Abstract
Inherent electrical properties of cells can be beneficial to characterize different cell lines and their response to experimental drugs. This paper presents a novel method to characterize the response of breast cancer cells to drug stimuli through use of off-chip passivated-electrode insulator-based dielectrophoresis (OπDEP) and the application of AC electric fields. This work is the first to demonstrate the ability of OπDEP to differentiate between two closely related breast cancer cell lines, LCC1 and LCC9 while assessing their drug sensitivity to an experimental anti-cancer agent, Obatoclax. Although both cell lines are derivatives of estrogen-responsive MCF-7 breast cancer cells, growth of LCC1 is estrogen independent and anti-estrogen responsive, while LCC9 is both estrogen-independent and anti-estrogen resistant. Under the same operating conditions, LCC1 and LCC9 had different DEP profiles. LCC1 cells had a trapping onset (crossover) frequency of 700 kHz and trapping efficiencies between 30–40%, while LCC9 cells had a lower crossover frequency (100 kHz) and showed higher trapping efficiencies of 40–60%. When exposed to the Obatoclax, both cell lines exhibited dose-dependent shifts in DEP crossover frequency and trapping efficiency. Here, DEP results supplemented with cell morphology and proliferation assays help us to understand the response of these breast cancer cells to Obatoclax.
Keywords: Breast cancer, Dielectrophoresis (DEP), Drug sensitivity, Estrogen receptor positive (ER+), GX15-070 (Obatoclax)
1 Introduction
At diagnosis, 70% of breast tumors express estrogen receptors (ER+) and the standard of care for these patients is endocrine therapy with antiestrogens or aromatase inhibitors [1]. These therapies are designed to deprive the tumor of growth promoting actions of estrogens and while they do have initial widespread clinical efficacy, 50% of women receiving endocrine therapy will show either de novo resistance or acquired resistance [2]. Therefore, endocrine resistance remains a major challenge in the clinic. In the laboratory, endocrine resistance has been studied at the molecular level using established breast cancer cell lines such as the ER+ MCF-7 derivatives, LCC1 and LCC9 [3]. MCF-7 cells have been used to model patients whose tumors are growth stimulated by estrogens and growth inhibited by anti-estrogen drugs [4]. LCC1 cells were selected from MCF-7 cells during in vivo propagation in mouse xenografts; although growth of LCC1 cells is estrogen independent, LCC1 grow is inhibited by antiestrogens. LCC9 cells are a derivative of LCC1; LCC9 cells were selected in vitro from LCC1 cells and are estrogen-growth independent and anti-estrogen resistant [3]. Thus, LCC1 and LCC9 cells represent forms of ER+ breast cancer that require new drug treatments are excellent models to screen new anti-cancer agents.
To provide patients with more targeted cancer treatments, cell lines must be further analyzed to characterize ER+ cells and their response to experimental anti-cancer agents. Conventional cell manipulation methods such fluorescent tagging with Annexin V [5], MTT assays, and the trypan blue assays can be used to study drug-induced growth inhibition and cell death. Such assays showed that LCC1 and LCC9 cell density decreased in response to GX, an anti-tumor BH3-mimetic drug (GX15-070) [6]. However, proliferation assays can be laborious, time consuming and fail to provide insight into drug-induced biophysical changes. Methods such as electrorotation have been utilized to characterize cells based on their dielectric properties [7]. Huang et al. demonstrated the ability of electrorotation to detect cell membrane responses to drug-induced apoptosis in HL-60 cells [8]. This technique was able to provide real-time membrane property measurements for single cells, however, due to its low throughput, it is very difficult to analyze the changes in cell populations.
One promising alternative for analyzing the drug response of cancer cells is dielectrophoresis (DEP). DEP can provide insight into changes occurring in cell biophysical attributes, including size, membrane capacitance, and cytoplasmic conductivity. DEP is an electrokinetic-based technique in which uncharged particles suspended in a fluid can be moved via polarization forces induced by an inhomogeneous electric field [9]. It is a highly versatile and customizable method which does not require biological markers [10] and can be used to manipulate bioparticles based on their dielectric properties and discriminate based on the cell’s physiological state [11]. Moreover, DEP based characterization can be further integrated into microfluidic platforms to provide separation of live and dead cells [5], to separate circulating tumors cells from blood samples [12], or provide real time analysis of drug induced cytotoxicity in cells [13]. Typical AC electric fields used in DEP have not shown adverse effects on cells when applied within certain frequency and voltage ranges [14]. Therefore, integrating microfluidics into DEP based characterization can provide Lab-On-Chip systems, which provide high throughput with real time analysis of live cells.
