Abstract
Allopolyploidy has played a major role in plant evolution but its impact on genome diversity and expression patterns remains to be understood. Some studies found important genomic and transcriptomic changes in allopolyploids, whereas others detected a strong parental legacy and more subtle changes. The allotetraploid C. bursa-pastoris originated around 100,000 years ago and one could expect the genetic polymorphism of the two subgenomes to follow similar trajectories and their transcriptomes to start functioning together. To test this hypothesis, we sequenced the genomes and the transcriptomes (three tissues) of allotetraploid C. bursa-pastoris and its parental species, the outcrossing C. grandiflora and the self-fertilizing C. orientalis. Comparison of the divergence in expression between subgenomes, on the one hand, and divergence in expression between the parental species, on the other hand, indicated a strong parental legacy with a majority of genes exhibiting a conserved pattern and cis-regulation. However, a large proportion of the genes that were differentially expressed between the two subgenomes, were also under trans-regulation reflecting the establishment of a new regulatory pattern. Parental dominance varied among tissues: expression in flowers was closer to that of C. orientalis and expression in root and leaf to that of C. grandiflora. Since deleterious mutations accumulated preferentially on the C. orientalis subgenome, the bias in expression towards C. orientalis observed in flowers indicates that expression changes could be adaptive and related to the selfing syndrome, while biases in the roots and leaves towards the C. grandiflora subgenome may be reflective of the differential genetic load.
Author summary
Most plant species have a polyploid at some stage of their ancestry. Polyploidy, genome doubling through either multiple copies of a single species or through genomes of different species coming into the same nucleus, is therefore a crucial step in plant evolution. Understanding its impact on basic biological functions is thus a matter of interest. Shepherd’s purse (Capsella bursa-pastoris) is a major weed that appeared about 100,000 years ago through hybridization of two diploid species of the same genus. In the present project, we measured genetic diversity and analyzed gene expression patterns in flowers, roots, and leaves of C. bursa-pastoris individuals as well as in its two parental species, the outcrossing C. grandiflora and the self-fertilizing C. orientalis. Our data shows that, after 100,000 generations of evolution, the origin of the two subgenomes can still be seen: the genome inherited from C. grandiflora still differs from the one inherited from self-fertilizing C. orientalis. However, there are also signs that the two genomes have started to work together and are jointly regulated, and the way expression pattern varied across the three tissues indicates that the evolution of gene expression was adaptive.
Introduction
Polyploidy, and in particular allopolyploidy, whereby a novel species is created by the merger of the genomes of two species, is considered to be a common mode of speciation in plants [1] as it induces an instant reproductive isolation, the difference in chromosome number impeding reproduction with the parental species. In the case of allopolyploidy, the daughter species thus has two divergent subgenomes at inception, one inherited from each parental species. Such an increase in genome copy number can be advantageous and could partly explain the apparent evolutionary success of allopolyploid species ([2, 3] but see [4]). For instance, genome doubling creates genetic redundancy, thereby increasing genetic diversity and allowing the masking of deleterious mutations through compensation. Genome doubling and initial redundancy also offer new possibilities for the evolution of genes over time: one copy can degenerate, both can be conserved by dosage compensation [5] or their pattern of expression can diverge and even lead to the evolution of new functions (see [6] and references therein). Gene redundancy also potentially allows tissue-specific expression of different gene copies [7, 8]. On the other hand, the evolutionary success of allopolyploids can also appear paradoxical since the birth of a new allopolyploid species will also be accompanied by numerous challenges [9–12]. These challenges are first associated with the initial hybridization between two divergent genomes, implying, among other things, potential changes of gene expression patterns [13].
The magnitude of gene expression changes has been reported to vary substantially across polyploid species, from minor modifications [14, 15] to so-called “transcriptomic shock” [8]. The balance in expression pattern between the two subgenomes also seems to be highly variable and ranges from the additivity of parental expression to extreme non-additivity. Several forms of non-additivity have been widely observed, such as homeologue expression bias, when the relative expression contributions from the two homeologues are altered, and expression level dominance, when the total expression level of both homeologues is similar to only one of the parental species [16, 17] (see [18] for definitions). These patterns also evolve through time. For example, in Mimulus peregrinus the genome-wide homeologue expression bias was established early on but also increased over successive generations [19]. However, the generality, timing, and causes of changes in expression pattern of the two parental genomes remain poorly known beyond a few case studies [17, 20] and may, to a large extent, depend on parental legacy because a part of the observed differences between the two subgenomes of the allopolyploid species may have already been present between the parental species [3].
Ultimately, changes in patterns of gene expression will follow from modifications in gene expression regulation. Differences in gene expression can be due to changes in cis- and trans-regulatory elements. Cis-regulatory elements alter allele-specific expression and are generally located close to the gene they regulate (e.g., promoters), whereas trans-regulatory elements can affect both alleles and can be located anywhere in the genome [21–24]. In the case of a newly formed allopolyploid species, one would expect the two copies of a gene to be under the influence of trans-regulatory elements inherited from both parents and its expression level to first move towards the mean expression of the two parental species. Retaining the parental pattern of expression in each subgenome would imply that only cis-regulation takes place, or there are forces opposing the establishment of cross trans-regulation. For instance, one could expect purifying selection to have a larger impact on trans-acting mutations than on cis-acting ones because the former have more pleiotropic expression than the latter. If so, the residual variants will mostly be cis-acting ([25] but see [26]). It was also shown that a gene is often under the influence of both trans- and cis-regulatory elements that act in opposite directions [24], leading to a cis-trans compensation that prevents overshooting optimal overall expression level. Such compensation between cis- and trans-regulatory elements is one of the predictions of the enhancer runaway (ER) model proposed by Fyon et al. [27]. Under the ER model, and especially in outcrossing species where heterozygotes are frequent, cis-regulatory variants facilitate the exposure of alleles to purifying selection. If the enhancer and the gene they regulate are linked then the up-regulating variants will hitch-hike with the allele carrying the lowest number of deleterious mutations, leading to an open-ended escalation in enhancer strength [27]. As selection on expression appears to be primarily stabilizing [24, 28, 29], at least at intermediate evolutionary timescales [30], a compensatory effect of expression in trans is predicted [27, 31]. The relative importance of cis- and trans-regulation can be examined by comparing the relative expression in the parental species with the relative expression of homeologous genes in the newly formed tetraploid [21, 32, 33].
Differential expression between the two genomes could result from a differential accumulation of deleterious or slightly deleterious mutations between the two subgenomes or, alternatively, be also related to phenotypic or adaptive changes associated to the differences between the two parental species. If the differential expression is only due to differential accumulations of deleterious mutations, we would expect to see the same differential expression pattern across different tissues, whereas if differential expression is related to phenotypic or adaptive changes then we may expect to see differences depending on the tissue considered.
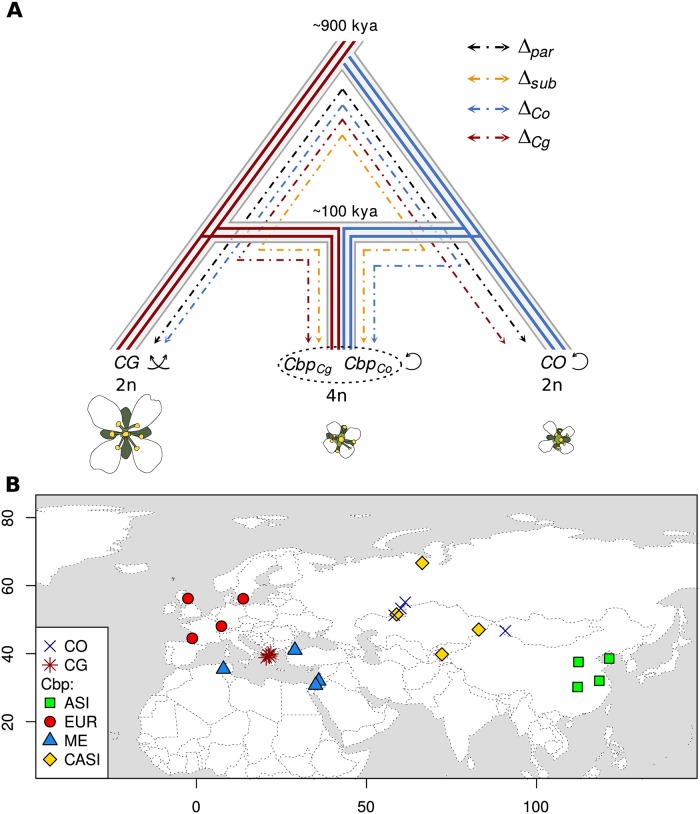
Shepherd’s purse, C. bursa-pastoris, is an allotetraploid selfing species that originated some 100-300 kya from the hybridization of the ancestors of C. orientalis and C. grandiflora [15] (Fig 1A). The two parental species are strikingly different: C. orientalis, a genetically depauperate selfer, occurs across the steppes of Central Asia and Eastern Europe [34], whereas C. grandiflora, an obligate outcrosser with a particularly high genetic diversity, is primarily confined to a tiny distribution range in the mountains of Northwest Greece and Albania [34] (Fig 1). Among Capsella species, only C. bursa-pastoris has a worldwide distribution [34], some of which might be due to extremely recent colonization events associated with human population movements [34, 35]. In Eurasia, the native range of C. bursa-pastoris is divided into three genetic clusters—Asia, Europe, and the Middle East (hereafter ASI, EUR and ME, respectively)—with low gene flow among them and strong differentiation both at the nucleotide and gene expression levels [35, 36]. Reconstruction of the colonization history suggested that C. bursa-pastoris spread from the Middle East towards Europe and then expanded into Eastern Asia. This colonization history resulted in a typical reduction of nucleotide diversity with the lowest diversity being found in the most recent Asian population [35]. It has been possible to phase the subgenomes by assigning each genome sequence (or transcript) to a parental species sequence [37]. In stark contrast to many other studies of allopolyploids, such as maize, cotton, Brassica, Xenopus laevis, [38–44], the phased data suggested that the differences in deleterious variants between the two subgenomes of C. bursa-pastoris are largely a legacy of the differences between the two parental species and that biased fractionation, the biased loss of ancestral genomes in an allopolyploid, is limited [15, 45].
Fig 1. Evolutionary history and sampling locations of the three Capsella species used in this study.
A Solid lines represent subgenomes segregation after the hybridization between C. grandiflora (CG) and C. orientalis (CO) ancestors. C. grandiflora and C. orientalis genetic backgrounds are marked with red and blue respectively. The ploidy levels (n) and the reproductive system are also indicated. Dashed and dotted lines represent the comparisons used to compute the gene expression convergence index (see Material and methods). B. CO, CG, ASI, EUR, ME, CASI correspond to C. orientalis, C. grandiflora, and four populations of C. bursa-pastoris, Cbp, (Asia, Europe, Middle East, and Central Asia) respectively. We shifted slightly population geographical coordinates when those overlapped to make all of them visible on the map.
