Abstract
Acinetobacter baumannii is a nosocomial pathogen that has emerged as a global threat because of high levels of resistance to many antibiotics, particularly those considered to be last-resort antibiotics, such as carbapenems. Although alterations in the efflux pump and outer membrane proteins can cause carbapenem resistance, the main mechanism is the acquisition of carbapenem-hydrolyzing oxacillinase-encoding genes. Of these, oxa23 is by far the most widespread in most countries, while oxa24 and oxa58 appear to be dominant in specific regions. Historically, much of the global spread of carbapenem resistance has been due to the dissemination of two major clones, known as global clones 1 and 2, although new lineages are now common in some parts of the world. The analysis of all publicly available genome sequences performed here indicates that ST2, ST1, ST79 and ST25 account for over 71 % of all genomes sequenced to date, with ST2 by far the most dominant type and oxa23 the most widespread carbapenem resistance determinant globally, regardless of clonal type. Whilst this highlights the global spread of ST1 and ST2, and the dominance of oxa23 in both clones, it could also be a result of preferential selection of carbapenem-resistant strains, which mainly belong to the two major clones. Furthermore, ~70 % of the sequenced strains have been isolated from five countries, namely the USA, PR China, Australia, Thailand and Pakistan, with only a limited number from other countries. These genomes are a vital resource, but it is currently difficult to draw an accurate global picture of this important superbug, highlighting the need for more comprehensive genome sequence data and genomic analysis.
Keywords: Acinetobacter baumannii, global clones, GC1, GC2, carbapenem resistance, oxa23, oxa58, oxa24, oxa235, Tn2006, Tn2008, Tn2009, AbaR4 and plasmid
Data Summary
1. Three thousand five hundred and seventy-five A. baumannii genomes were retrieved from the GenBank non-redundant and Whole Genome Shotgun (WGS) databases and analysed here. The full strain list and the ftp addresses used to retrieve the genomes are publicly available at https://www.ncbi.nlm.nih.gov/genome/?term=Acinetobacter+baumannii.
2. Variants of the beta-lactam resistance genes used for analyses were retrieved from the NCBI Antimicrobial Resistance Reference Gene database, which is publicly available at https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/.
Impact Statement.
Carbapenem antibiotics were once considered to be a last resort, but the rapid worldwide dissemination of multiply antibiotic-resistant (MAR) bacteria has made them the first, or only, treatment option left for many infections. However, organisms that are also resistant to carbapenems are now becoming commonplace. Diverse populations of carbapenem-resistant Acinetobacter baumannii (CRAB) have been observed worldwide, mainly driven by the spread of two MAR clonal lineages. Next-generation sequencing technologies have provided an unprecedented level of information to study the evolution and epidemiology of carbapenem resistance in this priority pathogen. Here, we delve into this rich resource to not only enhance what is known about the mechanisms and epidemiology of carbapenem resistance in A. baumannii , but also understand what is missing. Understanding the factors that lead CRAB to spread so successfully throughout the world is crucial to curtail its spread and prevent it from becoming universally untreatable.
Introduction
Antibiotic resistance has increased to dangerously high levels in bacterial strains recovered in all parts of the world, threatening our ability to treat common infectious diseases [1]. Acinetobacter baumannii is one such organism and a member of the ESKAPE group of six bacterial pathogens ( E nterococcus faecium , Staphylococcus aureus , Klebsiella pneumoniae , Acinetobacter baumannii , Pseudomonas aeruginosa and Enterobacter species) that are major causes of antibiotic-resistant infections [2]. A. baumannii is a Gram-negative opportunistic nosocomial pathogen that is most notably responsible for pneumonia, along with infections of burns and other wounds [3–5]. It can survive harsh environmental pressures, such as desiccation and pH extremes, making management of these infections particularly challenging in the intensive care and burns units of hospitals [6].
A. baumannii has been recognized as a threat since the 1970s [4, 7] due to the rapid development of resistance to a wide range of antibiotics, including last-resort treatments such as carbapenems [8–11]. Often, there are very limited or no remaining options to treat A. baumannii infections [3, 12]. In 2017, this prompted the World Health Organization (WHO) to recognize carbapenem-resistant A. baumannii (CRAB) as the critical, number 1 priority among a published list of 12 antibiotic-resistant bacteria that pose the greatest threat to modern medicine, underlining the clinical significance and global burden of infections caused by CRAB [13].
Here, we discuss the emergence, molecular mechanisms and global spread of CRAB. To develop a snapshot of the geographical distribution of genomes sequenced so far and their carbapenem resistance gene (CRG) repertoire, we explore over 3500 A. baumannii genomes deposited in the GenBank non-redundant and Whole Genome Shotgun (WGS) databases. We also examine the genomic context of CRGs in all 128 complete genomes to further understand the role of mobile genetic elements in the spread of CRGs in A. baumannii .
Global spread of carbapenem-resistant Acinetobacter baumannii
Carbapenem antibiotics such as meropenem and imipenem belong to the ß-lactam family and remain active against most ß-lactamase-producing organisms, including those with extended spectrum ß-lactamase enzymes [14]. Carbapenems are considered to be a front-line treatment for infections caused by multiply resistant bacteria [15], but carbapenem resistance is increasingly common in A. baumannii , imposing huge financial and healthcare burdens [8, 16–18].
The number of studies in PubMed reporting CRAB increased from a single report [19] in 2000 to over 266 in 2018, highlighting its global dissemination [20, 21]. This has been largely due to inter- and intra-hospital transfer of resistant strains over the last two decades [21–23]. One classic example involving both intra-hospital and international transfer was the dramatic increase in A. baumannii infections in soldiers injured in war zones in Iraq and Afghanistan between 2006–2008 [24–28]. These infections often were resistant to multiple antibiotics and one study showed that 37 % of isolates recovered from injured deployed military personnel were also resistant to carbapenems [29]. A subsequent study found that isolates recovered from injured soldiers were genetically related to those recovered on field hospital surfaces rather than pre-injury colonization or introduction at the time of injury [28]. It has been suggested that the return of soldiers from combat zones was an important factor that contributed to the epidemiology of A. baumannii infections in the USA [21].
Outbreaks caused by CRAB have been reported from civilian hospitals in the USA, Canada, South America, Europe, Africa, the Middle East, Southeast Asia, Australia and many more countries [11, 18, 23, 30–72]. Generally, these CRAB outbreaks have been caused by the spread of a few specific clones that were already resistant to a wide range of antibiotics [22, 41, 73]. Although it was initially thought that these clones were limited to Europe [43, 74], they have now been reported in different countries of all inhabited continents [16, 18, 75–86], raising widespread clinical concerns [9, 16, 56, 59, 78, 79, 83–90]. The two major clones responsible for most of these outbreaks are now commonly referred to as global clone 1 (GC1) and global clone 2 (GC2), but have also been referred to as international clones 1 and 2 [8, 16, 75, 78, 90, 91].
