Abstract
Analysis of gene expression can be challenging, especially if it involves genetically diverse populations that exhibit high variation in their individual expression profile. Despite this variation, it is conceivable that in the same individuals a high degree of coordination is maintained between transcripts that belong to the same signaling modules and are associated with related biological functions. To explore this further, we calculated the correlation in the expression levels between each of ATF4, CHOP (DDIT3), GRP94, DNAJB9 (ERdj4), DNAJ3C (P58IPK), and HSPA5 (BiP/GRP78) with the whole transcriptome in primary fibroblasts from deer mice following induction of endoplasmic reticulum (ER) stress. Since these genes are associated with different transducers of the unfolded protein response (UPR), we postulated that their profile, in terms of correlation of transcripts, reflects distinct UPR branches engaged, and therefore different biological processes. Standard gene ontology analysis was able to predict major functions associated with the corresponding transcript, and of the UPR arm related to that, namely regulation of the apoptotic response by ATF4 (PERK arm) and the ER stress-associated degradation for GRP94 (IRE1). BiP, being a global regulator of the UPR, was associated with activation of ER stress in a rather global manner. Pairwise comparison in the correlation coefficients for these genes' associated transcriptome showed the relevance of selected genes in terms of expression profiles. Conventional assessment of differential gene expression was incapable of providing meaningful information and pointed only to a generic association with stress. Collectively, this approach suggests that by evaluating the degree of coordination in gene expression, in genetically diverse biological specimens, may be useful in assigning genes in transcriptome networks, and more importantly in linking signaling nodules to specific biological functions and processes.
Keywords: chaperone, Peromyscus, ER stress
Introduction
Signaling networks respond to stimuli by modulating the expression of a multitude of genes. This activation proceeds by high quantitative variation and for certain transcripts, it can be high, while for others it can be minimal, yet both highly and marginally regulated transcripts are equally important for the production of the desired cellular response. This variation in the magnitude of the response increases if biological samples from genetically diverse populations are analyzed. Correlation clustering had been widely used for the discovery that the group of genes show similar expression pattern under different conditions (Ben-Dor et al., 1999; Ng et al., 2006). Such approaches have been used to define population structures and to identify variation in expression profiles between different groups (Wei et al., 2011; Brown et al., 2018), or to reveal disease-related genes (Cai et al., 2017; Tai et al., 2018).
The utilization of genetically diverse organisms allows applying such analysis to experiments performed on the same conditions where the diversity of the animals is projected to the diversity of the output evaluated. Despite the variation in the intensity of the response among individuals, it is conceivable that a high degree of coordination is maintained between targets that belong to the same signaling cascades (Komili and Silver, 2008). To that end, it is conceivable that co-regulated transcripts, due to their participation in the same networks, will exhibit higher coordination than with those associated with other signaling modules, and that identification and analysis of co-regulated transcripts may convey information regarding the function regulated by the corresponding signaling module.
Furthermore, by focusing on the degree of coordination, as opposed to fold induction, even minimal, although impactful, differences in gene expression should be unveiled and appreciated. In this study, we sought to exploit these hypotheses by testing if the degree of coordination in gene expression, rather than the magnitude of overexpression, bears information more valuable in assessing signal integration and biological function.
As a model for the transcriptional analyses, we used the induction of endoplasmic reticulum (ER) stress following exposure of fibroblasts to tunicamycin. ER stress is defined as the state of the cells at which protein production exceeds the capacity for protein folding and therefore results in the accumulation of misfolded and unfolded proteins (Walter and Ron, 2011; Han and Kaufman, 2017; Almanza et al., 2018). ER stress, for its resolution, inflicts the unfolded protein responses (UPRs), which is mediated by three major transducers, IRE1, ATF6 and PERK, each of which results in the activation of well-defined downstream targets (Szegezdi et al., 2006; Fu and Gao, 2014; Lemus and Goder, 2014; Hetz et al., 2015). Despite certain redundancies in the regulation of the downstream mediators, the activities and the molecular determinants of the corresponding three branches of UPR remain well defined, representing an appropriate system to study the integration of signals associated with different transcriptional nodules into an overall cascade of a well-orchestrated response.
The analyses were performed in primary cultures of fibroblasts from genetically diverse (outbred) Peromyscus (deer mice) (Havighorst et al., 2017). Recently, we reported that major UPR targets such as chaperones BiP, GRP94 and calnexin are highly coordinated in fibroblasts from outbred deer mice (P. maniculatus) (Havighorst et al., 2019). The utilization of an outbred species for this analysis allows assessment of the variation in gene expression in the context of the naturally existing diversity and within what should be considered physiological range.
Materials and Methods
Animals
Deer mouse, P. maniculatus bairdii (BW Stock), was closed colony bred in captivity since 1948 and descended from 40 ancestors wild-caught near Ann Arbor, Michigan. Sonoran deer mouse, P. maniculatus sonoriensis (SM2 Stock), was derived from about 50 animals wild-caught by Jack Hayes in 1995 near White Mountain Research Station, CA (Havighorst et al., 2017). In this study, we picked three outbred BWs, including two males and one female. They were 4-week old at weaning. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and the Department of Health and Human Services, Office of Laboratory Animal Welfare, University of South Carolina (Approval No. 2349-101211-041917).
Cell culture
Fresh ear punches were collected from deer mice during routine weaning and marking procedures. Ear punches were washed for 2 min in 70% ethanol and moved to Roswell Park Memorial Institute (RPMI)-1640 medium (HyClone) supplemented with 10% FBS (Gibco), 500 U/ml penicillin, 500 μl/ml streptomycin and 0.292 mg/ml L-glutamine (complete RPMI). Ear punches were minced into small pieces and then digested by collagenase I (Sigma, 2 mg/ml in RPMI-1640) for 1 h. Tissue debris from digested ear punches was removed once cells were visible. Cells were cultivated in complete RPMI at 37°C in a humidified incubator containing 5% CO2, and passaged when cells were at 90% confluency or above to 45% confluency or above. Cells were passaged no more than seven times before tunicamycin treatment. For tunicamycin treatment, cells were split into 6-well plates, 300,000 cells/well, and cultivated for 24 h. Then cells were treated with tunicamycin (5 μg/mL) for 5 h, immediately followed by RNA extraction. The analysis was performed in three pairs of fibroblasts, all obtained from different animals that resulted in six datasets (three treated and three untreated).
RNA sequencing
RNA and library preparation, sequencing, and postprocessing of the raw data and data analysis were performed by the USC CTT COBRE Functional Genomics Core. RNAs were extracted with a Qiagen RNeasy Plus Mini kit as per manufacturer's recommendations (Qiagen, Valencia, CA). RNA integrity was assessed using the Agilent Bioanalyzer and samples had a quality score >8.0. RNA libraries were prepared using established protocol with NEBNExt Ultra II Directional Library Prep Kit for Illumina (NEB, Lynn, MA). Each library was made with one of the TruSeq barcode index sequences and samples were sequenced across three lanes. The pools were clustered at 6.5 pM on a pair end read flow cell and sequenced for 300 cycles on an Illumina NextSeq.
Sequences were aligned to the P. maniculatus genome using STAR v2.6.1 (Dobin et al., 2013). Reads were counted using the featureCounts function of the Subreads package (Liao et al., 2013) in R using Gencode M6 GTF and summarized at exon, transcript, or gene level. Only reads that were mapped uniquely to the genome were used. Mapping quality minimum threshold was set at 10. Differential expression analysis was performed in R using the edgeR package (Robinson et al., 2010). The average read depth for the samples was 48 million reads and only genes with at least one count per million average depth were considered for differential expression analysis.
Raw counts were normalized using the trimmed mean of M values (TMM) method. Dispersion estimates were then calculated using the estimateGLMRobustDisp function (Zhou et al., 2008). The normalized read counts were then fitted to a generalized linear model using the function glmFit (McCarthy et al., 2012). Genewise tests for significant differential expression were performed using the function glmLRT. The p-value was then corrected for multiple testing using Benjamini-Hochberg's FDR (Benjamini and Hochberg, 1995). The results have been deposited in GEO (GSE131429).
