Abstract
Early life exposure to endocrine-disrupting chemicals (EDCs) is an emerging risk factor for the development of obesity and diabetes later in life. We previously showed that prenatal exposure to the EDC tributyltin (TBT) results in increased adiposity in the offspring. These effects linger into adulthood and are propagated through successive generations. TBT activates two nuclear receptors, the peroxisome proliferator–activated receptor (PPAR) γ and its heterodimeric partner retinoid X receptor (RXR), that promote adipogenesis in vivo and in vitro. We recently employed a mesenchymal stem cell (MSC) model to show that TBT promotes adipose lineage commitment by activating RXR, not PPARγ. This led us to consider the functional consequences of PPARγ vs RXR activation in developing adipocytes. We used a transcriptomal approach to characterize genome-wide differences in MSCs differentiated with the PPARγ agonist rosiglitazone (ROSI) or TBT. Pathway analysis suggested functional deficits in TBT-treated cells. We then compared adipocytes differentiated with ROSI, TBT, or a pure RXR agonist IRX4204 (4204). Our data show that RXR activators (“rexinoids,” 4204 and TBT) attenuate glucose uptake, blunt expression of the antidiabetic hormone adiponectin, and fail to downregulate proinflammatory and profibrotic transcripts, as does ROSI. Finally, 4204 and TBT treatment results in an inability to induce markers of adipocyte browning, in part due to sustained interferon signaling. Taken together, these data implicate rexinoids in the development of dysfunctional white adipose tissue that could potentially exacerbate obesity and/or diabetes risk in vivo. These data warrant further screening and characterization of EDCs that activate RXR.
The endocrine disruptor tributyltin activates retinoid X receptor during adipogenesis to develop a dysfunctional adipocyte, as compared with cells differentiated with the PPARγ agonist, rosiglitazone.
The economic burden of obesity (1) and diabetes (2) in the United States is colossal, with each pathology independently estimated to cost upwards of $200 billion annually. Moreover, rates of obesity and diabetes are on the rise globally, even in developing countries where prevalence has been historically low (3, 4). Obesity is by far the strongest predictor of type 2 diabetes (T2D) (3), which raises the stakes for identifying key drivers of the obesity epidemic beyond caloric intake and energy expenditure. Twin studies show that obesity is highly heritable (40% to 70%), yet genetic variants identified in large-scale genome-wide association studies account for only a small fraction (<3%) of the variance in body mass index (5, 6). This has led investigators to consider how environmental factors interact with the genome to contribute to the obesity and diabetes epidemics. In particular, researchers now understand that environmental factors in utero and in early life can program disease risk in adulthood (6, 7), collectively referred to as the developmental origins of health and disease paradigm (8). Maternal obesity, maternal weight gain, and gestational diabetes are all linked to the risk of obesity in the offspring later in life. Likewise, in utero exposure to hyperglycemia via maternal type 1, type 2, or gestational diabetes predicts diabetes risk in the offspring [reviewed in (7)]. Hence, researchers are examining a number of environmental factors, both maternal and paternal, that can influence metabolic disease risk in the offspring.
Emerging evidence links exposure to industrial chemicals with a number of noncommunicable diseases, including obesity and diabetes (9–12). Of the tens of thousands of chemicals used in industry, at least 1000 of them are considered endocrine-disrupting chemicals (EDCs) that can interfere with any aspect of hormone action (13). These chemicals can target endocrine and metabolic organs, including the hypothalamus, pituitary, thyroid, liver, pancreas and, of course, adipose tissue (10). The “obesogen hypothesis” postulated that developmental exposure to EDCs might promote obesity and metabolic disease later in life by reprogramming metabolic set points, promoting adipose hyperplasia and hypertrophy, or dysregulating hypothalamic and cortical circuits that control appetite, satiety, and reward (14, 15). In the ensuing decade, numerous obesogens have been identified in humans and animals and further characterized in vitro and in vivo (10).
Although the mechanisms of action of many obesogens remain elusive, many EDCs act through nuclear receptors such as peroxisome proliferator–activated receptor (PPAR) γ, estrogen receptor, and glucocorticoid receptor to stimulate adipogenesis (16–18). To study these chemicals mechanistically, many laboratories have employed the murine 3T3-L1 preadipocyte cell line as well as multipotent mesenchymal stem cells (MSCs, also known as mesenchymal stromal cells) from bone marrow, adipose tissue, and other organs (19, 20). We first showed that the fungicide tributyltin (TBT) activates PPARγ and its heterodimeric partner retinoid X receptor (RXR) to promote adipogenesis in 3T3-L1 cells and MSCs (14, 21, 22), results corroborated by independent studies in several laboratories (19, 23–25). Mice exposed to TBT in utero have increased fat mass and excess ectopic fat (14, 21), characteristics that persist into adulthood and across several generations (26, 27). We recently showed that TBT promotes adipose lineage commitment of undifferentiated MSCs through activation of RXR, but not PPARγ (28). This led us to question the implications of RXR vs PPARγ activation on the function of the induced adipocytes, especially because pharmaceutical PPARγ agonists are considered to improve adipocyte health (29) (in spite of important harmful side effects).
Adipocytes perform many functions essential to maintaining metabolic health. Principally, they are tasked with taking up glucose from the bloodstream in response to postprandial insulin (30). In obesity, disrupted insulin signaling through the Akt/protein kinase B (PKB) pathway can lead to insulin resistance and T2D; if unmanaged, the disease will eventually progress to pancreatic β-cell failure, requiring exogenous insulin (31). Adipose tissue is also an endocrine organ, releasing several “adipokines” that can improve or impair metabolic health (32). Chief among these is adiponectin, which inversely correlates with T2D risk and acts, in part, by suppressing gluconeogenesis and stimulating β-oxidation in the liver (33). Importantly, PPARγ agonists increase serum adiponectin levels through direct binding to and activation of its promoter in adipocytes (34). Two hallmarks of metabolically unhealthy obesity are inflammation and fibrosis (35, 36). Both are thought to be initiated by hypoxic signaling in hypertrophic adipocytes, which induces the expression of fibrotic (collagens, collagen crosslinking enzymes) and inflammatory (IL-6, TNFα, macrophage migration inhibitory factor) genes. The resultant increases in extracellular matrix (ECM) and cytokines stimulate adipocyte necrosis and recruit proinflammatory immune cells, such as M1-type macrophages, and a positive feedback loop ensues (35). Hence, functional adipocytes are characterized by (1) insulin sensitivity, (2) a healthy adipokine profile, and (3) small, normoxic cells that promote an anti-inflammatory and antifibrotic milieu (35, 36).
One final dimension of adipocyte function that has advanced greatly in recent years is the discovery that thermogenic brown adipose tissue (BAT) persists in adults, and white adipose tissue possesses the capability to “brown” into “beige/brite” fat (37, 38). These thermogenic adipocytes are characterized by their abundant mitochondria, which are able to uncouple cellular respiration from ATP synthesis and dissipate their proton gradient to generate heat. This is achieved by uncoupling protein 1 (Ucp1), whose activity is stimulated by lipolysed free fatty acids, but inhibited by purine nucleotides, such as ATP. Both cold exposure and dietary intake can stimulate lipolysis through β-adrenergic signaling, resulting in sufficient free fatty acids to promote thermogenesis. BAT has great therapeutic potential as a treatment of obesity and diabetes because it readily converts blood glucose and fats into heat (39). Interestingly, perinatal exposure to the insecticide DDT results in cold intolerance, reduced core temperature, and diminished energy expenditure in adult females (40). This was driven by diminished BAT expression of PPARγ coactivator 1α (Ppargc1a, also known as PGC-1α, a transcriptional master regulator of BAT genes) and iodothyronine deiodinase 2 (Dio2, which converts T4 to the more potent and thermogenic T3). Further studies are needed to clarify how DDT and other EDCs might disrupt adipose development to interfere with nonshivering thermogenesis in adulthood. Several reports in the literature show that brown/beige-like adipocytes can be differentiated from MSCs using genetic and pharmacologic approaches (41–43). Hence, these cells may serve as a platform to screen for EDCs that interfere with BAT function and to study the effects of these EDCs mechanistically.
RXR is a unique member of the nuclear receptor family in that it forms heterodimers with multiple other nuclear receptors (44). These partners are categorized either as “permissive,” where the heterodimer can be activated either by activating RXR with a rexinoid or the partner receptor with its cognate ligand, or as “nonpermissive,” in which RXR is unable to be activated and the heterodimer activated only by the partner receptor ligand. Given the multitude of permissive partners expressed in adipose tissue [e.g., liver X receptor (LXR), farnesoid X receptor, PPARα, PPARδ, PPARγ], rexinoids would be predicted to activate numerous, potentially interacting pathways in these cells. Interestingly, 3T3-L1 cells differentiated in the presence of the PPARγ agonist troglitazone or TBT were shown to have divergent expression of adipogenic transcripts and proteins and an altered glucose metabolism (23). Hence, we hypothesized that RXR activation in differentiating MSCs would result in a functionally distinct adipocyte compared with activating PPARγ. We employed the MSC model to differentiate adipocytes in the presence of a PPARγ agonist [rosiglitazone (ROSI)], an RXR agonist [IRX4204 (4204)], or the dual PPARγ/RXR activator TBT, and subsequently assessed the gene expression profiles and function of these cells. Despite comparable levels of lipid accumulation, MSCs differentiated in the presence of TBT had a strongly divergent transcriptome from ROSI-treated cells that suggested deficits in adipocyte function. Further analyses revealed that when compared with cells treated with ROSI, rexinoids reduced glucose uptake, ablated the secretion of adiponectin, sustained the expression of fibrotic and inflammatory genes, and repressed adipose browning. These data point to a deleterious role for rexinoids and RXR-disrupting chemicals on adipose tissue development and function.
Materials and Methods
Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated: dimethyl sulfoxide (Thermo Fisher Scientific, Waltham, MA), TBT, ROSI (Cayman Chemical, Ann Arbor, MI), 4204 (also known as AGN194204; a gift from Rosh Chandraratna, IO Therapeutics, Santa Ana, CA), T0070907 (Enzo Life Sciences, Farmingdale, NY), HX531 (a gift from Claes Bavik, Acucela, Inc., Seattle, WA), dexamethasone, human recombinant insulin, isobutylmethylxanthine, triiodothyronine, mouse recombinant interferon (IFN) α (PBL Assay Science, Piscataway, NJ), tofacitinib (TCN; Selleck Chemicals, Houston, TX), 2-deoxy-d-glucose, 2-deoxy-d-[3H]-glucose (American Radiolabeled Chemicals, St. Louis, MO), Nile Red, Hoechst 33342, and collagenase D.
Mesenchymal stem cell culture and differentiation
Mouse bone marrow–derived MSCs (OriCell, Cyagen Biosciences, Santa Clara, CA) from female C57BL/6 mice were stored at passage 9 in liquid N2. Cells were maintained and expanded at subconfluence as previously described (28, 45) in DMEM supplemented with 10% calf bovine serum (Sigma-Aldrich), 10 mM HEPES, 1 mM sodium pyruvate, 100 IU/mL penicillin, and 100 μg/mL streptomycin. To differentiate MSCs into adipocytes, cells were plated at 15,000 cells/cm2 and allowed to proliferate for 72 hours, at which point they were confluent and ready for adipose induction. Media were then switched to αMEM supplemented with 15% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA), 10 mM HEPES, 2 mM l-glutamine, 100 IU/mL penicillin, 100 μg/mL streptomycin, and an adipose induction cocktail (MDI) containing 1 μM dexamethasone, 5 μg/mL insulin, and 0.5 mM isobutylmethylxanthine; 10 nM T3 was added for brown adipose differentiation. Ligands were added in conjunction with differentiation media. Media was changed every 3 to 4 days for 14 days, at which point differentiated MSCs were ready for further analysis. For antagonist assays, the PPARγ antagonist T0070907 (1 μM) or the RXR antagonist HX531 (10 μM) was added during the last 7 days of differentiation. Owing to its short half-life, T0070907 was redosed every 12 hours (22). To assess the role of the IFN signaling, 1000 U/mL IFNα or 5 μM TCN was added during the last 7 days of differentiation. All cell culture experiments were carried out at 37°C with 5% CO2 unless otherwise indicated.
Stromal vascular fraction isolation and differentiation
Nine female C57BL/6J mice were purchased from The Jackson Laboratory (Sacramento, CA) and housed in a temperature-controlled room (25°C to 27°C) with a 12-hour light/dark cycle and unlimited access to food and water. All procedures were approved by the University of California, Irvine Institutional Animal Care and Use Committee. At 9 weeks of age, mice were euthanized and stromal vascular fraction (SVF) was harvested from inguinal fat pads as previously described (46). Inguinal fat pads were dissected, minced, pooled, and subsequently digested in a collagenase solution (100 mM HEPES, 120 mM NaCl, 50 mM KCl, 5 mM glucose, 1 mM CaCl2, 1.5% BSA, and 1 mg/mL collagenase D) at 37°C for 2 hours. Digested adipose tissue was passed through a 100-µm filter and the SVF was separated from adipocytes by centrifugation (5 minutes, 600g). SVF was resuspended in αMEM 15% FBS (see above), plated at high density in collagen-coated plates, and allowed to attach overnight. Media were changed daily to remove dead and nonadherent cells until adherent cells (preadipocytes, MSCs, fibroblasts) reached confluency. SVF was induced to differentiate into adipocytes in αMEM 15% FBS, MDI, T3, and ligands [dimethyl sulfoxide (DMSO), ROSI, 4204, TBT] as described above. After 48 hours, the induction cocktail was switched to insulin (5 µg/mL), T3, and ligands and differentiated for 4 more days.
