Abstract
Domestication of wild animals induces a set of phenotypic characteristics collectively known as the domestication syndrome. However, how this syndrome emerges is still not clear. Recently, the neural crest cell deficit hypothesis proposed that it is generated by a mildly disrupted neural crest cell developmental program, but clear support is lacking due to the difficulties of distinguishing pure domestication effects from preexisting genetic differences between farmed and wild mammals and birds. Here, we use a farmed fish as model to investigate the role of persistent changes in DNA methylation (epimutations) in the process of domestication. We show that early domesticates of sea bass, with no genetic differences with wild counterparts, contain epimutations in tissues with different embryonic origins. About one fifth of epimutations that persist into adulthood are established by the time of gastrulation and affect genes involved in developmental processes that are expressed in embryonic structures, including the neural crest. Some of these genes are differentially expressed in sea bass with lower jaw malformations, a key feature of domestication syndrome. Interestingly, these epimutations significantly overlap with cytosine-to-thymine polymorphisms after 25 years of selective breeding. Furthermore, epimutated genes coincide with genes under positive selection in other domesticates. We argue that the initial stages of domestication include dynamic alterations in DNA methylation of developmental genes that affect the neural crest. Our results indicate a role for epimutations during the beginning of domestication that could be fixed as genetic variants and suggest a conserved molecular process to explain Darwin’s domestication syndrome across vertebrates.
Keywords: developmental processes; animal domestication; epigenetics; neural crest; domestication syndrome; epimutation, transgenerational effects, glutamate receptors
Introduction
Domestication lies at the heart of human civilization and involves raising animals in an environment that differs substantially from their natural habitat. Darwin was the first to notice that domesticated animals evolve certain morphological, physiological, and behavioral traits (Darwin 1868), a phenomenon now known as the domestication syndrome (Wilkins et al. 2014). Research into the mechanisms underlying the domesticated phenotypes has traditionally focused on the detection of genes under positive selection in mammalian and avian domesticates, where domestication is mostly pronounced in behavioral traits such as tameness. However, this approach commonly involves interspecies comparisons, since most current domesticates have remained genetically isolated from their wild ancestors for thousands of years. Thus, the processes taking place during the initial stages of domestication, that is, the beginning of human-controlled rearing in a captive farming environment, have remained unclear and their contribution to the acquisition of a domesticated phenotype not well assessed (Wilkins 2017).
The first stages of domestication and adaption to a new environment likely involve epigenetic changes (Belyaev et al. 1981; Trut et al. 2009; Koch et al. 2016; Vogt 2017), that is, epimutations, defined as heritable changes in gene activity due to DNA modification, not mutation, occurring via mitotic transmission of epigenetic marks rather than DNA variants (Oey and Whitelaw 2014). Epigenetic regulatory mechanisms, such as DNA methylation, can respond to environmental change and, at the same time, be stable enough to be maintained throughout lifetime. Recently, there is mounting evidence in both plants and animals showing that epimutations can be transmitted across generations (Skinner et al. 2015; Chamorro-Garcia et al. 2017; Knecht et al. 2017; Leroux et al. 2017; Vogt 2017; Kamstra et al. 2018; Skjærven et al. 2018; Zhang Y-Y et al. 2018). To our knowledge, no studies have specifically addressed the involvement of epigenetic changes established during early development in the first steps of domestication prior to the appearance of major genetic differences. To determine whether such changes underlie the domestication syndrome, we reasoned that fish could provide a model in which to investigate the effects of the farming environment. Although the domestication syndrome refers originally to mammals, nevertheless domesticated fish exhibit many characteristics of domestication syndrome comparable to those of mammals, including jaw alterations, abnormal pigmentation, and behavioral changes (see examples in supplementary fig. 1, Supplementary Material online). Unlike domesticated mammals and birds, however, in fish wild relatives remain easily accessible. Consequently, it should be possible to make comparisons between genetically similar animals.
The European sea bass (Dicentrarchus labrax) is one of the main farmed species in all Mediterranean regions but selective breeding is not widespread. Thus, although some farms have carried out breeding for several generations others still rely on wild-caught fish for broodstock. A previous study identified genomic regions under positive selection in sea bass obtained from two independent Mediterranean hatcheries that had been carrying out selective breeding for 25 years (Bertolini et al. 2016). To confirm that sea bass would be a suitable model for analysis of epigenetic changes during domestication, we compared genes under positive selection in this species with those found in mammalian and avian domesticates (Carneiro et al. 2014; Montague et al. 2014; Schubert et al. 2014; Kukekova et al. 2018; Pendleton et al. 2018; Zhang Z et al. 2018) (supplementary fig. 2 and supplementary data set 1, Supplementary Material online). We found several genes in common, among them genes coding for glutamate receptors, which are found under positive selection in all the domesticates examined (O’Rourke and Boeckx 2018). This, along with the observation that farmed sea bass also shares some of the characteristics of the domestication syndrome, for example, changes in morphology (Arechavala-Lopez et al. 2012), jaw malformation (Babbucci et al. 2016), indicates that this species constitutes a suitable model to study the first steps of domestication in vertebrates.
Results and Discussion
Early Domesticates Differ in DNA Methylation from Wild Fish
To identify the underlying epigenetic processes operating during the early stages of domestication, we first compared wild and early domesticates of sea bass exhibiting no genetic differences, as confirmed by a low fixation index (mean FST = 0.0085) (supplementary fig. 3, Supplementary Material online; see Materials and Methods). Early domesticates were the offspring of a cross between a 3-year-old unselected broodstock that were hatchery raised for no more than one generation. We sampled adult tissues derived from all three embryonic layers—brain (ectoderm), muscle and testis (mesoderm), and liver (endoderm)—and analyzed DNA methylation by reduced representation bisulfite sequencing (RRBS). We chose to perform RRBS that effectively enriches for the informative parts of the genome, instead of whole genome bisulfite sequencing (WGBS), which produces 70–80% of uninformative reads for CpG dinucleotide methylation (Ziller et al. 2013). RRBS libraries were sequenced at an average 47-fold coverage per sample using 3 replicates per group and per tissue, that is, 12 samples for wild fish and 12 samples for early domesticates (technical details and data quality in supplementary table 1 and supplementary fig. 4, Supplementary Material online).
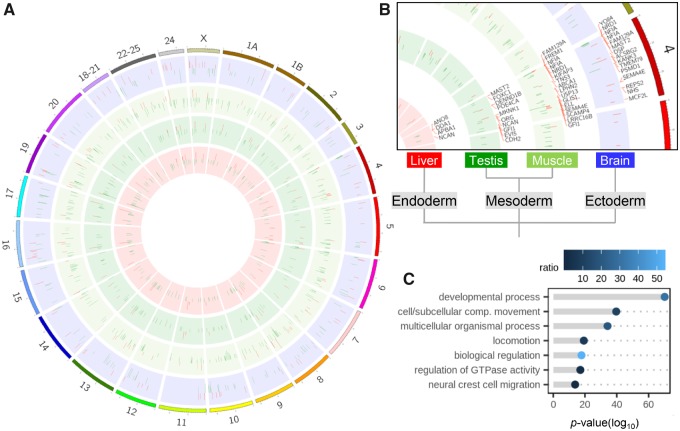
Differentially methylated regions (DMRs) were defined as having more than 15% methylation differences (False Discovery Rate, FDR <0.05) to reassure biologically meaningful results, between wild and early domesticates, kept under the same environmental conditions of temperature and photoperiod. DMRs were present in all four tissues examined and had a genome-wide distribution (fig. 1A and B). DMRs were preferentially located (z-score = 2.92, P value = 0.0023) inside gene bodies or within 5-kb upstream of the transcription start site or downstream of the transcription termination site (supplementary fig. 5a, Supplementary Material online) and overlapping CpG islands and/or shores (mean unique DMRs, 53.53%; supplementary fig. 5b, Supplementary Material online). Compared with wild fish, the number of hypomethylated DMRs in early domesticates (210 in brain, 476 in muscle, 275 in testis, and 52 in liver) was higher than the number of hypermethylated DMRs (148 in brain, 160 in muscle, 82 in testis, and 37 in liver; Fisher’s exact test for count data, odds ratio = 0.42, P value < 2.2e-16, supplementary fig. 5c, Supplementary Material online). The possibility that a fraction of the methylation differences may have a genetic basis cannot be excluded. However, considering the low FST between wild and early domesticates, a value even below the lowest value of the genetic differentiation of natural populations (FST = 0.011) (Loukovitis et al. 2015), the influence of the genetic background to epimutations can be considered minimal.
