Abstract
Background
The plant homeodomain (PHD) finger is a Cys4HisCys3-type zinc finger which promotes protein-protein interactions and binds to the cis-acting elements in the promoter regions of target genes. In Medicago truncatula, five PHD homologues with full-length sequence were identified. However, the detailed function of PHD genes was not fully addressed.
Results
In this study, we characterized the function of MtPHD6 during plant responses to drought stress. MtPHD6 was highly induced by drought stress. Ectopic expression of MtPHD6 in Arabidopsis enhanced tolerance to osmotic and drought stresses. MtPHD6 transgenic plants exhibited decreased water loss rate, MDA and ROS contents, and increased leaf water content and antioxidant enzyme activities under drought condition. Global transcriptomic analysis revealed that MtPHD6 reprogramed transcriptional networks in transgenic plants. Expression levels of ABA receptor PYR/PYLs, ZINC FINGER, AP2/EREBP and WRKY transcription factors were mainly up-regulated after transformation of MtPHD6. Interaction network analysis showed that ZINC FINGER, AP2/EREBP and WRKY interacted with each other and downstream stress induced proteins.
Conclusions
We proposed that ZINC FINGER, AP2/EREBP and WRKY transcription factors were activated through ABA dependent and independent pathways to increase drought tolerance of MtPHD6 transgenic plants.
Keywords: ABA, Drought stress, Interaction network, Medicago truncatula, PHD finger protein, Zinc finger protein
Background
The plant homeodomain (PHD) finger was named according to the Arabidopsis Histone acetyltransferases 3.1 (HAT3.1) [1]. As a common structural motif, it is found in all eukaryotic genomes and typically shows a Cys4HisCys3-type zinc finger signature [2, 3]. Along with promoting protein-protein interactions, the PHD-finger motif can also bind to the cis-acting elements in the promoter regions of target genes [4]. It has been widely reported that PHD-finger-containing proteins are localized in nucleus and most likely to be chromatin-mediated transcriptional regulators [3, 5]. In plants, many studies have shown that PHD-finger-containing proteins are involved in diverse physiological processes, including the epigenetic silencing, regulation of the flowering time, and the growth and development of roots [6–9].
Alfin1 from Medicago sativa (alfalfa) is a member of plant-specific PHD-finger protein subfamily. It was shown that alfin1 is a salt-inducible transcription factor and can regulate the expression of MsPRP2 gene, leading to enhanced salt tolerance [4, 10]. Alfin1-like (AL) proteins belonging to a small group of PHD-finger proteins were originally discovered to be a kind of transcription factor family in alfalfa [4]. Recently, more and more AL genes have been reported in many different plant species, including Arabidopsis thaliana, Oryza sativa, Glycine max, Brassica rapa, Brassica oleracea, Zea mays and Atriplex hortensis [5, 7, 11–15]. The expression of these genes is stress-responsive and varies due to stress types [7, 12]. Six GmPHD genes were identified from soybean and encoded Alfin1-type PHD finger proteins, which could response to diverse abiotic stresses. Ectopic expression of GmPHD2 in Arabidopsis improved salt tolerance of transgenic plants with decreased reactive oxygen species (ROS) [5]. GmPHD5 was capable of regulating histone crosstalk between methylated H3K4 and acetylated H3K14 in response to salinity stress, and could also recruit chromatin remodeling factors and salt stress induced transcription factors such as GmRD22 and GmGST to regulate their expression levels [16]. PHD fingers in ING (inhibitor of growth) homologues and AL proteins in Arabidopsis could bind to H3K4me3/2, the active histone markers in plants [6, 17]. AtAL6 played crucial roles in phosphate deficiency-induced root hair formation by binding to H3K4me3 of ETC1 through the PHD domain [6]. T-DNA insertion mutant and overexpression of AtAL7 exhibited a negative role in salt tolerance of A. thaliana [15]. AtAL5–overexpressing plants showed improved tolerance to salt, drought and cold stress [18]. Ectopic expression of AhAL1 contributed to improving survival rates of transgenic Arabidopsis plants under salt and drought conditions [13]. Above evidences suggested that AL/PHD genes play important roles in regulating plant responses to abiotic stresses by changing transcription and reading epigenetic histone modifications.
Forages are key components of sustainable agriculture. As a model plant of forage legume species, M. truncatula has been widely studied at the molecular level [19]; and has a close relationship to alfalfa which is the world’s most important forage legume [20]. However, forages are often grown in relatively severe environmental conditions, resulting in evolution of complicated protective mechanisms for survival [21]. Drought is the main environmental factor that limits plant growth, development and productivity in arid and semiarid regions [22]. Hence, the tolerance of drought is a vital breeding trait for forage legume. To date, the function of several transcription factors from M. truncatula has been dissected in plant responses to drought stress, including WXP1 and SPL8 [19, 23]. Overexpression of MtWXP1 in alfalfa facilitated plants to accumulate cuticular wax and improved the tolerance of drought [19]. However, the detailed function of MtPHDs in response to drought stress remains elusive.
In the present study, expression of MtPHD6 by drought stress was investigated. Drought tolerance of MtPHD6 transgenic Arabidopsis was identified in the physiological level. Water loss, oxidative damage and antioxidant enzyme activities were assayed. Genome-wide transcriptomic analysis of MtPHD6 transgenic plants under drought stress was performed to identify MtPHD6 mediated genes. Additionally, protein interaction networks modulated by MtPHD6 transgene were characterized. Our results provide further insights into the roles and the molecular mechanisms of MtPHD6 in regulating plant responses to drought stress.
Results
Characterization of MtPHDs family in M. truncatula
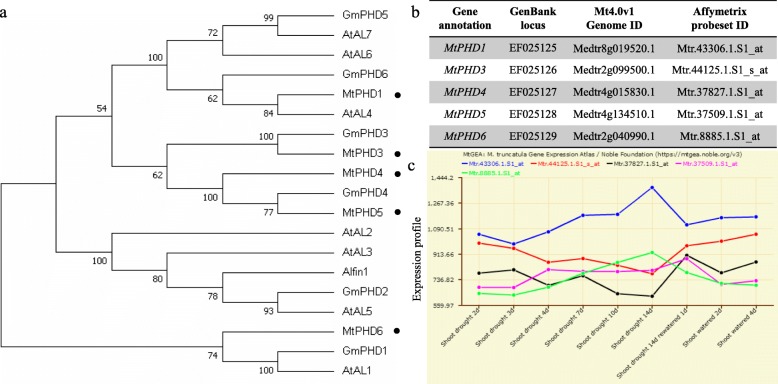
In M. truncatula, seven PHD homologues were identified and five full-length sequences were termed MtPHD1 and 3–6 (EF025125, EF025126, EF025127, EF025128, and EF025129) [5], among which MtPHD5 was almost identical to Alfin1 of M. sativa [4, 24]. Phylogenetic analysis of these five MtPHDs with ALs/PHDs from other plant species showed that MtPHDs were clustered into five different clades. However, MtPHD6 (EF025129/Medtr2g040990) was most distantly related to the other members of the MtPHDs family, and was more closely related to GmPHD1 and AtAL1 (Fig. 1a), which were significantly induced by PEG/drought and NaCl treatments [5, 18]. The detailed information of MtPHDs with Mt4.0 V1 genome (http://www.medicagogenome.org/home) and Affymetrix probeset IDs were presented (Fig. 1b). Affymetrix microarray data showed that MtPHD6 (Mtr.8885.1.S1_at) increased upon drought stress treatment, but decreased after re-watering (Fig. 1c).
Fig. 1.
