Abstract
Pluripotent stem cells (PSCs) harbor constitutive DNA replication stress during their rapid proliferation and the consequent genome instability hampers their applications in regenerative medicine. It is therefore important to understand the regulatory mechanisms of replication stress response in PSCs. Here, we report that mouse embryonic stem cells (ESCs) are superior to differentiated cells in resolving replication stress. Specifically, ESCs utilize a unique Filia-Floped protein complex-dependent mechanism to efficiently promote the restart of stalled replication forks, therefore maintaining genomic stability. The ESC-specific Filia-Floped complex resides on replication forks under normal conditions. Replication stress stimulates their recruitment to stalling forks and the serine 151 residue of Filia is phosphorylated in an ATR-dependent manner. This modification enables the Filia-Floped complex to act as a functional scaffold, which then promotes the stalling fork restart through a dual mechanism: both enhancing recruitment of the replication fork restart protein, Blm, and stimulating ATR kinase activation. In the Blm pathway, the scaffolds recruit the E3 ubiquitin ligase, Trim25, to the stalled replication forks, and in turn Trim25 tethers and concentrates Blm at stalled replication forks through ubiquitination. In differentiated cells, the recruitment of the Trim25-Blm complex to replication forks and the activation of ATR signaling are much less robust due to lack of the ESC-specific Filia-Floped scaffold. Thus, our study reveals that ESCs utilize an additional and unique regulatory layer to efficiently promote the stalled fork restart and maintain genomic stability.
Keywords: pluripotent stem cells, Filia, Floped, DNA replication stress, genomic stability
Introduction
During mammalian development, epiblast cells in the inner cell mass of a blastocyst are pluripotent and give rise to the whole embryo proper. The equivalent embryonic stem cells (ESCs) can be derived from the inner cell mass of a blastocyst and maintained in vitro 1. The pluripotent epiblast cells and ESCs have similar molecular signatures and biological features including rapid proliferation, unique cell cycle composition, aberrant cell cycle checkpoints and, in particular, a high competence to maintain genomic stability as required for their specific functions2,3,4,5. Mouse ESCs (mESCs) in culture display a 1 000-fold lower mutation rate than their isogenic mouse embryonic fibroblasts (MEFs)6. However, the underlying molecular mechanisms remain largely elusive. It has been shown that ESCs employ specific strategies to counter DNA insults and safeguard their genome stability5,7. For instance, ESCs preferentially utilize the error-free homologous recombination (HR)-mediated pathway to efficiently and accurately repair DNA double-strand breaks (DSBs)8. ESCs also display higher mitochondria priming and keep Bax constitutively active at the Golgi to ensure hyper-sensitivity to apoptosis9,10. Moreover, ESCs use specific proteins to efficiently regulate their DNA damage response (DDR)11.
In eukaryotic cells, DNA replication machinery needs to accurately duplicate the genome in each round of cell division. However, numerous endogenous and exogenous factors impede replication fork progression, causing fork slowing or stalling. This phenomenon is defined as DNA replication stress, a major source of endogenous DNA damage and genome instability12. Notably, DNA replication stress and the associated replication errors are implicated in the generation of nonrecurrent copy number variations (CNVs)13, a major type of genome instability in pluripotent stem cells (PSCs)14. When replication fork progression encounters obstacles such as base damage and dNTP depletion, which cause the physical uncoupling of DNA polymerases from the replicative helicase, an aberrant single-stranded DNA (ssDNA) structure arises due to the continuous unwinding of parental DNA by the replicative helicase after the polymerase has stalled. ssDNA is rapidly coated by the ssDNA-binding protein RPA (replication protein A), and the RPA-coated ssDNA then ignites the replication stress response primarily by triggering the activation of ATR (ATM- and Rad3-related) kinase. ATR activates downstream signaling pathways to stabilize the stalled forks and facilitate fork restart12. Any failure to restart the stalled replication forks leads to DNA DSBs and genomic instability12,15,16,17.
ESCs and the equivalent epiblast cells proliferate rapidly18 and harbor a high degree of replication stress19,20. However, compared to differentiated cells, ESCs have relatively low mutation rates. This suggests that ESCs and the equivalent epiblast cells may be superior to the differentiated cells in resolving replication stress. One recent study reported that more dormant origins are fired in ESCs to ensure the completion of DNA replication under replication stress21. Whether ESCs are unique in restarting stalled replication forks has never been explored. Here we show that mESCs are superior to other somatic cells in restarting stalled replication forks to prevent endogenous DNA damage, and this relies on an ESC-specific Filia-Floped protein complex-dependent mechanism.
Results
ESCs are superior to differentiated cells in stabilizing and restarting the stalled replication forks
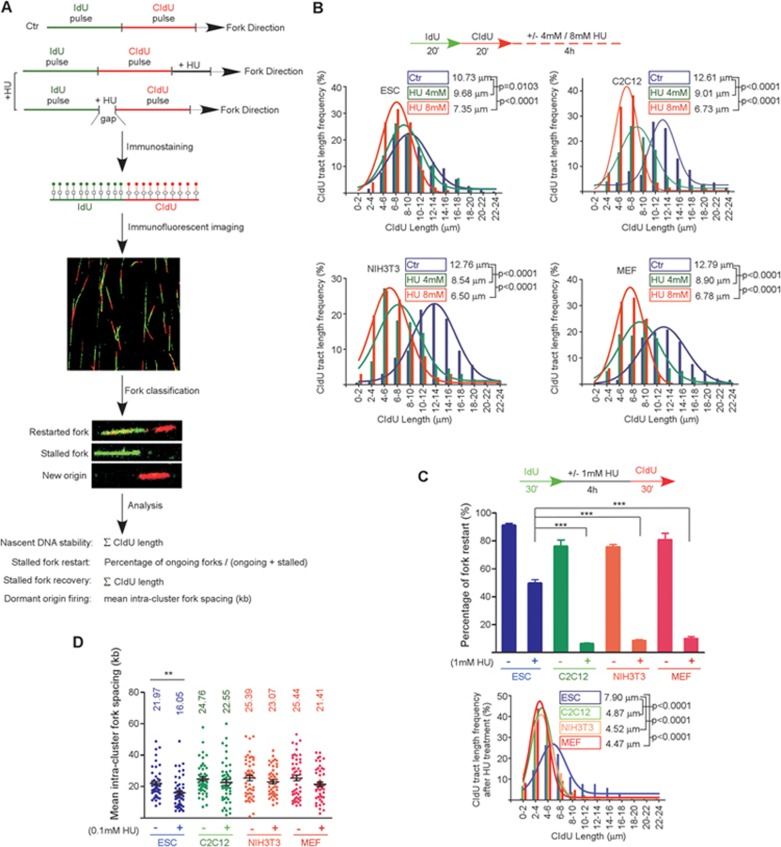
To test whether the ESCs are superior to tissue progenitor cells or terminally differentiated cells in stabilizing and restarting stalled replication forks, we compared the response of mESCs, C2C12 mouse myoblast cells, MEFs and NIH3T3 cells (spontaneously immortalized MEFs) to hydroxyurea (HU)-induced DNA replication stress by DNA fiber assay. In this assay, newly replicated DNA is substituted with nucleotide derivatives, such as bromodeoxyuridine (BrdU), chlorodeoxyuridine (CldU) or iododeoxyuridine (IdU), and is visualized by indirect immunofluorescence staining using specific antibodies after DNA combing22. Cells were temporarily treated with HU, which depletes the cellular dNTP pool and stalls replication forks17,23,24. Nascent DNA before and/or after HU treatment was differentially labeled with IdU (green fluorescent tract) and CIdU (red fluorescent tract), respectively (Figure 1A). We examined the abilities of cells to protect nascent DNA from degradation, to restart stalled replication forks, and to fire new replication origins in response to HU-induced fork stalling. To evaluate nascent DNA degradation, DNA was consecutively labeled with IdU and CIdU, and cells were subsequently cultured with or without HU treatment (Figure 1B). The degradation of nascent DNA was determined by the changes in the length of CIdU tracts with or without HU treatment25. In C2C12, MEF and NIH3T3 cells, HU treatment caused a drastic decrease of CIdU tract length in a dose-dependent manner. In contrast, a high dose of HU was required to achieve a much milder decrease of CIdU tract length in mESCs (Figure 1B). This suggests that ESCs are better at protecting nascent DNA upon replication fork stalling. To assess the fork restart ability of ESCs, HU treatment was performed between the two pulse labels (Figure 1C), and the fork restart was quantified as the percentages of restarted forks (IdU and CIdU labeled replication tracks) related to the total number of labeled forks (all IdU-labeled replication tracks). During the HU treatment, replication fork can move certain distance before complete arrest, which generates a gap between the IdU and CIdU tracts after fork restart (Figure 1A). To distinguish restarted CIdU tracts from newly initiated CIdU tracts, we included CIdU during the HU treatment to determine the average distance that the stalling forks can travel (gap length) (Supplementary information, Table S1). CIdU tracts shorter in distance than the average gap length from IdU tracts were then determined as restarted replication forks as previously reported25. In C2C12, MEF and NIH3T3 cells, the proportion of restarted forks decreased from about 75% under unimpeded conditions to ∼8% after HU treatment, whereas the decrease was significantly less in mESCs (from about 90% to 50%). Moreover, the restarted CIdU tract length in ESCs was significantly longer than in other cell types (Figure 1C). These data demonstrate that ESCs are more efficient than tissue progenitor cells and differentiated cells in protecting and restarting stalled forks under replication stress. In addition, more dormant replication origins were fired in ESCs than in the other cell types in response to HU treatment (Figure 1D), consistent with a recent report21.
Figure 1.
PSCs are superior in resolving DNA replication stress. (A) Scheme of DNA fiber analysis. (B) After fork stalling by HU, CIdU tract length in C2C12, NIH3T3 and MEF cells, but not ESCs, decreased dramatically in a dose-dependent manner. (C) After HU treatment, ESCs were better than C2C12, NIH3T3 and MEF cells at restarting stalled replication forks (upper panel). Moreover, the average CIdU tract length was less affected in ESCs than in C2C12, NIH3T3 and MEF cells (lower panel). (D) The frequency of dormant replication origin firing was evaluated as the mean intra-cluster fork spacing. Distributions of the mean intra-cluster fork spacing of 50 replication fork clusters are shown. Data are represented as mean ± SEM. **P < 0.01, ***P< 0.001.
