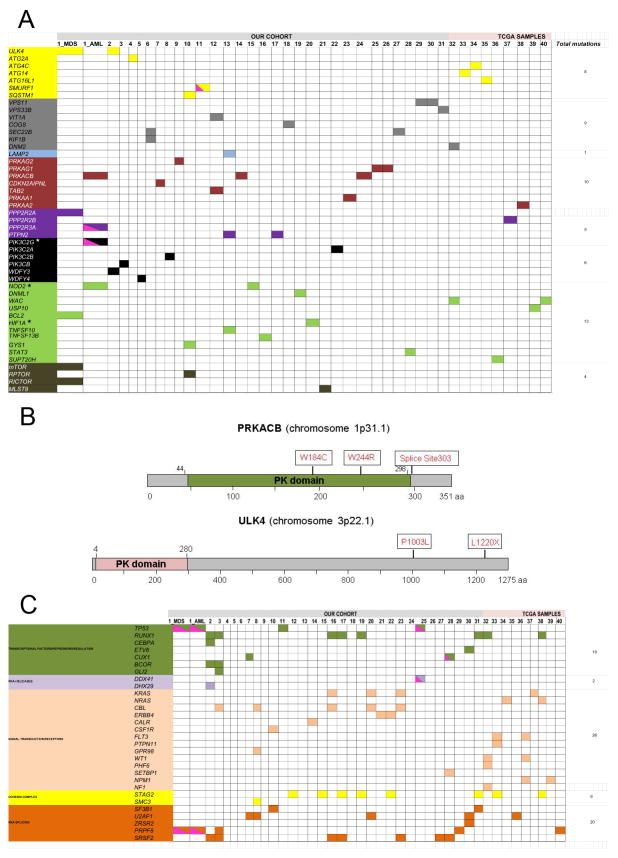
Autophagy is an evolutionarily conserved mechanism of bulk protein degradation. Aberrant autophagic activity contributes to the pathogenesis of many diseases including cancer.1, 2 Although allelic loss of key autophagy genes such as ATG6/BECN1 has been specifically associated with malignancies, the prevalence of mutations in the autophagy network as a whole has never been comprehensively studied in a diverse spectrum of patients with myeloid neoplasms.3 We conducted a cross-sectional analysis of 180 autophagy genes and their interactions by analyzing the results of whole exome sequencing (WES) derived from a cohort of 223 cases with myeloid neoplasms [(N=223; myelodysplastic syndromes (MDS)=120, acute myeloid leukemia (AML)=46, MDS/myeloproliferative neoplasms (MPN)=45, MPN=5, and others (paroxysmal nocturnal hemoglobinuria (PNH), aplastic anemia/PNH =7)]. We also reviewed the TCGA data of patients with primary AML (N=202). In total, we identified missense mutations and copy number alterations in the following genes of the autophagy network (ATG2A, ATG4C, ATG14, ATG16L1, BCL2, CDKN2AIPNL, COG8, DNM1L, DNM2, GYS1, HIF1A, KIF1B, LAMP2, MLST8, MTOR, NOD2, PIK3C2G, PIK3C2A/B, PIK3CB, PPP2R2A/B, PPP2R3A, PRKACB, PRKAA1/2, PRKAG1/G2, PTPN2, RICTOR, RPTOR, SEC22B, SMURF1, SQSTM1, STAT3, SUPT20H, TAB2, TNFSF10/13B, ULK4, USP10, VPS11/33B, VTI1A, WDFY3/4, WAC) in 40 patients (MDS=14, AML=15, MPN and MDS/MPN=9, Others=2). Bioinformatic analyses detected genetic alterations in at least one relevant gene in 31/223 (14%) of patients with available WES and in 9/202 (4.4%) of patients from the TCGA cohort (Table 1). Table S1 summarizes the clinical characteristics of the patients carrying mutations in autophagy-related genes [pt#1-31 (our cohort) and 32–40 (TCGA cohort)]. The analysis of matched CD3+ lymphocytes confirmed the somatic nature of each alteration. Mutations were: non-synonymous (45), stop-gain (5), frameshift insertions/deletions (3/1), non-frameshift deletion (1), and splice site (3); 32/40 patients had a sole mutation, 4 patients carried 2 mutations each (ULK4, WDFY3), (KIF1B, SEC22B), (VIT1A, TAB2), (DNM2, WAC). Two patients harbored mutations in 3 genes each [pt#10 (RPTOR, SQSTM1, GYS1) and pt#13 (TNFSF10, LAMP2, PTPN2)]. One patient (pt#15) carried 2 mutations in the NOD2 gene [stopgain, (13.5%) and a missense (8.2%)]. One patient (pt#1) had >3 mutations. We also identified recurrently