Abstract
In Streptomyces clavuligerus, the gene cluster involved in the biosynthesis of the clinically used β-lactamase inhibitor clavulanic acid contains a gene (orf12 or cpe) encoding a protein with a C-terminal class A β-lactamase-like domain. The cpe gene is essential for clavulanic acid production, and the recent crystal structure of its product (Cpe) was shown to also contain an N-terminal isomerase/cyclase-like domain, but the function of the protein remains unknown. In the current study, we show that Cpe is a cytoplasmic protein and that both its N- and C-terminal domains are required for in vivo clavulanic acid production in S. clavuligerus. Our results along with those from previous studies allude towards a biosynthetic role for Cpe during the later stages of clavulanic acid production in S. clavuligerus. Amino acids from Cpe essential for biosynthesis were also identified, including one (Lys89) from the recently described N-terminal isomerase-like domain of unknown function. Homologues of Cpe from other clavulanic acid-producing Streptomyces spp. were shown to be functionally equivalent to the S. clavuligerus protein, whereas those from non-producers containing clavulanic acid-like gene clusters were not. The suggested in vivo involvement of an isomerase-like domain recruited by an ancestral β-lactamase related protein, supports a previous hypothesis that Cpe could be involved in a step requiring the opening and modification of the clavulanic acid core during its biosynthesis from 5S precursors.
Introduction
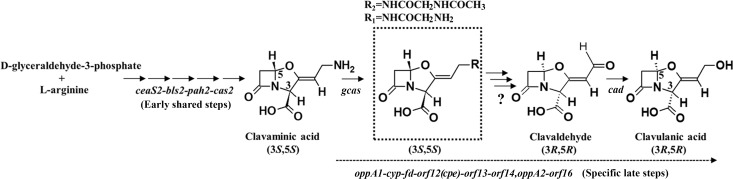
The β-lactam class of antibiotics have broad-spectrum activity and include some of the most commonly prescribed agents used for treating bacterial infections [1–3]. They have a long history of use in medicine, but as with other antibiotics, the emergence of resistance is a major problem [3–5]. There are several mechanisms responsible for β-lactam resistance, which include the production of secreted β-lactamases, enzymes that hydrolyze and inactivate certain members of this antibiotic class [6, 7]. Combinations of β-lactamase inhibitors such as clavulanic acid along with β-lactam antibiotics are often used as a strategy for treating some infections caused by β-lactamase-producing antibiotic resistant bacteria [8, 9]. Clavulanic acid belongs to the clavam family of specialized metabolites and it irreversibly inhibits class A β-lactamases, thereby restoring the activity of β-lactam antibiotics against target organisms in such combinations [10, 11]. The activity of clavulanic acid is attributed in part to its 3R,5R stereochemistry, as other naturally occurring clavams have a 5S configuration (collectively referred to as the 5S clavams) and do not inhibit β-lactamases [8, 12]. Commercial production of clavulanic acid is achieved by fermenting Streptomyces clavuligerus, and a cluster of ∼18 genes referred to as the clavulanic acid biosynthetic gene cluster (CA-BGC) encodes components of the core biosynthetic pathway [13]. It has previously been reported that Streptomyces jumonjinensis and Streptomyces katsurahamanus also produce clavulanic acid, but the sequences of their respective CA-BGCs are not available [12, 14]. On the other hand, the genome sequences of organisms such as Streptomyces flavogriseus (ATCC 33331, also known as S. pratensis) and Saccharomonospora viridis (DSM 43017) contain gene clusters closely resembling the S. clavuligerus CA-BGC, but neither has been shown to produce the metabolite to date [13, 15]. In addition, S. clavuligerus is somewhat unique among clavulanic acid producers as it also produces certain 5S clavams as products of a pathway related to clavulanic acid [13, 16]. Clavulanic acid and the 5S clavams have common biosynthetic origins and the pathway involved in their production can be roughly divided into two parts in S. clavuligerus (Fig 1). The “early” steps leading up to the intermediate clavaminic acid are shared during the production of both types of metabolites, with all intermediates possessing 5S configuration [17]. Beyond clavaminic acid (also a 5S clavam) the pathway diverges into specific “late” steps leading to either the 5S clavams or to clavulanic acid (Fig 1) [18].
Fig 1. Diagrammatic representation of the partial clavulanic acid biosynthetic pathway from Streptomyces clavuligerus.
Genes encoding enzymes known to be involved in the “early” shared stages of 5S clavam and clavulanic acid production, and those predicted to encode proteins involved exclusively in the biosynthesis of clavulanic acid (“late” steps) are indicated. In addition, genes encoding enzymes with known biosynthetic functions are shown next to arrows representing the respective reactions catalyzed by them, and the question mark indicates the unknown protein(s) responsible for the 5S to 5R epimerization and side chain modification of clavam intermediates during clavulanic acid biosynthesis. The two 5S clavam intermediates related to clavaminic acid (R1 = N-glycyl and R2 = N-acetyl-glycyl, respectively), which accumulate in the orf15 and orf16 gene mutants are shown in the dashed box.
The early shared portion of the pathway has been well characterized along with the genes involved in the process [19], but specific reactions involved in the production of each type of metabolite are yet to be elucidated [13]. It is currently hypothesized that during clavulanic acid production, the intermediate clavaminic acid undergoes oxidative deamination and ring inversion leading to clavaldehyde (Fig 1), which has 5R stereochemistry and is the immediate precursor of clavulanic acid [20]. The enzymes responsible for clavaldehyde formation are not known, but the products of orf10-17 from the CA-BGC are thought to play a role in the process [13, 17]. Previous reports have shown that the disruption of individual genes from the orf10-17 region abolishes or reduces clavulanic acid production without affecting 5S clavam levels [21–23]. Under certain conditions, the concomitant accumulation of acylated clavaminic acid derivatives was also observed in the orf15-16 mutants [23, 24], suggesting that the respective metabolites are intermediates from the clavulanic acid arm of the biosynthetic pathway (Fig 1). Because of the clinical applications of clavulanic acid, there is considerable interest in understanding how the metabolite is produced in S. clavuligerus.
Of particular relevance to the current study is the product of orf12 (SCLAV_4187) from the CA-BGC of S. clavuligerus, which resembles class A β-lactamases and also contains similar SXXK, SDN and KAG amino acid motifs [25]. orf12 is co-transcribed with orf13, which encodes a putative membrane transport protein (Fig 2A), and their relative arrangement also suggests possible translational coupling [23]. Due to the bioactivities of specialized metabolites (especially when the product is an antibacterial), producer organisms often employ self-resistance strategies for protection [26, 27]. Intrinsic resistance in β-lactam-producing organisms is often attributed to the presence of altered penicillin-binding proteins (PBPs, the targets of β-lactam antibiotics) with reduced binding affinities for endogenously-produced antibiotics [28], but BGCs from such organisms also contain genes encoding β-lactamases and efflux transporters [29, 30]. Studies have shown that orf12 is required for clavulanic acid, but not 5S clavam production [23] and that the encoded protein lacks any detectible β-lactamase activity. Instead, heterologously expressed and purified Orf12 was shown to function as a cephalosporin esterase under in vitro conditions [25], due to which it is henceforth referred to as Cpe (for cephalosporin esterase).
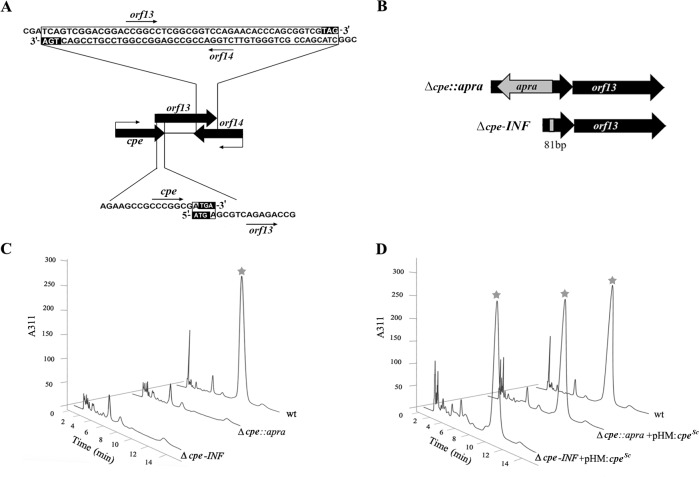
Fig 2. Preparation and analysis of S. clavuligerus cpe deletion mutants.
(A and B) The thick arrows depict genes with arrowheads indicating the direction of transcription. (A) The relative arrangement of genes from the chromosomal locus surrounding cpe in S. clavuligerus is shown, and the bent arrows represent the known promoters for the different transcriptional units. The DNA sequences of the overlapping regions between cpe-orf13 (bottom) and orf13-orf14 (top) are indicated in open boxes, whereas the respective start and stop codons are shown in filled boxes. (B) Diagrammatic representation of the Δcpe::apra and Δcpe-INF mutants, which were prepared such that the 5' and 3' ends of cpe were retained and intervening DNA sequences were replaced by an apramycin resistance cassette or an in-frame 81-bp sequence in the respective mutants. (C and D) HPLC analysis of 96 hour wt S. clavuligerus and different cpe mutant SA culture supernatants for assessing clavulanic acid production using the phosphate buffer system [32]. Peaks corresponding to imidazole-derivatized clavulanic acid are indicated by the star symbol, which were detected at 311nm.
The recently solved crystal structure of Cpe showed that in addition to a β-lactamase-like domain located in its C-terminus (residues 128–458), the protein also contains a previously unrecognized N-terminal domain (residues 1–127) resembling those found in steroid isomerases and polyketide cyclases [25]. In addition, two molecules of clavulanic acid were found to be bound non-covalently to Cpe when crystals of the protein were soaked in a solution of the metabolite during structural studies [25]. The first molecule (CA-1) was positioned in an active site pocket lined by residues (His88, Ser173, Thr209, Ser234, Ser278,Met383, Phe374, Ala376 and Phe385) from both the N- and the C-terminal domains, whereas the second molecule (CA-2) bound to a mostly hydrophobic cleft at the interface of the two domains via weak ionic interactions [25]. It was also shown that apart from Ser173, Ser234 and Ser378, other residues from Cpe or its N-terminal domain are not essential for its in vitro esterase activity. The ability of Cpe to bind clavulanic acid non-covalently under in vitro conditions is intriguing [25], as bona fide class A β-lactamases form irreversible covalent suicide adducts with the inhibitor [9]. In addition, β-lactamases are secreted out of the cell to inactive their antibiotic substrates [7], but the cellular location of Cpe in S. clavuligerus is not known. It is also not clear if Cpe undergoes post-translational processing in S. clavuligerus, or if both of its N- and C-terminal domains and associated amino acid residues are required for clavulanic acid production in the native host. Therefore, questions regarding the actual in vivo role of the Cpe gene product still remain unanswered, many of which are examined in the current study.
