Abstract
Cartilage homeostasis is governed by articular chondrocytes via their ability to modulate extracellular matrix production and degradation. In turn, chondrocyte activity is regulated by growth factors such as those of the transforming growth factor β (TGFβ) family. Members of this family include the TGFβs, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs). Signaling by this protein family uniquely activates SMAD-dependent signaling and transcription but also activates SMAD-independent signaling via MAPKs such as ERK and TAK1. This review will address the pivotal role of the TGFβ family in cartilage biology by listing several TGFβ family members and describing their signaling and importance for cartilage maintenance. In addition, it is discussed how (pathological) processes such as aging, mechanical stress, and inflammation contribute to altered TGFβ family signaling, leading to disturbed cartilage metabolism and disease.
Keywords: transforming growth factor β, bone morphogenetic proteins, osteoarthritis, cartilage, SMADs, aging, joint loading, inflammation, linker modifications
1. Introduction
The transforming growth factor β (TGFβ) family of polypeptide growth factors controls development and homeostasis of many tissues, including articular cartilage. Articular cartilage is the connective tissue covering joint surfaces and is a type of hyaline cartilage. This tissue is key in facilitating movement with its smooth lubricated surface, and it functions as a shock absorber to disperse forces acting upon movement with its physical properties. Articular cartilage is of mesodermal origin and, remarkably for a tissue, is neither innervated nor does it contain blood vessels to supply oxygen or nutrients [1,2]. The only cell type present in this tissue is the chondrocyte, which is surrounded by a large amount (up to 98% of cartilage volume) of extracellular matrix (ECM) [1,3]. The ECM is a highly organized network of hyaluronan, proteoglycans, and collagens. These collagens provide (tensile) strength and shape to the tissue [4], and they mainly are (up to 90%) collagen type II (COL2A1) fibrils [5]. Hyaluronan is a glycosaminoglycan (GAG) (i.e., a long, unbranched polysaccharide), which forms spontaneously aggregating networks to which other biomolecules such as proteoglycans can bind [6]. Proteoglycans are proteins glycosylated with sulfated GAGs. The most abundant proteoglycan (in mass) in articular cartilage is aggrecan (ACAN), and this proteoglycan forms large aggregates with hyaluronan. Because ACAN is heavily glycosylated with negatively charged sulfated GAGs, these aggregates generate a (static) charge density. This charge density generates the force required to counteract compressive forces during movement via attraction of solutes, which generate osmotic swelling pressure, and via electrostatic repulsion if aggrecan molecules come in close proximity [7].
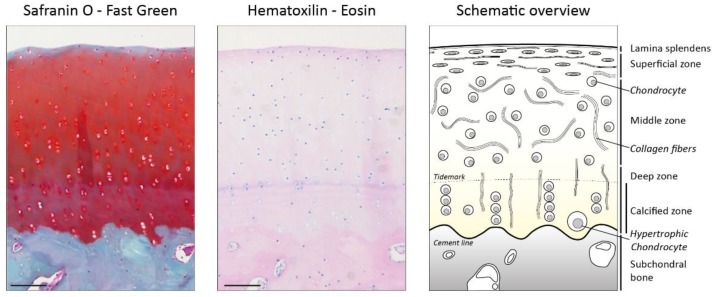
Articular cartilage is a highly structured tissue in which form fits function. Five distinct zones can be observed: the lamina splendens, the superficial zone, the middle zone, the deep zone, and the calcified zone (Figure 1). The lamina splendens consists of collagen fibrils oriented in parallel to the surface and high levels of hyaluronan and the lubricating proteoglycan lubricin (PRG4), but it contains no cells [8,9,10]. These characteristics provide a smooth surface with a low friction coefficient. Below the lamina splendens lies the superficial zone, which contains a relatively high number of cells with flattened morphology oriented in parallel to the surface, which produce high amounts of hyaluronan and PRG4 [8,10]. The collagen fibrils here are tightly packed and also oriented in parallel to the surface. This orientation counteracts osmotic swelling pressure of the tissue below [9]. Beneath the superficial zone lies the middle zone, which represents 40%–60% of articular cartilage [2]. Chondrocytes in this zone are rounded but sparse. The collagen fibers are oriented isotropically to the cartilage surface [9], which allows for efficient collapse of the tissue upon compression to dissipate the energy of the impact. The collapse of this layer is accompanied by displacement and loss of water. The more this layer is compacted, the more resistance to further collapse is increased [1]. This resistance to further compression is due to an increase in electrostatic repulsion between proteoglycans and also to an increase in hyaluronan viscosity because of an increase in concentration of this molecule [11]. Underneath the middle zone lies the deep zone (±30%). In the deep zone, chondrocytes are organized in columns placed between collagen fibrils oriented perpendicular to the cartilage surface [6,9]. Furthermore, this zone contains relatively the highest levels of aggrecan of all zones [1]. The high levels of aggrecan restrict water flow and thereby increase the compressive stiffness of cartilage. The collagen fibers are oriented to withstand the swelling pressure generated by these proteoglycans and to withstand sheer stresses generated by compression of the layers above. Part of the deep zone, below the tidemark, contains calcium salts in the ECM and is, thus, called the calcified zone [2]. The calcified zone forms the interface between bone and cartilage and anchors cartilage to the bone. This zone contains hypertrophic-like chondrocytes which resemble chondrocytes undergoing endochondral ossification [12].
Figure 1.
Appearance and structure of adult (bovine) articular cartilage. Note that the orientation of collagen fibrils differs per zone, from parallel in the superficial zone to perpendicular in the deep zone. Scale bar = 100 µm.
The importance of articular cartilage for joint function is illustrated by the effects of its loss, which leads to disability and pain, for example, in the word’s most common joint disease osteoarthritis (OA). Degeneration and loss of articular cartilage are the consequence of an imbalance in anabolic (e.g., ECM production) and catabolic (e.g., ECM degradation) processes. Chondrocytes, as the only cell type present, are essential for balancing these processes. Therefore, chondrocyte metabolism is a key regulator of cartilage homeostasis and joint health. Many factors can regulate chondrocyte homeostasis, like hydrostatic pressure, tensile strain, and proinflammatory cytokines, but various growth factors, including those of the TGFβ family, play a key role in this. Growth factors modulate chondrocyte metabolism, differentiation, proliferation, and survival, and they regulate ECM production and turnover. This review will address the pivotal role of the TGFβ family in chondrocyte (and thus cartilage) biology by listing several TGFβ family members and describing their importance. We will discuss how these members signal and how this signaling impacts cartilage. Furthermore, we will address how (pathological) processes such as aging, mechanical stress, and inflammation influence TGFβ signaling in chondrocytes, leading to an altered metabolism and disease.
2. The Transforming Growth Factor Β (TGFβ) Family and Its Signaling
The TGFβ family consists of over 30 members, including the TGFβs, activins, bone morphogenetic proteins (BMPs), and growth/differentiation factors (GDFs). These factors share a characteristic quaternary dimer structure [13] and signal via formation of heteromeric complexes between two types (type I and type II) of serine threonine-protein kinase receptors [14].
There are seven type I and five type II receptors (Figure 2). Because every growth factor recruits a specific, but not necessarily unique, receptor complex, this choice in receptors allows for differences in growth factor sensitivity. In addition, multiple type III receptors have been characterized that facilitate the interaction between ligand and receptor and can stabilize receptor complexes. For example, for TGFβ betaglycan [15,16] and endoglin [17], and for BMPs the repulsive guidance molecule (RGM) family members, RGM domain family member A (RGMA), dragon, and hemojuvelin have been identified as co-receptors [18]. Also, inhibitory (pseudo) receptors have been identified, which bind ligands but do not initiate signaling (e.g., CD109 for TGFβ signaling [19] and BMP and activin membrane bound inhibitor (BAMBI) for BMP and activin signaling [20]). After formation of functional receptor complexes, TGFβ family members can activate multiple intracellular signaling pathways, which are categorized as either receptor SMAD (R-SMAD)-dependent or -independent, but R-SMAD-dependent signaling is regarded as the unique and canonical signaling pathway of TGFβ family members.
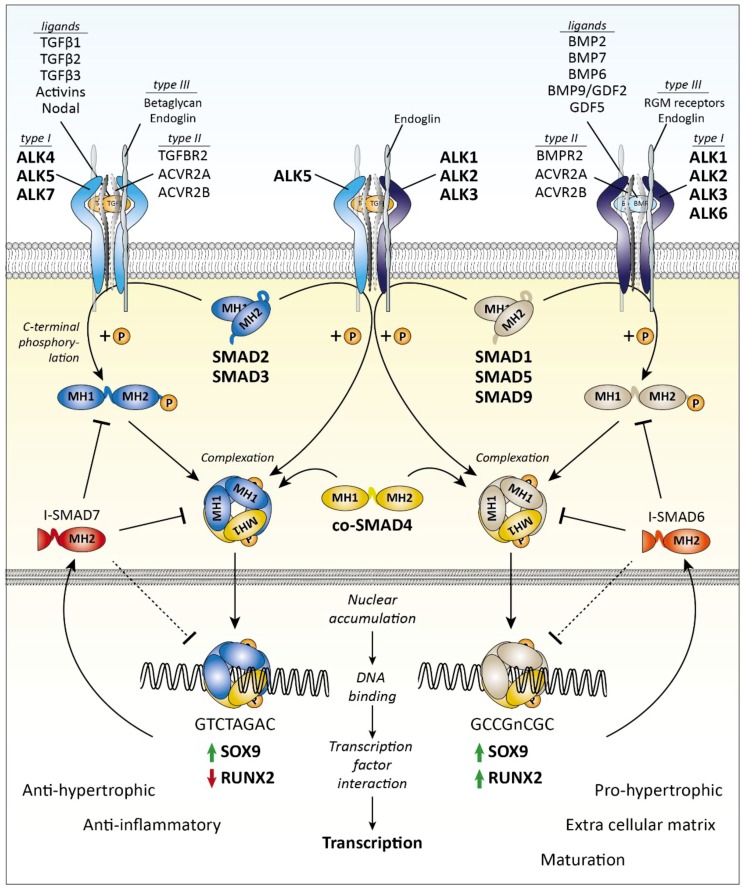
Figure 2.
Transforming growth factor β (TGFβ) family SMAD-dependent signaling. Ligand binding induces the formation of a receptor complex containing a type I and type II receptors. Type III receptors stabilize and facilitate the binding of TGFβ to the receptor complex, which recruits a receptor SMAD (R-SMAD). The R-SMAD is subsequently phosphorylated on its C-terminal domain, and a complex is formed with the common SMAD4 (co-SMAD4). This complex translocates to the nucleus where it can bind transcription factors, like SOX9 and RUNX2, and activates transcription. The activation of SMAD2/3 signaling results in anti-hypertrophic and anti-inflammatory function, whereas activation of SMAD1/5/9 signaling is associated with pro-hypertrophic regulation of the extracellular matrix and maturation of the cartilage. Other important target genes are the inhibitory SMADs, SMAD6 and SMAD7, whose expression provides the cell with a negative feedback mechanism. ACVR2 = Activin type-2 receptor, ALK = ALK tyrosine kinase receptor, BMP = Bone morphogenetic protein, BMPR = Bone morphogenetic protein receptor, GDF2 = Growth differentiation factor 2, MH(1/2) = MAD homology (1/2) domain, RGM = Repulsive guidance molecule, RUNX2 = Runt-related transcription factor 2, SOX9 = Transcription factor SOX9, TGFBR = TGFβ receptor
2.1. R-SMAD-Dependent Signaling
R-SMADs are proteins approximately 50 kDa in size that, upon receptor-mediated activation by carboxy terminal phosphorylation, migrate to the nucleus and function as transcription factors. Two groups of R-SMADs can be distinguished based upon structural and functional homology: one group consisting of SMAD1, SMAD5, and SMAD9 (also known as SMAD8) and one group containing SMAD2 and SMAD3 [14]. Upon receptor type I activation, these R-SMADs are directly phosphorylated on two serines at their C-terminal SXS motif. Specific ALKs phosphorylate specific SMADs: ALK1, 2, 3, and 6 phosphorylate SMAD1, 5, and 9, whereas ALK4, 5, and 7 phosphorylate SMAD2 and SMAD3. This phosphorylation facilitates complexation of R-SMADs with the common SMAD (co-SMAD), SMAD4, which promotes nuclear entry and accumulation. Inside the nucleus, this SMAD complex directly binds DNA, although with low affinity, and recruits transcription factors for \high-affinity interactions with DNA. The location where SMAD complexes bind DNA is dependent on characteristic SMAD-binding motifs present in the DNA sequence, which differ for SMAD3/4 and SMAD1/5/9 (e.g., GTCTAGAC for SMAD2/3/4 complexes [21] and GCCGnCGC for SMAD1/5/9/4 complexes [22]). Furthermore, the interaction between SMADs and DNA is regulated by the presence of master regulator transcription factors [23]. Master regulators vary per tissue, and an important master regulator of chondrocyte phenotype is SRY-box9 (SOX9), for example, which closely interacts with SMAD3/4 on the collagen type II promotor [24].
Two important target genes of R-SMAD signaling are the inhibitory-SMADs (I-SMAD): SMAD6 and SMAD7 [25,26]. These I-SMADs function as inhibitors of R-SMAD signaling on multiple levels by inhibiting C-terminal R-SMAD phosphorylation by inducing receptor dephosphorylation and degradation, by inhibiting R-SMAD DNA binding, and by inhibiting complex formation between SMAD4 and R-SMADs. In this way, SMAD6 predominantly inhibits SMAD1/5/9, and SMAD7 predominantly inhibits SMAD2/3 signaling, and both I-SMADs provide cells with an important negative feedback mechanism to inactivate R-SMAD signaling.
A (simplified) overview of the SMAD-dependent signaling pathway is depicted in Figure 2.
2.2. SMAD-Dependent Signaling in Chondrocytes and Cartilage Biology
SMAD signaling is essential for the formation and maintenance of healthy cartilage. Both in vivo and in vitro experiments point towards essential, but distinct, roles for SMAD2/3 versus SMAD1/5/9 signaling in chondrocyte biology.
In humans, genetic variation in SMAD3 is associated with hip and knee OA and total burden of the disease [27,28]. Dominant nonsense, missense, or frameshift mutations in SMAD3 lead to aneurysms-OA syndrome, a severe condition characterized by cardiovascular anomalies and early onset of OA in multiple joints [29,30]. Mice with a Smad3 null mutation (Smad3Δexon8/Δexon8) develop normal articular cartilage, but this starts to rapidly degrade one month after birth, resulting in severe OA [31]. This rapid degradation is linked to an increased number of hypertrophic chondrocytes present in both articular and epiphyseal cartilage [31]. These hypertrophic chondrocytes are unable to maintain a healthy articular cartilage matrix and produce collagen type 10 (COL10A1) and matrix metalloprotease 13 (MMP13), a potent cartilage-degrading enzyme. Increased chondrocyte hypertrophy and accelerated cartilage degeneration are also observed in chondrocyte-specific Smad3-null mice (Smad3Δexon2 + 3/Δexon2 + 3), which have been made using Col2-Cre and floxed Smad3 to circumvent systemic effects of Smad3 deletion, confirming the direct role of SMAD3 in these processes [32]. In vitro studies using overexpression or knockout of SMAD3 also support a role for SMAD3 in inhibiting chondrocyte terminal differentiation, by showing an inhibitory or stimulating effect on chondrocyte hypertrophy, respectively [33,34,35,36]. The anti-hypertrophic effect of SMAD3 is attributed to its interaction with core-binding factor subunit α 1 (CBFA1), also known as runt-related transcription factor 2 (RUNX2) [32,37,38]. RUNX2 is a potent regulator of chondrocyte maturation [39]. For example, overexpression of Runx2 in the murine teratoma-derived ATDC5 chondrocyte-like cell line induces chondrocyte hypertrophy, whereas inhibition of RUNX2 achieves the opposite [32,38,40,41]. SMAD3 counteracts RUNX2 function by directly binding this transcription factor and recruiting silencing histone class II deacetylases to RUNX2 responsive genes [37] such as MMP13, COL10A1, and alkaline phosphatase (ALPL).