Various DEP based methods have been used to study the effect of drugs on cells. In a study by Wang et al., the effectiveness of DEP characterization as a rapid method superior to DNA fragmentation for the detection of drug-induced apoptosis in human myelogenous HL-60 cells was demonstrated [15]. In addition, DEP has been utilized to complement flow cytometry to understand the underlying mechanism in multi-drug resistance reversal in human leukemic cells [16] and human breast cancer cells [17]. Henslee et al. used the 3DEP™ system to show DEP results were comparable to standard assays such as flow cytometry, trypan blue, and MTT assay whilst being faster and label-free [18]. The 3DEP™ system was also used to complement flow cytometry assessments of drug-induced apoptosis progression in Jurkat T-cells [19]. Similar studies have been carried out using DEP-based techniques to study the drug-induced apoptosis in lung cancer cells [20]. These studies illustrate the value of DEP in drug response analysis using electrodes in direct contact with cell samples. Such methods pose limitations such as sample contamination, electrolysis, and delamination of electrodes [21].
In contrast, the off-chip passivated-electrode insulator-based dielectrophoresis (OπDEP) uses a unique DEP design in which AC electric fields are capacitively coupled into the microchannel to alleviate some of the commonly known limitations in electrode-based DEP. This technique generates necessary electric field gradients through micro-pillars embedded within the microfluidic channel while utilizing reusable electrodes positioned on a separate detachable substrate for AC electric field generation. This platform has been previously used to isolate live and dead bacteria [22]. However, this is the first demonstration of the applicability of this technique for breast cancer cell drug analysis and the first use of insulator-based DEP for analyzing closely related drug sensitive and drug resistant breast cancer cells.
In this study, we employ the OπDEP platform to characterize two ER+ breast cancer cell lines, LCC1 and LCC9, and their DEP response to 100 nM, 500 nM, and 1 µM treatments of the anti-tumor drug GX. Scanning electron microscope (SEM) imaging was performed to compare the DEP results with cell morphological changes. Cell proliferation assays were performed to compare the DEP findings with a conventional drug assessment method.
2 Materials and methods
2.1 Device fabrication
Fabrication of OπDEP devices used in this study followed processes previously presented [22]. Electrodes were fabricated on a glass substrate through photolithography and deposition of chrome/gold by physical vapor deposition. A lift-off process was then employed to create a pair of electrodes with dimensions of 1000 and 600 µm in width and horizontal spacing, respectively, on a 500 µm thick glass substrate. After, a silicon wafer is patterned with photoresist and dry etched to create a negative silicon master.
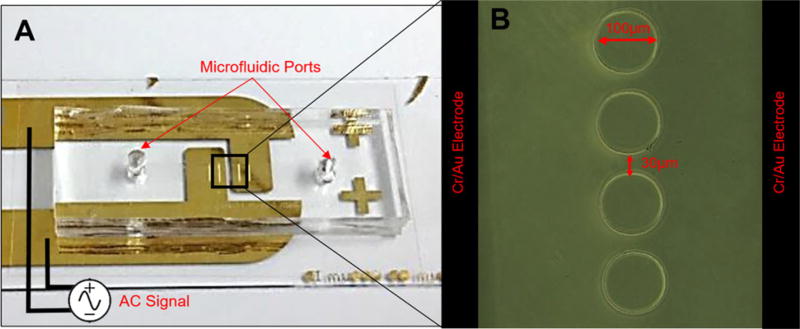
Polydimethlsiloxanne (PDMS) (Dow Corning, MI, USA) microfluidic channels with an array of insulator pillars were fabricated using the negative silicon wafer as the mold. Inlets and outlets were punched on each chip and then bonded to a 100 µm thick #0 cover glass slide (Electron Microscopy Sciences, PA, USA) which separates the microfluidic chip from the electrodes. The microfluidic component was placed directly above and between the electrodes with minimal alignment and attached using tape. Figure 1 shows an optical image of the OπDEP chip with a close-up of the insulating pillars which are 100 µm in diameter with edge-to-edge 30 µm spacing with a depth size 40 µm for the entire microfluidic channel. Overall size of the entire chip is 3 cm × 2.5 cm.
Figure 1.
(A) Top view of the fabricated microfluidic chip and electrodes. The image shows the alignments of the microfluidic device with the DEP pillars positions directly above and in-between the reusable (detachable) electrodes spaced 400 µm. (B) Close-up of the DEP pillars within the microfluidic chip and electrodes that lie below the microfluidic chip and surround the DEP pillars.