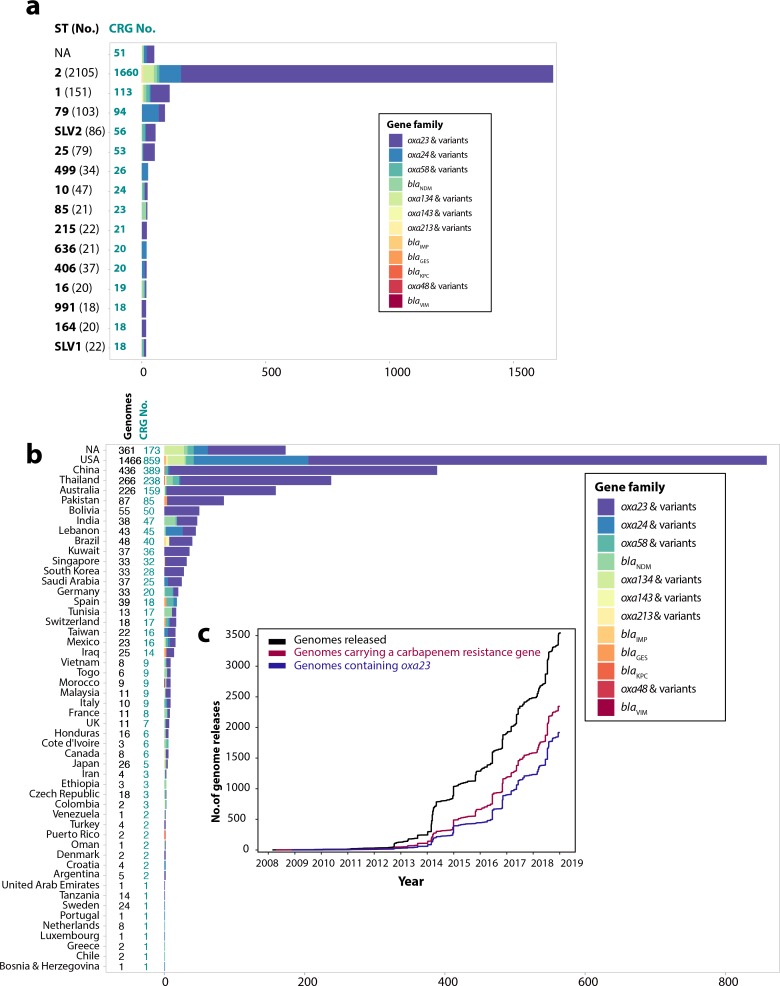
As of early April 2019, there were 3609 A. baumannii genomes available in the GenBank non-redundant and WGS databases (https://www.ncbi.nlm.nih.gov/genome/?term=Acinetobacter+baumannii). Here, these genomes were downloaded, and MLST types were determined in silico using MLST v2.16.1 (https://github.com/tseemann/mlst) followed by screening for antibiotic resistance genes using Abricate v0.8.10 (https://github.com/tseemann/abricate). These data were combined with the metadata available for each genome using R v3.5.2. Thirty-four duplicate, or passaged, isolates were removed from the analysis. Of the 3575 remaining genomes analysed here, 2364 (66 %) were members of GC1 (173 genomes) and GC2 (2191 genomes). These clones are defined here as ST1, representing GC1s according to the Institut Pasteur MLST scheme [92], and ST2, representing GC2s, along with their single-locus variants (ST1, SLV1, ST2 and SLV2 in Fig. 1a). However, ST2 itself is by far the dominant type, with 2105 genomes (59 %) among the available complete and draft genomes (Fig. 1a). This is also consistent with a large number of previous publications that continue to report outbreaks due to these two global clones, with GC2s accounting for the bulk of CRAB outbreaks [41, 43, 56, 77, 80, 81, 84, 85, 87, 93–96]. The global distribution of CRAB has been heavily influenced by the spread of GC2 isolates, with 1678 GC2 isolates also carrying at least 1 CRG. Only 109 of the 3575 genomes were GC1 isolates that harboured at least 1 CRG. Two other lineages contributed almost as much as GC1, with 91 and 53 belonging to ST79 and ST25, respectively.
Fig. 1.
Distribution of carbapenem resistance genes and trend of A. baumannii genomes released. (a) Distribution of carbapenem resistance genes in the 15 most prevalent sequence types (STs; according to the Institut Pasteur MLST scheme). https://github.com/tseemann/abricate.Numbers coloured turquoise indicate carbapenem resistance genes and black numbers show STs. SLV1 and SLV2 indicate single-locus variants of ST1 and ST2, respectively. All STs are based on the Institut Pasteur MLST scheme. (b) Geographical distribution of CRGs in A. baumannii genomes publicly available in the GenBank non-redundant and WGS databases (only countries with ≥1 CRG-containing genome are shown). Countries are shown on the y-axis and the numbers on x-axis indicate the number of CRGs. (c) Acinetobacter genomes released between 2008 and early April 2019. Black indicates total genome releases, red shows genomes with a carbapenem resistance gene and dark purple indicates genomes carrying the oxa23 gene. These figures were drawn using the ggplot2 package in R v3.5.2.
Molecular mechanisms of carbapenem resistance in A. baumannii
Many carbapenem resistance mechanisms have been described in A. baumannii , including alterations or loss of outer membrane proteins such as CarO [97, 98] and modifications of the AdeABC resistance nodulation division (RND) efflux pump [99, 100]. Although efflux modifications contribute to carbapenem resistance in A. baumannii , on their own they are are not sufficient to cause clinically relevant resistance [100]. Carbapenem resistance in A. baumannii is largely due to the horizontal acquisition of genes that encode carbapenem-hydrolyzing enzymes belonging to either Ambler class D (oxacillinases) or class B (metallo-ß-lactamases) [101–103].
Oxacillinases
Oxacillinase enzymes (OXAs) are a heterogeneous family [104] and, to date, several groups of carbapenem-hydrolyzing oxacillinases have been described in A. baumannii , most notably OXA-23, OXA-24, OXA-58, OXA-143, OXA-235 and an intrinsic OXA [105, 106], designated OXA-Ab for simplicity [89]. Genes encoding these acquired carbapenem-hydrolyzing enzymes are the main cause of carbapenem resistance in A. baumannii [107]. Generally, oxacillinases only hydrolyze carbapenems weakly and are often poorly expressed, hence they cannot cause clinically relevant levels of resistance on their own [108]. However, their expression is often enhanced by the insertion of an upstream IS, which enhances expression by providing a strong promoter, causing high resistance levels [109–111]. Reports of the prevalence of these genes vary by geographical distribution, but oxa23 is the most frequently described [34, 35, 47, 49, 50, 55, 58, 63, 66, 67, 75, 77, 106, 112, 113].
The intrinsic oxaAb
The oxaAb gene (also known as bla OXA51-like) occurs naturally in A. baumannii and is used as a marker for speciation [114]. So far, over 180 oxaAb variants (https://www.lahey.org/studies/) have been identified in A. baumannii strains [115–117]. The most common variants are oxa69 and oxa66, which are associated with members of GC1 and GC2, respectively [78, 89, 90]. It has been suggested that strains with an ISAba1 upstream of oxaAb can be carbapenem-resistant [109], but more than just overexpression of oxaAb is needed for significant levels of carbapenem resistance [118]. However, further work is required to understand the contribution of oxaAb overexpression in different genetic backgrounds and whether specific amino acid alterations in OXA-Ab also play a significant role.
Acquired oxacillinases
The oxa23 gene was first characterized in an A. baumannii strain recovered in Scotland in 1985, shortly after the introduction of carbapenems as therapeutic agents [119, 120]. Later, it was shown to have originated from the chromosome of Acinetobacter radioresistens , where it was mobilized into A. baumannii by ISAba1 [121]. To date, more than 25 variants of oxa23 have been identified (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/BETA-LACTAM).
The oxa58 gene was first identified on a plasmid from a multiply antibiotic-resistant A. baumannii recovered in France in 2003 [122], and to date six further variants have been found. The oxa58 gene has been associated with hospital outbreaks in Europe, the USA, South America, Australia and Africa [37, 39, 104, 105, 107, 123].
The oxa24 gene was originally identified in the chromosome of a CRAB isolate recovered in Spain [105]. However, now CRAB strains carrying the oxa24 gene, and its variants, often on small plasmids, have been recovered in hospital outbreaks worldwide [19, 53, 65, 105, 124–128]. This oxacillinase group consists of eight close relatives and amongst them OXA-25, OXA-26, OXA-40/OXA-24 (a sequencing error initially misclassified these as different) and OXA-72 are the most prevalent variants [129].
Other class D carbapenemases, such as those belonging to the OXA-143 and OXA-235 families, have also been associated with CRAB outbreaks in several countries [130], although they are reported less frequently and are generally considered to be minor causes for carbapenem resistance in A. baumannii .
Other carbapenemases
Other CRGs, such as those encoding metallo-ß-lactamases (MBL), bla VIM, bla IMP and bla NDM, or class A carbapenemases, bla KPC and bla GES-11, are also seen in A. baumannii [45]. However, unlike Enterobacteriaceae [131], they are not common in A. baumannii [45]. The bla NDM gene is often located in a mobile 10 kb ISAba125-bounded composite transposon called Tn125 [132], which is commonly seen on conjugative plasmids [133, 134].
