Abstract
Background
Redox metabolism is often considered a potential target for cancer treatment, but a systematic examination of redox responses in hepatocellular carcinoma (HCC) is missing.
Methods
Here, we employed systems biology and biological network analyses to reveal key roles of genes associated with redox metabolism in HCC by integrating multi-omics data.
Findings
We found that several redox genes, including 25 novel potential prognostic genes, are significantly co-expressed with liver-specific genes and genes associated with immunity and inflammation. Based on an integrative analysis, we found that HCC tumors display antagonistic behaviors in redox responses. The two HCC groups are associated with altered fatty acid, amino acid, drug and hormone metabolism, differentiation, proliferation, and NADPH-independent vs -dependent antioxidant defenses. Redox behavior varies with known tumor subtypes and progression, affecting patient survival. These antagonistic responses are also displayed at the protein and metabolite level and were validated in several independent cohorts. We finally showed the differential redox behavior using mice transcriptomics in HCC and noncancerous tissues and associated with hypoxic features of the two redox gene groups.
Interpretation
Our integrative approaches highlighted mechanistic differences among tumors and allowed the identification of a survival signature and several potential therapeutic targets for the treatment of HCC.
Keywords: Hepatocellular carcinoma, Redox metabolism, Systems biology, Precision medicine, Cancer, Transcriptomics, Liver cancer
Research in context.
Evidence before this study
Imbalances in redox metabolism are prevalent features in cancer, influencing tumorigenesis and proliferation. Because of this, several recent efforts have sought to target redox and antioxidant metabolism in cancer. However, a systematic examination and characterization of redox metabolism has never been performed in hepatocellular carcinoma, hindering our understanding of redox behavior and its relationship with metabolism, signaling, or patient clinical data.
Added value of this study
We performed a systems level multi-omics analysis of redox metabolism and identified two major opposing clusters of genes and their associations with central metabolism, differential metabolic responses, immunity and inflammation. Tumors stratified according to redox gene clusters display substantial progression and survival differences and subtype-specific potential therapeutic targets. Experiments with mouse models validated the antagonistic behavior and highlighted differential hypoxic associations for redox genes. We finally identified and validated a survival signature derived based on genes associated with redox metabolism.
Implications of all the available evidence
Our observations highlighted, for the first time, the antagonistic relationships between key genes in redox metabolism. Furthermore, while redox genes are heterogeneously expressed, HCC tumors may be stratified into two groups with distinct phenotypes, potential therapeutic targets, and associated patient survival.
Alt-text: Unlabelled Box
1. Introduction
Hepatocellular carcinoma (HCC) is a prevalent form of primary liver cancer and represents the second leading cause of worldwide cancer mortality [1,2]. Its prevalence is predicted to increase in the next two decades, and the prognosis is usually poor, with most diagnosed cases resulting in death [3]. Due to the high inter- and intra-tumor heterogeneity, it is challenging to establish a robust HCC classification and develop targeted therapies [4]. Hence, there is an urgent need to stratify patients to reveal the underlying molecular mechanisms of the disease that may be used in the development of targeted and effective treatment strategies.
Reactive oxygen species (ROS) such as hydrogen peroxide and superoxide are central players in redox metabolism and have important cellular functions in which they control signaling, cell cycle progression, proliferation, inflammation and immune responses [5]. However, the deregulation of redox and ROS metabolism may be toxic for cells, disrupting pathways and promoting mutagenesis and apoptosis [6,7], which are important features in cancer [8]. Due to their pivotal cellular functions, recent efforts have sought to devise ROS-based cancer treatments [9]. However, effective redox-based HCC therapeutics are currently lacking due to our incomplete understanding of the underlying redox mechanisms of the disease in different patient groups, and the extensive crosstalk between redox and central metabolism [5]. Systems level analyses have previously been employed to unravel cancer heterogeneity, to identify novel prognostic genes, therapeutic targets, and drug candidates and to enable patient stratification [[10], [11], [12], [13], [14]], including for HCC [[15], [16], [17]].
Here we used systems biology approaches (Fig. S1A) to reveal the heterogeneous redox responses in HCC combining transcriptomics (RNA-seq and an independent set of microarray data), proteomics and metabolomics data. We first identified several patterns of co-expression among genes involved in redox metabolism, liver-specific genes, and genes associated with immunity and inflammation that have prognostic properties based on integrative network analyses. Second, we identified several associations between the genes involved in redox and central metabolism. Next, we identified specific functional patterns between important ROS scavengers and how they are coupled to the central metabolism of the tumors. Tumor stratification based on the expression of redox genes and analysis of clinical data allowed the discovery of potential markers for effective treatment strategies of HCC patients and novel survival signatures based on the expression of key redox genes. Finally, we validated our results by performing experiments using HCC mouse models and analyzing independent human HCC tumors.
2. Materials & methods
2.1. Identification of redox genes
We identified 132 genes directly involved in redox metabolism (Dataset 1). This list included genes encoding for proteins 1. with antioxidant and/or ROS-dependent signaling activities (e.g., superoxide dismutases, catalase, peroxidases, peroxiredoxins, thioredoxins); 2. involved in reactions coupled with antioxidant defense or ROS-mediated signaling transduction (e.g., thioredoxin and glutathione reductases); 3. involved in producing compounds with relevant redox roles (e.g., glutathione synthetase, glutamate-cysteine ligase); 4. enzymes involved in reducing equivalent biosynthesis from pentose phosphate pathway (e.g., glucose- and hexose-6-phosphate dehydrogenases), folate metabolism (e.g., tetrahydrofolate synthase, bifunctional methylenetetrahydrofolate dehydrogenase), malate metabolism (malic enzymes), and other processes (e.g., NAD(P)H dehydrogenase, NAD(P) transhydrogenase, aldehyde and glutamate dehydrogenases); 5. transcription factors (e.g., hypoxia-inducible factor 1-alpha, nuclear factor erythroid 2-related factor 2) that coordinate responses to oxidative stress. This list was used to perform the initial hierarchical clustering, and co-expression analysis with all the genes in the genome. Due to the high co-expression between redox genes and those involved in central metabolic pathways, we expanded the list to 174 redox genes (Dataset 1), including those indirectly involved in redox metabolism with known importance for cancer and/or that control or are regulated by the above pathways (e.g., glycolytic enzymes).
2.2. Data selection and gene expression analysis
RNA-seq gene expression data for primary liver hepatocellular carcinoma in 360 subjects and 50 noncancerous samples were retrieved from NCI's Genome Data Commons [18] as raw counts or Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Only one sample was considered per patient, and samples belonging to control or normal types were excluded. Patient survival, expressed in Living Days, was identified by considering age at diagnosis for deceased patients, or days to last follow up for those alive. Unless otherwise stated, all analyses were performed based on FPKM data, and genes displaying a median (FPKM) < 1 across patients were considered as not detected.
In the cancer progression analyses, we considered grade and not stage classification of tumors because the former is a histological classification of the tumor and the latter takes into account other features that are not related to the primary tumor. In total, 55 Grade 1, 175 Grade 2, 118 Grade 3 and 12 Grade 4 patients were considered and divided according to early (G1), intermediate (G2) and late (G3 + G4) cancer.
For a smaller subset of the tumor samples (186), more detailed clinical information was available [13] and used to assess relationships with redox behavior.
Liver-specific and enriched genes, and group-enriched genes, were attained from ref. 25. Genes involved in the immune system (adaptive and innate immunity, and cytokine signaling in immune system) and inflammation (CLEC7A/inflammasome pathway, DEx/H-box helicases activate type I IFN and inflammatory cytokines production, inflammasomes) were identified using REACTOME [64].
2.3. Hierarchical clustering, differential expression and co-expression analysis
Hierarchical clustering was performed in R considering Ward.D2 and the Euclidean distance. Whenever clustering was performed based on gene expression matrices, genes were Z-normalized before clustering. Differential expression analyses were performed based on raw counts through R using the DESeq2 package [65] using default methods and considering genes with absolute log fold changes higher than 1 by setting the results function with arguments altHypothesis = “greaterAbs” and lfcThreshold = 1 and the DEseq function with argument betaPrior = FALSE. An FDR of 5% was set during the differential expression. Although the filtering based on the log fold change and FDR may be performed a posteriori, specifying these arguments upon performing the differential expression analysis is recommended for independent gene filtering and significance of the Wald testing [65]. Differential expression analysis was performed by partitioning subjects according to survival (in days), or between tumors displaying high vs low expression levels of redox genes. Because gene expression expressed in raw counts could not be compared across different samples, we sorted subjects according to the FPKM expression of the target gene and then identified the two groups that displayed highest and lowest gene expression, each with 50 individuals. Genes displaying median FPKM < 1 across individuals were ignored.
Spearman rank correlations were computed for gene vs gene expression across tumors, and Q values were used to find co-expressed genes. Genes with expression that was not statistically correlated (Q > 0.01) with at least one gene in the genome were disregarded.
2.4. Gene set enrichment analysis and reporter metabolite analysis
GSEA was performed through PIANO [66] using the whole-genome log2-fold changes and adjusted P values attained from DESeq2 as gene-level statistics, with geneSetStat = reporter, and nPerm = 1000. Gene Ontology biological processes were downloaded from MSigDB [67]. Gene Ontology processes were considered as enriched with an FDR of 5% and with clear direction (i.e., non-mixed). Additionally, we ignored Gene Ontology processes related to tissues/organs of different embryonic origin (e.g., brain, bone, hair). Reporter metabolites [28] were identified through PIANO using the same input as above through the function runGSA with default arguments and based on fold changes. Gene set collections were determined by assigning ensemble ids to each reaction's metabolites using the iHCC model [15]. Only those metabolites with clear direction were considered. The concept of reporter metabolites and their application in the determination of gene-set statistics and associated P values was detailed by Refs. [28,66].
After selection of liver-specific genes, as well as genes involved in the immune system and inflammation co-expressed with redox genes, gene set enrichment analysis was performed with BINGO in Cytoscape [68], and the significantly enriched biological processes (Q < 0.05) were reported.
2.5. Tumor stratification and comparison
Tumor stratification was carried out through Consensus Clustering [29,31] for those genes displaying a median FPKM > 1 across all samples after row-normalization of gene expression. This unsupervised method permits the determination of an optimum (i.e., stable) number of non-overlapping clusters. Briefly, the data were resampled 1000 times by considering 80% sample (i.e., tumor) and feature (genes) resampling to achieve robust clustering. Resampled data were then transformed into a similarity matrix, the consensus matrix. Agglomerative hierarchical clustering was performed to stratify tumors using the consensus matrix based on Pearson correlation distances through the R package ConsensusClusterPlus [31]. The optimum cluster number was determined by testing 2 to 10 clusters, and based on CDF and Δ(K), the area that increased in the cumulative distribution function increased as the number of clusters.
2.6. Genome-scale metabolic modeling and essentiality analysis
RNA expression data were integrated into the iHCC models to generate personalized genome-scale metabolic models using tINIT [15] and RAVEN [69]. The following thresholds for gene levels were considered: no expression (FPKM < 1), low expression (1 ≤ FPKM < 10), medium expression (10 ≤ FPKM < 50), and high expression (FPKM ≥ 50). Upper and lower bounds of the exchange reactions considered were based on Ref. [70].
To model HCC progression or the differences between tumor clusters, we used MADE [34] and TIGER [35] and used as input the gene-specific fold changes and Q values. MADE integrates expression data and a metabolic model without the prior definition of activity thresholds. MADE utilizes an optimization-based approach to generate a sequence of expression states reflecting the most significant changes in the series of gene expression measurements, simultaneously solving the Flux Balance Analysis problem for each condition, leading to a sequence of functioning models. To implement MADE, we used TIGER, which uses a custom MATLAB class CMPI to solve mathematical programming problems. The Common Mathematical Programming Interface defines a consistent structure for mixed-integer linear programming and mixed-integer quadratic programming problems, providing independence from the underlying mixed-integer linear programming solver software, and MADE is formulated as a single mixed-integer linear programming problem. CPLEX solver was also employed. In this study, genes were considered multilevel instead of in a binary state during the implementation of MADE. Using MADE and TIGER, we reconstructed grade-specific genome-scale metabolic models on the basis of their corresponding gene expression data. We integrated the expression data achieved from DESeq2 [65] and considered the upper and lower bounds for exchange reactions in GEMs based on experimental liver data [70].
