Abstract
Purpose:
The perturbation of metabolic pathways in high-grade BLCA has not been investigated. We aimed to identify a metabolic signature in high-grade BLCA by integrating unbiased metabolomics, lipidomics, and transcriptomics to predict patient survival and to discover novel therapeutic targets.
Experimental design:
We performed high-resolution liquid chromatography mass spectrometry (LC-MS) and bioinformatic analysis to determine the global metabolome and lipidome in high-grade BLCA. We further investigated the effects of impaired metabolic pathways using in vitro and in vivo models.
Results:
We identified 519 differential metabolites and 19 lipids that were differentially expressed between low-grade and high-grade BLCA using the NIST MS metabolomics compendium and lipidblast MS/MS libraries, respectively. Pathway analysis revealed a unique set of biochemical pathways that are highly deregulated in high-grade BLCA. Integromics analysis identified a molecular gene signature associated with poor patient survival in BLCA. Low expression of CPT1B in high-grade tumors was associated with low FAO and low acyl carnitine levels in high-grade BLCA which were confirmed using tissue microarrays. Ectopic expression of the CPT1B in high-grade BLCA cells led to reduced EMT in in vitro, and reduced cell proliferation, EMT, and metastasis in vivo.
Conclusions:
Our study demonstrates a novel approach for the integration of metabolomics, lipidomics, and transcriptomics data, and identifies a common gene signature associated with poor survival in BLCA patients. Our data also suggest that impairment of FAO due to down-regulation of CPT1B plays an important role in the progression towards high-grade BLCA and provide potential targets for therapeutic intervention.
Keywords: Multi-omics integration analysis, Metabolomics and Lipidomics, CPT1B, Fatty acid oxidation
Introduction
Bladder cancer (BLCA) causes significant morbidity and mortality worldwide. In the United States, there were about 79,000 cases and 17,000 deaths due to BLCA in 20171. In the past few decades advances in cancer genomics, transcriptomics, proteomics, and metabolomics resulted in the discovery of potential biomarkers for cancer2,3. Unfortunately, most of the biomarkers have failed to demonstrate superior performance characteristics compared with existing clinical tests. Recent advancements in OMICs technologies have ushered in a new era of targeted cancer therapies by improving our understanding of molecular carcinogenesis. OMICs studies have identified therapeutic targets and resulted in the successful development of new drugs to treat various solid tumors like breast cancer and lung cancer4,5. For BLCA, prognostication and treatment still depend mainly on pathological and clinical characteristics6. Patients diagnosed with high-grade invasive disease are difficult to treat effectively and have a relatively low life-expectancy despite available multimodal therapies. Therefore, novel prognostic and therapeutic targets against BLCA are needed.
Tumor progression relies on the reprogramming of cellular metabolism7. Our previous metabolomic and lipidomic studies highlighted the significance of altered xenobiotic and fatty acid metabolism in BLCA development8–10. However, we still know little about the altered metabolic pathways during the transition from low grade to high grade BLCA. The integration of OMICS facilitates the study of interactions among all classes of biomolecules that can occur in cell, which in turn, determines the cellular physiology and behavior. Treatment for high-grade muscle invasive bladder cancer (MIBC) has not advanced beyond cisplatin-based combination chemotherapy in the last three decades and very few drugs for the disease have been approved recently11. The identification of markers to predict poor outcomes among patients with BLCA will improve our ability to identify patients who might benefit from adjuvant therapy. Furthermore, a greater understanding of the metabolic molecular mechanisms of BLCA progression and the identification of novel therapeutic targets will improve outcomes for patients with high-grade BLCA.
We used a robust mass spectrometry (MS) platform to conduct global unbiased metabolomic and lipidomic analyses to identify critical alterations that may contribute to BLCA progression. We mapped the altered metabolites and lipids to corresponding genes using the Human Metabolome Database (HMDB) and then a pathway analysis to discover the ways in which metabolic pathways are perturbed in high-grade BLCA. We integrated the relevant genes with publicly available transcriptomic data and generated an integrated gene signature to predict the survival of patients with BLCA. Furthermore, we found that carnitine palmitoyl transferase 1B (CPT1B) is significantly downregulated in high-grade BLCA, which in turn leads to low fatty acid β-oxidation (FAO). Ectopic expression of CPT1B in high grade BLCA cells increased FAO which in turn reduced epithelial mesenchymal transition (EMT). Finally, overexpression of CPT1B in high grade BLCA cells reduced cell proliferation and liver metastasis in a chick chorioallantoic membrane (CAM) in vivo model. Our results clearly indicate that CPT1B plays an important role in BLCA tumor progression by affecting fatty acid metabolism. CPT1B might therefore have prognostic value and provide a target for therapeutic intervention in high-grade BLCA.
Materials and Methods
Reagents and internal standards
High-performance liquid chromatography (HPLC)-grade ammonium acetate, acetonitrile, methanol, chloroform and water were procured from Burdick & Jackson (Morristown, NJ). MS grade formic acid, standards and internal standards, N-acetyl aspartic acid-d3, tryptophan-15N2, sarcosine-d3, glutamic acid-d5, thymine-d4, gibberellic acid, trans-zeatin, jasmonic acid, anthranilic acid-15N, and testosterone-d3, were purchased from Sigma-Aldrich (St. Louis, MO). Mixture of LPC 17:0/0:0, PG 17:0/17:0, PE 17:0/17:0, PC 17:0/17:0, TAG 17:0/17:0/17:0, SM 18:1/17:0, MAG 17:0, DAG 16:0/18:1, CE 17:0, ceramide 18:1/17:0, PA 17:0, PI 17:0/20:4, and PS 17:0/17:0. were obtained from Avanti polar lipids (USA) (Supplementary Table S1).
Cell lines
RT4, T24, UMUC3, J82, and TCCSUP were procured from American Type Culture Collection (ATCC) and maintained as per ATCC instructions. All cell lines were verified using short-tandem repeat (STR) DNA fingerprinting at the MD Anderson Cancer Center and were tested for mycoplasma contamination every three months. All the BLCA cell lines were procured from the ATCC less than two years before starting the experiments.
Sample preparation for metabolomics and lipidomics
All the BLCA specimens were procured by a prior written informed consent under institute review board (IRB) approved protocols. Metabolites were extracted from BLCA tissues, and mouse liver pool was used as a quality control and followed the extraction procedure described8,12–14. Briefly, 10mg of tissue was used for the metabolic extraction. The extraction step starts with addition of 750μL ice-cold methanol: water (4:1) containing 20μL spiked internal standards (ISTDs). After homogenization, ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 4:3:2 methanol:chloroform:water. The organic and aqueous layers were collected, dried, and resuspended in methanol: water (1:1). The extract was deproteinized using a 3kDa molecular filter and the filtrate was dried under vacuum. The dried extracts were re-suspended in 100μL of injection solvent composed of 1:1 methanol: water and subjected to LC-MS.
Lipids were extracted using a modified Bligh-Dyer method15. The extraction was carried out using 2:2:2 ratio of water: methanol: dichloromethane at room temparature after ISTDs into tissues and quality control pool12. After homogenization of the samples, the organic layer was collected and completely dried under vacuum. Before MS analysis, the dried extract was resuspended in 100μL of buffer containing 10mM NH4Ac and subjected to LC/MS. The lipidome was separated using reverse-phase (RP) chromatography. To monitor the lipid extraction process, we used a standard pool of tissue samples from aliquots of the same samples.
