Abstract
The eukaryotic ribosome is assembled through a complex process involving more than 200 factors. As preribosomal RNA is transcribed, assembly factors bind the nascent pre-rRNA and guide its correct folding, modification, and cleavage. While these early events in the assembly of the small ribosomal subunit have been relatively well characterized, assembly of the large subunit precursors, or pre-60S, is less well understood. Recent structures of nucleolar intermediates of large subunit assembly have shed light on the role of many early large subunit assembly factors, but how these particles emerge is still unknown. Here, we use the expression and purification of truncated pre-rRNAs to examine the initial assembly of pre-60S particles. Using this approach, we can recapitulate the early recruitment of large subunit assembly factors mainly to the domains I, II, and VI of the assembling 25S rRNA.
Keywords: RNA, biochemistry, ribosome biogenesis
INTRODUCTION
Ribosome assembly in eukaryotes is a complex and intricate process that requires more than 200 nonribosomal factors in yeast (Woolford and Baserga 2013). Ribosome biogenesis begins in the nucleolus where RNA polymerase I transcribes the 35S pre-rRNA, a polycistronic RNA, which contains the 18S, 5.8S, and 25S rRNAs. These rRNAs are flanked by 5′ and 3′ external transcribed spacers (ETS) and two internal transcribed spacers (ITS), ITS1 and ITS2. The 5S rRNA is transcribed separately by RNA polymerase III as a pre-5S RNA species. As RNA polymerase I transcribes the 35S pre-rRNA, ribosome assembly factors bind the nascent pre-rRNAs, forming large multicomponent complexes, which represent the earliest precursors of the small and large ribosomal subunit. Cleavage of the pre-rRNA at the sites A2 and A3, located within ITS1, separate small and large subunit pre-rRNA and thereby small and large subunit assembly.
Recent structures of nucleolar pre-60S particles have revealed some of the folding pathways that large subunit pre-rRNA follows (Kater et al. 2017; Sanghai et al. 2018; Zhou et al. 2018) showing that domains I, II, and VI of the 25S rRNA, along with the 5.8S rRNA, fold first into a near-mature conformation. Domains III–V remain unfolded in the early stages of large subunit assembly.
Despite the wealth of information provided by these new structures, it remains unclear how nucleolar pre-60S particles emerge. The recruitment of ribosome assembly factors on pre-rRNA has been elucidated for the first part of the pre-rRNA using the expression of truncated pre-rRNA to recapitulate this process (Chaker-Margot et al. 2015; Zhang et al. 2016). This has led to the description of the assembly of the earliest precursor of the small subunit (SSU). More recently, another group has used the same approach to recapitulate the assembly of large subunit precursors onto pre-rRNA (Chen et al. 2017). Here, using a similar technique, we described our model for early large subunit assembly.
RESULTS AND DISCUSSION
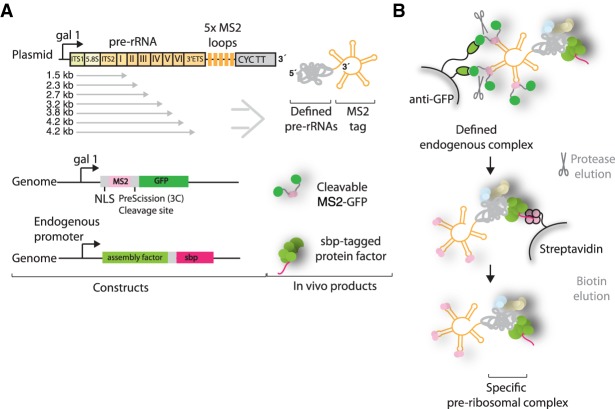
We set out to determine the order in which large ribosomal subunit assembly factors are recruited to the second half of the nascent pre-rRNA. For this purpose, purification of preribosomal complexes relies on the expression of tagged truncated pre-rRNAs, which are then bound by endogenous ribosome assembly factors and isolated in a two-step affinity purification, as described previously (Chaker-Margot et al. 2015). We coexpressed the truncated pre-rRNAs with five repeats of the MS2 aptamers at the 3′ end together with a protease-cleavable MS2-GFP fusion (Fig. 1A). The MS2 protein is fused to a nuclear localization signal (NLS) to promote its association with the truncated RNA in the cell. The expressed and truncated pre-rRNAs are recognized by the endogenous ribosome assembly machinery and isolated in a two-step purification (Fig. 1B).