Here we considered three processes as important cellular objectives by considering the following fake reactions as objective functions for maximization: biomass production, NADPH production and H2O2 scavenging (see below). These objective functions maximize the fluxes of NADPH oxidation and H2O2 production, such that the optimization method redirects the cell's resources towards reducing NADP+ and decomposing H2O2.
| => Biomass |
| NADPH[c] + NADPH[m] + NADPH[p] + NADPH[r] => => NADP+[c] + NADP+[m] + NADP+[p] + NADP+[r] + H+[c] + H+[m] + H+[p] + H+[r] |
| 2 H2O[c] + 2 H2O[m] + 2 H2O[p] + O2[c] + O2[m] + O2[p] => => 2 H2O2[c] + 2 H2O2[m] + 2 H2O2[p] |
2.7. Survival analysis and signature identification
Kaplan-Meier survival analysis was performed to compare two groups of stratified tumors and based on the 10-year survival data for all 360 patients in R through the package survival [71].
To determine the best survival signatures, we generated all possible signatures consisting of 3 or 4 genes by taking the average expression of the selected genes in tumors from the hALDH2 cluster. Then, all 360 patients were stratified based on the correlation between their expression of selected genes and the signature using a cutoff that was maximally selected similarly to our previous work [27]. The minimum group size considered was 20% of the total sample size. The signature that presented the lowest log-rank P value after multiple hypothesis correction among all the investigated gene combinations was then selected as the best one. KM analysis had been performed for single gene predictions [27], and was corrected for multiple hypothesis testing considering all redox genes.
2.8. Microarray and metabolomic analyses
Three separate microarray cohorts were used to validate the co-expression and gene expression patterns observed using transcriptomics. The microarray for 61 HCC samples was downloaded from GEO with accession number GSE76297 (Ref. [14]) for the Affymetrix Human Transcriptome Array 2.0 platform. Row data were normalized using the Robust Multi-array Average followed by quantile normalization. Log2-transformed data was then analyzed in R, with probes annotated through the hta20sttranscriptcluster.db package. Normalization of the second cohort of 91 HCC samples with accession GSE1898 was carried out as previously indicated [32,33]. The final microarray cohort of 221 hepatocellular carcinoma patients was retrieved from GSE14520 and tumor normalized data was used for all analyses [42,43].
The metabolomics data of the 127 most variable metabolites between HCC and intrahepatic cholangiocarcinoma, after ignoring unidentified metabolites, were analyzed for the 54 HCC samples with both gene expression and metabolomics data [14]. Correlations between gene vs gene, and gene vs metabolite were computed across the 61 and 54 HCC samples, respectively.
2.9. mRNA expression of redox genes in murine HCC samples
Total RNA was isolated from tumor and noncancerous liver tissues from six sets of DEN-treated (HCC) CB1R+/+ mice and CB1R−/− mice [46]. Next, 100 ng/sample of rRNA-depleted RNA was treated with RNase III to generate 100- to 200-nt fragments, which were pooled and processed for RNA sequencing. All data were normalized based on housekeeping genes through the Program CLC Genomics Workbench (version 5.1, CLC Bio). Absolute numbers were extracted from the reads, and the data were adjusted to non-HCC biopsies for each gene.
2.10. Statistical analyses
Statistical approaches were chosen according to the diverse data types in this study and indicated throughout. Hypothesis testing was performed by considering the null hypothesis of the absence of an association between the compared variables. The association with clinical features was tested according to the nature of the data: continuous vs continuous (Spearman rank correlation test); continuous vs categorical (Kruskal-Wallis test); categorical vs categorical (Fisher's exact test for 2 × 2 tables, or Chi-squared test). Q values were computed from the P values (false discovery rate with Benjamini–Hochberg procedure, family-wise error rate 0.05).
2.11. Data availability
The GEMs used in this work are found in SBML format (Computer Code 1). The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files, or otherwise clearly point to the online resources where they were obtained.
3. Results
3.1. Redox genes are co-expressed with liver-enriched, immune and inflammation genes, and differentiation markers
To clarify the role of genes involved in redox metabolism, we identified a panel of 132 protein-coding genes directly involved in this process through a literature search (Methods, Dataset 1). Specifically, we selected genes involved in i) ROS production, scavenging, and metabolism; ii) the metabolism of reducing equivalents and iii) genes that are essential for the response to oxidative stress using a generic human Genome-scale Metabolic Model (GEM) [15] and other publicly available biochemical reaction databases. We retrieved the gene expression data of 360 HCC tumors together with patient metadata from NIH's TCGA [18] and found that HCC tumors display highly heterogeneous expression of genes that are directly involved in redox metabolism (Fig. 1A).
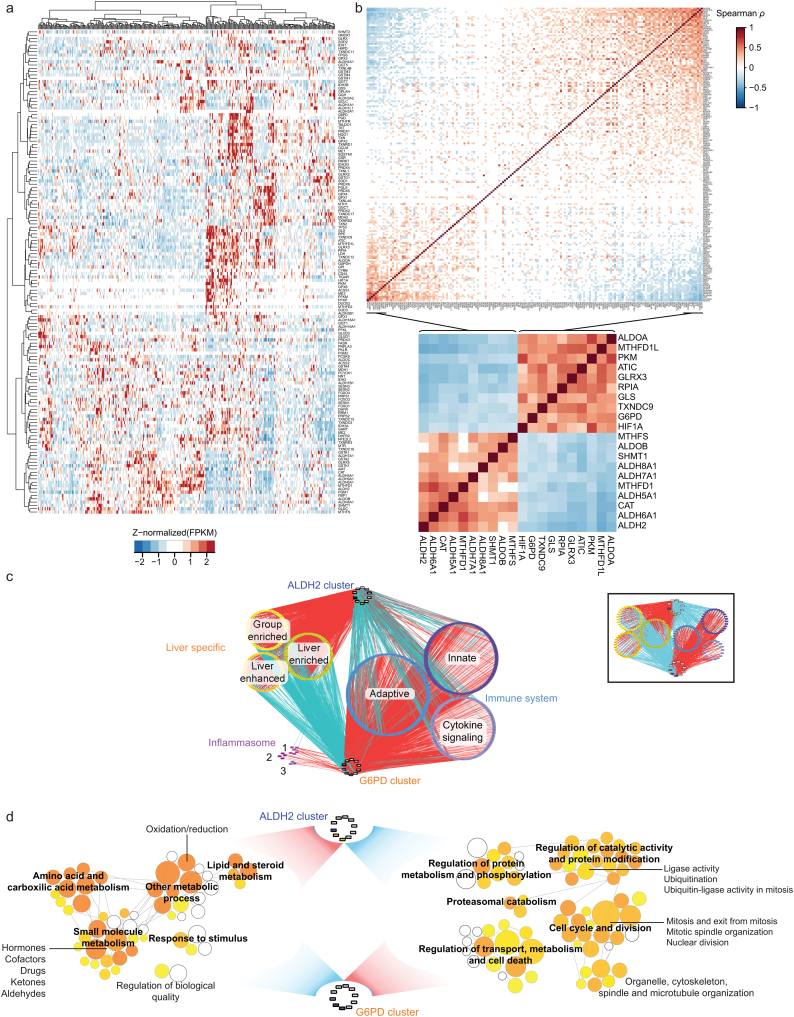
Fig. 1.
Highly heterogeneously expressed redox genes comprise two major clusters of co-expressed redox genes, highly co-expressed with genes that are liver-specific, involved in immune system or inflammation. A. Hierarchical clustering of 174 genes involved in redox metabolism across 360 HCC subjects showed highly heterogeneous redox responses. Genes (rows, mean FPKM > 1) were Z-normalized and clustered. B. Two redox gene clusters were highly positively co-expressed with genes within the clusters (red) but negatively co-expressed with genes of the opposing cluster (blue). Only statistically significant (Q < 0.01) correlations are presented, ordered according to first principal component. The inset displays the 10 genes in each cluster with the highest correlation coefficients. C. Genes in G6PD and ALDH2 clusters were strongly co-expressed with liver-specific genes and genes involved in inflammation (Q < 10−20, absolute Spearman ρ > 0.5). Inset: genes that were simultaneously co-expressed positively and negatively with >5 genes in ALDH2 and G6PD clusters and at least one gene in each cluster. Red edges indicate positive co-expression, blue edges indicate negative co-expression. Inflammasome categories: 1. Inflammasome; 2. CLEC7A/Inflammasome pathway; 3. DEx/H-box helicases that activate type I IFN and inflammatory cytokine production. D. Gene set enrichment analysis performed for liver-specific and immune system genes co-expressed with >5 genes in the ALDH2 and G6PD clusters (C inset). All colored nodes are statistically enriched functional terms (Q < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Gene co-expression analysis is a useful approach to reveal functional relationships [19,20]. We calculated pairwise Spearman rank correlations between these 132 genes and all other genes expressed in HCC tumors [21]. This unsupervised clustering approach identified genes showing coordinated co-expression and are likely co-regulated or functionally associated. The 132 genes were co-expressed with each other and with many other genes in the genome (Dataset 2), including genes involved in carbohydrate, lipid, nucleotide and amino acid metabolism. For instance, aldolases (ALDOA and ALDOB), pyruvate kinases (PKM, PKLR) and phosphofructokinases (PFKP) were highly co-expressed with these 132 genes (highest Spearman's ρ 0.71, Q < 10−50). Importantly, some enzymes involved in central metabolism (e.g., pyruvate kinases, isocitrate dehydrogenases, glutaminases) are not only important players in cancer but are also indirectly involved in redox metabolism [22]. Aiming to explore the relationships between genes directly or indirectly involved in redox metabolism, we expanded the list to 174 genes by considering those involved in the metabolism of carbohydrates, lipids or other compounds that are closely associated with redox metabolism (Dataset 1). This 174 highly heterogeneous gene set (Fig. 1A, Dataset 1) is hereafter referred to as “redox genes”.
Among the significantly correlated redox genes, we identified two major clusters with positively co-expressed genes in the same cluster but negatively co-expressed with those of the opposite cluster (Q < 0.01, Fig. 1B, Dataset 3). The first cluster of the top 10 correlated genes (Fig. 1B inset) comprised genes involved in the pentose phosphate pathway (PPP) (G6PD, RPIA), glycolysis (PKM, ALDOA), differentiation (TXNDC9), response to hypoxia (HIF1A), and metabolism of folate (MTHFD1L), glutathione (GLRX3), purines (ATIC), and amino acids (GLS) and is hereafter referred as the G6PD cluster. The second cluster also comprised genes involved in glycolysis and the PPP (ALDOB), metabolism of folate (MTHFD1, MTHFS, SHMT1), amino acids (ALDH5A1, ALDH6A1, ALDH7A1) or other compounds (ALDH2, ALDH8A1), and responses to oxidative stress (CAT) and is hereafter referred to as the ALDH2 cluster. Glucose 6 phosphate dehydrogenase (G6PD) controls the flux into the PPP and provides reducing power and ribose phosphates to maintain the redox balance and biosynthesis of nucleotides and lipids. These reducing equivalents may be used towards peroxidase-catalyzed antioxidant defense, which is linked to apoptosis, angiogenesis, and the efficacy of anti-cancer therapy and is identified as a promising target in cancer therapy [23]. ALDH2 is involved in alcohol metabolism and redox homeostasis, and it has recently been associated with HCC progression and a poor prognosis in mice [24].
