Abstract
DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase complex composed of a catalytic subunit (DNA-PKcs) and KU70/80 heterodimer bound to DNA. DNA-PK holoenzyme plays a critical role in non-homologous end joining (NHEJ), the major DNA repair pathway. Here, we determined cryo-electron microscopy structure of human DNA-PK holoenzyme at 6.6 Å resolution. In the complex structure, DNA-PKcs, KU70, KU80 and DNA duplex form a 650-kDa heterotetramer with 1:1:1:1 stoichiometry. The N-terminal α-solenoid (∼2 800 residues) of DNA-PKcs adopts a double-ring fold and connects the catalytic core domain of DNA-PKcs and KU70/80-DNA. DNA-PKcs and KU70/80 together form a DNA-binding tunnel, which cradles ∼30-bp DNA and prevents sliding inward of DNA-PKcs along with DNA duplex, suggesting a mechanism by which the broken DNA end is protected from unnecessary processing. Structural and biochemical analyses indicate that KU70/80 and DNA coordinately induce conformational changes of DNA-PKcs and allosterically stimulate its kinase activity. We propose a model for activation of DNA-PKcs in which allosteric signals are generated upon DNA-PK holoenzyme formation and transmitted to the kinase domain through N-terminal HEAT repeats and FAT domain of DNA-PKcs. Our studies suggest a mechanism for recognition and protection of broken DNA ends and provide a structural basis for understanding the activation of DNA-PKcs and DNA-PK-mediated NHEJ pathway.
Keywords: Cryo-EM structure, DNA-PK, DNA-PKcs, activation, NHEJ
Introduction
DNA double-strand breaks (DSBs) can be induced by intrinsic or extrinsic stimuli and may lead to genomic instability, tumorigenesis and other cellular abnormalities1. In mammalian cells, DSBs are primarily repaired by non-homologous end joining (NHEJ) and homologous recombination (HR)2. HR repairs DSBs by using a homologous sister chromatid to guide repair of the broken DNA strand, and is primarily active during the S and G2 phases of the cell cycle3. In contrast, NHEJ functions across cell cycle and during variable, diversity and joining (V(D)J) recombination4,5. NHEJ does not require a DNA template, but instead mediates direct re-ligation of the two free DNA ends of DSBs. In animal models and human diseases, NHEJ defects result in growth retardation, immune deficiency, embryonic lethality, etc.6,7,8,9.
When DSBs are present, NHEJ is initiated through the recognition of DSB ends by the KU70/80 heterodimer, which subsequently recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to the DNA damage sites5. The complex consisting of DNA, KU70/80, and DNA-PKcs is referred to as the “DNA-PK” complex or holoenzyme10,11,12,13,14. DNA-PK is essential for further recruitment and regulation of other NHEJ factors, including DNA ligase IV, X-ray cross complementing protein 4 (XRCC4), XRCC4-like factor (XLF), endonuclease Artemis, and other factors that function in the processing and ligation of broken DNA ends5. DNA-PK has been proposed to prompt the DNA broken ends to form a synapsis complex for further processing and joining15,16,17.
DNA-PKcs is a giant protein kinase that belongs to phosphoinositide-3-kinase-related kinase (PIKK) family13. Representative PIKK family members include two master regulators of DNA damage responses, ataxia-telangiectasia mutated (ATM), and ATM- and Rad3-related (ATR), as well as a key regulator for cell growth, mechanistic target of rapamycin (mTOR)18. Like other PIKK family members, DNA-PKcs is composed of a large N-terminal α-solenoid, a FAT (FRAP, ATM, TRRAP) domain, and a kinase domain (KD), followed by FATC, a C-terminal short segment19,20 (Figure 1A).
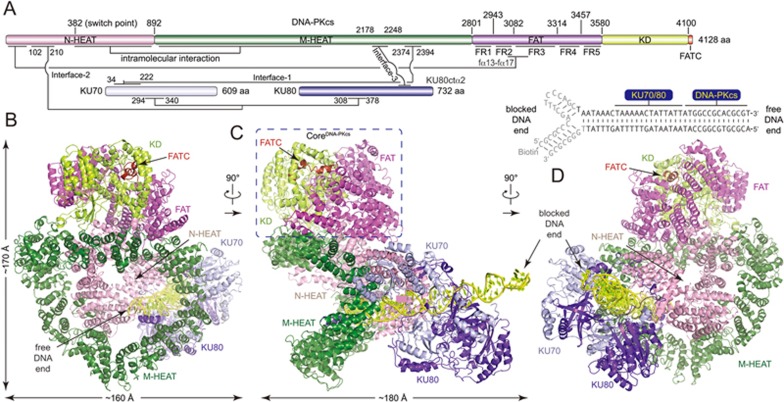
Figure 1.
Overall structure of the DNA-PK complex. (A) Color-coded domain architecture of human DNA-PKcs, KU70 and KU80. The same color scheme is used in all of the structure figures if not otherwise specified. The secondary structure of the biotinylated DNA is shown with the free and blocked DNA ends as indicated. The bases that were not built into the structural model are colored in gray. The inter- and intra-molecular contacts are shown as connected lines. Three major contacts (Interface-1 to Interface-3) between DNA-PKcs and KU70/80 are indicated. (B-D) Ribbon representations of the DNA-PK complex in three different views. The CoreDNA-PKcs (composed of the FAT, KD and FATC) is indicated with dashed rectangle. The DNA is colored in yellow. The bases of DNA are shown in stick representations. FAT, FRAP, ATM, TRRAP domain; FATC, FAT C-terminal domain; KD, kinase domain; M-HEAT, middle HEAT repeats; N-HEAT, N-terminal HEAT repeats.
