Abstract
Background
Although the nucleolus involves two major functions: pre-rRNA processing and ribosome biogenesis/assembly, increasing evidence indicates that it also plays important roles in response to abiotic stress. However, the possible regulatory mechanisms underlying the nucleolar proteins responsive to abiotic stress are largely unknown. High salinity is one of the major abiotic stresses, which hinders plant growth and productivity. Here, genetic screening approach was used to identify a salt hypersensitive mutant 9 (sahy9) mutant, also known as apum23, in Arabidopsis thaliana. Functional characterization of SAHY9/APUM23 through analyses of gene/protein expression profiles and metabolites was performed to decipher the possible regulatory mechanisms of the nucleolar protein SAHY9/APUM23 in response to salt stress.
Results
Seedlings of the sahy9/apum23 mutant displayed postgermination developmental arrest and then became bleached after prolonged culture under various salt stresses. Transcriptomic and proteomic analyses of salt-treated sahy9/apum23 and wild-type seedlings revealed differential expression of genes/proteins that have similar functional categories of biological processes, primarily those involved in cellular and metabolic processes as well as abiotic and biotic stress responses. However, the consistency of differential gene expression at both the transcript and protein levels was low (~ 12%), which suggests the involvement of posttranscriptional processing during the salt response. Furthermore, the altered expression of genes and proteins mediated by SAHY9/APUM23 regarding salt sensitivity involves abscisic acid (ABA) biosynthesis and signaling, abiotic stress responses, and ribosome biogenesis-related genes. Importantly, NCED3, ABI2, PP2CA, and major ABA-responsive marker genes, such as RD20 and RD29B, were down-regulated at both the transcript and protein levels in conjunction with lower contents of ABA and changes in the expression of a subset of LEA proteins in sahy9/apum23 mutants under salt stress. Moreover, the salt hypersensitivity of the sahy9/apum23 mutant was largely rescued by the exogenous application of ABA during salt stress.
Conclusion
Our results revealed that SAHY9/APUM23 regulated the expression of ribosome biogenesis-related genes and proteins, which further affected the ribosome composition and abundance, and potential posttranscriptional regulation. The salt hypersensitivity of sahy9/apum23 is associated with the ABA-mediated signaling pathway and the downstream stress-responsive network of this pathway.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1255-z) contains supplementary material, which is available to authorized users.
Keywords: ABA, APUM23, Arabidopsis thaliana, Nucleolus, Proteome, Salt stress, Transcriptome
Background
Salt stress is a major environmental factor, which limits plant growth, development, and productivity. In plants, high salinity generates both ionic toxicity and osmotic stress, which impairs new shoot growth and enhances the senescence of mature leaves, respectively [1]. Plants are sessile organisms that frequently suffer from deleterious environmental stimuli. To cope with abiotic stress, plants can reprogram their gene expression profile, change their signaling pathways, and adjust their metabolic pathways to better suit these adverse conditions. Plants have evolved sophisticated strategies to tolerate high salinity stress, such as increasing osmotic stress tolerance, excluding Na+ from leaf blades, and tolerating ion accumulation in tissues [2–4]. Numerous gene functions in salt stress responses and tolerance are induced through complex signal transduction pathways. These pathways include the salt overly sensitive (SOS)-mediated pathway [1, 5, 6], abscisic acid (ABA) biosynthesis and signaling [7], secondary signals (such as reactive oxygen species [ROS] and Ca2+) [8–10], the production of osmotic solutes such as proline [11], and small compatible molecules such as Late Embryogenesis Abundant (LEA) proteins [12]. Although great progress has been made in understanding the salt stress response in plants, the detailed regulatory mechanisms largely remain uncharacterized.
Gene expression is regulated at both the transcriptional and posttranscriptional levels, and these levels of regulation are essential for plant growth, development and stress responses [13, 14]. The posttranscriptional regulation of gene expression includes RNA processing, intron splicing, RNA transport and decay, and translation; in general, these processes are collectively referred to as RNA metabolism [14]. RNA-binding proteins (RBPs) are important factors involved in RNA metabolism. Studies of protein structures and functional characterization indicate that diverse organisms exhibit a number of conserved motifs and domains in RBPs, including an RNA-recognition motif (RRM), a zinc finger motif, a K homology (KH) domain, a glycine-rich region, an arginine-rich region, arginine/aspartic acid (RD) repeats, and serine/arginine (SR) repeats [15, 16]. Several nuclear-encoded chloroplast- and mitochondria-targeted RBPs are involved in RNA metabolism in organelles [17–19].
In addition to the aforementioned RBPs, Pumilio proteins are a class of RBP proteins that contain Puf domains, which are conserved across eukaryotes [20]. Puf domains contain multiple tandem repeats, and each repeat is composed of 35–39 amino acids that recognize one RNA base. Pumilio proteins are unique proteins that mostly direct binding to the 3′-untranslated region (UTR) [21]. Pumilio proteins have multifunctional roles, which include involvement in cytoplasmic deadenylation, translational repression, germline stem cell identity maintenance, mitochondria motility and biogenesis, translation initiation, rRNA processing and ribosome biogenesis [20, 22]. There are 25 members of Arabidopsis Pumilio (APUM) proteins identified to date. The expression of these APUM genes is tissue-specific and differential. Subcellular localization indicates that these proteins reside in distinct subcellular compartments, including the chloroplast, cytoplasm, and nucleolus, which indicates organelle-specific functions of APUMs [20].
APUM23, a nucleolar protein involved in pre-rRNA processing and regulation of ribosomal gene expression, is constitutively expressed in Arabidopsis, particularly in metabolically active and cell-division tissues. Mutation of Arabidopsis APUM23 results in slow growth, pointed leaves, and defects in venation patterns and leaf structure [23]. The nucleolus is the prominent substructure of the nucleus and functions in rRNA production and ribosome biogenesis/assembly. Moreover, the nucleolus may have multiple functions that are conserved across organisms. For instance, the nucleolus regulates the cell cycle, stress responses, telomerase activity, and aging [24–26]. Compelling evidence suggests that the nucleolus plays a critical role in sensing and responding to stresses. In mammals, diverse stresses can alter the nucleolar structure and protein composition [25]. However, little is known about the involvement of the nucleolus and nucleolar regulatory mechanisms in response to salt stress in higher plants.
To better understand the components involved in the salt-stress response and the affected pathways, we genetically isolated several salt hypersensitive (sahy) mutants; of which, sahy9 was identified as a new allele of the apum23 mutant. Although APUM23 is involved in pre-rRNA processing and ribosome biogenesis [23], the regulatory mechanisms of this protein in response to salt stress remain elusive. In this study, genome-wide analyses of gene expression profiles at both the transcriptomic and proteomic levels revealed that mutation of SAHY9/APUM23 altered the expression of both gene transcripts and proteins that had similar functional classifications based on Gene Ontology (GO) annotation for biological processes. Nevertheless, the consistency of gene expression at both the transcript and protein levels in sahy9/apum23 mutants is low under salt stress. These results suggest that posttranscriptional regulation is involved in the SAHY9/APUM23-mediated salt response. The sahy9/apum23 mutants contained lower ABA contents than did the wild type, and the salt hypersensitivity of the mutants under salt stress was largely recovered by exogenous ABA application. Thus, the present study provided molecular, protein, and metabolic data indicating that the nucleolar protein SAHY9/APUM23 regulates salt sensitivity through a mechanism involving the ABA signaling pathway and the downstream stress-responsive or tolerance genes of this pathway.
Results
Salt hypersensitive mutant 9 (sahy9) is a new allele of apum23
To identify novel components involved in the salt stress response or signaling, a genetic approach was used to screen transfer-DNA (T-DNA) insertion seed pools [27] on agar plates supplemented with 150 mM NaCl; at this concentration, wild-type seeds can grow, but salt-hypersensitive seeds display postgermination developmental arrest and yield bleached seedlings at subsequent growth stages. Ten salt hypersensitive mutants, referred to as sahys, were isolated. Of which, the sahy9 mutant exhibited striking phenotypes consisting of a small plant size, short roots, and serrated and scrunched leaves (Fig. 1a and Additional file 1: Figure S1b). Amplification of T-DNA-inserted flanking sequences indicated that the sahy9 mutant had a T-DNA insertion site at the 8th exon of APUM23 (Additional file 1: Figure S1a), denoted sahy9/apum23 hereafter. To further confirm whether the sahy9/apum23 mutant phenotype was due to mutation of the APUM23 gene, we requested another allelic mutant line, SALK_052992, also known as apum23–2 [23], from the Arabidopsis Biological Resource Center (ABRC, OH). Both sahy9/apum23 and apum23–2 mutant plants had similar phenotypes consisting of a small plant stature and altered leaves when grown on agar plates or in soil (Fig. 1a and Additional file 1: Figure S1b). Furthermore, an RT-PCR assay revealed no detectable APUM23 transcript in both mutant lines, reflecting that they were knockout mutants (Additional file 1: Figure S1c). These results suggest that the sahy9 mutant phenotypes are due to defects in the APUM23 gene.
Fig. 1.
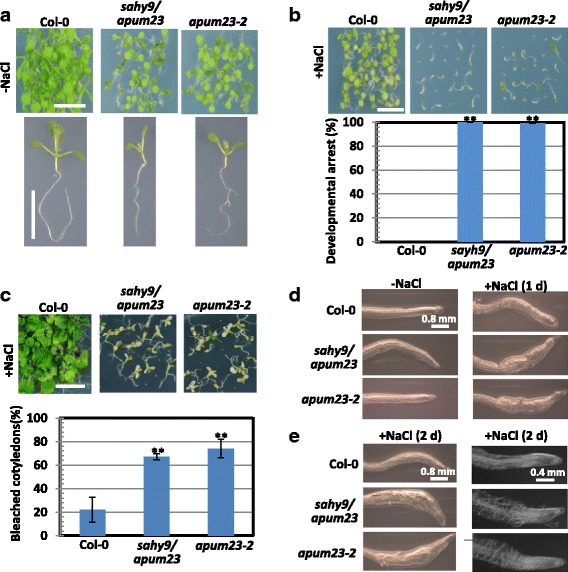
The sahy9/apum23 mutants show salt hypersensitivity. a-b Phenotypic comparison of plants under normal and salt stress conditions. Seedlings were grown for 10 days on basal medium without (a) or with 150 mM NaCl (b). The values indicate the means ± SD of three independent experiments, each with 100–150 seeds. **, P < 0.01, Student’s t-test. Scale = 1 cm. c Salt sensitivity. Plants were grown for 30 days on medium supplemented with 150 mM NaCl. The values indicate the means ± SD of three independent experiments, each with 100–150 seeds. **, P < 0.01, Student’s t-test. Scale = 1 cm. d-e: Comparison of root tips. The seedlings were grown on basal medium for eight or 9 days, and then transferred to medium supplemented with or without 150 mM NaCl for one (d) or two (e) days. The images were taken using a confocal microscope (e, right panel)
Mutation of SAHY9/APUM23 alters sensitivity to various salts
Seedlings of sahy9/apum23 and apum23–2 were smaller and had shorter roots than wild-type seedlings when grown for 10 days on agar plates composed of half-strength MS medium supplemented with 1% sucrose (referred to as basal medium) (Fig. 1a); however, these mutants showed postgermination developmental arrest when they grew directly for 10 days on basal medium supplemented with 150 mM NaCl (Fig. 1b). After culture for 30 days on agar plates containing salt, the majority of the mutant seedlings became bleached (died), but the wild-type seedlings largely exhibited steady growth (Fig. 1c). Transferring eight- or nine-day-old mutant seedlings to medium containing salt for one or 2 days resulted in the root tips of mutant plants to appear swollen (Fig. 1d, e), which is associated with irregular cell shapes in the epidermal and cortex cell layers, as observed using a confocal microscope (Fig. 1e, right panel). To further examine whether the salt hypersensitivity observed in sahy9/apum23 and apum23–2 mutants was specific to NaCl, we grew these mutant seedlings under various salt stress conditions. The results indicated that the sahy9/apum23 and apum23–2 seedlings were also sensitive to KCl (150 mM), NaNO3 (150 mM), and LiCl (15 mM), because the mutants showed postgermination developmental arrest at day 10 (Fig. 2a-c) and more bleached cotyledons at subsequent stages (Fig. 2d-f) compared with wild-type plants. However, developmental arrest and bleached cotyledons were not observed in the sahy9/apum23 mutants grown on 4% or 6% mannitol-containing media (Additional file 2: Figure S2). Nevertheless, a substantial proportion of the mutant seedlings showed a small plant size and no true leaf development, reflecting that osmotic stress may affect the growth and development of the sahy9/apum23 mutants. Collectively, these data suggest that the sahy9/apum23 mutants are sensitive to various salt or ion and osmotic stresses.