Assessment of lipid accumulation
MSCs were fixed in 3.7% buffered formaldehyde; neutral lipids were stained with Nile Red (1 μg/mL), and nucleic acid was stained with Hoechst 33342 (1 μg/mL). Relative fluorescence units (RFUs) were measured in a SpectraMax Gemini XS spectrofluorometer (Molecular Devices, Sunnyvale, CA) using SoftMax Pro (Molecular Devices). Lipid accumulation was calculated as Nile Red RFU normalized to Hoechst RFU.
Insulin challenge and glucose uptake
To assess insulin response, MSCs were differentiated for 12 days and then maintained in media (αMEM 15% FBS) without MDI or ligands for the final 2 days of differentiation (day 12 to 14). Cells were washed with Krebs–Ringer–HEPES buffer (KRH) containing 0.1% BSA and subsequently serum starved overnight in the same buffer supplemented with 5.5 mM glucose. MSCs were then washed with glucose-free KRH 0.1% BSA prior to insulin challenge in the same buffer. For protein and gene expression analysis, cells were collected at baseline or after 15 minutes (protein) or 4 hours (RNA) of 100 nM insulin treatment. Glucose uptake was measured as previously described (47). In brief, cells were treated with insulin (100 nM) or vehicle control (DMSO) in glucose-free KRH 0.1% BSA for 30 minutes. Wells were then spiked with 0.5 μCi of 2-deoxy-d-[3H]-glucose in 6.5 mM unlabeled 2-deoxy-d-glucose. After 5 minutes cells were thoroughly washed in ice-cold KRH 0.1% BSA and subsequently lysed in 0.05 N NaOH. Lysates were transferred into scintillation vials and radioactivity was measured in a Beckman LS6500 liquid scintillation counter (Beckman Coulter, Brea, CA).
Measurement of secreted adiponectin
Differentiated MSCs (14 days) were maintained for 72 hours in differentiation media lacking MDI or ligands (αMEM 15% FBS). Mouse adiponectin protein levels in conditioned media were measured by ELISA according to the manufacturer’s instructions (Thermo Fisher Scientific).
Lipolysis assay
Differentiated MSCs (14 days) were washed and subsequently incubated in the presence or absence of the β-adrenergic agonist isoproterenol (100 nM) for 4 hours. Free glycerol in the media was measured by a colorimetric method according to the manufacturer’s protocol (Abcam, Cambridge, United Kingdom). All assay buffers and chemicals were supplied by Abcam. Cells were fixed and stained with Nile Red to quantify lipid accumulation as described above. Free glycerol was normalized to total lipid (Nile Red RFU) for each well.
Mitochondrial stress test
Mitochondrial function was evaluated using a Seahorse XF24 analyzer (Agilent Technologies, Santa Clara, CA). MSCs were plated in 24-well Seahorse plates at 3000 cells per well (∼11,000 cells/cm2) in 100 µL of DMEM 10% calf bovine serum. Media volume was brought up to 200 µL the following day, and this volume was used for the remainder of the assay. Cells were allowed to grow to confluence and differentiated as described above in the presence of T3. The assay was carried out on two plates with five replicates from each treatment group on each plate (n = 10 total per group). After 2 weeks of differentiation, cells were washed and then incubated in buffer-free Seahorse XF media supplemented with 5.55 mM glucose, 1 mM sodium pyruvate, and 2 mM l-glutamine (identical to concentrations in αMEM) for 1 hour at 37°C without CO2. Plates were then loaded into the Seahorse XF analyzer and subjected to a Mito Stress Test. Cells were sequentially exposed to a complex V inhibitor (1.5 µM oligomycin), a mitochondrial uncoupler [1 µM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone], and a combination of complex I (1 µM rotenone) and complex III (1 µM antimycin A) inhibitors. Oxygen consumption rate (OCR; pmol/min) is measured repeatedly at baseline and following exposure to each compound. At the end of the assay, cells were immediately fixed and stained with Hoechst 33342 as described above; OCR measurements were normalized to Hoechst RFU for each well. ATP production, maximal respiration, and spare respiratory capacity were extrapolated from changes in normalized OCR in response to compounds.
Immunoblotting
To isolate protein, cells were washed twice in ice-cold PBS and then lysed in RIPA buffer containing protease (Research Products International, Mount Prospect, IL) and phosphatase (Thermo Fisher Scientific) inhibitors. Lysates were shaken at 4°C for 1 hour and then sheared with a 27-gauge needle prior to cold centrifugation. Supernatants were collected and protein concentrations were measured using the DC protein assay (Bio-Rad, Hercules, CA). Twenty micrograms of SDS denatured protein was run on 4% to 20% Tris-glycine polyacrylamide gels (Bio-Rad) and transferred to a polyvinylidene difluoride membrane (Millipore, Burlington, MA) by semidry transfer (Bio-Rad). Blots were blocked, washed, and incubated with primary antibodies according to product specifications [phospho-Akt Ser473, Abcam (48); pan-Akt, Abcam (49); GAPDH, Santa Cruz Biotechnology, Dallas, TX (50)]. Following incubation with HRP-conjugated secondary antibody (GE Healthcare Life Sciences, Marlborough, MA), blots were incubated with chemiluminescent substrate (Thermo Fisher Scientific) and visualized in a ChemiDoc imaging system (Bio-Rad). Blots were stripped in acidic glycine (pH 2.2, 0.1% SDS, 1% Tween 20) and reprobed as needed.
RNA/DNA isolation, reverse transcription, quantitative PCR
Cells were lysed in TRIzol (Thermo Fisher Scientific) and RNA was isolated by phenol/chloroform extraction. RNA concentrations were measured on a NanoDrop spectrophotometer (Thermo Fisher Scientific) and RNA quality was confirmed by gel electrophoresis. cDNA was generated from 1 μg of total RNA using Transcriptor reverse transcription (Roche, Basel, Switzerland) according to the manufacturer’s protocol. For DNA isolation, cells were collected in lysis buffer [10 mM Tris-HCl (pH 8.0), 400 mM NaCl, 2 mM EDTA, 1% SDS] and subsequently treated with proteinase K (0.25 mg/mL, 2 hours, 55°C). DNA was isolated by phenol/chloroform/isoamyl alcohol extraction and concentrations were measured on a NanoDrop. Quantitative RT-PCR was conducted as previously described (28, 45) using intron-spanning primers (Supplemental Table 1).
Deep sequencing
RNA for sequencing was isolated using a miRNeasy micro kit (Qiagen, Hilden, Germany). Intact RNA integrity >9.0 of total RNA samples was confirmed using the Agilent RNA 6000 Nano Kit for Bioanalyzer (Agilent Technologies). After ribosomal RNA removal using the RiboMinus eukaryote system v2 kit (Life Technologies), nondirectional RNA sequencing (RNA-seq) deep sequencing libraries for the ABI/SOLiD platform were constructed using the SOLiD total RNA-seq kit and EZ Bead (Life Technologies). Deep sequencing was performed using the SOLiD 5500XL deep sequencer (50-nt, single nondirectional reads; Life Technologies), and the XSQ-format raw data were converted to the csfasta/QV.qual format using the XSQ tool Linux script provided by Life Technologies.
Deep sequencing data analysis
The csfasta/QV.qual sequences were aligned to the NCBI37/mm9 mouse genome reference sequence to obtain the bam format aligned read data using the NovoAlignCS color-space aligner (Novocraft Technologies, Selangor, Malaysia) with the following command line: “novoalignCS -d Mouse.mm9.Exons.index -f csfastafile.csfasta -H -k -oSAM -t 110 -v 0 0 70 [>]([^:]*) -F CSFASTAnQV -r All 10 -oSAM.” The bam-format data of mapped reads were examined using the fastQC deep-sequencing reads quality control tool (Simon Andrews, Babraham Institute) to exclude poor-quality reads from the study. Quality control–passed mapped reads were subjected to extraction of uniquely mapped reads using samtools (51). Uniquely mapped reads were assigned to the mm9 gene model, and on-exon reads were counted using the Bioconductor package Rsubread (52). The assigned read counts were normalized using Bioconductor package DESeq2 (53) using the regularized logarithm (rlog) transformation. Prior to clustering analysis, normalized counts were filtered based on variance (standard deviation > 0.2) and expression level (minimum four replicates with more than four regularized logarithm normalized counts). Hierarchical and k-means clustering were performed on the 4770 genes that met the indicated criteria in cluster 3.0; dendrograms were visualized in Java TreeView. For gene ontology term and pathway analysis, differentially expressed genes were converted to HUGO gene symbols, then tested for enrichment in MSigDB (Broad Institute, Cambridge, MA) pathway (C2) gene sets by a hypergeometric test in R. P values were corrected for multiple testing using the Benjamini–Hochberg method. Differential expression was assessed in DESeq2 using the DESeq function with α = 0.01. Differentially expressed genes were defined by Benjamini–Hochberg corrected P values <0.01 and fold change [absolute value of log2(fold change) > log2(1.2)].
All sequencing data are available on the Gene Expression Omnibus (GSE115946).
Statistical analysis
Data visualization and statistical analyses were conducted in Prism 7 (GraphPad Software, La Jolla, CA), excluding RNA-seq analysis, which was completed in R and other software packages as noted above. Four to six biological replicates were used for all experiments except the Seahorse assay, which used 10 replicates. The primary endpoint of our study was to compare the function of terminally differentiated adipocytes with similar levels of lipid accumulation (ROSI, 4204, and TBT groups). Vehicle controls (DMSO) are presented but were excluded from most statistical tests, because these cells form far fewer adipocytes than do ligand-treated MSCs. However, because comparisons to vehicle control are useful to evaluate differentiation, we have included these data as a supplement (Supplemental Table 2). Standard propagation of error was used where appropriate (54). A P value of ≤0.05 was considered statistically significant for all assays other than RNA-seq analysis (see above).
Results
Differentiation of MSCs with ROSI and TBT forms adipocytes with distinct transcriptomes
Mouse bone marrow MSCs were differentiated into adipocytes using a standard adipogenic cocktail (insulin, dexamethasone, and isobutylmethylxanthine; also known as MDI) (55). Cells were additionally treated with the PPARγ agonist ROSI, the endocrine disruptor TBT, or vehicle control (0.1% DMSO). Doses of ROSI (500 nM) and TBT (50 nM) were chosen to achieve similar levels of lipid accumulation based on previously published reporter assays and adipogenesis assays (14, 21, 22, 45, 56, 57). After 2 weeks of differentiation, mature adipocytes were assessed for lipid accumulation and genome-wide transcription (Supplemental Fig. 1A). ROSI and TBT treatment resulted in comparable levels of lipid accumulation over DMSO control (Supplemental Fig. 1B). The transcriptomes of differentiated MSCs were analyzed by hierarchical clustering (Supplemental Fig. 1C) and principal component analysis (Supplemental Fig. 1D) of normalized transcript counts. Despite achieving similar levels of lipid accumulation, TBT and ROSI transcriptomes were distinct from one another (Supplemental Fig. 1C–1F). In fact, TBT-differentiated MSCs clustered closer to vehicle controls than to ROSI samples (Supplemental Fig. 1C). Genes differentially expressed by TBT and ROSI samples as compared with DMSO controls were largely distinct from one another (Supplemental Fig. 1E; Supplemental Table 3) and the overall correlation between TBT and ROSI expression changes over DMSO was modest (R2 = 0.3426; Supplemental Fig. 1F).
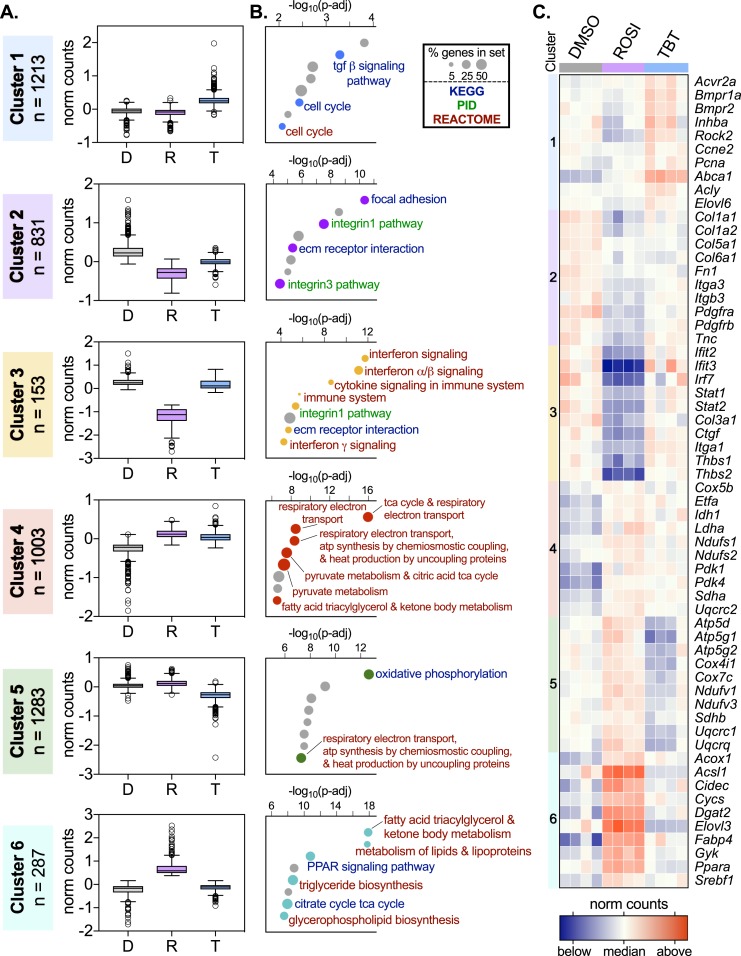
To better assess differences in gene expression patterns between TBT and ROSI-differentiated MSCs, we performed k-means clustering on 4770 transcripts with the highest variance and meaningful expression levels (Fig. 1A; Supplemental Table 4; see “Materials and Methods”). These transcripts were binned into six clusters that were subsequently evaluated for enrichment in the Molecular Signaling Database’s curated gene sets (C2 collection, MSigDB; Broad Institute). From this analysis we focused on enrichments in the Kyoto Encyplopedia of Genes and Genomes (Kyoto University, Kyoto, Japan), Pathway Interaction Database (National Cancer Institute, Bethesda, MD), and BioCarta (Allele Biotechnology, San Diego, CA) gene sets (Fig. 1B; Supplemental Table 5). Broadly, these clusters can be grouped into three categories. In the first group, TBT expression is distinct from both DMSO and ROSI (clusters 1 and 5, Fig. 1A). In the second group, ROSI expression is distinct from both DMSO and TBT (clusters 3 and 6, Fig. 1A). Although these clusters are the smallest in number (n = 153 and 287, respectively), they represent a failure by TBT to target a subset of genes effectively altered by ROSI. The final group contains genes congruently altered by ROSI and TBT treatments over DMSO controls (clusters 2 and 4, Fig. 1A), likely representing adipocyte differentiation induced by either ligand.
