Bacteria in biofilms are protected from killing by antibiotics, and this reduced susceptibility contributes to the persistence of infections such as those in the cystic fibrosis lung and chronic wounds. A generalized conceptual model of biofilm antimicrobial tolerance with the following mechanistic steps is proposed: (i) establishment of concentration gradients in metabolic substrates and products; (ii) active biological responses to these changes in the local chemical microenvironment; (iii) entry of biofilm cells into a spectrum of states involving alternative metabolisms, stress responses, slow growth, cessation of growth, or dormancy (all prior to antibiotic treatment); (iv) adaptive responses to antibiotic exposure; and (v) reduced susceptibility of microbial cells to antimicrobial challenges in some of the physiological states accessed through these changes.
KEYWORDS: antibiotic resistance, biofilms, physiology, starvation, stress response
ABSTRACT
Transcriptomic, metabolomic, physiological, and computational modeling approaches were integrated to gain insight into the mechanisms of antibiotic tolerance in an in vitro biofilm system. Pseudomonas aeruginosa biofilms were grown in drip flow reactors on a medium composed to mimic the exudate from a chronic wound. After 4 days, the biofilm was 114 μm thick with 9.45 log10 CFU cm−2. These biofilms exhibited tolerance, relative to exponential-phase planktonic cells, to subsequent treatment with ciprofloxacin. The specific growth rate of the biofilm was estimated via elemental balances to be approximately 0.37 h−1 and with a reaction-diffusion model to be 0.32 h−1, or one-third of the maximum specific growth rate for planktonic cells. Global analysis of gene expression indicated lower transcription of ribosomal genes and genes for other anabolic functions in biofilms than in exponential-phase planktonic cells and revealed the induction of multiple stress responses in biofilm cells, including those associated with growth arrest, zinc limitation, hypoxia, and acyl-homoserine lactone quorum sensing. Metabolic pathways for phenazine biosynthesis and denitrification were transcriptionally activated in biofilms. A customized reaction-diffusion model predicted that steep oxygen concentration gradients will form when these biofilms are thicker than about 40 μm. Mutant strains that were deficient in Psl polysaccharide synthesis, the stringent response, the stationary-phase response, and the membrane stress response exhibited increased ciprofloxacin susceptibility when cultured in biofilms. These results support a sequence of phenomena leading to biofilm antibiotic tolerance, involving oxygen limitation, electron acceptor starvation and growth arrest, induction of associated stress responses, and differentiation into protected cell states.
IMPORTANCE Bacteria in biofilms are protected from killing by antibiotics, and this reduced susceptibility contributes to the persistence of infections such as those in the cystic fibrosis lung and chronic wounds. A generalized conceptual model of biofilm antimicrobial tolerance with the following mechanistic steps is proposed: (i) establishment of concentration gradients in metabolic substrates and products; (ii) active biological responses to these changes in the local chemical microenvironment; (iii) entry of biofilm cells into a spectrum of states involving alternative metabolisms, stress responses, slow growth, cessation of growth, or dormancy (all prior to antibiotic treatment); (iv) adaptive responses to antibiotic exposure; and (v) reduced susceptibility of microbial cells to antimicrobial challenges in some of the physiological states accessed through these changes.
INTRODUCTION
What happens when microorganisms form a biofilm that renders them less susceptible to killing by antimicrobial agents? Among the answers we have considered are the following: (i) the antimicrobial agent fails to penetrate into the biofilm structure; (ii) biofilm cells deploy active (energy-dependent) stress responses (e.g., expression of drug efflux pumps, SOS response) that confer protection from the antimicrobial agent; (iii) some of the cells in the biofilm shift to alternative metabolisms and in these altered metabolic states are less susceptible; (iv) some of the microbial cells in the biofilm stop growing altogether and become less susceptible by virtue of their inactivity; (v) some biofilm cells enter dormant or persister states in which specific protective functions have been expressed (this state is conceptually distinct from the previous state in that dormant cells have actively remodeled the cell to afford protection, in contrast to simple cessation of activity); and (vi) microorganisms actively adapt once exposed to the antibiotic. A combination of these factors may also cause biofilm cells to be less susceptible to antibiotic treatments than planktonic cells.
We seek a mechanistic sequence of physical, chemical, and biological events that results in reduced biofilm susceptibility to antimicrobial agents (Fig. 1). In the particular case of ciprofloxacin and a Pseudomonas aeruginosa biofilm, experimental measurements from independent laboratories using varied techniques have consistently demonstrated rapid and effective penetration of fluoroquinolone antibiotics throughout the biofilm (1–5). We therefore regarded poor penetration as an unlikely mechanism and focused our attention on physiological changes in biofilm bacteria. Thus, the following steps are proposed for increased tolerance of biofilms to antibiotics: (i) establishment of concentration gradients in metabolic substrates and products through reaction-diffusion interactions (this term refers to the diffusive transport of oxygen [for example] and its simultaneous consumption [reaction] by microbial respiration) (6); (ii) active biological responses to these changes in the local chemical microenvironment through shifts in gene expression or alterations of enzyme activity; (iii) entry of biofilm cells into a spectrum of states involving alternative metabolisms, stress responses, slow growth, cessation of growth, and dormancy (7, 8); (iv) deployment of adaptive responses subsequent to antibiotic exposure (9, 10); and (v) reduced susceptibility of microbial cells to antimicrobial challenges in some of the resultant physiological states.
FIG 1.
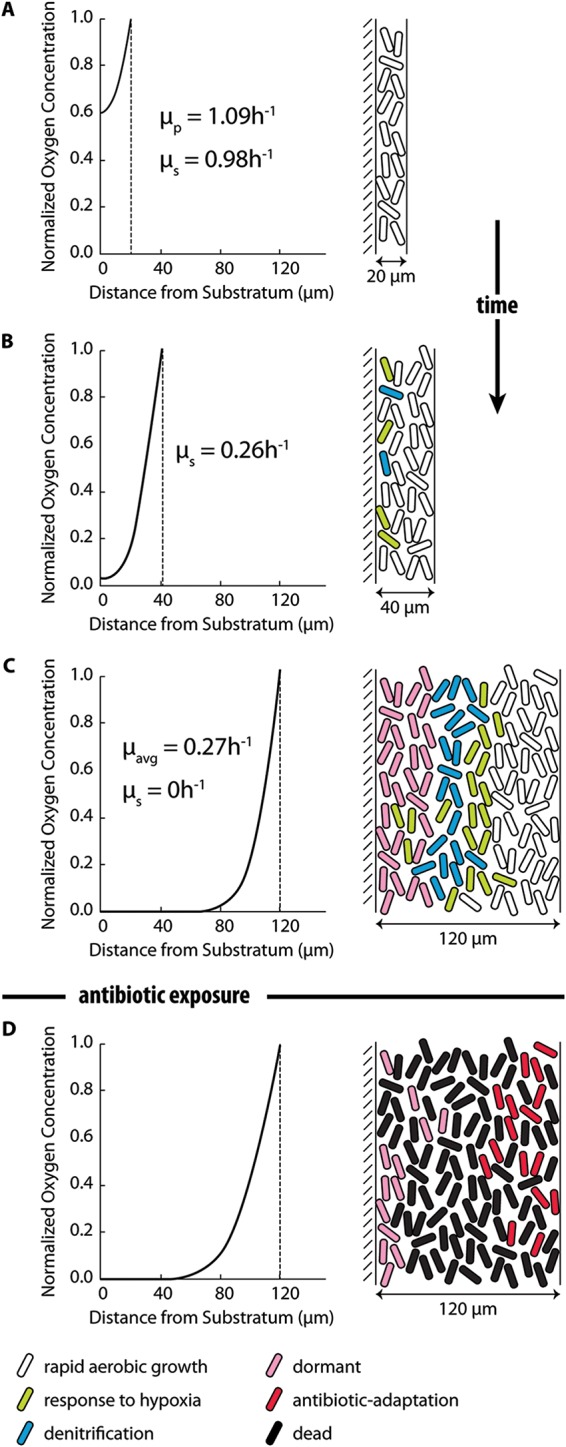
Hypothesized steps in the development of antibiotic tolerance in a P. aeruginosa biofilm. (A) Early on, there is little change in the chemical microenvironment within the biofilm. The biofilm is well permeated by oxygen and is characterized by rapid growth and relatively high antibiotic susceptibility. (B) Oxygen depletion first manifests when the biofilm reaches a thickness of approximately 40 μm, leading to the induction of genetic and metabolic responses to hypoxia and the onset of growth rate limitation. Anaerobic respiration of nitrate and nitrite may ensue, depending on the availability of these alternate electron acceptors. (C) As the biofilm attains its mature thickness, reaction-diffusion interactions lead to zones of decreased concentrations of metabolic substrates (e.g., oxygen, nitrite, zinc) and increased concentrations of metabolic products (e.g., homoserine lactone quorum-sensing molecules, phenazines) within the biofilm. Sustained changes in the chemical microenvironment subsequently modulate gene expression, enzyme activity, and metabolic fluxes and, over time, result in altered metabolisms, deployment of stress responses, anabolic inactivity, and dormancy, each of which can contribute to antibiotic tolerance. All of these hypothesized changes, which may have alternative sequences or parallel progress, occur prior to antibiotic exposure. (D) Finally, antibiotic treatment induces adaptive responses that may further enhance antibiotic tolerance. The oxygen gradients shown on the left were calculated with the reaction-diffusion model as explained in the text. Specific growth rates (μ) were determined from the calculated oxygen concentrations: μp, growth rate under planktonic conditions; μs, predicted growth rate at the substratum; μavg, average growth rate in the biofilm. Physiological states, shown on the right, are color coded as follows: white, rapidly growing cells, oxygen replete, physiologically similar to exponential-phase planktonic cells; green, cells deploying hypoxic stress, starvation, or quorum-sensing responses; blue, denitrifying cells; pink, dormant cells; red, cells deploying adaptive responses to antibiotic exposure; black, dead cells.
Prior work informs this hypothesized sequence for P. aeruginosa biofilms in particular. Depletion of oxygen in the interior of P. aeruginosa biofilms has been demonstrated through microelectrode measurements of oxygen concentration profiles (11–15). Increased concentrations (relative to the concentration outside the biofilm) of a metabolic product, pyocyanin, have been measured proximal to a biofilm cluster (16). Prior transcriptomic comparisons of gene expression between planktonic and biofilm P. aeruginosa bacteria have suggested responses to hypoxia (17), iron limitation (18), nutrient limitation (17), and upregulation of phenazine biosynthesis (17) and the denitrification pathway (18, 19). Characterization of the spatial distribution of anabolic activity within P. aeruginosa biofilms has revealed stratified patterns with activity focused near the source of oxygen and putative regions of nongrowing bacteria distal from the oxygen-supplied interface (4, 11, 13, 19). Data from the literature on the killing activity of ciprofloxacin against planktonic and biofilm P. aeruginosa PAO1 (12, 19–23) confirm the reduced susceptibility of bacteria in the biofilm state to the antibiotic (Fig. 2). Other reports describe the contributions of specific genes or regulatory systems to antibiotic tolerance in P. aeruginosa biofilms (21, 24–34).
FIG 2.
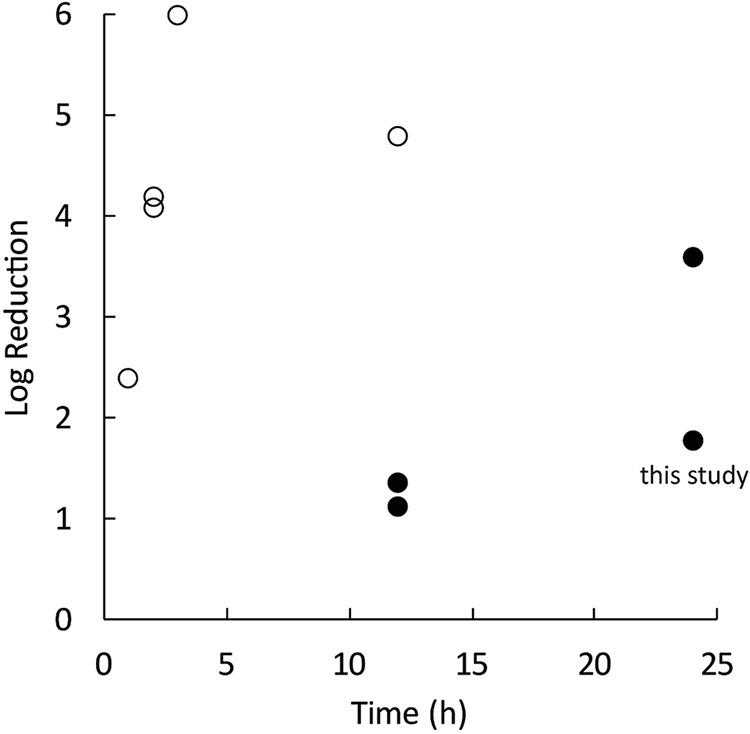
Planktonic P. aeruginosa bacteria (open circles) are killed more quickly by ciprofloxacin than are bacteria in mature biofilms (filled circles). The duration of antibiotic exposure is shown along the x axis. Data are from references 9, 15, and 17, to ,19 and this study.
