Significance
Identifying causal relationships within a web of human genotype–phenotype correlations is a substantial challenge. Using the largest genetic study to date of comeasured expression and DNA methylation (DNAme) in skeletal muscle, we identify correlations among expression, DNAme, and physiological traits. Leveraging Mendelian randomization (MR) and mediation techniques, we identify 213 putative causal relationships between expression and DNAme. We further use MR to prioritize hundreds of genes and thousands of DNAme sites that may drive genetic associations for diseases and quantitative traits. Our study integrates genetic, diverse -omics, and physiological measurements—a challenge of increasing importance in the field of human genomics.
Keywords: DNA methylation, gene expression, eQTL, mQTL, skeletal muscle
Abstract
We integrate comeasured gene expression and DNA methylation (DNAme) in 265 human skeletal muscle biopsies from the FUSION study with >7 million genetic variants and eight physiological traits: height, waist, weight, waist–hip ratio, body mass index, fasting serum insulin, fasting plasma glucose, and type 2 diabetes. We find hundreds of genes and DNAme sites associated with fasting insulin, waist, and body mass index, as well as thousands of DNAme sites associated with gene expression (eQTM). We find that controlling for heterogeneity in tissue/muscle fiber type reduces the number of physiological trait associations, and that long-range eQTMs (>1 Mb) are reduced when controlling for tissue/muscle fiber type or latent factors. We map genetic regulators (quantitative trait loci; QTLs) of expression (eQTLs) and DNAme (mQTLs). Using Mendelian randomization (MR) and mediation techniques, we leverage these genetic maps to predict 213 causal relationships between expression and DNAme, approximately two-thirds of which predict methylation to causally influence expression. We use MR to integrate FUSION mQTLs, FUSION eQTLs, and GTEx eQTLs for 48 tissues with genetic associations for 534 diseases and quantitative traits. We identify hundreds of genes and thousands of DNAme sites that may drive the reported disease/quantitative trait genetic associations. We identify 300 gene expression MR associations that are present in both FUSION and GTEx skeletal muscle and that show stronger evidence of MR association in skeletal muscle than other tissues, which may partially reflect differences in power across tissues. As one example, we find that increased RXRA muscle expression may decrease lean tissue mass.
Understanding the interplay between genetic inheritance and environmental exposure is critical to developing a full picture of human health and disease. However, this interplay cannot be revealed without a detailed understanding of the molecular events taking place in cells within multiple human tissues. Histone marks, transcription factor binding, and chemical modifications of DNA can actively influence or passively reflect gene expression programs, which are translated into action by proteins that carry out the actual work of the cell through molecularsignaling events. A critical challenge for genomic medicine is to understand how the molecular features within this dynamic landscape not only correlate but causally relate to one another, ultimately driving physiological traits or the development of disease. Such knowledge is crucial, as it can inform efficacious therapies, interventions, and disease diagnostics.
In recent years, common single nucleotide variant (SNV) genome-wide association studies (GWASs) have led to the identification of regions of the genome (and in some instances, specific genes) that influence diverse physiological and molecular traits (reviewed in refs. 1–3). These SNVs can also be remarkably useful statistical instruments for untangling causality, as for the vast majority of loci, the genotype is constant over the lifespan of an individual, regardless of disease or physiological or molecular trait changes. Because of the invariance of SNVs, they can be used as “anchors” to ground causal predictions—the basic insight behind Mendelian randomization (MR) (4, 5).
In this study, we present the largest integrative -omics analysis to date of human skeletal muscle, spanning gene expression, DNA methylation (DNAme), physiological traits, and genotype information (Fig. S1) derived from 318 Finnish participants (265 with all measurements; Table S1). Using skeletal muscle gene expression and DNAme, we identify associations with eight physiological traits and demonstrate that traits with a large number of associated genes also tend to be associated with many DNAme sites. We subsequently use the comeasured molecular traits to identify associations between gene expression and DNAme. By considering the role of tissue/cell composition and muscle fiber type, we document the importance of accounting for tissue/cell type heterogeneity when analyzing molecular trait readouts from bulk tissue. Finally, we map genetic regulators of expression and DNAme, and use these genetic variants to disentangle correlation from causation. Using MR and mediation techniques, we help unravel the complex web of associations between gene expression and DNAme, triangulating on a small number of loci where we predict a causal relationship (i.e., DNAme driving gene expression or vice versa). We then use MR to itemize genes and DNAme sites that may underlie 534 disease/quantitative traits (using “disease/quantitative traits” to distinguish the GWAS-based traits from eight physiological traits measured in our samples). We provide summary statistics from analyses as a publicly available resource (Methods). Collectively, these data represent the largest analysis to date of multiple molecular and physiological traits in skeletal muscle, and illustrate the challenges and potential combined approaches to narrow in on causal relationships.
Results
Molecular Trait Associations with Tissue/Fiber Types and Physiological Traits.
We previously described the signature of type 2 diabetes (T2D), body mass index (BMI), fasting serum insulin, and fasting plasma glucose in the transcriptome of skeletal muscle (6). We build on that study by (i) expanding our set of physiological traits to include waist, weight, waist–hip ratio (WHR), and height; (ii) measuring skeletal muscle DNAme, using the EPIC array on separate pieces of tissue from the same biopsies; (iii) exploring the effects of tissue/cell type heterogeneity on the levels of gene expression and DNAme; and (iv) identifying associations of DNAme with physiological traits. To prioritize sets of results for analysis, we use a false discovery rate (FDR) of ≤1% throughout this work as a pragmatic threshold to identify biologically relevant signals (7).