Figure 1.
Differentiation of MSCs with ROSI and TBT forms adipocytes with distinct transcriptomes. MSCs were differentiated for 2 wk in the presence of ROSI (500 nM), TBT (50 nM), or vehicle control (0.1% DMSO) and an adipose induction cocktail (MDI). After 2 wk, RNA was isolated and prepared for deep sequencing. Reads were aligned to the mouse genome (mm9), counted (Rsubread, R/Bioconductor), normalized (DESeq2, R/Bioconductor), and clustered using the k-means method (Cluster 3.0) as described in “Materials and Methods.” (A) Median-adjusted normalized counts for all genes within each of the six clusters are presented as Tukey box plots. (B) Pathway analysis of genes within each k-means cluster was performed using the MSigDB C2 gene sets. Results from Kyoto Encyplopedia of Genes and Genomes (KEGG; blue text), Pathway Interaction Database (PID; green text), and BioCarta (brown text) gene sets are displayed. Pathways were ranked by the Benjamini–Hochberg adjusted P value (p-adj). The number of genes from a given cluster within a particular gene set divided by the total number of genes in the set, as a percentage, is indicated by the size of the circle. Pathways not relevant to adipocyte biology are presented in gray but are listed in Supplemental Table 5. (C) Heat map of median-adjusted normalized counts for 10 representative genes from each of the 6 clusters are presented for each sample replicate.
Pathway analysis reveals that cluster 1 is enriched for the profibrotic TGF-β signaling pathway, suggesting that TBT may push developing MSCs toward a myofibroblast-like phenotype. Clusters 2 and 3 are enriched for cell adhesion and ECM gene sets (Fig. 1B; Supplemental Fig. 2B). Genes driving these enrichments include transcripts coding for collagens (Col1a1, Col1a2, Col3a1, Col5a1, Col6a1), integrins (Itga1, Itga3, Itgb3), thrombospondins (Thbs1, Thbs2), and fibronectin (Fn1) (Fig. 1C). Remarkably, cluster 3 is highly enriched for inflammatory pathways, particularly IFN signaling (Fig. 1B; Supplemental Fig. 3). These data reveal that TBT is unable to repress fibrotic and inflammatory transcripts that are effectively downregulated by ROSI. Furthermore, TBT activates the TGF-β pathway, which is known to promote adipose tissue fibrosis and dysfunction.
Genes from clusters 4 to 6 are highly enriched for metabolic pathways, particularly cellular respiration and lipid metabolism (Fig. 1B). Enrichment for cellular respiration terms was primarily found in clusters 4 and 5, driven by genes coding members of the electron transport chain (ETC; Supplemental Fig. 2A). The unique repression of nuclear-encoded ETC genes (cluster 5) implies respiratory deficits in TBT-differentiated cells. Lipid metabolism enrichment was principally seen in cluster 6, driven by classic adipose markers such as acyl-CoA oxidase 1 (Acox1), cell death–inducing DFFA-like effector C (Cidec, also known as Fsp27), fatty acid binding protein 4 (Fabp4), and sterol regulatory element binding transcription factor 1 (Srebf1) (Fig. 1C). The inability of TBT to effectively activate these genes suggests an inability to activate PPARγ targets as effectively as ROSI, despite comparable lipid accumulation. Taken together, these data demonstrate that energy metabolism is disrupted in TBT-treated MSCs.
Treatment with a selective RXR agonist mirrors TBT-induced changes in gene expression
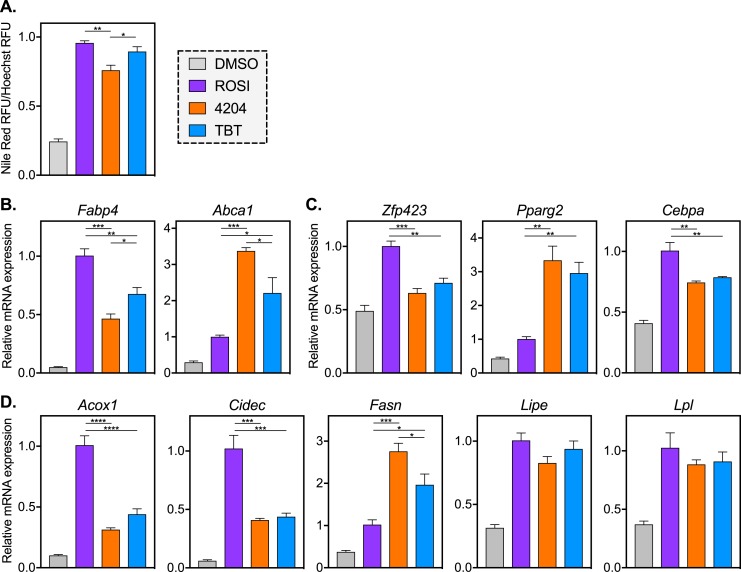
Because TBT is a dual PPARγ/RXR activator, we hypothesized that differences in the ROSI and TBT transcriptomes might be RXR-dependent. To explore this, MSCs were differentiated in the presence of ROSI, TBT, or the selective RXR agonist 4204 (58). After 2 weeks of differentiation, all three treatment groups accumulated similar levels of intracellular lipid compared with vehicle control (Fig. 2A), although 4204 produced slightly less lipid accumulation at the dose used in this study (100 nM).
Figure 2.
Treatment with a selective RXR agonist mirrors TBT-induced changes in gene expression. MSCs were differentiated into adipocytes with MDI and ROSI (500 nM), 4204 (100 nM), TBT (50 nM), or vehicle control (0.1% DMSO) for 2 wk. Cells were fixed and stained to assess (A) lipid accumulation or (B–D) RNA was extracted for analysis of gene expression. (A) Fixed MSCs were stained for neutral lipids (Nile Red) and nuclei (Hoechst 33342). Nile Red fluorescence was normalized to Hoechst for each well. Quantitative PCR analyses of (B) canonical PPARγ and RXR targets, (C) early adipose lineage markers, and (D) late markers of adipocyte function are shown. All data are represented as the mean ± SEM. For ROSI, 4204, and TBT samples, a one-way ANOVA and Tukey multiple comparison test were used: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We investigated gene expression in 4204 samples to compare with ROSI and TBT treatments. This began with the canonical PPARγ and RXR target genes Fabp4 and ATP-binding cassette transporter 1 (Abca1), respectively (Fig. 2B). As expected, expression of the PPARγ target Fabp4 was highest in ROSI-treated samples, whereas expression of the RXR target, Abca1, was highest in 4204-treated samples; TBT-induced expression levels fell between ROSI and 4204 (Fig. 2B). The transcriptional regulators of adipogenesis zinc finger protein 423 (Zfp423) and CCAAT/enhancer binding protein α (Cebpa) have higher expression in ROSI samples, whereas Pparg2 expression is highest with 4204 treatment (Fig. 2C), consistent with the ability of rexinoids to commit cells to the adipogenic lineage (28). For all three of these genes, TBT-induced expression levels resemble 4204 more so than ROSI (Fig. 2C). Genes critical for adipocyte function such as Acox1 and Cidec are preferentially targeted by the PPARγ agonist ROSI, whereas fatty acid synthase (Fasn) is better induced by rexinoids. The lipases lipase E (Lipe, also known as hormone-sensitive lipase) and lipoprotein lipase (Lpl) are expressed similarly between ligand treatment groups (Fig. 2D). These data show a clear rift in gene expression between MSCs differentiated with PPARγ and RXR agonists, and they suggest that the differences observed in ROSI and TBT transcriptomes might be driven by RXR.
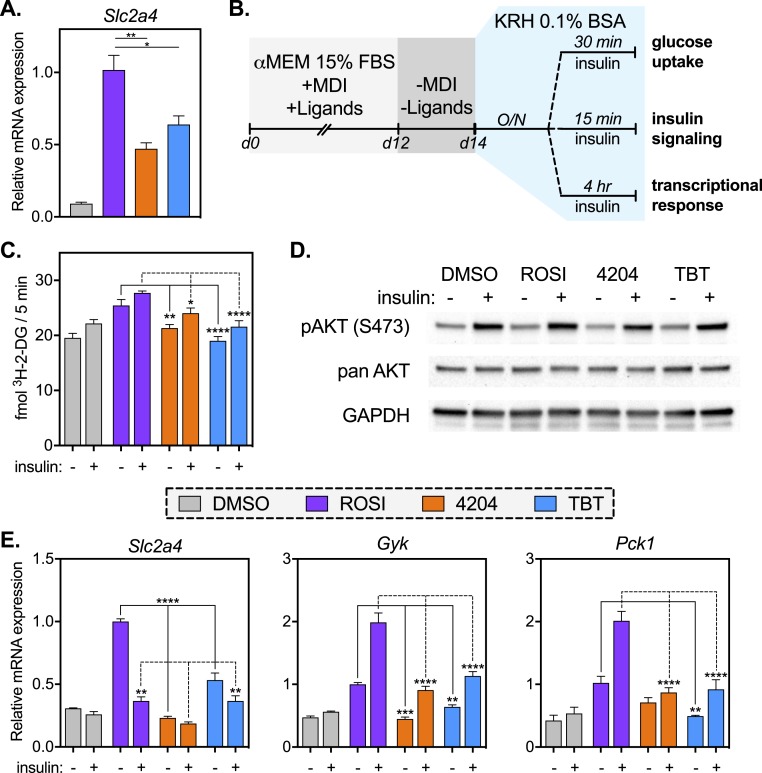
Rexinoid-differentiated MSCs have impaired glucose uptake but intact insulin signaling
Chief among the functions of a mature adipocyte is the ability to take up glucose in response to insulin. Transcript levels of the insulin-dependent glucose transporter solute carrier family 2 member 4 (Slc2a4, also known as GLUT4) are attenuated in 4204 and TBT samples when compared with ROSI (Fig. 3A). To test whether altered expression was reflected in glucose uptake, MSCs were differentiated with MDI and ligands as previously described (Supplemental Fig. 1A) for 12 days and then maintained in basal media for 2 days and finally starved overnight in a serum-free buffer. In this way, cells were starved first of the insulin in MDI and, subsequently, of insulinomimetic compounds in serum (Fig. 3B). Following serum starvation, cells were challenged with insulin and assessed for their ability to take up glucose (Fig. 3C), for signaling through the insulin pathway (Fig. 3D), and altered expression of insulin-response genes (Fig. 3E). Both basal and insulin-stimulated glucose uptake were highest in ROSI-differentiated MSCs compared with 4204 and TBT, as assessed by uptake of radiolabeled 2-deoxyglucose (Fig. 3C). Akt/PKB signaling was assessed by detecting phosphorylation of Akt at Ser473 (Fig. 3D). No discernable differences were seen between samples, suggesting that insulin signaling was intact across treatments (Fig. 3D; Supplemental Fig. 4).
Figure 3.
Rexinoid-differentiated MSCs have impaired glucose uptake but intact insulin signaling. (A) Quantitative PCR (qPCR) analysis of Slc2a4 (GLUT4) from material described in Fig. 2. For ROSI, 4204, and TBT samples, one-way ANOVA and Tukey multiple comparison test were used: *P < 0.05; **P < 0.01. (B) For insulin challenges, MSCs were differentiated for 12 d in the presence of MDI and ligands, then tapered off MDI, ligands, and serum as described in “Materials and Methods.” Cells were starved overnight in serum-free buffer prior to insulin challenge and measurement of (C) glucose uptake, (D) Akt/PKB signaling, or (E) transcriptional response. (C) Glucose uptake was measured using radiolabeled 2-deoxy-d-glucose as described in “Materials and Methods.” For ROSI, 4204, and TBT samples, a two-way ANOVA and Tukey multiple comparison test between treatment groups were used: *P < 0.05; **P < 0.01; ****P < 0.0001. (D) Protein lysates were prepared from samples at baseline or following insulin treatment (100 nM) for 15 min as described in “Materials and Methods.” Samples were immunoblotted for phosphorylation of AKT at serine 473 (pAKT S473), panAKT, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; loading control). (E) RNA was collected at baseline or after 4 h of insulin treatment (100 nM). Gene expression of insulin-responsive transcripts was assessed by qPCR. For ROSI, 4204, and TBT samples, a two-way ANOVA and Tukey multiple comparison test between treatment groups were used: **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are represented as the mean ± SEM. 3H-2-DG, 2-deoxy-d-[3H]-glucose.
Finally, the transcriptional response to insulin was assayed after 4 hours of insulin treatment. Basal Slc2a4 expression remained high in ROSI-differentiated MSCs, even after removal of MDI, ligands, and serum (Fig. 3E). Insulin treatment strongly repressed Slc2a4 in ROSI samples, mildly so in TBT treatment, but not in 4204-differentiated cells (Fig. 3E). Glycerol kinase (Gyk), an insulin target expressed in brown but not white adipose tissue, was induced by insulin in all treatments (Fig. 3E). Its basal expression, however, was highest in ROSI-differentiated samples (Fig. 3E). The gluconeogenic phosphoenolpyruvate carboxykinase 1 (Pck1, also known as PEPCK) was induced in ROSI-treated MSCs, whereas expression remained low in 4204 and TBT samples (Fig. 3E). Hence, the transcriptional response to insulin is altered in rexinoid-differentiated adipocytes, as is observed in human obesity (59).