Materials and methods
Bacterial strains, plasmids and culture conditions
Dehydrated media components and reagents were purchased from VWR International, Fisher Scientific or Sigma-Aldrich (Canada). Details of bacterial strains and plasmids used in the current study are described in Tables 1 and 2, respectively. Escherichia coli and S. clavuligerus cultures were grown and manipulated as described previously [31, 32]. Other Streptomyces species were cultured using tryptic soy broth (TSB) or ISP4 media, whereas S. viridis was grown in nutrient broth (BD, Canada). Unless otherwise specified, all E. coli, Streptomyces and S. viridis cultures were grown at 37, 28 and 42°C, respectively. Appropriate antibiotics were included in the media when required [31, 33], and liquid cultures were agitated at 200 rpm. For assessment of metabolite production, S. clavuligerus strains were grown in duplicate in starch asparagine (SA) or soy fermentation media as described previously [19]. All production phenotypes were verified using at least two independent fermentations.
Table 1. Bacterial strains used in the current study.
| Bacterial strain | Antibiotic resistance marker(s)a | Description | Source/Referenceb |
|---|---|---|---|
| Escherichia coli strains | |||
| E. coli NEB5α | NA | DH5α derived cloning host | NEB |
| E. coli BL21(DE3) | NA | Host for protein expression | NEB |
| E. coli ET12567(pUZ8002) | CamR, KanR | DNA methylation deficient conjugation host containing the plasmid pUZ8002 | [33] |
| E. coli BW25118 (pIJ790) | CamR, KanR | Host containing the plasmid pIJ790 for λ RED mediated ReDirect PCR targeting of genes | [34] |
| E. coli DH5α (BT340) | AmpR, CamR | Strain containing plasmid BT340 used for expressing the FLP recombinase | [35] |
| Streptomyces and other strains | |||
| Streptomyces clavuligerus NRRL 3585 | NA | Wild type; cephamycin and clavulanic acid producer | NRRL |
| Streptomyces clavuligerus Δcpe::apra | AprR | cpe deletion mutant; gene replaced by disruption cassette from plasmid pIJ773 | This study |
| Streptomyces clavuligerus Δcpe-INF | NA | cpe deletion mutant; gene replaced by 81bp marker less in-frame scar sequence | This study |
| Streptomyces flavogriseus ATCC 33331 | NA | Wild type; clavulanic acid non-producer | ATCC |
| Saccharomonospora viridis ATCC 15386 | NA | Wild type; clavulanic acid non-producer | ATCC |
| Streptomyces katsurahamanus | NA | Wild type; cephamycin and clavulanic acid producer | [12] |
| Streptomyces jumonjinensis | NA | Wild type; cephamycin and clavulanic acid producer | [12] |
| Klebsiella pneumoniae ATCC 15380 | NA | Indicator organism for clavulanic acid bioassays | [32] |
a AmpR, ampicillin resistance; AprR, apramycin resistance; CamR, chloramphenicol resistance; KanR, kanamycin resistance; NA, Not applicable.
b ATCC, American Type Culture Collection; NEB, New England Biolabs; NRRL, Northern Regional Research Laboratory.
Table 2. Plasmids and cosmids used in the current study.
| Plasmid/cosmid | Antibiotic resistance marker(s)a | Description | Source/Reference |
|---|---|---|---|
| pGEMT-Easy | AmpR | General E. coli cloning vector | Promega |
| pET30b | KanR | E. coli protein expression vector | Novagen |
| pHM11a | HygR | Integrative Streptomyces expression vector containing the constitutive ermEp* | [36] |
| pSET152 | AprR | Integrative Streptomyces cloning vector | [37] |
| pIJ773 | AprR | Template plasmid for preparing the ReDirect apra disruption cassette | [34] |
| pIJ10700 | HygR | Template plasmid for preparing the ReDirect hyg disruption cassette | [34] |
| pET30b-cpeSc | KanR | Plasmid vector used to express C-terminal 6×His-tagged Cpe in E. coli for purification | This study |
| 12B8 | AmpR, KanR | Cosmid clone containing the clavulanic acid biosynthetic gene cluster from S. clavuligerus | [19] |
| 12B8-Δcpe::apra | AprR, AmpR, KanR | Mutant cosmid 12B8 in which cpe has been replaced by the disruption cassette from plasmid pIJ773 using the ReDirect system | This study |
| 12B8-Δcpe-INF | AmpR, KanR | Mutant cosmid 12B8 in which cpe has been replaced by the 81-bp in-frame “scar” sequence using the ReDirect system | This study |
| 12B8-Δcpe-INF-Δamp::hyg | HygR, KanR | Cosmid 12B8-Δcpe-INF in which ampicillin resistance gene replaced by the hyg cassette from plasmid pIJ10700 using the ReDirect system | This study |
| pHM:cpeSc | HygR | Expression plasmid pHM11a containing cpe from S. clavuligerus | This study |
| pHM:cpeSf | HygR | Expression plasmid pHM11a containing cpe from S. flavogriseus | This study |
| pHM:cpeSv | HygR | Expression plasmid pHM11a containing cpe from S. viridis | This study |
| pHM:cpeSj | HygR | Expression plasmid pHM11a containing cpe from S. jumonjinensis | This study |
| pHM:cpeSk | HygR | Expression plasmid pHM11a containing cpe from S. katsurahamanus | This study |
| pHM:blipFLAG | HygR | Expression plasmid pHM11a containing blip from S. clavuligerus with a C-terminal FLAG tag | This study |
| pHM:ccaRFLAG | HygR | Expression plasmid pHM11a containing ccaR from S. clavuligerus with a C-terminal FLAG tag | This study |
| pHM:cpeSc-FLAG | HygR | Expression plasmid pHM11a containing cpe from S. clavuligerus with a C-terminal FLAG tag | This study |
| pHM:cpeSc-6×his | HygR | Expression plasmid pHM11a containing cpe from S. clavuligerus with a C-terminal 6×His tag | This study |
| pHM:cpeCt | HygR | Expression plasmid pHM11a containing the C-terminal domain of cpe from S. clavuligerus | This study |
| pHM:cpeNt | HygR | Expression plasmid pHM11a containing the N-terminal domain of cpe from S. clavuligerus | This study |
| pHM-cpeCt+Nt | HygR | Expression plasmid pHM11a containing the N-terminal and C- terminal domains of cpe from S. clavuligerus, each expressed independently under the control of the ermEp* | This study |
| pSET:cpeSc | AprR | Plasmid pSET152 containing the S. clavuligerus cpe gene along with ermEp* from pHM11a was used as template to prepare all described cpeSc site directed mutants | This study |
aAmpR, ampicillin resistance; AprR, apramycin resistance; KanR, kanamycin resistance, HygR, hygromycin resistance.
DNA isolation, manipulation and analysis
All oligonucleotide primers used in the current study were purchased from Integrated DNA Technologies (USA) and are listed in S1, S2 and S3 Tables. Standard techniques were used to introduce, isolate, manipulate and analyze plasmid DNA from E. coli (35). Restriction enzymes used in the study were purchased from New England Biolabs Ltd. (Canada). Chromosomal DNA was isolated from Streptomyces and S. viridis cultures using the QIAamp DNA Mini Kit (QIAGEN, Canada) and a SpeedMill PLUS Bead Homogenizer (Analytik Jena, Germany), which was also used in all subsequent bead-beating purposes. PCR was performed using either the Fisher BioReagents Taq DNA polymerase or the Phusion High-Fidelity DNA Polymerase kits (Fisher Scientific, Canada) according to the manufacturer’s recommendations, except that 5% DMSO was included in problematic reactions. DNA fragments were purified after standard TBE agarose gel electrophoresis using the EZ-10 Spin Column DNA Gel Extraction Kit according to the manufacturer’s instructions (Bio Basic Canada Inc.). Unless otherwise specified, all PCR products were cloned into the pGEM-T Easy vector (Promega, USA) and the DNA sequences of all inserts were determined at the Centre for Applied Genomics, University of Toronto, Canada. Plasmid and cosmid constructs were introduced into S. clavuligerus through intergeneric conjugation using E. coli ET12567/pUZ8002 as described previously [19, 33].
Preparation of the S. clavuligerus Δcpe::apra and Δcpe-INF mutants
The pWE15 vector based cosmid clone 12B8 (Table 2) containing the entire clavulanic acid gene cluster was used to prepare the S. clavuligerus Δcpe mutants according to the previously described ReDirect PCR-Targeting method [19, 34]. Specific oligonucleotide primers (S1 Table) along with pIJ773 as template were used to amplify a PCR product containing the apramycin resistance cassette (apra) to target cpe in 12B8. This led to the replacement of an internal fragment of cpe by the apra disruption cassette to give the mutant cosmid 12B8-Δcpe::apra. In addition, the apra cassette comprising the aac3(IV) gene and RK2 oriT flanked by FLP recombinase target sites (FRT), was inserted in the direction opposite to cpe transcription in the mutant cosmid. 12B8-Δcpe::apra was then introduced into wt S. clavuligerus for double homologous recombination and isolation of the apramycin resistant, Δcpe::apra mutant.
In order to prepare the in-frame (INF) marker-less Δcpe-INF mutant, cosmid 12B8-Δcpe::apra from above was introduced in E. coli DH5α/BT340, which expresses the FLP recombinase [35]. FLP caused the excision of the FRT-flanked apra cassette in 12B8-Δcpe::apra, leaving an 81-bp in-frame DNA sequence (“scar”) in its place in the mutant cosmid 12B8-Δcpe-INF (Table 2). Since oriT is part of the apra cassette, it was also lost, and 12B8-Δcpe-INF could not be transferred to S. clavuligerus via conjugation. Therefore, an oriT was introduced into 12B8-Δcpe-INF using a second round of ReDirect PCR-Targeting [34]. Specified primers (S1 Table) were used along with pIJ10700 as a template to amplify a PCR product containing the hygromycin resistance cassette (hyg) to target the ampicillin resistance gene present on the pWE15 vector backbone of 12B8-Δcpe-INF. The resulting cosmid 12B8-Δcpe-INF-Δamp::hyg (Table 2), containing the hyg cassette (which in turn contains an oriT) in place of the ampicillin resistance gene, was transferred to the S. clavuligerus Δcpe::apra mutant by conjugation with E. coli. Hygromycin-resistant colonies that arose were then made to undergo sporulation without any antibiotic selection to isolate the apramycin and hygromycin sensitive S. clavuligerus Δcpe-INF mutant. The replacement of the wt cpe gene with Δcpe::apra and Δcpe-INF in the respective S. clavuligerus mutants was confirmed by genomic DNA PCR and sequencing of products using specific primers (S1 Table).
