Abstract
Streptococcus pyogenes is a strict human pathogen responsible for more than 700 million infections annually worldwide. Strains of serotype M28 S. pyogenes are typically among the five more abundant types causing invasive infections and pharyngitis in adults and children. Type M28 strains also have an unusual propensity to cause puerperal sepsis and neonatal disease. We recently discovered that a one-nucleotide indel in an intergenic homopolymeric tract located between genes Spy1336/R28 and Spy1337 altered virulence in a mouse model of infection. In the present study, we analyzed size variation in this homopolymeric tract and determined the extent of heterogeneity in the number of tandemly-repeated 79-amino acid domains in the coding region of Spy1336/R28 in large samples of strains recovered from humans with invasive infections. Both repeat sequence elements are highly polymorphic in natural populations of M28 strains. Variation in the homopolymeric tract results in (i) changes in transcript levels of Spy1336/R28 and Spy1337 in vitro, (ii) differences in virulence in a mouse model of necrotizing myositis, and (iii) global transcriptome changes as shown by RNAseq analysis of isogenic mutant strains. Variation in the number of tandem repeats in the coding sequence of Spy1336/R28 is responsible for size variation of R28 protein in natural populations. Isogenic mutant strains in which genes encoding R28 or transcriptional regulator Spy1337 are inactivated are significantly less virulent in a nonhuman primate model of necrotizing myositis. Our findings provide impetus for additional studies addressing the role of R28 and Spy1337 variation in pathogen-host interactions.
Introduction
Streptococcus pyogenes (group A streptococcus, GAS) is a strict human pathogen responsible for >700 million infections and ~517,000 deaths annually worldwide [1]. Human infections range in severity from relatively mild conditions such as pharyngitis to life-threatening septicemia and necrotizing fasciitis/myositis [2]. GAS also causes skin infections such as impetigo and erysipelas [3] and post-infection sequelae, including rheumatic fever [4], rheumatic heart disease [5], and glomerulonephritis [6].
GAS strains are commonly classified based on serologic diversity in M protein, an antiphagocytic cell-surface virulence factor, or allelic variation in the 5’-end of the emm gene that encodes this protein [7, 8]. More than 250 emm types have been identified, but the majority of infections in many countries are caused by a relatively small number of prevalent emm types: emm1, emm3, and emm12 [9–12]. Strains of emm28 (serotype M28) GAS are of special importance because they are among the top five emm types causing invasive infections in the USA [11, 13] and several European countries [14–16]. They also have an unusual propensity to cause puerperal sepsis (childbed fever) and neonatal infections [17–21]. The molecular mechanisms contributing to the ability of emm28 strains to cause devastating peripartum infections are poorly understood.
The R28 protein is a surface-associated virulence factor made by emm28 strains and has been studied as a vaccine candidate [22–24]. R28 originally was described by Lancefield and colleagues based on serologic studies [25–27]. The gene (Spy1336/R28) encoding R protein in emm28 GAS strains is nearly identical to the alp3 gene encoding the Alp3 protein in group B streptococcus (GBS), a common cause of neonatal sepsis, pneumonia, and meningitis [24, 28, 29]. Genome sequencing of emm28 GAS strain MGAS6180 discovered that the gene encoding the R28 protein (Spy1336/R28) is located on an integrative-conjugative element (ICE)-like element originally designated as region of difference 2 (RD2) [30, 31]. In the genome of reference strain MGAS6180 [31], RD2 is a 37.4 Kb segment of DNA that has 34 annotated genes. RD2 is present in a small number of other GAS emm types and is >99% identical to a region present in the chromosome of the majority of GBS strains [31]. These characteristics suggest that RD2 has been disseminated into different streptococcal strains and species by horizontal gene transfer and recombination [32, 33]; published data support this idea [33]. The presence of RD2 in GBS and emm28 GAS strains causing infections associated with the female genital tract also suggests that genes located on this ICE element are causally involved in this clinical phenotype.
The Spy1337 gene is adjacent to, and divergently transcribed from, Spy1336/R28 in GAS. The Spy1337 protein is a member of the AraC family of prokaryotic transcriptional regulators. Members of this family of regulators commonly have two domains, a conserved C-terminal domain that defines the members of the family, and a variable N-terminal domain that differs among family members. The C-terminal domain comprises approximately 100 amino acids and has two helix-turn-helix (HTH) DNA-binding motifs. The N-terminal domain can be bifunctional, mediating effector-binding and multimerization of the regulator [34, 35]. Genes encoding Spy1337/AraC-like proteins are frequently found adjacent to genes encoding surface antigens containing the YSIRK signal sequence motif {(YF)SIRKxxxGxxS}, which is responsible for localized secretion at the division septum [36, 37]. This motif is present in the N-terminal domain of R28 at amino acid positions 19 through 30.
Recently, we proposed that the Spy1337 protein is a positive transcriptional regulator of both Spy1336/R28 and Spy1337, and that by regulating expression of Spy1336/R28 and other genes, Spy1337 is involved in emm28 GAS virulence [38]. To positively regulate virulence gene expression, AraC-like transcriptional regulators usually either bind to chemical effectors present at the site of infection to cause a conformational change favoring DNA-binding to their cognate gene targets [39, 40], or they bind to AraC negative regulators (ANRs) that inhibits such interactions [41]. In addition, they can regulate their own expression [42–46].
The R28 protein is a member of a family of streptococcal surface proteins termed alpha-like proteins (Alps), characterized by the presence of (i) a conserved amino-terminal domain (pfam08829), (ii) long tandem repeat (TR) elements (pfam08428), and (iii) a carboxy-terminal domain (pfam00746) containing the LPXTG consensus sequence that serves to attach these proteins to the cell wall [24, 28, 32, 47, 48] (Fig 1A). Alps participate in adhesion [49–51], can be immunogenic [52, 53], and have been studied in the context of vaccine development [23, 54]. The best-studied Alp family proteins are alpha C protein (bca) [55], Alp1 (alp1) [56], Alp2 (alp2) and Alp3 (alp3) [28], Alp4 (alp4) [57], and Rib (rib) [58], made by GBS strains A909 (bca), NCTC12906 (alp1), CJB110 (alp2), JM9-130013 (alp3), NCTC 9828 (alp4), and BM110 (rib). Compared to the wild-type parental strain, an isogenic alpha C gene deletion mutant of GBS is less virulent in a mouse model of sepsis [59], demonstrating the role of Alp proteins in virulence. It has been shown that changing the number of tandemly repeated domains in the alpha C protein in GBS altered antigenicity, host immune recognition, and virulence when immunized with alpha C protein.
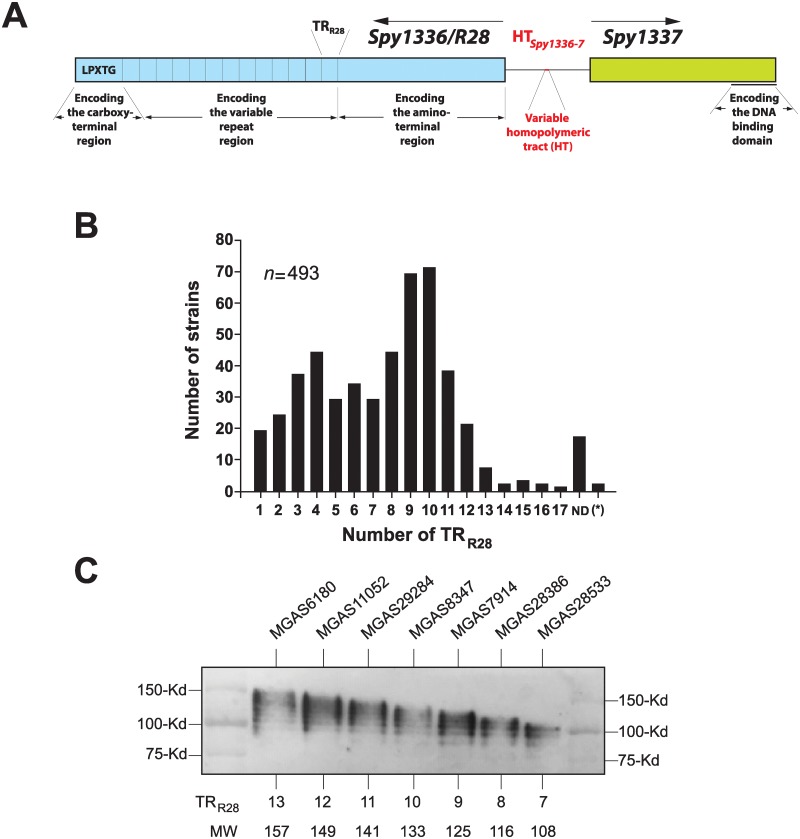
Fig 1. Analysis of the number of tandem repeats in the R28 protein.
(A) Schematic of Spy1336/R28, encoding R28, and Spy1337. HTSpy1336-7 refers to the variable homopolymeric tract. TRR28 refers to one 79-amino acid tandem repeat. LPXTG is the consensus sequence for peptidoglycan attachment. Regions encoding the amino-terminal, carboxy-terminal, and variable repeat regions are indicated. DNA binding domain refers to two predicted helix-turn-helix DNA-binding motifs. Schematic is not drawn to scale. (B) Data shown correspond to 493 strains. ND, not determined. (*), two strains studied did not contain the RD2 mobile genetic element, as confirmed by inspection of bam files using TABLET [78]. (C) Western immunoblot of strains with R28 proteins containing different numbers of TRR28, inferred based on gene sequence data. All strains analyzed had an HTSpy1336-7 with 10Ts because 9T strains do not produce detectable R28 protein. TRR28, number of tandem repeats per strain. MW, inferred molecular weight of the R28 protein.
The molecular mechanism(s) underlying how R28 or Alp proteins contribute to virulence is poorly understood. R28 adheres to human epithelial cells in vitro [24] via binding of the N-terminal domain to human integrin receptors α3β1, α6β1, and α6β4 [60], which in turn bind to laminin, an extracellular matrix (ECM) protein [61].