Correlation clustering
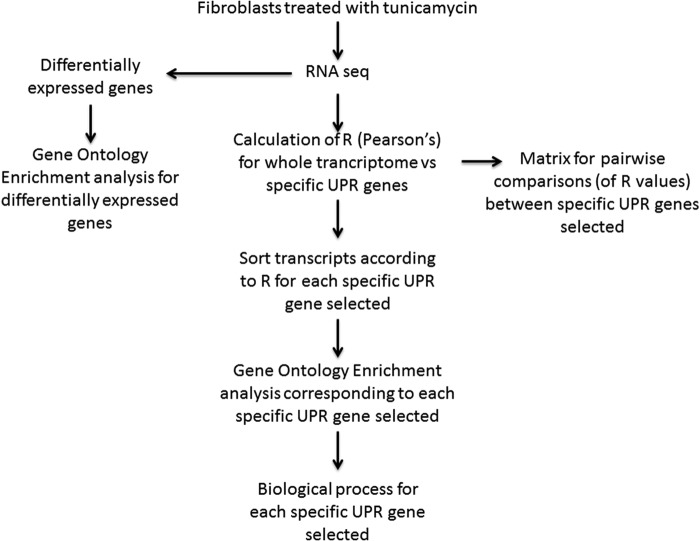
The Pearson's correlation was calculated between the whole transcriptome as obtained by the RNAseq analysis, and the UPR-associated transcripts indicated, by using Excel. Subsequently, all transcripts were sorted according to their R value with the given UPR genes and all exhibiting p < 0.05 (Pearson's) were introduced in the Gene Ontology Enrichment analysis. Both transcripts showing positive correlation only (Table 1) or positive and negative correlation (Supplementary Table S1) were analyzed. The process and analysis followed are shown in Figure 1.
Table 1.
Gene Ontology Enrichment Analysis for All Transcripts That Exhibited Positively Correlated Expression (p < 0.05, Pearson's) with Each of ATF4, CHOP, GRP94, BiP, ERdj4, and P58IPK (Top 6) or Were Upregulated According to Standard Fold Induction Analysis (Bottom)
| GO term | Description | p | FDR q-value | Enrichment (N, B, n, b) |
|---|---|---|---|---|
| Correlation with ATF4; 1198 genes with Pearson's p < 0.05 | ||||
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 2.46E-6 | 1.41E-2 | 23.19 (1002, 8, 27, 5) |
| GO:0043555 | Regulation of translation in response to stress | 2.76E-6 | 7.92E-3 | 39.29 (1002, 6, 17, 4) |
| GO:0048585 | Negative regulation of response to stimulus | 4.37E-6 | 8.36E-3 | 2.90 (1002, 93, 93, 25) |
| GO:1903573 | Negative regulation of response to ER stress | 1.01E-5 | 1.45E-2 | 9.87 (1002, 7, 87, 6) |
| GO:0032056 | Positive regulation of translation in response to stress | 1.22E-5 | 1.4E-2 | 58.94 (1002, 3, 17, 3) |
| GO:1900102 | Negative regulation of ER UPR | 1.35E-5 | 1.29E-2 | 18.56 (1002, 4, 54, 4) |
| GO:0097190 | Apoptotic signaling pathway | 1.49E-5 | 1.22E-2 | 10.82 (1002, 24, 27, 7) |
| GO:0006446 | Regulation of translational initiation | 1.98E-5 | 1.42E-2 | 29.47 (1002, 8, 17, 4) |
| GO:0034976 | Response to ER stress | 2.12E-5 | 1.35E-2 | 7.26 (1002, 46, 27, 9) |
| GO:0043558 | Regulation of translational initiation in response to stress | 3.94E-5 | 2.26E-2 | 44.21 (1002, 4, 17, 3) |
| GO:0045948 | Positive regulation of translational initiation | 3.94E-5 | 2.06E-2 | 44.21 (1002, 4, 17, 3) |
| GO:0097193 | Intrinsic apoptotic signaling pathway | 4.95E-5 | 2.37E-2 | 11.72 (1002, 19, 27, 6) |
| GO:0009968 | Negative regulation of signal transduction | 5.92E-5 | 2.61E-2 | 2.86 (1002, 79, 93, 21) |
| GO:0036493 | Positive regulation of translation in response to ER stress | 1.12E-4 | 4.58E-2 | 125.25 (1002, 2, 8, 2) |
| GO:1903912 | Negative regulation of ER stress-induced eIF2 alpha phosphorylation | 1.12E-4 | 4.27E-2 | 125.25 (1002, 2, 8, 2) |
| GO:0010648 | Negative regulation of cell communication | 1.15E-4 | 4.14E-2 | 2.76 (1002, 82, 93, 21) |
| GO:0080135 | Regulation of cellular response to stress | 1.24E-4 | 4.2E-2 | 5.96 (1002, 42, 36, 9) |
| GO:1903897 | Regulation of PERK-mediated UPR | 1.43E-4 | 4.56E-2 | 28.90 (1002, 4, 26, 3) |
| GO:0023057 | Negative regulation of signaling | 1.43E-4 | 4.32E-2 | 2.73 (1002, 83, 93, 21) |
| GO:0042493 | Response to drug | 1.9E-4 | 5.47E-2 | 5.69 (1002, 44, 36, 9) |
| GO:0007165 | Signal transduction | 3.18E-4 | 8.68E-2 | 1.91 (1002, 209, 93, 37) |
| GO:0036499 | PERK-mediated UPR | 3.22E-4 | 8.39E-2 | 8.52 (1002, 7, 84, 5) |
| GO:0032269 | Negative regulation of cellular protein metabolic process | 3.31E-4 | 8.26E-2 | 8.71 (1002, 69, 10, 6) |
| GO:0051248 | Negative regulation of protein metabolic process | 4.04E-4 | 9.66E-2 | 8.47 (1002, 71, 10, 6) |
| GO:1903898 | Negative regulation of PERK-mediated UPR | 5.42E-4 | 1.25E-1 | 58.94 (1002, 2, 17, 2) |
| GO:0032058 | Positive regulation of translational initiation in response to stress | 5.42E-4 | 1.2E-1 | 58.94 (1002, 2, 17, 2) |
| GO:0030968 | ER UPR | 6.76E-4 | 1.44E-1 | 4.51 (1002, 23, 87, 9) |
| GO:2001233 | Regulation of apoptotic signaling pathway | 6.93E-4 | 1.42E-1 | 1.28 (1002, 40, 783, 40) |
| GO:0071550 | Death-inducing signaling complex assembly | 7E-4 | 1.39E-1 | 37.11 (1002, 2, 27, 2) |
| GO:0043496 | Regulation of protein homodimerization activity | 7E-4 | 1.34E-1 | 37.11 (1002, 2, 27, 2) |
| GO:1990440 | Positive regulation of transcription from RNA polymerase II promoter in response to ER stress | 7.64E-4 | 1.41E-1 | 9.54 (1002, 5, 84, 4) |
| GO:0080134 | Regulation of response to stress | 8.06E-4 | 1.45E-1 | 2.80 (1002, 70, 87, 17) |
| GO:0036490 | Regulation of translation in response to ER stress | 8.49E-4 | 1.48E-1 | 62.63 (1002, 4, 8, 2) |
| GO:1905897 | Regulation of response to ER stress | 9.21E-4 | 1.55E-1 | 5.37 (1002, 15, 87, 7) |
| GO:0043620 | Regulation of DNA-templated transcription in response to stress | 9.45E-4 | 1.55E-1 | 6.21 (1002, 11, 88, 6) |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 9.45E-4 | 1.51E-1 | 6.21 (1002, 11, 88, 6) |
| GO:0035556 | Intracellular signal transduction | 9.49E-4 | 1.47E-1 | 1.53 (1002, 106, 347, 56) |
| GO:0010966 | Regulation of phosphate transport | 9.98E-4 | 1.51E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:0043267 | Negative regulation of potassium ion transport | 9.98E-4 | 1.47E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:0061395 | Positive regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance | 9.98E-4 | 1.43E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:2000118 | Regulation of sodium-dependent phosphate transport | 9.98E-4 | 1.4E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:2000120 | Positive regulation of sodium-dependent phosphate transport | 9.98E-4 | 1.36E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:1903793 | Positive regulation of anion transport | 9.98E-4 | 1.33E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:1905459 | Regulation of vascular associated smooth muscle cell apoptotic process | 9.98E-4 | 1.3E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:1903203 | Regulation of oxidative stress-induced neuron death | 9.98E-4 | 1.27E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:1903204 | Negative regulation of oxidative stress-induced neuron death | 9.98E-4 | 1.25E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:1905461 | Positive regulation of vascular associated smooth muscle cell apoptotic process | 9.98E-4 | 1.22E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:0044070 | Regulation of anion transport | 9.98E-4 | 1.19E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:0090649 | Response to oxygen-glucose deprivation | 9.98E-4 | 1.17E-1 | 1002.00 (1002, 1, 1, 1) |