To limit the potential effects of variations in genetic background and to further confirm the differential response to replication stress between ESCs and the differentiated cells, we removed LIF from the culture medium to induce ESC differentiation. At the earliest differentiation time-point when ESCs lost pluripotency but retained active proliferation (Supplementary information, Figure S1A), we compared the response to HU treatment between ESCs and their isogenic differentiated progeny as described above. Consistently, the ESCs displayed a higher competence in resolving replication stress (Supplementary information, Figure S1B-S1D). Because C2C12 cells and the isogenic differentiated progeny have comparable proliferation rates with mESCs (Supplementary information, Figure S1A and S1E), the superior ability of ESCs to resolve DNA replication stress is not related to cell proliferation rate. In addition, we examined the possible influence of the cellular dNTP level on stalled fork restart by culturing NIH3T3 cells in medium supplied with or without nucleosides after HU treatment. Increasing the cellular dNTP level by supplying exogenous nucleosides26 had no impact on stalled fork restart (Supplementary information, Figure S1F). Thus, the higher competence in restarting stalled forks in mESCs is irrelevant to cellular dNTP levels. Collectively, these data support the notion that ESCs are more efficient than differentiated cells in resolving DNA replication stress and this is an ESC-specific property.
Identification of Filia-Floped as a novel ESC-specific replication fork protein complex
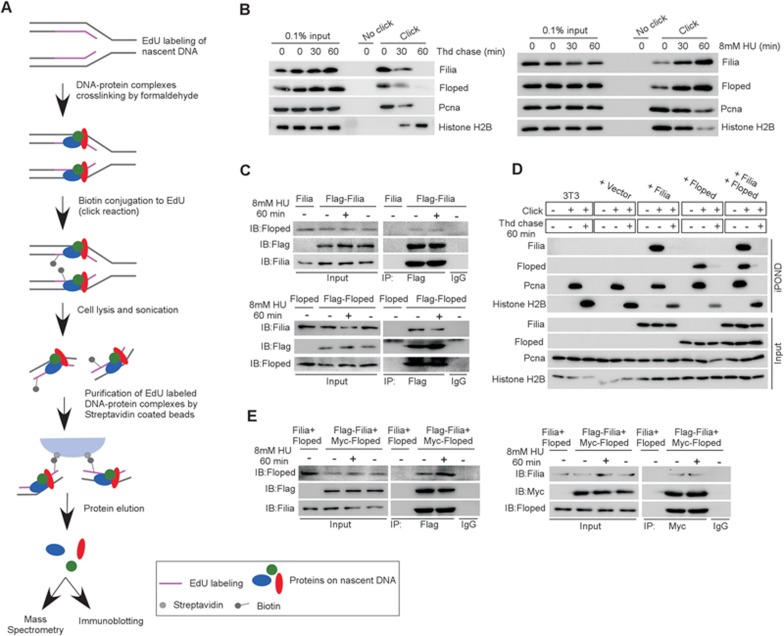
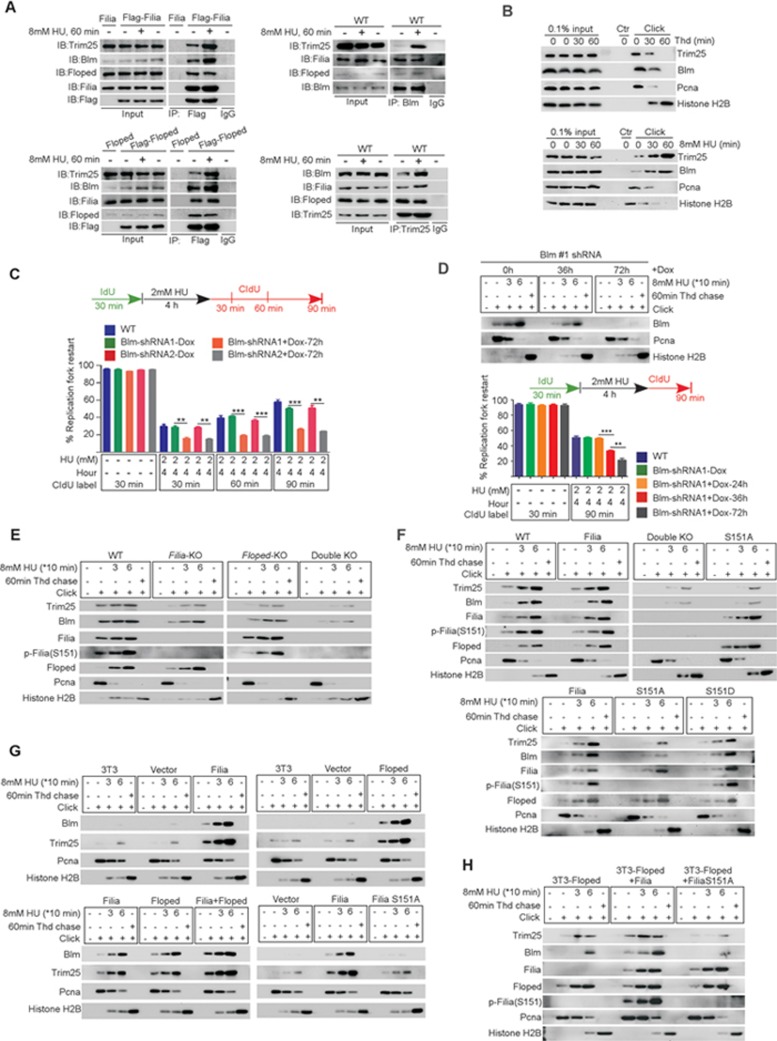
To understand the molecular basis underlying the observed superior replication stress response in ESCs, we performed iPOND (isolate proteins on nascent DNA)27 combined with an unbiased mass spectrometry analysis in three replicates to identify proteins that bind to replication forks in mESCs (Figure 2A). In one replicate we were able to detect an ESC-specific protein Filia, a protein that we had shown in a previous study to play critical roles in maintaining genome stability of mESCs by regulating DDR11 (Supplementary information, Figure S2A). In addition, Filia interacts with Floped28, which is also ESC specific (Supplementary information, Figure S2B). We therefore tested whether ESC-specific Filia and Floped proteins localize on replication forks. Binding of the two proteins to nascent DNA (or replication forks) under the normal culture condition was validated by immunoblotting analysis of iPOND captured samples (Figure 2B). In addition, we labeled the nascent DNA with BrdU for 5 min and showed by immunostaining that Filia and Floped co-localized with BrdU (Supplementary information, Figure S2C). Interestingly, iPOND analysis revealed that the loading of Filia and Floped on replication forks was enhanced by HU treatment (Figure 2B), suggesting their potential involvement in the replication stress response. Next, we examined whether chromatin-bound Filia and Floped interact on replication forks in mESCs. Co-immunoprecipitation of nuclear fractions of mESCs revealed that anti-Filia antibodies pulled down Floped, and vice versa. However, their association was not affected by HU treatment (Figure 2C). To further validate that Filia and Floped form a protein complex that localizes on replication forks, we ectopically expressed Filia and/or Floped in NIH3T3 cells (Supplementary information, Figure S2D) and performed iPOND and co-immunoprecipitation analyses. Indeed, iPOND could detect the presence of Filia and/or Floped proteins on replication forks of NIH3T3 cells ectopically expressing individual or both proteins (Figure 2D). Consistently, Filia and Floped formed a protein complex in nuclear fractions of NIH3T3 cells ectopically expressing both proteins (Figure 2E). Triple immunostaining of Filia, Floped and BrdU also validated their co-localization in mESCs and in NIH3T3 cells ectopically expressing both Filia and Floped (Supplementary information, Figure S2E). Together, we identified Filia-Floped as an ESC-specific replication fork protein complex.
Figure 2.
Filia-Floped is a novel ESC-specific replication fork protein complex. (A) Scheme of iPOND. (B) iPOND analysis confirmed the presence of Filia and Floped at replication forks in mESCs under normal (left panel) and HU treatment (right panel) conditions. HU treatment enhanced the retention of Filia and Floped on replication forks. (C) Filia and Floped reciprocally pulled down each other in ESCs treated with or without HU. (D) Filia and/or Floped proteins were ectopically expressed in NIH3T3 cells. iPOND analysis revealed their localization on replication forks. Thymidine (Thd) chased samples were included as negative controls. (E) Filia and Floped physically interacted in NIH3T3 cells co-expressing tagged Filia and Floped with or without HU treatment.
Filia and Floped function synergistically to promote efficient replication fork restart
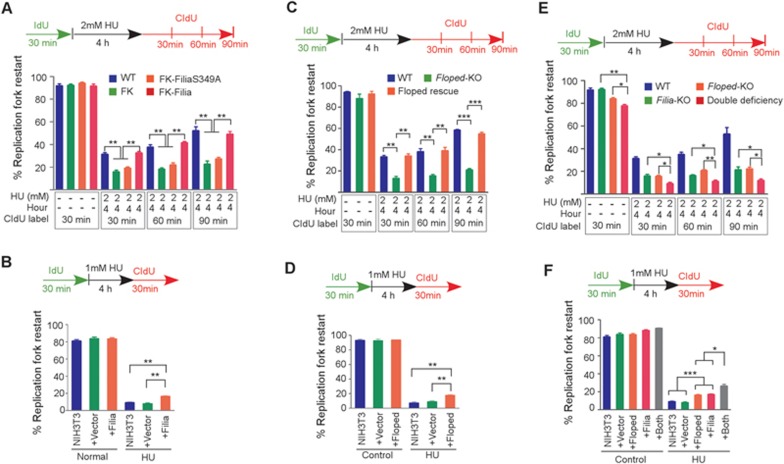
To determine whether this newly discovered ESC-specific replication fork protein complex is implicated in the HU-induced DNA replication stress response, we investigated the influence of Filia or Floped deficiency on nascent DNA degradation, replication fork restart and new replication origin firing by DNA fiber analysis. Filia−/− ESCs, Filia−/− ESCs expressing FiliaS349A, a mutant that does not undergo nuclear import of Filia, and Filia−/− ESCs complemented with wild-type (WT) Filia have been previously established11. Floped−/− ESCs were generated from Floped mutant mice28,29, and were complemented with Myc-Floped or Flag-Floped by stable transformation to establish Floped-rescued ESCs (Supplementary information, Figure S3A). Upon HU treatment, there was a decrease in the CIdU tract length in all four cell lines (WT, Filia-rescued, Filia−/− and FiliaS349A). However, the fold changes of CIdU tract length before and after treatment were not affected by depleting Filia from replication forks (Supplementary information, Figure S3B). This result suggests that Filia is not implicated in protecting nascent DNA from degradation upon fork stalling. Likewise, depletion of Filia from the replication forks did not compromise dormant origin firing in response to HU treatment (Supplementary information, Figure S3C). However, the stalled fork recovery was substantially impaired by Filia depletion, as shown by the lower rates of fork restart in Filia−/− and FiliaS349A ESCs compared with WT or Filia-rescued ESCs (Figure 3A). To further validate the functions of Filia on stalled replication forks, we performed the similar analyses on NIH3T3 cells with or without the ectopic expression of Filia (Filia-NIH3T3 and NIH3T3, respectively). Indeed, ectopic expression of Filia enhanced the stalled fork restart after HU treatment (Figure 3B), but had no effects on nascent DNA degradation or dormant replication origin firing (Supplementary information, Figure S3D and S3E). These data support the notion that Filia is implicated in regulating the restart of stalled replication forks. Consistently, Filia−/− and FiliaS349A-complemented ESCs display a range of genomic instability described in our previous study11.
