Abstract
Background
Colorectal cancer (CRC), the most common malignancy worldwide, causes inflammation. We explored the inflammatory pathophysiology of CRC by assessing the peripheral blood parameters.
Methods
The differences in gene expression profiles of whole blood cells and cell subpopulations between CRC patients and healthy controls were analyzed using DNA microarray. Serum cytokine/chemokine concentrations in CRC patients and healthy controls were measured via multiplex detection immunoassays. In addition, we explored correlations between the expression levels of certain genes of peripheral CD4+ cells and serum chemokine concentrations.
Results
The gene expression profiles of peripheral CD4+ cells of CRC patients differed from those of healthy controls, but this was not true of CD8+ cells, CD14+ cells, CD15+ cells, or CD19+ cells. Serum IL-8 and eotaxin-1 levels were significantly elevated in CRC patients, and the levels substantially correlated with the expression levels of certain genes of CD4+ cells. Interestingly, the relationships between gene expression levels in peripheral CD4+ cells and serum IL-8 and eotaxin-1 levels resembled those of monocytes/macrophages, not T cells.
Conclusions
Serum IL-8 and eotaxin-1 concentrations increased and were associated with changes in the gene expression of peripheral CD4+ cells in CRC patients.
1. Introduction
Colorectal cancer (CRC) is the third most common fatal malignancy worldwide [1]. It is important to detect CRC early to improve prognosis [2, 3]. Currently, the fecal occult blood test (FOBT) for in vitro diagnostic use is used to screen for CRC; however, the positive predictive rate is poor [3]. Colonoscopy is superior, but this is invasive and associated with multiple complications including perforation, pain, and discomfort [4]. Thus, alternative noninvasive diagnostic tests are required. To this end, it is necessary to understand the pathological features, including the immune status, of CRC patients.
We previously reported that the gene expression profiles of peripheral blood cells from patients with cancers of the digestive system differed from those of noncancerous controls [5]. Peripheral blood contains many types of immune cells including neutrophils, monocytes, and macrophages [6]. Changes in gene expression are hypothesized to reflect the reactions of the immune system to cancer, because cancer is frequently associated with the appearance of various types of inflammatory cells [7]. These include helper T cells and cytotoxic T lymphocytes [8], which inhibit cancer progression, and myeloid-derived suppressor cells [9], regulatory T cells [10], and programmed cell death 1 (PD-1) expressing T cells [11], which promote cancer development. We previously reported that immune response-eliciting or immunosuppressive molecules mediated interactions between circulating peripheral blood cells and local cancer tissues in patients with pancreatic ductal adenocarcinomas [12] and hepatocellular carcinomas [13, 14]. In contrast, the features of immune pathophysiology reflected in peripheral blood of colorectal cancer have yet to be investigated.
Here, we observed that the gene expression profiles of peripheral CD4+ cells and whole blood cells of CRC patients differed from those of healthy controls. The serum concentrations of IL-8 and eotaxin-1 were elevated in CRC patients compared to healthy controls.
2. Methods
2.1. CRC Patients and Healthy Controls
Blood was drawn from CRC patients prior to treatment and from healthy controls. A total of 30 CRC patients and 28 healthy controls (Supplemental Table 1) provided serum samples for cytokine and chemokine analyses. CRC was clinically staged using the tumor, node, and metastasis staging system of the Union of International Cancer Control (8th edition). Five CRC patients and seven healthy volunteers donated peripheral blood for gene expression analyses (Supplemental Table 2). Serum cytokine/chemokine levels were measured in four CRC patients and five healthy volunteers (Supplemental Table 2). Written informed consent was obtained from all participants. This study was approved by our Institutional Review Board and was performed in accordance with all relevant tenets of the Declaration of Helsinki.
2.2. Serum Cytokine and Chemokine Analyses
Serum cytokine and chemokine levels were measured using a Bio-Plex human cytokine 27-plex panel (Bio-Rad, Tokyo, Japan) according to the manufacturer's instructions. This kit was used to detect interleukin- (IL-) 1β, IL-ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17, fibroblast growth factor-basic, eotaxin-1, G-CSF, granulocyte-colony stimulating factor, IFN-γ, IP-10, monocyte chemotactic protein-1 (MCP-1), macrophage inflammatory protein- (MIP-) 1α, MIP-1β, platelet-derived growth factor-BB, tumor necrosis factor- (TNF-) α, and vascular endothelial growth factor.
2.3. Isolation of Peripheral Blood Mononuclear Cells
Peripheral blood mononuclear cells were isolated from heparinized venous blood via Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO, USA) density gradient centrifugation, as previously described [13]. Then the cells were incubated with bead-labeled anti-CD4, anti-CD8, anti-CD14, or anti-CD15 antibodies (Miltenyi, Cologne, Germany) and isolated using a magnet.
2.4. DNA Microarray and Data Analysis
PAXgene® Blood RNA Tubes (PreAnalytiX GmbH, Germany) were used to collect samples for mRNA extraction. Total RNA was isolated from subfractionated peripheral blood cells using a microRNA isolation kit (Stratagene, La Jolla, CA, USA). Isolated RNA was labeled with Cy3 using the Quick-Amp Labeling Kit (Agilent Technologies, Palo Alto, CA, USA) and hybridized to the Whole Human Genome Microarray kit, 4x44K (Agilent Technologies). The slides were scanned using a microarray scanner (Model G2505B; Agilent Technologies), and gene expression analyses were performed using the BRB array tools (NCI, http://linus.nci.nih.gov/BRB-ArrayTools.html). Hierarchical clustering of gene expression data was used to identify differentially expressed genes. Biological processes and networks were analyzed with the aid of the MetaCore® software suite (GeneGo, Carlsbad, CA, USA).
