Abstract
The autism spectrum disorders (ASDs) are a collection of human neurological disorders with heterogeneous etiologies. Hyperactivity of E3 ubiquitin (Ub) ligase UBE3A, stemming from 15q11-q13 copy number variations, accounts for 1%-3% of ASD cases worldwide, but the underlying mechanisms remain incompletely characterized. Here we report that the functionality of ALDH1A2, the rate-limiting enzyme of retinoic acid (RA) synthesis, is negatively regulated by UBE3A in a ubiquitylation-dependent manner. Excessive UBE3A dosage was found to impair RA-mediated neuronal homeostatic synaptic plasticity. ASD-like symptoms were recapitulated in mice by overexpressing UBE3A in the prefrontal cortex or by administration of an ALDH1A antagonist, whereas RA supplements significantly alleviated excessive UBE3A dosage-induced ASD-like phenotypes. By identifying reduced RA signaling as an underlying mechanism in ASD phenotypes linked to UBE3A hyperactivities, our findings introduce a new vista of ASD etiology and facilitate a mode of therapeutic development against this increasingly prevalent disease.
Keywords: ASD, UBE3A, ubiquitylation, ALDH1A2, retinoic acid, RA signaling, synaptic plasticity
Introduction
The autism spectrum disorders (ASDs) are a heterogeneous group of neurodevelopmental diseases characterized by a triad of core defects in social communication, reduced social interactions and stereotyped or repetitive behaviors1,2. The underlying genetic risk factors identified so far include de novo point mutations or copy number variations (CNVs) in large sets of genes that participate in diverse functions across many biological pathways3,4,5,6,7,8,9. Remarkably, despite such a complicated genetic landscape, ASD patients manifest similar core phenotypic deficits with typical perturbation in synaptic homeostasis, suggesting that there might exist certain common underlying mechanisms10,11. It is thus both intriguing and of impelling importance to elucidate the mechanisms underlying causal genetic variations and perturbed synaptic homeostasis in ASD.
The maternal 15q11-q13 duplication and isodicentric chromosome 15 (idic 15) have been identified in 1%-3% ASD cases worldwide3,4,12. The UBE3A gene, an E3 Ub ligase, has been validated as the major contributor for 15q11-q13 CNV symptoms in many population-based studies4,13,14,15, while transgenic mice expressing UBE3A at increased dosages also manifested three core autistic behavioral traits16, thus identifying UBE3A hyperactivities as one of the best defined genetic causes of ASD subtypes. More recently, an autism-linked mutation was found to disrupt protein kinase A (PKA)-mediated phosphorylation of UBE3A and result in excessive UBE3A activity and abnormal synapse formation17. In eukaryotes, E3 Ub ligases regulate a myriad of fundamental biological processes, mainly through conjugating single or multiple Ub moieties to their specific substrates in dynamic and spatiotemporally controlled manners4,18,19,20,21,22. Thus far, many proteins have been identified as E3 Ub ligase substrates for UBE3A, but few of these substrates have offered any potential ASD intervention opportunities. Because of the possible existence of yet unknown UBE3A substrate(s) whose functionalities might be dysregulated in ASD brains23, it is of critical importance to screen for UBE3A substrates of closer relevance to neuronal activities.
Retinol (vitamin A), whose deficiency is the leading cause of preventable childhood blindness (www.unicef.org), is one of the key nutrients that mammals cannot make and must take in principally from food and drink. In the cell, vitamin A is converted into retinoic acid (RA) through oxidation steps sequentially catalyzed by enzymes including: (1) the retinol dehydrogenases (mainly RDH10) that metabolize retinol to retinaldehyde; and (2) the ALDH1A family proteins (ALDH1A1-3, also named RALDH1-3), the rate-limiting retinaldehyde dehydrogenases that oxidize retinaldehyde into RA, the major active form of retinol24. RA is a key signaling molecule that regulates and mediates many fundamental biological processes, through its nuclear receptor, as indicated by genetic studies that eliminate RA biosynthesis, and perhaps extra-nuclear mechanisms, although these have not been demonstrated genetically25,26,27,28. In the nervous system, dynamically controlled RA homeostasis is not only critical for neuron differentiation, brain development and patterning27, but also essential for brain function, particularly in the maintenance of synaptic plasticity28. For example, RA synthesis is rapidly induced by loss of synaptic activity, followed by the direct activation of the local translation of AMPA receptors through an extra-nuclear RARα-dependent mechanism28,29,30. Exactly how the functionalities of ALDH1A family proteins as well as RA homeostasis might be dynamically regulated in neural systems and contribute to human neurological disorders such as ASD is still incompletely understood.
Here we report that UBE3A interacts with and ubiquitylates ALDH1A2, as well as the other members of the ALDH1A family of proteins, in a non-proteolytic manner, thereby inhibiting RA biogenesis and disrupting cellular RA homeostasis. Patch-clamp analyses with cultured primary neurons reveal that RA production in response to neuronal activity blockade is inhibited in the presence of UBE3A hyperactivities, subsequently interrupting RA-mediated neuronal synaptic scaling. Furthermore, ASD-like behavioral traits recapitulated in mice overexpressing UBE3A could be significantly alleviated by supplementing RA. Moreover, administration of disulfiram (DSF), an ALDH1A antagonist31, was sufficient to cause ASD-like phenotypes in mice. Our results might have thus uncovered perturbed RA homeostasis as a key mechanism underlying UBE3A hyperactivities-linked ASD-like behaviors, with potential implications for clinical interventions.
Results
Identification of ALDH1A family proteins as novel interacting partners for human UBE3A
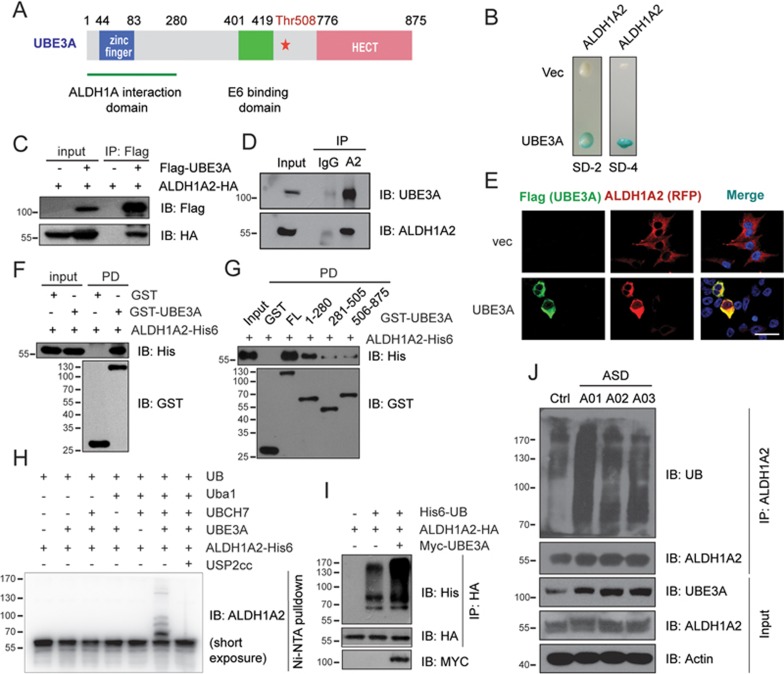
Little mechanistic insight into the link between ASD pathogenesis and excessive UBE3A dosage has been revealed due to few known UBE3A substrates4,23. We therefore carried out an initial yeast two-hybrid (Y2H) screening using the longest isoform of human UBE3A as the bait to find interacting partners (Figure 1A). Multiple positive colonies were found to harbor the full-length cDNA for human ALDH1A2, a rate-limiting enzyme in the cellular biogenesis of RA (Figure 1B). A co-immunoprecipitation assay indicated that the UBE3A and ALDH1A2 proteins, both endogenous and ectopically expressed, could form a complex in HEK-293FT cells (Figure 1C and 1D). We next expressed Flag-UBE3A and ALDH1A2-RFP ectopically in SH-SY5Y neuroblastoma cells and found that the two proteins significantly co-localized in the cytosol by immunofluorescence (Figure 1E). Furthermore, when recombinant GST-fused UBE3A protein was incubated with HA-tagged ALDH1A2 protein, ALDH1A2 protein was efficiently pulled down by the full-length or N-terminal fragment (amino acid 1-280) of GST-UBE3A, which was immobilized on glutathione-agarose beads (Figure 1A, 1F and 1G; Supplementary information, Figure S1A). These data validate a direct interaction between ALDH1A2 and UBE3A, mainly involving the N-terminal 1-280 residues of UBE3A. As human ALDH1A family proteins share high sequence homologies (Supplementary information, Figure S2), we examined the interaction of UBE3A with other family members; we found that UBE3A also directly bound to ALDH1A1 and ALDH1A3 in vitro and in cells (Supplementary information, Figure S1B-S1E). Consistent with the high sequence homology among ALDH1A family proteins, UBE3A bound to all three members via the same ALDH1A interaction domain (amino acids 1-280 of UBE3A; Figure 1A, Supplementary information, Figure S1F and S1G). Taken together, our results identify novel interactions between UBE3A and all members of the ALDH1A family of proteins, the rate-limiting enzymes for RA biogenesis.
Figure 1.
Identification of ALDH1A2 as a substrate of ASD-related UBE3A for ubiquitylation. (A) Schematic domain structure of human UBE3A protein. (B) Interaction between human UBE3A and ALDH1A2 in Y2H assay, indicated by both survival of the harboring colonies in SD-4 medium (deficient in Ura, His, Leu and Trp) and the blue color in X-gal assay. (C, D) Co-immunoprecipitation (co-IP) assay to identify the interaction of ectopically expressed Flag-UBE3A and ALDH1A2-HA proteins (C), or the endogenous partner proteins (D) in HEK-293FT cells. A2, ALDH1A2. (E) Immunofluorescence microscopy analysis indicated the co-localization of Flag-UBE3A with ALDH1A2-RFP in SH-SY5Y cells, with cell nucleus stained with DAPI. Scale bar, 30 μm. (F) GST pull-down assay was performed to validate the direct interaction between recombinant UBE3A and ALDH1A2 proteins in vitro. (G) Mapping the ALDH1A2-interacting regions in UBE3A. Purified ALDH1A2 protein was incubated with glutathione-agarose beads-bound GST-fused full-length UBE3A, the indicated fragments or the GST-only control proteins, respectively. The pulled down proteins were probed in immunoblotting. (H) UBE3A conjugated poly-Ub onto ALDH1A2 in an in vitro ubiquitylation assay. USP2cc: the catalytic core domain of human deubiquitylase USP2. (I) UBE3A promotes ALDH1A2 ubiquitylation in intact cells. HEK-293FT cells were co-transfected with His6-UB, ALDH1A2-HA and Myc-UBE3A constructs as indicated. Ubiquitylation-conjugated proteins were enriched using anti-HA antibody, and subjected to immunoblotting using anti-His antibody. (J) The ubiquitylation of endogenous ALDH1A2 increased in dependence of the dosages of UBE3A proteins in ASD patients-derived lymphocytes. All experiments were conducted at least three times.
Human UBE3A conjugates Ub onto ALDH1A family proteins
We next tested whether UBE3A might also conjugate ubiquitin onto ALDH1A2. To this end, an E. coli-based ubiquitylation system with or without E3 Ub ligase UBE3A was reconstituted as described before32 (see Materials and Methods for details), through co-expression of ALDH1A2 and the ubiquitylation system in the bacterial cells. Human ALDH1A2 was markedly ubiquitylated only when UBE3A was present; this ubiquitylation could be reversed upon treatment with USP2cc, the catalytic core of human deubiquitylating enzyme USP2, suggesting the covalent conjugation of Ub by UBE3A (Supplementary information, Figure S1H). We also assembled an in vitro ubiquitylation system that consisted of E1 Ub-activating enzyme (Uba1), E2 Ub-conjugating enzyme (UBCH7), E3 Ub ligase (UBE3A), the substrate (ALDH1A2) and ATP. Consistent with the above result using the E. coli-based system, ALDH1A2 protein was markedly ubiquitylated in vitro in the presence of the reaction components (Figure 1H). Ectopic expression of UBE3A also promoted the ubiquitylation of ALDH1A2 in HEK-293FT cells (Figure 1I) and in several other cell types (data not shown). This ubiquitylation was detectable even under denaturing conditions, excluding the potential interference through auto-ubiquitylation of UBE3A (Supplementary information, Figure S1I).