Data acquisition for metabolomics and lipidomics analysis
For metabolomics, 5μL of the metabolite extract was injected into a 3.5μm particle 4.6×150mm X bridge amide column which heats to 60°C. 0.1% formic acid in water as solvent-A and acetonitrile as solvent-B in positive ionization and 20mM ammonium acetate in 95% water; 5% acetonitrile and 100% acetonitrile as solvent-B in negative ionization mode. For lipidomics, 10μL of the lipid extract was injected into a 1.8μm particle 50 × 2.1mm column (Acquity HSS UPLC T3) which heats to 55°C. Acetonitrile: water (40:60, v/v) with 10mM ammonium acetate was solvent-A and acetonitrile: water: isopropanol (10:5:85 v/v) with 10mM ammonium acetate was solvent-B. Chromatographic elution used a linear gradient over a 20min total runtime, with 60% solvent-A and 40% solvent-B gradient in the first 10 minutes, Then the gradient was ramped in a linear fashion to 100% solvent-B which was maintained for 7 minutes. After that, the system was switched back to 60% solvent-B and 40% solvent-A for 3 minutes. The flow rate used for these experiments was 0.4mL/min. The data acquisition of each sample was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a Turbo VTM ion source. The instrument performed one TOF MS survey scan (150ms) and 15 MS/MS scans with a total duty cycle time of 2.4s. The mass range in both modes was 50–1200 m/z. We controlled the acquisition in both MS and MS/MS spectra by data-dependent acquisition function of the Analyst TF software (AB Sciex, Concord, Canada). Rolling collision energy spread was set whereby the software calculated the collision energy value to be applied as a function of m/z. Mass accuracy was maintained by the use of an automated calibrant delivery system interfaced to the second inlet of the Duo Spray source.
Metabolomics and lipidomics data Processing
The raw data (wiff) of metabolomics and lipidomics from ABsciex is converted to “mgf” data format using proteoWizard software16. We used the NIST MS Pep to search the converted files against NIST14 libraries17,18 for metabolomics and LipidBlast libraries for lipidomics. The m/z width was determined by the mass accuracy of ISTDs and was set 0.001 for positive mode and 0.005 for a negative mode with an overall mass error of less than 2 parts per million. The minimum match factor used was set to 400 score. The MS/MS identification results from all the files were combined using an in-house software tool to create a library for quantification. All raw data files were searched against this library of identified metabolites with mass and retention time using Multiquant 1.1.0.26 (AB Sciex, USA). We used MS/MS data as an intermediary step to help with identification, but quantification was done using MS1 data only. Relative abundance of peak spectra was used for the analyses. The metabolites identified in both positive and negative ion modes were initially analyzed separately for their relationship with the outcome to ensure persistent results. Identified metabolites were quantified by normalizing against their respective ISTDs. Quality control samples were used to monitor the overall quality of the metabolite extraction and MS analysis.
Statistical analysis for unbiased metabolomics and Lipidomics
After data acquisition, the missing values for metabolites/lipids were imputed using the k-nearest neighbors method. Then the data were log2 transformed followed by normalization using the median interquantile range normalization. The compound-by-compound t-test was applied, followed by the Benjamin-Hochberg procedure for FDR correction accounting for multiple comparisons. Significance was achieved for FDR<0.25.
Quantification of overall β-oxidation activity by using the Biocrates AbsoluteIDQ kit p180
Absolute concentrations of metabolites were quantified by AbsoluteIDQ p180 kit (Biocrates Life Science AG, Austria) using QTRAP 6500 LC/MS/MS system (AB Sciex, USA) equipped with an electrospray ionization (ESI) source, an Agilent G1367B autosampler and the Analyst 1.51 software (AB Sciex, USA). 10μL of the homogenized tissue sample, quality control samples, blank, and calibration standards were added to the appropriate wells. The plate was dried and the samples were derivatized with phenylisothiocyanate and dried again. The samples were eluted with 5mM ammonium acetate in methanol and then were diluted with running solvent provided in kit for flow injection analysis. 20μL of the sample was directly injected into the MS at a flow of 30μL/min. Concentrations were calculated and evaluated in the Analyst/MetIQ software by comparing measured analytes in a defined extracted ion count section to those of specific labeled internal standards or non-labeled, non-physiological standards (semi-quantitative) provided by Biocrates. Measurement of overall β-oxidation activity was derived by calculating the ratio of short-chain acylcarnitines to free carnitines.
Pathway analysis
Enriched pathways were determined by using the hypergeometric distribution against the Kyoto encyclopedia of genes and genomes (KEGG) pathway database, as compiled by the MSigDB gene set compendium; significance was achieved for p<0.05. Pathway networks were visualized using the Cytoscape scientific visualization platform.
Integration of unbiased metabolomics, unbiased lipidomics, and transcriptomics
Metabolites and lipids were mapped into associated genes/proteins using the HMDB ID. Next, we obtained the transcriptome fingerprint between high and low grade BLCA using the Kim cohort as described below. We generated an integrated metabolomics/lipidomics/transcriptomics gene signature, by intersecting the all three gene sets.
Gene expression analysis
To identify the gene signature of high vs low grade BLCA, we downloaded the publicly available data from the Kim cohort. Gene significance was assessed by using the student’s t-test; significance was achieved for p<0.05 and fold change either above 1.25 or below 0.8. To identify genes associated with low vs high CPT1B gene expression, we used the gene expression data from five independent public cohorts: The cancer genomic atlas (TCGA), Kim (GSE13507), Sjodahl (GSE32894), Lindgren (GSE32548), and Riester (GSE31684). For each cohort, we first stratified the patients by CPT1B, into the top 50% and bottom 50%. Next, we generated a rank file for each expressed gene by the log2 fold change of low CPT1B samples over high CPT1B samples. We ran Gene Set Enrichment Analysis (GSEA) against the HALLMARK pathway collection, as compiled by Molecular Signature Database (MSigDB). Significance was achieved for false discovery rate (FDR)-adjusted Q-value<0.25.
Analysis of prognostic power of the integrated gene signature in BLCA
Significant association with survival of an integrated BLCA metabolomics/lipidomics/transcriptomic gene signature was evaluated in four gene expression datasets of BLCA cohorts from TCGA, Kim, Sjodahl, and Lindgren for which clinical outcomes have been reported. For each gene in the signature and for each specimen, we computed the z-score for its expression within the cohort, as described previously9 and computed the sum z-score for each specimen. Specifically, the z-scores of genes repressed in the integrated signature were subtracted from the z-scores of genes induced in the integrated signature, resulting in a corresponding signature activity score for each specimen. In each cohort, the specimens were ranked according to their integrated signature activity score, and association with survival was evaluated via the log-rank test between the bottom 50% of the patients and the top 50% of the patients. Survival significance was assessed by employing the package survival in the R statistical system.
