Summary
Improving genetic resistance is a preferred method to manage Verticillium wilt of cotton and other hosts. Identifying host resistance is difficult because of the dearth of resistance genes against this pathogen. Previously, a novel candidate gene involved in Verticillium wilt resistance was identified by a genome‐wide association study using a panel of Gossypium hirsutum accessions. In this study, we cloned the candidate resistance gene from cotton that encodes a protein sharing homology with the TIR‐NBS‐LRR receptor‐like defence protein DSC1 in Arabidopsis thaliana (hereafter named GhDSC1). GhDSC1 expressed at higher levels in response to Verticillium wilt and jasmonic acid (JA) treatment in resistant cotton cultivars as compared to susceptible cultivars and its product was localized to nucleus. The transfer of GhDSC1 to Arabidopsis conferred Verticillium resistance in an A. thaliana dsc1 mutant. This resistance response was associated with reactive oxygen species (ROS) accumulation and increased expression of JA‐signalling‐related genes. Furthermore, the expression of GhDSC1 in response to Verticillium wilt and JA signalling in A. thaliana displayed expression patterns similar to GhCAMTA3 in cotton under identical conditions, suggesting a coordinated DSC1 and CAMTA3 response in A. thaliana to Verticillium wilt. Analyses of GhDSC1 sequence polymorphism revealed a single nucleotide polymorphism (SNP) difference between resistant and susceptible cotton accessions, within the P‐loop motif encoded by GhDSC1. This SNP difference causes ineffective activation of defence response in susceptible cultivars. These results demonstrated that GhDSC1 confers Verticillium resistance in the model plant system of A. thaliana, and therefore represents a suitable candidate for the genetic engineering of Verticillium wilt resistance in cotton.
Keywords: calmodulin binding transcription activator (CAMTA), Gossypium hirsutum, nonsynonymous mutation, TIR‐NBS‐LRR gene, Verticillium wilt
Introduction
Resistance (R) genes are key components of genetic interactions between plants and pathogens and are known to activate immunity/resistance responses against pathogen invasion (Chisholm et al., 2006; Dodds and Rathjen, 2010; Jones and Dangl, 2006). The common motifs of R gene products include a nucleotide‐blinding site (NBS), leucine‐rich repeat (LRR), Drosophila Toll domain, the mammalian interleukin‐1 receptor (TIR), coiled‐coil structure (CC), transmembrane domain (TM), and serine/threonine protein kinase domain (PK), which can be grouped into several typical families of TIR‐NBS‐LRRs (TNL), CC‐NBS‐LRR (CNL), nucleotide‐binding site leucine‐rich repeat (NBS‐LRR), LRR‐TM, LRR‐TM‐PK. (Joshi and Nayak, 2011; Martin et al., 2003; McHale et al., 2006). Most disease resistance genes in plants encode NBS‐LRR proteins. The genes encoding NBS‐LRR proteins can be subdivided into the functionally distinct TIR‐domain‐containing (TNL) and CC‐domain‐containing (CNL) subfamilies (McHale et al., 2006). For example, 149 NBS‐LRR proteins are encoded in the genome of Arabidopsis thaliana (Meyers et al., 2003).
To date, numerous NBS‐LRR proteins with roles in mediating plant disease resistance have been identified (Anderson et al., 1997; Ellis et al., 1999; Feuillet et al., 2003; Hinsch and Staskawicz, 1996; Li et al., 2017; Periyannan et al., 2013; Sanseverino et al., 2012; Shen et al., 2007; Wang et al., 2015; Whitham et al., 1994; Zhu et al., 2017). NBS‐LRR proteins generally are composed of tripartite domain architectures, an N‐terminal response domain involved in downstream signalling (CC or TIR are examples), a central molecular switch domain (NB‐ARC, a nucleotide‐binding adaptor shared by the mammalian apoptosis regulator Apaf1, and the Caenorhabditis elegans apoptosis regulator CED4), and a C‐terminal sensor domain‐containing LRRs (Collier and Moffett, 2009; Van der Biezen and Jones, 1998; Maekawa et al., 2011; Meyers et al., 2003; Qi and Innes, 2013).
The simplest model for NBS‐LRR protein function is that they act as receptors that bind effector proteins secreted by pathogens, but only a few such direct interactions have been characterized (Deslandes et al., 2003; Jia et al., 2000). In an alternative model, the ‘guard hypothesis’ predicts that NBS‐LRR proteins act by monitoring the status of plant proteins targeted by pathogen effectors, and that modification of this target by the effector results in the activation of the R protein, which triggers disease resistance in the host (Dangl and Jones, 2001; Van der Biezen and Jones, 1998). For instance, RPM1 (CC‐NBS‐LRR) detects phosphorylation of RPM1‐Interacting Protein 4 (RIN4) by the pathogen effectors from Pseudomonas syringae, and this modification elicits the resistance response (Mackey et al., 2002). In the majority of these interactions, it is the N‐terminus of the TIR or CC domain that is primarily responsible for the interaction with the downstream signalling partner, while the NBS is mainly involved in adenosine triphosphate (ATP) hydrolysis (the ADP [adenosine diphosphate] bound state represents the ‘off’ and the ATP the ‘on’ state) and release of the signalling. The LRR domain is responsible for interaction with signalling partners for the activation or interaction with the upstream activator (Belkhadir et al., 2004; McHale et al., 2006).
In Gossypium spp., R‐genes have been predicted and systematically compared using common motifs (Chen et al., 2015). The diploid Gossypium raimondii genome encodes more than 1000 resistance gene analogues (RGAs) and most of these genes cluster in homology groups based on high levels of protein sequence similarity (Chen et al., 2015). Amongst these, more than 300 G. raimondii RGAs encode NBS domains, largely of the CC‐NBS and CC‐NBS‐LRR subgroups (Paterson et al., 2012; Wei et al., 2013). Systematic analysis and comparison of NBS domain‐containing proteins in G. raimondii revealed 163 NBS genes that contain all five conserved motifs (P‐loop, Kinase2, Kinase3, GLPL and MHDL) (Paterson et al., 2012; Wei et al., 2013), and the disease resistance QTL (quantitative trait loci) were adjacent to the NBS‐encoding genes (Wei et al., 2013).
Verticillium wilt of cotton, caused by the soil‐borne fungus Verticillium dahliae, is a devastating disease that results in major losses in yield and boll quality (Xu et al., 2011b). Developing resistance in cotton cultivars is considered the optimal method to manage Verticillium wilt, which makes identifying Verticillium wilt resistance genes in cotton germplasm and incorporating them into elite cultivars a priority. The Ve1 gene (encoding a receptor‐like protein, LRR‐TM) mediates defence against V. dahliae race 1 strains in tomato (Fradin et al., 2009), and several similar genes have been identified in cotton using candidate homologues, including GbVe, GbVe1, Gbvdr5, GbaVd1 and GbaVd2 (Chen et al., 2017; Yang et al., 2015; Zhang et al., 2011, 2012). However, the Ve1 homologue does not provide adequate resistance in cotton since most of the V. dahliae strains from cotton lack the corresponding Ave1 effector (Song et al., 2018). Several other types of genes have been characterized that contribute to defence responses against Verticillium wilt, including GbCAD1 and GbSSI2 (Gao et al., 2013), GbRLK (Zhao et al., 2013), GbSTK (Zhang et al., 2013b), GbTLP1 (Munis et al., 2010), GbSBT1 (Duan et al., 2016), GhPAO (Mo et al., 2015) and GbNRX1 (Li et al., 2016). However, NBS domain‐containing proteins involved in Verticillium wilt resistance have rarely been reported.
Comparative genomics suggested that expansion and contraction in the numbers of NBS‐encoding genes has altered Verticillium wilt resistance in G. raimondii (nearly immune to the V. dahliae) and Gossypium arboretum (highly susceptible to V. dahliae) (Li et al., 2014). Transcriptome analysis revealed that the NBS‐encoding genes were significantly up‐regulated during infection by V. dahliae (Chen et al., 2015; Li et al., 2014; Xu et al., 2011b; Zhang et al., 2013a). Moreover, the NBS‐encoding genes GbRVd and GbaNA1 contribute to defence responses against Verticillium wilt (Li et al., 2018a; Yang et al., 2016). In our previous study, a Verticillium wilt resistance locus was determined by genome‐wide association study (GWAS) using a panel of 299 G. hirsutum accessions and identified a novel candidate gene CG02 that encodes a TIR‐NBS‐LRR protein (Li et al., 2017), which also shares affinity to the known resistance gene DSC1 from A. thaliana (Lolle et al., 2017) (and the gene is henceforth referred to as GhDSC1 in this study). Since the most widely deployed cultivar of G. hirsutum appears to lack genetic resistance against V. dahliae, and few of NBS‐encoding genes have been identified to confer Verticillium wilt resistance (Cai et al., 2009; Zhang et al., 2011), the pool of these candidate genes could be an important resource to develop resistance in cotton.
The main objectives of the current study were to: 1) investigate the conserved structure of GhDSC1‐encoded proteins and their subcellular localization; 2) explore the relationship between plant hormones and defence responses mediated by GhDSC1; 3) study the role of the GhDSC1 in Verticillium wilt resistance of A. thaliana transgenic lines using the DSC1 orthologous mutants of A. thaliana; 4) explore the defence responses mediated by GhDSC1; and 5) investigate the allelic divergence of GhDSC1 between resistant and susceptible accessions of G. hirsutum.
Results
GhDSC1 encodes a TIR‐NBS‐LRR protein
We previously identified a candidate gene, CG02 that encodes a NBS‐LRR protein, contributing to Verticillium wilt resistance by virus‐induced gene silencing (VIGS) in upland cotton (Li et al., 2017) and was subsequently named GhDSC1. To further investigate the role of GhDSC1 in Verticillium wilt resistance, DNA or cDNA sequences were identified from the resistant G. hirsutum cv. Zhongzhimian No. 2 at the genomic and transcriptional levels by Polymerase Chain Reaction (PCR). Sequencing results revealed that the full‐length, 3234 bp GhDSC1 cDNA (Accession No.:Gh_A10G2076) encodes a protein of 1077 amino acids (aa), and that there is a single intron of 85 bp in the genomic GhDSC1 sequence (Fig. S1). Prediction of the protein sequence structure by the web‐based programme SMART showed that GhDSC1 is a TIR‐NBS‐LRR protein that contains TIR, NBS and LRR domains (Fig. 1A). Phylogenetic analysis with known NBS‐LRR family members showed that CG02 is related to the known resistance gene DSC1 from A. thaliana and NgN from Nicotiana glutinosa, based on their clustering into an independent branch of the TIR‐NBS‐LRR family (Fig. 1B). Amino acid sequence alignments consisting of those from DSC1 and NgN showed that several distinctive motifs were present in the GhDSC1‐encoded protein, including P‐loop, RNBS‐A, Kinase 2, RNBS‐B and RNBS‐C, RNBS‐D and MHD motifs (Figs 1C and S2). Furthermore, analysis of the protein sequence by the web‐based programme LRRfinder (Offord and Werling, 2013) showed that the GhDSC1 structure includes four typical LRR domains, and two are leucine‐rich‐repeat C‐terminal (LRRCT) domains (Fig. 1C). Together, these results revealed that the cotton Verticillium wilt resistance candidate gene GhDSC1 encodes a typical TIR‐NBS‐LRR protein structure.
Figure 1.