Fig. 2.
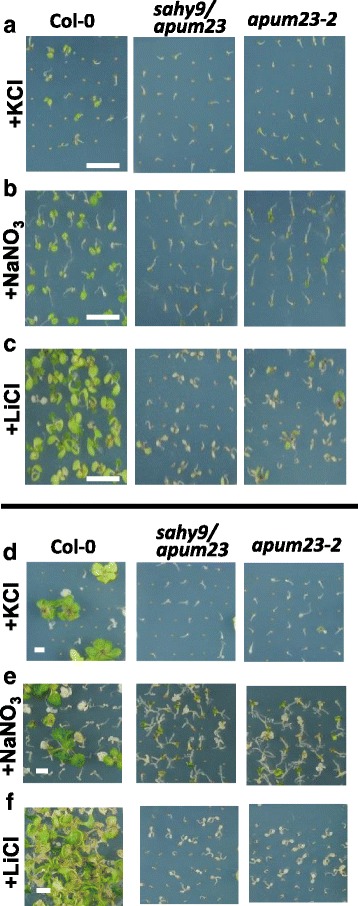
The sahy9/apum23 mutants are sensitive to various salt stresses. a-c Developmental arrest of the sahy9 mutant seedlings. Col-0, sahy9/apum23 and apum23–2 were grown for 10 days on basal medium supplemented with 150 mM KCl (a), 150 mM NaNO3 (b), or 15 mM LiCl (c). d-f Bleached cotyledons of the sahy9/apum23 mutants. Seeds were grown on basal medium supplemented with 150 mM KCl (d), 150 mM NaNO3 (e), and 15 mM LiCl (f) for 30, 30, and 20 days, respectively. Three biological repeats, each with approximately 100 seeds, were performed, and consistent results were obtained. Scale = 1 cm
Transcriptomic analysis reveals differential expression of genes involved in ABA and stress responses in sahy9/apum23 mutants under salt stress
To better understand the SAHY9-mediated gene expression profiles under salt stress conditions, ten-day-old wild-type and sahy9/apum23 seedlings grown on basal medium agar plates were transferred to agar plates containing 150 mM NaCl for 1 day. The salt-treated seedlings were then harvested and subjected to an Agilent Arabidopsis microarray (Agilent Technology, USA) analysis. After normalization to the wild-type levels, approximately 607 genes (319 up- and 288 down-regulated) were differentially expressed, with a signal fold change ≥3. The analysis of biological processes according to The Arabidopsis Information Resource (TAIR) GO annotations (http://www.arabidopsis.org/tools/index.jsp) indicated that these differentially expressed genes were primarily involved in four functional categorizations: cellular and metabolic processes as well as abiotic and biotic stress responses (Fig. 3a). The genes in these four functional groups constituted approximately 63.3% of the total number of differentially expressed genes, and stress-responsive genes constituted approximately one-fourth (25.1% = 13.3% + 11.8%) of the total number of differentially expressed genes. To gain insight into the GO categories of the differentially expressed genes, a GO enrichment analysis was performed using agriGO (http://systemsbiology.cau.edu.cn/agriGOv2/) [28]. Among the up-regulated genes, the sole GO category was lipid localization [GO:0010876 (2.57e-07)] with input (2.26%) vs. background (0.06%). The major GO category of these differentially expressed genes was response to stimulus, which includes several subcategories, such as response to hormone stimulus and abiotic stress (Fig. 3b). Among these differentially expressed genes, at least 57 were involved in ABA and abiotic stress responses (Table 1), particularly in the salt stress response. For instance, the expression of NCED3, a key gene involved in ABA biosynthesis, was decreased 4.4-fold in sahy9/apum23 compared with the wild type. Similarly, the expression of several protein phosphatase genes that participate in the ABA signaling pathway, such as ABI2, PP2CA, HAI2, and HAI1, was also down-regulated in sahy9/apum23. Several stress response marker genes, such as RD29A, COR15A, RD29B, and RD20, were also down-regulated, particularly during salt stress. The expression of 12 genes was verified by quantitative real-time PCR (qRT-PCR) and the results largely showed consistent expression patterns (Fig. 4). Under osmotic stress such as salt stress, plants in deleterious environments can induce the expression of LEA proteins. In this study, LEA genes were over-represented in the GO category of post-embryonic development (Fig. 3b). In sahy9/apum23 mutants, LEA genes, such as ABR (AT3G02480), EM1/LEA1 (AT3G51810), LEA4–5 (AT5G06760), AT2G03850, COR15A, and AT2G18340, were down-regulated in sahy9/apum23 compared with the wild type (Fig. 4 and Table 1). This finding further supports the salt hypersensitivity of sahy9/apum23. Although the proline biosynthesis gene P5CS1 (AT2G39800) was down-regulated in sahy9/apum23 under salt stress, the proline contents were not decreased in the mutants compared with the wild type (Additional file 3: Figure S3a). The net product of proline is likely determined by the balance between biosynthesis and catabolism. In addition, no reduction in the P5CS1 protein was observed in our proteomic analysis (see the isobaric tags for relative and absolute quantitation [iTRAQ] data below). Moreover, the expression of two major ABA-independent transcription factors, DREB2A and DREB2B, showed no significant change in sahy9/apum23 compared with the wild type (Additional file 3: Figure S3b). These data suggest that the mutation of SAHY9/APUM23 altered the ABA signaling pathway and the expression of the downstream stress-responsive genes of this pathway.
Fig. 3.
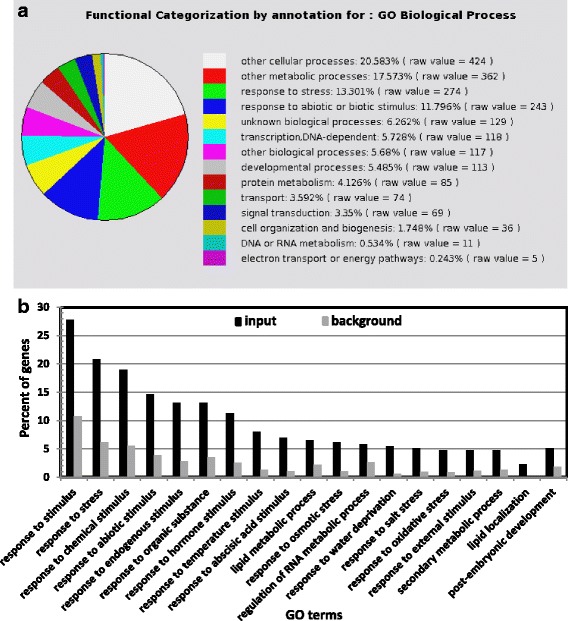
Functional categorization of differentially expressed genes by annotation for GO biological processes. a Genes showing altered expression patterns were identified through microarray analysis, and their functions were classified by an analysis of GO biological processes in TAIR. b GO enrichment analysis of the differentially expressed genes using agriGO, P value < 0.005
Table 1.