Biofilm tolerance to antimicrobial action refers to a comparison between the susceptibilities of the same microorganism tested in a biofilm model and in a planktonic assay. The biofilm susceptibility measured depends strongly on the biofilm method used (35). So, too, does planktonic susceptibility depend on the specific conditions of the test, including the medium, growth phase, aeration, and cell density. In this work, we have chosen to use aerated, exponential-phase planktonic cells for the planktonic reference state for metabolomic and transcriptomic comparisons to biofilm cells. One might reasonably argue that stationary-phase planktonic cells would be a more appropriate comparator. Cells in stationary phase may resemble cells in the nutrient-depleted regions of a biofilm. Stationary phase is, however, chemically and temporally ill defined and therefore makes a poor reference state. Exponential-phase planktonic cells can be assigned a single, quantitative specific growth rate, and this growth rate remains unchanged over multiple generations, establishing that the chemical environment of the culture is not changing in a way that impacts the physiology of the cells. The chemical environment is essentially given by the composition of the fresh medium, which is known. Stationary phase, despite the implication of stasis in this name, is neither fixed nor clearly defined. It is reasonable to anticipate that cells in a shaken batch culture that have just entered stationary phase at 12 h after inoculation will differ physiologically from cells in the same flask 2 days later. The chemical conditions in a stationary-phase culture have changed in multiple, uncharacterized ways from those in the initial medium, and the growth-limiting substrate (or inhibitive product) is not usually known. Thus, the conditions in a stationary-phase culture flask do not necessarily mimic those inside a biofilm. For these reasons, stationary phase is not an adequate model for biofilm physiology and, because of the lack of chemical definition, is a problematic reference state for dissecting biochemical mechanisms.
The assumption implicit in our conceptual model of biofilm antimicrobial tolerance is that susceptibility is dictated by the physiological state of the cell, whether that cell is embedded within a biofilm or is floating in an aqueous medium. There is no intrinsic difference between a cell in a biofilm and cell in a free aqueous suspension—only differences in their physiological states that arise from different environmental histories. By this logic, “biofilm tolerance” may be an artifact of comparing microorganisms in biofilms that have aged, often for a period of days, through transitions of microenvironmental chemistry with organisms in planktonic culture that are usually challenged soon after inoculation and are still growing rapidly. In this article, we seek to understand more clearly the physiological state of bacteria in a biofilm system and to explain these states mechanistically.
In the present study, we undertook an investigation of the model proposed in Fig. 1 using a single-species P. aeruginosa biofilm, a medium designed to mimic the milieu in a human chronic wound, and the antibiotic ciprofloxacin. An in vitro continuous-flow reactor system was used to investigate in detail changes in the chemistry, metabolism, gene expression, and antibiotic susceptibility of the biofilm bacteria and to gain insight into specific features of these possible protective mechanisms.
RESULTS
Reaction-diffusion modeling predicts oxygen concentration gradients in biofilms.
A reaction-diffusion model was solved to characterize the distribution of oxygen within the P. aeruginosa biofilm grown in the drip flow reactor (DFR). The purpose of this modeling was to demonstrate that changes in the chemical microenvironment within the biofilm, which determine subsequent alterations in physiology, derive from a fundamental chemical-physical interaction. Using parameter values representative of this reactor system (see Table S1 in the supplemental material), the model predicted that steep oxygen concentration gradients form in biofilms that are thicker than about 40 μm (Fig. 1). The concentration of oxygen at the substratum of the biofilm was calculated to decrease geometrically with increasing biofilm thickness (see Fig. S1 in the supplemental material). These calculations reinforce the plausibility of oxygen limitation as an important chemical microenvironmental condition for these biofilms.
The specific growth rate is reduced in biofilms.
In planktonic culture in an artificial chronic wound exudate (ACWE) medium at 33°C, P. aeruginosa PAO1 grew with an exponential-phase maximum specific growth rate of 1.09 ± 0.18 h−1. This medium and temperature were chosen to mimic the chemical and physical conditions in a human chronic wound. Material balances were used to estimate the average specific growth rate of the same bacteria when growing as a biofilm in the drip flow reactor in the same medium and at the same temperature. When the biofilm was 4 days old, the biofilm specific growth rate was calculated to be 0.37 h−1, which corresponds to 34% of the planktonic maximum specific growth rate. This growth rate estimate represents an average for all of the biomass in the biofilm. In addition, the reaction-diffusion model enables a theoretical determination of the biofilm average specific growth rate by converting the local oxygen concentration into a local growth rate. When calculated for a mature-biofilm thickness of 114 μm, the mean growth rate by this analysis was 0.32 h−1. The difference between these biofilm and exponential-phase planktonic growth rate values is consistent with growth rate limitation and slower growth in the biofilm.
The expression of genes associated with anabolism and ATP metabolism is reduced in biofilms.
When gene expression levels determined with microarrays were analyzed using the gene ontology tool DAVID, a general pattern of downregulation of anabolic and ATP-dependent activities in biofilms relative to exponential-phase planktonic cells was observed (Table 1). No metabolic pathways were identified as upregulated in biofilms by this analysis using the default DAVID parameters. The 17 gene ontology groups that were statistically significantly downregulated in the biofilm relative to exponential-phase planktonic conditions included ribosome components; ATP binding, ATP synthase, and ATP synthesis coupled electron transport activities; and multiple biosynthetic pathways (Table 1).
TABLE 1.
Expression of mRNAs associated with anabolic activity and ATP metabolism is reduced in P. aeruginosa biofilms relative to that in exponential-phase planktonic culturesa
| Gene ontology term | P value |
|---|---|
| Structural constituent of ribosome | 1 × 10−24 |
| ATP binding | 2 × 10−10 |
| Biosynthesis of secondary metabolites | 7 × 10−5 |
| Pyridoxal phosphate binding | 0.0004 |
| Aminoacyl-tRNA biosynthesis | 0.0007 |
| Proton-transporting ATP synthase activity, rotational mechanism | 0.0007 |
| Oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | 0.0016 |
| Histidine biosynthetic process | 0.0041 |
| Fatty acid biosynthetic process | 0.0104 |
| Peptidyl-prolyl cis-trans isomerase activity | 0.0111 |
| DNA topological change | 0.0132 |
| GTP binding | 0.0195 |
| Cobalamin/porphyrin biosynthetic process | 0.0221 |
| Arginine biosynthetic process | 0.0230 |
| Lysine biosynthetic process via diaminopimelate | 0.0285 |
| ATP synthesis coupled electron transport | 0.0304 |
| Pyridoxine biosynthetic process | 0.0350 |
Functional annotation clustering of P. aeruginosa genes downregulated in biofilms relative to planktonic growth (>2-fold change; P < 0.05) is based on gene enrichment analysis of gene ontology groups using DAVID. There were no statistically significant clusters for genes upregulated in biofilms using these parameters.
Multiple stress responses are activated in biofilms.
A customized gene set enrichment analysis comparing expression in exponential-phase planktonic cells, untreated biofilms, and ciprofloxacin-treated biofilms revealed the induction of multiple stress responses in biofilm cells (Table 2; see also Data Sets S1 to S3 in the supplemental material), even those not exposed to the antibiotic. Among the gene sets with statistically significantly higher expression in untreated biofilms than in exponential-phase planktonic cells were those associated with the stationary phase, zinc limitation, oxygen downshift, oxygen limitation, acyl-homoserine lactone quorum sensing, and osmotic stress. Metabolic pathways for phenazine biosynthesis and denitrification were transcriptionally activated in biofilms as well.
TABLE 2.
Stress responses are activated in P. aeruginosa biofilms relative to those in exponential-phase planktonic culturesa
| Activityb |
P value for association with differential gene expression in biofilms |
|
|---|---|---|
| Upregulation | Downregulation | |
| Stationary phase | 10−15 | 1.00 |
| Zn limitation | 10−15 | 1.00 |
| Oxygen downshift | 10−14 | 0.16 |
| Oxygen limitation | 10−13 | 0.98 |
| HSL quorum sensing | 10−10 | 0.42 |
| Denitrification | 10−6 | 1.00 |
| Phenazine biosynthesis | 10−4 | 1.00 |
| Virulence factors | 10−3 | 1.00 |
| Osmotic stress | 10−3 | 1.00 |
| Ammonia release | 0.05 | 10−6 |
Based on gene set enrichment analysis (2-fold change).
HSL, homoserine lactone.
P. aeruginosa is capable of synthesizing three extracellular polysaccharides (Psl, Pel, and alginate), which are known to be involved in biofilm formation depending on the strain and environmental conditions (36). The biosynthetic genes involved in producing these polymers were not expressed at higher levels in biofilms than in planktonic cells (P = 1.0). As a group, genes associated with drug efflux pumps (P = 0.21) and oxidative stress (P = 0.33) also were not differentially expressed under biofilm and exponential-phase planktonic conditions.
Regulon analyses indicated activation of the RpoS, MvfR, and VqsM regulons in biofilms relative to exponential-phase planktonic cells (Table 3). The RpoS and MvfR regulons are known to be expressed at high cell densities. Gene sets positively regulated by Anr or RsmA were not upregulated in biofilms with statistical significance. AmgRS-regulated genes were not differentially expressed under biofilm and planktonic conditions.
TABLE 3.
Regulons of RpoS and MvfR are activated in biofilms relative to exponential-phase planktonic culturesa
| Activityb |
P value for association with differential gene expression in biofilms |
|
|---|---|---|
| Upregulation | Downregulation | |
| RpoS+ | 10−15 | 1.00 |
| RpoS– | 10−6 | 0.61 |
| MvfR+ | 10−15 | 0.22 |
| MvfR– | 0.05 | 1.00 |
| VqsM+ | 10−4 | 10−15 |
| VqsM– | 10−7 | 1.00 |
| Anr+ | 0.09 | 10−4 |
| Crc+ | 1.00 | 10−5 |
| Crc– | 0.33 | 10−7 |
| RsaL+ | 1.00 | 10−5 |
| RsaL– | 10−15 | 1.00 |
| RsmA+ | 0.01 | 1.00 |
| RsmA– | 0.19 | 0.29 |
| AmgR+ | 0.69 | 0.48 |
Based on gene set enrichment analysis.
+ or –, positive or negative regulation.
Treatment of biofilms with ciprofloxacin induced responses to oxidative stress and DNA damage, the SOS response (37) in particular (Table 4; see also Data Set S3 in the supplemental material). Many of the stress response gene sets identified as upregulated in untreated biofilms relative to planktonic cells (Table 2) were downregulated in antibiotic-treated biofilms relative to untreated biofilms.
TABLE 4.
Oxidative stress and the SOS response are activated in P. aeruginosa biofilms treated with ciprofloxacin relative to untreated biofilmsa
| Activityb |
P value for association with differential gene expression in biofilms |
|
|---|---|---|
| Upregulation | Downregulation | |
| Peroxide stress | 10−14 | 0.33 |
| SOS response | 10−5 | 1.00 |
| Stationary phase | 1.00 | 10−15 |
| HSL quorum sensing | 1.00 | 10−15 |
| Virulence factors | 1.00 | 10−14 |
| Zn limitation | 1.00 | 10−13 |
| Oxygen limitation | 1.00 | 10−6 |
| Phenazine biosynthesis | 1.00 | 10−6 |
| Fe limitation | 0.94 | 10−5 |
Based on gene set enrichment analysis.
HSL, homoserine lactone.
Biofilm cells consume more glucose and lactate and a lower proportion of amino acids than planktonic cells.
Carbon sources in the ACWE medium that could potentially support bacterial growth included lactic acid, glucose, and all 20 amino acids. These compounds together provided 100 mM carbon. With the exception of three amino acids (cysteine, arginine, and asparagine) that were not detectable in the gas chromatography-mass spectrometry (GC-MS) analysis, these carbon sources were analyzed in fresh medium, exponential-phase planktonic cultures, and biofilm reactor effluents. The detectable analytes represented 98% of the usable carbon in the medium. By comparing the patterns of carbon source utilization by biofilm and exponential-phase planktonic cells, it can be seen that biofilm cells used more glucose and lactic acid and a lower proportion of amino acids than did planktonic cells (Fig. 3; see also Data Set S4 in the supplemental material). Biofilm bacteria excreted acetate (1.42 mM), whereas this metabolic product was not detected in planktonic-culture supernatants.