Genome sequencing; opportunities and challenges
In recent years, whole-genome sequencing (WGS) technologies and advances in bioinformatic tools have revolutionized the study of bacterial pathogens, enabling gene screening and phylogenomic studies of outbreak strains with unprecedented resolution [135–137]. The first A. baumannii genome was sequenced in 2006, ATCC 17978 [138], followed by an epidemic GC1 strain in France, a non-clonal strain from human body lice [139] and a carbapenem-resistant GC2 strain recovered in Italy [140]. A few additional strains, including three CRAB GC1 strains (AB0057, AB056 and AB059) recovered from military patients at Walter Reed Army Medical Center, were also sequenced in the USA between 2008–2010 [9, 16]. However, as short-read sequencing technologies became more affordable and accessible, the number of genomes sequenced exponentially increased from 2014 onwards, and by early April 2019, over 3500 A. baumannii genomes were available (Fig. 1c).
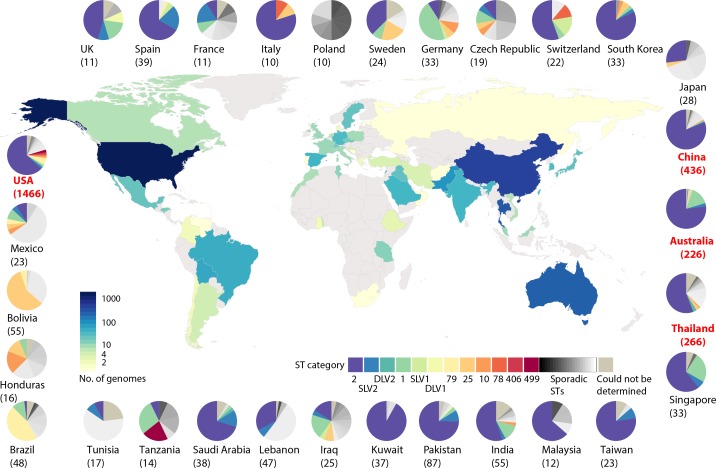
Amongst the 3575 non-redundant genome sequences studied here, 2345 (66 %) contained at least one CRG and of these, 1918 genomes (82 %) carry at least 1 copy of oxa23 (Fig. 1b), consistent with worldwide reports that oxa23 is overwhelmingly predominant in A. baumannii [34, 47, 49, 50, 55, 58, 63, 66, 77]. However, these sequences have a skewed geographical distribution, with 69 % (2481/3575) of all sequenced strains isolated from only five countries, namely the USA, PR China, Thailand, Australia and now Pakistan (Fig. 2). Notably, a total of 57 % (838/1466), 89 % (387/436), 84 % (224/266), 70 % (159/226) and 93 % (81/87) of genomes sequenced from the USA, PR China, Thailand, Australia and Pakistan, respectively, carry a CRG of one kind (Fig. 1b). However, it is unclear to what extent these CRG proportions reflect true population trends in these countries or whether they are a result of preferential selection of CRAB isolates for large-scale WGS projects, highlighting the need to sequence all A. baumannii that cause infection, regardless of their resistance phenotype. Hence, it is difficult to draw a clear picture of the true global A. baumannii population, including CRAB, unless more representative strains from Europe, the Middle East, Russia and Africa are sequenced and made publicly available.
Fig. 2.
Geographical distribution of A. baumannii genomes released. Countries are colour coded according to the number of genomes available as of April 2019. Countries with no genome available are coloured grey. Pie charts indicate the distribution of STs in each country. Sequence types (STs) were determined according to the Institut Pasteur MLST scheme.https://github.com/tseemann/mlst.
Furthermore, whilst GC1 and GC2 are the most common clones in many countries, this may not always be true. For instance, GC1 and GC2 strains do not seem to be the dominant types in South American countries (n=10/70), Tunisia (n=1/13), Tanzania (n=5/14), Poland (n=0/10) and Japan (n=7/26) (Fig. 2), although more genomes are needed to confirm this. Although GC2 appears to be the dominant type in some countries, for example Spain, the genomes show that oxa24 and oxa58 are the dominant CRGs rather than oxa23 (Fig. 2). However, given the relative paucity of genome data from these countries, caution needs to be exercised when drawing such conclusions.
Studying the genetic environment of antibiotic resistance genes, including CRGs, often provides valuable information on the origin, emergence, evolution and spread of resistance throughout bacterial populations [141]. Most of the currently available A. baumannii genomes have been sequenced using short-read technologies such as Illumina HiSeq or MiSeq. Although the data produced by these methods are sufficient to identify antibiotic resistance genes or draw phylogenetic trees, they lack the power to resolve complex resistance regions, which are often made up of numerous repeated elements [8, 142]. These regions tend to compromise assembly and can only be resolved manually via PCR and Sanger sequencing or by using long-read sequencing technologies such as Oxford Nanopore Technology (ONT) or Pacific Biosciences (PacBio) [143, 144]. Indeed, only 128 (4 %) genomes have been fully assembled and the majority of these were sequenced with PacBio (data not shown).
Genomic contexts and the role of mobile genetic elements in the spread of carbapenem resistance genes
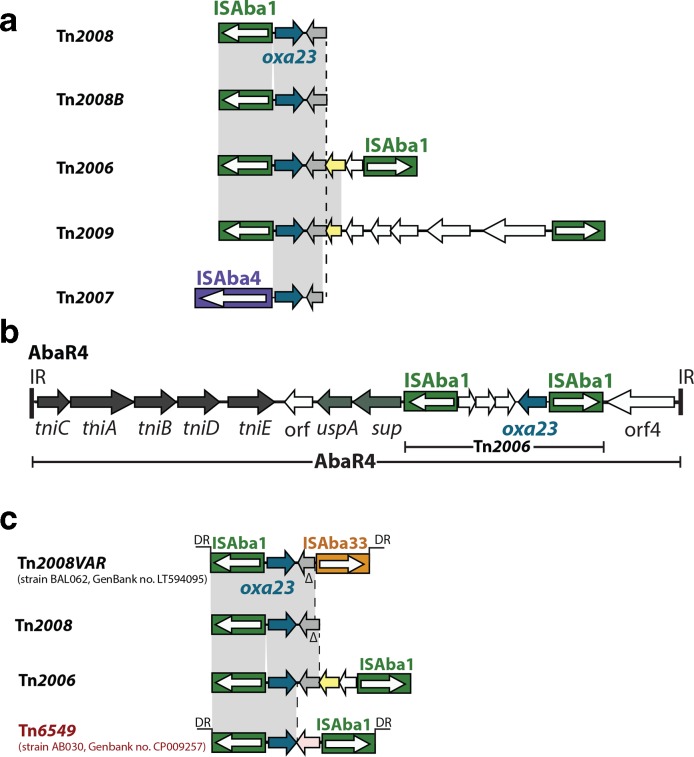
The oxa23 gene has moved into chromosomes and plasmids, on multiple occasions, via the transposons Tn2006, Tn2007, Tn2008, Tn2008B, Tn2009 and AbaR4 (Fig. 3a) [8, 9, 18, 112, 113, 145–148]. The oxa23-containing Tn2006 is the most commonly found transposon, and hence the most important in CRAB [110, 112]. It is a 4.8 kb class I transposon that consists of a central 2445 bp segment bounded by two inversely oriented copies of ISAba1 and generates a 9 bp target site duplication (TSD) upon insertion [110], characteristic of ISAba1 transposition [111]. Tn2006 can move independently and is found in many different chromosomal and plasmid contexts in distantly related A. baumannii strains [113]. In members of a distinct clade within GC1 lineage 1, Tn2006 has been found in a specific chromosomal location [8, 9, 18]. Tn2006 within AbaR4 is also located in the chromosomal comM gene in a member of another GC1 lineage, lineage 2 [70], where the A. baumannii Resistance Island (AbaR) is often present [149]. In GC2 isolates, oxa23 is often found in Tn2006 alone or in derivatives of AbaR4 as components of A. baumannii Genomic Resistance Island (AbGRI), which resides in the same location in comM as AbaR does in GC1s [11, 112]. Tn2008, Tn2008B and Tn2009 (Fig. 3 and Table S1, available in the online version of this article) are also seen in several chromosomal positions and are not associated with genomic islands [112].
Fig. 3.
Structure of transposons carrying the oxa23 gene. (a) Genes and open reading frames are shown using arrows. Filled boxes are insertion sequences (ISs) with ISAba1 coloured green, ISAba33 coloured dark orange and ISAba2 coloured dark purple. Arrows inside the boxes indicate the direction of transposition gene expression. The oxa23 gene is shown in dark blue and open reading frames encoding hypothetical proteins are shown in white. (b) Vertical bars marked as IR indicate inverted repeats of AbaR4. (c) DRs indicates direct repeats. Arrows located in the central segments of Tn2006, Tn2008 and Tn6549, coloured grey, yellow and pink, respectively, indicate open reading frames that encode unrelated hypothetical proteins.