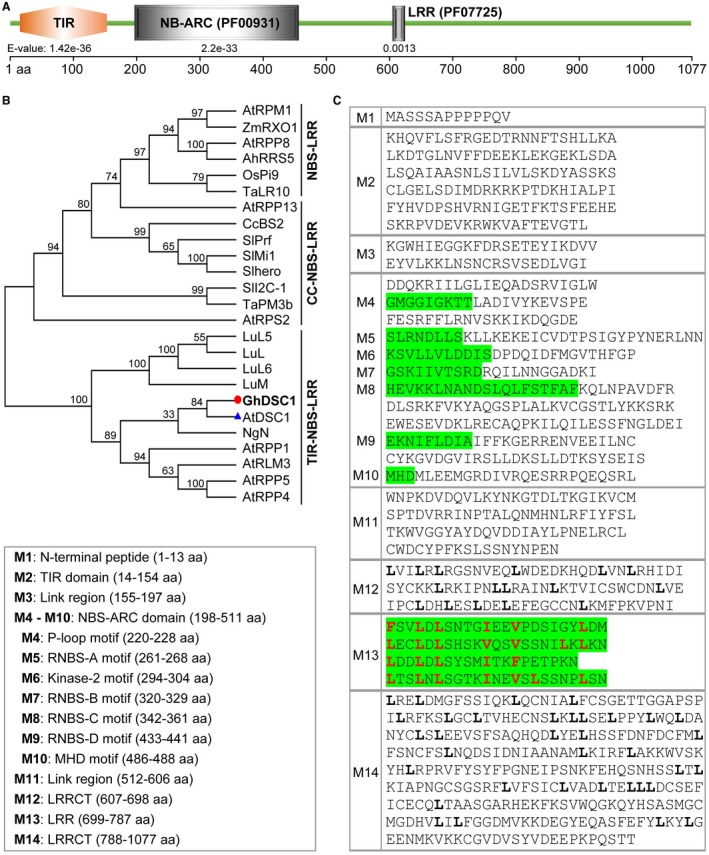
GhDSC1 from Gossypium hirsutum encodes a TIR‐NBS‐LRR protein. (A) Peptide domain prediction in GhDSC1. Conserved domains of GhDSC1 were predicted using the web‐based programme SMART (http://smart.embl-heidelberg.de/). TIR, Toll ‐ interleukin 1 ‐ resistance; NB‐ARC, nucleotide‐binding adaptor shared APAF‐1, R proteins and CED‐4; LRR, leucine‐rich repeat. E‐value represents the confidence of the predicted domains. (B) Phylogenetic tree constructed using GhDSC1 and known NBS‐LRR resistant proteins. The phylogeny was constructed by Mega 6.0, using maximum‐likelihood (Parameters: 1000 bootstraps, Jones‐Taylor‐Thornton model). GhDSC1 and the closest orthologue DSC1 (named AtDSC1 in this figure) from A. thaliana were labelled with a red dot and blue triangle, respectively. The known NBS‐LRR proteins include A. thaliana AtRPM1 (GeneBank: AGC12590.1), AtRPP13 (GeneBank: AAF42831.1), AtRPP8 (GeneBank: BAC67706.1), AtDSC1 (GeneBank: NP_192938.1), AtRPP1 (GeneBank: NP_190034.2), AtRLM3 (GeneBank: AEE83835.1), AtRPP4 (GeneBank: AAM18462.1), AtRPP5 (GeneBank: AAF08790.1) AtRPS2 (GeneBank: AAM90858.1); Solanum lycopersicum SlPrf (GeneBank: AAF76312.1), SlMi1 (GeneBank: AAC97933.1), SlI2C‐1 (GeneBank: AAB63274.1), Slhero (GeneBank: CAD29728.1); Linum usitatissimum LuL6 (GeneBank: AAA91022.1), LuL (GeneBank: AAD25969.1), LuM (GeneBank: AAB47618.1), LuL5 (GeneBank: AAD25972.1); Nicotiana glutinosa NgN (GeneBank: AAA50763.1); Capsicum chacoense CcBS2 (GeneBank: AAF09256.1); Triticum aestivum TaPM3b (GeneBank: AAQ96158.1), TaLR10 (GeneBank: AAQ01784.1); Zea mays ZmRXO1 (GeneBank: AAX31149.1); Oryza sativa OsPi9 (GeneBank: ABB88855.1); Arac hishypogaea AhRRS5 (Zhang et al., 2017). (C) Analysis of GhDSC1 sequence characteristics. Sequence characteristics were drawn by the multiple sequence alignment of the GhDSC1 to known TIR‐NBS‐LRR proteins. The amino acids represented in green indicate conserved motifs (labelled in M1–M14); search of LRRs (L, M and N regions) was conducted using the web‐based programme LRRfinder (http://www.lrrfinder.com/lrrfinder.php), L and N regions were predicted as LRRCT (PF01463: Leucine‐rich‐repeat C‐terminal domain). The leucine (L) residues were marked in bold font. Four LRRs were predicted and the L residues and similar hydrophobic amino acid residues were marked in red font.
GhDSC1 localizes to the cell nucleus
To gain insight into the function of GhDSC1, the subcellular localization was analysed bioinformatically and experimentally by transient expression of a GhDSC1‐green fluorescent protein (GFP) fusion. Prediction of subcellular localization by the web‐based programme Wolf‐Psort (Horton et al., 2007) suggested that GhDSC1 localizes to the cell nucleus (score of nucleus, chloroplast, and plasma is 6, 5 and 1, respectively). According to the prediction information from the web‐based programme cNLS Mapper (Kosugi, et al., 2009), GhDSC1 contains two nuclear localization signals (NLS1 and NLS2) (Figs 2A and S3), indicating that GhDSC1 may localize to the cell nucleus. The subcellular location of the GhDSC1‐GFP fusion examined by transient expression in tobacco showed that GhDSC1 was clearly localized to cell nucleus, in contrast to the fluorescence signal of GFP proteins alone, which was prevalent throughout the foliar cells in tobacco (Fig. 2B). To further confirm the role of NLS1 and NLS2 in localization, individual deletion of NLS1 (GhDSC1DNLS1) or NLS2 (GhDSC1DNLS2) and double‐deletion (GhDSC1DNLS1+2) mutants were constructed for transient expression (Fig. 2A). Interestingly, the effects of the nuclear localization of GhDSC1 could still be observed in individual NLS1 or NLS2 deletion mutants but failed to localize to cell nucleus in NLS1 and NLS2 double‐deletion mutants (Fig. 2B). These results suggested that GhDSC1 is localized to the cell nucleus, and the signal from NLS1 or NLS2 was sufficient for nuclear localization.
Figure 2.
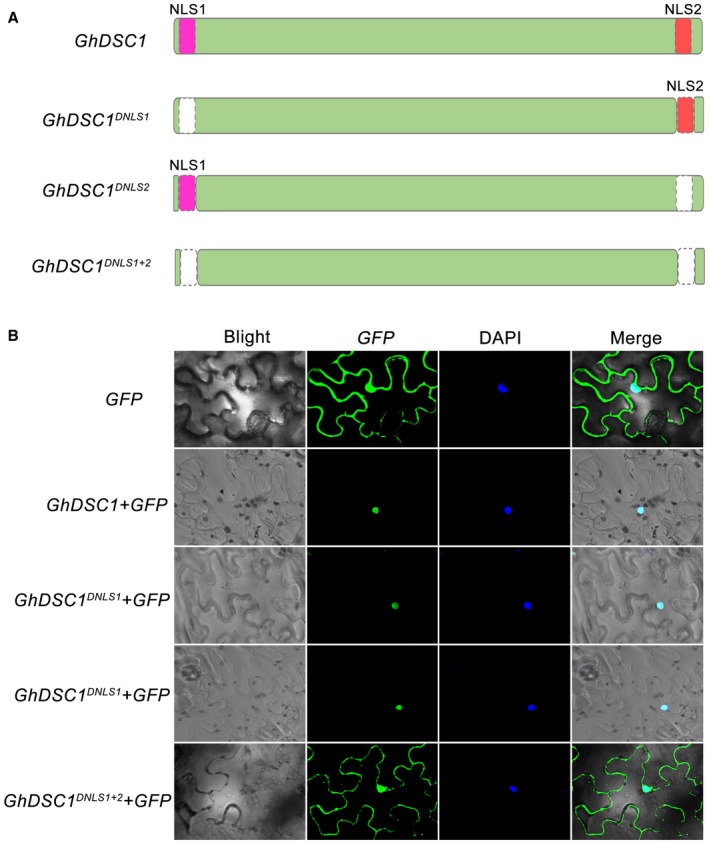
Subcellular localization of GhDSC1 in Nicotiana benthamiana. (A) Structure of nuclear localization signals (NLS) and mutations in GhDSC1. NLS1 and NLS2 represent two nuclear localization signals in GhDSC1. GhDSC1NLS1, GhDSC1NLS2, GhDSC1NLS1+2 represent mutation of NLS1, NLS2 and deletion of NLS1 and NLS2 together, respectively. (B) Subcellular localization of GhDSC1 and the mutant alleles were determined by transient expression of the C‐terminally green fluorescent protein (GFP)‐tagged proteins in N. benthamiana leaves. The fluorescence was scanned by a Leica TCS SP8 confocal microscopy system using × 200 magnification with a excitation at 488 nm and emission at 510 nm. The empty vector 35S::GFP (GFP) was a negative control. Nuclei were stained using 4',6‐diamidino‐2‐phenylindole (DAPI).
GhDSC1 expression is up‐regulated in response to Verticillium wilt and JA signalling in cotton
To test whether GhDSC1 expression correlates with Verticillium wilt resistance, expression patterns of GhDSC1 in resistant and susceptible cotton cultivars were determined during infection by V. dahliae. The expression of GhDSC1 was significantly up‐regulated in the two resistant cultivars, cv. Zhongzhimian No. 2 and cv. AA085, during the early infection stages (especially 6 h–24 h after inoculation) (Fig. 3A). Conversely, in the susceptible cotton cv. Junmian No. 1 and cv. Jimian No. 11, the transcript levels of GhDSC1 did not significantly change until 120 h after inoculation with V. dahliae (Fig. 3A), suggesting that the expression of GhDSC1 positively correlated with Verticillium wilt resistance in cotton. The involvement of GhDSC1 in Verticillium wilt resistance was also evident in the expression pattern of GhDSC1 in different cotton tissues at the adult‐plant stage, which was significantly up‐regulated in root, stem and petiole tissues compared with the expression in leaf at 72 h after flooding with V. dahliae conidial suspension (Fig. S4). To identify signalling pathway(s) linked with GhDSC1, the expression pattern of GhDSC1 was examined following treatment with salicylic acid (SA), ethephon (ETH), methyljasmonate (MeJA) and abscisic acid (ABA), respectively. Interestingly, the expression pattern of GhDSC1 was affected after application of MeJA, but not SA, ETH or ABA (Fig. 3B,C,D,E). These results suggested that GhDSC1 expression is mediated by JA signalling.
Figure 3.
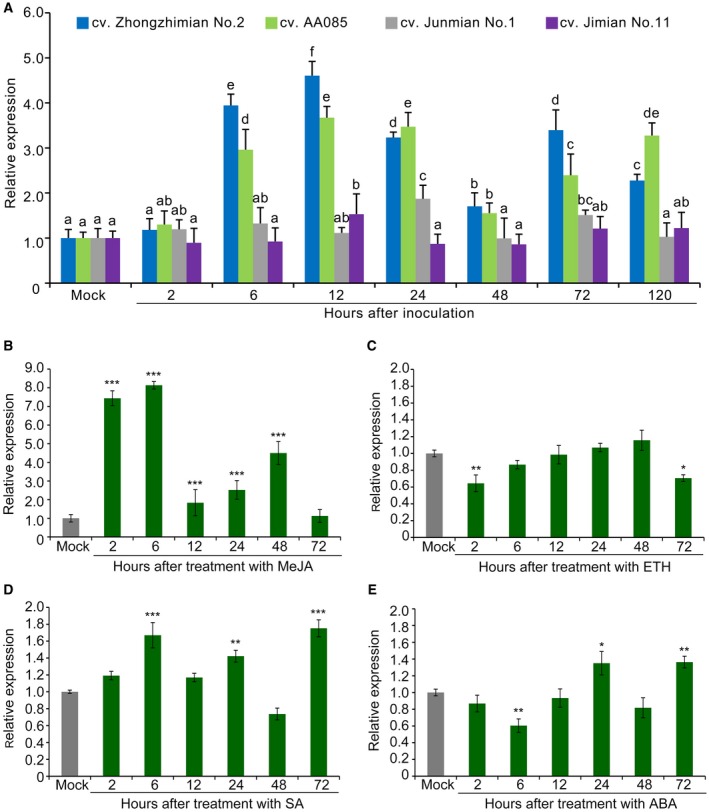
Expression of GhDSC1 in response to Verticillium dahliae infection and hormone signalling in cotton. (A) Expression analysis of GhDSC1 in four cotton cultivars over time after inoculation with V. dahliae strain Vd991 by Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR). Three‐week‐old cotton plants, including two resistance cultivars (cv. Zhongzhimian No. 2 and cv. AA085) and two susceptible cultivars (cv. Junmian No. 1 and cv.Jimian No. 11), were inoculated with conidial suspension (5 × 106 conidia/mL) and harvested at the respective time points. Different letters indicate significant differences at P < 0.01 based on Tukey's HSD. (B–D) GhDSC1 expression in response to the four hormone treatments. The transcript levels of GhDSC1 were detected in 3‐week‐old cotton plants (cv. Zhongzhimian No. 2) treated with the (B) MeJA, (C) ETH, (D) SA and (E) ABA. Relative expression analyses of GhDSC1 by RT‐qPCR was performed using the cotton 18S gene as a reference using the comparative threshold 2‐ΔΔCT method, and relative expression was compared with expression levels in cotton plants that were treated with sterile water (Mock). The values shown represent averages of three independent biological replicates of three plants each. Error bars were calculated based on three biological replicates using standard deviation; asterisks (∗) and double asterisks (∗∗) represents statistical significance of P < 0.05 and P < 0.01, respectively, according to an unpaired Student's t‐tests of each of treatment groups compared with control (Mock).
GhDSC1 enhances resistance to Verticillium wilt in Arabidopsis thaliana
To investigate the role of GhDSC1 in the defence against V. dahliae, GhDSC1 was heterologously expressed in A. thaliana. The GhDSC1 expression construct, in which GhDSC1 expression was driven by the CaMV35S (35S) promoter (P35S::GhDSC1), was transferred into A. thaliana (ecotype Col‐0) via Agrobacterium tumefaciens‐mediated transformation. Positive transgenic lines were verified by PCR and the expression of GhDSC1 was confirmed by Reverse Transcription (RT)‐PCR (Fig. S5A). Six independent GhDSC1‐transgenic lines (T3 generation) were obtained (Fig. S5B). Verticillium wilt resistance was evaluated using the highly virulent V. dahliae strain Vd991 on 4‐week‐old seedlings of three OE transgenic lines (OE1–OE3) that were arbitrarily selected. The results showed that the GhDSC1‐overexpressing lines exhibited significantly enhanced resistance to V. dahliae Vd991, as indicated by reductions in leaf chlorosis and withering compared to the wild‐type Col‐0 (Fig. 4A). Furthermore, real‐time quantitative PCR (qPCR) demonstrated that the GhDSC1‐transgenic lines developed significantly less fungal biomass in planta than the wild‐type A. thaliana plants (Fig. 4B). Thus, GhDSC1 conferred resistance to V. dahliae even after the interfamily transfer into A. thaliana.
