SUMMARY
In this article, we present the cloning of Hscm1, a gene for chorismate mutase (CM) from the beet cyst nematode Heterodera schachtii. CM is a key branch‐point enzyme of the shikimate pathway, and secondary metabolites that arise from this pathway control developmental programmes and defence responses of the plant. By manipulating the plant's endogenous shikimate pathway, the nematode can influence the plant physiology for its own benefit. Hscm1 is a member of the CM gene family and is expressed during the pre‐parasitic and parasitic stages of the nematode's life cycle. In situ mRNA hybridization reveals an expression pattern specific to the subventral and dorsal pharyngeal glands. The predicted protein has a signal peptide for secretion in addition to two domains. The N‐terminal domain of the mature protein, which is only found in cyst nematodes, contains six conserved cysteine residues, which may reflect the importance of disulphide bond formation for protein stabilization. The C‐terminal domain holds a single catalytic site and has similarity to secreted CMs of pathogenic bacteria, classifying HsCM1 as an AroQγ enzyme. The presumed catalytic residues are discussed in detail, and genetic complementation experiments indicate that the C‐terminal domain is essential for enzyme activity. Finally, we show how the modular design of the protein is mirrored in the genomic sequence by the intron/exon organization, suggesting exon shuffling as a mechanism for the evolutionary assembly of this protein.
INTRODUCTION
The beet cyst nematode Heterodera schachtii, like other plant‐parasitic nematodes, has a biotrophic parasitic strategy. After location of a suitable host, it penetrates the root and migrates through the plant tissue towards the vascular cylinder. There, a feeding site or syncytium is formed by local cell wall degradation and subsequent fusion of neighbouring protoplasts. For the rest of the nematode's life cycle, this syncytium will be its sole source of nutrients (Jones and Northcote, 1972).
A comprehensive analysis has revealed that proteins produced in the pharyngeal glands and secreted by the nematode are crucial for parasitism (Vanholme et al., 2004). For example, the nematode secretes a complex cocktail of cell wall‐degrading enzymes that facilitate its migration through the root tissue by softening the plant cell wall (Popeijus et al., 2000; Smant et al., 1998). Although these enzymes enable the parasite to cross a major defence barrier of its host, they are unlikely to be involved in the induction of the syncytium. The nematode factors responsible for this cellular transformation are still unknown, but many candidate parasitism genes have been identified. One of the corresponding gene products shows similarity to chorismate mutase (CM; EC 5.4.99.5) (Lambert et al., 1999), an enzyme that catalyses the conversion of chorismate into prephenate (Fig. 1A). Chorismate is the final product of the shikimate pathway and the last common intermediate for an extensive range of primary and secondary metabolites, including the aromatic amino acids phenylalanine, tyrosine and tryptophan and their derivatives. Several of the chorismate‐derived compounds have important functions in plant physiology, and include salicylic acid, indole‐3‐acetic acid (IAA), phytoalexins, (iso)flavonoids and lignin (Knaggs, 2003; Schmid and Amrhein, 1995). Changes in the concentration of some of these compounds, most notably IAA and flavonoids, have been implicated in the development of the feeding site or as part of the plant defence response (Goverse et al., 2000; Hutangura et al., 1999; Jones et al., 2007).
Figure 1.
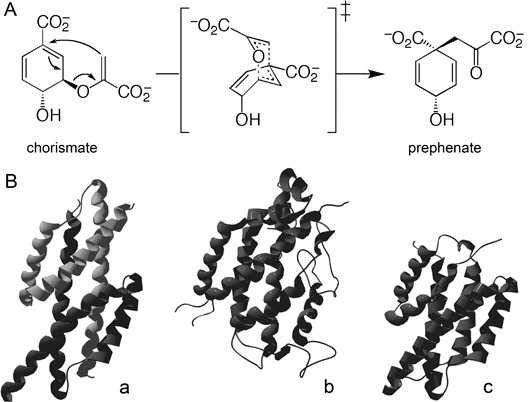
Biochemical function and different structures of AroQ chorismate mutases (CMs). (A) CM reaction. The proposed transition state structure for both the uncatalysed and enzyme‐catalysed rearrangements is shown in brackets. (B) Tertiary structure of the founding members of the three established AroQ subclasses. (a) EcCM of Escherichia coli representing the AroQα subclass. An active enzyme complex containing two active sites is formed by dimerization of two monomers (PDB entry: 1ECM). (b) ScCM of Saccharomyces cerevisiae representing the AroQβ subclass (PDB entry: 1CSM). (c) *MtCM of Mycobacterium tuberculosis representing the AroQγ subclass (PDB entry: 2FP2). (Adapted from Ökvist et al., 2006).
Immunolocalization of MjCM1, a CM of the root‐knot nematode Meloidogyne javanica, has revealed that the protein is released by the nematode into the surrounding root tissue during the early stages of the host–parasite interaction (Doyle and Lambert, 2003). Despite a lack of experimental evidence, it is thought that the protein is injected into the cytoplasm of the initial feeding cell. In this way, the nematode may change the concentration of chorismate‐derived compounds for its own benefit by altering the level of this key branch‐point enzyme. Expression of the gene encoding MjCM1 in soybean hairy root cultures resulted in altered root morphology, with a decrease in lateral roots and an inhibition of vascular tissue development (Doyle and Lambert, 2003). This phenotype was rescued by the addition of IAA, indicating that CM reduces IAA levels, possibly by draining the pool of the IAA precursor chorismate. This may help the nematode to alter normal plant development and to establish a successful parasitic interaction.
On the basis of the similar primary structure and the predicted secondary structure, nematode CMs were catalogued to the AroQ class (Calhoun et al., 2001), grouping all α‐helical bundle CMs. Recently, the nomenclature of the AroQ classification was refined into the subclasses now established as AroQα, AroQβ and AroQγ (Fig. 1B) (Ökvist et al., 2006). A fourth subclass (AroQδ), which was postulated from a set of unusual active site features (Ökvist et al., 2006), is the subject of recent (Kim et al., 2008) and ongoing investigations (P. Kast, unpublished results). The first subclass (AroQα) holds mainly small CMs composed of three α‐helices per subunit. These proteins dimerize to form a functional entity with two active sites. EcCM, the CM domain of the CM‐prephenate dehydratase of Escherichia coli, is the founding member of this group (Lee et al., 1995a). The AroQβ subclass catalogues more complex structures, such as the CM of Saccharomyces cerevisiae (ScCM), which is composed of 12 α‐helices (Xue and Lipscomb, 1995). Finally, the AroQγ (also known as *AroQ) subgroup encompasses the secreted CMs initially found in some pathogenic bacteria (Ökvist et al., 2006; Sasso et al., 2005; Xia et al., 1993). These proteins consist of two regions, each composed of three α‐helices. Structural studies have revealed that both regions interact to form a functional enzyme with a single active site (Kim et al., 2006; Ökvist et al., 2006; Qamra et al., 2006). Despite the different helix compositions and topologies of proteins belonging to different subclasses, their catalytic sites superimpose well and consist essentially of a four‐helix bundle.