| GO:0090650 | Cellular response to oxygen-glucose deprivation | 9.98E-4 | 1.15E-1 | 1002.00 (1002, 1, 1, 1) |
| Correlation with CHOP; 1010 genes with Pearson's p < 0.05 | ||||
| GO:0080090 | Regulation of primary metabolic process | 2.22E-4 | 1E-0 | 1.57 (849, 319, 95, 56) |
| GO:1901605 | Alpha-amino acid metabolic process | 3.27E-4 | 8.95E-1 | 2.74 (849, 12, 284, 11) |
| GO:0051171 | Regulation of nitrogen compound metabolic process | 3.88E-4 | 7.09E-1 | 1.57 (849, 314, 91, 53) |
| GO:0060255 | Regulation of macromolecule metabolic process | 6.05E-4 | 8.29E-1 | 1.54 (849, 327, 91, 54) |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 6.41E-4 | 7.03E-1 | 3.49 (849, 7, 243, 7) |
| GO:0035556 | Intracellular signal transduction | 9.82E-4 | 8.97E-1 | 2.39 (849, 82, 91, 21) |
| GO:0006520 | Cellular amino acid metabolic process | 9.84E-4 | 7.7E-1 | 2.01 (849, 30, 296, 21) |
| Correlation with GRP94; 1178 genes with Pearson's p < 0.05 | ||||
| GO:0034976 | Response to ER stress | 1.61E-5 | 9.34E-2 | 4.32 (986, 45, 71, 14) |
| GO:0010498 | Proteasomal protein catabolic process | 5.57E-5 | 1.62E-1 | 11.98 (986, 38, 13, 6) |
| GO:0036503 | ERAD pathway | 6.55E-5 | 1.27E-1 | 25.61 (986, 14, 11, 4) |
| GO:0030163 | Protein catabolic process | 1.49E-4 | 2.16E-1 | 10.34 (986, 44, 13, 6) |
| GO:0051603 | Proteolysis involved in cellular protein catabolic process | 2.23E-4 | 2.59E-1 | 7.70 (986, 56, 16, 7) |
| GO:0043620 | Regulation of DNA-templated transcription in response to stress | 2.46E-4 | 2.38E-1 | 6.05 (986, 12, 95, 7) |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 2.46E-4 | 2.04E-1 | 6.05 (986, 12, 95, 7) |
| GO:0034620 | Cellular response to unfolded protein | 2.46E-4 | 1.79E-1 | 93.90 (986, 3, 7, 2) |
| GO:0035967 | Cellular response to topologically incorrect protein | 2.46E-4 | 1.59E-1 | 93.90 (986, 3, 7, 2) |
| GO:0034975 | Protein folding in ER | 3.03E-4 | 1.76E-1 | 98.60 (986, 4, 5, 2) |
| GO:0036500 | ATF6-mediated UPR | 3.03E-4 | 1.6E-1 | 98.60 (986, 4, 5, 2) |
| GO:1901565 | Organonitrogen compound catabolic process | 3.16E-4 | 1.53E-1 | 4.81 (986, 57, 36, 10) |
| GO:0051402 | Neuron apoptotic process | 3.21E-4 | 1.44E-1 | 75.85 (986, 2, 13, 2) |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 3.63E-4 | 1.51E-1 | 6.21 (986, 8, 119, 6) |
| GO:1900102 | Negative regulation of ER UPR | 3.68E-4 | 1.43E-1 | 11.11 (986, 5, 71, 4) |
| GO:1903573 | Negative regulation of response to ER stress | 3.8E-4 | 1.38E-1 | 14.94 (986, 8, 33, 4) |
| GO:1903897 | Regulation of PERK-mediated UPR | 6.75E-4 | 2.31E-1 | 13.89 (986, 3, 71, 3) |
| GO:0043161 | Proteasome-mediated ubiquitin-dependent protein catabolic process | 8.36E-4 | 2.7E-1 | 10.53 (986, 36, 13, 5) |
| GO:0070997 | Neuron death | 9.22E-4 | 2.82E-1 | 50.56 (986, 3, 13, 2) |
| Correlation with BiP; 1182 genes with Pearson's p < 0.05 | ||||
| GO:0034976 | Response to ER stress | 1.75E-10 | 1.01E-6 | 11.93 (981, 47, 21, 12) |
| GO:0048585 | Negative regulation of response to stimulus | 2.11E-8 | 6.08E-5 | 6.02 (981, 94, 26, 15) |
| GO:1903573 | Negative regulation of response to ER stress | 3.44E-8 | 6.6E-5 | 43.79 (981, 7, 16, 5) |
| GO:1900102 | Negative regulation of ER UPR | 5.16E-8 | 7.43E-5 | 81.75 (981, 4, 12, 4) |
| GO:0009968 | Negative regulation of signal transduction | 5.21E-8 | 6E-5 | 7.56 (981, 82, 19, 12) |
| GO:0010648 | Negative regulation of cell communication | 8.57E-8 | 8.22E-5 | 7.29 (981, 85, 19, 12) |
| GO:0023057 | Negative regulation of signaling | 8.57E-8 | 7.05E-5 | 7.29 (981, 85, 19, 12) |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 1.38E-7 | 9.94E-5 | 35.04 (981, 7, 20, 5) |
| GO:0030968 | ER UPR | 1.78E-6 | 1.14E-3 | 14.22 (981, 23, 21, 7) |
| GO:0006984 | ER-nucleus signaling pathway | 3.48E-6 | 2.01E-3 | 23.36 (981, 10, 21, 5) |
| GO:1900101 | Regulation of ER UPR | 4.27E-6 | 2.24E-3 | 40.87 (981, 8, 12, 4) |
| GO:0036499 | PERK-mediated UPR | 6.01E-6 | 2.89E-3 | 32.70 (981, 6, 20, 4) |
| GO:0060548 | Negative regulation of cell death | 7.15E-6 | 3.17E-3 | 6.10 (981, 68, 26, 11) |
| GO:1903897 | Regulation of PERK-mediated UPR | 9.53E-6 | 3.92E-3 | 61.31 (981, 4, 12, 3) |
| GO:0097190 | Apoptotic signaling pathway | 1.35E-5 | 5.19E-3 | 21.33 (981, 23, 10, 5) |
| GO:0007165 | Signal transduction | 1.51E-5 | 5.44E-3 | 3.56 (981, 197, 21, 15) |
| GO:0033554 | Cellular response to stress | 2.63E-5 | 8.91E-3 | 4.16 (981, 146, 21, 13) |
| GO:1905897 | Regulation of response to ER stress | 4.44E-5 | 1.42E-2 | 17.03 (981, 18, 16, 5) |
| GO:0042981 | Regulation of apoptotic process | 4.94E-5 | 1.5E-2 | 4.16 (981, 118, 26, 13) |
| GO:0051716 | Cellular response to stimulus | 5.01E-5 | 1.44E-2 | 3.33 (981, 201, 22, 15) |
| GO:0043067 | Regulation of programmed cell death | 6.1E-5 | 1.67E-2 | 4.09 (981, 120, 26, 13) |
| GO:0006950 | Response to stress | 6.27E-5 | 1.64E-2 | 3.52 (981, 186, 21, 14) |
| GO:0080135 | Regulation of cellular response to stress | 7.55E-5 | 1.89E-2 | 6.31 (981, 56, 25, 9) |
| GO:0032056 | Positive regulation of translation in response to stress | 8.98E-5 | 2.16E-2 | 25.82 (981, 3, 38, 3) |
| GO:0043555 | Regulation of translation in response to stress | 9.03E-5 | 2.08E-2 | 17.21 (981, 6, 38, 4) |
| GO:0010941 | Regulation of cell death | 9.93E-5 | 2.2E-2 | 3.92 (981, 125, 26, 13) |
| GO:0097193 | Intrinsic apoptotic signaling pathway | 1.07E-4 | 2.28E-2 | 14.43 (981, 17, 20, 5) |
| GO:0080134 | Regulation of response to stress | 1.14E-4 | 2.35E-2 | 4.69 (981, 92, 25, 11) |
| GO:0071550 | Death-inducing signaling complex assembly | 1.16E-4 | 2.31E-2 | 122.62 (981, 2, 8, 2) |
| GO:0042493 | Response to drug | 1.72E-4 | 3.29E-2 | 6.68 (981, 47, 25, 8) |
| GO:0048523 | Negative regulation of cellular process | 1.96E-4 | 3.64E-2 | 2.72 (981, 275, 21, 16) |
| GO:0071496 | Cellular response to external stimulus | 2.22E-4 | 4E-2 | 9.67 (981, 29, 21, 6) |
