Abstract
Conventional wisdom holds that PCR amplification for sequencing should employ pooled replicate reactions to reduce bias due to jackpot effects and chimera formation. However, modern amplicon data analysis employs methods that may be less sensitive to such artifacts. Here we directly compare results from single vs. triplicate reactions for 16S amplicon sequencing and find no significant impact of adopting a less labor-intensive single reaction protocol.
Keywords: 16S rRNA gene amplicon sequencing, PCR, microbiome , replicate PCR reactions
Method Summary
We compared single PCR reactions to pooled triplicate PCR reactions for 16S rRNA gene amplicon sequencing on nearly 400 samples from a diverse range of environments across three independent laboratories.
For decades, 16S rRNA gene sequencing has been performed by pooling replicate PCR reactions, usually in triplicate. The primary benefit is to reduce “jackpotting”: the stochastic nature of PCR means that some molecules are amplified earlier than others, and exponential amplification in subsequent rounds of PCR substantially distort the frequencies of different molecules in heterogeneous pools of target genes[1]. This phenomenon is particularly important in environmental DNA sequencing where the goal is an accurate, or at least consistent, readout of the different gene targets matching a primer set.
However, since the guideline that PCR should be performed in triplicate was introduced[1], there have been substantial improvements in the processivity and fidelity of DNA polymerases. Therefore, triplicate PCR may no longer provide the benefits it once did, although performing single PCR reactions instead of triplicate would provide significant time and cost savings. Several studies have tested single versus triplicate PCR for 16S rRNA sequencing in limited settings with a small number of input samples (e.g. 18 soil samples[2], 2 soil and 2 stool samples[3], 3 soil samples[4]). However, it has never been tested across the wide range of samples and settings that would be needed to justify a general recommendation for change in protocol. We used the availability of standardized sample sets such as those from MBQC, the Microbiome Quality Control project[5], and from our previous technology testing to answer this question definitively across three different laboratories. In total, we tested the effects of replicate PCR pooling in 3 independent experiments containing nearly 373 samples from a diverse range of environments.
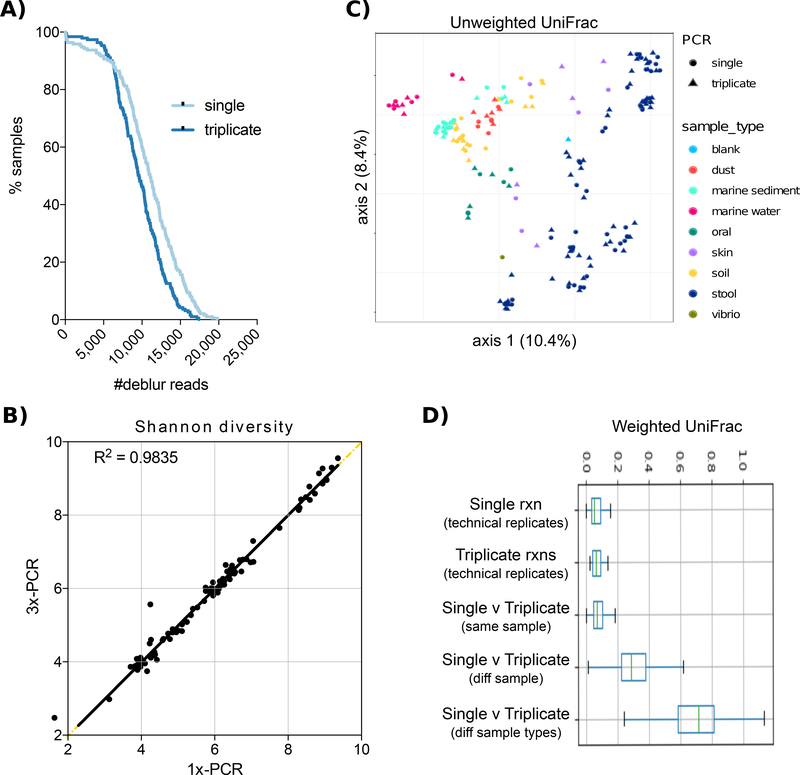
