Abstract
Oligodendrocytes are the critical cell types giving rise to the myelin nerve sheath enabling efficient nerve transmission in the central nervous system (CNS). Oligodendrocyte precursor cells differentiate into mature oligodendrocytes and are maintained throughout life. Deficits in the generation, proliferation, or differentiation of these cells or their maintenance have been linked to neurological disorders ranging from developmental disorders to neurodegenerative diseases and limit repair after CNS injury. Understanding the regulation of these processes is critical for achieving proper myelination during development, preventing disease, or recovering from injury. Many of the key factors underlying these processes are epigenetic regulators that enable the fine tuning or reprogramming of gene expression during development and regeneration in response to changes in the local microenvironment. These include chromatin remodelers, histone-modifying enzymes, covalent modifiers of DNA methylation, and RNA modification–mediated mechanisms. In this review, we will discuss the key components in each of these classes which are responsible for generating and maintaining oligodendrocyte myelination as well as potential targeted approaches to stimulate the regenerative program in developmental disorders and neurodegenerative diseases.
Keywords: epigenetics, developmental disorders, neurodegenerative disease, oligodendrocyte, myelination, myelin repair, multiple sclerosis, chromatin remodelers, histone-modifying enzymes, DNA methylation, RNA modification
Introduction
Oligodendrocytes are the specialized glial cells of the central nervous system (CNS) that produce the myelin sheaths surrounding axons and enabling salutatory conduction as well as providing metabolic support to axons 1. Defects in the myelination process have been associated with developmental disorders such as autism 2– 5 and coloboma, heart disease, atresia choanae, retarded growth and development, genital hypoplasia, and ear abnormalities (CHARGE) syndrome 6, 7 as well as neurodegenerative diseases such as the demyelinating disease multiple sclerosis (MS) and various leukodystrophies 8. The late-onset neurodegenerative diseases may also stem from subtle dysregulation of early developmental processes. In addition, dysregulation of the processes controlling proliferation and differentiation in the oligodendrocyte lineage has been linked to the development of various brain cancers 9. Understanding developmental myelination and remyelination processes will have impacts for the development of treatments to improve functional recovery after injury or disease 10– 13.
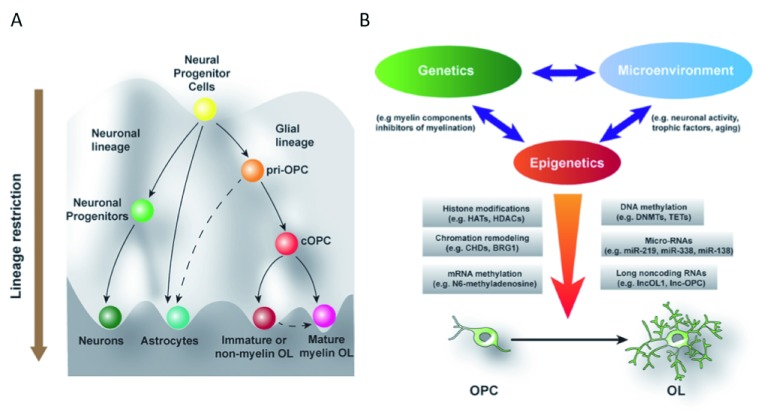
The oligodendrocyte lineage originates from multi-potent neural progenitor cells (NPCs). Early NPC divisions result predominantly in neurons before switching to a primarily glial progeny later in development 14– 16. First, NPCs become primitive oligodendrocyte progenitor cells (pri-OPCs or pre-OPCs) expressing Olig1/2, then committed OPCs (PDGFRα +/NG2 +), which persist in the CNS throughout life 17. OPCs can further proliferate and differentiate into mature myelinating oligodendrocytes 18– 21. The transition into each of these stages requires the coordination of intrinsic and extra-cellular cues where transcriptional regulatory events are closely interconnected and function together to safeguard the oligodendrocyte identity and prevent alternative cell fates such as astrocytes or neurons ( Figure 1A).
Figure 1. Differentiation of progenitor cells is a highly choreographed process.
( A) A diagram depicts an epigenetic landscape of cellular fate decision-making during oligodendrocyte development from neural progenitor cells. Beginning with neural progenitors, cell differentiation occurs along multiple potential pathways with cells taking on neuronal, astrocyte, or oligodendrocyte lineages. This differentiation from a common progenitor population involves the fine tuning of gene expression and turning on and off of lineage-specific genes and their epigenetic regulators. ( B) Many modulators of gene expression are through epigenetic mechanisms, which alter gene expression on the basis of local environmental factors. These mediators include covalent modifications to DNA or histones, RNA-mediated regulation of gene expression, or the enzymes responsible for mediating the effects of these modifications. BRG1, Brahma-related 1; CHD, chromodomain helicase DNA-binding; cOPC, committed oligodendrocyte progenitor cell; DNMT, DNA methyltransferase; HAT, histone acetyltransferase; HDAC, histone deacetylase; OL, oligodendrocyte; pri-OPC, primitive oligodendrocyte progenitor cell; TET, ten-eleven translocation.
The oligodendrocyte lineage is highly responsive to environmental cues. For example, activity or experience can promote myelination of axons by newly formed oligodendrocytes and even induce the proliferation of OPCs 22– 29. Additionally, there exist critical periods during oligodendrocyte development and myelination 30, 31 when oligodendrocytes are highly receptive and adaptive to environmental cues such as neuronal activity 32. The plasticity of myelinating oligodendrocytes and adaptive myelination are important for normal neural circuit function and cognition 33. Epigenetic regulation is likely the process through which the effects of these kinds of stimuli are carried out. At present, how epigenetic mechanisms mediate the environmental cues for oligodendrocyte myelination and remyelination remains poorly defined.
In recent years, the importance of epigenetic mechanisms and their non-genetic regulation of gene expression and cell states has been increasingly recognized 18, 20, 34, 35 ( Table 1). Epigenetic regulation of gene expression occurs through a variety of mechanisms, including covalent modifications of chromatin to regulate stearic access to DNA, ATP-dependent nucleosome remodeling, DNA methylation, non-coding RNAs, and RNA modifications 21, 78. All of these processes can modulate large-scale genetic programs to alter and maintain cell states during oligodendrocyte progenitor proliferation and maturation ( Figure 1B). Epigenetic modifications are often reversible and provide the necessary plasticity for progenitor cells to respond to environmental cues. Such pathways are amenable to pharmacological intervention and could be targeted to promote myelin growth or repair.
Table 1. Epigenetic pathways in oligodendrocyte development and myelination.
| Epigenetic regulators | Component | Description | Function in oligodendrocytes |
|---|---|---|---|
|
ATP-dependent
chromatin remodelers |
BRG1 (also known as
Smarca4) |
A key helicase subunit of
the SWI/SNF Family |
Stage-dependent promotion of OPC differentiation
but not required for OPC survival 36, 37. |
| CHD7 | Member of the chromo
helicase domain family |
CHD7 is required for oligodendrocyte differentiation
and remyelination in the spinal cord 6, 38, 39. |
|
| CHD8 | Member of the chromo
helicase domain family |
CHD8 has been linked to autism disorder with
white matter defects. CHD8 knockout in the oligodendrocyte lineage leads to myelination defects 40, 41. |
|
|
Histone acetylation
modifiers |
EP300 (also known as p300) | Histone acetyltransferase | Associated with Rubinstein–Taybi syndrome
42,
43 and
regulates oligodendrocyte differentiation 44. |
| EP400 (E1A Binding Protein
P400) |
Key subunit of TIP60
histone acetyltransferase complex |
Deletion in CNP
+ oligodendrocytes leads to defects
in terminal differentiation and hypomyelination 45. |
|
| HDAC1 | Class I histone
deacetylase (HDAC) |
Regulates oligodendrocyte differentiation via
co-repressor complexes 46, 47, 48 and has non– histone-dependent effects in oligodendrocyte differentiation 49. |
|
| HDAC2 | Class I HDAC | Functionally redundant regulation of
oligodendrocyte differentiation with HDAC1 46. |
|
| HDAC3 | Class I HDAC
Complexes with co- repressors NCOR/SMRT |
Regulates the fate choice of primitive OPCs
between astrocytic and oligodendrocytic fates and myelination 44. |
|
| SIRT1 | Class III NAD + HDAC | Stage-dependent effects on OPC proliferation.