Figure 3.
Filia and Floped cooperatively regulate replication fork restart. (A) Rates of stalled replication fork restart in wild-type (WT), Filia-knockout (FK), FiliaS349A-rescued (FK-FiliaS349A) and WT Filia-rescued (FK-Filia) ESCs. (B) Rates of stalled fork restart in NIH3T3 cells with or without ectopic Filia expression. (C) Rates of stalled fork restart in WT, Floped-knockout (Floped-KO) and Floped-rescued ESCs. (D) Rates of stalled fork restart in NIH3T3 cells with or without ectopic expression of Floped. (E) Rates of fork restart in WT, Filia-knockout (Filia-KO), Floped-knockout (Floped-KO), and double deficient ESCs with or without HU treatment. (F) Rates of fork restart in NIH3T3 cells expressing Filia and/or Floped. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We tested whether loss of Filia in epiblast cells affects post-implantation embryo development. We mated the Filia−/− females, in which the maternal Filia protein and its effects were completely eliminated, with Filia−/− or WT males. Mating to Filia−/− males generated a significantly higher rate of abnormal embryonic structures, in which the embryo proper was lost (Supplementary information, Figure S3F). Thus, depletion of Filia from epiblast cells compromised post-implantation embryo development in vivo.
We next examined the involvement of Floped in the DNA replication stress response using WT, Floped−/−, and Floped-rescued ESCs. Floped deficiency impaired stalled fork restart, which was rescued by re-introduction of Floped (Figure 3C). However, Floped depletion had no effects on nascent DNA degradation (Supplementary information, Figure S3G), or dormant origin firing (Supplementary information, Figure S3H). Similar results were obtained in NIH3T3 cells ectopically expressing Myc-Floped (Figure 3D and Supplementary information, Figure S3I and S3J). Thus, Floped plays a role in regulating stalled fork restart in response to HU treatment. Consequently, Floped−/− ESCs displayed a range of defects including genomic instability (Supplementary information, Figure S4A-S4D), tumorigenicity (Supplementary information, Figure S4E-S4G) and hypersensitivity to HU treatment (Supplementary information, Figure S4H).
To determine whether Filia and Floped have an additive effect in promoting the restart of stalled forks, we simultaneously depleted Filia and Floped by knocking down Filia in Floped−/− ESCs using short hairpin RNA (shRNA)11 (Supplementary information, Figure S5A). Compared with the individual depletion of Filia or Floped, loss of the two proteins further reduced the fork restart rate (Figure 3E). Consequently, Filia and Floped double knockdown ESCs (Supplementary information, Figure S5B) displayed a greater degree of DNA damage (Supplementary information, Figure S5C) and a higher level of apoptosis compared with the ESCs with individual knockdown of either protein (Supplementary information, Figure 5D). Accordingly, co-expressing Filia and Floped in NIH3T3 cells promoted fork restart better than expression of either individual protein (Figure 3F). Thus, Filia and Floped function in parallel to give synergistic regulation of efficient replication fork restart. Intriguingly, ESCs depleted of both proteins still had a higher competence to restart arrested forks than NIH3T3 cells. Likewise, NIH3T3 cells ectopically expressing both proteins displayed lower fork restart rates than mESCs (Supplementary information, Figure S5E). This result suggested that additional specific factors are involved in regulating stalled fork restart in ESCs.
ATR-dependent phosphorylation of serine 151 of Filia is required to promote stalled fork restart
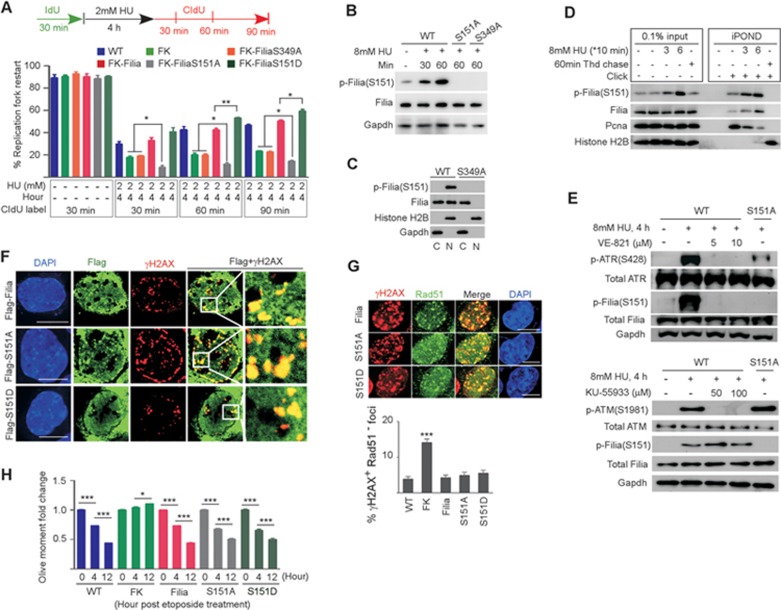
The replication stress response is orchestrated by ATR kinase12,23,30. As the Filia-Floped complex regulates the replication stress response, we examined whether Filia and/or Floped were subject to ATR-dependent phosphorylation. Mass spectrometry analysis identified a phosphorylation site on serine 151 of Filia. To validate this finding and to explore its relevance to replication stress response, we mutated serine to alanine (FiliaS151A) to mimic the non-phosphorylated status and to aspartic acid (FiliaS151D) to mimic constitutive phosphorylation. FiliaS151A or FiliaS151D was introduced into Filia−/− ESCs to establish stable complementing ESC lines (Supplementary information, Figure S6A). Neither mutation impaired Filia's normal distribution at replication forks (Supplementary information, Figure S6B and S6C). Moreover, FiliaS151A had no significant influence on nascent DNA degradation after HU treatment (Supplementary information, Figure S6D). However, compared with Filia-depleted cells, restart of stalled forks was further suppressed by FiliaS151A (Figure 4A). This finding suggests a dominant negative effect of FiliaS151A. In contrast, FiliaS151D functions as efficiently as the WT Filia (Figure 4A and Supplementary information, Figure S6D). Notably, FiliaS151A-complemented cells displayed a higher level of DNA DSBs than Filia−/− cells as determined by the neutral comet assay (Supplementary information, Figure S6E). These observations suggest that S151 of Filia is subject to phosphorylation that modulates Filia's roles in regulating stalled fork restart.
Figure 4.
ATR-dependent phosphorylation of the S151 residue on Filia is required for the restart of replication forks. (A) Rates of replication fork restart in WT, Filia-knockout (FK), FiliaS349A-rescued (FK-FiliaS349A), Filia-rescued (FK-Filia), FiliaS151A-rescued (FK-FiliaS151A) and FiliaS151D-rescued (FK-FiliaS151D) ESCs. (B) Phosphorylation of S151 on Filia (p-(FiliaS151)) was induced by HU treatment in WT ESCs, and no signal was detected in FiliaS151A- or FiliaS349A-complemented ESCs. (C) p-Filia(S151) was present in the nuclear fraction (N) but not the cytoplasmic fraction (C) of the WT ESCs. (D) iPOND confirmed that p-Filia(S151) localized at replication forks under normal and HU-treated conditions in ESCs. (E) Inhibition of the ATR kinase activity in ESCs suppressed the phosphorylation of FiliaS151 in response to HU treatment (upper panel), whereas inhibition of ATM kinase activity had no effect (lower panel). (F) Upon etoposide treatment of ESCs, Flag-tagged Filia, FiliaS151A and FiliaS151D were equally efficient at relocalizing to DNA damage sites. (G) Compared to WT ESCs, Filia-knockout (FK) ESCs were impaired in recruiting Rad51 proteins to DNA damage sites. WT Filia (Filia), FiliaS151A (S151A) and FiliaS151D (S151D) displayed equal ability in rescuing this defect. (H) Filia-knockout (FK) ESCs displayed lower DNA repair ability than WT cells after etoposide treatment. WT Filia, FiliaS151A and FiliaS151D mutants had a similar ability to rescue this defect. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar, 10 μm.
To further investigate this phosphorylation, we generated an antibody which specifically recognizes the phosphorylated S151 in Filia (p-Filia(S151)). S151 phosphorylation was robustly stimulated by HU treatment (Figure 4B). Moreover, the phosphorylation occurred in nucleus, because FiliaS349A mutation that blocks nuclear localization11 also eliminated S151 phosphorylation (Figure 4B). Indeed, cell fractionation showed that p-Filia(S151) is exclusively nuclear (Figure 4C). Immunoblotting analysis of iPOND captured samples further demonstrated the localization of p-Filia(S151) at replication forks and its enhanced retention at forks after HU treatment (Figure 4D). As ATR activation orchestrates the replication stress response12,23, we examined whether S151 phosphorylation is ATR dependent. Inhibition of ATR activity using a specific inhibitor, VE-82131, abrogated the phosphorylation of S151. As a control, inhibiting ATM activity with KU5593332 did not affect this modification (Figure 4E). Thus, the phosphorylation of S151 is dependent on ATR activity. However, S151 does not conform to the typical ATR substrate SQ/TQ motif, suggesting that ATR may regulate S151 phosphorylation indirectly via its downstream kinases or in a noncanonical manner. Intriguingly, the S151A mutation significantly decreased ATR but not ATM phosphorylation (Figure 4E), suggesting that Filia may also regulate ATR activation via a feedback loop.
