Summary
Genome-wide association studies have revealed numerous risk loci associated with diverse diseases. However, identification of disease-causing variants within association loci remains a major challenge. Divergence in gene expression due to cis-regulatory variants in noncoding regions is central to disease susceptibility. We show that integrative computational analysis of phylogenetic conservation with a complexity assessment of co-occurring transcription factor binding sites (TFBS) can identify cis-regulatory variants and elucidate their mechanistic role in disease. Analysis of established type 2 diabetes risk loci revealed a striking clustering of distinct homeo-box TFBS. We identified the PRRX1 homeobox factor as a repressor of PPARG2 expression in adipose cells and demonstrate its adverse effect on lipid metabolism and systemic insulin sensitivity, dependent on the rs4684847 risk allele that triggers PRRX1 binding. Thus, cross-species conservation analysis at the level of co-occurring TFBS provides a valuable contribution to the translation of genetic association signals to disease-related molecular mechanisms.
Introduction
Recent advances in genome-wide association studies (GWAS) have yielded a plethora of loci associated with diverse human diseases and traits (Hindorff et al., 2009). However, signals emerging from GWAS, which involve typically dozens of variants in linkage disequilibrium (LD), have rarely been traced to the disease-causing variants and even more rarely to the mechanisms by which they may increase disease risk (Califano et al., 2012). The majority of common genetic variants are located in noncoding regions (1000 Genomes Project Consortium et al., 2012), and disease-associated loci are enriched for expression quantitative trait loci (eQTLs) (Nica et al., 2010), DNase I hypersensitive sites sequencing (DHSseq) peaks, and chromatin immunoprecipitation sequencing (ChIP-seq) peaks (Maurano et al., 2012; ENCODE Project Consortium et al., 2012), suggesting that variants modulating gene regulation are major contributors to common disease risk.
Experimental DHS, RNA, and ChIP-seq approaches have been used to prioritize candidate cis-regulatory variants (Maurano et al., 2012; ENCODE Project Consortium et al., 2012; Ward and Kellis, 2012b). However, such functional approaches require access to appropriate human tissues and are further hampered by the spatial, temporal, environmental, and epigenetic complexity of gene regulation. These limitations emphasize the need for bioinformatics approaches that reliably assess the regulatory role of noncoding variants. So far, phylogenetic conservation has been a common denominator in the search for noncoding regulatory regions (Waterston et al., 2002; Pennacchio et al., 2006; ENCODE Project Consortium et al., 2007, 2012; Visel et al., 2009b; Blow et al., 2010; Lindblad-Toh et al., 2011). However, intra- and cross-species differences in gene expression are often driven by changes in transcription factor binding sites (TFBS), and their rapid evolutionary turnover results in lineage-specific regulatory regions that are functionally conserved but have low phylogenetic conservation (Ward and Kellis, 2012a), thus challenging the use of these algorithms. Importantly, gene regulatory regions in eukaryotes tend to be organized in cis-regulatory modules (CRMs), comprising complex patterns of co-occurring TFBS for combinatorial binding of transcription factors (TFs) (Arnone and Davidson, 1997; Pennacchio et al., 2006; Visel et al., 2013). CRMs integrate upstream signals to regulate expression of coordinated gene sets, making them a prime target to achieve phenotypic changes as a result of adaptive evolution (Junion et al., 2012). Despite the critical importance of CRMs, no algorithms have so far been developed to harness the potential power of conserved TFBS patterns within CRMs to predict regulatory variants in disease genetics.
We show that cross-species conservation at the level of the CRMs—rather than at the level of the regulatory sequence that comprises them—identifies cis-regulatory variants within disease-associated GWAS loci. Exploiting phylogenetic conservation of TFBS co-occurrences, we found homeobox TFBS as a cis-regulatory feature of type 2 diabetes (T2D) risk loci, for which the specific causal variants have rarely been pinpointed (Stitzel et al., 2010). Detailed analysis at the PPARG risk locus revealed the rs4684847 risk allele and, by changing binding of the homeo-box TF PRRX1, its genotype-dependent effect on PPARG2 expression and insulin sensitivity.
