Abstract
Mouse embryonic stem cells (mESCs) are key components in generating mouse models for human diseases and performing basic research on pluripotency, yet the number of genes essential for mESCs is still unknown. We performed a genome-wide screen for essential genes in mESCs and compared it to screens in human cells. We found that essential genes are enriched for basic cellular functions, are highly expressed in mESCs, and tend to lack paralog genes. We discovered that genes that are essential specifically in mESCs play a role in pathways associated with their pluripotent state. We show that 29.5% of human genes intolerant to loss-of-function mutations are essential in mouse or human ESCs, and that the human phenotypes most significantly associated with genes essential for ESCs are neurodevelopmental. Our results provide insights into essential genes in the mouse, the pathways which govern pluripotency, and suggest that many genes associated with neurodevelopmental disorders are essential at very early embryonic stages.
Essential genes are required for organism survival or development. Recent advances in CRISPR technology have enabled the identification of essential genes in multiple human cancer cell lines (Wang et al. 2015, 2017; Tzelepis et al. 2016) and in human embryonic stem cells (hESCs) (Yilmaz et al. 2018). A gene is considered human essential when a mutation in such gene will likely be either completely lethal or lead to a severe disorder in young age. Since essential genes are under severe negative (purifying) selection, they are expected to show reduced genetic variability. Thus, assessment of human essential genes can be based on population genetic data, including the identification of genes intolerant to mutations (Samocha et al. 2014; Lek et al. 2016). While intolerance to loss-of-function (LoF) mutations does not identify all essential genes, it is the best proxy for human in vivo essential genes (Bartha et al. 2018). However, one should note that measures of intolerance to mutations reflect the strength of selection acting on heterozygotes (Fuller et al. 2019). Previous studies found that intolerant genes are associated with neurodevelopmental risk genes (Samocha et al. 2014; Shohat et al. 2017) but not with other extensively studied disease gene, such as Type 2 diabetes, early-onset myocardial infarction, inflammatory bowel disease, ulcerative colitis, or Crohn's disease (Ganna et al. 2018).
Until now, efforts to detect essential genes in the mouse genome focused on generating and characterizing the phenotypes of knockout mice. These efforts generated knockouts for 4969 genes, of which 1187 were found to be essential (Dickinson et al. 2016). However, these studies still do not cover the entire mouse genome and, for many of the essential genes, do not provide precise information regarding the developmental stage or cell type affected by the essential gene.
Here, we report a genome-wide screen for genes essential in mouse embryonic stem cells (mESCs), which we compared to similar screens in human cells and genes essential in vivo. Our analysis reveals the cellular pathways which are globally essential and those that are specific to mESCs.
Results
CRISPR screen in mouse embryonic stem cells
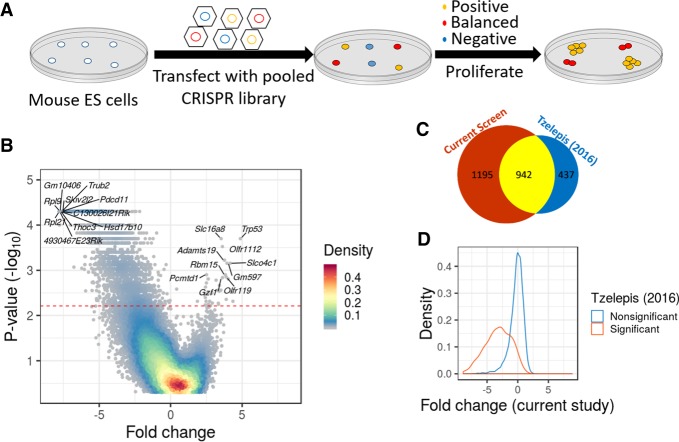
We performed a loss-of-function genome-wide screen to detect genes essential for the survival and proliferation of mESCs (Fig. 1A). We used a pooled knockout CRISPR library that consists of 77,637 guide RNAs (gRNAs) targeting 19,674 coding genes (four gRNAs per gene) and 1000 nongene targeting control gRNAs (Doench et al. 2016). Sequencing of the pretransfected plasmid library revealed the presence of 99.82% of gRNAs (143 gRNAs were missing) and that all genes had at least 97 reads from gene-targeting gRNAs (a mean of 1087 reads per gene) (Supplemental Fig. S1A,B; Supplemental Table S1). We transfected Cas9-expressing mESCs with lentiviruses containing the library and allowed the cells to proliferate for 18 d. Cells were collected on days 8, 11, 15, and 18 posttransfection, and the abundance of gRNAs was determined by sequencing. In order to assess which genes are under significant negative selection, we developed an approach that is based on a simulation of randomly selected control gRNAs (see Methods). This approach allows detection of negative and positive selection in the presence of random changes in gRNA abundance (random drift) (Supplemental Fig. S1C,D). Using this method, we were able to detect 2379 genes which are essential for survival or proliferation of mESCs (Fig. 1B). A CRISPR score calculated across all time points for each gene is available in Supplemental Table S2.
Figure 1.
A genome-wide CRISPR screen to identify genes essential in mESCs. (A) Schematic overview of the genome-wide CRISPR screen. (B) A volcano plot of the screen results. Significance (−log10 of the P-value) across all days as a function of the average fold change for each gene at the end of the screen. Colors indicate the density of the points. The red line represents a corrected P < 0.05. (C) Venn diagram presenting the overlap between essential genes in the current screen and essential genes in a previous study (Tzelepis et al. 2016). (D) The distribution of the average fold change per gene (day 15) was drawn separately for genes with significant evidence for negative selection in a previous study (Tzelepis et al. 2016) (blue) and genes with no significant evidence for selection (orange).