Fig. 1.
DNA methylation changes in early domesticate versus wild European sea bass. (A) Genome-wide distribution of DMRs; red, hypermethylation; green, hypomethylation. (B) Details of annotated genes on chromosome 4 containing DMRs in tissues derived from the ectoderm (brain; blue), paraxial mesoderm (muscle; light green), intermediate mesoderm (testis; green), and endoderm (liver; red). (C) GO term enrichment of genes that contain DMRs. The length of the bar indicates the log10-transformed P value of the enrichment and the color scale indicates the amount (in percent) of genes associated with the specific GO term as compared with the total number of genes with DMRs.
Independent assessment of DNA methylation levels was carried out by multiplex bisulfite sequencing (MBS) (Anastasiadi, Vandeputte, et al. 2018), targeting the detected DMRs of six genes (supplementary fig. 6, Supplementary Material online). MBS results fully validated our genome-wide approach with RRBS (Pearson’s correlation coefficient ρ = 0.89, P value < 2e-16; supplementary fig. 6, Supplementary Material online). In the muscle of hatchery-reared versus wild salmon, DMRs were found in the gene body or regulatory regions of 52 genes (Le Luyer et al. 2017). Fifteen genes of the sea bass showed similarities to the genes affected in salmon and these included five protocadherins (pcdh18 and fat3), three dual specificity phosphatases and one homeobox protein (supplementary data set 2, Supplementary Material online). Protocadherins and homeobox genes have also been found differentially methylated between wild and hatchery-reared steelhead trout (Gavery et al. 2018). Thus, DNA methylation differences between wild and domesticated fish are not exclusive to the sea bass.
To gain insight into the functions of the genes with DMRs, we performed enrichment tests for the associated Gene Ontology (GO) terms. The most significantly enriched biological processes GO terms were related to developmental process (GO:0032502), such as anatomical structure development (GO:0048856) and regulation of embryonic development (GO:0045995). Among the developmental processes affected by the farming environment, a large portion was associated with the nervous system and neural crest cell (NCC) migration (GO:0001755; fig. 1C; full list of genes with DMRs in supplementary data set 3, Supplementary Material online, and list of GO term enrichment in supplementary data set 4, Supplementary Material online). Genes that are responsible for NCC migration during early development are not necessarily expressed in differentiated, adult tissues. Thus, differences found in adults suggest that these genes became epimutated by the farming environment at the time they were expressed in the early domesticates.
Gene Expression Is Altered in Early Domesticates
It has been shown that just one generation of domestication can heritably alter gene expression (Christie et al. 2016). The possible effects of DNA methylation differences on gene expression in early domesticated sea bass were investigated by RNA sequencing (see quality of RNA-seq data in supplementary fig. 7, Supplementary Material online, and RNA-seq statistics in supplementary table 2, Supplementary Material online). The number of differentially expressed genes (DEGs; P adjusted <0.05 and absolute log2 fold change > 1; supplementary fig. 8, Supplementary Material online) ranged from 248 in the testis to 2,416 in the liver, with an approximately equal number of upregulated and downregulated genes in early domesticates versus wild sea bass (see full list of DEGs in supplementary data set 5, Supplementary Material online). RNA-seq results were validated by qPCR in four genes (Pearson’s correlation coefficient ρ = 0.60, P value = 0.00016; supplementary fig. 9, Supplementary Material online). Overall, DEGs were related to the defense response, including regulation of cytokine production and immune system process (supplementary fig. 10a, Supplementary Material online; see a full list of GO terms enriched in supplementary data set 6, Supplementary Material online). The number of DEGs that also contained DMRs was between 5 (testis) and 28 (muscle) (supplementary fig. 10b–e, Supplementary Material online). This low number is likely due to differences in the temporal and spatial expression of these genes, which should be affected during development rather than in adulthood, since most DMRs were found in developmental genes (fig. 1C). Changes in the expression of stress- and immune-related genes have been previously reported in farmed fish (Tymchuk et al. 2009; Bicskei et al. 2016; Wellenreuther et al. 2019). Therefore, our gene expression results are consistent with the phenotypic responses to the farming environment observed in other fish species.
DNA Methylation Changes in Developmental Genes Are Established during Gastrulation
The observation that DMRs were mostly found in development-related genes led us to hypothesize that the farming environment induces epimutations during early development, and that these are responsible for mild developmental deficits that are maintained through adulthood. Therefore, it was necessary to discriminate between epimutations present in adult tissues from those established already during early development, which have more potential for long-term effects since they are more persistent and may propagate consistently in a variety of differentiated tissues, including the germ line (Faulk and Dolinoy 2011). Since epimutations of the latter type may affect directly the germ line, they are likely more susceptible to be involved in transgenerational effects. Even if the extent to which DNA modifications can be inherited through meiotic divisions is still under debate, several cases of environmentally induced epimutations even in somatic tissues have been reported in the last years to be transgenerationally transmitted (Knecht et al. 2017; Leroux et al. 2017; Kamstra et al. 2018; Norouzitallab et al. 2019). In fish, additionally and in contrast to the situation in mammals, there seems to be little or no epigenetic reprograming after fertilization, providing further means for transgenerational epigenetic inheritance since epigenetic marks present in the genome of the sperm or of the eggs would not have been reprogramed or “erased” in the genome of the zygote (Jiang et al. 2013; Potok et al. 2013). Even though the detailed mechanisms of soma-to-germline transfer of epigenetic patterns remain mostly an unresolved question, recent studies point to possible ways of soma-to-germline transfer of information (Cossetti et al. 2014; Devanapally et al. 2015; Eaton et al. 2015; Reilly et al. 2016), including mediation by exosomes, circulating double-stranded RNAs, and transposable elements (Devanapally et al. 2015; Sharma 2017; Danchin et al. 2019; Norouzitallab et al. 2019; Perez and Lehner 2019).
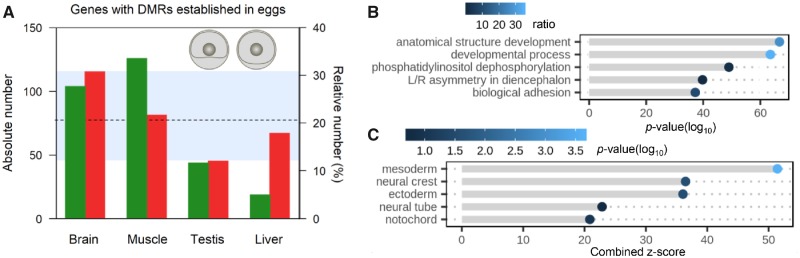
To determine whether epimutations were indeed established during early development, we therefore collected hatchery-reared sea bass embryos at 60–90% epiboly (∼30-h postfertilization), when the embryo remains a homogeneous cell mass and sequenced three RRBS libraries (see Principal Component Analysis in supplementary fig. 11, Supplementary Material online). This sampling point was chosen to allow enough time for epimutations to accumulate due to the farming environment, but not long enough for tissue- or organ-specific epimutations to have arisen. In this way, we also removed all noise due to differences in the environment (food, habitat, etc.) of early domesticates and wild fish. An average of 20.6% of loci harboring DMRs in adult tissues had similar levels of methylation already present in the embryos (defined as no more than ±10% differences in at least 50% of the CpGs within the DMRs), with a minimum of 12.1% in the testis and a maximum of 30.8% in the brain (fig. 2A). Thus, roughly one fifth of the DMRs present in adult tissues of farmed fish were already established within the first ∼30 h of life. Significantly, despite eliminating on average ∼80% of the DMRs present in adults after filtering, developmental processes (GO:0032502) such as anatomical structure development (GO:0048856) were still the most enriched terms (fig. 2B; see a full list of genes with DMRs established in embryos in supplementary data set 7, Supplementary Material online, and a list of GO terms enriched in supplementary data set 8, Supplementary Material online). More specifically, pathway analysis (Reactome database 2016) showed that these genes participate in several aspects of extracellular matrix (ECM) organization (e.g., nonintegrin membrane–ECM interactions, laminin and integrin cell-surface interactions, and collagen degradation), as well as glutamate binding, adrenoreceptors and synaptic plasticity (supplementary table 3, Supplementary Material online). Interestingly, genes harboring epimutations early in development are predominantly expressed in the mesoderm, the neural crest and the ectoderm (adjusted P value < 0.05) or similar structures, such as the neural tube and the notochord (fig. 2C; full list in supplementary table 4, Supplementary Material online). Taken together, these results suggest that the first steps in domestication involve epimutations established in genes related to developmental processes and expressed in the embryonic germ layers, including the neural crest.
