Abstract
Changes in gene copy number are important in the setting of precision medicine. Recent studies have established that copy number alterations (CNAs) can be detected in sequencing libraries prepared by hybridization-capture, but there has been comparatively little attention given to CNA assessment in amplicon-based libraries prepared by PCR. In this study, we developed an algorithm for detecting CNAs in amplicon-based sequencing data. CNAs determined from the algorithm mirrored those from a hybridization-capture library. In addition, analysis of 14 pairs of matched normal and breast carcinoma tissues revealed that sequence data pooled from normal samples could be substituted for a matched normal tissue without affecting the detection of clinically relevant CNAs (>|2| copies). Comparison of CNAs identified by array comparative genomic hybridization and amplicon-based libraries across 10 breast carcinoma samples showed an excellent correlation. The CNA algorithm also compared favorably with fluorescence in situ hybridization, with agreement in 33 of 38 assessments across four different genes. Factors that influenced the detection of CNAs included the number of amplicons per gene, the average read depth, and, most important, the proportion of tumor within the sample. Our results show that CNAs can be identified in amplicon-based targeted sequencing data, and that their detection can be optimized by ensuring adequate tumor content and read coverage.
The identification of molecular aberrations present in a tumor sample is becoming important in delivering precision cancer care. Targeted sequencing using next-generation technologies is effective in identifying the single-nucleotide substitutions and short indels that may help guide treatment decisions.1, 2, 3, 4, 5, 6, 7, 8, 9 Two widely used enrichment strategies for targeted sequencing are hybridization-capture, in which oligonucleotide baits complementary to the regions of interest are hybridized with fragmented genomic DNA,3 and PCR, in which a pool of primers is used to generate target-specific amplicons.4, 10 Both of these approaches work well on DNA purified from formalin-fixed, paraffin-embedded (FFPE) tumor tissue, and they require only small amounts of input DNA (10 to 100 ng).
Copy number alterations (CNAs) are also important in personalized cancer diagnostics. ERBB2 amplification is routinely screened in breast carcinomas to determine whether HER2-targeted therapies should be included in a patient's treatment plan. Similarly, amplifications of FGFR1, EGFR, MET, and PIK3CA are all being targeted in ongoing clinical trials. There are a variety of technologies that can be used to measure CNAs in tumor DNA, including genome-wide approaches such as array comparative genomic hybridization (aCGH) and whole-genome sequencing, as well as targeted approaches, such as whole-exome sequencing, single-nucleotide polymorphism (SNP) arrays, quantitative PCR, and fluorescence in situ hybridization (FISH).11, 12, 13, 14, 15, 16, 17 Among these methods, those based on next-generation sequencing (NGS) are gaining in popularity, because information on CNAs can be derived from the same data used to detect sequence alterations. Algorithms for assessing CNAs have been developed for NGS protocols that are based on hybridization-capture, whether in the setting of whole-exome sequencing12 or targeted sequencing.1, 3, 5, 8 In contrast, little work has been done on CNA assessment in NGS data from amplicon-based libraries.
Herein, we developed and validated an algorithm for assessing CNAs in NGS data derived from amplicon-based libraries of FFPE tumor DNA. We compared the results with CNAs assessed in a hybrid-capture library, as well as with CNAs determined from aCGH data, and from FISH for specific genes. In addition, we systematically examined several factors that can influence CNA detection, including tumor purity, the number of amplicons per gene, and the number of reads per amplicon. Our results show that CNAs are readily detected in amplicon-based libraries and correlate well with other methods. However, the sensitivity for CNAs is influenced by several parameters that should be taken into account in both the design of targeted panels and the interpretation of the NGS data that they yield.
Materials and Methods
Tumor Specimens and DNA Preparation
This study was conducted in accordance with federal and institutional guidelines. For all samples, excluding WA25 (see below), blocks of FFPE tumor or unstained sections of FFPE tissue were obtained from the pathology archives of Oregon Health and Science University (Portland, OR). The diagnosis in each case was confirmed by a board-certified pathologist (C.L.C.). Tumor-rich areas (20% to 90%) were macrodissected from unstained sections (5 μm thick) by comparison with a hematoxylin and eosin (H&E)–stained slide, and genomic DNA was extracted using a Macherey-Nagel NucleoSpin Tissue Kit (Clontech, Mountain View, CA). For 14 of the breast tumor samples, morphologically normal areas were identified and used as a source for matched normal DNA; these samples also served in the generation of a pool of data from normal DNA. Genomic DNA (20 ng) was used for library preparations from the tumor samples and from the matched normal samples.
Preparation of Amplicon Libraries
A custom Ion AmpliSeq (Ion Torrent, Carlsbad, CA) solid tumor panel was used to generate target amplicon libraries. This panel covers some or all of the coding exons of 37 genes known to play a role in cancer: AKT1, AKT2, AKT3, ALK, BRAF, CDK4, CDKN2A, DDR2, EGFR, ERBB2, FGFR1, FGFR3, GNA11, GNAQ, GNAS, KDR, KIT, KRAS, MAP2K1, MET, HRAS, NF1, NOTCH1, NRAS, NTRK2, NTRK3, PIK3CA, PIK3R1, PTEN, RAC1, RB1, RET, STK11, TSC1, TSC2, TP53, and VHL. The number of amplicons per gene in the panel varies from 1 to 145. DNA derived from FFPE tissue (20 ng) was amplified by PCR using the premixed AmpliSeq primer pools and AmpliSeq HiFi master mix (Ion AmpliSeq kit version 2.0). Primer sequences were manufactured specifically for use with the Ion AmpliSeq kits and contained proprietary modifications. The resulting 1164 multiplexed amplicons were treated with FuPa reagent (Ion Torrent) to partially digest primer sequences and phosphorylate the amplicons. The amplicons were then ligated to Ion Xpress bar-coded adapters, according to the manufacturer's instructions (Ion Torrent). The Ion Library Quantitation Kit was used to determine the library concentration.
Emulsion PCR and Sequencing
Multiplexed bar-coded libraries were amplified for 20 cycles by emulsion PCR on Ion Sphere particles (ISPs) at a 1:2 ratio of total library molecules/ISPs (280 × 106 molecules per reaction) (Ion Xpress Template kit version 2.0; Ion Torrent). The templated ISPs were recovered from the emulsion, and the ratio of templated ISPs/empty ISPs was determined by a fluorometric assay using fluorescently labeled oligonucleotides complementary to adapter sequences. The optimal templated signal ratio was determined to be between 10% and 40%. Positive templated ISPs were biotinylated during the emulsion PCR process so that the samples with an optimal templated signal ratio were then enriched with Dynabeads MyOne streptavidin C1 beads (Life Technologies/Thermo Fisher, Carlsbad, CA). Eight bar-coded samples were multiplexed on an Ion 318 chip. Sequencing was performed on a Personal Genome Machine (PGM) sequencer (Ion Torrent) using the Ion PGM 200 sequencing kit 2.0, according to the manufacturer's instructions. Torrent Suite software version 4.0 (Ion Torrent) was used to parse bar-coded reads, to align reads to the reference genome, and to generate run metrics, including chip loading efficiency and total read counts and quality. The total reads per run and the average number of reads per amplicon are listed in Supplemental Table S1.
CNAs in Castration-Resistant Prostate Cancer Sample WA25
WA25 was obtained from a rapid autopsy performed at the University of Michigan Health Systems (Ann Arbor, MI) on a patient who died of castration-resistant prostate cancer. This sample was collected under prior informed consent of the patient and previous University of Michigan Institutional Review Board approval. H&E-stained sections from FFPE blocks were reviewed by a board-certified pathologist (S.A.T.), and a representative section with >50% tumor content and a benign tissue section were identified. Three sections (10 μm thick) were cut from each block, and the tumor sections were macrodissected to enrich tumor content.