Figure 4.
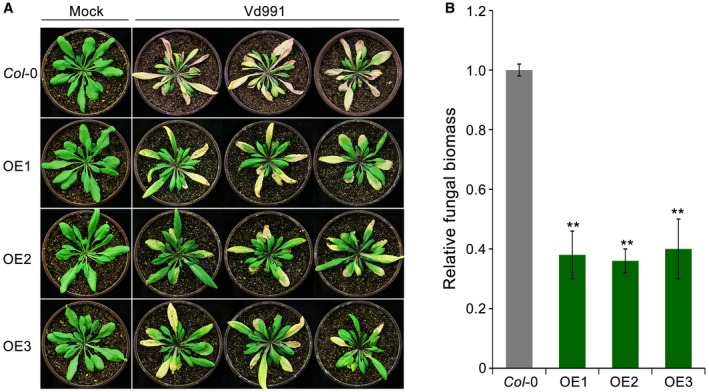
Transgenic expression of GhDSC1 enhances Verticillium wilt resistance in Arabidopsis thaliana. (A) Identification of Verticillium wilt resistance after interfamily transfer of GhDSC1 in A. thaliana. Three‐week‐old seedlings of homozygote transgenic A. thaliana (T3) were inoculated with 5 mL of Verticillium dahliae conidial suspension (5 × 106 conidia/mL). The Verticillium wilt phenotypes were determined and photographed 3 weeks after inoculation. Mock, inoculation with sterile water; OE1/OE2/OE3, GhDSC1 overexpressing transgenic plants. (B) Quantification of V. dahliae biomass in GhDSC1 transgenic A. thaliana plants (OE1, OE2 and OE3) compared to the wild‐type (Col‐0). Genomic DNA was extracted from three whole plants at 21 days after inoculation, and the relative fungal biomass was determined using quantitative Polymerase Chain Reaction (qPCR). V. dahliae elongation factor 1‐α (EF‐1α) was used to quantify fungal colonization, and A. thaliana UBQ5 was used as endogenous plant control. Error bars represent standard errors of three biological replicates, asterisks (∗∗) indicates statistical significance (P < 0.01), according to unpaired Student's t‐tests of plants of each OE compared to the wild‐type (Col‐0).
GhDSC1 can restore Verticillium wilt resistance in the Arabidopsis thaliana dsc1 mutant background
The A. thaliana DSC1 is the orthologue of GhDSC1 (Gh_A10G2076) in G. hirsutum (Zhang et al., 2015). BLASTp analysis using GhDSC1 as a query against A. thaliana proteins returned DSC1 (AT4G12010.1) as the best hit (amino acid identities = 354/1167, 30%; positives = 560/1167, 47%), which also has the typical TIR‐NBS‐LRR motif (Fig. S2), and the closest phylogenetic relationship (Fig. 1B). Because we had identified a role for GhDSC1 in Verticillium wilt resistance, we hypothesized that its orthologue, DSC1, also confers Verticillium wilt resistance in A. thaliana. To test this hypothesis, the sensitivity of DSC1 homozygosis mutant (dsc1, Stock ID in TAIR:SALK_014299) to V. dahliae was examined using the root‐dip inoculation method. The results showed that the dsc1 mutant grew normally as the wild‐type Col‐0 ecotype after disruption of DSC1 in A. thaliana but was more sensitive to V. dahliae compared with the wild‐type Col‐0 ecotype, showing significant leaf chlorosis and wilting 2 weeks after inoculation (Fig. 5A). Investigation of the fungal biomass by qPCR revealed rapid V. dahliae multiplication in the dsc1 lines relative to the wild‐type Col‐0 ecotype (Fig. 5C). These results suggested that the orthologue gene DSC1 is also involved in Verticillium wilt resistance in A. thaliana. To further confirm that the orthologue gene DSC1 is involved in Verticillium wilt resistance, the sensitivity to V. dahliae was assessed in the mutant dsc1 of A. thaliana following the complementation of GhDSC1 driven by the 35S promoter. As expected, inoculation of three separate dsc1 transgenic lines complemented with GhDSC1 displayed significantly less chlorosis and wilting compared with the dsc1 mutants (Fig. 5B). Correspondingly, the fungal biomass was significantly less in the GhDSC1‐recepient dsc1 mutants (Fig. 5C). Therefore, our results showed that both GhDSC1 and its orthologous gene (DSC1) share a common function in contributing to Verticillium wilt resistance in A. thaliana. The heterologously expressed GhDSC1 could also compensate for the Verticillium wilt sensitivity in the A. thaliana dsc1 mutant, providing confirmation that GhDSC1 confers Verticillium wilt resistance.
Figure 5.
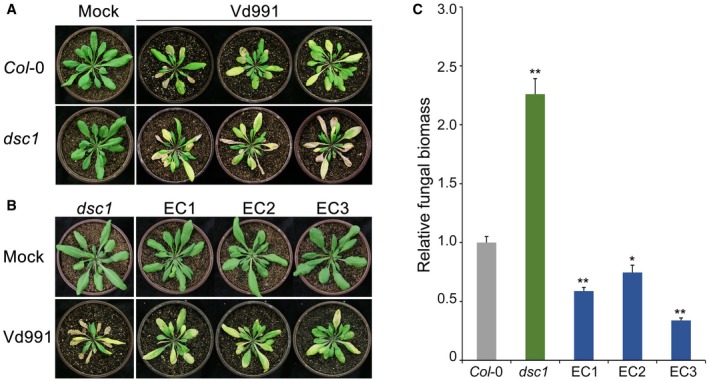
GhDSC1 complements Verticillium wilt resistance in an Arabidopsis thaliana dsc1 mutant. (A) Verticillium wilt phenotype A. thaliana line dsc1, a T‐DNA mutant of DSC1, which is orthologous to GhDSC1. (B) Identification of Verticillium wilt resistance of ectopic transformants in which GhDSC1 was introduced into the dsc1 mutant. Three‐week‐old transgenic (complement transformants) lines (EC1, EC2 and EC3) were subjected to a root‐dip inoculation in a suspension of 5 × 106 conidia/mL of Verticillium dahliae, strain Vd991. Verticillium wilt symptoms were assessed 21 days after inoculation. Sterile water was used in controls (Mock). (C) Quantification of V. dahliae biomass in transgenic GhDSC1 lines of the dsc1 mutants by quantitative Polymerase Chain Reaction (qPCR). Error bars represent standard errors of three biological replicates, asterisks (∗) and double asterisks (∗∗) represents statistical significance of P < 0.05 and P < 0.01, respectively, according to unpaired Student's t‐tests between dsc1 mutants and EC plants compared with the wild‐type (Col‐0).
GhDSC1 modulates defence responses of ROS accumulation and expression of JA‐regulated defence genes
To explore the potential mechanisms of GhDSC1‐mediated Verticillium wilt resistance in A. thaliana, ROS accumulation was assessed in GhDSC1‐transgenic lines during V. dahliae infection. ROS accumulation was assessed in leaves of A. thaliana ecotype Col‐0, transgenic lines overexpressing GhDSC1, dsc1 mutants and in the GhDSC1‐receipient dsc1 mutants following infiltration of a conidial suspension of V. dahliae strain Vd991. Leaves of both the wild‐type Col‐0 and the GhDSC1‐transgenic lines registered an enhanced ROS accumulation around the infiltration sites (indicated by dark brown deposits visible in leaves) 12 h after conidial infiltration, compared with leaves infiltrated with sterile water (Fig. 6A,C). In contrast to the wild type, however, the ROS accumulation in the GhDSC1‐transgenic lines was significantly enhanced 12 h after inoculation with V. dahliae (Fig. 6A). Furthermore, the dsc1 mutants displayed relatively lower ROS accumulation compared to the wild‐type Col‐0 because of the disruption of DSC1 in A. thaliana (Fig. 6A,B). Again, ROS was significantly up‐regulated 12 h after V. dahliae inoculation in the GhDSC1‐recipient dsc1 mutant (Fig. 6B,C). Expression analysis showed that the transcript levels of GhDSC1 were similar in the recipient Col‐0 and dsc1 mutant, which corresponded to the similar ROS accumulation between transgenic lines overexpressing GhDSC1 and the GhDSC1‐receipient dsc1 mutants (Figs 6C and S6). These results suggested that GhDSC1 activates ROS accumulation to enhance Verticillium wilt resistance.
Figure 6.
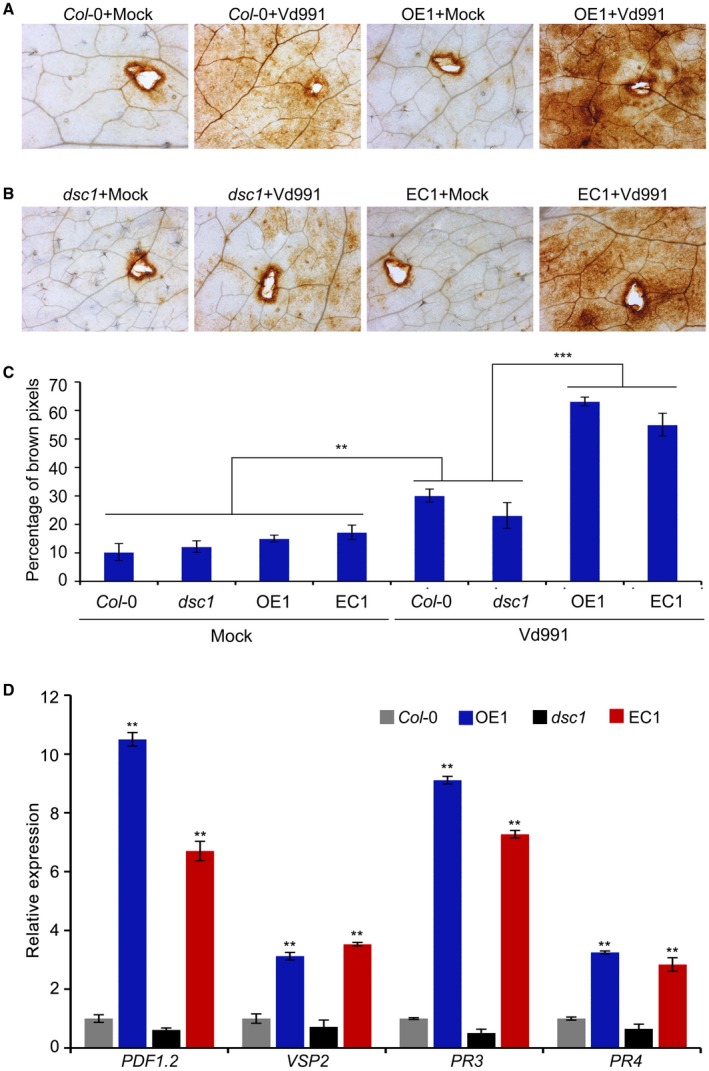
Identification of GhDSC1‐mediated defence responses in Arabidopsis thaliana. (A) Diaminobenzidine staining of ROS accumulation in A. thaliana transgenic line that overexpressed GhDSC1. ROS accumulation was assessed in GhDSC1 transgenic A. thaliana and wide type (Col‐0) leaves from 3‐week‐old plants 12 h after infiltration with a 10 μL suspension (5 × 106 conidia/mL) of Verticillium dahliae strain Vd991. Sterile water treatment was used as a control (Mock). ROS accumulation was captured by the microscopy with 13.5 × amplification under the stereomicroscope. (B) Detection of ROS‐inducing activities of A. thaliana dsc1 mutants and dsc1 mutants after introduction of GhDSC1. (C) The percentages of brown pixels of transgenic plants inoculated with V. dahliae strain Vd991. These included the GhDSC1 overexpression transgenic A. thaliana (OE1), A. thaliana dsc1 mutant that received GhDSC1 (EC1), wild‐type (Col‐0) and the A. thaliana dsc1 mutant. Values are means ± SD from three independent experiments. Asterisks (**) and (***) indicate a significant difference (P < 0.01) and (P < 0.005) relative to the control with sterile water (Mock) based on unpaired Student's t‐test. (D) Identification of the JA signalling‐associated gene expression mediated by GhDSC1 in A. thaliana. Overexpression transgenic line (OE1), dsc1 mutant, transgenic lines of the dsc1 mutant that introduced GhDSC1 (EC1), and wild‐type (Col‐0) were inoculated with a conidial suspension of 5 × 106 conidia/mL of V. dahliae strain Vd991 using a root‐dip method. Leaf samples were collected 24 h after inoculation. Relative expression was assessed by Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) using the comparative threshold 2‐∆∆CT method and A. thaliana UBQ5 as a reference. Values represent averages of three independent biological replicates. Error bars represent standard errors. Double asterisks (∗∗) represent statistical significance of P < 0.01, according to an unpaired Student's t‐tests of each dsc1 mutant, EC1 and OE1 plants compared with the wild‐type (Col‐0).