2.5. Statistical Analysis
The unpaired Student's t-test was used to assess differences between groups, and a p < 0.05 was considered statistically significant. Pearson correlations between IL-8 and eotaxin-1 levels and clinical parameters were calculated. Spearman correlations were derived to explore associations between changes in chemokine concentrations and genes that were differentially expressed in peripheral CD4+ cells of CRC patients and healthy volunteers.
3. Results
3.1. Serum IL-8 and Eotaxin-1 Levels in CRC Patients
First, we measured cytokine and chemokine levels in 30 CRC patients and 28 healthy controls using a bead-based multiplex immunoassay (Supplemental Table 1). The serum concentrations of IL-8 and eotaxin-1 were significantly elevated in CRC patients (n=30) compared to healthy controls (n=28) (Figures 1(a) and 1(b)). However, we did not observe an increase of the other proinflammatory cytokines such as TNF-α, IFN-γ, and IL-12, of a decrease of anti-inflammatory cytokines such as IL-10 (Supplemental Fig. 2).
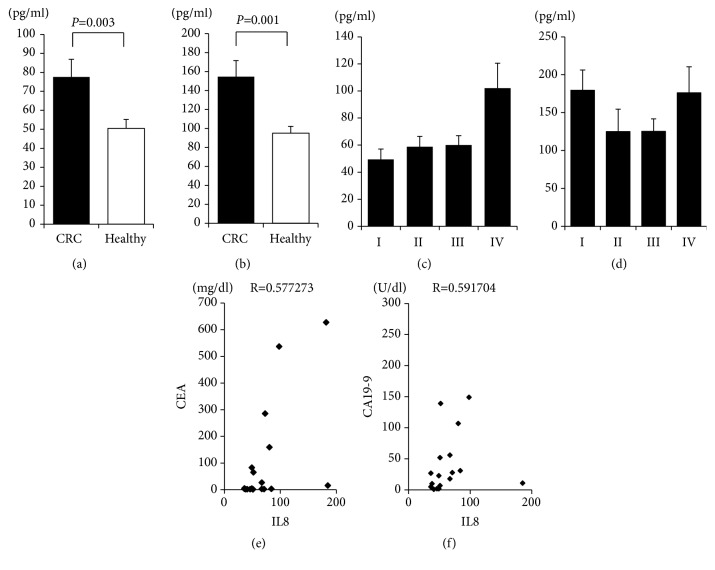
Figure 1.
Serum concentrations of eotaxin-1 and IL-8. Sera were obtained from CRC patients (n=30) prior to treatment and healthy volunteers (n=28). The serum concentrations of cytokines and chemokines were measured using a multiplex bead immunoassay system. (a) IL-8 levels; (b) eotaxin-1 levels. IL-8 levels in CRC patients by clinical stage (c) and eotaxin-1 levels by clinical stage (d). Correlations between IL-8 and CEA levels in CRC patients (e) and between IL-8 and CA19-9 levels (f).
IL-8 concentrations were only elevated in patients of advanced clinical stage (Stage IV; Figure 1(c)); eotaxin-1 levels did not differ by clinical stage (Figure 1(d)). IL-8 concentrations correlated with those of CEA (Figure 1(e)) and CA19-9 (Figure 1(f)) (r=0.577273 and r=0.591704, respectively), whereas eotaxin-1 concentrations did not (r=0.008045 and r=-0.06421, respectively) (data not shown). No correlation between the level of any other serum cytokine/chemokine and stage or level of tumor marker CEA or CA19-9 was apparent (data not shown).
3.2. Gene Expression Profiling of Peripheral Blood Cells from CRC Patients
Next, we used DNA microarray to determine if gene expression was altered in peripheral blood cells of five CRC patients and seven healthy volunteers (Supplemental Table 2). Unsupervised clustering analyses revealed a difference in the gene expression patterns of whole blood (Figure 2(a)) and CD4+ cells (Figure 2(b)), but not of CD8+ (Figure 2(c)), CD14+ (Figure 2(d)), or CD15+ cells (Figure 2(e)).
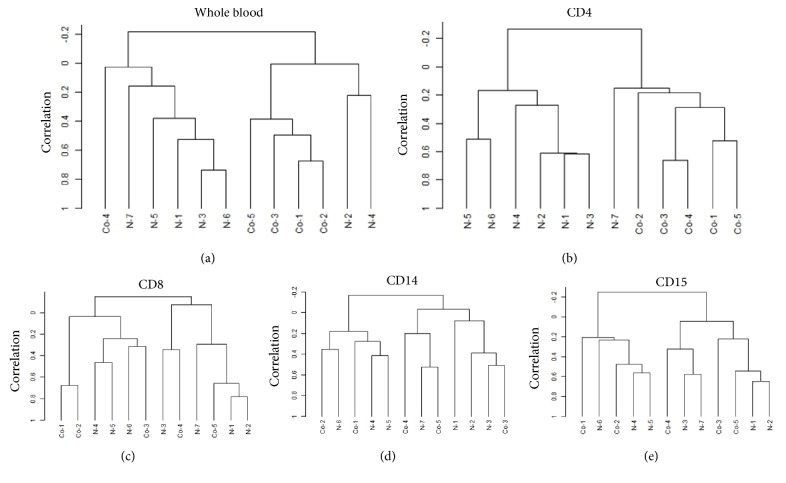
Figure 2.