To generate an experimental system that limited potential interference from endogenous proteins, we first surveyed cell lines for their expression levels of the ALDH1A family proteins. From all cell lines surveyed, the endogenous levels of ALDH1A family proteins appeared to be undetectable in H1299 cells33 (Supplementary information, Figure S3A). CRISPR/Cas9 was then employed to genetically ablate UBE3A in H1299 cells. This led to greatly reduced ubiquitylation of ectopically expressed ALDH1A2 protein (Supplementary information, Figures S3B, S3C and S4A). Accordingly, compared to the wild-type control, ubiquitylation of endogenous Aldh1a2 was much lower in mouse embryonic fibroblast (MEF) cells derived from Ube3a-knockout (Ube3a−/−) mice34 (Supplementary information, Figure S4B), suggesting that UBE3A might be a predominant E3 Ub ligase for ALDH1A2 in mammalian cells. Certainly, since ALDH1A2 protein was still ubiquitylated in mouse Ube3a−/− or human UBE3A−/− cells, there should exist other yet unknown E3 Ub ligase(s) that could also ubiquitylate ALDH1A2. Taken together, these results clearly indicate that ALDH1A2 is a bona fide substrate for UBE3A in mammalian cells.
As excessive UBE3A dosage is closely associated with ASD and naturally leads to the hyperactivity of UBE3A as an E3 Ub ligase, we further examined whether excess UBE3A resulted in increased ALDH1A2 ubiquitylation in cells. In HEK-293FT cells, the dosage of UBE3A positively correlated with ALDH1A2 ubiquitylation (Supplementary information, Figure S4C), suggesting that increased ALDH1A2 ubiquitylation may occur in ASD subtypes with excess UBE3A.
We next attempted to directly assess ALDH1A2 ubiquitylation in individuals with ASD with excess UBE3A dosage. Common amplifications in chromosome 15q regions3 were found in three Chinese probands with clinically evident ASD symptoms (A01 and A02 with maternal isodicentric 15q duplications; A03 with an interstitial 15q duplication), as validated by BeadChip microarray and karyotype analysis (Supplementary information, Figure S5A-S5D, Table S1). Gene amplifications found in this region implicated a battery of genes including the UBE3A gene. Among them, excessive dosage of UBE3A has been shown to have an established role in contributing to 15q duplication symptoms3 (Supplementary information, Figure S5C and S5D). Lymphocytes from patients in these three ASD probands were immortalized and directly assessed to have UBE3A hyperactivities in comparison with the healthy control (Figure 1J). Together with our additional finding that ubiquitylation of endogenous ALDH1A2 protein in immortal lymphocytes from the ASD patients was markedly higher than that from the healthy control (Figure 1J), our results demonstrate that human ASD-linked increased dosage of UBE3A apparently leads to increased conjugation of Ub onto endogenous ALDH1A2 protein.
Previously, residue Thr508 (T508) in human UBE3A protein was shown to undergo PKA-mediated phosphorylation that suppressed the Ub ligase activity of UBE3A, while an ASD-linked T508A mutation disrupted the inhibitory phosphorylation and led to hyperactive Ub ligase activity of UBE3A in autistic patients17. Conversely, a phospho-mimetic T508E mutation was found to abolish the E3 Ub ligase activity of UBE3A, resulting in a ligase-dead mutant of UBE3A. Using a ubiquitylation assay with Flag-tagged ALDH1A2 recovered from HEK-293FT cells, we found that ectopic expression of UBE3AT508E did not lead to any increase in ALDH1A2 ubiquitylation, whereas the overexpression of wild-type UBE3A significantly increased ALDH1A2 ubiquitylation, and the ubiquitylation was increased even more markedly by overexpression of the hyperactive T508A mutant of UBE3A (Supplementary information, Figure S4D). These data strongly suggest that UBE3AT508E lacks E3 Ub ligase activity, at least with ALDH1A2 as substrate. We therefore used UBE3AT508E as a suitable negative control for the E3 Ub ligase function of UBE3A in the rest of this study. Taken together, our findings indicate that ubiquitylation of ALDH1A2 correlates well with excessive UBE3A dosage in multiple experimental cell lines, in accord with UBE3A duplication or hyperactive T508A mutation in autistic cases.
Three isoforms of human UBE3A that result from alternative splicing contain distinct N-terminal residues35, and exhibit differential cellular distributions (Supplementary information, Figure S4E). Compared to isoforms 1 and 3, which mainly exist as puncta in the cell nucleus, isoform 2 of UBE3A co-localized with ALDH1A2 in the cytosol, manifesting the highest E3 Ub ligase activity toward ALDH1A2 (Supplementary information, Figure S4F).
The high sequence homologies among the human ALDH1A family proteins and their confirmed interaction with UBE3A (Supplementary information, Figures S1B-S1E and S2) led us to show that UBE3A also ubiquitylated ALDH1A1 and ALDH1A3, the other two members of the ALDH1A family proteins in both bacterial and mammalian cells (Supplementary information, Figure S1J-S1L). Therefore, UBE3A can ubiquitylate the whole family of human ALDH1A proteins, the only dehydrogenase family proteins that convert retinaldehyde into RA. Thus, fluctuations in UBE3A dosage or its activity might conceivably impact RA homeostasis in mammalian cells.
UBE3A ubiquitylates ALDH1A2 in non-proteolytic Ub lysine-linkages
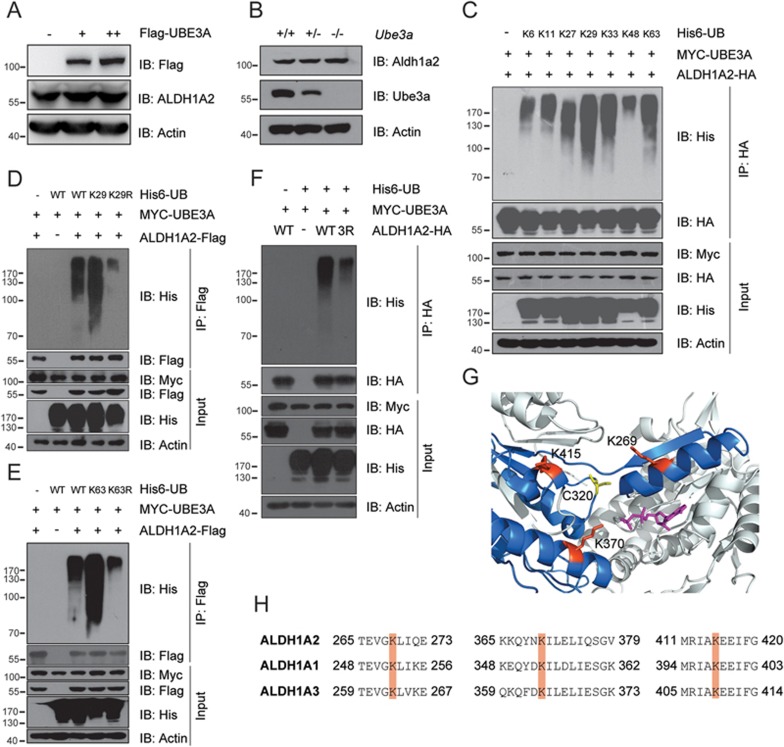
Previously, HHR23A36, PML37, RING1B38 and Tkv39 were found to be ubiquitylated by UBE3A and targeted for proteasome-dependent degradation in the absence of the human papillomavirus oncogene E6, whereas p53 proteins were ubiquitylated only by the E6-UBE3A complex40,41. Surprisingly, the static level of endogenous ALDH1A2 protein was not changed despite increasing dosage of UBE3A in HEK-293FT cells (Figure 2A). Similarly, the static level of Aldh1a2 protein remained unchanged in mouse MEF cells in disregard of the genetic ablation of Ube3a gene (Figure 2B). In addition, the protein level of ALDH1A2 was almost identical in all the immortalized lymphocytes from both ASD probands and the healthy control, despite the varying UBE3A dosages (Figure 1J). These data clearly indicated that, unlike other previously reported substrates, UBE3A-mediated ubiquitylation did not target ALDH1A2 for proteasomal degradation.
Figure 2.
Human UBE3A mediates ubiquitylation of ALDH1A2 with non-proteolytic Ub linkages. (A, B) The static level of endogenous ALDH1A2 protein remained unchanged in HEK-293FT cells ectopically expressing Flag-UBE3A in increasing dosages (A), or in MEF cells with wild-type Ube3a, deficient or genetically ablated (Ube3a +/+, +/− or −/−) (B). (C-E) UBE3A-mediated ubiquitylation of ALDH1A2 in K29- and K63-linked poly-Ub chains. H1299 cells were co-transfected with Myc-UBE3A, ALDH1A2-HA (or -Flag) and His6-Ub mutants that each retained only one Lys at the indicated position (C), or the K-to-R substitution only at K29 (D) or K63 (E). HA- or Flag-tagged ALDH1A2 proteins were enriched from the cells, and probed with the appropriate antibodies, respectively. (F) Identification of the sites for UBE3A-mediated ubiquitylation in human ALDH1A2 protein. H1299 cells were co-transfected with His6-Ub, Myc-UBE3A and ALDH1A2-HA mutant carrying K-to-R substitutions simultaneously at the indicated sites. ALDH1A2-HA proteins were enriched and followed by immunoblotting to detect ubiquitylation. 3R: ALDH1A2-HA mutant bearing K-to-R substitutions at K269, K370 and K415 sites. (G) The crystal structure of monomeric human ALDH1A2 in which the three major ubiquitylation sites (shown in red) residing close to the active center (Cys320, highlighted in yellow), with the co-factor NAD+ shown in violet. The structure was adopted from Protein Data Bank (PDB: 4X2Q; doi:10.2210/pdb4x2q/pdb). (H) The alignment of peptide sequences flanking the identified ubiquitylation sites in ALDH1A family proteins, with ubiquitylation sites shown in orange. All experiments were conducted at least three times.
As Ub has seven Lys (K) residues, the side chain of any of these lysines, together with the α amino group of the N-terminal Met of Ub, could be conjugation sites for additional Ub molecules to form poly-ubiquitin (poly-Ub) chains of different K-Ub linkages18,19,20,21,22. By retaining one Lys at a specific position and mutating the remaining Lys into Arg (K-to-R substitution), it is possible to characterize the Lys linkage types of poly-Ub chains. As shown in Figure 2C, Ub molecules with intact K29 or K63 were much more efficiently conjugated to ALDH1A2 than any other Ub mutants. Moreover, mass spectrometry analysis of the enriched proteins from both transfected HEK-293FT cells and the reconstituted bacterial strains also identified the K63 Ub linkage (Supplementary information, Figure S4G). Furthermore, only K-to-R substitution at residues K29 or K63 significantly compromised UBE3A-mediated ubiquitylation on ALDH1A2 (Figure 2D and 2E). These data strongly suggest that UBE3A might preferentially conjugate poly-Ub to ALDH1A2 in K29 or K63 Ub linkages or mixed linkages, which typically do not target the substrates for proteasome-dependent degradation.