DNA-PKcs phosphorylates a number of protein substrates, including the heat shock protein 90 (HSP90)21, the transcription factors p5310, specificity protein 1 (Sp1)22 and MYC23, and a majority of NHEJ factors24. The function of DNA-PKcs is regulated by extensive phosphorylation, autophosphorylation, and other protein partners24,25. The kinase activity of DNA-PKcs is essential for NHEJ26,27,28, although the exact role of its kinase activity is not fully understood. Sibanda et al. previously reported the crystal structure of DNA-PKcs in complex with C-terminal fragment of KU80 (residues 539-732, designated KU80ct194) at 6.6 Å resolution19 and recently pushed the resolution to 4.3 Å20. Sharif et al.29 recently reported a cryo-electron microscopy (cryo-EM) structure obtained from purified endogenous DNA-PK holoenzyme. However, no KU70/80-DNA was built into the structural model and very limited structural insights were revealed beyond the previously reported low-resolution cryo-EM structure30.
Results
Structure determination of the DNA-PK complex
The DNA fragment for structural study contains a 35-bp DNA duplex with a three-way junction formed by intra-strand annealing within 13 nucleotides (Figure 1A). Thus, the DNA contains a blocked DNA end so that the other end (free DNA end) mimics the broken DNA end in vivo and forces KU70/80 or DNA-PKcs-KU70/80 to adopt a single DNA-binding mode30,31. We purified the full-length human DNA-PKcs (the apo form) and the KU70/80 heterodimer (Supplementary information, Figure S1). KU70/80-DNA complex was pre-assembled and purified to homogeneity. DNA-PK (the holo form) was then assembled by mixing DNA-PKcs with KU70/80-DNA, followed by gel filtration. Purified DNA-PKcs, KU70/80, KU70/80-DNA, and DNA-PK complex were used for structural and biochemical studies.
The cryo-EM structure of DNA-PK was determined using single-particle reconstruction (Supplementary information, Figure S2, Table S1 and Movie S1). The overall structure of DNA-PK was determined at 6.6 Å resolution. We built structural model according to the cryo-EM map using the available structures of DNA-PKcs20 and KU70/80-DNA31 as template models (Supplementary information, Figure S3). In the DNA-PK structure, DNA-PKcs, KU70, KU80 and DNA duplex form a 650-kDa heterotetramer with 1:1:1:1 stoichiometry (Figure 1B-1D; Supplementary information, Movie S2), which is consistent with the gel filtration data (Supplementary information, Figure S1). The complex is composed of a compact core (the FAT, KD and FATC domains of DNA-PKcs, designated CoreDNA-PKcs), a “body” (N-terminal α-solenoid), and a “tail” (KU70/80-DNA), with overall dimensions of ∼160 × 180 × 170 Å3. The DNA is embedded within the complex with the free DNA end inserted into the “body” and the blocked DNA end flanking outside of KU70/80 (Figure 1C).
Structure of DNA-PK and intermolecular interactions
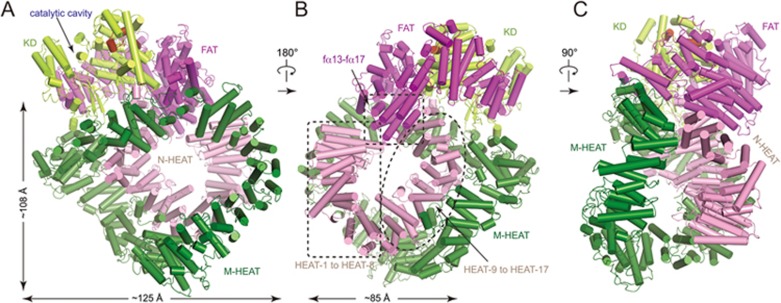
As in mTOR32, the N-terminal α-solenoid of DNA-PKcs contains an N-terminal (residues 1-892) and a middle (residues 893-2 801) HEAT (Huntingtin, elongation factor 3, PP2A and TOR1) repeats, referred to as “N-HEAT” and “M-HEAT”, respectively (Figure 1A). The M-HEAT forms a curved elliptical ring and serves as a scaffold to maintain the integrity of the whole complex (Figure 2A and Supplementary information, Movie S2). The N-HEAT forms an open ring (diameter of ∼80 Å) with a gap filled by five α-helices (FAT helices fα13-fα17 corresponding to residues 3 056-3 174) of the FAT domain (Figure 2B). The C-terminal part (HEAT9-HEAT17 corresponding to residues 383-892) of the N-HEAT makes extensive contacts with the M-HEAT. The N-HEAT and M-HEAT pack against each other and form a hollow double-ring structure (Figure 2C).
Figure 2.
Structure of DNA-PKcs in the DNA-PK complex. (A-C) Ribbon representations of the DNA-PKcs in three different views. The domains are indicated and colored as in Figure 1A. The sizes of the large ring (M-HEAT) and the small ring (N-HEAT and part of FAT) are indicated. Note that the catalytic cavity faces outward (A) and the M-HEAT primarily makes contacts with N-HEAT on repeats HEAT-9 to HEAT-17 (B).