Phylogenic tree of PHD-finger proteins from different species and expression analysis of MtPHDs under drought stress. a: The phylogenic tree was constructed using the neighbor-joining method in MEGA 7.0 software with 1000 bootstrap. Numbers on the figure are bootstrap values. The protein sequences are from Medicago truncatula (MtPHDs); Medicago sativa (Alfin1); Glycine max (GmPHDs); Arabidopsis thaliana (AtALs). Accession numbers were listed in Additional file 1: Table S1. b: Information of MtPHDs. c: Expression changes of MtPHDs under drought stress condition. The data are available from the M. truncatula Gene Expression Atlas (MtGEA) web server at https://mtgea.noble.org/v3/
MtPHD6-overexpressing plants showed increased tolerance to osmotic stress
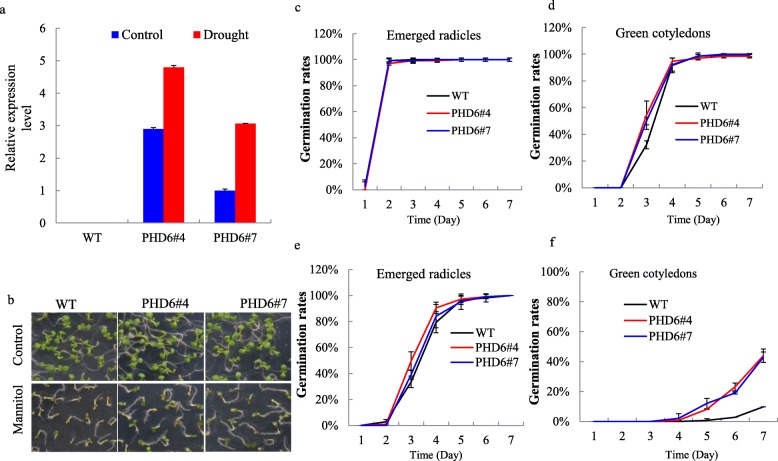
To test the function of MtPHD6 in response to abiotic stresses, MtPHD6 gene was cloned from M. truncatula using specific primers listed in Additional file 2: Table S2 and transgenic Arabidopsis plants over-expressing MtPHD6 were constructed. Drought treatment indeed significantly up-regulated the MtPHD6 expression in two transgenic lines (PHD6#4 and PHD6#7) when compared to control condition (Fig. 2a). These results showed that MtPHD6 is stress-inducible and might play roles in plant responses to drought stress.
Fig. 2.
Expression changes of MtPHD6 under drought stress and osmotic stress tolerance of MtPHD6 transgenic Arabidopsis. a: The relative expression level of MtPHD6 after drought 7 d for 12-day-old transgenic seedlings through qRT-PCR; b: Phenotypic changes after 300 mM mannitol treatment; c, e, Emerged radicles under control and osmotic stress conditions, respectively; d, f: Green cotyledons under control and osmotic stress conditions, respectively
Under 300 mM mannitol condition, MtPHD6 transgenic plants exhibited more vigorous growth than WT (Fig. 2b). There were no significant differences of emerged radicles and green cotyledons between transgenic lines and WT under normal growth condition (Fig. 2c, d). After 300 mM mannitol treatment for 7 d, the transgenic lines showed significantly higher percentages of green cotyledons (Fig. 2f), although emerged radicles between transgenic lines and WT had no significant differences (Fig. 2e). The results indicated that MtPHD6 might play a positive role in response to osmotic stress during seed germination stage.
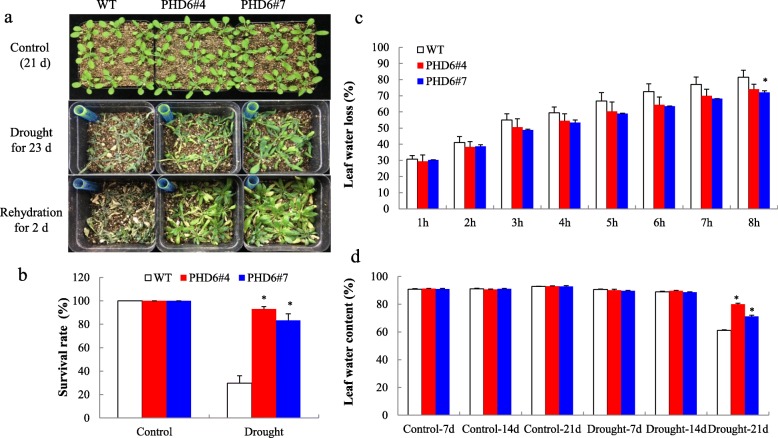
In order to further investigate drought tolerance, 12-day-old plants from normal growth condition were water-withheld for 23 d, and then the plants were re-watered for 2 d. The results showed that WT plants suffered more serious withering and damage from drought treatment (Fig. 3a). After rehydration, fewer than 30% of WT plants were alive whereas more than 80% of transgenic plants survived (Fig. 3b). Under control condition, the leaf water loss of transgenic lines was lower than that of WT (Fig. 3c). When subjected to drought treatment, transgenic plants showed significantly higher leaf water content than WT plants at drought 21 d (Fig. 3d). Taken together, these results suggested that MtPHD6 transgenic plants showed improved tolerance to drought stress through the regulation of water loss.
Fig. 3.
Drought stress tolerance of MtPHD6 transgenic lines. a: Phenotypic changes after drought treatment. 12-day-old seedlings were withheld water for 23 days. Photos were taken at 2 d after rehydration; b: Survival rate of MtPHD6 transgenic lines and WT after 2 d rehydration; c: Leaf water loss of MtPHD6 transgenic lines and WT. 21-day-old leaves were detached and air-dried for up to 8 h. d: Leaf water content of MtPHD6 transgenic lines and WT after 7, 14, 21 d drought treatment
Transcriptomic profiling analysis of MtPHD6 ectopic expression plants after drought treatment
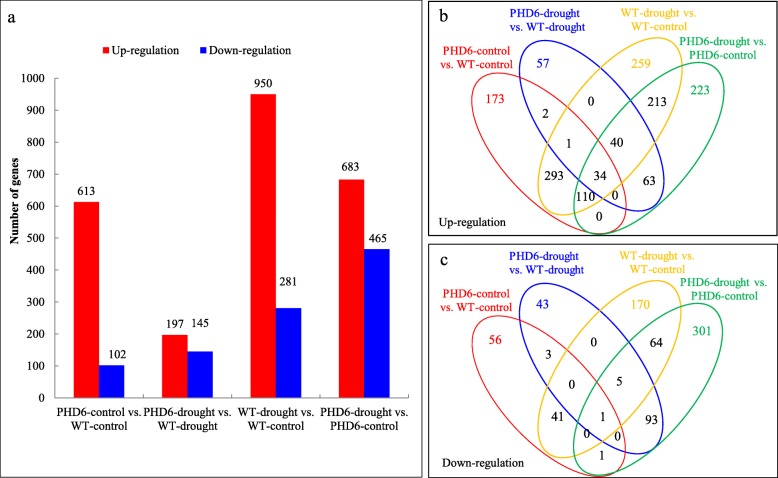
To characterize molecular mechanisms of MtPHD6-modulated drought tolerance, we performed RNA-sequencing to identify DEGs affected by the MtPHD6 transgene and drought treatment. In this study, twelve samples with three biological replicates per genotype/treatment combination were used. Each sample with at least 6 G clean data was obtained. In total, 2044 genes were transcriptionally affected by the MtPHD6 transgene or drought treatment (Additional file 3: Table S3). Quantitative real-time PCR qRT-PCR analysis showed that the trends of both up-regulated and down-regulated expression measured by RNA-seq and by qRT-PCR were similar (Additional file 7: Figure S1), indicating RNA-sequencing data were reliable.
Bioinformatics analysis showed that MtPHD6 transgene affected expression level of 715 and 342 genes under control and drought stress conditions, respectively. In WT plants, drought stress treatment modulated expression of 1231 genes, including 950 up-regulated and 281 down-regulated genes. Comparatively, significantly fewer DEGs were identified by drought stress treatment in MtPHD6 OE lines, with 683 up-regulated genes and 465 down-regulated genes (Fig. 4a). Overlapping analysis indicated that there were 397 up-regulated and 70 down-regulated genes which were commonly regulated in both MtPHD6 OE lines and WT plants after drought treatment (PHD6-drought vs. PHD6-control; WT-drought vs. WT-control), respectively. Moreover, 438 up-regulated and 42 down-regulated genes were commonly regulated by MtPHD6 transgene under control condition and drought stress condition (PHD6-control vs. WT-control; WT-drought vs. WT-control) (Fig. 4b, c).
Fig. 4.