First, we benchmarked single versus pooled-triplicate PCR across a broad range of sample types. In our previous study on comparison of DNA extraction methods[6] we assembled a set of 96 samples spanning a broad range of environments, including 48 fecal samples, 12 soil samples, 12 marine sediment samples, 6 seawater samples, 5 skin samples, 5 oral samples, and 6 mattress dust samples. We used the DNA from this previous study, extracted using the Earth Microbiome Project protocol[7] on the Kingfisher instrument, for this study. 16S rRNA gene amplification was performed according to the Earth Microbiome Project (EMP) protocol and is detailed in the supplemental file. We quantified amplicons by PicoGreen™ and pooled 240 ng of each for sequencing. We ran the entire sample set four times: twice with single PCR and twice with pooled-triplicate PCR. The pooled library was sequenced on the Illumina MiSeq sequencing platform with a MiSeq Reagent Kit v2 and paired-end 150 cycles. All data were processed and analyzed using the QIIME2 software suite[8] and Deblur[9]. Counterintuitively, single PCR reactions yielded significantly more reads than triplicate PCR reactions (mean ± SEM: 10,821 ± 298 versus 10,029 ± 262, respectively, paired T-test p=0.0003), and fewer dropouts (Fig. 1A). We saw no significant difference in alpha diversity, regardless of environment (Fig. 1B). Beta diversity analysis with Unweighted UniFrac demonstrates that samples cluster by sample type and not number of PCR reactions (Fig. 1C). The Weighted UniFrac distances are significantly larger among samples from different environments than among biological replicates, and distances among biological replicates are significantly greater than technical replicates, with both single and triplicate PCR reactions (Fig. 1D). Negligible taxonomic changes between single and triplicate reactions were observed (97.8% shared taxonomy at the species level, genus 98.4%, and phylum 100%, Fig. S1A and Fig. S2).
Figure 1. Effect of 16S PCR reaction number across a broad range of sample types.
A) The sequencing dropout rate of all samples run with either single or triplicate PCR reactions. B) Shannon diversity index is nearly identical between single and triplicate PCR reactions of the same sample. C) Unweighted UniFrac PCoA plot shows that samples cluster by sample type (color) and not number of PCR reactions (shape). D) Weighted UniFrac distance among technical replicates (same sample) run with either single or triplicate PCR reactions are smaller than the distance between samples of the same type (diff samples) or among samples from different environments (types).
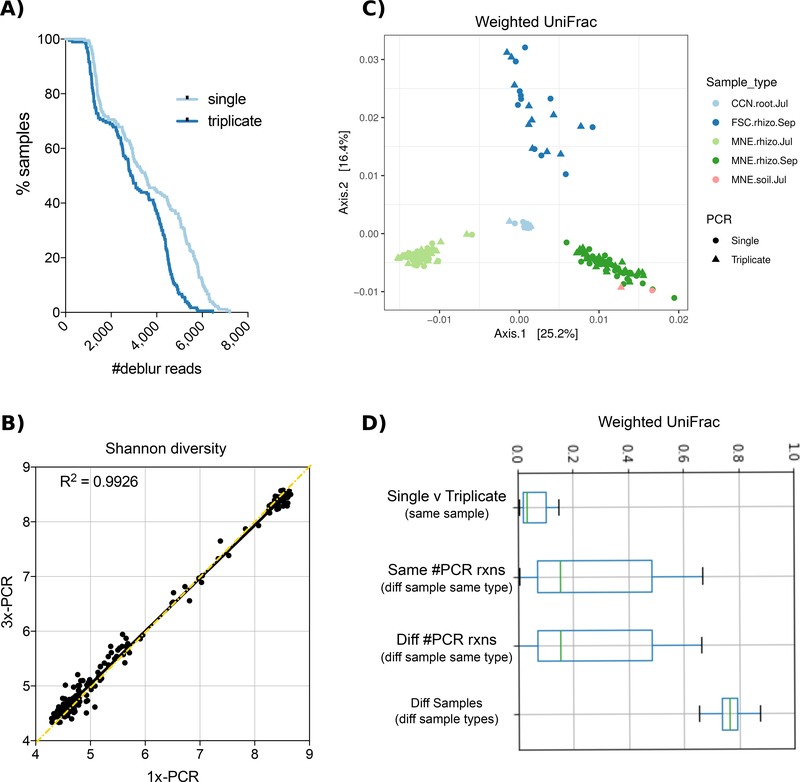
