Abstract
The small nucleolar RNAs (snoRNAs) are an abundant class of trans-acting RNAs that function in ribosome biogenesis in the eukaryotic nucleolus. Elegant work has revealed that most known snoRNAs guide modification of pre-ribosomal RNA (pre-rRNA) by base pairing near target sites. Other snoRNAs are involved in cleavage of pre-rRNA by mechanisms that have not yet been detailed. Moreover, our appreciation of the cellular roles of the snoRNAs is expanding with new evidence that snoRNAs also target modification of small nuclear RNAs and messenger RNAs. Many snoRNAs are produced by unorthodox modes of biogenesis including salvage from introns of pre-mRNAs. The recent discovery that homologs of snoRNAs as well as associated proteins exist in the domain Archaea indicates that the RNA-guided RNA modification system is of ancient evolutionary origin. In addition, it has become clear that the RNA component of vertebrate telomerase (an enzyme implicated in cancer and cellular senescence) is related to snoRNAs. During its evolution, vertebrate telomerase RNA appears to have co-opted a snoRNA domain that is essential for the function of telomerase RNA in vivo. The unique properties of snoRNAs are now being harnessed for basic research and therapeutic applications.
Keywords: Small nucleolar RNA, Nucleolus, RNP, Telomerase, Archaea, RNA transport, Ribosome, RNA modification, RNA processing, Cajal body, Ribozyme
SMALL NUCLEOLAR RNAs AND RIBOSOME BIOGENESIS
snoRNAs are essential for production of ribosomal RNA. Recent work has revealed that many snoRNAs base pair with precursor rRNA marking individual nucleotides for modification. Other snoRNAs are required for cleavages of precursor rRNA.
Despite the tremendous diversity of life on earth, all extant organisms depend upon structurally and functionally similar ribosomes. Furthermore, the major steps in the generation of ribosomes are generally similar in all organisms. For example, precursor rRNAs are typically synthesized, cleaved, and post-transcriptionally modified to yield small and large subunit rRNAs. Among the most abundant of the rRNA modifications are a specific sugar modification (methylation of the 2′ hydroxyl group of ribose) and a particular base modification (conversion of uridine to pseudouridine) (117,125,151). The fully processed and modified rRNAs assemble with numerous ribo-somal proteins to form small and large ribosomal subunits that together comprise the translational machinery of all cells.
While the basic steps of ribosome biogenesis have been studied for many decades, it is only in the last decade or so that we have come to discover the role of a collection of trans-acting RNAs known as the snoRNAs [reviewed in (6,53,64,93,123,158,188,201,214,224)]. snoRNAs constitute a very large family of RNAs found in diverse eukaryotic organisms. It is estimated that —150 different snoRNA species exist in each human cell (87). The known functions of snoRNAs include roles in both pre-rRNA processing (endonucleolytic cleavages) and pre-rRNA modification (ribose methylation and pseudouridylation).
Two snoRNA Classes
Based on the presence of conserved sequence elements and common secondary structures, snoRNAs fall into two major classes: box C/D snoRNAs and box H/ACA snoRNAs. Exceptions to this general rule include the RNA component of the MRP endo-nuclease (106,136) and a recently identified snoRNA species (U85) that contains both box C/D and box H/ ACA motifs (80). A few members in each of the two major snoRNA classes are involved in site-specific pre-rRNA cleavages (123,188,201,224). However, the vast majority of snoRNAs function as rRNA modification guide RNAs, directing site-specific nucleotide modifications of rRNA (6,151,201,224). The specification of the site of modification is achieved by base pairing of the snoRNA with rRNA, and the type of rRNA modification directed depends on the class of the snoRNA. Box C/D snoRNAs guide addition of a methyl group to the 2′ hydroxyl of specified rRNA ribose moieties (33,87,143,200,206). Box H/ ACA snoRNAs direct conversion of targeted uridines to pseudouridines (58,142). A given modification guide snoRNA may guide one or, less frequently, two rRNA modifications. In human cells, snoRNAs appear to be responsible for guiding ∼ 100 2′-0-methyl-ations and—100 pseudouridylations of rRNA (117). Thus, an elaborate RNA-guided rRNA modification system is present in eukaryotes.
Box C/D snoRNAs Guide 2′-O-Methylation of rRNA
The function of box C/D snoRNAs depends on the signature box C/D motif and regions of complementarity to rRNA (Fig. 1A). The box C/D motif is comprised of a conserved box C sequence element (RUGANGA, typically near the 5′ end of the RNA), a box D element (CUGA, typically near the 3′ end of the RNA), and a terminal stem. Sequences adjacent to the box C and box D elements base pair to form the terminal stem, which brings the two box elements together across from one another in an internal loop in the majority of the RNAs. In the mature snoRNAs that lack canonical 5′-3′ terminal stems, internal stems or stem structures that form transiently on precursor snoRNA molecules ensure the juxtaposition-ing of box C and D sequences (41,216). snoRNAs can have a second pair of box C-like and box D-like elements [called box C and box D′ (87)] that are located internally and are typically close to one another in the primary structure or brought together via a neighboring stem. Each methylation guide RNA contains one or two regions of extensive complementarity to rRNA (10–21 nucleotides in length) called “guide sequences” that mediate base pairing with specific rRNA regions.
Figure 1.
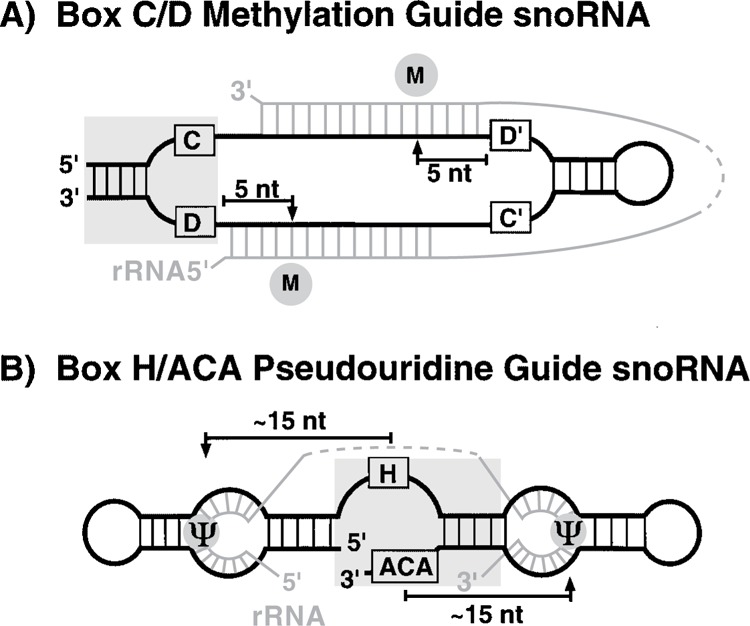
snoRNAs mark rRNA sites for modification. (A) Box C/D guide RNAs direct site-specific rRNA 2′-O-ribose methylation. A box C/ D snoRNA (black) base pairs with one or two complementary rRNA sequences (gray) via complementary sequences (guide sequences) found upstream of box D and/or box D′. The 2′-O-methyl group (M) is always added to the rRNA nucleotide that is paired to the fifth residue upstream of the conserved box D and/or box D′ element. The box C/D motif, a common feature of box C/D snoRNAs, is shaded. (B) Box H/ACA guide RNAs direct the conversion of uridines to pseudouridines. A box H/ACA snoRNA (black) base pairs with complementary rRNA sequences (gray) in one or two loop regions (“pseudouridine pockets”). The target uridine(s) (Ψ) remain unpaired. Pseudouridine formation takes place∼15 nucleotides from the conserved box H or box ACA element. The box H/ACA motif, a common feature of box H/ACA snoRNAs, is shaded.
The box D (or box D′) element and guide sequence(s) of the snoRNA play key roles in the meth-ylation reaction (Fig. 1A). The snoRNA guide sequences are always located immediately upstream of box D (and/or box D′). The rRNA nucleotide found across the snoRNA/rRNA duplex precisely five nucleotides upstream of box D is targeted for modification (6,7,87). There is no base specificity to the reaction (A, C, G, and U are all methylated) and the length of the snoRNA/rRNA duplex appears to be flexible (the observed range is 10–21 nucleotides). The “box D-plus-five” nucleotide selection rule, initially inferred from snoRNA sequences and mapped sites of methylation, was strengthened by the observation that alteration of the spacing between the guide sequence and box D results in a corresponding shift in the modification site to a new site precisely five nucleotides from box D (87). Furthermore, novel sites of methylation can be targeted in vivo simply by engineering new guide sequences in snoRNAs [(31,33,107) and W. Speckmann, R. M. Terns, and M. P. Terns, unpublished data].
Box H/ACA snoRNAs Guide Pseudouridylation of rRNA
Members of the box H/ACA class of snoRNAs exhibit a common “hairpin–hinge–hairpin–tail” secondary structure (Fig. 1B) and contain positionally conserved box H (ANANNA, located in “hinge”) and box ACA (ACANNN, located at the 3′ terminus) sequence elements (9,58,59). Box H and box ACA are tethered by a neighboring stem creating the box H/ ACA motif (17,138). The guide sequences of box H/ ACA snoRNAs are located in “pseudouridine pockets,” internal loops found in the 5′ and 3′ hairpins. The rRNA uridine to be modified remains unpaired amid flanking sequences that are base paired with the snoRNA guide sequences, and is located within a fixed range approximately 15 nucleotides from box H or box ACA (58).
Molecular Basis of snoRNA Function
The function of snoRNAs involved in both cleavage and modification appears to rely on transient, site-specific base pairing with pre-rRNA sites, but the ensuing molecular events are not understood in detail. For the few box C/D and H/ACA snoRNAs involved in endonucleolytic cleavage, a chaperone function has been proposed by Steitz and Tycowski whereby base pairing of the snoRNAs prepares the pre-rRNA substrate for action by site-specific protein endonucleases (193). On the other hand, the unrelated snoRNA, MRP (which is related to the RNA component of RNAse P), appears to directly cleave rRNA as a site-specific ribozyme (106,113,136,171). For box C/D and H/ACA snoRNAs that direct site-specific modification of rRNA, duplex formation appears to be a requirement for recognition of the pre-rRNA target site by recruited modifying enzymes (i.e., ribose methylase or pseudouridine synthase).
The precise function of the rRNA nucleotide modifications mediated by snoRNAs also remains obscure, but several observations indicate their biological significance. For example, the modifications occur on the newly transcribed rRNAs prior to cleavage, but are not found in the external and internal transcribed spacer sequences (117). Furthermore, the modified nucleotides cluster to functionally important domains of the mature rRNA including the peptidyl transferase and decoding centers (6,21,116,151). Nucleotide modification may alter intramolecular rRNA interactions and the three-dimensional structure of rRNA, and/or intermolecular rRNA interactions with ribosomal proteins. Pseudouridine has increased potential for hydrogen bonding relative to uridine, and methylation of ribose increases the hydrophobic character of the residues and is predicted to protect specific phosphodiester bonds against nucleolytic cleavage. Most individual modified nucleotides of rRNA are not required for cell viability (93,168,201). Thus, it is likely that the modifications perform a collective function in ribosome biogenesis and/or protein translation.
EXPANDING ROLES FOR snoRNAs
More recent work has revealed that snoRNAs also direct modification of snRNAs and perhaps mRNAs. In addition, during the course of evolution vertebrate telomerase RNA appears to have appropriated a snoRNA domain that provides stability and compart-mentalization within the nucleus. Finally, researchers are manipulating snoRNAs to provide vehicles for controlled subcellular delivery of RNAs including ri-bozymes, and to serve as agents for targeted modification of cellular RNAs.
New Targets for Modification
In addition to their well-established role in the 2′-O-methylation and pseudouridylation of RNA polymerase (Pol) I-transcribed ribosomal RNAs, the targets for snoRNA-directed modification have expanded to include Pol II- and Pol III-generated RNAs as well. For example, three box C/D snoRNAs have been found to guide 2′-O-methylation of U6 spliceosomal snRNA (a Pol III transcript) (60,209). The eight ribose methylations and three pseudouridylations of U6 are likely guided by dedicated snoRNPs within the nucleolus (60). U6 snRNA transiently travels through nucleoli where it presumably encounters the snoRNPs (101,152,160). A recent study indicates that internal modification of U2 snRNA (a Pol II transcript) is very likely also mediated by snoRNAs within the nucleolus or perhaps Cajal bodies (232). In this study, we demonstrated that ribose methylation and pseudouridylation of U2 and U2 RNA variants correlated with the nucleolar localization of the RNAs (232). Furthermore, modification could be rescued by appending an exogenous nucleolar localization signal (derived from a box C/D snoRNA) to U2 variants (232). Internal modification of U5 snRNA (also a Pol II transcript) is guided by an intriguing snoRNA found by Jady and Kiss (80). This RNA (U85) is the first example of a snoRNA that contains both box C/D and box H/ACA motifs and guides both 2′-O-methylation and pseudouridylation (80).