The catalytic pocket of the KD faces outward, presenting an open catalytic cavity for substrate phosphorylation (Figure 2A and Supplementary information, Figure S4). The FAT domain adopts a “C”-shaped conformation and wraps around the KD for about 2/3 of a turn (Supplementary information, Figure S4). The FAT domain contains five short α-solenoids: FAT region 1 (FR1) to FR5. FR1-FR4 wrap around the C-lobe of the KD. FR5 is followed by the FKBP12-rapamycin-binding (FRB) domain and packs against the characteristic five-stranded β-sheet of the KD.
The DNA-PKcs KD adopts an overall fold similar to that of mTOR33 with a root-mean-squared deviation (RMSD) of 3.01 Å for 325 aligned Cα (Supplementary information, Figure S5). Unlike mTOR32,33, DNA-PKcs has a three-helix insertion (Hairpin-2), which covers the FATC and the activation loop. The FRB and mLST8-binding elements (LBE) form two sides of the substrate entry groove. Substrate access to the active site of mTOR is proposed to be restricted by the helix kα9b (Supplementary information, Figure S5B), although deletion of this region did not increase its kinase activity33. The equivalent of kα9b in DNA-PKcs is a two-helix α-hairpin (Hairpin-1) that is stacked between the FRB domain and the catalytic loop (Supplementary information, Figure S5A).
While we used the full-length KU70 and KU80 for structural analysis, neither the C-terminus (residues 535-609) of KU70 nor the C-terminus (residues 542-732) of KU80 was built into the model, owing to a lack of density in the EM map. KU70/80 in the DNA-PK complex adopts an overall fold similar to that in the KU70/80-DNA structure (PDB: 1JEY), with an RMSD of 2.44 Å for 838 aligned Cα (Supplementary information, Figure S6). Structural comparison indicates that the two KU80 molecules are essentially similar with an RMSD of 1.88 Å for 507 aligned Cα, whereas the two KU70 molecules display distinct conformations with an RMSD of 3.21 Å for 377 aligned Cα. As observed in the KU70/80-DNA structure31, KU70 is proximal, and KU80 is distal, to the free DNA end. KU70 and KU80 share a similar fold and together form a pseudo-symmetrical ring that encircles the DNA duplex.
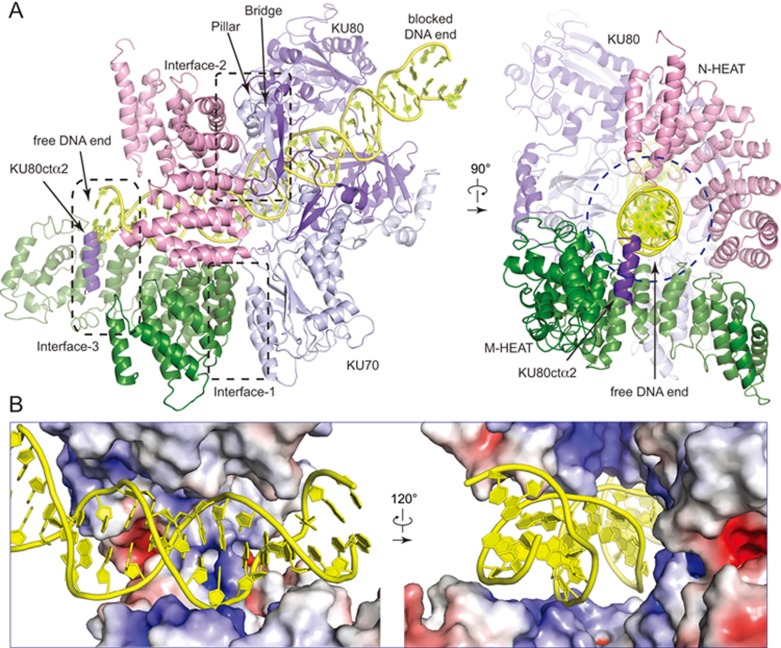
We observed three major intermolecular interfaces between DNA-PKcs and KU70/80 heterodimer (Figure 3A and Supplementary information, Figure S6). (1) The α/β domain of KU70 makes contacts (Interface-1) with the convex surface of the M-HEAT. (2) The convex surface of the N-HEAT interacts with the “bridge” formed by two anti-parallel strands of KU70/KU80 and the “pillar” of KU70 (Interface-2). Structural comparison indicates that DNA-PK complex formation forces the α/β domain of KU70 to move by as much as 12 Å, yet results in few changes on other regions (Supplementary information, Figure S6A). (3) Previous studies have indicated that the C-terminal region of KU80 is important for interaction between KU70/80 and DNA-PKcs34,35,36, and three α-helices of KU80ct194 bind to the M-HEAT in the DNA-PKcs-KU80ct194 structure20. In the DNA-PK structure, a 4-turn helix (equivalent to the second α-helix of KU80ct194, designated KU80ctα2) is located close to the free DNA end and binds to the concave side of the M-HEAT of DNA-PKcs (Interface-3). The other two helices were not observed in the DNA-PK complex structure.
Figure 3.
Intermolecular interactions within the DNA-PK complex. (A) Close-up views of intermolecular interactions within DNA-PK in two different views. Ribbon representations are shown with three major contact interfaces indicated. Note that a helix (KU80ctα2) binds to DNA-PKcs and obscures the free DNA end. The DNA-binding tunnel forces DNA duplex to undergo a 30° kink. (B) Close-up views of the electrostatic potential surface of DNA-PKcs in two different views. The inner surface of DNA-binding tunnel within DNA-PKcs is rich in positively-charged (blue) residues. KU80ctα2 is omitted for clarity.
