Abstract
Photosynthetic organisms provide food and energy for nearly all life on Earth, yet half of their protein-coding genes remain uncharacterized1,2. Characterization of these genes could be greatly accelerated by new genetic resources for unicellular organisms. Here, we generated a genome-wide, indexed library of mapped insertion mutants for the unicellular alga Chlamydomonas reinhardtii. The 62,389 mutants in the library, covering 83% of nuclear, protein-coding genes, are available to the community. Each mutant contains unique DNA barcodes, allowing the collection to be screened as a pool. We performed a genome-wide survey of genes required for photosynthesis, which identified 303 candidate genes. Characterization of one of these genes, the conserved predicted phosphatase-encoding gene CPL3, showed it is important for accumulation of multiple photosynthetic protein complexes. Notably, 21 of the 43 highest-confidence genes are novel, opening new opportunities for advances in our understanding of this biogeochemically fundamental process. This library will accelerate the characterization of thousands of genes in algae, plants and animals.
Editorial summary:
Generation of a library of 62,389 mapped insertion mutants for the unicellular alga Chlamydomonas reinhardtii enables screening for genes required for photosynthesis and the identification of 303 candidate genes.
The green alga Chlamydomonas has long been employed for genetic studies of eukaryotic photosynthesis because of its rare ability to grow in the absence of photosynthetic function3. In addition, it has made extensive contributions to our basic understanding of light signaling, stress acclimation, and metabolism of carbohydrates, lipids, and pigments (Fig. 1a)4-6. Moreover, Chlamydomonas retained many genes from the plant-animal common ancestor, which contributed to the understanding of fundamental aspects of the structure and function of cilia and basal bodies7,8. Like Saccharomyces cerevisiae, Chlamydomonas can grow as a haploid, facilitating genetic studies. However, until now, the value of Chlamydomonas has been limited by the lack of mutants in most of its nuclear genes.
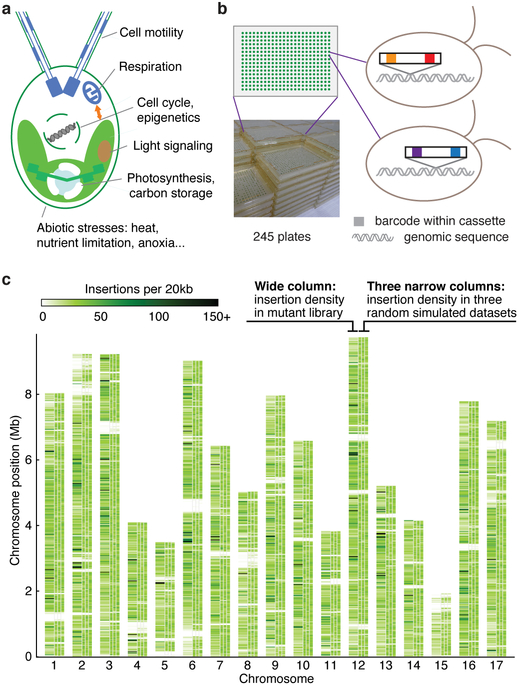
Fig. 1 ∣. A genome-wide library of Chlamydomonas mutants was generated by random insertion of barcoded cassettes and mapping of insertion sites.
a, Chlamydomonas reinhardtii is used for studies of various cellular processes and organism-environment interactions. b, Our library contains 62,389 insertional mutants maintained as 245 plates of 384-colony arrays. Each mutant contains at least one insertion cassette at a random site in its genome; each insertion cassette contains one unique barcode at each end (Supplementary Fig. 1a-c). c, The insertion density is largely random over the majority of the genome. This panel compares the observed insertion density over the genome (the left column above each chromosome number) to three simulations with insertions randomly distributed over all mappable positions in the genome (the three narrow columns to the right for each chromosome). Areas that are white throughout all columns represent regions where insertions cannot be mapped to a unique genomic position due to highly repetitive sequence. See also Supplementary Fig. 4.
In the present study, we sought to generate a genome-wide collection of Chlamydomonas mutants with known gene disruptions to provide mutants in genes of interest for the scientific community, and then to leverage this collection to identify genes with roles in photosynthesis. To reach the necessary scale, we chose to use random insertional mutagenesis and built on advances in insertion mapping and mutant propagation from our pilot study9. To enable mapping of insertion sites and screening pools of mutants on a much larger scale, we developed new tools leveraging unique DNA barcodes in each transforming cassette.
We generated mutants by transforming haploid cells with DNA cassettes that randomly insert into the genome and inactivate the genes they insert into. We maintained the mutants as indexed colony arrays on agar media containing acetate as a carbon and energy source to allow recovery of mutants with defects in photosynthesis. Each DNA cassette contained two unique barcodes, one on each side of the cassette (Supplementary Fig. 1a-d). For each mutant, the barcode and genomic flanking sequences on each side of the cassette were initially unknown (Supplementary Fig. 1e). We determined the sequence of the barcode(s) in each mutant colony by combinatorial pooling and deep sequencing (Supplementary Fig. 1f, Supplementary Fig. 2). We then mapped each insertion by pooling all mutants and amplifying all flanking sequences together with their corresponding barcodes followed by deep sequencing (Supplementary Fig. 1g). The combination of these datasets identified the insertion site(s) in each mutant. This procedure yielded 62,389 mutants on 245 plates, with a total of 74,923 insertions that were largely randomly distributed over the chromosomes (Fig. 1b,c; Supplementary Fig. 3, 4; Supplementary Table 5).
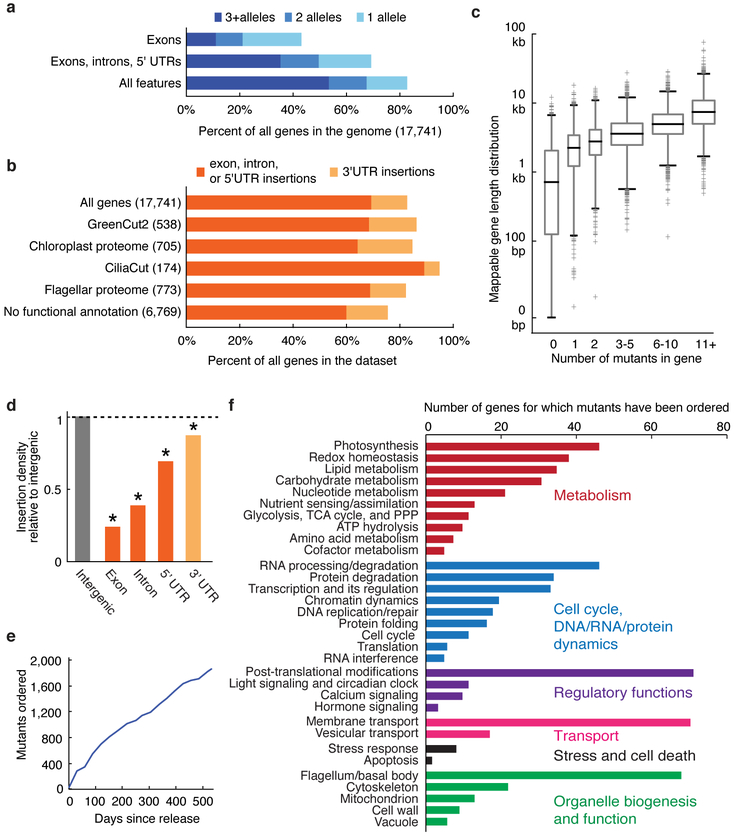
This library provides mutants for ~83% of all nuclear genes (Fig. 2a-d). Approximately 69% of genes are represented by an insertion in a 5’ UTR, an exon or an intron – regions most likely to cause an altered phenotype when disrupted. Many gene sets of interest to the research community are well represented, including genes encoding proteins phylogenetically associated with the plant lineage (GreenCut2)1, proteins that localize to the chloroplast10, or those associated with the structure and function of flagella or basal bodies11,12 (Fig. 2b). Mutants in this collection are available through the CLiP website (see URLs). Over 1,800 mutants have already been distributed to over 200 laboratories worldwide in the first 18 months of pre-publication distribution (Fig. 2e). These mutants are facilitating genetic investigation of a broad range of processes, ranging from photosynthesis and metabolism to cilia structure and function (Fig. 2f).
