Abstract
The potential for cardiotoxicity is carefully evaluated for pharmaceuticals, as it is a major safety liability. However, environmental chemicals are seldom tested for their cardiotoxic potential. Moreover, there is a large variability in both baseline and drug-induced cardiovascular risk in humans, but data are lacking on the degree to which susceptibility to chemically-induced cardiotoxicity may also vary. Human induced pluripotent stem cell (iPSC)-derived cardiomyocytes have become an important in vitro model for drug screening. Thus, we hypothesized that a population-based model of iPSC-derived cardiomyocytes from a diverse set of individuals can be used to assess potential hazard and inter-individual variability in chemical effects on these cells. We conducted concentration-response screening of 134 chemicals (pharmaceuticals, industrial and environmental chemicals and food constituents) in iPSC-derived cardiomyocytes from 43 individuals, comprising both sexes and diverse ancestry. We measured kinetic calcium flux and conducted high-content imaging following chemical exposure, and utilized a panel of functional and cytotoxicity parameters in concentration-response for each chemical and donor. We show reproducible inter-individual variability in both baseline and chemical-induced effects on iPSC-derived cardiomyocytes. Further, chemical-specific variability in potency and degree of population variability were quantified. This study shows the feasibility of using an organotypic population-based human in vitro model to quantitatively assess chemicals for which little cardiotoxicity information is available. Ultimately, these results advance in vitro toxicity testing methodologies by providing an innovative tool for population-based cardiotoxicity screening, contributing to the paradigm shift from traditional animal models of toxicity to in vitro toxicity testing methods.
Keywords: iPSC, In vitro, Alternative methods, Cardiotoxicity, High-content screening, Environmental chemicals
1. Introduction
Cardiovascular disease remains the leading cause of mortality and morbidity in the United States, and it is generally acknowledged that cardiovascular disease is the result of a complex interplay of genetic and environmental factors (Bhatnagar, 2017). For pharmaceuticals, the risk of cardiovascular adverse effects are assessed at all phases of drug development through a series of screening methods that include in silico, in vitro, non-clinical animal models, human clinical trials, and post-marketing surveillance (Arrigoni and Crivori, 2007; Laverty et al., 2011; Wallis et al., 2018). In addition, it is well established that people may vary greatly in their susceptibility to cardiovascular effects of drugs, and there has been considerable interest in using patient-specific induced pluripotent stem cell (iPSC)-derived in vitro cardiomyocytes with the promise of personalized drug safety evaluation (Yang et al., 2015; Sharma et al., 2017; Takasuna et al., 2017; Grimm et al., 2018).
Conversely, there is no routine testing for potential cardiovascular adverse effects of non-pharmaceutical chemicals, even though the potential for environmental chemicals to affect beating parameters of human iPSC-derived cardiomyocytes has been recently demonstrated (Sirenko et al., 2017). The evidence for the environmental contributions to cardiovascular disease is largely from epidemiological studies, most notably on air pollution, in which exposures and effects are already occurring in the population (Pruss-Ustun and Corvalan, 2006; Sacks et al., 2011). The extent to which the chemicals in commerce and the environment contribute to human cardiovascular disease morbidity is essentially unknown. Given the large number of chemicals in commerce and the expense of established in vivo animal models for cardiovascular effects (e.g., beagle dogs), in vitro screening is a sensible alternative approach to address this gap.
Human iPSC-derived cardiomyocytes have emerged as an attractive organotypic in vitro model for cardiotoxicity testing because they demonstrate biological relevance, high sensitivity and specificity, and consistent reproducibility (Burridge et al., 2016; Blinova et al., 2017; Kawatou et al., 2017; Sharma et al., 2017; Strauss et al., 2017). This human organotypic in vitro model has also been shown to replicate inter-individual variability in sensitivity to cardioactive and cardiotoxic drugs, thus offering a possibility for studies of the interaction of population genetics and chemical effects (Kilpinen et al., 2017; Panopoulos et al., 2017; Grimm et al., 2018). Therefore, we hypothesized that genetic variability across iPSC-derived cardiomyocytes can be used to characterize inter-individual variability in responses to chemicals. Moreover, we posit that this model can be used for population-based in vitro screening on environmental chemicals, being an organotypic model with more direct phenotypic correspondence to in vivo effects than the only other available population-based screening model, human lymphoblast cell lines (Abdo et al., 2015; Chiu et al., 2017).
Indeed, we have recently demonstrated (i) the feasibility of using a human iPSC-derived cardiomyocyte model to identify cardiotoxicity hazards of environmental chemicals (Sirenko et al., 2013b; Sirenko et al., 2017); (ii) the reproducibility of inter-individual variability in baseline and drug-induced phenotypes when using iPSC-derived cardiomyocytes from multiple donors (Grimm et al., 2018); and (iii) the qualitative and quantitative accuracy of this population-based in vitro cardiotoxicity testing model for predicting the concentration-response relationships for known pro-arrhythmic drugs (Blanchette et al., 2019). In this study, we simultaneously evaluated cardiotoxicity hazard, concentration-response, and population variability of over 130 chemicals using high-throughput screening in a population-based organotypic iPSC-derived cardiomyocyte model from over 40 human donors, considerably extending existing body of knowledge by demonstrating the utility of this in vitro human cell-based model to screening of a large number of drugs and chemicals.
2. Materials and methods
2.1. Chemicals and biologicals
Cardiomyocyte plating and maintenance media were obtained from Cellular Dynamics International (CDI, Madison, WI). Tissue-culture treated 384-well microplates were obtained from Corning (Product # 3764, Kennebunk, ME). Gelatin from porcine skin (CAS: 9000-70-8), penicillin/streptomycin (Product # P4333), tissue culture grade dimethyl sulfoxide (DMSO, CAS: 67–68–5), and cisapride monohydrate (CAS: 260779-88-2) were obtained from MilliporeSigma (St. Louis, MO). Trypan Blue 0.4% Solution (Product # 15250–061), Hoechst 33342 (Product # H3570), and MitoTracker Orange (Product # M7510) were obtained from Life Technologies (Grand Island, NY and Eugene, OR). Isoproterenol (CAS: 7683-59-2) and propranolol (CAS: 525-66-6) were included in the EarlyTox Cardiotoxicity Kit (Product # R8211) obtained from Molecular Devices (San Jose, CA). Test chemicals (N = 134, see Supplemental Table 1 for CAS, catalog number, and source) were provided for this study by the US Environmental Protection Agency, National Center for Computational Toxicology (Research Triangle Park, NC). Chemicals were selected to encompass a broad range of chemical classes, including pharmaceuticals, industrial chemicals, environmental chemicals such as pesticides and PAHs, and food constituents. Drugs of different risk classifications for Torsade de Pointes (high, intermediate, and low) listed in the Comprehensive In Vitro Pro-arrhythmia Assay (CiPA) initiative were also included in the panel of test compounds. Chemicals were divided into two plates for screening: plate 1 contained chemicals 1–68, and plate 2 contained chemicals 69–134.
2.2. iPSC-derived cardiomyocytes
De-identified iPSC-derived cardiomyocyte products from 43 human donors were obtained commercially from Fujifilm Cellular Dynamics (Madison, WI). These included the standard iCell cardiomyocyte product (Cat. No. CMC-100-010-001) and 42 distinct MyCell cardiomyocyte products (individual donor IDs and characteristics are listed in Supplemental Table 2). iPSC-derived cardiomyocytes were engineered from donor plasma samples from the repository of induced pluripotent stem cells (MyCell products) at Fujifilm Cellular Dynamics. Cardiomyocytes were derived from donors with no known cardiovascular disease or familial history of cardiovascular disease, and thus were deemed to represent “non-diseased” individuals.
2.3. Cell culture
All iPSC-derived cardiomyocytes were cultured under identical conditions in multiple batches using an established protocol as previously reported (Grimm et al., 2018; Blanchette et al., 2019). Cell screening was conducted in batches. Supplemental Table 3 lists the dates of testing and the donors tested in each batch, as well as the donors and batches for which replicate plates were tested. Briefly, tissue-culture treated 384-well microplates were gelatinized with 25 μL/well 0.1% (w/v) gelatin solution (gelatin from porcine skin in 18 MΩ sterile water) and incubated at standard conditions (37 °C and 5% CO2) for 2 h. Cells were then removed from liquid nitrogen storage and thawed in a 37 °C waterbath for 4 min. Cells were then incrementally resuspended in cardiomyocyte plating medium containing 1:500 penicillin/streptomycin solution to a final concentration of 2 × 105 cells/mL, based on manufacturer viability estimates. Live cell count and suspension density were confirmed using the trypan blue exclusion test prior to plating. Immediately prior to plating cells, gelatin solution was decanted from each plate. Cell suspension (25 μL) was then dispensed into each well of the microplates, resulting in a plating density of approximately 5000 cells/well. The microplates were allowed to rest at room temperature for 30 min to minimize edge effect prior to incubation at standard conditions for 48 h. At 48 h post-plating, 17.5 μL plating medium was exchanged with 32.5 μL cardiomyocyte maintenance medium containing 1:500 penicillin/streptomycin solution, to a final volume of 40 μL/well. Microplates were then incubated at standard conditions for 13 days post-plating, with 25 μL/well maintenance medium exchanged every 48–72 h. On day 13 post-plating, the entire volume in each well was aspirated and replaced with 25 μL maintenance medium. On day 14 post-plating, assays were performed.