Results
Cross-Species Analysis of TFBS Modularity Discovers cis-Regulatory SNPs at T2D Risk Loci
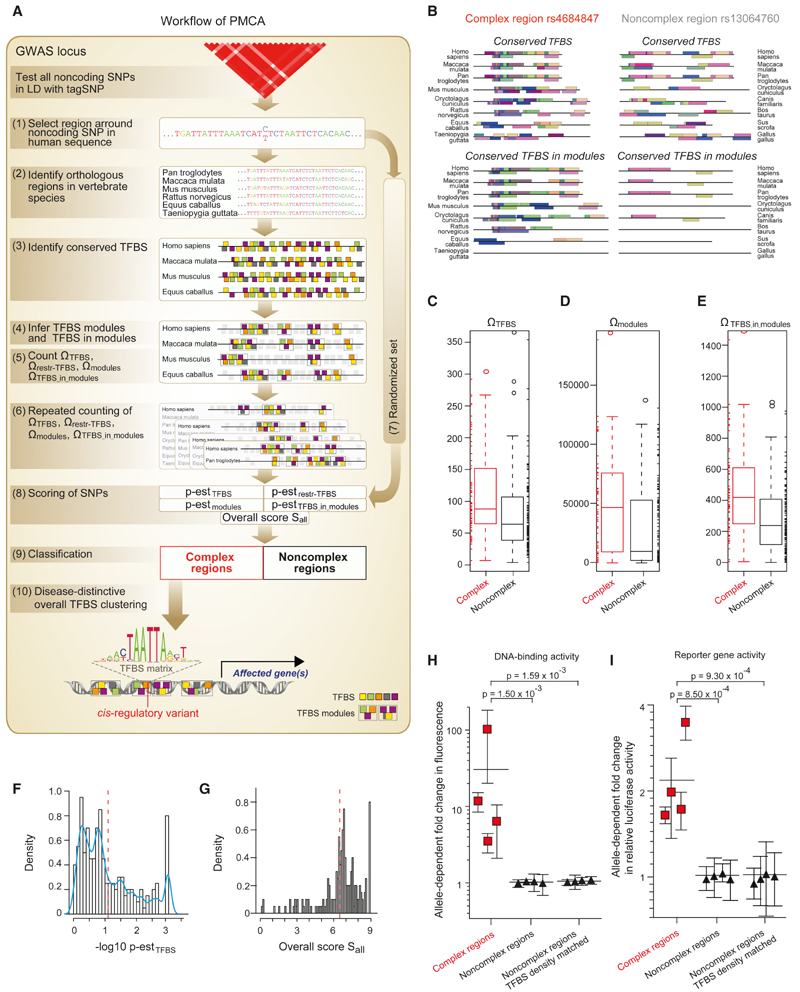
We developed a method, phylogenetic module complexity analysis (PMCA), which leverages conserved co-occurring TFBS patterns within CRMs to predict cis-regulatory variants, i.e., variants affecting gene expression (Figure 1A; Extended Experimental Procedures available online). To systematically identify cis-regulatory variants at GWAS risk loci, we extracted GWAS tagSNPs and consequently all noncoding (nc) SNPs that are in high LD with these tagSNPs. PMCA individually tests each nc variant by analyzing the flanking region for cross-species conserved TFBS patterns, regardless of global sequence conservation. This requires first the extraction of the region surrounding an nc SNP (±60 bp) from the human genome and consequent identification of orthologous regions in 15 vertebrate species. Within each SNP-specific set of orthologous regions, phylogenetically conserved TFBS, TFBS modules (a cross-species conserved pattern of two or more TFBS occurring in the same order and in a certain distance range), and TFBS in those TFBS modules were identified and then counted. SNP-flanking regions with a significant enrichment of phylogenetically conserved TFBS modules are classified as complex regions, as compared to noncomplex regions (example in Figure 1B) wherein the occurrence of TFBS modules does not exceed expectation by chance. To compute this enrichment weestimate background probabilities using randomizations of orthologous sets (details on scoring cut-offs in Extended Experimental Procedures).
Figure 1. Discovery of cis-Regulatory Diabetes SNPs.
(A) Workflow of the PMCA methodology: (1) the flanking region of a noncoding SNP is extracted from the human reference genome; (2) orthologous regions are searched in the genomes of 15 vertebrate species; (3) TFBS are identified in each orthologous sequence; (4) TFBS modules are identified in the set of orthologous sequences (TFBS modules defined as all, two or more TFBS occurring in the same order and in certain distance range in all or a subset of the orthologous sequences); (5) phylogenetically conserved TFBS ΩTFBS, TFBS modules Ωmodules, and occurrences of TFBS in TFBS modules ΩTFBS_in_modules are counted; (6) repeated counting for different numbers of input sequences weighs the degree of cross-species conservation and the number of TFBS in modules; computation of conserved TFBS with more restricted parameters Ωrestr_TFBS accounts for genomic regions with low numbers of orthologs; (7) steps 3-6 are repeated using randomized input sequences (randomization of sequences is done using local shuffling in order to conserve local nucleotide frequency distributions) to estimate; (8) the probability p-est of observing a given ΩTFBS, Ωrestr_TFBS, Ωmodules, and ΩTFBS_in_modules and to calculate the overall scoring criterion; (9) input sequences are classified as complex and noncomplex regions; and (10) complex regions harboring a trait-related TFBS at SNP position are selected for functional evaluation (trait-related TFBS are drawn from overall TFBS clustering analysis as described in text related to Figure 3). See also the Extended Experimental Procedures.
(B) Representative complex region (rs4684847) and noncomplex region (rs13064760). Conserved TFBS and conserved TFBS in modules occurring in more than two vertebrate species are shown to illustrate TFBS modularity across species.
(C-G) Classification of SNP regions for a set of eight T2D risk loci (Table S1; Figure S1). Box-whisker plots (IQR 50%) show the counts of conserved TFBS ΩTFBS (C), conserved TFBS modules Ωmodules (D) and occurrences of TFBS in TFBS modules ΩTFBS_in_modules (E) for complex regions (red lines) and noncomplex regions (black lines). Data points covered by the interquartile range (IQR) and the whiskers values were added as rug at the sides of the plot. Note that values vary over a large range with higher median for complex regions for all criteria (at 47 T2D loci we find a median of 354.5/470.46 and 310/382.35 for ΩTFBS_in_modules in complex/noncomplex regions). Scoring of SNP regions is illustrated by histograms showing the probability p-est of observing ΩTFBS across species (F) and showing the overall scoring criterion Sall (G). Blue curve: empirical density function of the histogram data. Red dashed line: cut-off scores separating complex from noncomplex regions (— log10 p-estTFBS = 1.12, Sall = 6.5); SNP regions with a value to the left of the red line were defined as noncomplex.