Fig. 2 ∣. The library covers 83% of Chlamydomonas genes.
a, 83% of all Chlamydomonas genes have one or more insertions in the library. b, In various functional groups, more than 75% of genes are represented by insertions in the library. c, The number of insertions per gene is roughly correlated with gene length. Box heights represent quartiles, whiskers represent 1st and 99th percentiles, and outliers are plotted as crosses. Box widths are proportional to the number of genes in each bin. d, Insertion density varies among different gene features, with the lowest density in exons. e, More than 1,800 mutants were distributed to approximately 200 laboratories around the world during the first 18 months of its availability. f, Distributed mutants are being used to study a variety of biological processes. Only genes with some functional annotation are shown.
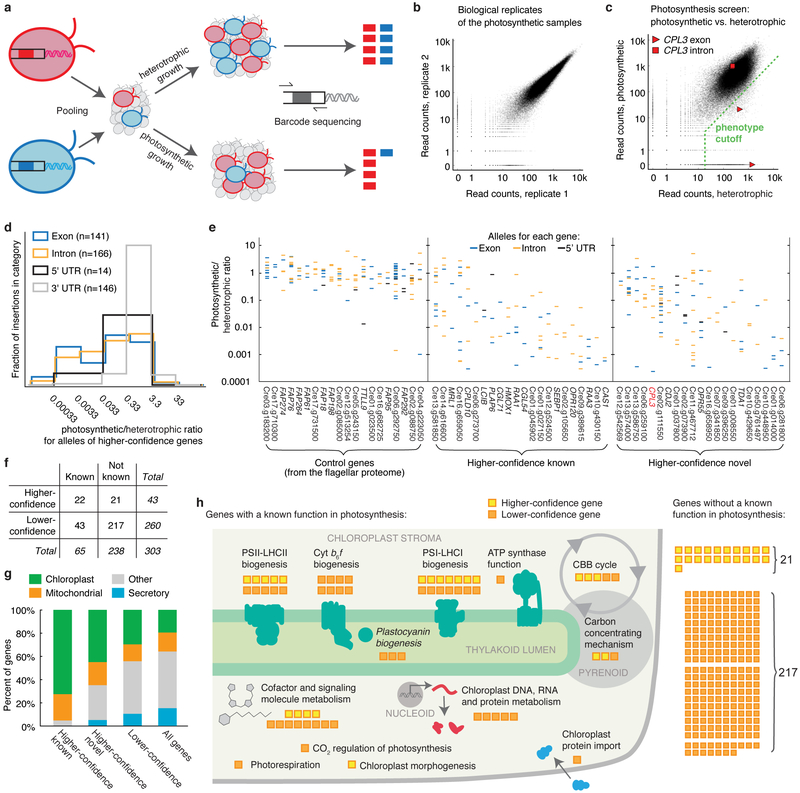
To identify genes required for photosynthesis, we screened our library for mutants deficient in photosynthetic growth. Rather than phenotyping each strain individually, we pooled the entire library into one culture and leveraged the unique barcodes present in each strain to track its abundance after growth under different conditions. This feature enables genome-wide screening with speed and depth unprecedented in photosynthetic eukaryotes. We grew a pool of mutants photosynthetically in light in minimal Tris-Phosphate (TP) medium with CO2 as the sole source of carbon, and heterotrophically in the dark in Tris-Acetate-Phosphate (TAP) medium, where acetate provides fixed carbon and energy3 (Fig. 3a). To quantify mutant growth under each condition, we amplified and deep sequenced the barcodes from the final cell populations. We then compared the ability of each mutant to grow under photosynthetic and heterotrophic conditions by comparing the read counts of each barcode from each condition (Supplementary Table 10; Supplementary Note). Mutant phenotypes were highly reproducible (Fig. 3b and Supplementary Fig. 5, a and b). We identified 3,109 mutants deficient in photosynthetic growth (Fig. 3c; Supplementary Note).
Fig. 3 ∣. A high-throughput screen using the library identifies many genes with known roles in photosynthesis and many novel components.
a, Unique barcodes allow screening mutants in a pool. Mutants deficient in photosynthesis can be identified because their barcodes will be less abundant after photosynthetic growth relative to after heterotrophic growth. b, Biological replicates were highly reproducible, with a Spearman’s correlation of 0.982. Each dot represents one barcode. See also Supplementary Fig. 5. c, The phenotype of each insertion was determined by comparing its read count under photosynthetic and heterotrophic conditions. Insertions that fell below the phenotype cutoff were considered to show a defect in photosynthesis. cpl3 alleles are highlighted. d, Exon and intron insertions are most likely to show strong phenotypes, while 3’UTR insertions rarely do. The plot is based on all insertions for the 43 higher-confidence genes. e, The photosynthetic/heterotrophic ratio of all the alleles are shown for hit and control genes. Each column is a gene; each horizontal bar is an allele. f, The 303 candidate genes were categorized based on statistical confidence in this screen and based on whether the genes had a previously known function in photosynthesis (see Supplementary Note). g, Known higher-confidence genes, novel higher-confidence genes, and lower-confidence genes are all enriched in predicted chloroplast-targeted proteins (P < 0.011). h, A schematic summary illustrates the numbers of candidate genes in each category (panel f) and the specific functions of the genes with a known role in processes related to photosynthesis.
To identify genes with roles in photosynthesis, we developed a statistical analysis framework that leverages the presence of multiple alleles for many genes. This framework allows us to overcome several sources of false positives that have been difficult to account for with previous methods, including cases where the phenotype is not caused by the mapped disruption. For each gene, we counted the number of mutant alleles with and without a phenotype, and evaluated the likelihood of obtaining these numbers by chance given the total number of mutants in the library that exhibit the phenotype (Supplementary Table 11; Supplementary Note).
We identified 303 candidate photosynthesis genes based on our statistical analysis above. These genes are enriched for membership in a diurnally regulated photosynthesis-related transcriptional cluster13 (P < 10−11), are enriched for upregulation upon dark-to-light transitions14 (P < 0.003), and encode proteins enriched for predicted chloroplast localization (P < 10−8). As expected15, the candidate genes also encode a disproportionate number of GreenCut2 proteins (P < 10−8), which are conserved among photosynthetic organisms but absent from non-photosynthetic organisms1: 32 GreenCut2 proteins are encoded by the 303 candidate genes (11%), compared to ~3% in the entire genome.
Photosynthesis occurs in two stages: the light reactions and carbon fixation. The light reactions convert solar energy into chemical energy, and require coordinated action of Photosystem II (PSII), Cytochrome b6f, Photosystem I (PSI), ATP synthase complexes, a plastocyanin or cytochrome c6 metalloprotein, as well as small molecule cofactors16. PSII and PSI are each assisted by peripheral light-harvesting complexes (LHCs) known as LHCII and LHCI, respectively. Carbon fixation is performed by enzymes in the Calvin-Benson-Bassham cycle, including the CO2-fixing enzyme Rubisco. In addition, most eukaryotic algae have a mechanism to concentrate CO2 around Rubisco to enhance its activity17.
