Abstract
Background: A subpopulation of cancer stem cells (CSCs) with capacity for self-renewal is believed to drive initiation, progression, and relapse of breast tumors.
Methods: Since the thyroid hormone receptor β (TRβ) appears to suppress breast tumor growth and metastasis, we have analyzed the possibility that TRβ could affect the CSC population using MCF-7 cells grown under adherent conditions or as mammospheres, as well as inoculation into immunodeficient mice.
Results: Treatment of TRβ-expressing MCF-7 cells (MCF7-TRβ cells) with the thyroid hormone triiodothyronine (T3) decreased significantly CD44+/CD24− and ALDH+ cell subpopulations, the efficiency of mammosphere formation, the self-renewal capacity of CSCs in limiting dilution assays, the expression of the pluripotency factors in the mammospheres, and tumor initiating capacity in immunodeficient mice, indicating that the hormone reduces the CSC population present within the bulk MCF7-TRβ cultures. T3 also decreased migration and invasion, a hallmark of CSCs. Transcriptome analysis showed downregulation of the estrogen receptor alpha (ERα) and ER-responsive genes by T3. Furthermore, among the T3-repressed genes, there was an enrichment in genes containing binding sites for transcription factors that are key determinants of luminal-type breast cancers and are required for ER binding to chromatin.
Conclusion: We demonstrate a novel role of TRβ in the biology of CSCs that may be related to its action as a tumor suppressor in breast cancer.
Keywords: thyroid hormone receptor β, breast cancer stem cells, self-renewal, estrogen receptors
Introduction
There is strong evidence that breast tumors are driven by a subpopulation of cells that display stem cell properties (cancer stem cells [CSCs]; also called tumor-initiating cells [TICs]) (1–4). CSCs appear to be required for sustained tumor progression, are resistant to radiation and chemotherapy (5,6), and may be responsible for cancer recurrence and metastasis (7). CSCs are enriched within cell subpopulations with a CD24−/CD44+ surface marker profile (8) and positivity for aldehyde dehydrogenase (ALDH+) (9). In contrast to other cancer cells, cells expressing these CSC markers have the ability to form colonies, or “mammospheres,” under low-adherence conditions and to initiate tumors in immunodeficient mice (8,9).
More than 70% of breast cancers are estrogen receptor alpha (ERα) positive (ER+). This receptor is a key driver of luminal breast tumor growth and is the most important therapeutic target in these tumors. Models of luminal human breast cancer have been based primarily on MCF-7 cells. Tumor initiation by this cell line appears to rely on estrogen. Thus, MCF-7 cells do not form tumors in ovariectomized immunodeficient mice, while estrogen supplementation allows MCF-7 cells to form tumors, indicating that estrogen could expand the population of MCF-7 CSCs (10). Although ERα levels are low in CSCs (10,11), estrogen induces paracrine fibroblast growth factor 9 (FGF9)/fibroblast growth factor receptor (FGFR) signaling in the non-CSC compartment that ultimately increases the CSC population (10,12).
The thyroid hormone receptors (TRs) mediate the biological actions of the thyroid hormone triiodothyronine (T3). Different studies have suggested that the TRβ isoform could have a tumor suppressor activity (13). In human breast tumors, TRβ mutations, anomalous subcellular localization, and biallelic inactivation of the TRβ gene by promoter methylation have been found (14).
Experimental evidence to further support the tumor suppressor role of TRβ in breast cancer came from studies showing that mice bearing a dominant negative mutant of TRβ are more susceptible than normal mice to the development of mammary tumors (15). In addition, in the presence of hormone, the expression of TRβ in MCF-7 cells leads to decreased proliferation and to inhibition of tumor development in xenograft experiments (16). However, the possibility that regulation of the number and/or function of breast CSCs could be related with the antitumorigenic effects of TRβ has not yet been explored.
Here we provide evidence that T3 is able to deplete the breast CSC population in estrogen-dependent MCF-7 cells expressing TRβ. T3 reduces the number of cells expressing CSC markers and inhibits mammosphere formation. These actions of T3 are linked to ERα downregulation and to an enrichment in repressed genes containing binding sites for transcription factors that are key determinants of luminal-type breast cancers and are required for ER binding to chromatin. Although the importance of these observations in clinically relevant human mammary cancers remains to be established, our results indicate a novel role of hormone-bound TRβ in the biology of CSCs that may be related to its action as a tumor suppressor in breast cancer.
Materials and Methods
Adherent cells
MCF-7 cells transduced with TRβ1 (MCF7-TRβ) or only with the selector Neo gene have been previously described (16). Cells were grown with Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% l-glutamine (Gibco, Waltham, MA), 1% penicillin/streptomycin (Gibco), and 10% fetal bovine serum (FBS; Sigma, St. Louis, Mo). BT474 cells were grown in Roswell Park Memorial Institute medium with the same supplements. Cells were transiently transfected (500 ng/mL) with an empty vector or with the same amount of a TRβ1 vector. Cell lines were mycoplasma free and were authenticated analyzing short tandem repeats of specific loci of the Human Genome with the StemElite ID system (Promega, Madison, WI).
Mammosphere culture
For the formation of primary mammospheres, cells were dispersed into single cells by trypsin digestion and plated in ultralow attachment plates (Corning, New York, NY) at a density of 15,000 cells/mL. Cells were grown in DMEM/F-12 GlutaMAX supplement medium (Gibco) with 1% penicillin/streptomycin, 2% B27 (Gibco), 10 ng/mL fibroblast growth factor b (PeproTech, London), and 20 ng/mL epidermal growth factor (PeproTech). Mammospheres were separated by gentle centrifugation, dissociated with trypsin, and mechanically with a micropipette. Cells were counted and plated at a concentration of 20,000 cells/mL for the development of second-generation mammospheres. Growth factors, and when indicated T3, were added every three days. Parallel cultures of adherent cells were plated at 125,000 cells/mL in 6-well plates and passaged every three days in the presence and absence of T3. Unless otherwise indicated in the figures, cells were incubated with 25 nM T3.
Limiting dilution assays
Single dissociated cells were plated at a concentration of 5 cells/well in 96-well plates in a mammosphere growing medium. The number of mammospheres was scored after 14 days.
Proliferation assays
Cell viability was determined by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H tetrazolium) using the CellTiter One-Solution-Assay (Promega) in 96-well plates inoculated with 1500 cells/well in the presence and absence of T3 for the times indicated following the manufacturer's recommendations. MTS (20 μL) was added to the wells and absorbance was read at 490 nm.
Cell staining for flow cytometry
Adherent or mammosphere cultures were trypsinized into single cells. Cells (1 × 106) were collected in 70 μL of flow cytometry buffer (1% ethylenediaminetetraacetic acid 0.5 M and 2% FBS in phosphate-buffered saline) and incubated with the primary antibodies for 20 min. The antibodies used for flow cytometry were the mouse FITC anti-CD24 antibody (clone ML5; BD) and the mouse PE/Cy7 anti-CD44 antibody (clone G44-26; BD). As a negative control, cells were incubated with isotype mouse FITC IgG2a κ (clone G155-178; BD) and mouse PE/Cy7 IgG2b κ (clone MPC-11; BD) nonspecific antibodies to establish the flow cytometer voltage settings, and single-color positive controls were used to adjust compensation. Side scatter and forward scatter were used to discard debris.
The flow cytometry data were acquired with an FACS Canto II and analyzed with FACS Diva or FlowJo software (BD). Cell cycle analysis was determined by incubating cells with propidium iodide and 0.1 mg/mL RNase for 30 min at 37°C before analysis. CD44+/CD24− cells were isolated by sorting in an FACSAria III cell sorter (BD Biosciences, Franklin Lakes, NJ), using the same antibodies.
ALDEFLUOR assays
ALDEFLUOR assays (StemCell Technologies, Vancouver) were used to determine ALDH activity following the manufacturer's instructions. Cells were incubated for 30 min at 37°C with the ALDEFLUOR reagent. For each sample, an aliquot of cells was stained under identical conditions with diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor, as an ALDH− control. Cells were analyzed by flow cytometry.
Western blotting
Proteins from cell lysates (20–30 μg) were separated in sodium dodecyl sulfate/polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Protran Whatman; Perkin Elmer, Waltham, MA) that were blocked for one hour at room temperature with 5% bovine serum albumin or nonfat milk. Incubation with primary antibodies was performed overnight at 4°C and with the secondary antibody for one hour at room temperature. Blots were visualized with enhanced chemoluminiscence (BioRad, Hercules, CA). The antibodies used are listed in the Supplementary Materials.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from cells using TRIzol (Sigma) or RNeasy Mini Kit (Qiagen, Hilden, Germany). mRNA levels were analyzed in technical triplicate by quantitative real-time polymerase chain reaction (RT-PCR), following specifications of SuperScript™ First-Strand Synthesis System (Invitrogen, Carlsbad, CA) and Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Data analysis was done using the comparative threshold cycle method and data were corrected with 36B4 mRNA levels. Primers used in this study are listed in the Supplementary Materials.
Transfection and reporter assays
Cells were plated in DMEM with 10% FBS in 24-well plates. After 24 h, cells were transfected in serum-free medium with 100 ng of luciferase reporter plasmids containing consensus thyroid hormone response elements (TRE) or estrogen response elements (ERE) and 10 ng of pRL-TK-Renilla (Promega) as a normalizer control, using Lipofectamine 2000 (Invitrogen) (17,18). After six hours, cells were transferred to medium supplemented with 10% thyroid hormone-stripped serum by treatment with AG1-X8 (Bio-Rad) resin. Cells were incubated in the presence of 25 nM T3 and/or 10 nM estradiol (E2) for 36 h or with increasing concentrations of T3 as indicated in the figures.
