Abstract
This review discusses Gō models broadly used in biomolecular simulations. I start with a brief description of the original lattice model study by Nobuhiro Gō. Then, the theory of protein folding behind Gō model, free energy approaches, and off-lattice Gō models are reviewed. I also mention a stringent test for the assumption in Gō models given from all-atom molecular dynamics simulations. Subsequently, I move to application of Gō models to protein dynamical functions. Various extension of Gō models is also reviewed. Finally, some publicly available tools to use Gō models are listed.
Keywords: Gō model, structure-based model, protein folding, funnel energy landscape, coarse-grained simulation
Significance.
Since the very first paper by Taketomi, Ueda, and Gō in 1975, the so-called Gō model has been broadly used in various biomolecular simulations. A brief history, a stringent test, various extensions, and broad range of applications are reviewed.
Since the very first paper by Taketomi, Ueda, and Gō in 1975 [1], the so-called Gō model has long been used to broad range of biomolecular simulations, primarily for, but not limited to, protein folding studies. In this review article, I start with a brief description of the original work by Gō, which is followed by subsequent developments, more recent application of Gō model, and discussion of future directions.
While not many, in the last 20 years, the Gō model has been reviewed in a few articles [2–4], which would complement this review.
Lattice model by Gō
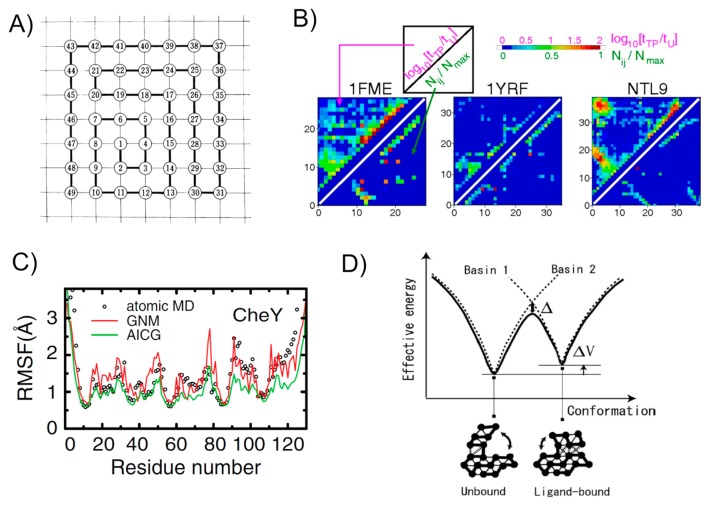
Teketomi, Ueda, and Gō, for the first time, introduced a lattice model to protein folding study [1]. In their model, each monomer in “protein” is placed on a lattice point in two dimension and is connected by bonds that have the unit length of the lattice (Fig. 1A). Starting from random configurations, the Metropolis Monte Carlo simulation was used to fold this model protein. Monomers that are separate by the unit length and not connected by a bond have non-local contact interactions. For the contact interactions, they considered three cases, a weak limit of specificity, an intermediate specificity, and a strong limit of specificity. It is in the third class that turned out to be called Gō-model later; two beads have negative contact interaction energy only when these pairs are in contact at the pre-defined native structure. Otherwise, the two beads have no contact energy. The Monte Carlo simulations resulted in complete folding to the native structure only for the strong limit of specificity, but not for the other two cases. A subsequent paper by Gō and Taketomi introduced the specificity to the local potential, a negative energy only when the bond angle is the same as that in the predefined native one, studying the balance between local and non-local specific interactions [5]. In these and other studies, Gō and his collaborators pursued statistical physics of the strong limit of specificity, finding many qualitatively consistent results with experiments, as summarized in the seminal review [6]. In their lattice model, the protein folds to the native configuration below the melting temperature, above which the protein unfolds. The transition is fairly cooperative, showing a clear peak in the heat capacity. The folding occurs faster when the local preference energy is larger, relative to the non-local contact energy, whereas the folding transition is more cooperative when the non-local contact contribution is larger. All these are very robust physical properties well supported by biophysical experiments and shared by more accurate protein simulations performed to date.
Figure 1.
Various studies with Gō models. A) One of the original two-dimensional lattice models used by Gō. The figure taken from [5]. B) A stringent test of the assumption in Gō models. For contact i and j, the log ratio of lifetime in the transition-path tTP to lifetime in the unfolded state tU is plotted by color in upper-left triangle, while the native contact map is depicted in the lower-right triangle. Results for three proteins are drawn here. The figure taken from [35]. C) Comparison of the root mean square fluctuation in the native basin for a test protein CheY. Results from all-atom MD (black circles), an elastic network model (GNM) (red curve), and a Gō model (green curve) are compared. The figure taken from [30]. D) A schematic plot for the multiple-basin Gō model that is based on two single Gō models. The picture taken from [41].
