Abstract
Machine learning techniques have been implemented to reveal brain features that distinguish people with autism spectrum disorders (ASDs) from typically developing (TD) peers. However, it remains unknown whether different neuroimaging modalities are equally informative for diagnostic classification. We combined anatomical magnetic resonance imaging (aMRI), diffusion weighted imaging (DWI), and functional connectivity MRI (fcMRI) using conditional random forest (CRF) for supervised learning to compare how informative each modality was in diagnostic classification. In-house data (N = 93) included 47 TD and 46 ASD participants, matched on age, motion, and nonverbal IQ. Four main analyses consistently indicated that fcMRI variables were significantly more informative than anatomical variables from aMRI and DWI. This was found (1) when the top 100 variables from CRF (run separately in each modality) were combined for multimodal CRF; (2) when only 19 top variables reaching >67% accuracy in each modality were combined in multimodal CRF; and (3) when the large number of initial variables (before dimension reduction) potentially biasing comparisons in favor of fcMRI was reduced using a less granular region of interest scheme. Consistent superiority of fcMRI was even found (4) when 100 variables per modality were randomly selected, removing any such potential bias. Greater informative value of functional than anatomical modalities may relate to the nature of fcMRI data, reflecting more closely behavioral condition, which is also the basis of diagnosis, whereas brain anatomy may be more reflective of neurodevelopmental history.
Keywords: autism, diagnostic classification, machine learning, MRI, multimodal imaging
Introduction
Autism Spectrum Disorders (ASDs) comprise a set of developmental disorders diagnosed on the basis of sociocommunicative deficits, and restricted and repetitive behaviors (American Psychiatric Association, 2013). Increasing prevalence, with recent estimates around 2% (Baio et al., 2018), and the need for lifelong services for most affected individuals make ASDs an urgent public health issue. Although ASDs are considered neurological disorders, no diagnostic brain markers are available. Instead, diagnosis of “idiopathic” or nonsyndromic ASDs solely relies on behavioral evaluations administered by clinicians. In the past decade, much of the focus in the search for brain substrates of ASDs has been directed at the systems level, with growing understanding that no single brain region can account for the observed complex patterns of behavioral anomalies in ASDs. Numerous findings of atypical network connectivity have been reported, but no consensus on crucial brain characteristics has emerged (Hull et al., 2017).
In the absence of definitive findings from traditional hypothesis-driven studies, several groups have begun to implement data-driven and machine learning (ML) techniques using different imaging modalities in an attempt to identify brain features that may distinctly separate individuals with ASDs from typically developing (TD) peers. Aside from a few electrophysiological studies (Bosl et al., 2011; Jamal et al., 2014; Stahl et al., 2012), most ML studies for diagnostic prediction (ASD vs. TD) have used magnetic resonance imaging (MRI). Early investigations by Ecker and colleagues (2010b) and Jiao and colleagues (2010) used anatomical MRI (aMRI) data, such as regional cortical volume and thickness, and applied support vector machines (SVMs) as well as functional trees and logistic model trees, achieving diagnostic prediction accuracies near 85%. A more recent study by Hazlett and colleagues (2017) in 106 infants considered “high-risk” (because they had an older sibling with ASD) showed that atypical expansion of cortical surface area in the first year of life strongly predicted an eventual ASD diagnosis, using deep learning. While aMRI variables have been used in other ML studies (Moradi et al., 2017; Xiao et al., 2017), relatively few studies have included diffusion weighted imaging (DWI) data. Two studies using fractional anisotropy (FA) and mean diffusivity (MD) features extracted from white matter regions of interest (ROIs) and tracts in infants (Jin et al., 2015) and school-age children (Ingalhalikar et al., 2011) implemented SVM and reached cross-validation accuracies up to 80%.
The majority of ML diagnostic classification studies have adopted functional MRI (fMRI) mostly using functional connectivity MRI (fcMRI) matrices from resting-state (rs) data (Abraham et al., 2017; Bi et al., 2018; Emerson et al., 2017; Iidaka, 2015; Nielsen et al., 2013). Among the very first, Anderson and colleagues (2011), using a very large matrix of over 26 million rs-fMRI connectivities, achieved an overall classification accuracy of 79%, although accuracy was lower (71%) in a small replication sample. More recently, Yahata and colleagues (2016) reported an 85% leave-one-out cross-validation (LOOCV) accuracy for a small set of 16 functional connections, with 75% accuracy in an independent validation sample from the Autism Brain Imaging Data Exchange (ABIDE) (Di Martino et al., 2014). Chen and colleagues (2015) and Jahedi and colleagues (2017) achieved accuracies of 91% and 99% in out-of-bag (OOB) validation samples, using random forest (RF) classifiers including 100 and 308 features, respectively; however, accuracies were ∼70% or lower in novel validation data sets. Overall, findings suggest that classifiers do not perform well in entirely new data sets (not included in training), probably due to clinical and neurobiological heterogeneity of the disorder (Tordjman et al., 2018).
ML studies have, in the past, commonly relied on a single imaging modality. However, information from fMRI or aMRI on their own may not be rich enough for a robust classifier because distinctive features of ASDs may affect different aspects of brain structure and function that can be captured only by combined use of imaging modalities. Yet, few multimodal ML studies of ASDs are currently available. Libero and colleagues (2015) included aMRI, DWI, and magnetic resonance spectroscopy data from small ASD and TD samples, reaching 92% LOOCV accuracy with a decision tree classifier. Ghiassian and colleagues (2016) used fMRI and aMRI data from ABIDE, achieving more modest accuracies <70%. However, neither of these studies specifically examined whether imaging modalities were differentially informative for diagnostic classification.