Within a muscle biopsy, gene expression and DNAme levels vary with the tissue/cell type composition, and therefore have the potential to strongly influence and/or confound conclusions with other molecular traits or phenotypes. To estimate the proportion of different tissue/cell types within our muscle biopsies, we compared gene expression signatures from our biopsies with signatures from four tissue/cell types from the GTEx study (8): skeletal muscle, subcutaneous adipose, whole blood, and Epstein–Barr virus–transformed lymphocytes. All our samples had >87% estimated muscle tissue and up to 13% estimated adipose tissue (Fig. S2A). The estimated proportion of muscle and adipose were positively (Pearson’s r = 0.76) and negatively (r = −0.76) correlated, respectively, with the first principal component of gene expression (Methods), suggesting we are capturing a large portion of tissue variability with our estimates. In addition, for six samples, we repeated the estimation with a second piece of tissue from the same biopsy stock and saw high correlation (r > 0.88) between first and second sample estimates (Fig. S2B). Because the FUSION DNAme data were also obtained from the same biopsy stock and a comprehensive tissue reference panel does not exist for DNAme, we used the gene expression-based tissue type proportions for both gene expression and DNAme analyses.
We found that estimated tissue type proportion was associated with 10,079 (48%) of 20,952 tested genes and 126 (0.0173%) of 727,141 DNAme sites. We also examined the association between eight physiological traits and tissue composition (Table S2A); only fasting serum insulin showed an association with tissue composition (P = 0.0069).
Gene expression and DNAme levels also vary by skeletal muscle fiber type composition. Muscle is composed of slow twitch type 1 fiber (oxidative), fast twitch 2A fiber (intermediate oxidative and glycolytic), and fast twitch type 2X fiber (glycolytic) (9). Each of the three main fiber types expresses a unique myosin heavy chain. As an mRNA-based fiber type proxy, we estimated the proportion of mRNA from each of the three MYH mRNA levels (estimated fiber type proportion; Methods). We observed substantial variability in estimated fiber type proportion across individuals (Fig. S3A), and an association with the third principal component of adjusted expression (rtype1 = 0.59; rtype2 = −0.13; rtype3 = −0.54). We repeated the estimation for the six replicates and saw very strong correlation (r > 0.98) for estimated fiber type proportions (Fig. S3B).
We found both that estimated fiber type proportion was associated with expression for 5,483 (26.2%) genes and DNAme at 13,582 (1.9%) DNAme sites (Dataset S1), and that coefficients of gene expression or DNAme for fiber type 2A were typically intermediate to those for type 1 and type 2X, consistent with the 2A fiber having both oxidative and glycolytic components.
Of the eight tested physiological traits, we found that fiber type was associated with fasting serum insulin, BMI, weight, waist, and WHR (P ≤ 1.4 × 10−4), but not with the other three traits (P > 0.12). Higher levels of these five physiological traits were associated with higher proportions of type 2X fiber type (Table S2B).
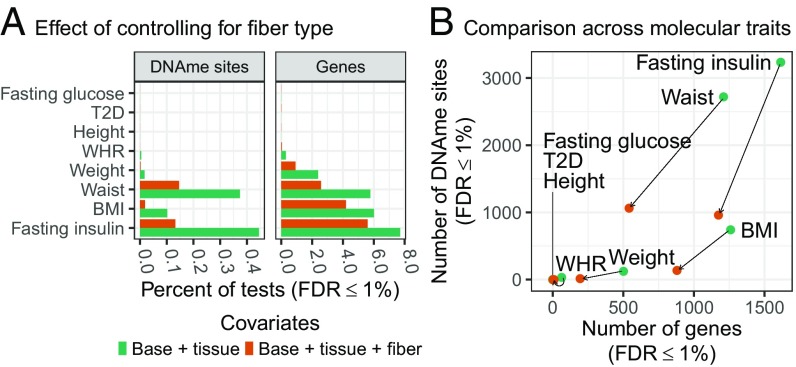
We tested for association of the eight physiological traits with gene expression and DNAme. Using a base set of covariates (technical covariates, smoking status, age, and sex), we found that >5% of genes and >0.1% of tested DNAme sites were associated with fasting serum insulin, BMI, and waist (Fig. S4A). To assess the effects of confounding of association by tissue or fiber type heterogeneity, we ran the physiological trait association analysis controlling for tissue composition, fiber type, or tissue composition and fiber type (Fig. S4A). Tissue composition had little effect on most traits, but increased the number of DNAme sites detected for fasting serum insulin and waist (Fig. S4B). In contrast, controlling for tissue composition and fiber type substantially reduced the number of genes and DNAme sites associated with fasting serum insulin, BMI, waist, and WHR (FDR ≤ 1%) relative to the base model (Fig. S4C) or tissue composition alone (Fig. 1), suggesting that many of the associations were driven by muscle fiber type.
Fig. 1.