Fig. 2.
Epimutations established during early development. (A) Absolute (green) and relative (red) number of genes with DMRs established during gastrulation, the latter shown as a percentage of the total number of genes with DMRs in adults. The dotted line indicates the mean of the relative number and the shaded area includes the range. (B) GO term enrichment of genes that contain DMRs. The most significantly enriched GO terms, related to development, are ranked according to the decreasing order of −log10-transformed P value of the enrichment and colored according to the percentage of genes that contain DMRs and that are members of each GO category. (C) Enrichment of tissues where the genes with DMRs are expressed. The most significantly enriched tissues are ranked according to the combined z-score as calculated by the Enrichr tool and colored according to the −log10-transformed adjusted P value of the enrichment.
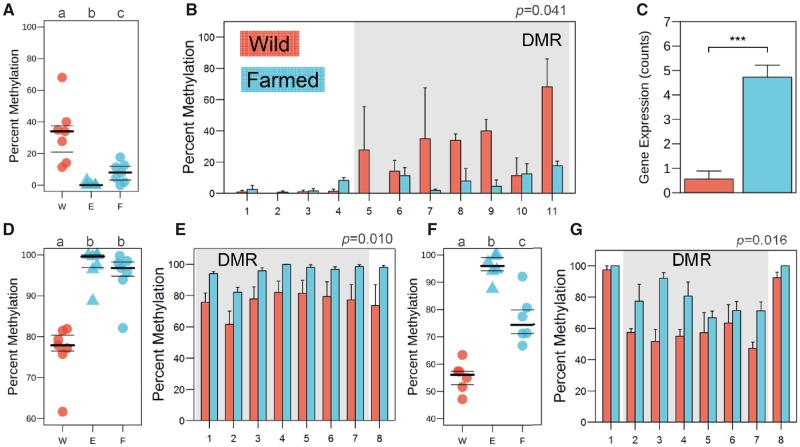
During NCC migration, cells separate from the surrounding tissues. This process, called delamination, requires the excretion of integrins, laminins and collagens such as the ones detected in this study (supplementary table 3, Supplementary Material online). For example, ADAMTS9, a matrix metalloproteinase whose function is conserved in vertebrates (Christian et al. 2013; Desanlis et al. 2018), is hypomethylated in embryos of early domesticate sea bass (fig. 3A) and this is maintained in adult muscle (fig. 3B), resulting in higher adamts9 expression levels (fig. 3C). Interestingly, homologs of ADAMTS9 are under positive selection in several domesticates, including rabbit (Carneiro et al. 2014), fox (Kukekova et al. 2018), and duck (Zhang Z et al. 2018). Collagens are also under positive selection in the genomes of rabbit (Carneiro et al. 2014), dog (Pendleton et al. 2018), fox (Kukekova et al. 2018), and horse (Schubert et al. 2014), and we found in sea bass domesticates that col18a1a contained a hypermethylated DMR in the testis (fig. 3E), with methylation patterns established already during gastrulation (fig. 3D). Another collagen, col14a1a, contained a hypermethylated DMR in the brain (supplementary fig. 12a, Supplementary Material online). The protocadherin γ-a11-like, which also belongs to a gene family involved in delamination, was found to carry a DMR in liver (supplementary fig. 12b, Supplementary Material online). This is consistent with previous work showing that cadherins and protocadherins are under positive selection in cat (Montague et al. 2014), dog (Pendleton et al. 2018), and duck (Zhang Z et al. 2018) and exhibit DMRs in hatchery-reared salmon (Le Luyer et al. 2017). DMRs in early domesticates versus wild sea bass were also observed for phldb1 in brain (supplementary fig. 12c, Supplementary Material online), foxc1 in testis (supplementary fig. 12d, Supplementary Material online) and mapk8a in muscle (supplementary fig. 12e, Supplementary Material online). The finding that many genes with epimutations established early in development are similar to genes under positive selection in domesticates from a range of species, and that these are also linked to processes of NCC migration, suggests that our observations in sea bass could reflect a general process underlying the onset of the domestication syndrome.
Fig. 3.
Influence of the farming environment on genes interacting with the ECM and neurotransmitters. Distribution of CpG methylation in wild (red, W) and farmed (blue) sea bass during gastrulation (E) and in adult tissues (F) inside the DMRs of adamts9 (A), col18a1 (D), and gria4a (F). Mean methylation of CpGs around the DMRs of adamts9 in muscle (B), col18a1 in testis (E), and gria4a in brain (G). Differential gene expression of adamts9 is also shown (C). The extent of DMRs is highlighted with gray shading and the CpGs are arbitrarily numbered. Differences in expression are shown with the following equivalence: ***P adjusted <0.001.
Members of the glutamate receptor family deserve particular attention because they have been found under positive selection in the genomes of all domesticated species studied without exception (O’Rourke and Boeckx 2018), and including the sea bass after 25 years of selective breeding (Bertolini et al. 2016) (supplementary fig. 2, Supplementary Material online). In this study, the ionotropic glutamate receptor gria4a contained a hypermethylated DMR (fig. 3G) in the brain of early domesticate sea bass and displayed similar methylation patterns during gastrulation (fig. 3F). Likewise, another glutamate receptor, grik4, exhibited a hypermethylated DMR in the muscle (supplementary data set S7, Supplementary Material online). In addition, the expression of grik5 was downregulated in the testis and liver of early domesticate sea bass, and the same pattern was observed for grm2, a glutamate metabotropic receptor in the liver (supplementary data set S5, Supplementary Material online). Among the most easily recognizable features of domestication syndrome are changes in the nozzle and associated jaw deformities. This can be explained in the NCC-deficit hypothesis by the fact that the craniofacial bones, including the jaws (upper maxilla and mandibula), are derived from the neural crest, whereas other bones are mesodermal in origin (Wilkins et al. 2014). In this context, it is interesting to note that glutamate receptors were also found differentially expressed in the lower jaw of European sea bass with mandibular prognathism (Babbucci et al. 2016).
Epimutations Associate with Genetic Variants after Generations of Rearing in Captivity
Experimental evidence in Atlantic salmon (Salmo salar) and delta smelt (Hypomesus transpacificus) suggest the possibility of rapid genetic adaptation to captivity (Finger et al. 2018; Horreo et al. 2018). It has been argued that epimutations, particularly DNA methylation changes, can eventually become permanently integrated into the genome by promoting mutation of methylated CpGs or through the generation of copy number variants or genome instability (Skinner et al. 2015; Vogt 2017). Indeed, it is well accepted that methylated cytosines tend to mutate 10–50 times faster than unmethylated ones (Chen et al. 2014; He et al. 2015). However, the role of epimutations in rapid genetic adaptation had not been shown.