In the present study, we used a multimodal in-house data set including aMRI, DWI, and fcMRI data to assess whether different modalities contribute equally to diagnostic prediction or whether some modalities may be more informative than others.
Methods
Participants
We recruited 164 participants, aged 7–18 years, from the community and local clinical collaborations. TD participants did not have a family or personal history of any neurological, developmental, or psychiatric disorders. ASD participants had diagnoses verified using the Autism Diagnostic Observation Schedule (ADOS-2) (Lord et al., 2012) and the Autism Diagnostic Interview-Revised (ADI-R) (Rutter et al., 2003). A total of 71 participants had to be excluded: 23 for insufficient quality of DWI data, 12 for poor aMRI quality, 18 for excessive motion during fMRI, 3 for failed or incomplete data acquisition, 4 for not meeting full ASD diagnostic criteria, and 5 for neurological or psychiatric findings that excluded them from the TD group. Six further participants were removed for optimal group matching on age, motion, and nonverbal IQ (Table 1). This yielded a final sample of 47 TD and 46 ASD participants.
Table 1.
Participant Information
| ASD, M ± SD (range) | TD, M ± SD (range) | p | |
|---|---|---|---|
| N (female) | 46 (7) | 47 (9) | 0.62 |
| Age (years) | 13.63 ± 2.81 (7.40–17.98) | 13.44 ± 2.76 (8.02–17.70) | 0.74 |
| Handedness (right:left) | 40:6 | 40:7 | 0.80 |
| Nonverbal IQ | 105.89 ± 17.28 (53.00–140.00) | 105.74 ± 14.20 (62.00–137.00) | 0.96 |
| fMRI head motion (RMSD) | 0.08 ± 0.04 (0.02–0.22) | 0.08 ± 0.06 (0.02–0.30) | 0.99 |
| DWI average translation (mm) | 0.83 ± 0.23 (1.82–0.44) | 0.86 ± 0.25 (1.71–0.46) | 0.58 |
| DWI average rotation (rad; × 10−3) | 4.65 ± 1.91 (0.01–0.00) | 4.64 ± 1.91 (0.01–0.00) | 0.96 |
| DWI % bad slices | 0.03 ± 0.18 (1.22–0.00) | 0.02 ± 0.10 (0.67–0.00) | 0.75 |
| DWI average dropout score | 1.07 ± 0.17 (1.66–1.00) | 1.05 ± 0.12 (1.41–1.00) | 0.40 |
ASD, autism spectrum disorder; DWI, diffusion weighted imaging; fMRI, functional magnetic resonance imaging; RMSD, root-mean-squared-difference; SD, standard deviation; TD, typically developing.
The study was approved by the institutional review boards of San Diego State University and University of California San Diego. Assent and informed consent were obtained from all participants and their caregivers.
Data acquisition
Imaging data were acquired at the University of California San Diego Center for Functional Magnetic Resonance Imaging (CFMRI) on a General Electric 3T Discovery MR750 scanner with an eight-channel head coil. Anatomical images were collected using a standard fast spoiled gradient-echo (FSPGR) T1-weighted sequence (172 slices; repetition time [TR] = 8.136 ms; echo time [TE] = 3.172 ms; field of view [FOV] = 256 mm; 256 × 256 matrix; flip angle = 8°; 1 mm3 resolution). Diffusion weighted images were collected using an echo planar imaging (EPI) pulse sequence, encoded for 61 noncollinear diffusion directions at b = 1000 s/mm2, and 1 at b = 0 s/mm2 (2D EPI; TR = 8500 ms; TE = 84.9 ms; flip angle = 90°; FOV = 240 mm; 1.875 × 1.875 × 2 mm3 resolution; 68 slices). Functional T2*-weighted images were obtained using a single-shot gradient-recalled EPI sequence of 180 whole-brain volumes (TR = 2000 ms; TE = 30 ms; FOV = 220 mm; flip angle = 90°; 64 × 64 matrix; 3.4 mm3 resolution; 42 axial slices covering the whole brain). For all EPI images, field maps were collected with the same spatial resolution to correct for field inhomogeneities. For the 6-min resting-state scan, participants were shown a white crosshair centered on a black background and instructed to fixate on the crosshair, relax, and to stay awake. Wakefulness and compliance were tracked with in-bore video monitoring.
Imaging variables
The aMRI, DWI, and fMRI data were preprocessed using standard procedures. Anatomical processing was performed using FreeSurfer version 5.3.0 for semiautomated cortical reconstruction (Dale et al., 1999). Images with persistent inaccuracies in surfaces or excessive artifacts, such as ghosting or ringing, were excluded. The anatomical data yielded five cortical features: cortical surface area, mean curvature, cortical thickness, cortical volume, and a local gyrification index (lGI). LGI, measured at each surface vertex using a FreeSurfer add-on (Schaer et al., 2008), is a 3D surface-based method that calculates the ratio of cortical surface area within the sulcal folds relative to the amount of cortex on the cortical hull. This calculation was made within a sphere of 25 mm radius around the pial surface vertex, resulting in an lGI value for each vertex on the mesh. This automated reconstruction feature has been validated against histological analysis and manual measurement and has been demonstrated as a reliable measure of gyrification (Schaer et al., 2008). The average value for the five cortical variables was extracted from 34 ROIs per hemisphere (Desikan et al., 2006), yielding 340 variables. Fifty-seven additional volume measures, including subcortical structures, white matter regions, and cerebrospinal fluid spaces, were added for a total of 397 anatomical variables.
