Abstract
Although CD4+ T cells likely play key roles in antitumor immune responses, most immuno-oncology studies have been limited to CD8+ T-cell responses due to multiple technical barriers and a lack of shared antigens across patients. Merkel cell carcinoma (MCC) is an aggressive skin cancer caused by Merkel cell polyomavirus (MCPyV) oncoproteins in 80% of cases. Because MCPyV oncoproteins are shared across most patients with MCC, it is unusually feasible to identify, characterize, and potentially augment tumor-specific CD4+ T cells. Here, we report the identification of CD4+ T-cell responses against six MCPyV epitopes, one of which included a conserved, essential viral oncogenic domain that binds/disables the cellular retinoblastoma (Rb) tumor suppressor. We found that this epitope (WEDLT209–228) could be presented by three population-prevalent HLA class II alleles, making it a relevant target in 64% of virus-positive MCC patients. Cellular staining with a WEDLT209–228–HLA-DRB1*0401 tetramer indicated that specific CD4+ T cells were detectable in 78% (14 of 18) of evaluable MCC patients, were 250-fold enriched within MCC tumors relative to peripheral blood, and had diverse T-cell receptor sequences. We also identified a modification of this domain that still allowed recognition by these CD4+ T cells, but disabled binding to the Rb tumor suppressor, a key step in the detoxification of a possible therapeutic vaccine. The use of these new tools for deeper study of MCPyV-specific CD4+ T cells may provide a broader insight into cancer-specific CD4+ T-cell responses.
Keywords: Immunotherapy, skin cancer, Merkel cell polyomavirus, Merkel cell carcinoma, CD4+ T cells
INTRODUCTION
Merkel cell carcinoma (MCC) is a rare but deadly skin cancer with a relative mortality rate of 46%, making it approximately three times as deadly as malignant melanoma on a per-case basis (1). The current annual US incidence of ~2,500 cases per year is projected to climb to ~3,200 by the year 2025 (2). This predicted increase is due to the unusually strong association of age with MCC and the aging of the “Baby Boomer” generation (2). In the United States, the majority of MCCs (~80%) are etiologically linked to the Merkel cell polyomavirus (MCPyV) (3), a small (~5.4 kilobase genome) double-stranded DNA virus that encodes oncogenic T-antigens, including large T (LT) and small T (sT) (3). Importantly, elimination of the C-terminal region of LT (necessary for viral DNA replication) is an invariant requirement for oncogenesis. In contrast, MCCs retain the N-terminal region, which promotes cell cycle progression and contains the ‘LxCxE motif’ (4,5). The LxCxE-binding motif is conserved in LT among human polyomaviruses and several other DNA viruses, suggesting that manipulation of retinoblastoma (Rb) biology is critical for viral pathogenesis (6). Interestingly, the LxCxE domain in several DNA viruses was recently shown to mediate direct binding to STING and, thus, antagonize the cyclic GMP-AMP synthase (cGAS) innate antiviral interferon pathway (7), which may further increase the selective pressure to maintain this sequence. Consequently, the MCPyV T-antigens, which are mechanistically involved in tumor formation, are both attractive targets for the study of tumor-specific T-cell responses and rational candidates for specific immunotherapy (8,9).
The importance of immune cell function in MCC is highlighted by the fact that MCC patients exhibit high response rates to PD-1 blocking agents (10–12). Robust CD8+ T-cell intratumoral infiltration is associated with 100% disease-specific survival, independent of stage at diagnosis (13,14). However, this favorable pattern is only observed in 4–18% of patients (13,14). In light of this link between local CD8+ T cells and MCC survival, we sought to explore mechanisms that could regulate the CD8+ T-cell response, including the adequacy of CD4+ T-cell help. CD4+ T cells are known to provide crucial support for CD8+ T-cell function in a variety of settings. Cellular therapies utilizing CD4+ CAR T-cell products (15), autologous CD4+ T cells (16), or TCR-transgenic CD4+ T cells (17,18) have mediated cancer regression. Therapeutic cancer vaccines primarily targeting CD4+ T cells can induce epitope spreading of CD8+ T-cell responses (19) and have shown promise in treating melanoma (20,21). These findings suggest that CD4+ T cells significantly contribute to cancer immune-based therapies. However, the low frequencies of antigen-specific CD4+ T cells within peripheral blood (several logs lower than antigen-specific CD8+ T cells) combined with a lack of specific tools to identify these cells have hindered their study. Consequently, we optimized and employed a suite of complementary methods to identify MCPyV-specific CD4+ T cells with the goal of enabling detailed tetramer-based characterization of CD4++ T-cell responses against MCPyV and of contributing to the development of novel therapeutic strategies for MCC patients.
METHODS
Subjects and specimens
Studies were approved by the Fred Hutchinson Cancer Research Center (FHCRC) and the Benaroya Research Institute (BRI) Institutional Review Boards and conducted according to the Declaration of Helsinki principles. Informed written consent was received from all participants. Subjects were HLA class II typed via polymerase chain reaction (PCR)-based methods at Bloodworks Northwest (Seattle, WA), by high-throughput next-generation sequencing at Scisco Genetics (FHCRC) as described (22), or by real-time PCR at BRI as described (23). We screened 89 MCC patients for appropriate HLA types from our repository of over 1400 patients. Of those, 5 patients with available tumor material had HLA types we could study. Peripheral mononuclear cells (PBMCs) were obtained from heparinized blood from MCC patients and healthy donors with Lymphocyte Separation Medium (Corning, Tewksbury MA) and cryopreserved in freezing medium [50% human serum (Valley Biomedical, Winchester VA), 40% RPMI (ThermoFisher, Waltham MA), and 10% DMSO (ThermoFisher, Waltham MA)]. Fresh MCC tumor material and punch biopsies (>1 cm3) were processed into single-cell digests by mincing them into small pieces with sterile forceps and scissors, followed by incubation in 20 mL of digestion medium composed of RPMI plus 0.002g DNase (Worthington Biochemical, Lakewood NJ), 0.008g collagenase (Worthington Biochemical, Lakewood NJ), and 0.002 g hyaluronidase (Worthington Biochemical, Lakewood NJ) in a 10 cm dish at 37°C with frequent, gentle swirling. After 3 hours of digestion, cells were strained through a 70 μm filter, centrifuged, resuspended in freezing medium (as described above), and cryopreserved. Tumor infiltrating lymphocytes (TILs) were expanded for 2 weeks as described (24) before analysis. Briefly, minced tumor biopsies were cultured in T-cell medium (TCM) [RPMI, 8% human serum, 200 nM L-glutamine and penicillin-streptomycin (100 U/mL; ThermoFisher, Waltham MA)] as described (25). Lymphoblastoid cell lines (LCL) were derived from PBMCs and maintained in LCL medium [RPMI, 10% fetal bovine serum, 200nM L-glutamine, penicillin-streptomycin (100 U/mL), 55 μM 2-Mercaptoethanol (ThermoFisher, Waltham MA), and 1 mM sodium pyruvate (ThermoFisher, Waltham MA)], as described (24).
Cell lines
MCC cell lines WaGa, MKL-1, and MCC-13 were cultured in RPMI-1640 supplemented with 10% Fetal bovine serum (Atlantic Biologicals, Miami FL), 200nM L-glutamine, and penicillin-streptomycin (100 U/mL). WaGa cells were a gift from Dr. Juergen Becker, German Cancer Research Center (8), MKL-1 cells from Masa Shuda (26), and MCC-13 cells from Helen Leonard (27). All cells, except COS-7, were confirmed authentic with short-tandem repeat analysis (STR) (ATCC, Manassas VA) and were mycoplasma negative via PlasmoTest (InvivoGen, San Diego CA). COS-7 cells (ATCC, CRL-1651, 2005) were maintained in DMEM supplemented with 10% FBS, 200 nM L-glutamine, and penicillin–streptomycin (100 U/mL) and were free of mycoplasmsa.
Tumor cell line reactivity determination
To examine the reactivity with intact tumor antigen, cell lysates of MCPyV-positive cell lines WaGa, MKL-1, and of the MCPyV-negative MCC cell line (MCC-13) were used. The resulting supernatants were used as antigen (or control non-antigen) in the IFNγ intracellular cytokine staining assays described below.
Epitope determination
MCPyV oncoprotein CD4+ T cell epitopes were identified using three approaches: Tetramer-guided epitope mapping (TGEM), intracellular cytokine staining (ICS), and carboxyfluorescein succinimidyl ester (CFSE) dilution.
