Abstract
During DNA virus infections, detection of cytosolic DNA by the cGAS-STING pathway leads to activation of IFN-β. Kaposi’s Sarcoma Herpesvirus (KSHV), an oncogenic DNA virus, is the etiological agent of Kaposi’s Sarcoma, an endothelial cell (EC)-based tumor. To investigate the role of STING during KSHV infection of primary ECs we identified a primary lymphatic EC sample that is defective for STING activation and we also knocked out STING in blood ECs. Ablation of STING in EC does not increase susceptibility to KSHV latent infection nor does it increase KSHV spread after lytic reactivation indicating STING signaling does not restrict KSHV. In contrast, STING ablation increases Adenovirus spread at low MOI, but STING is dispensable for blocking replication. These experiments reveal that the importance of STING depends on the DNA virus and that STING appears more important for restricting spread to bystander cells than for inhibition of viral replication.
Keywords: KSHV, Innate immunity, STING, Kaposi’s sarcoma, Endothelial cells, Interferons
1. Introduction
Innate immunity is critical for the detection and response to invading microbes. Detection relies on the sensing of pathogen-associated molecular patterns (PAMPs), which are structurally conserved molecules that are present or produced during infection (Takeuchi and Akira, 2010). PAMPs are detected by pattern recognition receptors (PRRs), proteins that then signal to activate production of antimicrobial factors, including cytokines such as type I interferon (IFN) (Akira et al., 2006). During viral infections, a major PAMP is nucleic acids, including double-stranded (ds)RNA and cytosolic DNA (Thompson et al., 2011). Therefore, cells express a variety of PRRs dedicated to the detection of nucleic acids. For detection of dsRNA, major sensors include retinoic acid induced gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (Kell and Gale, 2015). For mislocalized and foreign DNA, major sensors include absent in melanoma 2, interferon gamma inducible protein 16, and cyclic GMP-AMP synthase (cGAS) (Unterholzner, 2013; Keating et al., 2011). While several DNA sensors have been described, cGAS is likely the primary PRR for sensing cytosolic DNA during DNA virus infections in most situations (Sun et al., 2013; Wu et al., 2013; Gray et al., 2016). Upon binding to DNA, cGAS synthesizes the cyclic dinucleotide 2′3′ cyclic GMP-AMP (cGAMP), which binds to the ER adaptor stimulator of interferon genes (STING), leading to the ubiquitination of STING and recruitment of TANK-binding kinase 1 (TBK1) (Zhang et al., 2013; Ablasser et al., 2013; Diner et al., 2013; Ni et al., 2017; Wang et al., 2014; Ishikawa and Barber, 2008; Ishikawa et al., 2009). Activation of TBK1 leads to autophosphorylation and subsequent phosphorylation of STING and interferon regulator factor 3 (IRF3) (Liu et al., 2015; Zhong et al., 2008). Phosphorylated IRF3 then translocates to the nucleus where it acts as a transcription factor to induce expression of IFN-β and subsequent downstream IFN-stimulated genes (ISGs) (Tanaka and Chen, 2012). Despite extensive studies of these pathways, less is known about how STING affects the lifecycles of different DNA viruses.
Kaposi’s Sarcoma-associated herpes virus (KSHV) is a gammaherpesvirus, with a large double-stranded DNA (dsDNA) genome (Mesri et al., 2010). KSHV, like all herpesviruses, undergoes both lytic and latent replication, with latency being the predominant state in cell culture and its associated tumors (Lagunoff et al., 2002; Staskus et al., 1997). KSHV is the etiological agent of Kaposi’s Sarcoma, a common tumor of AIDS patients and one of the most common tumors in parts of sub-Saharan Africa (Mesri et al., 2010; Wabinga et al., 2014; Chokunonga et al., 2000). KS is a highly vascularized tumor composed primarily of spindle cells that express markers of endothelium (DiMaio and Lagunoff, 2012). Endothelial cells are divided into two main categories: blood endothelial cells (BECs), which line blood vessels and lymphatic endothelial cells (LECs), which make up lymphatic vessels (Dejana et al., 2017; Choi et al., 2012). While KS spindle cells express markers of both BECs and LECs, the expression profile of spindle cells from KS tumors aligns most closely with KSHV-infected LECs (DiMaio and Lagunoff, 2012; Wang et al., 2004; Weninger et al., 1999).
KSHV infects both BECs and LECs in cell culture and has been shown to both activate and antagonize innate immune pathways in these cells (Aresté and Blackbourn, 2009; Lee et al., 2016). Recent studies have focused on KSHV genes that antagonize the cGAS-STING pathway. It has been reported that ORF52 encodes a tegument protein that directly antagonizes cGAS during initial infection (Wu et al., 2015). In addition, KSHV encodes at least two genes that are expressed during lytic replication that antagonize either cGAS or STING. Cytoplasmic isoforms of the latency-associated nuclear antigen (LANA) are reported to accumulate during lytic replication and block cGAS signaling (Zhang et al., 2016). In addition, viral interferon regulatory factor 1 (vIRF1) is also expressed during lytic replication and antagonizes STING signaling by blocking its interaction with TBK1 (Ma et al., 2015). However, the role of STING during KSHV de novo infection remains unclear (Wu et al., 2015; Zhang et al., 2016; Ma et al., 2015). Additionally, while some studies have suggested a role for STING in restricting lytic reactivation, it is unknown if STING is important for controlling this process in primary endothelial cells (Zhang et al., 2016; Ma et al., 2017). In addition, while numerous studies have reported on the importance of STING in controlling DNA virus infections, it is unclear whether STING plays a role in blocking initial infection or replication of DNA viruses (Wu et al., 2015; Zhang et al., 2016; Ma et al., 2015, 2017; Paijo et al., 2016; Lio et al., 2016; Li et al., 2013; Schoggins et al., 2014).
Interestingly, we recently discovered a single-patient batch of primary neonatal LECs (LEC4) that are defective for activation of innate immune pathways following infection by DNA viruses, including KSHV and Adenovirus (Adv). We further characterized this defect and found that LEC4 are intrinsically defective in sensing cytosolic DNA and cGAMP, indicating a block in the cGAS-STING pathway. Further characterization reveals that STING, TBK1 and IRF3 are not phosphorylated in LEC4 following transfection with cGAS-STING agonists. Additionally, STING degradation, a hallmark of activation, fails to occur in LEC4. We show that pre-treatment of LEC4 with cGAMP fails to decrease susceptibility to KSHV while pre-treatment with IFN-β nearly completely blocks KSHV infection, thus demonstrating the importance of a fully functional DNA sensing pathway during viral infection. Because mutations that inactivate the pathway are common across human populations (Patel and Jin, 2019), we sought to better understand the role of STING in KSHV infection. We investigated the role of STING during initial viral infection and spread. We first found that KSHV induces minimal IFN-β expression in BECs and STING competent LECs and does not detectably activate the STING-TBK1-IRF3 axis. To further interrogate the role of STING during viral infection, we used CRISPR/Cas9 to create STING knockout BECs. Surprisingly, genetic ablation of STING does not increase susceptibility to infection by KSHV nor does it enhance the ability of KSHV to spread through the culture following lytic reactivation. Additionally, STING is not required for the induction of IFN-β during KSHV infection. However, the role of STING during Adv infection is more nuanced, with STING restricting spread at low MOI but playing no role in blocking replication at high MOI. Overall, these experiments reveal that while we have identified a LEC isolate that is defective for STING, STING appears to be dispensable during infection with KSHV. STING is important for blocking infection in bystander cells for a different dsDNA virus, Adv, but not for controlling replication following initial infection by Adv. Therefore, it appears that KSHV maintains high-level control of the activation of STING-related DNA sensing pathways.
