Abstract Abstract
Diaporthe species have often been reported as important plant pathogens, saprobes and endophytes on a wide range of plant hosts. Although several Diaporthe species have been recorded in China, little is known about species able to infect forest trees. Therefore, extensive surveys were recently conducted in Beijing, Heilongjiang, Jiangsu, Jiangxi, Shaanxi and Zhejiang Provinces. The current results emphasised on 15 species from 42 representative isolates involving 16 host genera using comparisons of DNA sequence data for the nuclear ribosomal internal transcribed spacer (ITS), calmodulin (cal), histone H3 (his3), partial translation elongation factor-1α (tef1) and β-tubulin (tub2) gene regions, as well as their morphological features. Three known species, D.biguttulata, D.eres and D.unshiuensis, were identified. In addition, twelve novel taxa were collected and are described as D.acerigena, D.alangii, D.betulina, D.caryae, D.cercidis, D.chensiensis, D.cinnamomi, D.conica, D.fraxinicola, D.kadsurae, D.padina and D.ukurunduensis. The current study improves the understanding of species causing diebacks on ecological and economic forest trees and provides useful information for the effective disease management of these hosts in China.
Keywords: Dieback, DNA phylogeny, Systematics, Taxonomy
Introduction
The genus Diaporthe Nitschke represents a cosmopolitan group of fungi occupying diverse ecological behaviour as plant pathogens, endophytes and saprobes (Muralli et al. 2006, Rossman et al. 2007, Garcia-Reyne et al. 2011, Udayanga et al. 2011, 2012a, b, 2014a, b, 2015, Gomes et al. 2013, Fan et al. 2015, Du et al. 2016, Dissanayake et al. 2017b, Guarnaccia and Crous 2017, Yang et al. 2017a, b, 2018, Guarnaccia et al. 2018, Marin-Felix et al. 2018). Diaporthe species are responsible for diseases on a wide range of plant hosts, including agricultural crops, forest trees and ornamentals, some of which are economically important. Several symptoms such as root and fruit rots, dieback, stem cankers, leaf spots, leaf and pod blights and seed decay are caused by Diaporthe spp. (Uecker 1988, Rehner and Uecker 1994, Mostert et al. 2001, Santos et al. 2011, Thompson et al. 2011, Udayanga et al. 2011). For example, D.ampelina, the causal agent of Phomopsis cane and leaf spot, is known as a severe pathogen of grapevines (Hewitt and Pearson 1988), infecting all green tissues and causing yield reductions of up to 30% in temperate regions (Erincik et al. 2001). Diaporthecitri is another well-known pathogen exclusively found on Citrus spp. causing melanose, stem-end rot and gummosis in all the citrus production areas except Europe (Mondal et al. 2007, Udayanga et al. 2014a, Guarnaccia and Crous 2017, 2018). Similarly, stem canker, attributed to several Diaporthe spp., is one of the most important diseases of sunflower (Helianthusannuus) worldwide (Muntañola-Cvetković et al. 1981, Thompson et al. 2011).
Several species of Diaporthe include a broad number of endophytes associated with hosts present in temperate and tropical regions (Udayanga et al. 2011). Gomes et al. (2013) considered that D.endophytica is a sterile endophyte on Schinusterebinthifolius and Maytenusilicifolia based on molecular phylogeny. Huang et al. (2015) distinguished seven undescribed Diaporthe species associated with citrus in China. Moreover, some endophytes have been shown to act as opportunistic plant pathogens. For instance, D.foeniculina has been found as both endophyte and opportunistic pathogen on various herbaceous weeds, ornamentals and fruit trees (Udayanga et al. 2014a, Guarnaccia et al. 2016).
The genus Diaporthe (syn. Phomopsis) was established by Nitschke (1870). Species identification criteria in Diaporthe were originally based on host association, morphology and culture characteristics (Mostert et al. 2001, Santos and Phillips 2009, Udayanga et al. 2012). As a consequence, a broad increase in the number of proposed Diaporthe species occurred. More than 1000 epithets for Diaporthe and 950 for Phomopsis were listed in Index Fungorum (2018) (http://www.indexfungorum.org/) (accessed 1 March 2018). The abolishment of the dual nomenclature system for pleomorphic fungi raised the question about which generic name to use. Given that both names are well known amongst plant pathologists and have been equally used, Rossman et al. (2015) proposed that the name Diaporthe (Nitschke 1870) has priority over Phomopsis (Saccardo and Roumeguère 1884) and has been adopted as the generic name in recent major studies (Gomes et al. 2013, Udayanga et al. 2014a, b, 2015, Fan et al. 2015, Huang et al. 2015, Du et al. 2016, Gao et al. 2017, Yang et al. 2017a, b, c, 2018).
The sexual morph of Diaporthe is characterised by immersed ascomata and an erumpent pseudostroma with elongated perithecial necks. Asci are unitunicate, clavate to cylindrical. Ascospores are fusoid, ellipsoid to cylindrical, hyaline, biseriate to uniseriate in the ascus and sometimes with appendages (Udayanga et al. 2011). The asexual morph is characterised by ostiolate conidiomata, with cylindrical phialides producing three types of hyaline, aseptate conidia (Udayanga et al. 2011). Previously, species identification of Diaporthe was largely referred to the assumption of host-specificity, leading to the proliferation of names (Gomes et al. 2013). More than one species of Diaporthe can colonise a single host, while one species can be associated with different hosts (Santos and Phillips 2009, Diogo et al. 2010, Santos et al. 2011, Gomes et al. 2013). In addition, considerable variability of the phenotype characters is present within a species (Rehner and Uecker 1994, Mostert et al. 2001, Santos et al. 2010, Udayanga et al. 2011, 2012a). Species identification is essential for understanding the epidemiology and plant diseases management and to guide the implementation of phytosanitary measures (Santos and Phillips 2009, Udayanga et al. 2011, Santos et al. 2017). Thus, molecular data are necessary to resolve Diaporthe taxonomy and, during the recent years, many species have been described through a polyphasic approach together with morphology (Gomes et al. 2013, Udayanga et al. 2014a, b, 2015, Huang et al. 2015, Gao et al. 2017, Guarnaccia and Crous 2017, Yang et al. 2018). Santos et al. (2017) revealed that the use of a five-loci dataset (ITS-cal-his3-tef1-tub2) is the optimal combination for species delimitation, showing the ribosomal ITS locus as the least informative, which is contrary to the result of Santos et al. (2010).
Although the classification of Diaporthe has been on-going, species are currently being identified based on a combination of morphological, cultural, phytopathological and phylogenetical analyses (Gomes et al. 2013, Huang et al. 2013, 2015, Udayanga et al. 2014a, b, 2015, Fan et al. 2015, Du et al. 2016, Gao et al. 2016, 2017, Guarnaccia and Crous 2017, Hyde et al. 2017, 2018, Guarnaccia et al. 2018, Jayawardena et al. 2018, Perera et al. 2018a, b, Tibpromma et al. 2018, Wanasinghe et al. 2018). However, fungi isolated from forest trees in China were recorded in old fungal literature without any living culture and molecular data (Teng 1963, Tai 1979, Wei 1979). The current study aimed to investigate the major ecological or economic trees in China by large-scale sampling and to identify isolates via morphology and multi-locus phylogeny based on modern taxonomic concepts. From 2015 to 2017, several surveys were conducted in six Provinces representing 16 host genera. The objectives of the present study were (i) to provide a multi-gene phylogeny for the genus Diaporthe based on a large set of freshly collected specimens in China; (ii) to identify Diaporthe taxa associated with disease symptoms or non-symptomatic tissues of various host genera distributed over six Provinces in China; (iii) to define the species limits of D.eres and closely related species based on multi-gene genealogies.
Materials and methods
Isolates
From 2015 to 2017, fresh specimens of Diaporthe were collected from symptomatic or non-symptomatic twigs or branches from Beijing, Heilongjiang, Jiangsu, Jiangxi, Shaanxi and Zhejiang Provinces in China (Table 1). A total of 105 isolates were obtained by removing a mucoid spore mass from conidiomata and spreading the suspension on the surface of 1.8% potato dextrose agar (PDA) in a Petri dish and incubating at 25 °C for up to 24 h. Single germinating conidia were transferred on to fresh PDA plates. Forty-two representative Diaporthe strains were selected based on cultural characteristics on PDA, conidia morphology and ITS sequence data. Specimens were deposited in the Museum of the Beijing Forestry University (BJFC). Axenic cultures are maintained in the China Forestry Culture Collection Centre (CFCC).
Table 1.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaporthe.
| Species | Isolate | Host | Location | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | cal | his3 | tef1 | tub2 | ||||
| D. acaciarum | CBS 138862 | Acacia tortilis | Tanzania | KP004460 | N/Aa | N/Aa | N/Aa | KP004509 |
| D. acaciigena | CBS 129521 | Acacia retinodes | Australia | KC343005 | KC343247 | KC343489 | KC343731 | KC343973 |
| D. acericola | MFLUCC 17-0956 | Acer negundo | Italy | KY964224 | KY964137 | N/Aa | KY964180 | KY964074 |
| D. acerigena | CFCC 52554 | Acer tataricum | China | MH121489 | MH121413 | MH121449 | MH121531 | N/Aa |
| CFCC 52555 | Acer tataricum | China | MH121490 | MH121414 | MH121450 | MH121532 | N/Aa | |
| D. acutispora | CGMCC 3.18285 | Coffea sp. | China | KX986764 | KX999274 | N/Aa | KX999155 | KX999195 |
| D. alangii | CFCC 52556 | Alangium kurzii | China | MH121491 | MH121415 | MH121451 | MH121533 | MH121573 |
| CFCC 52557 | Alangium kurzii | China | MH121492 | MH121416 | MH121452 | MH121534 | MH121574 | |
| CFCC 52558 | Alangium kurzii | China | MH121493 | MH121417 | MH121453 | MH121535 | MH121575 | |
| CFCC 52559 | Alangium kurzii | China | MH121494 | MH121418 | MH121454 | MH121536 | MH121576 | |
| D. alleghaniensis | CBS 495.72 | Betula alleghaniensis | Canada | KC343007 | KC343249 | KC343491 | KC343733 | KC343975 |
| D. alnea | CBS 146.46 | Alnus sp. | Netherlands | KC343008 | KC343250 | KC343492 | KC343734 | KC343976 |
| D. ambigua | CBS 114015 | Pyrus communis | South Africa | KC343010 | KC343252 | KC343494 | KC343736 | KC343978 |
| D. ampelina | STEU2660 | Vitis vinifera | France | AF230751 | AY745026 | N/Aa | AY745056 | JX275452 |
| D. amygdali | CBS 126679 | Prunus dulcis | Portugal | KC343022 | KC343264 | KC343506 | AY343748 | KC343990 |
| D. anacardii | CBS 720.97 | Anacardium occidentale | East Africa | KC343024 | KC343266 | KC343508 | KC343750 | KC343992 |
| D. angelicae | CBS 111592 | Heracleum sphondylium | Austria | KC343027 | KC343269 | KC343511 | KC343753 | KC343995 |
| D. apiculatum | CGMCC 3.17533 | Camellia sinensis | China | KP267896 | N/Aa | N/Aa | KP267970 | KP293476 |
| D. aquatica | IFRDCC 3051 | Aquatic habitat | China | JQ797437 | N/Aa | N/Aa | N/Aa | N/Aa |
| D. arctii | CBS 139280 | Arctium lappa | Austria | KJ590736 | KJ612133 | KJ659218 | KJ590776 | KJ610891 |
| D. arecae | CBS 161.64 | Areca catechu | India | KC343032 | KC343274 | KC343516 | KC343758 | KC344000 |
| D. arengae | CBS 114979 | Arenga enngleri | Hong Kong | KC343034 | KC343276 | KC343518 | KC343760 | KC344002 |
| D. aseana | MFLUCC 12-0299a | Unknown dead leaf | Thailand | KT459414 | KT459464 | N/Aa | KT459448 | KT459432 |
| D. asheicola | CBS 136967 | Vaccinium ashei | Chile | KJ160562 | KJ160542 | N/Aa | KJ160594 | KJ160518 |
| D. aspalathi | CBS 117169 | Aspalathus linearis | South Africa | KC343036 | KC343278 | KC343520 | KC343762 | KC344004 |
| D. australafricana | CBS 111886 | Vitis vinifera | Australia | KC343038 | KC343280 | KC343522 | KC343764 | KC344006 |
| D. baccae | CBS 136972 | Vaccinium corymbosum | Italy | KJ160565 | N/Aa | MF418264 | KJ160597 | N/Aa |
| D. batatas | CBS 122.21 | Ipomoea batatas | USA | KC343040 | KC343282 | N/Aa | KC343766 | KC344008 |
| D. beilharziae | BRIP 54792 | Indigofera australis | Australia | JX862529 | N/Aa | N/Aa | JX862535 | KF170921 |
| D. benedicti | BPI 893190 | Salix sp. | USA | KM669929 | KM669862 | N/Aa | KM669785 | N/Aa |
| D. betulae | CFCC 50469 | Betula platyphylla | China | KT732950 | KT732997 | KT732999 | KT733016 | KT733020 |
| CFCC 50470 | Betula platyphylla | China | KT732951 | KT732998 | KT733000 | KT733017 | KT733021 | |
| D. betulicola | CFCC 51128 | Betula albo-sinensis | China | KX024653 | KX024659 | KX024661 | KX024655 | KX024657 |
| CFCC 51129 | Betula albo-sinensis | China | KX024654 | KX024660 | KX024662 | KX024656 | KX024658 | |
| D. betulina | CFCC 52560 | Betula albo-sinensis | China | MH121495 | MH121419 | MH121455 | MH121537 | MH121577 |
| CFCC 52561 | Betula costata | China | MH121496 | MH121420 | MH121456 | MH121538 | MH121578 | |
| CFCC 52562 | Betula platyphylla | China | MH121497 | MH121421 | MH121457 | MH121539 | MH121579 | |
| D. bicincta | CBS 121004 | Juglans sp. | USA | KC343134 | KC343376 | KC343618 | KC343860 | KC344102 |
| D. biconispora | CGMCC 3.17252 | Citrus grandis | China | KJ490597 | KJ490539 | KJ490539 | KJ490476 | KJ490418 |
| D. biguttulata | CGMCC 3.17248 | Citrus limon | China | KJ490582 | N/Aa | KJ490524 | KJ490461 | KJ490403 |
| CFCC 52584 | Juglans regia | China | MH121519 | MH121437 | MH121477 | MH121561 | MH121598 | |
| CFCC 52585 | Juglans regia | China | MH121520 | MH121438 | MH121478 | MH121562 | MH121599 | |
| D. biguttusis | CGMCC 3.17081 | Lithocarpus glabra | China | KF576282 | N/Aa | N/Aa | KF576257 | KF576306 |
| D. bohemiae | CPC 28222 | Vitis vinifera | Czech Republic | MG281015 | MG281710 | MG281361 | MG281536 | MG281188 |
| D. brasiliensis | CBS 133183 | Aspidosperma tomentosum | Brazil | KC343042 | KC343284 | KC343526 | KC343768 | KC344010 |
| D. caatingaensis | CBS 141542 | Tacinga inamoena | Brazil | KY085927 | N/Aa | N/Aa | KY115603 | KY115600 |
| D. camptothecicola | CFCC 51632 | Camptotheca acuminata | China | KY203726 | KY228877 | KY228881 | KY228887 | KY228893 |
| D. canthii | CBS 132533 | Canthium inerme | South Africa | JX069864 | KC843174 | N/Aa | KC843120 | KC843230 |
| D. caryae | CFCC 52563 | Carya illinoensis | China | MH121498 | MH121422 | MH121458 | MH121540 | MH121580 |
| CFCC 52564 | Carya illinoensis | China | MH121499 | MH121423 | MH121459 | MH121541 | MH121581 | |
| D. cassines | CPC 21916 | Cassine peragua | South Africa | KF777155 | N/Aa | N/Aa | KF777244 | N/Aa |
| D. caulivora | CBS 127268 | Glycine max | Croatia | KC343045 | KC343287 | N/Aa | KC343771 | KC344013 |
| D. celeris | CPC 28262 | Vitis vinifera | Czech Republic | MG281017 | MG281712 | MG281363 | MG281538 | MG281190 |
| D. celastrina | CBS 139.27 | Celastrus sp. | USA | KC343047 | KC343289 | KC343531 | KC343773 | KC344015 |
| D. cercidis | CFCC 52565 | Cercis chinensis | China | MH121500 | MH121424 | MH121460 | MH121542 | MH121582 |
| CFCC 52566 | Cercis chinensis | China | MH121501 | MH121425 | MH121461 | MH121543 | MH121583 | |
| D. chamaeropis | CBS 454.81 | Chamaerops humilis | Greece | KC343048 | KC343290 | KC343532 | KC343774 | KC344016 |
| D. charlesworthii | BRIP 54884m | Rapistrum rugostrum | Australia | KJ197288 | N/Aa | N/Aa | KJ197250 | KJ197268 |
| D. chensiensis | CFCC 52567 | Abies chensiensis | China | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| CFCC 52568 | Abies chensiensis | China | MH121503 | MH121427 | MH121463 | MH121545 | MH121585 | |
| D. cichorii | MFLUCC 17-1023 | Cichorium intybus | Italy | KY964220 | KY964133 | N/Aa | KY964176 | KY964104 |
| D. cinnamomi | CFCC 52569 | Cinnamomum sp. | China | MH121504 | N/Aa | MH121464 | MH121546 | MH121586 |
| CFCC 52570 | Cinnamomum sp. | China | MH121505 | N/Aa | MH121465 | MH121547 | MH121587 | |
| D. cissampeli | CBS 141331 | Cissampelos capensis | South Africa | KX228273 | N/Aa | KX228366 | N/Aa | KX228384 |
| D. citri | AR 3405 | Citrus sp. | USA | KC843311 | KC843157 | N/Aa | KC843071 | KC843187 |
| D. citriasiana | CGMCC 3.15224 | Citrus unshiu | China | JQ954645 | KC357491 | KJ490515 | JQ954663 | KC357459 |
| D.citrichinensis | CGMCC 3.15225 | Citrus sp. | China | JQ954648 | KC357494 | N/Aa | JQ954666 | N/Aa |
| D. collariana | MFLU 17-2770 | Magnolia champaca | Thailand | MG806115 | MG783042 | N/Aa | MG783040 | MG783041 |
| D. compacta | CGMCC 3.17536 | Camellia sinensis | China | KP267854 | N/Aa | KP293508 | KP267928 | KP293434 |
| D. conica | CFCC 52571 | Alangium chinense | China | MH121506 | MH121428 | MH121466 | MH121548 | MH121588 |
| CFCC 52572 | Alangium chinense | China | MH121507 | MH121429 | MH121467 | MH121549 | MH121589 | |
| CFCC 52573 | Alangium chinense | China | MH121508 | MH121430 | MH121468 | MH121550 | MH121590 | |
| CFCC 52574 | Alangium chinense | China | MH121509 | MH121431 | MH121469 | MH121551 | MH121591 | |
| D. convolvuli | CBS 124654 | Convolvulus arvensis | Turkey | KC343054 | KC343296 | KC343538 | KC343780 | KC344022 |
| D. crotalariae | CBS 162.33 | Crotalaria spectabilis | USA | KC343056 | KC343298 | KC343540 | KC343782 | KC344024 |
| D. cucurbitae | CBS 136.25 | Arctium sp. | Unknown | KC343031 | KC343273 | KC343515 | KC343757 | KC343999 |
| D. cuppatea | CBS 117499 | Aspalathus linearis | South Africa | KC343057 | KC343299 | KC343541 | KC343783 | KC344025 |
| D. cynaroidis | CBS 122676 | Protea cynaroides | South Africa | KC343058 | KC343300 | KC343542 | KC343784 | KC344026 |
| D. cytosporella | FAU461 | Citrus limon | Italy | KC843307 | KC843141 | N/Aa | KC843116 | KC843221 |
| D. diospyricola | CPC 21169 | Diospyros whyteana | South Africa | KF777156 | N/Aa | N/Aa | N/Aa | N/Aa |
| D. discoidispora | ZJUD89 | Citrus unshiu | China | KJ490624 | N/Aa | KJ490566 | KJ490503 | KJ490445 |
| D. dorycnii | MFLUCC 17-1015 | Dorycnium hirsutum | Italy | KY964215 | N/Aa | N/Aa | KY964171 | KY964099 |
| D. elaeagni-glabrae | CGMCC 3.18287 | Elaeagnus glabra | China | KX986779 | KX999281 | KX999251 | KX999171 | KX999212 |
| D. ellipicola | CGMCC 3.17084 | Lithocarpus glabra | China | KF576270 | N/Aa | N/Aa | KF576245 | KF576291 |
| D. endophytica | CBS 133811 | Schinus terebinthifolius | Brazil | KC343065 | KC343307 | KC343549 | KC343791 | KC343065 |
| D. eres | AR5193 | Ulmus sp. | Germany | KJ210529 | KJ434999 | KJ420850 | KJ210550 | KJ420799 |
| CFCC 52575 | Castanea mollissima | China | MH121510 | N/Aa | MH121470 | MH121552 | MH121592 | |
| CFCC 52576 | Castanea mollissima | China | MH121511 | MH121432 | MH121471 | MH121553 | MH121593 | |
| CFCC 52577 | Acanthopanax senticosus | China | MH121512 | MH121433 | MH121472 | MH121554 | MH121594 | |
| CFCC 52578 | Sorbus sp. | China | MH121513 | MH121434 | MH121473 | MH121555 | MH121595 | |
| CFCC 52579 | Juglans regia | China | MH121514 | N/Aa | MH121474 | MH121556 | N/Aa | |
| CFCC 52580 | Melia azedarace | China | MH121515 | N/Aa | MH121475 | MH121557 | MH121596 | |
| CFCC 52581 | Rhododendron simsii | China | MH121516 | N/Aa | MH121476 | MH121558 | MH121597 | |
| D. eucalyptorum | CBS 132525 | Eucalyptus sp. | Australia | NR120157 | N/Aa | N/Aa | N/Aa | N/Aa |
| D. foeniculacea | CBS 123208 | Foeniculum vulgare | Portugal | KC343104 | KC343346 | KC343588 | KC343830 | KC344072 |
| D. fraxini-angustifoliae | BRIP 54781 | Fraxinus angustifolia | Australia | JX862528 | N/Aa | N/Aa | JX862534 | KF170920 |
| D. fraxinicola | CFCC 52582 | Fraxinus chinensis | China | MH121517 | MH121435 | N/Aa | MH121559 | N/Aa |
| CFCC 52583 | Fraxinus chinensis | China | MH121518 | MH121436 | N/Aa | MH121560 | N/Aa | |
| D. fukushii | MAFF 625034 | Pyrus pyrifolia | Japan | JQ807469 | N/Aa | N/Aa | JQ807418 | N/Aa |
| D. fusicola | CGMCC 3.17087 | Lithocarpus glabra | China | KF576281 | KF576233 | N/Aa | KF576256 | KF576305 |
| D. ganjae | CBS 180.91 | Cannabis sativa | USA | KC343112 | KC343354 | KC343596 | KC343838 | KC344080 |
| D. garethjonesii | MFLUCC 12-0542a | Unknown dead leaf | Thailand | KT459423 | KT459470 | N/Aa | KT459457 | KT459441 |
| D. goulteri | BRIP 55657a | Helianthus annuus | Australia | KJ197290 | N/Aa | N/Aa | KJ197252 | KJ197270 |
| D. gulyae | BRIP 54025 | Helianthus annuus | Australia | JF431299 | N/Aa | N/Aa | KJ197271 | JN645803 |
| D. helianthi | CBS 592.81 | Helianthus annuus | Serbia | KC343115 | KC343357 | KC343599 | KC343841 | KC344083 |
| D. helicis | AR5211 | Hedera helix | France | KJ210538 | KJ435043 | KJ420875 | KJ210559 | KJ420828 |
| D. heterophyllae | CBS 143769 | Acacia heterohpylla | France | MG600222 | MG600218 | MG600220 | MG600224 | MG600226 |
| D. hickoriae | CBS 145.26 | Carya glabra | USA | KC343118 | KC343360 | KC343602 | KC343844 | KC344086 |
| D. hispaniae | CPC 30321 | Vitis vinifera | Spain | MG281123 | MG281820 | MG281471 | MG281644 | MG281296 |
| D. hongkongensis | CBS 115448 | Dichroa febrífuga | China | KC343119 | KC343361 | KC343603 | KC343845 | KC344087 |
| D. incompleta | CGMCC 3.18288 | Camellia sinensis | China | KX986794 | KX999289 | KX999265 | KX999186 | KX999226 |
| D. inconspicua | CBS 133813 | Maytenus ilicifolia | Brazil | KC343123 | KC343365 | KC343607 | KC343849 | KC344091 |
| D. infecunda | CBS 133812 | Schinus terebinthifolius | Brazil | KC343126 | KC343368 | KC343610 | KC343852 | KC344094 |
| D. isoberliniae | CPC 22549 | Isoberlinia angolensis | Zambia | KJ869133 | N/Aa | N/Aa | N/Aa | KJ869245 |
| D. juglandicola | CFCC 51134 | Juglans mandshurica | China | KU985101 | KX024616 | KX024622 | KX024628 | KX024634 |
| CFCC 51135 | Juglans mandshurica | China | KU985102 | KX024617 | KX024623 | KX024629 | KX024635 | |
| D. kadsurae | CFCC 52586 | Kadsura longipedunculata | China | MH121521 | MH121439 | MH121479 | MH121563 | MH121600 |
| CFCC 52587 | Kadsura longipedunculata | China | MH121522 | MH121440 | MH121480 | MH121564 | MH121601 | |
| CFCC 52588 | Acer sp. | China | MH121523 | MH121441 | MH121481 | MH121565 | MH121602 | |
| CFCC 52589 | Acer sp. | China | MH121524 | MH121442 | MH121482 | MH121566 | MH121603 | |
| D. kochmanii | BRIP 54033 | Helianthus annuus | Australia | JF431295 | N/Aa | N/Aa | JN645809 | N/Aa |
| D. kongii | BRIP 54031 | Portulaca grandiflora | Australia | JF431301 | N/Aa | N/Aa | JN645797 | KJ197272 |
| D. litchicola | BRIP 54900 | Litchi chinensis | Australia | JX862533 | N/Aa | N/Aa | JX862539 | KF170925 |
| D. lithocarpus | CGMCC 3.15175 | Lithocarpus glabra | China | KC153104 | KF576235 | N/Aa | KC153095 | KF576311 |
| D. longicicola | CGMCC 3.17089 | Lithocarpus glabra | China | KF576267 | N/Aa | N/Aa | KF576242 | KF576291 |
| D. longicolla | ATCC 60325 | Glycine max | USA | KJ590728 | N/Aa | KJ659188 | KJ590767 | KJ610883 |
| D. longispora | CBS 194.36 | Ribes sp. | Canada | KC343135 | KC343377 | KC343619 | KC343861 | KC344103 |
| D. lonicerae | MFLUCC 17-0963 | Lonicera sp. | Italy | KY964190 | KY964116 | N/Aa | KY964146 | KY964073 |
| D. lusitanicae | CBS 123212 | Foeniculum vulgare | Portugal | KC343136 | KC343378 | KC343620 | KC343862 | KC344104 |
| D. macinthoshii | BRIP 55064a | Rapistrum rugostrum | Australia | KJ197289 | N/Aa | N/Aa | KJ197251 | KJ197269 |
| D. mahothocarpus | CGMCC 3.15181 | Lithocarpus glabra | China | KC153096 | N/Aa | N/Aa | KC153087 | KF576312 |
| D. malorum | CAA734 | Malus domestica | Portugal | KY435638 | KY435658 | KY435648 | KY435627 | KY435668 |
| D. maritima | DAOMC 250563 | Picea rubens | Canada | N/Aa | N/Aa | N/Aa | N/Aa | KU574616 |
| D. masirevicii | BRIP 57892a | Helianthus annuus | Australia | KJ197277 | N/Aa | N/Aa | KJ197239 | KJ197257 |
| D. mayteni | CBS 133185 | Maytenus ilicifolia | Brazil | KC343139 | KC343381 | KC343623 | KC343865 | KC344107 |
| D. maytenicola | CPC 21896* | Maytenus acuminata | South Africa | KF777157 | N/Aa | N/Aa | N/Aa | KF777250 |
| D. melonis | CBS 507.78 | Cucumis melo | USA | KC343142 | KC343384 | KC343626 | KC343868 | KC344110 |
| D. middletonii | BRIP 54884e | Rapistrum rugostrum | Australia | KJ197286 | N/Aa | N/Aa | KJ197248 | KJ197266 |
| D. miriciae | BRIP 54736j | Helianthus annuus | Australia | KJ197282 | N/Aa | N/Aa | KJ197244 | KJ197262 |
| D. momicola | MFLUCC 16-0113 | Prunus persica | China | KU557563 | KU557611 | N/Aa | KU557631 | KU55758 |
| D. multigutullata | ZJUD98 | Citrus grandis | China | KJ490633 | N/Aa | KJ490575 | KJ490512 | KJ490454 |
| D. musigena | CBS 129519 | Musa sp. | Australia | KC343143 | KC343385 | KC343627 | KC343869 | KC344111 |
| D. neilliae | CBS 144.27 | Spiraea sp. | USA | KC343144 | KC343386 | KC343628 | KC343870 | KC344112 |
| D. neoarctii | CBS 109490 | Ambrosia trifida | USA | KC343145 | KC343387 | KC343629 | KC343871 | KC344113 |
| D. neoraonikayaporum | MFLUCC 14-1136 | Tectona grandis | Thailand | KU712449 | KU749356 | N/Aa | KU749369 | KU743988 |
| D. nobilis | CBS 113470 | Castanea sativa | Korea | KC343146 | KC343388 | KC343630 | KC343872 | KC344114 |
| D. nothofagi | BRIP 54801 | Nothofagus cunninghamii | Australia | JX862530 | N/Aa | N/Aa | JX862536 | KF170922 |
| D. novem | CBS 127270 | Glycine max | Croatia | KC343155 | KC343397 | KC343640 | KC343881 | KC344123 |
| D. ocoteae | CBS 141330 | Ocotea obtusata | France | KX228293 | N/Aa | N/Aa | N/Aa | KX228388 |
| D. oraccinii | CGMCC 3.17531 | Camellia sinensis | China | KP267863 | N/Aa | KP293517 | KP267937 | KP293443 |
| D. ovalispora | ICMP20659 | Citrus limon | China | KJ490628 | N/Aa | KJ490570 | KJ490507 | KJ490449 |
| D. ovoicicola | CGMCC 3.17093 | Citrus sp. | China | KF576265 | KF576223 | N/Aa | KF576240 | KF576289 |
| D. oxe | CBS 133186 | Maytenus ilicifolia | Brazil | KC343164 | KC343406 | KC343648 | KC343890 | KC344132 |
| D. padina | CFCC 52590 | Padus racemosa | China | MH121525 | MH121443 | MH121483 | MH121567 | MH121604 |
| CFCC 52591 | Padus racemosa | China | MH121526 | MH121444 | MH121484 | MH121568 | MH121605 | |
| D. pandanicola | MFLU 18-0006 | Pandanus sp. | Thailand | MG646974 | N/Aa | N/Aa | N/Aa | MG646930 |
| D. paranensis | CBS 133184 | Maytenus ilicifolia | Brazil | KC343171 | KC343413 | KC343655 | KC343897 | KC344139 |
| D. parapterocarpi | CPC 22729 | Pterocarpus brenanii | Zambia | KJ869138 | N/Aa | N/Aa | N/Aa | KJ869248 |
| D. pascoei | BRIP 54847 | Persea americana | Australia | JX862532 | N/Aa | N/Aa | JX862538 | KF170924 |
| D. passiflorae | CBS 132527 | Passiflora edulis | South America | JX069860 | N/Aa | KY435654 | N/Aa | N/Aa |
| D. passifloricola | CBS 141329 | Passiflora foetida | Malaysia | KX228292 | N/Aa | KX228367 | N/Aa | KX228387 |
| D. penetriteum | CGMCC 3.17532 | Camellia sinensis | China | KP714505 | N/Aa | KP714493 | KP714517 | KP714529 |
| D. perjuncta | CBS 109745 | Ulmus glabra | Austria | KC343172 | KC343414 | KC343656 | KC343898 | KC344140 |
| D. perseae | CBS 151.73 | Persea gratissima | Netherlands | KC343173 | KC343415 | KC343657 | KC343899 | KC344141 |
| D. pescicola | MFLUCC 16-0105 | Prunus persica | China | KU557555 | KU557603 | N/Aa | KU557623 | KU557579 |
| D. phaseolorum | AR4203 | Phaseolus vulgaris | USA | KJ590738 | N/Aa | KJ659220 | N/Aa | KP004507 |
| D. podocarpi-macrophylli | CGMCC 3.18281 | Podocarpus macrophyllus | China | KX986774 | KX999278 | KX999246 | KX999167 | KX999207 |
| D. pseudomangiferae | CBS 101339 | Mangifera indica | Dominican Republic | KC343181 | KC343423 | KC343665 | KC343907 | KC344149 |
| D. pseudophoenicicola | CBS 462.69 | Phoenix dactylifera | Spain | KC343184 | KC343426 | KC343668 | KC343910 | KC344152 |
| D. pseudotsugae | MFLU 15-3228 | Pseudotsuga menziesii | Italy | KY964225 | KY964138 | N/Aa | KY964181 | KY964108 |
| D. psoraleae | CBS 136412 | Psoralea pinnata | South Africa | KF777158 | N/Aa | N/Aa | KF777245 | KF777251 |
| D. psoraleae-pinnatae | CBS 136413 | Psoralea pinnata | South Africa | KF777159 | N/Aa | N/Aa | N/Aa | KF777252 |
| D. pterocarpi | MFLUCC 10-0571 | Pterocarpus indicus | Thailand | JQ619899 | JX197451 | N/Aa | JX275416 | JX275460 |
| D. pterocarpicola | MFLUCC 10-0580a | Pterocarpus indicus | Thailand | JQ619887 | JX197433 | N/Aa | JX275403 | JX275441 |
| D. pulla | CBS 338.89 | Hedera helix | Yugoslavia | KC343152 | KC343394 | KC343636 | KC343878 | KC344120 |
| D. pyracanthae | CAA483 | Pyracantha coccinea | Portugal | KY435635 | KY435656 | KY435645 | KY435625 | KY435666 |
| D. racemosae | CBS 143770 | Euclea racemosa | South Africa | MG600223 | MG600219 | MG600221 | MG600225 | MG600227 |
| D. raonikayaporum | CBS 133182 | Spondias mombin | Brazil | KC343188 | KC343430 | KC343672 | KC343914 | KC344156 |
| D. ravennica | MFLUCC 15-0479 | Tamarix sp. | Italy | KU900335 | N/Aa | N/Aa | KX365197 | KX432254 |
| D. rhusicola | CBS 129528 | Rhus pendulina | South Africa | JF951146 | KC843124 | N/Aa | KC843100 | KC843205 |
| D. rosae | MFLU 17-1550 | Rosa sp. | Thailand | MG828894 | N/Aa | N/Aa | N/Aa | MG843878 |
| D. rosicola | MFLU 17-0646 | Rosa sp. | UK | MG828895 | N/Aa | N/Aa | MG829270 | MG843877 |
| D. rostrata | CFCC 50062 | Juglans mandshurica | China | KP208847 | KP208849 | KP208851 | KP208853 | KP208855 |
| CFCC 50063 | Juglans mandshurica | China | KP208848 | KP208850 | KP208852 | KP208854 | KP208856 | |
| D. rudis | AR3422 | Laburnum anagyroides | Austria | KC843331 | KC843146 | N/Aa | KC843090 | KC843177 |
| D. saccarata | CBS 116311 | Protea repens | South Africa | KC343190 | KC343432 | KC343674 | KC343916 | KC344158 |
| D. sackstonii | BRIP 54669b | Helianthus annuus | Australia | KJ197287 | N/Aa | N/Aa | KJ197249 | KJ197267 |
| D. salicicola | BRIP 54825 | Salix purpurea | Australia | JX862531 | N/Aa | N/Aa | JX862537 | JX862531 |
| D. sambucusii | CFCC 51986 | Sambucus williamsii | China | KY852495 | KY852499 | KY852503 | KY852507 | KY852511 |
| CFCC 51987 | Sambucus williamsii | China | KY852496 | KY852500 | KY852504 | KY852508 | KY852512 | |
| D. schini | CBS 133181 | Schinus terebinthifolius | Brazil | KC343191 | KC343433 | KC343675 | KC343917 | KC344159 |
| D. schisandrae | CFCC 51988 | Schisandra chinensis | China | KY852497 | KY852501 | KY852505 | KY852509 | KY852513 |
| CFCC 51989 | Schisandra chinensis | China | KY852498 | KY852502 | KY852506 | KY852510 | KY852514 | |
| D. schoeni | MFLU 15-1279 | Schoenus nigricans | Italy | KY964226 | KY964139 | N/Aa | KY964182 | KY964109 |
| D. sclerotioides | CBS 296.67 | Cucumis sativus | Netherlands | KC343193 | KC343435 | KC343677 | KC343919 | KC344161 |
| D. sennae | CFCC 51636 | Senna bicapsularis | China | KY203724 | KY228875 | N/Aa | KY228885 | KY228891 |
| CFCC 51637 | Senna bicapsularis | China | KY203725 | KY228876 | N/Aa | KY228886 | KY228892 | |
| D. sennicola | CFCC 51634 | Senna bicapsularis | China | KY203722 | KY228873 | KY228879 | KY228883 | KY228889 |
| CFCC 51635 | Senna bicapsularis | China | KY203723 | KY228874 | KY228880 | KY228884 | KY228890 | |
| D. serafiniae | BRIP 55665a | Helianthus annuus | Australia | KJ197274 | N/Aa | N/Aa | KJ197236 | KJ197254 |
| D. siamensis | MFLUCC 10-573a | Dasymaschalon sp. | Thailand | JQ619879 | N/Aa | N/Aa | JX275393 | JX275429 |
| D. sojae | FAU635 | Glycine max | USA | KJ590719 | KJ612116 | KJ659208 | KJ590762 | KJ610875 |
| D. spartinicola | CBS 140003 | Spartium junceum | Spain | KR611879 | N/Aa | KR857696 | N/Aa | KR857695 |
| D. sterilis | CBS 136969 | Vaccinium corymbosum | Italy | KJ160579 | KJ160548 | MF418350 | KJ160611 | KJ160528 |
| D. stictica | CBS 370.54 | Buxus sampervirens | Italy | KC343212 | KC343454 | KC343696 | KC343938 | KC344180 |
| D. subclavata | ICMP20663 | Citrus unshiu | China | KJ490587 | N/Aa | KJ490529 | KJ490466 | KJ490408 |
| D. subcylindrospora | MFLU 17-1195 | Salix sp. | China | MG746629 | N/Aa | N/Aa | MG746630 | MG746631 |
| D. subellipicola | MFLU 17-1197 | on dead wood | China | MG746632 | N/Aa | N/Aa | MG746633 | MG746634 |
| D. subordinaria | CBS 464.90 | Plantago lanceolata | New Zealand | KC343214 | KC343456 | KC343698 | KC343940 | KC344182 |
| D. taoicola | MFLUCC 16-0117 | Prunus persica | China | KU557567 | N/Aa | N/Aa | KU557635 | KU557591 |
| D. tectonae | MFLUCC 12-0777 | Tectona grandis | China | KU712430 | KU749345 | N/Aa | KU749359 | KU743977 |
| D. tectonendophytica | MFLUCC 13-0471 | Tectona grandis | China | KU712439 | KU749354 | N/Aa | KU749367 | KU749354 |
| D. tectonigena | MFLUCC 12-0767 | Tectona grandis | China | KU712429 | KU749358 | N/Aa | KU749371 | KU743976 |
| D. terebinthifolii | CBS 133180 | Schinus terebinthifolius | Brazil | KC343216 | KC343458 | KC343700 | KC343942 | KC344184 |
| D. thunbergii | MFLUCC 10-576a | Thunbergia laurifolia | Thailand | JQ619893 | JX197440 | N/Aa | JX275409 | JX275449 |
| D. thunbergiicola | MFLUCC 12-0033 | Thunbergia laurifolia | Thailand | KP715097 | N/Aa | N/Aa | KP715098 | N/Aa |
| D. tibetensis | CFCC 51999 | Juglandis regia | China | MF279843 | MF279888 | MF279828 | MF279858 | MF279873 |
| CFCC 52000 | Juglandis regia | China | MF279844 | MF279889 | MF279829 | MF279859 | MF279874 | |
| D. torilicola | MFLUCC 17-1051 | Torilis arvensis | Italy | KY964212 | KY964127 | N/Aa | KY964168 | KY964096 |
| D. toxica | CBS 534.93 | Lupinus angustifolius | Australia | KC343220 | KC343462 | C343704 | KC343946 | KC344188 |
| D. tulliensis | BRIP 62248a | Theobromacacao fruit | Australia | KR936130 | N/Aa | N/Aa | KR936133 | KR936132 |
| D. ueckerae | FAU656 | Cucumis melo | USA | KJ590726 | KJ612122 | KJ659215 | KJ590747 | KJ610881 |
| D. ukurunduensis | CFCC 52592 | Acer ukurunduense | China | MH121527 | MH121445 | MH121485 | MH121569 | N/Aa |
| CFCC 52593 | Acer ukurunduense | China | MH121528 | MH121446 | MH121486 | MH121570 | N/Aa | |
| D. undulata | CGMCC 3.18293 | Leaf of unknown host | China-Laos border | KX986798 | N/Aa | KX999269 | KX999190 | KX999230 |
| D. unshiuensis | CGMCC 3.17569 | Citrus unshiu | China | KJ490587 | N/Aa | KJ490529 | KJ490408 | KJ490466 |
| CFCC 52594 | Carya illinoensis | China | MH121529 | MH121447 | MH121487 | MH121571 | MH121606 | |
| CFCC 52595 | Carya illinoensis | China | MH121530 | MH121448 | MH121488 | MH121572 | MH121607 | |
| D. vaccinii | CBS 160.32 | Oxycoccus macrocarpos | USA | KC343228 | KC343470 | KC343712 | KC343954 | KC344196 |
| D. vangueriae | CPC 22703 | Vangueria infausta | Zambia | KJ869137 | N/Aa | N/Aa | N/Aa | KJ869247 |
| D. vawdreyi | BRIP 57887a | Psidium guajava | Australia | KR936126 | N/Aa | N/Aa | KR936129 | KR936128 |
| D. velutina | CGMCC 3.18286 | Neolitsea sp. | China | KX986790 | N/Aa | KX999261 | KX999182 | KX999223 |
| D. virgiliae | CMW40748 | Virgilia oroboides | South Africa | KP247566 | N/Aa | N/Aa | N/Aa | KP247575 |
| D. xishuangbanica | CGMCC 3.18282 | Camellia sinensis | China | KX986783 | N/Aa | KX999255 | KX999175 | KX999216 |
| D. yunnanensis | CGMCC 3.18289 | Coffea sp. | China | KX986796 | KX999290 | KX999267 | KX999188 | KX999228 |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
Newly sequenced material is indicated in bold type.
Morphological analysis
Agar plugs (6 mm diam.) were taken from the edge of actively growing cultures on PDA and transferred on to the centre of 9 cm diam Petri dishes containing 2% tap water agar supplemented with sterile pine needles (PNA; Smith et al. 1996) and potato dextrose agar (PDA) and incubated at 20–21 °C under a 12 h near-ultraviolet light/12 h dark cycle to induce sporulation as described in recent studies (Gomes et al. 2013, Lombard et al. 2014). Colony characters and pigment production on PNA and PDA were noted after 10 d. Colony colours were rated according to Rayner (1970). Cultures were examined periodically for the development of ascomata and conidiomata. The morphological characteristics were examined by mounting fungal structures in clear lactic acid and 30 measurements at 1000× magnification were determined for each isolate using a Leica compound microscope (DM 2500) with interference contrast (DIC) optics. Descriptions, nomenclature and illustrations of taxonomic novelties are deposited in MycoBank (www.MycoBank.org; Crous et al. 2004b).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from colonies grown on cellophane-covered PDA using a modified CTAB [cetyltrimethylammonium bromide] method (Doyle and Doyle 1990). DNA was estimated by electrophoresis in 1% agarose gel and the quality was measured using the NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA), following the user manual (Desjardins et al. 2009). PCR amplifications were performed in a DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The primer sets ITS1/ITS4 (White et al. 1990) were used to amplify the ITS region. The primer pair CAL228F/CAL737R (Carbone and Kohn 1999) were used to amplify the calmodulin gene (cal) and the primer pair CYLH4F (Crous et al. 2004a) and H3-1b (Glass and Donaldson 1995) were used to amplify part of the histone H3 (his3) gene. The primer pair EF1-728F/EF1-986R (Carbone and Kohn 1999) were used to amplify a partial fragment of the translation elongation factor 1-α gene (tef1). The primer sets T1 (O’Donnell and Cigelnik 1997) and Bt2b (Glass and Donaldson 1995) were used to amplify the beta-tubulin gene (tub2); the additional combination of Bt2a/Bt2b (Glass and Donaldson 1995) was used in case of amplification failure of the T1/Bt2b primer pair. Amplifications of different loci were performed under different conditions (Table 2). PCR amplification products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM® 3730XL DNA Analyser with a BigDye Terminater Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Table 2.
Genes used in this study with PCR primers, process and references.
| Gene | PCR primers (forward/reverse) | PCR: thermal cycles: (Annealing temp. in bold) | References of primers used |
|---|---|---|---|
| ITS | ITS1/ITS4 | (95 °C: 30 s, 51 °C: 30 s, 72 °C: 1 min) × 35 cycles | White et al. 1990 |
| cal | CAL228F/CAL737R | (95 °C: 15 s, 55 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| his3 | CYLH4F/H3-1b | (95 °C: 30 s, 58 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass and Donaldson 1995, Crous et al. 2004a |
| tef1 | EF1-728F/EF1-986R | (9 °C: 15 s, 55 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| tub2 | T1(Bt2a)/Bt2b | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass and Donaldson 1995, Glass and Donaldson 1995 |
Phylogenetic analyses
DNA generated sequences were used to obtain consensus sequences using SeqMan v.7.1.0 DNASTAR Lasergene Core Suite software programme (DNASTAR Inc., Madison, WI, USA). Sequences were aligned using MAFFT v.6 (Katoh and Toh 2010) and edited manually using MEGA6 (Tamura et al. 2013). Two different datasets were employed to estimate two phylogenetic analyses: one for Diaporthe species and one for Diaportheeres complex. The first analysis was undertaken to infer the interspecific relationships in Diaporthe. All the Diaporthe isolates recovered from samples collected during this study and additional reference sequences of Diaporthe species were included in the dataset of combined ITS, cal, his3, tef1, and tub2 regions (Table 1), with Diaporthellacorylina (CBS 121124) as outgroup. The second analysis focused on the Diaportheeres complex based on cal, tef1 and tub2 loci (Table 3) according to recent publications (Gao et al. 2014, 2015, 2016, Udayanga et al. 2014b, Tanney et al. 2016, Fan et al. 2018), with Diaporthecitri (AR3405) as outgroup. Maximum Parsimony analysis was performed by a heuristic search option of 1000 random-addition sequences with a tree bisection and reconnection (TBR) algorithm. Maxtrees were set to 5000, branches of zero length were collapsed and all equally parsimonious trees were saved. Other calculated parsimony scores were tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency (RC). Maximum Likelihood analysis was performed with a GTR site substitution model (Guindon et al. 2010). Branch support was evaluated with a bootstrapping (BS) method of 1000 replicates (Hillis and Bull 1993).
Table 3.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaportheeres complex.
| Species | Isolate/culture collection | Host | Location | GenBank accession numbers | ||
|---|---|---|---|---|---|---|
| CAL | TEF1-α | TUB | ||||
| D. alleghaniensis | CBS 495.72 | Betula alleghaniensis | Canada | KC343249 | GQ250298 | KC843228 |
| D. alnea | CBS 146.46 | Alnus sp. | Netherlands | KC343250 | KC343734 | KC343976 |
| CBS 159.47 | Alnus sp. | Netherlands | KC343251 | KC343735 | KC343977 | |
| LCM22b.02a | Alnus sp. | USA | KJ435020 | KJ210557 | KJ420825 | |
| LCM22b.02b | Alnus sp. | USA | KJ435021 | KJ210558 | KJ420826 | |
| D. betulina | CFCC 52560 | Betula albo-sinensis | China | MH121419 | MH121537 | MH121577 |
| CFCC 52561 | Betula costata | China | MH121420 | MH121538 | MH121578 | |
| CFCC 52562 | Betula platyphylla | China | MH121421 | MH121539 | MH121579 | |
| D. bicincta | CBS 121004 | Juglans sp. | USA | KC343376 | KC343860 | KC344102 |
| D. biguttusis | CGMCC 3.17081 | Lithocarpus glabra | China | N/Aa | KF576257 | KF576306 |
| D. camptothecicola | CFCC 51632 | Camptotheca acuminata | China | KY228881 | KY228887 | KY228893 |
| D. celastrina | CBS 139.27 | Celastrus sp. | USA | KC343289 | KC343773 | KC344015 |
| D. chensiensis | CFCC 52567 | Abies chensiensis | China | MH121426 | MH121544 | MH121584 |
| CFCC 52568 | Abies chensiensis | China | MH121427 | MH121545 | MH121585 | |
| D. citri | AR3405 | Citrus sp. | USA | KC843157 | KC843071 | KC843187 |
| D. citrichinensis | ZJUD034 | Citrus sp. | China | KC843234 | KC843071 | KC843187 |
| ZJUD034B | Citrus sp. | China | KJ435042 | KJ210562 | KJ420829 | |
| D. ellipicola | CGMCC 3.17084 | Lithocarpus glabra | China | N/Aa | KF576245 | KF576291 |
| D. eres | AR5193 | Ulmus laevis | Germany | KJ434999 | KJ210550 | KJ420799 |
| AR5196 | Ulmus laevis | Germany | KJ435006 | KJ210554 | KJ420817 | |
| DP0438 | Ulmus minor | Austria | KJ435016 | KJ210553 | KJ420816 | |
| LCM114.01a | Ulmus sp. | USA | KJ435027 | KJ210545 | KJ420787 | |
| LCM114.01b | Ulmus sp. | USA | KJ435026 | KJ210544 | KJ420786 | |
| FAU483 | Malus sp. | Netherlands | KJ435022 | JQ807422 | KJ420827 | |
| DAN001A | Daphne laureola | France | KJ434994 | KJ210540 | KJ420781 | |
| DAN001B | Daphne laureola | France | KJ434995 | KJ210541 | KJ420782 | |
| AR5197 | Rhododendron sp. | Germany | KJ435014 | KJ210552 | KJ420812 | |
| CBS 439.82 | Cotoneaster sp. | UK | JX197429 | GQ250341 | JX275437 | |
| AR3519 | Corylus avellana | Austria | KJ435008 | KJ210547 | KJ420789 | |
| FAU506 | Cornus florida | USA | KJ435012 | JQ807403 | KJ420792 | |
| FAU570 | Oxydendrum arboreum | USA | KJ435025 | JQ807410 | KJ420794 | |
| AR3723 | Rubus fruticosus | Austria | KJ435024 | JQ807354 | KJ420793 | |
| FAU522 | Sassafras albida | USA | KJ435010 | JQ807406 | KJ420791 | |
| DP0666 | Juglans cinerea | USA | KJ435007 | KJ210546 | KJ420788 | |
| DP0667 | Juglans cinerea | USA | KC843155 | KC843121 | KC843229 | |
| AR3560 | Viburnum sp. | Austria | KJ435011 | JQ807351 | KJ420795 | |
| AR5224 | Hedera helix | Germany | KJ435036 | KJ210551 | KJ420802 | |
| AR5231 | Hedera helix | Germany | KJ435038 | KJ210555 | KJ420818 | |
| AR5223 | Acer nugundo | Germany | KJ435000 | KJ210549 | KJ420830 | |
| CBS 109767 | Acer sp. | Austria | KC343317 | KC343801 | KC344043 | |
| DLR12a | Vitis vinifera | France | KJ434996 | KJ210542 | KJ420783 | |
| DLR12b | Vitis vinifera | France | KJ434997 | KJ210543 | KJ420784 | |
| AR4347 | Vitis vinifera | Korea | KJ435030 | JQ807356 | KJ420805 | |
| AR4355 | Prunus sp. | Korea | KJ435035 | JQ807359 | KJ420797 | |
| AR4367 | Prunus sp. | Korea | KJ435019 | JQ807364 | KJ420824 | |
| AR4346 | Prunus mume | Korea | KJ435003 | JQ807355 | KJ420823 | |
| AR4348 | Prunus persici | Korea | KJ435004 | JQ807357 | JQ807357 | |
| AR3669 | Pyrus pyrifolia | Japan | KJ435002 | JQ807415 | KJ420808 | |
| D. eres | AR3670 | Pyrus pyrifolia | Japan | KJ435001 | JQ807416 | KJ420807 |
| AR3671 | Pyrus pyrifolia | Japan | KJ435017 | JQ807417 | KJ420814 | |
| AR3672 | Pyrus pyrifolia | Japan | KJ435023 | JQ807418 | KJ420819 | |
| DP0591 | Pyrus pyrifolia | New Zealand | KJ435018 | JQ807395 | KJ420821 | |
| AR4369 | Pyrus pyrifolia | Korea | KJ435005 | JQ807366 | KJ420813 | |
| DP0180 | Pyrus pyrifolia | New Zealand | KJ435029 | JQ807384 | KJ420804 | |
| DP0179 | Pyrus pyrifolia | New Zealand | KJ435028 | JQ807383 | KJ420803 | |
| DP0590 | Pyrus pyrifolia | New Zealand | KJ435037 | JQ807394 | KJ420810 | |
| AR4373 | Ziziphus jujuba | Korea | KJ435013 | JQ807368 | KJ420798 | |
| AR4374 | Ziziphus jujuba | Korea | KJ434998 | JQ807369 | KJ420785 | |
| AR4357 | Ziziphus jujuba | Korea | KJ435031 | JQ807360 | KJ420806 | |
| AR4371 | Malus pumila | Korea | KJ435034 | JQ807367 | KJ420796 | |
| FAU532 | Chamaecyparis thyoides | USA | KJ435015 | JQ807408 | KJ435015 | |
| CBS 113470 | Castanea sativa | Australia | KC343388 | KC343872 | KC344114 | |
| AR4349 | Vitis vinifera | Korea | KJ435032 | JQ807358 | KJ420822 | |
| AR4363 | Malus sp. | Korea | KJ435033 | JQ807362 | KJ420809 | |
| CFCC 52575 | Castanea mollissima | China | N/Aa | MH121552 | MH121592 | |
| CFCC 52576 | Castanea mollissima | China | MH121432 | MH121553 | MH121593 | |
| CFCC 52577 | Acanthopanax senticosus | China | MH121433 | MH121554 | MH121594 | |
| CFCC 52578 | Sorbus sp. | China | MH121434 | MH121555 | MH121595 | |
| CFCC 52579 | Juglans regia | China | N/Aa | MH121556 | N/Aa | |
| CFCC 52580 | Melia azedarace | China | N/Aa | MH121557 | MH121596 | |
| CFCC 52581 | Rhododendron simsii | China | N/Aa | MH121558 | MH121597 | |
| D. helicis | AR5211 | Hedera helix | France | KJ435043 | KJ210559 | KJ420828 |
| D. longicicola | CGMCC 3.17089 | Lithocarpus glabra | China | N/Aa | KF576242 | KF576291 |
| D. mahothocarpus | CGMCC 3.15181 | Lithocarpus glabra | China | N/Aa | KC153087 | KF576312 |
| D. maritima | DAOMC 250563 | Picea rubens | Canada | N/Aa | N/Aa | KU574616 |
| D. momicola | MFLUCC 16-0113 | Prunus persica | China | N/Aa | KU557631 | KU55758 |
| D. neilliae | CBS 144. 27 | Spiraea sp. | USA | KC343386 | KC343870 | KC344112 |
| D. padina | CFCC 52590 | Padus racemosa | China | MH121443 | MH121567 | MH121604 |
| CFCC 52591 | Padus racemosa | China | MH121444 | MH121568 | MH121605 | |
| D. phragmitis | CBS 138897 | Phragmites australis | China | N/Aa | N/Aa | KP004507 |
| D. pulla | CBS 338.89 | Hedera helix | Yugoslavia | KC343394 | KC343878 | KC344120 |
| D. vaccinii | DF5032 | Vaccinium corymbosum | USA | KC849457 | JQ807380 | KC843225 |
| FAU633 | Vaccinium macrocarpon | USA | KC849456 | JQ807413 | KC843226 | |
| FAU446 | Vaccinium macrocarpon | USA | KC849455 | JQ807398 | KC843224 | |
| CBS 160.32 | Vaccinium macrocarpon | USA | KC343470 | GQ250326 | JX270436 | |
| FAU 468 | Vaccinium macrocarpon | USA | KC849458 | JQ807399 | KC843227 | |
Newly sequenced material is indicated in bold type.
Bayesian inference (BI) analysis, employing a Markov chain Monte Carlo (MCMC) algorithm, was performed (Rannala and Yang 1996). MrModeltest v. 2.3 was used to estimate the best-fit model of nucleotide substitution model settings for each gene (Posada and Crandall 1998). Two MCMC chains started from random trees for 1,000,000 generations and trees were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as the burn-in phase of each analysis. Branches with significant Bayesian posterior probabilities (BPP) were estimated in the remaining 7500 trees.
Sequences data were deposited in GenBank (Table 1). The multilocus sequence alignments were deposited in TreeBASE (www.treebase.org) as accession S22702 and S22703. The taxonomic novelties were deposited in MycoBank (Crous et al. 2004b).
Results
Collection of Diaporthe strains
Forty-two representative Diaporthe strains were isolated from 16 different host genera (Table 1) collected from six Provinces (Beijing, Heilongjiang, Jiangsu, Jiangxi, Shaanxi and Zhejiang) in China. All of these strains were isolated from symptomatic or non-symptomatic branches or twigs and preserved in the China Forestry Culture Collection Centre (CFCC).
Phylogenetic analyses
The first sequences dataset for the ITS, cal, his3, tef1, and tub2 was analysed in combination to infer the interspecific relationships within Diaporthe. The combined species phylogeny of the Diaporthe isolates consisted of 236 sequences, including the outgroup sequences of Diaporthellacorylina (culture CBS 121124). A total of 2948 characters including gaps (516 for ITS, 568 for cal, 520 for his3, 486 for tef1 and 858 for tub2) were included in the phylogenetic analysis. The maximum likelihood tree, conducted by the GTR model, confirmed the tree topology and posterior probabilities of the Bayesian consensus tree. For the Bayesian analyses, MrModeltest suggested that all partitions should be analysed with dirichlet state frequency distributions. The following models were recommended by MrModeltest and used: GTR+I+G for ITS, cal and his3, HKY+I+G for tef1 and tub2. The topology and branching order of ML were similar to BI analyses (Fig. 1). Based on the multi-locus phylogeny and morphology, 42 strains were assigned to 15 species, including 12 taxa which we describe here as new (Fig. 1).
Figure 1.
Phylogram of Diaporthe from a maximum likelihood analysis based on combined ITS, cal, his3, tef1 and tub2. Values above the branches indicate maximum likelihood bootstrap (left, ML BP ≥ 50%) and bayesian probabilities (right, BI PP ≥ 0.70). The tree is rooted with Diaporthellacorylina. Strains in the current study are in blue.
The second dataset with cal, tef1 and tub2 sequences were analysed to focus on the Diaportheeres complex. The alignment included 86 taxa, including the outgroup sequences of Diaporthecitri (Table 3). The aligned three-locus datasets included 1148 characters. Of these, 881 characters were constant, 105 variable characters were parsimony-uninformative and 162 characters were parsimony informative. The heuristic search using maximum parsimony (MP) generated 105 parsimonious trees (TL = 438, CI = 0.669, RI = 0.883, RC = 0.591), from which one was selected (Fig. 2). Based on the multi-locus phylogeny and morphology, seven strains were identified as D.eres, seven strains formed three distinct clades embedded in the D.eres complex, i.e. D.betulina, D.chensiensis and D.padina. MP and ML bootstrap support values above 50% are shown as first and second position, respectively. The branches with significant Bayesian posterior probability (≥ 0.70) in Bayesian analyses were thickened in the phylogenetic tree. The current results, based on the three genes (cal, tef1 and tub2), suggest that D.eres clade could be separated from other species in this complex (Fig. 2). However, D.biguttusis (CGMCC 3.17081), D.camptothecicola (CFCC 51632), D.ellipicola (CGMCC 3.17084), D.longicicola (CGMCC 3.17089), D.mahothocarpus (CGMCC 3.15181) and D.momicola (MFLUCC 16-0113) were clustered in D.eres clade and thus treated as the synonyms of D.eres in the current study.
Figure 2.
Phylogram of Diaportheeres complex based on combined cal, tef1 and tub2. Values above the branches indicate maximum parsimony bootstrap (left, MP BP ≥ 50%) and maximum likelihood bootstrap (right, ML BP ≥ 50%). Values below branches represent posterior probabilities (BI PP ≥ 0.70) from Bayesian inference. The tree is rooted with Diaporthecitri. Strains in the current study are in blue. The ex-type/ex-epitype culture is in bold.
Taxonomy
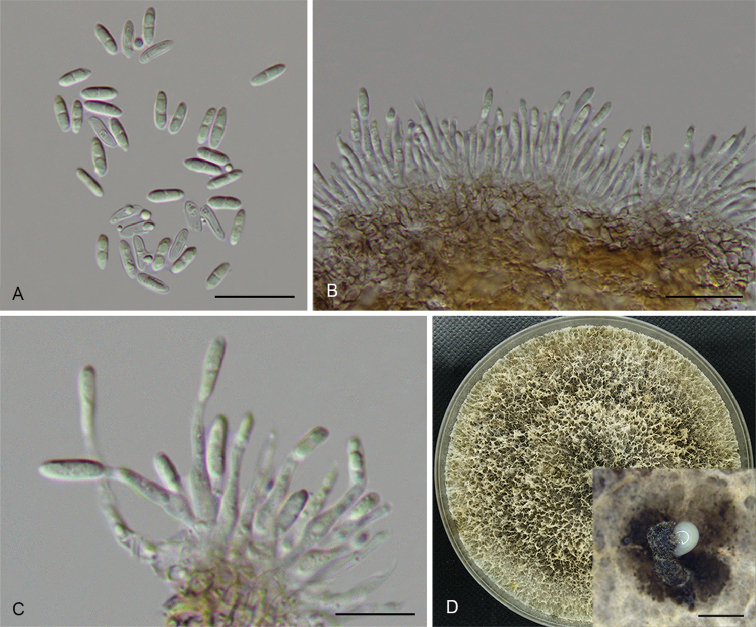
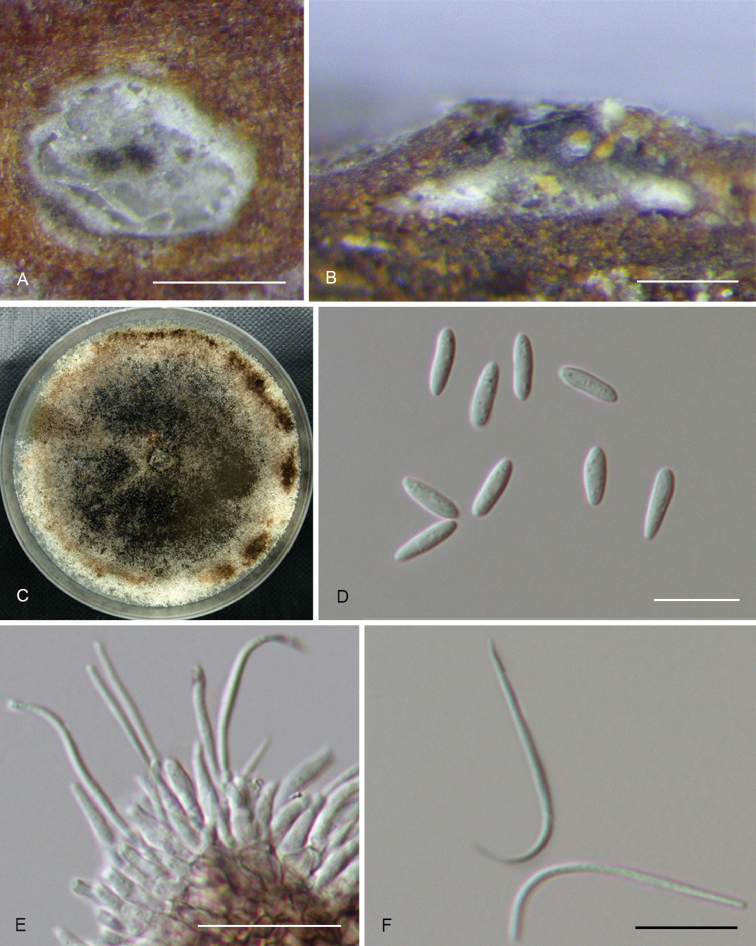
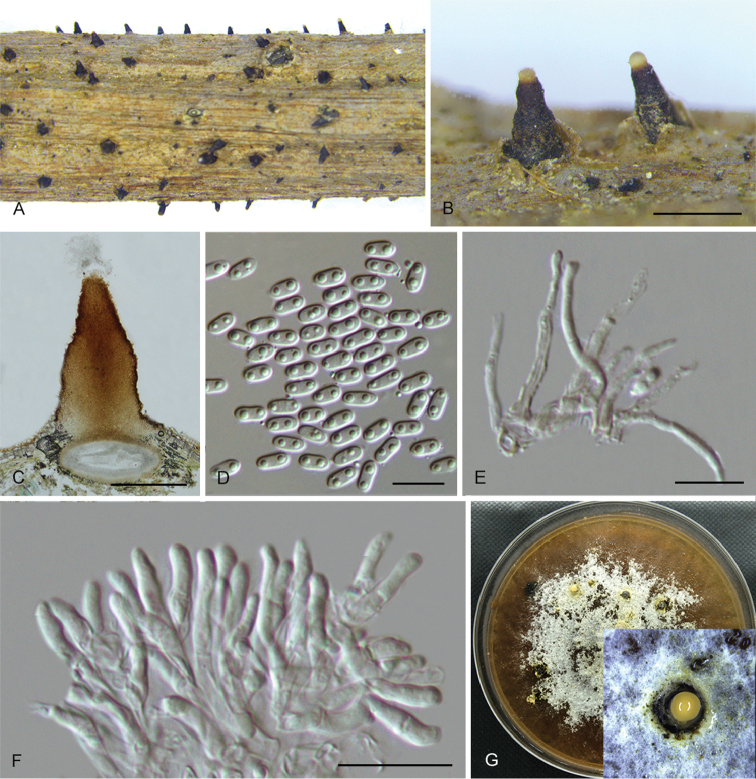
Diaporthe acerigena
C.M. Tian & Q. Yang sp. nov.
MB824703
Figure 3.
Diaportheacerigena (CFCC 52554) A Alpha conidia B–C Conidiophores D Culture on PDA and conidiomata. Scale bars: 20 μm (A–C), 200 μm (D).
Diagnosis.
Diaportheacerigena can be distinguished from the phylogenetically closely related species D.oraccinii in larger alpha conidia.
Holotype.
CHINA. Shaanxi Province: Qinling Mountain, on symptomatic twigs of Acertataricum, 27 June 2017, N. Jiang (holotype: BJFC-S1466; ex-type culture: CFCC 52554).
Etymology.
Named after the host genus on which it was collected, Acer.
Description.
On PDA: Conidiomata pycnidial, globose, solitary or aggregated, deeply embedded in the medium, erumpent, dark brown to black, 185–270 μm diam, whitish translucent to cream conidial drops exuding from the ostioles. Conidiophores 14.5–17 × 1.4–2.9 μm, cylindrical, hyaline, phiailidic, branched, straight to sinuous. Alpha conidia 7–10 × 2.1–2.9 μm (av. = 8.6 × 2.5 μm, n = 30), aseptate, hyaline, ellipsoidal, rounded at one end, slightly apex at the other end, usually with two-guttulate. Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony at first white, becoming dark brown in the centre with age. Aerial mycelium white, dense, fluffy, with cream conidial drops exuding from the ostioles.
Additional specimens examined.
CHINA. Shaanxi Province: Qinling Mountain, on symptomatic twigs of Acertataricum, 27 June 2017, N. Jiang, living culture CFCC 52555 (BJFC-S1467).
Notes.
Two strains representing D.acerigena cluster in a well-supported clade and appear most closely related to D.oraccinii. Diaportheacerigena can be distinguished from D.oraccinii based on ITS, his3, tef1 and tub2 loci (5/469 in ITS, 8/429 in his3, 8/326 in tef1 and 5/358 in tub2). Morphologically, D.acerigena differs from D.oraccinii in the longer and larger alpha conidia (8.6 × 2.5 vs. 6.6 × 1.9 μm) (Gao et al. 2016).
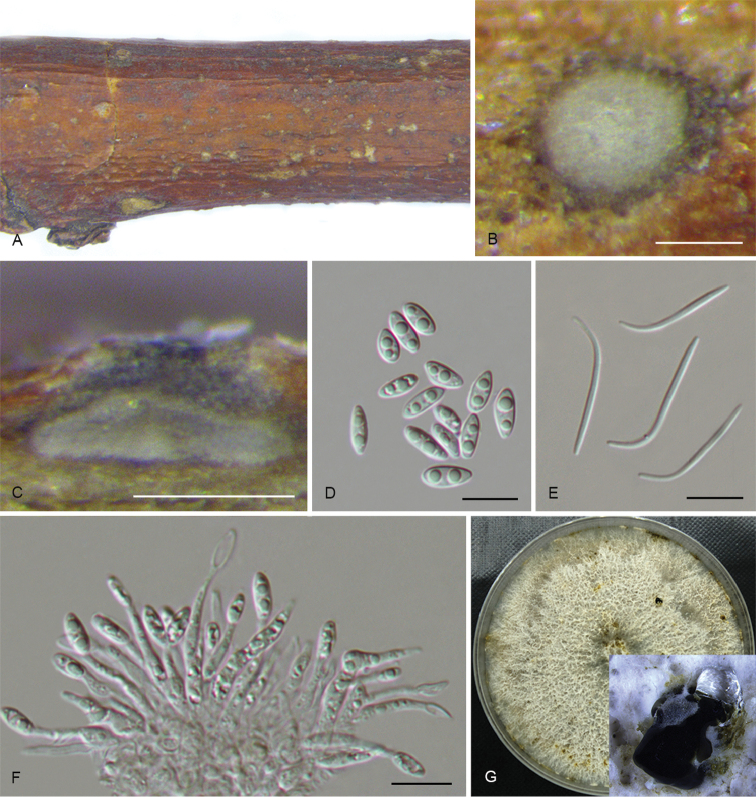
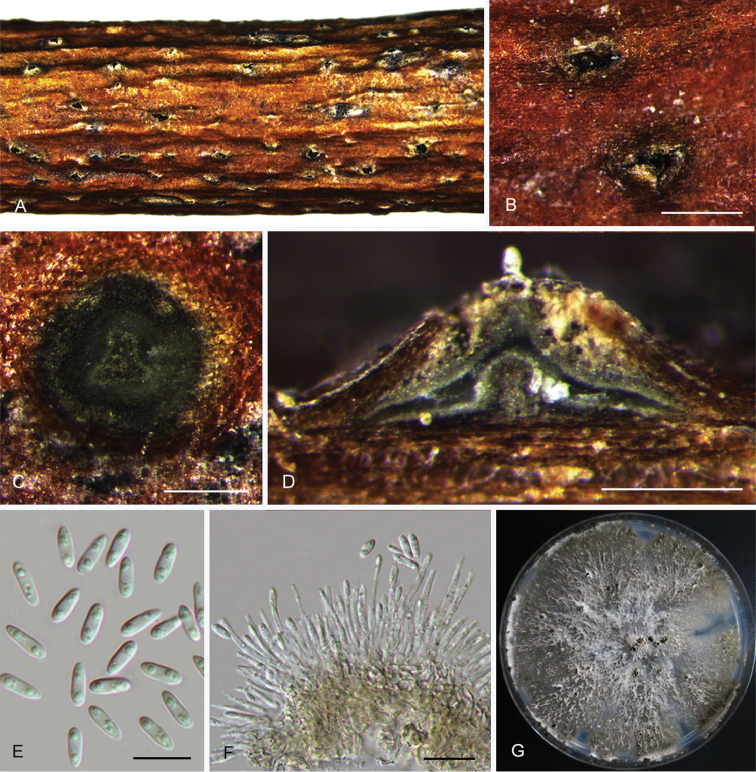
Diaporthe alangii
C.M. Tian & Q. Yang sp. nov.
MB824704
Figure 4.
Diaporthealangii (CFCC 52556) A Habit of conidiomata on branches B Transverse section of conidioma C Longitudinal section of conidioma D Alpha conidia E Conidiophores F Culture on PDA. Scale bars: 200 μm (B–C), 10 μm (D–E).
Diagnosis.
Diaporthealangii can be distinguished from the phylogenetically closely related species D.tectonae and D.tulliensis by the size of conidiophores and alpha conidia.
Holotype.
CHINA. Zhejiang Province: Tianmu Mountain, on symptomatic branches of Alangiumkurzii, 19 Apr. 2017, Q. Yang (holotype: BJFC-S1468; ex-type culture: CFCC 52556).
Etymology.
Named after the host genus on which it was collected, Alangium.
Description.
Conidiomata pycnidial, immersed in bark, scattered, erumpent through the bark surface, discoid, with a solitary undivided locule. Ectostromatic disc black, one ostiole per disc, 135–330 μm diam. Locule circular, undivided, 290–445 μm diam. Conidiophores 6–12 × 1.4–2 μm, cylindrical, hyaline, phiailidic, unbranched, straight. Alpha conidia 6.5–8 × 2 μm (av. = 7 × 2 μm, n = 30), aseptate, hyaline, ellipsoidal, biguttulate, mostly with one end obtuse and the other acute, occasionally submedian constriction. Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony initially white, producing beige pigment after 7–10 d. The colony is flat, felty with a thick texture at the centre and marginal area, with thin texture in the middle, lacking aerial mycelium, conidiomata absent.
Additional specimens examined.
CHINA. Zhejiang Province: Tianmu Mountain, on symptomatic branches of Alangiumkurzii, 19 Apr. 2017, Q. Yang, living culture CFCC 52557 (BJFC-S1469); ibid. living culture CFCC 52558 (BJFC-S1470); ibid. living culture CFCC 52559 (BJFC-S1471).
Notes.
Four isolates clustered in a clade distinct from its closest phylogenetic neighbour, D.tectonae and D.tulliensis. Diaporthealangii can be distinguished from D.tectonae in cal, tef1 and tub2 loci (6/458 in cal, 4/308 in tef1 and 11/407 in tub2); from D.tulliensis in ITS, tef1 and tub2 loci (6/462 in ITS, 8/308 in tef1 and 10/701 in tub2). Morphologically, D.alangii differs from D.tectonae in shorter conidiophores (6–12 vs. 11–18 μm) and longer alpha conidia (6.5–8 vs. 5.5–6 μm); from D.tulliensis in shorter conidiophores (6–12 vs. 15–20 μm) (Crous et al. 2015, Doilom et al. 2017).
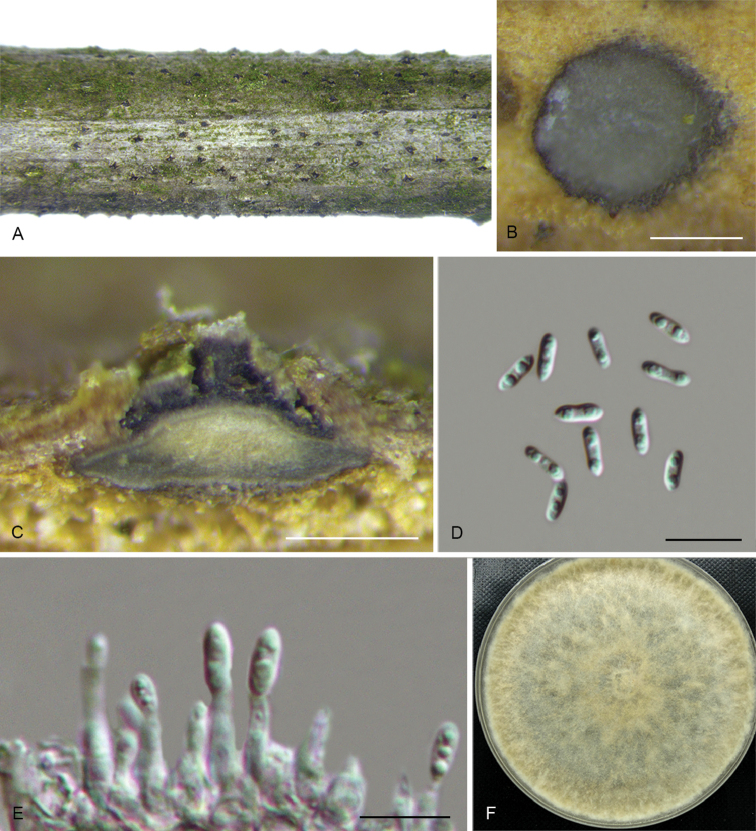
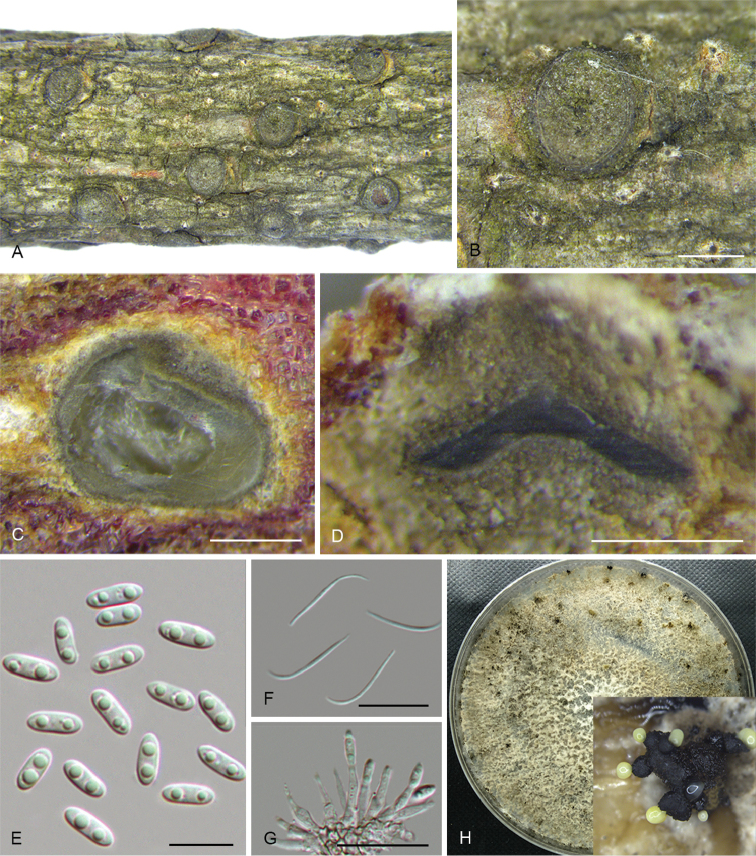
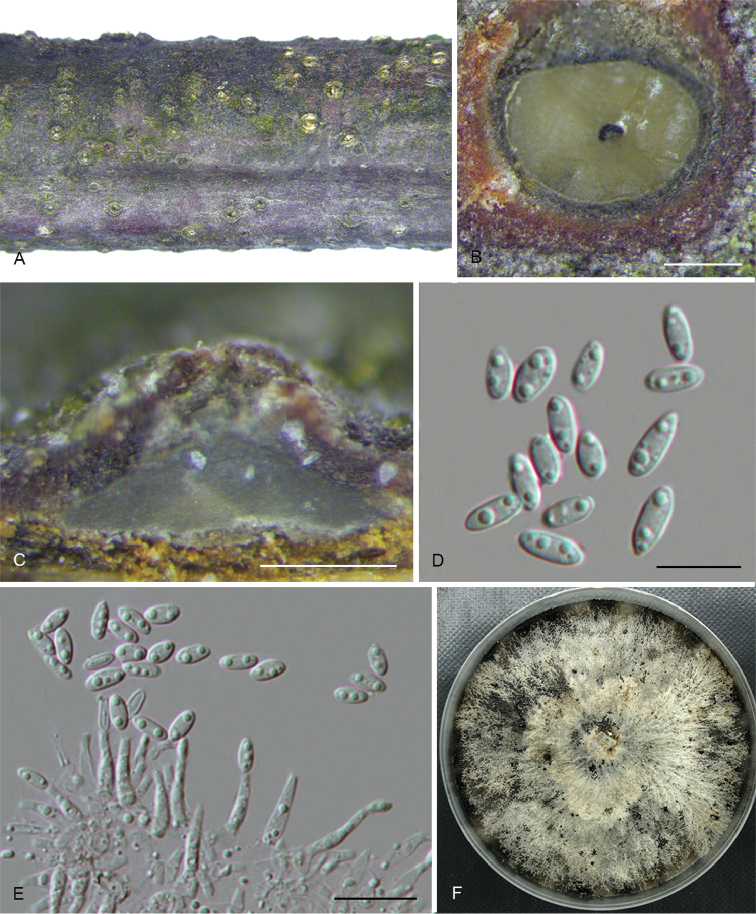
Diaporthe betulina
C.M. Tian & Q. Yang sp. nov.
MB824705
Figure 5.
Diaporthebetulina (CFCC 52562) A Habit of conidiomata on branches B Transverse section of conidioma C Longitudinal section of conidioma D Conidiophores E Alpha conidia F Beta conidia G Culture on PDA and conidiomata. Scale bars: 500 μm (A–C), 10 μm (D–F).
Diagnosis.
Diaporthebetulina can be distinguished from the phylogenetically closely related species D.betulae in smaller locule and wider alpha conidia.
Holotype.
CHINA. Heilongjiang Province: Yichun city, on symptomatic branches of Betulaplatyphylla, 27 July 2016, Q. Yang (holotype: BJFC-S1472; ex-type culture: CFCC 52562).
Etymology.
Named after the host genus on which it was collected, Betula.
Description.
Conidiomata pycnidial, conical, immersed in bark, scattered, erumpent through the bark surface, with a solitary undivided locule. Ectostromatic disc brown to black, one ostiole per disc, 290–645 μm diam. Ostiole medium black, up to the level of disc. Locule undivided, 670–905 μm diam. Conidiophores 12.5–17.5 × 1.5–2 μm, cylindrical, hyaline, phiailidic, branched, straight or slightly curved. Alpha conidia hyaline, aseptate, ellipsoidal to fusiform, 0–2-guttulate, sometimes acute at both ends, 8–10 × 2.5–3 μm (av. = 9 × 2.6 μm, n = 30). Beta conidia hyaline, aseptate, filiform, straight or hamate, eguttulate, base subtruncate, tapering towards one apex, 26–32.5 × 1 µm (av. = 30 × 1 µm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony flat with white felty aerial mycelium, turning white to dark brown aerial mycelium, conidiomata irregularly distributed on the agar surface.
Additional specimens examined.
CHINA. Heilongjiang Province: Yichun city, on symptomatic branches of Betulaalbo-sinensis, 27 July 2016, Q. Yang, living culture CFCC 52560 (BJFC-S1473); on symptomatic branches of Betulacostata, 27 July 2016, Q. Yang, living culture CFCC 52561 (BJFC-S1474).
Notes.
Diaporthebetulina was isolated from Betula spp. cankers in Heilongjiang Province. Three strains representing D.betulina cluster in a well-supported clade and appear most closely related to D.betulae, which was also isolated from Betulaplatyphylla in Sichuang Province (Du et al. 2016). Diaporthebetulina can be distinguished based on ITS, his3, tef1 and tub2 loci from D.betulae (11/461 in ITS, 9/453 in his3, 12/336 in tef1 and 7/695 in tub2). Morphologically, D.betulina differs from D.betulae in smaller locule (470–945 vs. 600–1250 μm) and wider alpha conidia (3–4 vs. 2.5–3 μm) (Du et al. 2016).
Diaporthe biguttulata
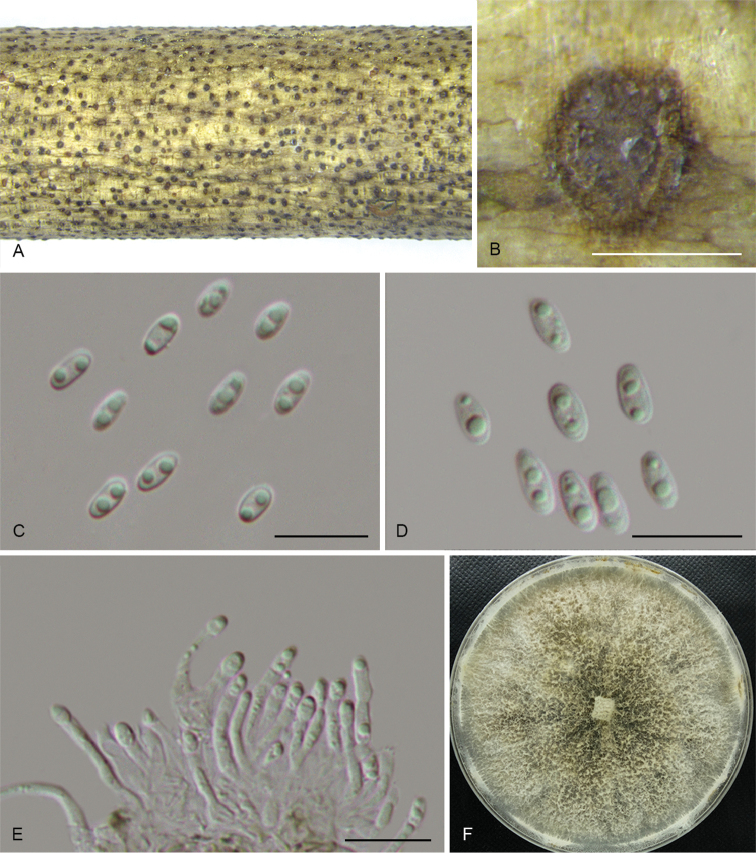
F. Huang, K.D. Hyde & H.Y. Li, 2015
Figure 6.
Diaporthebiguttulata (CFCC 52584) A Habit of conidiomata on branches B Transverse section of conidioma C Longitudinal section of conidioma D Alpha conidia E Conidiophores F Culture on PDA. Scale bars: 200 μm (B–C), 10 μm (D–E).
Description.
Conidiomata pycnidial, immersed in bark, scattered, erumpent through the bark surface, discoid, with a single locule. Ectostromatic disc dark brown, one ostiole per disc, 160–320 μm diam. Locule undivided, 235–350 μm diam. Conidiophores 8.5–11 × 1.5 μm, cylindrical, hyaline, branched, straight or slightly curved, tapering towards the apex. Alpha conidia hyaline, aseptate, ellipsoidal to oval, 2-guttulate, usually rounded at both ends, occasionally with one end acute, 7–8.5 × 1.5–2 μm (av. = 6.5 × 2.6 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white aerial mycelium, becoming pale grey, with dense aerial mycelium in the centre and sparse aerial mycelium at the marginal area, conidiomata absent.
Specimens examined.
CHINA. Zhejiang Province: Tianmu Mountain, on symptomatic branches of Juglansregia, 20 Apr. 2017, Q. Yang, living culture CFCC 52584 and CFCC 52585 (BJFC-S1504).
Notes.
Diaporthebiguttulata was originally described from a healthy branch of Citruslimon in Yunnan Province, China (Huang et al. 2015). In the present study, two isolates (CFCC 52584 and CFCC 52585) from symptomatic branches of Juglansregia were congruent with D.biguttulata based on morphology and DNA sequences data (Fig. 1). We therefore describe D.biguttulata as a known species for this clade.
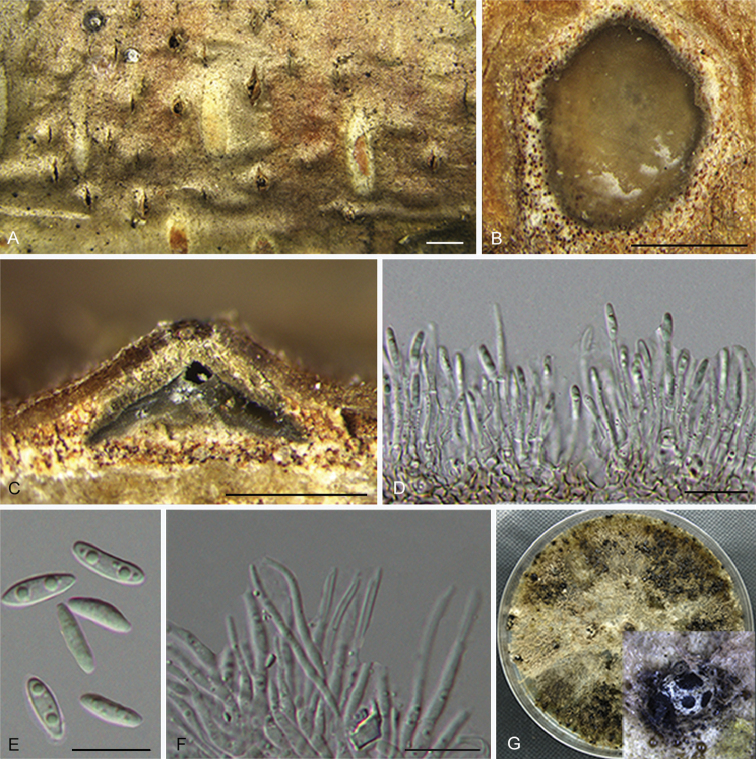
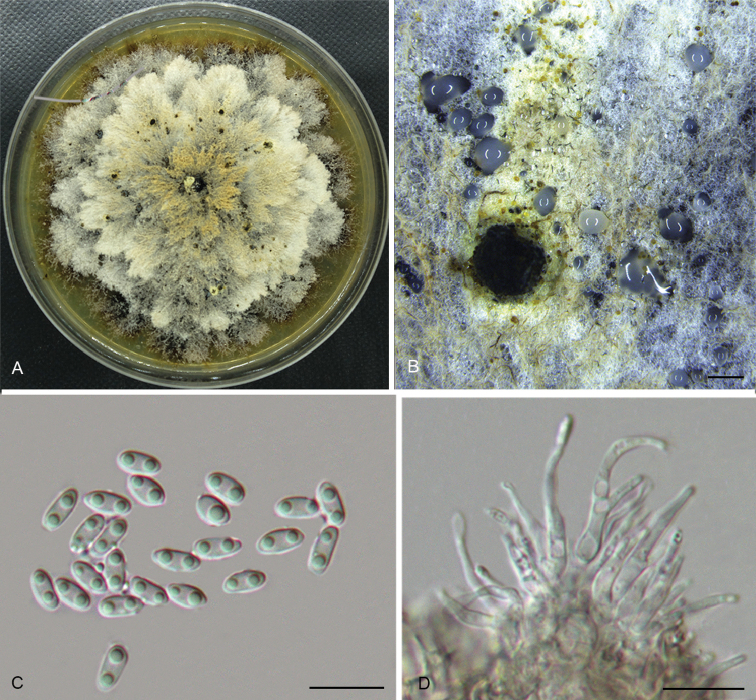
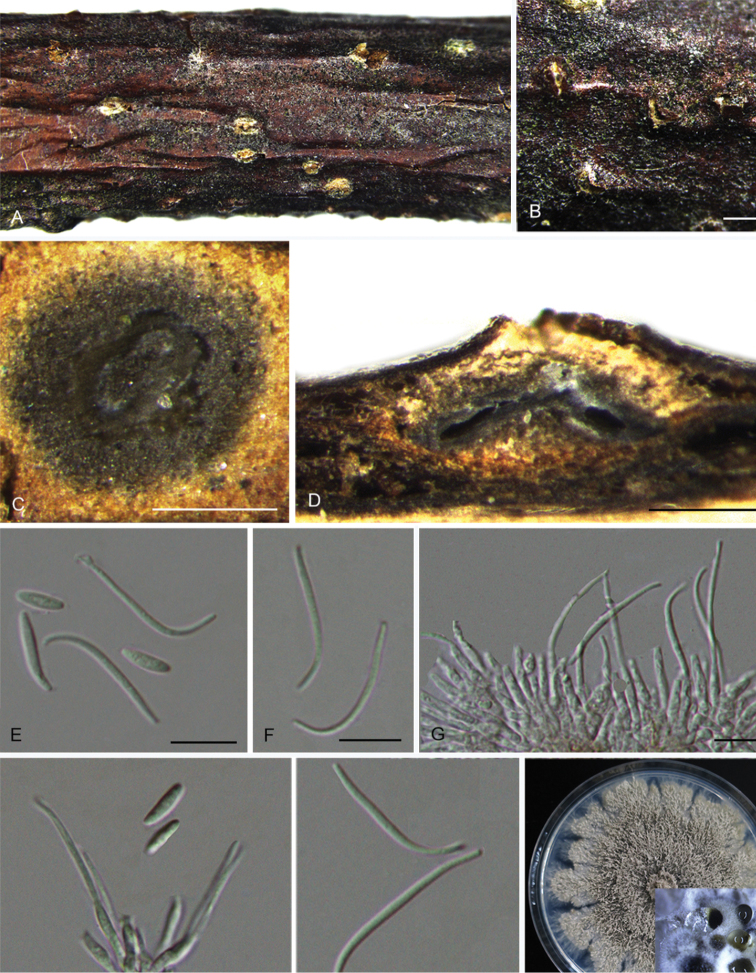
Diaporthe caryae
C.M. Tian & Q. Yang sp. nov.
MB824706
Figure 7.
Diaporthecaryae (CFCC 52563) A Transverse section of conidioma B Longitudinal section of conidioma C Culture on PDAD Alpha conidia E Conidiophores F Beta conidia. Scale bars: 200 μm (A), 100 μm (B), 10 μm (D, F), 20 μm (E).
Diagnosis.
Diaporthecaryae differs from its closest phylogenetic neighbour, D.charlesworthii and D.sackstonii, in ITS, tef1 and tub2 loci based on the alignments deposited in TreeBASE.
Holotype.
CHINA. Jiangsu Province: Nanjing city, on symptomatic twigs of Caryaillinoensis, 10 Nov. 2015, Q. Yang (holotype: BJFC-S1476; ex-type culture: CFCC 52563).
Etymology.
Named after the host genus on which it was collected, Carya.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, nearly flat, discoid, with a solitary undivided locule. Ectostromatic disc brown to black, one ostiole per disc. Locule undivided, 310–325 μm diam. Conidiophores 7–11 × 1.4–2.2 μm, cylindrical, phialidic, unbranched, sometimes inflated. Alpha conidia hyaline, aseptate, ellipsoidal or fusiform, eguttulate, obtuse at both ends, 7–8.5 × 2.1–2.5 μm (av. = 8 × 2.3 μm, n = 30). Beta conidia hyaline, aseptate, filiform, straight or hamate, eguttulate, base subtruncate, tapering towards one apex, 15.5–34 × 1.1–1.4 µm (av. = 27.5 × 1.2 µm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony at first flat with white felty mycelium, becoming black in the centre and black at the marginal area with age, conidiomata not observed.
Additional specimens examined.
CHINA. Jiangsu Province: Nanjing city, on symptomatic twigs of Caryaillinoensis, 10 Nov. 2015, Q. Yang, living culture CFCC 52564 (BJFC-S1477).
Notes.
Two strains representing D.caryae cluster in a well-supported clade and appear closely related to D.charlesworthii and D.sackstonii. Diaporthecaryae can be distinguished based on ITS, tef1 and tub2 loci from D.charlesworthii (50/468 in ITS, 107/338 in tef1 and 90/707 in tub2); from D.sackstonii (4/440 in ITS, 13/340 in tef1 and 23/701 in tub2). Morphologically, D.caryae can be distinguished from D.charlesworthii by its shorter conidiophores (7–11 vs. 15–35 μm); from D.sackstonii by its longer alpha conidia (7–8.5 vs. 6–7 μm) (Thompson et al. 2015).
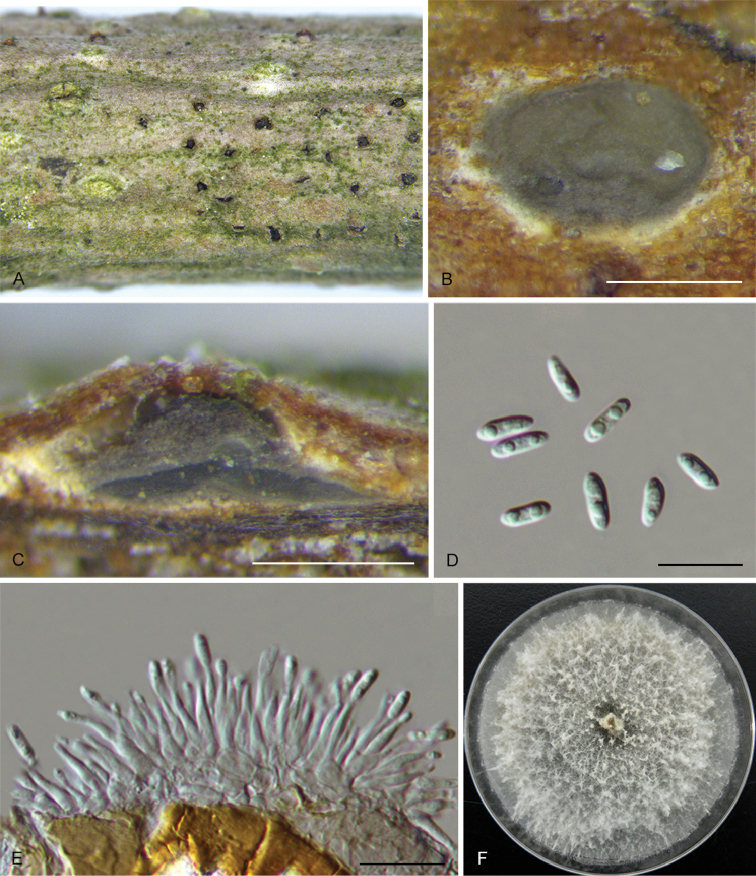
Diaporthe cercidis
C.M. Tian & Q. Yang sp. nov.
MB824707
Figure 8.
Diaporthecercidis (CFCC 52565) A Habit of conidiomata on branches B Transverse section of conidioma C Longitudinal section of conidioma D Alpha conidia E Beta conidia F Conidiophores G Culture on PDA and conidiomata. Scale bars: 100 μm (B–C), 10 μm (D–F).
Diagnosis.
Diaporthecercidis can be distinguished from the phylogenetically closely related species D.pescicola in larger alpha conidia.
Holotype.
CHINA. Jiangsu Province: Nanjing city, on twigs and branches of Cercischinensis, 11 Nov. 2015, Q. Yang (holotype: BJFC-S1478; ex-type culture: CFCC 52565).
Etymology.
Named after the host genus on which it was collected, Cercis.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, nearly flat, discoid, with a solitary undivided locule. Ectostromatic disc grey to brown, one ostiole per disc. Locule circular, undivided, 135–200 μm diam. Conidiophores 7–17 × 1.4–2.1 μm, phialidic, unbranched, straight or slightly curved, tapering towards the apex. Alpha conidia hyaline, aseptate, fusiform to oval, biguttulate, 6.5–10 × 3–3.5 μm (av. = 8.6 × 3.3 μm, n = 30). Beta conidia hyaline, aseptate, filiform, straight or hamate, eguttulate, 20–28.5 × 1–1.3 µm (av. = 25.5 × 1.2 µm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness showed colony at first white, becoming pale brown with yellowish dots with age, flat, with dense and felted mycelium, with visible solitary or aggregated conidiomata at maturity.
Additional specimens examined.
CHINA. Jiangsu Province: Yangzhou city, on twigs and branches of Ginkgobiloba, 11 Nov. 2015, N. Jiang, living culture CFCC 52566 (BJFC-S1479).
Notes.
Diaporthecercidis is distinguished from D.pescicola in the ITS, cal and tef1 loci (13/458 in ITS, 47/442 in cal and 6/328 in tef1). Morphologically, D.cercidis differs from D.pescicola in shorter conidiophores (7–17 vs. 21–35 μm) and larger alpha conidia (6.5–10 × 3–3.5 vs. 6–8.5 × 2–3 μm) (Dissanayake et al. 2017a).
Diaporthe chensiensis
C.M. Tian & Q. Yang sp. nov.
MB824708
Figure 9.
Diaporthechensiensis (CFCC 52567) A–B Habit of conidiomata on branches C Transverse section of conidioma D Longitudinal section of conidioma E Alpha conidia F Beta conidia G Conidiophores H Culture on PDA and conidiomata. Scale bars: 500 μm (B), 200 μm (C–D), 10 μm (E), 20 μm (F).
Diagnosis.
Diaporthechensiensis differs from its closest phylogenetic neighbour, D.vaccinii, in ITS, cal, his3 and tef1 loci based on the alignments deposited in TreeBASE.
Holotype.
CHINA. Shaanxi Province: Ningshan County, Huoditang forest farm, on symptomatic twigs of Abieschensiensis, 5 July 2017, Q. Yang (holotype: BJFC-S1480; ex-type culture: CFCC 52567).
Etymology.
Named after the host species on which it was collected, chensiensis.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, discoid, with a single locule. Ectostromatic disc white to brown, one ostiole per disc, 200–325 μm diam. Locule undivided, 385–540 μm diam. Conidiophores 8.5–13 × 2–3 μm, cylindrical, hyaline, phiailidic, unbranched, straight or slightly curved, tapering towards the apex. Alpha conidia hyaline, aseptate, smooth, ellipsoidal, biguttulate, rounded at both ends, 6.5–11 × 2–2.2 μm (av. = 8.5 × 2.1 μm, n = 30). Beta conidia present on the host, hyaline, eguttulate, smooth, filiform, hamate, 21–28.5 × 0.8–1.1 μm (av. = 25 × 1 μm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white felted aerial mycelium, becoming light brown mycelium due to pigment formation, conidiomata irregularly distributed over agar surface, with yellowish conidial drops exuding from the ostioles.
Additional specimens examined.
CHINA. Shaanxi Province: Ningshan County, Huoditang forest farm, on symptomatic twigs of Abieschensiensis, 5 July 2017, Q. Yang, living culture CFCC 52568 (BJFC-S1481).
Notes.
Diaporthechensiensis occurs in an independent clade (Fig. 1) and is phylogenetically distinct from D.vaccinii. Diaporhechensiensis can be distinguished from D.vaccinii by 57 nucleotides in concatenated alignment, in which 14 were distinct in the ITS region, 13 in the cal region, 10 in the his3 region, 15 in the tef1 region and 15 in the tub2 region. Although this species belongs to the D.eres complex, it is, however, distinct from the known species within the complex (Fig. 2).
Diaporthe cinnamomi
C.M. Tian & Q. Yang sp. nov.
MB824709
Figure 10.
Diaporthecinnamomi (CFCC 52569) A Culture on PDAB Conidiomata C Alpha conidia D Conidiophores. Scale bars: 200 μm (B), 10 μm (C–D).
Diagnosis.
Diaporthecinnamomi differs from its closest phylogenetic species D.discoidispora in ITS, his3 and tef1 loci based on the alignments deposited in TreeBASE.
Holotype.
CHINA. Zhejiang Province: Linan city, on symptomatic twigs of Cinnamomum sp., 22 Apr. 2017, Q. Yang (holotype: BJFC-S1482; ex-type culture: CFCC 52569).
Etymology.
Named after the host genus on which it was collected, Cinnamomum.
Description.
On PDA: Conidiomata pycnidial, globose, solitary or aggregated, deeply embedded in the substrate, erumpent, dark brown to black, 170–235 μm diam., whitish translucent to cream conidial drops exuding from the ostioles. Conidiophores 11–25 × 1.5–2 μm, cylindrical, hyaline, branched, straight or curved, tapering towards the apex. Alpha conidia hyaline, aseptate, ellipsoidal to oval, biguttulate, rounded at both ends, 5–7 × 2.5–3 μm (av. = 6 × 2.9 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness showed colony originally flat with white felty mycelium, developing petaloid mycelium after 7–10 d and turning yellowish at the centre and brownish at the marginal area after 15 d. Conidiomata erumpent at maturity.
Additional material examined.
CHINA. Zhejiang Province: Linan city, on symptomatic twigs of Cinnamomum sp., 22 Apr. 2017, Q. Yang, living culture CFCC 52570 (BJFC-S1483).
Notes.
Diaporthecinnamomi comprises strains CFCC 52569 and CFCC 52570 closely related to D.discoidispora in the combined phylogenetic tree (Fig. 1). Diaporthecinnamomi can be distinguished based on ITS, his3 and tef1 loci from D.discoidispora (4/460 in ITS, 17/448 in his3 and 38/339 in tef1).
Diaporthe conica
C.M. Tian & Q. Yang sp. nov.
MB824710
Figure 11.
Diaportheconica (CFCC 52571) A–B Habit of conidiomata on branches C Longitudinal section of conidioma D Alpha conidia E–F Conidiophores G Culture on PDA and conidiomata. Scale bars: 300 μm (B–C), 10 μm (D–F).
Diagnosis.
Diaportheconica is phylogenetically and morphologically distinct from D.rostrata, in smaller locule and alpha conidia.
Holotype.
CHINA. Zhejiang Province: Tianmu Mountain, on symptomatic branches of Alangiumchinense, 20 Apr. 2017, Q. Yang (holotype: BJFC-S1484; ex-type culture: CFCC 52571).
Etymology.
Named after the conical conidiomata.
Description.
Conidiomata pycnidial, 420–580 μm diam., solitary and with single necks erumpent through the host bark. Tissue around the neck is conical. Locule oval, undivided, 385–435 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells unbranched, straight or sinuous, apical or base sometimes swelling, 19–23.5 × 2.8 μm. Alpha conidia hyaline, aseptate, ellipsoidal, biguttulate, 5.5–7 × 2.3–3 μm (av. = 6.5 × 2.6 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony white to yellowish, with dense and felted mycelium, lacking aerial mycelium, with maize-coloured conidial drops exuding from the ostioles.
Additional material examined.
CHINA. Zhejiang Province: Tianmu Mountain, on symptomatic branches of Alangiumchinense, 20 Apr. 2017, Q. Yang, living culture CFCC 52572 (BJFC-S1485); ibid. living culture CFCC 52573 (BJFC-S1486); ibid. living culture CFCC 52574 (BJFC-S1487).
Notes.
Four isolates clustered in a clade distinct from further Diaporthe species based on DNA sequence data. Morphologically, this species is characterised by conical conidiomata, which is similar with D.rostrata from Juglansmandshurica. However, D.conica differs from D.rostrata by having smaller locule and alpha conidia (310–385 vs. 620–1100 μm in locule; 5.5–7 × 2.3–3 vs. 8.5–11.5 × 4–5 μm in alpha conidia) (Fan et al. 2015).
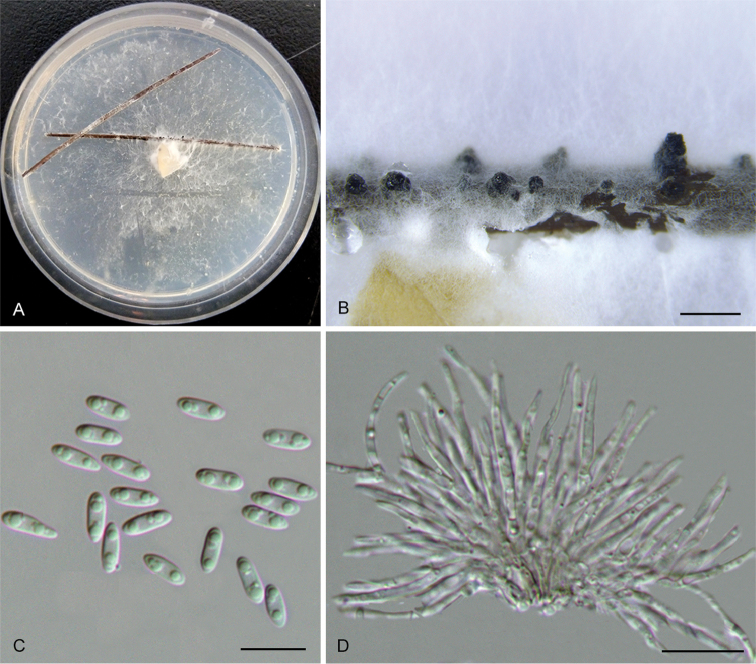
Diaporthe eres
Nitschke, 1870
Figure 12.
Diaportheeres (CFCC 52575) A–B Habit of conidiomata on branches C Transverse section of conidioma D Longitudinal section of conidioma E Alpha conidia F Conidiophores G Culture on PDA and conidiomata. Scale bars: 500 μm (B), 200 μm (C–D), 10 μm (E–F).
= Diaporthebiguttusis Y.H. Gao & L. Cai, 2015.
= Diaporthecamptothecicola C.M. Tian & Qin Yang, 2017.
= Diaportheellipicola Y.H. Gao & L. Cai, 2015.
= Diaporthelongicicola Y.H. Gao & L. Cai, 2015
= Diaporthemahothocarpus (Y.H. Gao, W. Sun & L. Cai) Y.H. Gao & L. Cai, 2015.
= Diaporthemomicola Dissan., J.Y. Yan, Xing H. Li & K.D. Hyde, 2017.
Description.
Conidiomata pycnidial, immersed in bark, erumpent through the bark surface, serried, with a single locule. Ectostromatic disc obviously, brown to black, with one ostiole per disc, 245–572 μm diam. Ostiole medium black, up to the level of disc. Locule circular, undivided, 335–450 μm diam. Conidiophores 10.5–19 × 1–1.5 μm, cylindrical, hyaline, unbranched, straight or slightly sinuous. Conidiogenous cells phialidic, cylindrical, terminal. Alpha conidia hyaline, aseptate, ellipsoidal to lanceolate, one guttulate at each end, 6–7.5 × 1.5–2.5 μm (av. = 6.5 × 2 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures on PDA incubated at 25 °C in darkness. Colony with white felty aerial mycelium, becoming white felted aerial mycelium in the centre and grey-brown mycelium at the marginal area, conidiomata irregularly distributed over agar surface.
Specimens examined.
CHINA. Beijing: Pinggu district, on symptomatic branches of Castaneamollissima, 1 Nov. 2016, N. Jiang, living culture CFCC 52576 (BJFC-S1489); ibid. living culture CFCC 52577 (BJFC-S1490). Heilongjiang Province: Liangshui Nature Reserve, on symptomatic twigs of Acanthopanaxsenticosus, 29 July 2016, Q. Yang, living culture CFCC 52580 (BJFC-S1493). Heilongjiang Province: Harbin city, Botanical garden, on symptomatic twigs of Sorbus sp., 2 Aug. 2016, Q. Yang, living culture CFCC 52575 (BJFC-S1488). Shaanxi Province: Zhashui County, on symptomatic branches of Juglansregia, 29 July 2016, Q. Yang, living culture CFCC 52579 (BJFC-S1492). Zhejiang Province: Yangzhou city, on symptomatic twigs of Meliaazedarace, 8 July 2017, N. Jiang, living culture CFCC 52578 (BJFC-S1491). Zhejiang Province: Tianmu Mountain, on symptomatic twigs of Rhododendronsimsii, 20 Apr. 2017, Q. Yang, living culture CFCC 52581 (BJFC-S1494).
Notes.
Diaportheeres, the type species of the genus, was described by Nitschke (1870) on Ulmus sp. collected in Germany, which has a widespread distribution and a broad host range as a pathogen, endophyte or saprobe causing leaf spots, stem cankers and diseases of woody plants (Udayanga et al. 2014b). Fan et al. (2018) indicated that D.biguttusis, D.ellipicola, D.longicicola and D.mahothocarpus should be treated as synonyms of D.eres using cal, tef1 and tub2 gene regions. In this study, we extended the work presented in Fan et al. (2018) and found seven additional strains belonging to D.eres. Additionally, the phylogenetic tree demonstrated that D.camptothecicola and D.momicola should also be treated as synonyms of D.eres (Fig. 2). Diaporthecamptothecicola from Camptothecaacuminate and D.momicola from Prunuspersica are described and illustrated based on the combined ITS, cal, his3, tef1 and tub2 regions (Dissanayake et al. 2017a, Yang et al. 2017c). Both of the two species are embedded in the D.eres complex. However, ITS analysis resulted in an unresolved phylogenetic tree without definitive bootstrap at the internodes, highly discordant to the trees resulting from the other four genes (Udayanga et al. 2014b). Therefore, the ITS region was not used in the combined analysis in the current study. To further investigate this complex, a second set of four (cal, his3, tef1 and tub2), three (cal, tef1 and tub2), two (tef1 and tub2) and one (tef1) data matrices were performed following Santos et al. (2017) and Fan et al. (2018). The results showed that the three genes analyses (cal, tef1 and tub2) appeared to be a better species recognition (Fig. 2). When it comes to this species complex, sequences supported by Udayanga et al. (2014b) are necessary to perform a more robust phylogenetic tree, clarifying the real species boundaries in this group in the future work.
Diaporthe fraxinicola
C.M. Tian & Q. Yang sp. nov.
MB824711
Figure 13.
Diaporthefraxinicola (CFCC 52582) A–B Habit of conidiomata on branches C Transverse section of conidioma D Longitudinal section of conidioma E Alpha conidia F Beta conidia G Culture on PDA and conidiomata. Scale bars: 500 μm (B), 200 μm (C), 100 μm (D), 10 μm (E–F).
Diagnosis.
Diaporthefraxinicola can be distinguished from the closely related species D.oraccinii and D.acerigena (described above) based on ITS, tef1 and tub2 loci. Diaporthefraxinicola differs from D.oraccinii in larger alpha conidia and from D.acerigena in wider alpha conidia.
Holotype.
CHINA. Shaanxi Province: Zhashui city, Niubeiliang Reserve, on symptomatic twigs of Fraxinuschinensis, 7 July 2017, Q. Yang (holotype: BJFC-S1495; ex-type culture: CFCC 52582).
Etymology.
Named after the host genus on which it was collected, Fraxinus.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, nearly flat, discoid, with a single locule. Ectostromatic disc grey to dark brown, circular to ovoid, one ostiole per disc, 150–325 μm diam. Locule circular, undivided, 275–480 μm diam. Conidiophores 10.5–17.5 × 2.1–3.2 μm, hyaline, branched, cylindrical to clavate, straight, tapering towards the apex. Alpha conidia hyaline, aseptate, ellipsoidal to oval, 2–3-guttulate, rounded at both ends, 7–10 × 2.9–3.2 μm (av. = 8.5 × 3 μm, n = 30). Beta conidia hyaline, filiform, straight or hamate, eguttulate, aseptate, base subtruncate, tapering towards one apex, 19–29.5 × 1.4 µm (av. = 24.5 × 1.4 µm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white aerial mycelium, becoming yellowish, dense and felted aerial mycelium with age, with visible solitary or aggregated conidiomata at maturity.
Additional material examined.
CHINA. Shaanxi Province: Zhashui city, Niubeiliang Reserve, on symptomatic twigs of Fraxinuschinensis, 7 July 2017, Q. Yang, living culture CFCC 52583 (BJFC-S1496).
Notes.
This new species is introduced as molecular data, shows it to be a distinct clade with high support (ML/BI=100/1) and it appears most closely related to D.oraccinii and D.acerigena. Diaporthefraxinicola can be distinguished from D.oraccinii by 22 nucleotides in concatenated alignment, in which 6 were distinct in the ITS region, 8 in the tef1 region and 8 in the tub2 region; from D.acerigena by 27 nucleotides in concatenated alignment, in which 11 were distinct in the ITS region, 3 in the tef1 region and 13 in the tub2 region. Morphologically, D.fraxinicola differs from D.oraccinii in longer and larger alpha conidia (7–10 × 2.9–3.2 vs. 5.5–7.5 × 0.5–2 μm); differs from D.acerigena in larger alpha conidia (2.9–3.2 vs. 2.1–2.9 μm) (Gao et al. 2016).
Diaporthe kadsurae
C.M. Tian & Q. Yang sp. nov.
MB824713
Figure 14.
Diaporthekadsurae (CFCC 52586) A Habit of conidiomata on branches B Transverse section of conidioma C Longitudinal section of conidioma D Alpha conidia E Conidiophores F Culture on PDA. Scale bars: 200 μm (B–C), 10 μm (D–E).
Diagnosis.
Diaporthekadsurae differs from its closest phylogenetic species D.fusicola and D.ovoicicola in ITS, cal and tef1 loci based on the alignments deposited in TreeBASE.
Holotype.
CHINA. Jiangxi Province: Shangrao city, Sanqing Mountain, on symptomatic branches of Kadsuralongipedunculata, 1 Apr. 2017, B. Cao, Y.M. Liang & C.M. Tian (holotype: BJFC-S1497; ex-type culture: CFCC 52586).
Etymology.
Named after the host genus on which it was collected, Kadsura.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, nearly flat, discoid, with a single locule. Ectostromatic disc obviously, brown to black, one ostiole per disc. Locule undivided, 475–525 μm diam. Conidiophores 7–11 × 1.8–2.9 μm, cylindrical, hyaline, unbranched, straight or slightly curved, tapering towards the apex. Alpha conidia hyaline, aseptate, oval or fusoid, biguttulate, 5.5–7.5 × 2.1–2.9 μm (av. = 6.5 × 2.5 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white aerial mycelium, becoming dense and felted aerial mycelium in the centre and grey to black mycelium at the marginal area with solitary conidiomata at maturity.
Additional specimens examined.
CHINA. Jiangxi Province: Shangrao city, Sanqing Mountain, on symptomatic branches of Kadsuralongipedunculata, 1 Apr. 2017, B. Cao, Y.M. Liang & C.M. Tian, living culture CFCC 52587 (BJFC-S1498); Yunbifeng National Forest Park, on symptomatic twigs of Acer sp., 31 Mar. 2017, B. Cao, Y.M. Liang & C.M. Tian, living culture CFCC 52588 (BJFC-S1499); ibid. living culture CFCC 52589 (BJFC-S1500).
Notes.
This new species is introduced as molecular data show it to be a distinct clade with high support (ML/BI=100/1) and it appears most closely related to D.fusicola and D.ovoicicola. Diaporthekadsurae can be distinguished from D.fusicola by 11 nucleotides in concatenated alignment, in which 4 were distinct in the ITS region and 7 in the cal region; from D.ovoicicola by 25 nucleotides in concatenated alignment, in which 12 were distinct in the ITS region, 6 in the cal region and 7 in the tef1 region. Morphologically, D.kadsurae differs from D.fusicola and D.ovoicicola in shorter conidiophores (7–11 μm in D.kadsurae vs. 11–24.1 μm in D.fusicola; 7–11 μm in D.kadsurae vs. 14.2–23.6 μm in D.ovoicicola) (Gao et al. 2014).
Diaporthe padina
C.M. Tian & Q. Yang sp. nov.
MB824714
Figure 15.
Diaporthepadina (CFCC 52590) A–B Habit of conidiomata on branches C Transverse section of conidioma D Longitudinal section of conidioma E Alpha and beta conidia F, I Beta conidia G–H Conidiophores J Culture on PDA and conidiomata. Scale bars: 500 μm (B), 200 μm (C–D), 10 μm (E–I).
Diagnosis.
Diaporthepadina can be distinguished from the phylogenetically closely related species D.betulae in smaller conidiomata and alpha conidia.
Holotype.
CHINA. Heilongjiang Province: Liangshui Nature Reserve, on symptomatic twigs of Padusracemosa, 31 July 2016, Q. Yang (holotype: BJFC-S1501; ex-type culture: CFCC 52590).
Etymology.
Named after the host genus on which it was collected, Padus.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through the bark surface, discoid, with a single locule. Ectostromatic disc light brown, one ostiole per disc, 330–520 μm diam. Locule circular, undivided, 250–550 μm diam. Conidiophores 5.5–12.5 × 1–1.5 μm, hyaline, unbranched, cylindrical, straight or slightly curved. Alpha conidia hyaline, aseptate, ellipsoidal to fusiform, eguttulate, 7–8 × 1.5–2 μm (av. = 7.5 × 1.8 μm, n = 30). Beta conidia hyaline, filiform, straight or hamate, eguttulate, aseptate, base truncate, 21–24 × 1 µm (av. = 22 × 1 µm, n = 30).
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white aerial mycelium, becoming grey to brown in the centre, with pale grey, felted, valviform mycelium at the marginal area and aggregated conidiomata at maturity.
Additional material examined.
CHINA. Heilongjiang Province: Liangshui Nature Reserve, on symptomatic twigs of Padusracemosa, 31 July 2016, Q. Yang, living culture CFCC 52591 (BJFC-S1502).
Notes.
Four strains representing D.padina cluster in a well-supported clade and appear closely related to D.betulae. This species is phylogenetically closely related to, but clearly differentiated from, D.betulae by 40 different unique fixed alleles in ITS, cal, his3, tef1 and tub2 loci (4, 7, 10, 13 and 6 respectively) based on the alignments deposited in TreeBASE. Morphologically, D.padina differs from D.betulae in smaller conidiomata and alpha conidia (250–550 vs. 600–1250 μm in conidiomata; 7–8 × 1.5–2 vs. 8.5–11 × 3–4 μm in alpha conidia) (Du et al. 2016).
Diaporthe ukurunduensis
C.M. Tian & Q. Yang sp. nov.
MB824715
Figure 16.
Diaportheukurunduensis (CFCC 52592) A Habit of conidiomata on branches B Transverse section of conidioma C–D Alpha conidia E Conidiophores F Culture on PDA. Scale bars: 200 μm (B), 10 μm (C–E).
Diagnosis.
Diaportheukurunduensis can be distinguished from the phylogenetically closely related species D.citrichinensis in longer conidiophores and shorter alpha conidia.
Holotype.
CHINA. Shaanxi Province: Qinling Mountain, on symptomatic twigs of Acerukurunduense, 27 June 2017, Q. Yang (holotype: BJFC-S1503; ex-type culture: CFCC 52592).
Etymology.
Named after the host species on which it was collected, Acerukurunduense.
Description.
Conidiomata pycnidial, immersed in bark, serried, slightly erumpent through the bark surface, nearly flat, discoid, with a single locule. Ectostromatic disc dark brown to black, one ostiole per disc. Locule circular, undivided, 165–215 μm diam. Conidiophores 11.5–18 × 1.5 μm, hyaline, branched, cylindrical, straight or curved. Alpha conidia hyaline, aseptate, ellipsoidal to oval, biguttulate, 5–6 × 2.1–2.9 μm (av. = 5.5 × 2.5 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in darkness. Colony originally flat with white aerial mycelium, becoming brown to pale black in the centre, dense, felted, conidiomata not observed.
Additional specimens examined.
CHINA. Shaanxi Province: Qinling Mountain, on symptomatic twigs of Acerukurunduense, 27 June 2017, Q. Yang, living culture CFCC 52593 (BJFC-S1503).
Notes.
Diaportheukurunduensis comprises strains CFCC 52592 and CFCC 52593 closely related to D.citrichinensis in the combined phylogenetic tree (Fig. 1). Diaportheukurunduensis can be distinguished from D.citrichinensis based on ITS and tef1 loci (10/470 in ITS and 4/336 in tef1).
Diaporthe unshiuensis
F. Huang, K.D. Hyde & H.Y. Li, 2015
Figure 17.
Diaportheunshiuensis (CFCC 52594) A Culture on PNA B Conidiomata C Alpha conidia D Conidiophores. Scale bars: 500 μm (B), 10 μm (C–D).
Description.
On PNA: Conidiomata pycnidial, globose or rostrated, black, erumpent in tissue, erumpent at maturity, 260–500 μm diam, often with translucent conidial drops exuding from the ostioles. Conidiophores 18–28.5 × 1.4–2.1 μm, cylindrical, hyaline, branched, septate, straight or curved, tapering towards the apex. Alpha conidia abundant in culture, hyaline, aseptate, ellipsoidal to fusiform, biguttulate, sometimes with one end obtuse and the other acute, 6.5–8.5 × 2.1–2.5 μm (av. = 7.8 × 2.3 μm, n = 30). Beta conidia not observed.
Culture characters.
Cultures incubated on PNA at 25 °C in darkness. Colony entirely white at surface, reverse with pale brown pigmentation, white, fluffy aerial mycelium.
Specimens examined.
CHINA. Jiangsu Province: Nanjing city, on non-symptomatic twigs of Caryaillinoensis, 10 Nov. 2015, Q. Yang, living culture CFCC 52594 and CFCC 52595 (BJFC-S1476).
Notes.
Diaportheunshiuensis was originally described from twigs of non-symptomatic Fortunellamargarita in Zhejiang Province, China (Huang et al. 2015). In the present study, two isolates from twigs of asymptomatic Caryaillinoensis were congruent with D.unshiuensis based on morphology and DNA sequences data (Fig. 1). We therefore describe D.unshiuensis as a known species for this clade.
Discussion
The current study described 15 Diaporthe species from 42 strains based on a large set of freshly collected specimens. It includes 12 new species and 3 known species, which were sampled from 16 host genera distributed over six Provinces of China (Table 1). In this study, 194 reference sequences (including outgroup) were selected based on BLAST searches of NCBIs GenBank nucleotide database and included in the phylogenetic analyses (Table 1). Phylogenetic analyses based on five combined loci (ITS, cal, his3, tef1 and tub2), as well as morphological characters, revealed the diversity of Diaporthe species in China, mainly focusing on diebacks from major ecological or economic forest trees.
Several studies have been conducted associated with various hosts in China. For instance, the research conducted by Huang et al. (2015) revealed seven apparently undescribed endophytic Diaporthe species on Citrus. Gao et al. (2016) demonstrated that Diaporthe isolates, associated with Camellia spp., could be assigned to seven species and two species complexes. Recently, Diaporthe has been revealed as paraphyletic by Gao et al. (2017), showing that Ophiodiaporthe, Pustulomyces, Phaeocytostroma and Stenocarpella embed in Diaporthes. lat. and eight new species of Diaporthe were introduced from leaves of several hosts. However, the identification of Diaporthe species associated with dieback of forest trees has rarely been studied, thus a large-scale investigation of Diaporthe spp. was conducted from 2015 to 2017. This study provides the first molecular phylogenetic frame of Diaporthe diversity associated with dieback in China, combined with morphological descriptions.
Diaportheeres, the type species of the genus, was initially described by Nitschke (1870), from Ulmus sp. collected in Germany. The major problem with this generic type was the lack of an ex-type culture or ex-epitype culture, although a broad species concept has historically been associated with D.eres (Udayanga et al. 2014b). Udayanga et al. (2014b) designed strain AR5193 as the epitype of D.eres and provided the phylogram of this complex using seven loci (ITS, act, Apn2, cal, his3, FG1093, tef1 and tub2), amongst which the tef1, Apn2 and his3 genes were recognised as the best markers for defining species in the D.eres complex. Moreover, they showed that poorly supported non-monophyletic grouping was observed when ITS sequences were included in the combined analysis. In this study, although we conducted phylogenetic analysis as performed in previous studies on Diaporthe species (Santos et al. 2017), much confusion has, however, occurred in species separation of the D.eres complex (Fig. 1). Especially, the ITS region could lead to a confused taxonomic situation within this species complex. We found the three-gene analysis, excluding the ITS and his3 regions, resulted in a more robust tree congruent with Udayanga et al. (2014b) and resolved the species boundaries within the D.eres species complex. The isolates, clustering with D.eres in this study, occur on multiple hosts from many different geographic locations. This study revealed three new species belonging to the D.eres complex, i.e. D.betulina, D.chensiensis and D.padina. It also shows D.biguttusis, D.camptothecicola, D.ellipicola, D.longicicola, D.mahothocarpus and D.momicola were clustered in D.eres and should be treated as synonyms of D.eres, which is in conformity with Fan et al. (2018).
The initial species concept of Diaporthe, based on the assumption of host-specificity, resulted in the introduction of more than 1000 taxa (http://www.indexfungorum.org/). Thus, during the past decade, a polyphasic approach, employing multi-locus DNA data together with morphology and ecology, has been employed for species boundaries in the genus (Crous et al. 2012, Udayanga et al. 2014a, b, Huang et al. 2015, Gao et al. 2016, 2017, Guarnaccia and Crous 2017, 2018, Hyde et al. 2017, 2018, Yang et al. 2017a, b, 2018, Guarnaccia et al. 2018, Jayawardena et al. 2018, Perera et al. 2018a, b, Tibpromma et al. 2018, Wanasinghe et al. 2018).
Further studies are required in order to conduct an extensive collection of Diaporthe isolates, to resolve taxonomic questions and to redefine species boundaries. Multiple strains from different locations should also be subjected to multi-gene phylogenetic analysis to determine intraspecific variation. The descriptions and molecular data of Diaporthe species provided in this study represent a resource for plant pathologists, plant quarantine officials and taxonomists for identification of Diaporthe.
Supplementary Material
Acknowledgements
This study is financed by National Natural Science Foundation of China (Project No.: 31670647). We are grateful to Chungen Piao, Minwei Guo (China Forestry Culture Collection Center (CFCC), Chinese Academy of Forestry, Beijing.
Citation
Yang Q, Fan X-L, Guarnaccia V, Tian C-M (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. https://doi.org/10.3897/mycokeys.39.26914
References
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3: 553–556. 10.2307/3761358 [DOI] [Google Scholar]
- Chi PK, Jiang ZD, Xiang MM. (2007) Flora Fungorum Sinicorum. Vol.34. Phomopsis. Science Press, Beijing.
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004a) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004b) Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BW, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GEStJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ. (2012) Fungal Planet description sheets: 107–127. Persoonia: Molecular Phylogeny and Evolution of Fungi 28: 138–182. 10.3767/003158512X652633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CM, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, de Jesús Yáñez-Morales M, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ. (2015) Fungal Planet description sheets: 371–399. Persoonia: Molecular Phylogeny and Evolution of Fungi 35: 264. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed]
- Desjardins P, Hansen JB, Allen M. (2009) Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. Journal of visualized experiments: JoVE 33: 1–3. 10.3791/1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo E, Santos JM, Phillips AJ. (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. 10.1007/s13225-010-0057-x [DOI] [Google Scholar]
- Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH. (2017b) The current status of species in Diaporthe. Mycosphere 8(5): 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- Dissanayake AJ, Zhang W, Liu M, Hyde KD, Zhao WS, Li XH, Yan JY. (2017a) Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere 8: 533–549. 10.5943/mycosphere/8/5/3 [DOI] [Google Scholar]
- Doilom M, Dissanayake AJ, Wanasinghe DN, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EHC, Hyde KD. (2017) Microfungi on Tectonagrandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. 10.1007/s13225-016-0368-7 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Du Z, Fan XL, Hyde KD, Yang Q, Liang YM, Tian CM. (2016) Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. in China. Phytotaxa 269: 90–102. 10.11646/phytotaxa.269.2.2 [DOI] [Google Scholar]
- Erincik O, Madden L, Ferree D, Ellis M. (2001) Effect of growth stage on susceptibility of grape berry and rachis tissues to infection by Phomopsisviticola. Plant Disease 85: 517–520. 10.1094/PDIS.2001.85.5.517 [DOI] [PubMed] [Google Scholar]
- Fan XL, Hyde KD, Udayanga D, Wu XY, Tian CM. (2015) Diaportherostrata, a novel ascomycete from Juglansmandshurica associated with walnut dieback. Mycological Progress 14: 1–8. 10.1007/s11557-015-1104-5 [DOI] [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, Alvarez LV, Tian CM. (2018) Diaporthe from walnut tree (Juglansregia) in China, with insight of Diaportheeres complex. Mycological Progress: 1–13. 10.1007/s11557-018-1395-4 [DOI]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA fungus 8: 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Su YY, Sun W, Cai L. (2015) Diaporthe species occurring on Lithocarpusglabra in China, with descriptions of five new species. Fungal Biology 115: 295–309. 10.1016/j.funbio.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai L. (2014) Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13: 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Garcia-Reyne A, López-Medrano F, Morales JM, Esteban CG, Martín I, Eraña I, Meije Y, Lalueza A, Alastruey‐Izquierdo A, Rodríguez‐Tudela JL, Aguado JM. (2011) Cutaneous infection by Phomopsislongicolla in a renal transplant recipient from Guinea: first report of human infection by this fungus. Transplant Infectious Disease 13: 204–207. 10.1111/j.1399-3062.2010.00570.x [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2018) Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathologia Mediterranea. 10.14601/Phytopathol_Mediterr-23254 [DOI]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia: Molecular Phylogeny and Evolution of Fungi 40: 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Vitale A, Cirvilleri G, Aiello D, Susca A, Epifani F, Perrone G, Polizzi G. (2016) Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. European Journal of Plant Pathology 146: 963–976. 10.1007/s10658-016-0973-z [DOI] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hewitt W, Pearson R. (1988) Phomopsis cane and leaf spot. Compendium of grape diseases: 17–18. APS Press, St Paul, Minnesota.
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Huang F, Hou X, Dewdney MM, Fu Y, Chen G, Hyde KD, Li HY. (2013) Diaporthe species occurring on citrus in China. Fungal Diversity 61: 237–250. 10.1007/s13225-013-0245-6 [DOI] [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva NI, Dissanayake AJ, Ekanayaka AH, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Jiang HB, Karunarathna A, Lin CG, Liu JK, Liu NG, Lu YZ, Luo ZL, Maharachchimbura SSN, Manawasinghe IS, Pem D, Perera RH, Phukhamsakda C, Samarakoon MC, Senwanna C, Shang QJ, Tennakoon DS, Thambugala KM, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Zhang JF, Zhang SN, Bulgakov TS, Bhat DJ, Cheewangkoon R, Goh TK, Jones EBG, Kang JC, Jeewon R, Liu ZY, Lumyong S, Kuo CH, McKenzie EHC, Wen TC, Yan JY, Zhao Q. (2018) Mycosphere notes 169–224. Mycosphere 9(2): 271–430. 10.5943/mycosphere/9/2/8 [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KT, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He MQ, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kušan I, Lee H, Li J, Lin CG, Liu NG, Lu YZ, Luo ZL, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Abdel-Aziz FA, Li WJ, Senanayake IC, Shang QJ, Daranagama DA, de Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones EBG, Kang JC, Karunarathna SC, Lim YW, Liu JK, Liu ZY, Plautz Jr. HL, Lumyong S, Maharachchikumbura SSN, Matočec N, McKenzie EHC, Mešić A, Miller D, Pawłowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su HY, Suetrong S, Tkalčec Z, Vizzini A, Wen TC, Wisitrassameewong K, Wrzosek M, Xu JC, Zhao Q, Zhao RL, Mortimer PE. (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87(1): 1–235. 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- Jayawardena RS, Hyde KD, Chethana KWT, Daranagama DA, Dissanayake AJ, Goonasekara ID, Manawasinghe IS, Mapook A, Jayasiri SC, Karunarathna A, Li CG, Phukhamsakda C, Senanayake IC, Wanasinghe DN, Camporesi E, Bulgakov TS, Li XH, Liu M, Zhang W, Yan JY. (2018) Mycosphere Notes 102–168: Saprotrophic fungi on Vitis in China, Italy, Russia and Thailand. Mycosphere 9(1): 1–114. 10.5943/mycosphere/9/1/1 [DOI] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Leeuwen GCM, Guarnaccia V, Polizzi G, Van Rijswick PC, Karin Rosendahl KC, Gabler J, Crous PW. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. 10.14601/Phytopathol_Mediterr-14034 [DOI] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsa-ard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW. (2018) Genera of phytopathogenic fungi: GOPHY2. Studies in Mycology. 10.1016/j.simyco.2018.04.002. [DOI] [PMC free article] [PubMed]
- Mostert L, Crous PW, Kang JC, Phillips AJ. (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167. 10.2307/3761612 [DOI] [Google Scholar]
- Mondal SN, Vincent A, Reis RF, Timmer LW. (2007) Saprophytic colonization of citrus twigs by Diaporthecitri and factors affecting pycnidial production and conidial survival. Plant Disease 91: 387–392. 10.1094/PDIS-91-4-0387 [DOI] [PubMed] [Google Scholar]
- Muntañola-Cvetković M, Mihaljčević M, Petrov M. (1981) On the identity of the causative agent of a serious Phomopsis-Diaporthe disease in sunflower plants. Nova Hedwigia 34: 417–435. [Google Scholar]
- Muralli TS, Suryanarayanan TS, Geeta R. (2006) Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52: 673–680. 10.1139/w06-020 [DOI] [PubMed] [Google Scholar]
- Nitschke T. (1870) Pyrenomycetes Germanici 2. Eduard Trewendt, Breslau, 245.
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Perera RH, Hyde KD, Dissanayake AJ, Jones EB, Liu JK, Wei D, Liu ZY. (2018b) Diaporthecollariana sp. nov., with prominent collarettes associated with Magnoliachampaca fruits in Thailand. Studies in Fungi 3(1): 141–151. 10.5943/sif/3/1/16 [DOI] [Google Scholar]
- Perera RH, Hyde KD, Peršoh D, Jones EBG, Liu JK, Liu ZY. (2018a) Additions to wild seed and fruit fungi 1: The sexual morph of Diaportherosae on Magnoliachampaca and Sennasiamea fruits in Thailand. Mycosphere 9(2): 256–270. 10.5943/mycosphere/9/2/7 [DOI] [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew.
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, Voglmayr H, Walker DM. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6: 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/s10267-007-0347-7 [DOI] [Google Scholar]
- Saccardo PA, Roumeguère C. (1884) Reliquiae mycologicae Libertianae. IV. Revue Mycologique, Toulouse 6: 26–39. [Google Scholar]
- Santos JM, Correia VG, Phillips AJL. (2010) Primers for mating-type diagnosis in Diaporthe and Phomopsis, their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114: 255–270. 10.1016/j.funbio.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculumvulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandečić K, Ćosić J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia: Molecular Phylogeny and Evolution of Fungi 27: 9–19. 10.3767/003158511X603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Alves A, Alves R. (2017) Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe PeerJ 5: e3120. 10.7717/peerj.3120 [DOI] [PMC free article] [PubMed]
- Smith H, Wingfeld MJ, Coutinho TA, Crous PW. (1996) Sphaeropsissapinea and Botryosphaeriadothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. 10.1016/S0254-6299(15)30596-2 [DOI] [Google Scholar]
- Tai FL. (1979) Sylloge fungorum sinicorum. Science press, 1527 pp.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Mcmullin DR, Green BD, Miller JD, Seifert KA. (2016) Production of antifungal and antiinsectan metabolites by the Picea endophyte Diaporthemaritima sp. nov. Fungal Biology 120: 1448–1457. 10.1016/j.funbio.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Teng SC. (1963) Fungi of China. Science press, 808 pp.
- Thompson SM, Tan YP, Shivas RG, Neate SM, Morin L, Bissett A, Aitken EAB. (2015) Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia: Molecular Phylogeny and Evolution of Fungi 35: 39. 10.3767/003158515X687506 [DOI] [PMC free article] [PubMed]
- Thompson SM, Tan YP, Young AJ, Neate SM, Aitken EAB, Shivas RG. (2011) Stem cankers on sunflower (Helianthusannuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia: Molecular Phylogeny and Evolution of Fungi 27: 80. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed]
- Tibpromma S, Hyde KD, Bhat JD, Mortimer PE, Xu J, Promputtha I, Mingkwan D, Yang JB, Tang AMC, Karunarathna SC. (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33: 25. 10.3897/mycokeys.33.23670 [DOI] [PMC free article] [PubMed]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014b) Insights into the genus Diaporthe: phylogenetic species delimitation in the D.eres species complex. Fungal Diversity 67: 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthesojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014a) Species limits in Diaporthe: molecular re-assessment of D.citri, D.cytosporella, D.foeniculina and D.rudis. Persoonia: Molecular Phylogeny and Evolution of Fungi 32: 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Liu X, Crous PW, McKenzie EH, Chukeatirote E, Chukeatirote E, Hyde KD. (2012a) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56: 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Hyde KD. (2012b) Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie, Mycologie 33: 295–309. 10.7872/crym.v33.iss3.2012.295 [DOI] [Google Scholar]
- Uecker FA. (1988) A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycological Memoirs 13: 1–231. [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Jones EG, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li WJ, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu JC, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89(1): 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- Wei JC. (1979) Identification of Fungus Handbook. Science press, 780 pp.
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. [Google Scholar]
- Yang Q, Du Z, Tian CM. (2018) Phylogeny and morphology reveal two new species of Diaporthe from Traditional Chinese Medicine in Northeast China. Phytotaxa 336: 159–170. 10.11646/phytotaxa.336.2.3 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Liang YM, Tian CM. (2017c) Diaporthecamptothecicola sp. nov. on Camptothecaacuminata in China. Mycotaxon 132: 591–601. 10.5248/132.591 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017a) Diaporthe species occurring on Sennabicapsularis in southern China, with descriptions of two new species. Phytotaxa 302: 145–155. 10.11646/phytotaxa.302.2.4 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017b) Diaporthejuglandicola sp. nov. (Diaporthales, Ascomycetes), evidenced by morphological characters and phylogenetic analysis. Mycosphere 8: 817–826. 10.5943/mycosphere/8/5/3 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.