The aim of the present study was to address questions on the evolution of gene expression patterns of the two subgenomes of the allotetraploid shepherd’s purse C. bursa-pastoris since they derived from the two parental species. We focused on two main questions. First, has the relative contribution of cis- and trans-regulation been altered by polyploidization? Second, could differential expression between the two subgenomes only results from a differential accumulation of deleterious/slightly deleterious mutations (nearly neutral hypothesis) or is it also related to phenotypic differences between the two parents (adaptive hypothesis)? One parent is outcrossing (C. grandiflora) and has large flowers as it needs to attract pollinators while the other parent is self-fertilizing (C. orientalis) and has tiny flowers. Hence one may expect differential expression in flower tissues of selfing C. bursa-pastoris to be biased towards the C. orientalis expression levels under the adaptive hypothesis whereas tissues that have not experienced adaptive specialization might show an expression bias towards C. grandiflora.
To address these questions and, more generally, to characterize the expression pattern of C. bursa-pastoris, we analyzed the genomes and the transcriptomes of three tissues (flowers, leaves, and roots) of 16 accessions coming from different populations of the C. bursa-pastoris natural range and compared them with those of the parental lineages C. grandiflora and C. orientalis (four accessions each) (Fig 1). In total, 24 transcriptomes in three tissues and 24 genomes were analyzed.
One hundred thousand generations after its inception, C. bursa-pastoris does not show any sign of a transcriptomic shock. Instead, our data revealed highly concerted changes with the expression levels of the two subgenomes converging towards an intermediate value. This was achieved by a balance between cis-and trans-regulation and a strong parental legacy that was also observed for the accumulation of deleterious mutations over the two subgenomes. While the differential accumulation of deleterious mutations between subgenomes could explain part of the differential expression between them, there were also significant tissue-specific differences in subgenome dominance and convergence indicating that adaptive changes may also have contributed to the evolution of the expression patterns of the two subgenomes.
Results
Population genetic structure
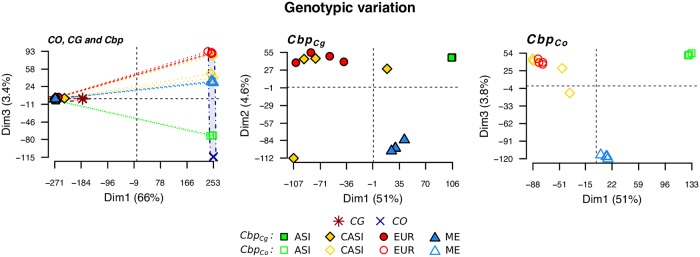
In order to assess the relationship of the newly obtained Central Asian samples with other populations, we analyzed the population structure of our samples. A SNP-based PCA (670K genomic SNPs without any missing data) confirmed the phylogenetic relationships between C. grandiflora (CG), C. orientalis (CO), and C. bursa-pastoris (Cbp) described in [35–37]. The first principal component (Dim1) explained the majority of the variance (66%) and clearly discriminated CG and the CbpCg subgenome from CO and the CbpCo subgenome (Fig 2, left panel). To investigate further population structure within C. bursa-pastoris, we then focused on genetic variation in each subgenome (Fig 2, middle and right, respectively for CbpCg and CbpCo). In both cases, there were three main clusters gathering accessions from Europe (EUR), Asia (ASI), and the Middle East (ME), respectively. Accessions from Central Asia (CASI) tended to cluster with European accessions for both subgenomes, even if they were more scattered. A phylogenetic analysis also confirmed that the new samples from Central Asia were most similar to the European genetic cluster and showed that they did not form a separate genetic cluster (S1 Fig).
Fig 2. Genomic variation patterns in three Capsella species.
Variation was visualized with principal component analyses based on the SNPs of C. grandiflora (CG), C. orientalis (CO), and four populations of C. bursa-pastoris (Cbp) (Asia (ASI), Central Asia (CASI), Europ (EUR), and Middle East (ME)). The left plot shows variation in the three species with lines connecting subgenomes of corresponding Cbp accessions and the dash-dotted circles highlighting two subgenomes of Cbp. The middle and right plots show only the variation within the subgenomes of C. bursa-pastoris (CbpCg and CbpCo).
Global variation in gene expression reflects genetic relationships
Given that the gene expression patterns in homeologue-specific and total expression can produce different results [46], we performed a differential gene expression analyses on both the unphased and phased data. Pairwise comparisons of a number of differentially expressed (DE) genes between species in unphased data (16,039 genes) showed that patterns of expression varied across tissues. First, the number of differentially expressed genes between parental species was the highest in flower tissues, while leaf tissues were the least differentiated (S2 Table). Second, in flowers, overall gene expression of C. bursa-pastoris was the closest to C. orientalis, while in the two other tissues it was the closest to C. grandiflora (S2 Table). At the population level, no clear pattern appeared: for instance, ME accessions were the closest to C. grandiflora in roots, while ASI accessions were the closest to C. grandiflora in leaves and CASI accessions in flowers (S3 Table).
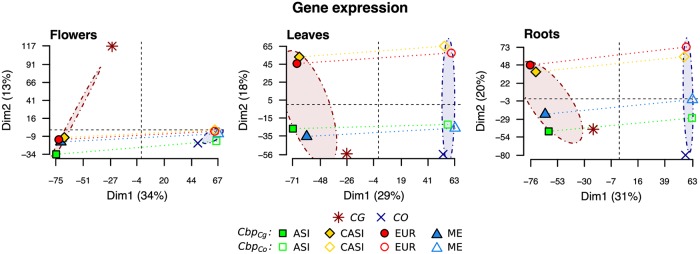
Gene expression variation was then surveyed in 11,931 genes for which phased expression of the two subgenomes was available in all populations of C. bursa-pastoris. Clustering of population/species mean expression values confirmed that the main difference in overall expression variation was between tissues (S2 Fig). The principal component analyses of the three tissues separately (Fig 3) revealed that the global variation pattern in gene expression reflected phylogenetic relationships (Fig 3 and S3 Fig). The two subgenomes of C. bursa-pastoris were most similar to their corresponding parental genomes along the first principal component, Dim1, i.e. expression in the CbpCg subgenome grouped with C. grandiflora, and the CbpCo subgenome grouped with C. orientalis. The second principal component, Dim2, reflected population structure; here again CASI accessions grouped with EUR accessions.
Fig 3. Transcriptomic variation patterns in three Capsella species.
Variation was visualized with principal component analyses of phased gene expression data (11,931 genes) for the three different tissues. CO, CG, ASI, EUR, ME, and CASI correspond to C. orientalis, C. grandiflora, and four populations of C. bursa-pastoris, Cbp, (Asia, Europe, Middle East, and Central Asia), respectively. The dash-dotted circles highlight the two different subgenomes of Cbp.
Testing for homeologue-specific expression (HSE) in C. bursa-pastoris showed that on average 4,096 genes (∼34%) per sample were significantly differentially expressed between the two subgenomes (FDR < 0.05). The expression ratio between subgenomes (defined as ) was on average 0.496 across all genes and 0.493 across genes with significant HSE indicating no strong bias towards one of the subgenomes (S4 Table). The ratio in DNA reads was 0.497 and thus there was no strong mapping bias towards either subgenome. Analyses of differential expression revealed no bias in the number of differentially expressed genes toward one subgenome either when comparing tissues (S5A Table, flowers and leaves being the most differentiated tissues and leaves and roots the least) or Cbp populations (S5B Table, Middle East and Asia being the most distant, except for CbpCo in flowers, while Europe and Central Asia are the closest).
Strong parental legacy and both cis- and trans-regulatory changes
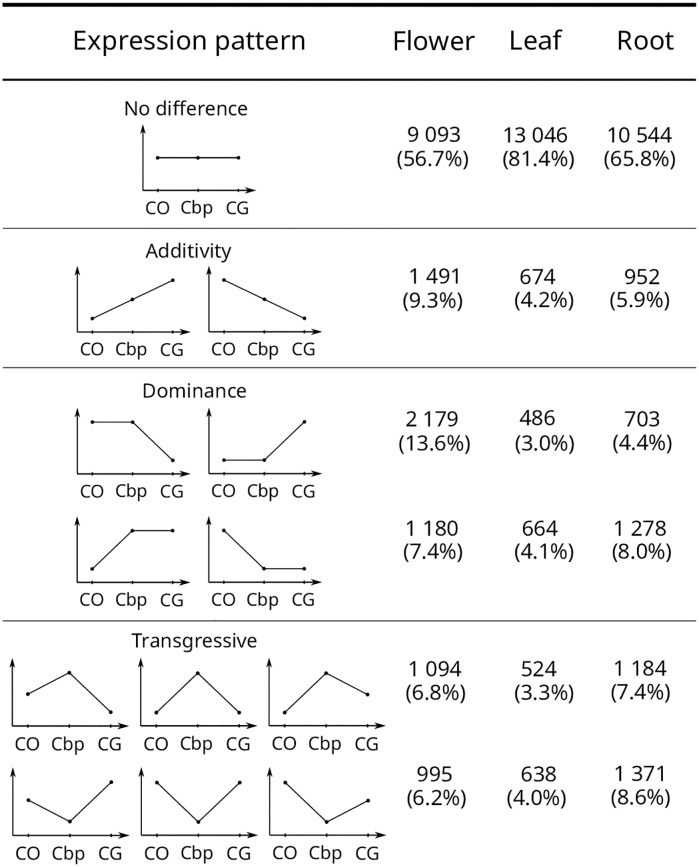
In order to investigate the total expression level changes in C. bursa-pastoris after C. grandiflora and C. orientalis hybridization, expression patterns of unphased data across the three species were classified into four categories: No difference, Intermediate/Additivity, Dominance and Transgressive (Fig 4). Up to 55-80% of the genes in C. bursa-pastoris were expressed at the same total level as in the parental species and 5 to 10% showed levels of expression intermediate to that of parental species. The dominance of one parental species over the other was most evident in flowers and roots. In flowers, ∼14% of C. bursa-pastoris genes were expressed at the same level as in C. orientalis but differed significantly from C. grandiflora, and ∼8% were expressed at the same level as in C. grandiflora but at a different level than in C. orientalis. The opposite dominance pattern was detected in the root tissue. Finally, a transgressive expression pattern, when expression levels in C. bursa-pastoris exceeded or were lower than the expression level of both parents, was detected in 8-16% of genes.
Fig 4. Levels of gene expression in C. bursa-pastoris relative to its parental species.
CO, CG, and Cbp correspond to C. orientalis, C. grandiflora, and C. bursa-pastoris, respectively. The y-axis indicates the level of expression. Expression levels were considered significantly different for the FDR < 0.05. In total, 16,032 genes were analyzed.