2. Materials and methods
2.1. Antibodies and reagents
Antibodies were purchased from the indicated source: anti-cGAS (catalog number 15102; Cell Signaling Technology), anti-STING (catalog number abl81125; Abcam) (catalog number 13647; Cell Signaling Technology), anti-p-STING (catalog number 85735; Cell Signaling Technology), anti-p-TBK1 (catalog number 5483; Cell Signaling, Technology), anti-TBK1 (catalog 3504; Cell Signaling, Technology), anti-p-IRF3 (catalog number 4947; Cell Signaling Technology), anti-IRF3 (catalog number 4302; Cell Signaling Technology), and anti-β-actin (catalog number A5441; Sigma Aldrich). The anti-LANA antibody was obtained from Don Ganem (University California, San Francisco). Recombinant IFN-β was purchased from EMD Millipore (catalogue number: IF014).
2.2. CRSIPR/Cas9 gene targeting of STING (TMEM173)
We obtained a pRRL plasmid expressing a gRNA targeting STING and Cas9-T2A cassette from Daniel Stetson (University of Washington), described here (Gray et al., 2016). Lentivirus targeting STING or the non-targeting scramble control was generated by co-transfection of 293T cells with the plasmid and two packaging plasmids, pMD2.G and psPAX2 (Addgene) into HEK293T cells using Transit 293 transfection reagent (Mirus) per the manufacturer’s instructions. For stable targeting of STING, BEC1-3 were transduced with the lentivirus for 6 h and selected with puromycin (1 μg/mL) (VWR Scientific) for 3 days.
2.3. Viruses and cells
Primary human neonatal microvascular BECs and LECs were obtained from Lonza and cultured in EBM-2 (Lonza) supplemented with a bullet kit containing 5% FBS, vascular endothelial growth factor, basic fibroblast growth factor, insulin-like growth factor 3, epidermal growth, and hydrocortisone. Numbers indicate that primary cells are from different donors (BEC1, BEC2, BEC3, LEC4, LEC6, and LEC8). HEK293 Integrin (IN)-β cells were cultured in DMEM (Corning) supplemented with 10% FBS, sodium pyruvate, l-glutamine, and penicillin/streptomycin. Cells were cultured at 37 °C, 5% CO2. KSHV-BAC16 was generated as previously described by inducing iSLK cells with doxycycline and harvesting the virus from the cellular supernatant by centrifugation (Brulois et al., 2012). BCBL-l-derived KSHV (WT-KSHV) was generated as previously described (Punjabi et al., 2007). A gutted helper-dependent Adenovirus expressing the KSHV lytic and replication activator (HD-Ad-RTA) was obtained from D. Ganem (Bechtel et al., 2003) and generated as previously described (Sanchez et al., 2017).
A BAC vector (pKSB2HAdV64) containing the entire genome of HAdV-64 was created as follows: 1) the first 1475 bp and the last 1008 bp of the HAdV-64 genome (Zhou et al., 2012) flanking a galK ORF from pgalK (Warming et al., 2005) were subcloned into pUC19; 2) the insert was then subcloned into Eco RI and Hin dIII digested pBluescript II SK (+); 3) the insert was then subcloned into Not I and Hin dIII digested pKSB2 (Sirena et al., 2005); 4) the resulting vector was used to transform SW102 cells (Warming et al., 2005); 5) HAdV-64 genomic DNA was isolated from virus and recombineered in place of the galK ORF (Warming et al., 2005). This vector was further modified by recombineering to replace the E3 region (bp 26215 to 30789) with a PCR product containing the CMV-eGFP ORF (bp 4709 to 1398) from pEGFP-N1 (Clontech Laboratories, Inc.) to create pKSB2HAdV64.eGFP. The fidelity of this construct was verified by Sanger sequencing of the recombineered region and by restriction digest of the entire BAC vector. Release of the viral genome from pKSB2HAdV64.eGFP by flanking Pac I sites was followed by transfection of 293β5 (Nguyen et al., 2010) cells to produce virus. Genomic DNA was isolated from purified virus, and the fidelity of the entire genome was verified by Illumina sequencing.
2.4. Nucleic acid and cGAMP transfection
E. coli dsDNA (dsDNA-EC) (2.5 μg/mL) (Invivogen), high molecular weight Polyinosinic-polycytidylic acid [Poly(I:C)] (0.5 μg/mL or 1 μg/mL) (Invivogen), interferon stimulatory DNA (ISD) 100mer (10 μg/mL) [synthesized by heating equimolar sense and antisense oligonucleotides (sequences in Table 1) to 95 °C and annealing at room temperature for 1 h], calf thymus DNA (1 μg/mL) (Invitrogen), and 2′3′ cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) (5 μg/mL) (Invivogen) were transfected into BECs or LECs using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol.
Table 1.
Oligonucleotide sequences for RT-qPCR and ISD 100mer.
| Gene/oligo name | Sense (S) | Antisense (AS) |
|---|---|---|
| IFN-β | AAACTCATGAGCAGTCTGCA | AGGAGATCTTCAGTTTCGGAGG |
| Mx1 | GACATTCGGCTGTTTACC | CTTCCAGTGCCTTGATTT |
| Mx2 | ACCGCCATTCGGCACAGT | TGCCCTTGGTTGGCTCCT |
| ISG15 | TGGACAAATGCGACGAACC | CCCGCTCACTTGCTGCTT |
| IFIT1 | CACCCACTTCTGTCTTACT | ACATTCTTGCCAGGTCTA |
| IFITM1 | GGATTTCGGCTTGTCCCGAG | CCATGTGGAAGGGAGGGCTC |
| K8.1 | AAAGCGTCCAGGCCACCACAG | GGCAGAAAATGGCACACGGTT |
| ORF10 | GTCCTGTCCCGCTCTCTTTTTTG | CAATAAGGTGTTCGTGCTTGCCC |
| Tubulin | TCCAGATTGGCAATGCCTG | GGCCATCGGGCTGGAT |
| ISD 100mer | GGATGAGTCCATGTCTAGATAATCACTA | ACATCTAGTACATGTCTAGTCAG |
| GATACTGACTAGACATGTACTAGATGTATGTCT | TATCTAGTGATTATCTAGACATACATCTAGTACATG | |
| AGATAATCACTAGATACTGACTAGACATGTACTAGATGT | TCTAGTCAGTATCTAGTGATTATCTAGACATGGACTCATCC |
2.5. Virus infection
Infections with KSHV were done as described (Punjabi et al., 2007). Briefly, virus stocks were diluted in serum free media containing 8 μg/mL of polybrene and incubated with cells for 4 h with gentle rocking. The inoculum was then replaced with complete media and incubated for the indicated time post infection. For KSHV IFN-β induction experiments, cells were infected with KSHV-BAC16 at a MOI such that > 75% of the cells were infected as indicated by immunofluorescence staining with an antibody to the KSHV LANA protein. For immunoblotting for p-STING, p-TBK1 and p-IRF3 following KSHV infection, cells were infected with WT-KSHV such that > 80% of the cells were infected. For comparing infection rates after the establishment of latency in ΔSTING and scramble BECs, cells were infected with varying concentrations of KSHV-BAC16. All KSHV infections were done in serum-free media with polybrene. For inducing lytic reactivation, cells were infected with either WT-KSHV or KSHV-BAC16 as previously described. After 4 h, cells were rinsed with PBS and superinfected with HD-Ad-RTA for 1 h in serum-free media, poly L:lysine (1μ/mL) and sodium butyrate (1 μM). Cells were then rinsed twice with PBS and incubated in complete media for 24 h. For HAdV-64 replication titer, cells were infected at 100 genomes per cell for 45 min at 4 °C, then rinsed with PBS and incubated for the indicated times. At 5 days post infection (dpi), cells were scraped, and the supernatant was frozen in liquid nitrogen. To titer HAdV-64 virions, the supernatant was then thawed and re-frozen in liquid nitrogen 3 more times and then added to 293T IN-β cells. Cells were incubated for 24 h and then fluorescence was quantified using Typhoon fluorescent scanner. For measuring Adv spread, cells were infected with HAdV-64 at 10 genomes per cell and cells were incubated for 5 days. Plates were scanned on the Typhoon fluorescent scanner.