(H and I) cis-Regulatory activity of SNP regions. Noncomplex regions include regions matched for TFBS density of complex regions (TFBS median = 88). The allele-dependent change in DNA-binding activity from EMSAs (n = 4) (H) and luciferase reporter activity (n = 10) (I) is shown for each SNP. Mean ± SD, p from linear mixed-effects model.
We applied PMCA to a set of eight GWAS T2D risk loci (MTNR1B, TCF7L2, PPARG, CENTD2, FTO, GCK, CAMK1D, and KLF14) (Dupuis et al., 2010; Voight et al., 2010) covering strong and weaker GWAS signals and reflecting the different T2D features, i.e., insulin resistance and impaired insulin secretion (Doria et al., 2008). Using noncoding sequence data, we defined 200 SNPs in LD with the tagSNPs (r2 ≥ 0.7, 1000 Genomes) (1000 Genomes Project Consortium et al., 2012) (Figure S1A). PMCA predicted 64 complex and 136 noncomplex regions (Figures 1C–1G; Table S1). We ranked complex regions based on the count of TFBS in conserved TFBS modules (Table S2) and examined the allele-dependent cis-regulatory potential of the 25% highest scoring SNPs using in vitro electrophoretic mobility shift assay (EMSA) and reporter assays. As predicted, SNPs in complex regions significantly differed in allele-dependent cis-regulatory activity compared to noncomplex regions (Figures 1H and 1I; Table S3). Indeed, the regulatory variants revealed effects ranging from 3.1-to 101-fold change in DNA-protein binding and 1.3- to 3.5-fold change in reporter activity. Moreover, the identified variants operated in a cell type-specific manner (Figure S1B).
To examine if the identified cis-regulatory variants in complex regions associate with T2D in vivo, we performed look-ups in the MAGIC and DIAGRAM cohorts (Dupuis et al., 2010; Voight et al., 2010). The variants in complex regions revealed a similar or stronger association compared to the initial GWAS signal (Table S4), and a look-up in a recent fine-mapping study (Maller et al., 2012) confirmed that our cis-regulatory SNPs belong to the predicted T2D-disease SNP set. GWAS signals are enriched for regulatory variants (Nica et al., 2010). Comparing random SNPs from the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2012) to a limited representation of GWAS signals for 19 human diseases (Hindorff et al., 2009) (Table S5A), we found a 1.12-fold overall enrichment of SNPs in complex regions (p = 1.9 × 10−4, binomial distribution) (Table S5B and S5C), reflecting disease-conferring and low effect cis-regulatory variants. Finally, we applied PMCA on reported cis-regulatory SNPs associated with diverse disease-related traits, including cancer, myocardial infarction, thyroid hormone resistance, hypercholesterolemia, and adiponectin levels (MYC, Pomerantz et al., 2009; MDM2, Post et al., 2010; PSMA6, Ozaki et al., 2006; THRB, Alberobello et al., 2011; SORT1, Musunuru et al., 2010; APM2, Laumen et al., 2009). Consistent with the reported functional proof, our analysis informed on all but one of the cis-regulatory SNPs (Table S6). The highest scores inferred from PMCA predicted the myocardial infarction risk variant shown to regulate hepatic SORT1 expression (Musunuru et al., 2010). Together, these results demonstrate the utility of cross-species TFBS modularity information within CRMs to elucidate functionality of GWAS signals in the noncoding genome.