2.4. Genotyping
Cells used in this study were genotyped using the Illumina (San Diego, CA) Infinium Global Screening Array-24 v2.0 Kit (48 samples, Cat. No. 20024444) as detailed in the manufacturer’s instructions.
2.5. Cardiotoxicity screening with Ca2+ flux assay
To evaluate functional effects of chemicals on cells, the Ca2+ flux assay (Early Cardiotoxicity Kit, Molecular Devices) was performed at multiple time points (baseline and up to 90 min post-chemical transfer), using an established protocol (Grimm et al., 2015; Grimm et al., 2018). Briefly, the Ca2+ flux assay was performed as follows. Ca2+ dye reagent was first prepared and equilibrated to 37 °C. Ca2+ dye reagent (25 μL) was then dispensed into each well of the microplates, resulting in a total volume of 50 μL/well, and microplates were incubated at standard conditions for 2 h. Following 2 h of equilibration with Ca2+ dye reagent, baseline intracellular Ca2+ flux was simultaneously recorded in all wells of the microplate using the FLIPR Tetra Cellular Screening System (Molecular Devices). Ca2+ flux was recorded every 0.125 s for 100 s (n = 800 total reads) with stage temperature = 37 °C, λexc = 470–495 nm, λem = 515–575 nm, gain = 2000, and exposure time = 0.05 s.
Following the baseline Ca2+ flux read, 12.5 μL 5× concentrated test chemicals (in 2.5% v/v DMSO and cardiomyocyte maintenance medium) were simultaneously added to all wells of the microplate using the internal liquid handler of the FLIPR Tetra. FLIPR Tetra settings were optimized to mix and then transfer test chemicals (in final concentrations of 0.1, 1, 10, or 100 μM) to the microplates at height = 40 μL, speed = 1 μL/s, and removal speed = 6 mm/s, for a total volume of 62.5 μL/well with 0.5% v/v DMSO in cardiomyocyte maintenance medium (vehicle). Additionally, multiple replicates of negative controls (vehicle-only and media-only wells) and positive controls (10 μM isoproterenol, 0.5 μM propranolol, and 0.1 μM cisapride monohydrate treatments) were included. A representative plate map is shown in Supplemental Fig. 1. Following chemical transfer, cells were incubated at standard conditions until Ca2+ flux was again simultaneously recorded in all wells of the microplate at various time intervals for up to 90 min post-chemical transfer. Data from the 90-min timepoint was analyzed further in this study, based on optimized protocols (Sirenko et al., 2017; Grimm et al., 2018). Data analysis was performed as detailed below and in (Blanchette et al., 2019). Four beating parameters were selected for further quantitative analysis (Table 1): peak frequency (beats per minute, BPM), ratio of decay time (time from top of peak to baseline) to rise time (time from baseline to top of peak), and coefficients of variation (CV) for peak amplitude and peak spacing based on our previous studies (Sirenko et al., 2017). The functional phenotypes selected for further analysis were largely non-redundant (Supplemental Fig. 2).
Table 1:
Cardiotoxicity screening phenotypes evaluated in this study.
| Phenotype | Description |
|---|---|
| Functional | |
| Peak frequency (beats per minute) | The average peak frequency, in beats per minute (BPM) |
| Decay To rise time ratio | The average ratio between the decay time and the rise time of the calcium flux (used as the in vitro surrogate for the QT interval) |
| Peak amplitude CV% | The coefficient of variation between the peak amplitudes |
| Peak spacing CV% | The coefficient of variation between the spacing of the peaks (used as a measure of irregular beating) |
| Cytotoxicity | |
| Total cells | Total number of nuclei (cell count) |
| All nuclei mean integrated intensity | The total pixel intensity of the nuclear stain over the nuclear area, divided by the total number of cells |
| Positive W2 mean stain area | The average stained area (nucleus, cymiddlelasm, or both as defined in the W2 settings) for all cells scored as positive for W2 (in μm2) |
| Positive W2 mean stain Integrated intensity | The total pixel intensity of the W2 stain over the stained area in W2 positive cells, divided by the number of cells positive for W2 |
| Positive W2 mean stain area/total cells | The average stained area (nucleus, cymiddlelasm, or both as defined in the W2 settings) for all cells scored as positive for W2, divided by the total number of cells |
2.6. Cardiotoxicity screening using high-content cell imaging
Cell viability-related phenotypes was quantitatively assessed by high-content imaging after conclusion of the Ca2+ flux measurements at 90 min post chemical exposure using an established protocol in the ImageXpress Micro Confocal Cellular Imaging System (Molecular Devices) as described previously (Grimm et al., 2015; Sirenko et al., 2017; Grimm et al., 2018). First, the total volume of cell culture medium containing Ca2+ dye was aspirated and replaced with 25 μL/well pre-warmed fluorescent staining solution (2.2 μg/mL Hoechst 33342 and 0.2 μM MitoTracker Orange in cardiomyocyte maintenance medium with 1:500 penicillin/streptomycin). Microplates were then incubated at standard conditions for 15 min. The staining solution was then aspirated and washed with 25 μL/well pre-warmed cardiomyocyte maintenance medium before proceeding to image acquisition. Images of each well at 10× magnification were acquired in succession using DAPI, TRITC, and FITC filters for Hoechst 33342, MitoTracker Orange, and Ca2+ dye, respectively. Image processing and quantification was performed using the multi-wavelength cell scoring module in the instrument-specific MetaXpress software package (Molecular Devices). Five parameters were selected for further quantitative analysis (Table 1): total cell count, nuclei mean integrated intensity, and mitochondrial integrity descriptors mitochondrial mean stain area, mitochondrial mean stain area divided by total cell count, and mitochondrial mean stain integrated intensity (Sirenko et al., 2017). The cytotoxicity phenotypes selected for further analysis were largely non-redundant (Supplemental Fig. 2).
2.7. Image processing and concentration-response analysis
Ca2+ flux data were processed and quantified using a custom script (Blanchette et al., 2019) in R Statistical Computing Software (R version3.5.1). Raw data were used for concentration-response modeling and derivation of the points of departure (POD) as previously described (Sirenko et al., 2017). Data were normalized to the phenotype-specific vehicle control (0.5% DMSO) mean prior to concentration-response modeling (four-parameter Hill model) for each phenotype, chemical, donor, and replicate (if available). The POD was defined as the concentration at which the concentration-response curve changed more than one standard deviation from the fit for vehicle controls.
2.8. Population variability analysis
Each of these parameters was analyzed to determine the contribution and reproducibility of inter-individual variability. For baseline inter-individual variability, data from vehicle- and media-only controls, in addition to replicate plates, were analyzed using a robust linear mixed effects model to evaluate the contribution of biological (i.e., donor) and technical (e.g., inter-plate, and media vs. vehicle) variability to overall variability, using the rlmer function from the R package robustlmm (Koller, 2016). The robust linear mixed effects model was used because the residuals from the usual linear mixed effects model had substantial outliers and were not normally distributed. We characterized reproducibility with treatment through intra-plate analyses of response to positive control compounds present on all plates and inter-plate analyses of 13 cell lines in which replicates were performed. Additionally, we characterized treatment-related inter-individual and inter-chemical variation. We also separately analyzed whether sex and/or race was a modifier of treatment-related effects using a linear mixed effects model, utilizing the lmer function from the R package lme4 (Bates et al., 2015).