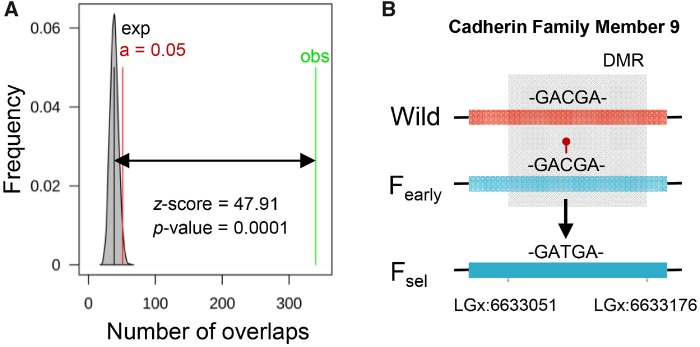
To test whether differential methylation due to early domestication is correlated with genetic variants and could therefore be genetically assimilated after several generations, we performed association analysis of genomic regions. We found that differentially methylated cytosines (DMCs) of early domesticates associated ∼10 times more than expected by chance (z-score = 47.91, P value = 0.0001, 10,000 permutations) with cytosine-to-thymine (C-T) single-nucleotide polymorphisms (SNPs) in two independent populations of sea bass after 25 years of selective breeding (Bertolini et al. 2016) (fig. 4A). This occurred, for example, in cadherin family member 9 (fig. 4B). These results do not provide a causal link between epigenetic and genetic changes and it could be possible that they represent only a correlation and a case of convergent evolution. Although we cannot exclude this possibility, our results conform to previous studies regarding the high mutability of methylated CpGs (Chen et al. 2014; He et al. 2015; Skinner et al. 2015) to highlight a significant nonrandom association of genomic sites. A recent study in domesticated chickens and their closest ancestors found that DNA methylation, depending on its levels and interindividual variability, can lead to biases toward SNPs or copy number variation mutations, promoting genomic specificity or flexibility, respectively (Pértille et al. 2019). Thus, these and our results suggest that epimutations occurring during the early stages of domestication can become integrated into the genome as genetic changes after just several generations.
Fig. 4.
Epimutations as a consequence of the farming environment associate with genetic changes in the sea bass after 25 years of domestication. (A) DMCs in early domesticate versus wild sea bass overlap with on-the-spot SNPs in two sea bass populations after 25 years of selective breeding (Bertolini et al. 2016). The number of overlaps of the two genomic sites is shown and tested via permutations. The shaded grey area shows the number of overlaps of randomized regions with the mean represented by the black bar. The green line represents the actual number of overlaps of SNPs with DMCs and the double arrow its distance from the significance limit in red. The significance of the association is indicated by the z-score and the P value. (B) Schematic representation inside the DMR of cadherin family member 9 of a CpG in wild sea bass that was found hypermethylated (red lollipop) in early domesticates (Fearly) and converted into a TG after 25 years of selective breeding (Fsel).
Conclusions
Recent research to elucidate the process of domestication has heavily relied on comparative genomics (Wilkins 2017). This is due to the complexity of the study of the domestication, which combines a polygenic basis, on one hand, with the “conditions of living,” according to Darwin, on the other. Domestication results in widespread common phenotypic characteristics (Darwin 1868; Wilkins 2017). Here, we not only used comparative genomics but integrated epigenomic and transcriptomic data of adult early domesticates and during gastrulation and made comparisons with their closest wild counterparts. One experimental aspect that we could not address concerns the epigenetic patterns of farmed fish reared in the wild environment. However, this would involve releasing freshly fertilized eggs from farmed progenitors into the ocean and recapturing the same fish when adults, which is an essentially impossible experiment in a fish like the sea bass. Likewise, functional experiments involving, for example, knocking-down specific genes among the ones discussed in this study or the DNA-methyltransferases, for example, would likely result in a drastically affected phenotype. Such type of strong effects would not provide reliable answers because the domestication syndrome is, by definition, thought to rise from mild developmental deficits, not strong ones. In vivo hybridization experiments of gene expression could be employed for specific phenotypic traits, such as the jaw deformities, whereas gene methylation editing technology could be used to functionally test the impact of few specific genes identified here. However, this technology is not yet applicable to marine fish with small pelagic eggs. Therefore, in this context, our design and data should be considered a good empirical demonstration of the role of epigenetic processes in the rise of the domesticated phenotype. The general NCC model for domestication, independently of the specific genes found here, could be, nevertheless, further tested in other early domesticated fish species for which wild counterparts are available and show signs of the domestication syndrome or in the experimentally domesticated foxes.
In summary, our findings show that the farming environment influences the genome of sea bass in the initial stages of domestication by inducing epimutations. These epimutations are established already during early life and involve genes related to developmental processes that are expressed in embryonic structures including the neural crest. The epimutations persist until adulthood in different tissues and some result in measurable changes in gene expression when compared with wild counterparts. Epimutated genes include those found with altered expression in sea bass with phenotypes associated with the domestication syndrome, such as jaw deformities. Epimutations significantly associate with SNPs, suggesting that they can be integrated into the genome as genetic polymorphisms due to the hypermutability of the CpGs and its neighboring sequences. Our results constitute, to the best of our knowledge, the first empirical demonstration incorporating epigenetic mechanisms in support of the NCC-deficit hypothesis to explain the emergence of Darwin’s domestication syndrome and suggest a conserved process that could be operating across vertebrates.
Materials and Methods
Animals and Rearing Conditions
Wild adult European sea bass (Dicentrarchus labrax L.) were captured around the Medes Islands Natural Reserve (Western Mediterranean, NE of Spain) and farmed sea bass were obtained from a farm located in the same area (St. Pere Pescador, Girona, Catalonia, Spain) (supplementary fig. 3, Supplementary Material online). Farmed sea bass were the offspring of a cross of 3-year-old unselected broodstock that were hatchery raised for no more than one generation. Farmed fish were, thus, at the early stages of domestication and are referred to as early domesticates. Analysis of 93,369 SNPs (see details in the Genetic Analysis section) showed that there were no major genetic differences between the wild and early domesticates used in this study, as confirmed by a mean FST = 0.0085, a value even below the lowest value of the genetic differentiation of natural populations (FST = 0.011) (Loukovitis et al. 2015).
Five days postfertilization (dpf) larvae of early domesticates were transported from the commercial hatchery to the aquarium facilities of the Institute of Marine Sciences. The fish were reared under standard raising conditions for this species as described previously (Díaz et al. 2013) and were maintained under natural conditions of photoperiod and temperature. Fish were fed pelleted food of the appropriate size (YM858, Efico, BioMar). The composition of this diet is considered conservative and is used as the control diet in studies of fish meal substitution (Simó-Mirabet et al. 2018) since among diets it has a composition closest to the diet encountered in the wild.
The aquarium facilities are authorized for experimentations with animals by the Ministry of Agriculture and Fisheries (certificate number 08039–46–A) according to Spanish law (R.D. 223 of March 1988). Fish treatments were in accordance with the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes (ETS Nu 123, 126, January 1, 1991). The experimental protocol was also authorized by the Ethics Committee of the Spanish National Research Council (CSIC) within projects AGL2013–41047–R and AGL2016–78710–R.
Sampling
Tissues from wild fish were dissected immediately upon capture, transported in RNAlater (ThermoFisher Scientific) and stored at −80 °C. Early domesticates were sacrificed using an overdose of 2-phenoxyethanol, tissues were immediately frozen in liquid nitrogen following dissection and stored at −80 °C. At sampling, both wild fish and early domesticates were more than 3 years old (mean weight 773.88 g; mean length 35.44 cm) to reduce possible effects due to differences in age or size. Both wild fish and early domesticates were sampled in the nonreproductive season (June) to prevent any effect due to differences in the annual reproductive cycle, especially in reproductive tissues. Three replicates per group and tissue were used for DNA methylation analysis and five replicates per group and tissue were used for gene expression analysis.
Fertilized eggs of early domesticates, the product of a natural spawning, were collected at 27–30-h postfertilization (60–90% epiboly). Eggs were briefly rinsed with water and were snap frozen in liquid nitrogen and stored at −80 °C until analysis. Three pools of five eggs each were used for DNA methylation analysis.