Second, because high-level conclusions crossing environment types might obscure relationships in particular sample types, we tested whether the conclusions held for a separate set of agricultural samples. We sampled root and rhizosphere samples from 3 different sites across 2 seasons. A variety of roots including crown, seminal, and primary roots were excavated and shaken for 1–2 min in 35 mL phosphate buffer and maintained on ice. In the laboratory, roots were surface sterilized by rinsing 30 seconds in 5.25% sodium hypochlorite + 0.01% Tween 20, followed by a 30 seconds rinse in 70% ethanol, followed by three rinses in sterile ultrapure water. Roots were blotted dry on a clean paper towel, placed in a 15 mL tube, frozen at −80° C and then ground in liquid nitrogen prior to DNA extraction. The rhizosphere samples were filtered through a sterile 100 μm mesh filter, pelleted at 3000 x g for 10 minutes, washed with 1.5 mL phosphate buffer, and re-pelleted by spinning for 5 minutes at full speed. The supernatant was drained off and the rhizosphere soil pellet was stored at −80° C until DNA extraction. DNA was extracted from soil, rhizosphere, and root samples using DNeasy PowerSoil HTP 96 Kit and quantified with the Quantifluor dsDNA reagent. Each sample was amplified both with a single PCR reaction and with pooled-triplicate reactions. The single PCR reactions yielded significantly more reads than triplicate PCR reactions (mean ± SEM: 3,631 ± 139 versus 3,000 ± 113, respectively, paired T-test p<0.0001), but had a similar dropout rate (Fig. 2A). Alpha diversity was not significantly different with single versus triplicate PCR (Fig. 2B), and as with the cross-environment comparison shown in Fig. 1, Weighted UniFrac analysis shows that the primary clustering is by sample type and the distances among samples does not differ in single versus triplicate PCRs (Fig. 2C,D). Negligible taxonomic changes between single and triplicate reactions were observed (99.3% shared taxonomy at the species level, genus 99.2%, and phylum 100%, Fig. S1B and Fig. S3).
Figure 2. Effect of 16S PCR reaction number across agricultural samples.
A). The sequencing dropout number of all samples run with either single or triplicate PCR reactions. B) Shannon diversity index of each sample is similar between single and triplicate PCR reactions. C) Weighted UniFrac PCoA plot shows that samples cluster by sample type (color) and not number of PCR reactions (shape). D) Weighted Unifrac distances between single or triplicate PCR reactions of the same sample are smaller than the distance between different samples of the same type run with either single or triplicate PCR reactions, and both are smaller than the distance between samples from different environments.
