Abstract
Background: Little knowledge about long non-coding RNAs(lncRNAs) in nasopharyngeal carcinoma (NPC) has been acquired.
Methods: Next-generation sequencing was applied in 7 cases of NPC tissues and 7 cases of normal tissues in nasopharynx. PLEX, CNCI and CPAT soft-wares were used to predict novel lncRNAs. Real-time Quantitative PCR (qPCR) further validated the data in 20 cases of NPC tissues and 14 cases of normal tissues. Then the cis-regulators and trans-regulators and potential biological functions together with pathways were predicted by Bioinformatics.
Results: Totally, 4248 novel lncRNAs were found to be expressed in our samples. And 2192 lncRNAs and 23342 mRNAs were considered to be differentially expressed in NPC. Among the results, 306 lncRNAs and 4599 mRNAs were significantly up-regulated, whereas 204 lncRNAs and 2059 mRNAs were significantly down-regulated, respectively. Moreover, 62 lncRNAs trans-regulated genes were involved in Epstein-Barr virus (EBV) infection pathway in our study. Jun proto-oncogene (JUN), which was related to a cis-regulator lncRNA RP4-794H19.1, was enriched in cancers and involved in Tumor Necrosis Factor (TNF) signaling pathway, might play a key role in NPC.
Conclusion: These findings broadened the lncRNAs landscape of NPC tissues and shed light on the roles of these lncRNAs, which might be conducive to the comprehensive management of NPC.
Keywords: Nasopharyngeal carcinoma, RNA sequencing, Long non-coding RNAs, Bioinformatics analysis, QPCR
Introduction
Globally, about 86,500 cases of nasopharyngeal carcinoma (NPC) were reported in 2012. NPC was a mortal disease prevailing in South-east Asia and North Africa 1. In China, the standardized mortality rate was 0.78 /105 issued in 20132. NPC arose as a result of its various genetic susceptibility, environmental or random factors and Epstein-Barr virus (EBV) infection3. Albeit researchers were at full tilt unraveling the tumorigenesis and the pathways, we seldom knew them yet.
Long non-coding RNAs(lncRNAs) previously were considered as “transcriptional noise”, which accounted for at least 75% of DNA transcripts4. Generally speaking, lncRNAs are longer than 200 nucleotides and short of protein-coding capacity, with lacking of open reading frame (ORF)5, 6. Encyclopedia of DNA Elements (ENCODE) Project Consortium issued at least 120,000 transcripts that served as lncRNAs in the human genome7. Now lncRNAs have been validated that it played a gorgeous role in regulating gene expression at various levels to affect cellular homeostasis, including proliferation, survival, migration and genomic stability4. Up-to-date evidences laid bare that the aberrantly expressed lncRNAs always were relative to the development and progression of cancers8-11. Given the eagerness to better understand NPC, many researchers took the cutting-edge lncRNAs to the spotlight.
The lncRNAs research in NPC usually resorted to Microarray and lncRNA-protein-based relationship technology 12-16, or at cell level17. Microarray was a high-throughput sequencing, but it was limited by countable numbers of lncRNAs, and all the probes were already known, which was the reason for refraining the technology from discovering novel lncRNAs. Despite LncRNA-protein-based relationship technology as RNA Immunoprecipitation (RIP)-chip and RIP-sequencing, its antibody specificity made the research focus on one point. The next-generation deep sequencing took its advantages in discovering massive exiting and novel lncRNAs18. Here, we applied the next-generation deep sequencing to provide a comprehensive view for insights into NPC tissues.
Materials and Methods
Samples collection
There were 20 cases of NPC tissues and 14 cases of normal tissues collected by the second Xiangya Hospital of Central South University. All sample tissues were confirmed by histopathological examination in the second Xiangya Hospital of Central South University. This study was approved by the Ethical Committee of Xiangya Hospital, Central South University. All patients had voluntarily signed a written consent before samples were collected. The acquired tissue samples were placed into liquid nitrogen immediately and then were transferred to a -80°C refrigerator.
RNA extraction
The abstraction of total RNAs derived from 20 cases of NPC tissues and 14 cases of normal tissues, using TRIzol (Life Technology, California, USA) according to the guideline protocol. The sample tissues were placed in 2ml RNA-free EP tube that grinded with four 4mm steel balls in TissueLyser LT (QIAGEN, Dusseldorf, Germany). The purity and concentration of RNAs were measured by OD260/280 readings shown on DeNovix DS-11+ (DeNovix, Delaware, USA), which ranging from 1.8-2.2 were kept. RNAs integrity was qualified by standard denaturing agarose gel electrophoresis.
cDNA synthesizing and Real-time Quantitative PCR(qPCR) analysis
The first strand of cDNA was synthesized by Promega GoScript (Promega, Madison, USA) with 2 μg total RNA. The qPCR setting 40 cycles of amplification was performed on GoTaq qPCR Master Mix (Promega, Madison, USA) on LightCycler 480 Ⅱ QPCR System (Roch, Basel, Switzerland). β-actin served as a reference, and relative-expression levels of lncRNAs were quantified through 2(-∆∆Ct) methods.