Univariate and multivariate survival analysis
Survival analysis was performed using the survival library in the R statistical system. Univariate analysis used 27 gene signature or the CPT1B with survival, after stratifying patients in top and bottom 50%. For multivariate models, we tested the gene signature and CPT1B, while controlling for other demographic and clinical factors, specifically age, sex, and tumor stage. Hazard ratios (HRs) derived from Cox proportional hazards models are expressed with 95% confidence intervals (CIs) and p-values. For all analyses and each variable, significant association with survival was achieved for p-value <0.05.
Chorio allantoic Membrane (CAM) Assay
The CAM was assessed as previously described19 on embryonic day seven, the eggs were inoculated with 5×10^5 UMUC3 vector control and CPT1B over expressing cells per egg. In vivo luminescence imaging was conducted on embryonic days 10, 13, and 15 (3, 5, and 7 days after seeding the tumor cells) by using in vivo imaging system (IVIS). Exposure time was 1 second, 5 minutes after addition of 100μL of 15mg/ml D-luciferin, and region of interest was quantitated by the Lumina software. Seventh day after inoculation, the eggs were euthanized as per the AVMA guidelines. The CAM tumors were fixed in 10% formalin and embedded in paraffin for immunohistochemistry (IHC). IHC was performed for CPT1B, E-cad, N-cad, and vimentin to check the expression levels. IHC stained slides were scanned and analyzed by Aperio CS2 system (Leica Biosystems) and number of positive cells/total cells were plotted.
To analyze metastasis of the tumor into the chick embryo, we harvested embryo visceral organs (i.e. lung, liver, bone, and brain) on day 15–18 post fertilization. To achieve this, the eggshell opening was widened to allow excision of the CAM tumor. After the tumor excision, the embryo was humanely euthanized by decapitation with a scissors and the body incised to dissect visceral organs including lung, liver, bone, and brain. Once dissected, the tissues were collected, flash-frozen, and stored at −80°C until further analysis. Quantitative assessment of tumor metastasis into visceral organs was achieved by quantification of human DNA from the metastasized cells in the chicken DNA by performing RT-PCR for human-specific Alu sequence.
To quantify the amount of human DNA from tumor cells that had metastasized to the chick embryo tissues, a standard curve was generated by amplification of genomic DNA isolated from a serial dilution of BLCA cells mixed with individual chick visceral tissue DNA (modified from published methods20,21). To detect the human Alu sequences in the chicken DNA background, human Alu-detecting primers and a probe were used (Supplementary Table S2). The standard curve was generated using serial dilutions (0.1ng-1μg) of UMUC3 DNA mixed with constant amount of chicken DNA. The RT-PCR was performed in triplicate for each standard as well as the experimental samples and analyzed the DNA content in control and CPT1B overexpression groups.
Results
Identification of altered metabolites and lipids in low-grade and high-grade BLCA
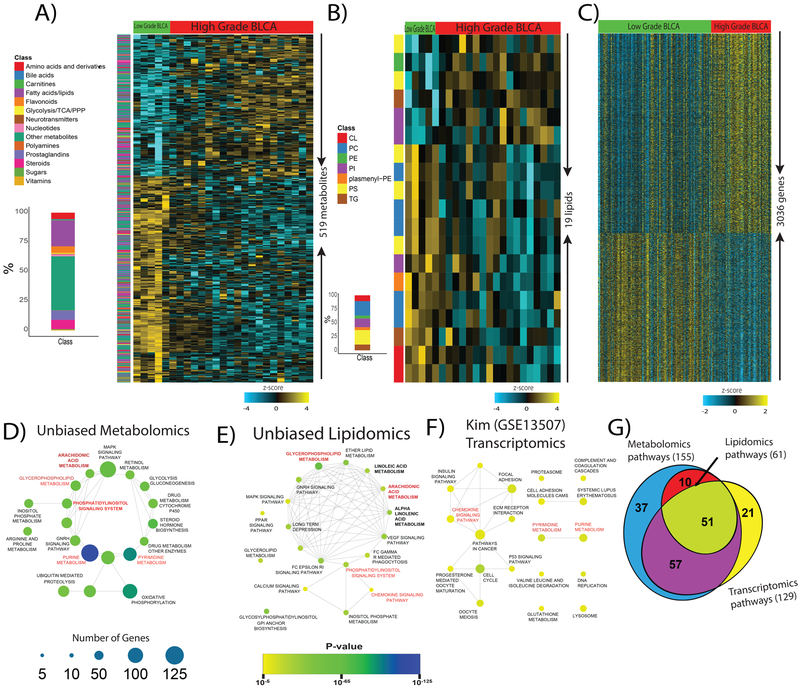
We characterized the global metabolome and lipidome of low-grade (n=5) and high-grade (n=20) BLCA tissues using high-resolution LC-MS and clinical information for the patients is shown in Supplementary Table S3. A cocktail of 10 internal standards along with a repetitive analysis of pooled liver extracts served as controls to ascertain the reproducibility and robustness of the profiling platform for metabolomics (Supplementary Figure S1A, S2A) and lipidomics (Supplementary Figure S1B, S2B). An extensive NIST database and lipid blast search analysis of the LC-MS data identified a total of ~2000 metabolites and 786 lipids in positive and negative ionization modes (Supplementary Figure S3). An overview of the study is shown in Figure 1. A total of 519 metabolites (14 different classes) were significantly altered (FDR<0.25) between low-grade and high-grade BLCA (Figure 2A and Supplementary Table S4). These include significantly altered levels of metabolites in multiple pathways (Figure 2D). Likewise, a total of 19 lipids (7 different classes) were significantly altered (FDR<0.25) (Figure 2B and Supplementary Table S5), that were involved in many pathways (Figure 2E). Using well annotated, publicly available Kim transcriptomics data we found 3036 genes that had significantly altered (p<0.05) between low-grade and high-grade BLCA (Figure 2C). Like the altered lipids and metabolites the altered genes were involved in multiple pathways (Figure 2F) and 51 common pathways were implicated in all three platforms (Figure 2G).
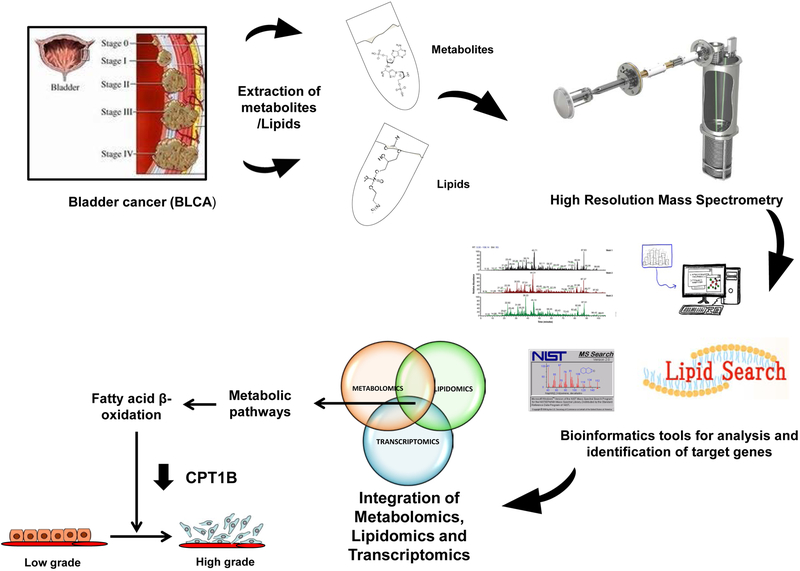
Figure 1. Overview of the strategy used to profile the global metabolome and lipidome and integrated them with the transcriptome to characterize low-grade and high-grade BLCA.