Differential expression of genes involved in ABA and abiotic stress responses in sahy9/apum23a under salt stress
| Locus | Gene name | Fold changeb | Biological/molecular function |
|---|---|---|---|
| AT2G18190 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | 43.97 | Response to salt stress |
| AT3G28580 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | 7.85 | Response to ABA |
| AT4G04490 | CRK36, CYSTEINE-RICH RLK 36 | 6.89 | Response to ABA |
| AT2G38340 | DREB19 | 5.32 | ABA-activated signaling pathway |
| AT4G12480 | EARLI1, EARLY ARABIDOPSIS ALUMINUM-INDUCED 1 | 4.99 | Response to salt stress |
| AT2G15390 | FUT4, FUCOSYLTRANSFERASE 4 | 4.15 | Response to salt stress |
| AT1G67760 | TCP-1/cpn60 chaperonin family protein | 4.12 | Response to salt stress |
| AT4G28950 | ROP9, RHO-RELATED PROTEIN FROM PLANTS 9 | 4.01 | ABA-activated signaling pathway |
| AT4G11890 | Encodes a receptor-like cytosolic kinase, ARCK1 | 3.96 | Response to ABA and salt |
| AT1G01680 | PUB54, PLANT U-Box 54 | 3.39 | Response to stress |
| AT5G01550 | LECRKA4.2 | 3.35 | ABA-activated signaling pathway |
| AT1G08910 | PIAL1, PROTEIN INHIBITOR OF ACTIVATED STAT LIKE 1 | 3.32 | Response to stress |
| AT4G23260 | CRK18, CYSTEINE-RICH RLK 18 | 3.28 | Response to ABA |
| AT5G01560 | LECRKA4.3, LECTIN RECEPTOR KINASE A4.3 | 3.18 | ABA-activated signaling pathway |
| AT1G43910 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | 3.10 | Response to ABA |
| AT2G40340 | DREB2C | 3.01 | ABA-activated signaling pathway |
| AT5G17490 | RGL3, RGA-LIKE PROTEIN 3 | −3.02 | Response to ABA |
| AT4G01060 | CPL3, CAPRICE-LIKE MYB3 | −3.05 | Response to ABA and salt stress |
| AT2G39800 | P5CS1 | −3.08 | Response ABA and salt stress |
| AT5G17460 | Unknown protein | −3.13 | Response to salt stress |
| AT2G47180 | GolS1, GALACTINOL SYNTHASE 1 | −3.22 | Response to salt stress |
| AT1G01520 | ASG4, ALTERED SEED GERMINATION 4 | −3.25 | Response to salt stress, |
| AT5G57050 | ABI2 | −3.37 | ABA-activated signaling pathway |
| AT3G63060 | EDL3, EID1-LIKE 3 | −3.39 | Response to ABA and salt stress |
| AT4G27410 | RD26 | −3.50 | Response to ABA |
| AT4G25480 | DREB1A, CBF3 | −3.52 | Response to cold and drought |
| AT5G10230 | ANNAT7, ANNEXIN 7 | −3.55 | Response to salt stress |
| AT4G05100 | MYB74 | −3.58 | Response to salt stress |
| AT3G02480 | ABR, ABA-RESPONSE PROTEIN; an LEA | −3.71 | Response to ABA |
| AT4G21440 | MYB102 | −3.85 | Response to ABA and salt stress |
| AT2G20880 | ERF53, ERF DOMAIN 53 | −3.96 | Response to salt stress |
| AT1G43160 | RAP2.6, RELATED TO AP2 6 | −4.04 | Response to ABA and salt stress |
| AT3G11410 | PP2CA | − 4.14 | ABA signaling pathway |
| AT1G56600 | GOLS2, GALACTINOL SYNTHASE 2 | −4.16 | Response to salt stress |
| AT1G54160 | NF-YA5, NUCLEAR FACTOR Y, SUBUNIT A5 | −4.21 | ABA-activated signaling pathway |
| AT1G69260 | AFP1, ABI FIVE BINDING PROTEIN | −4.27 | ABA-activated signaling pathway |
| AT3G28270 | AFL1, AT14A-LIKE1 | −4.31 | Response to water deprivation |
| AT3G14440 | NCED3 | −4.37 | ABA biosynthetic process |
| AT3G51810 | EM1, LEA 1 | −4.40 | Response to ABA |
| AT4G25000 | AMY1, ALPHA-AMYLASE-LIKE | −4.55 | Response to ABA |
| AT5G06760 | LEA4–5 | −4.57 | Response to osmotic stress |
| AT5G52310 | RD29A, RESPONSIVE TO DESICCATION 29A | −4.59 | Response to ABA and salt stress |
| AT4G23600 | CORI3, CORONATINE INDUCED 1 | −4.63 | Response to ABA and salt stress |
| AT1G07430 | HAI2, HIGHLY ABA-INDUCED PP2C GENE 2 | −4.63 | ABA signaling pathway |
| AT2G03850 | LEA family protein | −4.69 | |
| AT5G59220 | HAI1 | −4.82 | ABA signaling pathway |
| AT3G61890 | HB-12, HOMEOBOX 12 | −5.12 | Response to ABA and salt stress |
| AT2G47770 | TSPO | −5.48 | Response to ABA and salt stress |
| AT5G47450 | TIP2;3 | −5.50 | Response to salt stress |
| AT5G51760 | AHG1, ABA-HYPERSENSITIVE GERMINATION 1 | −5.74 | Response to ABA |
| AT2G42540 | COR15A, an LEA protein | −5.91 | Response to ABA and salt stress |
| AT5G52300 | RD29B, RESPONSIVE TO DESICCATION 29B | −6.83 | Response ABA and salt stress |
| AT2G33380 | RD20 | −7.22 | Response ABA and salt stress |
| AT2G18340 | LEA domain-containing protein | −7.26 | |
| AT2G16005 | ROSY1, INTERACTOR OF SYNAPTOTAGMIN1 | −8.56 | Response to salt stress |
| AT1G29395 | COR413IM1 | −9.36 | Response to ABA and cold |
| AT5G24770 | VSP2, VEGETATIVE STORAGE PROTEIN 2 | −9.81 | Response to salt stress |
aPlants were grown vertically on half-strength MS medium for 10 days and then transferred to fresh medium supplemented with or without 150 mM NaCl for one day. bThe fold change in sahy9/apum23 was normalized against the wild type. The genes in bold font were verified by qRT-PCR and are listed in Fig. 4. The raw data are available at the GEO database under Accession No. GSE99664
Fig. 4.
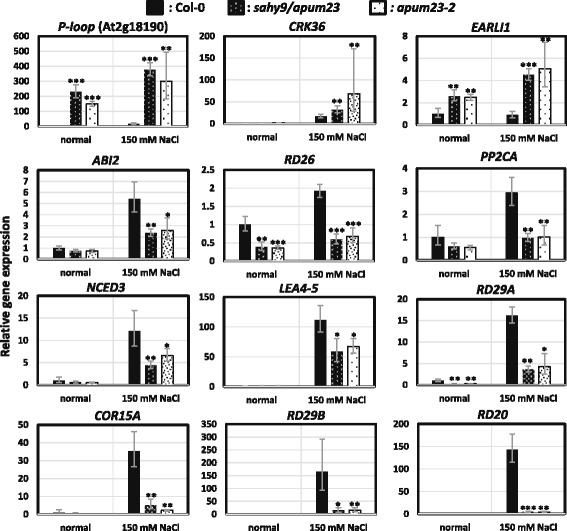
Validation of genes involved in ABA and salt stress responses. The genes used for quantitative real-time PCR (qRT-PCR) were derived from Table 1
Mutation of SAHY9/APUM23 alters the expression of genes involved in ribosome biogenesis and ribosome abundance under high salinity conditions
APUM23 is involved in pre-rRNA processing and regulates the expression of ribosomal protein (RP) and ribosome biogenesis factor (RBF) genes under soil-grown conditions [23]. However, the response of the APUM23-mediated expression of RP and RBF genes to salt stress remains unknown. In this study, microarray datasets indicated that at least 20 genes involved in ribosome biogenesis were differentially expressed in sahy9/apum23 compared with the wild type under salt stress (Table 2), and 17 of these differentially expressed genes were up-regulated. APUMs belong to a gene family composed of 25 members. The level of APUM23 transcript in the sahy9/apum23 mutant was 7.7-fold lower than that of the wild type, confirming a defect of the APUM23 transcript in this mutant. Mutation of APUM23 resulted in induced expression of other homologous members, such as APUM7, APUM8, and APUM12, suggesting possible genetic redundancy in this gene family. Interestingly, the expression of RBL19B was up-regulated in apum23 mutant plants grown in soil [23], whereas its expression was found to be highly suppressed under salt stress conditions in this study. These results suggest that RP or RBF expression patterns can change under different growth conditions. The expression of six up-regulated genes was verified by qRT-PCR and the results were largely consistent (Additional file 4: Figure S4). In addition, the expression of the verified genes was also up-regulated under normal growth conditions and exhibited a certain degree of difference from the salt treatment, as found for APUM8 and NUC-L2. Furthermore, ribosome profile assays revealed that 40S ribosome abundance decreased, but 80S ribosome levels increased in sahy9/apum23 under both normal and stress conditions (Fig. 5). These results support the important role of SAHY9/APUM23 in ribosome biogenesis/assembly.
Table 2.
Differential expression of genes involved in ribosome biogenesis in sahy9/apum23a under salt stress
| Locus | Gene name | Fold changeb | Biological/molecular function |
|---|---|---|---|
| AT2G03130 | Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein | 131.75 | translation |
| AT2G18720 | Translation elongation factor EF1A/initiation factor IF2 gamma family protein | 39.04 | translational elongation |
| AT3G22860 | EIF3C-2, EUKARYOTIC INITIATION FACTOR 3C-2 | 38.10 | translational initiation |
| AT1G22240 | APUM8 | 30.13 | regulation of translation |
| AT1G78160 | APUM7 | 22.36 | regulation of translation |
| AT3G18610 | NUC-L2, NUCLEOLIN LIKE 2 | 19.76 | rRNA processing |
| AT1G02830 | Ribosomal L22e protein family | 14.24 | structural constituent of ribosome |
| AT1G71770 | PAB5, POLY(A)-BINDING PROTEIN 5 | 12.07 | translational initiation |
| AT3G28500 | 60S acidic ribosomal protein family | 11.41 | structural constituent of ribosome |
| AT5G40040 | 60S acidic ribosomal protein family | 8.98 | structural constituent of ribosome |
| AT3G09680 | Ribosomal protein S12/S23 family protein | 8.92 | translation/ structural constituent of ribosome |
| AT5G56510 | APUM12 | 7.35 | regulation of translation |
| AT4G31520 | SDA1 family protein | 4.49 | ribosomal large subunit biogenesis |
| AT2G40010 | Ribosomal protein L10 family protein | 4.33 | structural constituent of ribosome |
| AT5G08600 | U3 ribonucleoprotein (Utp) family protein | 3.59 | rRNA processing |
| AT5G59240 | Ribosomal protein S8e family protein | 3.58 | ribosome biogenesis, translation |
| AT5G39850 | Ribosomal protein S4 | 3.03 | structural constituent of ribosome |
| AT1G72340 | NagB/RpiA/CoA transferase-like superfamily protein | −3.53 | translational initiation (Chloroplast) |
| AT1G72320 | APUM23 | −7.7 | regulation of translation |
| AT3G16780 | RPL19B, RIBSOMAL PROTEIN LIKE 19B | −4001.88 | ribosome biogenesis (60S) |
aPlants were grown vertically on half-strength MS medium for 10 days and then transferred to fresh media supplemented with or without 150 mM NaCl for 1 day. bThe fold change in sahy9/apum23 was normalized against the wild type. The genes in bold font were verified by qRT-PCR and are shown in Additional file 4: Figure S4. The raw data are available at the GEO database under Accession No. GSE99664
Fig. 5.
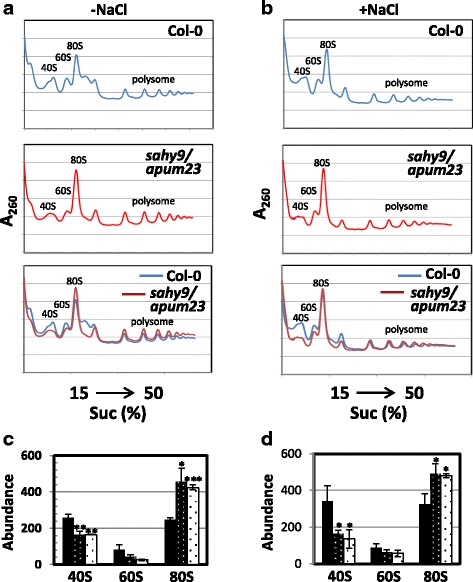
Changes in the ribosome subunit profile in the sahy9/apum23 mutant. a-b Analysis of the ribosome subunit profile using a sucrose gradient. c-d Quantified ribosome abundance derived from (a) and (b), respectively. Plants were grown vertically for 11 days on basal medium with or without 150 mM NaCl for 1 day. The values in (c) and (d) are the means ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t-test
Changes in the protein expression profile of the sahy9/apum23 mutant
Because ribosomes participate in protein translation, alteration in ribosome biogenesis and abundance might affect the function of ribosomes in protein translation. Thus, we expected proteomic changes in the sahy9/apum23 mutant. To examine the protein expression profile of the sahy9/apum23 mutant, an iTRAQ analysis was performed to compare the global protein expression profile of 11-day-old sahy9/apum23 mutants and wild-type plants grown on basal medium with or without salt treatment for 1 day. Proteins that were detected in at least two biological replicates of each genotype were selected for expression. Accordingly, 10,550 and 8830 proteins were expressed in sahy9/apum23 under normal and salt stress conditions, respectively. Proteins with differential expression in sahy9/apum23 was defined by proteins normalized to those of the wild type and having a fold change greater than 1.54 or less than 0.67 (P < 0.05, Z test). Based on these criteria, the sahy9/apum23 mutant had 528 (294 up-regulated and 234 down-regulated) and 534 (298 up-regulated and 236 down-regulated) differentially expressed proteins under normal and salt stress conditions, respectively (Fig. 6a). Of these proteins, approximately 314 (171 up-regulated and 143 down-regulated) proteins were differentially expressed exclusively under salt stress conditions, and approximately 220 overlapping proteins were expressed under both normal and salt stress conditions. This finding suggests that the expression of approximately 58.8% (314/534) of these proteins was specifically altered in the sahy9/apum23 mutant seedlings under salt stress conditions. A comparison of the transcriptome and proteome indicated that 68 genes showed differential expression at both the transcriptional and protein levels in sahy9/apum23 mutants under salt stress (Fig. 6b and Additional file 5: Table S1). These genes are largely involved in ribosome biogenesis, ABA and stress responses, carbohydrate metabolism, and lipid metabolism and transport. This low consistency of gene expression (~ 12%) at both the transcript and protein levels suggests an important role of posttranscriptional regulation in SAHY9/APUM23-mediated gene expression in response to salt stress.