Functional Conservation beyond Sequence Conservation
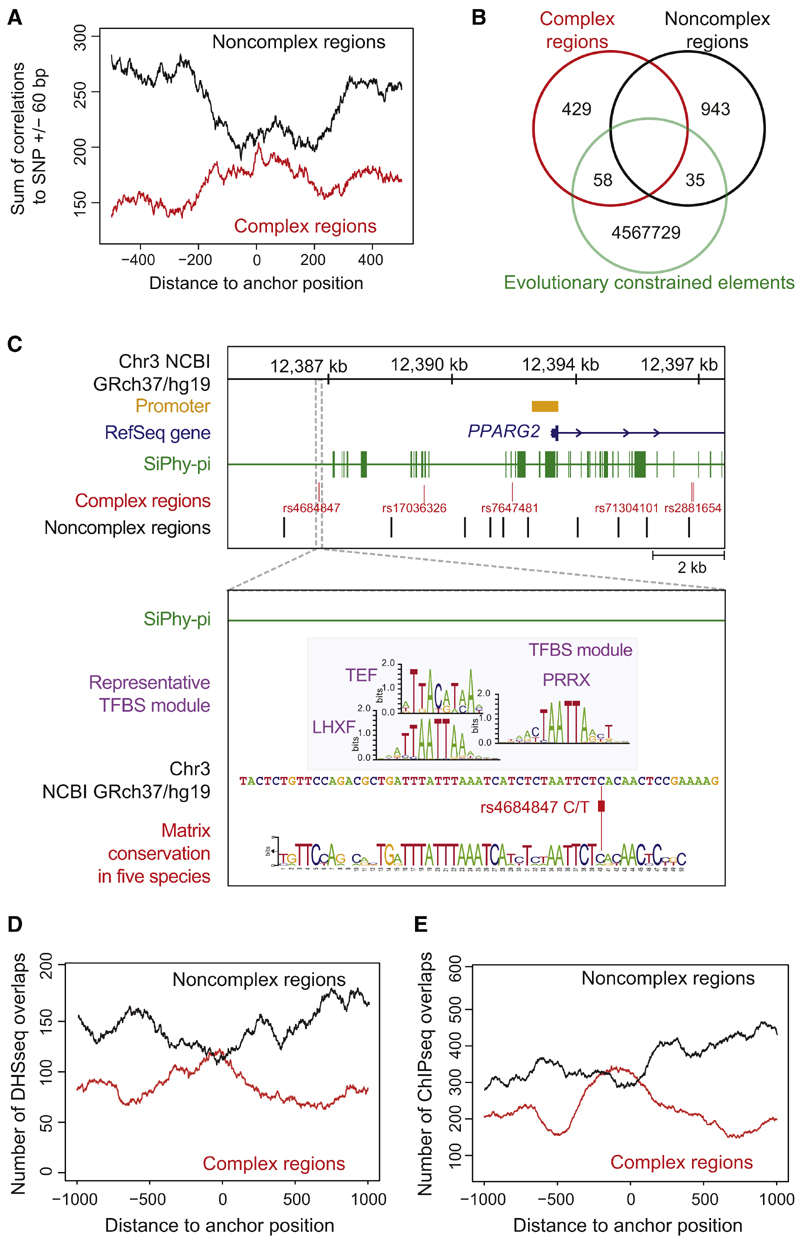
Given that TFBS turnover is characteristic of CRM evolution (Blow et al., 2010; Ward and Kellis, 2012a), the utility of sequence conservation in deciphering cis-regulatory variants may be limited. To assess the power of harnessing TFBS patterns beyond sequence conservation, allowing for sequence variability, we tested complex and noncomplex regions for correlations with evolutionary constrained elements detected by the SiPhy-Tc-method (Lindblad-Toh et al., 2011). For this analysis, we extended our initial PMCA analysis of eight T2D loci to a set of 47 T2D risk loci comprising all GWAS-reported autosomal variants (Hindorff et al., 2009) including 487 complex and 978 noncomplex regions (Figure S2; Table S7). Noncomplex regions were depleted of constrained elements in their close proximity (Figure 2A). Conversely, complex regions were enriched for nearby constrained elements, consistent with a 1.37-fold enrichment of GWAS SNPs relative to HapMap SNPs (Lindblad-Toh et al., 2011). Although complex regions overlapped 1.88-fold more with constrained elements than noncomplex regions (p = 2.4 × 10−9, hypergeometric distribution, right sided), strikingly the majority of complex regions lacked an overlap with constrained elements (Figure 2B; Table S8). This lack of overlap was true for all variants that we experimentally characterized as cis-regulatory (example in Figure 2C). In essence, considering sequence conservation helps to prioritize genomic regions that harbor potential causal variants, yet seems insufficient to pinpoint them. This underscores the importance of exploiting conservation in terms of a complexity assessment of co-occurring TFBS, in the search for cis-regulatory variants involved in human diseases.
Figure 2. Correlations of cis-Regulatory Predictions at 47 T2D Risk Loci with Evolutionary Constrained Elements and Functionally Annotated Genomic Regions.
(A) Correlation of PMCA results with evolutionary constrained regions. The occurrences of 487 complex and 978 noncomplex T2D-associated regions within constrained regions from SiPhy-p algorithm (Lindblad-Toh et al., 2011). Localization of SNPs relative to transcription start site in Figures S2A and S2B.
(B) Venn diagram illustrates the number of complex and noncomplex regions that directly map to a constrained element (overlap).
(C) Complex regions at the PPARG locus (Figure 4E) lack an overlap with constrained regions. Zoom-in: the rs4684847 cis-regulatory region does not map to a constrained region (393 bp upstream of nearest constrained element). A representative TFBS module (UTFBS_in_module = 3) is shown and its TFBS module conservation for a given quorum of five species is visualized by a sequence logo.
(D and E) Correlation of complex (red line) and noncomplex (black line) T2D-associated SNP regions to DHSseq (D) and ChIP-seq (E) peaks. From the midpoint of 487 complex and 978 noncomplex regions, 1,000 bp in both directions were scanned for DHSseq and ChIP-seq peaks (Extended Experimental Procedure). For each position, the sum of occurrences was plotted. T2D complex regions were significantly enriched for overlaps with DHSseq and ChIP-seq regions, displayed as a central peak (correlations with Crohn’s-associ-ated regions in Figures S2C and S2D).