All hitherto cloned nematode CMs have been described as multidomain proteins (Bekal et al., 2003; Doyle and Lambert, 2003). On the basis of the sequence similarity to EcCM, one specific domain was considered to be the catalytic domain. This led to the hypothesis that a functional CM with two catalytic sites was most probably formed by dimerization of two monomers (Lambert et al., 1999), in analogy with AroQα proteins (Lee et al., 1995b). The overall similarity to the recently described AroQγ proteins (Fig. 1B) indicated that this idea needed to be revised (Ökvist et al., 2006). According to the presented results, the so‐called catalytic domain is not active as such, but interacts with the C‐terminal region to form a functional enzyme with a single catalytic centre.
RESULTS
Hscm1: a CM‐encoding gene of H. schachtii
Primers were designed on the basis of the conserved sequences of various nematode CMs available in the database. Two highly similar CM fragments (93% identity) were amplified from the cDNA of H. schachtii. The genes corresponding to these fragments were named Hscm1 and Hscm2. Attempts to clone the 5′‐end of Hscm2 were unsuccessful; therefore, we decided to focus on Hscm1. This sequence contains a predicted open reading frame (ORF) of 825 bp; neither a spliced leader sequence commonly found in nematode transcripts nor a consensus polyadenylation signal (AAUAAA) was present. Polymerase chain reaction (PCR) amplification using genomic DNA as template resulted in the amplification of two fragments (796 and 1167bp). Cloning and sequencing of the fragments revealed that each contained a mixture of at least two highly similar sequences. All sequences contained two introns at conserved positions and the length difference between the fragments was a result of the size of the second intron (59 bp vs. 427 bp). A genomic clone corresponding to Hscm1 was found among the 1167‐bp fragment. Additional sequences obtained by genome walking revealed a third intron (246 bp) in the 5′‐region of the sequence. The introns of Hscm1 have the consensus GT‐AG splice junctions and are enriched in AT by 20% relative to the exons. During the cloning process, at least five different CM gene sequences were found, implying that Hscm1 belongs to a gene family. This was confirmed by Southern blot analysis, where multiple bands were observed using a CM probe (Fig. 2).
Figure 2.
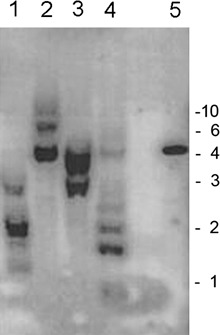
Southern blot of Heterodera schachtii DNA. Genomic DNA digested with HindIII/HincII (lane 1), HincII (lane 2), HindIII (lane 3) and EcoRI (lane 4). The blot was probed with a radioactively labelled chorismate mutase (CM) fragment. Positive control: Hscm1 fragment cloned in pGEM‐T (lane 5). Fragment sizes derived from a standard are indicated on the right in kilobase pairs.
Related sequences in pro‐ and eukaryotes
A blastp search with the putative protein of Hscm1 as query retrieved the secreted CMs of the AroQγ class of pathogenic bacteria, such as Burkholderia sp., Mycobacterium sp. and Pseudomonas sp. When the search was restricted to eukaryotic proteins, only the sequences of plant‐parasitic nematode species were retained (21 hits corresponding to 13 different proteins; E‐value cut‐off 10− 3). Interestingly, a successive blast search against dbEST retained two expressed sequence tags (ESTs) (EW751727 and EW749410) of the Arctic springtail Onychiurus arcticus, suggesting that CMs are not restricted to plant‐parasitic nematodes in the animal kingdom. However, both sequences were incomplete and did not include a convincing signal peptide processing site. To check the reliability of the origin of these sequences, an analysis was performed on all Onychiurus arcticus tags downloaded from the National Center for Biotechnology Information (NCBI) (n = 16 379). Seqclean trashed only one sequence, leaving 16 378 valid ESTs to work with, pointing to an overall high quality of the tags. Clustering of the valid ESTs resulted in 9739 unigenes (6954 singletons and 2785 contigs). For each unigene, a blast search was performed. The first 50 hits were retained, and the corresponding organism of each hit was determined. For 117 unigenes, all of the 50 hits were derived exclusively from Burkholderia species. This result points to a possible prokaryotic contamination of the EST library. As Burkholderia is known to possess CMs, a eukaryotic origin of the mentioned ESTs is uncertain, and it was therefore decided not to include these sequences in further analyses.
An unrooted phylogenetic tree, constructed over a conserved region of different blast hits using a maximum parsimony algorithm, revealed how HsCM1 clustered with CMs of cyst nematodes (Fig. 3). The CMs of root‐knot nematodes formed a separate cluster. Nematode sequences constituted a monophyletic group separated from prokaryotic sequences. A similar topology was obtained by neighbour joining in Treecon, as well as with Bayesian analyses (data not shown).
Figure 3.
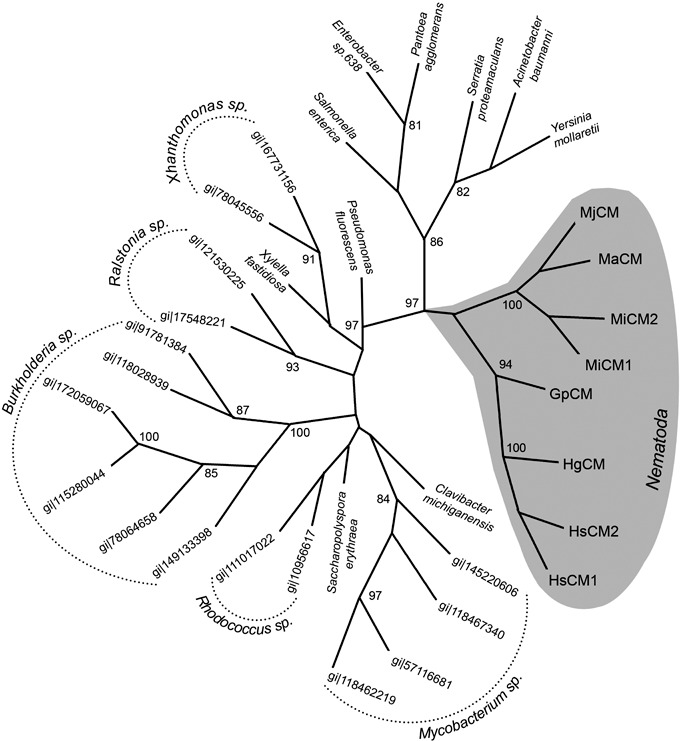
Unrooted evolutionary tree of AroQγ chorismate mutases (CMs) constructed with paup* 4.0. Nucleotide sequences were selected using TBLASTN searches with HsCM1 as query. gi|77799300 (Meloidogyne arenaria) was not included in the phylogenetic analysis as the corresponding protein lacked most active site residues, indicating that it is deduced from a pseudogene accumulating random mutations. Bootstrap values > 80 are indicated at each node; they were calculated from 1000 repetitions. The monophyletic cluster of nematode sequences is shaded in grey. Accession numbers and species names are listed as appearing in the tree (clockwise starting from the nematode sequences). Sequences derived from different species of one genus are listed by their accession number, otherwise the species name is given. MjCM, gi|5353513 Meloidogyne javanica; MaCM, gi|82621736 Meloidogyne arenaria; MiCM2, gi|40037061 Meloidogyne incognita; MiCM1, gi|40037053 Meloidogyne incognita; GpCM, gi|20330094 Globodera pallida; HgCM, gi|65431227 Heterodera glycines; HsCM2, gi|74422700 Heterodera schachtii; HsCM1, gi|74422698 Heterodera schachtii; gi|148271178, Clavibacter michiganensis; gi|145220606, Mycobacterium gilvum; gi|118467340, Mycobacterium smegmatis; gi|57116681, (*MtCM) Mycobacterium tuberculosis; gi|118462219, Mycobacterium avium; gi|134096620, Saccharopolyspora erythraea; gi|10956617, Rhodococcus equi; gi|111017022, Rhodococcus sp. RHA1; gi|149133398, Burkholderia ubonensis; gi|78064658, Burkholderia sp. 383; gi|115280044, Burkholderia ambifaria; gi|172059067, Burkholderia ambifaria; gi|118028939, Burkholderia phymatum; gi|91781384, Burkholderia xenovorans; gi|17548221, Ralstonia solanacearum; gi|121530225, Ralstonia pickettii; gi|71276019, Xylella fastidiosa; gi|78045556, Xanthomonas campestris; gi|167731156, Xanthomonas campestris; gi|77456228, Pseudomonas fluorescens; gi|153901725, Salmonella enterica; gi|146309667, Enterobacter sp. 638; gi|404036, Pantoea agglomerans; gi|157368249, Serratia proteamaculans; gi|126640115, Acinetobacter baumannii; gi|77963051, Yersinia mollaretii.
