Abstract
The genus Elizabethkingia is genetically heterogeneous, and the phenotypic similarities between recognized species pose challenges in correct identification of clinically derived isolates. In addition to the type species Elizabethkingia meningoseptica, and more recently proposed Elizabethkingia miricola, Elizabethkingia anophelis and Elizabethkingia endophytica, four genomospecies have long been recognized. By comparing historic DNA–DNA hybridization results with whole genome sequences, optical maps, and MALDI-TOF mass spectra on a large and diverse set of strains, we propose a comprehensive taxonomic revision of this genus. Genomospecies 1 and 2 contain the type strains E. anophelis and E. miricola, respectively. Genomospecies 3 and 4 are herein proposed as novel species named as Eliza-bethkingia bruuniana sp. nov. (type strain, G0146T = DSM 2975T = CCUG 69503T = CIP 111191T) and Elizabethkingia ursingii sp. nov. (type strain, G4122T = DSM 2974T = CCUG 69496T = CIP 111192T), respectively. Finally, the new species Elizabethkingia occulta sp. nov. (type strain G4070T = DSM 2976T = CCUG 69505T = CIP 111193T), is proposed.
Keywords: AAI, ANI, Elizabethkingia, MALDI-TOF, SNPs, Taxonomy
Introduction
First observed as a causative agent of neonatal meningitis by King (1959), Elizabethkingia infections can cause a variety of conditions including necrotizing fasciitis (Lee et al. 2006), endophthalmitis (Young et al. 2014), pneumonia (da Silva and Pereira 2013), and sepsis (Green et al. 2008; Ramanan and Razonable 2013). Elizabethkingia infections are most commonly observed in immunocompromised patients, mechanically ventilated patients, and neonates, but have been reported to cause meningitis in an immunocompetent adult (Hayek et al. 2013). Once Elizabethkingia infections occur, they have a high mortality rate, with reports of 25% for patients undergoing dialysis (Ratnamani and Rao 2013) and up to 57% for neonates with meningitis (Bloch et al. 1997). A review of 118 patients with Elizabethkingia bacteremia found an overall 14-day mortality rate of 23.4%, and approximately a five-fold increase in incidence per 100,000 admissions over an eight-year period (Hsu et al. 2011).
While hospital outbreaks have usually been attributed to Elizabethkingia meningoseptica, there have been recent reports of Elizabethkingia anophelis causing outbreaks in Intensive Care Units (Teo et al. 2014). A review of cases of bacteremia in Hong Kong hospitals that were caused by Elizabethkingia found that E. anophelis was frequently the causative agent, with an associated high degree of morbidity and mortality (Lau et al. 2016). The largest recognized outbreak to date of E. anophelis occurred in the spring of 2016, sickening 64 people in Wisconsin and nearby states of the United States (Perrin et al. 2017).
DNA–DNA hybridization was initially used to describe five distinct groups of Elizabethkingia strains (E. meningoseptica and genomospecies 1 through 4) (Ursing and Bruun 1987), but there are no known consistent phenotypic characteristics that define the various genomospecies of Elizabethkingia (Bruun and Ursing 1987). Additional Elizabethkingia species were later described with no comparison to the genomospecies reference strains, and each species was defined based on the description of a single strain: Elizabethkingia miricola was described in 2003 as Chryseobacterium miricola, and moved to the newly-formed Elizabethkingia genus in 2005, followed by E. anophelis in 2011 and Elizabethkingia endophytica in 2015 (Kim et al. 2005; Kampfer et al. 2011, 2015; Li et al. 2003). E. endophytica was subsequently recognized as a later subjective synonym of E. anophelis (Doijad et al. 2016), based on whole genome sequence analysis. Taxonomic correspondence of these recently named species with the genomospecies previously defined by DNA–DNA hybridization has not been formally addressed. In this paper, we have analyzed historical strains that had originally been assigned to genomospecies 1–4, modern type strains, and isolates recently obtained from clinical sources using whole genome sequencing (WGS) and optical mapping, and explored the use of MALDI-TOF mass spectrometry and targeted gene sequencing as identification methods.
Materials and methods
Strain selection and phenotypic testing
Traditional DNA–DNA (tDDH) hybridization values for Elizabethkingia strains that had been used to define the five genomospecies (E. meningoseptica and genomospecies 1 through 4) (Holmes et al. 2013) were reviewed, and strains representing the widest array of tDDH values were selected for WGS. This set of 17 strains will be referred to as the “historic strains” throughout this manuscript. During the course of the 2015–2016 Wisconsin Outbreak investigation, we requested that states send us all recently-collected Elizabethkingia isolates, and determined their optical maps using the OpGen optical mapping platform (see below); a subset of 21 strains was selected from these, based on the diversity of their optical maps. Ten additional strains were selected as potentially informative from the CDC (two strains) and Institut Pasteur (eight strains) strain collections, based on preliminary WGS data which again showed that they contained maximal diversity. Sixteen strains with whole genome sequences in the public domain were selected; strains that had been previously shown to have whole genome sequences that were essentially identical to other strains in the public domain were excluded and type strains of each of the validly published species were obtained, resulting in a total of 65 strains. Table 1 shows the BioSample identifier of each, along with the accession number for the draft and complete (if available) genomes of each. Our strain collection dates back to the 1960’s and contains 297 isolates that were previously designated as E. meningoseptica, or one of its earlier names (Flavobacterium meningosepticum, Chryseobacterium meningosepticum). Aggregate phenotypic data and MALDI-TOF mass spectra were examined for these, but only strains with WGS data are listed on Table 1.
Table 1.
Strains used in this study
| Strain | Taxon | Taxon originally described as | Acc # contigs | Acc # complete | BioSample | ENA sample id |
|---|---|---|---|---|---|---|
| FMS-007 | E. anophelis | Elizabethkingia meningoseptica | CP006576 | |||
| 0422 (H) | E. anophelis | Elizabethkingia genomosp. 1 | LNOG00000000 | CP016370 | SAMN05290454 | |
| 3375 (H) | E. anophelis | Elizabethkingia genomosp. 1 | MAHM00000000 | CP016373 | SAMN05273152 | |
| 502 | E. anophelis | Elizabethkingia meningoseptica | AVCQ00000000 | |||
| B2D | E. anophelis | Elizabethkingia meningoseptica | JNCG00000000 | |||
| CIP108654 | E. anophelis | Elizabethkingia anophelis | FTPG00000000 | SAMEA4026800 | ERS1197910 | |
| CIP111046 | E. anophelis | FTRB00000000 | SAMEA50777668 | ERS1505818 | ||
| CIP111067 | E. anophelis | FTQZ00000000 | SAMEA50774668 | ERS1505814 | ||
| CIP60.58 | E. anophelis | FTQY00000000 | SAMEA50775418 | ERS1505815 | ||
| CSID_3000516074 | E. anophelis | MAHA00000000 | SAMN05255124 | |||
| CSID_3000516810 | E. anophelis | MAHH00000000 | SAMN05256530 | |||
| CSID_3000516978 | E. anophelis | MAHJ00000000 | SAMN05256551 | |||
| CSID_3000517066 | E. anophelis | MAHL00000000 | SAMN05256625 | |||
| CSID_3015183678 | E. anophelis | MAFY00000000 | CP014805 | SAMN04567744 | ||
| CSID_3015183679 | E. anophelis | MAHO00000000 | SAMN05275358 | |||
| CSID_3015183680 | E. anophelis | MAHP00000000 | SAMN05275369 | |||
| CSID_3015183686 | E. anophelis | MAHR00000000 | SAMN05277596 | |||
| DSM_23781 (T) | E. anophelis | MAHN00000000 | SAMN02470677 | |||
| E6809 (H) | E. anophelis | Elizabethkingia genomosp. 1 | MAHS00000000 | CP014339 | SAMN05277610 | |
| Endophthalmitis | E. anophelis | Elizabethkingia meningoseptica | JSAA00000000 | |||
| F3201 (H) | E. anophelis | Elizabethkingia genomosp. 1 | MAHU00000000 | CP016374 | SAMN05277779 | |
