Abstract
Cobamides (Cbas) are coenzymes used by cells from all domains of life, but made de novo only by some bacteria and archaea. The last steps of the cobamide biosynthetic pathway activate the corrin ring and the lower ligand base, condense the activated intermediates, and dephosphorylate the product prior to the release of the biologically active coenzyme. In bacteria, a phosphoribosyltransferase (PRTase) enyzme activates the base into its α-mononucleotide. The enzyme from Salmonella enterica (SeCobT) has been extensively biochemically and structurally characterized. The crystal structure of the putative PRTase from the archaeum Methanocaldococcus jannaschii (MjCobT) is known but its function has not been validated. Here we report the in vivo and in vitro characterization of MjCobT. In vivo, in vitro, and phylogenetic data reported here show that MjCobT belongs to a new class of NaMN-dependent PRTase. We also show that the Synechococcus sp. WH7803 CobT protein has PRTase activity in vivo. Lastly, results of isothermal titration calorimetry and analytical ultracentrifugation analysis show that the biologically active form of MjCobT is a dimer, not a trimer, as suggested by its crystal structure.
Keywords: CobT phosphoribosyltransferase, cyanobacteria, B12 biosynthesis, cobamides, Methanocaldococcus jannaschii, archaeal PRTase
Graphical Abstract
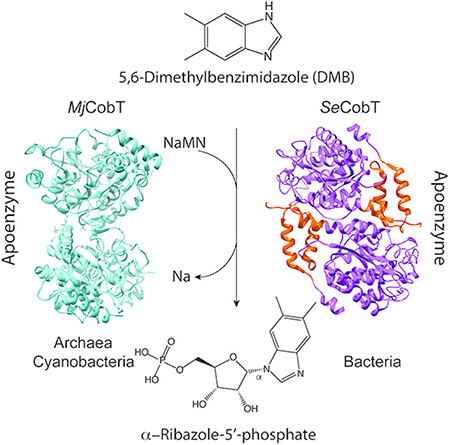
The activation of the lower ligand base of cobalamin is catalyzed by a new class of phosphoribosyltransferase found in archaea and cyanobacteria. The enzyme from the methanogenic archaeum Methanocaldococcus jannaschii (MjCobT) uses the same substrates but it is structurally different than the best studied bacterial enzyme from Salmonella enterica (SeCobT).
INTRODUCTION
Cobamides (Cba) are cobalt-containing cyclic tetrapyrroles used by cells from all domains of life but are produced de novo only by some bacteria and archaea [reviewed in1]. Cbas are involved in enzymatic carbon skeleton rearrangements, methyl-group transfers, reductive dehalogenation, and elimination reactions2–6. Additionally, cobamide-dependence is common amongst radical SAM enzymes7. Recent work has identified a role for Cbas in gene regulation as photoreceptors (reviewed in8).
Structurally, Cbas are characterized by a central cobalt ion equatorially coordinated by the nitrogens of the pyrrole rings. Unlike other cyclic tetrapyrroles (e.g., heme, factor F430, bacteriochlorophylls), most Cbas contain an upper and a lower ligand. The coenzymic form of the Cba known as cobalamin (Cbl) has a 5’-deoxyadenosyl (Ado) group as its upper ligand and 5,6-dimethylbenzimidazole (DMB) as the lower ligand (Fig. 1A). In addition to DMB, other purines and benzimidazole homologues can serve as the lower ligand and form a coordination bond with the central cobalt ion9 (Fig. 1B,C). Some organisms (e.g., Sporomusa ovata) can also incorporate phenolics as bases into Cbas (Fig. 1D), but such bases cannot form coordination bonds with the cobalt ion of the ring.
Figure 1. Adenosylcobamide (AdoCba) structure and nucleotide base diversity.
A. The nucleotide loop is boxed. B. Purines and purine analogues found in Cbas: [1], purine; [2]. hypoxanthine; [3], adenine; [4], 2-methyladenine; [5], 2-methylmercaptoadenine; [6], 2-methylsulfinyladenine; [7], 2-methylsulfonyladenine; [8], guanine. C. Benzimidazole and its analogues: [1], benzimidazole; [2], 5-methylbenzimidazole; [3], 5,6-dimethylbenzimidazole; [4], 5-hydroxybenzimidazole; [5], 5-methoxybenzimidazole; [6], 5-methoxy-6-methylbenzimidazole; [7], naphthimidazole. D. Phenolics: [1], phenol; [2], p-cresol.
Vitamin B12 (cyanocobalamin, CNCbl) is a Cba whose upper ligand is a cyano (CN) group, and its lower ligand is DMB. As shown in figure 1, the diversity of bases found in Cbas is broad4, 9. For example, Salmonella enterica produces three Cbas with varying nucleotide bases: one with DMB (a.k.a. cobalamin, Cbl), one with adenine (a.k.a. pseudocobalamin, psCbl), and one with 2-methyladenine (a.k.a. Factor A)10–12.
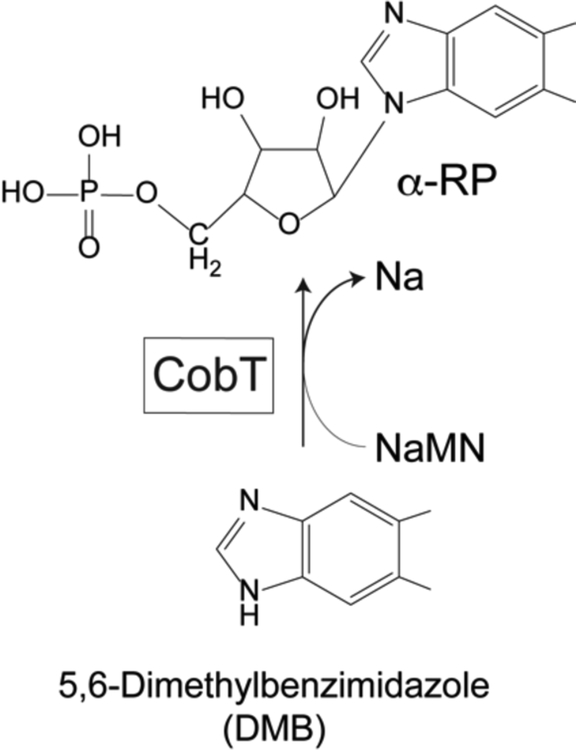
Lower ligand bases require activation to an α-ribotide prior to their incorporation into the final product of the pathway. In Salmonella enterica, the CobT enzyme (EC 2.4.2.21), a nicotinate mononucleotide (NaMN):Base phosphoribosyltransferase (PRTase), activates the lower ligand base. The product of this reaction is the intermediate known as α-ribazole-5’-phosphate (α-RP) (Fig. 2)13.
Figure 2. Base Activation by CobT.
The activation of 5,6-dimethylbenzimidazole (DMB) to its α-ribotide by CobT PRTase is shown. NaMN, nicotinate mononucleotide, Na, nicotinic acid, α-RP, alpha-ribazole-phosphate, CobT: NaMN:DMB phosboribosyltransferase (EC 2.4.2.21)
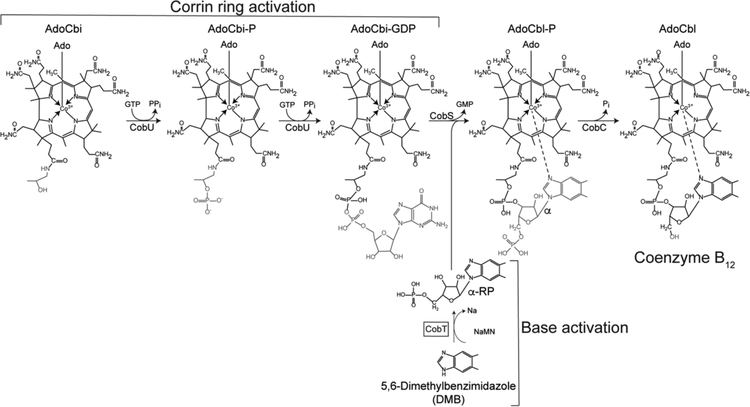
The last steps of AdoCba biosynthetic pathways activate the corrin ring and the lower ligand base, condense the activated intermediates into an AdoCba-phosphate (AdoCba-P), and, finally, removes the phosphate to yield the final product of the pathway (Fig. 3).
Figure 3. Late steps of coenzyme B12 biosynthesis.
This figure schematizes the two branches of the nucleotide loop assembly (NLA) pathway that yields coenzyme B12 in S. enterica. This bacterium can assimilate exogenous, incomplete corrinoids such as cobinamide (Cbi) converting it to AdoCbi and activating it to AdoCbi-GDP via AdoCbi-P using the bifunctional kinase/guanylyltransferase (CobU) enzyme. The base shown in this scheme is 5,6-dimethylbenzimidazole (DMB), which is activated to its α-ribotide by the CobT PRTase (shown inside the box). Condensation of the activated intermediates followed by dephosphorylation yields biologically active coenzyme B12.
Previous in vitro studies of SeCobT showed that the enzyme has a broad range of substrate specificity and can activate purine and benzimidazole analogues (Fig. 1)13, 14. This lack of substrate specificity is shared by S. enterica CobT (SeCobT) homologues in other microorganisms15. At present, we know little about the function of archaeal SeCobT homologues. The crystal structure of the putative apo NaMN:Base PRTase from the methanogenic archaeum Methanocaldococcus jannaschii (ORF MJ_RS08515) has been reported, but evidence supporting the annotated activity has not been published.