Increased OPC differentiation when knocked out in OPCs 50. |
|
| SIRT2 | Class III NAD + HDAC | Highly expressed in mature oligodendrocytes. Its
level is positively correlated with oligodendrocyte differentiation 51. |
|
| HDAC6 | Class II HDAC | Regulates oligodendrocyte differentiation via is
acetylation of tubulin in the cytoskeleton 52. |
|
| HDAC10 | Class II HDAC | No clear role, likely due to functional redundancy
with other HDACs in its regulation of OLIG1 nuclear localization 49. |
|
| HDAC11 | Class IV HDAC | Regulates oligodendrocyte differentiation possibly
via modulating regulatory elements of myelin- related genes 53, 54, 55. |
|
|
Histone methyl-
transferases |
COMPASS-like complex | Major subunits include
SETD1A, MLL1, and MLL2 (KMT2A) |
MLL2 works with CHD8 to deposit H3K4me3
at active promotors of oligodendrocyte lineage genes 41. |
| PRC2 complexes | Major subunits include
EZH2, EED, and SUZ12 |
Responsible for H3K27me3 deposition. Promotes
oligodendrogenesis and OPC differentiation 56, 57. |
|
| PRMT1 | Catalyzes histone arginine
methylation |
Required for proper OPC differentiation resulting in
hypomyelination defects 58. |
|
| PRMT5 | Catalyzes histone arginine
methylation |
Required for proper OPC differentiation resulting in
hypomyelination defects 59– 61. |
|
|
DNA methyl-
transferases and demethylases |
DNMT1 | DNA methyltransferase | Knockout early development impairs OPC
differentiation and results in hypomyelination 62. Has no effect on myelin repair 55. |
| DNMT3a | DNA methyltransferase | Plays a role in myelin repair after injury but not early
development of the oligodendrocyte lineage 55. |
|
| TET1–3
(ten-eleven translocation) |
DNA demethylases that
catalyze the conversion of 5mC to 5hmC |
Differentially regulated at different stages during OL
development. Tet1 is required for OL differentiation in vitro 63. |
|
| microRNAs | Dicer | Enzyme responsible for
processing microRNAs into mature form |
Required for OPC differentiation, myelination, and
myelin maintenance 64– 66. |
| miR-219 | miR-219 is necessary and sufficient to induce
differentiation 65, 66. Also required for remyelination after lysophosphatidylcholine (LPC)-induced demyelination 67. |
||
| miR-338 | miR-338 is dispensable for OPC differentiation
or myelination in vivo but has synergistic with miR-219 67. |
||
| miR-212 | Negatively regulates common oligodendrocyte and
myelin-related genes by miR-212 68. |
||
| miR-125a-3p | Upregulated in cerebrospinal fluid from multiple
sclerosis patients with active demyelinating lesions. Negatively regulates oligodendrocyte differentiation 69. |
||
|
Long non-coding
RNAs |
lncOL1 | LncOL1 positively regulates OPC differentiation
while having no effect on OPC formation. Affects timing of myelinogenesis but not the maintenance of myelin 56. |
|
| Lnc-OPC | Knockdown of lnc-OPC in NPCs limited their
differentiation into OPCs without affecting NPC proliferation 70. |
||
| Pcdh17it | A marker of the immature premyelinating
oligodendrocyte population 71. |
||
| SOX8OT | Regulates oligodendrocyte differentiation through
targeting SOX8 72, 73. |
||
| Neat1 | Knockout reduces the number of oligodendrocytes
in the frontal cortex 74. |
||
| Lnc158 | Correlates with oligodendrocyte differentiation-
associated gene expression 75. |
||
|
N
6-methyl-adenosine
(m 6A) modifiers |
METTL14 | m6A RNA writer | Required for OPC differentiation and proper
myelination 76. |
| PRRC2A | An m6A RNA binding
protein |
Highly expressed in OPCs and white matter
tracks. Required for normal OPC proliferation and differentiation 77. |
|
| FTO | m6A RNA demethylase
(alpha-ketoglutarate– dependent dioxygenase) |
Knockout mimics the effects of PRRC2A
overexpression increasing Olig2 expression 77. |
CHD, chromodomain helicase DNA-binding; HDAC, histone deacetylase; OL, oligodendrocyte cell line; OPC, oligodendrocyte progenitor cell; NPC, neural progenitor cell.
ATP-dependent chromatin remodelers in oligodendrocyte lineage progression and regeneration
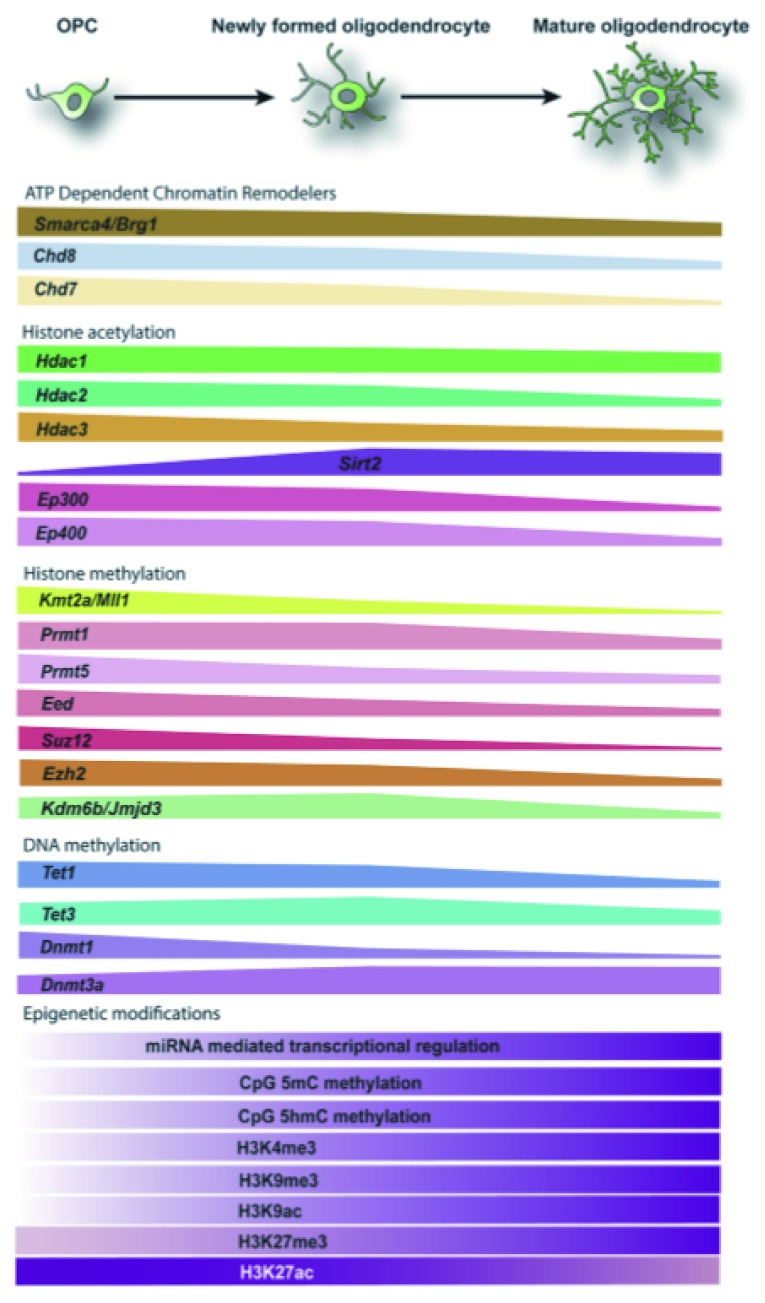
ATP-dependent chromatin remodeling uses ATP to remodel the nucleosome, opening up areas for enhancing transcription, and is critical for neural cell growth and differentiation 79, 80. Early work in cell cultures showed that OPCs differentiating into mature oligodendrocytes underwent substantial chromatin reorganization within the nucleus 81. The chromatin remodelers consist of several multi-subunit complexes which fall into four major families: the SWI/SNF family with the major ATPase subunits Brahma-related 1 (BRG1, also known as Smarca4) and Brahma (BRM, also known as SMARCA2), the INO80 family which includes the ATPases INO80 and SRCAP, the ISWI family with ATPase subunits SNF2L and SNF2H, and the chromodomain helicase DNA-binding (CHD) family consisting of CHD1–9 80, 82, 83. Of these, the SWI/SNF family and the CHD family members are dynamically regulated over the course of OPC specification and differentiation and have been implicated in oligodendrocyte development and myelination ( Figure 2).
Figure 2. Global expression levels of key epigenetic regulators during oligodendrocyte differentiation from progenitor cells.
Epigenetic modifiers, including ATP-dependent chromatin remodelers, histone acetyltransferases and deacetylases, histone methyltransferases, and demethylases, are critical components of the differentiation process, according to the data from a bulk RNA sequencing dataset 92. The change of epigenetic modifiers across oligodendrocyte differentiation is depicted. The global changes of expression levels in the epigenetic modifications themselves are based on the studies 41, 44, 62, 63, 66, 93. OPC, oligodendrocyte progenitor cell.
SWI/SNF family members
The SWI/SNF family of ATPase dependent chromatin remodelers have been shown to play critical roles in the development of the oligodendrocyte lineage. Deletion of Brg1/Smarca4, the core helicase component of the SWI/SNF family, in NPCs inhibits oligodendrocyte and astrocyte lineage development while increasing neuronal differentiation in the ventricular zone of the developing brain 80, 84. A lineage-specific transcription factor, OLIG2, can recruit the BRG1/SWI/SNF complex to the promoters and enhancers of oligodendrocyte lineage genes such as Sox10 to activate their transcription. BRG1 is also necessary for OPC differentiation. BRG1 expression increases after induction of rat OPC differentiation with T3 thyroid hormone 36. These increasing levels are critical for OPC differentiation as conditional knockout in Olig1-expressing oligodendrocyte progenitors and PDGFRa-expressing OPCs in vivo leads to oligodendrocyte differentiation defects and profound dysmyelination defects 36 (JW and QL, unpublished). Of note, the loss of Brg1 does not affect OPC survival in culture or in vivo 36. However, Brg1 knockout in later OPCs, such as NG2 + or CNP +, committed or post-mitotic OPCs, respectively, had progressively less severe effects on differentiation 37, suggesting that BRG1 effects are stage-dependent. This stage-dependent severity suggests that BRG1 activates early pro-differentiation factors, such as SOX10, that can continue to mediate downstream genetic programs in oligodendrocyte lineage progression despite the upstream loss of BRG1 36, 37. In addition, other chromatin remodelers such as CHD8 or CHD7 (discussed below) potentially compensate for the loss of BRG1 at later stages.