2.6. Immunofluorescence staining
To determine viral titers for comparing infection rates, aliquots of KSHV-BAC16-infected BECs and LECs were seeded on 4-well chamber slides and fixed with 4% (vol/vol) paraformaldehyde-phosphate-buffered saline. Infection rates were assessed using antibodies against the latent KSHV protein LANA (a kind gift from A. Polson and D. Ganem) as described previously (Lagunoff et al., 2002). Cells were incubated with fluorophore-conjugated secondary antibodies goat anti-rabbit Alexa Fluor 488 (Molecular Probes/Invitrogen). Cells were mounted in medium containing DAPI (4′,6′-diamidino-2-phenylindole) before being viewed under a Nikon Eclipse E400 fluorescence microscope (Nikon, Inc.).
2.7. Quantitative RT-PCR
Total RNA was extracted using RNA isolation kits (Macherey-Nagel). RNA was reverse-transcribed using iScript Reverse Transcription Supermix (Biorad). Gene expression was measured by qPCR using SsoAdvanced Universal SYBR Green supermix (Biorad). Primers used are shown in Table 1. The relative levels of each transcript were normalized by the delta threshold cycle method to the abundance of Tubulin mRNA.
2.8. Immunoblot
Cells were lysed in either RIPA lysis buffer or NP-40 lysis buffer supplemented with 1 mM sodium orthovanadate (Sigma Aldrich), 1 mM sodium fluoride (Sigma Aldrich), 40 mM 2-glycerophosphate (Sigma Aldrich), benzonase nuclease (Sigma Aldrich), and protease inhibitor cocktail tablets (Roche). Protein was quantified using bicinchoninic acid (BCA) assay (Pierce). Lysate was run on a gradient 4–20% SDS-PAGE gel (Biorad) and then transferred to an Immobilon F polyvinylidene difluoride membrane (Millipore). The membranes were blocked using Odyssey blocking buffer (Li-COR) for at least 1 h and then probed with the indicated primary antibodies overnight at 4 °C with rocking. Blots were washed 3 times with Tris-buffered saline with Tween 20 (TBST) and then probed with IRDye secondary antibody (Li-COR) diluted in Li-COR buffer for at least 1 h at room temperature. Blots were washed 3 times in TBST and then scanned with an Odyssey CLx infrared imaging system (Li-COR) for fluorescent blots.
2.9. Flow cytometry
ΔSTING and scramble BEC3 were infected with KSHV-BAC16 at low MOI as previously described. Immediately following infection, cells were treated with PMA (200 ng/mL) for 5 days. Cells were fixed in PFA and sorted by GFP fluorescence.
2.10. RNA sequencing of STING (TMEM173)
RNA was extracted from LEC4 and submitted for RNA sequencing to the Benaroya Genomics Core Facility and the results were analyzed by the Fred Hutchinson Cancer Research Center Genomics Shared Resource. LEC4 TMEM173 transcripts were aligned to human reference genome 38 and human STING1 (TMEM173) transcript variant 1 (Accession NM_198282.4) using Integrative Genomics Viewer.
2.11. Statistical analysis
Data are represented as the mean ± standard error of the mean (SEM) where indicated, and either Student’s t-test or Two-way ANOVA with Sidak posthoc test was used for statistical analyses with GraphPad Prism software.
3. Results
3.1. DNA viruses fail to activate innate immune signaling in LEC4
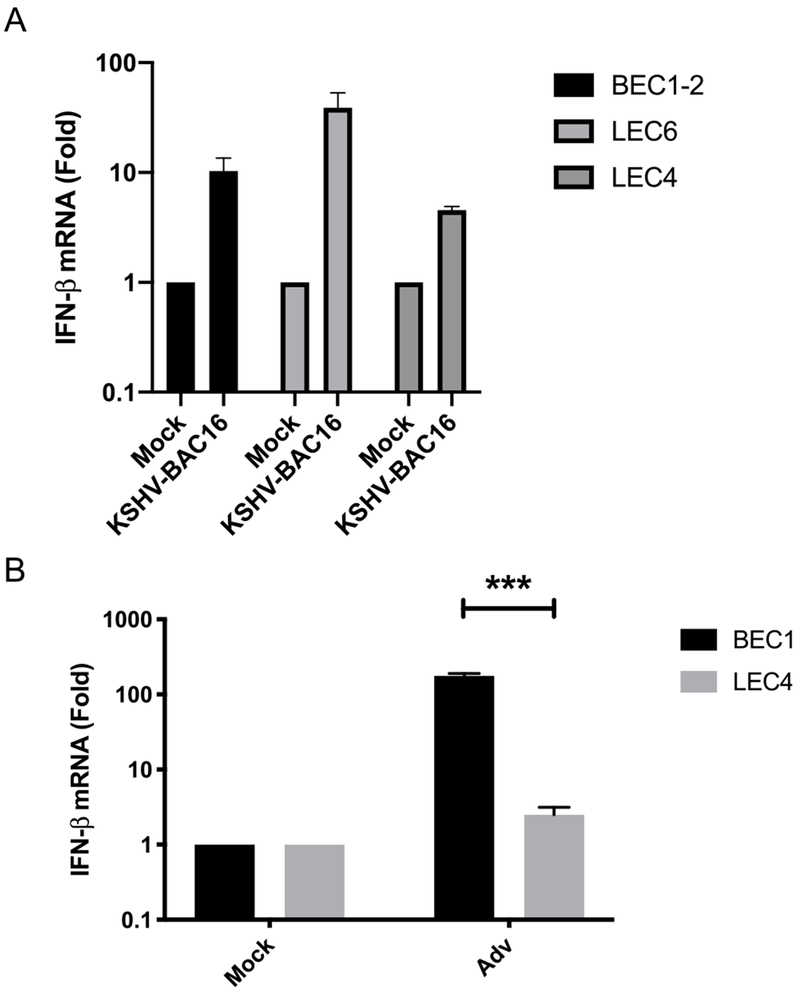
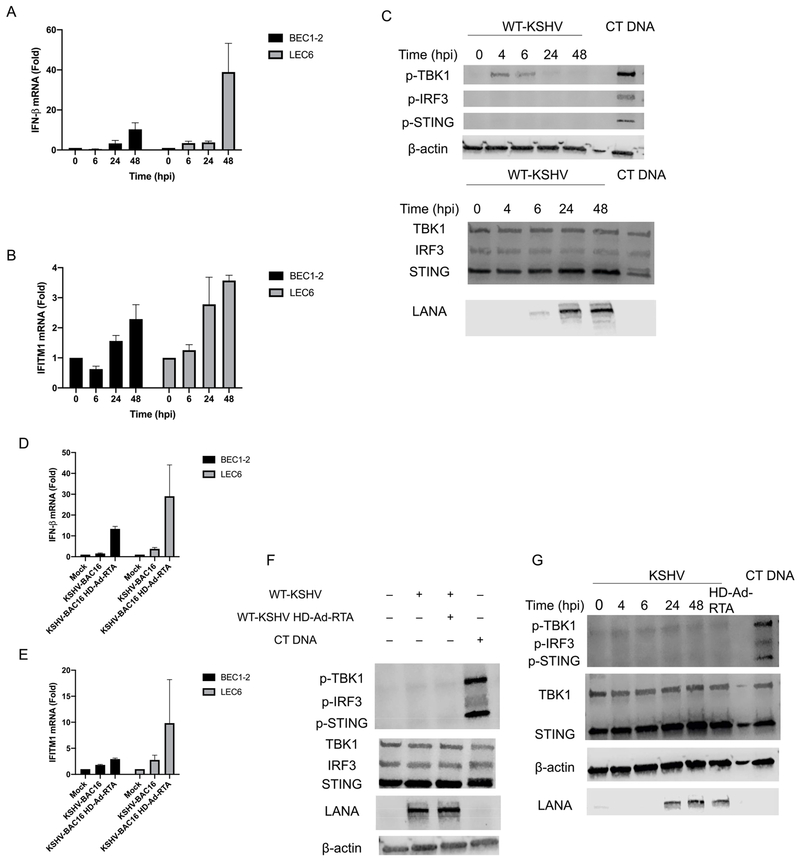
To determine if there are differences in the innate immune response to KSHV, we infected primary blood (BEC) and lymphatic (LEC) endothelial cells from different patients, BEC1-2, LEC4 and LEC6, with KSHV-BAC16 and measured the induction of IFN-β. Interestingly, IFN-β transcripts were slightly elevated in all 3 batches of cells following KSHV infection, albeit to varying degrees, with the greatest induction observed in LEC6, and the smallest increase in LEC4 (Fig. 1A). Since latency is the primary state during KSHV infection, we wanted to determine if DNA virus replication would similarly stimulate a differential IFN-β response in BECs and LEC4. Adv is a DNA virus that undergoes lytic replication in a wide variety of cell types. When infected with Adv, IFN-β was induced robustly in BEC1, but not in LEC4 (Fig. 1B). Overall, these results indicate modest increases in IFN-β expression following KSHV infection in cells from all donors tested, but defective innate immune activation to Adv viruses in LEC4.