2.9. Genome-wide association scanning (GWAS)
PLINK (Slifer, 2018) (v1.90b6.9 64-bit (4 Mar 2019)) was used to recode the 436,870 SNPs to counts of minor alleles (0, 1, 2) and to select a subset of 6965 SNPs for ancestry control with minor allele frequency (MAF) > 0.45. We required high MAF for ancestry-control SNPs due to the fact that we had a limited sample size in comparison to many GWAS studies. A five-level ancestry control factor variable was computed by comparison with 1000 Genomes data as described below, under the rationale that using this approach (as opposed to principal component control) would enable effective control without losing an excessive number of degrees of freedom for association testing. For association testing, the SNPs were filtered for MAF > 0.10 across the 41 individual cell lines for which genotyping data were available. For robust testing, each phenotype was subjected to a rank-inverse normal transformation, producing normally distributed phenotypes, except for ties in the data due to POD values censored at the maximum concentration. The resulting 177,908 SNPs were analyzed using linear regression for the transformed phenotypes for each of the 1242 chemical/phenotype combinations, with ancestry and sex as covariates. Thus, each chemical/phenotype was subjected to a Bonferroni threshold p-value = 2.81e-7 to control at alpha = 0.05, and the experiment wide threshold p-value = 2.26e-10 (for 220 million tests).
3. Results
3.1. A population-based iPSC-derived cardiomyocyte in vitro model
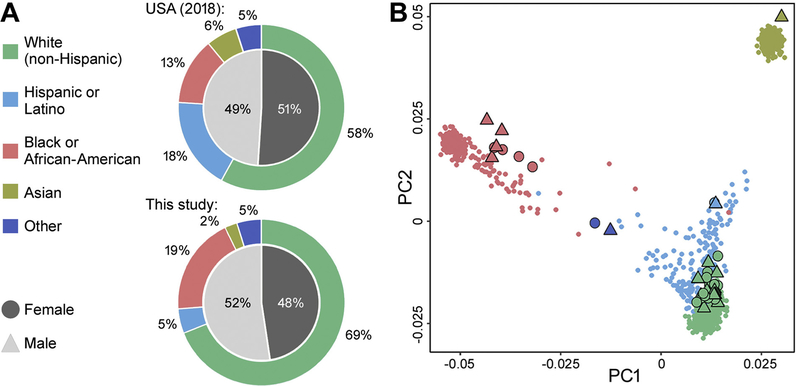
This study utilized iPSC-derived cardiomyocytes from 43 donors without known personal or familial history of cardiovascular disease (Supplemental Table 2). These cells are commercially available from FujiFilm Cellular Dynamics and represent a model that is publicly available and accessible. These cells passed stringent quality control procedures established by the manufacturer; only a fraction (~20%) of the iPSC clones from non-diseased human donors could be successfully differentiated into cardiomyocyte-like cells by the manufacturer (B. Anson, personal communication). To confirm ancestry and sex of each cell, Illumina Infinium Human Global Screening Array-based geno-typing was performed on 41 donors for which cell lysates were available for genotyping. Fig. 1A illustrates the distribution of population characteristics of the cells used in this study as compared to the ancestry/sex distribution in the USA in 2018. Fig. 1B shows a display of principal components for the donors used in this study overlaid with those from the 1000 Genomes/HapMap (Genomes Project et al., 2015) populations, labelled by population source. The donors used in this study included individuals of White (non-Hispanic) (69%), Black or African-American (19%), Hispanic or Latino (5%), and Asian (2%) ancestry, assigned by comparison with 1000 Genomes. Balanced sex distribution consisting of both male (52%) and female (48%) donors was also achieved.
Fig. 1.
Distribution of population characteristics for the donor pool of 41 iPSC-derived cardiomyocytes. (A) Distribution of population characteristics in the USA in 2018 and in this study, in terms of ancestry (outer ring) and sex (inner circle). (B) Scatter plot of the 1st and 2nd principal components for genotypes across donors, with colors and shapes representing the different ancestral backgrounds and sexes, respectively, presented in (A). Large symbols represent principal components for our population of 41 donors, and small symbols represent principal components for ~1000 human lymphoblastoid cell lines from the 1000 Genomes project.
3.2. Donor- and chemical-specific responses in the in vitro population-based cardiotoxicity model
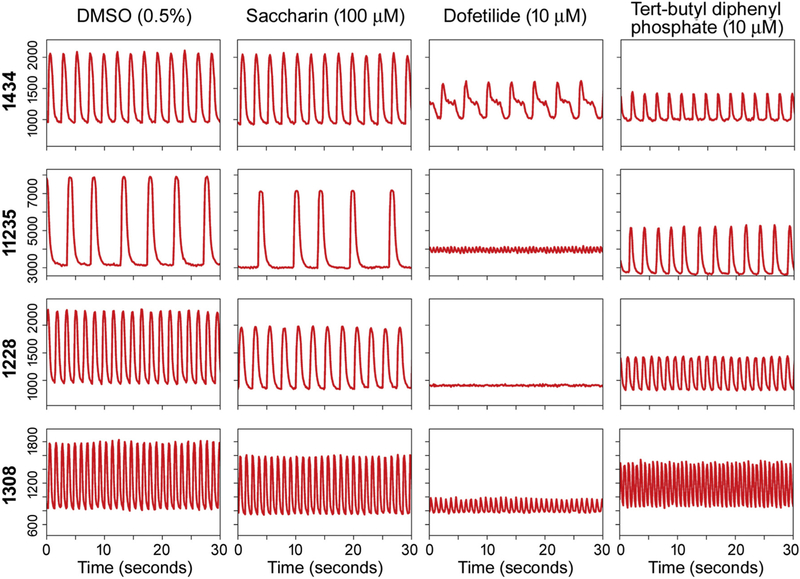
Kinetic changes in Ca2+ flux was used as a main readout for evaluating functional effects of the chemicals screened in this study (Sirenko et al., 2013b; Grimm et al., 2015). All test compounds were evaluated in a concentration-response (0.1, 1, 10, and 100 μM). Fig. 2 shows representative Ca2+ flux traces of four individual donors, both at baseline (vehicle, 0.5% DMSO) and upon treatment. Donor 1434 is the “standard” iPSC-derived cardiomyocyte FujiFilm Cellular Dynamics product that is widely used. Three other donors were selected to represent a range of baseline beating parameters. Treatment with 100 μM saccharin, a food constituent, generally did not affect peak frequency across donors. Treatment with 10 μM dofetilide, a class III antiar-rhythmic pharmaceutical, induced donor-specific cardiotoxic effects, such as formation of the in vitro “notch” phenotype associated with clinical QT-prolongation (donor 1434) and rapid beating with significantly decreased peak amplitude (donors 11,235, 1228, and 1308), an effect that is indicative of torsade de pointes (TdP), a type of arrhythmia that can result in sudden cardiac death. Treatment with 10 μM tert-butylphenyl diphenyl phosphate, a flame retardant, increased peak frequency in a number of donors. Importantly, despite differences in the spontaneous beat rate among cell lines, effects were observed upon treatment were similar to the observations reported previously (Grimm et al., 2018).
Fig. 2.
Representative Ca2= flux traces for iPSC-derived cardiomyocytes derived from 4 individual donors. Rows represent 4 individual donors, and columns represent different chemical treatments. The first column represents a vehicle control well (DMSO, 0.5%). The second column represents treatment with saccharin, a food constituent (100 μM). The third column represents treatment with dofetilide, a class III antiarrhythmic pharmaceutical (10 μM). The last column represents treatment with tert-butylphenyl diphenyl phosphate, a flame retardant (10 μM). The X-axis of each Ca2+ flux trace represents time in seconds, and the Y-axis represents relative fluorescence units (RFU) of the Ca2+ flux measured within each donor.
3.3. Basal and chemical-induced effects on iPSC-derived cardiomyocytes from multiple donors are highly reproducible
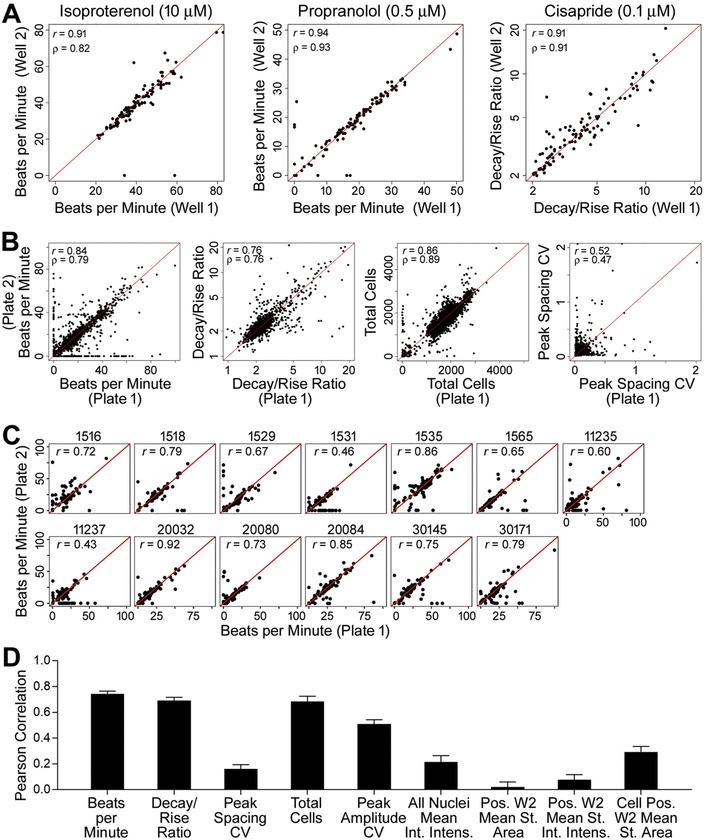
Our previous study of a 27 human iPSC-derived cardiomyocytes demonstrated high reproducibility of basal beating parameters and effects of three classic cardio-active drugs (Grimm et al., 2018). Here, a larger population of cell lines (i.e., individuals) and a far larger number of chemicals were tested. Our study design allowed for the evaluation of both intra- and inter-plate reproducibility. Fig. 3A demonstrates high intra-plate replicability across donors following treatment with the known cardio-active compounds isoproterenol, propranolol, and cisapride. For isoproterenol (β-adrenergic receptor agonist) and propranolol (β-adrenergic receptor antagonist), beats per minute (BPM) phenotype is shown as these compounds are known to affect the beat rate. For cisapride, a serotonin 5-HT₄ receptor agonist known to induce clinical prolongation of the QT interval, decay/rise ratio was the phenotype of interest, as this parameter is the in vitro surrogate for QT prolongation (Blanchette et al., 2019). Intra-plate correlation, was extremely high (> 0.90) as evidenced by both Pearson (r) and Spearman (ρ) coefficients.