mutated genes: PRKACB [pt#1 (AML stage), 14 and 24], ULK4 [pt#1 (MDS stage), #2], NOD2 [pt#1 (AML stage), #15], PTPN2 (pt#13, 17), PRKAG1 (pt#25, 26), VPS11 (pt#29, 30), and WAC (pt#32, 40). The same mutation (p.T192P; c.A574C) in the v-SNARE coiled-coil homology domain of SEC22B gene was also detected in pts#6 and #27. We designed a mutational diagram by clustering all the genes based on their function in the autophagy pathway (Figure 1a) and classified the mutations in the following autophagy interactome components: apoptotic effectors/inducers (13/55; 24%), protein kinases and dependent kinases (10/55; 18%), vesicle transport (9/55; 16%), early-stage autophagy (8/55; 15%) and phosphatidylinositol-3 phosphatase family (6/55; 11%; Figure S1, panel a). In the cohort of mutant patients, mutations were observed in more than 10% of all myeloid disease types with the majority of mutant patients being high-risk MDS (12/40; 30%) and AML patients (15/40; 37.5%) (Figure S1, panel b). A schematic diagram depicts 2 genes (PRKACB, ULK4) that were found mutated in 3 and 2 patients, respectively (Figure 1b). Of note, the association of mutations in the autophagy pathway with higher grade rather than lower grade MDS disease is emphasized by the analysis of consecutive samples. Indeed, we performed WES of patients #1 and 13 in paired specimens obtained at the stage of MDS and after progression to AML. We found a high mutation level in early stage autophagy (ULK4; 12.5%) genes and in the mTOR pathway [MTOR (10%) and RICTOR (16.7%)] at the stage of MDS in patient #1. Mutations in apoptotic effectors and inducers [BCL2; 14.2% (MDS stage) vs. NOD2; 63% (AML stage)] and in the protein kinase phosphatase pathway [PPP2R2A; 7.1% (MDS stage) vs. PPP2R3A; 10% (AML stage)] were detected at both the MDS and AML stages. This patient had multiple adverse features including a complex karyotype, TP53 and PRPF8 mutations, a rapid increase in bone marrow blasts with disease progression (3% vs. 35% in 3 months) and no response to hypomethylating therapy. In patient #13, we observed the emergence of mutations in autophagy-related genes such as TNFSF10 (7.6%), LAMP2 (7.7%) and PTPN2 (8%) only at the AML stage and not at the MDS stage. These characteristics suggest that alterations in autophagy-related genes may be a cooperative event associated with disease progression and that cells with autophagic defects may be more susceptible to clonal evolution.
Table 1.