DWI images were processed using FMRIB Software Library (FSL) (v5.0.8) (Smith et al., 2004) and AFNI (v17.3.0; AFNI) (Cox, 1996). Susceptibility distortions were corrected with field maps (FSLs prelude, fugue), and eddy currents and motion with eddy correct. Diffusivity measures were calculated using dtifit. The standard tract-based spatial statistics (Smith et al., 2006) processing pipeline was followed to produce spatially normalized metrics of diffusion (FA, MD, axial diffusivity, radial diffusivity), not confounded by partial volume effects or variations in regional volumes. White matter skeletons were then overlaid with 48 white matter regions provided by the Johns Hopkins University atlas (Mori et al., 2005) to extract average values for each of the 4 DWI metrics by region. These metrics contributed 192 features to the model.
FMRI data were processed and analyzed using methods identical to those in the study by Chen and colleagues (2015), using the AFNI and FSL software. The first five time points were discarded to allow for T1 equilibration. The remaining 180 time points were motion, slice-time, and field-map corrected. Functional data were aligned to anatomical images using FMRIB's Linear Image Regression Tool with six degrees of freedom, resampled to 3.0 mm isotropic voxels using sinc interpolation, and standardized to the MNI-152 template, using the FSLs nonlinear registration tool, all in a single transformation step. Data were spatially blurred to a full-width at half-maximum of 6 mm. Time series were band-pass filtered (0.008 < f < 0.08 Hz) using a second-order Butterworth filter (Power et al., 2013; Satterthwaite et al., 2013). In-scanner head motion was estimated using root-mean-squared-difference calculated from six motion parameters (three translational and three rotational). Average time series from trimmed white matter and ventricles (from FSL segmentation) as well as their first derivatives were regressed from the data. All nuisance regressors (including motion regressors) were band-pass filtered using the same procedures as for blood oxygen level dependent (BOLD) time series (Hallquist et al., 2013). A head motion threshold of 1 mm was applied. Any time points above the threshold, and the subsequent two time-points, were censored. If <10 time points remained between two blocks of censored time series, those time points were also removed.
We used two ROI schemes. First, we extracted time series from 220 of the 264 ROIs in the study by Power and colleagues (2011), excluding 44 ROIs with missing signal in >2 participants. Data from each participant were organized in a 220 × 219 matrix of Fisher-transformed Pearson correlation coefficients, with each cell in the matrix representing the time series correlation (functional connectivity) between two ROIs. There were thus 24,090 (220 × 219/2) unique features for each participant. To reduce the very large difference between modalities in the number of features, a second set with a smaller number of larger ROIs from the Harvard-Oxford atlas (Desikan et al., 2006) was used in additional analyses. Time series from 101 ROIs were extracted (excluding 35 ROIs with missing signal in >2 participants). Connectivity matrices from this ROI scheme included 5050 (101 × 100/2) unique features for each participant.
ML algorithm
Conditional random forest (CRF) is a variant of RF, which employs an ensemble method often used for classification, with the goal for a group of weak learners (decision trees) to combine into a strong learner (the forest). In a built-in training method, about 66% of the data are randomly chosen with replacement to create binary decision trees, and the remaining data are used as testing set—termed OOB sample. Breiman (2001) argued that OOB estimates can be used to estimate the generalization error, eliminating the need for a set-aside testing set. The same procedures are followed in CRF, except for variables being chosen without replacement (see CRF implementation section).
At each node of the tree, a certain fixed number, m, of predictor variables (typically the square root of the number of all predictor variables) is randomly chosen for splitting. After the forest is constructed, the majority vote determines the classifications in terminal nodes of each tree and is then averaged over all trees. Misclassifications, represented as the OOB error rate, are also averaged over all trees in the forest and provide the level of accuracy of the model.
Variable importance measures for feature selection are provided by RF and are attained by calculating the average relative difference between OOB prediction accuracy before and after the permutation of a variable. Strobl and colleagues (2008) showed that RF variable importance is biased when dealing with highly correlated and continuous variables—a likely concern for MRI data. To address this bias in CRF, variables are permuted in a conditional manner to measure variable importance, which reflects the true impact of each predictor variable more reliably (Strobl et al., 2008) and is calculated as the average difference in prediction accuracy or the mean decrease in accuracy (MDA).
CRF implementation
Four analyses using a supervised CRF algorithm were implemented. In the first analysis, each modality was run independently, and the top 100 variables with the highest MDAs from the first run (one dimension reduction) were selected from each modality. CRF was then run on this combined set of 300 variables and MDAs were recorded.
Analyses 2–3 were designed to reduce any potential bias favoring the fcMRI modality, for which much larger numbers of variables were available before feature selection. Analysis 2 permitted for more informative anatomical and DWI features to be selected. Instead of using a predetermined number of top variables from each modality (such as the top 100 variables in analysis 1), the performance of each modality was assessed during separate CRF runs for each modality, with accuracy, sensitivity, and specificity rates determined for each dimension reduction. The number of top variables for each modality was determined based on a minimum 67% accuracy criterion applied to each modality separately. Use of the top 19 variables fulfilled this criterion (>67% accuracy) in each modality (regardless of modality-specific accuracy peaks; Supplementary Fig. S1). CRF was run on the 57 variables (combined from the 3 modalities), and MDAs were recorded.