To analyze mothers against decapentaplegic (SMAD) activity, the reporter plasmid p3TP-lux bearing a fragment of the plasminogen activator inhibitor-1 promoter containing SMAD binding elements (SBEs) was cotransfected with expression vectors for SMAD3 (150 ng) and SMAD4 (200 ng) or with the appropriate amount of empty noncoding vectors (18). Cells transfected with a reporter plasmid containing consensus nuclear factor (NF)-κB binding sites (19) were incubated with 25 nM T3 for 36 h in the presence and absence of tumor necrosis factor-alpha (TNFα) (10 ng/mL), for the last 6 h. Luciferase activity was determined with the Dual Luciferase Assay System (Promega). Experiments were performed in triplicate and were repeated at least two times. Data are expressed as fold induction over the values obtained in the untreated cells as mean ± SD.
Invasion and migration assays
Matrigel invasion chambers, with cell culture inserts (Falcon, Pittsburgh, PA) containing an 8-μm-pore size polyethylene terephthalate membrane that has been treated with Matrigel matrix (Corning), were used for the invasion assays. A total of 140,000 cells in medium containing 0.5% FBS were inoculated in the upper chamber, and a medium containing 50% serum was added to the lower chamber. Cells were incubated in the presence and absence of T3 for 48 h. Noninvading cells were gently removed with a cotton swap, and invading cells were fixed and stained with Diff-Quik stain kit (Dade Behring, Deerfield, IL), photographed, and counted. Experiments were performed in triplicate. Migration assays were performed using 70,000 cells by the same procedure in the absence of Matrigel.
Microarray analysis
Analyses were carried out with triplicate cultures of adherent cells or secondary mammospheres (seven-day mammospheres generated from three-day primary mammospheres) treated with and without T3. Total RNA was extracted with the RNeasy Mini Kit, DNA was digested, and RNA integrity checked on an Agilent 2100 Bioanalyzer. cDNAs from total RNA (12 ng) were generated, fragmented, biotinylated, and hybridized to the GeneChip Human Transcriptome Array 2.0 (HTA 2.0) (Affymetrix; Thermo fisher, Waltham, MA). The arrays were washed and stained on a GeneChip Fluidics Station 450 (Affymetrix); scanning was carried out with the GeneChip Scanner 3000 7G; and image analysis with the Affymetrix GeneChip Command Console software.
Expression data were normalized, background and batch corrected using the Signal Space Transformation-Robust Multi-Chip Analysis implemented in the Transcriptome Analysis Console software version 4.0 (TAC4.0). Data have been deposited in GEO GSE117951 (adherent cells: GSE117948 and mammospheres: GSE117950). Gene Ontology and Chip Enrichment Analysis were performed using the Enrich web tool (20), and Gene Set Enrichment Analysis (GSEA) was performed using the Molecular Signatures Database (MSigDB) (1,21,22). Venn diagrams were drawn using the Bioinformatics and Evolutionary Genomics web tool.
In vivo tumorigenesis assays
Xenograft experiments were carried out in the animal facility of the Instituto de Investigaciones Biomédicas in compliance with the European Community Law (210/63/UE) and the Spanish law (R.D. 53/2013), with approval of the Ethics Committee of the Consejo Superior de Investigaciones Científicas and Comunidad de Madrid (PROEX 053/15). Mice were housed in a pathogen-free condition, in a 12-h light/12-h dark cycle, with water and normal diet food available ad libitum. Following ethical approval, mice were treated with 8 μg/mL of 17β-estradiol in the drinking water to supplement the estrogens requirement for MCF-7 cell proliferation. A week later, 200 or 2500 MCF-7 and MCF7-TRβ cells in 50% Matrigel were inoculated orthotopically into the fat pad of the bottom right abdominal mammary gland of female nu/nu mice, aged six to eight weeks, and allowed to form tumors. Tumor volume (8–9 tumors/group) was measured with a caliper and mice of all groups were sacrificed 38 days after inoculation. Estradiol treatment was continued during the whole period.
Statistical analysis
Statistical significance of data was determined by applying a two-tailed Student's t-test or analysis of variance (ANOVA) followed by the Bonferroni test for experiments with more than two experimental groups. Results are expressed as mean ± SD. Significance of ANOVA post-test or the Student's t-test between T3-treated and untreated groups is shown as *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
T3 inhibits mammosphere formation efficiency in TRβ-expressing MCF-7 cells
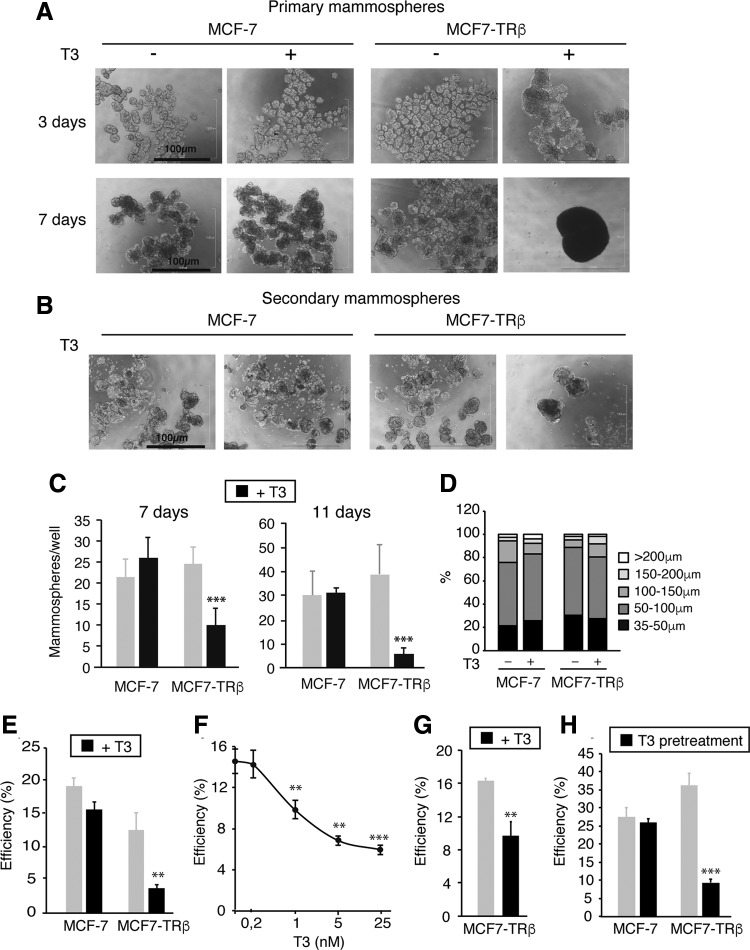
To elucidate the role of TRβ in mammary stem cell self-renewal, we used MCF-7 cells. As these cells express TRβ at undetectable levels by Western blot and present very low T3-dependent transcriptional activity in luciferase assays (Supplementary Fig. S1), we used MCF-7 cells expressing the TRβ1 isoform in a stable manner (MCF7-TRβ cells) or the corresponding empty vector (16). Figure 1 shows that three days after plating, both parental and MCF7-TRβ cells formed primary mammospheres in the presence and absence of T3. After six to seven days, mammospheres formed by MCF7-TRβ cells incubated with 25 nM T3 merged into very large and compact structures, a phenomenon never observed in the absence of hormone (Fig. 1A).
FIG. 1.
T3 inhibits mammosphere formation and expression of pluripotency markers in TRβ-expressing MCF-7 cells. (A) Representative optical microscope images of MCF7-TRβ and MCF-7 cells growing under nonadherent conditions for three and seven days in the presence and absence of 25 nM T3. (B) Images of 11-day second-generation mammospheres obtained after dispersion of 3-day primary mammospheres. Scale bars = 100 μM. (C) Quantification of 7- and 11-day secondary mammospheres grown with and without T3. (D) Size distribution of seven-day secondary mammospheres. (E) Cells were plated at a limiting dilution in 96-well plates under nonadherent conditions, and the efficiency of mammosphere formation was scored after 14 days in the presence and absence of T3. (F) MCF7-TRβ cells were inoculated at a limiting dilution and grown under nonadherent conditions in the presence of the indicated concentrations of T3. The number of mammospheres generated was scored after 14 days. Data are mean ± SD of three independent 96-well plates, and significance with respect to the untreated cells is shown with asterisks. (G) MCF7-TRβ cells were grown as primary mammospheres for three days with and without T3, dispersed, and plated at a limiting dilution in 96-well plates. The efficiency of mammosphere formation is shown. (H) Adherent 6-well cultures were incubated with and without T3 for 10 days, dispersed, inoculated at a limiting dilution in 96-well plates, and grown under nonadherent conditions. Mammospheres were counted after 14 days of incubation in the absence of T3. Data are mean ± SD of three independent plates. Asterisks denote the existence of statistically significant differences between T3-treated and untreated cells. **p < 0.01 and ***p < 0.001. T3, triiodothyronine; TR, thyroid hormone receptor.
Mammospheres can be serially passaged and contain a small number of stem cells capable of self-renewal, as well as multipotent progenitor cells (3). Thus, we next dissociated the three-day primary mammospheres, before aggregation was observed, and performed secondary mammosphere assays. No formation of compact structures was observed after T3 treatment (Fig. 1B), and the hormone very significantly reduced the efficiency of secondary mammosphere formation by MCF7-TRβ cells (Fig. 1C). Despite this, secondary mammosphere size distribution was similar in all groups (Fig. 1D).
To analyze if the reduction in mammosphere generation could reflect a T3-dependent inhibitory effect in cell proliferation and/or an increase in cell death, we next analyzed cell viability and cell cycle changes. Proliferation of adherent MCF7-TRβ cells was moderately blunted, and an increase in the percentage of cells in the G0/G1 phase of the cell cycle was observed after six days of T3 treatment (Supplementary Fig. S2A, B). In contrast, T3 did not increase the number of adherent subG1 cells, suggesting that it does not induce cell death. After seven days of T3 treatment, the number of subG1 MCF7-TRβ cells increased in primary mammospheres, most likely due to the observed cell aggregation (Supplementary Fig. S2C). However, no increase in the sub-G1 cell population was induced by T3 in secondary MCF7-TRβ mammospheres derived from three-day primary mammospheres (Supplementary Fig. S2D), indicating that T3 did not inhibit mammosphere formation by inducing cell death.
