Significance
Many microbial communities of ecological and medical importance reside in complex and heterogeneous environments, such as soils or intestinal tracts. While many studies consider the effects of flow or chemical gradients in structuring these communities, how the physical structure of the environment shapes community dynamics and outcomes remains poorly understood. Using simulations of competitive microbial communities, we show that stability and dynamics qualitatively shift in environments with complex surface structures compared with open isotropic environments. Therefore, in addition to biochemical interactions between species, our work suggests that the physical structure of the environment is an equally important determinant of dynamics and stability in microbial communities, in a manner dependent on the specific patterns of interactions within that community.
Keywords: interaction networks, anisotropic environments, microbial communities, reaction−diffusion models, microbial ecology
Abstract
The dynamics and stability of ecological communities are intimately linked with the specific interactions—like cooperation or predation—between constituent species. In microbial communities, like those found in soils or the mammalian gut, physical anisotropies produced by fluid flow and chemical gradients impact community structure and ecological dynamics, even in structurally isotropic environments. Although natural communities existing in physically unstructured environments are rare, the role of environmental structure in determining community dynamics and stability remains poorly studied. To address this gap, we used modified Lotka−Volterra simulations of competitive microbial communities to characterize the effects of surface structure on community dynamics. We find that environmental structure has profound effects on communities, in a manner dependent on the specific pattern of interactions between community members. For two mutually competing species, eventual extinction of one competitor is effectively guaranteed in isotropic environments. However, addition of environmental structure enables long-term coexistence of both species via local “pinning” of competition interfaces, even when one species has a significant competitive advantage. In contrast, while three species competing in an intransitive loop (as in a game of rock−paper−scissors) coexist stably in isotropic environments, structural anisotropy disrupts the spatial patterns on which coexistence depends, causing chaotic population fluctuations and subsequent extinction cascades. These results indicate that the stability of microbial communities strongly depends on the structural environment in which they reside. Therefore, a more complete ecological understanding, including effective manipulation and interventions in natural communities of interest, must account for the physical structure of the environment.
From the scale of large metazoans down to microbes, natural environments are replete with multispecies communities that compete for resources and space, and, in many cases, actively predate other species within their environment. Within complex ecosystems, the topology and type of interactions between constituent species are thought to be primary determinants of ecosystem dynamics and stability. Typical pairwise interactions, like competition, cooperation, or predation, form the building blocks for constructing multispecies interactions and can be used to predict dynamics and stability in “well-mixed” environments where spatial distributions are uniform (1, 2). Interaction topology plays a particularly important role in species coexistence. For instance, in three-species intransitive competition (as in the classic rock−paper−scissors game), extinction of any species results in extinction cascades that favor dominance of a single species. Microbial systems present a particularly salient manifestation of these concepts, not only because complex communities of microbes are found in a wide array of industrial- and health-relevant environments, like soils and the mammalian gut, but also because the ability to genetically recapitulate and manipulate specific pairwise interactions biochemically makes microbial systems particularly well suited for testing our understanding of fundamental mechanisms underlying ecosystem dynamics.
Characterization of interactions within ecological networks, and their corresponding biochemical mechanisms, often focuses on microbial communities in which the spatial distribution of actors can significantly impact the type and magnitude of those interactions, and the resulting population dynamics. For example, spatially localized clonal domains that result from competition between mutually killing isolates of Vibrio cholerae may facilitate emergence of cooperative behaviors like public good secretion (3). Large clonal domains stabilized three-way intransitive competition within a consortium of Escherichia coli strains (4), which was unstable in well-mixed environments; similar mechanisms were found to stabilize competing cancer cell lineages when comparing spatial vs. nonspatial models of tumorigenesis (5). Reversing the causative arrow, ecological interactions can also dictate spatial arrangements of genotypes: In simulated three-species intransitive consortia with mobile individuals, lack of a single dominant competitor leads to population waves that continually migrate throughout the environment (6), thereby ensuring dynamic and long-term stability in species representation. Conversely, in competition between two mutual killers, coarsening of clonal domains guarantees the eventual extinction of one of the species (3), unless additional interaction mechanisms are present (7). Therefore, in contrast to dynamics that play out in well-mixed environments, it is clear that the spatial distribution of organisms is an important determinant of community dynamics and long-term ecological outcomes.
A common condition imposed on simulations of spatially explicit ecological systems is environmental isotropy, defined by the system having the same chemical and physical properties in all directions [for example, a homogeneous 2D plane (3, 6, 8)]. While such simplifications are essential in building fundamental understanding of system dynamics, they do not reflect salient environmental anisotropies found in most natural systems, such as chemical gradients, fluid flow, and complex surfaces. Despite relevance to natural communities, examination of the mechanisms by which environmental anisotropies affect ecological communities is sparse. In single-species populations, colonizing complex environments can result in drastic changes in spatial distributions. For instance, using microfluidic devices, Drescher et al. (9) showed that surface morphology and fluid shear forces interact to drive formation of novel biofilm structures in Pseudomonas aeruginosa, while, in V. cholera, cell adhesiveness and fluid flow interact to dictate the cluster size of competing genotypes (10). Biofilm formation can also disrupt fluid flow in a microfluidic mimic of soil environments, which, in turn, allows for coexistence of competing cheater and cooperator phenotypes of P. aeruginosa that are otherwise unstable under well-mixed conditions (11); similar outcomes were observed for competing cooperative and cheating lineages of E. coli in heterogeneous nutrient landscapes (12). Complex environments may interact with bacterial social behaviors as well: When colonizing maze-like microfluidic environments, chemoattractant production by E. coli caused heterogeneous population structures to form due to concentration of such attractants in the “dead ends” of the maze (13). Importantly, these perturbations to population structure are commensurate with length scales at which mixing occurs for in vivo communities such as the mammalian (14, 15) and fish (16) guts, or in dental plaque (17). Theoretical investigations indicate that similar environmental perturbations are likely to affect multispecies communities; for example, turbulent flow (18) or directional motility (19) can disrupt spatial patterning of intransitive three-species communities and thus increase the risk of extinction cascades, while graph-theoretic approaches suggest that random perturbations to spatial lattices result in similar community destabilization (20). Together, these results suggest not only that spatial distributions of organisms influence ecological dynamics but that the magnitude of these effects depends strongly on the specific nature of anisotropies within the environment.