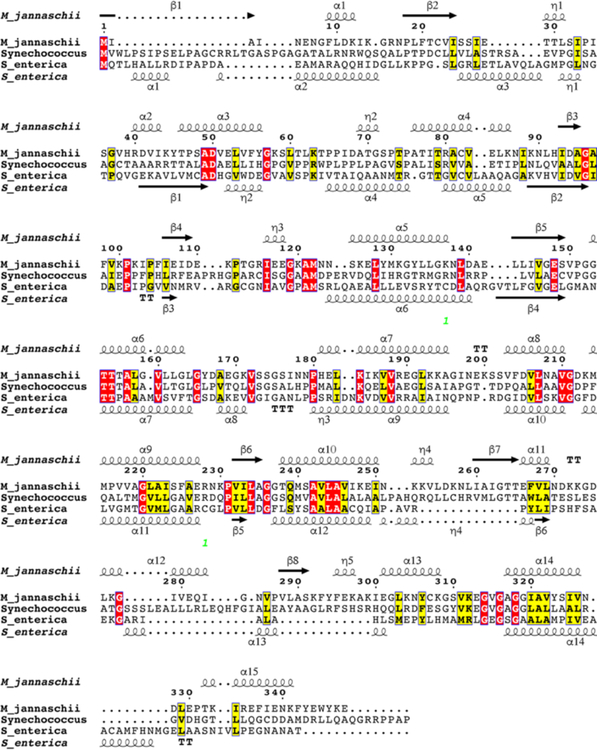
Bioinformatics analyses of the MJ_RS08515 amino acid sequence compared with that of the SeCobT enzyme showed limited identity (18%) and similarity (32%) (Fig. 4). These enzymes also differed in their oligomeric state, with the archaeal enzyme forming trimers (PDB 3L0Z), according to the structure deposited into the Research Collaboratory for Structural Bioinformatics Protein Database (RCSB PDB) (unpublished work), while the bacterial enzyme forms dimers13 (PDB 1L4B).
Figure 4. Protein sequence similarities and identities among MjCobT, SeCobT, and Synechococcus CobT.
The alignment was generated using Clustal Omega and visualized with ESPript 3.0 web-based program. Alpha helices and beta sheets for MjCobT (PDB 3L0Z) and SeCobT (PDB 1L4B) obtained from the reported crystal apo structures are shown. The structure of Synechococcus CobT has not been reported.
We note that previous studies by other investigators suggested that cyanobacteria solely produce psCbl. Additionally, these studies showed that 121 of the 123 cyanobacterial genomes searched lacked cobT16, 17. In contrast to these findings, our bioinformatics analysis identified CobT homologues in many cyanobacterial genomes. These homologues were similar in amino acid sequence composition to MJ_RS08515.
The work reported here shows that M. jannaschii CobT (MjCobT) belongs to a new class of base-activating PRTase enzymes. We determined that MjCobT represents a new class of base-activating PRTase enzymes based on both structural and taxonomic differences from the extensively characterized SeCobT. In this study we used S. enterica as a heterologous complementation system to show that MjCobT has bona fide NaMN:DMB PRTase activity in vivo. Consistent with our in vivo results, MjCobT exhibited robust enzymatic activity in vitro at pH 7. We found that under the conditions used to assay MjCobT activity, the SeCobT enzyme also displayed strong activity. This finding was of interest, because in the past we had only measured strong SeCobT activity at pH 10. We identify potassium phosphate (KPi) as the ingredient of the reaction mixture needed by SeCobT to be active at pH 7. We also report that MjCobT has broad substrate specificity and high affinity for its NaMN substrate. Finally, our data support the idea that MjCobT is biologically active as a dimer.
MATERIALS AND METHODS
Culture media and growth conditions.
All chemicals were reagent grade and were purchased from Sigma-Aldrich unless otherwise stated. Purchased chemicals were used without further purifications. All strains used in this study were grown at 37°C in no-carbon energy (NCE) minimal medium18 supplemented with ribose (20 mM) as a carbon and energy source. Minimal medium was also supplemented with MgSO4 (1 mM) and trace minerals19. When used, ampicillin, kanamycin, or chloramphenicol were added at 100 μg/mL, 50 μg/mL or 25 μg/mL, respectively. For all growth experiments, two μL of overnight cultures (12–16 h old) grown in nutrient broth (NB, Difco) was used to inoculate 198 μL of minimal medium with supplements (1% inoculum, v/v). Strains were grown in triplicate in 96-well microtiter plates for growth experiments. Growth was monitored using a computer-controlled BioTek plate reader (Model Eon). Optical density at 630 nm was measured every 30 min for a total time of 24 h; the microtiter dish was shaken between measurements. Data were analyzed using GraphPad Prism v6 software. Doubling times were calculated using the exponential growth equation included with the GraphPad Prism v6 software.
Strain constructions.
All strains used in in this study were derivatives of Salmonella enterica enterica sv Typhimurium (hereafter S. enterica) LT2 or Escherichia coli BL21. Strain genotypes are described in Table S1. Plasmids were introduced into S. enterica by electroporation20 and into E.coli by heat-shock transformation21. Deletions in S. enterica were constructed and resolved with pCP20 as described elsewhere22.
Plasmid constructions.
All primers used during the course of this work are described in Table S2. Restriction enzymes were purchased from Fermentas. BspQI restriction enzyme was purchased from New England BioLabs. Synechococcus sp. WH7803 cobT gene was codon optimized for expression in E. coli by GenScript (Fig. S1).
Plasmid pMjCobT1.
This plasmid was constructed using the polymerase incomplete primer extension (PIPE) method23. The Methanocaldococcus jannaschii wildtype allele of ORF MJ_RS08515 was amplified from M. jannaschii genomic DNA (a gift from W. B. Whitman, University of Georgia) using the primers Mj_1598_cobT_PIPE_f and Mj_1598_cobT_PIPE_r. Vector pBAD2424 was amplified using primers pBAD24-PIPE-5’ and pBAD24-PIPE-3’. Amplified fragments were cut with DpnI (ThermoFisher) overnight at 37°C. Fragments were mixed in equal volumes and transformed in chemically competent E. coli DH5a21. The completed plasmid contained ORF MJ_RS08515 coding sequence inserted between KpnI and SalI sites of pBAD24. The plasmid was used for complementation analyses and generation of cobamides in S. enterica. Plasmid pMjCobT1 is 5,601 bp long and encodes resistance to ampicillin.
Plasmid pMjCobT2.
This plasmid was also constructed using the PIPE method as described above using primers Mj_1598_TEV5_PIPE_f and Mj_1598_TEV5_PIPE_r to amplify ORF MJ_RS08515 and primers pTEV5-PIPE-5’ and pTEV5-PIPE-3’ to amplify the pTEV5 vector25. pMjCobT2 contained the wildtype MJ_RS08515 coding sequence inserted between NheI and XhoI sites of pTEV5. pMjCobT2 was used for protein overproduction. The plasmid is 6,381 bp long and encodes resistance to ampicillin. Transcription of MJ_RS08515 was induced by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG), which triggered the synthesis of T7 polymerase, the enzyme responsible for the transcription of MJ_RS08515.
Plasmid pCOBT140.
The construction of this vector has been described26. The plasmid encodes S. enterica cobT+, is ~5.6 kb long and encodes ampicillin resistance.
Plasmid pMjCobT3.
This plasmid encoded MjCobTE315A and was constructed using overlapping primers MjCobT_sdm_E315A_F and MjCobT_sdm_E315A_R with plasmid pMjCobT1 DNA as the template. Briefly, the primers indicated encoding E315A changes were used to amplify the ORF MJ_RS08515 on pMjCobT1. Amplified products were cut with DpnI (ThermoFisher) overnight at 37°C. The amplification product was then transformed in E. coli DH5a and its nucleotide sequence verified.
Plasmid pMjCobT4.
This plasmid encoded variant MjCobTE150A E315A and was constructed using overlapping primers MjCobT_sdm_E150A_F and MjCobT_sdm_E150A_R with plasmid pMjCobT3 DNA as the template. Briefly, the primers indicated encoding E150A changes were used to amplify the ORF MJ_RS08515 with the E315A change on pMjCobT3. Amplified products were cut with DpnI (ThermoFisher) overnight at 37°C. The amplification product was then transformed in E. coli DH5a and its nucleotide sequence verified.
Plasmid pMjCobT5.
This plasmid encoded MjCobTE150A and was constructed using overlapping primers MjCobT_sdm_E150A_F and MjCobT_sdm_E150A_R with plasmid pMjCobT1 DNA as the template. Briefly, the primers encoding E150A changes were used to amplify the ORF MJ_RS08515 on pMjCobT1. Amplified products were cut with DpnI (ThermoFisher) overnight at 37°C. The amplification product was then transformed in E. coli DH5a and its nucleotide sequence verified.
Plasmid pSynCobT2.
This plasmid was constructed using the BspQI high-efficiency cloning method outlined elsewhere27 using primers SynCobT pCV1 F and SynCobT pCV1 R to amplify the codon optimized Synechococcus sp. WH7803 CobT (SYNWH7803_RS09275). The completed plasmid contained codon optimized SYNWH7803_RS09275 coding sequence inserted between the pair of BspQI sites of pCV1. The plasmid was used for complementation analyses in S. enterica. Plasmid pSynCobT2 is 5,724 bp long and encodes resistance to ampicillin.
Cobamide extraction and high-performance liquid chromatography (HPLC) analysis.