Plasmids also play a crucial role in the spread of multiple carbapenem resistance genes in A. baumannii [45, 112, 130, 150]. For instance, large conjugative plasmids (80–130 kbp) encoding the RepAci6 replication initiation protein [71] are implicated in the spread of oxa23 in GC1 and GC2, as well as strains that do not belong to these clones [71, 91, 151]. To date, several oxa23 transposons have been found in different locations of related RepAci6 plasmids [71, 91, 146] . Moreover, oxa23 in Tn2006 was recently found in a RepAci1 plasmid where it was shown to be mobilized by a RepAci6 plasmid [152], further emphasizing the role of plasmids encoding RepAci6 in spreading and now in the mobilization of carbapenem resistance between disparate strains.
Currently, there are 128 completed A. baumannii genomes in GenBank, of which 91 carry at least one CRG. Examining these 91 genomes shows that 9 of the 11 oxa23-containing GC1s (Table S1) carry oxa23, either in Tn2006 in the chromosome or, in one case, Tn2006 in AbaR4 in comM [70]. In several strains, AbaR4 is found in a RepAci6 plasmid (Table S1). Members of GC2 often carry Tn2006 in the chromosome as part of AbGRI variants, with several strains carrying two oxa23 copies in this island (Table S1). Four strains, two from the Republic of Korea and two from Pakistan (Table S1), carried Tn2008B (Fig. 3a) in different chromosomal positions flanked by different TSDs. This suggests that Tn2008B is still quite active, as it was chromosomally incorporated on multiple occasions. Other GC2s carry either Tn2006 or Tn2008 in variants of RepAci6 plasmids (Table S1). Tn2009 appears most commonly in GC2 isolates from PR China (n=17) and the Republic of Korea (n=19), some with multiple chromosomal copies in tandem (Table S1). In non-GC1 or GC2 strains, chromosomal Tn2006 appears most often. Interestingly, 39 complete genomes harbour more than 1 copy of oxa23. Most often, there are multiple copies in the chromosome, although some also have oxa23 on a plasmid. This raises the question of whether these isolates, or those that harbour multiple families of CRG, have a selective advantage compared to those with fewer copies of oxa23, and warrants further investigation.
New oxa23-containing structures are still being identified, such as an ISAba1- and ISAba33-flanked transposon (Fig. 3c) described in 2016 [153]. Indeed, during the course of this work we found oxa23 in yet another novel structure with the features of a composite transposon, in the chromosome of a Canadian strain (BA30 in Fig. 3c). This 3902 bp transposon contains oxa23 flanked by directly oriented copies of ISAba1. Two copies of this transposon were found at different chromosomal locations (bases 3303155–3307056 and 4295797–4299698 in CP009257), with each copy flanked by novel 9 bp TSDs, providing evidence that it moves independently. Hence, we named this transposon Tn6549 (Fig. 3c). Tn6549 appears to be a derivative of Tn2008, rather than Tn2008B, as there is 27 bp between the start of the oxa23 gene and the ISAba1 sequence, which is indicative of Tn2008 [112]. The central segment of Tn6549 contains oxa23 and an open reading frame of unknown function (orf in Fig. 3c). A sequence identical to this open reading frame was found in the chromosome of Acinetobacter sp. strain ACNIH1 (GenBank accession no. CP026420) and fragments of this open reading frame are in several Acinetobacter lwoffii and Acinetobacter haemolyticus plasmids (e.g. GenBank accession nos CP038010 and CP032112).
The oxa58 gene is often embedded in the ISAba3::ISAba2-oxa58-ISAba3 structure, and carried on non-conjugative plasmids encoding both RepAci1 and RepAci10 [107, 140, 154]. This entire structure is now known to be surrounded by short inversely oriented inverted repeats similar to chromosomal dif sites, now referred to as pdif, targeted by XerC–XerD site-specific recombinases [154–156]. This dif module containing oxa58 is found in different plasmid backgrounds, indicating that it is a discrete mobile element that is responsible for the movement of oxa58, rather than the ISs that surround it [154–156]. Analysis of the completed genomes also indicated that oxa58 is mainly associated with dif modules often carried by similar small plasmids encoding RepAci1 and RepAci10 (Table S1).
The oxa24 gene is commonly seen in 8–12 kb plasmids that encode RepAci1 or RepAci2, as part of discrete dif modules flanked by pdif sites [155, 157–159]. This was also the case in all complete genomes with oxa24, (Table S1), adding further evidence that small plasmids, particularly those encoding RepAci1 and RepAci2, are a major force behind the global spread of oxa58 and oxa24.
The oxa253 gene, a variant of oxa143, may also occur in pdif modules, as it has been found near a single pdif site in a context similar to oxa24 in a RepAci2 plasmid [160]. The oxa235 gene, and its variants oxa236 and oxa237, are often found in single-nucleotide variants of a 5.2 kb ISAba1-bounded composite transposon called Tn6252, which has been found in chromosomes and plasmids [161–163].
Conclusions
Antibiotic resistance is on the rise and we are already running out of antibiotics to treat CRAB, which are unfortunately most commonly resistant to a wide range of additional antibiotics. Members of GC1 and GC2 are responsible for the bulk of globally disseminated multi-resistant A. baumannii , including CRAB. Although the current publicly available genomes provide an invaluable snapshot of the evolution and spread of CRAB throughout much of the world, the paucity of publicly available genome sequence data from regions such as Europe, the Middle East, Russia, Africa and South America has made it difficult to draw an accurate global picture of the spread of A. baumannii clones, CRGs and their phylogeny. Notably, whilst oxa23 is predominant globally, this is not always the case, particularly in countries such as Spain, Germany or Tunisia (Fig. 1b), further emphasizing the need for more sequencing coverage to understand why these different CRGs dominate in different regions. Sequencing strains from diverse regions is vital in understanding the evolutionary trajectory of the two major global clones, as well as other emerging clones, such as ST79 or ST25.
Expanding the use of long-read sequencing will facilitate a better understanding of the mobile elements responsible for moving CRGs and their broader contexts, while also enabling the characterization of further novel transposons and conjugative and mobilizable plasmids. It is now clear that alternative methods of horizontal gene transfer, such as dif modules and homologous recombination, play a larger role in the dissemination of CRGs than previously thought. Completing genome and plasmid assemblies will provide further knowledge regarding how widespread and important these mechanisms truly are. Indeed, this understanding will be vital in curtailing the future spread of CRGs in A. baumannii , as the issues of CRG spread via successful strains or clones and the spread of CRGs to susceptible strains via HGT are distinct problems that require different solutions. This will be crucial to identify molecular and epidemiological diagnostic markers to help identify resistant clones and track their spread. A more geographically uniform distribution of genome sequence data is also needed to further monitor plasmid movement and identify the true proportion of A. baumannii harbouring conjugative plasmids carrying bla NDM and other CRGs that are common in other species. With the lack of new antibiotics to treat CRAB, and the uncertainty about whether new drugs would even be effective, infection control policy and practice built upon the framework of these phylogenetic and epidemiological analyses are vital in stopping the spread of CRAB. Without such interventions, we will enter an era where common infections and minor injuries caused by CRAB can once again kill.
Data bibliography
1. GenBank non-redundant and Whole Genome Shotgun (WGS) databases. Complete ftp addresses used to retrieved the genomes are publicly available at https://www.ncbi.nlm.nih.gov/genome/?term=Acinetobacter+baumannii.
2. National Centre for Biotechnology Information (NCBI) Antimicrobial Resistance Reference Gene database, publicly available at https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/.
Supplementary Data
Funding information
This work and M.H. are supported by the University of Technology Sydney Chancellor’s Postdoctoral Research Fellowship (CPDRF PRO17-4005).
Acknowledgements
We would like to express our sincere gratitude to Professor Mark D. Adams of The Jackson Laboratory of Genomics for his support and helpful discussions.