Fig. 3.
High intra- and inter-plate reproducibility in treatment-related effects. (A) Intra-plate reproducibility (replicate treatment wells on the same plate) across all donors following treatment with control compounds isoproterenol (10 μM), propranolol (0.5 μM), and cisapride (0.1 μM), for the chemical-specific phenotype of interest. Pearson (r) and Spearman (ρ) correlation coefficients are shown in the upper left corner of each graph. (B) Inter-plate reproducibility (corresponding wells on replicate plates for the same donor, concentration, and cardiotoxicity parameter) following treatment with test chemicals for four main phenotypes, across 13 donors with replicate plates. Pearson (r) and Spearman (ρ) correlation coefficients are shown. (C) Inter-plate reproducibility following treatment with test chemicals for the phenotype beats per minute (BPM), shown individually for each donor with replicate plates. Pearson (r) correlation coefficients are shown. (D) Inter-batch reproducibility (all pairwise comparisons across 8 batches of the standard donor ID 1434), summarized by Pearson correlation coefficients, for all 9 phenotypes.
We also examined inter-plate reproducibility by comparing concordance of the readouts in each well across duplicate plates. For thirteen cell lines, replicate plates were available for analysis, and the outcome of concordance analysis of the wells where test chemicals were added is shown in Fig. 3B. High concordance was observed for both functional (e.g., beats per minute and decay/rise ratio) and viability (e.g., total cells) phenotypes where chemical effects were observed. For the phenotypes that were largely unaffected by treatments (e.g., peak spacing CV), the low correlation was consistent with a lack of the effect on both plates (i.e., comparisons between plates are consistent with intra-plate variability). High correlations for phenotypes affected by treatment were also observed for the replicate plates of the individual donors (Fig. 3C). In addition, experiments on the “standard” donor 1434 cells were repeated in a number of batches and data were analyzed to examine the concordance of the results when the experiment is repeated on different days. Fig. 3D shows the average correlation coefficients for all phenotypes for all pair-wise comparisons among batches. Similar to our findings with inter-plate variability, we find high correlations for the functional and viability phenotypes that are affected by treatment. Specifically, beats per minute, decay/rise ratio and total cells were highly correlated across batches, with a median Pearson’s r of 0.77, 0.71, and 0.78, respectively. Supplemental Fig. 2 shows pairwise correlations for all phenotypes and batches.
3.4. iPSC-derived cardiomyocytes from multiple donors exhibit reproducible inter-individual variability in baseline phenotypes
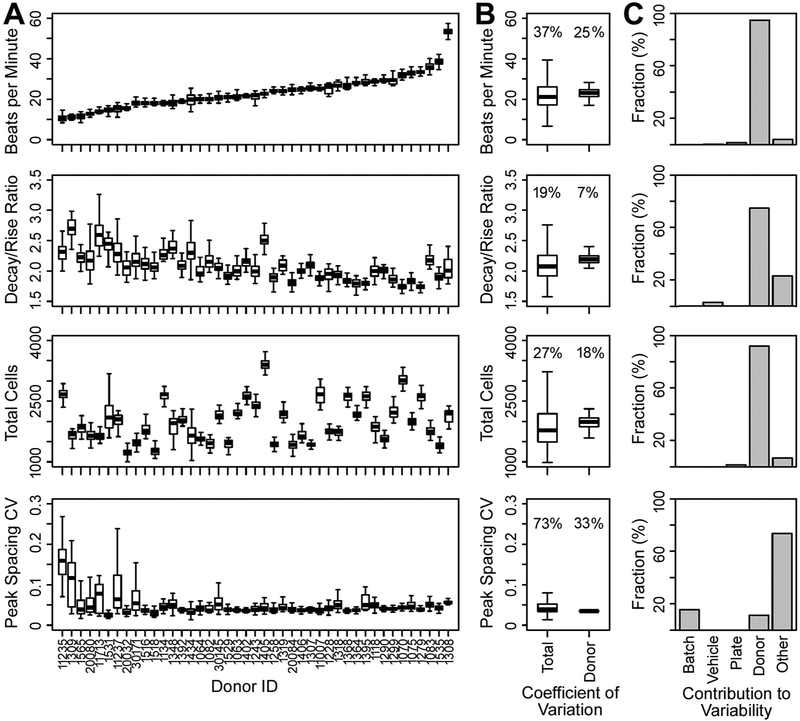
Inter-individual variability in baseline (examined from vehicle control – 0.5% DMSO) parameters was examined across the entire compendium of 43 iPSC-derived cardiomyocyte cell lines (Fig. 4A and Supplemental Fig. 4). The spontaneous beating of iPSC-derived cardiomyocytes was highly regular for most cell lines, with the coefficient of variation for peak spacing, a phenotype indicative of beating monotonicity, being < 7% in most cases. For the beats per minute phenotype, median values across donors ranged reproducibly from 10 to 54. For decay/rise ratio, median values across donors ranged reproducibly from 1.8 to 2.7. Data collected in these experiments were used to conduct multivariate analysis of variance to determine the contribution of the major factors, such as batch, plate, treatment (difference between untreated and DMSO-treated wells), donor and other (Fig. 4B and C). We found that inter-individual (donor to donor) variability was the primary contributor to total observed variability for most phenotypes, exceeding 80% of the total variance for many phenotypes. Batch, plate, and vehicle contributions were low or non-existent, except for phenotypes such as peak spacing CV where some individual donors had high intra-individual variation.
Fig. 4.
iPSC-derived cardiomyocytes exhibit cell line (i.e., inter-individual) variability in baseline beating parameters, largely driven by donor. (A) Boxplots showing baseline values for the phenotypes beats per minute, decay/rise ratio, total cells, and peak spacing CV for each donor, ordered by increasing baseline peak frequency (beats per minute). Boxplots for each donor are comprised of the median, interquartile range (IQR), and 95% confidence interval (CI) values. (B) Boxplots showing the distribution of the coefficient of variation for total and donor contributions to the total variability of each phenotype, based on robust linear mixed effects modeling. Total contribution accounts for technical and biological (i.e., donor) sources of variability, whereas donor variability accounts only for biological variability. (C) Histograms showing the fraction of technical (batch, plate, vehicle, other) and biological (donor) contributions to the total variability of each phenotype, based on robust linear mixed effects modeling. Sources of variability considered account for batch (screening batch dates) effects, plate (inter-plate) effects, vehicle (0.5% DMSO vehicle vs. cell culture media) effects, donor (biological) effects, and other residual contributions.