Overnight culture of strain JE18114 (ΔmetE ΔcobT ΔcobB / pMjCobT1) and strain JE18115 (ΔmetE ΔcobT ΔcobB / pCOBT140) grown in nutrient broth (NB) supplemented with ampicillin (100 μg/mL), kanamycin (50 μg/mL), and chloramphenicol (25 μg/mL) was used to inoculate NCE minimal medium supplemented with ribose (20 mM) as a carbon and energy source. Minimal medium also contained trace minerals, magnesium sulfate (1 mM), ampicillin (100 μg/mL) arabinose (500 μM), and dicyanocobinamide [(CN)2Cbi] (300 nM). Where indicated, DMB, adenine, 5-methoxybenzimidazole (5-MeO-Bza), or 5-hydroxybenzimidazole (5-HO-Bza) was provided at a final concentration of 200 μM. Cultures (250 mL) were grown for >20 h at 37°C in an Innova 44R gyratory incubator (New Brunswick Scientific) shaking at 180 rpm. Cells were harvested by centrifugation at 4°C for 15 min at 6,000 × g using an Avanti J20-XPI refrigerated centrifuge (Beckman) equipped with JLA-8.1000 rotor. Pelleted cells were re-suspended in ammonium acetate buffer (100 mM, pH 4.5) containing KCN (10 mM), and stored at −20 °C. Cobamides were extracted using methods described elsewhere28, 29. Cell suspensions were thawed on ice and subsequently incubated at 70°C with shaking at 180 rpm in an Innova 44R gyratory incubator (New Brunswick Scientific) for 2 h. After incubation, cell suspensions were centrifuged in an Avanti J-251 equipped with a JA 25.25 rotor (Beckman) at 43,000 × g for 40 min at 4°C. Clarified cell-free extract was filtered through a syringe filter unit (0.45 μm) and mixed with Amberlite XAD4 resin (Rohm-Haas). Cell-free extract and resin mixtures were incubated at 37°C with shaking at 180 rpm in an Innova 43 gyratory incubator (New Brunswick Scientific) for 16 h. After allowing the resin to settle, free liquid was removed, taking care not to disturb the resin. Resin was washed thrice with four bed volumes of deionized water (Millipore) allowing the resin to settle for 20 min between washes. After the final wash, two bed volumes of methanol (100%) were added to the resin and the solution incubated 16 h at room temperature. After incubation, methanol was removed from the resin, collected in a microfuge tube and subjected to vacuum centrifugation in an Eppendorf Vacufuge Plus set at 60 °C until dry. Dried pellets were re-suspended in a 1:4 ratio of buffer B [KH2PO4 (100 mM), KCN (10 mM), pH 8 + 50% (v/v) acetonitrile] and buffer C [KH2PO4 (100 mM), KCN (10 mM), pH 6.5] in preparation for separation by HPLC. Cbas were resolved by RP-HPLC using a Shimadzu Prominence UFLC SPD-M30A equipped with a Phenomenex Syngergi 4μ hydro-RP80A 150 × 4.6 mm LC column as described30, 31 with some modifications as outlined in26. Cbas were detected at 367 nm and 525 nm.
Mass spectrometry of corrinoids.
Corrinoids with retention times of 12, 13, and 16 min were collected, desalted using C18 SepPaks (Waters) and dried under vacuum centrifugation. The molecular mass of each corrinoid was determined by MALDI-TOF mass spectrometry at the PAMS facility of the University of Georgia.
Protein purification.
Wild-type MjCobT protein was overproduced from plasmid pMjCobT2 in strain JE6663 (E. coli C41) in 2-L cultures of Terrific Broth32. Protein synthesis was induced by the addition of IPTG at a final concentration of 500 μM in mid-log phase cultures (OD600 ~0.6) growing at 37°C with shaking at 180 rpm in an Innova44 (New Brunswick Scientific) gyratory incubator. After induction, cultures were grown for 17 h at 30°C with shaking at 180 rpm. Cultures were harvested by centrifugation at 4°C for 15 min at 6,000 × g in an Avanti J20-XPI refrigerated centrifuge equipped with JLA-8.1000 rotor. Pelleted cells were stored at −80°C until used. Frozen cells were thawed on ice and re-suspended in Tris-HCl buffer (25 mM, pH 7.5) containing NaCl (0.5 M) and imidazole (20 mM) at a rate of 20% cell weight to buffer volume. Lysozyme (1μg/mL) and DNaseI (25μg/mL) were added to the cell suspension and incubated on ice for 10 min. Cells were lysed by cell press in a cell disruptor (Constant Systems) set at 1.72e8 Pa. Phenylmethylsulfonyl fluoride (PMSF) was added to the cell lysate at a final concentration of 0.5 mM. The cell lysate was incubated for 30 min at 70°C to precipitate E. coli proteins. Cell debris and precipitated proteins were removed by centrifugation at room temperature for 30 min at 40,000 × g in an Avanti J-251 centrifuge (Beckman/Coulter) equipped with a JA 25.25 rotor. Clarified extract was filtered using a 0.45-μm syringe filter unit and applied to a 4-mL HisPur nickel-nitrilotriacetic acid (Ni-NTA) affinity column (ThermoFisher Scientific). The column was washed with 10 column volumes of Tris-HCl buffer (25 mM, pH 7.5) containing NaCl ((0.5 M) and imidazole (20 mM) and six column volumes of Tris-HC buffer (25 mM, pH 7.5) containing NaCl (0.5 M)) and imidazole (40 mM). H6-MjCobT was eluted with six column volumes of Tris-HCl buffer (25 mM, pH 7.5) containing NaCl (0.5 M) and imidazole (0.5 M). Fractions were collected throughout the wash and elution steps and H6-MjCobT purification was monitored by SDS-PAGE gel compared to Precision Plus Protein Standards (BioRad). Fractions containing H6-MjCobT were pooled and dialyzed against Tris-HCl buffer (25 mM, pH 7.5) containing NaCl (0.5 M) to remove imidazole. N-terminally tagged, recombinant tobacco etch virus protease (H7-rTEV) was purified as described elsewhere33 and mixed with H6-MjCobT at a ratio of 1:10 H6-MjCobT:H7-rTEV. The mixture was incubated at 34°C for 3 h to remove the H6 tag on MjCobT. Cleaved MjCobT was purified away from the tag and H7-rTEV using the Ni-NTA affinity purification method outlined above and tag-less protein was dialyzed against Tris-HCl buffer (25 mM, pH 7.5) in steps with decreasing concentrations of NaCl down to 150 mM. Purified MjCobT was flash frozen in liquid nitrogen and stored at −80°C until used. Protein concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific) using the molecular mass of 37600 Da and ε280 of 20400 M−1 cm−1 as determined by ProtParam33.
Analytical ultracentrifugation.
MjCobT was dialyzed against 50 mM Na/K phosphate (pH 7.5) and 100 mM NaCl and diluted to a final concentration of 1 mg/ml. Quantification was performed on an Agilent 8453 UV/vis and used an ε280 of 20400 M−1 cm−1 as determined by ProtParam34. The protein sample was loaded with the dialysis buffer to a final concentration of 1 mg/ml before being loaded into cells with 12 mm double-sector Epon centerpieces. The loaded cells were allowed to equilibrate in the rotor for 1 h at 20 °C. Sedimentation velocity data were collected at 50,000 rpm at 20 °C in an Optima XLA analytical ultracentrifuge. Data were recorded at 280 nm in radial step sizes of 0.003 cm. SEDNTERP35 was used to calculate the partial specific volume of MjCobT (0.756018 mL/g) as well as36,35,35 the density (1.0093 g/mL) and viscosity coefficient (0.010291) of the buffer. SEDFIT37 was used to analyze the raw sedimentation data. Data were modeled as a continuous sedimentation coefficient distribution [c(s)], and were fit using the baseline, meniscus, frictional coefficient, and systematic time-invariant and radial-invariant noise. Fit data for the experiment had a root mean square deviations of (r.m.s.d) < 0.004 AU. The theoretical sedimentation coefficient (s) values were calculated from the atomic coordinates of MjCobT(PDB ID 3L0Z) using HYDROPRO2038.
In vitro NaMN:DMB phosphoribosyltransferase assay.
NaMN:DMB PRTase activity was assayed as described elsewhere with minor changes to the reaction conditions detailed in each of the following sections13. Asterisks shown represent the following p-values: * denotes a p-value <0.05, ** denotes a p-value <0.01, *** denotes a p-value <0.001.
Specific activity measurements.
To determine the specific activity of MjCobT, the protein was diluted in potassium phosphate buffer (0.25 M, pH 8.0). Reaction mixtures contained phosphate buffer (0.25 M, pH 8.0), DMB (1.5 nmol), [2-14C]-DMB (0.5 nmol) and MjCobT (190 ng) in a final volume of 20 μL. NaMN was provided in the following amounts: 20, 15, 10, and 5 nmoles. Reactions mixtures were incubated at 37°C. Samples were removed after 0, 3, 5, 10, and 15 min incubations and immediately spotted (5 μL) onto a silica gel thin-layer chromatography (TLC) plate (Whatman PE SIL G/UV). TLCs were developed for 1.5 h in a TLC chamber pre-equilibrated with a 3:2 (v/v) chloroform:methanol mobile phase. After drying in a fume hood, plates were exposed to a phosphor screen for 18 h. Distribution of radioactivity on TLC plates was visualized using a Typhoon Trio+ Variable Mode Imager (GE Life Sciences) with ImageQuant v5.2. Pixel density of TLC plates was analyzed using 1D gel analysis in TotalLab TL100 (Nonlinear Dynamics) and the resulting data were analyzed using Prism v6 (GraphPad). Each reaction was performed in triplicate.
Effect of pH on MjCobT activity.
Reactions were conducted as outlined above with NaMN (1 mM) and buffer (25 mM) at the following pHs: sodium malate, pH 5, sodium succinate, pH 5.5, 4- morpholineethansulfonic acid (MES) pH 6, Bis-Tris pH 6.5, imidazole pH 7, Tris-HCl, pH 7, Tris-HCl pH 8, potassium phosphate pH 8, Tris-HCl pH 9, 2-(cyclohexylamino)ethanesulfonic acid (CHES) pH 9, CHES pH 9.5, glycine pH 10, and glycine pH 10.5. Reaction mixtures were incubated for 15 min at 37°C.