In this work, we systematically characterized the effects of structural anisotropy on multispecies population dynamics and spatial distributions within in silico ecological communities. The structural attributes of these simulations are intended to capture the primary spatial structure found in natural environments, like the packing of steric soil particles or the contents and epithelial structure of the mammalian gut. Using reaction−diffusion models, we simulated asymmetrically competing two-species and intransitively competing three-species ecological networks in the presence of steric barriers arranged in a lattice within the environment. These networks and the corresponding simulations were chosen for direct comparison with previous work (3, 6) that provides clear expectations for spatial distributions and community dynamics in homogeneous environments, and which we discuss within Results. We find that the addition of environmental structure fundamentally alters community dynamics in both two- and three-species competitive systems. In the two-species case, coarsening of genetic domains that would otherwise lead to extinction of one competitor is arrested due to “pinning” of competition interfaces between barriers, resulting in long-term coexistence of both species. This effect is strongly linked to the geometry of the steric barriers, and is robust to asymmetry in competitive fitness. For intransitive three-species competition, steric barriers cause interference between traveling population waves, inducing chaotic fluctuations in the abundances and spatial distributions of species and a concomitant increase in the probability of extinction cascades. Our results affirm that the trajectories, stability, and spatial structure of ecological communities are drastically altered by the arrangement and length scale of structural perturbations in the environment.
Results
Competition Model.
We model interspecies interactions using a modified version of the Lotka−Volterra (LV) competition framework. In the classic LV model, interaction mechanisms and fitness differences are combined into a single parameter that characterizes competition as an effective carrying capacity for the focal species relative to the density of a competitor; hence there is no differentiation between, e.g., competition for resources and toxin-mediated killing. Here, we extend the classic LV framework to reflect “active competition,” where passive competition for space and nutrients (affecting carrying capacity) is decoupled from active competition mechanisms that directly impact growth rate, such as Type VI secretion system mediated killing or bacteriocin production (21, 22), giving the system of partial differential equations (PDEs)
Here Ai is the local concentration of focal species i, Ak is the concentration of the active competitor species for Ai, and the sum is over all species passively competing for space and nutrients. The primary dispersal mechanism is through diffusion characterized by D, basal growth rate is given by r, and carrying capacity for each species is given by Cj. Active competition is characterized by the concentration parameter Pki, which is the concentration of Ak above which Ai is killed, and hence lower values of Pki indicate more potent active competition (i.e., lower concentrations of the active competitor are required to cause death). This framework has the capacity to model passive fitness differences (through species-specific values of Cj) and anticompetitor mechanisms (through Pki), thereby capturing two basal and distinct mechanisms of microbial competition. This model is appropriate for describing diffusively mediated local competitive interactions, like Type VI secretion contact-mediated killing or local killing by secreted toxins such as colicins (23). Additional PDEs would be required to describe highly motile cells, exogenous chemical gradients, or the production, potency, transport, and decay of rapidly diffusing secreted toxins. This set of PDEs establishes a baseline set of assumptions and corresponding phenomena from which to build more complex models.
In this work, we focus specifically on the competitive effects, and assume equal basal growth rates r, diffusion D, and carrying capacities C for all species in the community. This simplification allows the population density to be scaled by carrying capacity and the time to be scaled by the growth rate, which reduces the parameter space of the model, leaving the dimensionless version of Pki (i.e., Pki/C) as the single free parameter that dictates the strength of active interspecies competition,
Here time is in units of r−1, length is in units of , and organismal concentrations Ai (i.e., number per unit area) are in units of C; therefore . The natural length scale is proportional to the root-mean-squared distance an organism will move over a single doubling time. Finally, we employed a minimum concentration cutoff Amin, such that, if Ai < Amin, then Ai → 0. This is meant to approximate the fact that arbitrarily small organismal concentrations—compared with the carrying capacity—do not reflect the reality that single discrete organisms correspond to finite levels of organismal concentration.
We used this nondimensionalized model to simulate communities in a 2D environment into which we introduce structural anisotropy via a lattice of steric pillars (Fig. 1, and see Fig. 3). Like a grain in soil or tissue in a gut, these pillars do not allow free transport through them, nor allow microbes to occupy them; their perimeter is a reflecting boundary condition. Structural perturbations were explored by introducing a triangular lattice of steric circular pillars, with each lattice fully characterized by the radii of the pillars R and the center-to-center spacing of the pillars Δx, with each simulation evolving in a square domain of side length L. These parameters (pillar radius R, pillar spacing Δx, and simulation size L) are reported in units of λ. We then characterized the impact of these perturbations on the spatial distribution and dynamics of in silico communities across structural length scales by monitoring the distributions and abundances of resident community members as we varied the radius and density of pillars within the simulation environment.
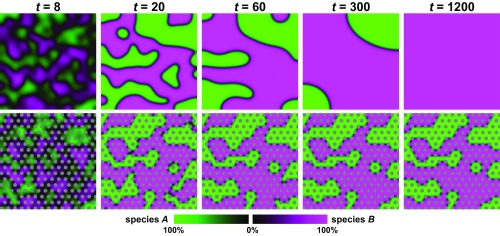
Fig. 1.
Structurally anisotropic environments arrest genetic phase separation in two-species systems, resulting in long-term coexistence. Panels depict snapshots from simulations of two-species competition in structurally isotropic (Top) and anisotropic (Bottom) environments, with color intensity reflecting species abundance, and pillars shown in gray. Time is measured in doubling times. Under isotropic conditions, domain coarsening robustly leads to extinction of one of the species. Anisotropic environments, however, allow for local pinning of competition interfaces, resulting in arrest of domain coarsening and thereby sustained coexistence. Simulation parameters are L/(1.29 λ) = 100, P = 0.1, and Amin = 0.001, with R/(1.29 λ) = 2 and Δx = 3.5 R for the anisotropic case.
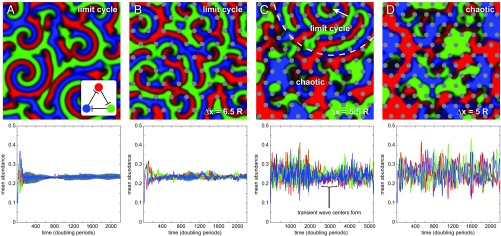
Fig. 3.
Structurally anisotropic environments disrupt spatial patterns and cyclic dynamics in intransitively competing three-species communities. (Top) Snapshots of spatial distributions from representative simulations, with (Bottom) corresponding abundance dynamics. (A) Following a brief “grow-in” period, isotropic conditions result in spiral waves and cyclic abundance dynamics with corresponding stable coexistence. The Inset illustrates the intransitive competitive network topology, with flat-headed arrows indicating predation. (B) Introducing pillars spaced far apart compared with the natural length scale of the system slightly perturbs wave center formation and dynamics, but the system still robustly exhibits spiral waves and stable coexistence. (C) Moving pillars closer disrupts cyclic pattern formation, leading to irregular spatial distributions and large fluctuations in species abundance. However, in this example, transient wave centers form (white arrow) and produce locally stable cyclic dynamics, indicated by the dashed boundary in Top. Note the longer time scale on this plot. (D) More densely packed pillars hinder transition to a limit cycle, resulting in sustained large fluctuations in abundance and irregular species distributions that exhibit extreme sensitivity to perturbations and corresponding chaotic dynamics. See SI Appendix for demonstration of positive Lyapunov exponents. Movies S7–S10 correspond to A through D, respectively. Simulation parameters are L/(1.29 λ) = 150, P = 0.1, Amin = 0, and, for simulations including pillars, R/(1.29 λ) = 3 and Δx as indicated.