Spatial and developmental expression analysis of Hscm1
In situ mRNA hybridization was used to analyse the spatial expression pattern of CM genes in pre‐parasitic juveniles of H. schachtii. Hybridization of an antisense probe from Hscm1 resulted in a signal in the three pharyngeal glands, whereas no staining could be observed using a sense probe (Fig. 4A). Gene‐specific primers for Hscm1 were used on cDNA to analyse the expression in different nematode life stages. It was found that Hscm1 was expressed during pre‐parasitic stages, during parasitic stages 4 and 7 days after inoculation and in young females (Fig. 4B). Expression was not observed in developing eggs and adult males. The successful amplification of an actin fragment in all samples confirmed that cDNA was obtained from the different life stages.
Figure 4.
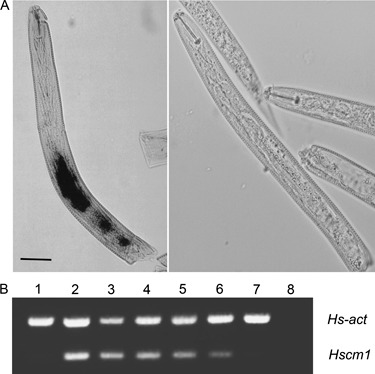
Spatial and temporal expression analysis of Hscm1. (A) Localization of chorismate mutase (CM) gene expression using whole‐mount in situ hybridization on second‐stage pre‐parasitic juveniles (J2s). Expression of Heterodera schachtii CM genes was observed in both subventral and dorsal glands of hatched J2s (left). No signal was detected using a sense probe as control (right). (B) Reverse transcriptase‐polymerase chain reaction (RT‐PCR) showing the expression of Hscm1 in different life stages of H. schachtii. The actin gene (Hs‐act) was used as an internal control. The different stages were as follows: developing eggs (lane 1); pre‐parasitic juveniles soaked (lane 2) or not soaked (lane 3) in cabbage root exudate; parasitic juveniles 4 days (lane 4) and 7 days (lane 5) after inoculation; adult females (lane 6); adult males (lane 7). In the negative control (lane 8), water was used instead of cDNA as template. Hscm1 mRNA is present in pre‐parasitic and parasitic stages, but not in developing eggs and mature males.
Features of the CM protein sequence
Hscm1 encodes a protein of 275 amino acids. The first 23 amino acids form a putative signal peptide for secretion according to SignalP set for eukaryotic sequences (Bendtsen et al., 2004). The predicted mature protein has a theoretical molecular mass of 27.9 kDa, a theoretical isoelectric point (pI) of 7.66 and is composed of two domains (Fig. 5A). Proteins deduced from Hscm2, as well as from the additional genomic sequences, have a similar modular structure. The N‐terminal domain of the mature protein is characterized by six highly conserved cysteine (Cys) residues (Fig. 5B), and was consequently named the Cys‐rich domain (CR domain; 72 amino acids). This domain is only found in CMs of cyst nematodes. The C‐terminal domain (180 amino acids) shows similarity to the secreted CMs of pathogenic bacteria and is, for this reason, referred to as the CM domain. It consists of two regions that were previously considered to be separate domains (Bekal et al., 2003; Lambert et al., 1999). The first region (82 amino acids) is similar to the CM sequence of E. coli and is separated from the second region (87 amino acids) by a proline‐rich motif (11 amino acids). Both regions, as well as the motif, are conserved among secreted CMs of the AroQγ class, although the sequence similarity over the second region is significantly lower. Interestingly, the domains and the signal peptide of HsCM1 are roughly separated by introns in the genomic sequence. In addition, the CM domain itself is encoded by two exons, corresponding to the two regions of this domain (Fig. 5A).
Figure 5.
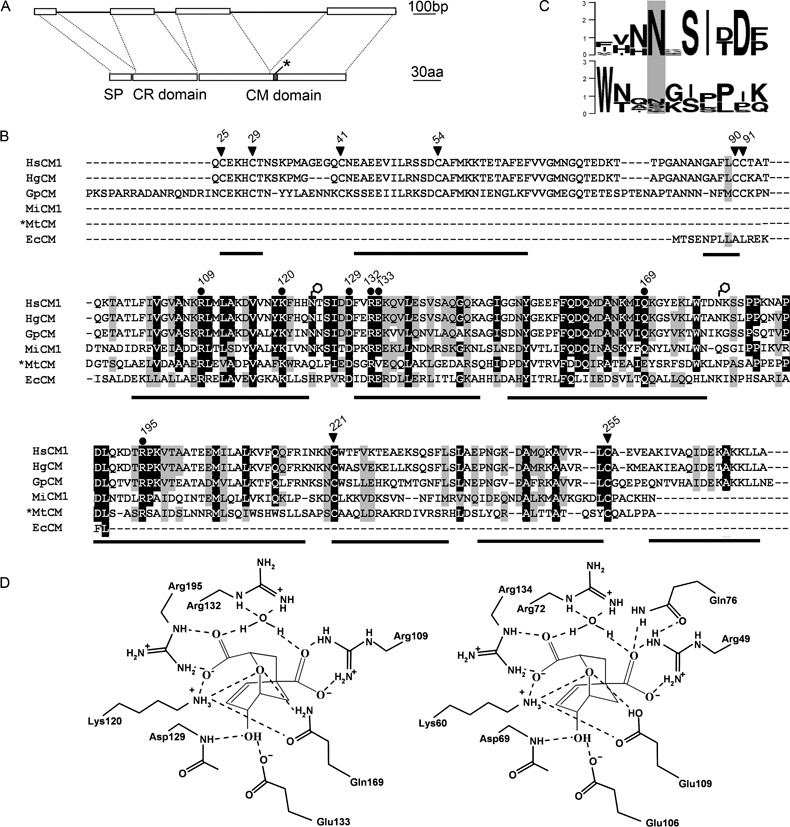
Primary and secondary structure of HsCM1. (A) Schematic representation of the modular structure of HsCM1. Top: exon (boxes) and intron (lines) structure of the genomic sequence. Bottom: protein domain structure. SP, signal peptide (23 amino acids); CR domain, cysteine‐rich domain (72 amino acids); CM domain, chorismate mutase domain (180 amino acids); grey box marked by an asterisk, proline‐rich motif. (B) Multiple sequence alignment of HsCM1 with chorismate mutases (CMs) of other plant‐parasitic nematodes and bacteria. Putative signal sequences were removed from the sequences. Filled dots above the alignment indicate active site residues in *MtCM or EcCM, which are conserved in nematodes. Conserved cysteine residues are marked with arrowheads. Numbers correspond to the amino acid position in HsCM1. Putative N‐glycosylation sites of HsCM1 are marked with open hexagons. The predicted α‐helices of HsCM1 are indicated below the alignment as black bars. Shading indicates identical (black) or similar (grey) amino acids. The species and accession numbers of the aligned proteins are as follows: HsCM1, Heterodera schachtii ABA06538; HgCM, Heterodera glycines AAM74953; GpCM, Globodera pallida CAD29887; MiCM1, Meloidogyne incognita AAR37372; *MtCM, Mycobacterium tuberculosis NP_216401; EcCM, Escherichia coli ZP_00708880. (C) WebLogo of the putative N‐glycosylation sites over the different nematode CMs currently in the database. Top: region surrounding asparagine‐124 (Asn124). Bottom: region surrounding Asn179. The overall height of a stack at a specific position reflects the sequence conservation at that position measured in bits. (D) Schematic representation of the putative active site of HsCM1 (left) in a complex with a transition state analogue. The drawing was made in analogy with the active site of *MtCM (right), as described previously (Ökvist et al., 2006).
