Abstract
Premise
Despite myriad examples of local adaptation, the phenotypes and genetic variants underlying such adaptive differentiation are seldom known. Recent work on freezing tolerance and local adaptation in ecotypes of Arabidopsis thaliana from Italy and Sweden provides an essential foundation for uncovering the genotype–phenotype–fitness map for an adaptive response to a key environmental stress.
Methods
We examined the consequences of a naturally occurring loss‐of‐function (LOF) mutation in an Italian allele of the gene that encodes the transcription factor CBF2, which underlies a major freezing‐tolerance locus. We used four lines with a Swedish genetic background, each containing a LOF CBF2 allele. Two lines had introgression segments containing the Italian CBF2 allele, and two contained deletions created using CRISPR‐Cas9. We used a growth chamber experiment to quantify freezing tolerance and gene expression before and after cold acclimation.
Results
Freezing tolerance was lower in the Italian (11%) compared to the Swedish (72%) ecotype, and all four experimental CBF2 LOF lines had reduced freezing tolerance compared to the Swedish ecotype. Differential expression analyses identified 10 genes for which all CBF2 LOF lines, and the IT ecotype had similar patterns of reduced cold responsive expression compared to the SW ecotype.
Conclusions
We identified 10 genes that are at least partially regulated by CBF2 that may contribute to the differences in cold‐acclimated freezing tolerance between the Italian and Swedish ecotypes. These results provide novel insight into the molecular and physiological mechanisms connecting a naturally occurring sequence polymorphism to an adaptive response to freezing conditions.
Keywords: adaptive phenotypic plasticity, Arabidopsis, CBF2, CRISPR‐Cas9, cold acclimation, freezing tolerance, genotype–phenotype mapping, local adaptation, RNAseq, winter annual
Adaptation to local environments, particularly those where stress imposes strong selection, should involve phenotypic differentiation for ecologically important traits. Such locally adaptive traits are unlikely to be beneficial in contrasting environments (Clausen et al., 1940). More generally, fitness trade‐offs across environments are thought to drive biological diversification at multiple scales (MacArthur, 1972; Futuyma and Moreno, 1988; Whitlock, 1996; Hereford, 2009). Despite many dozens of empirical studies of local adaptation (reviewed by Hereford, 2009) and a growing number of examples mapping the genetic basis of fitness in ancestral environments (Lowry et al., 2009; Hall et al., 2010; Ågren et al., 2013, 2017; Anderson et al., 2013; Leinonen et al., 2013; Postma and Ågren, 2016), there are still very few cases where the traits underlying local adaptation have been identified and experimentally confirmed. Rarer still are examples of the molecular and physiological mechanisms of local adaptation (Anderson et al., 2011; Savolainen et al., 2013; Tiffin and Ross‐Ibarra, 2014; VanWallendael et al., 2019).
Identifying the genes that underlie local adaptation may provide critical data to bear, for example, on a longstanding debate about the importance of moderate‐ to large‐effect alleles in adaptation (Fisher, 1930; Kimura, 1983; Orr, 1998, 2005; Rockman, 2012; Lee et al., 2014; Rausher and Delph, 2015; Remington, 2015; Dittmar et al., 2016). While quantitative trait loci (QTL) detected in crosses between populations and species suggest that large effect alleles may commonly contribute to variation in ecologically important traits (Rausher and Delph, 2015; Remington, 2015), identifying the causal genes is important for ruling out the possibility these QTL actually represent many linked genes of small effect (c.f., Rockman, 2012).
Approximately two thirds of the land on earth experiences freezing temperatures at least occasionally during a given year (Larcher, 1980); therefore, freezing tolerance is one trait that is likely to have broad adaptive significance for many plant species. Freezing tolerance typically requires a period of cold acclimation, an extended period of cold, but nonfreezing temperatures (Thomashow, 1999, 2010; Preston and Sandve, 2013; Barrero‐Gil and Salinas, 2018), which induces major changes in gene expression, metabolism, and physiology. Typical changes during acclimation include increased production of soluble sugars and other compounds that decrease the freezing point of the cell as well as proteins and metabolites to stabilize membranes, reduce or resist ice re‐crystallization in extracellular spaces, and resist desiccation (Thomashow, 1999, 2010; Preston and Sandve, 2013; Barrero‐Gil and Salinas, 2018; Zuther et al., 2018). Thus, cold‐acclimated freezing tolerance is an example of adaptive phenotypic plasticity, in which cold temperatures trigger an inducible physiological mechanism to increase survival through subsequent freezing periods.
Connecting the causal chain between sequence polymorphism, molecular phenotypes, organismal phenotypes, and ultimately fitness in contrasting environments is not an easy task. It is well beyond the scope of any individual study to provide all the necessary information. One clear path toward linking sequence polymorphism to ecologically relevant traits, and ultimately to fitness, is to conduct detailed studies of the molecular and physiological mechanisms of adaptive traits in study systems for which local adaptation has already been demonstrated (Tonsor et al., 2005). Indeed, in their recent review on stress response networks in plant local adaptation, VanWallendael et al. (2019) highlight that “integrating field‐based studies of local adaptation with mechanistic physiological and molecular biology promises advances in multiple areas of plant science.”
Differences in freezing tolerance between locally adapted (Ågren and Schemske, 2012; Ågren et al., 2013; Oakley et al., 2014) ecotypes of Arabidopsis thaliana (hereafter Arabidopsis) from Sweden (SW) and Italy (IT) represent one such opportunity to uncover the genetic and physiological mechanisms of plant interactions with a stressful environment in the context of local adaptation. In over 8 years of field experiments, QTL for local adaptation in a cross between SW and IT have been mapped (Ågren et al., 2013; Postma and Ågren, 2016; C. G. Oakley and J. Ågren, unpublished data). In field and laboratory studies, our group has identified freezing as a major selective agent in SW (Ågren and Schemske, 2012; Ågren et al., 2013; Oakley et al., 2014) and found large‐effect freezing tolerance QTL in the same genomic regions as the QTL for local adaptation and fitness trade‐offs (Oakley et al., 2014).
The causal variant underlying the largest‐effect freezing‐tolerance locus between the SW and IT ecotypes has been identified as a loss‐of‐function mutation in the IT allele of the gene encoding the transcription factor CBF2 (Gehan et al., 2015). The causal nature of CBF2 for this locus has been functionally validated using electrolyte‐leakage assays for freezing tolerance in both transgenic and CRISPR mutant lines (Gehan et al., 2015; Park et al., 2018). CBF2 is well known to be a major regulator of freezing tolerance in both the common Col‐0 ecotype and in other natural accessions of Arabidopsis (Thomashow, 1999, 2010; Alonso‐Blanco et al., 2005; Park et al., 2015; Barrero‐Gil and Salinas, 2018). The CBF genes generally, and CBF2 in particular, have been shown to mediate large scale changes in gene expression in response to even short‐term cold acclimation (Hannah et al., 2006; Gehan et al., 2015; Park et al., 2015, 2018; Jia et al., 2016; Zhao et al., 2016; Shi et al., 2017).
Recent studies have primarily focused on the effects of loss‐of‐function mutations of all three CBF genes on freezing tolerance and global gene expression and typically identify over a hundred genes with differential expression after cold acclimation (Park et al., 2015, 2018; Jia et al., 2016; Zhao et al., 2016). Here we are specifically interested in the effects of CBF2 on differential cold‐acclimated gene expression and survival through freezing between the IT and SW ecotypes because of direct implications of loss of function of CBF2 for local adaptation. We hypothesized that CBF2 is a key regulator of adaptive phenotypic plasticity in the SW ecotype of Arabidopsis. By identifying the target genes specifically regulated by CBF2 during cold acclimation and examining what is known about the function of those genes, we aimed to gain a greater understanding of the genetic and physiological mechanisms through which CBF2 mediates freezing tolerance in the SW ecotype.
In this study, we used a growth chamber freezing assay and RNAseq to investigate the role of CBF2 in differential cold‐acclimated freezing tolerance between the SW and IT ecotypes. We quantified freezing tolerance as survival through freezing and also quantified differential gene expression before and after cold acclimation using two independent CBF2 loss‐of‐function mutant lines (produced using CRISPR‐Cas9) in the SW genetic background, as well as two near‐isogenic lines (NILs) where we have introgressed a small part of the IT genome including the nonfunctional CBF2 allele into the SW genetic background. We asked the following questions: (1) What proportion of the difference in freezing tolerance between the SW and IT ecotypes can be explained by a CBF2 loss‐of‐function mutation? (2) Which of the genes that exhibit differences in cold‐responsive expression between SW and IT are downstream targets of CBF2? (3) For these genes, how much of the differential cold‐acclimated expression between SW and IT is regulated by CBF2, and what is known about their molecular and physiological roles in freezing tolerance?