Preparation of cpe complementation plasmids
Specific oligonucleotide primers (S1 Table) with engineered NdeI and HindIII/BamHI restriction sites were used to PCR amplify DNA fragments containing the cpe genes from S. clavuligerus (Sc), S. jumonjinensis (Sj), S. katsurahamanus (Sk), S. flavogriseus (Sf) and S. viridis (Sv) for complementation studies. Since the sequences of cpe from S. jumonjinensis and S. katsurahamanus were not know, degenerate oligonucleotide primers with engineered restriction sites were designed based on known cpe DNA sequences from the three other species. After PCR amplification, the DNA fragments were directly cloned into the NdeI and HindIII/BamHI sites of the Streptomyces expression plasmid pHM11a [36] to give pHM:cpeSj, pHM:cpeSk, pHM:cpeSf and pHM:cpeSv (Table 2). The DNA sequences of all inserts were also verified/determined for comparison using custom primers (S1 Table).
To examine the in vivo roles of the N- and C-terminal domains of CpeSc, custom oligonucleotide primers were used to amplify DNA fragments containing each domain separately (S1 Table). The respective PCR fragments were cloned into pHM11a at NdeI and BamHI after their sequences had been verified to give pHM:cpeNt and pHM:cpeCt, which functioned as the CpeSc N- and C-terminal domain expression constructs, respectively (Table 2). To prepare a construct that could express the two domains separately at the same time from a single plasmid, the insert from pHM:cpeCt was released as a BglII-BamHI fragment and ligated to BamHI-digested pHM:cpeNt. This led to the plasmid pHM:cpeNt+Ct, in which the expression of each domain (not as part of the same protein) was driven independently by ermEp* (Table 2). Plasmid constructs were introduced into either the S. clavuligerus Δcpe::apra and/or Δcpe-INF mutants for complementation studies.
Detection and localization of CpeSc in S. clavuligerus
Engineered oligonucleotide primers were used to add C-terminal FLAG tags onto Cpe, CcaR and Blip (S1 Table). PCR fragments containing the three respective genes (cpeSc-FLAG, ccaRFLAG and blipFLAG) were cloned into pHM11a and introduced into wt S. clavuligerus for localization studies (Table 2). One hundred milliliters S. clavuligerus SA cultures expressing each protein were separately grown for 48 hours, after which the cultures were subjected to centrifugation and the mycelial pellets were separated from the supernatants. Cell pellets were resuspended in 5 ml of lysis buffer (150 mM HEPES and 150 mM NaCl) and were sonicated on ice using a 5/64-inch probe (VWR International, Canada). The lysates were centrifuged at high-speed (27,000 × g) for 15 minutes to clarify the cytoplasmic fraction contained in the supernatants for subsequent use. Approximately 87 ml of culture supernatant (separated from the above mycelial pellet in the first step) was centrifuged at 27,000 × g for 15 minutes and was then filtered through 0.2 μm vacuum membranes (VWR International, Canada) to remove any residual particulate or insoluble material. To precipitate secreted proteins, 44.9 g of ammonium sulfate was added gradually to 500 ml of the filtered supernatant (final volume is made up by using lysis buffer) with constant stirring at 4°C to give 80% saturation. Precipitated protein fractions were collected by high-speed centrifugation as described above, after which the supernatant was discarded, and the protein pellet was left to air dry for 10 minutes. The pellet was then resuspended in 500 μl of 1M phosphate buffer (sodium phosphate, pH-7.0) for future analysis.
C-terminal 6×His tagged protein (CpeSc-6×His) was also expressed in S. clavuligerus and E. coli. Engineered oligonucleotide primers were used to introduce a C-terminal 6×His tag during the amplification of cpeSc (S1 Table), which was cloned into pHM11a for expression in S. clavuligerus. For expressing CpeSc-6×His in E. coli, the gene was PCR amplified using primers listed in S1 Table, was cloned into pET30b for expression at 15°C for 24 hours. CpeSc-6×His protein was purified using Ni-NTA resin as per the manufacturer’s instructions (Qiagen, USA) and was stored in 20 mM Tris-HCl, 150 mM NaCl (pH 7.6) + 20% (v/v) glycerol.
For western analysis, 20–50 μg of cell-free extract or 0.5–1 μg of purified CpeSc-6×His was subjected to standard 12% SDS-PAGE before being transferred to Immobilon-P PVDF membranes according to the manufacturer’s recommendations (Millipore, Canada). Membranes were washed with TBS-T buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, and 0.5% v/v Tween-20) and were blocked overnight at 4°C in blocking buffer (TBS-T with 10% w/v non-fat milk). The membranes were probed using anti-FLAG or anti-6×His antibodies (Thermo Scientific Pierce, USA) at 1:500 final dilutions before being washed several times with TBS-T buffer. The secondary antibody (Thermo Scientific Pierce, USA) was added at 1:400 dilution in TBS-T buffer and the membranes were processed using the ECL Western Blot Substrate (Promega, USA) for imaging using a GE ImageQuant LAS 4000 Digital Imaging System (GE Healthcare, USA).
RNA isolation and RT-PCR analysis
S. clavuligerus wt and Δcpe-INF strains were used to isolate RNA after 48 hours of growth in SA medium using the innuSPEED Bacteria/Fungi RNA Kit and a bead beater as per the manufacturer’s instructions (Analytik Jena, Germany). The cDNA was synthesized using 500 ng of DNaseI-treated RNA using random hexameric primers provided with the SuperScript II reverse transcriptase (RT) kit as per the manufacturer’s recommendations (Invitrogen, USA). PCR was performed using 2.5μl of the RT product from above in a final volume of 20μl using the GoTaq DNA Polymerase (Promega, Canada). Thirty cycle PCR was performed to detect ceaS2, oat2, oppA1, claR, car, cyp, cpe (orf12), orf13, orf14, oppA2, orf16, gcas, pbpA, and hrdB cDNA using gene-specific primers (S2 Table). Control reactions contained DNaseI-treated RNA preparations without reverse transcription for each reaction.
Site-directed mutagenesis of CpeSc
The cpeSc gene along with the ermEp* from pHM11a was isolated as a BglII/BamHI fragment and inserted into the BamHI site of pSET152 (45) to prepare a smaller expression plasmid (pSET:cpeSc), which would be more amenable for site-directed mutagenesis (Table 2). The QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, USA) along with mutagenic oligonucleotide primers (S3 Table) and pSET:cpeSc as template was to prepare selected single amino acid variants of CpeSc according to the manufacturer’s instructions. All introduced mutations were verified by DNA sequencing, and plasmids expressing CpeSc variants (Table 2) were introduced into the S. clavuligerus Δcpe-INF mutant for complementation studies.
Metabolite detection and analysis
S. clavuligerus strains were grown for fermentation studies and culture supernatants were assessed for clavulanic acid production using bioassays as described previously [32]. High-performance liquid chromatography analysis of imidazole-derivatized culture supernatants was performed using a 1260 Infinity system (Agilent Technologies, USA) and a Bondclone C18 (100×8mm, 10μm, 148Å) column (Phenomenex, USA) [23]. Selected supernatants were also analyzed by liquid chromatography-mass spectrometry on an LC-MS-Trap system (1100 LC-MS Agilent Technologies, USA) as previously described [23, 38], with the exception that an Xterra (2.1×150 mm, 3.5μm, 125Å) column (Waters Scientific, USA) was used in the analysis.
Results
Preparation and complementation of the S. clavuligerus Δcpe::apra and Δcpe-INF deletion mutants
In S. clavuligerus, cpe (orf12) and orf13 are transcribed together as a polycistronic mRNA and the stop codon of cpe also overlaps with the start codon of orf13 [23]. In addition, there is a 48 bp overlap between the 3' ends of orf13 and orf14, which are encoded on opposite DNA strands (Fig 2A). Therefore, there is potential for polar effects on the expression (transcription and/or translation) of orf13 in a cpe gene mutant, depending on how it was prepared. To test this hypothesis, two different cpe mutants (Table 1) were prepared using the ReDirect two-step protocol [34]. In the first mutant, the apramycin (apra) cassette flanked by FLP recombinase target (FRT) sites from the plasmid pIJ773 was used to delete an internal region of cpe (39 bp from the 5' end to 39 bp from the 3' end), leading to the S. clavuligerus Δcpe::apra mutant (Fig 2B). The apra gene was inserted in the orientation opposite to cpe transcription to maximize the potential for polar effects on the expression of downstream genes. For preparing the second mutant, the apra cassette was excised from the Δcpe::apra mutant and replaced with an 81 bp scar sequence in the correct reading frame to give the S. clavuligerus Δcpe-INF (in frame deletion) mutant (Fig 2B), which has the least potential for producing polar effects on the expression of the downstream genes. The prepared mutants were verified by genomic DNA PCR and were complemented using the cpe gene from S. clavuligerus (cpeSc) expressed under the control of the constitutive ermE* promoter (ermEp*) in the plasmid pHM11a (Table 2). Wild-type and cpe mutant strains of S. clavuligerus containing either pHM11a (control) or pHM:cpeSc were grown in SA medium for up to 120 hours to assess for clavulanic acid production. Bioassays and HPLC analysis of culture supernatants demonstrated that both the S. clavuligerus Δcpe::apra and Δcpe-INF mutants were completely blocked in clavulanic acid production when compared to the wt strain (Fig 2C). Introduction of pHM:cpeSc restored clavulanic acid production to 60%-70% of wt levels in both mutants (Fig 2D), suggesting that the cpe disruption(s) was not associated with any significant polarity. The marker-less S. clavuligerus Δcpe-INF mutant was chosen for further analysis in the current study.