2.2 Cell culture and treatments
LCC1 and LCC9 cells lines were obtained from Tissue Culture Shared Resources of the Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC. As recommended by the supplier, cells were maintained in T-25 flasks in Minimum Essential Medium (MEM Richter’s modification) (Life Technologies, NY, USA) containing 5% Donor Calf Serum Charcoal Stripped (Valley Biomedicals, VA, USA) and antibiotics, 100 U/mL penicillin and 100 µg/mL streptomycin (Mediatech, VA, USA), at 37°C in a humidified 5% CO2 incubator.
LCC1 and LCC9 cells were treated with GX (Selleckchem, TX, USA) for 12 h. Stock solutions of GX were prepared in dimethyl sulfoxide (DMSO) (Life Technologies, NY, USA), stored at −80°C, and diluted in culture media to the desired concentrations. A vehicle control treatment using 0.1% DMSO was also performed on both cell lines.
Immediately prior to DEP analysis, cells were harvested and fluorescently tagged with 5 µM 5-chloromethyl fluorescein diacetate (Cell Tracker Green CMFDA Dye) (Life Technologies, NY, USA) in a low conductivity DEP buffer prepared by adding 8.5 g sucrose (Sigma Aldrich, MO, USA) and 0.725 mL MEM (Life Technologies, NY, USA) in 100 mL deionized water [23]. The conductivity of the DEP buffer was found to be 113 ± 0.54 µS/cm as measured using Conductivity Meter SG7 (Mettler Toledo, Scherzenbach, Switzerland). Cells were washed two times with DEP buffer to remove residual drug and CMFDA and avoid alterations in the solution conductivity. The final cell concentration in the samples was adjusted to 2 × 106 cells/mL for all experiments.
2.3 Experimental setup
For experimentation, a 1 mL syringe containing the fluorescently tagged cell solution was connected to the chip and the solution was driven through the channel at a constant flow rate of 10, 20, or 50 µL/hr based on the experiment of interest, using a Pump11 Elite syringe pump (Harvard Apparatus, MA, USA). Experiments were conducted at frequencies 50 kHz and100 kHz–1 MHz at 100 kHz intervals. Voltages were applied over this frequency range to the electrodes by using a 50 MHz function generator (4079 50 MHz Dual-Channel Arbitrary Waveform/Function Generator, B&K Precision, CA, USA) and a voltage amplifier (A800DI Voltage Amplifier, FLC Electronics AB, Sweden) to initiate DEP forces induced by the application of the AC electric field causing trapping around the pillars. Trapping of the cells was observed using a Zeiss Axio Observer.Z1 inverted epifluorescence microscope (Germany) with a 10× objective lens and imaged using a Zeiss AxioCam MRc camera (Germany).
Trials were observed for 25 s to account for 20 s DEP excitation, cell release and to allow cells to return to a continuous flow (see supporting information 1 and 2 for videos of trapping). Trapping efficiencies were calculated as the ratio of trapped cells to the total number of cells in each run. Crossover frequencies were determined by experimentally sweeping the voltage over the 50 kHz–1 MHz range to determine the frequency at which the cell switch from being repelled by the highest gradient areas in the electric field (negative DEP) to being attracted to the highest gradient areas in the electric field (positive DEP). Only values for positive DEP are reported. Trials were operated using 20 µL/h flow rate and 80 Vpeak-to-peak applied voltage. These two values were the optimal values for this study, see Supporting Information 3 for further information.
2.4 Scanning electron microscope sample preparation
Cells were grown with and without drug treatment on individual glass slides and fixed using a 4% formaldehyde solution in phosphate-buffered saline (PBS, Baker, PA, USA). Samples were post-fixed in 1% osmium tetraoxide, serially dehydrated in a graded ethanol series (15, 30, 50, 70, 95, 100%), and critical point dried. After sputter coating the samples with gold, SEM images were acquired using a Carl Zeiss EVO 40 SEM (Germany).
2.5 Cell size measurement
Cell size in suspension was measured using images captured by the epifluorescence microscope and analyzed using the image processing software, Zen Pro Blue (Germany). Fifty cell sizes were measured for LCC1 and LCC9 vehicle and GX-treated trials. Final cell size was reported as the mean ± standard deviation, as shown in Table 1.
Table 1.