| F3543 (H) | E. anophelis | Elizabethkingia genomosp. 1 | MAHT00000000 | CP014340 | SAMN05277758 | |
| JM-87 | E. anophelis | E. endophytica | MAGY00000000 | CP016372 | SAMN05255122 | |
| NUH4 | E. anophelis | Elizabethkingia anophelis | ASYI00000000 | |||
| NUH6 | E. anophelis | Elizabethkingia anophelis | ASYJ00000000 | |||
| NUHP1 | E. anophelis | Elizabethkingia anophelis | CP007547 | |||
| PW2809 | E. anophelis | Elizabethkingia anophelis | CBYE00000000 | |||
| R26 (T) | E. anophelis | Elizabethkingia anophelis | MAHN00000000 | |||
| V0378064 | E. anophelis | Elizabethkingia anophelis | CCAB00000000 | |||
| ATCC_33958 | E. bruuniana | Elizabethkingia miricola | JRFN00000000 | |||
| BM10 | E. bruuniana | Elizabethkingia sp. | CP011059 | |||
| CSID_3000516589 | E. bruuniana | MAHG00000000 | SAMN05256516 | |||
| CSID_3015183685 | E. bruuniana | MAHQ00000000 | SAMN05277593 | |||
| G0146 (H) (T) | E. bruuniana | Elizabethkingia genomosp. 3 | MAHV00000000 | CP014337 | SAMN05277827 | |
| G0153 (H) | E. bruuniana | Elizabethkingia genomosp. 3 | MAHW00000000 | SAMN05277862 | ||
| G4075 (H) | E. bruuniana | Elizabethkingia genomosp. 3 | LNOJ00000000 | SAMN04254558 | ||
| CIP111048 | E. meningoseptica | FTPF00000000 | SAMEA50779918 | ERS1505821 | ||
| CIP80.33 | E. meningoseptica | FTRA00000000 | SAMEA4026802 | ERS1197912 | ||
| CSID_3000515919 | E. meningoseptica | MAGZ00000000 | SAMN05255123 | |||
| CSID_3000516359 | E. meningoseptica | MAHC00000000 | SAMN05255848 | |||
| CSID_3000516465 | E. meningoseptica | MAHE00000000 | SAMN05256018 | |||
| CSID_3000516535 | E. meningoseptica | MAHF00000000 | SAMN05256158 | |||
| CSID_3000516977 | E. meningoseptica | MAHI00000000 | SAMN05256543 | |||
| G4076 (H) | E. meningoseptica | E. meningoseptica | MAHZ00000000 | CP016376 | SAMN05281841 | |
| G4120 (H) | E. meningoseptica | E. meningoseptica | MAIA00000000 | CP016378 | SAMN05281842 | |
| KC1913 (H) (T) | E. meningoseptica | E. meningoseptica | LNOH00000000 | CP014338 | SAMN04254555 | |
| NBRC 12535 (T) | E. meningoseptica | Elizabethkingia meningoseptica | BARD00000000 | |||
| CIP108653 | E. miricola | FTRC00000000 | SAMEA4026803 | ERS1197913 | ||
| CIP111047 | E. miricola | FTQX00000000 | SAMEA50780668 | ERS1505822 | ||
| CSID_3000516464 | E. miricola | MAHD00000000 | SAMN05255998 | |||
| CSID_3000516998 | E. miricola | MAHK00000000 | SAMN05256599 | |||
| CSID_3000517120 | E. miricola | MAGX00000000 | SAMN05254999 | |||
| DSM_14571 (T) | E. miricola | E. miricola | LSGQ00000000 | SAMD00016624 | ||
| EM-CHUV | E. miricola | E. miricola | LIQC00000000 | |||
| G4071 (H) | E. miricola | Elizabethkingia genomosp. 2 | LNOI00000000 | SAMN04254557 | ||
| G4074 (H) | E. miricola | Elizabethkingia genomosp. 2 | MAHY00000000 | SAMN05277881 | ||
| G4121 (H) | E. miricola | Elizabethkingia genomosp. 2 | MAIB00000000 | SAMN05281843 | ||
| F8124 | E. occulta | MBDR00000000 | SAMN05334989 | |||
| G4070 (H) (T) | E. occulta | Elizabethkingia genomosp. 4 | MAHX00000000 | SAMN05277871 | ||
| C1558 | E. ursingii | MBDS00000000 | SAMN05335199 | |||
| CSID_3000516135 | E. ursingii | MAHB00000000 | SAMN05255125 | |||
| G4122 (H) (T) | E. ursingii | Elizabethkingia genomosp. 4 | LNOK00000000 | SAMN04254563 | ||
| G4123 (H) | E. ursingii | Elizabethkingia genomosp. 4 | MAIC00000000 | CP016377 | SAMN05281858 | |
| ATCC_33861 (T) | Sphingobacterium spiritivorum | ACHA00000000 | ||||
| ATCC_35910 (T) | Chryseobacterium gleum | ACKQ00000000 |
Strains that were originally used to define the distinct Elizabethkingia genomes, referred to as historic strains in this manuscript, are denoted with (H). Names of strains that are the type strain of their respective species are followed by (T)
Phenotypic testing was performed using conventional biochemical tests as previously described (Holmes et al. 2013; Bruun 1982; Bruun and Ursing 1987).
Genome sequencing and assembly
At CDC, strains were grown on heart infusion agar according to manufacturer instructions (Difco) and supplemented with 5% rabbit blood (Hemostat Laboratories) at 35 °C. DNA extraction for WGS was performed using the CTAB protocol provided by the Department of Energy’s Joint Genome Institute (JGI Bacterial DNA Isolation CTAB Protocol), libraries were prepared using the Illumina TruSeq DNA sample prep kit, and genomes were sequenced on an Illumina MiSeq using a 2 × 250 paired-end protocol as described previously (Nicholson et al. 2016). Using CLC Genomics Workbench version 7.51. (CLCbio, Aarhus, Denmark) adapters were removed and reads were trimmed based on quality (limit = 0.02), then the resulting reads were assembled using the de Bruijn graph method of de novo assembly. Contigs ≥500 bp that had an average mapping coverage ≥50× were selected for further analysis. Contigs were split at the positions of any ambiguous (“N”) nucleotides in the assembly. Selected genomes were closed based on orientation of the contigs as determined by optical mapping (see below), with the exact sequence of contig joins informed by read mapping.
At the Collection of Institut Pasteur (CIP), strains were cultivated on trypticase soy agar (Bio-Rad) at 30 °C and DNA was extracted using the MagNA Pure 96 robotic System with the MagNA Pure 96 DNA and Viral Nucleic Acid small volume kit (Roche Diagnostics). Libraries were constructed using the Nextera XT DNA Library Preparation kit (Illumina, Inc., San Diego, CA) and sequencing done on a NextSeq-500 instrument using a 2 × 150 paired-end protocol. Read trimming and clipping was performed with AlienTrimmer v.0.4.0 (Criscuolo and Brisse 2013), followed by sequencing error correction with Musket v.1.1 (Liu et al. 2013), and next by coverage homogenization with khmer v.1.3 (Crusoe et al. 2015). Processed reads were finally used to perform de novo assembly with SPAdes v3.6.2 (Bankevich et al. 2012).
rpoB sequencing
Positions 1939-3629 in the 3825 bp rpoB gene sequence were sequenced as described previously (Shewmaker et al. 2011), with minor modifications: Elizabethkingia-specific PCR primers were designed (EK_rpoB_fwd: 5′-ATGGGATCTAACATGAT-3′ and EK_rpoB_rev: 5′-GCCCAAACCTCCATCTC-3′), and the amplicon was sequenced using these primers plus two additional primers (EKrpoB1154F: 5′-GGGGATAAAATGGCRGG-3′ and EKrpoB11 54R 5′-CCYGCCATTTTATCCCC-3′). To compare rpoB for all genomes used in this analysis, the sequence of each predicted rpoB PCR product was located using BLAST, and aligned within CLC genomics workbench. Maximum likelihood (ML) trees were generated using MEGA v6 (Tamura et al. 2013).
In silico genome comparisons
The average nucleotide identity BLASTN (ANIb) method was described by Goris et al. and has been implemented in the Jspecies software package (Goris et al. 2007; Richter and Rossello-Mora 2009). Two-way average amino acid identity (AAI) scores were calculated, and percentage of conserved proteins (POCP) scores were calculated as described by Qin et al. (2014). Proteomes from each genome were generated by Prodigal v2.6.2 (Hyatt et al. 2010). For each pairwise comparison, an all-versus-all search of all proteins was carried out using BLASTp v2.4.0+ (Altschul et al. 1997) in both directions. If both directions of BLASTp searches resulted in the same protein match (pair) and exceeded 40% in amino acid identity and 50% in coverage length, we included the protein sequences for computing the arithmetic mean sequence identity. In silico genome comparisons based on calculating genome-to-genome distances as described by (Auch et al. 2010a, b; Meier-Kolthoff et al. 2013), were determined using their Genome-to-Genome Distance Calculator tool (GGDC) (http://ggdc.dsmz.de/distcalc2.php), and rounded to the nearest integer. Single nucleotide polymorphism (SNP) trees were generated using the HarvestTools (Treangen et al. 2014), and exported Newick files were edited with MEGA v6 (Tamura et al. 2013). Additional data visualizations were produced using JMP v11 (SAS Institute Inc., Cary, NC).
