Abstract
Cellular senescence is a mechanism that virtually irreversibly suppresses the proliferative capacity of cells in response to various stress signals. This includes the expression of activated oncogenes, which causes Oncogene-Induced Senescence (OIS). A body of evidence points to the involvement in OIS of chromatin reorganization, including the formation of senescence-associated heterochromatic foci (SAHF). The nuclear lamina (NL) is an important contributor to genome organization and has been implicated in cellular senescence and organismal aging. It interacts with multiple regions of the genome called lamina-associated domains (LADs). Some LADs are cell-type specific, whereas others are conserved between cell types and are referred to as constitutive LADs (cLADs). Here, we used DamID to investigate the changes in genome–NL interactions in a model of OIS triggered by the expression of the common BRAFV600E oncogene. We found that OIS cells lose most of their cLADS, suggesting the loss of a specific mechanism that targets cLADs to the NL. In addition, multiple genes relocated to the NL. Unexpectedly, they were not repressed, implying the abrogation of the repressive activity of the NL during OIS. Finally, OIS cells displayed an increased association of telomeres with the NL. Our study reveals that senescent cells acquire a new type of LAD organization and suggests the existence of as yet unknown mechanisms that tether cLADs to the NL and repress gene expression at the NL.
Cellular senescence is a virtually irreversible form of cell cycle arrest that occurs in response to diverse stress signals including telomere shortening, DNA damage, and oncogene expression. The latter is called Oncogene-Induced Senescence (OIS). OIS was first observed for an activated form of RAS, a cytoplasmic transducer of mitogenic signals (Serrano et al. 1997). Subsequently, other members of the RAS signaling pathway, like Raf-1, BRAF, and MEK (Lin et al. 1998; Zhu et al. 1998), were shown to cause senescence when overexpressed or expressed as oncogenic forms.
Work from several laboratories, including ours, demonstrated that this phenomenon, which was initially identified and characterized in vitro, acts as a robust tumor suppressive mechanism in vivo. For instance, we found that human melanocytic nevi (moles) harboring oncogenic mutant BRAFV600E display several hallmarks of senescence: durable lack of proliferation, increased expression of the tumor suppressor p16INK4a, and elevated senescence-associated β-galactosidase activity (Michaloglou et al. 2005). Concomitantly, OIS was demonstrated to occur in vivo also in response to a variety of other oncogenic mutations, an inactivated tumor suppressor, and in several types of premalignant lesions in human and different mouse models (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005). Together, these and many subsequent studies (Kuilman et al. 2010) demonstrated that OIS can effectively suppress progression of incipient cancer cells toward the malignant stage. Given the importance of OIS in limiting the tumorigenesis of human cancers, there is a crucial need to understand the mechanisms underlying this program.
Several studies pointed to a role for chromatin reorganization. For example, relocation of entire chromosomes relative to the nuclear periphery was observed in senescent cells (Bridger et al. 2000). Moreover, OIS is commonly accompanied by the accumulation of senescence-associated heterochromatic foci (SAHF), which correspond to condensed individual chromosomes (Narita et al. 2003; Zhang et al. 2007). These foci contain histone modifications and associated proteins characteristic of heterochromatin. They are thought to contribute to the onset of senescence by repressing the expression of proliferation-associated genes (Narita et al. 2003; Zhang et al. 2007). A detailed immunofluorescence microscopy analysis revealed that SAHF adopt a concentric organization with a central core enriched for compacted chromatin and H3K9me3, as well as a peripheral ring containing a more relaxed chromatin and an H3K27me3 mark (Chandra et al. 2012; Chandra and Narita 2013). The functional significance of this organization is unknown, however.
In recent studies, genome-wide approaches were applied to map different features of the senescent cell epigenome. Mapping of histone mark distribution identified large domains enriched for H3K4me3 and H3K27me3 in replicative senescent cells (Shah et al. 2013). In another study, FAIRE-seq analysis revealed a widespread change in the distribution of open and closed chromatin (De Cecco et al. 2013). Also, bisulfite-sequencing analysis identified large domains of hypomethylation and focal hypermethylation events in senescent cells, which resemble the methylome changes seen in cancer (Cruickshanks et al. 2013). Finally, a Hi-C study revealed a global change in the pattern of local chromatin interactions (Chandra et al. 2015). All these observations suggest that widespread changes occur in chromatin composition and 3D organization during senescence. However, it is still unclear whether and how these changes contribute to the establishment and maintenance of the senescent state.
Genome–nuclear lamina interactions are another important aspect of the spatial organization of the genome. The nuclear lamina (NL) is a fibrous multiprotein network lining the nucleoplasmic face of the inner nuclear membrane. Using the DamID technique, we have previously shown that genomes of human cells are organized into large lamina associated domains (LADs) (Guelen et al. 2008). The genes in LADs are generally expressed at low levels, and LADs are depleted for active histone marks such as acetylation of H3 and H4 and methylation of H3K4. Therefore, the NL is considered to be a repressive chromatin environment (Guelen et al. 2008; Kind and van Steensel 2010). We also previously described two classes of LADs—those that interact with the NL in all cell types, designated as constitutive LADs; and LADs that are cell-type specific, called facultative LADs (Meuleman et al. 2013).
Whether and how changes in LAD organization occur during senescence is largely unknown. We have addressed this question by using the DamID technology to generate and compare maps of genome–NL interactions of human proliferating, quiescent, and OIS cells.
Results
Massive changes in genome–nuclear lamina interactions occur during OIS
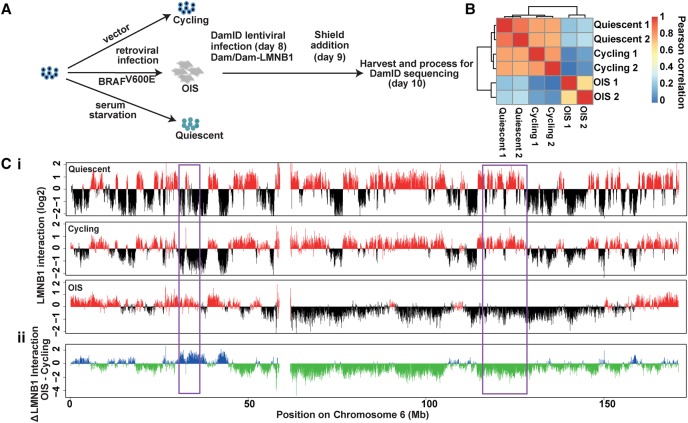
To map changes in LADs during OIS, we performed DamID for lamin B1 (LMNB1), a major constituent of the nuclear lamina (Peric-Hupkes et al. 2010). For this analysis, we used hTERT-immortalized Tig3 fibroblasts (Tig3ET), which were induced to senesce by ectopic expression of BRAFV600E. This is a well-defined OIS model that we used previously in several studies (Michaloglou et al. 2005; Kuilman et al. 2008; Kaplon et al. 2013). To express the Dam fusion proteins, we used the “ProteoTuner” inducible system, which is based on the fusion to a destabilization domain (DD). Dam-lamin B1 expression is induced by a small molecule (“Shield1”) that binds the DD, thereby preventing the targeting of Dam-lamin B1 for proteosomal degradation (Kind et al. 2013). Tig3ET cells were first transduced with retrovirus expressing either BRAFV600E or control empty vector. After a period of culturing to allow the cells to enter senescence, they were transduced with lentivirus encoding the Dam-lamin B1 fusion or unfused Dam and subsequently induced to express these proteins for 24 h. Genomic DNA was then subjected to DamID in combination with high-throughput sequencing (DamID-sequencing [DamID-seq]) (Fig. 1A).