Nucleic Acid Isolation
Total RNA was isolated from brain, muscle, testis and liver of adult wild fish and early domesticates using TRIzol Reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. Genomic DNA was isolated by phenol/chloroform/isoamyl alcohol extraction from brain, muscle, testis and liver of adult fish and from three pools of five eggs. In brief, cells were lysed by incubation in TENS buffer (0.1 M NaCl, 10 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid pH 8, 0.5% Sodium dodecyl sulfate, [SDS]) overnight with 1 μg of proteinase K (Sigma-Aldrich) for protein digestion. RNA was digested with 0.5 μg of ribonuclease A (PureLink RNase A; Life Technologies) and DNA was precipitated with 95% ethanol. Samples from tissues stored in RNAlater were further cleaned with 2× AMPure XP beads (Beckman Coulter).
RNA-seq
A total of 42 RNA-seq libraries were constructed from the four tissues of wild fish and early domesticates (supplementary table 2, Supplementary Material online). RNA quality was measured with the Agilent RNA 6000 Nano Kit (Agilent) and only samples with an RNA integrity number higher than 8 were further used for RNA-seq library preparation. The mRNA-seq libraries were constructed according to the manufacturer’s instructions using the TruSeqStranded mRNA LT Sample Prep Kit (Illumina Inc., Rev.E, October 2013). A total of 20 libraries for muscle and testis were sequenced using the TruSeq SBS Kit v3-HS and 20 libraries for liver and brain were sequenced using the TruSeq SBS Kit v4 on an Illumina HiSeq 2000 platform in paired-end mode with a read length of 76 bp. Image analysis, base calling and quality scoring of the runs were processed using the manufacturer’s software Real Time Analysis and followed by generation of FASTQ sequence files by CASAVA. Decontamination was performed with the BBDuk package (v. 37.61) from the BBTools Suite of the Joint Genome Institute, which employs kmers. The parameters set for adapter trimming were ktrim = r, k = 23, mink = 11, hdist = 1 tpe tbo, and for quality trimming they were qtrim = r, trimq = 30, minlen = 23. The HISAT2 (Kim et al. 2015) (v. 2.1.0) program, which uses a graph-based alignment, was employed for the alignment of the reads. The European sea bass genome (dicLab1 v1.0c, July 2012) (Tine et al. 2014) was indexed using the hisat2-index wrapper function and the trimmed reads were aligned using the core hisat2 function with default parameters. The reads were counted at the gene level with feature Counts (Liao et al. 2014) (v. 1.6.2) from the Subread (Liao et al. 2013) package. After performing clustering and principal component analysis, one sample of muscle and one sample of brain from wild fish were excluded from further analysis since they were clear outliers (supplementary fig. 7, Supplementary Material online). Differential expression was estimated using the default pipeline of the DESeq2 (Love et al. 2014) package (v. 1.20.0). The log2 fold changes were shrunk according to the original DESeq2 shrinkage estimators.
Quantitative Real-Time PCR
RNA extracted from the muscle of eight fish was used for validation of RNA-seq results using quantitative real-time PCR. Five-hundred nanograms of RNA were treated with 0.5 units of amplification grade DNAse I (ThermoFisher Scientific, 18068015) and reverse transcribed to cDNA using SuperScript III Reverse Transcriptase (ThermoFisher Scientific, 18080085) with 100 μM of random hexamers according to the manufacturer’s instructions. Primers were designed using Primer3Plus software targeting a short region covering two exons. To calculate the efficiency of the primers, we performed amplification of a pool from all samples and serial dilutions with cDNA concentration of 1, 0.2, 0.1, 0.02, 0.01, and 0.002. Then, we calculated the slope from the log-linear regression of the calibration curve and the efficiency as E = 10(−1/slope). Two reference genes were used previously validated in sea bass (Mitter et al. 2009). The reactions were performed in triplicate and negative controls without cDNA were included for each primer. Details of the primers used in qPCR reactions can be found in supplementary table 5, Supplementary Material online. The EvaGreen Dye (Biotium) was used in the qRT-PCR reactions performed on a QuantStudio 12K Flex (ThermoFisher Scientific). The Cq values were used to calculate the normalized values of each target gene after subtracting the geometric mean of the two reference genes.
Reduced Representation Bisulfite Sequencing
A total of 27 RRBS libraries were constructed as previously described (Anastasiadi, Esteve-Codina, et al. 2018). Briefly, 20 units of MspI (NEB) were used to digest 100 ng of genomic DNA at 37 °C overnight. In the same reaction, five units of Klenow fragment (3′ → 5′exo-; NEB) were added along with dNTPs at a final concentration of 300 μM dATP, 30 μM dCTP, and 30 μM dGTP. Incubation at 30 °C for 20 min was performed for end repair, followed by a 37 °C incubation for 20 min to account for A-tailing and 20 min incubation at 75 °C to inactivate the enzyme. Illumina TruSeq Adapters v2 were added to the fragments using the Quick Ligase (NEB) and incubating at 25 °C for 20 min. Size selection was performed using a ratio of 0.75× AMPure XP beads (1:5 diluted). The libraries were then quantified by qPCR and pooled according to the qPCR values. The pooled samples were further cleaned up using 2.5× AMPure XP beads (1:5 diluted). Bisulfite conversion was performed using the EZ DNA Methylation-Direct kit (Zymo Research) with 0.9× CT Conversion Reagent and 30 min of desulphonation. qPCRs were used to determine the optimal number of cycles per sample in the enrichment reaction. Enrichment of the libraries was performed using the PfuTurbo Cx HotStart Polymerase (Agilent Technologies) with the following cycling program: 95 °C for 2 min, determined number of cycles of 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 45 s, and a last step at 72 °C for 7 min. AMPure XP beads at 1× were used to clean up the final product. The RRBS libraries of liver, brain, and eggs were constructed using the Premium RRBS kit (Diagenode, Cat. No. C02030032) which compiles the exact manual protocol and which was followed according to the instructions of the manufacturer. Qubit High Sensitivity assays (ThermoFisher Scientific) were used to quantify the final RRBS libraries. Their quality was checked using the Experion DNA 1 k assays (BioRad) in the case of muscle and testis and with the DNA High Sensitivity assay (Agilent) for liver, brain and eggs. RRBS libraries were sequenced on an Illumina HiSeq 2000 instrument in single-end mode with a length of 50 bp.
Data analysis was performed in parallel exactly as described in Anastasiadi, Esteve-Codina, et al. (2018). The Trimmomatic software (Bolger et al. 2014) (v.0.32) was used for quality trimming with the following parameters: sliding window of 4, Phred score > 15, adaptive quality trimming with largest length of 20, and strictness of 0.50 and 18 as minimum read length. Reads were then aligned with the European sea bass genome using BSMAP (Xi and Li 2009; Xi et al. 2012) (v. 2.90) and restricting the parameters to RRBS mode and at least 5 reads coverage. The methratio.py script of the BSMAP package was used for methylation extraction. The bsrate script from the MethPipe pipeline (Song et al. 2013) (v. 3.4.3) was used to calculate the efficiency of bisulfite conversion.