Core genome phylogeny
The core orthologous genome of Elizabethkingia was calculated from Prodigal-generated (v2.6.2) (Hyatt et al. 2010) sequences of each Elizabethkingia (n = 63) isolate used as input for Roary v3.6.8 (Page et al. 2015). Highly related homologs were initially identified with CD-HIT v4.6 (Fu et al. 2012) by clustering sequences with five iterations beginning at 100% and going as low as 98% identity (0.5% decrement steps). Subsequent sequences were aligned to each other in an all-against-all fashion with BLASTp v2.4.0+ and a minimum identity of 40% was required. Sequence clusters were then identified with the mcl v14-137 algorithm (Enright et al. 2002) and paralogous sequences were discarded. The set of core orthologous genes were individually aligned with the codon-aware PRANK v.140603 software (Loytynoja and Goldman 2005). Concatenated alignments of these 2259 genes were filtered for invariant sites and the resulting 10,49,915 sites per isolate were analyzed to determine the most appropriate evolutionary model, using jModelTest v2.1.10 (Guindon and Gascuel 2003; Darriba et al. 2012). The JC69 model was then used in RAxML v8.2.9 (Stamatakis 2014) to generate 100 ML pseudoreplicate topologies, and 100 bootstraps provided convergence according to the extended majority—rule consensus tree criterion (Pattengale et al. 2010). The resulting tree was edited with MEGA v6 (Tamura et al. 2013).
Whole genome optical mapping
22 strains of Elizabethkingia, including at least two representatives for each proposed species, were compared by application of the OpGen optical mapping platform (OpGen, Inc., Gaithersburg, Maryland). High molecular weight genomic DNA from overnight grown bacterial cells was purified with Argus HMW DNA Isolation Kit (OpGen, Inc.) and examined for quality and concentration using the ARGUS QCards. The Enzyme Chooser function of MapManager version 1.3 (OpGen, Inc.) identified NcoI restriction endonuclease to be optimal for optical map production because its cleavage of reference genomes would result in fragments that average 6–12 kilobase pairs (kbp) in size, with no fragments larger than 80 kbp. Individual genomic DNA fragments were loaded onto a glass surface of a MapCard (OpGen, Inc.) using the microfluidic device, washed and then digested with NcoI, stained with JOJO-1 through the ARGUS MapCard Processor (OpGen, Inc.). Map cards after-ward were scanned and analyzed by automated fluorescent microscopy using the ARGUS Whole Genome Mapper v3.2.4 (OpGen, Inc.). The single molecule restriction map collections were then tiled according to overlapping fragment patterns to produce a consensus whole genome map. This map was imported into MapSolver v3.2 (OpGen, Inc.) along with predicted in silico maps of contigs derived from WGS, using the same restriction enzyme for ordering and orientation of contigs during genome circularization. In silico predicted optical maps of complete genomes were scaled according to the size of sequenced genomes. Final alignments were clustered in MapSolver v3.2 using a nearest neighbor algorithm to evaluate a similarity among Elizabethkingia strains.
MALDI-TOF
Matrix-Assisted Laser Desorption/Ionization-Time Of Flight (MALDI-TOF) mass spectrometry was performed using the BioTyper (Bruker, Germany). Main Spectrum Profiles (MSPs) were created to represent each genomospecies, using the historic strains and type strains of each species. For each MSP, cells were extracted using Bruker’s Formic Acid/Acetonitrile Procedure and overlaid with HCCA Matrix. Spectra were obtained using Bruker’s Flex Control and MALDI Biotyper 3 Software. The reproducibility of these spectra was confirmed using a whole-cell direct transfer, overlaid by HCCA matrix, on the same strains as well as additional strains with sequenced genomes. MSP’s (spectral profiles) are publically available using CDC’s MicrobeNet—a free, curated reference tool. (https://microbenet.cdc.gov/; see supplemental text). Real-Time Classification was performed using Bruker RTC Software.
Results and discussion
Criteria for determination of species among Elizabethkingia strains
The disadvantages of tDDH, and the need for microbial taxonomy to embrace the use of WGS data for species delineation have been widely discussed (Varghese et al. 2015; Thompson et al. 2015; Auch et al. 2010b; Goris et al. 2007; Moore et al. 2010; Coenye et al. 2005; Schleifer et al. 2015; Rossello-Mora and Amann 2015), and prominent prokaryotic systematists have been calling for the recognition that WGS provides sufficient information for species delimitation (Sutcliffe 2015; Hedlund et al. 2015; Whitman 2015). tDDH hybridization results at 70 °C (Holmes et al. 2013) were compared with results from each of the in silico methods used here (Supplemental Fig. 1, and Table 2). Consistent with previous reports (Meier-Kolthoff et al. 2013), GGDC formula 2 was the most highly correlated with tDDH. Comparing the in silico methods to each other, there was strong correlation between all of the methods, with the exception of GGDC formula 2, which is non-linear (Supplemental Fig. 2). The ANIb 95% cutoff-value for species delimitation (Goris et al. 2007) would therefore be equivalent to a predicted DDH value of slightly less than 65%, lower than the tDDH value of approximately 70% which has long been used for species delimitation (Wayne et al. 1987). The results of ANIb and predicted DDH analysis of all strains, as compared to all other strains, is summarized in Fig. 1. We followed the advice of Christensen et al., that multiple strains be used in describing a species (Christensen et al. 2001).
Table 2.
Traditional DNA–DNA hybridization (tDDH) results from representatives of all Elizabethkingia species (Holmes et al. 2013) correlate with in silico methods
| Method | tDDH at 70 °C | Two-way AAI | GGDC: formula 1 | GGDC: formula 2 | GGDC: formula 3 |
|---|---|---|---|---|---|
| Two-way AAI | 0.641 | ||||
| GGDC: formula 1 | 0.598 | 0.981 | |||
| GGDC: formula 2 | 0.912 | 0.812 | 0.757 | ||
| GGDC: formula 3 | 0.737 | 0.985 | 0.983 | 0.858 | |
| ANIb (Jspecies) | 0.751 | 0.986 | 0.956 | 0.876 | 0.986 |
Of the three formulae used to calculate Genome-to-Genome distance (Auch et al. 2010a), formula 2 was expected to best approximate tDDH results, and our results are an independent confirmation of this
Fig. 1.
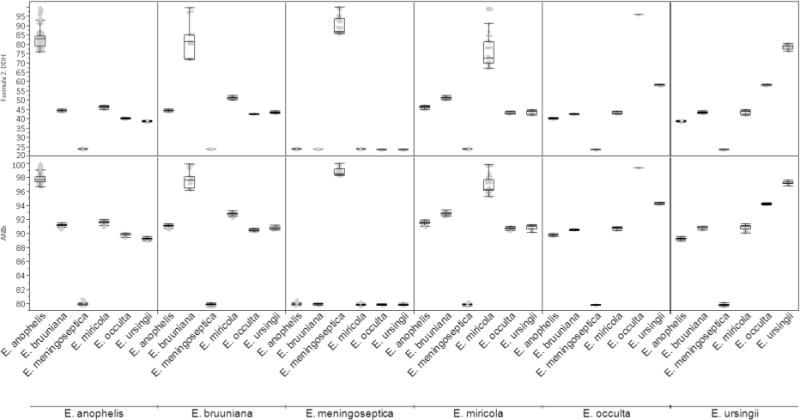
The variability of GGDC predicted DDH (top panel) and ANIb (lower panel) for pairwise comparisons is displayed, categorized based on the taxonomy assignments of each strain as described in this manuscript. Box-plots show the range and median of data for each comparison
WGS contigs have been shown for other species to produce ANIb and GGDC formula 2 predicted DDH (henceforth referred to simply as “predicted DDH”) results indistinguishable from those produced using complete circularized genomes (Richter and Rossello-Mora 2009; Auch et al. 2010a), and we confirmed that this was also the case for Elizabethkingia strains (see Supplemental text); WGS contig sets were used for all subsequent analyses.