Pattern of molecular mutations in the autophagy network
| # | Gene | Type of mutation | RefSeq Gene | AA change | cDNA change | Genomic location (start) | Protein Location/Interaction |
|---|---|---|---|---|---|---|---|
| 1 (MDS stage) | BCL2 | nonsynonymous substitution | NM_000633 | p.S105P | c.T313C | 60985587 | |
| 1 (MDS stage) | BCL2 | nonsynonymous substitution | NM_000633 | p.P57L | c.C170T | 60985730 | |
| 1 (MDS stage) | ULK4 | stopgain | NM_017886 | p.L1220X | c.T3659A | 41439589 | Repeat (Heat 5) |
| 1 (MDS stage) | PPP2R2A | nonsynonymous substitution | NM_002717 | p.A423D | c.C1268A | 26227853 | Repeat (WD 7) |
| 1 (MDS stage) | RICTOR | nonsynonymous substitution | NM_152756 | p.W295C | c.G885T | 38975643 | |
| 1 (MDS stage) | MTOR | nonsynonymous substitution | NM_004958 | p.K1471N | c.G4413T | 11217265 | Repeat (TPR 4) |
| 1 (AML stage) | PPP2R3A | nonsynonymous substitution | NM_002718 | p.Q88K | c.C262A | 135720602 | |
| 1 (AML stage) | PIK3C2G | nonsynonymous substitution | NM_004570 | p.A899D | c.C2696A | 18649021 | |
| 1 (AML stage) | PRKACB | splice site | NM_182948 | e9-1 | 84679835 | ||
| 1 (AML stage) | NOD2 | nonsynonymous substitution | NM_022162 | p.K118R | c.A353G | 50733678 | Domain (CARD 1) |
| 2 | WDFY3 | nonsynonymous substitution | NM_014991 | p.G3362V | c.G10085T | 85600134 | Region of interaction with SQSTM1 and ATG5 and LIR motif |
| 2 | ULK4 | nonsynonymous substitution | NM_017886 | p.P1003L | c.C3008T | 41705161 | |
| 3 | PIK3CB | nonsynonymous substitution | NM_006219 | p.V349E | c.T1046A | 138452207 | |
| 4 | ATG2A | nonsynonymous substitution | NM_015104 | p.A1533P | c.G4597C | 64666182 | |
| 5 | WDFY4 | splice site | NM_020945 | e48+2 | 50149895 | ||
| 6 | KIF1B | nonsynonymous substitution | NM_015074 | p.E1460K | c.G4378A | 10425470 | |
| 6 | SEC22B | nonsynonymous substitution | NM_004892 | p.T192P | c.A574C | 145115815 | Domain (v-SNARE coiled-coil homology) |
| 7 | CDKN2AIPNL | nonsynonymous substitution | NM_080656 | p.D100A | c.A299C | 133745634 | |
| 8 | PIK3C2B | non-frameshift deletion | NM_002646 | p.232_232del | c.694_696del | 204438235 | Region: of interaction with GRB2 |
| 9 | PRKAG2 | nonsynonymous substitution | NM_016203 | p.V533G | c.A1598C | 151257690 | Domain (CBS 4) |
| 10 | RPTOR | nonsynonymous substitution | NM_001163034 | p.A61D | c.C182A | 78599510 | |
| 10 | SQSTM1 | nonsynonymous substitution | NM_003900 | p.D347Y | c.G1039T | 179260656 | Proximal region to the LIR motif (amino-acid 336-341) |
| 10 | GYS1 | nonsynonymous substitution | NM_001161587 | p.G400V | c.G1199T | 49477908 | |
| 11 | SMURF1 | nonsynonymous substitution | NM_020429 | p.R564L | c.G1691T | 98636086 | Domain (HECT) |
| 12 | VTI1A | stopgain | NM_145206 | p.E53X | c.G157T | 114224309 | Region (Coiled coiled) |
| 12 | TAB2 | nonsynonymous substitution | NM_015093 | p.A152S | c.G454T | 149699505 | |
| 13 | TNFSF10 | nonsynonymous substitution | NM_003810 | p.A32D | c.C95A | 172241080 | |