CHD family members
CHD7 is highly enriched in the oligodendrocyte lineage, especially in differentiating oligodendrocytes. CHD7 mutations result in a series of birth defects called CHARGE syndrome, which exhibits impaired white matter development and myelination in addition to other congenital developmental abnormalities 85, 86. CHD7, like BRG1 above, does not affect OPC formation but instead causes defects in OPC differentiation 6, 38, 39. In fact, Chd7 is a direct target of the OLIG2/BRG1 complex and its expression is greatly increased by the binding of this complex at its promoter 6. CHD7 can complex with SOX10 to activate downstream regulators of oligodendrocyte differentiation. CHD7 activates the expression of OPC pro-differentiation regulators, including SOX10 and NKX2-2 39, as well as other oligodendrocyte-expressing transcription factors such as Osterix/Sp7 and Creb3l2 6, 39. Intriguingly, deletion of Chd7 in PDGFRa + OPCs appears to impair OPC survival via p53 upregulation 39. CHD7 binds to the p53 promotor in OPCs and limits p53 expression to maintain the survival of OPCs 39.
CHD7 is also required for remyelination after lysolecithin-induced demyelination 6 or spinal cord laminectomy, wherein it interacts with SOX2 to drive OPC differentiation 38. Chd7 deletion impairs OPC proliferation after spinal cord injury 38 but not in the developing brain 6, 39, suggesting a context-dependent CHD7 regulation of OPC proliferation. However, CHD7 appears to be dispensable for the maturation of oligodendrocytes, possibly due to compensation by other CHD members such as CHD8 39, which has been shown to work together with CHD7 to regulate OPC survival and maturation 39.
Another CHD family member, CHD8, is also critical for proper oligodendrocyte development. CHD8 has been linked to a subset of autism disorders, which exhibit a defect in white matter tracts and myelination 40, 41, 87– 89. Chd8 knockout in Olig1 + progenitors causes defects in CNS myelination, particularly in the spinal cord because of severe reductions in PDGFRα-expressing OPCs in this region 41, suggesting a region-specific role of CHD8 in OPC survival and differentiation. Deletion of Chd8 at the post-natal stages with an inducible PDGFRα-CreER driver also blocks OPC differentiation. The defects in oligodendrocyte differentiation are due to the cell-specific loss of Chd8 in the oligodendrocyte lineage as there are no defects seen after Chd8 knockout in post-mitotic neurons 41. This suggests that the myelination defects seen in CHD8 mutant patients are cell-autonomous defects due to the loss of CHD8. Remyelination after lysophosphatidylcholine (LPC)-induced demyelinating lesions in the spinal cord is also dependent on CHD8 expression 41. CHD8 dysregulation may be an important factor for white matter pathogenesis and remyelination failure given the critical role of CHD8 for OPC replenishment and remyelination in demyelinating lesions.
CHD7 and CHD8 have similar structures and can bind many of the same targets. However, they target different gene regions during oligodendrocyte differentiation. CHD7 predominantly binds to promotor regions in OPCs but switches to enhancer regions in oligodendrocytes 39. CHD8, in contrast, binds predominantly to promotor elements near transcription start sites marked by an activating histone mark H3K4me3 in OPCs and oligodendrocytes where it recruits an H3K4 activating histone methyltransferase MLL2 (mixed lineage leukemia 2) complex to drive expression of oligodendrocyte lineage genes 41. MLL2–4 and other family members can form a macromolecular complex called COMPASS (complex of proteins associated with Set1) to methylate H3K4 and regulate gene transcription 90. Strikingly, blocking lysine demethylase KDM5, an enzyme that erases methylation on H3K4 91, with a pan-KDM5 inhibitor CPI-455 rescues the differentiation defects in Chd8 mutant OPCs 41, suggesting that targeting this eraser to enhance H3K4me3 levels might facilitate the restoration of myelination defects caused by CHD8 defects.
The chromatin remodelers may all work together to regulate oligodendrocyte development but each has its own preferences for regulatory elements and mechanisms to control expression of specific sets of targeted genes. Of these, CHD8 appears to turn on the earliest, eventually promoting BRG1 expression which in turn induces CHD7 expression 6. This successive signaling cascade enables the progression from OPCs to mature myelinating oligodendrocytes and likely forms convergent points upon which other checkpoints and regulatory mechanisms act to facilitate this development. Nonetheless, these chromatin remodelers could operate simultaneously in a non-linear fashion to promote oligodendrocyte lineage progression.
Histone acetylation control of cell fates and differentiation in the oligodendrocyte lineage
Histone acetylation, in particular, has been strongly implicated in the regulation of oligodendrocyte development. The addition and elimination of acetyl groups are balanced through the competing work of histone acetyltransferases (HATs) 94 and histone deacetylases (HDACs).
HATs
The activity of HATs is responsible for the acetylation of histones, leading to relaxed chromatin coiling and increased gene expression. The acetylation of histone H3 on lysine 27 (H3K27ac) is often deposited at active regulatory elements such as enhancers and promoters and is positively correlated with the activation of gene transcription 95. H3K27 acetylation is catalyzed by multiple HATs, including p300 (also known as EP300), CREB-binding protein (CBP), TIP60, and PCAF 96, 97. Histone acetylation status can be further recognized by bromo-, PHD-, Tudor-, or WD40-domain-containing transcription activating regulators, which further modulate target gene expression 95, 98– 100.
In line with a critical role for histone acetylation in regulating the oligodendrocyte lineage, the loss of multiple HATs can lead to defects in the myelination process. Deletion of EP400, a key subunit of the TIP60 HAT complex, in Cnp-expressing oligodendroglial cells results in a defect in oligodendrocyte terminal differentiation, leading to profound hypomyelination 45. A genetic disorder, Rubinstein–Taybi syndrome, is associated with mutations in p300 (also known as EP300), which is characterized in part by hypoplasia of the corpus collosum and congenital hypomyelination 42, 43. The role of p300 in controlling oligodendrocyte development is still being explored, but p300 has been shown to interact with HDAC3 (discussed in more detail below) partly facilitating the role of HDAC3 in promoting oligodendrocyte as opposed to astrocytic cell lineages during early differentiation from NPCs via its promotion of Olig2 expression 101. Also, inhibition of p300 activity itself can lead to pronounced defects in OPC differentiation (JW and QL, unpublished).
HDACs
Histone acetylation status can be reversed by HDACs. Mammals possess four classes of HDACs. Class I contains HDACs 1–3 and 8, class II contains HDACs 4–7 and 9 and 10, class III are NAD-dependent HDACs (also known as sirtuins, encompassing SIRT1–7), and finally class IV contains one HDAC, HDAC11 102, 103. Pharmacological studies using HDAC inhibitors have indicated the importance of HDACs in oligodendrocyte fate specification and differentiation. Treating rat NPCs with valproic acid inhibited oligodendrogenesis and astrogenesis while promoting neurogenesis likely through NeuroD1 upregulation following the inhibition of HDAC activity 104. Blocking HDAC activity with pan inhibitors also disrupts the differentiation of OPCs into mature oligodendrocytes 105. The timing of this treatment appears to be critical. Treating cells with pan-HDAC inhibitors after the differentiation process has been shown to have minimal effect on oligodendrocyte differentiation 106. These studies indicate that HDACs play various roles at different stages during OPC differentiation and subsequent myelination. However, classic pan-HDAC inhibitors are non-specific and target HDACs across multiple classes. Genetic manipulation can specifically target individual HDACs to define their specific functions during oligodendrocyte lineage progression.
Class I HDACs
The expression levels and functions of class I HDACs are important for oligodendrocyte fate specification and differentiation ( Figure 2). HDAC1 and HDAC2, when knocked out individually in the oligodendrocyte lineage, have no obvious effects on OPC formation, proliferation, or differentiation 46. However, the double-knockout animals die shortly after birth, and analysis revealed a severe defect in OPC proliferation or differentiation in these animals, suggesting that HDAC1 and HDAC2 can functionally compensate for the loss of the other in oligodendrocyte lineage determination 46. Another HDAC class I family member, HDAC3, has been implicated in the control of oligodendrocyte lineage specification 44 but differs from those effects observed in HDAC1. HDAC3 deletion at the same stages as HDAC1 and 2 above results in a switch from oligodendrocyte to astrocytic fates, suggesting that HDAC3 regulates the fate choice of primitive OPCs between astrocytic and oligodendrocytic cell lineages 44.
HDACs have been shown to exhibit non–histone dependent functions during oligodendrocyte development. HDAC1/2 co-repressor complexes can compete with β-catenin for binding to TCF7L2 (TCF4), a member of the TCF transcription factor family, resulting in the disinhibition of TCF7L2, which then is free to promote OPC differentiation 46, 47, 48. This is an example of how non-enzymatic activity of HDACs through protein–protein interaction in addition to the deacetylase activity can function to regulate genetic expression. HDAC1 can also be recruited by YY1 transcription factor to the promotor of oligodendrocyte differentiation inhibitory genes such as Id4 to reduce their expression 107, 108. In addition, HDAC1 and HDAC3 can deacetylate the OLIG1 transcription factor, increasing its likelihood of nuclear translocation and ultimate promotion of OPC differentiation 49.
