Abstract
Restriction–modification (RM) systems are composed of genes that encode a restriction enzyme and a modification methylase. RM systems sometimes behave as discrete units of life, like viruses and transposons. RM complexes attack invading DNA that has not been properly modified and thus may serve as a tool of defense for bacterial cells. However, any threat to their maintenance, such as a challenge by a competing genetic element (an incompatible plasmid or an allelic homologous stretch of DNA, for example) can lead to cell death through restriction breakage in the genome. This post-segregational or post-disturbance cell killing may provide the RM complexes (and any DNA linked with them) with a competitive advantage. There is evidence that they have undergone extensive horizontal transfer between genomes, as inferred from their sequence homology, codon usage bias and GC content difference. They are often linked with mobile genetic elements such as plasmids, viruses, transposons and integrons. The comparison of closely related bacterial genomes also suggests that, at times, RM genes themselves behave as mobile elements and cause genome rearrangements. Indeed some bacterial genomes that survived post-disturbance attack by an RM gene complex in the laboratory have experienced genome rearrangements. The avoidance of some restriction sites by bacterial genomes may result from selection by past restriction attacks. Both bacteriophages and bacteria also appear to use homologous recombination to cope with the selfish behavior of RM systems. RM systems compete with each other in several ways. One is competition for recognition sequences in post-segregational killing. Another is super-infection exclusion, that is, the killing of the cell carrying an RM system when it is infected with another RM system of the same regulatory specificity but of a different sequence specificity. The capacity of RM systems to act as selfish, mobile genetic elements may underlie the structure and function of RM enzymes.
INTRODUCTION
Restriction enzymes and modification enzymes may not be merely sequence-specific DNA nucleases and methylases. Rather, they may represent one of the simplest forms of life, similar to viruses, transposons and homing endonucleases. As will be shown in this review, they can increase their relative frequency within a cell population by three strategies. First, they defend themselves (and the host bacterium) from invaders by attacking ‘non-self’ DNAs. Secondly, they kill cells that have eliminated them (Fig. 1A) due, for example, to the acquisition of another genetic element (Fig. 1B). Thirdly, they move between genomes. While such restriction–modification (RM) systems can help protect the cell from foreign DNAs, their behavior appears to reflect a primarily ‘selfish’ purpose, namely, to promote their survival. The purpose of this article is to review the evidence that supports this ‘selfish gene’ hypothesis.
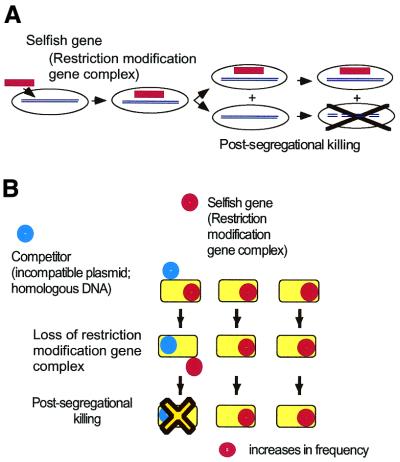
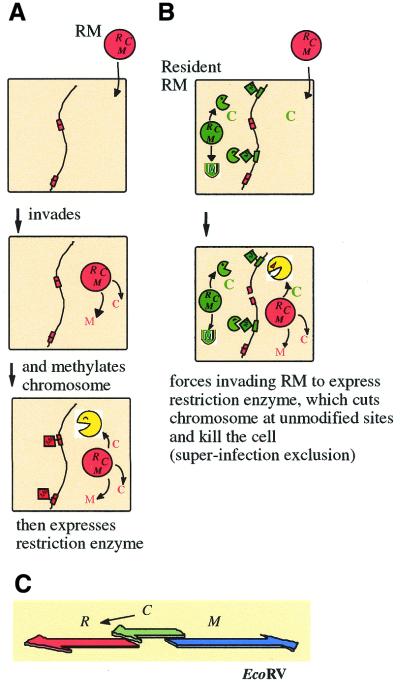
Figure 1.
Behavior of RM systems and other selfish genetic elements. (A) Parasitic behavior. Once established in a cell clone, the RM gene complex is difficult to eliminate because its loss leads to cell death (post-segregational cell killing). (B) Competitive advantage of post-segregational cell killing. A selfish genetic element (an RM gene complex) (red circle) is maintained in a cell clone. Invasion by a competing genetic element (such as an incompatible plasmid or allelic DNA) (blue circle) leads to competitive exclusion between the two elements. The cell where the resident genetic element is disturbed by the incoming element dies by post-segregational killing (see Fig. 2C). The incoming genetic element is thus eliminated together with the host cell. Thus, the resident genetic element maintains its frequency in the clonal cell population. [This scenario also operates in the super-infection exclusion between RM elements (Fig. 5).]
After a brief historical introduction, I will discuss recent evidence, particularly based on bacterial genome analysis, that suggests that some RM gene complexes behave as mobile genetic elements that can shape bacterial genomes. In the third section, I will introduce the parasitic life cycle of RM gene complexes with emphasis on their competition with each other. In the fourth section, the mechanisms used by bacteria and bacteriophages to protect themselves from the parasitic behavior of the RM complexes are described. It will be evident that the host–parasite-type interactions between bacteria and RM systems make a significant contribution to genome evolution.
The biochemistry, structural biology and biological and molecular evolution of RM systems have been reviewed from various points of view (1–7). The reference list here has been limited to recent sources that are immediately relevant to the concept of RM gene complexes as a form of life. Other references, representing many printed and online information sources, are easily accessible in the databases, MEDLINE/GenBank (NCBI), REBASE (New England Biolabs) and TIGR Microbial Database, among others, using keywords scattered throughout this article.
HYPOTHESES ABOUT THE EVOLUTION OF RM GENE COMPLEXES
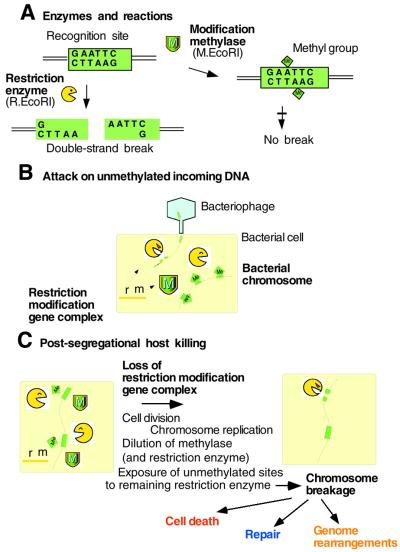
A restriction (R) endonuclease recognizes a specific DNA sequence and introduces a double-strand break (Fig. 2A), while a cognate modification (M) enzyme can methylate the same sequence and protect it from cleavage. The two together form a RM system. As each restriction gene is usually tightly associated with a cognate modification gene(s), these two (or more) genes can be termed an RM gene complex. There are several types of RM systems (2,7). A typical type II RM gene complex, such as EcoRI, contains one gene, R, for sequence recognition and restriction, and one gene, M, for sequence recognition and modification (Fig. 2A) (7). There are three, contrasting but not mutually exclusive hypotheses that explain the maintenance of RM systems in evolution.
Figure 2.
Action of type II RM systems. (A) Enzymes and reactions. (B) Direct attack on invading DNA. (C) Post-segregational killing.
RM systems may participate in cellular defense
Restriction enzymes will cleave incoming DNA if it has not been modified by a cognate or other appropriate methylase (Fig. 2B). It is thus widely believed that RM systems have been maintained by bacteria as tools for defense against infection by viral, plasmid and other foreign DNA. This may be called the cellular defense hypothesis.
RM complexes act as selfish genetic elements
Despite their function in cellular defense, there is increasing evidence that some RM systems operate on a primarily ‘selfish’ level. That is, the measures taken to maintain themselves in a cell population can lead to adverse consequences for their host cell. Some RM complexes resist being lost from a cell. For example, a plasmid carrying an RM gene complex cannot be readily displaced by an ‘incompatible’ plasmid carrying a different RM gene complex (8,9). Similarly, an RM gene complex on the bacterial chromosome cannot be easily replaced by a homologous stretch of DNA (a transducing fragment) (10). This resistance is manifested as cell death that occurs after an RM gene complex is lost (8,11–13).
Plausible steps involved in this process are illustrated in Figure 2C. If an RM gene complex is lost through an interaction with a competitor gene, for example, the cell’s descendants will contain fewer and fewer molecules of the modification enzyme. Eventually, the enzyme’s capacity to protect recognition sites on newly replicated chromosomes from the remaining pool of restriction enzymes will become inadequate and chromosomal DNA will be cleaved by the restriction enzyme, leading to the cell’s death (11,12). Naturally, the restriction enzyme molecules will also become increasingly limited in number. However, there is asymmetry between the roles of the methylase and the restriction enzyme—the asymmetry between life and death. For the restriction enzyme to kill the cell, a single break on the chromosome may well be sufficient (unless repaired). For the methylase to keep the host alive, all (or almost all) of the hundreds of recognition sites along the chromosome need to be methylated, and this will require a substantial pool of methylases to be present.
Evidence that supports this model was obtained in experiments in which an RM gene complex (PaeR7I, EcoRI and EcoRV) on a temperature-sensitive (ts) plasmid was lost from Escherichia coli cells after a temperature shift (8,11–14). This halted the increase in viable cell counts and resulted in loss of cell viability. Many cells formed long filaments, some of which were multinucleated and others anucleated (referring to those regions stained by DNA-staining dyes as nuclei). Accumulation of long linear chromosomal forms followed by extensive DNA degradation was observed by pulsed-field gel electrophoresis and conventional gel electrophoresis. The cells induced the SOS response.