To further investigate whether T3 reduces the self-renewal capability of CSCs, we plated cells into 96-well plates at a clonal density, using limiting dilution, to ensure that each of the resulting mammospheres represents the progeny of a single stem cell. Again, T3 significantly reduced the efficiency of mammosphere generation in MCF7-TRβ cells (Fig. 1E). This reduction showed a dose dependence similar to the T3-dependent transactivation in luciferase assays (Supplementary Fig. S1C), indicating that physiological amounts of T3 are able to inhibit self-renewal (Fig. 1F). In addition, the number of secondary mammospheres formed under limiting dilution conditions was also reduced in MCF7-TRβ cells treated with T3 (Fig. 1G).
Together, these results strongly suggest that TRβ expression results in a hormone-dependent decrease in the CSC population. This was further supported by the finding that prior incubation of adherent cells with T3 was sufficient to inhibit mammosphere generation under clonal conditions in the absence of hormone (Fig. 1H).
To further analyze the effect of TRβ and T3 on the self-renewal capacity of breast CSCs, we conducted mammosphere assays in another ER+ cell line, BT474 cells. These cells were transfected with an empty vector or with a vector coding for TRβ, which strongly increased TRβ expression and enhanced TRE-dependent transactivation (Supplementary Fig. S3A–C). As shown in Supplementary Figure S3D, T3 treatment caused a small but significant decrease of mammosphere formation after three and seven days in parental BT474 cells, and this decrease was stronger, and similar to that found in MCF7-TRβ cells after transient expression of TRβ. Therefore, the effect of T3 on breast CSCs does not appear to be limited to the MCF-7 cell line.
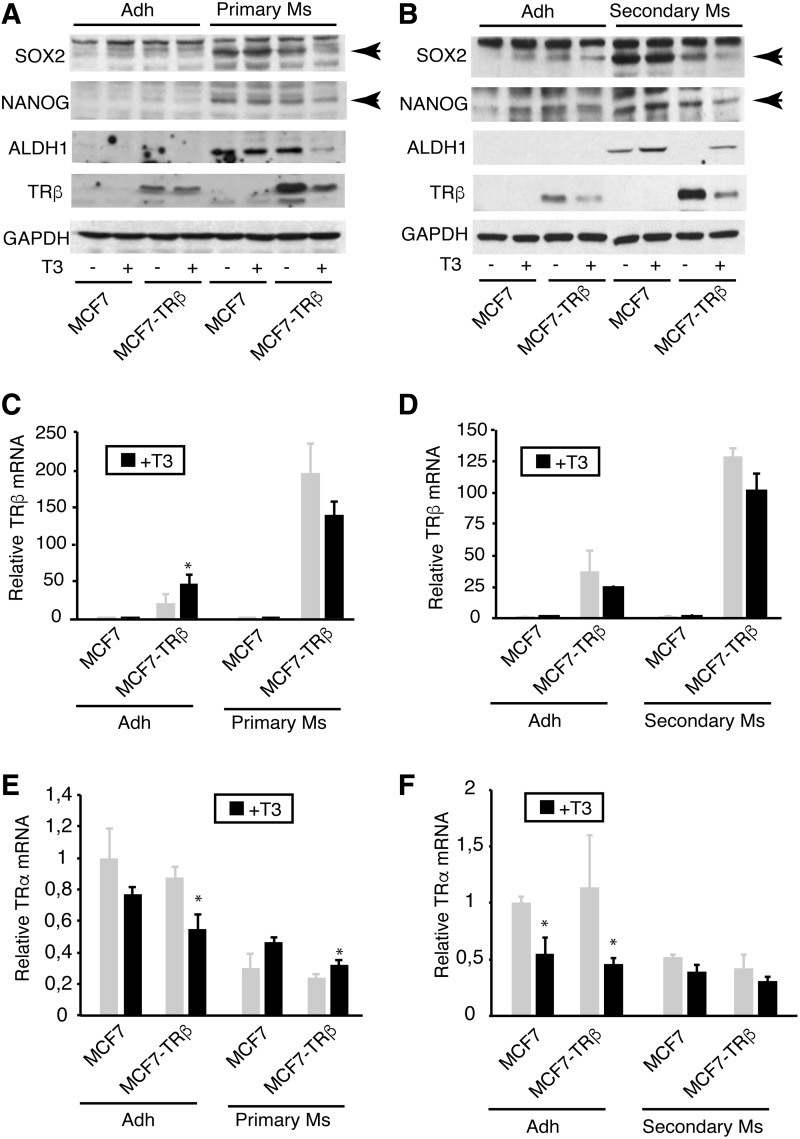
The pluripotency cell markers sex determining region Y-box 2 (SOX2) and NANOG, as well as the stem cell marker ALDH1, are expressed at high levels in breast tumor stem cells and their expression is reduced during cell differentiation (9,23). Thus, we next compared the levels of these proteins in primary and secondary mammospheres with those present in the adherent cultures of MCF-7 cells. There was an important increase in expression of SOX2, NANOG, and ALDH1 both in primary (Fig. 2A) and secondary mammospheres (Fig. 2B), confirming their enrichment in CSCs. Significantly, incubation of MCF7-TRβ mammospheres with T3 resulted in a very marked inhibition in the levels of pluripotency markers.
FIG. 2.
T3 inhibits expression of pluripotency markers. (A) The levels of SOX2, NANOG, ALDH1, and TRβ were determined by Western blotting in MCF-7 and MCF7-TRβ cells grown under adherent conditions (adh) or as primary mammospheres (Ms) for three days in the presence and absence of T3. GAPDH was used as a loading control. (B) Similar experiment performed in adherent cultures and seven-day secondary mammospheres derived from three-day primary mammospheres grown with and without T3. Arrows indicate the specific bands. (C) TRβ mRNA levels in untreated and T3-treated adherent cultures and in primary mammospheres. (D) TRβ mRNA levels in adherent cultures and secondary mammospheres incubated in the presence and absence of hormone. (E) TRα mRNA levels in untreated and T3-treated adherent cultures and primary mammospheres. (F) TRα transcripts in secondary mammospheres and adherent cells treated with and without T3. Data are mean ± SD. Statistically significant differences between T3-treated and untreated cells are shown by asterisks. *p < 0.05. ALDH1, aldehyde dehydrogenase 1; GAPDH, glyceraldehyde 3-phophate dehydrogenase; NANOG; SOX2, sex determining region Y-box 2.
We also analyzed TRβ expression, observing that its levels were higher in mammospheres than in their corresponding adherent cultures of MCF7-TRβ cells. In addition, TRβ levels were reduced after T3 treatment, since ligand binding can induce proteasome-mediated receptor degradation (24). Increased levels of TRβ mRNA in mammospheres with respect to adherent cultures were also observed (Fig. 2C, D). This suggests that either the transfected gene is more strongly expressed under nonadherent culture conditions or that mammospheres are enriched in cells that express higher levels of TRβ.
We also analyzed the endogenous levels of TRα mRNA in parental MCF7-TRβ cells grown as mammospheres and under adherent conditions (Fig. 2E, F). TRα transcripts were similar in adherent MCF-7 and MCF7-TRβ cells, but they decreased in the TRβ-expressing cells upon T3 treatment. Contrasting with the expression levels of TRβ, the abundance of TRα mRNA was lower both in primary and secondary mammospheres, which are enriched in CSCs, than in the cells grown as adherent cultures, and T3 did not decrease its levels.
Although the contribution of epithelial to mesenchymal transition (EMT) in the biology of breast CSCs is still under debate, previous studies have shown that mammospheres are enriched in EMT genes (25–28). Therefore, we next studied the effect of T3 on the expression of the epithelial marker E-cadherin and the mesenchymal markers vimentin, Slug, and Snail 1. The levels of E-cadherin were rather similar in adherent cells and mammospheres and were not significantly altered in the presence of T3 (Supplementary Fig. S4). As expected, the levels of mesenchymal proteins were generally higher in primary and secondary mammospheres than in their correspondent adherent cells. T3 did not alter expression of these proteins in parental MCF-7 mammospheres and had a variable effect in MCF7-TRβ mammospheres, since it reduced the expression of vimentin and Slug, while increasing the expression of Snail 1.
T3 reduces the CD44+/CD24− and ALDH+ stem cell populations
Both parental and TRβ-expressing MCF-7 cells used in this study showed a significant number of CD44+/CD24− cells, although they express much lower levels of CD44 and higher CD24 levels than basal carcinoma MDA-MB-231 cells (Supplementary Fig. S5), which is in agreement with the finding that luminal cells express less CD44 and more CD24 than basal breast cancer cell subtypes (29).
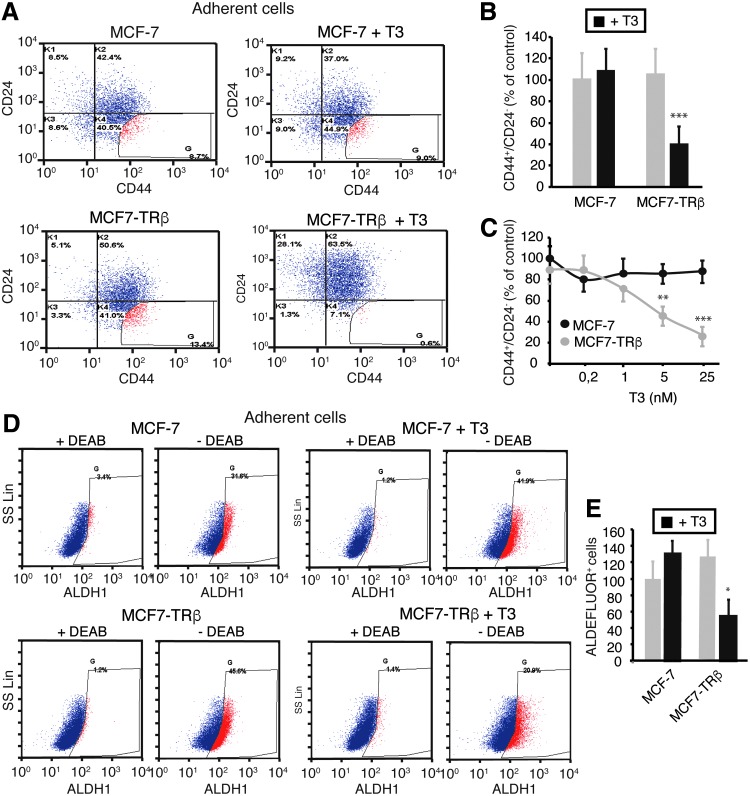
T3 at a 25 nM concentration did not alter the expression profile of these CSC markers in parental MCF-7 cells (Fig. 3A), but following T3 incubation for 72 h, most MCF7-TRβ cells became more positive for CD24 and displayed a decrease in CD44 expression, so that the proportion of gated CD44+/CD24− cells was very significantly reduced (Fig. 3B). As the optimal staining thresholds for identifying tumorigenic CD44+/CD24− cells are not well determined (30), we also gated the analysis to include CD24− cells with the highest CD44 expression, and under these conditions, we observed an almost total depletion of this cell population after T3 treatment (Fig. 3A).