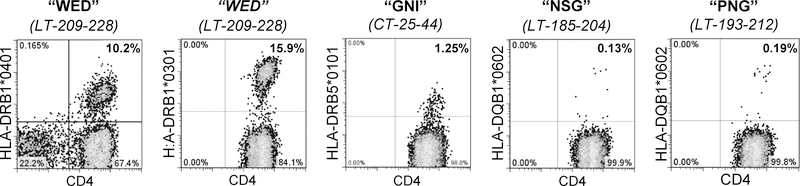
For TGEM studies (28), PBMCs obtained from 24 healthy donors expressing at least one of eight discrete HLA class-II allele types were cultured in TCM in the presence of pools of five MCPyV 20-mer peptides (Genscript; Supplementary Table S1) at 1 μg/mL final concentration of each peptide for 10–14 days. Subsequently, expanded PBMCs were subjected to two rounds of tetramer staining (first by tetramers loaded with a pool of five consecutive overlapping peptides and then with individual peptide loaded tetramers corresponding to positive pools) as previously described (28). Tetramers were generated at the BRI for the following alleles: HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1*0404, HLA- HLA-DRB1*0701, HLA-DRB1*1501, DRB5*0101, and HLA-DQB1*0602. Flow cytometry stains identified live CD4+ (Biolegend, #344618) cells containing tetramer pool-positive cells and excluded CD8+ (Invitrogen, MHCD0805), CD14+ (Biolegend, #301828), and CD19+ (Biolegend, #302232) cells. Cultures were then stained with single tetramers containing relevant peptides within the positive pool (Table 1). Four HLA alleles yielded MCPyV epitopes (HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB5*0101 and HLA-DQB1*0602), and for each allele type, four individuals were tested (Figure 1). The fraction of patients with a given allele in which tetramer-positive cells were detected are denoted in Table 1.
TABLE I:
NEWLY IDENTIFIED CD4 T CELL RESPONSES AGAINST MCPYV ONCOPROTEIN PEPTIDES
| Name of Peptide (first 3 A.A.) | Peptide sequence (core sequence underlined) | Location in MCPyV1 | HLA Restriction | Tet available? | T cell source | Detected in |
Method(s) of identification | ||
|---|---|---|---|---|---|---|---|---|---|
| Healthy donors (expanded4) | Healthy donors (ex vivo) | MCC pts | |||||||
| “MDL” | MDLVLNRKEREAL | CT-1–13 | Undetermined | No2 | PBMC | ND | ND | 1/1 | ICS |
| “GNI” | GNIPLMKAAFKRSCLKHHPD | CT-25–44 | DRB5*0101 | Yes | PBMC | 1/4 | ND | 1/1 | TGEM, ICS |
| “PVI” | PVIMMELNTLWSK | CT-49–61 | Undetermined | No2 | TIL | ND | ND | 1/1 | ICS |
| “NSG” | NSGRESSTPNGTSVPRNSSR | LT-185–204 | DQB1*0602 | Yes | PBMC | 1/4 | ND | 1/1 | TGEM, ICS |
| “PNG” | PNGTSVPRNSSRTYGTWEDL | LT-193–212 | |||||||
| “WED” | WEDLFCDESLSSP | LT-209–221 | DQB1*0301 | No2 | TIL/PBMC | ND | ND | 1/1 | ICS |
| “WED” | WEDLFCDESLSSPEPPSSSE | LT-209–228 | DRB1*0401 | Yes | TIL/PBMC | 4/4 | 8/10 | 14/18 | TGEM, ICS |
| “WED” | WEDLFCDESLSSPEPPSSSE | LT-209–228 | DRB1*0301 | Yes | PBMC | 3/4 | ND | 1/4 | TGEM |
| “CIS” | CISCKLSRQHCSLKTLKQKN | sT-121–140 | DRB1*0401 | No3 | PBMC | ND | ND | 1/1 | CFSE dilution, ICS |
The first 78 amino acids of LT and sT are shared and are referred to at ‘common T’ or CT.
Tetramer synthesis not attempted due to lack of specific HLA restriction or unavailability of specific HLA allele type.
Tetramer generation attempted but unsuccessful.
Cells were expanded in the presence of 10 peptide pools (20mers) for 10–14 days prior to staining.
Figure 1: TGEM identified four peptides presented by four population-prevalent HLA class II alleles.
TGEM was performed as outlined in the Methods testing seven allele types. PBMCs were cultured in the presence of pools of five MCPyV 20-mer peptides (WED, GNI, NSG, PNG; amino acid sequence indicated) for 10–14 days, followed by staining for pools of tetrameric complexes of 10 peptides (composed of two combined pools) bound to HLA class II molecules. After identification of CD4+tetramer+ T cells, the same cultures were stained with single tetramers containing each relevant peptide within the positive pool. Representative flow plots are shown for each peptide. For each epitope, four individuals were tested, and one to four persons were positive as detailed in column 7 of Table 1. The percentage of viable CD4+tetramer+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted.
For ICS assays focusing on IFNγ synthesis, peptide pools composed of 17–27 peptides [13-mers] at final concentrations of 1 μg/mL of each peptide were incubated with 2.5–5×105 TILs and an equal number of autologous PBMCs as antigen presenting cells (APCs) for 16 hours as previously described (24). Data were acquired on a Canto RUO cytometer (Becton Dickinson). Flow cytometry analysis evaluated the percentage of live IFNγ+ (BD Pharmingen, 554701) CD4+ cells and excluded dead (LIVE/DEAD Fixable Violet Dead Cell Stain, Invitrogen, #L34955), CD8+, CD14+, and CD19+ cells.
For CFSE dilution (29), PBMCs from three MCC patients were stained with CFSE (Vybrant CFDA SE cell tracer kit, Invitrogen Cat. # V-12883) following the manufacturer’s instructions and incubated in RPMI-1640 containing 10% human serum, 2 mM L-glutamine (Hyclone), and 1% penicillin/streptomycin (Hyclone) at a concentration of 4 × 106 cells per well in a 24-well plate. Two peptide pools composed of 24 peptides (20-mers overlapping by 12-amino acids; Genscript; Supplemental Table S1) were incubated with CFSE-labeled PBMCs at final concentrations of 1 μg/mL for each peptide for 5 days. Flow cytometry analysis evaluated the percentage of CFSE dilute among CD3+ (ECD; Beckman Coulter, Cat #: IM2705U) CD4+ cells (PE; Biolegend Cat #: 357404), excluding dead (7-AAD+; BD Biosciences Cat #: 559925).
In each of these methods, positive signals using peptide pools were confirmed through follow-up assays using individual peptides to define epitopes. The MCPyV CD4 epitopes discovered and validated through these complementary methods are summarized in Table 1.
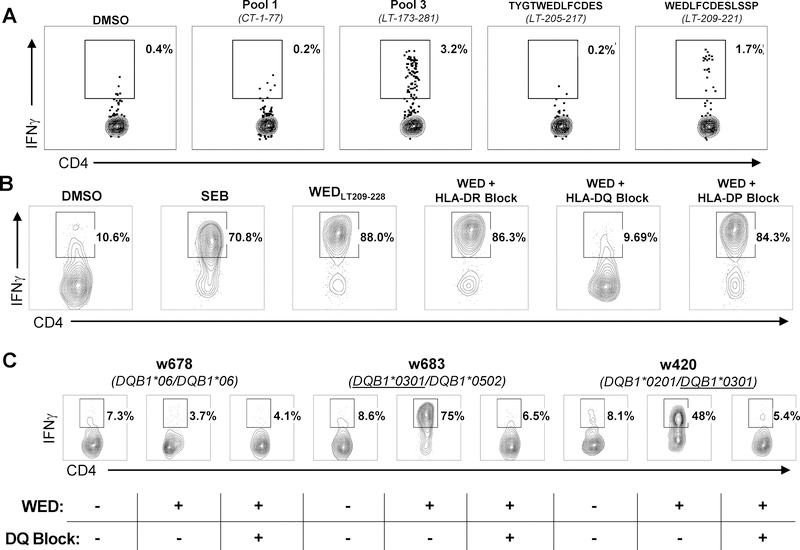
For one patient in whom WEDLT209–228-specificity was based on IFNγ secretion (Figure 2A), generated T-cell clones (described in the next section) were then tested for production of IFNγ following exposure to antigen in the presence of HLA class II locus-specific blocking monoclonal antibodies (30) in order to determine HLA class-II allele restriction (Figure 2B). The IFNγ flow cytometry release assay was the same protocol as described above, with the exception that APCs were incubated for 10 minutes at room temperature with 125 μL of HLA blocking antibodies, L243 (HLA-DR) and sPVL3 (HLA-DQ), prior to the addition of responder cells with antigenic peptide. Antibodies pan-specific for blocking all HLA DR (clone L243), HLA DP (clone B7.21), or HLA DQ (clone SPVL3) allelic variants were generated from murine hybridoma cell-lines (31) and stored as crude supernatants at −80 °C. Supernatants were used at a final 1:8 dilution for blocking.