Author contributions
Conceptualization: M.H. Data curation: M.H. and S.J.N. Formal analysis: M.H. and S.J.N. Funding acquisition: M.H. Investigation: M.H. and S.J.N. Methodology: M.H. and S.J.N. Visualization: M.H. and S.J.N. Writing – original draft: M.H. Writing – review and editing: M.H. and S.J.N.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CDS, coding sequence; CRG, carbapenem resistance gene; GC1, global clone 1; GC2, global clone 2; IS, insertion sequence; MAR, multiply-antibiotic resistant; MLST, multi-locus sequence typing; ONT, Oxford Nanopore Technology; PacBio, Pacific Biosciences; ST, sequence type; Tn, transposon; TSD, target site duplication.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table is available with the online version of this article.
References
- 1.Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Dis Clin North Am. 2016;30:313–322. doi: 10.1016/j.idc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 3.Towner KJ. Clinical importance and antibiotic resistance of Acinetobacter spp. Proceedings of a symposium held on 4-5 November 1996 at Eilat, Israel. J Med Microbiol. 1997;46:721–746. doi: 10.1099/00222615-46-9-721. [DOI] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 6.Cerqueira GM, Peleg AY. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life. 2011;63:1055–1060. doi: 10.1002/iub.533. [DOI] [PubMed] [Google Scholar]
- 7.Devaud M, Kayser FH, Bächi B. Transposon-mediated multiple antibiotic resistance in Acinetobacter strains. Antimicrob Agents Chemother. 1982;22:323–329. doi: 10.1128/AAC.22.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, et al. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom. 2016;2:e000052. doi: 10.1099/mgen.0.000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii . J Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi Y, Murray GL, Peleg AY. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med. 2015;36:85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nigro SJ, Hall RM. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother. 2012;67:1342–1346. doi: 10.1093/jac/dks037. [DOI] [PubMed] [Google Scholar]
- 12.Visca P, Seifert H, Towner KJ. Acinetobacter infection--an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 13.WHO Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organisation. 2017 [Google Scholar]
- 14.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Codjoe F, Donkor E. Carbapenem resistance: a review. Med Sci. 2017;6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii . Antimicrob Agents Chemother. 2010;54:3569–3577. doi: 10.1128/AAC.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez F, Ponce-Terashima R, Adams MD, Bonomo RA. Are we closing in on an "elusive enemy"? The current status of our battle with Acinetobacter baumannii . Virulence. 2011;2:86–90. doi: 10.4161/viru.2.2.15748. [DOI] [PubMed] [Google Scholar]
- 18.Hamidian M, Hawkey J, Wick R, Holt KE, Hall RM. Evolution of a clade of Acinetobacter baumannii global clone 1, lineage 1 via acquisition of carbapenem- and aminoglycoside-resistance genes and dispersion of ISAba1. Microb Genom. 2019;5:0.000242. doi: 10.1099/mgen.0.000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afzal-Shah M, Woodford N, Livermore DM. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii . Antimicrob Agents Chemother. 2001;45:583–588. doi: 10.1128/AAC.45.2.583-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peleg AY, Paterson DL. Multidrug-resistant Acinetobacter: a threat to the antibiotic era. Intern Med J. 2006;36:479–482. doi: 10.1111/j.1445-5994.2006.01130.x. [DOI] [PubMed] [Google Scholar]
- 21.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarrilli R, Crispino M, Bagattini M, Barretta E, Di Popolo A, et al. Molecular epidemiology of sequential outbreaks of Acinetobacter baumannii in an intensive care unit shows the emergence of carbapenem resistance. J Clin Microbiol. 2004;42:946–953. doi: 10.1128/JCM.42.3.946-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Broek PJ, Arends J, Bernards AT, De Brauwer E, Mascini EM, et al. Epidemiology of multiple Acinetobacter outbreaks in the Netherlands during the period 1999-2001. Clin Microbiol Infect. 2006;12:837–843. doi: 10.1111/j.1469-0691.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention (CDC) Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004. MMWR Morb Mortal Wkly Rep. 2004;53:1063–1066. [PubMed] [Google Scholar]
- 25.Calhoun JH, Murray CK, Manring MM. Multidrug-Resistant organisms in military wounds from Iraq and Afghanistan. Clin Orthop Relat Res. 2008;466:1356–1362. doi: 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith ME, Lazarus DR, Mann PB, Boger JA, Hospenthal DR, et al. Acinetobacter skin carriage among US army soldiers deployed in Iraq. Infect Control Hosp Epidemiol. 2007;28:720–722. doi: 10.1086/518966. [DOI] [PubMed] [Google Scholar]
- 27.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the walter reed army medical center. Antimicrob Agents Chemother. 2006;50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott P, Deye G, Srinivasan A, Murray C, Moran K, et al. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis. 2007;44:1577–1584. doi: 10.1086/518170. [DOI] [PubMed] [Google Scholar]
- 29.Davis KA, Moran KA, McAllister CK, Gray PJ. Multidrug-Resistant Acinetobacter extremity infections in soldiers. Emerg Infect Dis. 2005;11:1218–1224. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lolans K, Rice TW, Munoz-Price LS, Quinn JP. Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob Agents Chemother. 2006;50:2941–2945. doi: 10.1128/AAC.00116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dy ME, Nord JA, LaBombardi VJ, Kislak JW. The emergence of resistant strains of Acinetobacter baumannii: clinical and infection control implications. Infect Control Hosp Epidemiol. 1999;20:565–567. doi: 10.1086/501673. [DOI] [PubMed] [Google Scholar]
- 32.Go ES, Urban C, Burns J, Kreiswirth B, Eisner W, et al. Clinical and molecular epidemiology of Acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet. 1994;344:1329–1332. doi: 10.1016/s0140-6736(94)90694-7. [DOI] [PubMed] [Google Scholar]
- 33.Boyd DA, Mataseje LF, Pelude L, Mitchell R, Bryce E, et al. Results from the Canadian nosocomial infection surveillance program for detection of carbapenemase-producing Acinetobacter spp. in Canadian hospitals, 2010-16. J Antimicrob Chemother. 2019;74:315–320. doi: 10.1093/jac/dky416. [DOI] [PubMed] [Google Scholar]
- 34.Villegas MV, Kattan JN, Correa A, Lolans K, Guzman AM, et al. Dissemination of Acinetobacter baumannii clones with OXA-23 carbapenemase in Colombian hospitals. Antimicrob Agents Chemother. 2007;51:2001–2004. doi: 10.1128/AAC.00226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva KE, Maciel WG, Croda J, Cayô R, Ramos AC, et al. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PLoS One. 2018;13:e0209367. doi: 10.1371/journal.pone.0209367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavares LCB, de Vasconcellos FM, de Sousa WV, Rocchetti TT, Mondelli AL, et al. Emergence and persistence of high-risk clones among MDR and XDR A. baumannii at a Brazilian Teaching Hospital. Front Microbiol. 2018;9:2898. doi: 10.3389/fmicb.2018.02898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merkier AK, Catalano M, Ramírez MS, Quiroga C, Orman B, et al. Polyclonal spread of bla OXA-23 and bla OXA-58 in Acinetobacter baumannii isolates from Argentina. J Infect Dev Ctries. 2008;2:235–240. doi: 10.3855/jidc.269. [DOI] [PubMed] [Google Scholar]
- 38.Opazo-Capurro A, San Martín I, Quezada-Aguiluz M, Morales-León F, Domínguez-Yévenes M, et al. Evolutionary dynamics of carbapenem-resistant Acinetobacter baumannii circulating in Chilean hospitals. Infect Genet Evol. 2019;73:93–97. doi: 10.1016/j.meegid.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 39.Sevillano E, Fernández E, Bustamante Z, Zabalaga S, Rosales I, et al. Emergence and clonal dissemination of carbapenem-hydrolysing OXA-58-producing Acinetobacter baumannii isolates in bolivia. J Med Microbiol. 2012;61:80–84. doi: 10.1099/jmm.0.032722-0. [DOI] [PubMed] [Google Scholar]
- 40.Schulte B, Goerke C, Weyrich P, Gröbner S, Bahrs C, et al. Clonal spread of meropenem-resistant Acinetobacter baumannii strains in hospitals in the Mediterranean region and transmission to south-west Germany. J Hosp Infect. 2005;61:356–357. doi: 10.1016/j.jhin.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Coelho JM, Turton JF, Kaufmann ME, Glover J, Woodford N, et al. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J Clin Microbiol. 2006;44:3623–3627. doi: 10.1128/JCM.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wybo I, Blommaert L, De Beer T, Soetens O, De Regt J, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in a Belgian university hospital after transfer of patients from Greece. J Hosp Infect. 2007;67:374–380. doi: 10.1016/j.jhin.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Nemec A, Krízová L, Maixnerová M, Diancourt L, van der Reijden TJK, et al. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J Antimicrob Chemother. 2008;62:484–489. doi: 10.1093/jac/dkn205. [DOI] [PubMed] [Google Scholar]