Library preparation for lncRNA sequencing
There were 7 cases of NPC tissues and 7 cases of normal tissues for RNA sequencing. Total RNAs were abstracted as previously, strand-specific RNA-seq Libraries were generated with TruSeq® RNA LT Sample Prep Kit v2 (Illumina, CA, USA) following the handbook, subsequently the Ribo-Zero Gold kit (Illumina, CA, USA) was used for the removing of rRNA, and then the fragmentations of RNAs were harvested. RNA fragmentations were used to synthesize double-stranded cDNA with substituting dUTPs for dTTPs in the second strand cDNA synthesis. Next, double-stranded cDNA were added to A-tailed and ligated to adaptors. Finally, PCR amplification was performed to enrich cDNA libraries. PCR products were qualified by Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technology, MA, USA) for RNA sequencing.
RNA sequencing
Sequencing was performed on an Illumina Hiseq 2500 instrument (Illumina, CA, USA) with the TruSeq PE Cluster Kit v3-cBot-HS (Illumina, CA, USA).
Results
The prediction of novel lncRNAs
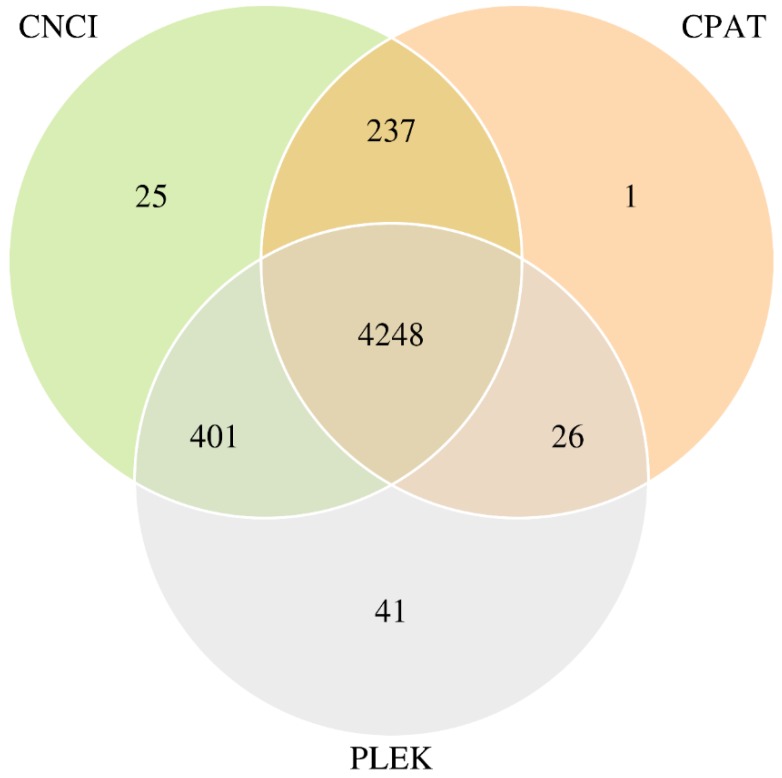
Transcripts were aligned with databases, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups (COG) and Swissprot Database, then the mapped transcripts (mRNA) were dropped. Subsequently, transcripts annotated in the lncRNA library (Ensemble) were also cast away. The remaining transcripts were inputted into three Coding Potential Calculator soft-wares (PLEX, CNCI and CPAT) to predict coding and non-coding sequences. We kept the 4248 transcripts which overlapped in three calculation methods, serving as novel lncRNAs. CNCI-CPAT, CPAT-PLEK and PLEK-CNCI overlapped numbers were 237, 26 and 401, respectively. Venn Diagram of predicted novel lncRNAs was shown in Figure 1.
Figure 1.
Venn Diagram of predicted novel lncRNAs with three the Coding Potential Calculator soft-wares.