The expression patterns of GhDSC1 in cotton (Fig. 3A) suggested the involvement of JA signalling. To test this hypothesis, the relative expression of four JA pathway‐regulated genes (PDF1.2, VSP2, PR3 and PR4) were examined in the background of A. thaliana plants in which GhDSC1 was absent or overexpressed. Compared to the transcript levels observed in the wild‐type Col‐0, all of the JA‐regulated genes were significantly up‐regulated in transgenic lines overexpressing GhDSC1. Additionally, these genes were significantly down‐regulated in the dsc1 mutant (Fig. 6D). Therefore, in addition to the activation of ROS accumulation, these results indicated that GhDSC1 activates defence responses through JA signalling to confer Verticillium wilt resistance.
Similar expression patterns of GhCAMTA3 and GhDSC1 in response to Verticillium wilt and JA signalling in cotton
Previous studies had shown that DSC1 functions in part through its association with the Calmodulin Binding Transcription Activator 3 (CAMTA3), which acts as a negative regulator of immunity to inhibit DCS1‐induced autoimmunity (Lolle et al., 2017). Comparative genomics revealed that one gene encodes CAMTA3 in the cotton (G. hirsutum) genome (hereafter referred to as GhCAMTA3) (Zhang et al., 2015). The expression of GhCAMTA3, as affected by GhDSC1, was detected in A. thaliana transgenic lines. The results showed that the transcript levels of GhCAMTA3 were significantly up‐regulated in the GhDSC1‐transgenic line compared with the wild‐type Col‐0, and similar results were recorded in the dsc1 mutant and the GhDSC1‐recipient dsc1 transgenic line, (Fig. 7A). To further explore the relationship between the expression patterns of GhCAMTA3 and GhDSC1, the transcript levels of GhCAMTA3 and GhDSC1 were detected following the same treatments. The expression levels of both GhCAMTA3 and GhDSC1 were also significantly enhanced after application of the MeJA at 6 h to 72 h (Fig. 7B), but not following treatments with ETH, SA and ABA (Fig. S7). Similar to the increased expression of GhDSC1 observed following V. dahliae inoculation, the expression of GhCAMTA3 in resistant plants (cv. AA085 and cv. Zhongzhimian No. 2) was induced 2 h–120 h following V. dahliae inoculation, when compared with the susceptible plants (cv. Junmian No. 1 and cv. Jimian No. 11) (Fig. 7C). The expression pattern of GhCAMTA3 was strikingly similar to patterns observed for GhDSC1, in response to multiple treatments that stimulate plant defence.
Figure 7.
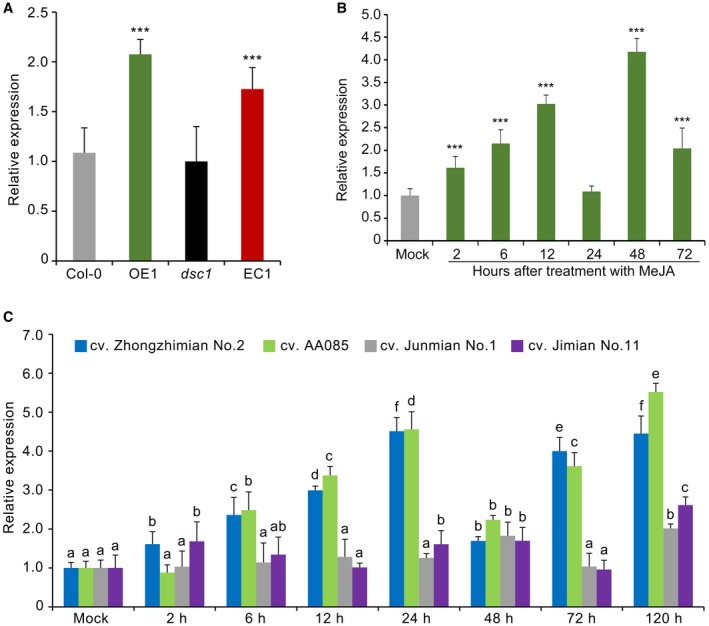
GhDSC1 and GhCAMTA3 show similar expression patterns in cotton. (A) Expression analysis of GhCAMTA3 in the GhDSC1 overexpression transgenic line (OE1), the dsc1 mutant, transgenic lines of dsc1 mutant in which GhDSC1 was introduced (EC1), and wild‐type Col‐0. The respective plants were inoculated with a conidial suspension of 5 × 106 conidia/mL of Verticillium dahliae (strain Vd991) using a root‐dip method. Leaf samples were collected 72 h after inoculation. The transcript relative expression was assessed by Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) using the comparative threshold 2‐∆∆CT method and the Arabidopsis thaliana UBQ5 as a reference. Values represent averages of three independent biological replicates. Error bars represent standard errors. Double asterisks (∗∗) represent statistical significance of P < 0.01, according to an unpaired Student's t‐tests between dsc1 mutants, EC1 and OE1 plants compared with the wild‐type (Col‐0). (B) Expression of GhCAMTA3 in response to MeJA treatment. Transcript levels of GhCAMTA3 were detected in RNA samples from 3‐week‐old cotton plants (cv. Zhongzhimian No. 2) treated with 10 mM MeJA. Asterisks (∗∗) and (***) represent statistical significance at P < 0.01 and P < 0.005, respectively, according to unpaired Student's t‐tests between treatment groups compared with the control group (Mock). (C) Expression analysis of GhCAMTA3 in four cotton cultivars after inoculation with V. dahliae strain Vd991 by RT‐qPCR. The samples of four cotton cultivars were treated as in detection of the expression of GhDSC1. Relative expression analyses of GhDSC1 by RT‐qPCR were performed using the cotton 18S gene as reference using the comparative threshold 2‐ΔΔCT method. Values represent the averages of three independent biological replicates of three plants each. Error bars represent standard errors. Different letters indicate significant differences at P < 0.01 based on Tukey's HSD.
A nonsynonymous mutation in the P‐loop motif of GhDSC1 differentiates resistance and susceptible cotton cultivars
A GWAS revealed that GhDSC1 (CG02) was located in the Verticillium wilt resistance locus in cotton (Li et al., 2017), suggesting that GhDSC1 may be conserved amongst the resistant cotton germplasm. To explore the genetic divergence of GhDSC1 in resistant and susceptible cultivars, nine typical resistant and susceptible G. hirsutum germplasm accessions were selected for sequence analyses (Table S1). The Verticillium wilt resistant germplasm accessions were analysed by both a disease index in a diseased field nursery and in a greenhouse after inoculations with V. dahliae. The average disease indices of the resistant germplasm was 20 compared with 60 for the susceptible germplasm (Fig. 8A,B). Furthermore, open reading frames (ORF) of GhDSC1 homologues were PCR‐amplified and sequenced from all 18 cotton germplasm accessions. Alignments of the ORF sequences showed that the GhDSC1 homologues were highly conserved amongst the 18 cotton accessions, except for 11 single nucleotide polymorphisms (SNPs) (Fig. S8). Of these SNPs, the polymorphism at 673 bp was a guanine (G) in resistant accessions but was cytosine (C) in susceptible accessions (Fig. 8C). This change represented the one nonsynonymous SNP (GGC >> CGC) that resulted in amino acid sequence divergence (225 aa, G >> R) (Fig. 8D). Interestingly, sequence analyses further revealed that this nonsynonymous mutation occurred in the P‐loop motif (Fig. 8D). The P‐loop motif plays a critical role in the functioning of TIR‐NBS‐LRR proteins (Hishida et al., 1999; Traut, 1994). These results indicated that Verticillium wilt resistance function of GhDSC1 may be distinguished by the nonsynonymous mutation between the resistant and susceptible cotton GhDSC1 sequences.
Figure 8.
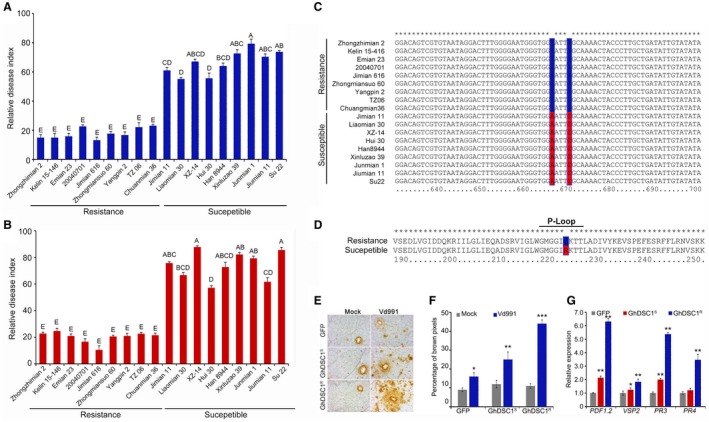
GhDSC1 sequence divergence in resistant and susceptible cotton cultivars and introduction of a P‐loop mutation. (A and B) Relative disease index of the Gossypium hirsutum germplasm accessions in response to Verticillium dahliae. Eighteen cotton germplasm accessions (nine are resistance phenotype) were selected for sequence divergence analysis; the relative disease index of Verticillium wilt was deployed previously (Li et al., 2017), and investigated in field (A) and greenhouse (B) experiments by a root‐dip inoculation method. Bars in green and red colour represent the relative disease index in the field and greenhouse, respectively. Error bars represent standard errors in a sample of three plants. Different letters indicate significant differences at P < 0.01 as calculated by one‐way analysis of variance (ANOVA) with Tukey's HSD used for mean comparisons.(C) Partial alignment of GhDSC1 homologues isolated from all G. hirsutum germplasm accessions. The partial alignment represents the only nonsynonymous mutation site (673 bp, GGC >> CGC) in GhDSC1 amongst G. hirsutum germplasm accessions, the resistant and susceptible nucleotide bases were labelled in green and red colour, respectively. Asterisks (∗) represent nucleotide base conservation. (D) Amino acid alignment of GhDSC1 between resistant and susceptible cotton cultivars. The genotype of GhDSC1 has a nonsynonymous mutation (225 aa, G >> R) in the P‐loop motif, and the amino acid residues of resistant and susceptible cultivars were marked in green and red colour, respectively. (E) Comparison of the ROS accumulation by transient expression between the resistant genotype GhDSC1R and susceptible genotype GhDSC1S. Nicotiana benthamiana leaves from 4‐week‐old plants were agroinfiltrated with GhDSC1R and GhDSC1S, respectively. Two days later, plants were inoculated with a conidial suspension of 5 × 106 conidia/mL of V. dahliae or sterile water (Mock). ROS accumulation stained with DAB solution was detected 2 days after inoculation. Agro‐infiltration of GFP served as a control. (F) The percentages of brown pixels of N. benthamiana after introduction of GFP, GhDSC1R and GhDSC1S. Asterisks (∗), (∗∗) and (∗∗∗) represents statistical significance at P < 0.05, P < 0.01 and P < 0.005 according to unpaired Student's t‐tests of each of the treatment groups compared to the control group (Mock). (G) Expression of JA signalling‐associated genes 2 days after transient expression of GFP, GhDSC1R and GhDSC1S in N. benthamiana. Asterisks (∗) and double asterisks (∗∗) represent statistical significance at P < 0.05 and P < 0.01, respectively, according to unpaired Student's t‐tests of each of the treatment groups compared to the control group.
To determine the functional divergence of GhDSC1, differences in defence response activation were analysed following transient expression of the GhDSC1 resistant genotype (GhDSC1R) and the susceptible genotype (GhDSC1S) and in Nicotiana benthamiana. ROS accumulation was similar 2 days after transient expression of GhDSC1R and GhDSC1S following treatment with sterile water but was significantly higher following inoculations with V. dahliae in the GhDSC1R compared to the GhDSC1S plants (Fig. 8E,F). The transcript levels of four JA‐regulated genes (PDF1.2, VSP2, PR3 and PR4) were significantly up‐regulated after transient expression of GhDSC1R compared to the GhDSC1S (Fig. 8G), further suggesting that the GhDSC1 may efficiently activate the defence response. These results suggested that GhDSC1 is associated with the Verticillium wilt resistance in cotton, and that the sequence divergence causing the P‐loop mutation determines the resistance or susceptibility of cotton to Verticillium wilt.