Cellular localization of CpeSc and its influence on the expression of other genes from the clavulanic acid biosynthetic gene cluster of S. clavuligerus
The CpeSc protein shares many sequence and structural similarities with class A β-lactamases, but it has been shown to lack any detectable β-lactamase activity under in vitro conditions [25]. Most bona fide β-lactamases are secreted proteins that inactivate β-lactam antibiotics in the periplasm, the site of peptidoglycan biosynthesis and crosslinking [39]. However, the predicted amino acid sequence of CpeSc does not contain any detectable secretion signals, warranting further investigation into its exact cellular location in S. clavuligerus. C-terminal FLAG (CpeSc-FLAG) and 6×His (CpeSc-6×His) epitope-tagged copies of the protein were separately expressed in wt S. clavuligerus using the constitutive ermEp* from plasmid pHM11a (Table 2). As controls for protein localization studies, S. clavuligerus strains expressing C-terminal FLAG-tagged copies of known cytoplasmic (CcaRFLAG) [40] and secreted (BlipFLAG) [41] proteins were also prepared separately (Table 2). S. clavuligerus strains expressing FLAG-tagged copies of the respective proteins were grown in SA medium for 48 hours for isolating different cellular protein fractions. Mycelial pellets were used to obtain cytoplasmic and cell wall-associated fractions, whereas enriched secreted fractions were prepared by using salt to precipitate soluble proteins from culture supernatants. Western blot analysis of different cellular fractions using anti-FLAG polyclonal antibodies demonstrated that CpeSc-FLAG was only detected in the cytoplasmic fraction (Fig 3). As expected, the CcarRFLAG and BlipFLAG controls were detected in cytoplasmic and secreted fractions, respectively (Fig 3). In addition, cultures of S. clavuligerus expressing CpeSc-6×His were also used for isolating fractions for western analysis, which confirmed that CpeSc is a cytoplasmic protein (S1 Fig). During the described western blot analysis, the size of epitope tagged CpeSc was determined to be ~54 kDa based on the signal obtained using anti-FLAG and anti-6×His antibodies (S1 Fig). This corresponded to the size of 6×His-tagged CpeSc heterologously expressed and purified from E. coli, which was used as a control (S1 Fig).
Fig 3. Cellular localization of Cpe in S. clavuligerus.
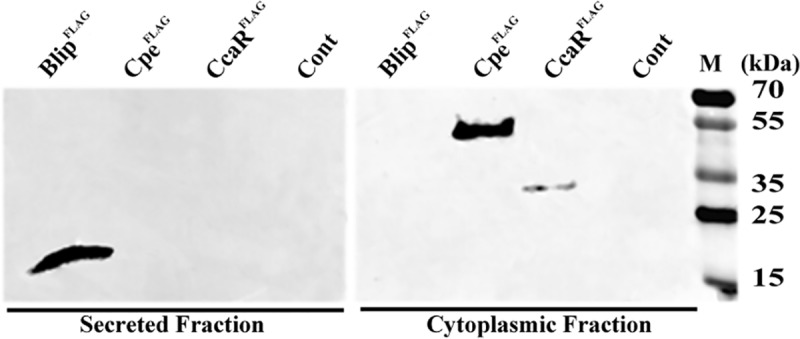
C-terminal FLAG-tagged copies of Cpe, secreted Blip and cytoplasmic CcaR were expressed in wt S. clavuligerus separately for western blot analysis. Cultures were used for isolating secreted (left panel) and cell/cytoplasmic (right panel) fractions, which were probed using anti-FLAG antibodies. The analysis of protein fractions from S. clavuligerus strains containing plasmids pHM:blipFLAG (expressing BlipFLAG), pHM:cpeFLAG (expressing CpeFLAG), pHM:ccaRFLAG (expressing CcaRFLAG) or pHM11a (Cont, empty vector) is shown. Lane M contains the PageRuler Plus Prestained Protein Ladder, which functioned as the molecular weight marker for resolving protein samples during 12% SDS-PAGE.
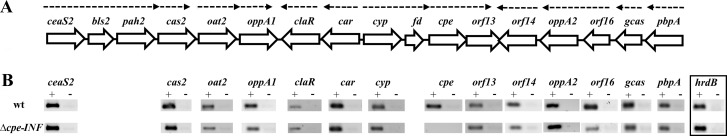
Clavulanic acid has been shown to bind non-covalently with CpeSc under in vitro conditions [25], but the relevance of this interaction is still not clear as the protein did not catalyze any associated reaction. In addition, CpeSc is located in the cytoplasm of S. clavuligerus (Fig 3), and cpe mutants are completely blocked in clavulanic acid production (Fig 2C). This raised the possibility that the protein could have a role in functioning as a cytoplasmic sensor/receptor for clavulanic acid or related metabolites to indirectly regulate production under in vivo conditions. To test this hypothesis, we analyzed the expression level of the first gene from each transcriptional unit (Fig 4A) from the clavulanic acid gene cluster of S. clavuligerus in the Δcpe-INF mutant and compared it with that from the wt strain (Fig 4B). RT-PCR analysis showed that only expression of the cpe gene was altered in the comparison, which was expected (Fig 4B). The analysis also demonstrated that the Δcpe-INF mutation is not associated with any transcriptional polarity as the expression of orf13 was unaffected in the strain. Therefore, it is clear that the deletion of cpe does not in any way influence the expression of other genes from the clavulanic acid gene cluster in S. clavuligerus.
Fig 4. Transcriptional analysis of genes from the clavulanic acid biosynthetic gene cluster (BGC) in wt S. clavuligerus and the Δcpe-INF mutant.
(A) The overall architecture of the BGC is shown with each hollow arrow representing a gene and the arrowhead its orientation. The known transcriptional units are also indicated, and the broken lines represent transcripts (B) The first gene from each transcriptional unit in (A) was selected for analysis to determine its comparative expression level in the two respective strains. RNA isolated from wt S. clavuligerus and the Δcpe-INF mutant after 48 hours of growth in SA medium was used for RT-PCR (+) analysis. As controls, treated RNA samples were used directly in PCR without RT or cDNA synthesis (-). The expression of the constitutively expressed hrdB gene (extreme right boxed panel) was used as internal control to normalize expression levels between different samples/strains.
Assessing the requirement of the N- and C-terminal domains of CpeSc for clavulanic acid production in S. clavuligerus
The crystal structure of heterologously expressed CpeSc from E. coli demonstrated that it contains distinct N-terminal (residues 1–127) and C-terminal (residues 128–458) domains resembling ketosteroid isomerases/polyketide cyclases and β-lactamases, respectively [25]. It was also shown that the C-terminal domain was responsible for the observed in vitro cephalosporin esterase activity of CpeSc, but a function or phenotype could not be assigned at the time for the N-terminal domain based on the assays used [25]. Results from the western blot analysis described above indicate that CpeSc does not undergo posttranslational processed in S. clavuligerus, but it is not known if the N-terminal domain is required for the activity of the protein in the native host. To investigate the in vivo roles of the N- and C-terminal domains during clavulanic acid biosynthesis, three additional CpeSc expression constructs were prepared for analysis. The N- and C-terminal domains were expressed separately or together (as separate polypeptides, Table 2) in complementation studies using the S. clavuligerus Δcpe-INF mutant. Analysis of SA and soy culture supernatants showed that except for full-length CpeSc, none of the other expression plasmids restored clavulanic acid production in the Δcpe-INF mutant, suggesting that both domains need to be part of a single polypeptide for biosynthesis to occur (Fig 5A and S2 Fig). Since the 5S clavams are not produced by wt S. clavuligerus when grown in SA medium [42], soy cultures were included in the analysis. Results showed that none of the strains accumulated any of the known intermediates from the clavulanic acid arm of the pathway (Fig 1 and S4 Table), and production of the 5S clavams was also unaffected in all of them when cultured in soy medium (S5 Table).
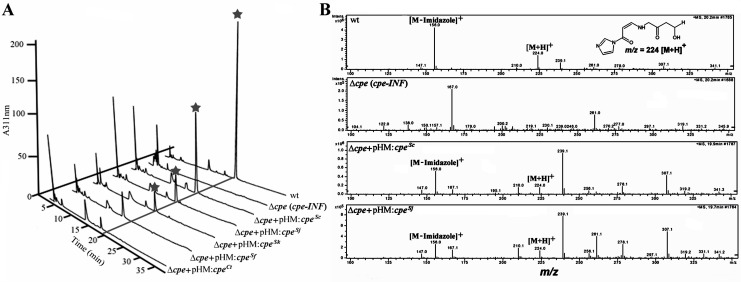
Fig 5. Functional analysis of different domains of CpeSc and its homologues during clavulanic acid production in S. clavuligerus.
(A and B) LC-MS analysis of 96 hour SA culture supernatants after imidazole derivatization using the ammonium bicarbonate buffer system [38]. Cultures of wt S. clavuligerus or the Δcpe-INF mutant expressing Cpe from S. clavuligerus (pHM:cpeSc), S. flavogriseus (pHM:cpeSf), S. jumonjinensis/S. katsurahamanus (pHM:cpeSj/Sk) or the C-terminal domain of CpeSc (pHM:cpeCt) were used in the analysis. (A) Liquid chromatography profiles showing the elution of the peaks corresponding to imidazole-derivatized clavulanic acid (indicated by the star symbol). (B) Mass spectra of the major peaks corresponding imidazole-derivatized clavulanic acid [M+H]+ (m/z = 224) and the fragmented product [M-imidazole]+ (m/z = 156), which were only detected in supernatants from clavulanic acid producing strains shown in (A).
Examination of the ability of other cpe homologues to support clavulanic acid production in the S. clavuligerus Δcpe-INF mutant
The S. flavogriseus and S. viridis genome sequences revealed that they encode clavulanic-like BGCs [13], which are thought to be “silent or cryptic” as the two organisms are not known to produce any clavam metabolites [15]. Whereas other studies have shown that S. jumonjinensis and S. katsurahamanus can also produce clavulanic acid [12], but details regarding the sequences of their respective gene clusters are unavailable [14]. Therefore, we amplified the cpe homologues from the four organisms using genomic DNA as a template for complementation studies, and we also determined the complete sequence of the genes from S. jumonjinensis and S. katsurahamanus (S3 Fig). The predicted amino acid sequences of the Cpe proteins from S. jumonjinensis (CpeSj) and S. katsurahamanus (CpeSk) share ~68% identity with Cpe from S. clavuligerus (CpeSc) (S4 Fig). In comparison, the predicted sequences of the proteins from S. flavogriseus (CpeSf) and S. viridis (CpeSv) showed 58.8% and 48.8% identity to CpeSc, respectively. The predicted C-terminal domains of all four proteins contain the characteristic class A β-lactamase SXXK and SDN catalytic motifs, whereas the KTG motif was replaced by KGG in the non-producers (S. flavogriseus and S. viridis) and KAG in the producers (S. clavuligerus, S. jumonjinensis and S. katsurahamanus), respectively (S4 Fig). In addition, all proteins also contained an extra N-terminal domain resembling that of CpeSc to different extents.