There is now evidence that snoRNAs may also direct the modification of certain messenger RNAs with fascinating potential consequences. A box C/D snoRNA that contains an 18-nucleotide, phylogeneti-cally conserved complementarity to the brain-specific serotonin 2C receptor mRNA has been identified in both mouse and human (32,52). The BII-52 snoRNA (called MBII-52 or HBII-52 in mouse and human, respectively) is predicted to target 2′-O-methylation of a particular adenosine residue within the serotonin 2C receptor mRNA that otherwise undergoes adeno-sine to inosine (A to I) editing (25,145). A to I editing of the serotonin 2C receptor mRNA (at up to five sites) can lead to structurally and functionally distinct serotonin receptor molecules (25,32,145). Furthermore, A to I editing is markedly inhibited by 2′-O-methylation in vitro (231). Taken together, the findings raise the intriguing possibility that this snoRNA (as well as others) may regulate A to I editing that occurs in brain (36,157).
The BII-52 box C/D RNA is expressed specifically in the brain. Two additional box C/D snoRNAs (named MBII-13/HBII-13 and MBII-85/HBII-85 in mouse/human) and one box H/ACA snoRNA (MBI-36/HBI-36) have been identified whose expression is limited to brain tissue (32,42,126). No obvious sequence complementarities between these snoRNAs and other cellular RNAs have been identified and it remains to be determined if these snoRNAs function in nucleotide modification. However, the genomic organization and expression patterns of these RNAs indicate their functional importance. For example, the genes encoding all three brain-specific box C/D snoRNAs map to a region of human chromosome 15 implicated in the neurogenetic disease Prader-Willi syndrome (PWS) (32,42,126). PWS results from a deficiency in paternal gene expression. The expression of the box C/D RNAs was not detected in the brains of PWS patients or PWS model mice (32). These findings indicate that expression of the snoRNAs is subject to paternal imprinting, and suggest a link between snoRNA expression and this disease. The gene encoding the brain-specific box H/ ACA snoRNA is located within the intron of the serotonin 2C receptor mRNA (the presumed target of ribose methylation by BII-52, see above).
In summary, it is becoming clear that snoRNAs act on a diverse repertoire of substrate RNAs. snoRNAs that lack significant sequence complementarity to rRNA or other known targets have been reported (32,79). It will be exciting to learn the nature of the target RNAs recognized by these “orphan snoRNAs.”
Essential Role of a snoRNA Domain in Telomerase RNA
Telomerase RNA is a core component of the telomerase enzyme, which is required for the synthesis of telomeres at chromosome ends and implicated in tumor progression and cellular senescence (22,40,67,68,74,210). Recent studies have identified a conserved box H/ACA snoRNA domain present at the 3′ termini of vertebrate telomerase RNAs (37,132) and revealed that vertebrate telomerase RNA associates with the known box H/ACA snoRNP proteins including GAR1, dyskerin (Cbf5p homolog), NOP10, and NHP2 (43,45,134,165). The snoRNA domain of telomerase RNA provides properties that are essential to the function of the RNA. The 3′ snoRNA domain of telomerase RNA is required for telomerase activity in human cells and has been shown by Mitchell et al. to be important for the stability and proper 3′ end formation of the RNA (134). Furthermore, we have found that telomerase RNA is retained in the nucleus (i.e., does not undergo nuclear export) due to the box H/ACA snoRNA motif (112a). A portion of endogenous telomerase RNA biochemically fractionates with nucleoli (134) and injected telomerase RNA becomes targeted to the dense fibrillar component of Xenopus oocyte nucleoli (138). The nucleolar localization of telomerase RNA is also mediated by the box H/ACA snoRNA motif (138). Our data demonstrate that the 3′ snoRNA domain of telomerase RNA, which is dispensable for telomerase activity in vitro (4,5,12,133,199), functions in intranuclear targeting of telomerase RNA in vivo. Interestingly, we also observed a stable association of telomerase RNAs with nucleoplasmic structures known as Cajal (coiled) bodies (112a).
It is not yet clear why telomerase RNA associates with nucleoli or Cajal bodies. While telomerase RNA resembles a box H/ACA snoRNA, it does not likely guide pseudouridylation of rRNA (37). Modification of telomerase RNA or the assembly of telomerase enzyme might take place within nucleoli and/or Cajal bodies. Moreover, because the precise intracellular location of telomere synthesis has not been determined in vertebrate cells, it is formally possible that telomeres become transiently associated with nucleoli and/or Cajal bodies during their synthesis (i.e., that te-lomerase functions within these structures). Alternatively, telomerase RNA localization studies in ciliates suggest a role for subnuclear structures (such as nucle-oli or Cajal bodies in vertebrates) in the compartmen-talization of telomerase RNA away from chromosomes when it is not involved in telomere synthesis (51).
Use of snoRNAs in Applied Research
Small Nucleolar RNAs as Vehicles for Intracellular Targeting of Ribozymes and Other RNAs
Ribozymes are naturally occurring enzymatic RNAs that catalyze the site-specific cleavage of other RNA molecules. Ribozymes can be engineered to cleave chosen target RNA molecules and inhibit expression of specific genes (81,163,173), and they are being developed to capitalize on their potential in therapeutic, biotechnological, and basic research applications. A major limitation in the application of ribozymes is that ribozymes that demonstrate good activity in vitro are often found to be ineffective when introduced into living cells (175). While the basis of the discrepancy between the in vitro and cellular activities of ribozymes is generally unknown, the efficacy of a ribozyme in vivo is certainly limited by access of the ribozyme to the target RNA; clearly, the ribo-zyme must encounter the target RNA in the cell for a productive interaction to take place.
Recently, a chimeric snoRNA-ribozyme (dubbed “snorbozyme” by Samarsky et al.) has been shown to perform with an unprecedented, near 100% efficiency against a colocalized RNA substrate in yeast [(179); reviewed in (174)]. In this model study, a hammerhead ribozyme embedded within U3 snoRNA sequences was coexpressed with a second RNA consisting of the ribozyme target sequences also embedded within U3 snoRNA. The near-perfect efficacy of the ribozyme in vivo was largely attributed to the demonstrated colocalization of the ribozyme and substrate in the cell nucleolus.
Another chimeric snoRNA-ribozyme has been shown to suppress HIV-1 infection of human cells (131). A hammerhead ribozyme directed against a conserved region of HIV-1 (the causative agent of human acquired immune deficiency syndrome, AIDS) RNA was embedded into U16 snoRNA sequences, thereby targeting the ribozyme to nucleoli. The efficacy of the chimeric snoRNA-ribozyme in targeting the destruction of HIV RNA provided insight that HIV-1 RNA trafficks through nucleoli en route to the cytoplasm. In a similar study, an HIV-1 Rev protein binding site was inserted into the U16 snoRNA sequence and targeted to nucleoli (24). Subsequent expression of Rev protein resulted in export of the chimeric RNA to the cytoplasm, indicating that the HIV-1 Rev protein is present and can function within nucleoli. It will be interesting to see whether the chimeric anti-Rev-snoRNA can function as a Rev decoy to effectively inhibit replication of the HIV-1 virus.
Small Nucleolar RNAs as Tools for Site-Specific Modification of RNA Targets
The ability of snoRNAs to guide site-selective nucleotide modification is also beginning to be exploited for RNA functional mapping studies (107,141). In this approach, snoRNAs are engineered to direct the modification of an RNA at targeted sites, and assays with the altered RNAs then test whether modification at a particular nucleotide interferes with function. In essence, the strategy is akin to modification interference assays used to determine the importance of particular nucle-otides in a process in vitro; however, snoRNA-guided modification offers a means to obtain the information in vivo. The feasibility of the approach has recently been demonstrated by Liu et al. with the identification of specific novel methylations in the peptidyl transferase center of rRNA that result in growth defects in yeast (107).
MULTIPLE MECHANISMS FOR THE GENERATION OF snoRNAs
The many individual snoRNAs present in cells have diverse and interesting genomic origins. Recent work has helped define the cellular machinery that harvests the snoRNAs from precursors.
Several unusual strategies are used to synthesize snoRNAs in eukaryotes. Many snoRNA genes are present within introns of protein coding genes, and functional snoRNAs are salvaged from introns excised by pre-mRNA splicing reactions (53,123,190,201,224). Interestingly, such intron-encoded snoRNAs reside in messenger RNAs encoding proteins involved in the synthesis, structure, or function of ribosomes, indicating that snoRNA production and ribosome production are coordinated events. Still other snoRNAs are encoded within the introns of host genes that do not appear to encode proteins; processing of the nonprotein coding transcripts can liberate up to 10 different snoRNA species (18,102,161,189,207,208). Other snoRNAs are more conventionally synthesized as the sole product of a transcriptional unit (123,205). These independently transcribed snoRNAs are synthesized with 5′ m7G caps that undergo hypermethylation within the nucleus to form a trimethylated m2,2,7G cap structure (192,196,197). Finally, in plants and yeast there are examples of multiple snoRNAs encoded as polycistrons that are liberated from their nonprotein encoding transcripts via a splicing-independent mechanism (34,35,102,162,168).
Considerable progress has been made in deciphering the mechanisms that generate mature snoRNA species from precursor molecules, and in identifying the cellular components responsible for the processing events. Whether the snoRNA is encoded within pre-mRNA introns or synthesized as a mono-or polycistronic transcript, biogenesis of mature snoRNA molecules generally involves the concerted action of endonuclease and exonuclease activities. For intron-encoded snoRNAs, the spliceosome provides the endonuclease activity responsible for releasing most, but not all (27,217), snoRNAs from longer precursor molecules. Subsequently, a debranching activity (provided by the Dbr1p protein in yeast) linearizes the released snoRNA/intron-lariat intermediates (154,162). An RNase III-like endonuclease (called Rnt1p in yeast) functions to liberate snoRNAs from independently transcribed units (34,35,168). The cleavage enzyme that generates the 3′ end of poly-adenylated mRNAs may also help release some pre-snoRNA precursor molecules (50). In all cases, the endonucleolytic cleavage events generate pre-snoRNA molecules with 5′ leader and 3′ trailer sequences that are subsequently removed by exo-nucleases to generate mature snoRNAs. In yeast, 5′ end trimming reactions are carried out by 5′–3′ exo-nucleases called Rat1p and Xrn1p (162) while 3′ end trimming is performed by a complex of 3′–5′ exo-nucleases known as the exosome (1,135,212,213). The generation of precise 5′ and 3′ termini of mature snoRNAs requires the prior association of snoRNP proteins, which protect the sequences within the body of the snoRNAs against exonuclease attack (28,220).
INTRANUCLEAR TRAFFICKING OF snoRNAs
The mechanisms that control the transport and localization of cellular RNAs are generally not well understood. The snoRNAs have provided a good model system to begin to dissect the means by which RNAs can be compartmentalized and targeted to cellular structures.
snoRNAs Are Retained in the Nucleus
After synthesis, most cellular RNAs (including mRNA, tRNA, rRNA, and snRNA) are exported from the nucleus to the cytoplasm (77,122,137,195). In contrast, snoRNAs remain within the nucleus where they are matured and function (196,197). Studies performed in the Xenopus oocyte system demonstrate that snoRNAs are actively retained in the nucleus by a mechanism that is saturable, sequence specific, and factor mediated (191,196,197). In vivo competition experiments have demonstrated that box C/D snoRNAs compete for a limiting component that is specifically involved in retaining these RNAs in the nucleus (197).
Extensive mutational analysis was used to dissect the cis-acting sequences essential for retaining U3 snoRNA within the nucleus (191). Interestingly, this work revealed that the nuclear retention of U3 snoRNA does not simply reflect its nucleolar localization. The retention of U3 in the nucleus can be independently mediated by both the box C′/D (common to all box C/D snoRNAs) and box B/C (U3-specific) motifs, which function as redundant nuclear retention elements (Fig. 2A). However, only the box C′/D motif targets U3 to nucleoli (139). Thus, when the box C′/D motif of U3 is disrupted, the RNA is retained in the nucleus but not localized to nucleoli (139,191). It is not known whether there are redundant sequence elements that mediate the retention of other box C/D RNAs, which do not contain a box B/ C motif. However, disruption of the box C/D motif in U8 and U14 results in mislocalization to the cytoplasm (197), indicating that U3 may be unique in containing two nuclear retention elements. Finally, the cis-acting RNA sequences and trans-acting proteins that mediate the nuclear retention of box H/ ACA snoRNAs are unknown.