FIG. 3.
TRβ mediates a T3-dependent decrease of the CD44+CD24− and ALDH+ cell subpopulations. (A) Flow cytometry analysis of the expression of CD44 and CD24 in adherent cultures of parental and TRβ-expressing MCF-7 cells treated with or without 25 nM T3 for 72 h. The threshold lines were set according to the isotype control. Gating of the more extreme CD44+CD24− cell population shows its disappearance in T3-treated MCF7-TRβ cells. (B) Quantification of the CD44+CD24− cell population obtained in four independent experiments. Data are expressed as the percentage of the value obtained in the parental control cells without T3 treatment and are mean ± SD. (C) Quantification of CD44+CD24− cells after 72-h incubation of MCF-7 and MCF7-TRβ cells, with the concentrations of T3 indicated. Data (mean ± SD) were obtained from two independent experiments performed in duplicate and are shown as the percentage of the results obtained in the untreated MCF-7 cells. Significant differences between MCF-7 and MCF7-TRβ cells are shown with asterisks. (D) ALDH enzymatic activity as assessed by ALDEFLUOR assays and flow cytometry in adherent cultures of parental and TRβ-expressing MCF-7 cells incubated in the presence and absence of 25 nM T3 for 72 h. To determine the percentage of ALDEFLUOR+ cells, assays were performed in the presence and absence of the ALDH-specific inhibitor DEAB. (E) Relative percentage of ALDEFLUOR+ cells (mean ± SD) obtained from three independent experiments. Significant differences between T3-treated and the corresponding untreated cells are shown with asterisks. *p < 0.05, **p < 0.01, and ***p < 0.001. DEAB, diethylaminobenzaldehyde.
Incubation of parental MCF-7 cells with increasing doses of T3 for 72 h did not alter the CD44+/CD24− cell population, but the hormone caused a dose-dependent decrease of this population in MCF7-TRβ cells, detectable at concentrations higher than 1 nM (Fig. 3C). A reduction of CD44+/CD24− cells was also observed after a longer treatment of adherent MCF7-TRβ cells with T3 (Supplementary Fig. S6A). In addition, expression of CD44 was markedly shifted to the right in secondary mammospheres with respect to the adherent cells, likely reflecting their enrichment in cancer initiating cells and the reduction caused by T3 was less marked under these conditions, suggesting a similar enrichment in putative stem cells in the mammospheres (Supplementary Fig. S6).
ALDEFLUOR assays also showed a significant reduction of the ALDH+ cell population after incubation of adherent MCF7-TRβ cells with T3 (Fig. 3D, E). As in the case of the CD44+/CD24− cells, the ALDH+ cell population in secondary mammospheres was not depleted in the presence of T3 (Supplementary Fig. S7). The decrease in the expression of the CSC markers observed in the T3-treated adherent cultures is consistent with the observed T3-mediated reduction in mammosphere formation by MCF7-TRβ cells, indicating again that the hormone reduces the population of CSCs with capacity for self-renewal.
T3 reduces migration and invasion
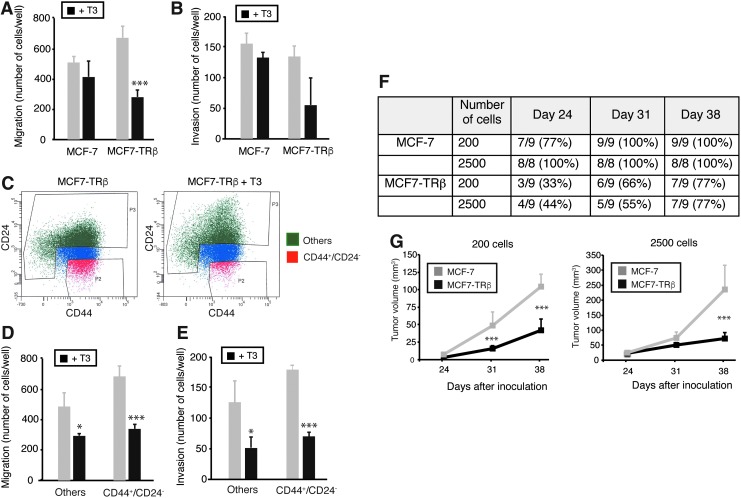
As CSCs appear to display an enhanced ability to disseminate and grow at metastatic sites, we next examined the effect of T3 on the migratory and invasive capacity of breast cancer cells. Consistent with a lower percentage of stem cells following incubation of MCF7-TRβ cells with T3, this treatment reduced the ability of the cells to migrate in Transwell assays (Fig. 4A) and to invade through Matrigel (Fig. 4B).
FIG. 4.
TRβ reduces migration, invasion, and tumor initiation. (A) Adherent cultures of MCF-7 and MCF7-TRβ cells were treated with and without T3 for 72 h. Cells were dispersed, inoculated into the upper chamber of 24-well plates, and allowed to migrate during 48 h in the presence and absence of hormone. (B) Invasion assays performed with the same experimental groups in Matrigel-coated chambers. Invasion proceeded for 48 h in the presence and absence of T3. (C) MCF7-TRβ cell cultures were treated with and without T3 for 72 h and FACS sorted to isolate the CD44+CD24− cell population (in red) and other cells (in green), discarding cells with intermediate levels of expression (in blue). (D) CD44+CD24− cells and other cells were subjected to migration assays for 48 h in the presence and absence of T3. (E) Invasion assays performed with the same groups in Matrigel-coated chambers. Data are mean ± SD, and asterisks denote the existence of statistically significant differences between T3-treated and untreated groups. (F) Tumor incidence was evaluated at the times indicated in estrogen-treated immunodeficient mice inoculated orthotopically into the mammary fat pad with MCF-7 and MCF7-TRβ cells at two different concentrations (200 and 2500 cells/injection). (G) Tumor volume measured at the indicated days after inoculation. Significant differences between MCF-7 and MCF7-TRβ tumors are shown with asterisks. *p < 0.05 and ***p < 0.001. FACS, fluorescence-activated cell sorting.
We next sorted the CD44+/CD24− MCF7-TRβ cells from the remaining cells (Fig. 4C) and subjected both populations to Transwell and Matrigel assays. As shown in Figure 4D and E, CD44+/CD24− cells showed increased migration and invasion with respect to other cells, but the hormone was able to significantly reduce the migratory and invasive properties of both populations, suggesting that not only the bulk of the breast cancer cells but also the putative stem cell populations are sensitive to this action of T3.
TRβ reduces tumor initiation and growth
The best criterion for assessing the number of TICs is to determine their ability to generate tumors in immunodeficient mice at limiting dilutions. Thus, we next compared the tumorigenicity of the parental and TRβ-expressing cells by injecting them into the fat pad of mammary glands of immunodeficient mice at two different dilutions (200 and 2500 cells). Since tumor initiation by the MCF7 cells appears to rely on ERα signaling, mice were treated with estrogen.
Figure 4F shows that 200 parental cells were sufficient to induce tumors in all mice one month after injection, while tumors originated by MCF7-TRβ cells appeared later and with a lower incidence. Furthermore, the volume of the tumors originated by TRβ-expressing cells was strongly decreased when compared with the parental tumors (Fig. 4G). These results are compatible with a reduction in the number of CSCs caused by binding of the endogenous circulating thyroid hormones of the mice to the receptor in the MCF7-TRβ cells.