MtPHD6 transgene- and drought stress-modulated genes through RNA-seq analysis. a: Number of genes changed by MtPHD6 transgene or drought stress. b and c: Overlapping analysis of up- and down-regulated genes by MtPHD6 transgene or drought stress. Arabidopsis seedlings at 12-day-old were subjected to drought treatment for 7 days. Leaves of transgenic lines and WT plants were harvested for RNA-seq analysis. The original data were presented in Additional file 3: Table S3
Cluster, GO term enrichment and MapMAN pathway analyses
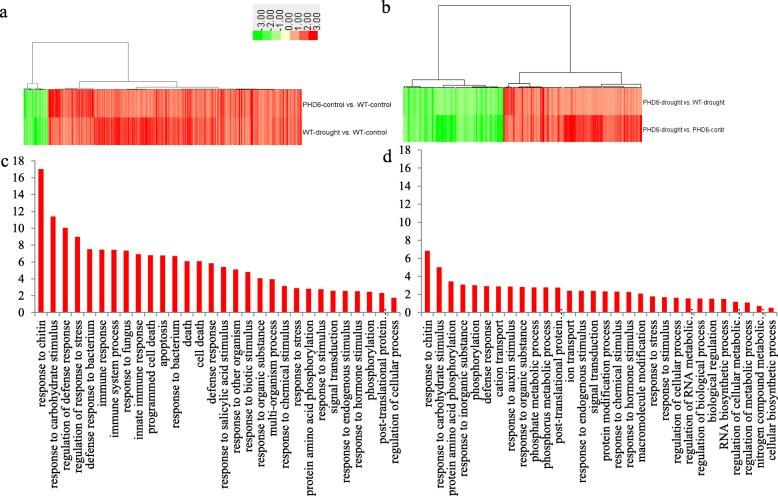
Totally, 480 genes were commonly regulated by MtPHD6 transgene under control condition (PHD6-control vs. WT-control) and drought stress on WT (WT-drought vs. WT-control), while 236 genes were commonly modulated by MtPHD6 transgene under drought condition (PHD6-drought vs. WT-drought) and drought stress on MtPHD6 OE lines (PHD6-drought vs. PHD6-control) (Fig. 5a, b). Further analysis showed that stress, carbohydrate metabolism, protein modification, signal transduction and hormone related GO terms were significantly enriched (Fig. 5c, d).
Fig. 5.
Cluster and GO term enrichment analyses of genes commonly regulated by MtPHD6 transgene and drought stress. In total 480 genes were commonly modulated by PHD6-control vs. WT-control and WT-drought vs. WT-control (a), while 236 genes were commonly modulated by PHD6-drought vs. WT-drought and PHD6-drought vs. PHD6-control (b). The lists of genes were submitted to agriGO (http://bioinfo.cau.edu.cn/agriGO/) with the analysis tool “Singular Enrichment Analysis (SEA)”. Top 15 significantly enriched GO terms with lowest P-value were listed (c and d). The original data were presented in Additional file 4: Table S4
For all DEGs modulated by MtPHD6 transgene or drought stress, cluster analysis further revealed that the majority of MtPHD6 transgene modulated genes were also changed by drought stress (Fig. 6a, columns 1 and 3). Pathway analysis exhibited that many AP2/EREBP, WRKY, and ZINC FINGER transcription factors were regulated by MtPHD6 transgene or drought stress (Fig. 6b, c). DEGs modulated by the MtPHD6 transgene or drought stress treatment were selected for GO term enrichment analysis. For biological process GO terms, response to stress, signal transduction, response to abiotic or biotic stimulus and other biological processes were significantly enriched (Additional file 8: Figure S2a). For molecular function GO terms, receptor binding or activity, transcription factor activity and kinase activity were significantly enriched (Additional file 8: Figure S2b). For cellular component GO terms, cell wall, plasma membrane and extracellular were significantly enriched (Additional file 8: Figure S2c).
Fig. 6.
Gene cluster analysis and transcription factors changed byMtPHD6 transgene or drought treatment. a: Cluster analysis of all genes modulated by MtPHD6 transgene or drought stress; b and c: Transcription factors regulated by MtPHD6 transgene or drought stress. Red squares represent up-regulated genes; blue squares mean down-regulated genes; and white squares mean no significant changes. The original data were presented in Additional file 6: Table S6
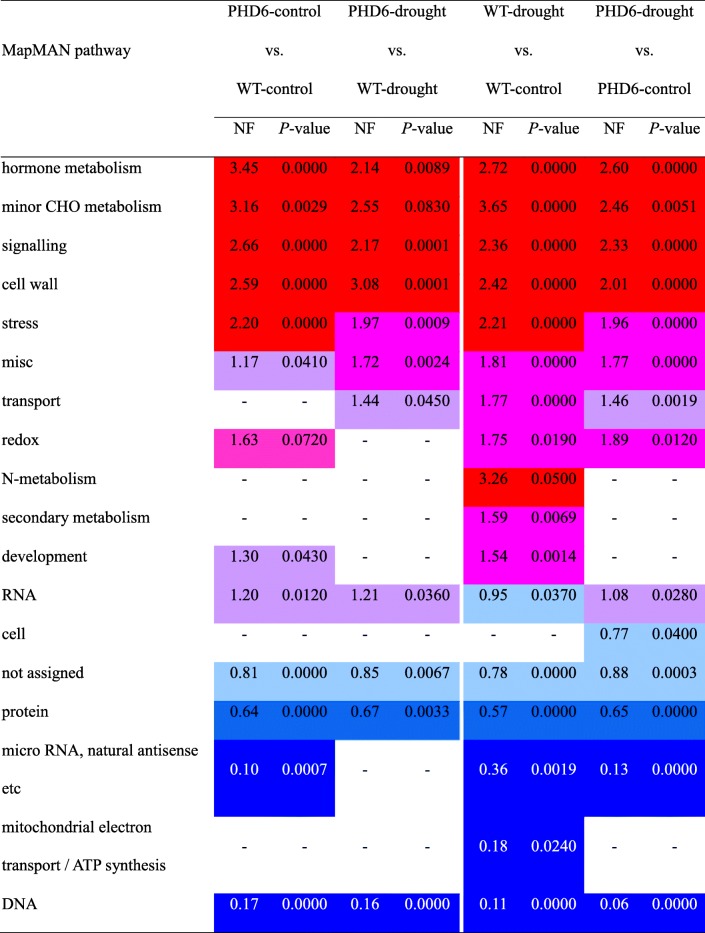
In addition, pathway enrichment analysis was performed for genes affected by MtPHD6 transgene or drought stress using MapMAN software. The results showed that five pathways were overrepresented by all comparisons, including hormone metabolism, minor CHO metabolism, signaling, cell wall, and stress. Pathway of redox was enriched after drought treatment in both WT and MtPHD6 transgenic lines, and by MtPHD6 transgene under control condition (P = 0.072) (Table 1). These data indicated that drought stress re-programed transcriptional networks in Arabidopsis and ectopic expression of MtPHD6 also extensively altered transcriptional networks in Arabidopsis.
Table 1.
