Abstract
Wheat is grown on more land than any other crop in the world. Current estimates suggest that yields will have to increase sixty percent by 2050 to meet the demand of an ever‐increasing human population; however, recent wheat yield gains have lagged behind other major crops such as rice and maize. One of the reasons suggested for the lag in yield potential is the lack of a robust hybrid system to harness the potential yield gains associated with heterosis, also known as hybrid vigor. Here, we set out to identify candidate genes for a genic hybrid system in wheat and characterize their function in wheat using RNASeq on stamens and carpels undergoing meiosis. Twelve genes were identified as potentially playing a role in pollen viability. CalS5‐ and RPG1‐like genes were identified as pre‐ and post‐meiotic genes for further characterization and to determine their role in pollen viability. It appears that all three homoeologues of both CalS5 and RPG1 are functional in wheat as all three homoeologues need to be knocked out in order to cause male sterility. However, one functional homoeologue is sufficient to maintain male fertility in wheat.
Keywords: callose synthase, gene editing, hybrid wheat, pollen fertility, ruptured pollen grain‐1
Significance Statement.
We report the identification and characterization of CalS5‐ and RPG1‐like genes as pre‐ and post‐meiotic candidate genes for a genic hybrid system in wheat. CRISPR/Cas9 editing of all six alleles for each gene in several lines results in aberrant pollen morphology and infertility—however, the presence of one allele of TaCalS5 or TaRPG1 is sufficient for fertility, as plants with only one non‐mutated allele were fertile.
1. INTRODUCTION
Wheat is grown on more land than any other crop in the world, with an estimated 240 million hectares grown in 2017 (FAO, 2017). Wheat production is second in the world, after maize, in terms of total tonnage created with nearly 750 million tons produced in 2016 (FAO, 2017). It has been estimated that to meet future demand of an ever‐increasing human population, yields will have to increase sixty percent by 2050 (Tilman, Balzer, Hill, & Befort, 2011). However, yield gains in wheat over the past few years have lagged behind other major crops such as rice and maize (Ray, Mueller, West, & Foley, 2013). One of the main reasons suggested for the lag in yield gains is the lack of a robust hybrid system to harness the potential yield gains associated with heterosis, also known as hybrid vigor. A second potential reason is the lack of focused wheat breeding resources compared with hybrid maize (Whitford et al., 2013). Hybrid systems are thought to offer two advantages: The first is heterotic yield, with gains of over 10% in many crops species easily obtainable (Longin et al., 2013; Longin & Würschum, 2014; Mühleisen, Piepho, Maurer, Longin, & Reif, 2014), and the second is as a driver for private investment in new and elite germplasm to further drive yield gains in prebreeding. However, due to the complex nature of wheat, with a large genome (17 Gb) and allohexaploid nature, the identification of genes involved in male sterility has been hard to establish. This is mainly due to the fact that the wheat genome is approximately forty times larger than rice (0.430 Gb) and seven times larger than maize (2.5 Gb). The added layer of redundancy that polyploids exhibit has meant that mutant screens have produced very few candidate genes involved in male sterility. Therefore, until the recent release of a reference wheat genome, little progress could be made with single mutations or mutant screens (Chapman et al., 2015; Clavijo et al., 2017; Mayer et al., 2014; Zimin et al., 2017).
Wheat being a species which regularly self‐fertilizes is considered as a cleistogamous pollinator, meaning it fertilizes its own stigma before the flower opens. Overcoming the cleistogamy to force outcrossing requires the modification of either timing of pollen release or a mutation to cause sterility (De Vries, 1971). The main mode of action for male sterility usually involves the disruption of the pollen cell wall. The pollen cell wall consists of a simple inner intine layer which is mainly composed of pectin and cellulose, and an intricate outer exine layer composed of sporopollenin, which is made up of fatty acids, phenylpropanoids, phenolics, and carotenoids. In diploid species such as Arabidopsis and rice, it has been found that many of the genes involved in exine formation are highly conserved and play the same roles in a number of plant species (Gómez, Talle, & Wilson, 2015). The two major components of the exine are callose and fatty acids. Callose, which is synthesized at the plasma membrane by an enzyme called callose synthase 5 (CalS5), plays multiple roles in pollen development, as it is involved in covering microspores as they form independent exine layers, but is also a major constituent of pollen tubes (Taylor & Hepler, 1997). The loss of CalS5 causes male sterility in Arabidopsis due to the degeneration of microspores in the mutant due to the failure of normal development of the bacula and tectum, which make up the sexine portion of the exine. Plant growth and development in cals5 mutants are otherwise normal, and female fertility is not affected (Dong, Hong, Sivaramakrishnan, Mahfouz, & Verma, 2005). CalS5 is thought mainly to be a pre‐meiotic gene as sterility from the lack of function is caused before the tetrad is formed (Nishikawa, Zinkl, Swanson, Maruyama, & Preuss, 2005). Other genes known to play a role in exine formation and targets for genic hybrid systems are DEX1 (Paxson‐Sowders, Dodrill, Owen, & Makaroff, 2001), NEF1 (Ariizumi et al., 2004), NPU (Chang et al., 2012), and RPG1 (Guan et al., 2008). The dex1 mutant has reduced and delayed exine formation, and the membrane undulation fails to form in dex1 mutants (Paxson‐Sowders et al., 2001). In the nef1 mutant, exine appears more coarse than that of the wild type, and nef1 microspore membrane undulation is abnormal (Ariizumi et al., 2004). In npu mutant, primexine is completely absent and no microspore plasma membrane undulation is observed (Chang et al., 2012). The RPG1 gene, a member of the MtN3+/saliva gene family, is important for primexine formation, although its detailed function is not currently clear.