This situation is reminiscent of ‘post-segregational killing’, which is recognized as a plasmid stabilization mechanism (Table 1) (15,16). The mechanism consists of a pair of genes, a killer and an anti-killer, that interact either at the protein level or at the RNA level. Loss of the gene pair from a cell brings about the lethal action of the killer element. Indeed, insertion of an RM gene complex into a plasmid increases the stability with which the plasmid is maintained in a culture of bacterial cells (8,11–14,17,18). In the case of the EcoRII RM gene complex on the ts plasmid described above, the temperature shift led to a decrease in the viable cell count instead of simply preventing its increase (14; Y.Naito, N.Handa, N.T.Kobayashi and I.Kobayashi, unpublished). The simplest explanation for this is that the RM system killed the cells when its copy number had decreased even before the complete loss of the gene complex from the cell. ‘Post-disturbance cell killing’ might be a more appropriate description of this behavior than post-segregational cell killing. The resistance of chromosomal RM gene complexes to replacement as described above (10) implies that the RM gene complex, rather than the plasmid, is the unit of post-segregational killing or selection.
Table 1. Selfish genetic elements programming host deaths.
| |
Type |
Subtype |
Example |
Killer/ anti-killer |
Mechanism |
Reference |
| I-1 | Post-segregational killing | |||||
| I-1-A | Proteic plasmid stabilization systems | Ccd on F plasmid | CcdB/CcdA | Inhibition of DNA gyrase | 15 | |
| I-1-B | Anti-sense RNA regulated plasmid systems | Hok/Sok on R1 plasmid | Hok RNA/Sok anti-sense RNA | Damaging cell membrane | 115 | |
| I-1-C | Restriction modification systems | EcoRI | R.EcoRI/ M.EcoRI | Restriction enzyme’s attack on chromosome | ||
| I-2 | Bacteriocin | ColE9 | Colicin E9/Immunity protein | DNase attack on chromosome | 116 | |
| I-3 | Meiotic drive | Segregation distorter | 19,117 | |||
| II-1 | Phage exclusion (abortive infection) | Prr | RNase on tRNALys | 118 | ||
| II-2 | Super-infection exclusion between RM systems | BamHI | Restriction enzyme’s attack on chromosome (restriction enzyme is coded by incoming RM gene complex) | 18 |
Given that RM systems will kill their host cell when they are eliminated from the cell, they could be called selfish genes in the sense that the word is used in genetics, evolutionary biology and in behavioral ecology (Table 1, I; Table 2) (19–21). The competition between an RM gene complex and a competing genetic element (Fig. 1B), which results in the host cell being killed, serves to maintain the presence of the RM gene complex in the neighboring clonal cells (Fig. 1B). While on a superficial level the cell death resembles altruistic suicide with respect to the other cells in the host cell clone, the cell death is actually programmed by a genetic element (RM) whose interests are not necessarily the same as those of the genome where it resides. Indeed, it will kill the cell in a fight with a competing genetic element. It is hypothesized that this competitive behavior has, at least in part, assured the maintenance of some RM systems (8,13,20,22).
Table 2. Selfish gene hypothesis for RM systems.
| Phenomenon |
Explanation |
| Occurrence | |
| Abundance in the genome of some eubacteria and archeae, especially in the chromosome of naturally competent (transformable) bacteria (26–28,119–121) | (i) Resistance to loss by host-killing, especially in competition with other genes(ii) Defense against invading DNAs by cleavage of their DNA |
| Potential mobility (see Table 3) | Mobility in accord with the selfish nature (infectious agents) |
| Gene organization | |
| Tight linkage of R and M genes | (i) Unit of selection and mobility(ii) Simultaneous loss essential to the post-segregational killing (Fig. 2) |
| Restriction and modification activities on separate, unrelated molecules (type II, unlike type I) | (i) Allows post-segregational cleavage of the host chromosome by restriction enzyme molecule(ii) Pairing of killer and anti-killer of independent origins |
| Individual specificity and collective diversity in sequence recognition | (i) Competition for recognition sequences among RM elements in post-segregational killing(ii) Effective defense of bacteria and RM genes against invading DNAs |
| Presence of rare cutters | Chromosome, rather than invading DNA, as a target |
| Regulatory system on R gene expression (such as C genes) | (i) Delays premature restriction in invasion to a new host cell(ii) Super-infection exclusion between RM gene complexes of different sequence specificities |
| Diversity of R gene sequences | Early specialization to specific sequences followed by sequence divergence and horizontal transfer (as in viruses and transposons) |
| Putative anti-RM adaptation | |
| Paucity of some potential RM sites (restriction avoidance) in some bacterial genomes | Protection of host chromosomes against attack of some RM systems |
| Orphan methylases | Protection of bacterial chromosome against some restriction modification systems (molecular vaccination) |
| Methylated DNA-specific endonuclease | Suicidal defense against invasion of RMs |
| Homologous recombination | (i) (Bacterial) Repair of host chromosomes (and destruction of invading DNA) after cleavage by restriction enzymes and genome diversification(ii) (Bacteriophages) Repair of phage DNA breaks made by restriction enzymes and genome diversification |
See text for details and references.
Other similarly selfish genes include bacteriocin operons and meiotic drive genes (Table 1, I). This mode of cell death—apparently altruistic cell death programmed by a resident genetic element upon invasion of its competitor genetic element—is also seen in phage exclusion (Table 1, II). We will see below that RM systems behave similarly.
Several type II systems have shown this type of self-stabilization and/or host killing, including PaeR7I (8), EcoRI (13), Bsp6I (17), EcoRV (18), PvuII (Y.Nakayama and I.Kobayashi, unpublished), EcoRII (Y.Naito, N.Handa, I.Kobayashi, unpublished; 14) and SsoII (14). The effect varies between type II systems as it is stronger for some (EcoRII, for example) and weaker for others (SsoII, for example) under the experimental conditions examined. Such selfish behavior has not yet been reported for other RM system types. The EcoKI system does not resist its loss and is readily replaced by alleles conferring different specificities (23). Similarly, the EcoR124I genes can be lost without killing the host cell (24). However, it would not be surprising if further work revealed a greater variation in the virulence of RM systems, similar to what we find in viruses and transposons.
The variation hypothesis
The third hypothesis about why RM systems are maintained in evolution is the variation hypothesis. This hypothesis suggests that RM systems participate in generating bacterial diversity by promoting homologous recombination (e.g. 25). This hypothesis needs another ad hoc hypothesis to explain why the diversity is advantageous. On the other hand, the selfish gene hypothesis can explain why homologous recombination is advantageous, as we see below.
RM GENE COMPLEXES ARE MOBILE AND CAN AFFECT GENOME EVOLUTION
RM gene complexes can act selfishly if they are independent from the rest of the genome (their host). This independence is facilitated by their movement between genomes. There is increasing evidence that supports the notion of RM independence and mobility, notably from genome analysis (Table 3).
Table 3. Evidence for mobility of restriction modification gene complexes.
| Type | Examples (comments and/or references) |
| Linkage with mobility-related elements | |
| Plasmid | EcoRI (122) |
| Bacteriophage | HindIII on Phi-flue (40); Sau42I RM on phi-42 (accession no. X94423); EcoO109I on P4-like prophage (42) |
| Transposon/ transposase | Rle39BI RM flanked by IS inverted repeats (44); AccI RM next to site-specific recombinase and transposase homologs (123) |
| Integron | XbaI (GenBank accession no. AF051092); M.Vch01I0 (REBASE, GenBank accession no. X64097) |
| Site-specific recombinase | PaeR7I (124) |
| [Abundance in the chromosome of naturally competent (transformable) bacteria] | (An RM element can move without a linked mobile element) |
| Genome sequence comparison | |
| Insertion/substitution (Fig. 3A and E) | HaeII on H.aegyptius (45); H.pylori (28,49,50) |
| Insertion with a long target duplication (Fig.3B and E) | H.pylori (49,50) |
| Linkage with inversion (Fig. 3C and E) | H.pylori (28); Pyrococcus (34) |
| Transposition (Fig.3D and E) | H.pylori (28); Pyrococcus (34) |
| Evolutionary/informatics analysis | |
| Occurrence of close homologs in distantly-related bacterial groups | Relatively frequent horizontal transfer (29) |
| Incongruence of phylogenetic trees with trees of the other genes | 29,36 |
| GC content difference | H.pylori (28,29); N.meningitidis (120) |
| Codon usage bias | 29,38,39 |
See text for details.
RM elements are abundant in some genomes
Some of the sequenced bacterial genomes, such as those of Haemophilus influenzae, Methanococcus jannaschii, Helicobacter pylori, Neisseria meningitidis, Neisseria gonorhoeae and Xylella fastidiosa, are impressively rich in RM genes (or their homologs) (see REBASE). Some of these genomes share the capacity for natural transformation, whereby chromosomal genes would be frequently replaced by incoming homologous DNA stretches. The RM gene complexes may resist this by killing their host or by cleaving the invading DNAs, thus assuring their continued presence in the cell population. However, other sequenced genomes, such as those of Rickettsia prowazekii, Treponema pallidum, Chlamydia and Buchnera, lack or almost lack open reading frames with homology to known RM genes. With regard to R.prowazekii, Chlamydia and Buchnera, one plausible explanation for their lack of RM genes is that their genomes are isolated from incoming genes because they occupy an intracellular niche within the cells that they infect.
RM elements are polymorphic
Considerable sequence diversity in the DNA sequences coding for RM enzyme homologs within a given species and within a given genome has been observed. Examples are found in E.coli, N.gonorrhoeae and H.pylori (REBASE) (26–28). Thus, a comparison of the M homologs from two completely sequenced strains of H.pylori, 26695 and J99, showed that, while many pairs were very similar to each other, there were also other M homologs that were specific to each individual strain (28,29). Another comparison between two strains revealed similar polymorphisms: among the gene clones identified as being present in a third strain of H.pylori but absent from the strain 26695, several were RM homologs (30). This diversity was verified by activity measurements (31–33). Polymorphisms of RM homologs have also been found between the genomes of two species of the genus Pyrococcus (34) and of two N.meningitidis strains (35).