Transcriptome analysis
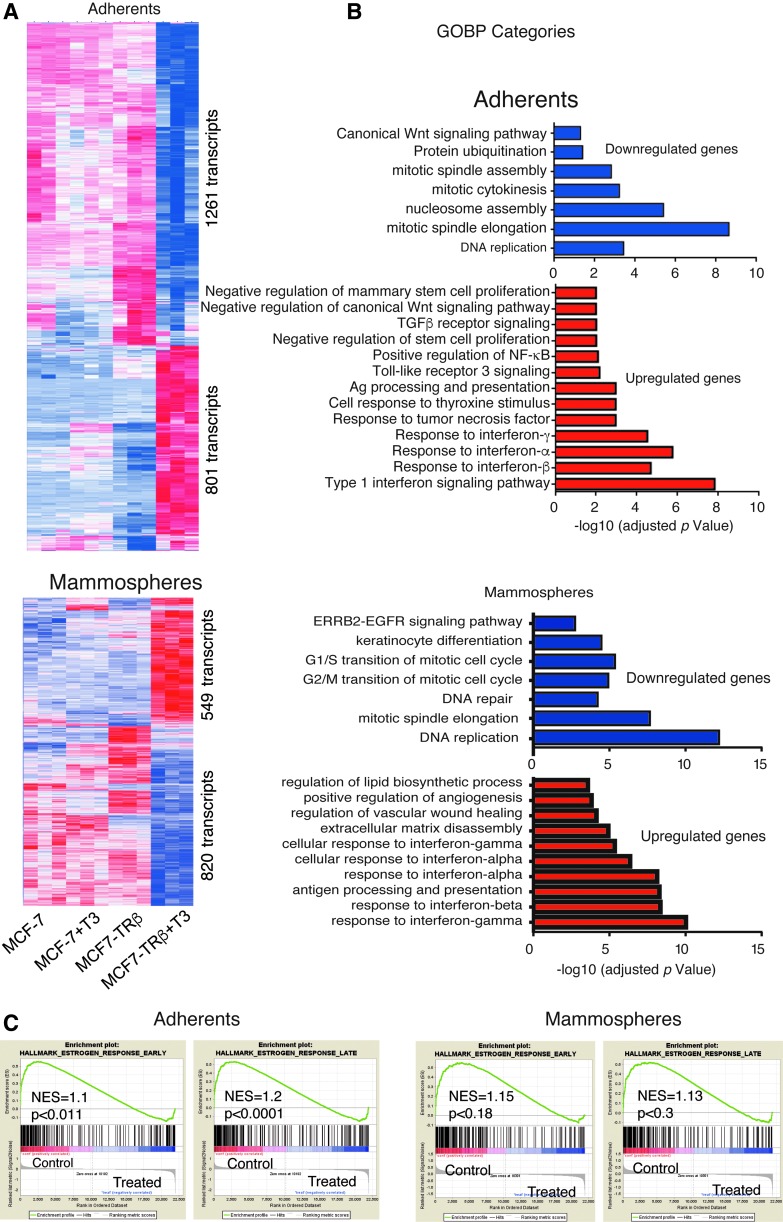
To comprehensively analyze the genes and pathways regulated by TRβ and T3, we next conducted microarray assays. Comparison of the expression profiles of parental and TRβ-expressing cells did not show substantial differences in the absence of T3. However, in MCF7-TRβ cells, T3 treatment caused widespread changes in gene expression (Fig. 5A and Supplementary Table S1). Functional annotation of the upregulated genes showed that the most enriched Gene Ontology of Biological Process (GOBP) term was the response to interferons (IFNs), while the downregulated GOBP terms among the T3-repressed genes included DNA replication and mitosis (Fig. 5B), consistent with the reduced proliferation observed in T3-treated cells. Similar processes were identified when considering the common set of genes that were regulated by T3 both in adherent cells and mammospheres (Supplementary Fig. S8).
FIG. 5.
Transcriptional profiling of adherent cells and mammospheres. (A) Heatmap showing differential gene expression between adherent MCF7 and MCF7-TRβ cells untreated and treated with T3 for 10 days (upper panel). Heatmap obtained in untreated and treated seven-day secondary mammospheres obtained from three-day primary mammospheres (lower panel). The number of transcripts downregulated and upregulated by T3 in MCF7-TRβ cells is indicated. (B) Top overrepresented GOBP categories with significant reduction (upper panel) or enrichment (lower panel) based in adjusted p-values in the T3-treated with respect to the untreated MCF7-TRβ cells, grown as adherent cultures or mammospheres. (C) Enrichment plots from GSEA showing the downregulation of Hallmark Estrogen Response Early and Late gene sets in T3-treated MCF7-TRβ cells. GOBP, Gene Ontology of Biological Process; GSEA, gene set enrichment analysis.
Furthermore, ranked GSEA revealed that the genes downregulated by T3 treatment of MCF7-TRβ cells were significantly enriched in both the Hallmark Estrogen Response Early and Late gene sets (Fig. 5C). Other top scoring gene sets enriched in the downregulated genes were Myc and E2F Hallmarks, also related with cell cycle control (Supplementary Fig. S9A), while IFN response and interleukin (IL)-6/STAT3 signaling response were found in the upregulated genes (Supplementary Fig. S9B), in agreement with the GOBP analysis.
GOBP and GSEA also showed that MCF7-TRβ cells treated with T3 undergo a negative regulation of mammary stem cell gene sets, in accordance with the observed depletion of the CSC population (Supplementary Fig. S10). Moreover, the absent or limited overlap between previously identified stem cell gene sets (1,21), and genes involved in the cell cycle, indicates that the decrease in the capacity for self-renewal is not just due to the inhibition of cell proliferation (Supplementary Fig. S10).
T3 inhibits ERα signaling in MCF7-TRβ cells
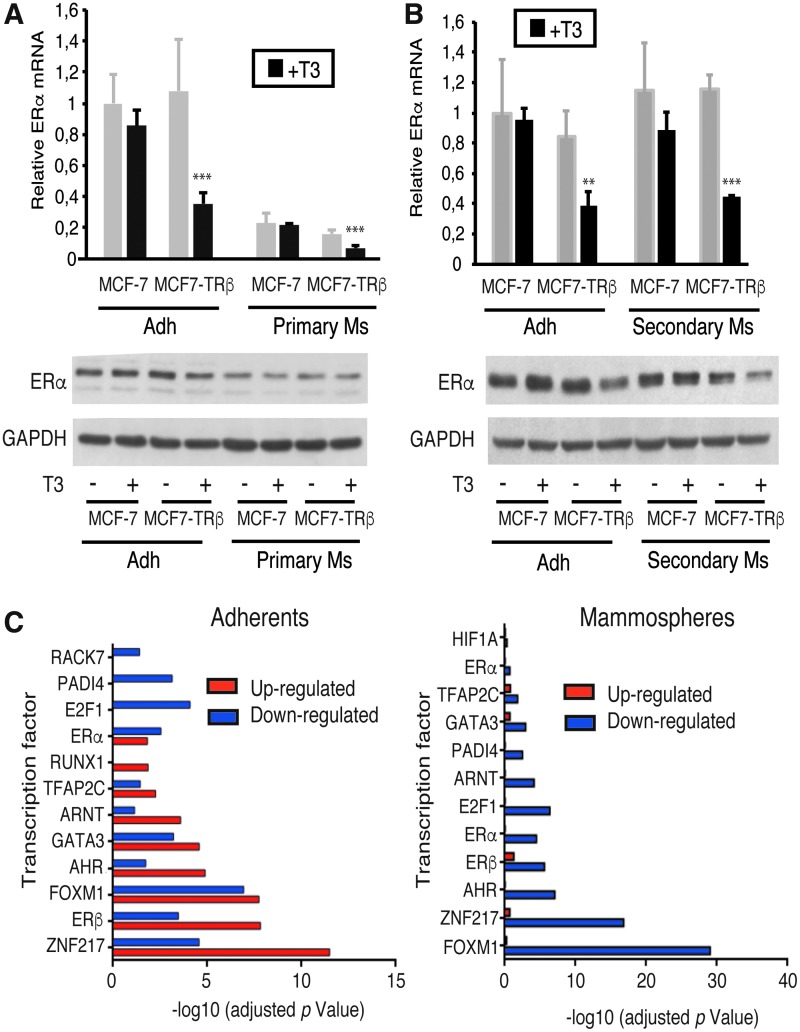
In agreement with the GSEA, ERα, the driving transcription factor in luminal breast cancer cells, was among the transcripts more strongly downregulated by T3 in MCF7-TRβ cells both under adherent and nonadherent conditions. These results were confirmed by RT-quantitative PCR (qPCR) and at the protein level in adherent cells and in primary and secondary mammospheres (Fig. 6A, B).
FIG. 6.
T3 inhibits ERα signaling in MCF7-TRβ cells. (A) ERα mRNA levels in adherent cultures (Adh) and primary mammospheres (primary Ms) of MCF-7 and MCF7-TRβ cells grown for three days in the presence and absence of T3. Data are mean ± SD and are represented relative to the values obtained in the untreated parental cells. **p < 0.01, ***p < 0.001. The lower panel shows a representative Western blot of ERα under the same experimental conditions. GAPDH was used as a loading control. (B) Similar experiments in adherent and seven-day second-generation mammospheres (secondary Ms) obtained from three-day primary mammospheres. (C) Summary of ChEA data showing the presence of binding sites for the indicated transcription factors in genes upregulated or downregulated by T3 in MCF7-TRβ cells. ChEA, chromatin immunoprecipitation enrichment analysis; ERα, estrogen receptor alpha.
The decrease in ERα expression led to reduced E2-dependent transactivation in transient transfection assays in the presence of T3 (Supplementary Fig. S11A), which was also dose dependent (Supplementary Fig. S11B). In addition, RT-qPCR of the two well-known ERα target genes PR (progesterone receptor) and TFF1 (trifolium factor 1) in MCF7-TRβ cells grown under both adherent and nonadherent conditions was very significantly reduced by the hormone (Supplementary Fig. S11C, D), confirming that T3 reduced ERα-dependent transcription.
T3 was also able to inhibit ERE-dependent transactivation (Supplementary Fig. S12A), as well as TFF1 and PR mRNA expression in both parental and TRβ-expressing BT474 cells (Supplementary Fig. S12B), further suggesting that inhibition of ERα signaling is an important mechanism by which T3 can modulate tumorigenesis of luminal breast cancer cells.
We also examined potential transcription factors involved in the observed gene deregulation by chromatin immunoprecipitation enrichment analysis (ChEA) of the transcriptome. This analysis revealed that the downregulated genes in T3-treated MCF7-TRβ cells (Fig. 6C) were not only enriched in binding motifs for ERs but also for ZNF217, FOXM1, or GATA3, among others. These transcription factors are key determinants of luminal-type breast cancers and are required for ERα binding to chromatin, confirming that T3 can be a key regulator of ERα signaling.