HsCM1 contains two putative N‐glycosylation sites (Asn124 and Asn179; Asn, asparagine), both located in the CM domain (Fig. 5B). The first site is conserved over the different nematode CM proteins in the database (ABB02655 being the only exception; however, the lack of most active site residues indicates that this protein could be deduced from a pseudogene). The second position is less conserved among different sequences. For example, it is predicted in the closest homologue from the related nematode H. glycines (AAM74953), but absent in the paralogous sequence HsCM2. Sequence logos over both putative N‐glycosylation sites of nematode CMs currently in the database are given in Fig. 5C.
Secondary and tertiary structure of HsCM1
Secondary structure predictions revealed that HsCM1 is composed predominantly of α‐helices. Thirty‐nine of the 72 amino acids of the N‐terminal CR domain are incorporated into three helices connected by two loop segments. The CM domain is composed of seven α‐helices, the first six of which have counterparts in AroQγ enzymes of bacteria. The last helix of this domain seems to be specific for CMs of cyst nematodes (Fig. 5B). A blastp search against all AroQγ proteins present in the Protein Data Bank (PDB) revealed a CM of Mycobacterium tuberculosis as the closest bacterial homologue (*MtCM; bit score, 37.8; sequence identity, 29%). The structure of this protein was determined independently by three different research groups, and can be accessed with PDB codes 2F6L (Kim et al., 2006), 2FP1/2FP2 (Ökvist et al., 2006) and 2AO2 (Qamra et al., 2006). Although the overall similarity with this protein is relatively modest, especially in the second half of the CM domain, most active site residues described for *MtCM are retained in HsCM1 (Fig. 5B,D). One interesting exception is glutamic acid‐106 (Glu106), which interacts with the 4‐hydroxyl group of chorismate in *MtCM (Ökvist et al., 2006). In HsCM1, this position is occupied by lysine‐166 (Lys166), which has different physicochemical properties. However, the hydroxyl group of the ligand is most probably bound by the conserved Glu133 in HsCM1. In EcCM, a homologous glutamic acid (Glu52) plays the same role (Lee et al., 1995a), whereas *MtCM lacks this residue (Fig. 5B,D).
The tertiary structure of the CM domain of HsCM1 [residues glutamine‐95 (Gln95) to alanine‐275 (Ala275)] was modelled using *MtCM as template (Fig. 6). The active site residues cluster in one half of the protein in a core formed by helices 1, 2, 3 and 4 of the CM domain. The proline‐rich motif, separating both CM domain regions, lies on the protein surface. At the same side of the protein, and exposed to the solvent, are the two putative glycosylation sites of HsCM1. At the opposite side, two conserved Cys residues (Cys221 and Cys255) are in close proximity, so that they could form an intramolecular disulphide bond. A similar cysteine bond is present in *MtCM, adding to the overall stability of this protein (Ökvist et al., 2006; Sasso et al., 2005).
Figure 6.
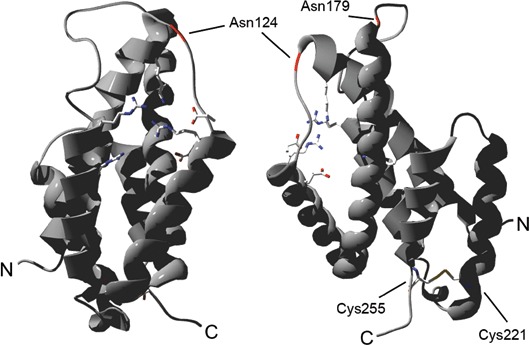
Tertiary structure of the chorismate mutase (CM) domain of HsCM1 [glutamine‐72 (Gln72) to alanine‐252 (Ala252)] modelled using *MtCM as template. Left: front view of active site residues (in stick representation) clustered between helices 1, 2, 3 and 4 of the CM domain. Right: side view. The positions of putative N‐glycosylation sites [asparagine‐124 (Asn124) and Asn179] are indicated in red. The presumed disulphide bond between cysteine‐221 (Cys221) and Cys255 is shown. N‐ and C‐termini are labelled.
Functional complementation
In order to perform a functional complementation assay, different deletion constructs of Hscm1 were made (Fig. 7A) and expressed in a CM‐deficient E. coli strain. The expression of the constructs was confirmed by Western blot (Fig. 7B), and the observed molecular masses matched well with the theoretical values predicted from the sequence (HsCMΔ1, 30.1 kDa; HsCMΔ2, 22.4 kDa; HsCMΔ3, 12.0 kDa). Notably, the bacteria transformed with the full‐length construct (HsCMΔ1, corresponding to the mature protein lacking the signal peptide) grew at a reduced rate and were even difficult to maintain on enriched medium plates. As a result, these cells required a longer incubation time at the appropriate temperature to reach the stationary phase (data not shown). The impaired growth suggests that the expression of this construct reduced the general fitness of the bacteria, possibly as a result of the intracellular toxicity of HsCMΔ1. In addition, on selective medium lacking phenylalanine (M9cY), the cells producing HsCMΔ1 were not able to grow (Fig. 7C). Transformants expressing the other deletion constructs grew well on agar plates with Luria–Bertani (LB) medium and on minimal medium supplemented with phenylalanine (M9cY + F; Fig. 7C). When phenylalanine was absent, only the transformant carrying a construct lacking the CR domain (HsCMΔ2) grew as well as the positive control. This indicates that the CR domain is not required for enzymatic activity, and may even be responsible for the toxic effect discussed above. Expression of a truncated CM domain (HsCMΔ3) failed to complement the CM deficiency (Fig. 7C).
Figure 7.
