Abstract
MicroRNAs (miRNAs) are short single-stranded RNAs, measuring 21 to 23 nucleotides in length and regulate gene expression at the post-transcriptional level through mRNA destabilization or repressing protein synthesis. Dysregulation of miRNAs can lead to tumorigenesis through changes in regulation of key cellular processes such as cell proliferation, cell survival, and apoptosis. miR-125a-5p has been implicated as a tumor suppressor miRNA in malignancies such as non-small cell lung cancer and colon cancer. However, the role of miR-125a-5p has not been fully investigated in head and neck squamous cell carcinoma (HNSCC). We performed microRNA microarray profiling of HNSCC tumor samples obtained from a prospective clinical trial evaluating the role of postoperative radiotherapy in head and neck cancer. We also mined through The Cancer Genome Atlas to evaluate expression and survival data. Biological experiments, including cell proliferation, flow cytometry, cell migration and invasion, clonogenic survival, and fluorescent microscopy, were conducted using HN5 and UM-SCC-22B cell lines. miR-125a-5p downregulation was associated with recurrent disease in a panel of high-risk HNSCC and then confirmed poor survival associated with low expression in HNSCC via the Cancer Genome Atlas, suggesting that miR-125a-5p acts as a tumor suppressor miRNA. We then demonstrated that miR-125a-5p regulates cell proliferation through cell cycle regulation at the G1/S transition. We also show that miR-125a-5p can alter cell migration and modulate sensitivity to ionizing radiation. Finally, we identified putative mRNA targets of miR-125a-5p, including ERBB2, EIF4EBP1, and TXNRD1, which support the tumor suppressive mechanism of miR-125a-5p. Functional validation of ERBB2 suggests that miR-125a-5p affects cell proliferation and sensitivity to ionizing radiation, in part, through ERBB2. Our data suggests that miR-125a-5p acts as a tumor suppressor miRNA, has potential as a diagnostic tool and may be a potential therapeutic target for the management and treatment of squamous cell carcinoma of the head and neck.
Abbreviations: MTT, 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide; DAPI, 4′,6-diamidino-2-phenylindole; 4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; BrdU, 5-bromo-2′-deoxyuridine; CO2, carbon dioxide; cDNA, complementary DNA; DMSO, dimethylsulfoxide; DNA, deoxyribonucleic acid; ECL, electrochemiluminescence; EDTA, ethylenediaminetetraacetic acid; EGFR, epidermal growth factor receptor; ERBB2, erythroblastic oncogene B 2; FBS, fetal bovine serum; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; LNA, locked nucleic acid; mRNA, messenger RNA; miRNA, microRNA; PARP, poly(ADP-ribose) polymerase; mTORC1, mTOR complex 1; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction; redox, reduction–oxidation; S6K1, S6 kinase 1; snRNA, small nuclear RNA; SDS, sodium dodecyl sulfate; TCGA, The Cancer Genome Atlas; TXNRD1, thioredoxin reductase 1
Introduction
Head and neck squamous cell carcinoma (HNSCC), encompassing an estimated 51,540 new cases in the United States in 2018 [1], represents an aggressive group of malignancies arising from various subsites in the head and neck region. Management of these cancers usually entails primary surgery followed by (chemo)radiotherapy or definitive (chemo)radiotherapy. In recent years, human papillomavirus (HPV) infection has been shown to be a strong, independent risk factor for oropharyngeal cancer [2], creating great interest in de-escalating therapy for these favorable group of patients [3]. However, it remains that a significant proportion of patients with HNSCC, especially those with HPV-negative disease, succumb to their disease, mandating a better understanding of the molecular mechanisms that can inform the development of better diagnostic and therapeutic modalities and improving clinical outcomes.
MicroRNAs (miRNA) are short, endogenous non-coding RNA, measuring approximately 21 to 23 nucleotides in length, that can regulate gene expression at the post-transcriptional level through the interaction of the miRNA with its cognate target mRNA leading to decreased protein expression through translational repression or mRNA destabilization [4]. In the context of cancer, dysregulation of miRNA expression can lead to alteration in expression of oncogenes or tumor suppressor protein coding genes, leading to tumorigenesis or potentiation of a cancer-related phenotype [5]. Given the importance of miRNAs in cancer, there has been a rapid development of novel laboratory techniques and computational methodologies for the comprehensive study of miRNAs and their function [6].
We have previously performed miRNA expression analysis of five patients with HNSCC of their tumor and adjacent normal tissue [7] that were obtained in a prospective trial evaluating pathologic risk features, total combined treatment duration, and post-operative radiation therapy [8]. miR-125a-5p was identified as a miRNA that had decreased expression in tumor tissue, compared to the adjacent normal tissue, with a log ratio of −0.83, suggesting that miR-125a-5p may act as a tumor suppressor miRNA in HNSCC. In this report, we sought to better elucidate the role of miR-125a-5p as a tumor suppressor miRNA and marker of poor prognosis in head and neck squamous cell carcinoma.
Material and Methods
MicroRNA Microarray Analysis
Total RNA was extracted from HNSCC patient samples treated in a prospective clinical trial evaluating the role of postoperative radiation therapy at MD Anderson Cancer Center [8]. Total RNA was extracted using the Qiagen RNeasy Plus kit (Qiagen). MicroRNA array profiling was done using Exiqon platform (Qiagen), using the Exiqon miRCURY LNA microRNA Hi-Power Labeling kit (Qiagen) with the miRCURY LNA microRNA Spike-in miRNA kit (Qiagen), used as a control. Then, labeled cRNA was hybridized on miRCURY LNA microRNA array (Qiagen), 7th generation slide, using Agilent chambers (Agilent Technologies, Santa Clara, CA). The hybridized slides were then scanned using the Tecan PowerScanner (Tecan US), and features were extracted using Arrayit ImaGene 9 (Arrayit Corporation) software for further analysis.
Survival Analysis of miR-125a-5p in The Cancer Genome Atlas
The OncoLnc tool (http://www.oncolnc.org) [9] was utilized to correlate miR-125a-5p expression in head and neck squamous cell carcinoma in The Cancer Genome Atlas and with the survival, using a 50th percentile cutoff was used to dichotomize the expression level of miR-125a-5p. Data was visualized using GraphPad Prism (GraphPad Software).
Target mRNA Expression Analysis in The Cancer Genome Atlas
The UALCAN tool (http://ualcan.path.uab.edu) [10] was utilized to assist in performing subgroup analysis of RNA-seq expression from the head and neck studies in The Cancer Genome Atlas. Expression levels of ERBB2, TXNRD1, and EIF4EBP1 were evaluated based on individual cancer stages, comparing mRNA expression levels to that of normal tissue. Data was visualized using GraphPad Prism (GraphPad).
microRNA Binding Site Analysis
To analyze the potential miRNA binding site of putative mRNA targets of miR-125a-5p, publically available miRNA prediction web tools were utilized, including DIANA-microT [11], miRANDA-miRSVR [12], MirTarget2 [13], and TargetScan [14].
Cell Culture
HN5 and UM-SCC-22B squamous cell carcinoma were cultured in Dulbecco's Modified Eagle Medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Atlas Biologicals) and penicillin/streptomycin (Sigma-Aldrich). Cells were cultured in a humidified atmosphere of 5% CO2 at 37 °C.
microRNA Transfection
Cells were transiently transfected with 25 nM of control miRNA, miR-125a-5p mimics, or miR-125a-5p antisense inhibitors (ThermoFisher) using Lipofectamine RNAiMAX (Invitrogen) using the reverse transfection protocol and were harvested after 72 hours for analysis.
Plasmid DNA Transfection
pcDNA3 vector (ThermoFisher) and pcDNA3-ERBB2 (Addgene) were transfected into cells using Lipofectamine 2000 transfection reagent (ThermoFisher). The pcDNA3-ERBB2 construct was previously constructed and described [15].
mRNA Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using the TRIzol reagent, per protocol (Invitrogen) and resuspended in nuclease-free water (Ambion). Complementary DNA (cDNA) was synthesized using Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR amplification was performed using the Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad), with TaqMan primer/probes sets from Applied Biosystems (Supplementary File 1). TaqMan Fast Advanced Master Mix (Applied Biosystems) was utilized for real-time PCR amplification. The data was analyzed using the 2-ΔΔCt method [16], using the ACTB mRNA as an endogenous control.
microRNA Quantitative Real-Time Polymerase Chain Reaction
microRNA transfection levels were assessed using the miRCURY LNA miRNA PCR System (Qiagen), per manufacturer's directions. The real-time PCR amplification was performed, using the PCR primer sets (Supplementary File 1) for hsa-miR-125a-5p and U6 snRNA, as an internal control. Real-time PCR amplification was performed on the Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad). The data was analyzed using the 2-ΔΔCt method [16], using the U6 snRNA as an endogenous control.
miRNA Target Validation Assay
To validate putative mRNA targets of miR-125a-5p, we utilized an affinity purification that has been previously described [17]. In brief, synthetic microRNAs of miR-125a-5p and miR-551a, used as a non-specific control, was biotinylated at the 3′ end of the RNA. Then, the biotinylated miRNAs were transfected into HN5 cells at a concentration of 50 nM for 48 hours. After lysis, streptavidin-sepharose beads were used to bind and recover biotinylated miRNA:mRNA complexes, and RNA was recovered using TRIzol reagent (Invitrogen). mRNA binding to biotinylated miRNA was then quantified using qRT-PCR.