While only one male fertility/sterility candidate gene from a model species has been translated to wheat or other major crops, a few loci in wheat have been identified. The single gene translated from a model species to wheat (ms1) was originally found in wheat almost 60 years ago as part of the identification of a number of male‐sterile mutants in wheat (Pugsley & Oram, 1959). Which is part of a suite of stable genic male sterility (GMS) loci (MS1–MS5) identified thus far in wheat (Driscoll, 1977; Fossati & Ingold, 1970; Pugsley & Oram, 1959; Sasakuma, Maan, & Williams, 1978; Zhou, Wang, Feng, Ji, & Wang, 2008), the five mutants identified contained ms1 and ms5 which are recessive mutants (Klindworth, Williams, & Maan, 2002; Sasakuma et al., 1978), and Ms2, Ms3, and Ms4 which are dominant mutants (Maan, Carlson, Williams, & Yang, 1987; Maan & Kianian, 2001; Qi & Gill, 2001). Currently, only one dominant gene, Ms2, and one recessive mutant have been cloned in wheat (Ni et al., 2017; Tucker, Baumann, & Kouidri, 2017; Wang et al., 2017; Xia et al., 2017), with Ms2 mutants being widely used for wheat breeding and potentially for hybrid wheat breeding (Ni et al., 2017). Other genes involved in male sterility in wheat include the genes Ms26 and Ms45 which are involved in the formation of the pollen cell wall similar to ms1 which is a transcription factor and regulates the post‐meiotic development of the anther (Dong et al., 2005; Singh, Kumar, Thilges, Cho, & Cigan, 2017; Tucker et al., 2017; Wang et al., 2017). Here, we set out to identify other genes involved in pollen viability in wheat and show their possible use as genes involved in a genic male sterility system. Two genes in particular, one gene from the literature listed as potentially a pre‐meiotic gene (TaCalS5) and one potential post‐meiotic gene (TaRPG1), are characterized for their role in wheat pollen development and viability (Dong et al., 2005; Guan et al., 2008).
2. MATERIALS AND METHODS
2.1. RNASeq
RNASeq was performed on total RNA extracted from stamen and pistil samples isolated from immature flowers at meiosis. Total RNA was extracted from three biologically replicated samples of developing stamens and carpels of wheat (Triticum aestivum) cultivar Fielder. Tissues were selected and dissected from wheat ears between the Zadok stages 41 and 49, and total RNA was isolated using Qiagen's RNeasy® Kit. Samples were then treated with DNAse to remove any further genomic contamination and purified using RNeasy MinElute® columns. Six RNASeq libraries (three from stamens and three from pistils) were generated for using Illumina HiSeq 2,500 paired‐end reads. The cDNA libraries were treated with the enzyme Ribo‐Zero (Illumina) to reduce the abundance of ribosomal RNAs before the libraries were run on the Illumina HiSeq2500. Sequencing was performed by Eurofins Genomics.
Reads obtained from the six libraries were analyzed using bioinformatics software tool “fastQC” to identify adapter contamination. Adapter contamination was removed from the reads using “cutadapt” software, and trimmed sequences were again run through fastQC to assure adapters had been removed. Trimmed reads were aligned to the TGAC cDNAs using Salmon (Patro, Duggal, Love, Irizarry, & Kingsford, 2017), and reads were quantified using EdgeR after removal of and cDNAs which had less than 10 total mapped reads total across the six libraries (McCarthy, Chen, & Smyth, 2012). A maximum FDR of 0.05 was used as a test of significance.
Validation of the RNASeq results was measured from two more biological replicated the same as above. cDNA synthesis was conducted on 500 ng of total RNA using Omniscript RT Kit (Qiagen). The cDNA was diluted 1:2 with water, and 0.5 µl was used as template in each RT‐PCR. Transcripts levels were quantified using SYBR Green JumpStartTaq ReadyMix (SIGMA) with the standard run conditions for the ABI 7900 HT. Four technical replicates were performed on each of the biological replicates. Two reference genes TaUbiquitin and TaEF1α were used for the normalization. The sequence of primers used in QPCR assays is shown in Table S9.
2.2. Bioinformatic comparison
Putative gene targets were identified from the RNASeq dataset, and identifiers were found in ENSEMBL plants (http://plants.ensembl.org/index.html) and the BioMart option of the site was used to identify conserved PFam domains listed under the transcript portion of a fertility candidate. The populated list of putative genes with the family‐specific PFam domain was then exported as either a fasta format to compare sequences information or .xls format for other data manipulation of the dataset. Splice variants of the same unique identifier in the wheat genome were then removed so only one variant was used for comparison. In most cases, if the gene had no prior characterization, the first splice variant was used for simplicity in the comparison.
RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐8675.
2.3. CRISPR‐mediated knockout
To produce plants with targeted mutations in TaRPG1 and TaCalS5‐like genes, we used a CRISPR Cas9 system with a wheat codon‐optimized Cas9 to introduce mutations in wheat plants (Howells, Craze, Bowden, & Wallington, 2018). We targeted TaRPG1‐ and TaCalS5‐like genes with four guide RNAs for each set of homoeologues. To identify the target sequences in these genes, we used the publicly available program DREG (http://emboss.sourceforge.net/apps/cvs/emboss/apps/dreg.html) to find sequences that match either 5’‐A(N)20GG‐3’ or 5’‐G(N)20GG‐3’ in both orientations of the Chinese Spring genomic sequence. We then selected four guides based on the following criteria: that the target sequence was conserved in all three homoeologues, that it was (at least partially) in an exon of TaRPG1‐ and TaCalS5‐like genes, and that it had a restriction enzyme site near the site of the protospacer associated motif (PAM) but in the sequence of the guide RNA and prioritized guides near the start of the coding sequences of each gene. The guide sequences selected are shown in Table S10. For targeting TaCalS5, one guide was driven by the OsU3, OsU6, TaU3, and TaU6 promoters with a total of four guides targeting TaCalS5 genes. For targeting the TaRPG1‐like gene, we duplicated the TaU6 promoter as we were unable to find a sequence in the TaRPG1 gene that could fulfill all our criteria for quality guides. These two promoter‐guide constructs were then synthesized (GenScript) and subsequently cloned into an intermediate vector containing attL1 attR5 flanking sites prior to a 2‐way MultiSite Gateway recombination with a wheat codon‐optimized cas9 sequence driven by the maize ubiquitin promoter flanked by attL5 and attL2 sites, into the final binary vector (Figures S4 and S5). Completed constructs were verified by restriction digest and sequencing before being electro‐transformed into Agrobacterium tumefaciens. Plasmids were re‐isolated from Agrobacterium cultures and verified by restriction digest prior to use in wheat experiments (Bates, Craze, & Wallington, 2017).