UBE3A polyubiquitylates human ALDH1A family proteins at conserved sites
We then mapped the sites for UBE3A-mediated ubiquitylation on ALDH1A2 protein, through proteomic analysis of ALDH1A2 protein recovered from both transfected HEK-293FT cells and reconstituted bacterial ubiquitylation system. This identified K269, K370 and K415 as the convergent sites for UBE3A-mediated polyubiquitylation on ALDH1A2 protein recovered in both systems (Supplementary information, Figure S4G). A cell-based ubiquitylation assay was then performed, where K-to-R substitutions were introduced on each of these potential polyubiquitylation sites in ALDH1A2 to examine its ubiquitylation status in human H1299 cells. As shown in Figure 2F, the ubiquitylation level of ALDH1A2 was dramatically reduced when K-to-R mutations were simultaneously introduced on all three Lys residues (K269, K370 and K415). Importantly, a close structural analysis revealed that these three validated polyubiquitylation sites are highly conserved in all ALDH1A family proteins and located in proximity to the active sites for their dehydrogenase activities42 (also Protein Data Bank, PDB: 4X2Q) (Figure 2G and 2H). Therefore, UBE3A polyubiquitylates human ALDH1A family proteins predominantly at the conserved sites close to the active centers for their dehydrogenase activities.
UBE3A-mediated ubiquitylation inhibits the dehydrogenase activity of ALDH1A2 in vitro
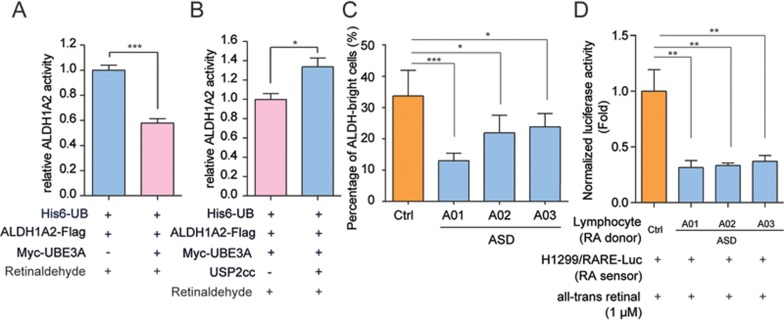
We next examined the effect of UBE3A-mediated ubiquitylation on the dehydrogenase activities of ALDH1A2. As shown in Figure 3A (also Supplementary information, Figure S6A and S6B), ALDH1A2 protein recovered from HEK-293FT cells over-expressing UBE3A, was highly ubiquitylated, and the total retinaldehyde dehydrogenase activity was approximately only half of that from the cells singly expressing endogenous UBE3A. We found that such inhibition was efficiently reversed after USP2cc treatment, which removes the poly-Ub chains on ALDH1A2 (Figure 3B). These data clearly suggest that UBE3A-mediated ubiquitylation of ALDH1A2 might significantly compromise its dehydrogenase activity toward retinaldehyde. Likewise, the enzymatic activity of ALDH1A2 in oxidizing propionaldehyde, another known substrate of its dehydrogenase, was also markedly decreased after ubiquitylation catalyzed by UBE3A, and this could also be reversed after USP2cc treatment (Supplementary information, Figure S6C and S6D). Moreover, since K-to-R substitutions at the three ubiquitylation sites in ALDH1A2 (K269, K370 and K415) led to a complete loss of its activity in oxidizing both aldehydes (Supplementary information, Figure S6E; data not shown), these three lysine residues seemed to be critically required for the dehydrogenase activity of ALDH1A2, and probably the whole ALDH1A family proteins (Figure 2H and Supplementary information, Figure S6F).
Figure 3.
UBE3A-mediated ubiquitylation suppresses the retinaldehyde dehydrogenase activities of ALDH1A. (A, B) UBE3A-conjugated poly-Ub chains on ALDH1A2 reduced its dehydrogenase activity toward retinaldehyde. ALDH1A2-Flag protein was enriched from HEK-293FT cells (A), followed by USP2cc treatment or not to remove the poly-Ub chains (B). The enriched ALDH1A2 protein was then subjected to dehydrogenase activity assays using all-trans retinaldehyde as the substrate. Relative dehydrogenase activity of polyubiquitylated ALDH1A2 protein was shown after normalization to that of the control group (A: n = 4; B: n = 3). Data were shown in means ± SEM. (C, D) Total cellular ALDH1A activities of the immortal lymphocytes from the autistic groups were significantly lower than that of the healthy control (multiple healthy lymphocytes cell lines), according to AldeFluor assay (C) or RARE-luciferase reporter assay (D). (C) The percentages of ALDH-bright cells were recorded after pre-setting DEAB (N,N-diethylaminobenzaldehyde)-treated cells for negative gating in flow cytometry. Values were expressed as means of quintuplicates of each individual ± SEM. (D) RA production in the immortal lymphocytes derived from the healthy control or autistic probands was assayed by co-culturing them (RA donor cells) with H1299 cells that bore RARE-luciferase reporter constructs (RA sensor cells). Cellular luciferase activities were assayed after 1 μM all-trans retinal treatment for 8 h, and normalized to control group (healthy lymphocytes). Values were expressed as means of quadruplicates of each individual ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t-test (A, B), one-way ANOVA with Dunnett's post hoc test (C, D).
UBE3A-mediated ubiquitylation inhibits the dehydrogenase activities of ALDH1A family proteins in cells
A flow cytometry-based aldehyde dehydrogenase (ALDH)-dependent fluorophore (Aldefluor) assay is commonly used to assess the cellular ALDH activities43 by assessing how much BAAA (BODIPY-aminoacetaldehyde) is converted into BAA (BODIPY-aminoacetate) by ALDHs (see Materials ang Methods for details). As shown in Figure 3C, the percentages of the ALDH-bright cells in the immortal lymphocytes from autistic patients were ∼20%-60% less than those from the healthy control, suggesting that ASD-linked increased dosage of UBE3A might lead to significantly lower cellular ALDH activities in patient-derived cells.
To further confirm that the specific ALDH1A enzyme activities were indeed compromised in cells from the autistic patients, a co-culture system was also constructed, in which the cellular level of all-trans RA (ATRA) from one cell population (RA-donor cells) was assayed by the uptake of ATRA and its activation of the RARE (RA-responsive element)-luciferase transcription reporter within the receiving cells (RA-sensor cells)26,27. As H1299 cells expressed endogenous ALDH1A family proteins at an undetectable level (Supplementary information, Figure S3A), they were considered to be free of potential interference from endogenous ALDHs33, and used to construct RA sensor cells stably harboring RARE-luciferase reporters. H1299-based RA sensor cells were then co-cultured with the tested RA-donor cells, which were the immortalized lymphocytes from either the autistic patients or the healthy control, at a cell number ratio of ∼1:1, before retinaldehyde was supplemented (see Materials and Methods for details). As shown in Figure 3D, RARE-based luciferase activities in co-cultures with RA donor cells derived from the autistic groups were decreased by ∼60%-70% compared with the healthy control, further supporting the results from the above two different assays.
Together, these data strongly suggest that ASD-associated UBE3A hyperactivity inhibits RA biogenesis through increased ubiquitylation of the ALDH1A family proteins and thus disrupts overall cellular RA homeostasis.
Excessive UBE3A dosage impairs RA production and disrupts synaptic transmission homeostasis
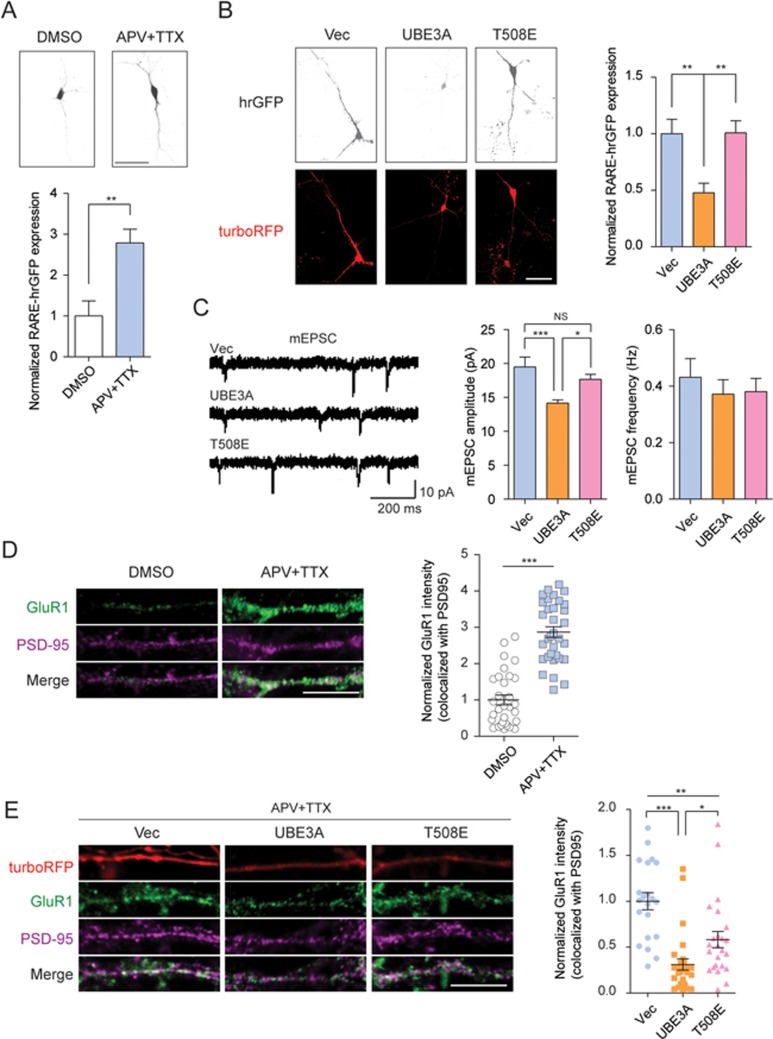
In the adult nervous system, RA has been increasingly recognized as a key player in regulating homeostatic synaptic plasticity25,28. When synaptic transmission is blocked with a concomitant reduction of dendritic calcium levels, local RA synthesis can be activated by inducing ALDH1A enzymatic activity in neurons, through translational control28,29. RA binds to dendritic-localized RARα, releasing the RARα-repressed translation of mRNAs for certain proteins, including AMPA receptor, to up-scale synaptic transmission29,30. Consistent with previous findings in hippocampus neurons28,29, inhibition of synaptic activity by 𝒟-(−)-2-amino-5-phosphonopentanoic acid (D-APV) and tetrodotoxin (TTX) treatment induced a surge in the cellular level of RA in primary neurons isolated from rat prefrontal cortex (PFC), as the RARE-driven expression of the humanized Renilla GFP (hrGFP) reporter was significantly activated, reaching a level up to 2-fold higher than in untreated cells (Figure 4A). However, in neurons co-transfected with both UBE3A-IRES-turboRFP and RARE-hrGFP reporter plasmids, APV and TTX treatment blocked synaptic activity, but no increase in hrGFP signal was observed. Remarkably, in neurons co-transfected with plasmids for RARE-hrGFP reporter and Ub ligase-dead mutant UBE3AT508E, the same treatment induced the expression of hrGFP to the level comparable to that of the control group (Figure 4B). These data provide the first evidence that the hyperactive E3 Ub ligase activity of UBE3A impaired RA production upon synaptic transmission blockade in neurons.
Figure 4.
UBE3A hyperactivity disrupts synaptic transmission scaling. (A) The fluorescence intensities of RARE-hrGFP were significantly increased in primary PFC neurons upon neuronal activity blockade with 1 μM APV and 100 μM TTX (24 h). Representative figures were shown in the upper panel (Scale bar, 50 μm); in the lower panel, fluorescence intensities were quantitated and normalized to DMSO group. DMSO, n = 8; APV/TTX, n = 11. (B) Introduction of UBE3A-IRES-turboRFP into primary neurons significantly reduced the RARE-hrGFP signals upon APV/TTX treatment for 24 h, compared with vector or the ligase-dead T508E mutant. The relative fluorescence intensities were presented in the right. Vector, n = 11; UBE3A, n = 14; T508E, n = 12. Scale bar, 40 μm. (C) Introduction of UBE3A into primary neurons reduced mEPSC amplitudes upon APV/TTX treatment (24 h), compared to that of the vector or T508E groups. Left, representative mEPSC traces; middle and right, quantitations of amplitudes and frequencies (vector, n = 14; UBE3A, n = 17; T508E, n = 15). (D) Surface staining of GluR1 and PSD-95 in primary PFC neurons upon treatment of DMSO or APV/TTX for 24 h. Left, representative images (Scale bar, 10 μm); right, quantitated intensities for GluR1 co-localized with PSD-95 (DMSO, n = 31; APV/TTX, n = 33). (E) Surface staining of GluR1 and PSD-95 in primary PFC neurons expressing wild-type or T508E mutant of UBE3A upon treatment of APV/TTX for 24 h. Left, representative images (Scale bar, 10 μm); right, relative intensities of co-localized GluR1 and PSD-95 in turboRFP positive dendrites (vector, n = 21; UBE3A, n = 28; T508E, n = 24). Data were shown as means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t-test (A, D), one-way ANOVA with Bonferroni post hoc test (B, C, E).