About 15 years later from the first paper by Gō, the lattice model of protein folding got popularity, notably by the studies of Dill, Shakhnovich, and Onuchic and Wolynes [7–10]. Importantly, in this second generation of the lattice protein model, monomer contact energies were not biased to their pre-defined native structure, but are purely sequence-based. So, these models are not a sort of Gō model. But, sometimes, the Gō model was mentioned as a control, which served as the limit of strong specificity. It was in this context that people started to call this type of models as “Gō model” [7,8]. Retrospectively, due to its strong geometrical constraint, the sequence-based, i.e., non-Gō type lattice models may have pronounced ruggedness, which might have overestimated the ruggedness of the energy landscape, comparing with nowadays-available all-atom molecular dynamics (MD) simulations for folding.
Theory behind
Through series of lattice model simulations and lessons from many X-ray crystallographic structures, Nobuhiro Gō proposed the consistency principle in the 1983 review [6]. In their native structures, proteins individually take the optimal local interactions, akin to the secondary-structure, and non-local interactions, i.e., the tertiary structure. This dual optimality was attained via evolutionary selections of the amino acid sequence. The review also mentioned about the kinetic aspect of the consistency principle; the balance between the local and non-local specificity, which is necessary for the well-behaved cooperative folding-unfolding transition. The consistency principle serves as a theoretical basis to use the Gō model. The consistency principle, and thus the Gō model, were considered as an ideal limit, whereas real proteins must have other restraints, such as functional restraints.
Some years later, extending Gō’s perspective, Wolynes and his coworkers formulated a statistical physical theory of protein folding and elucidated global picture of energy landscape of proteins [11,12]. The theory clarified that, for a given temperature, the energy bias to the native structure has to be large enough, relative to the average ruggedness of the energy surface (frustration) for the protein being able to fold to the native structure avoiding glassy slow dynamics. Here, the energy bias is linked to the thermodynamic stability T < Tm, where T is the physiological temperature and Tm is the melting temperature. The average ruggedness is correlated with the characteristic glass-transition temperature Tg for the onset of slow dynamics; thus, as the kinetic condition, T > Tg is to be satisfied. Together, Tg < T < Tm gives the foldability condition. For fast folding of proteins, the frustration at the native basin and on the route to it has to be small enough, which is termed the principle of minimum frustration.
While the consistency principle of Gō and the principle of minimum frustration apparently have overlap, they also differ in some respects. First, the consistency principle is a concept stated primarily by words, while the principle of minimum frustration gives mathematical expressions. Secondly, the consistency principle primarily states on the structural aspect at the native state and thus on the thermodynamic stability, in addition to the balance between the local and non-local specificities for experimentally observed cooperative folding-unfolding transitions. The Wolynes theory defines the foldability as the combination of the stability and kinetics; T < Tm from the stability and T > Tg from the kinetics.
Subsequent computer simulations by Onuchic and others pointed out importance of the network of folding pathways [13]; the folding is fast and efficient when there are multiple parallel pathways that are linked to the native state, while it is slow when there is a bottleneck where only few routes exist. This ends up with the concept of protein folding funnel; fast folding proteins have funnel-like energy landscapes [14]. The solvent-averaged energy of a fast-folding protein, on average, decreases as the protein approaches to the native state, which correspond to the bottom of the funnel. If the effective energy monotonically decreases, how does the folding free energy barrier arise? As the folding reaction proceeds, the number of available conformations, and thus the conformational entropy decreases, which opposes the folding. It is thus the energy-entropy compensation that gives rise to the free energy barrier along the folding reaction coordinate. Conformational bottleneck corresponds to a large conformational entropy loss, which increases the free energy at the bottleneck.
Gō model can be considered as a concise mean to realize the consistency principle, or the perfect funnel landscape. Assumed in Gō model is that only natively-contacting pairs (native contacts) have crucial contribution to the folding, but other pair interaction (non-native contacts) can be ignored.