Association of FUSION physiological traits with skeletal muscle gene expression and DNAme, controlling for estimated fiber type proportions. Analysis performed with base covariates (sex, age, sample collection site, smoking status, and molecular-trait specific technical covariates), plus tissue type (base+tissue, green bars and points), or base+tissue plus fiber type covariates (base+tissue+fiber, orange bars and points). (A) Percentage of DNAme sites or genes (x axis) associated with each physiological trait (y axis; FDR ≤ 1%). (B) Scatter plot of the number of DNAme sites (y axis) and number of genes (x-axis) associated with each physiological trait adjusting for base+tissue or for base+tissue+fiber covariates (results for a given trait connected with black line).
To assess the overall biological relationship of gene expression with physiological traits, we performed gene ontology term enrichment analysis. We found enrichment for lower expression of cellular respiration genes for T2D (Fig. S5) and for lower expression of proteins targeted to the endoplasmic reticulum for higher fasting serum insulin, BMI, and WHR (Figs. S6–S8), consistent with our previous analysis (6).
Potential Causal Relationships Between Gene Expression and DNAme.
A long-standing scientific challenge is to understand the molecular wiring of tissues/cells across diverse environmental contexts and molecular traits. In contribution to these efforts, we identified (i) DNAme sites whose DNAme level is correlated with gene expression, termed expression quantitative trait methylation (eQTM); (ii) genetic regulators of expression and DNAme; and (iii) potential causal relationships between gene expression and DNAme.
We identified eQTM by testing all DNAme sites at a variety of distances, up to 10 Mb from the transcription start site (TSS) of the target gene (Methods). At distances between 1 and 10 Mb from the TSS, we observed a low but constant discovery rate of eQTM that was attenuated by including tissue and fiber type proportions as covariates (Figs. S13 and S14), suggesting very little signal at distances >1 Mb. For our primary analysis of eQTMs, we used a 1-Mb window and controlled for additional variation, using latent factors learned from the gene expression and DNAme data (Methods), which captured tissue/fiber type, technical, and additional latent variation (Figs. S15 and S16). In total, we identified 37,464 eQTMs (FDR ≤ 1%; 38% positive effect and 62% negative effect) for 7,539 (36%) of 20,953 genes and 27,403 (3.8%) of 727,141 DNAme sites.
Using 7,128,878 autosomal SNVs, we mapped expression quantitative trait loci (eQTLs) and methylation quantitative trait loci (mQTLs), testing all SNVs within 1 Mb of the gene or DNAme site, while controlling for genetic population structure by using genotype principal components and for batch/tissue composition effects by using latent factors (Methods). We identified 10,154 (48%) of 20,953 genes and 149,543 (21%) of 727,141 DNAme sites with at least one QTL (FDR ≤ 1%; Fig. S17).
We used these genetic associations to infer potential causal relationships between gene expression and DNAme at the eQTMs (e.g., DNAme driving changes in gene expression or vice versa) by applying MR and mediation techniques.
MR is a statistical framework that uses a genetic association (“the instrument”) for one trait (“the exposure”) to test for a causal influence on another trait (“the outcome”). An MR result, in which the exposure instrument is associated with the outcome, can arise under four distinct models (10, 11): (i) there is no causal relationship, but a SNV that influences the outcome is in linkage disequilibrium (LD) with a SNV that influences the exposure; (ii) the exposure causally influences the outcome; (iii) the outcome causally influences the exposure (a reverse causal relationship); or (iv) the exposure and outcome are not causally related but share a SNV that influences both the exposure and the outcome independently (horizontal pleiotropy). To distinguish among these models, we use four complementary tests (defined here; Fig. S18): an MR test (consistent with all models), a colocalization test (distinguishing model i from models ii–iv; Note S1), the MR Steiger test (distinguishing model ii from model iii), and the causal inference test (CIT; distinguishing among models ii–iv).
Of the 37,464 eQTMs, 31,578 had an eQTL and/or mQTL (FDR ≤ 1%). For these 31,578 eQTMs, we modeled both expression and DNAme as an exposure, using the most strongly associated SNV for the respective molecular trait to perform an MR test (Methods). We identified 22,843 gene–DNAme site pairs with a putative MR association (FDR ≤ 1%) for which the results could be consistent with any of the four models described. Next, we removed pairs with evidence of being driven by two different SNVs in LD (distinguishing model i from ii–iv) by using the “heterogeneity in dependent instruments” (HEIDI) test (12) to identify 16,122 gene–DNAme site pairs (3,851 genes, 12,787 DNAme sites) with potentially colocalized eQTL and mQTL signals (PHEIDI > 0.05).
Having identified eQTMs with potentially colocalized genetic signals, we sought to both distinguish the direction of causality (model ii from iii; M → E or E → M) and distinguish between a causal and independent model (models ii–iv; Note S2). We used a recently developed MR extension, MR Steiger (13), that predicts the direction of causality by comparing the variance in gene expression explained by the SNV to the variance in DNAme explained by the SNV. We identified 7,952 of the 16,122 gene–DNAme site pairs with a predicted causal direction from the MR Steiger test (FDR ≤ 1%; Methods).