HDAC activity can be modulated through co-regulators or covalent modifications 109. For example, casein kinase 2 (CK2) phosphorylates HDAC3 to activate its activity while phosphatase 4 dephosphorylates it 110. Of interest, in vitro experiments revealed that the CK2 kinase, which activates HDAC3, also elevates expression of OLIG2, a critical transcription factor for initiating oligodendroglial cell fate 111. HDAC3 also forms protein complexes with co-repressor complexes such as NCOR and SMRT to regulate its activity 112. NCOR has been shown to negatively regulate astrogenesis through inhibiting JAK-STAT signaling, activation of which leads to astrocyte differentiation 113. In addition, HDAC3/NCOR can deacetylate and inactivate astrocyte-promoting factor STAT3 and therefore promote oligodendrogenesis while inhibiting the astroglial fate 44. HDAC3 also forms complexes with the HAT p300, to regulate OLIG2 expression levels during OPC specification 44. This interaction likely indicates that the coordinated activity of two opposing factors is required for oligodendrocyte and astrocytic lineage fate decisions.
HDAC3 functions not only as a transcriptional co-repressor, as one may assume from its histone deacetylation activity, but also as a transcriptional co-activator as in its role in the activation of retinoic acid response elements 114, 115. It is worth noting that HDAC3 deacetylase activity may not be vital for oligodendrocyte development. HDAC3 requires NCOR and SMRT to promote its deacetylation activity. Deleting the deacetylase-activating domains (DADs) in NCOR and SMRT abrogates the deacetylase activity of HDAC3. However, the DAD deletion mice survive to adulthood and exhibit normal myelination whereas the ablation of HDAC3 is embryonic lethal 116. Although the function of HDAC3 catalytic site mutants remains to be determined, the current data suggest that HDAC3 may serve as a scaffold for multi-component transcriptional regulatory complexes vital for oligodendrocyte myelination.
Other HDAC classes
Among class II HDACs, HDAC6 has been shown in rat oligodendrocyte cultures to acetylate the microtubule-associated protein tau and α-tubulin, both of which are required for normal oligodendrocyte development 52. HDAC10, along with HDAC1 and HDAC3, has also been shown to regulate the nuclear localization of OLIG1 for oligodendrocyte maturation 49. It is worth noting that the enzymatic activity of class II HDACs is dependent on the HDAC3/SMRT/N-CoR complex 117.
The class III HDACs SIRT1 and SIRT2 have been shown to regulate early oligodendrocyte lineage determination 50, 51, 118. SIRT2, in particular, is highly expressed in mature oligodendrocytes 119 and regulates the differentiation of oligodendrocytes as blocking its activity or overexpressing it prevents or promotes differentiation of CG4 oligodendroglial cells, respectively 120– 122. This class of HDACs also relies on NAD as a co-factor for deacetylase activity 123. The loss of NAMPT, the rate-limiting enzyme for NAD biosynthesis in mammals, leads to defective oligodendrocyte development 118, 124. Like those of many other epigenetic factors, the effects that SIRT1 and SIRT2 have on development are stage-dependent. For example, Sirt1 knockout in NPCs increases OPC proliferation 50 while Sirt1 knockout in PDGFRa + OPCs promotes cell cycle exit and OPC differentiation 125. Notably, SIRT2 is depleted in myelin sheathes of PLP-deficient oligodendrocytes, a model for spastic paraplegia, suggesting that SIRT2 might have a role in myelin sheath maintenance and provide trophic support of axons 126.
Finally, the class IV HDAC, HDAC11 has been shown to regulate H3K9 and H3K14 acetylation and expression levels of Mbp and Plp genes 53, 54. HDAC11 overexpression enhances the maturation of an oligodendrocyte cell line (OL-1) in vitro 53, 54, suggesting a potential role of HDAC11 in regulating myelin gene expression. At present, how the function of each HAT and HDAC is controlled, individually and coordinately, on a system-wide level to regulate the complex processes of oligodendrocyte development and myelination remains to be defined. This is of particular importance given the reiterative involvement of many HAT and HDAC enzymes in the gene regulatory network during CNS development and regeneration.
Histone methylation regulates oligodendrocyte differentiation
Histone methylation can be linked to either gene activation or gene repression. The activating histone mark H3K4 trimethylation (H3K4me3) is deposited mainly at promoter elements and associated with gene transcription 127. The COMPASS-like complex, consisting of SETD1A and MLL1/2, is a major enzyme responsible for H3K4me3 deposition 128, 129, although its function in oligodendrocyte development has not been fully defined.
During differentiation from a more plastic state to a more differentiated state, the level of repressive histone marks, for example, H3K27me3 and H3K9me3 increases across many different cell types 130, 131, including oligodendrocyte lineage cells 56, 93. The histone methyltransferases mediating the deposition of these marks are critical in the differentiation of oligodendrocytes. Inhibition of H3K9 histone methyltransferases in cell culture via pharmacological inhibitors or shRNAs suggested a role of H3K9 deposition in the progression of the OL lineage and the suppression of neuronal gene programs 93. However, the in vivo role of these H3K9 histone methyltransferases in oligodendrocyte development remains to be defined.
The importance of H3K27 trimethylation (H3K27me3) in oligodendrocyte development is more defined. Polycomb repressive complex 2 (PRC2), consisting of EZH2, EED, and SUZ12, is the sole enzyme responsible for H3K27me3 in mammals 132– 134. Expression of PRC2 complex components exhibits a spatiotemporal-specific pattern 135– 137, suggesting that individual PRC2 subunits may play distinct functions during oligodendrocyte development and myelination. EZH2, the core catalytic subunit of PRC2 mediating its methyltransferase activity, promotes oligodendrogenesis from neural stem cells as opposed to astrocyte formation in a dose-dependent manner 57. In addition, the loss of Ezh2 at later stages in Olig1-expressing progenitors prevents OPC differentiation, decreasing the number of mature oligodendrocytes in vivo 56. These observations suggest that elevation of H3K27me3 levels is required for oligodendrocyte differentiation. Of note, mutations in the histone such as H3.3K27M precludes PRC2-mediated H3K27me3 138, 139. This mutation limits the capacity for OPC differentiation and is a major factor contributing to the development of malignant diffuse intrinsic pontine glioma (DIPG) 138, 140, 141. OPCs or pri-OPCs have been implicated as the tumor cells of origin for H3K27M midline gliomas 142, 143, highlighting the critical nature of this epigenetic mechanism in regulating the development of the oligodendrocyte lineage.
Another histone methyltransferase family that catalyzes arginine instead of lysine methylation, PRMTs 144, is also implicated in OPC differentiation. PRMT1 58 and PRMT5 59– 61 have both been shown to be required for proper differentiation of OPCs into mature oligodendrocytes, and loss-of-function mutants develop hypomyelination phenotypes. The function of other PRMT family members in oligodendrocyte myelination remains to be further defined. Overall, these studies demonstrate that the balance of histone methyltransferases and histone demethylases is likely critically important for the regulation of oligodendrocyte development and remyelination.
DNA methylation and demethylation in oligodendrocyte development
DNA methylation is an epigenetic regulatory mechanism where cytosines, specifically those preceding guanine in so-called CpG islands, are methylated. CpG islands are preferentially found in the 5′ promotor region of genes and their methylation state can inhibit or promote the expression of the relevant gene 145. The methylation status of these sites is regulated by the coordinated activity of DNA methyltransferases (DNMTs), which add methyl groups to convert cytosine into 5-methylcytosine, and ten-eleven translocation (TET) proteins or DNA demethylases, which catalyze the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (5hmC), beginning the process of converting 5-methylcytosine back to cytosine 146. The expression of individual DNMTs and TETs varies across the OL lineage, suggesting a potential stage-specific role of DNMTs and TETs for oligodendrocyte development, myelination, and remyelination. In line with these observations, there was also a significant increase in DNA methylation during OL maturation 62.
DMNT1 is downregulated during oligodendrocyte differentiation where other DMNT family members had no change 62. Deletion of Dmnt1 early in the oligodendrocyte lineage had a profound effect on oligodendrocyte differentiation, resulting in hypomyelination in vivo 62. This effect was not due to the upregulation of normally methylated genes alone; defects in alternative splicing mediated by DNA methylation were also attributed to the failure in myelination 62. In contrast to the Dmnt1 knockout, Dmnt3a knockout in NPCs had no effect 62. However, after lysolecithin-induced demyelination, tamoxifen-inducible Dnmt3a deletion in mature oligodendrocytes using a PLP-CreERT2 driver line impaired remyelination whereas the inducible Dnmt1 knockout had no effect 147. Taken together, these results suggest that in some cases remyelination in adulthood does not fully recapitulate the developmental program.
TET1, TET2, and TET3 have all been implicated in the differentiation of oligodendrocytes in vitro 63. However, they each have different structures and their expression and subcellular localizations differ 63, suggesting that they may play different roles in regulating oligodendrocyte differentiation. TET1 is downregulated in mature oligodendrocytes, TET2 translocates from the cytoplasm to the nucleus during OPC differentiation, and TET3 is seen only in the nucleus of maturing oligodendrocytes 63. Of these, TET1 appears to show the strongest effect in regulating the oligodendrocyte lineage where the knockout impairs oligodendrocyte development and remyelination after lysolecithin-induced demyelination 148, 149.