To further support PMCA predictions at T2D risk loci, we merged our analysis with functional genomics data from The ENCODE Project Consortium (2011) (chromatin state and TF binding). We found complex regions highly enriched for both DHSseq peaks (p = 3.52 × 10−10) (Figure 2D) and ChIP-seq peaks (p = 4.68 × 10−6) (Figure 2E; Table S9). Additionally, crossing our regulatory predictions for T2D risk SNPs with Reg-ulomeDB, a data repository of multiple types of functional ENCODE data (Schaub et al., 2012), confirmed that complex regions are significantly enriched for functional annotations (p = 3 × 10−24, hypergeometric distribution, right-sided) (Table S10).
Clustering of Distinct Homeobox TFBS Is a Specific Feature of T2D-Related Complex Regions
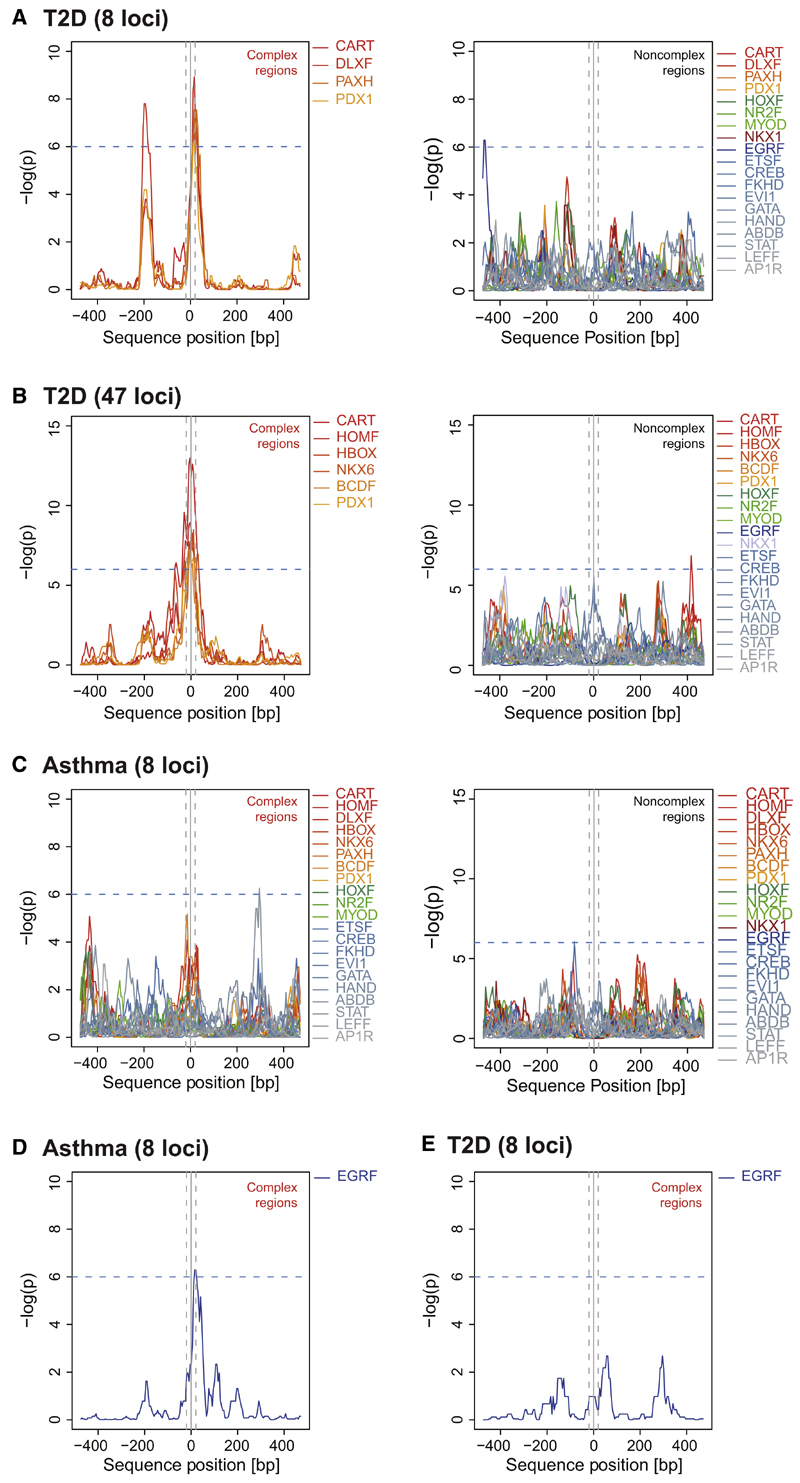
TFBS clustering relative to transcription start sites indicates biological significance (FitzGerald et al., 2004), and TFBS combination coupled with the TFs recruited to a CRM determines CRM function (Zinzen et al., 2009). Thus, we sought evidence for a discerning T2D functional feature by exploring TFBS characteristics in evolutionary conserved complex regions at T2D risk loci. Given a SNP genomic region we used positional bias analysis, scanning 1,000 bp with the SNP at midposition for the occurrence of putative TF binding sequences (883 TFBS matrices grouped in 192 TFBS matrix families) (Table S11). First, for the set of eight T2D risk loci selected for in-depth analysis above, we observed a significant positional bias for distinct TFBS families (−log10(p) > 6) exactly at SNP positions of complex contrary to noncomplex regions (Figure 3A). This striking SNP-directed overrepresentation in T2D complex regions was restricted to specific TFBS in the homeobox superfamily, including the matrix families CART (−log10(p) = 6.52) and PDX1 (−log10(p) = 6.18) (Table S12A). To test whether these findings could be retrieved in a larger set of T2D-associated variants, we extended TFBS clustering analysis to the set of 47 GWAS T2D risk loci (Hindorff et al., 2009). Indeed, this comprehensive analysis reproduced colocalization of T2D risk SNPs exclusively with homeobox TFBS matrices in complex regions as opposed to noncomplex regions (Figure 3B; Table S12B). We again found specific clustering of the CART (−log10(p) = 13.00) and PDX1 families (−log10(p) = 6.78) together with the homeobox matrix families NKX6 (−log10(p) = 8.50), HOMF (−log10(p) = 8.94), HBOX (−log10(p) = 8.54), and BCDF (−log10(p) = 7.24). No other TFBS matrices showed a significant peak in the bias profile at SNP positions. Importantly, when applying PMCA on risk loci of T2D nonrelated traits, asthma, and Crohn’s disease (Moffatt et al., 2010; Schaub et al., 2012) (Figures S3B and S3C; Table S13), we observed disease-distinctive TFBS at SNP positions (Table S12C and S12D). Both complex and noncomplex regions lacked a clustering of homeobox TFBS at asthma risk SNPs (Figure 3C). The specific clustering of the early growth response factor matrix family (EGRF) for asthma risk SNPs in complex regions (−log10(p) = 8.50; Figure 3D) was in strong contrast to T2D (−log10(p) = 3.97; Figure 2E) and Crohn’s (−log10(p) = 2.07; Figure S3D). Of note, the EGRF-binding factor EGR1 regulates asthma-related IL13-induced inflammation (Cho et al., 2006).
Figure 3. Positional Bias of Distinct Homeo-box TFBS Families at T2D Risk SNPs.
Distribution of TFBS matrices relative to SNP positions (SNP ± 500 bp) at T2D compared to asthma risk loci, calculated using positional bias analysis. One thousand base pair genomic regions with SNPs at midposition were scanned for the occurrence of TFBS matches for 192 TFBS matrix families (sliding 50 bp windows, p from binomial distribution model, Extended Experimental Procedures).