Fig. 1. KSHV induces minimal IFN-β expression in primary endothelial cells but LEC4 are defective for innate immune activation during Adv infection.
A) IFN-β mRNA was measured by RT-qPCR from BEC1-2, LEC4 and LEC6 that were infected with KSHV-BAC16 for 48 h. The relative amount of mRNA was normalized to tubulin mRNA in each sample, and fold change relative to mock was calculated (ΔΔct). B) IFN-β was measured by RT-qPCR from BEC1 and LEC4 that were infected with Adv for 48 h. The relative amount of mRNA was normalized as in (A). Data are shown as mean ± SEM from at least 3 biological replicates. ***P < 0.001; (Student’s t-test).
3.2. cGAS-STING pathway agonists fail to activate innate immune signaling in LEC4
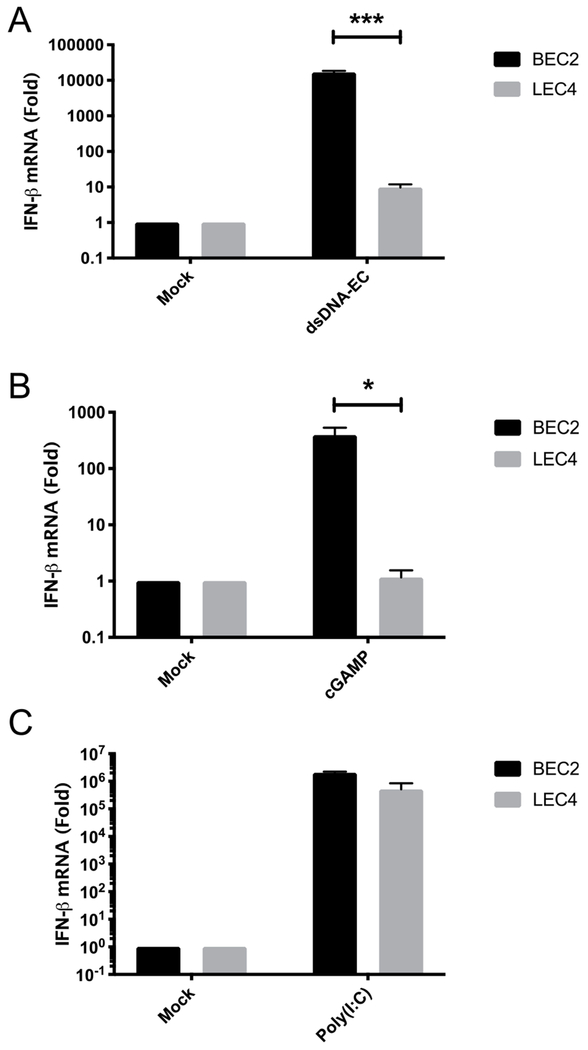
Since induction of IFN-β was minimal following infections with KSHV and Adv in LEC4, we next sought to determine if LEC4 are defective for innate immune activation of DNA sensing pathways. To test this, we transfected BEC2 and LEC4 with E. coli dsDNA (dsDNA-EC) and measured the induction of IFN-β. Surprisingly, IFN-β induction was completely absent in LEC4 but induced robustly in BEC2 (Fig. 2A). This trend was also observed for all other ISGs tested, including Mx1, Mx2, ISG15 and IFIT1 (Figs. S1A–D). Similarly, when cells were transfected with cGAMP, IFN-β induction was absent in LEC4 but robust in BEC2 (Fig. 2B). Examination of the expression levels of other ISGs followed a similar trend, including Mx1 and ISG15, with strong induction to cGAMP in BEC2, but no increase in expression in LEC4 (Figs. S1E and D). To determine if the defect in innate immune activation is a general feature of LECs, we transfected either dsDNA-EC or cGAMP into adult LECs as well as corresponding adult BECs and found that induction of IFN-β was robust in these cells, indicating that this defect is unique to LEC4 (Figs. S2A and B). To determine if this defect is a trait shared by other neonatal LECs, we transfected calf thymus (CT) DNA and cGAMP into two other primary neonatal LEC isolates (LEC6 and LEC8), and found a strong increase in expression of IFN-β in both LEC6 (Fig. S2C) and LEC8 (Fig. S2D) and were able to detect phosphorylated STING, TBK1 and IRF3 in both LEC6 (Fig. S2E) and LEC8 (Fig. S2F). Overall, these experiments show that other LECs patient isolates have intact cGAS-STING signaling. The cGAS-STING pathway utilizes TBK1, which is also critical for signaling in dsRNA sensing pathways, including RIG-1/MDA5-MAVS. In order to determine if the defect in innate immune signaling in LEC4 is upstream of TBK1, we transfected BEC2 and LEC4 with the dsRNA mimic, Polyinosinic-polycytidylic acid [Poly(I:C)], and found Poly(I:C) induced robust expression of IFN-β in both BEC2 and LEC4 (Fig. 2C), indicating that RNA sensing pathways are intact and that TBK1 is functional in LEC4. Overall, these experiments reveal that LEC4 have a unique defect in DNA sensing pathways leading to the induction of IFN-β not shared by other LEC isolates but RNA sensing pathways are functional.
Fig. 2. cGAS-STING pathway agonists fail to activate innate immune signaling in LEC4.
IFN-β mRNA was measured by RT-qPCR from BEC2 and LEC4 that were transfected with (A) 2.5μg/mL, dsDNA-EC, (B) 5 μg/mL, 2′3′ cGAMP or (C) 0.5 μg/mL, high molecular weight Poly(I:C) for 4 h. The relative amount of mRNA was normalized as in Fig. 1. Data are shown as mean ± SEM from at least 3 biological replicates. *P < 0.05; ***P < 0.001; (Student’s t-test).