MapMAN pathway enrichment analysis of genes affected by MtPHD6 transgene or drought stress. DEGs (fold change ≥2 and P-value ≤0.05) were annotated using the Classification SuperViewer Tool. MapMAN was used as the classification source. The original data were presented in Additional file 5: Table S5.The scales of normalized frequency (NF) are as follows 
Transcription factors modulated by MtPHD6 transgene and drought treatment
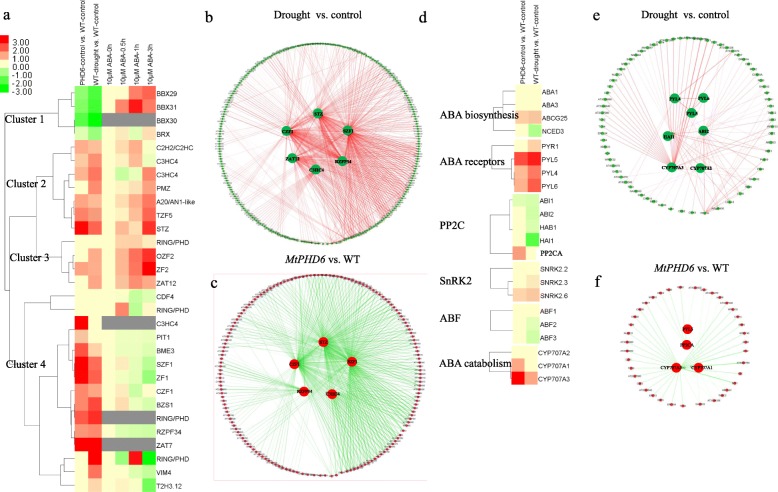
To identify transcription factors affected by MtPHD6 transgene and drought stress, we further performed MapMAN pathway analysis. The results showed that MtPHD6 transgene or drought stress modulated expression of many AP2/EREBP (AP2, RAV and CBF/DREB2/ERF), WRKY, and ZINC FINGER transcription factors (Fig. 6b, c), which have been reported to be involved in plant stress responses [25–27]. In this study, both drought stress and MtPHD6 transgene modulated expression of several ZINC FINGER transcription factors (STZ, SZF1, CZF1, etc) (Fig. 7). Cluster analysis showed that TZF5, STZ, OZF2, ZF2, ZAT12 identified in this study were also upregulated after ABA treatment (Fig. 7a, Clusters 2 and 3). Meanwhile, BBX29 and BBX31 were inhibited by drought stress and MtPHD6 transgene, but were highly induced after ABA treatment (Fig. 7a, Cluster 1). Interaction network analysis revealed that there were 249 nodes/proteins and 513 edges for drought, and 163 nodes/proteins and 388 edges for MtPHD6 transgene modulated ZINC FINGER transcription factors (Fig. 7b, c), including CPK, MPK, ERF, WRKY, DREB/CBF and NAC proteins.
Fig. 7.
Cluster and network analyses of zinc finger transcription factors and ABA pathway related genes. a: Cluster analysis of zinc finger transcription factors affected by MtPHD6 transgene, drought and ABA. b and c: Networks of zinc finger transcription factors modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively; d: Cluster analysis of ABA signaling pathway related genes affected by MtPHD6 transgene and drought. e and f: Networks of ABA signaling pathway related genes modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively. The original data were presented in Additional file 6: Table S6. ABA treatment expression data were downloaded from BAR Expression Browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi). The grey color in cluster analysis indicates the genes are missing in BAR database
Drought treatment and MtPHD6 transgene activated expression of the majority of AP2/EREBP transcription factors (Additional file 9: Figure S3a). AP2/EREBP transcription factors in clusters 1 and 2 were mainly downregulated by MtPHD6 transgene, but upregulated by ABA treatment. However, AP2/EREBP transcription factors in clusters 4 and 5 were upregulated by MtPHD6 transgene, but mainly downregulated by ABA treatment (Additional file 9: Figure S3a). These data indicated that AP2/EREBP transcription factors showed contrasting changes by MtPHD6 transgene and ABA. Interaction network analysis showed that there were 194 nodes/proteins and 445 edges for drought, and 155 nodes/proteins and 424 edges for MtPHD6 transgene modulated AP2/EREBP transcription factors (Additional file 9: Figure S3b, c), including WRKY, CML, MYB and ZINC FINGER proteins. WRKY transcription factors were mainly induced by MtPHD6 transgene and drought, but merely by ABA treatment (Additional file 9: Figure S3d). Interaction network analysis showed that there were 279 nodes/proteins and 807 edges for drought, and 196 nodes/proteins and 630 edges for MtPHD6 transgene modulated transcription factors (Additional file 9: Figure S3e, f), including MPK, MYB, AP2/ERF and ZINC FINGER proteins.
ABA pathway regulated by MtPHD6 transgene and drought treatment
Since zinc finger and AP2/EREBP transcription factors were modulated by ABA treatment based on publicly available microarray data analysis (Fig. 7a), we further investigated the gene expressions of ABA signaling transduction pathway. The results exhibited that several ABA receptor genes were highly induced by drought and MtPHD6 transgene, especially PYL4–6. Meanwhile, MtPHD6 transgene also activated expression of PP2CA and CYP707A1 and A3, while drought stress inhibited expressions of ABA1, ABA2, HAB1 and HAI1, and induced CYP707A3 expression (Fig. 7d). To better understand the effect of MtPHD6 transgene on ABA pathway, the possible interaction networks were also constructed and co-expressions of ABA pathway responsive genes were identified using STRING and Cytoscape software. The results showed that drought stress modulated PYLs (PYL4–5) and PP2C (HAI1, ABI2, PP2CA) interacted with several stress responsive genes, including ERFs, DREB1/CBF, LTI, COR, RD, LEA, WRKY, NAC and ZINC FINGER transcription factors (STZ, RHL41/ZAT12) (Fig. 7e). In MtPHD6 transgenic lines, PYLs and PP2Cs interact with ERF, DREB/CBF, RD, and ZINC FINGER transcription factors (STZ, SZF1, CZF1) (Fig. 7f). These results indicated that MtPHD6 transgene might activate ZINC FINGER and AP2/EREBP transcription factors through ABA dependent pathway, and induced WRKY transcription factors through ABA independent pathway.
Metabolism pathways regulated by MtPHD6 transgene and drought stress
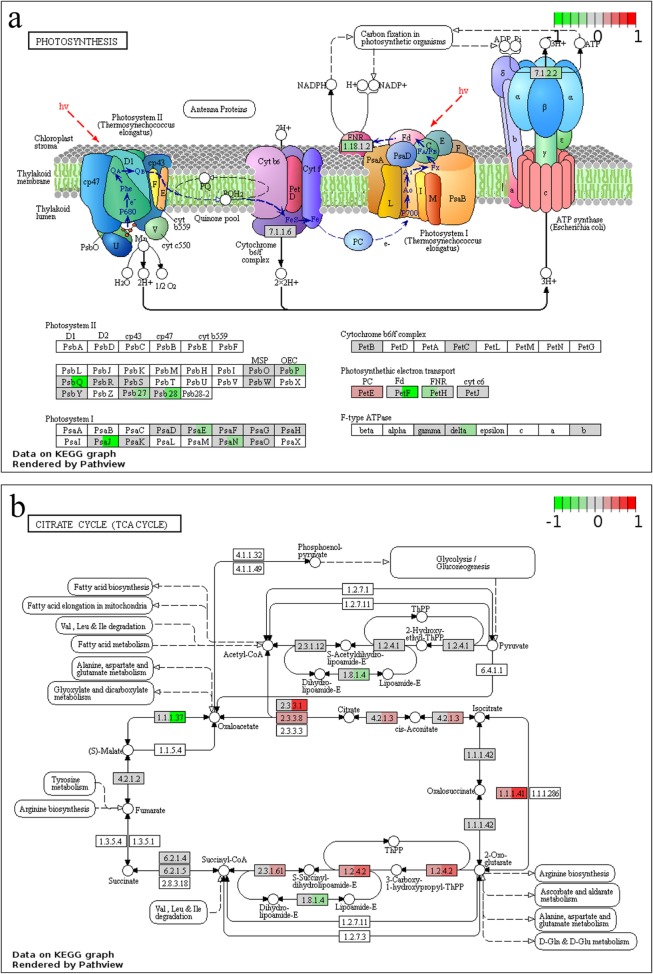
In the study, drought stress inhibited expression of several genes involved in photosystem I, II and photosynthetic electron transport pathways (Fig. 8a). However, few of photosynthesis related genes were regulated by MtPHD6 transgene, except plastocyanin (PETE) gene which was induced (Fig. 8a). Moreover, many genes encoding citrate cycle (TCA cycle) related enzymes were upregulated by drought, but barely by MtPHD6 transgene (Fig. 8b). These results showed that drought stress had more extensive effect on photosynthesis and TCA cycle than MtPHD6 transgene.
Fig. 8.