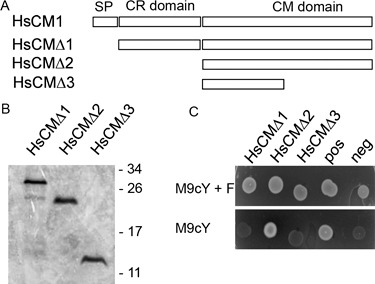
Complementation of the chorismate mutase (CM)‐deficient Escherichia coli strain KA12/pKIMP‐UAUC by HsCM1. (A) Schematic representation of the different deletion constructs used for the complementation test. For the nomenclature of the protein domains, see Fig. 5. (B) Western blot analysis of HsCM1 deletion constructs. Proteins were fractionated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and blotted on to a poly(vinylidene difluoride) (PVDF) membrane. HsCM1 derivatives were visualized using a polyhistidine (poly‐His) probe conjugated to horseradish peroxidase (HRP). Molecular masses from the size standard are indicated in kilodaltons on the right. (C) CM complementation assay. Different deletion constructs of Hscm1 were transformed into the CM‐deficient E. coli strain KA12/pKIMP‐UAUC (Kast et al., 1996). As a positive control, a construct expressing a CM gene of Bacillus subtilis was used (Sasso et al., 2005). KA12/pKIMP‐UAUC cells transformed with an empty pQE‐30 UA vector were used as negative control. Bacteria were grown for 3 days at 30 °C on minimal medium plates supplemented with or without phenylalanine (M9cY + F and M9cY, respectively).
DISCUSSION
The identification of CM in plant‐parasitic nematodes by Lambert et al. (1999) was remarkable, as this enzyme had not been found previously in animals. Based on the similarity to bacterial CMs, the authors posed the question of whether the gene could have been obtained by a horizontal gene transfer event as an alternative to inheritance through common ancestry. It is, however, extremely difficult to prove such hypothesis. Reconstruction of the evolutionary history by means of dense phylogenetic trees is one of the better approaches, but its validity depends on the availability of relevant sequence information (Stanhope et al., 2001). In the case of CMs, two ESTs suggestive of CM of the AroQγ subclass in the Arctic springtail Onychiurus arcticus were recently submitted to NCBI. The discovery of CM sequences in an arthropod could shed light on the evolutionary origin of animal CMs. However, subsequent analysis revealed that the springtail EST library in question is most likely contaminated, as a considerable number of the tags were probably derived from Burkholderia. These bacteria are known symbionts of arthropods (Kikuchi et al., 2005) and possess CMs belonging to the AroQγ subclass. Although this issue needs further clarification, it illustrates that publicly available sequences (especially ESTs) should be used with caution. Leaving the dubious EST sequences out of consideration, the topology of the constructed evolutionary tree does not speak against a horizontal gene transfer event between bacteria and nematodes.
Hscm1 is the first CM gene described from H. schachtii. In addition to this full‐length gene, we also obtained fragmentary sequence information pointing to multiple similar sequences, suggesting that the cloned gene is a member of a gene family. This is in agreement with observations from other nematode species, such as M. javanica (Lambert et al., 1999), M. incognita (Huang et al., 2005) and H. glycines (Bekal et al., 2003). Southern blotting experiments confirmed this, although fewer bands were observed than the number of sequences cloned. This can be a consequence of allelic polymorphism. Alternatively, it may suggest a specific genomic organization, in which different genes are organized in blocks of tandem repeats. This pattern of organization has been described for CM genes of H. glycines (Lambert et al., 2005).
HsCM1 has a modular architecture that is mirrored in the genome by four exons roughly corresponding to the different protein regions. A similar modular intron/exon architecture is found in other CMs of cyst and root‐knot nematodes (Bekal et al., 2003; Huang et al., 2005), and may have important evolutionary consequences as it allows the assembly of new proteins by exon shuffling. In line with this hypothesis, Ökvist et al. (2006) suggested that the two regions of the CM domain evolved from a duplication event. As the N‐terminal region is more conserved than the C‐terminal region, both evolved at distinct evolutionary rates. A possible explanation for this is the fact that the former region contributes more than the latter to the active site (Fig. 6), making it more vulnerable to mutations. The CR domain is unique to sequences from cyst nematodes. The fact that CMs of root‐knot nematodes lack this domain, but are functional, suggests that this domain is not crucial for enzymatic activity. This was confirmed in our complementation study, and is in agreement with experiments using H. glycines CM (Bekal et al., 2003). The function of this domain remains to be elucidated, but the six highly conserved Cys residues of this domain could be important in forming inter‐ or intra‐molecular disulphide bonds. Similar motifs have been found in other proteins, where they contribute to remarkable stability (Gelly et al., 2004). It is not known why root‐knot nematode CMs lack this CR domain, or whether the resulting difference in protein architecture is linked to the differences in parasitic behaviour or host range of root‐knot and cyst nematodes.
The CM domain holding the catalytic site was further studied by modelling its tertiary structure. We expect this model, which is based on *MtCM as a template, to be quite accurate, as structures of secreted CMs of the AroQγ subclass available in PDB superimpose extremely well, even though their sequence identity is low. For instance, on comparing *YpCM and *MtCM, which are only 23% identical, the active site residues are strongly conserved in identity and spatial positioning, and the average root‐mean‐square deviation over all Cα atoms is less than 0.8 Å (Kim et al., 2008). Thus, the model of the CM domain of HsCM1, which exhibits 29% sequence identity to *MtCM, should represent the actual structure reasonably well. Finally, two putative glycosylation sites were detected. Although one position was found to be conserved among nematode CMs, any putative glycosylations are not essential for catalytic activity as the protein is functional in a prokaryotic expression system. Nevertheless, glycosylation can still play a role in the natural host, for instance, to stabilize the protein.
Temporal expression analysis using reverse transcriptase (RT)‐PCR showed the expression of Hscm1 during the onset of the parasitic interaction and in the subsequent feeding stages of the nematode. Similar expression patterns during the parasitic stages were found for CMs of other plant‐parasitic nematodes using comparable techniques (Jones et al., 2003), whereas more detailed studies using quantitative RT‐PCR showed highest expression during the onset of parasitism (Painter and Lambert, 2003). In situ hybridization revealed that the CM gene is expressed in subventral and dorsal glands. This is in agreement with the observations of CM expression in H. glycines (Bekal et al., 2003). As a result of the high degree of similarity, a gene‐specific probe to discriminate between the different members of the CM gene family could not be constructed. As such, we were unable to conclude whether the signal corresponded to the expression of Hscm1 alone, or whether we were visualizing the expression of several genes. The specific expression pattern in the secretory glands during the onset of parasitism is in agreement with the general idea that CM is involved in the parasitic interaction of the nematode with its host.
The exact function of the secreted CMs is still a matter of debate (Sasso et al., 2005). As this CM is the only enzyme of the phenylalanine and tyrosine biosynthetic pathway detected in plant‐parasitic nematodes, and as long as there is no evidence for the presence of chorismate‐producing or prephenate‐utilizing enzymes, it appears to be unlikely that the protein functions as a CM in the basic metabolism within the nematode, but may point towards a role of the protein in the plant, where a functional shikimate pathway is present. A molecular pathway, which could explain the role of CM in the specific differentiation of nematode feeding cells, has been suggested by Doyle and Lambert (2003). It was proposed that the release of CM into the plant cytosol could influence auxin levels in the plant. Apart from these data, it is remarkable that similar enzymes are secreted by human pathogenic bacteria (Sasso et al., 2005). As mammals do not possess the shikimate pathway, this observation questions the fundamental role of nematode CMs in feeding cell formation. More general roles in the defence response during the host–pathogen interaction, as suggested previously (Jones et al., 2007), or in communication with co‐infecting pathogens could be interesting alternative hypotheses.