Figure 1.
Massive changes in genome–nuclear lamina interactions occur during OIS. (A) Experimental setup. The time points indicated in the brackets correspond to days after BRAFV600E retroviral transduction or serum starvation. (B) Heat map representation of correlations of Dam-LMNB1 methylation ratios between the two biological replicates each for quiescent, cycling, and OIS cells. (C, i). Genome–NL interaction profile along Chromosome 6 for quiescent cells after 10 d of serum starvation, and cycling and OIS cells 10 d after retroviral transduction. The y-axis represents log2 of Dam-LMNB1 over Dam-only methylation ratio. (ii) Changes in LMNB1 interaction in OIS cells for Chromosome 6. Some regions showing changes in NL interactions are highlighted in purple boxes.
At the time of harvesting, cells displayed typical features of senescence, including proliferation arrest, induction of senescence-associated-β-galactosidase activity, and elevated p16INK4a expression (Supplemental Fig. S1A–C). Western blot analysis confirmed Dam-lamin B1 expression in BRAFV600E-senescent cells, albeit at a lower level compared to cycling cells (Supplemental Fig. S1D). In these conditions of expression, we were able to identify a substantial number of LADs in OIS cell (see below). We previously demonstrated that Dam-lamin B1 can be used to map NL interactions in both cycling and noncycling cells (Peric-Hupkes et al. 2010), and immunofluorescence confocal microscopy confirmed that DD-Dam-lamin B1 was correctly localized at the nuclear periphery both in cycling and OIS cells (Supplemental Fig. S1E).
Two independent DamID-seq experiments were sequenced at a combined depth of 49.9 million reads (with an average of 6.2 million reads/sample). The Pearson correlation coefficient between the biological replicates was 0.79 for cycling cells and 0.64 for OIS cells (Fig. 1B). Consistent with previous DamID data generated in Tig3 cells (Guelen et al. 2008), cycling Tig3ET cells display large chromosomal domains of high NL association that alternate with regions of low-level NL association. However, the pattern of Dam-lamin B1 interaction was dramatically altered in OIS cells, with some regions showing a loss of interaction and some a gain (Fig. 1C). Such changes occurred on all chromosomes (Supplemental Fig. S2). Therefore, OIS cells profoundly change their chromosome domain organization with respect to the NL. These changes cannot be accounted for by the occurrence of nuclear blebs and/or micronuclei, because we found the frequencies of these structures to be similar between cycling and OIS cells (Supplemental Fig. S3A,B). The observed changes in DamID profile can also not be caused by systematic karyotype changes, because the Dam-lamin B1 data are normalized to Dam-only data. Moreover, systematic changes in karyotype are unlikely to accumulate, considering the rapid cell cycle arrest that accompanies OIS.
We also generated a map of genome–NL interactions in quiescent cells. Remarkably, this profile was highly similar to that of proliferative cells (Fig. 1C; Supplemental Fig. S2). This high degree of similarity was also reflected by unsupervised hierarchical clustering, which grouped proliferative and quiescent cells in one major branch and OIS cells in another (Fig. 1B). This indicates that most of the changes seen in OIS cells are not explained by cell cycle exit per se, but are specific for the senescent state.
Validation of the DamID-seq profile of OIS cells
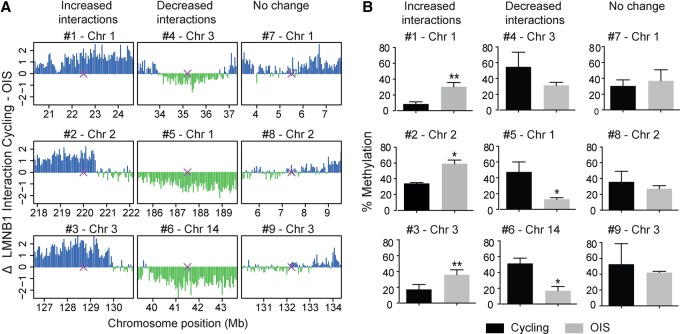
As DamID-seq provides only a relative estimate of the levels of methylation generated by Dam-LMNB1, we used the DpnII protection assay to quantify absolute differences in the level of Dam-LMNB1 mediated methylation at several loci (Kind et al. 2013). Briefly, we digested genomic DNA from cycling and OIS cells infected with Dam-lamin B1 with the methylation-sensitive enzyme DpnII, which cuts unmethylated but not Dam-methylated GATC sequences. This digest was assayed by real-time quantitative PCR with primers flanking sequences containing GATC. We selected nine loci, for which the DamID-seq results showed either an increase, decrease, or no change in NL interactions (Fig. 2A). The DpnII assay confirmed these results (Fig. 2B), indicating that increased and decreased DamID-seq signals may be interpreted as increased and decreased NL interactions, respectively.
Figure 2.
Validation of the changes in NL interactions at individual loci by DpnII assay. (A) Genomic regions showing either increased, decreased, or unchanged NL interactions in OIS cells selected for the DpnII assay. Purple crosses represent the location of the designed PCR primers. (B) Quantification by real-time PCR of Dam-LMNB1-mediated methylation at the loci presented in A in cycling and OIS cells. Results were generated from two independent replicates and are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01, based on a two-tailed paired t-test.