We previously reported the involvement of Filia in HR-mediated DNA DSB repair11, which plays essential roles in facilitating the recovery of stalled forks33. We determined whether the beneficial role of Filia in replication fork restart relied on its HR-mediated DNA repair function. To this end, we treated the FiliaS151A-complemented ESCs with etoposide to induce DNA DSBs34 and examined whether FiliaS151A affects HR repair. Notably, FiliaS151A mutant protein (Figure 4F) and the key HR recombinase Rad51 (Figure 4G) were normally recruited to DSB sites labeled with γH2AX. Moreover, FiliaS151A-complemented cells displayed efficient repair of DSBs as determined by neutral comet assay (Figure 4H). Therefore, Filia S151 phosphorylation is not implicated in its function in HR repair of DNA DSBs and thus Filia's role in replication fork restart is mechanistically distinct from its function in HR-mediated DNA DSB repair.
Filia and Floped interact with Blm and promote the recruitment of Blm to replication forks
To understand how the Filia-Floped complex regulates replication fork restart, we performed co-immunoprecipitation in Flag-Filia11 complemented ESCs. Unbiased mass spectrometry analysis identified the Bloom syndrome helicase Blm (Supplementary information, Figure S7A), which plays multiple critical roles in regulating stalled fork restart35,36,37, as a potential binding partner of Filia. These three proteins reciprocally pulled down each other in mESCs (Figure 5A) as well as in NIH3T3 cells co-expressing Flag-Filia and Myc-Floped (Supplementary information, Figure S7B). Moreover, iPOND analysis validated the localization of Blm on replication forks and its increased retention after HU treatment in ESCs and in NIH3T3 cells expressing Filia and Floped (Figure 5B and Supplementary information, Figure S7C). These results suggest that Filia, Floped and Blm form a protein complex at the replication forks of mESCs.
Figure 5.
Filia-Floped interact with Blm and promote its recruitment to replication forks. (A) Physical association of Filia, Floped, Blm and Trim25 in ESCs with or without HU treatment. (B) Blm and Trim25 localized at replication forks in ESCs under the normal (upper panel) and HU treatment (lower panel) conditions. (C) Blm knockdown impaired stalled fork restart in ESCs after HU treatment. (D) Blm regulated stalled fork restart in a protein dosage-dependent manner. (E) Depletion of Filia (Filia-KO) or Floped (Floped-KO) from ESCs decreased the recruitment of Blm and Trim25 to replication forks, double depletion (Double KO) resulted in a more severe phenotype. (F) FiliaS151A was as defective as Filia-Floped double deficiency in recruiting Trim25 and Blm to replication forks, whereas FiliaS151D functioned as effectively as WT Filia. (G) Ectopic expression of WT Filia and/or Floped in NIH3T3 cells stimulated the recruitment of Trim25 and Blm to replication forks. However, expression of FiliaS151A had no effect. (H) Ectopic expression of FiliaS151A in NIH3T3 cells impaired Blm recruitment induced by Floped. Data are represented as mean ± SEM. **P < 0.01, ***P < 0.001.
Blm is a key player in promoting stalled fork restart36,38. We therefore assessed Blm's roles in mESCs. Similar to Filia/Floped deficiency, the efficient knockdown of Blm by two independent shRNAs (Supplementary information Figure S7D) impaired the replication fork restart in mESCs (Figure 5C). Consequently, Blm-deficient ESCs showed an increase in DNA DSBs (Supplementary information, Figure S7E) and hypersensitivity to HU treatment (Supplementary information, Figure S7F). To further assess the relevance of Blm protein level to fork restart ability, we examined stalled fork restart following Doxycycline (Dox)-induced Blm knockdown at various time-points (Supplementary information, Figure S7G). We found that partial reduction of Blm on replication forks significantly impaired restart of stalling forks (Figure 5D). This result suggested that Blm regulates stalled replication fork restart in a dose-dependent manner.
We next examined whether Blm's accumulation on replication forks requires Filia and/or Floped. iPOND analysis revealed that depletion of Filia or Floped led to decreased retention of Blm on replication forks under normal and HU treatment conditions. Double depletion of Filia-Floped further reduced the amount of Blm on replication forks (Figure 5E). In addition, the FiliaS151A mutation resulted in a reduction of Blm on replication forks similar to that observed in Filia-Floped double depleted cells (Figure 5F), although this mutation impaired neither the retention of Filia and Floped on replication forks (Figure 5F) nor their association (Supplementary information, Figure S7H). In contrast, FiliaS151D was as effective as WT Filia in rescuing these defects (Figure 5F). Because the protein expression of Blm was not influenced by deficiency of Filia and/or Floped or the S151A mutation (Supplementary information, Figure S7I), we concluded that the formation of Filia-Floped complex and the phosphorylation of Filia at S151 are essential for the efficient recruitment of Blm to replication forks. Intriguingly, although Filia and Floped interact with each other, their loading onto the replication forks does not rely on their interaction (Figure 5E). This result further supports the notion that Filia and Floped function in parallel to synergistically regulate response to replication stress.
To further validate these findings, we performed a similar iPOND analysis on WT NIH3T3 cells, and NIH3T3 cells expressing Filia, FiliaS151A, Floped, Filia and Floped or FiliaS151A and Floped. The total protein levels of Blm were comparable among these cells (Supplementary information, Figure S7J). However, ectopic expression of Filia or Floped enhanced the recruitment of Blm to replication forks. Co-expressing both proteins further increased the amount of Blm on forks. However, expression of FiliaS151A had no effect on Blm recruitment (Figure 5G). Instead, expression of FiliaS151A impaired the recruitment of Blm induced by Floped (Figure 5H). This result suggests that FiliaS151A functions in a dominant-negative manner to inhibit Floped-dependent Blm recruitment. Collectively, these data demonstrate that the ESC-specific replication fork protein complex Filia-Floped promotes the recruitment of Blm to replication forks in response to replication stress and this function requires the ATR-dependent phosphorylation of S151 on Filia.
Trim25 regulates the recruitment of Blm to replication forks downstream of the Filia-Floped scaffold
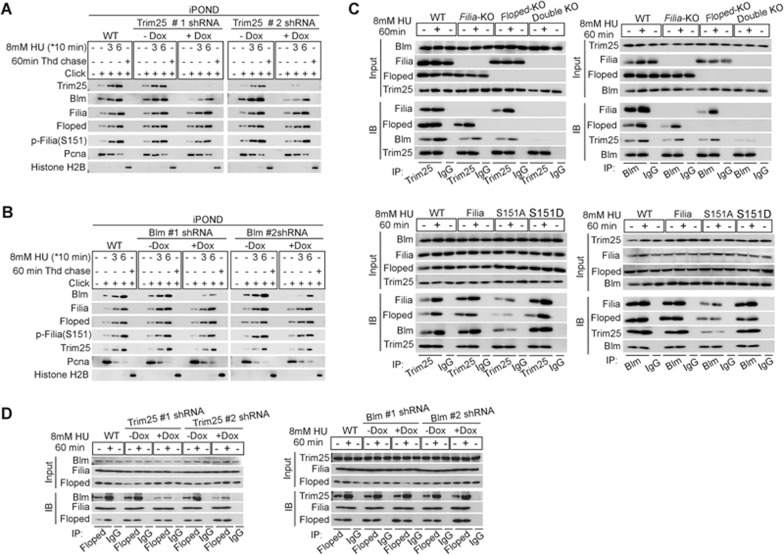
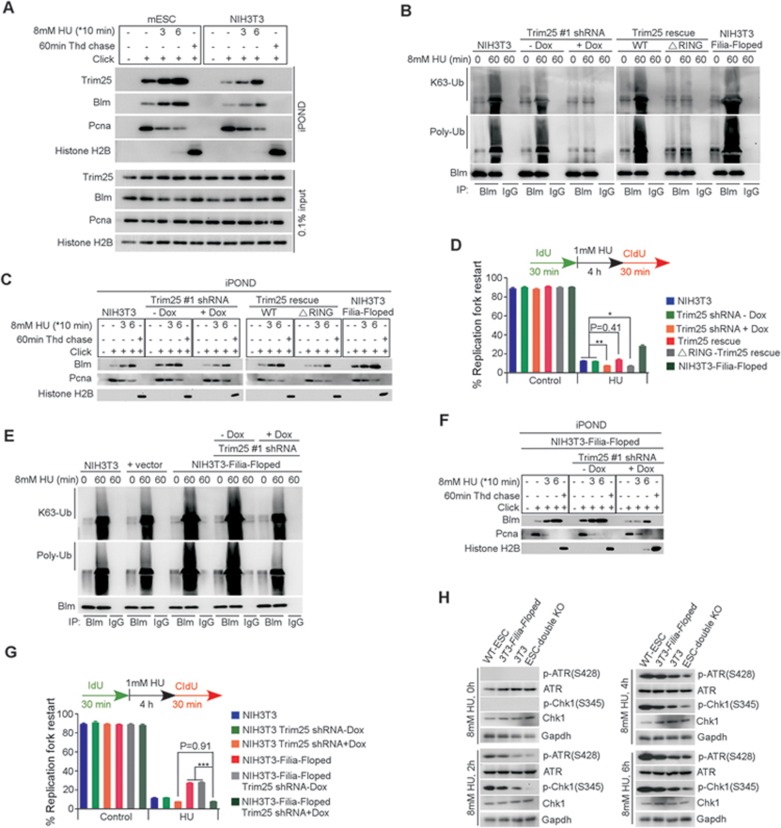
We next determined how the Filia-Floped protein complex promotes the recruitment of Blm to replication forks in response to replication stress. Blm's dynamic translocation and functions are modulated by post translational modifications such as ubiquitination39. Our co-immunoprecipitation/mass spectrometry analysis identified a potential Filia-interacting protein Trim25 (Supplementary information, Figure S7A), an E3 ubiquitin ligase that catalyzes the Lysine (K)63-linked ubiquitination40. The association of the four proteins, namely Filia-Floped-Trim25-Blm, was validated by reciprocal co-immunoprecipitation analysis in mESCs (Figure 5A) as well as in NIH3T3 cells co-expressing Flag-Filia and Myc-Floped (Supplementary information, Figure S7B). Moreover, iPOND analysis showed that Trim25 was localized on replication forks and the retention was similarly stimulated by HU treatment (Figure 5B and Supplementary information, Figure S7C). To examine whether Trim25 regulates the recruitment of Blm to replication forks, we knocked down Trim25 in mESCs with two individual shRNAs (Supplementary information, Figure S8A). We found that the amount of Blm protein on replication forks was significantly reduced (Figure 6A), although the total Blm level was not changed (Supplementary information, Figure S8B). Thus, Filia, Floped and Trim25 are all necessary for the efficient recruitment of Blm to the replication forks. Of note, the efficient recruitment of Trim25 to replication forks relied on the presence of Filia-Floped and the phosphorylation of Filia S151 in mESCs (Figure 5E and 5F) and in NIH3T3 cells expressing Filia and/or Floped (Figure 5G and 5H). However, the recruitment of Filia and Floped to replication forks and the phosphorylation of Filia S151 did not require Trim25 (Figure 6A). Thus, Trim25 functions downstream of Filia-Floped to regulate the recruitment of Blm. Conversely, Blm deficiency had no impact on the relocation of Filia, Floped and Trim25 to replication forks or Filia S151 phosphorylation (Figure 6B). Therefore, we propose a hierarchy in which Filia-Floped acts as a scaffold upon S151 phosphorylation to facilitate the efficient recruitment of E3 ubiquitin ligase Trim25, which then interacts with Blm and regulates its retention on replication forks. Supporting this hierarchy, depletion of Filia and/or Floped or mutation of FiliaS151A in mESCs significantly reduced the physical interaction of Trim25 and Blm (Figure 6C), whereas knockdown of Trim25 had no influence on the interaction between Filia and Floped, but compromised the association of Blm with Filia-Floped (Figure 6D). In addition, Blm depletion did not affect the assembly of the Filia-Floped-Trim25 protein complex (Figure 6D). In line with these observations in ESCs, the ectopic expression of WT Filia and/or Floped in NIH3T3 cells promoted the interaction of Trim25 and Blm (Supplementary information, Figure S8C) without affecting their expression (Supplementary information, Figure S7J).