Since results from a single screen might be influenced by the specific CRISPR library used or by other factors, we compared the results to data from another mESC line transfected with a different CRISPR library (Tzelepis et al. 2016). Despite the different experimental conditions, we found a highly significant overlap (odds ratio [OR] = 27, P < 10−16) (Fig. 1C). Quantitatively, we observed that the distribution of fold changes for gRNAs negatively selected in one screen was significantly shifted in the second screen, in the same direction (P < 10−16) (Fig. 1D). Since the two screens showed significant overlap, we combined the evidence of the two independent screens and generated a consensus list of genes under selection in mESCs, with 2164 genes under negative selection (henceforth, mESC-essential genes) (Supplemental Table S3).
mESC-essential genes are enriched for fundamental cellular processes
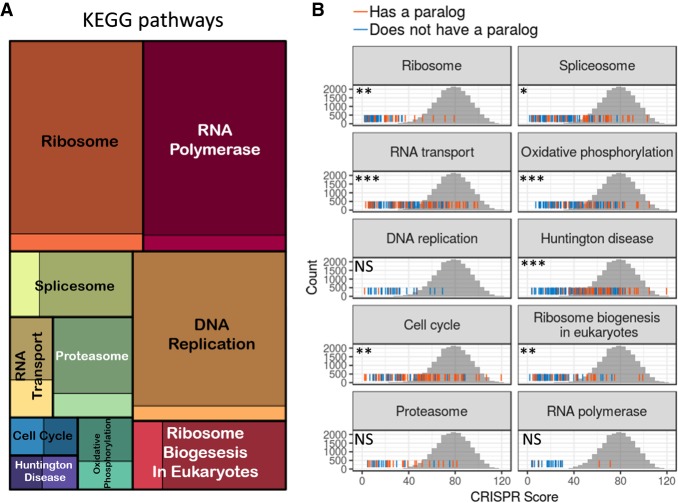
We next wanted to characterize the list of essential genes and test in which biological process they are involved. Using Gene Ontology (GO) and KEGG pathways enrichment analysis, we found that the essential genes are associated with fundamental cellular processes such as ribosome biogenesis, RNA processing, translation, DNA metabolism, and the cell cycle (Fig. 2A; Supplemental Fig. S2A; Supplemental Table S4). For some KEGG pathways (ribosome, DNA replication, and RNA polymerase), nearly all genes (>90%) in the pathway were found to be essential (Fig. 2A). The fact that not all genes in the top enriched KEGG pathways (“essential pathways”) emerged as essential in our screen could be a result of statistical power, but it also could be that some genes within the pathways are robust to mutations. One option for this robustness is functional redundancy, when an alternative gene or pathway, usually a paralog gene, can compensate for the mutated gene. To investigate this possibility, we tested within each essential pathway if genes with a paralog are less essential. For seven out of the 10 essential pathways, genes with paralogs were significantly less essential compared to genes without paralogs (Fig. 2B).
Figure 2.
Essential genes belong to basic cellular pathways and tend to lack paralogs. (A) A treemap of the top 10 most enriched KEGG pathways (“essential pathways”). The square size is proportional to the enrichment strength, and the color intensity indicates the proportion of essential (dark color) and nonessential genes (bright colors) in the pathway. (B) For each essential pathway, the rug plot displays the distribution of CRISPR scores (sum of ranks across days) for all genes in the pathway. Orange lines indicate genes with a paralog, and blue lines indicate genes without a paralog. The gray histogram behind is the empirical null distribution of CRISPR scores based on control gRNAs. Significance indicates the differences in scores between genes with and without a paralog. (NS) Nonsignificant, P > 0.05; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
We found 115 genes (5% of the essential genes) in our screen that are essential but currently not associated with any GO term or pathway. To exclude the possibility that these genes are false positives, we tested their expression in mESCs. We found that essential genes lacking a GO term show significantly higher expression in mESCs than nonessential genes (P < 10−16) (Supplemental Fig. S2B). This indicates that there is a large list of uncharacterized genes with unknown functions that are essential in mESCs and probably essential for the survival of early-stage embryos.