Interestingly, genes in the ALDH2 and G6PD clusters were co-expressed (absolute Spearman ρ > 0.5, Q < 10−20) with 1) 14 genes involved in inflammation; 2) 696 genes involved in immune mechanisms, including innate and adaptive immunity; and 3) 60 liver-enhanced and 89 liver-enriched genes [25]. Many of these were co-expressed with >5 genes in the ALDH2 and G6PD clusters but negatively and positively with respect to each cluster (Fig. 1C inset). For instance, IL1, TNFα and NFκB, key genes involved in inflammation, were negatively co-expressed with the ALDH2 cluster (Q < 10−4) but positively co-expressed with the G6PD cluster (Q < 0.001). In turn, liver-specific genes were negatively co-expressed with genes in the G6PD cluster but positively co-expressed with the ALDH2 cluster, in which we identified the liver-specific gene ALDOB [25]. This finding suggests that the G6PD cluster is associated with a poorly differentiated tumor phenotype, as opposed to the ALDH2 cluster. For instance, the G6PD cluster gene MTHFD2 has been previously identified as a marker of poor differentiation in other cancers [26]. Indeed, vimentin, a marker of poor differentiation and mesenchymal transition, was positively co-expressed with 9 genes in the G6PD cluster and negatively co-expressed with 7 genes in the ALDH2 cluster (absolute Spearman's ρ > 0.2, Q < 10−5, Dataset 4). Alpha-fetoprotein, also a marker of undifferentiated tumor cells, was positively co-expressed with 7 genes in the G6PD cluster and negatively co-expressed with 9 genes in the ALDH2 cluster (absolute Spearman's ρ > 0.2, Q < 10−4). Similar observations were obtained with respect to other markers of differentiation such as cytokeratins (e.g., KRT8, KRT19).
Gene set enrichment analysis (GSEA) for liver-specific or immune genes (Fig. 1D, Dataset 5) indicated that the liver-specific genes were involved in lipid, steroid, small molecule and amino acid metabolism; small molecule metabolism, including hormones, cofactors and ketones; and oxidation/reduction processes. In turn, the immune system genes were involved in cell cycle and division; regulation of transport, metabolism and cell death; ubiquitination, regulation of protein metabolism and phosphorylation; proteasomal-mediated protein catabolism, cytoskeleton and organelle organization. Finally, we observed that several genes were co-expressed with 10 or more genes in both clusters: liver-specific genes, including ABAT, CDO1, CYP4F3, F11, SEC14L2, SPDYC, and TTC36, were positively and negatively co-expressed with genes in the ALDH2 and G6PD clusters, respectively. Genes associated with the adaptive immune system, including PPP2R1A, UBE2L3, VASP, and YWHAZ, and innate immune system, including ARPC2 – 4, CFL1, GYG1, IKBKE, LIMK1, RHOG, and STK11IP, were negatively co-expressed with genes in the ALDH2 cluster but positively co-expressed with those in the G6PD cluster.
3.2. Several redox genes are associated with survival outcome in HCC patients
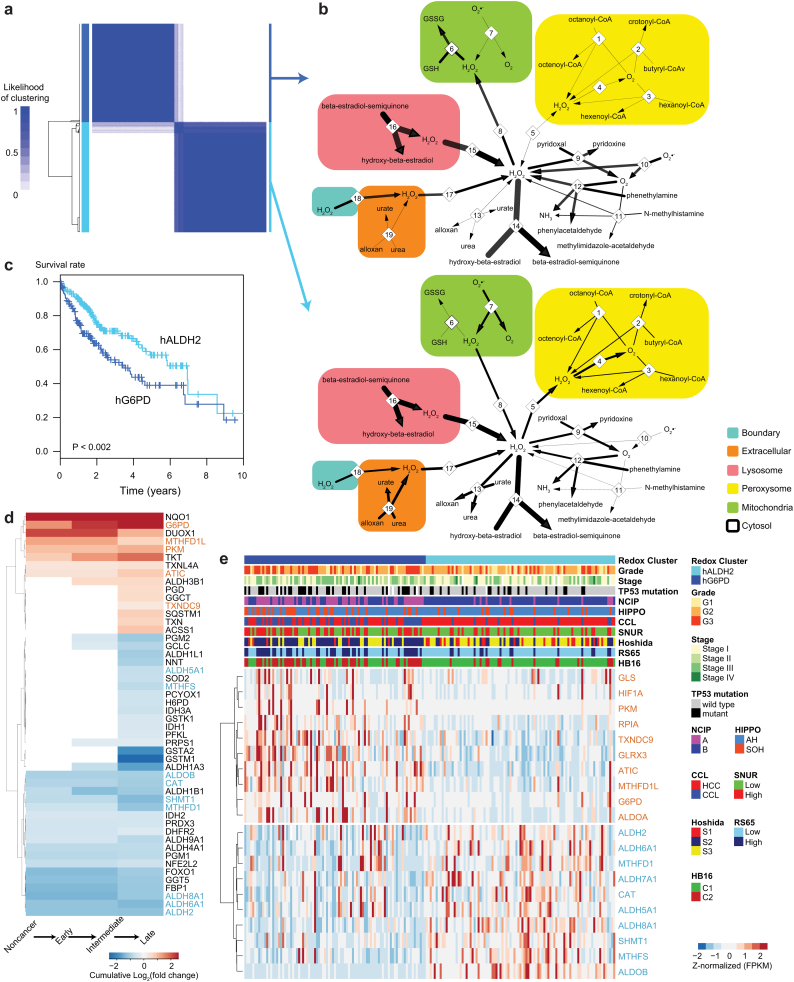
To reveal the association between the expression of redox genes and patient survival, we selected 50 subjects with the lowest and highest survival based on patient metadata and performed a differential expression analysis between the two groups (Methods, Dataset 6). We observed that 11 and 14 redox genes (Table 1) were significantly (Q < 0.05) up- and downregulated between patients in the low vs high survival groups, respectively. For instance, TXNRD1 and GSR were upregulated in the low-survival group and found to encode NADP-dependent reductases involved in peroxidase-dependent H2O2 defense. G6PD, which was upregulated in the low-survival group, encodes glucose-6-phosphate dehydrogenase, one of the main NADPH sources in cancer cells. In addition, many redox genes have been identified as potential prognostic genes (Table 1) [27]. Here, ALDH2 was identified as a potential prognostic gene since its downregulation in the low survival group is in agreement with its identification as a favorable prognostic gene in liver cancer [27]. These observations indicated that several genes involved in redox metabolism were significantly correlated with survival outcome in HCC patients.
Table 1.
Favorable and unfavorable survival and prognostic genes in HCC involved in redox metabolism. Significantly altered redox genes between 50 high vs 50 bottom survival subjects are displayed, where fold changes > 1 indicate an upregulated gene among the poor survival group, and prognostic genes in liver cancer [27] are shown. Favorable and unfavorable prognostic genes are those respectively up- and downregulated in the high survival group. CAT was identified as a prognostic gene in liver cancer in agreement with its expression variation between survival groups, despite being marginally not statistically differentially expressed.
| Module | Gene | Fold Change | P value | Q value | Prognostic genes |
|---|---|---|---|---|---|
| General ROSresponse | SQSTM1 | 1.74 | 0.00032 | 0.0065 | Unfavorable |
| HIF1A | 1.49 | 0.0026 | 0.026 | Unfavorable | |
| FOXO1 | 0.56 | 9.8 × 10−7 | 0.00012 | Favorable | |
| Catalase | CAT | 0.72 | 0.0074 | 0.052 | Favorable |
| Glutathione | GSR | 1.44 | 0.0006 | 0.01 | Unfavorable |
| GCLM | 1.52 | 0.0022 | 0.024 | Unfavorable | |
| GGT5 | 0.5 | 0.0002 | 0.0048 | Favorable | |
| GSTK1 | 0.75 | 0.0051 | 0.041 | Favorable | |
| GSTO1 | 0.71 | 0.0055 | 0.043 | Favorable | |
| Thioredoxin | TXNRD1 | 1.94 | 8.1 × 10−6 | 0.00049 | Unfavorable |
| TXNDC11 | 0.77 | 0.00099 | 0.014 | Favorable | |
| NAD(P)H | ALDH2 | 0.62 | 0.00064 | 0.01 | Favorable |
| ME1 | 1.27 | 0.0059 | 0.045 | Unfavorable | |
| G6PD | 2.68 | 4.9 × 10−9 | 2.6 × 10−6 | Unfavorable | |
| NNT | 0.62 | 0.00027 | 0.0059 | Favorable | |
| ALDH5A1 | 0.68 | 0.0043 | 0.036 | Favorable | |
| PPP | RPIA | 1.26 | 0.0023 | 0.024 | Unfavorable |
| GPI | 0.72 | 0.0037 | 0.033 | Favorable | |
| PGM1 | 0.74 | 0.0045 | 0.038 | Favorable | |
| Folate | FPGS | 0.73 | 0.00182 | 0.021 | Favorable |
| MTHFS | 0.62 | 0.00013 | 0.0037 | Favorable | |
| GART | 1.19 | 0.005 | 0.04 | Unfavorable | |
| Amino acid metabolism | SHMT1 | 0.63 | 0.0034 | 0.032 | Favorable |
| XCT | 2.05 | 0.00075 | 0.012 | Unfavorable | |
| Glycolysis | ALDOB | 0.45 | 0.0001 | 0.003 | Favorable |
| PFKP | 1.96 | 0.00028 | 0.006 | Unfavorable |
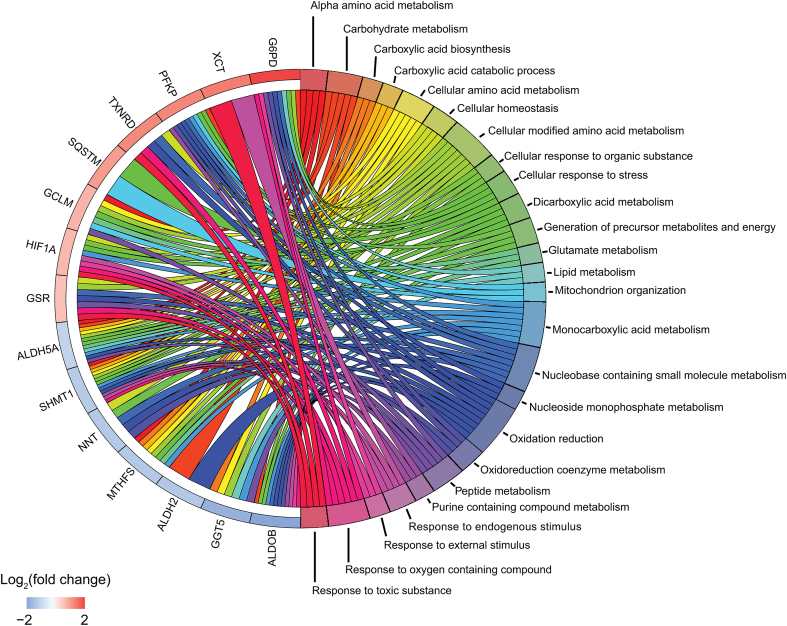
GSEA between low- and high-survival patients (Q < 0.05, Dataset 6) indicated that subjects with low survival tended to over represent genes involved in cell cycle regulation and related processes (e.g., chromatid/chromosome segregation and regulation, DNA replication, initiation and integrity checkpoint, spindle organization and recombination, regulation of cell division). In turn, these patients underrepresented processes such as metabolic and energy regulation (e.g., electron transport chain, aerobic respiration, oxidative phosphorylation, metabolism of amino acids, fatty acid β oxidation and catabolism, lipid oxidation). Only two upregulated redox genes (GART and HIF1A) were associated with overrepresented processes (Fig. 2, Dataset 7). However, several downregulated genes were associated with underrepresented processes, including energy and reducing equivalent metabolism (ALDH5A1, NNT), folate and amino acid metabolism (MTHFS, SHMT1), and detoxification (GSTO1, GSTK1), among other metabolic processes (Dataset 7). These observations suggested that the biological differences displayed between subjects with low vs high survival involved several redox genes associated with metabolic processes that are important in the occurrence of HCC.