3.5. Population variability in chemical-specific effects on iPSC-derived cardiomyocytes
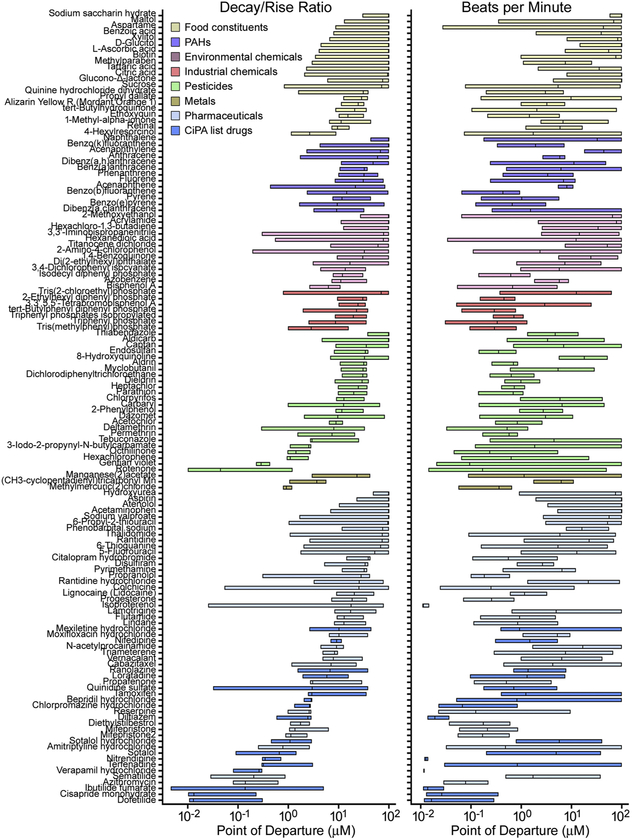
We next examined variability in the effects of all screened chemicals across the entire population of iPSC-derived cardiomyocytes (Fig. 5). Concentration-response modeling was conducted to derive quantitative estimates of the effects of each chemical in each cell line. We derived point-of-departure (POD) values and plotted the inter-quartile range in population variability. Compounds were grouped by chemical class and then sorted by the median POD across the population. The degree of population variability is indicated by spread of the POD interquartile range, with a smaller POD range representing lower population variability. For example, for the phenotype decay/rise ratio, dofetilide (a class III antiarrhythmic pharmaceutical) and cisapride (a serotonin 5-HT4 receptor agonist) exhibited high potency in affecting this phenotype (i.e., low median POD values across donors, log(POD) = −1.89 and − 1.88 μM, respectively). In contrast, hydroxyurea and thiabendazole, drugs without known cardiotoxicity liability, and sodium saccharin hydrate, a food constituent, were largely without effect (i.e., high median POD values across donors and relatively lower population variability). Other chemicals exhibited varied POD values. Generally, compounds with Comprehensive in vitro Pro-arrhythmia Assay (CiPA) classifications fell at the lower end of the POD range for decay/rise ratio, with compounds of higher relative risk classifications exhibiting higher population variability. Food constituents generally exhibited high median POD values and pharmaceuticals generally exhibited lower median POD values. Environmental chemicals (plasticizers, flame retardants, PAHs) varied in both potency of effect and degree of population variability.
Fig. 5.
Chemical-specific variability in potency and degree of population variability for decay/rise ratio and beats per minute. Box plots show median and inter-quartile range of the point of departure values across 43 donors, sorted within chemical class. Chemical names are along with left axis, and different colored boxes represent chemical classes as shown in the inset.
For the beats per minute phenotype, chemicals also exhibited a range of potency values and varied degrees of population variability, with compounds like isoproterenol, a β-adrenergic receptor agonist, exhibiting low median POD (log(POD) = −1.95μM) and low population variability. Again, food constituents generally exhibited high median POD values and environmental chemicals varied in both potency and degree of population variability.
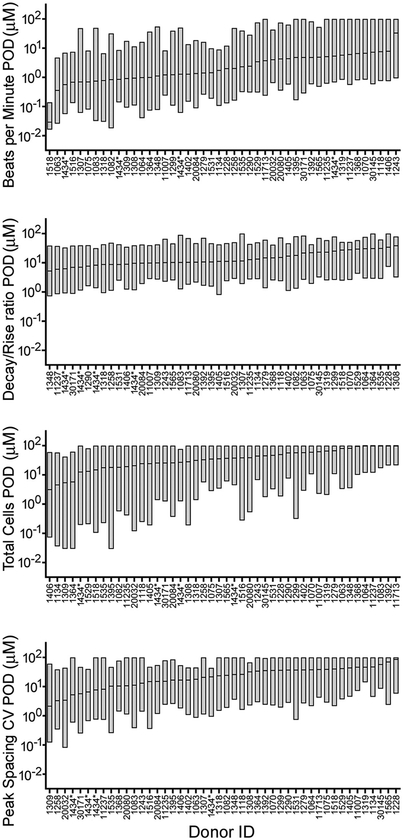
3.6. iPSC-derived cardiomyocyte cell line-specific variability in chemical-specific effects
We determined whether certain cell lines (i.e., donors) were more or less susceptible to the effects of all chemicals tested in this study. Variability in treatment-related effects on each iPSC-derived cardiomyocyte cell line, for each phenotype, is shown in Fig. 6 and Supplemental Fig. 5. Cell lines are sorted by the median POD and interquartile ranges are shown as box plots. For example, for the phenotype beats per minute, the median POD across the 134 tested chemicals was the lowest for donor 1518 (log(POD) = −1.53μM) and highest for donor 1243 (log(POD) = 1.52μM), with other donors exhibiting varied POD values within this range. Further, across 4 separate batches of standard donor 1434, the median POD value was less than an order of magnitude. For the phenotype decay/rise ratio, the median POD value across the tested chemicals was lowest for donor 1348 (log(POD) = 0.72μM) and highest for donor 1308 (log(POD = 1.59μM), but the extent of inter-individual variability in POD values was much smaller than that for beats per minute. Again, patterns across 4 separate batches of standard donor 1434 were generally identical.
Fig. 6.
iPSC-derived cardiomyocytes exhibit inter-individual variability in treatment-related points of departure (PODs) for phenotypes beats per minute, decay/rise ratio, total cells, and peak spacing CV. Box plots show median and interquartile range across 134 chemicals, and are ordered by the median POD. Donors more sensitive to chemical-induced effects are indicated by lower median POD across chemicals, and donors more resistant to effects are indicated by higher median POD.
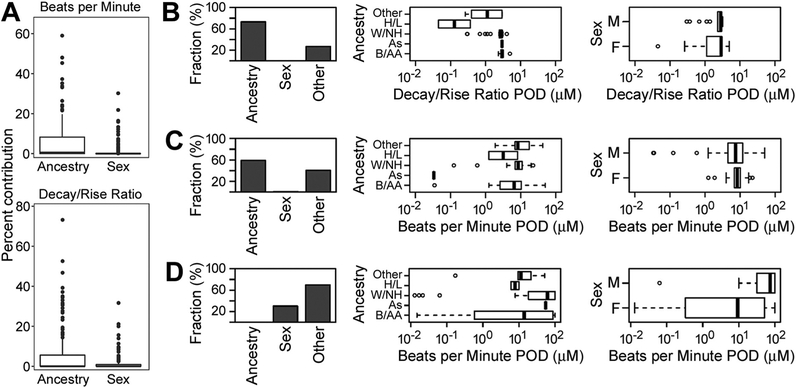
3.7. Sex- and ancestry-specific effects of chemicals on iPSC-derived cardiomyocytes
Because the population studied herein contained cells from both sexes and multiple race/ethnicity groups, we examined sex- and ancestry-specific effects of each chemical on each phenotype. For most chemicals tested, the relative contributions (regression R2) of sex and/or ancestry were low (Fig. 7 and Supplemental Fig. 6), with the contribution of ancestral origin to the overall variability in responses across cell lines being slightly higher than that of sex. Examples of several chemicals with high contribution of ancestry (bepridil and acenaphthene) and sex (titanocene) are shown in Fig. 7B–D. For the phenotype beats per minute, the ancestral contribution to variability ranged from 0 to 59%, with a median of 0%, whereas the sex contribution ranged from 0% to 30%, with a median of 0%. For the phenotype decay/rise ratio, the ancestral contribution to variability ranged from 0 to 73%, with a median of 0%, and the sex contribution ranged from 0 to 32%, with a median of 0%.
Fig. 7.
Summary of analysis of sex and ancestry effects on points of departure (PODs), based on linear mixed effect modeling. (A) Box plots showing the percent contribution of ancestry and sex across chemicals for the phenotypes Beats per Minute and Decay/Rise ratio. (B) – (D) Summary of contribution of ancestry, sex, and other to variation in PODs for three compounds: bepridil (B), acenaphthene (C), and titanocene (D). Included are the percent contributions to variations, and boxplots of PODs grouped by ancestry (Other; H/L, Hispanic/Latino; W/NH, White/non-Hispanic; As, Asian, B/AA, Black/African American), and by sex (M, male; F, female). See Supplemental Fig. 6 for complete analysis of all compounds.