2.3.1. Wheat transformation
Wheat variety Fielder plants were grown in controlled environment chambers (Conviron) at 20°C day/15°C night with a 16‐hr day photoperiod (approximately 400 μE m−2 s−1). Immature seeds were harvested for transformation experiments at 14–20 days post‐anthesis (dpa). Isolated immature wheat embryos were co‐cultivated with Agrobacterium tumefaciens for 2 days in the dark (Ishida, Tsunashima, Hiei, & Komari, 2015).
Subsequent removal of the embryonic axis and tissue culture was performed as described by Risacher, Craze, Bowden, Paul, and Barsby (2009). Individual plantlets were hardened off following transfer to Jiffy‐7 pellets (LBS Horticulture), potted up into 9‐cm plant pots containing M2 and grown on to maturity, and seed harvest in controlled environment chambers, as above.
2.3.2. DNA analysis of transformed wheat plants
Plantlets which regenerated under G418 selection in tissue culture and transferred to Jiffy‐7 pellets were validated using an nptII copy number assay relative to a single‐copy wheat gene amplicon, GaMyb, normalized to a known single‐copy wheat line (Milner et al., 2018). Plants were then screened for mutations using a PCR‐based method where the PCR products were designed to be homoeologue‐specific and covering all four guides or all three homoeologues containing a pair of guides.
2.4. Plant rescue of male sterility after CRISPR knockouts
Plants which appeared sterile were then fertilized with Wt Fielder pollen to rescue the male sterility and generate T1 seed. All sterile plants had two ears left unfertilized to check male sterility was not tiller specific.
3. RESULTS
3.1. RNASeq to identify pollen‐specific expressed genes
To identify candidate genes for use in a genic male sterility system, a differential expression analysis was performed on immature stamen and pistil tissues from wheat (cv. Fielder) inflorescences harvested between Zadok stages 41 and 49 (the “booting” stage of wheat development when meiosis takes place). Reads were mapped to the current wheat transcriptome (ref_seq v1.1). From this analysis, a total of 19,490 genes were found to be differentially expressed between the two tissues. A total of 10,474 were more highly expressed in stamens (Table S1), whereas 9,016 genes were more highly expressed in pistils (Table S2). GO analysis did not reveal informative GO terms associated with gene expressed in the stamens compared with pistil tissues (Tables S3 and S4).
Ten genes, which could be inferred to known male‐sterile phenotypes in other plant species or were members of the Sugars Will Eventually be Exported Transporter (SWEET) family and highly expressed in the stamens, were prioritized as potential stamen‐specific candidates for knockdown by RNAi silencing. Clear orthologs could be found in the genome sequence publicly available at that time (2015), and three homoeologues were easily identified (Table 1). Five potential candidates, which met these criteria and which showed differential expression based on the RNASeq data, were further confirmed by qRT‐PCR with two new biological samples. All five of the genes chosen were confirmed to be differentially expressed using qRT‐PCR although some of the values were greater or less than measured by RNASeq (Figure S1).
Table 1.
Candidate genes which could be used in a genic male sterility hybrid system identified from differential expression of developing wheat stamens and carpels
| Potential candidate | BLAST hit | TGAC v1 gene model | TGAC v1 homeologues | Significant homoeologues |
|---|---|---|---|---|
| 1 | RPG1 (Ruptured Pollen Grain1) like | TRIAE_CS42_7DL_TGACv1_603435_AA1983700 | TRIAE_CS42_7AL_TGACv1_556969_AA1774370; TRIAE_CS42_7BL_TGACv1_580455_AA1914070 | ABD |
| 2 | Callose synthase 5 | TRIAE_CS42_7BS_TGACv1_593715_AA1953990 | TRIAE_CS42_7AS_TGACv1_569258_AA1811650; TRIAE_CS42_7DS_TGACv1_622598_AA2042310 | ABD |
| 3 | Aborted microspore 1 like | TRIAE_CS42_6AS_TGACv1_486918_AA1566480 | TRIAE_CS42_6BS_TGACv1_514404_AA1659330; TRIAE_CS42_U_TGACv1_643846_AA2135420 | ABD |
| 4 | RPG1 (Ruptured Pollen Grain1) like | TRIAE_CS42_5BS_TGACv1_423307_AA1373980; | TRIAE_CS42_5AS_TGACv1_393366_AA1271880; TRIAE_CS42_5DS_TGACv1_457788_AA1489840 | AB |
| 5 | bHLH91 | TRIAE_CS42_2AL_TGACv1_094707_AA0301850 | TRIAE_CS42_2BL_TGACv1_129925_AA0399500; TRIAE_CS42_2DL_TGACv1_158620_AA0523420 | ABD |
| 6 | GAMYB (AtMYB101) | TRIAE_CS42_6AS_TGACv1_485682_AA1550030 | TRIAE_CS42_6DS_TGACv1_543879_AA1744870 | AD |
| 7 | Hothead | TRIAE_CS42_4BL_TGACv1_320326_AA1035360 | TRIAE_CS42_4DL_TGACv1_343496_AA1135340; TRIAE_CS42_5AL_TGACv1_375593_AA1224180 | ABD |
| 8 | Hothead | TRIAE_CS42_6DL_TGACv1_527115_AA1698830 | TRIAE_CS42_6AL_TGACv1_470984_AA1500160; TRIAE_CS42_6BL_TGACv1_500863_AA1610910 | ABD |
| 9 | Member of the sweet family | TRIAE_CS42_2DS_TGACv1_177708_AA0582810 | TRIAE_CS42_2AS_TGACv1_113352_AA0354890; TRIAE_CS42_2BS_TGACv1_149844_AA0497680 | ABD |
| 10 | member of the sweet family | TRIAE_CS42_7AS_TGACv1_570345_AA1834200 | TRIAE_CS42_7BS_TGACv1_591914_AA1925470 | A |
| 11 | Similar to OsSweet7e | TRIAE_CS42_U_TGACv1_640821_AA2075730 | no strong hit | U |
| 12 | Sweet4 | TRIAE_CS42_1DL_TGACv1_065128_AA0236610 | TRIAE_CS42_1AL_TGACv1_002319_AA0040790; TRIAE_CS42_1BL_TGACv1_030610_AA0095680 | ABD |
| 13 | Hothead | TRIAE_CS42_1DL_TGACv1_063432_AA0227210 | TRIAE_CS42_1AL_TGACv1_001690_AA0034080; TRIAE_CS42_1BL_TGACv1_032570_AA0131570 | ABD |
| 14 | Ms26 | TRIAE_CS42_4AS_TGACv1_308399_AA1027760.1 | TRIAE_CS42_4BL_TGACv1_321123_AA1055760; TRIAE_CS42_4DL_TGACv1_345634_AA1154040 | ABD |
NB Most of the above genes are shown in Table 2 of patent filing PCT/US2017/043009; they are designated Mfw genes cross‐referenced to the above Blast Hits, etc. A FDR cutoff of 0.05 was used for significance. Shown are the TGAC v1 representative gene models along with putative homeologues and whether the homoeologue(s) were also significantly differentially expressed.