3.3. Downstream STING signaling is blocked in LEC4 cells following treatment with cGAS-STING agonists
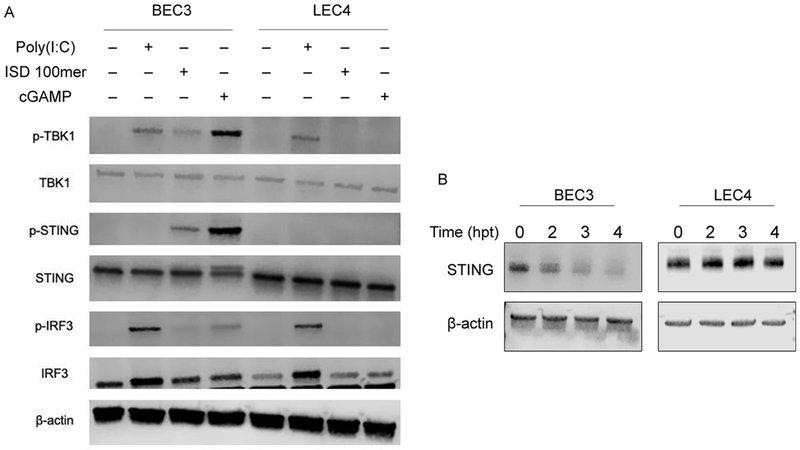
We next sought to characterize the block in STING-mediated innate immune activation. After activation by cGAMP, STING recruits TBK1, which autophosphorylates, and then phosphorylates STING, leading to the recruitment and phosphorylation of IRF3. To determine if TBK1, STING and IRF3 undergo phosphorylation in LEC4, we transfected BEC3 and LEC4 with interferon-stimulatory DNA (ISD) 100mer, cGAMP or Poly(I:C) and immunoblotted whole cell lysates. While Poly(I:C) led to the phosphorylation of TBK1 and IRF3 in BEC3 and LEC4, TBK1, STING and IRF3 were not phosphorylated in LEC4 in response to either ISD 100mer or cGAMP (Fig. 3A). In contrast, TBK1, STING and IRF3 were all phosphorylated following either ISD 100mer or cGAMP transfection in BEC3. Negative regulation of STING signaling is critical for controlling excessive and pathological innate immune signaling and inflammation (Li et al., 2017). In order to achieve this, STING undergoes K48-linked polyubiquitination and phosphorylation following activation, leading to degradation of STING by the proteasome (Li et al., 2018). To determine if STING is degraded in LEC4, we transfected BEC3 and LEC4 with cGAMP and immunoblotted for STING. Interestingly, while STING was rapidly degraded in BEC3, no such degradation occurred in LEC4 (Fig. 3B). STING protein expression remained stable through 4 h post transfection. STING cDNA from LEC4s was sequenced and determined to have a wild-type sequence, indicating that LEC4s have an unknown defect in the STING signaling pathway (Fig. S3). Together, these results indicate that downstream signaling events following STING activation are blocked in LEC4, preventing the phosphorylation of STING, TBK1 and IRF3 and the degradation of STING.
Fig. 3. Downstream STING signaling is blocked in LEC4 cells.
(A) BEC3 and LEC4 were transfected with either 1 μg/mL, Poly I:C, 10 μg/mL ISD 100mer or 5 μg/mL, 2′3′ cGAMP for 3 h and whole cell lysates were immunoblotted with the indicated phospho-specific or total protein antibodies. (B) BEC3 and LEC4 were transfected with 5 μg/mL 2′3′ cGAMP and cells were harvested at the indicated time points and whole cell lysates were immunoblotted in native (nonreducing) sample buffer for the indicated antibodies.
3.4. Pre-treatment with cGAMP decreases susceptibility to infection in BEC1, not LEC4
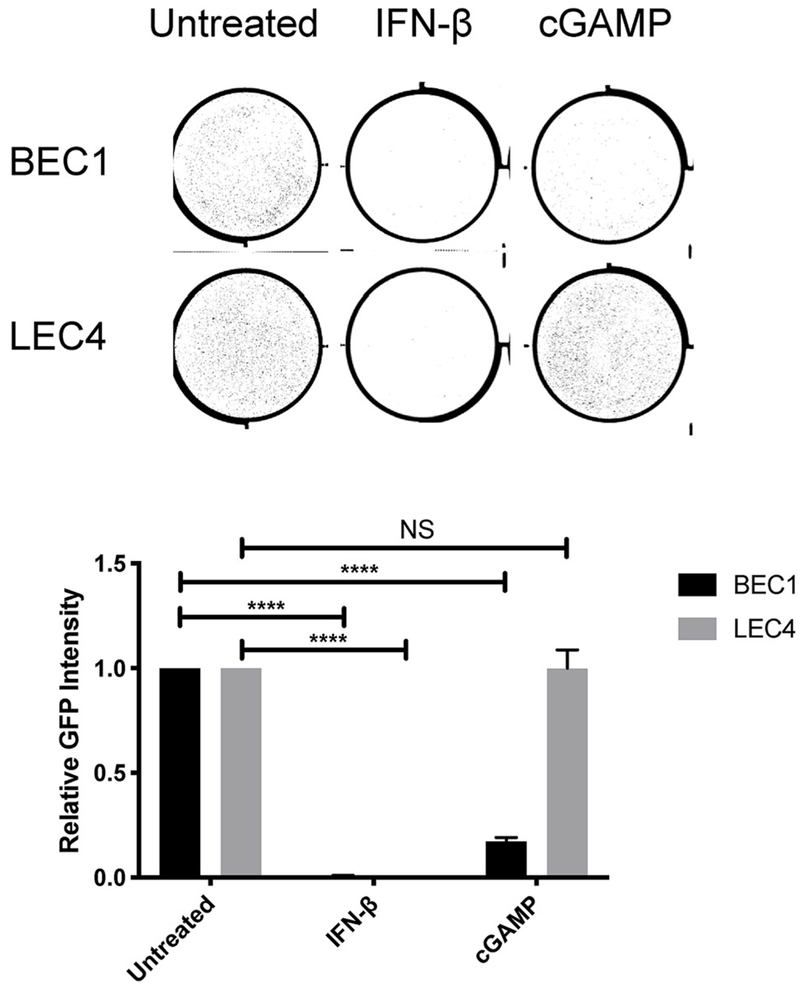
In addition to activating IRF3, STING also activates STAT6, NF-κB and autophagy pathways, which may have antiviral effects (Abe and Barber, 2014; Chen et al., 2011; Gui et al., 2019; Liu et al., 2018). To determine if STING is activating other antiviral pathways in LEC4, we pre-treated BEC1 and LEC4 with either IFN-β or cGAMP and infected with KSHV-BAC16 24 h later. Pre-treatment with cGAMP led to significantly decreased infection of BEC1 with KSHV, as measured by GFP intensity (Fig. 4, upper panel shows representative raw plate images, lower panel shows quantification). However, there was no change in infection of LEC4s following the pre-treatment. However, pre-treatment of both BEC1 and LEC4 with IFN-β protected both from infection by KSHV, with reduced susceptibility to infection relative to the untreated controls. Therefore, these results indicate STING is not able to activate other antiviral pathways in LEC4 and that activation of downstream pathways can rescue cells from the defect in STING signaling.
Fig. 4. cGAMP pre-treatment fails to reduce susceptibility to KSHV infection in LEC4.
BEC1 and LEC4 were transfected with 5 μg/mL 2′3′ cGAMP or treated with 1000 IU/mL IFN-β for 24 h. Cells were infected with KSHV-BAC16 and the integrated density of the fluorescence (relative GFP intensity) was quantified by Typhoon 48 h post infection (hpi). The upper panel show representative raw plate staining and the lower panel shows the mean ± SEM from at least 3 biological replicates. ****P < 0.0001 (Two-way ANOVA with Sidak posthoc test).