| 13 | LAMP2 | nonsynonymous substitution | NM_002294 | p.A314S | c.G940T | 119575738 | Region (Second lumenal domain) |
| 13 | PTPN2 | nonsynonymous substitution | NM_080423 | p.Q264L | c.A791T | 12814269 | Domain (Tyrosine-protein phosphatase) |
| 14 | PRKACB | nonsynonymous substitution | NM_001242862 | p.W184C | c.G552T | 84663456 | |
| 15 | NOD2 | stopgain | NM_022162 | p.E1008X | c.G3022T | 50763784 | |
| 15 | NOD2 | nonsynonymous substitution | NM_022162 | p.E1008G | c.A3023G | 50763785 | |
| 16 | TNFSF13B | nonsynonymous substitution | NM_001145645 | p.N164D | c.A490G | NA | |
| 17 | PTPN2 | stopgain | NM_001207013 | p.R26X | c.C76T | NA | |
| 18 | COG8 | nonsynonymous substitution | NM_032382 | p.P369S | c.C1105T | NA | |
| 19 | DNM1L | nonsynonymous substitution | NM_005690 | p.G362S | c.G1084A | 32883952 | |
| 20 | HIF1A | nonsynonymous substitution | NM_181054 | p.P429S | c.C1285T | NA | |
| 21 | MLST8 | nonsynonymous substitution | NM_001199173 | p.R221S | c.C661A | 2258298 | Repeat (WD 6) |
| 22 | PIK3C2A | nonsynonymous substitution | NM_002645 | p.P723S | c.C2167T | 17153527 | Domain (C2 PI3K-type) |
| 23 | PRKAA1 | nonsynonymous substitution | NM_206907 | p.A573V | c.C1718T | NA | |
| 24 | PRKACB | nonsynonymous substitution | NM_182948 | W244R | c.T730A | 84663454 | |
| 25 | PRKAG1 | nonsynonymous substitution | NM_001206709 | p.N15S | c.A44G | 49406859 | |
| 26 | PRKAG1 | nonsynonymous substitution | NM_002733 | p.R152G | c.A454G | NA | Domain (CBS 2) |
| 27 | SEC22B | nonsynonymous substitution | NM_004892 | p.T192P | c.A574C | 145115815 | Domain (v-SNARE coiled-coil homology) |
| 28 | STAT3 | nonsynonymous substitution | NM_003150 | p.G618R | c.G1852C | 40475058 | |
| 29 | VPS11 | frameshift insertion | NM_021729 | p.S74fs | 118939937 | ||
| 30 | VPS11 | nonsynonymous substitution | NM_021729 | p.S74P | c.T220C | 118939939 | |
| 31 | VPS33B | nonsynonymous substitution | NM_018668 | p.E128Q | c.G382C | 91553052 | |
| 32 (TCGA3009) | DNM2 | frameshift insertion | NM_001005360.1 | p.T471fs*11 | c.1411_1412insGCCATCGT | 10770237 | Domain (Pleckstrin homology) |
| 32 (TCGA3009) | WAC | frameshift insertion | NM_016628.3 | p.E87fs | ->AGGGC | 28864676 | |
| 33 (TCGA2871) | ATG14 | nonsynonymous substitution | NM_014924.3 | p.T213M | G>A | 54922339 | |
| 34 (TCGA2870) | ATG4C | nonsynonymous substitution | NM_032852.2 | p.D354Y | G>T | 63073082 | |
| 35 (TCGA2821) | ATG16L1 | nonsynonymous substitution | NM_030803.3 | p.L558Q | c.T1673A | 233866685 | |
| 36 (TCGA2919) | SUPT20H | nonsynonymous substitution | NM_001014286.2 | p.L635V | G>C | 36489471 | |
| 37 (TCGA2864) | PPP2R2B | nonsynonymous substitution | NM_181676.1 | p.R374C | G>A | 145949924 | Repeat (WD6) |
| 38 (TCGA2978) | PRKAA2 | stopgain | NM_006252.3 | p.R227* | C>T | 56934311 | Domain (Protein kinase) |
| 39 (TCGA2859) | USP10 | frameshift deletion | NM_005153.2 | p.P386fs | C>- | 83336744 | |
| 40 (TCGA3001) | WAC | splice site | NM_016628.3 | e12+1 | G>A | 28945298 |
Figure 1.