Figure 6.
Trim25 regulates the recruitment of Blm to replication forks downstream of Filia-Floped. (A) Depletion of Trim25 in ESCs impaired the recruitment of Blm, but not Filia, p-Filia(S151) or Floped, to replication forks. (B) Depletion of Blm in ESCs had no influence on the recruitment of Filia, p-Filia(S151), Floped or Trim25 to replication forks. (C) Depletion of Filia and/or Floped, or FiliaS151A mutation impaired the physical interaction of Trim25 and Blm as well as their association with Floped and/or Filia in ESCs. (D) Depletion of Blm in ESCs did not affect the assembly of Filia-Floped-Trim25 complex (right panel). Likewise, depletion of Trim25 had no effect on the interaction between Filia and Floped, but reduced the association of Blm with Filia-Floped (left panel).
Trim25 ubiquitinates Blm to promote its recruitment to replication forks
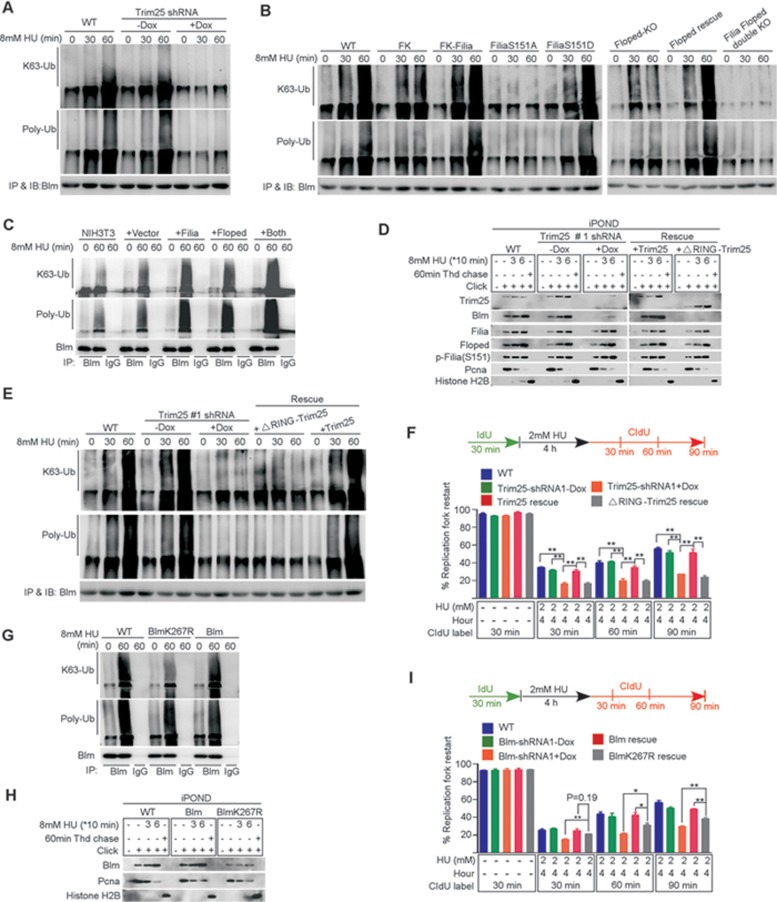
We next determined whether Trim25 ubiquitinates Blm to facilitate its retention on replication forks. Toward this goal, we found that poly-ubiquitination of Blm was robustly stimulated by HU treatment in WT ESCs, but was significantly suppressed in Trim25-knockdown cells (Figure 7A). This suggested that Trim25 was responsible for Blm ubiquitination. Consistently, in other mutant ESCs (Filia−/−, Floped−/−, Filia and Floped double depletion, or FiliaS151A-complemented cells), in which the loading of Trim25 and Blm on replication forks was reduced (Figure 5E and 5F) and their physical association was compromised (Figure 6C), the Blm ubiquitination level was not increased in response to HU treatment. However, the defect was rescued by re-introduction of WT Filia, FiliaS151D, or Floped (Figure 7B). In addition, the Blm ubiquitination level was increased in Filia- and/or Floped-expressing NIH3T3 cells (Figure 7C), in which the recruitment of Trim25 and Blm to replication forks was enhanced (Figure 5G) and their physical association was stimulated (Supplementary information, Figure S8C). These data together support the notion that Trim25 regulates the ubiquitination of Blm. Trim25 catalyzes K63-linked ubiquitination and this type of ubiquitin modification on Blm was validated by immunoblotting using an antibody recognizing K63-linked ubiquitin (Figure 7A-7C). To further confirm the enzyme-substrate relationship between Trim25 and Blm, we deleted the RING domain necessary for RING-finger protein E3 activity (ΔRING-Trim25)41,42,43. WT Trim25 and the ΔRING-Trim25 mutant were separately introduced into Trim25-knockdown ESCs to establish stable cell lines (Supplementary information, Figure S8D). ΔRING-Trim25 mutant protein was able to localize on replication forks (Figure 7D) and associate with Filia-Floped complex (Supplementary information, Figure S8E). However, it failed to restore Blm ubiquitination (Figure 7E) and retention of Blm on replication forks (Figure 7D), without affecting the total Blm level (Supplementary information, Figure S8F). Likewise, the association of ΔRING-Trim25 with its substrate Blm was impaired (Supplementary information, Figure S8E). As a result, ΔRING-Trim25-complemented cells had a reduced capacity for the stalled replication fork restart compared with WT Trim25-rescued cells (Figure 7F). Together, these lines of evidence support the notion that Trim25 ubiquitinates Blm and this modification is required for retention of Blm on replication forks.
Figure 7.
Trim25 ubiquitinates Blm to promote the recruitment of Blm to replication forks. (A) Blm ubiquitination was significantly increased by the HU treatment in WT ESCs. However, knockdown of Trim25 significantly suppressed this modification. (B) In Filia-knockout (FK), Floped-knockout or double knockout ESCs, the ubiquitination level of Blm was lower than that in WT ESCs. Re-expression of WT Filia, Floped or FiliaS151D restored the Blm ubiquitination. However, expression of FiliaS151A into FK cells failed to rescue the defect. (C) Ectopic expression of Filia, Floped or Filia plus Floped in NIH3T3 cells increased the Blm ubiquitination level. (D) ΔRING-Trim25 mutant protein was normally localized on replication forks. However, it could not recruit Blm to replication forks. (E) ΔRING-Trim25 failed to regulate the ubiquitination of Blm. (F) ΔRING-Trim25-expressing ESCs were as defective as Trim25-knockdown ESCs in restarting stalled replication forks. (G) BlmK267R mutation impaired the K63-linked poly-ubiquitination of Blm in response to HU treatment in mESCs. (H) BlmK267R mutation compromised Blm recruitment to replication forks. (I) The ability to restart replication forks in BlmK267R-complemented ESCs was lower than in WT Blm-rescued cells, but higher than in Blm-knockdown ESCs. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01.
The UbPreb program (http://www.ubpred.org/) predicts K267 residue with the highest confidence score (0.99) as a potential ubiquitination site on Blm. A Blm mutant with K267 converted to arginine was expressed in Blm-knockdown ESCs (Supplementary information, Figure S8G). Compared with WT Blm, the BlmK267R mutant showed a significant decrease in K63-linked poly-ubiquitination in response to HU treatment (Figure 7G). Consistently, the recruitment of Blm to replication forks (Figure 7H) and the restart of stalled forks (Figure 7I) were significantly impaired by the BlmK267R mutation compared with WT Blm in the rescue experiments. However, the fork restart rates in BlmK267R-complemented ESCs were higher than those in Blm-knockdown cells (Figure 7I), indicating that additional Trim25 ubiquitination sites may also contribute to Blm recruitment. Together, these data support the notion that K267 ubiquitination by Trim25 plays an important role in Blm recruitment to stalled replication forks.