Recognition and protection of broken DNA ends
According to the EM map, a 34-bp DNA duplex occurred in the model but no three-way junction was visible, possibly due to a lack of stabilization by protein-DNA contacts (Supplementary information, Figure S3). DNA-PKcs and KU70/80 together form a DNA-binding tunnel. DNA-PKcs is relatively proximal, and KU70/80 is distal, to the free DNA end. DNA-PKcs and KU70/80 both wrap around one and a half turn of the DNA duplex with the blocked DNA end flanking outside of the complex (Figure 3A and Supplementary information, Movie S2). KU70/80 binds to the dsDNA in a manner similar to that observed in the KU70/80-DNA structure (PDB: 1JEY; Supplementary information, Figure S6), wherein KU70/80 was proposed to sterically define the position of the DNA helix to pass through the protein ring31.
The DNA duplex fills the gap between the two ridges of the N-HEAT and the M-HEAT of DNA-PKcs (Figure 3B). The inner surface of the DNA-binding tunnel within DNA-PKcs is rich in positively-charged residues, in agreement with its DNA-binding affinity. Intriguingly, we did not observe extensive protein-DNA interaction, which may reflect limitations of the EM map at 6.6 Å resolution. Alternatively, the apparent lack of protein-DNA contact and the opened DNA-binding tunnel may allow DNA-PKcs to accommodate a wide spectrum of DNA ends to facilitate NHEJ. This observation is supported by the in vitro pull-down assay, in which DNA-PKcs alone barely binds to DNA but strongly binds to DNA in the presence of KU70/80 (Supplementary information, Figure S7), suggesting that DNA-PKcs-DNA interaction would not be stably maintained in the absence of KU70/80.
The free DNA end makes contacts with helix KU80ctα2 and a region that was not traced because the corresponding EM map was insufficiently resolved for model building (Figure 3A and Supplementary information, Figures S3 and S6). The existence of these structural elements likely provides steric hindrance to prevent inward sliding of DNA-PKcs along with the DNA duplex, and may also obscure the free DNA end to avoid unnecessary processing. Thus, DNA-PKcs and KU80 together provide a “block” for the DNA-binding tunnel proximal to the free DNA end. Such a striking structure agrees with previous studies, which show that DNA-PKcs-KU70/80 recognizes and protects DNA ends from unnecessary processing37,38. Although DNA-PK does not form a dimer of tetramer in solution (Supplementary information, Figure S1), two DNA-PK complexes might be bridged together by other NHEJ factors so that two free DNA ends (broken DNA ends in vivo) are brought into close proximity. Notably, the free DNA end is located within an open cradle (50 Å high, 50 Å wide, and 40 Å deep; Figure 1B and 1C) that is large enough to easily accommodate NHEJ factors such as DNA ligase IV and DNA polymerase μ39,40. It is tempting to speculate that some unidentified factors free the protected DNA end, allowing downstream NHEJ processes to occur under precise control.
Allosteric activation of DNA-PKcs
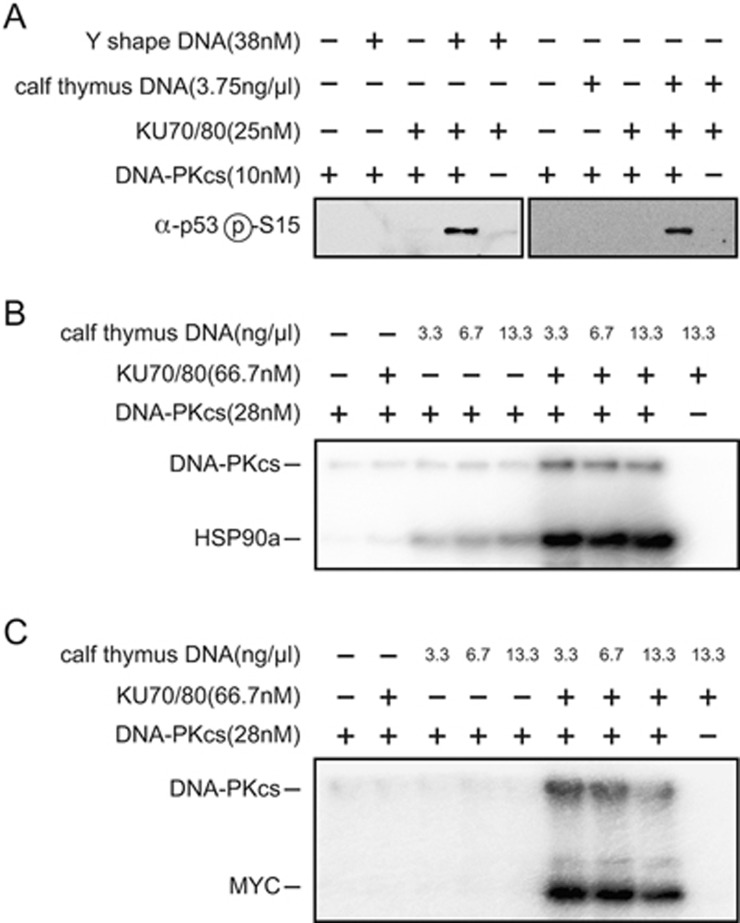