Unsupervised clustering analysis of the gene expression profiles of subfractionated peripheral blood cells. The dendrogram used for clustering using employed both correlations and average linkage for unsupervised analysis of the gene expression profile of peripheral blood. Significant up- or downregulated changes in gene expression (≥1.5-fold) in whole blood and CD4+ cells of CRC patients (compared to healthy volunteers) were observed for 3,243 and 2,459 genes at P-values <0.05, respectively, but few such changes were observed in CD8+ cells, CD14+ cells, or CD15+ cells (1,475, 128, and 333 genes; P <0.05, respectively).
3.3. Elevated Serum IL-8 and Eotaxin-1 Concentrations Were Significantly Correlated with Genes Expression, the Levels of Which Were Altered in the Peripheral CD4+ Cells of CRC Patients
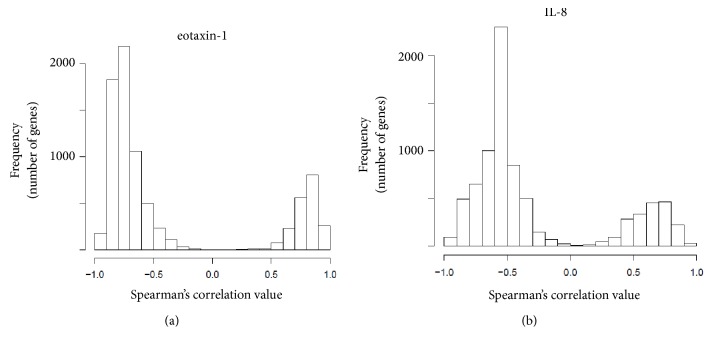
The data described above suggested that expression of the humoral chemokines, eotaxin-1 and IL-8, played a role in the inflammation of CRC patients. In addition, both peripheral CD4+ cells and whole blood cells were affected. Therefore, we derived Spearman's correlations between the serum concentrations of IL-8 and eotaxin-1 and the expression levels of 8,061 genes, the levels of which were altered (FDR<0.05) in CD4+ cells of four CRC patients compared to five healthy volunteers (Supplemental Table 2). We also confirmed that the serum concentrations of IL-8 (Supplemental Fig. 1A), eotaxin-1 (Supplemental Fig. 1B), MIP-1a (Supplemental Fig. 1C), and MCP-1 (Supplemental Fig. 1D) were significantly increased in the sera of CRC patients compared to healthy controls. The distribution frequencies of the 8,061 genes in terms of their Spearman correlations with eotaxin-1 levels are shown in Figure 3(a). Notably, the expression levels of all 8,061 genes correlated with serum eotaxin-1 concentrations. A total of 1,063 of these genes were involved in cell adhesion, inflammation, and the immune response (e.g., MHC, CD1d, TLR4, IL-15, Fc gamma, and Hck; Table 1). A total of 974 genes, the expression levels of which were negatively correlated with eotaxin-1 concentrations, were involved in proteolysis, development, and reproduction (Table 2). These biological processes are characteristics of monocytes and macrophages rather than T cells. The distribution frequencies of the 8,061 genes in terms of their Spearman correlations with serum IL-8 concentrations are shown in Figure 3(b). Almost all genes were so correlated. A total of 250 genes expressed in peripheral CD4+ cells were positively correlated with the serum IL-8 concentration, the genes played roles in cell adhesion, inflammation, the immune response, cytoskeletal processes, and development (Table 3). The expression levels of 586 genes were negatively correlated with serum IL-8 concentration; these genes were involved in cell proliferation, development, and reproduction (Table 4). These biological processes were also characteristic of monocytes and macrophages, rather than T cells. Overall, the serum levels of eotaxin-1 and IL-8 in CRC patients substantially correlated with the expression levels of certain genes in peripheral CD4+ cells compared to healthy controls.
Figure 3.
Spearman correlation coefficients between genes expressed by CD4+ cells. Genes expressed by CD4+ cells of CRC patients and healthy volunteers, with two-sided p-values, (a) eotaxin-1 and (b) IL-8 levels.
Table 1.
Biological process networks for 1063 genes whose expression in peripheral CD4+ cells was positively correlated with serum eotaxin concentration.