Western Blot
Whole cell extracts were extracted using standard RIPA (radioimmunoprecipitation assay) buffer, containing SigmaFAST protease inhibitor cocktail tablet (Sigma-Aldrich). Laemmli 6× sodium dodecyl sulfate (SDS) protein loading buffer, containing 5% β-mercaptoethanol, was used for sample preparation followed by boiling the sample to a temperature of 95° Celsius for 5 minutes. A 4–20% Mini-PROTEAN TGX precast protein gel was used for electrophoresis, in Tris-glycine-SDS running buffer followed by a semi-dry transfer to PVDF membrane (Bio-Rad) with the Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). The membrane was probed using antibodies obtained from Cell Signaling Technology as listed in Supplementary File 1. Peroxidase-linked secondary antibodies targeted against rabbit (catalog number NA934) or mouse (catalog number NA931) antibodies were obtained from GE Healthcare Life Sciences. Electrochemiluminescence was generated with the Pierce Electrochemiluminescence (ECL) Western Blotting Substrate (ThermoFisher) and detected and quantitated on the FluorChem M system (ProteinSimple).
Cell Proliferation and Viability
Transfected cells were harvested after 72 hours of transfection in 24-well tissue culture-grade plate. For cell proliferation, cell counts were quantified using the Z2 Coulter Counter Analyzer (Beckman Coulter). For cell viability, transfected cells were harvested after 72 hours of transfection, and cell viability was assayed using the CellTiter-Glo reagent, according to manufacturer's instructions (Promega). Luminescence was measured using the POLARstar OPTIMA multi-mode plate reader (BMG Labtech).
Cell Cycle Analysis
Cell cycle was analyzed by propidium iodide staining of DNA content. In brief, cells were harvested and resuspended in phosphate-buffered solution (PBS) (Sigma-Aldrich). Fixation of cells were performed by added resuspended cells dropwise into 200 proof ethanol and incubated on ice for 30 minutes. The cells were then pelleted by centrifugation at 1000 rpm for five minutes. The pelleted cells were then resuspended in a PBS solution, containing 20 μg/mL of RNase A, 0.1% (vol/vol) of Triton X-100, and 20 μg/mL of propidium iodide. The cells were then incubated at room temperature for 15 minutes. The cells were then analyzed on the FlowSight Imaging Flow Cytometer and IDEAS software package (Amnis Corporation).
In vitro Scratch Assay
Cells were grown and transfected in a 24-well tissue culture-grade plate to approximately 75% confluence. After 72 hours of transfection, a scratch was made using a P200 pipet tip, with an outer diameter of 1.2 mm. After 48 hours, the cells were fixed and stained with a solution of 6.0% (vol/vol) glutaraldehyde and 0.5% crystal violet (wt/vol) in water. Microscopy and measurement of the wound closure was performed with the Evos XL Cell Imaging System (ThermoFisher).
In Vitro Invasion Assay
Cells were transfected in 60 mm tissue culture plates for 24 hours. Harvested cells were counted on the Z2 Coulter Counter Analyzer (Beckman Coulter). Cells were then resuspended in Dulbecco's Modified Eagle Medium (Sigma-Aldrich) without serum. The resuspended cells were then pipetted into a Corning BioCoat Matrigel Invasion Chamber (ThermoFisher), 8 μm pore size, in 24-well tissue culture-grade plates. Dulbecco's Modified Eagle Medium (Sigma-Aldrich) supplemented with 10% FBS (Atlas Biologicals) was placed in the well of the plate as a chemo-attractant. After 24 hours, non-invading cells were removed using a cotton-tipped swab. Invasive cells, embedded into the Matrigel, was fixed using 70% ethanol then stained with a 0.5% crystal violet solution. Microscopy was performed with the Evos XL Cell Imaging System (ThermoFisher). Cells were counted in five fields of view at 10× magnification and then averaged as cells per 10× field.
Clonogenic Assay
Cells were transfected in 60 mm tissue culture plates for 24 hours. Cells were then harvested with trypsin–EDTA solution (Sigma-Aldrich) then counted on the Z2 Coulter Counter Analyzer (Beckman Coulter). Cells were diluted to the appropriate concentration, and then cells were plated on 60 mm tissue culture plates. Ionizing radiation was delivered using the Mark II 137Cs irradiator (JL Shepherd & Associates) at a dose rate of 3.47 Gy per minute. The dosimetry was measured using an ion chamber, calibrated with a National Institute of Standards and Technology traceable source. After 2 weeks, cells were fixed and stained with a solution of 6.0% (vol/vol) glutaraldehyde and 0.5% crystal violet (wt/vol) in water. Surviving fraction was calculated with the following equation:
Survival curves were generated using GraphPad Prism (GraphPad Software) on a log-linear graph with surviving fractions as a function of ionizing radiation dose, in Gray.
Immunofluorescence of γH2AX and 53BP1 foci
Cells were reverse transfected onto 8-well chamber slides (Nunc) for 24 hours. Ionizing radiation was delivered using the Mark II 137Cs irradiator (JL Shepherd & Associates) at a dose rate of 3.47 Gy per minute for a dose of 0, 1, or 2 Gy. Fixation was performed with 4% paraformaldehyde in PBS for 20 minutes at room temperature. The cells were permeabilized with PBS containing 0.5% Triton X-100 on ice for 10 minutes followed by blocking with 5% goat serum in PBS overnight at 4 °C. Rabbit anti-53BP1 antibody and mouse anti-γH2AX antibody (Supplementary File 1) were incubated, at 1:500 dilution in 5% goat serum (ThermoFisher) in PBS, with the cells for 2 hours at room temperature. Then, the cells were incubated with Alexa Fluor 488 goat anti-rabbit secondary antibody (Invitrogen, catalog number: A-11008) and Alexa Fluor 555 goat anti-mouse secondary antibody (Invitrogen, catalog number: A-21422) diluted at a concentration of 1:500 diluted in 1% bovine serum albumin and 2.5% goat serum in PBS and incubated for 1 hour in the dark. PBS was used for washing between each step. Fluoroshield mounting medium (Sigma-Aldrich) containing 4′,6-diamidino-2-phenylindole (DAPI) was used for mounting. Colocalized foci was visualized on the Zeiss Axio Imager 2 microscope (Carl Zeiss Microscopy) and analyzed using the ImageJ image processing program (National Institutes of Health).
BrdU Immunofluorescence Assay
Cells were reverse transfected onto 8-well chamber slides (Nunc) for 24 hours. Fresh 5-bromo-2′-deoxyuridine (BrdU) stock solution (BD Biosciences) was made by diluting BrdU into water for a final concentration of 10 mM. BrdU was further diluted into fresh growth medium at a final concentration of 10 μM and then filtered through a 0.2 μm filter under sterile conditions. The existing cell culture medium was then replaced with the BrdU-containing growth medium for 30 minutes at 37 °C in a CO2 incubator. The BrdU-containing growth medium was then removed, and the cells were immediately washed PBS. The cells were fixed with 4% paraformaldehyde in PBS for 20 minutes at room temperature. The cells were permeabilized with PBS containing 0.5% Triton X-100 on ice for 10 minutes then washed with PBS. DNA hydrolysis was performed by incubating the cells with a 2.5 M solution of hydrochloric acid for 1 hour at room temperature. Blocking was performed using 5% goat serum in PBS for 1 hour at room temperature. Mouse anti-BrdU antibody (BD Biosciences, clone B44) were diluted by mixing 20 μL of the antibody with 50 μL of PBS containing 0.5% Tween-20 and were incubated with the cells for 1 hour at room temperature. Then, the cells were incubated with Alexa Fluor 488 goat anti-mouse secondary antibody diluted at a concentration of 1:500 in 1% bovine serum albumin in PBS for 1 hour in the dark at room temperature. PBS was used for washing between each step. Cells were then mounted in Fluoroshield mounting medium (Sigma-Aldrich) containing 4′,6-diamidino-2-phenylindole (DAPI). 5-bromo-2′-deoxyuridine-positive cells were visualized and analyzed as described above.
Results
Decreased Levels of miR-125a-5p Is Associated With Locoregional Recurrence in Advanced Head and Neck Squamous Cell Carcinoma
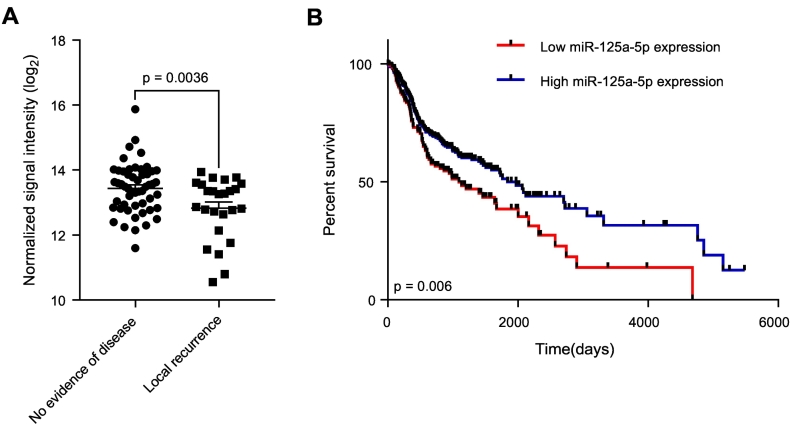
We first utilized microRNA profiling using microarrays to identify expression of miRNAs that are associated with local recurrence using patient samples obtained from a prospective study evaluating pathologic risk features, total combined treatment duration, and post-operative radiation therapy, which 288 registered patients with high-risk, locally advanced disease [8]. Our results reveal that in patients who developed a local recurrence, there was an associated decreased in miR-125a-5p expression, compared to patients who did not have evidence of disease (P = .0036), out of a total of 77 evaluable patient samples analyzed (Fig. 1A). Clinicopathologic characteristics of this patient cohort is shown in Supplementary Table 1. This results suggests that miR-125a-5p is a potential marker associating low expression levels with local recurrence. We then performed chi-squared analysis of our patient cohorts, comparing clinicopathologic risk factors such as age, tumor subsite, T stage, N stage, extracapsular extension, and surgical margin status (Supplementary Table 2). We did not observe any differences in between patients with no evidence of disease and patients with locoregional recurrence with the aforementioned risk factors, suggesting that miR-125a-5p expression is the only analyzed risk factor that is associated with locoregional recurrence.