Figure 2.
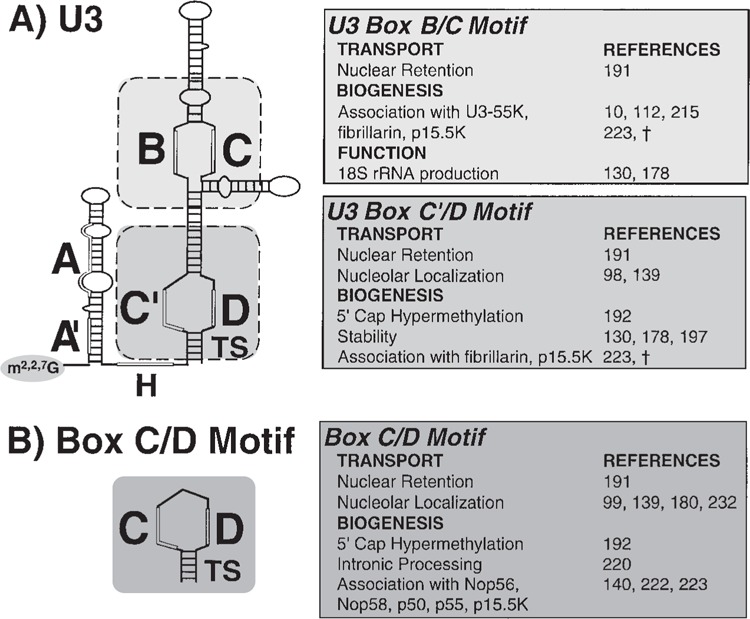
Structure–function analysis of box C/D snoRNAs. (A) U3 snoRNA (the most extensively analyzed box C/D snoRNA) contains five conserved sequence elements known as boxes A′, A, C′, B, C, and D as well as a “hinge” region (H). Box A′, box A, and the hinge each participate in U3 snoRNA/rRNA base pairing. Boxes B and C, and boxes C′ and D form functional units called the box B/C motif (U3 specific) and the box C′/D motif (common to other box C/D snoRNAs). A terminal stem (TS) is a critical component of the box C′/D motif of U3. The functions of the box B/C motif and the box C′/D motif elucidated by mutational analysis are indicated (see text for details). (B) The box C/D motif is a common feature of box C/D RNAs. The indicated properties of the box C/D motif have been inferred from studies of minimal RNAs comprised essentially of the box C/D motif (box C and box D separated by intervening sequences and linked by a terminal stem). †W. Speckmann, R. M. Terns, and M. P. Terns, unpublished data.
Motifs That Target snoRNAs to Nucleoli
To identify the cis-acting sequences that target snoRNAs to nucleoli, localization studies have been performed by a number of research groups in diverse experimental systems including Xenopus oocytes, mammalian cells, and yeast (78,97–100,138,139,176,180). Results from our laboratory indicate that in both the box C/D and box H/ACA family of snoRNAs, the signature motif, comprised of the common box elements and an adjacent tethering structure (Fig. 1), targets the RNAs to the nucleolus (138,139). Thus, we find that box C (C′ in U3, see Fig. 2A), box D, and an adjacent stem (but not other conserved elements) are required for nucleolar localization of box C/D RNAs (139). In addition, our laboratory and others have demonstrated that minimal RNAs consisting of the box C/D motif are localized to nucleoli, indicating that the box C/D motif is sufficient for nucleolar localization (99,139,180). Furthermore, we have found that addition of an exogenous box C/D motif is sufficient to efficiently target U2sm- (an snRNA variant that is normally exported from the nucleus) to nucleoli (232).
It is important to note that some studies are not in agreement with the box C/D RNA nucleolar localization motif delineated above. Some studies have suggested that structural elements of the box C/D motif (e.g., adjacent stems) are not important in nucleolar targeting of box C/D RNAs (97–99) and that an element of the box B/C motif is also essential for nucle-olar localization of U3 RNA (98). Some of the differences appear to be simply in interpretation of results; for example, use of alternate U8 RNA secondary structure models or interpretation of experiments in which structural elements were deleted (leaving short sequences that we would propose may function like a stem to tether the box elements) (97). However, in other cases very different results were obtained in very similar experiments. For example, Lange et al. found that disruption of the terminal stem of U14, or of the terminal stem or box C′ of U3 did not prevent nucleolar targeting of the RNAs (98,99). In these instances the sources of the differences are more likely experimental. We have found that injection of RNAs both in moderate amounts and in a solution containing dextrans is important to eliminate false-positive signals that appear to result from rapid, nonspecific, heterogeneous interaction of injected RNAs with nucleoli (A. Narayanan, R. M. Terns, and M. P. Terns, unpublished data).
With striking analogy to the box C/D RNAs, we have found that box H, box ACA, and the adjoining stem structure are each important for nucleolar localization of box H/ACA snoRNAs (138). However, our studies indicate that these three elements are not sufficient for localization, but that localization also requires at least one hairpin structure including an internal loop (138). Thus, box H and box ACA tethered by a single hairpin are targeted to nucleoli (138). On the other hand, other studies have concluded that the stem of the box H/ACA motif (176) and the hairpin structures (100) are not required for targeting, and that box H and box ACA function redundantly or additively in targeting (100). Again, in some cases contrasting conclusions may be a result of differences in the interpretation of experiments with deletion mutants, and contradictory results may reflect whether a dextran-blocking solution was used in microinjections.
Taken together, the results indicate that the nucleolar localization of box C/D snoRNAs and box H/ ACA snoRNAs requires cis-acting sequences and structural features that are common to the members of each family of RNAs. Furthermore, base pairing of snoRNAs to pre-rRNA is not essential for nucleolar localization/retention of snoRNAs because snoRNA variants devoid of all known rRNA complementarities are targeted to nucleoli (97–99,138,139,180). Instead, in each family, the conserved sequence elements (i.e., box C and box D, or box H and box ACA) are brought adjacent to one another as the result of formation of one or more nearby stems to form a core motif that serves as a nucleolar localization signal. These results indicate that the —200 identified snoRNA species utilize one of two distinct nucleolar targeting mechanisms.
snoRNAs Traverse Cajal Bodies En Route to Nucleoli
While the nucleolus is the functional destination of snoRNAs, biogenesis and maturation of the RNAs likely take place at distinct subnuclear sites. Box C/ D snoRNAs have been observed in nuclear structures called Cajal (coiled) bodies in mammalian and plant cells under steady-state expression conditions (11,84,180,186). More recently, microinjection experiments revealed that box C/D snoRNAs transiently associate with Cajal bodies prior to nucleoli (139). Strikingly, variant snoRNAs that failed to localize to nucleoli accumulated at Cajal bodies (139). These observations indicate that box C/D snoRNAs move through Cajal bodies en route to nucleoli. In a similar study, box H/ACA snoRNAs were not detected in Cajal bodies; however, the result may simply reflect more rapid transport through the Cajal bodies or the need to study precursor RNAs (138). Indeed, proteins associated with box H/ACA as well as box C/D snoRNAs are present in Cajal bodies (57,128,169). Other observations are also consistent with the hypothesis that Cajal bodies play an important role in snoRNP biogenesis and trafficking. For example, Cajal bodies have been observed in close association both with genes that encode snoRNAs (61,121,184) and with nucleoli (15,19,115,119,149,187).
THE PROTEIN PARTNERS OF snoRNAs
snoRNAs exist as small nucleolar ribonucleoprotein particles (snoRNPs) in the cell. Protein association with snoRNAs is likely dynamic, mediating the biogenesis, transport, and function of the RNAs.
A distinct set of proteins is found associated with each the box C/D and the box H/ACA snoRNA classes (Table 1). Additional proteins that are uniquely associated with a particular RNA have been identified for some of the snoRNAs involved in rRNA processing, including U3 and U8 (Table 2). Thus, it appears that a common set of proteins mediates the function of each of the modification guide snoRNAs, while snoRNAs involved in processing may require additional protein factors. The exact roles of each of the protein components in snoRNA biogenesis and rRNA modification or processing are not known. However, while it has not been formally demonstrated, the enzymes that catalyze the modification reactions (i.e., the methylase and pseudouridine synthase) appear to be among the integral components of the snoRNPs.
TABLE 1.
THE COMMON PROTEIN COMPONENTS OF THE BOX C/D snoRNPs AND THE BOX H/ACA snoRNPs
| Vertebrate Homolog | Yeast Homolog | Structural Motifs | Known or Predicted Function(s) | References |
|---|---|---|---|---|
| Box C/D snoRNPs | ||||
| Fibrillarin | Nop1p | GAR, RRM-like, SAM-methylase | 2-O-methylase?, RNA binding, 18S rRNA processing, ribosome assembly | 3,49,148,182,202,203 |
| Nop56 | Nop56p | Homology to Nop58, KKE/D repeats | snoRNA stability, 18S rRNA processing | 63,94 |
| Nop58 | Nop58p/Nop5p | Homology to Nop56, KKE/D repeats | 18S rRNA processing | 63,94,114,229 |
| p50 | Rvb2/Tih2 | Homology to p55, Walker A/B motifs | RNA helicase?, snoRNP biogenesis?, 18S rRNA processing | 140 |
| p55 | Rvb1 | Homology to p50, Walker A/B motifs | RNA helicase?, snoRNP biogenesis?, 18S rRNA processing | 140 |
| p15.5kD | Snu13p | Similarity to NHP2 | RNA binding (C/D motif), snoRNA stability | 223 |
| Box H/ACA snoRNPs | ||||
| GAR1 | Gar1p | GAR (two) | RNA binding, snoRNP/pre-rRNA interaction | 8,20,45,66,72,221 |
| Dyskerin | Cbf5p | KKE/D repeats | Pseudouridine synthase?, snoRNA stability | 26,43,45,72,134,221,233 |
| NOP10 | Nop10p | snoRNA stability | 43,45,72,165 | |
| NHP2 | Nhp2p | Similarity to P15.5kD | RNA binding? snoRNA stability | 43,45,72,165,221 |
TABLE 2.
PROTEINS ASSOCIATED WITH SPECIFIC EUKARYOTIC snoRNAs
| Vertebrate Homolog | Yeast Homolog | Structural Motifs | Known or Predicted Function(s) | References |
|---|---|---|---|---|
| U3 Specific | ||||
| U3-55k | Rrp9p | WD repeats | RNA binding?, Nuclear retention?, 18S rRNA processing | 111,112,164,215 |
| Sof1 | Sof1p | WD repeats | 18S rRNA processing | 82 |
| Mpp10 | Mpp10p | Phosphoprotein, Charged regions | 18S rRNA processing | 46,103,225 |
| Imp3p | Similarity to S4 family ribosomal proteins | RNA binding?, 18S rRNA processing | 104 | |
| Imp4p | 18S rRNA processing | 104 | ||
| Lcp5p | Charged regions | 18S rRNA processing | 226 | |
| Dhr1p | DEAH-Box | RNA helicase?, 18S rRNA processing | 39 | |
| Rcl1p | 3′-phosphate cyclase-like | 18S rRNA processing | 14 | |
| La | Lhp1p | RRM, PEST, ATP | RNA binding, pre-U3 biogenesis | 91 |
| Nucleolin | Gar2p, Nsr1p | Phosphoprotein RRM (2-4), GAR | 18S rRNA processing, Multiple cellular roles | 65 |
| U8 Specific | ||||
| X29 | ? | Sequence not determined | 5.8S and 28S rRNA processing? | 204 |
Common Components of box C/D snoRNPs
The box C/D snoRNAs that are involved in both rRNA cleavage and 2′-O-methylation are associated with a common set of at least six proteins: fibrillarin/Nop1p (3,148,182), Nop56 (63,94), Nop58/Nop5p (63,96,229), p50 (85a,140), p55 (85a,140), and p15.5kD/Snu13p (223) (see Table 1). Each of these proteins has been shown to be highly conserved and essential for yeast viability.
Fibrillarin is likely the 2′-O-methylase that acts in concert with the box C/D snoRNAs. Fibrillarin is required for rRNA 2′-O-methylation in yeast (203) and contains all of the hallmark structural features of known S-adenosylmethionine (SAM)-dependent methyltransferases (144,218). However, fibrillarin is known to associate with box C/D snoRNAs involved in both 2′-O-methylation and pre-rRNA processing. Furthermore, specific alleles of fibrillarin affect either rRNA processing, ribosome biogenesis, or 2′-O-methylation, indicating that fibrillarin plays independent roles in these processes (203). Fibrillarin does not appear to be required for the nucleolar localization or stability of most box C/D snoRNAs [(202); W. Speckmann, R. M. Terns, and M. P. Terns, unpublished data], indicating that it may not be essential for snoRNP formation.