The Spy1336/R28 gene has a centrally-located long TR motif (referred to herein as TRR28), characteristic of genes encoding Alp family proteins. Due to the homologous nature of repetitive DNA sequence, regions having TRs frequently vary in size as a consequence of mutational events involving either unequal crossover or intramolecular recombination [62–65]. The R28 protein made by GAS emm28 reference strain MGAS6180 [31] has 13 identical TRR28s of 79 amino acids (aa) each.
In a recent study of 492 M28 invasive isolates of GAS for which whole genome sequence and transcriptome data were available, we used machine learning to determine that a variable-length T-nucleotide homopolymeric tract (HT, referred to herein as HTSpy1336-7) in the intergenic region of Spy1336/R28 and Spy1337 was associated with differences in transcript levels of these two genes [38]. HTs are commonly characterized by rapid length variation [66]. When present in promoter regions, HTs can modify transcript levels by altering the distance between promoter elements [67] or changing the binding of transcription factors [68, 69]. In ~94% of the strains we studied, HTSpy1336-7 had either nine or ten T residues. Comparison of human infection isolates found that strains with 9Ts had significantly lower transcript levels of the Spy1336/R28 and Spy1337 genes compared to strains with 10Ts. Compared to a parental strain with 9Ts, an isogenic mutant strain with 10Ts in the HTSpy1336-7 had significantly increased transcript levels of Spy1336/R28 and Spy1337 and was significantly more virulent in a mouse necrotizing myositis infection model. In addition, compared to the isogenic strain with 9Ts, the strain with 10Ts was significantly more resistant to killing by human polymorphonuclear leukocytes ex vivo and produced more R28 protein. Thus, a one-nucleotide indel in the HTSpy1336-7 caused altered level of these two transcripts and changed the virulence phenotype [38]. Of note, there is an HT comprised of 8–15 T residues located in the regulatory region 99 nucleotides (nts) upstream of the bca gene encoding the GBS alpha C Alp protein [68].
In the present study, we determined the extent of heterogeneity in the number of TRR28s in Spy1336/R28 in a subset of 493 M28 invasive clinical isolates for which we had whole genome and transcriptome data, including the reference strain MGAS6180 [38]. In addition, we analyzed size variation of the HTSpy1336-7 region located upstream of Spy1336/R28 in >2,000 strains of emm28 GAS cultured from invasive human infections and compared transcriptome changes in isogenic strains containing variable lengths of HTSpy1336-7. Finally, we used isogenic mutant strains to determine the contributions of Spy1336/R28 and Spy1337 to global gene expression and virulence.
Materials and methods
Ethics statement
The clinical isolates strains used in this study were collected as part of comprehensive population-based public health surveillance studies of emm28 S. pyogenes infections conducted in 11 states in the United States, Canada (ON), the Faroe Islands in Denmark, Finland, Iceland, and Norway [38]. Consent for collection of these strains was waived and all data were fully anonymized.
Bacterial strains and growth conditions
GAS strains were grown at 37°C in Todd-Hewitt broth (Bacto Todd-Hewitt broth; Becton Dickinson and Co.) supplemented with 0.2% yeast extract (THY medium). THY medium was supplemented with chloramphenicol (20 μg ml-1) as needed. Trypticase soy agar supplemented with 5% sheep blood (Becton Dickinson and Co.) was used as required. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C, unless indicated otherwise. LB medium was supplemented with chloramphenicol (Acros Organics; 20 μg ml-1) as needed.
DNA manipulation
Standard protocols or manufacturer’s instructions were used to isolate plasmid DNA, and conduct restriction endonuclease, DNA ligase, PCR, and other enzymatic treatments of plasmids and DNA fragments. Enzymes were purchased from New England Biolabs, Inc (NEB). Q5 high-fidelity DNA polymerase (NEB) was used. Oligonucleotides were purchased from Sigma Aldrich.
Chromosomal DNA extraction and PCR amplification of the Spy1336/R28 repeat region
Chromosomal DNA extraction was performed as described [70], using Fast-Prep lysing Matrix B beads in 2-ml tubes (M P Biomedicals), or the DNeasy blood and tissue kit (Qiagen). The primer sequences used for PCR-based size determination of the Spy1336/R28 repeat region are shown in S5 Table. Three different primer sets were used. All primers were designed to bind to conserved regions located upstream, in the case of the forward (FWD) primers, or downstream, for the reverse (REV) primers, from the DNA sequence of the Spy1336/R28 gene encoding the repeat region. Primer set 1 comprised primers JE433 (FWD), binding 60-nt upstream of the repeat region, and JE431 (REV), binding 226-nt downstream of the repeat region. Primer set 2 included primers JE432 (FWD), binding 255-nt upstream of the repeat region, and JE431 (REV). Primer set 3 was comprised of primers JE410 (FWD), binding 2,264-nt upstream of the repeat region, and JE412 (REV), binding 1,098-nt downstream of the repeat region. The extension time used for the PCR reactions was adjusted to accommodate anticipated PCR fragment length. Typically primer set 1 was used first to obtain the desired PCR product and the other 2 primer sets were used if primer set 1 failed to yield an amplified product. Two different DNA ladders were used to determine the size of the PCR products, including the exACTGene 100 bp-10,000 bp DNA ladder (Fisher Scientific), and the 100-bp DNA step ladder (Promega). Both ladders were loaded at least 3 times in every agarose gel and used as reference to determine DNA fragment length (S1 Table).
Analysis of the number of T residues in the HT1336-7
A blastable database of SPADES [71] assemblies was made for 2,095 emm28 genomes [38]. Using two adjacent DNA sequences of 20-nts that flank HT1336-7, the DNA region surrounding these two DNA sequences, including the two sequences, was extracted from each strain, and the number of T nucleotides counted. In addition, we analyzed the number of T nucleotides present in HTSpy1336-7 in the 2,095 emm28 genomes with the command-line program Jellyfish [72], which uses k-mers, and counted the occurrences of each HTSpy1336-7 variant. Using these combined approaches we identified the length of HTSpy1336-7 in 2,074 (~99%) of the original 2,095 strains, corresponding to 30 different alleles (S2 and S3 Tables). Of these alleles, six contained indels in HTSpy1336-7 exclusively, which resulted in changes in the number of consecutive T nucleotides, with no additional polymorphisms. These six alleles were present in 2,020 (~97%) strains.
Western immunoblot analysis
Bacteria grown in THY were collected at OD600 = ~0.6 (ME), centrifuged at 16,100g for 1 min, and pellets were resuspended in PBS. The Western immunoblot procedure used has been described [38], with the following modifications: (i) protein transfer to nitrocellulose membranes was done for 80 (Fig 6) or 45 min (Fig 2C) at 120 volts, and (ii) the anti-R28 antibody [38] was diluted to 1:1,250 in PBS-T with 5% nonfat dry milk, whereas the HRP-conjugated anti-rabbit secondary antibody was used at 1:13,500 dilution.
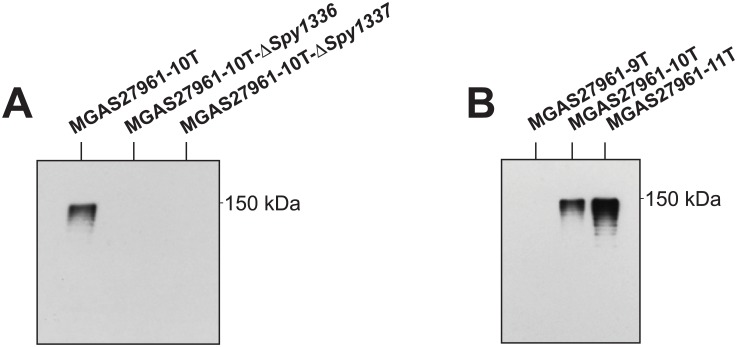
Fig 6. Western immunoblot analysis of Spy1336/R28.
Isogenic strains were collected at mid-exponential phase (OD600 = ~0.6). Whole cells were assayed for the presence of immunoreactive R28 protein with an anti-R28 polyclonal antibody [38]. (A) Isogenic deletion mutants assayed with anti-R28. (B) Isogenic HTSpy1336-7 size variant mutants assayed with anti-R28. 10% polyacrylamide gels were used.
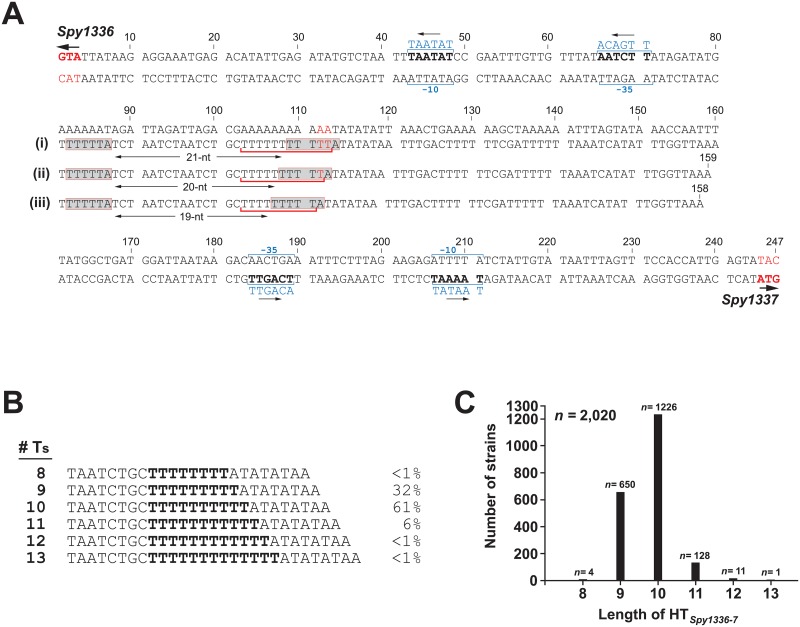
Fig 2. Heterogeneity in the homopolymeric tract located between Spy1336/R28 and Spy1337.