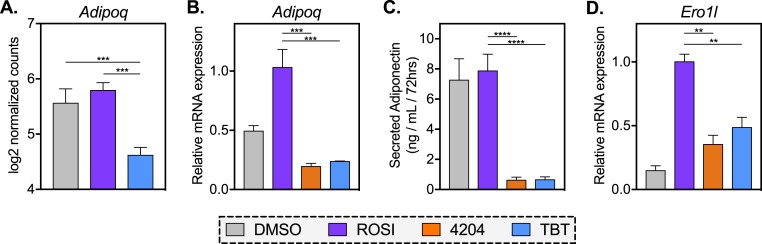
RXR agonists blunt the expression and secretion of adiponectin compared with ROSI-treated cells
A major role of adipose tissue in health and disease is the secretion of adipokines, some of which are insulin-sensitizing and anti-inflammatory whereas others promote insulin resistance and inflammation (32). Adiponectin belongs to the former group, acting on the liver, skeletal muscle, and pancreas to ameliorate glycemic control. In analyzing our RNA-seq data, we noted that adiponectin (Adipoq) was one of the most differentially expressed genes between ROSI and TBT (Fig. 4A; Supplemental Table 3). In agreement with previous work in human and mouse MSCs (21), we see minimal to modest induction of Adipoq in ROSI treatment over MDI plus vehicle controls. Diminished expression of Adipoq by TBT (and 4204) relative to ROSI was confirmed by quantitative PCR (qPCR; Fig. 4B). Notably, Adipoq transcript was repressed by 4204 (60%) and TBT (52%) as compared with vehicle control (DMSO), although this only reached statistical significance in the 4204 group (Supplemental Table 2). To assess whether these differences resulted in diminished secretion of adiponectin protein, we measured levels of adiponectin in cell media. ROSI and control cells secreted substantial levels of adiponectin during 3 days, but the adipokine was nearly undetectable in 4204 and TBT-conditioned media (Fig. 4C; Supplemental Table 2). In fact, diminished protein secretion by rexinoids relative to ROSI was greater than their effect on mRNA expression (92% vs 78%, respectively), suggesting that RXR activators may also inhibit adiponectin export. Therefore, we assayed transcript levels of endoplasmic reticulum oxidoreductase 1–like (Ero1l), which regulates the secretion of adiponectin from adipocytes (60). Indeed, expression of this gene in rexinoid treatments was less than half of what was detected in ROSI samples (Fig. 4D), suggesting that RXR activators not only produce less adiponectin mRNA, but they may either fail to induce or interfere with protein export compared with the ROSI group.
Figure 4.
RXR agonists blunt the expression and secretion of adiponectin compared with ROSI-treated cells. (A) Normalized read counts of Adipoq transcript were analyzed as described in “Materials and Methods.” (B) Quantitative PCR (qPCR) analysis of Adipoq expression in differentiated MSCs treated with ROSI (500 nM), 4204 (100 nM), TBT (50 nM), or vehicle control (0.1% DMSO). (C) MSCs were differentiated with MDI and ligands for 2 wk, washed, and allowed to incubate in media for 72 h. Adiponectin protein in conditioned media was measured by ELISA. (D) qPCR analysis of Ero1l in MSCs differentiated as described in (B). All data are represented as the mean ± SEM. For ROSI, 4204, and TBT samples, a one-way ANOVA and Tukey multiple comparison test were used: **P < 0.01; ***P < 0.001; ****P < 0.0001.
Rexinoids promote or fail to silence fibrotic and inflammatory genes that are downregulated by ROSI
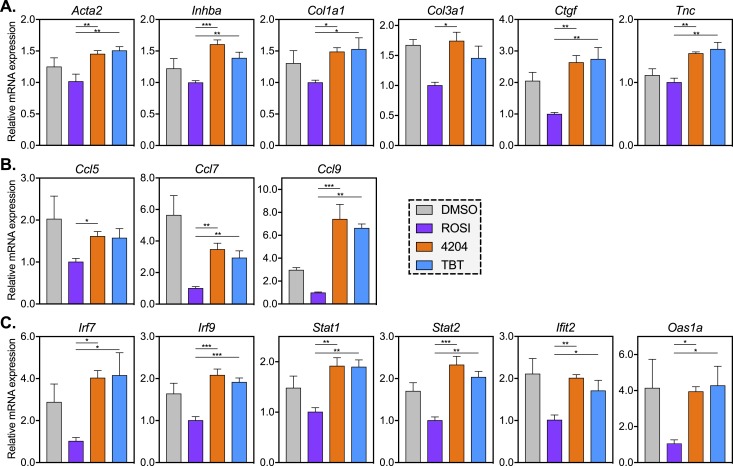
As in the liver, adipose fibrosis is promoted by TGF-β signaling, which recruits wound-healing myofibroblasts and elevates expression of profibrotic ECM genes such as collagens, fibronectin, and connective tissue growth factor (Ctgf) (61). In analyzing our RNA-seq data, we noted an enrichment of ECM terms in clusters 1 to 3 (where expression is higher in TBT than in ROSI groups). A comprehensive list of ECM genes expressed in adipose tissue or adipose progenitors (62) revealed 59 genes within our k-means cluster, 45 of which (76%) were found in clusters 1 to 3 (Supplemental Fig. 2B). TGF-β signaling was chiefly enriched in the TBT-induced cluster 1, driven by genes such as activin A receptor type 2A (Acvr2a), bone morphogenetic protein receptor type 1a and 2 (Bmpr1a, Bmpr2), inhibin A (Inhba), and Rho associated coiled-coil containing protein kinase 2 (Rock2). qPCR confirmed downregulation of fibrosis-associated genes by ROSI, but not 4204 or TBT (Fig. 5A). This included the myofibroblast marker actin, α2, smooth muscle, aorta (Acta2, also known as αSMA), the TGF-β marker Inhba, as well as several ECM genes such as Col1a1, Col3a1, and Ctgf, whereas the proinflammatory glycoprotein tenascin C (Tnc) was induced by rexinoids (Fig. 5A; Supplemental Table 2).
Figure 5.
Rexinoids promote or fail to silence the expression of fibrotic and inflammatory genes that are downregulated by ROSI. RNA was collected from MSCs differentiated with MDI and ROSI (500 nM), 4204 (100 nM), TBT (50 nM), or vehicle control (0.1% DMSO). qPCR analysis of gene expression was conducted for (A) genes involved in adipose tissue fibrosis, (B) chemokines, and (C) IFN genes. All data are represented as the mean ± SEM. For ROSI, 4204, and TBT samples, a one-way ANOVA and Tukey multiple comparison test were used: *P < 0.05; **P < 0.01; ***P < 0.001.
RNA-seq analysis also revealed an enrichment of inflammatory signaling pathways in cluster 3 (Fig. 1B). Whereas expression of classic markers of adipose inflammation such as IL-6, TNF-α, and macrophage migration inhibitory factor were not altered by TBT, levels of mRNAs encoding the macrophage recruiting C-C motif chemokine ligand 5, 7, and 9 (Ccl5, Ccl7, Ccl9) were all expressed at higher levels than ROSI either due to incomplete repression (Ccl5, Ccl7) or an RXR-mediated induction (Ccl9) (Fig. 5B; Supplemental Table 2). Genes involved in IFN signaling were among the most upregulated transcripts in TBT-differentiated MSCs when compared with ROSI (Supplemental Fig. 3A and 3B). qPCR confirmed high levels of interferon regulatory factors (IRFs) 7 and 9 (Irf7, Irf9) as well as the IFN-stimulated genes (ISGs) signal transducer and activator of transcription 1 and 2 (Stat1, Stat2), interferon induced protein with tetratricopeptide repeats 2 (Ifit2), and 2-5′ oligoadenylate synthetase 1a (Oas1a) in MSCs differentiated in the presence of rexinoids (Fig. 5C). Hence, rexinoid treatment promotes a proinflammatory, profibrotic gene expression profile in differentiated MSCs either by inducing (cluster 1) or failing to silence these genes (clusters 2 and 3) during adipogenesis.
Rexinoid-differentiated MSCs fail to induce the expression of BAT markers and respire less oxygen in the uncoupled state
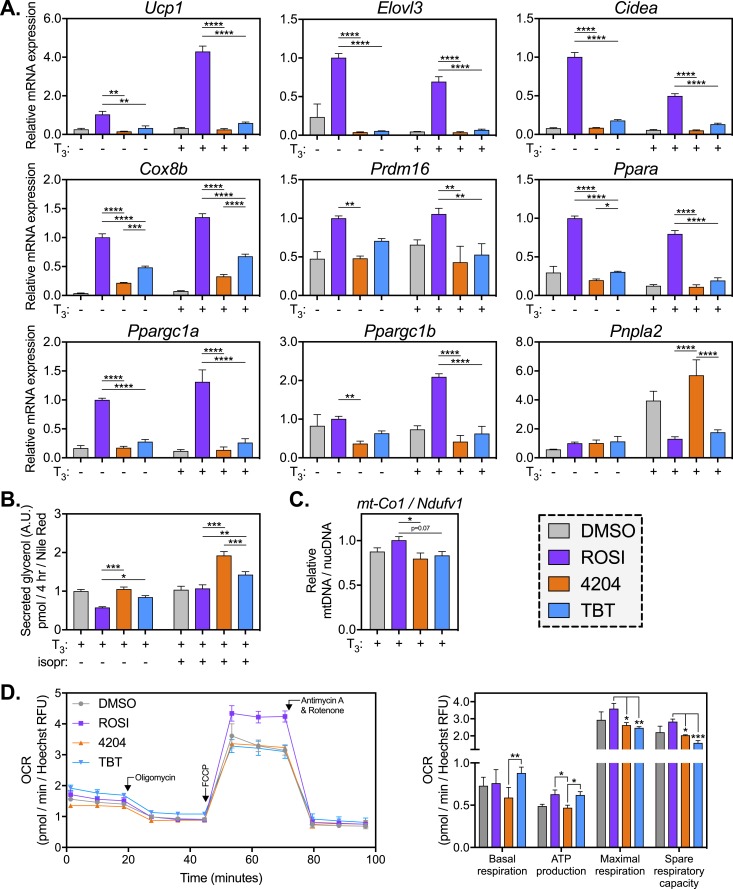
Analysis of clusters 4 and 5 revealed a pronounced enrichment of genes coding subunits of the ETC (pomplex I to IV and ATP synthase). Of the 75 ETC genes within our k-means cluster, 72 of them (96%) were in clusters 4 to 6 (Supplemental Fig. 2A). Along with mitochondrial uncoupling and lipid catabolism, elevated respiration is a key feature that distinguishes thermogenic brown or beige adipose tissue from white adipose tissue. Recent literature shows that bone marrow–derived MSCs can be differentiated into brown/beige-like adipocytes in vitro (41). Therefore, we hypothesized that rexinoids were inhibiting a brown/beige adipocyte phenotype in our MSCs. To test this hypothesis, MSCs were differentiated with MDI and ligands (as in Supplemental Fig. 1A) in the presence or absence of thyroid hormone (T3) to encourage browning (41). At the end of differentiation, MSCs were assessed for expression of brown/beige markers, including Ucp1, elongation of very-long-chain fatty acids (FEN1/Elo2, SUR4)/Elo3, yeast–like 3 (Elovl3), cell death–inducing DFFA-like effector A (Cidea), cytochrome c oxidase subunit VIIIb (Cox8b), PR/SET domain 16 (Prdm16), PPARα (Ppara), and PPARγ coactivator 1α and 1β (Ppargc1a and Ppargc1b, also known as PGC-1α, PGC-1β) (Fig. 6A). T3 effectively induced Ucp1 (4.3-fold in ROSI samples; Fig. 6A), although several other BAT markers (Elovl3, Cidea, and Ppara) were attenuated by T3 treatment. Importantly, 4204- and TBT-treated samples failed to upregulate most BAT markers over vehicle controls, revealing an inability to activate the brown/beige transcriptional program (Fig. 6A).
Figure 6.
Rexinoid-differentiated MSCs fail to induce the expression of BAT markers and respire less oxygen in the uncoupled state. MSCs were differentiated into adipocytes with MDI and ligands (500 nM ROSI, 100 nM 4204, 50 nM TBT, 0.1% DMSO) in the presence or absence of T3. (A) RNA was collected at the end of differentiation and analyzed for expression of BAT markers. For ROSI, 4204, and TBT samples, a two-way ANOVA and Tukey multiple comparison test were used: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (B) Differentiated MSCs were exposed to the β-adrenergic agonist isoproterenol (100 nM) or vehicle control (0.1% DMSO) for 4 h. Lipolysis was measured by assaying free glycerol in the media and normalizing to lipid stain as described in “Materials and Methods.” For ROSI, 4204, and TBT samples, a two-way ANOVA and Tukey multiple comparison test between treatment groups were used: *P < 0.05; **P < 0.01; ***P < 0.001. (C) DNA was isolated as described in “Materials and Methods.” Mitochondrial content was assessed by qPCR as the ratio of mitochondrial DNA (mt-Co1) to nuclear DNA (Ndufv1). For ROSI, 4204, and TBT samples, a one-way ANOVA and Tukey multiple comparison test were used: *P < 0.05. (D) MSCs were differentiated into adipocytes in the presence of MDI, T3, and ligands as described in “Materials and Methods.” After 2 wk, MSCs were starved of buffer and oxygen for 1 h prior to analysis of mitochondrial function by the Mito Stress Test (see “Materials and Methods”). OCR was normalized to cell number (Hoechst 33342 RFU) for each well. Parameters of mitochondrial function were extrapolated from normalized OCRs. For ROSI, 4204, and TBT samples, a one-way ANOVA and Tukey multiple comparison test were used: *P < 0.05; **P < 0.01; ***P < 0.001. All data are represented as the mean ± SEM.