Figure 2: The WEDLT209–228 epitope is presented by HLA-DRB1*0401, HLA-DRB1*0301, and HLA-DQB1*0301 alleles.
A: ICS screening of expanded TILs from an MCC patient biopsy incubated with four MCPyV T-antigen 13-mer peptide pools (two shown). Reactive pool 3 was broken down into individual 13-mer peptides (two shown). The percentage of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. Negative control: TILs stimulated with DMSO and the peptide ‘TYGWEDLFCDES’, which is 4 amino acids N-terminal to the ‘WED” peptide. Positive control: TILs stimulated with SEB. B: A WEDLT209–228-specific clone ± anti-HLA-class II locus-specific blocking monoclonal antibodies established HLA-DQ as the restricting locus. The percentage of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. Negative control: DMSO-stimulated; Positive control: SEB-stimulated. C: HLA-DQB1*0301 restriction was established using LCLs from three different patients (“w678”, “w683,” and “w420”) with defined HLA-DQ genotype with a WEDLT209–228-specific CD4+ T-cell clone, peptide (“WED”), and inclusion of anti–HLA-DQ mAb (“DQ-Block”). The percent of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. DMSO and SEB were used as negative and positive controls, respectively. Plots shown represent one of two replicates.
T-cell sorting
WEDLT209–228-specific CD4+ T cells were sorted using tetramer-based single-cell sorting, IFNγ capture, or CFSE dilution staining. Clones were derived from either expanded TIL cultures or peptide-stimulated PBMCs expanded as previously described (24,28).
For tetramer-based sorting, PBMCs or TILs (5–20 X 106) were thawed, phosphate-buffered saline (PBS)-washed, and incubated with 100 nM dasatinib (SelleckChem, #S1021) for 10 minutes at 37°C in T-cell medium (TCM; RPMI, 8% human serum, 200 nM L-glutamine and 100 U/ml penicillin-streptomycin). Cells were washed and resuspended in 50 μL TCM, stained with 2 μL of DRB1*0401-WEDLT209–228-PE tetramer (BRI) and incubated for 1 hour in the dark at room temperature (RT). Cells were washed with 3 mL of 4°C, 1% BSA in PBS (PBS-A), collected by centrifugation, and resuspended in 500 μL PBS-A and enriched immunomagnetically for PE-positive cells following manufacturer’s instructions (StemCell Technologies, #18551). Enriched cells were washed in 2 mL 4°C PBS-A, resuspended in 50 μL of 1:2000 diluted LIVE/DEAD Fixable Violet Dead Cell Stain (Invitrogen, #L34955) and incubated at 25°C for 20 minutes. Cells were then washed in 2 mL of 4°C PBS-A, resuspended in 95 μL of Fc block (StemCell Technologies, #18551) and stained with anti-CD4–Alexafluor 488 (Biolegend, #344618), anti-CD8–APC (Invitrogen, MHCD0805), anti-CD14–PacBlue (Biolegend, #301828), and anti-CD19–PacBlue (Biolegend, #302232). Following a 25-minute incubation on ice, cells were washed 2x with ice-cold PBS-A and resuspended in 150 μL of 2% human serum in RPMI. Viable CD4+tetramer+ cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region were sorted into 96-well plates containing 100 μL TCM. For single-cell sorting using IFNγ-capture, the IFNγ secretion enrichment kit (Miltenyi Biotec, #130–054-201) was used as per manufacturer’s instructions. T cells were sorted on an Aria II cell sorter (Becton Dickinson), and data were collected using Becton Dickinson-FACS Diva software (v6.1.1). For sorting using CFSE dilution, cells were stained and stimulated with peptide as described above.
T-cell expansion
Cells sorted using one of the three methods described above were expanded for 2 weeks in 96-well plates with allogeneic irradiated feeder PBMCs (150,000 cells/well) and phytohemagglutinin (PHA)(1.6 μg/mL) as described (32). After 24 hours, rIL15 (20 ng/mL; R&D Systems, #247-ILB-025) and natural IL2 (32 U/mL, Hemagen Diagnostics, #906011) were added. After 2 weeks, microcultures with visible growth were screened for specificity via ELISA or tetramer staining (described subsequently). Confirmed peptide-specific clones were further expanded in the presence of irradiated feeder cells, rIL2 (50 IU/ml; R&D Systems, #202-IL-010), and OKT3 (30 ng/mL; Miltenyi Biotec, #130–093-387) as described (24) plus rIL15 (20 ng/mL). Clones generated for these experiments are listed in Supplementary Table S2.
IFNγ and proliferation T-cell activation assays
The responsiveness of MCC patient WEDLT209–228-specific CD4+ T-cell clones were evaluated as previously described (24). Briefly, T-cell clones from 5 individuals (listed in Supplementary Table S2) were stimulated with antigen (1 μg/mL of the WEDLT209–228 20mer peptide or 1:100 dilution of tumor cell line lysates) using autologous PBMCs from the same 5 individuals as APCs that were CFSE-labeled to allow their exclusion from analysis (24). Cultures stimulated with dimethyl sulfoxide (DMSO) or staphylococcal enterotoxin B (SEB; 1 μg/mL; ThermoFisher, NC0333616) were included as a negative and a positive control, respectively. Cells were stained for IFNγ and analyzed with FlowJo (version 10.4.0, FlowJo LLC), and viable IFNγ+CD4+ cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region were designated as responder cells from the lymphocyte population. Some functional assays used cloned CD4+ T-cell responders and 3H-thymidine-based proliferation measurement as previously described (31).
Minimal epitope determination
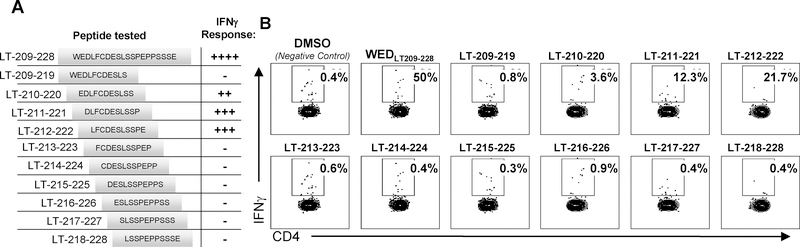
To determine the minimal antigenic motif for positive 20-mers, 11-mer peptides overlapping by 10 amino acids (AA) were synthesized spanning the entire 20-mer (listed in Figure 3; Genscript). ICS was performed as described previously (24). Briefly, autologous PBMCs were CFSE-labeled to allow dump-gating, and cocultured with responder T cells and peptide, as described above. Flow cytometric analysis done on viable IFNγ+CD4+ cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region were designated as responder cells in ICS assays.
Figure 3: The ‘WED’ minimal epitope presented by HLA-DRB1*0401 encompasses the Rb-binding motif.
A: Schematic of peptides synthesized for testing minimal epitope. B: A ‘WED’-specific CD4+ T-cell clone (wb688 clone 1,3) generated from a healthy donor, following ex vivo peptide-stimulated PBMC expansion, was incubated with an HLA-DRB1*0401 LCLs and stimulated with DMSO vehicle control, ‘WED’ 20-mer (positive control), or internal 11-mer peptides overlapping by 10 amino acids within the ‘WED’ 20-mer. The absence of responses to some 11-mer peptides provides an addition, internal negative control. Peptides were tested at 1 μg/mL and IFNγ responses measured by ICS assay. The percent of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. These data were representative of DRB1*0401-restricted WEDLT209–228 -specific CD4+ T-cell clones generated from three individuals as detailed using a secreted IFNγ readout in Supplemental Figure S5A.