Previous studies have indicated that purified endogenous DNA-PKcs-KU70/80 is activated by DNA10,12,21,41,42, and that DNA and KU70/80 heterodimer can independently stimulate the kinase activity of DNA-PKcs12,13,43. To investigate mechanism for the activation of DNA-PKcs, we performed in vitro kinase activity assays using purified human p53, HSP90a, and MYC as substrates. KU70/80 heterodimer showed undetectable activation of DNA-PKcs kinase activity toward the three substrates. DNA slightly enhanced the DNA-PKcs kinase activity toward HSP90a, but showed no detectable activation in the reaction using p53 or MYC as substrate (Figure 4). Robust activation of DNA-PKcs kinase activity, with substrate phosphorylation (Figure 4A and lower bands in Figure 4B and 4C) and autophosphorylation (upper bands in Figure 4B and 4C), was only observed in the reactions when both DNA and KU70/80 were present. Consistently, neither DNA nor KU70/80 exhibited binding to DNA-PKcs, whereas DNA and KU70/80 together obviously bound to DNA-PKcs under our experimental conditions (Supplementary information, Figure S7). This also agrees with multiple intermolecular contacts between DNA-PKcs, KU70/80, and dsDNA observed in the DNA-PK complex structure (Figures 1C and 3A). These results indicate that KU70/80 heterodimer and DNA together bind to DNA-PKcs and coordinately stimulate its kinase activity. Intriguingly, KU70/80 and KD bind to distinct surfaces of the N-terminal α-solenoid of DNA-PKcs, and the two structural units have no direct contact with each other (Figure 1D), suggesting that KU70/80-DNA allosterically stimulates the kinase activity of DNA-PKcs.
Figure 4.
Kinase activity assays of DNA-PKcs. Phosphorylation of p53 (A), HSP90a (B), or MYC (C) by DNA-PKcs in the presence of KU70/80 and/or increasing amounts of DNA. Y-shaped DNA employed in the holoenzyme assembly or sonicated calf thymus DNA is used as indicated. Purified proteins used for the assays are indicated in Supplementary information, Figure S1. Protein and DNA concentrations used in the assay are indicated. Substrate phosphorylation and/or DNA-PKcs autophosphorylation were detected by antibody (A) or autoradiography (B-C).
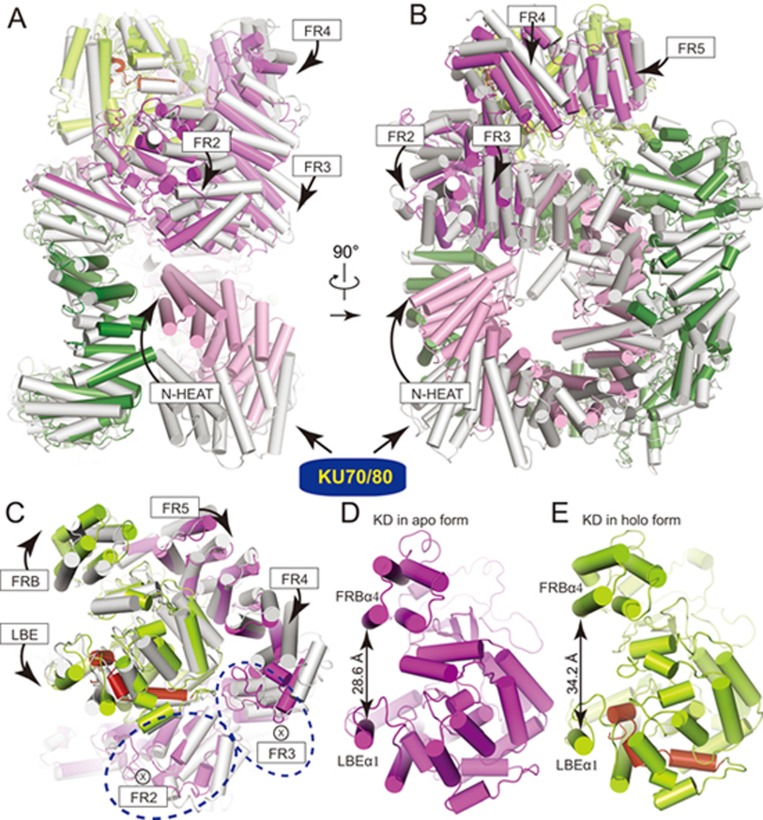
We next compared the structures of the apo and holo forms of DNA-PKcs (Figure 5A-5C). Note that in the DNA-PKcs-KU80ct194 structure (apo form), molecule B was used for comparison since it is slightly more concealed than the other (molecule A) in the asymmetric unit of the crystals20. The two M-HEAT domains are superimposed well with an RMSD of 2.60 Å for 839 aligned Cα. The most significant structural difference exists at the N-terminus (HEAT1-HEAT8 corresponding to residues 1-382) of the N-HEAT, which adopts an extended fold in the apo form but folds inward to join with the M-HEAT and FAT in the holo form (Figure 5A and 5B). DNA entry would be prohibited by this region in the apo form. Moreover, structural analyses indicate that intramolecular contact between FAT and KD-FATC is ∼3 500 Å2 in the apo form and ∼4 100 Å2 in the holo form, suggesting that the FAT domain may transmit KU70/80-DNA-triggered conformational changes to the KD and lead to the activation of DNA-PKcs. This hypothesis is supported by previous studies that the KD of DNA-PKcs is active and its kinase activity is inhibited by the N-terminal regions44,45.