Figure 1.
Decreased expression of miR-125a-5p is associated with local recurrence and poor prognosis in advanced head and neck cancer. (A) MicroRNA microarray analysis from patient tumor samples collected from a prospective trial of postoperative radiation therapy [8], comprising of 77 evaluable patient samples, was performed, revealing an association of local recurrence (n = 25) with decreased miR-125a-5p expression, compared to patients with no evidence of disease (n = 52) (P = .0036). The data were analyzed with the Student's t-test and is presented as the mean ± standard error of the mean. (B) Kaplan–Meier estimates of survival based on miR-125a-5p expression level in head and neck cancer, based on miRNA expression analysis in The Cancer Genome Atlas. Low levels of miR-125a-5p in head and neck squamous cell carcinoma confers worse survival than high expression of miR-125a-5p (P = .006). Data was analyzed using OncoLnc (www.oncolnc.org) [9]. The data was analyzed using a log rank test.
Decreased levels of miR-125a-5p in head and neck squamous cell carcinoma is associated with a poor prognosis. Using OncoLnc [9], which links RNA-seq expression data with survival in The Cancer Genome Atlas [18] (https://cancergenome.nih.gov), we find that patients with low levels of miR-125a-5p is associated with a worse overall survival than patients with high levels of miR-125a-5p (P = .006) (Figure 1B). This suggests that miR-125a-5p may act as a tumor suppressor miRNA and that decreased miR-125a-5p levels can lead to a more aggressive phenotype and clinical outcome. Univariable Cox regression analysis did not identify any other variable in our cohort that affected survival (Supplementary Table 3), suggesting that decreased miR-125a-5p expression is associated with poor prognosis.
miR-125a-5p Modulates Cell Proliferation by Regulating the G1/S Transition of the Cell Cycle
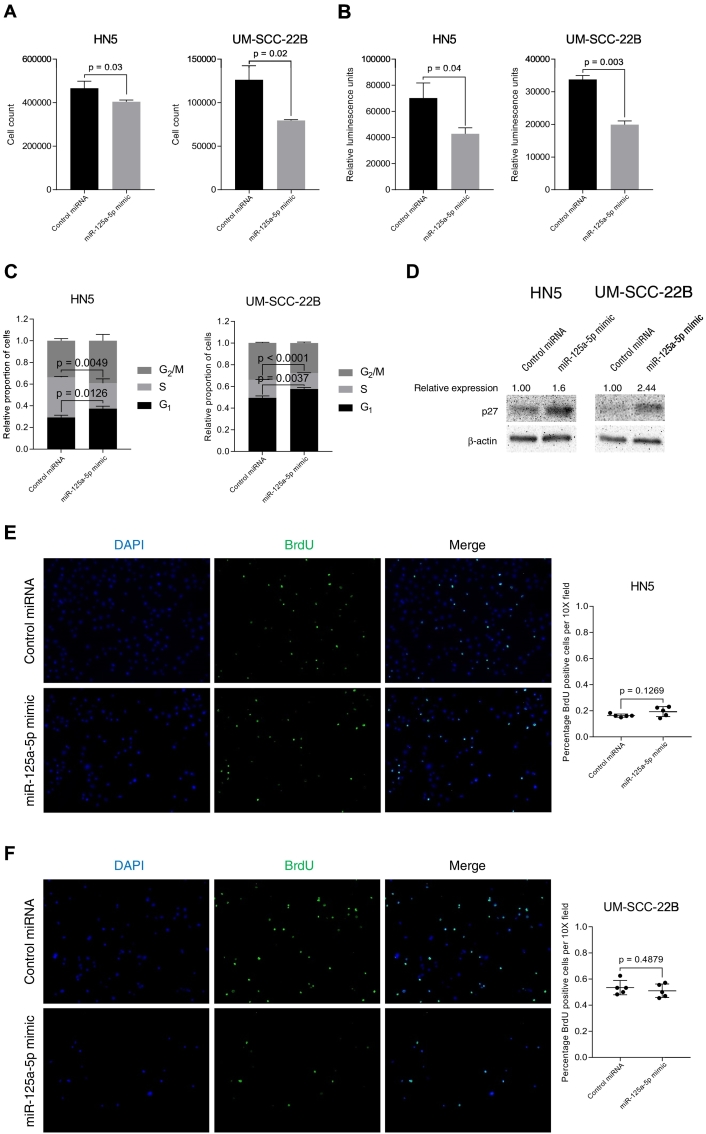
We hypothesize that miR-125a-5p functions as a tumor suppressor miRNA, and to test this hypothesis, we studied the effect of miR-125a-5p on cell proliferation in vitro. We transfected miR-125a-5p mimics into HN5 and UM-SCC-22B head and neck squamous cell carcinoma cell lines. We confirmed the transfection of miRNA mimics into each cell line (Supplementary Fig. 1). We observed that there is decreased cell proliferation in both HN5 and UM-SCC-22B cell lines when miR-125a-5p mimics are transfected, compared to the negative control miRNA, using an in vitro cell counting assay (Figure 2A). We corroborated the result by performing a cell viability assay with the CellTiter-Glo luminescent reagent, which showed decreased cell viability with transfection of miR-125a-5p, compared to cells transfected with control miRNA (Figure 2B).
Figure 2.
miR-125a-5p modulates cell proliferation in vitro via regulating the cell cycle at the G1/S transition by increasing p27 expression. (A) Transfection of miR-125a-5p into HN5 and UM-SCC-22B cell lines results in decreased cell counts, compared to transfection of a control miRNA. (B) Transfection of miR-125a-5p into HN5 and UM-SCC-22B cell lines results in luminenscence, using the CellTiter-Glo Assay (Promega), compared to transfection of a control miRNA. (C) Transfection of miR-125a-5p into HN5 and UM-SCC-22B cell lines results in increased proportion of cells in the G1 phase with a concomitant decrease in percentage of cells in S phase, compared to control miRNA transfection, as determined by the propidium iodide cell cycle assay. Cell cycle analysis was performed on the FlowSight Imaging Flow Cytometer (Amnis). (D) Western blotting of p27 protein shows increased expression of p27 after transfection of miR-125a-5p mimic, as compared to control miRNA. 5-bromo-2′-deoxyuridine (BrdU) assay reveals that miR-125a-5p does not affect DNA synthesis in HN5 (E) and UM-SCC-22B (F) cell lines. Data were analyzed with the Student's t-test and are presented as the mean ± standard deviation. Experiment was carried out in triplicate with the BrdU DNA synthesis assay performed in quintuplicate.
We then performed cell cycle analysis to further investigate the effect of miR-125a-5p on cell proliferation and cell cycle regulation. In both HN5 and UM-SCC-22B cell lines, there was an increase in the proportion of cells in the G1 phase with transfection of miR-125a-5p, with a concomitant proportion of cells decreased in S phase, in HN5 and UM-SCC-22B cells with miR-125a-5p mimic transfected, compared to controls (Figure 2C). To further investigate the role of miR-125a-5p on cell cycle progression, we found that there was an increase in p27 protein expression with transfection of miR-125a-5p mimics, compared to control cells, in both tested cell lines (Figure 2D). We also performed a BrdU incorporation immunofluorescence assay that did not show any changes with DNA synthesis with transfection of miR-125a-5p in either cell lines (Figure 2, E and F). We also did not see any effects from apoptosis or autophagy that can affect cell proliferation assays (Supplementary Fig. 2). Our data suggests that miR-125a-5p affects cell proliferation by regulating transit through the G1/S transition through p27 but does not affect DNA synthesis.
miR-125a-5p Regulates Cell Migration and Invasion In Vitro
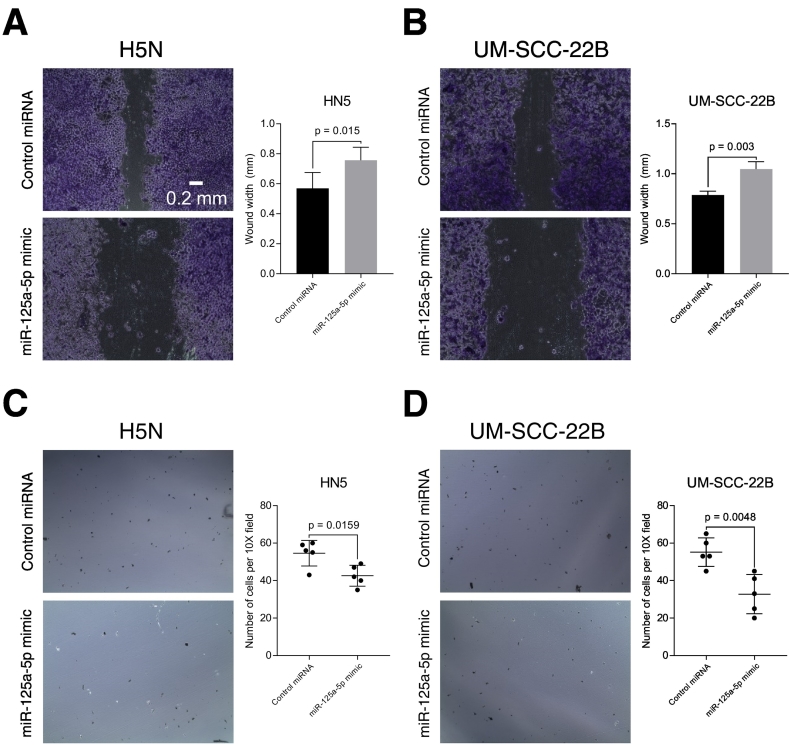
Head and neck squamous cell carcinoma are aggressive malignancies that are locally destructive with high rates of metastatic spread to regional lymph nodes. Histologically, worse pattern of invasion is an independent predictor of locoregional recurrence, in patients with low-stage oral cavity squamous cell carcinoma [19]. We performed an in vitro cell migration scratch assay and found that transfection of miR-125a-5p into both HN5 and UM-SCC-22B cell lines resulted in decreased cell migratory ability, as compared to the control miRNA (Figure 3, A and B). We then performed an in vitro transwell invasion assay using Matrigel extracellular matrix. We observed that there was decreased in vitro cell invasion with transfection of miR-125a-5p, compared to the control miRNA (Figure 3, C and D).