The third analysis followed the same methods as analyses 1–2, but the number of initially included fcMRI variables was reduced from 24,090 to 5,050 by using the Harvard-Oxford ROI scheme, as described above. This served to reduce the large difference in number of variables between fcMRI and the other two modalities. However, to further eliminate potential bias favoring the fcMRI modality, another analysis was implemented with no feature selection and randomly chosen variables. In this fourth analysis, 100 variables were randomly chosen from each modality and were combined to form a set of 300 variables. This set was run through CRF, and all variables within the set that had negative MDAs were removed. Also, only sets with an OOB error rate <0.4 were kept for further analysis to ensure that the retained sets performed above chance level. This process was repeated 1 million times to ensure that all variables from each modality were being chosen.
Results
Analysis 1
This analysis used the top 100 variables from each modality (cf. Supplementary Fig. S1). For the anatomical modality, white matter hypointensities and right parahippocampal cortical thickness stood out as features with highest MDA. However, MDAs were generally low (all <0.001). For the DWI modality, MDA levels were overall slightly higher, and one variable, FA of the right anterior limb of the internal capsule, stood out with an MDA of 0.0054 that was ≥3 times higher than for other DWI variables. Only 72 DWI variables had positive MDAs, suggesting that all others were noninformative. For the fcMRI modality, MDAs were generally higher, with all top 100 variables in a range from 0.0049 to 0.0152. The top four variables, with MDA >0.01, involved visual ROIs. Three were connections with ROIs in the default mode network, and one was a connection within the visual network. Visual and default mode networks also predominated among the top 100 features, with 57.5% of all ROIs participating in these connections (Supplementary Fig. S2).
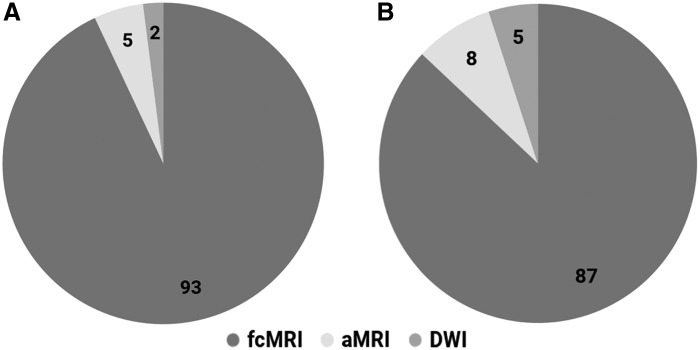
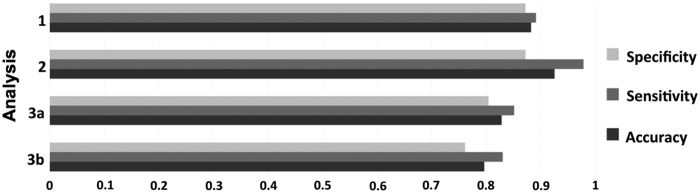
After combining the top 100 variables from each modality (300 top variables) and applying the CRF algorithm, 93 of the 100 most informative features belonged to the fcMRI modality (Fig. 1A). The accuracy of this run was at 88.2%, with 89.1% sensitivity and 87.2% specificity (Fig. 2).
FIG. 1.
Pie chart showing modality distribution of the top 100 variables from analysis 1 (A) and analysis 3a (B).
FIG. 2.
Accuracy, sensitivity, and specificity rates for each analysis.
Analysis 2
In this analysis, a smaller number of variables were selected from each modality and the criterion applied was a prediction accuracy of >67% when CRF was performed separately in each modality. CRF results for dimension reduction are shown for each modality in Supplementary Figure S3. For the anatomical data set, the highest accuracy rate of 69.9% was achieved with 19 variables as well as 3 variables. The highest accuracy rate for the DTI data set was at 71.0% with three variables. The fcMRI data set reached peak accuracy of 93.5% with 424 features. In all 3 modalities, the top 19 variables satisfied the criterion of a minimum of 67% accuracy.
CRF combining the top 19 variables from each modality yielded overall similar results to analysis 1. The accuracy for these 57 variables was 92.5%, with 97.8% sensitivity and 87.2% specificity (Fig. 2). The 19 most informative variables with the highest MDAs belonged to the fcMRI modality, again predominated by connections with participation of visual and default mode regions, which accounted for 65.8% of all connectivity participations (Supplementary Figs. S4 and S5). Only a single DWI variable (FA of the right anterior limb of the internal capsule) had an MDA >0.001.
Analysis 3
This analysis followed the same steps as the first and second analyses but implemented the Harvard-Oxford ROIs to reduce the very large number of fcMRI variables. Anatomical and DWI variables and their MDAs were identical to those in analyses 1–2. In analysis 3a, the pattern for the top 100 fcMRI variables differed, however, with cingulo-opercular, sensorimotor hand, auditory, dorsal attention, and visual networks accounting for 64% of ROI participations (Supplementary Fig. S6A, B). Results also showed that 87% of the most informative features belonged to the fcMRI modality (Fig. 1B). The accuracy of this run was at 82.8%, with 85.1% sensitivity and 80.4% specificity (Fig. 2).