For targeted sequencing, DNA was isolated using the Qiagen (Germantown, MD) Allprep FFPE DNA/RNA kit, according to the manufacturer's instructions, except with additional xylene/ethanol washes. DNA was quantified using the Qubit fluorometer (Life Technologies/Thermo Fisher). Bar-coded libraries were generated from 40 ng DNA using the Comprehensive Cancer Panel (CCP) and the Ion Ampliseq Library Kit version 2.0 according to the manufacturer's instructions, essentially as described above. The CCP contains multiplexed PCR primers for approximately 16,000 amplicons assessing all coding exons in 409 cancer genes. The PCR was done for 16 cycles. Templates were prepared using the Ion PGM Template OT2 Kit version 2 on the Ion One Touch 2, according to the manufacturer's instructions. Sequencing of multiplexed templates was performed on an Ion 318 chip using the Ion PGM 200 Sequencing Kit version 2 according to the manufacturer's instructions. Analysis was performed in Torrent Suite version 3.6, with alignment by TMAP version 3.6, using default parameters.
Whole-exome sequencing analysis of WA25 was previously reported.18 From the exome library, copy number aberrations were quantified and reported for each gene using the segmented normalized log2-transformed exon coverage ratios between each tumor sample and its matched normal sample, as previously described.12
FISH Data
For all probes used in the current study, an H&E-stained slide was marked by a board-certified pathologist (C.L.C.) to aid in the identification of the tumor cells by the Cytogenetics technologist. Sections were deparaffinized through heat treatment and pretreated for hybridization using an automated protocol (VP2000; Abbott, Abbott Park, IL), with the exception of ERBB2 slides, which were pretreated by hand, according to the manufacturer's guidelines (Abbott). FISH was performed per probe manufacturer's guidelines. A total of 50 to 100 cells were scored for each probe, with counts split between two qualified scorers. For ERBB2, 25 cells were scored (per American Society of Clinical Oncology guidelines, 2013 revision), and the slide and scores were reviewed by a pathologist before the results were reported.
aCGH Data
Chromosomal microarray using a custom exon-centric microarray was performed using DNA extracted from FFPE specimens. DNA (0.5 to 1 μg) was used per reaction. Probe labeling and hybridization conditions were performed per manufacturer's guidelines (Oxford Genome Technologies, Oxfordshire, UK) without modifications. Images were scanned using the Agilent SureScan Scanner (Agilent, Santa Clara, CA) and aligned for comparison using CytoSure Interpret Software version 4.5.3 (Oxford Genome Technologies). The exon-specific array is a custom design that has a minimum of three oligonucleotide probes per exon across all included genes, with a 60,000 oligonucleotide backbone based on the International Standards for Cytogenomic Arrays consortium design. To calculate Log2(copy number ratios) for each gene, we considered only probes that had Control Type equal to 0. We took the Log2 of the ratio of the green/red signal after correction (gProcessedSignal/rProcessedSignal) for each probe. Finally, for each gene, we took the average of the Log2(copy number ratios) over a window extending ±50 kb around that gene. These windows encompassed from 8 to 1390 probes, depending on the gene.
Results
An Algorithm for Detecting CNAs in Targeted Amplicon-Based NGS Data
The goal of this study was to develop, test, and validate an algorithm for detecting somatic variations in gene copy number in NGS data generated from amplicon-based libraries through comparisons to hybrid-capture library-based sequencing, FISH, and aCGH. In addition, amplicon characteristics that might influence the detection of CNAs were examined in detail.
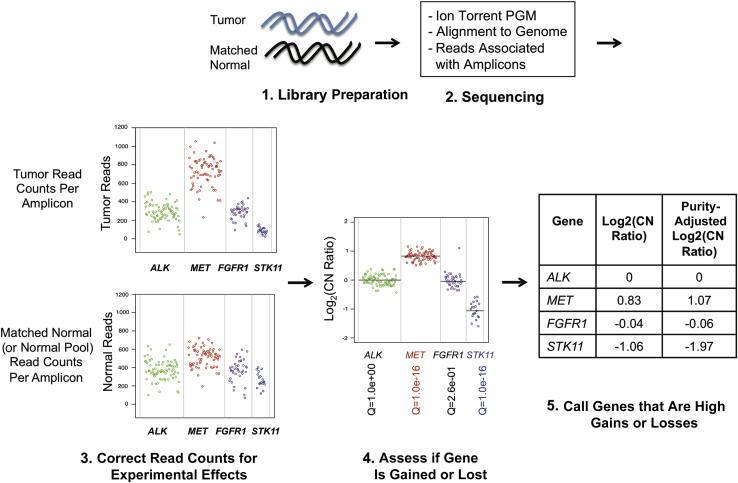
Figure 1 shows a schematic view of the algorithm. The approach is similar to that described by Lonigro et al12 for identifying CNAs in exome data, replacing average coverage of exon pull-down regions with read counts per amplicon. Amplicon-based libraries are prepared from tumor and matched normal DNA and sequenced on a semiconductor-based sequencing platform (Ion Torrent PGM). Next, reads are aligned to the genome and the number of reads per amplicon is tallied, with reads covering more than one amplicon being assigned to the amplicon that is most covered by that read. The amplicon-level read counts are normalized by dividing by the total number of reads from the sample to correct for sample-to-sample variation in total reads (Supplemental Figure S1). After this, the normalized tumor read counts are divided by the normalized reads from a matched normal sample (note: substitution with a normal pool is addressed below). This reduces amplicon-level effects, such as variability in mapping and primer efficiency (amplicon representation bias) (Supplemental Figure S2A).
Figure 1.
Overview of copy number analysis algorithm. (1) Library Preparation: tumor and matched normal genomic DNA are turned into targeted amplicon-based libraries. (2) Sequencing: the libraries are sequenced on the Ion Torrent PGM, and the reads are aligned to the human genome and associated with the amplicon targets. (3) Correct read counts for experimental effects: the tumor read counts for each amplicon are normalized for the total number of reads in the sample, and then divided by the normalized matched normal (or normal pool) read counts, and then GC content corrected to determine the copy number ratio for each amplicon. (4) Assess if gene is gained or lost: the weighted averages of the amplicon copy number ratios to generate a Log2(copy number ratio) for each gene are taken and then the variability of amplicons within the gene (and comparison with pooled normal samples, when available) is used to determine the q-value for the gene being gained or lost. (5) Call genes that are high gains or losses: the Log2(copy number ratio) for tumor purity is adjusted and then cutoffs of >0.58 for high gains and <−1 for high losses are applied. In this illustration, the amplicons for four genes are shown: ALK (green), MET (red), FGFR1 (purple), and STK11 (blue). The final weighted average for each gene is shown as a black line. A table shows the Log2(copy number ratios), both before and after the tumor purity correction. On the basis of the cutoffs for high gains and losses, MET and ALK are called.
The resulting Log2(raw copy number ratios) are corrected for the GC content in each amplicon, as previously described (Supplemental Figure S2B),19 to adjust for the observation that GC- and AT-rich fragments are underrepresented in sequencing due to the unimodal effect that GC content has on DNA melting temperature. This correction is standard for most sequencing-based CNA detection approaches. As can be seen in Supplemental Figure S2B, adjusting for GC content makes the Log2(copy number ratio) for most genes 0, as expected.
For a set of 14 normal samples and their matched breast tumors (Supplemental Table S2), amplicons emit reads with variance consistent with a Poisson process, once corrected for the total number of reads in the sample, the matched normal, and the GC content (Supplemental Figure S3). These data indicate that there are no unexpected, nonlinear effects in the PCR process that underlies the amplicon-based targeted sequencing method. Thus, the amplicon-based library approach allows for the use of a Poisson model for downstream statistics, similar to other NGS methods, and indicates that PCR duplicates do not need to be removed.