MATERIALS AND METHODS
Study system
Arabidopsis thaliana is a small selfing (Abbott and Gomes, 1989) annual with a wide native range in Europe, Asia, and Africa (Koornneef et al., 2004; Beck et al., 2008; Durvasula et al., 2017), where many populations exhibit a winter annual life history (Montesinos et al., 2009; Ågren and Schemske, 2012; Burghardt et al., 2016). At the IT site (Castelnuovo; 42°07′N, 12°29′E), seeds germinate in October and November, and plants experience cold but nonfreezing temperatures throughout the winter as rosettes (Ågren and Schemske, 2012). At the SW site (Rödåsen; 62°48′N, 18°12′E), seeds germinate in August and September, and seedlings experience low temperatures in the autumn before overwintering as rosettes (Ågren and Schemske, 2012). Both populations experience temperatures that trigger cold acclimation. The range of conditions over which cold acclimation is induced is poorly known and is likely to vary across taxa, but 4°C has been shown to induce cold acclimation in Arabidopsis thaliana (Alonso‐Blanco et al., 2005; Hannah et al., 2006; Zhen and Ungerer, 2008) and winter wheat (Zhu et al., 2014; Skinner, 2015). In winter at the SW site, soil temperatures are usually below freezing for more than 80 days, and temperatures can reach as low as −11°C, with air temperatures even colder (Oakley et al., 2014). Relative survival of the IT ecotype in Sweden has been shown to be positively correlated with winter minimum soil temperature across years (Ågren and Schemske, 2012), indicating strong temporal variation in the strength of selection imposed by freezing.
CRISPR and NIL construction
To mimic the loss‐of‐function mutation in CBF2 found in IT, we utilized the CRISPR/Cas9 system to generate two independent CBF2 loss‐of‐function mutant lines in the SW genetic background. We followed a multigenerational approach to create the CRISPR lines (Feng et al., 2014). Briefly, the 19‐bp oligonucleotides designed to target the coding region of CBF2 under control of the AtU6 promoter were cloned to a single binary vector (pCambia1300): CBF2, 5′‐TCGCCGCCATAGCTCTCCG‐3′. Seeds generated after a floral dip were exposed to an antibiotic medium to select the first generation of transformed seeds (T1), which were sequenced to confirm the CBF2 mutation. Transgenic plants with the CBF2 mutation were self‐pollinated for two generations to obtain T3 lines homozygous for the CBF2 loss‐of‐function mutations. The T3 lines were then backcrossed to SW to remove any possible insertional effects by the T‐DNA containing the CRISPR/Cas9 transgene. Two lines were produced (Appendix S1): SW:cbf2 a, which is the same line with a 19‐bp deletion in the coding region of CBF2 of Park et al. (2018), and SW:cbf2 b, with a 13‐bp deletion in the coding region of CBF2.
We also produced two independent NILs for the CBF2 region by crossing recombinant inbred lines with IT introgression segments including CBF2 to the SW ecotype. The backcrossed lines were then selfed for several generations, and lines of interest were genotyped using a combination of 2b‐RAD (Wang et al., 2012) and PCR‐based genotyping strategies. Two NILs were ultimately generated and used in this experiment, both with introgression segments that include CBF2: NIL R37, which has a 2.4‐Mb segment, and NIL R38, which has a 6.8‐Mb segment. Our use of both CRISPR lines and NILs in this experiment was motivated by a desire to link these results with field‐based estimates of survival and reproduction for plants with functional and nonfunctional CBF2 alleles. The inclusion of NILs in addition to the CRISPR lines here allows us to compare the effects of the native IT loss‐of‐function allele with those of experimental mutations. Having replicate lines of both types dramatically increases our confidence that the effects we observe in the CRISPR mutants are due to the loss of function of CBF2 and not to off‐target genes.
Freezing assay
To quantify the effect of the loss‐of‐function mutation in CBF2 on freezing tolerance, we exposed seedlings from six different lines (IT, SW, the two SW background NILs, and two SW background CRISPRs CBF2 loss‐of‐function lines) to a freezing assay in which seedlings in a growth chamber were subjected to a period of cold acclimation followed by freezing. The experiment was randomized in a stratified fashion in a complete block design. Each block consisted of two quartered petri dishes (i.e., eight cells total), and 12 individual seeds of each line were sown in one cell. There were 60 blocks in total, divided evenly among 10 trays to facilitate randomization within the growth chamber. This entire experimental design was repeated three times, with each temporally separated growth chamber experiment referred to as a batch.
The freezing assay protocol follows that of Oakley et al. (2014). The temperature and photoperiod conditions were loosely based on site‐level soil and air temperature data from the SW site and day length data from the U.S. Naval Observatory (Ågren and Schemske, 2012; Oakley et al., 2014). Briefly, seeds were sterilized using a 30% v/v bleach and 0.04% v/v Tween 20 solution for 10 min and suspended in 0.1% w/v Phytoblend agar (Caisson Laboratories, Smithfield, UT, USA) overnight in the dark at 4°C before sowing. All seeds were sown on autoclaved Gamborg's B‐5 basal salts (without sucrose) and Phytoblend agar and poured into sterilized petri dishes. The petri dishes were cold‐stratified in the dark at 4°C for 5 days to synchronize germination. This cold‐stratification period alone is insufficient to induce significant freezing tolerance (C. G. Oakley, unpublished data). This was followed by germination and early growth for 8 days in a growth chamber at 22°C, 16‐h day (16L:8D) with a photosynthetically active radiation (PAR) of 125 μmol photons m−2 s−1. After this period, we put lids on the trays to reduce drying of the agar medium and moved the trays to a chamber capable of freezing temperatures to initiate 10 days of cold acclimation (4°C, 10L:14D, 50 PAR). We next reduced the temperature to −2°C for 24 h and added shaved ice to each cell to facilitate ice nucleation (Smallwood and Bowles, 2002).
For the freezing period, plants were exposed to −7°C for 8 days, which represents a minimum soil temperature that plants might experience in a cold year in Sweden; previous work has shown that freezing at this temperature results in realistic (based on field survival, see below) and repeatable differences between the two ecotypes (Oakley et al., 2014). The duration of the freezing period was chosen based on practical constraints (Oakley et al., 2014). During this freezing period, the petri dishes were kept in the dark, as done previously (Alonso‐Blanco et al., 2005; Zhen and Ungerer, 2008; Kang et al., 2013; Oakley et al., 2014). Plants at the SW site are often covered in snow during freezing temperatures, which should minimize the exposure to light and thus photoperiod. To mitigate temperature variation within the chamber, we used supplemental fans and rotated trays twice a day.
After the freezing period, we brought the chamber up to 4°C for 24 h to gradually thaw the plants, followed by 48 h at 22°C before scoring. Although this protocol does not replicate natural conditions due to practical constraints, the relative survival of the IT and SW ecotypes after exposure to this protocol closely resembles overwinter survival at the SW site in cold years (Ågren and Schemske, 2012; Oakley et al., 2014), suggesting that we capture the essential elements of field conditions relevant to freezing tolerance.
We quantified freezing tolerance per cell as mean percentage survival after the freezing period. Some cells were not included in the freezing tolerance assay because the plants were sacrificed to collect RNA samples (see below). We excluded seedlings that did not develop true leaves, as preliminary results indicated that seedlings of this size are not freezing tolerant regardless of genotype. Plants included in the analysis typically had at least four true leaves and were assayed at a small size because freezing occurs early in the life history at the SW site (Oakley et al., 2014). Preliminary experiments with larger plants resulted in similar differences in freezing tolerance between the two ecotypes (D. W. Schemske, unpublished data). Of the total 942 cells included in the freezing assay, we excluded 97 cells because they contained fewer than four individual plants of sufficient size to collect freezing‐tolerance data. In the final data set, freezing tolerance was estimated for an average of 140.8 cells per line (range = 122–158), each containing an average of 8.26 individual plants for a grand total of 7005 individuals.
Freezing tolerance was analyzed with an analysis of variance with line as a fixed effect. Because of the limited number of batches (3), this factor was treated as a fixed effect. Block nested within batch was treated as a random effect, and significance was tested with a likelihood ratio test. We were primarily interested in the reduction in freezing tolerance resulting from a nonfunctional CBF2 allele; therefore, we limited pairwise comparisons to those involving the SW ecotype and tested these with a priori linear contrasts. With the exception of IT, which had about a 5‐fold greater number of zero values for cell mean freezing tolerance compared with the other lines (Appendix S2), the residuals of this model were approximately normally distributed with minimal heteroscedasticity. Reanalysis of a model excluding IT yielded qualitatively similar results for the overall effect of line and the pairwise contrasts to SW (not shown), so we proceeded with the full model. All statistics were performed in JMP v. 13 (JMP, 1989–2019).
RNA extraction
We randomly selected six blocks in the first batch to be completely harvested for RNA sequencing, and these blocks were excluded from the freezing‐tolerance assay. We harvested all available plant tissue (roots and leaves), 4 h after the lights came on to minimize the effects of circadian rhythm (Dong et al., 2011). We used the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) for RNA extraction using three replicates of each line at both pre‐acclimation (22°C) and post‐acclimation (4°C for 10 days) conditions. Total RNA was quantified and checked for quality using a Qubit Fluorometer (Life Technologies Holdings, Singapore) and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) at the RTSF Genomics Core at Michigan State University.