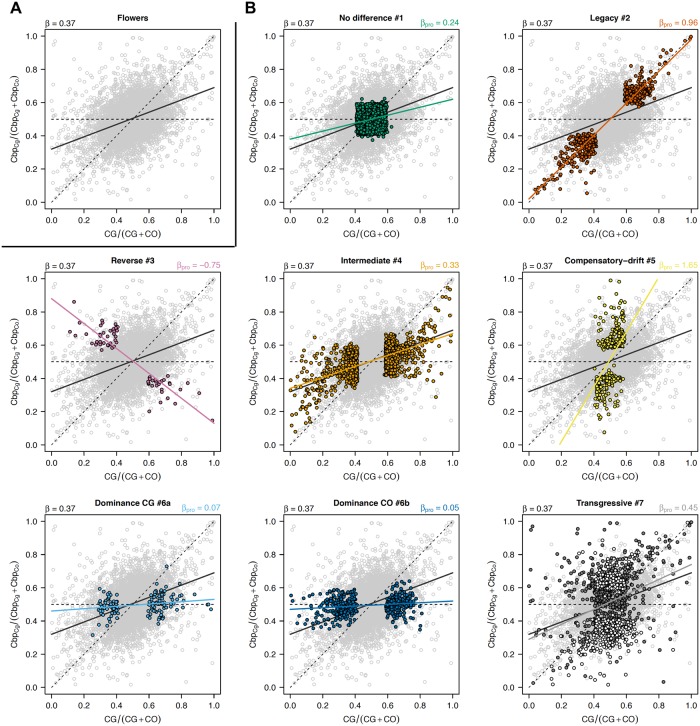
Gene expression in C. bursa-pastoris was further investigated by assessing the relative importance of cis- and trans-regulatory elements. The expression ratio of the two subgenomes was compared to the expression ratio between the two parental species (Fig 5A). For a given gene, if its expression in the homeologous genes of C. bursa-pastoris is only regulated by cis-regulatory changes, it should be completely explained by the divergence between the parental species (the diagonal line in Fig 5A). On the other hand, if homeologous genes are equally expressed in C. bursa-pastoris but not in the parental species, this means that Cbp expression is mainly controlled by trans-regulatory elements (the horizontal line in Fig 5A) [21]. First, the relationship between expression ratios in C. bursa-pastoris and parental species was positive and highly significant for all three tissues (p < 0.001), and the slope was intermediate between what would be expected if there were either only cis-(β = 1) or only trans-regulatory (β = 0) changes (β = 0.37, 0.42 and 0.46, respectively for flowers, leaves and roots). This indicates a strong parental legacy effect in expression of the two subgenomes of C. bursa-pastoris and suggests a joint effect of cis- and trans-regulation. Second, the variance of the expression ratio between subgenomes was significantly smaller than the variance of the expression ratio between parental genomes (Fisher’s variance test, all p < 0.001), indicating that the two subgenomes are closer to each other than the parental genomes are, therefore supporting a co-regulation of the two subgenomes through a mixture of trans- and cis-regulation [21, 32]. Finally, the slope of the regression between the two expression ratios was the weakest in flowers, suggesting a slightly stronger trans-regulation and a higher level of constraints in this tissue than in roots and leaves [32].
Fig 5. Relationships between the relative expression of the C. bursa-pastoris subgenomes and the relative expression of parental species.
The figure shows expression in flower as an example. A. Top-left panel is for all transcripts (11,931). B. Transcripts belonging to a specific category. The diagonal dashed lines indicate 100% cis-regulation divergence while the horizontal dashed lines indicate 100% trans-regulation. The solid lines give the slopes of the linear regressions between both ratios either for all transcript (black) or for transcript belonging to a specific category. β is the slope of the corresponding regression. For Transgressive category (bottom right panel), dark gray corresponds to categories #7a and b, light grey is for category #7c (see Fig 6).
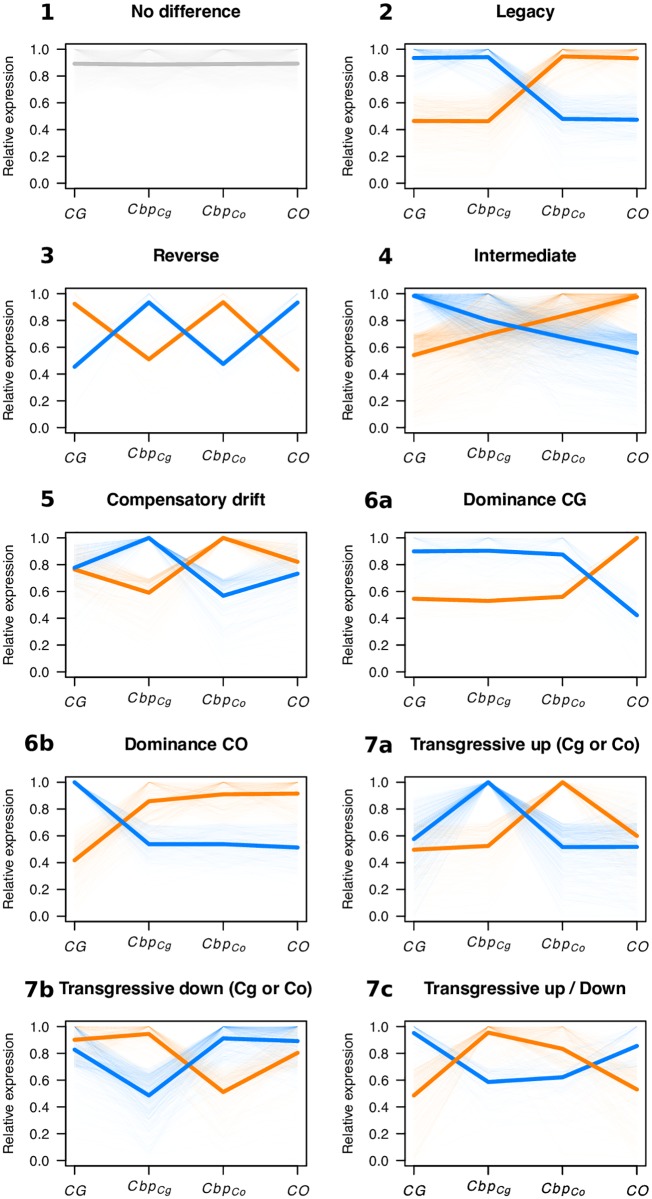
Classification of expression patterns
As mentioned above, subgenome expression level relative to parental species expression can help to disentangle the role of cis- and trans- components on overall gene regulation. However, the comparison between the ratios of expression in the tetraploid and in the parents is not sufficient to distinguish all possible patterns. We thus classified the expression patterns at the equivalent developmental stage in genes from the two subgenomes and the parental species in seven main categories by comparing the four expression levels (see Fig 6 for an example with flower tissues). The majority of the transcripts was not differentially expressed between parental genomes and subgenomes (No difference category), ranging from ∼60% in flowers to ∼78% in leaves (Table 1). However, the slope of the regression between relative expression of subgenomes and parental species clearly indicated that, even if the expression levels were not significantly different between parental species and C. bursa-pastoris subgenomes, crossed trans-regulation tended to make the two subgenomes expression closer to each other than to either parental species (Fig 5B “No difference” and Table 1). About 9% of genes had an Intermediate/Additive expression, i.e., the expression of both subgenomes being in between the expression of the two parental species. As expected this pattern was due to a combination of both cis- and trans-regulation (β ≃ 0.3 − 0.4). Only 3% showed a strict legacy of parental species expression which is primarily due to cis-regulation (β ≃ 1). About 4% of the genes showed a Dominance pattern of either CG or CO parental genetic background (i.e., both subgenome expression are similar to that of one parental species, categories 6a and 6b, Fig 6). However, within transcripts showing a Dominance pattern, 76% of the transcripts showed a dominance of CO in flowers, while there were only 45% and 34% in leaf and root tissues (Table 1). The Dominance pattern seems to be due to a dominance of transcription factors from one subgenome over the other (β ≃ 0.05 − 0.2); in favour of CO parental genetic background in flowers and of CG parental genetic background in leaves and roots (Fig 5B and Table 1). Finally, 3% of the genes had a Compensatory-drift profile (parental species expressions are similar but subgenome expressions diverge), a mere 0.4% showed a Reverse profile (each subgenome expression is similar to the opposite parental species) and about 10% of the transcripts showed a Transgressive pattern, either because of one (categories 7a and 7b) or of both subgenomes expression (category 7c) (Fig 5B and Table 1). These last profiles are less straightforward to interpret in terms of cis- and trans-regulation pattern as they involve more complex post-hybridization regulation processes.
Fig 6. Main categories of expression variation of C. bursa-pastoris subgenomes relative to expression in parental species.
The figure shows expression in flower as an example. Each transcript was assigned to one of seven main categories defined from the relative expression pattern of Cbp subgenomes (CbpCg and CbpCo) and parental species (CG and CO). For each category, dashed lines correspond to single transcript relative expression to the maximal expression of this transcript in parental genomes or subgenomes. Solid lines indicate the average expression for each genome or subgenome. Colors discriminate alternative patterns in the same category.
Table 1. Expression variation of C. bursa-pastoris subgenomes relative to expression in parental species across different tissues.
The percentage of transcripts within each category is given for all genes or only differentially expressed genes (i.e, without No difference category). The slope of the regression of relative expression between subgenomes and relative expression between parental species for all genes per category is also provided (β, see Fig 5). The percentages of transcripts showing a dominance of either CbpCg or CbpCo are given in parenthesis.
| Categories | Flowers | Leaves | Roots | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transcripts (%) | Transcripts (%) | Transcripts (%) | ||||||||
| All | DE only | β | All | DE only | β | All | DE only | β | ||
| No diff. | 1 | 60.4 | - | 0.24 | 78.4 | - | 0.32 | 67.6 | - | 0.32 |
| Legacy | 2 | 4.5 | 11.4 | 0.96 | 2.3 | 10.6 | 0.94 | 3.2 | 9.9 | 0.96 |
| Reverse | 3 | 0.5 | 1.3 | -0.75 | 0.2 | 0.9 | -0.78 | 0.4 | 1.2 | -0.78 |
| Intermediate | 4 | 12.6 | 31.9 | 0.33 | 6.8 | 31.5 | 0.44 | 8.8 | 27.3 | 0.41 |
| Comp. drift | 5 | 3.8 | 9.7 | 1.65 | 1.5 | 6.9 | 1.52 | 2.8 | 8.7 | 1.81 |
| Dominance | 6a | 1.2 | 3.0 (24) | 0.07 | 1.7 | 7.9 (54) | 0.18 | 2.7 | 8.4 (66) | 0.14 |
| 6b | 3.8 | 9.6 (76) | 0.05 | 1.4 | 6.5 (45) | 0.11 | 1.4 | 4.3 (34) | 0.1 | |
| Transgressive | 7a | 5.1 | 12.9 | - | 2.1 | 9.7 | - | 4.2 | 13 | - |
| 7b | 5.2 | 13.2 | - | 2.6 | 12 | - | 4.4 | 13.7 | - | |
| 7c | 2.8 | 7.1 | 0.45 | 3 | 13.9 | 0.55 | 4.3 | 13.4 | 0.61 | |
| Total | 100 | 100 | 0.37 | 100 | 100 | 0.42 | 100 | 0.46 | ||
Finally, although the relative proportions of the different categories were globally conserved across tissues (Table 1), expression patterns of individual genes were strongly tissue-specific. In our data, only half of the genes showed the same expression pattern in all three tissues. The most conserved category was No difference, 77%, and the least conserved one was Compensatory-drift, 3%. Pairwise comparisons between tissues revealed that the number of genes for which the expression pattern changed from one tissue to another was the largest between flowers and roots tissues (42%) and the smallest between leaf and root tissues (33%).