Non-coding RNAs in oligodendrocyte development and myelination
Non-coding RNAs such as microRNAs (miRNAs) or long-non-coding RNAs (lncRNAs) play regulatory roles in oligodendrocyte development, myelination, and remyelination. miRNAs are short RNA sequences that bind to homologous sequences on mRNA transcripts to inhibit translation into proteins. These miRNAs are processed into their mature active form by the enzyme Dicer. Conditional deletions of Dicer in OPCs and mature oligodendrocytes have all resulted in defects in myelination. Despite the myelin defects, the population of proliferating OPCs is increased in these animals, indicating a critical role for miRNAs in balancing OPC proliferation and differentiation 64– 66. Post-natal Dicer1 ablation in mature oligodendrocytes results in demyelination and oxidative damage, leading to neuronal degeneration and inflammatory astrogliosis and microgliosis in the brain 64, suggesting a critical role of Dicer and thus miRNAs in myelin lipid maintenance and redox homeostasis.
miRNAs
Comparisons between OPCs and immature and mature oligodendrocytes revealed a set of miRNAs enriched during oligodendrocyte differentiation, including miR-219, miR-138, and miR-338 64– 66, 150. miR-219 is necessary and sufficient to induce differentiation and can even partially rescue the Dicer knockout phenotype 65, 66. Knockout of miR-219–encoding genes ( miR-219-1 and miR-219-2) led to reduced myelination throughout the CNS 67. In contrast to miR-219, miR-338 is dispensable for OPC differentiation or myelination in vivo. However, there was a synergistic effect on the myelination defects in miR-219 and miR-338 double-conditional knockout mice. miR-219 is also required for remyelination after LPC-induced demyelination. Overexpression of miR-219 in OPCs increased oligodendrocyte differentiation and could even promote repair when overexpressed genetically or administered with intrathecal injections of miR-219 mimics 67. miR-219 likely functions by repressing inhibitors of OPC differentiation, including Lingo1 and Etv5 67. miR-219 has been suggested in zebrafish to regulate oligodendrocyte lineage specification from NPCs 151. However, there were no defects of OPC specification in the miR-219-1/2 double-null animals 67, suggesting a species-specific effect. Other miRNAs have been associated with oligodendrocyte development (reviewed in more detail in 152), but of these miR-219 exhibits the strongest effects.
A set of miRNAs have been identified to negatively regulate oligodendrocyte differentiation. One of these, miR-212 was found to be downregulated in oligodendrocytes after spinal cord injuries in rats, where it appears to repress expression of differentiation-associated genes 68. Another such miRNA, miR-125a-3p, was enriched in cerebrospinal fluid from MS patients with active demyelinating lesions. miR-125a-3p overexpression impaired oligodendrocyte differentiation whereas knocking it down promoted differentiation 69. Similarly, overexpression of miR-27a inhibits oligodendrocyte differentiation and myelination by activating the Wnt/beta-catenin signaling pathway 153. Such negative regulatory miRNAs are potential targets for enhancing remyelination.
LncRNAs
LncRNAs are long RNA sequences (more than 200 nucleotides) that are highly conserved across species but have no coding potential 154. LncRNAs have been implicated in the regulation of both normal development 155, 156 and diseases 157, 158. LncRNAs can be very specifically expressed in the oligodendrocyte lineage; for instance, lncOL1 and Pcdh17it were recently identified as specific markers for oligodendrocytes and immature premyelinating oligodendrocytes, respectively 56, 71. Gene-chip microarrays were initially used to identify lncRNAs such as SOX8OT (SOX8 opposite transcript) in cultured OPCs. SOX8OT might have a role in regulating oligodendrocyte differentiation through its regulation of SOX8 72, 73.
Combining RNA sequencing and chromatin mapping across oligodendrocyte lineage stages revealed several lncRNAs that are actively transcribed and restricted to this lineage 56. Of these, lncOL1 was identified as a top candidate on the basis of its abundance, regulation during oligodendrocyte differentiation, and preliminary screening for effects on myelin-associated gene expression. lncOL1 overexpression led to precocious oligodendrocyte differentiation in mouse embryos, and lncOL1 knockout led to defects in OPC differentiation while having no effect on OPC formation. Interestingly, these myelination defects were seen only during development but not in adulthood, suggesting a role of lncOL1 in regulating the timing of myelinogenesis and not the maintenance of myelin. lncOL1 mediates this effect in part by its interaction with SUZ12, a member of the PRC2 complex which mediates histone methylation through EZH2. lncOL1 directs the PRC2 complex to silence the expression of OPC-associated genes via H3K27me3 deposition 56. In contrast to lncOL1, lnc-OPC, another lncRNA found in the oligodendrocyte lineage, is enriched in OPCs and regulated by OLIG2 70. Knockdown of lnc-OPC in cultured NPCs limited their differentiation into OPCs without affecting NPC proliferation 70. In similar fashion, lnc158 expression directly correlated with oligodendrocyte-associated protein expression and differentiation along the oligodendrocyte lineage 75. In addition, another lncRNA, Neat1, was downregulated in schizophrenia. Neat1 knockout mice exhibited a reduction in the number of oligodendrocytes in the frontal cortex because of a failure in the retention of oligodendrocyte transcription factors in the nucleus 74. These studies indicate that lncRNAs regulate oligodendrocyte development and myelination via various processes such as controlling mRNA transcripts, nuclear localization of transcription factors, or interactions with chromatin remodelers.
m 6A RNA modification in oligodendrocyte progression and homeostasis
N 6-methyladenosine (m 6A) is the most abundant internal modification of mRNA in eukaryotes. A methyl group can be added to the N 6 position of adenosines in specific sequences by m 6A methyltransferases (m 6A writers) such as METTL3 and METTL14 or removed by demethylases (m 6A erasers) such as FTO and ALKBH5. The effects of m 6A methylation on translation and RNA stability is mediated by m 6A-specific binding proteins (m 6A readers) including YTH-domain containing family proteins, hnRNP proteins, PRRC2A, and IGF2BP 159– 161.
Recent studies have revealed a differential m 6A methylation of core oligodendrocyte lineage genes during OPC differentiation, suggesting an important role for this process in OL differentiation 76. Deleting the METTL14 led to defects in OPC differentiation and hypomyelination at least in part by regulating alternative mRNA splicing in OL-expressing genes, including the paranodal protein NF155, which is critical for the establishment and maintenance of nodes of Ranvier 76. In addition, the m 6A RNA binding protein PRRC2A is highly expressed in OPCs during development in the white matter tracks. Both knockout and knockdown of PRRC2A in NPCs via Nestin-Cre or Olig2-Cre + oligodendrocyte lineage cells led to hypomyelination due to defects in OPC proliferation and differentiation 77. PRRC2A was shown to bind to the Olig2 mRNA and further stabilize the expression of Olig2 transcript in an m 6A-dependent manner. Knocking out the RNA demethylase FTO mimicked the effects of PRRC2A overexpression 77 and led to increased Olig2 expression levels. These studies suggest a critical role for m 6A modification in the OL myelination process. The function of other mRNA modification enzymes remains to be determined in myelination and remyelination in the CNS.
Conclusion and perspectives
Chromatin modifications and epigenetic regulation are crucial for oligodendrocyte fate specification, OPC proliferation, and oligodendrocyte differentiation ( Figure 1). Many members of chromatin modifiers discussed above have not yet been examined in the context of oligodendrocyte development and regeneration. In particular, although the major mediators of these developmental processes are being identified, the environmental influences that modulate the epigenetic mechanisms are very poorly understood. A better understanding of the mechanisms underlying the windows of epigenetic engagement will facilitate oligodendrocyte regeneration and remyelination.
Targeting epigenetic factors to influence OPC differentiation as a means to promote myelin regeneration after nerve injury or in demyelinating diseases is an exciting potential therapeutic avenue. In MS, for example, many demyelinating plaques still contain OPCs; however, these cells fail to differentiate to replace those lost. Additionally, oligodendrocyte loss and the subsequent loss of myelin sheaths have been implicated in Alzheimer’s disease 55, 92, 162, 163. Stimulating OPCs to proliferate and differentiate would be an exciting treatment option in slowing the disease progression.