(A) Spy1336/R28-Spy1337 regulatory region from the reference strain MGAS6180 [31]. (i), (ii), and (iii) refer to HTSpy1336-7 containing 11, 10, or 9 T nucleotides, respectively, and the change in the spacing between two ATTTTT (shaded) direct repeats. The HTSpy1336-7 variants are underlined in red. Inferred promoters are shown. (B) DNA sequence of the prevalent HTSpy1336-7 alleles differing by one T nucleotide. Numbers at right refer to percent of all isolates analyzed with the designated number of Ts. (C) Frequency distribution of the most common HTSpy1336-7 alleles in the emm28 population. HTSpy1336-7 containing between 8 and 13 T nucleotides are shown with the corresponding number of strains (n) in which they are present. The total number of strains represented is 2,020.
Construction of isogenic mutant strains
All isogenic mutant strains used in this study are listed in S6 Table. Isogenic mutant strain MGAS27961-11T containing an 11T HTSpy1336-7 was generated using allelic exchange as described previously [73]. Briefly, primers HPN-1 and HPN-2 [38] were used to amplify a ~2,690-bp fragment using genomic DNA of MGAS11108, an emm28 clinical isolate with the naturally occurring 11T nucleotide region. The amplicon encompasses HTSpy1336-7. The resulting PCR product was cloned into suicide plasmid pBBL740 and transformed into parental strain MGAS27961-9T. The plasmid integrant was used for allelic exchange as described previously [73]. To identify strains putatively containing the allelic replacement region encompassing the expected polymorphism (11 T nucleotides), we sequenced the Spy1336/R28 upstream region using primer HPN-seq (S5 Table).
Isogenic mutant strain MGAS27961-10T-ΔSpy1336 was constructed using MGAS27961-10T genomic DNA as template for amplification. All primers are listed in S5 Table. Primer sets 1336–1 and -2 and 1336–3 and -4 were used to amplify two fragments upstream and downstream, respectively, of Spy1336/R28. The two PCR fragments were merged by combinatorial PCR and ligated into the BamHI site of suicide vector pBBL740. The recombinant plasmid containing a Spy1336/R28 deletion encompassing the entire gene was transformed into strain 27961-10T to replace the native Spy1336/R28 via allelic exchange. Isogenic mutant strains MGAS27961-10T-ΔSpy1337 and the MGAS27961-10T-ΔSpy1336/ΔSpy1337 double mutant were constructed with methods analogous to those described above. Primer sets 1337–1 and -2 and 1337–3 and -4 were used to generate MGAS27961-10T-ΔSpy1337, and 1336-1337-1 and -2 and 1336-1337-3 and -4 were used to generate the double mutant. Whole genome sequence analysis on the isogenic mutant strains confirmed the absence of spurious mutations.
RNAseq library preparation, sequencing, and analysis
Isogenic emm28 strains were grown in triplicate in THY and harvested at mid-exponential (ME; OD600 = 0.46–0.52) and early-stationary (ES; OD600 = 1.65–1.7) phases of growth. Bacteria (2 ml) from the ME phase and ES phase (1 ml) were added to 4 ml and 2 ml of RNAprotect Bacteria Reagent (Qiagen), respectively, incubated at room temperature for 20 min, and centrifuged at 4,000 rpm for 15 min. The supernatant was discarded, and the bacterial pellet was frozen in liquid nitrogen and stored at -80°C. The RNeasy kit (Qiagen) was used for total RNA isolation, and the quality of the total RNA was evaluated with RNA Nano chips (Agilent Technologies) and an Agilent 2100 Bioanalyzer. RNA extraction for all emm28 isogenic strains was performed as described previously [38, 70, 74]. The rRNA was depleted with the Ribo-Zero rRNA removal kit for Gram-positive bacteria (Illumina). The quality of the rRNA-depleted RNA was evaluated with RNA Pico chips (Agilent Technologies) and an Agilent 2100 Bioanalyzer. NEBNext Ultra II DNA library prep kit (NEB) was used to prepare the cDNA libraries, according to the manufacturer’s instructions. The quality of the cDNA libraries was evaluated with High-Sensitivity DNA chips (Agilent Technologies) and an Agilent 2100 Bioanalyzer. The cDNA library concentration was measured fluorometrically with Qubit dsDNA BR and HS assay kits (Invitrogen). Analysis of the RNAseq data was performed as described previously [38].
Necrotizing myositis infection models
A mouse model of necrotizing myositis was used to compare virulence of the 9T, 10T and 11T isogenic strains as previously described [38]. Briefly, 120 CD1 mice from Envigo were kept in cases containing 5 animals each, provided with chow pellets and acidified water ad libitum, and corn cob bedding with nesting material. They were inoculated in the right hindlimb with 5x108 CFU (n = 40 mice per strain) and followed for 7 days. They were euthanized with an overdose of isoflurane (primary), followed by cervical dislocation (secondary).
A well-described NHP model of necrotizing myositis was used to compare the virulence of the wildtype and isogenic MGAS27961-10T-ΔSpy1336/R28 and MGAS27961-10T-ΔSpy1337 strains [70, 75]. Three cynomolgus macaques (2–3 years, 2–4 kg) were used. Animals were randomly assigned to different strain treatment groups and inoculated with 5x109 CFU/kg of one strain in the right limb and a different strain in the left limb. Each strain was tested in triplicate. The animals were observed continuously and necropsied at 24 h post-inoculation. Lesions (necrotic tissue) were excised, measured in three dimensions, and volume was calculated using the formula for an ellipsoid. A full-thickness section of tissue taken from the inoculation site was fixed in 10% phosphate buffered formalin and embedded in paraffin using standard automated instruments. Histology of the three sections taken from each limb was scored by a pathologist blinded to the strain treatment groups [75, 76]. To obtain the quantitative CFU data, diseased tissue recovered from the inoculation site was weighed, homogenized (Omni International) in 1 mL PBS, and CFUs were determined by plating serial dilutions of the homogenate. Statistical differences between strain groups were determined with the Mann-Whitney test. Animal studies were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute (protocol numbers AUP-1217-0058 and AUP-0318-0016).
The humane endpoints used to determine when animals should be euthanized were for animals to be sacrificed if they either become immobile, reach lame scale = 4, reach body condition <2, develop an injection site abscess > 1 cm diameter, the injection site abscess ruptures, a metastatic (site other than injection site) abscess forms, lose >10% weight, or demonstrate other features of severe distress. Since none of the above criteria applied, the duration of the experiment was 7 days for mice and 24h for NHPs. Animals were euthanized immediately after the 7 day or 24h deadlines. The numbers of animals used was 120 (mice) and 3 (NHPs). They were all euthanized, and none was found dead. Animal care and handling was provided by the Comparative medicine core. Animal health and behavior were monitored at least once daily. All animal welfare considerations taken, including efforts to minimize suffering and distress, use of analgesics or anaesthetics, or special housing conditions were in accordance with guidelines specified by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute (protocol numbers AUP-1217-0058 and AUP-0318-0016).
Mice were housed in groups of 5 animals per individually ventilated techniplast cage. Cages are furnished with autoclaved quarter inch corncob bedding and a pulped virgin cotton fiber nestlet. Irradiated Teklad Global Diet 2920 pellets and acidified reverse osmosis water were provided ad libitum. Animals were examined at least once daily by the veterinary staff, attending veterinarian and investigator. Similarly, nonhuman primates were housed individually in squeeze back cages with a perch and provided chow, fresh fruit and vegetables, and water ad libitum. An enrichment program is maintained by the comparative medicine program.
Neutrophil bactericidal activity assays
Neutrophil bactericidal activity assays were performed in accordance with protocol 01-I-N055, approved by the Institutional Review Board for human subjects, National Institute of Allergy and Infectious Diseases. All volunteers gave written informed consent prior to participation in the study. Human neutrophils were isolated from the venous blood of healthy volunteers using a standard method [77]. Killing of S. pyogenes by human neutrophils was performed as described previously [38], except assay tubes were rotated for 3 h at 37°C.
Statistical analysis
Unless otherwise stated, error bars represent standard deviation (SD), and P values were calculated using either Kruskal-Wallis, or log-rank tests. Differential expression analysis was performed using DESeq2 1.16.1. Genes were considered differentially expressed if the fold-change was greater than 1.5-fold and associated with adjusted P value (Bonferroni corrected) < 0.05. For mouse survival studies, results were graphed as Kaplan-Meier curves and data were analyzed using the log-rank test with P < 0.05 considered to be significant. For the NHP virulence studies, lesion volume and CFU data were graphed as mean +/- SEM and analyzed using the Kruskal-Wallis test with P < 0.05 considered to be significant.
Results
Heterogeneity in the number of ALP-family long tandem repeats in the Spy1336/R28 gene and protein
The R28 protein has 3 domains: the amino-terminal and carboxy-terminal domains flank the size-variable central TRR28 domain. The amino-terminal domain is composed of 424 aa and it contains a secretion signal sequence with the YSIRK motif. The carboxy-terminal domain has 46 aa and it contains a cell-wall anchoring sequence with the LPXTG motif characteristic of surface-attached proteins (Fig 1A). Inasmuch as the number of TRs can vary by recombination or other mechanisms [62–65] we hypothesized that the large number of emm28 GAS strains we studied could have different size variants of the Spy1336/R28 gene as a consequence of having different numbers of TRR28s. Our previously reported Illumina paired-end 150nt read length whole genome sequence data [38] could not be used to accurately assemble and determine the number of repeats in the TRR28 region, as the sequence reads are of insufficient length to span the 237nt repeated motif. Thus, to determine the extent of size variation in TRR28s in the Spy1336/R28 gene, we used PCR analysis, as described in the Materials and Methods, and studied the 493 strains analyzed previously by RNAseq [38]. Overall, we identified TRR28s size variants ranging from 1 to 17 copies of the repeat (Fig 1B and S1 Table). The most common numbers of TRR28s identified was ten (n = 71, 14.4%) then nine (n = 69, 14.0%). The inferred molecular weight of several R28 variants was confirmed by Western immunoblot analysis (Fig 1C).