Further analyses were performed with the R (R Core Team 2015) and Bioconductor (Gentleman et al. 2004) packages using Rstudio (RStudio Team 2015), unless stated otherwise. The methylKit package (Akalin et al. 2012) (v. 1.6.3) was used for estimation of methylation values and for differential analysis for each tissue. CpGs with <10 reads coverage or with coverage higher than 99.9% of the distribution of read counts were filtered out. Coverage was then normalized across samples for each tissue and only CpGs covered in all samples of one tissue were maintained for further analysis. Eggs were analyzed in batches with all tissues from farmed fish and the methylation percentage for each CpG was extracted. To detect DMCs between wild and farmed fish, a logistic regression model was applied with correction for overdispersion as proposed by Feng et al. (2014) and McCullagh and Nelder (1989), and the χ2 test was used to determine methylation differences. We chose this method after testing the logistic regression with and without correction for overdispersion using the χ2 and the F-test to determine methylation differences, the beta-binomial model with parameter shrinkage as implemented by the DSS package (Feng et al. 2014), and the binomial mixed model integrating genetic relatedness data derived from the sequencing as implemented in MACAU (Lea et al. 2015). This method is characterized by high specificity without compromising its sensitivity (Wreczycka et al. 2017). The SLIM method was used to correct P values after multiple testing. The DMCs detected by methylKit were used in the edmr package (Li et al. 2013) (v. 0.6.2) to identify empirical DMRs based on bimodal normal distribution model and a weighted cost function to optimize regional methylation. The implementation of the algorithm followed the default settings and DMRs were further defined as regions containing a minimum of 4 CpGs, 2 DMCs with methylation difference >15% and q-value < 0.1 (as used previously [Lea et al. 2016; Ingerslev et al. 2018]), total methylation difference >15% and q-value < 0.05. The combination of this threshold, sample size, and depth coverage ensures detection of biologically meaningful differences. The COMBINED ANNOTATION track of the European sea bass genome (dicLab v1.0c, July 2012) was used for annotation procedures. Regions 5,000-bp upstream of the transcription start site and 5,000-bp downstream the 3′ UTR were included together with the full gene bodies to determine whether a DMR was present in a gene. The genomic association of DMRs with gene bodies and regulatory regions was tested using the Bioconductor package regioneR (Gel et al. 2016) (v. 1.12.0), which offers a permutation framework to evaluate statistically the association of genomic regions. We evaluated whether the number of overlaps of DMRs and gene bodies ±5 kb could be attributed to chance alone using a randomization strategy of creating a new set of the same number of regions with the same width, randomly placing all regions along the genome and performing 10,000 permutations.
Genetic Analysis
SNP data were extracted in parallel with DNA methylation data using BisSNP (Liu et al. 2012) (v. 0.82.2) from the four tissues of each individual per group separately and using the SNVs ILLUMINA track of the European sea bass as SNP reference. The VCFtool (Danecek et al. 2011) (v. 0.1.15) was used to prepare the files and calculate the Weir and Cockerham (Weir and Cockerham 1984) FST population estimates using only diploid sites per tissue, and then the FST population estimates were averaged.
SNPs were also extracted from the sequenced genomes of sea bass populations from two commercial hatcheries after 25 years of selective breeding for production traits from a previous study (Bertolini et al. 2016). Only SNPs that had a quality between 20 and 40, a minimum depth of 4 and were biallelic were retained for further analysis using samtools and bcftools (Li et al. 2009) (v. 1.9).
The genomic association of SNPs in selected populations of sea bass with the DMCs was tested using regioneR (Gel et al. 2016). Only SNPs from C to T, or from G to A, to take into account the reverse strand, were used to test whether their overlap with the DMCs was significantly higher than expected by chance. We evaluated the number of overlaps of SNPs and DMCs using the same strategy as before and performing 10,000 permutations.
Multiplex Bisulfite Sequencing
MBS (Anastasiadi, Vandeputte, et al. 2018) libraries were constructed to validate the RRBS results. Six genes were selected as candidates for validation and eight fish samples were used for each tissue. The genes selected had to follow three criteria: 1) contain DMRs between wild versus early domesticate fish, 2) the DMRs had to be already established in the eggs, and 3) genes of the same family had to under positive selection in at least one mammalian or avian domesticate. We targeted the identified DMRs of gria4a and phldb1 in the brain, adamts9 and mapk8a in the muscle, foxc1 in the testis, and protocadherin γ-a11-like in the liver (supplementary table 6, Supplementary Material online). Primers were designed using MethPrimer (Li and Dahiya 2002) and in silico validated using Primer3Plus (Untergasser et al. 2012, p. 3) against the bisulfite-converted amplicon sequence. The overhang adapter sequences (Illumina 18S Metagenomic protocol) were added to the 5′ of the locus-specific primers.
Five-hundred ng of genomic DNA were converted using the EZ DNA Methylation-Direct Kit (Zymo Research) extending the desulphonation time to 30 min and eluting in 40 μl of elution buffer. Two microliters of the bisulfite-converted DNA were used to amplify the targeted regions with 2.5 units of GoTaq G2 Hot Start polymerase (Promega) in a reaction that contained a final concentration of 4 mM of MgCl2, 0.8 mM dNTPs, and 0.8 μM of primers. The amplification was performed under the following conditions: 7 min at 95 °C, 40 cycles of 95 °C for 1 min, Tm specific for each primer pair (supplementary table 6, Supplementary Material online) for 2 min, 65 °C for 2 min, and finally 10 min at 65 °C. Tm was determined after gradient PCR, amplification was visually confirmed by gel electrophoresis and its specificity by Sanger sequencing using both the forward and the reverse adapter.
Bead-based size selection and normalization (Hosomichi et al. 2014) was performed according to Anastasiadi, Vandeputte, et al. (2018) using a home-made solution of Sera-mag SpeedBeads (Fisher 099981123) (Rohland and Reich 2012). The size selection followed a double incubation with beads, starting with 0.4× beads (2×) and followed by a second incubation of the supernatant with 0.6× beads (2×). The normalization was performed by incubating a 3:1 volume of 20-fold 2× diluted beads with the size-selected DNA. Two microliters of each of the six amplicons were pooled per sample. Indices from the Nextera XT index Kit SetA (Illumina FC-131-2001) were added using 1 unit of PCRBio HiFi Polymerase and 10 μl of the pooled amplicons under the following conditions: 3 min at 95 °C, 8 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and finally 5 min at 72 °C. Clean up of the reactions was performed using 0.4× beads (2×). The DNA was then quantified with a Qubit High Sensitivity assay (ThermoFisher Scientific) and these values were used to calculate the volumes used for the final pooling in a single tube which was cleaned up using 1× beads (2×). DNA High Sensitivity assays (Agilent) were performed separately for the 16 samples and for the final MBS library. The MBS library was quantified using the Kapa system and sequenced on a MiSeq (Illumina) instrument using the paired-end 300-bp protocol.
Demultiplexed raw reads were trimmed for adapter contamination and quality using the Trimmomatic (Bolger et al. 2014) (v. 0.32) and the same parameters used for RRBS data. FastQC software (Andrews 2010) (v. 0.11.4) was used for quality controls before and after trimming. The Bismark aligner (Krueger and Andrews 2011) (v. 0.14.4) was used for aligning the paired reads against the in silico converted genome using the following parameter: score_min L, 0,-0.6. Methylation was extracted using the bismark_methylation_extractor. The detailed statistics of the MBS libraries are found in supplementary table 7, Supplementary Material online.
Gene List Enrichment Analysis
The coding sequences of the European sea bass genome were used for annotation of transcripts and GO term (Ashburner et al. 2000) annotation using the go_enrichment (https://github.com/enormandeau/go_enrichment; last accessed July 05, 2019) suite. The nr and the Swissprot databases were downloaded from the blast databases of NCBI in August 2018. The sequences were blasted against the Swissprot database and annotated from Uniprot. The GOATOOLS python library (Klopfenstein et al. 2018) (v. 0.8.4) was used to test over- and under-representation of GO terms in gene lists using the GO database (August 3, 2018). The background set consisted of all annotated genes of the European sea bass genome (26,719 genes), which constitutes the most conservative approach for enrichment testing. Adjustment of P values was avoided since an FDR procedure can result in very conservative P values and interesting terms may be lost. Furthermore, an enrichment analysis includes many previous steps, so the sole number of GO terms may not be appropriate for multiple corrections, whereas the algorithms that take into account the terms’ topology perform dependent tests (Alexa and Rahnenfuhrer 2009). GO term enrichment was, nevertheless, performed by other means: using the GOATOOLS against a background set consisting only of feasible genes with RRBS (genes that within their gene body, 5-kb upstream the transcription start site or 5-kb downstream the 3′ UTR contained at least one CpG in the sample with the highest number of CpGs covered more than five times), with and without multiple correction and using the PANTHER tool (Mi et al. 2016) with human or zebrafish genes as reference with multiple testing correction. The chosen method was found to show overall consistency of the most important GO terms between pipelines without losing interesting information on gene set functions. For visualizations, REViGO software (Supek et al. 2011) was used to summarize the GO term lists by removing redundant GO terms. For further gene set enrichment analysis, the Enrichr tool (Chen et al. 2013; Kuleshov et al. 2016) was used, and results from the Reactome pathway database 2016 (Fabregat et al. 2018) and the Tissue expression database (Santos et al. 2015; Palasca et al. 2018) were presented.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Oscar Sagué and Àlex Lorente for assistance with the capture of the wild fish, Dr Antonio Mateos for providing the farmed fish, Dr Noelia Díaz and Sílvia Joly for help with the samplings, Dr Christoph Bock, Dr Matthias Farlik, Dr Paul Datlinger, and Dr Johanna Klughammer for training on RRBS libraries preparation, Dr Anna Esteve-Codina for advice on RNA-seq, Dr Diego Robledo for advice on SNP data, and Dr Lior David, Dr Robert H. Devlin, Dr Roderick N. Finn, Dr Ian Patten, and two anonymous reviewers for helpful comments on the manuscript. Funding: Research supported by the Spanish Ministry of Economy and Competitiveness (MINECO) grants “Epifarm” (ref. AGL2013–41047–R) and “Epimark” (ref. AGL2016–78710–R) to F.P. D.A. was supported by a PhD scholarship from the Spanish Government (BES–2011–044860) and an Epimark contract.