Discussion
Several genes in cotton that contribute to defence responses against Verticillium wilt have been characterized (Duan et al., 2016; Gao et al., 2013; Li et al., 2016, 2018c, 2016, 2018c; Mo et al., 2015; Munis et al., 2010; Yang et al., 2015, 2016, 2015, 2016; Zhang et al., 2011; 2012; 2013b; 2015), and studies on these genes provide an ever greater understanding of the bases for disease resistance in cotton. In most of these studies, however, candidate genes were identified in an arbitrary model such as conserved homologue cloning that lacks a genetic basis. Genetic methods to identify those genes that play a role in resistance are effective and practical, and also valuable for improved genetic selection by molecular breeding, but the complexity of allotetraploid genome has hitherto prevented such studies in cotton. In our previous study (Li et al., 2017), a GWAS was performed using a population of 299 cotton (G. hirsutum) germplasm accessions and a Verticillium wilt candidate resistance gene, GhDSC1, was identified from the associated locus. In this study, the role of GhDSC1 in Verticillium wilt resistance was examined from several different angles, including its expression and localization in cotton and its heterologous expression and phenotypic characterization representing resistant and susceptible responses in A. thaliana. We also observed that ROS activation and JA signalling were associated with GhDSC1‐mediated Verticillium wilt resistance in cotton.
Most of the disease resistance genes characterized in plants that encode NBS‐LRR proteins play key roles in pathogen resistance by activating defence responses (DeYoung and Innes, 2006). Generally, NBS‐LRR proteins are involved in the recognition of specialized pathogen effectors to activate the innate immunity (defence response) against pathogen invasion in two main mechanisms: detection through direct interaction of plant NBS‐LRR proteins and pathogen‐derived molecules or detection indirectly through the action of their effectors (guard model), which allows the plant to monitor a limited number of key targets of pathogenesis, and responds when those targets are perturbed (Chisholm et al., 2006; Van der Biezen and Jones, 1998; DeYoung and Innes, 2006). The NBS‐LRR family is encoded by hundreds of diverse genes per genome and can be subdivided into two functionally distinct subfamilies of TNL and CNL proteins (McHale et al., 2006), and many of these contribute to resistance (Belkhadir et al., 2004; Joshi and Nayak, 2011). In cotton, the genome encodes a large number of NBS‐LRR proteins (Chen et al., 2015; Khan et al., 2016), which have been a focus of attention for their respective functions in disease resistance, especially for Verticillium wilt.
Comparative genomic analysis showed that the expansion and contraction in the numbers of NBS‐encoding genes in different cotton species alter their resistance to V. dahliae (Li et al., 2014), and many of them are also involved in host responses during infection (Chen et al., 2015; Li et al., 2014; Xu et al., 2011b; Zhang et al., 2013b). However, except for GbRVd and GbaNA1 (Li et al., 2018a; Yang et al., 2016), few have been definitively characterized as contributing to Verticillium wilt resistance. We had previously identified GhDSC1 (typical TIR‐NBS‐LRR) (Fig. 1) by mining the GWAS using a population of G. hirsutum accessions (Li et al., 2017). Following overexpression in A. thaliana, GhDSC1 conferred Verticillium wilt resistance, and also restored resistance in the A. thaliana dsc1 mutant (Figs 4 and 5). This is perhaps the first NBS‐LRR gene associated with Verticillium wilt resistance that was identified and cloned using the genetic screen employed. This demonstrates the utility of such an approach to uncover mechanisms of Verticillium wilt resistance and augment molecular breeding strategies.
Plants have developed complex defence systems against diverse pathogens, systems, which comprise various responses to prevent infection (Caplan et al., 2008; DeYoung and Innes, 2006; Elmore et al., 2011; van Loon et al., 2006). In cotton, the chief defence mechanisms depend on pre‐formed defence structures including a thick cuticle, synthesis of phenolic compounds and delaying the invader through reinforcement of cell walls, accumulation of ROS, and release of phytoalexins (Shaban et al., 2018). For instance, a thioredoxin (GbNRX1) that scavenges apoplastic ROS following the ROS burst upon recognition of V. dahliae is critical for the apoplastic immune response (Li et al., 2016). Similarly, the defence responses mediated by overexpressed GhDSC1 also resulted in the ROS accumulation in A. thaliana (Fig. 6A,C) and following complementation with GhDSC1 in the A. thaliana dsc1 mutant (Fig. 6B,C). ROS accumulation as a Verticillium wilt resistance response also has been reported in association with several other candidate genes, including GbaNA1 (Li et al., 2018a), Gh‐LYK1 and Gh‐LYK2 (Gu et al., 2017) and GbRVd (Yang et al., 2016), suggesting that ROS accumulation plays a critical role in cotton resistance to V. dahliae. Furthermore, hormone‐mediated signalling is one of the most important aspects of this defence mechanism, (Fujita et al., 2006), and SA, JA and ETH are three main hormones contributing to defence against V. dahliae (Duan et al., 2016; Gao et al., 2013; Guo et al., 2016; He et al., 2017; Li et al., 2014, 2018b, 2014, 2018b; Mo et al., 2015; Parkhi et al., 2010; Sun et al., 2014; Wang et al., 2017; Xu et al., 2014; Yang et al., 2015; Zhang et al., 2013b; Zuo et al., 2007). Several functional studies have explored the roles of novel genes implicated in cotton defence, and some of these modulate JA signalling, such as GbSBT1 (Duan et al., 2016), GbSSN (Sun et al., 2014), GhNINJA (Wang et al., 2017), GhJAZ2 and GhbHLH171 (He et al., 2017) and GbSSI1 (Gao et al., 2013). In this study, we found that the expression of GhDSC1 was significantly up‐regulated after treatment with MeJA (Fig. 3B), and marker genes of JA signalling displayed a positive correlation with the presence of GhDSC1 (transgenic lines) (Fig. 6D). Interestingly, the expression levels of GhDSC1 were not altered following treatment with ETH (Fig. 3C), but ETH and JA are usually considered to act synergistically against V. dahliae (Xu et al., 2011a). Thus, these results indicated that GhDSC1‐associated defence responses to Verticillium wilt are mediated via JA signalling in cotton.
Analysis of the characteristic structural features within the translated GhDSC1 sequence revealed characteristics of a typical TIR‐NBS‐LRR protein, orthologous to DSC1 in A. thaliana. The A. thaliana DSC1 encodes a typical TIR‐NBS‐LRR, part of an NLR pair with the TIR‐NBS‐LRR At4g12020 (DSC2), similar to the RPS4 and RRS1 pair in A. thaliana (Narusaka et al., 2009). Interestingly, the GhDSC1 locus encodes another typical TIR‐NBS‐LRR protein (Gh_A10G2077, CG03) that also appears to be a part of an NLR head‐to‐head pair together with GhDSC1 (Li et al., 2017). In A. thaliana, the expression analysis of the majority of NBS‐LRR‐encoding genes showed that DSC1 is affected by SA or flg22 (Meyers et al., 2003). DSC1 is responsible for immunity in N. benthamiana since Agrobacterium expressing DSC1 resulted in HR, and the immunity can be suppressed by CAMTA3 as demonstrated by the co‐inoculation with CAMTA3 that inhibited the DSC1‐induced cell death. This suggests that DSC1 and CAMTA3 represent a guard/guardee pair as proposed by Lolle et al. (2017). Interestingly, the expression patterns of GhCAMTA3 (Gh_D12G0791, CAMTA3 orthologue gene in cotton) and GhDSC1 displayed similarities after inoculation with V. dahliae or treatment with JA (Fig. 7A,B), and the transcript levels of DSC1 were also enhanced after overexpressing GhDSC1 in wild‐type Col‐0 and the dsc1 mutant (Fig. 7B). GhDSC1 was further localized to the cell nucleus, corresponding to the findings of the localization of CAMTA3 and DSC1 that interact together in the cell nucleus (Lolle et al., 2017). However, GhDSC1 and GhCAMTA3 did not display interaction in a yeast two‐hybrid analysis (Fig. S9), suggesting that the interaction between GhDSC1 and GhCAMTA3 and the defence response mediated by both are different in cotton compared to those in A. thaliana. These results indicated that the function of GhDSC1 may be coupled with GhCAMTA3 through modulation of JA signalling.
Interestingly, identification of the sequence polymorphism of GhDSC1 between the resistant and susceptible cotton germplasm accessions showed that a single SNP was responsible for the nonsynonymous mutation in P‐loop motif in GhDSC1 that, resulted in the conserved glycine residue in resistant germplasm accessions and arginine residue in susceptible germplasm accessions. The NBS domain is mainly involved in ATP hydrolysis (the ADP bound state represents the ‘off’ and the ATP the ‘on’ state) and release of signalling, and the binding activity depends on a functional P‐loop motif, which is a glycine‐rich flexible loop containing a highly conserved lysine residue interacting with the phosphates of the nucleotide and with a magnesium cation that coordinates the β‐ and γ‐phosphates (Belkhadir et al., 2004; DeYoung and Innes, 2006; McHale et al., 2006; Qi and Innes, 2013). Previous studies showed that mutation of the glycine residue in the P‐loop resulted in the loss of function of several NBS‐LRR genes, such as RPM1 from A. thaliana and N from tobacco (Dinesh‐Kumar and Baker, 2000; Tornero et al., 2002). In our study, the mutation genotype from susceptible germplasm accessions also showed a reduction in the level of defence responses, including ROS accumulation and regulation of JA signalling‐related genes (Fig. 8E,F,G). In A. thaliana, introducing a P‐loop mutation of glycine in multiple A. thaliana NBS‐LRR proteins (like the DSC1) as the mutation in the susceptible genotype GhDSC1 in cotton also could disrupt their function in disease resistance (Lolle et al., 2017). The mutation in the P‐loop motif may thus underlie the functional divergence of GhDSC1 between the resistant and susceptible cotton accessions and suggests that the TIR‐NBS‐LRR protein encoded by GhDSC1 plays a critical role in Verticillium wilt resistance in cotton.
In conclusion, our study confirmed that GhDSC1 isolated by the genetic methods, encodes a typical TIR‐NBS‐LRR protein that confers Verticillium wilt resistance by modulating the ROS accumulation and JA signalling‐related genes. In addition, sequence divergence of GhDSC1 in G. hirsutum displayed a nonsynonymous mutation that determines susceptibility or resistance in cotton germplasm accessions. Taken together, our study demonstrated that GhDSC1 confers Verticillium wilt resistance, and hence is a valuable candidate for breeding wilt resistance in cotton.
Experimental Procedures
Plant and fungal culture conditions
The highly virulent V. dahliae strain Vd991 (Chen et al., 2018) (used in all experiments) was cultured in complete medium (CM) at 25 °C for 5 days on a shaker. Conidia were harvested by centrifugation and washed with sterile water; the final concentration was adjusted to 5 × 106 conidia/mL using a hemocytometer. A. thaliana seedlings were grown in pots with potting soil (PINDSTRUP, Denmark) including 20% vermiculite in a greenhouse maintained at 24 °C, 60%–70% relative humidity, and under a 16 h/8 h light/dark photoperiod. Cotton plants were grown and maintained in a greenhouse at 28 °C under a 16 h/8 h light/dark photoperiod. N. benthamiana plants were grown at 25 °C for 4 weeks prior to pathogenicity assay and transient expression, under a 14 h/10 h, light/dark regime in greenhouse.
Gene cloning
To clone GhDSC1, 3‐week‐old cotton seedlings of cv. Zhongzhimian No. 2 were inoculated with 5 mL of 5 × 106 conidia/mL conidial suspension, and root samples were collected at 24 h after inoculation. Total RNA was extracted using a Plant RNA Purification Kit (Tiangen, Beijing, China), and cDNA was synthesized by using a RevertAidTM First Strand cDNA Synthesis Kit from MBI (Fermentas, Glen Burnie, Maryland, MA, USA). Primers were designed according to the full ORF of the gene Gh_A10G2076 in the G. hirsutum reference genome (Zhang et al., 2015) (Table S2). Primers were used to amplify the target fragment from genomic DNA and cDNA. The PCR conditions consisted of an initial 95 °C denaturation step for 10 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 45 s, and 72 °C for 3 min. PCR products were cloned into the pGEM‐T‐Easy vector (Promega, Madison, WI, USA), transformed into Escherichia coli DH5α, and confirmed by sequencing.
Sequence analyses
The ORFs of GhDSC1 were determined using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The conserved domains of GhDSC1 were predicted using the web‐based programme SMART (Simple Modular Architecture Research Tool, (http://smart.embl.de) (Letunic and Bork, 2018). A phylogenetic tree was constructed using GhDSC1 and the sequences of other known NBS‐LRR resistance associated proteins by Mega 6.0 with Jones‐Taylor‐Thornton model, using maximum‐likelihood with 1000 bootstrap replicates (Tamura et al., 2013). Sequence characteristics of known TIR, NB‐ARC and LRR domains in GhDSC1 were analysed by the multiple sequence alignment of the GhDSC1 to known TIR‐NBS‐LRR proteins using the ClustalX 1.83 software (Thompson et al., 1997). LRR (L, M and N regions) searches were conducted using the web‐based programme LRRfinder (http://www.lrrfinder.com/lrrfinder.php). The potential subcellular localization of GhDSC1 was deduced using the web‐based programmes of WolfPsort (https://wolfpsort.hgc.jp/), Signal4.1 (http://www.cbs.dtu.dk/services/SignalP/), THHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi).