The landscape of differential expressions of lncRNAs and mRNAs sequencing in NPC and normal tissues
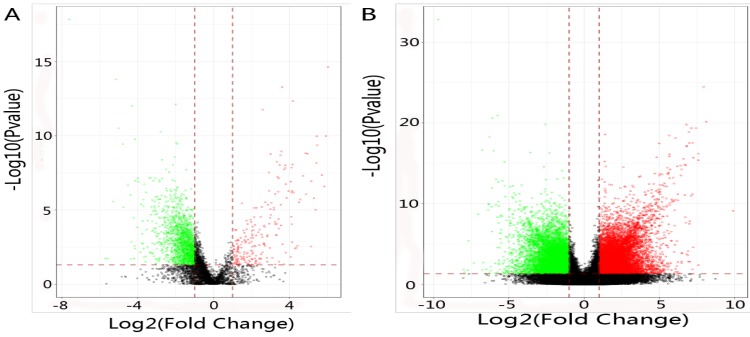
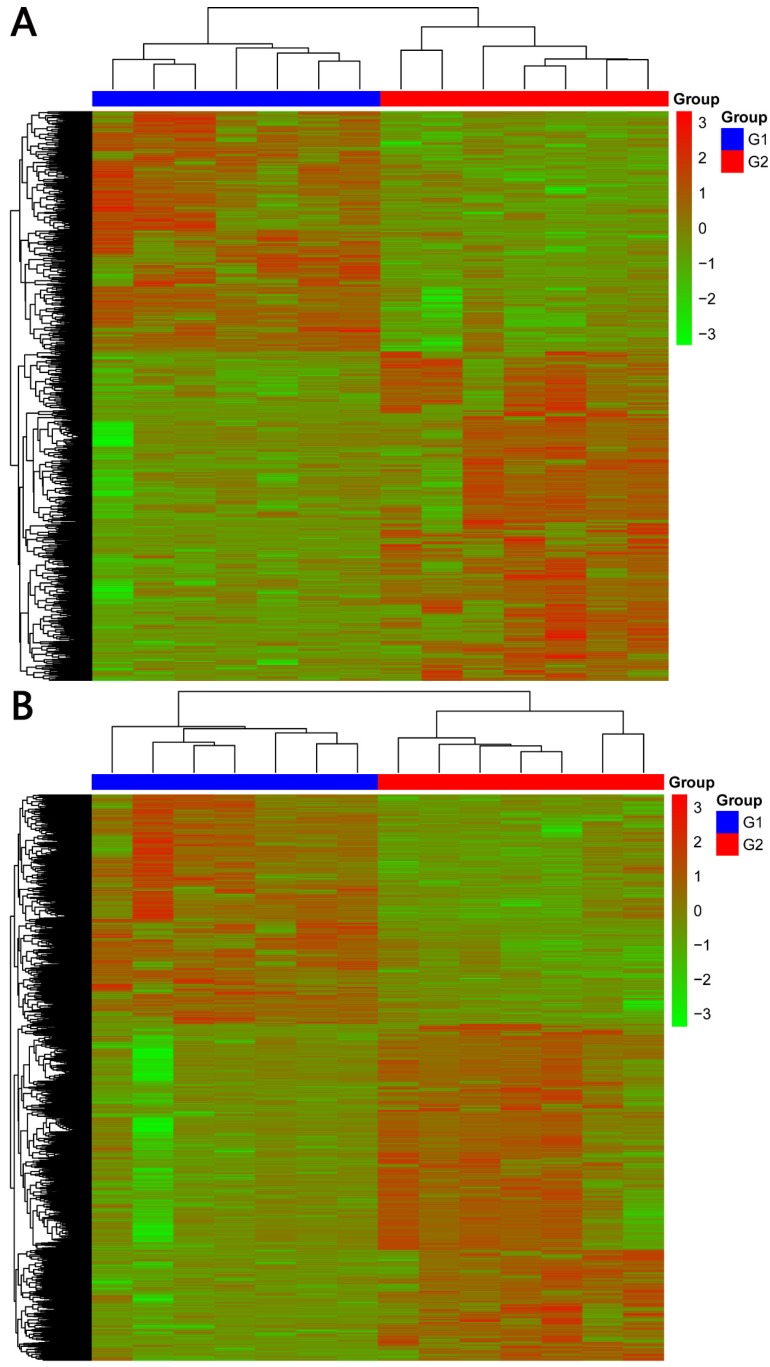
In total, 2192 lncRNAs and 23342 mRNAs were considered to be differentially expressed in NPC. Among the results, 204 lncRNAs and 2059 mRNAs were significantly down-regulated, and 306 lncRNAs and 4599 mRNAs were significantly up-regulated (≥2 fold change, FDR<0.05). Compared with normal tissues, OSER1-AS1 (fold change: -4.54, FDR<0.05) and MUC4 (fold change: -8.00, FDR<0.05) were the most down-regulated lncRNA and mRNA, as opposed to the most up-regulated lncRNA and mRNA, which were RP11-624L4.1 (fold change: 4.35, FDR<0.05) and XLOC.42541.1 (fold change:13.20, FDR<0.05), respectively. The volcano plot of differential expressions of lncRNAs and mRNAs were displayed in Figure 2. Heat maps of differential expressions and hierarchical clustering of lncRNAs and mRNAs in NPC and normal tissues were demonstrated in Figure 3.
Figure 2.
(A) The volcano plot of differential expressions of lncRNAs in NPC and normal tissues. Dark spot: log2(fold change)<1, P value>0.05; Red spot: log2(fold change)≥1, P value≤0.05; Green spot: log2(fold change)≤-1, P value≤0.05. (B) The volcano plot of differential expressions of mRNAs in NPC and normal tissues. Dark spot: log2(fold change)<1, P value>0.05; Red spot: log2(fold change)≥1, P value≤0.05; Green spot: log2(fold change)≤-1, P value≤0.05.
Figure 3.
(A) Heat maps of differential expressions of and hierarchical clustering of lncRNAsin NPC and normal tissues. (B) Heat maps of differential expressions of and hierarchical clustering of mRNAs in NPC and normal tissues.