| GO:1903895 | Negative regulation of IRE1-mediated UPR | 2.29E-4 | 3.99E-2 | 89.18 (981, 2, 11, 2) |
| GO:0043496 | Regulation of protein homodimerization activity | 2.5E-4 | 4.23E-2 | 93.43 (981, 3, 7, 2) |
| GO:1903898 | Negative regulation of PERK-mediated UPR | 2.75E-4 | 4.52E-2 | 81.75 (981, 2, 12, 2) |
| GO:0043066 | Negative regulation of apoptotic process | 3.01E-4 | 4.81E-2 | 5.39 (981, 63, 26, 9) |
| GO:0030433 | Ubiquitin-dependent ERAD pathway | 3.09E-4 | 4.8E-2 | 16.99 (981, 11, 21, 4) |
| GO:0043069 | Negative regulation of programmed cell death | 3.53E-4 | 5.35E-2 | 5.31 (981, 64, 26, 9) |
| GO:1901698 | Response to nitrogen compound | 3.66E-4 | 5.41E-2 | 5.27 (981, 67, 25, 9) |
| GO:0043620 | Regulation of DNA-templated transcription in response to stress | 4.22E-4 | 6.07E-2 | 16.35 (981, 12, 20, 4) |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 4.22E-4 | 5.93E-2 | 16.35 (981, 12, 20, 4) |
| GO:1901565 | Organonitrogen compound catabolic process | 4.56E-4 | 6.25E-2 | 5.92 (981, 51, 26, 8) |
| GO:0043558 | Regulation of translational initiation in response to stress | 5.54E-4 | 7.43E-2 | 19.36 (981, 4, 38, 3) |
| GO:0045948 | Positive regulation of translational initiation | 5.54E-4 | 7.26E-2 | 19.36 (981, 4, 38, 3) |
| GO:1990440 | Positive regulation of transcription from RNA polymerase II promoter in response to ER stress | 5.55E-4 | 7.11E-2 | 24.52 (981, 6, 20, 3) |
| GO:0030163 | Protein catabolic process | 5.74E-4 | 7.18E-2 | 6.77 (981, 39, 26, 7) |
| GO:0006446 | Regulation of translational initiation | 6.28E-4 | 7.7E-2 | 12.91 (981, 8, 38, 4) |
| GO:1903894 | Regulation of IRE1-mediated UPR | 6.54E-4 | 7.85E-2 | 59.45 (981, 3, 11, 2) |
| GO:0048519 | Negative regulation of biological process | 7.17E-4 | 8.43E-2 | 2.47 (981, 302, 21, 16) |
| GO:0048583 | Regulation of response to stimulus | 7.73E-4 | 8.9E-2 | 2.87 (981, 228, 21, 14) |
| GO:0061394 | Regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance | 7.91E-4 | 8.93E-2 | 49.05 (981, 2, 20, 2) |
| GO:2001235 | Positive regulation of apoptotic signaling pathway | 8.48E-4 | 9.39E-2 | 10.33 (981, 25, 19, 5) |
| GO:0031668 | Cellular response to extracellular stimulus | 8.49E-4 | 9.22E-2 | 10.16 (981, 23, 21, 5) |
| GO:0031669 | Cellular response to nutrient levels | 8.49E-4 | 9.05E-2 | 10.16 (981, 23, 21, 5) |
| GO:0007264 | Small GTPase mediated signal transduction | 9.1E-4 | 9.53E-2 | 3.67 (981, 10, 214, 8) |
| Correlation with ERdj4; 1250 genes with Pearson's p < 0.05 | ||||
| GO:1903573 | Negative regulation of response to ER stress | 2.24E-6 | 1.35E-2 | 25.70 (1028, 10, 20, 5) |
| GO:1900102 | Negative regulation of ER UPR | 5.01E-6 | 1.5E-2 | 34.27 (1028, 6, 20, 4) |
| GO:0034976 | Response to ER stress | 2.34E-5 | 4.68E-2 | 3.74 (1028, 53, 83, 16) |
| GO:0043555 | Regulation of translation in response to stress | 2.49E-5 | 3.73E-2 | 12.24 (1028, 6, 70, 5) |
| GO:0009968 | Negative regulation of signal transduction | 3.75E-5 | 4.5E-2 | 5.78 (1028, 89, 20, 10) |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 4.06E-5 | 4.07E-2 | 13.67 (1028, 8, 47, 5) |
| GO:0010648 | Negative regulation of cell communication | 5.15E-5 | 4.42E-2 | 5.59 (1028, 92, 20, 10) |
| GO:0023057 | Negative regulation of signaling | 5.15E-5 | 3.87E-2 | 5.59 (1028, 92, 20, 10) |
| GO:0048585 | Negative regulation of response to stimulus | 7.96E-5 | 5.32E-2 | 4.44 (1028, 103, 27, 12) |
| GO:1902842 | Negative regulation of netrin-activated signaling pathway | 1.04E-4 | 6.26E-2 | 93.45 (1028, 2, 11, 2) |
| GO:1902841 | Regulation of netrin-activated signaling pathway | 1.04E-4 | 5.69E-2 | 93.45 (1028, 2, 11, 2) |
| GO:1900101 | Regulation of ER UPR | 1.24E-4 | 6.21E-2 | 20.56 (1028, 10, 20, 4) |
| GO:0036499 | PERK-mediated UPR | 2.13E-4 | 9.84E-2 | 14.58 (1028, 6, 47, 4) |
| GO:1903897 | Regulation of PERK-mediated UPR | 2.25E-4 | 9.64E-2 | 30.84 (1028, 5, 20, 3) |
| GO:2001233 | Regulation of apoptotic signaling pathway | 2.48E-4 | 9.95E-2 | 7.66 (1028, 47, 20, 7) |
| GO:0032056 | Positive regulation of translation in response to stress | 2.58E-4 | 9.71E-2 | 18.69 (1028, 3, 55, 3) |
| GO:1903895 | Negative regulation of IRE1-mediated UPR | 2.9E-4 | 1.02E-1 | 57.11 (1028, 2, 18, 2) |
| GO:0071550 | Death-inducing signaling complex assembly | 3.45E-4 | 1.15E-1 | 73.43 (1028, 2, 14, 2) |
| GO:0043620 | Regulation of DNA-templated transcription in response to stress | 3.93E-4 | 1.24E-1 | 7.34 (1028, 12, 70, 6) |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 3.93E-4 | 1.18E-1 | 7.34 (1028, 12, 70, 6) |
| GO:1905897 | Regulation of response to ER stress | 4.3E-4 | 1.23E-1 | 11.68 (1028, 22, 20, 5) |
| GO:0043619 | Regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 5.29E-4 | 1.45E-1 | 14.69 (1028, 3, 70, 3) |
| GO:0030968 | ER UPR | 6.19E-4 | 1.62E-1 | 10.98 (1028, 26, 18, 5) |
| GO:0030163 | Protein catabolic process | 6.84E-4 | 1.71E-1 | 10.87 (1028, 43, 11, 5) |
| GO:2001235 | Positive regulation of apoptotic signaling pathway | 7.18E-4 | 1.73E-1 | 7.96 (1028, 25, 31, 6) |
| GO:0006520 | Cellular amino acid metabolic process | 8.59E-4 | 1.98E-1 | 4.20 (1028, 36, 68, 10) |
| GO:0002377 | Immunoglobulin production | 9.73E-4 | 2.16E-1 | 1028.00 (1028, 1, 1, 1) |
| Correlation with P58IPK; 1202 genes with Pearson's p < 0.05 | ||||
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 6.83E-7 | 3.88E-3 | 26.48 (1001, 7, 27, 5) |
| GO:0036493 | Positive regulation of translation in response to ER stress | 4E-6 | 1.14E-2 | 500.50 (1001, 2, 2, 2) |
| GO:1903912 | Negative regulation of ER stress-induced eIF2 alpha phosphorylation | 4E-6 | 7.57E-3 | 500.50 (1001, 2, 2, 2) |
| GO:0043555 | Regulation of translation in response to stress | 4.43E-6 | 6.3E-3 | 100.10 (1001, 6, 5, 3) |
| GO:0032056 | Positive regulation of translation in response to stress | 5.81E-6 | 6.61E-3 | 52.68 (1001, 3, 19, 3) |
| GO:1900102 | Negative regulation of ER UPR | 1.06E-5 | 1E-2 | 20.02 (1001, 4, 50, 4) |
| GO:0006446 | Regulation of translational initiation | 1.73E-5 | 1.4E-2 | 75.08 (1001, 8, 5, 3) |
| GO:0097193 | Intrinsic apoptotic signaling pathway | 2.2E-5 | 1.57E-2 | 13.08 (1001, 17, 27, 6) |