Essential genes with slower gRNA depletion are less essential but are more likely to be associated with human recessive mutations
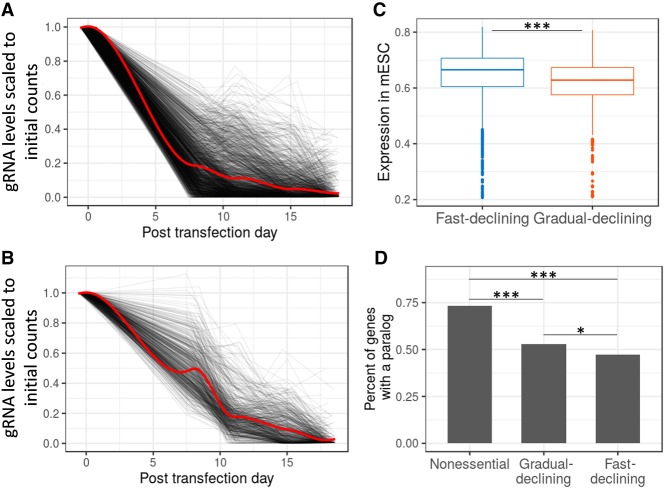
Our screen includes multiple time points, allowing us to quantify the kinetics of the negative selection. When fold change across time points was used in hierarchical clustering, most genes were mapped into two main clusters (Supplemental Fig. S3A): a fast-declining cluster (55%) that had a rapid reduction in gRNA representation in the first 8 d (Fig. 3A), and a gradual-declining group of genes (17%) with a linear decline of gRNAs across all days (Fig. 3B). To explain this phenomenon, we first tested if the gradual-declining group code for more stable proteins. Since data on protein stability in mESCs were unavailable, we used data from six other human or mouse cell lines. We observed no significant difference in protein half-life between the groups (Supplemental Fig. S3B). Another possibility is that essentiality is not an all or nothing phenomenon but a quantitative property. Supporting the possibility that the gradual-declining group is quantitatively less essential, we found that the expression of those genes is significantly lower in mESCs (P = 1.0 × 10−5) (Fig. 3C) and they have significantly more paralogs relative to the fast-declining group (P = 0.027) (Fig. 3D). When examining the enrichment of cellular processes in the two gene groups, we found that the fast-declining genes showed a stronger enrichment for ribosome biogenesis, transcription, and RNA processing terms. In contrast, the gradual-declining genes showed a higher enrichment for mitochondrial terms and terms related to modifications of DNA and proteins (Supplemental Fig. S3C–F). These results suggest that the difference in the selection rates mostly reflects differences in the quantitative level of essentiality, but it is also possible that genes in the fast-declining cluster cause cell lethality, while mutations in the gradual-declining genes tend to reduce the relative fitness of the cells. Genes that cause cell lethality will lead to early embryonic lethality and thus will less likely be associated with postnatal diseases. Consistent with this hypothesis, we found that the gradual-declining group contains a significantly higher proportion of genes associated with human recessive diseases (18.6% vs. 12.5%, P = 0.00088) and a significantly higher proportion of genes associated with postnatal lethality or abnormal growth in mice (34.6% vs. 26.2%, P = 0.013).
Figure 3.
Gene essentiality as a quantitative phenotype. (A,B) Two different dynamics of gRNA depletion rates (see Supplemental Fig. S3A for clustering results). (A) Fast-declining cluster, and (B) gradual-declining cluster. For each gene the gRNA with the strongest decline is shown (black lines). The red line indicates the trend of all genes in the cluster. (C) Expression in mESCs of genes in the fast- and gradual-declining groups. Expression values are probe signal intensities (log10) from microarray data. (D) Percentage of genes with at least one paralog gene. (*) P < 0.05; (***) P < 0.001.
Genes essential specifically in mESCs are associated with the early pluripotent state
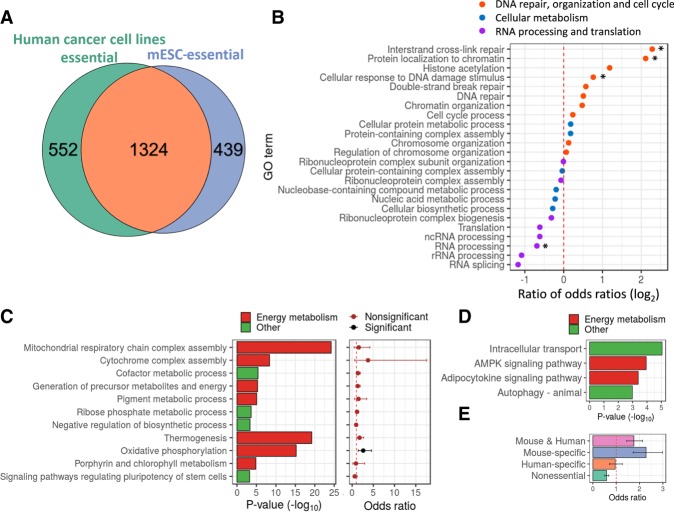
Essential genes in mESCs are enriched for the most basic cellular functions, similar to findings in other cell types (Wang et al. 2017; Yilmaz et al. 2018). We were interested to discover if there are genes that are essential specifically in mESCs. To this end, we compared the list of essential genes in mESCs to human cancer cell lines (Meyers et al. 2017) (n = 563) and two human ESCs (diploid and haploid hESCs). We found that 70% of essential genes in mESCs are also essential in >90% of the human cancer cell lines (OR = 62.6, P < 10−16) (Fig. 4A). When compared to hESCs, 66% of the essential genes in mESCs were also essential in at least one of the hESCs (OR = 24.2, P < 10−16) (Supplemental Fig. S4A). The overlap between results obtained in mESCs and haploid hESCs was stronger (OR = 31.7) than between the two hESCs (OR = 12.2, P < 10−5) (Supplemental Fig. S4B).
Figure 4.
Analysis of genes essential specifically in mESCs reveals pathways associated with the pluripotent state. (A) Venn diagram presenting the overlap between essential genes in mESCs and in human cancer cell lines (OR = 62.6, P < 10−16). (B) The ratio between odds ratios obtained for GO terms enrichment analysis for essential genes specific in ESCs relative to all essential genes in mESCs. (C) Significant enrichment of GO terms and KEGG pathways for genes essential specifically in mESCs (left plot), and corresponding association level of those terms with genes significantly up-regulated in mESCs relative to EpiSCs (right plot). Values are odds ratio ± 95% confidence interval. (D) Significant enrichment of GO terms and KEGG pathways for genes essential specifically in hESCs. (E) Association analysis between genes up-regulated in mESCs and genes essential specifically in (1) both human and mouse ESCs, (2) mESCs, (3) hESCs, and (4) nonessential genes. Values are odds ratio ± 95% confidence interval. (*) P < 0.05.
Although most essential genes are common across cell types, some are specific to ESCs (both human and mouse) and some are unique to mESCs. Genes essential specifically in ESCs were significantly more enriched for GO terms related to DNA repair organization and cell cycle (P = 0.010) and less enriched for RNA processing and translation (P = 0.0030) (Fig. 4B; Supplemental Fig. S4C). Genes specific to mESCs were enriched for mitochondria and other energy metabolism terms (Fig. 4C, left panel). Genes unique to hESCs showed enrichment for the AMPK and adipocytokine signaling pathways, which are part of the regulation on cellular energy state (Fig. 4D).