Sequence analysis showed that the Cpe proteins from clavulanic acid producers are more closely related to each other as compared to those from the non-producers (S4 and S5 Figs). To determine the significance of this finding, the respective cpe genes from different sources were expressed under the control of ermEp* in the S. clavuligerus Δcpe-INF mutant for complementation studies (Table 2). It was found that Cpe from S. jumonjinensis and S. katsurahamanus restored clavulanic acid production in the S. clavuligerus Δcpe-INF mutant to varying degrees (Fig 5A and 5B), whereas no complementation was observed in the case of CpeSv and CpeSf. Therefore, only the cpe genes from clavulanic acid producers (CpeSj and CpeSk) seem to be functionally equivalent to the known homologue from S. clavuligerus during clavulanic acid biosynthesis.
Identification of amino acid residues from CpeSc required for clavulanic acid production in S. clavuligerus
The crystal structure of CpeSc revealed that two molecules of clavulanic acid (CA-1 and CA-2, respectively) bind to the monomeric protein [25]. CA-1 binds to an active site pocket to form hydrogen bonds with Lys89, Tyr359 and Arg418 and its C2 side chain carboxylate is positioned deep in the active site of CpeSc, where it interacts with Lys375 [25]. Lys375 is part of the CpeSc KTG motif, where the equivalent catalytic residues in PBPs/β-lactamases also interact with the analogous carboxylates from penicillin and cephalosporin substrates, respectively [43]. In comparison, binding of CA-2 occurs in a mostly hydrophobic cleft comprised of Trp91, Leu362, Leu415, Arg418 and Ala422 at the interface of the N- and C-terminal domains [25]. The Trp91 and Arg418 residues are also highly conserved in the other predicted Cpe proteins (CpeSj, CpeSk, CpeSf, CpeSv), where Arg418 from CpeSc is also involved in binding to CA-1 (S4 Fig). Therefore, Lys89, Trp91 Tyr359, Lys375 and Arg418 from CpeSc were selected for mutagenesis studies to examine their in vivo contributions during the clavulanic acid production in S. clavuligerus. The cpeSc gene along with the ermEp* was transferred from pHM:cpeSc to pSET152 and subjected to site-directed mutagenesis, and the prepared cpeSc variants were assessed for their ability to complement the S. clavuligerus Δcpe-INF mutant. Replacement of Lys89, Tyr359, Lys375 or Arg418 with Ala individually in CpeSc led to a complete loss in clavulanic acid production (Table 3 and S6 Fig). However, when Lys375 was replaced with arginine (both being basic amino acids), clavulanic acid production was restored to 40% production levels in the Δcpe-INF mutant as compared to the wt strain (Table 3 and S6 Fig). As well, partial complementation was also observed in the case of the CpeSc Trp91Ala variant (Table 3).
Table 3. Clavulanic acid production in wild type (wt) S. clavuligerus and the Δcpe-INF mutant expressing different variants of CpeSc.
| S. clavuligerus straina | CpeSc protein variantb (cpeSc codon substitution) |
Bioactivityc | |
|---|---|---|---|
| SA | soy | ||
| wt | NA | ++++ | +++++ |
| Δcpe-INF | NA | - | - |
| Δcpe-INF (pSET-152) | NA | - | - |
| Δcpe-INF (pSET:cpeSc) | wt (none) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc- Ser27Ala) | Ser27Ala (TCC→GCC) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Lys89Ala) | Lys89Ala (AAG→GCG) | - | - |
| Δcpe-INF (pSET:cpeSc-Trp91Ala) | Trp91Ala (TGG→GCG) | ++ | +++ |
| Δcpe-INF (pSET:cpeSc-Arg115Ala) | Arg115Ala (CGC→GCC) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Ser173Ala) | Ser173Ala (TCG→GCG) | - | - |
| Δcpe-INF (pSET:cpeSc-Lys176Ala) | Lys176Ala (AAG→GCG) | - | - |
| Δcpe-INF (pSET:cpeSc-Ser206Ala) | Ser206Ala (AGC→GCC) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Ser234Ala) | Ser234Ala (AGC→GCC) | - | - |
| Δcpe-INF (pSET:cpeSc-Arg311Ala) | Arg311Ala (CGC→GCC) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Gln321Ala) | Gln321Ala (CAG→GCG) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Trp326Ala) | Trp326Ala (TGG→GCG) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Arg346Ala) | Arg346Ala (CGG→GCG) | +++ | ++++ |
| Δcpe-INF (pSET:cpeSc-Tyr359Ala) | Tyr359Ala (TAC→GCC) | - | - |
| Δcpe-INF (pSET:cpeSc-Lys375Ala) | Lys375Ala (AAG→GCG) | - | - |
| Δcpe-INF (pSET:cpeSc-Lys375Arg) | Lys375Arg (AAG→AGG) | ++ | +++ |
| Δcpe-INF (pSET:cpeSc-Ser378Ala) | Ser378Ala (TCC→GCC) | - | - |
| Δcpe-INF (pSET:cpeSc-Arg418Ala) | Arg418Ala (CGC→GCC) | - | - |
a Strains of S. clavuligerus were fermented in either SA or soy media for 96 hours and culture supernatants were used in bioassays for detecting clavulanic acid production.
b Single amino acid variants of CpeSc used in the analysis are shown and the corresponding codon changes in cpeSc leading to the respective substitutions are indicated in parenthesis; NA, Not applicable.
c Zones of inhibition relative the wt strain grown in each media are indicated, where (+) indicates clavulanic acid production and (-) indicates the lack of production, respectively.
Other amino acids from CpeSc have also been shown to interact with clavulanic acid, some of which contributed to its in vitro cephalosporin esterase activity [25]. These include residues from the SXXK (Ser173) and SDN (Ser234) motifs comprising the catalytic tetrad (Ser173/Lys176/Ser234/Lys375), which is conserved in all four Cpe protein sequences described above (S4 Fig). Valegård, et al. reported that the CpeSc Ser173Ala mutant showed a 100-fold reduction in esterase activity, whereas the Ser234Ala and Ser378A mutants were not affected to the same extent [25]. Since the roles of the respective amino acids during clavulanic acid are not known, Ser173, Lys176, Ser234, Ser378 were also individually substituted with Ala in CpeSc for in vivo analysis. All four variants were unable to complement the Δcpe-INF mutant, demonstrating that they essential for clavulanic acid production in S. clavuligerus (Table 3 and S6 Fig).
In the current analysis, amino acids were also identified that are either highly and/or partially conserved in all five Cpe proteins (S4 Fig). These include the Ser27, Arg115, Ser206, Arg311, Gln321, Trp326 and Arg346 from CpeSc, which are not part of any conserved motif and do not interact with clavulanic acid directly based on the reported crystal structure of the protein (29). When each of these residues was replaced with Ala, the respective CpeSc variants restored clavulanic acid production in the S. clavuligerus Δcpe-INF mutant to varying degrees (Table 3), demonstrating that they are not essential for production. Overall, a detailed set of residues were identified in CpeSc, some of which contribute to both the in vitro and in vivo activities of the protein, whereas others are only relevant during the latter process. These are important findings as they allude to the actual biochemical role/function of the protein, which occurs during in vivo clavulanic acid production in S. clavuligerus
Discussion
In the current study, we examined the function of cpe from the CA-BGC of S. clavuligerus, starting with the significance of the relative arrangement of neighboring genes located in its immediate vicinity. Polycistronic mRNAs often allow for the concerted expression of gene products involved in related biosynthetic pathways [44, 45], and gene knockout studies have implicated cpe as being essential for clavulanic acid production in S. clavuligerus [21, 23]. cpe is transcribed as part of a bicistronic operon along with orf13, the start codon of which also overlaps with the stop codon of cpe (Fig 2A), suggesting potential co-translation [46–48]. In addition, the 3′ ends of orf13 and orf14 overlap (Fig 2A), which is unusual in bacteria [49]. It can be challenging to decipher the precise roles of genes located within operons, particularly in cases where co-translation is involved [50–53]. The disruption of genes located in the 5′ regions of operons can influence the expression of downstream genes and also impact the relative stoichiometry of encoded gene products, thereby leading to polar effects [47, 51, 54]. Therefore, we prepared an in-frame S. clavuligerus cpe deletion mutant for use in the current study, while maintaining its stop codon and context with orf13 to minimize the potential for polar effects. During the process, we also prepared the S. clavuligerus Δcpe::apra deletion mutant in which a disruption cassette was inserted in the opposite orientation to cpe transcription. It was noted that both the in-frame and the insertional mutant could be successfully complemented to restore clavulanic acid production using a plasmid-borne copy of cpe, demonstrating that polar effects were not associated with either of them. Therefore, it seems that despite their relative organization, alternate mechanisms exist to facilitate the translation of Orf13 in the cpe mutants, enabling us to use the in-frame mutant for more detailed in vivo studies.
The Cpe protein resembles class A β-lactamases [25], proteins which are secreted to inactivate β-lactam antibiotics before they can inhibit peptidoglycan crosslinking on the outer surface of the cytoplasmic membrane. In addition, when cpe was first sequenced it was reported to share some similarity with the LpqF lipoprotein from Mycobacterium tuberculosis, [25], the function of which is still unknown [55]. Most β-lactamases are secreted using the Sec pathway [39], however, some are translocated by the Tat system in mycobacteria [56]. In addition, certain proteins unrelated to β-lactamases have been reported to be secreted in the absence of any recognizable N-terminal signal sequences [57]. To narrow down its biological function, we investigated the cellular location of CpeSc in its native host and showed that it is a cytoplasmic protein, with no evidence of any association with the cell wall or secreted fractions. Therefore, it can be inferred that unlike β-lactamases, the in vivo role of CpeSc lies in the cytoplasm, which is also the site of clavulanic acid biosynthesis in S. clavuligerus.