3.5. KSHV does not activate STING-TBK1-IRF3 signaling to induce minimal innate immune signaling in primary endothelial cells
STING is reported to play an important role during KSHV infection (Wu et al., 2015; Zhang et al., 2016; Ma et al., 2015, 2017). However, it is unknown if KSHV activates the cGAS-STING pathway in endothelial cells. To determine if KSHV de novo infection of primary cells activates the cGAS-STING pathway, we infected primary BEC1-2 and LEC6 with KSHV-BAC16 and measured the induction of IFN-β over time. Interestingly, IFN-β expression peaks at 48 h post infection (hpi), with minimal induction at 6 and 24 hpi (Fig. 5A). In addition, there was slight up regulation of the ISG IFITM1 at 48 hpi, following a similar trend as IFN-β (Fig. 5B). To determine if KSHV induces IFN-β through STING, we immunoblotted for p-STING, p-TBK1 and p-IRF3 at various time points up to 48 hpi with WT-KSHV. Interestingly, neither phosphorylated STING nor phosphorylated IRF3 was detected following KSHV infection, while phosphorylated TBK1 was detected up to 6 hpi but not at 24 or 48 hpi in BEC1 (Fig. 5C). However, when BEC1 were infected with KSHV first, then transfected with cGAMP, the pathway was activated as indicated by the presence of p-STING, p-TBK1 and p-IRF3 (Fig. S4). We next sought to determine if KSHV lytic replication activates innate immune signaling. To do this, we superinfected KSHV-BAC16-infected BEC1-2 and LEC6 with HD-Ad-RTA. This is a helper dependent adenovirus vector that expresses the KSHV lytic switch gene but no Adenovirus genes. While there was slight induction of IFN-β (Fig. 5D) and IFITM1 (Fig. 5E) in BEC1-2 and LEC6, the expression is far lower than what is observed during Adv infection (Fig. 1B). In addition, KSHV lytic reactivation did not activate STING signaling in BEC1, as induction of lytic WT-KSHV did not lead to detectable levels of p-STING, p-TBK1 or p-IRF3 (Fig. 5F). We confirmed lytic reactivation was occurring by measuring expression of the early gene K8.1 and late gene ORF10 and found robust induction in the KSHV + HD-Ad-RTA cells compared to KSHV-infected alone cells (Fig. S5A and B). We next investigated the activation of STING signaling in LEC6. Similar to BECs, we were unable to detect p-STING, p-TBK1 or p-IRF3 following de novo infection and lytic reactivation with WT-KSHV (Fig. 5G). Overall, these experiments reveal that while KSHV induces some expression of IFN-β in primary endothelial cells, the induction is minimal and correlates with undetectable or low levels of phosphorylated STING, TBK1 and IRF3.
Fig. 5. KSHV induces minimal innate immune signaling and does not strongly activate STING-TBK1-IRF3 in primary endothelial cells.
(A) IFN-β or (B) IFITM1 mRNA was measured by RT-qPCR from BEC1-2 and LEC6 that were infected with KSHV-BAC16 and harvested at the indicated timepoints. The relative amount of mRNA for each gene was normalized as in Fig. 1. (C) BEC1 were either infected with KSHV or transfected with 1 μg/mL CT DNA and whole cell lysates were harvested at the indicated timepoints (3 h post transfection for cells transfected with CT DNA) and immunoblotted with the indicated antibodies. The top and bottom panels are the same lysate, run on different gels. (D) IFN-β or (E) IFITM1 mRNA was measured by RT-qPCR from BEC1-2 and LEC6 that were infected with either KSHV-BAC16 or KSHV-BAC16 + HD-Ad-RTA and harvested at 24 hpi. The relative amount of mRNA for each gene was normalized as in Fig. 1. (F) BEC1 cells were infected with WT-KSHV alone or WT-KSHV + HD-Ad-RTA and harvested at 24 hpi or transfected with 1 μg/mL CT DNA (3-h transfection). Whole cell lysates were immunoblotted with the indicated antibodies. (G) LEC6 were infected with WT-KSHV, WT-KSHV + HD-Ad-RTA or transfected with CT DNA and whole cell lysates were harvested at the indicated time points (24 h for WT-KSHV + HD-Ad-RTA cells and 3 h for CT DNA transfection) and immunoblotted for the indicated antibodies. Data are shown as mean ± SEM from at least 3 biological replicates.
3.6. STING is dispensable during KSHV infection
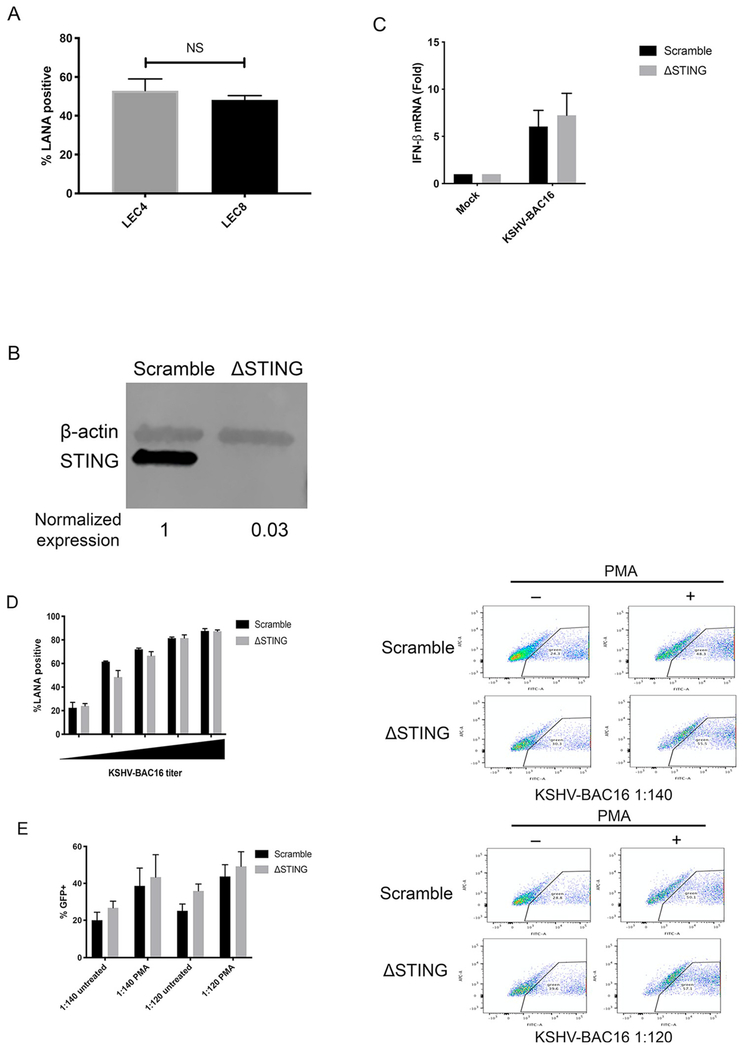
Due to the importance of cGAS-STING signaling during DNA virus infection and the presence of STING response variants in the human population, we next sought to further determine the role of STING during KSHV infection. To determine if the defect in STING signaling in LEC4 increases susceptibility to infection by KSHV, we infected LEC4 and LEC8 with KSHV-BAC16 and measured infection rates by immunofluorescence microscopy and counting of LANA-positive cells. Surprisingly, we found no difference in infection rates when comparing LEC4 to LEC8, indicating that STING may not restrict initial infection (Fig. 6A). We next wanted to further characterize the role of STING during KSHV infection by genetic ablation of STING. To study this, we used a lentivirus expressing CRISPR/Cas9 to create STING knockout (ΔSTING) BEC1-3 (Fig. 6B). To determine if STING signaling is required for the induction of IFN-β following KSHV infection, we infected ΔSTING and scramble BEC3 with KSHV-BAC16 and measured IFN-β transcripts at 48 hpi. Interestingly, ablation of STING did not reduce IFN-β expression in ΔSTING BEC3 as transcript levels were equivalent when compared with scramble BEC3 (Fig. 6C). As described in the discussion, the induction of IFN-β without activation of STING could be due to low level, undetectable activation of STING or possibly from viral dsRNA activating IFN through RNA-dependent pathways. To determine if ablation of STING would increase susceptibility to KSHV, we infected ΔSTING BEC1-3 with KSHV-BAC16 and measured infection rates as described above. Consistent with our experiments comparing susceptibility of LEC4 to LEC8, when we compared infection rates of ΔSTING BEC1-3 to the scramble control with increasing doses of KSHV-BAC16, we found no difference in susceptibility to KSHV (Fig. 6D). STING is reported to restrict lytic reactivation of KSHV (Zhang et al., 2016; Ma et al., 2017). However, this role has not been demonstrated yet in primary endothelial cells and the importance of STING in preventing KSHV spread through the culture is unclear. To examine the role of STING in restricting KSHV spread following lytic reactivation, we infected ΔSTING and scramble BEC3 with KSHV-BAC16 at different multiplicities of infection and induced lytic reactivation with PMA. We then sorted by GFP at 5 dpi and found that while PMA increased the proportion of GFP + cells in both ΔSTING and scramble BEC3, there was no difference in the increase in GFP + cells in the ΔSTING cells compared to the scramble at either dilution of virus (6E). Therefore, STING has no effect on the spread of KSHV through a culture following lytic induction. Overall, these experiments reveal that STING is dispensable during infection by KSHV, playing little or no role in reducing susceptibility to infection or restricting virus spread through the culture.