Photosynthesis (a) and TCA cycle (b) related genes changed by MtPHD6 transgene or drought. KEGG pathway changes were analyzed using Pathview (https://pathview.uncc.edu/home). Each rectangle was divided into 2 subsections, which represent genes modulated by MtPHD6 transgene (MtPHD6-control vs. WT-control) and drought (WT-drought vs. WT-control), respectively. Red means upregulation and green means downregulation
The effect of MtPHD6 transgene on oxidative damage under drought condition
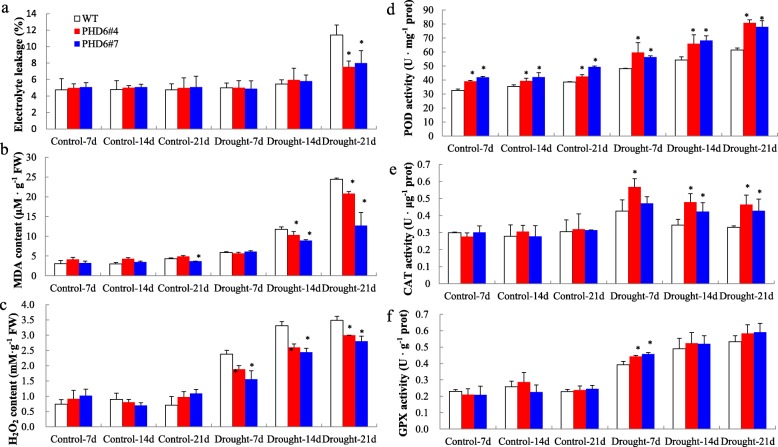
Drought stress can induce membrane lipid peroxidation and ROS overproduction, resulting in changes of plasma membrane permeability. In the study, the electrolyte leakage (EL) of transgenic and WT plants showed significant increase after 21 d of drought treatment. However, transgenic plants suffered less membrane damage and exhibited significantly lower EL than WT plants at 21 d of drought stress (Fig. 9a). Malondialdehyde (MDA) content in WT plants was significantly higher than that in transgenic plants at drought stress 14 d and 21 d, respectively (Fig. 9b). Moreover, significantly lower hydrogen peroxide (H2O2) content was observed in transgenic plants compared with WT plants after drought stress (Fig. 9c).
Fig. 9.
Physiological changes of MtPHD6 transgenic lines after drought stress treatment. 12-day-old seedlings were withheld water and leaf tissues were collected at 7, 14 and 21 d after drought treatment. a: Electrolyte leakage (EL); b: Malondialdehyde (MDA) content; c: Hydrogen peroxide (H2O2) content; d: Peroxidase (POD) activity; e: Catalase (CAT) activity; f: Glutathione peroxidase (GPX) activity
To avoid oxidative damage, plants activate antioxidant enzymatic defense system to keep redox homeostasis. Drought treatment triggered the activities of peroxidase (POD), catalase (CAT) and glutathione peroxidase (GPX) in WT and transgenic plants when compared to control condition. The POD activity in transgenic plants was significantly higher than that in WT plants under both control and drought conditions (Fig. 9d). Furthermore, transgenic plants showed significantly higher CAT (14 d, 21 d) and GPX (7 d) activities than WT plants under drought stress (Fig. 9e, f). These results showed that transgenic plants could effectively alleviate oxidative damage when exposed to drought stress.
Discussion
It is well known that drought stress constrains plant productivity and distribution. Through long-time evolution, plants have developed various responsive strategies to adapt to drought stress at the morphological, physiological, biochemical and molecular levels [28]. With rapidly developing analytical chemistry technologies, a large number of genes involved in drought tolerance have been excavated and functionally identified in various plant species [29, 30]. Among these genes, transcription factors play critical roles in multifarious signaling pathways that regulate plant responses to harsh environmental stresses [31–33]. So far, plenty of transcription factor families have been reported to be induced by abiotic stress, such as WRKY, bZIP, DREB, MYB/MYC, ERF, ZINC FINGER and Alfin1-like (AL) families [5, 13].
Alfin1-type PHD-finger-containing proteins are involved in plant abiotic stresses apart from their roles in developmental processes [12, 34]. In this study, we found that the expression of MtPHD6 was induced by drought stress (Figs. 1c, 2a). MtPHD6 transgenic lines showed improved tolerance to mannitol stress during seed germination (Fig. 2). Ectopic expression of MtPHD6 plants possessed enhanced tolerance to drought stress with significantly higher survival rates and antioxidant enzyme activities when compared to WT plants (Figs. 3b, 9). In Arabidopsis, AL proteins were found to bind to G-box elements and overexpression of AL5 improved drought tolerance [18]. Ectopic expression of soybean GmPHD2 increased salt tolerance in Arabidopsis probably by scavenging ROS [5]. The data in this study suggested that MtPHD6 could function as a positive regulator in response to drought stress.
In M. sativa, Alfin1 binds to G-rich elements in the promoter of MsPRP2, a salt-inducible and root-specific gene [4, 10]. Moreover, GmPHD5 binds to the promoter and coding region of salt stress responsive genes and acts as a positive regulator in response to salt stress [16]. Through large scale transcriptome analysis, we found that genes involving in stress related GO terms were enriched by drought stress and MtPHD6 transgene (Fig. 5, Additional file 8: Figure S2a), including ABA signaling pathway (Fig. 7d). In rice (O. sativa), five OsPHD genes were significantly induced while two inhibited by ABA [35]. In soybean, expression of GmPHD 1–6 were induced after ABA and drought treatment, and GmPHD2 transgene upregulated expression of ABA responsive bZIP transcription factor ABI5 [5]. In this study, MtPHD6 transgene regulated different subset of ABA responsive genes like PYL4–6 (Fig. 7d). As the key hormone during plant responses to drought stress, ABA can activate PYR/PYL-PP2C-SnRK2-ABF signaling pathway, which plays vital roles in plant drought tolerance. ABA pathways might function as the upstream signaling in MtPHD6 transgenic Arabidopsis in response to drought stress.
Similar to RING finger and LIM domain, the PHD finger is characterized as a conserved Cys4HisCys3-type zinc finger type. In Arabidopsis, ZINC FINGER and WRKY type transcription factors were identified as AL5-regulated genes by ChIP-Seq [18]. Indeed, several zinc finger transcription factors were modulated by MtPHD6 transgene and part of these genes were also induced by drought stress or ABA treatment (Fig. 7a). Arabidopsis zinc finger proteins ZAT6 and ZAT18 were transcriptionally induced by drought, salt, cold and heat stresses [26, 27]. Overexpression of Arabidopsis ZAT6, ZAT18 and pepper ZFP1 increased the expression level of stress induced genes and ABA signaling pathway related genes, resulting in enhanced drought tolerance [26, 27, 36]. AtZAT7 overexpression inhibited plant growth and enhanced salt tolerance of transgenic Arabidopsis [37]. Under stress condition, AtAZF1 and AtAZF2 had a negative effect on the expression of auxin-inducible and ABA repressive genes [31]. Moreover, a Cys2His2-type zinc finger transcription factor from Glyycine soja, GsZFP1, could be induced by abiotic stresses and ABA treatment. Ectopic expression of GsZFP1 in Arabidopsis activated the expression of ABA biosynthesis related genes and increased tolerance to drought stress [38]. These data showed that zinc finger transcription factors can function as positive regulators in response to drought stress. PHD domain transcription factors including Alfin1 encode novel members of zinc finger family proteins [5, 24]. MtPHD6 transgene upregulated the expressions of RZPF34, ZAT7, STZ/ZAT10, SZF1, ZF1, and several C3HC4 zinc finger transcription factors in Arabidopsis, which were induced after ABA treatment (Fig. 7a). These results indicated that several zinc finger proteins might function as ABA downstream regulators and play important roles in drought tolerance of MtPHD6 transgenic plants.