Loss of NL interactions during OIS affects primarily constitutive LADs
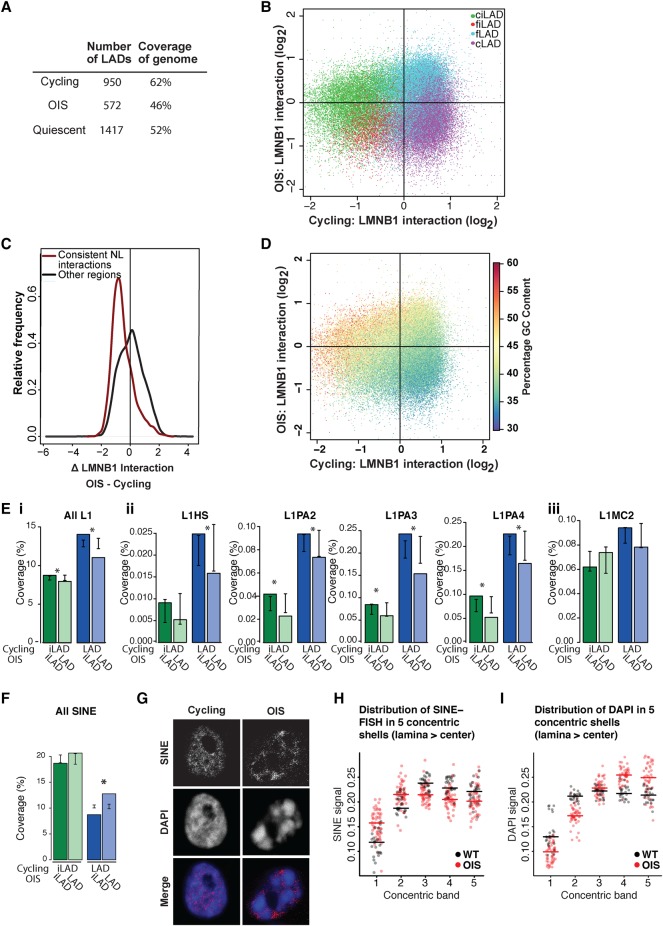
Inspection of the chromosomal profiles revealed that OIS cells exhibited very large regions losing NL interactions (Fig. 1A). Quantification showed that OIS cells displayed substantial decreases both in LAD number and in proportion of the genome covered with LADs, indicating an overall reduction in genome–NL interactions (Fig. 3A). We therefore investigated whether there were specific characteristics associated with the domains that were detaching from the NL in OIS cells.
Figure 3.
Loss of NL interactions during OIS affects primarily constitutive LADs. (A) LAD statistics in cycling, OIS cells, and quiescent cells. (B) Scatter plot representing the ratio of Dam-lamin B1 interaction within 50-kb windows for cycling (x-axis) and OIS cells (y-axis). The circles are colored according to their cLAD/ciLAD/fLAD/fiLAD status. cLADs (purple) show decreased NL binding in OIS cells. (C) Density plot of the change in LMNB1 interaction for consistent NL interactions compared to regions without consistent interactions. (D) Scatter plot showing LMNB1 binding ratio in cycling (x-axis) versus LMNB1 binding ratio in OIS cells (y-axis) in 50-kb windows. The circles are colored according to the percentage of GC content. (E,F) Repeat element coverage in LADs or iLADs in cycling cells after OIS: (green) repeat element coverage of iLADs in cycling cells that are still in iLADs in OIS cells (dark green) or moved to the NL in OIS cells (light green); (blue) repeat element coverage of LADs in cycling cells that detach from the NL in OIS cells (dark blue) or are still at the NL in OIS cells (light blue). (E) LINE-1 element coverage in LADs and iLADs: (i) all L1 elements; (ii) human- and primate-specific L1 repeats; (iii) L1MC2, a mammalian L1 repeat. Error bars indicate two standard deviations around mean L1 density for OIS LADs and iLADs randomly assigned to LAD or iLAD in OIS cells by circular permutations. Differences in iLAD and LAD L1 element density in OIS cells that was significantly different ([*] P < 0.05) to the expected density generated from random assignment of the LADs and iLADs in cycling cells. (F) SINE element coverage in LADs and iLADs. (G) Single z-section example of confocal images of cycling and OIS cells stained with a SINE probe (Alexa Fluor-555, grayscale). DNA was stained with DAPI. (H,I) Quantification of SINE (H) and DAPI (I) signal intensities in confocal sections, in five concentric shells having equal surface areas (shell 1 is closest to the NL, shell 5 is at the nuclear center). Each dot represents one nucleus; horizontal lines depict medians.
We previously reported that LADs can be divided into constitutive LADs (cLADs) that are conserved across cell types and facultative LADs (fLADs) that contact the NL in a cell-type–specific manner (Meuleman et al. 2013). We investigated whether there was any relationship between the cLAD/fLAD status and LAD reorganization during OIS. Because a cLAD/fLAD map is currently only available for mouse cells (Meuleman et al. 2013), we constructed one for human cells using previously published data from human ES cells, HT1080 fibrosarcoma cells (Kind et al. 2013), KBM7 chronic myelogenous leukemia cell line (Kind et al. 2015), and cycling Tig3 cells (this study). After identifying LADs in each of these four cell types, we divided them into two classes: cLADs that are NL-associated in all four cell types, and fLADs that are NL-associated in Tig3 cells, but do not associate with the NL in at least one of the other cell types. Likewise, we divided iLADs into ciLADs, which are not at the NL in any of the four cell types; and fiLADs, which are detached from the NL in Tig3 cells, but are NL-associated in at least one of the three other cell types.
We observed that the two classes of LADs behaved differently in OIS cells: whereas the majority of the cLADs had detached from the NL in OIS cells (79% of regions covered with cLADs move away from the NL after OIS), the fLADs mostly remained at the NL (72% of regions covered with fLADs remain at the NL after OIS) (Fig. 3B). We conclude from these observations that OIS cells undergo a preferential loss of constitutive genome–NL interactions.
Using single-cell DamID, we recently identified a class of LADs that are at the NL in almost every single cell. These stable LADs largely overlap with constitutive LADs (Kind et al. 2015). We found that 80% of these regions with consistent NL interactions in cycling cells had moved further away from the NL upon OIS compared to 52% of regions without consistent NL interactions (Fig. 3C). Therefore, the LADs that are not only invariant across cell types but also between individual cells appear to be the ones that preferentially detach from the NL during OIS.
DNA in cLADs has a low GC content (high AT content) (Meuleman et al. 2013). Accordingly, we found that the most pronounced loss of NL interaction in OIS cells occurred in regions with low GC content, whereas the regions with the strongest gain in NL interactions tended to be GC-rich (Fig. 3D). Our findings are consistent with, and extend, the previously reported correlation between GC content and LAD changes in the context of RASV12-induced senescence (Sadaie et al. 2013).