(A and B) TFBS family distribution in a set of eight and an extended set of 47 T2D risk loci. Complex regions reveal clustering of distinct homeobox TFBS matrix families at T2D risk SNP positions (±20 bp, gray dashed lines). All TFBS families displayed equal distributions within T2D non-complex regions (a subset of representative TFBS families is shown).
(C) TFBS family distribution in a set of eight asthma risk loci. Asthma complex and noncomplex regions lack a positional bias at SNP positions for the homeobox TFBS matrix families clustering in complex regions at T2D risk SNPs (see Figure S3 for details on Crohn’s).
(D and E) TFBS family distribution in asthma risk loci revealed a specific EGRF matrix family clustering in complex regions at asthma risk SNPs (D). T2D complex regions lack a clustering of EGRF matrices at SNP positions (E).
Homeobox TFs are known to be involved in tissue developmental processes including β-cell development (Jørgensen et al., 2007). However, except for the MODY gene PDX1 (Fajans et al., 2001) and the common T2D-associated loci HHEX1 and ALX4 (Sladek et al., 2007), the PMCA-inferred homeobox factors have not been implicated in T2D pathogenesis. T2D is marked by insulin resistance and impaired insulin secretion (Doria et al., 2008). To evaluate a functional role of the homeobox TFBS matrix families in T2D pathogenesis, we extracted data for insulin resistance (HOMA-IR) and impaired insulin secretion (HOMA-B) (Dupuis et al., 2010), to compute the enrichment of predicted cis-regulatory T2D risk SNPs that localize in close proximity to those homeobox TFBS (±20 bp, permutations on the phenotypes, n = 1,000, 95% confidence interval [CI]; Extended Experimental Procedures). We verified a significant enrichment of SNPs that localize ± 20 bp at inferred homeobox TFBS for both insulin resistance (mean = 1.09 × 10−6; 95% CI: 9.59 × 10−7-9.51 × 10−3, p = 3.28 × 10−4; mean permutation background) and impaired insulin secretion (mean = 9.45 × 10−4; 95% CI: 5.37 × 10−4-1.34 × 10−2, p = 1.29 × 10−7). Furthermore, we elucidated a potential effect of their binding TFs on impaired insulin secretion. Assessing mRNA levels in human islets from 51 healthy and eight T2D deceased donors by RNA-seq (L.G., unpublished data), we found a marked expression difference for RAX, PRRX2, BARX1, PITX1, EMX2, NKX6-3, BARX2, MSX2, and PDX1 in islets from T2D patients compared to healthy controls (7.28 × 10−9 < p < 4.02 × 10−4, false discovery rate [FDR] < 1%) (Table S14). By genome-wide coexpression analysis we found significantly coregulated gene sets (p < 5.02 × 10−3; FDR < 5%, n = 51 healthy donors) (Table S15). Except for the gene set coregulated with PITX1, we found metabolic pathways among the top five significantly enriched pathways (hypergeometric test, FDR corrected p < 0.05) (Figure S3E). Other top five enriched pathways included insulin signaling, MAPK signaling, notch signaling, calcium signaling, and pancreatic secretion. Knockdown of each candidate homeobox TF in pancreatic INS-1 β-cells significantly perturbed glucose-stimulated insulin secretion (Figure S3F). Moreover, except for PDX1 and MSX2 (corrected FDR, p = 0.96 and p = 0.89), all PMCA-inferred homeobox TFs were significantly coexpressed with the insulin gene in islets of 26 hyperglycemic individuals (hemoglobin A1C [HbA1C] > 6) (Table S16). Although the result for PDX1 was borderline nonsignificant, it is a well-known regulator of insulin expression (Brissova et al., 2002). The other TFs can be regarded as candidates for regulation of proinsulin production.