Heterogeneity in a homopolymeric tract in the intergenic region between Spy1336/R28 and Spy1337
We previously discovered that a single nucleotide indel located in an HT in the intergenic region between the divergently transcribed Spy1336/R28 and Spy1337 genes (Figs 1A and 2A) significantly altered the transcript levels of the two genes [38]. Specifically, strains with 9Ts in the HT produced little or no detectable transcript of these two genes, whereas organisms with 10Ts in this tract produced abundant and significantly increased levels of transcripts [38]. Moreover, increased transcript levels of Spy1336/R28 and Spy1337 resulted in increased production of the R28 virulence factor and increased virulence in a mouse necrotizing myositis infection model [38]. We reported that approximately two-thirds of 493 strains had the 10T variant of the HTSpy1336-7 region, whereas one-third of strains had a 9T variant [38].
Taken together, these observations provided the impetus to expand our study of heterogeneity in the HTSpy1336-7 region to include the entire previously described cohort of 2,095 emm28 clinical isolates recovered from invasive human infections collected in six countries over a 26-year period [38]. The HTSpy1336-7 region was analyzed in three ways, as described in detail in Materials and Methods. First, the contigs from genome assemblies generated with SPAdes [71] for all strains were searched with sequences of 20-nt flanking HTSpy1336-7 on each side using nucleotide Basic Local Alignment Search Tool (BLASTn). The identified HTSpy1336-7 target regions were retrieved, binned by alleles, and alleles were enumerated. Subsequently, the number of T nucleotides in the HTSpy1336-7 for each allele was counted. This method yielded results for the great majority of strains (~89%). As a second assembly method, we interrogated the Illumina sequencing reads for each strain using a set of eight probes of 31-nts in length corresponding to HTSpy1336-7 alleles with 6 to 13 Ts. In the aggregate, six alleles of HTSpy1336-7 were identified that differed from one another only by the number of T residues, varying in length from 8 to 13Ts (Fig 2B; S2 and S3 Tables). This analysis identified the same approximate frequency distribution for strains containing 9T or 10T nucleotides as described in our previous study [38], namely ~1/3 (n = 650) and ~2/3 (n = 1,226), respectively. We also discovered that ~6% (n = 128) of the strains had 11Ts in the HTSpy1336-7 sequence (Fig 2C). A small number of strains had 8Ts, 12Ts, or 13Ts in the HTSpy1336-7 sequence (S2 and S3 Tables). For any strain with discrepant or no results, we visually inspected read alignment, i.e. Binary Alignment Map (BAM) files corresponding to the Spy1336/R28-Spy1337 intergenic region using TABLET [78].
Construction and growth characteristics of isogenic mutant strains
We previously compared the global transcriptomes of isogenic mutant strains (MGAS27961-9T and MGAS27961-10T) that differ only in the number of T residues in the HTSpy1336-7 region [38]. In view of our finding that 6% of strains have 11Ts in this region, we constructed isogenic mutant strain MGAS27961-11T. We also constructed isogenic mutant strains MGAS27961-ΔSpy1336/R28, MGAS27961-ΔSpy1337, and MGAS27961-ΔSpy1336/ΔSpy1337 in which the target genes were deleted in a parental strain with 10Ts in the HTSpy1336-7 region. The goal of generating these strains was to perform comparative transcriptome and virulence analyses. All strains had a very similar growth curve under the laboratory conditions tested (S1 Fig).
Transcriptome analysis of clinical strains
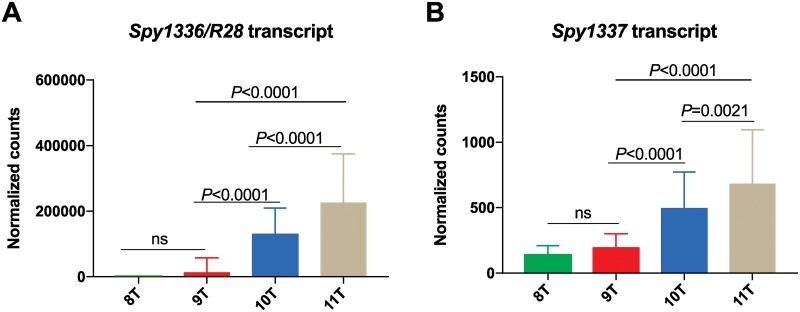
We examined the transcriptome data for 442 M28 clinical isolates [38] to determine if variation in the length of HTSpy1336-7 (Fig 2B) altered the level of transcripts for Spy1336/R28 and Spy1337. Among these 442 clinical isolates, 423 strains had alleles exclusively containing indels in HTSpy1336-7. All HT variants were represented in these 423 strains. We restricted examination of the transcriptome data to strains with 8 to 11Ts in the HTSpy1336-7 region because the 12T and 13T variants were represented by only one strain each. Strains with 8 and 9Ts in the HTSpy1336-7 region had low transcript levels of Spy1336/R28 and Spy1337 genes (Fig 3). In contrast, strains with the 10T variant had significantly higher transcript levels of Spy1336/R28 and Spy1337 compared to strains with the 9T variants (P<0.0001). Strains with the 11T variant (n = 25) had transcript levels for Spy1336/R28 (P<0.0001) and Spy1337 (P = 0.0021) that are significantly higher than either 10T or 9T strains (Fig 3).
Fig 3. Transcript levels of Spy1336/R28 and Spy1337 in 421 M28 clinical isolates.
RNAseq was performed on the isolates grown to early stationary phase, as described previously [38]. Transcript levels (normalized counts) of (A) Spy1336/R28 and (B) Spy1337 for strains with 8Ts (n = 4), 9Ts (n = 137), 10Ts (n = 255) or 11Ts(n = 25) in the HTSpy1336-7 region. Mean expression levels are plotted and error bars represent standard deviation. P values were calculated using one-way ANOVA with Tukey’s multiple comparison test. ns, not significant.
Transcriptome analysis of isogenic mutant strains
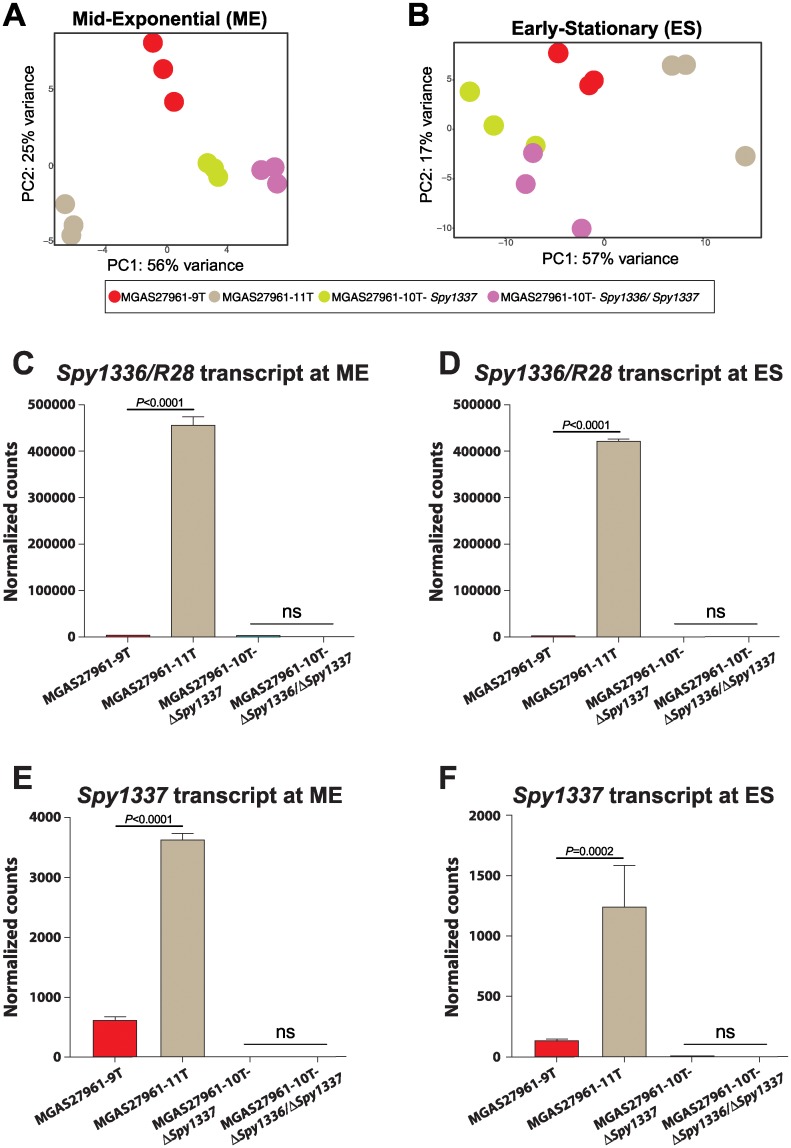
We next used RNAseq to test the hypothesis that, when compared to a parental strain with 9Ts (MGAS27961-9T) in the HTSpy1336-7 region, an isogenic mutant strain with 11Ts (MGAS27961-11T) had an altered transcriptome. RNAseq analysis confirmed that the gene expression profile of MGAS27961-11T is modestly altered compared to MGAS27961-9T. Principal component analysis supported these findings (Fig 4A and 4B). Moreover, compared to strain MGAS27961-9T, isogenic strain MGAS27961-11T had differential expression of 4.7% (3.2% upregulated and 1.5% downregulated at ME) and 6% (0.7% upregulated and 5.4% downregulated at ES) of the GAS genome (S2A Fig). We note that at both phases of growth, transcript levels of Spy1336/R28 in MGAS27961-11T were significantly higher compared to the MGAS27961-9T strain values (Fig 4C and 4D, and S2A Fig). As expected, no detectable transcript expression of Spy1336/R28 and Spy1337 was observed in the isogenic deletion mutant strains, MGAS27961-ΔSpy1337 and MGAS27961-ΔSpy1336/ΔSpy1337 (Fig 4C–4F). Virulence genes significantly upregulated, using a 1.5-fold cut-off, included: nga, encoding an NADase cytotoxin; slo, encoding the cytolytic protein streptolysin O; the sag operon encoding streptolysin S; mga, a positive transcriptional regulator of multiple virulence genes; emm28 encoding M protein; and sof, encoding serum opacity factor (S2 Fig).
Fig 4. Transcriptome analysis of parental and isogenic mutant strains.