Elizabethkingia meningoseptica is phylogenetically distinct from other Elizabethkingia species
Maximum Likelihood analysis of the core genome (Fig. 2) and UPGMA of the optical maps (Fig. 3) show that strains closely related to the E. meningoseptica type strain cluster at the end of a long branch, with E. anophelis strains in a sub-group distinct from the remaining strains. A SNP tree prepared from genomic sequence of all strains (Supplemental Fig. 3) had essentially the same topology as the core genome ML tree. This subdivision into three main phylogenetic groups is consistent with previous genome-based phylogenetic analyses (Breurec et al. 2016; Perrin et al. 2017).
Fig. 2.
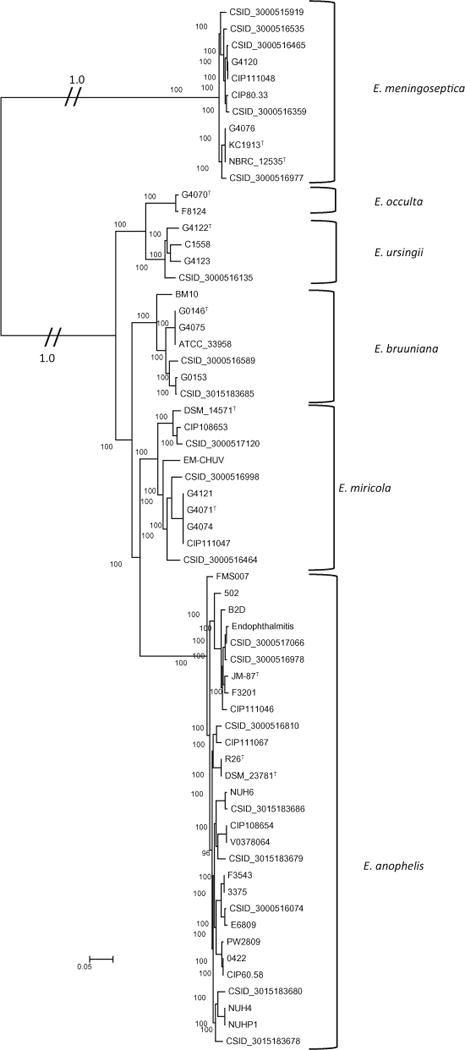
Core genome ML phylogeny of 1,049,915 variable nucleotide sites from 2259 genes. Only bootstrap values ≥70% are displayed
Fig. 3.
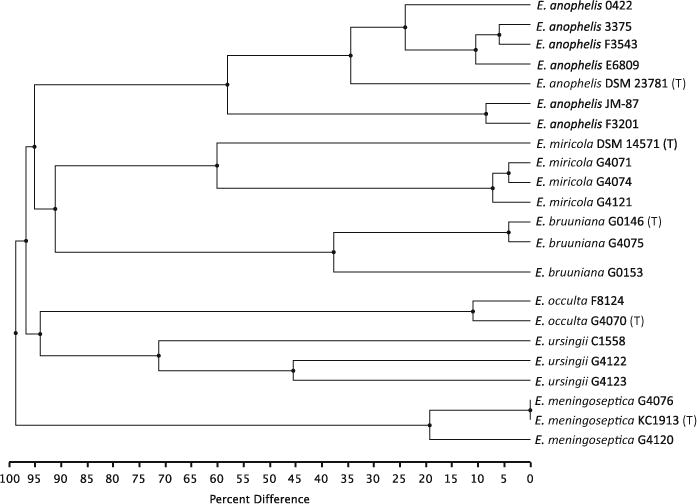
UPGMA based on optical maps. The percentage of restriction sites in common between optical maps of each isolate are indicated under the tree
The relatively large phylogenetic distance between E. meningoseptica strains and strains from other Elizabethkingia species raised the question of whether they really do belong to the same genus.
To examine this, we used the percentage of conserved proteins (POCP), calculated as described by Qin et al. (2014), which described that species in the same genus generally had a POCP value of ≥50%, with inter-species and inter-genus average POCP values varying considerably. The POCP values for pairwise comparisons of each of the type strains discussed in this manuscript are shown in Table 3, and the complete set of POCP values for all isolates can be found in Supplemental Table 1. All of the Elizabethkingia isolates had POCP values ≥88.3% when compared to any other Elizabethkingia isolate, confirming that they are in the same genus.
Table 3.
Percentage of Conserved Proteins (POCP) between type strains of Elizabethkingia anophelis (DSM 23781), Elizabethkingia bruuniana (G0146), Elizabethkingia meningoseptica (KC1913), Elizabethkingia miricola (DSM 14571), Elizabethkingia occulta (G4070), Elizabethkingia ursingii (G4122), Chryseobacterium gleum (ATCC 35910), and Sphingobacterium spiritivorum (ATCC 33861)
| Taxon | Strain | DSM_23781 | G0146 | KC1913 | DSM_14571 | G4070 | G4122 | ATCC_35910 | ATCC_33861 |
|---|---|---|---|---|---|---|---|---|---|
| Elizabethkingia anophelis | DSM_23781 | 100 | 90.9 | 91.7 | 90.9 | 93 | 91.7 | 79.1 | 72.7 |
| Elizabethkingia bruuniana | G0146 | 90.9 | 100 | 90.5 | 92.9 | 92.5 | 93 | 80.1 | 73.8 |
| Elizabethkingia meningoseptica | KC1913 | 91.7 | 90.5 | 100 | 91.1 | 91.8 | 91.8 | 78.1 | 73.9 |
| Elizabethkingia miricola | DSM_14571 | 90.9 | 92.9 | 91.1 | 100 | 92.2 | 93 | 77.9 | 72.7 |
| Elizabethkingia occulta | G4070 | 93 | 92.5 | 91.8 | 92.2 | 100 | 93 | 79.8 | 73.4 |
| Elizabethkingia ursingii | G4122 | 91.7 | 93 | 91.8 | 93 | 93 | 100 | 79.2 | 73.8 |
| Chryseobacterium gleum | ATCC_35910 | 79.1 | 80.1 | 78.1 | 77.9 | 79.8 | 79.2 | 100 | 71.4 |
| Sphingobacterium spiritivorum | ATCC_33861 | 72.7 | 73.8 | 73.9 | 72.7 | 73.4 | 73.8 | 71.4 | 100 |
E. anophelis strains constitute genomospecies 1 and Elizabethkingia miricola strains constitute genomospecies 2
Our earlier report describing WGS data for each genomospecies noted that the genome sequence of the type strain of Elizabethkingia anophelis was consistent with it belonging to genomospecies 1, and that the 16S rRNA gene of JM-87 (the type strain of “E. endophytica”) was identical to that of the historic strain F3201, which tDDH also classified as member of genomospecies 1 (Holmes et al. 2013). This similarity was borne out by comparisons of the whole genome sequences of both strains, which had an ANIb >98.70%, and a predicted DDH of 92%. Strain JM-87 had an ANIb ≥97%, and a predicted DDH ≥88%, compared to all of the genomospecies 1 strains, including the type strain of E. anophelis (DSM 23781), consistent with the recognition (Doijad et al. 2016; Perrin et al. 2017) that strain JM-87 is an Elizabethkingia anophelis strain.
The type strain of E. miricola (DSM 14571T) is most similar to the historic genomospecies 2 strains, with a predicted DDH of 70%, and an ANIb value slightly above 96%. We identified two additional strains that had marked similarity with the E. miricola type strain, and several others that were more closely related to the historic genomospecies 2 strains, as evidenced both by their phylogenetic proximity and their predicted DDH values. Strain EM-CHUV in the public domain (Opota et al. 2016), and strains CSID_3000516464 and CSID_3000516998 from this work, were similarly predicted to be E. miricola by ANIb. Several of the strains that were considered to be E. miricola (based on ANIb of ≥95%) had a predicted DDH of slightly less than 70%. A predicted DDH of at least 65% was determined to be sufficient for inclusion of the strains in the E. miricola species since the 9%.
Proposed nomenclature for the historically recognized genomospecies 3 and 4
Both ANIb and predicted DDH provide quantitative confirmation of the earlier DNA–DNA hybridization results that genomospecies 3 and 4 strains are species that are distinct from E. miricola and from each other. Core genome phylogenetic analysis and a SNP analysis of all strains that were not E. meningoseptica or E. anophelis (Supplemental Fig. 4) produced results consistent with ANIb and predicted DDH. Strain ATCC 33958 from the public domain was found to belong to genomospecies 3, as was strain BM10, by these methods. Modern strains were identified that belong to either genomospecies 3 or 4, as were strains retrieved from the CDC and CIP collections. This large set of well-characterized and historically recognized strains, which could not be provided with validly published names using pre-genomic technology, can now be named based on their complete genome sequences. In recognition of the foundational work done by Jan Ursing and Brita Bruun investigating the genus Elizabethkingia, we propose to name genomospecies 3 as Elizabethkingia bruuniana sp. nov. (bruun.i.a’na. N.L. fem. adj. bruuniana, named in honour of Brita Bruun), and genomospecies 4 as Elizabethkingia ursingii sp. nov. (ur.sing’i.i. N.L. gen. n. ursingii, of Ursing, named in honour of Jan Ursing).