We next examined the effect of excessive UBE3A dosage on RA-mediated synaptic homeostasis in neurons in response to a neuronal activity blockade. Upon treatment with APV and TTX, both the miniature excitatory post-synaptic current (mEPSC) amplitudes and frequencies significantly increased in normal primary PFC neurons (Supplementary information, Figure S7A). This suggested that the compensatory increases in excitatory synaptic strengths occurred, possibly via both pre-synaptic and post-synaptic mechanisms, in normal PFC neurons. However, in those primary neurons overexpressing wild-type UBE3A, the same neuronal activity deprivation elicited a significantly reduced (30% less) synaptic upscaling of mEPSC amplitudes, compared to control cells or cells expressing the ligase-dead T508E (Figure 4C). In addition, overexpression of wild-type UBE3A seemed not to alter the mEPSC frequencies upon neuronal activity blockade by APV/TTX, compared to that of control or T508E mutant (Figure 4C). Taken together, these data suggest that, upon neuronal activity blockade, excessive UBE3A dosage might impair RA-regulated compensation of synaptic homeostasis in an E3 Ub ligase activity-dependent manner, most likely through a post-synaptic rather than pre-synaptic mechanism.
It has been established that synaptic scaling via post-synaptic mechanisms involves de novo translation of dendritic receptors to augment their numbers for excitatory transmission44. In consistence with previous findings29, for two subtypes of AMPA receptor in dendrites, local translation of GluR1 (also known as Gria1), but not GluR2 (Gria2), was increased upon neural activity blockade in a transcription-independent manner (Figure 4D; Supplementary information, Figure S7B and S7C). However, in PFC neurons overexpressing wild-type UBE3A, neuronal activity blockade caused significantly less increase in post-synaptic GluR1 protein level compared to cells expressing the vector only (Figure 4E). In contrast, in PFC neurons overexpressing the ligase-dead T508E mutant, a similar neuronal activity blockade induced much higher levels of GluR1 expression than in cells overexpressing wild-type UBE3A, although the GluR1 expression level was still less than that of the vector control group (Figure 4E). These results strongly indicate that the excessive E3 Ub ligase activity of UBE3A disrupts homeostatic synaptic plasticity in the PFC neurons, most likely through impairing RA-mediated upregulation of post-synaptic local protein translation.
Ectopic expression of UBE3A in PFC region phenocopies ASD symptoms in mice
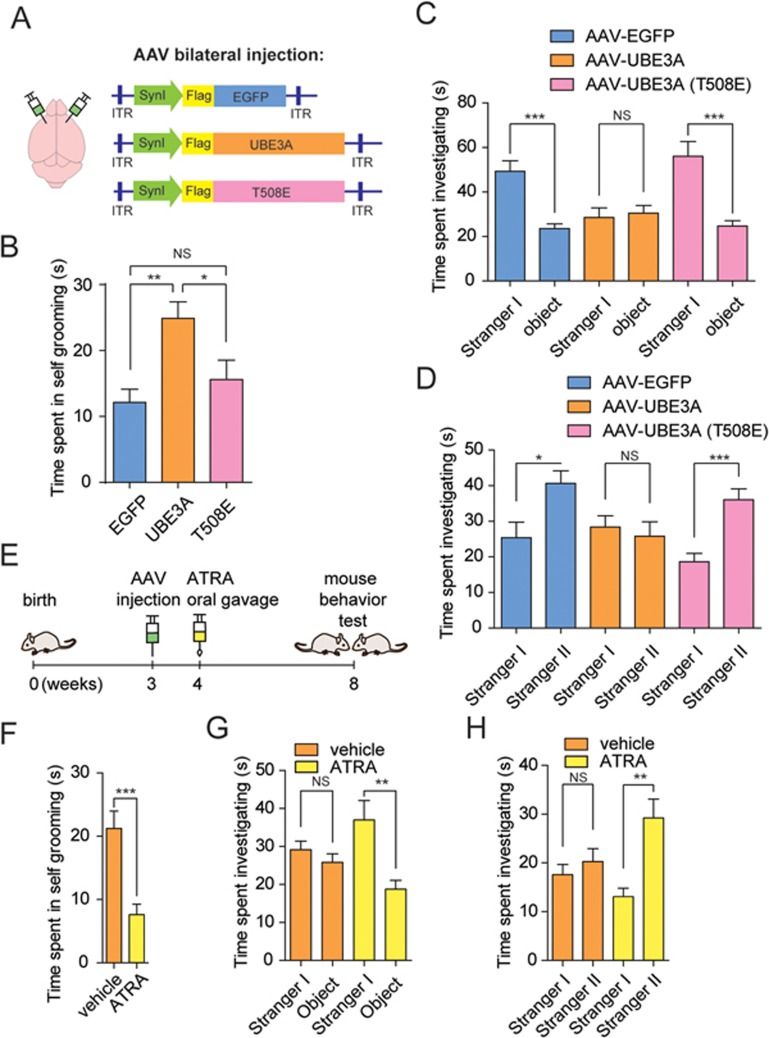
The PFC region controls many executive functions and higher-order cognitive processes of the brain, such as decision-making, cognitive movements, social behavior, learning and communication45,46. Recently, abnormal anatomic structures of the PFC region and/or its connections to the other regions of the brain are widely observed within ASD patients and these are considered to be closely associated with ASD etiology46,47,48,49. In addition, we took advantage of available data from the ALLEN mouse brain atlas50 and found that Ube3a and Aldh1a2 are expressed throughout the PFC region. To investigate the causative role of excessive UBE3A dosage in development of autistic behaviors, adeno-associated viruses (AAVs) encoding EGFP, UBE3A or UBE3AT508E under human Synapsin I (SynI) promoter were stereotaxically delivered into the medial PFC regions of the mouse brains, respectively (Figure 5A and Supplementary information, Figure S8A). Immunofluorescence analysis with the brain slices confirmed that UBE3A or UBE3AT508E proteins were expressed at almost identical levels in PFC regions from randomly chosen mice from each group (Supplementary information, Figure S8B).
Figure 5.
Increased dosages of UBE3A impair RA homeostasis and phenocopy ASD-like behaviors in mice. (A) The schematic brain map with indicated stereotaxic injection sites in PFC regions (left). Shown on the right were the constructs for hSynI (human Synapsin I)-promoter-driven expression of Flag-EGFP, Flag-UBE3A or Flag-T508E, respectively. (B) Time spent in self-grooming was recorded for mice of the groups stereotaxically injected with AAV virus-carried EGFP (n = 11), wild-type UBE3A (n = 11) or UBE3A-T508E (n = 9). (C) Time spent investigating between Stranger I and object was compared among three indicated groups. EGFP, n = 12; UBE3A, n = 13; T508E, n = 11. (D) Time spent investigating between Stranger I and Stranger II was compared among mice from the groups expressing EGFP (n = 10), wild-type UBE3A (n = 14) or T508E (n = 11). (E) Schematic processes of AAV stereotaxic injections and ATRA rescue experiments in mouse model. (F) Time spent in self-grooming was tested in AAV-infected mice in vehicle (olive oil, n = 9) or ATRA (3 mg/kg, dissolved in olive oil, n = 10) groups. (G, H) Time spent investigating in Stranger I versus Object (G), or Stranger I versus Stranger II (H) was compared among vehicle or ATRA group from AAV-infected mice. Vehicle group (n = 9), ATRA group (n = 10). Data were shown in means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t-test (C, D, F-H), one-way ANOVA with Bonferroni post hoc test (B). ITR, inverted terminal repeat; NS, not significant.
Four weeks after the stereotaxical injections, the mice were subjected to behavior tests. In a self-grooming test, mice overexpressing UBE3A spent almost 100% more time in repetitive grooming compared with control mice, while UBE3AT508E-expressing mice exhibited no significant differences compared with the control group (Figure 5B). These observations clearly demonstrated a strong tendency towards repetitive behavior in mice harboring hyperactive UBE3A. We also recorded the time that mice spent on investigating a social animal or a non-social object among each group in a three-chamber social test. Unlike the control or T508E group that spent roughly 100% more time (∼50 s) with the stranger than with the object (∼23 s), UBE3A-overexpressing mice spent an almost equal amount of time (∼30 s) on interacting with the stranger and the object (Figure 5C), indicating severe deficits in social interactions among the mice with hyperactive UBE3A. Meanwhile, time spent on the familiar social animal (Stranger I) over the novel social animal (Stranger II) was also compared among three groups. While control and T508E-overexpressing mice showed more interest in Stranger II and spent ∼60%-100% longer time interacting with them, UBE3A-expressing mice spent roughly equal amounts of time with Stranger I and Stranger II (Figure 5D), indicating that overexpression of UBE3A compromised the recognition of social novelty. Therefore, UBE3A hyperactivity, due to excessive wild-type UBE3A in the PFC region of the mouse brains, seemed sufficient to cause core ASD-like behavioral traits in mice, including increased repetitive behavior and deficits in social interaction and recognition of social novelty.
To further characterize the phenotypical changes caused by overexpression of wild-type UBE3A or the ligase-dead mutant T508E, the mice of each group were also subjected to other standard behavioral tests. It seemed that mice from all three groups showed similar levels of thigmotaxis in an open-field test, and locomotor activity in a rotarod test (Supplementary information, Figure S9A-S9C).
Repletion of RA homeostasis alleviates ASD-like phenotypes in mice caused by excessive UBE3A dosage
To further pinpoint the causative roles of impaired RA synthesis during the onset of autistic phenotypes in mice with UBE3A hyperactivity, mice overexpressing wild-type UBE3A in PFC were randomly assigned into groups receiving oral administration of the vehicle (olive oil) or ATRA (3 mg/kg, 5 consecutive days per week) for 4 weeks, before being subjected to a set of mouse behavioral tests (Figure 5E). In the self-grooming test, the ATRA-treated group spent a significantly reduced time in repetitive grooming, compared to untreated UBE3A-overexpressing mice (Figure 5F). Also in comparison with the vehicle group, deficits in social interaction were almost fully restored in the ATRA-treated group, as the treated mice spent ∼100% more time investigating Stranger I over a non-social object (Figure 5G) or interacting with Stranger II over Stranger I (Figure 5H). Therefore, an intervention of 4 weeks of oral ATRA administration seemed sufficient to alleviate the core traits of ASD-like behaviors in this mouse model of UBE3A hyperactivity.
Of note, in the open-field test, mice administered with ATRA seemed to have spent less time and explored less distances in the center, suggesting that ATRA administration might cause a certain level of anxiety in UBE3A-overexpressing mice (Supplementary information, Figure S9D and S9E). However, locomotor activities of all the mice overexpressing UBE3A seemed unaltered upon treatment with either vesicle or ATRA, according to the rotarod test (Supplementary information, Figure S9F).
To further investigate whether 4 weeks of ATRA treatment itself might cause side effects such as anxiety symptoms in wild-type mice, wild-type mice (n = 9-10) treated with ATRA or vehicle were subjected to behavioral tests and found to behave similarly in the self-grooming, social three-chamber, open-field or rotarod tests (Supplementary information, Figure S9G-S9I). These observations indicated that ATRA treatment might have caused slight anxiety specifically in mice overexpressing UBE3A in PFC but not in the wild-type mice. This suggests that ATRA-associated side effects might need to be taken into consideration if ATRA administration was ever adopted to alleviate ASD-like traits in future studies.