The role of SMAD2 in cartilage biology is less clear than that of SMAD3. In humans, there is no known association between genetic variation in SMAD2 and OA. Smad2 knockout animals or hypomorphs are not viable and die early during embryogenesis (≤embryonic day 9 [42,43,44,45]), before the formation of articular joints [46], and are thus not usable to study the role of SMAD2 in articular cartilage. Animals with only one genetic copy of Smad2 (Smad2+/−) can be viable [42,47], but no cartilage phenotype has been reported. Cartilage-specific Smad2 knockout mice (Smad2fx/fx;Col2a1Cre) have recently been studied [48], and such mice have an elongated hypertrophic zone in their growth plate. This last observation indicates a similar anti-hypertrophic effect of SMAD2 as SMAD3. An in vitro study supports a role for SMAD2 similar to that of SMAD3, because overexpression of a dominant negative variant of Smad2 enhances, whereas overexpression of wildtype Smad2 protects against chondrocyte hypertrophy, although less potent than SMAD3 [33].
SMAD1 and SMAD5 seem to have an important, but interchangeable, role in cartilage biology. In contrast, SMAD9 seems to be of little importance for cartilage formation or maintenance. In humans, no association has been reported between genetic variations in SMAD1, SMAD5, or SMAD9 and OA, possibly due to redundant roles in cartilage. Like Smad2 knockout mice, Smad1 [49] or Smad5 [50] knockout mice die before the formation of articular joints. Knockout of Smad9 is not embryonically lethal yet does not seem to affect (epiphyseal) cartilage biology [51]. To study SMAD1 and SMAD5 in chondrogenesis, cartilage-specific knockouts have been made using Col2-cre and floxed Smad1 or Smad5 [51]. Cartilage-specific removal of either Smad1 or Smad5 does not result in a phenotype, nor in combination with whole body Smad9 knockout. Only when both Smad1 and Smad5 are ablated, severe chondrodysplasia can be observed [51]. Additional removal of Smad9 does not add much to this severe phenotype. Together, these observations indicate that SMAD1 and SMAD5 have an important, but redundant, role in cartilage formation, whereas the role of SMAD9 seems limited [51]. The severe chondrodysplasia is, in part, due to a lack of hypertrophic chondrocytes in the growth plates, suggesting that SMAD1/5 signaling regulates chondrocyte maturation. In vitro studies support a stimulatory role of SMAD1/5 in chondrocyte maturation. For example, overexpression of Smad6, an inhibitor of SMAD1/5/9, results in inhibited chondrogenesis in the ATDC5 cell line [52] and delayed chondrocyte hypertrophy [53]. The stimulatory effects of SMAD1/5 on chondrocyte maturation most likely are due to a stimulation of RUNX2 function. Such a stimulatory role of SMAD1/5 on RUNX2 function has been described to be essential for BMP2-induced osteoblast differentiation [54,55,56,57]. Furthermore, in pre-hypertrophic chondrocytes, SMAD1/5-RUNX2 interaction induces the activation of a reporter construct based on the Col10a1 promoter, the expression of which is an important phenotypic marker of hypertrophy [38,58]. How exactly SMAD1 and SMAD5 stimulate RUNX2 function is yet unclear.
2.3. SMAD-Independent Signaling
As mentioned, TGFβ family members can also activate R-SMAD-independent pathways. Two examples are signaling via mitogen-activated protein kinase (MAPK) pathways involving TGFβ-activated kinase 1 (TAK1) or extracellular signal-regulated kinase 1 and 2 (ERK1/2, also known as MAPK3/1). The SMAD-independent signaling pathways are depicted in Figure 2. Of note, these pathways are not unique for TGFβ family signaling but are important signaling mediators of many other growth factors and cytokines as well.
Activation of MAPKs by TGFβ family members relies on adaptor proteins to bridge receptor activity to pathway activation—for example, SHC adaptor protein 1 (SHC1) for ERK [59] and tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) for TAK1 activation [60,61]. Receptor-mediated phosphorylation of SHC1 triggers a MAPK signaling cascade via GRB2/SOS, RAS, RAF, and MAP2K1, resulting in activation of ERK1/2 [59]. TRAF6 is activated differently: upon receptor type I/type II complex formation, TRAF6 oligomerization and auto-ubiquitination occurs, triggering complex formation between TAK1 and TGFβ-activated kinase 1/MAP3K7 binding protein 1 (TAB1) via poly-ubiquitination, which allows TAK1 auto-phosphorylation and activity [61]. Alternatively, TAK1 can be activated by E3 ubiquitin-protein ligase XIAP (in complex with TAB1), the expression of which is stabilized by interaction with BMP (and TGFβ) receptors [62,63]. Ultimately, activation of MAPK signaling by TGFβ family members results in activation of JNK and p38 kinases, which directly regulate gene expression by modulating transcription factors such as Jun proto-oncogene (c-JUN), MYC proto-oncogene (c-MYC), activating transcription factor 2 (ATF2), and CCAAT/enhancer-binding protein β (C/EBP-β) [64].
Both ERK and TAK1 contribute to TGFβ family signaling in chondrocytes. Functional inhibition of ERK1 in ATDC5 cells via inhibition of its activating kinase MAP2K1 with the inhibitor U0126 reduces TGFβ-induced Acan expression [65]. Functional inhibition of ERK1 in rat chondrocytes with the use of the MAP2K1 inhibitor PD98059 reduces TGFβ-induced proliferation and Acan and Col2a1 expression [66,67]. Furthermore, in both human and bovine primary chondrocytes, use of PD98059 lowers TGFβ-induced mRNA and protein expression of the metallopeptidase inhibitor TIMP3 [68], indicating that ERK activity is a cartilage-protective component of TGFβ signaling in mature chondrocytes. In contrast, during TGFβ-induced chondrogenesis in vitro, use of PD98059 increases Col2a1 and Sox9 expression and proteoglycan synthesis in rat, chicken, and human mesenchymal stem cells [69,70,71], indicating that ERK1/2 is an inhibitory component of TGFβ signaling during TGFβ-induced chondrogenesis.
Functional inhibition of TAK1 using a dominant negative form of Tak1 greatly diminishes the ability of TGFβ and BMP2 to induce Col2a1 expression in rabbit articular chondrocytes [72]. In Col2-Cre;Tak1fl/fl mice, Tak1 deletion in Col2a1-expressing cells during embryogenesis results in runting and severe chondrodysplasia because of the reduced chondrocyte proliferation and delayed maturation [73], and these mice die shortly before [73] or after birth [74]. This severe phenotype resembles the phenotype of mice lacking BMP receptor/SMAD(1/5) signaling. Indeed, these mice show impaired BMP-induced R-SMAD and p38 MAPK/JNK/ERK signaling [73,74], confirming that TAK1 is an important component of BMP signaling in cartilage. Postnatal deletion of Tak1 using inducible Col2a1-CreERT2;Tak1fl/fl mice also results in growth retardation, and these mice have profoundly decreased proteoglycan and COL2A1 production in their articular cartilage [75]. This last observation shows that the importance of TAK1 in cartilage biology is not limited to embryogenesis.
A possible role of TAK1 signaling in chondrocytes is the regulation of SOX9 production. In chondrocytes, deletion of Tak1 inhibits endogenous and BMP2-induced Sox9 mRNA expression and protein production, whereas overexpression of Tak1 enhances SOX9 levels [75]. This observation is reflected in vivo: both articular and epiphyseal chondrocytes from Col2a1-CreERT2;Tak1fl/fl mice show lower SOX9 protein production after deletion of Tak1 compared to control animals [75]. The induction of Sox9 expression by TAK1 involves binding of activating transcription factor 2 (ATF2), a downstream target of TAK1 via p38 MAPK, to the Sox9 promoter [75]. Furthermore, p38 MAPK signaling has also been shown to stabilize SOX9 mRNA in human chondrocytes, further strengthening the importance of this MAPK pathway in SOX9 regulation [76].
TGFβ family members can also signal via phosphoinositide 3-kinase (PI3K) or via the GTPase RhoA [64], but it is unclear how and to what extent this occurs in chondrocytes. Activation of PI3Ks leads to phosphorylation and activation of AKT serine/threonine kinase 1/2/3 (AKT1/2/3) and of mechanistic target of rapamycin (MTOR) [77]. Recently, it has been shown that mechanistic target of rapamycin complex 1 (mTORC1) activity is required for chondroblast growth and proliferation in the epiphyseal plate of mice, but that its inactivation is required for chondrocyte differentiation [78]. In articular cartilage, MTOR is upregulated during OA [79] and is an inhibitor of chondroprotective autophagy [79,80,81]. In vivo, intra-articular or intra-peritoneal injection of rapamycin (i.e., an inhibitor of MTOR) increases autophagy and reduces cartilage damage in the DMM (surgical destabilization of the medial meniscus) model of OA [80,81]. Furthermore, cartilage-specific Mtor knockout mice (Col2-rt-TA-Cre; Mtorfl/fl) are protected against DMM-induced OA [79], confirming that MTOR signaling is deleterious in OA conditions.
In chondrocytes, RhoA activates rho-associated coiled-coil containing protein kinase 1 (ROCK1) [82,83]. In primary human chondrocytes, ROCK inhibition induces SOX9, COL2A1, and ACAN mRNA expressions [76], while in primary murine chondrocytes, ROCK inhibition enhances the output of a SOX9-responsive luciferase assay along with Col2a1 and Acan mRNA expression [83]. Therefore, these studies indicate a negative effect of RhoA/ROCK signaling on chondrocyte phenotype. However, in murine mesenchymal cells during chondrogenic differentiation and chondrocytes cultured in 3D, ROCK1 inhibition negatively affects SOX9 function [83]. This effect has been attributed to direct phosphorylation of SOX9 by ROCK1, leading to nuclear accumulation and transcriptional activity of SOX9 [84]; thus, in contrast to the previous studies, this indicates a positive effect of Rhoa/ROCK signaling on chondrocyte phenotype.
A (simplified) overview of the SMAD-independent signaling pathways is depicted in Figure 3.
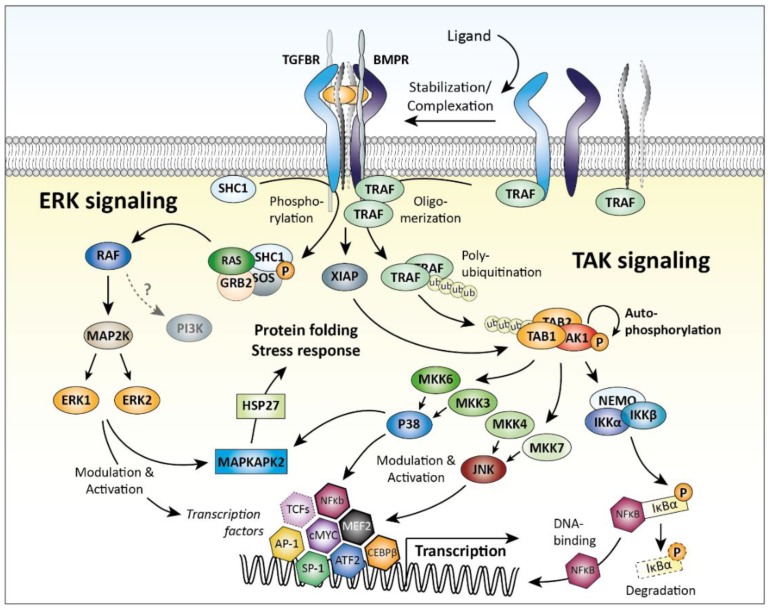
Figure 3.
Transforming growth factor β (TGFβ) family SMAD-independent signaling. Ligand binding induces the formation of a receptor complex. This complex can initiate SMAD-independent signaling via activation of adaptor proteins, e.g., SHC1 and TRAF6. This activation results in initiation of the ERK and TAK signaling cascades, culminating in the activation of MAPKAPK2, p38, JNK, and NFκb. These pathways induce transcription and translation and regulate diverse cellular processes. AKT = RAC-alpha serine/threonine-protein kinase, AP-1 = Activator protein 1, ATF2 = Cyclic AMP-dependent transcription factor ATF-2, CEBPβ = CCAAT/enhancer-binding protein β, ERK = Mitogen-activated protein kinase 3, GRB2 = Growth factor receptor-bound protein 2, HSP27 = Heat shock protein beta-1, IkBa = NF-kappa-B inhibitor alpha, IKK = Inhibitor of nuclear factor kappa-B kinase, JNK = c-Jun N-terminal kinase, MAP2K = Dual specificity mitogen-activated protein kinase kinase 1, MAPKAPK = MAP kinase-activated protein kinase, MEF2 = Myocyte-specific enhancer factor 2A, MKK = Dual specificity mitogen-activated protein kinase kinase, cMYC = Myc proto-oncogene protein, NEMO = NF-kappa-B essential modulator, NFκb = nuclear factor kappa-light-chain-enhancer of activated B cells, p38 = p38 mitogen-activated protein kinases, PI3K = Phosphatidylinositol 3-kinase, RAF = RAF proto-oncogene serine/threonine-protein kinase, RAS = GTPase HRas, SHC1 = SHC-transforming protein 1, SOS = Son of sevenless homolog 1, SP-1 = Sp1 transcription factor, TAB = TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, TAK1 = TGFβ-activated kinase 1, TCFs = ternary complex factors, TRAF6 = TNF receptor-associated factor 6, and XIAP = E3 ubiquitin-protein ligase XIAP.
3. TGFβ Family Members in Cartilage
3.1. TGFβ1, TGFβ2, and TGFβ3
The importance of TGFβ in cartilage biology was first observed in 1985 when two proteins were extracted from bone that were capable of inducing chondrogenesis in rat embryonic mesenchymal cells, and therefore were called cartilage inducing factor A and B [85]. Later research revealed that these proteins were identical to TGFβ1 and TGFβ2 [86,87].
In mammals, three TGFβs exist—TGFβ1, TGFβ2, and TGFβ3—encoded by three different genes—TGFB1, TGFB2 and TGFB3—and these homologs share a high percentage of amino acid identity (71%–79%). All three TGFβs are produced in inactive forms as homodimers (or more rarely as heterodimers [88]) bound by latency-associated peptide (LAP) and latent TGFβ binding protein (LTBP) [89]. This production in the inactive form separates secretion from activity, an important concept in TGFβ biology. All three forms of TGFβ are produced by chondrocytes [90,91,92] and a large amount (60–200 ng/mL) of inactive TGFβ is bound to the ECM of cartilage [93,94]. Because TGFβ is produced in the inactive form, activation is a crucial step in TGFβ signaling. For this, the noncovalent bond between LAP and TGFβ has to be disrupted. This can occur enzymatically by degradation of LAP by, for example, MMP3 [95], or nonenzymatically by a conformational change in the tertiary structure of LAP. Shearing stress [96] or chemical modification of LAP by reactive oxygen species [97] can induce such a conformational change. In addition, mechanical force as produced by compressive loading was also shown to be an important physiological activator of ECM-bound TGFβ in cartilage [98].
In chondrocytes, TGFβ induces both SMAD2/3 and SMAD1/5/9-dependent signaling via ALK5 and ALK1, respectively, in addition to SMAD-independent signaling [99,100]. Which pathway is activated by TGFβ depends on receptor level expression, co-receptor expression (i.e., endoglin enhances signaling via ALK1), and dose of TGFβ. In chondrocytes, a low dose of TGFβ predominantly signals via pSMAD2/3, whereas at high dosages, pSMAD1/5 signaling becomes more pronounced. Importantly, both pathways have been described to antagonize each other in chondrocytes [99,100].
TGFβ signaling is associated with cartilage ECM production and maintenance. In mice, a single intra-articular injection of rhTGFβ1 or rhTGFβ2 increases proteoglycan synthesis twofold after four days, as measured by 35S-sulfate incorporation in patellar articular cartilage [101,102]. Also in vitro, TGFβ enhances proteoglycan production in both chondrocytes and cartilage explants on both mRNA and protein level [103,104,105,106], together with production of other ECM components like COL2A1 [106,107], cartilage oligomeric matrix protein (COMP) [108,109], perlecan [110], fibronectin [111], and hyaluronan [112]. Furthermore, TGFβ is a potent inducer of lubricin (PRG4) in chondrocytes [113], a key lubricating component of synovial fluid. However, in contrast to the previously mentioned studies, inhibitory effects of TGFβ on chondrocyte proliferation, proteoglycan synthesis, and COL2A1 production have also been described both in vitro and in vivo [107,114,115,116,117,118,119]. Possibly, different receptor signals play a role in these apparently contradictory observations.