Sixty-five of the genes we identified encode proteins that were previously shown to play a role in photosynthesis or chloroplast function in Chlamydomonas or vascular plants (Fig. 3f). These include three PSII-LHCII subunits (PSBP1, PSBP2, and PSB27) and seven PSII-LHCII biogenesis factors (CGL54, CPLD10, HCF136, LPA1, MBB1, TBC2, and Cre02.g105650), two cytochrome b6f complex subunits (PETC and PETM) and six cytochrome b6f biogenesis factors (CCB2, CCS5, CPLD43, CPLD49, MCD1, and MCG1), five PSI-LHCI subunits (LHCA3, LHCA7, PSAD, PSAE, and PSAL) and nine PSI-LHCI biogenesis factors (CGL71, CPLD46, OPR120, RAA1, RAA2, RAA3, RAT2, Cre01.g045902, and Cre09.g389615), one protein required for ATP synthase function (PHT3), plastocyanin (PCY1) and two plastocyanin biogenesis factors (CTP2 and PCC1), 12 proteins involved in the metabolism of photosynthesis cofactors or signaling molecules (CHLD, CTH1, CYP745A1, DVR1, HMOX1, HPD2, MTF1, PLAP6, UROD3, Cre08.g358538, Cre13.g581850, and Cre16.g659050), three Calvin-Benson-Bassham Cycle enzymes (FBP1, PRK1, and SEBP1), two Rubisco biogenesis factors (MRL1 and RMT2), three proteins involved in the algal carbon concentrating mechanism (CAH3, CAS1, and LCIB), as well as proteins that play a role in photorespiration (GSF1), CO2 regulation of photosynthesis (Cre02.g146851), chloroplast morphogenesis (Cre14.g616600), chloroplast protein import (SDR17), and chloroplast DNA, RNA, and protein metabolism (DEG9, MSH1, MSRA1, TSM2, and Cre01.g010864) (Fig. 3h, Supplementary Table 12). We caution that not all genes previously demonstrated to be required for photosynthetic growth are detectable by this approach, especially the ones with paralogous genes in the genome, such as RBCS1 and RBCS2 that encode the small subunit of Rubisco18. Nonetheless, the large number of known factors recovered in our screen is a testament to the power of this approach.
In addition to recovering these 65 genes with known roles in photosynthesis, our analysis identified 238 candidate genes with no previously reported role in photosynthesis. These 238 genes represent a rich set of targets to better understand photosynthesis. Because our screen likely yielded some false positives, we divided all genes into “higher-confidence” (P < 0.0011; false discovery rate (FDR) < 0.27) and “lower-confidence” genes based on the number of alleles that supported each gene’s involvement in photosynthesis (Fig. 3d-f; Tables 1 and 2; Supplementary Note). The 21 higher-confidence genes with no previously reported role in photosynthesis are enriched in chloroplast localization (9/21, P < 0.011; Fig. 3g) and transcriptional upregulation during dark to light transition (5/21, P < 0.005), similar to the known photosynthesis genes. Thus, these 21 higher-confidence genes are particularly high-priority targets for the field to pursue.
Table 1 ∣.
Higher-confidence genes from the photosynthesis screen that had a previously known role in photosynthesis.
| Category | Gene | Defline/ description in Phytozome12 |
Pred Algoa |
Alleles in two replicates |
At homologe | Reference and the corresponding organism(s) |
||
|---|---|---|---|---|---|---|---|---|
| +b | −c | FDRd | ||||||
| Calvin-Benson-Bassham cycle | Cre03.g185550 (SEBP1, SBP1) | Sedoheptulose-1,7-bisphosphatase | C | 3 | 0 | 0.021 | AT3G55800.1 (SBPASE) | Arabidopsis29 |
| 3 | 0 | 0.018 | ||||||
| Cre12.g524500 (RMT2) | Rubisco small subunit N-methyltransferase | O | 3 | 0 | 0.021 | AT3G07670.1 | Pisum30 | |
| 3 | 0 | 0.018 | ||||||
| Cre06.g298300 (MRL1, PPR2) | Pentatricopeptide repeat protein, stabilizes rbcL mRNA | C | 1 | 1 | 1.000 | AT4G34830.1 (MRL1) | Chlamydomonas and Arabidopsis31 | |
| 2 | 0 | 0.239 | ||||||
| Carbon concentrating mechanism | Cre12.g497300 (CAS1, TEF2) | Rhodanese-like Ca-sensing receptor | C | 2 | 0 | 0.260 | AT5G23060.1 (CaS) | Chlamydomonas32 |
| 2 | 0 | 0.239 | ||||||
| Cre10.g452800 (LCIB) | Low-CO2-inducible protein | C | 2 | 0 | 0.260 | - | Chlamydomonas33 | |
| 1 | 1 | 1.000 | ||||||
| Chloroplast and thylakoid morphogenesis | Cre14.g616600 | - | M | 4 | 3 | 0.021 | AT1G03160.1 (FZL) | Arabidopsis34 |
| 4 | 3 | 0.018 | ||||||
| Cofactor and signaling molecule metabolism | Cre13.g581850 | - | M | 5 | 5 | 0.010 | AT4G31390.1 | Arabidopsis35 |
| 2 | 8 | 1.000 | ||||||
| Cre10.g423500 (HMOX1, HMO1) | Heme oxygenase | C | 3 | 0 | 0.021 | AT1G69720.1 (HO3) | Chlamydomonas14 | |
| 3 | 0 | 0.018 | ||||||
| Cre03.g188700 (PLAP6, PLP6) | Plastid lipid associated protein, Fibrillin | C | 3 | 1 | 0.070 | AT5G09820.2 | Arabidopsis36 | |
| 3 | 1 | 0.056 | ||||||
| Cre16.g659050 | - | C | 4 | 6 | 0.098 | AT1G68890.1 | Chlamydomonas37 | |
| 4 | 6 | 0.075 | ||||||
| PSI protein synthesis and assembly | Cre12.g524300 (CGL71) | Predicted protein | C | 2 | 0 | 0.260 | AT1G22700.1 | Synechocystis38; Arabidopsis39; Chlamydomonas40 |
| 2 | 0 | 0.239 | ||||||
| Cre01.g045902 | - | C | 1 | 1 | 1.000 | AT3G24430.1 (HCF101) | Arabidopsis41,42 | |
| 2 | 0 | 0.239 | ||||||
| PSI RNA splicing and stabilization | Cre09.g389615 | - | M | 5 | 0 | 0.0002 | AT3G17040.1 (HCF107) | Chlamydomonas43; Arabidopsis42,44,f |
| 5 | 0 | 0.0002 | ||||||
| Cre01.g027150 (CPLD46, HEL5) | DEAD/DEAH-box helicase | M | 5 | 1 | 0.0004 | AT1G70070.1 (EMB25, ISE2, PDE317) | Arabidopsis45 | |
| 5 | 1 | 0.0003 | ||||||
| Cre09.g394150 (RAA1) | - | M | 5 | 1 | 0.0004 | - | Chlamydomonas46 | |
| 5 | 1 | 0.0003 | ||||||
| Cre12.g531050 (RAA3) | PsaA mRNA maturation factor 3 | C | 3 | 0 | 0.021 | - | Chlamydomonas47 | |
| 3 | 0 | 0.018 | ||||||
| Cre10.g440000 (OPR120) | - | C | 2 | 0 | 0.260 | - | Chlamydomonas48,49 | |
| 2 | 0 | 0.239 | ||||||
| PSII protein synthesis and assembly | Cre13.g578650 (CPLD10, NUOAF5) | Similar to complex I intermediate-associated protein 30 | C | 3 | 3 | 0.260 | AT1G16720.1 (HCF173) | Arabidopsis42,50,51 |
| 3 | 3 | 0.208 | ||||||
| Cre02.g073850 (CGL54) | Predicted protein | C | 2 | 0 | 0.260 | AT1G05385.1 (LPA19, Psb27-H1) | Arabidopsis52 | |
| 2 | 0 | 0.239 | ||||||
| Cre02.g105650 | - | C | 2 | 0 | 0.260 | AT5G51545.1 (LPA2) | Arabidopsis53 | |
| 2 | 0 | 0.239 | ||||||
| Cre06.g273700 (HCF136) | - | C | 2 | 0 | 0.260 | AT5G23120.1 (HCF136) | Arabidopsis42; Synechocystis54 | |
| 1 | 1 | 1.000 | ||||||
| Cre10.g430150 (LPA1, REP27) | - | C | 2 | 0 | 0.260 | AT1G02910.1 (LPA1) | Arabidopsis55 | |
| 1 | 1 | 1.000 | ||||||
Prediction of protein localization by PredAlgo56: C = chloroplast, M = mitochondrion, SP = secretory pathway, O = other.
The number of exon/intron/5’UTR mutant alleles for that gene that satisfy our requirement of minimum 50 reads and showed at least 10X fewer normalized reads in the TP-light sample compared to the TAP-dark sample.
The number exon/intron/5’UTR mutant alleles for that gene that satisfy our minimum read count requirement but did not satisfy the at least 10X depletion in TP-light criterion.
the FDR for that gene compared to all alleles for all genes (see Supplementary Note).
Arabidopsis homolog, obtained from the “best_arabidopsis_TAIR10_hit_name” field in Phytozome12.
AT3G17040.1 is required for functional PSII in Arabidopsis whereas Cre09.g389615 was shown to be involved in PSI accumulation in Chlamydomonas.
Table 2 ∣.
Higher-confidence genes from the photosynthesis screen with no previously known role in photosynthesis.
| Gene | Defline/description in Phytozome |
PredAlgo | Alleles in two replicates |
At homolog | ||
|---|---|---|---|---|---|---|
| + | − | FDR | ||||
| Cre01.g008550 | Serine/threonine kinase-related | O | 2 | 0 | 0.260 | AT1G73450.1 |
| 1 | 1 | 1.000 | ||||
| Cre01.g014000 | - | C | 3 | 0 | 0.021 | - |
| 3 | 0 | 0.018 | ||||
| Cre01.g037800 (TRX21) | ATP binding protein; thioredoxin domain | O | 3 | 3 | 0.260 | AT2G18990.1 (TXND9) |
| 1 | 5 | 1.000 | ||||
| Cre02.g073900 | All-trans-10'-apo-beta-carotenal 13,14-cleaving dioxygenase | C | 3 | 1 | 0.070 | AT4G32810.1 (ATCCD8, CCD8, MAX4) |
| 3 | 1 | 0.056 | ||||
| Cre02.g111550 | Serine/threonine kinase-related | SP | 10 | 8 | < 10−6 | AT4G24480.1 |
| 6 | 12 | 0.015 | ||||
| Cre03.g185200 (CPL3, MPA6) | Metallophosphoesterase/metallodependent phosphatase | C | 3 | 4 | 0.260 | AT1G07010.1 |
| 3 | 4 | 0.239 | ||||
| Cre06.g259100 | - | C | 1 | 4 | 1.000 | - |
| 3 | 2 | 0.117 | ||||
| Cre06.g281800 | Domain of unknown function (DUF1995) | C | 3 | 0 | 0.021 | - |
| 3 | 0 | 0.018 | ||||
| Cre07.g316050 (CDJ2) | Chloroplast DnaJ-like protein | M | 2 | 0 | 0.260 | AT5G59610.1 |
| 1 | 1 | 1.000 | ||||
| Cre07.g341850 (EIF2, INFB) | Translation initiation factor IF-2, chloroplastic | C | 2 | 0 | 0.260 | AT1G17220.1 (FUG1) |
| 2 | 0 | 0.239 | ||||
| Cre08.g358350 (TDA1, OPR34) | Fast leu-rich domain-containinga | C | 3 | 2 | 0.152 | - |
| 3 | 2 | 0.117 | ||||
| Cre09.g396250 (VTE5) | Phosphatidate cytidylyltransferase | SP | 2 | 0 | 0.260 | AT5G04490.1 (VTE5) |
| 1 | 1 | 1.000 | ||||
| Cre10.g429650 | Alpha/beta hydrolase family (Abhydrolase_5) | O | 2 | 0 | 0.260 | - |
| 1 | 1 | 1.000 | ||||
| Cre10.g448950 | Nocturnin | C | 1 | 1 | 1.000 | AT3G58560.1 |
| 2 | 0 | 0.239 | ||||
| Cre11.g467712 | Structural maintenance of chromosomes smc family member; starch-binding domain | M | 7 | 7 | 0.0003 | AT5G05180.1 |
| 7 | 7 | 0.0003 | ||||
| Cre12.g542569 | Ionotropic glutamate receptor | O | 0 | 2 | 1.000 | AT1G05200.1 (ATGLR3.4, GLR3.4, GLUR3) |
| 2 | 0 | 0.239 | ||||
| Cre13.g566400 (OPR55) | Fast leu-rich domain-containinga | M | 4 | 2 | 0.018 | - |
| 4 | 2 | 0.015 | ||||
| Cre13.g574000 (GEF1, CLV1) | Voltage-gated chloride channel | O | 1 | 11 | 1.000 | AT5G26240.1 (ATCLC-D, CLC-D) |
| 4 | 8 | 0.144 | ||||
| Cre13.g586750 | Transportin 3 and importin | O | 3 | 4 | 0.260 | AT5G62600.1 |
| 2 | 5 | 1.000 | ||||
| Cre16.g658950 | - | C | 2 | 2 | 0.909 | - |
| 3 | 1 | 0.056 | ||||
| Cre50.g761497 | Magnesium transporter mrs2 homolog, mitochondrial | M | 2 | 0 | 0.260 | AT5G22830.1 (ATMGT10, GMN10, MGT10, MRS2-11) |
| 2 | 0 | 0.239 | ||||
The annotation of “fast leu-rich domain-containing” cannot be confirmed by BLASTp analysis at NCBI57.
Functional annotations for 15 of the 21 higher-confidence genes suggest that these genes could play roles in regulation of photosynthesis, photosynthetic metabolism, and biosynthesis of the photosynthetic machinery. Seven of the genes likely play roles in regulation of photosynthesis: GEF1 encodes a voltage-gated channel, Cre01.g008550 and Cre02.g111550 encode putative protein kinases, CPL3 encodes a predicted protein phosphatase, TRX21 contains a thioredoxin domain, Cre12.g542569 encodes a putative glutamate receptor, and Cre13.g586750 contains a predicted nuclear importin domain. Six of the genes are likely involved in photosynthetic metabolism: the Arabidopsis homolog of Cre10.g448950 modulates sucrose and starch accumulation19, Cre11.g467712 contains a starch-binding domain, Cre02.g073900 encodes a putative carotenoid dioxygenase, VTE5 encodes a putative phosphatidate cytidylyltransferase, Cre10.g429650 encodes a putative alpha/beta hydrolase, and Cre50.g761497 contains a magnesium transporter domain. Finally, two of the genes are likely to play roles in the biogenesis and function of photosynthesis machinery: EIF2 has a translation initiation factor domain, and CDJ2 has a chloroplast DnaJ domain. Future characterization of these genes by the community is likely to yield fundamental insights into our understanding of photosynthesis.
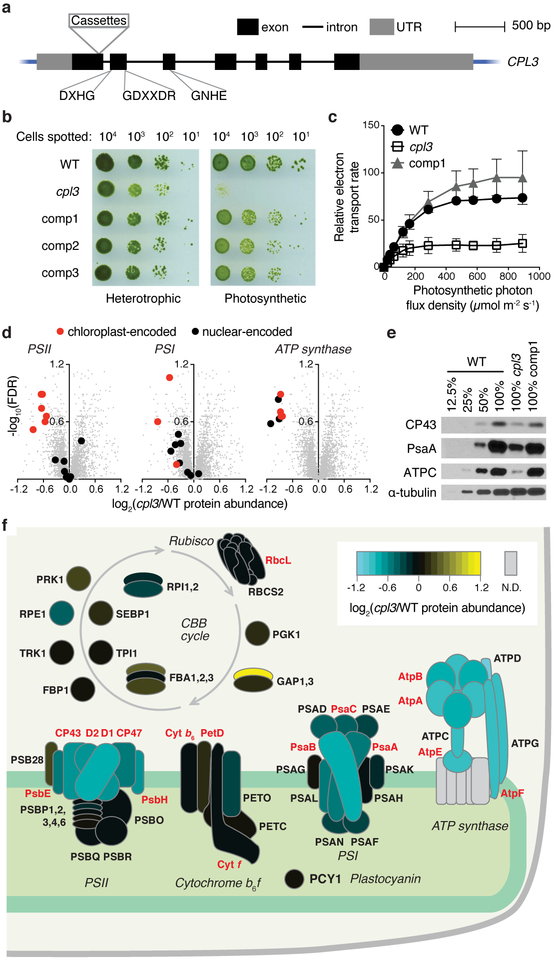
As an illustration of the value of genes identified in this screen, we sought to explore the specific function of one of the higher-confidence candidate genes, CPL3 (Conserved in Plant Lineage 3, Cre03.g185200, also known as MPA6), which encodes a putative protein phosphatase (Fig. 4a, Supplementary Fig. 6). Many proteins in the photosynthetic apparatus are phosphorylated, but the role and regulation of these phosphorylations are poorly understood20. An insertion junction mapped to the 3’ UTR of CPL3 was previously found in a collection of acetate-requiring mutants, although it was not determined if this mutation caused the phenotype15. In our screen, three mutants with insertion junctions in CPL3 exons or introns exhibited a deficiency in photosynthetic growth (Fig. 3c, Supplementary Table 13). We chose to examine one allele (LMJ.RY0402.153647, referred to hereafter as cpl3; Fig. 4a, Supplementary Fig. 6a) for phenotypic confirmation, genetic complementation, and further studies.
Fig. 4 ∣. CPL3 is required for photosynthetic growth and accumulation of photosynthetic protein complexes in the thylakoid membranes.
a, The cpl3 mutant contains cassettes inserted in the first exon of CPL3. The locations of conserved protein phosphatase motifs are indicated (see Supplementary Fig. 6e). b, cpl3 is deficient in growth under photosynthetic conditions and can be rescued upon complementation with the wild-type CPL3 gene (comp1–3 represent three independent complemented lines). c, cpl3 has a lower relative photosynthetic electron transport rate than the wild-type strain (WT) and comp1. Error bars indicate standard deviations (n = 3 for WT and comp1; n = 7 for cpl3). d, Whole-cell proteomics (Supplementary Table 14) indicate that cpl3 is deficient in accumulation of PSII, PSI, and the chloroplast ATP synthase. Each gray dot represents one Chlamydomonas protein. PSII, PSI and ATP synthase subunits are highlighted as black or red symbols. e, Western blots show that CPL3 is required for normal accumulation of the PSII subunit CP43, the PSI subunit PsaA, and the chloroplast ATP synthase subunit ATPC. α-tubulin was used as a loading control. See also Supplementary Fig. 7c. f, A heatmap of the protein abundance of subunits in the light reactions protein complexes or enzymes in the CBB cycle in cpl3 relative to the wild type based on proteomics data. Depicted subunits that were not detected by proteomics are filled with gray. Nuclear- and chloroplast-encoded proteins are labeled in black and red fonts respectively. A stack of horizontal ovals indicates different isoforms for the same enzyme, such as FBA1, FBA2, and FBA3.
Consistent with the pooled growth data, cpl3 showed a severe defect in photosynthetic growth on agar, which was rescued under heterotrophic conditions (Fig. 4b). We confirmed that the CPL3 gene is disrupted in the cpl3 mutant and found that complementation with a wild-type copy of the CPL3 gene rescues the phenotype, demonstrating that the mutation in CPL3 is the cause of the growth defect of the mutant (Supplementary Note, Supplementary Fig. 6a-d).
We then examined the photosynthetic performance, morphology of the chloroplast, and the composition of photosynthetic pigments and proteins in cpl3. Photosynthetic electron transport rate was decreased under all light intensities, suggesting a defect in the photosynthetic machinery (Fig. 4c). The chloroplast morphology of cpl3 appeared similar to the wild type based on chlorophyll fluorescence microscopy (Supplementary Fig. 7a). However, we observed a lower chlorophyll a/b ratio in cpl3 than in the wild type (Supplementary Fig. 7b), which suggests a defect in the accumulation or composition of the protein-pigment complexes involved in the light reactions21. Using whole-cell proteomics, we found that cpl3 was deficient in accumulation of all detectable subunits of the chloroplast ATP synthase (ATPC, ATPD, ATPG, AtpA, AtpB, AtpE, AtpF), some subunits of PSII (D1, D2, CP43, CP47, PsbE, PsbH), and some subunits of PSI (PsaA and PsaB) (FDR < 0.31 for each subunit, Fig. 4d,f; Supplementary Table 14). We confirmed these findings by western blots on CP43, PsaA, and ATPC (Fig. 4e; Supplementary Fig. 7c). Our results indicate that CPL3 is required for normal accumulation of thylakoid protein complexes (PSII, PSI, and ATP synthase) involved in the light reactions of photosynthesis.
Our finding that 21/43 of the higher-confidence photosynthesis hit genes were uncharacterized suggests that nearly half of the genes required for photosynthesis remain to be characterized. This finding is notable, considering that genetic studies on photosynthesis extend back to the 1950s22. Our validation of CPL3’s role in photosynthesis illustrates the value of the uncharacterized hit genes identified in this study as a rich set of candidates for the community to pursue.
More broadly, it is our hope that the mutant resource presented here will serve as a powerful complement to newly developed gene editing techniques23-28, and that together these tools will help the research community generate fundamental insights in a wide range of fields, from organelle biogenesis and function to organism-environment interactions.
Methods
Generation of the indexed and barcoded mutant library.
A three-step pipeline was developed for the generation of an indexed, barcoded library of insertional mutants in Chlamydomonas (Fig. 1b, Supplementary Fig. 1).
To generated mutants, CC-453358 (“wild type” in text and figures) cells were transformed with DNA cassettes that randomly insert into the genome, confer paromomycin resistance for selection, and inactivate the genes they insert into. Each cassette contained two unique 22 nucleotide barcodes, one at each end of the cassette (Supplementary Fig. 1a-d; Supplementary Note). Transformants were arrayed on agar plates and each insertion in a transformant would contain two barcodes. The barcode sequences as well as the insertion site were initially unknown (Supplementary Fig. 1e).
To determine the sequences of the barcodes in each colony, combinatorial pools of the individual mutants were generated, with DNA extracted, and barcodes amplified and deep-sequenced. The combinatorial pooling patterns were designed so that each colony was included in a different combination of pools, allowing us to determine the barcode sequences associated with individual colonies based on which pools the sequences were found in (Supplementary Fig. 1f and Supplementary Fig. 2a-e; Supplementary Note). This procedure was similar in concept to the approach we used in our pilot study9, but it consumed significantly less time because we used a simple PCR amplifying only the barcodes instead of a multi-step flanking sequence extraction protocol (ChlaMmeSeq58) on each combinatorial pool.
To determine the insertion site associated with each barcode, the library was pooled into a single sample or six separate samples. The barcodes and their flanking genomic DNA were PCR amplified using LEAP-Seq9 (Supplementary Fig. 1g and Supplementary Fig. 2f-j; Supplementary Note). The flanking sequences associated with each barcode were obtained by paired-end deep sequencing59,60. The final product is an indexed library in which each colony has known flanking sequences that identify the genomic insertion site, and barcode sequences that facilitate pooled screens in which individual mutants can be tracked by deep sequencing (Fig. 3a).
Insertion verification PCR.
The PCR reactions were performed in two steps to verify the insertion site9 (Supplementary Table 6): (1) Genomic locus amplification: genomic primers that are ~1 kb away from the flanking genomic sequence reported by LEAP-Seq were used to amplify the genomic locus around the flanking sequence. If wild type produced the expected PCR band but the mutant did not produce it or produced a much larger product, this indicated that the genomic locus reported by LEAP-Seq may be disrupted by the insertional cassette and we proceeded to the second step; (2) Genome-cassette junction amplification: one primer binding to the cassette (oMJ913 for the 5’ side and oMJ944 for the 3’ side, Supplementary Table 6) and the other primer binding to flanking Chlamydomonas genomic DNA (one of the genomic primers from the first step) were used to amplify the genome-cassette junction. If the mutant produced a PCR band with expected size that was confirmed by sequencing but wild type did not produce the expected PCR band, we categorized this insertion as “confirmed.” In some mutants, genomic primers surrounding the site of insertion did not yield any PCR products in wild type or the mutant even after several trials, possibly due to incorrect reference genome sequence or local PCR amplification difficulties. These cases were grouped as “failed PCR” and were not further analyzed.
72 mutants (24 insertions each for confidence levels 1 and 2, confidence level 3 and confidence level 4) were chosen randomly from the library and tested. The genomic DNA template was prepared from a single colony of each mutant using the DNeasy Plant Mini Kit (69106, Qiagen). The PCRs were performed using the Taq PCR core kit (201225, Qiagen) as described before58. PCR products of the expected size were verified by Sanger sequencing.
Southern blotting.
Southern blotting was performed as previously described in detail9. Genomic DNA was digested with StuI enzyme (R0187L, New England Biolabs) and separated on a 0.7% Tris-borate-EDTA (TBE) agarose gel. The DNA in the gel was depurinated in 0.25 M HCl, denatured in a bath of 0.5 M NaOH, 1M NaCl, neutralized in a bath of 1.5 M Tris-HCl, pH 7.4, 1.5 M NaCl, and finally transferred onto a Zeta-probe membrane (1620159, Bio-Rad) overnight using the alkaline transfer protocol given in the manual accompanying the membrane. On the next day, the membrane was gently washed with saline-sodium citrate (2xSSC: 0.3 M NaCl, 0.03 M sodium citrate), dried with paper towel, and UV cross-linked twice using the Stratalinker1800 (Stratagene). For probe generation, the AphVIII gene on CIB1 was amplified using primers oMJ588 and oMJ589 (Supplementary Table 1). The PCR product was purified and labeled according to the protocol of Amersham Gene Images AlkPhos Direct Labeling and Detection System (RPN3690, GE Healthcare). The membrane was hybridized at 60°C overnight with10 ng probe/mL hybridization buffer. On the next day, the membrane was washed with primary and secondary wash buffers and then visualized using a CL-XPosure film (34093, Thermo Fisher).
Analyses of insertion distribution and identification of hot/cold spots.
A mappability metric was defined to quantify the fraction of all possible flanking sequences from any genomic region that can be uniquely mapped to that region58. Calculation of mappability, hot/cold spot analysis and simulations of random insertions were performed as described previously58, except that a 30 bp flanking sequence lengths instead of a mix of 20 bp and 21 bp was used (because we now use 30 bp flanking sequence data derived from LEAP-Seq, rather than 20/21 bp ChlaMmeSeq sequences), and the v5.5 Chlamydomonas genome instead of the v5.3 genome was used12. This analysis was done on the original full set of mapped insertions, to avoid introducing bias from the choice of mutants into the consolidated set. The hot/cold spot analysis was performed on confidence level 1 insertions only, to avoid introducing bias caused by junk fragments and their imperfect correction. The full list of statistically significant hot/cold spots is provided in Supplementary Table 7.
Identification of underrepresented gene ontology (GO) terms.
For each GO category, we calculated the total number of insertions in all genes annotated with the GO term and the total mappable (mappability defined in a Supplementary Note) length of all such genes, and compared them to the total number of insertions in and total mappable length of the set of flagellar proteome genes11. We compared these numbers using Fisher’s exact test, and did correction for multiple comparisons61 to obtain the false discovery rate (FDR). This analysis was done on the original full set of mapped insertions to avoid introducing bias from the choice of mutants into the consolidated set. We decided to use the flagellar proteome as the comparison set because flagellar genes are very unlikely to be essential; we did not use intergenic insertions or the entire genome because we know that the overall insertion density differs between genes and intergenic regions. The statistically significant results are listed in Supplementary Table 8.
Prediction of essential genes.
To predict essential genes in Chlamydomonas, we sought to generate a list of genes that have fewer insertions than would be expected randomly was generated. Among them, those with 0 insertion are considered candidate essential genes.
To achieve these, for each gene, we calculated the total number of insertions in that gene and the total mappable length of that gene, and compared them to the total number of insertions in and total mappable length of the set of flagellar proteome genes11, as what we have performed on each GO category. The resulting list of genes with statistically significantly fewer insertions than expected is discussed in a Supplementary Note and shown in Supplementary Table 9: this includes 203 genes with no insertions, and 558 genes with at least one insertion. However, only genes 5 kb or longer yield a false discovery rate (FDR) of 0.05 or less when they have no insertions - our overall density of insertions is not high enough to detect smaller essential genes.
Pooled Screens.
Library plates that were replicated once every four weeks onto fresh medium were switched to a 2-week replication interval to support uniform colony growth before pooling. Cells were pooled from 5-days-old library plates: first, for each set of eight agar plates, cells were scraped using the blunt side of a razor blade (55411–050, VWR) and resuspended in 40 mL liquid TAP medium in 50-mL conical tubes. Second, cells clumps were broken up by pipetting, using a P200 pipette tip attached to a 10-mL serological pipette. In addition, cells were pipetted through a 100 µm cell strainer (431752, Corning). Third, these sub-pools were combined as the master pool representing the full library.
The master pool was washed with TP, and resuspended in TP. Multiple aliquots of 2 × 108 cells were pelleted by centrifugation (1,000g, 5 min, room temperature) and the supernatant was removed by decanting. Some aliquots were used for inoculation of pooled cultures, whereas other aliquots were frozen at −80 °C as initial pool samples for later barcode extraction to enable analysis of reproducibility between technical replicates. For pooled growth, 20 L TAP or TP in transparent Carboy containers (2251–0050, Nalgene) were inoculated with the initial pool to a final concentration of 2× 104 cells/mL. Cultures were grown under 22°C, mixed using a conventional magnetic stir bar and aerated with air filtered using a 1 µm bacterial air venting filter (4308, Pall Laboratory). The TAP culture was grown in dark. For the two replicate TP cultures, the light intensity measured at the surface of the growth container was initially 100 µmol photons m−2 s−1, and then increased to 500 µmol photons m−2 s−1 after the culture reached ~2× 105 cells/mL. When the culture reached the final cell density of 2× 106 cells/mL after 7 doublings, 2× 108 cells were pelleted by centrifugation (1,000g, 5 min, room temperature) for DNA extraction and barcode sequencing.
Molecular characterization of the cpl3 mutant.
Mutant genotyping PCRs were performed as previously described9. To complement the cpl3 mutant, the wild-type CPL3 gene was PCR amplified and cloned into the vector pRAM118 vector that contains the aph7’’ gene62, which confers resistance to hygromycin B. In this construct, the expression of CPL3 is under the control of the PSAD promoter. The construct was linearized before being transformed into the cpl3 mutant. Transformants were robotically arrayed and assayed in colony sizes in the presence and absence of acetate respectively (Supplementary Fig. 6, c and d). Three representative lines that showed rescued photosynthetic growth were used in further phenotypic analyses (Fig. 4).
Analyses of growth, chlorophyll, and photosynthetic electron transport.
For all physiological and biochemical characterizations of cpl3 below, we grew cells heterotrophically in the dark to minimize secondary phenotypes due to defects in photosynthesis.
For spot assays, cells were grown in TAP medium in dark to log phase to around 106 cells per mL. Cells were washed in TP and spotted onto solid TAP medium and TP medium respectively. The TAP plates were incubated in dark for 12 d before being imaged. The TP plates were incubated under 30 µmol photons m−2 s−1 light for 1 d, 100 µmol photons m−2 s1 light for 1 d, and then 500 µmol photons m−2 s−1 light for 4 d.
Chlorophyll a and b concentrations were measured as previously described63 using TAP-dark grown cells. We used TAP-dark-grown instead of TP-light-grown cells for chlorophyll analyses, photosynthetic performance analyses, microscopy, proteomics, and western blots (below) to avoid observing secondary effects due to the photosynthetic defects of the cpl3 mutants.
To measure photosynthetic electron transport rate, TAP-dark grown cells were collected, re-suspended in fresh TAP medium, and dark acclimated for 20 min. Cells were then measured in chlorophyll fluorescence under a series of increasing light intensities using the “Light Curve” function on a DUAL-PAM-100 fluorometer (Walz). PSII quantum yield (ΦPSII) was quantified as previously described64. Relative electron transport rate (rETR) was calculated according to the following equation rETR = ΦPSII x I. I represents the emitted irradiance.
Proteomics.
TAP-dark-grown cells were collected by centrifugation and flash-frozen. Proteins were extracted from the frozen pellets by resuspension in lysis buffer (6M guandium Hydrochloride, 10mM tris(2-carboxyethyl)phosphine, 40mM chloroacetamide, 100mM Tris pH8.5, 1x MS-Safe protease inhibitor, 1x Phosphatase inhibitor cocktail II), grinding with liquid nitrogen, followed by sonication. Protein lysates were then digested with trypsin (Promega) into peptides. Three biological replicates were processed for each strain.
The samples were labeled with tandem mass tags (TMTs), multiplexed and then fractionated before tandem mass spectrometry analyses. Briefly, each sample was labeled with the TMT labeling reagent (Thermo Fisher) according to the manufacturer’s instructions. The samples were then mixed in equimolar amounts and desalted using C18-stage tips65. The dried peptide mix was then separated using strong cation exchange (SCX) stage-tips66 into four fractions. Each of the four fractions were then diluted with 1% trifluoroacetic acid (TFA) and separated into three fractions using SDB-RPS stage tips. This procedure initially resulted in a total of 12 fractions. Fractions 1–3 (the children of the first SCX fraction) were pooled together yielding 10 final fractions. Each final fraction was diluted and injected per run using an Easy-nLC 1200 UPLC system (Thermo Fisher). Samples were loaded onto a nano capillary column packed with 1.9 µm C18-AQ (Dr. Maisch) mated to metal emitter in-line with a Fusion Lumos (Thermo Fisher). Samples were eluted using a split gradient of 10–20% solution B (80% ACN with 0.1% FA) in 32 min and 20–40% solution B in 92 min followed column wash at 100% solution B for 10 min. The mass spectrometer was operated in a data-dependent mode with the 60,000 resolution MS1 scan (380–1500 m/z), AGC target of 4e5 and max injection time of 50ms. Peptides above threshold 5e3 and charges 2–7 were selected for fragmentation with dynamic exclusion after 1 time for 60 s and 10 ppm tolerance. MS1 isolation windows of 1.6m/z, MS2 isolation windows 2 and HCD NCE of 55% were selected. MS3 fragments were detected in the Orbitrap at 50,000 resolution in the mass range of 120–500 with AGC 5e4 and max injection time of 86 ms. The total duty cycle was set to 3.0 sec.
Raw files were searched with MaxQuant67, using default settings for MS3 reporter TMT 10-plex data. Files were searched against sequences of nuclear, mitochondrial, and chloroplast-encoded Chlamydomonas proteins supplemented with common contaminants12,68,69. Raw files were also analyzed within the Proteome Discoverer (Thermo Fisher) using the Byonic70 search node (Protein Metrics). Data from Maxquant and Proteome Discoverer were combined in Scaffold Q+ (Proteome Software Inc.), which was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 96.0% probability and contained at least 2 identified peptides. Scaffold Q+ un-normalized data were exported in the format of the log2 value of the reporter ion intensities, which reflect the relative abundances of the same protein among different samples multiplexed. Each sample was then normalized to a median of 0 (by subtracting the original median from the raw values, since the values are log2). For each gene, for each pair of samples, the normalized log2 intensity values from the three replicates of one sample were compared against those for the other sample using a standard t-test. The resulting P values were adjusted for multiple testing61, yielding a false discovery rate (FDR) for each gene in each pair of samples. We note that our calculation of FDR does not take into account the spectral count of each protein (provided in Supplementary Table 14), which is related to the absolute abundance of the protein and impacts the accuracy of proteomic measurements. Specifically, proteins with a low spectral count are likely of low abundance in cells and often exhibit a large variation in the intensity value between the biological replicates.
Western blotting.
TAP-dark grown cells were pelleted by centrifugation, resuspended in an extraction buffer containing 5 mM HEPES-KOH, pH 7.5, 100 mM dithiothreitol, 100 mM Na2CO3, 2% (w/v) SDS, and 12% (w/v) sucrose, and lysed by boiling for 1 min. Extracted proteins were separated on SDS-PAGE (12% precast polyacrylamide gels, Bio-Rad) using tubulin as a loading and normalization control. Polypeptides were transferred onto polyvinylidene difluoride membranes using a semidry blotting apparatus (Bio-Rad) at 15 volts for 30 minutes. For western blot analyses, membranes were blocked for 1 h at room temperature in Tris-buffered saline-0.1% (v/v) Tween containing 5% powdered milk followed by a 1 h incubation of the membranes at room temperature with the primary antibodies in Tris-buffered saline-0.1% (v/v) Tween containing powdered milk (3% [w/v]). Primary antibodies were diluted according to the manufacturer’s recommendations. All antibodies were from Agrisera and the catalog numbers for the antibodies against CP43, PsaA, ATPC, and α-tubulin were AS11–1787, AS06–172-100, AS08–312, and AS10–680, respectively. Proteins were detected by enhanced chemiluminescence (K-12045-D20, Advansta) and imaged on a medical film processor (Konica) as previously described9.
Additional methods.
Additional method details are provided in a Supplementary Note.
Statistical analyses.
Statistical methods and tests used are provided throughout the manuscript. Fisher’s exact test with Benjamini-Hochberg correction61 for multiple comparisons was used to identify underrepresented gene ontology terms, essential genes, hit genes in the photosynthesis screen, and for the analysis of candidate gene enrichment. The binomial test with Benjamini-Hochberg correction for multiple comparisons was used for the hot/cold spot analysis. A t-test with Benjamini-Hochberg correction for multiple comparisons was used for analysis of the proteomics data. Please see the corresponding Methods or Supplementary Note section for details on each analysis.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary linked to this paper.
Code availability.
All programs written for this work are deposited at GitHub (see URLs).
Data availability.
Mutants’ insertion details and distribution information are available through the CLiP website: https://www.chlamylibrary.org/. The mass spectrometry proteomics data on cpl3 have been deposited to the ProteomeXchange Consortium via the PRIDE71 partner repository with the dataset identifier PXD012560. Other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank O.Vallon for helpful discussions; M.Cahn and Garret Huntress for developing and improving the CLiP website; X.Ji at the Stanford Functional Genomics Facility and Z. Weng at the Stanford Center for Genomics and Personalized Medicine for deep sequencing services; A. Itakura for help in library pooling; S. Ghosh, K. Mendoza, M. LaVoie, L. Galhardo, X. Li, Y. Wang, and Q. Chen for technical assistance; K. Barton, W. Briggs, and Z.-Y. Wang for providing lab space; J. Ecker, L. Freeman Rosenzweig and M. Kafri for constructive suggestions on the manuscript; and the Princeton Mass Spectrometry Facility for proteomics services. This project was supported by a grant from the National Science Foundation (MCB-1146621) awarded to M.C.J. and A.R.G., grants from the National Institutes of Health (DP2-GM-119137) and the Simons Foundation and HHMI (55108535) awarded to M.C.J., a German Academic Exchange Service (DAAD) research fellowship to F.F., Simons Foundation fellowships of the Life Sciences Research Foundation to R.E.J. and J.V.-B., EMBO long term fellowship (ALTF 1450–2014 and ALTF 563–2013) to J.V.-B and S.R., a Swiss National Science Foundation Advanced PostDoc Mobility Fellowship (P2GEP3_148531) to S.R, and the Westlake University startup fund to X.L.
Footnotes
URLs
CLiP website for mutant distribution, https://www.chlamylibrary.org/. Jonikas Lab GitHub repositories of scripts, https://github.com/Jonikas-Lab/Li-Patena-2019/.
Competing interests
The authors declare no competing interests.
References
- 1.Karpowicz SJ, Prochnik SE, Grossman AR & Merchant SS The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286, 21427–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnakumar V et al. Araport: the Arabidopsis information portal. Nucleic Acids Res 43, D1003–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine RP Genetic Control of Photosynthesis in Chlamydomonas Reinhardi. Proc Natl Acad Sci U S A 46, 972–8 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutman BL & Niyogi KK Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol 135, 607–10 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris EH, Stern DB & Witman GB The Chlamydomonas Sourcebook, (Academic Press, 2009). [Google Scholar]
- 6.Rochaix JD Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet 29, 209–30 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Li JB et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–52 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Silflow CD & Lefebvre PA Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol 127, 1500–7 (2001). [PMC free article] [PubMed] [Google Scholar]
- 9.Li X et al. An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terashima M, Specht M & Hippler M The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57, 151–68 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Pazour GJ, Agrin N, Leszyk J & Witman GB Proteomic analysis of a eukaryotic cilium. J Cell Biol 170, 103–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merchant SS et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–251 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zones JM, Blaby IK, Merchant SS & Umen JG High-Resolution Profiling of a Synchronized Diurnal Transcriptome from Chlamydomonas reinhardtii Reveals Continuous Cell and Metabolic Differentiation. Plant Cell (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duanmu D et al. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc Natl Acad Sci U S A 110, 3621–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent RM et al. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J 82, 337–351 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Allen JF, de Paula WB, Puthiyaveetil S & Nield J A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci 16, 645–55 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Giordano M, Beardall J & Raven JA CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56, 99–131 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Goldschmidt-Clermont M & Rahire M Sequence, evolution and differential expression of the two genes encoding variant small subunits of ribulose bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. J Mol Biol 191, 421–32 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Suzuki Y, Arae T, Green PJ, Yamaguchi J & Chiba Y AtCCR4a and AtCCR4b are Involved in Determining the Poly(A) Length of Granule-bound starch synthase 1 Transcript and Modulating Sucrose and Starch Metabolism in Arabidopsis thaliana. Plant Cell Physiol 56, 863–74 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Wang H et al. The global phosphoproteome of Chlamydomonas reinhardtii reveals complex organellar phosphorylation in the flagella and thylakoid membrane. Mol Cell Proteomics 13, 2337–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassi R, Soen SY, Frank G, Zuber H & Rochaix JD Characterization of Chlorophyll-a/B Proteins of Photosystem-I from Chlamydomonas-Reinhardtii. J Biol Chem 267, 25714–25721 (1992). [PubMed] [Google Scholar]
- 22.Sager R & Zalokar M Pigments and photosynthesis in a carotenoid-deficient mutant of Chlamydomonas. Nature 182, 98–100 (1958). [DOI] [PubMed] [Google Scholar]
- 23.Baek K et al. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep 6, 30620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Brueggeman AJ, Horken KM, Plucinak TM & Weeks DP Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell 13, 1465–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin SE et al. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep 6, 27810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slaninová M, Hroššová D, Vlček D & Wolfgang W Is it possible to improve homologous recombination in Chlamydomonas reinhardtii? Biologia 63, 941–46 (2008). [Google Scholar]
- 27.Greiner A et al. Targeting of Photoreceptor Genes in Chlamydomonas reinhardtii via Zinc-finger Nucleases and CRISPR/Cas9. Plant Cell (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferenczi A, Pyott DE, Xipnitou A & Molnar A Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Natl Acad Sci U S A (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu XL, Yu HD, Guan Y, Li JK & Guo FQ Carbonylation and loss-of-function analyses of SBPase reveal its metabolic interface role in oxidative stress, carbon assimilation, and multiple aspects of growth and development in Arabidopsis. Mol Plant 5, 1082–99 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Klein RR & Houtz RL Cloning and developmental expression of pea ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol Biol 27, 249–61 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Johnson X et al. MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22, 234–48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L et al. Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 113, 12586–12591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y & Spalding MH An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 103, 10110–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao H, Sage TL & Osteryoung KW FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc Natl Acad Sci U S A 103, 6759–64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinis J et al. ABC1K1/PGR6 kinase: a regulatory link between photosynthetic activity and chloroplast metabolism. Plant J 77, 269–83 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Kim EH, Lee Y & Kim HU Fibrillin 5 Is Essential for Plastoquinone-9 Biosynthesis by Binding to Solanesyl Diphosphate Synthases in Arabidopsis. Plant Cell 27, 2956–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre-Legendre L et al. Loss of phylloquinone in Chlamydomonas affects plastoquinone pool size and photosystem II synthesis. J Biol Chem 282, 13250–63 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Wilde A, Lunser K, Ossenbuhl F, Nickelsen J & Borner T Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem J 357, 211–6 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockel J, Bennewitz S, Hein P & Oelmuller R The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol 141, 870–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinnickel M et al. Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci U S A 113, 2774–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lezhneva L, Amann K & Meurer J The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J 37, 174–85 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Meurer J, Meierhoff K & Westhoff P Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198, 385–96 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Douchi D et al. A Nucleus-Encoded Chloroplast Phosphoprotein Governs Expression of the Photosystem I Subunit PsaC in Chlamydomonas reinhardtii. Plant Cell 28, 1182–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felder S et al. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13, 2127–41 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlotto N et al. The chloroplastic DEVH-box RNA helicase INCREASED SIZE EXCLUSION LIMIT 2 involved in plasmodesmata regulation is required for group II intron splicing. Plant Cell Environ 39, 165–73 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Perron K, Goldschmidt-Clermont M & Rochaix JD A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J 18, 6481–90 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivier C, Goldschmidt-Clermont M & Rochaix JD Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J 20, 1765–73 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobs J et al. Identification of a chloroplast ribonucleoprotein complex containing trans-splicing factors, intron RNA, and novel components. Mol Cell Proteomics 12, 1912–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marx C, Wunsch C & Kuck U The Octatricopeptide Repeat Protein Raa8 Is Required for Chloroplast trans Splicing. Eukaryot Cell 14, 998–1005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Link S, Engelmann K, Meierhoff K & Westhoff P The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol 160, 2202–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schult K et al. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19, 1329–46 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei L et al. LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem 285, 21391–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma J et al. LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19, 1980–93 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Komenda J et al. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283, 22390–9 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Peng L et al. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18, 955–69 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tardif M et al. PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29, 3625–39 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang R et al. High-Throughput Genotyping of Green Algal Mutants Reveals Random Distribution of Mutagenic Insertion Sites and Endonucleolytic Cleavage of Transforming DNA. Plant Cell 26, 1398–1409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubin BE et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci U S A 112, E6634–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wetmore KM et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995). [Google Scholar]
- 62.Berthold P, Schmitt R & Mages W An engineered Streptomyces hygroscopicus aph 7” gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153, 401–12 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Porra RJ, Thompson WA & Kriedemann PE Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) - Bioenergetics 975, 384–94 (1989). [Google Scholar]
- 64.Saroussi SI, Wittkopp TM & Grossman AR The Type II NADPH Dehydrogenase Facilitates Cyclic Electron Flow, Energy-Dependent Quenching, and Chlororespiratory Metabolism during Acclimation of Chlamydomonas reinhardtii to Nitrogen Deprivation. Plant Physiol 170, 1975–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rappsilber J, Ishihama Y & Mann M Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75, 663–70 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Kulak NA, Pichler G, Paron I, Nagaraj N & Mann M Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–24 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–72 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Maul JE et al. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14, 2659–79 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michaelis G, Vahrenholz C & Pratje E Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol Gen Genet 223, 211–6 (1990). [DOI] [PubMed] [Google Scholar]
- 70.Bern M, Kil YJ & Becker C Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics Chapter 13, Unit13 20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mutants’ insertion details and distribution information are available through the CLiP website: https://www.chlamylibrary.org/. The mass spectrometry proteomics data on cpl3 have been deposited to the ProteomeXchange Consortium via the PRIDE71 partner repository with the dataset identifier PXD012560. Other data that support the findings of this study are available from the corresponding author upon request.