Measured axial length cell size for vehicle and GX-treated LCC1 and LCC9 cells
| Treatment | LCC1 (µm) | LCC9 (µm) |
|---|---|---|
| Vehicle | 13.0 ± 2.0 | 15.0 ± 1.0 |
| GX 100 nM 12 h | 13.5 ± 1.0 | 14.5 ± 1.5 |
| GX 500 nM 12 h | 14.5 ± 1.5 | 11.0 ± 1.5 |
| GX 1 µM 12 h | 15.5 ± 1.5 | 10.0 ± 1.0 |
2.6 Cell proliferation assay
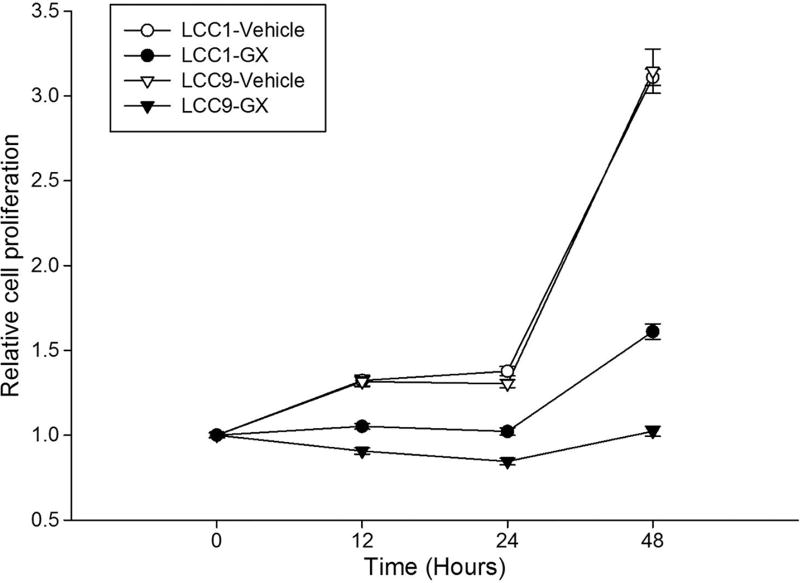
Cell proliferation assays were performed to assess the time course of response to GX. Cell proliferation was determined using the crystal violet assay. Cells were seeded at a proliferation of 5 × 103 per well in 96-well plates. Twenty-four hours later, the indicated concentration of GX or vehicle control was added for 12, 24 or 48 h treatments. Following treatment, cells were stained with a crystal violet staining solution as previously described [6]. Sodium citrate buffer was used to extract the dye, and an index of cell proliferation was obtained using the absorbance at 550 nm using a microplate reader (Bio-Rad, CA, USA). Results at 12, 24 and 48 h are shown in Fig. 2. LCC9 cells are significantly more sensitive to 500 nM GX than LCC1 cells, confirming an earlier report by Schwartz et al [6].
Figure 2.
Proliferation assay results for untreated and 500 nM GX-treated LCC1 and LLC9 cells under control conditions (DMSO vehicle alone) at 12, 24, and 48 h.
2.7 Statistical significance analysis
For multiple group comparisons, Student–Newman–Keuls method comparison test was applied following ANOVA. Results were considered to be significantly different at p < 0.05. Statistical analysis was carried out using the Sigma Plot software (Jandel Scientific, SPSS, Chicago, IL, USA).
3 Results
3.1 DEP characterization of breast cancer drug response
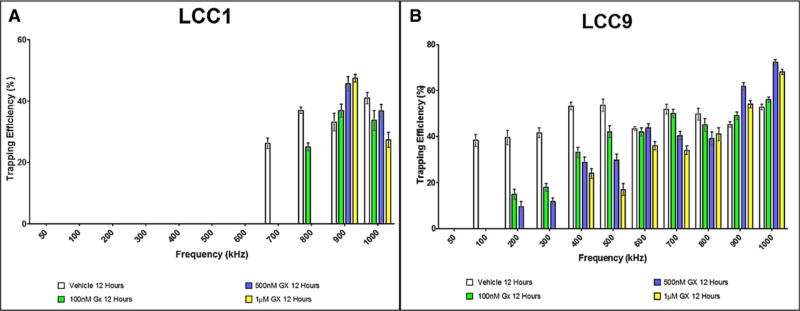
While discerning among different cell types is an established application of DEP-based technologies, drug response characterization is another area which can benefit from DEP characterization methods and has not been explored to a great extent. Therefore, a drug sensitivity study of LCC1 and LCC9 cells to the drug GX was performed using the OπDEP platform. Figure 3 shows the response of LCC1 and LCC9 cells to GX treatment at 100 nM, 500 nM and 1 µM compared to their vehicle counterparts.
Figure 3.