Figure 3.
miR-125a-5p regulates cell migratory and invasive ability in vitro. Transfection of miR-125a-5p into HN5 (A) and UM-SCC-22B (B) cell lines results in decreased wound healing, compared to control miRNA, in an in vitro wound healing assay. Cells were fixed in glutaraldehyde and stained with crystal violet. Brightfield microscopy images were taken with a 10x objective. Cell invasion was assessed with a Matrigel invasive chamber. Cells were transfected with miRNA mimics and then placed into the chamber for a period of 24 hours. The cells that were embedded in the Matrigel were stained using crystal violet and counted using brightfield microscopy with a 10x objective. There were decreased cells invading into the Matrigel with transfection of miR-125a-5p mimic, compared to the control miRNA, in both HN5 (C) and UM-SCC-22B (D) cells. Data were analyzed with the Student's t-test and are presented as the mean ± standard deviation. The in vitro scratch assay was carried out in triplicate. Five 10x fields were analyzed for the in vitro invasion assay.
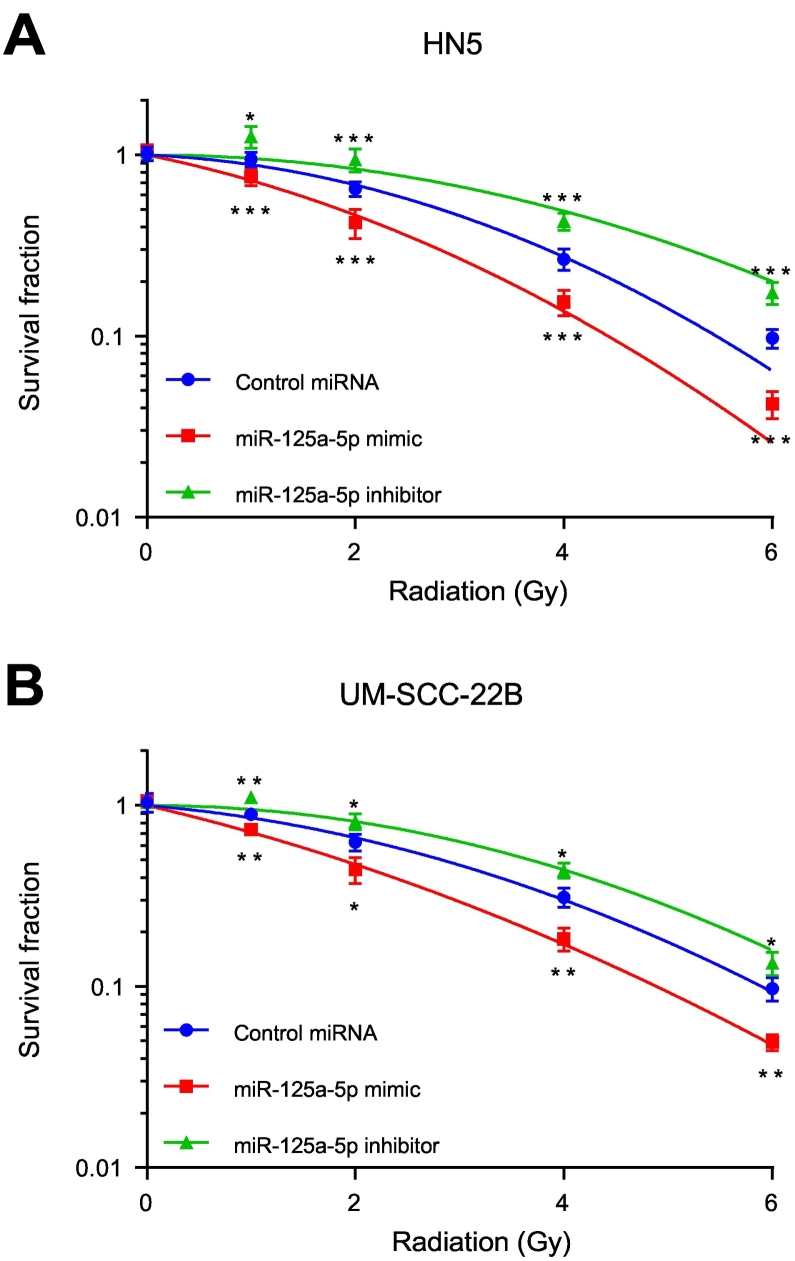
miR-125a-5p Modulates Sensitivity to Ionizing Radiation
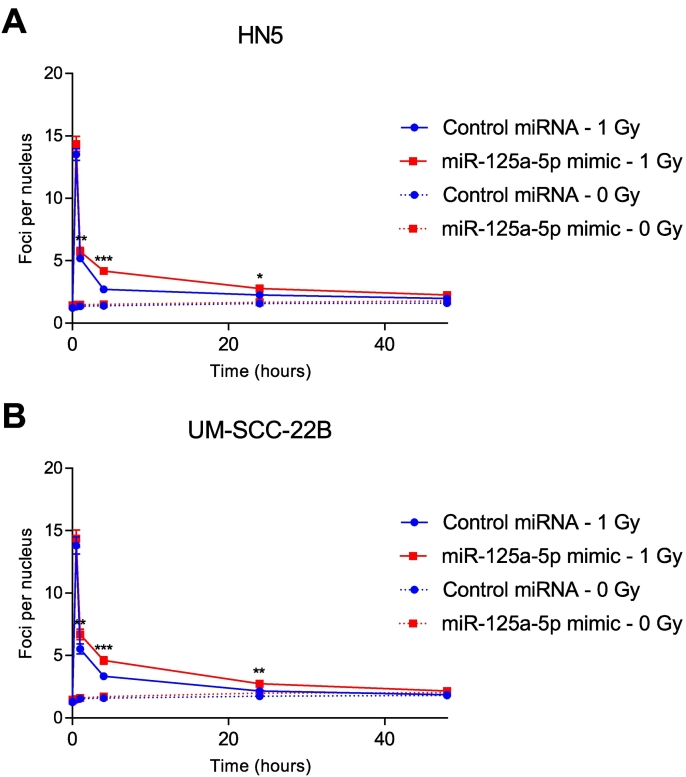
Radiation therapy is a key modality for the management of head and neck squamous cell carcinoma, either in the definitive, adjuvant, or recurrent setting. To evaluate the effect of miR-125a-5p on response to ionizing radiation, an in vitro clonogenic assay was performed. With transfection of miR-125a-5p mimics, we observed a decrease in surviving fraction in response to ionizing radiation, compared to the control miRNA, in HN5 (Figure 4A) and UM-SCC-22B cells (Figure 4B). Similarly, when miR-125a-5p is inhibited using antisense inhibitors, the cells are rendered for resistant to ionizing radiation. A γH2AX and 53BP1 foci formation assay was then performed to determine the effect of miR-125a-5p on double-stranded DNA break repair kinetics in response to ionizing radiation, in HN5 and UM-SCC-22B cell lines. After 1 Gy of ionizing radiation, there was an appreciable increase in γH2AX and 53BP1 colocalized foci at 1, 4, and 24 hours (Figure 5 and Supplementary Fig. 3), in both cell lines with miR-125a-5p, compared to the control miRNA. There was no difference in the number of colocalized 53BP1 and γH2AX foci at 48 hours, with the number of foci per nucleus returning to baseline levels. There was no increase in double-stranded DNA break formation without any ionizing radiation, in response to transfection of miR-125a-5p, suggesting that there is no contribution by miR-125a-5p by itself to double-stranded DNA damage (Figure 5 and Supplementary Fig. 4). This effect is also seen in response to 2 Gy of ionizing radiation (Supplementary Fig. 5, Supplementary Fig. 6). Our data suggests that miR-125a-5p has a role in modulating sensitivity to ionizing radiation in head and neck squamous cell carcinoma through delaying resolution of double-stranded DNA breaks.
Figure 4.
miR-125a-5p modulates sensitivity to ionizing radiation. HN5 (A) and UM-SCC-22B (B) cell lines were transfected with miR-125a-5p mimic, miR-125a-5p inhibitors, or control miRNA. Cells were then harvested and plated. Four hours after plating, cells were then treated increasing doses of ionizing radiation using a Cesium-137 irradiator. After two weeks of growth, colonies, consisting of 50 cells or more, were fixed with a solution of glutaraldehyde and stained with crystal violet. Colonies were then counted and plotted with surviving fraction as a function of radiation dose. Transfection of miR-125a-5p mimics results in increased sensitivity to ionizing radiation, compared to the cells transfected with control miRNA, while transfection of miR-125a-5p antisense inhibitors rendered the cells more resistant to ionizing radiation. Experiment was carried out in triplicate. Data were analyzed with the Student's t-test at each ionizing radiation dose level and are presented as the mean ± standard deviation with * signifying P ≤ .05, ** signifying P ≤ .01, and *** signifying P ≤ .001.