Previous studies have shown that energy metabolism is one of the key pathways that alters during transition between pluripotent states and that mESCs are found in an earlier pluripotent sate in comparison to hESCs (Brons et al. 2007; Tesar et al. 2007; Zhou et al. 2012). Therefore, the enrichment for energy metabolism-related terms implies that genes essential specifically in mESCs might be related to the pluripotent state of mESCs and not to the difference between mouse and human. To test this hypothesis, we identified genes differentially expressed (DE) between mESCs (early pluripotent) and mouse epiblast stem cells (EpiSCs) (representing primed pluripotent cells) (Brons et al. 2007; Tesar et al. 2007) and tested if terms or pathways associated with genes essential specifically in mESCs are significantly enriched for DE genes. Out of the 11 pathways and terms, only “oxidative phosphorylation” was significantly enriched for genes up-regulated in the early pluripotent state (Fig. 4C, right panel). When considering all essential genes (without grouping them into pathways), we found that genes essential specifically in mESCs are significantly associated with genes up-regulated in the early pluripotent state (OR = 1.7, P = 1.1 × 10−7) (Fig. 4E). This was also true when genes essential in both human and mouse ESCs were analyzed together (OR = 2.0, P = 5 × 10−9). In contrast, genes essential specifically in hESCs showed no significant association with genes up-regulated in the early pluripotent state (OR = 0.85, P = 0.53) (Fig. 4E).
Essential genes in mESCs are intolerant to heterozygous and homozygous mutations
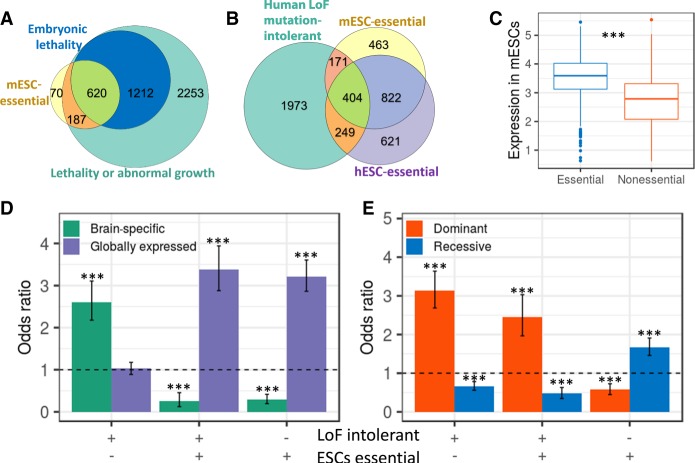
Our screen is based on mESCs proliferating in vitro, and thus the essential genes may not all be required in vivo. To test if the genes identified in the screen are known to be essential in vivo, we first compared the list with genes previously associated with embryonic lethality in mice. We found that 71% of the essential genes in mESCs (with phenotypic information) are known to be embryonic lethal (OR = 13.05, P < 10−16), and an additional 21% are known to be postnatal lethal or cause abnormal growth (Fig. 5A). Based on preferential gene expression in mESCs, the 8% of genes without developmental phenotypes are likely to be true essential genes (Supplemental Fig. S5A). Next, we tested the overlap of genes essential in mESCs with human genes intolerant to LoF mutations (Lek et al. 2016). We found that 30.9% of essential genes in mESCs are LoF mutation-intolerant genes (P < 10−16) (Fig. 5B). The relatively low overlap with LoF mutation-intolerant genes may be due to the differences between mouse and human. However, the percentage of overlap between hESC-essential genes and LoF mutation-intolerant genes was not significantly higher (31.2%, P = 0.69) (Fig. 5B). Given the similar level of magnitude in overlap with LoF mutation-intolerant genes, we combined the two lists of essential genes in mESCs and hESCs into one list of ESC-essential genes. Overall, 29.5% of the LoF mutation-intolerant genes are essential in ESCs (mESCs or hESCs).
Figure 5.
Overlap between essential genes in mESCs and genes essential in vivo in mouse or human. (A) Overlap between essential genes in mESCs and embryonic lethal genes in mice (odds ratio = 12.84, P < 10−16) and with genes known to be lethal or cause abnormal growth (odds ratio = 13.94, P < 10−16). (B) Overlap between human LoF mutation-intolerant genes, essential genes in mESCs (odds ratio = 1.88, P < 10−16), and essential genes in hESCs (odds ratio = 2.33, P < 10−16). (C) Expression levels in mESCs for genes intolerant to LoF mutations in human divided into essential and nonessential genes in ESCs. Expression values are signal intensities (log10) from microarray data. (D) Association between the pattern of expression (brain-specific genes vs. globally expressed genes) and being essential in ESCs and/or intolerant to LoF mutations in human. Values are odds ratio ± 95% confidence interval. (E) Association between dominant and recessive inheritance and being essential in ESCs and/or intolerant to LoF mutations in humans. Values are odds ratio ± 95% confidence interval. (***) P < 0.001.