Furthermore, TGFβ signaling has important anti-inflammatory effects in cartilage. TGFβ1 counteracts proinflammatory IL-1 signaling in vivo [120,121,122] and in vitro [123], and it helps cartilage proteoglycan content recover after inflammation induced depletion [90,124]. How TGFβ exerts its anti-inflammatory effects is not fully elucidated yet, but in primary rabbit articular chondrocytes, both TGFβ1 and TGFβ3 downregulate IL-1 receptor (IL1R1) expression approximately 50% on protein and mRNA level [123,125]. Furthermore, proinflammatory IL-6 signaling is also counteracted by TGFβ by downregulation of IL-6 receptor levels in human and bovine chondrocytes [126]. In macrophages, TGFβ signaling can also stabilize NFκB inhibitor alpha (NFKBIA, also known as IκBα), which is a potent inhibitor of NFκB, but this mechanism has not been described for chondrocytes yet [127]. Another possible mechanism is the competition between SMAD3 and NFκB for co-activating transcription factors like CREB binding protein, as has been described in endothelial cells [128]. Additionally, many inflammatory genes counteracted by TGFβ contain both SMAD3 and NFκb binding sites, leaving room for epigenetic regulation as a mechanism.
An especially important role for TGFβ signaling lies in regulation of chondrocyte hypertrophy. TGFβ-induced pSMAD2/3 signaling blocks hypertrophy and terminal differentiation of chondrocytes [31,33,35,100,129]. In contrast, TGFβ-induced pSMAD1/5 via ALK1 is associated with chondrocyte hypertrophy [99,100]. Both effects of TGFβ on hypertrophy can be observed in vivo. Removal of TGFβ signaling in cartilage of two-week-old Col2-CreER;Tgfbr2flox/flox mice results in an OA-like phenotype within 6 months via RUNX2-mediated Mmp13 and Adamts5 expression [130]. Furthermore, expression of a kinase-deficient dominant negative form of Tgfbr2 in mice induces chondrocyte hypertrophy and osteoarthritis [131]. In contrast, removal of TGFβ signaling in the hypertrophic chondrocytes of Col10a1-Cre;Tgfbr2flox/flox mice delays terminal differentiation in the epiphyseal plate [132], and removal of TGFβ signaling in cartilage of eight-week-old AgcCreERT2+/−;Tgfbr2flox/flox mice results in less hypertrophic chondrocytes after 12 months [133]. Because efficient removal of TGFBR2 was demonstrated in both the Col2-CreER;Tgfbr2flox/flox and AgcCreERT2+/−;Tgfbr2flox/flox models, their different outcomes illustrate that age of TGFβ signaling depletion (2 vs 8 weeks) and, thus, chondrocyte maturation status greatly affect the outcome and impact of TGFβ signaling in chondrocytes.
Other family members which can induce (chondro-protective) SMAD2/3 signaling in chondrocytes are the activins. However, little is known regarding their role and importance. Activin A, activin AB, and activin B signal via ACVR2A/ACVR2B and ALK4/ALK7 to induce SMAD2/3 [134]. Expression of the activin A subunit inhibin β A (INHBA) and activin A itself are upregulated in OA cartilage [135,136] but also expression of its inhibitor follistatin is upregulated [137,138]. A possible role, which has been demonstrated in vitro, is the inhibition of IL-1-induced ADAMTS4/5 activity [136], which would indicate an anti-inflammatory role similar to the TGFβs. However, a lack of response of adult cartilage tissue to activin A has been observed in multiple studies [109,139,140]; for example, activin A slightly induces proteoglycan synthesis (i.e., 35S-incorporation) in immature (1–3 months old) but not in mature (>18 months old) bovine articular cartilage explants [140]. Even less is known regarding the role in cartilage of the activin antagonists, the inhibins, and only one study reports that inhibin inhibits both proteoglycan synthesis (i.e., 35S-incorporation) and DNA synthesis in bovine chondrocytes [140], which would indicate that activin signaling contributes to both these processes, again similar to the TGFβs.
3.2. Bone Morphogenetic Proteins (BMPs)
Multiple BMPs play an important role in chondrocyte biology, including BMP2, BMP7 (OP1), BMP4, BMP6, BMP9 (GDF2), and BMP14 (GDF5) [141,142]. Chondrocytes express the BMP type I receptors ALK1, ALK2, ALK3, and ALK6 and the type II receptors BMPR2, ACVR2A, and ACVR2B. Genetic variation in either of these factors is not yet associated with OA. Like the TGFβs, BMPs are produced as a dimer, but not only as homodimers; heterodimers of BMP2/6, BMP2/7, and BMP4/7 have been described both in vitro and in vivo [143]. In contrast to TGFβ activity, BMP activity is not thought to be controlled via reassembly of BMPs with their prodomain to form a latency complex [143]. BMPs do reassemble with their prodomain to facilitate ECM binding, but this does not seem to confer latency because it does not impair receptor type I and II binding [89,143,144]. Instead, antagonistic scavenger proteins (e.g., gremlin, noggin, and sclerostin) and decoy receptors like BAMBI play an important role in modulating BMP activity, all of which are produced by chondrocytes. Additionally, BMPs can antagonize each other via competition for receptor type II binding [145].
In both healthy and OA cartilage, Bmp2 mRNA expression can be detected [146]. BMP2 signaling induces ECM production and proliferation. In ex vivo juvenile cartilage, BMP2 induces proteoglycan content and COL2A1 production, as measured by 3H-proline incorporation assays [147], whereas in vivo intra-articular injection of BMP2 in murine knee joints enhances proteoglycan synthesis 2.5-fold (i.e., 35S-incorporation) in patellar cartilage within two days [102]. Furthermore, intra-articular overexpression of BMP2 in the murine knee joint using adenoviruses also enhances cartilage proteoglycan content [148]. In in vitro mesenchymal stromal cells and chondrocyte-like cell lines, and in in vivo epiphyseal chondrocytes, BMP2 is a well-known inducer of chondrocyte maturation and hypertrophy [149]. However, in mice with cartilage-specific, tamoxifen-inducible overexpression of BMP2 (Col2 rTA; TRE-Bmp2), increased hypertrophy is not observed in articular cartilage after six weeks of prolonged exposure to tamoxifen [150]. Furthermore, in the DMM model of experimental OA, the presence of excessive BMP2 signaling does not negatively, nor positively, affect cartilage damage but does induce very extensive osteophyte formation [150]. These observations might indicate that, in vivo, articular cartilage is not very sensitive to BMP2 signaling compared to osteophyte-forming tissues like periosteum. Possibly, signaling of hyaluronan, an important structural component of cartilage, via CD44 plays a role in this, as it has been demonstrated that this inhibits BMP2-induced pSMAD1/5 signaling in the murine bone marrow-derived ST2 stromal cell line [151]. Alternatively, GDF5 has recently been shown to be a context-dependent inhibitor of BMP2 signaling, via a yet unknown mechanism, possibly involving receptor competition [152], and GDF5 is present in cartilage [153,154,155].
BMP7 mRNA and protein expression can be detected in both immature and mature articular cartilage. Immunohistochemically, mature BMP7 can predominantly be detected in chondrocytes of the superficial zone, whereas its pro-form is mainly detected in deep zone chondrocytes [156,157]. To signal, BMP7 can use ALK2, ALK3, and ALK6, and the type II receptors BMPR2, ACVR2A, and ACVR2B, to induce SMAD1/5 phosphorylation and SMAD-independent signaling [143]. In contrast to BMP2, a positive interaction of CD44 with BMP7 signaling has been described. Without CD44, chondrocytes are not able to induce pSMAD1/5 in response to BMP7 [158,159,160]. In chondrocytes, BMP7 signaling induces ECM production. Both proteoglycan and COL2A1 production are induced in vitro in juvenile and adult human primary chondrocytes [139] and ex vivo in juvenile bovine explants [147]. Furthermore, inhibition of endogenous BMP7 production with antisense oligonucleotides in cartilage explants lowers aggrecan production [161]. In vitro, BMP7 also induces the expression of cartilage oligomeric matrix protein [109] and hyaluronan synthase 2, resulting in more hyaluronan production by chondrocytes, possibly facilitating deposition of aggrecan in the ECM [162]. In addition, BMP7 can counteract an IL-1- or LPS-induced inhibition of proteoglycan synthesis in chondrocytes [163,164,165], and partly inhibit IL-1-induced MMP13 expression, especially in the presence of IGF1 [166]. The positive effects of BMP7 in vitro are reflected in vivo: conditional deletion of Bmp7 from limb mesenchyme in Prx-Cre;Bmp7flox/flox mice results in proteoglycan loss (at 8 weeks old) and enhanced Mmp13 expression (at 24 weeks old) [167]. Of note, joint and cartilage formation were not detectably affected in these mice, but Bmp7 deletion did result in synovial inflammation, making it difficult to exclude indirect effects. Administration of BMP7 is beneficial for cartilage in OA [168,169]. Osteoarthritic cartilage is still responsive to BMP7 [155], and weekly injections of BMP7 diminish cartilage damage in the rabbit ACLT model of OA [170]. Furthermore, BMP7 increases the quantity and quality of cartilage repair tissue in rabbits with a full thickness cartilage defect [171]. Remarkably, although BMP7 can induce ectopic bone formation [172], excessive osteophyte formation was not observed in these studies, nor was excessive osteophyte formation observed in humans after a single injection of BMP7 in a Phase 1 study done in symptomatic knee OA patients [173]. Although BMP7 induces SMAD1/5 phosphorylation, its signaling is not associated with chondrocyte hypertrophy. Bovine articular chondrocytes do not undergo hypertrophy when cultured with BMP7 [174], and BMP7 blocks hypertrophy in the ATDC5 cell line [175]. In long bone formation ex vivo, BMP7 does stimulate the formation of pre-hypertrophic chondrocytes in the epiphyseal plate but blocks the transition towards hypertrophy [176]. This anti-hypertrophic effect of BMP7 has contributed to its ability to induce the transcription factor BAPX-1/NKX-3.2, a potent modulator of chondrocyte hypertrophy [177]. However, BMP7 does induce ALPL activity and osteogenesis in primary fetal chondrocytes [178], and chick sternum chondrocytes do undergo hypertrophy when cultured with BMP7 [179], indicating that BMP7 can induce chondrocyte hypertrophy, but, like TGFβ, it is in a context-dependent manner.
Two other BMPs well known for their positive effect on chondrogenesis and cartilage formation are BMP4 and BMP6, which both signal via ALK2 and ALK3 to induce pSMAD1/5 and SMAD-independent signaling. However, very little is known regarding their role in mature articular cartilage. In bovine chondrocytes and cartilage explants, BMP4 enhances proteoglycan synthesis, COL2A1 production, and proliferation [140], and BMP4 has similar effects in human chondrocytes cultured in alginate [164]. BMP6 mRNA and protein are expressed in both healthy and OA articular cartilage [141]. In human chondrocytes, BMP6 induces proteoglycan synthesis [164,180], but with advancing age, chondrocyte response to BMP6 decreases [180]. However, in the epiphyseal plate, BMP6 is especially expressed in the hypertrophic zone [181] where it has a stimulatory role on chondrocyte maturation. Furthermore, a pro-hypertrophic effect of BMP6 has also been observed in ATDC5 cells [182]. These studies indicate that BMP6 signaling could be detrimental for articular cartilage
A BMP that has more recently come to attention is BMP9, also known as GDF2. This BMP signals via type II receptors ACVR2A/ACVR2B and BMPR2 and type I receptors ALK1, and/or ALK2 to induce SMAD1/5 and SMAD-independent signaling. Production of BMP9 could not be detected in chondrocytes [141], but BMP9 circulates in high levels in blood [183]. In juvenile bovine cartilage, BMP9 enhances the production of proteoglycans and does so more potently than BMP2 [147]. However, BMP9 can also induce ECM mineralization when chondrocytes are seeded in biodegradable polyglycolic acid (PGA) scaffolds [184]. Additionally, also in mesenchymal progenitor cells, BMP9 is a potent inducer of chondrogenesis accompanied by hypertrophy [185]. Therefore, BMP9 possibly has a detrimental effect on chondrocyte homeostasis. Indeed, increased hypertrophy was observed in primary bovine chondrocytes stimulated with this BMP, but remarkably, this was efficiently blocked via a synergy with TGFβ1 on SMAD2/3 phosphorylation [186].
BMP14, better known as GDF5, is a very important initiator of joint formation and patterning. The importance of GDF5 in cartilage biology is illustrated by the observations that mutations in GDF5 can lead to chondrodysplasias, and that a genetic SNP in the core promotor of GDF5, resulting in reduced GDF5 protein levels, is associated with OA development [187]. However, regarding the role of GDF5 in mature cartilage, relatively little is known. In chondrocytes, GDF5 predominantly signals via ALK6 [178] but can also signal via ALK2 and ALK3 [152] to induce SMAD1/5 phosphorylation and SMAD-independent signaling [188]. GDF5 is expressed in (im)mature cartilage, but its expression is not affected by OA [141,154,155]. In cartilage, GDF5 induces proteoglycan synthesis [154,178] and COMP expression [109] but neither COL2A1 synthesis nor chondrocyte proliferation [154]. Notably, OA chondrocytes do not respond consistently to GDF5 [188]; that is, large variations can be observed between OA patients on gene expression of targets like ACAN and SOX9 after addition of GDF5. Possibly, a yet undiscovered co-receptor plays a role in this. Such a co-receptor has been postulated to explain how GDF5 can function as a downstream BMP2 antagonist in ATDC5 cells even though it uses the same type I and type II receptors [152]. Identification of such a co-receptor would greatly help in understanding GDF5 signaling in chondrocytes and how it differs from BMP signaling using the same receptors.
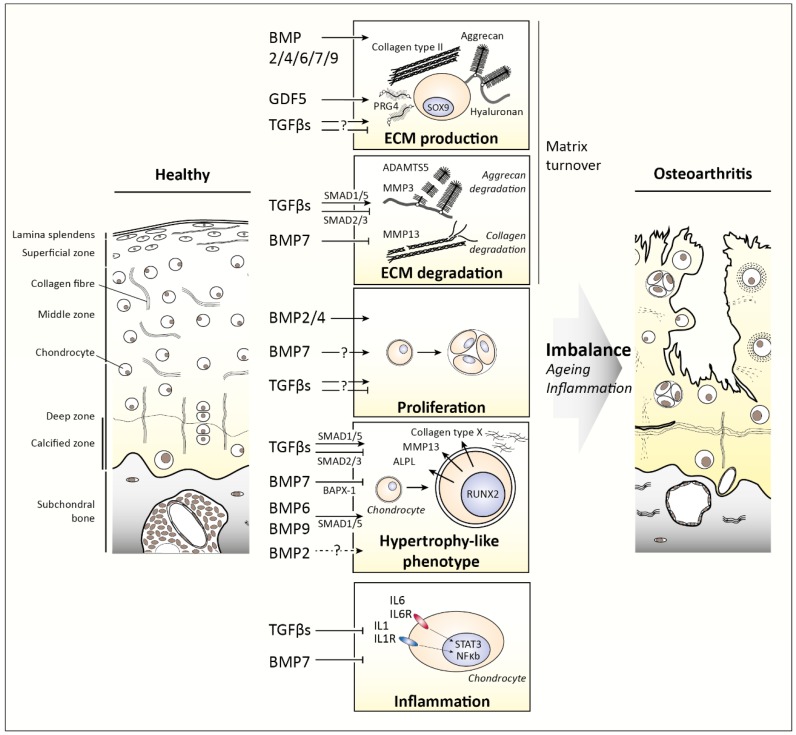
Figure 4 summarizes the effects of the different TGFβ family members on cartilage biology and OA development.
Figure 4.
Effects of the different Transforming Growth Factor β (TGFβ) family members on articular cartilage biology and osteoarthritis (OA) development. On the left healthy articular cartilage is depicted, on the right osteoarthritic cartilage. The middle depicts how TGFβ family members regulate matrix turnover, chondrocyte proliferation, hypertrophy-like differentiation, and inflammation. An imbalance in these effects of TGFβ family members because of age- and/or inflammation-related changes contributes to OA development. ADAMTS5 = A disintegrin and metalloproteinase with thrombospondin motifs 5, ALPL = Alkaline phosphatase tissue-nonspecific isozyme, BAPX-1 = Homeobox protein Nkx-3.2, BMP = Bone morphogenetic protein, GDF = Growth differentiation factor, IL = interleukin, MMP = matrix metalloproteinase, NFκb = nuclear factor kappa-light-chain-enhancer of activated B cells, PRG4 = proteoglycan 4, RUNX2 = Runt-related transcription factor 2, SOX9 = Transcription factor SOX-9, and STAT3 = Signal transducer and activator of transcription 3.