Figure 2. Identification of altered metabolites, lipids, and transcripts and their respective pathways in high-grade BLCA.
A) Heat map of hierarchical clustering of 519 differentially expressed metabolites from different classes of biomolecules detected across low-grade and high-grade BLCA samples (FDR<0.25). Each class of metabolites is represented in a different color. B) Heat map of hierarchical clustering of 19 differentially expressed lipids from various classes detected across low-grade and high-grade BLCA samples (FDR<0.25). Each class of lipids is represented in a different color. C) Heat map of hierarchical clustering of 3036 differentially expressed genes from the Kim dataset (GSE13507) across low-grade and high-grade BLCA samples (p<0.05). Columns represent individual tissue samples; rows refer to distinct metabolites, lipids, and genes. Shades of yellow indicate different levels of increase in expression; shades of blue indicate different levels of decrease in expression relative to the median levels. D, E, and F) Pathway enrichment analysis of differentially expressed metabolites, lipids, and genes shows the significantly altered pathways in high-grade BLCA (p<0.05). The node size indicates the number of genes involved in the pathway (deregulated pathways common to the metabolomics, lipidomics, and transcriptomics data sets are highlighted in red). G) Venn diagram of the deregulated metabolic pathways common to the metabolomics, lipidomics, and transcriptomics data sets.
Integration of metabolomics, lipidomics, and transcriptomics data identified a gene signature with prognostic value in BLCA
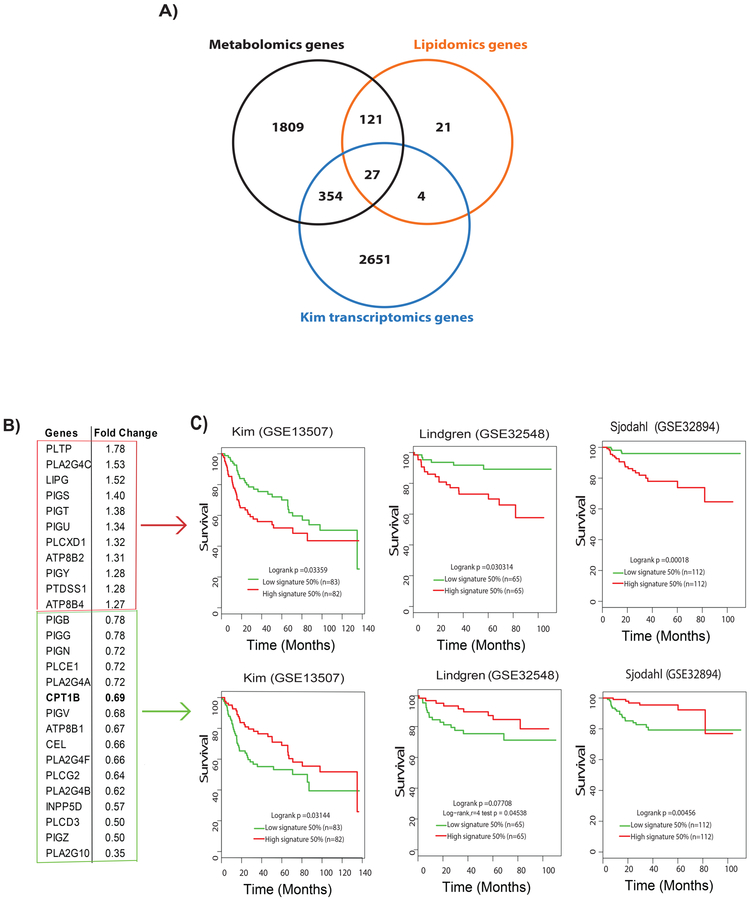
We mapped the panels of altered metabolites, to their corresponding genes using HMDB10,22, resulting in the identification of 2311 genes. A similar analysis of altered lipids led to the identification of 173 genes. Analysis of transcriptomics public data from the Kim cohort resulted in the identification of 3036 genes that were differentially expressed between low-grade and high-grade BLCA. Integration of these three platforms led to 27 intersecting genes between low-grade and high-grade BLCA (Figure 3A).
Figure 3. Metabolomic-lipidomic–transcriptomic integration strategies to identify potential prognostic gene signature for BLCA.
A) The integration of a significant genes derived from metabolomics, lipidomics, and transcriptomics data comparing low-grade and high-grade BLCA resulted in a set of 27 common genes. B) Eleven upregulated and 16 down-regulated genes in high-grade BLCA in the Kim cohert (GSE13507) with their fold change. C) Survival analysis shows poor survival associated with the upregulated gene signature (Kim, log rank p =0.03359;, Lindgren (GSE32584), log-rank p=0.030314;, Sjodahl (GSE32894), log-rank p=0.00018) and downregulated gene signature (Kim, log-rank p=0.03144;, Lindgren, log-rank p=0.07708; and Sjodahl, log-rank p=0.00456.
We next examined the expression of these 27 genes using Kim transcriptomics data. Eleven of these genes were upregulated while the other 16 were downregulated in BLCA (Figure 3B). We examined the association between the 27 genes and survival using publicly available BLCA cohorts (Kim23, Lindgren24, and Sjodahl25). We used the log-rank test to determine patient outcomes. In all three cohorts, high expression levels of the 11 genes that were upregulated in BLCA was associated with poor survival. Similarly, low expression levels of the genes that were downregulated in BLCA was associated with poor survival in all three cohorts (Figure 3C). Interestingly, 11 upregulated gene signature that were upregulated in BLCA in the Kim cohort were associated with poor survival in Lindgren and TCGA BLCA cohorts (Supplementary Figure S4).
We analyzed univariate and multivariate Cox proportional hazard regression models using the Kim, Lindgren, Sjodahl, TCGA data sets to assess the associations between the 27 gene signature and the time-to-death. Using multiivariate models, we tested the 27 gene signature (top 50% vs. bottom 50%) (Supplementary Table S6A–D) while controlling for sex, age, and tumor stage. Univariate Cox proportional hazard regression analysis showed that the gene signature was significantly associated with risk of death, with a hazard ratio (HR) above 1 in the Kim, Lindgren, Sjodahl and TCGA cohorts. Despite that, a multivariate analysis showed that although the HR was greater than 1 in the Kim, Lindgren and TCGA, the 27 gene signature was not associated with survival independently of the other clinical variables considered. The tumor stage was associated with survival in all four cohorts, while age was associated with survival in Kim, Lindgren and TCGA cohorts (Supplementary Table S6A–D).
To obtain additional insights into the 27 genes, we performed survival analysis based on the individual genes in the same publicly available cohorts. We found that six upregulated genes (PLA2G4C LIPG, PIGS, PIGU, ATP8B2 and ATP8B4) had a significant clinical association with survival (Supplementary Figure S5). High expression of PLA2G4C and ATP8B2 high expression was associated with poor survival in multiple cohorts (Kim, Lindgren, Sjodahl, and TCGA26) (Supplementary Figure S5). Eight of the downregulated genes (CPT1B, PIGB, PLA2G4A, PIGV, INPP5D, PLCD3, PIGZ, and PLA2G10) were associated with poor survival (Supplementary Figure S6).