Sequencing
Samples were prepared using the Illumina TruSeq Stranded mRNA Library preparation kit (Illumina, San Diego, CA, USA) on a Perkin Elmer Sciclone NGS (Perkin Elmer, Waltham MA, USA). Completed libraries were quality checked and quantified using a combination of Qubit dsDNA HS and Caliper LabChipGX HS DNA (Perkin Elmer) assays. Libraries were pooled for multiplexed sequencing. Sequencing was carried out in a 1 × 50‐bp single‐end format using Illumina HiSeq 4000 SBS reagents. Base calling was done by Illumina Real Time Analysis (RTA) v2.7.6, and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.18.0.
RNA‐sequencing analyses
Quality control and mapping
Remaining adapter sequences were removed, bases with quality scores less than 5 were trimmed, and reads smaller than 33 bp were excluded using cutadapt v. 1.8.1 (Martin, 2011). Quality of the remaining reads was inspected using FastQC (Andrews, 2010). RNA‐sequencing (RNA‐seq) reads from the different genotypes were mapped to the Arabidopsis thaliana reference genome (TAIR10) using TopHat v. 2.1.0 (Kim et al., 2013). TopHat was run in default mode with the following exceptions: the minimum intron length was set to 10 and maximum to 15,000 bp. A GTF file (TAIR10) was used to assist in the mapping of known junctions. Read counts for each gene were obtained using HTSeq 0.6.1 (EMBL, Heidelberg, Germany) using the intersection‐noempty option to only include counts for reads mapping to one unique gene.
Differential gene expression
Differential expression analysis was implemented in R version 3.0.1 (R Core Team, 2011) with the edgeR package v. 3.22.3 (Robinson et al., 2010). Because estimates of differential gene expression can be inflated by weakly expressed genes, we included only genes with more than one read per million (>1 CPM) in at least two samples. We used the trimmed mean of M‐values (TMM) as our normalization method (Robinson et al., 2010).
Ultimately, we were interested in differential gene expression as an interaction between CBF2 alleles and the cold‐acclimation treatment (pre‐ vs. post‐cold acclimation), and the extent to which loss‐of‐function mutations in CBF2 can explain differential expression between IT and SW in response to cold acclimation. The one thing in common among the IT ecotype and the four total NIL and CRISPR lines (in a SW background) is a nonfunctional CBF2 allele. We therefore used five separate generalized linear models to test for an interaction between genotype and the cold‐acclimation treatment on gene expression. SW was included in all five comparisons (see Appendix S3: Eqs. 1–5 for the log2 fold‐change calculations that correspond to the hypotheses tested in these models). We first identified the genes with differential cold‐responsive expression between IT and SW, because it is only this set of genes that can explain differential freezing‐tolerance between these ecotypes (Appendix S3: Eq. 1). For this set, we consider only genes for which there was a significant (at a Benjamini‐Hochberg FDR corrected P value [hereafter PFDR] ≤ 0.05) interaction between genotype and cold acclimation treatment in the SW vs. IT comparison.
We then narrowed the set of genes that were differentially cold responsive between SW and IT (above) to identify only those genes that are regulated by CBF2 in response to cold acclimation. Each of the SW vs. NIL/CRISPR line comparisons is a measure of the effect of a loss‐of‐function mutation in CBF2 on cold‐responsive gene expression (Appendix S3: Eqs. 2–5). Because we have multiple independent comparisons, which is in and of itself an approach to reducing false positives, we took a modified approach to adjusting P values for multiple comparisons to minimize false negatives. We therefore considered genes as potential targets of CBF2 if they were identified as significant in the comparison between SW and IT (above) and also had a significant genotype × treatment interaction in all four of the pairwise comparisons of SW vs. NIL/CRISPR lines at an uncorrected P value < 0.05. These independent comparisons comprise stringent criteria for eliminating false positives without resorting to log2 fold‐change thresholds commonly employed in studies of differential gene expression, allowing us to potentially identify target genes of CBF2 with subtle cold responsiveness.
To assess the extent to which CBF2 is responsible for differential cold responsiveness between SW and IT of each of these genes, we used the values for the log2 fold‐change (LFC) of the pairwise contrasts from edgeR for each of the genes that met both of the significance criteria (see above). Using the difference in cold responsive gene expression between SW and IT as a frame of reference (Appendix S3: Eq. 1), we calculated the average proportion of the differential cold responsiveness between SW and IT (among Appendix S3: Eqs. 2–5) explained by the four CBF2 loss‐of‐function lines. For a given target gene, this value represents the contribution of CBF2 in regulating differential cold‐responsive expression between the SW and IT ecotypes. This approach to using the difference between two ecotypes as the frame of reference, rather than simply the background genotype, is novel and allows us to more directly relate these patterns to local adaptation.
We used RT‐qPCR to confirm the results of our RNAseq experiment for two genes that exhibited significant genotype × cold acclimation treatment interactions. RNA was extracted as described above, and RT‐qPCR was performed using the Luna Universal Onestep RT‐qPCR kit (New England Biolabs, Ipswich, MA, USA) on a Bio‐Rad CFX Connect Real‐Time PCR Detection System (Bio‐Rad Laboratories, Hercules, CA, USA). The threshold quantification cycle (Cq) was determined using Bio‐Rad CFX Manager version 3.1. Relative expression ratios were quantified based on the corresponding efficiency of the primers for each gene and the deviation of Cq values for each sample from the mean Cq values of the pre‐acclimation samples for each gene (Pfaffl, 2001), in relation to the housekeeping gene ACT2.
Gene ontology
To assess the function of genes that exhibited significant genotype by environment interactions in the SW vs. IT comparison, we performed a gene ontology (GO) enrichment analysis using the PANTHER v. 14 overrepresentation test, performed with the GO biological processes complete annotations for the complete Arabidopsis thaliana gene database (Mi et al., 2019). Fisher exact tests were used to estimate the GO term enrichment P values, and a false discovery rate adjustment of P values was calculated to correct for the large number of comparisons.
RESULTS
Freezing assay
Overall freezing tolerance for the 845 cells (see Materials and Methods) was 50.2% (SD = 34.5%), and ranged from 0% to 100% (Appendix S4). Genotype had a highly significant effect on freezing tolerance (F 5, 689 = 109.5, P < 0.001). This strong signal of genetically based differences in freezing tolerance was apparent even with significant variation among batches (F 2, 156 = 32.5, P < 0.001), and among blocks nested within batch (χ 2 = 124.8, df = 1, P < 0.001). Least square mean freezing tolerance for IT was 11.4%, which was significantly lower than that of SW of 71.9% (Table 1, Fig. 1). These differences in freezing tolerance between IT and SW are similar to differences in overwinter survival at the SW site in cold years (Ågren and Schemske, 2012; Oakley et al., 2014), so differences reported here are reflective of differences in a key fitness component in nature. All four lines with a nonfunctional CBF2 in the SW genetic background had significantly and substantially reduced freezing tolerance compared to SW (Table 1, Fig. 1). Absolute reductions in mean freezing tolerance compared to SW for these four lines ranged from 13.1% to 25.7%, explaining roughly 1/5 to 2/5 of the difference between SW and IT in mean freezing tolerance (Table 1, Fig. 1). Much of the variation among these four lines is attributable to the somewhat higher freezing tolerance of NIL R37 compared to the other 3 lines (Table 1, Fig. 1).
Table 1.
Least square mean freezing tolerance (FrzTol) for each of the six lines in the study. Also given is the reduction in FrzTol of each line compared to SW and the results of the linear contrast from the ANOVA test of the significance of these differences. The final column gives the reduction in freezing tolerance for each CBF2 loss‐of‐function line expressed as a proportion of the difference in freezing tolerance between SW and IT.
| Line | FrzTol (%) | Reduction compared to SW | Linear contrast compared to SW | Proportion of difference between SW and IT explained |
|---|---|---|---|---|
| SW | 71.9 | n/a | n/a | n/a |
| NIL R37 | 58.8 | 13.1 | F 1,688 = 24.8, P < 0.001 | 0.22 |
| NIL R38 | 50.4 | 21.5 | F 1,687 = 66.5, P < 0.001 | 0.36 |
| SW:cbf2 b | 51.1 | 20.8 | F 1,696 = 54.1, P < 0.001 | 0.34 |
| SW:cbf2 a | 46.2 | 25.7 | F 1,695 = 85.7, P < 0.001 | 0.43 |
| IT | 11.4 | 60.5 | F 1,690 = 490.5, P < 0.001 | n/a |
Figure 1.
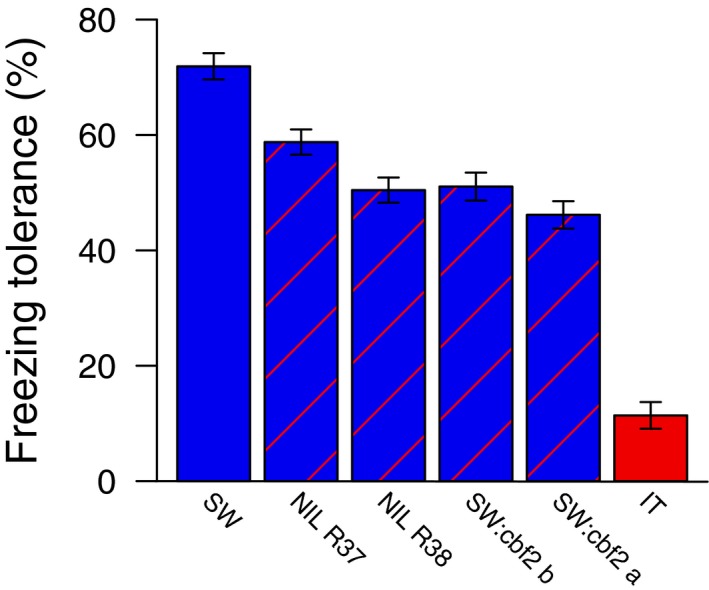
Mean freezing tolerance of the SW and IT ecotypes and the two NILs and two CRISPR mutant lines containing CBF2 loss‐of‐function alleles in the SW background. Error bars are 1 SE. Linear contrasts comparing SW to each of the other lines were all highly significant (Table 1).