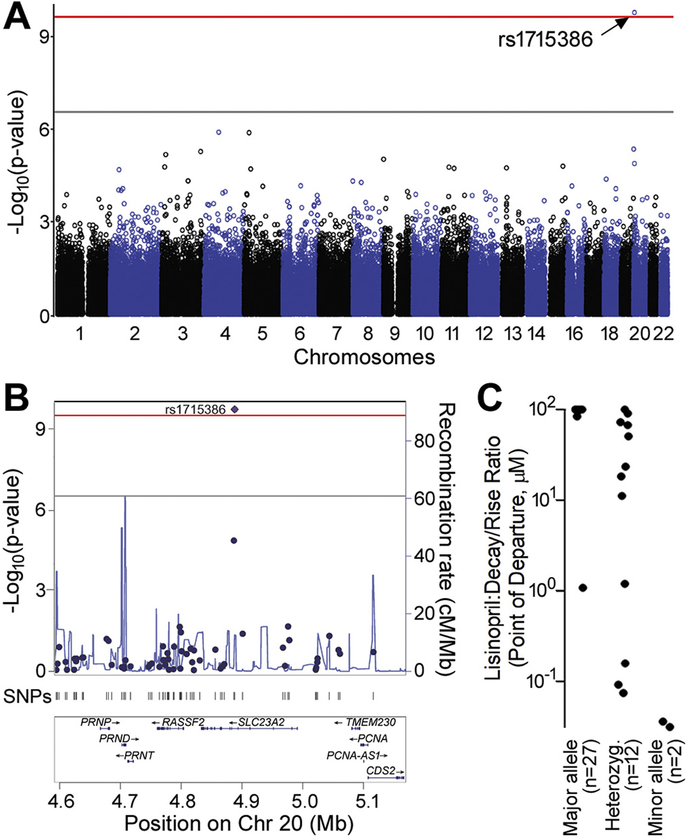
3.8. Exploring suggestive genetic contribution to the inter-individual variability in the effects of chemicals on iPSC-derived cardiomyocytes
While the number of individuals tested in this study is small, we performed exploratory genetic association mapping using low-density genotyping data, originally collected primarily to assign the ethnicity/race and sex to each cell line (Fig. 1). A noteworthy observation from this analysis is shown in Fig. 8 for lisinopril, an angiotensin-converting enzyme inhibitor used to treat life-threatening ventricular arrhythmias in patients with congestive heart failure and ventricular tachycardia, and decay/rise ratio phenotype. After correcting for the number of experiment-wide tests (177,908 SNPs × 1242 chemicals:phenotypes) and transformation to ensure robust analysis, an experiment-wide significant association was observed for sensitivity to decay/rise ratio disruptions with lisinopril and rs1715386 (p = 1.67e-10), a SNP [Chr20:4,887,900] in SLC23A2, a sodium-dependent vitamin C transporter of the SLC23 family. The distribution of allele-specific PODs for this drug/phenotype combination are shown in Fig. 8C.
Fig. 8.
Genome-wide association analysis. (A) Manhattan plot showing all analyzed SNPs and highlighting rs1715386 in SLC23A2 for associations with point of departure in the decay:rise ratio after treatment with lisinopril (adjusted for ancestry and sex) with minimum p = 2.26e-10. The grey line represents the −log10(p-value) = 0.05 significance threshold applied for a single tested phenotype. The red line represents the −log10(p-value) = 0.05 significance threshold for all ~220 million SNP × phenotype tests. (B) A locus zoom plot of the 500 kb region surrounding rs1715386 on chromosome 20. (C) Distribution of the decay/rise ratio POD of 41 iPSC lines treated with lisinopril. X-axis values for points have been slightly jittered in order to be distinguished. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we demonstrate the application of a population-based in vitro model using a panel of human iPSC-derived cardiomyocytes to test for potential cardiotoxicity hazard, dose-response, and toxicodynamic population variability of drugs and chemicals. Ca2+ flux monitoring and high-content imaging were conducted to quantitatively characterize baseline and treatment-related functional cardiomyocyte performance, as a function of 9 different phenotypes. A panel of over one hundred diverse chemicals, including pharmaceuticals, food constituents, and environmental chemicals (encompassing industrial chemicals, flame retardants, pesticides, and PAH chemical classes) were tested for treatment-related effects. No studies to date have used a population-based approach to assess the cardiotoxicity hazard and variability of environmental chemicals, and our results suggest that using a population of iPSC-derived cardiomyocytes can be a sensible approach for examining the potential for both the cardiotoxicity hazard of environmental chemicals, as well as the extent of toxicodynamic variability in these effects, if any.
Previous studies of a large number of environmental chemicals and drugs, conducted in one “standard” iPSC-derived cardiomyocyte cell line (Sirenko et al., 2017), showed that known cardio-toxic drugs show effects at concentrations comparable to blood concentrations. Even though it was found that many environmental chemicals may also affect human cardiomyocytes, effective concentrations were over 100-fold higher than expected blood concentrations from environmental exposures. The studies reported herein further extend the body of knowledge on the potential use of iPSC-derived cardiomyocytes to probe the potential cardiotoxicity of environmental chemicals, especially in the context of population variability. The panel of donors was representative of four ancestral backgrounds (European, African-American, Hispanic/Latino, and Asian) and both sexes, at distributions roughly similar to the population distribution of the United States in 2018. Moreover, the approach enabled detection of inter-individual differences in baseline beating parameters, as well as in treatment-related effects.
Importantly, we find that iPSC-derived cardiomyocytes exhibit high intra- and inter-plate reproducibility in treatment-related effects, in the individual donors and across treatment-related phenotypes. This finding corroborates previous studies demonstrating high reproducibility in treatment-related effects of control compounds in iPSC-derived cardiomyocytes (Grimm et al., 2018). Furthermore, our analysis shows that donor-attributable variability in the effects of chemicals on both functional and toxicity phenotypes is most pronounced as compared to other population/experimental factors. Collectively, these findings are noteworthy as they demonstrate that the in vitro population-based model can be used to test a large number of compounds and generate data that is comparable across batches and experiments. Because the cells used in these studies are commercially available, experimental protocols and culture conditions are standardized, and iPSC-derived cells from multiple donors are a renewable resource, this model can also be used by different laboratories to replicate responses in the same genetic backgrounds and experimental conditions.
Another key outcome of this study is the observation of chemical-specific variability in potency and degree of population variability. First, we find that the effects of known cardioactive or cardiotoxic drugs (e.g., cisapride, dofetilide, isoproterenol, etc.) corroborate the observations from studies of the “standard” donor of iPSC-derived cardiomyocytes (Sirenko et al., 2013a; Sirenko et al., 2013b). Furthermore, compounds included in the Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative (Fermini et al., 2016) were generally at the lower end of the POD range for the most relevant phenotype – the decay/rise ratio, with compounds listed with higher relative risk for torsades de pointes exhibiting wider population variability in effects. Because our goal is to address cardiac safety, our focus has been on population variation potency as expressed by a POD at a fixed magnitude of effect, rather than efficacy in terms of population variation in the magnitude of effect itself. Nonetheless, we note that there is also chemical-specific variability in the maximal effect size across donors (Supplemental Fig. 7), though the range of variability is smaller than for potency. Interestingly, potency and magnitude of effect are poorly correlated across individuals and chemicals (Supplemental Fig. 8).
Interestingly, we find that no single cell line was sensitive or resistant to the chemicals tested herein. This observation argues for the value of the population-wide approach rather than testing in one or several donor-specific cells that have been pre-selected for a particular response. Much work is done using donor-specific iPSC-derived cardiomyocytes to demonstrate the power of in vitro human iPSC-derived cardiomyocyte model in the context of personalized medicine (Burridge et al., 2016; Sayed et al., 2016; Sharma et al., 2017); however, our findings caution against the use of the selected lines to represent population variability. While the number of cell lines tested herein was relatively small, this population size is comparable to that used in human clinical trials that test for the potential liability of torsades de pointes (Vicente et al., 2018). Also, our previous study showing that an in vitro population as small as 27 individuals can achieve high concordance with the results of human clinical trials (Blanchette et al., 2019) further demonstrates the need for a population-wide approach to testing of chemicals in a compendium of human cell lines. The analysis of an optimal size of the population is needed, however, using the data reported herein to determine whether a smaller number of cell lines may be sufficient for larger screening campaigns (Chiu et al., 2017).