Fig. 6. STING does not reduce susceptibility during de novo infection or restrict spread of KSHV during lytic replication.
(A) LEC4 and LEC8 cells were infected with KSHV-BAC16 and infection rates were measured by immunofluorescence microscopy and quantifying the percent of LANA + cells in the culture at 48 hpi. (B) BEC1-3 were transduced with a lentivirus expressing Cas9 and a guide RNA targeting STING (ΔSTING) or a nontargeting (scramble) control and whole cell lysates were immunoblotted with the indicated antibodies. (C) Scramble and ΔSTING BEC3 from (B) were infected with KSHV-BAC16 and IFN-β mRNA was measured by RT-qPCR at 48 hpi as described earlier. (D) Scramble and ΔSTING BEC1-3 were infected with KSHV-BAC16 at different dilutions of virus and infection rates were measured by immunofluorescence microscopy and quantifying the percent of LANA + cells in the culture at 48 hpi. (E) Scramble and ΔSTING BEC3 were infected with KSHV-BAC16 at 2 different dilutions (1:140 and 1:120) for 4 h. Immediately following infection, cells were treated with PMA for 5 days. Cells were sorted by GFP by flow cytometry. Data are shown as mean ± SEM from at least 2 biological replicates.
3.7. STING is required for restricting Adv spread
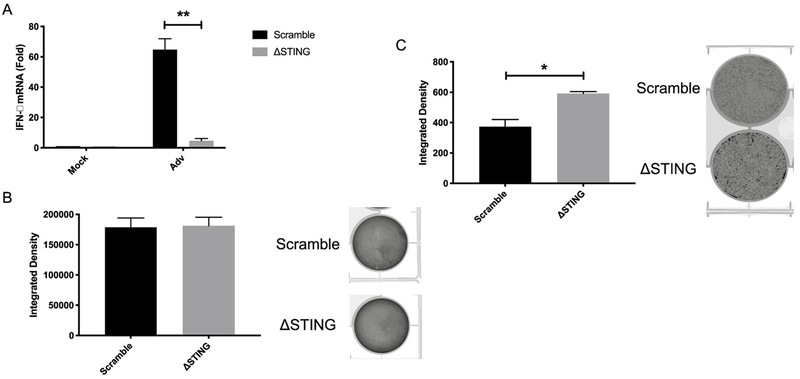
To determine if the cGAS-STING pathway is important for controlling other DNA viruses, we tested the role of defective STING signaling during Adv infection. To do this, we infected scramble and ΔSTING BEC3 with Adv and found that induction of IFN-β was nearly completely ablated in the STING knockouts (Fig. 7A). We next sought to examine the role of STING during Adv infection by infecting at high and low MOI. We reasoned that infection at high MOI would investigate the role of STING in restricting initial infection and replication whereas low MOI infections would interrogate whether STING is important for blocking spread from cell to cell. At high MOI, when infectious Adv progeny from ΔSTING and scramble control BEC3 were titered on 293T IN-β cells, there was no difference in viral titer harvested from the two cells types (Fig. 7B). However, at low MOI, Adv was able to spread through the culture more readily in the ΔSTING BEC3 compared to the scramble, as seen by an increase in integrated density (GFP intensity) in the ΔSTING BEC3 compared to the scramble control (Fig. 7C). Together, these results indicate that STING is required for restricting Adv spread through the culture but not replication.
Fig. 7. STING restricts Adv spread at low MOI.
(A) IFN-β mRNA was measured by RT-qPCR from scramble and ΔSTING BEC3 that were infected with Adv for 48 h. The relative amount of mRNA was normalized as in Fig. 1. (B) Scramble and ΔSTING BEC3 were infected with Adv at 100 genomes/cell and supernatant and cells were harvested at 5 days post infection (dpi). Viral progeny were titered on HER 293T-Integrin-β cells and integrated density of the fluorescent signal was measured by Typhoon. Representative raw plate staining is shown to the right (C) Scramble and ΔSTING BEC3 were infected with Adv at 10 genomes/cell and integrated density of the fluorescent signal was measured by Typhoon at 5 dpi. Representative raw plate staining is shown to the right. Data are shown as mean ± SEM from at least 3 biological replicates. *P < 0.05; **P < 0.01; (Student’s t-test).
4. Discussion
KSHV encodes multiple proteins that antagonize innate immune activation, including three that target cGAS or STING directly (Wu et al., 2015; Zhang et al., 2016; Ma et al., 2015). We wanted to determine if the cGAS-STING pathway was relevant for KSHV infection and the establishment of latency or if these inhibitors obviated the need for STING activation during infection. While studying the role of cGAS-STING in a relevant cell type for KS spindle cells, we fortuitously identified a lot of primary dermal microvascular lymphatic endothelial cells that were deficient for IFN-β activation by dsDNA or cGAMP. This primary cell lot did not induce significant IFN-β levels in response to KSHV infection. We found there were not significant differences in infection rates between LEC4 and other lots of primary LEC cells indicating that the ability to activate STING in response to infection is not likely to be important for initial KSHV infection. To further analyze the role of STING in KSHV infection, we used Crispr/Cas9 to knock out STING in primary dermal blood microvascular endothelial cells. Again, there was no difference between infection rates of cells with functional cGAS-STING pathway and cells where STING was knocked out. Therefore, we conclude that STING is not important for initial infection of endothelial cells and the establishment of latency.
Spread of KSHV through the culture is also not inhibited by the presence of STING. Following lytic induction of wild-type and knockout blood endothelial cells infected with KSHV, we were able to identify spread of KSHV through the culture. There were no differences in the increases in KSHV-positive cells in the culture following lytic induction in the wild-type or STING knockout endothelial cells, indicating that STING signaling is not critical for controlling KSHV lytic replication and spread to new cells. Interestingly, while replication following infection of the endothelial cells with Adv is also not altered in the absence of STING, spread though the culture at low MOI is. This result is consistent with a previous report regarding infection, although the finding that STING restricts Adv spread has not been reported previously (Lam and Falck-Pedersen, 2014). Pre-treatment of endothelial cells with activators of the cGAS/STING pathway nearly completely blocked infection with KSHV. Since pre-treatment with IFN-β blocks initial infection but STING has little to no effect on KSHV spread, KSHV spread through the culture must rely upon efficient inhibition of the cGAS-STING pathway by KSHV. Thus, the KSHV latent and lytic proteins that are able to inhibit the activation of the cGAS-STING pathway obviate the viral inhibition by the STING signaling pathway during lytic infection and spread. However, not all DNA viruses efficiently block the effects of STING activation as knockout of STING allowed more efficient spread of Adenovirus through the culture.
At 48 h post infection, KSHV induced small amounts of IFN-β. However, we could still not detect activation of STING at this time point. It is possible that there are simply low levels of activation of STING. However, we showed that even in the STING pathway deficient cells, dsRNA was still able to induce interferon. Therefore, at 48 h post infection there could be some activation of interferon through the formation of viral dsRNA. Alternatively, there could be other pathways that activate small amounts of interferon induction independent of the cGAS-STING pathways. While the induction of interferon was detectable in the KSHV-infected endothelial cells, it was greatly muted as compared to Adv infection or induction with dsDNA or cGAMP and it was only detected at 48 h post infection. Therefore, we believe that this amount was not significant enough to prevent spread of KSHV through the culture.