We also assessed expression of patatin-like phospholipase domain containing 2 [Pnpla2, also known as adipose triglyceride lipase (ATGL)], because intracellular lipolysis is thought to be critical for BAT thermogenesis (63). Interestingly, T3 increased expression of ATGL, and this increase was blocked in the ROSI and TBT groups, but not 4204 (Fig. 6A). We then assessed lipolysis in our differentiated cells, as measured by free media glycerol normalized to lipid accumulation, both under basal conditions and upon β-adrenergic stimulation with isoproterenol (Fig. 6B). In concordance with the ATGL expression data, 4204-treated cells lipolysed the most fat, whereas TBT treatment resulted in slightly more lipolysis than in the ROSI group (Fig. 6B). We infer from these data that thermogenesis in 4204- and TBT-differentiated adipocytes would not be limited by their ability to lipolyse intracellular fat. Consistent with this idea, recent in vivo work suggests that circulating, rather than intracellular, free fatty acids drive BAT thermogenesis (64).
Because BAT is, in part, defined by an abundance of mitochondria, we assessed whether rexinoid treatment reduced mitochondrial number by quantifying relative abundance of mitochondrial DNA. Compared with ROSI-differentiated MSCs, 4204- and TBT-treated cells had an ∼20% reduction in mitochondrial DNA, although this only reached statistical significance in the 4204 group (Fig. 6C). We then subjected differentiated MSCs to a Seahorse Mito Stress Test (see “Materials and Methods”) to assess mitochondrial function. Basal OCR was significantly higher in the TBT group as compared with 4204, suggesting an RXR-independent regulation of basal respiration by TBT (Fig. 6D). We also found that ATP production in the ROSI and TBT groups was higher than in 4204-treated cells. However, when respiration was uncoupled from ATP synthesis, ROSI-differentiated cells were the best equipped to respire oxygen (Fig. 6D). Hence, under basal conditions, TBT-differentiated MSCs maintain functional mitochondria that respire oxygen and produce ATP at the same levels seen in the ROSI group. Under stressed conditions (uncoupling) the rexinoid groups are unable to respire at the rate achieved by ROSI, again suggesting adipocyte dysfunction.
Multiple studies have now shown that long-term cold exposure induces the proliferation of beige adipocytes in inguinal fat depots from vascular progenitors (65, 66). Hence, we isolated the SVF from inguinal fat depots and differentiated them in the presence of MDI, T3, and ligands. Interestingly, rexinoids were not as effective as ROSI in inducing adipogenesis in SVF (Supplemental Fig. 5A), likely due to the fact that most nonhematopoietic cells in SVF are committed preadipocytes (28, 67). On average, DMSO-, 4204-, and TBT-treated SVF reached 47%, 66%, and 75% of the lipid accumulation achieved in ROSI samples, respectively (Supplemental Fig. 5A). In contrast, differences in gene expression of PPARγ target genes (Fabp4, GLUT4, Adipoq) were far more dramatic than the lipid accumulation data would suggest (Supplemental Fig. 5A and 5B). This suggests that ROSI induces expression of its target genes to levels beyond their maximal effect on adipose turnover, and it sheds light on how rexinoid-treated MSCs achieve the same level of lipid accumulation in spite of lower expression of PPARγ targets. The most striking difference in gene expression was seen in BAT markers, particularly Ucp1, whose expression was 1000- and 100-fold lower than ROSI in 4204 and TBT groups, respectively (Supplemental Fig. 5D). Interestingly, we do not see repression of adiponectin by rexinoids relative to the DMSO control as was observed in MSCs (Supplemental Fig. 5C; Supplemental Table 2), implying that this may occur during the lineage commitment process where RXR is known to play an important role (28). We additionally observe activation, not repression, of IFN genes by ROSI (and rexinoids to a lesser extent) (Supplemental Fig. 5E), again suggesting regulation during the commitment process or a depot-specific difference between the bone marrow and inguinal compartments.
Chemical antagonists reveal PPARγ- and RXR-dependent differences in ROSI- vs rexinoid-differentiated MSCs
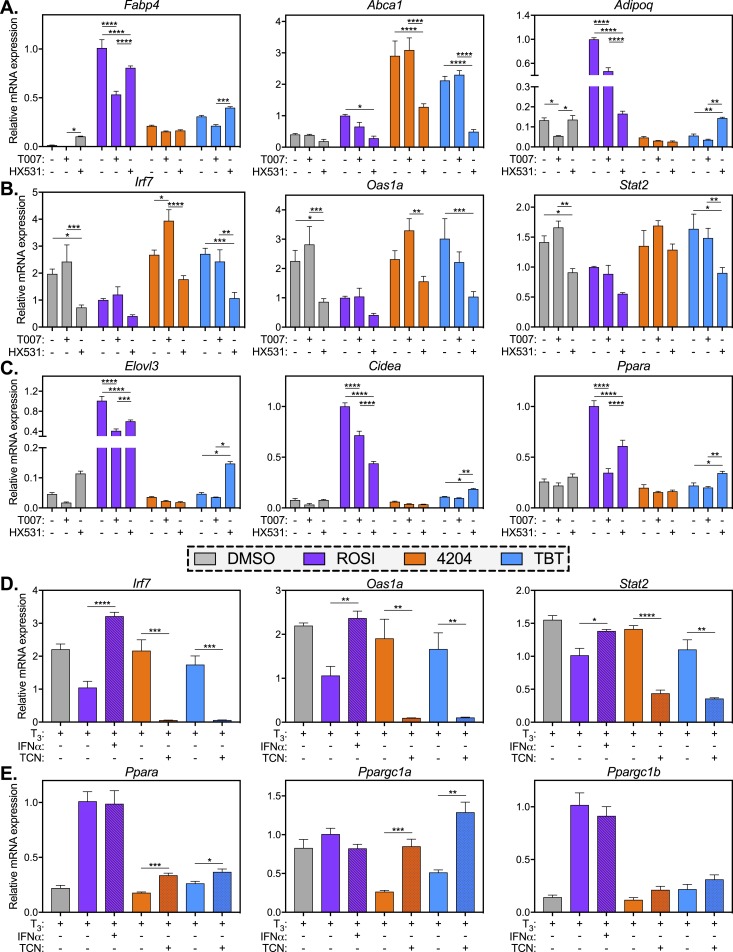
To evaluate the PPARγ/RXR dependence of our observed phenotypes, we differentiated MSCs into adipocytes for 7 days with MDI and ligands as before, then added in chemical antagonists of PPARγ (T0070907, also known as T007) and RXR (HX531) for the final 7 days of treatment (Supplemental Fig. 6A). In this way, we did not completely block the formation of adipocytes by inhibiting PPARγ/RXR early in adipogenesis (day 0 to 7), but we could still assess the functions of these receptors in committed preadipocytes and in mature adipocytes. As expected, Fabp4 was strongly induced by ROSI and this induction was blunted by T007 and, to a lesser extent, HX531 (Fig. 7A). Fabp4 induction was lower in both rexinoid treatments; however, in TBT-treated cells its expression was elevated by HX531 (Fig. 7A, left). Abca1 also behaved as predicted, showing induction by both rexinoid treatments that was inhibited by HX531, but not T007 (Fig. 7A, center). Interestingly, ROSI induction of the PPARγ target Adipoq was more effectively blocked by HX531 than T007, whereas RXR agonists 4204 and TBT strongly repressed Adipoq expression (Fig. 7A, right). Similar to what was observed with Fabp4, HX531 could only rescue Adipoq expression levels in TBT-treated samples. The ISGs Irf7, Oas1a, and Stat2 all showed expression patterns that suggested RXR dependence, with HX531 inhibiting ISG expression in TBT-treated samples (Fig. 7B). The BAT markers Elovl3, Cidea, and Ppara all showed a PPARγ-dependent expression pattern reminiscent of Fabp4 or Adipoq (Fig. 7C). BAT gene expression in 4204 samples was nearly unchanged by T007 or HX531 treatment, whereas, once again, expression in TBT samples was elevated in the presence of HX531.
Figure 7.
Chemical antagonists reveal PPARγ- and RXR-dependent differences in ROSI- vs rexinoid-differentiated MSCs. (A–C) MSCs were differentiated into adipocytes in the presence of MDI and ligands (500 nM ROSI, 100 nM 4204, 50 nM TBT, 0.1% DMSO). On day 7 of differentiation, chemical antagonists of PPARγ (T0070907, 1 μM) or RXR (HX531, 10 μM) were added to the culture medium through the end of differentiation (see Supplemental Fig. 6A). RNA was collected and analyzed by qPCR for gene expression of (A) classic PPARγ and RXR targets, (B) IFN genes, or (C) BAT markers. A two-way ANOVA and a Tukey multiple comparison within each treatment group were used: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (D and E) MSCs were differentiated for 7 d in the presence of MDI and ligands as in (A)–(C). On day 7, mouse recombinant IFNα (1000 U/mL) was added to ROSI samples and the JAK inhibitor TCN (5 μM) was added to 4204 and TBT samples (see Supplemental Fig. 6B). Gene expression of (D) IFN genes and (E) BAT markers was analyzed by qPCR. By an unpaired t test: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are represented as the mean ± SEM.
Finally, we asked whether repression of BAT genes in rexinoid-treated cells might be due to the sustained expression of ISGs, as was reported in a recent study by Seale and colleagues (68). To test this, we differentiated MSCs for 1 week in the presence of MDI and ligands, and then modulated IFN signaling by adding either mouse recombinant IFNα to ROSI-treated cells or the Janus kinase (JAK) inhibitor TCN to 4204- and TBT-treated cells. TCN was previously shown to induce browning in human induced pluripotent stem cell–derived adipocytes through JAK1/3 inhibition and, ultimately, by suppressing the expression of ISGs (69). IFNα treatment was able to reactivate the expression of ISGs in ROSI samples, whereas TCN strongly inhibited their expression in 4204/TBT samples (Fig. 7D). Interestingly, IFNα had no effect on the expression of BAT markers in ROSI-differentiated MSCs (Fig. 7E; Supplemental Fig. 6C). Alternatively, TCN significantly increased the expression of PGC-1α in 4204- and TBT-differentiated cells, and slightly elevated the expression of Ppara and Elovl3 (Fig. 7E; Supplemental Fig. 6C . These data are consistent with the possibility that persistent IFN signaling in rexinoid-treated samples suppresses the critical BAT regulator PGC-1α. Taken together, our data show that rexinoids fail to silence inflammatory signals in developing adipocytes that can interfere with the ability of these cells to acquire a brown/beige phenotype.
Discussion
In this study, we demonstrated that although rexinoids are potent inducers of lipid accumulation in differentiating MSCs, the resulting cells are dysfunctional when compared with MSCs differentiated with the thiazolidinedione ROSI. MSCs differentiated into adipocytes were assessed for four basic functions: (1) sensitivity to insulin and subsequent glucose uptake, (2) adipokine secretion, (3) expression of profibrotic and proinflammatory markers, and (4) the acquisition of a brown/beige fat phenotype. In all of these functions, rexinoids programmed poorly functioning adipocytes that displayed diminished glucose uptake (Fig. 3) and ablated adiponectin secretion (Fig. 4), persistent proinflammatory and profibrotic gene expression (Fig. 5), and a limited ability to respire oxygen in response to mitochondrial uncoupling (Fig. 6). These data shed light on a key question raised by our previous publications and within the EDC field: if pharmacologic PPARγ agonists improve adipocyte function, then why do environmental PPARγ activators such as TBT not produce a corresponding effect? For the prototypical obesogen TBT, the culprit appears to be its ability to activate the promiscuous nuclear receptor heterodimer RXR. Of course, developmental exposure (during gestation or early life) to any PPARγ agonist bears the risk of increasing total adipocyte number, a strong predictor of fat mass in adults (70, 71). However, our data highlight the potential for RXR activators to promote both adipose hyperplasia (28) and adipocyte dysfunction.
We recently showed that TBT exposure of MSCs prior to differentiation promotes adipose lineage commitment in an RXR-dependent manner (28). Transcriptomal analyses of undifferentiated MSCs revealed a distinct RXR-dependent gene expression profile in TBT-treated cells, whereas ROSI exposure had little effect, likely due to minimal PPARγ2 isoform expression in MSCs prior to adipose induction (28, 72). This led us to consider the potential role of RXR as a target of TBT in mature adipocytes. In our present study, MSCs differentiated in the presence of ROSI have a significantly different transcriptome from that of TBT-treated cells (Fig. 1; Supplemental Fig. 1). Unlike our previous work in undifferentiated cells (28), ROSI treatment altered the expression of numerous genes (Supplemental Fig. 1C–1F), some of which were congruently regulated by TBT (Supplemental Fig. 1E and 1F). This result is expected, because PPARγ2 is highly expressed in mature adipocytes and because TBT can activate PPARγ gene targets via either half of the RXR/PPARγ heterodimer. The consequence of RXR promiscuity, however, is that rexinoids will act not only on targets of RXR-PPARγ, but also on gene targets of other permissive RXR partners such as LXR, farnesoid X receptor, PPARα, and PPARδ, as well as targets of RXR homodimers (44). Hence, it is not surprising that the overlap between ROSI- and TBT-induced expression changes is relatively poor (Supplemental Fig. 1E and 1F). Subsequent pathway analysis revealed that transcriptomal differences between ROSI and TBT suggest functional deficits and inappropriate development of TBT-treated MSCs into adipocytes (Fig. 1).