Competition Between Two Mutual Killers.
Structured environments arrest genetic phase separation.
For an actively competing two-species community in an isotropic environment, recent theoretical and experimental work indicates that species phase separate according to genotype, with the eventual extinction of one species via domain coarsening (3). Our model produces results consistent with those findings, but, in contrast, we find that, when spatial structure is introduced into the environment, genetic phase separation is arrested, resulting in stable coexistence of mutually killing genotypes (Fig. 1 and Movie S1). Arrest occurs by pinning of competition interfaces between steric barriers (i.e., pillars). In both isotropic and anisotropic environments, coarsening of genetic domains is driven by the curvature of competition interfaces. If competition is symmetric, a flat interface will not move, whereas a curved interface will translate toward the center of the circumscribing circle. In isotropic conditions, stable interfaces are the exception, only found in the rare case where a single flat interface bisects the entire environment, which is itself increasingly unlikely in larger environments. Thus, all domains enclosed by a competitor will eventually be consumed, and one of the competitors will go extinct. In contrast, we find that flat competition interfaces are stabilized between steric barriers, resulting in the arrest of domain coarsening and subsequent long-term coexistence of both species (Fig. 1). Importantly, for symmetric competition, we observed that the size and/or density of pillars had little effect on community stabilization (left edge of Fig. 2A), suggesting that, for well-matched competitors, even slight structural perturbations that allow for interface pinning may be sufficient to foster coexistence.
Fig. 2.
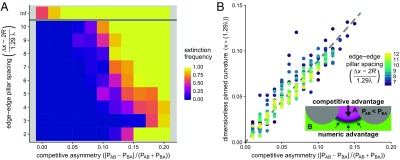
Coexistence of asymmetric competitors is maintained by stabilizing curved competition interfaces. (A) Extinction frequency from 30 replicate simulations per coordinate over 2,000 doubling times as a function of competition asymmetry and pillar spacing. Here, the ratio Δx/R is held constant at 3, while varying the lattice constant Δx relative to the natural length scale λ. Data for isotropic environmental conditions (no pillars) are depicted above the gray line; note that some isotropic simulations were not observed to go extinct, due to insufficient simulation duration; with more time, all isotropic simulations would go extinct. Higher pillar density stabilizes coexistence between strains with larger competitive asymmetries, up to the point at which the environment cannot sustain sufficiently large domains to stabilize the competition interface; this produces the increase in extinction frequency at bottom right (Movie S2). Simulation parameters are L/(1.29 λ) = 100, Amin = 0.001, and the average competition strength (PAB + PBA)/2 is held constant at 0.1. (B) Stable interface curvature () as a function of competitive asymmetry (SI Appendix). Edge-to-edge pillar spacing is indicated by point color. Pinning and stable coexistence was observed for competitive asymmetries greater than 0.15 (in A), but were omitted in B because pillar spacings were too small for reliable curvature estimation. Inset schematically depicts stable curved interfaces, where the numerical advantage of the weaker competitor species (green) balances the advantage of the stronger competitor species (magenta). See Methods for description of curvature calculation.
Pinning of genetic domain interfaces is robust to asymmetric competition.
When one species is a more potent competitor (e.g., PAB > PBA), even the symmetry of an environment fully bisected by a linear competition interface will result in extinction of the weaker competitor. While flat interfaces balance symmetric competition, they are not stable when one species has a competitive advantage, and instead will translate through space. Likewise, when competition is asymmetric in an isotropic environment, over an ensemble of random initial conditions, the dominant competitor will drive the weaker competitor to extinction in the overwhelming majority of cases. We wanted to know whether structural perturbations could stabilize coexistence even when competition was asymmetric. Thus, we performed simulations identical to those described above, but varied the difference in the competition parameters, |PAB − PBA|, while holding their mean constant. We observed that stable coexistence via interface pinning was robust to asymmetric competition within certain regimes of the lattice parameters (Fig. 2 and Movie S2). The mechanism, however, was somewhat counterintuitive: For a given degree of competition asymmetry, |PAB − PBA|, there exists some critical interface curvature that balances the numeric advantage of the weaker species against the competitive advantage of the more potent species (Fig. 2B, Inset). This is true regardless of the presence of environmental structure; however, in isotropic conditions, this competitive equilibrium is unstable, and any perturbation of domain curvature from the critical value will result in interface translation and eventual extinction. We found that structural perturbations stabilize the equilibrium created by curved competitive interfaces if the spatial structure of the environment can support the critical curvature between two steric surfaces (SI Appendix); only then will phase separation halt and coexistence be maintained. Otherwise, the dominant competitor will drive the weaker species to extinction (Fig. 2A), albeit with slower dynamics than isotropic conditions.
Unlike symmetric competition, where coexistence is fully determined by flat competition interfaces, the curved interfaces required to stabilize asymmetric competition also impose a minimum domain size on the competitively disadvantaged species that depends on the lattice parameters. This is because a sufficiently large population of weak competitors is required to compensate for competitive losses at the interface through growth and diffusion (note the increased levels of extinction with the smallest pillar spacings in Fig. 2A, and the dissolution of domains in Movie S2 that were stable under the symmetric competition of Movie S1). Furthermore, using geometric and scaling arguments (SI Appendix), we predicted that the critical curvature should be, to leading order, a linear function of the competitive asymmetry and confirmed this with our simulations (Fig. 2B).
Finally, we speculated that, because the stabilization of asymmetric competition was largely governed by interactions between pillar geometry and curved competition interfaces, structurally induced coexistence of competitors should be a general feature of models with sufficiently sharp competition interfaces. We tested this by simulating a classic competitive LV model with structural perturbations across the same range of competitive asymmetry and pillar spacing as Fig. 2A. We found that pinning and coexistence were robust features in the classic LV framework and generated quantitatively similar results to our modified framework from Fig. 2A (SI Appendix, Figs. S7 and S8).
Three Species Intransitive Competition.
Environmental structure disrupts three-species dynamics.