IFNγ ELISA Assays
WEDLT209–228-specific T-cell functional avidity was determined by co-incubating HLA-DRB1*0401-restricted WEDLT209–228 T-cell clones and autologous PBMCs (1:1 ratio, 2 × 105 cells/well each) in 200 μL TCM with 10-fold serial-diluted antigenic peptides at final concentrations of 10−6 to 10−14 g/mL for 16 hours. Cross-reactivity against homologous regions of 12 additional human polyomaviruses was assessed by incubating HLA-DRB1*0401-restricted WEDLT209–228 T-cell clones and autologous PBMC (1:1 ratio, 2 × 105 cells/well each) in 200 μl TCM with 13-mer peptides (Supplementary Table S3) at a final concentration of 1 μg/mL for 16 hours. To determine the reactivity of HLA-DRB1*0401-restricted WEDLT209–228 T-cell clones against wild-type versus S220-phosphorylated WEDLT209–228, HLA-DRB1*0401-restricted WEDLT209–228 T-cell clones were incubated with autologous PBMCs (1:1 ratio, 2 × 105 cells/well each) in 200 μL TCM with 20-mer wild-type WEDLT209–228 or S220-phosphorylated WEDLT209–228 (Genscript) peptides at a final concentration of 1 μg/mL for 16 hours. Concentrations of secreted IFNγ in cell culture supernatants were determined by ELISA (Affymetrix, #88–7316-76). To calculate EC50, IFNγ concentrations vs. peptide concentrations were analyzed via nonlinear regression using Prism version 7.0 (Graph-Pad Software, Inc.).
Large T-antigen production and site-directed mutagenesis
The expression plasmid for the Large T (LT) antigen fusion protein (pDEST103-GFP-LT) was created using Gateway recombination cloning technology (ThermoFisher Scientific) to insert LT from pCMV-MCV156 into pDEST103 (33). The resulting plasmid expresses the eGFP-LT fusion protein under the control of the CMV promoter. Site-directed mutagenesis (New England Biolabs, #E0554S) was performed to generate LT S220A and LT E216K mutants with the following primer sets: forward primer 5’-GCGATGAATCACTTTCCGCTCCTGAGCCTC-3’ and reverse primer 5’-AGAAGAGATCCTCCCAGGTGCC-3’ for S220A; forward primer 5’-CGACAAGTCACTTCCTCCCCTGAG-3’ and reverse primer 5’-CAGAACAGATCCTCCCAGGTGCCATC-3’ for E216K. Mutations at the specific sites were confirmed by sequencing. Protein was expressed by COS-7 cells (ATCC, CRL-1651, 2005). COS-7 cells were plated the day before transfection at 75,000 cells/well in 12-well plates in 0.5 mL of DMEM + 10% FBS, 200 nM L-glutamine, and penicillin–streptomycin (100 U/mL), and pDEST103-GFP-LT was transfected into COS-7 cells using FuGENE HD (Promega, #E2311) as per the manufacturer’s instructions. Seventy-two hours after transfection, eGFP-positive cells were sorted, and lysates were generated as described above. Clarified supernatants were used as the antigen in ICS assays as described above.
T-cell receptor beta sequencing and analysis of dextramer-sorted WEDLT209–228-specific cells
At least 3 million cells derived from fresh tumor digest of three HLA DRB1*0401-positive MCC patients were stained with HLA-DRB1*0401-WEDLT209–228-PE dextramer (Immundex, Denmark), and the same monoclonal antibody cocktail as described for tetramer sorting plus LIVE/DEAD Fixable Violet (Invitrogen, #L34955). Viable, dextramer+CD4+ cells (negative for CD14, CD19, and CD8) were bulk-sorted using an Aria II cell sorter, flash frozen, and submitted to Adaptive Biotechnologies (Seattle, WA) for genomic DNA extraction. Next-generation survey-level sequencing was used to determine T-cell receptor beta locus (TRB) CDR3 sequences with read normalization performed as described (33). All TRB CDR3 sequences detected at least twice were categorized as dextramer-positive clonotypes. Shannon entropy was calculated on the estimated number of genomes found twice or more for all productive TRB CDR3 reads and normalized by dividing the log2 of the number of unique productive sequences in each sample. Clonality was calculated as 1 minus normalized entropy. Sequences are available through Adaptive ImmuneACCESS (https://clients.adaptivebiotech.com/pub/longino-2019-cir).
Statistical analysis
Analyses were completed using Prism software, version 7 with a statistical significance threshold of 0.05. Specific tests used for each analysis are denoted in figure legends. Flow-cytometry data was repeated with biological replicates, typically from the same subject and the same source of PBMCs or tumor specimens. Biological replicates were assessed on different assay dates. Therefore, statistical analyses across these experiments was not deemed appropriate.
RESULTS
Identification of six CD4 epitopes within the MCPyV T-antigen oncoproteins
Our previously reported screens of expanded tumor-infiltrating lymphocytes (TILs) and overlapping 13-mer peptides spanning the MCPyV T-antigens identified MCPyV-specific CD4+ T cells responsive to TLWSKFQQNIHKL, located within the common T region (amino acids 57–69; TLWCT57–69) shared by LT and sT (24). Using this same method, we subsequently identified two additional CD4 epitopes, WEDLFCDESLSSP (WEDLT209–221; Figure 2A) and PVIMMELNTLWSK (PVICT49–61; Supplementary Figure S1A). Histologic examination has previously revealed CD4+ T cell infiltration into or surrounding many MCPyV-positive MCC tumors (34), therefore, we suspected that many additional CD4 MCPyV epitopes were present but remained undetected via this initial approach. Consequently, we pursued complementary strategies for epitope discovery. We synthesized 20-mer peptides overlapping by 12 amino acids (AA) spanning MCPyV LT and sT (Supplementary Table S1) and used tetramer-guided epitope mapping (TGEM)(28), IFNγ staining (24,35), and CFSE dilution (29) following MCPyV peptide stimulation of PBMCs. Using these approaches, we identified four additional epitopes and verified reactivity to WEDLT209–228 in additional patients (Table 1, Supplementary Figures S1B, S2–4).
T cells that recognized WEDLT209–228 were detected by TGEM using both HLA-DRB1*0401 and HLA-DRB1*0301 tetramers (Figure 1), indicating that the same epitope could be presented in the context of these two HLA class II alleles. To enhance detection of antigen-specific T cells, we generated an HLA-DRB1*0401-WEDLT209–228 dextramer, as dextramers are reported to have improved sensitivity compared to tetramers (36). Screening of peripheral blood samples using this dextramer indicated that WEDLT209–228 -specific CD4+ T cells could be visualized directly ex vivo in the majority of HLA-DRB1*0401 patients (14 of 18, 77.8%) and healthy donors (8 of 10, 80%)(Table 1).
The initial discovery of WEDLT209–228 using TILs from an HLA-DRB1*0401- and HLA-DRB1*0301-negative patient indicated that this peptide could also be presented by a non-HLA-DRB1*0401/DRB1*0301 allele (Figure 2A). To determine this additional allele, PBMCs from the TIL donor in which WEDLT209–228 was initially identified were expanded in the presence of WEDLT209–228 peptide and sorted using IFNγ capture to generate clones (37). After confirming WEDLT209–228-specificity, T-cell clones were tested for production of IFNγ following exposure to antigen in the presence of HLA class II locus-specific blocking monoclonal antibodies (30) (Figure 2B). Only anti-HLA-DQ blocked responses, indicating HLA-DQ restriction. The subject in which the WEDLT209–228 response was initially identified is heterozygous for HLA-DQB1*0301 and HLA-DQB1*0501. LCLs from HLA-genotyped donors (Figure 2C) partially matched to the restricting DQB1 alleles were used as antigen-presenting cells (APCs). Only APCs bearing HLA-DQB1*0301 induced IFNγ responses from T-cell clones, and responses were blocked by anti-HLA-DQ. These data collectively indicated that WEDLT209–228 could be presented by DRB1*0401, DRB1*0301, and DQB1*0301. Among the MCC patients in our cohort for which we have class II HLA typing data (DRB1 locus n=131; DQB1 locus n=53), 64% expressed at least one of these three alleles. Consequently, this epitope is immunologically relevant for the majority of patients with MCPyV-positive tumors.
