Abstract
Bone metastasis from breast and prostate carcinomas is facilitated by activation of bone-resorbing osteoclasts. Using proteomics approaches, we have identified peroxiredoxin-4 (PRDX4) as a cancer-secreted mediator of osteoclastogenesis. We now report characterization of L-plastin in the conditioned media (CM) of MDA-MB-231 human breast cancer cells using immunoblotting and mass spectrometry. The osteoclastogenic potential of MDA-MB-231 CM with siRNA-silenced L-plastin was significantly reduced. L-plastin was detected in cancer-derived exosomes, and inhibition of exosomal release significantly decreased the osteoclastogenic capacity of MDA-MB-231 CM. When added to osteoclast precursors primed with RANKL for 2 days, recombinant L-plastin induced calcium/NFATc1-mediated osteoclastogenesis to the levels similar to continuous treatment with RANKL. Using shRNA, we generated MDA-MB-231 cells lacking L-plastin, PRDX4, or both and injected these cell populations intratibially in CD-1 immunodeficient mice. Micro-CT and histomorphometric analysis demonstrated a complete loss of osteolysis when MDA-MB-231 cells lacking both L-plastin and PRDX4 were injected. A meta-analysis established an increase in L-plastin and PRDX4 mRNA expression in numerous human cancers, including breast and prostate carcinomas. This study demonstrates that secreted L-plastin and PRDX4 mediate osteoclast activation by human breast cancer cells.
Introduction
Bone is the common organ for metastasis from multiple tumors, including breast, prostate, lung, and kidney carcinomas [1]. Osteoclasts, bone cells that specialize in bone destruction, play a critical role in establishment of both osteolytic and osteoblastic bone metastases. Osteoclasts are formed by fusion from monocytic precursors of hematopoietic origin. Receptor activator of nuclear factor kB (RANK) and its ligand (RANKL) are critical for induction of osteoclast formation [2]. RANK signaling includes activation of the transcription factors NF-kB and AP-1 and protein kinases JNK, ERK, and p38 [3], leading to expression and activation of the osteoclastogenic transcription factor nuclear factor of activated T cells (NFAT) c1 [4], [5]. Inactive hyperphosphorylated NFATc1 is maintained in the cytosol. Calcium signaling, in particular, calcium oscillations observed in osteoclast precursors, stimulates calcineurin, a phosphatase that dephosphorylates NFATc1 allowing its nuclear translocation [5], [6]. In addition, signals through co-stimulatory immunoglobulin-like receptors, such as osteoclast-associated receptor (OSCAR) [7], [8] and triggering receptor expressed in myeloid cells-2 (TREM-2) [8], [9], are critical for stimulation of calcium/NFATc1 signaling and osteoclastogenesis.
Previously, we showed that while RANKL is required during early stages of osteoclastogenesis, osteoclast formation from late precursors can be stimulated independent of RANKL by factors produced by actively proliferating breast [10], [11] or prostate [12] cancer cells, which activated ERK and calcium/NFATc1 signaling pathways in osteoclast precursors. Since co-stimulatory immunoglobulin-like receptors are critical for inducing calcium oscillations and sustained NFATc1 activation [7], [9], we examined if the relevant mediators may be immunoprecipitated with IgG. We confirmed that removal of IgG-bound factors from media conditioned by MDA-MB-231 breast carcinoma cells significantly reduced its osteoclastogenic potential and identified one of the factors as peroxiredoxin-4 (PRDX4) [13]. In this study, we report L-plastin as an additional osteoclastogenic factor released by breast cancer cells and investigate the cooperative actions of L-plastin and PRDX4 in stimulating osteolysis in an experimental breast cancer bone metastasis model.
L-plastin belongs to a family of actin-binding proteins that consists of three mammalian isoforms sharing 75%-80% homology: I-plastin (plastin-1, fimbrin) expressed in the small intestine, colon, and kidney; L-plastin (plastin-2, LCP1) expressed mainly in hematopoietic cells; and T-plastin (plastin-3, T-fimbrin) expressed in solid tissues [14], [15]. All plastins have been implicated in the cross-linking of actin filaments [15], [16]. Physiologically, L-plastin is mainly expressed in hematopoietic cells [15]; however, many types of malignant human cells of non-hemapoietic origin express L-plastin [15], [17], [18], [19]. No extracellular function of L-plastin has been described to date; however, it has been detected in extracellular fluids [20].
In this study, we report identification of L-plastin as a novel soluble factor secreted from MDA-MB-231 human breast cancer cells. We examined the route of L-plastin secretion and explored the mechanism through which it stimulates osteoclastogenesis. An in vivo model of experimental bone metastases was used to assess the contribution of L-plastin together with PRDX4 to cancer-induced osteolysis. Finally, the importance of L-plastin and PRDX4 as a diagnostic and prognostic factor for the progression of different types of cancer was validated using publicly available datasets of differential gene expression in cancer patients.
Materials and Methods
This study was carried out in accordance with the recommendations of the Canadian Council on Animal Care. The protocol was approved by the McGill University Animal Care Committee.
Cell Cultures
The MDA-MB-231 breast cancer cell line was provided by Dr. Peter Siegel (McGill University, Montreal) and cultured as previously described [11]. Mouse bone marrow cells were collected as previously described [21]. Mouse bone marrow cells were collected from 6-week-old C57BL6/J mice (Charles River). Cells were cultured in 75-cm2 tissue culture flasks (1.5 × 107 cells per flask) with human recombinant macrophage-colony stimulating factor (M-CSF, 25 ng/ml, 300-25, PeproTech Inc.) for 24 hours, and then nonadherent cells were collected and plated at 5 × 104 cells/cm2 in α-MEM medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum, M-CSF (50 ng/ml), and recombinant GST-RANKL (100 ng/ml). Medium was changed every other day. On day 5, cell cultures were fixed using 10% formalin (23-245-685, Fisher) and stained for tartrate-resistant acid phosphatase (TRAP, Sigma-Aldrich, and 387A-KT). Osteoclasts were identified as multinucleated (more than three nuclei) TRAP-positive cells and were further characterized by image analysis using PixeLINK Capture SE software (PixeLINK) and Image J. RAW 264.7 cells (TIB-71, American Type Culture Collection) were cultured in DMEM supplemented with L-glutamine, 1 mM pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. RAW 264.7 cells were plated at 5 × 103 cells/cm2, and 24 hours later (day 1), recombinant GST-RANKL (50 ng/ml) was added. On days 2-3, cells were supplemented with fresh media with or without RANKL (50 ng/ml) or recombinant L-plastin (rP2, 2.5-25 μg/ml) +/− a [Ca2+]i chelator BAPTA-acetoxymethyl ester (6-50 μM BAPTA, Invitrogen, B6769) for 10 minutes as previously described [22], washed, treated with recombinant L-plastin (rP2, 2.5-25 μg/ml), cultured for 2 days, fixed, and stained for TRAP. L-plastin was provided by Dr. Jan Gettemans (University of Ghent, Belgium) [23].
Cell Culture Reagents
Fetal bovine serum (FBS) was from HyClone (SH 30396-03). Dulbecco's modified Eagle's medium (DMEM), Alpha MEM (αMEM, 310-022-CL), Opti-MEM Reduced Serum Medium (Gibco, Thermo Fisher, 31985070), sodium pyruvate (600-110-EL), L-glutamine (609-065-EL), penicillin/streptomycin (450-201-EL), and trypsin/ethylenediaminetetraacetic acid (T/E, 325-042-EL) were from Wisent Inc. Recombinant human M-CSF (300-25) was from Peprotech Inc. Recombinant glutathione S-transferase-soluble RANKL (GST-RANKL) was purified from clones kindly provided by Dr. M.F. Manolson (University of Toronto).
Preparation of Conditioned Medium
Parental or stably transfected MDA-MB-231 cells were cultured in 75-cm2 flasks to 80% confluence and rinsed twice with PBS, 10 ml of serum free medium was added, and cells were cultured for additional 24 hours. The conditioned medium was collected and centrifuged (100×g, 5 minutes), and then supernatant was filtered (0.2-μm filter) and stored (−80°C).
Mass Spectrometry
MDA-MB-231 cells were cultured in serum-free medium for 24 hours. The medium was collected, filtered (0.2-μm filter), and incubated with 10 μg/ml normal rabbit IgG overnight at 4°C then with immobilized Protein A/G-Sepharose overnight at 4°C. After centrifugation, the beads were collected and washed twice with 50 mM Tris-HCl pH 8.0; 40 mM NaCl; 5 mM EDTA; 1% NP-40 then with 10 mM Tris-HCl pH 6.8. Immunoprecipitates were eluted with 0.2 M glycine pH 2.5; the samples were boiled in 1× SDS-PAGE buffer with 10 mM DTT and separated on a gradient 7%-15% SDS-PAGE gel. The resulting peptides were identified using mass spectrometry (Proteomics service platform at the Centre d'Innovation Genome Québec).