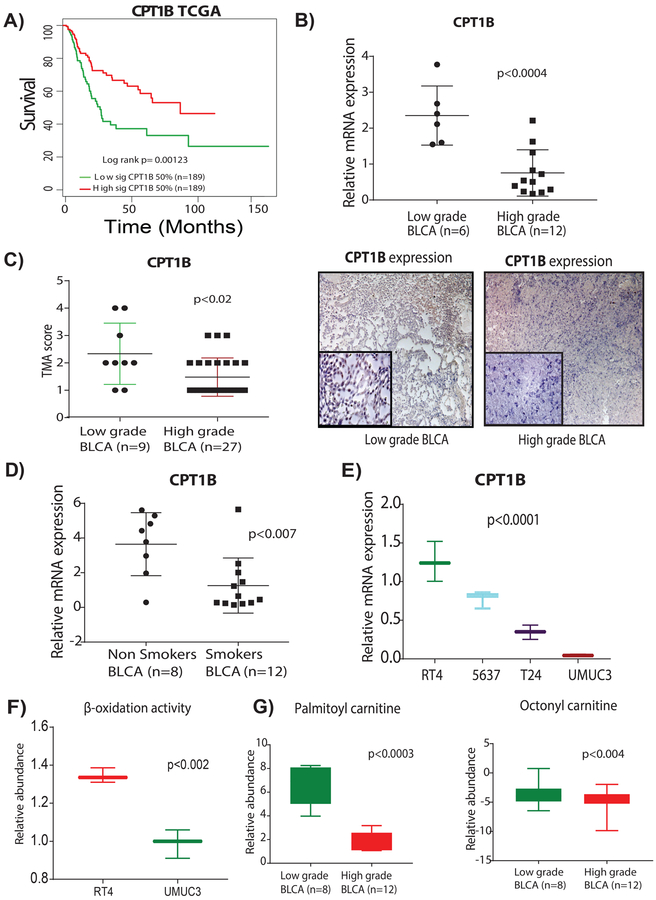
Remarkably, low expression of CPT1B was associated with poor survival in TCGA, (Figure 4A) Lindgren, and Sjodahl cohorts (Supplementary Figure S6), whereas in the Kim cohort there was no association between CPT1B and survival. However, the expression of CPT1B was significantly lower in high grade BLCA tumors in all three cohorts (Supplementary Figure S7). We further tested the univariate and multivariate models for CPT1B (top 50% vs. bottom 50%) by considering for sex, age, and tumor stage using Kim, Lindgren, Sjodahl, and TCGA data sets. Univariate Cox proportional hazard regression analysis showed that CPT1B had an HR below 1 in TCGA, Lindgren, and Sjodahl cohorts and was significantly associated with survival only in the TCGA cohort (Supplementary Table S7A–D). In a multivariate Cox proportional hazard regression model adjusted for sex, age and tumor stage showed that CPT1B had an HR below 1 and was significantly associated with survival in the TCGA cohort (Supplementary Table S7A–D). Pathway analysis also indicated that fatty acid metabolism was significantly increased in high-grade BLCA. CPT1B is involved in fatty acid transport and acts as a rate-limiting enzyme for FAO, thereby altering the fate of the cancer cells27. Recent studies showed that cancer metastatic potential is a consequence of an alteration in lipid metabolic pathways28–30, which led to our hypothesis that CPT1B plays a crucial role in BLCA progression.
Figure 4. Suppression of CPT1B is associated with poor survival, low β-oxidation, and low levels of acylcarnitines in high-grade BLCA.
A) Low expression of CPT1B was associated with poor survival in TCGA cohort (log-rank p= 0.00123). B) Low CPT1B mRNA expression demonstrates the low CPT1B expression in high-grade BLCA (n=6 and n=12 low-grade and high-grades tissues respectively; p<0.0004. C) TMA analysis of BLCA patient tissues (n=9 and n=27, low-grade and high-grade respectively) shows a trend for lower intensity in the high-grade tissues (p<0.02). Immunohistochemistry (IHC) analysis shows the low CPT1B protein levels in high-grade BLCA; on a scoring scale of 1–4, one is low intensity and 4 is the highest intensity of the CPT1B expression. D) mRNA expression analysis shows the low CPT1B expression in smokers with BLCA (n=8 and n=12 non-smokers and smokers respectively; p<0.007). E) mRNA expression analysis shows a gradual decrease of CPT1B levels with an increased grades of the BLCA in cell lines (p<0.0001). For all the mRNA measurements CPT1B levels were normalized to a GAPDH internal control. F) LC-MS based analysis using Biocrates AbsoluteIDQ p180 Kit shows the low β-oxidation activity in high-grade BLCA cell lines (p<0.02). G) LC-MS based analysis reveals low levels of palmitoyl (p<0.0003) and octanoyl (p<0.004) carnitines in patients with high-grade BLCA.
Suppression of CPT1B impairs the FAO in high-grade BLCA
To better understand the role of CPT1B in high-grade BLCA, we examined the expression of CPT1B in low-grade and high-grade BLCA. CPT1B expression was significantly lower in high-grade BLCA than in low-grade BLCA (Figure 4B) in an independent validation cohort (Supplementary Table S8). Histochemical analysis of a tissue micro array (TMA) (Figure 4C) confirmed the findings of the transcriptomic cohorts analyses (Supplementary Figure S7). It is known smoking is associated with high-grade BLCA31. In comparison with non-smokers, smokers with BLCA had reduced expression of CPT1B (Figure 4D). Likewise, the levels of CPT1B were also reduced in high-grade compared to low-grade BLCA cell lnes (Figure 4E). Consistent with the CPT1B expression, high-grade BLCA cell lines displayed low FAO activity compared to low-grade BLCA cell lines (Figure 4F). We measured the levels of acylcarnitines, which are indicators of FAO, and found that the levels of palmitoyl and octanoyl carnitines were significantly lower in high-grade BLCA patients (Figure 4G). Taken together, our results demonstrated the co-occurrence of CPT1B downregulation and low FAO levels in high-grade BLCA.