Gene expression
Differences between SW and IT
There were 249 genes that were differentially cold responsive between SW and IT (genotype by treatment interaction) at P FDR ≤ 0.05 (Appendices S5, S6). These genes are involved in genetic pathways including response to stress (54/3157 annotated GO terms, P FDR < 0.001), response to abiotic stimulus (38/2099, P FDR = 0.033), and response to water deprivation (14/346, P FDR = 0.007), among others (Appendix S7).
Potential role of targets of CBF2 in mediating cold acclimation
The 249 genes described above likely include a set of genes directly involved in differential freezing tolerance between SW and IT. To isolate only the genes that are differentially regulated by CBF2 in response to cold acclimation, we examined patterns of cold‐responsive gene expression in the CBF2 loss‐of‐function lines. For all six lines, expression of CBF2 before cold acclimation was very low (<0.3 CPM) and was very high after cold acclimation (range = 17–33 CPM). For the pairwise comparisons of SW to NILs, 36 genes for NIL R37 and 43 for NIL R38 differed significantly in cold‐responsive expression between SW and IT (P FDR ≤ 0.05) and further had a significant genotype × cold acclimation treatment interaction at an uncorrected P < 0.05 (Fig. 2; Appendices S8, S9). There were 21 genes meeting both criteria in common between both NILs. For the pairwise comparisons of SW to CRISPR lines, there were 38 genes for SW:cbf2 a and 29 for SW:cbf2 b met both criteria described above (Fig. 2; Appendices S10, S11). There were 17 genes meeting both criteria in common between both CRISPR lines. There were only 10 cold‐responsive genes meeting both criteria in common among the four NILs and CRISPR lines (Fig. 2; Appendix S12). The 10 genes had annotations with significantly enriched gene ontology terms such as response to abiotic stimulus (GO:0009628), response to stress (GO:0006950), and response to water (GO:0009415).
Figure 2.
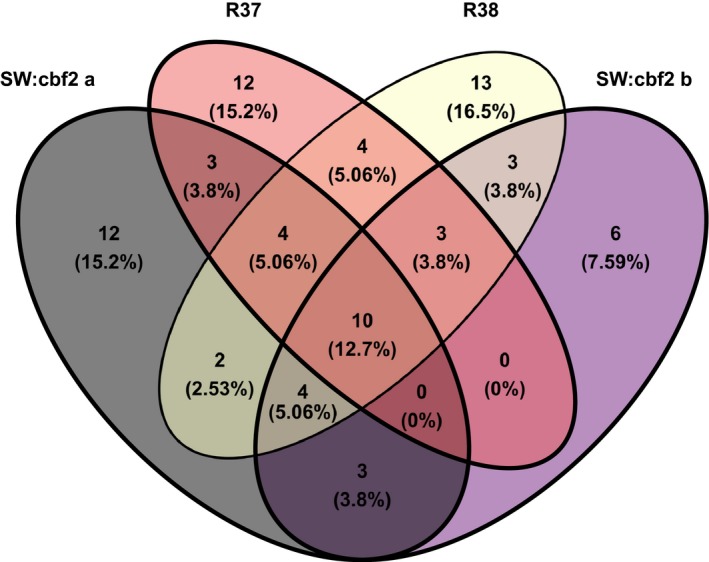
Venn diagram of the 79 genes that differ in cold‐responsive gene expression between SW and IT (of the 249 genes with a significant genotype by treatment interaction, PFDR ≤ 0.05) that also differ for at least one pairwise comparison of the NILs and CRISPR lines to SW (genotype × treatment interaction, P < 0.05). The plot was generated using the venn.diagram function in the R package Venn Diagram (Chen, 2018).
We further categorized these 10 genes based on the magnitude of differential expression between pre‐ and post‐cold acclimation treatments in SW and the average percentage of the difference between IT and SW that is attributable to CBF2 (Table 2). In other words, we categorized genes first based on how cold responsive they were in SW, then we quantified how much of the difference in cold responsiveness between SW and IT can be explained on average by loss‐of‐function mutations in CBF2 (see Appendix S3 for a full description of these calculations). The first category of genes comprises those that are very highly cold responsive (in terms of the log2 fold‐change in response to cold) in SW, GolS3 (LFCcaSW = 6.36) and AT4G30830 (LFCcaSW = 4.15; Table 2, Fig. 3; Appendix S12). GolS3 exhibited a striking pattern of cold acclimated gene expression where all four lines with loss‐of‐function mutations in CBF2 had nearly identical patterns of expression to IT (explaining on average 86% of the difference between SW and IT), suggesting that CBF2 almost completely mediates the difference between SW and IT in cold‐acclimated gene expression of GolS3. The relative expression patterns that we observed using RT‐qPCR for GolS3 were consistent with the results we obtained using RNAseq (Appendix S13). For AT4G30830, CBF2 could explain on average 44% of the log‐fold difference between SW and IT.
Table 2.
Differences in cold‐responsive gene expression between SW and IT and the average effect of CBF2 loss‐of‐function (LOF) mutations on cold‐responsive expression for the 10 candidate genes. Differences in cold responsiveness between SW and IT were calculated using Eq. 1 in Appendix S3. Average reductions in cold responsiveness due to CBF2 LOF were calculated as the average using Eqs. 2–5 in Appendix S3.
| Alias | Gene | Difference in cold responsiveness between SW and IT (log2 fold‐change) | Average reduction in cold responsiveness due to CBF2 LOF (log2 fold‐change) | Difference between SW and IT explained by CBF2 (%) |
|---|---|---|---|---|
| GolS3 | AT1G09350 | 2.53 | 2.16 | 86 |
| n/a | AT4G30830 | 2.71 | 1.18 | 44 |
| LEA14 | AT1G01470 | 0.91 | 0.75 | 81 |
| CCT2 | AT4G15130 | 0.85 | 0.59 | 69 |
| COR413‐PM1 | AT2G15970 | 0.98 | 0.52 | 53 |
| ERD10 | AT1G20450 | 1.53 | 0.75 | 49 |
| COR47 | AT1G20440 | 1.79 | 0.63 | 35 |
| ERD7 | AT2G17840 | 1.51 | 0.51 | 34 |
| n/a | AT3G55760 | 1.53 | 0.99 | 65 |
| DEAR3 | AT2G23340 | 0.86 | 0.73 | 84 |
Figure 3.
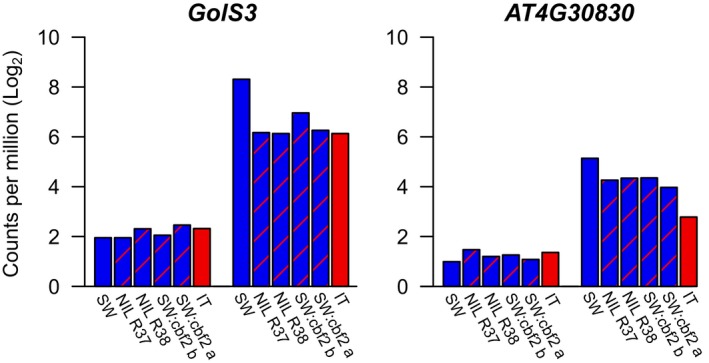
Log2 counts per million for the most highly cold‐responsive genes of the 10 candidates before (left group of bars) and after (right group of bars) cold acclimation.
The second category represents highly cold‐responsive genes (LFCcaSW between 1.91 and 2.70) in SW and included six genes LEA14, CCT2, COR‐413PM1, ERD10, COR47, and ERD7 (Table 2, Fig 4; Appendix S12). Among these, CBF2 explained the greatest difference in log2‐fold cold‐responsive gene expression between SW and IT for LEA14 (81%) and CCT2 (69%), with lower values for COR‐413PM1 (53%), ERD10 (49%), and even lower values for COR47 (35%) and ERD7 (34%). Some of these genes are therefore predominantly regulated by CBF2, whereas for others, CBF2 plays an important, but not predominant role in regulation. The relative expression patterns that we observed using RT‐qPCR for COR413‐PM1 were consistent with the results we obtained using RNAseq (Appendix S13).
Figure 4.