RNAseq was performed on isogenic mutant strains MGAS27961-9T, MGAS27961-11T, MGAS27961-ΔSpy1337, and MGAS27961-10T-ΔSpy1336/ΔSpy1337 using three biological replicates per strain at two phases of growth: mid-exponential (ME) and early-stationary (ES). Principal component analysis shows distinct clustering of isogenic mutant strains at the (A) ME and (B) ES phases of growth. Strain MGAS27961-11T clusters away from the isogenic strains with mutations that decrease (MGAS27961-9T) or eliminate expression of Spy1336/R28 and/or Spy1337 (MGAS27961-ΔSpy1337 and MGAS27961-10T-ΔSpy1336/ΔSpy1337). (C and D) At both ME and ES, transcript levels of Spy1336/R28 for strain MGAS27961-11T were significantly higher compared to strain MGAS27961-9T. At both phases of growth, no detectable Spy1336/R28 transcript was observed in the isogenic knockout strains (MGAS27961-ΔSpy1337 and MGAS27961-10T-ΔSpy1336/ΔSpy1337. (E and F) Similarly, at both ME and ES, transcript levels of the gene Spy1337 were significantly higher in MGAS27961-11T compared to strain MGAS27961-9T. Mean transcript levels are plotted and error bars represent standard deviation. P values were calculated using one-way ANOVA with Tukey’s multiple comparison test.
Heterogeneity in the HTSpy1336-7 region significantly affects virulence in a mouse model of necrotizing myositis
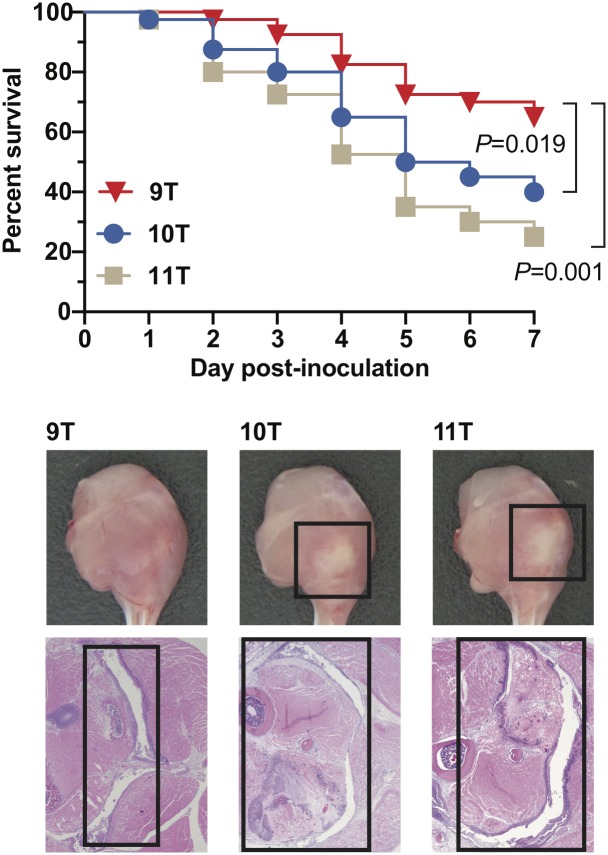
To test the hypothesis that the number of T nucleotides in HTSpy1336-7 region contributes to GAS virulence, we inoculated mice intramuscularly with either the parental strain with 9Ts in HTSpy1336-7 or an isogenic mutant strain with 10Ts or 11Ts. Compared to the parental 9T strain, the isogenic 10T and 11T strains each caused significantly greater mortality and larger lesions with more tissue destruction (Fig 5). Taken together, the data support the hypothesis that the number of T nucleotides in HTSpy1336-7 significantly affects virulence in this infection model, especially when comparing the 9T to either the 10T or 11T isogenic strains.
Fig 5. Virulence assessed in a mouse model of necrotizing myositis.
(A) Virulence of 9T, 10T, and 11T isogenic strains in a mouse model of necrotizing myositis (n = 40 mice per strain). The treatment groups’ size was based on a power calculation from results of a pilot study. Significantly increased near-mortality was observed for the 10T and 11T strains compared with the 9T strain. Significance was determined using the log-rank test. (B) Representative gross and microscopic pathology images of the hindlimb lesions from mice (n = 5 mice per strain). Boxed areas demarcate areas with major lesions.
Analysis of the R28 protein made by the isogenic mutant strains
In most bacteria, gene transcript levels typically correlate with amounts of the encoded proteins made, but this is not always the case [79, 80]. We reported previously that the R28 protein is produced and detected by Western immunoblot in whole cell extracts and supernatants derived from MGAS27961-10T, but not from the isogenic strain MGAS27961-9T. This finding is consistent with RNAseq data showing that the transcript level of Spy1336/R28 was higher in MGAS27961-10T compared to MGAS27961-9T [38].
To build on our previous observations, we used Western immunoblot analysis to assess R28 production by a strain with an HTSpy1336-7 11T variant and isogenic strains MGAS27961-11T, MGAS27961-10T-ΔSpy1336, and MGAS27961-10T-ΔSpy1337 (Fig 6A). As anticipated, R28 protein was undetectable in strain MGAS27961-10T-ΔSpy1336/R28, and production by strain MGAS27961-10T-ΔSpy1337 was also not detected under the conditions tested (Fig 6A). The lack of discernible R28 production by the MGAS27961-10T-ΔSpy1337 strain is consistent with our hypothesis that Spy1337 is a positive transcriptional regulator of Spy1336/R28 [38].
Next, we analyzed R28 protein production by parental and isogenic mutant strains containing the three different length variants of HTSpy1336-7 (Fig 6B). Consistent with previous data, we did not detect production of R28 by parental strain MGAS27961-9T, whereas immunoreactive R28 was produced by isogenic mutant strain MGAS27961-10T [38]. Also consistent with our hypothesis and RNAseq data, isogenic mutant strain MGAS27961-11T produced greater amounts of immunoreactive R28 compared to strain MGAS27961-10T and parental strain MGAS27961-9T (Fig 6B).
Spy1336/R28 and Spy1337 contribute significantly to virulence in a non-human primate (NHP) model of necrotizing myositis
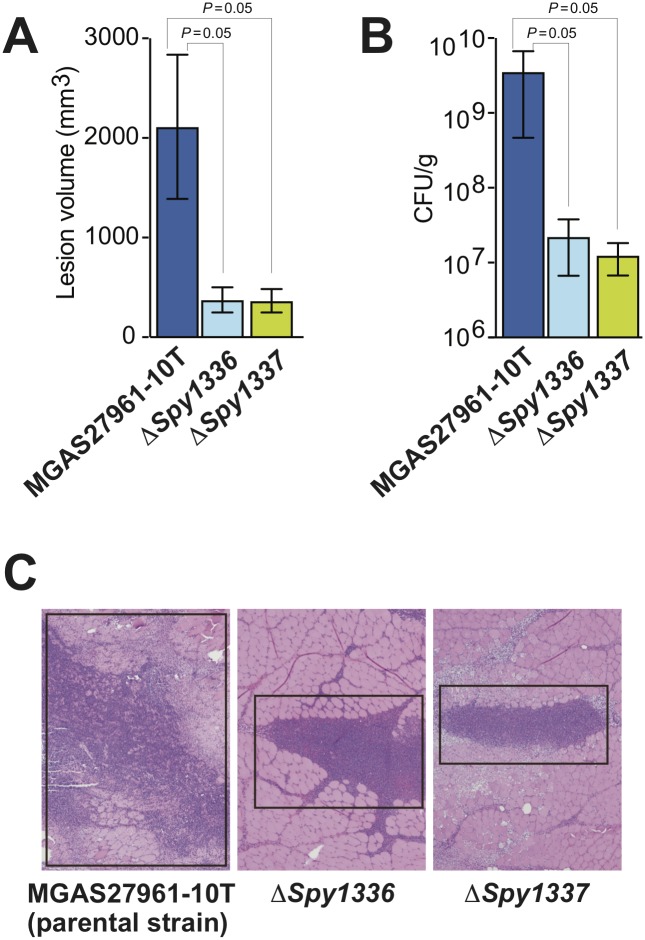
To test the hypothesis that Spy1336/R28 and Spy1337 contribute to GAS virulence we used NHPs, a well-studied animal model that closely resembles human hosts in terms of physiology and immune response [70, 81–83]. We inoculated NHPs with parental strain MGAS27961-10T and isogenic mutant strains MGAS27961-10T-ΔSpy1336/R28 or MGAS27961-10T-ΔSpy1337. Compared to the parental strain, each of the two isogenic deletion-mutant strains caused significantly smaller lesions characterized by less tissue destruction (Fig 7A and 7C). In addition, compared to the parental strain, significantly fewer CFUs of each isogenic mutant strain were recovered from the inoculation site (Fig 7B). Taken together, the data support the hypothesis that Spy1336/R28 and Spy1337 contribute to virulence in this infection model.
Fig 7. Virulence assessed in a nonhuman primate model of necrotizing myositis.
Compared to the parental wild-type strain, deletion of Spy1336/R28 (ΔSpy1336) or Spy1337 (ΔSpy1337) significantly decreases (A) lesion volume, (B) CFU recovery, and (C) tissue destruction. Shown in panel C are representative microscopic images of the necrotic lesion centered at the GAS injection site (black box); hematoxylin and eosin stain, original magnification 4X. P = 0.05, Mann-Whitney Test. n = 3 NHPs/GAS strain.
Discussion
S. pyogenes, a leading cause of human morbidity, mortality, and healthcare costs globally, produces a large number of extracellular virulence factors. Strains of serotype M28 S. pyogenes are repeatedly associated with puerperal sepsis (childbed fever) and are a prominent cause of pharyngitis in many countries. The molecular mechanisms responsible for puerperal sepsis, other invasive infections, and pharyngitis caused by serotype M28 strains are poorly understood. Several lines of evidence indicate that the Spy1336/R28 gene encodes a cell surface-anchored virulence factor [24, 84] that is involved in S. pyogenes pathogenesis. We analyzed the Spy1336/R28 gene encoding the R28 protein in approximately 2,000 emm28 invasive strains. We found DNA sequences located in the upstream regulatory region and repeat sequences in its coding sequence that make Spy1336/R28 highly polymorphic when compared across our population sample; importantly, these polymorphisms affect virulence.