Figure 5.
miR-125a-5p reduces rate of resolution of double-stranded DNA break in response to ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 1 and 0 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 and Alexa Fluor 555, respectively. Colocalized foci were then counted and displayed as the mean ± standard error of the mean. The data at each time point was analyzed using the Student's t-test, with * signifying P ≤ .05 and ** signifying P ≤ .01. There was a significant increase in colocalized 53BP1 and γH2AX foci with miR-125a-5p, compared to the control miRNA, at 4 hours after irradiation with 1 Gy that persisted at 24 hours.
miR-125a-5p Targets the mRNA of ERBB2, EIF4EBP1, and TXNRD1 in Head and Neck Squamous Cell Carcinoma
We hypothesized that miR-125a-5p acts as a tumor suppressor miRNA by targeting oncogenic mRNAs, and that through loss or downregulation of miR-125a-5p, there is a resultant increase in the expression of various oncogenic proteins. Therefore, to better understand how miR-125a-5p exerts its effects, we sought to identify mRNA targets of miR-125a-5p.
In order to identify putative mRNA targets of miR-125a-5p, we identified mRNAs that were upregulated in an mRNA expression microarray dataset that was generated from patient tumor samples that were collected in a prospective trial evaluating pathologic risk features, total combined treatment duration, and post-operative radiation therapy [8]. Using a cut-off of two-fold increase in mRNA expression levels, we identified 1249 mRNAs that were upregulated in tumor samples, compared to normal tissue. We then utilized four miRNA prediction algorithms, DIANA-microT [11], miRANDA-miRSVR [12], MirTarget2 [13], and TargetScan [14], to identify subsets of the upregulated mRNAs that contain well-conserved, low-binding energy binding sites for miR-125a-5p. We focused on mRNAs that were predicted in at least two prediction algorithms. In the end, we identified 41 mRNAs that were upregulated in tumor samples that contained a binding site for miR-125a-5p, as predicted by at least two prediction algorithms (Supplementary Table 2). From this list of 29 putative target mRNAs, we selected three mRNAs for further study as they have been previously implicated in head and neck squamous cell carcinoma, ERBB2 [20], EIF4EBP1 [21], and TXNRD1 [22].
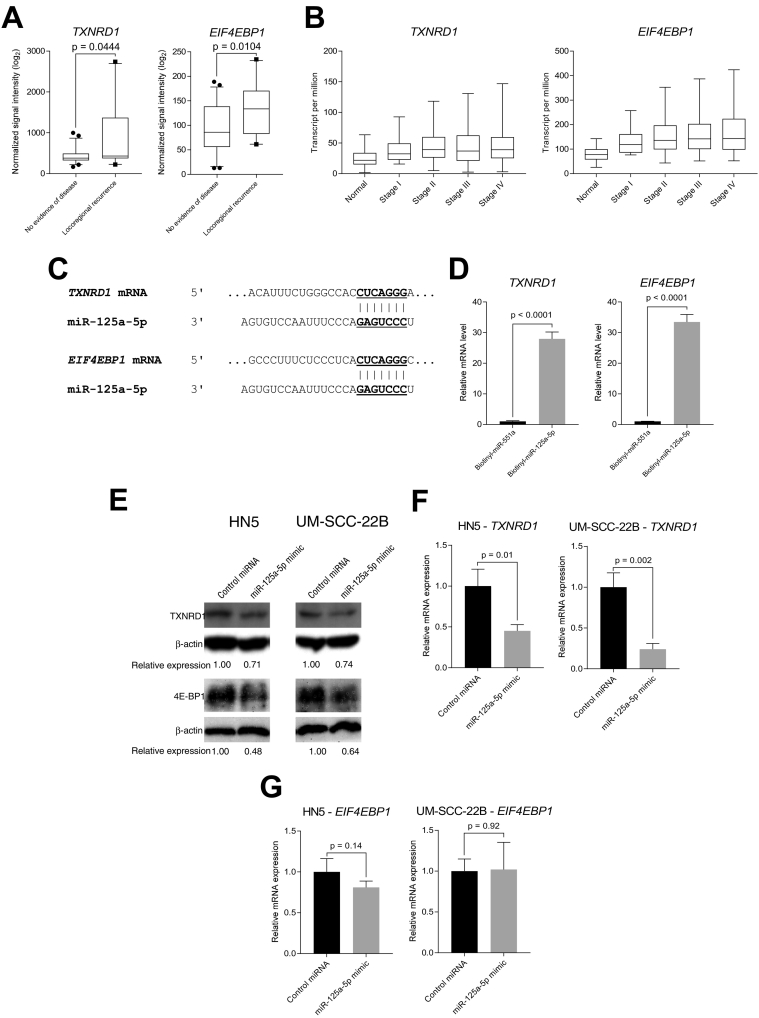
We then mined through our microarray profiling data and observed statistically significant increase in mRNA expression levels of these target mRNAs in our microarray profiled patients with locoregional recurrence, compared to patients with no evidence of disease (Figure 6A), the inverse of what was observed with miR-125a-5p expression (Figure 1). We then mined through The Cancer Genome Atlas, using the UALCAN web tool (ualcan.path.uab.edu) and found that that head and neck squamous cell carcinoma exhibited increasing mRNA levels of TXNRD1 and EIF4EBP1 with more advanced clinical stage of disease presentation (Figure 6B and Supplementary Table 5). Analysis of the 3′ untranslated region of TXNRD1 and EIF4EBP1 mRNA reveals putative, canonical binding sites for miR-125a-5p. (Figure 6C). To evaluate whether the mRNA of TXNRD1 and EIF4EBP1 physically binds to miR-125a-5p, we performed a biotin-streptavidin pulldown assay [17] to confirm the targeting of EIF4EBP1 and TXNRD1 by miR-125a-5p, using a biotinylated synthetic construct of miR-125a-5p. We found that both putative mRNA targets are bound to biotinylated miR-125a-5p, as measured by qRT-PCR, but not biotinylated miR-551a, which was used as a negative, nonspecific control, confirming the in silico prediction of the binding of the target mRNA to miR-125a-5p (Figure 6D). We then transfected miR-125a-5p into HN5 and UM-SCC-22B cell lines and then blotted for 4E-BP1 and TXNRD1 protein. We found that miR-125a-5p was able to repress protein levels of TXNRD1 and EIF4EBP1 mRNA targets (Figure 6E). We also measured mRNA levels using qRT-PCR and found that the mRNA levels of TXNRD1 were decreased after miR-125a-5p transfection, as compared to the control miRNA, suggesting that miR-125a-5p, at least in part, controls expression of TXNRD1, through mRNA destabilization (Figure 6F). However, when we evaluated the mRNA expression of EIF4EBP1, there was no statistically difference between the miR-125a-5p mimic and control miRNA (Figure 6G), signifying that decrease in protein level as seen in the immunoblotting analysis is mainly through translational repression, rather than mRNA destabilization.
Figure 6.
Putative mRNA targets of miR-125a-5p are associated with locoregional recurrence, are upregulated in advanced squamous cell carcinoma of the head and neck, compared to normal tissue and directly targets TXNRD1 and EIF4EBP1 mRNA to downregulate protein expression. (A) Messenger RNA microarray analysis from patient tumor samples collected from a prospective trial of postoperative radiation therapy was performed, revealing an association of local recurrence with increased mRNA expression of putative target mRNAs, TXNRD1 and EIF4EBP1, compared to patients with no evidence of disease. The data were analyzed with the Student's t-test and is presented as the mean ± standard error of the mean. (B) mRNA expression levels for EIF4EBP1, which encodes for the eukaryotic translation initiation factor 4E-binding protein 1, and TXNRD1, which encodes for thioredoxin reductase 1, from The Cancer Genome Atlas were analyzed according to mRNA expression levels, determined by RNA-seq, and analyzed based on cancer stage. UALCAN (ualcan.path.uab.edu) [10] was utilized for data acquisition and analysis, which combines mRNA expression data with clinical data. (C) Predicted binding sites of miR-125a-5p on the 3′ untranslated region of TXNRD1 and EIF4EBP1 mRNA. The TargetScan (www.targetscan.org) [14] was utilized to predict for the binding site based on conservation of the mRNA binding site to the miRNA seed sequence. Vertical lines show Watson-Crick base pairing between the mRNA and miRNA. (D) In vitro pulldown assay with a synthetically biotinylated miR-125a-5p were transfected into HN5, using biotinylated miR-551a as a negative control, and then recovered using streptavidin-agarose beads, which were then washed. Recovered RNAs were analyzed using quantitative RT-PCR, which revealed that TXNRD1 and EIF4EBP1 mRNAs are directly bound to miR-125a-5p, compared to the RNA species bound to miR-551a. (E) Transfection of miR-125a-5p results in decreased TXNRD1 and EIF4EBP1 protein expression, compared to control miRNA, in HN5 and UM-SCC-22B cell lines. (F) Transfection of miR-125a-5p results in decreased TXNRD1 mRNA expression, compared to control miRNA. (G) Transfection of miR-125a-5p does not change the levels of the EIF4EBP1 transcript, suggesting that translational repression is main mechanism of gene regulation for EIF4EBP1. Data were analyzed with the Student's t-test and are presented as the mean ± standard deviation. Experiment was carried out in triplicate.