RNA sequencing data validation by qPCR
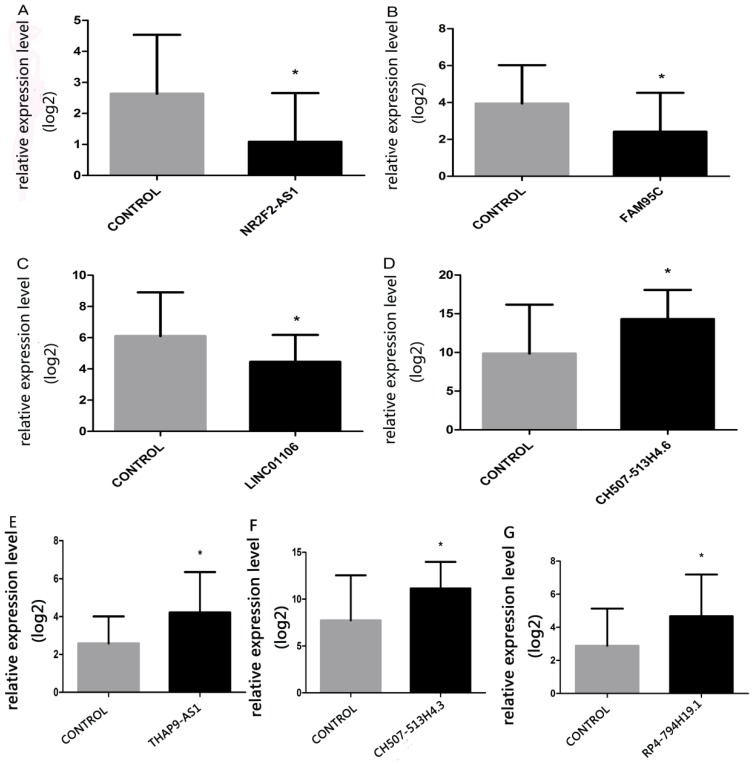
Seven lncRNAs were sifted through on the basis of differentially expressed lncRNAs prediction for validating RNA sequencing data, which were lncRNA-NR2F2 antisense RNA1(NR2F2-AS1) (fold change:-3.2), lncRNA-family with sequence similarity 95 member C(FAM95C) (fold change:-3.3), lncRNA- long intergenic non-protein coding RNA 1106 (LINC01106) (fold change:-3.7), lncRNA-CH507- 513H4.6 (fold change:3.8), lncRNA-THAP9 antisense RNA 1(THAP9-AS1) (fold change:3.7), lncRNA- CH507-513H4.3 (fold change:3.8) and lncRNA-RP4- 794H19.1 (fold change:1.8). QPCR results were consistent with RNA sequencing data(Table 1). The lncRNAs in NPC reported in former research also credited our RNA-seq data(Table 2). QPCR results were shown in Figure 4.
Table 1.
Primers used for qPCR analysis of lncRNA levels.
| LncRNA | Primer | Expression | Product Length (bp) |
|---|---|---|---|
| NR2F2-AS1 | F: GGCTTGGTTGGACCGTACTC R:ACTGCCTGCCACTCTTGTGA |
Down | 121 |
| FAM95C | F: CCTCATCCAGAGCAGGACAA R: GCCTGCATTAGCATCTGTGC |
Down | 128 |
| LINC01106 | F:GAGAGAGGAATGGCTGCATGT R:TCAGGAGGTTGCAGGTTGAC |
Down | 121 |
| CH507-513H4.6 | F: CATGCCAGAGTCTCGTTCGTT R: GGACACGGACAGGATTGACA |
Up | 125 |
| THAP9-AS1 | F: GAAGGTCCACAGCCAGTTCC R: CGCATTGCTGTAGGCATAGG |
Up | 121 |
| CH507-513H4.3 | F:TTGCTCAATCTCGGGTGGCT R:TGGTGGAGCGATTTGTCTGG |
Up | 138 |
| RP4-794H19.1 | F:CCTTCCGCCTTGGCAAAGAT R:TGTGGGGTGAAGGTCTGGTT |
UP | 120 |
Table 2.
LncRNAs reported in NPC in former research.
Figure 4.
The relative-expression levels of seven lncRNAs (A, NR2F2-AS1; B, FAM95C; C, LINC01106; D, CH507-513H4.6; E, THAP9-AS1, F, CH507-513H4.3; G, RP4-794H19.1) in NPC and normal tissues. Kruskal-Wallis H test was applied, *p<0.05.
Differential expressions of lncRNAs and Cis-regulation and trans-regulation genes co-expression networks
We defined the chromosomal genes (co-expression mRNA mapped), which were 100 kb upstream or downstream of differentially expressed lncRNAs, as potential lncRNA cis-regulatory genes. Totally, there were 1895 differentially expressed lncRNAs with 4848 cis-regulatory genes. Among them, LA16c-313D11.9 had 231 cis-regulatory genes(the most). One differentially expressed lncRNA for one cis-regulatory gene number was 59. One cis-regulatory gene for one differentially expressed lncRNA was 566.