EXPERIMENTAL PROCEDURES
Biological material
Large batches of pre‐parasitic stages of H. schachtii were purified from glasshouse‐grown cabbage (Brassica oleracea cv. Ramosa), as described previously (De Meutter et al., 2001). The nematodes were either used directly for in situ hybridization experiments or were stored at −80 °C for DNA extraction. The different developmental stages for RT‐PCR were dissected from sterile infected Arabidopsis thaliana roots, as described previously (Tytgat et al., 2004).
Extraction of DNA and RNA, and synthesis of cDNA
Genomic DNA was isolated from H. schachtii secondary‐stage juveniles, as described previously (Bolla et al., 1988). mRNA for RT‐PCR was isolated from different stages of H. schachtii with TRIzol reagent (Invitrogen, San Diego, CA, USA), and converted to cDNA using the SMART™‐cDNA synthesis system (BD Biosciences, Heidelberg, Germany), following the manufacturer's instructions. Large‐scale RNA purification from approximately 7 × 106 H. schachtii secondary‐stage juveniles for library construction was performed by LiCl precipitation (Sambrook et al., 1989). A cDNA library from H. schachtii pre‐parasitic juveniles was constructed on the basis of a modified literature procedure (Gubler and Hoffman, 1983).
Oligonucleotides
The following oligonucleotides were synthesized by Invitrogen or Isogen Life Science (Utrecht, The Netherlands): chor1, 5′‐GCAACTCCGACTGTGCCTTCATG‐3′; chor4, 5′‐TCGTCAATTTGTGCCACAATTTTCGC‐3′; CM1F, 5′‐GTGGACCGACAACAAATCGTCGCCTCCAA‐3′; CM1R, 5′‐AAGCTTTGCGACTTTTCGGCCTCCGTT‐3′; SP6+, 5′‐ATTTAGGTGACACTATAGAATACTCAAGC‐3′; T7+, 5′‐TAATACGACTCACTATAGGGCGAATTGG‐3′; 1224, 5′‐CGCCAGGGTTTTCCCAGTCACGAC‐3′; 1233, 5′‐AGCGGATAACAATTTCACACAGGA‐3′; AP1, 5′‐GTAATACGACTCACTATAGGGC‐3′; AP2, 5′‐ACTATAGGGCACGCGTGGT‐3′; hsactF, 5′‐ACTTCATGATCGAGTTGTAGGTGGACTCG‐3′; hsactR, 5′‐GACCTCACTGACTACCTGATGAAGATTC‐3′; CM‐FLF, 5′‐CAATGCGAGAAACATTGCACT‐3′; CM‐CATF, 5′‐AAAACCGCGACACTTTTCATC‐3′; CM‐CtF, 5′‐ACCGACAACAAATCGTCG‐3′; CM‐FLR, 5′‐TCCTCAGGCCAACAATTTCTT‐3′; CM‐CATR, 5′‐TCACGACGATTTGTTGTCGGT‐3′.
Cloning of the cDNA sequences
The 5′‐ and 3′‐terminal fragments of the coding sequences were obtained by PCR using 150 ng of cDNA library plasmid as template and a vector‐specific primer (1224 and 1233 for 5′‐ and 3′‐fragments, respectively) in combination with a CM‐specific primer (chor1 and chor4 for 3′‐ and 5′‐fragments, respectively). The PCR mixture was heated to 94 °C for 60 s, followed by 35 cycles of 94 °C (25 s), 58 °C (30 s) and 72 °C (90 s). PCR products were run on a 1% Tris‐acetate EDTA agarose gel and visualized by ethidium bromide staining. Fragments of interest were purified from the gel with the Qiaquick gel extraction kit (Qiagen, Crawlwy, UK) and cloned into the pGEM‐T vector (Promega, Madison, WI, USA) according to the standard T/A‐cloning protocol. Sequence analysis was performed using the chain termination method of Sanger et al. (1977) with the Thermo Sequenase Cycle Sequencing Kit (USB Corporation, Cleveland, OH, USA). Sequences can be found at GenBank under the accession number DQ176596 for the Hscml cDNA and DQ176597 for the Hscm2 cDNA.
Cloning of the genomic sequences
Gene fragments were obtained by PCR using 130 ng of genomic DNA as template in combination with the CM primers chor1 and chor4. The PCR conditions were as follows: 94 °C for 60 s, followed by 35 cycles of 94 °C (15 s), 58 °C (25 s) and 72 °C (120 s). PCR products and fragments of interest were processed and analysed as described above. Final sequencing was performed at the VIB Genetic Service Facility (Antwerp, Belgium). Gene‐specific primers were employed to amplify up‐ and downstream sequences using the Universal Genome Walker kit (BD Biosciences). Genomic sequences can be found at GenBank under the accession numbers DQ176598, DQ176599, DQ176600 and DQ176601.
Whole‐mount in situ hybridization
In situ hybridization was performed as described previously (Vanholme et al., 2002). Digoxigenin‐labelled single‐stranded DNA probes were generated using the gene‐specific primer chor1 (sense probe) and the vector primer 1224 (antisense probe).
RT‐PCR analysis
Temporal expression analysis was performed as described previously (Tytgat et al., 2004). Gene‐specific primers were constructed against Hscm1 (CM1F/CM1R). The PCR conditions were as follows: 94 °C for 60 s, followed by 23 cycles of 94 °C (15 s), 60 °C (25 s) and 72 °C (45 s). Intron‐spanning primers (hsactF/hsactR) complementary to the constitutively expressed actin gene were used in all PCRs as an internal control and to exclude the presence of contaminating genomic DNA. As a negative control, water was added instead of the cDNA template.
Software tools
Sequence similarity searches were performed with different blast programs (Altschul et al., 1990) as available on the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST), and alignments were generated using the BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Protein sequences were screened for putative N‐glycosylation sites using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc), and signal peptide sequences were predicted with the SignalP 3.0 program (Bendtsen et al., 2004; http://www.cbs.dtu.dk/services/SignalP). Putative intron splice sites were predicted using fsplice on the Softberry server (http://www.softberry.com). ESTs were cleaned using SeqClean (http://www.tigr.org) and clustered using the TIGR Gene Indices Clustering Tool (tgicl) (Pertea et al., 2003). For the prediction of the secondary structure, the Predict Protein package (Rost and Sander, 1995) or psipred (McGuffin et al., 2000; bioinf.cs.ucl.ac.uk/psipred) was used. Three‐dimensional homology modelling was performed with SWISS‐MODEL using the alignment mode. The protein structures were visualized using DeepView v3.7 (Guex and Peitsch, 1997), followed by POV‐Ray 3.6. Amino acid sequence alignments were generated with clustal‐w (Thompson et al., 1994). The phylogenetic tree was constructed based on a manually adjusted alignment, using the maximum parsimony method in paup* 4.0 with heuristic search (Swofford, 2003). The consensus tree was visualized with TreeView (Page, 1996). Bayesian analyses were performed by running 2 000 000 generations using MrBayes 3.1.2. Current trees were saved every 100 generations. Sequence logos were created using WebLogo (Crooks et al., 2004).