Effect of ionic strength on MjCobT activity.
Varying concentrations of salt tested as outlined above with NaMN (1 mM) and either potassium chloride or sodium chloride at the following concentrations: 0.1, 0.2, 0.3, 0.4 and 0.5 M. Reaction mixtures were incubated for 15 min at 37 °C.
Effect of temperature on MjCobT activity.
MjCobT activity as a function of incubation temperature was tested as outlined above with NaMN (1 mM). Reactions were incubated for 10 min at 40, 50, 70, or 80°C.
Effect of phosphate on MjCobT activity.
MjCobT activity as a function of phosphate concentration was measured as described above with NaMN (1 mM) and phosphate at either 0.025, 0.05, 0.075, 0.1, 0.2 or 0.4 M phosphate salts adjusted to pH 7.0. Reaction mixtures were incubated for 15 min at 80°C. Equivalent reactions were prepared with sodium salts of sulfate, vanadate, selenate, or molybate in lieu of potassium phosphate for comparison.
Determination of kinetic parameters.
Pseudo-first order kinetic data were obtained at a range of NaMN concentrations (0.075 to 1 mM) with a saturating concentration of DMB (100 μM). The data were fit to the Michaelis-Menten equation to determine the Km and maximum velocity (Vmax):
The turnover constant (kcat) was obtained with the following equation where E represents enzyme concentration:
Catalytic efficiency was determined by the following: kcat/Km.
Isothermal titration calorimetry.
Binding assays of MjCobT and NaMN substrate were performed using a Nano ITC isothermal titration calorimeter (TA instruments). MjCobT was dialyzed against Tris-HCl buffer (25 mM, pH 7.5) containing NaCl (150 mM); NaMN was solubilized in the final protein dialysate. Samples were degassed for 20 min at 25°C before use. MjCobT protein was present in the sample cell at 33 μM under constant stirring at 350 rpm, and NaMN was present in the injection syringe at 1 mM. Every 5 min, injections (2.4 μL) were made into the sample cell and heat released was recorded. Experiments were conducted at 25°C. Data were analyzed using NanoAnalyze software (TA instruments).
Phylogenetic analysis.
We searched for MjCobT homologues using NCBI BLASTP with CobT query sequences from M. jannaschii (accession: WP_064496984.1), S. enterica (accession: NP_460961.1), P. denitrificans (accession: WP_015478315.1), and B. megaterium (accession: YP_003563500.1)39. Query sequences were used to search cyanobacteria (taxid: 1117), thaumarchaeota (taxid: 651137), methanogens class I (taxid: 2283794), and methanogens class II (taxid: 224756). Results with a minimum 85% query coverage and e-value cutoff of <10−40 were included in the supplementary tables. Results were screened using the NCBI conserved domain database (CDD) to confirm DMB-PRT_CobT superfamily (accession: cl11435, PSSM Id: 325029) placement40. The percent identity and percent positives are also provided. Results from “uncultured marine thaumarchaeotes” were omitted from the tables.
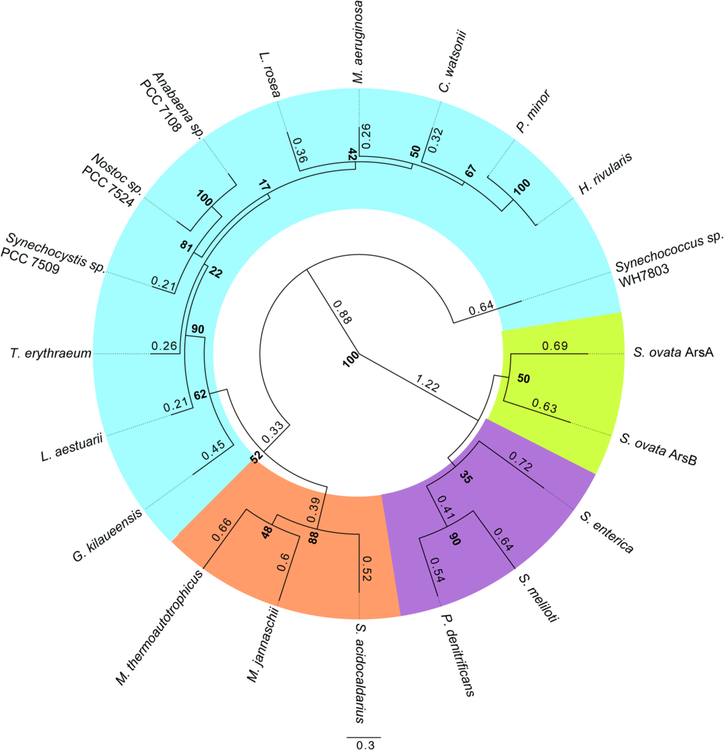
The phylogenetic tree (Fig. 14) was constructed using SeaView software41. CobT homologue sequences from the following organisms were used to generate a MUSCLE alignment: M. jannaschii, M. thermoautotrophicus, S. acidocaldarius, P. denitrificans, S. enterica, S. ovata (ArsAB), Synechococcus sp. WH7803, G. kilaueensis, L. rosea, Anabaena sp. PCC 7108, Nostoc sp. PCC 7524, Synechocystis sp. PCC 7509, T. erythraeum, L. aestuarii, H. rivularis, P. minor, C. watsonii, S. meliloti and M. aeruginosa. The tree was then constructed using the PhyML algorithm (100 bootstraps) with BLOSUM 62 as the model. The tree was modified for publication using the FigTree1.4.3 program available online (http://tree.bio.ed.ac.uk/software/figtree/).
Figure 14. Cyanobacterial CobT homologues are of archaeal origin.
Phylogenetic relationship of CobT homologues from diverse archaea and bacteria are shown. The tree was built based on a MUSCLE alignment using PhyML algorithm with BLOSUM62 model (100 bootstraps). Bootstrap values are presented in bold at the nodes and branch times are shown to 2 significant figures. Branch times below 0.2 were omitted from the tree. Organism highlighting is as follows: cyanobacteria in blue, archaea in orange, proteobacteria in purple, and Firmicutes in green.
RESULTS & DISCUSSION
In vivo evidence that M. jannaschii CobT (MjCobT) is a functional enzyme.
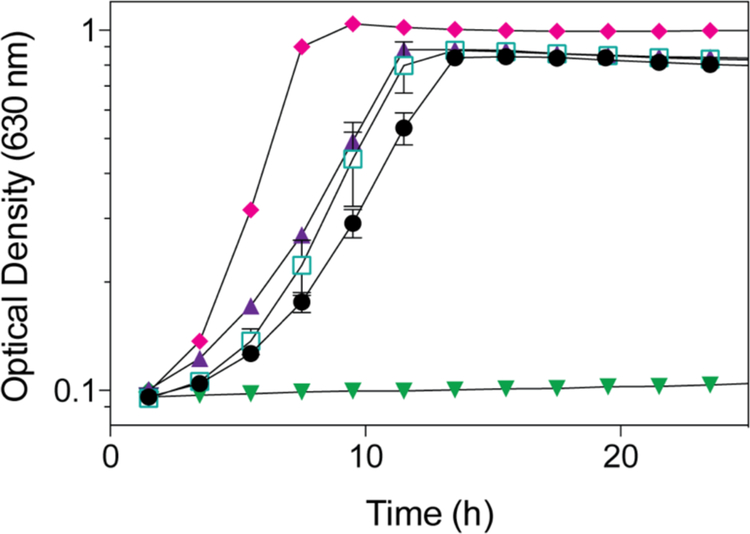
We used an in vivo approach to determine whether MjCobT had NaMN:DMB phoshoribosyltransferase (PRTase) activity. For this purpose, we introduced a plasmid encoding M. jannaschii cobT+ into a S. enterica ΔcobT strain to try to restore AdoCbl synthesis in the absence of SeCobT function. ORF MJ_RS08515 (referred to as M. jannaschii cobT+) was placed under the control of an arabinose-inducible promoter in plasmid pBAD2424 to yield pMjCobT1, which was moved into strain JE12893 (ΔcobT1379 ΔcobB1374) yielding strain JE18114 (Table S1).
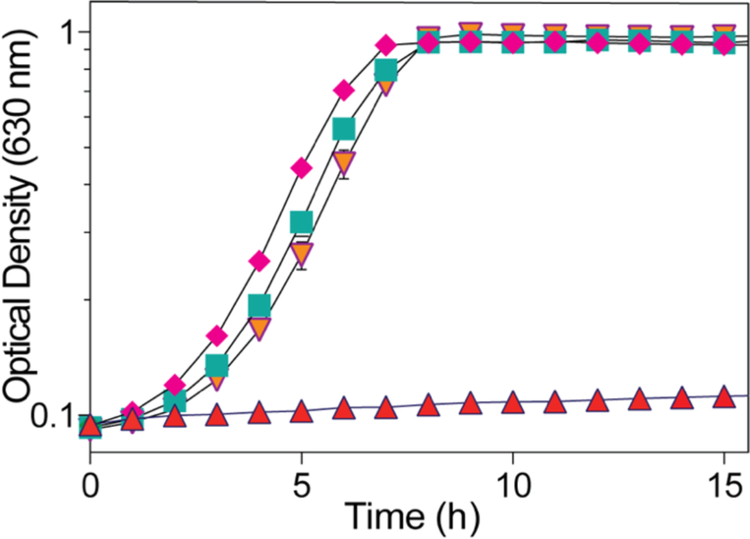
Strain JE12893 failed to grow without exogenous Cbl (Fig. 5, triangles), but grew when Cbl was present in the medium (Fig. 5, diamonds). Ectopic expression of M. janaschii cobT+ allowed strain JE18114 to grow when Cbi was present in the medium (Fig. 5, inverted triangles). This result indicated that MjCobT had SeCobT-like activity. Strain JE18115 (ΔcobT ΔcobB / pCOBT140) served as positive control for CobT function (Fig. 5, squares).