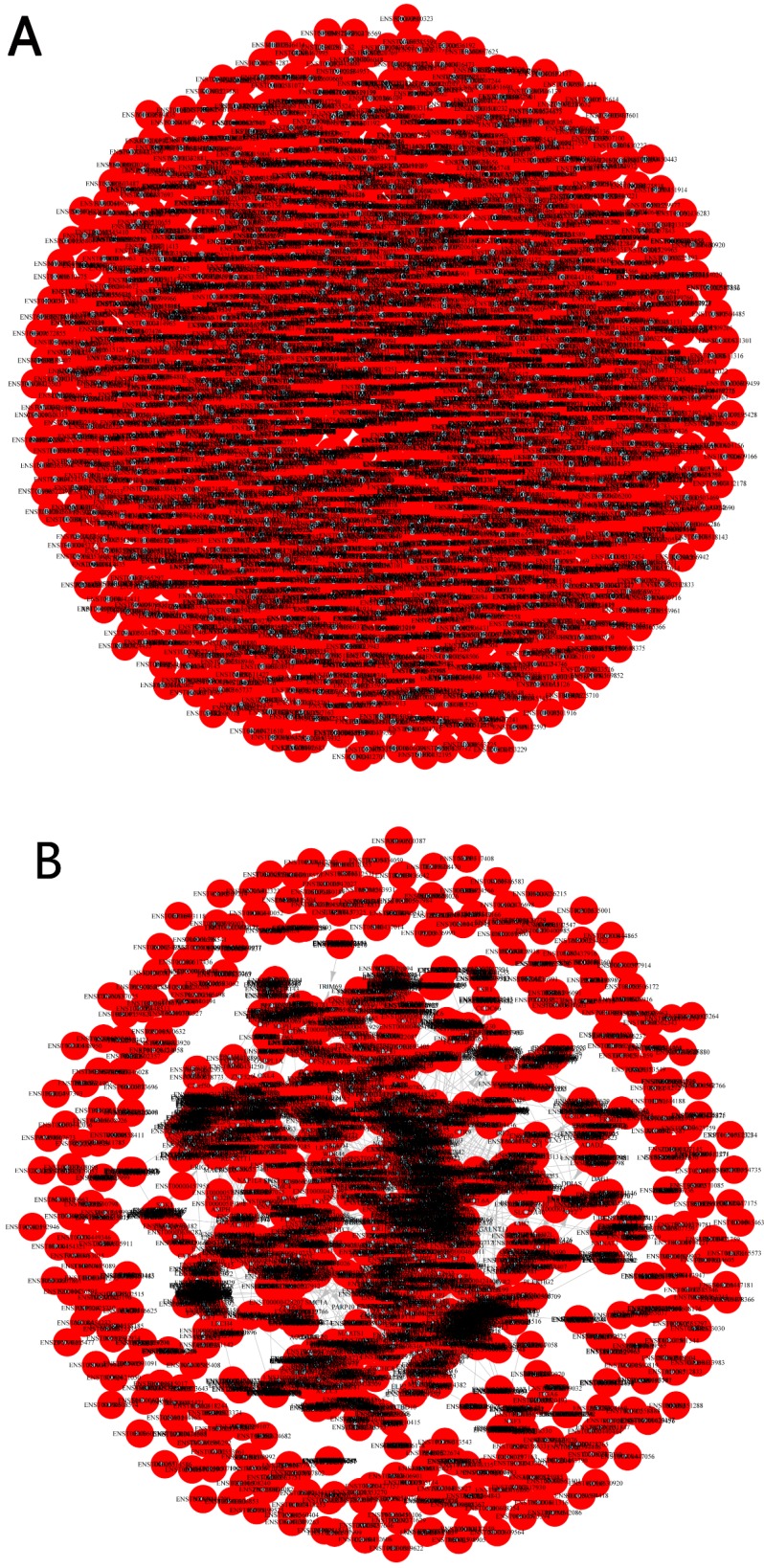
Pearson correlation coefficient(R2≥0.95) was applied to predict trans-regulatory genes. In total, there were 1284 differentially expressed lncRNAs with 6437 trans-regulatory genes. Among them, AC000123.4 had 577 trans-regulatory genes(the most). One differentially expressed lncRNA for one trans- regulatory gene number was 2263. One trans- regulatory genes for one differentially expressed lncRNA was 566. The networks of cis-regulation and trans-regulation were illuminated in Figure 5.
Figure 5.
(A) Cis-regulation of the networks. Red spot represented differential expression of lncRNA. Blue spot represented cis-regulation gene. (B) Trans-regulation of the networks. Red spot represented differential expression of lncRNA. Blue spot represented trans-regulation gene.