Complementation assay
The different fragments of Hscm1 were amplified by PCR using the cloned full‐length sequence as template. The fragments obtained were gel purified (Qiagen), cloned according to the standard T/A‐cloning protocol into the pQE‐30 UA vector (Qiagen) downstream of the strong T5 promoter, and transformed into Escherichia coli M15 cells. Transformants were selected on LB plates supplemented with 100 mg/L carbenicillin and 25 mg/L kanamycin. Plasmids were propagated, sequenced and transformed into E. coli KA12/pKIMP‐UAUC cells (Kast et al., 1996). Transformed cells were selected on LB plates supplemented with 100 mg/L carbenicillin and 30 mg/L chloramphenicol. Colonies were re‐streaked onto M9c‐type minimal medium agar plates (Gamper et al., 2000). M9cY medium consists of Na2HPO4 (6 g/L), KH2PO4 (3 g/L), NH4Cl (1 g/L), NaCl (0.5 g/L), d‐(+)‐glucose (2 g/L), MgSO4 (1 mm), 4‐hydroxybenzoic acid (5 mg/L), 4‐aminobenzoic acid (5 mg/L), 2,3‐dihydroxybenzoic acid (1.6 mg/L), CaCl2 (0.1 mm), thiamine‐HCl (5 mg/L), l‐tryptophan (20 mg/L) and l‐tyrosine (20 mg/L). M9cY + F plates were additionally supplemented with 20 mg/L of l‐phenylalanine; 133 µm isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) was always included to ensure full induction of the introduced CM gene variants as well as the auxiliary functions of the selection system itself (Sasso et al., 2005). For plates, 17 g/L agar was added. Bacterial growth was examined over several days of incubation at 30 °C. As a positive control, a transformant of KA12/pKIMP‐UAUC with plasmid pMG212H‐W (Gamper et al., 2000), encoding wild‐type Bacillus subtilis CM, was used. KA12/pKIMP‐UAUC cells transformed with an empty pQE‐30 UA vector were used as negative control.
Western blot
E. coli strains were grown (37 °C, 185 r.p.m., 16 h) in 5 mL of LB medium containing 30 µg/mL chloramphenicol, 100 µg/mL carbenicillin and 1 mm IPTG. The pellet from 2 mL of liquid culture was boiled in 40 µL of 2 × sodium dodecylsulphate (SDS) sample buffer supplemented with 130 mm dithiothreitol (DTT). Before loading, samples were centrifuged to pellet insoluble fractions. An aliquot of 5 µL of the supernatant was run on a 15% SDS‐polyacrylamide gel in a Mini‐PROTEAN Tetra electrophoresis system (Bio‐Rad, Hercules, CA, USA) according to the method of Laemmli (1970). Proteins were transferred to a pre‐wetted poly(vinylidene difluoride) (PVDF) membrane (Pierce, Rockford, IL, USA) using the Mini Trans‐Blot electrophoretic transfer cell (Bio‐Rad). Prestained PageRuler (Fermentas, Vilnius, Lithuania) was used as a marker to determine the molecular mass of the proteins and to estimate the efficiency of protein transfer. The membrane was rinsed in tris(hydroxymethyl)aminomethane (Tris)‐buffered saline (TBS: 0.1 m Tris‐HCl/0.15 m NaCl/0.1% Triton X‐100; pH 7.6) and incubated in blocking buffer [1% blocking reagent (Roche, Mannheim, Germany) in TBS] for 1 h. The blot was rinsed several times with TBS and incubated in TBS to which horseradish peroxidase‐conjugated histidine (HRP‐His) probe (dilution 1 : 1000; Pierce) had been added. After several rinses in TBS, the signal was visualized colorimetrically using 3,3′‐diaminobenzidine (DAB)/H2O2.
ACKNOWLEDGEMENTS
We would like to thank John Jones (Scottish Crop Research Institute, Dundee, UK) for help with the first draft of the manuscript. This work was supported by the fund for Scientific Research‐Flanders (5G.0078.97 and 3G.0031.08) and a grant from Ghent University (GOA 01G00805). AH is in receipt of a grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT‐Vlaanderen) and JJ a grant from Ghent University (Bijzonder Onderzoeksfonds, BOF). BV is a postdoctoral researcher at Ghent University (BOF).
REFERENCES
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bekal, S. , Niblack, T.L. and Lambert, K.N. (2003) A chorismate mutase from the soybean cyst nematode Heterodera glycines shows polymorphisms that correlate with virulence. Mol. Plant–Microbe Interact. 16, 439–446. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , Von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Bolla, R.I. , Weaver, C. and Winter, R.E.K. (1988) Genomic differences among pathotypes of Bursaphelenchus xylophilus . J. Nematol. 20, 309–316. [PMC free article] [PubMed] [Google Scholar]
- Calhoun, D.H. , Bonner, C.A. , Gu, W. , Xie, G. and Jensen, R.A. (2001) The emerging periplasm‐localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa . Genome Biol. 2, 301–30.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E. , Hon, G. , Chandonia, J.M. and Brenner, S.E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meutter, J. , Vanholme, B. , Bauw, G. , Tytgat, T. , Gheysen, G. and Gheysen, G. (2001) Preparation and sequencing of secreted proteins from the pharyngeal glands of the plant parasitic nematode Heterodera schachtii . Mol. Plant Pathol. 2, 297–301. [DOI] [PubMed] [Google Scholar]
- Doyle, E.A. and Lambert, K.N. (2003) Meloidogyne javanica chorismate mutase 1 alters plant cell development. Mol. Plant–Microbe Interact. 16, 123–131. [DOI] [PubMed] [Google Scholar]
- Gamper, M. , Hilvert, D. and Kast, P. (2000) Probing the role of the C‐terminus of Bacillus subtilis chorismate mutase by a novel random protein‐termination strategy. Biochemistry, 39, 14 087–14 094. [DOI] [PubMed] [Google Scholar]
- Gelly, J.C. , Gracy, J. , Kaas, Q. , Le Nguyen, D. , Heitz, A. and Chiche, L. (2004) The KNOTTIN website and database: a new information system dedicated to the knottin scaffold. Nucleic Acids Res. 32, D156–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverse, A. , Overmars, H. , Engelbertink, J. , Schots, A. , Bakker, J. and Helder, J. (2000) Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant–Microbe Interact. 13, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Gubler, U. and Hoffman, B.J. (1983) A simple and very efficient method for generating cDNA libraries. Gene, 25, 263–269. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. (1997) SWISS‐MODEL and the Swiss‐PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Huang, G.Z. , Dong, R.H. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2005) Two chorismate mutase genes from the root‐knot nematode Meloidogyne incognita . Mol. Plant Pathol. 6, 23–30. [DOI] [PubMed] [Google Scholar]
- Hutangura, P. , Mathesius, U. , Jones, M.G.K. and Rolfe, B.G. (1999) Auxin induction is a trigger for root gall formation caused by root‐knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Aust. J. Plant Physiol. 26, 221–231. [Google Scholar]
- Jones, J.T. , Furlanetto, C. , Bakker, E. , Banks, B. , Blok, V. , Chen, Q. , Phillips, M. and Prior, A . (2003) Characterization of a chorismate mutase from the potato cyst nematode Globodera pallida . Mol. Plant Pathol. 4, 43–50. [DOI] [PubMed] [Google Scholar]