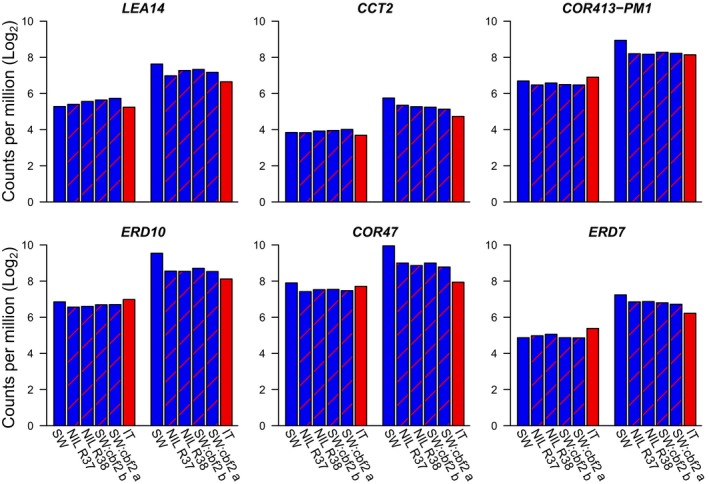
Log2 counts per million for the remaining highly cold‐responsive genes of the 10 candidates before (left group of bars) and after (right group of bars) cold acclimation.
The final category of genes comprises those that are modestly (AT3G55760, LFCcaSW = 1.18) or weakly (DEAR3, LFCcaSW = 0.43) cold responsive in SW (Table 2; Appendices S12, S14). Despite the limited cold responsiveness of these genes in SW, CBF2 could explain a large proportion of differential log2‐fold cold responsiveness between SW and IT, 65% for AT3G55760 and 84% for DEAR3.
DISCUSSION
Freezing tolerance is likely to be a key adaptation to stressful environments for many plants, and because freezing tolerance requires cold acclimation, it likely represents adaptive phenotypic plasticity. We found that a single mutation in CBF2 drives major differences between the SW and IT ecotypes in adaptive phenotypic plasticity in the form of cold acclimation, explaining one third of the differential freezing tolerance between the SW and IT ecotypes. Our approach using four independent genetic lines with loss‐of‐function mutations in CBF2 and explicitly testing for genotype × treatment interactions for gene expression identified a remarkably short list of 10 candidate target genes that may play an important role in cold‐acclimated freezing tolerance mediated by CBF2.
Loss of function of CBF2 has a large effect on freezing tolerance
The reduction in freezing tolerance of the NILs and CRISPR lines relative to SW provides direct evidence that the naturally occurring loss‐of‐function mutation in the IT allele of CBF2 drives approximately one third of the differential survival through freezing between SW and IT. The absolute effect size observed here is somewhat lower than a previous estimate for a QTL containing CBF2 (Oakley et al., 2014), but the proportional difference between SW and IT explained by CBF2 is similar to the QTL‐based estimate (Oakley et al., 2014). The difference in freezing tolerance between the NILs (59% in NIL R37 compared to 50% in NIL R38) is difficult to explain, but the overall reduction in freezing tolerance for both lines follows the expected direction given the loss of function of CBF2. Regardless, these results suggest that local adaptation between SW and IT is in part driven by a large‐effect allele of CBF2 in IT. Broad surveys of natural variation in CBF2 haplotypes in Arabidopsis have found that presumably functional variation in CBF2 only occurs in warmer climates (Zhen and Ungerer, 2008; Monroe et al., 2016). This suggests that CBF2 is likely important for regulating cold‐acclimated freezing tolerance across the northern portion of the species’ range and that CBF2 polymorphisms beyond that observed between SW and IT may contribute to regional climatic adaptation.
Large‐effect mutations have been thought to be unlikely to contribute to adaptive evolution (Fisher, 1930; Rockman, 2012) because of assumed negative pleiotropic effects. Despite these expectations, our results add to a growing body of literature that suggest large‐effect alleles contribute substantially to variation in physiological and life history traits (Stinchcombe et al., 2004; Chiang et al., 2012; Xie et al., 2015). The IT allele of CBF2 has a deletion that results in a loss of function, and similar loss‐of‐function mutations in genes that encode proteins involved in DNA or protein interactions have been found to drive large differences in flowering time and germination (Stinchcombe et al., 2004; Chiang et al., 2012), trichome production (Bloomer et al., 2012), growth and reproductive timing (Brock et al., 2010), and circadian timing and cold and salt tolerance (Xie et al., 2015). Taken together, our results as well as those of previous studies suggest that loss‐of‐function mutations in regulatory genes can have important roles in driving differentiation among populations for ecologically important quantitative traits.
Gene expression
Differences between SW and IT
We identified 249 genes with a significant genotype (SW vs. IT) × cold acclimation treatment interaction. In a recent study, 5200 genes in the SW and IT ecotypes were identified as differentially expressed in response to cold (Fig. 1D; Gehan et al., 2015), though that study did not explicitly test for genotype × environment interactions. The genes identified in that study include 145 of the 249 genes we identified as differentially cold‐responsive between the SW and IT ecotypes, including nine of our 10 candidate genes (Appendix S15). The 104 genes from our study that are not included in the Gehan et al. (2015) study include DEAR3, which exhibited slight but significant differences in cold‐responsive expression in all pairwise comparisons (see below; Appendices S14, S16).
Potential targets of CBF2‐mediated cold acclimation
There has been considerable recent interest in assessing the effects of loss‐of‐function mutations in CBF on cold‐acclimated gene expression and freezing tolerance (Jia et al., 2016; Zhao et al., 2016; Park et al., 2018). The focus of most of these studies is on the combined effects of loss of function in all three CBF genes to determine what is referred to as the “CBF regulon” for a given accession, rather than studying the effects of natural variation in individual CBF genes. Our work builds upon the recent study by Park et al. (2018), who used CRISPR‐Cas9 to produce mutations in CBF genes in the SW genetic background. Here we used a CBF2 null mutant from Park et al. (2018), an additional independent CBF2 null mutant, and two NILs with IT loss‐of‐function mutations in a SW genetic background to hone in on the downstream targets of CBF2 that are in part responsible for differences in cold‐acclimated freezing tolerance between IT and SW.
Examining the subset of genes that were differentially cold responsive between the SW and IT ecotypes that were also differentially cold responsive in all four lines with loss‐of‐function mutations in CBF2 narrowed the list of candidates to just 10 genes. Because these genes were identified as significant in all of our independent comparisons between SW and lines with nonfunctional CBF2, we have confidence that these are important candidate genes for downstream targets of CBF2. While not completely regulated by CBF2, these genes are likely responsible for most of the differences in freezing tolerance between SW and IT caused by the loss‐of‐function mutation in the IT CBF2 allele and, thus, are also candidates for contributing to local adaptation between these ecotypes.
Two of the 10 candidates were very highly cold responsive in SW, galactinol synthase 3 and AT4G30830 (Fig. 3). Galactinol synthase 3 (GolS3) was the most cold responsive of the 10 candidate genes in SW (Table 2), and CBF2 could explain almost all of the differences in cold‐acclimated GolS3 expression between SW and IT. GolS3 has been shown to be cold responsive in a number of studies (Maruyama et al., 2009; Park et al., 2015; Zhao et al., 2016), including those using the SW and IT ecotypes (Gehan et al., 2015; Park et al., 2018). This gene is also known to be induced by cold stress and plays a key role in biosynthesis of raffinose, an important osmoprotectant during cold and water stress (Taji et al., 2002). The other very highly cold‐responsive gene in SW was AT4G30830, but the expression differences between SW and IT explained by CBF2 for this gene were more modest (~44%). AT4G30830, which is a myosin‐like protein of unknown function (Krishnakumar et al., 2014) that has been described as cold responsive in other studies (Gehan et al., 2015; Park et al., 2018). Future studies to identify what role AT4G30830 plays in cold‐acclimated freezing tolerance would be worthwhile.
The next category of candidates included six genes that were all highly cold responsive in the SW ecotype (Fig. 4). Of these genes, ERD10, COR413‐PM1, and CCT2 were previously identified as part of the CBF regulon in a few accessions (Appendix S17). The amount of the differential cold‐responsive gene expression between SW and IT explained by CBF2 varied from 34–81% (Table 2), indicating variation in the extent to which these genes are regulated by CBF2. LEA14 encodes a late embryogenesis abundant protein, a class of proteins that is induced by cellular stress, particularly desiccation (Singh et al., 2005), and thought to contribute to biomolecule and membrane stability (Candat et al., 2014). CCT2 encodes a phosphorylcholine cytidylyltransferase, which acts to increase cellular phosphorylcholine content, an important component of biological membranes, in response to cold (Inatsugi et al., 2009). COR413‐PM1 encodes a multispanning transmembrane protein localized to the plasma membrane that is correlated with freezing tolerance in Arabidopsis and cereal crops (Breton et al., 2003) and may play a role in maintaining membrane fluidity under cold temperatures (Su et al., 2018). ERD10 and COR47 encode dehydrin family proteins, thought to play an important role in cellular desiccation resistance, and both have been shown to increase freezing tolerance (Puhakainen et al., 2004). ERD7 is a drought inducible gene that has been shown to be cold responsive (Kimura et al., 2003).
The final two genes in our list of 10 candidates were those with only modest or low cold responsiveness in SW. In spite of their relatively small cold responsiveness on an absolute scale, much of the differences in expression between SW and IT for these genes could be attributed to CBF2 (65% for AT3G55760 and 84% for DEAR3). Neither of these genes have been previously described as part of the CBF regulon for SW (Park et al., 2018), but both have been identified as part of regulons from other genetic backgrounds (Appendix S17). AT3G55760 is located in the chloroplast stroma and is involved in starch metabolism (Feike et al., 2016). DEAR3 is a member of the DREB subfamily ERF/AP2 transcription factors (Sazegari et al., 2015), which is the same subfamily of transcription factors as CBF2.
CONCLUSIONS
A more comprehensive understanding of local adaptation to stressful environments requires identifying the genetic and physiological changes that confer phenotypic variation, as well as the fitness consequences of such variation in contrasting environments. Detailed molecular studies of traits that have been established as contributing to local adaptation in large multi‐year field experiments is perhaps the best approach to connect sequence polymorphism to molecular and organismal phenotypes and ultimately fitness in contrasting environments. Using a novel approach of examining genotype × environment interactions in gene expression using replicate lines that either simulate (CRISPR) or contain (NILs) the loss‐of‐function mutation in CBF2 found in the IT ecotype, we narrowed the list of candidate targets regulated by CBF2 during cold acclimation to just 10 genes. These 10 genes are excellent candidates for further study of the genetic and physiological changes that underlie the differences in freezing tolerance and local adaptation in these natural populations. As a group, the 10 candidates have likely roles in desiccation resistance, sugar biosynthesis or starch metabolism, membrane structure and transport, and regulation of transcription, while some of the functions of these genes are unknown or poorly known. The interaction between the CBF2 protein and downstream targets on cold acclimation and local adaptation will be addressed with future growth chamber and field studies to quantify fitness, gene expression, and metabolite production for a set of NILs with pairwise combinations of IT alleles of CBF2 and each of these 10 genes.
AUTHOR CONTRIBUTIONS
C.G.O., D.W.S, and M.F.T. conceived the study; C.G.O. and S.P. designed the experiment; S.P. and C.G.O. developed the CRISPR and NIL lines, respectively; M.I.J., S.P., J.C.K, and C.G.O. carried out the experiment; B.J.S, C.G.O., and S.P. analyzed the data and produced the figures; B.J.S. and C.G.O. drafted the manuscript with help from S.P. and M.I.J.; and all authors contributed to revising the manuscript.
Supporting information
APPENDIX S1. Three transcription factors encoding CBF genes in tandem array in the SW and IT ecotypes, and two lines with CRISPR‐induced mutations in the CBF2 gene. Open boxes indicate CBF1, CBF2, and CBF3 coding regions (shaded portion indicates DNA binding domain) in the order they are arranged in the genome. The green lines indicate the transcription activation domains. The filled triangle indicates the site of the naturally occurring 13‐bp deletion in the IT CBF2 gene. Open triangles indicate the two independent CRISPR induced deletions. SW:cbf2 a is the same line used by Park et al. (2018).
APPENDIX S2. Distribution of mean freezing‐tolerance values per cell. The distribution of values for all six lines combined are given in black, and the distribution of values for just IT are given in red.
APPENDIX S3. Supplemental methods for calculating differences in cold‐responsiveness gene expression.
APPENDIX S4. Raw data for the freezing‐tolerance assay. Batch describes the three temporally separated replicates of the assay, within each of which there were 60 replicated blocks of lines. The number of plants dead and alive after the freezing period are represented by n_Dead and n_Alive, respectively, and freezing tolerance is expressed as a percentage.
APPENDIX S5. Heat map of expression differences for the 249 genes that were identified as having a significant genotype × treatment interaction (P FDR ≤ 0.05 for the comparison of IT to SW). The value of each cell represents the log2‐fold‐change in gene expression before and after cold acclimation (counts per million reads). Yellow‐red represents genes that are more highly expressed after cold acclimation; black‐purple represents genes that are more highly expressed before cold acclimation. The plot was generated using the heatmap.2 function in the R package plots (Warnes et al., 2019).
APPENDIX S6. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05). F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S7. Gene ontology enrichment for the 249 genes with significant genotype × environment interactions for the pairwise comparison of IT to SW (P FDR ≤ 0.05).
APPENDIX S8. List of genes with significant differences in cold‐responsive expression between SW and NIL R37 (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S9. List of genes with significant differences in cold‐responsive expression between SW and NIL R38 (P < 0.05) and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S10. List of genes with significant differences in cold‐responsive expression between SW and CRISPR line SW:cbf2 a (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S11. List of genes with significant differences in cold‐responsive expression between SW and CRISPR line SW:cbf2 b (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S12. Heat map of expression differences for the 10 genes that were identified as having a significant genotype × treatment interaction for all pairwise comparisons to SW. The value of each cell represents the log2 fold‐change in gene expression before and after cold acclimation (counts per million reads). Yellow‐red represents genes that are more highly expressed after cold acclimation; black‐purple represents genes that are more highly expressed before cold acclimation. The plot was generated using the heatmap.2 function in the R package plots (Warnes et al., 2019).
APPENDIX S13. Relative expression for GolS3 and COR413‐PM1 quantified by RT‐qPCR, normalized to expression of the housekeeping gene ACT2. PRE and POST refer to pre‐acclimation and post‐acclimation tissue collections, respectively. Each biological replicate was run in triplicate for three technical replicates. Points are means of three biological replicates; error bars are the standard error of those means. Primer sequences: GolS3 F, 5‐TGTGCCAAAGCTCCATCCGC‐3; GolS3 R, 5‐TGGTGTTGACAAGAACCTCGCT‐3; COR413‐PM1 F, 5‐TGCTGGCACATTCAGAGACAG‐3; COR413‐PM1 R, 5‐CAGACGGGGAAGACGACGAGA‐3; ACT2 F, 5‐CTGGATCGGTGGTTCCATTC‐3; ACT2 R, 5‐CCTGGACCTGCCTCATCATAC‐3.
APPENDIX S14. Log2 counts per million for the least responsive genes of the 10 candidates before (left group of bars) and after (right group of bars) cold acclimation.
APPENDIX S15. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05) that are included among those identified by Gehan et al. (2015).
APPENDIX S16. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05), which were not identified by Gehan et al. (2015).
APPENDIX S17. Comparison of genes included in Appendix S12 with previously published CBF regulons from four genetic backgrounds. SW and IT from Park et al. (2018), Col‐0 from Zhao et al. (2016), and WS from Park et al. (2015).
ACKNOWLEDGMENTS
The authors would like to thank A. Babbit, J. Eilers, and M. Kargul, who helped to implement the freezing tolerance assays, and the RTSF Genomics Core at MSU for sequencing and assistance. We are grateful to Dr. Jian‐Kang Zhu, Purdue University, for providing the plasmid used to generate the CRISPR lines. We thank A. Berardi, R. Deater, N. Mano, R. Watson, and members of the Zhang Lab at Purdue for assistance with qPCR. We thank members of the Aime, McAdam, McNickle, and Mickelbart labs at Purdue for helpful comments on a draft of the manuscript. We also thank R. Baucom and two anonymous reviewers for insightful comments on this manuscript. Funding was provided by NSF DEB grant (1743273) to C.G.O., D.W.S., and M.F.T. and by AgBioResearch at Michigan State University to M.F.T.
Sanderson, B. J. , Park S., Jameel M. I., Kraft J. C., Thomashow M. F., Schemske D. W., and Oakley C. G.. 2020. Genetic and physiological mechanisms of freezing tolerance in locally adapted populations of a winter annual. American Journal of Botany 107(2): 250–261.
Data Availability
Raw freezing‐tolerance data is available as a part of this submission in Appendix S4, and raw RNAseq read data are available in the sequence read archive of NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA556985).
LITERATURE CITED
- Abbott, R. J. , and Gomes M. F.. 1989. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity 62: 411–418. [Google Scholar]
- Ågren, J. , and Schemske D. W.. 2012. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytologist 194: 1112–1122. [DOI] [PubMed] [Google Scholar]
- Ågren, J. , Oakley C. G., Mckay J. K., Lovell J. T., and Schemske D. W.. 2013. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 110: 21077–21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ågren, J. , Oakley C. G., Lundemo S., and Schemske D. W.. 2017. Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: estimates of selection and QTL mapping. Evolution 71: 550–564. [DOI] [PubMed] [Google Scholar]
- Alonso‐Blanco, C. , Gomez‐Mena C., Llorente F., Koornneef M., Salinas J., and Martínez‐Zapater J. M.. 2005. Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis . Plant Physiology 139: 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. T. , Willis J. H., and Mitchell‐Olds T.. 2011. Evolutionary genetics of plant adaptation. Trends in Genetics 27: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. T. , Lee C.‐R., Rushworth C. A., Colautti R. I., and Mitchell‐Olds T.. 2013. Genetic trade‐offs and conditional neutrality contribute to local adaptation. Molecular Ecology 22: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. Website http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Barrero‐Gil, J. , and Salinas J.. 2018. Gene regulatory networks mediating cold acclimation: the CBF pathway In Iwaya‐Inoue M., Sakurai M., and Uemura M. [eds.], Survival strageies in extreme cold and desiccation, 3–22. Springer Singapore, Singapore. [DOI] [PubMed] [Google Scholar]
- Beck, J. B. , Schmuths H., and Schaal B. A.. 2008. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Molecular Ecology 17: 902–915. [DOI] [PubMed] [Google Scholar]
- Bloomer, R. H. , Juenger T. E., and Symonds V. V.. 2012. Natural variation in GL1 and its effects on trichome density in Arabidopsis thaliana . Molecular Ecology 21: 3501–3515. [DOI] [PubMed] [Google Scholar]
- Breton, G. , Danyluk J., Charron J.‐B. F., and Sarhan F.. 2003. Expression profiling and bioinformatic analyses of a novel stress‐regulated multispanning transmembrane protein family from cereals and Arabidopsis . Plant Physiology 132: 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock, M. T. , Maloof J. N., and Weinig C.. 2010. Genes underlying quantitative variation in ecologically important traits: PIF4 (PHYTOCHROME INTERACTING FACTOR 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana . Molecular Ecology 19: 1187–1199. [DOI] [PubMed] [Google Scholar]
- Burghardt, L. T. , Edwards B. R., and Donohue K.. 2016. Multiple paths to similar germination behavior in Arabidopsis thaliana . New Phytologist 209: 1301–1312. [DOI] [PubMed] [Google Scholar]
- Candat, A. , Paszkiewicz G., Neveu M., Gautier R., Logan D. C., Avelange‐Macherel M.‐H., and Macherel D.. 2014. The ubiquitous distribution of late embryogenesis abundant protiens across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 26: 3148–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. 2018. VennDiagram: Generate high‐resolution Venn and Euler plots. R package version 1.6.20. Website https://CRAN.R-project.org/package=VennDiagram.
- Chiang, G. C. K. , Barua D., Dittmar E., Kramer E. M., De Casas R. R., and Donohue K.. 2012. Pleiotropy in the wild: the dormancy gene DOG1 exerts cascading control on life cycles. Evolution 67: 883–893. [DOI] [PubMed] [Google Scholar]
- Clausen, J. , Keck D. D., and Hiesey W. M.. 1940. Experimental studies on the nature of plant species. I. The effect of varied environments on western North American plants. Carnegie Institute of Washington, Washington, D.C., USA. [Google Scholar]
- Dittmar, E. L. , Oakley C. G., Conner J. K., Gould B. A., and Schemske D. W.. 2016. Factors influencing the effect size distribution of adaptive substitutions. Proceedings of the Royal Society, B, Biological Sciences 283: 20153065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, M. A. , Farre E. M., and Thomashow M. F.. 2011. CIRCADIAN CLOCK‐ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C‐REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis . Proceedings of the National Academy of Sciences, USA 108: 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula, A. , Fulgione A., Gutaker R. M., Alacakaptan S. I., Flood P. J., Neto C., Tsuchimatsu T., et al. 2017. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 114: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feike, D. , Seung D., Graf A., Bischof S., Ellick T., Coiro M., Soyk S., et al. 2016. The starch granule‐associated protein EARLY STARVATION1 is required for the control of starch degradation in Arabidopsis thaliana leaves. Plant Cell 28: 1472–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Mao Y., Xu N., Zhang B., Wei P., Yang D., Wang Z., et al. 2014. Multigenerational analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis . Proceedings of the National Academy of Sciences, USA 111: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A. 1930. The genetical theory of natural selection. Oxford University Press, Oxford, UK. [Google Scholar]
- Futuyma, D. J. , and Moreno G.. 1988. The evolution of ecological specialization. Annual Review of Ecology and Systematics 19: 207–233. [Google Scholar]
- Gehan, M. A. , Park S., Gilmour S. J., An C., Lee C., and Thomashow M. F.. 2015. Natural variation in the C‐repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant Journal 84: 682–693. [DOI] [PubMed] [Google Scholar]
- Hall, M. C. , Lowry D. B., and Willis J. H.. 2010. Is local adaptation in Mimulus guttatus caused by trade‐offs at individual loci? Molecular Ecology 19: 2739–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah, M. A. , Wiese D., Freund S., Fiehn O., Heyer A. G., and Hincha D. K.. 2006. Natural genetic variation of freezing tolerance in Arabidopsis . Plant Physiology 142: 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford, J. 2009. A quantitative survey of local adaptation and fitness trade‐offs. American Naturalist 173: 579–588. [DOI] [PubMed] [Google Scholar]
- Inatsugi, R. , Kawai H., Yamaoka Y., Yu Y., Sekiguchi A., Nakamura M., and Nishida I.. 2009. Isozyme‐specific modes of activation of CTP: phosphorylcholine cytidylyltransferase in Arabidopsis thaliana at low temperature. Plant Cell Physiology 50: 1727–1735. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , Ding Y., Shi Y., Zhang X., Gong Z., and Yang S.. 2016. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis . New Phytologist 212: 345–353. [DOI] [PubMed] [Google Scholar]
- Jmp . 1989‐2019. JMP version 13. SAS Institute, Cary, NC, USA. [Google Scholar]
- Kang, J. , Zhang H., Sun T., Shi Y., Wang J., Zhang B., Wang Z., et al. 2013. Natural variation of C‐repeat‐binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytologist 199: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. 1983. The neutral theory of molecular evolution, 384. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Kimura, M. , Yamamoto Y. Y., Seki M., Sakurai T., Sato M., Abe T., Yoshida S., et al. 2003. Identification of Arabidopsis genes regulated by high light‐stress using cDNA microarray. Photochemistry and Photobiology 77: 226–233. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. , Alonso‐Blanco C., and Vreugdenhil D.. 2004. Naturally occurring genetic variation in Arabidopsis thaliana . Annual Review of Plant Biology 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Krishnakumar, V. , Hanlon M. R., Contrino S., Ferlant E. S., Karamycheva S., Kim M., Rosen B. D., et al. 2014. Araport: The Arabidopsis information portal. Nucleic Acids Research 43: D1003–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher, W. 1980. Physiological plant ecology, 32–33. Srpinger‐Verlag, Berlin, Germany. [Google Scholar]
- Lee, Y. W. , Gould B. A., and Stinchcombe J. R.. 2014. Identifying the genes underlying quantitative traits: a rationale for the QTN programme. AoB Plants 6: plu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen, P. H. , Remmington D. L., Leppälä J., and Savolainen O.. 2013. Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata . Molecular Ecology 22: 709–723. [DOI] [PubMed] [Google Scholar]
- Lowry, D. B. , Hall M. C., Salt D. E., and Willis J. H.. 2009. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus . New Phytologist 183: 776–788. [DOI] [PubMed] [Google Scholar]
- MacArthur, R. H. 1972. Geographical ecology: patterns in the distribution of species. Princeton University Press, Princeton, NJ, USA. [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Maruyama, K. , Takeda M., Kidokoro S., Yamada K., Sakuma Y., Urano K., Fujita M., et al. 2009. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiology 150: 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan A., Ebert D., Huang X., and Thomas P. D.. 2019. PANTHER version 14: more genomes, a new PANTHER GO‐slim and improvements in enrichment analysis tools. Nucleic Acids Research 47: D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe, J. G. , McGovern C., Lasky J. R., Grogan K., Beck J., and McKay J. K.. 2016. Adaptation to warmer climates by parallel functional evolution of CBF genes in Arabidopsis thaliana . Molecular Ecology 25: 3632–3644. [DOI] [PubMed] [Google Scholar]
- Montesinos, A. , Tonsor S. J., Alonso‐Blanco C., and Pico F. X.. 2009. Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 4: e7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, C. G. , Ågren J., Atchison R. A., and Schemske D. W.. 2014. QTL mapping of freezing tolerance: links to fitness and adaptive trade‐offs. Molecular Ecology 23: 4304–4315. [DOI] [PubMed] [Google Scholar]
- Orr, H. A. 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr, H. A. 2005. The genetic theory of adaptation: a brief history. Nature Reviews Genetics 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Park, S. , Lee C.‐M., Doherty C. J., Gilmour S. J., Kim Y., and Thomashow M. F.. 2015. Regulation of the Arabidopsis CBF regulon by a complex low‐temperature regulatory network. Plant Journal 82: 193–207. [DOI] [PubMed] [Google Scholar]
- Park, S. , Gilmour S. J., Grumet R., and Thomashow M. F.. 2018. CBF‐dependent and CBF‐independent regulatory pathways contribute to the differences in freezing tolerance and cold‐regulated gene expression of two Arabidopsis ecotypes locally adapted to sites in Sweden and Italy. PLoS ONE 13: e0207723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real time RT‐PCR. Nucleic Acids Research 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, F. M. , and Ågren J.. 2016. Early life stages contribute strongly to local adaptation in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 113: 7590–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, J. C. , and Sandve S. R.. 2013. Adaptation to seasonality and the winter freeze. Frontiers in Plant Science 4: 10.3389/fpls.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhakainen, T. , Hess M. W., Makela P., Svensson J., Heino P., and Palva E. T.. 2004. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis . Plant Molecular Biology 54: 743–753. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rausher, M. D. , and Delph L. F.. 2015. Commentary: When does understanding phenotypic evolution require identification of the underlying genes? Evolution 69: 1655–1664. [DOI] [PubMed] [Google Scholar]