Figure 5.
Mechanism for activation of DNA-PKcs. (A-C) Superimposition of the DNA-PKcs in the apo form (PDB: 5LUQ, molecule B) and the holo form shown in three different views. Transition from the apo form (colored in gray) to the holo form (colored as in Figure 1A) indicates conformational changes of DNA-PKcs induced by KU70/80-DNA. The directions of movement of structural units are indicated with arrows. ⊗ represents the direction of movement that is downward perpendicular to plane of paper. (D-E) The kinase domain in the apo form (D) and the holo form (E) shown in similar orientations. The distance between FRBα4 and LBEα1, two α-helices on top of both sides of the groove, is shown for each kinase domain, indicating obviously distinct substrate entry grooves. The same conclusion can be obtained using molecule A (PDB: 5LUQ) for comparison (Supplementary information, Figure S8).
We here propose a model for activation of DNA-PKcs in which allosteric signals are transmitted to the KD through the following steps (Figure 5 and Supplementary information, Movie S3). (1) Formation of the DNA-PK complex forces the N-terminal region (HEAT1-HEAT8) of DNA-PKcs to twist and move (by as far as 30 Å) and leaves the rest of the N-HEAT and M-HEAT essentially unchanged. The switch point is located at the site between HEAT8 and HEAT9. As a result, the N-terminus of the N-HEAT is brought to close proximity to fα13-fα15 of the FAT domain (Figure 5A and 5B). (2) This intramolecular contact forces FR2 and FR3 to twist and move toward the N-HEAT, which makes FR1-FR3 more compact (Figure 5A). (3) As shown in Figure 5B, FR4 and FR5 move leftward and rotate to fill the gap generated by the movement of FR3. (4) The above movements collectively lead to coordinated conformational changes of the KD. Most strikingly, the potential substrate entry groove is wider in the holo conformation, in which the two parallel helices (FRBα4 and LBEα1) on top of both sides are 34.2 Å (distance between Cα of K3672 and Cα of P3835) apart from each other, compared to 28.6/29.3 Å in the apo conformation (A/B molecule in apo form; Figure 5C-5E and Supplementary information, Figure S8). The wider groove may facilitate substrate entry to the catalytic cavity and thus stimulate the kinase activity of DNA-PKcs. Conformational changes within the catalytic pocket may also lead to more favorable substrate recognition or efficient reaction, which could be evaluated with a higher resolution structure and/or molecular dynamics analysis.
Discussion
DNA DSBs could be induced by a wide range of stimuli exampled by ionizing radiation and chemical agents. It can also result from intrinsic factors such as replication errors and reactive oxygen species. Failure to repair DSBs would cause genome instability, cell cycle checkpoint abnormalities, and ultimately cell death46. NHEJ plays a vital role in repairing DSBs throughout the cell cycle and is required in physiological processes such as V(D)J recombination. Core NHEJ factors include KU70/80 heterodimer, DNA-PKcs, ligation complex DNA ligase IV-XRCC4-XLF, and other processing factors like endonuclease Artemis, DNA polymerases, polynucleotide kinase/phosphatase (PNKP), and aprataxin and PNKP related protein (APLF). In the currently proposed model, upon detection of DNA DSB site by KU70/80, DNA-PKcs is recruited and activated24,25. Although the function of DNA-PK has been extensively studied, a number of critical questions remain elusive. (1) How are DNA-PKcs and KU70/80 organized at the broken DNA ends and how are the broken DNA ends protected? (2) How is DNA-PKcs activated by KU70/80 and/or dsDNA? (3) How are the broken DNA ends brought to close proximity for ligation? Addressing these questions is important to fully understand this complicated process.
In this work, we report the cryo-EM structure of DNA-PK holoenzyme at 6.6 Å resolution and reveal the mechanism for assembly of this giant protein kinase complex. Within DNA-PK holoenzyme, the N-terminal α-solenoid (∼2 800 residues) of DNA-PKcs forms a double-ring structure, which connects the catalytic core domain of DNA-PKcs and KU70/80-DNA. DNA-PKcs and KU70/80 together form a DNA-binding tunnel, which cradles ∼30-bp DNA. Previous models assumed that the bound DNA is unbent20,30. However, the cryo-EM structure clearly shows that DNA-PKcs and KU70/80 both recognize DNA and together result in a 30° kink of the DNA duplex (Figure 3A). This unexpected observation suggests that the DNA-binding tunnel is relatively rigid to allow distortion of the DNA. Whether and how this DNA distortion plays a role in NHEJ remain further investigation.