During the biosynthesis of certain bioactive natural products, mechanisms exist to coordinate different stages involved in the production of the terminal metabolite [58]. The strategy is used to regulate the expression of specific genes, including those involved in export or self-resistance, when threshold concentrations of a specific intermediate(s) from the pathway is achieved in the cell [59, 60]. As well, feedback inhibition during natural product biosynthesis by end products is also well documented [61]. The cytoplasmic location of CpeSc and its ability to bind to certain cephalosporins and clavulanic acid raises the possibility that it could function as an intracellular receptor for sensing such metabolites to elicit an associated response directly or indirectly. For example, the membrane-associated sensor kinase BlaR from Staphylococcus aureus also contains a PBP-like domain (related to β-lactamases) that binds to β-lactams and triggers the proteolysis of the cytoplasmic BlaI repressor to activate the expression of the BlaZ β-lactamase [62]. To investigate this hypothesis, the expression of key transcriptional units (comprising ceaS2, oat2, oppA1, claR, car, cyp, cpe, orf13, orf14, orf16, gcas and pbpA) from the CA-BGC was analyzed in the S. clavuligerus Δcpe-INF mutant for comparison with the wt strain. Except for the expression of cpe itself, the transcription of all other analyzed genes was unaffected in the mutant (Fig 4B). Results also clearly demonstrated that the transcription of orf13 (and orf14) was not affected in the Δcpe-INF mutant, despite their complex transcriptional/transnational arrangement (Fig 2A). Therefore, based on the results, we can rule out any apparent sensory or regulatory role for CpeSc during clavulanic acid biosynthesis in S. clavuligerus.
Homologues of cpeSc are also present in related BGCs from other clavulanic acid producing (S. jumonjinensis and S. katsurahamanus) and non-producing (S. flavogriseus and S. viridis) organisms (S4 Fig). It was found that only expression of Cpe from producer species (CpeSj/CpeSk) could complement the S. clavuligerus Δcpe-INF mutant. The probability of two proteins having a similar biological function increases proportionally with the relatedness of their respective amino acid sequences [63]. This might explain the complementation phenotypes as CpeSj/CpeSk are more closely related to CpeSc than CpeSf/CpeSv (S4 and S5 Figs). The inability of the corresponding S. flavogriseus and S. viridis cpe homologues to complement the S. clavuligerus mutant suggests that portions of the respective clavulanic acid-like BGCs from the non-producers (including cpe) might also be defective in addition to being “silent” [15], a hypothesis that is currently being explored. Since the same plasmid(s) and constitutive promoter (ermEp*) was used to drive the expression of all cpe genes independently in the current study, the lack of observed complementation in some cases is unlikely due to differences in expression levels.
A previous study showed that the C-terminal domain of recombinant CpeSc displays in vitro O-acetyl cephalosporin esterase activity, but a function could not be assigned for its N-terminal domain [25]. It was also suggested that CpeSc might undergo in vivo post-translational processing in S. clavuligerus to separate the two domains, which could not be addressed at the time since the protein was heterologous expressed and purified from E. coli [25]. Therefore, we examined the requirement of the two CpeSc domains during in vivo clavulanic acid production in S. clavuligerus by using them to complement the Δcpe-INF mutant. Results suggest that both domains are required, and that they should be present to on a single peptide for production to take place (S2 Fig). It is also possible that the inclusion of the two domains on separate plasmid constructs could lead to reduced expression of the respective peptides or they could become unstable/misfolded, which might explain the lack of complementation. This seems unlikely, as the C-terminal domain of CpeSc (completely lacking the N-terminus) was previously expressed and purified from E. coli for biochemical and structural studies [25]. Therefore the inability of the CpeSc C-terminal domain to complement the S. clavuligerus Δcpe-INF mutant is most likely due to the missing region of the protein, which is the N-terminus isomerase like domain. We also show that CpeSc with specific amino acid substitutions (but not all) in either its N- or C-terminal domain is unable to complement the Δcpe-INF mutant and that only a single band corresponding to intact CpeSc was observed in protein fractions from S. clavuligerus during western analysis (Fig 3 and S1 Fig). Therefore, results clearly demonstrate that CpeSc does not undergo of in vivo proteolytic processing in S. clavuligerus and that its N-terminal isomerase-like domain is required for clavulanic acid production, which is the first direct evidence for its involvement in the process.
The crystal structure of CpeSc showed that the protein binds to two molecules of clavulanic acid [25]. The first molecule (CA-1) forms hydrogen bonds with Tyr359, Arg418 and Lys89 from the active site, which also contains Ser173 and Ser234 from the S173XXK176 and S234DN motifs, respectively. In addition, these 5 amino acids are conserved across all five Cpe homologues included in the current study (S4 Fig). Ser173, Ser234 and Ser378 from CpeSc are also important for the in vitro cephalosporin esterase activity of the protein [25], whereas in class A β-lactamases the equivalent residues (including Lys176) are required for the binding and acylation of β-lactam substrates during catalysis [64–66]. It has been reported that certain esterases also exhibit β-lactamase activity and that some PBPs can conversely function as esterases [67, 68], which is not a true representation of their actual physiological function(s). This could also be the case for CpeSc, where the protein can function as an esterase if given permissive substrates (25). In the current study, we demonstrated that Ser173, Lys176, Ser234 and Ser378 from CpeSc are essential for in vivo clavulanic acid production in S. clavuligerus, reminiscent of their catalytic roles in class A β-lactamases. The K234 (T/S)235G catalytic motifs of serine β-lactamases also contain a conserved Ser/Thr residue, which interacts with the carboxylates of corresponding β-lactam substrates [69, 70]. Substitution of this Ser/Thr by amino acids with non-hydroxylated side chains (such as Ala) significantly reduces β-lactamase activity, especially against cephalosporin substrates [70, 71]. It is interesting to note that this conserved Ser/Thr residue is replaced by Gly (KGG) or Ala (KAG) in Cpe from clavulanic acid non-producers and producers, respectively (S4 Fig), which might explain why CpeSc lacks any detectable in vitro β-lactamase activity. As well, substitution of Lys234 by Thr (but not Arg) in class A β-lactamases substantially reduces their catalytic activities and ability to bind to clavulanic acid for inhibition [72, 73]. We also show that the positively charged electrostatic feature at position 375 is essential for the in vivo functional activity of CpeSc, as replacement of Lys (K375TG) with Ala but not Arg in the protein completely abolished clavulanic acid production in S. clavuligerus (Table 3 and S6 Fig).
The in vivo role of amino acids from CpeSc that interact with the second molecule of clavulanic acid (CA-2) via weak electrostatic interactions was also examined. These include the Trp91 or Arg418 residues [25], the substitution of which with Ala either reduced or abolish clavulanic acid production (Table 3). The blocked phenotype of the Arg418 mutant is consistent with the role of Arg418 as part of the CA-1 binding active site described above. In contrast, substitution of all other residues from the CA-2 binding cleft in CpeSc did not significantly impact clavulanic acid production in S. clavuligerus (Table 3), suggesting that they do not contribute towards catalysis. Therefore, the role of the second clavulanic acid binding site in CpeSc remains unclear. It is possible that the site occupied by CA-2 binds some other ligand and/or is an artifact of co-crystallizing purified CpeSc with clavulanic acid during structural studies (86, 87), possibilities that warrant further examination. In addition, the substitution of other residues in CpeSc that are conserved across all five homologues (S4 Fig), but which do not interact with clavulanic acid and/or are not part of any recognizable motif, did not affect the in vivo activity of the protein in S. clavuligerus.
To summarize, in the current study specific residues from CpeSc and its two domains were shown to be essential for in vivo clavulanic acid production in S. clavuligerus (Table 3). These are novel finding and allude towards a biosynthetic role for the protein during production. The described Cpe proteins share many similarities with class A serine β-lactamases, but some crucial differences are also apparent. For example, class A β-lactamases possess the characteristic Ω loop containing residue(s) involved in deacylation and subsequent release of hydrolyzed substrates [74], which are missing in CpeSc. In comparison, class C and D serine β-lactamases also lack the Ω loop and are believed to use alternate mechanisms involving the SXXK and SDN motifs for deacylation instead [75]. The corresponding residues from CpeSc (S173XXK and S234DN) bind to CA-1 in the crystal structure of the protein [25], but it is also possible that they might interact with some other intermediate(s) from the clavulanic acid biosynthetic pathway. Such precursors would not be detected in co-crystals reconstituted using heterologously expressed CpeSc and purified clavulanic acid, as all other components of the biosynthetic pathway would be missing. Therefore, Ser173 and Ser234 could promote a nucleophilic attack on a still unknown substrate to form the primary Cpe-substrate complex, while other essential residues including some form the N-terminal domain (Lys89, Lys375 and Arg418) might be involved in stabilizing the intermediate followed by isomerization and product formation. The N-terminal domain of CpeSc (residues 1–127) is structurally similar to a putative ketosteroid isomerase from Shewanella frigidimarina, which also contains the equivalent Lys89 from CpeSc shown to interact with CA-1 [25]. Lys89 and some of its neighboring residues are conserved in all five Cpe homologues included in the current study (S4 Fig). For the first time, we show that the N-terminal domain of CpeSc and specifically the Lys89 residue from it plays an essential role during clavulanic acid production in S. clavuligerus (S2 Fig and Table 3). We did not detect precursors or shunt products from the clavulanic acid pathway in culture supernatants from different S. clavuligerus mutants (S1 Table), suggesting that reaction intermediates remain tightly/covalently associated with CpeSc during catalysis or are perhaps unstable [25]. Therefore, it is possible that CpeSc is involved in an “altered/modified” β-lactamase-derived reaction required for clavulanic acid production by itself or in combination with another proteins(s), a hypothesis that is currently under investigation. It has been previously suggested that Cpe could be involved in the epimerization of 5S precursors to the 5R configuration during clavulanic acid biosynthesis [25], but this has not been demosntrated. Such a role for CpeSc is conceivable based on the stereospecificity and reversible nature of enzyme-catalyzed reactions, but is not trivial to examine as the natural substrate(s) of Cpe are unknown [25]. It is intriguing that the biosynthetic pathway for a β-lactamase inhibitor (clavulanic acid) has recruited an enzyme for its production that is evolutionarily related to the very proteins that it inhibits. In the long term, deciphering the roles of different residues from CpeSc involved in catalysis can enable us to engineer protein variants with the ability to accept altered substrates. Such a strategy would allow for the production of clavulanic acid analogues for future studies and possible applications.