FIGURE 1.
Purification of preribosomal particles. (A) Plasmid expression of aptamer-tagged pre-rRNAs of different lengths in genetically modified yeast strains containing a streptavidin-binding peptide (sbp)-tagged ribosome assembly factor. Plasmids contain the gal1 promoter and CYC transcriptional terminator (CYC TT). Pre-rRNA domains, sbp-tagged ribosome assembly factor, and MS2-GFP protein are shown. (B) Purification strategy using GFP and sbp as sequential tags, followed by biotin elution.
The RNA constructs used here begin at the A3 site, thereby mimicking A3 cleaved pre-rRNAs. Previous studies have suggested that separation of the rDNA locus between the A2 and A3 sites, at nucleotides 2739–2747 of the 35S pre-rRNA, leads to viable ribosome production, suggesting that there is no essential link between small and large subunit assembly factors (Liang and Fournier 1997). Target RNAs span the second half of the 35S locus, ending at each of the structural domains of the 25S, I–VI (Fig. 1A). The final and longest construct spans the entirety of the 25S and ends at the B0 site. A “full-length” construct that ended at the native end of the pre-rRNA underwent B0 cleavage, thereby separating the 3′ MS2 tag from the rest of the RNA and making isolation impossible (data not shown). The ITS2-binding nucleolar factor Cic1 was chosen as protein bait for the second capture (Wu et al. 2016). By using Cic1 as protein bait, we ensured that ITS2 processing has not yet occurred, thereby narrowing the type of pre-60S particles that can be isolated to earlier species.
Upon induction, truncated pre-rRNAs are expressed and bound by MS2-GFP and the endogenous large subunit assembly machinery, including the streptavidin-binding peptide (sbp)-tagged Cic1. During the purification, the RNA molecules are captured by the anti-GFP resin (Fig. 1B). In the second step, the targeted preribosomal complex is enriched by streptavidin capture and subsequent elution with the addition of biotin.
The expressed truncated pre-rRNAs were analyzed in total RNA extracts by northern blotting using an MS2 probe (Supplemental Fig. 1A). RNA bands corresponding to their expected size were visualized for each sample but were weak for the domain VI and B0-truncated species. Surprisingly, the major bands for the domain VI and B0 species run lower than the domain V band. These bands could represent degradation or processing products of these species and have been observed previously (Chen et al. 2017). Since the MS2 loops are located at the 3′ end of the constructs, this allows mapping the approximate cleavage site to around nucleotide 1350 in domain II. The mechanism by which this cleavage happens is unknown.
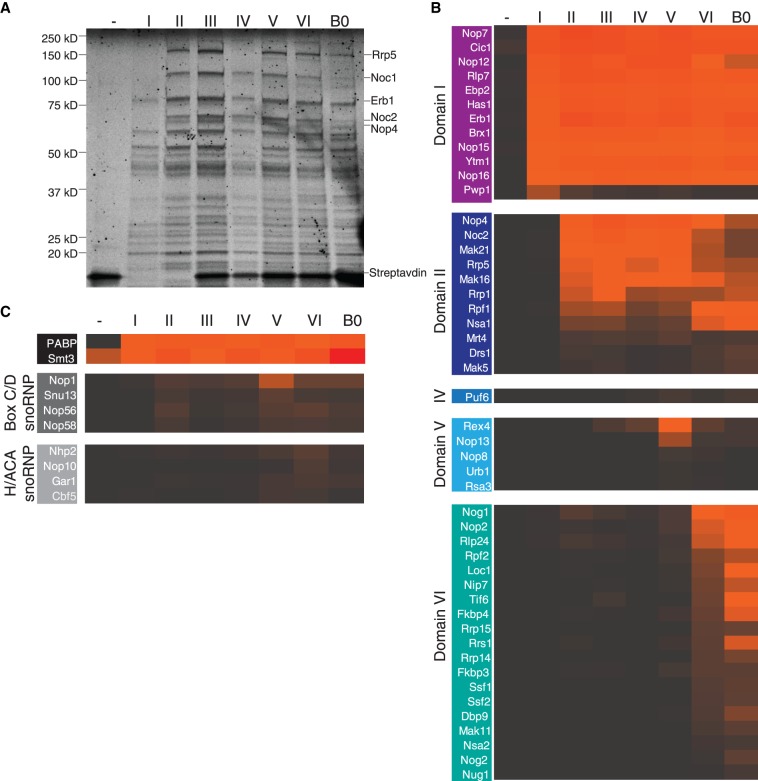
The use of our purification system has allowed us to recapitulate the assembly of the earliest large subunit assembly intermediates. The negative control, wherein only the MS2 loops were expressed, yielded an accordingly clean sample, except for the presence of streptavidin (Fig. 2A). The addition of the first section of pre-rRNA, spanning from the A3 site to the end of domain I of the 25S, led to the recruitment of approximately 10 factors. The subsequent addition of domains of the 25S induced the recruitment of more factors, although in a less dramatic fashion than what was observed