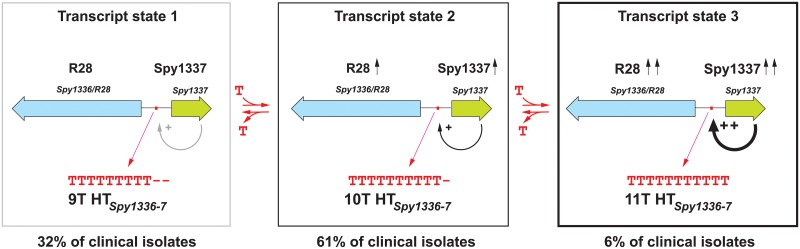
We previously reported a bimodal distribution in the transcripts made by Spy1336/R28 and Spy1337 in a study of 492 emm28 strains [38]. Namely, strains with 9Ts in HTSpy1336-7 located between Spy1336/R28 and Spy1337 produced low transcript levels (Figs 1A and 2A), whereas strains with 10Ts produced significantly greater transcript levels [38]. The current work confirmed and extended our observations and found that in the sample of ~2,000 strains, and when considering alleles exclusively containing indels in the HTSpy1336-7, (i) the number of T residues varied between 8 and 13, (ii) most strains (~90.4%) had 9T (32%) or 10T (61%) residues, (iii) an 11T variant was present in 6% of the strains (Figs 2B, 2C and 8), and (iv) non-isogenic clinical isolates and an isogenic strain with an 11T variant had significantly higher transcript levels of Spy1336/R28 and Spy1337, and produced more R28 protein than 9T and 10T variant isogenic strains (Figs 3 and 6, S2 and S3 Tables).
Fig 8. Frequency distribution of HTSpy1336-7 size variants in the emm28 strains studied and effect on Spy1336/R28 and Spy1337 transcript levels.
Addition or deletion of one T nucleotide in the homopolymeric tract results in a statistically significant change in transcript levels for Spy1336/R28 and Spy1337 and a consequent effect on virulence. There are three transcript states based on number of T residues in the HTSpy1336-7 region. In state 1 (9Ts, left), there are essentially no detectable transcripts of Spy1336/R28 and Spy1337. State 2 (10Ts, center) is characterized by increased transcripts of these two genes, and state 3 (11Ts, right) results in further increase in transcripts of Spy1336/R28 and Spy1337.
Size variants of the HTSpy1336-7 may arise because HTs, especially poly(T)-containing HTs such as HTSpy1336-7, are able to form transient mispaired regions leading to slipped-strand mispairing during DNA replication, resulting in expansion or contraction of the HT [85–90]. HTs located at the 5’ upstream untranslated regions of genes such as the HTSpy1336-7 can contribute to altered regulation of gene transcript expression [68, 91]. In this regard, HTs may be involved in phase variation [92] and bacterial adaptation [65, 93]. The finding that the vast majority of strains had a 9T or 10T genotype suggests that this system may influence GAS-human interaction in some settings. Under one scenario, lack of or very low transcript production (9T genotype) is advantageous to the organism in certain currently undefined physiologic conditions. Conversely, the significantly-higher transcript level conferred by the 10T genotype could be advantageous in other conditions, also currently not defined. Compared to the 9T strain, the isogenic mutant strain with the 11T genotype had a higher level of Spy1336/R28 transcript (Fig 4C and 4D). The 11T strain also had a higher level of production of R28 protein (Fig 6B), compared to both the 9T and 10T strains. We hypothesize that the increased R28 protein made by the 11T strain confers only slightly enhanced fitness to the organism (relative to the 10T genotype), an idea that is consistent with the observation that relatively few (6%) natural clinical isolates have this genotype, and substantiated by the fact that the isogenic mutant strain had only modestly increased virulence in the mouse model of necrotizing myositis. Alternatively, too much R28 protein could be detrimental during a natural course of infection in the human host by promoting too tight adherence, leading to decreased dissemination, or even promoting an increased immune response. Consistent with these ideas, the 10T and 11T isogenic mutant strains did not differ substantially in the number of differentially-expressed genes (S2 Fig, panels A and D), or in their resistance to killing by human PMNs ex vivo (Fig 9), although compared to the 9T wild-type parental strain, these two mutant strains had significantly enhanced resistance to killing by human PMN, and more differentially expressed genes (S2 Fig, panels A, B, and D). In terms of the small number of strains containing additional size variants of the HT region (8Ts, 12Ts, and 13Ts), strains with 8Ts produce very minimal levels of Spy1336/R28 transcripts (Fig 3A), and those with 12Ts and 13Ts might not further enhance pathogen fitness, or alternatively, global transcript changes occurring in strains with these genotypes in some way decrease fitness. Clearly, additional investigations are required to address these ideas.
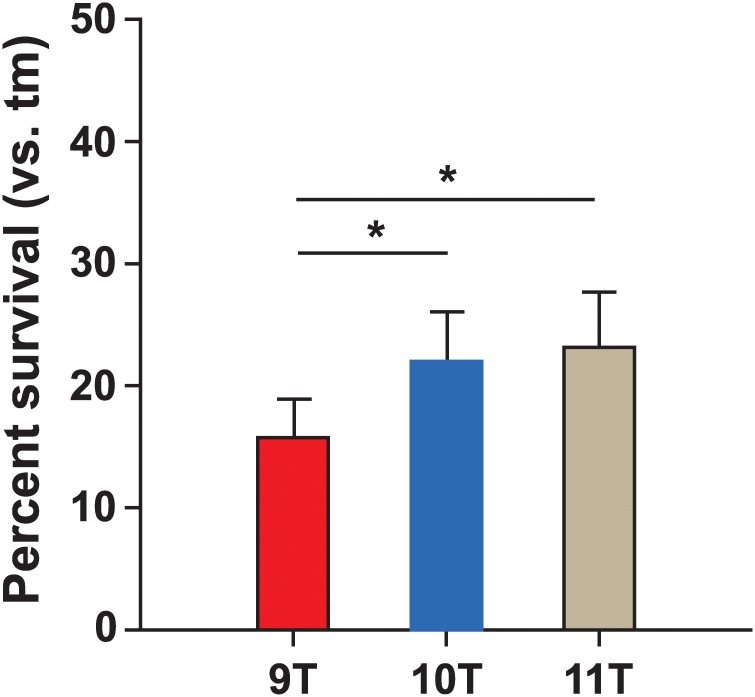
Fig 9. Resistance to killing by human PMNs.
Survival of 3 variant HTSpy1336-7 isogenic strains in the presence of purified human PMNs. Bacterial survival data are expressed as the mean ± SEM. *P < 0.05 using a repeated-measures one-way ANOVA and Tukey’s posttest (n = 7 experiments). tm, time-matched control lacking PMNs.
Puopolo and Madoff reported that deletion or expansion of a 5-nt repeat (AGATT) adjacent to the poly(T) tract is associated with a null phenotype for expression of the bca gene encoding the alpha C protein in GBS [68]. This region is highly conserved in GAS and GBS. However, only a small number of strains were studied. Here, analysis of 2,074 clinical isolates recovered from human patients with invasive disease identified only four strains with variation in this pentanucleotide motif (TCTAA in S3 Table), whereas most of the variation was found in HTSpy1336-7 region. Thus, in the natural populations of GAS we studied, variation in HTSpy1336-7 is the key driver of transcript level changes of Spy1336/R28 and Spy1337 [38].
Neither Spy1336/R28 transcript nor R28 protein production were detectable in an isogenic strain in which the Spy1337 transcription factor gene was deleted. In addition, deletion of either Spy1336/R28 or Spy1337 resulted in a significant decrease in virulence in an NHP model of necrotizing myositis (Fig 7). Thus, when taken together, our present and previous [38] data are consistent with a model in which the regulation of expression of the gene encoding the R28 virulence factor is partly dependent on a process whereby indels occurring in the HTSpy1336-7 affect binding of the Spy1337 transcriptional regulator to this DNA region by altering its consensus binding site, and/or changing the spacing, and therefore spatial orientation between two adjacent binding sites (Fig 2A). In this regard, the DNA sequence ATTTT present twice in the Spy1336/R28-Spy1337 regulatory region resembles part of the consensus binding site for the AraC transcriptional regulator ToxT [94]. In our proposed model, Spy1337 positively regulates the expression of both Spy1336/R28 and Spy1337 (Fig 8).
The R28 virulence factor binds through its N-terminal domain to integrin receptors α3β1, α6β1, and α6β4 [60], which in the human host bind to laminin, an ECM protein [61]. This constitutes an additional instance in which a pathogen binds to host integrins [95–98], a strategy described for other GAS proteins [99]. For example, GAS PrtF1 binds to α5β1 integrins and could trigger integrin clustering and internalization of α5β1 integrin, ultimately resulting in GAS uptake [100]. The second region in Spy1336/R28 containing repeat sequences is located in its coding sequence (Fig 1A). A 237-nt repetitive sequence, corresponding to one 79-aa TRR28, is present in Spy1336/R28 from 1 to 17 tandem copies (Fig 1B and S1 Table). Ten is the most prevalent number of TRR28 copies present in R28, followed by 9 (Fig 1B). Thus, emm28 GAS strains make different size variants of the R28 protein, likely arising through recombination [62–65, 101]. The function of TRs in virulence factors is not well understood. One possible function would be to extend the reach of a surface-anchored protein and expose its N-terminal domain at the bacterial surface without adding new mechanistic functions [102]. Variation in the number of TRs may alternatively produce antigenic variation. In this regard, variation in the conserved region of GAS M protein generates antigenic diversity [102], and similarly, variation in the GBS alpha C protein affects antigenicity and protective efficacy [52, 103]. TR number variation might decrease adhesion to and entry into host cells [104]. No correlation was found between the number of T nucleotides in HTSpy1336-7 and the number of TRR28 repeats (S3 Fig). Additional studies designed to address these ideas are warranted.