In the future, it may prove fruitful to further scrutinize in vivo models of demyelinating diseases for temporal changes in chromatin landscape, structure, occupancy, and activity in response to myelin-promoting stimuli or pharmacological treatments, such as those used in MS disease-modifying therapies. Future work of exploring these various family members of chromatin modifiers and identifying specific epigenetic modifiers responsible for CNS myelination and remyelination will facilitate the development of effective treatments for developmental disorders and neurodegenerative diseases.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Maria P Abbracchio, Department of Pharmacological and Biomolecular Sciences, University of Milan, Milan, Italy
Davide Marangon, Department of Pharmacological and Biomolecular Sciences, University of Milan, Milan, Italy
Robert H Miller, Department of Anatomy and Regenerative Biology, George Washington University School of Medicine and Health Sciences, Washington, DC, USA
Funding Statement
This study was funded in part by grants from the US National Institutes of Health (R01NS072427 and R01NS075243 to QRL) and the National Multiple Sclerosis Society (NMSS-1508 to QRL).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- 1. Simons M, Nave KA: Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb Perspect Biol. 2015;8(1):a020479. 10.1101/cshperspect.a020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boddaert N, Zilbovicius M, Philipe A, et al. : MRI findings in 77 children with non-syndromic autistic disorder. PLoS One. 2009;4(2):e4415. 10.1371/journal.pone.0004415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casanova MF: Neuropathological and genetic findings in autism: the significance of a putative minicolumnopathy. Neuroscientist. 2006;12(5):435–41. 10.1177/1073858406290375 [DOI] [PubMed] [Google Scholar]
- 4. Deoni SCL, Zinkstok JR, Daly E, et al. : White-matter relaxation time and myelin water fraction differences in young adults with autism. Psychol Med. 2015;45(4):795–805. 10.1017/S0033291714001858 [DOI] [PubMed] [Google Scholar]
- 5. Hardan AY, Fung LK, Frazier T, et al. : A proton spectroscopy study of white matter in children with autism. Prog Neuropsychopharmacol Biol Psychiatry. 2016;66:48–53. 10.1016/j.pnpbp.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He D, Marie C, Zhao C, et al. : Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat Neurosci. 2016;19(5):678–89. 10.1038/nn.4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gregory LC, Gevers EF, Baker J, et al. : Structural pituitary abnormalities associated with CHARGE syndrome. J Clin Endocrinol Metab. 2013;98(4):E737–43. 10.1210/jc.2012-3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Popescu BF, Lucchinetti CF: Pathology of demyelinating diseases. Annu Rev Pathol. 2012;7:185–217. 10.1146/annurev-pathol-011811-132443 [DOI] [PubMed] [Google Scholar]
- 9. Huse JT, Holland EC: Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–31. 10.1038/nrc2818 [DOI] [PubMed] [Google Scholar]
- 10. Bei F, Lee HHC, Liu X, et al. : Restoration of Visual Function by Enhancing Conduction in Regenerated Axons. Cell. 2016;164(1-2):219–32. 10.1016/j.cell.2015.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Franklin RJ, Gallo V: The translational biology of remyelination: Past, present, and future. Glia. 2014;62(11):1905–15. 10.1002/glia.22622 [DOI] [PubMed] [Google Scholar]
- 12. He X, Zhang L, Queme LF, et al. : A histone deacetylase 3-dependent pathway delimits peripheral myelin growth and functional regeneration. Nat Med. 2018;24(3):338–51. 10.1038/nm.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Toosy AT, Mason DF, Miller DH: Optic neuritis. The Lancet Neurology. 2014;13(1):83–99. 10.1016/S1474-4422(13)70259-X [DOI] [PubMed] [Google Scholar]
- 14. Lopez Juarez A, He D, Richard Lu Q: Oligodendrocyte progenitor programming and reprogramming: Toward myelin regeneration. Brain Res. 2016;1638(Pt B):209–20. 10.1016/j.brainres.2015.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldman SA, Kuypers NJ: How to make an oligodendrocyte. Development. 2015;142(23):3983–95. 10.1242/dev.126409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kessaris N, Fogarty M, Iannarelli P, et al. : Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9(2):173–9. 10.1038/nn1620 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Dawson MR, Polito A, Levine JM, et al. : NG2-expressing glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24(2):476–88. 10.1016/s1044-7431(03)00210-0 [DOI] [PubMed] [Google Scholar]
- 18. Zuchero JB, Barres BA: Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 2013;23(6):914–20. 10.1016/j.conb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emery B: Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20(5):601–7. 10.1016/j.conb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 20. Wegner M: A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35(1):3–12. 10.1007/s12031-007-9008-8 [DOI] [PubMed] [Google Scholar]
- 21. Bird A: Perceptions of epigenetics. Nature. 2007;447(7143):396–8. 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- 22. Gibson EM, Purger D, Mount CW, et al. : Neuronal Activity Promotes Oligodendrogenesis and Adaptive Myelination in the Mammalian Brain. Science. 2014;344(6183):1252304. 10.1126/science.1252304 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Zatorre RJ, Fields RD, Johansen-Berg H: Plasticity in gray and white: Neuroimaging changes in brain structure during learning. Nat Neurosci. 2012;15(4):528–36. 10.1038/nn.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scholz J, Klein MC, Behrens TE, et al. : Training induces changes in white-matter architecture. Nat Neurosci. 2009;12(11):1370–1. 10.1038/nn.2412 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Sampaio-Baptista C, Khrapitchev AA, Foxley S, et al. : Motor Skill Learning Induces Changes in White Matter Microstructure and Myelination. J Neurosci. 2013;33(50):19499–503. 10.1523/JNEUROSCI.3048-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. McKenzie IA, Ohayon D, Li H, et al. : Motor skill learning requires active central myelination. Science. 2014;346(6207):318–22. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Mangin JM, Li P, Scafidi J, et al. : Experience-dependent regulation of NG2 progenitors in the developing barrel cortex. Nat Neurosci. 2012;15(9):1192–4. 10.1038/nn.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bengtsson SL, Nagy Z, Skare S, et al. : Extensive piano practicing has regionally specific effects on white matter development. Nat Neurosci. 2005;8(9):1148–50. 10.1038/nn1516 [DOI] [PubMed] [Google Scholar]
- 29. Barrera K, Chu P, Abramowitz J, et al. : Organization of myelin in the mouse somatosensory barrel cortex and the effects of sensory deprivation. Dev Neurobiol. 2013;73(4):297–314. 10.1002/dneu.22060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makinodan M, Rosen KM, Ito S, et al. : A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science. 2012;337(6100):1357–60. 10.1126/science.1220845 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Liu J, Dietz K, DeLoyht JM, et al. : Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15(12):1621–3. 10.1038/nn.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Purger D, Gibson EM, Monje M: Myelin plasticity in the central nervous system. Neuropharmacology. 2016;110(Pt B):563–73. 10.1016/j.neuropharm.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 33. Monje M: Myelin Plasticity and Nervous System Function. Annu Rev Neurosci. 2018;41:61–76. 10.1146/annurev-neuro-080317-061853 [DOI] [PubMed] [Google Scholar]
- 34. Emery B, Lu QR: Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb Perspect Biol. 2015;7(9):a020461. 10.1101/cshperspect.a020461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun LO, Mulinyawe SB, Collins HY, et al. : Spatiotemporal Control of CNS Myelination by Oligodendrocyte Programmed Cell Death through the TFEB-PUMA Axis. Cell. 2018;175(7):1811–1826.e21. 10.1016/j.cell.2018.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Yu Y, Chen Y, Kim B, et al. : Olig2 Targets Chromatin Remodelers to Enhancers to Initiate Oligodendrocyte Differentiation. Cell. 2013;152(1–2):248–61. 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bischof M, Weider M, Küspert M, et al. : Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. J Neurosci. 2015;35(1):21–35. 10.1523/JNEUROSCI.1468-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doi T, Ogata T, Yamauchi J, et al. : Chd7 Collaborates with Sox2 to Regulate Activation of Oligodendrocyte Precursor Cells after Spinal Cord Injury. J Neurosci. 2017;37(43):10290–309. 10.1523/JNEUROSCI.1109-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Marie C, Clavairoly A, Frah M, et al. : Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc Natl Acad Sci U S A. 2018;115(35):E8246–E8255. 10.1073/pnas.1802620115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernier R, Golzio C, Xiong B, et al. : Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell. 2014;158(2):263–76. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Zhao C, Dong C, Frah M, et al. : Dual Requirement of CHD8 for Chromatin Landscape Establishment and Histone Methyltransferase Recruitment to Promote CNS Myelination and Repair. Dev Cell. 2018;45(6):753–768.e8. 10.1016/j.devcel.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Negri G, Milani D, Colapietro P, et al. : Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin Genet. 2015;87(2):148–54. 10.1111/cge.12348 [DOI] [PubMed] [Google Scholar]
- 43. Zimmermann N, Acosta AM, Kohlhase J, et al. : Confirmation of EP300 gene mutations as a rare cause of Rubinstein–Taybi syndrome. Eur J Hum Genet. 2007;15(8):837–42. 10.1038/sj.ejhg.5201791 [DOI] [PubMed] [Google Scholar]
- 44. Zhang L, He X, Liu L, et al. : Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev Cell. 2016;36(3):316–30. 10.1016/j.devcel.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elsesser O, Fröb F, Küspert M, et al. : Chromatin remodeler Ep400 ensures oligodendrocyte survival and is required for myelination in the vertebrate central nervous system. Nucleic Acids Res. 2019;47(2):6208–24. 10.1093/nar/gkz376 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Ye F, Chen Y, Hoang T, et al. : HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12(7):829–38. 10.1038/nn.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Zhao C, Deng Y, Liu L, et al. : Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. 2016;7:10883. 10.1038/ncomms10883 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Hammond E, Lang J, Maeda Y, et al. : The Wnt effector transcription factor 7-like 2 positively regulates oligodendrocyte differentiation in a manner independent of Wnt/β-catenin signaling. J Neurosci. 2015;35(12):5007–22. 10.1523/JNEUROSCI.4787-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dai J, Bercury KK, Jin W, et al. : Olig1 Acetylation and Nuclear Export Mediate Oligodendrocyte Development. J Neurosci. 2015;35(48):15875–93. 10.1523/JNEUROSCI.0882-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rafalski VA, Ho PP, Brett JO, et al. : Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol. 2013;15(6):614–24. 10.1038/ncb2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li W, Zhang B, Tang J, et al. : Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27(10):2606–16. 10.1523/JNEUROSCI.4181-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Noack M, Leyk J, Richter-Landsberg C: HDAC6 inhibition results in tau acetylation and modulates tau phosphorylation and degradation in oligodendrocytes. Glia. 2014;62(4):535–47. 10.1002/glia.22624 [DOI] [PubMed] [Google Scholar]