This study examines the role of endogenous cGAS-STING activity during de novo infection and lytic reactivation of KSHV in primary endothelial cells and shows that the cGAS-STING pathway is not efficiently activated during KSHV infection of endothelial cells. While many other studies have made important contributions to our understanding of KSHV-innate immune interactions and have pointed to a role for STING in restricting lytic replication (Zhang et al., 2016; Ma et al., 2017), we have performed wild-type infection studies in endothelial cells. We show that STING does not play a significant role in KSHV infection, replication or spread in endothelial cells. As KS spindle cells appear to be most closely aligned with endothelial cells, these studies show the relevance of STING in a cell type that potentially provides relevance to in vivo KSHV-related tumors.
Supplementary Material
Acknowledgements
We thank Daniel Stetson for the pRRL STING gRNA plasmid.
Grant information: DV was partially supported by the Viral Pathogenesis Training Grant (T32AI083203). These studies were supported by grants to ML from the National Cancer Institute (RO1 CA217788, RO1 CA097934 and R21CA240479). Additional funding was provided by the National Institute Of Allergy and Infectious Diseases under Award Number K22AI081870 (to J.G.S.) and by the Office Of The Director, National Institutes Of Health under Award Number S100D026741 (to J.G.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.11.012.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abe T, Barber GN, 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol 88, 5328–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Caviar T, Deimling T, Witte G, Röhl I, Hopfner K-P, Ludwig J, Hornung V, 2013. cGAS produces a 2’-5’,-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi 0, 2006. Pathogen recognition and innate immunity. Cell 124 (4), 783–801. [DOI] [PubMed] [Google Scholar]
- Aresté C, Blackbourn DJ, 2009. Modulation of the immune system by Kaposi’s sarcoma-associated herpesvirus. Trends Microbiol. 17 (3), 119–129. [DOI] [PubMed] [Google Scholar]
- Bechtel JT, Liang Y, Hvidding J, Ganem D, 2003. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol 77, 6474–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS-Y, Ensser A, Wong L-Y, Toth Z, Lee SH, Lee H-R, Myoung J, Ganem D, Oh T-K, Kim JF, Gao S-J, Jung JU, 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol 86, 9708–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z, 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147, 436–446. [DOI] [PubMed] [Google Scholar]
- Choi I, Lee S, Hong YK, 2012. The new era of the lymphatic system: No longer secondary to the blood vascular system. Cold Spring Harb. Perspect Med 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokunonga E, Levy LM, Bassett MT, Mauchaza BG, Thomas DB, Parkin DM, 2000. Cancer incidence in the African population of Harare, Zimbabwe: second results from the cancer registry 1993-1995. Int. J. Cancer 85, 54–59. [DOI] [PubMed] [Google Scholar]
- Dejana E, Hirschi KK, Simons M, 2017. The molecular basis of endothelial cell plasticity. Nat. Commun 8, 14361 (Nature Publishing Group). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio TA, Lagunoff M, 2012. KSHV induction of angiogenic and lymphangiogenic phenotypes. Front. Microbiol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner Elie J., Burdette Dara L., Wilson Stephen C., Monroe Kathryn M., Kellenberger Colleen A., Hyodo Mamoru, Hayakawa Yoshihiro, Hammond Ming C., Vance RE, Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE, 2013. The innate immune DNA sensor cGAS produces a non-canonical cyclic-di-nucleotide that activates human STING. Cell Rep. 3, 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB, 2016. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 45, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, Chen ZJ, 2019. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567 (7747), 262–266 (Nature Publishing Group). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN, 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN, 2009. STING regulates intracellular DNA-mediated , type I interferon-dependent innate immunity. Nature 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG, 2011. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32, 574–581. [DOI] [PubMed] [Google Scholar]
- Kell AM, Gale M, 2015. RIG-I in RNA virus recognition. Virology 479–480, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunoff M, Bechtel J, Venetsanakos E, Roy A, Abbey N, Herndier B, Mcmahon M, Ganem D, 2002. De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol 76, 2440–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Falck-Pedersen E, 2014. Unabated adenovirus replication following activation of the cGAS/STING-dependent antiviral response in human cells. J. Virol 88, 14426–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Choi UY, Hwang SW, Kim S, Jung JU, 2016. Viral inhibition of PRR-Mediated innate immune response: learning from KSHV evasion strategies. Mol. Cells 39 (11), 777–782 (Korean Society for Molecular and Cellular Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ, 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wilson HL, Kiss-Toth E, 2017. Regulating STING in health and disease. J. Inflamm 14, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lin L, Tong Y, Liu Y, Mou J, Wang X, Wang X, Gong Y, Zhao Y, Liu Y, Zhong B, Dai L, We YQ, Zhang H, Hu H, 2018. TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discov. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio C-WJ, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, Pham E, Benedict CA, Sharma S, 2016. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J. Virol 90, 7789–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIP induces IRF3 activation. Science 80, 347. [DOI] [PubMed] [Google Scholar]
- Liu D, Wu H, Wang C, Li Y, Tian H, Siraj S, Sehgal SA, Wang X, Wang J, Shang Y, Jiang Z, Liu L, Chen Q, 2018. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 26 (9), 1735–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B, 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. U. S. A 112, E4306–E4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Hopcraft SE, Yang F, Petrucelli A, Guo H, Ting JPY, Dittmer DP, Damania B, 2017. NLRX1 negatively modulates type I IFN to facilitate KSHV reactivation from latency. PLoS Pathog. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C, 2010. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen EK, Nemerow GR, Smith JG, 2010. Direct evidence from single-cell analysis that human {alpha}-defensins block adenovirus uncoating to neutralize infection. J. Virol 84, 4041–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G, Konno H, Barber GN, 2017. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijo J, Döring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U, 2016. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. 12, e1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Jin L, 2019. TMEM173 variants and potential importance to human biology and disease. Genes Immun. 20 (1), 82–89 (Nature Publishing Group). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjabi AS, Carroll PA, Chen L, Lagunoff M, 2007. Persistent activation of STAT3 by latent Kaposi’s sarcoma-associated herpesvirus infection of endothelial cells. J. Virol 81, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez EL, Pulliam TH, Dimaio TA, Thalhofer AB, Delgado T, Lagunoff M, 2017. Glycolysis, glutaminolysis, and fatty acid synthesis are required for distinct stages of Kaposi’s sarcoma-associated herpesvirus lytic replication. J. Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM, 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirena D, Ruzsics Z, Schaffner W, Greber UF, Hemmi S, 2005. The nucleotide sequence and a first generation gene transfer vector of species B human adenovirus serotype 3. Virology 343, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, Haase AT, 1997. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol 71, 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ, 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S, 2010. Pattern recognition receptors and inflammation. Cell 140, 805–820. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ, 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA, 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3, 920–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, 2013. The interferon response to intracellular DNA: why so many receptors? Immunobiology 218, 1312–1321. [DOI] [PubMed] [Google Scholar]
- Wabinga HR, Nambooze S, Amulen PM, Okello C, Mbus L, Parkin DM, 2014. Trends in the incidence of cancer in Kampala, Uganda 1991-2010. Int. J. Cancer 135, 432–439. [DOI] [PubMed] [Google Scholar]
- Wang H-W, Trotter MWB, Lagos D, Bourboulia D, Henderson S, Mäkinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C, 2004. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet 36, 687–693. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C, 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41, 919–933. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG, 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger W, Partanen TA, Breiteneder-Geleff S, Mayer C, Kowalski H, Mildner M, Pammer J, Stürzl M, Kerjaschki D, Alitalo K, Tschachler E, 1999. Expression of vascular endothelial growth factor receptor-3 and podoplanin suggests a lymphatic endothelial cell origin of Kaposi’s sarcoma tumor cells. Lab. Investig 79, 243–251. [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ, 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339 (80-), 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F, 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ, 2013. Cyclic GMP-AMP containing mixed Phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF, 2016. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. U. S. A 113, E1034–E1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang Y-Y, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu H-B, 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550. [DOI] [PubMed] [Google Scholar]
- Zhou X, Robinson CM, Rajaiya J, Dehghan S, Seto D, Jones MS, Dyer DW, Chodosh J, 2012. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Investig. Ophthalmol. Vis. Sci 53, 2804–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.