Trapping profile of vehicle and GX-treated at 100 nM, 500 nM, and 1 µM (A) LCC1 and (B) LCC9 cells, each bar represents mean ± standard error of mean for at least three experiments (n> = 3).
Vehicle control and GX-treated cells in both cases have notably different DEP profiles. In both cell lines, the crossover frequency shifted to a higher value in response to 1 µM GX exposure with LCC9 cells showing a larger shift in the crossover compared to LCC1 cells. As seen in Fig. 3, the LCC1 vehicle-treated cells had an initial crossover frequency of 700 kHz. After 100 nM GX treatment, the LCC1 cells shifted their crossover frequency to 800 kHz. When exposed to the 500 nM and 1 µM GX treatments, the cells reached a maximum in crossover frequency of 900 kHz.
Similarly, the LCC9 (vehicle) had a crossover of 100 kHz which shifted to a maximum of 400 kHz when exposed to GX. When exposed to 100 nM or 500 nM GX, the crossover frequency shifted to 200 kHz. However, when exposed to the 1 µM solution, the crossover frequency shifted to 400 kHz.
In addition to changes in the crossover frequency, GX-treatment resulted in alterations in the trapping efficiencies. For vehicle-treated LCC1 cells trapping efficiency at 900 kHz was 33% ± 2.8%. GX treatments of 100 nM, 500 nM and 1 µM GX caused graded increases in the trapping efficiency at 900 kHz to 38% ± 2.8%, 46% ± 2.3%, and 56% ± 1.2%, respectively. At the 1MHz frequency, the LCC1 vehicle-treated cells had a somewhat higher trapping efficiency (41% ± 1.8%) than the LCC1 GX-treated cells (36–27%).
In contrast, vehicle-treated LCC9 vehicle cells generally exhibited greater trapping efficiencies than GX-treated cells. The trend for 100–800 kHz was that with each concentration of GX, the trapping efficiency of the LCC9 cells decreased relative to the vehicle. However, at 900 kHz and 1 MHz the trapping efficiencies increased when exposed to GX. At 1 MHz the 100 nM, 500 nM, and 1 µM GX-treated cells had higher trapping efficiencies of 56% ± 1.1%, 72% ± 1.1%, and 68% ± 2.3%, respectively, versus 51% ± 1.2% for the vehicle.
These changes can be indicative of a change in the cell size and/or changes in the properties of the cell which relate to the electrical properties of the cell. Further experiments were carried out to understand the immediate effects of this drug on each cell line.
3.2 Morphology of vehicle and GX-treated LCC cells
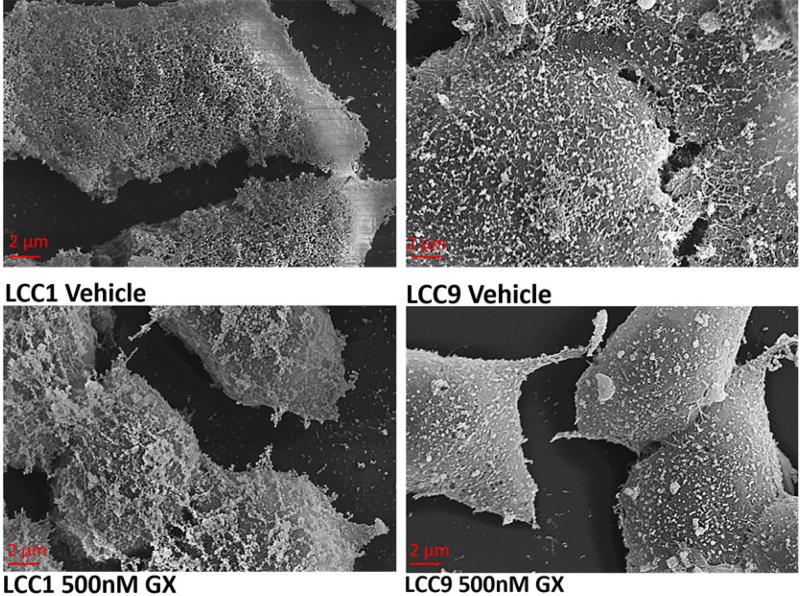
The cell morphology and surface properties are visible aspects of the cell which play a role in the DEP response. Hence, SEM images of vehicle and GX-treated LCC1 and LCC9 cells were acquired (Fig. 4). These images reveal changes in the cell membrane. The cell surface of LCC1 vehicle-treated cells appears generally smooth with many pores. However, the cells became ruffled with many protrusions after treatment with GX. In contrast, vehicle-treated LCC9 cells have a rough surface appearance with many surface protrusions. However, after treatment with GX, the LCC9 surfaces appear smoother with fewer surface protrusions.