Given gene expression data implying that TBT fails to activate PPARγ targets to levels achieved by ROSI, how then do these cells achieve the same levels of lipid accumulation? Notably, several lipogenic genes show higher levels of expression in TBT-treated samples, including Pparg2 (Fig. 2C), Fasn (Fig. 2D), ATP citrate lyase (Acly), and Elovl6 (Fig. 1C). ROSI treatment, however, highly induces the expression of many lipogenic genes found in cluster 6 (e.g., Srebf1 and Dgat2,Fig. 1C). What is more likely is that rexinoids induce the expression of enough adipogenic genes to a level sufficient for meaningful adipose differentiation, whereas ROSI promotes high levels of these transcripts such that they achieve their maximal adipogenic effect. Despite significant lipogenesis, rexinoids fail to silence genes that may belong to alternate mesenchymal lineages or preadipocytes, which may explain their dysfunction. Hence, these rexinoid-treated cells could also be viewed as immature or incompletely differentiated (i.e., dysfunctional) adipocytes, despite their ability to store and mobilize fat.
MSCs and 3T3-L1 cells have become common platforms to screen EDCs for obesogenic potential (19, 21, 45, 73, 74). Typically, the endpoints of these adipogenesis assays are intracellular lipid accumulation and gene expression of adipose lineage markers. These endpoints are then used to infer accelerated or inhibited commitment (MSCs only) and/or terminal differentiation (MSCs or 3T3-L1 cells). More recently, investigators have begun assessing the effects of EDCs on mature adipocyte function, primarily insulin sensitivity and adipokine secretion (10, 18, 75, 76). In vitro studies of adipocyte function using 3T3-L1 cells are nearly universal, with few exceptions utilizing primary adipose tissue explants or differentiated MSCs (77–79). This is despite literature showing that MSCs maintain lipolytic and endocrine functions when differentiated into adipocytes (80, 81).
Sargis and colleagues (23) investigated functional differences in 3T3-L1 cells differentiated with TBT or the thiazolidinedione troglitazone. These authors observed lower levels of adiponectin and CEBPα mRNA and intracellular protein in TBT-treated cells, as well as lower GLUT4 expression, but no differences in glucose uptake. They speculated that the observed differences may be RXR-dependent. In this study, we have used the primary MSC model to show that rexinoids fail to induce GLUT4 expression to levels achieved by ROSI, and therefore attenuate glucose uptake under basal and insulin-stimulated conditions (Fig. 3). Notably, we observed intact insulin signaling, suggesting that diminished glucose uptake is indeed due to lower expression of GLUT4.
There are multiple sources of ECM in adipose tissue fibrosis, although M1 macrophages and preadipocytes are major contributors. Recent work shows that TGF-β and PDGFRα signaling in preadipocytes induces a myofibroblast-like phenotype that results in ECM deposition and adipose fibrosis (82). ROSI-treated MSCs effectively downregulate the myofibroblast marker Acta2 and the TGF-β marker Inhba as well as several genes coding for ECM proteins, whereas rexinoid treatment sustains their expression at preadipocyte (vehicle control) levels (Figs. 1 and 5; Supplemental Fig. 2B). Unlike obese adipose tissue, fibrotic gene induction in our MSCs is not a product of hypoxia. Rather, it may reflect a developmental misprogramming by RXR whereby markers of other mesenchymal lineages (e.g., muscle, bone, cartilage) are either inappropriately activated or left unsilenced. This notion is supported by the fact that RXR is known to promote myogenic differentiation (83) and its heterodimeric partner retinoic acid receptor (RAR) is known to promote osteogenesis (84, 85). Whereas RAR is considered to be a nonpermissive RXR partner, previous studies report induction of RAR gene targets by rexinoids (86). This might occur through RXR homodimers or in concert with retinoids present in serum, because the RXR/RAR heterodimer is permissive when RAR is liganded (87). Notably, we observe markers of the myogenic [myosin heavy chain 11 (Myh11)] and osteogenic [carbonic anhydrase II (Car2), cytochrome P450 family 26 subfamily B member 1 (Cyp26b1)] lineages among the top genes upregulated by TBT in our RNA-seq data (Supplemental Table 3). Furthermore, many ECM genes are known to be expressed in preadipocytes but are downregulated during terminal differentiation (62). Given the diminished ability of rexinoids to induce PPARγ targets (cluster 6, Fig. 1B), it is likely that many of these transcripts reflect a failure by rexinoids to properly differentiate these cells into fully functional, mature adipocytes. Because adipose fibrosis is maintained through a positive feedback loop (35), aberrant rexinoid signaling in preadipocytes during adipogenesis could potentially initiate an inflammatory/fibrotic response in vivo.
In addition to a profibrotic transcripts, rexinoids also sustained the expression of proinflammatory genes in MSCs. The macrophage chemoattractants CCL5, CCL7, and CCL9 were highly expressed in MSCs treated with RXR activators when compared with ROSI (Fig. 5B). Importantly, these chemokines are upregulated in human obesity (88), with the exception of CCL9, which has no human ortholog but is upregulated in mouse models of obesity (89). Remarkably, rexinoids failed to repress many genes involved in IFN signaling (Figs. 1 and 5; Supplemental Fig. 3). The role of IFN signaling in adipose tissue is complex, as IFN and its effectors play roles in adipogenesis and in mature adipocyte function (90, 91). Importantly, IFNγ acts through STAT1 to induce insulin resistance and inhibit adipogenesis in human preadipocytes (92), and IFNγ-null mice on a high-fat diet have improved insulin sensitivity, smaller adipocytes, and an M2 shift in adipose tissue macrophages (93). Although the source of IFNs in obese adipose tissue is primarily T cells and natural killer cells, in this study we report the induction of IFN signaling within MSCs differentiated in the presence of RXR activators. Notably, IFN genes are strongly repressed by ROSI treatment but remain elevated in vehicle and TBT samples (Fig. 1A and 1B, cluster 3, Fig. 5B). Hence, the enrichment of IFN genes in rexinoid-treated samples probably reflects a failure to downregulate these transcripts via PPARγ, rather than an RXR-mediated induction. At the same time, chemical inhibition of RXR consistently repressed ISGs (Fig. 7B), suggesting some role for RXR in maintaining basal expression of these genes. Where in the IFN cascade PPARγ and RXR might act is unclear; however, recruiting or ousting IRFs to and from the genome is a plausible mechanism.
Research into the molecular regulation of brown and beige adipose tissue has exploded in the past decade (38), in part due to its potential as a pharmacologic target for the treatment of obesity and diabetes (39). Our data show that MSCs differentiated in the presence of rexinoids are unable to induce the expression of transcriptional and functional markers of brown and beige fat (Fig. 6A) despite uninhibited adipose turnover and T3 supplementation in the media. Interestingly, ATGL expression and lipolysis were induced by T3 in the 4204 group, but this was blunted by ROSI and TBT treatment (Fig. 6A and 6B), exposing a rare example of functional similarity between ROSI- and TBT-differentiated adipocytes. The other instance of congruence between ROSI and TBT can be seen in the basal respiration and ATP production rates measured in the Mito Stress Test (Fig. 6D), revealing that TBT-differentiated adipocytes are as functional as ROSI-differentiated cells under basal conditions. Only when respiration is uncoupled from ATP synthesis [carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone treatment] are the deficits of rexinoid-treated mitochondria apparent (Fig. 6D). Whether similar deficits would be seen in response to sympathetic activation (by isoproterenol or a β3 agonist) was not tested. Higher respiration in ROSI cells is likely due to higher expression of PGC-1α, a master regulator of mitochondrial biogenesis, and a subsequent increase in mitochondrial abundance (Fig. 6A and 6C). Our results in MSCs were mirrored in SVF differentiated into adipocytes, where rexinoid treatment resulted in slightly less adipose turnover, but a >100-fold decrease in Ucp1 expression compared with ROSI (Supplemental Fig. 5D).
Interestingly, recent work has highlighted the importance of IFN signaling in brown and beige adipose tissue. For example, IRF3 was shown to repress white adipose browning (94) whereas IRF4 is induced upon cold exposure and promotes thermogenesis (95). A recent high-throughput screen for potential browning agents using human pluripotent stem cell–derived adipocytes identified two JAK inhibitors (one of which was TCN) that were shown to act via repression of IFN signaling (69). Importantly, these pharmaceuticals increased mitochondrial content and basal lipolysis in these cells. Seale and colleagues (68) showed that the BAT transcriptional regulator PRDM16 represses type I IFN signaling in adipocytes to promote thermogenesis. PRDM16 was shown to negatively regulate ISGs in brown preadipocytes, including Irf7, Ifi44, Oas2, Oas3, Stat1, and Stat2, all genes that are highly upregulated in MDI-induced MSCs treated with vehicle or TBT compared with ROSI (Supplemental Fig. 3). Importantly, a downstream inhibitor of IFN signaling (TCN) was able to rescue expression of PGC-1α in 4204/TBT samples (Fig. 7E). Taken together, these data implicate persistent IFN signaling in rexinoid-treated MSCs as a repressor of adipose browning/beiging.
PPARs are permissive partners of RXR; hence, rexinoid treatment should induce the expression of PPARγ targets (44). To some extent, this is achieved in our experiments, because rexinoid-differentiated cells accumulate lipid and express many genes congruently with ROSI-treated MSCs (Supplemental Fig. 1). However, we observed a vastly divergent transcriptome between ROSI- and rexinoid-treated samples (Fig. 1; Supplemental Fig. 1). Our chemical antagonist assay sheds light on the complex mechanisms through which the effects of these ligands ultimately diverge (Fig. 7A–7C). Fabp4, for example, is effectively induced by ROSI, 4204, and TBT over vehicle, but this induction is much stronger in ROSI (Fig. 7A). Lower expression levels in rexinoid treatments might be due to dispersion of RXR among many permissive partners, thereby reducing the availability of RXR protein to partner with PPARγ. Interestingly, chemical inhibition of RXR in TBT-treated cells increases expression of Fabp4 and many other PPARγ targets (Adipoq, Elovl3, Cidea, Ppara) (Fig. 7A and 7C). This may be due to a “redirecting” of TBT toward its other nuclear receptor target, PPARγ. Alternatively, inhibition of RXR in the presence of rexinoids might increase RXR availability, and subsequent partnering with PPARγ is only unmasked in the presence of PPARγ agonist (i.e., TBT but not 4204). We also note that Adipoq mRNA levels are depressed by rexinoids relative to vehicle control, and this depression is only relieved by HX531 in TBT, but not 4204, samples (Fig. 7A). We hypothesize that RXR, likely as a PPARγ/RXR heterodimer, positively regulates the Adipoq promoter even in the absence of exogenous ligand (represented by the vehicle control group). Hence, ROSI treatment activates Adipoq, whereas the rexinoid-induced dispersal of RXR throughout the genome results in repression. Similar to Fabp4, this repression can be relieved by HX531 only in the presence of a PPARγ agonist, TBT (Fig. 7A). Taken together, we infer from these results that rexinoids are uniquely poised to disrupt normal adipocyte development and function due to the promiscuous nature of RXR. Notably, LXR has been implicated as a negative regulator of adiponectin (96) and adipose browning (97), making this permissive RXR partner suspect in mediating some of the adverse effects of rexinoids during adipogenesis.
Work in animals suggests that RXR agonists are, in fact, insulin-sensitizing and glucose-lowering agents, in spite of persistent hypertriglyceridemia and repression of the thyroid axis (98–101). Notably, every published animal study conducted on this class of drug used genetic models of obesity and/or diabetes in rodent models (ob/ob mouse, db/db mouse, or Zucker fatty rats). It is possible that rexinoids have unique effects in animals with these genetic backgrounds, and may in fact worsen glycemic control in wild-type animals or in dietary models of obesity. Diabetic patients taking the rexinoid bexarotene, indicated for cutaneous T-cell lymphoma, are warned that bexarotene might enhance the effect of insulin and to be wary of hypoglycemic crises (102). We find no evidence in the literature of such a case and presume that the warning was issued based on rodent data. We do find, however, a documented case of a pediatric patient developing insulin-dependent diabetes that persisted after discontinuation of bexarotene (103). Our data demonstrate that these pharmaceutical rexinoids stifle adipose browning, which, in combination with central repression of the thyroid axis, could result in deficits in nonshivering thermogenesis and energy expenditure that are worthy of further investigation. A recent paper identified bexarotene in a small molecule screen of browning agents in a muscle cell line (104), and subsequently showed increased browning and beiging in BAT and white adipose tissue, respectively, of adult mice fed a high-fat diet. The authors proposed that RXRs are master regulators of brown fat development and activation, more so than PPARγ. Importantly, doses of bexarotene used in this study (10 µM in vitro, 50 mg/kg/d in vivo) exceed the window of RXR selectivity for this drug (105) and likely activate RAR, which is a well-documented positive regulator of thermogenesis (106–108).
In conclusion, we have shown that RXR activation during the differentiation of MSCs promotes the formation of a dysfunctional and misprogrammed adipocyte in vitro. This comes on the heels of our recent work showing that rexinoids promote adipose lineage commitment in undifferentiated MSCs (28). Taken together, these data support a model in which RXR activators have the potential to increase the allocation of stem cells to the fat lineage in addition to programming a poorly functioning adipocyte. Given the focus within the endocrine disruption field on PPARγ activators, it would be prudent not to ignore EDCs that activate RXR, as they may prove more deleterious. Few of these chemicals have been identified to date (45, 109), and none, other than TBT, has been characterized in vivo or studied in human populations. Finally, we have shown, to our knowledge for the first time, that MSCs may provide a useful platform to screen and study EDCs that repress adipose browning. Given the high cost of endocrine disruption in the United States and Europe (110–112), the need to identify obesogens and characterize their mechanisms of action becomes increasingly critical.
Supplementary Material
Acknowledgments
We thank all members of the Blumberg and Shioda laboratories. We also acknowledge Dr. M. Cristina Kenney for making a Seahorse analyzer available to investigators at the University of California, Irvine, and Marilyn Chwa for providing time and expertise. We thank Rosh Chandraratna (IO Therapeutics, Santa Ana, CA) for a gift of IRX4204.