- Jones, J.T. , Furlanetto, C. and Phillips, M.S. (2007) The role of flavonoids produced in response to cyst nematode infection of Arabidopsis thaliana . Nematology, 9, 671–677. [Google Scholar]
- Jones, M.G.K. and Northcote, D.H. (1972) Nematode‐induced syncytium—a multinucleated transfer cell. J. Cell Sci. 10, 789–809. [DOI] [PubMed] [Google Scholar]
- Kast, P. , Asif‐Ullah, M. , Jiang, N. and Hilvert, D. (1996) Exploring the active site of chorismate mutase by combinatorial mutagenesis and selection: the importance of electrostatic catalysis. Proc. Natl. Acad. Sci. USA, 93, 5043–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Meng, X.Y. and Fukatsu, T. (2005) Gut symbiotic bacteria of the genus Burkholderia in the broad‐headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71, 4035–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.‐K. , Reddy, S.K. , Nelson, B.C. , Vasquez, G.B. , Davis, A. , Howard, A.J. , Patterson, S. , Gilliland, G.L. , Ladner, J . and Reddy, P.T . (2006) Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids. J. Bacteriol. 188, 8638–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.‐K. , Reddy, S.K. , Nelson, B.C. , Robinson, H. , Reddy, P.T. and Ladner, J.E. (2008) A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H37Rv and the secreted chorismate mutase (y2828) from Yersinia pestis . FEBS J. 275, 4824–4835. [DOI] [PubMed] [Google Scholar]
- Knaggs, A.R. (2003) The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 20, 119–136. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during assembly of the head of bacteriophage‐T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lambert, K.N. , Allen, K.D. and Sussex, I.M. (1999) Cloning and characterization of an esophageal‐gland‐specific chorismate mutase from the phytoparasitic nematode Meloidogyne javanica . Mol. Plant–Microbe Interact. 12, 328–336. [DOI] [PubMed] [Google Scholar]
- Lambert, K.N. , Bekal, S. , Domier, L.L. , Niblack, T.L. , Noel, G.R. and Smyth, C.A. (2005) Selection of Heterodera glycines chorismate mutase‐1 alleles on nematode‐resistant soybean. Mol. Plant–Microbe Interact. 18, 593–601. [DOI] [PubMed] [Google Scholar]
- Lee, A.Y. , Karplus, P.A. , Ganem, B. and Clardy, J. (1995a) Atomic‐structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 117, 3627–3628. [Google Scholar]
- Lee, A.Y. , Stewart, J.D. , Clardy, J. and Ganem, B. (1995b) New insight into the catalytic mechanism of chorismate mutases from structural studies. Chem. Biol. 2, 195–203. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J. , Bryson, K. and Jones, D.T. (2000) The PSIPRED protein structure prediction server. Bioinformatics, 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Ökvist, M. , Dey, R. , Sasso, S. , Grahn, E. , Kast, P. and Krengel, U. (2006) 1.6 Å crystal structure of the secreted chorismate mutase from Mycobacterium tuberculosis: novel fold topology revealed. J. Mol. Biol. 357, 1483–1499. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Painter, J.E. and Lambert, K.N. (2003) Meloidogyne javanica chorismate mutase transcript expression profile using real‐time quantitative RT‐PCR. J. Nematol. 35, 82–87. [PMC free article] [PubMed] [Google Scholar]
- Pertea, G. , Huang, X. , Liang, F. , Antonescu, V. , Sultana, R. , Karamycheva, S. , Lee, Y. , White, J. , Cheung, F. , Parvizi, B. , Tsai, J. and Quackenbush, J. . (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics, 19, 651–652. [DOI] [PubMed] [Google Scholar]
- Popeijus, H. , Overmars, H. , Jones, J. , Blok, V. , Goverse, A. , Helder, J. , Schots, A. , Bakker, J. and Smant, G . (2000) Degradation of plant cell walls by a nematode. Nature, 406, 36–37. [DOI] [PubMed] [Google Scholar]
- Qamra, R. , Prakash, P. , Aruna, B. , Hasnain, S.E. and Mande, S.C. (2006) The 2.15 Å crystal structure of Mycobacterium tuberculosis chorismate mutase reveals an unexpected gene duplication and suggests a role in host–pathogen interactions. Biochemistry, 45, 6997–7005. [DOI] [PubMed] [Google Scholar]
- Rost, B. and Sander, C. (1995) Progress of 1D protein‐structure prediction at last. Protein Struct. Funct. Genet. 23, 295–300. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sanger, F. , Nicklen, S. and Coulson, A.R. (1977) DNA sequencing with chain‐terminating inhibitors. Proc. Natl. Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso, S. , Ramakrishnan, C. , Gamper, M. , Hilvert, D. and Kast, P. (2005) Characterization of the secreted chorismate mutase from the pathogen Mycobacterium tuberculosis . FEBS J. 272, 375–389. [DOI] [PubMed] [Google Scholar]
- Schmid, J. and Amrhein, N. (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry, 39, 737–749. [Google Scholar]
- Smant, G. , Stokkermans, J.P.W.G. , Yan, Y.T. , De Boer, J.M. , Baum, T.J. , Wang, X.H. , Hussey, R.S. , Gommers, F.J. , Henrissat, B. , Davis, E.L. , Helder, J. , Schots, A. and Bakker, J. (1998) Endogenous cellulases in animals: isolation of beta‐1,4‐endoglucanase genes from two species of plant‐parasitic cyst nematodes. Proc. Natl. Acad. Sci. USA, 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope, M.J. , Lupas, A. , Italia, M.J. , Koretke, K.K. , Volker, C. and Brown, J.R. (2001) Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature, 411, 940–944. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4 beta 10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat, T. , Vanholme, B. , De Meutter, J. , Claeys, M. , Couvreur, M. , Vanhoutte, I. , Gheysen, G. , Van Criekinge, W. , Borgonie, G. , Coomans, A. and Gheysen, G . (2004) A new class of ubiquitin extension proteins secreted by the dorsal pharyngeal gland in plant parasitic cyst nematodes. Mol. Plant–Microbe Interact. 17, 846–852. [DOI] [PubMed] [Google Scholar]
- Vanholme, B. , De Meutter, J. , Tytgat, T. , Gheysen, G.D.C. , Vanhoutte, I. and Gheysen, G.D.R. (2002) An improved method for whole‐mount in situ hybridization of Heterodera schachtii juveniles. Parasitol. Res. 88, 731–733. [DOI] [PubMed] [Google Scholar]
- Vanholme, B. , De Meutter, J. , Tytgat, T. , Van Montagu, M. , Coomans, A. and Gheysen, G. (2004) Secretions of plant‐parasitic nematodes: a molecular update. Gene, 332, 13–27. [DOI] [PubMed] [Google Scholar]
- Xia, T. , Song, J. , Zhao, G. , Aldrich, H. and Jensen, R.A. (1993) The aroQ‐encoded monofunctional chorismate mutase (CM‐F) protein is a periplasmic enzyme in Erwinia herbicola . J. Bacteriol. 175, 4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y.F. and Lipscomb, W.N. (1995) Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc. Natl. Acad. Sci. USA, 92, 10 595–10 598. [DOI] [PMC free article] [PubMed] [Google Scholar]