Nop56 and Nop58 are very similar proteins that appear to have divergently evolved from a common ancestor (73,140), but these two proteins do not perform redundant functions because both gene products are essential in yeast (94,96,229). Both proteins are highly basic, a property that could be important in interaction with negatively charged snoRNAs. Depletion of Nop58 (but not Nop56) results in loss of box C/D RNAs from the cell (94,96).
A second pair of similar proteins, p50 and p55, was recently isolated as components of RNPs reconstituted in vitro on a model box C/D snoRNA substrate (140). Both p50 and p55 contain Walker A (ATP/GTP binding site) and Walker B (ATP hydrolysis) motifs, and may function as DNA/RNA heli-cases (118). Unlike the other common box C/D snoRNP proteins, p50 and p55 are concentrated in the nucleoplasm rather than nucleoli (140). Both proteins contribute to snoRNA stability (85a). p50 and p55 may be involved in the early steps of box C/D snoRNA production, snoRNP assembly, and/or intranuclear transport.
The most recently identified protein common to box C/D snoRNPs is p15.5kD (Snu13p in yeast) (223). This protein is required for box C/D snoRNA accumulation (223). Interestingly, p15.5kD had been previously identified as a protein that selectively interacted with the U4 spliceosomal snRNA (147). Evidence indicates that p15.5kD interacts directly with similar sequences present in both the 5′ stem loop of U4 and the box C/D motif of the snoRNAs (223). The occurrence of the protein in both snRNP and snoRNP complexes points to a possible significant connection between snRNP and snoRNP biogenesis and/or pre-mRNA and pre-rRNA metabolism.
The structural organization of box C/D snoRNP particles is unknown. The box C/D motif appears to be necessary for association of each of the six common proteins, but is not sufficient for binding of all of the proteins (10,29,222,223). p15.5kD interacts directly with box C and box D sequences in vitro, suggesting that this protein plays a crucial role in box C/D motif recognition and snoRNP assembly (223). Protein–protein interactions have been observed between fibrillarin, Nop56, and Nop58 in vivo (63,94). Association of Nop56 (but not Nop58) with box C/D snoRNAs is dependent on the presence of fibrillarin (94), suggesting that Nop56 interacts with box C/D snoRNAs indirectly via protein–protein interactions with fibrillarin. The association of fibrillarin with box C/D snoRNAs was thought to be indirect as well (111), but recent work suggests that fibrillarin is capable of weak but direct binding to U16 in a manner dependent on box C and box D (49). Ongoing work to define box C/D snoRNP composition including specific RNA–protein and protein–protein interactions will provide a better understanding of the roles of the various proteins in box C/D snoRNP biogenesis, transport, and function.
Common Components of Box H/ACA snoRNPs
Components of the box H/ACA snoRNPs have been identified by direct biochemical purification of native snoRNP particles from yeast (72,110,221). Four proteins, Gar1p, Cbf5p, Nhp2p, and Nop10p, associate with all tested box H/ACA snoRNAs (72,92,221) (see Table 1). Each of the four proteins is essential for growth and is required for both rRNA processing and pseudouridylation (20,66,72,92). All but Gar1p are required for box H/ACA snoRNA stability/accumulation in yeast (20,72,92). Depletion of Gar1p negatively affects the ability of box H/ACA snoRNPs to interact with pre-rRNA (20). Interestingly, depletion of either Cbf5p, Nhp2p, or Nop10p results in the codepletion of Gar1p, indicating that interaction of Gar1p with each of these components is required for the stability of the Gar1 protein (72,95). Each of the four core H/ACA snoRNA binding proteins also associates with human telomerase RNA (45,134,165), which contains a conserved box H/ACA snoRNA domain (37,132,138).
Cbf5p is very likely the catalytic component of the snoRNP responsible for rRNA pseudouridylation guided by the box H/ACA snoRNAs. Cbf5p exhibits significant sequence similarity to known pseudouri-dine synthases (89), and depletion of the protein or mutation of key residues results in a specific decrease in H/ACA snoRNA-mediated rRNA pseudouridyla-tion in Saccharomyces cerevisiae (92,233). Mutation of the human homolog of Cbf5p, dyskerin, can lead to a lethal X-linked bone marrow failure disease called dyskeratosis cogenita (71). While dyskerin associates with both telomerase RNA and box H/ACA snoRNAs, it is unclear why certain dyskerin mutations lead to a selective loss of telomerase RNA in human cells (134).
Box H/ACA snoRNP particles appear to contain two sets of the four core proteins; each set may be associated with one of the two box H/ACA snoRNA hairpin structures (221). However, the assembly of the box H/ACA snoRNPs is not well understood. Nhp2p may bind directly to H/ACA snoRNAs, as it contains a putative RNA binding motif (90,221). In addition, Gar1p has been reported to interact directly with box H/ACA snoRNAs in vitro (8). The glycine-and arginine-rich (GAR) domains, which are associated with RNA binding proteins and found in the amino- and carboxy-terminal sequences of Gar1p, are not essential for the in vitro RNA binding activity of Gar1p or for yeast viability (8,66). Nhp2p, Gar1p, Cbf5p, and Nop10p may each contact the RNA, as UV cross-linking products with corresponding molecular weights have been observed (45). There appears to be considerable flexibility in the requirements for protein assembly on different snoRNAs. Formation of a U17 snoRNP or the telomerase RNP in vitro requires only the 3′ hairpin and an intact box ACA, whereas the assembly of U19 and U64 snoRNPs seems to require the entire “hairpin–hinge– hairpin–tail” structure (45).
Proteins Uniquely Associated With Specific snoRNAs
U3 snoRNA interacts with at least eight proteins that have not been found in other snoRNP complexes (Table 2). U3 is a conserved, essential box C/D snoRNA required for 18S rRNA processing rather than 2′-O-methylation (13,16,47,75,85,181). In addition to the common box C/D proteins (see above), coimmunoprecipitation analysis indicates that U3 interacts with Sof1p (82), U3-55k/Rrp9p (111,112,164,215), Mpp10p (46,103,225), Imp3p (104), Imp4p (104), Lcp5p (226), Rcl1p (14), and Dhr1p (39). Like U3, each of these proteins is specifically required for the processing of mature 18S rRNA and is essential for cell viability (14,39,46,82,104,215,226). The observation that depletion of each protein leads to specific 18S rRNA processing defects but does not affect the stability of U3 RNA indicates the primary importance of the proteins in the function rather than biogenesis of the U3 snoRNP. In addition to these proteins, the nucleolar protein nucleolin selectively interacts with U3 snoRNPs and rRNA, and is required for 18S rRNA production (65). Finally, the La protein, which is known to associate with several RNAs containing U-rich 3′ termini (172,198), associates transiently with precursor U3 snoRNAs in yeast (91).
Interactions between the various protein components of the U3 snoRNP have been described. Mpp10p associates with Imp3p, Imp4p, and Dhr1p (a putative RNA helicase), and Dhr1p also associates with Rcl1p (a protein with similarities to 3′-phos-phate cyclases) in vivo (14,39,104). However, it is not known whether these proteins interact directly or indirectly with one another. Sof1p associates with fibrillarin in vivo, and allele-specific suppression of a fibrillarin mutant by Sof1 argues for a direct interaction between these two proteins (82). In cellular extracts, U3 snoRNA is found in both 12-15S and ∼80S ribonucleoprotein particles, which likely correspond to free U3 snoRNPs and U3 snoRNPs associated with pre-rRNA and other nucleolar components, respectively (14,48,86,205). Rclp cosediments with the complex associated with pre-rRNA processing but not with the free U3 snoRNP (14). Moreover, biochemically purified yeast U3 snoRNPs contain only one U3-specific protein (223), U3-55K/Rrp9p (112,215). It is likely that the interactions of proteins with the U3 snoRNP are dynamic and change during the course of U3 snoRNP biogenesis and function. It is not yet clear what role the individual proteins play in U3 snoRNP assembly and function.
The U3 snoRNA sequences required for protein association are found primarily in the 3′ domain of U3, which includes the conserved U3-specific box B/C motif and the common box C′/D motif (Fig. 2A) (70,83,130,156,178). The core box C/D snoRNA binding protein p15.5kD/Snu13p interacts directly with the structurally similar B/C and C′/D motifs of U3 (223). Similarly, in vivo coimmunoprecipitation experiments demonstrate that association of fibrillarin with U3 requires box B, box C, box C′, and box D [W. Speckmann, R. M. Terns, and M. P. Terns, unpublished data; see also (10)]. Finally, in both vertebrate and yeast cells, the binding site of the U3-specific protein, U3-55k/Rrp9p, has been mapped to the box B/C motif, with box C being of primary importance in the interaction (112,215).
Recently, a 29-kDa protein (X29) has been purified that exhibits the properties of a specific component of U8 snoRNPs (204). Like U3, the U8 snoRNA is a member of the box C/D snoRNA family. However, while U3 is involved in 18S rRNA processing, U8 is involved in the processing of 5.8S and 28S rRNAs (i.e., components of the large ribosomal subunit) in vertebrates (159). The gene encoding this U8-specific protein and further characterization of this protein have not yet been reported.
Putative snoRNP Assembly Factors
It has recently been proposed that the assembly of snoRNP complexes may be assisted by association with Nopp140 (230). Yang et al. have shown that Nopp140 specifically interacts with both box H/ACA snoRNPs and box C/D snoRNPs (Table 3) but appears not to be an integral component of either class of snoRNP particle (230). Nonetheless, the stability of several box H/ACA snoRNAs is dependent upon the function of the yeast Nopp140 homolog, Srp4p (230). Other properties of this interesting phospho-protein are consistent with the hypothesis. Nopp140 is present in quantities exceeding those of all known snoRNAs and is highly conserved (30,127,129,155). In addition, Nopp140 colocalizes with snoRNP components in both Cajal bodies and nucleoli, and is known to shuttle between these two nuclear structures (76). Further research is required to investigate the hypothesis that snoRNP assembly is regulated by transient interaction with Nopp140.
TABLE 3.
ADDITIONAL PROTEINS ASSOCIATED WITH BOX C/D AND/OR BOX H/ACA snoRNAs
| Vertebrate Homolog | Yeast Homolog | Structural Motifs | Known or Predicted Function(s) | References |
|---|---|---|---|---|
| Nopp140 | Srp40p | Phosphoprotein, Acidic/serine rich | snoRNP biogenesis/trafficking?, Interacts with C/D and H/ACA snoRNPs | 76,127,129,230 |
| SMN | Yab8p | Tudor domain | C/D snoRNP biogenesis?, Directly interacts with fibrillarin and GAR1 | 23,54,56,69,84a,108,161a,194 |
| Ssb1p | GAR, RRM | Interacts with snR10 > snR11 | 38 | |
| Sen1p | DEAD box, NTP | RNA helicase?, snoRNA maturation/stability | 170,211 |
The assembly of snRNPs, which function in pre-mRNA splicing, is chaperoned by protein complexes containing the survival motor neuron (SMN) protein (23,54,108). The SMN protein has been shown to interact directly with Sm proteins, the core components of the snRNPs (23,56,185). Interestingly, one of the most common inheritable and fatal human diseases, spinal muscular atrophy (SMA), results from loss of functional SMN protein and leads to progressive degradation of spinal motor neurons and muscle loss (105). We have recently found that SMN also interacts with fibrillarin (a core component of box C/D snoRNPs) in HeLa cells (Table 3) (84a). Interaction of recombinant SMN and fibrillarin in vitro indicates that the proteins interact directly, and we have determined that the interaction is mediated by the Tudor domain of SMN and the glycine/arginine-rich (GAR) domain of fibrillarin (84a). Moreover, single SMN missense mutations, including an SMA-causing mutation within the Tudor domain, impair the binding of SMN to fibrillarin (84a). Interestingly, we have also found an interaction between the core box H/ ACA snoRNP protein GAR1 and SMN (S. White-head, K. Jones, R. M. Terns, and M. P. Terns, in preparation). These results indicate that SMN may play a role in snoRNP biogenesis in a similar fashion to its known role as an assembly factor for snRNP biogenesis (198a), and suggest another cellular defect associated with spinal muscular atrophy.
ANCIENT EVOLUTIONARY ORIGINS
snoRNAs and snoRNP proteins appear to have arisen over 2 billion years ago and are present today in prokaryotes as well as eukaryotes. Gene duplications in eukaryotes appear to have increased the number of proteins associated with snoRNAs.