| Networks | Total | P value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Immune response_Phagocytosis | 222 | 2.27E-08 | 1.81E-06 | 33 | ITGB2, Syk, IL-15, RhoA, Myosin I, C/EBP, Dectin-1, Fc gamma RII beta, ILT4, MSN (moesin), ERM proteins, SHPS-1, Fc epsilon RI gamma, MSR1, MANR, Lyn, PLC-gamma 2, IL-15RA, Btk, ILT2, Hck, TLR4, MARCO, MARCKS, MLCK, PLC-gamma, gp91-phox, p40-phox, PAK1, p67-phox, FGR, Fc alpha receptor, Fc gamma RII alpha |
|
| |||||
| Cell adhesion_Platelet aggregation | 158 | 2.34E-08 | 1.81E-06 | 27 | ITGB2, Syk, GAB2, PLA2, RhoA, Thrombospondin 1, G-protein beta/gamma, COX-1 (PTGS1), Fc epsilon RI gamma, c-Src, ENP1, THAS, cPLA2, PTAFR, Lyn, PLC-gamma 2, PKA-reg (cAMP-dependent), GP-IX, P2Y12, Gab, G-protein alpha-i family, G-protein alpha-i2, P2X1, MLCK, PLC-gamma, CD36, VAV-2 |
|
| |||||
| Cell adhesion_Amyloid proteins | 195 | 6.02E-06 | 3.09E-04 | 26 | RhoA, FZD1, Nicastrin, NOTCH2, APLP2 active fragment, G-protein beta/gamma, Jagged1, Nidogen, p120-catenin, Tcf(Lef), c-Src, Presenilin 2, FZD2, FZD5, Notch, Presenilin 1, Alpha-catenin, APLP2 precursor, Cathepsin D, MALS, Frizzled, ADAM9, PKC, PAK1, Plakoglobin, Presenilin |
|
| |||||
| Immune response_Antigen presentation | 197 | 6.23E-05 | 2.40E-03 | 24 | CIITA, ICAM1, MHC class II beta chain, CD1b, HLADPA1, CD1d, HLA-DQA1, JAK2, HA2Z, Fc epsilon RI gamma, IP-30, HLA-DM, HLA-DRB1, CD1a, Cathepsin S, LFA-3, MHC class II, HLA-DQB1, HLA-DRA1, HLA-DPB1, HLA-DRB4, CD86, CD74, RING6 |
|
| |||||
| Proliferation_Positive regulation cell proliferation | 221 | 1.48E-04 | 4.56E-03 | 25 | p21, HGF, RhoG, Beta-arrestin1, GAB2, Fra-1, RhoA, Galpha(i)-specific peptide GPCRs, JAK2, MTG16 (CBFA2T3), c-Src, VEGF-A, RasGRP4, PKA-reg (cAMP-dependent), G-protein alpha-15, TCIRG1 (Atp6i), CCR1, G-protein alpha-i family, M-CSF receptor, G-protein alpha-i2, CSDA, MLCK, c-Fes, FLT3, PAK1 |
|
| |||||
| Chemotaxis | 137 | 2.00E-04 | 5.14E-03 | 18 | ITGB2, Syk, C5aR, Fra-1, GRO-2, Prokineticin 2, PD-ECGF (TdRPase), IL-1 beta, Galpha(i)-specific peptide GPCRs, PLAUR (uPAR), MIG, Integrin, VEGF-A, PTAFR, CCR1, G-protein alpha-i family, Galpha(q)-specific peptide GPCRs, PLD1 |
|
| |||||
| Inflammation_IFN-gamma signaling | 109 | 4.08E-04 | 8.97E-03 | 15 | CIITA, p21, ITGB2, IL-15, IL-18, ICAM1, PKC-delta, K12, JAK2, MIG, c-Src, PLC-gamma 2, TLR4, PLC-gamma, Fc alpha receptor |
|
| |||||
| Apoptosis_Anti-apoptosis mediated by external signals via NF-kB | 111 | 4.97E-04 | 9.57E-03 | 15 | IL-15, MyD88, G-protein beta/gamma, TNF-R2, VEGF-A, CD30(TNFRSF8), CSF2RA, PKA-reg (cAMP-dependent), IL-15RA, G-protein alpha-i family, TLR4, Bcl-3, TL1A(TNFSF15), APRIL(TNFSF13), BAFF(TNFSF13B) |
|
| |||||
| Inflammation_Neutrophil activation | 215 | 6.08E-04 | 1.02E-02 | 23 | ITGB2, C5aR, PLA2, ICAM1, GRO-2, RhoA, PKC-delta, G-protein beta/gamma, TNF-R2, Galpha(i)-specific peptide GPCRs, Syntaxin 7, cPLA2, Btk, G-protein alpha-15, G-protein alpha-i family, G-protein alpha-i2, PA24A, gp91-phox, ALOX5, p40-phox, PAK1, p67-phox, PLD1 |
|
| |||||
| Inflammation_IL-4 signaling | 115 | 7.27E-04 | 1.02E-02 | 15 | HLADPA1, HLA-DQA1, JAK2, MHC class II, Bax, HLA-DQB1, HLA-DRA1, HLA-DPB1, HLA-DRB4, CD86, CD74, c-Fes, CD13, IL13RA1, Fc gamma RII alpha |
Table 2.
Biological process networks for 974 genes whose expression was negatively correlated with CD4+ peripheral blood cells and Eotaxin.