Fig. 2.
Differentially expressed genes (left) and their associated biological processes (right) are significantly different between tumors of patients with low vs high survival (Q < 0.05). Differential expression analysis and GSEA were performed by comparing the 50 patients with the highest vs lowest survival. Differentially expressed redox genes and their respective biological processes were selected if genes and processes were simultaneously up- or downregulated. The full GSEA is indicated in Dataset 7.
3.3. Genes in ALDH2 and G6PD clusters are associated with opposing functional responses, prognostic genes, isoform-encoding genes, and hypoxia behavior
Based on our integrative analysis, we observed that HCC tumors displayed different gene expression patterns of redox genes in each cluster, which might translate into functional metabolic differences. To understand the key role of each redox gene in cancer, we classified HCC tumors based on expression and compared the global gene expression profiling of the 50 tumors with the highest and lowest expression of individual genes in each cluster (see Methods). Differential expression with respect to majority of the genes in the ALDH2 and G6PD clusters indicated similar expression patterns between redox genes that were distinct from those in the opposing cluster (Appendix Fig. S1B), in agreement with the above-described positive intra-cluster and negative inter-cluster correlations (Fig. 1B).
GSEA revealed underlying metabolic changes between tumors with high expression of genes in each cluster (Fig. 3). We found that tumors displaying high expression levels of genes in the ALDH2 cluster, or low expression of those in G6PD cluster, were enriched in several processes related to drug metabolism, lipid oxidation and metabolism, amino acid metabolism and biosynthesis, and carbohydrate metabolism. In turn, tumors with low expression levels of genes in the ALDH2 cluster, or high expression of those in the G6PD cluster, were enriched in several processes related to development, differentiation, and morphogenesis and, to a smaller extent, fatty acid biosynthesis (Dataset 8). No functional terms related to NADPH biosynthesis were downregulated with ALDH2 expression and upregulated with GPX and PRDX expression. Tumors enriched in genes co-expressed with those in the ALDH2 and G6PD clusters (Fig. 4) tended to display similarly antagonistic responses (Appendix Fig. S2). Similar observations were also identified in terms of CAT and G6PD, two important genes for defense against H2O2 (Doc S1, Appendix Fig. S3).
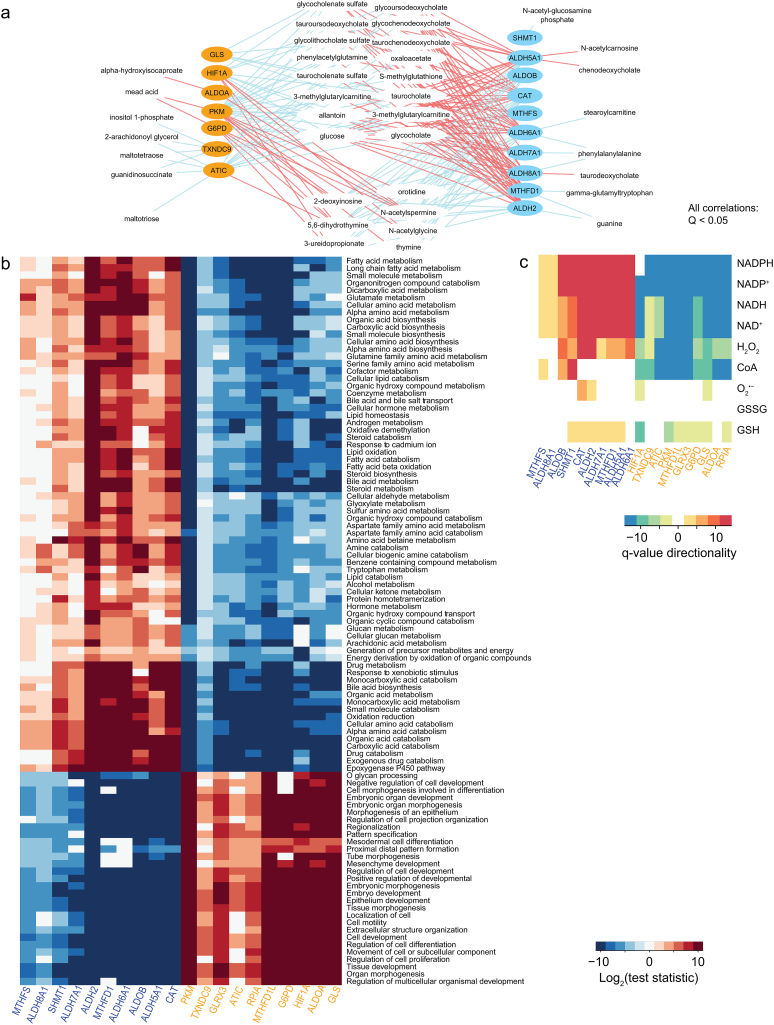
Fig. 3.
Opposite functional responses are observed in HCC tumors differentially expressing redox genes. A. Using metabolomics and gene expression data [14], we identified several metabolites with abundances that positively (red) or negatively (blue) correlated with the expression of genes in the ALDH2 and G6PD clusters (Q < 0.05, absolute Spearman's ρ > 0.4). Genes with expression that was not quantified by Ref. 14 are not indicated. B. GSEA for subjects expressing high and low redox genes in the ALDH2 (blue) and G6PD (orange) clusters. Only processes that were significant (Q < 0.05) in >4 columns are displayed (see Methods). Log2-normalized test statistics indicate the directionality of the processes (positive are upregulated, negative are downregulated, null are not statistically significant). C. Reporter metabolites were determined, and the minimum Q was computed for those with associated genes that were consistently up- or downregulated across compartments (Q < 0.05). Q values were normalized by computing −1 × log10(minimum Q), or log10(minimum Q), respectively, for up- or downregulated changes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
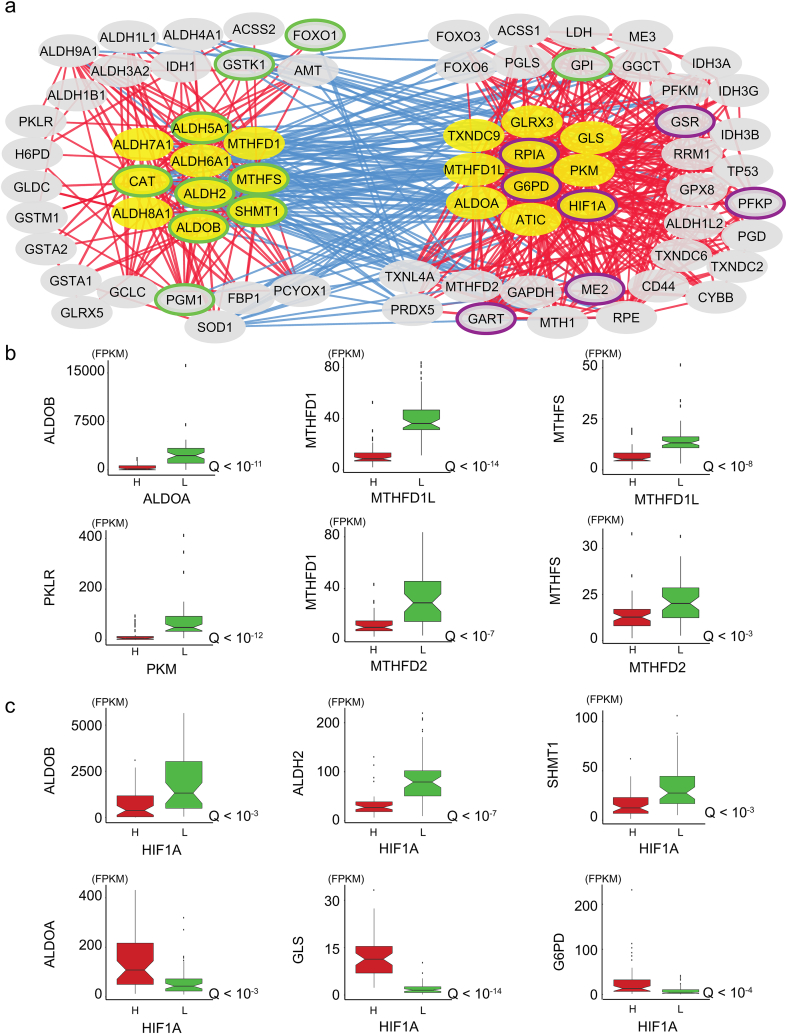
Fig. 4.
HCC tumors antagonistically co-express genes in ALDH2 and G6PD clusters, associated with opposite hypoxic environments. A. Genes in the two clusters (yellow) and their first neighbors (gray) are indicated. Redox genes that were positively co-expressed with genes in each cluster are displayed (absolute Spearman's ρ > 0.3, Q < 10−7). Green- and purple-circled nodes respectively indicate favorable and unfavorable prognostic genes (Table 1). Genes that were positively co-expressed with those in both clusters are not presented (e.g., NFE2L2, NNT, XCT, TXNRD1). Refer to Dataset 3 for the full list of correlations. B. Genes encoding enzyme isoforms or enzymes that catalyze alternative reactions in the same pathway were alternatively expressed. Boxplots display the expression of genes (FPKM) as a function of HCC samples displaying high (H) and low (L) expression of each gene (50 samples per group), and the respective Q values (Mann-Whitney U test). No significant difference (Q > 0.05) was observed for the other isoforms, with the exception of IDH1 as a function of IDH3A (Q < 0.005), for which high expression of the latter gene was associated with high expression of Idh1. Similar observations were obtained through differential expression analysis. C. Gene expression as a function of HIF1A expression. Note that among all genes in the ALDH2 and G6PD clusters, only the expression of MTHFD1, MTHFS and CAT did not significantly (Q < 0.05) differ between samples with high vs low HIF1A expression. For all boxplots, whiskers extended up to 1.5 times the interquartile range. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To reveal the detailed metabolic differences between tumors with high expression of genes in each cluster, we performed reporter metabolite analysis [28] using a generic human HCC GEM [15]. We found significant differences between reporter metabolites, with the most significant transcriptional changes observed in their associated enzymes, considering the full cellular metabolic network [28]. We identified 1999 reporter metabolites (out of 5134) that displayed the opposite behavior with respect to genes in ALDH2 and G6PD clusters (Q < 0.05, Dataset 9). Reporter metabolites identified in our study involved in redox metabolism included H2O2, NAD(P)H, NAD(P)+, glutathione, CoA, and O2•− (Dataset 9), as well as several metabolites involved in lipid metabolism (e.g., hydroperoxyeicosatetraenoic acids). These trends were consistently observed across cellular compartments, where 1566 reporter metabolites displayed similar antagonistic behaviors to those described above. For instance (Fig. 3B), genes involved in reactions involving NAD(P)H and NAD(P)+ were consistently upregulated in all compartments, with the exception of the nucleus, in subjects displaying high expression of genes in the ALDH2 cluster, or low expression of those in the G6PD cluster. Likewise, genes involved in H2O2-associated reactions were upregulated and downregulated in subjects displaying high ALDH2- and G6PD-co-expressed genes, respectively.