4. Changes in TGFβ Family Signaling as Cause for Disease
4.1. Age-Related Changes in TGFβ Signaling
In view of the aforementioned importance of TGFβ family members in chondrocyte and cartilage homeostasis, it is not surprising that alterations in their signaling affect cartilage biology and can lead to pathology. Alterations can occur because of various reasons, for example due to aging. Aging affects TGFβ family signaling on multiple levels in the signaling cascade. For example, ligand and receptor expression can be affected, just as intracellular signaling and regulation of gene transcription [90,121,189,190,191].
A prime example of how aging affects ligand expression is BMP7. In human cartilage, BMP7 gene and protein levels decline with advancing age. This decrease has been attributed to increased methylation of its promoter, resulting in silencing of its expression [192,193]. In contrast, mRNA expressions of BMP2, BMP6, and BMP9 do not seem to be affected by aging [141,146,156,190]. In mice, protein expression levels of TGFβ2 and TGFβ3 (but not TGFβ1) decreased with age [90], whereas in cows, a drop in Tgfb1 mRNA expression was observed [142].
Also, receptor expression is impacted by aging. Of type I receptors, a profound drop in ALK5 expression has been observed in humans, mice, guinea pigs, and cows on gene expression and protein level [100,142,194], indicating that this occurs consistently across species. In addition, on gene expression levels, loss of Alk2, Alk3, and Alk4 has also been observed in bovine cartilage [90,142]. Notably, loss of receptor expression can not only lead to loss of the growth factor response but also alters the signaling of ligands able to use multiple receptors. For example, this has been demonstrated for TGFβ signaling because aging affects Alk5 expression more than Alk1 expression, and TGFβ induces relatively more SMAD1/5 phosphorylation in old cartilage, with all due effects [142,194]. Next to these differences in type I receptor expression, an age-related decrease in Bmpr2 has also been observed [142]. This could possibly explain the age-related loss in the BMP7 response of chondrocytes that has been observed [190]. However, how age-related changes affect BMP signaling (besides BMP7) in chondrocyte homeostasis has not yet been sufficiently investigated.
Changes in receptor expression can affect intracellular signaling. As mentioned, an age-related change in receptor type I expression towards relatively more ALK1 shifts the balance from protective signaling (SMAD2/3) into deleterious signaling (SMAD1/5/9). The loss of protective SMAD2/3 signaling could explain the relation between aging and OA development [12,195]. Indeed, the changing ALK1/ALK5 ratio in aging correlates with Mmp13 expression and OA development [89]. Computational modeling supports a direct effect, especially TGFβ signaling via ALK1, which can explain Mmp13 expression and cartilage damage in aged cartilage [196]. Possibly, these effects run via decreased modulation of RUNX2 activity. SMADs interact with RUNX2 to modulate chondrocyte differentiation, where interaction of SMAD3 with RUNX2 inhibits RUNX2 functioning and SMAD1/5 stimulates it [36,53,197,198].
4.2. Joint Loading-Related Changes in TGFβ Signaling
Next to aging, another factor able to modulate TGFβ family signaling is (mechanical) loading. For example, mechanical joint loading has been shown to play a crucial role in activation of SMAD2/3 signaling [97,199,200]. In bovine articular cartilage explants, mechanical compression quickly induced SMAD2/3 signaling, most likely due to mechanical activation of latent TGFβ bound to the ECM of cartilage, as cartilage contains high levels of this. This loading-induced pSMAD2/3 signaling induced a positive feedback loop, by stimulating expression of Tgfb1 and Alk5. In contrast, unloading rapidly induced loss of SMAD2/3 signaling from articular cartilage (<2 h), possibly due to sequestering of available TGFβ [201]. As a consequence of this, prolonged (2 weeks) unloading of cartilage resulted in dramatically increased Col10a1 expression, a marker of chondrocyte hypertrophy. However, repeated loading of cartilage every third day efficiently blocked this increase in Col10a1 expression. These observations show the importance of cartilage loading for maintenance of SMAD2/3 signaling and cartilage homeostasis. In view of the effects of SMAD3 signaling in cartilage, this mechanism possibly also explains the observations that reduced joint loading contributes to cartilage degeneration and production of the proteolytic enzymes MMP8 and MMP13 [202,203]. Importantly, old cartilage, compared to young cartilage, has a strongly reduced capacity for this loading-mediated SMAD2/3 signaling, which possibly makes it more vulnerable to OA development [201,204]. Interestingly, a sedentary lifestyle was recently hypothesized to be the cause for the doubled OA prevalence in current populations compared to those from the 1800s to early 1900s after correction for age and BMI [205]; thus, possibly these observations are linked. Loading also induces BMP levels; BMP2, BMP4, BMP6, and BMPR2 are increased by moderate exercise and suppresses post-traumatic OA [206]. However, excessive loading has been shown to lead to collagen damage, proteoglycan loss, and OA progression, and these effects have been attributed to induction of gremlin-1, a BMP antagonist [207,208]. Together, these studies show the impact of loading on TGFβ family signaling.
4.3. Inflammation-Related Changes in TGFβ Signaling
Previously, cartilage breakdown during OA was considered to be a simple wear and tear process. However, nowadays, OA is regarded as a whole joint disease, with inflammation strongly implicated in its pathogenesis [209,210]. In over 50% of patients, synovitis (i.e., inflammation of the synovium or joint capsule) can be observed [211,212,213,214,215,216,217]. Important inflammatory mediators produced by this synovitis are IL-1, TNF-α, IL-8, and IL-6 [210,218,219,220,221]. These pro-inflammatory cytokines have a direct effect on cartilage homeostasis by upregulating MMPs and other cartilage-degrading enzymes [222,223,224,225] but, importantly, also have been described to modulate TGFβ family signaling [226,227].
For example, IL-1 can downregulate TGFBR2 on mRNA and protein expression in human articular chondrocytes [226]. Also, pro-inflammatory mediators produced by inflamed synovium, which was cultured in vitro (OA synovium conditioned medium), reduced Tgfbr2 expression in mechanically compressed bovine cartilage explants [228]. IL-1 has also been described to inhibit BMP7 signaling via downregulation of the ALK2 and ALK3 receptors and inhibition of SMAD1 expression [229]. But in contrast, IL-1 has also been described to induce BMP2 and BMP7 mRNA expression in chondrocytes, and a high dose of IL-1 leads to the activation of pro-BMP7 into the active form [230].
Next to modulation of receptor expression, inflammatory mediators can also affect SMAD-dependent signaling via various mechanisms. For instance, IL-1 can increase the expression of i-SMAD7 via NF-κB activation in chondrocytes, which inhibits SMAD2/3 signaling [231]. Furthermore, TNF-α and IL-1 have been described to be able to decrease DNA binding activity of SMAD3/4 complexes in human OA chondrocytes via unknown mechanisms but independent of i-SMAD7 [227]. In addition, inflammation-induced transcription factors can compete with SMADs for co-factors. For example, SMAD3 and NFκB (a transcription factor downstream of TAK1) can compete for their common co-transcription factor CREB binding protein (CBP) in endothelial cells [127].
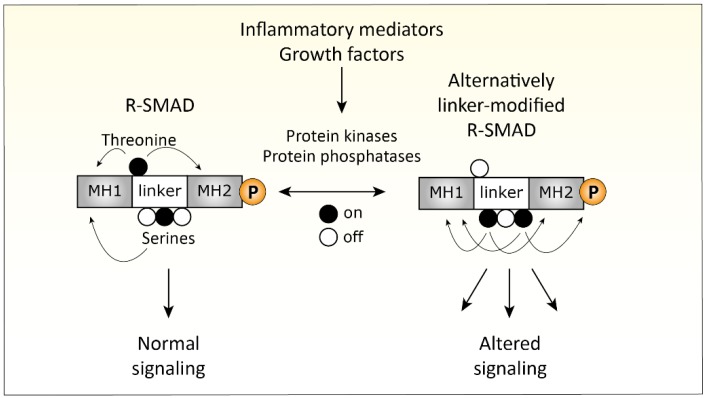
Recent insights provided another way of how pro-inflammatory pathways can interact with SMAD-dependent signaling, namely by post-translational modification of the linker region of SMAD proteins [232,233]. SMAD proteins consist of a N-terminal MH1 (missing in i-SMAD6 and i-SMAD7), important for DNA binding activity and nuclear importation, and a C-terminal MH2 domain, responsible for SMAD-receptor and SMAD–SMAD interactions and gene transcription activation [234,235,236,237]. These two conserved MH1 and MH2 domains are connected via a regulatory linker domain, which can be phosphorylated on specific serine and threonine residues. This phosphorylation of the linker region regulates nuclear entry, SMAD–protein interactions, and SMAD turnover [232,238,239]. For instance, mutation of the SMAD3 linker resulted in strongly enhanced TGFβ-induced responses in breast cancer cells and increased tumorigenesis during liver cancer [240,241]. Also, mutations in the linker of SMAD4, commonly found in colorectal and pancreatic cancers, result in rapid proteasomal degradation and loss of function [242,243]. These findings show that modification of the SMAD linkers greatly affect SMAD function (Figure 3).
Phosphorylation of the serine and threonine residues in the linker domain is dependent on kinases, including MAPKs, ERK, JNK, p38, AKT, CDKs, ROCK, and GSK-3 [233,238,239,244,245,246]. In addition, dephosphorylation of the SMAD linker, dependent on phosphatases [237,247,248], is just as important for regulating SMAD function. Notably, many of these kinases and phosphatases are induced in chondrocytes by inflammatory mediators [222,249,250,251,252] present in the OA joint. This makes it likely that inflammation-induced kinases can also affect SMAD function in cartilage, thereby affecting TGFβ and BMP signaling and, thus, a possible cause for OA. Still, the relative importance of these SMAD linker modifications in cartilage biology and OA pathogenesis is not well understood yet and warrants further research.
A (simplified) overview of SMAD modifications in the linker-domain that can alter TGFβ signaling is depicted in Figure 5.
Figure 5.
SMAD linker modifications alter Transforming growth factor β (TGFβ) signaling. The SMAD proteins consist of a MH1 and MH2 domain, connected with a non-conserved linker domain. Threonine and serine residues in the linker domain can be modulated by protein kinases and phosphatases like mitogen-activated protein kinases (MAPKs), c-Jun N-terminal kinases (JNK), and Glycogen synthase kinase 3 (GSK-3). These kinases can be activated by growth factors and inflammatory mediators. This process greatly affects SMAD function and leads to altered TGFβ signaling.
5. Future Perspectives
In conclusion, members of the TGFβ family are crucial for the homeostasis of chondrocytes and, thus, of cartilage. Signaling in cartilage goes via both SMAD-dependent and SMAD-independent signaling, although both pathways are more integrated than their names suggest. Cellular context plays a large role in TGFβ family signaling, greatly affecting its outcome, which is illustrated by, for example, the differential effects of TGFβ and BMP7 in immature versus mature chondrocytes. How aging or inflammation provide cellular context, which regulates TGFβ family signaling in articular cartilage, is still largely unknown. Gaining greater insights into this would help to better understand OA development and progression, and it would give leads for how to separate the beneficial effects of TGFβ family members from their detrimental effects. This topic, thus, definitely warrants further research. Of special interest might be the interaction of SMADs with other transcription factors. Such interactions control (efficient) targeting and transcription of (potential) target genes. Altered interactions, for example due to altered SMAD linker modifications, could explain context-dependent gain and loss of function. How, for instance, the SMAD–RUNX2 interaction is affected by aging or inflammation could be of great importance for cartilage degeneration. Closely related to this, it would be important to further study other SMAD binding partners involved in regulating chondrocyte hypertrophy such as myocyte enhancer-binding factor 2 (MEF2). This transcription factor has been shown to induce RUNX2 expression and hypertrophy in cartilage [253], and it has been described to interact and be inhibited by SMAD3 in muscle cells [254,255]. Identifying such interactions in articular chondrocytes can help us learn how cartilage degradation is regulated and would give leads on how to counteract this. Another topic of interest is preventing the negative impact of processes such as inflammation on TGFβ family signaling in cartilage. For this, a better understanding of the cellular processes driving these negative effects is needed, but this hardly has been investigated until now. Especially, the relevance of inflammation-driven SMAD linker post-translational modifications as causes for disturbed TGFβ and BMP signaling warrants attention, because this possibly will provide a new avenue of targeted therapy directed at the inhibition of cartilage degradation in OA by targeting the intracellular kinases and phosphatases responsible for these modifications.