To summarize, the R28 virulence factor in GAS is highly polymorphic in natural populations, both in level of transcript production and protein expression, and size. Differences in transcription levels are caused by variation in the number of Ts in a homopolymeric tract upstream of the Spy1336/R28 gene, whereas variance in R28 protein size is caused by variation in the number of identical TRR28 tandem repeats in the structural gene. Isogenic mutant strains in which genes encoding R28 or transcriptional regulator Spy1337 are inactivated are significantly less virulent in a NHP model of necrotizing myositis. These findings provide new information about the extent of natural genetic diversity and virulence in this two-gene virulence axis and set the stage for additional studies addressing the role of Spy1336/R28 and Spy1337 in pathogen-host interactions.
Supporting information
Strains were grown at 37°C in THY medium.
(PDF)
(A) Total number of differentially expressed genes using a fold cutoff ≥1.5. Upregulated virulence genes comparing 9T vs 10T (B), 9T vs 11T (C), and 10T vs 11T (D).
(PDF)
Number of TRR28 repeats in strains with HTSpy1336-7 containing 8Ts (A), 9Ts (B), 10Ts (C), and 11Ts (D). n = 490 strains.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Leslie Jenkins for veterinary assistance and Dr. Kathryn Stockbauer for critical comments to improve the manuscript.
Data Availability
Transcriptome data is available in the Gene Expression Omnibus under accession number GSE145621.
Funding Statement
This study was supported in part by the Fondren Foundation, Houston Methodist Hospital and Research Institute and National Institutes of Health grant AI139369-01A1 (to J.M.M.), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to F.R.D.). Fondren Foundation URL: https://fconline.foundationcenter.org/fdo-grantmaker-profile/?key=FOND00 NIH URL: https://www.nih.gov/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–94. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- 2.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, et al. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014;27(2):264–301. 10.1128/CMR.00101-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens DL, Bryant AE. Impetigo, Erysipelas and Cellulitis In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK) 2016. [Google Scholar]
- 4.Beaudoin A, Edison L, Introcaso CE, Goh L, Marrone J, Mejia A, et al. Acute rheumatic fever and rheumatic heart disease among children—American Samoa, 2011–2012. MMWR Morb Mortal Wkly Rep. 2015;64(20):555–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Carapetis JR, Beaton A, Cunningham MW, Guilherme L, Karthikeyan G, Mayosi BM, et al. Acute rheumatic fever and rheumatic heart disease. Nat Rev Dis Primers. 2016;2:15084 10.1038/nrdp.2015.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Iturbe B, Haas M. Post-Streptococcal Glomerulonephritis In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK) 2016. [Google Scholar]
- 7.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. Journal of clinical microbiology. 1996;34(4):953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferretti JJ, Stevens D. L., Fischetti V. A. Streptococcus pyogenes basic biology to clinical manifestations. Ferretti Joseph F. S DL, and Fischetti Vincent A., editor. Oklahoma City, OK: 2016. [PubMed] [Google Scholar]
- 9.Barnett TC, Bowen AC, Carapetis JR. The fall and rise of Group A Streptococcus diseases. Epidemiol Infect. 2018:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores AR, Chase McNeil J, Shah B, Van Beneden C, Shelburne SA 3rd. Capsule-Negative emm Types Are an Increasing Cause of Pediatric Group A Streptococcal Infections at a Large Pediatric Hospital in Texas. J Pediatric Infect Dis Soc. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson GE, Pondo T, Toews KA, Farley MM, Lindegren ML, Lynfield R, et al. Epidemiology of Invasive Group A Streptococcal Infections in the United States, 2005–2012. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2016;63(4):478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shea PR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, et al. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg Infect Dis. 2011;17(11):2010–7. 10.3201/eid1711.110159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherardi G, Vitali LA, Creti R. Prevalent emm Types among Invasive GAS in Europe and North America since Year 2000. Front Public Health. 2018;6(59):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naseer U, Steinbakk M, Blystad H, Caugant DA. Epidemiology of invasive group A streptococcal infections in Norway 2010–2014: A retrospective cohort study. Eur J Clin Microbiol Infect Dis. 2016;35(10):1639–48. 10.1007/s10096-016-2704-y [DOI] [PubMed] [Google Scholar]
- 15.Plainvert C, Doloy A, Loubinoux J, Lepoutre A, Collobert G, Touak G, et al. Invasive group A streptococcal infections in adults, France (2006–2010). Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18(7):702–10. [DOI] [PubMed] [Google Scholar]
- 16.Smit PW, Lindholm L, Lyytikainen O, Jalava J, Patari-Sampo A, Vuopio J. Epidemiology and emm types of invasive group A streptococcal infections in Finland, 2008–2013. Eur J Clin Microbiol Infect Dis. 2015;34(10):2131–6. 10.1007/s10096-015-2462-2 [DOI] [PubMed] [Google Scholar]
- 17.Ben Zakour NL, Venturini C, Beatson SA, Walker MJ. Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing. Journal of clinical microbiology. 2012;50(7):2224–8. 10.1128/JCM.00675-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang I, Van Beneden C, Beall B, Schuchat A. Population-based surveillance for postpartum invasive group a streptococcus infections, 1995–2000. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2002;35(6):665–70. [DOI] [PubMed] [Google Scholar]
- 19.Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980–1990 and their association with disease. J Med Microbiol. 1993;39(3):165–78. 10.1099/00222615-39-3-165 [DOI] [PubMed] [Google Scholar]
- 20.Gaworzewska E, Colman G. Changes in the pattern of infection caused by Streptococcus pyogenes. Epidemiol Infect. 1988;100(2):257–69. 10.1017/s095026880006739x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond J, Schlegel L, Garnier F, Bouvet A. Molecular characterization of Streptococcus pyogenes isolates to investigate an outbreak of puerperal sepsis. Infect Control Hosp Epidemiol. 2005;26(5):455–61. 10.1086/502567 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. 2006;24(2):191–7. 10.1038/nbt1179 [DOI] [PubMed] [Google Scholar]
- 23.Stalhammar-Carlemalm M, Stenberg L, Lindahl G. Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J Exp Med. 1993;177(6):1593–603. 10.1084/jem.177.6.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stalhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Molecular microbiology. 1999;33(1):208–19. 10.1046/j.1365-2958.1999.01470.x [DOI] [PubMed] [Google Scholar]
- 25.Lancefield RC. Studies on the Antigenic Composition of Group a Hemolytic Streptococci: I. Effects of Proteolytic Enzymes on Streptococcal Cells. J Exp Med. 1943;78(6):465–76. 10.1084/jem.78.6.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancefield RC, Perlmann GE. Preparation and properties of a protein (R antigen) occurring in streptococci of group A, type 28 and in certain streptococci of other serological groups. J Exp Med. 1952;96(1):83–97. 10.1084/jem.96.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lancefield RC. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med. 1957;106(4):525–44. 10.1084/jem.106.4.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lachenauer CS, Creti R, Michel JL, Madoff LC. Mosaicism in the alpha-like protein genes of group B streptococci. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(17):9630–5. 10.1073/pnas.97.17.9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. Early-onset neonatal sepsis. Clin Microbiol Rev. 2014;27(1):21–47. 10.1128/CMR.00031-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PloS one. 2007;2(8):e800 10.1371/journal.pone.0000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, et al. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. The Journal of infectious diseases. 2005;192(5):760–70. 10.1086/430618 [DOI] [PubMed] [Google Scholar]
- 32.Maeland JA, Afset JE, Lyng RV, Radtke A. Survey of immunological features of the alpha-like proteins of Streptococcus agalactiae. Clin Vaccine Immunol. 2015;22(2):153–9. 10.1128/CVI.00643-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC microbiology. 2011;11:65 10.1186/1471-2180-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egan SM. Growing repertoire of AraC/XylS activators. Journal of bacteriology. 2002;184(20):5529–32. 10.1128/JB.184.20.5529-5532.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61(4):393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. Journal of bacteriology. 2003;185(9):2910–9. 10.1128/JB.185.9.2910-2919.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442(7105):943–6. 10.1038/nature05021 [DOI] [PubMed] [Google Scholar]
- 38.Kachroo P, Eraso JM, Beres SB, Olsen RJ, Zhu L, Nasser W, et al. Integrated analysis of population genomics, transcriptomics and virulence provides novel insights into Streptococcus pyogenes pathogenesis. Nat Genet. 2019;51(3):548–59. 10.1038/s41588-018-0343-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):2860–5. 10.1073/pnas.0915021107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Tauschek M, Robins-Browne RM. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol. 2011;19(3):128–35. 10.1016/j.tim.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 41.Santiago AE, Yan MB, Tran M, Wright N, Luzader DH, Kendall MM, et al. A large family of anti-activators accompanying XylS/AraC family regulatory proteins. Molecular microbiology. 2016;101(2):314–32. 10.1111/mmi.13392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Laguna Y, Calva E, Puente JL. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Molecular microbiology. 1999;33(1):153–66. 10.1046/j.1365-2958.1999.01460.x [DOI] [PubMed] [Google Scholar]
- 43.Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol Med Microbiol. 2010;58(3):344–55. 10.1111/j.1574-695X.2010.00645.x [DOI] [PubMed] [Google Scholar]
- 44.Munson GP, Scott JR. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Molecular microbiology. 2000;36(6):1391–402. 10.1046/j.1365-2958.2000.01957.x [DOI] [PubMed] [Google Scholar]
- 45.Porter ME, Mitchell P, Roe AJ, Free A, Smith DG, Gally DL. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Molecular microbiology. 2004;54(4):1117–33. 10.1111/j.1365-2958.2004.04333.x [DOI] [PubMed] [Google Scholar]
- 46.Yu RR, DiRita VJ. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. Journal of bacteriology. 1999;181(8):2584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischetti VA, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Molecular microbiology. 1990;4(9):1603–5. 10.1111/j.1365-2958.1990.tb02072.x [DOI] [PubMed] [Google Scholar]