Homologs of snoRNAs are Found in a ”Third Domain of Life“
Bacteria do not appear to share the eukaryotic system of RNA-guided rRNA modification and processing. Studies performed mainly in Escherichia coli indicate that cis-acting sequences specify sites of pre-rRNA cleavage in bacteria, and each of the relatively few sites of rRNA 2′-O-methylation and pseudouridylation in bacteria appears to be recognized and modified by a dedicated enzyme (6,151). On the other hand, it has very recently become clear that homologs of both eukaryotic box C/D snoRNAs and snoRNA-associated proteins exist in the other prokaryotic domain, Archaea (formerly known as archaebacteria) (62,153). Archaea is a primary evolutionary lineage that is phylogenetically distinct from both Bacteria and Eukarya, comprising a third domain of life (44,227,228). The divergence of Archaea and Eukarya is thought to have occurred more than 2 billion years ago (55,177,183). The occurrence of methylation guide RNPs in both Archaea and Eukarya indicates that the RNA-guided modification system is of ancient evolutionary origin.
Discovery of Archaeal Box C/D RNAs
Archaea are unicellular prokaryotic organisms and, like bacteria, archaea lack nuclei and discernible nucleoli. It was therefore intriguing to find that archaeal genomes encoded proteins homologous to the essential eukaryotic box C/D snoRNA-associated proteins, fibrillarin and Nop56 and Nop58 (2,63,93). In addition, 67 2′-O-methylations were reported in the rRNA of the archaeon Sulfolobus solfataricus, a number much higher than has been found in bacterial species and similar to that observed in eukaryotic species (146). These early observations predicted the existence of trans-acting methylation guide RNAs (counterparts of eukaryotic box C/D snoRNAs) in organisms from the domain Archaea. This prediction was very recently borne out when directed computational screens of complete archaeal genome sequence databases systematically identified over 200 RNAs predicted to guide 2′-O-ribose methylation of rRNA in seven archaeal species (62,153).
The box C/D RNAs have presumably evolved independently in Archaea and Eukarya for many years. The sequences of individual archaeal box C/D RNAs are not significantly similar to eukaryotic box C/D RNAs; indeed, the only recognizable primary sequence similarities are the short box C and box D sequence elements. In this way, the archaeal RNAs appear to resemble the diverse individual paralogous box C/D RNAs in eukaryotes. However, the archaeal RNAs are more compact than the eukaryotic RNAs, on average only ∼60 nucleotides in length (versus, for example, ∼100 nucleotides for S. cerevisiae) (62,109,153). Interestingly, the regions of complementarity to rRNA are generally shorter in the archaeal RNAs (62,153). Furthermore, unlike most eukaryotic box C/D RNAs, the vast majority of archaeal RNAs appear to guide modification of two sites by means of two functional box C/box D/guide sequence units (62,88,109,153).
Biochemical experiments confirm that the computationally predicted box C/D RNAs exist in Archaea, and indicate that the RNAs are found as RNPs including fibrillarin and Nop56/58 protein homologs. At least 18 box C/D RNAs were coimmunoprecipi-tated from S. acidocaldarius using antibodies against fibrillarin and Nop56/58 (153). Primer extension reactions have confirmed the existence of predicted box C/D RNAs from four additional archaeal species: S. solfataricus (153), Methanococcus jannaschii (153), Pyrococcus abyssi (62), and Pyrococcus furiosus (S. Mattox, R. M. Terns, and M. P. Terns, unpublished data).
Some of the sites of rRNA modification predicted from box C/D RNA guide sequences (using the guide rule derived in eukaryotes) have been verified by primer extension assays in four archaeal species (62,153). These results indicate that the RNAs also function to guide 2′-O-ribose methylation in archaea by the same “box D-plus-five nucleotide” methyla-tion targeting rule observed with the eukaryotic box C/D snoRNAs (6), though this has not been demonstrated experimentally.
Archaea includes organisms that occupy extreme ecological niches including high salt, high pressure, extreme pH, and high temperature. It is interesting that the number of predicted modifications is much higher in extreme hyperthermophilic organisms than in mesophilic archaeal organisms (153). This observation suggests a role for increased ribose methyla-tion in the thermostability of rRNA or ribosomes in hyperthermophilic organisms. In addition, the number of experimentally detected rRNA methylation sites increases when the hyperthermophilic archaeon S. solfataricus is grown at progressively higher temperatures (146), suggesting that hyperthermophilic archaea may regulate rRNA methylation in response to elevated temperature.
Evidence for Expansion of the Number of Components of the Methylation Guide RNP in Eukarya
Homologs of each of the known common eukaryotic box C/D snoRNA binding proteins have been reported in archaeal genomes (2,63,140,153,218,223). However, with the exception of fibrillarin and Nop56/58, it is not known whether the homologous proteins interact with box C/D RNAs in archaea. There are interesting differences between the eukaryotic and putative archaeal box C/D proteins. For example, all of the predicted archaeal fibrillarin proteins lack the amino-terminal GAR domain present in eukaryotic fibrillarin homologs (2,73,218). However, the most notable difference between the systems may be the apparent expansion of individual box C/D RNP protein genes in archaea to pairs of essential paralogs in eukaryotes (Fig. 3). Two pairs of very similar proteins, Nop56p and Nop58p [36% identical in mouse (140)] and p50 and p55 [42% identical in mouse (140)], selectively interact with box C/D snoRNAs in eukaryotes (94,96,114,140,229). In both cases, archaeal genomes contain a single gene that is orthologous to the two eukaryotic genes (i.e., archaeal Nop56/58 and p50/55) (140). It seems likely that ancestral Nop56/58 and p50/55 genes underwent gene duplication during the evolution of eukaryotes, thereby expanding the number of core box C/D RNP proteins from perhaps four proteins to six proteins that are each essential in higher eukaryotes (Fig. 3).
Figure 3.
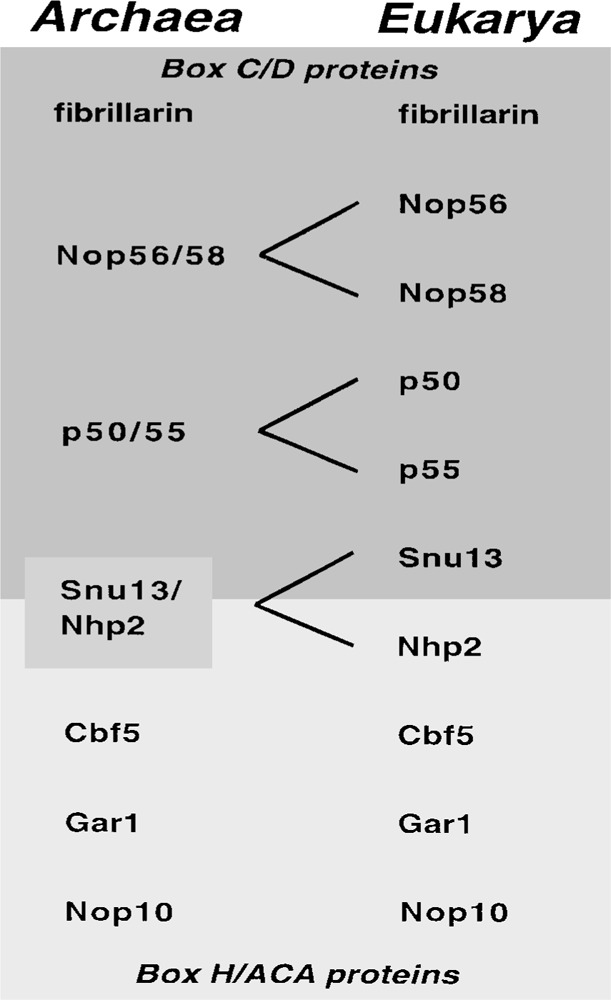
Relationships between the known protein components of box C/D and box H/ACA RNPs in eukaryotes, and homologous proteins encoded in archaeal genomes. Only fibrillarin and Nop56/ 58 have been experimentally determined to be associated with box C/D RNAs in archaea (153). Three pairs of eukaryotic snoRNP protein genes are represented by a single gene in archaea: Nop56 and Nop58 (140), p50 and p55 (140), and Snu13 and Nhp2 (R. M. Terns and M. P. Terns, see text). The archaeal Nop56/58 protein is also related to Prp31 (73). The archaeal Snu13/Nhp2 protein is also related to ribosomal protein RPL8 or L7a (R. M. Terns and M. P. Terns, see text). See text for additional details.
Another gene duplication event may have provided for the independent evolution of a box C/D protein and a box H/ACA protein in eukaryotes. Like Nop56p and Nop58p, and p50 and p55, the box C/D protein Snu13p and the box H/ACA protein Nhp2p are similar in eukaryotes (32% identical in yeast, R. M. Terns and M. P. Terns) and are represented by a single gene in each of eight archaeal genomes (R. M. Terns and M. P. Terns, data not shown). Interestingly, Snu13p and Nhp2p are also related (though to a lesser extent) to the ribosomal protein RPL8 or L7a (23% and 25% identical in yeast, R. M. Terns and M. P. Terns). The Snu13p, Nhp2p, and RPL8 proteins are each related to the archaeal ortholog to similar extents (e.g., the yeast proteins are 35%, 39%, and 35% identical to the Pyrococcus furiosus protein, R. M. Terns and M. P. Terns). Our analysis is consistent with the idea that Snu13, Nhp2, and RPL8 are paralogs that arose by two gene duplications early in the evolution of the eukaryotes. The eukaryotic Nhp2p and RPL8 proteins contain significant N-terminal (both) and C-terminal (RPL8) extensions not found in Snu13p and the archaeal ortholog. The function(s) of the Snu13/Nhp2/RPL8 ortholog in archaea remains to be determined. A similar relationship exists between eukaryotic Nop56, Nop58, and Prp31p and the orthologous archaeal protein Nop56/Nop58/ Prp31 (73). In this case, the archaeal protein has been found to associate with box C/D RNAs (153).
Despite the divergence, recent experiments in our laboratory indicate that the essential RNA/protein recognition elements of box C/D RNPs are remarkably well conserved between archaea and eukaryotes. We have microinjected box C/D RNAs from Pyrococcus furiosus into nuclei of oocytes from the aquatic frog Xenopus laevis, and found that the archaeal RNAs are retained in the nucleus, localize to nucleoli, interact with Xenopus fibrillarin, Nop56, and Nop58, and direct site-specific modification of rRNA (192a). This extraordinary recognition between two systems that have evolved independently for more than 2 billion years (55) may reflect the simple restriction imposed by the requirement for coevolution of the proteins with multiple RNAs. Once multiple guide RNAs became established in a common ancestor of Archaea and Eukarya, covariation of an individual RNA and common protein would require the unlikely simultaneous covariation of the other RNAs. Thus, many years later, the eukaryotic proteins recognize archaeal box C/D RNAs.
To date, none of the identified archaeal box C/D RNAs appears to represent homologs of eukaryotic snoRNAs that function in pre-rRNA cleavage events (e.g., U3 snoRNA). Furthermore, only one of the proteins associated specifically with U3 in eukaryotes, Imp4p, has been found in archaeal genomes [(124); R. M. Terns and M. P. Terns, data not shown]. However, the observation that in vitro rRNA processing in a cell-free system derived from Sulfolobus acidocaldarius appears to require one or more essential RNA cofactors indicates the potential involvement of trans-acting RNAs in rRNA cleavages in archaeal species (166,167).
Evidence Suggesting the Existence of Box H/ACA snoRNA Homologs in Archaea
The presence of box C/D RNPs in Archaea naturally begs the question as to whether box H/ACA snoRNPs will also be found in this domain. Archaeal ribosomal RNAs contain pseudouridine (120,146,150), which could be introduced by multiple pseudouridine synthases (as in bacteria) or via small guide RNAs that mark sites and recruit a common pseudouridine synthase (as in eukaryotes). The number of pseudouridine residues present in rRNA from archaea is much lower than that observed in eukaryotes, and is similar to that observed in bacteria (151). This observation might suggest that H/ACA snoRNA homologs do not exist in archaea. However, the recent realization that archaeal genomes encode proteins homologous to all of the known eukaryotic box H/ACA snoRNA-associated proteins (Fig. 3) (128,219,221) makes the question more interesting again. Watanabe and Gray have shown that the archaeal protein annotated as a TruB homolog is more similar to Cbf5p (the putative box H/ACA RNA-associated pseudouridine synthase) (219). However, the archaeal homolog of Nhp2p (219) is also related to Snu13p and RPL8/L7a (R. M. Terns and M. P. Terns, see above), and it is not clear that the protein would function as a box H/ACA protein in archaea. Computational search algorithms (similar to ones that identified the box C/D RNAs in yeast and archaea) have not yet been developed for box H/ACA RNAs.