After GC content correction, the Poisson model is applied so that gene-level CNAs can be determined from the weighted average of the amplicon-level copy number ratios, in which the weight for each amplicon is proportional to the number of reads on that amplicon in the matched normal sample (Supplemental Figure S2B). Genes are regarded as significantly different from 0 if, after converting the P value to a q-value using the Benjamini-Hochberg procedure for controlling false discovery rate, the q-value is <0.01. We define a high gain as a gene with a copy number ratio strictly >1.5 (3 total copies/2 total copies = 1.5 total copies) and a high loss as a gene with a copy number ratio strictly <0.5 (1 total copy/2 total copies = 0.5 total copies) (Figure 1). To reduce the number of false negatives resulting from sample-to-sample variation in tumor purity (estimated on microscope review), we apply the linear formula [(gene-level copy number ratio – 1)/tumor fraction + 1] before using these cutoffs.
In a previous study of amplicon-based sequencing, we observed that the normalized coverage for individual amplicons was highly reproducible across 45 different tumor samples.4 Herein, we ran each of the normal samples listed in Supplemental Table S1 twice. Again, there was a high level of correlation between technical replicates (Supplemental Figure S4, A and B).
Concordance of Fold Change Measurements Using Hybrid-Capture Whole-Exome Sequence Data and Amplicon-Based Targeted Sequence Data
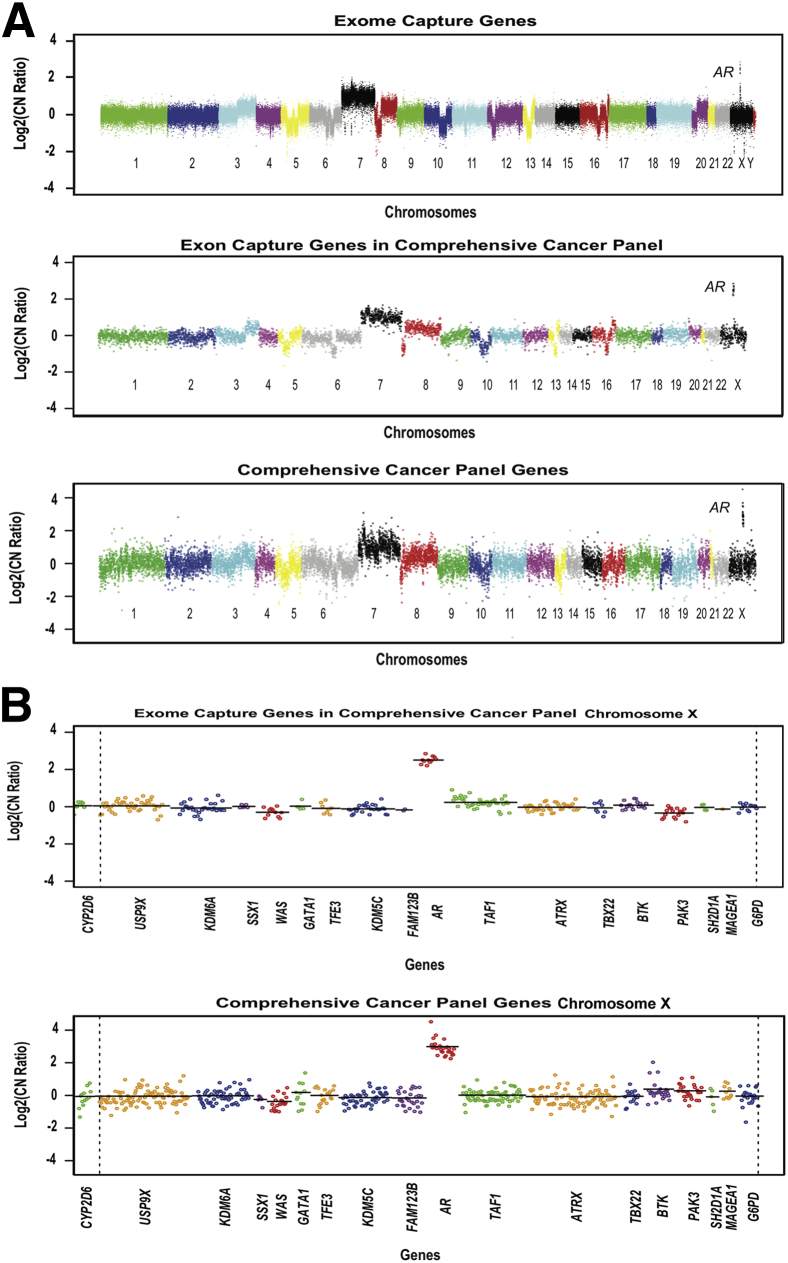
Whole-exome sequencing data were previously generated from a sample of fresh-frozen metastatic prostate tumor tissue and matching normal tissue using the Illumina (San Diego, CA) HiSeq 2000 and the Agilent Technologies (Santa Clara, CA) SureSelect Human All Exon Kit (approximately 18,000 genes and approximately 200,000 exons).18 DNAs from FFPE samples of the same tumor and normal tissue were subjected to targeted sequencing using the Ion Torrent CCP on the Ion Torrent PGM. Copy number plots from the exome capture and targeted sequencing approaches are compared in Figure 2A. When the full set of exons in the whole exome data was filtered down to the 409 genes on the CCP, the resulting Log2(copy number ratios) were highly concordant with those from the FFPE material run on the CCP panel. Large-scale amplifications and losses were similar in magnitude, including chromosome 3q gain, losses on chromosomes 5 and 6, gain of chromosome 7, loss in 8p, loss in the middle of chromosome 10, losses in chromosome 16, and an amplification on chromosome × focused on the AR gene (Figure 2B). Copy number ratios generated using exome capture data for the 409 genes in the CCP correlated well with those from the amplicon-derived data (R2 = 0.74 on a gene-by-gene level). There was somewhat greater variation in estimated copy number in the data from the amplicon-based library, possibly due to the effects of formalin fixation and paraffin embedding. Although this comparison was limited to only a single tumor-normal pair, it suggested that an amplicon-based library could yield copy number data comparable to a hybrid-capture library.
Figure 2.
Comparison of targeted sequencing to exome sequencing in detecting CNAs. A: Overall copy number across the genome is shown for metastatic prostate cancer sample WA25. Data from whole-exome sequencing of DNA from fresh-frozen tumor are compared with targeted sequencing of DNA, using the CCP, from FFPE tumor. The Log2(copy number ratio) for all exons in the exome capture and the exome sequencing restricted to the 409 genes present in the CCP targeted sequencing panel to facilitate comparison are shown. There is not necessarily an exact one-to-one match between amplicons and exon capture targets for each gene. Log2(copy number ratio) between tumor and matched normal tissue is shown on the vertical axis; each point represents the GC-content corrected, normalized, log-transformed ratio for targeted exon or amplicon, and ordered by genomic coordinates. B: Overall copy number across chromosome X for metastatic prostate sample WA25 by exome sequencing and targeted sequencing. Log2(copy number ratio) between the tumor and matched normal tissue is shown on the vertical axis; each point represents the GC-content corrected, normalized, log-transformed ratio for targeted exon or amplicon, and ordered by genomic coordinates. A line is drawn for the weighted average of the targeted exons or amplicons for each gene.
Using a Matched Normal versus a Normal Tissue Pool to Assess CNAs
To detect CNAs, a neutral copy number level must be established for each amplicon. As shown above, this can be done by comparison with a matched normal sample (Supplemental Figure S2A); however, this requires that the matched normal pool be sequenced, which effectively doubles the cost per sample. Moreover, a matched normal sample is not always available for each tumor sample. An alternative approach that is used in other high-throughput technologies, such as SNP arrays and aCGH, is to pool the amplicon-level read counts from multiple normal samples and use the resulting normal pool data set in place of reads from matched normal DNA during the data analysis. To establish a normal pool, we sequenced amplicon-based libraries from 14 normal breast tissue FFPE samples (Supplemental Table S1), along with 14 matched breast tumor FFPE samples using a 37 cancer-related gene panel. We found that the normalized reads from pairs of different normal samples showed a high level of correlation (Supplemental Figure S5A), and this high degree of agreement was consistent throughout the normal cohort (Supplemental Figure S5B). These data suggested that normalized read counts from unmatched samples might be substituted for those of a matched normal sample.