(a) Somatic mutations of the autophagy network in patients with myeloid neoplasms. Whole exome and targeted sequencing identified 55 somatic mutations in autophagy-related genes (a complete list of mutations is presented in Table 1). A schematic diagram illustrates mutations per patients (N=40; pts #1-31; our cohort; pts #32-40, TCGA cohort). For pt #1, exome sequencing was performed at the stage of MDS and AML. Different colors represent the specific pathways where genes were clustered based on their function in the autophagy pathway (yellow: early stage autophagy; gray: vesicles-mediated transport; light blue: lysosomal enzymes: red: protein kinases and dependent kinases family; purple: protein tyrosine phosphatase family; black: phosphatidylinositol-3 phosphatases; green: apoptotic inducers/inhibitors; dark green: insulin receptor/mTOR associated signaling. Clustering was conducted by reviewing previous published reports and by using the reactome pathway database (http://www.reactome.org/). Pink half square triangles represent patients who were hemizygous for specific mutations. Stars indicate genes for which germline variants were also detected. The total number of mutations per pathway is indicated in the right column. (b) Schematic diagrams of two mutated autophagy-related genes. Protein kinase cAMP-activated catalytic subunit beta (PRKACB) is a serine/threonine protein kinase composed by 351 amino acids which mediates signaling through cyclic AMP and it is involved in the induction of autophagy. The protein kinase (PK) domain spans amino-acid 44-244. Three patients in our cohort carried PRKACB mutations [p.W184C, p.W244R, Splice site_303 (exon 9-1)]. Unc-51 like kinase 4 (ULK4) is a serine/threonine protein kinase involved in autophagy. The protein is composed by 1275 amino acids with a kinase domain spanning amino acids 4-280. Two patients carried ULK4 mutations (p.L1220X, p.P1003L).
(c) Mutational spectrum of patients with myeloid neoplasms harboring mutations in the autophagy network. A schematic diagram illustrates the complete molecular profile of patients with myeloid malignancies (N=40; pts #1-31; our cohort; pts #32-40, TCGA cohort). For pt #1, exome sequencing was performed at the stage of MDS and AML. Data are a collection of exome sequencing, TruSeq analysis, and Sanger sequencing. Genes were clustered in pathway based on their function according to the literature (blue: DNA-methylation; light green: Chromatin modification; dark green: transcriptional factors/repressors/regulation; purple: RNA-helicases; light orange: signal transduction/receptors; yellow: cohesin complex; dark orange: RNA-splicing). Pink half square triangle represents patients who are hemizygous for specific mutations.