Two candidate genes were selected based on their characterization in other plant species as being either pre‐ or post‐meiotic developmentally sterile, and one candidate from each category was chosen for further study. Each gene had three clear homoeologues identified, plus significantly different expression in stamen and pistil tissues from the RNASeq dataset, which was also confirmed by qRT‐PCR analysis. The two candidates chosen for further study were a Ruptured Pollen Grain‐like (RPG1) and a Callose Synthase‐like (CalS5) gene which were selected for further analysis and to determine whether manipulation of these genes would result in male‐sterile wheat.
3.2. Bioinformatic analysis of TaCaLS5 and TaRPG1
To understand the conservation of the two candidate genes, public databases were used to ensure that the proper targets were identified. A conserved domain for each protein which helped to define the family was chosen to help identify all putative members of the family in wheat. For CalS5, searching for the PFam domain PF14288 in wheat returns 13 loci on the A genome, 14 loci on the B genome, 10 loci on the D genome, and one unanchored locus. A BLAST search of public EST databases showed that only one locus on A, B, and D genomes is expressed only in the anther or pollen and was likely to be homoeologous group loci. These loci TraesCS7A02G146200, TraesCS7B02G048700, and TraesCS7D02G147700 under current refseq v1.1 locations were all differentially expressed in our RNASeq dataset and thus would be the target of future characterization. The overall number of unique loci in the CalS family in wheat is similar to that of Arabidopsis which has 12 unique loci. A phylogenetic comparison of the 41 loci in wheat, listed in Table S5, relative to the AtCalS5 and OsCalS5 is shown in Figure S2.
The RPG1‐like genes which are members of the SWEET family of genes showed 108 loci in wheat with the PFam domain PF03083. Again there was an uneven distribution in the genome. There are 29 loci on the A genome, 39 on the B genome, 30 on the D genome, and 10 unanchored. Unlike the CalS5‐like candidates, there was not a clear RPG1‐like candidate as many different SWEET transporters were expressed in the pollen or stamens. Table S1 shows that there are different members of the SWEET family which are differentially expressed residing on chromosomes 1, 2, 3, 5, and 7. The three potential homoeologues on chromosome 7 were chosen for further analysis as these three were the most highly expressed of the potential candidate genes in a previous iteration of the genome. Although with current genome sequence a triplicate set of homoeologues can also be found on wheat chromosomes 1 and 2, for which the genes on chromosome 2 did match our cutoff (Table 1). However, since the 112 loci are more than three times the 17 members of the family found in Arabidopsis, redundancy and potentially miss‐assignment of orthologous genes may occur. A phylogenetic comparison of the 112 loci in wheat, listed in Table S6, relative to the AtRPG1 is shown in Figure S3.
3.3. CRISPR‐mediated knockout of TaRPG1 and TaCalS5
A construct was designed to reduce expression of the TaCalS5 and TaRPG1 genes simultaneously through RNAi silencing, with the potential to cause male sterility in wheat. Thirty‐nine positive transgenic plants were generated, of which two showed male sterility, with unfilled and collapsed pollen (data not shown). The two male‐sterile plants set seed only after crossing with wild‐type (Wt) pollen or pollen from another RNAi line.
To understand which, or both, pollen expressed genes were causing the sterility phenotype, four guides were designed to target all three homoeologues of either TaRPG1 or TaCalS5 (Figures S4 and S5). Wheat transformation experiments with two CRISPR/Cas9 constructs generated a total of 64 T0 plants with guides targeting TaRPG1 and 101 plants targeting TaCalS5. Nine of the 64 TaRPG1‐targeted plants (14%) were sterile, and 7 of the 101 TaCalS5‐targeted plants (7%) were sterile. Pollen from sterile plants was analyzed and showed the same phenotype as the RNAi lines, with unfilled and collapsed pollen grains (Figures 1, 2, 3, 4). All sterile plants from each of the CRISPR‐targeted genes were fertilized with Wt Fielder donor pollen to ensure sterility was indeed male specific. All sterile plants (9 TaRPG1 sterile plants and 7 TaCalS5 sterile plants) were rescued by Wt donor pollen, confirming the male‐sterile phenotype, and generating T1 seed for further analysis. No observable differences were detected in transformed plants compared with untransformed control Wt Fielder lines, except for the male sterility. The obvious phenotype of the male‐sterile plants was gaping phenotype of the flowers after anthesis (Figure 2b,c, and e) This suggests no deleterious effects from the loss of either of these genes on normal growth patterning or any other agronomic trait of interest (Figures 1, 2, 3, 4).