Because MR Steiger cannot distinguish between a causal and independent model, we next used the CIT (14), a mediation-based approach in which a causal chain from SNV to exposure to outcome is predicted using a series of conditional regression tests (Fig. S19). Of the 7,952 pairs, 214 pairs had a predicted causal direction from the CIT (Methods), of which 213 had concordant directions of effect with the MR Steiger prediction.
These 213 gene–DNAme site pairs (Dataset S2) are likely to be a conservative estimate, given we use fairly stringent criteria for identifying causal relationships and because measurement error can lead the CIT to predict independence for truly causal relationships (13). Within our 213 predicted causal relationships (115 genes, 190 DNAme sites), 137 (64%) predict methylation to causally influence expression (M → E) and 76 (36%) predict expression to causally influence methylation (E → M). DNAme sites were closer to the gene TSS for the M → E predictions than for E → M (P = 0.0082; Fig. S20A); however, we did not observe a substantial difference in chromatin state overlaps between M → E and E → M predictions (minimum Bonferroni P = 1; Fig. S20 B and C).
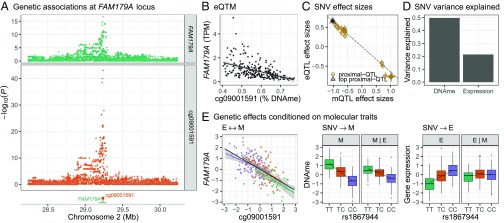
As an example with strong evidence from each causal test, we highlight the predicted M → E effect for cg09001591 DNAme and FAM179A expression. FAM179A expression and cg09001591 DNAme are strongly associated (P = 5.7 × 10−32; Fig. 2B), and they share the same top QTL SNV, rs1867944 (PFAM179A-eQTL = 2.1 × 10−17, Pcg09001591-mQTL = 9.5 × 10−44; Fig. 2A). Both the CIT and MR Steiger test predict a causal methylation to expression (M → E) relationship (Fig. 2 C–E). The cg09001591 DNAme site (chr2:29236578) lies in a skeletal muscle TSS between two skeletal muscle ATAC-seq peaks at the start of the most highly expressed FAM179A exons (Fig. S21). We analyzed the chromatin states of the six SNVs in strong LD with rs1867944 (r2 > 0.8) across a panel of tissues (Methods) and found both rs1867944 and cg09001591 lie in a TSS state unique to skeletal muscle and duodenal mucosa (Fig. S22). This TSS may explain why the strongest FAM179A-rs1867944 associations in GTEx (phs000424.v7.p2) are for skeletal muscle, stomach, and colon-sigmoid (P = 8.2 × 10−9, 3.3 × 10−7, and 3.0 × 10−6, respectively).
Fig. 2.
FAM179A-cg09001591 causal analysis. (A, Top) SNV association with cg09001591 (orange) or FAM179A (green). (A, Bottom facet) cg09001591 DNAme site (orange lollipop) and FAM179A gene body (green line). (B) Scatter plot of cg09001591percentage DNAme (x axis) and FAM179A transcripts per million (TPM; y axis). (C) Scatter plot of mQTL effect sizes with SEs (x axis) by eQTL effect sizes with SEs (y axis) for SNVs used in the HEIDI test. The black dashed line is the estimated MR effect based on the top QTL SNV (black triangle). (D) Percent DNAme and gene expression variance explained (y axis) by rs1867944 genotypes. (E) Scatter plot of residual cg09001591 DNAme (adjusted for PEER factors used in eQTM mapping; x axis) and residual FAM179A gene expression (adjusted for PEER factors; y axis). Linear regression line for eQTM association overall (black) and colored by the rs1867944 genotype (TT, green; TC orange; CC, purple; Left). Box plots and linear regression line (additive model) of residual cg09001591 DNAme by rs1867944 genotype (facet M). Box plot and regression line as for M, except with adjustment of residual cg09001591 DNAme by residual FAM179A gene expression (facet M|E). Box plots and linear regression line (additive model) of residual FAM179A gene expression by rs1867944 genotype (facet E). Box plot and regression line as for E except with adjustment of residual FAM179A gene expression by residual cg09001591 DNAme (facet E|M).
Candidate Links Between Disease/Quantitative Trait Genetic Signals: FUSION Skeletal Muscle Gene Expression and DNAme.
A primary goal motivating molecular trait genetics is to understand the molecular effects of noncoding genetic loci associated with disease. To integrate our genetic maps of molecular traits with genetic maps for disease/quantitative traits, we performed MR for FUSION skeletal muscle gene expression and DNAme, using GWAS summary statistics for 522 disease/quantitative traits from the UK Biobank and GWAS meta-analysis summary statistics for T2D and 11 T2D-related traits (a total of 534 disease/quantitative traits; Table S3 and Methods). Controlling for the number of tests performed across all 534 disease/quantitative traits (Benjamini Hochberg), we found 7,145 preliminary MR associations for gene expression and 79,444 for DNAme (FDR ≤ 1%, PGWAS ≤ 5 × 10−8), spanning 1,059 genes and 13,112 DNAme sites. We performed HEIDI colocalization analysis and identified 2,417 gene expression and 26,718 DNAme associations (560 genes, 6,722 DNAme sites) with potentially colocalized molecular and disease/quantitative trait genetic signals (FDR ≤ 1%, PHEIDI > 0.05, PGWAS ≤ 5 × 10−8), removing many associations that lacked evidence of shared genetic factors, consistent with previous studies (10, 12, 15). We refer to these potentially colocalized results, constituting pairs of disease/quantitative traits and genes or DNAme sites, as MR associations (FDR ≤ 1%, PHEIDI > 0.05, PGWAS ≤ 5 × 10−8). Assuming a low probability of a reverse causation (Note S2), these results are consistent with either a causal or independent (horizontal pleiotropy) model (12).