Evolutionary analyses suggest RM genes have undergone horizontal gene transfer
Comparisons of RM sequence alignments (often in the form of a phylogenetic tree) with sequence alignments of other genes, such as ribosomal RNA genes, in the genomes suggest that RM genes have undergone extensive lateral gene transfer (29,36,37). The GC content and/or codon usage of RM genes are often different from those of other genes in the genome (28,29,38,39). This is consistent with the notion that these RM genes joined the genome relatively recently, although it is difficult to estimate the time of their arrival.
RM elements are often linked to mobility-associated genes
As implied in the above discussion, any type of genetic element that is relatively independent from the remainder of the genome would benefit from carrying an RM gene complex, as the selfish behavior of this complex would help maintain the element in the cell population. In turn, the RM gene complex would enjoy increased mobility. Supporting this notion is that a variety of mobile genetic units are found to be linked with RM gene complexes (Table 3). Some plasmids are known to carry RM gene complexes. For example, the sequencing of the M.jannaschii genome identified a type I RM system on a large plasmid. Prophages are also known to bear RM genes. For example, the HindIII RM gene complex was found on a cryptic prophage identified in the H.influenzae Rd genome sequence (40). Other examples are the Sau42I RM, found on a Staphylococcus aureus bacteriophage (accession no. X94423) and linked with a phage integrase (41), and EcoO109I, present on a P4-like prophage (42). There are also RM gene homologs on a prophage in the sequenced Bacillus subtilis genome (43).
The sequencing of clones containing RM genes often reveals their linkage to homologs of mobility-related genes and elements. For example, the Rle39BI RM is flanked by IS repeats and could move as a composite transposon (44). In addition, the XbaI RM (GenBank accession no. AF051092) and M.Vch01I0 (REBASE, GenBank accession no. X64097) are flanked by repeat elements of integrons, and their sequences suggest that they could move as an integron cassette. Many more RM genes are linked with a site-specific recombinase homolog or a transposase homolog (see REBASE, GenBank and MEDLINE) (41).
RM elements are also found to be inserted into operons. When the HaeIII RM gene region in Haemophilus aegyptius was compared to that in H.influenzae, it appeared that the HaeIII RM had been inserted into an operon in an ancestor of H.aegyptius, replacing a short intergenic sequence (45). Similar insertion into an operon was found in Neisseria (46) and in Streptococcus (47). As an operon can be regarded as a unit of mobility on an evolutionary time scale (48), the association of RMs with operons may be essentially similar to the association of RMs with other mobile elements. The operon would enjoy stability, especially in competition with an operon of a similar function, while the RM unit would enjoy mobility. Another benefit of being inserted into an operon is that the host operon would allow the RM unit to be transcribed.
How often RM units are associated with a mobile genetic element is not clear as yet because cloned RM units are rarely sequenced outside of the coding regions. Thus, it may be that other RM gene units are also associated with mobile genetic elements. However, some RM gene complexes appear to behave as a mobile unit without being linked to another mobile unit. This can be inferred from the comparison between closely related genomes of naturally transformable bacteria. In naturally transformable bacteria, RM gene complexes are as mobile as any other DNA in the genome. They do not have to be linked with a mobile element.
Linkage of RM genes with large genome polymorphisms
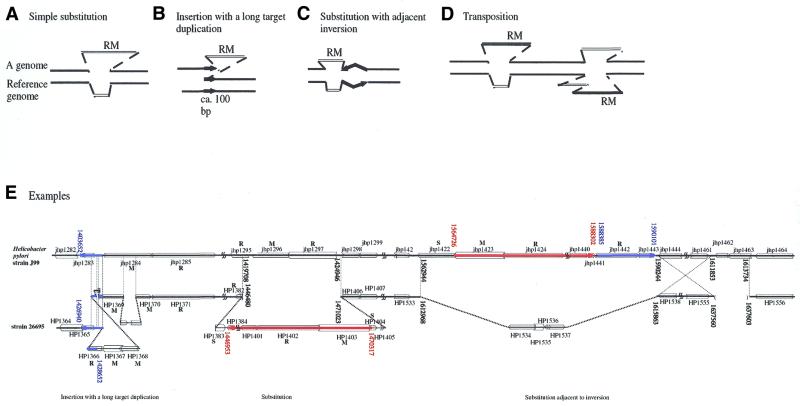
That RM elements have moved between genomes and are associated with genome rearrangements is evidenced by comparison of two closely related genomes. A simple-substitution-type polymorphism (Fig. 3A and E, middle) was detected when two strains of H.pylori (49) and two species of Pyrococcus (34) were compared. Also in H.pylori was observed the unique structure of ‘an insertion with a long target duplication’ (Fig. 3B and E, left) (49,50). This structure is formally similar to the classical transposon insertion except that the target duplications here are much longer. Analysis of the neighboring sequences suggested that this polymorphism reflects an ancient insertion event. This is also consistent with the possibility that later recombination between the duplicated region caused the RM gene complex to be deleted (49). Several RM homologs have also been found to be inserted next to an inversion polymorphism between two closely associated genomes both in H.pylori and Pyrococcus (Fig. 3C and E, right) (28,34,49). Figure 3E (right) illustrates a case involving type I RMS homologs.
Figure 3.
Large-scale genome polymorphisms associated with RM genes. The following RM-associated polymorphisms were detected when two closely related genomes were compared. (A) Simple substitution. (B) Insertion with a long target duplication (49,50). (C) Substitution adjacent to an inversion (28,34,49). (D) Translocation/transposition. Two homologous DNA segments carrying an RM gene complex are present at different locations in the two genomes (28,34,49). (E) Examples found when two strains of H.pylori were compared. (Left) A type IIS gene complex (HP1366/HP1367/HP1368) is inserted, with a long target duplication, upstream of a type III RM gene complex (HP1369/HP1370/HP1371 = jhp1284/jhp1285). The type III gene complex may have been inactivated following the insertion (49). (Middle) Simple substitution between a type III RM gene pair (jhp1296/jhp1297) and a long region including a type I RM gene complex (HP1402/HP1403/HP1404) at its right end. (Right) A long DNA region carrying multiple RMS homologs is present as a substitution adjacent to a large inversion. The long substitution region consists of (i) a (blue and striped) region (jhp1442/jhp1443) homologous to the type IIS R homolog region (HP1366/HP1365) in the left part of the figure and (ii) a (red and solid) region (jhp1423 – jhp1441) homologous to the substitution region in the middle part of the figure. The type I homologs (jhp1424/jhp1423/jhp1422) are at the end of the substitution. These two regions can be described as transposed. Analysis by A.Nobusato and I.Kobayashi (49,114) of the areas first analyzed by Alm et al. (28).
Comparison of the genome sequences of two H.pylori strains (28,29,49) and of two Pyrococcus species (34) also brought to light cases where a homologous segment containing RM complexes is present at different chromosomal loci in the two genomes (Fig. 3D and E). This strongly suggests that the DNA segments containing the RM complexes can move between different positions in a genome—a transposition or a translocation event. (The transposition/translocation could have occurred between these two loci or could have involved a third locus.) These examples (Fig. 3D and E) suggest that the unit of transposition for a particular RM gene complex can vary between events in that an RM gene complex may or may not move together with neighboring DNA.
Mechanisms generating genome rearrangements involving RM complexes
Although these polymorphisms involving RM gene complexes can be formally described as insertions, substitutions, inversions, translocations, transpositions etc., the molecular mechanisms generating them are not yet understood. In some cases, certain mechanisms can be ruled out through homology searches (34,49).
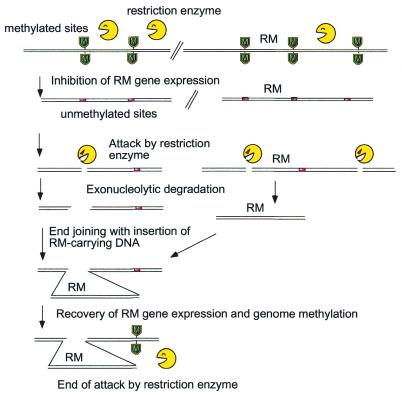
One hypothesis of how these polymorphisms arise suggests they are due to attacks by restriction enzymes on the host chromosome after disturbance of the RM gene complex (34,49). An example of how an RM unit could be transposed within a genome is shown in Figure 4. Here it is assumed that the balance between restriction enzyme and modification enzyme is somehow disturbed, for example, by the insertion of some genetic element in the neighborhood. The modification enzyme now fails to methylate its chromosomal recognition sites, and the remaining restriction enzyme molecules will introduce breaks into the chromosome. Degradation from the cut ends follows. The host, in an attempt to repair the chromosome, tries to rejoin the broken ends. These steps would generate a variety of rearranged genomes. A DNA fragment may be excised and inserted elsewhere in the genome. If the ends are properly closed with the insertion of a DNA fragment carrying the RM gene complex in question, the RM gene complex may be able to express the methylase again, and thus can protect chromosomal sites from its new locus. This would prevent further attack by the restriction enzyme. (Note that the RM unit on a DNA fragment, even if expressed, cannot be stably inherited by the progeny. Their descendants would be attacked by restriction enzyme after the loss of the DNA fragment.) The net result is the transposition of the RM gene complex to a new locus in this surviving clone.
Figure 4.
Selfish transposition model for genome rearrangements associated with RM gene complexes. Expression of an RM gene complex is disturbed. Unmethylated sites are exposed to attack by the restriction enzyme. The DNA may be further degraded in two directions from the cut ends. The host joins the broken ends. When a DNA fragment carrying the RM gene complex is properly inserted at the joint, the methylation can resume and the restriction attack will cease.