| GO:0048585 | Negative regulation of response to stimulus | 3.12E-5 | 1.97E-2 | 3.51 (1001, 97, 50, 17) |
| GO:0036490 | Regulation of translation in response to ER stress | 3.37E-5 | 1.91E-2 | 250.25 (1001, 4, 2, 2) |
| GO:0045948 | Positive regulation of translational initiation | 3.37E-5 | 1.74E-2 | 250.25 (1001, 4, 2, 2) |
| GO:0001933 | Negative regulation of protein phosphorylation | 5.03E-5 | 2.38E-2 | 12.26 (1001, 35, 14, 6) |
| GO:0043558 | Regulation of translational initiation in response to stress | 5.53E-5 | 2.42E-2 | 39.51 (1001, 4, 19, 3) |
| GO:1903573 | Negative regulation of response to ER stress | 6.43E-5 | 2.61E-2 | 12.51 (1001, 8, 50, 5) |
| GO:0097190 | Apoptotic signaling pathway | 8.61E-5 | 3.26E-2 | 8.38 (1001, 22, 38, 7) |
| GO:0042326 | Negative regulation of phosphorylation | 9.92E-5 | 3.52E-2 | 11.00 (1001, 39, 14, 6) |
| GO:0034976 | Response to ER stress | 1.42E-4 | 4.75E-2 | 4.69 (1001, 47, 50, 11) |
| GO:0032269 | Negative regulation of cellular protein metabolic process | 1.62E-4 | 5.12E-2 | 5.72 (1001, 75, 21, 9) |
| GO:0010563 | Negative regulation of phosphorus metabolic process | 1.73E-4 | 5.19E-2 | 8.02 (1001, 46, 19, 7) |
| GO:0045936 | Negative regulation of phosphate metabolic process | 1.73E-4 | 4.93E-2 | 8.02 (1001, 46, 19, 7) |
| GO:0031400 | Negative regulation of protein modification process | 1.73E-4 | 4.69E-2 | 8.02 (1001, 46, 19, 7) |
| GO:1902903 | Regulation of supramolecular fiber organization | 1.81E-4 | 4.68E-2 | 35.75 (1001, 6, 14, 3) |
| GO:0051704 | Multiorganism process | 1.94E-4 | 4.8E-2 | 17.88 (1001, 56, 4, 4) |
| GO:0051248 | Negative regulation of protein metabolic process | 2.05E-4 | 4.86E-2 | 5.57 (1001, 77, 21, 9) |
| GO:0031327 | Negative regulation of cellular biosynthetic process | 2.34E-4 | 5.31E-2 | 6.09 (1001, 94, 14, 8) |
| GO:0009890 | Negative regulation of biosynthetic process | 2.72E-4 | 5.94E-2 | 5.96 (1001, 96, 14, 8) |
| GO:0061394 | Regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance | 3.12E-4 | 6.56E-2 | 77.00 (1001, 2, 13, 2) |
| GO:0007015 | Actin filament organization | 3.56E-4 | 7.22E-2 | 30.64 (1001, 7, 14, 3) |
| GO:0032273 | Positive regulation of protein polymerization | 3.64E-4 | 7.12E-2 | 71.50 (1001, 2, 14, 2) |
| GO:0030838 | Positive regulation of actin filament polymerization | 3.64E-4 | 6.89E-2 | 71.50 (1001, 2, 14, 2) |
| GO:0036499 | PERK-mediated UPR | 5.26E-4 | 9.64E-2 | 12.71 (1001, 7, 45, 4) |
| GO:0042493 | Response to drug | 6.76E-4 | 1.2E-1 | 2.06 (1001, 42, 277, 24) |
| GO:0032058 | Positive regulation of translational initiation in response to stress | 6.83E-4 | 1.18E-1 | 52.68 (1001, 2, 19, 2) |
| GO:1903898 | Negative regulation of PERK-mediated UPR | 6.83E-4 | 1.14E-1 | 52.68 (1001, 2, 19, 2) |
| GO:0048511 | Rhythmic process | 7.36E-4 | 1.2E-1 | 2.86 (1001, 21, 217, 13) |
| GO:0001932 | Regulation of protein phosphorylation | 7.42E-4 | 1.17E-1 | 6.34 (1001, 79, 14, 7) |
| GO:0006418 | tRNA aminoacylation for protein translation | 7.76E-4 | 1.19E-1 | 9.53 (1001, 15, 35, 5) |
| GO:1990440 | Positive regulation of transcription from RNA polymerase II promoter in response to ER stress | 8.12E-4 | 1.21E-1 | 5.27 (1001, 5, 190, 5) |
| GO:1903897 | Regulation of PERK-mediated UPR | 8.32E-4 | 1.21E-1 | 16.68 (1001, 4, 45, 3) |
| GO:1900101 | Regulation of ER UPR | 8.38E-4 | 1.19E-1 | 11.44 (1001, 7, 50, 4) |
| GO:0051172 | Negative regulation of nitrogen compound metabolic process | 9.15E-4 | 1.27E-1 | 4.29 (1001, 150, 14, 9) |
| GO:0032535 | Regulation of cellular component size | 9.22E-4 | 1.25E-1 | 22.58 (1001, 7, 19, 3) |
| GO:0030182 | Neuron differentiation | 9.43E-4 | 1.25E-1 | 8.58 (1001, 11, 53, 5) |
| GO:0036494 | Positive regulation of translation initiation in response to ER stress | 9.99E-4 | 1.29E-1 | 1001.00 (1001, 1, 1, 1) |
| Conventional analysis with 310 upregulated genes (p < 0.05) | ||||
| GO:0048583 | Regulation of response to stimulus | 4.86E-9 | 1.65E-5 | 1.97 (306, 83, 88, 47) |
| GO:0009966 | Regulation of signal transduction | 1.55E-8 | 2.63E-5 | 2.02 (306, 74, 88, 43) |
| GO:0006950 | Response to stress | 1.68E-8 | 1.9E-5 | 2.08 (306, 71, 85, 41) |
| GO:0010033 | Response to organic substance | 2.07E-8 | 1.76E-5 | 2.40 (306, 51, 80, 32) |
| GO:0010646 | Regulation of cell communication | 3.15E-8 | 2.14E-5 | 1.99 (306, 75, 88, 43) |
| GO:0023051 | Regulation of signaling | 3.15E-8 | 1.79E-5 | 1.99 (306, 75, 88, 43) |
| GO:0065009 | Regulation of molecular function | 3.76E-8 | 1.83E-5 | 2.16 (306, 61, 86, 37) |
| GO:0007165 | Signal transduction | 1.54E-7 | 6.53E-5 | 1.71 (306, 83, 114, 53) |
| GO:0042221 | Response to chemical | 2.33E-7 | 8.82E-5 | 2.28 (306, 67, 64, 32) |
| GO:0080134 | Regulation of response to stress | 2.91E-7 | 9.9E-5 | 2.43 (306, 40, 85, 27) |
| GO:0050896 | Response to stimulus | 7.57E-7 | 2.34E-4 | 1.78 (306, 93, 87, 47) |
| GO:0051716 | Cellular response to stimulus | 9.21E-7 | 2.61E-4 | 1.92 (306, 75, 85, 40) |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to ER stress | 1.12E-6 | 2.93E-4 | 21.25 (306, 8, 9, 5) |
| GO:0034976 | Response to ER stress | 1.57E-6 | 3.8E-4 | 7.36 (306, 22, 17, 9) |
| GO:0042493 | Response to drug | 1.75E-6 | 3.96E-4 | 3.03 (306, 19, 85, 16) |
| GO:0043620 | Regulation of DNA-templated transcription in response to stress | 2.02E-6 | 4.3E-4 | 17.00 (306, 6, 15, 5) |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 2.02E-6 | 4.05E-4 | 17.00 (306, 6, 15, 5) |
| GO:0010941 | Regulation of cell death | 2.4E-6 | 4.53E-4 | 2.08 (306, 47, 97, 31) |
| GO:0009968 | Negative regulation of signal transduction | 2.45E-6 | 4.39E-4 | 3.22 (306, 38, 45, 18) |
| GO:0042981 | Regulation of apoptotic process | 3.02E-6 | 5.14E-4 | 2.10 (306, 45, 97, 30) |
| GO:0033554 | Cellular response to stress | 3.4E-6 | 5.51E-4 | 2.11 (306, 53, 85, 31) |
| GO:0048585 | Negative regulation of response to stimulus | 4.13E-6 | 6.39E-4 | 3.00 (306, 43, 45, 19) |
| GO:0050790 | Regulation of catalytic activity | 5.94E-6 | 8.8E-4 | 3.25 (306, 39, 41, 17) |
| GO:0043067 | Regulation of programmed cell death | 6.37E-6 | 9.04E-4 | 2.06 (306, 46, 97, 30) |
| GO:0010648 | Negative regulation of cell communication | 7.36E-6 | 1E-3 | 3.06 (306, 40, 45, 18) |