Fig. 6.
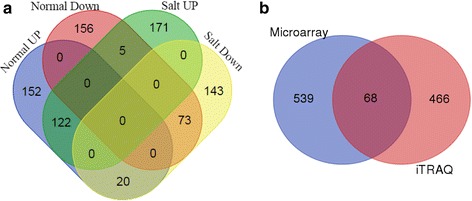
Overlap of identified transcripts and proteins in sahy9/apum23 under normal or salt stress conditions. a Venn diagram representing the overlap of the identified proteins under normal and salt stress conditions. b Venn diagram showing the overlap of the expression of transcripts and proteins under salt stress
The functional classification of these differentially expressed proteins through TAIR GO annotations revealed that these proteins were mostly involved in cellular and metabolic processes and stress response (abiotic and biotic) stimuli, as these proteins constituted 65.5% and 67.0% of all differentially expressed proteins under normal and salt stress conditions, respectively (Fig. 7a, b). To further evaluate the differentially expressed proteins, a GO enrichment analysis was performed. The results indicated that the enriched GO terms belonged to four main GO categories: response to stimulus, cellular component biogenesis, secondary metabolism, and post-embryonic development. These four categories and their prominent corresponding subcategories (or GO terms) are shown in Fig. 7c and d. In general, the enriched GO terms identified for sahy9/apum23 were similar between normal (Fig. 7c) and salt stress (Fig. 7d) conditions, but they differed somewhat in the input percentage.
Fig. 7.
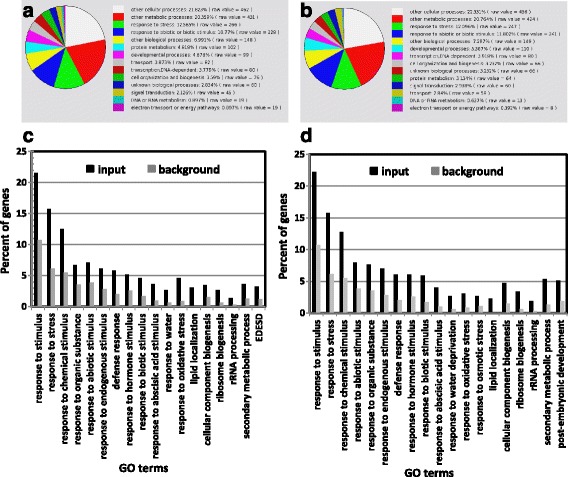
Functional categorization of differentially expressed proteins in sahy9/apum23 compared with the wild type. a-b Functional classification of the differentially expressed proteins in sahy9/apum23 under normal (a) and salt stress (b) conditions through analysis of GO biological processes in TAIR. c-d GO enrichment analysis of differentially expressed genes in sahy9/apum23. The seedlings were grown under normal (c) or salt stress (d) conditions. P value < 0.005
Differential expression of proteins involved in ribosome biogenesis in sahy9/apum23 mutants under normal and salt stress conditions
As mentioned above, one of the four major GO categories was cellular component biogenesis, which included the enriched GO terms “ribosome biogenesis” and “rRNA processing”. Among these differentially expressed proteins, approximately 45 participated in ribosome biogenesis under normal and/or salt stress conditions. Of which, 23 proteins (23/45, 51%) were differentially regulated under both normal and salt stress conditions (Table 3). In contrast, 22 proteins (22/45, 49%) were differentially expressed under either normal or salt stress conditions but not both. This suggests that approximately 50% of differentially expressed proteins involved in ribosome biogenesis can show changes in expression when the sahy9/apum23 mutant seedlings are shifted from normal growth conditions to high saline conditions for 1 day. Interestingly, the comparison of gene expression profiles (Tables 2 and 3) indicated that the expression of only AT5G40040 (60S acidic RP family protein) and AT3G16780 (ribosome protein L19e family protein) was detected at both the transcript and protein levels, reflecting an important role of posttranscriptional regulation in ribosome biogenesis under salt stress conditions.
Table 3.
Differential expression of proteins involved in ribosome biogenesis in sahy9/apum23a under normal and salt stress conditions
| Locus | Protein name | Biological/molecular function | Fold changeb (P value) normal cond. | Fold change (P value) salt stress |
|---|---|---|---|---|
| AT5G40040 | 60S acidic ribosomal protein family | Structural constituent of ribosome | 3.48 (2.59E-09) | 3.13 (1.52E-07) |
| AT4G25630 | FIB2, a fibrillarin | rRNA processing | 2.17 (0.0002) | 3.49 (8.5E-09) |
| AT5G15550 | ATPEIP2, Arabidopsis thaliana PESCADILLO ORTHOLOG | rRNA processing | 1.94 (0.0017) | 2.16 (0.00040) |
| AT5G18180 | H/ACA ribonucleoprotein complex | snoRNA binding | 1.93 (0.0019) | 2.55 (1.74E-05) |
| AT4G12600 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | Ribosome biogenesis | 1.79 (0.006) | 1.83 (0.0056) |
| AT4G15770 | RNA binding protein | Ribosome biogenesis/assembly | 1.79 (0.006) | 2.09 (0.0007) |
| AT1G48920 | NUC-L1, NUCLEOLIN-LIKE 1 | Ribosome biogenesis | 1.73 (0.01) | 2.01 (0.0013) |
| AT1G16280 | RH36, RNA HELICASE 36 | rRNA processing | 1.71 (0.011) | 1.81 (0.0069) |
| AT4G36420 | Ribosomal protein L12 family protein | Structural constituent of ribosome | 1.70 (0.012) | ns |
| AT3G03920 | H/ACA ribonucleoprotein complex | RNA binding, rRNA processing | 1.68 (0.014) | 2.16 (0.00042) |
| AT2G24500 | FZF, a C2H2 zinc finger protein | Ribosomal large subunit biogenesis | 1.68 (0.014) | 2.10 (0.00068) |
| AT5G08180 | Ribosomal L7Ae/L30e/S12e/Gadd45 family protein | RNA binding | 1.66 (0.017) | 1.66 (0.021) |
| AT5G61330 | rRNA processing protein-related | 1.62 (0.023) | 1.96 (0.0021) | |
| AT3G22660 | EBP2, rRNA processing protein-related | Ribosomal large subunit biogenesis | 1.61 (0.024) | 1.91 (0.0030) |
| AT3G55620 | EIF6A, EMBRYO DEFECTIVE 1624 | Ribosomal large subunit biogenesis | 1.60 (0.028) | 2.11 (0.00063) |
| AT5G66540 | U3 small nucleolar ribonucleoprotein | rRNA processing | 1.59 (0.029) | 1.74 (0.012) |
| AT1G63780 | IMP4, small nucleolar ribonucleoprotein protein | rRNA processing | 1.58 (0.032) | 1.62 (0.027) |
| AT5G20600 | rRNA processing-like protein | rRNA processing | 1.56 (0.038) | 1.77 (0.0088) |
| AT3G16810 | APUM24 | RNA binding | 1.53 (0.046) | 1.73 (0.013) |
| AT1G13160 | ARM repeat superfamily protein | Ribosomal large subunit biogenesis | 1.51 (0.051) | 2.63 (8.92E-06) |
| AT1G80750 | Ribosomal L30/L7 family protein | Structural constituent of ribosome | 1.51 (0.053) | 1.93 (0.0026) |
| AT2G37990 | Ribosome biogenesis regulatory (RRS1) family protein | Ribosome biogenesis | ns | 1.84 (0.0051) |
| AT4G25730 | FtsJ-like methyltransferase family protein | rRNA processing | ns | 2.06 (0.00089) |
| AT5G14520 | PES, PESCADILLO | rRNA processing and ribosome biogenesis | ns | 1.84 (0.0051) |
| AT2G20490 | EDA27, EMBRYO SAC DEVELOPMENT ARREST 27 | rRNA processing, ribosome biogenesis | ns | 1.66 (0.02) |
| AT5G62190 | PRH75, DEAD/DEAH box RNA helicase PRH75 | RNA metabolic process | ns | 1.74 (0.012) |
| AT3G19630 | Radical SAM superfamily protein | rRNA processing | ns | 1.89 (0.0037) |
| AT2G44860 | Ribosomal L24e family protein | Ribosome biogenesis | ns | 1.60 (0.033) |
| AT1G50920 | NOG1–1, nucleolar GTP-binding protein | Ribosome biogenesis | ns | 1.65 (0.021) |
| AT1G52930 | ATBRX1–2, ARABIDOPSIS HOMOLOGUE OF YEAST BRX1 2 | rRNA processing, ribosomal large subunit assembly | ns | 1.67 (0.019) |
| AT3G27180 | An SAM-dependent methyltransferase | RNA/rRNA methylation | ns | 1.74 (0.011) |
| AT2G40590 | Ribosomal S26e family protein | Structural constituent of ribosome | ns | 0.57 (0.0091) |
| AT3G15460 | ATBRX1 1, ARABIDOPSIS HOMOLOGUE OF YEAST BRX1 1 | rRNA processing, ribosomal large subunit assembly | ns | 1.86 (0.0043) |
| AT5G10360 | RPS6B, RIBOSOMAL PROTEIN SMALL SUBUNIT 6B | Ribosomal small subunit biogenesis | ns | 0.64 (0.028) |
| AT3G43980 | Ribosomal S14p/S29e family protein | Structural constituent of ribosome | 0.67 (0.046) | ns |
| AT5G04800 | Ribosomal S17 family protein | Structural constituent of ribosome | 0.66 (0.041) | ns |
| AT2G39390 | Ribosomal L29 family protein | Structural constituent of ribosome | 0.66 (0.040) | ns |
| AT3G46040 | RPS15AD, RIBOSOMAL PROTEIN S15A D | Structural constituent of ribosome | 0.66 (0.040) | 0.59 (0.012) |
| AT2G32220 | Ribosomal L27e protein family | Structural constituent of ribosome | 0.64 (0.0278) | ns |
| AT4G39200 | Ribosomal S25 family protein | Structural constituent of ribosome | 0.63 (0.024) | ns |
| AT3G48960 | Ribosomal L13e family protein | Structural constituent of ribosome | 0.63 (0.021) | ns |
| AT5G23740 | RPS11-BETA, RIBOSOMAL PROTEIN S11-BETA | Structural constituent of ribosome | 0.60 (0.012) | ns |
| AT1G61580 | RPL3B, RIBOSOMAL PROTEIN L3 B | Ribosomal large subunit assembly | 0.48 (0.00032) | ns |
| AT3G28900 | Ribosomal L34e superfamily protein | Structural constituent of ribosome | 0.45 (9.76E-05) | 0.60 (0.016) |
| AT3G16780 | Ribosomal L19e family protein | Ribosome biogenesis | 0.310 (1.17E-08) | 0.35 (9.07E-07) |
aPlants were grown vertically on half-strength MS medium for 10 days and then transferred to fresh medium supplemented with or without 150 mM NaCl for one day. bThe fold change in sahy9/apum23 was normalized against the wild type. Cond. conditions, ns no significance
Altered expression of proteins involved in ABA and stress responses in the sahy9/apum23 mutants
The GO enrichment analysis identified response to stimulus as a major enriched GO term, which included the subcategories: response to ABA and abiotic stress stimuli (Fig. 7c, d). At least 45 proteins involved in the ABA and stress responses were differentially expressed in the sahy9/apum23 mutants compared with wild-type plants grown under normal or salt stress conditions (Table 4). Of which, 20 proteins were significantly up- or down-regulated under both normal and salt stress conditions. These proteins have diverse functions, including in vacuolar storage (cruciferins), in lipid transfer and seed oil body biosynthesis (EARLI1 and OBAP1A), in the defense response (PR4), as a transcription factor (WRKY57), in ion or metal transport (TIP1;1; IRT1), in carbohydrate metabolism (BGLU21, BGLU22, and AMY1), and as a kinase (MPK11). However, 25 of the 45 proteins that show response to ABA and/or osmotic stress, particularly salt stress, were exclusively observed in sahy9/apum23 under salt stress conditions. These 25 proteins included the key ABA biosynthesis protein NCED3, protein phosphatases such as ABI1, ABI2, and PP2CA involved in the ABA signaling pathway, and marker genes of ABA signaling, such as KIN2, RD20, and NHL6. Although KIN1 and RD29B were up-regulated in sahy9/apum23 grown under normal conditions, the expression of these proteins was down-regulated in these plants under high salinity conditions compared with the wild type. This finding supports the salt hypersensitivity of the sahy9/apum23 mutant seedlings under salt stress conditions. Furthermore, the ABA contents in sahy9/apum23 and apum23–2 were significantly lower than those in the wild type under salt stress (Fig. 8a); these results are in accordance with the lower expression levels of NCED3 and the downstream ABA-responsive marker proteins. These data support the involvement of the ABA biosynthesis and signaling pathways in the salt hypersensitivity of the sahy9/apum23 mutants.