Previous in silico simulations of an intransitively competing three-species network (i.e., displaying a cyclic competitive hierarchy, as in the game rock−paper−scissors) within an isotropic environment resulted in the formation of striking spiral wave patterns, in which dense waves of species constantly migrate throughout the environment, with each species wave chasing its prey and being followed by its predator (see ref. 6, and recapitulated in our model in Fig. 3A). Despite constant flux of species at small length scales, the community robustly exhibited stable coexistence of all three species on ecological time scales (more than 104 generations) when provided with a sufficiently large environment relative to the natural length scale set by diffusion and growth. These findings agree with the earlier experimental results of Kerr et al. (4), albeit at different time and length scales. However, it should be noted that previous theoretical work indicates that fluctuations (24) or finite number effects (25) can force such systems into heteroclinic cycles that eventually lead to extinction cascades.
Given the drastic changes in ecological outcomes when structural perturbations were introduced in two-species competitive systems, we wanted to characterize how dynamics and outcomes changed in three-species competition when we included structural perturbations. We performed simulations using the same set of governing equations as in the two-species case, now accounting for the topology of a cyclic competitive hierarchy and imposing fully symmetric competition for simplicity (i.e., all Pki set equal). We found that the introduction of spatial structure into the environment significantly destabilizes wave patterns observed under isotropic conditions in a manner that strongly depends on the spacing and size of steric barriers and the value of the minimum concentration Amin. For example, while densely packed barriers prevent regular pattern formation and result in erratic fluctuations in species abundance (Fig. 3 C and D), increasing the space between pillars allows the system to reestablish wave patterns that dominate the environment and significantly reduce the magnitude of population fluctuations (Fig. 3B). Thus, whereas structure in the two-species competitive model promotes coexistence and long-term stability that is otherwise absent in isotropic conditions, three-species intransitive competition displays the opposite qualitative trend: Adding structure reduces coexistence and promotes extinction cascades. We therefore set out to characterize the complex dynamics arising from intransitive competition in structured environments, with special attention paid to transitions in population dynamics as a function of quantitative changes in environmental structure.
Introducing structural anisotropy leads to chaotic fluctuations in species abundance and extinction cascades.
To quantify how structural perturbations destabilize pattern formation and cyclic dynamics in our deterministic simulations, we examined the dynamic trajectories of multiple replicates of the same steric pillar array initialized with controlled, random differences in the initial distributions of the three species. We then compared the correlations in species distributions between replicate simulations as the system evolved. In contrast to limit cycle dynamics in isotropic environments, we found that increasing pillar density resulted in extreme sensitivity to perturbations of initial conditions, a hallmark of chaotic dynamics (26), with an exponential decay in pairwise replicate correlations through time (Fig. 4). Chaotic fluctuations were accompanied by transitions into extinction cascades (evident in Fig. 4, where correlation traces are truncated at the first extinction event among replicates), and we found that higher values of Amin favored faster transitions into extinction cascades; for all subsequent analyses, we used Amin = 0.01 to make these large-scale computations tractable. In the SI Appendix, we further examine the difference between limit cycle and chaotic behaviors, demonstrating that the latter has positive Lyapunov exponents. We also performed demonstrative simulations of intransitive three-species competition in a typical LV model, and found that, even in the classic LV framework, introducing environmental structure could elicit transitions from limit cycle to chaotic dynamics as pillar density increased (SI Appendix, Fig. S6).
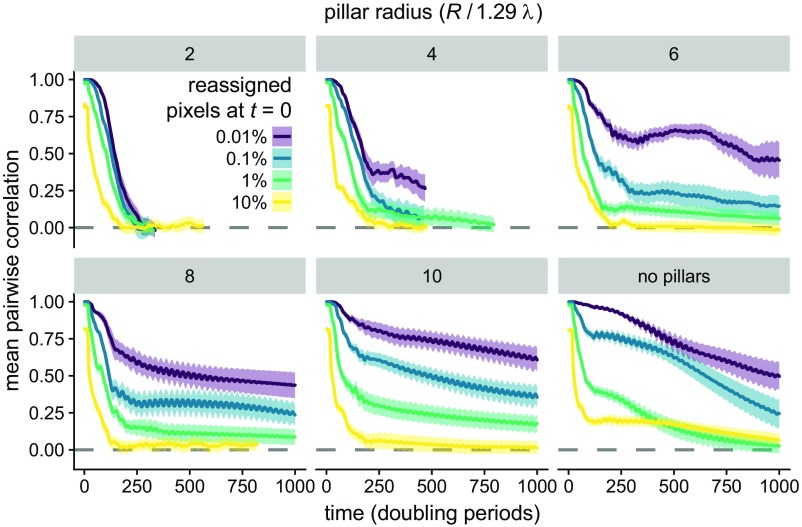
Fig. 4.
Ecological dynamics display extreme sensitivity to initial conditions depending on environmental structure. In each panel, 10 replicate simulations were identically initialized, and then a small percentage of grid locations in each replicate (indicated by line color) were randomly resampled, creating correlated initial conditions. Each panel shows the spatial (pixel-by-pixel) correlation over time between replicate simulations, averaged over all 45 unique pairwise comparisons. Shaded regions indicate SEM. As the pillar array becomes denser, rapid decorrelation among replicates results from minute perturbations to initial conditions, a hallmark of chaotic dynamics. In contrast, as pillar spacing increases, some fraction of simulations fall into a limit cycle and thus have nonzero correlations with similar initial conditions. Simulation parameters are L/(1.29 λ) = 100, P = 0.1, Amin = 0.01, and Δx = 4 R, with pillar size R indicated at the top of each panel.
In initial simulations, we noted that species distributions often exhibited dynamic transitions between patterns of spiral waves and chaotic fluctuations (Fig. 3C), and thus we sought to characterize overall system dynamics as a function of environmental structure. We performed simulations with uncorrelated initial conditions across a range of pillar sizes and spacings, and classified system dynamics as “limit cycle” or “chaotic” by calculating the temporal autocorrelation of the species distribution in space. If the spatiotemporal autocorrelation of all three species (minus steric barriers) at time t reached an autocorrelation value above a threshold value of 0.8 two or more times after t, we defined the dynamic state as cyclic at time t (Methods and SI Appendix, Fig. S12). With this definition, we classified the dynamics as a function of R and Δx into pseudophase diagrams for fraction of time spent in cyclic dynamics (Fig. 5A) and the extinction frequency over the simulation time scale (Fig. 5B). Example simulations are provided in Movies S3–S6. We found that smaller and more densely packed pillars lead to greater destabilization, with less time spent in limit cycle dynamics and higher rates of extinction. Intriguingly, however, with the smallest and most densely packed pillar structures, we observed a reduced extinction frequency, reversing the trend seen at larger pillar spacings (Fig. 5B, bottom). This appears to be specific to the mechanisms by which pillars destabilize the system. With large pillars and spacings, spiral waves develop in open areas and are largely unperturbed by the pillars, resulting in cyclic behavior and few extinctions (Movie S3). As pillar spacing decreases, open areas narrow to the point that spiral wave centers are destabilized, migrating erratically and eventually collapsing due to interference from other wave fronts (Movie S4). With smaller pillar radii, the pillars themselves often act as wave centers, and appear to be particularly vulnerable to disruption via interference (Movie S5). However, when small pillars are so densely packed that a pillar cannot serve as a wave center, the centers again migrate erratically between pillars, but the pillar density is high enough to “cage” the rapidly diffusing wave centers and prolong their existence in a chaotically fluctuating state (Movie S6). Thus, the prevalence of extinction cascades is a nonmonotonic function of pillar density, suggesting that intermediate scales of spatial structure produce the strongest destabilizing effects on intransitive communities. Finally, to ensure that the observed changes to system dynamics and corresponding destabilizing effects were not dependent on the symmetry of a triangular lattice, we performed a subset of simulations where pillar radii or spacing were independently and randomly perturbed, and no significant changes to system dynamics and ecological outcomes were observed (SI Appendix, Fig. S13).