Filia-Floped scaffold promotes ATR activation in parallel with, but independent of Trim25-Blm recruitment
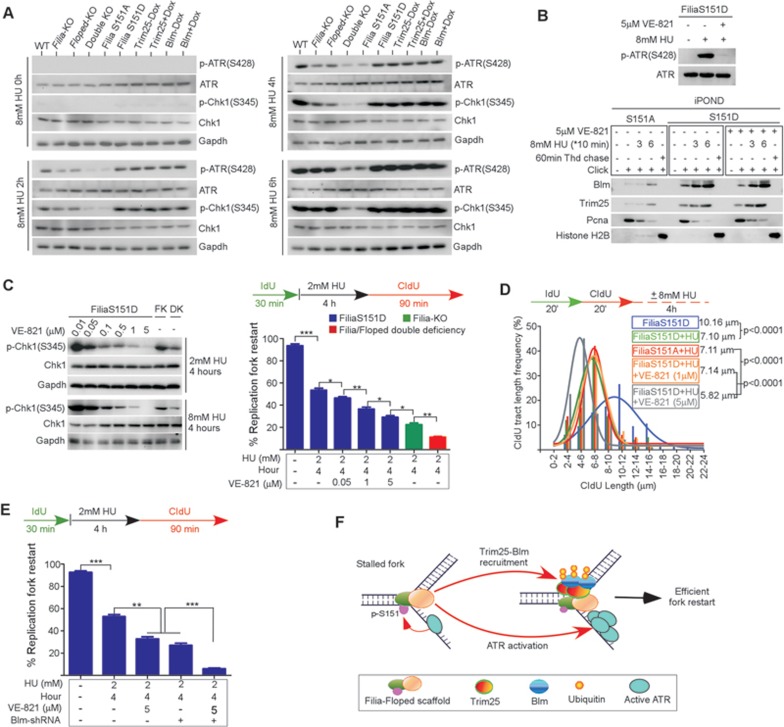
We observed that ATR phosphorylation at Ser428 was compromised by the FiliaS151A mutation in response to HU treatment (Figure 4E). This raised a question regarding whether the Filia-Floped scaffold is essential for optimal ATR activation in the replication stress response. To this end, we conducted a time-course analysis of ATR activation in WT ESCs and in ESCs deficient for Filia, Floped, Filia plus Floped, Trim25 or Blm, by examining the phosphorylation of ATR and its downstream target Chk1 (p-Chk1(S345)). Depletion of individual Filia or Floped protein compromised ATR activation, and the double deficiency or the FiliaS151A mutation similarly caused a further decrease, although total ATR protein levels were comparable (Figure 8A). This defect could be rescued by FiliaS151D (Figure 8A). Intriguingly, ATR activity was not affected by depletion of Trim25 or Blm (Figure 8A). These results suggest that the Filia-Floped scaffold, but not Trim25-Blm, facilitates the activation of ATR kinase in the replication stress response.
Figure 8.
Filia-Floped scaffolds promote ATR activation independent of the Trim25-Blm pathway to regulate stalled fork restart. (A) Time-course analysis of ATR activation in response to HU treatment. Deficiency of Filia or Floped impaired ATR activation, double deficiency or FiliaS151A mutation caused further inhibition. However, depletion of Trim25 or Blm had no influence on ATR activation. (B) Inhibition of ATR activity in FiliaS151D ESCs did not affect the recruitment of Trim25 and Blm to stalled replication forks. (C) ATR activity was suppressed to different extents in FiliaS151D ESCs to determine its influence on stalled fork restart and nascent DNA degradation. ATR signaling regulated stalled fork restart in a dose-dependent manner. Filia-knockout (FK) ESCs and Filia-Floped double deficient (DK) cells were used as controls. (D) However, nearly complete inhibition of ATR signaling was required for nascent DNA degradation. (E) Individual inhibition of ATR activation or Blm protein expression in FiliaS151D-complemented ESCs decreased stalled fork restart ability. Simultaneous inhibition of both pathways caused further suppression of fork restart. (F) Working model of Filia-Floped scaffold in promoting stalled fork restart in ESCs.
The Filia-Floped scaffold promotes ATR activation and Trim25-Blm recruitment. Because ATR kinase orchestrates many downstream events in the replication stress response, we wondered whether ATR signaling regulated the recruitment of Trim25-Blm to replication forks. To this end, we inhibited ATR activity in FiliaS151D-complemented ESCs, in which the Filia-Floped complex forms a functional scaffold. Without the ATR activity but with intact scaffolds, Trim25 and Blm were efficiently recruited to replication forks (Figure 8B). Thus, Trim25-Blm recruitment is not regulated by ATR signaling once the scaffolds have formed. Likewise, ATR activation does not require Trim25-Blm (Figure 8A). These observations together support the notion that the Filia-Floped scaffold independently regulates ATR activation and Trim25-Blm recruitment to stalling forks in response to replication stress.
Next, we investigated whether ATR signaling participates in the regulation of stalled fork restart in ESCs. To this end, we suppressed the ATR activity to different levels in FiliaS151D-complemented ESCs whose Trim25-Blm pathway remained intact. The results showed that ATR signaling stimulated stalled fork restart in a dose-dependent manner (Figure 8C). In contrast, inhibition of ATR activity to a level similar to that in Filia-Floped double deficient ESCs (1 μM VE-821 treatment) had no effect on nascent DNA degradation (Figure 8D). This is consistent with the notion that the Filia-Floped scaffold is not implicated in protecting nascent DNA from degradation. Taken together, these data suggest that the Filia-Floped scaffold amplifies ATR activity to promote stalled fork restart rather than nascent DNA protection.
Although nearly complete inhibition of ATR activity in FiliaS151D-rescued ESCs impaired stalled fork restart, the fork restart rate was still higher than that in ESCs depleted of the scaffold (Figure 8C). This result suggested that the ATR pathway and the Trim25-Blm pathway might have addictive effects on stalled fork restart. To test this hypothesis, we separately or simultaneously inhibited ATR activation and Blm protein expression in FiliaS151D-complemented ESCs and compared their effects. Suppression of individual pathways compromised fork restart ability, and simultaneous inhibition of the both led to a further significant decrease (Figure 8E). Thus, these two pathways have an addictive effect on stalled fork restart. Taken together, we proposed a working model in which the Filia-Floped scaffold promotes stalled fork restart through two independent but converging pathways (Figure 8F).
Trim25-Blm recruitment and ATR activation are less robust in terminally differentiated cells than in ESCs during the replication stress response
Since the Filia-Floped scaffold is ESC specific, we sought to determine the efficiency of differentiated cells to evoke Blm recruitment and ATR activation in the replication stress response. Compared with ESCs, the recruitment of Blm and Trim25 to replication forks were less robust in NIH3T3 cells under normal and HU-treated conditions (Figure 9A), although the total protein expression was comparable in both cell types (Supplementary information, Figure S9A). In NIH3T3 cells, Trim25 interacted with Blm (Supplementary information, Figure S8C), and knockdown of Trim25 via shRNA (Supplementary information, Figure S9B) impaired the K63-linked poly-ubiquitination of Blm (Figure 9B) and Blm retention on replication forks (Figure 9C) after HU treatment. These defects were rescued by WT Trim25 but not ΔRING-Trim25 (Figure 9B and 9C). Consequently, Trim25 deficiency in NIH3T3 cells compromised replication fork restart (Figure 9D) and caused DNA DSBs (Supplementary information, Figure S9C). Consistently, ectopic expression of Filia and Floped in NIH3T3 cells significantly enhanced the recruitment of Trim25 to replication forks (Figure 5G), Trim25-mediated Blm ubiquitination (Figure 9E and Supplementary information, Figure S9D) and retention on replication forks (Figure 9F), as well as stalled fork restart (Figure 9G). Thus, the Trim25-Blm pathway functions similarly in differentiated cells to regulate stalled fork restart, but with a lower efficiency compared to ESCs due to the lack of the Filia-Floped scaffold.
Figure 9.
Trim25-Blm recruitment and ATR activation are less robust in differentiated cells than in ESCs during replication stress response. (A) NIH3T3 cells had lower levels of Blm and Trim25 on replication forks than ESCs. (B) Knockdown of Trim25 in NIH3T3 cells impaired the ubiquitination of Blm. This defect could be rescued by WT Trim25, but not ΔRING-Trim25. (C) Knockdown of Trim25 in NIH3T3 cells impaired the recruitment of Blm to replication forks. This defect was rescued by WT Trim25, but not ΔRING-Trim25. (D) Knockdown of Trim25 in NIH3T3 cells impaired stalled fork restart. This defect was rescued by WT Trim25, but not ΔRING-Trim25. (E) Co-expression of Filia and Floped in NIH3T3 cells stimulated the ubiquitination of Blm upon HU treatment. Knockdown of Trim25 suppressed this modification. (F) In NIH3T3 cells co-expressing Filia and Floped, the recruitment of Blm to replication forks was decreased by depletion of Trim25. (G) Co-expression of Filia and Floped in NIH3T3 cells significantly increased the potential of stalled fork restart in a Trim25-dependent manner. (H) Time-course comparison of ATR activities among ESCs, ESCs depleted of Filia and Floped (ESC-double KO), NIH3T3 cells, and NIH3T3 cells expressing both Filia and Floped (3T3-Filia-Floped). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We then compared the strength of ATR activation in response to HU treatment between NIH3T3 cells and ESCs. Intriguingly, WT ESCs displayed higher ATR activation than NIH3T3 cells. Ectopic expression of Filia and Floped in NIH3T3 cells stimulated the ATR activation, but the level was still lower than that in WT ESCs (Figure 9H). This is in line with the observation that NIH3T3 cells with forced expression of both proteins displayed lower fork restart rates than WT ESCs (Supplementary information, Figure S5E).
Discussion
PSCs show rapid cell division and unique cell cycle composition, which subjects them to unusually high DNA replication stress20. Here, we systematically compared the DNA replication stress response in mouse ESCs and differentiated cells and found that ESCs are better at restarting stalled replication forks to maintain genome stability. Mechanistically, we identified an ESC-specific replication fork protein complex Filia-Floped that is implicated in promoting stalled fork restart. The Filia-Floped complex resides on replication forks under normal condition. Upon replication stress, binding of the complex to replication forks is stimulated, and the S151 residue of Filia is phosphorylated in an ATR-dependent manner. This phosphorylation enables Filia-Floped to become a functional scaffold, which then promotes fork restart through two independent pathways: increasing the abundance of Blm on stalled forks and promoting ATR activation. These findings support the notion that ESCs employ additional regulatory mechanisms to resolve higher replication stress to meet a higher demand of genomic stability than differentiated cells.
Of note, our findings in ESCs and NIH3T3 cells support that Filia and Floped function in parallel, yet synergistically, to provide the most efficient scaffolding when they interact. In ESCs, depletion of both proteins causes more severe defects than depletion of either individual protein. Similarly, ectopic expression of the two proteins in NIH3T3 cells shows more robust effects than the expression of each individual protein. Notably, the optimal scaffolding function of the Filia-Floped complex requires ATR kinase-dependent phosphorylation of Filia S151. S151 phosphorylation may induce a conformational change and generate suitable binding domains. The S151A mutation not only eliminates the scaffold role of Filia, but also interferes with the function of Floped, causing a complete loss of scaffold function. Indeed, FiliaS151A mutant ESCs displayed a similar level of defects to Filia-Floped double deficient ESCs. Thus, the phosphorylated Filia-Floped protein complex constitutes a unique molecular mechanism underlying the superior competence of fork restart in ESCs. However, it is intriguing to note that ESCs depleted of this scaffold still have a higher ability to restart stalled forks than differentiated cells. Likewise, ectopic expression of the scaffolds in NIH3T3 cells does not increase the fork restart ability to an extent similar to that in ESCs. These observations suggest that except for the Filia-Floped scaffold, additional ESC-specific molecules also participate in the regulation of efficient replication fork restart.