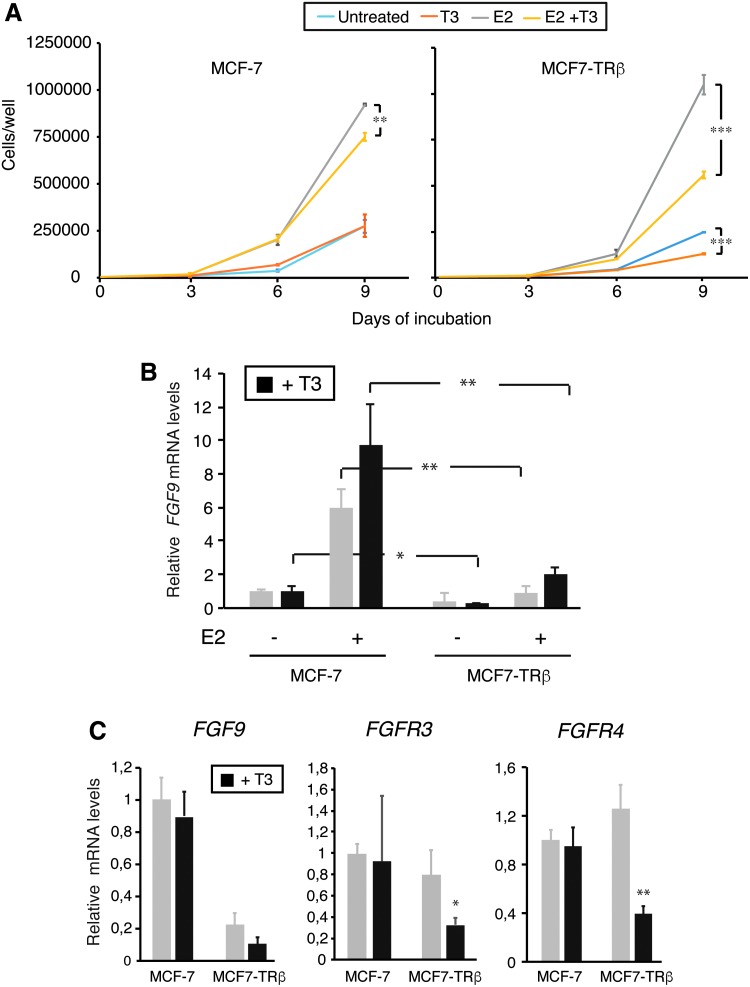
Since ERα target genes determine cell proliferation in luminal breast cancer cells, we also examined the effect of T3 on E2-dependent proliferation of MCF-7 and MCF7-TRβ cells. As shown in Figure 7A, E2 increased proliferation of both and T3 strongly reversed this response in MCF7-TRβ cells. In addition, it has been shown that E2 is able to induce the expression of FGF9, which has an important paracrine role in the expansion of the stem cell population (10). As shown in Figure 7B, FGF9 transcripts were markedly lower in the TRβ-expressing cells irrespectively of the presence of E2. Furthermore, there are several FGFRs, and MCF-7 cells express high levels of FGFR3, which binds with high affinity to FGF9 (10). Interestingly, transcripts for this receptor, as well as for FGFR4, were strongly reduced by T3 in MCF7-TRβ cells (Fig. 7C).
FIG. 7.
T3 inhibits E2-dependent proliferation and autocrine FGF induction. (A) MCF-7 and MCF7-TRβ cells were cultured under adherent conditions in the presence and absence of E2 and/or T3, as indicated in 6-well plates. Cells were counted three, six, and nine days after inoculation. Data are mean ± SD, and differences between T3-treated and T3-untreated cells are indicated with asterisks. (B) FGF9 transcripts in cells treated for nine days with E2 and/or T3 as indicated. Data are mean ± SD and are expressed relative to the levels obtained in MCF-7 untreated cells. Statistically significant differences between MCF-7 and MCF7-TRβ cells in each condition are shown. (C) FGF9, FGFR3, and FGFR4 mRNA levels were determined in cells treated with and without T3 for 72 h. Data (mean ± SD) are expressed relative to the levels obtained in the untreated cells, and differences between treated and untreated cells are indicated. *p < 0.05, **p < 0.01, and ***p < 0.001. FGF, fibroblast growth factor.
T3 alters the activity of signaling pathways involved in breast CSC self-renewal
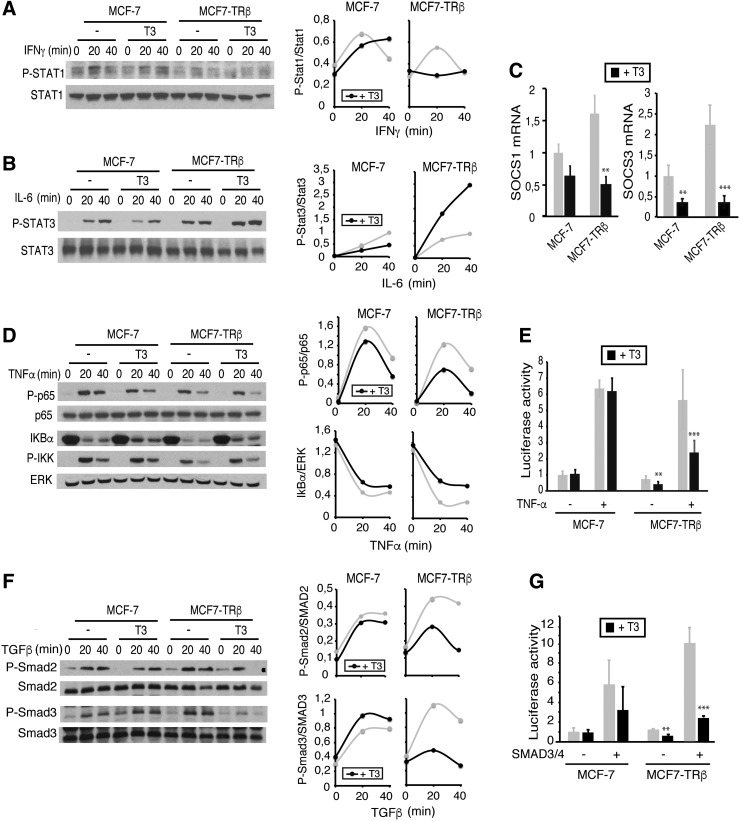
Signal transducer and activator of transcription (STAT) and NF-κB transcription factors are the main effectors in cytokine signaling. As the transcriptome analysis revealed that the response to IFNs and IL-6 appears to be among the most upregulated pathways by T3, we next analyzed the effect of the hormone on STAT-1 activation by IFNγ and STAT-3 activation by IL-6. As shown in Figure 8A and B, phosphorylation of STAT-1 was transiently induced by IFNγ in adherent MCF-7 and MCF7-TRβ cells and, surprisingly, this induction was blunted in TRβ-expressing cells that were pretreated with T3. In contrast, in MCF7-TRβ cells, the hormone induced a detectable increase in STAT-3 activation by IL-6 (Fig. 8B).
FIG. 8.
T3 regulates activation of signaling pathways essential for CSC function. (A) Western blot analysis of the total and phosphorylated STAT-1 in MCF-7 and MCF7-TRβ cells pretreated with 25 nM T3 for 48 h and then treated with 5 ng/mL IFNγ for the indicated time points. Quantification of the pSTAT-1/STAT-1 ratio with ImageJ from two experiments is shown at the right panel. (B) Total and phosphorylated STAT-3 in cells treated with 10 ng/mL IL-6 under the same conditions, and quantification of the pSTAT3/STAT3 ratio. (C) Transcript levels of SOCS1 and SOCS3 in MCF-7 and MCF7-TRβ cells incubated in the presence and absence of T3. Data (mean ± SD) are expressed relative to the levels obtained in the untreated cells. (D) Western blots of total and phosphorylated p65/NF-κB, IκBα, and phospho-IKK performed with extracts from cells incubated with T3 for 48 h and with 10 ng/mL TNFα for the indicated time periods. Total ERK levels were used as a loading control. The right panels show quantification of blots from two independent experiments. (E) Luciferase activity in MCF-7 and MCF7-TRβ cells transfected with a reporter plasmid containing consensus NF-κB motifs. Cells were treated with T3 for 36 h, and with TNFα during 6 h. (F) Western blot analysis of MCF-7 and MCF7-TRβ cell extracts with pSMAD2, pSMAD3, and total SMAD2 and SMAD3 antibodies after treatment with 25 nM T3 for 48 h and with 10 ng/mL TGFβ for the indicated time periods. Blots from two separate experiments were quantitated, and the pSMAD2/SMAD2 and pSMAD3/SMAD3 ratios are illustrated at the right panels. (G) Transient transfection assays in MCF-7 and MCF7-TRβ cells with an SBE-containing reporter plasmid. Cells were cotransfected with expression vectors for SMAD3 and SMAD4 or with an empty vector (−), and luciferase activity was determined after 36 h of incubation in the absence and presence of T3. All data (mean ± SD) are expressed as fold induction over the values obtained in the untreated cells and are representative of three independent experiments. Statistical significant differences between T3-treated and untreated cells are indicated. **p < 0.01, ***p < 0.001. CSC, cancer stem cell; IFN, interferon; IL, interleukin; NF, nuclear factor; pSMAD2, phospho-SMAD2; pSMAD3, phospho-SMAD3; SBEs, SMAD binding elements; SMAD, mothers against decapentaplegic; STAT, signal transducer and activator of transcription; TNFα, tumor necrosis factor-alpha.
Activation of STAT transcription factors is subjected to a negative feedback by the suppressor of cytokine signaling (SOCS) family of proteins (31). Interestingly, the transcriptome analysis showed that SOCS3 was included among the downregulated genes in T3-treated MCF7-TRβ cells (Supplementary Table S1). Indeed, not only SOCS3 but also SOCS1 transcript levels were strongly repressed after incubation with T3 (Fig. 8C).
To assess the effect of T3 on NF-κB activation, cells were treated with TNFα. This cytokine caused a detectable phosphorylation of p65 and a rapid disappearance of IκBα, and hormone reduced NF-κB activation in MCF7-TRβ cells, as shown by a lower p65 phosphorylation and a reduced degradation of IκBα (Fig. 8D). Furthermore, in transient transfection assays, TNFα stimulated the activity of a reporter plasmid bearing consensus NF-κB-binding elements and T3 significantly antagonized this response (Fig. 8E).
Cytokines that induce activation of SMAD transcription factors also play an important role in CSC self-renewal (27). Therefore, we also tested the effect of T3 on SMAD signaling. As shown in Figure 8F, T3 reduced SMAD2 and SMAD3 phosphorylation in response to TGFβ in MCF7-TRβ cells and also inhibited transcriptional stimulation of a reporter plasmid containing SBEs by SMAD factors in transient transfection assays (Fig. 8G).