The above-described antagonistic behavior (Fig. 1) extended to the genes co-expressed with those in the two clusters, which tended to be negatively co-expressed with those of the opposing cluster (Fig. 4A, Appendix Fig. S4). Importantly, the ALDH2 and G6PD clusters were co-expressed with the genes encoding for enzyme isoforms or enzymes that catalyze alternative reactions in the same pathway (Fig. 4B, Q < 10−3). For instance, among the genes in the G6PD cluster or their co-expressed genes (Fig. 4 right), we found that PKM, MTHFD2, MTHFD1L, and ALDOA, as opposed to genes in the ALDH2 cluster or their first neighbors PKLR, MTHFD1, and ALDOB (Fig. 4 left). This opposite behavior was not observed for GLRX3 vs GLRX5, IDH3ABG vs IDH1, or ACSS1 vs ACSS2, as previously observed [16]. Transcription factors and genes involved in the response to oxidative stress, TP53, HIF1A, and MTH1, were co-expressed with genes in the G6PD cluster, whereas FOXO1 was co-expressed with genes in the ALDH2 cluster.
The observation that HIF1A was part of the G6PD cluster suggested that HCC tumors and their redox behavior responded to hypoxia. We observed that PKM and HIF1A were co-expressed, consistent with the response of PKM to hypoxia [16]. Interestingly, we found that HIF1A expression reflected the antagonistic responses of ALDH2 and G6PD clusters observed herein (Fig. 4C, Q < 0.02), in which the only genes that showed no significant difference with respect to HIF1A expression were MTHFD1, MTHFS and CAT (Q > 0.05).
Additionally, we identified 7 unfavorable prognostic genes and favorable prognostic genes among the G6PD cluster and its co-expressed genes, and 9 favorable prognostic genes among the ALDH2 cluster and its co-expressed genes. Functional enrichment analysis with respect to tumors with high expression of each gene showed similar observations to those described above (Appendix Fig. S2).
3.4. Tumors stratified based on redox genes display substantial metabolic differences
The above observations indicated potentially important and opposite responses in HCC subjects differentially expressing redox genes. We then questioned whether HCC tumors could be stratified based on genes in ALDH2 and G6PD clusters. Hierarchical clustering of tumors has some pitfalls that have been overcome by recent methods that achieve robust stratification even considering small gene subsets [29,30], and does not permit a clear separation of tumors (Fig. 1A). Unsupervised clustering methods permit the identification of unknown groups in a sample based on intrinsic features without external information [31]. Here, we employed consensus clustering for partitioning patients into stable groups through repeated subsampling and clustering [29,31]. Briefly, considering genes in the ALDH2 and G6PD clusters (Fig. 1B inset), we determined an optimum number of clusters of 2 (Fig. 5A) by determining the highest area increase in the cumulative distribution function as the number of clusters increased.
Fig. 5.
HCC tumors stratified based on redox genes display substantial differences in the survival and metabolism of H2O2, purines and fatty acids. A. Tumors were stratified based on the expression of genes in the ALDH2 and G6PD clusters through consensus clustering into two clusters that respectively displayed high expression of genes in ALDH2 and G6PD clusters (cyan and blue horizontal bars, respectively, hALDH2 and hG6PD). Clustering displayed the likelihood that a tumor was found in that cluster. Similar partitioning was achieved if all 174 redox genes were considered, reinforcing the robustness of the clustering. Note that >94% of the samples were consistently clustered in the two clusters (likelihood > 0.9). B. Metabolic differences displayed by the two groups of tumors as determined through metabolic modeling. GEMs for hG6PD (top) and hALDH2 (bottom) tumors were integrated with the gene expression data. Cell compartments are indicated by colored boxes. Numbers indicate enzyme-catalyzed reactions or transport reactions, as indicated in Dataset 11. Thin arrows indicate reactions with low or null fluxes (B: 6, 10, 11; C: 1–5, 7, 13, 19). Bidirectional reactions (e.g., transport reactions) indicate the estimated direction based on the flux value. Common metabolites (e.g., H2O and H+) are not displayed. C. Kaplan-Meier survival plot for hALDH2 (cyan) and hG6PD (blue) clusters indicates substantially lower survival for patients in the latter cluster (respective median 5-year survival of 0.57 and 0.39, P ≈ 0.0018). D. Differential expression analysis showing the opposing behavior between ALDH2-co-expressed and G6PD-co-expressed genes. Cumulative log2(fold changes) were computed for the significantly differentially expressed redox genes (Q < 0.05, DESeq2) between late (G3 + G4) vs intermediate (G2) vs early (G1) HCC and noncancerous samples. Genes in the ALDH2 (cyan) and G6PD (blue) clusters (Fig. 1B, inset). The full output is displayed in Dataset 15. E. Expression of genes in ALDH2 and G6PD clusters and detailed clinical information for 186 tumor samples (Cancer Genome Atlas Research Network, 2017). Gene expression data (FPKM) were row-normalized and clustered. Columns were sorted according to the redox cluster. Clinical information is shown for grade, stage, TP53 mutation status, and known tumor subtypes (NCIP, HIPPO, CCL, SNUR, Hoshida, RS65, HB16). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We repeated these analyses using gene expression data in independent human HCC cohorts [14,32,33] and identified 2 tumor clusters (Appendix Fig. S5). The two clusters (Fig. 5A) comprised 190 and 170 patients with high-expression of ALDH2- and G6PD-co-expressed genes, respectively, and hereafter are referred to as the hALDH2 and hG6PD groups. Consensus clustering based on the full set of redox genes yielded similar clusters. Importantly, among the top differentially expressed genes between the two clusters, we identified ALDOB, PKM, G6PD and MTHFS (Appendix Fig. S6, Dataset 10).
3.5. Simulation of the metabolic differences between HCC tumor types
To reveal the metabolic differences between HCC subpopulations, we generated two HCC GEMs, one per patient cluster, using the MADE [34] and TIGER [35] algorithms by incorporating gene-specific fold changes and Q values. Considering tumor growth as an objective function, we observed that models for hG6PD and hALDH2 tumors (Fig. 5B top and bottom) displayed major differences in terms of H2O2 consumption. NADPH-independent and –dependent systems, i.e., reactions catalyzed by catalase but not by peroxidases, displayed fluxes in hALDH2 but not in hG6PD tumors (compare NADPH-independent reaction 4 with NADPH-dependent reaction 6). NADPH-production by glucose-6-phosphate dehydrogenase mirrored this trend, displaying low flux in hALDH2 but high flux in hG6PD (Dataset 11, Appendix Fig. S7). Additionally, mitochondrial superoxide dismutase-catalyzed H2O2 production (Fig. 5B, reaction 7) displayed flux and promoted H2O2 permeation to the cytoplasm (reaction 8) in hALDH2. This was not observed in hG6PD, where cytoplasmic superoxide dismutase produces H2O2 that enters mitochondria and is therein consumed by peroxidases (reaction 6). Finally, reactions involved in fatty acid β-oxidation (reactions 1–3), and purine metabolism (reactions 13 and 19) and that produce H2O2 displayed fluxes in hALDH2 but not in hG6PD. NADPH metabolism through the folate pathway displayed fluxes in hALDH2 but not in hG6PD (reactions 15, 25, 28 and 33, Appendix Fig. S7). In turn, reactions involved in mitochondrial but not cytoplasmic reduction of oxidized glutathione (reaction 27) displayed fluxes in hALDH2 but not in hG6PD, reflecting the increased peroxidase activity in the latter group of tumors. Similar observations were observed through patient-specific GEMs, reflecting the major redox patterns of the two stratified groups (Dataset 12). Reporter metabolite analysis performed between hALDH2 vs hG6PD qualitatively reproduces (Dataset 12) the observations found for gene-specific reporter metabolites (Fig. 3C).
To validate these observations, we repeated these analyses using gene expression data from an independent cohort [32,33]. Overall, these cohorts displayed metabolic features that were qualitatively similar to those observed above, including fluxes in glutathione-reductase driven H2O2 scavenging and glucose-6-phosphate dehydrogenase, and fatty acid biosynthesis but not β-oxidation in hG6PD, and not in hALDH2 (Dataset 13).
Essentiality analysis performed on each of the genes in the ALDH2 and G6PD clusters indicated that they were essential for hALDH2 and hG6PD tumors, respectively, but not the converse (with the only exception being GLRX3, which is not in the model).
3.6. Redox responses vary with tumor progression and subtype and identify survival signatures across HCC patients
The above observations indicated that HCC tumors display substantial metabolic differences, leading us to question how these differences relate to clinical properties of the patients. For instance, survival analysis indicated that hALDH2 patients displayed significantly higher survival than hG6PD patients (median 5-year survival of 0.57 and 0.39 respectively, log-rank test P = .0017, Fig. 5C). We also identified significantly altered redox gene expression between patients with different tumor grades. Cancer grades identify the dedifferentiation stage of HCC with respect to normal cells, and they include well (Grade 1), moderately (Grade 2), poorly (Grade 3) differentiated and undifferentiated (Grade 4) tumors. Here we identified differentially expressed genes throughout cancer progression between early (G1), intermediate (G2) and late (G3 + G4) HCC. We observed that few redox genes were significantly (Q < 0.05) different between early and intermediate cancer, but many of them were significantly differentially expressed between intermediate and late cancer (Fig. 5D, Dataset 14). Notably, the differences followed the shifts in gene expression observed between the opposing G6PD and ALDH2 clusters. Namely, expression of genes in the G6PD cluster was upregulated throughout cancer progression, whereas expression of genes in the ALDH2 cluster decreased with cancer progression. Metabolic modeling and accounting grade-specific gene expression for early and late HCC showed trends similar to those observed in hALDH2 and hG6PD tumors above (Fig. 5B). These observations indicated that through HCC progression, and particularly in more advanced cancer, there was a switch from NADPH-independent to NADPH-dependent H2O2 defense supported by glucose-6-phosphate dehydrogenase activity.
Detailed clinical information, including the tumor subtype, viral infection and inflammation status, is available for 186 patients [13]. Analysis of clinical data (Fig. 5E) indicated that the proportions of tumors in hALDH2 and hG6PD groups were not independent of clinical traits (Dataset 15) such as TP53 mutation incidence, histological grade and stage (Q < 0.05), tumor growth pattern (Q < 0.005), and molecular data including mRNA and microRNA expression and protein levels (Q < 0.01). The proportion of tumors in the two clusters also varied with the tumor subtype: NCI proliferation (NCIP) [32], cholangiocarcinoma-like (CCL) [36] and hepatoblastoma 16 gene (HB16) [37] signatures (Q < 10−5); Hoshida [38] and 65-gene recurrence risk score (RS65) [39] signatures (Q < 10−4); Seoul National University recurrence (SNUR) [40], and Hippo pathway [41] signatures (Q < 0.05). In contrast, the proportion of tumors in hALDH2 and hG6PD did not vary (Q > 0.2) with race, gender, alcohol liver disease or consumption, viral infection (hepatitis B and/or C), inflammation (including intra-tumoral), cirrhosis, fibrosis, tumor ploidy, purity, or DNA methylation. At the gene expression level (Fig. 5E, Dataset 15), we observed that expression of most genes in the ALDH2 and G6PD clusters (≥17 genes, Q < 10−4) varied with the tumor subtype (CCL, HB16, Hoshida, NCIP, SNUR, Hippo), RS65, and mRNA expression. The expression of many genes (≥11) also varied with miRNA expression and protein levels.
Negative correlations were identified between RS65 index score and the expression of genes in the ALDH2 cluster (ALDH2 and MTHFD1, ρ < −0.3, Q < 0.0005), but positive correlation were observed with those in the G6PD cluster (PKM, MTHFD1L, G6PD, ATIC, GLRX3, ALDOA, RPIA and TXNDC9, ρ > 0.3, Q < 0.0006). Only the expression of genes in the G6PD cluster (G6PD, RPIA, PKM, ALDOA, ATIC, and MTHFD1L) strongly varied with the TP53 mutation incidence (Q < 10−4), which was also observed to a lower extent for genes in the ALDH2 cluster (ALDH2 and ALDH5A1, Q < 0.01).