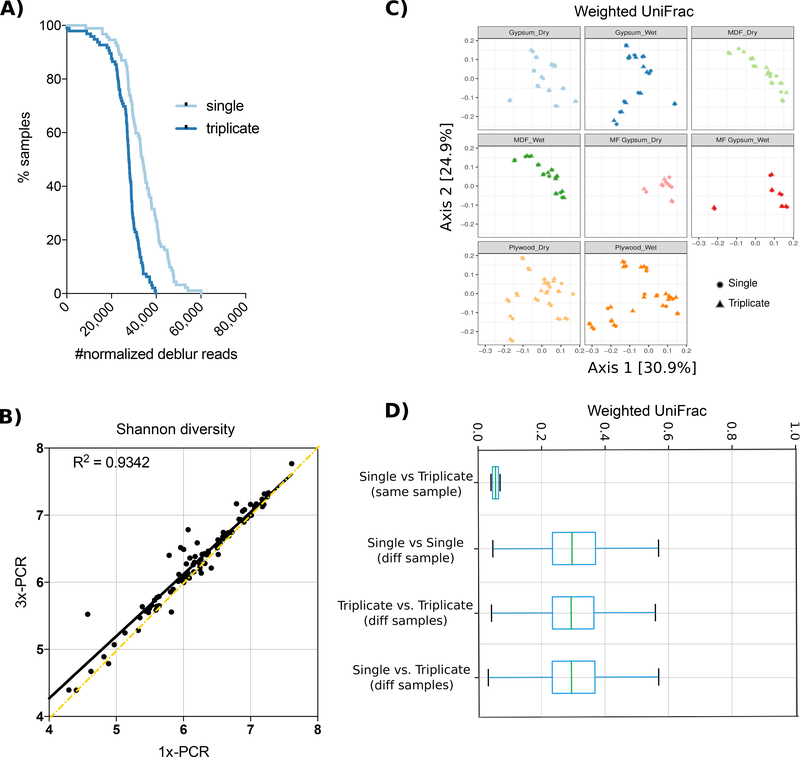
Finally, the microbiology of the built environment has been a rapidly expanding topic of interest over the past decade, but poses unique challenges for molecular analysis. In particular, samples tend to be contaminated with high levels of human DNA and have low bacterial biomass[10]. We used samples from a previous study that collected 96 samples longitudinally from four commonly used building materials maintained at a high relative humidity (~94%)[11]. Genomic DNA was extracted from environmental samples using the PowerSoil DNA isolation kit as previously described[12], and genomic DNA was amplified using the EMP protocol as detailed in the supplemental file. Samples were processed both with single PCR and pooled-triplicate PCR reactions, and sequenced on an Illumina MiSeq sequencing platform with a MiSeq Reagent Kit v2 and paired-end 150 cycles. Once again, yields were higher with single PCR than triplicate PCR (Fig. 3A), Shannon diversity was not affected by single versus triplicate PCR (Fig. 3B), and beta diversity was driven by biological parameters of the sample rather than by single versus triplicate PCR (Fig. 3C,D). Negligible taxonomic changes between single and triplicate reactions were observed (96.5% shared taxonomy at the species level, genus 95.8%, and phylum 100%, Fig. S1C and Fig. S4). All data from each of the three experiments are publicly available from the EBI under accession number ERP113817.
Figure 3. Effect of 16S PCR reaction number across building materials.
A) The sequencing dropout number of all samples run with either single or triplicate PCR reactions. B) Shannon diversity index is similar between single and triplicate PCR reactions. C) Weighted UniFrac PCoA plot shows that samples do not cluster by number of PCR reactions (shape). D) Weighted UniFrac distances between single or triplicate PCR reactions of the same sample are smaller than the distance between different samples with either single or triplicate PCR reactions.
Taken together, these results demonstrate that with modern methods pooling triplicate PCR reactions for 16S rRNA amplicon sequencing is more expensive and does not provide improvement over single PCR reactions. This result was confirmed in studies spanning three laboratories, hundreds of samples, and numerous distinct environment types. However, although these results hold true for the range of conditions tested here, there are so many variations in PCR techniques that this type of benchmarking effort should be validated for specific sample types and PCR protocols before a switch from established procedure is implemented for specialized protocols. For the general sample types tested here, we recommend using single PCRs rather than triplicate PCRs. Combined with other technical improvements in miniaturizing PCR reactions[13], this change in protocol will substantially reduce the cost and complexity of amplicon studies.
Supplementary Material
Figure S1. Taxonomic relative abundance profiles comparing single vs triplicate PCR reactions.
Bar charts comparing genera with greater than 1% relative abundance between single and triplicate reactions across a broad range of sample types (A), agricultural samples (B), and built environment samples (C).