Free energy function type Gō model
In the same spirit of Gō model, Wako and Saito proposed an Ising-like model to describe protein folding processes [15,16], which was, many-years later, reinvented and applied to many proteins by Munoz and Eaton [17]. In the model, each residue takes two state, n (native) and u (unfolded) so that a microstate of a protein is described as a sequence of n and u, such as uuuunnnnuu for a 10-residue case. The global native state N and unfolded state U correspond to the states that all the residues take n and u microstates, respectively. Only when both of natively interacting residues as well as all the residues in between take the n states, they get a certain stability, which basically is the same assumption as the Gō model. Non-native contacts are completely ignored. As residues change from the u state to the n state, the chain loses its conformational entropy. Thus, as the protein folds, it gains energetic stability and loses its conformational entropy. Since the folding transition is modeled as the energy-entropy compensation process, the optimal folding pathways are determined so as to minimize the entropy loss for a given energy gain. Under some assumptions, free energy of microstates can be analytically calculated by a transfer-matrix approach, which is a clear advantage of this type of modeling, compared to the lattice Gō model. This simple modeling turned out to have marked predictive power of folding transition ensembles and folding rates, when compared with experimental phi-value analysis and folding rates. Related free energy function approaches were developed by a few other studies, as well [2,18–20]. Conversely, this success strongly supports the consistency principle and the perfect funnel view. More recently, the Wako-Saito-Munoz-Eaton model and its extension have been broadly used in diverse folding studies [21,22].
Off-lattice Cα Gō model
While the lattice model was powerful to reveal conceptual aspects in protein folding, its highly restricted geometry precludes direct application to real protein structures. Especially, low-frequency and collective fluctuations around the native structures cannot easily be represented by lattice models. In this respect, another class of minimal protein models, called the elastic-network-model, was invented first by Tirion in 1996 [23], and further developed by Jernigan, Bahar, and others [24]. The elastic network model consists of elastic bonds between two monomers that are spatially close to each other at the native structures, with the natural lengths of the elastic bond being the lengths at the native structure. Albeit its ultra-simplicity, the elastic network model was shown to represent low-frequency fluctuations of proteins in the native state surprisingly well. As in the case of Gō model, the elastic network model directly uses and is biased towards the native structure by construction. However, because all the interactions are elastic, the elastic network model cannot approximate the unfolded state at all, and thus is not considered as a Gō model.
The lattice Gō model is a concise realization of protein folding, but not good for native fluctuation dynamics. On the other hand, the elastic network model is a simple and minimal model to approximate low-frequency native fluctuations, but does not take into consideration of unfolding. Soon after Tirion’s work, Clementi, Nymeyer, and Onuchic proposed an off-lattice Gō model that resulted in taking advantage of the two minimal models; the model represents a protein as a chain of Cα atoms of every amino acids and that has both local angle and non-local contact potentials biased towards the native structure. In the low temperature limit, the model converges to the elastic network model, whereas the protein unfolds cooperatively at a higher temperature. They applied this off-lattice Cα Gō model to small fast-folding proteins, directly comparing the folding pathways with experiments [25]. Several subsequent works together showed its predictive power for folding reaction mechanisms [26,27].
As is clear by now, “Gō model” does not mean a single model, but represents a class of models that share the concept in the original work by Gō. Although no one clearly defined it, in practice, Gō models share the two concepts. 1) They represent folding-unfolding transitions and 2) take into accounts pairwise contact energies only for the pairs that are in contact in the native structure. The Gō model is often called “structure-based model” as well since the energy function is explicitly dependent on the native structure. While the two names are used as synonym, they can be slightly different; e.g., the elastic network model of Tirion is clearly a structure-based model, but is not a sort of Gō model.
The first version of off-lattice Cα Gō model by Clementi, C., et al. is a minimal and concise model so that there is room to add some more details. Karanicolas and Brooks added sequence-dependent local terms, chemically-motivated energy scales for the contact energies, and a desolvation potential in non-local contacts, improving predictive power of folding transitions and pathways [28]. A similar desolvation energy was addressed by Kaya and Chan, as well [29]. Multiscale algorithms were utilized to reflect more sequence-and structure-specific interaction at atomic level within Cα Gō model [30,31]. Furthermore, the chemical denaturant effect was incorporated into an off-lattice Gō model [32].
Testing Gō model assumption via all-atom molecular simulations
A special-purpose supercomputer Anton and the associated MD software Desmond were used to simulate long-time folding dynamics of small proteins with an all-atom force-field and explicit water solvent, which successfully realized reversible folding and unfolding for more than 10 proteins [33,34]. They observed in the trajectories that, whereas nonnative secondary structures do form in the unfolded state ensemble for some proteins, they are transient and disappear either before initiation of folding events or early during the folding. This implies that these non-native contacts do not contribute much to the folding mechanisms, supporting the assumption behind Gō models.
