Abstract
Trichothecenes are a family of terpenoid toxins produced by multiple genera of fungi, including plant and insect pathogens. Some trichothecenes produced by the fungus Fusarium are among the mycotoxins of greatest concern to food and feed safety because of their toxicity and frequent occurrence in cereal crops, and trichothecene production contributes to pathogenesis of some Fusarium species on plants. Collectively, fungi produce over 150 trichothecene analogs: i.e., molecules that share the same core structure but differ in patterns of substituents attached to the core structure. Here, we carried out genomic, phylogenetic, gene-function, and analytical chemistry studies of strains from nine fungal genera to identify genetic variation responsible for trichothecene structural diversity and to gain insight into evolutionary processes that have contributed to the variation. The results indicate that structural diversity has resulted from gain, loss, and functional changes of trichothecene biosynthetic (TRI) genes. The results also indicate that the presence of some substituents has arisen independently in different fungi by gain of different genes with the same function. Variation in TRI gene duplication and number of TRI loci was also observed among the fungi examined, but there was no evidence that such genetic differences have contributed to trichothecene structural variation. We also inferred ancestral states of the TRI cluster and trichothecene biosynthetic pathway, and proposed scenarios for changes in trichothecene structures during divergence of TRI cluster homologs. Together, our findings provide insight into evolutionary processes responsible for structural diversification of toxins produced by pathogenic fungi.
Author summary
Toxins produced by pathogens can contribute to infection and/or colonization of hosts. Some toxins consist of a family of metabolites with similar but distinct chemical structures. This structural variation can affect biological activity, which in turn likely contributes to adaptation to different environments, including to different hosts. Trichothecene toxins consist of over 150 structurally distinct molecules produced by certain fungi, including some plant and insect pathogens. In multiple systems that have been examined, trichothecenes contribute to pathogenesis on plants. To elucidate the evolutionary processes that have given rise to trichothecene structural variation, we conducted comparative analyses of nine fungal genera, most of which produce different trichothecene structures. Using genomic, molecular biology, phylogenetic, and analytical chemistry approaches, we obtained evidence that trichothecene structural variation has arisen primarily from gain, loss, and functional changes of trichothecene biosynthetic genes. Our results also indicate that some structural changes have arisen independently in different fungi. Our findings provide insight into genetic and biochemical changes that can occur in toxin biosynthetic pathways as fungi with the pathways adapt to different environmental conditions.
Introduction
Secondary metabolites (SMs) are low-molecular-weight metabolites that are not required for growth or development, but instead provide ecological advantages under certain environmental conditions. Microbial SMs are diverse in chemical structure and biological activity; some are toxins, plant hormones, pigments, or antibiotics, and some have pharmaceutical properties. Many SMs contribute to host-pathogen interactions. Despite their structural diversity, most microbial SMs are derived from one of three classes of parent compounds: non-ribosomal peptides, polyketides, and terpenes [1]. SM structural diversity results from functional variation in enzymes that synthesize the parent compounds (i.e., non-ribosomal peptide synthetases, and polyketide and terpene synthases) as well as enzymes that catalyze modifications of the parent compound. The latter enzymes include acyltransferases, amino transferases, dehydrogenases, reductases, dioxygenases, methyltransferases, monooxygenases, and prenyltransferases. In fungi, genes encoding enzymes required for synthesis of the same SM are typically located adjacent to one another in a biosynthetic gene cluster [2]. Such clusters can also encode transport proteins that export SMs from cells, and transcription factors that activate expression of cluster genes. SMs often consist of families of analogs that share a core structure, but vary in the presence of substituents (functional groups) attached to the core structure. Structural variation among analogs of the same SM family typically results from the presence, absence, or differences in function of genes encoding modifying enzymes [2,3].
Trichothecenes are a family of toxic SMs produced by some, but not all, species in multiple fungal genera, including Fusarium, Isaria, Microcyclospora, Myrothecium, Peltaster, Spicellum, Stachybotrys, Trichoderma, and Trichothecium [4–8]. Most known trichothecene-producing fungi are plant pathogens, and one, Isaria tenuipes, is an insect pathogen [5]. In Fusarium, trichothecene production contributes to pathogenesis on multiple crop plants [9–11], and some Fusarium trichothecenes are among the mycotoxins of greatest concern to food and feed safety [12]. In addition, Stachybotrys trichothecenes have been implicated in negative health effects of mold growth in damp buildings [13]. In contrast, trichothecene production by Trichoderma arundinaceum contributes to its biological control activity against some plant pathogenic fungi [14].
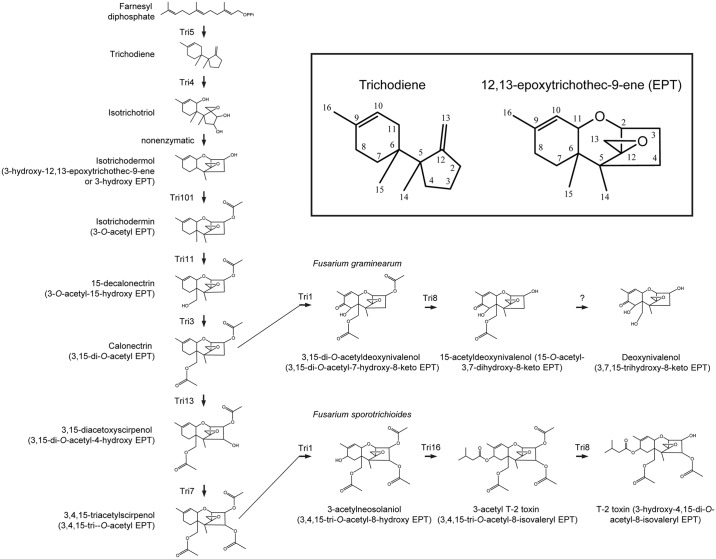
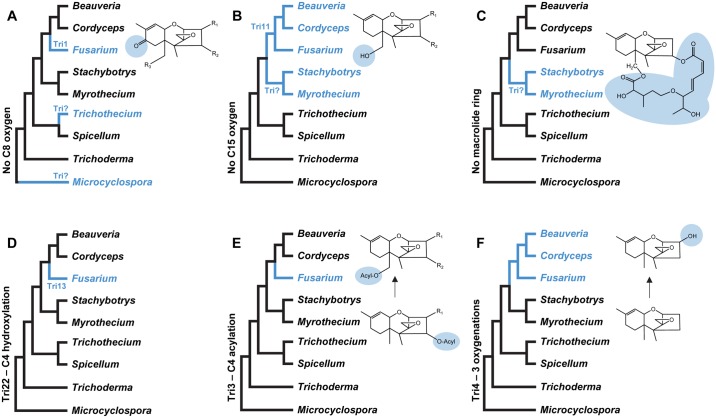
The core structure of trichothecenes consists of a three-ring molecule known as 12,13-epoxytrichothec-9-ene (EPT; Fig 1), and analogs of trichothecenes differ from one another in the patterns of substituents attached to EPT (Fig 2). One type of structural variation has resulted in classification of trichothecenes into two groups [15]. Analogs in the first group, macrocyclic trichothecenes, have a macrolide ring resulting from a 12 or 14-atom chain esterified via hydroxyl groups at carbon atoms 4 and 15 (C4 and C15) of EPT. Analogs in the second group, simple trichothecenes, lack a macrolide ring.
Fig 1. Biosynthetic pathways for the trichothecene analogs deoxynivalenol and T-2 toxin in Fusarium graminearum and F. sporotrichioides, respectively.
The inset at the top right shows the structure and numbering systems for trichodiene and 12,13-epoxytrichothec-9-ene (EPT).
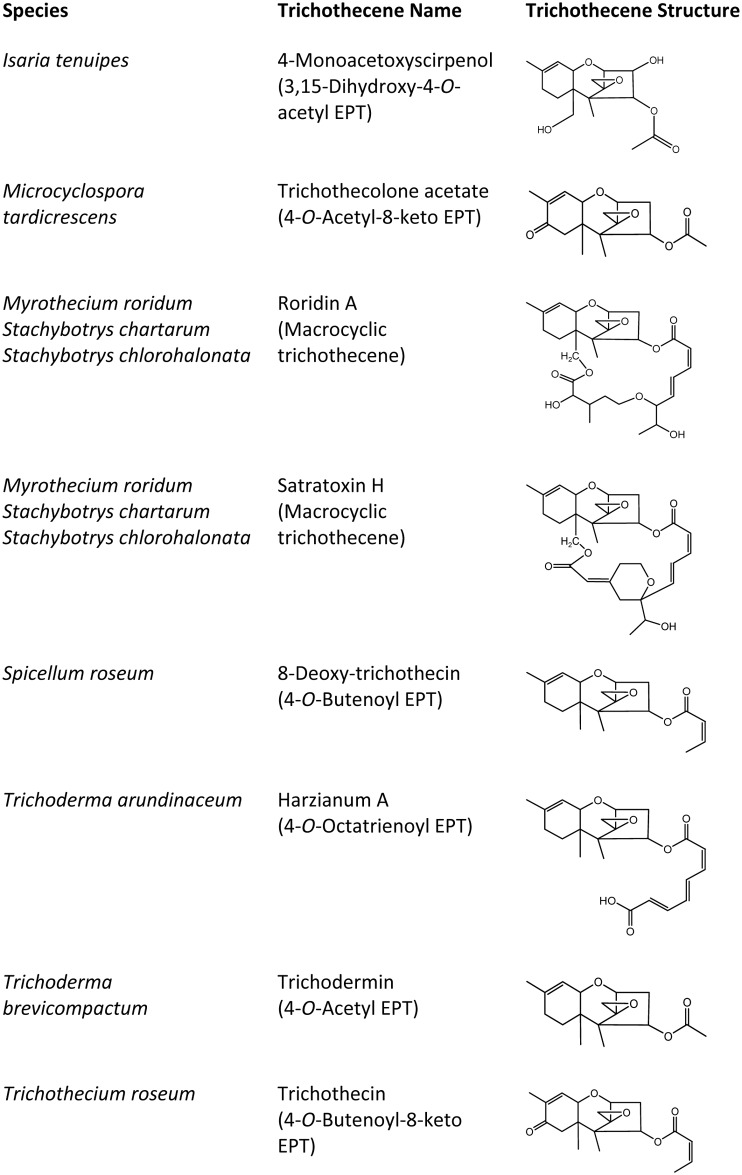
Fig 2. Representative diversity of trichothecene structures in fungi other than Fusarium.
See Fig 1 for examples of Fusarium trichothecenes.
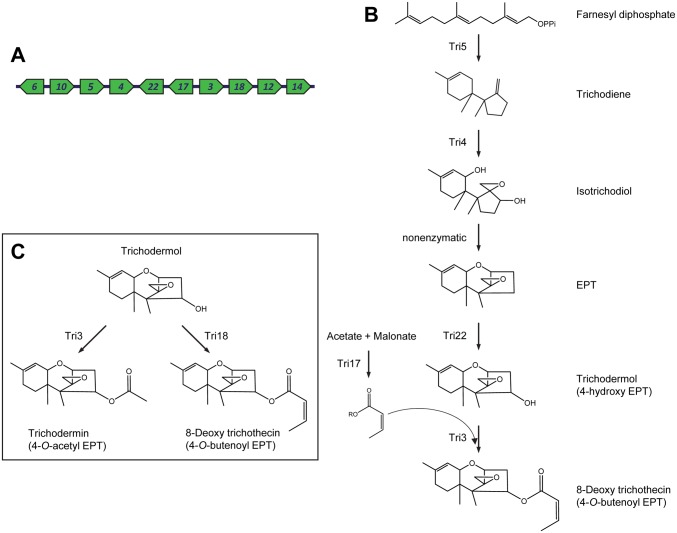
The genetics and biochemistry of trichothecene biosynthesis have been studied most extensively in Fusarium, and biosynthetic pathways for Fusarium trichothecenes that significantly impact agriculture have been elucidated (e.g., deoxynivalenol, nivalenol and T-2 toxin; Fig 1) [3,16]. Additional studies indicate that at least the initial steps in trichothecene biosynthesis are similar in Fusarium, Myrothecium and Trichoderma [17–20]. Trichothecene biosynthesis begins with the cyclization of the primary metabolite farnesyl diphosphate to form the terpene trichodiene. This reaction is catalyzed by a terpene synthase (trichodiene synthase). Subsequently, a cytochrome P450 monooxygenase (trichodiene oxygenase) catalyzes oxygenation of trichodiene at three or four positions to yield isotrichodiol or isotrichotriol, which can cyclize nonenzymatically to form EPT or 3-hydroxy EPT (isotrichodermol), respectively. These latter molecules undergo one or more additional oxygenations, acylations and sometimes other modifications to form all trichothecene analogs [4].
The trichothecene biosynthetic gene (TRI) cluster is one of the most studied SM gene clusters in fungi. Homologs of the TRI cluster have been identified in Fusarium, Stachybotrys, and Trichoderma [3,16,17,21]. In addition, sequence analysis of a Myrothecium roridum cosmid clone identified three adjacent TRI genes presumed to be part of a larger cluster [22], and RNAseq analysis of the fungus has identified homologs of six TRI genes [23]. The number of TRI genes per cluster varies among Fusarium, Stachybotrys, and Trichoderma and in some cases among species of the same genus. The Fusarium and Stachybotrys cluster homologs include the trichodiene synthase gene (TRI5), the trichodiene oxygenase gene (TRI4), two regulatory genes (TRI6 and TRI10), and other genes encoding enzymes that catalyze addition of substituents to the core EPT structure (Table 1). The Trichoderma TRI cluster differs in that it lacks TRI5, which is located elsewhere in the genome [17]. Fusarium and Stachybotrys also have TRI genes at loci other than the TRI cluster. In some Fusarium species, monooxygenase (TRI1) and acyltransferase (TRI16) genes are at a second locus, and an acetyltransferase gene (TRI101) is at a third locus [3]. In other Fusarium species, however, TRI1 and TRI101 are located in the cluster [24]. In Stachybotrys, TRI genes at loci other than the cluster are paralogs of genes in the cluster [21].
Table 1. Functions of trichothecene biosynthetic genes.
| Genea | Functional Categoryb | Function in trichothecene biosynthesisc |
|---|---|---|
| TRI3 | acetyl transferase | acetylation at C15 (Fusarium) |
| TRI4 | cytochrome P450 monooxygenase | oxygenation of trichodiene at C2, C11, and C13 (Myrothecium, Trichoderma), or C2, C3, C11, and C13 (Fusarium) |
| TRI5 | terpene synthase | cyclization of farnesyl pyrophosphate to trichodiene (Fusarium, Trichoderma) |
| TRI6 | Zn2His2 transcription factor | transcriptional regulation of TRI gene expression (Fusarium) |
| TRI7 | acetyl transferase | acetylation at C4 (Fusarium) |
| TRI8 | esterase | deacetylation at C3 or C15 (Fusarium) |
| TRI9 | unknown | unknown |
| TRI10 | transcriptional regulator | transcriptional regulation of TRI gene expression (Fusarium) |
| TRI11 | cytochrome P450 monooxygenase | hydroxylation at C15 (Fusarium) |
| TRI12 | major facilitator superfamily transporter | trichothecene efflux pump (Fusarium) |
| TRI13 | cytochrome P450 monooxygenase | hydroxylation at C4 (Fusarium) |
| TRI14 | unknown | unknown; not required for synthesis in culture (Fusarium) |
| TRI16 | acyl transferase | acylation at C8 (Fusarium) |
| TRI17 | polyketide synthase | synthesis of polyketide esterified at C4 (predicted in Stachybotrys) |
| TRI18 | acyl/acetyl transferase | unknown |
| TRI19 | terpene synthase | TRI5 paralog |
| TRI22d | cytochrome P450 monooxygenase | hydroxylation at C4 (Trichoderma) |
| TRI101 | acetyl transferase | acetylation at C3 (Fusarium) |
a In literature on trichothecene biosynthetic genes, wild-type genes are variously designated as TRI, Tri or tri. For consistency among fungi examined in the current study, we have used the same format for all fungi. Uppercase italicized letters are used to indicate wild-type genes (e.g., TRI5), lowercase italicized letters are used to indicate inactivated genes (e.g., tri5), and non-italicized letters, with the first letter uppercase and the second and third letters lowercase, are used to indicate proteins (e.g., Tri5).
b Functional categories are based on previously reported BLAST analyses.
c Functions in trichothecene biosynthesis have been determined by chemical analysis of fungal strains in which the gene has been inactivated or by heterologous expression [3,16,17,25]. The fungal genus names in parentheses indicate the origin of the TRI homologs used in functional analyses.
d In the initial characterization of TRI22, it was designated as TRI11. But here, we propose that it be re-designated as TRI22 (S1 Fig).