CONCLUDING REMARKS
Investigation of the snoRNAs has revealed a large family of trans-acting RNAs of great diversity and versatility. Recent work has shown that the targets of the snoRNAs may include snRNAs and mRNAs as well as rRNAs. The genomic origins of the snoRNAs are also varied and include salvage from mRNA introns. Furthermore, it has recently become clear that snoRNAs exist not only in eukaryotes but also in diverse organisms from the prokaryotic domain Archaea. These findings have resulted in an explosion in our comprehension of the numbers and functions of nonprotein-coding RNAs that may be encoded in the genomes of organisms.
snoRNA research is also providing inroads in other areas of biological importance. The unexpected realization that telomerase RNA is a hybrid snoRNA is contributing to our understanding of this key RNA. Furthermore, definition of the RNA motifs responsible for cellular trafficking of snoRNAs has provided new means to direct the localization of other RNAs including ribozymes. The impressive developments of the last few years provide exciting new research frontiers in the field of snoRNA biology.
ACKNOWLEDGMENTS
We thank Jean-Pierre Bachellerie, Witold Filipowicz, Skip Fournier, Tamas Kiss, Stu Maxwell, and Joan Steitz for providing results prior to publication. We are grateful to the National Institutes of Health, National Science Foundation, and the American Cancer Society for supporting our work.
REFERENCES
- 1. Allmang C.; Kufel J.; Chanfreau G.; Mitchell P.; Petfalski E.; Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399–5410; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amiri K. A. Fibrillarin-like proteins occur in the domain Archaea. J. Bacteriol. 176:2124–2127; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aris J. P.; Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA 88:931–935; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Artandi S. E.; DePinho R. A. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39–46; 2000. [DOI] [PubMed] [Google Scholar]
- 5. Autexier C.; Pruzan R.; Funk W. D.; Greider C. W. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 15:5928–5935; 1996. [PMC free article] [PubMed] [Google Scholar]
- 6. Bachellerie J.-P.; Cavaille J. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Grosjean H.; Benne B., eds. Modification and editing of RNA. Washington, DC: ASM Press; 1998:255–272. [Google Scholar]
- 7. Bachellerie J. P.; Cavaill J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257–261; 1997. [DOI] [PubMed] [Google Scholar]
- 8. Bagni C.; Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J. Biol. Chem. 273:10868–10873; 1998. [DOI] [PubMed] [Google Scholar]
- 9. Balakin A.; Smith L.; Fournier M. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86:823–834; 1996. [DOI] [PubMed] [Google Scholar]
- 10. Baserga S. J.; Yang X. D.; Steitz J. A. An intact box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 10:2645–2651; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bauer D.; Murphy C.; Wu Z.; Wu C.; Gall J. In vitro assembly of coiled bodies in Xenopus egg extract. Mol. Biol. Cell 5:633–644; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beattie T. L.; Zhou W.; Robinson M. O.; Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177–180; 1998. [DOI] [PubMed] [Google Scholar]
- 13. Beltrame M.; Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14:4350–4356; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Billy E.; Wegierski T.; Nasr F.; Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19:2115–2126; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bohmann K.; Ferreira J.; Lamond A. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J. Cell Biol. 131:817–831; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borovjagin A. V.; Gerbi S. A. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 286:1347–1363; 1999. [DOI] [PubMed] [Google Scholar]
- 17. Bortolin M. L.; Ganot P.; Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ri-bosomal RNAs. EMBO J. 18:457–469; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bortolin M. L.; Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4:445–454; 1998. [PMC free article] [PubMed] [Google Scholar]
- 19. Boudonck K.; Dolan L.; Shaw P. J. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol. Biol. Cell 10:2297–2307; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bousquet-Antonelli C.; Henry Y.; G’elugne J. P.; Caizergues-Ferrer M.; Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 16:4770–4776; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brimacombe R.; Mitchell P.; Osswald M.; Stade K.; Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 7:161–167; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Bryan T. M.; Cech T. R. Telomerase and the maintenance of chromosome ends. Curr. Opin. Cell. Biol. 11:318–324; 1999. [DOI] [PubMed] [Google Scholar]
- 23. Buhler D.; Raker V.; Luhrmann R.; Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: Implications for spinal muscular atrophy. Hum. Mol. Genet. 8:2351–2357; 1999. [DOI] [PubMed] [Google Scholar]
- 24. Buonomo S. B.; Michienzi A.; De Angelis F. G.; Bozzoni I. The Rev protein is able to transport to the cytoplasm small nucleolar RNAs containing a Rev binding element. RNA 5:993–1002; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burns C. M.; Chu H.; Rueter S. M.; Hutchinson L. K.; Canton H.; Sanders-Bush E.; Emeson R. B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308; 1997. [DOI] [PubMed] [Google Scholar]
- 26. Cadwell C.; Yoon H. J.; Zebarjadian Y.; Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 17:6175–6183; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caffarelli E.; Arese M.; Santoro B.; Fragapane P.; Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis . Mol. Cell. Biol. 14:2966–2974; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caffarelli E.; Fatica A.; Prislei S.; De Gregorio E.; Fragapane P.; Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15:1121–1131; 1996. [PMC free article] [PubMed] [Google Scholar]
- 29. Caffarelli E.; Losito M.; Giorgi C.; Fatica A.; Bozzoni I. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol. 18:1023–1028; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cairns C.; McStay B. Identification and cDNA cloning of a Xenopus nucleolar phosphoprotein, xNopp180, that is the homolog of the rat nucleolar protein Nopp140. J. Cell Sci. 108:3339–3347; 1995. [DOI] [PubMed] [Google Scholar]
- 31. Cavaillé J.; Bachellerie J. SnoRNA-guided ribose methylation of rRNA: Structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 26:1576–1587; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cavaillé J.; Buiting K.; Kiefmann M.; Lalande M.; Brannan C. I.; Horsthemke B.; Bachellerie J. P.; Brosius J.; Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97:14311–14316; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cavaillé J.; Nicoloso M.; Bachellerie J. P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732–735; 1996. [DOI] [PubMed] [Google Scholar]
- 34. Chanfreau G.; Legrain P.; Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975–988; 1998. [DOI] [PubMed] [Google Scholar]
- 35. Chanfreau G.; Rotondo G.; Legrain P.; Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726–3737; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen C. X.; Cho D. S.; Wang Q,; Lai F.; Carter K. C.; Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6:755–767; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J. L.; Blasco M. A.; Greider C. W. Secondary structure of vertebrate telomerase RNA. Cell 100:503–514; 2000. [DOI] [PubMed] [Google Scholar]
- 38. Clark M. W.; Yip M. L.; Campbell J.; Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J. Cell Biol. 111:1741–1751; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colley A.; Beggs J. D.; Tollervey D.; Lafontaine D. L. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol. Cell. Biol. 20:7238–7246; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Collins K. Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12:378–383; 2000. [DOI] [PubMed] [Google Scholar]
- 41. Darzacq X.; Kiss T. Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5′,3′-terminal stem structure. Mol. Cell. Biol. 20:4522–4531; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Los Santos T.; Schweizer J.; Rees C. A.; Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am. J. Hum. Genet. 67:1067–1082; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dez C.; Henras A.; Faucon B.; Lafontaine D. L.; Caizergues-Ferrer M.; Henry Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598–603; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doolittle W. F.; Brown J. R. Tempo, mode, the progenote, and the universal root. Proc. Natl. Acad. Sci. USA 91:6721–6728; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dragon F.; Pogacic V.; Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037–3048; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dunbar D.; Wormsley S.; Agentis T.; Baserga S. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17:5803–5812; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Enright C.; Maxwell E.; Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 2:1094–1099; 1996. [PMC free article] [PubMed] [Google Scholar]
- 48. Epstein P.; Reddy R.; Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry 23:5421–5425; 1984. [DOI] [PubMed] [Google Scholar]
- 49. Fatica A.; Galardi S.; Altieri F.; Bozzoni I. Fibrillarin binds directly and specifically to U16 box C/D snoRNA. RNA 6:88–95; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fatica A.; Morlando M.; Bozzoni I. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218–6229; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feng J.; Funk W. D.; Wang S. S.; Weinrich S. L.; Avilion A. A.; Chiu C. P.; Adams R. R.; Chang E.; Allsopp R. C.; Yu J.; et al. The RNA component of human telomerase. Science 269:1236–1241; 1995. [DOI] [PubMed] [Google Scholar]
- 52. Filipowicz W. Imprinted expression of small nucleolar RNAs in brain: Time for RNomics. Proc. Natl. Acad. Sci. USA 97:14035–14037; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Filipowicz W.; Pelczar P.; Pogacic V.; Dragon F. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 46:377–389; 1999. [PubMed] [Google Scholar]
- 54. Fischer U.; Liu Q.; Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023–1029; 1997. [DOI] [PubMed] [Google Scholar]
- 55. Fox G. E.; Stackebrandt E.; Hespell R. B.; Gibson J.; Maniloff J.; Dyer T. A.; Wolfe R. S.; Balch W. E.; Tanner R. S.; Magrum L. J.; Zablen L. B.; Blakemore R.; Gupta R.; Bonen L.; Lewis B. J.; Stahl D. A.; Luehrsen K. R.; Chen K. N.; Woese C. R. The phylogeny of prokaryotes. Science 209:457–463; 1980. [DOI] [PubMed] [Google Scholar]
- 56. Friesen W. J.; Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem. 275:26370–26375; 2000. [DOI] [PubMed] [Google Scholar]
- 57. Gall J. G.; Bellini M.; Wu Z.; Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10:4385–4402; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ganot P.; Bortolin M. L.; Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799–809; 1997. [DOI] [PubMed] [Google Scholar]
- 59. Ganot P.; Caizergues-Ferrer M.; Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941–956; 1997. [DOI] [PubMed] [Google Scholar]
- 60. Ganot P.; Jady B. E.; Bortolin M. L.; Darzacq X.; Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceoso-mal RNA. Mol. Cell. Biol. 19:6906–6917; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao L.; Frey M.; Matera A. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 25:4740–4747; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaspin C.; Cavaille J.; Erauso G.; Bachellerie J. P. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J. Mol. Biol. 297:895–906; 2000. [DOI] [PubMed] [Google Scholar]
- 63. Gautier T.; Berges T.; Tollervey D.; Hurt E. Nu-cleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088–7098; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gerbi S. Small nucleolar RNA. Biochem. Cell. Biol. 73:845–858; 1995. [DOI] [PubMed] [Google Scholar]
- 65. Ginisty H.; Amalric F.; Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476–1486; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Girard J. P.; Lehtonen H.; Caizergues-Ferrer M.; Amalric F.; Tollervey D.; Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 11:673–682; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Greider C. W. Telomerase activation. One step on the road to cancer? Trends Genet. 15:109–112; 1999. [DOI] [PubMed] [Google Scholar]
- 68. Greider C. W. Telomerase activity, cell proliferation, and cancer. Proc. Natl. Acad. Sci. USA 95:90–92; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hannus S.; Buhler D.; Romano M.; Seraphin B.; Fischer U. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 9:663–6674; 2000. [DOI] [PubMed] [Google Scholar]
- 70. Hartshorne T.; Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 22:3354–3364; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heiss N. S.; Knight S. W.; Vulliamy T. J.; Klauck S. M.; Wiemann S.; Mason P. J.; Poustka A.; Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19:32–38; 1998. [DOI] [PubMed] [Google Scholar]
- 72. Henras A.; Henry Y.; Bousquet-Antonelli C.; Noaillac-Depeyre J.; Gélugne J. P.; Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078–7090; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hickey A. J.; Macario A. J.; Conway de Macario E. Identification of genes in the genome of the archaeon Methanosarcina mazeii that code for homologs of nuclear eukaryotic molecules involved in RNA processing. Gene 253:77–85; 2000. [DOI] [PubMed] [Google Scholar]
- 74. Holt S. E.; Shay J. W. Role of telomerase in cellular proliferation and cancer. J. Cell. Physiol. 180:10–18; 1999. [DOI] [PubMed] [Google Scholar]
- 75. Hughes J.; Ares M. J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231–4239; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Isaac C.; Yang Y.; Meier U. T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell. Biol. 142:319–329; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Izaurralde E.; Mattaj I. W. RNA export. Cell 81:153–159; 1995. [DOI] [PubMed] [Google Scholar]