Figure 4.
SEM images of vehicle control and 500 nM GX-treated LCC1 and LCC9 cells with labeling below each image.
Cell size is another factor affecting the DEP force. The size of vehicle and GX-treated LCC1 and LCC9 cells were measured (Table 1). LCC9 GX-treated cells showed a significant decrease in cell size while LCC1 GX-treated cells were slightly larger than the control cells. Cell shrinkage also termed apoptotic volume decrease (AVD) is a hallmark of apoptosis and might well account for the reduced cell size observed here in response to GX [24].
4 Discussion
The DEP force acting upon a particle is a factor of various parameters as shown by Pohl [9]. The DEP profile (trapping efficiency in this work) can provide valuable information regarding the electrical properties of the cell. One such information can be extracted from the low crossover frequency which has been observed in the results presented in this work. Based on previously developed models [25], the low crossover frequency can be expressed as
| (1) |
where r is the particle radius, Cm is the membrane capacitance, and σs is the conductivity of the solution the cells are suspended in. Based on this equation, at constant solution conductivity, the crossover frequency has an inverse relationship with both the cell radius and membrane capacitance.
Based on the DEP profile, studies have demonstrated that DEP can be a useful tool for drug assessment for cancer cells, while also providing insight into the cell physiology [17]. Wang et al. cited membrane changes determined by DEP profiles could be used as an indication of apoptosis in HL-60 cells. The study suggests that these membrane variations are caused by changes in the membrane area, which is associated with cell size or surface ruffles, folds or microvilli alterations [15]. Variations in these parameters modifies fx1, as described by Eq. (1), such that larger membrane capacitances and particle radii result in decreased values. Other studies investigated multidrug resistance in breast cancer cells using DEP and a single-shell model to extract membrane and cytoplasm properties. Their findings suggested cytoplasmic features are more critical than membrane properties in determining drug sensitivity. These investigators found no significant change in the electrical nature of the membrane between the drug sensitive and multi-drug resistant cell lines tested [16, 17].
Based on the frequencies used for these DEP studies, these variations in cellular DEP profiles can be attributed to differing physical characteristics, such as cell size, membrane capacitance, morphology, and membrane passive conductivity [26]. As shown by Gascoyn et al., the membrane capacitance is directly related to the degree of membrane folding which is dictated by the membrane morphology [11]. In their work, a membrane morphological score (M) is defined as
| (2) |
where R, P, and F are roughness, projections, and flattening compared to a smooth sphere, respectively. Based on this definition, we can predict that membrane capacitance would be greater in the vehicle-treated LCC9 than LCC1 cells; additionally, the vehicle-treated LCC9 cells may be slightly larger than the LCC1 cells which again would predict a higher membrane capacitance. High number of protrusions and larger membrane capacitances translate to lower crossover frequencies, as seen by the DEP profiles.
The LCC9 are strongly growth inhibited by GX while LCC1 are relatively GX insensitive to GX. The DEP profiles displayed by LCC9 and LCC1 should reflect these distinct drug sensitivities. As mentioned previously the fx1 is inversely proportional to cell size and membrane capacitance, meaning an increase in either parameter results in a decrease in crossover frequency. The trapping efficiency has the opposite relationship where in increase in cell size or capacitance results in higher trapping values. In response to GX, LCC9 cells showed a noticeable decrease in roughness, slight increase in projection, no change in cell flattening after GX treatment, and a concentration-dependent decrease in cell size. These physical observations would be expected to increase crossover frequency and decrease trapping efficiency. In LCC1 cells, however, we see noticeable increase in cell surface roughness and slight increase in projections. However, the cells become significantly rounded compared to vehicle condition where they show a flat surface mostly. These characteristic changes for both cell lines translates to their shifts in the DEP profiles.
Assuming constant membrane permittivity and thickness, findings regarding LCC9 are consistent with the concepts that the smoother membrane and smaller cell size contributes to a higher crossover frequency. Results show that when exposed to GX, LCC9 cells decreased proportionally in size as the GX concentration was increased. In addition, the cells decreased in the number of surface protrusions when exposed to the 500 nM GX treatment. The LCC9 cell’s crossover frequency shifted from 100 kHz to a higher frequency of 400 kHz.