One possible explanation why many LoF mutation-intolerant genes are not essential in ESCs is that they are essential in later developmental stages. In accordance, we found that LoF mutation-intolerant genes not essential in ESCs express at relatively lower levels in mESCs (P < 10−16) (Fig. 5C). In addition, when testing which tissues are most influenced by these genes, we found that LoF mutation-intolerant genes not essential in ESCs preferentially express in the brain, while genes that are both LoF mutation-intolerant and essential in ESCs are broadly expressed across multiple tissues (Fig. 5D). The relatively low overlap between LoF mutation-intolerant genes and genes essential in ESCs also raises the question of why many genes that are essential in ESCs are not LoF mutation-intolerant. We hypothesized that this is because LoF mutation-intolerant genes represent genes sensitive to heterozygote mutations (Fuller et al. 2019), while CRISPR screens tend to create complete knockouts. Indeed, we found that essential genes that are LoF mutation-intolerant are more likely to be associated with dominant disorders, while essential genes that are tolerant to LoF mutations are more likely to be associated with recessive disorders (Fig. 5E).
Essential genes in ESCs are associated with neurodevelopmental phenotypes
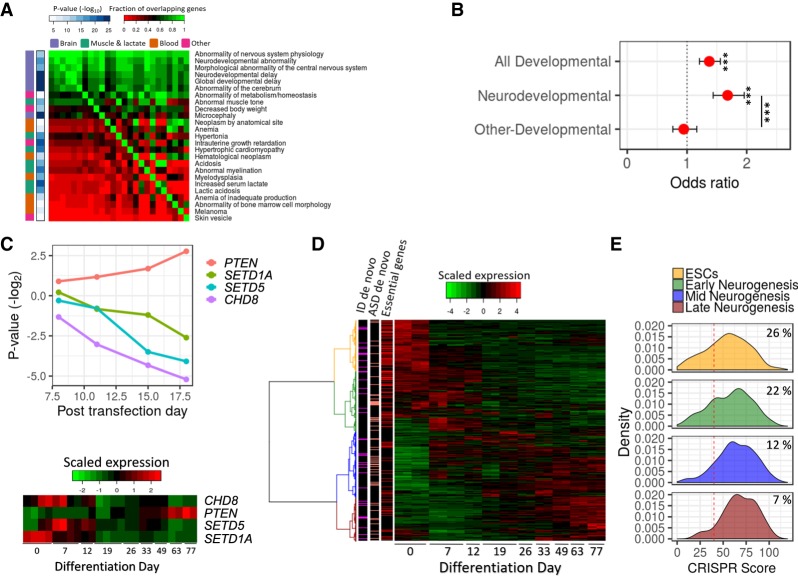
Since ESCs serve as a model for studying many human diseases (Zhu and Huangfu 2013), we tested what type of human phenotypes are associated with ESC-essential genes. When testing all human phenotypes in the Human Phenotype Ontology (HPO) (Köhler et al. 2019), we found that essential genes are enriched for abnormal phenotypes related to the brain, muscle, or blood systems (Fig. 6A). Notably, there is a large overlap between the list of genes belonging to different phenotypes, where most belong to more than one, and 88% are associated with the brain (Supplemental Fig. S5B).
Figure 6.
Essential genes in ESCs are associated with neurodevelopmental phenotypes. (A) Human phenotypes significantly enriched for essential genes. The phenotypes are ordered by the degree of overlap with other phenotypes. The color in the heat map corresponds to the degree of overlap in genes between phenotypes (red is low overlap; green is high overlap). The right-side bar indicates the significance level of the enrichment of each phenotype with essential genes. The left-side bar indicates the organs involved in the phenotypes. (B) Association between ESC-essential genes and risk genes for human developmental and neurodevelopmental disorders. Values are odds ratio ± 95% confidence interval. (***) Corrected P < 0.001. (C) (Top) Dynamic of gRNAs (average fold change) targeting four genes associated with neurodevelopmental disorders. (Bottom) The expression of the four genes during human in vitro corticogenesis. The colors of the heat map correspond to the normalized expression (red is high and green is low levels of expression). (D) A heat map of gene expression of risk genes for neurodevelopmental disorders during human in vitro corticogenesis. The colors of the heat map correspond to the normalized expression (red is high and green is low levels of expression). Side bars indicate ESC-essential genes (red), genes implicated in ASD by multiple de novo mutations (salmon), and genes implicated in ID by multiple de novo mutations (magenta). (E) Distribution of CRISPR scores for NDDs risk genes divided by expression patterns during human in vitro corticogenesis. The red line indicates a corrected P < 0.05. The numbers are the percentage of ESC-essential genes in each group.
The association between genes essential in ESCs and human phenotypes may be related to the role of these genes in regulating proliferation and differentiation during development, so we next checked the association between ESC-essential genes and human developmental disorders, including neurodevelopmental disorders (NDDs). We tested the overlap with genes previously identified as disrupted by rare mutations in individuals with different developmental phenotypes (Wright et al. 2015). Out of the list of essential genes in ESCs, 13% were previously identified to be mutated in individuals with a developmental phenotype (OR = 1.3, P = 3.6 × 10−6) (Fig. 6B). We separated the individuals into ones with neurodevelopmental phenotypes and ones without brain-related phenotypes and found that the genetic overlap with ESC-essential genes was driven only by the neurodevelopmental phenotypes (ORNDDs = 1.65, PNDDs = 4.1 × 10−10; ORnon-NDDs = 0.94, Pnon-NDDs = 0.56; NDDs vs. non-NDDs P = 2.7 × 10−5) (Fig. 6B). Among the top NDDs risk genes identified as ESC-essential are CHD8 (autism spectrum disorders [ASD]) (Bernier et al. 2014), SETD1A (schizophrenia) (Singh et al. 2016), and SETD5 (intellectual disability [ID]) (Fig. 6C; Grozeva et al. 2014).