Figure 5. MjCobT supports AdoCbl-dependent growth of S. enterica.
Strains were grown in minimal NCE medium supplemented with ribose (20 mM) as carbon and energy source. (CN)2Cbi (15 nM) was added to all cultures, except to the culture growing in the presence of CNCbl (diamonds). DMB (150 μM) was added to all cultures except for the culture to which CNCbl was added (diamonds).
MjCobT activates diverse bases to their corresponding α-ribotides.
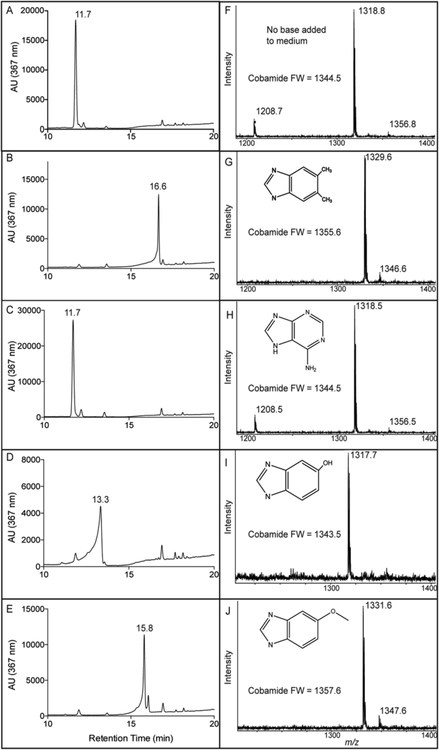
To gain insights into the substrate specificity of MjCobT, guided biosynthesis experiments were performed. For this purpose, Cbas were extracted from S. enterica ΔcobT ΔcobB / pMjCobT (strain JE18114) after growth in culture medium supplemented with Cbi alone or Cbi plus varying bases (i.e., 5,6-dimethylbenzimidazole (DMB), adenine, 5-methoxybenzimidazole, or 5-hydroxybenzimidazole). Reverse-phase, high-performance liquid chromatography (RP-HPLC) and MALDI-TOF mass spectrometry analyses of extracted Cbas showed that all the bases tested were incorporated into the final product (Fig. 6), indicating that MjCobT activated each base to its corresponding α-ribotide. Notably, when no base was provided, MjCobT activated endogenous adenine and the final product was psCbl. As a positive control, the same analyses were performed with Cbas extracted from the S. enterica ΔcobT ΔcobB / pSeCobT (strain JE18115) (data not shown).
Figure 6. MjCobT can activate different bases.
The figure shows RP-HPLC tracings (panels A-E) and MALDI-TOF mass spectra (panels F-J) of α-ribotides synthesized by MjCobT under conditions described under Materials and Methods. The substrates used were: 5,6-dimethylbenzymidazole (panels B, G), adenine (panels C, H), 5-hydroxybenzimidazole (panels D, I), and 5-methoxybenzimidazole (panels (E, J).
In vitro optimization of conditions for MjCobT NaMN:DMB PRTase activity.
NaMN:DMB PRTase activity was measured using thin layer chromatography (TLC) and [2-14C]-DMB substrate as described elsewhere42.
Effect of pH on the reaction.
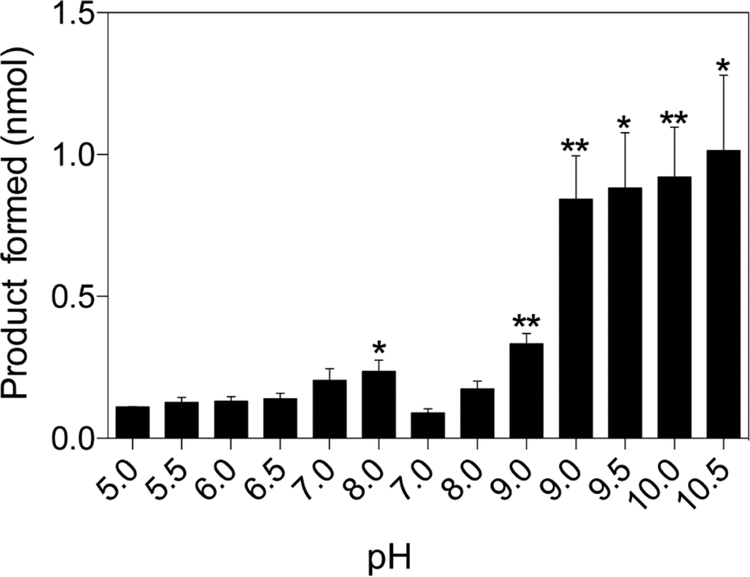
To determine optimal buffer and pH for MjCobT activity, activity level was assessed using multiple buffers over a range of pH (Fig. 7). MjCobT showed greatest activity at pH 10, similar to SeCobT42. Given that the internal pH of S. enterica is not 10, the buffer system with the highest activity within a physiologically relevant pH range was used for subsequent analyses. Subsequent experiments were performed with KPi buffer pH 8.
Figure 7. Effect of pH on MjCobT activity.
Conditions for the PRTase reaction are described under Materials and Methods. Product formation is displayed as a function of pH. Error bars represent the standard error of the mean. Asterisks represent significance determined by unpaired T test compared to pH 7. The following buffers were tested in order of appearance: sodium malate (pH 5), sodium succinate (pH 5.5), MES (pH 6), 2-[Bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol (Bis-Tris, pH 6.5), imidazole (pH 7), phosphate (pH 8), 2-amino-2-(hydroxymethyl)-1,3-propanediol chloride (Tris-HCl pH 7, 8, 9), N-cyclohexyl-2-aminoethanesulfonic acid (CHES pH 9, 9.5), and glycine (pH 10, 10.5).
Phosphate positively affects MjCobT activity.
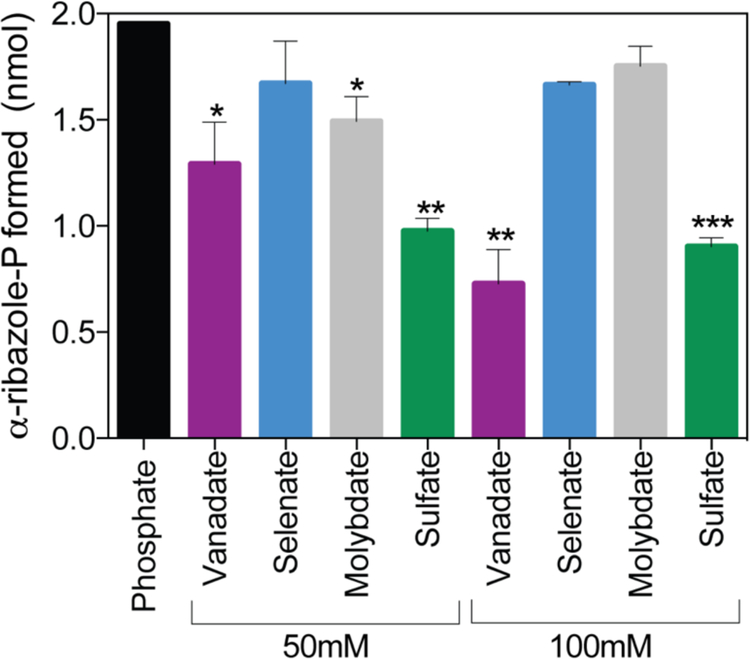
Given the precedent of positive effects of phosphate on the activity of another Cba biosynthetic enzyme from a hyperthermophile43, we varied the phosphate concentration in the reaction mixture to investigate the effect of phosphate on MjCobT activity. We varied the concentration of KPi in the reaction mixture (25–400 mM) and observed a modest increase in the amount of α-RP synthesized over a period of 15 min, from 1.0 to 1.8 nmol α-RP. To determine whether observed effect of activity was specific to phosphate, an equivalent experiment was performed with sulfate (Fig 8). The amount of DMB converted to α-RP by MjCobT was measured after incubation of enzyme (250 nM) with substrates at 80°C for 15 min in the presence of sulfate, phosphate or neither. In the absence of either sulfate or phosphate the enzyme converted 0.8 nmol of DMB (40% of the initial amount) to α-RP. To assess the effect of other oxyanions on the activity of MjCobT compared to phosphate, an equivalent experiment was performed with sodium salts of vanadate, selenate, or molybdate. Compared to phosphate (25 mM), none of the oxyanions tested stimulated MjCobT activity, even when tested at 50 mM or 100 mM (Fig. 8).
Figure 8. Phosphate positively affects MjCobT activity, vanadate, selenate, molybdate and sulfate do not.
Conditions for the PRTase reaction are described under Materials and Methods. Product formation is shown for each oxyanion provided at 50 mM and 100 mM compared to 25 mM phosphate. Error bars represent the standard error of the mean. Asterisks show significance determined by unpaired T test compared to phosphate condition.
Effect of ionic strength.
To determine the effect of salts on the activity of MjCobT, varying concentrations of sodium chloride and potassium chloride were tested (100–500 mM). Altering the ionic strength of the reaction system showed no significant change in the activity of MjCobT with either salt (data not shown).
Enzyme activity as a function of temperature.
We also probed the effect of temperature on the activity of MjCobT. As expected for an enzyme from a hyperthermophile, the activity of MjCobT increased as a function of temperature, exhibiting the highest activity (~4-fold increase from 37°C) at 80°C (data not shown).
