Abstract
The protein kinase (PK, kinome) family is one of the largest families in plants and regulates almost all aspects of plant processes, including plant development and stress responses. Despite their important functions, comprehensive functional classification, evolutionary analysis and expression patterns of the cotton PK gene family has yet to be performed on PK genes. In this study, we identified the cotton kinomes in the Gossypium raimondii, Gossypium arboretum, Gossypium hirsutum and Gossypium barbadense genomes and classified them into 7 groups and 122–24 subfamilies using software HMMER v3.0 scanning and neighbor-joining (NJ) phylogenetic analysis. Some conserved exon-intron structures were identified not only in cotton species but also in primitive plants, ferns and moss, suggesting the significant function and ancient origination of these PK genes. Collinearity analysis revealed that 16.6 million years ago (Mya) cotton-specific whole genome duplication (WGD) events may have played a partial role in the expansion of the cotton kinomes, whereas tandem duplication (TD) events mainly contributed to the expansion of the cotton RLK group. Synteny analysis revealed that tetraploidization of G. hirsutum and G. barbadense contributed to the expansion of G. hirsutum and G. barbadense PKs. Global expression analysis of cotton PKs revealed stress-specific and fiber development-related expression patterns, suggesting that many cotton PKs might be involved in the regulation of the stress response and fiber development processes. This study provides foundational information for further studies on the evolution and molecular function of cotton PKs.
Introduction
The protein kinase (PK, kinome) family is one of the largest families in plants and regulates many signaling pathways that are triggered during stress and development processes [1]. The common structural feature of PKs is their 250–300 amino acid catalytic domain, which uses the γ-phosphate of adenosine triphosphate (ATP) to phosphorylate serine, threonine or tyrosine residues of proteins [2]. Phylogenetic analyses of the PK family were initially conducted using the conserved feature of the catalytic domain [3]. Based on analysis of the hundreds of known kinase domain sequences at that moment, the PK family was classified into five major groups: (1) AGC, which mainly includes PKA (cyclic AMP-dependent protein kinase), PKG (cyclic GMP-dependent protein kinase) and PKC (protein kinase C); (2) CAMK, which mainly includes CDPK (calcium- and calmodulin-regulated kinase) and Snfl/AMPK (adenosine 5'-monophosphate (AMP)-activated protein kinase); (3) CMGC, which mainly includes CDKs (cyclin-dependent kinases), MAPK (mitogen-activated protein kinases), GSK3 (glycogen synthase kinase) and Clk (cyclin-dependent-like kinases); (4) PTK (protein-tyrosine kinase); (5) others [2]. In 2002, Manning analyzed the evolution of sequences from yeast to human and further divided Metazoan PKs into eight major groups: AGC, CAMK, CK1 (casein kinase 1), CMGC, STE (sterile), TK (tyrosine kinase), TKL (tyrosine kinase-like kinase) and others [4]. Recently, PKs from 25 plant genomes were identified and classified into nine main groups: AGC, CMGC, CAMK, CMGC, CK1, STE, TKL, RLK (receptor-like kinase) and others [5]. Arabidopsis PKs were identified and classified into six large groups: RLK (620), CMGC (65), CaMK (89), AGC (43), STE (67) and Raf (52) [6]. Gene duplication events and functional diversification of the Arabidopsis kinome were further studied [7]. Identification, functional classification and expression analysis of the soybean kinome were performed by Liu et al. [8]. They demonstrated that whole genome duplication (WGD) events might play a vital role in soybean PK expansion, whereas tandem duplication (TD) events might only contribute to the expansion of certain soybean PK subfamilies.
Numerous studies demonstrated that the expansion of plant PKs is mainly due to the expansions of certain RLK subfamilies [9–12]. Lehti-Shiu and Shiu also demonstrated that each flower plant kinome is significantly larger than other eukaryotes’ kinomes mainly due to the expansion of certain RLK subfamilies [5]. In 2001, Shiu and Bleecker performed a genome-wide identification and phylogenetic analysis of Arabidopsis RLKs [13]. Then, they analyzed the expansion mechanism of Arabidopsis RLKs and found that the RLKs ancient origin could be attributed to a more recent plant-specific expansion [9]. By comparing rice (Oryza sativa) RLKs with Arabidopsis RLKs, they found that OsRLKs involved in development have rarely undergone duplications after the rice-Arabidopsis split, whereas OsRLKs involved in defense resistance have undergone more duplications [10]. In 2008, Vij et al. performed identification and phylogenetic analysis of OsRLCKs (receptor-like cytoplasmic kinase, subfamily of RLKs) and analyzed the expression patterns of certain OsRLCKs involved in development and stress [14]. The expression analysis of rice RLKs was studied in different tissues, with the emphasis on seed development and abiotic stress response [11]. Evolutionary analysis indicated that the expansion of RLK family coincided with the establishment of land plants, and expression analysis of Arabidopsis RLKs supported the importance of RLKs in biotic stress response [15]. In addition, phylogenetic analysis and expression profile analysis of the LRRs (leucine-rich repeat, subfamily of RLKs) were performed in Populus trichocarpa [16], Platanus × acerifolia [17], Brassica rapa [18], Arabidopsis [19], tomato [20] and cotton [21]. Based on phylogenetic analysis of the RLK-LRR genes from 31 sequenced flower plants, Fischer et al. found that subgroup- and species-specific expansion rates differ significantly due to the complex history of expansion-retention-loss cycles and whole genome multiplication [22].
Functional characterization studies of cotton PKs indicated that cotton mitogen-activated protein kinases (MAPKs) are involved in abiotic and biotic stress responses [23–26], and cotton RLKs function in fiber development [27], anther development [28], disease resistance [29] and drought-stress tolerance [30]. The completion of cotton genome sequencing provides an opportunity to perform genome-wide research on cotton PKs. In this study, we identified the putative PK genes in Gossypium raimondii, Gossypium arboretum, Gossypium hirsutum and Gossypium barbadense and classified them into groups, families and subfamilies. Some conserved exon-intron structures were identified not only in cotton species but also in primitive plants fern and moss, suggesting that these subfamilies may play important and conserved roles during plant evolutionary processes. Analyses of chromosome location and collinearity events were combined to explore the relationship between expansion and duplication events (WGD, TD and synteny analysis). Expression profiles in developing fibers and under abiotic and biotic stresses were assessed in G. hirsutum and G. barbadense PKs. This work will help to understand the cotton PK evolution mechanism and provide a starting point for further experimental research.
Materials and methods
Identification and classification of cotton PKs
The genome and proteome sequences of G. raimondii were downloaded from Phytozome V10.0 (http://phytozome.jgi.doe.gov/pz/portal.html). The genome and proteome sequences of G. arboretum (V2.0) and G. hirsutum (V1.0) were downloaded from the Cotton Genome Project (CGP: http://cgp.genomics.org.cn/). The genome and proteome sequences of G. barbadense were downloaded from the website CHGC (http://database.chgc.sh.cn/cotton/index.html). HMMER v3.0 [31] in our local server and Pfam 28.0 in batch (http://pfam.xfam.org/) with an E-value of less than 0.01 were performed against all proteomes. The Pfam profiles PF00069 (Pkinase domain) and PF07714 (Pkinase_Tyr domain) were used in HMMER. After the initial screen, putative typical PKs were retained only if Pkinase or Pkinase_Tyr domain alignments covered greater than 50%, as previously described by Legti-Shiu and Shui [5]. Classifications of "typical" cotton PKs were performed using the HMM models developed by Legti-Shiu and Shui [5]. The classifications were further confirmed by phylogenetic analysis using the neighbor-joining (NJ) method. The truncated sequences of the Pkinase or Pkinase_Tyr domain were aligned using ClustalW (v2.0) [32]. The NJ phylogenetic analysis with the p-distance model and 1000 bootstrap repetitions was constructed by MEGAcc 7.0 [33] in our local server. The classification information of Vitis vinifera, Arabidopsis thaliana, Carica papaya, Zea mays and Oryza sativa PKs was extracted from the supplemental files of Legti-Shiu and Shui [5].
Chromosome location and synteny analysis of cotton PKs
The chromosome location information of G. raimondii, G. arboretum, G. hirsutum and G. barbadense was extracted from GFF files of Phytozome, CGP and CHGC using our perl script. A BLAST search was performed against all the cotton PKs with an E-value of 1e-100. Based on the BLAST and GFF results, the segmental/tandem duplication events of G. raimondii, G. arboretum, G. hirsutum and G. barbadense were determined using MCScanX software [34]. The "add_ka_and_ks_to_collinearity.pl" of MCScanX was used to calculate the synonymous (Ks) and non-synonymous substitution (Ka) rates. The collinearity visualization of duplicated PKs was performed using GenomePixelizer [35]. The chromosomal location of identified tandem PKs was mapped on each chromosome with Mapchart v2.3 (http://www.wageningenur.nl/en/show/mapchart.htm). The synteny blocks between G. arboretum and G. hirsutum At-subgenome, G. raimondii and G. hirsutum Dt-subgenome; G. arboretum and G. barbadense At-subgenome; G. raimondii and G. barbadense Dt-subgenome; and G. raimondii and G. arboretum were identified by MCScanX [34]. All paralogous and orthologous PK pairs were visualized using the Circos program [36].
Domain and exon-intron structure search of cotton PKs
The classifications of V. vinifera, A. thaliana, O. sativa, Selaginella moellendorffii, Physcomitrella patens and Chlamydomonas reinhardtii PKs were extracted from Legti-Shiu and Shui [5]. Exon-intron structure diagrams of these six plants and four cotton species were generated by our perl and R scripts based on extracting information from Phytozome, CGP and CHGC GFF files. The domain information was obtained from pfam 28.0 in batch (http://pfam.xfam.org/).
Microarray expression data analysis
Public cotton expression datasets were retrieved from the Gene Expression Omnibus (GEO) of NCBI. Microarray datasets for stress treatments and fiber development stages were selected from the Affymetrix Cotton Genome Array platform GPL8672. (1) Abiotic stresses: ① Five abiotic stresses: GSE50770, cotton microarray-based datasets under ABA, cold, drought, salinity and alkalinity treatments [37]. ② Heat stress: GSE41725, G. hirsutum (cultivar: Sicala 45 and Sicot 53) in heat tolerance (32°C and 42°C). ③ Waterlog and drought stresses: GSE16467, flooded-stress G. hirsutum root and leaf tissues [38]. GSE29810 (This SuperSeries is composed of GSE29566 and GSE29567), G. hirsutum under drought stress in leaf tissue and during fiber development stages (0, 5, 10, 15, and 20 dpa) [39]. (2) Biotic stresses: ① Alternaria alternata disease: GSE74412, G. hirsutum leaf tissues under A. alternata infected stress with or without chilling pre-treatment. ② Bollworm infection: GSE55511, G. hirsutum during boll development stages (0, 2, 5 and 10 dpa (days post anthesis) under bollworm-infected biotic stress. (3) Fiber development: GSE36228, cotton fiber at different developmental stages (0, 6, 9, 12, 19 and 25 dpa) from five G. hirsutum varieties (JKC 725 and JKC 777 were superior in fiber traits compared with JKC 703, JKC 737 and JKC 783) [40]. GSE36021, GhHD-1-silenced, GhHD-1-over-expression and wild-type cotton lines [41]. GSE38490, fiber bearing and fuzzless-lintless mutant G. hirsutum during fiber development stages (0, 5, 10, 15 and 20 dpa) [42]. The raw data were normalized using RMAexpress v1.2.0 (http://www.rmaexpress.bmbolstad.com/). The normalization of raw data and quality control were performed by RMAexpress v1.2.0 (http://www.rmaexpress.bmbolstad.com/). The expression levels (treatment vs. control, log2 value of fold change) of cotton PKs for stress treatments and fiber development were calculated using R software and R package limma. Heat maps of cotton PK expression levels were generated by Mev4.9 [43].
RNA-seq expression data analysis
Public cotton expression datasets were retrieved from the Sequence Read Archive (SRA) of NCBI. The RNA-seq data of two commercial cotton species, G. barbadense and G. hirsutum, in fiber developmental stages (10, 15, 18, 21, and 28 dpa) were used (PRJNA263926). The RNA-seq data of G. hirsutum Li1 mutant and wild type in leaf tissue and ovules of different fiber developmental stages (1, 3, and 8dpa) were used (PRJNA301536). Quality control assessment of raw data was performed using FastQC. High-quality RNA-seq reads were aligned to reference cotton genomes by software Hisat2. The counts of expression genes were performed using Samtools and HTseq software. The reads per kilobase of exon model per million mapped reads (RPKM) algorithm was used to normalize the data. The expression levels (PRJNA301536, Li1 mutant vs. wild type, log2 value of fold change) of cotton PKs for fiber development was calculated using R software and R package DESeq2. Heat maps of cotton PK expression levels were generated using Mev4.9 [43].
Results
Genome-wide identification and classification of cotton PKs
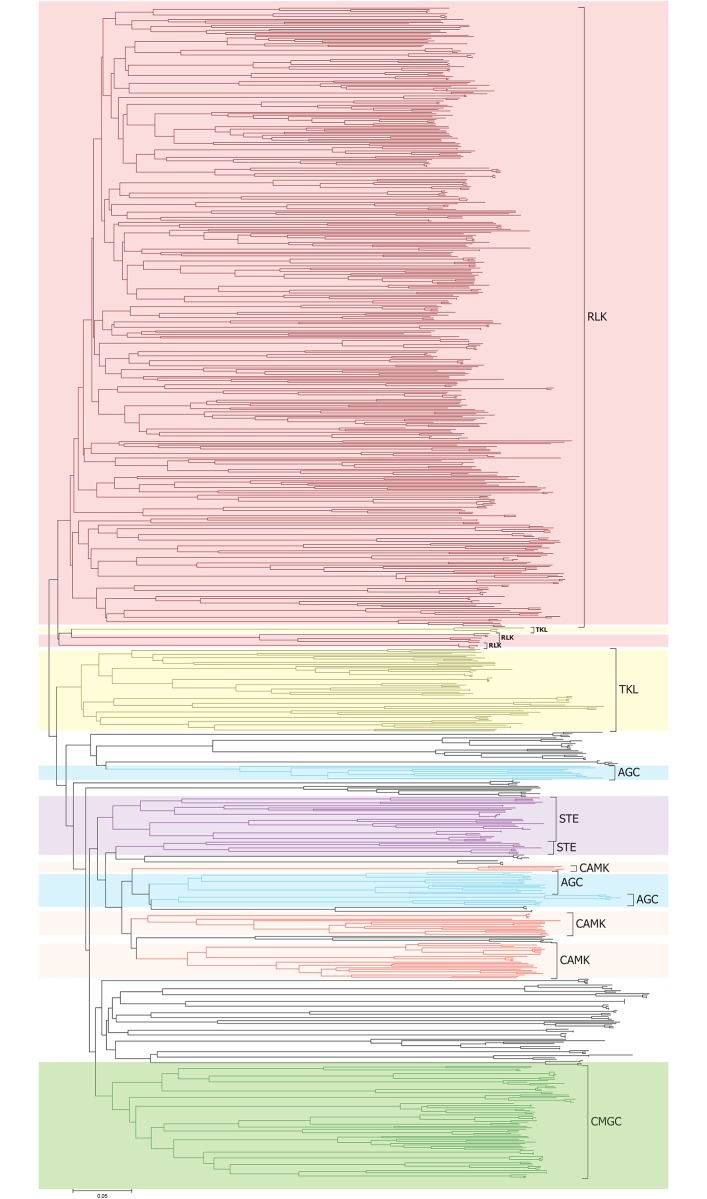
We identified 1517, 1407, 2508 and 2745 PK genes with typical kinase domains (S1 Table) (see Method) and excluded 70, 147, 363 and 390 sequences with atypical kinase domains (domain alignments covered less than 50% of Pfam domain models) in G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively (S2 Table). The classifications of these cotton PKs were performed against HMM models developed by Legti-Shiu and Shui [5]. These genes were classified into 122–124 subfamilies (S1 Table). To validate the classification by HMMER, we selected 1–3 random genes from each subfamily as representatives to construct a NJ phylogenetic tree using the truncated kinase domain sequences with the p-distance model and 1000 bootstraps (Fig 1, Figure A in S1 File). Interestingly, almost all the cotton PKs belonging to the same clade indicated the same subfamily classification as identified by HMMER. We also constructed the NJ tree based on all the cotton PK members (Figure B in S1 File). In summary, cotton PKs were classified into seven groups: RLK (G. raimondii 1019, G. arboretum 913, and G. hirsutum 1681, and G. barbadense 1855), AGC (52, 55, 81, 96), CAMK (114, 113, 189, 207), CMGC (105, 100, 180, 182), STE (60, 59, 103, 112), TKL (80, 80, 130, 143), and others (87, 87, 144, 150). These seven groups were further classified into 123, 123, 124 and 123 subfamilies in G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively. However, each subfamily member size was highly variable. The RLK group contained 57–58 subfamilies and formed two subgroup clusters, which are leucine-rich repeat (LRR) and receptor-like cytoplasmic kinase (RLCK). The RLK-DLSV subfamily contained the largest members (129, 93, 205, 225) among the cotton PK subfamilies.
Fig 1. Classification and phylogenetic relationships of the cotton PK subfamilies.
The Neighbor-Joining tree was built by the amino acid sequences of the kinase domain using the MEGAcc 7.0 with the p-distance model. Random representative PKs of each subfamily are selected by following criteria: members ≤ 6, 1 PK; 6 < members ≤ 30, 2 PKs; members > 30, 3 PKs. Six major groups are labeled with different colors, including AGC, CAMK, CMGC, RLK, STE, TKL. Detailed information is provided in S1A Fig.
To comprehensively study the cotton PK classifications, we compared the PK distributions of four cotton species with three eudicots (V. vinifera, A. thaliana, and C. papaya) and two monocots (Z. mays and O. sativa) (S3 Table). First, the "one-two-four" pattern was found to be existed in cotton PK subfamily member size comparing with grape. For instance, the RLK/Pelle_RLCK-XI subfamily of V. vinifera, G. raimondii, G. arboretum, G. hirsutum and G. barbadense contained 2, 4, 4, 8 and 6 members, respectively. Similar examples were also found in other PK subfamilies, such as AGC_Pl, AGC_RSK-2, CAMK_CAMKL-CHK1 and CAMK_CDPK. This expansion pattern could be explained by the report indicating that cotton has experienced only one cotton-specific WGD event after it split from the resemble-grape ancestor [44]. Moreover, tetraploidization of G. hirsutum and G. barbadense also contributed to the "one-two-four" expansion pattern [45–46]. Second, some species-specific expansions were identified in some PK subfamilies. For example, cotton RLK/Pelle_CrRLK1L-1 (G. raimondii 55, G. arboretum 56, G. hirsutum 93 and G. barbadense 103) contained more members compared with V. vinifera (9), suggesting that it potentially experienced cotton-specific expansion during the evolutionary process. Similar examples were also identified in RLK/Pelle_L-LEC, RLK/Pelle_LRK10L-2 and RLK/Pelle_WAK. One monocot-specific PK subfamily, CMGC_CDKL-Os, was absent in all the six investigated eudicots but was present in the two monocots (Z. mays and O. sativa). RLK/Pelle_URK-4 had only one member in V. vinifera, A. thaliana, and C. papaya, separately, but was absent in four cottons and two monocots (Z. mays and O. sativa). These results suggest that it might arise in an eudicot ancestor after the monocot-eudicot split but was lost in cotton evolution. RLK/Pelle_LRR-I-1 contained more members in in O. sativa (34) and A. thaliana (48) compared with other investigated plants (8–22), implying that it might experience rice- and Arabidopsis-specific expansions. RLK/Pelle_DLSV contained the largest members among PK subfamilies in all the nine investigated plants.
Conserved domains and intron-exon structures of cotton PKs
Based on the Pfam information, we diagrammed the domain distributions of all the putative cotton PKs (G. raimondii 1517, G. arboretum 1407, G. hirsutum 2508 and G. barbadense 2745) (S2 Fig). Our results indicated that approximately half of cotton PKs exclusively contained the kinase domain, including G. raimondii (662, 43.6%), G. arboretum (669, 47.5%), G. hirsutum (1217, 48.5%) and G. barbadense (1247 45.4%). The other PKs contained additional domains in addition to the kinase domain. We calculated the proportion of PKs with additional domains in PK groups, including AGC (G. raimondii 36.5%, G. arboretum 38.2%, G. hirsutum 33.3% and G. barbadense 38.5%); CAMK (G. raimondii 72.8%, G. arboretum 73.5%, G. hirsutum 74.6% and G. barbadense 69.6%); CMGC (G. raimondii 1.9%, G. arboretum 2%, G. hirsutum 1.7% and G. barbadense 10.4%); RLK-Pelle (G. raimondii 68.5%, G. arboretum 63.3%, G. hirsutum 61.3% and G. barbadense 64.4%); STE (G. raimondii 0%, G. arboretum 1.7%, G. hirsutum 1% and G. barbadense 3.6%) and TKL (G. raimondii 58.8%, G. arboretum 57.5%, G. hirsutum 58.5% and G. barbadense 53.8%). These results demonstrate that various subfamilies contained various domain compositions. By contrast, PKs of the same subfamily shared similar domain arrangements, suggesting that they might originate from a common ancestor through domain fusion between the kinase domain and additional domains. We identified some special PKs containing 2–4 PK kinase domains (G. raimondii 58, G. arboretum 63, G. hirsutum 115 and G. barbadense 175, S4 Table). Most of these genes belonged to the AGC_NDR, AGC_RSK-2, and CMGC_SRPK subfamilies and RLK group.
To gain further insights into the cotton PKs, we generated the exon-intron structures within the kinase domain in four cotton species (S3 Fig). Interestingly, cotton PKs belonging to the same subfamily contained similar exon-intron structures within the conserved exon phases, especially in the kinase domain. For example, AGC_ NDR sequences in G. raimondii (Gorai.011G175100.1), G. arboretum (Cotton_A_39141), G. hirsutum (CotAD_71296) and G. barbadense (GOBAR_DD23594) shared the same exon-intron structure within "0020–200" exon phases in the kinase domain.
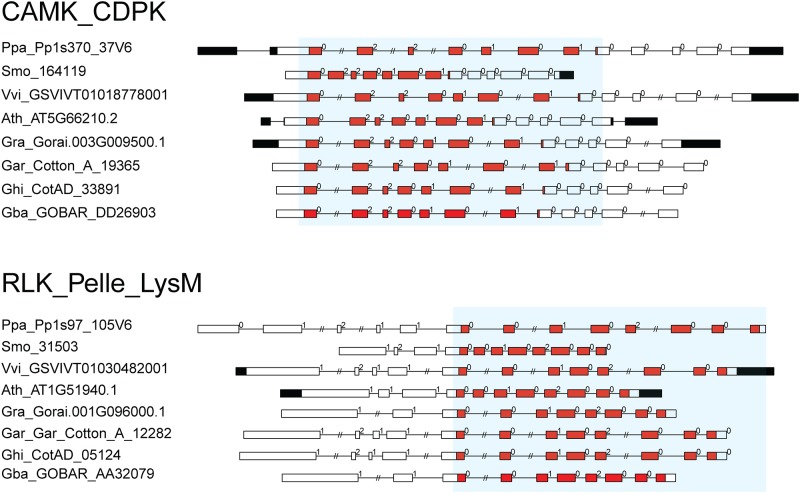
Our other research on wheat PKs showed that these exon-intron structures were conserved across the plant evolution [47]. To test it in cotton PKs, we also diagramed the PK exon-intron structures in V. vinifera (grape), A. thaliana, O. sativa (rice), S. moellendorffii (a fern), P. patens (moss) and C. reinhardtii (a green alga) (S3 Fig). Similarly, our results revealed that these conserved exon-intron structures were also identified in investigated plants, especially in the primitive plants S. moellendorffii and P. patens but not in C. reinhardtii. These findings suggest that these conserved exon-intron structures may have occurred during the emergence of Pteridophytes or Bryophytes. For instance, the CAMK_CDPK subfamily sequences from P. patens (Pp1s370_37V6), S. moellendorffii (164119), O. sativa (LOC_Os02g03410.1), V. vinifera (GSVIVT01018778001) A. thaliana (AT5G66210.2), G. raimondii (Gorai.003G009500.1), G. arboretum (Cotton_A_19365), G. hirsutum (CotAD_33891) and G. barbadense (GOBAR_DD26903) shared the same exon-intron structure of the kinase domain within the exon phases "022–0101" (Fig 2A). Similarly, RLK_Pelle_LysM members from moss to cotton also shared the conserved exon-intron structure with exon phases "001–0200" in the kinase domain (Fig 2B). Based on the above analysis, we summarized all the conserved exon-intron structures of some PK subfamilies in P. patens, S. moellendorffii, V. vinifera and G. raimondii (S4 Fig). These results suggested that these conserved exon-intron structures might have important biological functions for plants during the evolutionary process.
Fig 2. Two examples of conserved exon–intron structure.
This diagram indicated that conserved exon–intron structure with conserved exon phases existed in kinase domain. Filled boxes: red represents Pkinase or Pkinase_Tyr domain; black boxes: untranslated regions (UTRs); white boxes: other exon regions; lines: introns. Numbers 0, 1, and 2: exon phases. The lengths of the boxes and lines are scaled based on the lengths of genes. The long introns are shorted by “//”. (A) non-RLK example: CAMK_CDPK. (B) RLK example: RLK_Pelle_LysM.
Chromosome location and duplication events of cotton PKs
We mapped the 1501 G. raimondii PKs (Figure A in S5 File, excluding 16 genes from scaffold), 1407 G. arboretum PKs (Figure B in S5 File), 1893 G. hirsutum PKs (Figure C in S5 File, excluding 615 genes from scaffolds) and 2477 G. barbadense PKs (Figure D in S5 File, excluding 268 genes from scaffolds) to chromosome positions (S5 Table). The distributions of cotton PKs were assessed in G. raimondii, G. arboretum, G. hirsutum and G. barbadense (Table 1). The results showed that cotton PKs were unequally distributed across 13 or 26 chromosomes. G. raimondii chromosome 9 contained the most PKs (205), whereas G. arboretum chromosome 10, G. hirsutum chromosome 9Dt and G. barbadense chromosome 5At/5Dt contained the most PKs (145, 207, and 175, respectively).
Table 1. The PK distributions among the chromosomes in of G. raimondii, G. arboretum, G. hirsutum and G. barbadense.
| Chromosome | G. arboretum | G. hirsutum (At) | G. barbadense (At) | G. raimondii | G. hirsutum (Dt) | G. barbadense (Dt) |
|---|---|---|---|---|---|---|
| 1 | 127 | 71 | 72 | 104 | 105 | 89 |
| 2 | 59 | 56 | 78 | 120 | 34 | 110 |
| 3 | 73 | 59 | 60 | 63 | 32 | 45 |
| 4 | 135 | 8 | 46 | 79 | 15 | 60 |
| 5 | 73 | 43 | 175 | 115 | 99 | 175 |
| 6 | 122 | 71 | 91 | 134 | 116 | 108 |
| 7 | 123 | 44 | 88 | 171 | 102 | 90 |
| 8 | 119 | 67 | 60 | 102 | 78 | 67 |
| 9 | 113 | 183 | 108 | 205 | 207 | 113 |
| 10 | 145 | 63 | 119 | 85 | 70 | 131 |
| 11 | 130 | 78 | 125 | 155 | 101 | 141 |
| 12 | 98 | 42 | 91 | 81 | 49 | 87 |
| 13 | 90 | 43 | 68 | 88 | 57 | 80 |
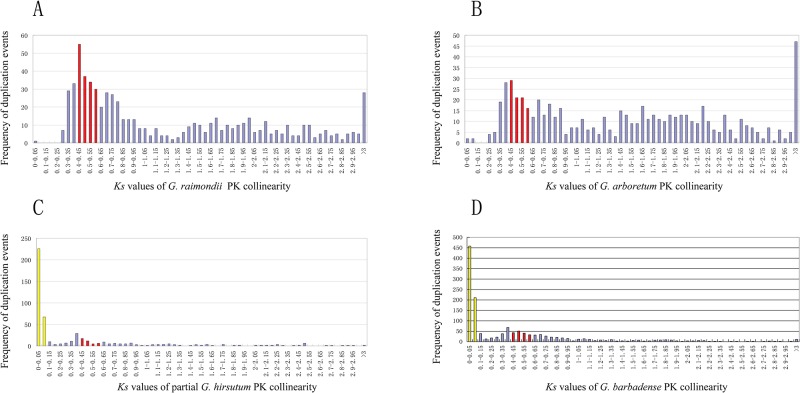
Gossypium lineages experienced two WGD events, which occurred 16.6 and 130.8 million years ago (Mya) [48–50]. Therefore, we decided to investigate the contributions of WGD events to cotton PK expansions. We identified 681, 643, 510 and 1391 collinearity events in G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively (S6 Table). These results suggest that segmental duplication events might play important roles in the cotton PK expansions. The WGD event appeared after cotton speciation 16.6 Mya, and its synonymous distance (Ks value) peak ranged from 0.4 to 0.6 [48]. Our results were consistent with a paper suggesting that cotton PK collinearity events also formed peaks of Ks 0.4–0.6 (Fig 3A–3D). We detected the chromosome positions of these collinearity events with Ks 0.4–0.6 in G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively (Figure A-N in S6 File). The results indicated that 271, 158, 72 and 264 PKs of G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively, were involved in the collinearity events (Ks 0.4–0.6). These results suggest that the cotton PK expansions might be due in part to WGD 16.6 Mya. We noticed that 440 G. hirsutum PKs and 1333 G. barbadense PKs were involved in the collinearity events (Ks 0.0–0.1) (Fig 3C and 3D, Figure G and K in S6 File), suggesting that tetraploidization of G. hirsutum and G. barbadense might play important roles in the PK expansion.
Fig 3. Ks values of PK collinearity in G. raimondii, G. arboretum, G. hirsutum and G. barbadense.
This diagram indicated that the Ks values of PK collinearity in G. raimondii (A), G. arboretum (B), G. hirsutum (C) and G. barbadense (D). The red bars denote the collinearity events resulting from 16.6 Mya WGD (Ks values 0.4–0.6) in G. raimondii, G. arboretum, G. hirsutum and G. barbadense. The yellow bars denote the collinearity events contributed by tetraploidization in G. hirsutum and G. barbadense. The blue bars denote the other collinearity events.
We identified 291, 246, 264 and 455 tandem cotton PKs in G. raimondii, G. arboretum, G. hirsutum and G. barbadense, respectively. The chromosome positions of these cotton PKs were detected in G. raimondii, G. arboretum, G. hirsutum and G. barbadense (S7 Table). They formed several tandem PK clusters among 13 or 26 cotton chromosomes. The G. raimondii tandem PKs formed 83 clusters that were distributed among the 13 chromosomes (Figure A in S7 File). The largest G. raimondii cluster contained 13 RLK-Pelle_LRR-XII-1 genes located on chromosome 11. The G. arboretum tandem PKs formed 87 clusters among the 13 chromosomes (Figure B in S7 File). The largest G. arboretum cluster was identified on chromosome 1 and contained 11 RLK-Pelle_CrRLK1L-1 genes. The G. hirsutum tandem PKs formed 89 clusters among 24 chromosomes (Figure C in S7 File). Tandem PKs were not detected on chromosome Dt2 and At4 of G. hirsutum perhaps due to its incomplete chromosome location information (scaffold). The largest G. hirsutum cluster contained 8 RLK-Pelle_CrRLK1L-1 genes located on chromosome Dt7. The G. barbadense tandem PKs formed 152 clusters among 26 chromosomes (Figure D in S7 File). The largest G. barbadense cluster contained 13 RLK-Pelle_LRR-XI-1 genes located on chromosome Dt9. Most of these cotton tandem PK clusters belonged to the RLK subfamilies, suggesting that TD mainly contributed to the expansion of the cotton RLK group.
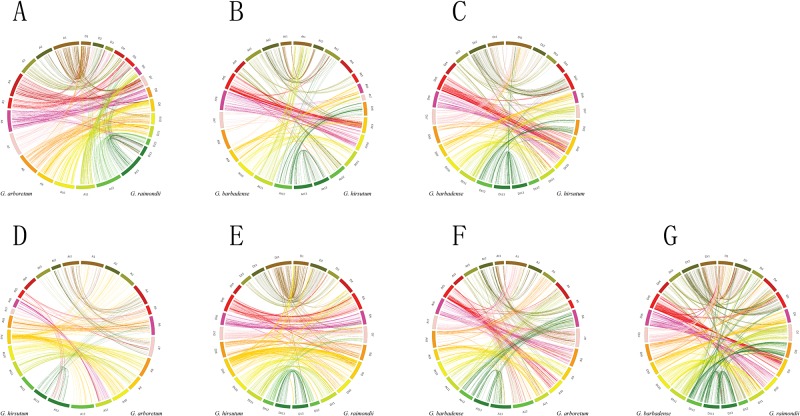
To further explore the relationship between PK expansions and cotton duplication events, we constructed the synteny analysis of cotton PKs between G. raimondii and G. arboretum (Fig 4A); At-subgenomes of G. hirsutum and G. barbadense (Fig 4B); and Dt-subgenomes of G. hirsutum and G. barbadense (Fig 4C). The similar synteny analysis of cotton PKs was also performed between G. arboretum and G. hirsutum At-subgenome (Fig 4D), G. raimondii and G. hirsutum Dt-subgenome (Fig 4E), G. arboretum and G. barbadense At-subgenome (Fig 4F), and G. raimondii and G. barbadense Dt-subgenome (Fig 4G). The related collinearity events were also demonstrated among G. raimondii, G. arboretum, G. hirsutum and G. barbadense (S8 Table). Our results demonstrated that approximately half of collinearity events exhibited single gene correspondence (S9 Table), indicating that these single gene pairs might exist in the common cotton ancestor and did not experience expansion after the split of cotton A- and D-subgenomes. We also noticed that some gene pairs from chromosome fragments did not locate on the corresponding chromosomes between subgenomes, suggesting that they might experience chromosome rearrangement after the split of cotton A- and D-subgenomes. For instance, eight gene pairs (from Cotton_A_10476-Gorai.013G272100.1 to Cotton_A_10556-Gorai.013G263500.1) are located in the A8 chromosome fragment of G. arboretum and D13 chromosome fragment of G. raimondii, separately (S8 Table).
Fig 4. Synteny analysis of PK genes.
(A) Synteny between G. raimondii (A1-A13) and G. arboretum (D1-D13). (B) Synteny between G. barbadense (At1-At13) and G. hirsutum (At1-At13). (C) Synteny between G. barbadense (Dt1-Dt13) and G. hirsutum (Dt1-Dt13). (D) Synteny between G. arboretum (A1-A13) and G. hirsutum (At1-At13). (E) Synteny between G. raimondii (D1-D13) and G. hirsutum (Dt1-Dt13). (F) Synteny between G. arboretum (A1-A13) and G. barbadense (At1-At13). (G) Synteny between G. raimondii (D1-D13) and G. barbadense (Dt1-Dt13). The different cotton chromosomes were labeled with different colors.
Expression pattern of cotton PKs in abiotic and biotic stresses
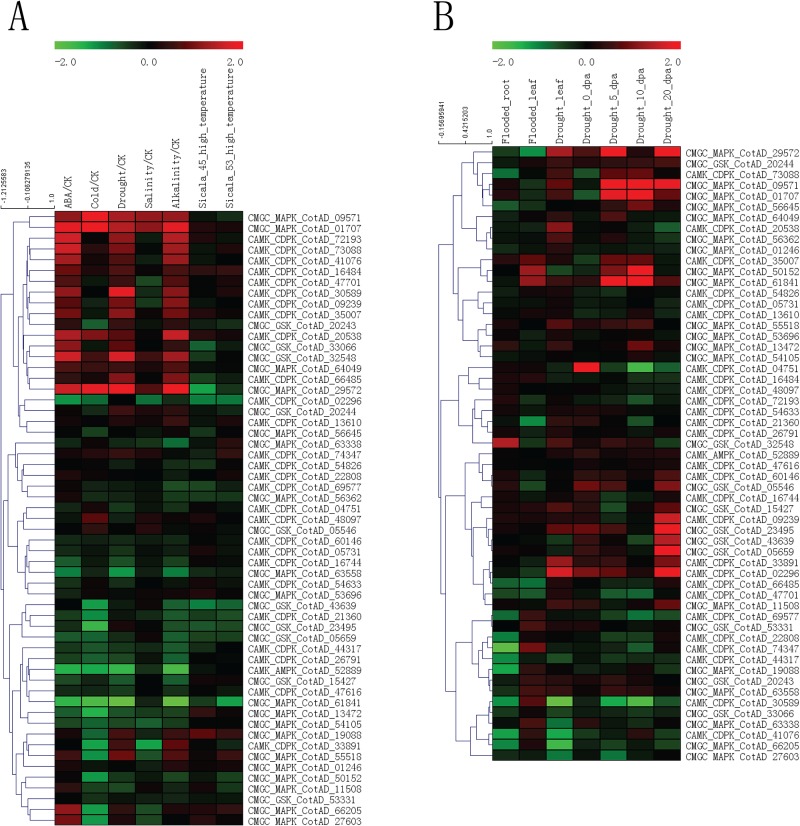
We analyzed the gene expression profiles of G. hirsutum PKs using 11 public datasets of the Affymetrix microarray GPL8672 platform. As a result, 564 of 2508 G. hirsutum PKs have probes in GPL8672. Based on the RLE and NUSE diagrams (S8 Fig), we performed quality control assessment and excluded 5 CEL files in the following analysis (S10 Table). The expression patterns of 564 G. hirsutum PKs were assessed under various abiotic stresses (S9 Fig and S11 Table). To identify PK genes differentially expressed under abiotic stresses, the PKs with |FC|>1.5 (fold change) and p<0.01 were retained (S12 Table). (1) Five stress conditions: We assessed genes in response to five abiotic stress conditions: ABA, cold, drought, salinity and alkalinity stresses. Some PKs, such as CotAD_10936 (CAMK_CAMKL-CHK1), CotAD_29572 (CMGC_MAPK) and CotAD_24426 (AGC-Pl), exhibited up-regulation in response to these five stresses. These results suggest that several common components of signaling pathways might be shared by these five abiotic stress response. In addition, certain PKs responded to particular stress treatments. For example, CotAD_51610 (CMGC_CDK-CRK7-CDK9) only exhibited up-regulation in response to ABA treatment but down-regulation to other four abiotic stress treatments. (2) Heat stress: Most PKs exhibited the same expression trend in both varieties "Sicala 45" and "Sicala 53" under high temperature treatment. However, some PKs exhibited two opposing expression trends between variety "Sicala 45" and "Sicala 53". For instance, CotAD_74347 (CAMK_CDPK) exhibited slight down-regulation in "Sicala 45" but up-regulation in "Sicala 53". (3) Waterlog and drought stresses (S10 Fig, S13 and S14 Tables): We observed that some PKs, such as CotAD_65513 (RLK-Pelle_RLCK-VIIa-2), CotAD_27962 (AGC-Pl) and CotAD_57424 (RLK-Pelle_DLSV), exhibited up-regulation in root under waterlog stress, whereas these PKs exhibited no change or down-regulation in leaf. The reason for these findings might be that the low oxygen of the cotton root influenced some energy metabolism. Under drought stress, some PKs, such as CotAD_09571 (CMGC_MAPK), CotAD_06862 (CAMK_CAMKL-CHK1) and CotAD_43021 (CAMK_OST1L), exhibited up-regulation at 5, 15 and 20 dpa (days post anthesis). Similarly, several PKs, such as CotAD_70308 (RLK-Pelle_LRK10L-2), CotAD_62253 (TKL-Pl-3) and CotAD_62987 (RLK-Pelle_DLSV), exhibited down-regulation at 5, 15 and 20 dpa. We also selected PKs of some known subfamilies, such as CMGC_MAPK, CAMK_CDPK and CAMK_AMPK, to investigate their expression patterns under abiotic stresses (Fig 5A and 5B).
Fig 5. Heat map of the expression patterns of G. hirsutum PK genes from known subfamilies under various abiotic stress conditions.
The expression patterns of PKs of known subfamilies, including CAMK_CDPK, CAMK_AMPK, CMGC_GSK and CMGC_MAPK. (A) The expression patterns under abiotic stress treatments. (B) The expression patterns under waterlog and drought stress treatments.
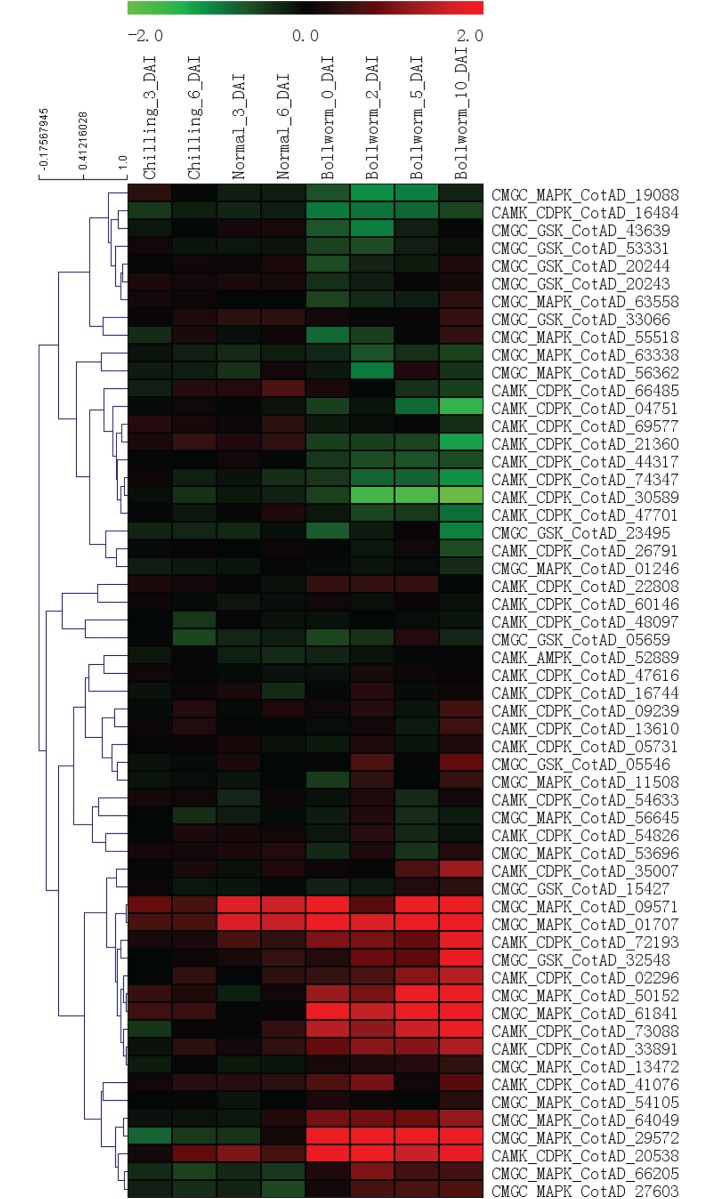
The expression patterns of 564 G. hirsutum PKs were also analyzed under biotic stresses (S11 Fig and S15 Table). The differential expression PKs with |FC|>1.5 and p<0.01 were also assessed under biotic stresses (S16 Table). (1) A. alternata disease: Some PKs, such as CotAD_29801 (WNK_NRBP), CotAD_07539 (RLK-Pelle_LRR-XI-1) and CotAD_37359 (RLK-Pelle_LRR-XII-1), exhibited up-regulation at 3 and 6 DAI (days after inoculation) under A. alternata-infected stress conditions with or without chilling pre-treatment, suggesting that these genes might participate in the A. alternata defense response. (2) Bollworm infection: Some PKs, such as CotAD_20538 (CAMK_CDPK), CotAD_75319 (RLK-Pelle_WAK) and CotAD_29572 (CMGC_MAPK), exhibited up-regulation at 0, 2, 5 and 10 dpa with Helicoverpa armigera infection. We also selected the same PKs of known subfamilies to investigate their expression patterns under biotic stresses (Fig 6).
Fig 6. Heat map of the expression patterns of G. hirsutum PK genes from known subfamilies under various biotic stress conditions.
The selected PKs of known subfamilies are same as Fig 5. The expression patterns under biotic stress treatments.
Expression pattern of cotton PKs during fiber development stages
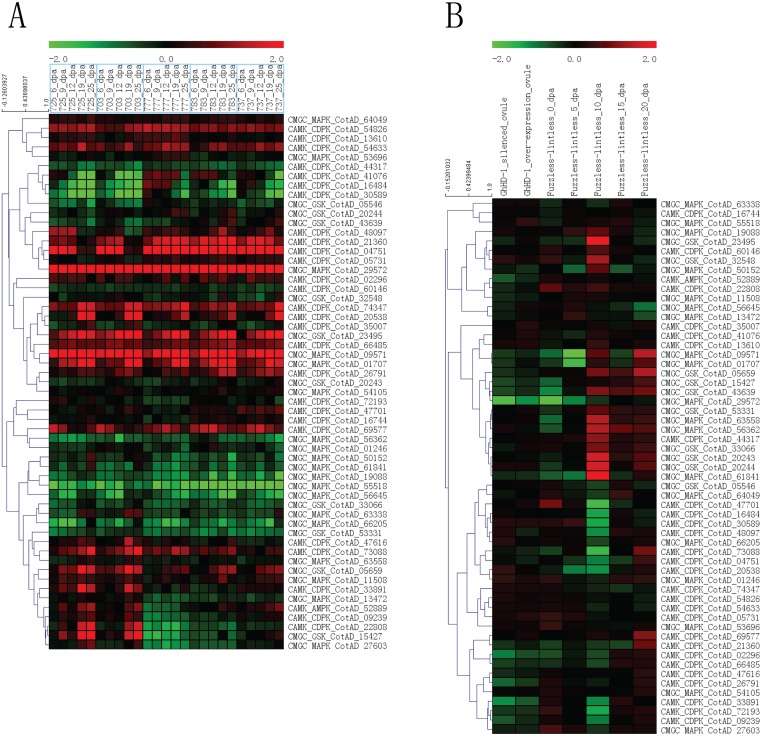
We assessed the expression pattern of cotton PK genes in five G. hirsutum varieties during fiber development stages (6, 9, 12, 19 and 25 dpa) (S12 Fig and S17 Table). Some PKs, such as CotAD_10084 (RLK-Pelle_RLCK-IXa), CotAD_70308 (RLK-Pelle_LRK10L-2) and CotAD_29572 (CMGC_MAPK), exhibited sustained up-regulation at 6–25 dpa among the investigated five varieties. Similarly, several PKs, such as CotAD_06790 (RLK-Pelle_RLCK-VIIa-2), CotAD_54619 (CAMK_OST1L) and CotAD_17222 (RLK-Pelle_LRR-XI-1), exhibited sustained down-regulation at 6–25 dpa among the five varieties. We also noticed that some PKs exhibited different expression patterns among these five cotton varieties. For example, a few PKs, including CotAD_22808 (CAMK_CDPK), CotAD_15427 (CMGC_GSK) and CotAD_61960 (TKL-Pl-4), exhibited peak up-regulation at 19 and 25 dpa in varieties "JKC 725" and "JKC 703", respectively, but remained at the same expression level or exhibited down-regulation in the other three cotton varieties. We also assessed PKs exhibiting differential expression (|FC|>1.5 and p<0.01) (S18 Table) and selected the PKs of known subfamilies to investigate their expression patterns during fiber development stages (Fig 7A).
Fig 7. Heat map of the expression patterns of G. hirsutum PK genes from known subfamilies during fiber development stages.
The selected PKs of known subfamilies are same as Fig 5. (A) The expression patterns for genotypes (725, 703, 777, 783 and 737) and fiber development stages (6, 9, 12, 19 and 25 dpa). (B) The expression patterns about GhHD-1 and fuzzless-lintless mutant during fiber development stages.
The cotton transcription factor GhHD-1 plays roles in cotton fiber initiation and is expressed in trichomes and early fibers [41]. The expression patterns of cotton PKs between GhHD-1 silenced and over-expressed lines were assessed during the early fiber development stage (S13 Fig and S19 Table). Our result revealed that a few PKs exhibited reverse expression trends between GhHD-1 silenced and over-expressed lines, hinting that they might be the downstream of GhHD-1 in the signaling pathway. For instance, CotAD_03134 (RLK-Pelle_LRR-XI-1) and CotAD_34129 (RLK-Pelle_CR4L) exhibited down-regulation in a GhHD-1-silenced line but up-regulation in a line exhibiting GhHD-1 over-expression.
Fuzzless-lintless cotton mutants represent ideal materials to identify genes involving in fiber development by comparison with fiber-bearing wild-type cottons [42]. Our result revealed that several cotton PKs exhibited peak up- or down-regulation at 10 dpa in the fuzzless-lintless cotton lines (S13 Fig and S19 Table). For example, CotAD_67858 (CMGC_CDK-CDK7), CotAD_10699 (AGC_NDR) and CotAD_32629 (CK1_CK1-Pl) exhibited peak up-regulation at 10 dpa. Similarly, some PKs, including CotAD_75319 (RLK-Pelle_WAK), CotAD_08639 (STE_STE11) and CotAD_16484 (CAMK_CDPK), exhibited down-regulation at 10 dpa. PKs exhibiting differential expression (|FC|>1.5 and p<0.01) (S20 Table) and PKs of known subfamilies (Fig 7B) were also assessed in GhHD-1 lines and fuzzless-lintless lines.
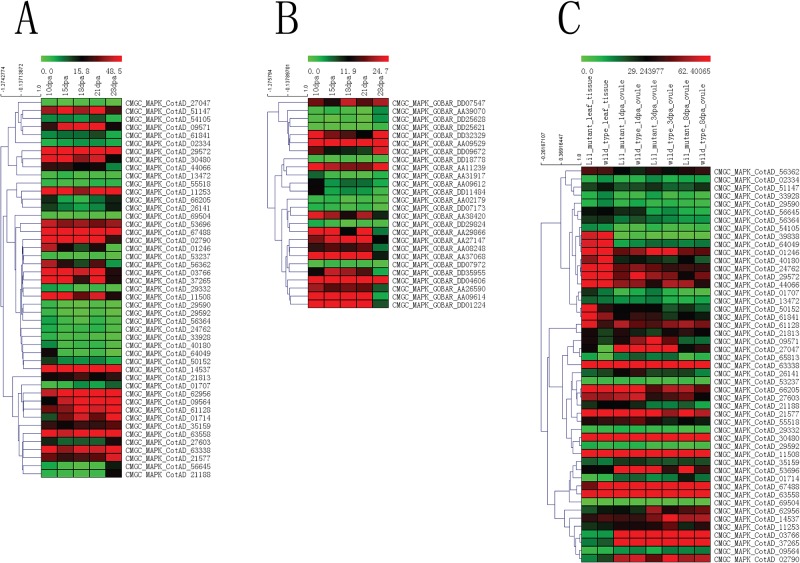
We also analyzed two public RNA-seq data of NCBI SRA about the fiber development of G. hirsutum and G. barbadense. Based on the results of FastQC, we performed quality control assessment and excluded 15 RUN files in the following analysis (S21 Table). 1619 G. hirsutum PKs and 963 G. barbadense PKs were identified in all the five runs of 10, 15, 18, 21 and 28 dpa (S14 and S15 Figs, and S22 Table). We noticed that some G. hirsutum PKs, such as CotAD_12382 (NEK) and CotAD_16078 (CAMK_CAMKL-CHK1), exhibited sustained high expression from 10 to 28 dpa. Similarly, some G. hirsutum PKs, such as CotAD_31836 (RLK-Pelle_LRR-XI-1) and CotAD_21804 (RLK-Pelle_DLSV), exhibited sustained low expression from 10 to 28 dpa. We also selected PKs of the known subfamily, CMGC_MAPK, to investigate their expression patterns of fiber development (Fig 8A and 8B).
Fig 8. Heat map of the expression patterns of G. hirsutum and G. barbadense PK genes from MAPK subfamily during fiber development stages.
(A) Normalized gene expression RPKM values of G. hirsutum MAPK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). (B) Normalized gene expression RPKM values of G. barbadense MAPK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). (C) Normalized gene expression RPKM values of G. hirsutum PK genes for genotypes (Li1 mutant and wild type) ahout leaf tissue and ovule tissues in fiber development stages (1, 3 and 8 dpa).
1749 G. hirsutum PKs of two genotypes (Li1 mutant and wild type) were identified in all the eight RUN files of leaf tissue and ovule tissues in fiber development stages (1, 3 and 8 dpa) (S16 Fig and S23 Table). Most PKs exhibited the similar expression levels between genotypes Li1 mutant and wild type. We also found that some PKs, such as CotAD_21607 (RLK-Pelle_DLSV) and CotAD_58349 (RLK-Pelle_RLCK-XII-1), exhibited different expression levels in leaf tissue between the two genotypes. In order to investigate the differential expression PKs between genotypes Li1 mutant and wild type, we also caculated the log2 Fold Change (Li1 mutant vs. wild type) and padj values (S24 Table). For instance, the log2 value of CotAD_27047 (CMGC_MAPK) in leaf tissue is 5.885973619 and its padj value is 0.000776909, suggesting that Li1 mutant might greatly influence the expression of CotAD_27047 (CMGC_MAPK) in leaf tissue. We also selected PKs of the known subfamily, CMGC_MAPK, to investigate their expression patterns of fiber development between the two genotypes (Fig 8C).
Discussion
Phylogeny of cotton PK family
The identification and analyses of A. thaliana, wheat, soybean and maize kinomes has been reported in recent years [7–8, 12, 47]. Diversity, classification and functions of 25 plant kinomes were also assessed to determine the evolutionary history of PK subfamilies [5]. In this study, we also identified, classified, and performed phylogenetic and expression pattern analyses of the PK family in four cotton species G. raimondii, G. arboretum, G. hirsutum and G. barbadense. As reported in the other plant kinomes, cotton PKs were also classified into seven groups: RLK, AGC, CAMK, CMGC, STE, TKL and others.
Our previous research on wheat PKs revealed that some conserved exon-intron structures were present from primitive plant moss to flowering plant wheat [47]. In this study, we also revealed that some conserved exon-intron structures with conserved exon phases in the kinase domain existed in cotton PKs (S4 Fig). For instance, the CAMK_AMPK subfamily in cotton (Gorai.007G080200.1), grape (GSVIVT01011467001), fern (80443) and moss (Pp1s3_550V6) shared the same exon-intron structure within the exon phase "1000" in the kinase domain. This finding suggested that these conserved exon-intron structures might play important roles in plant development and evolution. Indeed, previous functional studies of plant PKs indicated that these PK genes within conserved exon-intron structures function as core components of various signaling pathways that control multiple plant cellular processes. (1) Mitosis: Aur [51] and ULK_ULK4 [52] function in cell division in A. thaliana. The rice and Arabidopsis CMGC_CDK−CDK7 homologs phosphorylate both CDKs (cyclin-dependent protein kinases) and the CTD (carboxy-terminal domain) of RNA polymerase II [53–54]. (2) Microtubule: Arabidopsis CK1_CK1 affects cortical microtubules organization [55]. Arabidopsis NEKs form homo- or heterodimers to regulate microtubule organization during epidermal cell expansion [56]. (3) Metabolism: The Arabidopsis CAMK_AMPK homolog SnRK1 forms the SnRK1-ADK complex and plays significant roles in energy homeostasis [57]. The Arabidopsis CAMK_CAMKL−LKB homologs GRIK1 and GRIK2 specifically activate SnRK1, and the GRIK-SnRK1 cascade may coordinate metabolic requirements of rapidly growing cells [58]. The Arabidopsis CAMK_CAMKL−CHK1 homologs SOS2 (Salt Overly Sensitive2) and SOS3 maintain ion homeostasis and confer salt tolerance [59]. (4) MAPK signaling network: Plant MAPK cascades regulate several processes, including stress response, immunity and development [60]. STE_STE11 functions as a MAPK3K [60]. Similar to animal RSK−2, the plant AGC_RSK−2 homolog is also activated by PDK1 [61], which is involved in the MAPK signaling cascade [62]. (5) Stress response and development: The plant CMGC_GSK homolog GSK3 functions in floral organs, cell expansion, and abiotic and biotic stress responses [63]. Plant RLK/Pelle members play various roles in cell specification [64], pathogen recognition [65], stress response and development [66]. The soybean WNK_NRBP member GmWNK1 regulates root system architecture via ABA and osmotic signals [67] and modulates the osmotic stress response [68].
We observed that some cotton PKs contained two or more kinase domains (S4 Table). Interestingly, soybean and wheat also contained similar special PKs within two or three kinase domains [8, 47]. Comparing these special PKs within 2–4 kinase domains of cotton, soybean and wheat, we identified four overlapping PK subfamilies: AGC_NDR, AGC_RSK-2, CMGC_SRPK and RLK-Pelle_DLSV. Notably, the structure of AGC_RSK-2 and CMGC_SRPK within two kinase domains can be found in moss, fern, grape and cotton (S4 Fig), suggesting that the duplication event of the kinase domain in these PKs occurred during the emergence of land plants and these structures were subsequently retained during the evolutionary process. Given that some PKs form homo- or hetero-dimers [56, 69], we hypothesized that these structures within two or three kinase domains might be required for specific substrates to form PK dimers. This hypothesis is consistent with soybean PKs [8] and requires more functional research.
Evolution, duplication and expansion of cotton PKs
The cotton genome experienced two WGD events: a cotton-specific WGD event 16.6 Mya and a WGD that occurred 130.8 Mya and was shared by eudicots [48]. Our results revealed that the cotton-specific WGD that occurred 16.6 Mya partly contributed to the expansion of cotton PKs (Fig 3). Similar to the Ks peak (0.4–0.6) of the G. raimondii whole genome [48], G. raimondii PKs also formed a Ks peak (0.4–0.6) (Fig 3A). The paleo-hexaploidy of ancient eudicot occurred after the split of Monocotyledons and Dicotyledons [70]. Considering that the WGD that occurred 130.8 Mya is shared by eudicots [48], we proposed that most of the cotton PK collinearity events (Ks 0.6–3) potentially attributed to the 130.8 Mya WGD. However, we did not identify the second obvious cotton PK Ks peak, which corresponds to the WGD that occurred 130.8 Mya. We were potentially unable to identify this peak because these duplicated PKs might exhibit low retention after the WGD that occurred 130.8 Mya. Indeed, the TD after the paleo 130.8 Mya WGD caused dosage imbalance as the gene balance hypothesis claimed [71]. Tetraploidization of G. hirsutum occurred 1.5 Mya and corresponds to a Ks peak [50]. In our study, we also identified G. hirsutum and G. barbadense PK Ks peaks (0–0.1) (Fig 3C and 3D), indicating that the tetraploidization event of G. hirsutum and G. barbadense was important for PK expansion. We also identified several PK collinearity blocks among G. raimondii (D-genome), G. arboretum (A), G. hirsutum (At and Dt) and G. barbadense (At and Dt) (Fig 4).
Most tandem cotton PKs belonged to the RLK group, suggesting that TD partly contributed to the cotton PKs expansion (S7 Table). Our result was consistent with reports demonstrating that most soybean and wheat TD PKs belonged to the RLK group [8, 47]. An A-genome ancestor resembling G. arboretum and a D-genome ancestor resembling G. raimondii diverged from a common ancestor 5–10 Mya and subsequently reunited to produce an allotetraploid Gossypium species [50]. Our result demonstrated that the TD PK clusters of G. raimondii, G. arboretum, G. hirsutum and G. barbadense exhibited different chromosome locations and related subfamilies, suggesting that they might experience independent PK TDs or different chromosome rearrangements after the split of cotton A- and D-subgenomes. Indeed, the G. raimondii genome underwent large-scale rearrangement on chromosomes 2 and 3 compared with G. arboretum [49].
Similar to A. thaliana, rice and soybean [8–10], cotton RLK groups were also remarkably large, ranging from 913 to 1,855. The expansion of cotton RLKs was associated with specific subfamilies or subgroups, including LRR, RLCK and DLSV.
LRR contained the highest number of members among the RLK group in cottons (G. raimondii 347, G. arboretum 323, G. hirsutum 619, and G. barbadense 660) and formed a cluster of 23 subfamilies. RLK/Pelle_LRR-XI-1 contained the highest number in LRR subfamilies in G. raimondii (82), G. arboretum (80), G. hirsutum (162) and G. barbadense (155). This finding suggested that RLK-LRRs experienced remarkable expansions and play important roles in plant growth, development and defense response. Indeed, many RLK-LRRs enhance drought resistance [72], improve drought and salt stress tolerance [73], respond to diverse abiotic stresses [74], and regulate root meristem development [75]. RLCK contained the second highest number of members among the RLK group in cottons (G. raimondii 184, G. arboretum 178, G. hirsutum 310, and G. barbadense 346) and was grouped into 17–18 subfamilies. Without the extracellular domain, RLCK interacts with RLK to form the RLCK-RLK complex, which could mediate plant growth and immune responses [76]. DLSV contained the third highest number of members among the RLK group in cottons (G. raimondii 129, G. arboretum 93, G. hirsutum 205, and G. barbadense 225).
Expression pattern of cotton PKs
Our previous studies demonstrated that cotton PKs GhMKK1 [24], GhMKK3 [26] and GhMKK5 [23] are involved in drought stress resistance. In this study, we provided some candidate cotton PKs involved in drought resistance. For example, expression pattern analysis revealed that CotAD_10106 (STE_STE7) of G. hirsutum exhibited slight up-regulation (log2 value 0.3230955 in leaf) under drought stress (S13 Table), suggesting that it exerts a positive effect in drought tolerance. Using BLAST in NCBI, we found that cotton PK CotAD_10106 is a homolog of A. thaliana MKK3. This prediction is consistent with our laboratory research demonstrating that GhMKK3 positively regulates the drought stress response [26]. In addition, a cotton RLK GbRLK from Gossypium barbadense is involved in drought and high salinity stress pathways by participating or activating the ABA signaling pathway [30]. In this study, we provided some candidate cotton RLKs for drought resistance. For instance, CotAD_53902 (RLK-Pelle_SD-2b) and CotAD_15239 (RLK-Pelle_LRR-Xb-1) exhibited up-regulation at 5 and 10 dpa under drought stress (S13 Table).
Our previous research also demonstrated that several cotton PKs are involved in disease defense. GhMPK2 mediates defense responses to oxidative stress and pathogen infection [77]. GhMPK6a interacts with the upstream MAPK kinase GhMKK4 and negatively regulates bacterial infection and osmotic stress [78]. GhMPK7 plays a role in SA-regulated resistance to pathogen infection [79]. GhMPK16 displays significant resistance to fungi (Alternaria alternata and Colletotrichum nicotianae) and bacteria (Pseudomonas solanacearum) [80]. In this study, we also observed that some cotton MAPKs, such as CotAD_66205, CotAD_27603 and CotAD_29572, exhibited up-regulation in response to bollworm infection at 2–10 DAI (S11 Fig). Interestingly, CotAD_53696 (CMGC_MAPK, a homolog of A. thaliana MPK16) of G. hirsutum exhibited slight up-regulation at 3 and 6 DAI (log2 values 0.213498 and 0.296303333, respectively) after A. alternate infection without chilling pre-treatment (S15 Table), suggesting that it positively regulates the A. alternate defense response. This prediction is consistent with our laboratory research demonstrating that GhMPK16 regulates A. alternata resistance [80]. In addition, cotton RLKs are also involved in disease defense. Two cotton RLK genes, Gbve1 [29] and GbRLK [81], improve tolerance to Verticillium dahliae infection. In this study, we also found that some cotton RLKs, such as CotAD_75319 (RLK-Pelle_WAK), CotAD_53902 (RLK-Pelle_SD-2b) and CotAD_03142 (RLK-Pelle_WAK_LRK10L-1), exhibited up-regulation at 2–10 DAI after bollworm infection (S11 Fig).
Cotton PKs are also involved in fiber development. According to expression analysis, some cotton CrRLK1L proteins were predicted to be related to fiber development [21]. A cotton CDPK gene GhCPK1 [82] and a cotton CDK gene GhCDKA [83] were cloned and characterized to be associated with fiber development. In this study, we also found that some cotton RLKs (CotAD_49930, CotAD_10084 and CotAD_70308), MAPKs (CotAD_29572 and CotAD_09571), CDPKs (CotAD_54826 and CotAD_04751) and CDKs (CotAD_20875 and CotAD_75205) exhibited up-regulation during fiber development (S12 Fig).
Conclusion
In this study, we systematically identified cotton PKs and analyzed their classification, evolution and expression patterns. Some conserved exon-intron kinase domain structures were identified during plant evolution. WGD, TD and synteny PKs of three cotton species G. raimondii, G. arboretum and G. hirsutum were analyzed by MCScanX. These results suggest that cotton-specific WGD, TD and tetraploidization of G. hirsutu that occurred 16.6 Mya partially contributed to the expansion of cotton PKs. We also performed global expression pattern analysis under abiotic and biotic stress conditions and during fiber development, providing candidate PKs for further experimental research. Our results will provide clues for further research on the evolution and physiology of the cotton kinome.
Supporting information
(A) Representatives of each subfamily; (B) All members.
(PDF)
Filled boxes: red represents the Pkinase or Pkinase_Tyr domain; other colors represent various domains labeled in each page. The lengths of the boxes and lines are scaled based on the length of proteins.
(PDF)
The descriptions of domain and exon phases are the same as in Fig 2. The lengths of the boxes and lines are scaled based on the length of genes.
(PDF)
The descriptions of domain and exon phases are the same as in Fig 2. The lengths of the boxes and lines are scaled based on the length of genes.
(PDF)
Chromosomal locations of G. raimondii (A), G. arboretum (B), G. hirsutum (C) and G. barbadense (D) PKs. Yellow boxes denote PK genes.
(PDF)
(A) The collinearity events of G. raimondii PKs resulting from 16.6-Mya WGD. (B) The other collinearity events of G. raimondii PKs. (C) All of the collinearity events of G. raimondii PKs. (D) The collinearity events of G. arboretum PKs resulting from 16.6-Mya WGD. (E) The other collinearity events of G. arboretum PKs. (F) All of the collinearity events of G. arboretum PKs. (G) The collinearity events of G. hirsutum PKs contributed by tetraploidization. (H) The collinearity events of G. hirsutum PKs resulting from 16.6-Mya WGD. (I) The other collinearity events of G. hirsutum PKs. (J) All of the collinearity events of G. hirsutum PKs. (K) The collinearity events of G. barbadense PKs contributed by tetraploidization. (L) The collinearity events of G. barbadense PKs resulting from 16.6-Mya WGD. (M) The other collinearity events of G. barbadense PKs. (N) All of the collinearity events of G. barbadense PKs. Red lines denote the collinearity events resulting from 16.6-Mya WGD. Red lines denote the collinearity events resulting from 16.6-Mya WGD. Yellow lines denote the collinearity events contributed by tetraploidization. Blue lines denote other collinearity events.
(PDF)
(A) The 291 tandemly arrayed G. raimondii PK genes were grouped in 83 clusters distributed unevenly among the 13 chromosomes. (B) The 246 tandemly arrayed G. arboretum PK genes were grouped in 87 clusters distributed unevenly among the 13 chromosomes. (C) The 264 tandemly arrayed G. hirsutum PK genes were grouped in 91 clusters distributed unevenly among the 26 chromosomes. (D) The 455 tandemly arrayed G. barbadense PK genes were grouped in 152 clusters distributed unevenly among the 26 chromosomes. Gene IDs and subfamilies are labeled in the right of each chromosome, and the chromosomal location of each cluster is in the left of each chromosome. Genes in the same cluster are highlighted in the same color.
(PDF)
RLE (Relative log expression) and NUSE (Normalized unscaled standard errors) values of each GEO microarray dataset.
(PDF)
The expression patterns of 564 PK genes under abiotic stress treatments are presented. The heat maps were generated using MeV (Multiple Experiment Viewer) software, version 4.9. Red and green correspond to up-regulation and down-regulation, respectively. Normalized gene expression values are provided in Supplemental S11 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S13 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S15 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S17 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S19 Table.
(PDF)
Normalized gene expression RPKM values of G. hirsutum PK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). Normalized gene expression values are provided in Supplemental S22 Table.
(PDF)
Normalized gene expression RPKM values of G. barbadense PK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). Normalized gene expression values are provided in Supplemental S22 Table.
(PDF)
Normalized gene expression RPKM values of G. hirsutum PK genes for genotypes (Li1 mutant and wild type) ahout leaf tissue and ovule tissues in fiber development stages (1, 3 and 8 dpa). Normalized gene expression values are provided in Supplemental S23 Table.
(PNG)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense classifications are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense atypical kinases are provided in sheets 1–4, respectively.
(XLS)
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense protein kinases within 2–4 Pkinase or Pkinase_Tyr domains are provided in sheets 1–4, respectively.
(XLS)
Chromosome locations of PKs in G. raimondii, G. arboretum, G. hirsutum and G. barbadense are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense Ka/Ks values of protein kinases are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense tandemly arrayed PKs are provided in sheets 1–4, respectively.
(XLS)
Synteny analyses of PK genes bewteen G. arboretum A-genome and G. raimondii D-genome; bewteen G. hirsutum At-subgenome and G. barbadense At-subgenome; bewteen G. hirsutum Dt-subgenome and G. barbadense Dt-subgenome; bewteen G. hirsutum At-subgenome and G. arboretum A-genome; bwtween G. hirsutum Dt-subgenome and G. raimondii D-genome; between G. barbadense At-subgenome and G. arboretum A-genome; and between G. barbadense Dt-subgenome and G. raimondii D-genome are provided in sheets 1–7, respectively.
(XLS)
The descriptions of sheets 1–7 are same as S8 Table.
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
G. hirsutum and G. barbadense PK genes are provided in sheets 1–2, respectively.
(XLS)
(XLS)
Leaf tissue, ovule tissues in 1, 3 and 8 dpa are provided in sheets 1–4, respectively.
(XLS)
Acknowledgments
We would like to thank Dr. An-Yuan Guo (Huazhong University of Science and Technology) for advice on this study and Dr. Shizhong Zhang (Shandong Agricultural University) for manuscript revisions. This work was supported by the fund of National Natural Science Foundation of China (325–35238). This work was also supported by the funding of J.Y: Young Teacher Innovation Fund of Shandong Agricultural University (140–23848), First-class Discipline Fund, Key Cultivation Discipline Fund for NSFC, and Fund for Fostering Talents of Information College (xxxy201707). This work was also supported by the Natural Science Foundation of Shandong (ZR2016BB13).
Abbreviations
- AGC
PKA–PKG–PKC
- CAMK
calcium- and calmodulin-regulated kinase
- CDPK
calcium-dependent PKs
- CK1
casein kinase 1
- CMGC
cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinase and cyclin-dependent-like kinases
- LRR
leucine-rich repeat
- PK
protein kinase
- RLCK
receptor-like cytoplasmic kinase
- RLK
receptor-like kinase
- TK
tyrosine kinase
- TKL
tyrosine kinase-like kinase
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the fund of National Natural Science Foundation of China (325-35238). This work was also supported by the funding of J.Y: Young Teacher Innovation Fund of Shandong Agricultural University (140-23848), First-class Discipline Fund, Key Cultivation Discipline Fund for NSFC, and Fund for Fostering Talents of Information College (xxxy201707). This work was also supported by the Natural Science Foundation of Shandong (ZR2016BB13).
References
- 1.Singh A, Pandey GK. Protein phosphatases: a genomic outlook to understand their function in plants. Journal of Plant Biochemistry & Biotechnology. 2012;21(1):100–7. [Google Scholar]
- 2.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–96. Epub 1995/05/01. . [PubMed] [Google Scholar]
- 3.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. Epub 1988/07/01. . [DOI] [PubMed] [Google Scholar]
- 4.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27(10):514–20. Epub 2002/10/09. . [DOI] [PubMed] [Google Scholar]
- 5.Lehti-Shiu MD, Shiu SH. Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2619–39. Epub 2012/08/15. doi: 10.1098/rstb.2012.0003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champion A, Kreis M, Mockaitis K, Picaud A, Henry Y. Arabidopsis kinome: after the casting. Funct Integr Genomics. 2004;4(3):163–87. Epub 2004/01/24. doi: 10.1007/s10142-003-0096-4 . [DOI] [PubMed] [Google Scholar]
- 7.Zulawski M, Schulze G, Braginets R, Hartmann S, Schulze WX. The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genomics. 2014;15:548 Epub 2014/07/06. doi: 10.1186/1471-2164-15-548 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Chen N, Grant JN, Cheng ZM, Stewart CN Jr., Hewezi T. Soybean kinome: functional classification and gene expression patterns. J Exp Bot. 2015;66(7):1919–34. Epub 2015/01/24. doi: 10.1093/jxb/eru537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132(2):530–43. Epub 2003/06/14. doi: 10.1104/pp.103.021964 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16(5):1220–34. Epub 2004/04/24. doi: 10.1105/tpc.020834 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao LL, Xue HW. Global analysis of expression profiles of rice receptor-like kinase genes. Mol Plant. 2012;5(1):143–53. Epub 2011/07/19. doi: 10.1093/mp/ssr062 . [DOI] [PubMed] [Google Scholar]
- 12.Wei K, Wang Y, Xie D. Identification and expression profile analysis of the protein kinase gene superfamily in maize development. Molecular Breeding. 2014;33(1):155–72. [Google Scholar]
- 13.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 2001;98(19):10763–8. Epub 2001/08/30. doi: 10.1073/pnas.181141598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant. 2008;1(5):732–50. Epub 2009/10/15. doi: 10.1093/mp/ssn047 . [DOI] [PubMed] [Google Scholar]
- 15.Lehti-Shiu MD, Zou C, Hanada K, Shiu SH. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009;150(1):12–26. Epub 2009/03/27. doi: 10.1104/pp.108.134353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zan Y, Ji Y, Zhang Y, Yang S, Song Y, Wang J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genomics. 2013;14:318 Epub 2013/05/15. doi: 10.1186/1471-2164-14-318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilotti M, Brunetti A, Uva P, Lumia V, Tizzani L, Gervasi F, et al. Kinase domain-targeted isolation of defense-related receptor-like kinases (RLK/Pelle) in Platanus×acerifolia: phylogenetic and structural analysis. BMC Res Notes. 2014;7:884 Epub 2014/12/10. doi: 10.1186/1756-0500-7-884 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rameneni JJ, Lee Y, Dhandapani V, Yu X, Choi SR, Oh MH, et al. Genomic and Post-Translational Modification Analysis of Leucine-Rich-Repeat Receptor-Like Kinases in Brassica rapa. PLoS One. 2015;10(11):e0142255 Epub 2015/11/21. doi: 10.1371/journal.pone.0142255 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Xun Q, Guo Y, Zhang J, Cheng K, Shi T, et al. Genome-Wide Expression Pattern Analyses of the Arabidopsis Leucine-Rich Repeat Receptor-Like Kinases. Mol Plant. 2016;9(2):289–300. Epub 2015/12/30. doi: 10.1016/j.molp.2015.12.011 . [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto T, Deguchi M, Brustolini OJ, Santos AA, Silva FF, Fontes EP. The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 2012;12:229 Epub 2012/12/04. doi: 10.1186/1471-2229-12-229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu E, Cai C, Zheng Y, Shang X, Fang L, Guo W. Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol Genet Genomics. 2016;291(3):1137–54. Epub 2016/02/03. doi: 10.1007/s00438-016-1169-0 . [DOI] [PubMed] [Google Scholar]
- 22.Fischer I, Dievart A, Droc G, Dufayard JF, Chantret N. Evolutionary Dynamics of the Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK) Subfamily in Angiosperms. Plant Physiol. 2016;170(3):1595–610. Epub 2016/01/17. doi: 10.1104/pp.15.01470 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Li Y, Lu W, Meng F, Wu CA, Guo X. Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J Exp Bot. 2012;63(10):3935–51. Epub 2012/03/24. doi: 10.1093/jxb/ers086 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu W, Chu X, Li Y, Wang C, Guo X. Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS One. 2013;8(7):e68503 Epub 2013/07/12. doi: 10.1371/journal.pone.0068503 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhang L, Lu W, Wang X, Wu CA, Guo X. Overexpression of cotton GhMKK4 enhances disease susceptibility and affects abscisic acid, gibberellin and hydrogen peroxide signalling in transgenic Nicotiana benthamiana. Mol Plant Pathol. 2014;15(1):94–108. Epub 2013/08/29. doi: 10.1111/mpp.12067 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Lu W, He X, Wang F, Zhou Y, Guo X. The Cotton Mitogen-Activated Protein Kinase Kinase 3 Functions in Drought Tolerance by Regulating Stomatal Responses and Root Growth. Plant Cell Physiol. 2016;57(8):1629–42. Epub 2016/06/24. doi: 10.1093/pcp/pcw090 . [DOI] [PubMed] [Google Scholar]
- 27.Li YL, Sun J, Xia GX. Cloning and characterization of a gene for an LRR receptor-like protein kinase associated with cotton fiber development. Mol Genet Genomics. 2005;273(3):217–24. Epub 2005/05/20. doi: 10.1007/s00438-005-1115-z . [DOI] [PubMed] [Google Scholar]
- 28.Shi YL, Guo SD, Zhang R, Meng ZG, Ren MZ. The role of Somatic embryogenesis receptor-like kinase 1 in controlling pollen production of the Gossypium anther. Mol Biol Rep. 2014;41(1):411–22. Epub 2013/11/28. doi: 10.1007/s11033-013-2875-x . [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Yang Y, Chen T, Yu W, Liu T, Li H, et al. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One. 2012;7(12):e51091 Epub 2012/12/20. doi: 10.1371/journal.pone.0051091 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Gao Y, Zhang Z, Chen T, Guo W, Zhang T. A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biol. 2013;13:110 Epub 2013/08/07. doi: 10.1186/1471-2229-13-110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23(1):205–11. Epub 2010/02/25. . [PubMed] [Google Scholar]
- 32.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. Epub 2007/09/12. doi: 10.1093/bioinformatics/btm404 . [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. Epub 2016/03/24. doi: 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49 Epub 2012/01/06. doi: 10.1093/nar/gkr1293 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozik A, Kochetkova E, Michelmore R. GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics. 2002;18(2):335–6. Epub 2002/02/16. . [DOI] [PubMed] [Google Scholar]
- 36.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. Epub 2009/06/23. doi: 10.1101/gr.092759.109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu YN, Shi DQ, Ruan MB, Zhang LL, Meng ZH, Liu J, et al. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS One. 2013;8(11):e80218 Epub 2013/11/14. doi: 10.1371/journal.pone.0080218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010;51(1):21–37. Epub 2009/11/20. doi: 10.1093/pcp/pcp163 . [DOI] [PubMed] [Google Scholar]
- 39.Padmalatha KV, Dhandapani G, Kanakachari M, Kumar S, Dass A, Patil DP, et al. Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol Biol. 2012;78(3):223–46. Epub 2011/12/07. doi: 10.1007/s11103-011-9857-y . [DOI] [PubMed] [Google Scholar]
- 40.Nigam D, Kavita P, Tripathi RK, Ranjan A, Goel R, Asif M, et al. Transcriptome dynamics during fibre development in contrasting genotypes of Gossypium hirsutum L. Plant Biotechnol J. 2014;12(2):204–18. Epub 2013/10/15. doi: 10.1111/pbi.12129 . [DOI] [PubMed] [Google Scholar]
- 41.Walford SA, Wu Y, Llewellyn DJ, Dennis ES. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 2012;71(3):464–78. Epub 2012/03/27. doi: 10.1111/j.1365-313X.2012.05003.x . [DOI] [PubMed] [Google Scholar]
- 42.Padmalatha KV, Patil DP, Kumar K, Dhandapani G, Kanakachari M, Phanindra ML, et al. Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation. BMC Genomics. 2012;13:624 Epub 2012/11/16. doi: 10.1186/1471-2164-13-624 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. Epub 2003/03/05. . [DOI] [PubMed] [Google Scholar]
- 44.Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: a tale of duplications. Trends Plant Sci. 2009;14(12):680–8. Epub 2009/10/13. doi: 10.1016/j.tplants.2009.09.001 . [DOI] [PubMed] [Google Scholar]
- 45.Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:17662 Epub 2015/12/05. doi: 10.1038/srep17662 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–7. Epub 2015/04/22. doi: 10.1038/nbt.3207 . [DOI] [PubMed] [Google Scholar]
- 47.Yan J, Su P, Wei Z, Nevo E, Kong L. Genome-wide identification, classification, evolutionary analysis and gene expression patterns of the protein kinase gene family in wheat and Aegilops tauschii. Plant Mol Biol. 2017;95(3):227–42. Epub 2017/09/18. doi: 10.1007/s11103-017-0637-1 . [DOI] [PubMed] [Google Scholar]
- 48.Wang K, Wang Z, Li F, Ye W, Wang J, Song G, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–103. Epub 2012/08/28. doi: 10.1038/ng.2371 . [DOI] [PubMed] [Google Scholar]
- 49.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–72. Epub 2014/05/20. doi: 10.1038/ng.2987 . [DOI] [PubMed] [Google Scholar]
- 50.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–30. Epub 2015/04/22. doi: 10.1038/nbt.3208 . [DOI] [PubMed] [Google Scholar]
- 51.Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, et al. Arabidopsis alpha Aurora kinases function in formative cell division plane orientation. Plant Cell. 2011;23(11):4013–24. Epub 2011/11/03. doi: 10.1105/tpc.111.089565 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krupnova T, Sasabe M, Ghebreghiorghis L, Gruber CW, Hamada T, Dehmel V, et al. Microtubule-associated kinase-like protein RUNKEL needed [corrected] for cell plate expansion in Arabidopsis cytokinesis. Curr Biol. 2009;19(6):518–23. Epub 2009/03/10. doi: 10.1016/j.cub.2009.02.021 . [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi M, Umeda M, Uchimiya H. A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J. 1998;16(5):613–9. Epub 1999/02/26. . [DOI] [PubMed] [Google Scholar]
- 54.Shimotohno A, Matsubayashi S, Yamaguchi M, Uchimiya H, Umeda M. Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 2003;534(1–3):69–74. Epub 2003/01/16. . [DOI] [PubMed] [Google Scholar]
- 55.Ben-Nissan G, Cui W, Kim DJ, Yang Y, Yoo BC, Lee JY. Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 2008;148(4):1897–907. Epub 2008/10/24. doi: 10.1104/pp.108.129346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Motose H, Hamada T, Yoshimoto K, Murata T, Hasebe M, Watanabe Y, et al. NIMA-related kinases 6, 4, and 5 interact with each other to regulate microtubule organization during epidermal cell expansion in Arabidopsis thaliana. Plant J. 2011;67(6):993–1005. Epub 2011/05/25. doi: 10.1111/j.1365-313X.2011.04652.x . [DOI] [PubMed] [Google Scholar]
- 57.Mohannath G, Jackel JN, Lee YH, Buchmann RC, Wang H, Patil V, et al. A complex containing SNF1-related kinase (SnRK1) and adenosine kinase in Arabidopsis. PLoS One. 2014;9(1):e87592 Epub 2014/02/06. doi: 10.1371/journal.pone.0087592 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen W, Reyes MI, Hanley-Bowdoin L. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 2009;150(2):996–1005. Epub 2009/04/03. doi: 10.1104/pp.108.132787 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X. The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant. 2013;6(2):275–86. Epub 2013/01/29. doi: 10.1093/mp/sst017 . [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–49. Epub 2010/05/06. doi: 10.1146/annurev-arplant-042809-112252 . [DOI] [PubMed] [Google Scholar]
- 61.Hirt H, Garcia AV, Oelmuller R. AGC kinases in plant development and defense. Plant Signal Behav. 2011;6(7):1030–3. Epub 2011/10/19. doi: 10.4161/psb.6.7.15580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9(10):747–58. Epub 2008/09/25. doi: 10.1038/nrm2509 . [DOI] [PubMed] [Google Scholar]
- 63.Saidi Y, Hearn TJ, Coates JC. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012;17(1):39–46. Epub 2011/11/05. doi: 10.1016/j.tplants.2011.10.002 . [DOI] [PubMed] [Google Scholar]
- 64.De Smet I, Voss U, Jurgens G, Beeckman T. Receptor-like kinases shape the plant. Nat Cell Biol. 2009;11(10):1166–73. Epub 2009/10/02. doi: 10.1038/ncb1009-1166 . [DOI] [PubMed] [Google Scholar]
- 65.Afzal AJ, Wood AJ, Lightfoot DA. Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe Interact. 2008;21(5):507–17. Epub 2008/04/09. doi: 10.1094/MPMI-21-5-0507 . [DOI] [PubMed] [Google Scholar]
- 66.Vaid N, Macovei A, Tuteja N. Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol Plant. 2013;6(5):1405–18. Epub 2013/02/23. doi: 10.1093/mp/sst033 . [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Suo H, Zheng Y, Liu K, Zhuang C, Kahle KT, et al. The soybean root-specific protein kinase GmWNK1 regulates stress-responsive ABA signaling on the root system architecture. Plant J. 2010;64(2):230–42. Epub 2010/08/26. doi: 10.1111/j.1365-313X.2010.04320.x . [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Suo H, Zhuang C, Ma H, Yan X. Overexpression of the soybean GmWNK1 altered the sensitivity to salt and osmotic stress in Arabidopsis. J Plant Physiol. 2011;168(18):2260–7. Epub 2011/09/20. doi: 10.1016/j.jplph.2011.07.014 . [DOI] [PubMed] [Google Scholar]
- 69.Afzal AJ, Srour A, Goil A, Vasudaven S, Liu T, Samudrala R, et al. Homo-dimerization and ligand binding by the leucine-rich repeat domain at RHG1/RFS2 underlying resistance to two soybean pathogens. BMC Plant Biol. 2013;13:43 Epub 2013/03/19. doi: 10.1186/1471-2229-13-43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449(7161):463–7. Epub 2007/08/28. doi: 10.1038/nature06148 . [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Wang X, Paterson AH. Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci. 2012;1256:1–14. Epub 2012/01/20. doi: 10.1111/j.1749-6632.2011.06384.x . [DOI] [PubMed] [Google Scholar]
- 72.Xing HT, Guo P, Xia XL, Yin WL. PdERECTA, a leucine-rich repeat receptor-like kinase of poplar, confers enhanced water use efficiency in Arabidopsis. Planta. 2011;234(2):229–41. Epub 2011/03/15. doi: 10.1007/s00425-011-1389-9 . [DOI] [PubMed] [Google Scholar]
- 73.Ouyang SQ, Liu YF, Liu P, Lei G, He SJ, Ma B, et al. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010;62(2):316–29. Epub 2010/02/05. doi: 10.1111/j.1365-313X.2010.04146.x . [DOI] [PubMed] [Google Scholar]
- 74.Wu T, Tian Z, Liu J, Xie C. A novel leucine-rich repeat receptor-like kinase gene in potato, StLRPK1, is involved in response to diverse stresses. Mol Biol Rep. 2009;36(8):2365–74. Epub 2009/02/14. doi: 10.1007/s11033-009-9459-9 . [DOI] [PubMed] [Google Scholar]
- 75.Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 2016;26(6):686–98. Epub 2016/05/28. doi: 10.1038/cr.2016.63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin W, Ma X, Shan L, He P. Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol. 2013;55(12):1188–97. Epub 2013/05/29. doi: 10.1111/jipb.12071 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Xi D, Luo L, Meng F, Li Y, Wu CA, et al. Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 2011;278(8):1367–78. Epub 2011/02/23. doi: 10.1111/j.1742-4658.2011.08056.x . [DOI] [PubMed] [Google Scholar]
- 78.Li Y, Zhang L, Wang X, Zhang W, Hao L, Chu X, et al. Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J. 2013;280(20):5128–44. Epub 2013/08/21. doi: 10.1111/febs.12488 . [DOI] [PubMed] [Google Scholar]
- 79.Shi J, An HL, Zhang L, Gao Z, Guo XQ. GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol Biol. 2010;74(1–2):1–17. Epub 2010/07/06. doi: 10.1007/s11103-010-9661-0 . [DOI] [PubMed] [Google Scholar]
- 80.Shi J, Zhang L, An H, Wu C, Guo X. GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol Biol. 2011;12:22 Epub 2011/05/18. doi: 10.1186/1471-2199-12-22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jun Z, Zhang Z, Gao Y, Zhou L, Fang L, Chen X, et al. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci Rep. 2015;5:15048 Epub 2015/10/09. doi: 10.1038/srep15048 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang QS, Wang HY, Gao P, Wang GY, Xia GX. Cloning and characterization of a calcium dependent protein kinase gene associated with cotton fiber development. Plant Cell Rep. 2008;27(12):1869–75. Epub 2008/09/11. doi: 10.1007/s00299-008-0603-0 . [DOI] [PubMed] [Google Scholar]
- 83.Gao W, Saha S, Ma DP, Guo Y, Jenkins JN, Stelly DM. A cotton-fiber-associated cyclin-dependent kinase a gene: characterization and chromosomal location. Int J Plant Genomics. 2012;2012:613812 Epub 2012/06/30. doi: 10.1155/2012/613812 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representatives of each subfamily; (B) All members.
(PDF)
Filled boxes: red represents the Pkinase or Pkinase_Tyr domain; other colors represent various domains labeled in each page. The lengths of the boxes and lines are scaled based on the length of proteins.
(PDF)
The descriptions of domain and exon phases are the same as in Fig 2. The lengths of the boxes and lines are scaled based on the length of genes.
(PDF)
The descriptions of domain and exon phases are the same as in Fig 2. The lengths of the boxes and lines are scaled based on the length of genes.
(PDF)
Chromosomal locations of G. raimondii (A), G. arboretum (B), G. hirsutum (C) and G. barbadense (D) PKs. Yellow boxes denote PK genes.
(PDF)
(A) The collinearity events of G. raimondii PKs resulting from 16.6-Mya WGD. (B) The other collinearity events of G. raimondii PKs. (C) All of the collinearity events of G. raimondii PKs. (D) The collinearity events of G. arboretum PKs resulting from 16.6-Mya WGD. (E) The other collinearity events of G. arboretum PKs. (F) All of the collinearity events of G. arboretum PKs. (G) The collinearity events of G. hirsutum PKs contributed by tetraploidization. (H) The collinearity events of G. hirsutum PKs resulting from 16.6-Mya WGD. (I) The other collinearity events of G. hirsutum PKs. (J) All of the collinearity events of G. hirsutum PKs. (K) The collinearity events of G. barbadense PKs contributed by tetraploidization. (L) The collinearity events of G. barbadense PKs resulting from 16.6-Mya WGD. (M) The other collinearity events of G. barbadense PKs. (N) All of the collinearity events of G. barbadense PKs. Red lines denote the collinearity events resulting from 16.6-Mya WGD. Red lines denote the collinearity events resulting from 16.6-Mya WGD. Yellow lines denote the collinearity events contributed by tetraploidization. Blue lines denote other collinearity events.
(PDF)
(A) The 291 tandemly arrayed G. raimondii PK genes were grouped in 83 clusters distributed unevenly among the 13 chromosomes. (B) The 246 tandemly arrayed G. arboretum PK genes were grouped in 87 clusters distributed unevenly among the 13 chromosomes. (C) The 264 tandemly arrayed G. hirsutum PK genes were grouped in 91 clusters distributed unevenly among the 26 chromosomes. (D) The 455 tandemly arrayed G. barbadense PK genes were grouped in 152 clusters distributed unevenly among the 26 chromosomes. Gene IDs and subfamilies are labeled in the right of each chromosome, and the chromosomal location of each cluster is in the left of each chromosome. Genes in the same cluster are highlighted in the same color.
(PDF)
RLE (Relative log expression) and NUSE (Normalized unscaled standard errors) values of each GEO microarray dataset.
(PDF)
The expression patterns of 564 PK genes under abiotic stress treatments are presented. The heat maps were generated using MeV (Multiple Experiment Viewer) software, version 4.9. Red and green correspond to up-regulation and down-regulation, respectively. Normalized gene expression values are provided in Supplemental S11 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S13 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S15 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S17 Table.
(PDF)
Normalized gene expression values are provided in Supplemental S19 Table.
(PDF)
Normalized gene expression RPKM values of G. hirsutum PK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). Normalized gene expression values are provided in Supplemental S22 Table.
(PDF)
Normalized gene expression RPKM values of G. barbadense PK genes in fiber development stages (10, 15, 18, 21 and 28 dpa). Normalized gene expression values are provided in Supplemental S22 Table.
(PDF)
Normalized gene expression RPKM values of G. hirsutum PK genes for genotypes (Li1 mutant and wild type) ahout leaf tissue and ovule tissues in fiber development stages (1, 3 and 8 dpa). Normalized gene expression values are provided in Supplemental S23 Table.
(PNG)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense classifications are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense atypical kinases are provided in sheets 1–4, respectively.
(XLS)
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense protein kinases within 2–4 Pkinase or Pkinase_Tyr domains are provided in sheets 1–4, respectively.
(XLS)
Chromosome locations of PKs in G. raimondii, G. arboretum, G. hirsutum and G. barbadense are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense Ka/Ks values of protein kinases are provided in sheets 1–4, respectively.
(XLS)
G. raimondii, G. arboretum, G. hirsutum and G. barbadense tandemly arrayed PKs are provided in sheets 1–4, respectively.
(XLS)
Synteny analyses of PK genes bewteen G. arboretum A-genome and G. raimondii D-genome; bewteen G. hirsutum At-subgenome and G. barbadense At-subgenome; bewteen G. hirsutum Dt-subgenome and G. barbadense Dt-subgenome; bewteen G. hirsutum At-subgenome and G. arboretum A-genome; bwtween G. hirsutum Dt-subgenome and G. raimondii D-genome; between G. barbadense At-subgenome and G. arboretum A-genome; and between G. barbadense Dt-subgenome and G. raimondii D-genome are provided in sheets 1–7, respectively.
(XLS)
The descriptions of sheets 1–7 are same as S8 Table.
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
G. hirsutum and G. barbadense PK genes are provided in sheets 1–2, respectively.
(XLS)
(XLS)
Leaf tissue, ovule tissues in 1, 3 and 8 dpa are provided in sheets 1–4, respectively.
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.