For gene expression, the disease/quantitative traits with the most MR associations were height (standing or sitting: 140 genes), bioimpedance-derived traits excluding fat mass/percentage traits (43–71 genes), and weight (40 genes; Fig. S23 and Dataset S3). We observed a similar pattern for DNAme MR associations (Fig. S24 and Dataset S4) and a strong correlation between the number of gene expression- and DNAme-based MR associations per trait (r = 0.99; Fig. S25).
We looked for MR associations in which the same SNV had an expression MR association and DNAme MR association for the same disease/quantitative trait, identifying 593 trait–gene–DNAme site sets that spanned 171 unique gene–DNAme site pairs and 89 unique SNVs. Within the 171 gene–DNAme site pairs, 86 (50%) were eQTM (FDR ≤ 1%); for the remaining 85 pairs, the eQTM P value distribution was markedly skewed toward smaller P values (Fig. S26), suggesting many additional eQTM associations that did not reach the FDR ≤ 1% threshold. None of the 86 gene–DNAme site pairs that passed the eQTM FDR threshold were in our set of 213 causal gene–DNAme site predictions (0.006% of the 37,464 considered gene–DNAme pairs that passed the eQTM FDR threshold of 1%); thus, we did not identify instances of predicted causal pathways from gene expression to DNAme to disease/quantitative trait or from DNAme to gene expression to disease/quantitative trait. We note, however, that given the stringent thresholds of our causal predictions, this result does not prove horizontal pleiotropy at these loci.
Candidate Links Between Disease/Quantitative Trait Genetic Signals: FUSION Skeletal Muscle and GTEx Tissue Gene Expression.
We compared the FUSION skeletal muscle gene expression MR associations with those for GTEx skeletal muscle and put them in context of the other 47 GTEx study tissues (8). For each GTEx tissue, we performed MR for the 534 disease/quantitative traits, using eQTL summary statistics from the GTEx study (Methods).
Overall, the number of GTEx MR associations scaled roughly with tissue sample size (Fig. S27); both GTEx (n = 491) and FUSION (n = 301) skeletal muscle had similar numbers of MR associations, at 2,229 and 2,417, respectively. The number of MR associations per disease/quantitative trait was strongly correlated between FUSION and GTEx skeletal muscle (r = 0.98), but also between FUSION skeletal muscle and other GTEx tissues (Fig. S28). The number of MR associations was positively associated with the number of GWAS trait SNVs with P < 5 × 10−8 (Fig. S29), suggesting that for this set of traits, the number of MR associations for each disease/quantitative trait is more strongly influenced by the number of tissue samples and number of trait GWAS signals than the biology underlying a specific tissue and disease/quantitative trait combination. However, within a given trait, we observed a stronger correlation between the FUSION and GTEx skeletal muscle gene-specific MR association strengths (Fig. S30, shown for trunk predicted mass; r = 0.62) than between FUSION and other GTEx tissues (maximum r = 0.45). This observation is consistent with either different genes potentially influencing a disease/quantitative trait in different tissues and/or with different levels of power to detect eQTLs in different tissues.
As an example, for T2D, we saw five MR associations with FUSION skeletal muscle, three for GTEx skeletal muscle, four for subcutaneous adipose, and associations with other tissues (e.g., pancreas, brain; Fig. S31). All these tissues are known to play a role in T2D etiology (16); however, we cannot draw strong conclusions about T2D tissue specificity from these results due to the lack of other important T2D tissues in GTEx (e.g., pancreatic islets), the small number of MR associations, and the small sample sizes for most GTEx tissues.
Of the 2,417 FUSION skeletal muscle gene–disease/quantitative trait MR associations, 921 were also observed in GTEx skeletal muscle (FDR ≤ 1%, PHEIDI > 0.05, PGWAS ≤ 5 × 10−8 with the FUSION and/or GTEx eQTL SNV). For 300 of these pairs, the GTEx muscle MR association was stronger (smaller P value) than that for all other GTEx tissues (Dataset S5). These genes were more highly expressed in muscle (P = 7.1 × 10−6), and showed increased muscle specificity (P = 0.0021; Fig. S32); thus, some of the signals may reflect skeletal muscle-specific biology. However, as more and stronger MR associations are seen in GTEx tissues with larger sample sizes (of which muscle is the largest; Fig. S27), some top skeletal muscle signals are likely due to greater power rather than muscle specificity.