Funding
This research was funded by the Dutch Arthritis Foundation (ReumaNederland), grant number 16-1-401. The APC was funded by the Dutch Arthritis Foundation (ReumaNederland) and Radboud University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sophia Fox A.J., Bedi A., Rodeo S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health. 2009;1:461–468. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mescher A.L., Junqueira L.C.U.A.B.H. Junqueira’s Basic Histology: Text and Atlas. 12th ed. McGraw-Hill Medical; New York, NY, USA: London, UK: 2010. [Google Scholar]
- 3.Pearle A.D., Warren R.F., Rodeo S.A. Basic science of articular cartilage and osteoarthritis. Clin. Sports Med. 2005;24:1–12. doi: 10.1016/j.csm.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Shoulders M.D., Raines R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruckner P., van der Rest M. Structure and function of cartilage collagens. Microsc. Res. Tech. 1994;28:378–384. doi: 10.1002/jemt.1070280504. [DOI] [PubMed] [Google Scholar]
- 6.Fraser J.R., Laurent T.C., Laurent U.B. Hyaluronan: Its nature, distribution, functions and turnover. J. Intern. Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 7.Kiani C., Chen L., Wu Y.J., Yee A.J., Yang B.B. Structure and function of aggrecan. Cell Res. 2002;12:19–32. doi: 10.1038/sj.cr.7290106. [DOI] [PubMed] [Google Scholar]
- 8.Teshima R., Otsuka T., Takasu N., Yamagata N., Yamamoto K. Structure of the most superficial layer of articular cartilage. J. Bone Jt. Surg. Br. 1995;77:460–464. doi: 10.1302/0301-620X.77B3.7744937. [DOI] [PubMed] [Google Scholar]
- 9.Jeffery A.K., Blunn G.W., Archer C.W., Bentley G. Three-dimensional collagen architecture in bovine articular cartilage. J. Bone Jt. Surg. Br. 1991;73:795–801. doi: 10.1302/0301-620X.73B5.1894669. [DOI] [PubMed] [Google Scholar]
- 10.Rhee D.K., Marcelino J., Baker M., Gong Y., Smits P., Lefebvre V., Jay G.D., Stewart M., Wang H., Warman M.L., et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J. Clin. Investig. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowman M.K., Schmidt T.A., Raghavan P., Stecco A. Viscoelastic Properties of Hyaluronan in Physiological Conditions. F1000Res. 2015;4:622. doi: 10.12688/f1000research.6885.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kraan P.M., van den Berg W.B. Chondrocyte hypertrophy and osteoarthritis: Role in initiation and progression of cartilage degeneration? Osteoarthr. Cartil. 2012;20:223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Mueller T.D., Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012;586:1846–1859. doi: 10.1016/j.febslet.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 14.Heldin C.H., Miyazono K., ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 15.Wang X.F., Lin H.Y., Ng-Eaton E., Downward J., Lodish H.F., Weinberg R.A. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Casillas F., Cheifetz S., Doody J., Andres J.L., Lane W.S., Massague J. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-beta receptor system. Cell. 1991;67:785–795. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 17.Cheifetz S., Bellon T., Cales C., Vera S., Bernabeu C., Massague J., Letarte M. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J. Biol. Chem. 1992;267:19027–19030. [PubMed] [Google Scholar]
- 18.Siebold C., Yamashita T., Monnier P.P., Mueller B.K., Pasterkamp R.J. RGMs: Structural Insights, Molecular Regulation, and Downstream Signaling. Trends Cell Biol. 2017;27:365–378. doi: 10.1016/j.tcb.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finnson K.W., Tam B.Y., Liu K., Marcoux A., Lepage P., Roy S., Bizet A.A., Philip A. Identification of CD109 as part of the TGF-beta receptor system in human keratinocytes. FASEB J. 2006;20:1525–1527. doi: 10.1096/fj.05-5229fje. [DOI] [PubMed] [Google Scholar]
- 20.Onichtchouk D., Chen Y.G., Dosch R., Gawantka V., Delius H., Massague J., Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 21.Zawel L., Dai J.L., Buckhaults P., Zhou S., Kinzler K.W., Vogelstein B., Kern S.E. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell. 1998;1:611–617. doi: 10.1016/S1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 22.Kusanagi K., Inoue H., Ishidou Y., Mishima H.K., Kawabata M., Miyazono K. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell. 2000;11:555–565. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaarenstroom T., Hill C.S. TGF-beta signaling to chromatin: How Smads regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 2014;32:107–118. doi: 10.1016/j.semcdb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Furumatsu T., Tsuda M., Taniguchi N., Tajima Y., Asahara H. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J. Biol. Chem. 2005;280:8343–8350. doi: 10.1074/jbc.M413913200. [DOI] [PubMed] [Google Scholar]
- 25.Nakao A., Afrakhte M., Moren A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 26.Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 27.Valdes A.M., Spector T.D., Tamm A., Kisand K., Doherty S.A., Dennison E.M., Mangino M., Tamm A., Kerna I., Hart D.J., et al. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62:2347–2352. doi: 10.1002/art.27530. [DOI] [PubMed] [Google Scholar]
- 28.Aref-Eshghi E., Zhang Y., Hart D., Valdes A.M., Furey A., Martin G., Sun G., Rahman P., Arden N., Spector T.D., et al. SMAD3 is associated with the total burden of radiographic osteoarthritis: The Chingford study. PLoS ONE. 2014;9:e97786. doi: 10.1371/journal.pone.0097786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Laar I.M., van der Linde D., Oei E.H., Bos P.K., Bessems J.H., Bierma-Zeinstra S.M., van Meer B.L., Pals G., Oldenburg R.A., Bekkers J.A., et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J. Med. Genet. 2012;49:47–57. doi: 10.1136/jmedgenet-2011-100382. [DOI] [PubMed] [Google Scholar]
- 30.van de Laar I.M., Oldenburg R.A., Pals G., Roos-Hesselink J.W., de Graaf B.M., Verhagen J.M., Hoedemaekers Y.M., Willemsen R., Severijnen L.A., Venselaar H., et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 31.Yang X., Chen L., Xu X., Li C., Huang C., Deng C.X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C.G., Thuillier D., Chin E.N., Alliston T. Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 2012;64:3278–3289. doi: 10.1002/art.34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson C.M., Schwarz E.M., Reynolds P.R., Puzas J.E., Rosier R.N., O’Keefe R.J. Smad2 and 3 mediate transforming growth factor-beta1-induced inhibition of chondrocyte maturation. Endocrinology. 2000;141:4728–4735. doi: 10.1210/endo.141.12.7848. [DOI] [PubMed] [Google Scholar]
- 34.Ionescu A.M., Schwarz E.M., Zuscik M.J., Drissi H., Puzas J.E., Rosier R.N., O’Keefe R.J. ATF-2 cooperates with Smad3 to mediate TGF-beta effects on chondrocyte maturation. Exp. Cell Res. 2003;288:198–207. doi: 10.1016/S0014-4827(03)00181-2. [DOI] [PubMed] [Google Scholar]
- 35.Kim K.O., Sampson E.R., Maynard R.D., O’Keefe R.J., Chen D., Drissi H., Rosier R.N., Hilton M.J., Zuscik M.J. Ski inhibits TGF-beta/phospho-Smad3 signaling and accelerates hypertrophic differentiation in chondrocytes. J. Cell Biochem. 2012;113:2156–2166. doi: 10.1002/jcb.24089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T.F., Gao L., Sheu T.J., Sampson E.R., Flick L.M., Konttinen Y.T., Chen D., Schwarz E.M., Zuscik M.J., Jonason J.H., et al. Aberrant hypertrophy in Smad3-deficient murine chondrocytes is rescued by restoring transforming growth factor beta-activated kinase 1/activating transcription factor 2 signaling: a potential clinical implication for osteoarthritis. Arthritis Rheum. 2010;62:2359–2369. doi: 10.1002/art.27537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang J.S., Alliston T., Delston R., Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leboy P., Grasso-Knight G., D’Angelo M., Volk S.W., Lian J.V., Drissi H., Stein G.S., Adams S.L. Smad-Runx interactions during chondrocyte maturation. (Pt 1)J. Bone Jt. Surg. Am. 2001;83-A Suppl 1:S15–S22. doi: 10.2106/00004623-200100001-00003. [DOI] [PubMed] [Google Scholar]
- 39.Takeda S., Bonnamy J.P., Owen M.J., Ducy P., Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enomoto H., Enomoto-Iwamoto M., Iwamoto M., Nomura S., Himeno M., Kitamura Y., Kishimoto T., Komori T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 2000;275:8695–8702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- 41.Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 42.Nomura M., Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 43.Waldrip W.R., Bikoff E.K., Hoodless P.A., Wrana J.L., Robertson E.J. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/S0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 44.Weinstein M., Yang X., Li C., Xu X., Gotay J., Deng C.X. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc. Natl. Acad. Sci. USA. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vivian J.L., Chen Y., Yee D., Schneider E., Magnuson T. An allelic series of mutations in Smad2 and Smad4 identified in a genotype-based screen of N-ethyl-N- nitrosourea-mutagenized mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2002;99:15542–15547. doi: 10.1073/pnas.242474199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decker R.S., Koyama E., Pacifici M. Genesis and morphogenesis of limb synovial joints and articular cartilage. Matrix Biol. 2014;39:5–10. doi: 10.1016/j.matbio.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamamoto T., Beppu H., Okada H., Kawabata M., Kitamura T., Miyazono K., Kato M. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 2002;62:5955–5961. [PubMed] [Google Scholar]
- 48.Wang W., Song B., Anbarchian T., Shirazyan A., Sadik J.E., Lyons K.M. Smad2 and Smad3 Regulate Chondrocyte Proliferation and Differentiation in the Growth Plate. PLoS Genet. 2016;12:e1006352. doi: 10.1371/journal.pgen.1006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lechleider R.J., Ryan J.L., Garrett L., Eng C., Deng C., Wynshaw-Boris A., Roberts A.B. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Dev. Biol. 2001;240:157–167. doi: 10.1006/dbio.2001.0469. [DOI] [PubMed] [Google Scholar]
- 50.Chang H., Huylebroeck D., Verschueren K., Guo Q., Matzuk M.M., Zwijsen A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126:1631–1642. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- 51.Retting K.N., Song B., Yoon B.S., Lyons K.M. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136:1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujii M., Takeda K., Imamura T., Aoki H., Sampath T.K., Enomoto S., Kawabata M., Kato M., Ichijo H., Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell. 1999;10:3801–3813. doi: 10.1091/mbc.10.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horiki M., Imamura T., Okamoto M., Hayashi M., Murai J., Myoui A., Ochi T., Miyazono K., Yoshikawa H., Tsumaki N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J. Cell Biol. 2004;165:433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Javed A., Afzal F., Bae J.S., Gutierrez S., Zaidi K., Pratap J., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs. 2009;189:133–137. doi: 10.1159/000151719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Javed A., Bae J.S., Afzal F., Gutierrez S., Pratap J., Zaidi S.K., Lou Y., van Wijnen A.J., Stein J.L., Stein G.S., et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J. Biol. Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phimphilai M., Zhao Z., Boules H., Roca H., Franceschi R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 2006;21:637–646. doi: 10.1359/jbmr.060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bae J.S., Gutierrez S., Narla R., Pratap J., Devados R., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B., Javed A. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J. Cell Biochem. 2007;100:434–449. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]
- 58.Kielty C.M., Kwan A.P., Holmes D.F., Schor S.L., Grant M.E. Type X collagen, a product of hypertrophic chondrocytes. Biochem. J. 1985;227:545–554. doi: 10.1042/bj2270545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee M.K., Pardoux C., Hall M.C., Lee P.S., Warburton D., Qing J., Smith S.M., Derynck R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y.E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landstrom M. The TAK1-TRAF6 signalling pathway. Int. J. Biochem. Cell Biol. 2010;42:585–589. doi: 10.1016/j.biocel.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi K., Nagai S., Ninomiya-Tsuji J., Nishita M., Tamai K., Irie K., Ueno N., Nishida E., Shibuya H., Matsumoto K. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galban S., Duckett C.S. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ. 2010;17:54–60. doi: 10.1038/cdd.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe H., de Caestecker M.P., Yamada Y. Transcriptional cross-talk between Smad, ERK1/2, and p38 mitogen-activated protein kinase pathways regulates transforming growth factor-beta-induced aggrecan gene expression in chondrogenic ATDC5 cells. J. Biol. Chem. 2001;276:14466–14473. doi: 10.1074/jbc.M005724200. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y., Tao H., Jin C., Liu Y., Lu X., Hu X., Wang X. Transforming growth factor-beta1 induces type II collagen and aggrecan expression via activation of extracellular signal-regulated kinase 1/2 and Smad2/3 signaling pathways. Mol. Med. Rep. 2015;12:5573–5579. doi: 10.3892/mmr.2015.4068. [DOI] [PubMed] [Google Scholar]
- 67.Miyazaki Y., Tsukazaki T., Hirota Y., Yonekura A., Osaki M., Shindo H., Yamashita S. Dexamethasone inhibition of TGF beta-induced cell growth and type II collagen mRNA expression through ERK-integrated AP-1 activity in cultured rat articular chondrocytes. Osteoarthr. Cartil. 2000;8:378–385. doi: 10.1053/joca.1999.0313. [DOI] [PubMed] [Google Scholar]
- 68.Qureshi H.Y., Sylvester J., El Mabrouk M., Zafarullah M. TGF-beta-induced expression of tissue inhibitor of metalloproteinases-3 gene in chondrocytes is mediated by extracellular signal-regulated kinase pathway and Sp1 transcription factor. J. Cell Physiol. 2005;203:345–352. doi: 10.1002/jcp.20228. [DOI] [PubMed] [Google Scholar]
- 69.Tuli R., Tuli S., Nandi S., Huang X., Manner P.A., Hozack W.J., Danielson K.G., Hall D.J., Tuan R.S. Transforming growth factor-beta-mediated chondrogenesis of human mesenchymal progenitor cells involves N-cadherin and mitogen-activated protein kinase and Wnt signaling cross-talk. J. Biol. Chem. 2003;278:41227–41236. doi: 10.1074/jbc.M305312200. [DOI] [PubMed] [Google Scholar]
- 70.Li J., Zhao Z., Liu J., Huang N., Long D., Wang J., Li X., Liu Y. MEK/ERK and p38 MAPK regulate chondrogenesis of rat bone marrow mesenchymal stem cells through delicate interaction with TGF-beta1/Smads pathway. Cell Prolif. 2010;43:333–343. doi: 10.1111/j.1365-2184.2010.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bobick B.E., Kulyk W.M. The MEK-ERK signaling pathway is a negative regulator of cartilage-specific gene expression in embryonic limb mesenchyme. J. Biol. Chem. 2004;279:4588–4595. doi: 10.1074/jbc.M309805200. [DOI] [PubMed] [Google Scholar]
- 72.Qiao B., Padilla S.R., Benya P.D. Transforming growth factor (TGF)-beta-activated kinase 1 mimics and mediates TGF-beta-induced stimulation of type II collagen synthesis in chondrocytes independent of Col2a1 transcription and Smad3 signaling. J. Biol. Chem. 2005;280:17562–17571. doi: 10.1074/jbc.M500646200. [DOI] [PubMed] [Google Scholar]
- 73.Gunnell L.M., Jonason J.H., Loiselle A.E., Kohn A., Schwarz E.M., Hilton M.J., O’Keefe R.J. TAK1 regulates cartilage and joint development via the MAPK and BMP signaling pathways. J. Bone Miner. Res. 2010;25:1784–1797. doi: 10.1002/jbmr.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shim J.H., Greenblatt M.B., Xie M., Schneider M.D., Zou W., Zhai B., Gygi S., Glimcher L.H. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28:2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao L., Sheu T.J., Dong Y., Hoak D.M., Zuscik M.J., Schwarz E.M., Hilton M.J., O’Keefe R.J., Jonason J.H. TAK1 regulates SOX9 expression in chondrocytes and is essential for postnatal development of the growth plate and articular cartilages. J. Cell Sci. 2013;126:5704–5713. doi: 10.1242/jcs.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tew S.R., Hardingham T.E. Regulation of SOX9 mRNA in human articular chondrocytes involving p38 MAPK activation and mRNA stabilization. J. Biol. Chem. 2006;281:39471–39479. doi: 10.1074/jbc.M604322200. [DOI] [PubMed] [Google Scholar]