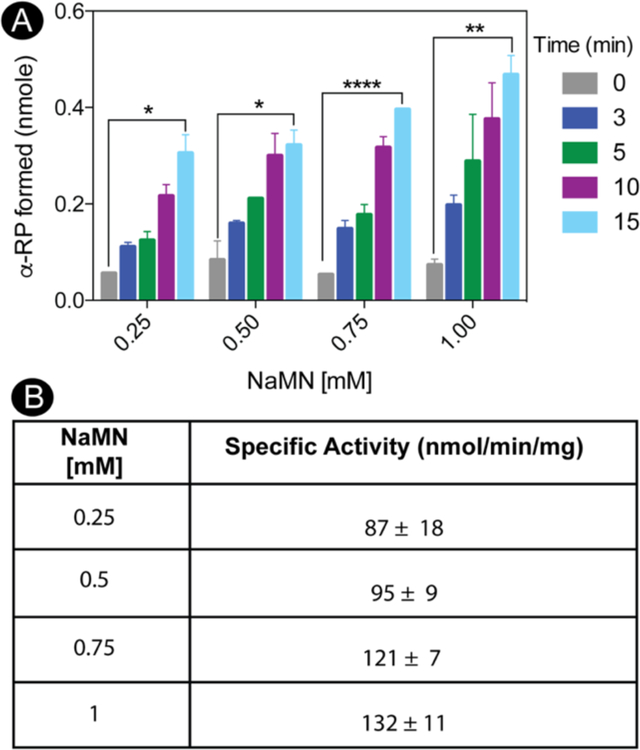
Enzyme activity as a function of NaMN.
As shown in figure 9A, the amount of MjCobT-generated α-RP is displayed as a function of time for each concentration of NaMN tested. The specific activity of MjCobT at each level of NaMN is shown in Figure 9B. MjCobT shows NaMN:DMB PRTase activity at a physiological pH, where SeCobT activity was previously undetectable42.
Figure 9. MjCobT activates 5,6- dimethylbenzimidazole to α-ribazole- phosphate (α-RP).
Conditions for the PRTase reaction are described under Materials and Methods. In panel A, product formation is shown as a function of time across varying NaMN concentrations. Product formation was quantified as a function of time. Error bars represent standard error of the mean. Asterisks shown significance determined by unpaired T test comparing time 0 to time 15. Panel B shows average specific activities with standard deviations for each concentration of NaMN tested.
Effect of salting out agents.
To assess the effect of crowding agents on the activity of MjCobT, varying amounts of ammonium sulfate (75–750 mM) were added to the reaction, but no significant changes in activity were observed (data not shown).
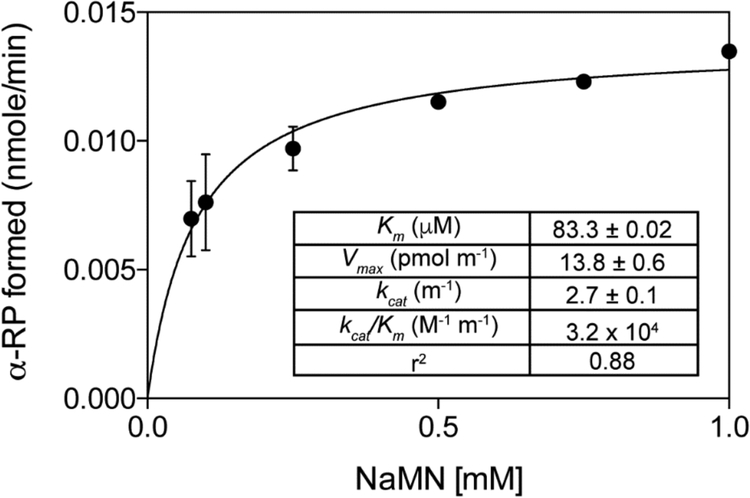
Pseudo-first order kinetics analysis of MjCobT.
Kinetic parameters for the reaction catalyzed by MjCobT were obtained with NaMN when DMB was provided at saturating levels (100μM). The rate of the reaction as a function of NaMN concentration is shown in figure 10 with the corresponding summary of apparent kinetic parameters shown in the inset. Kinetic parameters of MjCobT for DMB were not obtained because of the limit of product detection of the assay. The plot in figure 10 is missing points below 50 μM NaMN because below that concentration product formation was not detectable. Of note, the apparent Km of MjCobT for NaMN was 5-fold lower than that of SeCobT44.
Figure 10. Pseudo-first order kinetics of the MjCobT reaction as a function of NaMN.
In all reaction mixtures DMB was present at saturating levels (100 μM). Details of the protocol used can be found under Materials and Methods. Apparent kinetic parameters are shown in the inset.
Biologically active MjCobT is a dimer.
The oligomeric state of MjCobT was determined by analytical ultracentrifugation. MjCobT protein used for this analysis was 99% homogenous (Fig. 11A). Sedimentation velocity analysis showed that 27 μM MjCobT existed primarily as a 4.4 S (81.3%) species that was consistent with the expected size of a dimer (Fig. 11B). Examining the crystal structure with PISA identified a dimeric assembly with a reasonable buried interface (940 Å2) and a favorable P-value of 0.164, which is good evidence of a specific, stable dimer interface45 (Fig. 11C). The identified dimer gives a predicted sedimentation value of 4.4 S, which matches the experimentally observed species. Additional evidence supporting our identification of the dimer can be seen in the crystal structure of the distant homologue, CobT from Pyrococcus horikoshii (PDB 3U4G), which contains the same dimeric assembly despite a sequence identity of 37% (Fig. 11C).
Figure 11. Oligomeric state and purity of MyOobT.
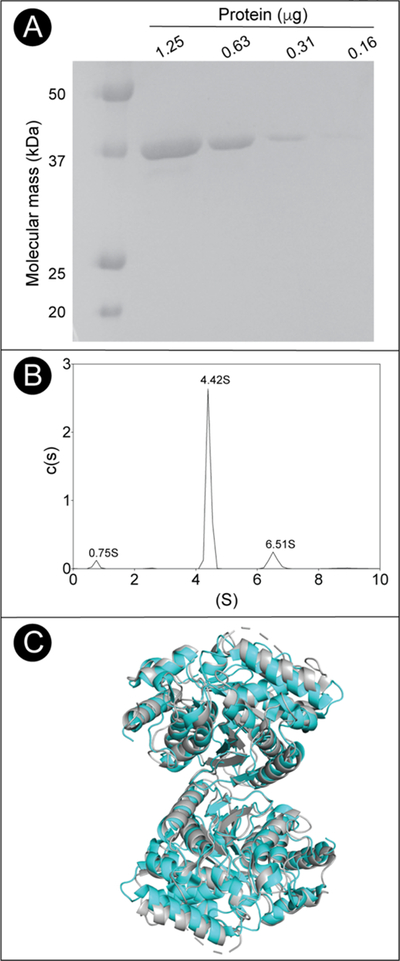
A. SDS-PAGE behavior of MjCobT compared to BioRad Precision Plus Protein Standards. Based on the data shown in panel B, MjCobT was estimated to be >99% homogeneous. B. Sedimentation velocity c(s) distribution of MjCobT at pH7.5 showing three species at 0.75 S, 4.42 S, and 6.51 S C. Cartoon model of the MjCobT dimer (cyan) identified in the crystal structure (PDB ID 3L0Z) superimposed onto the homologous CobT from Pyrococcus horikoshii dimer in grey (PDB ID 3U4G).
The sedimentation distribution also revealed a small amount of a 6.5 S species (12.7%), which was consistent with a predicted tetramer (6.7 S) formed from the association of two of the dimers. We believe that this species is most likely caused by aggregation of the protein, with the 0.2 difference in S value being the result of the aggregate forming a less compact structure than the modeled tetramer. Finally, there was a minor peak representing 3.8% of the distribution at 0.75S. Based on the small size, this is likely a minor contaminant in the sample.
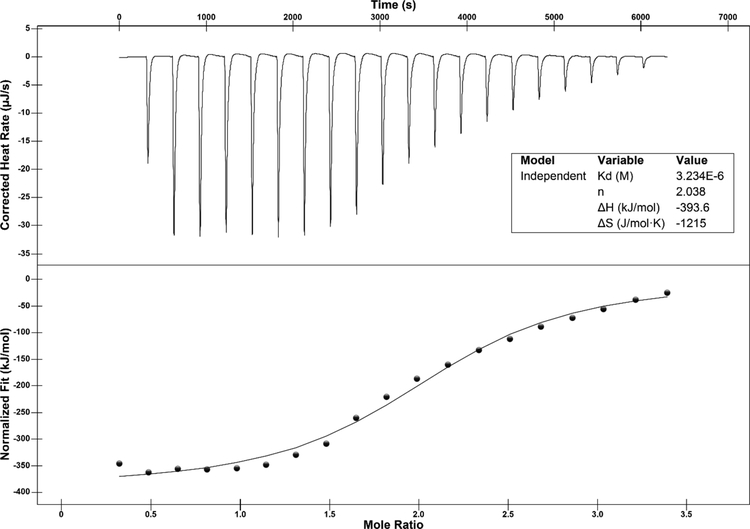
MjCobT has a high affinity for NaMN substrate.
Isothermal titration calorimetry (ITC) was used to determine the affinity of MjCobT for the NaMN substrate. The calculated dissociation constant (Kd) of the MjCobT/NaMN complex was 3.2 μM, indicating a high affinity for the substrate (Fig. 12). Given the low concentration of NaMN in the cell46, a high affinity for NaMN would be consistent with our expectations. ITC experiments aimed at determining the Kd of MjCobT for DMB were not performed due to incompatibility of the solvent used to solubilize DMB (DMSO) with the ITC instrument. The molar ratio of ~2 was consistent with two active sites per MjCobT dimer.