Our results suggest that many genes associated with NDDs are essential as early as the ESC stage. We predicted that those NDD genes will be preferentially expressed in ESCs, since generally genes essential in ESCs were preferentially expressed in human and mouse ESCs (Supplemental Fig. S5C). To study NDDs genes, we tested the expression patterns of NDDs risk genes and specifically ASD and ID risk genes during human and mouse in vitro corticogenesis (23% of ASD and ID risk genes are ESC-essential). While ASD and ID risk genes are preferentially expressed across multiple differentiation stages (Fig. 6D; Supplemental Fig. S5D), the genes that show the highest expression (in both human and mouse) at the embryonic stem cell stages have a significant propensity to be essential in ESCs (Fig. 6E; Supplemental Fig. S5E).
Discussion
CRISPR screens have been used to identify essential genes mainly in human cancer cells but less commonly in other types of cells and organisms. We have shown here that a screen in mESCs can identify not only mouse essential genes and genes involved in pluripotency but also genes related to human disorders. Our results raise questions about the quantitative definition of essential genes and about the biological insights that can be attained by comparing essential genes identified in vivo and in vitro.
Our analysis shows that essential genes are less likely to have a paralog. This phenomenon was observed in yeast (Keane et al. 2014) and Caenorhabditis elegans (Kamath et al. 2003) but was debated in the mouse (Liao and Zhang 2007; Makino et al. 2009; White et al. 2013). This debate was based on a limited number of genes and biased screens. Our analysis, which covers the vast majority of mouse genes, confirms that essential genes are less likely to have a paralog across the eukaryotic tree.
The screen we performed spans several time points, which allowed us to quantify the gRNA decline rate and cluster the essential genes into two groups. We found that genes in the fast-declining group are more highly expressed, less likely to have a paralog, and are enriched for fundamental cellular functions, all of which are general properties of essential genes (White et al. 2013; Rancati et al. 2018). These findings imply that genes in the gradual-declining group might have a milder effect on cell viability than genes in the fast-declining group—for example, a reduction in proliferation rate or partial lethality versus complete lethality in the fast-declining group. This is consistent with the higher propensity of genes in the gradual-declining group to be associated with human recessive diseases.
Our results imply that the differences between essential genes in human and mouse ESCs are mostly related to differences in their pluripotent state and not to the organism. We show that genes essential specifically in mESCs are in pathways involved in the pluripotent state, such as mitochondria organization and oxidative phosphorylation (Brons et al. 2007; Zhou et al. 2012). Genes essential specifically in hESCs showed enrichment in terms related to the regulation of cellular energy, such as the AMPK pathway (Herzig and Shaw 2018).
Our study provides an important new resource for studying human disease. Genes known to be intolerant to LoF mutations in humans are mainly dominant acting essential genes. For most of those LoF mutation-intolerant genes, the developmental period affected is still unknown. Our screen provides a list of essential genes that includes recessive and dominant inheritance and shows that 29% of human LoF mutation-intolerant genes are essential already at the ESCs stage. The remaining 71% show lower expression in ESCs and are likely essential in later stages of development. When testing which human disorders are associated with essential genes in ESCs, we found that brain developmental disorders are the most significantly enriched. Since human genes intolerant to LoF mutations are also associated with neurodevelopmental disorders (Samocha et al. 2014; Shohat et al. 2017), a question is why essential genes are not linked to diseases affecting other important organs. We suggest that this could be related to the inheritance pattern, since mutations disrupting both copies of essential genes will frequently lead to embryonic lethality and therefore will not be linked to any human disease. However, it is possible that during development the brain is the most sensitive organ to heterozygote LoF mutations in essential genes, leading to deficits in proliferation and differentiation (Courchesne et al. 2003; Stephenson et al. 2011; Ernst 2016).
In summary, we provide a map of genes essential for mESC proliferation and survival. These data greatly expands our knowledge about genes essential in the mouse and for pluripotency and can help researchers determine how and which human disorders can be modeled in the mouse. Noteworthy in this regard is that 5% of the essential genes are of unknown function, and many others have very limited functional information. These genes will be of great interest for further study.
Methods
Screen for essential genes using pooled CRISPR library
CAS9-expressing mESCs were transfected with lentiviruses containing the Brie pooled library (see Supplemental Methods). Cells were passaged at days 8, 11, 15, and 18 posttransfection. For each passage, a minimum number of 31 million cells was retained for sequencing, and 31 million additional cells were replated, allowing for a maintenance of adequate library representation (an average of ∼400 cells per gRNA).
Identification of essential genes
Genomic DNA extracted from the cells at the different time points was amplified with primers targeting the gRNAs and sequenced (see Supplemental Methods). For all samples, library sizes were normalized using the calcNormFactors function in edgeR (Robinson et al. 2010; McCarthy et al. 2012). This normalization corrects for underestimation of gRNAs abundances due to the presence of a few highly represented gRNAs. Following the normalization, the log fold change of each gRNA in each proliferation day was calculated relative to the initial counts in the plasmid library. A simulation-based approach was used to detect genes with significant negative or positive selection. For each proliferation day, we ranked all gRNAs by their representation fold change relative to the library. For each gene, we calculated the sum of ranks of its gRNA across all days. This score was compared to an empirical null distribution generated by randomly selecting four control gRNAs and calculating their sum of ranks (10,000 simulation). P-values were corrected for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) procedure. CRISPR scores were defined as the sum of ranks for each gene divided by 104.
Overlap of essential genes with a previous published data set
The overlap between genes found to be under significant negative selection (FDR corrected P < 0.05) in the screens was tested using Fisher's exact test. Welch's t-test was used to test the significance for the quantitative difference in the fold change (day 15 posttransfection) between genes found to be under significant or nonsignificant negative selection in a previous screen (Tzelepis et al. 2016). A combined P-value for the two screens were obtained using the sum z (Stouffer's) method (Stouffer et al. 1949; Whitlock 2005).