Samples containing 50 μg of proteins were loaded onto a 10% SDS-PAGE followed by staining and destaining. The desired band was excised from the gel and reduced with DTT, alkylated with iodoacetic acid, and digested with trypsin. The lyophilized peptides were resolubilized in 0.1% aqueous formic acid/2% acetonitrile and loaded onto a Thermo Acclaim Pepmap (Thermo, 75 μM ID × 2 cm C18 3-μM beads) precolumn and then onto an Acclaim Pepmap Easyspray (Thermo, 75 μM × 15 cm with 2-μM C18 beads) analytical column separation using a Dionex Ultimate 3000 μHPLC at 220 nl/min with a 2%-35% organic gradient (0.1% formic acid in acetonitrile) over 2 hours. Peptides were analyzed using a Thermo Orbitrap Fusion mass spectrometer operating at 120,000 resolutions (FWHM in MS1, 15,000 for MS/MS) with HCD sequencing all peptides with a charge of 2+ or greater. Raw data were converted into *.mgf format (Mascot generic format) and searched using Mascot 2.3 against human sequences (Swissprot). Database search results were loaded onto Scaffold Q+ Scaffold_4.4.8 (Proteome Sciences) for spectral counting, statistical treatment, and data visualization.
siRNA
MDA-MB-231 cells (150,000 cells/ml) were transfected in suspension (according to manufacturer's suggestion) as follows: handling control (untreated cells that underwent all the procedures without treatments), lipofectamine-treated only (Invitrogen), AllStars negative control siRNA target sequence 5′-CAGTCTTATTCTGTAATGTAA-3′ (Qiagen, 1027280, 10 nM) or Hs_LCP1_5 target sequence 5′-AACTGCGTTATGAAGAGCTAA-3′ (Qiagen, SI04197375, 10 nM). Cells were plated in 6-well plates at 15,000 cells/cm2, and after 6 hours, the medium was changed to culture medium (DMEM). Twenty-four hours later, a second transfection was performed in Opti-MEM followed by a medium change after 6 hours. Forty-eight hours after the first transfection, the cells were washed twice with PBS, and serum-free medium was added. Twenty-four hours later, CM was collected (0.2-μm filter), aliquoted and stored (−80°C).
shRNA
All lentiviral shRNA vectors were retrieved from the arrayed Mission TRC genome-wide shRNA collections purchased from Sigma-Aldrich Corporation [24]. Additional information describing the shRNA vectors can be found at http://www.sigmaaldrich.com/life-science/functional-genomics-and-rnai/shrna/library-information.html or http://www.broad.mit.edu/genome_bio/trc/rnai.html using the TRCN number. The following lentiviral shRNA vectors were used: shhumanLCP1, TRCN0000056494 and TRCN0000056497 and shhumanPRDX4, TRCN0000064819. Lentiviral supernatants were generated as described at http://www.broadinstitute.org/rnai/public/resources/protocols. Pooled stable populations were maintained under antibiotic selections of 2 μg/ml pyromycin ±7.5 μg/ml blasticidin.
Intratibial Injection of Female Nude CD-1 Mice
For in vivo studies, nude CD-1 mice (Charles River) were maintained under sterile conditions in ventilated cages and racks with a 12-hour light/12-hour dark cycle. At 6 weeks of age, female mice were randomized into six groups: vehicle (sham) (n = 4); parental MDA-MB-231 cells (n = 5); luciferase shRNA control vector (Luc, n = 5); shRNA for L-plastin (shL, n = 7, shRNA for peroxiredoxin-4 (shP, n = 7); shRNA for L-plastin and PRDX4 (shLP, n = 6). MDA-MB-231 cells (1 × 105 cells/mouse) in 20 μl of PBS were inoculated in the left tibia [25]. Mice were euthanized 2 weeks after the injection.
Tissue Preparation, μCT, and Histomorphometry
Left tibiae were dissected and immediately fixed in 10% formalin for 24 hours at 4°C. Bones were transferred to 70% ethanol and kept at 4°C until analysis. Micro–computed tomography (μCT) was performed using a SkyScan 1272 high-resolution μCT scanner connected to a Hamamatsu 20-megapixel camera (Hamamatsu L11871). Images were captured at 61 kV, 142 μA, using a detection pixel of 6 μm and a 0.5-mm aluminum filter. Integration time was set at 1400 milliseconds, and 3 images were captured every 0.30° through 180° rotation. Reconstruction was performed using SkyScan Recon software and analyzed using SkyScan CT Analysis software. Tibiae metaphyseal trabecular parameters were measured 100 μm below the growth plate, and a volume of interest of 1.50 mm (250 slides) was analyzed from images acquired at an isotropic resolution of 6 μm. For histological staining and dynamic histomorphometry, fixed tibiae were decalcified in PBS/EDTA and embedded in paraffin. Five micrometer-thick cross-sectional sections and tibial longitudinal sections were collected using a rotary microtome (Leica Microsystems, Germany). Sections were stained for H&E and TRAP, and images were taken with a Leica DMRB light microscope. TRAP-stained images were taken with a Polyvar light microscope at 40× 100 μm below the growth plate. Osteoclasts were counted and measured using the osteometric software (Osteometrics Inc.). Embedding, sectioning, and histomorphometry were performed by the histology platform of the Bone and Periodontal Research Center of McGill University.
Nanoparticle Tracking Analysis (NTA)
Extracellular vesicle profiling entailed measurements of size and numbers of particles secreted from cells cultured in serum-depleted medium using the NS500 nanoparticle tracking analysis system (Nanosight, Amesbury, UK) as described previously [26].
Isolation of Exosomes
Exosomes were purified from MDA-MB-231 conditioned medium originating from untreated and shRNA-treated MDA-MB-231 breast cancer cells according to the procedure by Vlassov et al. [27]. Briefly, MDA-MB-231 breast cancer cells were cultured under standard conditions. When they reached 80% confluency, the cells were washed twice with PBS, and serum-free medium was added to the culture. After 24 hours, the conditioned medium was collected and centrifuged at 100×g for 10 minutes to remove cell debris. The supernatant was further centrifuged at 2500×g for 15 minutes and then filtered by a 0.22-μm filter (Millipore) before a final ultracentrifugation at 100,000×g for 2 hours. The supernatant was discarded, and the exosome pellet was washed twice with PBS and intermittent ultracentrifugation at 100,000×g for 2 hours.
Test Compounds
Cells were incubated with [Ca2+]i chelator BAPTA-acetoxymethyl ester (25 μM, Invitrogen, B6769) for 10 minutes as described previously [22], washed, and treated with 10 μg/ml of recombinant plastin I, L, or T-plastin. Recombinant human L-plastin was a kind gift from Dr. Jan Gettemans, Ghent University, Belgium [23]. Before use, the neutral sphingomyelinase inhibitor GW4869 (used to inhibit exosome production) was resuspended in DMSO (7.5 mM) and aliquoted and stored at −70°C. Upon use, GW4869 was solubilized in 1/20 volume of 5% methanesulfonic acid (working solution 10 μM) [28].
Protein Extraction, Immunoblotting, and Immunofluorescence
For CM, 1 ml of CM was TCA-precipitated and applied on a 10% SDS-PAGE gel followed by immunoblotting. For cell lysates, extraction was performed in RIPA lysis buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 1 mg/ml aprotinin, 2 mg/ml leupeptin, 0.1 mM phenylmethylsulfonyl fluoride, 20 mM sodium fluoride, and 0.5 mM sodium orthovanadate. Samples were centrifuged at 12,000×g for 10 minutes at 4°C, supernatant was collected, and protein was measured using a Quant-iT protein assay kit (Invitrogen, Q33211). Ten to 20 μg of lysates were separated on a 10% SDS-PAGE and transferred to a nitrocellulose membrane (0.45 μm, 162-0115, Bio-Rad) using 10 mM sodium borate buffer. The membranes were blocked in 5% milk for 1 hour at room temperature followed by overnight incubation at 4°C with primary antibodies: anti–plastin-1 (1:300, NBP1-56555, Novus), anti–L-plastin (1:300, MA5-11921, Thermo Fisher), anti–plastin-3 (1:300, T-plastin, A-3, SC-16620B), anti-calnexin (1:1000, NB100-1965, Novus), anti–α-tubulin (1:5000, T9026, Sigma), anti-CD9 (1:150, ab92726, Abcam), and anti-TSG101 (1:150, ab83, Cedarlane). The blots were washed, incubated with horseradish peroxidase–conjugated secondary antibodies (1:1500, anti-mouse, 170-5047; 1:1500, anti-rabbit, 170-5046; Bio-Rad), and visualized with a chemiluminescence system (Super signal West Pico; 34,080, Pierce). Immunoblots were quantified using densitometry with ImageJ. For immunofluorescence, cells plated on glass coverslips were fixed with 10% formalin, and immunostaining was performed as described previously [10] using anti-NFATc-1 (1:100, 7A6, Santa Cruz Biotechnology) and Alexa Fluor 488–conjugated secondary antibody (Invitrogen, A32723). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). Five random images per experimental condition were collected each containing 10-15 precursors. Cells were rated positive for nuclear localization of NFAT if fluorescence intensity of nuclei exceeded that of the cytoplasm.
Transmission Electron Microscopy (TEM)
Purified exosomes were washed by ultracentrifugation (100,000×g, 90 minutes, and 4°C) in 0.2 M hydroxyethyl piperazineethanesulfonic acid (HEPES) pH 7.25 solution. All supernatant was removed, and pellet was resuspended in 2.5% glutaraldehyde in 0.1 M HEPES pH 7.25 solution. Five microliters of fixed exosome solutions were absorbed on each glow-discharged carbon-coated nickel grid and allowed to air dry for 5 minutes. Uranyl formate solutions were prepared by dissolving 7.5 μg uranyl formate in 500 μl boiling water and subsequently adding 1.25 μl 10 N NaOH and vigorous shaking. Solutions were then centrifuged at 21,000×g for 5 minutes to remove debris, and supernatants were collected and used. The grids were negatively stained with 5 μl of uranyl formate solution and allowed to air-dry for 15 minutes. Exosomes were imaged using a Philips TECNAI 12 electron transmission microscope. Microscope tension was set to 120 kV, and grid was inserted to the sample holder. After ensuring that vacuum was maintained, the height of the beam was adjusted using the wobbler function. Intensity, focus, and magnification were adjusted accordingly, and AMT camera was used to capture images.
Expression Clones and Recombinant Protein Purification
The ORF expression clones for human Plastin-1 (EX-Z6971-B31) and Plastin-3 (Ex-S0326-B1) were from Gene Copoeia Inc. and expressed in DH5α cells according to manufacturer's recommendation. The recombinant plastin-1 and -3 were purified using nickel magnetic beads for his-tag protein purification (B23601) from BioTools Inc. Recombinant human L-plastin was a kind gift from Dr. Jan Gettemans, Ghent University, Belgium [23]. The GST-tagged L-plastin was purified on a glutathione sepharose 4B column (17075602, GE healthcare).
Microspectrofluorometry
Raw 264.7 cells were plated onto glass-bottom 35-mm dishes (MatTek Corp.). RANKL-primed cells were washed twice with DMEM containing 10 mM HEPES and labeled with Fura-2-AM (F1221, Invitrogen) at room temperature for 40 minutes. Cells were washed twice and imaged using a fluorescence inverted microscope (T2000, Nikon), cooled charge-coupled device camera (Hamamatsu) connected to the image analysis software (Volocity, Improvision), which recorded fluorescence emission at 510 nm, following excitation at 340 and 380 nm alternated by a high-speed wavelength switching device (Lambda DG-4, Quorum Technologies). Vehicle and 10 μg L-plastin were added to the bath. Changes in [Ca2+]i were measured for 2 minutes at baseline and after the addition of compounds.