A third novel Elizabethkingia species is proposed as Elizabethkingia occulta
The strain G4070 had been originally identified as belonging to genomospecies 4, but its optical map showed that it was unlike the other genomospecies 4 strains, and both ANIb and GGDC put it outside of that genomospecies. This suggested that strain G4070 was not actually a member of genomospecies 4, but instead a representative of its own novel genomospecies. MALDI-TOF mass spectra of strains from the CDC strain collection were reviewed to locate strains with spectra similar to G4070, and a subset of these had their rpoB sequenced. Strain F8124 was thereby identified as potentially belonging to the same genomospecies as G4070, and this similarity was confirmed by whole genome sequencing with an ANIb of 99.4% and a predicted DDH of 96. Strain F8124 (CL50/86 = CCUG 15909 = GIFU 2120) had been originally published as a founding member of the combination Sphingobacterium mizutae (Yabuuchi et al. 1983) due to its phenotypic similarities. Later studies based on tDDH showed that it was distinct from S. mizutae strains and it was instead assigned to the F. meningosepticum species (Holmes et al. 1988), but not included in the experiments that delineated the Elizabethkingia genomospecies. Here we propose that both strains belong to a novel species that we name Elizabethkingia occulta sp. nov. (oc.cul’ta. L. fem. adj. occulta hidden), to reflect that it was hiding in plain sight.
Elizabethkingia species identification by MALDI-TOF mass spectrometry and target gene sequencing
Using an expanded spectrum database (https://microbenet.cdc.gov/), analysis by MALDI-TOF mass spectrometry can reliably identify E. anophelis and E. meningoseptica, but cannot distinguish between the remaining species. 274 Elizabethkingia strains from the CDC collection of mostly clinical isolates were analyzed using MALDI-TOF mass spectrometry, and only 23 (8%) were found to be E. meningoseptica. 210 (71%) were E. anophelis, and 41 (14%) were one of the other Elizabethkingia species.
Both the 16S rRNA and rpoB genes were evaluated as target genes for Elizabethkingia species identification. The five copies of the 16S rRNA gene present in all Elizabethkingia genomes can be quite different from each other. The most extreme example of this among the strains described here was E. ursingii strain G4123, which contained three distinct variants of the 16S rRNA gene, one being most similar to all five from the E. ursingii type strain, two being most similar to E. bruuniana strains, and two matching each other but otherwise unique. This confounds the use of 16S rRNA sequence comparisons for Elizabethkingia species identification.
A Maximum Likelihood tree was generated based on the single-copy rpoB gene sequence for all strains (Fig. 4). E. meningoseptica strains clustered separately from other Elizabethkingia strains, and all E. anophelis strains clustered within a distinct subgroup of the remaining strains, consistent with the core-genome phylogeny. The topology of the section of the rpoB tree containing all strains except those assigned to E. meningoseptica or E. anophelis is consistent with the species designations determined by whole genome sequencing, despite E. bruuniana strains forming two separate clusters and a slightly ambiguous positioning of E. miricola strain EM-CHUV. A laboratory that has only Sanger sequencing capacity should now be able to correctly identify Elizabethkingia strains to the species level by constructing a phylogenetic tree based on this alignment (available in the supplemental material) with their rpoB sequences included. This confirms the power of gene sequencing for Elizabethkingia species identification (Breurec et al. 2016) and adds rpoB gene sequencing to existing molecular identification methods (http://bigsdb.pasteur.fr/elizabethkingia).
Fig. 4.
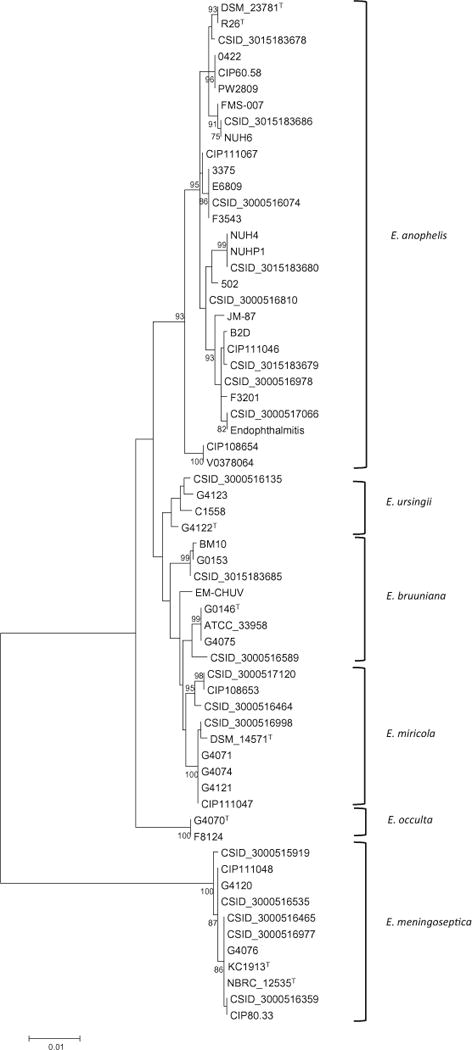
Molecular phylogenetic analysis of positions 1939-3629 from the rpoB gene. The evolutionary history was inferred by using the Maximum Likelihood method based on the JC69 model. The tree with the highest log likelihood is shown, and the percentage of trees in which the associated taxa clustered together is shown next to the branches, based on 100 bootstrap replicates. Bootstrap values greater than 70% are displayed. Branch lengths show the number of substitutions per site. The analysis involved 63 nucleotide sequences, and gaps were eliminated, yielding a total of 1690 nucleotide positions
Phenotypic testing cannot reliably distinguish Elizabethkingia strains
As Ursing and Bruun reported (Bruun and Ursing 1987), there is a great deal of phenotypic variability among Elizabethkingia strains, even those belonging to the same genomospecies. Results of our testing of the historic strains are shown in Supplemental Table 2. When the Elizabethkingia genus was first published, urease was the only biochemical test that was consistently different between E. meningoseptica strains (negative) and the type strain of E. miricola (positive), but our tests show that all of the genomo-species 1 strains and some of the genomospecies 3 and 4 strains were also positive for urease, while genomo-species 2 strains G4071 and G4121 were negative. Similarly, “E. endophytica” was described as being negative for acid production from cellobiose as compared to E. anophelis, which was positive. While we confirmed these results for the strains described, three of the four genomospecies 1 strains most closely related to E. anophelis strain R26 were negative in the cellobiose assay, while the strain that was most closely related to the “E. endophytica” strain JM-87 (F3201) was positive. We did not perform cellular fatty acid testing on these strains, as all Elizabethkingia have been previously described as having polar lipid profiles that are very similar to each other, and to species of the genus Chryseobacterium (Kampfer et al. 2015).
For many decades, CDC’s Special Bacteriology Reference Laboratory performed a series of standard phenotypic tests on all strains that were added to the collection, and these results were reviewed in hope of discovering any consistent phenotypic difference between the three groups (E. meningoseptica, E. anophelis, and all others) that could be distinguished by MALDI-TOF mass spectrometry. Supplemental Table 3 summarizes this review, showing that they cannot be reliably distinguished by phenotype alone. Certain characteristics were found to be almost entirely strain-dependent (variable within a species), and not at all useful in species determination; these were urease, acid production from lactose, and growth on Simmons’ citrate (citrate as a sole carbon source).
Although is it likely that labs will rely on DNA sequence analysis and/or MALDI-TOF mass spectrometry when classifying Elizabethkingia strains, phenotypic descriptions have been published for Elizabethkingia species previously. We have combined all available information to update the existing descriptions, and provide the traditional phenotypic descriptions of the newly named species.
Emended description of the genus Elizabethkingia
Elizabethkingia (E.liz.a.beth.kin′gi.a. N.L. fem. n. Elizabethkingia in honour of Elizabeth O. King, who first described the bacteria associated with infant meningitis as [Flavobacterium] meningosepticum in 1959).