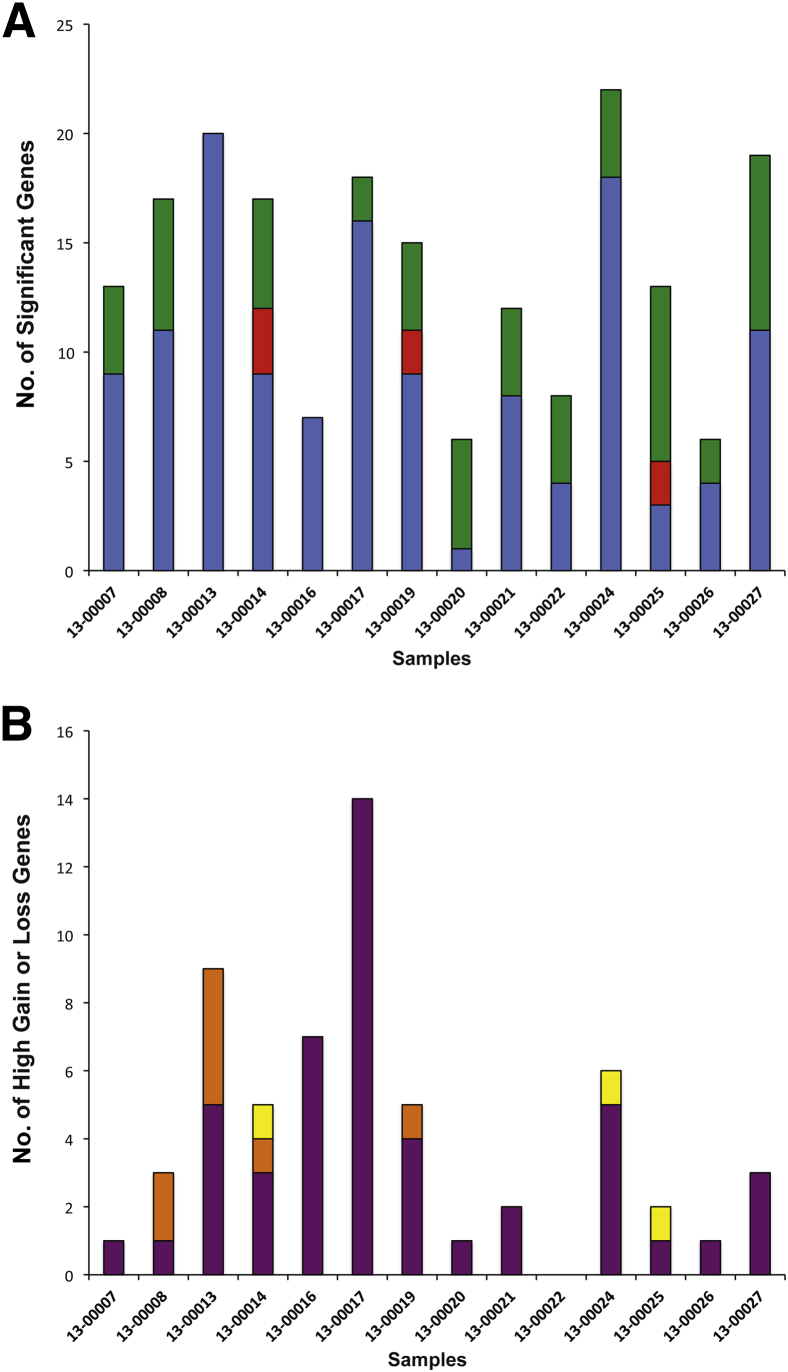
We next examined the CNA calls made for the 14 breast tumor samples, comparing (after normalization for read counts) each tumor analyzed against its matched normal pool versus each tumor analyzed against a pool of 13 unmatched normal controls. This was done to ensure that an individual tumor sample was compared only to normal controls from other subjects. There were a total of 193 significant (q < 0.01) CNA calls among the 14 breast tumors. Of these, 130 (67%) were significant using both the matched and pooled normal controls, 56 (29%) were significant only versus the matched normal controls, and 7 (4%) were significant only versus the pooled normal controls (Figure 3A). Closer examination revealed that among the 56 CNA calls made (only versus the matched normal controls), 45 fell within 3 SDs of the observed distribution of the Log2(copy number ratios) across all of the normal controls (Supplemental Figure S5C). This suggests that using a normal pool actually improves the significance assessment because it eliminates the noise that is inherent within a single matched normal sample. There were 18 CNAs for which significance calls differed between the two approaches; all of these fell between −0.75 and 1.33 copy number ratio (corrected for tumor purity), so they would not be called high gains or losses. A total of 59 genes were called as high gains and losses across the breast tumor samples by either or both approaches. Among these, there was agreement on 48 (Figure 3B). Of the 11 that did not agree (8 were called versus the normal pool and 3 were called versus the matched normal), all were close to the cutoff for high gain or the cutoff for high loss (Supplemental Figure S5D). These findings suggest that a normal pool can be used in place of matched normal pool for assessing high gains and losses, but that alterations of smaller magnitude are less reliably detected regardless of which approach is used.
Figure 3.
Comparison of copy number calls using both pooled and matched normal samples. A: Comparison of significant calls made using the matched normal sample or the normal pool. The bar plot shows the number of CNA calls made across the 14 matched breast tumor samples, indicating the number of significant calls that were made using both the matched normal and the normal pool (blue bars), as compared with those made only when using the matched normal pool (green bars) and those made only when using the normal pool (red bars). B: Comparison of high copy number gain or loss calls made using the matched normal sample or the normal pool. The bar plot shows the number of CNA calls made across the 14 matched breast tumor samples, indicating the number of high copy number gain or loss calls that were made using both the matched normal and the normal pool (purple bars), as compared with those made only when using the matched normal (yellow bars) and those made only when using the normal pool (orange bars).
Amplicon Characteristics that Influence the Assessment of Gene CNAs
Our study focused on a targeted 37 cancer-related gene panel designed to detect point mutations and indels in genes commonly mutated in solid tumors. To use this panel for CNA detection, we first had to determine whether any of the included amplicons did not consistently assess gene coverage, measured as the weighted average of the amplicon-level Log2(copy number ratio). In other words, we wanted to identify any amplicons that behaved significantly different from the others for a particular gene. For each amplicon, we looked at its GC-corrected copy number ratio across the sequencing data from 14 normal breast tissue samples, with each normal sample being compared with the pool of the other normal controls. There was a clear downward trend, indicating that amplicons yielding greater average read depths produced more consistent copy number estimates, as expected if the underlying emission of reads from amplicons is a Poisson process (Supplemental Figure S3A). Similarly, we looked at the GC-corrected copy number ratio of each amplicon across the 14 matched tumor samples (Supplemental Figure S3B), comparing each sample with the pool of normals with the sample's own matched normal removed. In this case, we normalized the copy number ratios for all amplicons for a gene by the estimated copy number ratio of that gene to correct for actual copy number changes. As with the normal samples, higher average coverage resulted in more consistent copy number estimates (Supplemental Figure S3B). More important, none of the amplicons was an outlier with respect to how consistently it measured a gene's copy number, indicating that no amplicons needed to be excluded from further calculations.
Next, we assessed whether particular genes needed to be excluded from consideration for CNA assessment as a result of having too few amplicons. At the time the panel was originally designed, CNA assessment was not a priority and the number of amplicons per gene on the panel ranged from 1 to 145. As expected, the variance in the copy number estimate decreased with increasing numbers of amplicons (Supplemental Figure S6). We selected four amplicons as a cutoff for this study, because there was a notable increase in variation lower than this level. We recognize that choosing a higher number of amplicons per gene would have somewhat further reduced the variance in CNA assessments, but it would have also restricted the list of assessable genes.