GO terms and KEGG pathway enrichment analysis
GO terms enrichment was performed using the GOrilla tool (Eden et al. 2009), with all genes tested in the screens as background. GO terms significant at a FDR corrected value P < 0.05 were summarized using REVIGO (Supek et al. 2011; see Supplemental Methods). Comparison of GO term enrichment between essential genes in the fast- and gradual-declining group, and between genes essential specifically in mESCs or in hESCs relative to genes essential in both, was performed using Fisher's exact test. Comparison of GO term enrichment between all mESC-essential genes and ESCs specific genes was performed on all the terms that were significant for ESC-essential genes and the top 10 terms enriched for all essential genes. P-values were calculated using a permutation test by sampling 187 genes (the number of ESC-essential genes) from the mESC-essential gene list and testing their enrichment for each GO term. P-values were corrected for multiple testing using an FDR procedure. Analysis of KEGG pathways was performed using the clusterProfiler R package (Yu et al. 2012). Comparison of KEGG pathways between essential genes in the fast- and gradual-declining groups and between genes essential specifically in mESCs or hESCs relative to genes essential in both was performed using Fisher's exact test.
Analysis of paralog genes
Paralog genes were identified using Ensembl BioMart (Smedley et al. 2015) and TreeFam (Ruan et al. 2008) databases. The significance of the difference between the CRISPR score distribution of genes with and without a paralog in the top KEGG pathways was tested using a Mann–Whitney U test. The enrichment for genes without a paralog for the fast- and gradual-declining clusters and for nonessential genes was tested using a Fisher's exact test.
Cluster identification based on gRNA kinetics
Clustering of essential genes in mESCs was performed based on the correlation between all essential genes in depletion rates. The correlation matrix was then used for hierarchical clustering using the R (R Core Team 2019) hclust function with default settings. The dendrogram branches were cut to obtain two main clusters.
Gene expression in ESCs
Gene expression in mESCs and hESCs was obtained from previous studies (Tesar et al. 2007; van de Leemput et al. 2014; see Supplemental Methods). To test for significant differences in gene expression between groups, we used the Welch's t-test for two groups and Tukey's test for three groups.
Difference in mean half-life between genes in the gradual- and fast-declining groups
Data on protein half-life were obtained from previous studies (Schwanhäusser et al. 2011; Mathieson et al. 2018). The difference in the mean log10 half-life for genes in the fast- and gradual-declining groups was determined using Welch's t-test.
Comparison of essential genes between mESCs, hESCs, and cancer cell lines
Data on essential genes in haploid hESCs grown on feeder cells were from Yilmaz et al. (2018), on essential genes in diploid hESCs grown on feeder cells was from Mair et al. (2019), and on essential genes in human cancer cell lines were from the Achilles project (Meyers et al. 2017). Genes essential in cancer cell lines were defined as genes in the top ranked essential genes in >90% of cell lines. Human mouse orthologs were identified using BioMart (Ensembl release 97) (Smedley et al. 2015). Genes without direct 1:1 orthologs were filtered out and were not used in the human-mouse comparison. We defined ESC-specific essential genes as genes essential in mESCs and in at least one of the hESCs (FDR corrected P < 0.05) but not essential in the human cancer cells. Genes essential specifically in mESCs were defined as genes essential in mESCs but not in both hESCs lines. Similarly, genes essential specifically in hESCs were defined as genes essential in at least one of the hESCs lines (FDR corrected P < 0.05) but not in mESCs.
Differential expression analysis between mESCs and EpiSCs
Microarray gene expression data for three mESCs samples and three EpiSCs samples were obtained from Zhou et al. (2012). The data were normalized by quintile normalization, and differential expression was performed using limma (Ritchie et al. 2015). Association with genes significantly up-regulated in mESCs relative to EpiSCs was determined using Fisher's exact test.
Overlap with mouse embryonic lethal genes and human LoF mutation-intolerant genes
A list of genes leading to pre- or postnatal lethality or abnormal survival phenotype in knockout mice was obtained from the Mouse Genome Informatics (MGI) database (Supplemental Table S5; Bult et al. 2019). The overlap between genes essential in mESCs and genes associated with growth or lethality in mice was tested only for genes with knockout phenotypic data at the MGI database. The list of human LoF mutation-intolerant genes was from Lek et al. (2016). Significance of the overlaps was based on Fisher's exact test.
Association of essential genes with human phenotypes
Analysis of human phenotypes for genes essential in ESCs was based on the Human Phenotype Ontology (Köhler et al. 2019). A phenotype that was significant but had more than 90% overlap of genes with a more significant phenotype was filtered out. Association with developmental and neurodevelopmental phenotypes for genes essential in ESCs was based on the DDG2P data set from the Deciphering Developmental Disorders project (Wright et al. 2015). Neurodevelopmental phenotypes were defined as any developmental phenotype involving the brain. The significance of the association was calculated by Fisher's exact test, and P-values were corrected for multiple testing by FDR procedure.
Gene expression during in vitro human and mouse corticogenesis
Neurodevelopmental disorders risk genes, as defined by DDG2P (Wright et al. 2015) or genes present in the developmental brain disorders database (tires 1 & 2) (Gonzalez-Mantilla et al. 2016), were clustered according to their expression patterns during in vitro corticogenesis of mouse (Hubbard et al. 2013) or human (van de Leemput et al. 2014) cells. Clustering was performed using the R hclust function with the default settings.