In contrast, GX-treated LCC1 cells, showed a higher proliferation of surface protrusions, while also shifting their crossover frequency from 700 kHz to a higher value of 900 kHz. The cell size increase was negligible in response to GX which is consistent with lack of apoptosis in this line (Table 1). This indicates that factors other than cell size or surface roughness contribute to the DEP profile. Because the crossover frequency for LCC1 is relatively high, changes in the cytoplasm as demonstrated by other studies [18] or membrane passive conductance [26] might be responsible.
Beyond crossover frequency, we observed that for GX-treated LCC9 cells, the overall trapping efficiency at lower frequencies (200–500 kHz) decreased compared to the vehicle-treated cells. In the more GX-sensitive LCC9 line, that the trapping efficiency over this range decreased with increasing concentrations of GX. This change is consistent with membrane morphology and cell size as LCC9 GX-treated cells have fewer protrusions and are smaller in size than the vehicle cells (Table 1). However, at frequencies beyond 500 kHz, the trapping efficiencies became more similar to the vehicle cells, suggesting minimal or no changes in internal dielectric properties.
The LCC1 vehicle and GX-treated cells also have relatively similar trapping efficiencies for high frequencies. This may imply that the cell size and surface morphology is no longer playing as large of a role in the DEP profile as the frequencies are becoming larger. Again, this suggests that the internal cell components such as the cytoplasm are predominant influences on the DEP profile in this cell line. Supporting this are the findings that GX elicits cytoplasmic autophagosomes in LCC1 cells and that compared with LCC9 cells, autophagy plays a more prominent role than apoptosis in the toxicity of GX in this cell line [6].
We have shown the OπDEP platform was able to detect changes in the DEP profile after 12 h GX treatment, while a traditional drug assessment method such as proliferation assays require seeding and at least 24 h incubation prior to treatment. Similarly, DEP has shown to detect drug response in cells (after 2 h of treatment) faster than other assessment methods such as flow cytometry which typical requires 24 h [20]. Along with this advantage, the OπDEP platform eliminates certain common drawbacks associated with other DEP platforms. The benefits of OπDEP include reusable electrodes, passivation from cell solution (preventing interactions between the cells and electrodes) and localization of electric fields, making this a cost-effective for tool for rapid drug assessment.
Nevertheless, we acknowledge the OπDEP platform is limited by the thick passivation layer (100 µm) which causes the system to require large electric field magnitudes. This translates to limited frequency ranges due to equipment restrictions and large electronics. Further limitations of the platform are discussed in Zellner et al. [22]. In addition, as this study evaluated frequencies at 100 kHz intervals, the DEP profiles generated are only discrete values. In order to produce a more continuous profile, additional data points should be added for future studies.
Further studies need to be performed to better define the relationships which link DEP characteristics and the cellular alterations which lead to each kind of profile. Additionally, to evaluate the predictive nature of the trapping efficiency and crossover frequency DEP analyses with unknown cell samples should be conducted. Future directions for this platform encompass the fabrication of multiple channels on a single chip in order to enable simultaneous differential measurements for various trials, leading to more robust results and reduced experimental time.
5 Conclusion
By utilizing the previously established OπDEP platform, we have shown that different cell lines exhibit different and distinctive trapping profiles. When the cells were exposed to a chemical stimulus, the initial trapping frequency and the trapping efficiencies vary. The treated cells are clearly distinguished from their vehicle counterparts based on shifts in their DEP profiles, exhibiting the applicability of this method for drug sensitivity studies. These shifts in the DEP profiles can be further examined to determine the underlying biophysical changes due to drug exposure. Thus, this method of analysis provides greater insight into the biophysical changes of cells in a rapid, label-free assay. Additionally, the inherent benefits of the OπDEP platform such as reusable electrodes and separation between the electrodes and sample shows the platform’s tremendous potential for drug sensitivity studies.
Supplementary Material
Acknowledgments
This work was supported primarily by the National Science Foundation under award number ECCS-1310090.
Abbreviations
- ER+
estrogen receptor positive
- GX
GX15-070 (Obatoclax)
- OπDEP
off-chip passivated-electrode insulator-based dielectrophoresis
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
The authors have declared no conflict of interest.