- 77.Rokutanda S., Fujita T., Kanatani N., Yoshida C.A., Komori H., Liu W., Mizuno A., Komori T. Akt regulates skeletal development through GSK3, mTOR, and FoxOs. Dev. Biol. 2009;328:78–93. doi: 10.1016/j.ydbio.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 78.Yan B., Zhang Z., Jin D., Cai C., Jia C., Liu W., Wang T., Li S., Zhang H., Huang B., et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat. Commun. 2016;7:11151. doi: 10.1038/ncomms11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Vasheghani F., Li Y.-h., Blati M., Simeone K., Fahmi H., Lussier B., Roughley P., Lagares D., Pelletier J.-P., et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann. Rheum. Dis. 2015;74:1432–1440. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 80.Takayama K., Kawakami Y., Kobayashi M., Greco N., Cummins J.H., Matsushita T., Kuroda R., Kurosaka M., Fu F.H., Huard J. Local intra-articular injection of rapamycin delays articular cartilage degeneration in a murine model of osteoarthritis. Arthritis Res. Ther. 2014;16:482. doi: 10.1186/s13075-014-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carames B., Hasegawa A., Taniguchi N., Miyaki S., Blanco F.J., Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann. Rheum. Dis. 2012;71:575–581. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woods A., Wang G., Beier F. RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem. 2005;280:11626–11634. doi: 10.1074/jbc.M409158200. [DOI] [PubMed] [Google Scholar]
- 83.Woods A., Beier F. RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J. Biol. Chem. 2006;281:13134–13140. doi: 10.1074/jbc.M509433200. [DOI] [PubMed] [Google Scholar]
- 84.Haudenschild D.R., Chen J., Pang N., Lotz M.K., D’Lima D.D. Rho kinase-dependent activation of SOX9 in chondrocytes. Arthritis Rheum. 2010;62:191–200. doi: 10.1002/art.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seyedin S.M., Thomas T.C., Thompson A.Y., Rosen D.M., Piez K.A. Purification and characterization of two cartilage-inducing factors from bovine demineralized bone. Proc. Natl. Acad. Sci. USA. 1985;82:2267–2271. doi: 10.1073/pnas.82.8.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seyedin S.M., Thompson A.Y., Bentz H., Rosen D.M., McPherson J.M., Conti A., Siegel N.R., Galluppi G.R., Piez K.A. Cartilage-inducing factor-A. Apparent identity to transforming growth factor-beta. J. Biol. Chem. 1986;261:5693–5695. [PubMed] [Google Scholar]
- 87.Segarini P.R., Roberts A.B., Rosen D.M., Seyedin S.M. Membrane binding characteristics of two forms of transforming growth factor-beta. J. Biol. Chem. 1987;262:14655–14662. [PubMed] [Google Scholar]
- 88.Ogawa Y., Schmidt D.K., Dasch J.R., Chang R.J., Glaser C.B. Purification and characterization of transforming growth factor-beta 2.3 and -beta 1.2 heterodimers from bovine bone. J. Biol. Chem. 1992;267:2325–2328. [PubMed] [Google Scholar]
- 89.Sengle G., Ono R.N., Sasaki T., Sakai L.Y. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J. Biol. Chem. 2011;286:5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blaney Davidson E.N., Scharstuhl A., Vitters E.L., van der Kraan P.M., van den Berg W.B. Reduced transforming growth factor-beta signaling in cartilage of old mice: Role in impaired repair capacity. Arthritis Res. Ther. 2005;7:R1338–R1347. doi: 10.1186/ar1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Villiger P.M., Lotz M. Differential expression of TGF beta isoforms by human articular chondrocytes in response to growth factors. J. Cell Physiol. 1992;151:318–325. doi: 10.1002/jcp.1041510213. [DOI] [PubMed] [Google Scholar]
- 92.Pombo-Suarez M., Castano-Oreja M.T., Calaza M., Gomez-Reino J., Gonzalez A. Differential upregulation of the three transforming growth factor beta isoforms in human osteoarthritic cartilage. Ann. Rheum. Dis. 2009;68:568–571. doi: 10.1136/ard.2008.090217. [DOI] [PubMed] [Google Scholar]
- 93.Morales T.I., Joyce M.E., Sobel M.E., Danielpour D., Roberts A.B. Transforming growth factor-beta in calf articular cartilage organ cultures: Synthesis and distribution. Arch. Biochem. Biophys. 1991;288:397–405. doi: 10.1016/0003-9861(91)90212-2. [DOI] [PubMed] [Google Scholar]
- 94.Albro M.B., Nims R.J., Cigan A.D., Yeroushalmi K.J., Shim J.J., Hung C.T., Ateshian G.A. Dynamic mechanical compression of devitalized articular cartilage does not activate latent TGF-beta. J. Biomech. 2013;46:1433–1439. doi: 10.1016/j.jbiomech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maeda S., Dean D.D., Gay I., Schwartz Z., Boyan B.D. Activation of latent transforming growth factor beta1 by stromelysin 1 in extracts of growth plate chondrocyte-derived matrix vesicles. J. Bone Miner. Res. 2001;16:1281–1290. doi: 10.1359/jbmr.2001.16.7.1281. [DOI] [PubMed] [Google Scholar]
- 96.Albro M.B., Cigan A.D., Nims R.J., Yeroushalmi K.J., Oungoulian S.R., Hung C.T., Ateshian G.A. Shearing of synovial fluid activates latent TGF-beta. Osteoarthr. Cartil. 2012;20:1374–1382. doi: 10.1016/j.joca.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jobling M.F., Mott J.D., Finnegan M.T., Jurukovski V., Erickson A.C., Walian P.J., Taylor S.E., Ledbetter S., Lawrence C.M., Rifkin D.B., et al. Isoform-specific activation of latent transforming growth factor beta (LTGF-beta) by reactive oxygen species. Radiat. Res. 2006;166:839–848. doi: 10.1667/RR0695.1. [DOI] [PubMed] [Google Scholar]
- 98.Madej W., van Caam A., Blaney Davidson E.N., van der Kraan P.M., Buma P. Physiological and excessive mechanical compression of articular cartilage activates Smad2/3P signaling. Osteoarthr. Cartil. 2014;22:1018–1025. doi: 10.1016/j.joca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 99.Finnson K.W., Parker W.L., ten Dijke P., Thorikay M., Philip A. ALK1 opposes ALK5/Smad3 signaling and expression of extracellular matrix components in human chondrocytes. J. Bone Miner. Res. 2008;23:896–906. doi: 10.1359/jbmr.080209. [DOI] [PubMed] [Google Scholar]
- 100.Blaney Davidson E.N., Remst D.F., Vitters E.L., van Beuningen H.M., Blom A.B., Goumans M.J., van den Berg W.B., van der Kraan P.M. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J. Immunol. 2009;182:7937–7945. doi: 10.4049/jimmunol.0803991. [DOI] [PubMed] [Google Scholar]
- 101.van Beuningen H.M., van der Kraan P.M., Arntz O.J., van den Berg W.B. Transforming growth factor-beta 1 stimulates articular chondrocyte proteoglycan synthesis and induces osteophyte formation in the murine knee joint. Lab. Investig. 1994;71:279–290. [PubMed] [Google Scholar]
- 102.van Beuningen H.M., Glansbeek H.L., van der Kraan P.M., van den Berg W.B. Differential effects of local application of BMP-2 or TGF-beta 1 on both articular cartilage composition and osteophyte formation. Osteoarthr. Cartil. 1998;6:306–317. doi: 10.1053/joca.1998.0129. [DOI] [PubMed] [Google Scholar]
- 103.Morales T.I., Roberts A.B. Transforming growth factor beta regulates the metabolism of proteoglycans in bovine cartilage organ cultures. J. Biol. Chem. 1988;263:12828–12831. [PubMed] [Google Scholar]
- 104.Malemud C.J., Killeen W., Hering T.M., Purchio A.F. Enhanced sulfated-proteoglycan core protein synthesis by incubation of rabbit chondrocytes with recombinant transforming growth factor-beta 1. J. Cell Physiol. 1991;149:152–159. doi: 10.1002/jcp.1041490119. [DOI] [PubMed] [Google Scholar]
- 105.van der Kraan P., Vitters E., van den Berg W. Differential effect of transforming growth factor beta on freshly isolated and cultured articular chondrocytes. J. Rheumatol. 1992;19:140–145. [PubMed] [Google Scholar]
- 106.Redini F., Galera P., Mauviel A., Loyau G., Pujol J.P. Transforming growth factor beta stimulates collagen and glycosaminoglycan biosynthesis in cultured rabbit articular chondrocytes. FEBS Lett. 1988;234:172–176. doi: 10.1016/0014-5793(88)81327-9. [DOI] [PubMed] [Google Scholar]
- 107.Galera P., Vivien D., Pronost S., Bonaventure J., Redini F., Loyau G., Pujol J.P. Transforming growth factor-beta 1 (TGF-beta 1) up-regulation of collagen type II in primary cultures of rabbit articular chondrocytes (RAC) involves increased mRNA levels without affecting mRNA stability and procollagen processing. J. Cell Physiol. 1992;153:596–606. doi: 10.1002/jcp.1041530322. [DOI] [PubMed] [Google Scholar]
- 108.Recklies A.D., Baillargeon L., White C. Regulation of cartilage oligomeric matrix protein synthesis in human synovial cells and articular chondrocytes. Arthritis Rheum. 1998;41:997–1006. doi: 10.1002/1529-0131(199806)41:6<997::AID-ART6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 109.Motaung S.C., Di Cesare P.E., Reddi A.H. Differential response of cartilage oligomeric matrix protein (COMP) to morphogens of bone morphogenetic protein/transforming growth factor-beta family in the surface, middle and deep zones of articular cartilage. J. Tissue Eng. Regen. Med. 2011;5:e87–e96. doi: 10.1002/term.358. [DOI] [PubMed] [Google Scholar]
- 110.Iozzo R.V., Pillarisetti J., Sharma B., Murdoch A.D., Danielson K.G., Uitto J., Mauviel A. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming growth factor-beta via a nuclear factor 1-binding element. J. Biol. Chem. 1997;272:5219–5228. doi: 10.1074/jbc.272.8.5219. [DOI] [PubMed] [Google Scholar]
- 111.Burton-Wurster N., Lust G. Fibronectin and proteoglycan synthesis in long term cultures of cartilage explants in Ham’s F12 supplemented with insulin and calcium: effects of the addition of TGF-beta. Arch. Biochem. Biophys. 1990;283:27–33. doi: 10.1016/0003-9861(90)90607-Z. [DOI] [PubMed] [Google Scholar]
- 112.Morales T.I. Transforming growth factor-beta 1 stimulates synthesis of proteoglycan aggregates in calf articular cartilage organ cultures. Arch. Biochem. Biophys. 1991;286:99–106. doi: 10.1016/0003-9861(91)90013-9. [DOI] [PubMed] [Google Scholar]
- 113.Niikura T., Reddi A.H. Differential regulation of lubricin/superficial zone protein by transforming growth factor beta/bone morphogenetic protein superfamily members in articular chondrocytes and synoviocytes. Arthritis Rheum. 2007;56:2312–2321. doi: 10.1002/art.22659. [DOI] [PubMed] [Google Scholar]
- 114.van der Kraan P.M., Vitters E.L., van den Berg W.B. Inhibition of proteoglycan synthesis by transforming growth factor beta in anatomically intact articular cartilage of murine patellae. Ann. Rheum. Dis. 1992;51:643–647. doi: 10.1136/ard.51.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Glansbeek H.L., van der Kraan P.M., Vitters E.L., van den Berg W.B. Correlation of the size of type II transforming growth factor beta (TGF-beta) receptor with TGF-beta responses of isolated bovine articular chondrocytes. Ann. Rheum. Dis. 1993;52:812–816. doi: 10.1136/ard.52.11.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chadjichristos C., Ghayor C., Herrouin J.F., Ala-Kokko L., Suske G., Pujol J.P., Galera P. Down-regulation of human type II collagen gene expression by transforming growth factor-beta 1 (TGF-beta 1) in articular chondrocytes involves SP3/SP1 ratio. J. Biol. Chem. 2002;277:43903–43917. doi: 10.1074/jbc.M206111200. [DOI] [PubMed] [Google Scholar]
- 117.O’Keefe R.J., Puzas J.E., Brand J.S., Rosier R.N. Effects of transforming growth factor-beta on matrix synthesis by chick growth plate chondrocytes. Endocrinology. 1988;122:2953–2961. doi: 10.1210/endo-122-6-2953. [DOI] [PubMed] [Google Scholar]
- 118.Vivien D., Galera P., Lebrun E., Loyau G., Pujol J.P. Differential effects of transforming growth factor-beta and epidermal growth factor on the cell cycle of cultured rabbit articular chondrocytes. J. Cell Physiol. 1990;143:534–545. doi: 10.1002/jcp.1041430319. [DOI] [PubMed] [Google Scholar]
- 119.van Beuningen H.M., Glansbeek H.L., van der Kraan P.M., van den Berg W.B. Osteoarthritis-like changes in the murine knee joint resulting from intra-articular transforming growth factor-beta injections. Osteoarthr. Cartil. 2000;8:25–33. doi: 10.1053/joca.1999.0267. [DOI] [PubMed] [Google Scholar]
- 120.van Beuningen H.M., van der Kraan P.M., Arntz O.J., van den Berg W.B. Protection from interleukin 1 induced destruction of articular cartilage by transforming growth factor beta: studies in anatomically intact cartilage in vitro and in vivo. Ann. Rheum. Dis. 1993;52:185–191. doi: 10.1136/ard.52.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van Beuningen H.M., van der Kraan P.M., Arntz O.J., van den Berg W.B. In vivo protection against interleukin-1-induced articular cartilage damage by transforming growth factor-beta 1: age-related differences. Ann. Rheum. Dis. 1994;53:593–600. doi: 10.1136/ard.53.9.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takahashi N., Rieneck K., van der Kraan P.M., van Beuningen H.M., Vitters E.L., Bendtzen K., van den Berg W.B. Elucidation of IL-1/TGF-beta interactions in mouse chondrocyte cell line by genome-wide gene expression. Osteoarthr. Cartil. 2005;13:426–438. doi: 10.1016/j.joca.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 123.Redini F., Mauviel A., Pronost S., Loyau G., Pujol J.P. Transforming growth factor beta exerts opposite effects from interleukin-1 beta on cultured rabbit articular chondrocytes through reduction of interleukin-1 receptor expression. Arthritis Rheum. 1993;36:44–50. doi: 10.1002/art.1780360108. [DOI] [PubMed] [Google Scholar]
- 124.Glansbeek H.L., van Beuningen H.M., Vitters E.L., van der Kraan P.M., van den Berg W.B. Stimulation of articular cartilage repair in established arthritis by local administration of transforming growth factor-beta into murine knee joints. Lab. Investig. 1998;78:133–142. [PubMed] [Google Scholar]
- 125.Pronost S., Segond N., Macro M., Redini F., Penfornis H., Jullienne A., Moukhtar M.S., Pujol J.P. Modulation of interleukin-1 receptor expression by transforming growth factor-beta in cultured rabbit articular chondrocytes: analysis by reverse transcription-polymerase chain reaction. Osteoarthr. Cartil. 1995;3:147–155. doi: 10.1016/S1063-4584(05)80049-4. [DOI] [PubMed] [Google Scholar]
- 126.Wiegertjes R., van Caam A., van Beuningen H., Koenders M., van Lent P., van der Kraan P., van de Loo F., Blaney Davidson E. TGF-beta dampens IL-6 signaling in articular chondrocytes by decreasing IL-6 receptor expression. Osteoarthr. Cartil. 2019 doi: 10.1016/j.joca.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 127.Shiou S.R., Yu Y., Guo Y., Westerhoff M., Lu L., Petrof E.O., Sun J., Claud E.C. Oral administration of transforming growth factor-beta1 (TGF-beta1) protects the immature gut from injury via Smad protein-dependent suppression of epithelial nuclear factor kappaB (NF-kappaB) signaling and proinflammatory cytokine production. J. Biol. Chem. 2013;288:34757–34766. doi: 10.1074/jbc.M113.503946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.DiChiara M.R., Kiely J.M., Gimbrone M.A., Jr., Lee M.E., Perrella M.A., Topper J.N. Inhibition of E-selectin gene expression by transforming growth factor beta in endothelial cells involves coactivator integration of Smad and nuclear factor kappaB-mediated signals. J. Exp. Med. 2000;192:695–704. doi: 10.1084/jem.192.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li T.F., Darowish M., Zuscik M.J., Chen D., Schwarz E.M., Rosier R.N., Drissi H., O’Keefe R.J. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J. Bone Miner. Res. 2006;21:4–16. doi: 10.1359/JBMR.050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen J., Li J., Wang B., Jin H., Wang M., Zhang Y., Yang Y., Im H.J., O’Keefe R., Chen D. Deletion of the transforming growth factor beta receptor type II gene in articular chondrocytes leads to a progressive osteoarthritis-like phenotype in mice. Arthritis Rheum. 2013;65:3107–3119. doi: 10.1002/art.38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Serra R., Johnson M., Filvaroff E.H., LaBorde J., Sheehan D.M., Derynck R., Moses H.L. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J. Cell Biol. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sueyoshi T., Yamamoto K., Akiyama H. Conditional deletion of Tgfbr2 in hypertrophic chondrocytes delays terminal chondrocyte differentiation. Matrix Biol. 2012;31:352–359. doi: 10.1016/j.matbio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 133.Chen R., Mian M., Fu M., Zhao J.Y., Yang L., Li Y., Xu L. Attenuation of the progression of articular cartilage degeneration by inhibition of TGF-beta1 signaling in a mouse model of osteoarthritis. Am. J. Pathol. 2015;185:2875–2885. doi: 10.1016/j.ajpath.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mitsugi S., Ariyoshi W., Okinaga T., Kaneuji T., Kataoka Y., Takahashi T., Nishihara T. Mechanisms involved in inhibition of chondrogenesis by activin-A. Biochem. Biophys. Res. Commun. 2012;420:380–384. doi: 10.1016/j.bbrc.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 135.Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 2004;279:43514–43521. doi: 10.1074/jbc.M407041200. [DOI] [PubMed] [Google Scholar]