Discussion
TRβ is widely accepted to be a hormone-dependent tumor suppressor (13) and binding of T3 to this receptor has been shown to inhibit proliferation of a variety of cell types (32). However, the effect of TRβ on a CSC-like population has not been previously examined. Using TRβ-expressing MCF-7 cells, we show here that T3 can be an important modulator of breast CSC properties both in vitro and in vivo.
We found that the proportion of CD44+/CD24− and ALDH+ stem-like cells was significantly decreased by T3 in TRβ-expressing MCF-7 cells, suggesting that they display a lower CSC content than the untreated cultures. If so, this should be reflected in a reduced ability of the treated cells to generate mammospheres, a widely used surrogate assay to assess CSC self-renewal that enriches for the presence of progenitor cells (8). Indeed, we found that T3 significantly inhibited the formation of mammospheres. Furthermore, mammospheres are enriched in stem and early progenitor cells expressing pluripotency stem cell markers (9), and the expression of these markers was reduced in mammospheres generated in the presence of T3, suggesting that they contain cells with a more differentiated genotype.
Although it has been described that T3 can either stimulate (33,34) or inhibit (35) proliferation of breast cancer cell lines, including MCF-7 cells, we did not find a significant effect of T3 on MCF-7 cell proliferation, unless TRβ was transfected. Confirming previous observations (16), we found that T3 reduces the proliferation of MCF7-TRβ cells, but depletion of the putative CSC population does not appear to reflect the generic growth inhibitory properties of T3, since the decrease of the CD44+/CD24− and ALDH+ stem-like cells is observed well before a detectable decrease of cell proliferation.
In agreement with these results, there is a reduction of the breast CSC signature in T3-treated MCF7-TRβ cells (1,21), while there is little overlap between genes involved in breast cancer stemness and proliferation, strongly suggesting that T3 decreases stemness by a mechanism independent of the decrease in cell proliferation. Importantly, T3 was able to reduce the self-renewal capacity of the MCF7-TRβ cells at physiological concentrations, with a dose dependence similar to that required for hormone-dependent transactivation.
One limitation of our study with MCF7-TRβ cells is that they overexpress TRβ. However, we have confirmed that mammosphere generation was also reduced by T3 in the BT474 mammary cancer cell line without exogenous TRβ expression, although this reduction was stronger after transient transfection with TRβ. Therefore, high receptor levels appear to be required to maximally diminish CSC content. Interestingly, TRβ levels, but not TRα levels, have been negatively associated with the histological grade of ER+ tumors and were also a predictor of improved distant metastasis-free survival in tamoxifen-treated patients (36). This is compatible with the hypothesis that TRβ could reduce the CSC population in clinically relevant mammary tumors, as CSCs appear to be responsible for resistance to therapy and tumor recurrence and metastasis (5–7).
CD44+/CD24− breast cancer cells exhibit high migratory capacity and enhanced invasive properties, an early step necessary for metastasis (7). Accordingly, we found that T3 was able to reduce migration and invasion in MCF7-TRβ cells, which is again compatible with a depletion of the CSC population. As CSCs are believed to drive initiation and progression of tumors, in vivo assays in xenotransplantation experiments are the gold standard for identifying CSCs (8). We observed that TRβ expression inhibits tumor incidence and tumor growth when cells were inoculated at limiting numbers in E2-treated immunodeficient mice, reinforcing the idea that T3 reduces the number of cells with tumor-initiating ability. The strongly decreased tumor initiating ability of MCF7-TRβ cells with respect to the parental cells in euthyroid mice again indicates that physiological levels of thyroid hormones are sufficient to deplete the putative CSC population in vivo.
Different studies have indicated a close association between breast CSCs and invasiveness and EMT (25,27), while others have proposed that these processes are independent of the expression of EMT-associated genes or that breast CSCs transition between epithelial and mesenchymal states (26,28). The reduced expression of CSC markers, as well as the decreased migratory and invasive capacity of T3-treated cells, could have been a consequence of a mesenchymal/epithelial transition (MET), typified by the acquisition of epithelial features and downregulation of mesenchymal characteristics. However, T3 did not increase E-cadherin expression, the main epithelial marker, and although it reduced vimentin levels, it also increased expression of Snail 1, a mesenchymal marker. Thus, the effect of T3 on CSC biology is not associated with induction of a clear MET phenotype. This could be related to the complex effects of T3 on the activity of critical signaling pathways responsible for self-renewal of CSCs and regulation of epithelial and mesenchymal characteristics.
It is known that cytokines, which may be released in an autocrine manner or by the tumor microenvironment, induce activation of SMAD (27,37), NF-κB (38), or STAT (39,40) transcription factors and play a key role in the generation of breast CSCs and tumorigenesis. Our results indicate that in MCF7-TRβ cells, T3 reduces activation of SMAD and NF-κB by TGFβ and TNFα, respectively, confirming previous results obtained in other cell types (18,19). This reduced response could be involved in the downregulation of the CSC population by T3, as these pathways govern mammosphere formation, stem cell expansion, and EMT.
Paradoxically, we found that STAT3 activation by IL-6, also very important for the acquisition of stem cell properties, was increased by T3. Furthermore, transcriptome analysis showed that the IL-6/STAT pathway is one of the main pathways activated by the thyroid hormone in MCF7-TRβ cells. This is in contrast with previous results showing that T3 reduces IL-6/STAT3-mediated transcription in hepatocarcinoma cells (18), and with the finding that STAT3 phosphorylation is strongly decreased in the xenografts generated by MCF7-TRβ cells (16), which suggests that during tumor growth in vivo, the activity of this pathway is downregulated by TRβ by unknown mechanisms, possibly involving the tumor microenvironment. A possible cause for the increased STAT3 activation shown by T3-treated MCF7-TRβ cells is the downregulation of the expression of the SOCS3 gene, the major negative regulator of STAT activity found in the microarrays and confirmed by RT-qPCR.
The increased response to IFNs found in the microarrays in T3-treated MCF7-TRβ cells is particularly interesting, as these molecules are implicated in tumor immune surveillance and induce the expression of immune checkpoint proteins (41). Thus, through modulating the response to IFN, TRβ could potentially regulate immunotherapeutic responses. However, in contrast with the increased STAT3 phosphorylation in response to IL-6, phosphorylation of STAT-1 in response to incubation with IFNγ was reduced by T3 in MCF7-TRβ cells, despite the finding that SOCS1 mRNA levels were decreased. Therefore, more studies will be required to analyze the role of the receptor in the immune response of breast cancer cells.
ERα is the distinctive feature of luminal breast cancers, where it regulates cell proliferation and tumor progression even after the development of resistance to antiestrogen therapy (42). Our results show that ERα transcripts were markedly reduced by T3 both in adherent and CSC-enriched mammosphere cultures of MCF7-TRβ cells and that, consequently, T3 dampens ER-dependent transcription. ER-dependent transcriptional activity was also reduced by T3 in BT474 cells expressing only endogenous TRβ, as indicated by the decreased abundance of the E2-responsive TFF1 and PR target genes. The finding that T3 reduces self-renewal and ER-dependent gene expression in unmanipulated mammary cancer cells again suggests that MCF7-TRβ cells could be a valid model to analyze the role of the receptor in CSC biology.
The reduction of E2-dependent signaling could be important for the observed effects of T3 on proliferation and is in concordance with the downregulation of genes involved in DNA replication and mitosis observed in the transcriptome analysis of MCF7-TRβ cells. Decreased ERα levels could also be related to the T3-dependent depletion of the CD44+/CD24− population and to the reduced efficiency of mammosphere formation, since estrogen-bound ERα downregulates CD24, while increasing CD44 (43) and enhances mammosphere generation in ER+ breast cancer cell lines (10).
Although ERα levels are low in CSCs (10,11), estrogen induces paracrine signals in the non-CSC compartment that ultimately increase the CSC population (10,12). This suggests that inhibition of ERα signaling by T3 could result in a reduction of this paracrine axis. In agreement with this hypothesis, expression of the FGF9 gene, as well as members of the family of receptors for this growth factor, is strongly diminished in MCF7-TRβ cells, which could contribute to the CSC depletion found in TRβ expressing cells.
Binding of ligand to ERα causes the recruitment of the receptor to its accessible binding motifs in chromatin. Remarkably, ChEA of T3-regulated genes in MCF7-TRβ cells unveils the enrichment of binding sites for the transcription factors GATA3, ZNF217, FOXM1, and RUNX1, as well as ERs. This enrichment was almost totally restricted to the downregulated genes in the mammospheres.
Importantly, GATA3 is a critical determinant of ER binding to its recognition motifs, acting as a pioneer factor required for genome-wide chromatin accessibility to ERα binding at sites that lack active histone modifications (44–46). In addition, ChIP-seq analysis shows clustering of ZNF217 (47) and FOXM1 (48) with FOXA1, another pioneer factor that determines ERα recruitment (44,49), GATA3 and ERα binding sites. On the contrary, RUNX1 is a transcription factor mutated in breast cancer, which controls the fate of ER+ mammary luminal cells and also influences binding of ER to chromatin (50,51).
This suggests that binding of T3 to TRβ could induce still undetermined epigenetic modifications creating a more compacted chromatin landscape, which could restrain binding of key luminal transcription factors to ERα target genes and consequently repress the master ER-dependent transcriptional program that dictates the hormone-responsive phenotype in luminal breast cancer.
In conclusion, our results show that TRβ mediates a ligand-dependent depletion of luminal breast CSCs, by modulating the activity of ERα and other key signaling pathways. These results indicate a novel role of TRβ in the biology of CSCs that may be related to its action as a tumor suppressor in ER+ breast cancer tumors. As CSCs appear to be responsible for resistance to therapy and tumor relapse, future studies are needed to underscore a potential role of TRβ as a positive prognostic marker of disease outcome and as a target for therapeutic interventions on the CSC population in luminal breast cancer.
Clinical studies have shown that elevated TRβ levels are also associated with a better prognosis and overall survival in other types of breast tumors (52,53). Furthermore, expression of TRβ in triple-negative breast cancer cells reduces tumor growth and has a strong inhibitory effect on invasion and metastasis formation in immunodeficient mice (54–57). Whether or not TRβ could regulate CSC biology by an ER-independent mechanism in other types of mammary tumors is an intriguing possibility that remains to be studied.