FIG 3.
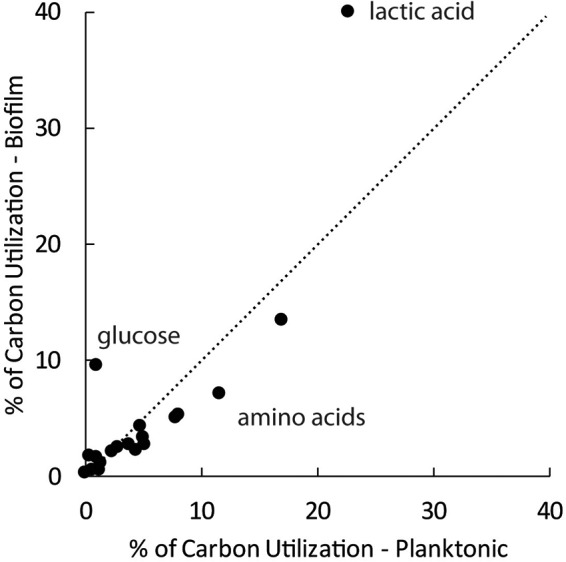
Biofilm cells consume more glucose and lactate and a lower proportion of amino acids than planktonic cells. The dotted line is the line of equality.
Biofilms treated with ciprofloxacin for 24 h continued to consume carbon sources at rates essentially unchanged from those in the pretreatment state. Untreated biofilms consumed a total of 49.7 mM carbon, while antibiotic-treated biofilms consumed 49.9 mM carbon. The profiles of carbon source usage were similar for untreated and treated biofilms (see Fig. S2 in the supplemental material). Ciprofloxacin-treated biofilms excreted less acetate (0.87 mM carbon) than untreated biofilms (2.83 mM carbon).
Additional metabolite measurements are consistent with an overall metabolic downshift in biofilms with activation of quorum sensing.
Analysis of intracellular metabolites in exponential-phase planktonic and biofilm cells detected thousands of molecular features corresponding to 352 tentatively identified metabolites (see Data Sets S5 and S6 in the supplemental material). In this group, 58 metabolites had significantly different abundances in exponential-phase planktonic and biofilm cells (P < 0.05). Of these, 57 were present at lower concentrations in biofilm samples than in planktonic cells, whereas just 1 was present at a higher concentration in biofilm cells (see Fig. S3 in the supplemental material). To take a specific group of metabolites, consider the four amino acids detected in both biofilm and planktonic cells for which a difference in concentration at a significance level (P) of <0.05 was determined. All four of these amino acids showed lower abundances in biofilm cells than in planktonic cells, with fold changes (FC) ranging from 9 to 63. Decreased concentrations of metabolic intermediates may reflect an overall decrease in the metabolic activity of biofilm cells. In foundational work by Frimmersdorf et al. (38), intracellular metabolites of P. aeruginosa showed a general pattern of lower concentrations in stationary-phase cells than in exponential-phase cells when grown on Casamino Acids, citrate, or succinate.
Two features corresponding to N-(3-oxo-dodecanoyl)-l-homoserine lactone were detected, indicating higher concentrations of this signaling molecule in biofilm cells than in exponential-phase planktonic cells (FC = 36.2 and 4.1 [P = 0.03 and 0.06] for the two features, respectively). The phenazine product pyocyanin was detected in the intracellular metabolite pool. Pyocyanin had a relatively small increase in biofilm cells relative to exponential-phase planktonic cells that was not statistically significant (FC = 2.3; P = 0.32). Four tricarboxylic acid (TCA) cycle metabolites were detected. Three of these did not differ in concentration between biofilm and exponential-phase planktonic cells (succinate, fumarate, and oxaloacetate [P = 0.20, 0.51, and 0.97, respectively]). α-Ketoglutarate levels were lower under biofilm conditions than under planktonic conditions (FC = 6.3; P = 0.05).
In examining the transcriptome data, we noticed a number of genes associated with the release of free ammonia in the breakdown of amino acids that were downregulated in the biofilm mode of growth (relative to the planktonic mode). We hypothesized that biofilm bacteria might be experiencing ammonia stress. Ammonia could come either from amino acid catabolism or from the hydrolysis of urea, an abundant constituent of the ACWE medium. To further investigate this possibility, assays for ammonium, urea, and pH were performed on biofilm reactor effluents. These measurements, summarized in Table S2 in the supplemental material, revealed no substantial pH change, urea hydrolysis, or ammonia accumulation in the biofilm system. When the wild-type (WT) strain was grown as a biofilm on a medium lacking urea, its susceptibility to ciprofloxacin (log reduction, 1.71; n = 1) was not different from that in the baseline experiment in complete medium (log reduction, 1.79).
Mutants deficient in Psl polysaccharide synthesis, the stringent response, the stationary-phase response, and the membrane stress response exhibit increased ciprofloxacin susceptibility in biofilms.
P. aeruginosa formed robust biofilms when grown on ACWE medium in a drip flow reactor (see Fig. S4 in the supplemental material). Untreated 4-day-old biofilms contained 9.45 ± 0.46 log10 CFU cm−2 (mean ± standard deviation; n = 17). When these biofilms were subsequently treated with 1 μg of ciprofloxacin ml−1 (the MIC for planktonic cells in ACWE medium was 0.06 μg ml−1) for 24 h, the resulting log reduction was 1.79 ± 0.43 (n = 15). Antibiotic-treated biofilms contained less biomass than untreated biofilms (see Table S3). The protein content of treated biofilms was 23% that of the untreated biofilm; the carbohydrate content of treated biofilms was 59% that of the untreated biofilm; and the DNA content of treated biofilms was 77% that of the untreated biofilm. The mean thickness (±standard deviation) of a single untreated biofilm (48 microscale measurements) was 114 ± 48 μm, whereas the mean thickness of a single biofilm after ciprofloxacin treatment (47 microscale measurements) was 157 ± 56 μm.
Selected mutant strains were grown as biofilms for 3 days and were then treated with ciprofloxacin. Most of these mutants (15 of 18), when cultured in biofilms, exhibited ciprofloxacin susceptibilities similar to that of the wild-type strain (Fig. 4). Four mutants exhibited higher susceptibilities than the WT when grown as biofilms: the psl, ΔrelA ΔspoT, rpoS, and amgR mutants. An isocitrate lyase (aceA) mutant exhibited decreased biofilm susceptibility to ciprofloxacin.
FIG 4.
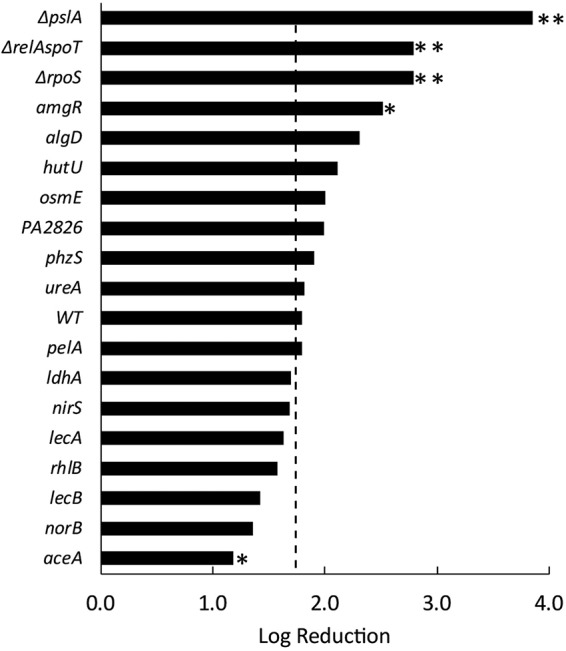
Mutants deficient in Psl biosynthesis (ΔpslA), the stringent response (ΔrelA ΔspoT), the membrane stress response (amgR), stationary-phase sigma factor regulation (ΔrpoS), and the glyoxylate shunt pathway (aceA) exhibit differential susceptibilities to ciprofloxacin in biofilms. The killing of wild-type (WT) and mutant P. aeruginosa strains in 3-day-old biofilms by ciprofloxacin was determined by viable plate counts. The dashed line delineates the killing of the WT bacteria, which resulted in a log reduction of 1.79 ± 0.43 (n = 15). Mutations that gave statistically significantly different log reductions are marked with asterisks (**, P < 0.01; *, P < 0.05).
It is known that the pretreatment cell density of a biofilm affects its antimicrobial susceptibility (35). Could the differential sensitivities of mutant strains result from differences in starting cell numbers? The viable areal cell densities of biofilms grown from mutant strains displaying a significant differential susceptibility phenotype, expressed as percentages of the mean wild-type density, were 63% for the psl mutant, 56% for the ΔrelA ΔspoT mutant, 68% for the amgR mutant, 205% for the rpoS mutant, and 115% for the aceA mutant. None of these differences between mutant and wild-type densities was statistically significant. To test for the hypothesized effect, we performed a linear regression of log reduction versus initial areal cell density for all mutant and wild-type data. No statistically significant correlation was found (R2 = 0.096; P = 0.47).
DISCUSSION
The goal of this project was to integrate the chemical, transcriptomic, metabolomic, and physiological data from an experimental biofilm system in a conceptual and mechanistic framework that provides a detailed characterization of how bacterial phenotypes change within a biofilm and which of these changes are associated with decreased antibiotic susceptibility. The narration that follows presents this integrated mechanism, the sequence of which is diagrammed in Fig. 1, with discussion of its successes and limitations.
Reaction-diffusion interactions change the chemical microenvironment inside a biofilm.
As a P. aeruginosa biofilm develops in the drip flow reactor, an oxygen concentration gradient manifests, and oxygen becomes limiting in the depths of the biofilm. Based on prior measurements with oxygen microelectrodes in similar biofilms, the onset of oxygen limitation at the base of the biofilm is anticipated by the time the biofilm is approximately 50 μm thick (11, 19). Mature biofilms composed of or containing P. aeruginosa, when probed with a microsensor, always exhibit steep oxygen concentration gradients and regions of very low oxygen concentration (4, 11–15, 19, 39, 40). Oxygen gradients and hypoxic zones have also been measured in vivo or ex vivo in humans or in animal models of chronic wound infections (15, 41, 42). Oxygen penetration into the biofilm grown in this investigation was calculated with a reaction-diffusion model to confirm this outcome using system-specific parameter values (Fig. 1). For a biofilm that is 100 μm thick, the oxygen concentration at the base of the biofilm (i.e., at the substratum) was calculated to be a millionfold less than the air saturation concentration. This result is in accord with a long history of reaction-diffusion modeling in the biofilm field (6, 43–45) and underscores the rigorous, quantitative physical-chemical basis for predicting and understanding such oxygen concentration profiles.
In contrast, carbon and nitrogen sources in the medium (lactic acid, glucose, and amino acids) are not predicted to be limiting within the biofilm. Once oxygen is locally depleted, the catabolism of these electron donors will largely cease. For example, for lactic acid, the concentration at the base of a 100-μm-thick biofilm was calculated to be 97.2% of the bulk fluid concentration. Thus, the microenvironment in the lower zones of the mature biofilm is expected to be replete with carbon- and nitrogen-containing substrates while anoxic.
Gene expression and enzyme activities are altered within the biofilm.
It is logical to anticipate that oxygen deprivation will trigger multiple biological consequences. The first of these may be the expression of genes that are induced under conditions of an oxygen downshift (a transfer from aerobic to anaerobic conditions, as described by Trunk et al., for example [46]) or under hypoxic conditions (as described by Alvarez-Ortega and Harwood, for example [47]). Indeed, highly significant overlap was found between both of these published gene sets and the genes upregulated in 96-h-old biofilms (Table 2). Of the 75 genes with ≥10-fold higher expression in biofilm cells than in exponential-phase planktonic cells, 23 appear on either the Trunk et al. list (46) or the Alvarez-Ortega and Harwood list (47). These include, for example, genes coding for enzymes in the denitrification pathway. Many of these 23 oxygen limitation-associated genes are annotated as hypothetical proteins (16 genes, or 70%), indicating that the response to oxygen limitation remains incompletely understood.