The RRBS and RNA-seq data that support the findings of this study have been deposited in the NCBI Gene Expression Omnibus and are available with the following accession codes: GSE104366 for muscle and testis of wild fish and GSE125124 for the rest of NGS libraries.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE.. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 1310:R87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J.. 2009. Gene set enrichment analysis with topGO. Bioconductor Improv, 27. https://bioconductor.org/packages/release/bioc/vignettes/topGO/inst/doc/topGO.pdf. last accessed July 05, 2019.
- Anastasiadi D, Esteve-Codina A, Piferrer F.. 2018. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet Chromatin 111:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadi D, Vandeputte M, Sánchez-Baizán N, Allal F, Piferrer F.. 2018. Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination. Epigenetics 139:988–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. last accessed July 05, 2019.
- Arechavala-Lopez P, Sanchez-Jerez P, Bayle-Sempere JT, Sfakianakis DG, Somarakis S.. 2012. Morphological differences between wild and farmed Mediterranean fish. Hydrobiologia 6791:217–231. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT.. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 251:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbucci M, Ferraresso S, Pauletto M, Franch R, Papetti C, Patarnello T, Carnier P, Bargelloni L.. 2016. An integrated genomic approach for the study of mandibular prognathism in the European seabass (Dicentrarchus labrax). Sci Rep. 61:38673.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev DK, Ruvinsky AO, Trut LN.. 1981. Inherited activation-inactivation of the star gene in foxes: its bearing on the problem of domestication. J Hered. 724:267–274. [DOI] [PubMed] [Google Scholar]
- Bertolini F, Geraci C, Schiavo G, Sardina MT, Chiofalo V, Fontanesi L.. 2016. Whole genome semiconductor based sequencing of farmed European sea bass (Dicentrarchus labrax) Mediterranean genetic stocks using a DNA pooling approach. Mar Genomics. 28:63–70. [DOI] [PubMed] [Google Scholar]
- Bicskei B, Taggart JB, Glover KA, Bron JE.. 2016. Comparing the transcriptomes of embryos from domesticated and wild Atlantic salmon (Salmo salar L.) stocks and examining factors that influence heritability of gene expression. Genet Sel Evol. 48:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Rubin C-J, Di Palma F, Albert FW, Alfoldi J, Barrio AM, Pielberg G, Rafati N, Sayyab S, Turner-Maier J, et al. 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 3456200:1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R, Diaz-Castillo C, Shoucri BM, Käch H, Leavitt R, Shioda T, Blumberg B.. 2017. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun. 81:2012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Miller BF, Furano AV.. 2014. Repair of naturally occurring mismatches can induce mutations in flanking DNA. eLife 3:e02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian L, Bahudhanapati H, Wei S.. 2013. Extracellular metalloproteinases in neural crest development and craniofacial morphogenesis. Crit Rev Biochem Mol Biol. 486:544–560. [DOI] [PubMed] [Google Scholar]
- Christie MR, Marine ML, Fox SE, French RA, Blouin MS.. 2016. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun. 71:10676.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossetti C, Lugini L, Astrologo L, Saggio I, Fais S, Spadafora C.. 2014. Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes. PLoS One 97:e101629.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin E, Pocheville A, Rey O, Pujol B, Blanchet S.. 2019. Epigenetically facilitated mutational assimilation: epigenetics as a hub within the inclusive evolutionary synthesis. Biol Rev. 941:259–282. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1868. The variation of animals and plants under domestication. London: J. Murray. [Google Scholar]
- Desanlis I, Felstead HL, Edwards DR, Wheeler GN.. 2018. ADAMTS9, a member of the ADAMTS family, in Xenopus development. Gene Expression Patterns 29:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanapally S, Ravikumar S, Jose AM.. 2015. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc Natl Acad Sci U S A. 1127:2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz N, Ribas L, Piferrer F.. 2013. The relationship between growth and sex differentiation in the European sea bass (Dicentrarchus labrax). Aquaculture 408–409:191–202. [Google Scholar]
- Eaton SA, Jayasooriah N, Buckland ME, Martin DI, Cropley JE, Suter CM.. 2015. Roll over Weismann: extracellular vesicles in the transgenerational transmission of environmental effects. Epigenomics 77:1165–1171. [DOI] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. 2018. The reactome pathway knowledgebase. Nucleic Acids Res. 46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Dolinoy DC.. 2011. Timing is everything. Epigenetics 67:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Conneely KN, Wu H.. 2014. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 428:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger AJ, Mahardja B, Fisch KM, Benjamin A, Lindberg J, Ellison L, Ghebremariam T, Hung T-C, May B.. 2018. A conservation hatchery population of delta smelt shows evidence of genetic adaptation to captivity after 9 generations. J Hered. 1096:689–699. [DOI] [PubMed] [Google Scholar]
- Gavery MR, Nichols KM, Goetz GW, Middleton MA, Swanson P.. 2018. Characterization of genetic and epigenetic variation in sperm and red blood cells from adult hatchery and natural-origin steelhead, Oncorhynchus mykiss. G3 (Bethesda) 811:3723–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, Malinverni R.. 2016. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 510:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tillo D, Vierstra J, Syed K-S, Deng C, Ray GJ, Stamatoyannopoulos J, FitzGerald PC, Vinson C.. 2015. Methylated cytosines mutate to transcription factor binding sites that drive tetrapod evolution. Genome Biol Evol. 711:3155–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horreo JL, Valiente AG, Ardura A, Blanco A, Garcia-Gonzalez C, Garcia-Vazquez E.. 2018. Nature versus nurture? Consequences of short captivity in early stages. Ecol Evol. 81:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosomichi K, Mitsunaga S, Nagasaki H, Inoue I.. 2014. A Bead-based Normalization for Uniform Sequencing depth (BeNUS) protocol for multi-samples sequencing exemplified by HLA-B. BMC Genomics. 151:645.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerslev LR, Donkin I, Fabre O, Versteyhe S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT, Barrès R.. 2018. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin Epigenet. 101:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang J-J, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. 2013. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 1534:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamstra JH, Hurem S, Martin LM, Lindeman LC, Legler J, Oughton D, Salbu B, Brede DA, Lyche JL, Aleström P.. 2018. Ionizing radiation induces transgenerational effects of DNA methylation in zebrafish. Sci Rep. 8: Article number: 15373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 124:357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Warwick Vesztrocy A, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, et al. 2018. GOATOOLS: a Python library for Gene Ontology analyses. Sci Rep. 81:10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht AL, Truong L, Marvel SW, Reif DM, Garcia A, Lu C, Simonich MT, Teeguarden JG, Tanguay RL.. 2017. Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol Appl Pharmacol. 329:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch IJ, Clark MM, Thompson MJ, Deere‐Machemer KA, Wang J, Duarte L, Gnanadesikan GE, McCoy EL, Rubbi L, Stahler DR, et al. 2016. The concerted impact of domestication and transposon insertions on methylation patterns between dogs and grey wolves. Mol Ecol. 25:1838–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR.. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2711:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukekova AV, Johnson JL, Xiang X, Feng S, Liu S, Rando HM, Kharlamova AV, Herbeck Y, Serdyukova NA, Xiong Z, et al. 2018. Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviours. Nat Ecol Evol. 29:1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Luyer J, Laporte M, Beacham TD, Kaukinen KH, Withler RE, Leong JS, Rondeau EB, Koop BF, Bernatchez L.. 2017. Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc Natl Acad Sci U S A. 11449:12964–12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Altmann J, Alberts SC, Tung J.. 2016. Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus). Mol Ecol. 258:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Tung J, Zhou X.. 2015. A flexible, efficient binomial mixed model for identifying differential DNA methylation in bisulfite sequencing data. PLoS Genet. 1111:e1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux S, Gourichon D, Leterrier C, Labrune Y, Coustham V, Rivière S, Zerjal T, Coville J-L, Morisson M, Minvielle F, et al. 2017. Embryonic environment and transgenerational effects in quail. Genet Sel Evol. 491:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L-C, Dahiya R.. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 1811:1427–1431. [DOI] [PubMed] [Google Scholar]