| GO:0023057 | Negative regulation of signaling | 7.36E-6 | 9.63E-4 | 3.06 (306, 40, 45, 18) |
| GO:0010243 | Response to organonitrogen compound | 8.44E-6 | 1.06E-3 | 2.91 (306, 21, 80, 16) |
| GO:0043085 | Positive regulation of catalytic activity | 9.88E-6 | 1.2E-3 | 2.67 (306, 24, 86, 18) |
| GO:0032502 | Developmental process | 1.2E-5 | 1.41E-3 | 1.74 (306, 86, 88, 43) |
| GO:0071310 | Cellular response to organic substance | 1.23E-5 | 1.39E-3 | 2.75 (306, 33, 64, 19) |
| GO:0032269 | Negative regulation of cellular protein metabolic process | 1.36E-5 | 1.49E-3 | 3.88 (306, 25, 41, 13) |
| GO:0009636 | Response to toxic substance | 1.78E-5 | 1.89E-3 | 3.81 (306, 11, 73, 10) |
| GO:0044093 | Positive regulation of molecular function | 1.84E-5 | 1.89E-3 | 2.41 (306, 31, 86, 21) |
| GO:0080135 | Regulation of cellular response to stress | 2.11E-5 | 2.11E-3 | 2.59 (306, 25, 85, 18) |
| GO:1901698 | Response to nitrogen compound | 2.3E-5 | 2.23E-3 | 2.66 (306, 23, 85, 17) |
| GO:0035556 | Intracellular signal transduction | 2.42E-5 | 2.29E-3 | 1.87 (306, 46, 114, 32) |
| GO:0051248 | Negative regulation of protein metabolic process | 2.45E-5 | 2.25E-3 | 3.73 (306, 26, 41, 13) |
| GO:0097193 | Intrinsic apoptotic signaling pathway | 2.61E-5 | 2.34E-3 | 14.17 (306, 12, 9, 5) |
| GO:0019752 | Carboxylic acid metabolic process | 3.28E-5 | 2.86E-3 | 3.27 (306, 23, 57, 14) |
| GO:0036499 | PERK-mediated UPR | 3.45E-5 | 2.93E-3 | 6.24 (306, 6, 49, 6) |
| GO:0042594 | Response to starvation | 3.83E-5 | 3.18E-3 | 4.59 (306, 12, 50, 9) |
| GO:1901216 | Positive regulation of neuron death | 4.22E-5 | 3.42E-3 | 8.27 (306, 5, 37, 5) |
| GO:0036003 | Positive regulation of transcription from RNA polymerase II promoter in response to stress | 4.22E-5 | 3.34E-3 | 16.32 (306, 5, 15, 4) |
| GO:0032268 | Regulation of cellular protein metabolic process | 5.14E-5 | 3.97E-3 | 1.88 (306, 61, 88, 33) |
| GO:0070887 | Cellular response to chemical stimulus | 5.43E-5 | 4.11E-3 | 2.55 (306, 48, 50, 20) |
| GO:0051246 | Regulation of protein metabolic process | 5.58E-5 | 4.13E-3 | 1.77 (306, 66, 97, 37) |
| GO:0043436 | Oxoacid metabolic process | 6.78E-5 | 4.91E-3 | 3.13 (306, 24, 57, 14) |
| GO:0006082 | Organic acid metabolic process | 6.78E-5 | 4.81E-3 | 3.13 (306, 24, 57, 14) |
| GO:1905897 | Regulation of response to ER stress | 8.45E-5 | 5.87E-3 | 4.95 (306, 11, 45, 8) |
| GO:0009605 | Response to external stimulus | 8.69E-5 | 5.91E-3 | 2.40 (306, 35, 73, 20) |
| GO:0009719 | Response to endogenous stimulus | 8.96E-5 | 5.98E-3 | 2.82 (306, 19, 80, 14) |
| GO:0071407 | Cellular response to organic cyclic compound | 1.02E-4 | 6.67E-3 | 3.68 (306, 13, 64, 10) |
| GO:0007154 | Cell communication | 1.16E-4 | 7.44E-3 | 3.51 (306, 16, 60, 11) |
| GO:0097190 | Apoptotic signaling pathway | 1.17E-4 | 7.35E-3 | 11.33 (306, 15, 9, 5) |
| GO:0051336 | Regulation of hydrolase activity | 1.2E-4 | 7.45E-3 | 4.15 (306, 18, 41, 10) |
| GO:0051188 | Cofactor biosynthetic process | 1.22E-4 | 7.44E-3 | 102.00 (306, 3, 2, 2) |
| GO:1900101 | Regulation of ER UPR | 1.35E-4 | 8.05E-3 | 6.80 (306, 5, 45, 5) |
| GO:1901700 | Response to oxygen-containing compound | 1.62E-4 | 9.5E-3 | 2.12 (306, 39, 85, 23) |
| GO:0051346 | Negative regulation of hydrolase activity | 1.68E-4 | 9.69E-3 | 7.97 (306, 6, 32, 5) |
| GO:0032270 | Positive regulation of cellular protein metabolic process | 1.73E-4 | 9.83E-3 | 2.10 (306, 38, 88, 23) |
| GO:0010469 | Regulation of signaling receptor activity | 1.84E-4 | 1.02E-2 | 3.44 (306, 10, 80, 9) |
| GO:0006986 | Response to unfolded protein | 1.85E-4 | 1.02E-2 | 5.83 (306, 7, 45, 6) |
| GO:0035966 | Response to topologically incorrect protein | 1.85E-4 | 1E-2 | 5.83 (306, 7, 45, 6) |
| GO:0045861 | Negative regulation of proteolysis | 1.92E-4 | 1.02E-2 | 9.56 (306, 4, 32, 4) |
| GO:0031399 | Regulation of protein modification process | 1.99E-4 | 1.04E-2 | 2.02 (306, 43, 88, 25) |
| GO:0040011 | Locomotion | 2.07E-4 | 1.07E-2 | 2.70 (306, 14, 97, 12) |
| GO:0048870 | Cell motility | 2.07E-4 | 1.05E-2 | 2.70 (306, 14, 97, 12) |
| GO:0010942 | Positive regulation of cell death | 2.1E-4 | 1.05E-2 | 2.27 (306, 25, 97, 18) |
| GO:0051247 | Positive regulation of protein metabolic process | 2.48E-4 | 1.22E-2 | 2.04 (306, 41, 88, 24) |
| GO:0014070 | Response to organic cyclic compound | 2.5E-4 | 1.22E-2 | 2.68 (306, 20, 80, 14) |
| GO:0006954 | Inflammatory response | 2.59E-4 | 1.24E-2 | 3.73 (306, 9, 73, 8) |
| GO:0051186 | Cofactor metabolic process | 2.72E-4 | 1.29E-2 | 8.69 (306, 8, 22, 5) |
| GO:0048519 | Negative regulation of biological process | 2.75E-4 | 1.28E-2 | 1.52 (306, 119, 81, 48) |
| GO:0030162 | Regulation of proteolysis | 2.81E-4 | 1.29E-2 | 4.20 (306, 16, 41, 9) |
| GO:0030154 | Cell differentiation | 3.02E-4 | 1.37E-2 | 2.28 (306, 40, 67, 20) |
| GO:0006984 | ER-nucleus signaling pathway | 3.08E-4 | 1.38E-2 | 5.35 (306, 7, 49, 6) |
| GO:0006928 | Movement of cell or subcellular component | 3.16E-4 | 1.4E-2 | 2.56 (306, 16, 97, 13) |
| GO:0043086 | Negative regulation of catalytic activity | 3.37E-4 | 1.47E-2 | 4.78 (306, 16, 32, 8) |
| GO:1902531 | Regulation of intracellular signal transduction | 3.67E-4 | 1.58E-2 | 1.91 (306, 43, 97, 26) |
| GO:0016579 | Protein deubiquitination | 4.09E-4 | 1.74E-2 | 3.60 (306, 7, 85, 7) |
| GO:0070646 | Protein modification by small protein removal | 4.09E-4 | 1.72E-2 | 3.60 (306, 7, 85, 7) |
| GO:0048869 | Cellular developmental process | 4.49E-4 | 1.86E-2 | 2.10 (306, 54, 62, 23) |
| GO:0065008 | Regulation of biological quality | 4.65E-4 | 1.91E-2 | 1.64 (306, 57, 118, 36) |
| GO:1901214 | Regulation of neuron death | 5.05E-4 | 2.05E-2 | 4.66 (306, 10, 46, 7) |
| GO:0006766 | Vitamin metabolic process | 5.06E-4 | 2.03E-2 | 13.91 (306, 3, 22, 3) |
| GO:0012501 | Programmed cell death | 5.81E-4 | 2.3E-2 | 2.60 (306, 18, 85, 13) |
| GO:0008219 | Cell death | 5.81E-4 | 2.27E-2 | 2.60 (306, 18, 85, 13) |
| GO:0035690 | Cellular response to drug | 5.87E-4 | 2.27E-2 | 2.83 (306, 14, 85, 11) |
| GO:0006915 | Apoptotic process | 6.03E-4 | 2.3E-2 | 2.77 (306, 17, 78, 12) |
| GO:0016477 | Cell migration | 6.41E-4 | 2.42E-2 | 2.67 (306, 13, 97, 11) |