The epitope within WEDLT209–228 encompasses the LxCxE motif
CD4+ T-cell epitopes typically range in size from 9–22 amino acids in length (38). We sought to determine the core sequence necessary for T-cell recognition within WEDLT209–228 in the context of HLA-DRB1*0401, the most prevalent allele of the three restriction elements in the general population that accommodated this epitope. HLA-DRB1*0401–expressing APCs were loaded with 11-mer peptides overlapping by 10 AA spanning WEDLT209–228 and incubated with WEDLT209–228-specific, HLA DRB1*0401-restricted CD4 clones from three individuals (Supplemental Table S2) to identify its minimal epitope (Figure 3). WEDLT209–228-specific clones responded to each 11-mer spanning LT-210–222, which share the 9 AA core LFCDESLSS (LT-212–220)(Figure 3). Stimulation with this 9-mer did not elicit a significant response in three DRB1*0401-restricted WEDLT209–228-specific clones from the three individuals (Supplementary Figure S5A), suggesting that flanking residues may stabilize and improve the strength of TCR stimulation and/or HLA binding as has been described (39). Importantly, this epitope encompasses the conserved LxCxE binding motif that is critical for MCPyV-LT binding to Rb. The minimal epitope presented by DQB1*0301 was found to be LT-210–219 (Supplementary Figure S5B), whereas the minimal epitope presented by DRB1*0301 was not determined. Because the LxCxE motif is conserved among many DNA viruses, including the other described human polyomaviruses, four HLA-DRB1*0401-restricted WEDLT209–228 T-cell clones were incubated overnight with 13-mer peptides spanning the LxCxE motif of 12 additional human polyomaviruses with homologous regions (Supplementary Table S3). IFNγ secretion from CD4+ T cells clones was detected against the MCPyV peptide (LT-210–222), as well as the homologous sequence from the human polyomavirus 9 (HuPyV9) for one clone and human polyomavirus 12 (HuPyV12) for three of the four clones (Supplementary Figure S5C). These data indicated that the T-cell receptors expressed by these three clones could tolerate different degrees of variance and that cross-reactivity against other polyomaviruses was possible.
CD4+ T cells recognize the WEDLT209–228 epitope in the context of MCC tumors
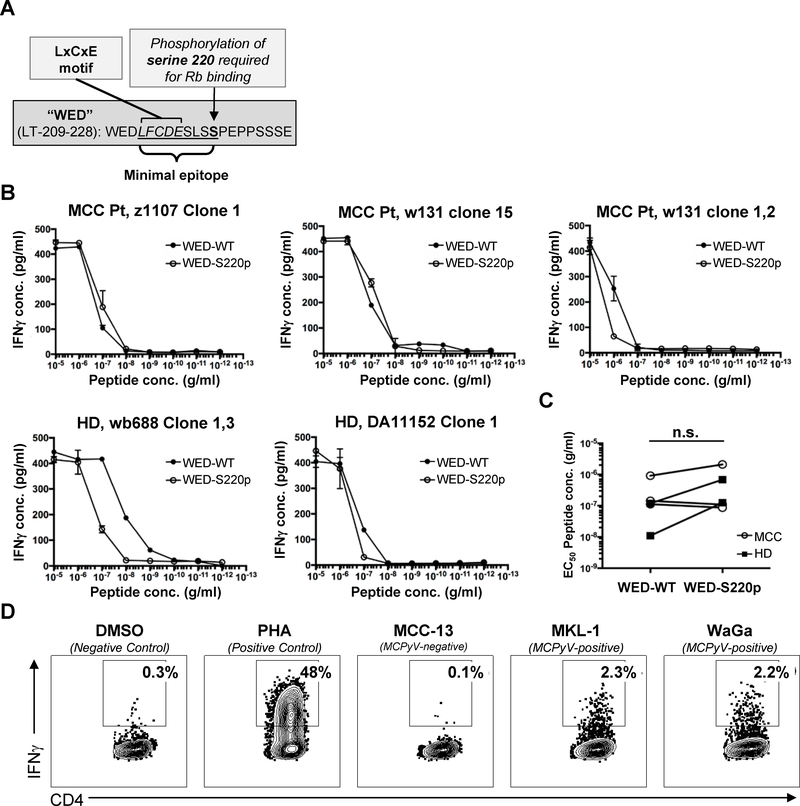
Phosphorylation of serine residue 220 adjacent to the MCPyV LT LxCxE motif is required for binding to Rb (40) (schematic, Figure 4A). To test whether WEDLT209–228 -specific T cells tolerated this post-translational modification, five HLA-DRB1*0401-restricted T-cell clones were assayed for recognition of wild-type (WED-WT) or phosphorylated-S220 (S220p) 20-mers (Figure 4B). Functional avidity, expressed as EC50, was calculated for the five DRB1*0401-restricted clones from four donors (two MCC patients and two healthy controls; Figure 4C). Functional avidities ranged over two orders of magnitude for WED-WT and one order of magnitude for S220p. Phosphorylation of S220 did not significantly change the EC50 for WEDLT209–228-specific CD4+ T-cell clones collectively. However, the two clones derived from healthy donors did show a decrease in functional avidity against WED-S220p relative to WED-WT. Evaluation of a much larger number of clones from additional subjects would be necessary to determine whether this trend is significant. To evaluate whether the WEDLT209–228 epitope can be processed from the LT protein in MCC tumor cells, APCs (autologous PBMCs) and DRB1*0401-restricted WEDLT209–228-specific CD4+ T clones (data from MCC patient z1107 clone 1 depicted, representing replicate data from three tested individuals) were incubated with lysates from two MCPyV-positive MCC cell lines, MKL-1 and WaGa, and a MCPyV-negative cell line MCC-13. WEDLT209–228-specific CD4+ T-cell clones responded specifically to the MCPyV-positive lysates (Figure 4D). These data indicated that the LT protein was present in an immunogenic context in virus-positive MCC cell lines and suggest that WEDLT209–228 is naturally processed in the virus-positive MCC tumor context in vivo.
Figure 4: HLA-DRB1*0401 WEDLT209–228-specific CD4+ T-cell clones can recognize LT presented in the context of MCC tumors.
A: Schematic of ‘WED’ peptide features. B: Dose-response curves depicted for five WEDLT209–228-specific clones stimulated with WEDLT209–228 (WED-WT) or WEDLT209–228-S220p peptides. IFNγ secretion was measured by ELISA, and samples were run in duplicate. Lines connect means. Error bars denote standard deviation. Of the five clones tested, three derived from MCC patients (MCC) and two were derived from healthy donors (HD) C: Dose-response curves were used to calculate the EC50 of five distinct ‘WED’-specific CD4+ T-cell clones stimulated with either WEDLT209–228 or WEDLT209–228-S220p peptides. Clones from healthy donors are filled squares, clones generated from MCC patients are open circles. Wilcoxon matched-pairs signed rank test was used to compare EC50 from clones stimulated with WED-WT or WED-S220p (n.s. not significant). D: WEDLT209–228 -specific clones were incubated with DRB1*0401-positive APCs and lysate from either MCPyV-negative (MCC-13) or MCPyV-positive (MKL-1 and WaGa) cell lines, DMSO (negative control), or PHA (positive control). The percent of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. Flow plots are representative data of four distinct TRB CDR3 clonotypes.
Virus-specific CD4 T cells are enriched within tumors and have diverse TCR repertoires
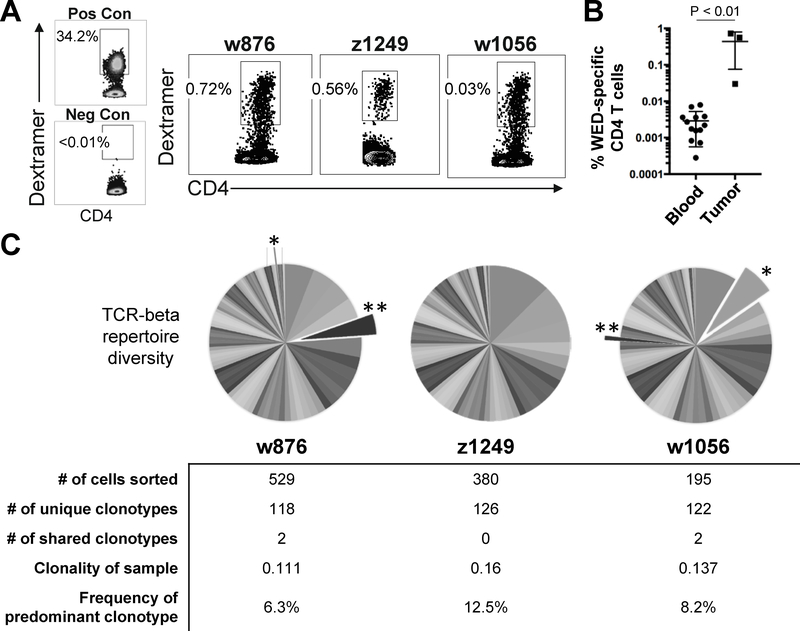
To determine if WEDLT209–228-specific cells infiltrate MCC tumors, we stained tumor digests from three MCC patients with the HLA-DRB1*0401–WEDLT209–228 dextramer. The percentage of WEDLT209–228-specific cells within the total tumor-infiltrating CD4+ T cells ranged from 0.03–0.72% (median 0.56%; Figure 5A). In contrast, PBMCs from 13 MCC patients were stained with the HLA-DRB1*0401-WEDLT209–228 dextramer, and the median frequency of WEDLT209–228 -specific CD4+ T cells within the periphery was significantly lower at 0.0022% of total CD4+ T cells (Figure 5B). These frequencies closely aligned with prior reports of peripheral virus-specific CD4+ T-cell frequencies (41). These results indicated an enrichment of WEDLT209–228-specific CD4+ T cells within MCC tumors compared to the periphery, suggesting active recruitment of these cells into the tumor microenvironment.