Hallmark cancer pathways are associated with low CPT1B expression
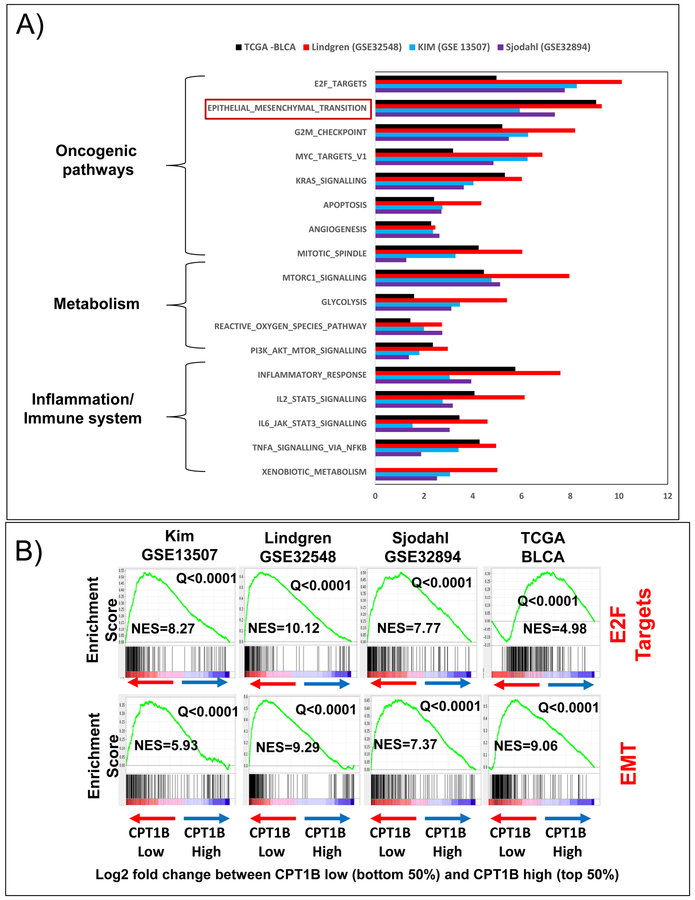
To obtain additional insights into the effects of CPT1B on metabolism in high-grade BLCA, an in-silico analysis was performed on data divided into CPT1B high vs CPT1B low signature using public gene expression data sets. For each transcriptomics cohort that profiled the CPT1B gene (TCGA, Kim, Sjodahl, and Lindgren), we stratified the patients based on CPT1B gene expression. Next, we divided the patients based on the median CPT1B value into top 50% and bottom 50%. We then computed a gene signature using a parametric t-test and a minimum difference in expression of 1.25 fold between the patients in the top 50% of CPT1B expression and bottom 50% of CPT1B expression. For each cohort, the signature consisted of the genes that were differentially expressed by at least 1.25 fold between the patients with high and low CPT1B expression, respectively. We referred the obtained signatures as “CPT1B high vs CPT1B low” signatures. We used GSEA method to determine the enriched pathways of the CPT1B high vs. CPT1B low signatures in each cohort. A total of 17 pathways were uniformly enriched (FDR-adjusted Q-value<0.25) in all five datasets (Figure 5A). These pathways include inflammation/immune system, metabolism, and oncogenic pathways (Figure 5A). Notably, E2F targets and the EMT pathway were enriched with a strict Q-value<0.0001, further supporting the biological relevance of CPT1B in BLCA progression (Figure 5B).
Figure 5. Low expression of CPT1B is associated with key oncogenic, metabolic pathways in high-grade BLCA.
A) Gene set enrichment analysis (GSEA) revealed that genes associated with CPT1B down-regulation are significantly enriched in oncogenic, metabolic, and immune pathways in the Kim, Lindgren, Sjodahl, and TCGA cohorts. B) CPT1B downregulation was highly associated with enrichment of E2F targets and EMT signatures in all cohorts. The values on the y-axis for each graph represent enrichment scores (corresponding to the magnitude of the enrichment for each analysis). For each graph, the Normalized Enrichment Score (NES, computed via the GSEA analysis) and the significance of the enrichment (q=false discovery rate, also computed via the GSEA analysis). The NES scores ranged from 4 to 11 (all q<0.0001).
Effects of CPT1B overexpression in high grade invasive BLCA cell lines
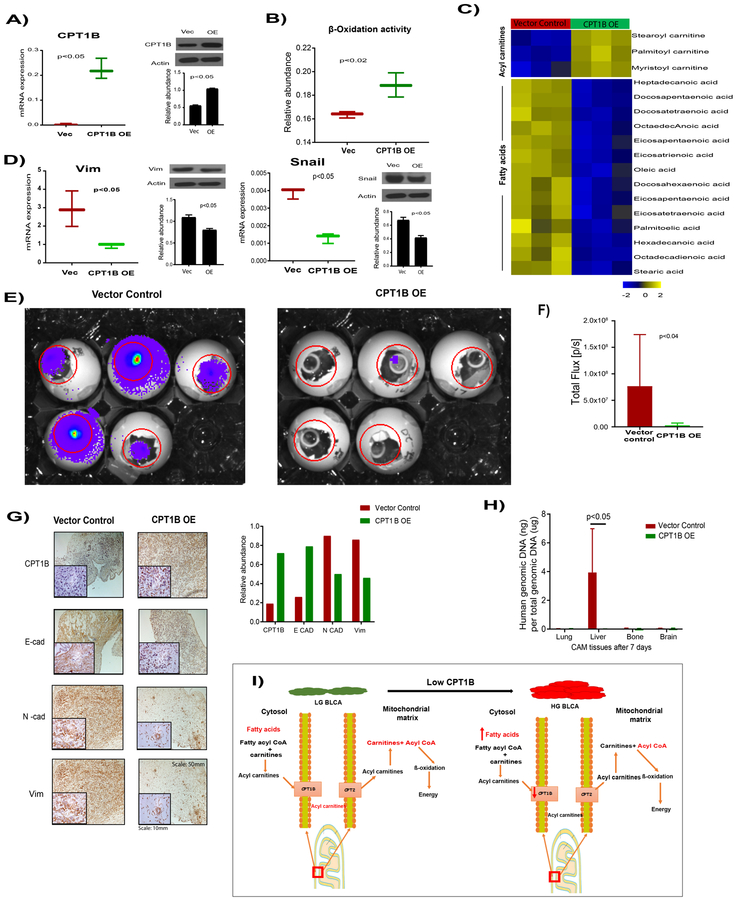
To determine the effects of CPT1B, in high grade BLCA, we ectopically expressed the CPT1B in high-grade UMUC3 BLCA cells (Figure 6A). The CPT1B overexpressed cells displayed increased FAO activity (Figure 6B), with increased levels of acyl carnitines and reduced levels of fatty acids compared to control cells (Figure 6C). EMT is one of the major hallmarks of the metastasis in high-grade cancer. The CPT1B overexpressed cells displayed decreased levels of the EMT markers vimentin and snail compare to vector control (Figure 6D).
Figure 6. Effects of CPT1B in high-grade BLCA cell lines.
A) Adenovirus-mediated overexpression of CPT1B in the high grade BLCA cell line UMUC3 compared with a vector control, confirmed by qPCR and western blot. B) CPT1B overexpression increases the β-oxidation in high grade BLCA cells. C) Heat map of increased acyl carnitine levels and reduced fatty acid levels in CPT1B overexpressed cells compared to control cells (false discovery rate-corrected p-value <0.05). D) Expression of the epithelial and mesenchymal transition (EMT) markers vimentin and snail in CPT1B overexpressed cells compared with that in vector control cells. E) CPT1B over expressed cells formed significantly smaller tumors on chicken chorioallontoic membranes (CAMs) as measured by total photon flux of the region of interest (Day 15; red circle) (F). G) CPT1B overexpression significantly increased the E-cad, reduced the N-cad and vimentin levels in tumors after 7 days of tumor growth on CAMs, as evidenced by IHC analysis and quantification. H) CPT1B overexpression reduced the metastasis of UMUC3 cells into the chick embryo liver tissue as measured by RT-PCR of a human-specific Alu sequence in total genomic DNA (gDNA). I) Graphical representation of the role of CPT1B in BLCA progression. OE represents overexpression.