| Networks | Total | P value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Proteolysis_ECM remodeling | 85 | 5.38E-05 | 7.21E-03 | 10 | Collagen XIV, Tenascin-C, NEPH2, MMP-16, Protein C inhibitor, Serpin B12, COL18A1, Kallikrein 2, Trypsin II, Aggrecanase-1 |
|
| |||||
| Neurophysiological process_Transmission of nerve impulse | 212 | 3.13E-04 | 2.10E-02 | 15 | L-type Ca(II) channel, alpha 1C subunit, GABA-A receptor gamma-2 subunit, KCC2, mGluR3, Galpha(i)-specific peptide GPCRs, mGluR1, Galpha(q)-specific metabotropic glutamate GPCRs, Ionotropic glutamate receptor, Galpha(i)-specific metabotropic glutamate GPCRs, GluR6, Galpha(i)-specific amine GPCRs, CHT1, RIN, G-protein alpha-s, Kainate receptor |
|
| |||||
| Reproduction_Gonadotropin regulation | 199 | 1.64E-03 | 5.29E-02 | 13 | L-type Ca(II) channel, alpha 1C subunit, GABA-A receptor gamma-2 subunit, mGluR3, mGluR1, Galpha(q)-specific metabotropic glutamate GPCRs, Ionotropic glutamate receptor, Galpha(i)-specific metabotropic glutamate GPCRs, Secretogranin 1, Protein kinase G1, Adenylate cyclase, G-protein alpha-s, Protein kinase G, Kainate receptor |
|
| |||||
| Development_Blood vessel morphogenesis | 228 | 1.97E-03 | 5.29E-02 | 14 | PDE, Galpha(i)-specific peptide GPCRs, PDE7A, Endomucin, Galpha(q)-specific amine GPCRs, Galpha(i)-specific amine GPCRs, Protein kinase G1, Galpha(q)-specific peptide GPCRs, COL18A1, Tissue kallikreins, Neuropilin-1, G-protein alpha-s, Protein kinase G, Transferrin |
|
| |||||
| Reproduction_Spermatogenesis, motility and copulation | 228 | 1.97E-03 | 5.29E-02 | 14 | PDGF receptor, MFGE8, IGF-1 receptor, Ropporin, MSK1, S5AR2, BBS2, Tissue kallikreins, BMP2, Kallikrein 2, SOX5, CREM (activators), ZFP37, PDGF-R-alpha |
|
| |||||
| Proteolysis_Connective tissue degradation | 119 | 3.23E-03 | 6.80E-02 | 9 | Trypsin, Tenascin-C, MMP-16, Protein C inhibitor, Serpin B12, Tissue kallikreins, Kallikrein 2, Trypsin II, Aggrecanase-1 |
|
| |||||
| Development_Neurogenesis in general | 192 | 3.55E-03 | 6.80E-02 | 12 | WNT4, RET, CHRM, Neuromodulin, WNT7A, WNT, Galpha(q)-specific amine GPCRs, Galpha(i)-specific amine GPCRs, HDAC7, ACM3, SOX8, SOX14 |
|
| |||||
| Development_Cartilage development | 66 | 6.45E-03 | 1.08E-01 | 6 | TR-alpha, Noggin, COL1A2, BMP2, SOX5, Aggrecanase-1 |
|
| |||||
| Reproduction_Male sex differentiation | 243 | 8.98E-03 | 1.30E-01 | 13 | AP-2A, PDGF receptor, Olfactory receptor, RET, IGF-1 receptor, MSK1, S5AR2, HSF2, BMP2, SOX5, CREM (activators), ZFP37, PDGF-R-alpha |
|
| |||||
| Reproduction_GnRH signaling pathway | 166 | 9.70E-03 | 1.30E-01 | 10 | GABA-A receptor gamma-2 subunit, mGluR3, mGluR1, Galpha(q)-specific metabotropic glutamate GPCRs, Ionotropic glutamate receptor, Galpha(i)-specific metabotropic glutamate GPCRs, Protein kinase G1, G-protein alpha-s, Protein kinase G, Kainate receptor |
Table 3.
Biological process networks for 250 genes whose expression in peripheral CD4+ cells was positively correlated with serum IL-8 concentration.
| Networks | Total | P value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Cytoskeleton_Actin filaments | 176 | 2.12E-05 | 2.31E-03 | 10 | Actin muscle, Talin, Tropomyosin, RhoA, Myosin I, CAPZA, MELC, TARA, Actin, CAPZA1 |
|
| |||||
| Cytoskeleton_Regulation of cytoskeleton rearrangement | 183 | 8.79E-04 | 3.67E-02 | 8 | Actin muscle, Talin, RhoA, CAPZA, MELC, TARA, Actin, CAPZA1 |
|
| |||||
| Development_Skeletal muscle development | 144 | 1.01E-03 | 3.67E-02 | 7 | ACTA2, Smooth muscle myosin, Actin muscle, Tropomyosin, RhoA, MELC, Actin |
|
| |||||
| Muscle contraction | 173 | 2.90E-03 | 6.54E-02 | 7 | ACTA2, Syntrophin B, Smooth muscle myosin, Actin muscle, Tropomyosin, MELC, Actin |
|
| |||||
| Immune response_Phagocytosis | 222 | 3.00E-03 | 6.54E-02 | 8 | ILT2, Talin, CD63, RhoA, Myosin I, MSR1, MELC, Actin |
|
| |||||
| Cell adhesion_Integrin priming | 110 | 7.22E-03 | 1.17E-01 | 5 | ACTA2, ITGA2B, Talin, Integrin, Actin |
|
| |||||
| Cell adhesion_Platelet aggregation | 158 | 7.84E-03 | 1.17E-01 | 6 | COX-1 (PTGS1), Talin, RhoA, ENP1, MELC, GP-IB beta |
|
| |||||
| Cell adhesion_Integrin-mediated cell-matrix adhesion | 214 | 9.17E-03 | 1.17E-01 | 7 | ITGA2B, ITGB5, Talin, Integrin, RhoA, MELC, Actin |
|
| |||||
| Inflammation_Amphoterin signaling | 118 | 9.64E-03 | 1.17E-01 | 5 | ITGAM, MyD88, RhoA, MELC, Actin |
|
| |||||
| Proteolysis_Proteolysis in cell cycle and apoptosis | 125 | 1.22E-02 | 1.33E-01 | 5 | Presenilin 2, Cathepsin C, FBX6, Pseudo-ICE, Presenilin |
Table 4.
Biological process networks for 586 genes whose expression in peripheral CD4+ cells was negatively correlated with serum IL-8 concentration.
| Networks | Total | P value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Muscle contraction | 173 | 2.69E-03 | 1.78E-01 | 7 | K(+) channel, subfamily J, Dystrophin, PKC-alpha, MyHC, PKC, Titin, cPKC (conventional) |
|
| |||||
| Development_Blood vessel morphogenesis | 228 | 3.26E-03 | 1.78E-01 | 8 | PDE3B, PKC-alpha, COL18A1, PDE, PDE9A, TERT, PDE7A, Endomucin |
|
| |||||
| Cardiac development_Wnt_beta-catenin, Notch, VEGF, IP3 and integrin signaling | 150 | 5.75E-03 | 2.09E-01 | 6 | PKC-alpha, Polycystin, CHIBBY, MyHC, Titin, LRP6 |
|
| |||||
| Cardiac development_FGF_ErbB signaling | 124 | 1.12E-02 | 3.05E-01 | 5 | PKC-alpha, Polycystin, MyHC, Titin, gp130 |
|
| |||||
| Development_Skeletal muscle development | 144 | 2.02E-02 | 4.41E-01 | 5 | Dystrophin, HDAC7, Histone deacetylase class II, MyHC, Titin |
|
| |||||
| Proliferation_Positive regulation cell proliferation | 221 | 3.32E-02 | 5.15E-01 | 6 | PUR-alpha, RASGRF1, COL18A1, IGF-1 receptor, IL-11 receptor, gp130 |
|
| |||||
| Development_Cartilage development | 66 | 3.51E-02 | 5.15E-01 | 3 | TR-alpha, Noggin, Aggrecanase-1 |
|
| |||||
| Reproduction_Spermatogenesis, motility and copulation | 228 | 3.78E-02 | 5.15E-01 | 6 | MSK1, MFGE8, Oct-3/4, IGF-1 receptor, PKC, ZFP37 |
|
| |||||
| Reproduction_Male sex differentiation | 243 | 4.90E-02 | 5.41E-01 | 6 | MSK1, Oct-3/4, IGF-1 receptor, PKC, ZFP37, PMEPA1 |
|
| |||||
| Transcription_Chromatin modification | 127 | 4.97E-02 | 5.41E-01 | 4 | SATB1, MSK1, HDAC7, Histone deacetylase class II |
4. Discussion
Based on our previous findings that the immune pathophysiology of digestive system cancers is reflected in peripheral blood, we investigated the inflammatory conditions of CRC patients by assessing cytokine/chemokine and performing gene expression analyses of peripheral blood using bead-based multiplex immunoassay and DNA microarray, respectively. Gene expression in peripheral CD4+ and whole blood cells differed between CRC patients and healthy controls [5]. The serum levels of eotaxin-1 and IL-8 were significantly elevated in CRC patients, and the levels significantly correlated with changes in the gene expression levels in CD4+ cells.
Cytokines/chemokines (humoral immunomodulators) regulate cancer-associated immune responses [15]. Eotaxin-1 (CCL11) controls both the eosinophil-mediated immune response [16] and other immune responses of Th2 cells [17]. We found that eotaxin-1 levels were elevated in the sera of CRC patients. The expression levels of certain other genes that significantly differed between peripheral CD4+ cells of CRC patients and those of healthy controls also correlated with serum eotaxin-1 concentrations. The upregulated genes, including those encoding MHC, CD1d, TLR4, IL-15, Fc gamma, and Hck [18], are normally expressed in monocytes/macrophages. In humans, both T cells and monocytes express CD4; however, the cell functions differ [19].
Serum IL-8 (CXCL8) levels were significantly elevated in CRC patients and those with other cancers [20–22]. IL-8 fosters CRC tumor growth, invasion, and metastasis [23, 24], promoting in vitro cell proliferation of human colon carcinoma cells via metalloproteinase-mediated cleavage [25]. Additionally, tumor-derived IL-8 induces the formation of immunosuppressive neutrophils and myeloid-derived suppressor cells in tumor microenvironments [26, 27]. Thus, elevated serum IL-8 levels in CRC patients may play an important role in cancer progression; indeed, attainment of an advanced clinical stage was associated with an increase in serum IL-8 concentration. Serum IL-8 levels correlated with changes in the expression levels of CD4+ cell genes compared to healthy controls; these changes also suggested that phagocytosis was in play.
Because immune-mediating cells are miscellaneous, including myeloid-derived cells such as neutrophils, monocytes, and lymphocytes, the interaction of these immune-mediating cells in CRC should be studied to further understand the immune pathophysiological features of CRC. The most frequent subpopulation of whole blood cells is neutrophils. We observed that the gene expression profile of whole blood cells and CD4+ cells was discernible between CRC patients and healthy volunteers; thus, the interaction between these two populations should particularly be investigated.
Collectively, this study showed that transcriptional alteration of peripheral blood, especially CD4+ cells, and elevation of humoral mediators were possibly reflection of immune pathophysiology of CRC, which are compatible to the recent other reports showing gene expression profile alteration [28–30] as well as alteration of concentration of humoral immune mediators [31] in peripheral blood. Humoral immune mediators and cellular immunity are interactive [32, 33]. As the immune system and its reaction are extremely complex, especially in cancers [34], each humoral mediator and cellular fraction should be further investigated to understand immune pathophysiology in detail. Despite these possible immune pathophysiological features being reflected by serum chemokines and peripheral CD4+ cells, further analysis in a larger cohort than that used in the current study should be performed to explore interactive features between chemokines, eotaxin-1 and IL-8, and CD4+ cells in peripheral blood of CRC patients.
In conclusion, we showed that CRC featured systemic inflammation, changes in the serum concentrations of eotaxin-1 and IL-8, and correlated changes in gene expression in peripheral blood CD4+ cells. Further studies exploring the roles played by chemokines and peripheral CD4+ cells in CRC patients are required. In addition, it should be explored how eotaxin-1 and IL-8 elevation is correlated with clinical outcome of CRC in terms of overall survival, therapeutic response after curative treatment with endoscopy or surgery, and relapse rate after complete cure.
Acknowledgments
This study was supported, in part, by a grant-in-aid from the Japanese Agency for Medical Research and Development.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
This study was approved by the Institutional Review Board and was performed in accordance with the Declaration of Helsinki.
Consent
Written informed consent was obtained from all participants.
Conflicts of Interest
The authors declare that they have no conflicts of interest in this paper.
Authors' Contributions
Takuya Komura and Masaaki Yano contributed to write a manuscript and analysis and interpretation of data. Akimitsu Miyake contributed to acquisition of data and interpretation of data. Hisashi Takabatake, Masaki Miyazawa, Norihiko Ogawa, and Akihiro Seki contributed to acquisition of data and analysis of data. Masao Honda and Takashi Wada contributed to analysis of data and interpretation of data. Shigeyuki Matsui and Shuichi Kaneko contributed to interpretation of data and supervision of the research. Yoshio Sakai contributed to interpretation of data and supervision of the research and wrote a manuscript. Takuya Komura and Masaaki Yano contributed equally to this study.
Supplementary Materials
Supplemental Table 1. Characteristics of serum chemokine study subjects.
Supplemental Table 2. Characteristics of study subjects.
Supplemental Fig. 1. Serum concentrations of cytokines and chemokines.
Supplemental Fig. 2. Serum concentrations of cytokines and chemokines.
References
- 1.Smith R. A., Manassaram-Baptiste D., Brooks D., et al. Cancer screening in the United States, 2014: A review of current American Cancer Society guidelines and current issues in cancer screening. CA: A Cancer Journal for Clinicians. 2014;64(1):30–51. doi: 10.3322/caac.21212. [DOI] [PubMed] [Google Scholar]
- 2.Bond J. H. Colorectal cancer screening: The potential role of virtual colonoscopy. Journal of Gastroenterology. 2002;37(13):92–96. doi: 10.1007/BF02990108. [DOI] [PubMed] [Google Scholar]
- 3.Bond J. H. Fecal occult blood test screening for colorectal cancer. Gastrointestinal Endoscopy Clinics of North America. 2002;12(1):11–21. doi: 10.1016/S1052-5157(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 4.Ramos M., Llagostera M., Esteva M., et al. Knowledge and attitudes of primary healthcare patients regarding population-based screening for colorectal cancer. BMC Cancer. 2011;11 doi: 10.1186/1471-2407-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda M., Sakai Y., Yamashita T., et al. Differential gene expression profiling in blood from patients with digestive system cancers. Biochemical and Biophysical Research Communications. 2010;400(1):7–15. doi: 10.1016/j.bbrc.2010.07.123. [DOI] [PubMed] [Google Scholar]
- 6.Pecht T., Gutman-Tirosh A., Bashan N., Rudich A. Peripheral blood leucocyte subclasses as potential biomarkers of adipose tissue inflammation and obesity subphenotypes in humans. Obesity Reviews. 2014;15(4):322–337. doi: 10.1111/obr.12133. [DOI] [PubMed] [Google Scholar]
- 7.Fridman W. H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nature Reviews Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 8.Zhang N., Bevan M. J. CD8+ T cells: foot soldiers of the immune system. Immunity. 2011;35(2):161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solito S., Marigo I., Pinton L., Damuzzo V., Mandruzzato S., Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Annals of the New York Academy of Sciences. 2014;1319(1):47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- 10.Facciabene A., Motz G. T., Coukos G. T-Regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Research. 2012;72(9):2162–2171. doi: 10.1158/0008-5472.can-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okazaki T., Chikuma S., Iwai Y., Fagarasan S., Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nature Immunology. 2013;14(12):1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 12.Komura T., Sakai Y., Harada K., et al. Inflammatory features of pancreatic cancer highlighted by monocytes/macrophages and CD4+ T cells with clinical impact. Cancer Science. 2015;106(6):672–686. doi: 10.1111/cas.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai Y., Honda M., Fujinaga H., et al. Common transcriptional signature of tumor-infiltrating mononuclear inflammatory cells and peripheral blood mononuclear cells in hepatocellular carcinoma patients. Cancer Research. 2008;68(24):10267–10279. doi: 10.1158/0008-5472.CAN-08-0911. [DOI] [PubMed] [Google Scholar]
- 14.Sakai Y., Tatsumi I., Higashimoto M., et al. Association of changes in the gene expression profile of blood cells with the local tumor inflammatory response in a murine tumor model. Biochemical and Biophysical Research Communications. 2012;428(1):36–43. doi: 10.1016/j.bbrc.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Pengjun Z., Xinyu W., Feng G., et al. Multiplexed cytokine profiling of serum for detection of colorectal cancer. Future Oncology. 2013;9(7):1017–1027. doi: 10.2217/fon.13.71. [DOI] [PubMed] [Google Scholar]
- 16.Kitaura M., Nakajima T., Imai T., et al. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3. The Journal of Biological Chemistry. 1996;271(13):7725–7730. doi: 10.1074/jbc.271.13.7725. [DOI] [PubMed] [Google Scholar]
- 17.Teran L. M., Mochizuki M., Bartels J., et al. Th1- and Th2-Type Cytokines Regulate the Expression and Production of Eotaxin and RANTES by Human Lung Fibroblasts. American Journal of Respiratory Cell and Molecular Biology. 1999;20(4):777–786. doi: 10.1165/ajrcmb.20.4.3508. [DOI] [PubMed] [Google Scholar]
- 18.Lynch G. W., Turville S., Carter B., et al. Marked differences in the structures and protein associations of lymphocyte and monocyte CD4: Resolution of a novel CD4 isoform. Immunology & Cell Biology. 2006;84(2):154–165. doi: 10.1111/j.1440-1711.2005.01403.x. [DOI] [PubMed] [Google Scholar]
- 19.Szabo G., Miller C. L., Kodys K. Antigen presentation by the CD4 positive monocyte subset. Journal of Leukocyte Biology. 1990;47(2):111–120. doi: 10.1002/jlb.47.2.111. [DOI] [PubMed] [Google Scholar]
- 20.Xie K. Interleukin-8 and human cancer biology. Cytokine & Growth Factor Reviews. 2001;12(4):375–391. doi: 10.1016/S1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 21.Waugh D. J., Wilson C. The interleukin-8 pathway in cancer. Clinical Cancer Research. 2008;14(21):6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 22.Jin W.-J., Xu J.-M., Xu W.-L., Gu D.-H., Li P.-W. Diagnostic value of interleukin-8 in colorectal cancer: A case-control study and meta- Analysis. World Journal of Gastroenterology. 2014;20(43):16334–16342. doi: 10.3748/wjg.v20.i43.16334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baggiolini M., Dewald B., Moser B. Human chemokines: an update. Annual Review of Immunology. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 24.Ning Y., Lenz H.-J. Targeting IL-8 in colorectal cancer. Expert Opinion on Therapeutic Targets. 2012;16(5):491–497. doi: 10.1517/14728222.2012.677440. [DOI] [PubMed] [Google Scholar]
- 25.Itoh Y., Joh T., Tanida S., et al. IL-8 promotes cell proliferation and migration through metalloproteinase- cleavage proHB-EGF in human colon carcinoma cells. Cytokine. 2005;29(6):275–282. doi: 10.1016/j.cyto.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Fridlender Z. G., Sun J., Mishalian I., et al. Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0031524.e31524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao H.-L., Chen J.-W., Li M., et al. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients' adverse prognosis. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0030806.e30806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciarloni L., Hosseinian S., Monnier-Benoit S., Imaizumi N., Dorta G., Ruegg C. Discovery of a 29-gene panel in peripheral blood mononuclear cells for the detection of colorectal cancer and adenomas using high throughput real-time PCR. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0123904.e0123904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Y.-T., Huang C.-S., Yao C.-T., et al. Gene expression profile of peripheral blood in colorectal cancer. World Journal of Gastroenterology. 2014;20(39):14463–14471. doi: 10.3748/wjg.v20.i39.14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichita C., Ciarloni L., Monnier-Benoit S., Hosseinian S., Dorta G., Rüegg C. A novel gene expression signature in peripheral blood mononuclear cells for early detection of colorectal cancer. Alimentary Pharmacology & Therapeutics. 2014;39(5):507–517. doi: 10.1111/apt.12618. [DOI] [PubMed] [Google Scholar]
- 31.Johdi N. A., Mazlan L., Sagap I., Jamal R. Profiling of cytokines, chemokines and other soluble proteins as a potential biomarker in colorectal cancer and polyps. Cytokine. 2017;99:35–42. doi: 10.1016/j.cyto.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Komura T., Takabatake H., Harada K., et al. Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Science. 2017;108(11):2122–2129. doi: 10.1111/cas.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardini G., Antonangeli F., Bonanni V., Santoni A. Dysregulation of chemokine/chemokine receptor axes and NK cell tissue localization during diseases. Frontiers in Immunology. 2016;7(402) doi: 10.3389/fimmu.2016.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandya P. H., Murray M. E., Pollok K. E., Renbarger J. L. The Immune System in Cancer Pathogenesis: Potential Therapeutic Approaches. Journal of Immunology Research. 2016;2016:13. doi: 10.1155/2016/4273943.4273943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Characteristics of serum chemokine study subjects.
Supplemental Table 2. Characteristics of study subjects.
Supplemental Fig. 1. Serum concentrations of cytokines and chemokines.
Supplemental Fig. 2. Serum concentrations of cytokines and chemokines.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.