In addition to oxygen limitation, there is a strong signature for Zn limitation (48) in the biofilm transcriptome (Table 2). No zinc was added to the medium as a distinct ingredient, so the zinc that was available must have been present as a contaminant in the water used to prepare the medium or in one or more of the reagents. Zinc limitation was not evident in the transcriptome of exponential-phase planktonic cultures. Zinc limitation within the biofilm could occur by the same reaction-diffusion interaction that leads to oxygen limitation. Another explanation for why zinc becomes limiting in the biofilm but not in planktonic cultures is that extracellular polymeric substances in the biofilm sequester zinc. If the biofilm matrix binds zinc, and biofilm cells synthesize more matrix, then the zinc requirement for building a biofilm would be greater than the zinc requirement for forming planktonic cells, and the biofilm would consequently be more vulnerable to experiencing limitation for this element. Recent transcriptomic and proteomic analyses of P. aeruginosa from cystic fibrosis (CF) sputum have reported data indicative of zinc limitation in vivo in this milieu (49, 50).
In contrast to metabolic substrates such as oxygen and zinc, which are found at lower concentrations inside a biofilm than outside, a metabolic product will be present at a higher concentration inside the biofilm. This is expected to be the case with quorum-sensing signal molecules such as the acyl-homoserine lactones and could explain the observed higher expression of certain quorum-sensing-regulated virulence factors in biofilm cells (Table 2). Oxo-dodecanoyl homoserine lactone was measured at higher intracellular concentrations in biofilm cells than in exponential-phase planktonic cells. Genes known to be specifically regulated by homoserine lactones, such as the elastase gene lasA (23-fold-higher expression in biofilms than in planktonic cultures), were upregulated in biofilms (Table 2).
Shifts in metabolism, deployment of stress responses, growth arrest, and entry into dormancy occur in biofilms prior to antibiotic exposure.
As oxygen limitation persists in the depths of the biofilm, one would expect to observe a shift from aerobic respiration to alternative metabolisms that do not depend on molecular oxygen: denitrification (51, 52), arginine fermentation (52), or pyruvate fermentation (53–55). Although evidence of activation of the denitrification pathway in biofilms is seen in the transcriptomic data, the flux through this pathway and its functional impact would be constrained by the low concentration of nitrite in the medium (58 μM). This concentration in ACWE medium was based on one published measurement of 65 μM combined nitrite and nitrate in chronic wound exudate (56). In the cystic fibrosis lung, considerably higher concentrations of nitrite and nitrate (approximately 350 μM combined) have been measured (57–59), and there is debate about whether anaerobic respiration contributes to P. aeruginosa fitness in vivo (47, 58, 60, 61). In our model system, we estimate the relative amount of ATP available through the use of nitrite as an alternate electron acceptor to be only 0.23% of that generated by aerobic respiration of the lactate consumed experimentally. Similarly, arginine fermentation could provide a mere 0.12% of the ATP generated aerobically.
One speculative explanation for the increased lactate consumption and acetate excretion observed in the biofilm system involves a metabolic sharing strategy first proposed by Schobert and Jahn (62). In this scheme, bacteria in the aerobic layer oxidize lactate to pyruvate, which is excreted. Pyruvate is then fermented by cells in the anoxic layer, resulting in the release of acetate. Although pyruvate fermentation generates insufficient ATP to enable growth, it does prolong cell survival under starvation conditions (53, 54) and may contribute to protection from antibiotics (63). Genes in this pathway (ldhA, ackA, and pta) have been independently shown to provide a fitness advantage to P. aeruginosa under oxygen-limiting growth (64). We observed strong expression of the pyruvate dehydrogenase complex PA3415 to -17 in biofilms relative to that in exponential-phase planktonic cells (mean FC, 19). Our metabolomic method was not able to measure pyruvate. In our experimental system, if all of the acetate generated by biofilm cells arose from pyruvate fermentation, the ATP generated would correspond to 1.90% of that available by aerobic respiration.
The basis for metabolic shifts observed in biofilm cells relative to exponential-phase planktonic cells—increased utilization of lactate and glucose and decreased utilization of amino acids—was not apparent in the transcriptomic comparison of these two growth modes. This result underscores the reality that it is not in general valid to infer metabolic activity directly from gene expression. We hypothesize that the increased consumption of glucose by biofilm cells is directed toward the synthesis of extracellular polysaccharide. This would be a particularly efficient strategy under conditions of oxygen limitation. Glucose is a much more oxygen-efficient substrate for making polysaccharide (Psl) than are the other constituents of the medium. The oxygen requirement for synthesizing Psl from glucose is >5 times less than that for synthesizing Psl from lactic acid or the average amino acid (see Table S4 in the supplemental material).
Prolonged oxygen limitation in the absence of alternate electron acceptors or fermentable substrates can be expected to trigger the expression of stress response gene sets associated with growth arrest, such as those related to the stationary phase and the stringent response (65). The stringent response regulon has not been mapped in P. aeruginosa, but the stationary-phase gene set (see Data Set S1 in the supplemental material) compiled on the basis of four published reports (47, 66–68) overlaps strongly with the genes upregulated in biofilms (Table 2). Of the 75 genes with ≥10-fold-higher expression in biofilm cells than in exponential-phase planktonic cells, 35 appear on the stationary-phase composite list. These genes include the stationary-phase sigma factor (rpoS) and rmf, the ribosome modulation factor, known to be associated with growth arrest and ribosome hibernation in Escherichia coli. Many of the other genes are annotated as hypothetical proteins (21 of 35 [60%]). The subset of genes positively regulated by RpoS was collectively upregulated in biofilms (Table 3), providing further evidence of the stationary-phase character of biofilm cells and a role for this regulator in biofilm physiology. RpoS has been shown to confer a fitness advantage on P. aeruginosa under conditions of energy-limited growth arrest (54). The RpoS protein has been shown previously to be present at higher levels in a continuously fed P. aeruginosa biofilm than in planktonic cells (69). Finally, the expression of RpoS has been shown to be spatially stratified, with predominant localization beneath the most metabolically active layer (69). Reduced abundance of ribosomal proteins was measured in the region of the biofilm associated with higher RpoS expression (70). We hypothesize that some of the genes controlled through the rpoS and relA spoT growth arrest stress responses facilitate the entry of bacterial cells into a protected dormant state. This hypothesis is consistent with prior reports of susceptibility phenotypes in these mutants (17, 26, 71, 72).
As oxygen starvation continues in the depths of the biofilm, one would also expect to observe diminished anabolic activity, such as lower overall rates of transcription and translation and, ultimately, reduced biomass growth rates. These outcomes are consistent with DAVID analysis comparing the transcriptomes of biofilm and planktonic cultures and quantitative estimates of the population-averaged specific growth rates in these two culture modes. Our finding of reduced expression of genes related to structural constituents of the ribosome in biofilms (Table 1) accords with recent transcriptomic and proteomic analyses that reported reduced ribosome biogenesis in human clinical CF sputum specimens (49, 50). The specific growth rate of biofilms was approximately one-third of the exponential-phase planktonic rate, which qualitatively agrees with other measurements of lowered growth rates in P. aeruginosa biofilms. Specific growth rates measured in mature P. aeruginosa colony biofilms have been reported to be approximately 2% of the planktonic cell growth rate (4, 12). In a prior investigation with drip flow reactor-grown biofilms, the specific growth rate was estimated at 10% of the planktonic rate (19). Single-cell estimates of the specific growth rate of P. aeruginosa in expectorated sputum from the lungs of individuals with CF returned heterogeneous measurements with a median value that was 18% of the planktonic growth rate (73).
Because it is founded on a reaction-diffusion interaction, our model predicts that anabolism and growth will be spatially stratified inside the biofilm (4, 11–13, 19, 45). We further predict that antimicrobial-sensitive cells will localize to the metabolically active region of the biofilm and antimicrobial-tolerant cells will localize to the metabolically inactive region of the biofilm. These predictions were not experimentally tested in the current investigation, but they are consistent with prior reports (70, 74–76). For example, ciprofloxacin killing of P. aeruginosa was spatially restricted to the metabolically active cells at the top of a biofilm (75).
In summary, much of what is observed in this biofilm system in terms of gene expression and overall anabolism follows logically as a consequence of reaction-diffusion limitation for oxygen. Although the prediction was not experimentally investigated in this study, we expect that the biofilm exhibits a stratification of anabolic activity with spatially heterogeneous gene expression. Previous studies of spatial patterns of anabolism in P. aeruginosa in varied experimental systems have revealed regions of active anabolism in the biofilm adjacent to the source of oxygen, with regions of inactivity and dormant cells below this layer (4, 11, 13, 18, 19). Figure 1 illustrates some possible patterns of heterogeneous physiology and antibiotic tolerance that are plausible in biofilms. These patterns are expected to change as the provision of electron acceptors and donors changes.
Biofilm antibiotic tolerance manifests and may be enhanced by adaptive responses post-antibiotic exposure.
A critical question in the investigation of biofilm tolerance to drugs is whether changes in biofilm gene expression can be used to predict antibiotic susceptibility. Our findings, summarized in Table 5, suggest multiple possibilities for the dependence of antibiotic susceptibility on genotype. For example, a straightforward prediction would be that a gene that is expressed more strongly in a biofilm than in cells in a susceptible exponential-phase culture could contribute to reduced biofilm susceptibility. However, it can be anticipated that not all of the genes induced in a biofilm (here a total of 449) play roles in modulating antimicrobial tolerance. For example, seven genes that were more highly expressed in biofilms than in exponential-phase planktonic cells (nirS, norB, rhlB, phzS, lecA, lecB, and osmE) displayed no differential susceptibility phenotype when the corresponding mutant biofilm was challenged with ciprofloxacin. Another six genes (pelA, algD, hutU, PA2826, ldhA, and ureA), whose expression did not differ between biofilm and exponential-phase planktonic cultures, also had no effect on biofilm susceptibility when mutated, conforming to the null hypothesis. Consider the specific example of the extracellular electron-shuttling molecule phenazine. Phenazines are redox-active compounds that balance the intracellular redox state of P. aeruginosa when growing in biofilms (77–79). We measured higher levels of transcription of phenazine biosynthetic genes in biofilms than in exponential-phase planktonic cells (Table 2) but found no statistically significant increase in intracellular pyocyanin levels and detected no differential ciprofloxacin susceptibility of a phzS mutant when grown in a biofilm (Fig. 4). Using stimulated Raman spectroscopy, Schiessl et al. (33) showed that phenazines are heterogeneously distributed in P. aeruginosa PA14 biofilms, with most of the phenazines localized to the hypoxic interface in the biofilms. This distribution presumably allows the cells to utilize the phenazines as alternative electron acceptors under hypoxic conditions, increasing the metabolic potential of cells in this zone of the biofilms. Schiessl et al. also demonstrated that phenazines promote the survival of P. aeruginosa in the presence of ciprofloxacin (33). Phenazines are also linked to biofilm formation through c-di-GMP. Okegbe et al. (80) showed that P. aeruginosa PA14 senses the local level of phenazines through a regulatory protein that they labeled RmcA. RmcA contains both Pas domains (which sense redox states) and domains for the production and degradation of c-di-GMP (GGDEF and EAL domains). Interaction with the redox-cycling compound phenazine stimulates the degradation of c-di-GMP, resulting in reduced extracellular polysaccharide production and reduced colony wrinkling. In summary, while the literature supports the potential for phenazine biosynthesis to contribute to biofilm antibiotic tolerance, measurements in our system failed to find either elevated pyocyanin production in the biofilm or a phenotype in the pyocyanin-affected phzS mutant.
TABLE 5.
Summary of hypothesized chemical triggers and of transcriptomic, metabolomic, and mutant susceptibility evidence for stress response or metabolic activities influencing ciprofloxacin tolerance in P. aeruginosa biofilmsa
| Chemical triggerb | Activityc | Gene overexpressiond | Mutant evidence |
|---|---|---|---|
| O2 depletion | O2 downshift | 38/117 (32) | Not tested |
| Hypoxic growth | 43/159 (27) | Not tested | |
| Anr+ regulon | Not significant | Not tested | |
| Denitrification | 7/9 (78) | nirS, norB | |
| Stationary phase | 148/250 (59) | Not tested | |
| Stringent response | Not mapped | ΔrelA ΔspoT | |
| RpoS+ regulon | 108/423 (26) | ΔrpoS | |
| Unknown | PSL biosynthesis | 0/12 (0) | ΔpslA |
| PEL biosynthesis | 0/7 (0) | pelA | |
| Alginate biosynthesis | 0/12 (0) | algD | |
| AHL accumulation | AHL quorum sensing | 25/72 (35) | Not tested |
| Rhamnolipid production | 6.7 FC | rhlB | |
| Lectin production | 29.7, 60.4 FC | lecA, lecB | |
| PQS accumulation | Pyocyanin biosynthesis | 7/17 (41) | phzS |
| MvfR+ regulon | 50/115 (43) | Not tested | |
| Zn depletion | Zn limitation | 26/44 (59) | Not tested |
| Osmotic stress | Osmotic stress response | 12/56 (21) | osmE |
| Unknown | Glyoxylate shunt | 1/2 (50) | aceA |
| Isocitrate lyase | 3.4 FC | aceA | |
| Malate synthase | 1.06 FC | Not tested | |
| Mistranslated proteins? | Membrane stress | 1/14 (7) | amgR |
Light shading indicates logical concordance with the identified chemical trigger, annotated activity, or effect on biofilm antimicrobial susceptibility, whereas dark shading indicates a contradiction of the hypothesis that the upregulated gene affords protection from killing by ciprofloxacin.