Supplementary Material
Author Disclosure Statement
No competing financial interests exist.
Funding Information
We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
This work was supported by grants BFU2014-53610-P (MINECO, AEI, FEDER, UE), CIBERONC CB/16/00228, and SAF2017-90604-REDT (Spanish Ministry of Economy and Competitiveness), and B2017/BMD-3724 (Comunidad de Madrid). The cost of this publication has been paid, in part, by FEDER funds.
Supplementary Material
References
- 1. Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. 2010. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140:62–73 [DOI] [PubMed] [Google Scholar]
- 2. Luo M, Clouthier SG, Deol Y, Liu S, Nagrath S, Azizi E, Wicha MS. 2015. Breast cancer stem cells: current advances and clinical implications. Methods Mol Biol 1293:1–49 [DOI] [PubMed] [Google Scholar]
- 3. Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17:1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clarke MF, Fuller M. 2006. Stem cells and cancer: two faces of eve. Cell 124:1111–1115 [DOI] [PubMed] [Google Scholar]
- 5. Zhang M, Atkinson RL, Rosen JM. 2010. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci U S A 107:3522–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. 2007. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A 104:618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr., Badve S, Nakshatri H. 2006. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 8:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. 2010. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci U S A 107:21737–21742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, Visvader JE. 2010. Control of mammary stem cell function by steroid hormone signalling. Nature 465:798–802 [DOI] [PubMed] [Google Scholar]
- 12. Simoes BM, Alferez DG, Howell SJ, Clarke RB. 2015. The role of steroid hormones in breast cancer stem cells. Endocr Relat Cancer 22:T177–T186 [DOI] [PubMed] [Google Scholar]
- 13. Aranda A, Martinez-Iglesias O, Ruiz-Llorente L, Garcia-Carpizo V, Zambrano A. 2009. Thyroid receptor: roles in cancer. Trends Endocrinol Metab 20:318–324 [DOI] [PubMed] [Google Scholar]
- 14. Li Z, Meng ZH, Chandrasekaran R, Kuo WL, Collins CC, Gray JW, Dairkee SH. 2002. Biallelic inactivation of the thyroid hormone receptor beta1 gene in early stage breast cancer. Cancer Res 62:1939–1943 [PubMed] [Google Scholar]
- 15. Guigon CJ, Kim DW, Willingham MC, Cheng SY. 2011. Mutation of thyroid hormone receptor-beta in mice predisposes to the development of mammary tumors. Oncogene 30:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park JW, Zhao L, Cheng SY. 2013. Inhibition of estrogen-dependent tumorigenesis by the thyroid hormone receptor beta in xenograft models. Am J Cancer Res 3:302–311 [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez L, Agullo-Ortuno MT, Garcia-Martinez JM, Calcabrini A, Gamallo C, Palacios J, Aranda A, Martin-Perez J. 2006. Role of c-Src in human MCF7 breast cancer cell tumorigenesis. J Biol Chem 281:20851–20864 [DOI] [PubMed] [Google Scholar]
- 18. Alonso-Merino E, Martin Orozco R, Ruiz-Llorente L, Martinez-Iglesias OA, Velasco-Martin JP, Montero-Pedrazuela A, Fanjul-Rodriguez L, Contreras-Jurado C, Regadera J, Aranda A. 2016. Thyroid hormones inhibit TGF-beta signaling and attenuate fibrotic responses. Proc Natl Acad Sci U S A 113:E3451–E3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lasa M, Gil-Araujo B, Palafox M, Aranda A. 2010. Thyroid hormone antagonizes tumor necrosis factor-alpha signaling in pituitary cells through the induction of dual specificity phosphatase 1. Mol Endocrinol 24:412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, Michalowska AM, Rosen JM. 2008. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res 68:4674–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dace A, Zhao L, Park KS, Furuno T, Takamura N, Nakanishi M, West BL, Hanover JA, Cheng S. 2000. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci U S A 97:8985–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, Yang J, Weinberg RA. 2017. Upholding a role for EMT in breast cancer metastasis. Nature 547:E1–E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sikandar SS, Kuo AH, Kalisky T, Cai S, Zabala M, Hsieh RW, Lobo NA, Scheeren FA, Sim S, Qian D, Dirbas FM, Somlo G, Quake SR, Clarke MF. 2017. Role of epithelial to mesenchymal transition associated genes in mammary gland regeneration and breast tumorigenesis. Nat Commun 8:1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schafer R, van Diest P, Voest E, van Oudenaarden A, Vrisekoop N, van Rheenen J. 2016. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep 14:2281–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fillmore CM, Kuperwasser C. 2008. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lawson JC, Blatch GL, Edkins AL. 2009. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res Treat 118:241–254 [DOI] [PubMed] [Google Scholar]
- 31. Kubo M, Hanada T, Yoshimura A. 2003. Suppressors of cytokine signaling and immunity. Nat Immunol 4:1169–1176 [DOI] [PubMed] [Google Scholar]
- 32. Pascual A, Aranda A. 2013. Thyroid hormone receptors, cell growth and differentiation. Biochim Biophys Acta 1830:3908–3916 [DOI] [PubMed] [Google Scholar]
- 33. Hall LC, Salazar EP, Kane SR, Liu N. 2008. Effects of thyroid hormones on human breast cancer cell proliferation. J Steroid Biochem Mol Biol 109:57–66 [DOI] [PubMed] [Google Scholar]
- 34. Dinda S, Sanchez A, Moudgil V. 2002. Estrogen-like effects of thyroid hormone on the regulation of tumor suppressor proteins, p53 and retinoblastoma, in breast cancer cells. Oncogene 21:761–768 [DOI] [PubMed] [Google Scholar]
- 35. Martinez MB, Ruan M, Fitzpatrick LA. 2000. Altered response to thyroid hormones by breast and ovarian cancer cells. Anticancer Res 20:4141–4146 [PubMed] [Google Scholar]
- 36. Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, Davis MJ, Clyne C, Funder JW, Simpson ER, Ragan MA, Kuczek E, Fuller PJ, Tilley WD, Leedman PJ, Clarke CL. 2013. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol 27:350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruna A, Greenwood W, Le Quesne J, Teschendorff A, Miranda-Saavedra D, Rueda OM, Sandoval JL, Vidakovic AT, Saadi A, Pharoah P, Stingl J, Caldas C. 2012. TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun 3:1055. [DOI] [PubMed] [Google Scholar]
- 38. Hinohara K, Kobayashi S, Kanauchi H, Shimizu S, Nishioka K, Tsuji E, Tada K, Umezawa K, Mori M, Ogawa T, Inoue J, Tojo A, Gotoh N. 2012. ErbB receptor tyrosine kinase/NF-kappaB signaling controls mammosphere formation in human breast cancer. Proc Natl Acad Sci U S A 109:6584–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafe M. 2007. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest 117:3988–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(-) stem cell-like breast cancer cells in human tumors. J Clin Invest 121:2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brockwell NK, Parker BS. 2019. Tumor inherent interferons: impact on immune reactivity and immunotherapy. Cytokine 118:42–47 [DOI] [PubMed] [Google Scholar]
- 42. Visvader JE. 2009. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 23:2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaipparettu BA, Malik S, Konduri SD, Liu W, Rokavec M, van der Kuip H, Hoppe R, Hammerich-Hille S, Fritz P, Schroth W, Abele I, Das GM, Oesterreich S, Brauch H. 2008. Estrogen-mediated downregulation of CD24 in breast cancer cells. Int J Cancer 123:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Theodorou V, Stark R, Menon S, Carroll JS. 2013. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 23:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67:6477–6483 [DOI] [PubMed] [Google Scholar]
- 46. Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9:201–209 [DOI] [PubMed] [Google Scholar]
- 47. Frietze S, O'Geen H, Littlepage LE, Simion C, Sweeney CA, Farnham PJ, Krig SR. 2014. Global analysis of ZNF217 chromatin occupancy in the breast cancer cell genome reveals an association with ERalpha. BMC Genomics 15:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. 2013. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol 14:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Bragt MP, Hu X, Xie Y, Li Z. 2014. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife 3:e03881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. 2010. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol 30:3943–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gu G, Gelsomino L, Covington KR, Beyer AR, Wang J, Rechoum Y, Huffman K, Carstens R, Ando S, Fuqua SA. 2015. Targeting thyroid hormone receptor beta in triple-negative breast cancer. Breast Cancer Res Treat 150:535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heublein S, Mayr D, Meindl A, Angele M, Gallwas J, Jeschke U, Ditsch N. 2015. Thyroid hormone receptors predict prognosis in brca1 associated breast cancer in opposing ways. PLoS One 10:e0127072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martinez-Iglesias OA, Alonso-Merino E, Gomez-Rey S, Velasco-Martin JP, Martin Orozco R, Luengo E, Garcia Martin R, Ibanez de Caceres I, Fernandez AF, Fraga MF, Gonzalez-Peramato P, Varona C, Palacios J, Regadera J, Aranda A. 2016. Autoregulatory loop of nuclear corepressor 1 expression controls invasion, tumor growth, and metastasis. Proc Natl Acad Sci U S A 113:E328–E337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martinez-Iglesias O, Olmeda D, Alonso-Merino E, Gomez-Rey S, Gonzalez-Lopez AM, Luengo E, Soengas MS, Palacios J, Regadera J, Aranda A. 2016. The nuclear corepressor 1 and the thyroid hormone receptor beta suppress breast tumor lymphangiogenesis. Oncotarget 7:78971–78984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martinez-Iglesias O, Garcia-Silva S, Tenbaum SP, Regadera J, Larcher F, Paramio JM, Vennstrom B, Aranda A. 2009. Thyroid hormone receptor beta1 acts as a potent suppressor of tumor invasiveness and metastasis. Cancer Res 69:501–509 [DOI] [PubMed] [Google Scholar]
- 57. Martinez-Iglesias O, Garcia-Silva S, Regadera J, Aranda A. 2009. Hypothyroidism enhances tumor invasiveness and metastasis development. PLoS One 4:e6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.