Analysis of the cytogenetic profiles of the mutant patients showed that 4 genes mapped to commonly deleted regions e.g., 5q (CDKN2AIPNL, SQSTM1) or 7q (SMURF1, PRKAG2) and coincided with haploinsufficient expression, while 3 genes had hemizygous configuration (SMURF1, PPP2R3A, PIK3C2G) (Table S2). Analysis of the variant types showed that loss-of-function mutations were observed in ULK4 (stopgain; p.L1220X), NOD2 (stopgain; p.E1008X), VTI1A (stopgain; p.E53X), PTPN2 (stopgain; p.R26X), PRKAA2 (stopgain; p.R227X), VPS11 (frameshift insertion; p.S64fs*), DNM2 (frameshift insertion; T471fs*), WAC (frameshift insertion; p.E87fs), and USP10 (frameshift deletion; p.P386fs*) genes. We also dissected the clonal hierarchy of autophagy gene mutations. Clonal hierarchy showed that autophagy mutations were predominantly secondary events, were ancestral events in 7 patients (ATG2A, SEC22B, STAT3, PRKACB, DNML1, PTPN2, PRKAG1) and co-dominant in 2 patients (NOD2, MLST8) When autophagy gene mutations were secondary, the most represented ancestral mutations were in splicing factors (N=9; SRSF2, PRPF8, U2AF1) and DNA methylation genes (N=4; TET2, DNMT3A). The characterization of ancestral vs. secondary events suggested to us that autophagy gene mutations are cooperative rather than initiating events in clonal evolution. We also observed that autophagy gene mutations are secondary mutations in several cases when ancestral mutations occurred in splicing factor genes. A recent article reported that autophagy genes may be implicated in the epigenetic mechanism of patients with MDS carrying U2AF1 mutations and may contribute to disease phenotypes. U2AF1S34 mutations have been described to possibly promote leukemogenesis by aberrant splicing of the autophagy gene ATG7.4 This observation led us to investigate whether epigenetic defects in autophagy genes are also commonly detected in MDS and, if present, whether they correlate with specific biological and clinical MDS phenotypes. We analyzed RNA sequencing of patients with MDS and found that the expression levels of several autophagy genes was increased in SF3B1MUT MDS compared to SF3B1WT MDS cells: early stage autophagy genes (ATG2A/B, FC=2; ATG4A, FC=2; ATG9A, FC=5; ATG4C, P=0.05; ATG18 (FC=4.8; P= 0.02), autophagy-initiating kinases (ULK1, FC=2; ULK3, FC=3.9; P=0.05), and late stage autophagy genes (CTSL1, FC=20; CTSD, FC=5.8; P=0.05; CTSB, FC=2.1; CTSE, FC=5.9; CTSD, FC=2; P=0.01). Of note, 2 genes (ATG2A, ATG4C) carried somatic mutations and increased expression levels. One of them, ATG2A was also an ancestral event when mutated. We originally reported a list of genes found differentially expressed in Sf3b1+/− BM cells compared to Sf3b1+/+ BM cells5 (see Table S1) by RNA sequencing analysis. Interestingly enough we merged the list of genes found mutated in our cohort of patients with the list of genes found modulated in Sf3b1+/− mice and found that 17 genes were up-regulated more than 2-fold change in our mouse model of MDS (ATG16L1, PIK3CB, GYS1, SEC22B, KIF1B, STAT3, VT1A, TNFSF10, PTPN2, DNML1, DNM2, HIF1A, MLST8, CDKN2AIPNL, WAC, BCL2, USP10). These observations suggested that the autophagy pathway can potentially be triggered in MDS cells for therapeutic benefit. Autophagy gene mutations were most prevalent in high-risk MDS (12/40; 30%) and almost equally prevalent in sAML and pAML patients (6/9). 21/37 (57%) had abnormal karyotype with 4 patients having complex karyotype including -17 and -7. Among the cases with abnormal karyotype, 4 cases had del (5q). 21 patients had a fatal outcome. Comparative analysis of Sanger sequencing, TruSeq targeted DNA sequencing, WES and TCGA data was used to assess the global mutational spectrum of the patients harboring mutations in the autophagy-network. We found that the most commonly mutated gene was TET2 (11/40; 27.5%), followed by RUNX1 and STAG2 (8/40; 20%), SRSF2 (7/40; 7.5%) and DNMT3A and ASXL1 (6/40; 15%). Hemizygosity due to somatic deletions for genes associated with poor prognosis was also noted (TP53: 2 patients; PRPF8, CUX1, DDX41: 1 patient each) (Figure 1c). Comparative analysis (Sanger, TruSeq, WES, TCGA) found that patients with autophagy-related genes are more susceptible to acquire mutations in signal transduction pathways (26 mutations), DNA methylation (24 mutations), and RNA-splicing (20 mutations) (Figure 1c and Figure S2). Twenty-five mutations had a cut-off more than 20% variant allele frequency (VAF) (Figure S3). Targeted gene panel sequencing was applied for 22 genes and confirmed the presence of high variant allele frequency mutations like NOD2 (63%), PIK3C2B (44%), PRKACB (36%) and STAT3 (38%). The majority of mutant patients had a dismal prognosis, suggesting that these patients had cumulative unfavorable mutations like RUNX1, ASXL1, DNMT3A and that these negative prognostic mutations6 represented driver mutations in these patients. Seven patients receiving hypomethylating agents had no response to therapy (Table S1). Comparison of mutant and wild type (WT) patients showed that mutant patients had lower survival trending toward significance compared to WT (14 mo vs. 20 mo; NMUT vs. NWT = 20 vs. 90; log-rank=0.09). Among disease subtypes, autophagy network mutations were associated with significantly inferior survival in patients with MDS (NMUT = 13, mean survival: 17 mo; NWT = 61, mean survival: 35 mo, log-rank: 0.0187) and MDS/MPN overlap syndromes (NMUT = 4, mean survival: 12 mo; NWT= 31, mean survival: 30 mo, log-rank: 0.0374).