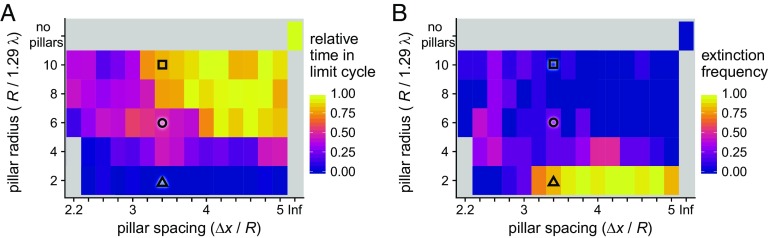
Fig. 5.
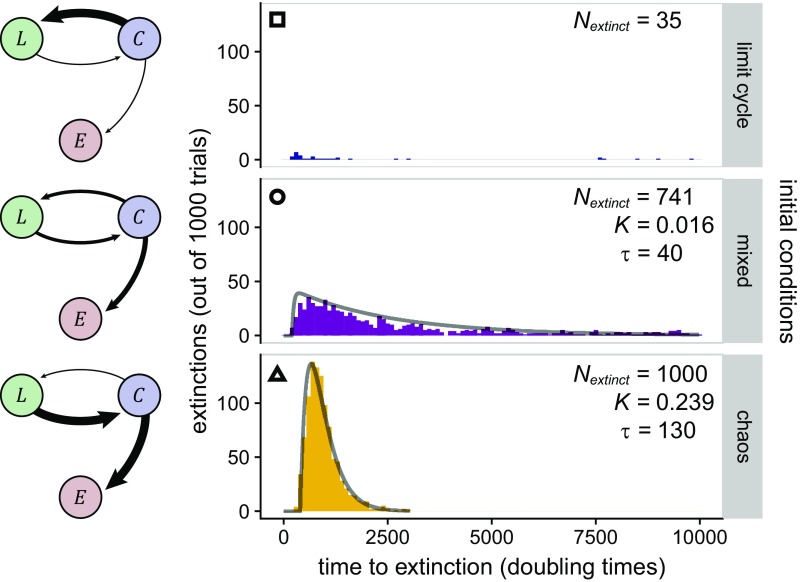
Severity of chaotic disruptions depend on structural characteristics of the environment. Over 1,000 doubling times, A shows the fraction of time the system displayed cyclic dynamics, and B shows extinction frequency, both as functions of the size and spacing of the pillar array. Ten simulations were performed for each grid point. For each simulation, the amount of time spent in a limit cycle was normalized by the average time that isotropic simulations were classified as cyclic; this adjusts for systematically acyclic periods such as the grow-in phase. Three primary dynamic regimes were identified: (i) stable cycles, with larger and widely spaced pillars (square symbol); (ii) a transitory region at intermediate pillar size and spacing, where communities tend to either relax into a limit cycle or collapse (circle); and, (iii) when pillars are small and densely packed, unstable chaos with rapid community collapse (triangle). Simulation parameters are L/(1.29 λ) = 100, P = 0.1, and Amin = 0.01, with pillar size and spacing as indicated in the figure. The simulations at R/(1.29 λ) = 2 and 4 with Δx/R = 2.2 were omitted because the pillar spacing did not allow for accurate simulation of diffusion. Black symbols correspond to simulation conditions whose extinction time distributions were analyzed in Fig. 6.
A three-state kinetic model describes coupling of dynamic transitions and extinction.
In our three-species simulations, we observed transitions from chaotic dynamics to limit cycles and back again, with many simulations ultimately making the transition from chaotic dynamics to the fully absorbing state of extinction. Although the simulations are deterministic, the ensemble of random initial conditions creates statistical variability in system dynamics. Thus, we wanted to characterize how the distribution of extinction times, and hence the time scale of coexistence, depended on environmental structure. We developed a three-state kinetic model to describe transitions between chaotic (C), limit cycle (L), and extinct (E) states, using three positive rate parameters to connect the states (kCL, kLC, and kCE). The closed-form solution to our model (SI Appendix) predicts that any system whose structural perturbations result in kLC > 0 and kCE > 0 will go extinct given enough time, which is consistent with previous work (24, 25). It also predicts that the rates of arrival to the extinct state depend on the dynamics fostered by the environmental structure. To test this, we took structural conditions whose initial dynamics were classified as either limit cycle, chaotic, or mixed for the first 1,000 doubling times (marked tiles in Fig. 5), and used the observed distribution of extinction times as a function of environmental structure to fit extinction probability densities predicted by the model over a period of 10,000 doubling times. We found that our model recapitulated observed distributions of extinction times (Fig. 6), and that, indeed, changes in environmental structure had significant effects on the distribution of extinction times. These results indicate that structurally induced destabilization results from a combination of decreased rates of transition from chaotic fluctuations to limit cycle dynamics and/or increased rates of transition from chaotic dynamics to extinction (see model diagrams in Fig. 6). Accordingly, systems that remained largely in a limit cycle had slower rates of extinction. The fitted model parameters were functions of multiple individual transition rates with complex mappings (SI Appendix, Fig. S10); hence direct inference of the effects of structural perturbations on individual transition rates (e.g., from limit cycle to chaos) was not possible with this model.
Fig. 6.