Gene ontology (GO) enrichment and pathway analysis
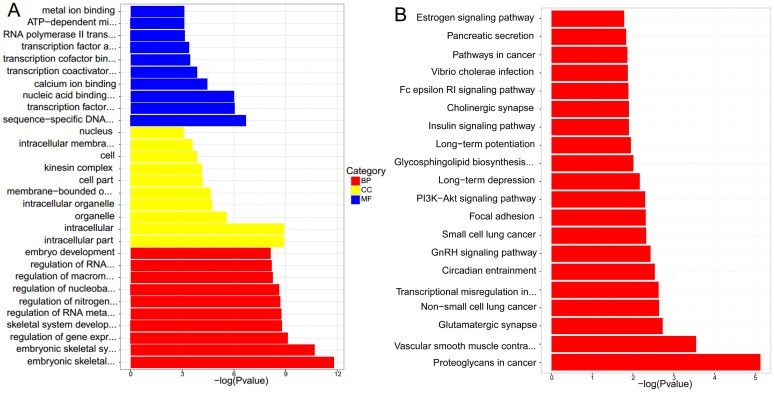
GO enrichment analysis was conducted by TopGO software, and associated with Fisher Exact Test. The results suggested that the significant functions mainly focused on embryonic skeletal system development, embryonic skeletal system morphogenesis, ATP-dependent microtubule motor activity and regulation of gene expression in lncRNAs cis-regulation genes between NPC and normal tissues. As for lncRNAs trans-regulation genes, they laid in intracellular part, intracellular and nuclear part. The detailed results were presented in Figure 6. A and Figure 7. A.
Figure 6.
(A) GO enrichmentanalysis of differential expressions of lncRNAs cis-regulation genes. (B) KEGG pathway analysis of differential expressions of lncRNAs cis-regulation genes.
Figure 7.
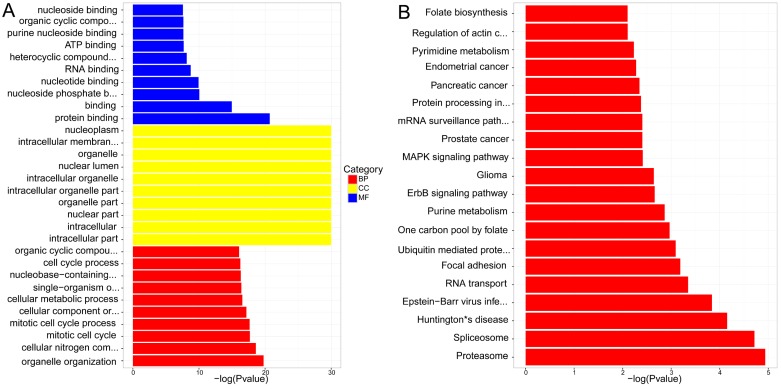
(A) GO enrichment of differential expressions of lncRNAs trans-regulation genes (B) KEGG pathway analysis of differential expressions of lncRNAs trans-regulation genes.
KEGG pathway analysis concluded that the most significant pathways of lncRNAs cis-regulation genes were Proteoglycans in cancer, Vascular smooth muscle contraction and Glutamatergic synapse, while Proteasome, Spliceosome and Huntington's disease were in lncRNAs trans-regulation genes. The rest were demonstrated in Figure 6. B and Figure 7. B.
Discussion
In 2015, there were sixty thousand new NPC cases reported in China19. For NPC patients usually were diagnosed at late stage, and it's a challenge to determine the disease at an early stage. Here, we resorted to the next-generation sequencing technology in 7 cases of NPC tissues and 7 cases of nasopharyngeal normal tissues. 2192 differentially expressed lncRNAs were found, which had the making of potential biomarkers for early-stage diagnosis, progression and prognosis20. Moreover, 4248 novel lncRNAs were obtained, and might tremendously enrich the human lncRNA pool.
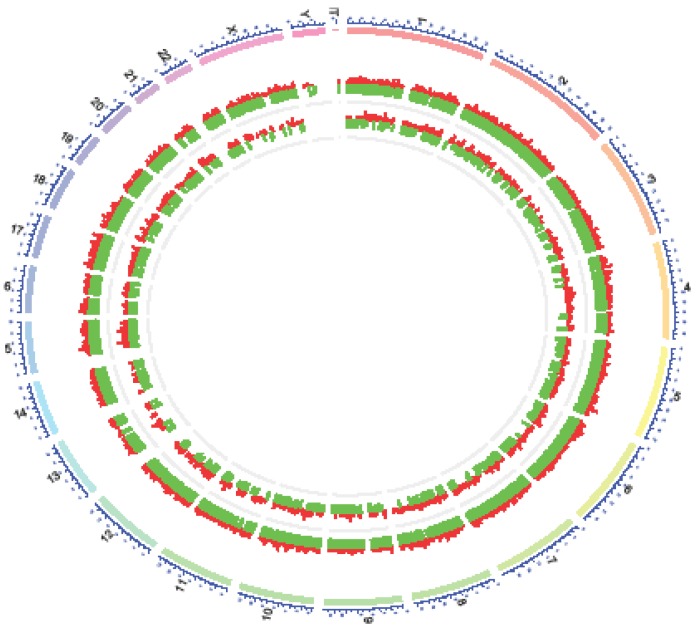
There was an interesting finding when we mapped differentially expressed lncRNAs to human chromosomes. Y- chromosome had no differentially expressed lncRNAs in NPC and normal tissues(Figure 8), meanwhile, no such studies have been issued. According to demographic trends, men were two to three fold more likely to acquire NPC than women were1. Maybe Y- chromosome was short of some suppressor lncRNAs inflicting a higher morbidity compared with women in NPC.