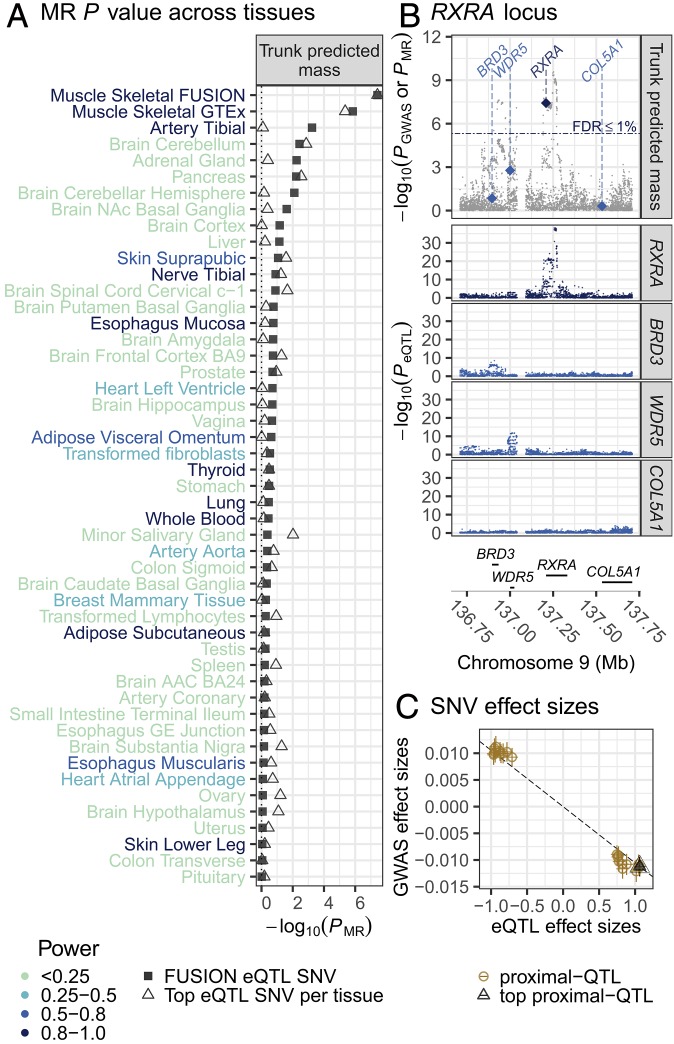
We found strong evidence in FUSION and GTEx skeletal muscle for overlap between genetic regulators of RXRA (retinoic acid receptor RXR-alpha) expression and a locus associated with four body mass/composition related traits derived from bioimpedance measures (Fig. 3 and Fig. S33; muscle expression specificity index = 0.55). The strongest association was for trunk predicted mass (PGWAS = 1.29 × 10−9, PMR-FUSION = 3.78 × 10−8, PMR-GTEx = 4.39 × 10−6), a measurement that approximates trunk lean tissue mass based on bioimpedance, weight, age, and height (17). Using rs6583658 as an instrument, our MR results suggest that increased RXRA expression may decrease lean tissue mass (Fig. 3C and Dataset S5), which is approximated by the four correlated bioimpedance-derived traits: trunk predicted mass, trunk fat-free mass, whole body fat-free mass, and whole body water mass. Analysis of LD identified 31 SNVs in LD with rs6583658 (r2 > 0.8), making the identification of candidate causal SNVs difficult, although several SNVs lie in muscle-specific flanking TSS chromatin states (Fig. S34).
Fig. 3.
MR association of UK Biobank GWAS of trunk predicted mass and RXRA-eQTL results in FUSION skeletal muscle and GTEx tissues. (A) MR association (x axis) for UK Biobank trunk predicted mass with the top RXRA-eQTL SNV from FUSION (rs6583658; square) or the top GTEx tissue-specific RXRA-eQTL SNV (triangle) across FUSION skeletal muscle and GTEx tissues (y axis). Power to detect an RXRA MR association (color of tissue name; Methods). ACC, anterior cingulate cortex; GE, gastroesophageal, NAc, nucleus accumbens basal. (B, Top) UK Biobank trunk predicted mass–SNV association (gray points); MR association P values for RXRA (dark blue diamond) and nearby, protein-coding genes (light blue diamond; diamonds drawn at the TSS of the gene). (Middle) FUSION SNV-gene expression association results for RXRA and other nearby genes. (Bottom) Genes in the region. (C) Scatter plot of FUSION RXRA-eQTL effect sizes and SEs (x axis) and trunk predicted mass GWAS effect sizes and SEs (y axis) for SNVs used in the HEIDI test. The black dashed line is the estimated MR effect based on the top QTL SNV (black triangle).
Muscle may have stronger RXRA MR associations for these traits than other GTEx tissues due to larger sample size. To address this issue, we estimated the power to detect an RXRA MR association for trunk predicted mass in other tissues, assuming the rs6583658 effect size observed in GTEx skeletal muscle was observed in the other GTEx tissues (Methods). GTEx muscle had an estimated 99% power to detect an MR association (FDR ≤ 1%), whereas of the 48 other GTEx tissues, only 8 tissues had >80% power and 35 tissues had <50% power to detect an MR association (Fig. 3A). These findings leave open the possibility that other tissues might have similar evidence for overlap of genetic associations with RXRA expression and trunk predicted mass.
We also investigated whether this RXRA MR association may be driven by height [lean tissue mass is known to be associated with height (18)], even though height is accounted for in the lean tissue mass calculations (17). Compared with lean tissue mass, we found weaker RXRA MR signals for height (standing height: PGWAS = 1.01 × 10−6, PMR-FUSION = 4.7 × 10−6, PMR-GTEx = 3.2 × 10−4; sitting height: PGWAS = 9.46 × 10−8, PMR-FUSION = 7.87 × 10−7, PMR-GTEx = 2.13 × 10−5), suggesting this result is not driven by height.