Figure 12. ITC analysis of NaMN binding to MjCobT.
Sample preparation for ITC analysis is described in detailed under Materials and Methods. Twenty 2.4-μL injections of 1 mM NaMN spaced every 5 min were made during the course of the experiment. The top panel shows the heat released per injection. The interaction between MjCobT and NaMN is shown as enthalpy per mol of NaMN injected as a function of the molar ratio of protein to ligand (bottom panel). The binding isotherm was obtained from the integrals of the peaks. Shown in the inset are the best-fit values for the dissociation constant (Kd), stoichiometry (n), enthalpic change (ΔH), and entropic change (ΔS).
MjCobT residues E150 and E315 are necessary for function.
In SeCobT, residues E174 and E317 are required for activity47. To investigate whether the equivalent residues in MjCobT, i.e., E150 and E315 (Fig. 4) were needed for activity, these residues were changed to alanine and the activity of the resulting variants was assessed in vivo. Concentration sweeps of DMB or adenine in the growth medium were performed to determine the minimum concentration of base necessary to confer growth of strains expressing variant proteins. Data from these experiments are shown in Table 1. In the presence of DMB (50 μM) the strain that synthesized variant MjCobTE150A had a doubling time similar to that of the strain that synthesized wild-type MjCobT (Table 1). The strain that synthesized variant MjCobTE150A failed to grow when adenine (1 mM) substituted for DMB in the medium. At present it is unclear whether the MjCobTE150A variant could not use adenine as substrate because it cannot bind it or because it is catalytically inactive. ITC experiments aimed at answering the question of binding could not be performed due to incompatibility of the solvent needed to keep adenine in solution.
Table 1.
Doubling times (h) of a S. enterica cobT cobB strain harboring plasmids encoding MjCobT proteins
| Variant (plasmid) | Cbi (15 nM) | Cbi + DMB | Cbi + Ade | Cbl (100 nM) |
| Vector (pBAD24) | NG | NG | NG | 1.2 ± 0.01[3 h lag] |
|
MjCobTWT (pMjCobT1) |
1.6 ± 0.01 [3 h lag] |
1.4 ± 0.01, [3 h lag] (10 nM) |
1.5 ± 0.01, [3 h lag] (10 nM) |
1.2 ± 0.01, [3 h lag] |
|
MjCobTE150A (pMjCobT5) |
NG | 2.0 ± 0.02, [3 h lag] (50 μM) 3.4 ± 0.12, [20 h lag] (1 μM) |
NG (1mM) | 1.5 ± 0.01, [4 h lag] |
|
MjCobTE315A (pMjCobT3) |
NG | 1.6 ± 0.01, [3 h lag] (5 μM) 2.0 ± 0.19[19 h lag] (50 nM) |
2.2 ± 0.03, [6 h lag] (0.5 mM) |
1.8 ± 0.03, [4 h lag] |
|
MjCobTE150A E315A (pMjCobT4) |
NG | NG | NG | 1.9 ± 0.17, [4 h lag] |
Cells were grown in NCE minimal medium supplemented with ribose (20mM) as the sole carbon and energy source. Doubling times are shown in hours with the standard error of the mean. Lag times are shown in brackets. Concentrations of added ring precursor or base are shown in parenthesis. NG, no growth; Cbi, dicyanocobinamide [(CN)2Cbi]; DMB, 5,6-dimethylbenzimidazole; Cbl, cyanocobalamin (CNCbl, vitamin B12); Ade, adenine
In contrast, the MjCobTE315A variant restored growth with 10-fold less DMB (5 μM) in the medium, suggesting that the E315A substitution did not negatively affect enzyme activity as much as the E150A substitution. Unlike MjCobTE150A, MjCobTE315A did support psCbl synthesis, even when the concentration of adenine in the medium was reduced to 0.5 mM (Table 1).
The strain that synthesized the double variant MjCobTE150A,E315A failed to synthesize AdoCbl or psCbl when inoculated into medium containing DMB or adenine, suggesting that in spite of their structural differences, SeCobT and MjCobT likely catalyze the reaction via similar mechanisms. It was previously suggested that in archaeal orthologues, an acidic residue at the position equivalent to SeCobT E174 might be necessary and sufficient for activity47.
This idea was based on analyses of structural alignments which showed a valine residue in place of SeCobT E317 in two archaeal orthologues, one of them being MjCobT. Here we show that the MjCobT E315 is in fact, the equivalent to SeCobT E317 and both acidic residues in the active site are necessary for function in vivo.
MjCobT homologues are commonly found in methanogenic archaea.
Using MjCobT as a query sequence, we identified many homologues in both class I and class II methanogens (Table S3). Homologues identified in class I methanogens showed a high level of identity and similarity to MjCobT, while class II methanogens showed a more divergent homologue. On the basis of phylogenetic analysis, MjCobT is likely representative of the class of NaMN:DMB phosphoribosyltransferases found in class I methanogenic archaea.
The cobT+ allele of the cyanobacterium Synechococcus sp. restores AdoCba biosynthesis in a CobT-deficient S. enterica strain when DMB is provided.
To determine whether Synechococcus sp. CobT had NaMN:Base PRTase activity in vivo, we introduced a plasmid encoding Synechococcus sp. WH7803 cobT+ into a S. enterica ΔcobT strain to attempt to restore AdoCbl synthesis in the absence of SeCobT function. ORF SYNWH7803_RS09275 (i.e., Synecococcus cobT+) placed under the control of an arabinose-inducible promoter in plasmid pCV127 was moved into strain JE12893 (ΔcobT ΔcobB) yielding strain JE24636 (ΔcobT ΔcobB / pSynCobT2) (Table S1). Strain JE12893 failed to grow without exogenous CNCbl (Fig. 13, inverted triangles), but grew when CNCbl was present in the medium (Fig. 13, diamonds). Ectopic expression of Synechococcus sp. cobT+ allowed strain JE24636 to grow when (CN)2Cbi and DMB were present in the medium. Strain JE24636 failed to grow when only (CN)2Cbi was provided. This result indicated Synechococcus sp. CobT supported SeCobT-like activity in vivo when DMB was provided (Fig. 13, triangles). Previously, cyanobacterial species were shown to solely produce psCbl. Our findings indicate Synechococcus sp. CobT can activate DMB, ultimately yielding AdoCbl. Strain JE18115 (ΔcobT ΔcobB / pCOBT140) served as positive control for CobT function (Fig. 13, squares).
Figure 13. Synechococcus CobT supports AdoCbl-dependent growth of S. enterica in the absence of cobT and cobB.
S. enterica strains were grown in minimal medium supplemented with ribose (20 mM). (CN)2Cbi (15 nM) and DMB (150 μM) were added to all cultures except to the one to which CNCbl was added (diamonds). Synechococcus CobT (triangles) and SeCobT (squares) were expressed in trans in a ΔcobT ΔcobB S. enterica strain. A cobT+ cobB+ S. enterica strain (circles) served as positive control. A S. enterica strain deficient in CobT and CobB activity served as negative control (inverted triangles).
Phylogenetic analysis suggests that CobT in cyanobacteria has an archaeal origin.
Previous studies have suggested that the majority of cyanobacterial genomes lack cobT and that none of the sequenced Synechococcus genomes encode cobT17. When searching for homologues using CobT from S. enterica, P. denitrificans, and B. megaterium, our findings were consistent with the data reported by Helliwell et. al (Table S4). With the identification of the archaeal cobT from M. jannaschii, we found numerous cobT homologues in the genomes of cyanobacteria, including Synechococcus (Table S5). The report by Helliwell et. al. showed that Synechococcus solely produced psCbl even when provided with DMB (1μM) to “guide” the biosynthesis of cobalamin. The sole synthesis of psCbl by Synechococcus may be attributed to a lack of DMB transport and may not indicate the exclusive production of psCbl. Our bioinformatics findings in combination with the complementation of Cbl-dependent growth by a Synechococcus CobT homologue suggest that cyanobacteria can activate DMB via the canonical NaMN:DMB phosphoribosyltransfer mechanism. These findings also suggest that cyanobacteria can produce Cbl in addition to psCbl.
Our phylogenetic analysis shows that the CobT homologues found in archaea and cyanobacteria cluster into a distinct clade separate from the homologues found in Firmicutes and proteobacteria (Fig 14). Within this clade, the majority of the cyanobacterial CobT homologues cluster tightly within a major clade and are distinctly separate from the archaeal CobTs which form another major clade. This likely suggests that the cyanobacterial CobTs are of archaeal origin and significantly differ from the CobTs found in Firmicutes and Proteobacteria. This difference is likely why previous studies did not identify CobT homologues in cyanobacteria.
Previous work has implicated thaumarchaeota as major Cbl producers in marine environments16. Previously reported genome analyses performed showed the absence of cobT from all thaumarchaeota and suggested that an alternative pathway for DMB activation existed in these microorganisms. Our identification of archaeal MjCobT prompted the search for homologues in thaumarchaeota. We found that cobT is commonly found in thaumarchaeotal genomes (Table S6). Given the prevalence of cobT in thaumarchaeotal genomes and the previously reported production of cobalamin, it is likely that thaumarchaeota activate DMB using a NaMN:DMB phosphoribosyltransferase enzyme.