Chemically impaired RA homeostasis is sufficient to elicit ASD-like phenotypes in wild-type mice
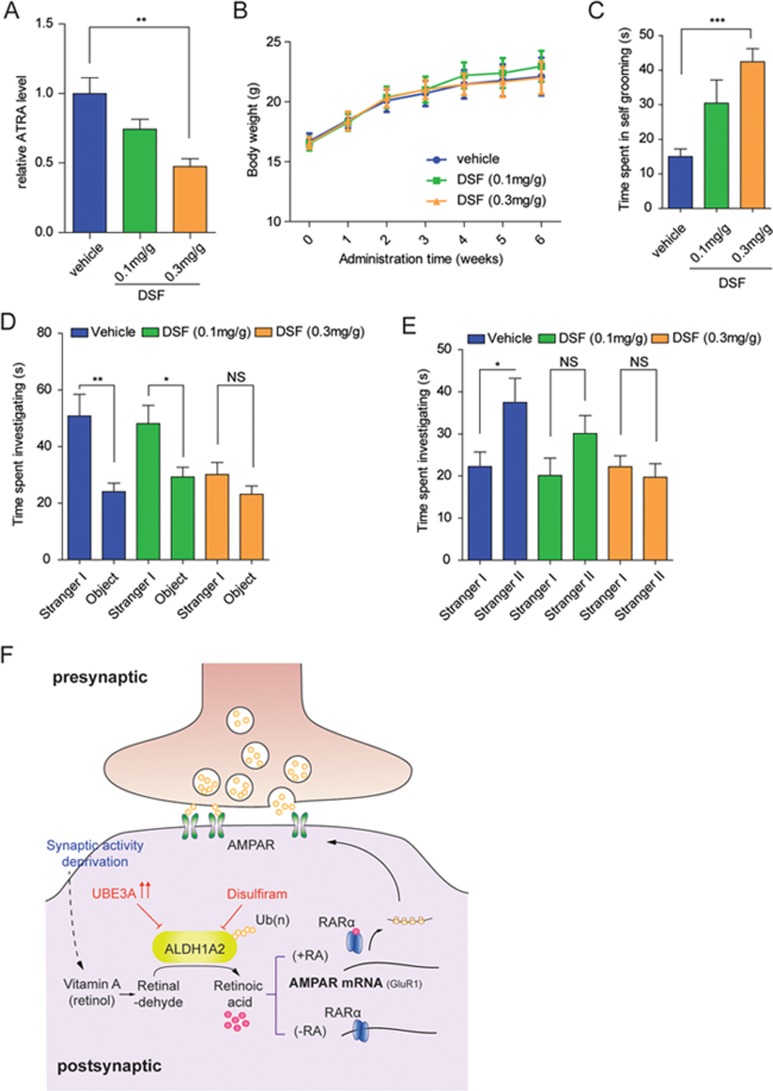
To further investigate whether repressed ALDH1A activities alone would be sufficient to cause ASD-like phenotypes in mice, DSF, an Aldh1a enzyme inhibitor31, was dissolved in olive oil and orally delivered into wild-type male C57BL/6 mice (at a single dose of 0.1 or 0.3 mg/g every other day, for 6 weeks) at the juvenile stage (∼4 weeks old) to chemically impair RA biosynthesis in vivo. After DSF administration was completed, the levels of ATRA in mouse brains were quantitated, using a previously reported LC-MS/MS (liquid chromatography tandem mass spectrometry) approach51,52. As shown in Figure 6A, after oral administration of DSF for 6 weeks, ATRA levels of DSF-treated mice decreased by 30% (at 0.1 mg/g dosage) or 60% (at 0.3 mg/g dosage) compared to mice receiving vehicle only, indicating that DSF administration can inhibit ATRA synthesis in mouse brains in a dose-dependent manner. Of note, body weights were not affected by DSF treatment (Figure 6B), suggesting that, at the dosages used, DSF administration itself might not be toxic to the mice.
Figure 6.
Oral administration of ALDH1A inhibitor DSF causes ASD-like behaviors in mice. (A) Relative ATRA levels within mouse brains (n = 6), dissected from the mice orally administered with either DSF (0.1 or 0.3 mg/g) or vehicle only for 6 weeks, were quantitated using HPLC-MS/MS method, and normalized to those of the vehicle groups. (B) Six-week administration of DSF did not cause significant differences in mouse body weights, compared to those of the control groups. Vehicle group (n = 10), DSF (0.1 mg/g, n = 9), DSF (0.3 mg/g, n = 12). (C) Time spent in self-grooming for the three indicated groups. (D) Time spent interacting with Stranger I or Object was compared among three indicated groups. (E) Time spent interacting with Stranger I or Stranger II was compared among indicated groups. (F) A working model depicts how excessive UBE3A dosage might disrupt post-synaptic transmission scaling through ubiquitylating ALDH1A family proteins and impairing RA homeostasis. Data were shown in means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t-test (D, E), one-way ANOVA with Bonferroni post hoc test (A-C). Data in C-E, significance were determined from the indicated biological replicates consisted of vehicle group (n = 15), DSF (0.1 mg/g, n = 17) and DSF (0.3 mg/g, n = 19). NS, not significant.
After 6-week administration of DSF or the vehicle only, mice were subjected to behavioral tests. The self-grooming test showed that DSF-treated mice spent on average 100% (∼30 s for mice in the 0.1 mg/g treatment group) or 200% (∼42 s for mice in the 0.3 mg/g group) longer in repetitive grooming than mice receiving the vehicle only (∼15 s) (Figure 6C). In the social interaction test, the mice in vehicle group or those receiving DSF at 0.1 mg/g did not exhibit obvious differences in social preference of a social animal over a non-social object (Figure 6D). However, the group of mice receiving 0.3 mg/g DSF manifested a distinct behavioral pattern from groups receiving the lower DSF dosage or the vehicle only; they lacked any significant preference of the social animals over non-social objects (Figure 6D). Furthermore, when assessed for their preference of social novelty, the mice treated with DSF at 0.3 mg/g spent a shorter time (∼30% less) interacting with stranger II than the vehicle-only group (Figure 6E). More specifically, DSF administration appeared to have shortened the time that the mice spent on interacting with Stranger II, in a DSF dose-dependent manner (Figure 6E). Taken together, it seems that, when RA biosynthesis is chemically disrupted through oral intake of DSF, a specific inhibitor for Aldh1a, ASD-like phenotypes including increased repetitive grooming and deficits in social interaction and recognition of social novelty could be robustly elicited in mice.
Other behavioral tests were also performed to evaluate the potential effects of DSF administration on anxiety, exploratory behavior and locomotor activity. Remarkably, among all three groups of mice, no significant differences were observed in the open-field, elevated plus maze, or rotarod tests (Supplementary information, Figure S10), suggesting that DSF treatment at the above indicated doses did not cause detectable changes in anxiety, exploratory behaviors, or motor activities in these animals. Data from these additional behavior tests as well as body weight measurements strongly suggested that the chemical toxicity of DSF might not contribute to the ASD-like phenotypes in mice following the indicated drug administration (Figure 6B). Thus, chemical disruption of RA homeostasis in mice appears sufficient to elicit ASD-like behavioral traits.
Collectively, these findings strongly support a model in which excessive UBE3A dosage might lead to increased ubiquitylation of the ALDH1A family members and decreased RA biosynthesis resulting in reduced RA activity, which might mechanistically underlie this subtype of human ASD (Figure 6F).
Discussion
ASDs have been defined as extremely heterogeneous and complex syndromes associated with diverse genetic factors1. Many subtypes of ASD are caused by abnormal dosages of the susceptibility genes, such as duplications in MECP253, SHANK354, or deletions of NRXN1, CNTNAP255. Together these all point to the contributory roles of synaptic dysfunctions in ASD2,5,56. In the subtype of ASD cases implicating UBE3A gene, excessive UBE3A dosages are usually attributable to duplications or triplications of the chromosomal regions3 (Supplementary information, Figure S5), or more rarely caused by mutations disrupting the site for inhibitory phosphorylation that typically leads to E3 Ub ligase hyperactivity17. Mouse models, in which overexpression of UBE3A successfully recapitulates ASD-like phenotypes16, substantiate the notion that UBE3A hyperactivity may critically contribute to subtypes of human ASD through mechanisms that are yet incompletely understood23.
Through an unbiased Y2H screen and subsequent characterization of the targets, we identified human retinaldehyde dehydrogenase ALDH1A2 and its family members as novel interacting partners for UBE3A. Subsequently, our work has provided evidence that excessive UBE3A leads to the increased non-proteolytic ubiquitylation of ALDH1A proteins at sites essential for their retinaldehyde dehydrogenase activity, which compromised RA biosynthesis and impaired the overall cellular RA homeostasis in multiple types of cells including neurons. In primary neurons overexpressing wild-type UBE3A but not the ligase-dead T508E mutant, of which the homeostatic levels of RA were disrupted, synaptic transmission scaling was markedly compromised upon blockade of neuronal activities, through reducing the up-scaled expression of GluR1. This is not only reminiscent of previous findings that impaired RA biosynthesis significantly decreased synaptic scaling28, but also offering mechanistic insights into the previously reported compromise in excitatory synaptic transmission in mice overexpressing Ube3a16.
We later demonstrated that the core traits of autistic behaviors could be successfully recapitulated in mice by stereotaxic overexpression of wild-type UBE3A, but not the control fluorescent protein or the ligase-dead T508E mutant in the mouse PFC regions. Moreover, oral administration of RA replenished RA homeostasis and alleviated autistic phenotypes in mice overexpressing UBE3A. Furthermore, impairing RA biogenesis through direct administration of DSF, an Aldh1a inhibitor, was sufficient to cause ASD-like phenotypes in mice (Figure 6F). This work has revealed, for the first time, the causal roles of impaired RA biosynthesis and reduced RA signaling in the onset of ASD-like behavior, such as increased repetitive behavior, lower interest in social interactions and deficits in recognizing social novelty.
In the broader literature, there is accumulating evidence that perturbation of RA homeostasis and/or RA signaling pathways might be associated with neurological or cognitive disorders. Deficit or loss of homeostatic synaptic plasticity was observed in either RARα-knockout or vitamin A-deficient (VAD) mice, underscoring a possible causal effect of defective RA signaling pathway in neuropsychiatric diseases10,25,57,58. In addition, deletions or functional mutations in the ALDH1A1-3 genes were clinically associated with neurological disorders, such as Parkinson's disease, schizophrenia59,60 and autism61. Taking ALDH1A2 as an example, our data now clearly indicate that ubiquitylation-led suppression of retinaldehyde dehydrogenase activity might critically contribute to the etiology of excessive UBE3A activity-caused ASD. Therefore, our data have unraveled, for the first time, a robust mechanistic link between impaired RA biosynthesis-caused deficient synaptic plasticity and human ASD associated with UBE3A hyperactivity.
It is essential to note that in this study ASD-like behavior could be fully recapitulated by oral administration of DSF, which is currently a drug in use for treating chronic alcoholism, and also with potential efficacy in treating cancer, HIV or cocaine addiction31. Given the current pandemic and rising incidence of ASD cases worldwide, it is both intriguing and critically important to investigate whether and how some drugs might contribute to etiologies of other ASD subtypes through perturbing RA signaling homeostasis in human brains. This possibility, if true, suggests that safeguarding these chemicals or drugs from entering the human body might help lower the future risk of some subtypes of ASD.
It is also important to note the limitations of our current study. Our experimental study does not rule out an alternative or additional mechanisms that could involve the nuclear role of RA given that RA can function through genomic regulation in brain development26,27. Moreover, we mainly focused on the effects exerted by the cytosol-located isoform of UBE3A in ASD etiology. Therefore, the roles of the two other nuclear isoforms of UBE3A would need to be clarified in follow-up studies to asses either E3 ubiquitin ligase activity or co-activator effects62,63.