| GO:0048523 | Negative regulation of cellular process | 6.64E-4 | 2.48E-2 | 1.58 (306, 109, 73, 41) |
| GO:0017144 | Drug metabolic process | 6.93E-4 | 2.56E-2 | 4.18 (306, 9, 57, 7) |
| GO:0051093 | Negative regulation of developmental process | 7.12E-4 | 2.6E-2 | 2.24 (306, 19, 108, 15) |
| GO:0051239 | Regulation of multicellular organismal process | 7.29E-4 | 2.64E-2 | 1.88 (306, 48, 88, 26) |
| GO:1902532 | Negative regulation of intracellular signal transduction | 7.35E-4 | 2.63E-2 | 3.92 (306, 19, 37, 9) |
| GO:0009991 | Response to extracellular stimulus | 7.76E-4 | 2.75E-2 | 3.40 (306, 18, 50, 10) |
| GO:0031667 | Response to nutrient levels | 7.76E-4 | 2.72E-2 | 3.40 (306, 18, 50, 10) |
| GO:0048856 | Anatomical structure development | 7.87E-4 | 2.73E-2 | 1.80 (306, 56, 88, 29) |
| GO:1990440 | Positive regulation of transcription from RNA polymerase II promoter in response to ER stress | 7.95E-4 | 2.73E-2 | 15.30 (306, 4, 15, 3) |
| GO:0071495 | Cellular response to endogenous stimulus | 8.04E-4 | 2.74E-2 | 3.67 (306, 15, 50, 9) |
| GO:0009267 | Cellular response to starvation | 8.31E-4 | 2.8E-2 | 4.37 (306, 10, 49, 7) |
| GO:0043555 | Regulation of translation in response to stress | 8.39E-4 | 2.8E-2 | 18.36 (306, 5, 10, 3) |
| GO:0006952 | Defense response | 8.54E-4 | 2.82E-2 | 2.84 (306, 15, 79, 11) |
| GO:0015804 | Neutral amino acid transport | 8.91E-4 | 2.92E-2 | 5.02 (306, 5, 61, 5) |
| GO:0010604 | Positive regulation of macromolecule metabolic process | 9.01E-4 | 2.92E-2 | 1.55 (306, 82, 101, 42) |
| GO:0070848 | Response to growth factor | 9.13E-4 | 2.93E-2 | 4.94 (306, 5, 62, 5) |
| GO:1901701 | Cellular response to oxygen-containing compound | 9.22E-4 | 2.93E-2 | 2.64 (306, 27, 60, 14) |
| GO:0043619 | Regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 9.64E-4 | 3.04E-2 | 30.60 (306, 2, 10, 2) |
| GO:1902882 | Regulation of response to oxidative stress | 9.64E-4 | 3.01E-2 | 30.60 (306, 2, 10, 2) |
| GO:1901522 | Positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 9.64E-4 | 2.98E-2 | 30.60 (306, 2, 10, 2) |
The number of genes that exhibited p < 0.05 in the coordination analysis is indicated next to the specific gene for which coordination was assessed. The major biological processes predicted by this analysis are shown. The list in the second column (description) indicates the corresponding biological process, as obtained by the enrichment analysis.
ER, endoplasmic reticulum; UPR, unfolded protein response.
FIG. 1.
Flowchart of the process and analysis followed.
Results and Discussion
By using three pairs of fibroblasts from different P. maniculatus animals that were treated with tunicamycin, we performed RNA sequencing that revealed expression of 14,159 transcripts. The upregulation of six selected UPR targets, as revealed by the RNAseq analysis, indicates the canonical activation of the UPR under these conditions (Supplementary Fig. S1). Subsequently, we determined the correlation of the 14,158 genes with each of ATF4, CHOP (DDIT3), GRP94, DNAJB9 (ERdj4), DNAJ3C (P58IPK), and HSPA5 (BiP/GRP78) and then calculated the correlation of the corresponding R (Pearson's) values for all pairwise comparisons.
These genes represent targets of different branches of the UPR: ATF4 is regulated by PERK (Harding et al., 2003), while CHOP by both PERK (in a manner that is ATF4 dependent) and ATF6 (Zinszner et al., 1998; Ye et al., 2000). GRP94 is not only regulated by IRE1 (Yoshida et al., 2001; Marzec et al., 2012) but also by ATF6 (Yamamoto et al., 2007; Shoulders et al., 2013). DNAJB9 and DNAJ3C have been recognized as XBP-1 specific targets (Lee et al., 2003), while BiP (HSPA5) is induced by ATF6, but then it plays a nodal role in orchestrating globally the UPR.
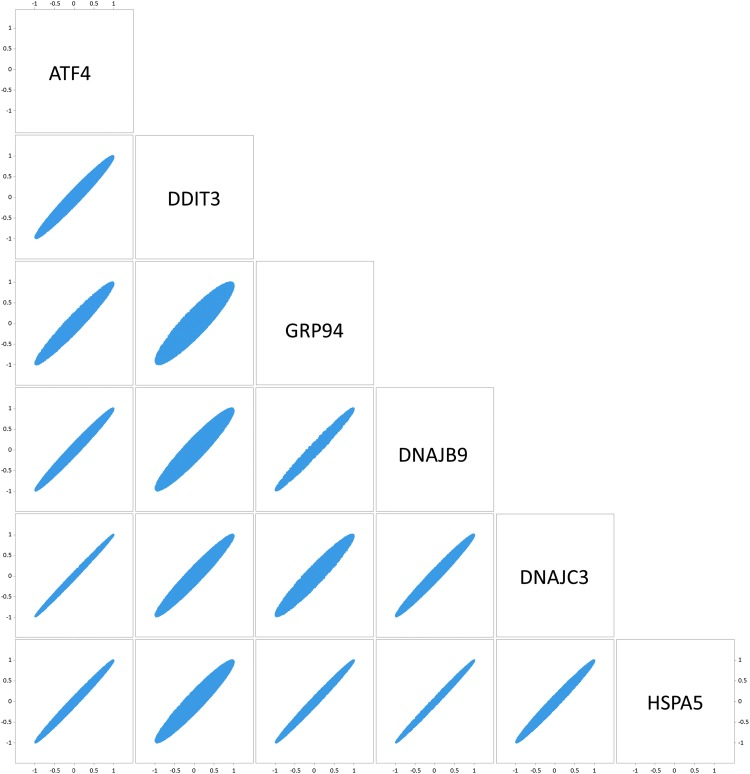
Calculation of the correlation of the transcriptome coordinated with each of the UPR-associated genes analyzed, in all pairwise comparisons, showed similar—yet distinct—profiles. BiP, being a global transducer of the UPR, was highly coordinated with all UPR targets, while CHOP exhibited the most deviant profile (Fig. 2). This is not surprising considering that CHOP is activated by several stress-inducing signals, beyond UPR (Jauhiainen et al., 2012). CHOP and ATF4 that are co-regulated following stimulation of the PERK branch of the UPR showed tight coordination with each other, exceeding that obtained by CHOP and the other UPR targets analyzed. DNAJB9 and DNAJC3 that are XBP1 targets showed high coordination with all UPR targets, and especially with BiP.
FIG. 2.
Pairwise scatterplots' matrix showing the R (Pearson's) for the transcripts correlated with each of the indicated pairwise comparisons. More narrow plots indicate tighter coordination.
These observations imply that interpretation of the degree of co-ordination between specific transcripts and the whole transcriptome may be useful in assigning genes into networks, signaling nodules, and biological processes. It is noted that UPR is a highly integrated response at which different branches of the UPR crosstalk and mask linear and causative associations between specific targets. For example, BiP that is an ATF6 target globally activates all branches of the UPR, while XBP1 is regulated by both IRE1 (at the level of splicing) and ATF6 (at the level of transcription). Yet, we hypothesized that the impact of particular targets and their association with specific biological processes may be revealed.
Based on these notions, we explored if the identification of the array of genes that are mostly correlated with each of these UPR-associated genes can predict biological processes that are known to reflect activation of corresponding UPR branches and reveal their corresponding impact in specific processes. The identification of signaling networks and associated biological processes was performed by using the gene ontology online platform (Eden et al., 2007, 2009) at which the list of genes that showed significance (p < 0.05, Pearson's) with GRP94, ATF4, CHOP, DNAJB9 (ERdj4), DNAJ3C (P58IPK), or HSPA5 (BiP/GRP78) were considered hierarchically, in a progressively decreasing manner.