Supporting information
Purified protein (E. coli) or cell free lysates (S. clavuligerus) were used in the analysis along with anti-6×His antibodies for detecting epitope-tagged Cpe. The lane labeled as “Mock prep” contains S. clavuligerus pHM11a empty vector lysate as control to account for any non-specific antibody binding. The size of the band corresponding to CpeSc-6×His in all lanes was approximately 50–55 kDa, and the prestained protein ladder (Marker) was used as a reference for estimating molecular weights during 12% SDS-PAGE.
(PDF)
The peak corresponding to imidazole-derivatized clavulanic acid (CA) is indicated and was only observed when the full-length protein was used in the analysis.
(PDF)
The complete gene sequences starting from initiation (ATG) to the stop (TGA) codon for each gene were determined as part of the current study and are reported.
(PDF)
Analysis was performed with Clustal Omega (ver 1.2.1) using translated Cpe amino acid sequences from S. clavuligerus (WP_003952519.1), S. flavogrisius (WP_014152684.1), S. viridis (WP_015787620), S. jumonjinensis and S. katsurahamanus. The DNA sequences of cpe from the latter two producers (S. jumonjinensis and S. katsurahamanus) were determined as part of the current study and are reported in S3 Fig. The boxes in black highlight the conserved SXXK, SDN and KTG/KAG motifs present in class A β-lactamses and the respective Cpe proteins, whereas the box in blue represents the N-terminus domain indentifed in CpeSc. The arrows indicate amino acids from CpeSc that were selected for mutagenesis and the ones highlighted in red were shown to be essential for in vivo clavulanic acid production in S. clavuligerus.
(PDF)
Multiple sequence alignments using the predicted amino acid sequences of Cpe proteins listed in S4 Fig. and class A β-lactamases including Bla (from the S. clavuligerus cephamycin C biosynthetic gene cluster, CAA90895.1) and TEM-1 (from E.coli, AMM70781.1) were used to prepare the tree, and bootstrap analyses were performed using 100 replicates. All positions containing gaps were eliminated during the analysis and the number next to each node represents the percentage of trees in which the respective topologies were observed.
(PDF)
Clavulanic acid (CA) production was monitored at 311nm following imidazole derivatization.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The described work was funded by an operating grant from the Natural Science and Engineering Research Council of Canada (NSERC: 386417–2010 and 2018–05949) to KT. KSK, NFA and CJM were also the recipients of NSERC graduate/undergraduate student awards. In addition, we acknowledge Memorial University of Newfoundland for providing graduate student support to KSK, NFA and BMP.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The described work was funded by an operating grant from the Natural Science and Engineering Research Council of Canada (NSERC: 386417-2010 and 2018-05949) to KT. KSK, NFA and CJM were also the recipients of NSERC graduate/undergraduate student awards. In addition, we acknowledge Memorial University of Newfoundland for providing graduate student support to KSK, NFA and BMP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tahlan K, Jensen SE. Origins of the β-lactam rings in natural products. J Antibiot (Tokyo). 2013;66(7):401–10. 10.1038/ja.2013.24 [DOI] [PubMed] [Google Scholar]
- 2.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the β-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol. 1979;33(1):113–37. 10.1146/annurev.mi.33.100179.000553 [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12(5):371–87. 10.1038/nrd3975 [DOI] [PubMed] [Google Scholar]
- 4.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5(3):175–86. 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- 5.Essack SY. The development of β-lactam antibiotics in response to the evolution of β-lactamases. Pharm Res. 2001;18(10):1391–9. [DOI] [PubMed] [Google Scholar]
- 6.Livermore DM. β-lactamase-mediated resistance and opportunities for its control. J Antimicrob Chemother. 1998;41 [DOI] [PubMed] [Google Scholar]
- 7.Heesemann J. Mechanisms of resistance to β-lactam antibiotics. Infection. 1993;21 . [DOI] [PubMed] [Google Scholar]
- 8.Reading C, Cole M. Clavulanic acid: a β-lactamase-inhibiting β-lactam from Streptomyces clavuligerus. Antimicrob Agents Chemother. 1977;11(5):852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti SN, Phillips OA, Micetich RG, Livermore DM. β-lactamase inhibitors: agents to overcome bacterial resistance. Curr Med Chem. 1998;5(6):441–56. [PubMed] [Google Scholar]
- 10.Matsuura M, Nakazawa H, Hashimoto T, Mitsuhashi S. Combined antibacterial activity of amoxicillin with clavulanic acid against ampicillin-resistant strains. Antimicrob Agents Chemother. 1980;17(6):908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd PA, Benfield P. Amoxicillin/clavulanic acid. An update of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs. 1990;39(2):264–307. 10.2165/00003495-199039020-00008 [DOI] [PubMed] [Google Scholar]
- 12.Jensen SE, Paradkar AS. Biosynthesis and molecular genetics of clavulanic acid. Antonie Van Leeuwenhoek. 1999;75(1–2):125–33. [DOI] [PubMed] [Google Scholar]
- 13.Jensen SE. Biosynthesis of clavam metabolites. J Ind Microbiol Biotechnol. 2012;39(10):1407–19. 10.1007/s10295-012-1191-0 [DOI] [PubMed] [Google Scholar]
- 14.Ward JM, Hodgson JE. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’in three Streptomyces. FEMS Microbiol Lett. 1993;110(2):239–42. 10.1111/j.1574-6968.1993.tb06326.x [DOI] [PubMed] [Google Scholar]
- 15.Álvarez-Álvarez R, Martínez-Burgo Y, Pérez-Redondo R, Braña A, Martín JF, Liras P. Expression of the endogenous and heterologous clavulanic acid cluster in Streptomyces flavogriseus: why a silent cluster is sleeping. Applied Microbiology and Biotechnology. 2013;97(21):9451–63. 10.1007/s00253-013-5148-7 [DOI] [PubMed] [Google Scholar]
- 16.Tahlan K, Anders C, Wong A, Mosher RH, Beatty PH, Brumlik MJ, et al. 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chemistry & Biology. 2007;14(2):131–42. 10.1016/j.chembiol.2006.11.012 [DOI] [PubMed] [Google Scholar]
- 17.Hamed RB, Gomez-Castellanos JR, Henry L, Ducho C, McDonough MA, Schofield CJ. The enzymes of β-lactam biosynthesis. Nat Prod Rep. 2013;30(1):21–107. 10.1039/c2np20065a [DOI] [PubMed] [Google Scholar]
- 18.Egan LA, Busby RW, Iwata-Reuyl D, Townsend CA. Probable role of clavaminic acid as the terminal intermediate in the common pathway to clavulanic acid and the antipodal clavam metabolites. 1997;119(10):2348–55. 10.1021/ja963107o [DOI] [Google Scholar]
- 19.Tahlan K, Park HU, Wong A, Beatty PH, Jensen SE. Two sets of paralogous genes encode the enzymes involved in the early stages of clavulanic acid and clavam metabolite biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother. 2004;48(3):930–9. 10.1128/AAC.48.3.930-939.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKenzie AK, Kershaw NJ, Hernandez H, Robinson CV, Schofield CJ, Andersson I. Clavulanic acid dehydrogenase: structural and biochemical analysis of the final step in the biosynthesis of the β-lactamase inhibitor clavulanic acid. Biochemistry. 2007;46(6):1523–33. 10.1021/bi061978x [DOI] [PubMed] [Google Scholar]
- 21.Li R, Khaleeli N, Townsend CA. Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Bacteriol. 2000;182(14):4087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellado E, Lorenzana LM, Rodriguez-Saiz M, Diez B, Liras P, Barredo JL. The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology. 2002;148(Pt 5):1427–38. 10.1099/00221287-148-5-1427 [DOI] [PubMed] [Google Scholar]
- 23.Jensen SE, Paradkar AS, Mosher RH, Anders C, Beatty PH, Brumlik MJ, et al. Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother. 2004;48(1):192–202. 10.1128/AAC.48.1.192-202.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Álvarez-Álvarez R, Rodríguez-García A, Martínez-Burgo Y, Martín J, Liras P. Transcriptional studies on Streptomyces clavuligerus oppA2-deleted mutant: N-acetylglycyl-clavaminic acid is an intermediate of clavulanic acid biosynthesis. 2018: Appl Environ Microbiol. 01701–18. 10.1128/AEM.01701-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valegård K, Iqbal A, Kershaw NJ, Ivison D, Genereux C, Dubus A, et al. Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway. Acta Crystallogr D Biol Crystallogr. 2013;69(8):1567–79. 10.1107/S0907444913011013 [DOI] [PubMed] [Google Scholar]
- 26.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43(1):207–33. 10.1146/annurev.mi.43.100189.001231 [DOI] [PubMed] [Google Scholar]
- 27.Hopwood DA. How do antibiotic‐producing bacteria ensure their self‐resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63(4):937–40. 10.1111/j.1365-2958.2006.05584.x [DOI] [PubMed] [Google Scholar]
- 28.Ogawara H. Self-resistance in Streptomyces, with special reference to β-Lactam antibiotics. Molecules. 2016;21(5):605 10.3390/molecules21050605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coque JJ, Liras P, Martin JF. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993;12(2):631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Llarena F, Martin JF, Galleni M, Coque JJ, Fuente JL, Frere JM, et al. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A β-lactamase of low enzymatic activity. J Bacteriol. 1997;179(19):6035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green MR, Sambrook J. Molecular cloning: A laboratory manual: Cold Spring Harbor Laboratory Press; 2012. www.cshlpress.com [Google Scholar]
- 32.Mosher RH, Paradkar AS, Anders C, Barton B, Jensen SE. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1999;43(5):1215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieser T. Practical streptomyces genetics: John Innes Foundation; 2000. www.jic.ac.uk [Google Scholar]
- 34.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100(4):1541–6. 10.1073/pnas.0337542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motamedi H, Shafiee A, Cai SJ. Integrative vectors for heterologous gene expression in Streptomyces spp. Gene. 1995;160(1):25–31. [DOI] [PubMed] [Google Scholar]
- 37.Bierman M, Logan R, O'brien K, Seno E, Rao RN, Schoner B. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–9. [DOI] [PubMed] [Google Scholar]