References
- 1.Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y, Hilakivi-Clarke LA. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 2.Thompson A, Brennan K, Cox A, Gee J, Harcourt D, Harris A, Harvie M, Holen I, Howell A, Nicholson R, Steel M, Streuli C. Breast Cancer Res. 2008;10:1–25. doi: 10.1186/bcr1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brünner N, Boysen B, Jirus S, Skaar TC, Holst-Hansen C, Lippman J, Frandsen T, Spang-Thomsen M, Fuqua SAW, Clarke R. Cancer Res. 1997;57:3486–3493. [PubMed] [Google Scholar]
- 4.Lee AV, Oesterreich S, Davidson NE. JNCI: J. Nat. Cancer Inst. 2015;107:djv073–djv073. doi: 10.1093/jnci/djv073. [DOI] [PubMed] [Google Scholar]
- 5.Lv Y, Zeng L, Zhang G, Xu Y, Lu Y, Mitchelson K, Cheng J, Xing W. Int. J. Nanomed. 2013;8:2333–2350. doi: 10.2147/IJN.S31678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz-Roberts JL, Shajahan AN, Cook KL, Warri A, Abu-Asab M, Clarke R. Mol. Cancer Ther. 2013;12:448–459. doi: 10.1158/1535-7163.MCT-12-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stylianou S, Clarke RB, Brennan K. Cancer Res. 2006;66:6840–6844. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 8.Huang HE, Chin SF, Ginestier C, Bardou VJ, Adélaïde J, Iyer NG, Garcia MJ, Pole JC, Callagy GM, Hewitt SM, Gullick WJ, Jacquemier J, Caldas C, Chaffanet M, Birnbaum D, Edwards PA. Cancer Res. 2004;64:1517–1525. doi: 10.1158/0008-5472.CAN-04-1762. [DOI] [PubMed] [Google Scholar]
- 9.Pohl HA. J. Appl. Phys. 1951;22:869–871. [Google Scholar]
- 10.Pethig R. Adv. Drug Del. Rev. 2013;65:1589–1599. doi: 10.1016/j.addr.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Gascoyne PRC, Shim S, Noshari J, Becker FF, Stemke-Hale K. Electrophoresis. 2013;34:1042–1050. doi: 10.1002/elps.201200496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gascoyne PR, Shim S. Cancers (Basel) 2014;6:545–579. doi: 10.3390/cancers6010545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshmanesh K, Akagi J, Nahavandi S, Skommer J, Baratchi S, Cooper JM, Kalantar-Zadeh K, Williams DE, Wlodkowic D. Anal. Chem. 2011;83:2133–2144. doi: 10.1021/ac1029456. [DOI] [PubMed] [Google Scholar]
- 14.Yusvana R, Headon DJ, Markx GH. Biotechnol. Bioeng. 2010;105:945–954. doi: 10.1002/bit.22615. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Becker FF, Gascoyne PR. Biochim. Biophys. Acta. 2002;1564:412–420. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labeed FH, Coley HM, Thomas H, Hughes MP. Biophys. J. 2003;85:2028–2034. doi: 10.1016/S0006-3495(03)74630-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coley HM, Labeed FH, Thomas H, Hughes MP. Biochim. Biophys. Acta. 2007;1770:601–608. doi: 10.1016/j.bbagen.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Henslee EA, TorcalSerrano RM, Labeed FH, Jabr RI, Fry CH, Hughes MP, Hoettges KF. Analyst. 2016;141:6408–6415. doi: 10.1039/c6an01596d. [DOI] [PubMed] [Google Scholar]
- 19.Mulhall HJ, Cardnell A, Hoettges KF, Labeed FH, Hughes MP. Integr. Biol. (Camb.) 2015;7:1396–1401. doi: 10.1039/c5ib00109a. [DOI] [PubMed] [Google Scholar]
- 20.Kumar RTK, Liu S, Minna JD, Prasad S. Biochim. Biophys. Acta. 2016;1860:1877– 1883. doi: 10.1016/j.bbagen.2016.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou C, Tegenfeldt JO, Bakajin O, Chan SS, Cox EC, Darnton N, Duke T, Austin RH. Biophys. J. 2003;83:2170–2179. doi: 10.1016/S0006-3495(02)73977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zellner P, Shake T, Sahari A, Behkam B, Agah M. Anal. Bioanal. Chem. 2013;405:6657–6666. doi: 10.1007/s00216-013-7123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan LA, Lu J, Wang L, Marchenko SA, Jeon NL, Lee AP, Monuki ES. Stem Cells. 2008;26:656–665. doi: 10.1634/stemcells.2007-0810. [DOI] [PubMed] [Google Scholar]
- 24.Bortner CD, Cidlowski JA. Phil. Trans. R. Soc. B. 2014;369:20130104. doi: 10.1098/rstb.2013.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pethig R, Talary MS. IET Nanobiotechnol. 2007;1:2–9. doi: 10.1049/iet-nbt:20060018. [DOI] [PubMed] [Google Scholar]
- 26.Pethig RR. Dielectrophoresis: Theory, Methodology and Biological Applications. John Wiley & Sons; Hoboken: 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.