Discussion
In this study, we integrate skeletal muscle gene expression, DNAme, estimates of tissue and muscle fiber type, physiological traits, SNVs, and external GWAS results for disease/quantitative traits. We use genetics to begin to untangle the complex web of correlations between molecular and physiological traits and identify putative causal relationships.
Within our data, we find that estimated tissue/cell type proportions are associated with a large proportion of genes (26.2–48%) and a small proportion of DNAme sites (0.017–1.9%). Although our estimated tissue/cell-type proportions do not capture the full variety of cell types present in muscle, the low proportion of associated DNAme sites is consistent with previous whole genome bisulfite sequencing studies that report only ∼15–21% of CpG sites showing tissue-specific DNAme patterns (reviewed in ref. 19)—most of which lie in enhancers, a class of regulatory elements poorly captured by the EPIC array (20). Even though proportionally fewer DNAme sites are associated with tissue/cell type heterogeneity, we find that controlling jointly for tissue and muscle fiber type decreases the number of genes and DNAme sites associated with physiological traits and substantially reduces the number of long-range (>1 Mb) associations between gene expression and DNAme. Overall, these results emphasize the importance of tissue/cell type composition as a component of physiological traits and the need for single cell data, either for the study of samples or as a source of cell type signatures for more accurate estimates of tissue composition.
Our study also demonstrates how putative causal predictions can be inferred using genetic associations through multilayered analysis, giving special consideration to the assumptions and biases of the models being used. Many MR approaches between molecular traits and disease/quantitative traits rely on the assumption that a proximal SNV association with a molecular trait is not mediated through the complex trait (Note S2). However, for MR tests of two molecular traits, such as gene expression and DNAme, there is less a priori information about how the SNV might affect the traits; thus, other methods must be used in attempt to infer the direction of causality, if present.
In our analysis, we use two methods (MR Steiger and CIT), leveraging the strengths of each method to identify a modest set of 213 possibly causal relationships. Roughly two-thirds of these gene expression–DNAme pairs have evidence of DNAme driving gene expression, and one third the reverse—highlighting that a model in which DNAme always drives changes in gene expression cannot be assumed. Our findings may represent true causal relationships of DNAme on expression or expression on DNAme, but are in themselves not proof. Of the 7,952 causal predictions from MR Steiger, 7,731 are predicted to be driven by independent genetic effects based on the CIT. This low level of CIT causal predictions is consistent with the findings from other studies (21, 22), as well as with limited power to detect causality due to modest sample sizes and noise (e.g., measurement error) within the data (13). If the many independent predictions are due to biological effects and not statistical issues, such results are consistent with a model in which SNVs influence the local regulatory environment (23), which then influences both gene expression and DNAme.
Finally, we integrate genetic maps for 534 disease/quantitative traits with genetic maps for FUSION skeletal muscle gene expression and DNAme, as well as GTEx gene expression from 48 diverse tissues. We use these data to identify variants that may work through (or be easier to detect in) muscle, highlighting RXRA as an example—although we also show that we have greater power to detect MR associations in muscle than other GTEx tissues.
RXRA belongs to the RXR transcription factor family of nuclear receptors that, in the context of skeletal muscle, has been linked to myoblast differentiation (24–26), insulin sensitivity, and glucose and fatty acid metabolism (reviewed in ref. 27). Although we observe a stronger MR association for skeletal muscle than other tissues, we cannot rule out the possibility of an independent model (horizontal pleiotropy) with action through other tissues, genes, or molecular traits. Nonetheless, our results suggest that increased RXRA muscle expression may contribute to decreased lean tissue mass, perhaps through long-term changes in muscle physiology.
With the increasing accessibility and affordability of molecular measurements on humans for both genetic loci and specific molecular traits (e.g., RNA-seq, DNAme), multilayered datasets will become commonplace across many tissues and diseases. As more comprehensive genome-wide QTL catalogs become available (i.e., distal/trans QTLs) and multi-instrument MR methods mature, it may become possible to better distinguish instances of horizontal pleiotropy (reviewed in ref. 11). MR approaches, when their assumptions can be verified, will help provide a way to cut through the Gordian knot of correlations to better understand the molecular underpinnings of disease.
Materials and Methods
The study was approved by the coordinating ethics committee of the Hospital District of Helsinki and Uusimaa. A written informed consent was obtained from each participant. A detailed description of computational and experimental analyses is provided in Supplementary Materials and Methods. Briefly, we conducted strand-specific mRNA-seq, DNAme assessment using the 850K EPIC chip, and dense array genotyping spanning 318 human skeletal muscle biopsies. We tested for associations of estimated tissue type and fiber type proportions, physiological traits, and proximal SNVs, with gene expression and DNAme. We tested for association of gene expression with DNAme and for evidence of a causal relationship between gene expression and DNAme, using MR, MR Steiger, and the CIT. We tested for MR associations of T2D, 11 T2D-related traits, and 522 traits measured in the UK Biobank (Table S3) with eQTLs from FUSION skeletal muscle and 48 GTEx tissues, and mQTLs from FUSION DNAme. EPIC methylation array blacklist probes and summary statistics of physiological trait associations, eQTMs, eQTLs, mQTLs, and disease/quantitative trait MR associations are publicly available at https://fusion.sph.umich.edu/public/tissue_biopsy/share/2018_muscle.