A kinetic model of dynamic transitions describes extinction time distributions for a range of environmental structures. From the lattice structures indicated by overlaid symbols in Fig. 5 (indicated here at top left in histogram plots), we performed 1,000 replicate simulations for 10,000 doubling times to measure the distribution of extinction times and compare them to our model. These conditions typify the three observed dynamic regimes [(Top) limit cycle, (Middle) transitory, and (Bottom) chaotic], and map to a three-state model of system dynamics with two correlated rate parameters that depend on structural characteristics (in this case, pillar grid parameters; see SI Appendix). The histograms were constructed from observed extinction times, and gray lines are fits to probability distributions predicted from the three-state model. Fitting was not attempted for the cyclic case (Top), as only 3.5% of simulations were observed to go extinct over the simulation period. The number of extinctions and (where applicable) the fit parameters are shown within the corresponding plots. (Left), Connections between the dynamical states of limit cycle (L), chaotic (C), and extinction (E) are depicted, with relative rates qualitatively indicated by the width of the arrows. Simulation parameters are L/(1.29 λ) = 100 and P = 0.1, with pillar size and spacing as indicated in Fig. 5.
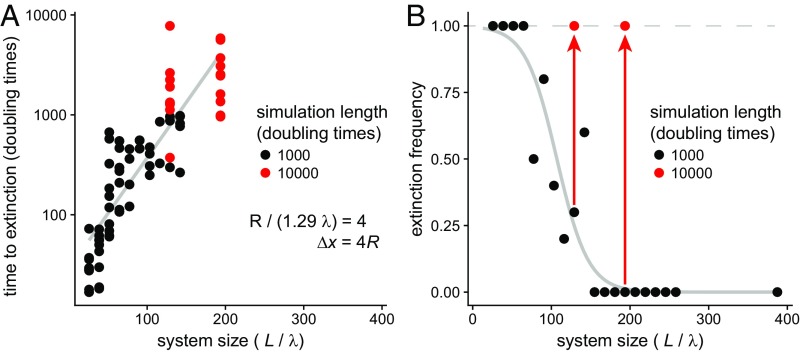
Larger systems prolong species coexistence despite chaotic fluctuations.
Lastly, we sought to characterize the effect of system size on community stability. Holding the structure of the pillar array constant, we observed that the mean time to an extinction cascade increased approximately exponentially with increasing system size (Fig. 7A). This suggests that, with sufficiently large systems relative to the natural length scale, communities can coexist for long periods despite continual chaotic fluctuations in individual species abundances and distributions. However, consistent with the predictions of our kinetic model (Fig. 6), larger systems cannot fully prevent extinctions, as evidenced by observed extinction frequencies when simulation times were extended. In Fig. 7B, we demonstrate this effect, showing that, for a given simulation duration, there is a system size above which the extinction frequency drops to nearly zero, but, by simply extending the simulation time, the extinction frequency increases to unity.
Fig. 7.
Larger system sizes delay but do not prevent extinction cascades. (A) Plotting time to extinction as a function of system size reveals an approximately exponential relationship, suggesting that larger systems can persist in a state of chaotic fluctuation for longer time periods. Each point is a single simulation, with 10 replicate simulations per system size L. (B) Extinction frequency as a function of system size. The black points and fitted gray logistic regression curve suggest a critical system size at which extinctions are no longer observed for the fixed number of simulated doubling times (here, 1,000). However, by increasing simulation duration, the observed extinction frequency saturates to ∼1 (red points and arrows), indicating that large systems delay but do not prevent extinction, consistent with our model, which predicts that all anisotropic environments will eventually end with an extinction cascade. Simulation parameters are R/(1.29 λ) = 4, Δx = 4 R, P = 0.1, and Amin = 0.01, with system size L as indicated in the figure.
Discussion
Using in silico simulations of ecological communities, we found that adding structural complexity to the environment results in fundamental changes to community dynamics and outcomes in a manner dependent on the interaction network topology. Specifically, we observed that, for two mutually competitive species, structured environments allowed for long-term coexistence between species with relatively large differences in competitive fitness, an outcome impossible in well-mixed or isotropic environments. Conversely, for a three-species intransitively competing community, which is expected to be stable under isotropic conditions (6), we found that environmental structure can disrupt the dynamic spatial patterns that stabilize these communities, resulting in chaotic fluctuations in species abundances and spatial distributions, and an increased frequency of extinction cascades. Together, these findings strongly suggest that the physical structure of the environment can interact significantly with the specific nature of interspecies interactions within resident communities to affect stability and dynamics, and more generally indicate that physical attributes of the environment must be considered when assessing the stability of resident communities.
Our results extend established findings that spatially structured communities maintain biodiversity by localizing interactions among community members (8, 27, 28). In particular, in the context of simple competition, the spatial bottlenecks that structurally complex environments provide impede competitive mechanisms to the point that only a small fraction of a given population is engaged in active competition, and hence fitness differences become less important relative to geometric advantages provided by specific localization within the environment. However, our findings also suggest that intransitive interaction networks are not a robust means of stabilizing communities, as has been theoretically postulated (29, 30). If deviation from isotropic conditions (which is found in virtually all natural environments) only serves to accelerate the frequency of extinction cascades within these networks, this work offers a mechanism as to why such networks are only rarely observed outside of the laboratory (31–34). More generally, developing experimental systems to directly assess the impact of spatial structure on microbial community dynamics will be essential in isolating and characterizing the mechanisms that stabilize or perturb communities. We suggest that derived systems, such as microfluidic environments, will be crucial in this respect, as finding suitable natural environments that vary in degree of spatial structuring and are not also confounded by factors like nutrient availability, phages, predation, or other physical anisotropies like fluid flow may be difficult. We note that the complementary approaches of Kerr et al. (4) and Kirkup and Riley (32), where the same experimental community was observed within in vitro and in vivo environments, respectively, may offer a possible alternative.
We speculate, based on scaling effects, that the increase in surface area-to-volume ratio going from 2D into 3D will enhance the stabilization of asymmetric competition between two species. Conversely, given the potential augmentation of structural complexity available in higher dimensions, we expect that, in dynamic intransitive communities, chaotic fluctuations would be a robust feature. We also expect that the shape of steric barriers could play a nontrivial role in ecosystem dynamics and stability; we chose circles for simplicity, as they are characterized by a single parameter (R). The spectrum of available interface curvatures within a particular environmental structure is a function of both overall spatial scale (e.g., here Δx) and the shape of the steric objects themselves. Rationally designed structures could therefore potentially be used to tune the range of competitive asymmetries and/or stochastic fluctuations that an environment can stably support, and to shift system dynamics and stability to favor particular species or interaction topologies.