Financial Support: This work was funded by National Institute of Environmental Health Sciences Grant R01ES023316 (to B.B. and T.S) and by Environmental Protection Agency Grant STAR FP917800 (to B.M.S.).
Author Contributions: B.M.S. and B.B. conceived the study and wrote the manuscript. B.M.S., T.S., and B.B. designed the experiments and secured funding. B.M.S., V.T.H., T.S., and R.C.-G. performed all experiments. B.M.S. and T.S. analyzed the data.
Disclosure Summary: B.B. is a named inventor on several patents related to PPARγ. The remaining authors have nothing to disclose.
Glossary
Abbreviations:
- 4204
IRX4204
- Abca1
ATP-binding cassette transporter 1
- ATGL
adipose triglyceride lipase
- Adipoq
adiponectin
- BAT
brown adipose tissue
- Cidea
cell death–inducing DFFA-like effector A
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- EDC
endocrine-disrupting chemical
- Elovl3
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)–like 3
- ETC
electron transport chain
- Fabp4
fatty acid binding protein 4
- FBS
fetal bovine serum
- IFN
interferon
- Inhba
inhibin A
- IRF
interferon regulatory factor
- ISG
interferon-stimulated gene
- JAK
Janus kinase
- KRH
Krebs–Ringer–HEPES buffer
- LXR
liver X receptor
- MSC
mesenchymal stem cell
- OCR
oxygen consumption rate
- PKB
protein kinase B
- PPAR
peroxisome proliferator–activated receptor
- Ppara
peroxisome proliferator–activated receptor α
- Ppargc1a
peroxisome proliferator–activated receptor γ coactivator 1α
- qPCR
quantitative PCR
- RAR
retinoic acid receptor
- RNA-seq
RNA sequencing
- RFU
relative fluorescence unit
- ROSI
rosiglitazone
- RXR
retinoid X receptor
- Slc2a4
solute carrier family 2 member 4
- Stat2
signal transducer and activator of transcription 2
- SVF
stromal vascular fraction
- T2D
type 2 diabetes
- TBT
tributyltin
- TCN
tofacitinib
- Ucp1
uncoupling protein 1
References
- 1. Cawley J, Meyerhoefer C. The medical care costs of obesity: an instrumental variables approach. J Health Econ. 2012;31(1):219–230. [DOI] [PubMed] [Google Scholar]
- 2. American Diabetes Association Economic costs of diabetes in the U.S. in 2012 [published correction apprears in Diabetes Care 2013;36(6):1797]. Diabetes Care. 2013;36(4):1033–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization Global Report on Diabetes. Geneva, Switzerland: World Health Organization; 2016. [Google Scholar]
- 4. Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, Abraham JP, Abu-Rmeileh NM, Achoki T, AlBuhairan FS, Alemu ZA, Alfonso R, Ali MK, Ali R, Guzman NA, Ammar W, Anwari P, Banerjee A, Barquera S, Basu S, Bennett DA, Bhutta Z, Blore J, Cabral N, Nonato IC, Chang JC, Chowdhury R, Courville KJ, Criqui MH, Cundiff DK, Dabhadkar KC, Dandona L, Davis A, Dayama A, Dharmaratne SD, Ding EL, Durrani AM, Esteghamati A, Farzadfar F, Fay DF, Feigin VL, Flaxman A, Forouzanfar MH, Goto A, Green MA, Gupta R, Hafezi-Nejad N, Hankey GJ, Harewood HC, Havmoeller R, Hay S, Hernandez L, Husseini A, Idrisov BT, Ikeda N, Islami F, Jahangir E, Jassal SK, Jee SH, Jeffreys M, Jonas JB, Kabagambe EK, Khalifa SE, Kengne AP, Khader YS, Khang YH, Kim D, Kimokoti RW, Kinge JM, Kokubo Y, Kosen S, Kwan G, Lai T, Leinsalu M, Li Y, Liang X, Liu S, Logroscino G, Lotufo PA, Lu Y, Ma J, Mainoo NK, Mensah GA, Merriman TR, Mokdad AH, Moschandreas J, Naghavi M, Naheed A, Nand D, Narayan KM, Nelson EL, Neuhouser ML, Nisar MI, Ohkubo T, Oti SO, Pedroza A, Prabhakaran D, Roy N, Sampson U, Seo H, Sepanlou SG, Shibuya K, Shiri R, Shiue I, Singh GM, Singh JA, Skirbekk V, Stapelberg NJ, Sturua L, Sykes BL, Tobias M, Tran BX, Trasande L, Toyoshima H, van de Vijver S, Vasankari TJ, Veerman JL, Velasquez-Melendez G, Vlassov VV, Vollset SE, Vos T, Wang C, Wang X, Weiderpass E, Werdecker A, Wright JL, Yang YC, Yatsuya H, Yoon J, Yoon SJ, Zhao Y, Zhou M, Zhu S, Lopez AD, Murray CJ, Gakidou E. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384(9945):766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman ÅK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen YI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van ’t Hooft FM, Vinkhuyzen AAE, Westra HJ, Zheng W, Zondervan KT, Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin MR, Jöckel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJP, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Marchand LL, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PAF, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O’Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJF, Speliotes EK; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herrera BM, Keildson S, Lindgren CM. Genetics and epigenetics of obesity. Maturitas. 2011;69(1):41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanson MA, Gluckman PD. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol Rev. 2014;94(4):1027–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305(5691):1733–1736. [DOI] [PubMed] [Google Scholar]
- 9. Heindel JJ, Skalla LA, Joubert BR, Dilworth CH, Gray KA. Review of developmental origins of health and disease publications in environmental epidemiology. Reprod Toxicol. 2017;68:34–48. [DOI] [PubMed] [Google Scholar]
- 10. Heindel JJ, Blumberg B, Cave M, Machtinger R, Mantovani A, Mendez MA, Nadal A, Palanza P, Panzica G, Sargis R, Vandenberg LN, Vom Saal F. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. 2017;68:3–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gore AC, Chappell VA, Fenton SE, Flaws JA, Nadal A, Prins GS, Toppari J, Zoeller RT. EDC-2: The Endocrine Society’s second scientific statement on endocrine-disrupting chemicals. Endocr Rev. 2015;36(6):E1–E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meeker JD. Exposure to environmental endocrine disruptors and child development. Arch Pediatr Adolesc Med. 2012;166(10):952–958. [PubMed] [Google Scholar]
- 13. Zoeller RT, Brown TR, Doan LL, Gore AC, Skakkebaek NE, Soto AM, Woodruff TJ, Vom Saal FS. Endocrine-disrupting chemicals and public health protection: a statement of principles from the Endocrine Society. Endocrinology. 2012;153(9):4097–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grün F, Watanabe H, Zamanian Z, Maeda L, Arima K, Cubacha R, Gardiner DM, Kanno J, Iguchi T, Blumberg B. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol. 2006;20(9):2141–2155. [DOI] [PubMed] [Google Scholar]
- 15. Grün F, Blumberg B. Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006;147(6, Suppl):S50–S55. [DOI] [PubMed] [Google Scholar]
- 16. Janesick A, Blumberg B. Endocrine disrupting chemicals and the developmental programming of adipogenesis and obesity. Birth Defects Res C Embryo Today. 2011;93(1):34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heindel JJ, Newbold R, Schug TT. Endocrine disruptors and obesity. Nat Rev Endocrinol. 2015;11(11):653–661. [DOI] [PubMed] [Google Scholar]
- 18. Mimoto MS, Nadal A, Sargis RM. Polluted pathways: mechanisms of metabolic disruption by endocrine disrupting chemicals. Curr Environ Health Rep. 2017;4(2):208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pereira-Fernandes A, Vanparys C, Hectors TL, Vergauwen L, Knapen D, Jorens PG, Blust R. Unraveling the mode of action of an obesogen: mechanistic analysis of the model obesogen tributyltin in the 3T3-L1 cell line. Mol Cell Endocrinol. 2013;370(1–2):52–64. [DOI] [PubMed] [Google Scholar]
- 20. Bateman ME, Strong AL, McLachlan JA, Burow ME, Bunnell BA. The effects of endocrine disruptors on adipogenesis and osteogenesis in mesenchymal stem cells: a review. Front Endocrinol (Lausanne). 2017;7:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirchner S, Kieu T, Chow C, Casey S, Blumberg B. Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol Endocrinol. 2010;24(3):526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Ycaza J, Blumberg B. The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J Steroid Biochem Mol Biol. 2011;127(1–2):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Regnier SM, El-Hashani E, Kamau W, Zhang X, Massad NL, Sargis RM. Tributyltin differentially promotes development of a phenotypically distinct adipocyte. Obesity (Silver Spring). 2015;23(9):1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker AH, Watt J, Huang CK, Gerstenfeld LC, Schlezinger JJ. Tributyltin engages multiple nuclear receptor pathways and suppresses osteogenesis in bone marrow multipotent stromal cells. Chem Res Toxicol. 2015;28(6):1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanayama T, Kobayashi N, Mamiya S, Nakanishi T, Nishikawa J. Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptor γ/retinoid X receptor pathway. Mol Pharmacol. 2005;67(3):766–774. [DOI] [PubMed] [Google Scholar]
- 26. Chamorro-Garcia R, Diaz-Castillo C, Shoucri BM, Käch H, Leavitt R, Shioda T, Blumberg B. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun. 2017;8(1):2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chamorro-García R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect. 2013;121(3):359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shoucri BM, Martinez ES, Abreo TJ, Hung VT, Moosova Z, Shioda T, Blumberg B. Retinoid X receptor activation alters the chromatin landscape to commit mesenchymal stem cells to the adipose lineage. Endocrinology. 2017;158(10):3109–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugii S, Olson P, Sears DD, Saberi M, Atkins AR, Barish GD, Hong SH, Castro GL, Yin YQ, Nelson MC, Hsiao G, Greaves DR, Downes M, Yu RT, Olefsky JM, Evans RM. PPARγ activation in adipocytes is sufficient for systemic insulin sensitization. Proc Natl Acad Sci USA. 2009;106(52):22504–22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. [DOI] [PubMed] [Google Scholar]
- 31. Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106(4):473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li S, Shin HJ, Ding EL, van Dam RM. Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2009;302(2):179–188. [DOI] [PubMed] [Google Scholar]
- 34. Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, Nagaretani H, Matsuda M, Komuro R, Ouchi N, Kuriyama H, Hotta K, Nakamura T, Shimomura I, Matsuzawa Y. PPARγ ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50(9):2094–2099. [DOI] [PubMed] [Google Scholar]
- 35. Sun K, Tordjman J, Clément K, Scherer PE. Fibrosis and adipose tissue dysfunction. Cell Metab. 2013;18(4):470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kusminski CM, Bickel PE, Scherer PE. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15(9):639–660. [DOI] [PubMed] [Google Scholar]
- 37. Nam M, Cooper MP. Role of energy metabolism in the brown fat gene program. Front Endocrinol (Lausanne). 2015;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76(1):225–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poher AL, Altirriba J, Veyrat-Durebex C, Rohner-Jeanrenaud F. Brown adipose tissue activity as a target for the treatment of obesity/insulin resistance. Front Physiol. 2015;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. La Merrill M, Karey E, Moshier E, Lindtner C, La Frano MR, Newman JW, Buettner C. Perinatal exposure of mice to the pesticide DDT impairs energy expenditure and metabolism in adult female offspring [published correction appears in PLoS One 2014;9(9):e107332]. PLoS One. 2014;9(7):e103337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang YL, Lin SP, Hsieh PC, Hung SC. Concomitant beige adipocyte differentiation upon induction of mesenchymal stem cells into brown adipocytes. Biochem Biophys Res Commun. 2016;478(2):689–695. [DOI] [PubMed] [Google Scholar]
- 42. Huang PI, Chen YC, Chen LH, Juan CC, Ku HH, Wang ST, Chiou SH, Chiou GY, Chi CW, Hsu CC, Lee HC, Chen LK, Kao CL. PGC-1α mediates differentiation of mesenchymal stem cells to brown adipose cells. J Atheroscler Thromb. 2011;18(11):966–980. [DOI] [PubMed] [Google Scholar]
- 43. Morganstein DL, Wu P, Mane MR, Fisk NM, White R, Parker MG. Human fetal mesenchymal stem cells differentiate into brown and white adipocytes: a role for ERRα in human UCP1 expression. Cell Res. 2010;20(4):434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014;157(1):255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Janesick AS, Dimastrogiovanni G, Vanek L, Boulos C, Chamorro-García R, Tang W, Blumberg B. On the utility of ToxCast™ and ToxPi as methods for edentifying new obesogens. Environ Health Perspect. 2016;124(8):1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, Seale P, Gupta RK. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab. 2016;23(6):1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamamoto N, Ueda-Wakagi M, Sato T, Kawasaki K, Sawada K, Kawabata K, Akagawa M, Ashida H.. Measurement of glucose uptake in cultured cells. Curr Protoc Pharmacol. 2015;71:12.141–26. [DOI] [PubMed] [Google Scholar]
- 48. RRID:AB_329825.
- 49. RRID:AB_1147620.
- 50. RRID:AB_10167668.
- 51. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. 3rd ed. Boston, MA: McGraw-Hill; 2003.