Soon after this all-atom folding simulations, these trajectories were further analyzed from the perspective of folding mechanisms by Best, Hummer, and Eaton [35]. Probably, this analysis provides the most convincing data to date that supports the native-contact centric view and thus the use of Gō model. For each residue pair, they compared the lifetime of that contact in the transition-path and the lifetime of the same contact in the non-native ensemble in the trajectories (the transition-path stands for a fragment of MD trajectory that departs from the unfolded basin and reaches at the native state) (Fig. 1B). If the former lifetime is much longer than the latter for a pair, this indicates the pair has significant role for folding. Quantifying this ratio, they found that the high scores are located only in the native contact pairs and their neighbors for all but one protein analyzed. Interestingly, one exception was a designed protein, of which sequence has not evolved naturally. For naturally-evolved proteins, not all the native contacts are equally resistant; the high scored regions correlate with the folding initiation sites. They also performed a Bayesian analysis finding similar tendency; native contacts are formed at higher probabilities in the transition-path than the non-native contacts. This analysis unambiguously shows that, in the successful folding and unfolding transitions, natively formed contacts play major roles, while non-native contacts do not contribute significantly; thus, this directly rationalizes the use of Gō models in predicting/revealing folding mechanisms for natural proteins. Furthermore, the Wako-Saito-Munoz-Eaton model was applied to the same set of proteins as those simulated with Anton, finding highly consistent results [36].
From folding to function with Gō model
It has been shown that off-lattice Gō models are useful not only for protein folding, but also for native-state dynamic simulations [30,37]. Somewhat surprisingly, comparing with the root-mean-square-fluctuations (RMSF) calculated by all-atom MD with explicit water, off-lattice Gō models agree better than the elastic network model, which indicates that even the native dynamics of proteins reflect some un-harmonic part of potentials that is linked to local unfolding (Fig. 1C). It was investigated that, relative to the elastic network model, local unfolding, or cracking, decreases the free energy barrier for conformational transition [38]. With a multiscale-calibrated version of Gō model, the correlation between the RMSF by the Gō model and that by all-atom MD with explicit water was as high as the correlation between the RMSF by all-atom MD with implicit water and that with explicit water [39].
Given that the native dynamics can be well captured by Gō models, we can go further simulating conformational transition between two or more distinct conformations relevant to allosteric regulations. When two distinct conformations, A and B, for a target protein are available from experiments, we can construct the two respective Gō models V(R|A), and V(R|B), each making funnel-like landscapes centered around A and B basins, respectively, where the vector R collectively represents all the particle coordinates. If we think of V(R) = Min (V(R|A), V(R|B)), this potential V(R) encodes two basins A and B in its energy landscape (Fig. 1D). To use it in MD simulations, we need to make it differentiable. There can be multiple means to do it. Best and Hummer proposed the so-called soft-min function, represented as a combination of logarithmic and exponential functions, describing conformational change of a protein [40]. Okazaki, K., et al. used the secular equation formalism for the purpose [41]. Notably, these multiple-Gō model formalisms create two basins in the global energy landscape, but do not make intermediate states between two conformations. As an opposite limit of cooperativity, one can put two local minima in every pair contact potentials, corresponding to the pair distances in conformations A and B [42,43]. While this formalism also encodes two basins A and B, it also allows to possess multiple intermediate states between two endpoint configurations. This locally multiple Gō contact formalism is useful to investigate detailed pathways for conformational change. There can be ways to put a cooperativity in between these two limits: One can use the global multiple-Gō modeling for certain fragments of a protein [44,45]. Alternatively, one can design an arbitrary range of cooperativity in the conformational transition [46].
With the framework of constructing multiple-basins in hand, one can treat the ligand binding dynamic process in a few different manners. For a minimal modeling of conformational transition dynamics, one may simply change the relative free energy between two basins mimicking the ligand binding [47]. As a second level, one can introduce an implicit ligand binding interaction potential which effectively stabilizes the residues neighboring the ligand. Then, the ligand binding and unbinding is mimicked by truing on and off of this implicit ligand binding potential. This turning on and off process can be realized by Monte Carlo process, thus making the entire simulation as the hybrid-MC/MD simulation. Namely, the ligand binding and unbinding, or even chemical reactions, can be treated by MC, whereas protein conformational motions are treated by MD [48]. This scheme has been applied to modeling whole enzymatic dynamics of adenylate kinase as well as FO part of F-type ATP synthase [44,49]. As a third and more microscopic view, one can treat ligand as an explicit molecule represented by a few beads [42].
To mimic an entire cycle of conformational transition for huge molecular complexes, such as molecular machines, one may favor even a simpler approach; sudden switch of the reference (native) structures in Gō models. For example, a protein may have an open conformation A in the apo state and a closed conformation B in the ligand-bound state. For mimicking the ligand-binding induced conformational change, during a MD simulation with a Gō model based on the A structure, one can suddenly (or gradually) switch the potential surface to a new Gō model based on the B structure. After the sudden (or gradual) switch, the protein conformation smoothly relaxes from the A basin to the new structure B, which can be thought as a proxy to the conformational change. It should be noted that the sudden switch of potential places the protein at quite a high energy state in the new potential surface so that the initial part of the relaxation process tends to contain more artefacts. This “switching Gō model” was applied to a rotary molecular motor, F1-ATPase [50], molecular chaperone [51], and, more recently, for ATP-dependent chromatin remodeler [52].