Finally, by supplementing RA within a therapeutically safe dose to restore RA homeostasis and alleviate ASD-like behaviors in UBE3A-overexpressing mice, our results suggest a prototypical strategy for treating human ASD of relevant etiology. This would apply specifically to ASD associated with amplifications of the 15q11-13 chromosomal region and UBE3A hyperactivity. Meanwhile, human ASD could also be re-categorized into subtypes according to whether RA homeostasis or RA signaling pathways are affected, which might entail a re-prioritization of future efforts for targeted therapy64. In an era when cure or effective treatments are yet unavailable for human ASD, our findings might facilitate development of mechanism-based therapeutics for the precise management of ASD in clinics64,65.
Materials and Methods
Human subjects and samples
All subjects were enrolled and evaluated according to protocols approved by the institutional review board of the State Key Laboratory of Medical Genetics, School of Life Sciences at Central South University, Changsha, China, firmly adhering to the tenets of the Declaration of Helsinki. The details of subject recruitment and diagnosis were followed as previously described8,66. The written informed consents have been received upon all blood sample collections.
CNV array and karyotype analysis
The CNVs in three Han Chinese patients (males of 3-6 years) were identified using the Human660W-Quad BeadChip microarray (Illumina). Microarray analysis was performed using GenomeStudio v2011.1 (Illumina) as previously reported67. CNVs were mapped using the hg18 human genome as a reference. Metaphase slides were prepared from peripheral blood lymphocytes from human subjects. G-banded chromosomes were analyzed at 400-550 band level using Genetix' CytoVision platform with a GSL-120 slide loader. The detailed analysis including culture and metaphase harvest of peripheral blood cells, chromosome slide preparation and Giemsa banding were performed following standard procedures68.
Bisulfite DNA sequencing
Methylation analysis of the SNRPN gene in ASD patients was performed as previously reported69. Briefly, genomic DNA was extracted from peripheral blood lymphocytes and treated using bisulfite modification reagents. The modified DNA was amplified with the respective primer sets that only recognized bisulfite-modified DNA and submitted to TA cloning followed by Sanger sequencing. The pair of primers was listed as below: 5′-TTAGGTTATTTCGGTGAGGGAG-3′, and 5′-ACACAACAACAAACCTCTAAACATT-3′.
Cell lines and transfection
The immortal lymphocytes were established by Epstein-Barr virus transformation using standard methods70 and cultured in RPMI-1640 (Gibco) supplemented with 20% FBS (Biochrom). HEK-293FT (Life Technologies), HEK-293 (ATCC), SH-SY5Y (ATCC), H1299 (ATCC) and A549 (ATCC) cell lines were all cultured in DMEM (Corning) supplemented with 10% FBS and penicillin/streptomycin (Life Technologies). Mouse MEF cells were prepared and established following the reported methods71. Primary neurons were isolated from the PFC region of rat brains (Sprague Dawley) at embryonic day 18 and cultured in DMEM/F12 medium (Gibco) supplemented with 10% FBS. On the next day, cells were switched to serum-free NeuroBasal medium (Gibco) supplemented with B-27 supplements (Gibco) and GlutaMAX (Gibco). Cells were maintained at 37 °C in a humidified 5% CO2 air incubator. All cell lines were routinely tested for mycoplasma contamination.
HEK-293FT cells were transfected with the indicated plasmids using polyethylenimine (Sigma), while transfection of SH-SY5Y and H1299 cells were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. H1299-based mono- or double-allele knockout cell lines were generated by genome editing using the CRISPR/Cas9 system72. A pair of sgRNAs targeting UBE3A was designed as previously reported72 and listed below (target sequences are lowercase):
(1) 5′-CACCGagcacaaaactcattcgtgc-3,
(2) 5′-AAACgcacgaatgagttttgtgctC-3′.
Primary neurons were transfected with the indicated plasmids using calcium phosphate transfection reagent (Beyotime) following the manufacturer's protocols.
Plasmid construction
Detailed information of all plasmids is listed in Supplementary information, Table S2. Briefly, restriction enzyme digestion and ligation reactions (NEB) were performed using traditional cloning methods. Plasmids used in Y2H screening were obtained using Gateway LR clonase reaction (Thermo Fisher) according to the manufacturer's protocols. Point mutations of the indicated plasmids were introduced by site-directed mutagenesis as previously reported73. In addition, plasmids consisting of several cassettes were constructed using Gibson assembly (NEB) following the manufacturer's instruction. In this work, the full-length human UBE3A isoform 2 (1-875 AA) was used as the template, and expression tags fused with UBE3A were inserted at its N-terminus.
Yeast two-hybrid screening
Y2H screening was performed as previously described74 using UBE3A as the bait protein. Both pDEST32-UBE3A and the human ORFeome library (pDEST22 backbone) were co-transformed into yeast strain Mav203 (Thermo Fisher). The positive colonies could both survive in SD-4 (deficient in Ura, His, Leu and Trp) medium (Clontech) and show a blue color in the presence of X-Gal (Sigma).
GST pull-down assay
GST, GST-UBE3A and GST-UBE3A-truncated proteins were expressed in BL21 competent cells and purified using Glutathione-agarose beads (GE Healthcare) according to the manufacturer's instructions. ALDH1A1-His6, ALDH1A2-His6 and ALDH1A3-His6 proteins were purified using Ni-NTA agarose beads (Qiagen) following the manufacturer's protocols. Purified ALDH1A2-His6 and GST-UBE3A proteins were incubated in pull-down buffer (50 mM Tris-Cl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM DTT, 10 mM MgCl2) for 2 h at 4 °C. The beads were washed four times with pull-down buffer, and analyzed by immunoblotting assay. The pull-down assays of other proteins followed the same procedures.
In vitro ubiquitylation assay
In vitro ubiquitylation was performed as previously described74. Briefly, 100 ng Uba1 (E1), 150 ng UBCH7 (E2), 500 ng UBE3A (E3), 2.5 μg ALDH1A2-His6 and 5 μg wild-type ubiquitin were added into ubiquitylation buffer (25 mM Tris-Cl, pH 7.6, 100 mM NaCl, 1 mM DTT, 5 mM MgCl2, supplemented with 2 mM ATP) with the final reaction volume of 50 μL, and incubated at 37 °C for 1 h. The ubiquitylation level of ALDH1A2 protein was examined by immunoblotting assay.
E. coli ubiquitylation system reconstitution
The entire components of the ubiquitylation system were cloned into the dual-expression backbone pACYCDuet-1 vector (Novagen) using traditional cloning methods. HA-UB, UBCH7 and Uba1 were assembled in the first multiple cloning sites (MCS) under T7 promoter/lac operator and ribosome-binding sites (RBS), each separated with Shine-Dalgarno sequences to form a polycistronic cassette. UBE3A was inserted into the second MCS, to generate the plasmid pACYC-HA-UB-UBCH7-Uba1-UBE3A. BL21 competent cells were co-transformed with pACYC and pET22b-ALDH1A2-His6 plasmids using electroporation, and selected with chloramphenicol and ampicillin antibiotics. The E. coli cell culture was induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; Sigma) at the OD600 of 0.8, and further cultured at 18 °C for 16 h. Cells were harvested and resuspended in RIPA buffer (150 mM NaCl, 50 mM Tris-Cl, pH 7.4, 1% NP-40, 0.1% SDS). The cells were then sonicated using the Vibra-Cell processors (SONICS) and pelleted by centrifugation. The supernatant was purified using Ni-NTA agarose beads. In USP2cc-treatment group, the purified protein binding on Ni-NTA beads was incubated with USP2cc enzyme overnight at 4 °C. The ubiquitylation level of purified ALDH1A2 protein was examined by immunoblotting assay.
Mass spectrometry analysis of ubiquitylation modification sites
The protein samples were obtained both from in vivo ubiquitylation assay in HEK-293FT cells and the E. coli reconsitituted system as described above. Procedures for MS analysis were as previously described74. Briefly, the protein pellet was dissolved in 8 M urea, 100 mM Tris-Cl (pH 8.5), followed by TCEP reduction, NEM alkylation and trypsin digestion. Peptides were separated by the EASY-nLC system (Thermo Fisher) and analyzed by the Q Exactive mass spectrometer (Thermo Fisher). Protein and ubiquitylation analysis were performed with Thermo Proteome Discoverer 2.1 (Thermo Fisher) and searched against Uniprot Human database (http://www.uniprot.org/).
Immunoprecipitation and immunoblotting
Specific endogenous or exogenous protein-expressing cells were lysed in IP buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol) supplemented with protease inhibitor cocktail (Roche), and sonicated using the Vibra-Cell processors. After centrifugation to remove the cell debris, the supernatant was incubated with specific antibodies and Protein G agarose beads (Merck Millipore) overnight at 4 °C. The primary antibodies were as follows: normal rabbit IgG (sc-2027, Santa Cruz), anti-Flag (F1804, Sigma), anti-ALDH1A2 (sc-367527, Santa Cruz), anti-HA (H6908, Sigma). The immunoprecipitates were enriched and denatured at 95 °C for 10 min in 2× SDS-PAGE loading buffer. The inputs, immunoprecipitates and other cell lysates were then subjected to SDS-PAGE, and transferred to PVDF membrane (Bio-Rad). The membranes were immunoblotted with the specified antibodies: anti-UBE3A (sc-166689, Santa Cruz, 1:500 dilution), anti-Flag (F1804, Sigma, 1:8 000 dilution), anti-HA (H6908, Sigma, 1:4 000 dilution), anti-ALDH1A2 (sc-367527, Santa Cruz, 1:500 dilution), anti-ALDH1A1 (15910-1-AP, Proteintech, 1:500 dilution), anti-ALDH1A3 (25167-1-AP, Proteintech, 1:500 dilution), anti-His (H1029, Sigma, 1:4 000 dilution), anti-GST (66001-1-lg, Proteintech, 1:5 000 dilution), anti-Myc (sc-40, Santa Cruz, 1:1 000 dilution), anti-actin (A2228, Sigma, 1:8 000 dilution) and anti-GAPDH (sc-32233, Santa Cruz, 1:4 000 dilution).
Cells expressing His6-tagged Ub, ALDH1A2-HA and Flag-UBE3A were lysed in 1.0% SDS denaturing buffer, followed by boiling at 95 °C for 10 min, then diluted by 10-fold in immunoprecipitation buffer. The Ub-conjugated ALDH1A2 protein was then specifically enriched using anti-HA antibody, and detected by immunoblot analysis.
Immunofluorescence
SH-SY5Y cells were transfected with the specific plasmids and cultured for at least 24 h before fixation in 4% paraformaldehyde, followed by staining of Flag-tagged UBE3A. The permeabilized cells were incubated with primary antibody (anti-Flag, F1804, Sigma) at 4 °C overnight, and then Alexa Fluor 488-conjugated secondary antibody (A11029, Thermo Fisher) at room temperature for 1 h. The cell nucleus was counterstained with 4′, 6-diamidino-2-phenylindole (DAPI, Thermo Fisher). Primary rat neurons at day 10 in vitro (DIV) were transfected with the indicated plasmids using a calcium phosphate transfection method and treated with DMSO (Sigma) or 1 μM TTX (Aladdin) and 100 μM 𝒟-(−)-2-amino-5-phosphonopentanoic acid (D-APV, Tocris) at DIV 12 for 24 h. The cells were fixed and stained following the above procedures but using primary antibodies, anti-GluR1 (sc-55509, Santa Cruz) and anti-PSD95 (ab18258, Abcam), and subsequently Alexa Fluor 488 (A11029, Thermo Fisher) and Alexa Fluor 647 (A21245, Thermo Fisher)-conjugated secondary antibodies were applied, respectively. Images were acquired on an Olympus FV1200 confocal microscope (Olympus) with sequential acquisition settings at 1024 × 1024 pixel resolution. Identical confocal settings were maintained during all scans in the same experiments. Fluorescence intensities were analyzed and quantitated with Image-Pro plus software (Media Cybernetics).