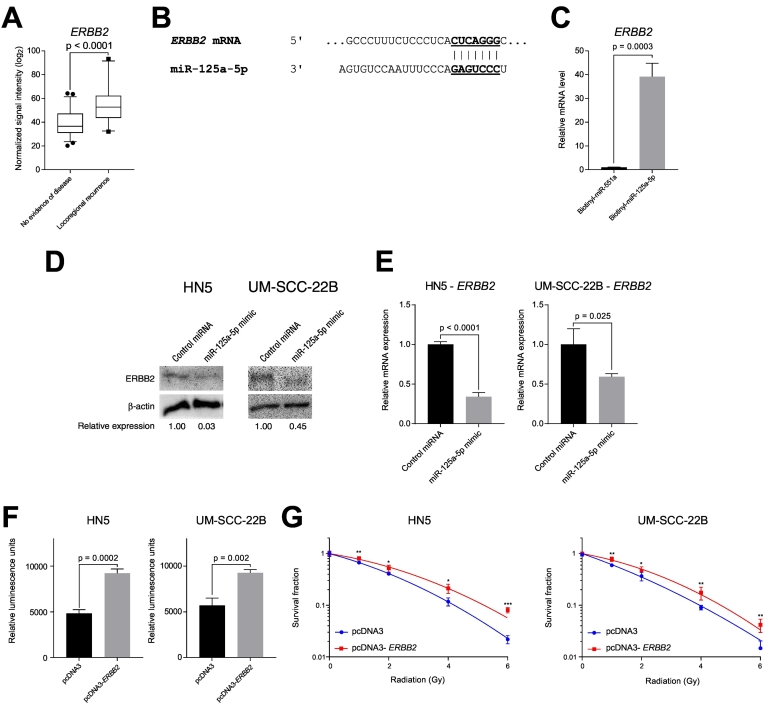
We also investigated ERBB2 mRNA as a target of miR-125a-5p, as it is an important signaling molecule in many cancers, including head and neck cancer. Similar to TXNRD1 and EIF4EBP1, we observed an increased in mRNA expression in patients who had a locoregional recurrence, compared to patients with no evidence of disease (Figure 7A). We then analyzed the 3′ untranslated region of ERBB2 and found that it contains a binding site for miR-125a-5p (Figure 7B). Using an in vitro pulldown assay, we found that the ERBB2 mRNA preferentially binds to biotinylated miR-125a-5p, compared to the nonspecific, biotinylated miR-551a (Figure 7C). Transfection of miR-125a-5p resulted in decreased ERBB2 protein expression, compared to control miRNA transfection (Figure 7D), likely in part to decreased ERBB2 mRNA expression (Figure 7E). In addition, overexpression of ERBB2 using an expression plasmid construct resulted in increased cell proliferation in HN5 and UM-SCC-22B cells, compared to the empty vector pcDNA3 control (Figure 7F). We observed increased PI3K-AKT signaling with a concomitant decrease in p27Kip1 protein expression (Supplementary Fig. 7), and we also found that overexpression of ERBB2 results in increased resistance to ionizing radiation, compared to transfection of a control cDNA plasmid, suggesting that miR-125a-5p modulates sensitivity to radiation therapy, in part, through ERBB2 (Figure 7G).
Figure 7.
The putative mRNA target of miR-125a-5p, ERBB2, is associated with locoregional recurrence and modulates sensitivity to ionizing radiation. (A) Messenger RNA microarray analysis from patient tumor samples collected from a prospective trial of postoperative radiation therapy was performed, revealing an association of local recurrence with increased mRNA expression of putative target mRNA, ERBB2, compared to patients with no evidence of disease. The data were analyzed with the Student's t-test and is presented as the mean ± standard error of the mean. (B) Predicted binding sites of miR-125a-5p on the 3′ untranslated region of ERBB2 mRNA. The TargetScan (www.targetscan.org) [14] was utilized to predict for the binding site based on conservation of the mRNA binding site to the miRNA seed sequence. Vertical lines show Watson-Crick base pairing between the mRNA and miRNA. (C) In vitro pulldown assay with a synthetically biotinylated miR-125a-5p were transfected into HN5, using biotinylated miR-551a as a negative control, and then recovered using streptavidin-agarose beads, which were then washed. Recovered RNAs were analyzed using quantitative RT-PCR, which revealed that ERBB2 mRNA binds directly to miR-125a-5p, compared to the RNA species bound to miR-551a. (D) Transfection of miR-125a-5p results in decreased ERBB2 protein expression, compared to control miRNA, in HN5 and UM-SCC-22B cell lines. (E) Transfection of miR-125a-5p results in decreased ERBB2 mRNA expression, compared to control miRNA. (F) Overexpression of ERBB2 by transfection of plasmid DNA containing the coding sequence of ERBB2 (pcDNA3-ERBB2) increases cell proliferation in HN5 and UM-SCC-22B, as measured using the CellTiter-Glo Assay (Promega), compared to transfection of an empty control plasmid (pcDNA3), measured 72 hours after transfection. The data were analyzed with the Student's t-test and is presented as the mean ± standard deviation. (G) ERBB2 overexpression increases resistance to ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with a plasmid containing an ERBB2 cDNA or control cDNA. Cells were then harvested and plated. Four hours after plating, cells were then treated increasing doses of ionizing radiation using a Cesium-137 irradiator. After 2 weeks of growth, colonies, consisting of 50 cells or more, were fixed with a solution of glutaraldehyde and stained with crystal violet. Colonies were then counted and plotted with surviving fraction as a function of radiation dose. Transfection of ERBB2 cDNA results in increased resistance to ionizing radiation, compared to the cells transfected with control cDNA plasmid. Data were analyzed with the Student's t-test at each ionizing radiation dose level and are presented as the mean ± standard deviation with * signifying P ≤ .05, ** signifying P ≤ .01, and *** signifying P ≤ .001.
Discussion
With the increasing amount of data accumulating from systems-based efforts such as next generation sequencing, microRNA in cancer has created a great amount of interest as its study can allow for interrogation of the underlying biology in cancer, revealing novel mechanisms in tumorigenesis, as well as the potential development of markers for diagnostic and therapeutic applications. However, given the amount of data created, a greater amount of effort is required to validate the significance of potential genes or non-coding RNAs, such as miRNAs, in cancer. Regulation of gene expression is finely regulated in normal cells, and aberrations in gene expression, such as in cancer, are dependent on the cell type and context. Specifically, the regulation of a single mRNA at the post-transcriptional level can be govern by changes in secondary structure, binding by RNA-binding proteins, and miRNAs, requiring genomic approaches and computational methods for study and analysis [6]. Nonetheless, it is very important to experimentally validate the significance of miRNAs and their posited roles in tumorigenesis. In our study, we establish a defined role of miR-125a-5p as a tumor suppressor miRNA in head and neck squamous cell carcinoma, whose basis was from a previous study from our group showing that miR-125a-5p is downregulated in HNSCC, as compared to normal tissue [7].
In our study, we demonstrate that miR-125a-5p functions as a tumor suppressor microRNA (miRNA) in head and neck squamous cell carcinoma (HNSCC) through regulating cell proliferation and mediating sensitivity to ionizing radiation. Our report suggests potential clinical implications as radiotherapy is the backbone of treatment for patients with head and neck cancer, commonly used either in the definitive or adjuvant setting. The tumor suppressive function of miR-125a-5p in HNSCC, as demonstrated by our results, is concordant with previous studies in other malignancies, such as lung cancer [23], breast cancer [24], multiple myeloma [25], gastric cancer [26], hepatocellular carcinoma [27], chronic lymphocytic leukemia [28], nasopharyngeal carcinoma [29], glioblastoma [30], colon cancer [31], prostate cancer [32], cervical carcinoma [33], and thyroid cancer [34], implicating miR-125-5p as a general tumor suppressor miRNA, across a broad spectrum of malignancies.
This report has also identified three mRNA targets of miR-125a-5p in head and neck squamous cell carcinoma, specifically, ERBB2, which encodes for the erbB-2, otherwise known as HER2/neu, receptor tyrosine kinase, TXNRD1, which encodes for thioredoxin reductase 1, and EIF4EBP1, which encodes for the 4E-BP1 translation initiation factor. ERBB2/HER2 is a receptor tyrosine kinase that belongs to the ERBB family and is commonly amplified in breast cancer [35] or gastric cancer [36], can result in homodimerization, enhancing downstream signaling, increasing cell proliferation, decreased apoptosis, and increased cell migratory and invasive abilities [37]. ERBB2 has also emerged a potent target of trastuzumab and pertuzumab in breast cancer [38]. Our functional assays evaluating cell proliferation and sensitivity to ionization radiation suggest that miR-125a-5p acts, in part, through ERBB2 [39], [40]. Thioredoxin reductase 1, a selenocysteine-containing protein, is a major component in the reduction–oxidation (redox) that is a critical system maintaining cellular homeostasis and normal function, including cell proliferation, protection from oxidative stress, and regulating the extracellular redox state [41]. Thioredoxin reductase 1 is found to be overexpressed in many human cancers [42], leading to resistance to ionizing radiation, likely through increased AP-1 DNA-binding activity, a transcription factor that contain redox-sensitive cysteine motifs [43]. Targeting thioredoxin reductase 1 with the plant polyphenol, curcumin, can induce radiosensitization in squamous carcinoma cells [44]. Another mRNA target of miR-125a-5p reported in our study is EIF4EBP1, which encodes for the eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1). Normally, 4E-BP1 is phosphorylated, alongside S6 kinase 1 (S6K1), by mTOR complex 1 (mTORC1), in response of upstream growth factor signals to stimulate protein synthesis [45]. 4E-BP1 can promote the translation of cap-independent mRNAs over cap-dependent mRNAs, especially in times of cellular stress, such as in the case of Bcl-2, to promote cellular survival [46]. Overexpression of 4E-BP1 occurs in many types of cancer, including breast cancer, colorectal cancer, kidney cancer, and lung cancer and is thought to acts a tumor promoter [47]. Our data suggests an additional mechanism for increased 4E-BP1 levels through a miR-125a-5p-mediated, post-transcriptional mechanism, in addition to the previously described mechanisms of gene amplification of the 11q13 chromosomal region [48]. Our study highlights the potential of understanding miRNA biology to reveal important targets and dissect influential mechanisms in head and neck cancer.