- 38.Zelyas NJ, Cai H, Kwong T, Jensen SE. Alanylclavam biosynthetic genes are clustered together with one group of clavulanic acid biosynthetic genes in Streptomyces clavuligerus. J Bacteriol. 2008;190(24):7957–65. 10.1128/JB.00698-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pradel N, Delmas J, Wu LF, Santini CL, Bonnet R. Sec- and Tat-dependent translocation of beta-lactamases across the Escherichia coli inner membrane. Antimicrob Agents Chemother. 2009;53(1):242–8. 10.1128/AAC.00642-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santamarta I, Rodriguez-Garcia A, Perez-Redondo R, Martin JF, Liras P. CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J Bacteriol. 2002;184(11):3106–13. 10.1128/JB.184.11.3106-3113.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thai W, Paradkar AS, Jensen SE. Construction and analysis of ss-lactamase-inhibitory protein (BLIP) non-producer mutants of Streptomyces clavuligerus. Microbiology. 2001;147 10.1099/00221287-147-2-325 [DOI] [PubMed] [Google Scholar]
- 42.Tahlan K, Anders C, Jensen SE. The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. J Bacteriol. 2004;186(18):6286–97. 10.1128/JB.186.18.6286-6297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghuysen JM. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45(1):37–67. 10.1146/annurev.mi.45.100191.000345 [DOI] [PubMed] [Google Scholar]
- 44.Laing E, Mersinias V, Smith CP, Hubbard SJ. Analysis of gene expression in operons of Streptomyces coelicolor. Genome Biol. 2006;7(6). 10.1186/gb-2006-7-6-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charaniya S, Mehra S, Lian W, Jayapal KP, Karypis G, Hu WS. Transcriptome dynamics-based operon prediction and verification in Streptomyces coelicolor. Nucleic Acids Res. 2007;35(21):7222–36. 10.1093/nar/gkm501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das A, Yanofsky C. Restoration of a translational stop-start overlap reinstates translational coupling in a mutant trpB'-trpA gene pair of the Escherichia coli tryptophan operon. Nucleic Acids Res. 1989;17(22):9333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khosla C, Ebert-Khosla S, Hopwood DA. Targeted gene replacements in a Streptomyces polyketide synthase gene cluster: role for the acyl carrier protein. Mol Microbiol. 1992;6(21):3237–49. [DOI] [PubMed] [Google Scholar]
- 48.Pradhan P, Li W, Kaur P. Translational coupling controls expression and function of the DrrAB drug efflux pump. J Mol Biol. 2009;385(3):831–42. 10.1016/j.jmb.2008.11.027 [DOI] [PubMed] [Google Scholar]
- 49.Fukuda Y, Nakayama Y, Tomita M. On dynamics of overlapping genes in bacterial genomes. Gene. 2003;323:181–7. [DOI] [PubMed] [Google Scholar]
- 50.Raynal A, Karray F, Tuphile K, Darbon-Rongere E, Pernodet JL. Excisable cassettes: new tools for functional analysis of Streptomyces genomes. Appl Environ Microbiol. 2006;72(7):4839–44. 10.1128/AEM.00167-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Løvdok L, Bentele K, Vladimirov N, Müller A, Pop FS, Lebiedz D, et al. Role of translational coupling in robustness of bacterial chemotaxis pathway. PLoS Biol. 2009;7(8):e1000171 10.1371/journal.pbio.1000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goh S, Hohmeier A, Stone TC, Offord V, Sarabia F, Garcia-Ruiz C, et al. Silencing of essential genes within a highly coordinated operon in Escherichia coli. Appl Environ Microbiol. 2015;81(16):5650–9. 10.1128/AEM.01444-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng C, Nair AD, Jaworski DC, Ganta RR. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One. 10.1371/journal.pone.0132657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G, Tian Y, Hu K, Feng C, Tan H. SCO3900, co-transcripted with three downstream genes, is involved in the differentiation of Streptomyces coelicolor. Curr Microbiol. 2010;60(4):268–73. 10.1007/s00284-009-9536-2 [DOI] [PubMed] [Google Scholar]
- 55.Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28(5):645–59. 10.1016/j.femsre.2004.06.002 [DOI] [PubMed] [Google Scholar]
- 56.McDonough JA, Hacker KE, Flores AR, Pavelka MS, Braunstein M. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial β-lactamases. J Bacteriol. 2005;187(22):7667–79. 10.1128/JB.187.22.7667-7679.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5(1):58 10.1186/1471-2180-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim FY, Keller NP. Spatial and temporal control of fungal natural product synthesis. Nat Prod Rep. 2014;31(10):1277–86. 10.1039/c4np00083h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tahlan K, Ahn SK, Sing A, Bodnaruk TD, Willems AR, Davidson AR, et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol. 2007;63(4):951–61. [DOI] [PubMed] [Google Scholar]
- 60.Tahlan K, Yu Z, Xu Y, Davidson AR, Nodwell JR. Ligand recognition by ActR, a TetR-like regulator of actinorhodin export. J Mol Biol. 2008;383(4):753–61. 10.1016/j.jmb.2008.08.081 [DOI] [PubMed] [Google Scholar]
- 61.Liu C, Hermann T, Miller PA. Feedback inhibition of the synthesis of an antibiotic: aurodox (X-5108). J Antibiot (Tokyo). 1977;30(3):244–51. [DOI] [PubMed] [Google Scholar]
- 62.Clarke SR, Dyke KG. The signal transducer (BlaRI) and the repressor (BlaI) of the Staphylococcus aureus β-lactamase operon are inducible. Microbiology. 2001;147(Pt 4):803–10. 10.1099/00221287-147-4-803 [DOI] [PubMed] [Google Scholar]
- 63.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8(12):995–1005. 10.1038/nrm2281 [DOI] [PubMed] [Google Scholar]
- 64.Matagne A, Frere JM. Contribution of mutant analysis to the understanding of enzyme catalysis: the case of class A β-lactamases. Biochim Biophys Acta. 1995;1246(2):109–27. [DOI] [PubMed] [Google Scholar]
- 65.Maveyraud L, Pratt RF, Samama JP. Crystal structure of an acylation transition-state analog of the TEM-1 β-lactamase. Mechanistic implications for class A β-lactamases. Biochemistry. 1998;37(8):2622–8. 10.1021/bi972501b [DOI] [PubMed] [Google Scholar]
- 66.Vandavasi VG, Weiss KL, Cooper JB, Erskine PT, Tomanicek SJ, Ostermann A, et al. Exploring the mechanism of β-lactam ring protonation in the class A β-lactamase acylation mechanism using neutron and X-ray crystallography. J Med Chem. 2015;59(1):474–9. 10.1021/acs.jmedchem.5b01215 [DOI] [PubMed] [Google Scholar]
- 67.Jones M, Page MI. An esterase with β-lactamase activity. J. Chem Soc Chem. Commun. 1991;(5):316–7. 10.1039/C39910000316 [DOI] [Google Scholar]
- 68.Ryu BH, Ngo TD, Yoo W, Lee S, Kim BY, Lee E, et al. Biochemical and structural analysis of a novel esterase from Caulobacter crescentus related to Penicillin-Binding Protein (PBP). Sci Rep. 2016;6:37978 10.1038/srep37978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubus A, Wilkin JM, Raquet X, Normark S, Frere JM. Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the Lys-Thr(Ser)-Gly triad. Biochem J. 1994;301 (Pt 2)(2):485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imtiaz U, Manavathu EK, Lerner SA, Mobashery S. Critical hydrogen bonding by serine-235 for cephalosporinase activity of TEM-1 β-lactamase. Antimicrob Agents Chemother. 1993;37(11):2438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lamotte-Brasseur J, Knox J, Kelly JA, Charlier P, Fonze E, Dideberg O, et al. The structures and catalytic mechanisms of active-site serine β-lactamases. Biotechnol Genet Eng Rev. 1994;12(1):189–230 [DOI] [PubMed] [Google Scholar]
- 72.Ellerby LM, Escobar WA, Fink AL, Mitchinson C, Wells JA. The role of lysine-234 in β-lactamase catalysis probed by site-directed mutagenesis. Biochemistry. 1990;29(24):5797–806. [DOI] [PubMed] [Google Scholar]
- 73.Lenfant F, Labia R, Masson JM. Replacement of lysine-234 affects transition state stabilization in the active site of β-lactamase TEM1. J Biol Chem. 1991;266(26):17187–94. [PubMed] [Google Scholar]
- 74.Banerjee S, Pieper U, Kapadia G, Pannell LK, Herzberg O. Role of the omega-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry. 1998;37(10):3286–96. 10.1021/bi972127f [DOI] [PubMed] [Google Scholar]
- 75.Medeiros AA. β-Lactamases: quality and resistance. Clin Microbiol Infect. 1997;3 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purified protein (E. coli) or cell free lysates (S. clavuligerus) were used in the analysis along with anti-6×His antibodies for detecting epitope-tagged Cpe. The lane labeled as “Mock prep” contains S. clavuligerus pHM11a empty vector lysate as control to account for any non-specific antibody binding. The size of the band corresponding to CpeSc-6×His in all lanes was approximately 50–55 kDa, and the prestained protein ladder (Marker) was used as a reference for estimating molecular weights during 12% SDS-PAGE.
(PDF)
The peak corresponding to imidazole-derivatized clavulanic acid (CA) is indicated and was only observed when the full-length protein was used in the analysis.
(PDF)
The complete gene sequences starting from initiation (ATG) to the stop (TGA) codon for each gene were determined as part of the current study and are reported.
(PDF)
Analysis was performed with Clustal Omega (ver 1.2.1) using translated Cpe amino acid sequences from S. clavuligerus (WP_003952519.1), S. flavogrisius (WP_014152684.1), S. viridis (WP_015787620), S. jumonjinensis and S. katsurahamanus. The DNA sequences of cpe from the latter two producers (S. jumonjinensis and S. katsurahamanus) were determined as part of the current study and are reported in S3 Fig. The boxes in black highlight the conserved SXXK, SDN and KTG/KAG motifs present in class A β-lactamses and the respective Cpe proteins, whereas the box in blue represents the N-terminus domain indentifed in CpeSc. The arrows indicate amino acids from CpeSc that were selected for mutagenesis and the ones highlighted in red were shown to be essential for in vivo clavulanic acid production in S. clavuligerus.
(PDF)
Multiple sequence alignments using the predicted amino acid sequences of Cpe proteins listed in S4 Fig. and class A β-lactamases including Bla (from the S. clavuligerus cephamycin C biosynthetic gene cluster, CAA90895.1) and TEM-1 (from E.coli, AMM70781.1) were used to prepare the tree, and bootstrap analyses were performed using 100 replicates. All positions containing gaps were eliminated during the analysis and the number next to each node represents the percentage of trees in which the respective topologies were observed.
(PDF)
Clavulanic acid (CA) production was monitored at 311nm following imidazole derivatization.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.