The T2D-Identified Variant rs4684847 Regulates PPARG2 Gene Expression
To establish the informative value of TFBS pattern analysis for pinpointing the cis-regulatory variant and binding TF underlying GWAS association signals, we chose the PPARG locus for detailed study. PPARg is crucial in adipogenesis, lipid metabolism, and systemic insulin sensitivity (Rosen et al., 1999; Medina-Gomez et al., 2005) and exists as two isoforms: PPARg1 (PPARG1, PPARG3 mRNA) and PPARg2 (PPARG2 mRNA) (Fajas et al., 1998), the latter mainly expressed in adipocytes (Tontonoz et al., 1994). There is a robust association of PPARG with T2D (Deeb et al., 1998; Heikkinen et al., 2009; Dupuis et al., 2010; Voight et al., 2010). The T2D GWAS association comes from an LD region mainly tagged by the coding missense mutation Pro12Ala (Figure 4A, upper panel). However, the minor 12Ala allele, associated with enhanced insulin sensitivity in humans, paradoxically blunts the transcriptional activity of the insulin-sensitizing PPARγ2 TF (Deeb et al., 1998). Hypothesizing that the elusive PPARG T2D signal instead arises from a regulatory variant that affects PPARG2 expression, we first confirmed—before analyzing variants at the PPARG locus with PMCA—a risk allele-dependent 3.8-fold decrease of PPARG2 mRNA in human adipose stromal cells (hASCs) (p = 1.0 × 10−3) (Figure 4B). This effect was specific for PPARG2, as there was no effect on PPARG1 expression (Figure 4C).
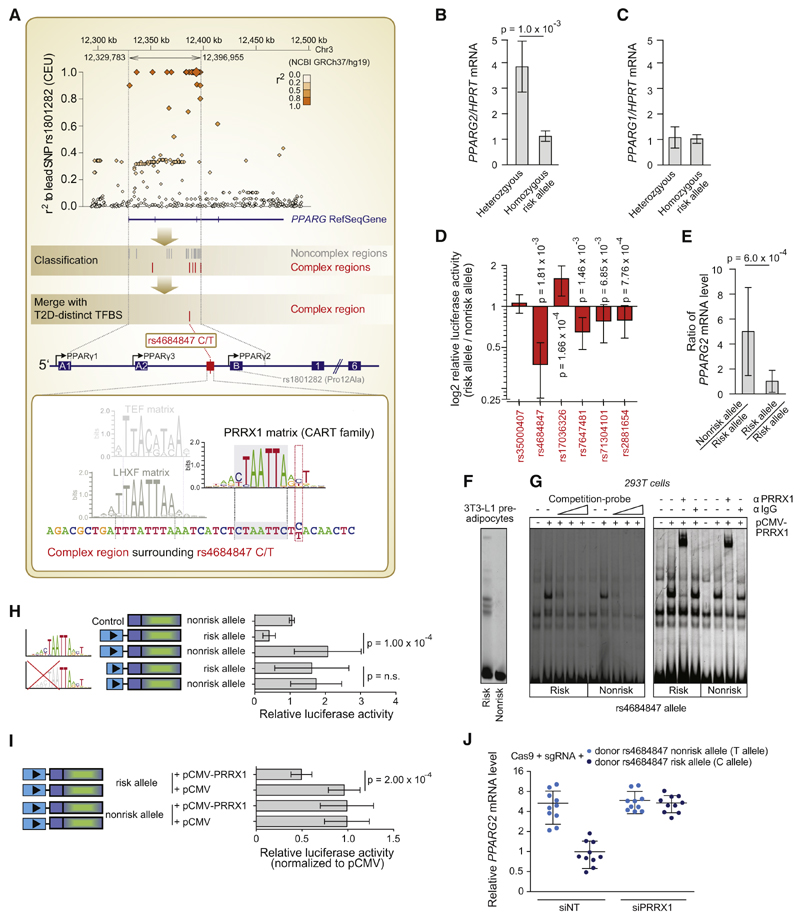
Figure 4. The Noncoding SNP rs4684847 by Binding the Homeobox Factor PRRX1, Represses PPARG2 Expression at the PPARG Diabetes Risk Locus.