For this analysis, we considered both genes exhibiting positive correlation only (Table 1 and Supplementary Figs. S2 and S3) and genes exhibiting positive and negative correlation (Supplementary Table S1 and Supplementary Figs. S4 and S5). With GRP94, 1178 genes exhibited significant positive correlation, 1010 with CHOP (DDIT3), 1198 with ATF4, 1182 with BiP, 1250 with ERdj4 and 1202 with P58IPK (for all p < 0.05, Pearson's). Relevant biological processes are shown in Table 1 at which all functions unveiled, along with the corresponding statistical significance indicated. Similar results were obtained when both positively and negatively transcripts were analyzed (Supplementary Table S1).
For BiP, as expected, the response to ER stress was predicted to be the major process unveiled, underscoring its role as the nodal activator of the UPR (Table 1). For GRP94, besides the cumulative response to ER stress, major functions that had been identified are also protein catabolism, ER-associated (ERAD) degradation, and transcriptional activation, which are established processes that are linked to the IRE1 arm of the UPR (Chiang et al., 2012). We note that earlier studies utilizing either qualitative changes in gene expression or chemical activators showed that in the absence of IRE1 and its target XBP1, but not in the absence of ATF6, MEFs can induce GRP94, which points to the important role of ATF6 in GRP94 expression (Yamamoto et al., 2007; Shoulders et al., 2013). This association we also noted since GRP94 was identified as an ATF6 target (Table 1).
Yet, with such qualitative alterations, the relative impact of each of these upstream regulators cannot be evaluated and adequately appreciated. This experimental setting, at which all branches are physiologically expressed, implies that GRP94 is associated with functions (as opposed to specific genes), previously established as IRE1-regulated functions, such as ERAD. This observation does not contradict the role of ATF6 in the regulation of GRP94, but rather underscores the biological relevance of IRE1 at conditions at which all branches of the UPR are physiologically expressed.
With ATF4, major processes revealed were the regulation of translation and regulation of apoptosis in response to stress, which are known processes that are mediated by PERK (Szegezdi et al., 2006).
For ERdj4 and P58IPK, no specific functions were revealed beyond their association with ER stress, with the exception of the fact that for both, a negative function was predicted. This probably implies their function in establishing negative feedback regulatory associations, consistent with the relief of stress following UPR activation.
An unexpected finding was the consistent association of CHOP with the regulation of metabolic processes. While CHOP is known to regulate metabolism, it is believed that this activity is produced indirectly, by competing with other cEBP family members. Indeed, negative regulation of cAMP-response element binding protein (CREB) has been revealed (Rutkowski et al., 2008). In addition, a major role of CHOP in regulating metabolism should also be considered.
Although this analysis possesses limitations in predicting linear association at the level of regulation, it possesses high power in identifying relevant biological processes and underscoring the relative impact of the transcripts of interest when simultaneously several such processes operate. The power of this approach in deciphering biologically relevant processes was reflected to the fact that by using only three primary cell lines, we were able to distinguish between well-established major functions for the different arms of the UPR and accurately predicting processes, such as ERAD and apoptosis for different transcripts. Similar gene ontology analysis by identifying transcripts significantly upregulated during ER stress and subjecting them, by an analogous hierarchical manner, to gene ontology analysis, was powerless to predict specific biological functions and only unveiled a generic and rather wide response to stress (Table 1).
Furthermore, while the proposed analysis allows the extraction of information with regard to the function of individual genes, conventional analysis focusing on relative expression can be useful only if genetic manipulation targeting the gene of interest is applied. The latter, beyond its methodological limitation related to that it is hypothesis driven, also possesses conceptual limitations since qualitative changes may be distinct from quantitative changes that occur in naturally existing populations. It is plausible that the power of the correlation analysis over the standard overexpression analysis is associated with the fact that different transcription modules exhibit different saturation levels at which the coordination in their activation is abolished. This can only be studied by using genetically diverse biological systems at which variation in expression has the capacity to reveal the presence of correlation in expression levels. When, however, only the degree of differential expression is evaluated, co-regulation and deviation from this cannot be assessed.
Collectively, this study illustrates the power of using outbred species in analyzing gene expression and suggests that evaluation of the degree of coordination as opposed to the magnitude of expression may be particularly valuable in assigning genes into transcriptional networks. It is plausible that by increasing the depth of RNA sequencing and concomitantly analyzing a large number of specimens and transcripts, more precise predictions will be made regarding signal integration. It is conceivable that this strategy can find application to the study of virtually all signaling networks at which variation in the response, accompanied by coordination in the expression of the corresponding transcripts, is anticipated. Whether in disease the coordination in particular signaling nodes is abolished, remains to be seen.
Supplementary Material
Funding Information
Peromyscus research in the Kiaris laboratory is supported by NSF (Award No. 1736150). The PGSC is supported by a grant from NSF (Award No. 1755670).
Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- Almanza A., Carlesso A., Chintha C., Creedican S., Doultsinos D., Leuzzi B., et al. (2018). Endoplasmic reticulum stress signalling—from basic mechanisms to clinical applications. FEBS J 286, 241–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dor A., Shamir R., and Yakhini Z. (1999). Clustering gene expression patterns. J Comput Biol 6, 281–297 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 57, 289–300 [Google Scholar]
- Brown B.C., Bray N.L., and Pachter L. (2018). Expression reflects population structure. PLoS Genet 14, e1007841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Li Q., Du Y., Yun J., Xie Y., DeBerardinis R.J., et al. (2017). Genomic regression analysis of coordinated expression. Nat Commun 8, 2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W.-C., Messah C., and Lin J.H. (2012). IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell 23, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Lipson D., Yogev S., and Yakhini Z. (2007). Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., and Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.L., and Gao D.S. (2014). Endoplasmic reticulum proteins quality control and the unfolded protein response: the regulative mechanism of organisms against stress injuries. Biofactors 40, 569–585 [DOI] [PubMed] [Google Scholar]
- Han J., and Kaufman R.J. (2017). Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev 31, 1417–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- Havighorst A., Crossland J., and Kiaris H. (2017). Peromyscus as a model of human disease. Semin Cell Dev Biol 61, 150–155 [DOI] [PubMed] [Google Scholar]
- Havighorst A., Zhang Y., Farmaki E., Kaza V., Chatzistamou I., and Kiaris H. (2019). Differential regulation of the unfolded protein response in outbred deer mice and susceptibility to metabolic disease. Dis Model Mech 12, dmm037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Chevet E., and Oakes S.A. (2015). Proteostasis control by the unfolded protein response. Nat Cell Biol 17, 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhiainen A., Thomsen C., Strömbom L., Grundevik P., Andersson C., Danielsson A., et al. (2012). Distinct cytoplasmic and nuclear functions of the stress induced protein DDIT3/CHOP/GADD153. PLoS One 7, e33208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S., and Silver P.A. (2008). Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet 9, 38–48 [DOI] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., and Glimcher L.H. (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemus L., and Goder V. (2014). Regulation of endoplasmic reticulum-associated protein degradation (ERAD) by Ubiquitin. Cells 3, 824–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M., Eletto D., and Argon Y. (2012). GRP94: an HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim Biophys Acta 1823, 774–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., and Smyth G.K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.K., McLachlan G.L., Wang K., Ben-Tovim Jones L., and Ng S.-W. (2006). A mixture model with random-effects components for clustering correlated gene-expression profiles. Bioinformatics 22, 1745–1752 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D.T., Wu J., Back S.H., Callaghan M.U., Ferris S.P., Iqbal J., et al. (2008). UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 15, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., et al. (2003). Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep 3, 1279–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., and Samali A. (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Y., Liu C., Yu S., Yang H., Sun J., Guo C., et al. (2018). Gene co-expression network analysis reveals coordinated regulation of three characteristic secondary biosynthetic pathways in tea plant (Camellia sinensis). BMC Genomics 19, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- Wei P., Milbauer L.C., Enenstein J., Nguyen J., Pan W., and Hebbel R.P. (2011). Differential endothelial cell gene expression by African Americans versus Caucasian Americans: a possible contribution to health disparity in vascular disease and cancer. BMC Med 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., et al. (2007). Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson R.B., Komuro R., Chen X., Davé UP, Prywes R., et al. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6, 1355–1364 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Shu N., Liu Y., Song M., Hao Y., Liu H., et al. (2008). Altered resting-state functional connectivity and anatomical connectivity of hippocampus in schizophrenia. Schizophr Res 100, 120–132 [DOI] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., et al. (1998). CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.