Functional analyses of TRI genes have elucidated the genetic bases for much of the structural diversity of trichothecene analogs produced by Fusarium [3,16,26]. However, the genetic bases for most of the structural diversity of trichothecenes produced by other fungi are not known. For example, T. arundinaceum produces harzianum A, a trichothecene with a polyketide-derived side chain [6,27]. The macrolide ring of macrocyclic trichothecenes is thought to be composed of both polyketide- and isoprenoid-derived moieties [15]. Although the genes responsible for formation of these substituents have not been identified, a polyketide synthase (PKS) gene is located in the TRI cluster of Stachybotrys species that produce macrocyclic trichothecenes [21].
The objective of the current study was to investigate variation of TRI genes among selected fungi in order to identify evolutionary processes that have likely contributed to structural diversity of trichothecene analogs produced by different fungi. To this end, we used genome sequencing to compare the gene content, arrangement, and sequences of TRI loci in selected species of nine genera. We also conducted additional functional analyses of selected TRI genes. The results indicate that gain, loss, and changes in function of genes are major contributors to structural diversity of trichothecenes. We used the results to infer an ancestral trichothecene biosynthetic pathway and to propose scenarios for gain and loss of trichothecene substituents during divergence of TRI cluster homologs. Together, our findings and inferences provide insights into the evolutionary processes that have given rise to biochemical diversity in plant pathogenic, entomopathogenic, and other fungi.
Results
Genomic analysis and TRI-gene content
We used genome sequence data to examine the content and arrangement of TRI genes in 20 fungal strains that included 14 species from nine genera (Table 2). Genome sequence data for 12 strains were generated during the current study, while data for eight strains were generated in previous studies. The strains represented fungi with a range of lifestyles, including saprophytism, endophytism, plant pathogenicity, and entomopathogenicity. The two entomopathogenic fungi, Beauveria bassiana and Cordyceps confragosa (Lecanicillium lecanii), have TRI genes but have not been reported to produce trichothecenes as far as we are aware. To assess TRI gene content in the fungi, we used coding region sequences of the 18 previously described TRI genes (Table 1) as queries in BLASTn and BLASTx analyses against genome sequence databases of the 20 fungi.
Table 2. Information on fungal strains and genome sequences examined in the current study.
| Species a | Strain | Lifestyle | GenBank Accession No.c |
Genome Size (Mb) |
No. Contigs | No. Genes | N50 |
|---|---|---|---|---|---|---|---|
| Beauveria bassiana | ARSEF 2860 | Insect pathogen/ endophyte |
ADAH00000000 | 33.7 | 1,229 | 10,364 | 84,720 |
| Cordyceps confragosa | RCEF 1005 | Insect pathogen | AZHF00000000 | 35.6 | 197 | 11,030 | 782,161 |
| Cordyceps confragosa | UM487 | Insect pathogen | LUKN00000000 | 32.6 | 8,204 | 8,126 | 9,866 |
| FIESC 12b | NRRL 13405 | Plant pathogen | PXXK00000000† | 39.6 | 1,073 | 13,092 | 112,688 |
| Fusarium graminearum | PH-1 | Plant pathogen | AACM00000000 | 36.6 | 435 | 13,313 | 184,591 |
| Fusarium longipes | NRRL 20695 | Plant pathogen | PXOG00000000† | 35.3 | 544 | 11,461 | 144,380 |
| Fusarium sporotrichioides | NRRL 3299 | Plant Pathogen | PXOF00000000† | 37.4 | 446 | 12,014 | 235,034 |
| Microcyclospora tardicrescens | HJS 1936 | Plant pathogen | PXOE00000000† | 26.2 | 1,069 | 10,375 | 169,506 |
| Myrothecium roridum | NRRL 2183 | Plant pathogen | PXOD00000000† | 45.1 | 2,310 | 14,215 | 54,581 |
| Spicellum ovalisporum | DAOM 186447 | Saprophyte | PXOC00000000† | 32.1 | 1,632 | 9,497 | 91,139 |
| Spicellum roseum | DAOM 209012 | Saprophyte | PXOB00000000† | 34.1 | 1,154 | 10,426 | 168,957 |
| Stachybotrys chartarum | IBT 7711 | Saprophyte | APIU00000000 | 36.9 | 3,848 | 11,530 | 55,709 |
| Stachybotrys chartarum | IBT 40288 | Saprophyte | AQPQ00000000 | 36.0 | 3,659 | 11,368 | 46,546 |
| Stachybotrys chartarum | IBT 40293 | Saprophyte | ASEQ00000000 | 36.5 | 4,267 | 11,453 | 60,116 |
| Stachybotrys chlorohalonata | IBT 40285 | Saprophyte | APWP00000000 | 34.4 | 5,591 | 10,706 | 47,022 |
| Trichoderma arundinaceum | IBT 40837 (Ta37) | Saprophyte | PXOA00000000† | 36.9 | 1,370 | 10,539 | 134,831 |
| Trichoderma brevicompactum | IBT 40841 (Tb41) | Saprophyte | PXNZ00000000† | 37.0 | 1,404 | 10,467 | 57,298 |
| Trichothecium roseum | DAOM 195227 | Saprophyte/Plant pathogen | PXNY00000000† | 33.9 | 1,466 | 9,759 | 77,986 |
| Trichothecium roseum | DAOM 197141 | Saprophyte/Plant pathogen | PXNX00000000† | 32.2 | 4,816 | 10,007 | 13,428 |
| Trichothecium roseum | K7-1 | Saprophyte/Plant pathogen | PXNW00000000† | 31.6 | 2,463 | 9,356 | 24,156 |
a Fungi were obtained from the following individuals and institutions: Beauveria—Richard Humber, Agriculture Research Service Collection of Entomopathogenic Fungal Cultures (ARSEF); Cordyceps—Timothy James, Biology Department at the University of Michigan; Fusarium and Myrothecium—Agriculture Research Service (NRRL) Culture Collection, National Center for Agricultural Utilization Research, U.S. Department of Agriculture; Microcyclospora—Hans Josef Schroers at Agricultural Institute of Slovenia; Spicellum and Trichothecium (DAOM strains)—Keith A. Seifert, Ottawa Research and Development Centre, Agriculture and Agri-Food Canada; Trichoderma—Ulf Thrane, IBT Culture Collection of Fungi, Mycology Group, Technical University of Denmark; Trichothecium (strain K7-1)—Anne E. Desjardins, National Center for Agricultural Utilization Research, U.S. Department of Agriculture.
b FIESC 12 is phylogenetic species 12 of the Fusarium incarnatum-equiseti species complex [28].
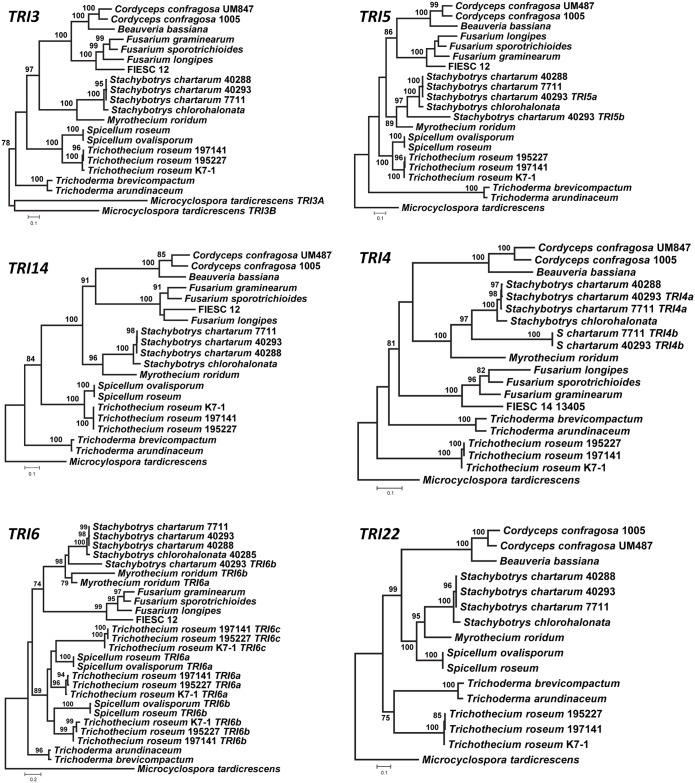
There was considerable variation in the presence and absence of TRI genes among the fungi examined (Fig 3, Table 3). The number of TRI genes per genome varied from six in B. bassiana and C. confragosa to 15 in Fusarium sporotrichioides and Stachybotrys chartarum strain 40293. The number of TRI genes varied within some genera and species as well. For example, S. chartarum had from nine to 15 TRI genes, because some TRI genes were duplicated in two strains [21]. TRI3, TRI5 and TRI14 were the only TRI genes that were present in all 20 fungi examined, while TRI4 was present in all the fungi except the two Spicellum strains (Table 3). In some cases, TRI-gene counts per genome included two or three paralogs of the same gene (Table 3). We identified paralogs of the structural genes TRI3, TRI4, TRI5, TRI17 and TRI18 and the regulatory genes TRI6 and TRI10 (Table 3). TRI6 had the largest number of paralogs; two each in Myrothecium, Spicellum, and S. chartarum 40293, and three in each Trichothecium strain (Table 3). S. chartarum 40293 had the largest number of TRI paralogs, with over half of the TRI genes in this strain being paralogs.
Fig 3. Gene content and arrangement at TRI loci in fungi examined in this study.
The phylogenetic tree to the left was inferred by maximum likelihood analysis of concatenated nucleotide sequences of TRI3, TRI5, and TRI14, the only three TRI genes common to all the fungi. Numbers near branch nodes are bootstrap values based on 1000 pseudoreplicates. The four colored blocks (labeled A, B, C and D) indicate four lineages of TRI genes. The diagrams to the right show the content and arrangement of genes at TRI loci. Green arrows represent homologs of previously described TRI genes, and numbers within arrows indicate TRI gene designations (e.g., 14 indicates TRI14). TRI22 was originally described as TRI11 in Trichoderma, but here we consider TRI11 and TRI22 to be functionally and phylogenetically distinct genes (S1 Fig). For the purposes of this study, paralogs are indicated by the lowercase letters a, b, and c; e.g., TRI6 paralogs are indicated as 6a, 6b, and 6c. Gray arrows represent genes present in the TRI cluster of only one genus; orange arrows represent genes unique to Beauveria and Cordyceps; and purple arrows represent genes unique to Stachybotrys and Myrothecium (S2 Fig). Arrows overlaid on the same line indicate genes on the same contig, whereas arrows overlaid on different lines indicate genes on different contigs. For Stachybotrys, the numbers 7711 and 40293 above TRI paralogs indicate strains in which the paralogs occur. TRI3b in S. chartarum strain 40293 is truncated relative to other TRI3 homologs and, as a result, is likely nonfunctional.
Table 3. TRI gene content of fungi examined in the current study.
A gray box indicates that a TRI gene is present in the genome of a fungus, while a white box indicates the gene was not detected. Numbers within boxes indicate the number of paralogs. The Greek letter ψ indicates that a large portion of the gene is present but that it is a pseudogene.
| Species | Strain | No. Locia |
No. TRI Genes |
TRI Gene | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 16 | 17 | 18 | 22b | 101 | ||||
| Beauveria bassiana | ARSEF 2860 | 1 | 6 | ||||||||||||||||||
| Cordyceps confragosa | RCEF 1005 | 1 | 6 | ||||||||||||||||||
| Cordyceps confragosa | UM487 | 1 | 6 | ||||||||||||||||||
| FIESC 12 | NRRL 13405 | 2 | 14 | ||||||||||||||||||
| Fusarium graminearum | PH-1 | 3 | 12 | Ψ | Ψ | Ψ | |||||||||||||||
| Fusarium longipes | NRRL 20695 | 3 | 15 | ||||||||||||||||||
| Fusarium sporotrichioides | NRRL 3299 | 3 | 15 | ||||||||||||||||||
| Microcylospora tardicrescens | HJS 1936 | 1 | 11 | 2 | |||||||||||||||||
| Myrothecium roridum | NRRL 2183 | 2 | 13 | 2 | |||||||||||||||||
| Spicellum ovalisporum | DAOM 186447 | nd | 10 | 2 | |||||||||||||||||
| Spicellum roseum | DAOM 209012 | nd | 10 | 2 | |||||||||||||||||
| Stachybotrys chartarum | IBT 7711 | 2 | 12 | 2 | 2 | 2 | |||||||||||||||
| Stachybotrys chartarum | IBT 40288 | 1 | 9 | ||||||||||||||||||
| Stachybotrys chartarum | IBT 40293 | 3 | 14 | 2 | 2 | 2 | 2 | 2 | |||||||||||||
| Stachybotrys chlorohalonata | IBT 40285 | 1 | 9 | ||||||||||||||||||
| Trichoderma arundinaceum | IBT 40837 | 3 | 10 | ||||||||||||||||||
| Trichoderma brevicompactum | IBT 40841 | 3 | 10 | ||||||||||||||||||
| Trichothecium roseum | DAOM 195227 | nd | 12 | 3 | |||||||||||||||||
| Trichothecium roseum | DAOM 197141 | nd | 12 | 3 | |||||||||||||||||
| Trichothecium roseum | K7-1 | nd | 12 | 3 | |||||||||||||||||
The sequence data also indicated that TRI genes can occur at one to as many as five distinct genomic locations (loci). In some fungi with multiple TRI loci, genes located at different loci were paralogs. For example, the Myrothecium and Stachybotrys TRI clusters include nine and ten known TRI genes, respectively, but the TRI genes at other loci were paralogs of genes in the cluster. In Fusarium and Trichoderma, by contrast, TRI genes occurred at two or three loci, but the gene at the same or different loci in these fungi were not paralogous.
In the Spicellum and Trichothecium strains examined, TRI genes were dispersed over five or six contigs (S2 Fig). Although this dispersion of TRI genes on different contigs was likely an artifact of the genome sequence assembly in some cases, in other cases it was not artefactual. In S. roseum, for example, TRI12 was near the middle of 243-kb contig and TRI3, TRI5, TRI6a, TRI10, and TRI14 were located adjacent to one another and near the middle of a 169-kb contig (S2 Fig). In both of these contigs, the TRI genes were flanked by multiple genes that were unlikely to be involved in trichothecene biosynthesis based on their predicted functions. Thus, like Fusarium, Myrothecium, Stachybotrys, and Trichoderma, TRI genes in Spicellum and Trichothecium occur at two or more loci (S2 Fig).
The apparent absence of TRI4 in the Spicellum genome sequences was unexpected, because TRI4 is required for essential steps that occur early in trichothecene biosynthesis in other fungi [3,16]. We used three approaches to determine whether the absence of TRI4 was a sequencing or assembly artifact: 1) generation of genome sequence data for both strains of Spicellum using two or three Methods (MiSeq, TruSeq, and Ion Torrent); 2) RNAseq analysis of S. roseum (strain 209012) grown under conditions that induced expression of other TRI genes; and 3) PCR analysis of the Spicellum strains using multiple primer pairs that amplified TRI4 fragments from Fusarium, Myrothecium and Trichothecium strains. None of these methods yielded evidence for a full-length TRI4 homolog in either Spicellum strain. However, BLASTx analysis of the TRI3-TRI6a intergenic region in S. roseum 209012 revealed a 558-nucleotide sequence that is likely a remnant of TRI4 (S3 Fig).
The absence of TRI4 in both Spicellum strains led us to predict that neither strain would produce trichothecenes. In Gas chromatography-mass spectrometry (GC-MS) analysis, we did not detect trichothecenes in culture extracts of S. ovalisporum, but we did detect them in culture extracts of S. roseum strain 209012 (S4 Fig). Consistent with a previous study [32], the most abundant trichothecene analog produced was 8-deoxy-trichothecin (4-O-butenoyl EPT). The absence of a TRI4 homolog in S. roseum suggests that it must have a gene(s) that encodes another trichodiene oxygenase. Attempts to identify such a gene by RNAseq analysis were not successful. That is, we did not find evidence for an oxygenase gene in S. roseum 209012 that exhibited a pattern of expression similar to those of known TRI genes.