Immunohistochemistry
Brains from mice that received stereotaxic injection were fixed in PBS containing 4% paraformaldehyde overnight at 4 °C, and immersed in PBS containing 30% sucrose (Sigma) until settling. Coronal brain sections (40 μm) were obtained using a Leica CM3050 S Research Cryostat (Leica Biosystems) and treated with PBST (0.3% Triton X-100 resolved in PBS) for 15 min at room temperature. The brain slices were then blocked with 3% normal goat serum (Boster, China), and incubated with primary antibody anti-Flag (14793, Cell Signaling, 1:800 dilution) overnight at 4 °C. The slides were then incubated with Cy3-conjugated secondary antibody (111-165-045, Jackson ImmunoResearch) for 1 h at room temperature. The brain sections were counterstained with DAPI for nuclear staining and coverslipped with Mowiol mounting solution (Sigma). Fluorescent images were obtained using an Olympus FV1200 confocal microscope (Olympus).
ALDH1A enzyme activity assay
ALDH1A2-Flag protein was ectopically expressed in the HEK-293FT cell line (otherwise indicated) with or without Myc-UBE3A expression, and immunoprecipitated using anti-Flag agarose beads (Sigma), followed by elution using Flag peptide in ALDH1A assay buffer (0.1 M sodium pyrophosphate, pH 8.0, 1.0 mM EDTA, 2.0 mM DTT). For both assays with or without de-ubiquitylation, USP2cc enzyme or bovine serum albumin (BSA) was incubated with the eluted protein overnight at 4 °C. The reaction mixture contained 2.5 mM NAD+, 20 mM DTT, 100 μM propionaldehyde (Sigma) in activity assay buffer, and the enzyme activity was measured on a BioTek Synergy Neo spectrophotometer at 340 nm with 3-min intervals at room temperature. The reaction was terminated when all values reached their plateau states. Using NADH as a standard, ALDH1A2 enzyme activity was calculated as: (amount of NADH generated (nmol) within total reaction time × sample dilution factor) / (reaction time × reaction volume). In measuring activity oxidizing substrate of all-trans retinaldehyde (Sigma), the ALDH activity assay kit (Cayman) was used through a fluorescence-based approach. Briefly, in the presence of 100 μM all-trans retinaldehyde, enzyme activity was assayed by monitoring the fluorescence at Ex = 530-540 nm, Em = 585-595 nm to minimize the interference from retinaldehyde at low absorbance wavelengths.
Aldefluor assay
The aldefluor assay was performed using an Aldefluor kit (Stemcell Technologies) according to the manufacturer's instructions. Briefly, 1 × 106 immortalized lymphocytes were incubated with the fluorescent substrate BODIPY-aminoacetaldehyde-diethyl acetate (BAAA-DA) (1.5 μM, Aldefluor assay buffer resolved) for 30 min at 37 °C. Each fraction of the cells was split into two halves: one half for the fluorescence assay, whereas the other half of the cells was pre-treated with 15 μM ALDH inhibitor, diethylamino-benzaldehyde (DEAB), to serve as the negative control during flow cytometry analyses.
RARE-luciferase reporter assay and the RA donor-sensor co-culture system
The plasmid pGL4.22-RARE-TK-luciferase was constructed by cloning 3× RARE (RA-responsive elements) into the pGL4.22 vector (Promega). The RA sensor cell line was established by transfecting H1299 cells with pGL4.22-RARE-TK-luciferase, followed by screening of the stable line with puromycin (1 μg/ml, Sigma). The RA sensor cells were co-cultured with the RA donor cells derived from immortal lymphocytes at ∼1:1 ratio (cell numbers) in VP-SFM medium (virus production serum-free medium, Gibco). After treatment with 1 μM all-trans retinaldehyde for 8 h, total luciferase activities of the cultured cells were assayed, following the manufacturer's instructions (Promega).
Electrophysiology
The primary neurons and post-transfected neurons at DIV12-14 were treated with 1 μM TTX and 100 μM D-APV for 24 h, and then patch clamp recordings were measured at room temperature. The recording pipettes were filled with an internal solution (20 mM KCl, 5 mM MgCl2, 20 mM HEPES, 110 mM K-gluconate, 0.6 mM EGTA, 2 mM Na2-ATP, 0.2 mM Na3-GTP, pH 7.3, 290 mOsm). The internal resistance was about 3-6 MΩ. Cell cultures were positioned in an external solution containing 129 mM NaCl, 5 mM KCl, 1 mM MgCl2, 25 mM HEPES, 2 mM CaCl2, 30 mM glucose, pH 7.3, 310 mOsm. mEPSC recordings were performed at −70 mV in the presence of 1 μM TTX and 100 μM picrotoxin (Tocris) in the external solution. Individual events were recorded and data were analyzed with the Mini Analysis Program (Synaptosoft).
Quantitative RT-PCR
An RNAsimple total RNA kit (Tiangen) was used to extract total RNAs from primary neurons treated with 1 μM TTX and 100 μM D-APV for 24 h or 0.5 μM ATRA for 8 h. Complementary DNA samples were synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo). qRT-PCR assay was performed with the CFX96 real-time PCR system (Bio-Rad) using SYBR Green Master Mix (Toyobo). The relative abundances of the transcripts of the indicated genes were normalized to that of Gapdh, using the ΔΔCt method. Sequences of the PCR primers used in this work are listed in Supplementary information, Table S3.
Surface protein biotinylation assay
The surface protein biotinylation assay was performed following a previously reported method29. Briefly, after treatment with DMSO or 1 μM TTX and 100 μM D-APV for 24 h, primary PFC neurons were washed with PBS buffer and incubated with biotin solution (1 mg/ml EZ-Link Sulfo-NHS-SS-Biotin, Pierce) for 2 h at 4 °C. The reaction was quenched by washing with 0.1 M glycine (prepared in PBS), followed by washing three times with PBS. The biotinylated cells were solubilized in cell lysis buffer (25 mM MgCl2, 1% NP-40, 1% Triton X-100 and 10% glycerol in PBS) supplemented with protease inhibitor cocktail (Roche). After centrifugation to remove the cell debris, the clarified supernatant was incubated with UltraLink Streptavidin resin (Pierce) with rotation at 4 °C overnight. Following centrifugation, the biotinylated proteins were pelleted and washed three times with cell lysis buffer. The biotinylated proteins were denatured with 2× SDS-PAGE loading buffer at 75 °C for 30 min and analyzed by immunoblotting assay using antibodies: anti-GluR1 (13185, Cell Signaling, 1:500 dilution) and anti-GluR2 (13607, Cell Signaling, 1:500 dilution).
Animals
Mice were housed in groups (n = 3-5) on a standard 12 h/12 h light-dark cycle, and food and water were provided ad libitum. All behavioral experiments were performed during the light cycle. All animal studies were performed in strict accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Shanghai Institute of Biochemistry and Cell Biology, CAS. Mice used for all experiments in this study were male, with C57BL/6 genetic background (SLAC, China).
Virus preparation and stereotaxic injection
Expression of all indicated genes was driven by the Synapsin I (SynI) promoter, in fusion with Flag tag at N-terminus. AAV vectors were serotyped with AAV2/9 coat protein and packaged by Obio Technology. Viral titers were approximately in 1.5 × 1013 genome copies per ml for the AAV-SynI-Flag-UBE3A and AAV-SynI-Flag-UBE3A-T508E, and 9.5 × 1012 genome copies per ml for the AAV-SynI-Flag-EGFP.
Three-week-old mice were anesthetized with sodium pentobarbital (50 mg/kg, i.p.) and placed in a stereotaxic apparatus (RWD Life Science) to infuse virus. PBS-diluted AAVs (1 μl) were bilaterally injected into mPFC regions of the brain at a constant rate of 0.2 μl/min using a syringe pump (Stoelting). The stereotaxic injection coordinates relative to bregma were: AP, +2 mm; ML, ±0.5 mm; DV, −1.3 mm. The injected viral particles for AAV-SynI-Flag-UBE3A and its mutant T508E were 3 × 109, and 1.5 × 109 for AAV-SynI-Flag-EGFP at each injected site. The needle was kept in place for another 3 min to prevent virus backflow. The mice were maintained on a 37 °C heat blanket until fully recovered from anesthesia. At post-surgery day 1-2, 0.5 mg/ml meloxicam (Sigma) was injected (2 mg/kg, i.p.) to help relieve pain. Four weeks after the surgery, behavioral tests were conducted. In the ATRA rescue experiments carried out 1 week after the injection of AAV-SynI-Flag-UBE3A virus, mice were administered daily (5 consecutive days per week) with ATRA (Sigma; dissolved in olive oil, at dosages of 3 mg/kg) or vehicle (olive oil) via oral gavage. Behavioral tests were started after 4 weeks of such treatment.
DSF administration
Four-week-old mice were administered a single dose of 0.1 mg/g, or 0.3 mg/g DSF (Sigma, dissolved in olive oil) via oral gavage every other day for 6 weeks. The control group received the vehicle only. Body weights of the mice were measured weekly. Behavioral tests of the mice were initiated after administration of DSF or vehicle for 6 weeks. Brains were dissected from the animals receiving DSF or vehicle only and flash-frozen immediately in liquid nitrogen, followed by tissue homogenization and ATRA quantitation using the HPLC-MS/MS approach described below.
Quantitation of ATRA using HPLC-MS/MS
Sample preparation, retinoid extraction and subsequent analysis using HPLC-MS/MS approach were performed as previously reported51,52. Briefly, each frozen brain was homogenized on ice with 2 ml of 0.9% saline under red light. 13-cis-RA-d5 (20 ng/ml, TRC) was added as the internal standard to the homogenates and mixed. 1.5 ml of 0.025 M KOH in ethanol was added to the homogenates. After addition of 7 ml hexane, the aqueous phase was extracted, and then neutralized with 120 μl of 4 M HCl. Next, the organic phase containing RA and polar retinoids was extracted with another 7 ml of hexane. The hexane extracts were evaporated under nitrogen gas, and resuspended in 50 μl of acetonitrile. For ATRA resolution in HPLC, the 2.1 × 100 mm Supelcosil ABZ+PLUS column (3 μm, Sigma) was employed, with the following running solvents: A, H2O with 0.1% formic acid; B, acetonitrile with 0.1% formic acid. Identity of the ATRA-containing fraction was determined with an AB Sciex 4000 QTRAP LC-MS/MS system equipped with APCI in positive ion mode. The amounts of ATRA in each fraction were determined using calibration curves generated from standard amounts of ATRA.
Self-grooming test
The mice used for the self-grooming test were firstly habituated in a cleaning chamber covered with beddings (the height was about 0.5-1 cm) for 10 min. After the habituation, the time of mice spent in grooming was recorded within 10-min intervals, using stop-watches by two observers who were blinded to the groupings of mice.
Three-chamber social test
The three-chamber social test was performed according to previously reported protocols75. Briefly, the apparatus consisted of a transparent acrylic box divided into three chambers of the same size, and removable doors in each partition. Two wire cups were placed with the opening down in the left and right chambers. Two days prior to the testing, two age- and gender-matched C57BL/6 mice, not littermates, were placed under both wire cups as stranger mice, and habituated for 1 h per day. Test mice, grouped in a blinded manner, were habituated to the test room for 1 h before the start of behavior tests. Each test mouse was introduced to the central chamber and allowed to explore for 10 min and then isolated in the central chamber with doors closed. In the first stage, a stranger mouse was placed randomly into the left or right wire cup, to avoid any side preference of the test mice; while an inanimate object was placed into the other wire cup. Each test mouse was allowed to explore with open access to all three chambers of the apparatus for 10 min. The time spent in interacting with the stranger mouse or an inanimate object was recorded manually. Following this stage, the test mouse remained in the stranger mouse chamber for extra 5 min. In the second stage, another stranger mouse was shifted to replace the inanimate object. Each test mouse was allowed to investigate for additional 10 min. The time spent in interacting between familiar or novel animals was measured manually. The observers were blinded to the groupings of test mice.