In analysis 3b, which combined the top 19 variables from each modality, the accuracy, sensitivity, and specificity rates for the CRF run within the fcMRI modality were generally lower than those in analysis 2. The highest accuracy rate achieved here was at 83.9% with seven variables (sensitivity 82.6%, specificity 85.1%; Supplementary Fig. S7). The top 19 fcMRI variables again fulfilled the 67% accuracy criterion. CRF results for the top 19 variables from each modality combined showed a general pattern consistent with analysis 2. The accuracy for this run was slightly lower at 79.6%, with sensitivity at 83.0% and specificity at 76.1% (Fig. 2). Among the 19 top variables with MDA >0.002, there were 18 from the fcMRI modality, predominated by connections in the sensorimotor hand, auditory, and cingulo-opercular regions accounting for 52% of ROI participations (Supplementary Fig. S8), and only a single DWI variable (again FA of the right anterior limb of internal capsule). Six more variables had MDA >0.001, five of which were anatomical and one fcMRI (Supplementary Fig. S9).
Analysis 4
In the final analysis, which consisted of 1 million iterations using 100 randomly chosen variables per modality combined, 2530 sets of 300 variables survived the 0.4 OOB error rate cutoff. These sets were further analyzed, and the mean MDAs for each modality were calculated: 4.35e-4 for fcMRI, 2.84e-4 for DWI, and 2.71e-4 for aMRI. The difference in MDAs between the modalities was significant (p < 0.001). Outliers in the box plot in Figure 3 show the wide range of MDAs for the fcMRI modality compared with the DWI and anatomical modalities, with many variables in the fcMRI modality attaining higher MDAs than any from the other two modalities.
FIG. 3.
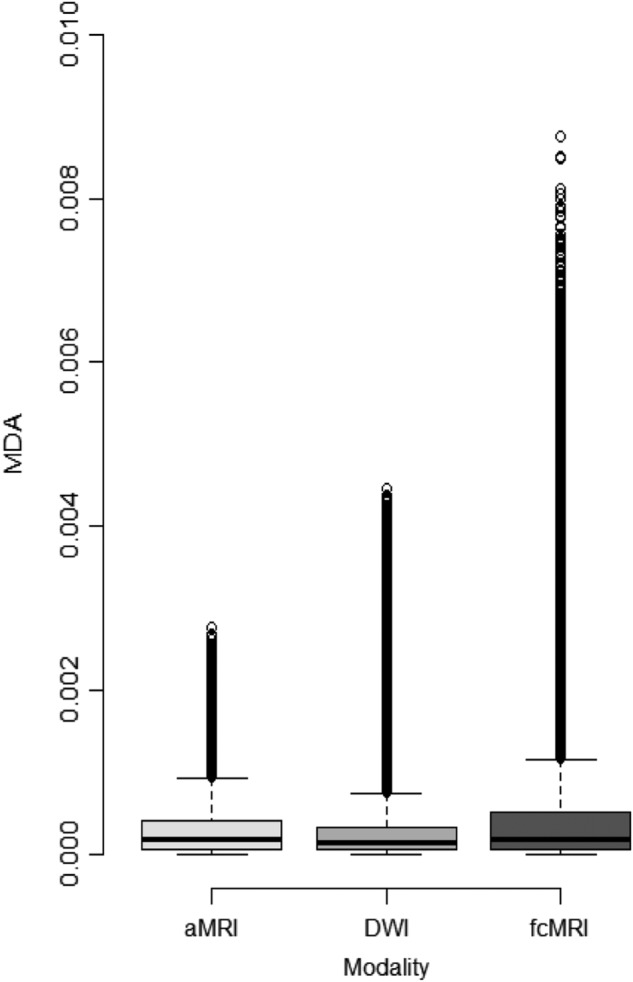
Box plot from analysis 4. The dots above each modality represent the outliers, which have higher MDA values when compared with the other two modalities. The mean MDA values for fcMRI, DWI, and aMRI are 4.35e-4, 2.84e-4, and 2.71e-4, respectively. The Kruskal–Wallis test showed differences between modalities at p < 0.001. Three pairwise Wilcoxon rank sum tests between modalities were also all p < 0.001. aMRI, anatomical magnetic resonance imaging; DWI, diffusion weighted imaging; fcMRI, functional connectivity MRI; MDA, mean decrease in accuracy.
Discussion
A growing number of studies have applied ML techniques for diagnostic classification of ASDs, using different functional and anatomical types of imaging data with varying level of success. However, there has been no systematic investigation assessing whether different imaging modalities are equally informative in this process. In four comparative analyses that included fcMRI, aMRI, and DWI data, we consistently found that fcMRI variables performed at superior levels compared with the other modalities.
Features supporting diagnostic classification
While not the focus of the current study, some region- and network-specific findings from the different CRF analyses are worth noting. When using the ROI scheme from the study by Power and colleagues (2011), visual and default mode networks stood out among fcMRI features that overall topped features from other modalities in MDA, with >57% of ROI participations among the 100 most informative fcMRI features (as detected in analysis 1). A similar pattern was seen in analysis 2 that included only the top 19 fcMRI features, with ∼66% participation from visual and default mode ROIs. Prominence of fcMRI variables of the default mode network has been noted in some previous diagnostic classification studies using multisite consortium data (Abraham et al., 2017; Nielsen et al., 2013), whereas connectivity of visual ROIs has been highlighted by few ML studies (Chen et al., 2015). However, prominence of visual features is not unexpected given extensive evidence of atypical visual processing (Dakin and Frith, 2005) and atypical task-related neural activity of visual cortices in ASDs (Samson et al., 2012). In analyses 3 and 4 using the less granular Harvard-Oxford ROI scheme (Desikan et al., 2006), visual and default mode networks appeared less informative, with higher percentages of top ROI participations for cingulo-opercular, somatosensory/motor hand, auditory, and dorsal attention regions. These differences illustrate how ML findings may differ depending on the selected ROI scheme, even when the exact same data set is used.