To conclude, only about 10% of the 11,931 transcripts had a transgressive or a reverse expression pattern. Expression patterns were poorly conserved between tissues except for the No difference category, indicating that the evolution of expression regulation is highly tissue-specific. Flower tissue differed the most from the two other tissues. In addition to a lower proportion of differentially expressed genes, flower tissues also had the lowest proportion of Transgressive category in the differentially expressed genes, indicating that when expression changes occurred, they either took place within the expression range of the parental species or they were compensated by the other subgenome (Compensatory-drift). This suggests a higher level of constraints on gene expression in flower tissues than in leaves and roots. Moreover, in flowers, the CO genetic background clearly dominates over the CG background, in striking contrast with the dominance of the CG genetic background in the other two tissues. Finally, expression profiles are more conserved between leaves and roots than between flowers and roots.
Expression similarity and convergence between subgenomes: Flowers differ from roots and leaves
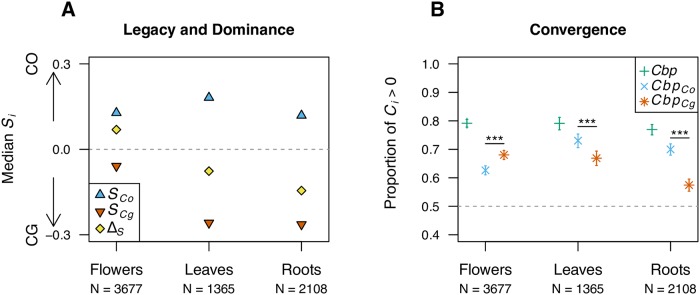
To understand better the joint dynamics of expression in the two subgenomes across tissues, and to avoid a priori classifications, we defined a new similarity index, S, that measures the similarity between mean expression level of each subgenome in each gene and the mean expression level in the parental species for the same gene (see Material and methods § Similarity and Convergence indices). This index is centered on 0, so that S < 0 means that the expression of a given transcript from a given subgenome is more similar to the expression of that transcript in CG, and S > 0 means that its expression is closer to that of CO. For all tissues, S indices of both subgenomes were biased towards the corresponding parental genome, i.e. CbpCg towards CG and CbpCo towards CO (binomial test, all p < 0.001). However, the strength of this bias differed between subgenomes and across tissues (Fig 7A). The distributions of S values for leaf and root tissues were more spread than the distribution for flowers, meaning that the relative expression in the two subgenomes was globally less constrained in these tissues than in the flower tissue (S4 Fig).
Fig 7. Similarity and convergence indices for differentially expressed genes between subgenomes of C. bursa-pastoris.
A. For each tissue and each subgenome, the median of similarity indices for each subgenome (SCo and SCg) are presented as well as the difference between the two indices (ΔS) that indicates the dominance of one parental genetic background. Grey dotted lines (S = 0) indicate level of no bias. B. The proportion of transcripts showing convergence (Ci > 0) is reported for the whole genome (green plus signs) or each subgenome (CbpCo, CbpCg). The significance of difference between the subgenome convergence indices is also depicted (binomial test,***, p < 0.001). The number of differentially expressed genes considered for each tissue are indicated with N.
As S index reflects the similarity between each subgenome expression and parental expression, the difference between S values for a given transcript (ΔS = |SCbpCo| − |SCbpCg|) can be viewed as the overall dominance of one parental genetic background over the other (ΔS < 0 means dominance of CG and ΔS > 0 means dominance of CO). In flowers, median S values for genes that showed significant differential expression between parental species (FDR < 0.05) showed dominance of the CO over CG genetic background (ΔS = 0.07), while the opposite pattern—i.e. dominance of CG back-ground over CO—was observed in leaves and roots (ΔS = -0.08 and -0.14, respectively; Fig 7A). This pattern was also observed when considering all genes, though it was less pronounced (S4 Fig). Such a dominance cannot only be due to the genes showing strict dominance of one genetic background (Dominance category, ∼3-5%), but rather indicate a more global dominance of trans-regulation of one genetic background. Indeed, even if S indices tended to show a large legacy of parental genome expression, positive correlations between SCg and SCo (Spearman’s ρ, all p < 0.001) confirmed that both subgenomes were co-regulated in the same direction (S4 Fig), towards C. orientalis in flower tissues and towards C. grandiflora in leaf and root tissues.
Finally, since subgenomes expression tended to converge, we defined a convergence index to measure the strength of the convergence of each subgenome expression toward the other (C index, see Material and methods § Similarity and Convergence indices). Indeed, a closer expression between subgenomes than between parental species can be due to a change in expression of both subgenomes toward an intermediate expression level or to a change in expression of only one subgenome toward the expression level of the other. In all tissues, most convergence indices were positive (Fig 7B and S5 Fig), indicating that the difference in gene expression between subgenomes was generally lower than the difference between parental species; also, the larger the difference in expression between parental species, the stronger the convergence between subgenomes, CCbp (Spearman’s ρ = 0.63, ρ = 0.74, ρ = 0.66, respectively for flowers, leaves and roots; all p < 0.001). One could expect that the expression patterns of homeologous genes were inherently more correlated because the RNA was extracted from the exact same pool of cells in C. bursa-pastoris while it was obviously not the case for the parental species. However, the way the analysis was carried out has likely attenuated this effect. First, the convergence index was computed from the average expression of each subgenome across all Cbp accessions, thereby partly breaking such an association. Second, to evaluate the strength of such a potential bias, we also estimated the C indices for subgenomes coming either from the same individuals or from different individuals S6 Fig. Although the convergence indices computed from the same individuals were stronger (closer to one) than the ones computed from different individuals, the overall pattern did not change: namely, the vast majority of the convergence indices are positive indicating that the subgenome expression levels converged. Although the overall degree of convergence was the same in the three tissues, the amount of convergence was not the same between the two subgenomes. In flowers, CbpCg expression tended to shift more towards CbpCo expression than the converse, while the opposite was true in the two other tissues (Fig 7B). This explains the dominance patterns observed through the S indices and confirms the role of unbalanced trans-regulation in the present system.
Genes showing converging expression patterns are enriched for specific functions
Regardless of the tissue considered, the expression profiles did not correspond to specific physical clusters along the genome with transcripts belonging to a given profile being spread across the genome: for each scaffold and each category, the average distance (bp) between two transcripts randomly sampled within a given category was not significantly different than that of two transcripts randomly sampled in different categories (Wilcoxon-Mann-Whitney’s test, all p > 0.05, S7 Fig). This suggests that the differential expression is not driven by large-scale epigenetic changes along chromosomes.
Gene ontology analyses revealed that the different expression profile categories (Fig 6) were enriched for different molecular functions (MF, average overlap between categories: 8.9, 9.0 and 8.6% for flowers, leaves and roots tissues, respectively, S6A Table) and biological processes (BP, average overlap, 5.4, 4.2 and 6.0%, S6B Table), though neither MF nor BP of a given category tended to cluster into specific networks. At the tissue level, the different expression profile categories were enriched for different MF and BP with a small average overlap between tissues (MF, 5.3% and BP, 4.8%, S7A and S7B Table), highlighting the specificity of expression regulation in different tissues.
We showed above that the main difference in expression between tissues was in the convergence of the two subgenomes: in flowers, CbpCg expression pattern converged toward that of CbpCo, while for the two other tissues convergence was in the opposite direction (CbpCo toward CbpCg). We tested whether the transcripts showing a convergence of CbpCg toward CbpCo (hereafter, ConvCo genes) or a convergence of CbpCo toward CbpCg (hereafter, ConvCg genes) were enriched for different molecular functions and biological processes. The two gene sets, ConvCo or ConvCg genes, were indeed enriched for GO terms belonging to different clusters (S8 Fig). For instance, in flower tissues, ConvCo genes are enriched for biological processes involved in the transition between vegetative and reproductive phases, the dormancy of floral meristems and male meiosis, while ConvCg genes were enriched for cell redox homeostasis and related biological processes (S8 and S9 Figs). As expected, underlying molecular functions also tended to group into distinct clusters corresponding to different functional networks (S8 and S9 Figs). There was also an enrichment for similar biological processes (e.g., drug transport in flowers, sucrose and carbohydrate metabolisms in leaves and roots) or molecular functions (e.g, RNA, nucleotide and GTP binding or MF related to transporter activity, S8 and S9 Figs) indicating some concerted changes of gene expression between the two subgenomes.
Deleterious mutations accumulate preferentially on the C. orientalis subgenome and are associated with the level of expression
Among the 11 million genomic sites segregating across the five genomes, about 3 million alleles were specific to the Capsella species, and 669,675 of these species-specific alleles were annotated for tolerated (TOL) and deleterious mutations (DEL) by SIFT4G with the A. thaliana database, and 432,354 of them were annotated with the C. rubella database.
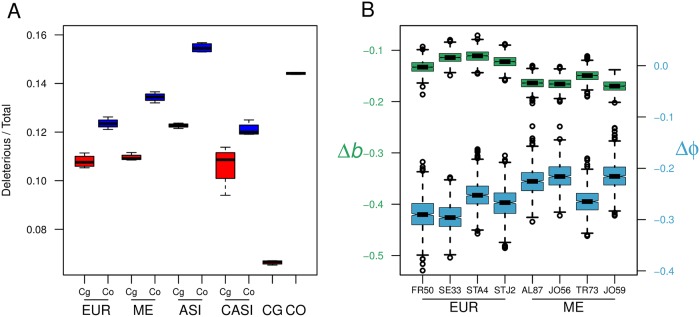
The estimated proportion of deleterious mutations among species and among the four populations of C. bursa-pastoris were similar independently of whether A. thaliana or C. rubella was used for SIFT4G annotation (Fig 8A and S10A Fig). Despite a lower number of accessions, the same pattern as in [37] was observed: i) the C. grandiflora genome had a lower proportion of deleterious mutations than C. orientalis or either subgenome of C. bursa-pastoris ii) within C. bursa-pastoris, the CbpCg subgenome always had a lower proportion of deleterious mutations than the CbpCo subgenome of the same population and iii) among the C. bursa-pastoris populations, both subgenomes of the Asian population had a higher proportion of deleterious mutations than the corresponding subgenomes in the other three populations, indicating a higher rate of mutation accumulation in this population. The proportion of deleterious mutations of the newly added CASI population was most similar to that of the EUR population with a larger variance of the proportion of deleterious mutations carried by CbpCg subgenome of CASI accessions (Fig 8A).