Blm is a key player regulating replication fork restart, probably by promoting fork regression (formation of “chicken-foot” intermediate structure)44,45,46. The amount of Blm proteins on stalled replication forks closely reflects fork restart ability. We reveal that Blm recruitment to stalled replication forks is regulated by Trim25-catalyzed ubiquitination in both ESCs and differentiated cells. Replication fork-localized E3 ubiquitin ligase Trim25 physically interacts with its substrate Blm and catalyzes the K63-linked ubiquitination. Depletion of Trim25, disruption of the Trim25-Blm interaction, or deletion of RING domain necessary for the E3 ubiquitin ligase activity of Trim25 all impair Blm ubiquitination. Trim25-catalyzed Blm ubiquitination does not cause protein degradation, but stimulates the retention of Blm on replication forks. Sustained localization of Trim25 to stalled replication forks is critical to allow for the efficient interaction between Trim25 and Blm. In differentiated cells, Trim25 anchoring to stalled forks might be stochastic and less efficient. However, Filia-Floped scaffolds can tether Trim25 to stalled forks and significantly increase its local concentration, thereby promoting Blm retention on stalled forks.
In addition to facilitating Trim25-Blm's regional accumulation on stalled forks, Filia-Floped scaffolds are also necessary for greater ATR activation in the replication stress response. Without functional scaffold in ESCs, the amplitude of ATR activation is significantly decreased. Likewise, ectopic expression of the Filia-Floped scaffold in NIH3T3 cells elevates ATR activity. ATR signaling is necessary to prevent fork collapse and promote fork restart in differentiated cells and ESCs47. In vertebrates, TopBP1 is a direct activator of ATR. The TopBP1-ATR interaction and reaction is enabled by ATRIP and the Rad9-Rad1-Hus1 protein complex (also called the 9-1-1 complex). ATRIP interacts with and recruits ATR to stalling forks through binding to ssDNA-coating RPAs, whereas the 9-1-1 complex binds to DNA ends adjacent to a stretch of RPA-ssDNA and functions as a clamp to bring TopBP1 to ATR47. It is unclear whether the Filia-Floped scaffold could increase the co-localization of ATR-ATRIP and 9-1-1-TopBP1 complexes at the stalled forks, thereby increasing their local concentration and promoting the activation of ATR. Recently, a new ATR activator ETAA1 was identified to function parallel to the TopBP1 pathway48,49,50. It warrants further investigation whether Filia-Floped regulates ATR activation through a novel mechanism independent of these known pathways.
Materials and Methods
Mice, cell lines and culture
Animal care and experimental procedures were conducted in compliance with the guidelines of the Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. The Filiatm/tm mice were kindly provided by Dr Jurrien Dean (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, USA)51. Mice were maintained in specific pathogen-free conditions.
MEFs were prepared and cultured in standard procedures52. Oct4-GFP ES cell line was a gift from Dr Qi Zhou (Institute of Zoology, Chinese Academy of Sciences). C2C12 cell line was obtained from Conservation genetics CAS Kunming Cell Bank. NIH3T3 cell line was kindly provided by Dr Bingyu Mao (Kunming Institute of Zoology, Chinese Academy of Sciences). Filia- and Floped-knockout ESCs were derived from Filiatm/tm and Flopedtm/tm blastocysts, respectively11,29. Maintenance of ESCs was performed as described11. ESC Differentiation was initiated following LIF withdrawal. All cells were tested and showed no mycoplasma contamination.
BrdU proliferation assay
The BrdU proliferation assay was performed as described53. Briefly, cells were incubated with media containing 10 mM BrdU for 40 min. Cells were fixed with 4% formaldehyde in PBS for 15 min and washed in PBS. Before permeabilization, cells were incubated for 1 h in 2 N HCl at room temperature followed by extensive washes in PBS. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and blocked with 1% PBS-BSA for 30 min at room temperature. Cells were then incubated overnight with anti-BrdU antibody (Novus) diluted in 1% PBS BSA, followed by 1-h incubation with secondary antibody conjugated with Alexa Fluor Cy3 at room temperature. The percentages of BrdU-positive cells were analyzed by FACS.
DNA fiber assay
The DNA fiber assay was performed as described25. Specifically, replicating DNA was first labeled with 25 μM 5-iodo-2′-deoxyuridine (IdU; Sigma, I7125) for 30 min, with or without HU (Selleck, S1896; Sigma, H8627) treatment. Cells were then subjected to the second DNA labeling with 250 μM 5-chloro-2′-deoxyuridine (CldU; Sigma, C6891). After labeling, 2.5 μl of the cell suspension (∼2 500 cells) were spotted onto one end of the glass slide, followed by addition of 7.5 μl of lysis buffer (50 mM EDTA, 0.5% SDS, 200 mM Tris-HCl, pH 7.5). After incubation for 8 min at room temperature, the slides were tilted to 15° to allow the DNA fibers to spread along the slide. DNA fibers were treated with 2.5 M hydrochloric acid and incubated with rat anti-BrdU (BU1/75) monoclonal antibody (Novus, NB500-169) that recognizes CIdU, but not IdU at 4 °C overnight, followed by an AlexaFluor cy3-conjugated goat anti-rat secondary antibody for 1.5 h at room temperature. The mouse anti-IdU monoclonal antibody (BD, 347580) that recognizes IdU but not CIdU (4 °C overnight) and AlexaFluor 488-conjugated goat anti-mouse secondary antibody (1.5 h at room temperature) were used to detect IdU. DNA fibers were analyzed on a Leica DM6000B microscope equipped with a CoolSNAP HQ CCD camera (Roper Scientifics). The lengths of CIdU (AF cy3, red) and IdU (AF 488, green) labeled patches were measured using the Image J software, and μm values were converted into kb using the conversion factor 1 μm = 2.59 kb. 200 fibers were analyzed for assessment of nascent DNA degradation and stalled fork restart.
New replication origin firing analysis
New replication origin firing analysis was performed as described21. Briefly, cells were labeled with BrdU for 10 min without HU or with 100 μM HU for 20 min to achieve similar replication fork length. 50 DNA fibers containing a replicon cluster of four BrdU-labeled forks were analyzed for measurement of the mean intra-cluster fork spacing.
iPOND
iPOND was performed as described54,55. Briefly, ESCs were cultured under normal conditions. NIH3T3 cells were arrested in S phase by treating with 2 mM thymidine (Sigma, T1895) for 15 h, followed by release into thymidine-free medium for 2.5 h. Logarithmically growing cells were incubated with 10 mM EdU (Life technologies, A10044) for 10 min. Following EdU labeling, cells were treated with or without HU, or treated with 10 μM thymidine for a chase. Cells were fixed in 1% formaldehyde (Sigma, F1635), followed by quenching with 0.125 M glycine (Sangon Biotech, A100167). Cells were then collected and washed three times in ice-cold PBS, and permeabilized by resuspending in ice-cold 0.25% Triton X-100/PBS at a concentration of 1 × 107 cells/ml followed by incubation at room temperature for 30 min. Before the click reaction, samples were washed once in 0.5% BSA/PBS and once in ice-cold PBS.
For the click reaction, cells were incubated in click reaction buffer for 1 h at room temperature containing 10 μM Biotin-azide (Life Technologies, B10184). The “no-click” reaction (negative control) contained DMSO instead of Biotin-azide. Following the Click reaction, cells were washed once in 0.5% BSA/PBS and once in ice-cold PBS. Cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 1% SDS) containing 1 μg/ml aprotinin (Sigma, A6103) and 1 μg/ml leupeptin (Sigma, L2884) and sonicated using a BioruptorTM UCD-200 for 60 cycles (30-s pulse/30-s pause). Samples were centrifuged at 16 100× g at 4 °C for 10 min and the supernatant was collected. The supernatant was filtered through a 90-micron nylon mesh and diluted 1:1 (v/v) with ice-cold PBS containing 1 μg/ml aprotinin and 1 μg/ml leupeptin. The input samples were collected.
Streptavidin-agarose beads (Millipore, 69203) were washed three times in lysis buffer containing aprotinin and leupeptin. 200 μl bead slurry was used per 1 × 108 cells. The streptavidin-agarose beads were added to the samples, which were then incubated at 4 °C for 16 h in dark. Following binding, the beads were washed with ice-cold lysis buffer, followed by one wash with 1 M NaCl and two washes with ice-cold lysis buffer. To elute proteins bound to nascent DNA, the 2× SDS Laemmli sample buffer (2× SB) mix (0.4 g SDS, 2 ml 100% Glycerol, 1.25 ml 1 M Tris, pH 6.8 and 0.01 g Bromophenol blue in 8 ml H2O) was added to packed beads (1:1; v/v). Samples were incubated at 95 °C for 25 min, followed by mass spectrometry or immunoblotting.
Immunoblotting
Immunoblotting was performed as described11. Images were captured using a ProteinSimple FluorChem system. In each case, experiments were carried out in triplicate and a representative blot is shown unless otherwise stated.
Immunostaining
Immunostaining was performed as described11. For co-localization analysis of Filia/Floped with BrdU-labeled replication foci, cells were labeled with 10 μM BrdU for 5 min, followed by treatment with 2.5 M HCl (Hydrochloric acid) overnight at 4 °C. Images were captured using a Leica TCS SP5 confocal microscope system (Leica Microsystems).
Immunoprecipitation and mass spectrometry assay
Cells were lysed with nuclear protein isolation buffer containing complete EDTA-free (Roche) inhibitors56. Lysates were treated in the presence of 10 units/ml DNase I (New England Biolabs, M0303). For the ubiquitination assay, cells were harvested in SDS lysis buffer (50 mM Tris-HCl, pH 6.8, 1.5% SDS) and boiled for 15 min. Immunoprecipitation was carried out using protein G Dynabeads (Novex, 10009D) according to the manufacturers' protocol. After immunoprecipitation, proteins were fractionated by SDS-PAGE gel followed by immunoblotting or silverQuest staining (Beyotime, P0017L). The protein bands of interest were manually excised and mass spectrometry analysis was performed by MASS core facility in Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
Antibody information
Pan-Filia antibody was generated as described11. Pan-Floped antibody was generated as described28. Serine 151 phospho-Filia antibody was generated by Abmart Inc. Synthetic peptide antigens (corresponding to CAPRPAS(p)VKV) were utilized to immunize rabbits. All other primary and secondary antibodies were obtained commercially (Supplementary information, Table S2).