Table 4.
Differential expression of proteins involved in ABA and abiotic stress responses in sahy9/apum23a under normal and salt stress conditions
| Locus | Protein name | Biological/molecular function | Fold changeb (P-value) Normal cond. | Fold change (P-value) salt stress |
|---|---|---|---|---|
| AT1G03880 | CRU2, CRUCIFERIN 2 | Response to ABA | 2.76 (1.26E-06) | 2.90 (9.79E-07) |
| AT5G44120 | CRU1, CRUCIFERINA | Response to ABA | 2.87 (4.76E-07) | 2.85 (1.41E-06) |
| AT3G15353 | MT3, METALLOTHIONEIN 3 | Response to salt stress | ns | 2.78 (2.47E-06) |
| AT4G28520 | CRU3, CRUCIFERIN 3 | Response to ABA | 2.23 (0.00014) | 2.30 (0.00013) |
| AT3G43700 | ATBPM6, BTB-POZ AND MATH DOMAIN 6 | Response to salt stress | ns | 2.28 (0.00016) |
| AT1G24120 | ARL1, ARG1-LIKE 1 | Response to ABA | ns | 2.09 (0.00071) |
| AT4G12480 | EARLI 1, EARLY ARABIDOPSIS ALUMINUM-INDUCED 1 | Response to ABA and salt stress | 2.10 (0.00042) | 1.98 (0.0017) |
| AT5G14920 | GASA14 | A-STIMULATED IN ARABIDOPSIS 14 | 1.73 (0.010) | 1.96 (0.0021) |
| AT3G23830 | RBGA4, RNA-BINDING GLYCINE-RICH PROTEIN A4 | Response to salt stress | ns | 1.77 (0.0092) |
| AT5G03740 | HD2C, HISTONE DEACETYLASE 3 | Response ABA and salt stress | ns | 1.76 (0.0099) |
| AT2G38310 | ATPYL4, PYR1-LIKE 4 | ABA-activated signaling pathway | ns | 1.72 (0.013) |
| AT1G05510 | OBAP1A, OIL BODY-ASSOCIATED PROTEIN1A | Response to ABA | 1.81 (0.0050) | 1.66 (0.021) |
| AT3G04720 | PR4, PATHOGENESIS-RELATED 4 | Defense and salt response | 1.65 (0.019) | 1.65 (0.023) |
| AT5G47450 | TIP2;3, TONOPLAST INTRINSIC PROTEIN 2;3 | Response to salt stress | ns | 1.63 (0.025) |
| AT4G12470 | AZI1, AZELAIC ACID INDUCED 1 | Response to cold | ns | 1.60 (0.033) |
| AT1G69310 | WRKY57, WRKY DNA-BINDING PROTEIN 57 | Response to salt stress | 1.52 (0.047) | 1.57 (0.040) |
| AT5G52310 | RD29A, RESPONSIVE TO DESICCATION 29A | Response to ABA and salt stress | 1.68 (0.014) | ns |
| AT5G15960 | KIN1 | Response to ABA and stress | 2.04 (0.00069) | 0.66 (0.050) |
| AT2G36830 | TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 | Response to salt stress | 0.65 (0.033) | 0.66 (0.049) |
| AT5G26751 | ATSK11, SHAGGY-RELATED KINASE 11 | Response to salt stress | ns | 0.66 (0.046) |
| AT1G66270 | BGLU21, a beta-glucosidase | Response to salt stress | 0.67 (0.047) | 0.65 (0.041) |
| AT4G14630 | GLP9, GERMIN-LIKE PROTEIN 9 | Response to salt stress | 0.60 (0.013) | 0.64 (0.033) |
| AT1G69260 | AFP1, ABI FIVE BINDING PROTEIN | ABA signaling pathway | ns | 0.63 (0.030) |
| AT1G54100 | ALDH7B4, ALDEHYDE DEHYDROGENASE 7B4 | Response to ABA and salt stress | ns | 0.63 (0.030) |
| AT5G66400 | ATD18, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8 | Response to ABA and stress | ns | 0.63 (0.030) |
| AT5G15970 | KIN2 | Response to ABA and stress | ns | 0.62 (0.026) |
| AT5G02020 | SIS, SALT-INDUCED SERINE RICH | Response to salt stress | ns | 0.62 (0.022) |
| AT1G65690 | NHL6 (NDR1/HIN1-like 6) | Response to ABA and salt stress | ns | 0.62 (0.023) |
| AT2G37770 | AKR4C9, ALDO-KETO REDUCTASE FAMILY 4 MEMBER C9 | Response to salt stress | ns | 0.60 (0.016) |
| AT3G50970 | LTI30, LOW TEMPERATURE-INDUCED 30 | Response to ABA and stress | ns | 0.58 (0.010) |
| AT1G01560 | MPK11, MAP KINASE 11 | Response to ABA | 0.62 (0.020) | 0.57 (0.0090) |
| AT4G26080 | ABI1, ABA INSENSITIVE 1 | Negative regulator of ABA signaling | ns | 0.57 (0.0080) |
| AT3G22231 | PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 | ABA and defense response | 0.34 (1.02E-07) | 0.57 (0.0076) |
| AT5G57050 | ABI2, ABA INSENSITIVE 2 | Negative regulator of ABA signaling | ns | 0.54 (0.0043) |
| AT2G47770 | TSPO, OUTER MEMBRANE TRYPTOPHAN-RICH SENSORY PROTEIN-RELATED | Response to ABA and osmotic stress | ns | 0.54 (0.0039) |
| AT3G22060 | Response to ABA | 0.65 (0.030) | 0.53 (0.0026) | |
| AT5G52300 | RD29B, RESPONSIVE TO DESICCATION 29B | Response to ABA and osmotic stress | 1.75 (0.0084) | 0.51 (0.0018) |
| AT3G11410 | PP2CA, PROTEIN PHOSPHATASE 2CA | Negative regulator of ABA signaling | ns | 0.50 (0.0011) |
| AT1G66280 | BGLU22, a beta-glucosidase | Response to salt stress | 0.60 (0.011) | 0.46 (0.00030) |
| AT2G33380 | RD20, RESPONSIVE TO DESICCATION 20 | Response to ABA and stress | ns | 0.45 (0.00019) |
| AT3G26830 | PAD3, PHYTOALEXIN DEFICIENT 3 | Response to ABA | ns | 0.45 (0.00016) |
| AT1G32350 | AOX1D, ALTERNATIVE OXIDASE 1D | Response to ABA | 0.64 (0.028) | 0.44 (0.00012) |
| AT3G14440 | NCED3, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 | Involved in ABA biosynthesis | ns | 0.41 (3.28E-05) |
| AT4G25000 | AMY1, ALPHA-AMYLASE-LIKE | Response to ABA | 0.44 (7.41E-05) | 0.33 (3.25E-07) |
| AT4G19690 | IRT1, IRON-REGULATED TRANSPORTER 1 | Response to ABA | 0.47 (0.00024) | 0.32 (1.17E-07) |
aPlants were grown vertically on half-strength MS medium for 10 days and then transferred to fresh medium supplemented with or without 150 mM NaCl for one day. bThe fold change in sahy9/apum23 was normalized against the wild type. Cond. conditions, ns no significance
Fig. 8.
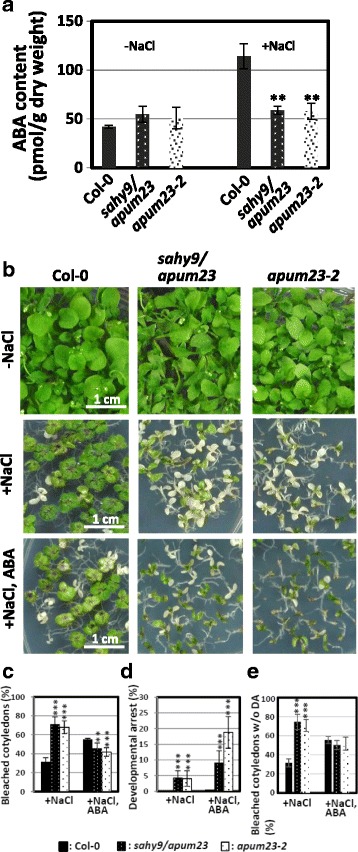
ABA contents and exogenous application of ABA in the sahy9/apum23 mutant and wild type. a ABA contents. Seedlings were grown on basal medium for 10 days, after which they were transferred to basal medium with or without 150 mM NaCl for 1 day. The values indicate the means ± SD of three independent experiments. **, P < 0.01, Student’s t-test. b-d Exogenous application of ABA rescues the salt hypersensitivity of sahy9/apum23 seedlings. Seedlings were grown on basal medium or medium supplemented with 150 mM NaCl and/or 50 nM ABA for 24 days (b). Salt hypersensitivity (c) and developmental arrest (d) were quantified. Salt hypersensitivity (e) was derived from the data shown in (c) by excluding the developmentally arrested (DA) seedlings from the denominator. The values indicate the means ± SD of three biological repeats, each with 100 seeds. **, P < 0.01; ***, P < 0.001, Student’s t-test
Another enriched GO term is embryo development ending in seed dormancy (EDESD; Fig. 7c) or post-embryonic development (Fig. 7d), which included a substantial number of proteins belonging to the LEA protein family. Under osmotic stress, plants can produce small molecules such as LEA proteins to protect larger molecules or cellular compartmental membranes from deleterious environmental effects. In this study, 12 LEA proteins, seven of which belong to the LEA 4 group, exhibited different expression patterns between normal and salt stress conditions (Table 5). Although the majority of these LEA proteins were up-regulated in sahy9/apum23 under normal growth conditions, most of them (eight proteins) were down-regulated under salt stress conditions. These results also support the salt hypersensitivity of the sahy9/apum23 mutant seedlings under salt stress conditions.