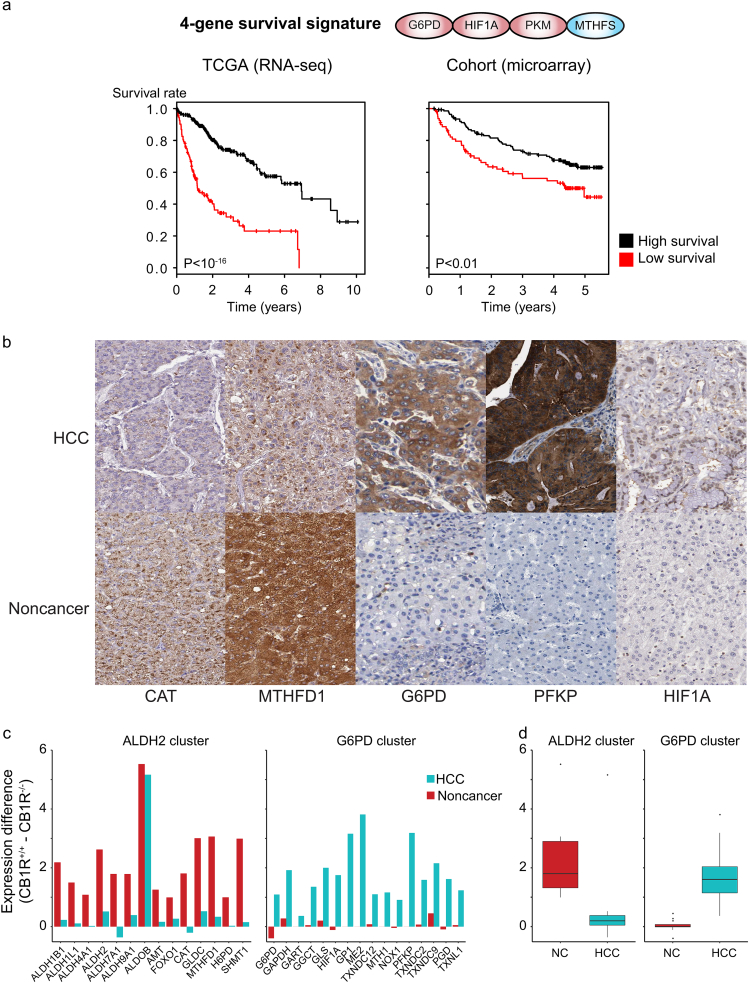
The above observations indicated that HCC tumors were successfully stratified into different groups with distinct redox behavior and survival (Fig. 5) and that several redox genes were potentially favorable and unfavorable prognostic genes (Table 1). We sought to determine a signature that would permit a more efficient prediction of patient survival by using a subset of genes in the redox clusters. We examined all 3 by 3 and 4 by 4 combinations of genes in the redox clusters and, through Kaplan-Meier analysis, identified PKM, G6PD, MTHFS, and HIF1A as achieving the best survival signature where the group size comprised made up at least 20% of the total sample (Fig. 6A, log-rank P < 10−16, Q < 10−16, Dataset 16). PKM, MTHFS and G6PD also presented the most significant 3 by 3 combination (log-rank P < 10−15, Q < 10−12) and were substantially better than any single gene prediction (log-rank P < 10−12, Q < 10−9, Appendix Fig. S8, Dataset 16, analysis based on ref. [27]). This 4-gene signature included genes from pentose phosphate pathway and folate metabolism, suggesting that the survival of HCC patients was associated with these pathways.
Fig. 6.
Predominant expression of G6PD- and ALDH2-clustered genes in mouse and human HCC and noncancerous cells. A. Kaplan-Meier plots for the 4-gene signature (G6PD, HIF1A, PKM, MTHFS) using RNA-seq data for 360 patients from TCGA (left), and from an independent microarray cohort with 222 patients [42,43]. B. Immunohistochemistry staining of G6PD, HIF1A, PFKP, CAT, and MTHFD1 through antibodies HPA000834, HPA001275, HPA018257, HPA055838, and HPA050052, respectively [25,27]. Protein expression is displayed in blue, with counterstaining in blue. C. Gene expression profile in murine samples revealed substantially higher expression of G6PD cluster genes in HCC than noncancer samples and higher expression in CB1R+/+ (hypoxia-associated) compared with CB1R−/− samples, compared with the expression of genes in the ALDH2 cluster. mRNA expression profile measured in six pooled wild-type HCC tumor and noncancerous samples using RNA-seq in CB1R+/+ and CB1R−/− six murine samples. For all boxplots, whiskers extended up to 1.5 times the interquartile range. D. The distribution of differences for genes in the two clusters in HCC and noncancerous (NC) murine samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We performed Kaplan-Meier analysis in independent cohorts [42,43] and validated the prognostic value of this gene signature (Fig. 6A, log-rank P < .01, Q ≈ 0.023). Taken together, these observations indicated that tumors displayed substantial differences in redox behavior that were related to the clinical and survival differences of their respective patients.
3.7. Validation of the redox response in independent human HCC cohorts
The above relationships were validated using several independent cohorts at gene expression protein and metabolite levels. First, the antagonistic behavior observed among co-expressed redox genes (Fig. 1) was also displayed at the gene expression level in 61 HCC tumors (Appendix Fig. S9A, Ref. [14]) and 91 HCC tumors [42,43] (Appendix Fig. S2A), and at the proteomic level in 19 HCC tumors [44] (Dataset 17).
Second, the above antagonistic behavior was observed in an independent microarray and metabolomics tumor dataset [14] (Fig. 3A). The authors used multi-omics data, including gene expression and metabolomic data that quantified 718 metabolites using LC/MS and GC/MS, to identify and identified molecular subtypes and driver genes shared by HCC and intrahepatic cholangiocarcinoma. The integrative approach employed by the authors permitted identifying allowed the identification of potential biomarkers and shared metabolic features in common in the two liver diseases despite their intrinsic heterogeneity. Using gene expression and metabolomics data from this cohort, we observed that the abundance of glycolytic/gluconeogenic compounds (e.g. glucose and oxaloacetate), pyrimidines (e.g. thymine) and other compounds was directly or inversely proportional to the expression of genes in the ALDH2 and G6PD clusters. Notably, we observed that the reporter metabolite analysis between the clusters was consistent with the observed gene-metabolite associations. For instance, we observed that 75% of the metabolites simultaneously found by the reporter metabolite analysis and gene-metabolite association analysis (Fig. 3) showed concordant behavior between the two independent analyses on the two datasets, i.e., positive correlations with ALDH2-cluster genes and upregulated reporter metabolites in hALDH2 tumors, and the same with respect to G6PD cluster/hG6PD. Analyses performed using the additional cohort supported these observations (Dataset 13). The metabolites identified included glucose, oxaloacetate, and cholate derivatives (positively correlated with ALDH2 cluster genes), and guanosine, thymidine and deoxyinosine (positively correlated with G6PD cluster genes). While the reporter metabolite analysis alone did not permit inferences of metabolite abundance, they pointed towards metabolic hotspots with likely increases or decreases in metabolic fluxes [28]. The observed metabolite-gene associations suggested that aside from representing these flux-alteration hotspots, the abundances of those metabolites simultaneously identified in the 2 analyses were associated with redox genes and thus might represent potential biomarkers in ALDH2 or G6PD clusters.
Third, we observed that patient stratification according to genes in ALDH2 and G6PD clusters was associated with differences in survival (Fig. 5C) and known HCC subtypes (e.g., NCIP, Fig. 5D), which were reproduced using an independent cohort (Appendix Fig. S9B) [32,33].
In addition to these observations, we have previously shown that similar antagonistic patterns are observed for other genes associated with the G6PD cluster (ACSS1, PKM) and ALDH2 cluster (ACSS2) [16,45]. Finally, we have identified the expression of G6PD, HIF1A, PFKP, CAT, and MTHFD1 using the antibodies HPA000834, HPA001275, HPA018257, HPA055838, and HPA050052, respectively [25,27], in human HCC and noncancerous samples (Fig. 6B). The G6PD-clustered genes HIF1A, G6PD and PFKP show moderate/high protein expression in HCC samples but low expression in noncancerous samples, whereas the ALDH2-clustered genes CAT and MTHFD1 show medium protein expression in HCC but high expression in noncancerous tissues.
3.8. Validation of redox responses in independent mouse HCC models
Retrospective analysis of murine and human samples [16,46] showed agreement between hypoxic behavior and progression for the 2 redox gene clusters. G6PD-clustered genes were associated with hypoxic behavior, a less favorable prognosis or accumulated throughout progression when compared with ALDH2-clustered genes. Our observations (Fig. 6C, D) indicated that most genes associated with the G6PD cluster displayed more marked expression increases between HCC vs noncancerous samples (e.g., Hif1a, G6pd, Pfkp, Gart, Me1) than those in the ALDH2 cluster (e.g., Aldh2, Cat, Shmt1, Mthfd1) in N-nitrosodiethylamine (DEN)-induced HCC in wild-type (wt) mice. These changes were more pronounced when comparing expression levels of HCC tumors in CB1R+/+ vs CB1R−/− mice since the wild-type mice were associated with hypoxic tumor responses when compared with the mutant littermates (Fig. 6C, D). Aldob presented as an exception to this behavior, possibly because no other aldolase was detected in either HCC or noncancerous samples. Overall, these observations indicate that the antagonistic redox behavior observed in human HCC is also displayed in a mouse model of HCC.
4. Discussion
Redox metabolism regulates cell proliferation and aberrant redox metabolism, and it is now recognized as one of the hallmarks of cancer [22,47]. Antioxidants scavenge ROS that may otherwise have toxic effects, such as promoting oxidative DNA damage and tumorigenesis. This phenomenon has led many researchers to propose the use of antioxidants as therapeutic agents against cancer [48,49], but several clinical trials have failed to demonstrate the beneficial effects of antioxidants and instead showed that they may increase cancer risk and promote progression and metastasis [5]. Due to the highly heterogeneous responses of HCC, including in terms of global antioxidant gene expression [5], the commonalities and relationships between redox metabolism, patient survival, and global metabolism had never been systematically investigated. Here we extended our previous pan-cancer analysis [27] to systematically analyze heterogeneous redox behavior across HCC tumors and used systems biology approaches to analyze transcriptomics, proteomics and metabolomics data. We considered genes directly and indirectly involved in redox metabolism, i.e., redox genes, and found that they were co-expressed with liver-specific genes and genes involved in the immune system and inflammation, all of which are involved in important processes associated with cancer, such as cell cycle progression, apoptosis, amino acid and lipid metabolism. The close relationships between redox metabolism, inflammation and immune defense are well known in other contexts [7,50], and the immune system is targeted for the treatment of HCC [51]. Our observations emphasized the necessity of jointly targeting immunity, inflammation and redox metabolism for the effective treatment of HCC.