- 44.Corbella X, Montero A, Pujol M, Domínguez MA, Ayats J, et al. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii . J Clin Microbiol. 2000;38:4086–4095. doi: 10.1128/jcm.38.11.4086-4095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Da Silva G, Domingues S. Insights on the horizontal gene transfer of carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii . Microorganisms. 2016;4:29. doi: 10.3390/microorganisms4030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeannot K, Diancourt L, Vaux S, Thouverez M, Ribeiro A, et al. Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS One. 2014;9:e115452. doi: 10.1371/journal.pone.0115452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoeva T, Higgins PG, Bojkova K, Seifert H. Clonal spread of carbapenem-resistant OXA-23-positive Acinetobacter baumannii in a Bulgarian university hospital. Clin Microbiol Infect. 2008;14:723–727. doi: 10.1111/j.1469-0691.2008.02018.x. [DOI] [PubMed] [Google Scholar]
- 48.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, et al. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000-09) J Antimicrob Chemother. 2011;66:2767–2772. doi: 10.1093/jac/dkr390. [DOI] [PubMed] [Google Scholar]
- 49.El Bannah AMS, Nawar NN, Hassan RMM, Salem STB. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a tertiary care hospital in Egypt: clonal spread of blaOXA-23 . Microb Drug Resist. 2018;24:269–277. doi: 10.1089/mdr.2017.0057. [DOI] [PubMed] [Google Scholar]
- 50.Cheikh HB, Domingues S, Silveira E, Kadri Y, Rosário N, et al. Molecular characterization of carbapenemases of clinical Acinetobacter baumannii-calcoaceticus complex isolates from a University Hospital in Tunisia. 3 Biotech. 2018;8:297. doi: 10.1007/s13205-018-1310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marais E, de Jong G, Ferraz V, Maloba B, Dusé AG. Interhospital transfer of pan-resistant Acinetobacter strains in johannesburg, South Africa. Am J Infect Control. 2004;32:278–281. doi: 10.1016/j.ajic.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Kumburu HH, Sonda T, van Zwetselaar M, Leekitcharoenphon P, Lukjancenko O, et al. Using WGS to identify antibiotic resistance genes and predict antimicrobial resistance phenotypes in MDR Acinetobacter baumannii in Tanzania. J Antimicrob Chemother. 2019;74:1484–1493. doi: 10.1093/jac/dkz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasiri MJ, Zamani S, Fardsanei F, Arshadi M, Bigverdi R, et al. Prevalence and mechanisms of carbapenem resistance in Acinetobacter baumannii: a comprehensive systematic review of cross-sectional studies from Iran. Microb Drug Resist. 2019;25:2018.0435. doi: 10.1089/mdr.2018.0435. [DOI] [PubMed] [Google Scholar]
- 54.Mugnier P, Poirel L, Pitout M, Nordmann P. Carbapenem-resistant and OXA-23-producing Acinetobacter baumannii isolates in the united arab emirates. Clin Microbiol Infect. 2008;14:879–882. doi: 10.1111/j.1469-0691.2008.02056.x. [DOI] [PubMed] [Google Scholar]
- 55.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf cooperation Council states: dominance of OXA-23-type producers. J Clin Microbiol. 2015;53:896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed SS, Alp E, Ulu-Kilic A, Dinc G, Aktas Z, et al. Spread of carbapenem-resistant international clones of Acinetobacter baumannii in turkey and azerbaijan: a collaborative study. Eur J Clin Microbiol Infect Dis. 2016;35:1463–1468. doi: 10.1007/s10096-016-2685-x. [DOI] [PubMed] [Google Scholar]
- 57.Marchaim D, Navon-Venezia S, Leavitt A, Chmelnitsky I, Schwaber MJ, et al. Molecular and epidemiologic study of polyclonal outbreaks of multidrug-resistant Acinetobacter baumannii infection in an Israeli Hospital. Infect Control Hosp Epidemiol. 2007;28:945–950. doi: 10.1086/518970. [DOI] [PubMed] [Google Scholar]
- 58.Al Atrouni A, Hamze M, Jisr T, Lemarié C, Eveillard M, et al. Wide spread of OXA-23-producing carbapenem-resistant Acinetobacter baumannii belonging to clonal complex II in different hospitals in Lebanon. Int J Infect Dis. 2016;52:29–36. doi: 10.1016/j.ijid.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 59.Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol. 2014;63:50–55. doi: 10.1099/jmm.0.063925-0. [DOI] [PubMed] [Google Scholar]
- 60.Ng DHL, Marimuthu K, Lee JJ, Khong WX, Ng OT, et al. Environmental colonization and onward clonal transmission of carbapenem-resistant Acinetobacter baumannii (CRAB) in a medical intensive care unit: the case for environmental hygiene. Antimicrob Resist Infect Control. 2018;7:51. doi: 10.1186/s13756-018-0343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan TY, Poh K, Ng SY. Molecular typing of imipenem-resistant Acinetobacter baumannii-calcoaceticus complex in a Singapore Hospital where carbapenem resistance is endemic. Infect Control Hosp Epidemiol. 2007;28:941–944. doi: 10.1086/518964. [DOI] [PubMed] [Google Scholar]
- 62.Gurung M, Rho JS, Lee YC, Kim HS, Moon SY, et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii sequence type 191 in a Korean hospital. Infect Genet Evol. 2013;19:219–222. doi: 10.1016/j.meegid.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 63.Jeon H, Kim S, Kim MH, Kim SY, Nam D, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates from a Korean hospital that carry blaOXA-23 . Infect Genet Evol. 2018;58:232–236. doi: 10.1016/j.meegid.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Lee Y, Kim YR, Kim J, Park YJ, Song W, et al. Increasing prevalence of bla OXA-23-carrying Acinetobacter baumannii and the emergence of bla OXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn Microbiol Infect Dis. 2013;77:160–163. doi: 10.1016/j.diagmicrobio.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 65.Kuo SC, Huang WC, Huang TW, Wang HY, Lai JF, et al. Molecular epidemiology of emerging blaOXA-23-Like and blaOXA-24-Like-carrying Acinetobacter baumannii in Taiwan. Antimicrob Agents Chemother. 2018;62:e01215–01217. doi: 10.1128/AAC.01215-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang TH, Leu YS, Wang NY, Liu CP, Yan TR. Prevalence of different carbapenemase genes among carbapenem-resistant Acinetobacter baumannii blood isolates in Taiwan. Antimicrob Resist Infect Control. 2018;7:123. doi: 10.1186/s13756-018-0410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y, Zhou J, Zhou H, Yang Q, Wei Z, et al. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J Antimicrob Chemother. 2010;65:644–650. doi: 10.1093/jac/dkq027. [DOI] [PubMed] [Google Scholar]
- 68.Alshahni MM, Asahara M, Kawakami S, Fujisaki R, Matsunaga N, et al. Genotyping of Acinetobacter baumannii strains isolated at a Japanese hospital over five years using targeted next-generation sequencing. J Infect Chemother. 2015;21:512–515. doi: 10.1016/j.jiac.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Teerawattanapong N, Panich P, Kulpokin D, Na Ranong S, Kongpakwattana K, et al. A systematic review of the burden of multidrug-resistant healthcare-associated infections among intensive care unit patients in Southeast Asia: the rise of multidrug-resistant Acinetobacter baumannii . Infect Control Hosp Epidemiol. 2018;39:525–533. doi: 10.1017/ice.2018.58. [DOI] [PubMed] [Google Scholar]
- 70.Hamidian M, Hall RM. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2011;66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 71.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. A conjugative plasmid carrying the carbapenem resistance gene bla OXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother. 2014;69:2625–2628. doi: 10.1093/jac/dku188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peleg AY, Franklin C, Bell JM, Spelman DW. Emergence of carbapenem resistance in Acinetobacter baumannii recovered from blood cultures in Australia. Infect Control Hosp Epidemiol. 2006;27:759–761. doi: 10.1086/507012. [DOI] [PubMed] [Google Scholar]
- 73.Towner KJ, Levi K, Vlassiadi M. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin Microbiol Infect. 2008;14:161–167. doi: 10.1111/j.1469-0691.2007.01911.x. [DOI] [PubMed] [Google Scholar]