Integrating with physical model
While the Gō model represents an ideal protein encoding the prefect funnel picture, real proteins do have some non-ideality. Functional restraints give rise to frustrations that are enriched near active sites as well as allosteric sites [30,53]. To model non-ideality on top of the funnel picture, it has been devised to integrate the Gō type bias with purely sequence-based and physico-chemical potentials. It should be noted that earlier off-lattice Cα Gō models such as [28] already took into account the sequence-based interaction to some extent. More thorough sequence-based interactions, i.e., non-native contact interactions, were also incorporated into Gō type modeling; for example, the AWSEM provides a spectrum of models from a purely sequence-based model for protein structure prediction to a Gō type model representing the perfect funnel landscape [54,55].
Within Cα models, one can approximate backbone hydrogen bonds and sidechain interactions using orientation-dependent potential functions. Hoang, T. X., et al. proposed to use three consecutive Cα positions to calculate orientations of backbone amide- and carbonyl- groups of the central residue so that the orientation-dependent backbone hydrogen bonds can be modeled as a function of 6 Cα coordinates, which was carefully calibrated later [56,57]. A similar approach was further developed to model the sidechain orientation in terms of three consecutive Cα positions [58,59]. This type of modeling is particularly useful to model intrinsically disorder proteins/regions that lack the native structure, as well as amyloid like higher-order structures. While these physico-chemical interactions alone cannot specific enough to fold a globular protein purely from sequence information, one can combine it with Gō type bias, whenever necessary.
Further spreading and future directions
While Gō models have originally been developed for and applied to proteins, one can extend the same idea to other macromolecules. The RNA folding and functional dynamics have been studied by some Gō models together with physico-chemical interactions [60–62]. For DNA, Gō type biases have been added to more physico-chemical interactions to stabilize B-type duplex form [63] although this bias was much weakened in more recent modeling. Notably, Gō type model has never been used for lipid, to my knowledge; lipid in biology stays in the liquid phase and thus it is not compatible with the idea of Gō model. In other words, the applicability of Gō models is linked to the availability of X-ray crystallographic structures; if a biomolecule crystalizes and its crystal diffracts X-ray well, that structure can be specific enough to rationalize the use of Gō models.
Gō models have also been used at the level of all-atom models [64]. Sanbonmatsu, Whitford, and Onuchic successfully applied it to reveal energy landscape of ribosome conformational dynamics and others [65]. The SMOG server provides a useful platform to run the all-atom, as well as coarse-grained, Gō models [66,67].
While Gō models were originally proposed as means to elucidate protein folding mechanisms, current and perhaps future usage of Gō models more often aims at fast conformation sampling. To characterize free energy landscape/pathways of some protein conformational change at atomic details, one often uses advanced sampling methods, such as Markov state models, and the string-method. These methods need initial and rough samples of relevant part of conformational space. Gō models are very efficient tools to prepare these samples.
Another interesting direction is structure-modeling; when high-resolution structures are available for most parts of a protein separately, but not the full-length of the protein, one can integrate them using a sort of Gō models [45]. Notably, most Gō models do not need information of Cartesian coordinates of the entire system. Instead, they need collections of many pairwise contact information. Thus, structure-information of many parts can be naturally merged into the full-length protein within Gō models
For many of these useful applications, when one uses Cα Gō model, one needs to reconstruct all-atom models that are compatible with the given Cα structure. This so-called back-mapping has been well developed and one can immediately reconstruct all-atom protein models given a reasonable Cα-model structure [68,69].
Publicly available tools for Gō models
There are several publicly available tools/servers to run Gō model simulations immediately although writing the code from scratch can also be straightforward. The SMOG server, a very useful web server (the SMOG can be used as a standalone program, as well), produces the necessary data for Gō model simulations with GROMACS [70], by which one can run MD [67,71]. It covers many of the variants described here, including Cα and all-atom Gō models, multiple-basin Gō model, as well as RNA Gō model. The MMTSB (Multiscale Modeling Tools for Structural Biology) tool set also offers a “Go Model Builder”, which provides necessary files for running Cα Gō model simulations with MD engines, such as CHARMM, GENESIS, and others [72–74]. The eSBMTools is a convenient python source-code package for various Gō model simulations, providing interface to run MD with GROMACS, somewhat similar to SMOG [75]. The CafeMol is a standalone software that implements a simple as well as a fine-tuned version of Cα Gō models, a multiple-basin Gō model, a RNA Gō model, together with an experiment-based coarse-grained DNA model and protein-DNA interactions [37]. The AWSEM-MD package offers a broad range of models from a purely physico-chemical and sequence-based interaction model, in one limit, to a Gō model in the other limit [55]. It uses a three-beads-per-amino-acid resolution. The AWSEM-MD produces an interface to run MD with the LAMMPS MD suite.