Whether our findings are robust when placed in the context of other physical and ecological phenomena will meaningfully comment on whether principles gleaned through idealized contexts transfer to complex natural systems. For example, how robust are pinned competition interfaces to stochastic spatial fluctuations caused either by finite organism size or other forms of motility (besides diffusion), tunable interaction strengths, such as with competition sensing (23, 35), or phenotypic differentiation (36)? Are chaotic fluctuations a dominant dynamic state when cells can respond to chemical gradients via chemotaxis? What are the effects of physical structure on species distributions for larger networks, where specific interaction motifs are embedded within a more complex ecological context? These extensions will pave the way toward future theoretical work, as well as generating specific hypotheses to be tested experimentally. Conversely, phenomena that were salient in this work, in particular the pinning phenomena that slowed or halted genetic coarsening, play important roles in other physical and biological systems, including domain wall stabilization in Ising-like systems due to pinning at random spatial impurities (37), pinning-induced transitions of supercooled liquids into glassy states (38), and arrest of lipid bilayer domain coarsening in the presence of biopolymers that impose structure on the bilayer (39). Likewise, other physical mechanisms, such as flow (40), have been found to slow or halt coarsening in phase-separating systems, suggesting that modulation of system dynamics and stability due to complexities within the physical environment is not the exception but rather the rule.
Finally, we note that the reductionist approach we take here is valuable toward unraveling the multitude of forces acting on microbial communities in complex environments. While we focus specifically on environmental structure, and others give similar focus to flow (10, 41) and chemical gradients (42) in structuring communities, all of these environmental features are intimately linked and, in combination, will modulate impacts on communities in important ways (11). Building a bottom-up understanding of how various features interact to drive community processes is therefore essential in determining the primary forces acting on a community in a given environmental context, paving the way toward the ultimate goals of understanding basal mechanisms of ecosystem dynamics and of targeted and robust engineering of microbial communities.
Methods
Two-Species Mutual Killer Simulations.
Simulations were randomly seeded with pink noise (43) at an average density of 10% of the carrying capacity, with each species represented by its own field matrix. Pillars were placed in a triangular lattice with the specified radius and spacing. Microbial density that coincided with pillar locations was removed from the simulation. The bounding box and pillar edges were modeled as reflecting boundary conditions. At each simulated time step (Δt = 0.01t, with t in doubling times), populations diffused via a symmetric and conservative Gaussian convolution filter with standard deviation set by . After the diffusion step, changes in population density (growth and death) were calculated using the equations given in Competition Model, and used to update the density of each species according to a forward Euler scheme. In combination with the small dimensionless time step and concentration-conserving convolution filter, hard upper and lower bounds (1 and Amin in units of carrying capacity, respectively) were enforced on each species field to ensure numerical stability of simulations; population densities outside this range were set to 1 and 0, respectively. For each set of lattice constants and competitive asymmetry values, 30 independently initialized replicates were simulated for 2,000 doubling times. Mean population abundances and images of the simulation were recorded every doubling time for the duration of the simulation. Extinction was defined as the mean population density of either species dropping below a threshold value of , where R is the pillar radius and A is the area of lattice points not obstructed by pillars, to account for surviving populations “trapped” between a pillar and the corner of the simulation box and therefore not in contact with the rest of the simulation.
Calculation of Pinned Curvature.
To obtain higher resolution of pinned curvature in asymmetric competition, two pillars of R/(1.29 λ) = 10 were put at two opposing edges of a simulation box, and in contact with the simulation boundary, leaving a single gap between the pillars. Two competing species were symmetrically and uniformly inoculated at 30% of the carrying capacity on either side of this gap, leaving a single flat interface spanning the distance between the two pillars. Simulations were then allowed to evolve as above until dynamics ceased due to either pinning or extinction. All combinations of the indicated competitive asymmetries were sampled, and pillar gap distances were sampled by varying the size of the simulation box. For simulations where pinning was observed, the interface location was defined as the boundary points where species A and B were of equal abundance. The interface curvature was calculated from three points along that boundary (the midpoint and the two points in contact with the pillars); this method was found to be more robust than other circle-fitting methods, especially for low curvatures and narrow pillar gaps.
Intransitive Three-Species Simulations.
Three-species intransitive simulations were carried out similarly to the two-species cases described above, with the competition terms in the model modified to reflect the intransitive interaction network topology. Simulations were inoculated randomly with 10 replicate simulations per structural condition. Unless otherwise indicated, simulations were evolved for 1,000 doubling times, with images written every 0.4t.
A schematic of the classification of simulation dynamics is given in SI Appendix, Fig. S12. Spatiotemporal autocorrelations were calculated for each simulation, where correlations at each time point were calculated from the concatenated vectorized simulation matrix of all nonpillar grid locations for each species; e.g., for the autocorrelation matrix in SI Appendix, Fig. S12A, each matrix entry represents the correlation of two 438,000 (three species multiplied by 146,000 unique nonpillar grid locations) length vectors at the indicated time points. Using the autocorrelation matrix, at every time point (i.e., starting from the matrix diagonal and moving forward in time), that time point was classified as exhibiting limit cycle dynamics if the autocorrelation rose above the threshold value of 0.8 for at least two cycles. This threshold was chosen empirically as the level at which isotropic simulations were reliably classified as limit cycles (excluding grow-in periods and final time points for which future dynamics were not observed). Extinction events were calculated as in the two-species cases.
To establish correlated initial conditions (Fig. 4), the following procedure was used: For each replicate set of simulations, a random initial inoculum at a density of 10% of the carrying capacity was generated using the same random seed (i.e., constructing 10 identical initial condition matrices). Then, for each individual replicate, a randomly selected percentage (as indicated in Fig. 4) of nonpillar grid locations were randomly resampled between 0% and 10% of the carrying capacity. Simulations were then allowed to evolve as described above. At each time point, each unique pairwise correlation (45 for the 10 replicates used) between vectorized simulation matrices was calculated, and the mean over all pairwise correlations was used to generate Fig. 4. Correlation traces were truncated upon the first observed extinction event among the replicates.
Kinetic Modeling.
For details on assumptions and analysis of the kinetic state model, and derivation of the closed-form solutions for the time-to-extinction distributions, see SI Appendix. Histograms in Fig. 6 were generated from randomly initialized simulations as described above, with 1,000 replicates per set of lattice constants, each over 10,000 doubling times. Model parameters K and were fit by minimizing the squared error between the empirical cumulative distribution function (CDF) from simulated data and the corresponding CDF predicted by the model; global minima in the parameter space were found using grid search. A temporal offset parameter was also fit to account for grow-in periods, effectively shifting the histogram along the time axis and setting extinction probability for to zero.
Data Availability.
Code to run simulations and analyses is available in Datasets S1–S12.
Supplementary Material
Acknowledgments
We thank Kerwyn Huang, Will Ratcliff, Kalin Vetsigian, Ajay Gopinathan, Raghu Parthasarathy, and Brendan Bohannan for helpful discussions and/or comments on the manuscript. We thank Rob Yelle and Michael Coleman for assistance with high-performance computing resources at the University of Oregon. Research reported in this publication was supported, in part, by the National Institute of General Medical Sciences of the National Institutes of Health under Award 1P50GM098911 (to T.U.) and the University of Oregon (T.U.). This work was performed, in part, at the Aspen Center for Physics, which is supported by National Science Foundation Grant PHY-1607611. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811887116/-/DCSupplemental.