CPT1B overexpression in in vivo reduces tumor growth, EMT, and liver metastasis in CAM model
Given that CPT1B loss and FAO disruption led to significantly increased activity in pathways associated with EMT and invasion (Figure 5), we hypothesized that CPT1B overexpression leads to a reduction in the metastatic capabilities of cancer cells. To functionally test that hypothesis, we performed in vivo CAM xenograft assay32 using CPT1B overexpressing cells. Compared to vector control UMUC3 cells, CPT1B overexpression cells demonstrated reduced tumor growth in the xenograft assays (Figure 6E and F). Immunohistochemistry confirmed that the CPT1B overexpression CAM tumors had high E-cad levels, but low N-cad and vimentin levels (Figure 6G). We tested the metastatic potential of the CPT1B overexpression cells by Alu PCR. There was decreased metastasis to liver of the chick embryo from CPT1B overexpression tumors than controls (Figure 6H). Overall, our results confirmed the role of CPT1B in tumor growth and metastasis in high-grade BLCA. Taken together our results suggested that CPT1B downregulation impairs FAO and increases the tumor growth and metastasis in BLCA (Figure 6I).
Discussion
Treatments for high-grade muscle-invasive and metastatic bladder cancer have not advanced beyond cisplatin-based combination chemotherapy33. The median survival time of patients with high-grade MIBC is only 14–15 months with cisplatin-based chemotherapy15, highlighting the need for novel therapeutic approaches. Genomic integrative studies showed that genes involved in the PI3-kinase/AKT/mTOR, RTK/RAS, and CDKN2A/CDK4/CCND1 pathways are associated with high-grade BLCA26. We used integrated approach combining unbiased metabolomics, lipidomics, and transcriptomics to investigate the progression of high-grade BLCA and to delineate opportunities for therapeutic and prognostic intervention.
Alteration of metabolism is one of the hallmarkers for cancer progression34. Metabolic alterations enable cancer cells to meet the increased energy demands for proliferation and survival in the nutrient-deprived tumor microenvironment35. Our metabolomics analysis identified 519 differentially metabolites that were differentially produced in high-grade BLCA, including glycolysis, TCA cycle, PPP pathway, polyamines, bile acids, carnitine, prostaglandins, and nucleotides indicating that metabolism is a highly dynamic process in BLCA. Unbiased lipidomics analysis identified 19 differentially produced lipids in high-grade BLCA, including triglycerides, phosphatidylcholines, phosphatidylethanolamines, and phosphatidyl inositols. The higher levels of triglycerides and low levels of phosphatidylcholines and phosphatidylethanolamines are in line with previous studies10. We used transcriptomics data from the Kim cohort26 to identify genes that were differentially expressed in high-grade BLCA. We mapped the altered metabolites and lipids to their corresponding genes and then to enriched pathways. The results from metabolomics, lipidomics, and transcriptomics suggest that there were common deregulated metabolic pathways implicated the arachidonic acid, phosphatidylinositol, and glycerophospholipid metabolism. These pathways are important in various cancers36,37,38,39 and will be focal points in future BLCA research. Clinical trials of drugs targeting these metabolic alterations in BLCA patients are warranted.
Our integrative multi-OMICs analysis identified 27 common genes that were implicated by metabolomics, lipidomics, and transcriptomics data. Each of these genes has a profound impact on the cellular metabolism, and particularly lipid metabolism highlighting the importance of fatty acids in cell proliferation and tumorigenesis40. The upregulated genes (PLTP, PLA2G4C, LIPG, PIGS, PIGT, PIGU, PLCXD1, ATP8B2, PIGY, PTDSS1, ATP8B4) and downregulated genes (PIGB, PIGG, PIGN, PLCE1, PLA2G4A, CPTIB,PIGV, ATP8B1, CEL, PLA2G4F, PLCG2, PLA2G4B, INPP5D, PLCD3, PIGZ, PLA2G10) provided a gene signature that was associated with poor survival in Lindgren, Kim, and Sjodhal cohorts. Further characterization and validation of this gene signature may provide a prognostic marker for high-grade BLCA. Some of these genes in the signature have been reported to be involved in the metastasis of other cancers; for example, PLTP, and LIPG upregulation in breast cancer41,42, ATP8B1 in colorectal cancer43, and PLCG2 in chronic lymphocytic leukemia44. So far, the role of these genes in high-grade BLCA remains unclear. Further characterization and functional validation of the genes could provide insights into the molecular events that are related to progression of BLCA from low-grade to high-grade.
Our integrated analysis of these OMICs datasets revealed the previously unappreciated importance of CPT1B in high-grade BLCA. CPT1B resides at the outer mitochondrial membrane and regulates intracellular lipid metabolism by transporting long-chain fatty acids into the mitochondria for β-oxidation45,46. Overexpression of fatty acid synthase (FAS)47, leads to greater accumulation of fatty acids and low expression of CPT1B in high-grade BLCA. That, in turn, supports the requirement of actively proliferating cells for membrane structural and signaling lipids. FAO and fatty acid synthesis are mutually antagonistic process48. The low abundance of palmitoyl and octanoyl carnitine and low FAO activity observed in patients with high-grade BLCA are consistent with low CPT1B expression observed in patient with high-grade BLCA and cell lines. GSEA of genes associated with low CPT1B expression in five distinct public BLCA cohorts revealed alterations of many immunological, metabolic, and oncogenic pathways that are known hallmarks of cancer. Low CPT1B expression was associated with high levels of EMT which regulates metastasis in cancer by conferring an invasive phenotype49, is the salient feature of high-grade BLCA. CPT1B over expression reduced the cell proliferation and EMT markers supporting the relevance of FAO in cellular transformation and BLCA progression towards high grade disease. Furthermore, CPT1B over expression in in vivo significantly reduced tumor growth, and metastasis, suggesting that a strategy to increase the CPT1B levels in high grade BLCA might help to inhibit tumor growth and metastasis.
In conclusion, for the first time, we identified critical alterations of metabolite and lipid profiles that contribute to BLCA progression by levaraging high resolution LC-MS based unbiased metabolomics, and lipidomics in combination with bioinformatics analysis. Our integrated study of low-grade versus high-grade BLCA provided numerous novel insights into disease biology and delineated pathways that provide potential opportunities for therapeutic intervention. The low levels of FAO and downregulation of CPT1B in high-grade BLCA indicate that the impairment of β-oxidation plays an important role in BLCA tumor progression. These results may have prognostic value and provide targets for therapeutic intervention in the future. Taken together, our results suggest that interventions that increase the CPT1B levels may have therapeutic value for high-grade BLCA patients that are notably dependent on FAO metabolism.
Supplementary Material
Translational Relevance.