- 53. Liu H, Hu Q, D'ercole AJ, et al. : Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57(1):1–12. 10.1002/glia.20729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Douvaras P, Rusielewicz T, Kim KH, et al. : Epigenetic Modulation of Human Induced Pluripotent Stem Cell Differentiation to Oligodendrocytes. Int J Mol Sci. 2016;17(14): pii: E614. 10.3390/ijms17040614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Behrendt G, Baer K, Buffo A, et al. : Dynamic changes in myelin aberrations and oligodendrocyte generation in chronic amyloidosis in mice and men. Glia. 2013;61(2):273–86. 10.1002/glia.22432 [DOI] [PubMed] [Google Scholar]
- 56. He D, Wang J, Lu Y, et al. : lncRNA Functional Networks in Oligodendrocytes Reveal Stage-Specific Myelination Control by an lncOL1/Suz12 Complex in the CNS. Neuron. 2017;93(2):362–78. 10.1016/j.neuron.2016.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Sher F, Rössler R, Brouwer N, et al. : Differentiation of neural stem cells into oligodendrocytes: Involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875–83. 10.1634/stemcells.2008-0121 [DOI] [PubMed] [Google Scholar]
- 58. Hashimoto M, Murata K, Ishida J, et al. : Severe Hypomyelination and Developmental Defects Are Caused in Mice Lacking Protein Arginine Methyltransferase 1 (PRMT1) in the Central Nervous System. J Biol Chem. 2016;291(5):2237–45. 10.1074/jbc.M115.684514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scaglione A, Patzig J, Liang J, et al. : PRMT5-mediated regulation of developmental myelination. Nat Commun. 2018;9(1): 2840. 10.1038/s41467-018-04863-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Calabretta S, Vogel G, Yu Z, et al. : Loss of PRMT5 Promotes PDGFRα Degradation during Oligodendrocyte Differentiation and Myelination. Dev Cell. 2018;46(4):426–440.e5. 10.1016/j.devcel.2018.06.025 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Huang J, Vogel G, Yu Z, et al. : Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J Biol Chem. 2011;286(52):44424–32. 10.1074/jbc.M111.277046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moyon S, Huynh JL, Dutta D, et al. : Functional Characterization of DNA Methylation in the Oligodendrocyte Lineage. Cell Rep. 2016;15(4):748–60. 10.1016/j.celrep.2016.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao X, Dai J, Ma Y, et al. : Dynamics of ten-eleven translocation hydroxylase family proteins and 5-hydroxymethylcytosine in oligodendrocyte differentiation. Glia. 2014;62(6):914–26. 10.1002/glia.22649 [DOI] [PubMed] [Google Scholar]
- 64. Shin D, Shin JY, McManus MT, et al. : Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66(6):843–57. 10.1002/ana.21927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dugas JC, Cuellar TL, Scholze A, et al. : Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. 10.1016/j.neuron.2010.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Zhao X, He X, Han X, et al. : MicroRNA-Mediated Control of Oligodendrocyte Differentiation. Neuron. 2010;65(5):612–26. 10.1016/j.neuron.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang H, Moyano AL, Ma Z, et al. : miR-219 Cooperates with miR-338 in Myelination and Promotes Myelin Repair in the CNS. Dev Cell. 2017;40(6):566–582.e5. 10.1016/j.devcel.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang CY, Deneen B, Tzeng SF: MicroRNA-212 inhibits oligodendrocytes during maturation by down-regulation of differentiation-associated gene expression. J Neurochem. 2017;143(1):112–25. 10.1111/jnc.14138 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Lecca D, Marangon D, Coppolino GT, et al. : MiR-125a-3p timely inhibits oligodendroglial maturation and is pathologically up-regulated in human multiple sclerosis. Sci Rep. 2016;6:34503. 10.1038/srep34503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dong X, Chen K, Cuevas-Diaz Duran R, et al. : Comprehensive Identification of Long Non-coding RNAs in Purified Cell Types from the Brain Reveals Functional LncRNA in OPC Fate Determination. PLoS Genet. 2015;11(12):e1005669. 10.1371/journal.pgen.1005669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kasuga Y, Fudge AD, Zhang Y, et al. : Characterization of a long noncoding RNA Pcdh17it as a novel marker for immature premyelinating oligodendrocytes. Glia. 2019;67(11):2166–77. 10.1002/glia.23684 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Stolt CC, Schmitt S, Lommes P, et al. : Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281(2):309–17. 10.1016/j.ydbio.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 73. Mercer TR, Qureshi IA, Gokhan S, et al. : Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. 10.1186/1471-2202-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Katsel P, Roussos P, Fam P, et al. : The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 2019;5(1):1856. 10.1038/s41537-019-0071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Li Y, Guo B, Yang R, et al. : A novel long noncoding RNA lnc158 promotes the differentiation of mouse neural precursor cells into oligodendrocytes by targeting nuclear factor-IB. Neuroreport. 2018;29(13):1121–8. 10.1097/WNR.0000000000001083 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Xu H, Dzhashiashvili Y, Shah A, et al. : m 6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron. 2020;105(2):293–309.e5. 10.1016/j.neuron.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Wu R, Li A, Sun B, et al. : A novel m 6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29(1):23–41. 10.1038/s41422-018-0113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Allis CD, Jenuwein T: The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Wilson BG, Roberts CWM: SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–92. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- 80. Yoo AS, Crabtree GR: ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19(2):120–6. 10.1016/j.conb.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nielsen JA, Hudson LD, Armstrong RC: Nuclear organization in differentiating oligodendrocytes. J Cell Sci. 2002;115(Pt 21):4071–9. 10.1242/jcs.00103 [DOI] [PubMed] [Google Scholar]
- 82. Runge JS, Raab JR, Magnuson T: Epigenetic Regulation by ATP-Dependent Chromatin-Remodeling Enzymes: SNF-ing Out Crosstalk. Curr Top Dev Biol. 2016;117:1–13. 10.1016/bs.ctdb.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. de La Serna IL, Ohkawa Y, Imbalzano AN: Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–73. 10.1038/nrg1882 [DOI] [PubMed] [Google Scholar]
- 84. Matsumoto S, Banine F, Feistel K, et al. : Brg1 directly regulates Olig2 transcription and is required for oligodendrocyte progenitor cell specification. Dev Biol. 2016;413(12):173–87. 10.1016/j.ydbio.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Martin DM: Chromatin remodeling in development and disease: Focus on CHD7. PLoS Genet. 2010;6(7):e1001010. 10.1371/journal.pgen.1001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bergman JEH, Janssen N, Hoefsloot LH, et al. : CHD7 mutations and CHARGE syndrome: The clinical implications of an expanding phenotype. J Med Genet. 2011;48(5):334–42. 10.1136/jmg.2010.087106 [DOI] [PubMed] [Google Scholar]
- 87. Stolerman ES, Smith B, Chaubey A, et al. : CHD8 intragenic deletion associated with autism spectrum disorder. Eur J Med Genet. 2016;59(4):189–94. 10.1016/j.ejmg.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 88. Cotney J, Muhle RA, Sanders SJ, et al. : The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. 10.1038/ncomms7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Katayama Y, Nishiyama M, Shoji H, et al. : CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537(7622):675–9. 10.1038/nature19357 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 90. Piunti A, Shilatifard A: Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352(6290):aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- 91. Rasmussen PB, Staller P: The KDM5 family of histone demethylases as targets in oncology drug discovery. Epigenomics. 2014;6(3):277–86. 10.2217/epi.14.14 [DOI] [PubMed] [Google Scholar]
- 92. Moyon S, Casaccia P: DNA methylation in oligodendroglial cells during developmental myelination and in disease. Neurogenesis (Austin). 2017;4(1):e1270381. 10.1080/23262133.2016.1270381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu J, Magri L, Zhang F, et al. : Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J Neurosci. 2015;35(1):352–65. 10.1523/JNEUROSCI.2606-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Marmorstein R, Zhou MM: Writers and Readers of Histone Acetylation: Structure, Mechanism, and Inhibition. Cold Spring Harb Perspect Biol. 2014;6(7):a018762–a018762. 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Raisner R, Kharbanda S, Jin L, et al. : Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep. 2018;24(7):1722–9. 10.1016/j.celrep.2018.07.041 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Lee KK, Workman JL: Histone acetyltransferase complexes: One size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8(4):284–95. 10.1038/nrm2145 [DOI] [PubMed] [Google Scholar]
- 97. Roth SY, Denu JM, Allis CD: Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. 10.1146/annurev.biochem.70.1.81 [DOI] [PubMed] [Google Scholar]
- 98. Chiang DY, Kongchan N, Beavers DL, et al. : Loss of MicroRNA-106b-25 Cluster Promotes Atrial Fibrillation by Enhancing Ryanodine Receptor Type-2 Expression and Calcium Release. Circ Arrhythm Electrophysiol. 2014;7(6):1214–22. 10.1161/CIRCEP.114.001973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yun M, Wu J, Workman JL, et al. : Readers of histone modifications. Cell Res. 2011;21(4):564–78. 10.1038/cr.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gacias M, Casaccia P: Epigenetic mechanisms in multiple sclerosis. Rev Esp Escler Mult. 2014;6(29):25–35. [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang L, He X, Liu L, et al. : Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev Cell. 2016;37(6):582. 10.1016/j.devcel.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 102. Glaser KB: HDAC inhibitors: clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74(5):659–71. 10.1016/j.bcp.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 103. Thiagalingam S, Cheng KH, Lee HJ, et al. : Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. 10.1111/j.1749-6632.2003.tb05964.x [DOI] [PubMed] [Google Scholar]