Figure 1.
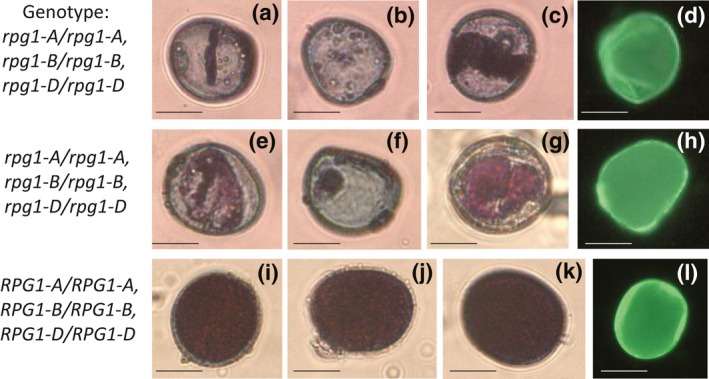
Disrupted pollen formation in TaRPG1. Mature pollen stained with either Alexander stain (a‐c, e‐g, i‐k) or Auramine‐O (d, h, l) from male‐sterile CRISPR lines RPG 12 with the genotype rpg1‐A/rpg1‐A, rpg1‐B/rpg1‐B, rpg1‐D/rpg1‐D (a‐d), and RPG 13 with the genotype rpg1‐A/rpg1‐A, rpg1‐B/rpg1‐B, rpg1‐D/rpg1‐D (e‐h), and Wt Fielder (i‐l) bar = 50 µm
Figure 2.

Plant growth and female fertility of TaRPG1 knockout lines 12 and 13. Images of whole plants (a), florets (b‐d), an ear (e‐f), and dissected flower (g‐h). Bar in A = 10cm, B‐H = 1mm
Figure 3.
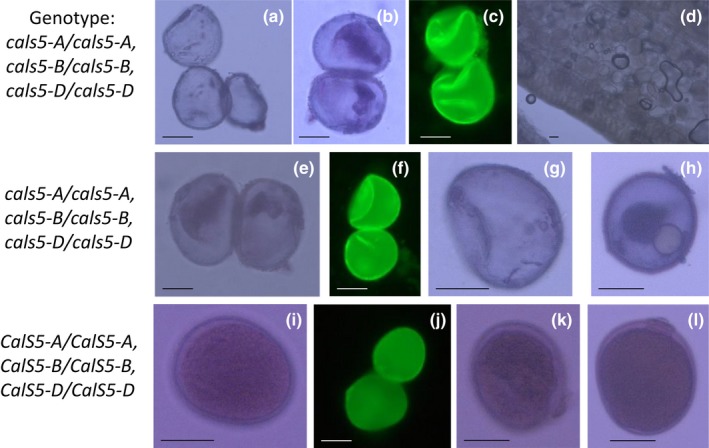
Disrupted pollen formation in TaCalS5. Mature pollen stained with either Alexander stain (a, b, d, e, g, h, i, k, and l) or Auramine‐O (c, f, j) from male‐sterile CRISPR lines CalS 5 with the genotype cals5‐A/cals5‐A, cals5‐B/cals5‐B, cals5‐D/cals5‐D, (a‐d), and CalS 17 with the genotype cals5‐A/cals5‐A, cals5‐B/cals5‐B, cals5‐D/cals5‐D, (e‐h), and Wt Fielder (i‐l) bar = 50 µm
Figure 4.

Plant growth and female fertility of TaCalS5 knockout lines 5 and 17. Images of dissected flower before fertilization (a) after fertilization could occur (b‐c), florets (d‐e), and the entire plant (f). Bar in a‐e = 1 mm, f = 10 cm
3.4. Identification of CRISPR‐mediated mutations in TaRPG1 and TaCalS5
To understand the basis of the sterility, the homoeologous regions targeted by the guides were sequenced in 20 plants per construct including both sterile and fertile plants. For guides targeting TaRPG1, two plants contained no mutations, 8 plants contained a range of mutations on either one or two of the homoeologues targeted, and two plants each contained a full knockout of all 6 alleles (Table S7). No plant with less than a mutation in all six RPG1 alleles showed a male‐sterile phenotype. All of the predicted changes to the coding sequences from Table S7 are shown in Figures [Link], [Link], [Link].
For TaCalS5‐targeted plants, only one plant contained no mutations at any of the four guide target sites sequenced. One plant (CalS 3) contained six different mutations but as one allele on the B genome contained a 3 bp mutation, resulting in 2 amino acids being substituted by one amino acid, with the rest of the protein conserved in frame, fertility was maintained. The range of deletions ranged from a deletion of 1,029 bases to the addition of 118 bases on one allele (Table S8). TaCalS5‐targeted plants required all six alleles to be mutated to cause male sterility; seven plants had a complete knockout of all six alleles and exhibited the resultant sterile phenotype. Again all of the predicted changes as a result of the mutations are shown in Table S8 and Figures [Link], [Link], [Link]. It should also be noted that neither of the OsU3 guides produced any mutations for either gene targeted. Of the male‐sterile plants identified for both TaRPG1 and TaCalS5 genes, every sterile plant had mutations in all six alleles. Among the fully fertile plants, two had only one Wt allele (with the other five being mutated): 1 TaRPG1 plant (RPG 11) and 1 TaCalS5 plant (CalS 3).
4. DISCUSSION
With an ever‐improving reference genome, identification of wheat orthologues involved in pollen development can now occur at greater speeds. This, along with the development of ever‐increasing public resources such as public expression databases and publicly available TILLING mutants, has led to a new dawn in the ability to finally translate some of the gains from other species such as Arabidopsis and rice to more complex genomes such as wheat. The release of the wheat genome data along with the rapid development of gene‐editing/genomic modification tools such as CRISPR‐Cas9 has resulted in a major step forward in the development of a hybrid wheat breeding system.