Long interspersed nuclear elements (LINEs) are enriched at the NL in murine cells (Meuleman et al. 2013). We also found that LINE-1 elements are enriched at the NL in cycling Tig3 cells (Fig. 3Ei). Along with the massive changes in genome localization occurring in OIS, we observed the opposite in this setting: the LADs from cycling cells detaching from the NL in OIS cells showed a significantly higher L1 density than those that remained at the NL, whereas iLADs from cycling cells remaining detached after OIS had a significantly higher L1 density than those that moved to the NL. We then looked more closely at the more recent L1 elements: L1HS (found specifically in humans) and L1PA2, L1PA3, and L1PA4 (primate-specific) (Fig. 3Eii). For each of these subtypes, LINE-1 elements in LADs in cycling cells were significantly more likely to detach from the nuclear lamina after OIS. We also observed that the iLADs in cycling cells were significantly more likely to remain detached from the NL after OIS in all of these L1 subtypes except L1HS. This appears to be specific to the primate- and human-specific L1 elements, because L1MC2, a mammalian L1 element, failed to show a significant shift away from the nuclear lamina in OIS cells.
This was paralleled by short interspersed nuclear elements (SINEs) moving in OIS cells in the opposite direction compared to LINEs (Fig. 3F–H). It thus seems that the constitutive iLADs move toward the nuclear periphery in OIS cells, which is in agreement with the DamID data and DAPI distribution. That this analysis is specific for the SINE signal is shown by a comparative analysis, based on the DAPI signal, which does not show this pattern (Fig. 3I).
Loss of gene repression at the NL during OIS
In addition to the widespread loss of NL interaction, we noted that some regions moved closer to the NL in OIS cells (Fig. 1C; Supplemental Fig. S2). This was the case for iLADs, which again displayed a differential behavior according to their constitutive or facultative status. Specifically, ciLADs displayed a strong trend toward increased NL association in OIS cells, in contrast to fiLADs, which mainly remained away from the lamina (Fig. 3B). Thus, regions that do not even associate with the NL in other (nonsenescent) cell types showed a preference to contact the NL in OIS cells. A correlation with GC content was apparent here also, with GC-rich regions showing a trend toward increased interactions (Fig. 3D).
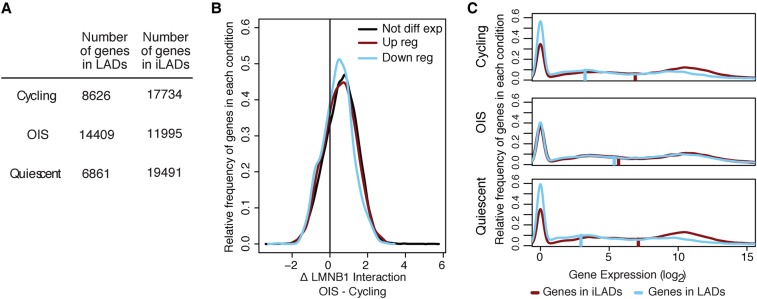
GC-rich regions are generally more gene dense (Mouchiroud et al. 1991; Zoubak et al. 1996). Therefore, we predicted that genic regions would move toward the NL in OIS cells. Indeed, as previously reported (Guelen et al. 2008), LADs in cycling cells tended to be depleted of genes, containing only a third of them (Fig. 4A). This was also the case for quiescent cells. In contrast, this proportion increased in OIS cells, such that more than half of all genes were at the NL.
Figure 4.
Loss of gene repression at the NL in OIS cells. (A) Quantification of the number of genes in LAD and iLADs in cycling, OIS cells, or quiescent cells. Genes were assigned to LAD or iLAD based on which state their transcription start site fell into for each cell condition. For each cell type, a small number of genes fell into regions we were unable to assign a state to; those genes were censored. (B) Distribution of log2 fold change in LMNB1 binding for genes showing up-regulation, down-regulation, or unchanged expression in OIS cells. Genes were considered not differentially expressed if they had an expression fold change <1 on a log2 scale. Genes were differentially expressed if they had a fold change >2 on a log2 scale and an adjusted P-value <0.05. (C) Density plots of expression levels of genes located within LADs or in inter-LADs (iLADs) in cycling, OIS cells, or quiescent cells. Gene expression data in the different conditions were obtained by RNA-sequencing (at day 10 post BRAFV600E retroviral introduction or serum starvation). The vertical bars on each plot represent the median gene expression level of each condition.
Given the previously documented role of the NL in gene repression (Guelen et al. 2008; Kind and van Steensel 2010), we next wished to explore a possible relationship between these altered gene–NL associations and changes in the transcriptome after OIS induction. For this, we generated gene expression profiles of cycling, OIS, and quiescent cells using RNA-sequencing. We first compared the distribution of NL interaction changes between up-, down-, and nonregulated genes in OIS cells. For all three gene groups, we observed a shift toward the NL of a similar magnitude (Fig. 4B). Thus, surprisingly, increased association of genes with the NL is not linked to changes in their expression status.
We then examined the expression level by LAD/iLAD status in the three conditions. Consistent with our prior work (Guelen et al. 2008), the genes outside LADs (i.e., those in iLADs) were more highly expressed than genes inside LADs in cycling cells (4.9-fold higher mean expression), as well as in quiescent cells (6.5-fold higher mean expression) (Fig. 4A). However, this difference in expression was virtually lost in OIS cells (1.2-fold increase in mean expression) (Fig. 4C). This indicates that the genes moving toward the NL in OIS cells do not become repressed. Therefore, there appears to be no global relationship between differential NL interactions and differential gene expression during OIS. This is in agreement with a previous study reporting lack of correlation between changes in lamin B1 interactions and changes in gene expression (Sadaie et al. 2013). In addition, the complete loss of the difference in gene expression between LADs and iLADs in OIS (Fig. 4B) suggests that the mechanisms responsible for gene repression at LADs are abolished in OIS cells.