Table 5.
Differential expression of LEA proteins in the sahy9/apum23 mutanta compared with the wild type
| Locus | Protein name | Biological/molecular function | Fold changeb (P value) normal conditions | Fold change (P value) salt stress |
|---|---|---|---|---|
| AT1G52690 | LEA7 (LEA_4)c | Embryo development ending in seed dormancy | 2.83 (7.23E-07) | 0.36 (2.04E-06) |
| AT4G21020 | (LEA_4) | Embryo development ending in seed dormancy | 2.68 (2.61E-06) | 2.22 (0.00026) |
| AT5G44310 | (LEA_4) | Embryo development ending in seed dormancy | 2.64 (3.73E-06) | 2.73 (3.92E-06) |
| AT3G17520 | (LEA_4) | Embryo development ending in seed dormancy | 2.60 (5.1E-06) | 0.54 (0.0041) |
| At2G42540 | COR15A, (LEA_4) | Response to ABA and cold | 2.33 (5.66E-05) | 0.60 (0.018) |
| AT5G53820 | LEA | 2.31 (7.05E-05) | ns | |
| AT4G02380 | ATLEA5 (LEA_3), SAG21 | Response to abscisic acid | 1.98 (0.0012) | ns |
| AT5G06760 | LEA4–5 (LEA_1) | Response to osmotic stress | 1.90 (0.0024) | 0.54 (0.0041) |
| AT3G02480 | (LEA_4) | ABR, ABA-RESPONSE PROTEIN | 1.86 (0.0033) | 0.61 (0.020) |
| AT3G15670 | LEA76 (LEA_4) | Embryo development ending in seed dormancy | 1.79 (0.0059) | 0.64 (0.035) |
| AT2G42560 | (LEA_4) | Embryo development ending in seed dormancy | 1.73 (0.010) | 0.33 (1.78E-07) |
| AT2G42530 | COR15B (LEA_4) | Response to ABA and cold | ns | 0.55 (0.0056) |
aPlants were grown vertically on half-strength MS medium for 10 days and then transferred to fresh medium supplemented with or without 150 mM NaCl for 1 day. bThe fold change in sahy9/apum23 was normalized against the wild type. ns no significance. cLEA proteins were classified into groups based on previous description [24]
Exogenous application of ABA largely rescues the salt hypersensitivity of sahy9/apum23 seedlings under salt stress conditions
As mentioned above, the expression of many proteins involved in both ABA and stress responses were changed in sahy9/apum23 under salt stress. To further confirm whether the salt hypersensitivity of sahy9/apum23 is associated with reductions in ABA and its signaling pathway constituents, exogenous ABA was applied to agar plates under salt stress. As shown in Fig. 8b-e, the salt hypersensitive phenotype of sahy9/apum23 was more intense than that of the wild type under salt stress conditions, even though the seedlings were smaller than the wild-type seedlings; however, exogenous application of ABA (50 nM) largely rescued the salt hypersensitivity of the sahy9/apum23 seedlings. Instead, the rate of bleached cotyledons in the sahy9/apum23 seedlings was slightly lower than that in the wild type under 150 mM NaCl + 50 nM ABA conditions (Fig. 8b, c). The lower rate of bleached cotyledons observed in the sahy9/apum23 mutants was likely due to the slight induction of post-germination developmental arrest (~ 13.4%) in these mutants under NaCl + ABA conditions (Fig. 8d). Thus, examination of only the percentage of expanded and bleached cotyledons in the wild type and mutants (i.e., excluding the developmentally arrested seedlings from the denominator), the rates of bleached cotyledons in the wild type and mutants showed no difference (Fig. 8e). The rate of bleached cotyledons was slightly higher in the wild-type seedlings grown in the presence of NaCl + ABA than in those grown in the presence of NaCl alone. It is likely that the exogenous application of ABA to NaCl-containing medium enhances the intensity of the stress and leads to a slight increase in the rate of bleached cotyledons in the wild-type plants. Furthermore, transcriptional analyses indicated that the expression of NCED3 was slightly higher in sahy9/apum23 than in the wild type under NaCl + ABA conditions (Additional file 6: Figure S5). Although transcripts of three PP2Cs (ABI1, ABI2, and PP2CA) and four stress-responsive genes (RD29A, COR15A, RD26, and RD20) were slightly increased in the mutants under NaCl + ABA conditions compared with NaCl conditions, their expression levels were still lower than those in the wild type. Interestingly, the expression of RD29B and three LEA genes (LEA4–5, LEA7, and At3g17520) were higher in the mutants than in the wild type under salt stress, and the induced expression of these genes became more pronounced in the mutants under NaCl + ABA conditions. Furthermore, although the ABA contents in sahy9/apum23 were lower than in the wild type under NaCl conditions, the ABA contents showed no difference between wild type and the mutants under NaCl + ABA conditions (Additional file 7: Figure S6). These data indicate that SAHY9/APUM23-mediated salt sensitivity is associated with the ABA signaling pathway together with its downstream responsive genes and that the small plant size of sahy9/apum23 mutants is likely due to the effects of other pathways.
Discussion
Changes in the expression of ribosome biogenesis-related genes and ribosome abundance in sahy9/apum23 under normal and salt stress conditions
SAHY9/APUM23 is a nucleolus-localized protein that functions in pre-rRNA processing and ribosome biogenesis [23]. In this study, GO enrichment analyses of the transcriptome and proteome datasets revealed that the differentially expressed genes identified in sahy9/apum23 were over-represented in one of the major GO categories: cellular component biogenesis. Within this category, the main sub-categories involved were the regulation of RNA metabolic processes (Fig. 3) and ribosome biogenesis and rRNA processing (Fig. 7). A transcriptomic analysis has indicated that at least 43 genes relative to ribosome biogenesis are differentially expressed in the apum23 mutants grown in the soil for 3 weeks [23]. However, in the present study, at least 20 genes involved in ribosome biogenesis were differentially expressed in the sahy9/apum23 seedlings grown on agar plates supplemented with NaCl. Of these 20 genes, eight (40%, 8/20) overlapped with those previously identified (Table 2 in this study vs. Table 1 of [23]). Moreover, proteomic analyses revealed that at least 45 proteins involved in ribosome biogenesis were differentially expressed in sahy9/apum23 seedlings grown on normal or NaCl-treated agar plates. Of which, approximately 50% of ribosome biogenesis-related genes can be activated or suppressed under normal or salt stress conditions. Changes in these ribosome biogenesis-related genes and/or proteins under distinct environments could alter ribosome biogenesis/assembly and lead to differences in ribosome subunit abundance (Fig. 5). Altered ribosome profiles have also been reported regarding the mutation of DIG6 (drought-inhibited growth of lateral roots), which encodes a large 60S subunit nuclear export GTPase1 involved in ribosome biogenesis [29]. Changes in ribosome biogenesis/assembly and abundance may further affect protein translation. The majority of the RP or RBF mutants exhibit auxin-mediated developmental defects in leaf morphology, venation patterning, and root growth (Fig. 1 and Additional file 1: Figure S1). These mutants are presumably due to changes in ribosome composition and further cause preferential translation or undertranslation of certain auxin-related genes, such as PINs [23, 30–32]. However, such changes in genes involved in auxin transport and perception are largely not detectable through transcriptomic and proteomic analyses (in this study; [29]). It is likely that the expression of these auxin-related genes is too low to be detected under these experimental conditions and that only abundant transcripts or proteins can be detected.
In addition to its two major roles in pre-rRNA and ribosome biogenesis/assembly, the nucleolus has been proposed to function in multiple processes, such as the cell cycle and stress responses [25, 26]. Our data provided evidence that the expression of ABA- or abiotic stress-related genes was up-regulated by salt stress (Fig. 4). However, the induction ratios of these genes were lower in sahy9/apum23 than in the wild type, which suggests that mutation of the nucleolar protein SAHY9/APUM23 affects transcriptional and posttranscriptional regulation both sensitively and widely. The multiple functions of the nucleolus are also reflected by its high heterogeneity, including modification of rRNA, variation in the RP composition and dynamic compositional changes in response to cellular cues or environmental stimuli [26, 33, 34]. In the Arabidopsis genome, each RP and its homologs form a multimember family. Additionally, different tissues, developmental stages, and environmental stimuli may activate the expression of distinct subsets of RPs (in this study and [35]). Thus, the data obtained from the present study may complement the previous report [23].
Genome-wide analyses of gene expression reveal similar functional categories but low consistency of transcript and protein profiles in sahy9/apum23 mutants under salt stress conditions
Transcriptomic analyses indicated that approximately 607 genes were differentially expressed under salt stress and that changes in the expression of approximately 534 proteins were detectable via iTRAQ analysis under the same growth conditions. Although these differentially expressed genes/proteins had similar functional classifications primarily consisting of cellular and metabolic processes as well as biotic and abiotic stress responses, the changes in transcript and protein profiles showed little overlap. Only 68 genes were differentially expressed at both the transcript and protein levels in sahy9/apum23 mutants under salt stress conditions. For instance, 20 genes and 36 proteins involved in ribosome biogenesis/assembly showed differential expression in sahy9/apum23 under salt stress, but only two genes, the 60S acidic RP (AT5G40040) and RPL19e (AT3G16780), were expressed at both the transcript and protein levels (Table 2 vs. Table 3). In addition, 57 transcripts (Table 1) and 45 proteins (Table 4) were differentially expressed and involved in ABA and abiotic stress responses in sahy9/apum23 under salt stress. Among these, only six genes, ABI2, PP2CA, RD20, NCED3, RD29B, and AMY1, were detected at both the transcript and protein levels. This low consistency in the changes in RNA and protein profiles might be due to the following reasons. First, low-abundance proteins, such as membrane-associated proteins, might not be detected under the applied experimental conditions, whereas the transcript levels of the corresponding proteins could show significant changes. Second, high-abundance proteins might exhibit low rates of protein degradation or important housekeeping functions, but their transcript levels may not have met the criterion of a three-fold change expression applied to the transcriptomic analysis. Finally, the differential expression of gene transcripts may not have been efficiently translated to the proteins due to the regulation of posttranscriptional processing. In addition to salt stress, as observed in the present study, a low congruency of transcript and protein profiles has also been detected during ion starvation [36] and light morphogenesis [37]. Thus, posttranscriptional processing or RNA metabolism plays important roles in gene regulation that predominantly affects plant growth and stress responses.