CONCLUDING REMARKS
This study reports a new class of NaMN:DMB phosphoribosyltransferases (CobT) enzymes involved in cobamide biosynthesis in archaea and in cyanobacteria, but not in bacteria. Structurally, there are significant differences between MjCobT and SeCobT, most notably the absence of the small domain in MjCobT which is essential in forming the active site of SeCobT. MjCobT groups with both the CobT homologues found in archaea and cyanobacteria and this group is highly divergent from the CobT homolgues found in bacteria. MjCobT sequence identity compared to SeCobT is too low to be detected by NCBI BLASTp. Additionally, we see a greater degree of amino acid sequence similarity between MjCobT and the cyanobacterial and archaeal CobT homologues than we see compared to bacterial CobT. Given this information, we feel MjCobT represents a class of NaMN:DMB phosphoribosyltransferase that is significantly different from bacterial CobT. In addition, this work corrects information regarding the oligomeric state of the PRTase from Methanocaldococcus jannaschii (PDB 3L0Z), whose crystal structure suggests it is a trimer. Our data indicate that in solution, the biologically active MjCobT enzyme is dimeric.
Supplementary Material
ACKNOWLEDGEMENTS
The authors have no conflict of interest to declare. This work was supported by NIH grant R37-GM040313 to J.C.E.-S.
Footnotes
REFERENCES
- [1].Mattes TA, Deery E, Warren MJ, and Escalante-Semerena JC (2017) Cobalamin Biosynthesis and Insertion, In Encyclopedia of Inorganic and Bioinorganic Chemistry (Scott RA, Ed.), pp 1–24, John Wiley & Sons, Ltd, Chichester, UK. [Google Scholar]
- [2].Kräutler B, Fieber W, Osterman S, Fasching M, Ongania K-H, Gruber K, Kratky C, Mikl C, Siebert A, and Diekert G (2003) The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is Norpseudo-B12, a new type of natural corrinoid., Helv. Chim. Acta 86, 3698–3716. [Google Scholar]
- [3].Marsh EN (1999) Coenzyme B12 (cobalamin)-dependent enzymes, Essays Biochem 34, 139–154. [DOI] [PubMed] [Google Scholar]
- [4].Yan J, Bi M, Bourdon AK, Farmer AT, Wang PH, Molenda O, Quaile AT, Jiang N, Yang Y, Yin Y, Simsir B, Campagna SR, Edwards EA, and Löffler FE (2018) Purinyl-cobamide is a native prosthetic group of reductive dehalogenases, Nat. Chem. Biol 14, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jones AR, Rentergent J, Scrutton NS, and Hay S (2015) Probing reversible chemistry in coenzyme B12 -dependent ethanolamine ammonia lyase with kinetic isotope effects, Chemistry (Easton) 21, 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bobik TA, Xu Y, Jeter RM, Otto KE, and Roth JR (1997) Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase., J. Bacteriol 179, 6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, and Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods., Nucleic Acids Res 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Padmanabhan S, Jost M, Drennan CL, and Elias-Arnanz M (2017) A new facet of vitamin B12: Gene regulation by cobalamin-based photoreceptors, Annu. Rev. Biochem 86, 485–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Renz P (1999) Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of other bases found in natural corrinoids, In Chemistry and Biochemistry of B12 (Banerjee R, Ed.), pp 557–575, John Wiley & Sons, Inc., New York. [Google Scholar]
- [10].Johnson MG, and Escalante-Semerena JC (1992) Identification of 5,6-dimethylbenzimidazole as the Coα ligand of the cobamide synthesized by Salmonella typhimurium. Nutritional characterization of mutants defective in biosynthesis of the imidazole ring, J. Biol. Chem 267, 13302–13305. [PubMed] [Google Scholar]
- [11].Keck B, and Renz P (2000) Salmonella typhimurium forms adenylcobamide and 2-methyladenylcobamide, but no detectable cobalamin during strictly anaerobic growth, Arch. Microbiol 173, 76–77. [DOI] [PubMed] [Google Scholar]
- [12].Anderson PJ, Lango J, Carkeet C, Britten A, Kräutler B, Hammock BD, and Roth JR (2008) One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica, J. Bacteriol 190, 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Trzebiatowski JR, and Escalante-Semerena JC (1997) Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2, J. Biol. Chem 272, 17662–17667. [DOI] [PubMed] [Google Scholar]
- [14].Cheong CG, Escalante-Semerena JC, and Rayment I (2001) Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica, J. Biol. Chem 276, 37612–37620. [DOI] [PubMed] [Google Scholar]
- [15].Hazra AB, Tran JL, Crofts TS, and Taga ME (2013) Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin B(12), Chem. Biol 20, 1275–1285. [DOI] [PubMed] [Google Scholar]
- [16].Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote-Maestas W, Hmelo LR, Moffett JW, Devol AH, Armbrust EV, Stahl DA, and Ingalls AE (2017) Two distinct pools of B12 analogs reveal community interdependencies in the ocean, Proc. Natl. Acad. Sci. U S A 114, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Krautler B, Scanlan DJ, Warren MJ, and Smith AG (2016) Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12, Curr. Biol 26, 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Berkowitz D, Hushon JM, Whitfield HJ Jr., Roth J, and Ames BN (1968) Procedure for identifying nonsense mutations, J. Bacteriol 96, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Balch WE, Fox GE, Magrum LJ, Woese CR, and Wolfe RS (1979) Methanogens: reevaluation of a unique biological group, Microbiol. Rev 43, 260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].O’Toole GA, Rondon MR, and Escalante-Semerena JC (1993) Analysis of mutants of defective in the synthesis of the nucleotide loop of cobalamin, J. Bacteriol 175, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Maniatis T, Fritsch EF, and Sambrook J (1982) Introduction of plasmid and bacteriophage lambda into Escherichia coli, In Molecular cloning: a laboratory manual (Maniatis T, Fritsch EF, and Sambrook J, Eds.), pp 250–251, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. [Google Scholar]
- [22].Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products., Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klock HE, Koesema EJ, Knuth MW, and Lesley SA (2008) Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts, Prot.-Struct. Funct. Bioinformat 71, 982–994. [DOI] [PubMed] [Google Scholar]
- [24].Guzman LM, Belin D, Carson MJ, and Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter, J. Bacteriol 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rocco CJ, Dennison KL, Klenchin VA, Rayment I, and Escalante-Semerena JC (2008) Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli, Plasmid 59, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mattes TA, and Escalante-Semerena JC (2017) Salmonella enterica synthesizes 5,6-dimethylbenzimidazolyl-(DMB)-alpha-riboside. Why some Firmicutes do not require the canonical DMB activation system to synthesize adenosylcobalamin, Mol. Microbiol 103, 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].VanDrisse CM, and Escalante-Semerena JC (2016) New high-cloning-efficiency vectors for complementation studies and recombinant protein overproduction in Escherichia coli and Salmonella enterica, Plasmid 86, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kittaka-Katsura H, Fujita T, Watanabe F, and Nakano Y (2002) Purification and characterization of a corrinoid compound from Chlorella tablets as an algal health food, J. Agric. Food Chem 50, 4994–4997. [DOI] [PubMed] [Google Scholar]
- [29].Keller S, Treder A, SH VR, Escalante-Semerena JC, and Schubert T (2016) The SMUL_1544 gene product governs norcobamide biosynthesis in the tetrachloroethenerespiring bacterium Sulfurospirillum multivorans, J. Bacteriol 198, 2236–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Blanche F, Thibaut D, Couder M, and Muller JC (1990) Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography, Anal. Biochem 189, 24–29. [DOI] [PubMed] [Google Scholar]
- [31].Chan CH, and Escalante-Semerena JC (2011) ArsAB, a novel enzyme from Sporomusa ovata activates phenolic bases for adenosylcobamide biosynthesis, Mol. Microbiol 81, 952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Second ed, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- [33].Blommel PG, and Fox BG (2007) A combined approach to improving large-scale production of tobacco etch virus protease, Protein Expr. Purif 55, 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, and Bairoch A (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis, Nucleic Acids Res 31, 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Laue TM, Shah BD, and Ridgeway TM (1992) Analytical ultracentrifugation in biochemistry and polymer science, R. Soc. Chem, 90–125. [Google Scholar]
- [36].Laue TM, Shah BD, Ridgeway TM, and Pelletier SL (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science, R. Soc. Chem, 90–125. [Google Scholar]
- [37].Schuck P (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation, Anal. Biochem 320, 104–124. [DOI] [PubMed] [Google Scholar]
- [38].Ortega A, Amoros D, and Garcia de la Torre J (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models, Biophys. J 101, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, and Bryant SH (2009) CDD: specific functional annotation with the Conserved Domain Database, Nucleic Acids Res 37, D205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gouy M, Guindon S, and Gascuel O (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building, Mol. Biol. Evol 27, 221–224. [DOI] [PubMed] [Google Scholar]
- [42].Claas KR, Parrish JR, Maggio-Hall LA, and Escalante-Semerena JC (2010) Functional analysis of the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) enzyme, involved in the late steps of coenzyme B12 biosynthesis in Salmonella enterica, J. Bacteriol 192, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Woodson JD, and Escalante-Semerena JC (2006) The cbiS gene of the archaeon Methanopyrus kandleri AV19 encodes a bifunctional enzyme with adenosylcobinamide amidohydrolase and alpha-ribazole-phosphate phosphatase activities, J. Bacteriol 188, 4227–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maggio-Hall LA, and Escalante-Semerena JC (2003) Alpha-5,6-Dimethylbenzimidazole adenine dinucleotide (alpha-DAD), a putative new intermediate of coenzyme B12 biosynthesis in Salmonella typhimurium, Microbiology 149, 983–990. [DOI] [PubMed] [Google Scholar]
- [45].Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- [46].Bochner BR, and Ames BN (1982) Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography, J. Biol. Chem 257, 9759–9769. [PubMed] [Google Scholar]
- [47].Chan CH, Newmister SA, Talyor K, Claas KR, Rayment I, and Escalante-Semerena JC (2014) Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica, Biochim. Biophys. Acta 1840, 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.