Acknowledgement
This work was supported by the MEXT as “Priority Issue on Post-K computer”, and by the Japan Science and Technology Agency (JST) grant JPMJCR1762.
Footnotes
Conflict of Interest
None
Author contribution
S. T. wrote the manuscript.
References
- 1.Taketomi H, Udea Y, Go N. Studies on protein folding, unfolding and fluctuations by computer simulation. I. The effect of specific amino acid sequence represented by specific inter-unit interactions. Int J Pept Protein Res. 1975;7:445–459. [PubMed] [Google Scholar]
- 2.Takada S. Go-ing for the prediction of protein folding mechanisms. Proc Natl Acad Sci USA. 1999;96:11698–11700. doi: 10.1073/pnas.96.21.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Hills R, Brooks C. Insights from Coarse-Grained Gō Models for Protein Folding and Dynamics. Int J Mol Sci. 2009;10:889–905. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Go N, Taketomi H. Respective roles of short-and long-range interactions in protein folding (mechanism of folding/range of interactions/lattice model/Monte Carlo method) Proc Nat Acad Sci USA. 1978;75:559–563. doi: 10.1073/pnas.75.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Go N. Theoretical studies of protein folding. Annu Rev Biophys Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- 7.Dill KA, Bromberg S, Yue K, Fiebig KM, Yee DP, Thomas PD, et al. Principles of protein folding―A perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leopold PE, Shakhnovich EI. Protein folding kinetics in the dense phase. [1993] Proceedings of the Twenty-sixth Hawaii International Conference on System Sciences; 1993.IEEE; pp. 726–735. [Google Scholar]
- 9.Šali A, Shakhnovich E, Karplus M. Kinetics of Protein Folding. A lattice model study of the requirements for folding to the native state. J Mol Biol. 1994;235:1614–1636. doi: 10.1006/jmbi.1994.1110. [DOI] [PubMed] [Google Scholar]
- 10.Socci ND, Onuchic JN, Wolynes PG. Diffusive dynamics of the reaction coordinate for protein folding funnels. J Chem Phys. 1996;104:5860–5868. [Google Scholar]
- 11.Bryngelson JD, Wolynes PG. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 13.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 15.Wako H, Saitô N. Statistical Mechanical Theory of the Protein Conformation. I. General Considerations and the Application to Homopolymers. J Physical Soc Japan. 1978;44:1931–1938. [Google Scholar]
- 16.Wako H, Saitô N. Statistical Mechanical Theory of the Protein Conformation. II. Folding Pathway for Protein. J Physical Soc Japan. 1978;44:1939–1945. [Google Scholar]
- 17.Muñoz V, Eaton WA. A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc Natl Acad Sci USA. 1999;96:11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portman JJ, Takada S, Wolynes PG. Variational Theory for Site Resolved Protein Folding Free Energy Surfaces. Phys Rev Lett. 1998;81:5237–5240. [Google Scholar]
- 19.Galzitskaya OV, Finkelstein AV. A theoretical search for folding/unfolding nuclei in three-dimensional protein structures. Proc Natl Acad Sci USA. 1999;96:11299–11304. doi: 10.1073/pnas.96.20.11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alm E, Baker D. Prediction of protein-folding mechanisms from free-energy landscapes derived from native structures. Proc Natl Acad Sci USA. 1999;96:11305–11310. doi: 10.1073/pnas.96.20.11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh K, Sasai M. Cooperativity, connectivity, and folding pathways of multidomain proteins. Proc Natl Acad Sci USA. 2008;105:13865–13870. doi: 10.1073/pnas.0804512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruscolini P, Naganathan AN. Quantitative Prediction of Protein Folding Behaviors from a Simple Statistical Model. J Am Chem Soc. 2011;133:5372–5379. doi: 10.1021/ja110884m. [DOI] [PubMed] [Google Scholar]
- 23.Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys Rev Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 24.Bahar I, Jernigan RL. Inter-residue potentials in globular proteins and the dominance of highly specific hydrophilic interactions at close separation. J Mol Biol. 1997;266:195–214. doi: 10.1006/jmbi.1996.0758. [DOI] [PubMed] [Google Scholar]
- 25.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 26.Hoang TX, Cieplak M. Sequencing of folding events in Go-type proteins. J Chem Phys. 2000;113:8319–8328. [Google Scholar]
- 27.Koga N, Takada S. Roles of native topology and chain-length scaling in protein folding: a simulation study with a Go-like model. J Mol Biol. 2001;313:171–180. doi: 10.1006/jmbi.2001.5037. [DOI] [PubMed] [Google Scholar]