The diversity of donors included in this study, with respect to sex and ancestry, allowed us to examine the relative contribution of these factors on the effects of chemicals. We observed some sex differences in treatment-related prolongation of the decay/rise ratio (correlating with clinical prolongation of the QT interval), findings that are in accord with previous reports showing that females are sensitive to treatment-induced effects of some drugs (Huo et al., 2019). We found that titanocene, an organotitanium compound that is a common reagent in organometallic and organic synthesis, showed the strongest sex effect on the beat frequency. This compound has no known cardiac effects largely because it has not been tested for such effects; thus, further studies are needed to fully characterize the contribution of sex to treatment-related variability on a population level of this and other compounds. With respect to the ancestral origin as a variable, we find that a greater number of compounds showed some contribution of ethnicity/race, and to a greater degree than sex, to the observed variance. Specifically, bepridil and acenaphthene were two compounds with most pronounced overall ancestry effects on decay/rise ratio and beat frequency, respectively. Bepridil is a calcium antagonist with direct negative chronotropic, dromotropic, inotropic and vasodilatory actions that is no longer marketed in the United States. It has been associated with rate-dependent prolongation of the QTc interval and development of torsade de pointes (Hollingshead et al., 1992). Thus, our findings of its effects on decay/rise ratio and its wide inter-individual variability are concordant with its known adverse effects in some patients. With respect to acenaphthene, no studies explored its potential effects on the heart rhythm, but other PAHs are known to be arrythmogenic (Zhang et al., 2013; Brown et al., 2015).
While this study was not designed to be suitable for discovery of the genomic loci that may be associated with inter-individual differences in the effects of tested chemicals, and cardiovascular diseases are known to be complex traits that require large study populations and replication (Mendez-Giraldez et al., 2017; Bapat et al., 2018), we performed exploratory genome-wide association analysis. The significance required for such analysis in the context of this experimental design (multiple testing-corrected p-values < 10−9), is very stringent. One finding, the association between the effects of lisinopril on decay/rise ratio, did achieve genome-wide multiple testing corrected significance for a SNP in SLC23A2. While vitamin C status has been shown to affect QT parameters in patients after myocardial infarction (Bednarz et al., 2003), the significance of our observation for the association between lisinopril, QT interval and vitamin C transport may be a chance observation that needs to be interpreted with caution. We do, however, note that proximity to cellular transport genes was a common finding for significant SNPs from our much larger study (Abdo et al., 2015; Eduati et al., 2015) of chemical-induced cytotoxicity response in over 1000 lymphoblastoid cell lines.
This experimental approach and our study have a number of limitations. Though the population of 43 iPSC-derived cardiomyocyte lines is the largest of those published to date, it is still limited by the availability of differentiated cells and challenges of establishing highly functional cardiomyocytes from a large number of donors (Hausburg et al., 2017). While studies using undifferentiated human iPSCs from hundreds of individuals have been conducted, consistent differentiation of iPSC-derived cells from multiple donors currently limits the use of iPSC-derived cells from large populations (Kilpinen et al., 2017; Panopoulos et al., 2017; Schwartzentruber et al., 2018). Therefore, the ability to observe extreme sensitivities in baseline characteristics and treatment-related responses within the population is limited. Additional analyses are needed to determine whether larger or smaller populations of iPSC-derived cardiomyocytes may be needed to characterize population variability in potential cardio effects of chemicals and drugs with reasonable confidence. In addition, as we have found with the exploratory genome-wide association analysis, the small size of the in vitro population also limits the ability to conduct discovery analyses of potential genomic loci that may underpin the variability in responses across cell lines.
Another limitation is that we studied the population of “healthy” individuals with no known cardiovascular disease. Therefore, individual susceptibilities contributed by disease state(s) have not been examined in this study, though these susceptibilities may contribute extensively to toxicodynamic variability within the population (Zeise et al., 2013). Because iPSC-derived cardiomyocytes from individuals with disease states (especially cardiovascular diseases) are available (Kilpinen et al., 2017; Panopoulos et al., 2017; Schwartzentruber et al., 2018), inclusion of these cells in future studies may be informative to further address the potential for cardiotoxicity hazard and variability in a population with more susceptible individuals.
Finally, our study used a large number of test compounds, but it is still small in comparison to the number of chemicals in commerce and the environment. Even though test compounds included pharmaceuticals, food constituents, and environmental chemicals (e.g., industrial chemicals, flame retardants, plasticizers, and PAHs), there are thousands more chemicals that are lacking data on their potential to elicit cardiotoxicity. Therefore, larger studies are needed to increase the number of environmental chemicals screened in iPSC-derived cardiomyocytes. To aid future designs, additional analyses of the minimal required size of the population are needed, as these experiments are quite laborious and cannot be easily scaled.
In conclusion, we have demonstrated that a population-based in vitro model consisting of a panel of 43 iPSC-derived cardiomyocyte cell lines has utility for evaluation of cardiotoxicity hazard, dose-response, and toxicodynamic population variability of a large number of chemicals. Specifically, Ca2+ flux monitoring and high-content imaging revealed reproducible qualitative and quantitative differences in functional performance both at baseline and in treatment-related effects, in a donor- and chemical-specific manner. We showed that inter-individual variability in cardiotoxicity hazard is present across a wide range of drugs and some environmental chemicals. This model is also highly reproducible and shows that donor-specific variability far exceeds technical and other factors that must be considered in complex experimental designs. Overall, as current pre-clinical and chemical safety testing provides little information on the potential population variability in response to drugs and chemicals, this model provides a sensible strategy for improving safety evaluation of both pharmaceutical and non-pharmaceutical compounds. Ultimately, this research expands the range of in vitro testing methods by providing an innovative tool for cardiotoxicity screening of chemicals with unknown toxicological profiles and contributes to the paradigm shift from traditional animal models of toxicity to new approach methods.
Supplementary Material
Acknowledgements
The authors thank Dr. Blake Anson (FujiFilm Cellular Dynamics) for kind cooperation in developing the panel of iPSC-derived cardiomyocytes. We are grateful to Dr. Andrew Hillhouse (Texas A&M University) for processing samples for genotyping and Dr. Oksana Sirenko (Molecular Devices) for useful discussions. We wish to acknowledge Drs. Ann Richard, Keith Houck and Rusty Thomas (US EPA) for providing chemicals used in these experiments.
Sources of funding
This work was supported, in part, by grants from US EPA (STAR RD83561202) and NIH (P42 ES027704 and T32 ES026568). Fabian Grimm was the recipient of the Society of Toxicology Colgate-Palmolive and Society of Toxicology Syngenta Fellowship Awards. The views expressed in this manuscript do not reflect those of the funding agencies. The use of specific commercial products in this work does not constitute endorsement by the funding agencies.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.taap.2019.114711.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abdo N, Xia M, Brown CC, Kosyk O, Huang R, Sakamuru S, Zhou YH, Jack JR, Gallins P, Xia K, Li Y, Chiu WA, Motsinger-Reif AA, Austin CP, Tice RR, Rusyn I, Wright FA, 2015. Population-based in vitro hazard and concentration-response assessment of chemicals: the 1000 genomes high-throughput screening study. Environ. Health Perspect 123, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni C, Crivori P, 2007. Assessment of QT liabilities in drug development. Cell Biol. Toxicol 23, 1–13. [DOI] [PubMed] [Google Scholar]
- Bapat A, Anderson CD, Ellinor PT, Lubitz SA, 2018. Genomic basis of atrial fibrillation. Heart 104, 201–206. [DOI] [PubMed] [Google Scholar]
- Bates D, Machler M, Bolker B, Walker S, 2015. Fitting linear mixed-effects models using lme4. J Stat Soft 67, 1–48. [Google Scholar]
- Bednarz B, Chamiec T, Ceremuzynski L, 2003. Antioxidant vitamins decrease exercise-induced QT dispersion after myocardial infarction. Kardiol. Pol 58, 375–379. [PubMed] [Google Scholar]