The Number of Reads Necessary to Detect CNAs
To assess how the total number of reads in a sample affects CNA calling, we performed the CNA analysis on breast tumor sample 13-00027 using all 610,000 reads (average number of reads per amplicon, 558) generated when the sample was run on the PGM (Supplemental Figure S7A). Then, we repeated the analysis after sequentially removing randomly selected read counts down to the level of 60,000 total reads (10-fold decrease; average number of reads per amplicon, 55.8) and 6000 total reads (100-fold decrease; average number of reads per amplicon, 5.58) (Supplemental Figure S7, B and C, respectively). The high-level gains and losses affecting DDR2, ERBB2, and RB1 were significant and called in all three analyses despite the increased noise in the samples with 10- and 100-fold reductions in reads. KRAS and MAP2K1 were falsely called for the sample with a 100-fold reduction, indicating that the amplicon-level noise will result in false calls as the number of reads decreases. These results indicate that the bias due to read depth per amplicon, shown in Supplemental Figure S3, is present, but only affects calling of high gains and losses at low numbers of reads that are not typically accepted in targeted sequencing runs.
Effect of Tumor Purity on the Sensitivity for CNA Detection
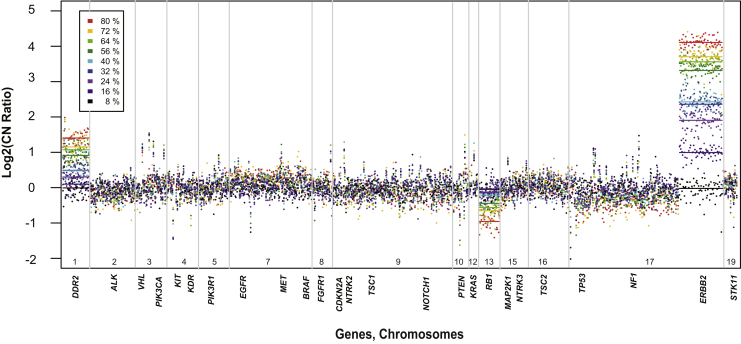
To assess the sensitivity for detecting CN alterations, we prepared sequencing libraries from breast tumor sample 27 (80% tumor nuclei on the basis of microscope estimation) diluted with varying amounts of its matched normal DNA, and sequenced them using the 37-gene panel. The three most significant CN changes (DDR2, ERBB2, and RB1) showed the expected decrease in CN ratio with increasing tumor dilution (Figure 4). Interestingly, the copy number for ERBB2, which by FISH was highly amplified in this tumor (ERBB2/Cen17 ratio, >5.55), correlated linearly with tumor dilution (R2 = 0.98) (Supplemental Figure S8A) and was significantly higher than other genes down to the level of 8% tumor. Loss of RB1 and gain of DDR2 were likewise linear across the dilution series (R2 = −0.97 and R2 = 0.97, respectively) (Supplemental Figure S8B), and was significant down to 16% tumor purity. Together, these data show that copy number ratios are linear in relation to tumor purity, making it possible to use the tumor purity to rescale the copy number ratios before applying cutoffs for high gains and losses. But, genes with greater CN changes can be detected at lower tumor purity than those with less significant changes.
Figure 4.
Serial dilution to test sensitivity of CNA detection in targeted sequencing. Visualization of dilutions of breast tumor 13-00027 with its matched normal DNA. Each dilution has a unique color. Lines represent the weighted average for copy number ratio for each gene.
Compensating for Tumor Purity by Increasing the Number of Amplicons
As discussed above, tumor purity has a profound effect on sensitivity for CNA assessment. One way to compensate for this might be to increase the number of amplicons covering a gene of interest. Revisiting breast tumor sample 27, the sample diluted with varying amounts of its matched normal DNA, we examined the ERBB2 gain while varying the number of ERBB2 amplicons included in the algorithm. For each tumor purity level, box-and-whisker plots were generated for both copy number ratios and Z-scores for 10 random samplings of the set of ERBB2 amplicons, varying the number of amplicons from 1 to a maximum of 71. Not surprisingly, the copy number ratio estimate became more accurate as the number of amplicons was increased (Supplemental Figure S9A). Z-score data derived from the same calculations confirmed that increasing the number of amplicons increased sensitivity; this was particularly evident at intermediate tumor purities (Supplemental Figure S9B). Data from RB1 (35 amplicons) yielded parallel results regarding copy number loss (Supplemental Figure S9, C and D). Thus, increasing the number of amplicons across a gene can partially compensate for tumor purity.
Comparison of CNA Calling with aCGH
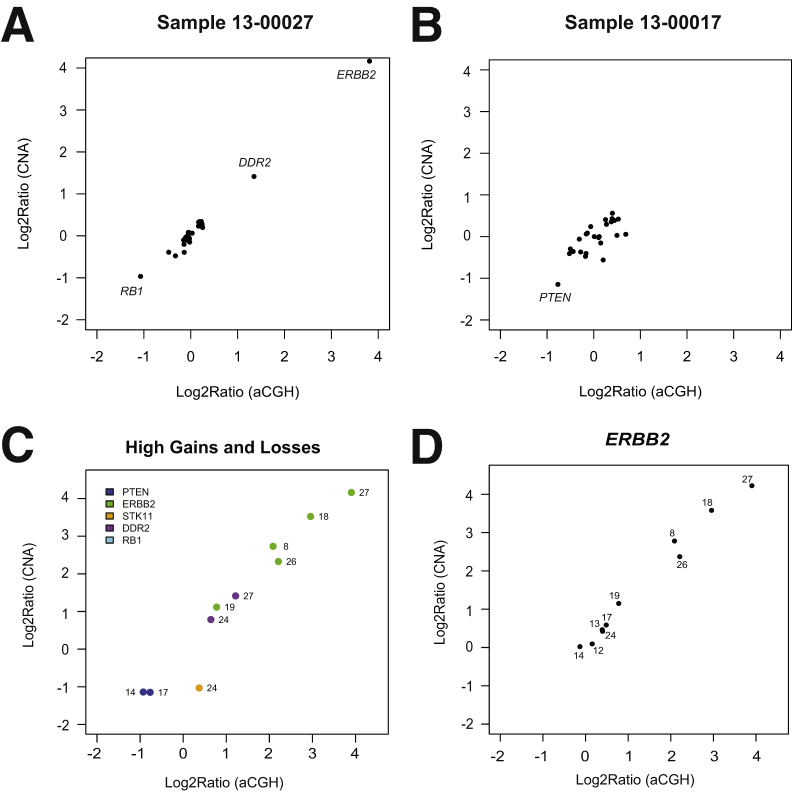
Gene-level fold-change assessments performed on whole-exome–capture sequencing data and amplicon-based sequencing data compared favorably (Figure 2). To further assess the accuracy of the gene-level fold changes called on amplicon-based sequencing data, we compared the results with aCGH data for 10 breast tumor samples. Figure 5, A and B, shows the linear relationship between the fold changes measured using aCGH and amplicon-level sequencing for samples 13-00027 and 13-00017, respectively. The two methods showed good concordance for all high gains and high losses across 10 breast tumor samples (Figure 5C). This concordance supports the accuracy of the targeted sequencing approach for the high gains and losses. There was a linear relationship between the fold changes measured using aCGH and amplicon-level sequencing for ERBB2 (Figure 5D). Thus, amplicon-based sequencing has sufficient dynamic range to accurately assess fold change over a large range of values.
Figure 5.
Comparison of fold change measurement with aCGH data. A and B: Log2(copy number ratios) for all genes from targeted amplicon-based sequencing approach are plotted versus Log2(copy number ratios) assessed using aCGH for sample 13-00027 (A) and sample 13-00017 (B). C: Log2(copy number ratios) from targeted amplicon-based sequencing are plotted versus Log2(copy number ratios) assessed using aCGH for observations that were high gains or losses (copy number ratio, >1.5 or <0.5) on the basis of the targeted sequencing approach among 10 breast tumor samples. D:ERBB2 Log2(copy number ratios) from targeted amplicon-based sequencing of 10 breast tumor samples are plotted versus Log2(copy number ratios) assessed using aCGH. Sample names have been abbreviated (eg, sample 13-00012 is denoted as 12).