AP2/EREBP and WRKY were shown to play key roles in plant abiotic stress responses. AP2/EREBP family is a large group of plant-specific transcription factors, and has four major subfamilies, including the AP2, DREB, ERF and RAV [39]. Totally 123 putative AP2/ERF genes were identified in M. truncatula genome. Further analysis showed that cold stress induced a cluster of DREB subfamily genes on chromosome 6 indicating the contribution of these genes in response to cold stress [40]. Overexpression of tomato DREB2 improved salt tolerance in transgenic tomato and Arabidopsis through activation of proline and polyamines biosynthesis and increase of K+/Na+ ratio [41]. Vitis CBF1 and CBF4 transgenic Arabidopsis showed improved cold and drought stress tolerances. Different subsets of stress responsive genes were modulated by VrCBF1 and VrCBF4 transgene, indicating that Vitis CBFs have different regulons [42]. Additionally, WRKY family also plays a crucial role in regulating plant responses to abiotic stresses, including drought, cold, salt and osmotic stresses [43]. Ectopic expression of sunflower HaWRKY76 increased drought and flood stress tolerances of transgenic Arabidopsis plants [44]. Wheat TaWRKY1 and TaWRKY33 conferred drought and heat stress tolerance in Arabidopsis [45]. Mutation of Arabidopsis WRKY transcription factor ABO3 resulted in decreased expression of ABF2/AREB1, RD29A and COR47, indicating the important role of WRKY in plant stress responses [46]. Ectopic expression of MtPHD6 increased the expression of AP2/EREBP and WRKY transcription factors in Arabidopsis (Fig. 6, Additional file 9: Figure S3). Therefore, MtPHD6 transgene modulated AP2/EREBP and WRKY transcription factors might contribute to enhanced plant drought tolerance.
Conclusions
Taken together, MtPHD6 transgene might activate stress responsive ZINC FINGER, AP2/EREBP and WRKY transcription factors through ABA dependent and independent pathways. These transcription factors interact with stress related proteins, regulate other stress responsive genes including NAC, CML, RD, LEA, LTI and MPK, and/or modulate accumulation of osmolites, photosynthesis, and ROS metabolism, resulting in improved plant drought stress tolerance (Fig. 10).
Fig. 10.
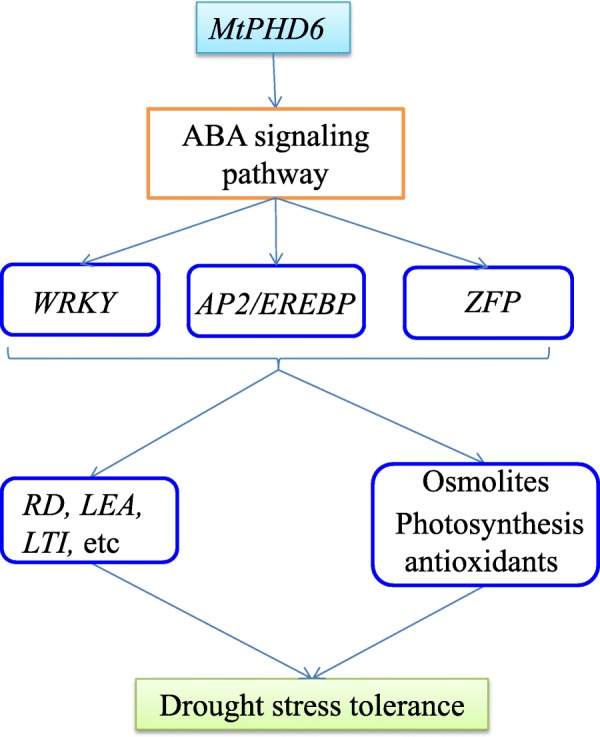
Model depicting roles of MtPHD6 in response to drought stress
Methods
Plant materials and growth conditions
In this study, M. truncatula Jemalong A17 and A. thaliana Columbia-0 were used and identified by Wuhan Botanic Garden. Seeds of M. truncatula Jemalong A17 were provided by Dr. Haiqing Wang from Chinese Academy of Sciences and seeds of A. thaliana Columbia-0 were self-propagated in the lab.
After surface-sterilization in 2% NaClO solution for 5 min, Arabidopsis seeds were stratified in deionized water at 4 °C for 3 d in darkness. Then the seeds were sown on full-strength Murashige and Skoog (MS) medium plates (pH 5.7), and were incubated in a growth chamber at 22 °C with 120 μmol m− 2 s− 1 irradiance, 60% relative humidity, and a light/ dark cycle of 16/8 h. After growth for 7 d, the seedlings, except those used in germination tests, were transferred into soil-filled containers and irrigated with nutrient solution every 3 days.
Cloning of MtPHD6 and gene transformation
The full-length coding sequence of MtPHD6 from M. truncatula was isolated by PCR amplification using MtPHD6-F and MtPHD6-R primers with KpnI and BamHI restriction sites, respectively (Additional file 2: Table S2). Then PCR products were purified and inserted into pCAMBIA1301S vectors to construct recombinant plasmids. The transformed A. thaliana plants were generated through introduction of Agrobacterium tumefaciens strain GV3101 containing the constructs using the floral lip method [47]. The positive transgenic plants were identified on MS medium with 25 μg ml− 1 hygromycin under the same growth condition. T3 homozygous lines were confirmed by PCR analyses and used for following experiments.
Germination tests and drought treatment
The sensitivity of MtPHD6-overexpressing seeds to mannitol was analyzed on MS medium plates at the germination stage. After vernalization, WT and transgenic seeds were sown on MS medium plates supplemented with or without 300 mM mannitol, respectively. It was considered as germinated seeds when radicles fully penetrated the seed coat. Germinated seeds were counted daily for 7 d.
For the 12-day-old soil-grown seedlings, water was withheld for 23 d before rehydration. Leaf samples under control and stress conditions were harvested at 7 d, 14 d and 21 d after drought treatment, respectively. The survival rate was calculated at 2 d after re-watering.
Analysis of water loss and leaf water content
To analyze water loss under control condition, detached leaves from 21-day-old plants were weighted at 1 h intervals for up to 8 h. The leaf samples at different intervals after drought stress were used for measurement of leaf water content (LWC). Fresh weight (FW) of leaf samples was immediately quantified, and dry weight (DW) was measured after 16 h at 80 °C oven. LWC (%) calculation was made as follows: (FW-DW)/FW × 100 [26].
Measurement of EL and MDA
EL was measured as described [48]. Briefly, detached leaves were placed in a tube with 15 mL deionized water on a shaker for 6 h. The Ci of leaf samples was determined before boiling for 20 min. After boiling and cooling to room temperature, the Cmax was measured and then the percentage of EL was computed based on the formula of (Ci/Cmax) × 100.
MDA determination was performed as described [27]. Briefly, 0.3 g leaf samples were ground with liquid nitrogen and 5% trichloroacetic acid (TCA; w/v) was added and mixed with fine leaf powder. Before adding 2 mL of 0.67% thiobarbituric acid (TBA, w/v), the supernatants were centrifuged at 12,000 g for 10 min. The solutions were mixed and boiled for 30 min. After centrifugation and cooling to room temperature, the concentration of MDA was computed according to the absorbance at 450, 532 and 600 nm, respectively.
Quantification of H2O2 content and antioxidant enzyme activities
For the assessment of H2O2 content and antioxidant enzyme activities, the supernatant from fresh leaves was prepared according to Quan et al. [49] and used for the following measurement.
The content of H2O2 was analyzed using H2O2 Assay Kit (A064–1-1, Nanjing Jiancheng Bioengineering Institute, China) based on the manufacturer’s instructions. The measurement of POD, CAT and GPX activities was performed with Plant POD Assay Kit (A084–3-1, Nanjing Jiancheng Bioengineering Institute, China), CAT Assay Kit (A007–1-1, Nanjing Jiancheng Bioengineering Institute, China) and GPX Assay Kit (A005–1-2, Nanjing Jiancheng Bioengineering Institute, China), respectively, as the introduction described.
RNA isolation and quantitative real-time PCR
Leaf samples of WT and MtPHD6 transgenic lines after 7 d of drought treatment were collected for RNA isolation. Using Trizol reagent (Invitrogen, Carlsbad, CA, USA), total RNA was extracted and then treated by DNase I (Promega, Madison, WI, USA) to remove the contamination of genomic DNA [48, 49]. RNA concentration and purity were measured by the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). The integrity of RNA was assessed with 1.5% (w/v) agarose gel electrophoresis. Totally 2 μg of above RNA was reverse-transcribed into cDNA using reverse transcriptase (TOYOBO, Ohtsu, Janpan) according to the manufacturer’s instructions.
qRT-PCR was executed by a CFX 96 Real Time System (Bio-Rad, California, USA) with SYBR green fluorescence. The expression levels of genes were standardized with ubiquitin 10 (AtUBQ10, AT4G05320) and calculated by the 2-ΔΔCT method [50]. Gene-specific primer sequences used for qRT-PCR are listed in Additional file 2: Table S2. The experiment had three technical and biological replicates.