Among aMRI and DWI variables, a few stood out as relatively informative. These were white matter hypointensities and cortical thickness of the right parahippocampal gyrus from aMRI, and FA in the right anterior limb of the internal capsule from DWI. However, MDAs were generally low for these two MRI modalities and did not reach levels seen for fcMRI variables mentioned above.
Functional variables are more informative than anatomical variables
All analyses consistently indicated that fcMRI variables were more informative for diagnostic prediction than anatomical variables from aMRI and DWI. This would not be expected from the literature, with good prediction accuracies in some early aMRI studies (Ecker et al., 2010a) and more modest prediction rates in some fcMRI studies (Nielsen et al., 2013). However, the literature does not permit direct comparison across studies that use data from different cohorts and apply different analysis techniques. In contrast, our findings using the same ML technique (CRF) in the identical sample of ASD and TD participants for each imaging modality robustly indicated superiority of the fcMRI modality for the specific purpose of diagnostic prediction.
One caveat relates to the almost unavoidable fact that fcMRI matrices generate much larger numbers of features than aMRI and DWI methods. We addressed this potential bias in multiple ways. In analysis 2, we strictly limited the number of variables, by selecting only the top 19 variables per modality for which CRF achieved a minimum accuracy of 67% when performed separately in each modality. Strikingly, when these 57 variables were combined into multimodal CRF, the 19 most informative variables (with top MDA) exclusively belonged to the fcMRI modality. In a second attempt, we reduced the number of fcMRI variables by using an ROI scheme with fewer but larger regions (i.e., by reducing the granularity of fcMRI measurements). The pattern of findings remained similar, with an overwhelming majority (≥87%) of the most informative features being fcMRI variables. However, even in this analysis, bias may not have been completely removed because the reduced number of initial fcMRI variables was still much higher than the number of aMRI and DWI variables. We therefore performed a fourth analysis that was fully protected from this confound, by randomly selecting 100 variables from each modality and running CRF numerous times on different randomizations. The resulting pattern was similar, with fcMRI variables reaching significantly higher mean MDA than variables from the other modalities. This finding may be considered particularly striking, as arguably analysis 4 was biased in favor of aMRI and DWI, given that each CRF run randomly included <2% of potentially informative fcMRI variables, but >25% of such variables in aMRI and DWI modalities.
Differential relations between imaging modalities and diagnosis
The very robust differences indicating superiority of fcMRI over aMRI and DWI in diagnostic prediction were unexpected. However, when considering the differential nature of functional versus anatomical data, they are interpretable. Diagnostic procedures in ASDs are exclusively behavioral (American Psychiatric Association, 2013). While research diagnostic instruments include information about behavioral history, starting with the first years of life (Rutter et al., 2003), the primary research diagnostic instrument, the ADOS-2 (Lord et al., 2012), exclusively focuses on observations of a child's current behavior. It can be assumed that fcMRI and matrices of BOLD correlations that indirectly reflect neuronal activity fluctuations during an MRI scan are comparatively close in relation to current behavioral state, whereas anatomical brain variables probably reflect developmental history to a greater extent. This implies that fcMRI measures may be more closely related to diagnostic measures, which is exactly what our analyses suggest.
Caveats should be considered. First, functional connectivity detected in resting-state fMRI is considered “intrinsic” (Van Dijk et al., 2010) and therefore thought to demonstrate the architecture of specialized brain networks. Specifically, intrinsic functional connectivity (iFC) has been attributed to history of coactivation and Hebbian principles of plasticity (Gordon et al., 2016; Lewis et al., 2009). However, it is also understood that the “resting state” during which fMRI data are acquired is cognitively demanding, rich, and variable (Buckner et al., 2013) and that BOLD signal correlations are likely affected by current state or by dynamic changes in current state [as seen in novel implementations of dynamic fcMRI in ASDs (Falahpour et al., 2016; Rashid et al., 2018)]. While iFC therefore probably reflects a mix of current online cognitive activity and history of coactivation, the latter may be mostly recent history, as suggested by findings of rapidly changing iFC patterns after brief periods of learning (Lewis et al., 2009). This is consistent with a relatively close relation to behavioral profiles that are the basis of diagnostic procedures.
Second, brain anatomy undergoes change throughout life and some of these changes reflect plasticity in response to environmental interactions. It is therefore too simple to attribute neuroanatomy entirely to early developmental history. For example, the recent finding of atypically increased gyrification in the vicinity of the—prenatally emerging—Sylvian fissure in children and adolescents with ASDs (Kohli et al., 2019) may contain traces of very early growth anomalies in ASDs, but cortical morphology will likely be affected by plastic changes relating to behavior and environmental interaction later in life. However, the unequal contribution to diagnostic prediction observed in the current study is fully compatible with a relative difference between functional and anatomical measurements, with fcMRI reflecting current behavioral states and dynamics more closely (and therefore being more directly linked to diagnostic measurements), but aMRI and DWI reflecting neurodevelopmental history more predominantly. The latter, speculatively, would suggest that neuroanatomical data may be more informative of etiological subtypes of the disorder that vary in developmental history, consistent with the notion of (anatomical) neuroimaging data providing “intermediate phenotypes” (Rasetti and Weinberger, 2011). It is furthermore possible that greater informative value of functional than anatomical features observed in our study may be age-specific and that anatomical variables may be more predictive early in life. This is supported by Hazlett and colleagues (2017) who found that cortical surface area in 6- to 12-month-old high-risk infants was strongly predictive of subsequent diagnosis of ASD.