Fig 8. Variation in deleterious mutations in the two subgenomes of C. bursa-pastoris.
A. Proportion of deleterious mutations in the subgenomes and in the parental species. CO, CG, ASI, EUR, ME, CASI correspond to C. orientalis, C. grandiflora, and four populations of C. bursa-pastoris, respectively. The two subgenomes are indicated with Co and Cg. Functional effects were annotated with the C. rubella SIFT database (the annotation with A. thaliana SIFT database is in the S10 Fig). B. Maximum likelihood estimates of parameters of the distribution of deleterious mutations on CbpCg genes. Each box represents the estimates for one accession, with 1000 bootstrap replicates. The estimates are presented as the difference between the estimated parameter for deleterious mutations, DEL, and the estimated parameter for synonymous mutations, SYN (Δb = bDEL − bSYN, Δφ = φDEL − φSYN). Notches represent the median and the 95% confidence interval. The left axis refers to Δb (green boxes), and the right axis refers to Δφ (blue boxes). The estimated parameters (b and φ) for DEL and SYN are shown separately in S11 Fig.
We then assessed the distribution of deleterious mutations between the two subgenomes of C. bursa-pastoris to test whether they accumulated (i) more in one gene copy than in the other at the homeologue level, as would be expected under a pseudogenization process, (ii) more in one subgenome than in the other as expected if one subgenome predominates (see Material and methods § Difference between species and subgenomes in deleterious mutations). Mutation accumulation pattern between the two subgenomes was thus investigated by estimating the mutation accumulation bias towards CbpCg, b, and the overdispersion parameter φ; a large value of φ indicates that mutations tend to accumulate preferentially in one of the two homeologous genes. b was positive for synonymous (SYN) mutations indicating a mapping bias towards CbpCg. b was also positive for DEL mutations in all accessions (S11 Fig), but much smaller than for SYN (bDEL < bSYN, Fig 8B). This indicates a general bias towards more DEL mutations in the CbpCo subgenome, despite the mapping bias toward CG. The same pattern was observed for φ (φDEL< φSYN, Fig 8B and S11 Fig). Hence, contrary to what is expected under a scenario of pseudogenization, the distribution of deleterious mutations was less over-dispersed than expected at random, suggesting that the accumulation of too many deleterious mutations per gene is prevented; a mechanism that might contribute to the maintenance of both homeologue copies. However, it should be noted that more silenced genes were observed in CbpCo than in CbpCg (S12 Fig).
Finally, the relationship between the number of deleterious mutations and the homeologue expressions was investigated by comparing, for each transcript, the difference in number of deleterious mutations () and the homeologue expression bias (). The categories where deleterious mutations and expression bias varied in the same direction (i.e., dDEL > 0 and e > 0 or dDEL and e < 0) were over-represented (Fisher’s exact test, all p < 0.001, S8 Table). This means that the homeologue copy carrying the highest number of deleterious mutations tends to show the lowest expression level. No such association was found when considering transcripts carrying only synonymous mutations (Fisher’s exact test, p = 0.57, 0.74 and 0.27 for flowers, leaves and roots tissues, respectively), confirming that the association between deleterious mutations and expression level was not the result of a mapping or annotation bias toward one of the two subgenomes (S8 Table).
Discussion
The events accompanying the birth of a polyploid species have often been described in rather dramatic terms, with expressions such as “transcriptomic shock” or “massive genome-wide transcriptomic response” often used (e.g. [8, 47, 48]). The early and formative years of a young polyploid might indeed be eventful, but what happens afterward may well be less dramatic, especially for tetraploid species with a disomic inheritance such as shepherd’s purse. In the present study, we compared some of the genomic and transcriptomic changes that occurred between C. bursa-pastoris and its two parental species C. grandiflora and C. orientalis. Overall, the emerging picture is one of an orderly and rather conservative transition towards a new “normal” state. A conservative transition, because after around 100,000 generations we can still detect a significant parental legacy effect on both the number of deleterious mutations accumulated and gene expression patterns. And an orderly one too, since the emerging pattern of expression involves a balance between cis- and trans-regulatory changes suggesting the emergence of coordinated functioning of the two subgenomes. This general impression of a non-stochastic transition process to polyploidy [49] is reinforced by the variation in patterns of gene expression across the three tissues: as one would expect, the expression of both subgenomes in selfing C. bursa-pastoris was biased towards the selfing parent C. orientalis in flower, whereas in leaf expression of the two subgenomes were mostly similar, and in roots expression was biased towards C. grandiflora. This expression bias towards the C. orientalis subgenome in flowers despite a higher accumulation of deleterious mutations in this subgenome suggests that the evolution of gene expression is not entirely random.
Demography and expression: A limited effect of introgression?
Previous studies have stressed the importance of population structure and demographic history in genomic and transcriptomic studies of C. bursa-pastoris [36, 37]; [37], for instance, showed a significant admixture between C. orientalis and Asian populations of C. bursa-pastoris. In the present study, we indeed showed that the overall gene expression pattern reflected the main phylogenetic relationships. Each subgenome was the closest to the parental species from which it was inherited and populations from close geographic areas tended to cluster together, except for Central Asian accessions (CASI), which clustered with European ones even though they were geographically closer to the Asian or Middle-East ones. Most likely these samples were recently introduced to Central Asia, as it was suggested for C. bursa-pastoris accessions with European ancestry inhabiting the Russian Far East [35].
When comparing the number of differentially expressed genes between C. bursa-pastoris and parental species, no specific trend was detected and Asian accessions were not the closest to C. orientalis as one would have expected because of introgression. In leaf and roots tissues, ASI was even closer to C. grandiflora than to C. orientalis. This can be explained by the fact that the vast majority of the genes (up to 80%) did not show any difference in expression (thus hiding a more subtle signal). Assessing the influence of introgression on expression pattern would require a more thorough investigation, for instance by focusing on genes for which introgression was actually characterized.
Transition to polyploidy: Compensatory cis-trans effects, and stabilizing selection
As mentioned above, in the case of a newly formed allopolyploid species one would expect the two homoeologous copies of a gene to be under the influence of trans-regulatory elements inherited from both parents and its expression level to first move towards the mean expression of the two parental species. However, different forces could lead to an excess of divergence in subgenome expression compared to what would be expected under a pure drift model. Polyploidy creates a large redundancy in gene function that should free one of the copies from purifying selection. Generally, the copy carrying more deleterious mutations is expected to degenerate, biasing the expression pattern toward one of the two parental species, even if sub- or neo-functionalization can still occur but to a much lower extent. This ought to be particularly true for C. bursa-pastoris as one of its parental species, C. orientalis, is a selfer that has accumulated more deleterious mutations than the other parent, the outcrossing C. grandiflora [37]. This process will be reinforced by the enhancer runways process [27], that should strengthen cis-acting elements from the CbpCg subgenome as the CbpCg subgenome has higher heterozygosity and lower genetic load than the CbpCo subgenome.
In our study, however, we did not observe any “transcriptomic shock” (as for instance in, [8, 47]) neither major homeologue expression remodeling and/or subgenome expression asymmetry (as in e.g. [18]). In contrast, our study, like some others before it [16, 49–52], instead suggests overall conservation of the expression pattern in polyploids and hybrids. And even if a “transcriptomic shock” did take place during the formation of the tetraploid, expression changes have stabilized since then. Some 100,000 years later parental legacy on subgenome expression is still detectable and the two subgenomes’ expression patterns are still closer to each other than that of parental species, clearly indicating that none of the subgenomes has degenerated; as expected, however, the CbpCo subgenome carries more silenced genes and a higher proportion of deleterious mutations than CbpCg.
Most of the genes were under both cis- and trans-acting elements; the No difference and Intermediate expression categories represented up to 70-80% of genes depending on the tissue considered, a percentage similar to that observed in F1 hybrids between A. thaliana and A.arenosa [52]. Only a small fraction (5 to 10%) of genes showed either almost pure cis- (Legacy category) or trans-regulation (Dominance category). While the former can be explained by the absence of crossed trans-regulation, the latter could be due to the dominance of transcription factor of one subgenome over the other; though, in both cases, post-hybridization mutations affecting either cis- or trans-acting elements or both could have evolved. The remaining fraction (up to 15%, Reverse, Compensatory-drift and Transgressive) showed a more complex pattern that is hard to assign to a simple factor but could be in part due to new intertwined cis- and trans-regulations across subgenomes. It should be noted that such patterns can naturally emerge after hybridization as a byproduct of stabilizing selection on diverging optima [53] for Transgressive profiles, on the overall amount of protein produced for Compensatory-drift profile, and on the intermediate level of expression for Reverse profile, without invoking additional specific processes. To address further this question, it would be interesting to compare auto- and allopolyploids to tease apart the effects of hybridization and genome doubling.
Even though this does not, in any way, alter the conclusion above, we also would like to note here that the classification of overall expression patterns in different categories used in Fig 6 and Table 1 is somewhat arbitrary as some expression patterns are ambiguous and could have been classified in different categories. It should also be pointed out that these classifications were dependent on the chosen False Discovery Rate (FDR). As a control, we reproduced the analysis based on unphased data of Cbp expression, with FDR < 0.01 and 0.1 (S9 Table). It indicated that the number of genes within the different categories can vary substantially with the different FDR level (mainly because of variation in No difference category), however, the main patterns were not altered. Moreover, the main pattern of variation we described was a change in dominance between tissue that is obviously not affected by the bias described before. In part to overcome the limitations inherent to any a priori classification, we developed the expression similarity and convergence indices, S and C, that confirmed our conclusions.
Level of expression dominance varies across tissues and functions
Allopolyploid species are often examined for unequal expression between homeologous genes because of their hybrid nature but other aspects of gene expression have been less extensively studied. For example, there might be no difference in the relative expression of subgenomes (balanced homeologue expression), but the total amount of transcripts can vary and reflect the dominance of the level of expression of one of the parents [18]. C. bursa-pastoris exhibits rather balanced homeologue expression, but the summed expression of the two homeologues shows differentiation across tissues with the dominance of C. orientalis expression level in flowers, and C. grandiflora level in leaves and roots. The genes with significant expression bias between subgenomes also show strong dominance of CbpCo expression over CbpCg in flower. However, a positive correlation between the expression deviation indices of the two subgenomes indicates that this dominance is not primarily caused by up-regulation or down-regulation of one parental copy, but rather unidirectional regulation of homeologous genes as it has been observed, for instance, in cotton and coffee [2, 32, 54]. This convergence could be possible because of the low divergence between the subgenomes of C. bursa-pastoris and, hence, the absence of barriers for trans-acting regulation of homeologous genes.