Figure 5: WEDLT209–228 -specific CD4+ T cells are enriched in MCC tumors and are diverse.
A: Fresh tumor digest samples were acquired from three HLA-DRB1*0401-positive patients and stained with DRB1*0401-WEDLT209–228 dextramer. A WEDLT209–228 -specific CD4 clone generated against the WEDLT209–228 epitope was spiked into PBMCs as a positive control. TILs from an MCPyV-positive, HLA DB1*0401-negative patient were used as a negative control. Frequency of CD4+dextramer+ T cells shown for the indicated clones. Positive dextramer staining was consistent between 2 replicate experiments. B: Peripheral blood samples (n=13) and fresh tumor digest samples (n=3) from HLA-DRB1*0401-positive MCC patients were stained with DRB1*0401-WEDLT209–228 dextramer as described in Materials and Methods. The percent of viable CD4+dextramer+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region were divided by the total CD4+ T-cell population (in the lymphocyte forward/side scatter region, viable CD4+CD8−CD14−CD19−). The median number of WEDLT209–228 -specific CD4+ T cells of the total CD4 population was 0.0022% in the periphery compared to 0.56% within tumors (p = 0.0036, Mann-Whitney Test). C: TCR beta chain sequencing of the variable CDR3 region was performed on sorted cells from panel A. Pie slices depict the frequency of an individual clonotype within the dextramer-positive, WEDLT209–228 -specific CD4+ T-cell population. Exploded and starred slices indicate two TRB sequences with shared amino acid sequences between patients w876 and w1056.
Next, we evaluated the clonal diversity of WEDLT209–228-specific CD4+ T cells within MCC tumors by next-generation sequencing of the T-cell receptor beta locus (TRB) from dextramer-sorted cells isolated from these tumor digests as previously described (33). Analysis of the TRB complementary determining region 3 (CDR3) sequences yielded 366 unique TRB amino acid sequences from the three tumors (Figure 5C). Of the 366, two sequences were shared between patients ‘w876’ and ‘w1056’ (exploded, starred pie slices; Figure 5C). These were identical at the amino acid but not nucleotide level, suggesting convergence. In order to estimate immunodominance within each tumor’s WEDLT209–228-specific CD4+ T-cell population, clonality scores were calculated (see Materials and Methods). The clonality score of the dextramer-sorted samples ranged from 0.111–0.137, indicating that although the TRB repertoires of infiltrating T cells within these MCC tumors were diverse, a few dominant clonotypes were expanded, with the top clonotype occupying 6.3% to 12.5% of the populations (Figure 5C).
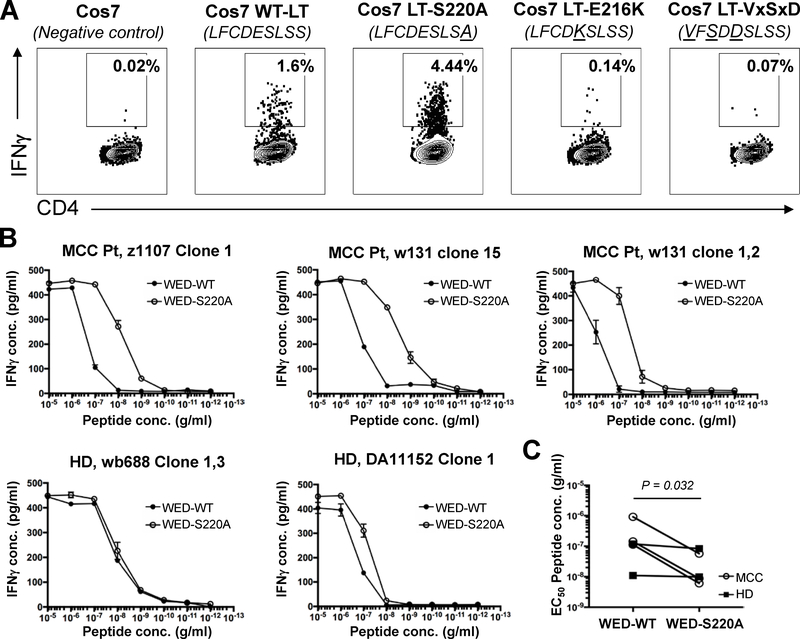
The S220A mutation within WEDLT209–228 retains immunogenicity
The data presented thus far suggests that the WEDLT209–228 epitope could be a therapeutically useful target, either for adoptive cellular therapy or in the context of a therapeutic cancer vaccine. However, inclusion of the LxCxE motif in a vaccine is not straightforward due to the Rb-binding, oncogenic activity of this domain. This issue has been addressed in HPV-vaccines by mutating the LxCxE motif to render the HPV-E7 oncoprotein non-oncogenic (42). Two mutations within the LxCxE domain of MCPyV LT have been reported to inhibit oncogenic activity in vitro, E216K (4) and S220A (40). To test whether immunogenicity of the WEDLT209–228 epitope could be retained with either of these mutations, we created an antigen by transfecting COS-7 cells with wild-type LT (LT-WT), or the LT-S220A or LT-E216K mutants (33). As an experimental control, we also tested a mutant that has all three critical LxCxE residues mutated (LT-VxSxD). Lysates from LT-expressing cells were tested as antigen with autologous PBMCs as APCs and a WEDLT209–228-specific, HLA DRB1*0401-restricted CD4+ T-cell clone (wb688 clone 1,3). WEDLT209–228-specific clones responded to wild-type and MCPyV-LT-S220A (Figure 6A) but not to the ‘detoxified’ E216K and VxSxD variants. To further compare the strength of response against LT-WT and LT-S220A, HLA-DRB1*0401-restricted CD4+ T-cell clones were stimulated with serial dilutions of WED-WT and WED-S220A peptides (20-mers). The S220A variant stimulated clones derived from MCC patients more potently than WED-WT, as measured by EC50 for each responder CD4 clone, whereas little change was observed in the measured EC50 of clones derived from healthy donors (Figure 6B). It will be important to test additional donors and MCC patients to validate whether this difference in response is consistently found between these two donor sources. However, these data do suggest that the LT-S220A mutant (reported not to bind Rb) retained and may even have strengthen immune recognition of this region in the context of HLA-DRB1*0401. These data suggest that this mutation may be clinically useful in the design of a therapeutic cancer vaccine.
Figure 6: WEDLT209–228 -specific CD4+ T-cell clones can recognize mutant S220A but not E216K.
A: A WEDLT209–228-specific clone (wb688 clone 1,3) was incubated with DRB1*0401-positive PBMCs (as APCs) and COS-7 cell lysates that were either untransfected (negative control), transfected with wild-type (WT) LT, or mutant LT-S220A, LT-E216K, or LT-VxSxD. Intracellular IFNγ was measured by ICS. Flow plots are representative data of four HLA DRB1*0401-restricted clones. The percent of viable CD4+IFNγ+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted. B: Dose-response curves of a WEDLT209–228-specific HLA DRB1*0401-restricted CD4 clones stimulated with WEDLT209–228 (WED-WT) or WEDLT209–228-S220A peptides. Clones were incubated with HLA-DRB1*0401-positive LCLs (as APCs) and peptides. IFNγ secretion was measured by ELISA. Of the five clones tested, three were from MCC patients (MCC) and two were from healthy donors (HD). Lines connect means. Error bars denote standard deviation. C: Dose-response curves were used to calculate the EC50 of five distinct WEDLT209–228-specific HLA DRB1*0401-restricted CD4+ T-cell clones for WED-WT or WED-S220A peptides. Wilcoxon matched-pairs signed rank test was used to compare EC50 values. P value shown.