- 55. Scott MA, Nguyen VT, Levi B, James AW. Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2011;20(10):1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li X, Pham HT, Janesick AS, Blumberg B. Triflumizole is an obesogen in mice that acts through peroxisome proliferator activated receptor gamma (PPARγ). Environ Health Perspect. 2012;120(12):1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chamorro-García R, Kirchner S, Li X, Janesick A, Casey SC, Chow C, Blumberg B. Bisphenol A diglycidyl ether induces adipogenic differentiation of multipotent stromal stem cells through a peroxisome proliferator-activated receptor γ-independent mechanism. Environ Health Perspect. 2012;120(7):984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vuligonda V, Thacher SM, Chandraratna RA. Enantioselective syntheses of potent retinoid X receptor ligands: differential biological activities of individual antipodes. J Med Chem. 2001;44(14):2298–2303. [DOI] [PubMed] [Google Scholar]
- 59. Rydén M, Hrydziuszko O, Mileti E, Raman A, Bornholdt J, Boyd M, Toft E, Qvist V, Näslund E, Thorell A, Andersson DP, Dahlman I, Gao H, Sandelin A, Daub CO, Arner P. The adipose transcriptional response to insulin is determined by obesity, not insulin sensitivity. Cell Reports. 2016;16(9):2317–2326. [DOI] [PubMed] [Google Scholar]
- 60. Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-Lα. Mol Cell Biol. 2007;27(13):4698–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors. 2011;29(5):196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mariman EC, Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell Mol Life Sci. 2010;67(8):1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. [DOI] [PubMed] [Google Scholar]
- 64. Schreiber R, Diwoky C, Schoiswohl G, Feiler U, Wongsiriroj N, Abdellatif M, Kolb D, Hoeks J, Kershaw EE, Sedej S, Schrauwen P, Haemmerle G, Zechner R.. Cold-induced thermogenesis depends on ATGL-mediated lipolysis in cardiac muscle, but not brown adipose tissue. Cell Metab. 2017;26(5):753–763.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berry DC, Jiang Y, Arpke RW, Close EL, Uchida A, Reading D, Berglund ED, Kyba M, Graff JM. Cellular aging contributes to failure of cold-induced beige adipocyte formation in old mice and humans [published correction appears in Cell Metab. 2017;25(2):481]. Cell Metab. 2017;25(1):166–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vishvanath L, MacPherson KA, Hepler C, Wang QA, Shao M, Spurgin SB, Wang MY, Kusminski CM, Morley TS, Gupta RK. Pdgfrβ+ mural preadipocytes contribute to adipocyte hyperplasia induced by high-fat-diet feeding and prolonged cold exposure in adult mice. Cell Metab. 2016;23(2):350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135(2):240–249. [DOI] [PubMed] [Google Scholar]
- 68. Kissig M, Ishibashi J, Harms MJ, Lim HW, Stine RR, Won KJ, Seale P. PRDM16 represses the type I interferon response in adipocytes to promote mitochondrial and thermogenic programing. EMBO J. 2017;36(11):1528–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moisan A, Lee YK, Zhang JD, Hudak CS, Meyer CA, Prummer M, Zoffmann S, Truong HH, Ebeling M, Kiialainen A, Gérard R, Xia F, Schinzel RT, Amrein KE, Cowan CA. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat Cell Biol. 2015;17(1):57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Janesick A, Blumberg B. Obesogens, stem cells and the developmental programming of obesity. Int J Androl. 2012;35(3):437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Näslund E, Britton T, Concha H, Hassan M, Rydén M, Frisén J, Arner P. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. [DOI] [PubMed] [Google Scholar]
- 72. Aprile M, Ambrosio MR, D'Esposito V, Beguinot F, Formisano P, Costa V, Ciccodicola A. PPARG in human adipogenesis: differential contribution of canonical transcripts and dominant negative isoforms. PPAR Res. 2014;2014:537865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pereira-Fernandes A, Vanparys C, Vergauwen L, Knapen D, Jorens PG, Blust R. Toxicogenomics in the 3T3-L1 cell line, a new approach for screening of obesogenic compounds. Toxicol Sci. 2014;140(2):352–363. [DOI] [PubMed] [Google Scholar]
- 74. Watt J, Schlezinger JJ. Structurally-diverse, PPARγ-activating environmental toxicants induce adipogenesis and suppress osteogenesis in bone marrow mesenchymal stromal cells. Toxicology. 2015;331:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nadal A, Quesada I, Tudurí E, Nogueiras R, Alonso-Magdalena P. Endocrine-disrupting chemicals and the regulation of energy balance. Nat Rev Endocrinol. 2017;13(9):536–546. [DOI] [PubMed] [Google Scholar]
- 76. Regnier SM, Sargis RM. Adipocytes under assault: environmental disruption of adipose physiology. Biochim Biophys Acta. 2014;1842(3):520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Valentino R, D’Esposito V, Passaretti F, Liotti A, Cabaro S, Longo M, Perruolo G, Oriente F, Beguinot F, Formisano P. Bisphenol-A impairs insulin action and up-regulates inflammatory pathways in human subcutaneous adipocytes and 3T3-L1 cells. PLoS One. 2013;8(12):e82099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hugo ER, Brandebourg TD, Woo JG, Loftus J, Alexander JW, Ben-Jonathan N. Bisphenol A at environmentally relevant doses inhibits adiponectin release from human adipose tissue explants and adipocytes. Environ Health Perspect. 2008;116(12):1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Steinmetz R, Brown NG, Allen DL, Bigsby RM, Ben-Jonathan N. The environmental estrogen bisphenol A stimulates prolactin release in vitro and in vivo. Endocrinology. 1997;138(5):1780–1786. [DOI] [PubMed] [Google Scholar]
- 80. Dicker A, Le Blanc K, Aström G, van Harmelen V, Götherström C, Blomqvist L, Arner P, Rydén M. Functional studies of mesenchymal stem cells derived from adult human adipose tissue. Exp Cell Res. 2005;308(2):283–290. [DOI] [PubMed] [Google Scholar]
- 81. Rydén M, Dicker A, Götherström C, Aström G, Tammik C, Arner P, Le Blanc K. Functional characterization of human mesenchymal stem cell-derived adipocytes. Biochem Biophys Res Commun. 2003;311(2):391–397. [DOI] [PubMed] [Google Scholar]
- 82. Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, Botbol Y, Ambrosini M, Fradet M, Rouault C, Hénégar C, Hulot JS, Poitou C, Torcivia A, Nail-Barthelemy R, Bichet JC, Gautier EL, Clément K. A PDGFRα-mediated switch toward CD9high adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metab. 2017;25(3):673–685. [DOI] [PubMed] [Google Scholar]
- 83. AlSudais H, Aabed K, Nicola W, Dixon K, Chen J, Li Q. Retinoid X receptor-selective signaling in the regulation of Akt/protein kinase B isoform-specific expression. J Biol Chem. 2016;291(6):3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Geng S, Zhou S, Bi Z, Glowacki J. Vitamin D metabolism in human bone marrow stromal (mesenchymal stem) cells. Metabolism. 2013;62(6):768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wan DC, Shi YY, Nacamuli RP, Quarto N, Lyons KM, Longaker MT. Osteogenic differentiation of mouse adipose-derived adult stromal cells requires retinoic acid and bone morphogenetic protein receptor type IB signaling. Proc Natl Acad Sci USA. 2006;103(33):12335–12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S, Gyuris T, Keirsse J, Gysemans C, Van Ginderachter JA, Balint BL, Evans RM, Barta E, Nagy L. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014;28(14):1562–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6(10):793–810. [DOI] [PubMed] [Google Scholar]
- 88. Huber J, Kiefer FW, Zeyda M, Ludvik B, Silberhumer GR, Prager G, Zlabinger GJ, Stulnig TM. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab. 2008;93(8):3215–3221. [DOI] [PubMed] [Google Scholar]
- 89. Jiao P, Chen Q, Shah S, Du J, Tao B, Tzameli I, Yan W, Xu H. Obesity-related upregulation of monocyte chemotactic factors in adipocytes: involvement of nuclear factor-κB and c-Jun NH2-terminal kinase pathways. Diabetes. 2009;58(1):104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Richard AJ, Stephens JM. The role of JAK-STAT signaling in adipose tissue function. Biochim Biophys Acta. 2014;1842(3):431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Eguchi J, Yan QW, Schones DE, Kamal M, Hsu CH, Zhang MQ, Crawford GE, Rosen ED. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7(1):86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. McGillicuddy FC, Chiquoine EH, Hinkle CC, Kim RJ, Shah R, Roche HM, Smyth EM, Reilly MP. Interferon γ attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem. 2009;284(46):31936–31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. O’Rourke RW, White AE, Metcalf MD, Winters BR, Diggs BS, Zhu X, Marks DL. Systemic inflammation and insulin sensitivity in obese IFN-γ knockout mice. Metabolism. 2012;61(8):1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kumari M, Wang X, Lantier L, Lyubetskaya A, Eguchi J, Kang S, Tenen D, Roh HC, Kong X, Kazak L, Ahmad R, Rosen ED. IRF3 promotes adipose inflammation and insulin resistance and represses browning. J Clin Invest. 2016;126(8):2839–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, Cypess AM, Xue R, Kleiner S, Kang S, Spiegelman BM, Rosen ED. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell. 2014;158(1):69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zheng F, Zhang S, Lu W, Wu F, Yin X, Yu D, Pan Q, Li H. Regulation of insulin resistance and adiponectin signaling in adipose tissue by liver X receptor activation highlights a cross-talk with PPARγ. PLoS One. 2014;9(6):e101269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Korach-André M, Archer A, Barros RP, Parini P, Gustafsson JA. Both liver-X receptor (LXR) isoforms control energy expenditure by regulating brown adipose tissue activity. Proc Natl Acad Sci USA. 2011;108(1):403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pinaire JA, Reifel-Miller A. Therapeutic potential of retinoid x receptor modulators for the treatment of the metabolic syndrome. PPAR Res. 2007;2007:94156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li X, Hansen PA, Xi L, Chandraratna RA, Burant CF. Distinct mechanisms of glucose lowering by specific agonists for peroxisomal proliferator activated receptor gamma and retinoic acid X receptors. J Biol Chem. 2005;280(46):38317–38327. [DOI] [PubMed] [Google Scholar]
- 100. Ogilvie KM, Saladin R, Nagy TR, Urcan MS, Heyman RA, Leibowitz MD. Activation of the retinoid X receptor suppresses appetite in the rat. Endocrinology. 2004;145(2):565–573. [DOI] [PubMed] [Google Scholar]
- 101. Macchia PE, Jiang P, Yuan YD, Chandarardna RA, Weiss RE, Chassande O, Samarut J, Refetoff S, Burant CF. RXR receptor agonist suppression of thyroid function: central effects in the absence of thyroid hormone receptor. Am J Physiol Endocrinol Metab. 2002;283(2):E326–E331. [DOI] [PubMed] [Google Scholar]
- 102. Wong SF. Oral bexarotene in the treatment of cutaneous T-cell lymphoma. Ann Pharmacother. 2001;35(9):1056–1065. [DOI] [PubMed] [Google Scholar]
- 103. Mehta N, Wayne AS, Kim YH, Hale GA, Alvarado CS, Myskowski P, Jaffe ES, Busam KJ, Pulitzer M, Zwerner J, Horwitz S. Bexarotene is active against subcutaneous panniculitis-like T-cell lymphoma in adult and pediatric populations. Clin Lymphoma Myeloma Leuk. 2012;12(1):20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nie B, Nie T, Hui X, Gu P, Mao L, Li K, Yuan R, Zheng J, Wang H, Li K, Tang S, Zhang Y, Xu T, Xu A, Wu D, Ding S. Brown adipogenic reprogramming induced by a small molecule. Cell Reports. 2017;18(3):624–635. [DOI] [PubMed] [Google Scholar]
- 105. Gottardis MM, Bischoff ED, Shirley MA, Wagoner MA, Lamph WW, Heyman RA. Chemoprevention of mammary carcinoma by LGD1069 (Targretin): an RXR-selective ligand. Cancer Res. 1996;56(24):5566–5570. [PubMed] [Google Scholar]
- 106. Wang B, Fu X, Liang X, Deavila JM, Wang Z, Zhao L, Tian Q, Zhao J, Gomez NA, Trombetta SC, Zhu MJ, Du M. Retinoic acid induces white adipose tissue browning by increasing adipose vascularity and inducing beige adipogenesis of PDGFRα+ adipose progenitors. Cell Discov. 2017;3:17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kiefer FW, Vernochet C, O’Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, Plutzky J. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012;18(6):918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Alvarez R, de Andrés J, Yubero P, Viñas O, Mampel T, Iglesias R, Giralt M, Villarroya F. A novel regulatory pathway of brown fat thermogenesis. Retinoic acid is a transcriptional activator of the mitochondrial uncoupling protein gene. J Biol Chem. 1995;270(10):5666–5673. [DOI] [PubMed] [Google Scholar]
- 109. Bowers RR, Temkin AM, Guillette LJ, Baatz JE, Spyropoulos DD. The commonly used nonionic surfactant Span 80 has RXRα transactivation activity, which likely increases the obesogenic potential of oil dispersants and food emulsifiers. Gen Comp Endocrinol. 2016;238:61–68. [DOI] [PubMed] [Google Scholar]
- 110. Attina TM, Hauser R, Sathyanarayana S, Hunt PA, Bourguignon JP, Myers JP, DiGangi J, Zoeller RT, Trasande L. Exposure to endocrine-disrupting chemicals in the USA: a population-based disease burden and cost analysis. Lancet Diabetes Endocrinol. 2016;4(12):996–1003. [DOI] [PubMed] [Google Scholar]
- 111. Trasande L, Zoeller RT, Hass U, Kortenkamp A, Grandjean P, Myers JP, DiGangi J, Bellanger M, Hauser R, Legler J, Skakkebaek NE, Heindel JJ. Estimating burden and disease costs of exposure to endocrine-disrupting chemicals in the European union. J Clin Endocrinol Metab. 2015;100(4):1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Legler J, Fletcher T, Govarts E, Porta M, Blumberg B, Heindel JJ, Trasande L. Obesity, diabetes, and associated costs of exposure to endocrine-disrupting chemicals in the European Union. J Clin Endocrinol Metab. 2015;100(4):1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.