An intuitive explanation of this bias in flower tissues could be that this simply reflects the fact that both C. orientalis and C. bursa-pastoris are selfing species with tiny flowers, in contrast to C. grandiflora, an outcrossing species that has large flowers. A way to test this hypothesis would be to compare C. orientalis with both C. grandiflora and C. rubella for the genes implicated in the bias towards C. orientalis using root tissues as a control. In contrast, in the non-reproductive leaf and root tissues, expression is biased towards the genome of the outcrossing C. grandiflora. Although this interpretation needs further validation, it stands against the genomic shock pattern that implies a disruption of expression patterns.
Finally, although the bias of expression observed between homeologous genes is not strongly shifted towards either subgenome, it is not random either: one subgenome can dominate over the other for a given function or pathway in a given tissue, suggesting constrained evolution in gene expression regulation at a tissue/function level. In many cases, it is not straightforward to explain why a particular subgenome dominates for a particular function, and this could simply be the result of coincidence in neutral evolution of gene regulation networks. In other cases such as flower tissues, however, the observed dominance makes biological sense.
Both subgenomes of C. bursa-pastoris are maintained, but they are not equal
Redundancy of polyploid genomes often assumes evolution of non-functionalization of duplicated genes [55–57] or even of a whole subgenome [38, 44, 58]. When one gene copy of a duplicated gene starts to degenerate, the purifying selection on that copy becomes weaker and the deleterious mutations accumulate further, while the other copy of the gene remains functional and under purifying selection. If non-functionalization is prevalent, deleterious mutations are expected to be more unevenly distributed between the homeologous genes and even between the two subgenomes. We indeed observed more deleterious load in the CbpCo subgenome with the absolute load comparison and with the estimated parameter b indicating its degeneration. However, the dispersion for deleterious mutations indicated that they tend to be more evenly distributed between the homeologous genes than expected at random. This suggests that CbpCo genes cannot degenerate further after a certain amount of genetic load is accumulated. Thus, although the amount of accumulated genetic load differs between subgenomes of C. bursa-pastoris, both subgenomes are maintained and there is no large-scale non-functionalization at the gene and subgenome levels.
One might expect the differences between homeologues in accumulation of deleterious mutations would lead to bias in gene expression. For example, Arabidopsis suecica, like C. bursa-pastoris, is an allopolyploid species with parents characterized by different mating systems: the outcrossing Arabidopsis arenosa, and the selfing Arabidopsis thaliana [59]. Chang et al. [60] observed a bias in expression in favor of the A. arenosa subgenome and, among other hypotheses, suggested that this bias could be due to the fact that mildly deleterious alleles are not purged as efficiently from the A. thaliana subgenome as from the A. arenosa subgenome. In C. bursa-pastoris, the CbpCo subgenome had a higher proportion of deleterious mutations than the CbpCg subgenome, but there was no strong bias in expression between subgenomes. However, when we paired the amount of derived deleterious mutations with the expression level of each gene and compared homeologous genes, we found that there was a significant association between deleterious mutation bias and expression bias (S8 Table). The homeologous gene with more deleterious mutations tends to have a lower expression level than the other one. Moreover, we also found that there are more silenced genes in CbpCo, which is the subgenome with a higher proportion of deleterious mutations. These results are in accordance with the hypothesis that the bias in expression is linked to the accumulation of deleterious mutations. Yet, it is worth noting that the expression bias may not necessarily be the result of the biased distribution of deleterious mutations. The homeologue expression bias could also be the cause of the observed deleterious mutation bias, especially considering that we have only investigated the deleterious mutations in coding regions. Purifying selection on the homeologue with lower expression can be weaker [61], therefore it is less efficient in eliminating deleterious mutations. At any rate, the fact that we have a relative dominance of expression of CbpCo in flowers and of CbpCg in other tissues, despite CbpCo subgenome having a higher proportion of deleterious mutations than CbpCg, suggests that parental legacy and functional constraints may also play a major role.
Conclusion
In 1929, George Shull, one of the most prominent geneticists of his time [62], wrote: “It is considered a matter of fundamental significance that the increase in a number of chromosomes in the bursa-pastoris group is correlated with greater variability, greater adaptability, greater vigor, and greater hardiness”. In the present study, the merging of the two parental genomes was not accompanied by major disruptions of the transcriptome. Instead, there was a strong parental legacy and the emergence of a shift in the subgenome expression pattern towards a new “equilibrium” state reflecting the composite nature of the new species. Hence, being a selfer like its C. orientalis parent, there was a shift in flower tissues of the expression pattern of the C. grandiflora subgenome towards that of C. orientalis. Similarly, it seems also possible that the dominance of the C. grandiflora inherited subgenome in roots and leaves contributed to the high competitive ability of C. bursa-pastoris, which was similar to that of C. grandiflora but much higher than that of C. orientalis and C. rubella, its two self-fertilizing congeners [63, 64]. It therefore seems that the present study, together with those more focused on fitness of C. bursa-pastoris [63, 64] contributed to better understanding of the causes of the correlation pointed out almost 100 years ago by Shull.
Material and methods
Samples, sequencing and data preparation
We obtained the whole genome and RNA-Seq data from flower, leaf and root tissues of (i) 16 accessions of C. bursa-pastoris coming from already characterized populations from Europe (EU), the Middle East (ME) and Eastern Asia (ASI) [35] and from hitherto unstudied Central Asian populations (CASI) and (ii) four accessions each of C. grandiflora and C. orientalis (Fig 1). The genomic data included both published and newly sequenced genomes (S1 Table). For newly sequenced genomes, DNA was extracted from leaves with the Qiagen DNeasy Plant Mini Kit, libraries were prepared using the TruSeq Nano DNA kit, and 150-bp paired-end reads were sequenced on Illumina HiSeqX platform (SciLife, Stockholm, Sweden). All 72 RNA-Seq libraries (24 accessions × three tissues) were sequenced in this study. For RNA sequencing, seeds were surface-sterilized and germinated as described in [36]. Seedlings were then transplanted into pots (10 × 10 × 10cm) filled with soil seven days after germination and cultivated in one growth chamber (22°C, 16:8h light/dark period, light intensity 150 μmol/m2/s). Seven days after the onset of flowering, we collected flower buds, leaves, and roots of visually similar developmental stage. Tissues were snap-frozen in liquid nitrogen, and stored at -80°C before extraction following manufacturer protocol (Plant Total RNA Kit (Spectrum) for flower buds and leaves, and RNeasy Plant Mini Kit (Qiagen) for roots). RNA sequencing libraries were prepared using the TruSeq stranded mRNA library preparation kit including polyA selection and sequenced for 125-bp paired-end reads on Illumina HiSeq 2500 platform (SciLife, Stockholm, Sweden). Sequencing of new samples yielded an average library size of 57 million reads for DNA sequencing and 59 million reads for RNA-Seq.
DNA and RNA-Seq reads were mapped to the C. rubella reference genome [65] with Stampy v1.0.22 [66]. To account for the divergence from the reference genome, the substitution rate was set to 0.025 for C. bursa-pastoris, 0.02 for C. grandiflora, and 0.04 for C. orientalis. On average, 85%, 90% and 85% of the DNA reads were successfully mapped for the corresponding three species and 98% in all species for RNA mapping. This yielded an average coverage of 51x and 52x for DNA and RNA data, respectively. Genotyping of DNA and RNA-Seq alignments were performed using HaplotypeCaller from the Genome Analysis Tool Kit (GATK) v3.5 [67] as described in [37]. The subgenomes of C. bursa-pastoris were phased with HapCUT version 0.7 [68] following the procedure by [37]. The quality of this phasing procedure was ascertained by comparing the phased subgenomes with the subgenome assembly obtained by [45]. The unphased expression data was generated for non-overlapping feature positions (option: -m union) using the htseq-count program from HTSeq v0.6.1 [69]. To compare the expression between the two subgenomes of C. bursa-pastoris, homeologue-specific counting of alleles was performed using ASEReadCounter from GATK and phased according to the phased genomic data. We analyzed only the counts of SNPs that showed no strong deviation from the 0.5 mapping ratio in DNA data defined with a statistical model developed by [70]. To correct for potential bias in homeologue count data due to the uneven density of SNPs and/or uneven coverage along the gene, we scaled the homeologue expression counts using the unphased data and the allelic ratio from the phased data.
Population structure
Principal component analyses were performed using the ade4 R package [71]. A neighbor-joining phylogenetic tree was reconstructed from the absolute genetic distance in genomic SNPs with the ape R package [72]. A hierarchical distance clustering with bootstrap support was perfromed in the pvclust R package, [73].
Gene expression analyses
Differential gene expression analyses were carried out in edgeR [74]. The TMM normalization for different library sizes [74] was used for differential gene expression analyses, while for all other analyses, we used the count per million (CPM) normalization (one was added to every gene count to bypass log-transformation of zero expression). Phased counts were normalized by the mean library size of the two subgenomes and only genes showing no strong mapping bias were retained (see below). For both datasets (unphased or phased), only genes with at least one sample having a non-zero expression in every population/species were kept.
Differences between the two subgenomes (homeologue-specific expression) were assessed with the integration of the information from both RNA and DNA data to exclude highly biased SNPs and to account for the noise in read counts due to statistical variability. The data were analyzed using the three-stage hierarchical Bayesian model for allelic read counts developed by [70]. The model was implemented using Markov chain Monte Carlo (MCMC) with 200,000 iterations with burn-in of 20,000 and thinning interval of 100. Each analysis was run three times to assess convergence. The significance of homeologue-specific expression (HSE) was defined from a Bayesian analog of the false discovery rate (FDR < 0.05).
Expression patterns in C. bursa-pastoris and its parental species were classified into categories based on significant and non-significant differential expression defined with edgeR [74]. We considered the four genomes/subgenomes (CG, CO, Cbpcg, and Cbpco) and three possibilities for each of the six pairwise comparisons (significantly over, under or equally expressed, FDR < 0.05), and grouped the resulting combinations into seven main categories: No difference, Intermediate, Legacy, Reverse, Dominance, Compensatory drift, and Transgressive (see the Results for categories description). We also performed similar analysis for the unphased total C. bursa-pastoris expression (thus considering only three pairwise comparisons) by classifying the expression patterns into four major categories: 1) no differential expression, when no significant differences are detected in any of the three pairwise comparisons, 2) intermediate, when the expression of C. bursa-pastoris (Cbp) is intermediate between C. grandiflora (CG) and C. orientalis (CO), 3) dominance of one of the parents over the other, when the mean expression of C. bursa-pastoris is equal to only one parental species and the two parents are significantly different, and finally 4) transgressive, when the mean expression of C. bursa-pastoris is outside the range of expression of both parents and statistically significantly different from the parental species with the closer level of expression.