Relocalization of telomeres and centromeres in OIS cells
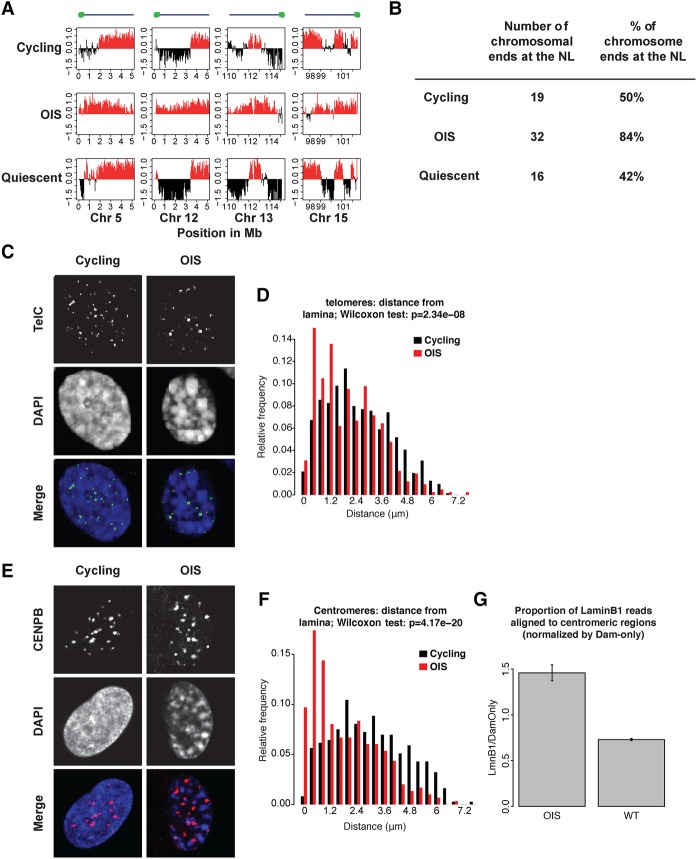
The DamID data reveal that chromosomal extremities often exhibit increased NL interactions (Fig. 5A). When considering the last 2 Mb of each chromosomal end, the DamID sequencing data revealed a dramatic increase in the percentage of chromosomal extremities associating with the NL (Fig. 5B). To confirm this result, we assessed telomere localization by telomere fluorescence in situ hybridization (FISH). In OIS cells compared to cycling cells, we detected an enrichment of telomere signals at the nuclear periphery (Fig. 5C,D). Of note, fixation and subsequent treatment of cells did not cause loss of cellular architecture by artificial flattening, as demonstrated by the appearance of a different spatial localization of telomeres throughout consecutive z-layers (Supplemental Fig. S4).
Figure 5.
Telomeric and centromeric regions shift toward the NL in OIS cells. (A) Increased nuclear lamina association of chromosomal ends in OIS cells compared to cycling and quiescent cells in the 2-Mb region closest to the end of the chromosome. The y-axis shows log2 Lamin B1 binding ratio for each graph. (B) Quantification of the frequency of interaction of chromosomal extremities as defined in A in cycling, OIS, and quiescent cells. Only the 38 chromosome ends for which a sequence assembly is available were analyzed. (C) Single z-section example of confocal images of cycling and OIS cells stained by FISH with a telomere probe (FITC, grayscale). DNA was stained with DAPI. (D) Distribution of distances of telomere signals to the NL in cycling and OIS cells. Data are from two independent experiments. Number of nuclei counted: 59 (OIS), 61 (cycling); total number of telomere signals counted: 417 (OIS), 740 (cycling). The difference in the distribution of the telomere distance between cycling and OIS cells was statistically significant (P-value = 2.34 × 10−8, Wilcoxon test). (E) Centromeres labeled with a CENP-B antibody (Alexa Fluor-568, grayscale). Single z-section examples of confocal images are shown. DNA was stained with DAPI. (F) Distribution of centromeres determined as in D. Number of nuclei counted: 55 (OIS), 49 (cycling); total number of centromere signals counted: 299 (OIS), 373 (cycling). The distributions differ statistically significantly (P-value = 4.17 × 10−20, Wilcoxon test). (G) Average DamID signals in centromere-proximal regions are stronger in OIS cells compared to cycling cells (P = 0.0067, Welch two-sample t-test; data based on two independent DamID experiments).
A similar analysis of nuclei labeled with a CENP-B antibody showed that centromeres in OIS cells also tend to move toward the NL (Fig. 5E,F); likewise, we observed that centromeric regions exhibit stronger DamID signals in OIS cells (Fig. 5G). Therefore, in addition to providing a microscopy validation of our DamID-seq data, these results indicate that the telomeres and centromeres shift toward a more peripheral localization in OIS cells.
Loss of LBR is not sufficient to induce OIS-like changes in NL interactions
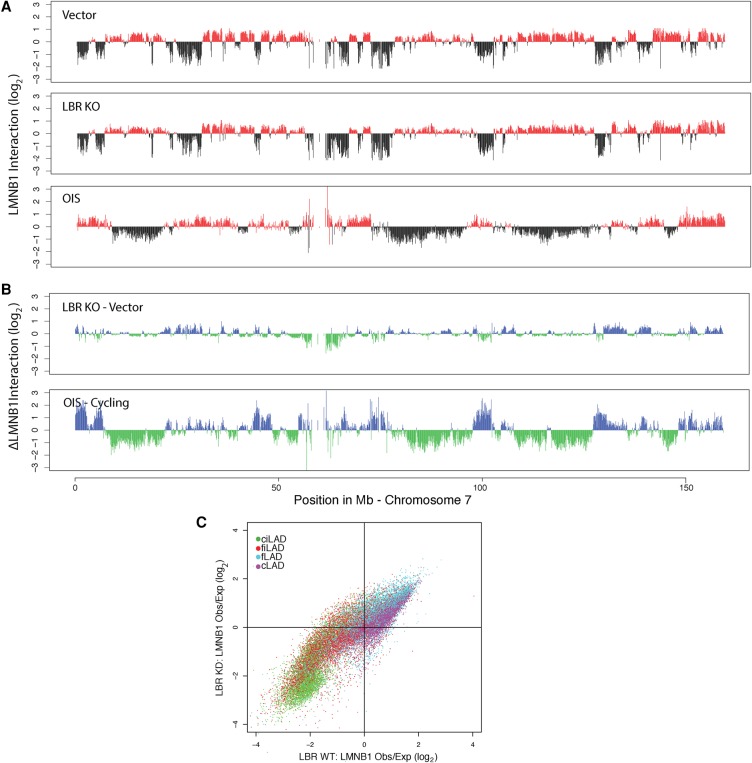
The lamin B receptor (LBR) has been implicated in the peripheral positioning of heterochromatin in mouse cells (Clowney et al. 2012; Solovei et al. 2013) and is down-regulated in OIS (Lenain et al. 2015). To test whether loss of LBR can account for the changes in NL interactions as observed by DamID, we created a LBR knockout (KO) cell line by CRISPR/Cas9 technology (Methods; Maresca et al. 2013). For this we chose human HAP1 cells, in which the haploid genome simplifies the engineering of KO cells (Burckstummer et al. 2013). Strikingly, DamID experiments revealed that the LBR KO cells exhibit a genome–NL interaction pattern that is comparable to wild-type cells (Fig. 6A), with essentially no detachment of cLADs from the NL (Fig. 6B). In particular, loss of LBR does not replicate the movement of the ciLADs toward the lamina that is observed in senescence (Figs. 3B, 6C). We conclude therefore that loss of LBR alone does not induce the massive alterations in NL interactions as we observe in OIS cells.
Figure 6.