Transcriptomic analysis
RNA-seq analysis was performed by the Novogene Corporation (Beijing, China) as described [27]. Totally 3 μg RNA for each sample was used for preparation of sequencing libraries using NEBNext® UltraTM RNA Library Prep Kit for Illumina®. Index codes were added to attribute sequences and the clustering of index-coded samples was performed on a cBot Cluster Generation System (Illumina). Library preparations were then sequenced on an Illumina Hiseq platform and paired-end reads with 125 bp/150 bp were generated. After removing reads containing poly-N, adapter and low quality reads from raw data, clean data (clean reads) were generated. Paired-end clean reads were aligned to the Arabidopsis genome using Hisat2 v2.0.5. The read numbers mapped to each gene were counted using featureCounts v1.5.0-p3, and the FPKM of each gene was computed according to the length of the gene and read counts mapped to this gene. Differential expression analysis of each treatment versus control was investigated with the DESeq2 R package (1.16.1). There were 12 samples with three biological replicates for each sample. DEGs were selected with a fold change > 2 and P-value < 0.05. The RNA-seq data have been submitted to NCBI Gene Expression Omnibus (GEO) with the accession number GSE134945.
Hierarchical cluster analysis
Cluster 3.0 program (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) was used for hierarchical cluster analysis with uncentered matrix and complete linkage method. The resulting tree figures were displayed with Java Treeview software (http://jtreeview.sourceforge.net/) [51].
GO term and pathway enrichment analysis
All DEGs from RNA-seq data were annotated using the Classification SuperViewer Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) for GO term and pathway enrichment analysis [52]. GO and MapMan were respectively chosen as classification source. The normalized frequency (NF) of each functional category was computed according to the following formula: NF = sample frequency in each sample/background frequency in Arabidopsis genome.
Transcriptional network analysis
Interaction networks of DEGs were analyzed using STRING database (https://string-db.org/) with high confidence (score > 0.7) [53]. Tab separated values from STRING were then analyzed with Cytoscape software (http://www.cytoscape.org/).
Statistical analysis
All experiments were repeated three times independently. Leaf samples used in each experiment were collected from 3 containers with at least 15 plants of each container. The results presented were the mean ± SD. The significant differences relative to WT were showed by asterisks above the columns of figures at P ≤ 0.05 level using Student’s t-test.
Supplementary information
Additional file 1: Table S1. Genbank accession numbers of AL/PHD proteins used for phylogenetic analysis
Additional file 2: Table S2. Primer sequences used for gene cloning and qRT-PCR experiments
Additional file 3: Table S3. Spreadsheet of genes changed by drought stress or MtPHD6 transgene.
Additional file 4: Table S4. Spreadsheet of genes commonly regulated by MtPHD6 transgene and drought stress
Additional file 5: Table S5. Spreadsheet of genes for MapMAN pathway analysis.
Additional file 6: Table S6. Spreadsheet of specific genes including ABA pathway related genes, zinc finger, AP2/ EREBP and WRKY transcription factors.
Additional file 7: Figure S1. Correlation expression analysis of selected genes by RNA-seq and qRT-PCR. Totally 11 genes co-regulated by MtPHD6 transgene and drought treatment were selected for qRT-PCR analysis.
Additional file 8: Figure S2. GO term enrichment analysis of genes changed by drought or MtPHD6 transgene in Arabidopsis.
Additional file 9: Figure S3. Cluster and network analyses of AP2/EREBP and WRKY transcription factors. For cluster analysis, the list of genes were analyzed using Cluster 3.0 and the resulting tree figure was shown by Java Treeview. Network analysis was performed using STRING (https://string-db.org) and Cytoscape 3.7.1. a: Cluster analysis of AP2/EREBP transcription factors affected by MtPHD6 transgene, drought and ABA. b and c: Networks of AP2/EREBP transcription factors modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively. d: Cluster analysis of WRKY transcription factors affected by MtPHD6 transgene, drought and ABA. e and f: Networks of WRKY transcription factors modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively. The original data were presented in Additional file 6: Table S6. ABA treatment expression data were downloaded from BAR Expression Browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi). The grey color in cluster analysis indicates the genes are missing in BAR database.
Acknowledgments
We are grateful to the editor and reviewers for their comments and suggestions. We would thank Dr. Haiqing Wang from Chinese Academy of Sciences for providing seeds of M. truncatula Jemalong A17.
Authors’ contributions
WQ and ZC planned and designed the research. WQ, XL, MY and LY performed experiments. WQ and ZC analyzed data and wrote the manuscript. LW revised the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0201305), Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (2016RC010), the National Natural Science Foundation of China (31872143) to Zhulong Chan, and the Youth Project of Science and Technology Department of Hubei Education Department (Q20182702) to Wenli Quan. Xun Liu acknowledges the support from the China Scholarship Council (2016–2020). The funding bodies had no roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Sequence data from this article can be found in GEO data library (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE134945. DEGs analysis is included in Additional files 3, 4, 5, 6, 7, 8 and 9.
Ethics approval and consent to participate
Plant materials were self-propagated for research use only. Sampling of plant material was performed in compliance with institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenli Quan, Email: quanwenli0426@163.com.
Xun Liu, Email: xunliu0123@hotmail.com.
Lihua Wang, Email: lwang@hbeu.edu.cn.
Mingzhu Yin, Email: baihemingzhu114@126.com.
Li Yang, Email: 935354092@qq.com.
Zhulong Chan, Email: zlchan@mail.hzau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6350-5.