Similarity and convergence indices
To quantify the similarity between each subgenome expression level and the expression level in the parental species, we developed a similarity index (S). For each transcript i and each subgenome j ∈ {CbpCg, CbpCo}, S was computed as the subgenome relative expression deviation from the mean expression level in the parental species, :
Where (Eij) is the average expression of a given transcript i in a given genetic background j (CO or CG for parental species, and CbpCg or CbpCo for subgenomes of C. bursa-pastoris). This index is centered on 0 and oriented (i.e, when ), so that if Sij < 0 or Sij > 0, the expression of a given transcript in a given subgenome is more similar to the expression of that transcript in CG or CO, respectively. The difference between the absolute values of the indices values for CbpCg and CbpCo, was used as a measure of dominance of one of the parental genetic background.
Finally, for each gene that was differentially expressed between the two parental species, a convergence index, C, was computed from the absolute difference in expression for:
subgenomes: Δsub = |EiCg − EiCo|
parental species: Δpar = |EiCG − EiCO|
each subgenome and the opposite parental species: ΔCg = |EiCg − EiCO| and ΔCo = |EiCo − EiCG|.
These differences correspond to the phylogenetic distances (Fig 1A). In principle, if the regulation of gene expression in CbpCg is independent of the regulation of gene expression in CbpCo, then the overall Δsub, Δpar, ΔCg and ΔCo are expected to be equal. To compare these quantities, for each transcript i, we used a convergence index (Ci):
So, measures the expression convergence of CbpCg toward CbpCo, measures the expression convergence of CbpCo toward CbpCg, and CCbp measures the overall subgenomes convergence within Cbp. Δx stands for either ΔCo, ΔCg or Δsub, respectively. Ci thus ranges from -1 to 1, with positive values indicating more similar expression between the subgenomes of C. bursa-pastoris than between parental species, and negative values indicating increased differences between subgenomes; the closer Ci to 0, the more similar are the expression patterns to parental species.
Gene ontology enrichment test
Gene ontology (GO) enrichment tests were performed using the topGO R package [75]. The GO term annotation was downloaded from PlantRegMap (http://plantregmap.cbi.pku.edu.cn) and we used a custom background list of genes that included only the expressed genes for which phasing was possible in the relevant tissue. Fisher’s exact-test procedure (weight algorithm) was performed to assess the enrichment (p < 0.05) for either molecular functions (MF) or biological processes (BP). Finally, the REViGO software [76] was used to remove GO terms redundancy and to cluster remaining terms in a two-dimensional space derived by applying multidimensional scaling to a matrix of the GO terms semantic similarities. Cytoscape v3.6.1 was used to visualize GO terms networks [77].
Difference between species and subgenomes in deleterious mutations
Mutations were classified into tolerated and deleterious (DEL) using SIFT4G [78]. We used C. rubella [37] and Arabidopsis thaliana (TAIR10.22) SIFT4G reference databases. This helps avoid reference bias towards C.rubella away from calling mutations to be deleterious in the C. grandiflora homeologue. We considered only the mutations that accumulated after speciation of C. bursa-pastoris and identified mutations specific to C. grandiflora, C. orientalis, the two subgenomes of C. bursa-pastoris, and Neslia paniculata that was used as an outgroup here. All estimates were relative to the total number of SIFT4G annotated sites to minimize the bias associated with variation in missing data as in [37]. Only the European and Middle Eastern populations were used in further analysis of the distribution of deleterious mutations, in order to exclude the effect of gene flow between C. orientalis and the Asian population of C. bursa-pastoris [37].
We assessed the distribution of deleterious mutations between the two subgenomes of C. bursa-pastoris to test whether they accumulated (i) more in one gene copy than in the other at the homeologue level, as would be expected under a pseudogenization process, (ii) more in one subgenome than in the other as expected if one subgenome predominates. Under the null hypothesis (random accumulation without subgenome bias) the distribution of deleterious mutations between the two subgenomes should follow a binomial distribution with mean 1/2. Under the first hypothesis, the distribution should be more dispersed with the same mean, which can be modeled by a Beta-binomial distribution. Under the second hypothesis, the mean should differ from 1/2. However, over-dispersion and bias can also occur because of missing data and sampling error, we thus used synonymous mutations (SYN) to control for this and built the correct null distribution. To do so, we developed a maximum likelihood method implemented in R [79] as follows. First, we identified a most likely probability distribution model by fitting four models to the SYN dataset, where nSYN is the sum of SYN mutations occurring on both homeologous genes and kSYN is the number of SYN mutations occurring on CbpCg genes. The four models are:
M1: kSYN ∼ B(nSYN, 0.5), a binomial distribution with no bias between CbpCg and CbpCo,
M2: kSYN ∼ B(nSYN, 0.5 + b), a binomial distribution with bias,
M3: kSYN ∼ BB(nSYN, 0, φ), a beta-binomial distribution with no bias,
M4: kSYN ∼ BB(nSYN, b, φ), a beta-binomial distribution with bias.
For convenience, the beta-binomial distribution:
was re-parameterized as:
where and [80, 81]. In this way, the parameter b was a measure of the bias towards the CbpCg genes, and φ was a measure of the variance of the probability that a mutation is found within the CbpCg homeologues, and can be interpreted as an index of overdispersion. A large value of φ indicates that mutations tend to accumulate preferentially in one of the two homeologous genes, and a small value of φ indicates that mutations are more evenly distributed between them. We calculated the likelihood of each model and chose the best-fitting model with a hierarchical likelihood ratio test (hLRTs). After choosing the beta-binomial distribution with bias as the most likely null distribution, we estimated the parameters b and φ. We introduced a new set of models to test for the specific features of the distribution of deleterious mutations:
The null model assumes that both parameters b and φ are the same for the SYN and DEL datasets, while the alternative models allow the DEL dataset to have different parameters from the SYN dataset: only bDEL, only φDEL, or both bDEL and φDEL were allowed to vary. We calculated the likelihood of each model, chose the best fitting model with hierarchical likelihood ratio tests (hLRTs) and estimated the parameters of the selected model. Bootstrap estimates of confidence intervals were estimated with 1000 bootstrap replicates.
Relationship between deleterious mutations and gene expression
The SIFT4G annotation of the C. rubella database was used to match the gene IDs of the mutation and expression data. For each tissue, the relationship between the bias in the number of deleterious mutations between subgenomes and the bias in homeologue expression was investigated by calculating, for gene i in accession j, the difference (dij) in the number of deleterious mutations (DEL) between homeologous gene pairs:
The expression ratio between the homeologues of genes with significant HSE was used as a measure of homeologue expression bias:
Genes were further classified into four categories according to the deleterious mutations bias, d, and homeologue expression bias, e:
d > 0 and e > 0.5;
d > 0 and e < 0.5;
d < 0 and e > 0.5;
d < 0 and e < 0.5.
Genes with no bias in the distribution of deleterious mutation (d = 0) or no significant HSE (FDR < 0.05) were removed from the analysis. Fisher’s exact test was then used to test for independence between the difference in the number of deleterious mutations (d) and homeologue expression bias (e). As a control, the whole analysis was reproduced with dij computed from the number of synonymous mutations in genes with no DEL mutations. In addition, we also compared the number of silenced genes (genes with zero expression values) of each subgenome of C. bursa-pastoris, to check if there was a relationship between genetic load and silenced genes.
Data access
SRA numbers of the previously published samples are listed in S1 Table. New sequences of the DNA and RNA samples are avaliable under the project PRJNA533007 at NCBI and their SRA numbers are also provided in S1 Table. Phased and unphased genomic and expression data are deposited to the Open Science Framework Repository (DOI 10.17605/OSF.IO/G6H57) [82].
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Karl Holm, Kerstin Dalman, and Kerstin Jeppsson for help in the lab. Most of the analyses were carried out at Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under the project b2016212.
Data Availability
SRA numbers of the previously published samples are listed in S1 Table. New sequences of the DNA and RNA samples will be deposited to NCBI and their SRA numbers will be added to S1 Table. Phased and unphased genomic and expression data will be deposited to the Dryad Digital Repository. All new data will be made available upon the acceptance of this manuscript.