Given the recent discovery of familial AML and MDS cases carrying pathogenic germ-line variants, 7, 8 we also evaluated whether the same autophagy genes we found to be somatically mutated may also be prone to germ-line alterations. An in house analysis of a database of 263,973 variants obtained from WES was conducted by testing different predictive tools assessing deleterious variants and by calling a variant as deleterious if present as deleterious in all Supplemental methods). We found 4 variants in 3 genes among 4 patients with possible deleterious exonic non-synonymous variants that were predicted to be significantly damaging by all the tested scores: PIK3C2G (rs377020826, c.T3083C, total allele frequency: 0.0001; rs201569993, c.C2938T, total allele frequency: 0.0007), NOD2 (rs104895423, c.T662G, total allele frequency: 0.0006), and HIF1A (c.A1666G; total allele frequency: undetermined) (Table S3). In total, PIK3C2G carried 1 somatic and 2 germ-line variants, NOD2 had 2 somatic and 1 germ-line variants, and HIF1A harbored 1 somatic and 1 germ-line variant. A complete table summarizes the somatic and germ-line status of the variants we identified in the autophagy pathway (Table S4).
As mentioned previously, many types of malignant cells have been reported to exhibit defective or dysregulated autophagy including mutations in BECN1 (17q21), which is monoallelically deleted in 40–75% of patients with breast, ovarian, and prostate cancer.9 In other tumors, autophagy genes such as SH3GLB1 have been associated with regions of loss of heterozygosity.10 In our study, we identified mutations in proximal co-factors of BECN1 such as PI3KCIII, vacuolar sorting proteins (Vps34, Vps11) and ATG14. We performed an analysis of the related protein sequences (http://www.uniprot.org/) of the mutated regions and found that several mutations occurred in essential domains/regions involved in protein-protein interactions, particularly those required for autophagosome formation (ATG14, BECN1, PI3KC3, SEC22B, VPS34/11) (Table 1). Several mutations were found in functional domains of the proteins [pt#32 (DNM2; pleckstrin homology domain), pt#38 (PRKAA2; protein kinase domain)]. Other mutations were detected in regions of interactions between genes highly relevant in the autophagy pathway such as WDFY3 mutation (p.G3362V; pt#2) maps in a region spanning the interaction with SQSTM1 (amino-acid 2586-3526), ATG5 (amino-acid 2981-3526) and the region containing the LC3-interacting region (LIR) motif, which allows the targeting of autophagy receptors to LC3 (or other ATG8 family proteins) as a crucial step in the autophagy cascade. SQSTM1 was also found mutated in our cohort (p.D347Y; pt#10) and the mutated region was proximal to the LIR motif (amino-acid 336-341). We also detected a mutation in a Beclin-1-associated autophagy key regulator (ATG14; pt#33), which has been shown to directly interact with Beclin-1 and increases phosphatidylinositol 3-phosphate kinase class III (PI3KC3) activity to initiate autophagosome membrane nucleation by targeting SNARE-associated proteins.11 We also reviewed extensively the literature to identify possible variants occurring also in other diseases (Table S5).