Deletion of LBR does not cause OIS-like changes in NL interactions in cycling human HAP1 cells. (A) LMNB1 DamID profiles along Chromosome 7 for wild-type and LBR KO cycling human HAP1 cells. (B) Changes in LMNB1 interaction after LBR KO (top). For comparison, the changes in NL interactions upon OIS (as in Fig. 1C) are shown (bottom). (C) Scatter plot of the ratio of Dam-LMNB1 interaction within 100 kb windows for wild-type (x-axis) and LBR KO cells (y-axis). The circles are colored according to their cLAD/ciLAD/fLAD/fiLAD status.
Discussion
Although several lines of evidence point to a spatial reorganization of the chromatin during senescence, it has remained unclear whether these changes are associated with alterations in genome–NL interactions. Using DamID-sequencing for lamin B1 in cells undergoing senescence following the expression of the common oncogene BRAFV600E, we found that OIS cells undergo a dramatic reorganization of genome–NL interactions. Our results are consistent with previous work reporting changes in lamin B1 genome-binding distribution during RASV12-induced senescence (Sadaie et al. 2013). This altered genome–NL interaction profile is likely to reflect a global spatial repositioning of the genome that is also manifested by SAHF formation (Chandra et al. 2012; Chandra and Narita 2013).
We found a widespread loss of NL contacts affecting preferentially the AT-rich, constitutive LAD compartment. These cLADs are also rich in LINE elements; at present, it is unknown whether AT-content or LINE elements play an active role in the positioning of cLADs. cLADs are likely to play an important role in genome function given the high degree of their conservation across species and cell types (Meuleman et al. 2013). Therefore, it is striking that almost all cLADs detached from the NL during OIS. This suggests that constitutive genome–NL interactions are governed by a (yet to be molecularly defined) distinct molecular mechanism that appears to be specifically lost during OIS.
It is remarkable that some of the epigenomic alterations previously identified in senescent cells, including large domains of H3K4me3 and H3K27me3 enrichment (Shah et al. 2013) and DNA hypomethylation (Cruickshanks et al. 2013), overlap largely with LADs in normal cells. This suggests that LADs are relatively more prone to undergo these epigenetic changes during the transition to the senescent state. It will be of interest to determine whether those domains that acquire a different chromatin composition correspond to the cLADs for which we observed detachment from the NL in OIS cells.
Another senescence-associated chromatin alteration that has been linked to LADs during senescence is a change in the pattern of interactions within topological domains. Indeed, a recent Hi-C study revealed that OIS cells exhibit loss of interactions within topologically associating domains (TADs) and an increased degree of interactions between TADs. This change was predominantly observed for LADs harboring a lower GC content (Chandra et al. 2015). Furthermore, it has been shown by microscopy that H3K9me3 and late-replicating DNA form the core of SAHFs (Chandra et al. 2012). This, together with our observation that cLADs detach from the NL, suggests that cLADs cluster together within the nuclear interior, probably contributing to SAHF formation.
We also show that the proportion of genes present at the NL significantly increases during OIS. Surprisingly, this does not appear to be accompanied by their repression. In normal cells, genes in LADs are generally repressed and for this reason, attachment to the NL is thought to generate a repressive environment (Guelen et al. 2008; Kind and van Steensel 2010). Therefore, it appears that the repressive mechanisms operating at the NL are no longer functional in OIS cells. The reason for this remains to be determined, but one possible explanation is an uncoupling between changes in NL interactions and those in chromatin composition. Indeed, previous work using ChIP-sequencing for multiple repressive histone marks failed to identify substantial changes in the overall pattern of their distribution in RASV12-induced senescent cells (Chandra et al. 2012), while major changes in lamin B1 interaction were found in the same setting (Sadaie et al. 2013).
The remodeling of nuclear architecture in OIS cells is remarkably reminiscent of the inverted chromatin organization described in retinal rod cells of nocturnal mammals (Solovei et al. 2009, 2013). In these cells, gene-poor, L1-rich heterochromatin coalesces into a large chromocenter in the nuclear interior, which can be paralleled with the clustering of the same regions into SAHF during OIS. Similarly to OIS cells, genes—whether silent or active—are localized at the nuclear periphery of rod cells, reflecting an uncoupling between transcriptional status and nuclear positioning (Solovei et al. 2009). Interestingly, this rod cell phenotype is associated with the absence of lamin A and LBR expression; concomitant deletion of the LMNA and LBR genes in nonrod cells results in chromatin inversion (Solovei et al. 2013). We recently showed that OIS cells down-regulate several NE proteins, including lamin A and LBR (Lenain et al. 2015). Our data in HAP1 cells indicate that loss of LBR alone is insufficient for the reshaping of NL interaction during OIS. An interesting possibility is that OIS and mouse rod cell differentiation use the same mechanism for spatial reorganization of the genome. One difference, however, is the positioning of centromeres. In rod cells, centromeres are preferentially located in the nuclear interior; in OIS, we observe them to move toward the nuclear periphery. At present, we cannot explain this difference, but it may be related to the fact that mouse chromosomes are acrocentric, whereas most human chromosomes are not.
Finally, we found that OIS cells display an altered positioning of the telomeres within the nucleus, with a shift toward the nuclear periphery. Telomere enrichment at the NL has been observed in two other physiological contexts also: in meiosis and immediately after mitosis (Ding et al. 2007; Crabbe et al. 2012). It remains to be determined whether a common mechanism drives peripheral telomere position in these different situations. The relocalization of telomeres to the NE could have an impact on telomere maintenance. This is supported by the observation that loss of lamin A in mouse cells results in telomere accumulation at the nuclear periphery that is accompanied by their shortening, a change in their chromatin composition and a reduction in telomeric noncoding RNA (TERRA) expression (Gonzalez-Suarez et al. 2009). Of note, it has been reported that oncogene expression, including BRAFV600E, results in DNA replication stress preferentially affecting telomeres and rapid telomere shortening (Suram et al. 2012). Future study should shed light on whether and how this oncogene-driven telomere dysfunction is mechanistically related to telomere relocation.
In conclusion, using DamID, we mapped the changes in genome–NL interactions during OIS. Our study reveals that OIS cells dramatically alter their chromosomal organization relative to the NL, consisting mostly of loss of constitutive interactions. Investigation of the molecular mechanisms driving these changes and their functional significance provides critical information about the principles of genome organization and function in normal and senescent cells, with implications for our understanding of aging and tumor suppression.
Methods
Plasmids and virus production
The retroviral vectors pBabe-puro and pBabe-puro-BRAFV600E were used to overexpress the oncogene BRAFV600E. The lentiviral plasmids pCCL-hPGK-DD-Dam-V5/Wpre (DD-Dam) and pCCL-hPGK-DD-Dam-V5-hLMNB1/Wpre (DD-Dam-LMNB1) that encode the Shield1-inducible unfused Dam or Dam-lamin B1 were used for DamID experiments. Retroviruses were produced in the Phoenix packaging cell line and lentiviruses in HEK293T using Polyethylenimine (PEI).