Funding Statement
The study was financed by grants from the Swedish Research Council (https://www.vr.se/), grant 2015-03797 to ML, and a grant from the Erik Philip-Sörensens Stiftelse (http://www.epss.se/) also to ML. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wood T, Takebayashi N, Barker M, Mayrose I, Greenspoon P, Rieseberg L. The frequency of polyploid speciation in vascular plants. Proc Nat Acad Sci. 2009;106:13875–13879. 10.1073/pnas.0811575106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoo MJ, Szadkowski E, Wendel JF. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 2013;110(2):171–180. 10.1038/hdy.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buggs RJA, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE. The legacy of diploid progenitors in allopolyploid gene expression patterns. Phil Trans R Soc B. 2014;369 (1648). 10.1098/rstb.2013.0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, et al. Recently formed polyploid plants diversify at lower rates. Science (New York, NY). 2011;333(6047):1257 10.1126/science.1207205 [DOI] [PubMed] [Google Scholar]
- 5. Thompson A, Zakon HH, Kirkpatrick M. Compensatory Drift and the Evolutionary Dynamics of Dosage-Sensitive Duplicate Genes. Genetics. 2016;202(2):765–774. 10.1534/genetics.115.178137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nature Reviews Genetics. 2010;11(2):97–108. 10.1038/nrg2689 [DOI] [PubMed] [Google Scholar]
- 7. Buggs RJ, Elliott NM, Zhang L, Koh J, Viccini LF, Soltis DE, et al. Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytologist. 2010;186(1):175–183. 10.1111/j.1469-8137.2010.03205.x [DOI] [PubMed] [Google Scholar]
- 8. Buggs RJA, Zhang L, Miles N, Tate JA, Gao L, Wei W, et al. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol. 2011;21(7):551–556. 10.1016/j.cub.2011.02.016 [DOI] [PubMed] [Google Scholar]
- 9. Hollister JD. Polyploidy: adaptation to the genomic environment. New Phyt. 2015;205(3):1034–1039. 10.1111/nph.12939 [DOI] [PubMed] [Google Scholar]
- 10. Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, et al. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol. 2013;23(21):2151–2156. 10.1016/j.cub.2013.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bomblies K, Higgins JD, Yant L. Meiosis evolves: adaptation to external and internal environments. New Phyt. 2015;208(2):306–323. 10.1111/nph.13499 [DOI] [PubMed] [Google Scholar]
- 12. Pelé A, Rousseau-Gueutin M, Chèvre AM. Speciation Success of Polyploid Plants Closely Relates to the Regulation of Meiotic Recombination. Frontiers in plant science. 2018;9:907 10.3389/fpls.2018.00907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role? Chromosoma. 2016;125(2):287–300. 10.1007/s00412-015-0571-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soltis DE, Misra BB, Shan S, Chen S, Soltis PS. Polyploidy and the proteome. Biochimica et biophysica acta. 2016;1864(8):896–907. 10.1016/j.bbapap.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 15. Douglas GM, Gos G, Steige KA, Salcedo A, Holm K, Josephs EB, et al. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc Nat Acad Sci. 2015;112(9):2806–2811. 10.1073/pnas.1412277112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE. Nonadditive gene expression in polyploids. Ann Rev Gen. 2014;48:485–517. 10.1146/annurev-genet-120213-092159 [DOI] [PubMed] [Google Scholar]
- 17. Wendel JF, Lisch D, Hu G, Mason AS. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Current opinion in genetics & development. 2018;49:1–7. 10.1016/j.gde.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 18. Grover C, Gallagher J, Szadkowski E, Yoo M, Flagel L, Wendel J. Homoeolog expression bias and expression level dominance in allopolyploids. New Phyt. 2012;196(4):966–971. 10.1111/j.1469-8137.2012.04365.x [DOI] [PubMed] [Google Scholar]
- 19. Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y, et al. Subgenome Dominance in an Interspecific Hybrid, Synthetic Allopolyploid, and a 140-Year-Old Naturally Established Neo-Allopolyploid Monkeyflower. The Plant cell. 2017;29(9):2150–2167. 10.1105/tpc.17.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bird KA, VanBuren R, Puzey JR, Edger PP. The causes and consequences of subgenome dominance in hybrids and recent polyploids. The New Phytologist. 2018;. 10.1111/nph.15256 [DOI] [PubMed] [Google Scholar]
- 21. Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430(6995):85 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- 22. Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature reviews Genetics. 2011;13(1):59–69. 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- 23. Buggs RJA, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE. The legacy of diploid progenitors in allopolyploid gene expression patterns. Philosophical Transactions Of The Royal Society Of London Series B-Biological Sciences. 2014;369 (1648). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Signor SA, Nuzhdin SV. The Evolution of Gene Expression in cis and trans. Trends In Genetics. 2018;34:532–544. 10.1016/j.tig.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Metzger BPH, Duveau F, Yuan DC, Tryban S, Yang B, Wittkopp PJ. Contrasting Frequencies and Effects of cis- and trans-Regulatory Mutations Affecting Gene Expression. Molecular Biology and Evolution. 2016;33(5):1131–1146. 10.1093/molbev/msw011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bell GDM, Kane NC, Rieseberg LH, Adams KL. RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome biology and evolution. 2013;5(7):1309–1323. 10.1093/gbe/evt072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fyon F, Cailleau A, Lenormand T. Enhancer Runaway and the Evolution of Diploid Gene Expression. PLoS genetics. 2015;11(11):e1005665 10.1371/journal.pgen.1005665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hodgins-Davis A, Rice DP, Townsend JP. Gene Expression Evolves under a House-of-Cards Model of Stabilizing Selection. Molecular Biology and Evolution. 2015;32(8):2130–2140. 10.1093/molbev/msv094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. Tempo and mode of regulatory evolution in Drosophila. Genome Research. 2014;24(5):797–808. 10.1101/gr.163014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nourmohammad A, Rambeau J, Held T, Kovacova V, Berg J, Lässig M. Adaptive Evolution of Gene Expression in Drosophila. Cell reports. 2017;20(6):1385–1395. 10.1016/j.celrep.2017.07.033 [DOI] [PubMed] [Google Scholar]
- 31. de Meaux J. Cis-regulatory variation in plant genomes and the impact of natural selection. American Journal of Botany. 2018;in press. 10.1002/ajb2.1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Combes MC, Dereeper A, Severac D, Bertrand B, Lashermes P. Contribution of subgenomes to the transcriptome and their intertwined regulation in the allopolyploid Coffea arabica grown at contrasted temperatures. New Phyt. 2013;200(1):251–260. 10.1111/nph.12371 [DOI] [PubMed] [Google Scholar]
- 33. Combes MC, Hueber Y, Dereeper A, Rialle S, Herrera JC, Lashermes P. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biology And Evolution. 2015;7(4):1110–1121. 10.1093/gbe/evv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hurka H, Friesen N, German DA, Franzke A, Neuffer B. ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae). Mol Ecol. 2012;21(5):1223–1238. 10.1111/j.1365-294X.2012.05460.x [DOI] [PubMed] [Google Scholar]
- 35. Cornille A, Salcedo A, Kryvokhyzha D, Glémin S, Holm K, Wright S, et al. Genomic signature of successful colonization of Eurasia by the allopolyploid shepherd’s purse (Capsella bursa-pastoris). Mol Ecol. 2016;25(2):616–629. 10.1111/mec.13491 [DOI] [PubMed] [Google Scholar]
- 36. Kryvokhyzha D, Holm K, Chen J, Cornille A, Glémin S, Wright SI, et al. The influence of population structure on gene expression and flowering time variation in the ubiquitous weed Capsella bursa-pastoris (Brassicaceae). Mol Ecol. 2016;25(5):1106–1121. 10.1111/mec.13537 [DOI] [PubMed] [Google Scholar]
- 37. Kryvokhyzha D, Salcedo A, Eriksson MC, Duan T, Tawari N, Chen J, et al. Parental legacy, demography, and admixture influenced the evolution of the two subgenomes of the tetraploid Capsella bursa-pastoris (Brassicaceae). PLOS Genetics. 2019;15(2):1–34. 10.1371/journal.pgen.1007949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Nat Acad Sci. 2011;108(10):4069–4074. 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature biotechnology. 2015;33(5):531 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- 40. Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proceedings of the National Academy of sciences. 2003;100(8):4649–4654. 10.1073/pnas.0630618100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature communications. 2014;5:3930 10.1038/ncomms4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. science. 2014;345(6199):950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- 43. Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genetics. 2011;43(10):1035 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 44. Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538(7625):336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kasianov AS, Klepikova AV, Kulakovskiy IV, Gerasimov ES, Fedotova AV, Besedina EG, et al. High-quality genome assembly of Capsella bursa-pastoris reveals asymmetry of regulatory elements at early stages of polyploid genome evolution. The Plant Journal. 2017;91(2):278–291. 10.1111/tpj.13563 [DOI] [PubMed] [Google Scholar]
- 46. Huang HR, Liu JJ, Xu Y, Lascoux M, Ge XJ, Wright SI. Homeologue-specific expression divergence in the recently formed tetraploid Capsella bursa-pastoris (Brassicaceae). New Phytologist. 2018;. 10.1111/nph.15299 [DOI] [PubMed] [Google Scholar]
- 47. Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr Biol. 2006;16(16):1652–1659. 10.1016/j.cub.2006.06.071 [DOI] [PubMed] [Google Scholar]
- 48. Hu G, Wendel JF. Cis-trans controls and regulatory novelty accompanying allopolyploidization. The New phytologist. 2018;. [DOI] [PubMed] [Google Scholar]
- 49. Akama S, Shimizu-Inatsugi R, Shimizu KK, Sese J. Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Research. 2014;42(6):e46 10.1093/nar/gkt1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, et al. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345(6194):1250091 10.1126/science.1250091 [DOI] [PubMed] [Google Scholar]
- 51. Göbel U, Arce AL, He F, Rico A, Schmitz G, de Meaux J. Robustness of Transposable Element Regulation but No Genomic Shock Observed in Interspecific Arabidopsis Hybrids. Genome biology and evolution. 2018;10(6):1403–1415. 10.1093/gbe/evy095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi X, Ng DWK, Zhang C, Comai L, Ye W, Chen ZJ. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nature communications. 2012;3:950 10.1038/ncomms1954 [DOI] [PubMed] [Google Scholar]
- 53. Chevin LM, Decorzent G, Lenormand T. Niche dimensionality and the genetics of ecological speciation. Evolution; international journal of organic evolution. 2014;68(5):1244–1256. 10.1111/evo.12346 [DOI] [PubMed] [Google Scholar]
- 54. Rambani A, Page JT, Udall JA. Polyploidy and the petal transcriptome of Gossypium. BMC Plant Biol. 2014;14(1):3 10.1186/1471-2229-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sankoff D, Zheng C, Zhu Q. The collapse of gene complement following whole genome duplication. BMC genomics. 2010;11(1):313 10.1186/1471-2164-11-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Force A, Lynch M, Pickett FB, Amores A, Yan Yl, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- 58. Marcet-Houben M, Gabaldón T. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast lineage. PLoS biology. 2015;13(8):e1002220 10.1371/journal.pbio.1002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Novikova PY, Tsuchimatsu T, Simon S, Nizhynska V, Voronin V, Burns R, et al. Genome Sequencing Reveals the Origin of the Allotetraploid Arabidopsis suecica. Mol Ecol Evol. 2017;34(4):957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang PL, Dilkes BP, McMahon M, Comai L, Nuzhdin SV. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 2010;11(12):R125 10.1186/gb-2010-11-12-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(10):4069–4074. 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shull GH. Species hybridizations among old and new species of shepherd’s purse. Proc Int Congr Pl Sci. 1929;1:837–888. [Google Scholar]
- 63. Petrone Mendoza S, Lascoux M, Glémin S. Competitive ability of Capsella species with different mating systems and ploidy levels. Annals of botany. 2018;121(6):1257–1264. 10.1093/aob/mcy014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang X, Lascoux M, Glémin S. Variation in competitive ability with mating system, ploidy and range expansion in four Capsella species. PCI Evolutionary Biology. 2018;. [Google Scholar]
- 65. Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45(7):831–835. 10.1038/ng.2669 [DOI] [PubMed] [Google Scholar]
- 66. Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–939. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24(16):i153–i159. 10.1093/bioinformatics/btn298 [DOI] [PubMed] [Google Scholar]
- 69. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 2011;21(10):1728–1737. 10.1101/gr.119784.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software. 2007;22(4):1–20. 10.18637/jss.v022.i04 [DOI] [Google Scholar]
- 72. Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 73. Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. 10.1093/bioinformatics/btl117 [DOI] [PubMed] [Google Scholar]
- 74. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R package version. 2010;2(0). [Google Scholar]
- 76. Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6(7):e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nature Prot. 2016;11(1):1–9. 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- 79.Team RC, et al. R: A language and environment for statistical computing; 2013. [Google Scholar]
- 80. Skellam J. A probability distribution derived from the binomial distribution by regarding the probability of success as variable between the sets of trials. J Royal Stat Soc B (Methodol). 1948;10(2):257–261. [Google Scholar]
- 81. Hughes G, USA LMP, 1993. Using the beta-binomial distribution to describe aggregated patterns of disease incidence. Phytopathology. 1993;83:759–763. 10.1094/Phyto-83-759 [DOI] [Google Scholar]
- 82.Kryvokhyzha D. Towards the new normal: Transcriptomic convergence and genomic legacy of the two subgenomes of an allopolyploid weed (Capsella bursa-pastoris)—DATA; 2019. Available from: osf.io/g6h57. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
SRA numbers of the previously published samples are listed in S1 Table. New sequences of the DNA and RNA samples will be deposited to NCBI and their SRA numbers will be added to S1 Table. Phased and unphased genomic and expression data will be deposited to the Dryad Digital Repository. All new data will be made available upon the acceptance of this manuscript.