- 78. Jacobson M.; Pederson T. A 7-methylguanosine cap commits U3 and U8 small nuclear RNAs to the nucleolar localization pathway. Nucleic Acids Res. 26:756–760; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jady B. E.; Kiss T. Characterisation of the U83 and U84 small nucleolar RNAs: Two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res. 28:1348–1354; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jady B. E.; Kiss T. A small nucleolar guide RNA functons both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosoal RNA. EMBO J. 20:541–551; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. James H. A.; Gibson I. The therapeutic potential of ribozymes. Blood 91:371–382; 1998. [PubMed] [Google Scholar]
- 82. Jansen R.; Tollervey D.; Hurt E. C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 12:2549–2558; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jeppesen C.; Stebbins B. B.; Gerbi S. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 16:2127–2148; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jimenez-Garcia L.; Segura-Valdez M.; Ochs R.; Rothblum L.; Hannan R.; Spector D. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell 5:955–966; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84a. Jones K. W.; Gorzynski K.; Hales C. M.; Fischer V.; Badbanch F.; Terns R. M.; Terns M. P. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarian. J. Biol. Chem. 276:38645–38651; 2001. [DOI] [PubMed] [Google Scholar]
- 85. Kass S.; Tyc K.; Steitz J. A.; Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60:897–908; 1990. [DOI] [PubMed] [Google Scholar]
- 85a. King T. H.; Decatur W. A.; Betrand E.; Maxwell E. S.; Fournier M. J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21:7731–7746; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kiss T.; Marshallsay C.; Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65:517–526; 1991. [DOI] [PubMed] [Google Scholar]
- 87. Kiss-László Z.; Henry Y.; Bachellerie J. P.; Caizergues-Ferrer M.; Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85:1077–1088; 1996. [DOI] [PubMed] [Google Scholar]
- 88. Kiss-László Z.; Henry Y.; Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17:797–807; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Koonin E. V. Pseudouridine synthases: Four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP de-aminases. Nucleic Acids Res. 24:2411–2415; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koonin E. V.; Bork P.; Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22:2166–2167; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kufel J.; Allmang C.; Chanfreau G.; Petfalski E.; Lafontaine D. L.; Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415–5424; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lafontaine D. L.; Tollervey D. Birth of the snoRNPs: The evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 23:383–388; 1998. [DOI] [PubMed] [Google Scholar]
- 94. Lafontaine D. L.; Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650–2659; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lafontaine D. L. J.; Bousquet-Antonelli C.; Henry Y.; Caizergues-Ferrer M.; Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527–537; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lafontaine D. L. J.; Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455–467; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lange T.; Borovjagin A.; Gerbi S. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA 4:789–800; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lange T.; Ezrokhi M.; Borovjagin A.; Rivera-Leon R.; North M.; Gerbi S. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol. Biol. Cell 9:2973–2985; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lange T. S.; Borovjagin A.; Maxwell E. S.; Gerbi S. A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 17:3176–3187; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lange T. S.; Ezrokhi M.; Amaldi F.; Gerbi S. A. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 10:3877–3890; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lange T. S.; Gerbi S. A. Transient nucleolar localization Of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell 11:2419–2428; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Leader D. J.; Clark G. P.; Watters J.; Beven A. F.; Shaw P. J.; Brown J. W. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 16:5742–5751; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lee S. J.; Baserga S. J. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl. Acad. Sci. USA 94:13536–13541; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lee S. J.; Baserga S. J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribo-nucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441–5452; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lefebvre S.; Burglen L.; Reboullet S.; Clermont O.; Burlet P.; Viollet L.; Benichou B.; Cruaud C.; Millasseau P.; Zeviani M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165; 1995. [DOI] [PubMed] [Google Scholar]
- 106. Lindahl L.; Zengel J. M. RNase MRP and rRNA processing. Mol. Biol. Rep. 22:69–73; 1995. [DOI] [PubMed] [Google Scholar]
- 107. Liu B.; Ni J.; Fournier M. J. Probing RNA in vivo with methylation guide snoRNAs. Methods 23:276–286; 2001. [DOI] [PubMed] [Google Scholar]
- 108. Liu Q.; Fischer U.; Wang F.; Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013–1021; 1997. [DOI] [PubMed] [Google Scholar]
- 109. Lowe T. M.; Eddy S. R. A computational screen for methylation guide snoRNAs in yeast. Science 283:1168–1171; 1999. [DOI] [PubMed] [Google Scholar]
- 110. Lubben B.; Fabrizio P.; Kastner B.; Luhrmann R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae . J. Biol. Chem. 270:11549–11554; 1995. [DOI] [PubMed] [Google Scholar]
- 111. Lubben B.; Marshallsay C.; Rottmann N.; Luhrmann R. Isolation of U3 snoRNP from CHO cells: A novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 21:5377–5385; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lukowiak A. A.; Granneman S.; Mattox S. A.; Speckmann W. A.; Jones K.; Pluk H.; Venrooij W. J.; Terns R. M.; Terns M. P. Interaction of the U3-55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55k. Nucleic Acids Res. 28:3462–3471; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112a. Lukowiak A. A.; Narayanan A.; Li Z.-H.; Terns R. M.; Terns M. P. The snoRNA domain of human telomerase RNA functions to localize the RNA within the nucleus. RNA (in press). [PMC free article] [PubMed] [Google Scholar]
- 113. Lygerou Z.; Allmang C.; Tollervey D.; Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268–270; 1996. [DOI] [PubMed] [Google Scholar]
- 114. Lyman S. K.; Gerace L.; Baserga S. J. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 5:1597–1604; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lyon C.; Bohmann K.; Sleeman J.; Lamond A. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 230:84–93; 1997. [DOI] [PubMed] [Google Scholar]
- 116. Maden B. E. Eukaryotic rRNA methylation: The calm before the Sno storm. Trends Biochem. Sci. 23:447–450; 1998. [DOI] [PubMed] [Google Scholar]
- 117. Maden B. E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39:241–303; 1990. [DOI] [PubMed] [Google Scholar]
- 118. Makino Y.; Kanemaki M.; Kurokawa Y.; Koji T.; Tamura T. A rat RuvB-like protein, TIP49a, is a germ cell-enriched novel DNA helicase. J. Biol. Chem. 274:15329–15335; 1999. [DOI] [PubMed] [Google Scholar]
- 119. Malatesta M.; Zancanaro C.; Martin T.; Chan E.; Amalric F.; Luhrmann R.; Vogel P.; Fakan S. Is the coiled body involved in nucleolar functions? Exp. Cell Res. 211:415–419; 1994. [DOI] [PubMed] [Google Scholar]
- 120. Massenet S.; Ansmant I.; Motorin Y.; Branlant C. The first determination of pseudouridine residues in 23S ribosomal RNA from hyperthermophilic Archaea Sulfolobus acidocaldarius . FEBS Lett. 462:94–100; 1999. [DOI] [PubMed] [Google Scholar]
- 121. Matera A. G. Of coiled bodies, gems, and salmon. J. Cell Biochem. 70:181–192; 1998. [PubMed] [Google Scholar]
- 122. Mattaj I. W.; Englmeier L. Nucleocytoplasmic transport: The soluble phase. Annu. Rev. Biochem. 67:265–306; 1998. [DOI] [PubMed] [Google Scholar]
- 123. Maxwell E. S.; Fournier M. J. The small nucleolar RNAs. Annu. Rev. Biochem. 35:897–933; 1995. [DOI] [PubMed] [Google Scholar]
- 124. Mayer C.; Suck D.; Poch O. The archaeal homolog of the Imp4 protein, a eukaryoticv U3 snoRNP component. Trends Biochem. Sci. 26:143–144; 2001. [DOI] [PubMed] [Google Scholar]
- 125. McCloskey J.; Crain P. The RNA modification database—1998. Nucleic Acids Res. 26:196–197; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Meguro M.; Mitsuya K.; Nomura N.; Kohda M.; Kashiwagi A.; Nishigaki R.; Yoshioka H.; Nakao M.; Oishi M.; Oshimura M. Large-scale evaluation of imprinting status in the Prader-Willi syndrome region: An imprinted direct repeat cluster resembling small nucleolar RNA genes. Hum. Mol. Genet. 10:383–394; 2001. [DOI] [PubMed] [Google Scholar]
- 127. Meier U. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 271:19376–19384; 1996. [PubMed] [Google Scholar]
- 128. Meier U.; Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 127:1505–1514; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Meier U. T.; Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 70:127–138; 1992. [DOI] [PubMed] [Google Scholar]
- 130. Mereau A.; Fournier R.; Gregoire A.; Mougin A.; Fabrizio P.; Luhrmann R.; Branlant C. An in vivo and in vitro structure–function analysis of the Saccharomyces cerevisiae U3A snoRNP: Protein–RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 273:552–571; 1997. [DOI] [PubMed] [Google Scholar]
- 131. Michienzi A.; Cagnon L.; Bahner I.; Rossi J. J. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 97:8955–8960; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mitchell J. R.; Cheng J.; Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567–576; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mitchell J. R.; Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell. 6:361–371; 2000. [DOI] [PubMed] [Google Scholar]
- 134. Mitchell J. R.; Wood E.; Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551–555; 1999. [DOI] [PubMed] [Google Scholar]
- 135. Mitchell P.; Tollervey D. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7:843–846; 2000. [DOI] [PubMed] [Google Scholar]
- 136. Morrissey J. P.; Tollervey D. Birth of the snoRNPs: The evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem. Sci. 20:78–82; 1995. [DOI] [PubMed] [Google Scholar]
- 137. Nakielny S.; Fischer U.; Michael W. M.; Dreyfuss G. RNA transport. Annu. Rev. Neurosci. 20:269–301; 1997. [DOI] [PubMed] [Google Scholar]
- 138. Narayanan A.; Lukowiak A..; Jady B. E.; Dragon F.; Kiss T.; Terns R. M.; Terns M. P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 18:5120–5130; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Narayanan A.; Speckmann W.; Terns R.; Terns M. Role of the box C/D motif in the localization of small nucleolar RNAs to nucleoli and coiled bodies. Mol. Biol. Cell 10:2131–2147; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Newman D. R.; Kuhn J. F.; Shanab G. M.; Maxwell E. S. Box C/D snoRNA-associated proteins: Two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6:861–879; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ni J.; Samarsky D. A.; Liu B.; Ferbeyre G.; Cedergren R.; Fournier M. J. SnoRNAs as tools for RNA cleavage and modification. Nucleic Acids Symp. Ser. 61–63; 1997. [PubMed] [Google Scholar]
- 142. Ni J.; Tien A.; Fournier M. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565–573; 1997. [DOI] [PubMed] [Google Scholar]
- 143. Nicoloso M.; Qu L. H.; Michot B.; Bachellerie J. P. Intron-encoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol. 260:178–195; 1996. [DOI] [PubMed] [Google Scholar]
- 144. Niewmierzycka A.; Clarke S. S-Adenosylmethio-nine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274:814–824; 1999. [DOI] [PubMed] [Google Scholar]
- 145. Niswender C. M.; Copeland S. C.; Herrick-Davis K.; Emeson R. B.; Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 274:9472–9478; 1999. [DOI] [PubMed] [Google Scholar]
- 146. Noon K. R.; Bruenger E.; McCloskey J. A. Post-transcriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus . J. Bacteriol. 180:2883–2888; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Nottrott S.; Hartmuth K.; Fabrizio P.; Urlaub H.; Vidovic I.; Ficner R.; Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119–6133; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ochs R. L.; Lischwe M. A.; Spohn W. H.; Busch H. Fibrillarin: A new protein of the nucleolus identified by autoimmune sera. Biol. Cell 54:123–133; 1985. [DOI] [PubMed] [Google Scholar]
- 149. Ochs R. L.; Stein T. W. Jr.; Tan E. M. Coiled bodies in the nucleolus of breast cancer cells. J. Cell Sci. 107:385–399; 1994. [DOI] [PubMed] [Google Scholar]
- 150. Ofengand J.; Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266:246–268; 1997. [DOI] [PubMed] [Google Scholar]
- 151. Ofengand J.; Fournier M. J. The pseudouridine residues of rRNA: Number, location, biosynthesis, and function. In: Grosjean H.; Benne B., eds. Modification and editing of RNA. Washington, DC: ASM Press; 1998:229–253. [Google Scholar]
- 152. Olson M. O.; Dundr M.; Szebeni A. The nucleolus: An old factory with unexpected capabilities. Trends Cell. Biol. 10:189–196; 2000. [DOI] [PubMed] [Google Scholar]
- 153. Omer A. D.; Lowe T. M.; Russell A. G.; Ebhardt H.; Eddy S. R.; Dennis P. P. Homologs of small nucleolar RNAs in Archaea. Science 288:517–522; 2000. [DOI] [PubMed] [Google Scholar]
- 154. Ooi S. L.; Samarsky D. A.; Fournier M. J.; Boeke J. D. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA 4:1096–1110; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pai C. Y.; Chen H. K.; Sheu H. L.; Yeh N. H. Cell-cycle-dependent alterations of a highly phosphorylated nucleolar protein p130 are associated with nucleologenesis. J. Cell Sci. 108:1911–1920; 1995. [DOI] [PubMed] [Google Scholar]