AHL, acyl-homoserine lactone; PQS, Pseudomonas quinolone signal.
PSL and PEL, polysaccharide synthesis locus and pellicle polysaccharides, respectively. Plus signs with regulons indicate positive regulation.
Given as the number of overexpressed genes on the current study list/number of genes on the target activity list (percentage of target list genes that are overexpressed in the current study) or as the fold change (FC) in expression. The current study list has a total of 449 genes.
Two loci that exhibited no discernible differential gene expression at the transcriptional level did produce a susceptibility phenotype (ΔpslA and amgR). This outcome could possibly be attributed to posttranscriptional regulation (81, 82). Studies have shown that Psl helps provide the cohesive structure of P. aeruginosa PAO1 biofilms, and a recent study showed that the degradation of Psl with the Psl glycoside hydrolase, PslG, improves antibiotic efficacy in biofilm models (83). Regulation of Psl production is complex and includes both transcriptional and posttranscriptional control mechanisms. The psl operon is induced in part by RpoS (84) and is negatively regulated by the alginate transcriptional activator, AmrZ, which plays a role in coordinating the expression of these two extracellular polysaccharides (85, 86). At the posttranscriptional level, the regulator RsmA binds to the hairpin loop structure of the psl operon 5ʹ untranslated region and represses psl translation (84). Psl may also stimulate its own expression at the posttranscriptional level during biofilm formation by stimulating the diguanylate cyclases SiaD and SadC to produce cyclic di-GMP, resulting in increased Psl production (87). This example illustrates the reality that transcriptional activity may not be a reliable indicator of metabolic activity.
Only one gene locus, rpoS, followed the hypothesized logical progression from higher expression in biofilms to increased susceptibility in a mutant strain. Because the stringent response regulon has not been mapped in P. aeruginosa, we were unable to test for correspondence between gene expression and phenotype for this regulatory mechanism. The phenotype of one other gene, aceA, contradicts logic. This gene was more highly expressed in biofilms (and also in CF sputum [49, 50]), but the biofilms grown from the aceA mutant were found to be less susceptible than those from the wild type.
Overall, these results suggest that only a fraction of the genes expressed at higher levels in a biofilm contribute significantly to protection against antimicrobial agents. This work also reinforces the potential role in antimicrobial tolerance of genes that are not transcriptionally activated in biofilms. Elaboration of Psl was the activity with the strongest phenotype for protection against ciprofloxacin in mutants tested in the current work, yet like others, we found no induction of the psl operon in biofilm cells (18, 19, 88–91).
We do not view these apparent discrepancies as a logical problem at all: they are simply observations of the messy reality. There is no reason to expect that all of the genes differentially expressed in a biofilm would also be involved in antibiotic tolerance. And it is completely reasonable that genes that are not differentially transcribed in a biofilm could contribute to biofilm protection. The scenario that seems likely is one in which overlapping regulons enable multiple stimuli to contribute to the activation of many genes, some likely with redundant functions, creating a robust, difficult-to-defeat defense.
We found evidence for oxygen gradients and induction of responses to hypoxia in the biofilm investigated in this work. Multiple prior studies have found biological evidence for hypoxia (15, 92, 93) and have reported that oxygen limitation reduces the killing of P. aeruginosa by antibiotics (12, 94–97). In agreement with this dependence, the provision of more oxygen to the biofilm through exposure to hyperbaric oxygen has recently been shown to enhance the killing of P. aeruginosa biofilms by ciprofloxacin (98).
Although the phenomenon was not tested experimentally in this study, it has been shown that bacteria in a biofilm can express specific protective systems that augment biofilm tolerance in response to antimicrobial treatment (73, 99). We found that in response to ciprofloxacin treatment, P. aeruginosa biofilms induced oxidative stress and SOS responses (Table 4). Work with Escherichia coli has established a role for the SOS response in enhancing biofilm tolerance to fluoroquinolone antibiotics (100). Likewise, prior work with P. aeruginosa and Streptococcus mutans has demonstrated a contribution of oxidative stress defenses to biofilm antibiotic tolerance in these bacteria (71, 101, 102).
We found that ciprofloxacin-treated biofilms continued to consume all carbon sources at rates similar to those measured for untreated biofilms. This result is seemingly at odds with the 99.9% reduction in viable cell counts measured for antibiotic-treated biofilms relative to untreated biofilms. A possible explanation for this observation is that antibiotic-damaged cells may continue to exhibit anabolic activity for some time, even as long as 24 h, although they are incapable of forming a visible colony on an agar plate. Other researchers have reported continued bacterial respiration following aggressive antimicrobial treatment (103–108).
A corollary of the hypothesis diagrammed in Fig. 1 is that a planktonic cell can acquire the same degree of antibiotic tolerance as a biofilm cell. If a planktonic cell experiences the same chemical microenvironmental history as a cell within a biofilm, it has the chance to enter a protected physiological state just like that of the cell in the biofilm. This conjecture is supported by experimental comparisons of killing of exponential-phase planktonic, stationary-phase planktonic, and biofilm cells that show similar levels of reduced susceptibility for bacteria from stationary-phase cultures and biofilms (27, 109; see also Fig. 12 in reference 110). A caveat to be aware of is that there are surely many possible variants of “stationary phase.” For example, the physiological state of bacteria in an overnight shake flask culture, aerated and grown on a rich medium, is probably significantly different from the oxygen-deprived state in the interior of a biofilm grown on ACWE medium for 3 days.
In summary, the proposed model (Fig. 1) is grounded in physical, chemical, and biological mechanisms and is consistent with a large, diverse body of evidence from the literature and the current work. That said, many details remain unclear. In particular, gene regulatory networks that govern gene expression in biofilms, posttranscriptional regulation, and posttranslational modification and activity are areas where additional research is needed.
MATERIALS AND METHODS
Bacterial strain and culture medium.
P. aeruginosa PAO1 was cultured in biofilms using a medium designed to mimic the exudate from a human chronic wound (55, 111–113). We refer to this medium as artificial chronic wound exudate (ACWE) medium. ACWE medium was composed of a basal salt solution of KCl (4.40 mM), NaCl (99.93 mM), Na2SO4 (300 μM), Na2HPO4 (1.27 mM), urea (9.60 mM), NaNO2 (58 μM), and Tris (413 μM). The salt solution was adjusted to pH 7.5 and was sterilized by autoclaving. Stock solutions of amino acids and carbon sources were filter sterilized using 0.2-μm-pore-size poly(ether sulfone) (PES) bottle-top filters (Fisher Scientific, Waltham, MA). The carbon sources were l-lactate (13.4 mM), glucose (1.8 mM), and amino acids. The concentrations of each amino acid were as follows: l-alanine, 786 μM; l-arginine, 88 μM; l-asparagine, 45 μM; l-aspartate, 451 μM; l-cysteine, 495 μM; l-glutamate, 460 μM; l-glutamine, 589 μM; l-glycine, 641 μM; l-histidine, 167 μM; l-isoleucine, 281 μM; l-leucine, 561 μM; l-lysine, 631 μM; l-methionine, 77 μM; l-phenylalanine, 280 μM; l-proline, 1,720 μM; l-serine, 317 μM; l-threonine, 291 μM; l-valine, 502 μM; l-tryptophan, 376 μM; l-tyrosine, 616 μM. Sterile MgCl2 (940 μM) and CaCl2 (557 μM) were added to the basal salt solution. A vitamin solution and an iron solution were added to give final concentrations of 7.5 μM thiamine, 16 μM nicotinic acid, and 0.36 μM FeSO4·7H2O. When used, ciprofloxacin was added to give a final concentration of 1 mg/liter, approximately 10-fold higher than the MIC of ciprofloxacin against P. aeruginosa growing in planktonic culture.
Determination of MICs.
An aliquot of an overnight culture of P. aeruginosa PAO1 (250 μl) incubated in ACWE medium was inoculated into 25 ml of ACWE medium in a 125-ml baffled Erlenmeyer flask. The culture was grown until it reached an optical density at 600 nm (OD600) of 0.27 (approximately 1.2 × 108 CFU/ml). The culture was then diluted 100-fold in ACWE medium and was used to inoculate a sterile, clear, 96-well flat-bottom microtiter plate (Evergreen Scientific, Los Angeles, CA) containing ACWE medium with ciprofloxacin at concentrations of 1.00, 0.50, 0.25, 0.13, 0.06, 0.03, 0.02, and 0.01 mg/liter, or without ciprofloxacin. Initially, each well contained approximately 1.2 × 105 cells. The microtiter plate was placed in a SpectraMax Paradigm plate reader (Molecular Devices LLC, Sunnyvale, CA) and was incubated at 33°C with shaking, and the OD600 was assayed every 30 min for 20 h. The minimum ciprofloxacin concentration that prevented growth for 16 h was defined as the MIC.
Planktonic culture conditions.
Three independent P. aeruginosa planktonic cultures were prepared by inoculating 1.0 ml of an overnight culture into 100 ml of ACWE medium in 500-ml baffled Erlenmeyer flasks. Cultures were incubated to early-exponential phase (OD, 0.250; 1 × 108 CFU) at 33°C (the approximate surface temperature of a dermal wound). Cultures were harvested by dividing into two 45-ml aliquots in 50-ml Falcon tubes (Corning Inc., Corning, NY), and centrifuging at 5,125 × g for 5 min at 4°C in an Allegra X-15R centrifuge (Beckman Coulter, Brea, CA). The supernatants were decanted. Both the supernatants and the cell pellets were frozen at −80°C for use in metabolomic or transcriptomic studies.
Biofilm growth conditions.
P. aeruginosa biofilms were grown on hydroxyapatite-coated glass slides (Clarkson Chromatography, South Williamsport, PA) in a drip flow reactor (DFR) designed by BioSurface Technologies Corp., Bozeman, MT (114). P. aeruginosa PAO1 was cultured overnight in ACWE medium at 33°C with shaking at 250 rpm. A 1:100 dilution of the overnight culture in ACWE medium was incubated until the culture reached an OD600 of 1.0. The culture was diluted 1:10 with ACWE medium, and 5 ml of this suspension was added to each channel of the DFR. The DFR was incubated for 1 h at 33°C to allow cell attachment. Following the batch phase, a Masterflex L/S peristaltic pump (model 7519-20; Cole-Parmer, Vernon Hills, IL) was started at a flow rate of 10 ml/h, and biofilms were cultivated at 33°C for 72 h (see Fig. S4 in the supplemental material). ACWE medium containing 1 mg/liter ciprofloxacin was then pumped through the ciprofloxacin treatment chambers, while ACWE medium without ciprofloxacin continued to flow through the untreated control chambers for 24 h. After a total of 96 h of growth, the P. aeruginosa PAO1 biofilms were harvested. The susceptibility of biofilms formed by defined P. aeruginosa mutants was measured in the same way using mutant strains obtained from the University of Washington (UW) transposon mutant collection (115) and other sources (see Table S5 in the supplemental material). Transposon insert locations in the UW mutant strains (ISphoA/hah or ISlacZ/hah) were confirmed with a modified dual-round PCR method using previously described primers (116–120). The P. aeruginosa PAO1 ΔrelA ΔspoT mutant, deficient in the stringent response (26), was graciously provided by Pradeep Singh, and the PAO1 ΔpslA mutant (WFPA60), which was impaired in the ability to produce the Psl polysaccharide (121), was kindly provided by Daniel Wozniak. San-Jin Suh supplied the ΔrpoS mutant (122). Mutant selection was made on the basis of experimentally observed transcriptional activation of related genes or pathways and was also guided by prior reports (21, 26, 123–131). If the first experiment challenging a mutant biofilm with ciprofloxacin resulted in an apparent difference from wild-type susceptibility, the experiment was repeated a total of three times.
Each slide was removed from its chamber using sterile forceps and was placed in a 50-ml Falcon tube (Corning Inc., Corning, NY) containing 10 ml phosphate-buffered saline (PBS). Slides were scraped using sterile 18-cm GeneMate cell lifters (BioExpress, Kaysville, UT). The cell suspension was homogenized by vortexing and passing slowly twice through an 18-gauge, 1.5-in-long PrecisionGlide needle and a 20-ml syringe (Becton Dickinson and Co., Franklin Lakes, NJ). Aliquots (1 ml) were collected and were centrifuged at 10,000 × g for 1 min. The supernatant was removed by pipetting and discarded. Cell pellets were placed in a −80°C freezer.