Cell culture
For all the senescence experiments, we used the human diploid fibroblast cell line Tig3 expressing the ecotropic receptor to allow infection with ecotropic retroviruses and hTERT to prevent replicative senescence (Tig3ET). Cells were propagated in DMEM (GIBCO) supplemented with 9% fetal bovine serum.
OIS was induced by infecting Tig3ET with retroviruses encoding oncogene BRAFV600E as described (Michaloglou et al. 2005). Tig3ET cells were made quiescent by culturing them in 0% FBS DMEM medium. Eight days after retroviral infection (a time point at which cells have entered senescence) or after serum starvation, cells were transduced with the Dam or Dam-lamin B1-encoding lentiviruses. Twenty-four hours after transduction, cells were refreshed with medium supplemented with 0.5 µM Shield (Clontech). After 24 h of induction, cells were harvested and processed for DamID sequencing. These experiments were repeated, resulting in n = 2 for each condition.
LBR knockout cell line
HAP1 cells were cultured in IMDM 1× (Gibco) supplemented with 10% fetal bovine serum (Thermo Scientific HyClone), 1% penicillin, and 1% streptomycin. We first established a HAP1 clone stably expressing a Shield1-inducible Dam-LMNB1 as well as the Fucci cell cycle reporters by simultaneous lentiviral transductions of the previously described hPGK-DD-Dam-LMNB1 and Fucci constructs (Sakaue-Sawano et al. 2008; Kind et al. 2015). Cells were transduced with both lentiviruses simultaneously in the presence of 8 µg/mL of polybrene and clonally selected based on the expression of both Fucci markers and the expression of the Dam-LMNB1 by screening for Dam methylation levels as determined by DpnII-qPCR assays (Kind et al. 2013). Based on this, we selected clone #5F5. Next, we established a LBR knockout subclone of #5F5 using CRISPR/Cas9 technology in combination with the ObLiGaRe system for selection (Maresca et al. 2013). In brief, a sgRNA expression vector targeting exon 1 of LBR was designed using an online CRISPR design tool (http://chopchop.cbu.uib.no). Oligonucleotides CACCGGCCGATGGTGAAGTGGTAAG and AAA CCTTACCACTTCACCATCGGCC were annealed and ligated into pX330 (Addgene plasmid #42230), yielding plasmid pX330-LBR2. This plasmid drives expression of the sgRNA as well as Cas9. We then transfected #5F5 cells in a 24-well plate with 3 µg plasmid pX330-LBR2, 1 µg 2A-Blas plasmid for insertion of the Blasticidin cassette, and 6 µL FuGENE6 (Promega #E2691) according to the manufacturer's protocol. After 48 h, the cells were plated on 10 cm dishes and selected for 14 d with 10 µg/mL Blasticidin. Colonies were picked and screened for the correct integration event by PCR using a primer binding in the Blasticidin cassette (GACATGGTGCTTGTTGTCCTC) together with an LBR-specific primer (GTAGCCTTTCTGGCCCTAAAAT). PCR products from the positive clones were sequence verified by Sanger sequencing. This screen yielded LBR knockout clone #5F5-LBR2-5. The sequence of the junction of the LBR gene and the inserted marker gene is: GGTTAATTATAGAAAATGCCAAGTAGGAAATTTGCCGATGGTGAAGTGGTTGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTG, with the first 50 nucleotides corresponding to Chr 1: 225611743–225611792 (hg19 coordinates; ending in exon 2 of LBR) and the remaining nucleotides corresponding to part of the inserted plasmid region. RT-qPCR with two sets of primers downstream from LBR exon 1 (set 1: CCGTGAATTAAACCCTCGAA and ACGCTTCCTCATTCCAGAGA; set 2: TTTGGGGTCTACCTCCTGTG and TCTACGCCCTGGAAGAGAGA) showed that expression of the remaining (mutated) mRNA in this clone was reduced to ∼3.5% of that in wild-type HAP1 cells. Haploidy and proper expression of the Fucci markers were confirmed in clone #5F5-LBR2-5 by FACS.
Cell proliferation and senescence assays
Cells were labeled for 3 h with BrdU (Roche), followed by fixation in 70% ethanol. After incubation with 0.1 mg/mL RNase A (Roche) and denaturation in 2 M HCL/0.2% Triton X-100, immunostaining for BrdU was performed using anti-BrdU antibody (DAKO) and FITC-conjugated anti-mouse antibody (DAKO). DNA was stained with propidium iodide. Samples were analyzed for fluorescein and propidium iodide on a BD Biosciences FACSCalibur flow cytometer. SA-β-galactosidase was detected using the Senescence Associated β-galactosidase staining kit (Cell Signaling) at pH6, following the manufacturer's instructions.
DamID microarray data
We generated new DamID maps of lamin B1 in cycling Tig3 cells with both microarray and sequencing technology. Using DamID sequencing, we also generated lamin B1 profiles of Tig3 cells in OIS and quiescence and LBR knockout and control profiles lamin B1 in HAP1 cells. Microarray samples were processed and hybridized to the human whole-genome design as described (Meuleman et al. 2013).
DamID sequencing
Genomic DNA was isolated using the using Gentra Puregene Cell Kit according to the manufacturer's instructions. To generate and amplify the Dam-methylated DNA fragments, we used a modified version of the procedure described (Vogel et al. 2007). Details are in Supplemental Methods; R code for processing DamID sequencing reads is available as Supplemental Code.
Assignment of NL interaction status
A hidden Markov model (HMM) was used to assign a state to each normalized bin using a two state model as described (Meuleman et al. 2013). State calls were derived through the Viterbi algorithm. Bins in the “bound” state were interpreted as overlapping LADs, and bins in the “unbound” state were interpreted as overlapping iLADs. The number of domains was calculated by the number of consecutive runs of the “bound” or “unbound” state, uninterrupted by bins without any data.
DpnII assay
The DpnII assay is a PCR-based method for quantifying the amount of A6-methylation in cells infected with the DamID constructs (Kind et al. 2013). Details are in Supplemental Methods.
Facultative and constitutive LAD definitions
Microarray LMNB1 DamID data from Tig3 cells, ESCs (Meuleman et al. 2013), HTC75 (a human HT1080 line which stably expresses the Tet-off) (van Steensel and de Lange 1997), and KBM7 (Kind et al. 2015) were used in our definition of facultative and constitutive LADs. Probes were averaged into the same windows as the OIS DamSeq data, and the HMM procedure as described above was used to find states for each window; those that were in perfect agreement were termed “constitutive” and the rest “facultative.” The facultative regions were split into two groups based on their status in our cell type of interest: cycling Tig3 cells (fLADs), those that were at the NL in Tig3 cells; and fiLADs, those that were away from the NL in Tig3 cells. We additionally developed a model of facultative LADs in the wild-type HAP1 cells used in the LBR knockout experiment, with the HMM procedure being applied to the observed to expected ratios of reads in each window. These states were combined with the microarray-based fLAD data to define the facultative LADs for the HAP1 genome.