- 156. Parker K.; Steitz J. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol. Cell. Biol. 7:2899–2913; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Paul M. S.; Bass B. L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17:1120–1127; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Peculis B. RNA processing: Pocket guides to ribosomal RNA. Curr. Biol. 7:R480–482; 1997. [DOI] [PubMed] [Google Scholar]
- 159. Peculis B. A.; Steitz J. A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73:1233–1245; 1993. [DOI] [PubMed] [Google Scholar]
- 160. Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871–3876; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Pelczar P.; Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol. 18:4509–4518; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161a. Pellizzoni L.; Baccon J.; Charroux B.; Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11:1079–1088; 2001. [DOI] [PubMed] [Google Scholar]
- 162. Petfalski E.; Dandekar T.; Henry Y.; Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181–1189; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Phylactou L. A.; Kilpatrick M. W.; Wood M. J. Ribozymes as therapeutic tools for genetic disease. Hum. Mol. Genet. 7:1649–1653; 1998. [DOI] [PubMed] [Google Scholar]
- 164. Pluk H.; Soffner J.; Luhrmann R.; Van V. W. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol. Cell. Biol. 18:488–498; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Pogacic V.; Dragon R.; Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20:9028–9040; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Potter S.; Durovic P.; Dennis P. P. Ribosomal RNA precursor processing by a eukaryotic U3 small nucleolar RNA-like molecule in an archaeon. Science 268:1056–1060; 1995. [Retracted by Russell, A. G.; de Sa M. M.; Dennis, P. P. Science 277(5330):1189; 1997] [DOI] [PubMed] [Google Scholar]
- 167. Potter S.; Durovic P.; Russell A.; Wang X.; de Jong-Wong D.; Dennis P. P. Preribosomal RNA processing in archaea: Characterization of the RNP endonuclease mediated processing of precursor 16S rRNA in the thermoacidophile Sulfolobus acidocal-darius . Biochem. Cell. Biol. 73:813–823; 1995. [DOI] [PubMed] [Google Scholar]
- 168. Qu L. H.; Henras A.; Lu Y. J.; Zhou H.; Zhou W. X.; Zhu Y. Q.; Zhao J.; Henry Y.; Caizergues-Ferrer M.; Bachellerie J. P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19:1144–1158; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Raska I.; Andrade L.; Ochs R.; Chan E.; Chang C.; Roos G.; Tan E. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 195:27–37; 1991. [DOI] [PubMed] [Google Scholar]
- 170. Rasmussen T. P.; Culbertson M. R. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 18:6885–6896; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Reilly T. H.; Schmitt M. E. The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Mol. Biol. Rep. 22:87–93; 1995. [DOI] [PubMed] [Google Scholar]
- 172. Rinke J.; Steitz J. A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell 29:149–159; 1982. [DOI] [PubMed] [Google Scholar]
- 173. Rossi J. J. Ribozyme therapy for HIV infection. Adv. Drug Deliv. Rev. 44:71–78; 2000. [DOI] [PubMed] [Google Scholar]
- 174. Rossi J. J. Ribozymes in the nucleolus. Science 285:1685; 1999. [DOI] [PubMed] [Google Scholar]
- 175. Rossi J. J. Therapeutic antisense and ribozymes. Br. Med. Bull. 51:217–225; 1995. [DOI] [PubMed] [Google Scholar]
- 176. Ruhl D. D.; Pusateri M. E.; Eliceiri G. L. Multiple conserved segments of E1 small nucleolar RNA are involved in the formation of a ribonucleoprotein particle in frog oocytes. Biochem. J. 348(Pt. 3):517–524; 2000. [PMC free article] [PubMed] [Google Scholar]
- 177. Runnegar B. Proterozoic eukaryotes: Evidence from biology and geology. In: Bengtson S., ed. Early life on earth. New York: Columbia University Press; 1994:287–297. [Google Scholar]
- 178. Samarsky D.; Fournier M. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae . Mol. Cell. Biol. 18:3431–444; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Samarsky D. A.; Ferbeyre G.; Bertrand E.; Singer R. H.; Cedergren R.; Fournier M. J. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. USA 96:6609–6614; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Samarsky D. A.; Fournier M. J.; Singer R. H.; Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17:3747–3757; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Savino R.; Gerbi S. A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9:2299–2308; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Schimmang T.; Tollervey D.; Kern H.; Frank R.; Hurt E. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015–4024; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Schopf J. W. Tracing the roots of the Universal Tree of Life. In: Brack A., ed. The molecular origins of life: Assembling pieces of the puzzle. Cambridge: Cambridge University Press; 1998:337–362. [Google Scholar]
- 184. Schul W.; Adelaar B.; van Driel R.; de Jong L. Coiled bodies are predisposed to a spatial association with genes that contain snoRNA sequences in their introns. J. Cell Biochem. 75:393–403; 1999. [PubMed] [Google Scholar]
- 185. Selenko P.; Sprangers R.; Stier G.; Buhler D.; Fischer U.; Sattler M. SMN Tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8:27–31; 2001. [DOI] [PubMed] [Google Scholar]
- 186. Shaw P.; Beven A.; Leader D.; Brown J. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J. Cell Sci. 111:2121–2128; 1998. [DOI] [PubMed] [Google Scholar]
- 187. Sleeman J.; Lyon C. E.; Platani M.; Kreivi J. P.; Lamond A. I. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 243:290–304; 1998. [DOI] [PubMed] [Google Scholar]
- 188. Smith C.; Steitz J. Sno storm in the nucleolus: New roles for myriad small RNPs. Cell 89:669–672; 1997. [DOI] [PubMed] [Google Scholar]
- 189. Smith C. M.; Steitz J. A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18:6897–6909; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Sollner-Webb B. Novel intron-encoded small nucle-olar RNAs. Cell 75:403–405; 1993. [DOI] [PubMed] [Google Scholar]
- 191. Speckmann W.; Narayanan A.; Terns R.; Terns M. P. Nuclear retention elements of U3 small nucleolar RNA. Mol. Cell. Biol. 19:8412–8421; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Speckmann W. A.; Terns R. M.; Terns M. P. The box C/D motif directs snoRNA 5′ cap hypermethylation. Nucleic Acids Res. 28:4467–4473; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192a. Speckmann W.; Li Z.-H.; Lowe T.; Eddy S.; Terns R. M.; Terns M. P. Archaeal guide RNAs function in rRNA modification in the eukaryotic nucleus. Curr. Biol. (in press). [DOI] [PubMed] [Google Scholar]
- 193. Steitz J.; Tycowski K. Small RNA chaperones for ribosome biogenesis. Science 270:1626–1627; 1995. [DOI] [PubMed] [Google Scholar]
- 194. Talbot K.; Ponting C. P.; Theodosiou A. M.; Rodrigues R. N.; Surtees R.; Mountford R.; Davies K. E. Missense mutation clustering in the survival motor neuron gene: A role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum. Mol. Genet. 6:497–500.; 1997. [DOI] [PubMed] [Google Scholar]
- 195. Terns M.; Goldfarb D. Nuclear transport of RNAs in microinjected Xenopus oocytes. Methods Cell Biol. 53:559–589; 1998. [DOI] [PubMed] [Google Scholar]
- 196. Terns M. P.; Dahlberg J. E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science 264:959–961; 1994. [DOI] [PubMed] [Google Scholar]
- 197. Terns M. P.; Grimm C.; Lund E.; Dahlberg J. E. A common maturation pathway for small nucleolar RNAs. EMBO J. 14:4860–4871; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Terns M. P.; Lund E.; Dahlberg J. E. 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol. 12:3032–3040; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198a. Terns M. P.; Terns R. M. Macromolecular complexes: SMN—the master assembler. Curr. Biol. 11:R862–R864; 2001. [DOI] [PubMed] [Google Scholar]
- 199. Tesmer V. M.; Ford L. P.; Holt S. E.; Frank B. C.; Yi X.; Aisner D. L.; Ouellette M.; Shay J. W.; Wright W. E. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hT-ERT) in vitro. Mol. Cell. Biol. 19:6207–6216; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tollervey D. Small nucleolar RNAs guide ribosomal RNA methylation. Science 273:1056–1057; 1996. [DOI] [PubMed] [Google Scholar]
- 201. Tollervey D.; Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell. Biol. 9:337–342; 1997. [DOI] [PubMed] [Google Scholar]
- 202. Tollervey D.; Lehtonen H.; Carmo-Fonseca M.; Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10:573–583; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Tollervey D.; Lehtonen H.; Jansen R.; Kern H.; Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443–457; 1993. [DOI] [PubMed] [Google Scholar]
- 204. Tomasevic N.; Peculis B. Identification of a U8 snoRNA-specific binding protein. J. Biol. Chem. 274:35914–35920; 1999. [DOI] [PubMed] [Google Scholar]
- 205. Tyc K.; Steitz J. A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113–3119; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Tycowski K.; Smith C.; Shu M.; Steitz J. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus . Proc. Natl. Acad. Sci. USA 93:14480–14485; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Tycowski K. T.; Shu M. D.; Steitz J. A. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379:464–466; 1996. [DOI] [PubMed] [Google Scholar]
- 208. Tycowski K. T.; Steitz J. A. Non-coding snoRNA host genes in Drosophila: Expression strategies for modification guide snoRNAs. Eur. J. Cell Biol. 80:119–125; 2001. [DOI] [PubMed] [Google Scholar]
- 209. Tycowski K. T.; You Z. H.; Graham P. J.; Steitz J. A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 2:629–638; 1998. [DOI] [PubMed] [Google Scholar]
- 210. Urquidi V.; Tarin D.; Goodison S. Role of telomerase in cell senescence and oncogenesis. Annu. Rev. Med. 51:65–79; 2000. [DOI] [PubMed] [Google Scholar]
- 211. Ursic D.; Himmel K. L.; Gurley K. A.; Webb F.; Culbertson M. R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 25:4778–4785; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. van Hoof A.; Lennertz P.; Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441–452; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. van Hoof A.; Parker R. The exosome: A proteasome for RNA? Cell 99:347–350; 1999. [DOI] [PubMed] [Google Scholar]
- 214. Venema J.; Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae . Annu. Rev. Genet. 33:261–311; 1999. [DOI] [PubMed] [Google Scholar]
- 215. Venema J.; Vos H. R.; Faber A. W.; van Venrooij W. J.; Raue H. A. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA 6:1660–1671; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Villa T.; Ceradini F.; Bozzoni I. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 20:1311–1320; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Villa T.; Ceradini F.; Presutti C.; Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol. 18:3376–3383; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Wang H.; Boisvert D.; Kim K. K.; Kim R.; Kim S. H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 19:317–323; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Watanabe Y.; Gray M. W. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 28:2342–2352; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Watkins N.; Leverette R.; Xia L.; Andrews M.; Maxwell E. Elements essential for processing in-tronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2:118–133; 1996. [PMC free article] [PubMed] [Google Scholar]
- 221. Watkins N. J.; Gottschalk A.; Neubauer G.; Kast-ner B.; Fabrizio P.; Mann M.; Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549–1568; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Watkins N. J.; Newman D. R.; Kuhn J. F.; Maxwell E. S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA 4:582–593; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Watkins N. J.; Segault V.; Charpentier B.; Not-trott S.; Fabrizio P.; Bachi A.; Wilm M.; Rosbash M.; Branlant C.; Luhrmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457–466; 2000. [DOI] [PubMed] [Google Scholar]
- 224. Weinstein L. B.; Steitz J. A. Guided tours: From precursor snoRNA to functional snoRNP. Curr. Opin. Cell. Biol. 11:378–384; 1999. [DOI] [PubMed] [Google Scholar]
- 225. Westendorf J.; Konstantinov K.; Wormsley S.; Shu M.; Matsumoto T. N.; Pirollet F.; Klier F.; Gerace L.; Baserga S. M phase phosphoprotein is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell 9:437–449; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Wiederkehr T.; Prétôt R. F.; Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4:1357–1372; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Woese C. R.; Kandler O.; Wheelis M. L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576–4579; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Woese C. R.; Magrum L. J.; Fox G. E. Archaebac-teria. J. Mol. Evol. 11:245–251; 1978. [DOI] [PubMed] [Google Scholar]
- 229. Wu P.; Brockenbrough J.; Metcalfe A.; Chen S.; Aris J. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem. 273:16453–16463; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Yang Y.; Isaac C.; Wang C.; Dragon F.; Pogacic V.; Meier U. T. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell 11:567–577; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Yi-Brunozzi H. Y.; Easterwood L. M.; Kamilar G. M.; Beal P. A. Synthetic substrate analogs for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res. 27:2912–2917; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Yu Y. T.; Shu M. D.; Narayanan A.; Terns R. M.; Terns M. P.; Steitz J. A. Internal modification of U2 snRNA occurs in nucleoli of Xenopus oocytes. J. Cell Biol. 152:1279–1288; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Zebarjadian Y.; King T.; Fournier M. J.; Clarke L.; Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461–7472; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