References
- 1.Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313X.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 2.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatinmediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/S0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 3.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Bastola DR, Pethe VV, Winicov I. Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol. 1998;38:1123–1135. doi: 10.1023/A:1006081926699. [DOI] [PubMed] [Google Scholar]
- 5.Wei W, Huang J, Hao YJ, Zou HF, Wang HW, Zhao JY, et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One. 2009;4:e7209. doi: 10.1371/journal.pone.0007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrika NNP, Sundaravelpandian K, Schmidt W. A PHD in histone language: on the role of histone methylation in plant responses to phosphate deficiency. Plant Signal Behav. 2013;8:e24381. doi: 10.4161/psb.24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayum MA, Park JI, Ahmed NU, Jung HJ, Saha G, Kang JG, et al. Characterization and stress-induced expression analysis of Alfin-like transcription factors in Brassica rapa. Mol Gen Genomics. 2015;290:1299–1311. doi: 10.1007/s00438-015-0993-y. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez MDLP, Gutierrez C. Arabidopsis ORC1 is a PHD containing H3K4me3 effector that regulates transcription. Proc Natl Acad Sci U S A. 2009;106:2065–2070. doi: 10.1073/pnas.0811093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung S, Schmitz RJ, Amasino RM. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Gene Develop. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winicov I, Valliyodan B, Xue L, Hoober JK. The MsPRP2 promoter enables strong heterologous gene expression in a root-specific manner and is enhanced by overexpression of Alfin 1. Planta. 2004;219:925–935. doi: 10.1007/s00425-004-1296-4. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol. 2005;59:191–203. doi: 10.1007/s11103-005-6503-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhou W, Wu J, Zheng Q, Jiang Y, Zhang M, Zhu S. Genome-wide identification and comparative analysis of Alfin-like transcription factors in maize. Genes Genom. 2017;39:261–275. doi: 10.1007/s13258-016-0491-6. [DOI] [Google Scholar]
- 13.Tao JJ, Wei W, Pan WJ, Lu L, Li QT, Ma JB, et al. An Alfin-like gene from Atriplex hortensis enhances salt and drought tolerance and abscisic acid response in transgenic Arabidopsis. Sci Rep. 2018;8:2707. doi: 10.1038/s41598-018-21148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu L, Zhu Y. Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol. 2006;9:544–549. doi: 10.1016/j.pbi.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Song Y, Gao J, Yang F, Kua CS, Liu J, Cannon CH. Molecular evolutionary analysis of the Alfin-like protein family in Arabidopsis lyrata, Arabidopsis thaliana, and Thellungiella halophila. PLoS One. 2013;8:e66838. doi: 10.1371/journal.pone.0066838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu T, Pi EX, Tsai SN, Lam HM, Sun SM, Kwan YW, et al. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 2011;11:178. doi: 10.1186/1471-2229-11-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee WY, Lee D, Chung WI, Kwon CS. Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 2009;58:511–524. doi: 10.1111/j.1365-313X.2009.03795.x. [DOI] [PubMed] [Google Scholar]
- 18.Wei W, Zhang YQ, Tao JJ, Chen HW, Li QT, Zhang WK, et al. The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J. 2015;81:871–883. doi: 10.1111/tpj.12773. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Summer LW, Wang ZY. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa) Plant J. 2005;42:689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- 20.May GD. From models to crops: integrated medicago genomics for alfalfa improvement. In: Hopkins A, Wang ZY, Mian R, Sledge M, Barker RE, editors. Molecular breeding of forage and turf. Developments in plant breeding. Dordrecht: Springer; 2004. [Google Scholar]
- 21.Wang ZY, Hopkins A, Mian R. Forage and turf grass biotechnology. Crit Rev Plant Sci. 2001;20:573–619. doi: 10.1080/20013591099281. [DOI] [Google Scholar]
- 22.Yu Y, Bi C, Wang Q, Ni Z. Overexpression of TaSIM provides increased drought stress tolerance in transgenic Arabidopsis. Biochem Bioph Res Co. 2019;512:66–71. doi: 10.1016/j.bbrc.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Gou J, Debnath S, Sun L, Flanagan A, Tang Y, Jiang Q, et al. From model to crop: functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotech J. 2018;16:951–962. doi: 10.1111/pbi.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winicov I, Bastola DR. Transgenic overexpression of the transcription factor Alfin 1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol. 1999;120:473–480. doi: 10.1104/pp.120.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina J, Catalá R, Salinas J. The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011;180:3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, et al. The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiol. 2014;165:1367–1379. doi: 10.1104/pp.114.242404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin M, Wang Y, Zhang L, Li J, Quan W, Yang L, et al. The Arabidopsis Cys2/His2 zinc-finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J Exp Bot. 2017;68:2991–3005. doi: 10.1093/jxb/erx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Y, Xu Y, Hettenhausen C, Lu C, Shen G, Zhang C, et al. Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 2018;18:35. doi: 10.1186/s12870-018-1250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, et al. The ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci U S A. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kodaira KS, Qin F, Tran LSP, Maruyama K, Kidokoro S, Fujita Y, et al. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011;157:742–756. doi: 10.1104/pp.111.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Y, Chen P, Yan Y, Bao C, Li X, Wang L, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018;218:201–218. doi: 10.1111/nph.14952. [DOI] [PubMed] [Google Scholar]
- 34.Mouriz A, López-González L, Jarillo JA, Piñeiro M. PHDs govern plant development. Plant Signal Behav. 2015;10:e993253. doi: 10.4161/15592324.2014.993253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun M, Jia B, Yang J, Cui N, Zhu Y, Sun X. Genome-wide identification of the PHD-finger family genes and their responses to environmental stresses in Oryza sativa L. Int J Mol Sci. 2017;18:2005. doi: 10.3390/ijms18092005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SH, Hong JK, Lee SC, Sohn KH, Jung HW, Hwang BK. CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol Biol. 2004;55:883–904. doi: 10.1007/s11103-005-2151-0. [DOI] [PubMed] [Google Scholar]
- 37.Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, et al. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282:9260–9268. doi: 10.1074/jbc.M611093200. [DOI] [PubMed] [Google Scholar]
- 38.Luo X, Bai X, Zhu D, Li Y, Ji W, Cai H, et al. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta. 2012;235:1141–1155. doi: 10.1007/s00425-011-1563-0. [DOI] [PubMed] [Google Scholar]
- 39.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul Mech. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Shu Y, Liu Y, Zhang J, Song L, Guo C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front Plant Sci. 2016;6:1247. doi: 10.3389/fpls.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hichri I, Muhovski Y, Clippe A, Žižková E, Dobrev PI, Motyka V, et al. SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and Arabidopsis. Plant Cell Environ. 2016;39:62–79. doi: 10.1111/pce.12591. [DOI] [PubMed] [Google Scholar]
- 42.Siddiqua M, Nassuth A. Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 2011;34:1345–1359. doi: 10.1111/j.1365-3040.2011.02334.x. [DOI] [PubMed] [Google Scholar]
- 43.Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2014;239:255–266. doi: 10.1007/s00425-013-1985-y. [DOI] [PubMed] [Google Scholar]
- 44.Raineri J, Ribichich KF, Chan RL. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 2015;34:2065–2080. doi: 10.1007/s00299-015-1852-3. [DOI] [PubMed] [Google Scholar]
- 45.He GH, Xu JY, Wang YX, Guo W, Zhang X. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2006;16:116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 48.Quan W, Liu X, Wang H, Chan Z. Comparative physiological and transcriptional analyses of two contrasting drought tolerant alfalfa varieties. Front Plant Sci. 2016;6:1256. doi: 10.3389/fpls.2015.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quan W, Liu X, Wang H, Chan Z. Physiological and transcriptional responses of contrasting alfalfa (Medicago sativa L.) varieties to salt stress. Plant cell tissue organ. Cult. 2016;126:105–115. [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time itative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Chan Z, Bigelow PJ, Loescher W, Grumet R. Comparison of salt stress resistance genes in transgenic Arabidopsis thaliana indicates that extent of transcriptomic change may not predict secondary phenotypic or fitness effects. Plant Biotech J. 2012;10:284–300. doi: 10.1111/j.1467-7652.2011.00661.x. [DOI] [PubMed] [Google Scholar]
- 52.Provart NJ, Zhu T. A browser-based functional classification superviewer for Arabidopsis genomics. Curr Comput Mol Biol. 2003;2003:271–272. [Google Scholar]
- 53.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:362–368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Genbank accession numbers of AL/PHD proteins used for phylogenetic analysis
Additional file 2: Table S2. Primer sequences used for gene cloning and qRT-PCR experiments
Additional file 3: Table S3. Spreadsheet of genes changed by drought stress or MtPHD6 transgene.
Additional file 4: Table S4. Spreadsheet of genes commonly regulated by MtPHD6 transgene and drought stress
Additional file 5: Table S5. Spreadsheet of genes for MapMAN pathway analysis.
Additional file 6: Table S6. Spreadsheet of specific genes including ABA pathway related genes, zinc finger, AP2/ EREBP and WRKY transcription factors.
Additional file 7: Figure S1. Correlation expression analysis of selected genes by RNA-seq and qRT-PCR. Totally 11 genes co-regulated by MtPHD6 transgene and drought treatment were selected for qRT-PCR analysis.
Additional file 8: Figure S2. GO term enrichment analysis of genes changed by drought or MtPHD6 transgene in Arabidopsis.
Additional file 9: Figure S3. Cluster and network analyses of AP2/EREBP and WRKY transcription factors. For cluster analysis, the list of genes were analyzed using Cluster 3.0 and the resulting tree figure was shown by Java Treeview. Network analysis was performed using STRING (https://string-db.org) and Cytoscape 3.7.1. a: Cluster analysis of AP2/EREBP transcription factors affected by MtPHD6 transgene, drought and ABA. b and c: Networks of AP2/EREBP transcription factors modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively. d: Cluster analysis of WRKY transcription factors affected by MtPHD6 transgene, drought and ABA. e and f: Networks of WRKY transcription factors modulated by drought stress (green node and red edge) and MtPHD6 transgene (red node and green edge), respectively. The original data were presented in Additional file 6: Table S6. ABA treatment expression data were downloaded from BAR Expression Browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi). The grey color in cluster analysis indicates the genes are missing in BAR database.
Data Availability Statement
Sequence data from this article can be found in GEO data library (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE134945. DEGs analysis is included in Additional files 3, 4, 5, 6, 7, 8 and 9.