Cells are Gram-negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.5 μm). Good growth is observed on TSA and nutrient agar at 28–37 °C, but no growth is observed at 5 or 42 °C. Colonies are white or yellow, semi-translucent, circular and shiny with entire edges. Catalase, phosphatase and β-galactosidase activities are positive. Indole is produced. Casein is hydrolysed, but starch is not. Malonate is not utilized. Acid is not produced from galactose, melezitose, raffinose, adonitol, dulcitol, sorbitol, or inositol, or salicin. The fatty acid profile consists largely of 15 : 0 iso, 17 : 0 iso 3-OH and summed feature 4 (15 : 0 iso 2-OH and/or 16 : 1 ω7 c/t). Menaquinone MK-6 is the predominant quinone. The G+C content of the DNA is 35.0–38.2 mol%.
The type species is E. meningoseptica. This emended genus description is represented in the Digital Protologue by taxon number GA00018.
Emended description of Elizabethkingia anophelis
Elizabethkingia anophelis (a.no.phe′lis. N.L. gen. n. anophelis of/from a mosquito of the genus Anopheles, as the type strain was isolated from the midgut of Anopheles gambiae).
Cells are aerobic Gram-reaction-negative, non-motile, non-spore-forming rods, approximately 1 μm in width and 2 μm in length. Catalase-positive. Good growth occurs after 48 h on NA, R2A agar and TSA (all Oxoid) at 11–36 °C. Growth on MacConkey agar (Oxoid) at 28 °C is strain-dependent. Unable to grow at temperatures below 10 °C or above 37 °C. Two growth optima are detected on LB medium: 30–31 °C with a doubling time of 50 min; and 37 °C with a doubling time of 42 min. Colonies on NA are smooth, yellowish, circular, translucent and shiny with entire edges. The non-diffusible and non-fluorescent yellow pigment is not of the flexirubin-type (KOH test-negative). Resistant to a number of antibiotics; MICs in LB medium are >400 μg ml−1 for ampicillin, >250 μg ml−1 for kanamycin, >250 μg ml−1 for streptomycin, >30 μg ml−1 for chloramphenicol and >10 μg ml−1 for tetracycline. Indole is produced. Acid is produced from trehalose. No acid is produced from adonitol, D-arabitol, dulcitol, erythritol, i-inositol, methyl α- D-glucoside, raffinose, salicin, or D-sorbitol. Acid production from D-melibiose, D-cellobiose, D-glucose, lactose, D-mannitol, maltose, D-mannitol, D-xylose, lactose, sucrose and arabinose is variable. Indole production from tryptophan and β-galactosidase activity (ONPG) are positive. Aesculin hydrolysis, nitrate reduction, urease and oxidase activity is variable; Hydrolysis of casein, starch, DNA and tyrosine, activity of arginine dihydrolase, lysine decarboxylase and ornithine decarboxylase, and utilization of malonate are negative. Hydrogen sulfide and gelatinase production is strain dependent. The following compounds are not utilized as sole sources of carbon: N-acetyl-D-galactosamine, N-acetyl-D-glucosamine, L-arabinose, L-arbutin, cellobiose, D-fructose, D-glucose, maltose, D-galactose, gluconate, glycerol, D-mannose, D-mannitol, maltitol, α-melibiose, L-rhamnose, D-ribose, sucrose, salicin, trehalose, D-xylose, adonitol, inositol, D-sorbitol, putrescine, acetate, propionate, cis-aconitate, trans-aconitate, 4-aminobutyrate, adipate, azelate, fumarate, glutarate, DL-3-hydroxybutyrate, itaconate, DL-lactate, 2-oxoglutarate, pyruvate, suberate, mesaconate, L-alanine, β-alanine, L-ornithine, L-phenylalanine, L-serine, L-aspartate, L-histidine, L-leucine, L-proline, L-tryptophan, 3-hydroxybenzoate, 4-hydroxybenzoate and phenylacetate. Utilization of citrate as the sole source of carbon is strain-dependent. The chromogenic substrates p-nitrophenyl (pNP)-β-D-glucopyranoside, pNP-β-D-galactopyranoside, pNP-α-D-glucopyranoside, bis-pNP-phosphate, bis-pNP-phenyl-phosphonate, bis-pNP-phosphoryl-choline, 2-deoxythymidine-2′-pNP-phosphate, L-alanine-p-nitroanilide (pNA), γ-L-glutamate-pNA and L-proline-pNA are hydrolysed but not pNP-β-D-xylopyranoside or pNP-β-D-glucuronide. Major cellular fatty acids are iso-C15: 0, iso-C17: 0 3-OH and summed feature 4 (iso-C15: 0 2-OH and/or C16: 1ω7 c/t). The only menaquinone is MK-6. The major polar lipids are diphosphatidylglycerol, phosphatidylinositol, a characteristic unknown phospholipid, and unknown polar lipids and glycolipids.
The type strain is R26T (= CCUG 60038T = CCM 7804T), isolated from the midgut of Anopheles gambiae G3, originating from McCarthy Island, The Gambia, and deposited by Dr William Collins at Malaria Research Reference Resource Centre. This emended species description is represented in the Digital protologue by taxon number TA00064.
Emended description of Elizabethkingia meningoseptica
Elizabethkingia meningoseptica (me.nin.go.sep′ti.ca. Gr. n. meninx, meningos meninges, membrane covering the brain; Gr. adj. septikos putrefactive; N.L. fem. adj. meningoseptica apparently referring to association of the bacterium with both meningitis and septicaemia, but not septic meningitis as the name implies).
Basonym Flavobacterium meningosepticum King (1959) (Approved Lists 1980).
Cells are Gram-negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.0 μm). Growth on MacConkey agar is strain-dependent. Oxidase, gelatinase, H2S and indole are produced. Aesculin is hydrolyzed. Acid is produced from ethanol, D-glucose, glycerol, lactose, D-maltose, D-mannitol and trehalose, but not from L-arabinose, D-cellobiose, raffinose, sucrose, salicin or rhamnose, D-xylose. Urea hydrolysis, use of citrate as a sole carbon source, and acid production from fructose is strain-dependent. The fatty acid profile consists largely of 15 : 0 iso (43.9 ± 2.0 %), 17 : 0 iso 3-OH (14.6 ± 1.0 %) and summed feature 4 (15 : 0 iso 2-OH and/or 16 : 1 ω7 c/t; 19.6 ± 1.0%). The G+C content of the DNA is 37.2 ± 0.6 mol% (37.1 mol% for the type strain).
The type strain is ATCC 13253T (= KC1913T = NCTC 10016T = LMG 12279T = CCUG 214T). This emended species description is represented in the Digital protologue by Taxon Number TA00060.
Emended description of Elizabethkingia miricola
Elizabethkingia miricola [mi.ri′co.la. N.L. neut. n. mirum derived from mir (peace) (name of Russian space station); L. suff.—cola from L. masc. or fem. n. incola inhabitant; N.L. masc. or fem. n. miricola inhabitant of the Mir space station].
Basonym Chryseobacterium miricola Li et al. (2003).
Cells are Gram-negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.5 μm). Hydrolysis of urea and gelatin, nitrate reduction, growth on MacConkey agar, H2S production and use of citrate as a sole carbon source are strain dependent. Indole is produced. Aesculin and oxidase are positive. Acid is produced from D-fructose, D-mannitol and trehalose, but not from L-arabinose, raffinose, sucrose, salicin, rhamnose, or D-xylose. Acid production from D-glucose, lactose, D-maltose, and D-cellobiose is strain dependent. The fatty acid profile consists largely of 15 : 0 iso (46.4 ± 2.2 %), 17 : 0 iso 3-OH (15.3 ± 0.2%) and summed feature 4 (15 : 0 iso 2-OH and/or 16 : 1 ω7 c/t, 17.0 ± 1.3%). The G+C content of the DNA is 35.3 ± 0.3 mol% (35.0 mol% for the type strain).
The type strain is DSM 14571T (=JCM 11413T = GTC 862T). This emended species description is represented in the Digital protologue by Taxon Number TA00061.
Description of Elizabethkingia bruuniana sp. nov
Elizabethkingia bruuniana (bruun.i.a’na. N.L. fem. adj. bruuniana, named in honour of Brita Bruun).
Cells are Gram-stain negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.5 μm). Good growth is observed on TSA and nutrient agar at 28–37 °C, but no growth is observed at 5 or 42 °C. Colonies are white or yellow, semi-translucent, circular and shiny with entire edges. Catalase, phosphatase, gelatinase, and β-galactosidase activities are positive. Nitrate is not produced. Oxidase, aesculin, H2S production, and ability to use citrate as a sole carbon source are strain-dependent. Indole is produced. Casein is hydrolysed, but starch is not. Malonate is not utilized. Acid is produced from D-mannitol, glucose, and maltose but not produced from arabinose, lactose, rhamnose, sucrose, xylose, galactose, melezitose, raffinose, sucrose, adonitol, dulcitol, sorbitol, inositol, or salicin. Acid production from cellobiose, fructose, mannitol, and trehalose is strain-dependent.