- 74.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii . Nat Rev Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 75.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 76.Saranathan R, Vasanth V, Vasanth T, Shabareesh PRV, Shashikala P, et al. Emergence of carbapenem non-susceptible multidrug resistant Acinetobacter baumannii strains of clonal complexes 103(B) and 92(B) harboring OXA-type carbapenemases and metallo-beta-lactamases in Southern India. Microbiol Immunol. 2015;59:277–284. doi: 10.1111/1348-0421.12252. [DOI] [PubMed] [Google Scholar]
- 77.Pournaras S, Dafopoulou K, Del Franco M, Zarkotou O, Dimitroulia E, et al. Predominance of international clone 2 OXA-23-producing- Acinetobacter baumannii clinical isolates in Greece, 2015: results of a nationwide study. Int J Antimicrob Agents. 2017;49:749–753. doi: 10.1016/j.ijantimicag.2017.01.028. [DOI] [PubMed] [Google Scholar]
- 78.Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii . J Antimicrob Chemother. 2010;65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 79.Peymani A, Higgins PG, Nahaei M-R, Farajnia S, Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int J Antimicrob Agents. 2012;39:526–528. doi: 10.1016/j.ijantimicag.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 80.Milan A, Furlanis L, Cian F, Bressan R, Luzzati R, et al. Epidemic dissemination of a carbapenem-resistant Acinetobacter baumannii clone carrying armA two years after its first isolation in an italian hospital. Microb Drug Resist. 2016;22:668–674. doi: 10.1089/mdr.2015.0167. [DOI] [PubMed] [Google Scholar]
- 81.Dortet L, Bonnin RA, Girlich D, Imanci D, Bernabeu S, et al. Whole-genome sequence of a european clone II and OXA-72-producing Acinetobacter baumannii strain from serbia. Genome Announc. 2015;3:e01390–15. doi: 10.1128/genomeA.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brahmi S, Touati A, Cadière A, Djahmi N, Pantel A, et al. First description of two sequence type 2 Acinetobacter baumannii isolates carrying OXA-23 carbapenemase in pagellus acarne fished from the mediterranean sea near bejaia, algeria. Antimicrob Agents Chemother. 2016;60:2513–2515. doi: 10.1128/AAC.02384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bakour S, Olaitan AO, Ammari H, Touati A, Saoudi S, et al. Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in algeria: first case report. Microb Drug Resist. 2015;21:279–285. doi: 10.1089/mdr.2014.0214. [DOI] [PubMed] [Google Scholar]
- 84.Al-Sultan AA, Evans BA, Aboulmagd E, Al-Qahtani AA, Bohol MFF, et al. Dissemination of multiple carbapenem-resistant clones of Acinetobacter baumannii in the Eastern District of Saudi Arabia. Front Microbiol. 2015;6:634. doi: 10.3389/fmicb.2015.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levy-Blitchtein S, Roca I, Plasencia-Rebata S, Vicente-Taboada W, Velásquez-Pomar J, et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg Microbes Infect. 2018;7:1–9. doi: 10.1038/s41426-018-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Correa A, Del Campo R, Escandón-Vargas K, Perenguez M, Rodríguez-Baños M, et al. Distinct genetic diversity of carbapenem-resistant Acinetobacter baumannii from colombian hospitals. Microb Drug Resist. 2018;24:48–54. doi: 10.1089/mdr.2016.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother. 2013;57:5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mathlouthi N, Ben Lamine Y, Somai R, Bouhalila-Besbes S, Bakour S, et al. Incidence of OXA-23 and OXA-58 carbapenemases coexpressed in clinical isolates of Acinetobacter baumannii in tunisia. Microb Drug Resist. 2018;24:136–141. doi: 10.1089/mdr.2016.0306. [DOI] [PubMed] [Google Scholar]
- 89.Nigro SJ, Post V, Hall RM. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother. 2011;66:1504–1509. doi: 10.1093/jac/dkr163. [DOI] [PubMed] [Google Scholar]
- 90.Post V, Hall RM. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii . Antimicrob Agents Chemother. 2009;53:2667–2671. doi: 10.1128/AAC.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, et al. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio. 2014;5:e00963–13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ko KS. Antibiotic-resistant clones in gram-negative pathogens: presence of global clones in Korea. J Microbiol. 2019;57:195–202. doi: 10.1007/s12275-019-8491-2. [DOI] [PubMed] [Google Scholar]
- 94.Matsui M, Suzuki M, Suzuki M, Yatsuyanagi J, Watahiki M, et al. Distribution and molecular characterization of Acinetobacter baumannii international clone II lineage in Japan. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.02190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giannouli M, Antunes LCS, Marchetti V, Triassi M, Visca P, et al. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282. doi: 10.1186/1471-2334-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karah N, Giske CG, Sundsfjord A, Samuelsen Ørjan. A diversity of OXA-carbapenemases and class 1 integrons among carbapenem-resistant Acinetobacter baumannii clinical isolates from Sweden belonging to different international clonal lineages. Microb Drug Resist. 2011;17:545–549. doi: 10.1089/mdr.2011.0089. [DOI] [PubMed] [Google Scholar]
- 97.Lee Y, Kim CK, Lee H, Jeong SH, Yong D, et al. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the blaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii . Antimicrob Agents Chemother. 2011;55:361–363. doi: 10.1128/AAC.01672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mussi MA, Limansky AS, Relling V, Ravasi P, Arakaki A, et al. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J Bacteriol. 2011;193:4736–4748. doi: 10.1128/JB.01533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coyne S, Courvalin P, Périchon B. Efflux-Mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naas T, Nordmann P. OXA-type beta-lactamases. Curr Pharm Des. 1999;5:865–879. [PubMed] [Google Scholar]
- 102.Ambler RP, Coulson AFW, Frère JM, Ghuysen JM, Joris B, et al. A standard numbering scheme for the class A β -lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 104.Poirel L, Mansour W, Bouallegue O, Nordmann P. Carbapenem-resistant Acinetobacter baumannii isolates from Tunisia producing the OXA-58-like carbapenem-hydrolyzing oxacillinase OXA-97. Antimicrob Agents Chemother. 2008;52:1613–1617. doi: 10.1128/AAC.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 107.Bertini A, Poirel L, Bernabeu S, Fortini D, Villa L, et al. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii . Antimicrob Agents Chemother. 2007;51:2324–2328. doi: 10.1128/AAC.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Héritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii . Antimicrob Agents Chemother. 2005;49:3198–3202. doi: 10.1128/AAC.49.8.3198-3202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii . FEMS Microbiol Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 110.Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii . Antimicrob Agents Chemother. 2007;51:1530–1533. doi: 10.1128/AAC.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii . J Bacteriol. 2009;191:2414–2418. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nigro SJ, Hall RM. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother. 2016;71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 113.Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii . Emerg Infect Dis. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Héritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, et al. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii . Antimicrob Agents Chemother. 2005;49:4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brown S, Amyes SGB. The sequences of seven class D beta-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin Microbiol Infect. 2005;11:326–329. doi: 10.1111/j.1469-0691.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 116.Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006;57:1–3. doi: 10.1093/jac/dki425. [DOI] [PubMed] [Google Scholar]
- 117.Evans BA, Hamouda A, Towner KJ, Amyes SGB. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii . Clin Microbiol Infect. 2008;14:268–275. doi: 10.1111/j.1469-0691.2007.01919.x. [DOI] [PubMed] [Google Scholar]
- 118.Nigro SJ, Hall RM. Does the intrinsic oxaAb (bla OXA-51-like) gene of Acinetobacter baumannii confer resistance to carbapenems when activated by ISAba1? J Antimicrob Chemother. 2018;73:3518–3520. doi: 10.1093/jac/dky334. [DOI] [PubMed] [Google Scholar]
- 119.Paton R, Miles RS, Hood J, Amyes SG, Miles RS, et al. Ari 1: beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii . Int J Antimicrob Agents. 1993;2:81–87. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 120.Scaife W, Young HK, Paton RH, Amyes SG. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36:585–586. doi: 10.1093/jac/36.3.585. [DOI] [PubMed] [Google Scholar]