Quantification of protein, carbohydrates, DNA, and thickness in biofilms.
Total protein in biofilm samples was quantified by the Bradford assay. Cell pellets were resuspended in RLT lysis buffer (Qiagen) and were heated at 65°C until complete lysis. Ten microliters of the lysed cell suspension was added to 90 μl of 0.85% NaCl, and 1 ml of Bradford reagent was added. Absorbance was measured at 595 nm. A standard curve was constructed using bovine serum albumin (BSA).
Total carbohydrates in biofilm samples were quantified by a phenol-sulfuric acid assay. Standards were made by diluting an 80-ng/ml stock solution of d-(+)-galactose (Sigma-Aldrich, St. Louis, MO). Samples were resuspended in 450 μl H2O, and 150 μl was transferred to new glass test tubes. Concentrated phenol was diluted 1:13 in H2O to yield a 5% (wt/vol) phenol solution. To each tube, 150 μl of the 5% (wt/vol) phenol solution was added. The tubes were vortexed. To each tube, 750 μl of concentrated sulfuric acid was added rapidly. The tubes were incubated for 10 min at room temperature before vortexing. Then the tubes were incubated for 30 min to develop color. Absorbance was measured at 480 nm and 490 nm.
DNA was isolated from biofilm samples using the QIAamp Miniprep kit (Qiagen), with the following modifications. Samples were resuspended in 180 μl Buffer ATL (Qiagen) by passing the solution three times through a 23-gauge, 1-in-long PrecisionGlide needle. A 20-μl volume of proteinase K was added, and samples were then incubated at 56°C for 1.5 h with periodic vortexing. DNA extraction then proceeded according to the QIAamp protocol, with the use of RNase A to obtain RNA-free genomic DNA. Total DNA was eluted with 200 μl water and was quantified on a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Biofilm thickness was measured by microscopy and image analysis of frozen cross sections of cryoembedded specimens (132).
Preparation of RNA.
Each cell pellet was resuspended in 100 μl of lysozyme–Tris-EDTA (TE) (5 mg/ml; pH 8.0). After incubation for 3 min at room temperature, 650 μl of Tri reagent (Zymo Research, Irvine, CA) was added. The sample was vortexed for 20 s and was then allowed to incubate at room temperature for 5 min. The lysed pellets were then placed in a Sorvall Legend Micro 21R tabletop microcentrifuge (Thermo Scientific, Waltham, MA) and were centrifuged at 13,000 × g for 4 min to pellet cell debris. Each supernatant, approximately 750 μl, was added to 750 μl of molecular-biology-grade pure ethanol (Sigma-Aldrich, St. Louis, MO). The mixtures were first vortexed and then processed according to the Direct-zol RNA MiniPrep RNA purification protocol. Total RNA was quantified on the NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and the quality was verified on a Bioanalyzer 2100 nanochip (Agilent, Santa Clara, CA). Purified RNA was diluted to a concentration of 500 ng/μl for Turbo DNase treatment. Treatment was performed in the presence of 1 μl of the RNase inhibitor RNasin (Promega, Madison, WI), according to a rigorous DNase treatment protocol (Ambion, Thermo Fisher Scientific, Waltham, MA).
Microarray analysis.
Samples were prepared for microarrays as described previously (13). Briefly, 8 μg of Turbo DNase-treated RNA was reverse transcribed, fragmented, and labeled according to the Affymetrix prokaryotic target labeling protocol (GeneChip expression analysis technical manual, November 2004). Labeled cDNA was then hybridized to Affymetrix P. aeruginosa microarrays (part 900339) for 16 h at 50°C with constant rotation. Microarrays were stained using a GCOS Fluidics Station 450 and were scanned with an Affymetrix 7G scanner. Affymetrix GCOS, v1.4, was used to generate CEL files, which were imported into FlexArray, v1.6.1, for quality control and data analysis.
Transcriptomic data analysis.
Microarray data from three independent biological replicates for each condition (planktonic, biofilm, and treated biofilm) were background corrected and normalized using the GC-RMA algorithm in FlexArray, v1.6.1. Genes with statistically significantly changes in expression (2-fold change at a P value of <0.05) as determined by analysis of variance (ANOVA) were uploaded into the Database for Annotation, Visualization and Integrated Discovery (DAVID), v6.8. The Functional Annotation Clustering Tool was used to find enriched terms so as to identify the most pertinent biological processes in the study. The most significant gene ontology term in each cluster was selected as the category identifier for use in Table 1. Each P value listed in Table 1 is the geometric mean of all the enrichment P values (Expression Analysis Systematic Explorer [EASE] scores) of each annotation term in the group. The EASE score was determined in DAVID with a modified Fisher exact test (log-transformed average P value). The formula P = 10–EASE was used to determine the P values in Table 1 (133). Gene set enrichment analysis was performed by determining the overlap between the gene lists from this study and activity gene lists compiled from the literature as described elsewhere (17) with reference to the Pseudomonas Genome Database (134). P values for assessing the statistical significance of gene set enrichment were calculated using a negative binomial distribution.
Analysis of intracellular metabolites by liquid chromatography-mass spectrometry (LC-MS).
Intracellular metabolite samples were prepared by thawing bacterial cell pellets on ice. The pellets were suspended in 300 μl H2O (Fisher Scientific, Waltham, MA) and were transferred to a 2-ml Lysing Matrix B tube (MP Biomedicals LLC, Santa Ana, CA). Lysing Matrix B contained 0.1-mm silica beads. The tubes were placed in a FastPrep FP120 reciprocating device (Thermo Electron Corporation, Thermo Fisher Scientific, Waltham, MA). The FastPrep system was run for 20 s at 6 m/s, and then the samples were placed on ice for 1 min. This procedure was repeated two additional times. To each sample, 300 μl of ice-cold methanol (Fisher Scientific, Waltham, MA) was added, and the mixture was allowed to incubate on ice for 3 min. The bead-beating procedure was followed as before. The lysing matrix was allowed to settle, and the top 300 μl of sample was added to 1.4 ml cold acetone (Fisher Scientific, Waltham, MA). Samples were placed in a –80°C freezer overnight to precipitate cell debris and protein, followed by centrifugation at 20,000 × g for 5 min at 4°C. The supernatant was collected and placed in a fresh tube. The precipitate was stored in a –80°C freezer. The supernatants were placed in a SpeedVac Plus SC110A evaporator (Thermo Fisher Scientific, Waltham, MA) and were dried down under a vacuum on the medium-speed setting. The samples were resuspended in 25 μl of H2O-methanol (50:50).
LC-MS was used to analyze intracellular metabolites. For reverse-phase analysis, a Kinetex C18 column (particle size, 1.7 μm; length, 150 mm; inside diameter [i.d.], 2.1 mm; Phenomenex, Torrance, CA) was kept at 50°C with a flow rate of 600 μl min−1. Solvent A consisted of 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile (ACN). The elution gradient was initially 2% solvent B for 2 min and was then ramped to 95% solvent B over 24 min, held at 95% for 2 min, and then returned to 2% for 2 min, with a total run time of 30 min, using an Agilent 1290 Infinity ultrahigh-performance liquid chromatography (UHPLC) system connected to an Agilent 6538 quadrupole time of flight (Q-TOF) mass spectrometer (both from Agilent, Santa Clara, CA).
Normal-phase analysis was carried out using a Cogent Diamond Hydride hydrophilic interaction chromatography (HILIC) column (length, 150 mm; i.d., 2.1 mm; MicroSolv, Eatontown, NJ) for LC separation with a flow rate of 600 μl min−1. Solvent A consisted of 0.1% formic acid in water, while solvent B consisted of 0.1% formic acid in ACN. The elution gradient consisted of 95% solvent B for 2 min and was then ramped to 50% solvent B over 24 min, held at 50% for 2 min, and then returned to 95% for 2 min, with a total run time of 30 min, using an Agilent 1290 Infinity UHPLC system connected to an Agilent 6538 Q-TOF mass spectrometer (both from Agilent, Santa Clara, CA).
The mass spectrometer was operated in positive-ion mode for both reverse-phase and normal-phase analysis, with a cone voltage of 3,500 V and a fragmentor voltage of 120 V. The drying gas temperature was 350°C with a flow rate of 12 liters min−1, and the nebulizer was set to 60 lb/in2. Spectra were collected at a rate of 2.52/s with a mass range of 50 to 1,000 m/z. The mass analyzer resolution was greater than 18,000, and the pre- and postcalibration tests had a mass accuracy of approximately 1 ppm.
Raw data files from the LC-MS were converted to .MZxml format using the MassHunter qualitative software provided with Agilent instruments (Agilent, Santa Clara, CA). Alignment, retention time (R/T) correction, peak picking, and identification were performed using the MZmine software package (v2.14). The procedures, together with the parameters used for the alignment of features and identification in MZmine, were as follows. LC-MS files were imported into MZmine, followed by data set filtering to remove the first minute of elution data for HILIC analysis and the first 2 min of elution data for reverse-phase analysis. A minimum intensity cutoff of 5,000 and a minimum elution time window of 0.1 min were used to create molecular feature lists. Additionally, lists were R/T adjusted with a tolerance of 0.2 min or less. These R/T-adjusted lists were then aligned into one mass list, which was gap-filled to add missing peaks not detected in all runs with an m/z tolerance of 15.0 ppm. The m/z peak intensities were normalized, adjusting the total ion intensity of each run to the most intense total ion intensity. Significance was determined using a two-tailed t test. Metabolites were putatively identified by generating a list of curated metabolites found in the BioCyc server for PAO1. Monoisotopic masses for each metabolite were calculated, and protonated (M + H), deprotonated (M – H), and sodiated (M + Na) adducts were added to each metabolite mass; these were then matched to features in the peak list using MZmine, with the error level set to 15 ppm.
Analysis of extracellular metabolites by gas chromatography-mass spectrometry (GC-MS).
Extracellular metabolite samples from biofilms were collected from DFR effluent lines into 15-ml Falcon tubes (Corning Inc., Corning, NY). Collection lines were unclamped and were allowed to drain for approximately 1 h. Samples were capped and placed in a –80°C freezer. After biofilms were harvested, stock medium and ciprofloxacin-containing medium were collected in 50-ml Falcon tubes and frozen at −80°C. Extracellular metabolites from planktonic cultures were collected by decanting and freezing the supernatant from the pelleting process. Extracellular metabolite samples were thawed, and 1-ml aliquots were dried under a nitrogen stream. Prior to analysis, samples were derivatized using the method described below, which enabled the consistent detection of 17 amino acids and 3 central carbon metabolites (135). Two additional central carbon metabolites, formate and acetate, were detected as underivatized compounds in aqueous solution, using a ZB-WAX column.
N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) was obtained from Sigma (CAS no. 77377-52-7; Sigma-Aldrich, Switzerland). ACN solvent was obtained from Fisher (CAS no. 75-05-8; Fisher Scientific, USA). Hexane solvent was obtained from Fisher (CAS no. 110-54-3; Fisher Scientific, USA). Formic acid and acetic acid were obtained from Fisher. The remaining central carbon metabolites were obtained from Sigma. All amino acids were purchased from Sigma.
Samples were analyzed using an Agilent 7890A GC oven system, coupled to an Agilent 5975C inert XL EI/CI MSD (electron ionization/chemical ionization mass selective detector) mass spectrometer with a Triple-Axis Detector. Sample organization and injection were performed with an Agilent 7693 autosampler.
For the analysis of amino acids and central carbon metabolites, aqueous solutions previously dried using nitrogen were resuspended in ACN and derivatized using MTBSTFA. Equal volumes of MTBSTFA and ACN (100 μl) were added to each sample in a glass GC vial, which was capped, vortexed briefly, and then incubated on a hot plate at 50°C for 30 min. Derivatized samples were transferred to new GC vials containing glass inserts for analysis. Analytes were separated with a Phenomenex Zebron ZB-5MS nonpolar column (length, 30 m; i.d., 0.25 mm; particle size, 0.25 μm) The GC temperature gradient for the MTBSTFA-derivatized samples was as follows: 60°C for 2 min, ramping to 120°C at a rate of 20°C/min (3-min ramp time), then to 155°C at a rate of 6°C/min (5.83-min ramp time), and finally at a rate of 14.5°C/min (10-min ramp time) to 300°C, where the temperature was held for 10 min. The total run time was 30.833 min. Helium gas was used as the carrier at a flow rate of 1.5 ml/min. Injections were performed at a volume of 1 μl, with a split ratio of 10:1 and an inlet temperature of 325°C. The interface temperature between the GC and the MS was set at 230°C. The volatile central carbon metabolites, formate and acetate, were analyzed underivatized in aqueous solution. Analysis was performed using a Phenomenex Zebron ZB-WAX column (length, 30 m; i.d., 0.25 mm; particle size, 0.50 μm). The oven temperature started at 75°C and was held for 1 min. It was then ramped at a rate of 6°C/min to 180°C (17.5-min ramp time) and next at 10°C/min to 230°C (5-min ramp time), where it was held for 5 min. The total run time was 28.5 min. Helium gas was used as a carrier at a flow rate of 3 ml/min. Injections were performed at a volume of 0.5 μl, splitless, with the inlet temperature set at 240°C and the interface temperature set at 280°C.