Neutral comet assay
The Neutral Comet assay was performed as described57. Comets were analyzed by Komet 7 comet assay software (Andor Technology). A total of 100 cells were counted per slide. Each experiment was independently repeated twice.
Karyotyping and telomere fluorescence in situ hybridization
Metaphase chromosome spreads were prepared as described11. Chromosomes were stained with KaryoMAXGiemsa Stain solution (Life Technologies; 10092-013) or DAPI (4′,6-Diamidine-2′-phenylindole dihydrochloride). Peptide-nucleic acid (PNA) telomere probes (Tel G-Alexa 488, PANAGENE, F1010-5) were used to detect telomeres58. Images were captured using the Genus System (Applied Imaging Corp, UK) with a CCCD camera mounted on a Zeiss Axioplan 2 microscope.
Micronuclei assay
WT and Floped-knockout ESCs were cultured under normal conditions. Cells were fixed with 4% (w/v) paraformaldehyde for 30 min on ice and subjected to DAPI staining. A total of 100 mitotic cells were analyzed per sample. Each experiment was independently repeated three times.
Teratoma formation and immunohistochemistry
The injection of ESCs and collection of teratomas was described previously11. Teratomas were fixed and cut at 8-10 μm. After heat-induced antigen retrieval, slides were incubated with GFP primary antibody at 4 °C overnight, followed by incubation with secondary antibody for 1 h at room temperature. DNA was counterstained with DAPI.
shRNA knockdown
shRNA sequences are listed in Supplementary information, Table S3. shRNA knockdown was conducted using pTRIPZ lentiviral tet-on inducible shRNAmir system according to the manufacture's instruction (Open Biosystems). shRNAmir expression vectors were co-transfected with packaging plasmids (psPAX2 and PMD2G) in 293T cells to package viruses. ESCs were cultured with 0.5 μg/ml puromycin on puromycin-resistant feeder layer for 48 h post viral transfection. To verify the knockdown efficiency, cells were treated with 2 μg/ml doxycycline for 48 h before harvest for immunoblotting.
Construction of lentiviral expression vectors
Primers used for cloning cDNA and point mutations are shown in Supplementary information, Tables S4 and S5, respectively. cDNAs fused with Myc or 3× Flag tag at its N-terminus were cloned into pTOMO-IRES-EGFP lentiviral expression vector. Point mutations were performed according to the manufacturer's instructions (QuikChange II Site-Directed Mutagenesis Kit, Agilent Technologies, Cat# 200524). Mutations were confirmed by sequencing. Virus was packaged in 293T cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. Virus-infected ESCs and NIH3T3 cells expressed GFP and were purified by FACS.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Fisher's exact test was performed. All reported P-values are two-sided and were considered statistically significant when < 0.05.
RT-qPCR
Total RNA was extracted using Trizol reagent (TIANGEN, Cat# DP419). Reverse transcription was performed with superscript II (Invitrogen, Cat# 18064014). qPCR was performed with SYBR premix (TaKaRa, Cat# RR8208). The primers are listed in Supplementary information, Table S6.
Author Contributions
BZ and WZ performed most of the experiments. YC analyzed the phenotypes of Floped−/− ESCs. JL and YL prepared some expressing vectors. JG, H Zhu and H Zhou conducted the mass spectrometry analysis. RZ provided critical comments and helped revise the manuscript. PZ supervised the study and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Dr Yingliang Duan and Yongqing Lv for making the FiliaS151A- and FiliaS151D-rescued ESC lines. This work is supported by the National Key Research and Development Program of China (Stem Cell and Translational Research, 2016YFA0100300), the Key Basic Research Project of Yunnan Province (2013FA049 to PZ), the project from Yunnan Province (2015HA038 to PZ), the Basic Research Project of Yunnan Province (2015FB178 to BZ), the Youth Innovation Promotion Association to BZ, and the West Light Foundation of the Chinese Academy of Sciences to BZ.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
The replication fork travelling distances during the HU treatment in different cells.
Antibody information.
Short hairpin RNA sequences used for mRNA knockdown.
Primers used for cDNA cloning.
Primers used for making FiliaS151A, FiliaS151D and BlmK267R mutation.
Primers used for qPCR analysis of Blm and Gapdh.
PSCs are superior at resolving DNA replication stress.
Filia-Floped is a novel ESC-specific replication fork protein complex.
Filia and Floped are not involved in nascent DNA protection or dormant replication fork firing under replication stress.
Loss of Floped leads to genomic instability in mouse ESCs.
Filia and Floped have additive effects on genomic stability and cell sensitivity to HU treatment.
Phosphorylation of serine 151 of Filia is required for the stalled fork restart but not nascent DNA protection.
Filia-Floped associates with Blm and promotes its recruitment to replication forks.
Trim25 promotes Blm recruitment to replication forks downstream of Filia-Floped scaffold.
Blm-Trim25 pathway operates in differentiated cells with relatively lower efficiency than in ESCs.
References
- Nichols J, Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol 2012; 4:a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Lombard P, et al. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell 2015; 35:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boheler KR. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J Cell Physiol 2009; 221:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PH, da Silva CL, Cabral JM. Concise review: genomic instability in human stem cells: current status and future challenges. Stem Cells 2014; 32:2824–2832. [DOI] [PubMed] [Google Scholar]
- Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell 2017; 66:306–319. [DOI] [PubMed] [Google Scholar]
- Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res 2008; 314:1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles SP, Brandt EB, Nelson TJ. Stem cells: the pursuit of genomic stability. Int J Mol Sci 2014; 15:20948–20967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Pillai R, Deng L, et al. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev 2010; 19:1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Guan X, Ryan JA, et al. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell 2013; 13:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru R, Gama V, Fagan BM, et al. Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis. Mol Cell 2012; 46:573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Zhang WD, Duan YL, et al. Filia is an ESC-specific regulator of DNA damage response and safeguards genomic stability. Cell Stem Cell 2015; 16:684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Wilson TE, Glover TW. Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev 2012; 22:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N, Schöler HR. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell 2016; 19:298–309. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res 2007; 35:7545–7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013; 494:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, Jirmanova L, Vitelli L, Samarut J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol 2002; 185:27–33. [DOI] [PubMed] [Google Scholar]
- Banath JP, Banuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 2009; 315:1505–1520. [DOI] [PubMed] [Google Scholar]
- Ahuja AK, Jodkowska K, Teloni F, et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat Commun 2016; 7:10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Han J, Cheng EC, et al. Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Reports 2015; 5:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techer H, Koundrioukoff S, Azar D, et al. Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol 2013; 425:4845–4855. [DOI] [PubMed] [Google Scholar]
- Berti M, Vindigni A. Replication stress: getting back on track. Nat Struct Mol Biol 2016; 23:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz S, Méndez J. DNA replication stress: from molecular mechanisms to human disease. Chromosoma 2017; 126:1–15. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011; 145:435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 2008; 15:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Cun YX, He XC, Zheng P. Maternal-effect Floped gene is essential for the derivation of embryonic stem cells in mice. Dongwuxue Yanjiu 2013; 34:E82–E86. [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair 2007; 6:953–966. [DOI] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011; 7:428–430. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- Syeda AH, Hawkins M, McGlynn P. Recombination and replication. Cold Spring Harb Perspect Biol 2014; 6:a016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 2009; 19:617–629. [DOI] [PubMed] [Google Scholar]
- Wu L. Role of the BLM helicase in replication fork management. DNA Repair 2007; 6:936–944. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 2007; 14:677–679. [DOI] [PubMed] [Google Scholar]
- Manthei KA, Keck JL. The BLM dissolvasome in DNA replication and repair. Cell Mol Life Sci 2013; 70:4067–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm S, Bernstein KA. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair 2014; 22:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007; 446:916–920. [DOI] [PubMed] [Google Scholar]
- Urano T, Saito T, Tsukui T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 2002; 417:871–875. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Nakamura N, Kashiwagi M, et al. RING finger, B-box, and coiled-coil (RBCC) protein expression in branchial epithelial cells of Japanese eel, Anguilla japonica. Eur J Biochem 2002; 269:6152–6161. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. Bioessays 2005; 27:1147–1157. [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 2006; 281:22839–22846. [DOI] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 2009; 37:3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 2013; 5:a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Zhou Q, Chen J, Yuan J. RPA-binding protein ETAA1 is an ATR activator involved in DNA replication stress response. Curr Biol 2016; 26:3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr P, Hoffmann S, Tollenaere MA, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- Bass TE, Luzwick JW, Kavanaugh G, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci USA 2009; 106:7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol 1999; 9:1107–1110. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 2012; 489:304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Gene Dev 2011; 25:1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Piccolo L, Bonaccorso R, Onorati MC. Nuclear and cytoplasmic soluble proteins extraction from a small quantity of Drosophila's whole larvae and tissues. Int J Mol Sci 2015; 16:12360–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain U, Subba Rao K. Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging. Mech Ageing Dev 2011; 132:374–381. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM, Verwoerd NP, van de Rijke FM, et al. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet 1996; 5:685–691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The replication fork travelling distances during the HU treatment in different cells.
Antibody information.
Short hairpin RNA sequences used for mRNA knockdown.
Primers used for cDNA cloning.
Primers used for making FiliaS151A, FiliaS151D and BlmK267R mutation.
Primers used for qPCR analysis of Blm and Gapdh.
PSCs are superior at resolving DNA replication stress.
Filia-Floped is a novel ESC-specific replication fork protein complex.
Filia and Floped are not involved in nascent DNA protection or dormant replication fork firing under replication stress.
Loss of Floped leads to genomic instability in mouse ESCs.
Filia and Floped have additive effects on genomic stability and cell sensitivity to HU treatment.
Phosphorylation of serine 151 of Filia is required for the stalled fork restart but not nascent DNA protection.
Filia-Floped associates with Blm and promotes its recruitment to replication forks.
Trim25 promotes Blm recruitment to replication forks downstream of Filia-Floped scaffold.
Blm-Trim25 pathway operates in differentiated cells with relatively lower efficiency than in ESCs.