- 104. Hsieh J, Nakashima K, Kuwabara T, et al. : Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101(47):16659–64. 10.1073/pnas.0407643101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marin-Husstege M, Muggironi M, Liu A, et al. : Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22(23):10333–45. 10.1523/JNEUROSCI.22-23-10333.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shen S, Li J, Casaccia-Bonnefil P: Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169(4):577–89. 10.1083/jcb.200412101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. He Y, Sandoval J, Casaccia-Bonnefil P: Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007;3(3):221–31. 10.1017/S1740925X08000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. He Y, Dupree J, Wang J, et al. : The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55(2):217–30. 10.1016/j.neuron.2007.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Seto E, Yoshida M: Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6(4):a018713–a018713. 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang X, Ozawa Y, Lee H, et al. : Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005;19(7):827–39. 10.1101/gad.1286005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhou J, Tien AC, Alberta JA, et al. : A Sequentially Priming Phosphorylation Cascade Activates the Gliomagenic Transcription Factor Olig2. Cell Rep. 2017;18(13):3167–77. 10.1016/j.celrep.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Guenther MG, Barak O, Lazar MA: The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21(18):6091–101. 10.1128/mcb.21.18.6091-6101.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hermanson O, Jepsen K, Rosenfeld MG: N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419(6910):934–9. 10.1038/nature01156 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Greer CB, Tanaka Y, Kim YJ, et al. : Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep. 2015;13(7):1444–55. 10.1016/j.celrep.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Jepsen K, Hermanson O, Onami TM, et al. : Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102(6):753–63. 10.1016/s0092-8674(00)00064-7 [DOI] [PubMed] [Google Scholar]
- 116. You SH, Lim HW, Sun Z, et al. : Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20(2):182–7. 10.1038/nsmb.2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fischle W, Dequiedt F, Hendzel MJ, et al. : Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9(1):45–57. 10.1016/s1097-2765(01)00429-4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 118. Stein LR, Imai S: Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014;33(12):1321–40. 10.1002/embj.201386917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dugas JC, Tai YC, Speed TP, et al. : Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26(43):10967–83. 10.1523/JNEUROSCI.2572-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Ji S, Doucette JR, Nazarali AJ: Sirt2 is a novel in vivo downstream target of Nkx2.2 and enhances oligodendroglial cell differentiation. J Mol Cell Biol. 2011;3(6):351–9. 10.1093/jmcb/mjr009 [DOI] [PubMed] [Google Scholar]
- 121. Thangaraj MP, Furber KL, Gan JK, et al. : RNA-binding Protein Quaking Stabilizes Sirt2 mRNA during Oligodendroglial Differentiation. J Biol Chem. 2017;292(13):5166–82. 10.1074/jbc.M117.775544 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 122. Zhu H, Zhao L, Wang E, et al. : The QKI-PLP pathway controls SIRT2 abundance in CNS myelin. Glia. 2012;60(1):69–82. 10.1002/glia.21248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Haigis MC, Sinclair DA: Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang P, Xu TY, Guan YF, et al. : Nicotinamide phosphoribosyltransferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Ann Neurol. 2011;69(2):360–74. 10.1002/ana.22236 [DOI] [PubMed] [Google Scholar]
- 125. Jablonska B, Gierdalski M, Chew LJ, et al. : Sirt1 regulates glial progenitor proliferation and regeneration in white matter after neonatal brain injury. Nat Commun. 2016;7:13866. 10.1038/ncomms13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Werner HB, Kuhlmann K, Shen S, et al. : Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27(29):7717–30. 10.1523/JNEUROSCI.1254-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Heintzman ND, Stuart RK, Hon G, et al. : Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 128. Schneider J, Wood A, Lee JS, et al. : Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19(6):849–56. 10.1016/j.molcel.2005.07.024 [DOI] [PubMed] [Google Scholar]
- 129. Miller T, Krogan NJ, Dover J, et al. : COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98(23):12902–7. 10.1073/pnas.231473398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mohn F, Weber M, Rebhan M, et al. : Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–66. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 131. Mikkelsen TS, Ku M, Jaffe DB, et al. : Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 132. Cao R, Wang L, Wang H, et al. : Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–43. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 133. Laugesen A, Højfeldt JW, Helin K: Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell. 2019;74(1):8–18. 10.1016/j.molcel.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yu JR, Lee CH, Oksuz O, et al. : PRC2 is high maintenance. Genes Dev. 2019;33(15–16):903–35. 10.1101/gad.325050.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ai S, Peng Y, Li C, et al. : EED orchestration of heart maturation through interaction with HDACs is H3K27me3-independent. eLife. 2017;6: pii: e24570. 10.7554/eLife.24570 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 136. Margueron R, Li G, Sarma K, et al. : Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol Cell. 2008;32(4):503–18. 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang M, Wang Y, Jones S, et al. : Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46(11):1170–2. 10.1038/ng.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 138. Lewis PW, Muller MM, Koletsky MS, et al. : Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013;340(6134):857–61. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 139. Bender S, Tang Y, Lindroth AM, et al. : Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell. 2013;24(5):660–72. 10.1016/j.ccr.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 140. Larson JD, Kasper LH, Paugh BS, et al. : Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell. 2019;35(1):140–155.e7. 10.1016/j.ccell.2018.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 141. Kim KH, Roberts CW: Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–34. 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Filbin MG, Tirosh I, Hovestadt V, et al. : Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–5. 10.1126/science.aao4750 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 143. Weng Q, Wang J, Wang J, et al. : Single-Cell Transcriptomics Uncovers Glial Progenitor Diversity and Cell Fate Determinants during Development and Gliomagenesis. Cell Stem Cell. 2019;24(5):707–723.e8. 10.1016/j.stem.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 144. Bedford MT: Arginine methylation at a glance. J Cell Sci. 2007;120(Pt 24):4243–6. 10.1242/jcs.019885 [DOI] [PubMed] [Google Scholar]
- 145. Saxonov S, Berg P, Brutlag DL: A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103(5):1412–7. 10.1073/pnas.0510310103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kohli RM, Zhang Y: TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Moyon S, Ma D, Huynh JL, et al. : Efficient Remyelination Requires DNA Methylation. eNeuro. 2017;4(2): pii: ENEURO.0336-16.2017. 10.1523/ENEURO.0336-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 148. Moyon S, Frawley R, Marshall-Phelps KL, et al. : TET1-mediated DNA hydroxy-methylation regulates adult remyelination. bioRxiv. 2019; 819995. 10.1101/819995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhang M, Wang J, Zhang K, et al. : TET1-mediated Oligodendrocyte Homeostasis Regulates Myelination and Synaptic Functions. bioRxiv. 2019; 821496. 10.1101/821496 [DOI] [Google Scholar]
- 150. Lau P, Verrier JD, Nielsen JA, et al. : Identification of Dynamically Regulated MicroRNA and mRNA Networks in Developing Oligodendrocytes. J Neurosci. 2008;28(45):11720–30. 10.1523/JNEUROSCI.1932-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 151. Hudish LI, Blasky AJ, Appel B: miR-219 Regulates Neural Precursor Differentiation by Direct Inhibition of Apical Par Polarity Proteins. Dev Cell. 2013;27(4):387–98. 10.1016/j.devcel.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 152. Galloway DA, Moore CS: miRNAs As Emerging Regulators of Oligodendrocyte Development and Differentiation. Front Cell Dev Biol. 2016;4:59. 10.3389/fcell.2016.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Tripathi A, Volsko C, Garcia JP, et al. : Oligodendrocyte Intrinsic miR-27a Controls Myelination and Remyelination. Cell Rep. 2019;29(4):904–919.e9. 10.1016/j.celrep.2019.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 154. Guttman M, Amit I, Garber M, et al. : Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 155. Batista PJ, Chang HY: Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Fatica A, Bozzoni I: Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 157. Bhan A, Mandal SS: Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9(9):1932–56. 10.1002/cmdc.201300534 [DOI] [PubMed] [Google Scholar]
- 158. Lalevée S, Feil R: Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7(6):877–9. 10.2217/epi.15.55 [DOI] [PubMed] [Google Scholar]
- 159. Liu J, Yue Y, Han D, et al. : A METTL3-METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Fu Y, Dominissini D, Rechavi G, et al. : Gene expression regulation mediated through reversible m 6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 161. Ji P, Wang X, Xie N, et al. : N6-Methyladenosine in RNA and DNA: An Epitranscriptomic and Epigenetic Player Implicated in Determination of Stem Cell Fate. Stem Cells Int. 2018;2018: 3256524. 10.1155/2018/3256524 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 162. Tse KH, Cheng A, Ma F, et al. : DNA damage-associated oligodendrocyte degeneration precedes amyloid pathology and contributes to Alzheimer's disease and dementia. Alzheimers Dement. 2018;14(5):664–79. 10.1016/j.jalz.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 163. Mathys H, Davila-Velderrain J, Peng Z, et al. : Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570(7761):332–7. 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation