Abstract
New therapeutic approaches are needed for gestational diabetes mellitus (GDM), but must show safety and efficacy in a historically understudied population. We studied associations between electronic medical record (EMR) phenotypes and genetic variants to uncover drugs currently considered safe in pregnancy that could treat or prevent GDM.
We identified 129 systemically active drugs considered safe in pregnancy targeting the proteins produced from 196 genes. We tested for associations between GDM and/or type 2 diabetes (DM2) and 306 SNPs in 130 genes represented on the Illumina Infinium Human Exome Bead Chip (DM2 was included due to shared pathophysiological features with GDM). In parallel, we tested the association between drugs and glucose tolerance during pregnancy as measured by the glucose recorded during a routine 50-gram glucose tolerance test (GTT).
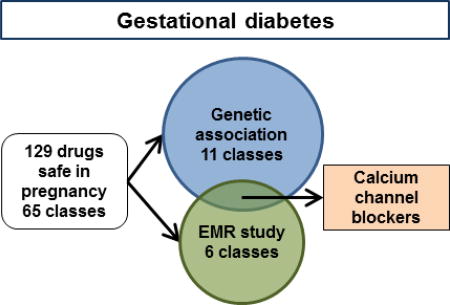
We found an association between GDM/DM2 and the genes targeted by 11 drug classes. In the EMR analysis, 6 drug classes were associated with changes in GTT. Two classes were identified in both analyses. L-type calcium channel blocking antihypertensives (CCBs), were associated with a 3.18mg/dL (95% CI −6.18 to −0.18) decrease in glucose during GTT, and Serotonin receptor type 3 (5HT-3) antagonist antinausea medications were associated with a 3.54mg/dL (95% CI 1.86 to 5.23) increase in glucose during GTT.
CCBs were identified as a class of drugs considered safe in pregnancy could have efficacy in treating or preventing GDM. 5HT-3 antagonists may be associated with worse glucose tolerance.
Graphical abstract
Introduction
Gestational diabetes (GDM) affects nearly 1 in 12 pregnancies in the United States, and is increasing in the developing world including up to 14% in Africa1,2. GDM is generally defined as elevated fasting glucose or elevated glucose in an oral glucose tolerance test3. GDM is caused by a combination of insulin resistance (similar to DM2) and inadequate insulin production to accommodate the fetus (despite elevated maternal insulin levels)4. It is associated with complications including macrosomia, shoulder dystocia, preterm birth, pre-eclampsia, and stillbirth5. Unlike other major pregnancy complications that occur late in gestation (e.g., preterm birth or pre-eclampsia), GDM is diagnosed in the second trimester, providing a relatively long window for potential intervention.
GDM is associated with an increased risk of subsequent GDM and DM2 in both mother and infant, raising the worrisome possibility of a vicious intergenerational cycle6,7. Current therapy for GDM relies on the use of insulin and two oral hypoglycemic medications, metformin and glyburide. There is good quality evidence that treatment of GDM normalizes blood sugar and results in lower birthweight and frequency of large for gestational age infants8. However, treatment does not reduce BMI Z-score, waist circumference, or fasting glucose in offspring at 8–10 years of follow-up9. Treatment of women with a history of GDM with metformin or intensive lifestyle modification reduces their risk of progression to DM2 to approximately that of women with similar baseline characteristics but no history of GDM10.However, there is no evidence that intrapartum therapy has any effect on progression to DM211. The identification of additional medications safe in pregnancy that treat GDM could improve outcomes and give providers more alternatives to oral or systemic glucose-lowering agents.
Although the need for new treatments is apparent, the usual target-driven path to drug discovery is especially cumbersome in pregnancy. The high cost of new, in-patent medications is particularly problematic for a disease with a growing burden in low and middle-income countries. An alternative strategy is drug repurposing using an FDA-approved drug applied to a novel disease, particularly those known to be safe in pregnancy. One example of successful repurposing is metformin for polycystic ovarian syndrome. In the maternal-fetal arena, drug repurposing has an additional motivation – because of the perceived medical, ethical and legal consequences of teratogenesis, new medications are essentially never tested on pregnant women12.
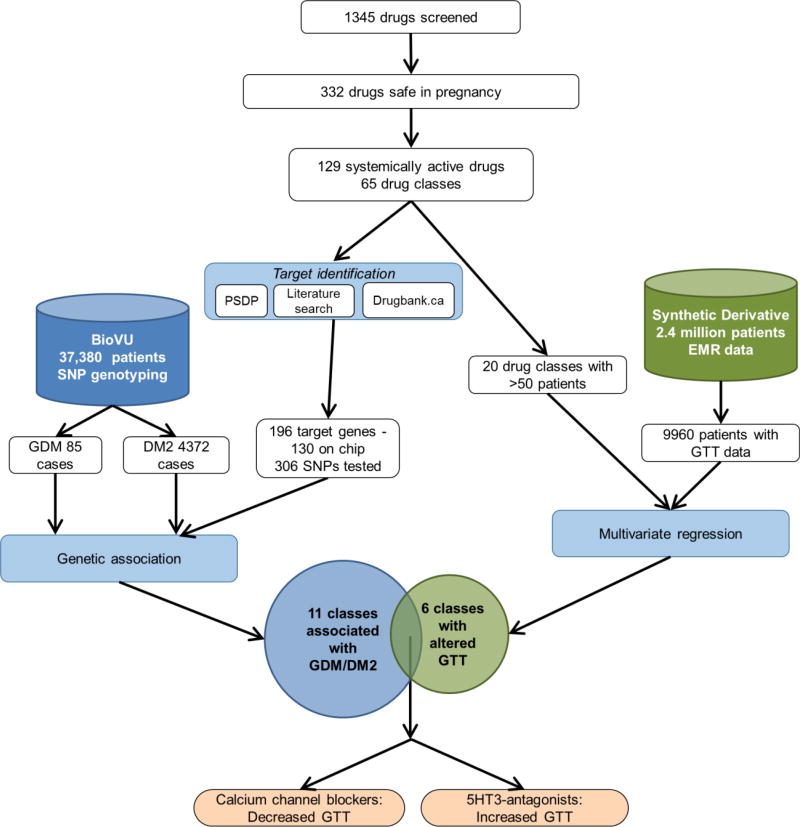
The purpose of this study was to use an informatics-based approaches as a hypothesis-generating method to identify drug repurposing candidates for GDM (Figure 1).
Figure 1. Experimental design.
We screened 1345 drugs and identified 332 considered safe in pregnancy. Of these, 129 drugs in 65 classes are systemically active, have identifiable human targets, and are not contraindicated by the US FDA. We pursued a two-pronged strategy. In the first (left side), we identified the drug target genes using publicly available data. Of the 196 target genes, 130 genes, represented by 306 SNPs, were present on our genotyping platform. We used SNP genotyping data from 37,380 patients with ICD9-level diagnostic information (BioVU). We examined the association between the genes targeted by our candidate drugs and GDM or DM2, combining our analyses using gene set analysis, which resulted in 11 drug classes genetically associated with GDM/DM2. In the second strategy (right side), we generated a cohort of patients with GTT data and examined the effect of drug exposure on GTT values in a multivariate regression. We identified 6 classes of drugs associated with changes in GTT. The overlap of these two independent strategies (bottom) are two drugs – 5HT-3 antagonists, which are associated with increased GTT, indicating worse glucose tolerance, and calcium channel blockers, which are associated with decreased GTT, indicating improved glucose tolerance.
Research Design and Methods
Drug and gene selection
Because we were unable to identify a comprehensive list of U.S. Food and Drug Administration (FDA) drug safety-in-pregnancy ratings, we compiled a list of drugs considered safe in pregnancy from the Australian Prescribing Medicines in Pregnancy database and two guides for U.S. practitioners13–15. Within the Australian database, we selected drugs rated A, indicating no teratogenicity in large human studies or B1, indicating no teratogenicity in small human studies and no teratogenicity in animal studies. The U.S. FDA has moved away from general pregnancy categorizations in favor of narrative description, however these roughly correspond to FDA categories A and B16. Where categories were given, e.g., “tricyclic antidepressants”, representative U.S. approved examples were included. This initial list consisted of 332 drugs. We selected systemically-active drugs delivered orally or subcutaneously. We excluded drugs that lacked a clear human protein target (e.g., many antibiotics), or are illicit, withdrawn, or contraindicated in pregnancy by the FDA regardless of other ratings. This left 129 candidate therapeutic agents in 65 mechanistic classes (Table S1a–b). We identified targets based on receptor profiling by the Psychoactive Drug Screening Program and “pharmacological action” targets listed in DrugBank.ca and mapped these to their respective genes (51, Figure 2). For drugs where targets were not identified, we performed a literature search using PubMed and Google Scholar to identify plausible targets. A total of 196 genes were identified, reflecting drug activity at many targets and multiple drugs with activity at a single protein.
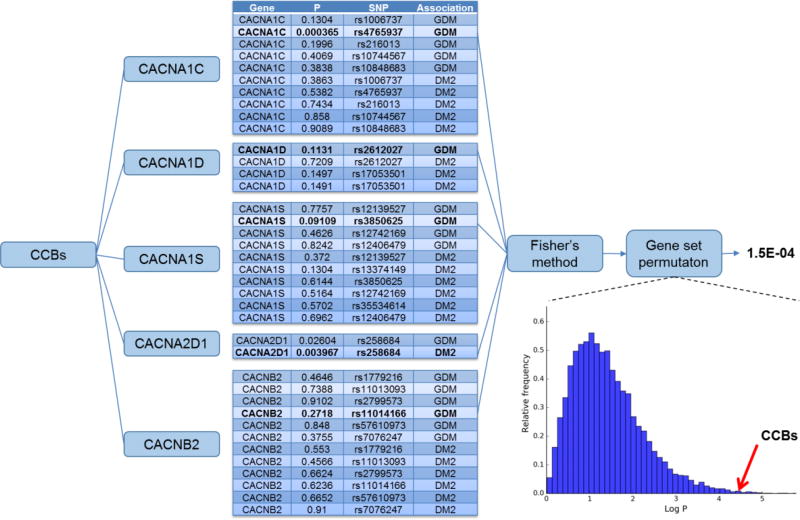
Figure 2. Example drug-gene set P-value calculation.
Each drug class (e.g. calcium channel blockers (CCBs)) acts by targeting one or more proteins (in this case CACNA1C, CACNA1D, CACNA1S, CACNA2D1, and CACNB2). Within the gene that codes for the protein are one or more SNPs. The association of each SNP with GDM or DM2 was tested. From each gene, we identified the SNP/disease association with the lowest P-value (e.g. rs4765937 with GDM in CACNA1C). We combined the lowest P-value from each gene using Fisher’s method and controlled the result using gene set permutation.
Patient population and genotyping
Patients were identified using the Synthetic Derivative (SD), a de-identified image of Vanderbilt University Medical Center’s electronic medical record (EMR). Diagnoses are abstracted in the SD according to the International Classification of Diseases, version 9 (ICD9). Within the SD, ICD9 codes are aggregated into Phecodes, reducing complexity and allowing for variation in coding over time17. BioVU is a subset of the SD that includes DNA samples and analyses, as previously described18–20.The findings reported here are based on the 37,380 BioVU subjects tested using the Illumina Infinium Human Exome Bead Chip. Of our 196 genes of interest, 130 are represented on the chip by 306 SNPs.
Identification of disease associated SNPs in GDM
We identified 1281 women with SNP genotyping aged 15–50 years with clinic visits for supervision of a normal or high-risk pregnancy (ICD9 V22.x or V23.x). We excluded women with a diagnosis of type I diabetes (Phecode 250.1). GDM shares pathophysiological features with DM2, including insulin resistance associated with inadequate insulin secretion4. Women with GDM are at increased risk for developing DM2, as opposed to DM1. Therefore, we retained women with a diagnosis of DM2 as they have the most severe disease. There is no single ICD9 code which maps to GDM. Therefore, we defined cases as patients meeting inclusion criteria those with at least one diagnosis of diabetes in pregnancy (ICD9 648.0x) or glucose intolerance in pregnancy (ICD9 648.8x). We included the latter cases because GDM is defined as glucose intolerance incident in pregnancy and because the preferred mapping of ICD10 (which does include codes for GDM) to ICD8 is 648.8x. Controls were other women in the cohort. Age at first prenatal visit was used as a covariate. This yielded 130 cases and 950 controls. Due to the low number of patients with non-European ancestry, we further restricted the analysis to those with European ancestry – yielding a final total of 85 GDM cases and 622 controls. The average age of the GDM GWAS cohort was 27.8+/−6.9 years. For controls, it was 27.6+/−7.0 years vs 29.3+/−6.5 years for cases (P = 0.002), consistent with the increased incidence of GDM in older patients. We performed logistic regression for the presence of GDM in plink version 1.9 using age as a covariate a minimum minor allele frequency of 0.0521.
Identification of disease associated SNPs in DM2
We reanalyzed data reported in our first description of phenome screening17. We selected cases as patients with the diagnosis of type II diabetes (Phecode 250.2), and compared their genetics to controls,22. For DM2, there were 4372 cases and 17646 controls. We performed logistic regression for the presence of DM2 in plink version 1.9 using age and current sex as covariates and a minimum minor allele frequency of 0.001. Due to the lower minimum minor allele frequency, we tested 305 SNP associations for DM2 versus 215 for GDM.
Replication of previously established SNP associations with GDM
For 9 SNPs previously established to be associated with GDM (ref 28), we searched our datasets for the corresponding SNPs and report the P-value. Because 2 of the 9 SNPs were not directly genotyped on the Illumina Infinium Human Exome Bead Chip, we identified proxy SNPs in high linkage disequilibrium with the published SNPs using SNAP23. Consistent with our previously reported methodology, we considered a SNP as successfully replicated if the analysis showed a P-value of < 0.05 (unadjusted)22.
Aggregation and statistics
We combined the SNPs and P-values for GDM and DM2 into a single dataset. Using the target genes for each drug as a gene set, we combined the strongest single SNP per gene in the gene set using Fisher’s method24,25, as implemented in Python version 3.6, pandas version 0.18, numpy version 1.13, and statsmodels 0.8.0 Figure 2. We corrected the resulting P-values using gene set permutations with 10,000 permutations per gene count25. The final P-value represents the probability that degree of association between the set of genes targeted by the drug and GDM/DM2 could have arisen if the genes were selected randomly.
Robustness testing
We used 3 approaches – 1) We replaced the custom drug-gene target associations above with the Pharmacologically Active set from Drugbank.ca and ran the same statistics, 2) We replaced the custom drug-gene target associations above with the complete set from Drugbank.ca and ran the same statistics, 3) We used the same custom drug-gene target associations as above but changed the analysis. Rather than using the strongest P-value SNP in each gene to represent that gene, we multiplied the P-value by the square root of the number of SNPs in that gene26. This penalizes genes with large numbers of SNPs which may have falsely low P-values due to chance.
Effect of drugs commonly used in pregnancy on glucose tolerance test
We sought to determine the extent to which any candidate drugs altered glucose tolerance in pregnancy as measured by the GTT. This test is generally administered during the 2nd trimester and involves women consuming a 50-gram glucose load with a blood glucose measurement 1 hour afterward. Higher blood glucose values indicate worse glucose tolerance. We chose to use the GTT as our outcome because, as compared to a diagnosis of GDM, GTT is more sensitive to small variations as it is a continuous as opposed to a dichotomous variable. We generated a cohort of patients with a GTT in pregnancy. In the SD, we selected females with CPT codes indicating a prenatal visit (0500F, 0502F) with a CPT code for a 50-gram GTT (82950) and a lab value for a 50-gram GTT (GTT50g), all on the same day. We excluded women with a diagnosis of pre-pregnancy diabetes (ICD10 O24.8x) or type 1 diabetes (ICD10 E10.x, ICD9 250.01, 250.03). This generated a cohort of 9960 patients. We had complete data for 6390 pregnancies. We identified patients with a history of exposure to drug classes based on mention in the EMR using generic and brand names. Because the SD does not provide accurate chronological information for medications, we could not reliably record the exact timing of exposure relative to gestation and treated patients as ‘exposed’ if the medication was mentioned in their record and otherwise ‘unexposed’. We excluded classes with fewer than 50 patients in the cohort (e.g. memantine, used to treat Alzheimer dementia), lack of FDA approval or a known FDA contraindication, over-the-counter (OTC) medications for which ascertainment is questionable, and drugs with significant potential for addiction and withdrawal such as opiates. Of the 65 medication classes, we examined 20 (Table S1c).
As use of particular drugs generally indicates a particular diagnosis we controlled for the effect of their usual indication, e.g. metoclopramide and 5HT-3-antagnoist antinausea medications in the context of hyperemesis gravidarum (HG, ICD10 O21.x, ICD9 643.x). We performed multivariate linear regression controlling for age, BMI at the time of GTT, race, ethnicity, and the indication (e.g., HG for antinausea medications) using statsmodels. We plotted mean and SEM of GTT for each drug by adjusting the reported GTT value for covariates as well as other drugs that the patient was exposed to that treat the same indication using matplotlib version 2.1.
This study was approved by the Vanderbilt University Institutional Review Board, #151121.
Results
Replication of prior SNP associations with GDM
As a check on the fidelity of our genetic analysis, we compared our results to previously published findings for GDM. Of the 9 SNPs most frequently replicated across data sets, the GDM analysis replicated 2 (28, Table S2). The DM2 analysis replicated 6 of 9, and in combination the two analyses replicated 7 of 9. Based on this finding, we combined the results for GDM and DM2 in subsequent studies.
Gene set analysis of candidate drugs
We tested whether the genes targeted by each drug, as a set, were associated with GDM/DM2 (Table 1, Figure 2). The strongest association was for calcium channel blockers (CCBs) with P = 0.00015. This finding remained significant after adjustment for multiple comparisons for the 65 drug classes tested at 0.00975 (0.00015 * 65). We tested the robustness of our result using 2 variant gene sets and one alternative statistical analysis. CCBs remained in the top 5 hits for all three, while 5-HT3 antagonists were lower ranked in the variant analyses (Table S3).
Table 1. Drug classes with target gene sets associated with GDM and DM2.
Of 129 drugs, representing 65 classes, 11 classes had adjusted P < 0.05 and are shown. L-type calcium channel blockers, represented by nifedipine, had the strongest P, which remained significant after adjusted for multiple comparisons. Examples and usual indication are shown. The number of genes is the number of genes targeted by the drug which had at least 1 SNP tested.
| Class | Example | Indication | Genes | P |
|---|---|---|---|---|
| L-type calcium channel blocker | nifedipine | Hypertension | 5 | 1.50E-04 |
| NMDA receptor blockers | memantine | Alzheimer dementia | 2 | 4.91E-04 |
| Alpha blockers | prazosin | Hypertension | 2 | 6.27E-03 |
| Alpha/beta blockers | carvedilol | Hypertension | 4 | 9.62E-03 |
| Beta blockers | labetalol | Hypertension | 4 | 9.62E-03 |
| Sodium channel blocker | mexiletine | Arrhythmia | 1 | 1.16E-3 |
| Phosphodiesterase inhibitors | sildenafil | Pulmonary hypertension | 1 | 1.18E-02 |
| 5HT-3 blockers | ondansetron | Nausea/vomiting | 4 | 1.57E-02 |
| PPARa activator | clofibrate | High cholesterol | 1 | 2.12E-02 |
| D2 blocker | metoclopramide | Nausea/vomiting | 8 | 2.60E-02 |
Glucose Tolerance Tests
We sought to identify what effect, if any, the drugs identified had on glucose tolerance during pregnancy. We grouped the drugs by their mechanistic class and examined the effect in a multivariate analysis that controlled for age at GTT, BMI at GTT, presence of the usual indication, self-identified white race, and self-identified Hispanic ethnicity. All, except white race, were significant in multivariate analysis. Each additional year of age was associated with a 1.00 mg/dL (95% CI 0.86 to 1.13 mg/dL, p < 0.001) increase in GTT, as expected. Each additional point of BMI was associated with an increase of 0.52 mg/dL (95% CI 0.41 to 0.63, p < 0.001). Hispanic ethnicity was associated with an increase of 6.69 mg/dL (95% CI 4.53 to 8.85 mg/dL, p < 0.001) as previously reported27. Demographics are described in Table S4.
We tested the reliability of our analysis by examining the effect of hypoglycemic drugs prescribed for GDM. As expected, biguanides, insulins, and sulfonylureas were all associated with large increases in glucose during GTT (Table 2)
Table 2. Multivariate regression of impact of drugs and their usual indication on GTT.
ΔGTT indicates the change in the value of the GTT attributable to the medication.
| Effect of drug | |||||
|---|---|---|---|---|---|
| Class | Example(s) | Indication | ΔGTT (mg/dL) |
95% CI (mg/dL) | P |
| L-type calcium channel blocker | nifedipine | Hypertension | −3.18 | −6.18 to −0.18 | 0.038 |
| Alpha 2 agonists | clonidine, methyldopa | Hypertension | −2.90 | −7.45 to 1.65 | 0.211 |
| Beta blockers | labetalol | Hypertension | 3.14 | −2.59 to 8.87 | 0.283 |
| Alpha/beta blockers | carvedilol | Hypertension | 0.72 | −8.25 to 9.69 | 0.876 |
| 5HT-3-antagonists | ondansetron | Nausea/vomiting | 3.54 | 1.86 to 5.23 | < 0.001 |
| D2 antagonist | metoclopramide | Nausea/vomiting | 0 | −2.38 to 2.38 | 0.998 |
| 5HT-1a partial agonist | buspirone | Anxiety | −6.48 | −11.89 to −1.07 | 0.019 |
| SNRIs | duloxetine | Depression/Anxiety | 1.24 | −2.75 to 5.23 | 0.543 |
| NDRI | bupropion | Depression | −2.06 | −5.32 to 1.21 | 0.217 |
| SARIs | trazodone | Depression/Anxiety | −1.45 | −5.79 to 2.89 | 0.512 |
| NASSA | mirtazapine | Depression/Anxiety | 6.13 | −1.57 to 13.83 | 0.118 |
| TCAs | amitriptyline | Depression/Anxiety | −0.31 | −3.69 to 3.07 | 0.858 |
| Biguanide | metformin | Diabetes | 5.65 | 2.49 to 8.81 | < 0.0001 |
| Insulins | insulin aspart | Diabetes | 19.62 | 15.93 to 23.31 | < 0.0001 |
| Sulfonylurea | glyburide | Diabetes | 40.78 | 37.76 to 43.81 | < 0.0001 |
Terms: GTT, 1 hour 50-gram glucose tolerance test; SNRI: serotonin-norepinephrine reuptake inhibitor, NDRI: norepinephrine-dopamine reuptake inhibitor, SARI: serotonin antagonist and reuptake inhibitor, NASSA: norepinephrine and selective serotonin agonist, TCA: tricyclic antidepressant.
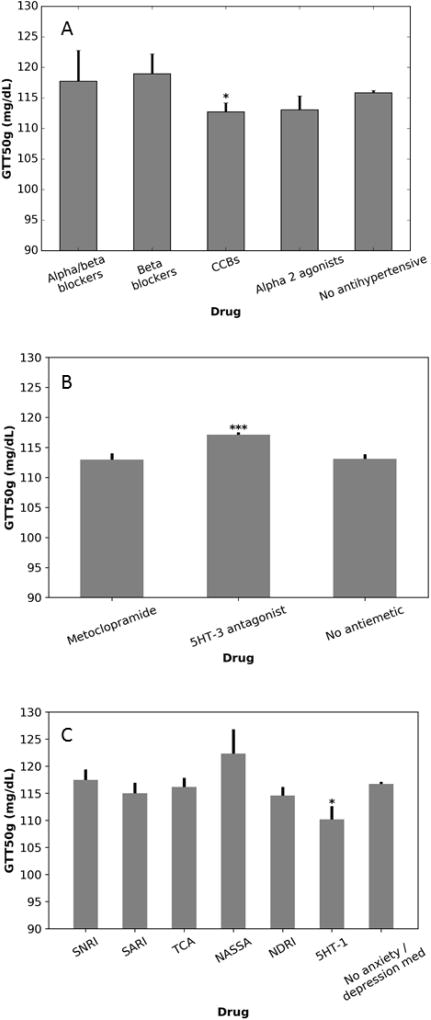
In multivariate analysis, CCB use was associated with a −3.18 mg/dL (95% CI −6.18 to −0.18 mg/dL, p = 0.038) decrease in blood glucose during the GTT, while blood pressure was associated with a 0.12 mg/dL (0.07 to 0.18mg/dL, p< 0.0001) increase per mmHg of mean arterial pressure (Figure 3, Table 2). Other antihypertensives had no effect. 5HT-3 antagonists were associated with a 3.54 mg/dL (95% CI 1.856 to 5.225, p < 0.001) increase in blood glucose during the GTT (Figure 3, Table 2). Hyperemesis gravidarum, the condition treated by 5HT-3 antagonists, was associated with a 6.58 mg/dL (95% CI −9.63 to −3.52 mg/dL, p < 0.001) decrease. D2 antagonist anti-nausea medications had no effect. 5HT-1a agonist anti-anxiety medications were also associated with a decrease in glucose during the GTT (−6.48 mg/dL, CI −11.89 to −1.07). Anxiety was not associated with significant changes in glucose tolerance, nor was depression, or treatment with other anti-anxiety or anti-depressant medications (Figure 3, Table 2).
Figure 3. Effect of various drugs on GTT.
Data are shown for antihypertensives (A), medications for nausea and vomiting (B), and medications for depression and/or anxiety (C). CCBs and 5HT-1a partial agonists are associated with decreased GTT while 5HT-3-antagonists are associated with an increase. Values are adjusted for demographics and indication as described in the methods. Mean +/− SEM is shown. *: P<0.05 versus no antihypertensive / no anxiety/depression medication, ***: P<0.001 versus no drug. P-values are not corrected for multiple comparisons. 5HT-1: 5HT-1a partial agonist, CCB: calcium channel blocker, SNRI: serotonin-norepinephrine reuptake inhibitor, SARI: serotonin antagonist and reuptake inhibitor, TCA: tricyclic antidepressant.
Conclusions
To our knowledge, this is the first study aimed at drug repurposing in GDM. The use of a parallel genetic and EMR-exposure based design increases the reliability of the results. We identified several drugs that could be related to GDM, most prominently an association and worse glucose tolerance with 5HT-3 antagonists and an association and improved glucose tolerance with pharmacologic calcium channel blockade and improved glucose tolerance with a 5HT-1a partial agonist.
Our genetic association study identified a strong link between the genes targeted by the CCBs and GDM/DM2. In our EMR-based study, calcium channel blockers were associated with a decrease in GTT while hypertension was associated with an increase. We did not observe a significant effect on GTT with other antihypertensives, pointing toward a mechanism-specific effect for calcium channel blockers. CCBs, as currently used to treat hypertension in pregnancy, are not associated with an increased risk of birth defects, nor with changes in uteroplacental flow in hypertensive emergency, representing a favorable safety profile28,29. In our sources, nifedipine was the only calcium channel blocker described as safe in pregnancy however, other CCBs may be safe in pregnancy and at least as effective in improving glucose tolerance, an area for future study14. Nifedipine is out of patent and widely available. Therefore, it represents an attractive option for use in low-resource settings.
Several clinical trials have examined the effect of CCBs on measures of glucose tolerance. Use of nifedipine to treat non-pregnant, non-diabetic patients with hypertension was reported to improve glucose tolerance30. In a study of diabetics with hypertension, CCBs were associated with lower fasting glucose31. Other studies have shown that beta blockers and diuretics are associated with worsening glucose tolerance, CCBs are neutral, and medications which interfere with the renin-angiotensin system are associated with improved glucose tolerance32. In a recent network meta-analysis of medications to prevent new-onset diabetes, diuretics were associated with increased risk of new-onset diabetes versus placebo, but beta blockers were not; CCBs were neutral33. In in vitro models of insulin secretion, inhibition of L-type calcium channels by nifedipine decreases glucose stimulated insulin secretion from islets, which would correlate with post-prandial hyperglycemia34. However, treatment of intact islets with nifedipine increases basal insulin secretion and decreases glucagon secretion35. By inhibiting calcium influx during hyperglycemia and hypoxia, nifedipine may also protect against ER stress and beta-cell apoptosis36,37.
SNPs in genes involved in serotonin synthesis (TPH1) and receptors (including 5HT-2A, 5HT-3B) are associated with multiple metabolic phenotypes, including metabolic syndrome38. We showed that HG was associated with decreased GTT while 5HT-3 antagonists, which treat HG are associated with an increase. It seems plausible that patients suffering from nausea and vomiting would consume less and therefore have a lower blood glucose before adding the 50 grams of glucose used in the GTT. Antiemetics, like 5HT-3 antagonists, could be expected to reverse this effect. However, a similar effect was not seen with the mechanistically unrelated D2 antagonists, which may favor a serotonin-specific effect. Serotonin inhibits glucose induced insulin secretion in vitro, and the 5HT-3 antagonist topisetron reverses this effect and potentiates insulin secretion from INS-1 cells39. However, mice with targeted mutations in HTR3A or TPH1 showed impaired glucose tolerance and decreased insulin secretion due to membrane hyperpolarization in islet cells40,41. The significance of these competing findings is unclear, but underlines the importance of serotonin and the 5HT-3 receptor in islet physiology.
We identified 9 other classes in the SNP arm. Of these, 4 could be tested for GTT and had no significant effect. We also identified one class, 5HT-1a partial agonists, which altered the GTT but were not identified in the SNP arm. The effect of 5HT-1a partial agonists on glucose homeostasis has not been well studied. In rats, 5HT-1a partial agonism prevented the hyperglycemic, renal, and cardiac manifestations of the streptozotocin -induced diabetes model42. Conversely, in a study of schizophrenic patients, adding on 5HT-1a partial agonist therapy to atypical antipsychotics was associated with an increased risk of glucose intolerance43. The relevance of these findings to the present study is unclear.
Limitations
This study is hypothesis generating and subject to several limitations. We chose the drugs based on their reputation for safety, which has often not been rigorously tested. In the genetic analysis, the small size of the GDM cohort reduced our power to identify associations. Admixing the association data for DM2 provided a much larger cohort, but reduced the specificity for our results. Our patient assignments were based upon ICD9 coding, which is unreliable. The lack of a precise ICD9 code for GDM exacerbates this issue. Additionally, we cannot guarantee that all patients were ascertained for GDM.
The GTT study complemented the SNP study, however it was also limited. There were many drug classes with insufficient patients. Medications may be taken at a random time not related to pregnancy, GTT, or GDM. This increases the probability of a type II error, and therefore we do not consider any non-associations between a drug and GTT value as meaningful. Conversely, associations that persist despite chronological uncertainty deserve further study. It is also difficult to separate the indication for medications from their effects. We approached this conundrum by examining the differential effects of medications with similar indications. For example, 5HT-3 antagonists were associated with a +3.54 mg/dL change (95% CI +1.86 to +5.23, p < 0.001) in GTT, while the D2 antagonist was not associated with any significant change. We conclude that the variable effects of medications used to treat the same indication might be attributable to differing mechanisms of action. We arrive at a similar conclusion in regards to anti-hypertensives and CCBs versus beta blockers and alpha-2 agonists. CCBs may also be used to treat premature labor. GDM is a risk factor for preterm labor, so this could cause a spurious association, however the chain of causation from GDM to preterm labor and then CCB treatment would result in CCBs being associated with an increased GTT, rather than a decrease. We did not include other complications of pregnancy (e.g. pre-eclampsia) or parity. Finally, an abnormal screening GTT does not define GDM. We chose to focus on the value of the GTT because a numerical value is more information-rich than a binary disease/no-disease. Because of the roughly random assortment of genotypes the genetic association study does not suffer from this problem.
Conclusion
GDM continues to be a major problem for pregnant women and their offspring. Current therapies improve hyperglycemia and prevent some complications, however more interventions are needed. This paper sought to identify candidate drugs for repurposing to treat GDM by starting with the subset of drugs considered safe in pregnancy. Genes targeted by 11 drug classes were significantly associated with GDM/DM2 in our SNP study, while 6 drug classes were associated with changes to the GTT in our EMR study. Notably, genes targeted by CCBs were significantly associated with the diagnosis of GDM/DM2 and exposure to CCBs improved GTT values, while 5HT-3 antagonists were associated with worse glucose tolerance. Prior research shows calcium channel blockers are associated with improved glucose control in non-pregnant populations. These findings support CCBs as a candidate for drug repurposing in GDM
Supplementary Material
Acknowledgments
The project was supported by NIH R01 LM 010685, and P50 GM115305. The dataset used in the analyses described were obtained from BioVU, which is supported by institutional funding and by the Vanderbilt CTSA grant ULTR000445 from NCATS/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
Study conception: JAG, DMR, JMP, DMA; Data acquisition: JCD, DMR, JMP; Data analysis: JAG, LAB; Data interpretation: JAG, LAB, JCD, DMR, JMP, DMA; Writing: JAG, LAB, JCD, DMR, JMP, DMA. Manuscript final approval: JAG, LAB, JCD, DMR, JMP, DMA; Accountability for completeness and accuracy: JAG, LAB, JCD, DMR, JMP, DMA.
Conflict of Interest
The authors state they have no conflicts of interest.
Conflicts of interest: none
References
- 1.Macaulay S, Dunger DB, Norris SA. Gestational diabetes mellitus in Africa: a systematic review. PloS One. 2014;9(6):e97871. doi: 10.1371/journal.pone.0097871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y, Zhang C. Prevalence of Gestational Diabetes and Risk of Progression to Type 2 Diabetes: a Global Perspective. Curr Diab Rep. 2016;16(1):7. doi: 10.1007/s11892-015-0699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Collaborating Centre for Women’s and Children’s Health (UK) Diabetes in Pregnancy: Management of Diabetes and Its Complications from Preconception to the Postnatal Period. London: RCOG Press; 2008. [Accessed May 14, 2016]. http://www.ncbi.nlm.nih.gov/books/NBK51920/ [PubMed] [Google Scholar]
- 4.Koning SH, Hoogenberg K, Lutgers HL, VAN DEN Berg PP, Wolffenbuttel BHR. GESTATIONAL DIABETES MELLITUS: current knowledge and unmet needs. J Diabetes. 2016 Apr; doi: 10.1111/1753-0407.12422. [DOI] [PubMed] [Google Scholar]
- 5.HAPO Study Cooperative Research Group. Metzger BE, Lowe LP, et al. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358(19):1991–2002. doi: 10.1056/NEJMoa0707943. [DOI] [PubMed] [Google Scholar]
- 6.Sweeting AN, Ross GP, Hyett J, et al. Gestational Diabetes Mellitus in Early Pregnancy: Evidence for Poor Pregnancy Outcomes Despite Treatment. Diabetes Care. 2016;39(1):75–81. doi: 10.2337/dc15-0433. [DOI] [PubMed] [Google Scholar]
- 7.Reece EA. The fetal and maternal consequences of gestational diabetes mellitus. J Matern-Fetal Neonatal Med Off J Eur Assoc Perinat Med Fed Asia Ocean Perinat Soc Int Soc Perinat Obstet. 2010;23(3):199–203. doi: 10.3109/14767050903550659. [DOI] [PubMed] [Google Scholar]
- 8.Landon MB, Spong CY, Thom E, et al. A multicenter, randomized trial of treatment for mild gestational diabetes. N Engl J Med. 2009;361(14):1339–1348. doi: 10.1056/NEJMoa0902430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landon MB, Rice MM, Varner MW, et al. Mild gestational diabetes mellitus and long-term child health. Diabetes Care. 2015;38(3):445–452. doi: 10.2337/dc14-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratner RE, Christophi CA, Metzger BE, et al. Prevention of diabetes in women with a history of gestational diabetes: effects of metformin and lifestyle interventions. J Clin Endocrinol Metab. 2008;93(12):4774–4779. doi: 10.1210/jc.2008-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton S, Kirkwood S, Thangaratinam S. Interventions to modify the progression to type 2 diabetes mellitus in women with gestational diabetes: a systematic review of literature. Curr Opin Obstet Gynecol. 2014;26(6):476–486. doi: 10.1097/GCO.0000000000000127. [DOI] [PubMed] [Google Scholar]
- 12.Malek A, Mattison DR. Drug development for use during pregnancy: impact of the placenta. Expert Rev Obstet Gynecol. 2010;5(4):437–454. doi: 10.1586/eog.10.29. [DOI] [Google Scholar]
- 13.Ratcliffe SD, editor. Family Medicine Obstetrics. 3. Philadephia: Mosby Elsevier; 2008. Drugs for common conditions in pregnancy; pp. 676–684. [Google Scholar]
- 14.Hamilton RJ. Tarascon Pocket Pharmacopoeia. Burlington, Mass.: Jones & Bartlett Learning; 2012. [Accessed May 14, 2016]. http://site.ebrary.com/id/10546129. [Google Scholar]
- 15. [Accessed May 14, 2016];Prescribing medicines in pregnancy database | Therapeutic Goods Administration (TGA) https://www.tga.gov.au/prescribing-medicines-pregnancy-database.