Open-field test
Mice, grouped in a blinded manner, were placed in the open-field apparatus (Med Associates) and allowed to explore for 30 min. The time spent in center (defined by the area from 1/3 to 2/3 of the length of each side) and distances traveled in center compared with total traveling distances were measured using Ethovision automated tracking software (Noldus).
Elevated plus maze
Mice were placed in the closed arm of elevated plus maze apparatus (Med Associates) and allowed to explore for 5 min. The movements of animals were filmed by an overhead camera, and the entries running into open arms and closed arms within the 5-min interval were analyzed using the ANY-maze software (Stoelting).
Rotarod test
The motor coordination and locomotor activity of blindly grouped mice were examined using a Rotamex rotarod apparatus (Columnbus Instruments). In a single day, the mice were tested on three trials, with each trial lasting 5 min with the rod rotation accelerated from 4 to 40 rpm. The intervals permitting rest between each trial were about 30 min. The time of mouse falling down the rod was automatically recorded by the infrared detection system and results of the last trial were compared.
Statistics
All statistical analyses were performed using GraphPad Prism (GraphPad Software). Data sets were analyzed for significance using Student's two-tailed t-test, one-way ANOVA with Bonferroni post hoc test, or Dunnett's post hoc test, respectively. All data were expressed as means ± SEM. Details for each statistical analysis were clarified in each figure legend. All data were collected from at least 3 independent replicates of experiments. No statistical methods were applied to pre-estimate sample size. All mice were randomly allocated to different experimental groups.
Accession number
The CNV data have been deposited in the Gene Expression Omnibus database at NCBI under accession number GSE93207.
Author Contributions
RH designed and supervised the whole UBE3A project. XX led the study; XX, CL and XG performed most of the experiments and analyzed data. CL performed Y2H and biochemistry experiments, and was the first to identify ALDH1A2 as a novel UBE3A substrate. XX, XG, ZH, LZ and CX performed animal study and behavior tests. YL carried out the electrophysiology experiments under supervision of HX. KX supervised the collection and analysis of ASD patient samples; HG contributed human data and immortalized the patient-derived cells. ZX provided Ube3a-knockout mice. ZX, ZQ, LM and KR provided advices on experiments. RH and XX wrote the manuscript with inputs from other authors. All authors read and approved the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Prof Qishui Lin (SIBCB, CAS) for invaluable advice. We appreciate Drs Xiang Yu, Jiulin Du, Bo Yuan (Institute of Neuroscience, SIBS, CAS), Donghong Cui, Han Li (Shanghai Mental Health Center) for providing technical assistance, and Zi Li (Institute of Nutritional Sciences, SIBS, CAS) for technical help in HPLC-MS/MS. We specially acknowledge the excellent support from proteomics facility at the National Center for Protein Science Shanghai, and the cell imaging center led by Dr Wei Bian at SIBCB. We thank all members of our laboratory for support and Ms Yalan Wu for assistance. We are also grateful to Dr ZeNan Chang (University of California, Los Angeles) for critical reading of the manuscript. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000 and XDA12040323), the National Natural Science Foundation of China (31470770 and 81525019 to RH; 81601203 to XX; 81330027 and 81525007 to KX; 31400919 and 31671114 to HG), the Ministry of Science and Technology of China (2013CB910900 to RH) and the China Postdoctoral Science Foundation (2016M591724 to XX).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
UBE3A interacts with, and ubiquitylates other members of human ALDH1A family.
The members of mammalian ALDH1A family proteins are highly conserved in both amino acid sequences and secondary structures.
Low abundances of endogenous ALDH1A family proteins in H1299 cell line and its derivative H1299 UBE3A−/− cells.
UBE3A ubiquitylates endogenous or exogenous ALDH1A2 protein in a dosage-dependent manner.
Copy number amplification identified in three autism patients.
Ubiquitylation of ALDH1A2 compromises its dehydrogenase activity, using propionaldehyde as substrate.
Neuronal activity blockade induced changes in synaptic homeostasis through translational control.
Stereotaxic injection of indicated AAVs into mouse PFC regions.
Mouse behavior tests for mice after stereotaxic injection of indicated AAVs or those receiving oral intake of ATRA.
Administration with disulfiram did not alter the mouse behaviors in open-field, elevated plus maze or rotarod tests.
Methylation analysis of SNRPN gene in ASD patients with 15q11-13 duplication.
Plasmids used in this study.
qRT-PCR primers used in this study.
References
- Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet 2013; 14:191–213. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 2012; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008; 9:341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009; 459:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 2015; 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Guo H, Xiong B, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun 2016; 7:13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan RN, Bae BI, Cubelos B, et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 2016; 167:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Konyukh M, Delorme R, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet 2010; 26:363–372. [DOI] [PubMed] [Google Scholar]
- Santini E, Klann E. Reciprocal signaling between translational control pathways and synaptic proteins in autism spectrum disorders. Sci Signal 2014; 7:re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet 2006; 15:R138–R150. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Bradford Y, Chen Y, et al. Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics 2001; 77:105–113. [DOI] [PubMed] [Google Scholar]
- Baron CA. Genomic and functional profiling of duplicated chromosome 15 cell lines reveal regulatory alterations in UBE3A-associated ubiquitin-proteasome pathway processes. Hum Mol Genet 2006; 15:853–869. [DOI] [PubMed] [Google Scholar]
- Noor A, Dupuis L, Mittal K, et al. 15q11.2 Duplication encompassing only the UBE3A gene is associated with developmental delay and neuropsychiatric phenotypes. Hum Mutat 2015; 36:689–693. [DOI] [PubMed] [Google Scholar]
- Smith SE, Zhou YD, Zhang G, et al. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med 2011; 3:103ra197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Berrios J, Newbern JM, et al. An autism-linked mutation disables phosphorylation control of UBE3A. Cell 2015; 162:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 2011; 12:295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med 2000; 6:1073–1081. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 2006; 22:159–180. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem 2012; 81:203–229. [DOI] [PubMed] [Google Scholar]
- Swatek KN, Komander D. Ubiquitin modifications. Cell Res 2016; 26:399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell GL, Margolis SS. From UBE3A to Angelman syndrome: a substrate perspective. Front Neurosci 2015; 9:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Duester G. SnapShot: retinoic acid signaling. Cell 2011; 147:1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer KD, Stoney PN, Morgan PJ, McCaffery PJ. A vitamin for the brain. Trends Neurosci 2012; 35:733–741. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dolle P. Retinoic acid signalling during development. Development 2012; 139:843–858. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol 2015; 16:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lau AG, Sarti F. Synaptic retinoic acid signaling and homeostatic synaptic plasticity. Neuropharmacology 2014; 78:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron 2008; 60:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Napoli JL. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARα. FASEB J 2008; 22:236–245. [DOI] [PubMed] [Google Scholar]
- Koppaka V, Thompson DC, Chen Y, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev 2012; 64:520–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Kaplan T, Attali I, Motamedchaboki K, et al. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J 2012; 31:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreb JS, Zucali JR, Ostmark B, Benson NA. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by Aldefluor flow cytometry-based assay. Cytometry B Clin Cytom 2007; 72:281–289. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 1998; 21:799–811. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics 1997; 41:263–266. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem 1999; 274:18785–18792. [DOI] [PubMed] [Google Scholar]
- Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, et al. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ 2009; 16:1156–1166. [DOI] [PubMed] [Google Scholar]
- Zaaroor-Regev D, de Bie P, Scheffner M, et al. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci USA 2010; 107:6788–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yao A, Zhi H, et al. Angelman syndrome protein Ube3a regulates synaptic growth and endocytosis by inhibiting BMP signaling in Drosophila. PLoS Genet 2016; 12:e1006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol 1993; 13:4918–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari T, Brimer N, Vande Pol SB. Peptide interactions stabilize and restructure human papillomavirus type 16 E6 to interact with p53. J Virol 2012; 86:11386–11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Li J, Donini S, et al. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci Rep 2016; 6:35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA 1999; 96:9118–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han EB, Stevens CF. Development regulates a switch between post- and presynaptic strengthening in response to activity deprivation. Proc Natl Acad Sci USA 2009; 106:10817–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicks LK, Koike H, Akbarian S, Morishita H. Prefrontal cortex and social cognition in mouse and man. Front Psychol 2015; 6:1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak B, Feng G. Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat Neurosci 2016; 19:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow ML, Pramparo T, Winn ME, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet 2012; 8:e1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med 2014; 370:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffney LJ, Zhong P, Wei J, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep 2015; 11:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445:168–176. [DOI] [PubMed] [Google Scholar]
- Kane MA, Napoli JL. Quantification of endogenous retinoids. Methods Mol Biol 2010; 652:1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JE, McCaffery P. HPLC/MS(N) analysis of retinoids. Methods Mol Biol 2010; 652:149–162. [DOI] [PubMed] [Google Scholar]
- Ramocki MB, Peters SU, Tavyev YJ, et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann Neurol 2009; 66:771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007; 39:25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet 2009; 85:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature 2008; 455:912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misner DL, Jacobs S, Shimizu Y, et al. Vitamin A deprivation results in reversible loss of hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA 2001; 98:11714–11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarti F, Schroeder J, Aoto J, Chen L. Conditional RARα knockout mice reveal acute requirement for retinoic acid and RARα in homeostatic plasticity. Front Mol Neurosci 2012; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Shi Y, Zhao X, et al. Positive association between ALDH1A2 and schizophrenia in the Chinese population. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33:1491–1495. [DOI] [PubMed] [Google Scholar]
- Galter D, Buervenich S, Carmine A, Anvret M, Olson L. ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson's disease and in the ventral tegmental area in schizophrenia. Neurobiol Dis 2003; 14:637–647. [DOI] [PubMed] [Google Scholar]
- Fares-Taie L, Gerber S, Chassaing N, et al. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet 2013; 92:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Nawaz Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl Recept Signal 2008; 6:e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Stoppel DC, Nong Y, et al. Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 2017; 543:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, Zoghbi HY. Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci 2016; 19:1408–1417. [DOI] [PubMed] [Google Scholar]
- Delorme R, Ey E, Toro R, et al. Progress toward treatments for synaptic defects in autism. Nat Med 2013; 19:685–694. [DOI] [PubMed] [Google Scholar]
- Xia K, Guo H, Hu Z, et al. Common genetic variants on 1p13.2 associate with risk of autism. Mol Psychiatry 2013; 19:1212–1219. [DOI] [PubMed] [Google Scholar]
- Nava C, Keren B, Mignot C, et al. Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. Eur J Hum Genet 2013; 22:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn P, Delach J. Human lymphocyte culture and chromosome analysis. CSH Protoc 2008; 2008:pdbprot5035. [DOI] [PubMed] [Google Scholar]
- Xu X, Tao Y, Gao X, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2016; 2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro 1984; 20:856–858. [DOI] [PubMed] [Google Scholar]
- Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol 2005; Chapter 28:Unit 28.1. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 2008; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen P, Gao H, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 2014; 26:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, Chen H-m, Swann JW, et al. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 2015; 528:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UBE3A interacts with, and ubiquitylates other members of human ALDH1A family.
The members of mammalian ALDH1A family proteins are highly conserved in both amino acid sequences and secondary structures.
Low abundances of endogenous ALDH1A family proteins in H1299 cell line and its derivative H1299 UBE3A−/− cells.
UBE3A ubiquitylates endogenous or exogenous ALDH1A2 protein in a dosage-dependent manner.
Copy number amplification identified in three autism patients.
Ubiquitylation of ALDH1A2 compromises its dehydrogenase activity, using propionaldehyde as substrate.
Neuronal activity blockade induced changes in synaptic homeostasis through translational control.
Stereotaxic injection of indicated AAVs into mouse PFC regions.
Mouse behavior tests for mice after stereotaxic injection of indicated AAVs or those receiving oral intake of ATRA.
Administration with disulfiram did not alter the mouse behaviors in open-field, elevated plus maze or rotarod tests.
Methylation analysis of SNRPN gene in ASD patients with 15q11-13 duplication.
Plasmids used in this study.
qRT-PCR primers used in this study.