- Remington, D. L. 2015. Alleles versus mutations: Understanding the evolution of genetic architecture requires a molecular perspective on allelic origins. Evolution 69: 3025–3038. [DOI] [PubMed] [Google Scholar]
- Robinson, M. D. , Mccarthy D. J., and Smyth G. K.. 2010. edgeR: a Bioconductor package for differential expression analyis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman, M. V. 2012. The QTN program and the alleles that matter for evolution: All that's gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen, O. , Lascoux M., and Merilä J.. 2013. Ecological genomics of local adaptation. Nature Reviews Genetics 14: 807–820. [DOI] [PubMed] [Google Scholar]
- Sazegari, S. , Niazi A., and Ahmadi F. S.. 2015. A study on the regulatory network with promoter analysis for Arabidopsis DREB‐genes. Bioinformation 11: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Huang J., Sun T., Wang X., Zhu C., Ai Y., and Gu H.. 2017. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana . Journal of Integrative Biology 59: 118–133. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Cornilescu C. C., Tyler R. C., Cornilescu G., Tonelli M., Lee M. S., and Markley J. L.. 2005. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress‐related protein. Protein Science 14: 2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, D. Z. 2015. Time and temperature interactions in freezing tolerance of winter wheat. Crop Science 54: 1523–1529. [Google Scholar]
- Smallwood, M. , and Bowles D. J.. 2002. Plants in a cold climate. Philosophical Transactions of the Royal Society, B, Biological Sciences 357: 831–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe, J. R. , Weinig C., Ungerer M., Olsen K. M., Mays C., Halldorsdottir S. S., Purugganan M. D., and Schmitt J.. 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA . Proceedings of the National Academy of Sciences, USA 101: 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, C. , Chen K., Ding Q., Mou Y., Yang R., Zhao M., Ma B., et al. 2018. Proteomic analysis of the function of a novel cold‐regulated multispanning transmembrane protein COR413‐PM1 in Arabidopsis . International Journal of Molecular Sciences 19: 2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji, T. , Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi‐Shinozaki K., and Shinozaki K.. 2002. Important roles of drought‐ and cold‐inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana . Plant Journal 29: 417–426. [DOI] [PubMed] [Google Scholar]
- Thomashow, M. F. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow, M. F. 2010. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiology 154: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin, P. , and Ross‐Ibarra J.. 2014. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution 29: 673–680. [DOI] [PubMed] [Google Scholar]
- Tonsor, S. J. , Alonso‐Blanco C., and Koornneef M.. 2005. Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana . Plant, Cell & Environment 28: 2–20. [Google Scholar]
- VanWallendael, A. , Soltani A., Emery N. C., Peixoto M. M., Olsen J., and Lowry D. B.. 2019. A molecular view of plant local adaptation: incorporating stress‐response networks. Annual Review of Plant Biology 70: 14.11–14.25. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Meyer E., Mckay J. K., and Matz M. V.. 2012. 2b‐RAD: a simple and flexible method for genome‐wide genotyping. Nature Methods 9: 808–812. [DOI] [PubMed] [Google Scholar]
- Warnes, G. R. , Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., et al. 2019. gplots: Various R programming tools for plotting data. R package version 3.0.1.1. Website https://CRAN.R-project.org/package=gplots. [Google Scholar]
- Whitlock, M. C. 1996. The red queen beats the jack‐of‐all‐trades: the limitations on the evolution of phenotypic plasticity and niche breadth. American Naturalist 148: XS65–XS77. [Google Scholar]
- Xie, Q. , Lou P., Hermand V., Aman R., Park H. J., Yun D.‐J., Kim W. Y., et al. 2015. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa . Proceedings of the National Academy of Sciences, USA 112: 3829–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Zhang Z., Xie S., Si T., Li Y., and Zhu J.‐K.. 2016. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis . Plant Physiology 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen, Y. , and Ungerer M. C.. 2008. Relaxed selection on the CBF/DREB1 regulatory genes and reduced freezing tolerance in the southern range of Arabidopsis thaliana . Molecular Biology and Evolution 25: 2547–2555. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Pearce S., Burke A., See D. R., Skinner D. Z., Dubcovsky J., and Garland‐Campbell K.. 2014. Copy number and haplotype variation at the VRN‐A1 and central FR‐A2 loci are associated with frost tolerance in hexaploid wheat. Theoretical and Applied Genetics 127: 1183–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther, E. , Lee Y. P., Erban A., Kopka J., and Hincha D. K.. 2018. Natural variation in freezing tolerance and acclimation response in Arabidopsis thaliana and related species In Iwaya‐Inoue M., Sakurai M., and Uemura M. [eds.], Survival strategies in extreme cold and dessication, 81–98. Springer Singapore, Singapore. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Three transcription factors encoding CBF genes in tandem array in the SW and IT ecotypes, and two lines with CRISPR‐induced mutations in the CBF2 gene. Open boxes indicate CBF1, CBF2, and CBF3 coding regions (shaded portion indicates DNA binding domain) in the order they are arranged in the genome. The green lines indicate the transcription activation domains. The filled triangle indicates the site of the naturally occurring 13‐bp deletion in the IT CBF2 gene. Open triangles indicate the two independent CRISPR induced deletions. SW:cbf2 a is the same line used by Park et al. (2018).
APPENDIX S2. Distribution of mean freezing‐tolerance values per cell. The distribution of values for all six lines combined are given in black, and the distribution of values for just IT are given in red.
APPENDIX S3. Supplemental methods for calculating differences in cold‐responsiveness gene expression.
APPENDIX S4. Raw data for the freezing‐tolerance assay. Batch describes the three temporally separated replicates of the assay, within each of which there were 60 replicated blocks of lines. The number of plants dead and alive after the freezing period are represented by n_Dead and n_Alive, respectively, and freezing tolerance is expressed as a percentage.
APPENDIX S5. Heat map of expression differences for the 249 genes that were identified as having a significant genotype × treatment interaction (P FDR ≤ 0.05 for the comparison of IT to SW). The value of each cell represents the log2‐fold‐change in gene expression before and after cold acclimation (counts per million reads). Yellow‐red represents genes that are more highly expressed after cold acclimation; black‐purple represents genes that are more highly expressed before cold acclimation. The plot was generated using the heatmap.2 function in the R package plots (Warnes et al., 2019).
APPENDIX S6. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05). F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S7. Gene ontology enrichment for the 249 genes with significant genotype × environment interactions for the pairwise comparison of IT to SW (P FDR ≤ 0.05).
APPENDIX S8. List of genes with significant differences in cold‐responsive expression between SW and NIL R37 (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S9. List of genes with significant differences in cold‐responsive expression between SW and NIL R38 (P < 0.05) and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S10. List of genes with significant differences in cold‐responsive expression between SW and CRISPR line SW:cbf2 a (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S11. List of genes with significant differences in cold‐responsive expression between SW and CRISPR line SW:cbf2 b (P < 0.05), and which are also in Appendix S6. F, P Value, and FDR refer to the significance test of the interaction term.
APPENDIX S12. Heat map of expression differences for the 10 genes that were identified as having a significant genotype × treatment interaction for all pairwise comparisons to SW. The value of each cell represents the log2 fold‐change in gene expression before and after cold acclimation (counts per million reads). Yellow‐red represents genes that are more highly expressed after cold acclimation; black‐purple represents genes that are more highly expressed before cold acclimation. The plot was generated using the heatmap.2 function in the R package plots (Warnes et al., 2019).
APPENDIX S13. Relative expression for GolS3 and COR413‐PM1 quantified by RT‐qPCR, normalized to expression of the housekeeping gene ACT2. PRE and POST refer to pre‐acclimation and post‐acclimation tissue collections, respectively. Each biological replicate was run in triplicate for three technical replicates. Points are means of three biological replicates; error bars are the standard error of those means. Primer sequences: GolS3 F, 5‐TGTGCCAAAGCTCCATCCGC‐3; GolS3 R, 5‐TGGTGTTGACAAGAACCTCGCT‐3; COR413‐PM1 F, 5‐TGCTGGCACATTCAGAGACAG‐3; COR413‐PM1 R, 5‐CAGACGGGGAAGACGACGAGA‐3; ACT2 F, 5‐CTGGATCGGTGGTTCCATTC‐3; ACT2 R, 5‐CCTGGACCTGCCTCATCATAC‐3.
APPENDIX S14. Log2 counts per million for the least responsive genes of the 10 candidates before (left group of bars) and after (right group of bars) cold acclimation.
APPENDIX S15. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05) that are included among those identified by Gehan et al. (2015).
APPENDIX S16. List of genes with significant gene × environment interactions in our study for the pairwise comparison of IT to SW (P FDR ≤ 0.05), which were not identified by Gehan et al. (2015).
APPENDIX S17. Comparison of genes included in Appendix S12 with previously published CBF regulons from four genetic backgrounds. SW and IT from Park et al. (2018), Col‐0 from Zhao et al. (2016), and WS from Park et al. (2015).
Data Availability Statement
Raw freezing‐tolerance data is available as a part of this submission in Appendix S4, and raw RNAseq read data are available in the sequence read archive of NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA556985).