Identifying Clinically Relevant CNA and Somatic Mutations
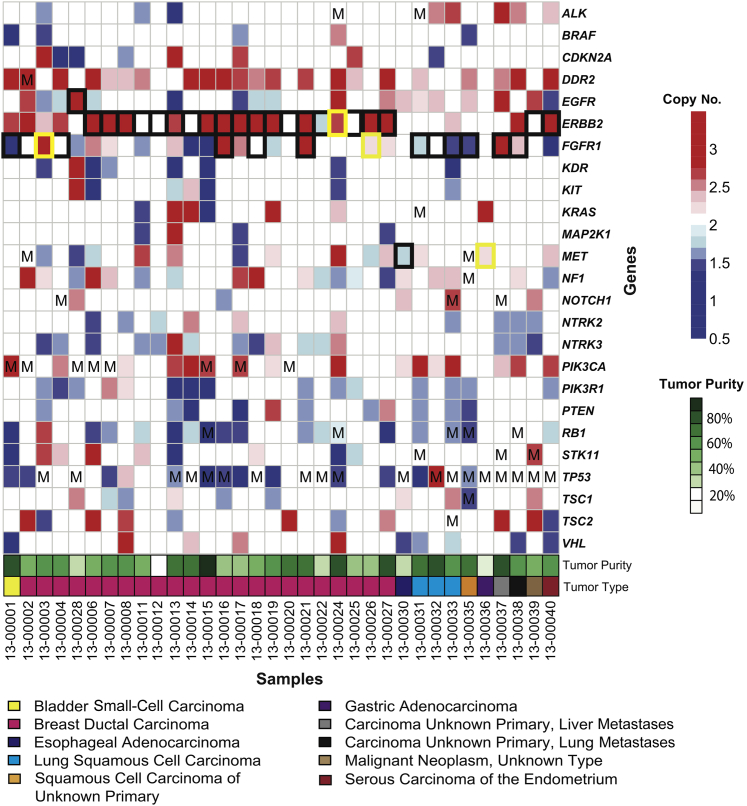
The 37-gene panel was run on 34 tumors of various types, including 23 breast carcinomas and 11 other carcinomas, with estimated tumor purity ranging from 15% to 80%. Mutations were identified in several genes that are typically altered in these malignancies (eg, TP53, PIK3CA, RB1, and STK11) (Figure 6). Equally important, there was a good consistency between high copy number gains determined by sequencing compared with FISH (Figure 6 and Supplemental Table S3): 33 of 38 FISH results agreed in total, including 20 of 21 for ERBB2, 11 of 14 for FGFR1, 1 of 2 for MET, and 1 of 1 for EGFR. Consistent with our results, all of the high copy number gain calls with high tumor purity-corrected copy number ratios were in agreement with the FISH results. This included 16 calls in which the tumor purity-corrected copy number ratio was >1.87. Notably, all but one of these would have been called without the tumor purity adjustment. When the tumor purity-adjusted copy number ratios were between 0.51 and 1.45 and a negative call was made, the results agreed with FISH. Among the 38 FISH calls, there were five that differed from the targeted sequencing algorithm. All of these were close to the 1.5 high gain cutoff, ranging from 1.23 to 1.44 for four that were false negatives, and 1.78 for one case that was a false positive. Inaccuracy in tumor purity estimates may have contributed to these discrepancies. Certainly, borderline cases would be worthy of further exploration.
Figure 6.
Summary of all significant CNAs and somatic mutations across 34 solid tumors. Significant (q < 0.01) gains and losses are indicated by red squares and blue squares, respectively, whereas white squares indicate the gene was not significantly gained or lost. FISH for ERBB2, FGFR1, and/or MET was performed on a subset of tumors and correlated with high gain or loss calls made using the targeted sequencing approach. Agreement is shown by a black border, whereas disagreement is shown by a yellow border. Mutations (Ms) were identified across several genes using the targeted sequencing approach.
One of the benefits of this method over non–sequencing-based platforms, such as FISH, is the ability to combine point mutation data with copy number data to make assessments. For example, we found that when the known tumor suppressors TP53, RB1, PTEN, and PIK3R1 harbored likely deleterious mutations, they nearly always showed decreased CN with the targeted sequencing algorithm (Figure 6).
Discussion
Alterations in gene copy number are of increasing interest in the era of precision medicine. ERBB2 amplification is a well-established biomarker for the treatment of breast and gastric carcinomas with trastuzumab. Similarly, amplifications of FGFR1, EGFR, MET, and PIK3CA are all being targeted in ongoing clinical trials. Most clinical laboratories assess these alterations using immunohistochemistry and/or FISH; however, these methods consume considerable amounts of tumor material while generally yielding results for only one gene at a time. The recent introduction of next-generation DNA sequencing into the laboratory presents the opportunity to screen for CNAs across many genes simultaneously. This has been done successfully using sequencing library protocols that are based on hybridization-capture approaches, but to date, there has been little effort focused on amplicon-based libraries.
Herein, we undertook a systematic assessment of CNA detection in sequencing data generated from amplicon-based libraries. We developed a relatively simple algorithm beginning with the assignment of reads to specific amplicons, followed by normalization for total reads, comparison to a matched normal, GC-content correction, a statistical analysis for the likelihood of true variation from the normal, adjustment for tumor purity, and a call of high gain or loss on the basis of a cutoff. We determined that this algorithm generated results that compared favorably with data from a hybrid-capture library. In addition, we found that averaged amplicon reads from a pool of normal DNA samples could substitute for reads from a matched normal sample. Indeed, use of the pooled normal reduced the number of statistical CNA calls that might otherwise be regarded as marginal.
We focused primarily on high gains and losses in this study (>1.5 or <0.5 copy number ratio). Although single-copy variation may be biologically important in some cases, and larger gains and losses occurring in subclonal populations might also affect tumor behavior, there are inherent limitations to a sequencing-based approach using data from only a few dozen genes. Sampling across a larger number of genes and analysis of SNPs may support more robust copy number analysis and the detection of loss of heterozygosity.20 Nevertheless, a relatively small panel of genes can still provide information on high gains and losses, which are being targeted in many ongoing clinical studies.
In examining the data from amplicon-based libraries, we looked for additional factors that might influence CNA calls. We found that the distribution of reads from an amplicon across many samples followed a Poisson model. No outlier amplicons were identified, indicating that standard statistics could be applied in our analyses and that no individual amplicons needed to be excluded from the calculation. On the basis of these results, we looked at the impact of amplicon number per gene and, as expected, observed that the fewer the number of amplicons, the larger the SE in measuring copy number. Although we selected four amplicons as the minimum in our study, large increases in copy number can still be detected with just one or two amplicons per gene. On the other hand, increasing the amplicon number to 10 or even greater would likely allow more subtle alterations to be picked up. These observations should be kept in mind when designing new amplicon-based sequencing panels.
Average read depths also had an impact on CNA detection, with increasing noise becoming apparent when reads were reduced by 10-fold or more. Nevertheless, significant CNAs were still readily detected. The 37 cancer-related gene panel used in our study is routinely sequenced to an average of 1000 reads per amplicon. A run yielding only 10% of this read depth would not be reported by our laboratory; however, our data suggest that CNAs can still be reliably identified even if the total read counts fall lower than expected levels.
One of the most important factors affecting CNA detection is tumor purity (ie, the fraction of the sample composed of tumor cells as opposed to stromal cells, lymphoid cells, and other normal cellular elements). We observed a linear correlation between copy number across tumor fractions ranging from 80% to 16%, indicating that a tumor purity correction on the basis of a simple rescaling of the copy number ratios is reasonable. More important, whether a gain (ERBB2) or loss (RB1) was detectable depended on both the degree of copy alteration and the tumor content in the specimen. Although a highly amplified gene might be detected at <20% tumor cells, observing a gene loss may require 50% or more tumor cells. Unfortunately, pathologists are not accurate in their estimates of tumor content. In one recent study comparing nine pathologists, 38% of samples containing <20% tumor content were inappropriately judged to be sufficient for testing (>20% tumor).21 Given the relative uncertainty surrounding H&E-based estimates of tumor content, there remains a risk for false-negative CNA calls, particularly in the setting of low tumor fraction and borderline alterations (close to 1.5 or 0.5 for copy number ratio). Such calls need to be interpreted carefully.