Figure 8.
Distribution of differential expressions of lncRNAs and mRNAs along each chromosome. The chromosome constitution (outer circle, depicted in blue), distribution of differential expressions of mRNAs (middle circle, depicted in red: up-regulation, green: down-regulation) and distribution of differential expressionsof lncRNAs (inner circle, depicted in red: up-regulation, green: down-regulation).
Seven qPCR results were credible based on our work, and several reported studies also supported it. Increased X inactive specific transcript (XIST) expression was associated with poor prognosis and stunted cell growth21. HOX transcript antisense RNA(Hotair) might play an oncogenic role in NPC progression, and its knockdown attenuated NPC cell growth and angiogenesis20. Both DNA copy number loss and aberrant promoter methylation led to maternally expressed 3(MEG3) inactivation. As a tumor suppressor, which could be significantly inhibited in cell proliferation, colony formation22. Overexpressed cancer susceptibility 9 (CASC9) promoted the glycolysis and tumorigenesis of NPC cells via activating hypoxia inducible factor 1 alpha subunit (HIF1alpha)23. Ewing sarcoma associated transcript 1(EWSAT1) could up-regulate the expressions of miR-326/330-5p clusters targeted gene cyclin D1 through a competitive 'sponge' of miR-326/330-5p clusters, which might promote the development of NPC24.
However, only few known lncRNAs had functional annotations. The methods for lncRNAs function prediction always investigated their correlated protein coding genes and related biological pathways25. It also had been proved that lncRNAs function through cis-regulation of nearby protein coding genes was a common mechanism26. Only a few lncRNAs were refined as cis-regulators, while the majority of functionally annotated ones were trans-regulators27. Therefore, we divided the differentially expressed lncRNAs cis-regulation or trans-regulation genes into two categories to explore the potential target genes, GO enrichment and KEGG pathways analysis. 62 lncRNAs trans-regulated genes were involved in the EBV infection pathway in our study (Figure 7B), as CD4428 and interleukin 1 receptor associated kinase 1(IRAK1)29 (Table S1). In our work, we defined 100kb distance as a criterion between lncRNAs and cis-regulated genes. Jun proto-oncogene(JUN), which was located 98542b downstream to LncRNA RP4-794H19.1, was enriched in cancers and involve in Tumor Necrosis Factor (TNF) signaling pathway. For example, the expression levels of JUN family members were positively correlated to the cell proliferation in diffuse large B-cell lymphomas30. Furthermore, NAG7 through the activity of JUN pathways could promote the invasion in NPC31. For good measure, the EBV-encoded EBNA1 regulated the JUN transcription factor pathway in NPC and increased angiogenesis in vitro32. Our results underpinned that lncRNA RP4-794H19.1 in relation to JUN might play a key role in NPC. However our work strongly depended on the theoretically computerized calculation, lacking further numerous practice validation. These findings broadened the lncRNAs landscape of NPC and shed light on the roles of these lncRNAs, which might be conducive to the comprehensive management of NPC.
Supplementary Material
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary table S4.
Supplementary table S5.
Supplementary table S6.
Supplementary table S7.
Supplementary table S8.
Supplementary table S9.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81372516), and Hunan Provincial Natural Science Foundation of China (No. 2016JJ5013).
Abbreviations
- lncRNAs
long non-coding RNAs
- qPCR
Real-time Quantitative PCR
- JUN
Jun proto-oncogene
- TNF
Tumor Necrosis Factor
- NPC
nasopharyngeal carcinoma
- EBV
Epstein-Barr virus
- ORF
open reading frame
- ENCODE
Encyclopedia of DNA Elements
- RIP RNA
Immunoprecipitation
- KEGG
the Kyoto Encyclopedia ofGenes and Genomes
- COG
Clusters of Orthologous Groups
- NR2F2-AS1
lncRNA-NR2F2antisense RNA 1
- FAM95C
lncRNA-family with sequencesimilarity 95 member C
- LINC01106
lncRNA- long intergenic nonprotein coding RNA 1106
- THAP9-AS1
lncRNA-THAP9 antisense RNA 1
- Gene ontology
GO
- XIST X
inactive specific transcript
- Hotair
HOX transcript antisense RNA
- MEG3
maternally expressed 3
- CASC9
Overexpressed cancer susceptibility 9
- HIF1alpha
hypoxia inducible factor 1 alphasubunit
- EWSAT1
Ewing sarcoma associated transcript 1
- IRAK1
interleukin 1 receptor associated kinase 1.