In this case (Fig. 4), the transposition results in substitution polymorphism of an RM segment (Fig. 3A). The case of ‘insertion with a long target duplication’ (Fig. 3B) (mentioned above) can be explained if unequal end joining between two restriction breaks on two sister chromosomes takes place together with the insertion of a DNA carrying the RM gene complex (49). The substitution adjacent to a large inversion (Fig. 3C) (28,34) can be explained by restriction breaks at two sites, possibly behind each of the two bi-directional replication forks. The fragment between the two breaks will thus be rejoined in the opposite orientation together with the DNA fragment carrying the RM gene complex (34).
In this ‘selfish transposition model’, the RM gene complex appears to take advantage of the host’s attempt to repair in order to transpose itself to a new locus. Supporting the notion that disturbance of the resident RM complex triggers the development of these polymorphisms are experiments demonstrating that an attempt to replace an RM gene complex by allelic DNA lead to genome rearrangements in the laboratory (see below) (10).
Linkage with a paralog cluster
In S.aureus, genes homologous to those of type IC RM systems have been found to be inserted into each of two tandem clusters of paralog family members (exotoxin cluster and serine protease cluster) (51). The exotoxin cluster is polymorphic in that the number of repeats vary among strains. The role, if any, of these RM genes in the maintenance or formation of this cluster is to be examined.
Phase variation of RM systems
Two types of phase variation of RM systems that involve changes in the primary structure of the RM genes have been documented.
One is site-specific recombination within the gene for the specificity subunit (S) of type I systems (52,53). This (i) changes the sequence specificity of the RM system by changing altering the primary structure of the specificity subunit and (ii) also switches gene expression on or off.
The second type of phase variation of RM systems has been predicted to occur in H.pylori, N.gonorrhoeae, H.influenzae and Pasteurella haemolytica (54,55) and is due to changes in the length of simple base pair repeats in either the R or the M gene. Changes in tract length have been demonstrated experimentally (28). How such changes affect RM activities remains to be elucidated, but a change in sequence specificity due to differences in repeat numbers has been reported for type I systems (Eco124 and R124/3) (56).
Allelic RM systems may show phase variation through intercellular DNA transfer (57), and the RM systems showing strain polymorphisms involving substitution and insertion (discussed above) are candidates for this.
Turning R and/or M genes on or off by phase variation could lead to cell death through restriction attacks on the genome. This has been hypothesized to be an altruistic cell death process that releases cellular DNA for its uptake by other cells in natural transformation (54).
Phase variation would be promoted when the genes making up one RM system are not linked together on the genome and are instead at separate locations (discussed further below). This could help facilitate their recombination and, possibly, also their phase variation.
RM pseudogenes
Genomic analysis of a variety of bacterial systems suggests that there are many RM homologs that appear to be inoperative because of insertions, deletions or point mutations (32,49 and REBASE). This is reminiscent of the defective transposons observed in some prokaryotic genomes and many eukaryotic genomes. In H.pylori genomes, there are indications of RM units that have been inserted into another RM unit. An example is HpyAII (= HP1366, HP1367, HP1368) (type IIS) that appears to have been inserted into a type III gene cluster (HP1369/HP1370, HP1371) (Fig. 3E, left). The resident RM unit appears to have been further destroyed by mutation, possibly after the insertion, because their homologs in another strain, J99, seem to be intact (jhp1284, jhp1285) (Fig. 3E, left). This is again reminiscent of transposon insertion into another transposon that results in the structures found in the genome of many eukaryotes.
In some cases, the R gene appears to be non-functional while the neighboring M gene seems to be functional (58). The M gene may serve to protect the genome against RM systems that recognize the same sequence (as described below for orphan methylases).
Tightness of linkage of the genes constituting an RM system may vary
In most of the RM systems identified so far, the constituent genes are tightly linked with each other. This cluster of genes confers the RM unit with mobility and the ability, upon its elimination from a cell, to kill the cell. The simultaneous loss of R and M genes assured by their tight linkage is particularly critical for their maintenance by post-segregational killing. In several cases, however, R, M or S homologs have been found isolated from each other in the genome. Such isolated genes may be due to (i) the decay of an RM cluster, (ii) their unique function as an orphan gene or (iii) their participation in an RM system composed of unlinked genes. Examples of the first possibility (i) are found in sequenced genomes, for example, H.pylori (32,49). Variability in operon structure among diverse genomes has been noticed earlier (59). This may be explained by the decay in the structure of the operon, which represents a unit of mobility (48). That isolated genes may play a unique function as orphan genes (ii) will be discussed later.
The third possibility (iii) is exemplified by an S gene on a plasmid that works together with a chromosomal RM gene cluster in a type I system (60). Similarly, the Mycoplasma pneumoniae genome carries a single type I gene cluster (SMR) together with several orphan S genes at different loci (61) although their activities have not been examined. Candidates for another example of unlinked RM genes are found in the two S.aureus genomes discussed above. In both of these genomes, an S and M gene pair is linked to a toxin gene cluster, while another S and M pair is linked with another, unlinked toxin gene cluster. An R homolog of the same family (type IC) is located elsewhere (unlinked) in the genome. The two M genes are highly homologous while the two S genes diverge in their putative target recognition domains (51). It is not known whether the two different SM pairs, together with the R gene, form two type I systems that differ in sequence specificity. The separation of genes for an RM system into different loci may facilitate their recombination and phase variation (mentioned above). It is possible that such unlinked RM systems may be more common than is currently believed, because the conventional methods of identifying RM genes (as opposed to the genome decoding strategy) may miss RM systems composed of unlinked genes.
PARASITIC/SYMBIOTIC LIFE CYCLE OF RM GENE COMPLEXES
Several of the phenomena observed with RM gene complexes and RM enzymes may be better understood if we assume that they are parasitic or symbiotic elements. One example of such parasitic behavior is the post-segregational killing discussed above, which appears to be a programmed process that serves to overcome competing genetic elements (Fig. 1). Other examples of how RM complexes can act in a parasitic or symbiotic manner are described below.
RM systems establish their parasitism/symbiosis within a new host by delaying R expression
When some RM gene complexes establish themselves in a cell, they delay expression of the restriction enzyme relative to the expression of the methylase to avoid the lethal restriction of the host genome (Fig. 5A). In this way, RM complexes behave as ‘smart’ parasites because they assure the survival of the host cell. There are several different ways in which this delay is achieved.
Figure 5.
Establishment of an RM system in a host cell and super-infection exclusion. (A) Establishment of an RM system in a host cell. An RM gene complex invades a cell. Its methylase and regulatory (C) genes are expressed first. After the modification enzyme has modified most of the chromosomal recognition sites, the accumulation of C protein induces the expression of the restriction enzyme. (B) Super-infection exclusion. A plasmid carrying an RM gene complex invades a cell that harbors another RM gene complex with a different sequence specificity but with the same C protein specificity. The resident C protein induces the expression of the incoming R gene, whose product then cleaves the unmodified chromosomal sites recognized by the incoming restriction enyzme and kills the cell. The resident RM gene complex thus promotes its survival in the clonal population over that of other RM systems (compared with Fig. 1B.) (C) The gene regulation circuit underlying the delay of restriction and super-infection exclusion. In some RM gene complexes, such as EcoRV, its C regulatory protein regulates the expression of its restriction enzyme.
Some RM systems (PvuII, BamHI, EcoRV, etc.) employ a regulatory gene, called C, to control the expression of R and/or the repression of M (Fig. 5C) (18,62–64). The C gene product binds the promoter region of the R gene (65), and either promotes its transcription (64) or stabilizes its mRNA (66). The C gene products for the PvuII and BamHI RM systems recognize the same sequences (65,67), while the C gene product for the EcoRV system does not share this specificity (18,67).
In some systems, the methylase itself either positively regulates R expression or negatively regulates M expression. M.EcoRII autoregulates its own expression (68). In the case of SsoII, the methylase binds the intergenic region of a divergent R and M gene pair, and thus regulates both M and R expression (69). Its N-terminus region is important in this regulation. DNA methylation at the CfrBI site in the promoter region of R.CfrBI is involved in transcriptional control of the CfrBI RM system (70).
The salIR and salIM genes constitute an operon that is mainly transcribed from sal-pR1, a promoter located upstream of salIR. Another promoter, sal-pM, is within the salIR coding region and allows the M gene to be expressed in the absence of sal-pR1. The sal-pM promoter might be involved in the establishment of modification prior to restriction endonuclease activity (71). The pvuIIW gene is a possible modulator of PvuII endonuclease subunit association, which delays restriction upon establishment (72).
Systems for avoiding lethal restriction upon entry into a new cell have been documented for type I and III RM systems (3,24,73,74).
Super-infection exclusion between RM systems
The presence of an RM gene complex in a cell would prevent another RM gene complex from establishing itself in the cell because the resident RM would attack the incoming RM if it was not properly methylated, just as it would attack any other DNA. However, should the incoming RM complex be properly methylated or lack the recognition sequence of the resident RM at all, the resident RM may still be able to abort the establishment of the incoming RM in the cell by eliciting host killing (Fig. 5B) (18,64). It takes place specifically between two RM systems that share the specificity in the regulatory system for establishment (see the previous section). It was demonstrated between two RM systems that are regulated by C gene products that bind the same sequence. The PvuII and BamHI RM systems have such cross-complementing C genes. The presence of the BamHI RM gene complex in the recipient cell reduced the transformation efficiency of a plasmid that carried the PvuII RM gene complex. A similar decrease in transformation efficiency was also observed when the recipient cell contained only the C regulatory gene of BamHI, and this decrease was dependent on the restriction gene of the incoming PvuII RM system (18). Thus, the regulatory C gene product of the resident RM element in the cell appears to force the expression of the incoming R gene, which leads to it cleaving its as-yet-unmodified recognition sites on the chromosome and killing the host cell (Fig. 5B). This mechanism should work only if the two RM systems involved have non-identical recognition sequences.