Supplementary Material
Acknowledgments
We thank Jian Yang for support using the SMR software, Joshua Millstein for support using the CIT R package, and Tingfen Yan for her comments and help. We thank the reviewers for excellent feedback and suggestions. This research was supported in part by National Institutes of Health Grants 1-ZIA-HG000024 (to F.S.C.), U01DK062370 (to M.B. and L.J.S.), and R00DK099240 (to S.C.J.P.); American Diabetes Association Pathway to Stop Diabetes Grant 1-14-INI-07 (to S.C.J.P.); Academy of Finland Grants 271961, 272741 (to M.L.), 258753 (to H.A.K.); European Molecular Biology Laboratory (to E.B.); UK Medical Research Council Grant MC_UU_00011/1 (to G.D.S.); and Wellcome Trust/Royal Society Grant 208806/Z/17/Z (to G.H.).
Footnotes
Conflict of interest statement: D.J.G. and E.B. are members of the Human Induced Pluripotent Stem Cell Initiative and coauthors on a 2017 research article.
Data deposition: Individual-level genotype, RNA-seq, and DNAme data from this study have been deposited to the database of Genotypes and Phenotypes (dbGaP; accession no. phs001048.v2.p1); data are available via the repository’s standard data access request procedures. EPIC methylation array blacklist probes and summary statistics of physiological trait associations, eQTMs, eQTLs, mQTLs, and disease/quantitative trait MR associations are publicly available at https://fusion.sph.umich.edu/public/tissue_biopsy/share/2018_muscle.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814263116/-/DCSupplemental.
References
- 1.Visscher PM, et al. (2017) 10 years of GWAS discovery: Biology, function, and translation. Am J Hum Genet 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaffney DJ. (2013) Global properties and functional complexity of human gene regulatory variation. PLoS Genet 9:e1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rockman MV, Kruglyak L (2006) Genetics of global gene expression. Nat Rev Genet 7:862–872. [DOI] [PubMed] [Google Scholar]
- 4.Davey Smith G, Ebrahim S (2003) ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32:1–22. [DOI] [PubMed] [Google Scholar]
- 5.Davey Smith G, Hemani G (2014) Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23:R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott LJ, et al. (2016) The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat Commun 7:11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasserstein RL, Lazar NA (2016) The ASA’s statement on p-values: Context, process, and purpose. Am Stat 70:129–133. [Google Scholar]
- 8.GTEx Consortium (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213, and erratum (2018) 553:530. [DOI] [PubMed] [Google Scholar]
- 9.Schiaffino S, Reggiani C (2011) Fiber types in mammalian skeletal muscles. Physiol Rev 91:1447–1531. [DOI] [PubMed] [Google Scholar]
- 10.Richardson TG, et al. (2017) Mendelian randomization analysis identifies CpG sites as putative mediators for genetic influences on cardiovascular disease risk. Am J Hum Genet 101:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemani G, Bowden J, Davey Smith G (2018) Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 27:R195–R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48:481–487. [DOI] [PubMed] [Google Scholar]
- 13.Hemani G, Tilling K, Davey Smith G (2017) Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millstein J, Zhang B, Zhu J, Schadt EE (2009) Disentangling molecular relationships with a causal inference test. BMC Genet 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannon E, et al. (2016) Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 19:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn SE, Hull RL, Utzschneider KM (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444:840–846. [DOI] [PubMed] [Google Scholar]
- 17.Tanita, Body composition analyzer BC-418 instruction manual Available at https://www.tanita.com/en/bc-418/. Accessed August 1, 2018.
- 18.Heymsfield SB, Heo M, Thomas D, Pietrobelli A (2011) Scaling of body composition to height: Relevance to height-normalized indexes. Am J Clin Nutr 93:736–740. [DOI] [PubMed] [Google Scholar]
- 19.Luo C, Hajkova P, Ecker JR (2018) Dynamic DNA methylation: In the right place at the right time. Science 361:1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pidsley R, et al. (2016) Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez-Arcelus M, et al. (2013) Passive and active DNA methylation and the interplay with genetic variation in gene regulation. elife 2:e00523, and erratum (2013) 2:e01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng B, et al. (2017) An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci 20:1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaneau O, et al. (2017) Intra- and inter-chromosomal chromatin interactions mediate genetic effects on regulatory networks. bioRxiv:10.1101/171694.
- 24.Le May M, et al. (2011) Contribution of retinoid X receptor signaling to the specification of skeletal muscle lineage. J Biol Chem 286:26806–26812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.AlSudais H, et al. (2016) Retinoid X receptor-selective signaling in the regulation of Akt/protein Kinase B isoform-specific expression. J Biol Chem 291:3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamed M, et al. (2017) Insights into interplay between rexinoid signaling and myogenic regulatory factor-associated chromatin state in myogenic differentiation. Nucleic Acids Res 45:11236–11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szanto A, et al. (2004) Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ 11(Suppl 2):S126–S143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.