SAHY9/APUM23 regulates salt sensitivity in association with the ABA signaling pathway and ABA-mediated downstream stress-responsive or tolerance genes
Although the APUM23 gene function has been well characterized [23], its response to salt stress, particularly as a nucleolar protein, remains unknown. Arabidopsis nucleolin 1 (NUC1) is also an RBP predominantly localized in the nucleolus and functions in pre-rRNA processing, ribosome biogenesis, and plant normal growth [38, 39]. Mutation of NUC1 (i.e., parl1) results in plants with slowed growth and auxin-mediated developmental defects [31]. Rice NUC1 (OsNUC1) is transcriptionally regulated by salt stress, and the overexpression of OsNUC1 in Arabidopsis or rice leads to salt stress tolerance [40]. Because the expression of several ABA biosynthesis and signaling genes, including NCED3, ABI1, ABF3, RD29A, and KIN1, is down-regulated in OsNUC1 overexpressors under salt stress conditions [41], the relationship between ABA-mediated gene expression and salt-resistant phenotypes remains to be illustrated. Salt stress increases the calcium ion (Ca2+) concentration in the cytosol of plant cells, which further results in activation of calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs). Subsequently, CBL-CIPK mediates the SOS pathway to increase the tolerance of plant cells to salt stress [42]. Although many genes that were identified as differentially expressed in sahy9/apum23 under salt stress in this study were classified as being involved in response to ABA or abiotic stress, the expression of the SOS components (SOS1, SOS2, and SOS3) in sahy9/apum23 did not differ from that of the wild type. In addition, although salt stress induces the production of the osmotic solute proline in plants, the proline contents in sahy9/apum23 also showed levels similar to those found in the wild type. Thus, the SOS pathway and proline content are not likely involved in SAHY9/APUM23-mediated salt sensitivity.
ABA is the major regulator of abiotic stress resistance and coordinates a complex regulatory network to adapt to osmotic stress [43, 44]. The core ABA signaling pathway is composed of three protein classes: PYR/PYL/RCAR receptors, protein phosphatase 2Cs (PP2Cs), and SNF1-related protein kinase 2 s (SnRK2s). PP2Cs are transcriptionally regulated by ABA [45]. In this study, the PP2C proteins ABI1, ABI2, and PP2CA were down-regulated in sahy9/apum23 under salt stress (Table 4). Because PP2Cs function as negative regulators [46, 47], a reduction in these PP2C proteins might presumably activate SnRK2s, further triggering downstream ABA-responsive gene expression. Instead, the expression of several ABA-responsive marker genes, such as KIN1, RD29A, RD29B and RD20, were reduced at the transcript and/or protein level in the mutant under salt stress conditions. A previous study proposed the existence of two ABA signaling pathways: the ABI1-dependent and ABI1-independent pathways [48]. Thus, our data support the notion that the SAHY9/APUM23-mediated salt response most likely occurs through an ABI1-independent pathway. Furthermore, several lines of evidence also support SAHY9/APUM23-mediated salt sensitivity through the ABA signaling pathway, including reduced expression levels of NCED3 and a subset of LEAs, and lower ABA contents in sahy9/apum23 compared with the wild type under salt stress. Moreover, exogenous ABA application largely rescued the salt-hypersensitive phenotype together with induction of NCED3 expression and ABA contents in the sahy9/apum23 mutants under NaCl + ABA conditions.
Because LEA structures are highly hydrophilic and natively unfolded, they may interact with other large molecules to stabilize them against deleterious stress conditions [12, 49]. Most LEAs, including dehydrin RAB18, can be induced by ABA and osmotic stress [50–52]. Furthermore, overexpression of LEAs in transgenic plants of rice, Arabidopsis, or mustard (Brassica juncea) confers tolerance to drought and/or salt stress [53–55]. Consistently, in this study, exogenous ABA application partially rescued the salt-hypersensitive phenotype of the sahy9/apum23 mutants and led to notably induced expression of three LEAs (LEA4–5, LEA7, and At3g17520) in the mutants relative to the wild type under NaCl + ABA conditions. Notably, the expression of LEA4–5 in the mutants was lower than that in the wild type after short-term (one-day) salt treatment. Nevertheless, long-term (24-day) salt treatment increased LEA4–5 expression to a level higher than that in the wild type. A similar expression pattern was also observed for LEA7 and At3g17520 (Table 5 vs. Additional file 6: Figure S5). These data suggest that LEA proteins might play an important role in adaptation to salt stress. Moreover, with the exception of RD29B, which was highly induced, several ABA-mediated stress responsive genes, such as RD29A, RD26, RD20 and COR15A, were only induced slightly under NaCl + ABA conditions. This differentially induced expression of the canonical stress marker genes also supports the involvement of the ABA-mediated ABI-independent signal pathway in the sahy9/apum23 mutants in response to salt stress.
Conclusions
In conclusion, analyses of gene expression profiles and metabolites were performed in the present study to characterize the possible regulatory mechanisms of the nucleolar protein SAHY9/APUM23 in response to salt stress. Gene/protein expression profiles revealed changes in the differential expression of genes or proteins primarily involved in ribosome biogenesis and ABA biosynthesis and signaling as well as changes in the differential expression of both stress-responsive marker genes and a subset of LEA proteins in sahy9/apum23 in response to salt stress. The altered expression of ribosome biogenesis-related genes or proteins in sahy9/apum23 might alter the composition and abundance of ribosomes, further affecting the translation of proteins with distinct functions and causing pleiotropic phenotypes. Among these phenotypes, the salt hypersensitivity of sahy9/apum23 is associated with the ABA-mediated signaling pathway and its downstream stress-responsive network.
Methods
Plant materials and growth conditions
A. thaliana ecotype Columbia (Col-0) plants were used in this study. Seeds were sterilized and subjected to cold pretreatment at 4 °C for 3 days in the dark, after which they were sowed on agar plates or in soil on the first day of germination. The basal medium used in this study was composed of half-strength MS basal salt [56], B5 organic compounds [57], 0.05% MES [2-(N-morpholino)ethanesulfonic acid monohydrate], and 1% sucrose. The medium was adjusted to pH 5.7, followed by the addition of 7 g/L phytoagar (Duchefa Biochemie, Haarlem, the Netherlands) prior to autoclaving. Unless stated otherwise, the seeds were germinated at 22 °C under a 16/8 h day/night photoperiod and a light intensity of approximately 80 μmol m− 2 s− 1. The plant materials used in this study was approved by the Academia Sinica Biosafety Review & Biomaterials and Lab Biosafety Information System.
Genetic isolation of sahy9
For the genetic screening of salt-responsive mutants, T-DNA insertion seed pools [27] consisting of more than 10,000 lines were requested from the ABRC. Approximately 400,000 seeds were grown on basal agar medium supplemented with 150 mM NaCl, a concentration in which wild-type plants can germinate and steadily grow. Seedlings that displayed postgermination developmental arrest at day 10 and subsequent bleaching of the cotyledons after three- or four-week culture were referred to as salt hypersensitive (sahy) mutants. Approximately 10 sahy mutants were isolated through this genetic approach. Of which, sahy9 exhibited a unique phenotype consisting of slow growth, small plant size, and pointed leaves. The sahy9 mutant was further characterized for its function.
Microarray analysis
Col-0 and sahy9/apum23 seedlings were grown on basal medium for 10 days, after which they were transferred to medium supplemented with 150 mM NaCl for 1 day. After salt treatment, the seedlings were harvested for total RNA extraction using the RNeasy Plant Mini Kit (Qiagen, Germany). After RNA labeling and hybridization, the GeneChip (Agilent, Arabidopsis 4 × 4.4 K V4) was scanned in accordance with the Agilent standard protocol. The resulting CEL files were analyzed using GeneSpring GX V11.5 software (Agilent). The data were normalized using MAS V5.0 and filtered based on expression levels, employing a raw signal value of 100 as the cutoff value, at least in the wild type or the sahy9/apum23 mutant. The filtered genes were statistically analyzed using the unpaired t-test (P-value cutoff of 0.05) and multiple testing corrections in accordance with the Benjamini-Hochberg false discovery rate (FDR) [58]. A three-fold signal change in gene expression was defined as differential expression. Two biological experiments were performed in this study. The raw data are available in the Gene Expression Omnibus (GEO) database under Accession No. GSE99664.
iTRAQ analysis
Seedlings harvested after 11 days of growth on agar plates with or without 150 mM NaCl treatment for 1 day were used for total protein extraction, which was followed by iTRAQ analysis. Protein sample preparation and iTRAQ analysis were performed as previously described [36]. The final proteomic data were derived from three biological experiments, each with three technical replicates.
Quantitative RT-PCR
Total RNA was extracted from 11-day-old seedlings grown on agar plates with or without 150 mM NaCl treatment for 1 day, using the RNeasy Plant Mini Kit (Qiagen). Subsequently, 2 μg of DNase I (Qiagen)-treated total RNA was reverse-transcribed with 2 μg of an oligo dT primer using Superscript III Reverse Transcriptase (Invitrogen). qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and an Applied Biosystems 7500 Real-Time PCR System. Primer Express v2.0 software (Applied Biosystems) was used to design the primers (Additional file 8: Table S2). The relative transcript levels of the genes were determined via the comparative threshold cycle (CT) method using PP2A (At1g13320) as the internal control. All experiments were performed in three biological replicates, each with three technical repeats.
Ribosome profile analyses
Ribosome profile analyses were performed as previously described [37]. In general, 0.3-g samples of 11-day-old seedlings grown on agar plates with or without 150 mM NaCl treatment for 1 day were used for ribosome or polysome extraction using a buffer composed of 200 mM Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) (pH 8.5), 50 mM KCl, 25 mM MgCl2, 100 μg/mL heparin, 50 μg/mL cycloheximide, 400 U/mL RNasin (Promega, Madison, WI, USA), 2% polyoxyethylene 10-tridecyl ether, and 1% deoxycholic acid. The mixture was incubated on ice for 5 min, followed by centrifugation at 15000 g for 5 min at 4 °C. Thereafter, the supernatant was collected and the pellet discarded. A total of 300 μL of the supernatant was loaded onto a 10-mL continuous sucrose gradient (15–50%) prepared with a gradient maker (ISCO, Lincoln, NE), after which the mixture was centrifuged at 35000 g for 3.5 h at 4 °C. The distribution of nucleic acids was detected based on the 254-nm UV absorbance profile (Brandel BR-188, Gaithersburg, MD, USA).
ABA and proline assays
For the ABA assays, eleven-day-old seedlings grown on agar plates with or without 150 mM NaCl treatment for 1 day were harvested and subsequently subjected to ABA extraction as described previously [59]. ABA was quantified using enzyme-linked immunosorbent assay (ELISA) (Phytodetek ABA kit; Agdia) in accordance with the manufacturer’s recommended protocol. The protocol for the proline assays followed the description provided in previous reports [60].
Additional files
Figure S1. SAHY9/APUM23 gene structure showing the T-DNA insertion sites. a: Exon-intron structure of SAHY9/APUM23 and T-DNA insertion sites in the mutant lines. b: Phenotypic comparison between the wild-type and mutant lines. Plants were grown in soil for 35 days. c: RT-PCR analysis of the APUM23 transcript in the wild-type and mutant plants. (PPTX 531 kb)
Figure S2. Effect of osmotic stress on sahy9/apum23 mutant plants. a-b: Seedlings were grown on basal medium supplemented with 4% (a) or 6% (b) mannitol for 10 or 30 days, respectively. (PPTX 482 kb)
Figure S3. qRT-PCR results of stress-responsive genes and proline contents. a: qRT-PCR of the P5CS1 gene and proline contents. b: qRT-PCR of stress-responsive genes. Plants were grown on basal medium for 10 days, followed by treatment with 150 mM NaCl for 1 day. *, P < 0.05; **, P < 0.01, Student’s t-test. (PPTX 2453 kb)
Figure S4. Validation of the expression of genes involved in ribosome biogenesis in sahy9/apum23 under salt stress conditions. The verified genes are listed in Table 2. (PPTX 12701 kb)
Table S1. Overlap of differential gene expression at both the transcript and protein levels in the sahy9/apum23 mutants under salt stress conditions. (DOCX 26 kb)
Figure S5. Expression of genes regulated by ABA and salt stress. Plants were grown on basal medium supplemented with 150 mM NaCl or 150 mM NaCl + 50 nM ABA for 24 days. qRT-PCR was performed for the detection of relative gene expression levels. (PPTX 29522 kb)
Figure S6. ABA contents. Seedlings were grown on basal medium supplemented with NaCl or NaCl + ABA for 24 days. The values indicate the means ± SD of four independent experiments. ***, P < 0.001, Student’s t-test. (PPTX 88 kb)
Table S2. Primer sequences used in this study. (DOCX 16 kb)
Acknowledgments
We thank the Arabidopsis Biological Research Center (ABRC, OH) for providing the T-DNA insertion mutant seeds. We are also grateful to the Core Labs of DNA Microarray and Proteomics at the Institute of Plant and Microbial Biology (IPMB, Academia Sinica, Taipei) for their services regarding the Agilent GeneChip and iTRAQ, respectively.