Glucose, urea, and ammonia levels were determined using enzymatic kits (product codes GAGO-20, KA-1652, and AA0100; Sigma, St. Louis, MO).
Calculation of biofilm specific growth rates by material balance.
Elemental balances on C, N, H, and O were derived to estimate the rate of biomass production and subsequently the average specific growth rate in the biofilm. The overall reaction analyzed had the following form:
where substrates included lactate, glucose, and 17 amino acids; acetate was the sole product. Experimental measurements of the concentrations of these solutes were used to determine net elemental input. In this reaction, C5H7O2N was the assumed composition of cellular biomass, and EPS refers to the extracellular polymeric substances, which were assumed to have the composition C6H12O6. There were six unknowns (a to f) and four elemental balances, so two additional equations were required to solve this system. The measured ratio of protein (0.362 mg/cm2) to carbohydrate (0.184 mg/cm2) in P. aeruginosa biofilms (Table S3 in the supplemental material) was used to estimate the ratio d/c. Assuming that biomass is 50% protein, d/c = 0.1595. The final piece of information was the production of ammonia, which was measured experimentally (Table S2). When this system of linear algebraic equations is solved, the biomass stoichiometric coefficient c has units of biomass concentration. Multiplying by the flow rate and dividing by the standing biomass in the biofilm gives a growth rate. Specifically, growth rate = [(c mmol/liter biomass) (113 g biomass/mol) (0.5 g protein/g biomass) (0.01 liter/h)]/[(0.362 mg protein/cm2) (18.75 cm2)]. The growth rate thus calculated is expressed per hour.
Oxygen requirement to synthesize Psl from different carbon sources.
Flux balance analysis (136) and a P. aeruginosa metabolic network model (137) (see the supplemental material) were used to compute the oxygen requirement to synthesize Psl using a single amino acid, lactate, or glucose as the sole carbon source. First, the maximum Psl yield from each carbon source was computed. Then the minimum oxygen consumption required to achieve the maximum Psl yield was computed for each carbon source.
Mathematical modeling of oxygen concentration gradients.
Reaction-diffusion modeling was performed with the biofilm accumulation model described previously (138, 139) and was based on the structure derived by Wanner and Gujer (140). Parameter values are given in Table S1 in the supplemental material.
Accession number(s).
The data derived from microarray analyses have been deposited in the Gene Expression Omnibus database with accession number GSE120760.
Supplementary Material
ACKNOWLEDGMENTS
This work was sponsored by the Military Operational Medicine Program Area Directorate of the U.S. Army Medical Research and Materiel Command, Fort Detrick, MD, and by the U.S. Department of Defense (DoD) Medical Research and Development Program. Funding for the Proteomics, Metabolomics and Mass Spectrometry Facility used for this publication was made possible in part by the MJ Murdock Charitable Trust and the National Institute of General Medical Sciences of the National Institutes of Health under award P20GM103474.
This was not an industry-supported study. We declare that we have no financial conflicts of interest. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or of the U.S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00307-19.
REFERENCES
- 1.Suci PA, Mittelman MW, Yu FP, Geesey GG. 1994. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38:2125–2133. doi: 10.1128/aac.38.9.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shigeta M, Tanaka G, Komatsuzawa H, Sugai M, Suginaka H, Usui T. 1997. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy 43:340–345. doi: 10.1159/000239587. [DOI] [PubMed] [Google Scholar]
- 3.Vrany JD, Stewart PS, Suci PA. 1997. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa biofilms displaying rapid-transport characteristics. Antimicrob Agents Chemother 41:1352–1358. doi: 10.1128/AAC.41.6.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters MC III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/aac.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol 15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart PS. 2003. Diffusion in biofilms. J Bacteriol 185:1485–1491. doi: 10.1128/jb.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 8.Morgan SJ, Lippman SI, Bautista GE, Harrison JJ, Harding CL, Gallagher LA, Cheng AC, Siehnel R, Ravishankar S, Usui ML, Olerud JE, Fleckman P, Wolcott RD, Manoil C, Singh PK. 2019. Bacterial fitness in chronic wounds appears to be mediated by the capacity for high-density growth, not virulence or biofilm functions. PLoS Pathog 15:e1007511. doi: 10.1371/journal.ppat.1007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szomolay B, Klapper I, Dockery J, Stewart PS. 2005. Adaptive responses to antimicrobial agents in biofilms. Environ Microbiol 7:1186–1191. doi: 10.1111/j.1462-2920.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- 10.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE. 2013. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Xu KD, Stewart PS, Xia F, Huang C-T, McFeters GA. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol 64:4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, Pitts B, Stewart PS. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James GA, Ge Zhao A, Usui M, Underwood RA, Nguyen H, Beyenal H, deLancey Pulcini E, Agostinho Hunt A, Bernstein HC, Fleckman P, Olerud J, Williamson KS, Franklin MJ, Stewart PS. 2016. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen 24:373–383. doi: 10.1111/wrr.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jo J, Cortez KL, Cornell WC, Price-Whelan A, Dietrich LE. 2017. An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6:e30205. doi: 10.7554/eLife.30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koley D, Ramsey MM, Bard AJ, Whiteley M. 2011. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc Natl Acad Sci U S A 108:19996–20001. doi: 10.1073/pnas.1117298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart PS, Franklin MJ, Williamson KS, Folsom JP, Boegli L, James GA. 2015. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 59:3838–3847. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentzer M, Eberl L, Givskov M. 2005. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2:37–62. doi: 10.1017/S1479050505001699. [DOI] [Google Scholar]
- 19.Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, Mazurie A, Stewart PS. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol 10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brochmann RP, Toft A, Ciofu O, Briales A, Kolpen M, Hempel C, Bjarnsholt T, Høiby N, Jensen PØ. 2014. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int J Antimicrob Agents 43:140–147. doi: 10.1016/j.ijantimicag.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Marques CNH, Nelson SM. 2019. Pharmacodynamics of ciprofloxacin against Pseudomonas aeruginosa planktonic and biofilm-derived cells. Lett Appl Microbiol 68:350–359. doi: 10.1111/lam.13126. [DOI] [PubMed] [Google Scholar]
- 24.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Mah T-F. 2008. Involvement of a novel efflux system in biofilm specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao J, Sauer K. 2012. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol 194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Chiang W-C, Gao Q, Givskov M, Tolker-Nielsen T, Yang L, Zhang G. 2012. The catabolite repression control protein Crc plays a role in the development of antimicrobial-tolerant subpopulations in Pseudomonas aeruginosa biofilms. Microbiology 158:3014–3019. doi: 10.1099/mic.0.061192-0. [DOI] [PubMed] [Google Scholar]
- 29.Liao J, Schurr MJ, Sauer K. 2013. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol 195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poudyal B, Sauer K. 2017. The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother 62:e01981-17. doi: 10.1128/AAC.01981-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poudyal B, Sauer K. 2018. The PA3177 gene encodes an active diguanylate cyclase that contributes to biofilm antimicrobial tolerance but not biofilm formation by Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e01049-18. doi: 10.1128/AAC.01049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dingemans J, Poudyal B, Sondermann H, Sauer K. 2018. The yin and yang of SagS: distinct residues in the HmsP domain of SagS independently regulate biofilm formation and biofilm drug tolerance. mSphere 3:e00192-18. doi: 10.1128/mSphere.00192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiessl KT, Hu F, Jo J, Nazia SZ, Wang B, Price-Whelan A, Min W, Dietrich L. 2019. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat Commun 10:762. doi: 10.1038/s41467-019-08733-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciofu O, Tolker-Nielsen T. 2019. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents—how P. aeruginosa can escape antibiotics. Front Microbiol 10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart PS, Parker AE. 2019. Measuring antimicrobial efficacy against biofilms: a meta-analysis. Antimicrob Agents Chemother 63:e00020-19. doi: 10.1128/AAC.00020-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maslowska KH, Makiela-Dzbenska K, Fijalkowska IJ. 2019. The SOS system: a complex and tightly regulated response to DNA damage. Environ Mol Mutagen 60:368–384. doi: 10.1002/em.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. 2010. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ Microbiol 12:1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen K, Lewandowski Z. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng 59:302–309. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol 171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 41.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Döring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. 2015. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio 6:e00767. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williamson K, McCarty PL. 1976. A model of substrate utilization by bacterial films. J Water Pollut Control Fed 48:9–24. [PubMed] [Google Scholar]
- 44.Stewart PS, Zhang T, Xu R, Pitts B, Walters MC III, Roe F, Kikhney J, Moter A. 2016. Reaction-diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. NPJ Biofilms Microbiomes 2:16012. doi: 10.1038/npjbiofilms.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T, Pabst B, Klapper I, Stewart PS. 2013. General theory for integrated analysis of growth, gene, and protein expression in biofilms. PLoS One 8:e83626. doi: 10.1371/journal.pone.0083626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trunk K, Benkert B, Quäck N, Münch R, Scheer M, Garbe J, Jänsch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 47.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pederick VG, Eijkelkamp BA, Begg SL, Ween MP, McAllister LJ, Paton JC, McDevitt CA. 2015. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci Rep 5:13139. doi: 10.1038/srep13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi E, Falcone M, Molin S, Johansen HK. 2018. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun 9:3459. doi: 10.1038/s41467-018-05944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Siehnel RJ, Garudathri J, Staudinger BJ, Hisert KB, Ozer EA, Hauser AR, Eng JK, Manoil C, Singh PK, Bruce JE. 2019. In vivo proteome of Pseudomonas aeruginosa in airways of cystic fibrosis patients. J Proteome Res 18:2601–2612. doi: 10.1021/acs.jproteome.9b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tata M, Wolfinger MT, Amman F, Roschanski N, Dötsch A, Sonnleitner E, Häussler S, Bläsi U. 2016. RNASeq based transcriptional profiling of Pseudomonas aeruginosa PA14 after short- and long-term anoxic cultivation in synthetic cystic fibrosis sputum medium. PLoS One 11:e0147811. doi: 10.1371/journal.pone.0147811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamath KS, Krisp C, Chick J, Pascovici D, Gygi SP, Molloy MP. 2017. Pseudomonas aeruginosa proteome under hypoxic stress conditions mimicking the cystic fibrosis lung. J Proteome Res 16:3917–3928. doi: 10.1021/acs.jproteome.7b00561. [DOI] [PubMed] [Google Scholar]
- 53.Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2012. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol 86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiber K, Boes N, Eschbach M, Jaensch L, Wehland J, Bjarnsholt T, Givskov M, Hentzer M, Schobert M. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J Bacteriol 188:659–668. doi: 10.1128/JB.188.2.659-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debats I, Booi D, Deutz NEP, Buurman WA, Boeckx WD, van der Hulst R. 2006. Infected chronic wounds show different local and systemic arginine conversion compared with acute wounds. J Surg Res 134:205–214. doi: 10.1016/j.jss.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Grasemann H, Ioannidis I, Tomkiewicz RP, de Groot H, Rubin BK, Ratjen F. 1998. Nitric oxide metabolites in cystic fibrosis lung disease. Arch Dis Child 78:49–53. doi: 10.1136/adc.78.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer KL, Brown SA, Whiteley M. 2007. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J Bacteriol 189:4449–4455. doi: 10.1128/JB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolpen M, Bjarnsholt T, Moser C, Hansen CR, Rickelt LF, Kühl M, Hempel C, Pressler T, Høiby N, Jensen PØ. 2014. Nitric oxide production by polymorphonuclear leucocytes in infected cystic fibrosis sputum consumes oxygen. Clin Exp Immunol 177:310–319. doi: 10.1111/cei.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, Burns JL, Stahl DA, Hassett DJ, Fang FC, Miller SI. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Line L, Alhede M, Kolpen M, Kühl M, Ciofu O, Bjarnsholt T, Moser C, Toyofuku M, Nomura N, Høiby N, Jensen PØ. 2014. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front Microbiol 5:554. doi: 10.3389/fmicb.2014.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol 300:549–556. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Goodwine J, Gil J, Doiron A, Valdes J, Solis M, Higa A, Davis S, Sauer K. 2019. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci Rep 9:3763. doi: 10.1038/s41598-019-40378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basta DW, Bergkessel M, Newman DK. 2017. Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. MBio 8:e01170-17. doi: 10.1128/mBio.01170-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain V, Kumar M, Chatterji D. 2006. ppGpp: stringent response and survival. J Microbiol 44:1–10. [PubMed] [Google Scholar]