Granularity
As mentioned above, the number of initial features (before feature selection) differed greatly between fcMRI versus aMRI and DWI. While we took multiple measures to limit any comparative bias related to these differences, the problem remains that granularity cannot be easily compared or equated across modalities. For example, a given ROI, such as a Freesurfer parcel, may generate a number of variables in aMRI (volume, surface area, cortical thickness, gyrification), whereas in fcMRI many more variables will be generated based on BOLD correlations (iFC) with each of the other ROIs in the selected ROI scheme, typically in the hundreds. However, this much larger number of features does not actually indicate higher granularity in fcMRI because the identical ROI scheme is used in both modalities. The question of what may be optimal granularity (i.e., the one generating most informative features) was not the focus of our study, but slightly higher accuracies in analyses 1–2 than analyses 3a–b suggest that for fcMRI, higher granularity of the ROI scheme from the study by Power and colleagues (2011) was superior to the lower granularity of an alternative scheme (Desikan et al., 2006). Generally, it can be assumed that there will be trade-offs between reduced informative value due to signal averaging (very low granularity; e.g., cortical thickness of an entire hemisphere) on the one hand, and due to noisy measurements (very high granularity, e.g., BOLD time series correlations between single voxels) on the other. Calibration of optimal granularity likely depends on data quality and acquisition protocols and will differ between MRI (and other imaging) modalities.
Limitations
The comparisons performed in this study required high-quality data in all three imaging modalities in each participant, which resulted in limited sample size compared with some previous single-modality diagnostic classification studies. In view of the known heterogeneity of ASDs (Lombardo et al., 2019), cohort effects in small samples cannot be ruled out. Specifically, the requirement of low-motion data limited participation largely to the high-functioning segment of the autism spectrum. It remains possible that brain features distinctive of low-functioning ASD, or of ASD variants not captured in our moderate sample, differ from the ones detected in our study (Gabrielsen et al., 2018; Reiter et al., 2019) and that informative contributions from various MRI modalities also differ. Finally, while we performed multiple comparative analyses, they all relied on CRF for classification. It cannot be ruled out that our findings were (to some extent) algorithm-specific.
Conclusions
Our findings suggest that neuroimaging modalities vary in their informative value for ML diagnostic classification of ASD. Specifically, resting-state functional connectivities were found to be overall more informative than anatomical measures from aMRI and DWI for classification of children and adolescents with ASD versus matched TD cohorts.
Supplementary Material
Acknowledgments
The authors thank the children and their parents for participating in this study.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by the National Institutes of Health R01 MH081023 (R.-A.M.), R01 MH101173 (R.-A.M.), and K01 MH097972 (Inna Fishman, Brain Development Imaging Laboratories, Department of Psychology, San Diego State University, San Diego, California), and by San Diego State University Graduate Fellowships to A.J. and J.S.K.
Supplementary Material
References
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G. 2017. Deriving reproducible biomarkers from multi-site resting-state data: an Autism-based example. Neuroimage 147:736–745 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. 2013. Diagnostic and Statistical Manual of Mental Disorders–5. Washington, DC: American Psychiatric Association [Google Scholar]
- Anderson JS, Nielsen JA, Froehlich AL, DuBray MB, Druzgal TJ, Cariello AN, et al. . 2011. Functional connectivity magnetic resonance imaging classification of autism. Brain 134:3742–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, et al. . 2018. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 Sites, United States, 2014. MMWR Surveill Summ 67:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi XA, Wang Y, Shu Q, Sun Q, Xu Q. 2018. Classification of autism spectrum disorder using random support vector machine cluster. Front Genet 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl W, Tierney A, Tager-Flusberg H, Nelson C. 2011. EEG complexity as a biomarker for autism spectrum disorder risk. BMC Med 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn 45:5–32 [Google Scholar]
- Buckner RL, Krienen FM, Yeo BT. 2013. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci 16:832–837 [DOI] [PubMed] [Google Scholar]
- Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA, Müller R-A. 2015. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. Neuroimage Clin 8:238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Dakin S, Frith U. 2005. Vagaries of visual perception in autism. Neuron 48:497–507 [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9:179–194 [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. . 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980 [DOI] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, et al. . 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourao-Miranda J, Johnston P, Daly EM, Brammer MJ, et al. . 2010a. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci 30:10612–10623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM, et al. . 2010b. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage 49:44–56 [DOI] [PubMed] [Google Scholar]