This form of competition between RM systems is reminiscent of the super-infection exclusion between viruses (bacteriophages) (and between a plasmid and a virus) whereby cell death is triggered by infection of another virus (Table 1, II). Thus, this form of competition has been designated as ‘super-infection exclusion’. It is another example of how resident selfish genetic elements can act in altruistic cell death strategies to protect a clonal cell population against invading genetic elements (Fig. 1B). By analogy to apoptosis in animal cells infected with viruses, I have called this phenomenon ‘apoptotic mutual exclusion’. The exclusion resembles immunity between prophages and incompatability between plasmids in that the identity of specificity in the regulatory network prevents propagation of a second element. The super-infection RM exclusion would be prominent during the establishment of an RM system and thus resembles surface exclusion between plasmids in this respect.
This type of exclusion constitutes another dimension to RM system classification. Thus, when the regulatory specificities of the two RM systems involved are identical, they constitute an incompatibility group. PvuII RM and BamHI RM consequently define one incompatibility group while the EcoRV RM, which is controlled by a C gene that recognizes different sequences, defines another (18). SsoII RM and EcoRII RM do not show such exclusion, probably because their restriction enzyme expression is regulated with different specificities (14). A bacterial cell can carry multiple RM systems of different super-infection exclusion specificities just as it may carry multiple plasmids of different incompatibility specificities.
RM systems compete with each other on the basis of their recognition sequences
Sequence recognition by RM systems is individually highly specific but collectively quite diverse. This has been explained by the existence of frequency-dependent selection in cellular defense (Fig. 2B). If the recognition sequence be rare, an RM system would be more likely to recognize invading DNAs as non-self.
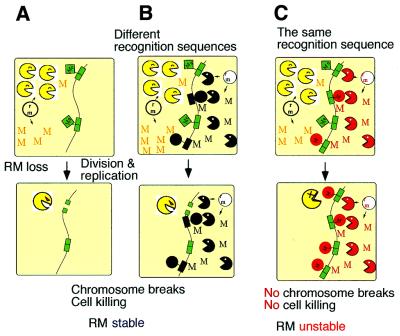
However, competition between two RM systems for a recognition sequence has also been demonstrated to occur in the absence of any invading DNA (Fig. 6) (13). Post-segregational host killing by an RM gene complex did not occur when a second RM gene complex within the cell shared the same sequence specificity (Fig. 6C). In other words two RM systems of the same specificity were unable to enjoy stabilization simultaneously. This type of ‘incompatibility’ or mutual exclusion implies competition for specific sequences by RM systems.
Figure 6.
Competition for recognition sequences between RM gene complexes. (A) The cell contains one plasmid with an RM gene complex. When the plasmid (and thus also the RM gene complex) is eliminated from the cell, the modification enzyme is diluted and this leads to cell death by post-segregational killing. (B) The cell contains two plasmids, each with an RM gene complex that has a different sequence specificity. When either of the plasmids is lost together with its RM gene complex, the cells die due to post-segregational cell killing. Both of the RM gene complexes can enjoy stabilization. The situation is stable. (C) The cell contains two plasmids, each with an RM gene complex that has the same sequence specificity. Loss of either of the plasmids together with its RM gene complex does not lead to cell killing because the other RM system modify the recognition sites on the host chromosome. The two RM gene complexes cannot simultaneously enjoy stabilization. The situation is unstable.
In addition, a less specific recognition site can provide an advantage. For example, when two RM systems, one recognizing CCNGG (N = A, T, G or C) and the other recognizing CCWGG (W = A or T), were present in the same cell, the former won the intracellular competition (14). The former was able to prevent host killing by the latter, while the latter was unable to prevent killing by the former. This one-sided incompatibility implies that there is a selective pressure for a less specific recognition sequence.
This direct competition between RM systems in the absence of any invading DNAs thus could cause RM systems to evolve, with respect to their sequence recognition, faster than they would only through selection by invaders.
BACTERIA PROTECT THEMSELVES FROM THE SELFISH BEHAVIOR OF RM GENE COMPLEXES AND THIS CAN AFFECT GENOME EVOLUTION
Bacteria may have evolved several means to defend their genome from attack by restriction enzymes. That restriction enzymes participate crucially in various types of DNA recombination has been known for a long time. Indeed, RM systems are sometimes regarded as a mechanism for generating variety in a genome (the variation hypothesis) as mentioned above (75). A contrasting view is that the various recombination systems are measures taken by the host to protect itself from the selfish behavior of RM systems (20). This hypothesis is described below together with other possible adaptation strategies. Bacterial strategies against type I RM systems have been analyzed (3,76). Our emphasis will be on type II RM systems.
Evidence that bacteria use homologous recombination to defend themselves from RM gene complexes
Interaction with RM gene complexes may explain the apparently contradictory behavior of a major homologous recombination pathway of E.coli (77). From a double-stranded break, the RecBCD enzyme begins exonucleolytic DNA degradation. When the enzyme encounters a specific sequence called Chi, the degradation is attenuated and followed by RecA-mediated recombination of this DNA with a homologous DNA. The recombinogenic double-strand breaks may be provided by a type II restriction enzyme in vivo (78). In a mutant defective in RecBCD exonuclease/recombinase, the symptoms of cell death following the loss of an RM gene complex are more severe (12). These include growth inhibition, loss of cell viability, cell filamentation, and loss of nuclei (the region stained by DAPI). Huge linear forms of the chromosome accumulate. Similar syndromes were observed with a recBCD mutant defective in Chi recognition, and growth inhibition is also severe in recA, ruvAB, ruvC, recG and recN mutants. The cells induce the SOS response in a RecBC-dependent manner. These observations strongly suggest that the bacterial cell death after the loss of an RM gene complex is caused by chromosome breakage, and that the bacterial RecBCD/RecA machinery helps the cells to survive by repairing the broken chromosomes. RecA/BC-mediated cell survival is also observed when a mutant restriction enzyme is activated or when a restriction enzyme is overproduced (79).
A plausible explanation for the dual nature of the RecBCD system may be the interactions that occur between three genetic elements (the RM system, invading alien DNA and the host RecBCD system) within one bacterial genome. Thus, the RM system will attack non-self DNA, marked by an unmethylated recognition site, whether it is on invading DNA or on the chromosome. After restriction breakage, the host RecBCD system destroys the invading non-self DNA, defined by the host as the absence of the Chi sequence. On the other hand, it allows the restoration of its own Chi-marked chromosomal DNA through recombination.
If an incoming DNA carries Chi, exonucleolytic degradation will be attenuated. Furthermore, parts of the remaining fragment will be incorporated into the chromosome if they survive the mismatch repair process during homologous pairing. This would result in the mosaic-type genome polymorphism observed within a group of bacteria sharing the identification marker such as Chi (80). Pairs consisting of a RecBCD homolog and a Chi equivalent have been identified in other bacteria such as H.influenzae and Lactococcus lactis (81). The RecBCD enzyme changes its sequence preference through mutation (82,83). The bacterial group defined by the RecBCD analog/Chi analog pair may be regarded as a type of species in the sense that it defines a unit of reproductive isolation. The above role of Chi equivalents is distinct from, but nonetheless analogous to, the role of the uptake signal sequence in naturally transformable bacteria (84).
Interaction between a selfish RM system and bacterial homologous recombination results in genome rearrangements
Genome comparison revealed that RM genes are sometimes associated with large genome polymorphisms as discussed above (in the section ‘linkage of RM genes with large genome polymorphisms’). We then introduced the hypothesis that RM gene complexes can cause genome rearrangements when their presence is threatened. Experiments have provided support for this hypothesis. When the PaeR7I gene complex on the E.coli chromosome was challenged through general transduction by a homologous stretch of PaeR7I-modified DNA, the replacement efficiency was lower than expected. Many of the resulting recombinant clones retained the recipient RM gene complex as well as the homologous donor DNA. Analysis of these clones suggested that multiple rounds of unequal homologous recombination between copies of a repetitive element (IS) generated large-scale duplication and inversion in the chromosome and that only one of the duplicated copies of the PaeR7I gene complex was replaced by the donor allele. The interaction between selfish attack by RM systems and defensive homologous recombination system of bacteria is likely responsible for these genome rearrangements (10).
Bacteriophages also use homologous recombination to protect themselves from RM systems
Of the many anti-restriction strategies used by bacteriophages and plasmids (5), the homologous recombination machinery carried by bacteriophages appears well adapted to counteracting attacks by RM systems (85). An example is bacteriophage lambda, which encodes the homologous recombination proteins Redalpha and Redbeta. Rac prophage, a lambdoid bacteriophage present in some E.coli strains, also encodes similar recombinases, RecE and RecT. Redalpha and RecE both show 5′–3′ exonuclease activity at a double-stranded DNA end and expose a 3′ single-stranded end while Redbeta and RecT proteins stimulate annealing of complementary single strands. DNA strand invasion activity has been demonstrated for RecT (86). The cooperation of Redalpha and Redbeta, or of RecE and RecT, can promote homologous recombination near a DNA double-strand break made by restriction enzymes. Two parental DNAs can give rise either to only one progeny DNA (non-conservative recombination) or to two progeny DNAs (conservative recombination). When it is conservative, the repair may or may not be accompanied by crossing-over of the flanking sequences (87,88). Lambda Redalpha exonuclease has structural similarity to restriction enzymes, its enemies, and may be related to them in evolutionary terms (89).