Here, we set out to identify genes which might be involved in pollen viability and show two such examples of how this can be applied to genic hybrid systems. From the original RNASeq dataset, we were able to identify fourteen candidate homologous gene sets as potentially being vital in the pollen fertility. From this list, two genes involved in cell wall formation were further characterized for their potential effects on creating sterility in wheat. As shown by CRISPR‐mediated knockout, both genes studied here are involved in maintaining the integrity of the pollen cell wall, while the bioinformatic comparison of the two genes characterized summarizes the potential transfer of knowledge from other plants to wheat. For example in the case of TaCalS5 there was a clear orthologous set of genes on chromosome 7; this along with the expression data from public databases clearly shows only one clear candidate showing about 77% homology to the Arabidopsis orthologue. This even carries through to the rough estimation of family size, as 12 genes were found in diploid Arabidopsis and 38 were found in hexaploid wheat. However, in the case of TaRPG1, the direct transfer of this knowledge was not straightforward as it was found that at least four separate homoeologue sets could potentially be involved in pollen development.
We identified 21 such members of the SWEET family of transporters which were differentially expressed in stamens versus pistils including homoeologous groups on chromosomes 1, 2, and 7. This is not surprising as in Arabidopsis at least three members of the family have already been shown to be important in pollen development or viability (AtSweet5, AtSweet8, and AtSweet13) (Engel, Holmes‐Davis, & Mccormick, 2005; Guan et al., 2008; Sun, Huang, Yang, Guan, & Yang, 2013). While we targeted the TaRPG1/Sweet8 member in our work presented here, there appears to be additional potential targets as genes for manipulation in a genic hybrid system. However, as our 21 loci were more highly expressed in stamen tissues, it would suggest that, at least in wheat, there may be more sugar transporters involved in pollen formation than in Arabidopsis and therefore greater potential for redundancy.
RNAi‐silencing, as an established technology, was used at the first stage to provide evidence of the genes involved in male fertility in wheat. However, its use in a commercial wheat hybrid system is of limited interest due to it having a “dominance” effect: The expression of one exogenous RNAi allele can silence all Wt alleles. In contrast, gene knockout technology, such as CRISPR‐Cas, creates mutant alleles which are usually recessive to wild‐type alleles—preferably to such an extent that just one Wt allele will suffice for expression. This has major attractions for a hybrid wheat system as an elite line (wild‐type male fertility) pollinator may be used to produce a fertile hybrid. This is why, when it became available, CRISPR‐mediated mutation was used in the second phase of our work. A workable model has been proposed by Wan et al. (2019) and has been slightly modified for this work highlighting the role of a genic male‐sterile system in wheat (Figure 5).
Figure 5.
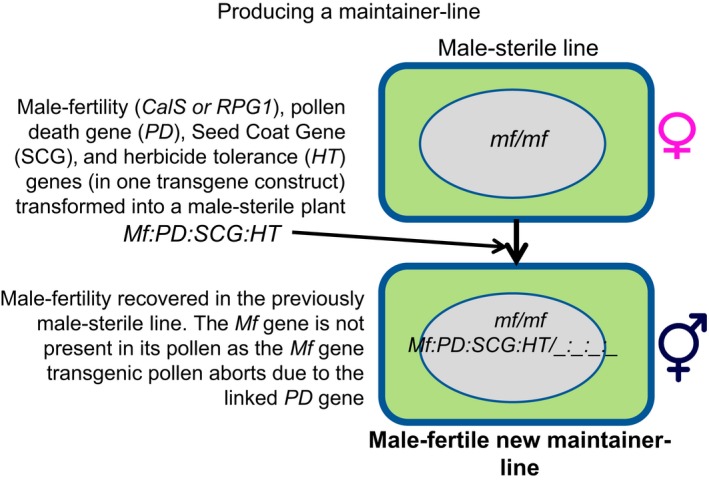
Scheme to produce a maintainer line using a genic male sterility gene to force outcrossing. Figure adapted from Wan et al., 2019. The system would also use other marker genes to prevent the release of the functional copy of the male sterility gene, and the selection for a heterozygote maintainer
Using CRISPR/Cas9 editing, we were fortunate in our ability to show a direct phenotype in the T0 gene‐edited plants; more work needs to be done to determine which homoeologue is the best candidate for a genic hybrid system with these two genes. Our data suggest that in contrast to Ms26 in wheat, one functional allele appears sufficient to allow normal seed set (Singh et al., 2017). We also show the power of combining multiple guides in one gene‐editing cassette as we achieved an overall success rate of knocking out all six alleles in 14% of the T0 plants in the case of TaRPG1‐targeted homoeologues. With the advances in guide prediction and a better understanding of promoters driving expression of the guides, this level of success may be increased to that of diploid species such as rice where it is routine to achieve mutations in 90% of the T0 plants regenerated (Miao et al., 2013). This can be illustrated in Figure 5 which shows how a genic male sterility system can be maintained using a genic male sterility gene (adapted from Wan et al., 2019).
This dataset can also be used for other future advancements in genic male fertility/sterility hybrid breeding systems as we identified 62 pentatricopeptide repeat‐containing proteins (PPR) which are the basis of a number of cytoplasmic male hybrid breeding systems in rice (Akagi et al., 2004; Dahan & Mireau, 2013; Hu et al., 2012; Kazama & Toriyama, 2003; Zhang, Lu, Bharaj, Virmani, & Huang, 1997). However, since the PPR family of proteins is a poorly conserved and complex family, identifying the direct orthologues of known rice genes is not a simple task (Dahan & Mireau, 2013). In our dataset, there were no clear orthologues of known PPRs which matched known rice Rf genes. A thorough understanding of the role played by these mitochondrial proteins and a route for their efficient manipulation for a functioning genic hybrid system to work are areas for future study and investment.
CONFLICT OF INTEREST
M.M. and A.K. have filed a patent application based on the results reported in this paper. All other authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
M.M., E.J.W., and A.K. designed the experiments and wrote the manuscript. M.M. conducted the experiments and analyzed the data. M.C., S.B., and R.B. generated the transgenic plants and assisted with microscopy.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Andy Greenland for his support and Nelzo Ereful for advice on analysis of RNASeq data. Elsoms Developments Ltd funded this work.
Milner MJ, Craze M, Bowden S, Bates R, Wallington EJ, Keeling A. Identification of genes involved in male sterility in wheat (Triticum aestivum L.) which could be used in a genic hybrid breeding system. Plant Direct. 2020;4:1–10. 10.1002/pld3.201
REFERENCES
- Akagi, H. , Nakamura, A. , Yokozeki‐Misono, Y. , Inagaki, A. , Takahashi, H. , Mori, K. , & Fujimura, T. (2004). Positional cloning of the rice Rf‐1 gene, a restorer of BT‐type cytoplasmic male sterility that encodes a mitochondria‐targeting PPR protein. Theoretical and Applied Genetics, 108, 1449–1457. 10.1007/s00122-004-1591-2 [DOI] [PubMed] [Google Scholar]
- Ariizumi, T. , Hatakeyama, K. , Hinata, K. , Inatsugi, R. , Nishida, I. , Sato, S. , … Toriyama, K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana . The Plant Journal, 39, 170–181. [DOI] [PubMed] [Google Scholar]
- Bates, R. , Craze, M. , & Wallington, E. J. (2017). Agrobacterium ‐mediated transformation of oilseed rape (Brassica napus). Current Protocols in Plant Biology, 2, 287–298. 10.1002/cppb.20060 [DOI] [PubMed] [Google Scholar]
- Chang, H.‐S. , Zhang, C. , Chang, Y.‐H. , Zhu, J. , Xu, X.‐F. , Shi, Z.‐H. , … Yang, Z.‐N. (2012). No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiology, 158, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, J. A. , Mascher, M. , Buluç, A. , Barry, K. , Georganas, E. , Session, A. , … Rokhsar, D. S. (2015). A whole‐genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biology, 16, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo, B. J. , Venturini, L. , Schudoma, C. , Accinelli, G. G. , Kaithakottil, G. , Wright, J. , … Clark, M. D. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Research, 27, 885–896. 10.1101/gr.217117.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan, J. , & Mireau, H. (2013). The Rf and Rf‐like PPR in higher plants, a fast‐evolving subclass of PPR genes. RNA Biology, 10, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. , Hong, Z. , Sivaramakrishnan, M. , Mahfouz, M. , & Verma, D. P. S. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. The Plant Journal, 42, 315–328. [DOI] [PubMed] [Google Scholar]
- Driscoll, C. J. (1977). Registration of cornerstone malesterile wheat Germplasm1 (Reg. No. GP 74). Crop Science, 17, 190. [Google Scholar]
- Engel, M. L. , Holmes‐Davis, R. , & Mccormick, S. (2005). Green sperm. Identification of male gamete promoters in arabidopsis. Plant Physiology, 138, 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2017). FAOSTAT (www.fao.com). Retrieved from http://www.fao.org/faostat/en/#home [Google Scholar]
- Fossati, A. , & Ingold, M. (1970). A male sterile mutant in Triticum aestivum. Wheat Information Service, 30, 8–10. [Google Scholar]
- Gómez, J. F. , Talle, B. , & Wilson, Z. A. (2015). Anther and pollen development: A conserved developmental pathway. Journal of Integrative Plant Biology, 57, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y.‐F. , Huang, X.‐Y. , Zhu, J. , Gao, J.‐F. , Zhang, H.‐X. , & Yang, Z.‐N. (2008). RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiology, 147, 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells, R. M. , Craze, M. , Bowden, S. , & Wallington, E. J. (2018). Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biology, 18, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Wang, K. , Huang, W. , Liu, G. , Gao, Y. A. , Wang, J. , … Zhu, Y. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in hong‐lian cytoplasmic male‐sterile lines via a complex with the glycine‐rich protein GRP162. The Plant Cell, 24, 109–122. 10.1105/tpc.111.093211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, Y. , Tsunashima, M. , Hiei, Y. , & Komari, T. (2015). Wheat (Triticum aestivum L.) transformation using immature embryos. Methods in Molecular Biology, 1223, 189–198. [DOI] [PubMed] [Google Scholar]
- Kazama, T. , & Toriyama, K. (2003). A pentatricopeptide repeat‐containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male‐sterile rice. FEBS Letters, 544, 99–102. [DOI] [PubMed] [Google Scholar]
- Klindworth, D. L. , Williams, N. D. , & Maan, S. S. (2002). Chromosomal location of genetic male sterility genes in four mutants of hexaploid wheat. Crop Science, 42, 1447–1450. [Google Scholar]
- Longin, C. F. H. , Gowda, M. , Mühleisen, J. , Ebmeyer, E. , Kazman, E. , Schachschneider, R. , … Reif, J. C. (2013). Hybrid wheat: Quantitative genetic parameters and consequences for the design of breeding programs. Theoretical and Applied Genetics, 126, 2791–2801. [DOI] [PubMed] [Google Scholar]
- Longin, C. F. H. , & Würschum, T. (2014). Genetic variability, heritability and correlation among agronomic and disease resistance traits in a diversity panel and elite breeding material of spelt wheat. Plant Breeding, 133, 459–464. [Google Scholar]
- Maan, S. S. , Carlson, K. M. , Williams, N. D. , & Yang, T. (1987). Chromosomal arm location and gene‐centromere distance of a dominant gene for male sterility in wheat 1. Crop Science, 27, 494–500. [Google Scholar]
- Maan, S. S. , & Kianian, S. F. (2001). Third dominant male sterility gene in common wheat. Wheat Information Service, 93, 27–31. [Google Scholar]