Data access
The read counts of gRNAs at different time points are available in Supplemental Table S1. The mean fold change, CRISPR scores, and P-values for all genes tested in the screen are available in Supplemental Table S2. The consensus list of genes under selection in mESCs and the combined P-values for negative and positive selection are available in Supplemental Table S3.
Supplementary Material
Acknowledgments
We thank Nissim Benvenisty, Alana Amelan, and Eran Meshorer for valuable comments on the manuscript. This research was supported by the Israel Science Foundation (grant no. 575/17) and by the Israel Science Foundation Broad Institute Joint Program (grant no. 2612/18).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.250019.119.
References
- Bartha I, di Iulio J, Venter JC, Telenti A. 2018. Human gene essentiality. Nat Rev Genet 19: 51–62. 10.1038/nrg.2017.75 [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, et al. 2014. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158: 263–276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Mouse Genome Database Group. 2019. Mouse Genome Database (MGD) 2019. Nucleic Acids Res 47: D801–D806. 10.1093/nar/gky1056/5165331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N. 2003. Evidence of brain overgrowth in the first year of life in autism. JAMA 290: 337–344. 10.1001/jama.290.3.337 [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C. 2016. Proliferation and differentiation deficits are a major convergence point for neurodevelopmental disorders. Trends Neurosci 39: 290–299. 10.1016/j.tins.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Berg JJ, Mostafavi H, Sella G, Przeworski M. 2019. Measuring intolerance to mutation in human genetics. Nat Genet 51: 772–776. 10.1038/s41588-019-0383-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, Alfoldi J, Martin AR, Havulinna AS, Byrnes A, et al. 2018. Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am J Hum Genet 102: 1204–1211. 10.1016/j.ajhg.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. 2016. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry 73: 275 10.1001/jamapsychiatry.2015.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park SM, Canham N, Holder SE, Wilson M, et al. 2014. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet 94: 618–624. 10.1016/j.ajhg.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ. 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19: 121–135. 10.1038/nrm.2017.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KS, Gut IM, Lyman ME, McNutt PM. 2013. Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000Res 2: 35 10.12688/f1000research.2-35.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Keane OM, Toft C, Carretero-Paulet L, Jones GW, Fares MA. 2014. Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Res 24: 1830–1841. 10.1101/gr.176792.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine J-P, Gargano M, Harris NL, Matentzoglu N, McMurry JA, et al. 2019. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 47: D1018–D1027. 10.1093/nar/gky1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J. 2007. Mouse duplicate genes are as essential as singletons. Trends Genet 23: 378–381. 10.1016/j.tig.2007.05.006 [DOI] [PubMed] [Google Scholar]
- Mair B, Tomic J, Masud SN, Tonge P, Weiss A, Usaj M, Tong AHY, Kwan JJ, Brown KR, Titus E, et al. 2019. Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Rep 27: 599–615.e12. 10.1016/j.celrep.2019.02.041 [DOI] [PubMed] [Google Scholar]
- Makino T, Hokamp K, McLysaght A. 2009. The complex relationship of gene duplication and essentiality. Trends Genet 25: 152–155. 10.1016/j.tig.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, Poeckel D, Ratnu VS, Schramm M, Becher I, et al. 2018. Systematic analysis of protein turnover in primary cells. Nat Commun 9: 689 10.1038/s41467-018-03106-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. 2017. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 49: 1779–1784. 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rancati G, Moffat J, Typas A, Pavelka N. 2018. Emerging and evolving concepts in gene essentiality. Nat Rev Genet 19: 34–49. 10.1038/nrg.2017.74 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJM, Guo Y, Hériché J-K, Hu Y, Kristiansen K, Li R, et al. 2008. TreeFam: 2008 update. Nucleic Acids Res 36: D735–D740. 10.1093/nar/gkm1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. 2014. A framework for the interpretation of de novo mutation in human disease. Nat Genet 46: 944–950. 10.1038/ng.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Shohat S, Ben-David E, Shifman S. 2017. Varying intolerance of gene pathways to mutational classes explain genetic convergence across neuropsychiatric disorders. Cell Rep 18: 2217–2227. 10.1016/j.celrep.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G, et al. 2016. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 19: 571–577. 10.1038/nn.4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43: W589–W598. 10.1093/nar/gkv350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson DT, O'Neill SM, Narayan S, Tiwari A, Arnold E, Samaroo HD, Du F, Ring RH, Campbell B, Pletcher M, et al. 2011. Histopathologic characterization of the BTBR mouse model of autistic-like behavior reveals selective changes in neurodevelopmental proteins and adult hippocampal neurogenesis. Mol Autism 2: 7 10.1186/2040-2392-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM Jr. 1949. The American soldier: adjustment during army life. (Studies in social psychology in World War II, Vol. 1). Princeton University Press, Princeton, NJ. [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, et al. 2016. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep 17: 1193–1205. 10.1016/j.celrep.2016.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, Lee C, Martinez RA, Levi BP, Thompson CL, et al. 2014. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 83: 51–68. 10.1016/j.neuron.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168: 890.– . 10.1016/j.cell.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin A-K, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. 10.1016/j.cell.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. 2005. Combining probability from independent tests: The weighted Z-method is superior to Fisher's approach. J Evol Biol 18: 1368–1373. 10.1111/j.1420-9101.2005.00917.x [DOI] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, et al. 2015. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 385: 1305–1314. 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N. 2018. Defining essential genes for human pluripotent stem cells by CRISPR–Cas9 screening in haploid cells. Nat Cell Biol 20: 610–619. 10.1038/s41556-018-0088-1 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. 2012. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J 31: 2103–2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Huangfu D. 2013. Human pluripotent stem cells: an emerging model in developmental biology. Development 140: 705–717. 10.1242/dev.086165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.