for the SSU processome (Chaker-Margot et al. 2015). Samples for all seven constructs, with termini at domains I–VI and B0 plus a negative control were purified in biological triplicates and analyzed by LC-MS. The median amounts (as quantified by areas) for each protein were then used for the data analysis (Fig. 2B,C). Only proteins with amounts within 100-fold of the protein bait Cic1, and with more than five unique peptides were considered for the analysis. The amounts of Cic1, which should stay similar throughout, were used to normalize the samples. As described previously (Chaker-Margot et al. 2015), ribosomal proteins have been excluded from this analysis, as signals of highly abundant peptides from contaminating mature ribosomes prevent an unambiguous assignment of ribosomal proteins to a particular stage of ribosome assembly in our system.
FIGURE 2.
Stage-specific assembly of nucleolar pre-60S particles. (A) SYPRO-Ruby-stained SDS-PAGE gel of preribosomal complexes corresponding to increasing length of large subunit pre-RNA. Ribosome assembly factors are indicated on the right. (B) LC-MS analysis of ribosome assembly factors, based on a triplicate of samples, normalized against Cic1 and classified by recruitment stage. Protein abundance is measured by peak area, and high abundance proteins are shown in orange/red while low abundance and absent proteins are shown in black/gray. (C) LC-MS analysis of the polyadenylate-binding protein (PABP), Smt3, and the H/ACA and box C/D snoRNP proteins.
This analysis has allowed us to determine the timing of recruitment of approximately 50 assembly factors on nascent pre-rRNA (Fig. 2B,C). The first construct (containing 5.8S rRNA, ITS2, and domain I) recruits 12 assembly factors, including several known binders of ITS2 (Cic1, Nop7, Nop15, and Rlp7) (Wu et al. 2016). The helicase Has1, which is involved in small and large subunit processing (Emery et al. 2004), is also incorporated in this particle. Strikingly, this first construct also induces the recruitment of the protein Pwp1, whose levels decrease drastically for longer constructs, indicating its transient role in early large subunit assembly (Talkish et al. 2014). High levels of the ubiquitin-like modifier SUMO (Smt3) were detected in all intermediates, suggesting a possible functional role in early large subunit assembly (Fig. 2C; Panse et al. 2006; Raman et al. 2016).
Extension of the pre-rRNA into domain II recruits another 11 factors. Importantly, and in contrast to previous data (Chen et al. 2017), these include the large protein Rrp5 and its known interaction partners, Noc1 and Noc2. Rrp5 is required for the assembly of the SSU while Noc1/Noc2 have been shown to coprecipitate with the SSU processome (Chaker-Margot et al. 2015), likely through their interaction with Rrp5 (Hierlmeier et al. 2013). In vivo cross-linking studies of Rrp5 have shown its proximity to several sites on the pre-rRNA, including the central domain of the 18S, ITS1, and domain II of the 25S, consistent with our data (Lebaron et al. 2013). Domain II also leads to the association of several factors completing the ring structure around domains I and II, which was described in the nucleolar pre-60S structure (Sanghai et al. 2018), composed of Nsa1, Rpf1, Mak16, Rrp1, and the already associated Brx1 and Ebp2.