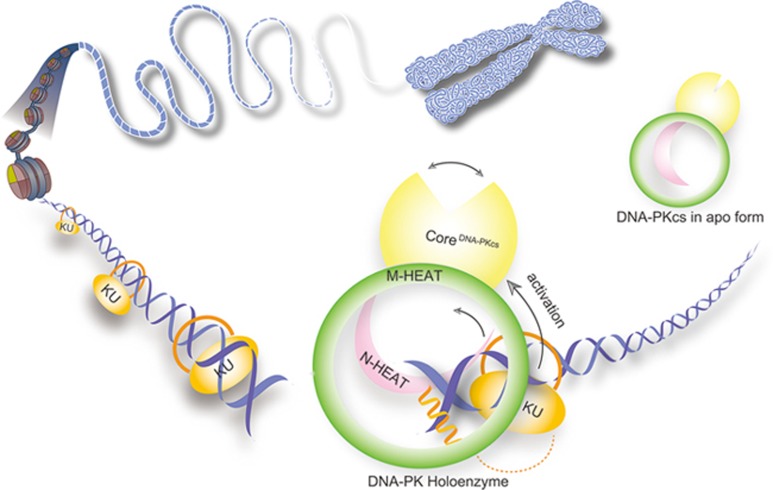
Intriguingly, the free DNA end inserts into the double-ring structure of DNA-PKcs and a helix of KU80 prevents sliding inward of DNA-PK along with DNA duplex (Figure 6). Such structural organization suggests that the broken DNA ends might be protected from unnecessary processing upon DNA-PK complex formation, and DNA-PK holoenzyme serves as a scaffold to allow further dynamic ligation processes to occur. The observation is consistent with previous studies, which show that DNA-PKcs and KU70/80 recognize and protect the free DNA ends from unnecessary processing37,38.
Figure 6.
A working model for DNA-PK complex assembly and activation. When DNA DSB occurs, KU70/80 heterodimer recognizes the broken DNA ends and binds to the DNA with the preformed ring structure (left). DNA-PKcs is then recruited to the DSB end and binds to KU70/80-DNA (right). KU70/80-DNA stimulates the kinase activity of DNA-PKcs through an allosteric activation mechanism. DNA-PKcs and KU70/80 together bind to DNA and prevent sliding inward of DNA-PKcs along with DNA duplex, suggesting a mechanism by which the broken DNA end is protected from unnecessary processing.
DNA-PK was initially purified and identified as a high-molecular weight protein kinase10,21,22. Subsequently, multiple lines of evidence converge on the conclusion that purified endogenous DNA-PK is strongly activated by linearized dsDNA and KU70/80, but the activation mechanism has been unknown over two decades47. In this work, by comparing structures of DNA-PKcs in apo form and DNA-PK holoenzyme, a significant conformational difference was observed, indicating that KU70/80-DNA induces conformational changes of DNA-PKcs and allosterically stimulates its kinase activity (Figure 6). We therefore propose a model for activation of DNA-PKcs in which allosteric signals are initiated by DNA-PK holoenzyme formation and then transmitted through the FAT domain to the KD. In the absence of KU70/80-DNA, DNA-PKcs possesses basal-level kinase activity to avoid non-specific phosphorylation. Once DNA-PKcs is recruited to the broken DNA ends by KU70/80, the kinase is activated in an impermissible genomic region.
In summary, our study reveals a structural basis for the complex assembly of DNA-PK and an allosteric mechanism for the activation of DNA-PKcs (Figure 6). The structure also provides foundation for the future design of allosteric activators/inhibitors that target DNA-PKcs for potential therapeutic or basic biological research applications. Finally, our structure motivated us to propose a mechanism for the recognition and positioning of broken DNA ends, thus shedding light on understanding the complicated processes of NHEJ.
Materials and Methods
Protein purification
Biotinylated Y-shaped dsDNA was synthesized and annealed as described previously (Supplementary information, Table S2)30. KU70/80-DNA was purified to homogeneity using Superose 6 (Increase 5/150 GL, GE healthcare). Purified DNA-PKcs and KU70/80-DNA were mixed and DNA-PK complex was obtained by gel filtration through Superose 6. Human p53, HSP90a and MYC were expressed in E. coli BL21(DE3) and purified by affinity purification followed by ion exchange chromatography.
Kinase activity assays
In vitro kinase assays were performed as described previously10. In brief, 1 μg HSP90a or MYC was incubated with 200 ng DNA-PKcs in 15 μl reaction buffer containing 50 mM HEPES-NaOH, pH 7.4, 100 mM KCl, 10 mM MgCl2, 2 mM EGTA, 0.1 mM EDTA, and 2 μl of [γ-32P]ATP (6 000 Ci/mmol, 10 mCi/ml) at 30 °C for 10 min. For assays using p53, the same condition was used except that 2.3 μg p53 was catalyzed by DNA-PKcs in 40 μl reaction buffer and the final ATP concentration is 1 mM. Sonicated calf thymus DNA was added as indicated. The assays were performed in the absence or presence of KU70/80. The reaction was stopped by adding SDS-PAGE loading buffer. The reaction product was analyzed by SDS-PAGE, and the gel was subjected to autoradiography (for MYC and HSP90a) or western blot using site-specific antibody (for p53).
Accession codes
The electron density map and corresponding atomic coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code: 5Y3R and EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code: EMD-6803.
Please refer to Supplementary information, Data S1 for EM data acquisition, image processing, model building, and other details on Materials and Methods.
Author Contributions
XY and ML contributed to the protein sample preparation. ML and YT carried out cryo-EM data collection. ML and JW analyzed the cryo-EM data and determined the structure. XY led the biochemical analyses and assay setup. YX performed structure-function analyses and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank staff members of National Center for Protein Science Shanghai (NCPSS), Institute of Biophysics (IBP), Chinese Academy of Sciences (CAS), and Biomedical Core Facility of Fudan University for their help on cryo-EM sample screening and data collection. We thank Minghong He from Prof Jinqiu Zhou's Lab at Shanghai Institute of Biochemistry and Cell Biology for his help on kinase activity assays. We thank Prof David Chen for kindly providing the cDNA of DNA-PKcs. This work was supported by the Ministry of Science and Technology of China (2016YFA0500700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08000000), the National Natural Science Foundation of China (U1432242, 31425008 and 91419301), and the Program of Shanghai Subject Chief Scientist (14XD1400500).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Complex preparation of DNA-PK.