- 136.Alexander S., Watt F., Sawaji Y., Hermansson M., Saklatvala J. Activin A is an anticatabolic autocrine cytokine in articular cartilage whose production is controlled by fibroblast growth factor 2 and NF-kappaB. Arthritis Rheum. 2007;56:3715–3725. doi: 10.1002/art.22953. [DOI] [PubMed] [Google Scholar]
- 137.Tardif G., Hum D., Pelletier J.P., Boileau C., Ranger P., Martel-Pelletier J. Differential gene expression and regulation of the bone morphogenetic protein antagonists follistatin and gremlin in normal and osteoarthritic human chondrocytes and synovial fibroblasts. Arthritis Rheum. 2004;50:2521–2530. doi: 10.1002/art.20441. [DOI] [PubMed] [Google Scholar]
- 138.Tardif G., Pelletier J.P., Boileau C., Martel-Pelletier J. The BMP antagonists follistatin and gremlin in normal and early osteoarthritic cartilage: An immunohistochemical study. Osteoarthr. Cartil. 2009;17:263–270. doi: 10.1016/j.joca.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 139.Flechtenmacher J., Huch K., Thonar E.J., Mollenhauer J.A., Davies S.R., Schmid T.M., Puhl W., Sampath T.K., Aydelotte M.B., Kuettner K.E. Recombinant human osteogenic protein 1 is a potent stimulator of the synthesis of cartilage proteoglycans and collagens by human articular chondrocytes. Arthritis Rheum. 1996;39:1896–1904. doi: 10.1002/art.1780391117. [DOI] [PubMed] [Google Scholar]
- 140.Luyten F.P., Chen P., Paralkar V., Reddi A.H. Recombinant bone morphogenetic protein-4, transforming growth factor-beta 1, and activin A enhance the cartilage phenotype of articular chondrocytes in vitro. Exp. Cell Res. 1994;210:224–229. doi: 10.1006/excr.1994.1033. [DOI] [PubMed] [Google Scholar]
- 141.Chen A.L., Fang C., Liu C., Leslie M.P., Chang E., Di Cesare P.E. Expression of bone morphogenetic proteins, receptors, and tissue inhibitors in human fetal, adult, and osteoarthritic articular cartilage. J. Orthop. Res. 2004;22:1188–1192. doi: 10.1016/j.orthres.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 142.van Caam A., Madej W., Thijssen E., Garcia de Vinuesa A., van den Berg W., Goumans M.J., Ten Dijke P., Blaney Davidson E., van der Kraan P.M. Expression of TGFbeta-family signalling components in ageing cartilage: Age-related loss of TGFbeta and BMP receptors. Osteoarthr. Cartil. 2016 doi: 10.1016/j.joca.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 143.Bragdon B., Moseychuk O., Saldanha S., King D., Julian J., Nohe A. Bone morphogenetic proteins: A critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 144.Sengle G., Ono R.N., Lyons K.M., Bachinger H.P., Sakai L.Y. A new model for growth factor activation: Type II receptors compete with the prodomain for BMP-7. J. Mol. Biol. 2008;381:1025–1039. doi: 10.1016/j.jmb.2008.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Aykul S., Martinez-Hackert E. Transforming Growth Factor-beta Family Ligands Can Function as Antagonists by Competing for Type II Receptor Binding. J. Biol. Chem. 2016;291:10792–10804. doi: 10.1074/jbc.M115.713487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pregizer S.K., Mortlock D.P. Dynamics and cellular localization of Bmp2, Bmp4, and Noggin transcription in the postnatal mouse skeleton. J. Bone Miner. Res. 2015;30:64–70. doi: 10.1002/jbmr.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hills R.L., Belanger L.M., Morris E.A. Bone morphogenetic protein 9 is a potent anabolic factor for juvenile bovine cartilage, but not adult cartilage. J. Orthop. Res. 2005;23:611–617. doi: 10.1016/j.orthres.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 148.Blaney Davidson E.N., Vitters E.L., van Lent P.L., van de Loo F.A., van den Berg W.B., van der Kraan P.M. Elevated extracellular matrix production and degradation upon bone morphogenetic protein-2 (BMP-2) stimulation point toward a role for BMP-2 in cartilage repair and remodeling. Arthritis Res. Ther. 2007;9:R102. doi: 10.1186/ar2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou N., Li Q., Lin X., Hu N., Liao J.Y., Lin L.B., Zhao C., Hu Z.M., Liang X., Xu W., et al. BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell Tissue Res. 2016 doi: 10.1007/s00441-016-2403-0. [DOI] [PubMed] [Google Scholar]
- 150.Blaney Davidson E.N., Vitters E.L., Bennink M.B., van Lent P.L., van Caam A.P., Blom A.B., van den Berg W.B., van de Loo F.A., van der Kraan P.M. Inducible chondrocyte-specific overexpression of BMP2 in young mice results in severe aggravation of osteophyte formation in experimental OA without altering cartilage damage. Ann. Rheum. Dis. 2015;74:1257–1264. doi: 10.1136/annrheumdis-2013-204528. [DOI] [PubMed] [Google Scholar]
- 151.Kaneko K., Higuchi C., Kunugiza Y., Yoshida K., Sakai T., Yoshikawa H., Nakata K. Hyaluronan inhibits BMP-induced osteoblast differentiation. FEBS Lett. 2015;589:447–454. doi: 10.1016/j.febslet.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 152.Klammert U., Mueller T.D., Hellmann T.V., Wuerzler K.K., Kotzsch A., Schliermann A., Schmitz W., Kuebler A.C., Sebald W., Nickel J. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol. 2015;13:77. doi: 10.1186/s12915-015-0183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chang S.C., Hoang B., Thomas J.T., Vukicevic S., Luyten F.P., Ryba N.J., Kozak C.A., Reddi A.H., Moos M., Jr. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J. Biol. Chem. 1994;269:28227–28234. [PubMed] [Google Scholar]
- 154.Erlacher L., Ng C.K., Ullrich R., Krieger S., Luyten F.P. Presence of cartilage-derived morphogenetic proteins in articular cartilage and enhancement of matrix replacement in vitro. Arthritis Rheum. 1998;41:263–273. doi: 10.1002/1529-0131(199802)41:2<263::AID-ART10>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 155.Bobacz K., Gruber R., Soleiman A., Graninger W.B., Luyten F.P., Erlacher L. Cartilage-derived morphogenetic protein-1 and -2 are endogenously expressed in healthy and osteoarthritic human articular chondrocytes and stimulate matrix synthesis. Osteoarthr. Cartil. 2002;10:394–401. doi: 10.1053/joca.2002.0522. [DOI] [PubMed] [Google Scholar]
- 156.Chubinskaya S., Merrihew C., Cs-Szabo G., Mollenhauer J., McCartney J., Rueger D.C., Kuettner K.E. Human articular chondrocytes express osteogenic protein-1. J. Histochem. Cytochem. 2000;48:239–250. doi: 10.1177/002215540004800209. [DOI] [PubMed] [Google Scholar]
- 157.Muehleman C., Kuettner K.E., Rueger D.C., Ten Dijke P., Chubinskaya S. Immunohistochemical localization of osteogenetic protein (OP-1) and its receptors in rabbit articular cartilage. J. Histochem. Cytochem. 2002;50:1341–1350. doi: 10.1177/002215540205001007. [DOI] [PubMed] [Google Scholar]
- 158.Peterson R.S., Andhare R.A., Rousche K.T., Knudson W., Wang W., Grossfield J.B., Thomas R.O., Hollingsworth R.E., Knudson C.B. CD44 modulates Smad1 activation in the BMP-7 signaling pathway. J. Cell Biol. 2004;166:1081–1091. doi: 10.1083/jcb.200402138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Andhare R.A., Takahashi N., Knudson W., Knudson C.B. Hyaluronan promotes the chondrocyte response to BMP-7. Osteoarthr. Cartil. 2009;17:906–916. doi: 10.1016/j.joca.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Luo N., Knudson W., Askew E.B., Veluci R., Knudson C.B. CD44 and hyaluronan promote the bone morphogenetic protein 7 signaling response in murine chondrocytes. Arthritis Rheumatol. 2014;66:1547–1558. doi: 10.1002/art.38388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Soder S., Hakimiyan A., Rueger D.C., Kuettner K.E., Aigner T., Chubinskaya S. Antisense inhibition of osteogenic protein 1 disturbs human articular cartilage integrity. Arthritis Rheum. 2005;52:468–478. doi: 10.1002/art.20856. [DOI] [PubMed] [Google Scholar]
- 162.Nishida Y., Knudson C.B., Eger W., Kuettner K.E., Knudson W. Osteogenic protein 1 stimulates cells-associated matrix assembly by normal human articular chondrocytes: up-regulation of hyaluronan synthase, CD44, and aggrecan. Arthritis Rheum. 2000;43:206–214. doi: 10.1002/1529-0131(200001)43:1<206::AID-ANR25>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 163.Huch K., Wilbrink B., Flechtenmacher J., Koepp H.E., Aydelotte M.B., Sampath T.K., Kuettner K.E., Mollenhauer J., Thonar E.J. Effects of recombinant human osteogenic protein 1 on the production of proteoglycan, prostaglandin E2, and interleukin-1 receptor antagonist by human articular chondrocytes cultured in the presence of interleukin-1beta. Arthritis Rheum. 1997;40:2157–2161. doi: 10.1002/art.1780401209. [DOI] [PubMed] [Google Scholar]
- 164.Chubinskaya S., Segalite D., Pikovsky D., Hakimiyan A.A., Rueger D.C. Effects induced by BMPS in cultures of human articular chondrocytes: Comparative studies. Growth Factors. 2008;26:275–283. doi: 10.1080/08977190802291733. [DOI] [PubMed] [Google Scholar]
- 165.Bobacz K., Sunk I.G., Hofstaetter J.G., Amoyo L., Toma C.D., Akira S., Weichhart T., Saemann M., Smolen J.S. Toll-like receptors and chondrocytes: The lipopolysaccharide-induced decrease in cartilage matrix synthesis is dependent on the presence of toll-like receptor 4 and antagonized by bone morphogenetic protein 7. Arthritis Rheum. 2007;56:1880–1893. doi: 10.1002/art.22637. [DOI] [PubMed] [Google Scholar]
- 166.Im H.J., Pacione C., Chubinskaya S., Van Wijnen A.J., Sun Y., Loeser R.F. Inhibitory effects of insulin-like growth factor-1 and osteogenic protein-1 on fibronectin fragment- and interleukin-1beta-stimulated matrix metalloproteinase-13 expression in human chondrocytes. J. Biol. Chem. 2003;278:25386–25394. doi: 10.1074/jbc.M302048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Abula K., Muneta T., Miyatake K., Yamada J., Matsukura Y., Inoue M., Sekiya I., Graf D., Economides A.N., Rosen V., et al. Elimination of BMP7 from the developing limb mesenchyme leads to articular cartilage degeneration and synovial inflammation with increased age. FEBS Lett. 2015;589:1240–1248. doi: 10.1016/j.febslet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 168.Hurtig M., Chubinskaya S., Dickey J., Rueger D. BMP-7 protects against progression of cartilage degeneration after impact injury. J. Orthop. Res. 2009;27:602–611. doi: 10.1002/jor.20787. [DOI] [PubMed] [Google Scholar]
- 169.Badlani N., Inoue A., Healey R., Coutts R., Amiel D. The protective effect of OP-1 on articular cartilage in the development of osteoarthritis. Osteoarthr. Cartil. 2008;16:600–606. doi: 10.1016/j.joca.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 170.Hayashi M., Muneta T., Ju Y.J., Mochizuki T., Sekiya I. Weekly intra-articular injections of bone morphogenetic protein-7 inhibits osteoarthritis progression. Arthritis Res. Ther. 2008;10:R118. doi: 10.1186/ar2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kuo A.C., Rodrigo J.J., Reddi A.H., Curtiss S., Grotkopp E., Chiu M. Microfracture and bone morphogenetic protein 7 (BMP-7) synergistically stimulate articular cartilage repair. Osteoarthr. Cartil. 2006;14:1126–1135. doi: 10.1016/j.joca.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 172.Spiro A.S., Beil F.T., Baranowsky A., Barvencik F., Schilling A.F., Nguyen K., Khadem S., Seitz S., Rueger J.M., Schinke T., et al. BMP-7-induced ectopic bone formation and fracture healing is impaired by systemic NSAID application in C57BL/6-mice. J. Orthop. Res. 2010;28:785–791. doi: 10.1002/jor.21044. [DOI] [PubMed] [Google Scholar]
- 173.Hunter D.J., Pike M.C., Jonas B.L., Kissin E., Krop J., McAlindon T. Phase 1 safety and tolerability study of BMP-7 in symptomatic knee osteoarthritis. BMC Musculoskelet Disord. 2010;11:232. doi: 10.1186/1471-2474-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen P., Vukicevic S., Sampath T.K., Luyten F.P. Bovine articular chondrocytes do not undergo hypertrophy when cultured in the presence of serum and osteogenic protein-1. Biochem. Biophys. Res. Commun. 1993;197:1253–1259. doi: 10.1006/bbrc.1993.2612. [DOI] [PubMed] [Google Scholar]
- 175.Caron M.M., Emans P.J., Cremers A., Surtel D.A., Coolsen M.M., van Rhijn L.W., Welting T.J. Hypertrophic differentiation during chondrogenic differentiation of progenitor cells is stimulated by BMP-2 but suppressed by BMP-7. Osteoarthr. Cartil. 2013;21:604–613. doi: 10.1016/j.joca.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 176.Haaijman A., D’Souza R.N., Bronckers A.L., Goei S.W., Burger E.H. OP-1 (BMP-7) affects mRNA expression of type I, II, X collagen, and matrix Gla protein in ossifying long bones in vitro. J. Bone Miner. Res. 1997;12:1815–1823. doi: 10.1359/jbmr.1997.12.11.1815. [DOI] [PubMed] [Google Scholar]
- 177.Caron M.M., Emans P.J., Surtel D.A., van der Kraan P.M., van Rhijn L.W., Welting T.J. BAPX-1/NKX-3.2 acts as a chondrocyte hypertrophy molecular switch in osteoarthritis. Arthritis Rheumatol. 2015;67:2944–2956. doi: 10.1002/art.39293. [DOI] [PubMed] [Google Scholar]
- 178.Erlacher L., McCartney J., Piek E., ten Dijke P., Yanagishita M., Oppermann H., Luyten F.P. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J. Bone Miner. Res. 1998;13:383–392. doi: 10.1359/jbmr.1998.13.3.383. [DOI] [PubMed] [Google Scholar]
- 179.Chen P., Vukicevic S., Sampath T.K., Luyten F.P. Osteogenic protein-1 promotes growth and maturation of chick sternal chondrocytes in serum-free cultures. Pt 1J. Cell Sci. 1995;108:105–114. doi: 10.1242/jcs.108.1.105. [DOI] [PubMed] [Google Scholar]
- 180.Bobacz K., Gruber R., Soleiman A., Erlacher L., Smolen J.S., Graninger W.B. Expression of bone morphogenetic protein 6 in healthy and osteoarthritic human articular chondrocytes and stimulation of matrix synthesis in vitro. Arthritis Rheum. 2003;48:2501–2508. doi: 10.1002/art.11248. [DOI] [PubMed] [Google Scholar]
- 181.Nilsson O., Parker E.A., Hegde A., Chau M., Barnes K.M., Baron J. Gradients in bone morphogenetic protein-related gene expression across the growth plate. J. Endocrinol. 2007;193:75–84. doi: 10.1677/joe.1.07099. [DOI] [PubMed] [Google Scholar]
- 182.Ito H., Akiyama H., Shigeno C., Nakamura T. Bone morphogenetic protein-6 and parathyroid hormone-related protein coordinately regulate the hypertrophic conversion in mouse clonal chondrogenic EC cells, ATDC5. Biochim. Biophys. Acta. 1999;1451:263–270. doi: 10.1016/S0167-4889(99)00100-7. [DOI] [PubMed] [Google Scholar]
- 183.Bidart M., Ricard N., Levet S., Samson M., Mallet C., David L., Subileau M., Tillet E., Feige J.J., Bailly S. BMP9 is produced by hepatocytes and circulates mainly in an active mature form complexed to its prodomain. Cell Mol. Life Sci. 2012;69:313–324. doi: 10.1007/s00018-011-0751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Blunk T., Sieminski A.L., Appel B., Croft C., Courter D.L., Chieh J.J., Goepferich A., Khurana J.S., Gooch K.J. Bone morphogenetic protein 9: A potent modulator of cartilage development in vitro. Growth Factors. 2003;21:71–77. doi: 10.1080/0897719031000148822. [DOI] [PubMed] [Google Scholar]
- 185.Luu H.H., Song W.X., Luo X., Manning D., Luo J., Deng Z.L., Sharff K.A., Montag A.G., Haydon R.C., He T.C. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J. Orthop. Res. 2007;25:665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 186.van Caam A., Blaney Davidson E., Garcia de Vinuesa A., van Geffen E., van den Berg W., Goumans M.J., Ten Dijke P., van der Kraan P. The high affinity ALK1-ligand BMP9 induces a hypertrophy-like state in chondrocytes that is antagonized by TGFbeta1. Osteoarthr. Cartil. 2015;23:985–995. doi: 10.1016/j.joca.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 187.Miyamoto Y., Mabuchi A., Shi D., Kubo T., Takatori Y., Saito S., Fujioka M., Sudo A., Uchida A., Yamamoto S., et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 188.Ratnayake M., Ploger F., Santibanez-Koref M., Loughlin J. Human chondrocytes respond discordantly to the protein encoded by the osteoarthritis susceptibility gene GDF5. PLoS ONE. 2014;9:e86590. doi: 10.1371/journal.pone.0086590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Martin J.A., Buckwalter J.A. Aging, articular cartilage chondrocyte senescence and osteoarthritis. Biogerontology. 2002;3:257–264. doi: 10.1023/A:1020185404126. [DOI] [PubMed] [Google Scholar]
- 190.Chubinskaya S., Kumar B., Merrihew C., Heretis K., Rueger D.C., Kuettner K.E. Age-related changes in cartilage endogenous osteogenic protein-1 (OP-1) Biochim. Biophys. Acta. 2002;1588:126–134. doi: 10.1016/S0925-4439(02)00158-8. [DOI] [PubMed] [Google Scholar]