- Li S, Garrett-Bakelman FE, Akalin A, Zumbo P, Levine R, To BL, Lewis ID, Brown AL, D’Andrea RJ, Melnick A, et al. 2013. An optimized algorithm for detecting and annotating regional differential methylation. BMC Bioinformatics 14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 4110:e108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 307:923–930. [DOI] [PubMed] [Google Scholar]
- Liu Y, Siegmund KD, Laird PW, Berman BP.. 2012. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 137:R61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukovitis D, Ioannidi B, Chatziplis D, Kotoulas G, Magoulas A, Tsigenopoulos CS.. 2015. Loss of genetic variation in Greek hatchery populations of the European sea bass (Dicentrarchus labrax L.) as revealed by microsatellite DNA analysis. Medit Mar Sci. 161:197–200. [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 1512:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA.. 1989. Generalized linear models. London; New York. Chapman and Hall/ CRC Press. [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD.. 2016. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter K, Kotoulas G, Magoulas A, Mulero V, Sepulcre P, Figueras A, Novoa B, Sarropoulou E.. 2009. Evaluation of candidate reference genes for QPCR during ontogenesis and of immune-relevant tissues of European seabass (Dicentrarchus labrax). Comp Biochem Physiol B Biochem Mol Biol. 153B:340–347. [DOI] [PubMed] [Google Scholar]
- Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, Minx P, Hillier LW, Koboldt DC, Davis BW, et al. 2014. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A. 11148:17230–17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzitallab P, Baruah K, Vanrompay D, Bossier P.. 2019. Can epigenetics translate environmental cues into phenotypes? Sci Total Environ. 647:1281–1293. [DOI] [PubMed] [Google Scholar]
- Oey H, Whitelaw E.. 2014. On the meaning of the word ‘epimutation.’ Trends Genet. 3012:519–520. [DOI] [PubMed] [Google Scholar]
- O’Rourke T, Boeckx C.. 2018. Converging roles of glutamate receptors in domestication and prosociality. bioRxiv. 439869. Available from: https://www.biorxiv.org/content/10.1101/439869v1.
- Palasca O, Santos A, Stolte C, Gorodkin J, Jensen LJ.. 2018. TISSUES 2.0: an integrative web resource on mammalian tissue expression. Database (Oxford) 2018:bay003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton AL, Shen F, Taravella AM, Emery S, Veeramah KR, Boyko AR, Kidd JM.. 2018. Comparison of village dog and wolf genomes highlights the role of the neural crest in dog domestication. BMC Biol. 16:64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MF, Lehner B.. 2019. Intergenerational and transgenerational epigenetic inheritance in animals. Nat Cell Biol. 212:143.. [DOI] [PubMed] [Google Scholar]
- Pértille F, Da Silva VH, Johansson AM, Lindström T, Wright D, Coutinho LL, Jensen P, Guerrero-Bosagna C.. 2019. Mutation dynamics of CpG dinucleotides during a recent event of vertebrate diversification. Epigenetics 147:685–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR.. 2013. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 1534:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: https://www.R-project.org, last accessed July 05, 2019. [Google Scholar]
- Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL, et al. 2016. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci Rep. 61:31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D.. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 225:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team. 2015. RStudio: integrated development environment for R. Boston (MA): RStudio, Inc; Available from: http://www.rstudio.com/; last accessed July 05, 2019. [Google Scholar]
- Santos A, Tsafou K, Stolte C, Pletscher-Frankild S, O’Donoghue SI, Jensen LJ.. 2015. Comprehensive comparison of large-scale tissue expression datasets. PeerJ 3:e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Jónsson H, Chang D, Der Sarkissian C, Ermini L, Ginolhac A, Albrechtsen A, Dupanloup I, Foucal A, Petersen B, et al. 2014. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci U S A. 11152:E5661–E5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. 2017. Transgenerational epigenetics: integrating soma to germline communication with gametic inheritance. Mech Ageing Dev. 163:15–22. [DOI] [PubMed] [Google Scholar]
- Simó-Mirabet P, Felip A, Estensoro I, Martos-Sitcha JA, de las Heras V, Calduch-Giner J, Puyalto M, Karalazos V, Sitjà-Bobadilla A, Pérez-Sánchez J.. 2018. Impact of low fish meal and fish oil diets on the performance, sex steroid profile and male-female sex reversal of gilthead sea bream (Sparus aurata) over a three-year production cycle. Aquaculture 490:64–74. [Google Scholar]
- Skinner MK, Guerrero-Bosagna C, Haque MM.. 2015. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 108:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjærven KH, Jakt LM, Fernandes JMO, Dahl JA, Adam A-C, Klughammer J, Bock C, Espe M.. 2018. Parental micronutrient deficiency distorts liver DNA methylation and expression of lipid genes associated with a fatty-liver-like phenotype in offspring. Sci Rep. 81:3055.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD.. 2013. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 812:e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 67:e21800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tine M, Kuhl H, Gagnaire P-A, Louro B, Desmarais E, Martins RST, Hecht J, Knaust F, Belkhir K, Klages S, et al. 2014. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun. 51:5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trut L, Oskina I, Kharlamova A.. 2009. Animal evolution during domestication: the domesticated fox as a model. BioEssays 313:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymchuk W, Sakhrani D, Devlin R.. 2009. Domestication causes large-scale effects on gene expression in rainbow trout: analysis of muscle, liver and brain transcriptomes. Gen Comp Endocrinol. 164(2-3):175–183. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG.. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 4015:e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G. 2017. Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: insights from clonal, invasive, polyploid, and domesticated animals. Environ Epigenet. 31:dvx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Luyer JL, Cook D, Ritchie PA, Bernatchez L.. 2019. Domestication and temperature modulate gene expression signatures and growth in the Australasian snapper Chrysophrys auratus. G3 (Bethesda) 9:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AS. 2017. Revisiting two hypotheses on the “domestication syndrome” in light of genomic data. Vestn Vogis 214:435–442. [Google Scholar]
- Wilkins AS, Wrangham RW, Fitch WT.. 2014. The “Domestication Syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 1973:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreczycka K, Gosdschan A, Yusuf D, Grüning B, Assenov Y, Akalin A.. 2017. Strategies for analyzing bisulfite sequencing data. J Biotechnol. 261:105–115. [DOI] [PubMed] [Google Scholar]
- Xi Y, Bock C, Muller F, Sun D, Meissner A, Li W.. 2012. RRBSMAP: a fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing. Bioinformatics 283:430–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W.. 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 101:232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-Y, Latzel V, Fischer M, Bossdorf O.. 2018. Understanding the evolutionary potential of epigenetic variation: a comparison of heritable phenotypic variation in epiRILs, RILs, and natural ecotypes of Arabidopsis thaliana. Heredity. 1213:257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jia Y, Almeida P, Mank JE, van Tuinen M, Wang Q, Jiang Z, Chen Y, Zhan K, Hou S, et al. 2018. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. GigaScience 74:giy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai L-Y, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 5007463:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.