Functional analyses of TRI genes
Trichoderma arundinaceum TRI3
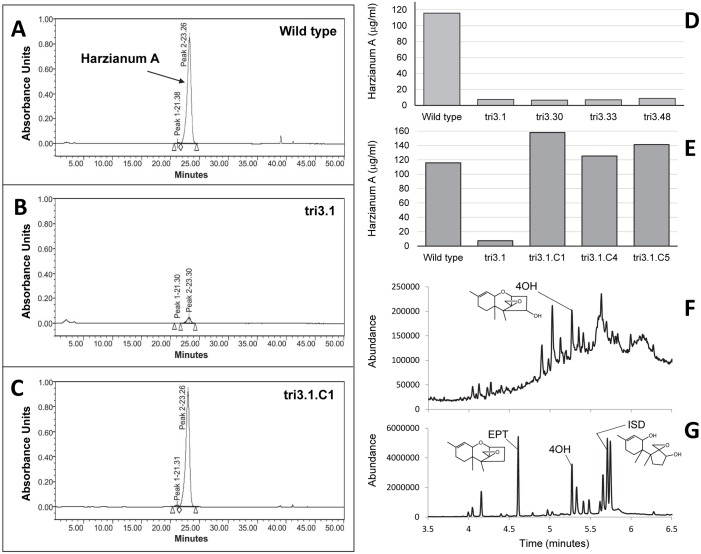
The presence of TRI3 in fungi that produce trichothecenes that lack a C15 ester suggests that the gene can have a function other than the C15 esterification function previously described in Fusarium [33]. To determine whether this is the case, we inactivated TRI3 in T. arundinaceum by gene deletion (S1 File). The most abundant trichothecene produced by wild-type strains of this fungal species is harzianum A, which consists of EPT with an eight-carbon side chain (octatrienedioate) esterified to a hydroxyl group at C4 (Fig 2). Harzianum A does not have an ester or hydroxyl group at C15. Analysis of T. arundinaceum tri3 deletion mutants indicated that harzianum A production was reduced by 92–94% compared to the wild-type progenitor strain (Fig 4A, 4B and 4D). However, GC-MS analysis revealed that the tri3 mutants produced relatively high levels of trichodermol (4-hydroxy EPT) and two other harzianum A precursors, EPT and its immediate precursor isotrichodiol (Fig 4G). In contrast, the wild-type progenitor strain produced only low levels of trichodermol (Fig 4F). Complementation of a tri3 mutant with a wild-type copy of TRI3 restored high levels of harzianum A production (Fig 4C and 4E). The trichothecene production phenotypes of the tri3 mutants and complemented mutant indicate that the T. arundinaceum Tri3 catalyzes esterification of octatrienedioate to the C4 hydroxyl group of 4-hydroxy EPT. Thus, the results also indicate that different homologs of Tri3 can have different functions: C4 esterification in T. arundinaceum and C15 esterification in Fusarium. Given that Microcyclospora tardicrescens, S. roseum and T. roseum can produce trichothecenes esterified at C4 but not C15, we propose that TRI3 functions in C4 esterification in these fungi as well.
Fig 4. Functional analysis of TRI3 in Trichoderma arundinaceum.
(A-C) High performance liquid chromatograms showing harzianum A (HA) production by: (A) wild-type progenitor strain Ta37; (B) tri3 mutant strain tri3.1; and (C) strain tri3.1.C1, tri3 mutant strain tri3.1 complemented with a wild-type copy of TRI3. (D) Quantification of HA production in the wild type and tri3 deletion mutant strains tri3.1, tri3.30, tri3.33, and tri3.48. (E) Quantification of HA production in the wild type, tri3 mutant strain tri3.1, and three tri3.1-derived strains that were complemented with a wild-type copy of TRI3 (strains tri3.1.C1, tri3.1.C4 and tri3.1.C5). (F and G) Total ion chromatograms from gas chromatography-mass spectrometry analysis of culture extracts of the (F) wild-type strain and (G) tri3 mutant strain tri3.1. The peaks labeled 4OH and ISD are for trichodermol (4-hydroxy EPT) and isotrichodiol, respectively.
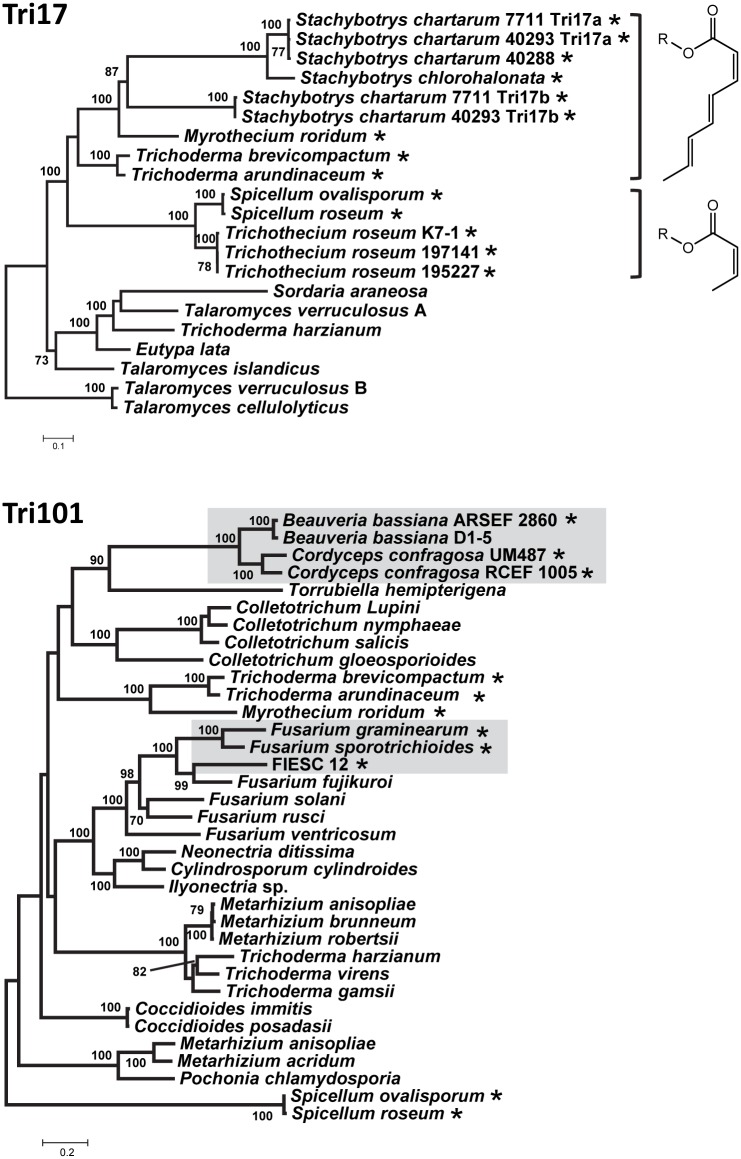
Trichoderma arundinaceum and Myrothecium roridum TRI17
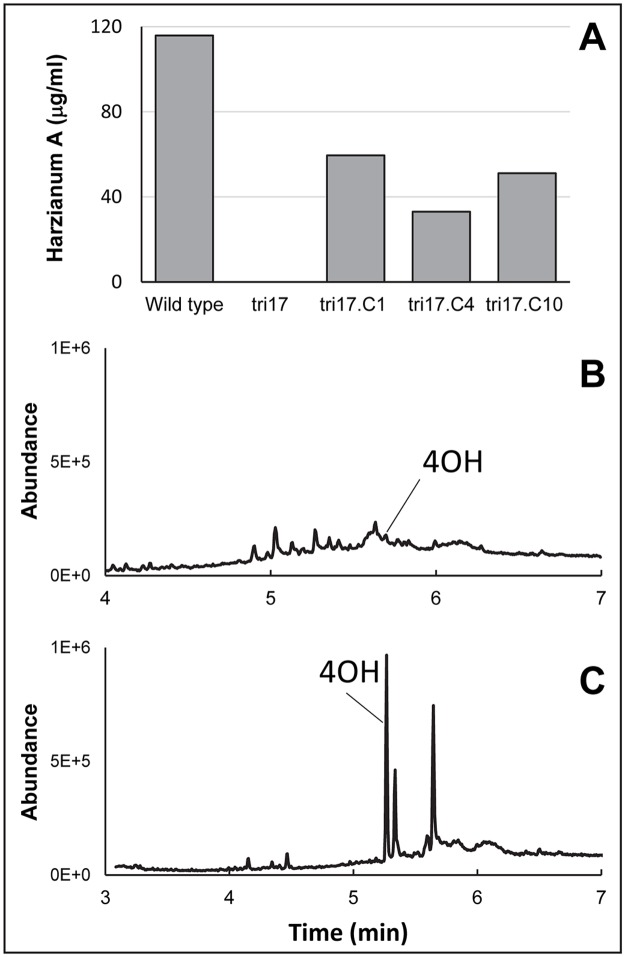
The octatrienedioate substituent in harzianum A is thought to be derived from a polyketide [6] and, therefore, likely requires a PKS gene for its synthesis. We detected homologs of the PKS gene TRI17 in Myrothecium, Spicellum, Trichoderma, and Trichothecium in addition to Stachybotrys, the fungus in which the gene was originally reported [21]. Its presence in the Stachybotrys TRI cluster led Semeiks et al. [21] to propose that Tri17 catalyzes synthesis of the polyketide portion of the macrolide ring of macrocyclic trichothecenes, but they did not confirm its role in trichothecene biosynthesis. Therefore, we used gene deletion and complementation analyses to determine whether TRI17 is required for synthesis of the polyketide side chain of harzianum A in T. arundinaceum (S1 File). If TRI17 is required for synthesis of the polyketide, deletion of the gene should block formation of harzianum A. tri17 deletion mutants did not produce detectable amounts of harzianum A, but did produce trichodermol (4-hydroxy EPT), and complementation of a mutant with a wild-type copy of TRI17 restored harzianum A production (Fig 5A, 5B and 5C). These results demonstrate that TRI17 is required for synthesis of harzianum A and are consistent with the hypothesis that Tri17 catalyzes synthesis of the polyketide precursor of octatrienedioate.
Fig 5. Functional analysis of TRI17 in Trichoderma arundinaceum.
(A) Quantitation of harzianum A production in the wild-type strain Ta37, tri17 mutant strain tri17.139, and three tri17.139-derived strains (tri17.C1, tri17.C4 and tri17.C10) that were complemented with a wild-type copy of T. arundinaceum TRI17. (B and C) Total ion chromatograms of culture extracts of (B) wild-type progenitor strain Ta37 and (C) tri17 deletion mutant tri17.139. The trichothecene biosynthetic intermediate trichodermol (4-hydroxy EPT) is indicated at 5.268 min. 4OH indicates trichodermol (4-hydroxy EPT).
Because the 6,7-dihydroxy-2,4-octadienoate substituent that occurs in the macrolide ring of some macrocyclic trichothecenes is similar in structure to octatrienedioate (Fig 2), it is possible that the TRI17 homolog from a macrocyclic trichothecene-producing fungus could complement the T. arundinaceum tri17 mutant. To test this hypothesis, we introduced a wild-type copy of M. roridum TRI17 into a T. arundinaceum tri17 mutant (S1 File). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis revealed that harzianum A production was restored in the resulting transformants and, thus, that M. roridum TRI17 can complement the T. arundinaceum tri17 mutant (S5 Fig). This finding indicates that biosynthesis of the polyketide-derived substituents of harzianum A and some macrocyclic trichothecenes (e.g., roridins and satratoxins) requires the same polyketide precursor.
Beauveria bassiana TRI genes
We attempted to induce trichothecene production in B. bassiana ARSEF 2860 and C. confragosa UM487 by growing each fungus under multiple conditions that induce trichothecene production in other fungi. However, trichothecenes were not detected in extracts from any of the resulting cultures. To obtain evidence as to whether the B. bassiana TRI cluster is functional, we heterologously expressed selected TRI genes from B. bassiana in either Fusarium verticillioides or Saccharomyces cerevisiae, two trichothecene-nonproducing fungi that have been used previously to determine TRI gene function [20,26,34]. We used F. verticillioides for analysis of TRI4 and TRI22, which have introns, and S. cerevisiae for analysis of TRI101, which does not have introns. In the heterologous expression experiments, a TRI gene was introduced into F. verticillioides or S. cerevisiae by standard transformation methods [26]; selected trichothecene biosynthetic intermediates were added to cultures of the resulting transformants; and the ability of the cultures to modify the intermediates was assessed by GC-MS.
In the first heterologous expression experiment, we introduced the B. bassiana TRI4 gene into F. verticillioides. In trichothecene-producing fungi, Tri4 catalyzes oxygenation at three or four positions of trichodiene leading to the formation of EPT or 3-hydroxy EPT, respectively. Therefore, we reasoned that expression of the B. bassiana TRI4 homolog in F. verticillioides would lead to the conversion of exogenously added trichodiene to EPT or 3-hydroxy EPT. Although F. verticillioides does not produce trichothecenes or have a TRI cluster, wild-type strains of the fungus can acetylate the C3 hydroxyl of trichothecenes and, as a result, can convert 3-hydroxy EPT to 3-O-acetyl EPT (isotrichodermin) [18]. Thus, if 3-hydroxy EPT were formed by expression of B. bassiana TRI4 in F. verticillioides, it would be converted to 3-O-acetyl EPT by the endogenous C3 acetylase activity. Indeed, addition of trichodiene to cultures of F. verticillioides expressing the B. bassiana TRI4 gene resulted in formation of 3-O-acetyl EPT (Fig 6A and 6B), thereby confirming that the B. bassiana TRI4 is functional, and that it is required for oxygenation of trichodiene at four positions to yield 3-hydroxy EPT, the same function reported in Fusarium [18,20].
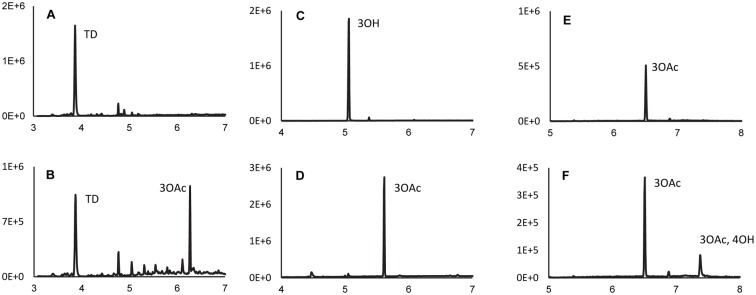
Fig 6. Total ion chromatograms from GC-MS analysis of cultures from heterologous expression of TRI genes from Beauveria bassiana strain ARSEF 2860.
(A) wild-type Fusarium verticillioides grown on YEPD medium containing trichodiene (TD); (B) F. verticillioides expressing the B. bassiana TRI4 grown on YEPD medium containing trichodiene; (C) ayt1 mutant of Saccharomyces cerevisiae grown on YG medium containing isotrichodermol (3-hydroxy EPT, 3OH); (D) ayt1 mutant of S. cerevisiae expressing the B. bassiana TRI101 grown on YG medium containing isotrichodermol; (E) wild-type F. verticillioides grown on YEPD medium containing isotrichodermin (3-O-acetyl EPT, 3OAc); (F) F. verticillioides expressing the B. bassiana TRI22 grown on YEPD medium containing isotrichodermin. In each chromatogram, the Y-axis is total ion abundance, and the X-axis is time in minutes. The peak labeled 3OAc,4OH indicates 4-hydroxy isotrichodermin (3-O-acetyl-4-hydroxy EPT).
In a second experiment, we introduced the B. bassiana TRI101 gene into S. cerevisiae. In trichothecene-producing fusaria, Tri101 catalyzes trichothecene C3 acetylation. The S. cerevisiae Ayt1 enzyme also catalyzes trichothecene C-3 acetylation. Therefore, in order to study the activity of B. bassiana Tri101 in this yeast species, we expressed TRI101 in an ayt1 deletion mutant. Addition of 3-hydroxy EPT (isotrichodermol) to cultures of a S. cerevisiae ayt1 mutant expressing the B. bassiana TRI101 homolog resulted in the formation of 3-O-acetyl EPT (isotrichodermin), thereby confirming that B. bassiana TRI101 confers trichothecene C3 acetylation (Fig 6C and 6D).
In a third experiment, we introduced the B. bassiana TRI22 homolog into F. verticillioides, and assessed whether expression of the gene conferred trichothecene C4 hydroxylation, the reaction catalyzed by Tri22 in T. arundinaceum [17]. Addition of 3-O-acetyl EPT to cultures of F. verticillioides expressing B. bassiana TRI22 resulted in formation of a novel trichothecene product that was isolated and identified as 3-O-acetyl-4-hydroxy EPT on the basis of mass spectral and NMR data (Fig 6E and 6F, S6 Fig). This result indicated that B. bassiana Tri22 catalyzes trichothecene C4 hydroxylation. Together, the heterologous expression experiments described above indicate that the B. bassiana TRI genes examined are functional. Given this, the rest of the B. bassiana TRI genes might also be functional. If this is the case, our inability to induce trichothecene production in cultures of B. bassiana in the current study was likely because we did not use suitable culture conditions.
Phylogenetic analysis of TRI genes
To gain insight into the variation of TRI gene homologs, we generated phylogenetic trees for individual TRI genes and for concatenated sequences of three of the genes. In preliminary analyses of individual TRI genes, we employed outgroup sequences of non-TRI genes from fungi that do not produce trichothecene and do not have a TRI cluster. Although distantly related, these outgroup sequences aligned to TRI sequences. In trees inferred from the resulting alignments, Microcyclospora homologs were consistently either the most or among the most basal lineages of TRI genes (S7 Fig). Given this and its distant relationships to the other trichothecene-producing fungi examined in this study, Microcyclospora homologs were used as the root in subsequent TRI gene trees that excluded a non-TRI-gene outgroup.
In trees inferred from homologs of individual TRI genes, relationships among more closely related homologs were generally well resolved (bootstrap values >70%), whereas relationships among more distantly related homologs were generally not well resolved (Figs 7 and 8, S8 Fig). In most single-TRI-gene trees, Myrothecium and Stachybotrys formed a well-supported clade, and with the exception of TRI22, Spicellum and Trichothecium formed a well-supported clade. In addition, Beauveria, Cordyceps and Fusarium also formed a well-supported clade in which Beauveria and Cordyceps had a sister relationship. Although branch conflicts were observed in comparisons of some TRI gene trees, most of the conflicts were not statistically supported by bootstrap analysis. We also performed Shimodaira-Hasegawa (SH) [35] and the Approximately Unbiased (AU) [36] tests to assess the significance of conflicting branches with bootstrap values > 70. According to the results of these tests, the conflicts were not significant, with one exception; in the TRI22 tree, Spicellum homologs grouped in a well-supported clade with Myrothecium and Stachybotrys rather than with Trichothecium (Fig 7). This result suggests that the evolutionary history of TRI22 differs from other TRI genes in Spicellum, a phenomenon that has been previously reported for some Fusarium TRI genes [37,38].
Fig 7. Maximum likelihood trees inferred from sequences of selected TRI genes: TRI3, TRI4, TRI5, TRI6, TRI14, and TRI22.
Trees for TRI10, TRI12 and TRI18 are shown in the Supporting Information (S8 Fig). Numbers near branch nodes are bootstrap values based on 1000 pseudoreplicates.
Fig 8. Maximum likelihood trees inferred from predicted amino acid sequences of Tri17 (top) and Tri101 (bottom) and related homologs from trichothecene-nonproducing fungi.
The chemical structures shown to the right of the Tri17 tree are predicted structures of polyketides synthesized by the different Tri17 homologs. Asterisks (*) indicate species/strains that produce trichothecenes or are predicted to produce trichothecene based the presence of TRI genes. In the TRI101 tree, the gray boxes indicate the strains/species that have a TRI101 gene that functions or is likely to function in trichothecene biosynthesis. Numbers near branch nodes are bootstrap values based on 1000 pseudoreplicates. Strain designations are shown only for species with two or three strains included in a tree.
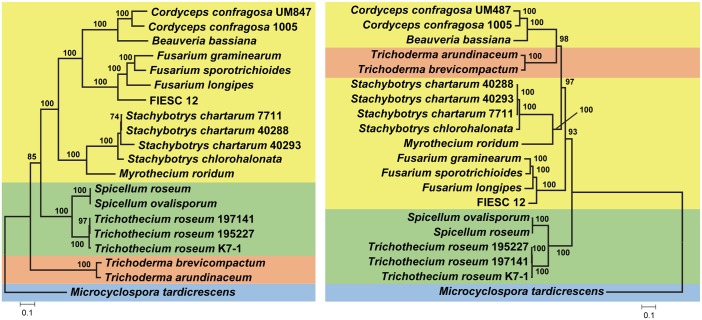
We surmised that trees inferred from multiple TRI genes would better reflect relationships of TRI-cluster homologs than single-TRI-gene trees. Therefore, we inferred a phylogenetic tree from concatenated sequences of TRI3, TRI5 and TRI14, the only three TRI genes that were common to all fungi that were the focus of this study (Table 3). Trees inferred from these three genes individually did not have any well-supported branches that conflicted and were not significantly different from one another according to the SH and AU tests. Although results of a partition homogeneity test indicated inclusion of F. graminearum sequences resulted in significant heterogeneity in the data, relationships between genera did not differ in the concatenated gene trees with or without inclusion of F. graminearum sequences. Therefore, F. graminearum sequences were included in the final concatenated dataset.
Some relationships among TRI gene homologs that were evident in single-TRI-gene trees were also evident in the concatenated gene tree. For example, Myrothecium and Stachybotrys formed a well-supported clade, as did Beauveria, Cordyceps, and Fusarium in the concatenated and most single-TRI-gene trees (Figs 7 and 9). The concatenated-TRI-gene tree had high bootstrap support for almost all branches, and therefore provided information for relationships of more distantly related TRI clusters. Based on the clades resolved in the concatenated-gene tree, we divided the cluster homologs into four lineages: lineage A consisted of the outgroup, M. tardicrescens; lineage B was the next most basal clade and consisted of Trichoderma sequences; lineage C consisted of Spicellum and Trichothecium sequences; and lineage D consisted of Beauveria, Cordyceps, Fusarium, Myrothecium and Stachybotrys sequences (Figs 3 and 9). Although there were no consistent differences in gene content of the different cluster lineages, lineages A-C occurred in fungi that produce less complex trichothecenes (i.e., with a hydroxyl or ester at C4 of EPT, a carbonyl at C8 in some cases, and a hydroxyl at C7 in one case), whereas lineage D cluster homologs occurred in fungi that can produce more complex trichothecenes (i.e., with carbonyl, hydroxyl or ester groups at up to five positions of EPT) (Fig 2) [7].
Fig 9. Comparison phylogenetic tree inferred from concatenated sequences of TRI3, TRI5 and TRI14 (left) and a species phylogeny inferred from concatenated sequences of 20 housekeeping genes (right).
Numbers near branch nodes are bootstrap values from 1000 pseudoreplicates. Only bootstrap values greater than 70% are shown. The housekeeping genes used in this analysis are listed in S3 File.
Visual inspection indicated that there was one or more well-supported branches (bootstrap value > 70) in the TRI10, TRI18, TRI22 and TRI101 trees that conflicted with branches in the concatenated-TRI3-5-14-gene tree. Results of SH and AU tests indicated that the conflicts for TRI22 and TRI101 were significant, but those for TRI10 and TRI18 were not (S2 File).
To compare phylogenetic relationships of TRI cluster homologs to the relationships of the fungi in which the homologs occur, we inferred a species tree from the concatenated sequences of 20 housekeeping genes. We analyzed trees inferred from each housekeeping gene individually and assessed whether conflicts between the single-gene trees affected the species tree inferred from all 20 genes. The results of these analyses are shown in S3 File and suggest that the 20-housekeeping-gene tree provides a reasonable estimate of the species phylogeny. The high bootstrap values for almost all branches in the species tree provided evidence for the hierarchical relationships of most of the genera examined (Fig 9). There were two notable conflicts in the topologies of the species tree and the concatenated-TRI-gene tree. First, the sister relationship of Beauveria-Cordyceps and Fusarium observed in the TRI tree did not exist in the housekeeping gene tree; and second, the sister relationship of Beauveria-Cordyceps and Trichoderma observed in the housekeeping gene tree did not exist in the TRI tree. We used SH and AU tests to assess the significance of the conflicts between the trees overall and between the branches noted above. In a first set of tests, the TRI tree was constrained to conform to the housekeeping-gene tree, and housekeeping-gene tree were constrained to conform to the TRI tree. In these reciprocal assessments, the constrained trees were significantly worse than the unconstrained trees (p < 0.05). In a second set of tests, the TRI tree was constrained to include a sister relationship of Beauveria-Cordyceps and Trichoderma, and the housekeeping-gene tree was constrained to include the sister relationship of Beauveria-Cordyceps and Fusarium. Both tests indicated that the conflicts were significant (p < 0.05). In addition, none of the trees inferred from individual housekeeping genes included a well-supported Beauveria-Cordyceps-Fusarium clade (S3 File), and none of the single-TRI-gene trees included a well-supported Beauveria-Cordyceps-Trichoderma clade.
Homologs of TRI101 have been identified in trichothecene-producing and nonproducing species of Fusarium and other fungal genera [39,40]. In the current study, BLAST analyses indicated that the Beauveria and Cordyceps TRI101 homologs were more similar to TRI101 homologs in some other genera of trichothecene-nonproducing fungi than they were to homologs in Fusarium. To further investigate sequence differences of TRI101 homologs from Beauveria, Cordyceps, and Fusarium, we conducted a phylogenetic analysis with TRI101 homologs from diverse Ascomycetes. The resulting tree suggests that TRI101 homologs from Beauveria and Cordyceps are more closely related to a homolog from the trichothecene-nonproducing fungus Torrubiella hemipterigena than to homologs from trichothecene-producing fusaria (Fig 8). The tree also suggests that TRI101 homologs in trichothecene-producing fusaria are more closely related to homologs in trichothecene-nonproducing species of Fusarium, Cylindrosporum, Ilyonectria, and Neonectria than they are to the homologs in Beauveria-Cordyceps. The relatively distant relationships of TRI101 homologs in Beauveria-Cordyceps and trichothecene-producing species of Fusarium were unexpected given the close relationships of other TRI genes in these fungi.
Discussion
In this study, a combination of genomic, phylogenetic, functional, and biochemical analyses has provided unprecedented insights into the evolutionary history of trichothecene biosynthesis in representatives of diverse genera of filamentous fungi. Our results indicate that structural diversity of trichothecenes produced by these fungi has arisen largely from gain, loss and changes in function of TRI genes during the collective evolutionary histories of TRI loci. Further, our phylogenetic analyses indicate that the evolutionary histories of TRI genes do not necessarily mirror the phylogenetic relationships of trichothecene-producing fungi, a phenomenon that has been observed for multiple fungal SM biosynthetic genes [24,37,38,41,42]. Below, we discuss the evidence for gain, loss and functional changes of TRI genes and consider the relationships between trichothecene structural changes and divergence of TRI cluster homologs.
TRI-gene gain
We consider gain of a TRI gene to be the addition of a gene to trichothecene biosynthesis that was not previously involved in the process. Gain is suggested by the absence of a TRI gene in the genomes of multiple trichothecene-producing fungi, particularly those with a basal TRI cluster (lineage A and B clusters), and the presence of the gene in the genome(s) of only one or a few fungi. TRI1, TRI7, TRI8, TRI11, and TRI13 are examples of such genes, because they were absent in all the fungi examined except Fusarium. Multiple mechanisms, including neofunctionalization, horizontal gene transfer (HGT), and horizontal chromosome transfer, have the potential to contribute to gain of a SM biosynthetic gene in fungi. The presence of TRI gene paralogs in several fungi suggested gain of some TRI genes might have resulted from neofunctionalization (i.e., the process of gene duplication and subsequent divergence in function of a resulting paralog). However, with the exception of the paralogs, known TRI genes are more closely related to non-TRI genes than they are to other TRI genes [21,24,37]. This suggests that neofunctionalization of TRI genes has not contributed to TRI-gene gain, but instead neofunctionalization of closely related non-TRI genes has contributed to gain.
TRI-gene gain may have also resulted when non-TRI genes changed function due to selection or other evolutionary processes to become involved in trichothecene biosynthesis. In fungi, SM biosynthetic gene clusters can degenerate such that some genes are pseudogenized or deleted and others remain intact [42–45]. If a TRI gene were gained by adaptation of a gene once involved in another process, the gene could have originated in a degenerating cluster. In our search for outgroups for phylogenetic analyses of individual TRI genes, BLASTx analysis of the fungal protein database in GenBank indicated that distantly related homologs of TRI genes occur in other fungi (Fig 8, S7 Fig). Furthermore, F. graminearum and F. sporotrichioides have genes that can partially compensate for the absence of TRI genes in deletion mutants [46,47]. Such genes encode enzymes that can modify trichothecene structures, and suggest another possible origin of gained TRI genes.
The discussion above indicates that multiple mechanisms could have contributed to gain of TRI genes, but that the mechanisms responsible for gain of specific genes are not evident from our analyses. The trichothecene C3 acetylation gene, TRI101, is a possible exception. Homologs of TRI101 are present in some trichothecene-producing fungi and in many trichothecene-nonproducing fungi [39,48]. In fact, all trichothecene-nonproducing species of Fusarium that have been examined have a TRI101 homolog, which is often designated as TRI201 [49]. All of the fungi examined here with a lineage-A–C TRI cluster and some fungi with a lineage-D cluster (i.e., Myrothecium and Stachybotrys) produce trichothecenes that lack a C3 acetyl group, and therefore do not require TRI101 for trichothecene biosynthesis. But, some trichothecene-producing fungi that do not require TRI101 for production have a TRI101 homolog (Fig 8) that is not located near other TRI genes. The presence of TRI101 in some trichothecene-nonproducing fungi and trichothecene-producing fungi that do not require C3-acetylation activity indicates that some TRI101 homologs have a function(s) other than trichothecene biosynthesis. In most trichothecene-producing fusaria that have been examined, TRI101 is not in the TRI cluster, but instead is located in the same genomic context as the homolog in some trichothecene-nonproducing species [40,49]. In addition, there is evidence that TRI101 has translocated into the TRI cluster rather than out of it during the evolutionary history of the Fusarium incarnatum-equiseti species complex (FIESC) [24]. These observations plus the knowledge that TRI101 functions in trichothecene C3 acetylation in both Beauveria and Fusarium suggest that TRI101 has become incorporated into trichothecene biosynthesis (i.e., gained) in Beauveria-Cordyceps and Fusarium. The presence of TRI101 in trichothecene-nonproducing fungi further suggests that TRI101 gain has involved its adaptation from another function. It has been proposed that in Fusarium TRI101 and TRI201 are paralogs [49]. If this is the case, gain of TRI101 could be a result of neofunctionalization, whereby the ancestral gene was and the TRI201 paralog is involved in a process other than trichothecene biosynthesis, and the TRI101 paralog diverged to function in trichothecene biosynthesis.
Results of the phylogenetic analysis of Tri101 homologs from trichothecene-producing and nonproducing fungi (Fig 8) suggests that the gain of TRI101 and, therefore, the evolution of the C3 acetylation in the trichothecene biosynthetic pathway occurred independently in Fusarium and in Beauveria-Cordyceps. Two other trichothecene structural modifications, C8 and C15 oxygenation, appear to have also evolved independently in different fungi. Some Fusarium, Microcyclospora, and Trichothecium trichothecenes have a C8 oxygen atom (Fig 2). Functional analyses of F. graminearum and F. sporotrichioides indicate that Tri1 catalyzes trichothecene C8 oxygenation in Fusarium [50–53]. The absence of TRI1 in the Microcyclospora and Trichothecium genome sequences indicates that an enzyme other than Tri1 catalyzes C8 oxygenation in these fungi, which in turn indicates that C8 oxygenation in Fusarium evolved independently of its evolution in Microcyclospora and Trichothecium. (Fig 10A). It is not known whether C8 oxygenation arose independently in Microcyclospora and Trichothecium.
Fig 10. Scenarios for changes in trichothecene biosynthesis during evolutionary divergence of TRI cluster homologs.
The scenarios presume the ancestral TRI cluster and trichothecene pathway presented in Fig 12. In each scenario, the tree is a simplified version of the tree inferred from concatenated sequences of TRI3, TRI5, and TRI14 (Fig 3). A change in biosynthesis from the ancestral state is indicated by a change in color of a branch from black to blue. A. acquisition of C8 oxygen by gain of Tri1 in Fusarium and an unknown enzyme(s) in Microcyclospora and Trichothecium; B. acquisition of C15 oxygen by gain of Tri11 in Fusarium, Beauveria and Cordyceps, and an unknown enzyme in Myrothecium and Stachybotrys; C. acquisition of macrolide ring by gain of unknown enzymes in Myrothecium and Stachybotrys; D. change in C4 hydroxylation enzyme from Tri22 to Tri13 during divergence of Fusarium TRI cluster; E. change in Tri3 function from C4 acylation to C15 acetylation during divergence of Fusarium TRI cluster; and F. change in Tri4 function from 3 to 4 oxygenations during divergence of TRI cluster lineage in Fusarium, Beauveria, and Cordyceps.
Among the fungi examined in this study, only Fusarium, Myrothecium and Stachybotrys are reported to produce trichothecenes with a C15 oxygen. In Fusarium, Tri11 catalyzes trichothecene C15 oxygenation [54]. The absence of a TRI11 homolog in the Myrothecium and Stachybotrys genome sequences (Table 3) indicates that a gene other than TRI11 is required for C15 oxygenation in these fungi, and therefore that trichothecene C15 oxygenation in Myrothecium and Stachybotrys arose independently of its evolution in Fusarium (Fig 10B). On the other hand, the presence of TRI11 homologs in Beauveria, Cordyceps and Fusarium suggests that gain of TRI11 occurred prior to divergence of the TRI cluster homologs in these fungi (Fig 10B). To our knowledge and with the exception of TRI17, genes required for synthesis of the macrolide rings of macrocyclic trichothecene have yet to be identified. Production of macrocyclic trichothecenes only by fungi with lineage-D TRI clusters suggests that the formation macrolide rings of these trichothecenes resulted from gain of genes in the Myrothecium-Stachybotrys lineage of trichothecene-producing fungi (Fig 10C).
TRI-gene loss
We consider that loss of a TRI gene results from pseudogenization or complete deletion of the gene such that a functional version of it is no longer present in a genome. Evidence for loss is the occurrence of a gene in multiple fungi with a more basal (i.e., lineage A and B) TRI cluster, but absence of the gene in one or more other fungi. TRI4, TRI6, TRI10, TRI12, TRI13, TRI17, and TRI22 are examples of genes that have likely been lost (Table 3). Gene loss is reported to contribute to structural variation of multiple fungal SMs [42,55,56]. Given the structural diversity of trichothecenes, TRI gene loss was expected to contribute to variation in gene content among the fungi examined, and indeed was previously reported from analyses of Fusarium and Stachybotrys [21,50,55]. However, absence of TRI4 in the Spicellum genomes was unexpected, because Tri4 catalyzes multiple reactions that are essential for formation of the EPT structure common to all trichothecenes (Fig 2) [18–20]. Production of trichothecenes by S. roseum 209012 (S4 Fig) indicates that the fungus has a gene(s) that compensates for the absence of TRI4. Furthermore, production of low levels of trichothecenes by T. arundinaceum tri4 deletion mutants indicates the existence of a gene that can partially compensate for the absence of TRI4 in this fungus [14]. The absence of TRI4 in Spicellum strains raises a question: what caused the loss of an enzyme that catalyzes multiple reactions essential for trichothecene biosynthesis and its replacement with another enzyme?
The absence of TRI6 and TRI10 in the B. bassiana and C. confragosa genomes was also unexpected given that these genes regulate expression of TRI genes in Fusarium [57–59]. Assuming B. bassiana and C. confragosa produce trichothecenes under some conditions, the absence of TRI6 and TRI10 indicates the existence of two fundamentally different regulatory systems for trichothecene biosynthesis in fungi. Among the fungi examined, B. bassiana and C. confragosa are the only insect pathogens. This raises a question: does the apparent change in regulation of TRI gene expression in B. bassiana and C. confragosa reflect an adaptation of trichothecene production for a lifestyle that includes insect pathogenesis?
The absence of the MFS transporter gene TRI12 was previously noted in analyses of the FIESC [24] and Stachybotrys species [21]. As a result, the absence of TRI12 in the B. bassiana and C. confragosa genomes was not surprising, but instead contributes to evidence that TRI12 is not essential for trichothecene production in fungi [60]. Presumably, another transporter(s) can compensate for the absence of Tri12 in trichothecene-producing fungi that lack TRI12. The presence of TRI12 in all fungi with a lineages A–C TRI cluster and its absence in some fungi with a lineage-D cluster (Fig 3) suggests that TRI12 was present in the ancestral TRI cluster. Further, the variable presence of TRI12 in lineage-D TRI clusters suggests three independent losses of the gene: once in Stachybotrys after divergence from Myrothecium; once in the Beauveria-Cordyceps clade after divergence from Fusarium; and once in FIESC after divergence from other fusaria.
TRI13 and TRI22: An unusual case of gain and loss
Except for F. graminearum, all known trichothecene-producing fungi examined here can produce trichothecenes that have a hydroxyl or ester group at C4 (Fig 2). This suggests that C4 oxygenation arose early in the evolutionary history of trichothecene biosynthesis, and therefore, that the common ancestor of extant TRI clusters likely encoded an enzyme that catalyzed this reaction. The results of the current and previous studies indicate that C4 hydroxylation is catalyzed by Tri22 in T. arundinaceum [17] and B. bassiana (Fig 6E and 6F) but by Tri13 in Fusarium [55]. Together, the presence of TRI22 in all the fungi examined here except Fusarium and the presence of TRI13 only in Fusarium (Table 3) suggest that TRI22 is the ancestral C4 hydroxylase gene, and that TRI13 was acquired after the Fusarium TRI cluster diverged from the cluster in other genera (Fig 10D). With the exception of the position of Spicellum and absence of Fusarium, the topology of the TRI22 tree is similar to the topology of the combined TRI3-TRI5-TRI14 tree, suggesting that the evolutionary history of TRI22 mirrors that of the TRI cluster to some extent. This, in turn, is consistent with the hypothesis that TRI22 is the ancestral C4 hydroxylase gene. If this hypothesis is correct, TRI22 would have been lost from and TRI13 would have been gained during the evolutionary divergence of the Fusarium cluster. Within Fusarium, production of trichothecenes that lack a C4 oxygen (e.g., deoxynivalenol) results from pseudogenization of TRI13 [55,61]. This observation suggests a possible scenario to explain how the gene conferring trichothecene C4 hydroxylation changed from TRI22 to TRI13. The scenario is based on the idea that if some extant fusaria produce trichothecenes that lack a C4 oxygen, ancestral trichothecene-producing fusaria could have produced trichothecenes that lack a C4 oxygen as well. In the scenario, TRI22 conferred C4 hydroxylation in ancestral trichothecene-producing fungi. Subsequently, during early divergence of the Fusarium TRI cluster, selection for production of trichothecenes with a C4 oxygen was relaxed and, as a result, TRI22 was lost. This gave rise to production of trichothecenes that lack a C4 oxygen. Subsequent to TRI22 loss, selection for production of trichothecene with a C4 oxygen was restored and, as a result, TRI13 was gained. Because all trichothecene-producing fusaria that have been examined to date have a functional or pseudogenized TRI13, we surmise that gain of TRI13 occurred early in divergence of the Fusarium TRI cluster [16,24,62]. After gain of TRI13, the gene was pseudogenized in one lineage of Fusarium, resulting in a mixed population in which some individuals produced trichothecenes that lacked C4 oxygen and others produced trichothecenes that have a C4 oxygen, a situation that still occurs in some lineages of Fusarium [37,63]. Thus, according to this scenario, trichothecene C4 hydroxylation has undergone a cycle whereby it existed in the ancestral trichothecene-producing fungus, was lost, then reacquired, and subsequently lost again.
Changes in TRI gene function
The results of the current and previous studies indicate that some trichothecene structural diversity has resulted from changes in function of TRI3 (Fig 4) and TRI4 (Fig 6A and 6B) [18–20,25]. Tri3 catalyzes C4 acylation in T. arundinaceum and C15 acylation (acetylation) in Fusarium. We propose that the C4 acylation activity is ancestral and C15 acylation is derived, because trichothecenes produced by fungi with a lineage A or B TRI cluster (i.e., Microcyclospora and Trichoderma) have an acyl group at C4 but not at C15 (Fig 10E). The proposed ancestral and derived activities of Tri3 are consistent with the acquisition of C15 oxygenation in fungi with a lineage C and D TRI cluster (Fig 10B), because a hydroxyl group at C15 is a prerequisite for C15 acetylation catalyzed by the Fusarium Tri3 [33,64]. If the proposed ancestral and derived activities of Tri3 are correct, the low level of C4 acetylation activity reported for recombinant F. graminearum Tri3 [64] indicates that some of the ancestral activity has been retained in this fungus.
The results of this and previous studies indicate Myrothecium and Trichoderma Tri4 homologs catalyze oxygenation at three positions of trichodiene, whereas Beauveria (Fig 6A and 6B) and Fusarium Tri4 homologs catalyze oxygenation at four positions [18–20,25]. The three oxygenations result in formation of trichothecenes that lack a C3 hydroxyl, while the four oxygenations result in formation of trichothecenes that have a C3 hydroxyl. Further, trichothecenes that lack a C3 hydroxyl are produced by all the fungi with a lineage-A–C TRI cluster and some fungi with a lineage-D cluster, whereas trichothecenes that have a C3 hydroxyl are produced only by some fungi with a lineage-D cluster (i.e., Fusarium and presumably Beauveria and Cordyceps). Based on this information, we propose that the ability to catalyze three oxygenations is the ancestral condition of Tri4, and the ability to catalyze four reactions is derived (Fig 10F). This hypothesis is consistent with the idea that C3 acetylation is also a derived condition, because an oxygen atom at C3 is a prerequisite for C3 acetylation catalyzed by Tri101 [3,16]. There is evidence for changes in functions of other TRI genes/enzymes as well, because Tri1 and Tri8 are reported to differ in function within and/or among Fusarium species [26,50–52].
Synthesis of polyketide-derived substituents of trichothecenes
Functional analyses of the T. arundinaceum and M. roridum TRI17 homologs indicate that Tri17 catalyzes synthesis of the polyketide precursors of the substituents esterified to C4 of some trichothecene analogs (Fig 5, S5 Fig). The polyketide-derived substituents of harzianum A and macrocyclic trichothecenes are made up of linear molecules that are either six (hexa-2,4-dienedioate) or eight (octatrienedioate and 6,7-dihydroxy-2,4-octadienoate) carbon atoms long (Fig 11A) [15,65]. The presence of TRI17 in Spicellum and Trichothecium suggests that the Tri17 homologs in these fungi catalyze synthesis of the four-carbon chain (2-butenoyl) that is esterified to the C4 oxygen of trichothecenes produced by these fungi (Fig 11A). The variable lengths of the polyketide-derived substituents in trichothecenes produced by Myrothecium, Stachybotrys, Spicellum, Trichoderma, and Trichothecium suggest that collectively, Tri17 homologs can catalyze synthesis of four, six or eight-carbon polyketides. Furthermore, single species of Myrothecium and Stachybotrys and even single isolates of some species can produce macrocyclic trichothecenes that have six- or eight-carbon polyketide-derived substituents [66,67]. Thus, it is likely that in some species, a single Tri17 homolog can catalyze synthesis of both six and eight-carbon polyketides.
Fig 11.
(A) Variation in the structure of polyketide-derived substituents in selected trichothecene analogs. The polyketide-derived substituent of each structure is highlighted with a gray background. Names below and/or to the right of highlighted areas are chemical or trivial names of the polyketide-derived substituents. Trichothecene names are indicated above each structure. (B) Proposed schemes for synthesis of octatrienedioate and 6,7-dihydroxy-2,4-octadienoate, the polyketide-derived substituents that occur in the structures of harzianum A and the macrocyclic trichothecene roridin A, respectively. In the proposed schemes, the parent compound of both substituents is 2,4,6-octatrienoate, an eight-carbon polyketide with three carbon-carbon double (enoyl) bonds. To form octatrienedioate, the polyketide would undergo a hydroxylation reaction followed by two oxidation reactions, first to form an aldehyde and second to form a carboxylic acid group. To form 6,7-dihydroxy-2,4-octadienoate, the polyketide would undergo two hydroxylation reactions during which an enoyl bond is lost. Formation of hexa-2,4-dienedioate, the polyketide-derived substituent in verrucarin A is not included in the figure, but would involve the same reactions as for synthesis of octatrienedioate, except that the parent polyketide would be a six-carbon polyketide with two enoyl bonds. It is not known whether the structural modifications of the polyketides occur before or after esterification to EPT. Thus, in the proposed schemes, R could be C4 of EPT, Co-enzyme A, or a hydrogen atom.
The Tri17 protein is predicted to include an enoyl reductase (ER) domain [21]. During polyketide biosynthesis, an ER domain catalyzes reduction of carbon-carbon double bonds to carbon-carbon single bonds [68]. PKSs that have the appropriate combination of other functional domains but lack a functional enoyl reductase domain catalyze synthesis of polyketides that have alternating double and single bonds. The polyketide-derived substituents in Trichoderma, Myrothecium, and Stachybotrys trichothecenes have such alternating double and single bonds. Thus, the Tri17 enoyl reductase domain is almost certainly nonfunctional.
In polyketide biosynthesis, carbon-chain length is controlled by the PKS enzyme [69]. In trichothecene biosynthesis, therefore, differences in polyketide-chain length (i.e., four carbons vs. six or eight carbons) likely result from differences in amino acid sequence of Tri17 homologs. In phylogenetic trees inferred from predicted amino acid sequences of Tri17 and related PKSs, Spicellum and Trichothecium Tri17 homologs, which likely catalyze synthesis of a four-carbon polyketide, form a clade basal to the Myrothecium, Stachybotrys and Trichoderma homologs, which likely catalyze synthesis of six- and eight-carbon polyketides (Fig 8). These phylogenetic relationships suggest that Tri17 homologs that catalyze synthesis of a four-carbon polyketide represent the ancestral Tri17 condition, whereas homologs that catalyze synthesis of six and/or eight-carbon polyketides represent a derived condition.
Given the predicted Tri17 functional domains and the structures of polyketide-derived substituents of harzianum A and macrocyclic trichothecenes, the polyketides precursors of these substituents are likely modified after release from Tri17. For example, the polyketide precursor of octatrienedioate is likely a linear, eight-carbon polyketide with alternating double and single carbon-carbon bonds, and one carboxylic acid group (Fig 11B). Because octatrienedioate has two carboxylic acid groups, its polyketide precursor likely undergoes modifications to form a second carboxylic acid group (Fig 11B). Likewise, the 6,7-dihydroxy-2,4-octadienoate substituent in some macrocyclic trichothecenes contains two adjacent hydroxyl groups. Because the polyketide precursor of this substituent is likely the same as the octatrienedioate precursor, formation of 6,7-dihydroxy-2,4-octadienoate would also require modifications of its polyketide precursor (Fig 11B).
Inference of an ancestral trichothecene biosynthetic pathway
The structural diversity of trichothecene analogs produced by the fungi examined in this study combined with information on the distribution, phylogenetic relationships and functions of TRI genes allow for inference of ancestral states of the TRI cluster and trichothecene biosynthetic pathway (Fig 12). The inferred ancestral cluster included the structural genes TRI3, TRI4, TRI5, TRI17, TRI18 and TRI22, the regulatory genes TRI6 and TRI10, the transporter gene TRI12, and TRI14 (Fig 12A). The presence of these 10 genes in the ancestral cluster is consistent with their presence in all fungi examined and/or their presence in all lineages of the TRI cluster (Fig 3). Based on observations discussed above, TRI4 in this ancestral cluster conferred the ability to catalyze three oxygenation reactions to yield EPT [18,19], and TRI3 conferred the ability to catalyze C4 rather than C15 acylation (Fig 12B). In addition to rationales described above, this role for TRI3 in the ancestral cluster is consistent with the observation that during trichothecene biosynthesis in T. arundinaceum, Tri22 and Tri3 function in tandem; Tri22 catalyzes C4 hydroxylation, and Tri3 catalyzes acylation of the resulting C4 hydroxyl (Fig 4) [17]. This tandem function of Tri22 and Tri3 is also consistent with the presence of both TRI22 and TRI3 in fungi that have a C4 but not C15 ester. As noted above, the ancestral Tri17 likely catalyzed synthesis of a four-carbon polyketide. Thus, the product of the inferred ancestral pathway would be 8-deoxy trichothecin (4-O-butenoyl EPT) (Fig 12B).
Fig 12.
(A) Proposed ancestral TRI cluster based on the distribution of TRI genes among fungi examined in the current study. (B) Proposed ancestral trichothecene biosynthetic pathway based on inferred gene content of the ancestral TRI cluster and knowledge of TRI gene functions. (C) The presence of two acetyl/acyl transferase genes in the proposed ancestral cluster raises the possibility of a branch at the end of the ancestral pathway that would result in the formation of two trichothecene analogs: 4-acetyl EPT and 4-butenoyl EPT.
We have included TRI18 in the inferred ancestral cluster because of its widespread distribution among trichothecene-producing fungi. However, we do not know its function in these fungi nor its likely function in the ancestral trichothecene biosynthetic pathway. In all fungi examined, when TRI17 is present, TRI18 is located adjacent to or near it (Fig 3). This consistent physical linkage of TRI17 and TRI18 suggests that the two genes could function together in biosynthesis. Given that TRI18 is predicted to encode an acyltransferase (Table 1), the most obvious possibility is that Tri18 catalyzes C4 esterification of the polyketide product of Tri17 to the C4 hydroxyl; i.e., Tri18 could have the same function as the proposed ancestral function of Tri3. Consistent with this idea is the evidence that there is a gene in T. arundinaceum that can partially compensate for TRI3 in tri3 deletion mutants of the fungus (Fig 4). Thus, one possibility is that in the ancestral trichothecene pathway, both Tri3 and Tri18 catalyzed esterification of the Tri17 product (butenoyl) to the hydroxyl group at C4. Another possibility is that both enzymes catalyzed trichothecene C4 esterification, but esterified different molecules to the C4 hydroxyl (e.g., acetyl and butenoyl). Such a difference in function of Tri3 and Tri18 would have caused a branch at the end of the ancestral pathway, with one branch leading to 8-deoxy trichothecin (4-O-butenoyl EPT) and the other leading to trichodermin (4-O-acetyl EPT) (Fig 12C). It is noteworthy that S. roseum can produce both of these metabolites (S4 Fig), and has homologs of both TRI3 and TRI18. Thus, the trichothecene products of the ancestral TRI cluster could be the same as those produced by an extant species of Spicellum.
We have not included C8 oxygenation in the proposed ancestral pathway. However, the presence and absence of a C8 oxygenation step in the ancestral pathway are both consistent with currently available data. Our rational for not including a C8 oxygenation step in the pathway was based on: 1) the hypothesis that the ancestral pathway would have been simpler than most extant pathways; and 2) the trichothecene-C8-oxygenase gene(s) in Microcyclospora and Trichothecium has not been identified, and therefore its distribution is not known. Given this, it is not possible to say whether the gene(s) was likely to have been present in the ancestral TRI cluster. If the ancestral pathway included a C8 oxygenation step, this ability would have to have been lost four or more times and re-acquired at least once to account for the distribution of the C8-oxygenation ability among trichothecene-producing fungi examined in this study. On the other hand, if C8 oxygenation was absent in the ancestral pathway, three independent gain events could account for the current distribution of the ability to produce C8-oxygenated trichothecenes (Fig 10A). Identification of gene(s) required for C8 oxygenation in Microcyclospora and Trichothecium should provide insight into the whether the ancestral pathway included this reaction.
Distribution and phylogenetic relationships of TRI genes
The sampling of fungi in the current study represents a majority of fungal genera in which trichothecene production has been reported [4–8]. The fungi are also phylogenetically diverse; M. tardicrescens is a member of the class Dothideomycetes, while the other fungi are members of five lineages within the class Sordariomycetes (order Hypocreales). Thus, the TRI cluster and trichothecene production are uncommon and discontinuously distributed among ascomycetous fungi. How the current distribution of the TRI cluster arose is not clear, but comparison of TRI-gene and species trees inferred in the current study suggest some possibilities (Fig 9). The trees suggest that Fusarium TRI genes are more closely related than expected to those of Beauveria and Cordyceps, whereas Trichoderma TRI genes are more distantly related than expected to those of Beauveria and Cordyceps. In other studies, similar conflicts among phylogenetic trees have been attributed to lineage sorting and HGT [37,38,41]. If the conflicts were caused by lineage sorting, the Beauveria-Cordyceps, Fusarium, and Trichoderma TRI clusters could represent ancestral alleles or ancient paralogs that have been differentially inherited by these fungi such that Beauveria-Cordyceps and Fusarium inherited one allele (or paralog), and Trichoderma inherited another allele (or paralog). On the other hand, the close relationship of TRI cluster homologs in Beauveria-Cordyceps and Fusarium could have resulted from a HGT event between these two fungal lineages. The distant relationship of Trichoderma and Beauveria-Cordyceps TRI clusters could have also resulted from HGT events in which these two fungal lineages were recipients of distantly related TRI clusters. It is also possible that the distribution and phylogenetic relationships of the TRI cluster among the fungi examined here are a product of lineage sorting in some cases and HGT in others. Future studies aimed at identification and analysis of additional TRI cluster homologs in phylogenetically more diverse fungi could provide more definitive evidence for processes that have contributed to the distribution of the cluster.
The conflict between the TRI22 tree (Fig 7) and the concatenated TRI-gene tree (Fig 9) with respect to the position of Spicellum suggests that the evolutionary history of TRI22 homologs has differed from other TRI genes in this fungal genus. This conflict could be attributed to lineage sorting or HGT, but either process would have involved TRI22 but not other extant TRI genes in Spicellum. Similar conflicts among TRI gene trees in closely related species of Fusarium were attributed to sorting of ancestral TRI-cluster alleles, which provide a mechanism to maintain production of two acetylated forms of deoxynivalenol [37]. It is unclear how a similar scenario could apply to TRI22 given that it confers C4 hydroxylation, and therefore, alleles of TRI22 are not likely to contribute to trichothecene structural diversity. It is noteworthy that the Spicellum strains were unusual among the fungi examined with respect to their position in the TRI22 tree as well as the absence of TRI4 in their genomes. Analyses of additional Spicellum species and their close relatives may provide insight into whether these two unusual features of Spicellum TRI genes resulted from related or independent events.
Conclusions
Together, the results of the current and previous studies provide insights into evolutionary processes that have given rise to trichothecene structural diversity in fungi. The findings of the current study have facilitated inference of an ancestral trichothecene biosynthetic pathway (Fig 12) that is consistent with extant pathways that collectively yield over 150 structurally diverse trichothecene analogs. Knowledge obtained from functional analyses of TRI genes in Fusarium and Trichoderma has contributed significantly to insights of the evolutionary history of trichothecene biosynthesis. However, it is likely that functional analyses of TRI homologs in other fungi will provide evidence that refines or disproves evolutionary scenarios proposed in this study. Functional studies in other fungi should also lead to identification of additional TRI genes responsible for structural diversity of trichothecene analogs, including genes responsible for: i) C8 oxygenation in Microcyclospora and Trichothecium; ii) structural modification of the polyketide precursors of macrocyclic trichothecenes and harzianum A; and iii) formation of macrolide rings of macrocyclic trichothecenes. Future studies that sample numerous strains of the same species should provide evidence for whether gains and losses of genes are consistent across species.
Trichothecene structural diversity appears to have arisen largely from gain, loss, and changes in function of TRI genes, evolutionary processes that have also attributed to structural diversity of ergot alkaloids produced by fungi of the family Clavicipitaceae [56]. Our results also indicate that the presence of some substituents of trichothecenes have evolved independently in different lineages of fungi through gain of different genes with the same function. In addition, at least one trichothecene modification (C4 oxygenation) appears to have been lost, reacquired, and subsequently lost again during divergence of the Fusarium TRI cluster. Structural diversity of trichothecene analogs likely reflects differences in selection experienced by fungi that produce the analogs [37]. Thus, the cycle of loss, reacquisition, and subsequent loss of C4-oxygenated trichothecenes likely reflects changes in selection for biological activity conferred by the analogs. Trichothecene production contributes to pathogenesis of some fusaria on some hosts [10], and there is evidence that trichothecenes structural diversity in one lineage of Fusarium has been maintained by balancing selection [37]. It is not clear whether trichothecene production contributes to the pathogenicity of other fungi as well, but adaptation to pathogenesis on different plants and insects could provide selection pressure that has driven structural diversification of trichothecenes.
Materials and methods
Genome sequence and RNAseq analyses
Genome sequences of B. bassiana [29], C. confragosa [30], F. graminearum [31] and Stachybotrys species [21] have been reported previously, and were downloaded from the National Center for Biotechnology Information (NCBI) database. Genome sequences for all other fungi were generated as part of the current study, primarily with a MiSeq Illumina platform (Illumina, Inc.). In the initial genome sequence assemblies for the Spicellum and Trichothecium strains, almost every TRI gene was on a different contig, most of which were less than 5 kb in length. We partially overcame this limitation by generating a single genome sequence assembly from sequence reads generated with MiSeq, TruSeq (Illumina, Inc.), and an Ion Torrent Ion Proton Sequencer (Thermo Fisher Scientific Inc.). In the resulting assemblies, TRI genes were present on only five or six contigs (Fig 3, S2 Fig). To prepare DNA for genome sequencing, fungal strains were grown in YEPD medium (0.1% yeast extract, 0.1% peptone, 2% glucose) for 2 days at room temperature with shaking at 200 rpm. The exception to this was M. tardicrescens HJS 1936, which was grown in liquid YMG medium (0.4% yeast extract, 1% malt extract, 0.4% glucose) for 10 days as previously described [7]. Mycelia were harvested by filtration, lyophilized, ground to a powder, and genomic DNA was extracted using the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research) or the chloroform-phenol method as previously described [70]. DNA sequencing libraries were prepared as follows. For the MiSeq platform, the Nextera XT DNA library Preparation Kit was used as specified by the manufacturer (Illumina). For the TruSeq platform, genomic DNA was first sonicated for four cycles with Diagenode Bioruptor system as specified by the manufacture to obtain 500-bp fragments (Diagenode). Sequencing libraries were prepared from the fragmented DNA with the TruSeq DNA LT Library Preparation Kit as specified by the manufacturer (Illumina). For the Ion Torrent Ion Proton Sequencer, the NEBNext Fast DNA Fragmentation & Library Prep Set for Ion Torrent was used as specified by the manufacturer (New England BioLabs).
Sequence reads obtained from each platform were processed and assembled using CLC Genomics Workbench (Qiagen Inc.). Gene predictions were done using the program Augustus [71] and FGENESH (Softberry, Inc., Mount Kisco, New York). All TRI and housekeeping genes obtained from the genome sequences and used in phylogenetic analyses were also manually annotated. The 18 TRI genes used as queries in BLAST analyses have been described in Fusarium [55], Trichoderma [17] and/or Stachybotrys [21]. BLASTx and BLASTn analyses were done against our in-house genome sequence database using CLC Genomics Workbench. Once contigs with TRI genes were identified in genome sequence assemblies, a portion of or the entire contig (depending on contig length) were subjected to gene prediction via FGENESH, and the resulting coding regions, as well as genomic sequences, were subjected to BLASTx analysis against the NCBI Non-redundant Protein Sequences database at NCBI to confirm the presence of TRI genes and to identify putative functions of other genes in the same region.
For RNAseq analysis, Spicellum and Trichothecium strains were grown in liquid YEPD medium for one, two, and three days, after which mycelia were harvested by filtration and lyophilized. RNA was isolated with the RNeasy method (Qiagen), and cDNA libraries were prepared with the MinElute Reaction Cleanup Kit (Qiagen). cDNA libraries were then sonicated for five cycles with Diagenode Bioruptor system (Diagenode) as specified by the manufacturer to obtain 100- to 300-bp fragments. Sequencing libraries were prepared from the sonicated DNA using a NEBNext Fast DNA Library Prep Set for Ion Torrent (New England BioLabs). The resulting library was then sequenced using Ion Torrent Ion Proton Sequencer platform (Thermo Fisher Scientific). The resulting sequence reads were analyzed using the RNA-Seq Analysis function in CLC Genomics Workbench.
TRI6 analysis
To confirm sequences of the three TRI6 paralogs in Trichothecium, we amplified each paralog from three strains of T. roseum by PCR and sequenced the amplicons via Sanger Sequencing. PCR primers used for these amplifications are shown in S1 Table. The DNA polymerase GoTaq was used for amplification, and the conditions were those recommended by the manufacturer (Promega). Amplicons were purified using standard agarose gel electrophoresis and the UltraClean protocol (Mo Bio Laboratories). Amplicons were sequenced using BigDye Terminator version 3.1 and BigDye Xterminator Purification reagents (Thermo Fisher Scientific), and sequences were determined with a 3739 DNA Analyzer (Thermo Fisher Scientific). Sequences were viewed and edited using Sequencher (Gene Codes Corporation).
Deletion and complementation of Trichoderma TRI genes
For deletion and complementation of T. arundinaceum TRI genes, previously described plasmid and protoplast-mediated transformation methods were used [17]. Deletion mutants and complemented deletion mutants were examined for their ability to produce harzianum A and other trichothecene analogs using the previously described two-step culture procedure [17].
TRI3 deletion was accomplished by transformation with plasmid pΔtri3 that had been linearized with ApaI prior to transformation (Fig A in S1 File). Transformants were selected using 100 μg hygromycin B per mL of selection medium as previously described [17,72]. Transformants were analyzed by PCR with oligonucleotides Tarun-TrpC3/Tarun-compT35F (S1 Table) for the presence of a fragment expected to result from replacement of the TRI3 coding region with the hygromycin resistance cassette (hygB). Transformants that yielded a PCR product were then analyzed by PCR to confirm the absence of TRI3. TRI3 deletion was confirmed by Southern blot analysis using four hybridization probes (Fig A in S1 File). Based on these analyses, we concluded that transformants tri3.1, tri3.30, tri3.33 and tri3.48 were tri3 deletion mutants (Fig A in S1 File).
tri3 deletion mutant tri3.1 was complemented by transformation with plasmid pTCtri3-ble linearized with EcoRI (Fig B in S1 File). Transformants were selected using 75 μg phleomycin per mL of selection medium. Five transformants were analyzed by PCR for the presence of TRI3 and the phleomycin resistance cassette (bleR) with oligonucleotides Tarun-T3int3/Tarun-T3int5 and Tarun-Phleo-3/Tarun-Phleo-4, respectively (S1 Table). Three transformants that yielded amplicons from both primer pairs were analyzed by Southern blot analysis with two hybridization probes to confirm the presence of TRI3 and bleR (Fig B in S1 File).
TRI17 deletion was accomplished by transformation with plasmid pΔtri17 linearized with XhoI prior to transformation (Fig C in S1 File). Transformants were selected with hygromycin as described above and analyzed by PCR with oligonucleotides Tarun-Db741/Tarun-Db742 to detect hygB, and with oligonucleotides Tarun-pks-F/Tarun-pks-R to test for the absence of TRI17 (S1 Table). Transformants that yielded the appropriate amplicons were analyzed further by PCR with oligonucleotides Tarun-5-disrT/Tarun-TtrpC-disrT (S1 Table) to test for a fragment expected to result from replacement of the TRI17 coding region with hygB. Selected transformants were also analyzed by Southern blot analysis to confirm deletion of TRI17 (Fig C in S1 File). Based on these analyses, we concluded that transformants tri17.96, tri17.109 and tri17.139 were tri17 deletion mutants.
tri17 deletion mutant tri17.139 was complemented by transformation with plasmid pTCtri17-ble linearized with NotI (Fig D in S1 File). Transformants were selected with 100 μg of phleomycin per mL of selection medium. Transformants were analyzed by PCR for the presence of TRI17 and ble with oligonucleotides Tarun-pks-F/Tarun-pks-R and Tarun-Phleo-3/Tarun-Phleo-4, respectively (S1 Table). A subset of six transformants that yielded amplicons from both primer pairs were subjected to Southern blot analysis with three hybridization probes to confirm the presence of TRI17 and bleR (Fig D in S1 File).
Tri17 deletion mutant tri17.139 was also complemented with a plasmid, pTCMrtri17-ble, carrying M. roridum TRI17 and that had been linearized with EcoRI (Fig E in S1 File). Transformants were selected as indicated above, and analyzed by PCR for the M. roridum TRI17 and bleR genes (Fig E in S1 File). Based in the PCR results, transformants tri17.MrT17.C3, .C4, .C5, and .C13 were selected for further studies.
Heterologous expression of B. bassiana TRI genes
To express TRI4 and TRI22 from B. bassiana in F. verticillioides, the coding region (with intron sequences intact) plus ~500-bp of the 3’ flanking region of each gene was fused to the promoter sequence of the translation elongation factor 1α (TEF1) gene from Aureobasidium pullulans. This was done using a previously described PCR-based fusion method [26]. The primers used for the PCR are shown in S1 Table, and DNA polymerases used were iProof High Fidelity DNA polymerase (Bio-Rad Laboratories) for TRI4 and Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific) for TRI22. For the fusion PCR, the A. pullulans TEF1 promoter was amplified from plasmid pTEFEGFP [73] with a reverse primer that consisted of approximately 22 nucleotides of the 3’ end of the TEF1 promoter and approximately 22 nucleotides of the 5’ end of the B. bassiana TRI4 (or TRI22) coding region. Likewise, the B. bassiana TRI4 (or TRI22) coding region and 3’ flanking region were amplified from genomic DNA of B. bassiana strain ARSEF 2860 using a forward primer that was essentially the reverse complement of the primer described above; that is, the primer included sequences of both the 3’ end of the A. pullulans TEF1 promoter and 5’ end of the B. bassiana TRI4 (or TRI22) coding region. The two amplicons were then annealed and amplified by PCR without primers to generate the chimeric TEF1 promoter::TRI4 (or TRI22) construct, which was then further amplified with nested primers as previously described [26]. The fusion amplicons were cloned into the PCR cloning vector pCR-XL1 TOPO (Thermo Fisher Scientific) and sequenced to confirm that PCR amplification did not introduce nucleotide errors. The geneticin resistance gene (genR) was then introduced into the resulting vector via NotI digestion using standard molecular biology protocols as described previously [26].
The TEF1 promoter::TRI4 (or TRI22)-genR vector was then introduced into F. verticillioides strain M-3125 via a protoplast-mediated transformation protocol as previously described [72]. The presence of the construct in F. verticillioides transformants was confirmed by PCR with primer combinations used to generate the TEF1 promoter::TRI constructs.
For precursor feeding experiments, F. verticillioides transformants were inoculated into liquid YEPD medium. Trichodiene or isotrichodermin (3-O-acetyl EPT), obtained from previous studies [74,75], were dissolved in acetone and then added to cultures at a final concentration of 250 μM. The final concentration of acetone was less than 1%. The cultures were incubated in the dark at 28 °C with shaking (200 rpm). After six days, cultures were extracted with ethyl acetate, and the extracts were analyzed by GC-MC as described below.
To express the B. bassiana TRI101 in S. cerevisiae, the TRI101 coding region was amplified from B. bassiana ARSEF 2860 genomic DNA using primers indicated in S1 Table and iProof High Fidelity DNA polymerase (Bio-Rad Laboratories) following the manufacturer’s recommendations. The PCR product was gel purified using the QIAEX II Gel Extraction Kit (Invitrogen), treated with 1000 units of Taq DNA polymerase (Qiagen) to add A overhangs to the amplicon 3’ ends, and then cloned into pYES2.1 using the TOPO TA Yeast Expression Kit (Invitrogen). In the resulting plasmid, the TRI101 coding region was fused to the GAL1 promoter and termination sequence. The cloned TRI101 was sequenced to confirm that PCR amplification did not introduce errors, and then the plasmid was introduced into an ayt1 mutant of S. cerevisiae (strain YLL063C; GE Healthcare Dharmacon) using the TRAFO protocol [76]. For feeding studies, yeast transformants were grown in the dark at 28 °C with shaking (200 rpm) on minimal medium [77], supplemented with leucine (1g/L), methionine (200 mg/L) and histidine (200 mg g/L. After 3 days, cultures were centrifuged and the pellet was re-suspended in YGal medium (1% yeast extract, 2% peptone, 2% galactose) to induce TRI101 expression. Isotrichodermol (3-hydroxy EPT) was added to the cultures at a final concentration of 250 μM. After an additional four-day incubation in the dark at 28 °C with shaking (200 rpm), cultures were extracted with ethyl acetate and analyzed with GC-MS as described below.
Chromatography and mass spectrometry
HPLC, LC-MS/MS and GC-MS were used to monitor trichothecenes and other metabolites produced by fungal strains. Harzianum A (HA), which is not detectable by GC-MS analysis, was detectable and quantified by HPLC and/or LC-MS/MS analysis. For HPLC analysis of harzianum A, T. arundinaceum cultures were extracted with an equal volume of ethyl acetate. The upper phase was recovered and evaporated to dryness in a rotary evaporator at room temperature, and then redissolved in acetonitrile at 10% of the original volume. After a final 1/5 dilution, a 20 μL aliquot of the resulting sample was used for HPLC analysis. The HPLC system consisted of a Waters 600 HPLC connected to a 996 Photodiode Array Detector (Waters Corporation) [17]. The column was a Waters YMC analytical column (150 mm length, 4.6 mm internal diameter). The initial mobile phase was 40:60 acetonitrile:water with 0.1% trifluoroacetic acid, and had a flow rate of 1 mL/min. After 30 min, the mobile phase was adjusted to 100% acetonitrile over 10 min, held for 5 min, and then returned to the initial condition [17].
For both GC-MS and LC-MS/MS analyses, 5-mL aliquots of liquid cultures (fungal biomass and medium) were combined with 2 mL ethyl acetate, and mixed vigorously for 5 min. The ethyl acetate phase was recovered and used directly in GC-MS and LC-MS/MS analyses. The GC-MS system consisted of a Hewlett Packard 6890 gas chromatograph fitted with a HP-5MS column (30 m length, 0.25 mm film thickness) and a 5973 mass detector (Hewlett Packard). The carrier gas was helium with a 20:1 split ratio and a 20 mL/min split flow. The column was held at 120 °C at injection, heated to 210 °C at 15 °C/min and held for 1 min, then heated to 260 °C at 5 °C/min and held for 8 min.
The LC-MS/MS system consisted of a ThermoDionex Ultimate 3000 UPLC fitted with a Phenomenex Kinetex F5 column (150 mm length, 2.1 mm diameter, 1.7 μm particle size) connected to the ionspray interface of an ABSciex Qtrap 3200 mass spectrometer operated in negative mode. The chromatographic separation was done with a 200 μL/min gradient flow of water and acetonitrile with 0.3% ammonium acetate. The separation utilized a gradient from 35 to 95% aqueous acetonitrile over 10 min. The column was held at 50 °C for the entire analysis.
In the analytical systems described above, identification of trichothecene analogs was confirmed by comparing their chromatographic retention times and, in MS analyses, molecular masses and mass spectra to those of standards. Novel trichothecenes were purified and their structures were determined by 1H and 13C NMR spectrometry as previously described [17,78].
Phylogenetic analyses
For phylogenetic analyses of TRI and housekeeping genes, coding region sequences were aligned via the computer program MUSCLE as implemented in MEGA7 [79]. In general, nucleotide sequences were translated to amino acid sequences, aligned, and then converted back to nucleotide sequences before further analysis. Aligned sequences were subjected to maximum likelihood analysis using the computer program IQ-Tree version 1.5.5 [80]. In analyses with IQ-Tree, the best substitution model was determined by the program prior to tree building. Some alignments (e.g., TRI6) were also subjected to maximum parsimony analysis using MEGA7, and for some genes (e.g., TRI17 and TRI101) alignments of deduced amino acid sequences were analyzed in addition to nucleotide sequences. For TRI genes, potential conflicts within sets of concatenated gene sequences were initially assessed using the Homogeneity Partition test as implemented in PAUP version 4.0a [81]. Bootstrap [80], SH [35], and AU [36] analyses, as implemented in IQ-Tree version 1.5.5, were also used to assess whether trees inferred from different sequences were significantly different. In preliminary analyses of individual TRI genes, it was not clear which strain/species should be used to root trees. To assess the most appropriate TRI-gene root in each tree, we conducted BLASTx analyses against the GenBank non-redundant database to identify genes that were distantly related to but would still align to TRI genes [82]. Four to six of the best BLAST hits were then used for alignment and tree building with the corresponding TRI gene, and thereby obtain information on which TRI gene homolog(s) was most basal.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
In the diagrams (above), green arrows represent known TRI genes, and gray arrows represent genes that are unique to a region in a particular genus. Numbers below arrows are locus tag numbers (with five-letter prefix, the underscore, and in some cases a zero omitted). The tables below the diagrams include predicted functions of genes based on Blast2Go analysis and supplemented with manual blast analysis in some cases. Tables also include information on contigs on which the genes occur. In the tables, genes above and below a double line are on different contigs. A. Beauveria bassiana; B. Cordyceps confragosa—orange arrows represent genes that are common to the TRI cluster locus in Beauveria and Cordyceps; C. Microcylospora tardicrescens; D. Myrothecium roridum—purple arrows represent genes that are common to TRI loci in Myrothecium and Stachybotrys chartarum; E. Spicellum ovalisporum; F. Spicellum roseum; G. Stachybotrys chartarum; H. Trichoderma arundinaceum; and I. Trichothecium roseum.
(PPTX)
The nucleotide sequence of the query is shown below the BLASTx results.
(PPTX)
Peaks corresponding to trichodermol (4-hydroxy EPT), trichodermin (4-O-acetyl EPT), and 8-deoxy trichothecin (4-O-butenoyl EPT) are shown. Based on mass spectral fragmentation patterns, the unlabeled peaks at 5 min and 6.1 min do not correspond to trichothecenes.
(PPTX)
Results are presented for the wild-type progenitor strain (blue trace) and complementation strains tri17.MrT17.C3 (black trace) and tri17.MrT17.C4 (red trace). The trace for tri17 mutant strain tri17.139 (purple trace) does not rise above the base line. Identity of harzianum A in samples was confirmed by comparison of mass spectra to the spectrum of a purified harzianum A standard, the identity of which was confirmed by 1H and 13C NMR. Strains were grown in YEPD medium for one week.
(PPTX)
Fig A: Total ion chromatogram of an ethyl acetate extract of the culture (above), and mass spectrum of 4-hydroxy isotrichodermin (below). Fig B: 13C (left) and 1H (right) NMR spectra of 4-hydroxy isotrichodermin.
(PPTX)
In the tree shown here, a distantly related homolog was selected for each of the three TRI genes. For TRI3, we selected a homolog of TRI18 as the outgroup. We searched GenBank by BLASTx [75] to identify non-TRI gene homologs to use at outgroups in the TRI5 and TRI14 trees; the outgroups are represented by their GenBank accession numbers. To generate trees, deduced amino acid sequences of each gene were aligned with Muscle as implemented in MEGA7 [79], and trees were inferred using the maximum likelihood method with ultrafast bootstrapping [83] as implemented in IQ-Tree [73]. Numbers near branches are bootstrap values based on 1000 pseudoreplicates.
(PPTX)
(PPTX)
(DOCX)
Acknowledgments
We are grateful for the technical assistance of Crystal Probyn, José Alvarez, Jennifer Teresi, Marcie Moore, Amy McGovern, Chris McGovern, Stephanie Folmar, Christine Hodges, and Nathane Orwig. We are also grateful to Todd J. Ward for helpful comments on phylogenetic analyses.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Data Availability
Genome sequence data generated as part of this study have been submitted to the GenBank database at the National Center for Biotechnology Information. The accession numbers for the genome sequences are as follows: PXOG00000000 Fusarium longipes NRRL 20695, PXOF00000000 Fusarium sporotrichioides NRRL, 3299 PXXK00000000 Fusarium sp. FIESC 12 NRRL 13405, PXOE00000000 Microcyclospora tardicrescens HJS1936, PXOD00000000 Myrothecium roridum NRRL 2183, PXOC00000000 Spicellum ovalisporum DAOM 186447, PXOB00000000 Spicellum roseum DAOM 209012, PXOA00000000 Trichoderma arundinaceum IBT 40837, PXNZ00000000 Trichoderma brevicompactum IBT 40841, PXNX00000000 Trichothecium roseum DAOM 197141, PXNY00000000 Trichothecium roseum DAOM 195227, PXNW00000000 Trichothecium roseum K7-1.
Funding Statement
SG received funding from the Spanish Ministry of Economy and Competitiveness (grant number MINECO-AGL2015-70671-C2-2-R). TL received funding from the National Institute of Agricultural Sciences, Rural Development Administration (grant number PJ00843203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24: 393–416. doi: 10.1039/b603084j [DOI] [PubMed] [Google Scholar]
- 2.Keller NP, Turner G, Bennett JW (2005) Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3: 937–947. doi: 10.1038/nrmicro1286 [DOI] [PubMed] [Google Scholar]
- 3.Alexander NJ, Proctor RH, McCormick SP (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev 28: 198–215. [Google Scholar]
- 4.Cole R, Jarvis BB, Schweikert MA (2003) Handbook of Secondary fungal Metabolites Volume III. San Diego: Academic Press. [Google Scholar]
- 5.Kikuchi H, Miyagawa Y, Sahashi Y, Inatomi S, Haganuma A, et al. (2004) Novel spirocyclic trichothecanes, spirotenuipesine A and B, isolated from entomopathogenic fungus, Paecilomyces tenuipes. J Org Chem 69: 352–356. doi: 10.1021/jo035137x [DOI] [PubMed] [Google Scholar]
- 6.McCormick SP, Stanley AM, Stover NA, Alexander NJ (2011) Trichothecenes: from simple to complex mycotoxins. Toxins (Basel) 3: 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surup F, Medjedović A, Szczygielski M, Schroers H-J, Stadler M (2014) Production of Trichothecenes by the Apple Sooty Blotch Fungus Microcyclospora tardicrescens. J Agricl Food Chem 62: 3525–3530. [DOI] [PubMed] [Google Scholar]
- 8.Venkatasubbaiah P, Sutton TB, Chilton WS (1995) The structure and biological properties of secondary metabolites produced by Peltaster fructicola, a fungus associated with apple sooty blotch disease. Plant Dis 79: 1157–1160. [Google Scholar]
- 9.Desjardins AE, Hohn TM, McCormick SP (1992) Effect of gene disruption of trichodiene synthase on the virulence of Gibberella pulicaris. Mol Plant Microbe Interact 5: 214–222. [DOI] [PubMed] [Google Scholar]
- 10.Desjardins AE, Proctor RH, Bai G, McCormick SP, Shaner G, et al. (1996) Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol Plant-Microbe Interact 9: 775–781. [Google Scholar]
- 11.Cuzick A, Urban M, Hammond-Kosack K (2008) Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. New Phytol 177: 990–1000. doi: 10.1111/j.1469-8137.2007.02333.x [DOI] [PubMed] [Google Scholar]
- 12.Desjardins AE (2006) Fusarium Mycotoxins Chemistry, Genetics and Biology. St. Paul: APS Press. [Google Scholar]
- 13.Straus DC (2009) Molds, mycotoxins, and sick building syndrome. Toxicol Ind Health 25: 617–635. doi: 10.1177/0748233709348287 [DOI] [PubMed] [Google Scholar]
- 14.Malmierca MG, Cardoza RE, Alexander NJ, McCormick SP, Hermosa R, et al. (2012) Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl Environ Microbiol 78: 4856–4868. doi: 10.1128/AEM.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamm C, Breitenstein W (1980) The biosynthesis of trichothecene Mycotoxins In: Steyn P, editor. The Biosynthesis of Mycotoxins: A Study in Secondary Metabolism. New York: Academic Press; pp. 69–104. [Google Scholar]
- 16.Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M (2007) Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem 71: 2105–2123. [DOI] [PubMed] [Google Scholar]
- 17.Cardoza RE, Malmierca MG, Hermosa MR, Alexander NJ, McCormick SP, et al. (2011) Identification of loci and functional characterization of trichothecene biosynthesis genes in filamentous fungi of the genus Trichoderma. Appl Environ Microbiol 77: 4867–4877. doi: 10.1128/AEM.00595-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick SP, Alexander NA, Proctor RH (2006) Fusarium Tri4 encodes a multifunctional oxygenase required for trichothecene biosynthesis. Canadian Journal of Microbiology 52: 636–642. doi: 10.1139/w06-011 [DOI] [PubMed] [Google Scholar]
- 19.McCormick SP, Alexander NJ (2007) Myrothecium roridum Tri4 encodes a multifunctional oxygenase required for three oxygenation steps. Can J Microbiol 53: 572–579. doi: 10.1139/W07-025 [DOI] [PubMed] [Google Scholar]
- 20.Tokai T, Koshino H, Takahashi-Ando N, Sato M, Fujimura M, et al. (2007) Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem Biophys Res Commun 353: 412–417. [DOI] [PubMed] [Google Scholar]
- 21.Semeiks J, Borek D, Otwinowski Z, Grishin NV (2014) Comparative genome sequencing reveals chemotype-specific gene clusters in the toxigenic black mold Stachybotrys. BMC Genomics 15: 590 doi: 10.1186/1471-2164-15-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapp SC, Hohn TM, McCormick SP, Jarvis BB (1998) Characterization of the macrocyclic trichothecene gene cluster in Myrothecium roridum. Mol Gen Genet 257: 421–432. [DOI] [PubMed] [Google Scholar]
- 23.Ye W, Liu T, Zhu M, Zhang W, Li H, et al. (2017) De novo transcriptome analysis of plant pathogenic fungus Myrothecium roridum and identification of genes associated with trichothecene mycotoxin biosynthesis. Int J Mol Sci 18: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor RH, McCormick SP, Alexander NJ, Desjardins AE (2009) Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol 74: 1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x [DOI] [PubMed] [Google Scholar]
- 25.Cardoza RE, McCormick SP, Malmierca MG, Olivera ER, Alexander NJ, et al. (2015) Effects of trichothecene production on the plant defense response and fungal physiology: overexpression of the Trichoderma arundinaceum tri4 gene in T. harzianum. Appl Environ Microbiol 81: 6355–6366. doi: 10.1128/AEM.01626-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander NJ, McCormick SP, Waalwijk C, van der Lee T, Proctor RH (2011) The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet Biol 48: 485–495. doi: 10.1016/j.fgb.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 27.Corley DG, Miller-Wideman M, Durley RC (1994) Isolation and structure of harzianum A: a new trichothecene from Trichoderma harzianum. J Nat Prod 57: 422–425. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, et al. (2009) Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium inacrnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol 47: 3851–3861. doi: 10.1128/JCM.01616-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao G, Ying SH, Zheng P, Wang ZL, Zhang S, et al. (2012) Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep 2: 483 doi: 10.1038/srep00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y, Xiao G, Zheng P, Cen K, Zhan S, et al. (2016) Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol 8: 1374–1387. doi: 10.1093/gbe/evw082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, et al. (2010) Comparative genomics reveal mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. doi: 10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seifert KA, Louis-Seize G, Savard ME (1997) The phylogenetic relationships of two trichothecene-producing hyphomycetes, Spicellum roseum and Trichothecium roseum. Mycologia 89: 250–257. [Google Scholar]
- 33.McCormick SP, Hohn TM, Desjardins AE (1996) Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl Environ Microbiol 62: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick SP, Alexander NJ, Proctor RH (2006) Heterologous expression of two trichothecene P450 genes in Fusarium verticillioides. Can J Microbiol 52: 220–226. doi: 10.1139/w05-124 [DOI] [PubMed] [Google Scholar]
- 35.Shimodaira H, Hasagawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16: 1114–1116. [Google Scholar]
- 36.Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51: 492–508. doi: 10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- 37.Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K (2002) Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci U S A 99: 9278–9283. doi: 10.1073/pnas.142307199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proctor RH, Van Hove F, Susca A, Stea G, Busman M, et al. (2013) Birth, death and horizontal transfer of the fumonisin biosynthetic gene cluster during the evolutionary diversification of Fusarium. Mol Microbiol 90: 290–306. doi: 10.1111/mmi.12362 [DOI] [PubMed] [Google Scholar]
- 39.Khatibi PA, Newmister SA, Rayment I, McCormick SP, Alexander NJ, et al. (2011) Bioprospecting for trichothecene 3-O-acetyltransferases in the fungal genus Fusarium yields functional enzymes with different abilities to modify the mycotoxin deoxynivalenol. Appl Environ Microbiol 77: 1162–1170. doi: 10.1128/AEM.01738-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokai T, Fujimura M, Inoue H, Aoki T, Ohta K, et al. (2005) Concordant evolution of trichothecene 3-O-acetyltransferase and rDNA species phylogeny of trichothecene-producing and nonproducing fusaria and other ascomycetous fungi. Microbiology 151: 509–519. doi: 10.1099/mic.0.27435-0 [DOI] [PubMed] [Google Scholar]
- 41.Campbell MA, Rokas A, Slot JC (2012) Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol 4: 289–293. doi: 10.1093/gbe/evs011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds H, Slot JC, Divon HH, Lysoe E, Proctor RH, et al. (2017) Differential retention of gene functions in a secondary metabolite cluster. Mol Biol Evol. 34: 2002–2015. doi: 10.1093/molbev/msx145 [DOI] [PubMed] [Google Scholar]
- 43.Susca A, Proctor RH, Morelli M, Haidukowski M, Gallo A, et al. (2016) Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae isolates from multiple crop and geographic origins. Front Microbiol 7: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus E-M, et al. (2013) Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog 9: e1003475 doi: 10.1371/journal.ppat.1003475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell MA, Staats M, van Kan JA, Rokas A, Slot JC (2013) Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 105: 1126–1134. doi: 10.3852/12-390 [DOI] [PubMed] [Google Scholar]
- 46.Maeda K, Tanaka A, Sugiura R, Koshino H, Tokai T, et al. (2016) Hydroxylations of trichothecene rings in the biosynthesis of Fusarium trichothecenes: evolution of alternative pathways in the nivalenol chemotype. Environ Microbiol 18: 3798–3811. doi: 10.1111/1462-2920.13338 [DOI] [PubMed] [Google Scholar]
- 47.McCormick SP, Hohn TM (1997) Accumulation of trichothecenes in liquid cultures of a Fusarium sporotrichioides mutant lacking a functional trichothecene C-15 hydroxylase. ApplEnvironMicrobiol 63: 1685–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura M, Matsumoto G, Shingu Y, Yoneyama K, Yamaguchi I (1998) The mystery of the trichothecene 3-O-acetyltransferase gene analysis of the region around Tri101 and characterization of its homologue from Fusarium sporotrichioides. FEBS Letters 435: 163–168. [DOI] [PubMed] [Google Scholar]
- 49.Kimura M, Tokai T, Matsumoto G, Fujimura M, Hamamoto H, et al. (2003) Trichothecene nonproducer Gibberella species have both functional and nonfunctional 3-O-actyltransferase genes. Genetics 163: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown DW, Proctor RH, Dyer RB, Plattner RD (2003) Characterization of a fusarium 2-gene cluster involved in trichothecene C-8 modification. J Agric Food Chem 51: 7936–7944. doi: 10.1021/jf030607+ [DOI] [PubMed] [Google Scholar]
- 51.McCormick SP, Harris LJ, Alexander NJ, Ouellet T, Saparno A, et al. (2004) Tri1 in Fusarium graminearum encodes a P450 oxygenase. ApplEnvironMicrobiol 70: 2044–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meek IB, Peplow AW, Ake C, Phillips TD, Beremand MN (2003) Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. ApplEnvironMicrobiol 69: 1607–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varga E, Wiesenberger G, Hametner C, Ward TJ, Dong Y, et al. (2015) New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ Microbiol 17: 2588–2600. doi: 10.1111/1462-2920.12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexander NJ, Hohn TM, McCormick SP (1998) The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl Environ Microbiol 64: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE (2002) Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet Biol 36: 224–233. [DOI] [PubMed] [Google Scholar]
- 56.Young CA, Schardl CL, Panaccione DG, Florea S, Takach JE, et al. (2015) Genetics, genomics and evolution of ergot alkaloid diversity. Toxins (Basel) 7: 1273–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasmith CG, Walkowiak S, Wang L, Leung WW, Gong Y, et al. (2011) Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum. PLoS Pathog 7: e1002266 doi: 10.1371/journal.ppat.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tag AG, Garifullina GF, Peplow AW, Ake C, Phillips TD, et al. (2001) A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. ApplEnvironMicrobiol 67: 5294–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seong KY, Pasquali M, Zhou X, Song J, Hilburn K, et al. (2009) Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol Microbiol 72: 354–367. doi: 10.1111/j.1365-2958.2009.06649.x [DOI] [PubMed] [Google Scholar]
- 60.Nakajima Y, Koseki N, Sugiura R, Tominaga N, Maeda K, et al. (2015) Effect of disrupting the trichothecene efflux pump encoded by FgTri12 in the nivalenol chemotype of Fusarium graminearum. J Gen Appl Microbiol 61: 93–96. doi: 10.2323/jgam.61.93 [DOI] [PubMed] [Google Scholar]
- 61.Lee T, Han YK, Kim KH, Yun SH, Lee YW (2002) Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl Environ Microbiol 68: 2148–2154. doi: 10.1128/AEM.68.5.2148-2154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelly A, Proctor RH, Belzile F, Chulze SN, Clear RM, et al. (2016) The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet Biol 95: 39–48. doi: 10.1016/j.fgb.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 63.Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, et al. (2007) Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol 44: 1191–1204. doi: 10.1016/j.fgb.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 64.Tokai T, Takahashi-Ando N, Izawa M, Kamakura T, Yoshida M, et al. (2008) 4-O-acetylation and 3-O-acetylation of trichothecenes by trichothecene 15-O-acetyltransferase encoded by Fusarium Tri3. Biosci Biotechnol Biochem 72: 2485–2489. doi: 10.1271/bbb.80501 [DOI] [PubMed] [Google Scholar]
- 65.Jarvis BB (1991) Macrocyclic trichothecenes In: Sharma RP, Salunkhe DK, editors. Mycotoxins and Phytoalexins. Boca Raton: CRC Press, Inc. pp. 361–421. [Google Scholar]
- 66.Mondol MA, Surovy MZ, Islam MT, Schuffler A, Laatsch H (2015) Macrocyclic trichothecenes from Myrothecium roridum strain M10 with motility inhibitory and zoosporicidal activities against Phytophthora nicotianae. J Agric Food Chem 63: 8777–8786. doi: 10.1021/acs.jafc.5b02366 [DOI] [PubMed] [Google Scholar]
- 67.Rosso M, Maier M, Bertoni M (2000) Macrocyclic trichothecene production by the fungus epibiont of Baccharis coridifolia. Molecules 5: 345–347. [Google Scholar]
- 68.Hopwood DA (1997) Genetic contributions to understanding polyketide synthases. Chem Rev 97: 2465–2498. [DOI] [PubMed] [Google Scholar]
- 69.Crawford JM, Townsend CA (2010) New insights into the formation of fungal aromatic polyketides. Nat Rev Micro 8: 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raeder U, Broda P (1885) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1: 17–20. [Google Scholar]
- 71.Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33: W465–467. doi: 10.1093/nar/gki458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Proctor RH, Desjardins AE, Plattner RD, Hohn TM (1999) A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet Biol 27: 100–112. doi: 10.1006/fgbi.1999.1141 [DOI] [PubMed] [Google Scholar]
- 73.Van den Wymelenberg AJ, Cullen D, Spear RN, Schoenike B, Andrews JH (1997) Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 23: 686–690. [DOI] [PubMed] [Google Scholar]
- 74.Hohn TM, Desjardins AE (1992) Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol Plant-Microbe Interact 5: 249–256. [DOI] [PubMed] [Google Scholar]
- 75.McCormick SP, Alexander NJ, Trapp SC, Hohn TM (1999) Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. ApplEnvironMicrobiol 65: 5252–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Birky CW Jr. (1975) Effects of glucose repression of the transmission and recombination of mitochondrial genes in yeast (Saccharomyces cerevisiae). Genetics 80: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Desjardins AE, McCormick SP, Appell M (2007) Structure-activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J Agric Food Chem 55: 6487–6492. doi: 10.1021/jf0709193 [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2014) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32: 268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10 ed Sunderland, MA: Sinauer Associates. [Google Scholar]
- 82.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minh BQ, Nguyen MA, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30: 1188–1195. doi: 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
In the diagrams (above), green arrows represent known TRI genes, and gray arrows represent genes that are unique to a region in a particular genus. Numbers below arrows are locus tag numbers (with five-letter prefix, the underscore, and in some cases a zero omitted). The tables below the diagrams include predicted functions of genes based on Blast2Go analysis and supplemented with manual blast analysis in some cases. Tables also include information on contigs on which the genes occur. In the tables, genes above and below a double line are on different contigs. A. Beauveria bassiana; B. Cordyceps confragosa—orange arrows represent genes that are common to the TRI cluster locus in Beauveria and Cordyceps; C. Microcylospora tardicrescens; D. Myrothecium roridum—purple arrows represent genes that are common to TRI loci in Myrothecium and Stachybotrys chartarum; E. Spicellum ovalisporum; F. Spicellum roseum; G. Stachybotrys chartarum; H. Trichoderma arundinaceum; and I. Trichothecium roseum.
(PPTX)
The nucleotide sequence of the query is shown below the BLASTx results.
(PPTX)
Peaks corresponding to trichodermol (4-hydroxy EPT), trichodermin (4-O-acetyl EPT), and 8-deoxy trichothecin (4-O-butenoyl EPT) are shown. Based on mass spectral fragmentation patterns, the unlabeled peaks at 5 min and 6.1 min do not correspond to trichothecenes.
(PPTX)
Results are presented for the wild-type progenitor strain (blue trace) and complementation strains tri17.MrT17.C3 (black trace) and tri17.MrT17.C4 (red trace). The trace for tri17 mutant strain tri17.139 (purple trace) does not rise above the base line. Identity of harzianum A in samples was confirmed by comparison of mass spectra to the spectrum of a purified harzianum A standard, the identity of which was confirmed by 1H and 13C NMR. Strains were grown in YEPD medium for one week.
(PPTX)
Fig A: Total ion chromatogram of an ethyl acetate extract of the culture (above), and mass spectrum of 4-hydroxy isotrichodermin (below). Fig B: 13C (left) and 1H (right) NMR spectra of 4-hydroxy isotrichodermin.
(PPTX)
In the tree shown here, a distantly related homolog was selected for each of the three TRI genes. For TRI3, we selected a homolog of TRI18 as the outgroup. We searched GenBank by BLASTx [75] to identify non-TRI gene homologs to use at outgroups in the TRI5 and TRI14 trees; the outgroups are represented by their GenBank accession numbers. To generate trees, deduced amino acid sequences of each gene were aligned with Muscle as implemented in MEGA7 [79], and trees were inferred using the maximum likelihood method with ultrafast bootstrapping [83] as implemented in IQ-Tree [73]. Numbers near branches are bootstrap values based on 1000 pseudoreplicates.
(PPTX)
(PPTX)
(DOCX)
Data Availability Statement
Genome sequence data generated as part of this study have been submitted to the GenBank database at the National Center for Biotechnology Information. The accession numbers for the genome sequences are as follows: PXOG00000000 Fusarium longipes NRRL 20695, PXOF00000000 Fusarium sporotrichioides NRRL, 3299 PXXK00000000 Fusarium sp. FIESC 12 NRRL 13405, PXOE00000000 Microcyclospora tardicrescens HJS1936, PXOD00000000 Myrothecium roridum NRRL 2183, PXOC00000000 Spicellum ovalisporum DAOM 186447, PXOB00000000 Spicellum roseum DAOM 209012, PXOA00000000 Trichoderma arundinaceum IBT 40837, PXNZ00000000 Trichoderma brevicompactum IBT 40841, PXNX00000000 Trichothecium roseum DAOM 197141, PXNY00000000 Trichothecium roseum DAOM 195227, PXNW00000000 Trichothecium roseum K7-1.