Subcellular localization of GhDSC1
To study the subcellular localization of GhDSC1 in planta, and whether the nuclear localization signals (NLS) affect their subcellular localization, the full‐length GhDSC1 coding region and also the sequences without one or two NLS peptide were inserted into the pBGFP4 vector to generate a C‐terminal fusion with the GFP sequence under the control of Cauliflower mosaic virus (CaMV) 35S promoter, respectively. Plasmids harbouring GFP alone (empty vector, p35S::GFP) were used as controls. These vectors were transiently expressed in N. benthamiana leaves using Agrobacterium infection (van der Hoorn et al., 2000). The subcellular localization of the above fusion protein was observed 2 days post‐agroinfiltration with a laser scanning confocal microscope (LSMT‐PMT) with excitation at 488 nm and emission at 510 nm. Nuclei were stained with 4',6‐diamidino‐2‐phenylindole (DAPI; Invitrogen, Carlsbad, CA, USA).
Generation and analysis of transgenic Arabidopsis thaliana
The ORF fragments from GhDSC1 were amplified with primers containing NcoI and SpeI enzyme sites and were integrated into the binary vector pCAMBIA1304 under the control of the CaMV35S promoter. The recombinant plasmid (pCAMBIA1304::GhDSC1) was transformed into A. tumefaciens (strain GV3101) and introduced into 4‐week‐old A. thaliana plants (ecotype Col‐0) using an Agrobacterium‐mediated floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 50 mg/L hygromycin, and the T3 homozygous transgenic plants were identified with PCR and RT‐PCR using genomic DNA and cDNA samples, respectively. The wild‐type gDNA and cDNA were used as controls. The amplification conditions consisted of an initial 95 °C denaturation step for 10 min, which was followed by 35 cycles of 95 °C for 45 s, 58 °C for 30 s, and 72 °C for 1 min; and the gene encoding ubiquitin extension protein 5 (UBQ5, NM_116090.3) was used as a control. GhDSC1 was also introduced into the A. thaliana GhDSC1 orthologue gene At4g12010.1 (DSC1) mutant (dsc1, SALK_014299) as described above.
Detection of transgenic plant resistance to Verticillium wilt
The phenotypes of transgenic A. thaliana plants resistant to V. dahliae Vd991 were assayed using a root‐dip method. Three‐week‐old A. thaliana plants were up‐rooted, and the roots were dipped in V. dahliae conidial suspension (5 × 106 conidia/mL) for 5 mins followed by replanting into vermiculite soil. The Verticillium wilt symptoms were recorded 3 weeks after inoculation.
For fungal biomass quantification, roots and stems of three inoculated plants were harvested at 21 days post‐inoculation. Quantitative PCR was performed using a SYBR Premix Ex Taq II kit (Takara, Japan) with primers for the V. dahliae elongation factor 1‐α (EF‐1α) and primers for A. thaliana UBQ5 as an endogenous control (Table S2).
Detection of ROS accumulation
ROS accumulation was detected in transgenic A. thaliana and wild‐type (Col‐0) leaves from 3‐week‐old plants 12 h after infiltration with 10 µL of a V. dahliae (strain Vd991) conidia suspension (2 × 106 conidia/mL) using 3'3‐diaminobenzidine (DAB) solution as previously described (Bindschedler et al., 2006; Thordal‐Christensen et al., 1997). A sterile water treatment was used as the control. For comparing the ROS accumulation after transient expression of the resistance genotype GhDSC1R and susceptible genotype GhDSC1S, each was cloned into a PVX vector pCHF3 and transformed into the A. tumefaciens strain GV3101. Agroinfiltration assays were performed on N. benthamiana plants expressing GFP as a negative control. Four‐week‐old N. benthamiana leaves were agroinfiltrated with 10 µL (OD = 0.8) of GhDSC1R and GhDSC1S A. tumefaciens strains, respectively, then conidial suspensions each of 5 × 106 conidia/mL of V. dahliae were inoculated 2 days later. ROS accumulation was stained with DAB solution for detection at 2 days after treatment. Briefly, the leaves were treated with 1 mg/mL DAB containing 0.05% v/v Tween 20 and 10 mM sodium phosphate buffer (pH 7.0). The leaves were incubated at 25 °C in the dark and infiltrated under gentle vacuum. The reaction was terminated at 10 h–12 h post‐inoculation and the DAB solution was removed with a distilled water rinse. Ethanol (75%) was added to the leaves to remove the chlorophyll and placed in 30% glycerol after the decolourization. Six leaves per treatment were included in each of the three replicates. Samples were observed using a SMZ18 stereo microscope (Nikon, Japan). The percentages of brown pixels were obtained in every image (1 cm2) from six leaves examined for each treatment, and replicates of the same size and resolution were included in calculations using ImageJ software (Rasband, 2012).
Relative gene expression analysis
For relative expression analysis of GhDSC1 and GhCAMTA3 in cotton plants (G. hirsutum cv. Zhongzhimian No. 2, cv. AA085, cv. Junmian No. 1, cv. Jimian No. 11), which differed in resistance to Verticillium wilt, the cotton plants were inoculated with a conidial suspension of 5 × 106 conidia/mL of V. dahliae (strain Vd991) using a root‐dip method upon the development of the first euphylla. The inoculated samples were collected at seven time points (2, 6, 12, 24, 48, 72 and 120 h) after treatment, with three seedlings for each sample. For the expression analysis of GhDSC1 and GhCAMTA3 in cotton after hormone treatment, 4‐week‐old seedlings of G. hirsutum cv. Zhongzhimian No. 2 with first euphylla were sprayed with 10 mM MeJA, 10 mM SA, 5 mM ETH, 100 μM ABA, respectively. The inoculated samples were collected at six time points (2, 6, 12, 24, 48 and 72 h) after treatment, with three seedlings for each sample. For detection of the expression of GhDSC1 in different tissues of G. hirsutum cv. Zhongzhimian No. 2, the different tissues (leaf, root, stem, petiole, flower, boll and seed) were collected 72 h after inoculation for RNA extraction. For analysis of MeJA signalling‐associated genes and GhCAMTA3 expression in different A. thaliana transgenic lines (ecotype Col‐0, GhDSC1 overexpression transgenic Col‐0 mutants, dsc1 mutants, and dsc1 mutants complemented with GhDSC1), each were inoculated with 5 × 106 conidia/mL of V. dahliae (strain Vd991) conidia suspension using a root‐dip method. Three root samples from each treatment were collected at 24 h after inoculation.
RT‐qPCR analyses were performed using the SYBR Premix Ex Taq kit (Takara, Kusatsu, Shiga, Japan) and a QuantStudio 6 Flex Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR cycling programme included an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 20 s. The A. thaliana UBQ5 (At3g62250, Paparella et al., 2014) and N. benthamiana actin (Gui et al., 2017) were amplified as endogenous controls using the primer pairs listed in Table S2. All assays were carried out with three independent biological replicates. The relative expression levels of genes were evaluated using the 2‐∆∆CT method (Livak and Schmittgen, 2001).
Supporting information
Fig. S1 Cloning of GhDSC1 from Gossypium hirsutum. (A) Amplification of GhDSC1 by Reverse Transcription‐Polymerase Chain Reaction (RT‐PCR). RNA was isolated from cotton roots of G. hirsutum cv. Zhongzhimian No. 2 24 h after inoculation with V. dahliae Vd991. GhDSC1 was amplified by RT‐PCR using the cDNA template (cDNA lane). DNA contamination in the RNA sample was assayed by PCR (RNA lane). The GhDSC1 structure was determined by amplification using the genomic DNA (DNA lane). (B) Exon and intron boundaries of GhDSC1 were obtained by comparison of the cDNA sequence to the genomic sequence of GhDSC1.
Fig. S2 Structure‐based multiple sequence alignment of the subdomains in GhDSC1 to known TIR‐NBS‐LRR proteins. The secondary structure assignments of the known TIR‐NBS‐LRR proteins are underlined. Conserved residues are marked by asterisks. N. glutinosa NgN (Genebank ID : AAA50763.1), and A. thaliana AtRPP5 (Genebank ID : AAF08790.1).
Fig. S3 Prediction of nuclear localization signals (NLS) in GhDSC1. Nuclear localization signals prediction of GhDSC1 was conducted using the web‐based programme cNLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). Peptide sequences in red colour represent two nuclear localization signals (NLS1 and NLS2) in GhDSC1.
Fig. S4 Expression analysis of GhDSC1 in different cotton tissues. Plants of 3‐week‐old cotton (cv. Zhongzhimian No. 2) were inoculated with a suspension of 5 × 106 conidia/mL of V. dahliae strain Vd991 using a root‐dip method. Different tissue samples (leaf, Root, Stem, Petiole, Flower, Boll and Seed) were collected 72 h after inoculation for RNA isolation and cDNA synthesis. Relative expression analysis of GhDSC1 was performed by quantitative Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) using the cotton 18S gene as a reference. Values represent the averages of three independent biological replicates of three plants each. Error bars represent standard errors. Asterisks (∗∗) represent statistical significance at P < 0.01, respectively, according to unpaired Student's t‐tests of each of the leaf samples used as control.
Fig. S5 Validation of positive transformants of GhDSC1 transgenic Arabidopsis thaliana lines. Polymerase Chain Reaction (PCR) products targeting a fragment of GhDSC1 amplified from DNA extracted from transgenic lines, (A) GhDSC1‐overexpressing transgenic lines of A. thaliana ecotype Col‐0 and (C) the GhDSC1‐recepient dsc1 mutants. Reverse transcription‐PCR amplification of GhDSC1 cDNA in the same transgenic A. thaliana, (A) GhDSC1‐overexpressing transgenic lines of A. thaliana ecotype Col‐0 and (C) the GhDSC1‐recepient dsc1 mutants, UBQ5 is shown as a control.
Fig. S6 Quantification of GhDSC1 expression in the transgenic line overexpressing GhDSC1 and the GhDSC1‐receipient dsc1 mutant. The transcript levels of GhDSC1 were detected in 3‐week‐old plants grown in Murashige‐Skoog medium. Relative expression analyses of GhDSC1 using Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) was performed using the comparative threshold 2‐ΔΔCT method, and relative expression was compared with expression levels in the transgenic lines overexpressing GhDSC1 compared to the GhDSC1‐receipient dsc1 mutant. Values represent averages of three independent biological replicates of three plants each. Error bars (standard errors of the mean) were calculated based on three biological replicates using standard deviation.
Fig. S7 Quantification of GhCAMTA3 expression in response to ethylene (ETH), salicylic acid (SA) and abscisic acid (ABA) treatment. The transcript levels of GhCAMTA3 were detected in 3‐week‐old cotton plants (cv. Zhongzhimian No. 2) that treated with the ETH, SA and ABA. Relative expression analyses of GhCAMTA3 using Reverse rTanscription‐quantitative Polymerase Chain Reaction (RT‐qPCR) was performed using the cotton 18S gene as a reference using the comparative threshold 2‐ΔΔCT method, and relative expression was compared with expression levels in cotton plants that were treated with sterile water (Mock). Values represent averages of three independent biological replicates of three plants each. Error bars were calculated based on three biological replicates using standard deviation; asterisks (∗) and (∗∗) represent statistical significance at P < 0.05 and P < 0.01, respectively, according to unpaired Student's t‐tests of each of the treatment groups compared to control group (Mock).
Fig. S8 Nucleotide sequence alignment of GhDSC1 in Gossypium hirsutum resistant and susceptible germplasm accessions. The alignment was performed by Clustal X2 with a GONNET 80 protein weight matrix. Only residues that deviate from the reference sequences are shown in the alignment; deletions are indicated by dashes (‐). The polymorphism positions are written vertically, i.e. the first polymorphism occurs at position 177 of the CDS. The position in orange colour (673 bp) represents the nonsynonymous mutation in GhDSC1.
Fig. S9 Yeast two‐hybrid assay of GhDSC1 and GhCAMTA3 proteins. SD/‐LWHA represents the selection medium lacking Leu, Trp, His and Ade. The interaction of murine p53 (p53) and SV40 large T‐antigen (T) was used as a positive control for the system, and human lamin C (lam) was used in the negative interaction control.
Table S1 Information of cotton varieties used in this study.
Table S2 Primers used in this study.
Acknowledgements
This work was supported by the Special Public Welfare Industry Research on Agriculture (201503109), the National Key Research and Development Program of China (2017YFD0201900, 2017YFD0200601), the National Natural Science Foundation of China (31671986, 31471759, 31772245, 31501600, 31671980), the Young Elite Scientists Sponsorship Program by Cast (2016QNRC001), an Agricultural Science and Technology Innovation Program grant to X.F.D, the Fundamental Research Funds for Central Non‐profit Scientific Institution (Y2016CG11, S2016JC05, S2016CG01), and the Outstanding Youth Fund of Jiangsu Province (BK20160016).
Contributor Information
Krishna V. Subbarao, Email: kvsubbarao@ucdavis.edu.
Jie‐Yin Chen, Email: chenjieyin@caas.cn.
Xiao‐Feng Dai, Email: daixiaofeng_caas@126.com.
References
- Anderson, P.A. , Lawrence, G.J. , Morrish, B.C. , Ayliffe, M.A. , Finnegan, E.J. and Ellis, J.G. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine‐rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y. , Subramaniam, R. and Dangl, J.L. (2004) Plant disease resistance protein signaling: NBS‐LRR proteins and their partners. Curr. Opin. Plant Bio. 7, 391–399. [DOI] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Dewdney, J. , Blee, K.A. , Stone, J.M. , Asai, T. , Plotnikov, J. , Denoux, C. , Hayes, T. , Gerrish, C. , Davies, D.R. , Ausubel, F.M. and Bolwell, G.P. (2006) Peroxidase‐dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y.F. , He, X.H. , Mo, J.C. , Sun, Q. , Yang, J.P. and Liu, J.G. (2009) Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: A review. Afr. J. Biotechnol. 8, 7363–7372. [Google Scholar]
- Caplan, J. , Padmanabhan, M. and Dinesh‐Kumar, S.P. (2008) Plant NB‐LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host & Microbe, 3, 126–135. [DOI] [PubMed] [Google Scholar]
- Chen, J.Y. , Huang, J.Q. , Li, N.Y. , Ma, X.F. , Wang, J.L. , Liu, C. , Liu, Y.F. , Liang, Y. , Bao, Y.M. and Dai, X.F. (2015) Genome‐wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.Y. , Li, N.Y. , Ma, X.F. , Gupta, V.K. , Zhang, D.D. , Li, T.G. and Dai, X.F. (2017) The ectopic overexpression of the cotton Ve1 and Ve2‐homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 8, 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.Y. , Liu, C. , Gui, Y.J. , Si, K.W. , Zhang, D.D. , Wang, J. , Short, D.P.G. , Huang, J.Q. , Li, N.Y. , Liang, Y. , Zhang, W.Q. , Yang, L. , Ma, X.F. , Li, T.G. , Zhou, L. , Wang, B.L. , Bao, Y.M. , Subbarao, K.V. , Zhang, G.Y. and Dai, X.F. (2018) Comparative genomics reveals cotton‐specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium . New Phytol. 217, 756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host‐microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collier, S.M. and Moffett, P. (2009) NB‐LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung, B.J. and Innes, R.W. (2006) Plant NBS‐LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh‐Kumar, S.P. and Baker, B.J. (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA, 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Duan, X.P. , Zhang, Z.D. , Wang, J. and Zuo, K.J. (2016) Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS ONE, 11, e153988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. , Luck, J.E. and Dodds, P.N. (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene‐for‐gene specificity. Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, J.M. , Lin, Z.J. and Coaker, G. (2011) Plant NB‐LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 14, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C. , Travella, S. , Stein, N. , Albar, L. , Nublat, A. and Keller, B. (2003) Map‐based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA, 100, 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Ayala, J.C.J. , Castroverde, C.D.M. , Nazar, R.N. , Robb, J. , Liu, C.M. and Thomma, B.P.H.J. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1 . Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Noutoshi, Y. , Takahashi, F. , Narusaka, Y. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. [DOI] [PubMed] [Google Scholar]
- Gao, W. , Long, L. , Zhu, L.F. , Xu, L. , Gao, W.H. , Sun, L.Q. , Liu, L.L. and Zhang, X.L. (2013) Proteomic and virus‐induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae . Mol. Cell. Proteomics, 12, 3690–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z.H. , Liu, T.L. , Ding, B. , Li, F.F. , Wang, Q. , Qian, S.S. , Ye, F. , Chen, T.Z. , Yang, Y.W. , Wang, J.Y. , Wang, G.L. , Zhang, B.L. and Zhou, X.P. (2017) Two lysin‐motif receptor kinases, Gh‐LYK1 and Gh‐LYK2, contribute to resistance against Verticillium wilt in upland cotton. Front. Plant Sci. 8, 2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, Y.J. , Chen, J.Y. , Zhang, D.D. , Li, N.Y. , Li, T.G. , Zhang, W.Q. , Wang, X.Y. , Short, D.P.G. , Li, L. , Guo, W. , Kong, Z.Q. , Bao, Y.M. , Subbarao, K.V. and Dai, X.F. (2017) Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate‐binding module 1. Environ. Microbiol. 19, 1914–1932. [DOI] [PubMed] [Google Scholar]
- Guo, W.F. , Li, J. , Miao, Y.H. , He, X. , Hu, Q. , Guo, K. , Zhu, L.F. and Zhang, X.L. (2016) An ethylene response‐related factor, GbERF1‐like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. [DOI] [PubMed] [Google Scholar]
- He, X. , Zhu, L.F. , Wassan, G.M. , Wang, Y.J. , Miao, Y.H. , Shaban, M. , Hu, H.Y. , Sun, H. and Zhang, X.L. (2017) GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171 . Mol. Plant Pathol. 19, 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch, M. and Staskawicz, B. (1996) Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi . Mol. Plant‐Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- Hishida, T. , Iwasaki, H. , Yagi, T. and Shinagawa, H. (1999) Role of walker motif A of RuvB protein in promoting branch migration of holliday junctions: walker motif A mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of RuvB. J. Biol. Inorg. Chem. 274, 25335–25342. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Laurent, F. , Roth, R. and de Wit, P.J.G.M. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf‐9‐induced and Avr4/Cf‐4‐induced necrosis. Mol. Plant‐Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y.L. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 15, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Joshi, R.K. and Nayak, S. (2011) Functional characterization and signal transduction ability of nucleotide‐binding site‐leucine‐rich repeat resistance genes in plants. Genet. Mol. Res. 10, 2637–2652. [DOI] [PubMed] [Google Scholar]
- Khan, A.M. , Khan, A.A. , Azhar, M.T. , Amrao, L. and Cheema, H.M. (2016) Comparative analysis of resistance gene analogues encoding NBS‐LRR domains in cotton. J. Sci. Food Agric. 96, 530–538. [DOI] [PubMed] [Google Scholar]
- Kosugi, S. , Hasebe, M. , Matsumura, N. , Takashima, H. , Miyamoto‐Sato, E. , Tomita, M. and Yanagawa, H. (2009) Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 284, 478–485. [DOI] [PubMed] [Google Scholar]
- Letunic, I. and Bork, P. (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F.G. , Fan, G.Y. , Wang, K.B. , Sun, F.M. , Yuan, Y.L. , Song, G.L. , Li, Q. , Ma, Z.Y. , Lu, C.R. , Zou, C.S. , Chen, W.B. , Liang, X.M. , Shang, H.H. , Liu, W.Q. , Shi, C.C. , Xiao, G.H. , Gou, C.Y. , Ye, W.W. , Xu, X. , Zhang, X.Y. , Wei, H.L. , Li, Z.F. , Zhang, G.Y. , Wang, J.Y. , Liu, K. , Kohel, R.J. , Percy, R.G. , Yu, J.Z. , Zhu, Y.X. , Wang, J. and Yu, S.X. (2014) Genome sequence of the cultivated cotton Gossypium arboreum . Nat. Genet. 3, 567–572. [DOI] [PubMed] [Google Scholar]
- Li, Y.B. , Han, L.B. , Wang, H.Y. , Zhang, J. , Sun, S.T. , Feng, D.Q. , Yang, C.L. , Sun, Y.D. , Zhong, N.Q. and Xia, G.X. (2016) The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T.G. , Ma, X.F. , Li, N.Y. , Zhou, L. , Liu, Z. , Han, H.Y. , Gui, Y.J. , Bao, Y.M. , Chen, J.Y. and Dai, X.F. (2017) Genome‐wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N.Y. , Ma, X.F. , Short, D.P.G. , Li, T.G. , Zhou, L. , Gui, Y.J. , Kong, Z.Q. , Zhang, D.D. , Zhang, W.Q. , Li, J.J. , Subbarao, K.V. , Chen, J.Y. and Dai, X.F. (2018a) The island cotton NBS‐LRR gene GbaNA1 confers resistance to the non‐race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N.Y. , Zhou, L. , Zhang, D.D. , Klosterman, S.J. , Li, T.G. , Gui, Y.J. , Kong, Z.Q. , Ma, X.F. , Short, D.P.G. , Zhang, W.Q. , Li, J.J. , Subbarao, K.V. , Chen, J.Y. and Dai, X.F. (2018b) Heterologous expression of the cotton NBS‐LRR gene GbaNA1 enhances Verticillium wilt resistance in Arabidopsis. Front. Plant Sci. 9, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T.G. , Zhang, D.D. , Zhou, L. , Kong, Z.Q. , Hussaini, A.S. , Wang, D. , Li, J.J. , Short, D.P.G. , Dhar, N. , Klosterman, S.J. , Wang, B.L. , Yin, C.M. , Subbarao, K.V. , Chen, J.Y. and Dai, X.F. (2018c) Genome‐wide identification and functional analyses of the CRK gene family in cotton reveals GbCRK18 confers Verticillium wilt resistance in Gossypium barbadense . Front. Plant Sci. 9, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lolle, S. , Greeff, C. , Petersen, K. , Roux, M. , Jensen, M.K. , Bressendorff, S. , Rodriguez, E. , Sømark, K. , Mundy, J. and Petersen, M. (2017) Matching NLR immune receptors to autoimmunity in camta3 mutants using antimorphic NLR alleles. Cell Host Microbe, 21, 518–529. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Geraats, B.P. and Linthorst, H.J. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. [DOI] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. III , Wiig, A. and Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis . Cell, 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Maekawa, T. , Cheng, W. , Spiridon, L.N. , Töller, A. , Lukasik, E. , Saijo, Y. , Liu, P. , Shen, Q.H. , Micluta, M.A. , Somssich, I.E. , Frank, L.W. , Takken, F.L.K. , Petrescu, A.J. , Chai, J.J. and Schulze‐Lefer, P. (2011) Coiled‐coil domain‐dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe, 9, 187–199. [DOI] [PubMed] [Google Scholar]
- Martin, G.B. , Bogdanove, A.J. and Sessa, G. (2003) Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- McHale, L. , Tan, X.P. , Koehl, P. and Michelmore, R.W. (2006) Plant NBS‐LRR proteins: adaptable guards. Genome Biol. 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C. , Kozik, A. , Griego, A. , Kuang, H. and Michelmore, R.W. (2003) Genome‐wide analysis of NBS‐LRR‐encoding genes in Arabidopsis. Plant Cell, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, H.J. , Wang, X.F. , Zhang, Y. , Zhang, G.Y. , Zhang, J.F. and Ma, Z.Y. (2015) Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae . Plant J. 83, 962–975. [DOI] [PubMed] [Google Scholar]
- Munis, M.F.H. , Tu, L.L. , Deng, F.L. , Tan, J.F. , Xu, L. , Xu, S.C. , Long, L. and Zhang, X.L. (2010) A thaumatin‐like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 393, 38–44. [DOI] [PubMed] [Google Scholar]
- Narusaka, M. , Shirasu, K. , Noutoshi, Y. , Kubo, Y. , Shiraishi, T. , Iwabuchi, M. and Narusaka, Y. (2009) RRS1 and RPS4 provide a dual Resistance‐gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Offord, V. and Werling, D. (2013) LRRfinder2.0: a webserver for the prediction of leucine‐rich repeats. Innate Immun. 19, 398–402. [DOI] [PubMed] [Google Scholar]
- Paparella, C. , Savatin, D.V. , Marti, L. , De Lorenzo, G. and Ferrari, S. (2014) The Arabidopsis LYSIN MOTIF‐CONTAINING RECEPTOR‐LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 165, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhi, V. , Kumar, V. , Campbell, L.M. , Bell, A.A. , Shah, J. and Rathore, K.S. (2010) Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1 . Transgenic Res. 19, 959–975. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H. , Wendel, J.F. , Gundlach, H. , Guo, H. , Jenkins, J. , Jin, D. , Llewellyn, D. , Showmaker, K.C. , Shu, S.Q. , Udall, J. , Yoo, M.J. , Byers, R. , Chen, W. , Doron‐Faigenboim, A. , Duke, M.V. , Gong, L. , Grimwood, J. , Grover, C. , Grupp, K. , Hu, G.J. , Lee, T.H. , Li, J.P. , Lin, L.F. , Liu, T. , Marler, B.S. , Page, J.T. , Roberts, A.W. , Romanel, E. , Sanders, W.S. , Szadkowski, E. , Tan, X. , Tang, H.B. , Xu, C.M. , Wang, J.P. , Wang, Z.N. , Zhang, D. , Zhang, L. , Ashrafi, H. , Bedon, F. , Bowers, J.E. , Brubaker, C.L. , Chee, P.W. , Das, S. , Gingle, A.R. , Haigler, C.H. , Harker, D. , Hoffmann, L.V. , Hovav, R. , Jones, D.C. , Lemke, C. , Mansoor, S. , Ur Rahman, M. , Rainville, L.N. , Rambani, A. , Reddy, U.K. , Rong, J.K. , Saranga, Y. , Scheffler, B.E. , Scheffler, J.A. , Stelly, D.M. , Triplett, B.A. , Van Deynze, A. , Vaslin, M.F. , Waghmare, V.N. , Walford, S.A. , Wright, R.J. , Zaki, E.A. , Zhang, T.Z. , Dennis, E.S. , Mayer, K.F. , Peterson, D.G. , Rokhsar, D.S. , Wang, X.Y. and Schmutz, J. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature, 492, 423–427. [DOI] [PubMed] [Google Scholar]
- Periyannan, S. , Moore, J. , Ayliffe, M. , Bansal, U. , Wang, X.J. , Huang, L. , Deal, K. , Luo, M.C. , Kong, X.Y. , Bariana, H. , Mago, R. , McIntosh, R. , Dodds, P. , Dvorak, J. and Lagudah, E. (2013) The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science, 341, 786–788. [DOI] [PubMed] [Google Scholar]
- Qi, D. and Innes, R.W. (2013) Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband, W.S. (2012) ImageJ: Image processing and analysis in Java. Astrophysics Source Code Library, 2, 378. [Google Scholar]
- Sanseverino, W. , Hermoso, A. , D'Alessandro, R. , Vlasova, A. , Andolfo, G. , Frusciante, L. , Lowy, E. , Roma, G. and Ercolano, M.R. (2012) PRGdb 2.0: towards a community‐based database model for the analysis of R‐genes in plants. Nucleic Acids Res. 41, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban, M. , Ahmed, M.M. , Sun, H. , Ullah, A. and Zhu, L.F. (2018) Genome‐wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics, 19, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.H. , Saijo, Y. , Mauch, S. , Biskup, C. , Bieri, S. , Keller, B. , Seki, H. , Ulker, B. , Somssich, I.E. and Schulze‐Lefert, P. (2007) Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Song, Y. , Liu, L.L. , Wang, Y.D. , Valkenburg, D.J. , Zhang, X.L. , Zhu, L.F. and Thomma, B.P.H.J. (2018) Transfer of tomato immune receptor Ve1 confers Ave‐dependent Verticillium resistance in tobacco and cotton. Plant Biotechnol. J. 16, 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L.Q. , Zhu, L.F. , Li, X. , Yuan, D.J. , Min, L. and Zhang, X.L. (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley‐powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tornero, P. , Chao, R.A. , Luthin, W.N. , Goff, S.A. and Dangl, J.L. (2002) Large‐scale structure‐function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell, 14, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, T.W. (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide binding sites. Eur. J. Biochem. 222, 9–19. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A. and Jones, J.D. (1998) Plant disease‐resistance proteins and the gene‐for‐gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Wang, G.F. , Ji, J.B. , Ei‐Kasmi, F. , Dangl, J.L. , Johal, G. and Balint‐Kurti, P.J. (2015) Molecular and functional analyses of a maize autoactive NB‐LRR protein identify precise structural requirements for activity. PLoS Pathog. 11, e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wu, S.M. , Zhu, Y. , Fan, Q. , Zhang, Z.N. , Hu, G. , Peng, Q.Z. and Wu, J.H. (2017) Functional characterization of a novel jasmonate ZIM‐domain interactor (NINJA) from upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 112, 152–160. [DOI] [PubMed] [Google Scholar]
- Wei, H.L. , Li, W. , Sun, X.W. , Zhu, S.J. and Zhu, J. (2013) Systematic analysis and comparison of nucleotide‐binding site disease resistance genes in a diploid cotton Gossypium raimondii . PLoS ONE, 8, e68435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S. , Dinesh‐Kumar, S.P. , Choi, D. , Hehl, R. , Corr, C. and Baker, B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Zhu, L.F. , Tu, L.L. , Guo, X.P. , Long, L. , Sun, L.Q. , Gao, W. and Zhang, X.L. (2011a) Differential gene expression in cotton defence response to Verticillium dahliae by SSH. J. Phytopathol. 159, 606–615. [Google Scholar]
- Xu, L. , Zhu, L.F. , Tu, L.L. , Liu, L.L. , Yuan, D.J. , Jin, L. , Long, L. and Zhang, X.L. (2011b) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA‐Seq‐dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhang, W.W. , He, X. , Liu, M. , Zhang, K. , Shaban, M. , Sun, L.Q. , Zhu, J.C. , Luo, Y.J. , Yuan, D.J. , Zhang, X.L. and Zhu, L.F. (2014) Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 65, 6679–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.W. , Ling, X.T. , Chen, T.Z. , Cai, L.W. , Liu, T.L. , Wang, J.Y. , Fan, X.H. , Ren, Y.Z. , Yuan, H.B. , Zhu, W. , Zhang, B.L. and Ma, D.P. (2015) A cotton Gbvdr5 gene encoding a leucine rich‐repeat receptor‐like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Rep. 33, 987–1001. [Google Scholar]
- Yang, J. , Ma, Q. , Zhang, Y. , Wang, X.F. , Zhang, G.Y. and Ma, Z.Y. (2016) Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene, 575, 687–694. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X.F. , Yang, S. , Chi, J.N. , Zhang, G.Y. and Ma, Z.Y. (2011) Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana . Plant Cell Rep. 30, 2085–2096. [DOI] [PubMed] [Google Scholar]
- Zhang, B.L. , Yang, Y.W. , Chen, T.Z. , Yu, W.G. , Liu, T.L. , Li, H.J. , Fan, X.H. , Ren, Y.Z. , Shen, D.Y. , Liu, L. , Dou, D.L. and Chang, Y.H. (2012) Island cotton Gbve1 gene encoding a receptor‐like protein confers resistance to both defoliating and non‐defoliating isolates of Verticillium dahliae . PLoS ONE, 7, e51091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X.F. , Ding, Z.G. , Ma, Q. , Zhang, G.R. , Zhang, S.L. , Li, Z.K. , Wu, L.Q. , Zhang, G.Y. and Ma, Z.Y. (2013a) Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics, 14, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, X.F. , Li, Y.Y. , Wu, L.Z. , Zhou, H.M. , Zhang, G.Y. and Ma, Z.Y. (2013b) Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 32, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zhang, T.Z. , Hu, Y. , Jiang, W.K. , Fang, L. , Guan, X.Y. , Chen, J.D. , Zhang, J.B. , Saski, C.A. , Scheffler, B.E. , Stelly, D.M. , Hulse‐Kemp, A.M. , Wan, Q. , Liu, B.L. , Liu, C.X. , Wang, S. , Pan, M.Q. , Wang, Y.K. , Wang, D.W. , Ye, W.X. , Chang, L.J. , Zhang, W.P. , Song, Q.X. , Kirkbride, R.C. , Chen, X.Y. , Dennis, E. , Llewellyn, D.J. , Peterson, D.G. , Thaxton, P. , Jones, D.C. , Wang, Q. , Xu, X.Y. , Zhang, H. , Wu, H.T. , Zhou, L. , Mei, G.F. , Chen, S.Q. , Tian, Y. , Xiang, D. , Li, X.H. , Ding, J. , Zuo, Q.Y. , Tao, L.N. , Liu, Y.C. , Li, J. , Lin, Y. , Hui, Y.Y. , Cao, Z.S. , Cai, C.P. , Zhu, X.F. , Jiang, Z. , Zhou, B.L. , Guo, W.Z. , Li, R.Q. and Chen, Z.J. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM‐1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Chen, H. , Cai, T.C. , Deng, Y. , Zhuang, R.R. , Zhang, N. , Zeng, Y.H. , Zheng, Y.X. , Tang, R.H. , Pan, R.L. and Zhuang, W.J. (2017) Overexpression of a novel peanut NBS‐LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Gao, Y.L. , Zhang, Z.Y. , Chen, T.Z. , Guo, W.Z. and Zhang, T.Z. (2013) A receptor like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought‐stress tolerance in Arabidopsis. BMC Plant Biol. 13, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Zhang, Z.Y. , Gao, Y.L. , Zhou, L. , Fang, L. , Chen, X.D. , Ning, Z.Y. , Chen, T.Z. , Guo, W.Z. and Zhang, T.Z. (2015) Overexpression of GbRLK, a putative receptor‐like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5, 15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.L. , Lu, C.G. , Du, L.P. , Ye, X.G. , Liu, X. , Coules, A. and Zhang, Z.Y. (2017) The wheat NB‐LRR gene TaRCR90 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 15, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, K.J. , Qin, J. , Zhao, J.Y. , Ling, H. , Zhang, L.D. , Cao, Y.F. and Tang, K.X. (2007) Over‐expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene, 391, 80–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Cloning of GhDSC1 from Gossypium hirsutum. (A) Amplification of GhDSC1 by Reverse Transcription‐Polymerase Chain Reaction (RT‐PCR). RNA was isolated from cotton roots of G. hirsutum cv. Zhongzhimian No. 2 24 h after inoculation with V. dahliae Vd991. GhDSC1 was amplified by RT‐PCR using the cDNA template (cDNA lane). DNA contamination in the RNA sample was assayed by PCR (RNA lane). The GhDSC1 structure was determined by amplification using the genomic DNA (DNA lane). (B) Exon and intron boundaries of GhDSC1 were obtained by comparison of the cDNA sequence to the genomic sequence of GhDSC1.
Fig. S2 Structure‐based multiple sequence alignment of the subdomains in GhDSC1 to known TIR‐NBS‐LRR proteins. The secondary structure assignments of the known TIR‐NBS‐LRR proteins are underlined. Conserved residues are marked by asterisks. N. glutinosa NgN (Genebank ID : AAA50763.1), and A. thaliana AtRPP5 (Genebank ID : AAF08790.1).
Fig. S3 Prediction of nuclear localization signals (NLS) in GhDSC1. Nuclear localization signals prediction of GhDSC1 was conducted using the web‐based programme cNLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). Peptide sequences in red colour represent two nuclear localization signals (NLS1 and NLS2) in GhDSC1.
Fig. S4 Expression analysis of GhDSC1 in different cotton tissues. Plants of 3‐week‐old cotton (cv. Zhongzhimian No. 2) were inoculated with a suspension of 5 × 106 conidia/mL of V. dahliae strain Vd991 using a root‐dip method. Different tissue samples (leaf, Root, Stem, Petiole, Flower, Boll and Seed) were collected 72 h after inoculation for RNA isolation and cDNA synthesis. Relative expression analysis of GhDSC1 was performed by quantitative Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) using the cotton 18S gene as a reference. Values represent the averages of three independent biological replicates of three plants each. Error bars represent standard errors. Asterisks (∗∗) represent statistical significance at P < 0.01, respectively, according to unpaired Student's t‐tests of each of the leaf samples used as control.
Fig. S5 Validation of positive transformants of GhDSC1 transgenic Arabidopsis thaliana lines. Polymerase Chain Reaction (PCR) products targeting a fragment of GhDSC1 amplified from DNA extracted from transgenic lines, (A) GhDSC1‐overexpressing transgenic lines of A. thaliana ecotype Col‐0 and (C) the GhDSC1‐recepient dsc1 mutants. Reverse transcription‐PCR amplification of GhDSC1 cDNA in the same transgenic A. thaliana, (A) GhDSC1‐overexpressing transgenic lines of A. thaliana ecotype Col‐0 and (C) the GhDSC1‐recepient dsc1 mutants, UBQ5 is shown as a control.
Fig. S6 Quantification of GhDSC1 expression in the transgenic line overexpressing GhDSC1 and the GhDSC1‐receipient dsc1 mutant. The transcript levels of GhDSC1 were detected in 3‐week‐old plants grown in Murashige‐Skoog medium. Relative expression analyses of GhDSC1 using Reverse Transcription‐quantitative Polymerase Chain Reaction (RT‐qPCR) was performed using the comparative threshold 2‐ΔΔCT method, and relative expression was compared with expression levels in the transgenic lines overexpressing GhDSC1 compared to the GhDSC1‐receipient dsc1 mutant. Values represent averages of three independent biological replicates of three plants each. Error bars (standard errors of the mean) were calculated based on three biological replicates using standard deviation.
Fig. S7 Quantification of GhCAMTA3 expression in response to ethylene (ETH), salicylic acid (SA) and abscisic acid (ABA) treatment. The transcript levels of GhCAMTA3 were detected in 3‐week‐old cotton plants (cv. Zhongzhimian No. 2) that treated with the ETH, SA and ABA. Relative expression analyses of GhCAMTA3 using Reverse rTanscription‐quantitative Polymerase Chain Reaction (RT‐qPCR) was performed using the cotton 18S gene as a reference using the comparative threshold 2‐ΔΔCT method, and relative expression was compared with expression levels in cotton plants that were treated with sterile water (Mock). Values represent averages of three independent biological replicates of three plants each. Error bars were calculated based on three biological replicates using standard deviation; asterisks (∗) and (∗∗) represent statistical significance at P < 0.05 and P < 0.01, respectively, according to unpaired Student's t‐tests of each of the treatment groups compared to control group (Mock).
Fig. S8 Nucleotide sequence alignment of GhDSC1 in Gossypium hirsutum resistant and susceptible germplasm accessions. The alignment was performed by Clustal X2 with a GONNET 80 protein weight matrix. Only residues that deviate from the reference sequences are shown in the alignment; deletions are indicated by dashes (‐). The polymorphism positions are written vertically, i.e. the first polymorphism occurs at position 177 of the CDS. The position in orange colour (673 bp) represents the nonsynonymous mutation in GhDSC1.
Fig. S9 Yeast two‐hybrid assay of GhDSC1 and GhCAMTA3 proteins. SD/‐LWHA represents the selection medium lacking Leu, Trp, His and Ade. The interaction of murine p53 (p53) and SV40 large T‐antigen (T) was used as a positive control for the system, and human lamin C (lam) was used in the negative interaction control.
Table S1 Information of cotton varieties used in this study.
Table S2 Primers used in this study.