Perhaps, the more intriguing aspects of studying miRNA in cancer is in its value for diagnostic and therapeutic applications. There has been great interest in using miRNAs as a therapeutic strategy. Recently, in a phase I study of liposomal miR-34a mimic, dubbed MRX34, many patients enrolled on the study experienced adverse effects, which was suspected to be due to infusion-related intolerability, limiting its potential clinical utility [49]. However, the potential for RNA interference-based therapeutics continues to exist through the recent Breakthrough Therapy Designation from the U.S. Food and Drug Administration for patisiran, an RNA interference therapeutic agent, for the treatment of hereditary transthyretin amyloidosis with polyneuropathy based on recent phase III study demonstrating improved neurological outcomes [50]. More interestingly, there has been increasing interesting in incorporating genomics into clinical practice, such as the use of gene expression profiling in the management of early stage breast cancer to guide the decision of use of adjuvant systemic therapy [51] or genomic classifiers to predict for metastatic disease in patients with prostate cancer post-prostatectomy [52]. In nasopharyngeal cancer, the detection of Epstein–Barr virus DNA after primary therapy with chemotherapy and radiotherapy was associated with worse survival than patients with undetectable levels of Epstein–Barr virus DNA [53], suggesting that there is a subset of patients that can potentially benefit from treatment escalation, which is the basis of the currently accruing NRG Oncology trial, NRG HN-001.
Our study suggests that miR-125a-5p has a molecularly pivotal role in head and neck cancer, and lower expression of miR-125a-5p is associated with increased risk of locoregional recurrence, proposing that miR-125a-5p expression can be utilized as a biomarker to individualize treatment for patients with head and neck cancer, especially those that are HPV-negative, a malignancy whose biology does not currently have any clinically defined and useful biomarker. These patients can potentially benefit from treatment escalation, for example, in the form of dose-escalated radiotherapy or use of novel and emerging systemic agents such as immunotherapies. In sum, the study of miRNAs in cancer can allow for the uncovering and discovery of novel targets in cancer as well as allow for the use biomarker-based risk stratification and response assessment to personalize cancer therapy, making the study of miRNAs even more important than before.
The following are the supplementary data related to this article.
Validating transfection of miRNA mimics, miR-125a-5p and control miRNA, by quantitative real-time polymerase chain reaction (qRT-PCR). HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p and control miRNA mimics for a period of 72 hours. The cells were then extracted with the TRIzol reagent then quantified using a the miRCURY LNA miRNA PCR System, using the U6 snRNA as an internal, housekeeping control. qRT-PCR reveals efficacy of transfection of miR-125a-5p mimic into HN5 and UM-SCC-22B cell lines, compared to the control miRNA. Data were analyzed with the Student’s t-test and are presented as the mean ± standard deviation. Experiment was carried out in triplicate.
miR-125a-5p does not alter cell death pathways of apoptosis or autophagy. miR-125a-5p mimics were transfected into HN5 and UM-SCC-22B cell lines. Protein was extracted, and Western blots were performed with the antibodies for poly-ADP ribose polymerase (PARP), LC3B, and β-actin, a loading control. No difference in PARP cleavage, a marker of apoptosis, or LC3B expression, a marker of autophagy. LC3B-II is the autophagosome-associating form of LC3B-I. However, there was a decrease in total, uncleaved PARP protein levels that was noted.
Representative microscope images of double-stranded DNA break formation after 1 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 1 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
Representative microscope images of double-stranded DNA break formation after 0 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then sham irradiated with 0 Gy. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary fluorescent antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
miR-125a-5p reduces rate of resolution of double-stranded DNA break in response to 2 Gy of ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 2 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 and Alexa Fluor 555, respectively. Colocalized foci were then counted and displayed as the mean ± standard error of the mean. The data at each time point was analyzed using the Student’s t-test with * signifying P ≤ .05. There was a significant increase in colocalized 53BP1 and γH2AX foci with miR-125a-5p, compared to the control miRNA, at 4 hours after irradiation with 2 Gy. At 24 hours, there was complete resolution of double-stranded DNA break as the number of colocalized foci formation returned to baseline levels.
Representative microscope images of double-stranded DNA break formation after 2 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 2 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
ERBB2 increases PI3K-AKT signaling. Transfection of pcDNA3-ERBB2 also increases phosphorylation of Akt at serine 473 while decreases expression of p27Kip1, compared to the control transfection of pcDNA3, as determined by Western blot.
Characteristics of the 77 patients who had miR-125a-5p expression analyzed. Patient characteristics included age, tumor subsite, T stage, N stage, presence of extracapsular extension, and surgical margin status.
chi-Squared analysis of clinicopathologic risk factors in 77 patients who had miR-125a-5p expression analyzed. Contingency tables were constructed and analyzed using chi-squared analysis. Risk factors that were analyzed included age, tumor subsite, T stage, N stage, extracapsular extension, and surgical margin status. NED denotes no evidence of disease, and LR denotes locoregional recurrence. No statistical difference was noted between any of the groups.
Univariable analysis of clinicopathologic risk factors of head and neck cancer patients in The Cancer Genome Atlas cohort. Univariable Cox regression analysis was performed for the following risk factors, including, race, ethnicity, and tumor subsite. No statistical difference was noted between each analyzed risk factor.
Complete list of putative mRNA targets of miR-125a-5p predicted in silico in locally advanced head and neck cancer.
Statistical analysis of comparisons between different stage groups in The Cancer Genome Atlas cohort for TXNRD1 and EIF4EBP1 mRNA expression. Comparisons between stage groupings were analyzed by Student’s t-test.
List of reagents, with catalog numbers, used in this study.
Footnotes
Competing interests: The authors declare that they have no competing interests with the contents of this article.
Acknowledgements: The authors would thank Brock Sishc, PhD and Elliot Fletcher, PhD for their assistance throughout this work.
Funding: Cancer Prevention and Research Institute of Texas Individual Research Award (CPRIT) to MDS, the David A. Pistenmaa Distinguished Chair in Radiation Oncology to MDS, National Cancer Institute (NCI) RO1 CA168484 to JVH, and an American College of Radiation Oncology (ACRO) Resident Seed Grant to DTV.