Funding
This work was supported by the Ministry of Science and Technology (MOST), Taipei, Taiwan (Grant No. MOST 103–2321-B-001-014 to W.-H. Cheng). The funding body did not play a role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript, but it just provided the financial support.
Availability of data and materials
The data sets generated or analyzed during this study are included in this published article and its additional files. The raw microarray data reported in this paper has been submitted to the GEO database under Accession No. GSE99664.
Abbreviations
- ABA
Abscisic acid
- ABI
Abscisic acid insensitive
- ABR
ABA-RESPONSE PROTEIN
- APUM
Arabidopsis Pumilio protein
- CBL
Calcineurin B-like protein
- CIPK
CBL-interacting protein kinase
- ELISA
Enzyme-linked immunosorbent assay
- GO
Gene Ontology
- iTRAQ
isobaric tags for relative and absolute quantitation
- LEA
Late Embryogenesis Abundant Protein
- NCED3
Nine-cis-epoxycarotenoid dioxygenase 3
- NUC
Nucleolin
- P5CS1
DELTA1-PYRROLINE-5-CARBOXYLATE SYNTHASE 1
- PP2CA
Protein phosphatase 2CA
- PYR/PYL/RCAR
Pyrabactin resistance/PYR1-like/regulatory components of the ABA receptor
- qRT-PCR
quantitative real-time polymerase chain reaction
- RBF
Ribosome biogenesis factor
- RBP
RNA-binding protein
- ROS
Reactive oxygen species
- RP
Ribosomal protein
- RRM
RNA-recognition motif
- SAHY
SALT HYPERSENSITIVE MUTANT
- SnRK
SNF1-related protein kinase
- SOS
Salt overly sensitive
- TAIR
The Arabidopsis Information Resource
- UTR
Untranslated region
Authors’ contributions
WHC conceived the study, supervised the experiments and wrote the manuscript. KCH and WCL performed the experiments. All of the authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1255-z) contains supplementary material, which is available to authorized users.
Contributor Information
Kai-Chau Huang, Email: d99b42009@ntu.edu.tw.
Wei-Chih Lin, Email: d01b42006@ntu.edu.tw.
Wan-Hsing Cheng, Email: whcheng@gate.sinica.edu.tw.
References
- 1.Munns R, Tester M. Mechanisms of salinity tolerance. Ann Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 2.Bartels D, Sunkar R. Drought and salt tolerance in plants. CRC Crit Rev Plant Sci. 2005;24:23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- 3.Munns R. Genes and salt tolerance bringing them together. New Phytol. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhu JK. Salt and drought signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Fu X, Koo YD, Zhu JK, Jenny FE, Jr, Adams MWW, et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cell Biol. 2007;27:5214–5224. doi: 10.1128/MCB.01989-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkelstein RR, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 8.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 9.Pardo JM. Biotechnology of water and salinity stress tolerance. Curr Opin Biotech. 2010;21:339–346. doi: 10.1016/j.copbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Seckin B, Turkan I, Sekmen AH, Ozfidan C. The role of antioxidant defense systems at differential salt tolerance of Hordeum marinum Huds. (sea barleygrass) and Hordeum vulgare L. (cultivated barley) Environ Exp Bot. 2010;69:76–85. doi: 10.1016/j.envexpbot.2010.02.013. [DOI] [Google Scholar]
- 11.Verslues PE, Sharma S. Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book. 2010;8:e0140. doi: 10.1199/tab.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 13.Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B. Post-transcriptional regulation of gene expression in plants during abiotic stress. Int J Mol Sci. 2009;10:3168–3185. doi: 10.3390/ijms10073168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- 15.Alba MM, Pages M. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 1998;3:15–21. doi: 10.1016/S1360-1385(97)01151-5. [DOI] [Google Scholar]
- 16.Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delvillani F, Papiani G, Dehò G, Briani F. S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res. 2011;39:7702–7715. doi: 10.1093/nar/gkr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammani K, Giege P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014;19:380–389. doi: 10.1016/j.tplants.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 20.Abbasi N, Park YI, Choi SB. Pumilio Puf domain RNA-binding proteins in Arabidopsis. Plant Signal Behav. 2011;6:364–368. doi: 10.4161/psb.6.3.14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spassov DS, Jurecic R. The PUF family of RNA binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 22.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 23.Abbasi N, Kim HB, Park NI, Kim YK, Park YI, Choi SB. APUM23, a nucleolar PUF domain RNA-binding protein, is required for pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64:960–976. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- 24.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 25.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw P, Brown J. Nucleoli: composition, function, and dynamics. Plant Physiol. 2012;158:44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 28.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic acid Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Lu S, Li R, Chen T, Zhang H, Cui P, et al. The Arabidopsis gene DIG6 encodes a large 60S subunit nuclear export GTPase 1 that is involved in ribosome biogenesis and affects multiple auxin-regulated development processes. J Exp Bot. 2015;66:6863–6875. doi: 10.1093/jxb/erv391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008;147:128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrica JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144:173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, et al. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010;22:143–158. doi: 10.1105/tpc.109.068320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, et al. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001;127:398–415. doi: 10.1104/pp.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll AJ. The Arabidopsis cytosolic ribosomal proteome: from form to function. Front Plant Sci. 2013;4:1–14. doi: 10.3389/fpls.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weis BL, Kovacevic J, Missbach S, Schleiff E. Plant-specific features of ribosome biogenesis. Trends Plant Sci. 2015;20:729–740. doi: 10.1016/j.tplants.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W. iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol. 2011;155:821–834. doi: 10.1104/pp.110.169508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu MJ, Wu SH, Chen HM, Wu SH. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Sys Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pontvianne F, Matía I, Douet J, Tourmente S, Medina FJ, Echeverria M, et al. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong CC, Reichler S, Blumenthal S, Balk J, Hsieh HL, Roux SJ. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 1997;114:643–652. doi: 10.1104/pp.114.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sripinyowanich S, Chamnanmanoontham N, Udomchalothorn T, Maneeprasopsuk S, Santawee P, Buaboocha T, et al. Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Sci. 2013;213:67–78. doi: 10.1016/j.plantsci.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Udomchalothorn T, Plaimas K, Sripinyowanich S, Boonchai C, Kojonna T, Chutimanukul P, et al. OsNucleolin1-L expression in Arabidopsis enhances photosynthesis via transcriptome modification under salt stress conditions. Plant Cell Physiol. 2017;58:717–734. doi: 10.1093/pcp/pcx024. [DOI] [PubMed] [Google Scholar]
- 42.Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 43.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 44.Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol. 2015;56:1043–1052. doi: 10.1093/pcp/pcv060. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 47.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci. 2002;115:4891–4900. doi: 10.1242/jcs.00175. [DOI] [PubMed] [Google Scholar]
- 49.Candat A, Paszkiewicz G, Neveu M, Gautier R, Logan DC, Avelange-Macherel M, et al. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell. 2014;26:3148–3166. doi: 10.1105/tpc.114.127316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalal M, Trayal D, Chinnusamy V, Bansal KC. Abiotic stress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotech. 2009;139:137–145. doi: 10.1016/j.jbiotec.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 51.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118–140. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welin BV, Olson Å, Nylander M, Palva ET. Characterization and differential expression of dhn/lea/rab-like genes during cold acclimation and drought stress in Arabidopsis thaliana. Plant Mol Biol. 1994;26:131–144. doi: 10.1007/BF00039526. [DOI] [PubMed] [Google Scholar]
- 53.Chen YS, Lo SF, Sun PK, Lu CA, Ho TH, Yu SM. A late embryogenesis abundant protein HAV1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotech J. 2015;13:105–116. doi: 10.1111/pbi.12241. [DOI] [PubMed] [Google Scholar]
- 54.Saha B, Mishra S, Awasthi JP, Sahoo L, Panda SK. Enhanced drought and salinity tolerance in transgenic mustard [Brassica juncea (L.) Czern & Coss.] overexpressing Arabidopsis group 4 late embryogenesis abundant gene (AtLEA4-1) Environ Exp Bot. 2016;128:99–111. doi: 10.1016/j.envexpbot.2016.04.010. [DOI] [Google Scholar]
- 55.Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 57.Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Statist Soc B. 1995;57:289–300. [Google Scholar]
- 59.Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007;143:745–758. doi: 10.1104/pp.106.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. SAHY9/APUM23 gene structure showing the T-DNA insertion sites. a: Exon-intron structure of SAHY9/APUM23 and T-DNA insertion sites in the mutant lines. b: Phenotypic comparison between the wild-type and mutant lines. Plants were grown in soil for 35 days. c: RT-PCR analysis of the APUM23 transcript in the wild-type and mutant plants. (PPTX 531 kb)
Figure S2. Effect of osmotic stress on sahy9/apum23 mutant plants. a-b: Seedlings were grown on basal medium supplemented with 4% (a) or 6% (b) mannitol for 10 or 30 days, respectively. (PPTX 482 kb)
Figure S3. qRT-PCR results of stress-responsive genes and proline contents. a: qRT-PCR of the P5CS1 gene and proline contents. b: qRT-PCR of stress-responsive genes. Plants were grown on basal medium for 10 days, followed by treatment with 150 mM NaCl for 1 day. *, P < 0.05; **, P < 0.01, Student’s t-test. (PPTX 2453 kb)
Figure S4. Validation of the expression of genes involved in ribosome biogenesis in sahy9/apum23 under salt stress conditions. The verified genes are listed in Table 2. (PPTX 12701 kb)
Table S1. Overlap of differential gene expression at both the transcript and protein levels in the sahy9/apum23 mutants under salt stress conditions. (DOCX 26 kb)
Figure S5. Expression of genes regulated by ABA and salt stress. Plants were grown on basal medium supplemented with 150 mM NaCl or 150 mM NaCl + 50 nM ABA for 24 days. qRT-PCR was performed for the detection of relative gene expression levels. (PPTX 29522 kb)
Figure S6. ABA contents. Seedlings were grown on basal medium supplemented with NaCl or NaCl + ABA for 24 days. The values indicate the means ± SD of four independent experiments. ***, P < 0.001, Student’s t-test. (PPTX 88 kb)
Table S2. Primer sequences used in this study. (DOCX 16 kb)
Data Availability Statement
The data sets generated or analyzed during this study are included in this published article and its additional files. The raw microarray data reported in this paper has been submitted to the GEO database under Accession No. GSE99664.