- Bhatnagar A, 2017. Environmental determinants of cardiovascular disease. Circ. Res 121, 162–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette AD, Grimm FA, Dalaijamts C, Hsieh NH, Ferguson K, Luo YS, Rusyn I, Chiu WA, 2019. Thorough QT/QTc in a dish: an in vitro human model that accurately predicts clinical concentration-QTc relationships. Clin. Pharmacol. Ther 105, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K, Stohlman J, Vicente J, Chan D, Johannesen L, Hortigon-Vinagre MP, Zamora V, Smith G, Crumb WJ, Pang L, Lyn-Cook B, Ross J, Brock M, Chvatal S, Millard D, Galeotti L, Stockbridge N, Strauss DG, 2017. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhythmias. Toxicol. Sci 155, 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Clark BW, Garner LV, Di Giulio RT, 2015. Zebrafish cardiotoxicity: the effects of CYP1A inhibition and AHR2 knockdown following exposure to weak aryl hydrocarbon receptor agonists. Environ. Sci. Pollut. Res. Int 22, 8329–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Diecke S, Matsa E, Sharma A, Wu H, Wu JC, 2016. Modeling cardiovascular diseases with patient-specific human pluripotent stem cell-derived cardiomyocytes. Methods Mol. Biol 1353, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA, Wright FA, Rusyn I, 2017. A tiered, Bayesian approach to estimating of population variability for regulatory decision-making. ALTEX 34, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduati F, Mangravite LM, Wang T, Tang H, Bare JC, Huang R, Norman T, Kellen M, Menden MP, Yang J, Zhan X, Zhong R, Xiao G, Xia M, Abdo N, Kosyk O, Friend S, Dearry A, Simeonov A, Tice RR, Rusyn I, Wright FA, Stolovitzky G, Xie Y, Saez-Rodriguez J, Collaboration N-N-UDT, 2015. Prediction of human population responses to toxic compounds by a collaborative competition. Nat. Biotechnol 33, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermini B, Hancox JC, Abi-Gerges N, Bridgland-Taylor M, Chaudhary KW, Colatsky T, Correll K, Crumb W, Damiano B, Erdemli G, Gintant G, Imredy J, Koerner J, Kramer J, Levesque P, Li Z, Lindqvist A, Obejero-Paz CA, Rampe D, Sawada K, Strauss DG, Vandenberg JI, 2016. A new perspective in the field of cardiac safety testing through the comprehensive in vitro proarrhythmia assay paradigm. J. Biomol. Screen 21, 1–11. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, 2015. A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Iwata Y, Sirenko O, Bittner M, Rusyn I, 2015. High-content assay multiplexing for toxicity screening in induced pluripotent stem cell-derived cardiomyocytes and hepatocytes. Assay Drug Dev Technol 13, 529–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Blanchette A, House JS, Ferguson K, Hsieh NH, Dalaijamts C, Wright AA, Anson B, Wright FA, Chiu WA, Rusyn I, 2018. A human population-based organotypic in vitro model for cardiotoxicity screening. ALTEX 35, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausburg F, Jung JJ, Hoch M, Wolfien M, Yavari A, Rimmbach C, David R, 2017. (Re-)programming of subtype specific cardiomyocytes. Adv. Drug Deliv. Rev 120, 142–167. [DOI] [PubMed] [Google Scholar]
- Hollingshead LM, Faulds D, Fitton A, 1992. Bepridil. A review of its pharmacological properties and therapeutic use in stable angina pectoris. Drugs 44, 835–857. [DOI] [PubMed] [Google Scholar]
- Huo J, Wei F, Cai C, Lyn-Cook B, Pang L, 2019. Sex-related differences in drug-induced QT prolongation and torsades de pointes: a new model system with human iPSC-CMs. Toxicol. Sci 167, 360–374. [DOI] [PubMed] [Google Scholar]
- Kawatou M, Masumoto H, Fukushima H, Morinaga G, Sakata R, Ashihara T, Yamashita JK, 2017. Modelling torsade de pointes arrhythmias in vitro in 3D human iPS cell-engineered heart tissue. Nat. Commun 8, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ, 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller M, 2016. An R package for robust estimation of linear mixed-effects models. J Stat Soft 75, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty H, Benson C, Cartwright E, Cross M, Garland C, Hammond T, Holloway C, McMahon N, Milligan J, Park B, Pirmohamed M, Pollard C, Radford J, Roome N, Sager P, Singh S, Suter T, Suter W, Trafford A, Volders P, Wallis R, Weaver R, York M, Valentin J, 2011. How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol 163, 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Giraldez R, Gogarten SM, Below JE, Yao J, Seyerle AA, Highland HM, Kooperberg C, Soliman EZ, Rotter JI, Kerr KF, Ryckman KK, Taylor KD, Petty LE, Shah SJ, Conomos MP, Sotoodehnia N, Cheng S, Heckbert SR, Sofer T, Guo X, Whitsel EA, Lin HJ, Hanis CL, Laurie CC, Avery CL, 2017. GWAS of the electrocardiographic QT interval in Hispanics/Latinos generalizes previously identified loci and identifies population-specific signals. Sci. Rep 7, 17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, D’Antonio M, Benaglio P, Williams R, Hashem SI, Schuldt BM, DeBoever C, Arias AD, Garcia M, Nelson BC, Harismendy O, Jakubosky DA, Donovan MKR, Greenwald WW, Farnam K, Cook M, Borja V, Miller CA, Grinstein JD, Drees F, Okubo J, Diffenderfer KE, Hishida Y, Modesto V, Dargitz CT, Feiring R, Zhao C, Aguirre A, McGarry TJ, Matsui H, Li H, Reyna J, Rao F, O’Connor DT, Yeo GW, Evans SM, Chi NC, Jepsen K, Nariai N, Muller FJ, Goldstein LSB, Izpisua Belmonte JC, Adler E, Loring JF, Berggren WT, D’Antonio-Chronowska A, Smith EN, Frazer KA, 2017. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports 8, 1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss-Ustun A, Corvalan C, 2006. Preventing Disease through Healthy Environments: Towards an Estimate of the Environmental Burden of Disease. World Health Organization, Geneva, Switzerland. [Google Scholar]
- Sacks JD, Stanek LW, Luben TJ, Johns DO, Buckley BJ, Brown JS, Ross M, 2011. Particulate matter-induced health effects: who is susceptible? Environ. Health Perspect 119, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed N, Liu C, Wu JC, 2016. Translation of human-induced pluripotent stem cells. From Clinical Trial in a Dish to Precision Medicine. J Am Coll Cardiol 67, 2161–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, Patel M, Goncalves A, Ferreira R, Benn CL, Wilbrey A, Bictash M, Impey E, Cao L, Lainez S, Loucif AJ, Whiting PJ, Consortium H, Gutteridge A, Gaffney DJ, 2018. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet 50, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmstrom A, Matsa E, Zhang Y, Kumar A, Fan AC, Del Alamo JC, Wu SM, Moslehi JJ, Mercola M, Wu JC, 2017. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med 9 (pii: eaaf2584). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O, Crittenden C, Callamaras N, Hesley J, Chen YW, Funes C, Rusyn I, Anson B, Cromwell EF, 2013a. Multiparameter in vitro assessment of compound effects on cardiomyocyte physiology using iPSC cells. J. Biomol. Screen 18, 39–53. [DOI] [PubMed] [Google Scholar]
- Sirenko O, Cromwell EF, Crittenden C, Wignall JA, Wright FA, Rusyn I, 2013b. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol. Appl. Pharmacol 273, 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O, Grimm FA, Ryan KR, Iwata Y, Chiu WA, Parham F, Wignall JA, Anson B, Cromwell EF, Behl M, Rusyn I, Tice RR, 2017. In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol. Appl. Pharmacol 322, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifer SH, 2018. PLINK: key functions for data analysis. Curr Protoc Hum Genet 97, e59. [DOI] [PubMed] [Google Scholar]
- Strauss DG, Vicente J, Johannesen L, Blinova K, Mason JW, Weeke P, Behr ER, Roden DM, Woosley R, Kosova G, Rosenberg MA, Newton-Cheh C, 2017. Common genetic variant risk score is associated with drug-induced QT prolongation and torsade de pointes risk: a pilot study. Circulation 135, 1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasuna K, Asakura K, Araki S, Ando H, Kazusa K, Kitaguchi T, Kunimatsu T, Suzuki S, Miyamoto N, 2017. Comprehensive in vitro cardiac safety assessment using human stem cell technology: overview of CSAHi HEART initiative. J. Pharmacol. Toxicol. Methods 83, 42–54. [DOI] [PubMed] [Google Scholar]
- Vicente J, Zusterzeel R, Johannesen L, Mason J, Sager P, Patel V, Matta MK, Li Z, Liu J, Garnett C, Stockbridge N, Zineh I, Strauss DG, 2018. Mechanistic model-informed Proarrhythmic risk assessment of drugs: review of the “CiPA” initiative and Design of a Prospective Clinical Validation Study. Clin. Pharmacol. Ther 103, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis R, Benson C, Darpo B, Gintant G, Kanda Y, Prasad K, Strauss DG, Valentin JP, 2018. CiPA challenges and opportunities from a non-clinical, clinical and regulatory perspectives. An overview of the safety pharmacology scientific discussion. J. Pharmacol. Toxicol. Methods 93, 15–25. [DOI] [PubMed] [Google Scholar]
- Yang C, Al-Aama J, Stojkovic M, Keavney B, Trafford A, Lako M, Armstrong L, 2015. Concise review: cardiac disease Modeling using induced pluripotent stem cells. Stem Cells 33, 2643–2651. [DOI] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ, 2013. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ. Health Perspect 121, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zuo Z, Chen Y, Wang C, 2013. Phenanthrene exposure causes cardiac arrhythmia in embryonic zebrafish via perturbing calcium handling. Aquat. Toxicol 142–143, 26–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.