Quantitative Real-Time PCR (qPCR)
Total RNA was isolated from primary cultures using the RNeasy mini kit and QIAshredder columns (Qiagen, 74104 and 79654). For real-time PCR, 1 μg of total RNA was reverse transcribed using a cDNA archive kit (Applied Biosystems, 74322171). Real-time PCR was performed using 7500 Applied Biosystems instrument using TaqMan probes with the universal PCR Master Mix (Life technologies, 4304437) in a total volume of 20 μl. The following probes were used: TaqMan: L-plastin, (LCP1): Hs00158701_m1 and glyceraldehyde dehydrogenase phosphate (GAPDH): Hs03929097_g1.
Meta-Analysis of LCP1 and PRDX4 Differential Expression in Human Cancers
Datasets for LCP1 and PRDX4 mRNA expression (expressed as transcripts per million; TPM) in normal tissues and tumors were extracted from The Cancer Genome Atlas (TCGA) database using TCGA Assembler 2 on July 2, 2018 [29], and data were log-transformed to establish normality. Differential expression of LCP1 and PRDX4 was determined as the standardized mean difference (SMD) between normal tissue and primary cancer sites. To determine the average differential expression of each gene, the pooled SMD across all tumor types was estimated using the meta-analytic toolbox MetaLab in MATLAB R2016b (MathWorks) [30], assuming a random effects model for which the variance of cancer-specific effects was determined using the DerSimonian-Laird estimator [31]. To investigate whether there was an association between the changes in LCP1 and PRDX4 expression, data were first partitioned into two groups in which LCP1 was either up- or downregulated at the primary cancer site, and the relationship between LCP1 and PRDX4 changes for each subgroup was analyzed by linear regression in MATLAB. The direction (positive or negative) and strength (R2) of association were reported. Where applicable, 95% confidence intervals (CIs) were estimated using critical values from the z-distribution (i.e., z = 1.96 for significance level α = .05), and results were deemed statistically significant if the 95% CI did not overlap with the null effect (i.e., SMD = 0). When specified, pairwise comparisons between normal tissue and primary tumor gene expression were conducted using two-tailed Student's t test, and P values were reported.
Statistics
Data are presented as means ± standard error of the mean (S.E.) with sample size (n) indicating the number of independent experiments, or as means ± standard deviation (S.D.) with sample size (n) indicating the number of samples. Differences were assessed by ANOVA or Student's t test and accepted as statistically significant at P < .05.
Results
Identification of L-Plastin as a Potential Osteoclast Stimulating Factor Secreted from Breast Cancer Cells
We have previously demonstrated that soluble factors produced by breast [10], [11] and prostate [12] cancer cells induce osteoclast differentiation and that peroxiredoxin-4 (PRDX4) is one the mediators of these osteoclastogenic effects [13]. Another protein identified by mass spectrometry in the osteoclastogenic fraction was plastin (Supplemental Figure 1A). The three plastin isoforms, I-plastin (PLS1), L-plastin (LCP1), and T-plastin (PLS3), are typically described as intracellular proteins. Since plastin isoforms have a high degree of homology, we first confirmed the specificity of the antibodies for different isoforms. Antibodies for T- and L-plastins were specific, while the I-plastin antibody exhibited high nonspecific binding at lower molecular weights; antibody against T-plastin identified a product in both T- and I-plastin lysates; however, the molecular weight was lower than expected (Supplemental Figure 1B). Next, we examined the protein levels of plastin isoforms in MDA-MB-231 cell lysates and conditioned media (CM) using immunoblotting. All plastin isoforms were present in the breast cancer cell lysates; however, only L-plastin was detected extracellularly in the MDA-MB-231 CM (Figure 1A). We dissected the electrophoresis band between 60 and 70 kDa and analyzed it with mass spectrometry. With a 95% probability, we identified 13 peptides that originated from L-plastin and 5 peptides from T-plastin. A transient siRNA knockdown of L-plastin in MDA-MB-231 cells (Figure 1B) resulted in a decrease in the ability of breast cancer cell to induce osteoclastogenesis (Figure 1C, D). Similarly, knockdown of L-plastin in mouse breast carcinoma 4T1 cells decreased their ability to stimulate osteoclast formation (Supplemental Figure 2). L-plastin was found in cell lysates (Figure 1E) and CM (Figure 1F) in the breast (MCF7, 4T1, and MDa-MB-231), prostate (PC3), and erythroleukemia (K562) cancer cells but not in normal skin fibroblasts (NSF). Of interest, we commonly observed two bands in some cell lysates and most CM samples, suggesting the possibility of splice variants or proteolytic products. Thus, we demonstrate that L-plastin is expressed and secreted by breast cancer cells and is potentially involved in stimulation of osteoclastogenesis.
Figure 1.
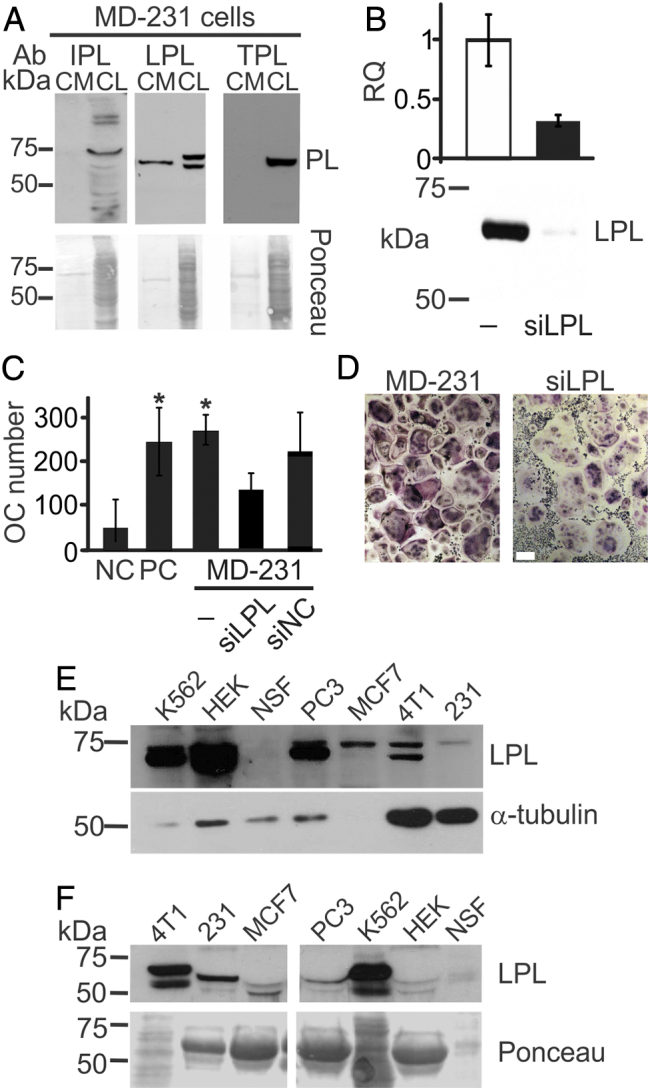
L-plastin is an osteoclastogenic factor secreted from MDA-MB-231 breast cancer cells. (A) Immunoblotting for I, L, and T-plastin (IPL, LPL, and TPL) in MDA-MB-231 cell lysates (CL) and conditioned medium (CM). Equal loading was visualized by Ponceau staining (lower gels). (B-D) L-plastin was transiently depleted in MDA-MB-231 cells (MD-231) using siRNA for L-plastin specific oligos (siLPL) or negative control (siNC). (B) L-plastin mRNA (top) and protein (bottom) expression in parental (−) and siLPL cells. (C, D) RAW 264.7 cells were primed with RANKL (50 ng/ml) for 2 days and then cultured for additional 2 days without treatment (negative control, NC), with RANKL (50 ng/ml, PC), or with 10% CM from siNC or siLPL MDA-MB-231. Average numbers (C) and representative images (D) of osteoclasts (OC) formed in indicated conditions. Scale bar of 100 μm applies to both images. Data are means ± SD, N = 2. (E, F) Protein expressions of L-plastin in whole cell lysates (E) and CM (F) in human erythroleukemia cells K562, human embryonic kidney cells 293 (HEK), normal skin fibroblasts (NSF), prostate carcinoma cells (PC3), human breast carcinoma cells MCF7 and MDA-MB-231 (231), and mouse breast carcinoma cells (4T1) were determined using immunoblotting. Protein loading was assessed using α-tubulin for cell lysates (E) and Ponceau stain for CM (F).
Exosomal Release of L-Plastin from MDA-MB-231 Cells
Plastins are intracellular proteins; therefore, we next examined the mechanism of their release from MDA-MB-231 cells. Since L-plastin was previously found in the vesicular compartment [32], [33], we isolated the vesicular fraction from MDA-MB-231 CM using differential centrifugation. The average vesicular size was 135 nm (Figure 2A), and their transmission electron microscopy appearance was circular with a central depression (Figure 2B), consistent with exosomes. Immunoblotting confirmed the presence of two specific exosome markers, CD9 and TSG101, as well as L-plastin in the vesicular fraction (Figure 2C). Nevertheless, protein expression of L-plastin was still evident in the conditioned medium after isolation of exosomes by ultracentrifugation (Figure 2D). The treatment of MDA-MB-231 cells with an exosome inhibitor GW4869 significantly reduced the osteoclastogenic potency of MDA-MB-231 CM (Figure 2E). We examined if PRDX4 was also released by exosomes; however, PRDX4 was not detected in purified exosomes but was present in MDA-MB-231 cell lysates and CM both before and after depletion of exosomes (Figure 2F). Thus, at least in part, breast cancer cells deliver L-plastin into the extracellular space through exosomal release.
Figure 2.
Exosomes in MDA-MB-231 conditioned medium contain L-plastin. (A) Exosomes were purified from MDA-MB-231 CM, and the distribution of particle sizes was analyzed with Nanosight. (B) Representative transmission electron microscopy image of exosomes purified from MDA-MB-231 CM. (C) Proteins from purified exosomes collected from 10 ml of MDA-MB-231 CM were immunoblotted against exosomal markers, CD9 and TSG101, and L-plastin (LPL). (D) Immunoblotting for L-plastin in MDA-MB-231 cell lysate (CL) or in CM before (+) and after (−) exosome depletion by differential centrifugation. Numbers above the lanes indicate relative density of L-plastin band compared to Ponceau (lower gels). (E) CM was collected from MDA-MB-231 cells cultured without or with exosome inhibitor GW4869 (10 μM, 48 hours). RANKL-primed osteoclast precursors were cultured for an additional 2 days without RANKL (negative control, NC), with RANKL (50 ng/ml, PC), or with 10% CM from untreated or GW4869-treated (white bar) MDA-MB-231. Data are means ± SEM, N = 3; *P < .05, compared to negative control, #P < .05 compared to no GW4869 treatment, assessed by Student's t test. (F) Immunoblotting for PRDX4 and exosomal marker TSG101 in MDA-MB-231 cell lysate (CL), in CM before (+) and after (−) exosome depletion, or in purified exosomes (Exo). Ponceau stain (lower gels) was used as loading control.
Induction of Osteoclast Formation by Recombinant Human L-Plastin Induces
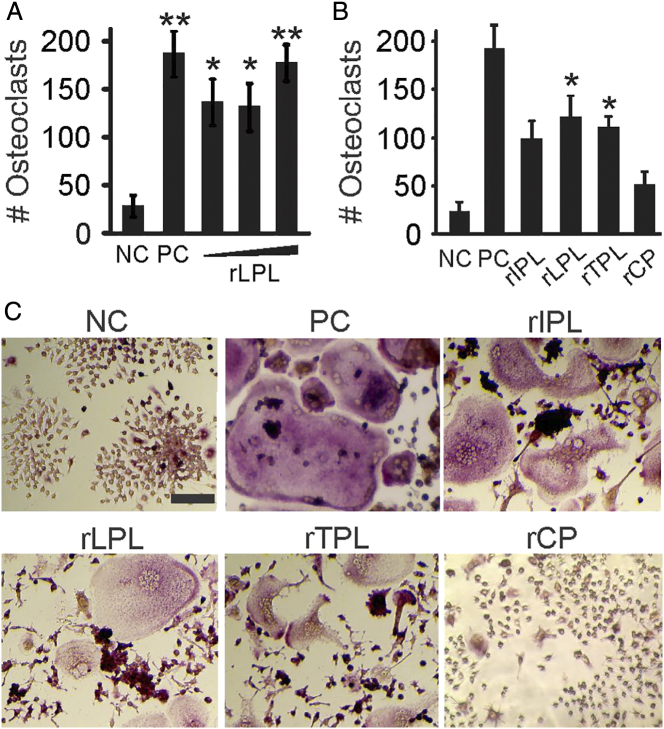
We next examined if L-plastin can directly induce osteoclast formation. We primed RAW 264.7 cells with RANKL for 2 days, washed the precursors, and cultured them with recombinant human L-plastin for an additional 2 days (Figure 3A). The effect of L-plastin on osteoclast formation was similar to the positive control (cells cultured with RANKL for 4 days) and resulted in a significant increase in osteoclast numbers compared to the negative control (cells cultured with RANKL for 2 days, then without additions for 2 days). Similarly, addition of recombinant T-plastin resulted in a significant increase in osteoclast number, while in the presence of I-plastin, increase in osteoclast numbers did not reach statistical significance (Figure 3, B-C). Addition of control proteins recombinantly produced in bacterial system did not affect osteoclast formation (Figure 3, B-C). Increase in the number of osteoclasts in recombinant plastin-treated cultures was accommodated with a corresponding increase in osteoclast size and nucleation (Supplemental Figure 3).
Figure 3.
Recombinant L-plastin induces osteoclast differentiation from RANKL-primed precursors. (A-C) RAW 264.7 cells were primed with RANKL (50 ng/ml) for 2 days and then cultured for an additional 2 days without RANKL treatment (negative control, NC), with RANKL (50 ng/ml, positive control, PC), with human recombinant L-plastin (rLPL) at different concentrations (5, 10, or 25 μg/ml) (A), or with human recombinant I-plastin-1 (rIPL), L-plastin (rLPL), T-plastin (rTPL), or control protein (rCP, an unrelated protein produced by the same process) (5-10 μg/ml), (B) and average osteoclast numbers were assessed. Data are means ± SEM., N = 3-11 independent experiments, ***P < .001 **P < .01, *P < .05 assessed by ANOVA and compared to NC by post hoc Bonferroni test. (C) Representative images of TRAP-stained osteoclast cultures formed in indicated conditions; 100-μm scale bar applies to all images.
We have previously shown that soluble factors produced by MDA-MB-231 cells stimulate osteoclastogenesis through the calcium-dependent NFATc1 nuclear translocation [10], [11]. We examined if L-plastin similarly affects Ca2+/NFATc1 pathway in osteoclast precursors (Figure 4). Addition of recombinant L-plastin to the RANKL-primed osteoclast precursors induced immediate oscillations in [Ca2+]i (Figure 4A), evident in a significant increase in the average and variability in [Ca2+]i levels (Figure 4B). Moreover, L-plastin induced a significant increase in the proportion of osteoclast precursors exhibiting nuclear localization of NFATc1 (Figure 4C). When calcium signaling was prevented using a calcium chelator BAPTA-AM, L-plastin–induced NFATc1 nuclear translocation was significantly decreased (Figure 4C), and the effect of L-plastin on osteoclast differentiation was significantly diminished (Figure 4D). Thus, recombinant human L-plastin induced osteoclast formation from RANKL-primed precursors through stimulation of Ca2+/NFATc1 pathway.
Figure 4.
L-plastin induces [Ca2+]i oscillations and sustained NFATc1 translocation in osteoclast precursors. (A-B) Fura2-loaded RANKL-primed RAW 264.7 cells were imaged at baseline, 50 μg/ml recombinant L-plastin (rLPL) or same amount of media, vehicle (Veh) was added to control for mechanical perturbations, and changes in [Ca2+]i were imaged for additional 60-120 seconds. (A) Top: representative [Ca2+]i recordings (top, 10- second bar indicates time scale) in seven individual cells per experimental condition. Bottom: For each cell, the average variation in [Ca2+]i, measured as a standard deviation of [Ca2+]i, levels was determined and presented in ascending order. (B) Average [Ca2+]i (top) and variation (standard deviations of [Ca2+]i level, bottom). Data are means ± SEM, N = 33-63 cells/condition, from four independent experiments, **P < .01, ***P < .001 compared to baseline, assessed by ANOVA and post hoc Bonferroni test. (C) NFATc1 localization was assessed by immunofluorescence (green); nuclei were stained with DAPI (blue). RAW264.7 cells cultured for 2 days untreated (−) or primed with RANKL (50 ng/ml). The cells were washed and incubated for 2 hours in the fresh medium without RANKL (2), with RANKL (50 ng/ml, 2+), with L-plastin (rLPL, 10 μg/ml), or with L-plastin (10 μg/ml) following pretreatment with 25 μM BAPTA-AM (rLPL+BA). Left: representative images, 10-μm scale bar applies to all images. Right: average percentage of cells with nuclear NFATc1 from total number of cells scored (>50 cells per condition). Data are means ± SEM, N = 2-4 independent experiments, *P < .05 compared to NC, assessed by ANOVA and post hoc Bonferroni test, #P < .05 for compared to rLPL vs. rLPL+BAPTA by Student's t test. (D) RAW 264.7 cells were cultured untreated or primed with RANKL (50 ng/ml) for 2 days and then cultured for additional 2 days without treatment, with RANKL (50 ng/ml), or pretreated with 25 μM of BAPTA or its vehicle and treated with L-plastin (rLPL). Average numbers of osteoclasts formed in indicated conditions (numbers 2 and 5 indicate for how many days cells were exposed to RANKL). Data are means ± SEM, N = 3-6 independent experiments, **P < .01, compared to NC, #P < .05 compared to rLPL by ANOVA.
Loss of Osteolytic Potential by L-Plastin and PRDX4-Deficient MDA-MB-231 Breast Cancer Cells
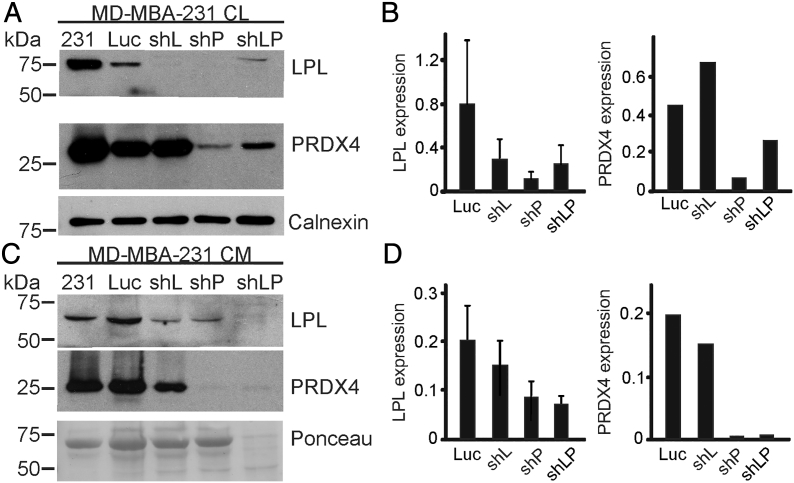
We generated MDA-MB-231 cells stably transfected with shRNAs against L-plastin (shL), peroxiredoxin-4 (shP), or the combination of both L-plastin and peroxiredoxin-4 targeting shRNAs (shLP) (Figure 5). The expression of L-plastin and PRDX4 in the cell lysates (Figure 5A, B) and CM (Figure 5C, D) of shRNA clones was compared to parental and empty vector (Luc) MDA-MB-231 cells. L-plastin expression in cell lysates and CM was reduced in all clones, including MDA-MB-231 cells harboring only the PRDX4-targeting shRNAs (shP). In contrast, PRDX4 expression was reduced only in the MDA-MB-231 clones that harbor PRDX4 targeting shRNAs (shP). These data suggest that L-plastin expression is downstream of PRDX4.
Figure 5.
Generation of L-plastin and PRDX4 targeting shRNA clones. (A-D) MDA-MB-231 cells (231) were transfected with shRNA targeting L-plastin (shL), peroxiredoxin-4 (shP), a combination of both L-plastin and PRDX4 (shLP), or an empty vector (Luc), and stable clones were generated. Expression of L-plastin and PRDX4 in cell lysates (CL, A, B) and conditioned medium (CM, C, D) was assessed by immunoblotting. Calnexin and Ponceau stains were used as loading controls for CL and CM, respectively. Shown are representative immunoblots (A, C) and their quantification relative to loading control (B, D). Data are means ± SEM, N = 3 independent experiments for L-plastin and N = 1 for PRDX4.
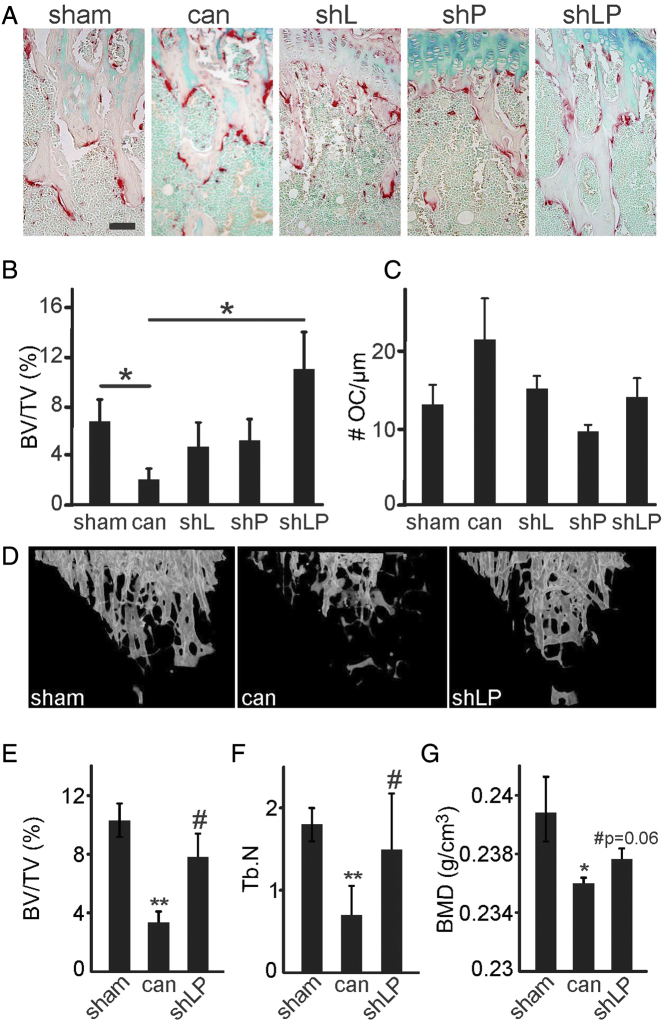
Next, female CD-1 immunodeficient mice were injected in the left tibia with i) PBS (sham); ii) human MDA-MB-231 cells, naïve or empty-vector transfected; iii) L-plastin deficient MDA-MB-231 cells (shL); iv) PRDX4 deficient MDA-MB-231 cells (shP); or v) MDA-MB-231 cells deficient for both L-plastin and PRDX4 (shLP), and bone loss was examined 2 weeks after surgery (Figure 6). We observed no significant difference between mice inoculated with parental or empty vector-transfected MDA-MB-231 cells and combined these groups for further analysis as a positive control group. Histological analysis demonstrated a significant decrease of bone volume per tissue volume (BV/TV) and a tendency for increase in osteoclast number per bone perimeter in positive control samples compared to sham (Figure 6A-C). In tibias injected with clones harboring shL, shP, and shLP targeting shRNAs, BV/TV was not significantly different from sham, while in tibias injected with clones harboring the shLP shRNAs, the BV/TV was also significantly higher than in positive control group (Figure 6B). No increase in osteoclast numbers compared to sham was evident in tibia injected with MDA-MB-231 clones harboring shL, shP, or shLP constructs, and clones deficient in PRDX4 demonstrated significant (shP) or trending (shLP, P = .1) decrease in osteoclast numbers compared to positive control (Figure 6C). μCT confirmed that MDA-MB-231 cells deficient in both L-plastin and PRDX4 (shLP) were unable to induce osteolysis, which is evident by higher BV/TV, trabecular number, and bone mineral density compared to positive control, cancer samples (Figure 6D-E). These data suggest that L-plastin and PRDX4 are involved in mediating breast cancer–induced osteolysis in vivo.
Figure 6.
Double knockdown of L-plastin and PRDX4 significantly attenuates osteolysis in vivo. Six-week-old female nude CD-1 mice were injected in the left tibiae with PBS (sham), 10,000 parental MDA-MB-231 cells, or MDA-MB-231 cells transfected with empty vector (Luc), L-plastin (shL), peroxiredoxin-4 (shP), or both L-plastin and PRDX4 (shLP). Data from MDA-MB-231 parental and Luc groups were not significantly different and were pooled as a single cancer group (can) for further analysis. Two weeks postsurgery, the mice were euthanized, and hind limbs were collected for histology and μCT. (A-C) Histological evaluation. (A) Representative images of upper tibial metaphyseal region stained for TRAP; 100-μm scale bar applies to all images. (B) The bone volume per tissue volume (BV/TV). (C) Number of osteoclasts per bone perimeter (# OC/μm). Data are means ± SEM, N = 4-11 mice/group, *P < .05 assessed by ANOVA followed by post hoc Bonferroni test. (D-G) μCT analysis. D) Representative 3D μCT reconstructions of the trabecular bone in the proximal tibia of mice that were sham-operated, or injected with parental MDA-MB-231 cells (cancer) or with MDA-MB-231 cells lacking L-plastin and PRDX4 (shLP). (E) Bone volume per tissue volume (BV/TV). (F) Trabecular number (Tb.N). (G) Bone mineral density. Data are means ± SEM, N = 3-7 mice per group, *P < .05, **P < .01 compared to sham, #P < .05 compared to cancer, assessed by ANOVA and then Bonferroni test.
Differential Gene Expression of LCP1 and PRDX4 in Primary Carcinomas
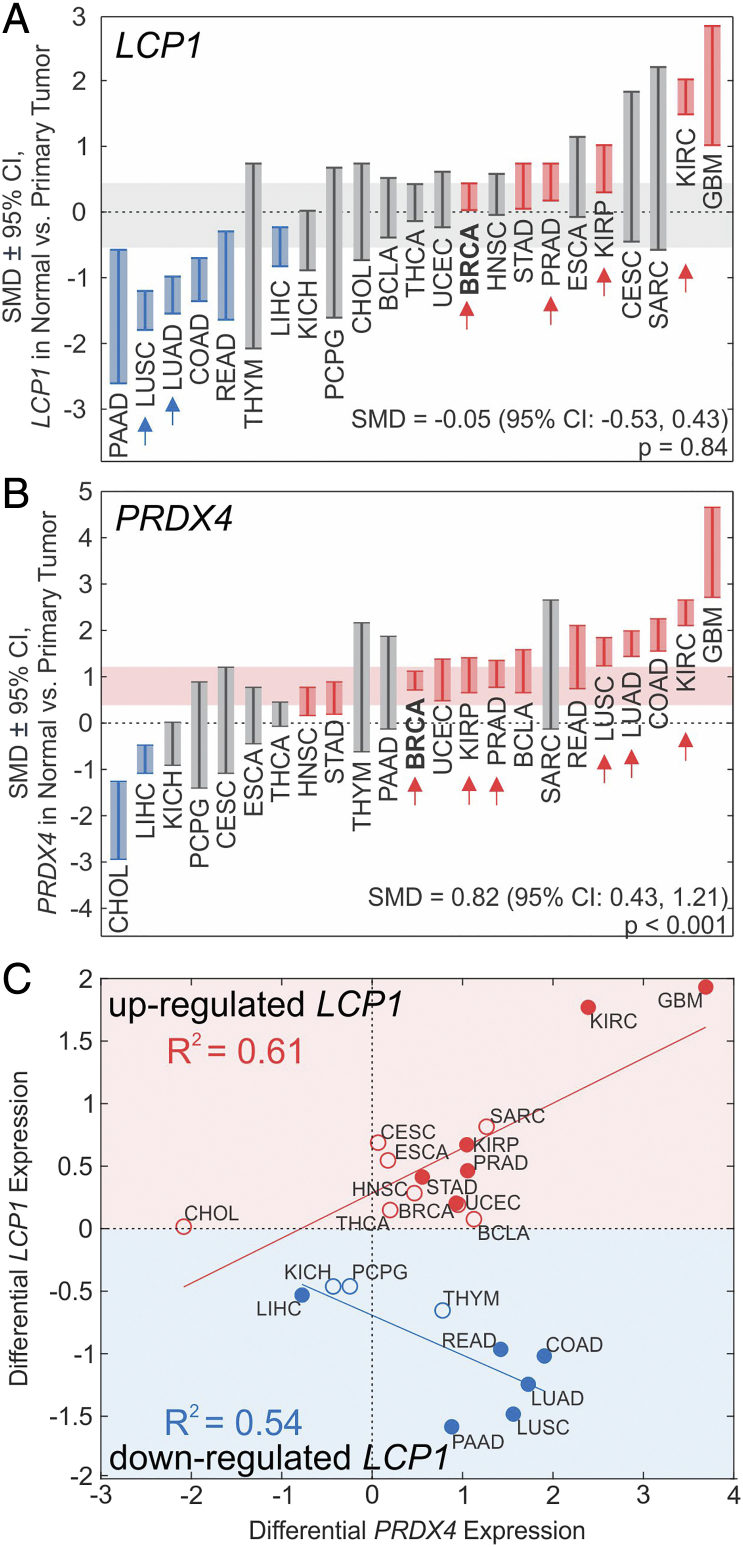
Using datasets from TCGA [29], we examined LCP1 and PRDX4 mRNA levels in normal human tissue and primary tumors; regrettably, the data available for LCP1 or PRDX4 expression at metastatic sites were scarce. We investigated the differential mRNA expression of LCP1 and PRDX4 in other primary cancer types (Figure 7, Table S1). Both LCP1 and PRDX4 were significantly elevated in primary invasive breast carcinoma (BRCA) and primary prostate adenocarcinoma (PRAD). In addition, LCP1 was significantly upregulated or demonstrated tendencies towards an increase in numerous cancer types, including kidney renal clear and papillary cell carcinomas, as well as glioblastoma multiform; and was significantly downregulated in several cancers, including liver hepatocellular carcinoma, pancreatic, colon, and rectum adenocarcinomas, as well as lung adeno- and squamous cell carcinomas (Figure 7A, Table S1). PRDX4 was significantly upregulated in the majority of cancers, including uterine, bladder, head and neck, and kidney carcinomas, and colon, rectum, lung, stomach adenocarcinomas, and significantly downregulated in liver and cholangiocarcinomas (Figure 7B). Of interest, cancers that commonly metastasize to bone (i.e., breast, prostate, lung, and renal cancers [1]) demonstrated significantly higher levels of PRDX4 and, except for lung cancers, LCP1. Since our in vivo data suggested that L-plastin and PRDX4 have cooperative roles in bone metastasis (Figure 6), we examined the relationship between changes in LCP1 and PRDX4 expression in cancer (Figure 7C). Cancer types in which L-plastin was upregulated were consistently characterized by a positive correlation between changes in L-plastin and PRDX4 (Figure 7C, red). In contrast, in cancer types in which LCP1 was downregulated, changes in LCP1 and PRDX4 were negatively correlated (Figure 7C, blue). These data support that LCP1 and PRDX4 are significantly upregulated in invasive breast carcinoma and prostate adenocarcinoma, as well as other cancers that commonly metastasize to bone, with changes in PRDX4 being more pronounced than those observed in LCP1.
Figure 7.
Differential gene expression of L-plastin (LCP1) and PRDX4 in cancer. (A, B) Standardized mean difference (SMD) ± 95% CI between LCP1 (A) and PRDX4 (B) expression in normal tissue and primary tumors for specified cancers. BLCA: bladder urothelial carcinoma, BRCA: breast invasive carcinoma, CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: cholangiocarcinoma, COAD: colon adenocarcinoma, ESCA: esophageal carcinoma, GBM: glioblastoma multiforme, HNSC: head and neck squamous cell carcinoma, KICH: kidney chromophobe, kidney renal clear cell carcinoma, KIRP: kidney renal papillary cell carcinoma, LIHC: liver hepatocellular carcinoma, LUAD: lung adenocarcinoma, LUSC: lung squamous cell carcinoma, PAAD: pancreatic adenocarcinoma, PCPG: pheochromocytoma and paraganglioma, PRAD: prostate adenocarcinoma, READ: rectum adenocarcinoma, SARC: sarcoma, STAD: stomach adenocarcinoma, THCA: thyroid carcinoma, THYM: thymoma, UCEC: uterine corpus endometrial carcinoma. Red: expression significantly higher than normal; blue: significantly lower than normal; gray: not significantly different from normal tissue. Horizontal shaded bands: random effects SMD estimate ± 95% CI of overall pooled SMD across all cancer types. Arrows: Cancer types commonly associated with bone metastases [31]. (C) Linear regression analysis of relationship between differential expression of LCP1 and PRDX4, grouped by cancer types in which LCP1 was either downregulated (blue) or upregulated (red). Filled markers: significant change in LCP1 expression, empty markers: nonsignificant change in LCP1 expression.
Discussion
In this study, we demonstrated that the cytosolic protein L-plastin is released from breast cancer cells with exosomes. Extracellular L-plastin stimulated osteoclast formation from late osteoclast precursors in the absence of RANKL. The osteoclastogenic effect of L-plastin was mediated through stimulation of calcium oscillations and nuclear translocation of NFATc1 transcription factor, supporting a receptor-mediated action of L-plastin. Using an experimental bone metastasis model, we demonstrated that L-plastin, together with PRDX4 which we described previously [13], mediated breast-cancer induced osteolysis. We validated the relevance of L-plastin and PRDX4 in human cancer biology by demonstrating that expression of LCP1 and PRDX4 was significantly increased in carcinoma samples from patients across different cancer types, including primary invasive breast carcinoma and primary prostate adenocarcinoma.
We have now reported that two proteins with established cytosolic function, L-plastin (current manuscript) and PRDX4 [13], are actively released from breast cancer cells and participate in the intercellular communications. While PRDX4 was previously shown to have a signal peptide allowing its secretion [34] and was found in the extracellular space [35], [36], [37], L-plastin was only reported to function as an actin binding protein involved in motility and invasion [16], [23], [38], [39]. To evaluate the route of L-plastin release from breast cancer cells, we considered that the cancer secretome consists of macromolecules released from cancer cells by all means, including secretion, shedding, and leaking through vesicular release [40], [41]. Many cytosolic proteins have been shown to be present within exosomes released from cancer cells [27], [42], [43]. We found that exosomes were important for breast cancer–induced osteoclast formation, as inhibition of exosomal release significantly reduced the osteoclastogenic potential of breast cancer–derived CM. We purified exosomes from the media conditioned by breast cancer cells and have demonstrated that L-plastin was present within the exosomal fraction, while PRDX4 was not. Detectable levels of L-plastin were also found in exosome-depleted CM, suggesting either the presence of an additional route of L-plastin release or L-plastin spillage from exosomes. In this regard, exosomes have been demonstrated to communicate with the target cells through several pathways, including i) interaction of ligand present on the outer surface of exosomal membrane with a cell surface receptor; ii) release of soluble ligands through proteolytic cleavage of exosomal membrane; or iii) release of exosomal content into the cytoplasm after uptake by target cells [44]. Our data demonstrate that in addition to the established prometastatic function of intracellular L-plastin known to enhance cancer invasiveness [23], [45], extracellular L-plastin contributes to cancer metastasis to bone by promoting a favorable osteolytic microenvironment.
The establishment of osteolytic bone metastases requires the action of bone-resorbing osteoclasts [46], [47]. RANKL is a key osteoclastogenic cytokine produced by bone-forming osteoblasts. Breast cancer cells are known to stimulate osteoblastic expression of RANKL, which in turn stimulates osteoclast formation [48], [49], [50]. We have previously determined that cancer cells also release factors that can serve as a substitute for RANKL and directly induce osteoclast formation from precursors deprived of RANKL after an initial period of RANKL-mediated priming [10], [11], [12]. We have found that breast cancer cells lacking L-plastin are ineffective in inducing osteoclast formation and that addition of recombinant L-plastin to RANKL-primed osteoclast precursors is sufficient to stimulate osteoclastogenesis. Moreover, we demonstrated that similar to breast cancer–derived factors, recombinant L-plastin induced calcium oscillations and NFATc1 nuclear translocation. This signaling signature is consistent with the function of co-stimulatory immunoglobulin-like receptors previously reported to be important for osteoclastogenesis, including osteoclast-associated receptor (OSCAR), triggering receptor expressed in myeloid cells 2 (TREM2), signal-regulatory protein β1 (SIRPβ1), and paired immunoglobulin-like receptor A (PIRA) [8], [51]. Our data suggest that secreted L-plastin is a novel osteoclast regulator potentially acting through one of the co-stimulatory immunoglobulin-like receptors to augment and partially substitute for RANKL in promoting osteoclastogenesis.
To confirm the relevance of L-plastin for bone metastasis in vivo, we used the experimental breast cancer bone metastasis model in which parental or shRNA-transfected human MDA-MB-231 cells were injected directly in the tibias of immunodeficient CD-1 mice. We found that L-plastin–deficient MDA-MB-231 cells exhibited a reduced capacity to stimulate osteolysis in vivo. Similarly, PRDX4-deficiency resulted in a moderate reduction in cancer-induced osteolysis in this model. Previously, we examined the role of PRDX4 in prostate cancer bone metastasis using a model of tibial injection of PRDX4-deficient human PC3 cells in CD-1 mice and demonstrated significant but partial inhibition of osteolysis [13]. Importantly, MDA-MB-231 cells deficient in both L-plastin and PRDX4 exhibited severe deficits in their ability to induce osteolysis, suggesting a potential cooperative mechanism of action for L-plastin and PRDX4. This model specifically targets the stage of direct interactions between the invading cancer cells and resident bone cells rather than the processes of selective homing, suggesting that the reduction in osteoclastogenic capacity of L-plastin- and PRDX4-deficient breast cancer cells—rather than changes in motility or homing—likely underlies the reduced osteolysis observed in vivo.
To examine the translational relevance of L-plastin and PRDX4, we used the publicly available data sets to examine differential gene expression in normal and tumor tissues of cancer patients (https://cancergenome.nih.gov/). We found that changes in LCP1 expression were cancer-type specific: LCP1 was significantly increased in invasive breast carcinoma and prostate adenocarcinoma, as well as in kidney cell carcinomas and glioblastoma, and significantly decreased in tumors originated in lower gastrointestinal tract. PRDX4 was significantly upregulated in the majority of cancer types (13 of 23) and significantly downregulated only in 2 cancer types: cholangio and liver carcinomas. Importantly, PRDX4 was significantly upregulated in all cancer types known for their high propensity to metastasize to bone—breast, prostate, lung, and kidney [52]—while LCP1 was upregulated in all except for lung cancers. These data strongly support the potential utility of LCP1 and PRDX4 as diagnostic markers and therapy targets. Our data are consistent with the previous studies of PRDX4, overexpression of which was identified in many cancer types [53], including breast [54] and prostate [52], [55]. Based on the L-plastin role in tumor cell invasiveness [17], [45], [56], [57], [58], [59], [60], it was proposed to be used as a diagnostic marker [19], [61]; prognostic marker [62]; or a therapeutic target [60], [63], [64]. Our data demonstrate that extracellular L-plastin and PRDX4 have a specific role in stimulating osteoclastogenesis and thus promoting osteolysis during cancer metastasis to bone and suggest that information regarding the expression of these proteins may be particularly useful in cancers with frequently bone metastasis.
The following are the supplementary data related to this article.
Identification of osteoclastogenic factors in MDA-MB-231 conditioned medium. (A) MDA-MB-231 CM-derived osteoclastogenic proteins were immunoprecipitated by IgG, separated on a gradient 7%-15% SDS-PAGE gel, divided into 10 sections, and digested with trypsin. Mass spectrometry identified PRDX4 (validated in the previous study) and plastin as highly expressed candidates. This panel is identical to that published as Figure 1C in Rafiei et al, Cancer Letters, 2015 Jun 1;361(2):262-70 (reused with permission). (B) Antibody validation for plastin isoforms. Recombinant human plastin-1, 2, and 3 (rhPL I, L, T) were immunoblotted with antibodies for I, L, and T-plastin (upper gels, dashed line: predicted MW of plastins); equal loading was visualized by Ponceau staining (lower gels).
Generation of L-plastin targeting shRNA 4T1 clones. 4T1 cells were transfected with shRNA targeting L-plastin with two independent shRNA expression vectors (KD1 and KD2), or an empty vector (LMP), and stable clones were generated. (A) Expression of Lcp1 in parental 4T1 cells and shRNA cells harboring control vector (LMP) or shRNA for L-plastin (KD1 and KD2) was assessed and normalized to beta-actin (TaqMan: Lcp1: Mn00786153_s1, Actb: Mn00607938_s1. (B) Expression of L-plastin in cell lysates (top) and conditioned medium (bottom) was assessed by immunoblotting. (C) RAW 264.7 cells were cultured untreated (RAW) or primed with RANKL (50 ng/ml) for 2 days and then cultured for additional 2 days without treatment (negative control, NC), with RANKL (50 ng/ml, PC), or with 10% CM with from parental 4T1 cells (4T1), shRNA control clone LMP, or shLCP1 clones KD1 and KD2. Average numbers (left) and representative images (right) of osteoclasts formed in indicated conditions. Scale bar of 50 μm applies to all images. Data are means ± SEM, N = 6; *P < .05, **P < .01 compared to negative control, #P < .05, ##P < .01 compared to 4T1, assessed by Student's t test.
RAW 264.7 cells were primed with RANKL (50 ng/ml) for 2 days and then cultured for an additional 2 days without RANKL treatment (negative control, NC); with RANKL (50 ng/ml, positive control, PC); or with human recombinant plastin-1 (rIPL), 2 (rLPL), and 3 (rTPL) at 10 μg/ml. (A) Average osteoclast planar area. (B) Average number of nuclei per osteoclast. Data are means ± SEM, N= 30-55 cells, *P < .05, **P < .01 compared to NC assessed by Student’s t test.
Differential Expression of LCP1 and PRDX4 in Normal Tissue and Primary Tumours for Different Cancer Types.
LCP1 and PRDX4 mRNA expression data were obtained from the TCGA database, and standardized mean differences (SMDs) between normal tissue and primary tumor expression were estimated along with corresponding standard errors (SEs) and 95% confidence intervals (loCI: lower limit, hiCI: upper limit). Additionally, random effects estimate of overall pooled SMD across all cancer types was determined. Nnorm: normal tissue sample size, Nprim: primary tumor site sample size. Red: significant upregulation, blue: significant downregulation, gray: nonsignificant difference.
Acknowledgments
Acknowledgements
We are grateful to Dr. Morris F. Manolson for providing GST-RANKL clones, the laboratory of Dr. Januzs Rak for the help with the Nanosight, Drs. Martin Pellicelli and Elizabeth Zimmermann for technical assistance with the SkyScan 1272 high-resolution μCT scanner, Ms. Manuella Widjaja for help with selection of shRNA clones, and Ms. Mariya Stavnichuk for help with cell culture experiments. We are also grateful to the animal facility staff at the Shriner's Hospital for Children for technical assistance with the in vivo study. This work was supported by the Canadian Institutes of Health Research (MOP-77643 to S.V.K.).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Appendix A
Supplementary meta-analysis data can be found under Supplemental Table 1.
References
- 1.Hernandez RK, Wade SW, Reich A, Pirolli M, Liede A, Lyman GH. Incidence of bone metastases in patients with solid tumors: analysis of oncology electronic medical records in the United States. BMC Cancer. 2018;18:44. doi: 10.1186/s12885-017-3922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DH, Kong YY, Penninger JM. Role of RANKL and RANK in bone loss and arthritis. Ann Rheum Dis. 2002;61(Suppl. 2):ii32–39. doi: 10.1136/ard.61.suppl_2.ii32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 4.Ishida N, Hayashi K, Hoshijima M, Ogawa T, Koga S, Miyatake Y, Kumegawa M, Kimura T, Takeya T. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J Biol Chem. 2002;277:41147–41156. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- 5.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 6.Hwang SY, Putney JW., Jr. Calcium signaling in osteoclasts. Biochim Biophys Acta. 2011;1813:979–983. doi: 10.1016/j.bbamcr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim N, Takami M, Rho J, Josien R, Choi Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J Exp Med. 2002;195:201–209. doi: 10.1084/jem.20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 9.Cella M, Buonsanti C, Strader C, Kondo T, Salmaggi A, Colonna M. Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J Exp Med. 2003;198:645–651. doi: 10.1084/jem.20022220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Tiedemann K, Khalil JA, Russo C, Siegel PM, Komarova SV. Osteoclast precursors acquire sensitivity to breast cancer derived factors early in differentiation. Bone. 2008;43:386–393. doi: 10.1016/j.bone.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Tiedemann K, Hussein O, Sadvakassova G, Guo Y, Siegel PM, Komarova SV. Breast cancer-derived factors stimulate osteoclastogenesis through the Ca2+/protein kinase C and transforming growth factor-beta/MAPK signaling pathways. J Biol Chem. 2009;284:33662–33670. doi: 10.1074/jbc.M109.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafiei S, Komarova SV. Molecular signaling pathways mediating osteoclastogenesis induced by prostate cancer cells. BMC Cancer. 2013;13:605. doi: 10.1186/1471-2407-13-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafiei S, Tiedemann K, Tabaries S, Siegel PM, Komarova SV. Peroxiredoxin 4: a novel secreted mediator of cancer induced osteoclastogenesis. Cancer Lett. 2015;361:262–270. doi: 10.1016/j.canlet.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Delanote V, Vandekerckhove J, Gettemans J. Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol Sin. 2005;26:769–779. doi: 10.1111/j.1745-7254.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 15.Shinomiya H. Plastin family of actin-bundling proteins: its functions in leukocytes, neurons, intestines, and cancer. Int J Cell Biol. 2012;2012:213492. doi: 10.1155/2012/213492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samstag Y, Klemke M. Ectopic expression of L-plastin in human tumor cells: diagnostic and therapeutic implications. Adv Enzyme Regul. 2007;47:118–126. doi: 10.1016/j.advenzreg.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Zheng J, Rudra-Ganguly N, Powell WC, Roy-Burman P. Suppression of prostate carcinoma cell invasion by expression of antisense L-plastin gene. Am J Pathol. 1999;155:115–122. doi: 10.1016/S0002-9440(10)65106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz DM, Bollner C, Thomas G, Atkinson M, Esposito I, Hofler H, Aubele M. Identification of differentially expressed proteins in triple-negative breast carcinomas using DIGE and mass spectrometry. J Proteome Res. 2009;8:3430–3438. doi: 10.1021/pr900071h. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Zhao R. Expression and clinical significance of L-plastin in colorectal carcinoma. J Gastrointest Surg. 2011;15:1982–1988. doi: 10.1007/s11605-011-1678-4. [DOI] [PubMed] [Google Scholar]
- 20.Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong S, Pereverzev A, Dixon SJ, Sims SM. Activation of P2X7 receptors causes isoform-specific translocation of protein kinase C in osteoclasts. J Cell Sci. 2009;122:136–144. doi: 10.1242/jcs.031534. [DOI] [PubMed] [Google Scholar]
- 22.Komarova SV, Pilkington MF, Weidema AF, Dixon SJ, Sims SM. RANK ligand-induced elevation of cytosolic Ca2+ accelerates nuclear translocation of nuclear factor kappa B in osteoclasts. J Biol Chem. 2003;278:8286–8293. doi: 10.1074/jbc.M206421200. [DOI] [PubMed] [Google Scholar]
- 23.Delanote V, Vanloo B, Catillon M, Friederich E, Vandekerckhove J, Gettemans J. An alpaca single-domain antibody blocks filopodia formation by obstructing L-plastin-mediated F-actin bundling. FASEB J. 2010;24:105–118. doi: 10.1096/fj.09-134304. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C, Prahallad A. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussein O, Tiedemann K, Murshed M, Komarova SV. Rapamycin inhibits osteolysis and improves survival in a model of experimental bone metastases. Cancer Lett. 2012;314:176–184. doi: 10.1016/j.canlet.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 29.Wei L, Jin Z, Yang S, Xu Y, Zhu Y, Ji Y. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34:1615–1617. doi: 10.1093/bioinformatics/btx812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikolajewicz N, Mohammed A, Morris M, Komarova SV. Meta-analysis of mechanically-stimulated ATP release from mammalian cells. J Cell Sci. 2018;131(22) doi: 10.1242/jcs.223354. [DOI] [PubMed] [Google Scholar]
- 31.Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Wiley; 2009. Introduction to Meta-Analysis. [Google Scholar]
- 32.Dubovsky JA, Chappell DL, Harrington BK, Agrawal K, Andritsos LA, Flynn JM, Jones JA, Paulaitis ME, Bolon B, Johnson AJ. Lymphocyte cytosolic protein 1 is a chronic lymphocytic leukemia membrane-associated antigen critical to niche homing. Blood. 2013;122:3308–3316. doi: 10.1182/blood-2013-05-504597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piehl LL, Fischman ML, Hellman U, Cisale H, Miranda PV. Boar seminal plasma exosomes: effect on sperm function and protein identification by sequencing. Theriogenology. 2013;79:1071–1082. doi: 10.1016/j.theriogenology.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Wong CM, Chun AC, Kok KH, Zhou Y, Fung PC, Kung HF, Jeang KT, Jin DY. Characterization of human and mouse peroxiredoxin IV: evidence for inhibition by Prx-IV of epidermal growth factor- and p53-induced reactive oxygen species. Antioxid Redox Signal. 2000;2:507–518. doi: 10.1089/15230860050192288. [DOI] [PubMed] [Google Scholar]