- 16. [Accessed March 29, 2017];Research C for DE and. Labeling - Pregnancy and Lactation Labeling (Drugs) Final Rule. https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/Labeling/ucm093307.htm.
- 17.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma Oxf Engl. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3(1):42–48. doi: 10.1111/j.1752-8062.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6(234):234cm3. doi: 10.1126/scitranslmed.3008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinforma Oxf Engl. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet TIG. 2014;30(9):390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mooney MA, Wilmot B. Gene set analysis: A step-by-step guide. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2015;168(7):517–527. doi: 10.1002/ajmg.b.32328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieffer EC. Maternal obesity and glucose intolerance during pregnancy among Mexican-Americans. Paediatr Perinat Epidemiol. 2000;14(1):14–19. doi: 10.1046/j.1365-3016.2000.00228.x. [DOI] [PubMed] [Google Scholar]
- 28.Papatsonis DN, Lok CA, Bos JM, Geijn HP, Dekker GA. Calcium channel blockers in the management of preterm labor and hypertension in pregnancy. Eur J Obstet Gynecol Reprod Biol. 2001;97(2):122–140. doi: 10.1016/s0301-2115(00)00548-0. [DOI] [PubMed] [Google Scholar]
- 29.Cornette J, Buijs EaB, Duvekot JJ, et al. Hemodynamic effects of intravenous nicardipine in severely pre-eclamptic women with a hypertensive crisis. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2016;47(1):89–95. doi: 10.1002/uog.14836. [DOI] [PubMed] [Google Scholar]
- 30.Koyama Y, Kodama K, Suzuki M, Harano Y. Improvement of insulin sensitivity by a long-acting nifedipine preparation (nifedipine-CR) in patients with essential hypertension. Am J Hypertens. 2002;15(11):927–931. doi: 10.1016/s0895-7061(02)03019-4. [DOI] [PubMed] [Google Scholar]
- 31.Khodneva Y, Shalev A, Frank SJ, Carson AP, Safford MM. Calcium channel blocker use is associated with lower fasting serum glucose among adults with diabetes from the REGARDS study. Diabetes Res Clin Pract. 2016;115:115–121. doi: 10.1016/j.diabres.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancia G, Grassi G, Zanchetti A. New-onset diabetes and antihypertensive drugs. J Hypertens. 2006;24(1):3–10. doi: 10.1097/01.hjh.0000194119.42722.21. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Xu H. Comparing six antihypertensive medication classes for preventing new-onset diabetes mellitus among hypertensive patients: a network meta-analysis. J Cell Mol Med. 2017;21(9):1742–1750. doi: 10.1111/jcmm.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran K, Peng X, Bokvist K, Stehno-Bittel L. Assessment of re-aggregated human pancreatic islets for secondary drug screening. Br J Pharmacol. 2014;171(12):3010–3022. doi: 10.1111/bph.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Marchand SJ, Piston DW. Glucose decouples intracellular Ca2+ activity from glucagon secretion in mouse pancreatic islet alpha-cells. PloS One. 2012;7(10):e47084. doi: 10.1371/journal.pone.0047084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Gao L, Li Y, Chen H, Sun Z. Nifedipine protects INS-1 β-cell from high glucose-induced ER stress and apoptosis. Int J Mol Sci. 2011;12(11):7569–7580. doi: 10.3390/ijms12117569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Z, Moruzzi N, Catrina S-B, et al. Preconditioning with associated blocking of Ca2+ inflow alleviates hypoxia-induced damage to pancreatic β-cells. PloS One. 2013;8(7):e67498. doi: 10.1371/journal.pone.0067498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh CM, Park S, Kim H. Serotonin as a New Therapeutic Target for Diabetes Mellitus and Obesity. Diabetes Metab J. 2016;40(2):89–98. doi: 10.4093/dmj.2016.40.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heimes K, Feistel B, Verspohl EJ. Impact of the 5-HT3 receptor channel system for insulin secretion and interaction of ginger extracts. Eur J Pharmacol. 2009;624(1–3):58–65. doi: 10.1016/j.ejphar.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 40.Ohara-Imaizumi M, Kim H, Yoshida M, et al. Serotonin regulates glucose-stimulated insulin secretion from pancreatic β cells during pregnancy. Proc Natl Acad Sci U S A. 2013;110(48):19420–19425. doi: 10.1073/pnas.1310953110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K, Oh C-M, Ohara-Imaizumi M, et al. Functional role of serotonin in insulin secretion in a dietinduced insulin-resistant state. Endocrinology. 2015;156(2):444–452. doi: 10.1210/en.2014-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghunathan S, Tank P, Bhadada S, Patel B. Evaluation of buspirone on streptozotocin induced type 1 diabetes and its associated complications. BioMed Res Int. 2014;2014:948427. doi: 10.1155/2014/948427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hedenmalm K, Hägg S, Ståhl M, Mortimer O, Spigset O. Glucose intolerance with atypical antipsychotics. Drug Saf. 2002;25(15):1107–1116. doi: 10.2165/00002018-200225150-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.