- 66.Lequette Y, Lee J-H, Ledgham F, Lazdunski A, Greenberg EP. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol 188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nalca Y, Jansch L, Bredenbruch F, Geffers R, Buer J, Häussler S. 2006. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Chemother 50:1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu KD, Franklin MJ, Park CH, McFeters GA, Stewart PS. 2001. Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously-fed Pseudomonas aeruginosa biofilms. FEMS Microbiol Lett 199:67–71. doi: 10.1111/j.1574-6968.2001.tb10652.x. [DOI] [PubMed] [Google Scholar]
- 70.Babin BM, Atangcho L, van Eldijk MB, Sweredoski MJ, Moradian A, Hess S, Tolker-Nielsen T, Newman DK, Tirrell DA. 2017. Selective proteomic analysis of antibiotic-tolerant cellular subpopulations in Pseudomonas aeruginosa biofilms. MBio 8:e01593-17. doi: 10.1128/mBio.01593-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kragh KN, Alhede M, Jensen PØ, Moser C, Scheike T, Jacobsen CS, Seier Poulsen S, Eickhardt-Sørensen SR, Trøstrup H, Christoffersen L, Hougen HP, Rickelt LF, Kühl M, Høiby N, Bjarnsholt T. 2014. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun 82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. 2012. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol 194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 76.Chiang WC, Pamp SJ, Nilsson M, Givskov M, Tolker-Nielsen T. 2012. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol Med Microbiol 65:245–256. doi: 10.1111/j.1574-695X.2012.00929.x. [DOI] [PubMed] [Google Scholar]
- 77.Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price-Whelan A, Dietrich LE, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol 189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Kern SE, Newman DK. 2010. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol 192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, Price-Whelan A, Stewart RC, Lee VT, Dietrich L. 2017. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci U S A 114:E5236–E5245. doi: 10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. 2012. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol 14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Babin BM, Bergkessel M, Sweredoski MJ, Moradian A, Hess S, Newman DK, Tirrell DA. 2016. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E597–E605. doi: 10.1073/pnas.1514412113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pestrak MJ, Baker P, Dellos-Nolan S, Hill PJ, Passos da Silva D, Silver H, Lacdao I, Raju D, Parsek MR, Wozniak DJ, Howell PL. 2019. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob Agents Chemother 63:e00234-19. doi: 10.1128/AAC.00234-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol 195:1637–1644. doi: 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu B, Soukup RJ, Jones CJ, Fishel R, Wozniak DJ. 2016. Pseudomonas aeruginosa AmrZ binds to four sites in the algD promoter, inducing DNA-AmrZ complex formation and transcriptional activation. J Bacteriol 198:2673–2681. doi: 10.1128/JB.00259-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mikkelsen H, Bond NJ, Skindersoe ME, Givskov M, Lilley KS, Welch M. 2009. Biofilms and type III secretion are not mutually exclusive in Pseudomonas aeruginosa. Microbiology 155:687–698. doi: 10.1099/mic.0.025551-0. [DOI] [PubMed] [Google Scholar]
- 89.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, Curtis MA. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 91.Bielecki P, Puchałka J, Wos-Oxley ML, Loessner H, Glik J, Kawecki M, Nowak M, Tümmler B, Weiss S, dos Santos VAPM. 2011. In-vivo expression profiling of Pseudomonas aeruginosa infections reveals niche-specific and strain-independent transcriptional programs. PLoS One 6:e24235. doi: 10.1371/journal.pone.0024235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich LE. 2014. Morphological optimization for access to dual oxidants in biofilms. Proc Natl Acad Sci U S A 111:208–213. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin YC, Cornell WC, Jo J, Price-Whelan A, Dietrich L. 2018. The Pseudomonas aeruginosa complement of lactate dehydrogenases enables use of d- and l-lactate and metabolic cross-feeding. MBio 9:e00961-18. doi: 10.1128/mBio.00961-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Field TR, White A, Elborn JS, Tunney MM. 2005. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur J Clin Microbiol Infect Dis 24:677–687. doi: 10.1007/s10096-005-0031-9. [DOI] [PubMed] [Google Scholar]
- 95.King P, Citron DM, Griffith DC, Lomovskaya O, Dudley MN. 2010. Effect of oxygen limitation on the in vitro activity of levofloxacin and other antibiotics administered by the aerosol route against Pseudomonas aeruginosa from cystic fibrosis patients. Diagn Microbiol Infect Dis 66:181–186. doi: 10.1016/j.diagmicrobio.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 96.Schaible B, Taylor CT, Schaffer K. 2012. Hypoxia increases antibiotic resistance in Pseudomonas aeruginosa through altering the composition of multidrug efflux pumps. Antimicrob Agents Chemother 56:2114–2118. doi: 10.1128/AAC.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gupta S, Laskar N, Kadouri DE. 2016. Evaluating the effect of oxygen concentrations on antibiotic sensitivity, growth, and biofilm formation of human pathogens. Microbiol Insights 9:37–46. doi: 10.4137/MBI.S40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kolpen M, Lerche CJ, Kragh KN, Sams T, Koren K, Jensen AS, Line L, Bjarnsholt T, Ciofu O, Moser C, Kühl M, Høiby N, Jensen PØ. 2017. Hyperbaric oxygen sensitizes anoxic Pseudomonas aeruginosa biofilm to ciprofloxacin. Antimicrob Agents Chemother 61:e01024-17. doi: 10.1128/AAC.01024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol 65:4594–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppée JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jensen PØ, Briales A, Brochmann RP, Wang H, Kragh KN, Kolpen M, Hempel C, Bjarnsholt T, Høiby N, Ciofu O. 2014. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog Dis 70:440–443. doi: 10.1111/2049-632X.12120. [DOI] [PubMed] [Google Scholar]
- 102.Nilsson M, Jakobsen TH, Givskov M, Twetman S, Tolker-Nielsen T. 2019. Oxidative stress response plays a role in antibiotic tolerance of Streptococcus mutans biofilms. Microbiology 165:334–342. doi: 10.1099/mic.0.000773. [DOI] [PubMed] [Google Scholar]
- 103.Stewart PS, Griebe T, Srinivasan R, Chen C-I, Yu FP, deBeer D, McFeters GA. 1994. Comparison of respiratory activity and culturability during monochloramine disinfection of binary population biofilms. Appl Environ Microbiol 60:1690–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.von Ohle C, Gieseke A, Nistico L, Decker EM, DeBeer D, Stoodley P. 2010. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl Environ Microbiol 76:2326–2334. doi: 10.1128/AEM.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiamco MM, Atci E, Mohamed A, Call DR, Beyenal H. 2017. Hyperosmotic agents and antibiotics affect dissolved oxygen and pH concentration gradients in Staphylococcus aureus biofilms. Appl Environ Microbiol 83:e02783-16. doi: 10.1128/AEM.02783-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kiamco MM, Mohamed A, Reardon PN, Marean-Reardon CL, Aframehr WM, Call DR, Beyenal H, Renslow RS. 2018. Structural and metabolic responses of Staphylococcus aureus biofilms to hyperosmotic and antibiotic stress. Biotechnol Bioeng 115:1594–1603. doi: 10.1002/bit.26572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin J, Wang Z, Zang Y, Zhang D, Xin Q. 2019. Detection of respiration changes inside biofilms with microelectrodes during exposure to antibiotics. J Environ Sci Health A Tox Hazard Subst Environ Eng 54:202–207. doi: 10.1080/10934529.2018.1544782. [DOI] [PubMed] [Google Scholar]
- 108.Simkins JW, Stewart PS, Codd SL, Seymour JD. 2019. Non-invasive imaging of oxygen concentration in a complex in vitro biofilm infection model using 19F MRI: persistence of an oxygen sink despite prolonged antibiotic therapy. Magn Reson Med 82:2248–2256. doi: 10.1002/mrm.27888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trengove NJ, Langton SR, Stacey MC. 1996. Biochemical analysis of wound fluid from nonhealing and healing chronic leg ulcers. Wound Repair Regen 4:234–239. doi: 10.1046/j.1524-475X.1996.40211.x. [DOI] [PubMed] [Google Scholar]
- 112.James TJ, Hughes MA, Cherry GW, Taylor RP. 2000. Simple biochemical markers to assess chronic wounds. Wound Repair Regen 8:264–269. doi: 10.1046/j.1524-475x.2000.00264.x. [DOI] [PubMed] [Google Scholar]
- 113.James TJ, Hughes MA, Cherry GW, Taylor RP. 2003. Evidence of oxidative stress in chronic venous ulcers. Wound Repair Regen 11:172–176. doi: 10.1046/j.1524-475X.2003.11304.x. [DOI] [PubMed] [Google Scholar]
- 114.Goeres DM, Hamilton MA, Beck NA, Buckingham-Meyer K, Hilyard JD, Loetterle LR, Lorenz LA, Walker DK, Stewart PS. 2009. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc 4:783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 115.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chun KT, Edenberg HJ, Kelley MR, Goebl MG. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233–240. doi:. [DOI] [PubMed] [Google Scholar]
- 117.Kulasekara HD. 2014. Transposon mutagenesis. Methods Mol Biol 1149:501–519. doi: 10.1007/978-1-4939-0473-0_39. [DOI] [PubMed] [Google Scholar]
- 118.Liberati NT, Urbach JM, Thurber TK, Wu G, Ausubel FM. 2008. Comparing insertion libraries in two Pseudomonas aeruginosa strains to assess gene essentiality. Methods Mol Biol 416:153–169. doi: 10.1007/978-1-59745-321-9_10. [DOI] [PubMed] [Google Scholar]
- 119.O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to study of biofilms. Methods Enzymol 310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 120.O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 121.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suh S-J, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol 181:3890–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahn S, Jung J, Jang IA, Madsen EL, Park W. 2016. Role of glyoxylate shunt in oxidative stress response. J Biol Chem 291:11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. 2017. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo Q, Wei Y, Xia B, Jin Y, Liu C, Pan X, Shi J, Zhu F, Li J, Qian L, Liu X, Cheng Z, Jin S, Lin J, Wu W. 2016. Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci Rep 11:19141. doi: 10.1038/srep19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol 92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/jb.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, Singh PK, Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci U S A 106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Murakami K, Ono T, Viducic D, Somiya Y, Kariyama R, Hori K, Amoh T, Hirota K, Kumon H, Parsek MR, Miyake Y. 2017. Role of psl genes in antibiotic tolerance of adherent Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e02587-16. doi: 10.1128/AAC.02587-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goltermann L, Tolker-Nielsen T. 2017. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob Agents Chemother 61:e02696-16. doi: 10.1128/AAC.02696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Murga R, Stewart PS, Daly D. 1995. Quantitative analysis of biofilm thickness variability. Biotechnol Bioeng 45:503–510. doi: 10.1002/bit.260450607. [DOI] [PubMed] [Google Scholar]
- 133.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 134.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas Genome Database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hamerly T, Tripet BP, Tigges M, Giannone RJ, Wurch L, Hettich RL, Podar M, Copié V, Bothner B. 2015. Untargeted metabolomics studies employing NMR and LC–MS reveal metabolic coupling between Nanoarcheum equitans and its archaeal host Ignicoccus hospitalis. Metabolomics 11:895–907. doi: 10.1007/s11306-014-0747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orth JD, Thiele I, Palsson BO. 2010. What is flux balance analysis? Nat Biotechnol 28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vital-Lopez FG, Reifman J, Wallqvist A. 2015. Biofilm formation mechanisms of Pseudomonas aeruginosa predicted via genome-scale kinetic models of bacterial metabolism. PLoS Comput Biol 11:e1004452. doi: 10.1371/journal.pcbi.1004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stewart PS. 1994. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38:1052–1058. doi: 10.1128/aac.38.5.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roberts ME, Stewart PS. 2004. Modeling antibiotic tolerance in biofilms by accounting for nutrient limitation. Antimicrob Agents Chemother 48:48–52. doi: 10.1128/aac.48.1.48-52.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wanner O, Gujer W. 1986. A multispecies biofilm model. Biotechnol Bioeng 28:314–328. doi: 10.1002/bit.260280304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