While the addition of domain III recruits no specific assembly factors, domain IV induces the binding of a single protein, Puf6. Domain V leads to the appearance of Nop13, a known nucleolar pre-60S factor and three members of the Npa/Urb subcomplex: Urb1, Nop8, and Rsa3. These proteins form a subcomplex, which also includes Urb2 and the helicase Dbp6, involved in the earliest steps of large subunit biogenesis (Rosado et al. 2007; Joret et al. 2018). While its role in large subunit assembly is still poorly understood, these data suggest that this complex associates with domain V.
Completion of the 25S rRNA induced the recruitment of many factors, including the GTPase Nog1, which remains associated with the pre-60S particle until its removal in the nucleus, as well as Tif6 and Rlp24, which are removed in the cytoplasm. Several nucleolar-specific factors also bound at this stage, such as Nop2, Rrp14, the Ssf1-Rrp15 dimer, and Nip7. Surprisingly, low levels of Nog2 were also detected in the A3-domain VI sample (Fig. 2B). Nog2 is a nuclear assembly factor whose binding site in the pre-60S particles overlaps with nucleolar factors such as Nip7, Sbp1, and Nop2 (Wu et al. 2016; Kater et al. 2017), which makes its association with nucleolar intermediates unlikely. This may be evidence that the A3-domain VI sample represents multiple pre-60S intermediates, which have matured to different extents rather than a specific large subunit precursor. Importantly, we did not detect the presence of Nop53, which is now known to have overlapping binding sites with Erb1 and is required for ITS2 processing at later stages of maturation (Thoms et al. 2015; Kater et al. 2017; Sanghai et al. 2018).
Maturation of large subunit precursors involves the recruitment of the 5S rRNA, as well as the modification of pre-rRNA guided by small nucleolar RNAs. We therefore tested the presence of such RNAs in each of the samples. Northern blotting analysis of the samples showed that the 5S rRNA appears with the addition of domain VI (Supplemental Fig. 1B). Consistent with this result, we observed that the chaperones of the 5S rRNA, Rpf2 and Rrs1, are also recruited at domain VI.
snR10, which is involved in the pseudouridylation of U2923, in domain V near domain VI (Ni et al. 1997), is also recruited to domain VI (Supplemental Fig. 1B). Correspondingly, there was a moderate increase in signal in the levels of box H/ACA proteins (Cbf5, Nhp2, Nop10, and Gar1) at domain VI, consistent with the presence of snR10. The presence of both 5S rRNA and snR10 is qualitative only, since the truncated pre-rRNAs could not be used as loading controls due to degradation (data not shown).
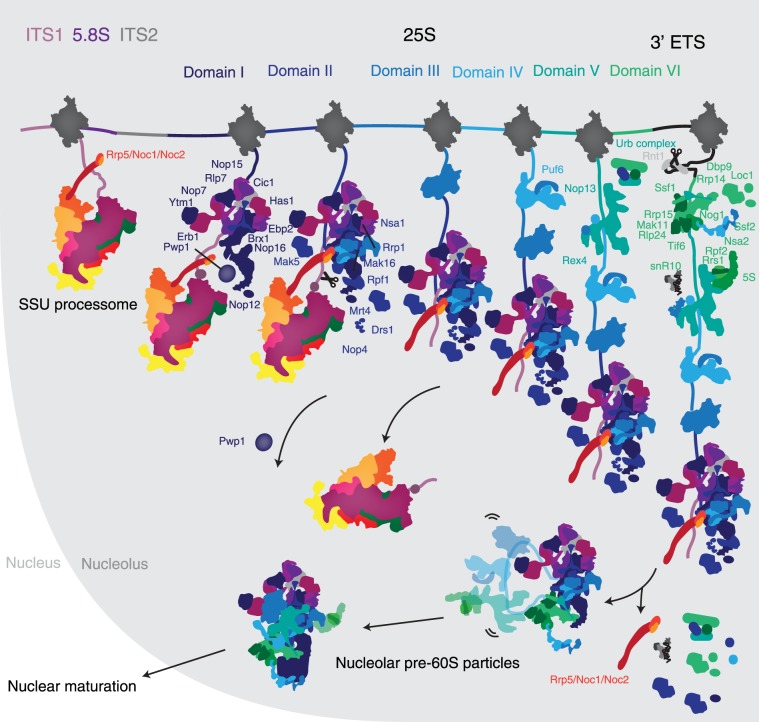
By expressing truncated pre-rRNAs, we have recapitulated the assembly of ribosome biogenesis factors on the second half of the 35S pre-rRNA. Large subunit assembly begins with the recruitment of many factors, which bind ITS2, 5.8S, and domain I. With the addition of domain II, the Rrp5-Noc1-Noc2 complex, which is also associated with the SSU processome, binds the forming large subunit, thereby bridging small and large subunit assembly (Fig. 3). Further transcription does not lead to the addition of many additional factors, since domains III–V are flexible in the early stages of pre-60S maturation (Supplemental Fig. 