In summary, we have developed and validated an approach for detecting CNAs in sequencing data from amplicon-based libraries. Use of the algorithm in the analysis of samples from previously diagnosed cases has already brought to light amplifications of known actionable genes that would have been otherwise missed. The approach nicely complements mutation detection and broadens the possible clinical utility of NGS.
Footnotes
Supported by Knight Cancer Institute research funds and the Department of Pathology, Oregon Health and Science University research funds.
Disclosures: K.R. and M.A. are employees of Ion Torrent, a division of ThermoFisher, and participate in the company stock plan. C.L.C. has received honoraria and travel support from Ion Torrent. Some reagents used in the study were provided by Ion Torrent/Life Technologies (ThermoFisher).
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2014.09.008.
Supplemental Data
Normalization of read counts by the total number of reads in the sample. For amplicon AMPL1128718 (length, 102 bp), A shows the total number of reads on the amplicon versus the total number of reads in the sample across the 14 breast normal samples, whereas B shows the amplicon read fraction, which is the total number of reads on the amplicon normalized by the total number of reads from the sample, across the 14 breast normal samples. The red line is the best fit for the amplicon read fraction.
Key algorithm steps illustrated for breast sample 13-00020. A: Normalization of the amplicon-level read counts in the tumor by the matched normal tissue. The amplicon-level read counts for each gene and the copy number ratios, resulting from dividing the tumor amplicon-level read counts by the normal amplicon-level read counts, are shown. B: GC-content correction of the copy number ratios for each amplicon. The uncorrected Log2(copy number ratios) for each amplicon and the GC-content corrected Log2(copy number ratios) for each amplicon are shown. The black lines indicate the weighted average of the amplicon-level copy number ratios. The amplicons from each gene are highlighted using alternating colors. Chromosomes are numbered along the bottom of each panel.
Assessment of how consistently each amplicon assesses the copy number of each gene. For the 14 breast normal samples and the 14 matched breast tumor samples, each amplicon's GC-corrected copy number ratio was divided by the estimated copy number of the gene. A: Plot shows the SD of each amplicon's relative copy number estimate among the 14 breast normal samples, versus the average reads from the amplicon over the same set of samples. B: Plot shows the SD of each amplicon's relative copy number estimate among the 14 matched breast tumor samples versus the average reads from the amplicon over the same set of samples. The shaded regions represent a range of Poisson models chosen so that 1% of the probes are lower than the lower limit (×1.34 and ×1.35 for the normal and tumor distributions, respectively) and 1% are higher than the upper limit (×5.92 and ×6.93); the blue curves represent the median Poisson model (×2.78 and ×3.07).
Similarity of technical replicates. A: Visualization of the similarity between two technical replicates of normal breast tissue sample 13-00017. The scatterplot shows the read counts per amplicon. B: Visualization of the similarity between all technical replicates for breast normal samples. The histogram shows the R2 for each of the technical replicate pairs.
Assessment of the feasibility of replacing the matched normal pool with a normal pool. A: Visualization of the similarity between two normal samples. The scatterplot shows the read counts per amplicon for two normal breast samples, 13-00017 and 13-00020. B: Visualization of the similarity between the full set of 14 breast normal samples. The histogram shows the distribution of R2 for the read counts per amplicon for each normal-to-normal comparison. C: For each gene assessed as significant in the matched normal pool, but not the normal pool, its Log2(copy number ratio) is shown beside the box-and-whisker plot, showing the variability of this value in the set of breast normal samples. Next to each box-and-whisker data point are whiskers showing where 3 SDs lie on the distribution. D: For each high copy gain or loss that was called differently when using the matched normal versus the normal pool, the copy number ratio estimate generated by each approach is shown. The red line shows the cutoff for a high gain (>1.5). The blue line shows the cutoff for a high loss (<0.5).
Amount of variance in copy number estimates relative to the number of amplicons per gene. By using sequencing data from the 14 normal breast samples, box-and-whisker plots were generated for the copy number estimate for each gene. The number of amplicons for each gene is shown in parentheses after the name of the gene, and the genes are sorted in order of increasing number of amplicons.
Total reads in the sample necessary to detect CNAs. For breast tumor sample 13-00027, the Log2(copy number ratio) is shown for each amplicon. The numbers at the bottom of each panel correspond to different chromosomes. There is a horizontal black line showing the weighted average of all of the amplicons for that gene. The gene names in red indicate that the gene is significantly increased relative to neutral, whereas the gene names in blue indicate that the gene is significantly decreased relative to neutral. The asterisks indicate genes that are high gains or losses. A: CNA analysis on the basis of 610,000 total reads (an average of 558 reads per amplicon) originally generated from the sample. B: CNA analysis on the basis of 61,000 total reads (an average of 55.8 reads per amplicon) from the sample. C: CNA analysis on the basis of 6100 total reads (an average of 5.58 reads per amplicon) from the sample.
Effect of tumor purity on CNA detection sensitivity in targeted sequencing. A: Plot of ERBB2 copy number ratio versus tumor purity (blue points; the slope of the black line is the correlation R2). B: Plot of RB1 copy number ratio versus tumor purity and DDR2 copy number ratio versus tumor purity (green and red points, respectively; the slope of the black line is the correlation R2). Points that are x's are not significant, whereas squares are significant.
Assessment of accuracy and sensitivity in calling ERBB2 gain and RB1 loss relative to tumor purity and the number of amplicons assessed. For ERBB2, for each tumor purity level, the box-and-whisker plot of the copy number ratio (A) or the Z-score (B) is shown for 10 random samplings of the set of ERBB2 amplicons. The box-and-whisker plots are shown in order of increasing number of amplicons, where the number of amplicons varies from 1 to 71. For RB1, for each tumor purity level, the box-and-whisker plot of the copy number ratio (C) and Z-score (D) is shown for 10 random samplings of the set of RB1 amplicons. The box-and-whisker plots are shown in order of increasing number of amplicons, ranging from 1 to 35.