Recombination stimulated by a restriction break may take place between co-infecting sister genomes in a clone. Recombination may also take place with a partner, possibly present as a prophage, from another phage clone possessing a divergent genome. Such out-crossing could confer several advantages in addition to the primary advantage of immediate restoration of the cleaved chromosome. If the template DNA were to lack the recognition site, recombination might result in a DNA region devoid of this particular restriction site. The crossing-over (or half crossing-over) of flanking sequences triggered by the break would confer a third kind of advantage: alleles at different locations, either sensitive or resistant to a particular restriction enzyme, would be recombined to generate rare combinatory genotypes. Some of these would be more resistant to attack by the present RM systems than the current combination, and they would increase in number. As a phage population encounters bacterial populations possessing various combinations of RM systems of diverse sequence specificities, the process of breakage, repair, gene conversion and crossing-over will continue.
End joining by bacteria in defense against RM and in genome rearrangements
Host bacterial cells also attempt to repair restriction breakage by rejoining the broken ends. Precise joining at a restriction break by ligase would restore intact DNA (90). Non-homologous or illegitimate joining of DNA ends resulting from further degradation is regarded as an unsatisfactory form of DNA repair, because the degraded genes are lost forever, even though the chromosome and the genome may be repaired. Non-homologous end joining at unequal loci in sister chromosomes might result in tandem duplication, but this would be preferable to joining the ends of a single chromosome, as it would not lead to the loss of genes and would increase the probability of genome survival. Insertion of RM gene complexes with a long target duplication (see above) (Fig. 3B) may be a strategy on the side of RM systems that takes advantage of this defense reaction by the host.
An interaction between non-homologous end joining, homologous recombination, and restriction has been identified in a special type of non-homologous recombination that is dependent upon homologous interaction and type I restriction (91). As expected from this in vivo work, a type I restriction enzyme was shown to cut DNA at a Holliday structure, an intermediate in homologous recombination (92). This could represent an attempt by a type I RM complex to abort homologous recombination involving unmodified non-self DNA.
Bacteria protect themselves from RM systems by orphan methylases and Vsr
Some DNA methylase genes are not linked to a restriction enzyme gene and are thus called orphans. One such gene is the dcm gene, present in several bacteria including E.coli, whose product methylates DNA to generate 5′ CmCWGG. A linked Vsr mismatch repair function prevents the C-to-T mutagenesis that is enhanced by this methylation but promotes other types of mutagenesis (93). Why the dcm–vsr gene pair is present has to date been an enigma. However, a clue to the function of dcm was provided by a study that blocked the EcoRII RM gene complex, which recognizes the same sequence as dcm, from replicating. This blockade led to chromosome degradation and severe loss of cell viability (14). However, when dcm was placed on the chromosome, the cell killing was attenuated, while when dcm was over-expressed, cell killing was completely abolished (Y.Naito, N.Handa, A.Chinen and I.Kobayashi, unpublished). Dcm thus can defend the genome against the parasitism of EcoRII.
Some RM gene complexes [HpaII (94), XorII (95), NaeI (96)] carry a Vsr homolog. (It is not known how often a C5 RM system is associated with a Vsr homolog/analog.) It is possible that the Vsr homologs may have evolved to prevent the loss of RM recognition sites, which are the target sites of their parasitism, from the host genome. Vsr has a structure similar to that of restriction enzymes, and is likely to be related to them phylogenetically (93,97).
Bacteria avoid attack by RM systems by eliminating their recognition sites in their genome
Some short palindromic sequences are rare in certain bacterial genomes (98,99). This has been described as ‘restriction avoidance’ as it may be an adaptation of these genomes to prevent attack by RM systems. A detailed analysis of completely sequenced genomes (100) showed that the sequences that would be attacked by the resident type II RM complex were amongst the rarest palindromes in the host cell’s genome. For example, EcoRI sites are quite rare in the E.coli chromosome (100). Palindromes recognized by restriction enzymes from other species were also avoided, albeit to a lesser extent.
When Rocha et al. (101) analyzed the oligonucleotide abundance in the complete genome of B.subtilis, they found that prophage-like elements embedded in the genome were more likely to contain several potential restriction sites than the rest of the genome. This suggests that, unlike the recently embedded prophages, the genomes themselves have evolved over past exposure to a variety of restriction enzymes to eliminate their recognition sites (101,102).
The strength of such selection would depend on the population substructure and the modes and frequency of horizontal gene transfer (103). For example, the E.coli population consists of few long-lasting clones and might be easily influenced by the attack by RM systems brought in by plasmids and bacteriophages. One can also imagine that the selection would be stronger for attack by resident RMs than for attack by invading RMs.
CONCLUSIONS AND PERSPECTIVES
I have reviewed here the increasing evidence that supports the hypothesis that some RM systems behave as a simple form of life. They increase their own frequency in the cell population by destroying non-self DNAs, whether these are invading DNA or their (ex-)host chromosome. The non-self DNA is recognized as such by the absence of methylation on a recognition sequence. The observations reviewed here also support the hypothesis that RM gene complexes are mobile genetic elements, in the broad sense of the term, that sometimes move between genomes, and occasionally cause evolutionary changes in these genomes.
Our knowledge of RM systems as a minimal form of life is still fragmentary. However, their life cycle and their interaction with the host (the genome) may be studied in much the same way as other genetic elements or genomic parasites have been studied. These include homing endonucleases (104), transposons, viruses, bacteria and parasites. Post-segregational killing, or addiction (105), is just one example of the principles that can be studied in the simple biological system consisting of RM and the bacterial genome. It might turn out to be a general principle that underlies the symbiosis of a variety of genetic elements in a genome.
Homologous recombination and several other cellular mechanisms appear to be well adapted to the selfish behavior of RM complexes. Several forms of genome polymorphism that were detected when closely related genome sequences were compared may have resulted from parasite–host-type interactions between RM elements and the rest of the genome. This interaction could be a major force for genome evolution in at least some bacteria. Laboratory studies could be designed to test the hypotheses emerging from the comparison of genome sequences. The immediate subjects of study could include genome changes involving RM systems and the host machinery of recombination, repair, replication and mutagenesis.
The behavior of RM gene complexes as genomic parasites or symbionts may have also affected the evolution of the structure and function of the RM enzymes themselves. An unexpected diversity of RM enzymes has been recognized at the levels of amino acid sequence (2,7) and three-dimensional structure (106–108). Structural analysis has revealed surprising diversity in the members of the restriction enzyme family (109). This family includes an exonuclease for homologous recombination (89), Vsr mismatch repair endonuclease (93,97) and MutH mismatch repair endonuclease (110), some of which have been discussed above in the context of their interaction with restriction enzymes. Other new findings include a resolvase of the Holliday intermediate in homologous recombination (111,112) and a transposase (113). More proteins of these families will undoubtedly be found in bacterial genomes, our human genomes and other genomes during the coming years of structural genomics or proteomics, with potentially exciting implications for biology, biotechnology and medicine.
Acknowledgments
ACKNOWLEDGEMENTS
I am grateful to my collaborators in my laboratory for data, figures, database survey and discussion. This work was supported by the MEXT (Ministry of Education, Culture, Sports, Science and Technology) of the Japanese government (Genome, Recombination), NEDO and the Uehara Memorial Foundation.
References
- 1.Fauman E.B., Blumenthal,R.M. and Chen,X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing Co., Singapore, pp. 1–38.
- 2.Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing Co., Singapore, pp. 283–340.
- 3.Murray N.E. (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev., 64, 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao D.N., Saha,S. and Krishnamurthy,V. (2000) ATP-dependent restriction enzymes. Prog. Nucleic Acid Res. Mol. Biol., 64, 1–63. [DOI] [PubMed] [Google Scholar]
- 5.Bickle T.A. and Kruger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X. and Roberts,R.J. (2001) AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res., this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naito T., Kusano,K. and Kobayashi,I. (1995) Selfish behavior of restriction-modification systems. Science, 267, 897–899. [DOI] [PubMed] [Google Scholar]
- 9.Naito Y., Naito,T. and Kobayashi,I. (1998) Selfish restriction modification genes: Resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol. Chem., 379, 429–436. [DOI] [PubMed] [Google Scholar]
- 10.Handa N., Nakayama,Y., Sadykov,M. and Kobayashi,I. (2001) Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction modification gene complex. Mol. Microbiol., 40, 932–940. [DOI] [PubMed] [Google Scholar]
- 11.Handa N. and Kobayashi,I. (1999) Post-segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie, 81, 931–938. [DOI] [PubMed] [Google Scholar]
- 12.Handa N., Ichige,A., Kusano,K. and Kobayashi,I. (2000) Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol ., 182, 2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusano K., Naito,T., Handa,N. and Kobayashi,I. (1995) Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl Acad. Sci. USA, 92, 11095–11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinen A., Naito,Y., Handa,N. and Kobayashi,I. (2000) Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol. Biol. Evol., 17, 1610–1619. [DOI] [PubMed] [Google Scholar]
- 15.Jensen R.B. and Gerdes,K. (1995) Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol. Microbiol., 17, 205–210. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes K., Ayora,S., Canosa,I., Ceglowski,P., Diaz,R., Franch,T., Gultyaev,A.P., Jensen,R.B., Kobayashi,I., Macpherson,C., Summers,D., Thomas,C. and Zielenkiewicz,U. (2000) Plasmid maintenance systems. In Thomas,C.M. (ed.), The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread. Harwood Academic Publishers, Amsterdam, The Netherlands, pp. 49–86.