Our collective findings define for the first time an important and perhaps so far underappreciated role for genetic aberrations in the autophagy network in the pathogenesis of myeloid neoplasms. Our data demonstrate that autophagy are autophagy-related gene mutations are more prevalent in MDS and AML, co-occur with unfavorable mutations (RUNX1, ASXL1, DNMT3A), and are associated with inferior survival. These findings support further investigation of genetic loss of autophagy regulation as a novel cooperative mechanism leading to leukemogenesis that can potentially be therapeutically targeted with synthetic lethal approaches. Results from high-throughput next generation sequencing collected from a larger population of genetically annotated patients with myeloid neoplasms and comprehensive clinical information from different institutional centers will help define the exact frequency of single gene mutations and the precise correlation with the clinical phenotypes.
Supplementary Material
Acknowledgments
This work was supported by the Scott Hamilton CARES (V.V) and 2016 ASH Scholar award (V.V), the National Cancer Institute grants R01CA172443 (J.S.C) and R01CA190789 (S.T.N). We thank The Cancer Genome Atlas (TCGA) data portal and cBioPortal for Cancer Genomics to allow us the retrieval of helpful information.
Footnotes
Conflicts of interest
J.M.P. (Celgene and Ra Pharmaceuticals)
Author contributions
V.V. designed the study, analyzed and interpreted data, and wrote the manuscript; B.P. analyzed and interpreted the data and edited the manuscript; Y.H. performed experiments and edited the manuscript; S.T.N. analyzed and interpreted data and edited the manuscript; S. T. collected clinical data; K.R.K. provided intellectual input; B.J.P. collected clinical information; C.H. analyzed the data; A.S.A., H.E.C., and M.A.S. contributed to data interpretation and manuscript preparation; J.M.P. contributed patient specimens and genomic data and participated in data analysis and manuscript preparation; J.S.C. designed the study, analyzed and interpreted data, and wrote the manuscript. All authors approved the manuscript before submission.
References
- 1.Kroemer G. Autophagy: a druggable process that is deregulated in aging and human disease. J Clin Invest. 2015;125(1):1–4. doi: 10.1172/JCI78652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85(6):830–8. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–6. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius CM, et al. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3’ End Formation. Mol Cell. 2016;62(4):479–90. doi: 10.1016/j.molcel.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visconte V, Tabarroki A, Zhang L, Parker Y, Hasrouni E, Mahfouz R, et al. Splicing factor 3b subunit 1 (Sf3b1) haploinsufficient mice display features of low risk Myelodysplastic syndromes with ring sideroblasts. J Hematol Oncol. 2014;7:89. doi: 10.1186/s13045-014-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harada H, Harada Y. Recent advances in myelodysplastic syndromes: Molecular pathogenesis and its implications for targeted therapies. Cancer Sci. 2015;106(4):329–36. doi: 10.1111/cas.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liew E, Owen C. Familial myelodysplastic syndromes: a review of the literature. Haematologica. 2011;96(10):1536–42. doi: 10.3324/haematol.2011.043422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–90. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C, Song E, Codogno P, Mehrpour M. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy. 2012;8(12):1853–5. doi: 10.4161/auto.21996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang C, Jung JU. Autophagy genes as tumor suppressors. Curr Opin Cell Biol. 2010;22(2):226–33. doi: 10.1016/j.ceb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HJ, Zhong Q, Sheng ZH, Yoshimori T, Liang C, Jung JU. Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J Cell Sci. 2012;125(Pt 20):4740–50. doi: 10.1242/jcs.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.