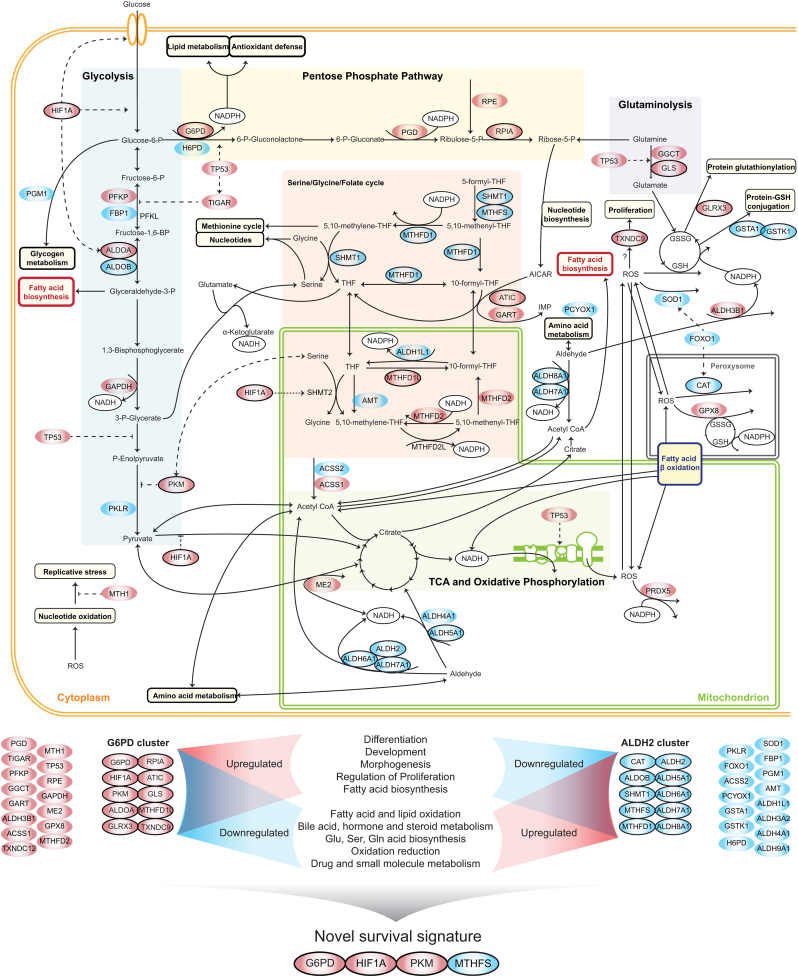
Importantly, our unsupervised approach highlights opposite behaviors among redox genes through two antagonistic and highly co-expressed groups of genes co-expressed with ALDH2 or G6PD. ALDH2 is a gene with important roles in redox homeostasis associated with survival and lower malignancy [24]. In turn, the role of G6PD in cancer is widely known [22]. Their co-expressed genes have known relationships in tumorigenesis. For instance, ALDOA is co-expressed with G6PD and has been previously associated with the response to hypoxia and with HIF1A [52,53], similarly to our observations in HCC. Tumors with high ALDH2-co-expressed genes displayed high fatty acid and lipid oxidation, catabolism, and lower lipid biosynthesis, whereas those with high G6PD-co-expressed genes (hG6PD) showed the opposite behavior. Our previous analyses indicated that the metabolic alterations in HCC promote NADPH biosynthesis through the PPP, lipid biosynthesis and decreased fatty acid oxidation [16]. Here we observed that HCC tumors differentially displayed the above metabolic changes and were closely related to redox behavior (Fig. 7). The correlation between the expression of genes in the ALDH2 cluster with alterations in lipid, amino acid and carbohydrate metabolism additionally suggested that tumors with high expression of those genes might be metabolically more active. In turn, tumors with high expression of G6PD-clustered genes, including HIF1A, were associated with several markers of dedifferentiation, poor prognosis and low survival, consistent with the higher aggressiveness of those tumors.
Fig. 7.
HCC tumors display antagonistic metabolic behavior associated with redox responses. Genes of ALDH2 and G6PD clusters (blue and red nodes) promoted several reactions and processes, as summarized at the bottom of the figure. Reducing equivalents (NADPH and NADH) are highlighted (black nodes). Dashed lines indicate regulatory processes. Genes in the G6PD and ALDH2 clusters (black-circled red and blue nodes) and some of their co-expressed genes (Fig. 4). The reaction catalyzed by H6PD occurred in the endoplasmic reticulum. Note the differences in ROS metabolism in tumors with high expression of ALDH2-clustered genes (blue) vs G6PD-clustered genes (red). Prdx5 was expressed by hALDH2 and hG6PD tumors, but mitochondrial ROS consumption by this gene was only active in hG6PD. Expression profiles of G6PD, HIF1A, PKM and MTHFS, genes in the two redox clusters, identified a signature that more effectively predicted patient survival than predictions from a single gene. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HCC tumors displayed antagonistic redox responses at the gene expression, protein and metabolite level. For instance, hG6PD tumors displayed lower levels of glycolytic metabolites (including oxaloacetate and glucose) and higher levels of nucleoside and nucleobases (thymine, deoxyinosine and orotidine) and amino acid intermediates (acetylglycine, phenylalanine, γ-glutamyl tryptophan). Although other important metabolites, such as NADP and ROS, were not available in the metabolomics data [14], our modeling analyses indicated that genes involved in the metabolism of NAD(P)H, H2O2, and, to a smaller extent, O2●-, glutathione, and CoA, were significantly upregulated in hALDH2 tumors but downregulated in hG6PD tumors. This finding suggested that tumors responded differently to reducing equivalents and ROS and implied that targeting redox metabolism might lead to different outcomes depending on whether the tumors display the hALDH2 or hG6PD subtypes. Using metabolic models and essentiality analysis for hALDH2 and hG6PD tumors, we observed that the former group was not sensitive to inhibition of genes in G6PD cluster but displayed no growth if ALDH2-clustered genes ALDH2 are inhibited, and vice versa.
It is generally considered that cancer cells rely on high antioxidant activity, together with promoted NADPH biosynthesis that is used towards peroxidase-catalyzed defense, enabling tumors to proliferate under sustained oxidative stress [54]. HCC displayed lower glutathione content, catalase and glutathione peroxidase activities in comparison with normal cells [55,56]. Our analyses showed that hALDH2 tumors displayed substantial catalase-mediated H2O2 scavenging, as opposed to hG6PD tumors that instead rely on glutathione/glutathione-peroxidase-mediated H2O2 scavenging. Additionally, we observed that mitochondria were a significant source of H2O2 in hALDH2 tumors, but not in hG6PD tumors, which displayed high mitochondrial glutathione-peroxidase activity. These observations highlighted important NADPH-independent and –dependent differences in ROS scavenging in HCC. Furthermore, they indicated that targeting antioxidants and redox metabolism for HCC treatment is highly dependent on the cell phenotype, i.e., hALDH2 or hG6PD, and thus requires subgroup-tailored drug choices.
Major metabolic and functional alterations that are hallmarks of malignant transformation and often linked to different cancer stages are associated with differential redox metabolism [8]. Expression and functional switches are associated with genes in the ALDH2 and G6PD clusters and thus hALDH2 and hG6PD phenotypes. Importantly, a switch between the two phenotypes was observed from early to late HCC, associated with two main features. First, genes in the ALDH2 and G6PD clusters tended to be respectively co-expressed with favorable and unfavorable prognostic genes. Second, liver-specific genes and markers of differentiation loss respectively were co-expressed positively and negatively with genes in the ALDH2 and G6PD clusters. These two features suggested a malignant association with the expression of genes in the G6PD cluster. They were consistent with the low survival of patients with hG6PD tumors and the observed inverse relationship between the expression of ALDOB, a liver-specific gene in the ALDH2 cluster [57], and ALDOA, a G6PD cluster gene repressed in adult liver and expressed in other tissues [25]. Furthermore, a decrease in serum aldolase B and malignant liver cancer has been previously observed [58], whereas a positive relationship between ALDOA and malignancy and progression has been observed in other cancers [53,59].
Importantly, genes in ALDH2 and G6PD clusters were co-expressed with genes encoding alternative enzymes but that catalyze the same or alternative reactions, sometimes exhibiting the same flux directions. This suggests a compensatory behavior in the metabolic switching of the hALDH2 to hG6PD phenotype. This phenomenon was observed for the expression of MTHFD1, ALDOB and PKLR, as well as all parts of the ALDH2 cluster or their co-expressed genes, which were replaced by MTHFD2, MTHFD1L, ALDOA and PKM, and all parts of the G6PD cluster or co-expressed genes. Such switching has been previously observed in acetate metabolism in HCC [16], and in glycolysis in rat hepatomas [60].
Tumor stratification based on the expression of genes in the ALDH2 and G6PD clusters, where ALDOB is the best stratifying gene, has shown not only differences at the functional and genomic levels but also at the clinical level, including TP53 mutation and known tumor subtypes [36]. Through patient metadata, we also identified several markers of favorable and unfavorable survival among genes involved in redox metabolism. The 4-gene signature herein introduced (PKM, G6PD, MTHFS and HIF1A) improved the commonly used single-gene survival markers and showed great potential for clinical use to predict the prognosis of HCC patients. These 4 genes and their individual association with redox metabolism and tumorigenesis are known in other cancers [22,[61], [62], [63]]. However, here we showed for the first time the relationship of their joint behavior with metabolic and functional responses in HCC and translation to survival differences.
The current study also revealed gene expression behaviors of ALDH2 and G6PD clusters in a DEN-induced HCC murine model. In human HCC, high expression of the hG6PD cluster resulted in low survival and was dominant in late-stage HCC. In mice, G6pd cluster genes were more robustly increased in HCC tumors than Aldh2 clusters in wild-type mice. A similar polarization trend between humans and mice with a dominance of the G6PD cluster increased the translational value of DEN-induced HCC murine models for studying the preclinical efficacy of pharmacological agents in HCC as metabolism mimicking the low-survival stratified cluster. Furthermore, this study also exemplified an effect of deletion of a druggable gene, Cnr1, on the regulation of G6PD cluster genes in DEN-induced HCC in mice. Overactivation of CB1R has been shown to promote HCC initiation and progression [46]. Deletion of the Cnr1 gene significantly attenuated the activation of G6PD cluster genes in HCC. This finding supports previous observations of the potential involvement of CB1R in the stratification of HCC patients [16] and also reinstates the therapeutic potential of peripheral CB1R antagonism in HCC [46].
In conclusion, our systems level analyses of multi-omics data [72] highlighted substantial differences in redox behavior in HCC tumors, relationships between redox genes, and genes that are liver specific, involved in the immune system, or inflammation. Different redox responses in subgroups of HCC tumors and their relationships with clinical outcome were also identified. We finally identified several novel markers of survival and prognosis and showed that they may be targeted for the treatment of HCC in specific patient groups, depending on the redox response.
Acknowledgments
Acknowledgements
This work was financially supported by the Knut and Alice Wallenberg Foundation and by intramural funds from the National Institute on Alcohol Abuse and Alcoholism, USA.
Declaration of interests
A.M., M.U., and J.B. are founders and shareholders of ScandiBio Therapeutics and ScandiEdge Therapeutics. All other authors have nothing to declare.
Author contributions
Conceptualization, R.B., G.B., A.M.; Formal Analysis, R.B., G.B., S.L., C.Z., M.A.; Resources, R.B., G.B., C.Z; Investigation, R.B., G.B., B.M., M.A., M.K.; Data Curation, R.B., G.B, B.M.; Writing – Original Draft, R.B., G.B., J.N., M.U., J.B., A.M.; Writing – Review & Editing R.B., G.B., B.M., M.K., R.C., C.Z., S.L., M.A., J.N., M.U., G.K., J.B., A.M.; Visualization, R.B., G.B.; Supervision – A.M.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.057.
Contributor Information
Rui Benfeitas, Email: rui.benfeitas@scilifelab.se.
Gholamreza Bidkhori, Email: gholamreza.bidkhori@scilifelab.se.
Bani Mukhopadhyay, Email: mukhopadhyayb@mail.nih.gov.
Martina Klevstig, Email: martina.klevstig@wlab.gu.se.
Muhammad Arif, Email: muhammad.arif@scilifelab.se.
Cheng Zhang, Email: cheng.zhang@scilifelab.se.
Sunjae Lee, Email: sunjae.lee@scilifelab.se.
Resat Cinar, Email: resat.cinar@nih.gov.
Jens Nielsen, Email: nielsenj@chalmers.se.
Mathias Uhlen, Email: mathias.uhlen@scilifelab.se.
Jan Boren, Email: jan.boren@wlab.gu.se.
George Kunos, Email: george.kunos@nih.gov.
Adil Mardinoglu, Email: adilm@scilifelab.se.
Appendix A. Supplementary data
Appendix Figures S1 – S9 and Doc S1.
Genome-scale metabolic model used in this work (SBML)
Genes involved in redox metabolism.