- 48.Lindahl G, Stalhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev. 2005;18(1):102–27. 10.1128/CMR.18.1.102-127.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolduc GR, Baron MJ, Gravekamp C, Lachenauer CS, Madoff LC. The alpha C protein mediates internalization of group B Streptococcus within human cervical epithelial cells. Cell Microbiol. 2002;4(11):751–8. 10.1046/j.1462-5822.2002.00227.x [DOI] [PubMed] [Google Scholar]
- 50.Courtney HS, Hasty DL, Dale JB. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann Med. 2002;34(2):77–87. 10.1080/07853890252953464 [DOI] [PubMed] [Google Scholar]
- 51.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73(3):407–50, Table of Contents. 10.1128/MMBR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gravekamp C, Horensky DS, Michel JL, Madoff LC. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infection and immunity. 1996;64(9):3576–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madoff LC, Michel JL, Gong EW, Kling DE, Kasper DL. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(9):4131–6. 10.1073/pnas.93.9.4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michel JL, Madoff LC, Kling DE, Kasper DL, Ausubel FM. Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infection and immunity. 1991;59(6):2023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michel JL, Madoff LC, Olson K, Kling DE, Kasper DL, Ausubel FM. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(21):10060–4. 10.1073/pnas.89.21.10060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kvam AI, Mavenyengwa RT, Radtke A, Maeland JA. Streptococcus agalactiae alpha-like protein 1 possesses both cross-reacting and Alp1-specific epitopes. Clin Vaccine Immunol. 2011;18(8):1365–70. 10.1128/CVI.05005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Creti R, Fabretti F, Orefici G, von Hunolstein C. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. Journal of clinical microbiology. 2004;42(3):1326–9. 10.1128/JCM.42.3.1326-1329.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wastfelt M, Stalhammar-Carlemalm M, Delisse AM, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. The Journal of biological chemistry. 1996;271(31):18892–7. 10.1074/jbc.271.31.18892 [DOI] [PubMed] [Google Scholar]
- 59.Li J, Kasper DL, Ausubel FM, Rosner B, Michel JL. Inactivation of the alpha C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(24):13251–6. 10.1073/pnas.94.24.13251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weckel A, Ahamada D, Bellais S, Mehats C, Plainvert C, Longo M, et al. The N-terminal domain of the R28 protein promotes emm28 group A Streptococcus adhesion to host cells via direct binding to three integrins. The Journal of biological chemistry. 2018;293(41):16006–18. 10.1074/jbc.RA118.004134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belkin AM, Stepp MA. Integrins as receptors for laminins. Microsc Res Tech. 2000;51(3):280–301. [DOI] [PubMed] [Google Scholar]
- 62.Bi X, Liu LF. A replicational model for DNA recombination between direct repeats. J Mol Biol. 1996;256(5):849–58. 10.1006/jmbi.1996.0131 [DOI] [PubMed] [Google Scholar]
- 63.Richard GF, Paques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000;1(2):122–6. 10.1093/embo-reports/kvd031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rocha EP. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 2003;13(6A):1123–32. 10.1101/gr.966203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2014;38(1):119–41. 10.1111/1574-6976.12036 [DOI] [PubMed] [Google Scholar]
- 66.van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17(3):581–611, table of contents. 10.1128/CMR.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Ende A, Hopman CT, Dankert J. Multiple mechanisms of phase variation of PorA in Neisseria meningitidis. Infection and immunity. 2000;68(12):6685–90. 10.1128/iai.68.12.6685-6690.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puopolo KM, Madoff LC. Upstream short sequence repeats regulate expression of the alpha C protein of group B Streptococcus. Molecular microbiology. 2003;50(3):977–91. 10.1046/j.1365-2958.2003.03745.x [DOI] [PubMed] [Google Scholar]
- 69.van Ham SM, van Alphen L, Mooi FR, van Putten JP. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73(6):1187–96. 10.1016/0092-8674(93)90647-9 [DOI] [PubMed] [Google Scholar]
- 70.Eraso JM, Olsen RJ, Beres SB, Kachroo P, Porter AR, Nasser W, et al. Genomic Landscape of Intrahost Variation in Group A Streptococcus: Repeated and Abundant Mutational Inactivation of the fabT Gene Encoding a Regulator of Fatty Acid Synthesis. Infection and immunity. 2016;84(12):3268–81. 10.1128/IAI.00608-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–70. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Molecular microbiology. 2010;78(6):1332–47. 10.1111/j.1365-2958.2010.07427.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beres SB, Kachroo P, Nasser W, Olsen RJ, Zhu L, Flores AR, et al. Transcriptome Remodeling Contributes to Epidemic Disease Caused by the Human Pathogen Streptococcus pyogenes. mBio. 2016;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu L, Olsen RJ, Nasser W, de la Riva Morales I, Musser JM. Trading Capsule for Increased Cytotoxin Production: Contribution to Virulence of a Newly Emerged Clade of emm89 Streptococcus pyogenes. mBio. 2015;6(5):e01378–15. 10.1128/mBio.01378-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6901–6. 10.1073/pnas.092148299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26(3):401–2. 10.1093/bioinformatics/btp666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eraso JM, Roh JH, Zeng X, Callister SJ, Lipton MS, Kaplan S. Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. Journal of bacteriology. 2008;190(14):4831–48. 10.1128/JB.00301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS letters. 2009;583(24):3966–73. 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 81.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(17):E1768–76. 10.1073/pnas.1403138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olsen RJ, Sitkiewicz I, Ayeras AA, Gonulal VE, Cantu C, Beres SB, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):888–93. 10.1073/pnas.0911811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu L, Olsen RJ, Beres SB, Eraso JM, Saavedra MO, Kubiak SL, et al. Gene fitness landscape of group A streptococcus during necrotizing myositis. J Clin Invest. 2019;129(2):887–901. 10.1172/JCI124994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stalhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. Cross-protection between group A and group B streptococci due to cross-reacting surface proteins. The Journal of infectious diseases. 2000;182(1):142–9. 10.1086/315693 [DOI] [PubMed] [Google Scholar]
- 85.Achaz G, Rocha EP, Netter P, Coissac E. Origin and fate of repeats in bacteria. Nucleic Acids Res. 2002;30(13):2987–94. 10.1093/nar/gkf391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Henderson IR, Owen P, Nataro JP. Molecular switches—the ON and OFF of bacterial phase variation. Molecular microbiology. 1999;33(5):919–32. 10.1046/j.1365-2958.1999.01555.x [DOI] [PubMed] [Google Scholar]
- 87.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4(3):203–21. 10.1093/oxfordjournals.molbev.a040442 [DOI] [PubMed] [Google Scholar]
- 88.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128(4):655–68. 10.1016/j.cell.2007.01.023 [DOI] [PubMed] [Google Scholar]
- 89.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72(4):686–727. 10.1128/MMBR.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62(2):275–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Orsi RH, Bowen BM, Wiedmann M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC genomics. 2010;11:102 10.1186/1471-2164-11-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Molecular microbiology. 2004;52(2):357–69. 10.1111/j.1365-2958.2004.03996.x [DOI] [PubMed] [Google Scholar]
- 93.Kassai-Jager E, Ortutay C, Toth G, Vellai T, Gaspari Z. Distribution and evolution of short tandem repeats in closely related bacterial genomes. Gene. 2008;410(1):18–25. 10.1016/j.gene.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 94.Withey JH, Dirita VJ. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. Journal of bacteriology. 2005;187(23):7890–900. 10.1128/JB.187.23.7890-7900.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gorrell RJ, Guan J, Xin Y, Tafreshi MA, Hutton ML, McGuckin MA, et al. A novel NOD1- and CagA-independent pathway of interleukin-8 induction mediated by the Helicobacter pylori type IV secretion system. Cell Microbiol. 2013;15(4):554–70. 10.1111/cmi.12055 [DOI] [PubMed] [Google Scholar]
- 96.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–6. 10.1038/nature06187 [DOI] [PubMed] [Google Scholar]
- 97.LaFoya B, Munroe JA, Miyamoto A, Detweiler MA, Crow JJ, Gazdik T, et al. Beyond the Matrix: The Many Non-ECM Ligands for Integrins. Int J Mol Sci. 2018;19(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tegtmeyer N, Hartig R, Delahay RM, Rohde M, Brandt S, Conradi J, et al. A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. The Journal of biological chemistry. 2010;285(30):23515–26. 10.1074/jbc.M109.096214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stockbauer KE, Magoun L, Liu M, Burns EH Jr., Gubba S, Renish S, et al. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(1):242–7. 10.1073/pnas.96.1.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ozeri V, Rosenshine I, Ben-Ze'Ev A, Bokoch GM, Jou TS, Hanski E. De novo formation of focal complex-like structures in host cells by invading Streptococci. Molecular microbiology. 2001;41(3):561–73. 10.1046/j.1365-2958.2001.02535.x [DOI] [PubMed] [Google Scholar]
- 101.Puopolo KM, Hollingshead SK, Carey VJ, Madoff LC. Tandem repeat deletion in the alpha C protein of group B streptococcus is recA independent. Infection and immunity. 2001;69(8):5037–45. 10.1128/IAI.69.8.5037-5045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jones KF, Hollingshead SK, Scott JR, Fischetti VA. Spontaneous M6 protein size mutants of group A streptococci display variation in antigenic and opsonogenic epitopes. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(21):8271–5. 10.1073/pnas.85.21.8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gravekamp C, Kasper DL, Michel JL, Kling DE, Carey V, Madoff LC. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infection and immunity. 1997;65(12):5216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gerlach RG, Jackel D, Stecher B, Wagner C, Lupas A, Hardt WD, et al. Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol. 2007;9(7):1834–50. 10.1111/j.1462-5822.2007.00919.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains were grown at 37°C in THY medium.
(PDF)
(A) Total number of differentially expressed genes using a fold cutoff ≥1.5. Upregulated virulence genes comparing 9T vs 10T (B), 9T vs 11T (C), and 10T vs 11T (D).
(PDF)
Number of TRR28 repeats in strains with HTSpy1336-7 containing 8Ts (A), 9Ts (B), 10Ts (C), and 11Ts (D). n = 490 strains.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Transcriptome data is available in the Gene Expression Omnibus under accession number GSE145621.