References
- 1.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1:109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 2.Faust K, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNally L, et al. Killing by type VI secretion drives genetic phase separation and correlates with increased cooperation. Nat Commun. 2017;8:14371. doi: 10.1038/ncomms14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 5.You L, et al. Spatial vs. non-spatial eco-evolutionary dynamics in a tumor growth model. J Theor Biol. 2017;435:78–97. doi: 10.1016/j.jtbi.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Reichenbach T, Mobilia M, Frey E. Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature. 2007;448:1046–1049. doi: 10.1038/nature06095. [DOI] [PubMed] [Google Scholar]
- 7.Xiong L, Cooper R, Tsimring LS. Coexistence and pattern formation in bacterial mixtures with contact-dependent killing. Biophys J. 2018;114:1741–1750. doi: 10.1016/j.bpj.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 9.Drescher K, Shen Y, Bassler BL, Stone HA. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci USA. 2013;110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-García R, Nadell CD, Hartmann R, Drescher K, Bonachela JA. Cell adhesion and fluid flow jointly initiate genotype spatial distribution in biofilms. PLoS Comput Biol. 2018;14:e1006094. doi: 10.1371/journal.pcbi.1006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadell CD, Ricaurte D, Yan J, Drescher K, Bassler BL. Flow environment and matrix structure interact to determine spatial competition in Pseudomonas aeruginosa biofilms. eLife. 2017;6:e21855. doi: 10.7554/eLife.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert G, Vyawahare S, Austin RH. Bacteria and game theory: The rise and fall of cooperation in spatially heterogeneous environments. Interface Focus. 2014;4:20140029. doi: 10.1098/rsfs.2014.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, et al. Influence of topology on bacterial social interaction. Proc Natl Acad Sci USA. 2003;100:13910–13915. doi: 10.1073/pnas.1935975100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: Connecting spatial organization to function. Cell Host Microbe. 2017;21:433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiles TJ, et al. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grošelj D, Jenko F, Frey E. How turbulence regulates biodiversity in systems with cyclic competition. Phys Rev E Stat Nonlin Soft Matter Phys. 2015;91:033009. doi: 10.1103/PhysRevE.91.033009. [DOI] [PubMed] [Google Scholar]
- 19.Avelino PP, et al. How directional mobility affects coexistence in rock-paper-scissors models. Phys Rev E. 2018;97:032415. doi: 10.1103/PhysRevE.97.032415. [DOI] [PubMed] [Google Scholar]
- 20.Szolnoki A, Szabó G. Phase transitions for rock-scissors-paper game on different networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:037102. doi: 10.1103/PhysRevE.70.037102. [DOI] [PubMed] [Google Scholar]
- 21.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: Poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drissi F, Buffet S, Raoult D, Merhej V. Common occurrence of antibacterial agents in human intestinal microbiota. Front Microbiol. 2015;6:441. doi: 10.3389/fmicb.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavridou DAI, Gonzalez D, Kim W, West SA, Foster KR. Bacteria use collective behavior to generate diverse combat strategies. Curr Biol. 2018;28:345–355.e4. doi: 10.1016/j.cub.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 24.May RM, Leonard WJ. Nonlinear aspects of competition between three species. SIAM J Appl Math. 1975;29:243–253. [Google Scholar]
- 25.Reichenbach T, Mobilia M, Frey E. Coexistence versus extinction in the stochastic cyclic Lotka-Volterra model. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:051907. doi: 10.1103/PhysRevE.74.051907. [DOI] [PubMed] [Google Scholar]
- 26.Strogatz SH. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry and Engineering. Westview Press; Boulder, CO: 2001. [Google Scholar]
- 27.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci USA. 2014;111:17941–17946. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine JM, Bascompte J, Adler PB, Allesina S. Beyond pairwise mechanisms of species coexistence in complex communities. Nature. 2017;546:56–64. doi: 10.1038/nature22898. [DOI] [PubMed] [Google Scholar]
- 30.Gallien L, Zimmermann NE, Levine JM, Adler PB. The effects of intransitive competition on coexistence. Ecol Lett. 2017;20:791–800. doi: 10.1111/ele.12775. [DOI] [PubMed] [Google Scholar]
- 31.Sinervo B, Lively CM. The rock-paper-scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. [Google Scholar]
- 32.Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- 33.Godoy O, Stouffer DB, Kraft NJB, Levine JM. Intransitivity is infrequent and fails to promote annual plant coexistence without pairwise niche differences. Ecology. 2017;98:1193–1200. doi: 10.1002/ecy.1782. [DOI] [PubMed] [Google Scholar]
- 34.Higgins LM, Friedman J, Shen H, Gore J. 2017. Co-occurring soil bacteria exhibit a robust competitive hierarchy and lack of non-transitive interactions. bioRxiv:175737.
- 35.Cornforth DM, Foster KR. Competition sensing: The social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 36.Lowery NV, McNally L, Ratcliff WC, Brown SP. Division of labor, bet hedging, and the evolution of mixed biofilm investment strategies. MBio. 2017;8:e00672-17. doi: 10.1128/mBio.00672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huse DA, Henley CL. Pinning and roughening of domain walls in Ising systems due to random impurities. Phys Rev Lett. 1985;54:2708–2711. doi: 10.1103/PhysRevLett.54.2708. [DOI] [PubMed] [Google Scholar]
- 38.Cammarota C, Biroli G. Ideal glass transitions by random pinning. Proc Natl Acad Sci USA. 2012;109:8850–8855. doi: 10.1073/pnas.1111582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arumugam S, Petrov EP, Schwille P. Cytoskeletal pinning controls phase separation in multicomponent lipid membranes. Biophys J. 2015;108:1104–1113. doi: 10.1016/j.bpj.2014.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu X, Cueto-Felgueroso L, Juanes R. Thermodynamic coarsening arrested by viscous fingering in partially miscible binary mixtures. Phys Rev E. 2016;94:033111. doi: 10.1103/PhysRevE.94.033111. [DOI] [PubMed] [Google Scholar]
- 41.Siryaporn A, Kim MK, Shen Y, Stone HA, Gitai Z. Colonization, competition, and dispersal of pathogens in fluid flow networks. Curr Biol. 2015;25:1201–1207. doi: 10.1016/j.cub.2015.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borer B, Tecon R, Or D. Spatial organization of bacterial populations in response to oxygen and carbon counter-gradients in pore networks. Nat Commun. 2018;9:769. doi: 10.1038/s41467-018-03187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milotti E. 2002. 1/F noise: A pedagogical review. arXiv:physics/0204033.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code to run simulations and analyses is available in Datasets S1–S12.