Cancer metabolism varies depending on the tumor grade. Currently, there is an absence of multi-OMICs data integration to predict bladder cancer (BLCA) survival. To fill in this gap, we performed unbiased metabolomics and lipidomics analyses of matched BLCA tissues and integrated them with BLCA transcriptomics analyses to generate an integrated gene signature that was associated with patient survival in multiple BLCA cohorts. Our results revealed altered metabolic differences between high-grade BLCA and low-grade BLCA and suggest that impaired fatty acid β-oxidation (FAO) due to the downregulation of CPT1B plays a crucial role in the progression of low grade to high grade BLCA. CPT1B overexpression in high-grade BLCA cell lines reduced cell proliferation, epithelial-mesenchymal transition, and metastasis to the liver in vivo by increasing FAO. Our metabolic-centered multi-OMICs based integrative analysis provides a system-level perspective on BLCA and potential targets for novel therapeutics against high-grade BLCA.
Acknowledgments
This research was fully supported by American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N.P.), NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.), partially supported by the following grants: NIH/NCI U01 CA167234 (A.S.K), CPRIT RP170295 (C.C.), as well as funds from Alkek Center for Molecular Discovery (A.S.K.). This project was also supported by the Agilent Technologies Center of Excellence (COE) in Mass Spectrometry at Baylor College of Medicine, Metabolomics Core, Human Tissue Acquisition and Pathology at Baylor College of Medicine with funding from the NIH (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility (N.P.), (RP170005), and Dan L. Duncan Cancer Center. CAM assay was supported by the Patient-Derived Xenograft and Advanced in vivo Models Core Facility at Baylor College of Medicine with funding from the Cancer Prevention and Research Institute of Texas (CPRIT) grant #170691. We would like thank the team of the Georgia Cancer Center Biorepository / BRAG-Onc for biospecimen collection and annotation.
Funding support: Supported by American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N.P.), NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.).
Footnotes
Conflict of interest: Authors do not have any conflict of interest.
Reference
- 1.Siegel RL, Miller KD & Jemal A Cancer Statistics, 2017. CA Cancer J Clin 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lin LL, Hsia CR, Hsu CL, Huang HC & Juan HF Integrating transcriptomics and proteomics to show that tanshinone IIA suppresses cell growth by blocking glucose metabolism in gastric cancer cells. BMC Genomics 16, 41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koplev S, Lin K, Dohlman AB & Ma’ayan A Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Comput Biol 14, e1005911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ettinger DS, et al. Non-Small Cell Lung Cancer, Version 6.2015. J Natl Compr Canc Netw 13, 515–524 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Gradishar WJ, et al. Breast Cancer Version 2.2015. J Natl Compr Canc Netw 13, 448–475 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Li R, Choi W, Ferguson Rd JE, Metcalfe MJ & Kamat AM New discoveries in the molecular landscape of bladder cancer. F1000Res 5, 2875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putluri N, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res 71, 7376–7386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Rundstedt FC, et al. Integrative Pathway Analysis of Metabolic Signature in Bladder Cancer: A Linkage to The Cancer Genome Atlas Project and Prediction of Survival. J Urol 195, 1911–1919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piyarathna DWB, et al. Distinct Lipidomic Landscapes Associated with Clinical Stages of Urothelial Cancer of the Bladder. Eur Urol Focus (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JC, Citrin DE, Agarwal PK & Apolo AB Multimodal management of muscle-invasive bladder cancer. Curr Probl Cancer 38, 80–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vantaku V, et al. Expression of ganglioside GD2, reprogram the lipid metabolism and EMT phenotype in bladder cancer. Oncotarget 8, 95620–95631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amara CS, et al. Serum metabolic profiling identified a distinct metabolic signature in Bladder Cancer Smokers: A key metabolic enzymes associated with patient survival. Cancer Epidemiol Biomarkers Prev (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vantaku V, et al. Large-scale profiling of serum metabolites in African American and European American patients with bladder cancer reveals metabolic pathways associated with patient survival. Cancer (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 16.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kind T, et al. Qualitative analysis of algal secretions with multiple mass spectrometric platforms. J Chromatogr A 1244, 139–147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ejsing CS, et al. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem 78, 6202–6214 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Li M, et al. The In Ovo Chick Chorioallantoic Membrane (CAM) Assay as an Efficient Xenograft Model of Hepatocellular Carcinoma. J Vis Exp (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quigley JP & Armstrong PB Tumor cell intravasation alu-cidated: the chick embryo opens the window. Cell 94, 281–284 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Zijlstra A, et al. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res 62, 7083–7092 (2002). [PubMed] [Google Scholar]
- 22.Wishart DS, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res 35, D521–526 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim WJ, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer 9, 3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindgren D, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One 7, e38863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjodahl G, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 18, 3377–3386 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong J Fatty Acid Oxidation in Cell Fate Determination. Trends Biochem Sci (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascual G, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Luo X, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer 16, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beloribi-Djefaflia S, Vasseur S & Guillaumond F Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin F, et al. Tobacco-Specific Carcinogens Induce Hypermethylation, DNA Adducts, and DNA Damage in Bladder Cancer. Cancer Prev Res (Phila) 10, 588–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Deryugina EI & Quigley JP Chick embryo chorioallantoic membrane model systems to study and visualize human tumor cell metastasis. Histochem Cell Biol 130, 1119–1130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiess PE, et al. Bladder Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 15, 1240–1267 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Cairns RA & Mak TW An Alternative Sugar Fuels AML. Cancer Cell 30, 660–662 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Liu P, Cheng H, Roberts TM & Zhao JJ Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8, 627–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolce V, Cappello AR, Lappano R & Maggiolini M Glycerophospholipid synthesis as a novel drug target against cancer. Curr Mol Pharmacol 4, 167–175 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Yang P, et al. Arachidonic acid metabolism in human prostate cancer. Int J Oncol 41, 1495–1503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudry AA, Wahle KW, McClinton S & Moffat LE Arachidonic acid metabolism in benign and malignant prostatic tissue in vitro: effects of fatty acids and cyclooxygenase inhibitors. Int J Cancer 57, 176–180 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Fritz V & Fajas L Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene 29, 4369–4377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallo LH, Ko J & Donoghue DJ The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle 16, 634–648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slebe F, et al. FoxA and LIPG endothelial lipase control the uptake of extracellular lipids for breast cancer growth. Nat Commun 7, 11199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aziz MA, et al. Integrated exon level expression analysis of driver genes explain their role in colorectal cancer. PLoS One 9, e110134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang SQ, Smith SM, Zhang SY & Lynn Wang Y Mechanisms of ibrutinib resistance in chronic lymphocytic leukaemia and non-Hodgkin lymphoma. Br J Haematol 170, 445–456 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Britton CH, et al. Human liver mitochondrial carnitine palmitoyltransferase I: characterization of its cDNA and chromosomal localization and partial analysis of the gene. Proc Natl Acad Sci U S A 92, 1984–1988 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis BC, Esser V, Foster DW & McGarry JD Rat heart expresses two forms of mitochondrial carnitine palmitoyltransferase I. The minor component is identical to the liver enzyme. J Biol Chem 269, 18712–18715 (1994). [PubMed] [Google Scholar]
- 47.Sugino T, et al. Overexpression of fatty acid synthase in human urinary bladder cancer and combined expression of the synthase and Ki-67 as a predictor of prognosis of cancer patients. Med Mol Morphol 44, 146–150 (2011). [DOI] [PubMed] [Google Scholar]
- 48.McGarry JD & Brown NF The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem 244, 1–14 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Davis FM, Stewart TA, Thompson EW & Monteith GR Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci 35, 479–488 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.