The type strain is G0146T (= DSM 2975T = CCUG 69503T = CIP 111191T). The species description is represented in the Digital protologue by taxon number TA00058.
Description of Elizabethkingia ursingii sp. nov
Elizabethkingia ursingii (ur.sing’i.i. N.L. gen. n. ursingii, of Ursing, named in honour of Jan Ursing).
Cells are Gram-stain negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.5 μm). Good growth is observed on TSA and nutrient agar at 28–37 °C, but no growth is observed at 5 or 42 °C. Colonies are white or yellow, semi-translucent, circular and shiny with entire edges. Aesculin, oxidase, catalase, phosphatase and β-galactosidase activities are positive. H2S and Indole are produced. Casein is hydrolysed, but starch is not. Malonate is not utilized and nitrate is not reduced. Gelatinase, urease activity, use of citrate as the sole carbon source and growth on MacConkey agar are strain dependent. Acid is produced from fructose, glucose, maltose, and mannitol but not produced from cellobiose, rhamnose, sucrose, xylose, galactose, melezitose, raffinose, sucrose, adonitol, dulcitol, sorbitol, or inositol, or salicin. Acid production from lactose is strain dependent.
The type strain is G4122T (=DSM 2974T = CCUG 69496T = CIP 111192T). The species description is represented in the Digital protologue by Taxon Number TA00059.
Description of Elizabethkingia occulta sp. nov
Elizabethkingia occulta (oc.cul’ta. L. fem. adj. occulta hidden, to reflect that it was hiding in plain sight, and had been previously masquerading as Sphingobacterium mizutae or Elizabethkingia genomospecies 4).
Cells are Gram-stain negative, non-motile, non-spore-forming rods (0.5 × 1.0–2.5 μm). Good growth is observed on MacConkey agar, TSA and nutrient agar at 28–37 °C, but no growth is observed at 5 or 42 °C. Colonies are white or yellow, semi-translucent, circular and shiny with entire edges. Aesculin, catalase, oxidase, phosphatase, urease, and β-galactosidase activities are positive. Indole is produced and nitrate is reduced. Casein is hydrolysed, but gelatin and starch are not. Malonate is not utilized, and citrate cannot be used as the sole carbon source. H2S production is strain-dependent. Acid is produced from cellobiose, glucose, lactose, maltose, mannitol, and trehalose, but not produced from arabinose, fructose, rhamnose, galactose, melezitose, raffinose, sucrose, adonitol, dulcitol, sorbitol, or inositol, or salicin. Acid production from xylose is strain-dependent.
The type strain is G4070T (= DSM 2976T = CCUG 69505T = CIP 111193T). The species description is represented in the Digital protologue by Taxon Number TA00062.
Supplementary Material
Acknowledgments
We thank John McInroy for providing strain JM-87, and the State Health Departments of Wisconsin, Minnesota, Illinois, Michigan, Florida, Arizona, Texas, South Carolina, and California in the USA for providing Elizabethkingia clinical specimens. We also thank Aharon Oren for nomenclature advice, Barry Holmes for supplying phenotypic data on some of the strains included in this study, and Aaron Villarma for technical assistance.
Funding CDC research was supported by the Advanced Molecular Detection (AMD) initiative, and work done at the Institut Pastuer was funded by the French government’s Investissement d’Avenir program Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (Grant ANR-10-LABX-62-IBEID).
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Electronic supplementary material The online version of this article (doi:10.1007/s10482-017-0926-3) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest All authors report that they have no conflicts of interest.
Contributor Information
Ainsley C. Nicholson, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA
Christopher A. Gulvik, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA
Anne M. Whitney, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA
Ben W. Humrighouse, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA
James Graziano, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Brian Emery, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Melissa Bell, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Vladimir Loparev, Division of Scientific Resources, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Phalasy Juieng, Division of Scientific Resources, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Jarrett Gartin, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA.
Chantal Bizet, Microbiology Department, Institut Pasteur, Collection de L’Institut Pasteur (CIP), Paris, France.
Dominique Clermont, Microbiology Department, Institut Pasteur, Collection de L’Institut Pasteur (CIP), Paris, France.
Alexis Criscuolo, Institut Pasteur – Bioinformatics and Biostatistics Hub – C3BI, USR 3756 IP CNRS, Paris, France.
Sylvain Brisse, Microbial Evolutionary Genomics, Institut Pasteur, Paris, France; CNRS, UMR 3525, Paris, France; Institut Pasteur, Biodiversity and Epidemiology of Bacterial Pathogens, Paris, France.
John R. McQuiston, Special Bacteriology Reference Laboratory, Bacterial Special Pathogens Branch, Division of High Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, Klenk HP, Goker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Gen Sci. 2010a;2(1):142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk HP, Goker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Gen Sci. 2010b;2(1):117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine. 1997;76(1):30–41. doi: 10.1097/00005792-199701000-00003. [DOI] [PubMed] [Google Scholar]
- Breurec S, Criscuolo A, Diancourt L, Rendueles O, Vanden-bogaert M, Passet V, Caro V, Rocha EP, Touchon M, Brisse S. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci Rep. 2016;6:30379. doi: 10.1038/srep30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun B. Studies on a collection of strains of the genus Flavobacterium. 1. Biochemical studies. Acta Pathol Microbiol Immunol Scand. 1982;90(6):415–421. doi: 10.1111/j.1699-0463.1982.tb00140.x. [DOI] [PubMed] [Google Scholar]
- Bruun B, Ursing J. Phenotypic characterization of Flavobacterium meningosepticum strains identified by DNA-DNA hybridization. Acta Pathol Microbiol Immunol Scand. 1987;95(1):41–47. doi: 10.1111/j.1699-0463.1987.tb03085.x. [DOI] [PubMed] [Google Scholar]
- Christensen H, Bisgaard M, Frederiksen W, Mutters R, Kuhnert P, Olsen JE. Is characterization of a single isolate sufficient for valid publication of a new genus or species? Proposal to modify recommendation 30b of the Bacteriological Code (1990 Revision) Int J Syst Evol Microbiol. 2001;51(Pt 6):2221–2225. doi: 10.1099/00207713-51-6-2221. [DOI] [PubMed] [Google Scholar]
- Coenye T, Gevers D, Van de Peer Y, Vandamme P, Swings J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol Rev. 2005;29(2):147–167. doi: 10.1016/j.femsre.2004.11.004. [DOI] [PubMed] [Google Scholar]
- JGI Bacterial DNA Isolation CTAB Protocol. http://1ofdmq2n8tc36m6i46scovo2e.wpengine.netdna-cdn.com/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf. http://1ofdmq2n8tc36m6i46scovo2e.wpengine.netdna-cdn.com/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf.
- Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102(5–6):500–506. doi: 10.1016/j.ygeno.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, Fenton J, Fenzl T, Fish J, Garcia-Gutierrez L, Garland P, Gluck J, Gonzalez I, Guermond S, Guo J, Gupta A, Herr JR, Howe A, Hyer A, Harpfer A, Irber L, Kidd R, Lin D, Lippi J, Mansour T, McA’Nulty P, McDonald E, Mizzi J, Murray KD, Nahum JR, Nanlohy K, Nederbragt AJ, Ortiz-Zuazaga H, Ory J, Pell J, Pepe-Ranney C, Russ ZN, Schwarz E, Scott C, Seaman J, Sievert S, Simpson J, Skennerton CT, Spencer J, Srinivasan R, Standage D, Stapleton JA, Steinman SR, Stein J, Taylor B, Trimble W, Wiencko HL, Wright M, Wyss B, Zhang Q, Zyme E, Brown CT. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva PS, Pereira GH. Elizabethkingia meningoseptica: emergent bacteria causing pneumonia in a critically ill child. Pediatr Int. 2013;55(2):231–234. doi: 10.1111/j.1442-200X.2012.03650.x. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doijad S, Ghosh H, Glaeser S, Kampfer P, Chakraborty T. Taxonomic reassessment of the genus Elizabethkingia using whole genome sequencing: Elizabethkingia endophytica Kampfer et al. 2015 is a later subjective synonym of Elizabethkingia anophelis Kampfer et al. 2011. Int J Syst Evol Microbiol. 2016 doi: 10.1099/ijsem.0.001390. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Green O, Murray P, Gea-Banacloche JC. Sepsis caused by Elizabethkingia miricola successfully treated with tigecycline and levofloxacin. Diagn Microbiol Infect Dis. 2008;62(4):430–432. doi: 10.1016/j.diagmicrobio.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hayek SS, Abd TT, Cribbs SK, Anderson AM, Melendez A, Kobayashi M, Polito C, Wayne Wang YF. Rare Elizabethkingia meningosepticum meningitis case in an immunocompetent adult. Emerg Microbes Infect. 2013;2(4):e17. doi: 10.1038/emi.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund BP, Dodsworth JA, Staley JT. The changing landscape of microbial biodiversity exploration and its implications for systematics. Syst Appl Microbiol. 2015;38(4):231–236. doi: 10.1016/j.syapm.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Holmes B, Weaver RE, Steigerwalt AG, Brenner DJ. A Taxonomic Study of Flavobacterium spiritivorum and Sphingobacterium mizutae: proposal of Flavobacterium yabuuchiae sp. nov. and Flavobacterium mizutaii comb. nov. Int J Syst Bacteriol. 1988;38(4):348–353. doi: 10.1099/00207713-38-4-348. [DOI] [Google Scholar]
- Holmes B, Steigerwalt AG, Nicholson AC. DNA-DNA hybridization study of strains of Chryseobacterium, Elizabethkingia and Empedobacter and of other usually indole-producing non-fermenters of CDC groups IIc, IIe, IIh and IIi, mostly from human clinical sources, and proposals of Chryseobacterium bernardetii sp. nov., Chryseobacterium carnis sp. nov., Chryseobacterium lactis sp. nov., Chryseobacterium nakagawai sp. nov. and Chryseobacterium taklimakanense comb. nov. Int J Syst Evol Microbiol. 2013;63(Pt 12):4639–4662. doi: 10.1099/ijs.0.054353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MS, Liao CH, Huang YT, Liu CY, Yang CJ, Kao KL, Hsueh PR. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999–2006. Eur J Clin Microbiol Infect Dis. 2011;30(10):1271–1278. doi: 10.1007/s10096-011-1223-0. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61(Pt 11):2670–2675. doi: 10.1099/ijs.0.026393-0. [DOI] [PubMed] [Google Scholar]
- Kampfer P, Busse HJ, McInroy JA, Glaeser SP. Elizabethkingia endophytica sp. nov., isolated from Zea mays and emended description of Elizabethkingia anophelis. Kampfer et al. 2011. Int J Syst Evol Microbiol. 2015;65(7):2187–2193. doi: 10.1099/ijs.0.000236. [DOI] [PubMed] [Google Scholar]
- Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol. 2005;55(Pt 3):1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- King EO. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am J Clin Pathol. 1959;31(3):241–247. doi: 10.1093/ajcp/31.3.241. [DOI] [PubMed] [Google Scholar]
- Lau SK, Chow WN, Foo CH, Curreem SO, Lo GC, Teng JL, Chen JH, Ng RH, Wu AK, Cheung IY, Chau SK, Lung DC, Lee RA, Tse CW, Fung KS, Que TL, Woo PC. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep. 2016;6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Chen PL, Wang LR, Lee HC, Chang CM, Lee NY, Wu CJ, Shih HI, Ko WC. Fatal case of community-acquired bacteremia and necrotizing fasciitis caused by Chryseobacterium meningosepticum: case report and review of the literature. J Clin Microbiol. 2006;44(3):1181–1183. doi: 10.1128/JCM.44.3.1181-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kawamura Y, Fujiwara N, Naka T, Liu H, Huang X, Kobayashi K, Ezaki T. Chryseobacterium miricola sp. nov., a novel species isolated from condensation water of space station Mir. Syst Appl Microbiol. 2003;26(4):523–528. doi: 10.1078/072320203770865828. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schroder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013;29(3):308–315. doi: 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102(30):10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore ER, Mihaylova SA, Vandamme P, Krichevsky MI, Dijkshoorn L. Microbial systematics and taxonomy: relevance for a microbial commons. Res Microbiol. 2010;161(6):430–438. doi: 10.1016/j.resmic.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Nicholson AC, Humrighouse BW, Graziano JC, Emery B, McQuiston JR. Draft genome sequences of strains representing each of the Elizabethkingia genomospecies previously determined by DNA-DNA hybridization. Genome Announc. 2016 doi: 10.1128/genomeA.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opota O, Diene SM, Bertelli C, Prod’hom G, Eckert P, Greub G. Genome of the carbapenemase-producing clinical isolate Elizabethkingia miricola EM_CHUV and comparative genomics with Elizabethkingia meningoseptica and Elizabethkingia anophelis: evidence for intrinsic multidrug resistance trait of emerging pathogens. Int J Antimicrob Agents. 2016 doi: 10.1016/j.ijantimicag.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol. 2010;17(3):337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Perrin A, Larsonneur E, Nicholson AC, Edwards DJ, Gundlach KM, Whitney AM, Gulvik CA, Bell ME, Rendueles O, Cury J, Hugon P, Clermont D, Enouf V, Loparev V, Juieng P, Monson T, Warshauer D, Elbadawi LI, Walters MS, Crist MB, Noble-Wang J, Borlaug G, Rocha EPC, Criscuolo A, Touchon M, Davis JP, Holt KE, McQuiston JR, Brisse S. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun. 2017;8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196(12):2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Razonable RR. Elizabethkingia species sepsis after lung transplantation: case report and literature review. Transpl Infect Dis. 2013;15(6):E229–E234. doi: 10.1111/tid.12146. [DOI] [PubMed] [Google Scholar]
- Ratnamani MS, Rao R. Elizabethkingia meningoseptica: emerging nosocomial pathogen in bedside hemodialysis patients. Indian J Crit Care Med. 2013;17(5):304–307. doi: 10.4103/0972-5229.120323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossello-Mora R, Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015;38(4):209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Amann R, Rossello-Mora R. Taxonomy in the age of genomics. Introduction Syst Appl Microbiol. 2015;38(4):207–208. doi: 10.1016/j.syapm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Shewmaker PL, Steigerwalt AG, Nicholson AC, Carvalho Mda G, Facklam RR, Whitney AM, Teixeira LM. Reevaluation of the taxonomic status of recently described species of Enterococcus: evidence that E. thailandicus is a senior subjective synonym of “E. sanguinicola” and confirmation of E. caccae as a species distinct from E. silesiacus. J Clin Microbiol. 2011;49(7):2676–2679. doi: 10.1128/JCM.00399-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe IC. Challenging the anthropocentric emphasis on phenotypic testing in prokaryotic species descriptions: rip it up and start again. Front Genet. 2015;6:218. doi: 10.3389/fgene.2015.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J, Tan SY, Liu Y, Tay M, Ding Y, Li Y, Kjelleberg S, Givskov M, Lin RT, Yang L. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol Evol. 2014;6(5):1158–1165. doi: 10.1093/gbe/evu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Amaral GR, Campeao M, Edwards RA, Polz MF, Dutilh BE, Ussery DW, Sawabe T, Swings J, Thompson FL. Microbial taxonomy in the post-genomic era: rebuilding from scratch? Arch Microbiol. 2015;197(3):359–370. doi: 10.1007/s00203-014-1071-2. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/PREACCEPT-2573980311437212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursing J, Bruun B. Genetic heterogeneity of Flavobacterium meningosepticum demonstrated by DNA-DNA hybridization. Acta Pathol Microbiol Immunol Scand. 1987;95(1):33–39. doi: 10.1111/j.1699-0463.1987.tb03084.x. [DOI] [PubMed] [Google Scholar]
- Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43(14):6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PA, Kandler O, Krichevsky MI, Moore LH, Moore WE, Murray R, Stackebrandt ES, Starr MP, Truper HG. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- Whitman WB. Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol. 2015;38(4):217–222. doi: 10.1016/j.syapm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Yabuuchi E, Kaneko T, Yano I, Moss CW, Miyoshi N. Sphingobacterium gen. nov., Sphingobacterium spiritivorum comb. nov., Sphingobacterium multivorum comb. nov., Sphingobacterium mizutae sp. nov., and Flavobacterium indologenes sp. nov.: glucose-Nonfermenting Gram-Negative Rods in CDC Groups IIK-2 and IIb. Int J Syst Bacteriol. 1983;33(3):580–598. doi: 10.1099/00207713-33-3-580. [DOI] [Google Scholar]
- Young SM, Lingam G, Tambyah PA. Elizabethkingia Meningoseptica Engodenous Endophthalmitis—a case report. Antimicrob Resist Infect Control. 2014;3(1):35. doi: 10.1186/2047-2994-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