- 28.Karanicolas J, Brooks CL. The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaya H, Chan HS, Solvation Effects. Driving Forces for Protein Thermodynamic and Kinetic Cooperativity: How Adequate is Native-centric Topological Modeling? J Mol Biol. 2003;326:911–931. doi: 10.1016/s0022-2836(02)01434-1. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Wolynes PG, Takada S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc Natl Acad Sci USA. 2011;108:3504–3509. doi: 10.1073/pnas.1018983108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Terakawa T, Wang W, Takada S. Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot. Proc Natl Acad Sci USA. 2012;109:17789–17794. doi: 10.1073/pnas.1201807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Reddy G, O’Brien EP, Thirumalai D. Collapse kinetics and chevron plots from simulations of denaturant-dependent folding of globular proteins. Proc Natl Acad Sci USA. 2011;108:7787–7792. doi: 10.1073/pnas.1019500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How Fast-Folding Proteins Fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 34.Piana S, Lindorff-Larsen K, Shaw DE. Atomic-level description of ubiquitin folding. Proc Natl Acad Sci USA. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best RB, Hummer G, Eaton WA. Native contacts determine protein folding mechanisms in atomistic simulations. Proc Natl Acad Sci USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry ER, Best RB, Eaton WA. Comparing a simple theoretical model for protein folding with all-atom molecular dynamics simulations. Proc Natl Acad Sci USA. 2013;110:17880–17885. doi: 10.1073/pnas.1317105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenzaki H, Koga N, Hori N, Kanada R, Li W, Okazaki KI, et al. CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. J Chem Theory Comput. 2011;7:1979–1989. doi: 10.1021/ct2001045. [DOI] [PubMed] [Google Scholar]
- 38.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takada S, Kanada R, Tan C, Terakawa T, Li W, Kenzaki H. Modeling Structural Dynamics of Biomolecular Complexes by Coarse-Grained Molecular Simulations. Acc Chem Res. 2015;48:3026–3035. doi: 10.1021/acs.accounts.5b00338. [DOI] [PubMed] [Google Scholar]
- 40.Best RB, Chen Y-G, Hummer G. Slow Protein Conformational Dynamics from Multiple Experimental Structures: The Helix/Sheet Transition of Arc Repressor. Structure. 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi F, Kikuchi M. Structural Change and Nucleotide Dissociation of Myosin Motor Domain: Dual Gō Model Simulation. Biophys J. 2007;93:3820–3827. doi: 10.1529/biophysj.106.103796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Q, Wang J. Single Molecule Conformational Dynamics of Adenylate Kinase: Energy Landscape, Structural Correlations, and Transition State Ensembles. J Am Chem Soc. 2008;130:4772–4783. doi: 10.1021/ja0780481. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Wang J, Zhang J, Takada S, Wang W. Overcoming the Bottleneck of the Enzymatic Cycle by Steric Frustration. Phys Rev Lett. 2019;122:238102. doi: 10.1103/PhysRevLett.122.238102. [DOI] [PubMed] [Google Scholar]
- 45.Kubo S, Li W, Takada S. Allosteric conformational change cascade in cytoplasmic dynein revealed by structure-based molecular simulations. PLOS Comput Biol. 2017;13:e1005748. doi: 10.1371/journal.pcbi.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terada TP, Kimura T, Sasai M. Entropic Mechanism of Allosteric Communication in Conformational Transitions of Dihydrofolate Reductase. J Phys Chem B. 2013;117:12864–12877. doi: 10.1021/jp402071m. [DOI] [PubMed] [Google Scholar]
- 47.Kanada R, Kuwata T, Kenzaki H, Takada S. Structure-based Molecular Simulations Reveal the Enhancement of Biased Brownian Motions in Single-headed Kinesin. PLoS Comput Biol. 2013;9:e1002907. doi: 10.1371/journal.pcbi.1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okazaki K, Takada S. Dynamic energy landscape view of coupled binding and protein conformational change: Inducedfit versus population-shift mechanisms. Proc Natl Acad Sci USA. 2008;105:11182–11187. doi: 10.1073/pnas.0802524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo S, Niina T, Takada S. Molecular dynamics simulation of proton-transfer coupled rotations in ATP synthase FO motor. bioRxiv. 2019 doi: 10.1038/s41598-020-65004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koga N, Takada S. Folding-based molecular simulations reveal mechanisms of the rotary motor F1-ATPase. Proc Natl Acad Sci USA. 2006;103:5367–5372. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyeon C, Lorimer GH, Thirumalai D. Dynamics of allosteric transitions in GroEL. Proc Natl Acad Sci USA. 2006;103:18939–18944. doi: 10.1073/pnas.0608759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brandani GB, Takada S. Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA. PLOS Comput Biol. 2018;14:e1006512. doi: 10.1371/journal.pcbi.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci USA. 2011;108:3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papoian GA, Ulander J, Eastwood MP, Luthey-Schulten Z, Wolynes PG. Water in protein structure prediction. Proc Natl Acad Sci USA. 2004;101:3352–3357. doi: 10.1073/pnas.0307851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davtyan A, Schafer NP, Zheng W, Clementi C, Wolynes PG, Papoian GA. AWSEM-MD: protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J Phys Chem B. 2012;116:8494–8503. doi: 10.1021/jp212541y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoang TX, Trovato A, Seno F, Banavar JR, Maritan A. Geometry and symmetry presculpt the free-energy landscape of proteins. Proc Natl Acad Sci USA. 2004;101:7960–7964. doi: 10.1073/pnas.0402525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enciso M, Rey A. A refined hydrogen bond potential for flexible protein models. J Chem Phys. 2010;132:235102. doi: 10.1063/1.3436723. [DOI] [PubMed] [Google Scholar]
- 58.Hung NB, Le D-M, Hoang TX. Sequence dependent aggregation of peptides and fibril formation. J Chem Phys. 2017;147:105102. doi: 10.1063/1.5001517. [DOI] [PubMed] [Google Scholar]
- 59.Mioduszewski Ł, Cieplak M. Disordered peptide chains in an α-C-based coarse-grained model. Phys Chem Chem Phys. 2018;20:19057–19070. doi: 10.1039/c8cp03309a. [DOI] [PubMed] [Google Scholar]
- 60.Hyeon C, Thirumalai D. Mechanical unfolding of RNA hairpins. Proc Nat Acad Sci USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denesyuk NA, Thirumalai D. Coarse-Grained Model for Predicting RNA Folding Thermodynamics. J Phys Chem B. 2013;117:4901–4911. doi: 10.1021/jp401087x. [DOI] [PubMed] [Google Scholar]
- 62.Hori N, Takada S. Coarse-grained structure-based model for RNA-protein complexes developed by fluctuation matching. J Chem Theory Comput. 2012;8:3384–3394. doi: 10.1021/ct300361j. [DOI] [PubMed] [Google Scholar]
- 63.Knotts TA, Rathore N, Schwartz DC, de Pablo JJ. A coarse grain model for DNA. J Chem Phys. 2007;126:084901. doi: 10.1063/1.2431804. [DOI] [PubMed] [Google Scholar]
- 64.Clementi C, García AE, Onuchic J. Interplay Among Tertiary Contacts, Secondary Structure Formation and Sidechain Packing in the Protein Folding Mechanism: All-atom Representation Study of Protein L. J Mol Biol. 2003;326:933–954. doi: 10.1016/s0022-2836(02)01379-7. [DOI] [PubMed] [Google Scholar]
- 65.Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN. SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 2010;38:W657–W661. doi: 10.1093/nar/gkq498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noel JK, Levi M, Raghunathan M, Lammert H, Hayes RL, Onuchic JN, Whitford, et al. 2016 SMOG 2: A Versatile Software Package for Generating Structure-Based Models. PLoS Comput Biol. 2016;12:e1004794. doi: 10.1371/journal.pcbi.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gront D, Kmiecik S, Kolinski A. Backbone building from quadrilaterals: a fast and accurate algorithm for protein backbone reconstruction from alpha carbon coordinates. J Comput Chem. 2007;28:1593–1597. doi: 10.1002/jcc.20624. [DOI] [PubMed] [Google Scholar]
- 69.Kmiecik S, Gront D, Kolinski M, Wieteska L, Dawid AE, Kolinski A. Coarse-Grained Protein Models and Their Applications. Chem Rev. 2016;116:7898–7936. doi: 10.1021/acs.chemrev.6b00163. [DOI] [PubMed] [Google Scholar]
- 70.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 71.Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75:430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feig M, Karanicolas J, Brooks CLMMTSB. Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Jung J, Mori T, Kobayashi C, Matsunaga Y, Yoda T, Feig M, et al. GENESIS: a hybrid-parallel and multi-scale molecular dynamics simulator with enhanced sampling algorithms for biomolecular and cellular simulations. Wiley Interdiscip Rev Comput Mol Sci. 2015;5:310–323. doi: 10.1002/wcms.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lutz B, Sinner C, Heuermann G, Verma A, Schug A. 2013 eSBMTools 1.0: enhanced native structure-based modeling tools. Bioinformatics. 2019;29:2795–2796. doi: 10.1093/bioinformatics/btt478. [DOI] [PubMed] [Google Scholar]