References
- 1.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerick M., Isau M., Timmermann B., Sultmann H., Herwig R., Krobitsch S., Schaefer G., Verdorfer I., Bartsch G., Klocker H., Lehrach H., Schweiger M.R. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68. doi: 10.1186/1755-8794-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagle N., Berger M.F., Davis M.J., Blumenstiel B., Defelice M., Pochanard P., Ducar M., Van H.P., Macconaill L.E., Hahn W.C., Meyerson M., Gabriel S.B., Garraway L.A. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beadling C., Neff T.L., Heinrich M.C., Rhodes K., Thornton M., Leamon J., Andersen M., Corless C.L. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mol Diagn. 2013;15:171–176. doi: 10.1016/j.jmoldx.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Won H.H., Scott S.N., Brannon A.R., Shah R.H., Berger M.F. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013;(80):e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Endris V., Penzel R., Warth A., Muckenhuber A., Schirmacher P., Stenzinger A., Weichert W. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: feasibility, costs, and performance compared with conventional sequencing. J Mol Diagn. 2013;15:765–775. doi: 10.1016/j.jmoldx.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard C.C., Salipante S.J., Koehler K., Smith C., Scroggins S., Wood B., Wu D., Lee M.K., Dintzis S., Adey A., Liu Y., Eaton K.D., Martins R., Stricker K., Margolin K.A., Hoffman N., Churpek J.E., Tait J.F., King M.C., Walsh T. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16:56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell C.E., Al-Kateb H., Bredemeyer A.J., Duncavage E.J., Spencer D.H., Abel H.J., Lockwood C.M., Hagemann I.S., O'Guin S.M., Burcea L.C., Sawyer C.S., Oschwald D.M., Stratman J.L., Sher D.A., Johnson M.R., Brown J.T., Cliften P.F., George B., McIntosh L.D., Shrivastava S., Nguyen T.T., Payton J.E., Watson M.A., Crosby S.D., Head R.D., Mitra R.D., Nagarajan R., Kulkarni S., Seibert K., Virgin H.W., Milbrandt J., Pfeifer J.D. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16:89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones M.A., Bhide S., Chin E., Ng B.G., Rhodenizer D., Zhang V.W., Sun J.J., Tanner A., Freeze H.H., Hegde M.R. Targeted polymerase chain reaction-based enrichment and next generation sequencing for diagnostic testing of congenital disorders of glycosylation. Genet Med. 2011;13:921–932. doi: 10.1097/GIM.0b013e318226fbf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B., Morrison C.D., Johnson C.S., Trump D.L., Qin M., Conroy J.C., Wang J., Liu S. Computational methods for detecting copy number variations in cancer genome using next generation sequencing: principles and challenges. Oncotarget. 2013;4:1868–1881. doi: 10.18632/oncotarget.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonigro R.J., Grasso C.S., Robinson D.R., Jing X., Wu Y.M., Cao X., Quist M.J., Tomlins S.A., Pienta K.J., Chinnaiyan A.M. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia. 2011;13:1019–1025. doi: 10.1593/neo.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Dairkee S.H., Ljung B.M., Gray J.W., Albertson D.G. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 14.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Oseroff V.V., Albertson D.G., Pinkel D., Eichler E.E. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A., Elliott A.L., Parkin M., Hubbell E., Webster T., Mei R., Veitch J., Collins P.J., Handsaker R., Lincoln S., Nizzari M., Blume J., Jones K.W., Rava R., Daly M.J., Gabriel S.B., Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 16.Chiang D.Y., Getz G., Jaffe D.B., O'Kelly M.J., Zhao X., Carter S.L., Russ C., Nusbaum C., Meyerson M., Lander E.S. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore D.A., Saldanha G., Ehdode A., Potter L., Dyall L., Bury D., Pringle J.H. Accurate detection of copy number changes in DNA extracted from formalin-fixed, paraffin-embedded melanoma tissue using duplex ratio tests. J Mol Diagn. 2013;15:687–694. doi: 10.1016/j.jmoldx.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., Asangani I.A., Ateeq B., Chun S.Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J.R., Palanisamy N., Ryslik G.A., Vandin F., Raphael B.J., Kunju L.P., Rhodes D.R., Pienta K.J., Chinnaiyan A.M., Tomlins S.A. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y., Speed T.P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho C.C., Mun K.S., Naidu R. SNP array technology: an array of hope in breast cancer research. Malays J Pathol. 2013;35:33–43. [PubMed] [Google Scholar]
- 21.Smits A.J., Kummer J.A., de Bruin P.C., Bol M., van den Tweel J.G., Seldenrijk K.A., Willems S.M., Offerhaus G.J., de Weger R.A., van Diest P.J., Vink A. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014;27:168–174. doi: 10.1038/modpathol.2013.134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalization of read counts by the total number of reads in the sample. For amplicon AMPL1128718 (length, 102 bp), A shows the total number of reads on the amplicon versus the total number of reads in the sample across the 14 breast normal samples, whereas B shows the amplicon read fraction, which is the total number of reads on the amplicon normalized by the total number of reads from the sample, across the 14 breast normal samples. The red line is the best fit for the amplicon read fraction.
Key algorithm steps illustrated for breast sample 13-00020. A: Normalization of the amplicon-level read counts in the tumor by the matched normal tissue. The amplicon-level read counts for each gene and the copy number ratios, resulting from dividing the tumor amplicon-level read counts by the normal amplicon-level read counts, are shown. B: GC-content correction of the copy number ratios for each amplicon. The uncorrected Log2(copy number ratios) for each amplicon and the GC-content corrected Log2(copy number ratios) for each amplicon are shown. The black lines indicate the weighted average of the amplicon-level copy number ratios. The amplicons from each gene are highlighted using alternating colors. Chromosomes are numbered along the bottom of each panel.
Assessment of how consistently each amplicon assesses the copy number of each gene. For the 14 breast normal samples and the 14 matched breast tumor samples, each amplicon's GC-corrected copy number ratio was divided by the estimated copy number of the gene. A: Plot shows the SD of each amplicon's relative copy number estimate among the 14 breast normal samples, versus the average reads from the amplicon over the same set of samples. B: Plot shows the SD of each amplicon's relative copy number estimate among the 14 matched breast tumor samples versus the average reads from the amplicon over the same set of samples. The shaded regions represent a range of Poisson models chosen so that 1% of the probes are lower than the lower limit (×1.34 and ×1.35 for the normal and tumor distributions, respectively) and 1% are higher than the upper limit (×5.92 and ×6.93); the blue curves represent the median Poisson model (×2.78 and ×3.07).
Similarity of technical replicates. A: Visualization of the similarity between two technical replicates of normal breast tissue sample 13-00017. The scatterplot shows the read counts per amplicon. B: Visualization of the similarity between all technical replicates for breast normal samples. The histogram shows the R2 for each of the technical replicate pairs.
Assessment of the feasibility of replacing the matched normal pool with a normal pool. A: Visualization of the similarity between two normal samples. The scatterplot shows the read counts per amplicon for two normal breast samples, 13-00017 and 13-00020. B: Visualization of the similarity between the full set of 14 breast normal samples. The histogram shows the distribution of R2 for the read counts per amplicon for each normal-to-normal comparison. C: For each gene assessed as significant in the matched normal pool, but not the normal pool, its Log2(copy number ratio) is shown beside the box-and-whisker plot, showing the variability of this value in the set of breast normal samples. Next to each box-and-whisker data point are whiskers showing where 3 SDs lie on the distribution. D: For each high copy gain or loss that was called differently when using the matched normal versus the normal pool, the copy number ratio estimate generated by each approach is shown. The red line shows the cutoff for a high gain (>1.5). The blue line shows the cutoff for a high loss (<0.5).
Amount of variance in copy number estimates relative to the number of amplicons per gene. By using sequencing data from the 14 normal breast samples, box-and-whisker plots were generated for the copy number estimate for each gene. The number of amplicons for each gene is shown in parentheses after the name of the gene, and the genes are sorted in order of increasing number of amplicons.
Total reads in the sample necessary to detect CNAs. For breast tumor sample 13-00027, the Log2(copy number ratio) is shown for each amplicon. The numbers at the bottom of each panel correspond to different chromosomes. There is a horizontal black line showing the weighted average of all of the amplicons for that gene. The gene names in red indicate that the gene is significantly increased relative to neutral, whereas the gene names in blue indicate that the gene is significantly decreased relative to neutral. The asterisks indicate genes that are high gains or losses. A: CNA analysis on the basis of 610,000 total reads (an average of 558 reads per amplicon) originally generated from the sample. B: CNA analysis on the basis of 61,000 total reads (an average of 55.8 reads per amplicon) from the sample. C: CNA analysis on the basis of 6100 total reads (an average of 5.58 reads per amplicon) from the sample.
Effect of tumor purity on CNA detection sensitivity in targeted sequencing. A: Plot of ERBB2 copy number ratio versus tumor purity (blue points; the slope of the black line is the correlation R2). B: Plot of RB1 copy number ratio versus tumor purity and DDR2 copy number ratio versus tumor purity (green and red points, respectively; the slope of the black line is the correlation R2). Points that are x's are not significant, whereas squares are significant.
Assessment of accuracy and sensitivity in calling ERBB2 gain and RB1 loss relative to tumor purity and the number of amplicons assessed. For ERBB2, for each tumor purity level, the box-and-whisker plot of the copy number ratio (A) or the Z-score (B) is shown for 10 random samplings of the set of ERBB2 amplicons. The box-and-whisker plots are shown in order of increasing number of amplicons, where the number of amplicons varies from 1 to 71. For RB1, for each tumor purity level, the box-and-whisker plot of the copy number ratio (C) and Z-score (D) is shown for 10 random samplings of the set of RB1 amplicons. The box-and-whisker plots are shown in order of increasing number of amplicons, ranging from 1 to 35.