DISCUSSION
Here, we report six MCPyV CD4+ T-cell epitopes, including a conserved and immunogenic WEDLT209–228 epitope within a key oncogenic region of the MCPyV Large T-antigen. The WEDLT209–228 epitope was recognized in the context of at least three discrete, population-prevalent HLA allelic variants. Within our HLA-typed MCC cohort, 64% expressed one or more of these alleles, indicating that this epitope was immunologically relevant in most MCC patients with MCPyV-positive tumors. Further supporting this notion, the WEDLT209–228 epitope encompasses the LxCxE motif, which is a critical binding site of the tumor suppressor Rb (4,40). Persistent expression of this region is required for MCC tumor development and growth (4), and this sequence is conserved in MCC tumors. Indeed, of 99 MCPyV tumor-associated LT sequences in GenBank, only one has a coding variation within LT-212–220 (S219F; Genbank KJ128376.1). WEDLT209–228-specific T cells infiltrated each of three tested MCC tumors from DRB1*0401-positive patients, and WEDLT209–228-specific T cells were 250-fold enriched within these tumor samples compared to blood, as assessed by MHC-peptide multimer staining. These WEDLT209–228-specific T cells exhibited diverse TCR repertoires that include expanded clonotypes, suggesting in vivo antigen recognition and, consequently, that these T cells could home appropriately into tumor tissue and have therapeutic potential.
To date, tumor immunology research has largely focused upon studying and improving CD8+ (rather than CD4+) T-cell responses. This is due to the lower relative frequencies of antigen-specific CD4+ T cells compared to CD8+ T cells (~100–1000 fold (41,43)), making detailed characterization of tumor-specific CD4+ T cells more difficult. The generation of HLA class II tetramers for the isolation and study of antigen-specific CD4+ T cells poses numerous technical challenges (44). However, a growing body of evidence suggests that harnessing tumor-specific CD4+ T cells can improve immune-based therapies and sustain CD8+ T-cell responses (44). Specifically, infusion of autologous TILs, which contain both CD4+ and CD8+ T cells, have shown higher response rates (50–70%) in malignant melanoma than have CD8+ T cells alone (45). A study has described the case of a stage IV acral melanoma patient who experienced a complete response after TIL therapy, which included CD4+ T cells specific for BRAF-V600E. These tumor-specific CD4+ T cells increased in frequency within the blood after therapy and persisted long-term, suggesting that these cells were of clinical significance and augmented the antitumor immune response (46). Administration of autologous cancer-specific CD4+ T cells has mediated regression of distant metastatic disease in epithelial cholangiocarcinoma (17) and complete clinical remission in another patient with metastatic melanoma (16). The use of genetically engineered CD4+ T cells targeting melanoma-associated antigen-A3 (MAGE-A3) yields objective responses among patients with esophageal cancer, urothelial cancer, and osteosarcoma (18), indicating that cellular therapy utilizing CD4+ T cells has powerful therapeutic potential in a wide array of solid cancers. Clinical trials utilizing neoantigen-based therapeutic vaccines for melanoma have yielded impressive clinical responses that correlate with robust CD4+ T-cell stimulation, particularly in the setting of combined PD-1 blockade therapy (20,21). Against this backdrop, our identification of CD4+ T-cell epitopes and the generation of four HLA class II multimers (tetramers/dextramers) facilitated characterization of MCPyV-specific CD4+ T-cell responses and the subsequent development of CD4-targeted therapies for MCC, including a therapeutic cancer vaccine.
Vaccination approaches targeting MCPyV oncoproteins have clinical appeal for several reasons. Viral antigens are shared among MCC patients and, therefore, could be used to generate ‘off-the-shelf’ vaccines that are not patient-specific. Indeed, MCPyV LT and sT are immunogenic, containing at least 35 known T-cell epitopes (28 CD8 and 7 CD4), including WEDLT209–228, and are restricted by diverse HLA class I and class II alleles, suggesting that an MCPyV oncoprotein vaccine has the potential to induce T-cell immunity in most MCC patients (24,47–50). To date, two groups have described potential MCPyV therapeutic vaccine strategies utilizing both in vivo models (50) and ex vivo (51). Both studies indicate the ability to induce MCPyV-specific immunity following vaccination with the MCPyV LT-antigen. However, additional work will be required to translate these findings to the clinic.
Importantly, vaccination with MCPyV T-antigens in humans raises safety concerns due to the known oncogenic activity of these proteins. Approaches to ‘detoxify’ oncogene-based cancer vaccines have shown promising safety profiles and efficacy in treating HPV-induced pre-malignant lesions (52). These ‘detoxified’ vaccines harbored mutations within the LxCxE motif of the HPV-E7 oncoprotein, suggesting that a similar approach in the context of MCPyV LT may be possible (52). One possible ‘detoxification’ strategy would be to simply delete the LxCxE region. However, this would result in loss of the immunogenic WEDLT209–228 CD4 epitope. Notably, two point mutations within the LxCxE region at residues 216 (E216K mutation) or 220 (S220A mutation) have been reported to prevent Rb binding, thereby blocking oncogenic activity at this site (4,5,40). However, we showed that of these two mutations, only S220A retained the relevant epitope that allowed recognition by HLA DRB1*0401-restricted WEDLT209–228 -specific T-cell clones. Collectively, these data indicated that the WEDLT209–228 epitope is immunogenic, expressed within MCC tumors, and could be ‘detoxified’ through mutating S220 without disrupting antigenicity.
There are several limitations of our studies. First, it is unlikely that we identified all relevant CD4 epitopes within the MCPyV oncoproteins. Study of additional MCC patients, as well as other methods of epitope identification, may yield additional disease-relevant epitopes (53). The peptide lengths used in our initial mapping studies were 13-mers, but minimal CD4+ T-cell epitopes can sometimes be longer and indeed, other groups have reported using different lengths or RNA-transfected APCs encoding the antigen of interest for CD4 epitope mapping studies (54). Consequently, alternative antigen formats could be utilized to more comprehensively map CD4 epitopes within the MCPyV T-antigens. The characterization of WEDLT209–228 reported here was largely carried forward in the context of the HLA-DRB1*0401 allele. Therefore, additional research is required to fully characterize the response to WEDLT209–228 in the context of HLA DRB1*0301 and HLA DQB1*0301.
The findings reported here have key implications for future investigation and clinical applications. The ability to deeply probe the immunobiology of this disease using the tools described herein provides a powerful opportunity to understand tumor-specific T-cell responses against a shared tumor antigen. Although checkpoint inhibitors have revolutionized cancer therapy, only half of MCC patients experience durable responses (10–12). Therefore, the development of novel therapies including cancer vaccines and/or CD4+ T-cell therapy may provide much needed adjunctive therapeutic strategies for MCC patients and cancer patients.
Supplementary Material
Acknowledgements
We would like to thank David Crispin and Martin McIntosh for their assistance in the sequencing of CD4+ T-cell receptors.
Funding Support: 1P01CA225517, 5K24CA139052-10; NIH T32ES007032, P30-CA01570.
Footnotes
Conflict of Interest: Consultant: EMD Serono (PN), Merck (PN), GSK (DMK). Sponsored research: Sanofi Pasteur (DMK), Admedus Vaccines, (DMK), Immune Design Corporation (DMK), EMD Serono (PN) and Bristol Myers Squibb (PN).