Co-expression analysis between genes directly involved in redox metabolism vs all protein-coding genes.
Co-expression analysis between redox genes.
Co-expression analysis between redox genes, and markers of differentiation.
Co-expression between redox genes, liver-specific genes, genes involved in the immune system/inflammation, and gene set enrichment with respect to liver-specific and immune system genes.
GSEA and associated differentially expressed redox genes in low vs high survival patients.
GSEA for genes in the ALDH2 and G6PD clusters.
Reporter metabolite analysis, GSEA, and metabolite-gene associations.
Differential expression analysis between hALDH2 and hG6PD tumors.
GEM modeling of hALDH2 or hG6PD tumors.
Tumor-specific GEM modeling and Reporter metabolites.
Cluster-specific GEM modeling and reporter metabolites performed on an independent cohort.
Differential expression analysis through HCC progression.
Comparison of clinical traits between groups of tumors and with respect to gene expression.
Kaplan-Meier analysis and survival signatures based on single and combinations of redox genes.
Co-expression analysis among redox proteins.
Patient clinical data (Sheet 2) and GSEA (Sheet 3) for differentially expressed genes between low- vs high-survival patients.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2001;5(1):87–107. doi: 10.1016/s1089-3261(05)70155-0. [vi] [DOI] [PubMed] [Google Scholar]
- 3.Altekruse S.F., McGlynn K.A., Reichman M.E. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friemel J., Rechsteiner M., Frick L. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21(8):1951–1961. doi: 10.1158/1078-0432.CCR-14-0122. [DOI] [PubMed] [Google Scholar]
- 5.Benfeitas R., Uhlen M., Nielsen J., Mardinoglu A. New challenges to study heterogeneity in cancer redox metabolism. Front Cell Dev Biol. 2017;5:65. doi: 10.3389/fcell.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown G.C., Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12(1):1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Nathan C., Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat Rev Immunol. 2013;13(5):349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornsveld M., Dansen T.B. The hallmarks of cancer from a redox perspective. Antioxid Redox Signal. 2016;25(6):300–325. doi: 10.1089/ars.2015.6580. [DOI] [PubMed] [Google Scholar]
- 9.Fulda S., Galluzzi L., Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 10.Yizhak K., Chaneton B., Gottlieb E., Ruppin E. Modeling cancer metabolism on a genome scale. Mol Syst Biol. 2015;11(6):817. doi: 10.15252/msb.20145307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mardinoglu A., Gatto F., Nielsen J. Genome-scale modeling of human metabolism - a systems biology approach. Biotechnol J. 2013;8(9):985–996. doi: 10.1002/biot.201200275. [DOI] [PubMed] [Google Scholar]
- 12.Mardinoglu A., Nielsen J. Systems medicine and metabolic modelling. J Intern Med. 2012;271(2):142–154. doi: 10.1111/j.1365-2796.2011.02493.x. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. [e23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaisaingmongkol J., Budhu A., Dang H. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell. 2017;32(1):57–70. doi: 10.1016/j.ccell.2017.05.009. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., Nielsen J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10(3):721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjornson E., Mukhopadhyay B., Asplund A. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 2015;13(9):2014–2026. doi: 10.1016/j.celrep.2015.10.045. [DOI] [PubMed] [Google Scholar]
- 17.Bidkhori G, Benfeitas R, Klevstig M, et al. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. P Natl Acad Sci USA in press. [DOI] [PMC free article] [PubMed]
- 18.Grossman R.L., Heath A.P., Ferretti V. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 20.Lee S., Zhang C., Arif M. TCSBN: a database of tissue and cancer specific biological networks. Nucleic Acids Res. 2018;46(D1):D595–D600. doi: 10.1093/nar/gkx994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Dam S., Vosa U., van der Graaf A., Franke L., de Magalhaes J.P. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2018;19(4):575–592. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C., Zhang Z., Zhu Y., Qin S. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med Chem. 2014;14(2):280–289. doi: 10.2174/18715206113136660337. [DOI] [PubMed] [Google Scholar]
- 24.Hou G., Chen L., Liu G. Aldehyde dehydrogenase-2 (ALDH2) opposes hepatocellular carcinoma progression by regulating AMP-activated protein kinase signaling in mice. Hepatology. 2017;65(5):1628–1644. doi: 10.1002/hep.29006. [DOI] [PubMed] [Google Scholar]
- 25.Uhlen M., Fagerberg L., Hallstrom B.M. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes D.R., Yu J., Shanker K. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M., Zhang C., Lee S. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352) doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 28.Patil K.R., Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci U S A. 2005;102(8):2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monti S., Tamayo P., Mesirov J., Golub T. Consensus clustering. Machine Learn. 2003;52(1):91–118. [Google Scholar]
- 30.Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Metagenes and molecular pattern discovery using matrix factorization. P Natl Acad Sci USA. 2004;101(12):4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J.S., Chu I.S., Heo J. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.S., Heo J., Libbrecht L. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12(4):410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 34.Jensen P.A., Papin J.A. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics. 2011;27(4):541–547. doi: 10.1093/bioinformatics/btq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen P.A., Lutz K.A., Papin J.A. TIGER: toolbox for integrating genome-scale metabolic models, expression data, and transcriptional regulatory networks. BMC Syst Biol. 2011;5:147. doi: 10.1186/1752-0509-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo H.G., Lee J.H., Yoon J.H. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70(8):3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cairo S., Armengol C., De Reynies A. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14(6):471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Hoshida Y., Nijman S.M., Kobayashi M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S.M., Leem S.H., Chu I.S. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55(5):1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo H.G., Park E.S., Cheon J.H. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14(7):2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 41.Sohn B.H., Shim J.J., Kim S.B. Inactivation of hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin Cancer Res. 2016;22(5):1256–1264. doi: 10.1158/1078-0432.CCR-15-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roessler S., Jia H.L., Budhu A. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roessler S., Long E.L., Budhu A. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(4):957–966. doi: 10.1053/j.gastro.2011.12.039. [e12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naboulsi W., Megger D.A., Bracht T. Quantitative tissue proteomics analysis reveals versican as potential biomarker for early-stage hepatocellular carcinoma. J Proteome Res. 2016;15(1):38–47. doi: 10.1021/acs.jproteome.5b00420. [DOI] [PubMed] [Google Scholar]
- 45.Lee S., Zhang C., Liu Z. Network analyses identify liver-specific targets for treating liver diseases. Mol Syst Biol. 2017;13(8):938. doi: 10.15252/msb.20177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay B., Schuebel K., Mukhopadhyay P. Cannabinoid receptor 1 promotes hepatocellular carcinoma initiation and progression through multiple mechanisms. Hepatology. 2015;61(5):1615–1626. doi: 10.1002/hep.27686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 48.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11(12):1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vafa O., Wade M., Kern S. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9(5):1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 50.Ben-Neriah Y., Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12(8):715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 51.Prieto J., Melero I., Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2015;12(12):681–700. doi: 10.1038/nrgastro.2015.173. [DOI] [PubMed] [Google Scholar]
- 52.Kawai K., Uemura M., Munakata K. Fructose-bisphosphate aldolase A is a key regulator of hypoxic adaptation in colorectal cancer cells and involved in treatment resistance and poor prognosis. Int J Oncol. 2017;50(2):525–534. doi: 10.3892/ijo.2016.3814. [DOI] [PubMed] [Google Scholar]
- 53.Ji S.R., Zhang B., Liu J. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett. 2016;374(1):127–135. doi: 10.1016/j.canlet.2016.01.054. [DOI] [PubMed] [Google Scholar]
- 54.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corrocher R., Casaril M., Bellisola G. Severe impairment of antioxidant system in human hepatoma. Cancer. 1986;58(8):1658–1662. doi: 10.1002/1097-0142(19861015)58:8<1658::aid-cncr2820580814>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 56.Bellisola G., Casaril M., Gabrielli G.B., Caraffi M., Corrocher R. Catalase activity in human hepatocellular carcinoma (HCC) Clin Biochem. 1987;20(6):415–417. doi: 10.1016/0009-9120(87)90007-5. [DOI] [PubMed] [Google Scholar]
- 57.Kampf C., Mardinoglu A., Fagerberg L. The human liver-specific proteome defined by transcriptomics and antibody-based profiling. FASEB J. 2014;28(7):2901–2914. doi: 10.1096/fj.14-250555. [DOI] [PubMed] [Google Scholar]
- 58.Asaka M., Kimura T., Nishikawa S., Miyazuki T., Alpert E. Decreased serum aldolase B levels in patients with malignant tumors. Cancer. 1988;62(12):2554–2557. doi: 10.1002/1097-0142(19881215)62:12<2554::aid-cncr2820621217>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 59.Chang Y.C., Chan Y.C., Chang W.M. Feedback regulation of ALDOA activates the HIF-1alpha/MMP9 axis to promote lung cancer progression. Cancer Lett. 2017;403:28–36. doi: 10.1016/j.canlet.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Nordmann Y. A shift of liver aldolase towards the muscle type in hepatoma: immunological study. Eur J Cancer. 1969;5(5):485–489. doi: 10.1016/0014-2964(69)90102-9. [DOI] [PubMed] [Google Scholar]
- 61.Baba Y., Nosho K., Shima K. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol. 2010;176(5):2292–2301. doi: 10.2353/ajpath.2010.090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z., Lu X.Y., Wang Z.C. Co-expression of PKM2 and TRIM35 predicts survival and recurrence in hepatocellular carcinoma. Oncotarget. 2015;6(4):2539–2548. doi: 10.18632/oncotarget.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matakidou A., el Galta R., Rudd M.F. Prognostic significance of folate metabolism polymorphisms for lung cancer. Brit J Cancer. 2007;97(2):247–252. doi: 10.1038/sj.bjc.6603830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabregat A., Sidiropoulos K., Garapati P. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varemo L., Nielsen J., Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41(8):4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 69.Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9(3) doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyotylainen T., Jerby L., Petaja E.M. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat Commun. 2016;7:8994. doi: 10.1038/ncomms9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Therneau T.M., Grambsch P.M. Springer Science & Business Media; 2013. Modeling survival data: Extending the Cox model. [Google Scholar]
- 72.Mardinoglu A., Boren J., Smith U., Uhlen M., Nielsen J. Systems biology in hepatology: approaches and applications. Nature Reviews Gastroenterology & Hepatology. 2018;15:365–377. doi: 10.1038/s41575-018-0007-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Figures S1 – S9 and Doc S1.
Genome-scale metabolic model used in this work (SBML)
Genes involved in redox metabolism.
Co-expression analysis between genes directly involved in redox metabolism vs all protein-coding genes.
Co-expression analysis between redox genes.
Co-expression analysis between redox genes, and markers of differentiation.
Co-expression between redox genes, liver-specific genes, genes involved in the immune system/inflammation, and gene set enrichment with respect to liver-specific and immune system genes.
GSEA and associated differentially expressed redox genes in low vs high survival patients.
GSEA for genes in the ALDH2 and G6PD clusters.
Reporter metabolite analysis, GSEA, and metabolite-gene associations.
Differential expression analysis between hALDH2 and hG6PD tumors.
GEM modeling of hALDH2 or hG6PD tumors.
Tumor-specific GEM modeling and Reporter metabolites.
Cluster-specific GEM modeling and reporter metabolites performed on an independent cohort.
Differential expression analysis through HCC progression.
Comparison of clinical traits between groups of tumors and with respect to gene expression.
Kaplan-Meier analysis and survival signatures based on single and combinations of redox genes.
Co-expression analysis among redox proteins.
Patient clinical data (Sheet 2) and GSEA (Sheet 3) for differentially expressed genes between low- vs high-survival patients.
Data Availability Statement
The GEMs used in this work are found in SBML format (Computer Code 1). The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files, or otherwise clearly point to the online resources where they were obtained.