References
- 1.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. The Lancet. 2016;387(10022):1012–24. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 2.Xu ZX, Lin ZX, Fang JY, Wu KS, Du PL, Zeng Y. et al. Mortality Characteristic and Prediction of Nasopharyngeal Carcinoma in China from 1991 to 2013. Asian Pacific Journal of Cancer Prevention. 2015;16(15):6729–34. doi: 10.7314/apjcp.2015.16.15.6729. [DOI] [PubMed] [Google Scholar]
- 3.He R, Hu Z, Wang Q, Luo W, Li J, Duan L. et al. The role of long non-coding RNAs in nasopharyngeal carcinoma: As systemic review. Oncotarget. 2017;8(9):16075–83. doi: 10.18632/oncotarget.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nature Medicine. 2015;21(11):1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jalali S, Gandhi S, Scaria V. Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Human Genomics. 2016;10(1):35. doi: 10.1186/s40246-016-0090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Clara E, Gourvest M, Ma H, Vergez F, Tosolini M, Dejean S. et al. Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica. 2017;102(10):1718–26. doi: 10.3324/haematol.2017.171645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malouf GG, Zhang J, Yuan Y, Comperat E, Roupret M, Cussenot O. et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Molecular Oncology. 2015;9(1):32–43. doi: 10.1016/j.molonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Zhang H, Li X, Chen W. Differential expression profile analysis of lncRNA UCA1alpha regulated mRNAs in bladder cancer. Journal of Cellular Biochemistry. 2017 doi: 10.1002/jcb.26345. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Qiu X, Jia B, Sun X, Hu W, Chu H, Xu S. et al. The Critical Role of Long Noncoding RNA in Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Biomed Res Int. 2017;2017:5045827. doi: 10.1155/2017/5045827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang QQ, Deng YF. Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology. 2015;66(7):1022–30. doi: 10.1111/his.12616. [DOI] [PubMed] [Google Scholar]
- 13.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y. et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–25. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Research. 2014;24(5):513–31. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K. et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(5):1273–8. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8(24):39001–11. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren S, Li G, Liu C, Cai T, Su Z, Wei M. et al. Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma. Oncology Reports. 2016;36(4):1861–7. doi: 10.3892/or.2016.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida K, Sanada M, Ogawa S. Deep sequencing in cancer research. Japanese Journal of Clinical Oncology. 2013;43(2):110–5. doi: 10.1093/jjco/hys206. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 20.Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang HD. et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7(4):4712–23. doi: 10.18632/oncotarget.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 22.Chak WP, Lung RW, Tong JH, Chan SY, Lun SW, Tsao SW. et al. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Molecular Carcinogenesis. 2017;56(3):1041–54. doi: 10.1002/mc.22569. [DOI] [PubMed] [Google Scholar]
- 23.Su X, Li G, Liu W. The Long Noncoding RNA Cancer Susceptibility Candidate 9 Promotes Nasopharyngeal Carcinogenesis via Stabilizing HIF1alpha. DNA and Cell Biology. 2017;36(5):394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 24.Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging. 2016;8(11):2948–60. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagel D, Vincendeau M, Eitelhuber AC, Krappmann D. Mechanisms and consequences of constitutive NF-kappaB activation in B-cell lymphoid malignancies. Oncogene. 2014;33(50):5655–65. doi: 10.1038/onc.2013.565. [DOI] [PubMed] [Google Scholar]
- 27.Young RM, Shaffer AL 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Seminars in Hematology. 2015;52(2):77–85. doi: 10.1053/j.seminhematol.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janisiewicz AM, Shin JH, Murillo-Sauca O, Kwok S, Le QT, Kong C. et al. CD44+cells have cancer stem cell-like properties in nasopharyngeal carcinoma. Int Forum Allergy Rh. 2012;2(6):465–70. doi: 10.1002/alr.21068. [DOI] [PubMed] [Google Scholar]
- 29.Luftig M, Prinarakis E Fau - Yasui T, Yasui T Fau - Tsichritzis T, Tsichritzis T Fau - Cahir-McFarland E, Cahir-McFarland E Fau - Inoue J-I, Inoue J Fau - Nakano H. et al. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA. 2003;200(26):15595–15600. doi: 10.1073/pnas.2136756100. (0027-8424 (Print)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papoudou-Bai A, Goussia A, Batistatou A, Stefanou D, Malamou-Mitsi V, Kanavaros P. The expression levels of JunB, JunD and p-c-Jun are positively correlated with tumor cell proliferation in diffuse large B-cell lymphomas. Leukemia & Lymphoma. 2016;57(1):143–50. doi: 10.3109/10428194.2015.1034704. [DOI] [PubMed] [Google Scholar]
- 31.Huang C, Wu MH, Tang YL, Li XL, Ouyang J, Xiao L. et al. NAG7 Promotes Human Nasopharyngeal Carcinoma Invasion Through Inhibition of Estrogen Receptor Alpha and Up-Regulation of JNK2/AP-1/MMP1 Pathways. J Cell Physiol. 2009;221(2):394–401. doi: 10.1002/jcp.21867. [DOI] [PubMed] [Google Scholar]
- 32.O'Neil JD, Owen TJ, Wood VH, Date KL, Valentine R, Chukwuma MB. et al. Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. The Journal of General Virology. 2008;89(Pt 11):2833–42. doi: 10.1099/vir.0.2008/003392-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary table S4.
Supplementary table S5.
Supplementary table S6.
Supplementary table S7.
Supplementary table S8.
Supplementary table S9.