Regions with consistent interactions with the NL
We used the regions identified in Kind et al. (2015) in 1N KBM7 cells (clone 14) with consistent interactions with the NL. These are 100-kb regions that have more interaction with LMNB1 than expected by chance in 80% of the single cells that were analyzed. We then binned our OIS DamID data into equivalent 100-kb windows to look at the change in LMNB1 interaction after OIS.
NL localization of LINE1 elements
LINE elements were obtained from the UCSC Table Browser. Coverage of LAD and iLAD regions was obtained by calculating the overlap of the 50-kb regions as defined previously with L1 elements. We split the genome into regions that were LADs and iLADs in cycling cells. Error bars for L1 element density in OIS cells were then calculated by performing 1000 random circular permutations to randomly assign these regions into two groups based on the observed distribution of the regions as LADs and iLADs in OIS cells, calculating the average L1 density and reporting two standard deviations. The significance of the difference between density of regions that ended up in LADs versus iLADs was calculated using the difference in the density of L1 elements assigned to LADs and iLADs in the random circular permutations.
SINE probe design and SINE FISH
SINE elements were PCR-amplified from human genomic DNA with primers based on the Alu consensus from http://bioinfo.ut.ee/cgi-bin/primer3-0.4.0/cat_humrep_and_simple.cgi, resulting in a ±230-bp PCR product. Details are in Supplemental Methods.
Distribution of SINEs and DAPI relative to nuclear lamina
SINEs are ubiquitous over the entire genome, and the SINE-FISH signal is present in the entire nucleus. We used the single confocal section through the approximate center of the nucleus to determine the distribution of SINEs. We identified the nuclear lamina as above. The midsection was divided into five concentric bands of equal area. We summed the SINE-FISH signal in each of the five bands and divided that by the total SINE-FISH signal in the section. For the distribution of DAPI signal, we used the same method.
RNA-sequencing
RNA-seq libraries on biological triplicate vector and BRAFV600E-transduced Tig3 cells and senescent Tig3 cells were generated from 1 µg total RNA using the Illumina TruSeq RNA Sample Preparation kit, following the manufacturer's instruction. Libraries were single-end sequenced on the Illumina HTSeq 2000 platform. The reads were aligned to the human genome build GTC37.75 using TopHat 2.012 (Kim et al. 2013). HTSeq-count (Anders et al. 2015) was used to determine expression per gene using the default union mode. Differentially expressed genes were determined in R using edgeR (Robinson et al. 2010) and voom (Law et al. 2014) and were considered differentially expressed if they had a P-value <0.05 after Benjamini-Hochberg adjustment (Hochberg and Benjamini 1990). Relative expression of fragments per kilobase per million reads (FPKM) of each gene was calculated by normalizing the reads for each gene-by-gene length (kb) and library size (million reads).
Telomere interaction with the lamina
Telomere locations were downloaded from the UCSC Table Browser. Two-megabase regions adjacent to telomeres were filtered to only those that were more than 2 Mb from a centromere and which were composed of more than 50% bins containing mappable reads. This left 38 telomeric regions. The average lamin B1 binding score across this region was calculated and positive scores were considered to be at the NL.
Data access
Genome-wide DamID and RNA-seq data from this study have been submitted to the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE76605.
Supplementary Material
Acknowledgments
We thank the NKI Genomic Core Facility for sequencing of the DamID and RNA samples, Anita Pfauth and Frank van Diepen in assisting with flow cytometry, and Lenny Brocks and Lauran Oomen for technical assistance in using the microscopy facility. This work was supported by a Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO-ALW) open program grant 822.02.001 (C.L., D.S.P.), a European Molecular Biology Organization (EMBO) Long Term Fellowship (C.A.dG.), a National Health and Medical Research Council (NHMRC) early career fellowship (C.A.dG.), and European Research Council (ERC) Advanced Grant 293662 (B.vS).
Author contributions: C.L. and C.A.dG. conceived and designed the study. C.L. performed most experiments, with additional data and reagents generated by N.L.V., M.dH., S.S.dV., and D.P.H. C.A.dG. performed the bioinformatics analyses of the DamID and RNA-sequencing data. L.P. carried out the analysis of microscopic and centromere DamID data. D.S.P. and B.vS. also conceived the study and analyzed the data along with the lead authors. C.L., C.A.dG., N.L.V., D.S.P., and B.vS. wrote the manuscript. All authors read and approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.225763.117.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol 10: 149–152. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Banning C, Hainzl P, Schobesberger R, Kerzendorfer C, Pauler FM, Chen D, Them N, Schischlik F, Rebsamen M, et al. 2013. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods 10: 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Narita M. 2013. High-order chromatin structure and the epigenome in SAHFs. Nucleus 4: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. 2012. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Ewels PA, Schoenfelder S, Furlan-Magaril M, Wingett SW, Kirschner K, Thuret JY, Andrews S, Fraser P, Reik W. 2015. Global reorganization of the nuclear landscape in senescent cells. Cell Rep 10: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. 2012. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Cesare AJ, Kasuboski JM, Fitzpatrick JA, Karlseder J. 2012. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep 2: 1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks HA, McBryan T, Nelson DM, Vanderkraats ND, Shah PP, van Tuyn J, Singh Rai T, Brock C, Donahue G, Dunican DS, et al. 2013. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol 15: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12: 863–872. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, et al. 2009. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J 28: 2414–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9: 811–818. [DOI] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. 2013. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498: 109–112. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome–nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. 2010. The essence of senescence. Genes Dev 24: 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenain C, Gusyatiner O, Douma S, van den Broek B, Peeper DS. 2015. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis 36: 1263–1274. [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12: 3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Lin VG, Guo N, Yang Y. 2013. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. 2013. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D, D'Onofrio G, Aïssani B, Macaya G, Gautier C, Bernardi G. 1991. The distribution of genes in the human genome. Gene 100: 181–187. [DOI] [PubMed] [Google Scholar]
- Narita M, Nuñez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Pérez-Mancera PA, Bennett DC, Chong H, et al. 2013. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602. [DOI] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. [DOI] [PubMed] [Google Scholar]
- Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL, et al. 2012. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J 31: 2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743. [DOI] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. 2007. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2: 1467–1478. [DOI] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2343–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 12: 2997–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubak S, Clay O, Bernardi G. 1996. The gene distribution of the human genome. Gene 174: 95–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.