- 17.Kulakauskas S., Lubys,A. and Ehrlich,S.D. (1995) DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol., 177, 3451–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama Y. and Kobayashi,I. (1998) Restriction-modification gene complexes as selfish gene entities: Roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl Acad. Sci. USA, 95, 6442–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst L.D., Atlan,A. and Bengtsson,B.O. (1996) Genetic conflicts. Q. Rev. Biol., 71, 317–364. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi I. (1998) Selfishness and death: raison d’être of restriction, recombination and mitochondria. Trends Genet., 14, 368–374. [DOI] [PubMed] [Google Scholar]
- 21.Haig D. (1997) The Social Gene. In Krebs,J. and Davies,N.B. (eds), Behavioural Ecology: An Evolutionary Approach, 4th Edn. Blackwell Science, Oxford, UK, pp. 284–304.
- 22.Kobayashi I. (1996) DNA modification and restriction: Selfish behavior of an epigenetic system. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 155–172.
- 23.O’Neill M., Chen,A. and Murray,N.E. (1997) The restriction-modification genes of Escherichia coli K-12 may not be selfish: they do not resist loss and are readily replaced by alleles conferring different specificities. Proc. Natl Acad. Sci. USA, 94, 14596–14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulik E.M. and Bickle,T.A. (1996) Regulation of the activity of the type IC EcoR124I restriction enzyme. J. Mol. Biol., 264, 891–906. [DOI] [PubMed] [Google Scholar]
- 25.Price C. and Bickle,T.A. (1986) A possible role for DNA restriction in bacterial evolution. Microbiol. Sci., 3, 296–299. [PubMed] [Google Scholar]
- 26.Stein D.C., Gunn,J.S., Radlinska,M. and Piekarowicz,A. (1995) Restriction and modification systems of Neisseria gonorrhoeae. Gene, 157, 19–22. [DOI] [PubMed] [Google Scholar]
- 27.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 28.Alm R.A., Ling,L.S., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., deJonge,B.L. et al. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 29.Nobusato A., Uchiyama,I. and Kobayashi,I. (2000) Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene, 259, 89–98. [DOI] [PubMed] [Google Scholar]
- 30.Akopyants N.S., Fradkov,A., Diatchenko,L., Hill,J.E., Siebert,P.D., Lukyanov,S.A., Sverdlov,E.D. and Berg,D.E. (1998) PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl Acad. Sci. USA, 95, 13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Q., Morgan,R.D., Roberts,R.J. and Blaser,M.J. (2000) Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc. Natl Acad. Sci. USA, 97, 9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong H., Lin,L.F., Porter,N., Stickel,S., Byrd,D., Posfai,J. and Roberts,R.J. (2000) Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res., 28, 3216–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L.F., Posfai,J., Roberts,R.J. and Kong,H. (2001) Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA, 98, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chinen A., Uchiyama,I. and Kobayashi,I. (2000) Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene, 259, 109–121. [DOI] [PubMed] [Google Scholar]
- 35.Bart A., Pannekoek,Y., Dankert,J. and van der Ende,A. (2001) NmeSI restriction-modification system identified by representational difference analysis of a hypervirulent Neisseria meningitidis strain. Infect. Immun., 69, 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeltsch A., Kroger,M. and Pingoud,A. (1995) Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene, 160, 7–16. [DOI] [PubMed] [Google Scholar]
- 37.Bujnicki J.M. and Radlinska,M. (1999) Molecular phylogenetics of DNA 5mC-methyltransferases. Acta. Microbiol. Pol., 48, 19–30. [PubMed] [Google Scholar]
- 38.Jeltsch A. and Pingoud,A. (1996) Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol., 42, 91–96. [DOI] [PubMed] [Google Scholar]
- 39.Mrázek J. and Karlin,S. (1999) Detecting alien genes in bacterial genomes. In Caporale,L.H. (ed.), Molecular Strategies in Biological Evolution. Annals of New York Academy of Sciences. New York Academy of Sciences, NY, Vol. 870, pp. 314–329. [DOI] [PubMed]
- 40.Hendrix R.W., Smith,M.C., Burns,R.N., Ford,M.E. and Hatfull,G.F. (1999) Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl Acad. Sci. USA, 96, 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi I., Nobusato,A., Kobayashi-Takahashi,N. and Uchiyama,I. (1999) Shaping the genome – restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev., 9, 649–656. [DOI] [PubMed] [Google Scholar]
- 42.Kita K., Tsuda,J., Okamoto,K., Yanase,H. and Tanaka,M. (1999) Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol ., 181, 6822–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 44.Rochepeau P., Selinger,L.B. and Hynes,M.F. (1997) Transposon-like structure of a new plasmid-encoded restriction-modification system in Rhizobium leguminosarum VF39SM. Mol. Gen. Genet., 256, 387–396. [DOI] [PubMed] [Google Scholar]
- 45.Stein D.C., Gunn,J.S. and Piekarowicz,A. (1998) Sequence similarities between the genes encoding the S.NgoI and HaeII restriction/modification systems. Biol. Chem., 379, 575–578. [PubMed] [Google Scholar]
- 46.Claus H., Friedrich,A., Frosch,M. and Vogel,U. (2000) Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J. Bacteriol., 182, 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekizaki T., Otani,Y., Osaki,M., Takamatsu,D. and Shimoji,Y. (2001) Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol ., 183, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence J. (1999) Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev., 9, 642–648. [DOI] [PubMed] [Google Scholar]
- 49.Nobusato A., Uchiyama,I., Ohashi,S. and Kobayashi,I. (2000) Insertion with long target duplications: A mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene, 259, 99–108. [DOI] [PubMed] [Google Scholar]
- 50.Nobusato A., Uchiyama,I., Ohashi,S. and Kobayashi,I. (1999) Insertion with long target duplication: a mechanism for bacterial gene mobility suggested from genome comparison. In Asai,K., Miyano,S. and Takagi,T. (ed.), Genome Informatics 1999. Universal Academy Press, Tokyo, Japan, pp. 346–347.
- 51.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kobayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 52.Dybvig K., Sitaraman,R. and French,C.T. (1998) A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl Acad. Sci. USA, 95, 13923–13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chambaud I., Heilig,R., Ferris,S., Barbe,V., Samson,D., Galisson,F., Moszer,I., Dybvig,K., Wroblewski,H., Viari,A., Rocha,E.P. and Blanchard,A. (2001) The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res., 29, 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunders N.J., Peden,J.F., Hood,D.W. and Moxon,E.R. (1998) Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol., 27, 1091–1098. [DOI] [PubMed] [Google Scholar]
- 55.Ryan K.A. and Lo,R.Y. (1999) Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res., 27, 1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price C., Lingner,J., Bickle,T.A., Firman,K. and Glover,S.W. (1989) Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J. Mol. Biol., 205, 115–125. [DOI] [PubMed] [Google Scholar]
- 57.Lacks S.A., Mannarelli,B.M., Springhorn,S.S. and Greenberg,B. (1986) Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell, 46, 993–1000. [DOI] [PubMed] [Google Scholar]
- 58.Ibanez M., Alvarez,I., Rodriguez-Pena,J.M. and Rotger,R. (1997) A ColE1-type plasmid from Salmonella enteritidis encodes a DNA cytosine methyltransferase. Gene, 196, 145–158. [DOI] [PubMed] [Google Scholar]
- 59.Itoh T., Takemoto,K., Mori,H. and Gojobori,T. (1999) Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol., 16, 332–346. [DOI] [PubMed] [Google Scholar]
- 60.Schouler C., Gautier,M., Ehrlich,S.D. and Chopin,M.C. (1998) Combinational variation of restriction modification specificities in Lactococcus lactis. Mol. Microbiol., 28, 169–178. [DOI] [PubMed] [Google Scholar]
- 61.Dandekar T., Huynen,M., Regula,J.T., Ueberle,B., Zimmermann,C.U., Andrade,M.A., Doerks,T., Sanchez-Pulido,L., Snel,B., Suyama,M., Yuan,Y.P., Herrmann,R. and Bork,P. (2000) Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res., 28, 3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks J.E., Nathan,P.D., Landry,D., Sznyter,L.A., Waite-Rees,P., Ives,C.L., Moran,L.S., Slatko,B.E. and Benner,J.S. (1991) Characterization of the cloned BamHI restriction modification system: its nucleotide sequence, properties of the methylase, and expression in heterologous hosts. Nucleic Acids Res., 19, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao T., Bourne,J.C. and Blumenthal,R.M. (1991) A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol ., 173, 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vijesurier R.M., Carlock,L., Blumenthal,R.M. and Dunbar,J.C. (2000) Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol ., 182, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sohail A., Ives,C.L. and Brooks,J.E. (1995) Purification and characterization of C.BamHI, a regulator of the BamHI restriction-modification system. Gene, 157, 227–228. [DOI] [PubMed] [Google Scholar]
- 66.O’Sullivan D.J. and Klaenhammer,T.R. (1998) Control of expression of LlaI restriction in Lactococcus lactis. Mol. Microbiol., 27, 1009–1020. [DOI] [PubMed] [Google Scholar]
- 67.Ives C.L., Sohail,A. and Brooks,J.E. (1995) The regulatory C proteins from different restriction-modification systems can cross-complement. J. Bacteriol ., 177, 6313–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friedman S. and Som,S. (1993) Induction of EcoRII methyltransferase: evidence for autogenous control. J. Bacteriol ., 175, 6293–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karyagina A., Shilov,I., Tashlitskii,V., Khodoun,M., Vasil’ev,S., Lau,P.C.K. and Nikolskaya,I. (1997) Specific binding of SsoII DNA methyltransferase to its promoter region provides the regulation of SsoII restriction-modification gene expression. Nucleic Acids Res., 25, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beletskaya I.V., Zakharova,M.V., Shlyapnikov,M.G., Semenova,L.M. and Solonin,A.S. (2000) DNA methylation at the CfrBI site is involved in expression control in the CfrBI restriction-modification system. Nucleic Acids Res., 28, 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alvarez M.A., Chater,K.F. and Rodicio,M.R. (1993) Complex transcription of an operon encoding the SalI restriction- modification system of Streptomyces albus G. Mol. Microbiol., 8, 243–252. [DOI] [PubMed] [Google Scholar]
- 72.Adams G.M. and Blumenthal,R.M. (1995) Gene pvuIIW: a possible modulator of PvuII endonuclease subunit association. Gene, 157, 193–199. [DOI] [PubMed] [Google Scholar]
- 73.Prakash-Cheng A. and Ryu,J. (1993) Delayed expression of in vivo restriction activity following conjugal transfer of Escherichia coli hsdK (restriction-modification) genes. J. Bacteriol ., 175, 4905–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Redaschi N. and Bickle,T.A. (1996) Posttranscriptional regulation of EcoP1I and EcoP15I restriction activity. J. Mol. Biol., 257, 790–803. [DOI] [PubMed] [Google Scholar]
- 75.Arber W. (1993) Evolution of prokaryotic genomes. Gene, 135, 49–56. [DOI] [PubMed] [Google Scholar]
- 76.Makovets S., Doronina,V.A. and Murray,N.E. (1999) Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc. Natl Acad. Sci. USA, 96, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 78.Stahl M.M., Kobayashi,I., Stahl,F.W. and Huntington,S.K. (1983) Activation of Chi, a recombinator, by the action of an endonuclease at a distant site. Proc. Natl Acad. Sci. USA, 80, 2310–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heitman J., Ivanenko,T. and Kiss,A. (1999) DNA nicks inflicted by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in Escherichia coli. Mol. Microbiol., 33, 1141–1151. [DOI] [PubMed] [Google Scholar]
- 80.McKane M. and Milkman,R. (1995) Transduction, restriction and recombination patterns in Escherichia coli. Genetics, 139, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.el Karoui M., Ehrlich,D. and Gruss,A. (1998) Identification of the lactococcal exonuclease/ recombinase and its modulation by the putative Chi sequence. Proc. Natl Acad. Sci. USA, 95, 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handa N., Ohashi,S., Kusano,K. and Kobayashi,I. (1997) Chi*, a chi-related 11-mer partially active in an E. coli recC* strain. Genes Cells, 2, 525–536. [DOI] [PubMed] [Google Scholar]
- 83.Arnold D.A., Handa,N., Kobayashi,I. and Kowalczykowski,S.C. (2000) A novel, 11-nucleotide variant of chi, chi*: one of a class of sequences defining the E. coli recombination hotspot, chi. J. Mol. Biol., 300, 469–479. [DOI] [PubMed] [Google Scholar]
- 84.Smith H.O., Gwinn,M.L. and Salzberg,S.L. (1999) DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol., 150, 603–616. [DOI] [PubMed] [Google Scholar]
- 85.Stahl F.W. (1998) Recombination in phage lambda: one geneticist’s historical perspective. Gene, 223, 95–102. [DOI] [PubMed] [Google Scholar]
- 86.Noirot P. and Kolodner,R.D. (1998) DNA strand invasion promoted by Escherichia coli RecT protein. J. Biol. Chem., 273, 12274–12280. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi N. and Kobayashi,I. (1990) Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc. Natl Acad. Sci. USA, 87, 2790–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi N.K., Sakagami,K., Kusano,K., Yamamoto,K., Yoshikura,H. and Kobayashi,I. (1997) Genetic recombination through double-strand break repair: shift from two-progeny mode to one-progeny mode by heterologous inserts. Genetics, 146, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kovall R.A. and Matthews,B.W. (1998) Structural, functional, and evolutionary relationships between lambda-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heitman J., Zinder,N.D. and Model,P. (1989) Repair of the Escherichia coli chromosome after in vivo scission the EcoRI endonuclease. Proc. Natl Acad. Sci. USA, 86, 2281–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kusano K., Sakagami,K., Yokochi,T., Naito,T., Tokinaga,Y., Ueda,E. and Kobayashi,I. (1997) A new type of illegitimate recombination is dependent on restriction and homologous interaction. J. Bacteriol ., 179, 5380–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janscak P., MacWilliams,M.P., Sandmeier,U., Nagaraja,V. and Bickle,T.A. (1999) DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J., 18, 2638–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsutakawa S.E. and Morikawa,K. (2001) The structural basis of damaged DNA recognition and endonucleolytic cleavage for very short patch repair endonuclease. Nucleic Acids Res., this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kulakauskas S., Barsomian,J.M., Lubys,A., Roberts,R.J. and Wilson,G.G. (1994) Organization and sequence of the HpaII restriction-modification system and adjacent genes. Gene, 142, 9–15. [DOI] [PubMed] [Google Scholar]
- 95.Choi S.H. and Leach,J.E. (1994) Identification of the XorII methyltransferase gene and a vsr homolog from Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet., 244, 383–390. [DOI] [PubMed] [Google Scholar]
- 96.Taron C.H., Van Cott,E.M., Wilson,G.G., Moran,L.S., Slatko,B.E., Hornstra,L.J., Benner,J.S., Kucera,R.B. and Guthrie,E.P. (1995) Cloning and expression of the NaeI restriction endonuclease-encoding gene and sequence analysis of the NaeI restriction-modification system. Gene, 155, 19–25. [DOI] [PubMed] [Google Scholar]
- 97.Tsutakawa S.E., Jingami,H. and Morikawa,K. (1999) Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell, 99, 615–623. [DOI] [PubMed] [Google Scholar]
- 98.Karlin S., Burge,C. and Campbell,A.M. (1992) Statistical analyses of counts and distributions of restriction sites in DNA sequences. Nucleic Acids Res., 20, 1363–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karlin S., Campbell,A.M. and Mrazek,J. (1998) Comparative DNA analysis across diverse genomes. Annu. Rev. Genet., 32, 185–225. [DOI] [PubMed] [Google Scholar]
- 100.Gelfand M.S. and Koonin,E.V. (1997) Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res., 25, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rocha E.P.C., Viari,A. and Danchin,A. (1998) Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res., 26, 2971–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rocha E.P., Danchin,A. and Viari,A. (2001) Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res., 11, 946–958. [DOI] [PubMed] [Google Scholar]
- 103.Hartl D.L. (1992) Population genetics of microbial organisms. Curr. Opin. Genet. Dev., 2, 937–942. [DOI] [PubMed] [Google Scholar]
- 104.Belfort M., Reaban,M.E., Coetzee,T. and Dalgaard,J.Z. (1995) Prokaryotic introns and inteins: a panoply of form and function. J. Bacteriol., 177, 3897–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yarmolinsky M.B. (1995) Programmed cell death in bacterial populations. Science, 267, 836–837. [DOI] [PubMed] [Google Scholar]
- 106.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Structure of the multimodular endonuclease FokI bound to DNA. Nature, 388, 97–100. [DOI] [PubMed] [Google Scholar]
- 107.Huai Q., Colandene,J.D., Chen,Y., Luo,F., Zhao,Y., Topal,M.D. and Ke,H. (2000) Crystal structure of NaeI — an evolutionary bridge between DNA endonuclease and topoisomerase EMBO J., 19, 3110–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 Å resolution. Nature Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 109.Bujnicki J.M. (2000) Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol., 50, 39–44. [DOI] [PubMed] [Google Scholar]
- 110.Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Daiyasu H., Komori,K., Sakae,S., Ishino,Y. and Toh,H. (2000) Hjc resolvase is a distantly related member of the type II restriction endonuclease family. Nucleic Acids Res., 28, 4540–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hickman A.B., Li,Y., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- 114.Nobusato A. (2001) Evidence from genome sequence comparison that restriction modification gene complexes are mobile genetic elements. PhD dissertation, University of Tokyo, Japan.
- 115.Gerdes K., Gultyaev,A.P., Franch,T., Pedersen,K. and Mikkelsen,N.D. (1997) Antisense RNA-regulated programmed cell death. Annu. Rev. Genet., 31, 1–31. [DOI] [PubMed] [Google Scholar]
- 116.Kleanthous C., James,R., Hemmings,A.M. and Moore,G.R. (1999) Protein antibiotics and their inhibitors. Biochem. Soc. Trans, 27, 63–67. [DOI] [PubMed] [Google Scholar]
- 117.Merrill C., Bayraktaroglu,L., Kusano,A. and Ganetzky,B. (1999) Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science, 283, 1742–1745. [DOI] [PubMed] [Google Scholar]
- 118.Snyder L. (1995) Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol., 15, 415–420. [DOI] [PubMed] [Google Scholar]
- 119.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 120.Parkhill J., Achtman,M., James,K.D., Bentley,S.D., Churcher,C., Klee,S.R., Morelli,G., Basham,D., Brown,D., Chillingworth,T. et al. (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature, 404, 502–506. [DOI] [PubMed] [Google Scholar]
- 121.Tettelin H., Saunders,N.J., Heidelberg,J., Jeffries,A.C., Nelson,K.E., Eisen,J.A., Ketchum,K.A., Hood,D.W., Peden,J.F., Dodson,R.J. et al. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science, 287, 1809–1815. [DOI] [PubMed] [Google Scholar]
- 122.Betlach M., Hershfield,V., Chow,L., Brown,W., Goodman,H. and Boyer,H.W. (1976) A restriction endonuclease analysis of the bacterial plasmid controlling the EcoRI restriction and modification of DNA. Fed. Proc., 35, 2037–2043. [PubMed] [Google Scholar]
- 123.Brassard S., Paquet,H. and Roy,P.H. (1995) A transposon-like sequence adjacent to the AccI restriction- modification operon. Gene, 157, 69–72. [DOI] [PubMed] [Google Scholar]
- 124.Vaisvila R., Vilkaitis,G. and Janulaitis,A. (1995) Identification of a gene encoding a DNA invertase-like enzyme adjacent to the PaeR7I restriction-modification system. Gene, 157, 81–84. [DOI] [PubMed] [Google Scholar]