2), as seen in the cryo-EM structure of nucleolar pre-60S particles (Kater et al. 2017; Sanghai et al. 2018; Zhou et al. 2018). Completion of domain VI induces the incorporation of almost 20 ribosome assembly factors, the 5S rRNA and association of snR10. Many of these factors will stay associated with the pre-60S particle until their removal in the nucleus or the cytoplasm. In contrast, the Rrp5-Noc1-Noc2 complex dissociates from these early precursors before their exit from the nucleolus (Fig. 3).
FIGURE 3.
Assembly of the nucleolar pre-60S particle. Updated model of the early assembly of large ribosomal subunit precursors. Stepwise recruitment of ribosome assembly factors is concluded by cleavage of the B0 site by Rnt1 and release of the emerging pre-60S. Rearrangement of this particle induces the departure of several factors and the formation of the nucleolar pre-60S that was observed previously.
Our approach relies on the uncoupling of small and large subunit assembly, which happen simultaneously in vivo. It is therefore possible that specific recruitment events, especially those involving early factors, occur in a different order when the assembly processes of both subunits are coupled.
The study of pre-rRNA structural transitions by chemical probing in the early stages of pre-60S maturation (Burlacu et al. 2017) provides interesting complementary data to our model. Comparative chemical probing of 35S, 27SA, and 27SB shows both early stabilization of the 5.8S and domain I–II, and flexibility of domain III–VI. These data are consistent with our model, whereby the early recruitment of factors to domains I and II lead to the stabilization of the first half of the large subunit pre-rRNA, and domains III–V remain flexible in the early stages of maturation. The observed flexibility in domain VI seems incompatible with the large number of factors recruited to that domain but may be explained by the 3′ processing of the pre-rRNA, which likely requires rearrangement of the pre-rRNA.
While our proposed model is in mostly good agreement with the data published in Chen et al. (2017), there remain significant differences. Most notably, their data suggests association of Rrp5-Noc1-Noc2 at domain I instead of domain II, and several factors such as Nip7, Nop2, Rrs1, and Rpf2 already recruited at domain V, instead of only later to domain VI. Furthermore, several factors are completely absent from their model, such as Mak5, Rpf1, Rrp14, Rrp14, Ssf1, Ssf2, and snR10, which we know are part of early nucleolar pre-60S particles. These differences may be attributable to a slightly different design of the expressed RNA—the MS2 aptamers are placed in ITS2 instead of at the 3′ end of the RNA—or different stringency during purification.
Maturation of large subunit precursors requires processing at the 5′ and 3′ of the precursor RNA, in ITS1 and the 3′ ETS. This processing involves cleavage and resection of the spacer RNAs by Rnt1 and Rex1 for the 3′ end and Rrp17 or Rat1 for the 5′ end (Henry et al. 1994; van Hoof 2000; Oeffinger et al. 2009). It is tempting to speculate that the removal of Rrp5-Noc1-Noc2 may be coupled to 5′ and 3′ processing of the pre-rRNA. Ongoing and future studies will elucidate the precise mechanisms by which these early precursors mature to form nucleolar pre-60S particles.
MATERIALS AND METHODS
Cloning of RNA sequences
Ribosomal RNA sequences were amplified directly for BY4741 yeast genomic DNA. Sequences corresponding to all constructs used here from Saccharomyces cerevisiae rDNA were cloned downstream from the Gal1 promoter of a derivative of pESC URA (Agilent Technologies) containing five copies of the MS2 aptamer tag at the 5′ or 3′ end.
Purification of preribosomal complexes assembled on truncated pre-rRNA
Large ribosomal subunit precursors were purified from yeast strains bearing one sbp tag on Cic1 transformed with the appropriate plasmid. The yeast was grown in fully synthetic medium supplement with 2% raffinose and induced with 2% galactose for 16 h. The cells were then harvested and cryogenically ground (Retsch PM100 CM). Cryo-ground yeast powder was resuspended in buffer A (50 mM Tris-HCl, pH 7.7 [20°C], 150 mM NaCl, 1 mM EDTA, 0.1% Triton-X100, PMSF, Pepstatin, E-64), and the lysate cleared by high-speed centrifugation. The cleared lysate was incubated with anti-GFP beads. After washes in buffer A, the immobilized complexes were incubated with TEV protease and the supernatant was applied to streptavidin beads. Beads were subsequently washed in buffer B (50 mM Tris-HCl pH 7.7 [20°C], 150 mM NaCl, 1 mM EDTA) and the complex was eluted in the same buffer, supplemented with 5 mM D-Biotin. Composition of the particles was analyzed on a 4%–12% SDS-PAGE gradient gel and by LC-MS.
Mass spectrometry-based quantitation of ribosome assembly factors
Following isolation of ∼5 µg of total protein, proteins were denatured in urea and reduced prior to alkylation of cysteine with iodoacetamide. Proteins were digested with LysC (Wako Chemicals) followed by trypsin (Promega) and desalted. Approximately 2 µg of each sample was injected for LC-MS/MS analysis (Q Exactive Plus, Thermo Scientific). Peptides were separated using a 12 cm × 75 µm C18 column (Nikkyo Technos Co., Ltd., Japan) at a flow rate of 200 nL/min, with a 5%–40% gradient over 100 min (buffer A 0.1% formic acid, buffer B 0.1% formic acid in acetonitrile).
Peptide/protein identification was conducted by searching a Swiss-Prot S. cerevisiae database (downloaded March 2016) with Mascot 2.5.1 (Matrix Science) using Proteome Discoverer 1.4 (Thermo Scientific) and filtered with <4 ppm error and Percolator at a 1% FDR. Search parameters were as follows: trypsin/P, allowing two missed cleavages, with oxidized methionine and protein amino-terminal acetylation for variable modifications and carbamidomethyl cysteine as a static modification. All the analyzed data are compiled in Supplemental File 1.
Northern blotting
RNA was extracted from either ∼0.1 g of yeast powder, or 200 µL of biotin eluates, which were resuspended in 1 mL of TRIzol (Life Technologies) and processed according to the manufacturer's instructions. Total RNA samples were separated on a denaturing 3.7% formaldehyde–1.2% agarose gel (SeaKem LE, Lonza). The RNA was subsequently transferred onto a cationized nylon membrane (Zeta-Probe GT, Bio-Rad) using downward capillary transfer. RNA was cross-linked to the membrane for northern blot analysis by UV irradiation with a UV Stratalinker 2400 (Stratagene). Cross-linked membranes were incubated with hybridization buffer (750 mM NaCl, 75 mM trisodium citrate, 1% [w/v] SDS, 10% [w/v] dextran sulfate, 25% [v/v] formamide) at 65°C for 30 min prior to addition of γ-32P-end-labeled DNA oligonucleotide probes.
To generate γ-32P-end-labeled DNA oligonucleotide probes, 1.5–2 µL of DNA oligonucleotide was incubated with 4 µL of γ-32P-ATP and 1 µL T4 PNK in 20 µL total T4 PNK buffer (NEB). The reaction was incubated at 37°C for 30 min and subsequently cleaned up by using the Illustra MicroSpin G-25 columns (GE Healthcare). Probes were allowed to hybridize at 65°C for 1 h and then at 37°C overnight. Membranes were washed once with wash buffer 1 (300 mM NaCl, 30 mM trisodium citrate, 1% [w/v] SDS) and once with wash buffer 2 (30 mM NaCl, 3 mM trisodium citrate, 1% [w/v] SDS) for 20 min each at 45°C. Radioactive signal was detected by exposure of the washed membranes to a storage phosphor screen which was scanned with a Typhoon 9400 variable-mode imager (GE Healthcare).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonas Baradun, Mirjam Hunziker, Zahra Sanghai, and Linamarie Miller for critical advice in designing the experiments. We also thank Henrik Molina, Milica Tesic Mark, and Brian D. Dill from the Rockefeller proteomics core facility for the processing of mass spectrometry samples. M.C-M. was supported by an NSERC PGS D fellowship. S.K. is supported by the Robertson Foundation, the Irma T. Hirschl Trust, the Alexandrine and Alexander L. Sinsheimer Fund, the Rita Allen Foundation, and a National Institutes of Health New Innovator Award (1DP2GM123459).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069799.118.
REFERENCES
- Burlacu E, Lackmann F, Aguilar LC, Belikov S, Van NR, Trahan C, Hector RD, Dominelli-Whiteley N, Cockroft SL, Wieslander L, et al. 2017. High-throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat Commun 8: 714 10.1038/s41467-017-00761-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. 2015. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22: 920–923. 10.1038/nsmb.3111 [DOI] [PubMed] [Google Scholar]
- Chen W, Xie Z, Yang F, Ye K. 2017. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res 45: 6837–6847. 10.1093/nar/gkx254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, De La Cruz J, Rocak S, Deloche O, Linder P. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol 52: 141–158. 10.1111/j.1365-2958.2003.03973.x [DOI] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463. 10.1002/j.1460-2075.1994.tb06530.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, Hamperl S, Ohmayer U, Rachel R, Jacob A, et al. 2013. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res 41: 1191–1210. 10.1093/nar/gks1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joret C, Capeyrou R, Belhabich-Baumas K, Plisson-Chastang C, Ghandour R, Humbert O, Fribourg S, Leulliot N, Lebaron S, Henras AK, et al. 2018. The Npa1p complex chaperones the assembly of the earliest eukaryotic large ribosomal subunit precursor. PLOS Genet 14: e1007597 10.1371/journal.pgen.1007597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I, et al. 2017. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171: 1599–1610.e13. 10.1016/j.cell.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe Å, French SL, Dudnakova T, de lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D. 2013. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol Cell 52: 707–719. 10.1016/j.molcel.2013.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W-Q, Fournier MJ. 1997. Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vivo rDNA system for dissecting ribosome biogenesis. Proc Natl Acad Sci 94: 2864–2868. 10.1073/pnas.94.7.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. 10.1016/S0092-8674(00)80238-X [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. 2009. Rrp17p Is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol Cell 36: 768–781. 10.1016/j.molcel.2009.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnädig M, Tollervey D, Hurt E. 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7: 1311–1321. 10.1111/j.1600-0854.2006.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman N, Weir E, Müller S. 2016. The AAA ATPase MDN1 acts as a SUMO-targeted regulator in mammalian pre-ribosome remodeling. Mol Cell 64: 607–615. 10.1016/j.molcel.2016.09.039 [DOI] [PubMed] [Google Scholar]
- Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. 2007. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol Cell Biol 27: 1207–1221. 10.1128/MCB.01523-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker-Margot M, Wang J, Chait BT, Klinge S. 2018. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556: 126–129. 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL. 2014. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 34: 1863–1877. 10.1128/MCB.01322-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. 2015. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162: 1029–1038. 10.1016/j.cell.2015.07.060 [DOI] [PubMed] [Google Scholar]
- van Hoof A. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365. 10.1093/emboj/19.6.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. 2016. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534: 133–137. 10.1038/nature17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G, Chen S, Ye K. 2016. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev 30: 718–732. 10.1101/gad.274688.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhu X, Zheng S, Tan D, Dong MQ, Ye K. 2018. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 10: 120–130. 10.1007/s13238-018-0526-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.