- Mayer, K. F. X. , Rogers, J. , Dole el, J. , Pozniak, C. , Eversole, K. , Feuillet, C. , … Praud, S. (2014) A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science, 345, 1251788. [DOI] [PubMed] [Google Scholar]
- McCarthy, D. J. , Chen, Y. , & Smyth, G. K. (2012). Differential expression analysis of multifactor RNA‐Seq experiments with respect to biological variation. Nucleic Acids Research, 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, J. , Guo, D. , Zhang, J. , Huang, Q. , Qin, G. , Zhang, X. , … Qu, L.‐J. (2013). Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Research, 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, M. J. , Howells, R. M. , Craze, M. , Bowden, S. , Graham, N. , & Wallington, E. J. (2018). A PSTOL‐like gene, TaPSTOL, controls a number of agronomically important traits in wheat. BMC Plant Biology, 18, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühleisen, J. , Piepho, H.‐P. , Maurer, H. P. , Longin, C. F. H. , & Reif, J. C. (2014). Yield stability of hybrids versus lines in wheat, barley, and triticale. Theoretical and Applied Genetics, 127, 309–316. [DOI] [PubMed] [Google Scholar]
- Ni, F. , Qi, J. , Hao, Q. , Luo, M. C. , Wang, Y. , Chen, F. , … Fu, D. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nature Communications, 8, 15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, S. , Zinkl, G. M. , Swanson, R. J. , Maruyama, D. , & Preuss, D. (2005). Callose (beta‐1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology, 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R. , Duggal, G. , Love, M. I. , Irizarry, R. A. , & Kingsford, C. (2017). Salmon provides fast and bias‐aware quantification of transcript expression. Nature Methods, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson‐Sowders, D. M. , Dodrill, C. H. , Owen, H. A. , Makaroff, C. A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiology, 127, 1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Pugsley, A. T. , & Oram, R. N. (1959). Genic male sterility in wheat. Australian Plant Breeding and Genetics Newsletter, 14, 10–11. [Google Scholar]
- Qi, L. L. , & Gill, B. S. (2001). High‐density physical maps reveal that the dominant male‐sterile gene Ms3 is located in a genomic region of low recombination in wheat and is not amenable to map‐based cloning. Theoretical and Applied Genetics, 103, 998–1006. 10.1007/s001220100699 [DOI] [Google Scholar]
- Ray, D. K. , Mueller, N. D. , West, P. C. , & Foley, J. A. (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE, 8, e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher, T. , Craze, M. , Bowden, S. , Paul, W. , & Barsby, T. (2009). Highly efficient agrobacterium‐mediated transformation of wheat via in planta inoculation. Methods Molecular Biology, 478, 115–124). [DOI] [PubMed] [Google Scholar]
- Sasakuma, T. , Maan, S. S. , & Williams, N. D. (1978). EMS‐induced male‐sterile mutants in euplasmic and alloplasmic common wheat 1. Crop Science, 18, 850–853. [Google Scholar]
- Singh, M. , Kumar, M. , Thilges, K. , Cho, M.‐J. , & Cigan, A. M. (2017) MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE, 12, e0177632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M.‐X. , Huang, X.‐Y. , Yang, J. , Guan, Y.‐F. , & Yang, Z.‐N. (2013). Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reproduction, 26, 83–91. 10.1007/s00497-012-0208-1 [DOI] [PubMed] [Google Scholar]
- Taylor, L. P. , & Hepler, P. K. (1997). Pollen germination and tube growth. Annual Review of Plant Physiology and Plant Molecular Biology, 48, 461–491. [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. , & Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences, 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, E. J. , Baumann, U. , Kouidri, A. et al (2017). Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nature Communications, 8, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries, A. P. D. (1971). Flowering biology of wheat, particularly in view of hybrid seed production ‐ A review. Euphytica, 20, 152–170. 10.1007/BF00056076 [DOI] [Google Scholar]
- Wan, X. , Wu, S. , Li, Z. , Dong, Z. , An, X. , Ma, B. , … Li, J. (2019). Maize genic male‐sterility genes and their applications in hybrid breeding: progress and perspectives. Molecular Plant, 12, 321–342. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Li, J. , Chen, S. , Heng, Y. , Chen, Z. , Yang, J. , … Ma, L. (2017). Poaceae‐specific MS1 encodes a phospholipid‐binding protein for male fertility in bread wheat. Proceedings of the National Academy of Sciences of the United States of America, 114, 12614–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford, R. , Fleury, D. , Reif, J. C. , Garcia, M. , Okada, T. , Korzun, V. , & Langridge, P. (2013). Hybrid breeding in wheat: Technologies to improve hybrid wheat seed production. Journal of Experimental Botany, 64, 5411–5428. 10.1093/jxb/ert333 [DOI] [PubMed] [Google Scholar]
- Xia, C. , Zhang, L. , Zou, C. , Gu, Y. , Duan, J. , Zhao, G. , … Kong, X. (2017). A TRIM insertion in the promoter of Ms2 causes male sterility in wheat. Nature Communications, 8, 15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Lu, Y. , Bharaj, T. S. , Virmani, S. S. , & Huang, N. (1997). Mapping of the Rf‐3 nuclear fertility‐restoring gene for WA cytoplasmic male sterility in rice using RAPD and RFLP markers. Theoretical and Applied Genetics, 94, 27–33. 10.1007/s001220050377 [DOI] [PubMed] [Google Scholar]
- Zhou, K. , Wang, S. , Feng, Y. , Ji, W. , & Wang, G. (2008). A new male sterile mutant LZ in wheat (Triticum aestivum L.). Euphytica, 159, 403–410. 10.1007/s10681-007-9551-y [DOI] [Google Scholar]
- Zimin, A. V. , Puiu, D. , Hall, R. , Kingan, S. , Clavijo, B. J. , & Salzberg, S. L. (2017). The first near‐complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience, 6, 1–7. 10.1093/gigascience/gix097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