Availability of data and materials: The data generated or analyzed during this study are included in this article, or if absent are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Conception and design: Dat Vo, Narasimha Kumar Karanam, and Michael Story; Acquisition of data: Dat Vo, Narasimha Kumar Karanam, Uma Giri and John Yordy; Analysis and interpretation of the data: all authors; Drafting the article: Dat Vo; Critically revising the article: all authors; Approved the final version of the manuscript on behalf of all authors: Dat Vo; Study supervision: Michael Story
References
- 1.Siegel RL, Miller KD. Jemal A (2018). Cancer statistics. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirghani H, Amen F, Blanchard P, Moreau F, Guigay J, Hartl DM, Lacau St Guily J. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: ongoing trials, critical issues and perspectives. Int J Cancer. 2015;136:1494–1503. doi: 10.1002/ijc.28847. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. Vol. 116. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function Cell; pp. 281–297. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahrami-Samani E, Vo DT, de Araujo PR, Vogel C, Smith AD, Penalva LO, Uren PJ. Computational challenges, tools, and resources for analyzing co- and post-transcriptional events in high throughput. Wiley Interdiscip Rev RNA. 2015;6:291–310. doi: 10.1002/wrna.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramdas L, Giri U, Ashorn CL, Coombes KR, El-Naggar A, Ang KK, Story MD. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck. 2009;31:642–654. doi: 10.1002/hed.21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang KK, Trotti A, Brown BW, Garden AS, Foote RL, Morrison WH, Geara FB, Klotch DW, Goepfert H, Peters LJ. Randomized trial addressing risk features and time factors of surgery plus radiotherapy in advanced head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51:571–578. doi: 10.1016/s0360-3016(01)01690-x. [DOI] [PubMed] [Google Scholar]
- 9.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2 [Google Scholar]
- 10.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YM, Pan Y, Wei Y, Cheng X, Zhou BP, Tan M, Zhou X, Xia W, Hortobagyi GN, Yu D. Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell. 2004;6:459–469. doi: 10.1016/j.ccr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method Methods25; pp. 402–408. [DOI] [PubMed] [Google Scholar]
- 17.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Bai S, Carroll W, Dayan D, Dort JC, Heller K, Jour G, Lau H, Penner C, Prystowsky M. Validation of the risk model: high-risk classification and tumor pattern of invasion predict outcome for patients with low-stage oral cavity squamous cell carcinoma. Head Neck Pathol. 2013;7:211–223. doi: 10.1007/s12105-012-0412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erjala K, Sundvall M, Junttila TT, Zhang N, Savisalo M, Mali P, Kulmala J, Pulkkinen J, Grenman R, Elenius K. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12:4103–4111. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- 21.Llanos S, Garcia-Pedrero JM, Morgado-Palacin L, Rodrigo JP, Serrano M. Stabilization of p21 by mTORC1/4E-BP1 predicts clinical outcome of head and neck cancers. Nat Commun. 2016;7 doi: 10.1038/ncomms10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin ZX, Hang W, Liu G, Wang YS, Shen XF, Sun QH, Li DD, Jian YP, Zhang YH, Quan CS. PARP-1 inhibitors sensitize HNSCC cells to APR-246 by inactivation of thioredoxin reductase 1 (TrxR1) and promotion of ROS accumulation. Oncotarget. 2018;9:1885–1897. doi: 10.18632/oncotarget.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY, Huang YY, Liu XG, Le HB, Zhang YK. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest. 2014;32:394–401. doi: 10.3109/07357907.2014.922569. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai CY, Hou MF, Lee JN, Wu DC, Wang SC, Tsai EM. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2015;6:494–509. doi: 10.18632/oncotarget.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S, Lombardi L. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 26.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 27.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 28.Rigolin GM, Saccenti E, Rizzotto L, Ferracin M, Martinelli S, Formigaro L, Cibien F, Cavallari M, Lista E, Daghia G. Genetic subclonal complexity and miR125a-5p down-regulation identify a subset of patients with inferior outcome in low-risk CLL patients. Oncotarget. 2014;5:140–149. doi: 10.18632/oncotarget.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y, Zhao Q, Luo F. MiRNA-125a-5p: a regulator and predictor of gefitinib's effect on nasopharyngeal carcinoma. Cancer Cell Int. 2014;14:24. doi: 10.1186/1475-2867-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan J, Xiao G, Peng G, Liu D, Wang Z, Liao Y, Liu Q, Wu M, Yuan X. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457:171–176. doi: 10.1016/j.bbrc.2014.12.078. [DOI] [PubMed] [Google Scholar]
- 31.Tong Z, Liu N, Lin L, Guo X, Yang D, Zhang Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 32.Fu Y, Cao F. MicroRNA-125a-5p regulates cancer cell proliferation and migration through NAIF1 in prostate carcinoma. Onco Targets Ther. 2015;8:3827–3835. doi: 10.2147/OTT.S92314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin X, Wan Y, Wang S, Xue M. MicroRNA-125a-5p modulates human cervical carcinoma proliferation and migration by targeting ABL2. Drug Des Devel Ther. 2016;10:71–79. doi: 10.2147/DDDT.S93104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang P, Mao LF, Zhang ZP, Lv WW, Feng XP, Liao HJ, Dong C, Kaluba B, Tang XF, Chang S. Down-Regulated miR-125a-5p Promotes the Reprogramming of Glucose Metabolism and Cell Malignancy by Increasing Levels of CD147 in Thyroid Cancer. Thyroid. 2018;28:613–623. doi: 10.1089/thy.2017.0401. [DOI] [PubMed] [Google Scholar]
- 35.Borg A, Baldetorp B, Ferno M, Killander D, Olsson H, Sigurdsson H. ERBB2 amplification in breast cancer with a high rate of proliferation. Oncogene. 1991;6:137–143. [PubMed] [Google Scholar]
- 36.Nakajima M, Sawada H, Yamada Y, Watanabe A, Tatsumi M, Yamashita J, Matsuda M, Sakaguchi T, Hirao T, Nakano H. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer. 1999;85:1894–1902. doi: 10.1002/(sici)1097-0142(19990501)85:9<1894::aid-cncr3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 37.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA, White FM. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Appert-Collin A, Hubert P, Cremel G, Bennasroune A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front Pharmacol. 2015;6:283. doi: 10.3389/fphar.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, Ciruelos E, Ferrero JM, Schneeweiss A, Heeson S. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duru N, Fan M, Candas D, Menaa C, Liu HC, Nantajit D, Wen Y, Xiao K, Eldridge A, Chromy BA. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res. 2012;18:6634–6647. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 42.Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. The thioredoxin-thioredoxin reductase system: over-expression in human cancer. Anticancer Res. 2003;23:2425–2433. [PubMed] [Google Scholar]
- 43.Karimpour S, Lou J, Lin LL, Rene LM, Lagunas L, Ma X, Karra S, Bradbury CM, Markovina S, Goswami PC. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene. 2002;21:6317–6327. doi: 10.1038/sj.onc.1205749. [DOI] [PubMed] [Google Scholar]
- 44.Javvadi P, Hertan L, Kosoff R, Datta T, Kolev J, Mick R, Tuttle SW, Koumenis C. Thioredoxin reductase-1 mediates curcumin-induced radiosensitization of squamous carcinoma cells. Cancer Res. 2010;70:1941–1950. doi: 10.1158/0008-5472.CAN-09-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 46.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 47.Musa J, Orth MF, Dallmayer M, Baldauf M, Pardo C, Rotblat B, Kirchner T, Leprivier G, Grunewald TG. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): a master regulator of mRNA translation involved in tumorigenesis. Oncogene. 2016;35:4675–4688. doi: 10.1038/onc.2015.515. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson E, Waltersson MA, Bostner J, Perez-Tenorio G, Olsson B, Hallbeck AL, Stal O. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. 2011;50:775–787. doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 49.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, et al. (2018). Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis N Engl J Med 379, 11–21. [DOI] [PubMed]
- 51.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC, Mennel RG. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34:1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM, Trock BJ, Zhang J, Glass AG, Dicker AP. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–1998. doi: 10.1200/JCO.2016.70.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, Jiang RS. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461–2470. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validating transfection of miRNA mimics, miR-125a-5p and control miRNA, by quantitative real-time polymerase chain reaction (qRT-PCR). HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p and control miRNA mimics for a period of 72 hours. The cells were then extracted with the TRIzol reagent then quantified using a the miRCURY LNA miRNA PCR System, using the U6 snRNA as an internal, housekeeping control. qRT-PCR reveals efficacy of transfection of miR-125a-5p mimic into HN5 and UM-SCC-22B cell lines, compared to the control miRNA. Data were analyzed with the Student’s t-test and are presented as the mean ± standard deviation. Experiment was carried out in triplicate.
miR-125a-5p does not alter cell death pathways of apoptosis or autophagy. miR-125a-5p mimics were transfected into HN5 and UM-SCC-22B cell lines. Protein was extracted, and Western blots were performed with the antibodies for poly-ADP ribose polymerase (PARP), LC3B, and β-actin, a loading control. No difference in PARP cleavage, a marker of apoptosis, or LC3B expression, a marker of autophagy. LC3B-II is the autophagosome-associating form of LC3B-I. However, there was a decrease in total, uncleaved PARP protein levels that was noted.
Representative microscope images of double-stranded DNA break formation after 1 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 1 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
Representative microscope images of double-stranded DNA break formation after 0 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then sham irradiated with 0 Gy. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary fluorescent antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
miR-125a-5p reduces rate of resolution of double-stranded DNA break in response to 2 Gy of ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 2 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 and Alexa Fluor 555, respectively. Colocalized foci were then counted and displayed as the mean ± standard error of the mean. The data at each time point was analyzed using the Student’s t-test with * signifying P ≤ .05. There was a significant increase in colocalized 53BP1 and γH2AX foci with miR-125a-5p, compared to the control miRNA, at 4 hours after irradiation with 2 Gy. At 24 hours, there was complete resolution of double-stranded DNA break as the number of colocalized foci formation returned to baseline levels.
Representative microscope images of double-stranded DNA break formation after 2 Gy ionizing radiation. HN5 and UM-SCC-22B cell lines were transfected with miR-125a-5p mimic or control miRNA then exposed to ionizing radiation at 2 Gy, using a Cesium-137 source. Cells were then stained at specific time points after irradiation and stained for 53BP1, γH2AX, and DAPI, a nuclear stain. Secondary antibodies were then used to fluorescently tag the bound antibodies to 53BP1 and γH2AX with Alexa Fluor 488 (green) and Alexa Fluor 555 (red), respectively. Colocalized foci, consisting of fluorescently tagged 53BP1 and γH2AX, were then counted. Microscope images were taken with a 63x objective with oil immersion.
ERBB2 increases PI3K-AKT signaling. Transfection of pcDNA3-ERBB2 also increases phosphorylation of Akt at serine 473 while decreases expression of p27Kip1, compared to the control transfection of pcDNA3, as determined by Western blot.
Characteristics of the 77 patients who had miR-125a-5p expression analyzed. Patient characteristics included age, tumor subsite, T stage, N stage, presence of extracapsular extension, and surgical margin status.
chi-Squared analysis of clinicopathologic risk factors in 77 patients who had miR-125a-5p expression analyzed. Contingency tables were constructed and analyzed using chi-squared analysis. Risk factors that were analyzed included age, tumor subsite, T stage, N stage, extracapsular extension, and surgical margin status. NED denotes no evidence of disease, and LR denotes locoregional recurrence. No statistical difference was noted between any of the groups.
Univariable analysis of clinicopathologic risk factors of head and neck cancer patients in The Cancer Genome Atlas cohort. Univariable Cox regression analysis was performed for the following risk factors, including, race, ethnicity, and tumor subsite. No statistical difference was noted between each analyzed risk factor.
Complete list of putative mRNA targets of miR-125a-5p predicted in silico in locally advanced head and neck cancer.
Statistical analysis of comparisons between different stage groups in The Cancer Genome Atlas cohort for TXNRD1 and EIF4EBP1 mRNA expression. Comparisons between stage groupings were analyzed by Student’s t-test.
List of reagents, with catalog numbers, used in this study.