Figure S2. Relative abundance between single and triplicate PCR is not biased by GC content across a broad range of sample types.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.
Figure S3. Relative abundance between single and triplicate PCR is not biased by GC content in agricultural samples.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.
Figure S4. Relative abundance between single and triplicate PCR is not biased by GC content in built environment samples.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.
Funding Acknowledgement
Data from JGI was collected with support from a grant from the Office of Science (BER), U.S. Department of Energy, Grant no DE-SC0014395 to Daniel Schachtman and Susannah Tringe. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231.
References
- 1.Polz MF, Cavanaugh CM. Bias in Template-to-Product Ratios in Multitemplate PCR [Internet]. Available from: http://aem.asm.org/. [DOI] [PMC free article] [PubMed]
- 2.Wen C, Wu L, Qin Y, et al. Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PLoS One [Internet]. 12(4), e0176716 (2017). Available from: https://dx.plos.org/10.1371/journal.pone.0176716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy K, Hall MW, Lynch MDJ, Moreno-Hagelsieb G, Neufeld JD. Evaluating Bias of Illumina-Based Bacterial 16S rRNA Gene Profiles. (2014). Available from: http://dx.doi.org/10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DP, Peay KG. Sequence Depth, Not PCR Replication, Improves Ecological Inference from Next Generation DNA Sequencing. PLoS One [Internet]. 9(2), e90234 (2014). Available from: https://dx.plos.org/10.1371/journal.pone.0090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha R, Abu-Ali G, Vogtmann E, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol. [Internet]. 35(11), 1077 (2017). Available from: http://www.nature.com/doifinder/10.1038/nbt.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, Knight R. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 62(6), 290–293 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Gilbert J, Jansson J, Knight R. The Earth Microbiome Project. (2010). Available from: http://www.earthmicrobiome.org/
- 8.Bolyen E, Rideout JR, Dillon MR, et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. (2018). Available from: https://peerj.com/preprints/27295/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir A, McDonald D, Navas-Molina JA, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems [Internet]. 2(2), e00191–16 (2017). Available from: https://msystems.asm.org/content/2/2/e00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley ST, Gilbert JA. Studying the microbiology of the indoor environment. Genome Biol. [Internet]. 14(2), 202 (2013). Available from: http://genomebiology.biomedcentral.com/articles/10.1186/gb-2013-14-2-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardona C, Lax S, Larsen P, et al. Environmental Sources of Bacteria Differentially Influence Host-Associated Microbial Dynamics. (2018). Available from: http://msystems.asm.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPherson MR, Wang P, Marsh EL, Mitchell RB, Schachtman DP. Isolation and Analysis of Microbial Communities in Soil, Rhizosphere, and Roots in Perennial Grass Experiments. J. Vis. Exp. [Internet]. (137) (2018). Available from: http://www.ncbi.nlm.nih.gov/pubmed/30102263. [DOI] [PMC free article] [PubMed]
- 13.Minich JJ, Humphrey G, Benitez RAS, et al. High-Throughput Miniaturized 16S rRNA Amplicon Library Preparation Reduces Costs while Preserving Microbiome Integrity. mSystems [Internet]. 3(6), e00166–18 (2018). Available from: http://msystems.asm.org/lookup/doi/10.1128/mSystems.00166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Taxonomic relative abundance profiles comparing single vs triplicate PCR reactions.
Bar charts comparing genera with greater than 1% relative abundance between single and triplicate reactions across a broad range of sample types (A), agricultural samples (B), and built environment samples (C).
Figure S2. Relative abundance between single and triplicate PCR is not biased by GC content across a broad range of sample types.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.
Figure S3. Relative abundance between single and triplicate PCR is not biased by GC content in agricultural samples.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.
Figure S4. Relative abundance between single and triplicate PCR is not biased by GC content in built environment samples.
Taxa were ranked according to their % GC content, and the highest 10% of genera (A) and lowest 10% of genera (B) are plotted to compare single vs triplicate PCR reactions.