- 35.Okado-Matsumoto A, Matsumoto A, Fujii J, Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem. 2000;127:493–501. doi: 10.1093/oxfordjournals.jbchem.a022632. [DOI] [PubMed] [Google Scholar]
- 36.Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123–130. doi: 10.1179/135100002125000352. [DOI] [PubMed] [Google Scholar]
- 37.Fujii J, Ikeda Y, Kurahashi T, Homma T. Physiological and pathological views of peroxiredoxin 4. Free Radic Biol Med. 2015;83:373–379. doi: 10.1016/j.freeradbiomed.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Lin CS, Park T, Chen ZP, Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J Biol Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- 39.Van Audenhove I, Denert M, Boucherie C, Pieters L, Cornelissen M, Gettemans J. Fascin Rigidity and L-plastin Flexibility Cooperate in Cancer Cell Invadopodia and Filopodia. J Biol Chem. 2016;291:9148–9160. doi: 10.1074/jbc.M115.706937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rak J. Microparticles in cancer. Semin Thromb Hemost. 2010;36:888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 41.Schaaij-Visser TB, de Wit M, Lam SW, Jimenez CR. The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context. Biochim Biophys Acta. 2013;1834:2242–2258. doi: 10.1016/j.bbapap.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Choi D, Lee TH, Spinelli C, Chennakrishnaiah S, D'Asti E, Rak J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin Cell Dev Biol. 2017;67:11–22. doi: 10.1016/j.semcdb.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Isola AL, Chen S. Exosomes: the messengers of health and disease. Curr Neuropharmacol. 2017;15:157–165. doi: 10.2174/1570159X14666160825160421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, Tancini B, Emiliani C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes. 2013;4:152–170. doi: 10.3390/genes4020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klemke M, Rafael MT, Wabnitz GH, Weschenfelder T, Konstandin MH, Garbi N, Autschbach F, Hartschuh W, Samstag Y. Phosphorylation of ectopically expressed L-plastin enhances invasiveness of human melanoma cells. Int J Cancer. 2007;120:2590–2599. doi: 10.1002/ijc.22589. [DOI] [PubMed] [Google Scholar]
- 46.Boyde A, Maconnachie E, Reid SA, Delling G, Mundy GR. Scanning electron microscopy in bone pathology: review of methods, potential and applications. Scan Electron Microsc. 1986:1537–1554. [PubMed] [Google Scholar]
- 47.Taube T, Elomaa I, Blomqvist C, Beneton MN, Kanis JA. Histomorphometric evidence for osteoclast-mediated bone resorption in metastatic breast cancer. Bone. 1994;15:161–166. doi: 10.1016/8756-3282(94)90703-x. [DOI] [PubMed] [Google Scholar]
- 48.Thomas RJ, Guise TA, Yin JJ, Elliott J, Horwood NJ, Martin TJ, Gillespie MT. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–4458. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 49.Ohshiba T, Miyaura C, Inada M, Ito A. Role of RANKL-induced osteoclast formation and MMP-dependent matrix degradation in bone destruction by breast cancer metastasis. Br J Cancer. 2003;88:1318–1326. doi: 10.1038/sj.bjc.6600858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan H, Tumber A, Hill PA. Breast cancer cells induce osteoclast formation by stimulating host IL-11 production and downregulating granulocyte/macrophage colony-stimulating factor. Int J Cancer. 2004;109:653–660. doi: 10.1002/ijc.20056. [DOI] [PubMed] [Google Scholar]
- 51.Takayanagi H. Interaction between the immune system and bone metabolism: an emerging field of osteoimmunology. Proc Jpn Acad Ser B Phys Biol Sci. 2007;83:136–143. doi: 10.2183/pjab.83.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saraon P, Cretu D, Musrap N, Karagiannis GS, Batruch I, Drabovich AP, van der Kwast T, Mizokami A, Morrissey C, Jarvi K. Quantitative proteomics reveals that enzymes of the ketogenic pathway are associated with prostate cancer progression. Mol Cell Proteomics. 2013;12:1589–1601. doi: 10.1074/mcp.M112.023887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolussi A, D'Inzeo S, Capalbo C, Giannini G, Coppa A. The role of peroxiredoxins in cancer. Mol Clin Oncol. 2017;6:139–153. doi: 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karihtala P, Mantyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9:3418–3424. [PubMed] [Google Scholar]
- 55.Basu A, Banerjee H, Rojas H, Martinez SR, Roy S, Jia Z, Lilly MB, De Leon M, Casiano CA. Differential expression of peroxiredoxins in prostate cancer: consistent upregulation of PRDX3 and PRDX4. Prostate. 2011;71:755–765. doi: 10.1002/pros.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foran E, McWilliam P, Kelleher D, Croke DT, Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int J Cancer. 2006;118:2098–2104. doi: 10.1002/ijc.21593. [DOI] [PubMed] [Google Scholar]
- 57.Chaijan S, Roytrakul S, Mutirangura A, Leelawat K. Matrigel induces L-plastin expression and promotes L-plastin-dependent invasion in human cholangiocarcinoma cells. Oncol Lett. 2014;8:993–1000. doi: 10.3892/ol.2014.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riplinger SM, Wabnitz GH, Kirchgessner H, Jahraus B, Lasitschka F, Schulte B, van der Pluijm G, van der Horst G, Hammerling GJ, Nakchbandi I. Metastasis of prostate cancer and melanoma cells in a preclinical in vivo mouse model is enhanced by L-plastin expression and phosphorylation. Mol Cancer. 2014;13:10. doi: 10.1186/1476-4598-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koide N, Kasamatsu A, Endo-Sakamoto Y, Ishida S, Shimizu T, Kimura Y, Miyamoto I, Yoshimura S, Shiiba M, Tanzawa H. Evidence for critical role of lymphocyte cytosolic protein 1 in oral cancer. Sci Rep. 2017;7 doi: 10.1038/srep43379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kell MJ, Riccio RE, Baumgartner EA, Compton ZJ, Pecorin PJ, Mitchell TA, Topczewski J, LeClair EE. Targeted deletion of the zebrafish actin-bundling protein L-plastin (lcp1) PLoS One. 2018;13 doi: 10.1371/journal.pone.0190353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueo H, Sugimachi K, Gorges TM, Bartkowiak K, Yokobori T, Muller V, Shinden Y, Ueda M, Ueo H, Mori M. Circulating tumour cell-derived plastin3 is a novel marker for predicting long-term prognosis in patients with breast cancer. Br J Cancer. 2015;112:1519–1526. doi: 10.1038/bjc.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otsuka M, Kato M, Yoshikawa T, Chen H, Brown EJ, Masuho Y, Omata M, Seki N. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- 63.Peng XY, Won JH, Rutherford T, Fujii T, Zelterman D, Pizzorno G, Sapi E, Leavitt J, Kacinski B, Crystal R. The use of the L-plastin promoter for adenoviral-mediated, tumor-specific gene expression in ovarian and bladder cancer cell lines. Cancer Res. 2001;61:4405–4413. [PubMed] [Google Scholar]
- 64.Chung I, Deisseroth AB. Recombinant adenoviral vector containing tumor-specific L-plastin promoter fused to cytosine deaminase gene as a transcription unit: generation and functional test. Arch Pharm Res. 2004;27:633–639. doi: 10.1007/BF02980163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of osteoclastogenic factors in MDA-MB-231 conditioned medium. (A) MDA-MB-231 CM-derived osteoclastogenic proteins were immunoprecipitated by IgG, separated on a gradient 7%-15% SDS-PAGE gel, divided into 10 sections, and digested with trypsin. Mass spectrometry identified PRDX4 (validated in the previous study) and plastin as highly expressed candidates. This panel is identical to that published as Figure 1C in Rafiei et al, Cancer Letters, 2015 Jun 1;361(2):262-70 (reused with permission). (B) Antibody validation for plastin isoforms. Recombinant human plastin-1, 2, and 3 (rhPL I, L, T) were immunoblotted with antibodies for I, L, and T-plastin (upper gels, dashed line: predicted MW of plastins); equal loading was visualized by Ponceau staining (lower gels).
Generation of L-plastin targeting shRNA 4T1 clones. 4T1 cells were transfected with shRNA targeting L-plastin with two independent shRNA expression vectors (KD1 and KD2), or an empty vector (LMP), and stable clones were generated. (A) Expression of Lcp1 in parental 4T1 cells and shRNA cells harboring control vector (LMP) or shRNA for L-plastin (KD1 and KD2) was assessed and normalized to beta-actin (TaqMan: Lcp1: Mn00786153_s1, Actb: Mn00607938_s1. (B) Expression of L-plastin in cell lysates (top) and conditioned medium (bottom) was assessed by immunoblotting. (C) RAW 264.7 cells were cultured untreated (RAW) or primed with RANKL (50 ng/ml) for 2 days and then cultured for additional 2 days without treatment (negative control, NC), with RANKL (50 ng/ml, PC), or with 10% CM with from parental 4T1 cells (4T1), shRNA control clone LMP, or shLCP1 clones KD1 and KD2. Average numbers (left) and representative images (right) of osteoclasts formed in indicated conditions. Scale bar of 50 μm applies to all images. Data are means ± SEM, N = 6; *P < .05, **P < .01 compared to negative control, #P < .05, ##P < .01 compared to 4T1, assessed by Student's t test.
RAW 264.7 cells were primed with RANKL (50 ng/ml) for 2 days and then cultured for an additional 2 days without RANKL treatment (negative control, NC); with RANKL (50 ng/ml, positive control, PC); or with human recombinant plastin-1 (rIPL), 2 (rLPL), and 3 (rTPL) at 10 μg/ml. (A) Average osteoclast planar area. (B) Average number of nuclei per osteoclast. Data are means ± SEM, N= 30-55 cells, *P < .05, **P < .01 compared to NC assessed by Student’s t test.
Differential Expression of LCP1 and PRDX4 in Normal Tissue and Primary Tumours for Different Cancer Types.
LCP1 and PRDX4 mRNA expression data were obtained from the TCGA database, and standardized mean differences (SMDs) between normal tissue and primary tumor expression were estimated along with corresponding standard errors (SEs) and 95% confidence intervals (loCI: lower limit, hiCI: upper limit). Additionally, random effects estimate of overall pooled SMD across all cancer types was determined. Nnorm: normal tissue sample size, Nprim: primary tumor site sample size. Red: significant upregulation, blue: significant downregulation, gray: nonsignificant difference.