References
- 1.Lemos BD, Storer BE, Iyer JG, Phillips JL, Bichakjian CK, Fang LC, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol 2010;63(5):751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson KG, Park SY, Vandeven NA, Lachance K, Thomas H, Chapuis AG, et al. Merkel cell carcinoma: Current US incidence and projected increases based on changing demographics. J Am Acad Dermatol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008;319(5866):1096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. PNAS 2008;105(42):16272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer 2012;130(4):847–56. [DOI] [PubMed] [Google Scholar]
- 6.Sobhy H A Review of Functional Motifs Utilized by Viruses. Proteomes 2016;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015;350(6260):568–71. [DOI] [PubMed] [Google Scholar]
- 8.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol 2010;84(14):7064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuda M, Chang Y, Moore PS. Merkel cell polyomavirus-positive Merkel cell carcinoma requires viral small T-antigen for cell proliferation. J Invest Dermatol 2014;134(5):1479–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Eng J Med 2016;374(26):2542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Angelo SP, Russell J, Lebbe C, Chmielowski B, Gambichler T, Grob JJ, et al. Efficacy and Safety of First-line Avelumab Treatment in Patients With Stage IV Metastatic Merkel Cell Carcinoma: A Preplanned Interim Analysis of a Clinical Trial. JAMA Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topalian S, Bhatia S, Hollebecque A, Awada A, De Boer J, Kudchadkar R, et al. Non-comparative, open-label, multiple cohort, phase 1/2 study to evaluate nivolumab (NIVO) in patients with virus-associated tumors (CheckMate 358): Efficacy and safety in Merkel cell carcinoma (MCC) American Association for Cancer Research Annual Meeting 2017;CT074. [Google Scholar]
- 13.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol 2011;29(12):1539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson KG, Iyer JG, Simonson WT, Blom A, Thibodeau RM, Schmidt M, et al. CD8+ lymphocyte intratumoral infiltration as a stage-independent predictor of Merkel cell carcinoma survival: a population-based study. Am J Clin Pathol 2014;142(4):452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 2016;126(6):2123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Eng J Med 2008;358(25):2698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344(6184):641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu YC, Parker LL, Lu T, Zheng Z, Toomey MA, White DE, et al. Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II-Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. J Clin Oncol 2017;35(29):3322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015;520(7549):692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547(7662):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547(7662):222–6. [DOI] [PubMed] [Google Scholar]
- 22.Zhao LP, Carlsson A, Larsson HE, Forsander G, Ivarsson SA, Kockum I, et al. Building and validating a prediction model for paediatric type 1 diabetes risk using next generation targeted sequencing of class II HLA genes. Diabetes Metab Res Rev 2017;33(8). [DOI] [PubMed] [Google Scholar]
- 23.Gersuk VH, Nepom GT. A real-time polymerase chain reaction assay for the rapid identification of the autoimmune disease-associated allele HLA-DQB1*0602. Tissue Antigens 2009;73(4):335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer JG, Afanasiev OK, McClurkan C, Paulson K, Nagase K, Jing L, et al. Merkel cell polyomavirus-specific CD8(+) and CD4(+) T-cell responses identified in Merkel cell carcinomas and blood. Clin Cancer Res 2011;17(21):6671–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afanasiev OK, Yelistratova L, Miller N, Nagase K, Paulson K, Iyer JG, et al. Merkel polyomavirus-specific T cells fluctuate with merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clin Cancer Res 2013;19(19):5351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosen ST, Gould VE, Salwen HR, Herst CV, Le Beau MM, Lee I, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab Invest 1987;56(3):302–12. [PubMed] [Google Scholar]
- 27.Leonard JH, Dash P, Holland P, Kearsley JH, Bell JR. Characterisation of four Merkel cell carcinoma adherent cell lines. Int J Cancer 1995;60(1):100–7. [DOI] [PubMed] [Google Scholar]
- 28.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, et al. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol 2001;166(11):6665–70. [DOI] [PubMed] [Google Scholar]
- 29.Cai G, Hafler DA. Multispecific responses by T cells expanded by endogenous self-peptide/MHC complexes. Eur J Immunol 2007;37(3):602–12. [DOI] [PubMed] [Google Scholar]
- 30.Pawelec G, Fernandez N, Brocker T, Schneider EM, Festenstein H, Wernet P. DY determinants, possibly associated with novel class II molecules, stimulate autoreactive CD4+ T cells with suppressive activity. J Exp Med 1988;167(2):243–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, et al. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J Virol 1994;68(5):2803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. Direct recovery of herpes simplex virus (HSV)-specific T lymphocyte clones from recurrent genital HSV-2 lesions. J Infect Dis 1994;169(5):956–61. [DOI] [PubMed] [Google Scholar]
- 33.Miller NJ, Church CD, Dong L, Crispin D, Fitzgibbon MP, Lachance K, et al. Tumor-Infiltrating Merkel Cell Polyomavirus-Specific T Cells Are Diverse and Associated with Improved Patient Survival. Cancer Immunol Res 2017;5(2):137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sihto H, Bohling T, Kavola H, Koljonen V, Salmi M, Jalkanen S, et al. Tumor infiltrating immune cells and outcome of Merkel cell carcinoma: a population-based study. Clin Cancer Res 2012;18(10):2872–81. [DOI] [PubMed] [Google Scholar]
- 35.Neller MA, Lai MH, Lanagan CM, O’Connor LE, Pritchard AL, Martinez NR, et al. High efficiency ex vivo cloning of antigen-specific human effector T cells. PloS one 2014;9(11):e110741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massilamany C, Gangaplara A, Jia T, Elowsky C, Kang G, Riethoven JJ, et al. Direct staining with major histocompatibility complex class II dextramers permits detection of antigen-specific, autoreactive CD4 T cells in situ. PloS one 2014;9(1):e87519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker C, Pohla H, Frankenberger B, Schuler T, Assenmacher M, Schendel DJ, et al. Adoptive tumor therapy with T lymphocytes enriched through an IFN-gamma capture assay. Nat Med 2001;7(10):1159–62. [DOI] [PubMed] [Google Scholar]
- 38.Oyarzun P, Ellis JJ, Boden M, Kobe B. PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity. BMC Bioinformatics 2013;14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland CJ, Cole DK, Godkin A. Re-Directing CD4(+) T Cell Responses with the Flanking Residues of MHC Class II-Bound Peptides: The Core is Not Enough. Front Immunol 2013;4:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrama D, Hesbacher S, Angermeyer S, Schlosser A, Haferkamp S, Aue A, et al. Serine 220 phosphorylation of the Merkel cell polyomavirus large T antigen crucially supports growth of Merkel cell carcinoma cells. Int J Cancer 2015. [DOI] [PubMed] [Google Scholar]
- 41.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 2013;38(2):373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res 2014;2(2):91–8. [DOI] [PubMed] [Google Scholar]
- 43.Bacher P, Scheffold A. Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A 2013;83(8):692–701. [DOI] [PubMed] [Google Scholar]
- 44.Muranski P, Restifo NP. Adoptive immunotherapy of cancer using CD4(+) T cells. Curr Opin Immunol 2009;21(2):200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol 2008;26(32):5233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veatch JR, Lee SM, Fitzgibbon M, Chow IT, Jesernig B, Schmitt T, et al. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J Clin Invest 2018;128(4):1563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapuis AG, Afanasiev OK, Iyer JG, Paulson KG, Parvathaneni U, Hwang JH, et al. Regression of metastatic Merkel cell carcinoma following transfer of polyomavirus-specific T cells and therapies capable of re-inducing HLA class-I. Cancer Immunol Res 2014;2(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyngaa R, Pedersen NW, Schrama D, Thrue CA, Ibrani D, Met O, et al. T-cell responses to oncogenic merkel cell polyomavirus proteins distinguish patients with merkel cell carcinoma from healthy donors. Clin Cancer Res 2014;20(7): 1768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibrani D, Iyer J, Miller N, Vandeven N, Afanasiev O, Koelle D, et al. Identifying Merkel polyomavirus-specific CD4+ and CD8+ T-cells in Merkel cell carcinoma patients’ tumor-infiltrating lymphocytes. Presented at the 2015 Society for Investigative Dermatology Annual Meeting May 6–9, 2015. Atlanta, GA 2015. [Google Scholar]
- 50.Zeng Q, Gomez BP, Viscidi RP, Peng S, He L, Ma B, et al. Development of a DNA vaccine targeting Merkel cell polyomavirus. Vaccine 2012;30(7):1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerer KF, Erdmann M, Hadrup SR, Lyngaa R, Martin LM, Voll RE, et al. Preclinical evaluation of NF-kappaB-triggered dendritic cells expressing the viral oncogenic driver of Merkel cell carcinoma for therapeutic vaccination. Ther Adv Med Oncol 2017;9(7):451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez RD, Huh WK, Bae S, Lamb LS Jr., Conner MG, Boyer J, et al. A pilot study of pNGVL4a-CRT/E7(detox) for the treatment of patients with HPV16+ cervical intraepithelial neoplasia 2/3 (CIN2/3). Gynecol Oncol 2016;140(2):245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Provenzano M, Panelli MC, Mocellin S, Bracci L, Sais G, Stroncek DF, et al. MHC-peptide specificity and T-cell epitope mapping: where immunotherapy starts. Trends Mol Med 2006;12(10):465–72. [DOI] [PubMed] [Google Scholar]
- 54.Simon P, Omokoko TA, Breitkreuz A, Hebich L, Kreiter S, Attig S, et al. Functional TCR retrieval from single antigen-specific human T cells reveals multiple novel epitopes. Cancer Immunol Res 2014;2(12):1230–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.