- 121.Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother. 2008;52:1252–1256. doi: 10.1128/AAC.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poirel L, Marqué S, Héritier C, Segonds C, Chabanon G, et al. OXA-58, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii . Antimicrob Agents Chemother. 2005;49:202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Héritier C, Dubouix A, Poirel L, Marty N, Nordmann P. A nosocomial outbreak of Acinetobacter baumannii isolates expressing the carbapenem-hydrolysing oxacillinase OXA-58. J Antimicrob Chemother. 2005;55:115–118. doi: 10.1093/jac/dkh500. [DOI] [PubMed] [Google Scholar]
- 124.Pailhoriès H, Kempf M, Belmonte O, Joly-Guillou ML, Eveillard M. First case of OXA-24-producing Acinetobacter baumannii in cattle from Reunion Island, France. Int J Antimicrob Agents. 2016;48:763–764. doi: 10.1016/j.ijantimicag.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Sarı AN, Biçmen M, Gülay Z. The first report on the outbreak of OXA-24/40-like carbapenemase-producing Acinetobacter baumannii in turkey. Jpn J Infect Dis. 2013;66:439–442. doi: 10.7883/yoken.66.439. [DOI] [PubMed] [Google Scholar]
- 126.Todorova B, Velinov T, Ivanov I, Dobreva E, Kantardjiev T. First detection of OXA-24 carbapenemase-producing Acinetobacter baumannii isolates in Bulgaria. World J Microbiol Biotechnol. 2014;30:1427–1430. doi: 10.1007/s11274-013-1562-3. [DOI] [PubMed] [Google Scholar]
- 127.Chen Y, Yang Y, Liu L, Qiu G, Han X, et al. High prevalence and clonal dissemination of OXA-72-producing Acinetobacter baumannii in a Chinese hospital: a cross sectional study. BMC Infect Dis. 2018;18:491. doi: 10.1186/s12879-018-3359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang L, Sun L, Yan Y. Clonal spread of carbapenem resistant Acinetobacter baumannii ST92 in a Chinese Hospital during a 6-year period. J Microbiol. 2013;51:113–117. doi: 10.1007/s12275-013-2341-4. [DOI] [PubMed] [Google Scholar]
- 129.Evans BA, Amyes SG. Oxa beta-lactamases. Clin Microbiol Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rodríguez CH, Nastro M, Famiglietti A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Revista Argentina de Microbiología. 2018;50:327–333. doi: 10.1016/j.ram.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 131.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae . Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bontron S, Nordmann P, Poirel L. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii . Antimicrob Agents Chemother. 2016;60:7245–7251. doi: 10.1128/AAC.01755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, Zhang Z, Hao Q, Wu J, Xiao J, et al. Complete Genome Sequence of Acinetobacter baumannii ZW85-1. Genome Announc. 2014;23:e01083–13. doi: 10.1128/genomeA.01083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, et al. Complete sequence of the bla (NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother. 2013;68:1681–1682. doi: 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 135.Quainoo S, Coolen JPM, van Hijum S, Huynen MA, Melchers WJG, et al. Whole-Genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Popovich KJ, Snitkin ES. Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr Infect Dis Rep. 2017;19:15. doi: 10.1007/s11908-017-0570-0. [DOI] [PubMed] [Google Scholar]
- 137.Schürch AC, van Schaik W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci. 2017;1388:108–120. doi: 10.1111/nyas.13310. [DOI] [PubMed] [Google Scholar]
- 138.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii . PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008;52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31 doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holt KE, Hamidian M, Kenyon JJ, Wynn MT, Hawkey J, et al. Genome sequence of Acinetobacter baumannii strain A1, an early example of antibiotic-resistant global clone 1. Genome Announc. 2015;3:e00032–15. doi: 10.1128/genomeA.00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wick RR, Judd LM, Gorrie CL, Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 2017;3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Greig DR, Dallman TJ, Hopkins KL, Jenkins C. MinION nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli . Microb Genom. 2018;4:0.000213. doi: 10.1099/mgen.0.000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gheorghe I, Novais Ângela, Grosso F, Rodrigues C, Chifiriuc MC, et al. Snapshot on carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in Bucharest hospitals reveals unusual clones and novel genetic surroundings for bla OXA-23 . J Antimicrob Chemother. 2015;70:1016–1020. doi: 10.1093/jac/dku527. [DOI] [PubMed] [Google Scholar]
- 146.Nigro SJ, Holt KE, Pickard D, Hall RM. Carbapenem and amikacin resistance on a large conjugative Acinetobacter baumannii plasmid. J Antimicrob Chemother. 2015;70:1259–1261. doi: 10.1093/jac/dku486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim DH, Park YK, Ko KS. Variations of AbaR4-type resistance islands in Acinetobacter baumannii isolates from South Korea. Antimicrob Agents Chemother. 2012;56:4544–4547. doi: 10.1128/AAC.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou H, Zhang T, Yu D, Pi B, Yang Q, et al. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother. 2011;55:4506–4512. doi: 10.1128/AAC.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hamidian M, Hall RM. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 - Structure, origin and evolution. Drug Resist Updat. 2018;41:26–39. doi: 10.1016/j.drup.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 150.Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, et al. Carbapenem-resistant Acinetobacter baumannii and enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev. 2017;30:1–22. doi: 10.1128/CMR.00042-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii . Antimicrob Agents Chemother. 2010;54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blackwell GA, Hall RM. Mobilisation of a small Acinetobacter plasmid carrying an oriT transfer origin by conjugative RepAci6 plasmids. Plasmid. 2019;103:36–44. doi: 10.1016/j.plasmid.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 153.Schultz MB, Pham Thanh D, Tran Do Hoan N, Wick RR, Ingle DJ, et al. Repeated local emergence of carbapenem-resistant Acinetobacter baumannii in a single hospital ward. Microb Genom. 2016;2:e000050. doi: 10.1099/mgen.0.000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Poirel L, Nordmann P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene bla OXA-58 in Acinetobacter baumannii . Antimicrob Agents Chemother. 2006;50:1442–1448. doi: 10.1128/AAC.50.4.1442-1448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Blackwell GA, Hall RM. The tet39 Determinant and the msrE-mphE Genes in Acinetobacter Plasmids Are Each Part of Discrete Modules Flanked by Inversely Oriented pdif (XerC-XerD) Sites. Antimicrob Agents Chemother. 2017;61:e00780–17. doi: 10.1128/AAC.00780-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cameranesi MM, Morán-Barrio J, Limansky AS, Repizo GD, Viale AM. Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a bla OXA-58- and TnaphA6-resistance module in Acinetobacter baumannii . Front Microbiol. 2018;9:66. doi: 10.3389/fmicb.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, et al. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii . Antimicrob Agents Chemother. 2009;53:3528–3533. doi: 10.1128/AAC.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Grosso F, Quinteira S, Poirel L, Novais Ângela, Peixe L. Role of common bla OXA-24/OXA-40 - carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob Agents Chemother. 2012;56:3969–3972. doi: 10.1128/AAC.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, et al. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob Agents Chemother. 2010;54:2724–2727. doi: 10.1128/AAC.01674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Girlich D, Damaceno QS, Oliveira AC, Nordmann P. OXA-253, a variant of the carbapenem-hydrolyzing class D β-lactamase OXA-143 in Acinetobacter baumannii . Antimicrob Agents Chemother. 2014;58:2976–2978. doi: 10.1128/AAC.02640-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hamidian M, Nigro SJ, Hartstein RM, Hall RM. RCH51, a multiply antibiotic-resistant Acinetobacter baumannii ST103IP isolate, carries resistance genes in three plasmids, including a novel potentially conjugative plasmid carrying oxa235 in transposon Tn6252. J Antimicrob Chemother. 2017;72:1907–1910. doi: 10.1093/jac/dkx069. [DOI] [PubMed] [Google Scholar]
- 162.Higgins PG, Pérez-Llarena FJ, Zander E, Fernández A, Bou G, et al. OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii . Antimicrob Agents Chemother. 2013;57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ou HY, Kuang SN, He X, Molgora BM, Ewing PJ, et al. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep. 2015;5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.