- Emerson RW, Adams C, Nishino T, Hazlett HC, Wolff JJ, Zwaigenbaum L, et al. . 2017. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci Transl Med 9:eaag2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahpour M, Thompson WK, Abbott AE, Jahedi A, Mulvey ME, Datko M, et al. . 2016. Underconnected, but not broken? Dynamic functional connectivity MRI shows underconnectivity in autism is linked to increased intra-individual variability across time. Brain Connect 6:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen TP, Anderson JS, Stephenson KG, Beck J, King JB, Kellems R, et al. . 2018. Functional MRI connectivity of children with autism and low verbal and cognitive performance. Mol Autism 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiassian S, Greiner R, Jin P, Brown MR. 2016. Using functional or structural magnetic resonance images and personal characteristic data to identify ADHD and autism. PLoS One 11:e0166934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. 2016. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex 26:288–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallquist MN, Hwang K, Luna B. 2013. The nuisance of nuisance regression: spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage 82:208–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, et al. . 2017. Early brain development in infants at high risk for autism spectrum disorder. Nature 542:348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JV, Jacokes ZJ, Torgerson CM, Irimia A, Van Horn JD. 2017. Resting-state functional connectivity in autism spectrum disorders: a review. Front Psychiatry 7:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iidaka T. 2015. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex 63:55–67 [DOI] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Bloy L, Roberts TP, Verma R. 2011. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. Neuroimage 57:918–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahedi A, Nasamran CA, Faires B, Fan J, Müller R-A. 2017. Distributed intrinsic functional connectivity patterns predict diagnostic status in large autism cohort. Brain Connect 7:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal W, Das S, Oprescu IA, Maharatna K, Apicella F, Sicca F. 2014. Classification of autism spectrum disorder using supervised learning of brain connectivity measures extracted from synchrostates. J Neural Eng 11:046019. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Chu K, Lu Z, Herskovits EH. 2010. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage 50:589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wee CY, Shi F, Thung KH, Ni D, Yap PT, Shen D. 2015. Identification of infants at high-risk for autism spectrum disorder using multiparameter multiscale white matter connectivity networks. Hum Brain Mapp 36:4880–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli JS, Kinnear MK, Fong CH, Fishman I, Carper RA, Müller R-A. 2019. Local cortical gyrification is increased in children with autism spectrum disorders, but decreases rapidly in adolescents. Cereb Cortex 29:2412–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Baldassarre A, Committeri G, Romani GL, Corbetta M. 2009. Learning sculpts the spontaneous activity of the resting human brain. Proc Natl Acad Sci U S A 106:17558–17563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libero LE, DeRamus TP, Lahti AC, Deshpande G, Kana RK. 2015. Multimodal neuroimaging based classification of autism spectrum disorder using anatomical, neurochemical, and white matter correlates. Cortex 66:46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Lai MC, Baron-Cohen S. 2019. Big data approaches to decomposing heterogeneity across the autism spectrum. Mol Psychiatry. [Epub ahead of print]; DOI: 10.1038/s41380-018-0321-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore P, Risi S, Gotham K, Bishop SL. 2012. Autism Diagnostic Observation Schedule–2. Torrance, CA: Western Psychological Services [Google Scholar]
- Moradi E, Khundrakpam B, Lewis JD, Evans AC, Tohka J. 2017. Predicting symptom severity in autism spectrum disorder based on cortical thickness measures in agglomerative data. Neuroimage 144:128–141 [DOI] [PubMed] [Google Scholar]
- Mori S, Wakana S, van Zijl PCM, Nagae-Poetscher LM. 2005. MRI Atlas of Human White Matter. Amsterdam: Elsevier Science [Google Scholar]
- Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED, et al. . 2013. Multisite functional connectivity MRI classification of autism: ABIDE results. Front Hum Neurosci 7:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2013. Steps toward optimizing motion artifact removal in functional connectivity MRI; a reply to Carp. Neuroimage 76:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. . 2011. Functional network organization of the human brain. Neuron 72:665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasetti R, Weinberger DR. 2011. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev 21:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B, Blanken LME, Muetzel RL, Miller R, Damaraju E, Arbabshirani MR, et al. . 2018. Connectivity dynamics in typical development and its relationship to autistic traits and autism spectrum disorder. Hum Brain Mapp 39:3127–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter MA, Mash LE, Linke AC, Fong CH, Fishman I, Müller R-A. 2019. Distinct patterns of atypical functional connectivity in lower-functioning autism. Biol Psychiatry Cogn Neurosci Neuroimaging 4:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, LeCouteur A, Lord C. 2003. Autism Diagnostic Interview–R. Los Angeles, CA: Wester Psychological Services [Google Scholar]
- Samson F, Mottron L, Soulieres I, Zeffiro TA. 2012. Enhanced visual functioning in autism: an ALE meta-analysis. Hum Brain Mapp 33:1553–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, et al. . 2013. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64:240–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer M, Cuadra MB, Tamarit L, Lazeyras F, Eliez S, Thiran J-P. 2008. A surface-based approach to quantify local cortical gyrification. IEEE Trans Med Imaging 27:161–170 [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, et al. . 2006. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31:1487–1505 [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. . 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23(Suppl 1):S208–S219 [DOI] [PubMed] [Google Scholar]
- Stahl D, Pickles A, Elsabbagh M, Johnson MH, Team B. 2012. Novel machine learning methods for ERP analysis: a validation from research on infants at risk for autism. Dev Neuropsychol 37:274–298 [DOI] [PubMed] [Google Scholar]
- Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A. 2008. Conditional variable importance for random forests. BMC Bioinformatics 9:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordjman S, Cohen D, Coulon N, Anderson GM, Botbol M, Roubertoux PL. 2018. Reprint of reframing autism as a behavioral syndrome and not a specific mental disorder: perspectives from a literature review. Neurosci Biobehav Rev 89:132–150 [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. 2010. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol 103:297–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Fang H, Wu J, Xiao C, Xiao T, Qian L, et al. . 2017. Diagnostic model generated by MRI-derived brain features in toddlers with autism spectrum disorder. Autism Res 10:620–630 [DOI] [PubMed] [Google Scholar]
- Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y, et al. . 2016. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat Commun 7:11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.