- 191.Iqbal J., Dudhia J., Bird J.L., Bayliss M.T. Age-related effects of TGF-beta on proteoglycan synthesis in equine articular cartilage. Biochem. Biophys. Res. Commun. 2000;274:467–471. doi: 10.1006/bbrc.2000.3167. [DOI] [PubMed] [Google Scholar]
- 192.Loeser R.F., Im H.J., Richardson B., Lu Q., Chubinskaya S. Methylation of the OP-1 promoter: Potential role in the age-related decline in OP-1 expression in cartilage. Osteoarthr. Cartil. 2009;17:513–517. doi: 10.1016/j.joca.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Loeser R.F., Gandhi U., Long D.L., Yin W., Chubinskaya S. Aging and oxidative stress reduce the response of human articular chondrocytes to insulin-like growth factor 1 and osteogenic protein 1. Arthritis Rheumatol. 2014;66:2201–2209. doi: 10.1002/art.38641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao W., Wang T., Luo Q., Chen Y., Leung V.Y., Wen C., Shah M.F., Pan H., Chiu K., Cao X., et al. Cartilage degeneration and excessive subchondral bone formation in spontaneous osteoarthritis involves altered TGF-beta signaling. J. Orthop. Res. 2016;34:763–770. doi: 10.1002/jor.23079. [DOI] [PubMed] [Google Scholar]
- 195.Bauge C., Girard N., Lhuissier E., Bazille C., Boumediene K. Regulation and Role of TGFbeta Signaling Pathway in Aging and Osteoarthritis Joints. Aging Dis. 2014;5:394–405. doi: 10.14336/AD.2014.0500394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hui W., Young D.A., Rowan A.D., Xu X., Cawston T.E., Proctor C.J. Oxidative changes and signalling pathways are pivotal in initiating age-related changes in articular cartilage. Ann. Rheum. Dis. 2016;75:449–458. doi: 10.1136/annrheumdis-2014-206295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hjelmeland A.B., Schilling S.H., Guo X., Quarles D., Wang X.F. Loss of Smad3-mediated negative regulation of Runx2 activity leads to an alteration in cell fate determination. Mol. Cell Biol. 2005;25:9460–9468. doi: 10.1128/MCB.25.21.9460-9468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zheng L., Baek H.J., Karsenty G., Justice M.J. Filamin B represses chondrocyte hypertrophy in a Runx2/Smad3-dependent manner. J. Cell Biol. 2007;178:121–128. doi: 10.1083/jcb.200703113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Responte D.J., Lee J.K., Hu J.C., Athanasiou K.A. Biomechanics-driven chondrogenesis: From embryo to adult. FASEB J. 2012;26:3614–3624. doi: 10.1096/fj.12-207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nam J., Perera P., Rath B., Agarwal S. Dynamic regulation of bone morphogenetic proteins in engineered osteochondral constructs by biomechanical stimulation. Tissue Eng. Part A. 2013;19:783–792. doi: 10.1089/ten.tea.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Madej W., van Caam A., Blaney Davidson E., Buma P., van der Kraan P.M. Unloading results in rapid loss of TGFbeta signaling in articular cartilage: Role of loading-induced TGFbeta signaling in maintenance of articular chondrocyte phenotype? Osteoarthr. Cartil. 2016;24:1807–1815. doi: 10.1016/j.joca.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 202.Hinterwimmer S., Krammer M., Krotz M., Glaser C., Baumgart R., Reiser M., Eckstein F. Cartilage atrophy in the knees of patients after seven weeks of partial load bearing. Arthritis Rheum. 2004;50:2516–2520. doi: 10.1002/art.20378. [DOI] [PubMed] [Google Scholar]
- 203.Nomura M., Sakitani N., Iwasawa H., Kohara Y., Takano S., Wakimoto Y., Kuroki H., Moriyama H. Thinning of articular cartilage after joint unloading or immobilization. An experimental investigation of the pathogenesis in mice. Osteoarthr. Cartil. 2017;25:727–736. doi: 10.1016/j.joca.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 204.Madej W., van Caam A., Davidson E.N., Hannink G., Buma P., van der Kraan P.M. Ageing is associated with reduction of mechanically-induced activation of Smad2/3P signaling in articular cartilage. Osteoarthr. Cartil. 2016;24:146–157. doi: 10.1016/j.joca.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 205.Wallace I.J., Worthington S., Felson D.T., Jurmain R.D., Wren K.T., Maijanen H., Woods R.J., Lieberman D.E. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. USA. 2017;114:9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Iijima H., Ito A., Nagai M., Tajino J., Yamaguchi S., Kiyan W., Nakahata A., Zhang J., Wang T., Aoyama T., et al. Physiological exercise loading suppresses post-traumatic osteoarthritis progression via an increase in bone morphogenetic proteins expression in an experimental rat knee model. Osteoarthr. Cartil. 2017;25:964–975. doi: 10.1016/j.joca.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 207.Chang S.H., Mori D., Kobayashi H., Mori Y., Nakamoto H., Okada K., Taniguchi Y., Sugita S., Yano F., Chung U.I., et al. Excessive mechanical loading promotes osteoarthritis through the gremlin-1-NF-kappaB pathway. Nat. Commun. 2019;10:1442. doi: 10.1038/s41467-019-09491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Henao-Murillo L., Ito K., van Donkelaar C.C. Collagen Damage Location in Articular Cartilage Differs if Damage is Caused by Excessive Loading Magnitude or Rate. Ann. Biomed. Eng. 2018;46:605–615. doi: 10.1007/s10439-018-1986-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Scanzello C.R., Goldring S.R. The role of synovitis in osteoarthritis pathogenesis. Bone. 2012;51:249–257. doi: 10.1016/j.bone.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Robinson W.H., Lepus C.M., Wang Q., Raghu H., Mao R., Lindstrom T.M., Sokolove J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.D’Agostino M.A., Conaghan P., Le Bars M., Baron G., Grassi W., Martin-Mola E., Wakefield R., Brasseur J.L., So A., Backhaus M., et al. EULAR report on the use of ultrasonography in painful knee osteoarthritis. Part 1: Prevalence of inflammation in osteoarthritis. Ann. Rheum. Dis. 2005;64:1703–1709. doi: 10.1136/ard.2005.037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Baker K., Grainger A., Niu J., Clancy M., Guermazi A., Crema M., Hughes L., Buckwalter J., Wooley A., Nevitt M., et al. Relation of synovitis to knee pain using contrast-enhanced MRIs. Ann. Rheum. Dis. 2010;69:1779–1783. doi: 10.1136/ard.2009.121426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sokolove J., Lepus C.M. Role of inflammation in the pathogenesis of osteoarthritis: Latest findings and interpretations. Ther. Adv. Musculoskelet Dis. 2013;5:77–94. doi: 10.1177/1759720X12467868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Guermazi A., Roemer F.W., Hayashi D., Crema M.D., Niu J., Zhang Y., Marra M.D., Katur A., Lynch J.A., El-Khoury G.Y., et al. Assessment of synovitis with contrast-enhanced MRI using a whole-joint semiquantitative scoring system in people with, or at high risk of, knee osteoarthritis: the MOST study. Ann. Rheum. Dis. 2011;70:805–811. doi: 10.1136/ard.2010.139618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pessler F., Dai L., Diaz-Torne C., Gomez-Vaquero C., Paessler M.E., Zheng D.H., Einhorn E., Range U., Scanzello C., Schumacher H.R. The synovitis of “non-inflammatory” orthopaedic arthropathies: A quantitative histological and immunohistochemical analysis. Ann. Rheum. Dis. 2008;67:1184–1187. doi: 10.1136/ard.2008.087775. [DOI] [PubMed] [Google Scholar]
- 216.Goldring M.B., Goldring S.R. Osteoarthritis. J. Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 217.Bondeson J., Blom A.B., Wainwright S., Hughes C., Caterson B., van den Berg W.B. The role of synovial macrophages and macrophage-produced mediators in driving inflammatory and destructive responses in osteoarthritis. Arthritis Rheum. 2010;62:647–657. doi: 10.1002/art.27290. [DOI] [PubMed] [Google Scholar]
- 218.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J.P., Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 219.Wojdasiewicz P., Poniatowski L.A., Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm. 2014;2014:561459. doi: 10.1155/2014/561459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sellam J., Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat. Rev. Rheumatol. 2010;6:625–635. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- 221.Laavola M., Leppanen T., Hamalainen M., Vuolteenaho K., Moilanen T., Nieminen R., Moilanen E. IL-6 in Osteoarthritis: Effects of Pine Stilbenoids. Molecules. 2018;24:109. doi: 10.3390/molecules24010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Legendre F., Bogdanowicz P., Boumediene K., Pujol J.P. Role of interleukin 6 (IL-6)/IL-6R-induced signal tranducers and activators of transcription and mitogen-activated protein kinase/extracellular. J. Rheumatol. 2005;32:1307–1316. [PubMed] [Google Scholar]
- 223.Ryu J.H., Yang S., Shin Y., Rhee J., Chun C.H., Chun J.S. Interleukin-6 plays an essential role in hypoxia-inducible factor 2alpha-induced experimental osteoarthritic cartilage destruction in mice. Arthritis Rheum. 2011;63:2732–2743. doi: 10.1002/art.30451. [DOI] [PubMed] [Google Scholar]
- 224.Cortial D., Gouttenoire J., Rousseau C.F., Ronziere M.C., Piccardi N., Msika P., Herbage D., Mallein-Gerin F., Freyria A.M. Activation by IL-1 of bovine articular chondrocytes in culture within a 3D collagen-based scaffold. An in vitro model to address the effect of compounds with therapeutic potential in osteoarthritis. Osteoarthr. Cartil. 2006;14:631–640. doi: 10.1016/j.joca.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 225.Fernandes J.C., Martel-Pelletier J., Pelletier J.P. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237–246. [PubMed] [Google Scholar]
- 226.Bauge C., Legendre F., Leclercq S., Elissalde J.M., Pujol J.P., Galera P., Boumediene K. Interleukin-1beta impairment of transforming growth factor beta1 signaling by down-regulation of transforming growth factor beta receptor type II and up-regulation of Smad7 in human articular chondrocytes. Arthritis Rheum. 2007;56:3020–3032. doi: 10.1002/art.22840. [DOI] [PubMed] [Google Scholar]
- 227.Roman-Blas J.A., Stokes D.G., Jimenez S.A. Modulation of TGF-beta signaling by proinflammatory cytokines in articular chondrocytes. Osteoarthr. Cartil. 2007;15:1367–1377. doi: 10.1016/j.joca.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Madej W., Buma P., van der Kraan P. Inflammatory conditions partly impair the mechanically mediated activation of Smad2/3 signaling in articular cartilage. Arthritis Res. Ther. 2016;18:146. doi: 10.1186/s13075-016-1038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Elshaier A.M., Hakimiyan A.A., Rappoport L., Rueger D.C., Chubinskaya S. Effect of interleukin-1beta on osteogenic protein 1-induced signaling in adult human articular chondrocytes. Arthritis Rheum. 2009;60:143–154. doi: 10.1002/art.24151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Merrihew C., Soeder S., Rueger D.C., Kuettner K.E., Chubinskaya S. Modulation of endogenous osteogenic protein-1 (OP-1) by interleukin-1 in adult human articular cartilage. J. Bone Jt. Surg. Am. 2003;85-A Suppl 3:67–74. doi: 10.2106/00004623-200300003-00012. [DOI] [PubMed] [Google Scholar]
- 231.Bauge C., Attia J., Leclercq S., Pujol J.P., Galera P., Boumediene K. Interleukin-1beta up-regulation of Smad7 via NF-kappaB activation in human chondrocytes. Arthritis Rheum. 2008;58:221–226. doi: 10.1002/art.23154. [DOI] [PubMed] [Google Scholar]
- 232.Matsuzaki K. Smad phospho-isoforms direct context-dependent TGF-beta signaling. Cytokine Growth Factor Rev. 2013;24:385–399. doi: 10.1016/j.cytogfr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 233.Kamato D., Burch M.L., Piva T.J., Rezaei H.B., Rostam M.A., Xu S., Zheng W., Little P.J., Osman N. Transforming growth factor-beta signalling: Role and consequences of Smad linker region phosphorylation. Cell Signal. 2013;25:2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 234.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 235.Shi Y. Structural insights on Smad function in TGFbeta signaling. Bioessays. 2001;23:223–232. doi: 10.1002/1521-1878(200103)23:3<223::AID-BIES1032>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 236.Hill C.S. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 237.Sapkota G., Knockaert M., Alarcon C., Montalvo E., Brivanlou A.H., Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- 238.Matsuzaki K. Smad phosphoisoform signaling specificity: The right place at the right time. Carcinogenesis. 2011;32:1578–1588. doi: 10.1093/carcin/bgr172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Millet C., Yamashita M., Heller M., Yu L.R., Veenstra T.D., Zhang Y.E. A negative feedback control of transforming growth factor-beta signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J. Biol. Chem. 2009;284:19808–19816. doi: 10.1074/jbc.M109.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bae E., Sato M., Kim R.J., Kwak M.K., Naka K., Gim J., Kadota M., Tang B., Flanders K.C., Kim T.A., et al. Definition of smad3 phosphorylation events that affect malignant and metastatic behaviors in breast cancer cells. Cancer Res. 2014;74:6139–6149. doi: 10.1158/0008-5472.CAN-14-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Murata M., Yoshida K., Yamaguchi T., Matsuzaki K. Linker phosphorylation of Smad3 promotes fibro-carcinogenesis in chronic viral hepatitis of hepatocellular carcinoma. World J. Gastroenterol. 2014;20:15018–15027. doi: 10.3748/wjg.v20.i41.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Demagny H., De Robertis E.M. Point mutations in the tumor suppressor Smad4/DPC4 enhance its phosphorylation by GSK3 and reversibly inactivate TGF-beta signaling. Mol. Cell Oncol. 2016;3:e1025181. doi: 10.1080/23723556.2015.1025181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Demagny H., Araki T., De Robertis E.M. The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-beta signaling. Cell Rep. 2014;9:688–700. doi: 10.1016/j.celrep.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 244.Heldin C.H., Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 245.Sekimoto G., Matsuzaki K., Yoshida K., Mori S., Murata M., Seki T., Matsui H., Fujisawa J., Okazaki K. Reversible Smad-dependent signaling between tumor suppression and oncogenesis. Cancer Res. 2007;67:5090–5096. doi: 10.1158/0008-5472.CAN-06-4629. [DOI] [PubMed] [Google Scholar]
- 246.Yu J.S., Ramasamy T.S., Murphy N., Holt M.K., Czapiewski R., Wei S.K., Cui W. PI3K/mTORC2 regulates TGF-beta/Activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nat. Commun. 2015;6:7212. doi: 10.1038/ncomms8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bruce D.L., Sapkota G.P. Phosphatases in SMAD regulation. FEBS Lett. 2012;586:1897–1905. doi: 10.1016/j.febslet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 248.Liu T., Feng X.H. Regulation of TGF-beta signalling by protein phosphatases. Biochem. J. 2010;430:191–198. doi: 10.1042/BJ20100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Saklatvala J. Inflammatory signaling in cartilage: MAPK and NF-kappaB pathways in chondrocytes and the use of inhibitors for research into pathogenesis and therapy of osteoarthritis. Curr. Drug Targets. 2007;8:305–313. doi: 10.2174/138945007779940115. [DOI] [PubMed] [Google Scholar]
- 250.Djouad F., Rackwitz L., Song Y., Janjanin S., Tuan R.S. ERK1/2 activation induced by inflammatory cytokines compromises effective host tissue integration of engineered cartilage. Tissue Eng. Part A. 2009;15:2825–2835. doi: 10.1089/ten.tea.2008.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Geng Y., Valbracht J., Lotz M. Selective activation of the mitogen-activated protein kinase subgroups c-Jun NH2 terminal kinase and p38 by IL-1 and TNF in human articular chondrocytes. J. Clin. Investig. 1996;98:2425–2430. doi: 10.1172/JCI119056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Manley G.C.A., Stokes C.A., Marsh E.K., Sabroe I., Parker L.C. DUSP10 Negatively Regulates the Inflammatory Response to Rhinovirus through Interleukin-1beta Signaling. J. Virol. 2019;93 doi: 10.1128/JVI.01659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Arnold M.A., Kim Y., Czubryt M.P., Phan J., McAnally D., Qi X., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 254.Quinn Z.A., Yang C.C., Wrana J.L., McDermott J.C. Smad proteins function as co-modulators for MEF2 transcriptional regulatory proteins. Nucleic Acids Res. 2001;29:732–742. doi: 10.1093/nar/29.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liu D., Kang J.S., Derynck R. TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004;23:1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]