Cryo-EM structure determination.
Cryo-EM map for the DNA-PK complex.
Structure of the CoreDNA-PKcs.
Structural comparison of the kinase domains of DNA-PKcs and mTOR.
Structures of KU70/80-DNA.
Interaction between DNA-PKcs, KU70/80 and DNA.
Structural comparison of the kinase domains of DNA-PKcs in apo form and holo form.
Statistics for data collection and model refinement of DNA-PK complex
Oligonucleotides used in this work
EM map and model building of DNA-PK complex.
Structural analysis of DNA-PK complex.
KU70/80-DNA induces conformational changes and activation of DNA-PKcs.
Materials and Methods
References
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2001; 2:196–206. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell 2007; 28:739–745. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem Sci 1998; 23:394–398. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Buck D, le Deist F, de Villartay JP. The repair of DNA damages/modifications during the maturation of the immune system: lessons from human primary immunodeficiency disorders and animal models. Adv Immunol 2005; 87:237–295. [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001; 105:177–186. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Cerosaletti KM, Girard PM, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell 2001; 8:1175–1185. [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval P, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006; 124:287–299. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol 1990; 10:6472–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AI, Hunt T, Jackson RJ, Anderson CW. Double-stranded DNA induces the phosphorylation of several proteins including the 90 000 mol. wt. heat-shock protein in animal cell extracts. EMBO J 1985; 4:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 1993; 72:131–142. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995; 82:849–856. [DOI] [PubMed] [Google Scholar]
- Suwa A, Hirakata M, Takeda Y, Jesch SA, Mimori T, Hardin JA. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA 1994; 91:6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak KS, Turchi JJ. A mechanism for DNA-PK activation requiring unique contributions from each strand of a DNA terminus and implications for microhomology-mediated nonhomologous DNA end joining. Nucleic Acids Res 2008; 36:4022–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu YP, Mahaney BL, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 2010; 285:1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 2002; 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretic D, Williams RL. PIKKs--the solenoid nest where partners and kinases meet. Curr Opin Struct Biol 2014; 29:134–142. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 2010; 463:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Ascher DB, Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 2017; 355:520–524. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Anderson CW. The human double-stranded DNA-activated protein kinase phosphorylates the 90-kDa heat-shock protein, hsp90α at two NH2-terminal threonine residues. J Biol Chem 1989; 264:17275–17280. [PubMed] [Google Scholar]
- Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell 1990; 63:155–165. [DOI] [PubMed] [Google Scholar]
- Iijima S, Teraoka H, Date T, Tsukada K. DNA-activated protein kinase in Raji Burkitt's lymphoma cells. Phosphorylation of c-Myc oncoprotein. Eur J Biochem 1992; 206:595–603. [DOI] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol 2015; 117:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienker LJ, Shin EK, Meek K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res 2000; 28:2752–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Wong N, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell 2009; 34:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimasa A, Kumano S, Boubnov NV, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol 1999; 19:3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H, Li Y, Dong Y, et al. Cryo-EM structure of the DNA-PK holoenzyme. Proc Natl Acad Sci USA 2017; 114:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 2006; 22:511–519. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001; 412:607–614. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang J, Liu M, et al. 4.4 Å Resolution cryo-EM structure of human mTOR Complex 1. Protein Cell 2016; 7:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature 2013; 497:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res 1999; 27:3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol 1999; 19:3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DS, Sears CR, Turchi JJ. Recognition of DNA termini by the C-terminal region of the Ku80 and the DNA-dependent protein kinase catalytic subunit. PLoS One 2015; 10:e0127321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JHJ, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res 2003; 31:7238–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Halbrook J, Nickoloff JA, Ullrich RL, Goodwin EH. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair 2004; 3:225–233. [DOI] [PubMed] [Google Scholar]
- Ochi T, Gu X, Blundell TL. Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure 2013; 21:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Pryor JM, Ramsden DA, Kunkel TA, Bebenek K, Pedersen LC. Sustained active site rigidity during synthesis by human DNA polymerase mu. Nat Struct Mol Biol 2014; 21:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol 1997; 7:588–598. [DOI] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J 1997; 16:5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic J, Rivera-Calzada A, Maman JD, et al. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J 2003; 22:5875–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Lee KJ, Chen DJ. The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation. J Biol Chem 2013; 288:7037–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Lees-Miller SP, Modesti M. N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res 2012; 40:2964–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene 2005; 24:949–961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complex preparation of DNA-PK.
Cryo-EM structure determination.
Cryo-EM map for the DNA-PK complex.
Structure of the CoreDNA-PKcs.
Structural comparison of the kinase domains of DNA-PKcs and mTOR.
Structures of KU70/80-DNA.
Interaction between DNA-PKcs, KU70/80 and DNA.
Structural comparison of the kinase domains of DNA-PKcs in apo form and holo form.
Statistics for data collection and model refinement of DNA-PK complex
Oligonucleotides used in this work
EM map and model building of DNA-PK complex.
Structural analysis of DNA-PK complex.
KU70/80-DNA induces conformational changes and activation of DNA-PKcs.
Materials and Methods