(A) Top panel: an LD regional plot of the PPARG locus. Diamonds, tagSNP Pro12Ala and pairwise correlation of SNPs in LD (MAF ≥ 1%) against genomic position; blue, PPARG gene and exons. Middle/lower panel: classification of SNPs in complex regions (red lines) and noncomplex regions (gray lines) (PMCA steps 1–9, Figure 1A). Scanning of PPARG complex regions for T2D-distinct homeobox TFBS matrix families (CART, HOMF, HBOX, NKX6, BCDF, PDX1; Figure 3B) pinpoints rs4684847 (C/T), based on its overlap with the CART binding matrix for PRRX1 (step 10, Figure 1A). Zoom-in, human PPARG gene; arrows, transcription start site (TSS) of PPARG1-3 mRNA isoforms; boxes, coding exons (filled) and untranslated exons (open); lines, introns. Second zoom-in, CRM at rs4684847; the PRRX1 matrix co-occurs with diverse TFBS matrices in consistent orientation and distance range across species, exemplarily illustrated by one conserved TFBS module (UTFBS_in_modules = 3; TFBS matrices: PRRX1, TEF, LHXF).
(B and C) Genotype-dependent mRNA expression in undifferentiated hASCs genotyped for Pro12Ala and rs4684847 (r2 = 1.0). qPCR of PPARG1 and PPARG2 mRNA isoforms (standardized to HPRT) homozygous CC risk (n = 9) and CT nonrisk allele carriers (n = 5) normalized to mean for CC. Mean ± SD, t test.
(D) Validation of cis-regulatory predictions for complex regions at the PPARG locus. Quantified change in reporter activity in 3T3-L1 adipocytes is shown for each SNP, using luciferase constructs harboring the risk or nonrisk alleles, representing an activating or repressing effect of the risk allele on transcriptional activity.
Mean ± SD, n = 3–14, paired t test.
(E) Allele-specific primer extension analysis in hASCs of heterozygous rs4684847 carriers (n = 6) normalized to mean risk allele levels (D). Mean ± SD, Mann-Whitney U test.
(F and G) Increased PRRX1 binding at the risk allele in EMSAs with rs4684847 allelic probes and 3T3-L1 preadipocyte nuclear extracts (F), confirmed by competition with cold PRRX1 probe (G, left panel) and PRRX1 antibody shift of protein-DNA complex in 293T with ectopically expressed PRRX1 (G, right panel).
(H) Reporter assays with constructs harboring the rs4684847 risk and nonrisk allele in 3T3-L1 preadipocytes. Truncation of the PRRX1 matrix without affecting rs4684847 reveals abrogated allelic cis-regulatory activity. Mean ± SD, n = 9, paired t test.
(I) Inhibition of reporter activity (normalized to pCMV control) at the rs4684847 risk allele by ectopic expression of PRRX1 in 3T3-L1 preadipocytes. Mean ± SD; n = 9, paired t test.
(J) Regulation of PPARG2 mRNA expression in SGBS adipocytes with the CC risk allele, or TT nonrisk allele introduced by CRISPR/Cas9 genome editing approach. siPRRX1 and siNT transfection concurrent with induction of differentiation, PPARG2 mRNA assessed by quantitative RT-PCR (qRT-PCR), standardized to HPRT. Mean ± SD, n = 12, t test. siNT, nontargeting siRNA.
First, to narrow-down the variants that could explain the decrease in PPARG2 expression and thereby the underlying T2D association, we applied PMCA to each of the 23 correlated noncoding variants at the PPARG locus (r2 ≥ 0.7, 1000 Genomes) (1000 Genomes Project Consortium et al., 2012) (Figure 4A). Seventeen variants were ruled out being located in noncomplex regions (Figure S4A; Table S17). Among the six variants in complex regions, five had either activating or repressing cis-regulatory activity (Figure 4D), which may reflect gene regulatory dependency on the tissue/cell-type and the spatial, temporal, environmental, and epigenetic context. In fact, while the quantitative PCR (qPCR) data in undifferentiated hASCs showed a suppressive effect specific for the PPARG2 mRNA isoform, adipose tissue eQTL data showed an upregulation of total PPARG mRNA in risk allele carriers (p = 0.01) (Figure S4B).
Second, to pinpoint the functional variants that may explain the GWAS-reported T2D association, we scrutinized the complex regions for those TFBS showing a clustering at T2D risk SNP positions (drawn from the overall TFBS clustering analysis in complex regions; Figure 3), pursuing the variants overlapping a TFBS matrix in the disease-distinctive cluster. As shown above, our comprehensive cross-species TFBS pattern analysis of 47 T2D risk loci unveiled a clustering of specific homeobox TFBS families as a characteristic feature of T2D risk SNPs (Figure 3B). Among the six noncoding variants at the PPARG locus, only one variant, rs4684847 (C/T), overlaps with the T2D-distinct clustering of the homeobox TFBS matrix. The TFBS matrix overlapping with rs4684847 belongs to the CART matrix family (−log10(p) = 13.00, the highest score among TFBS matrix families), and is predicted to bind the homeobox TF PRRX1. The other five noncoding variants showed no homeobox TFBS matrix match (Figure 4A, lower panel).
Third—as an independent approach to confirm rs4684847 mediating the PPARG2 suppression—we examined the cellular context of genotype-dependent PPARG2 suppression and epigenomic profiling data that allow for temporal chromatin state-dependent regulatory functional annotations. By allele-specific primer extension analysis in heterozygous undifferentiated hASCs genotyped for rs4684847, where each allele serves as an internal control for the other, we first confirmed a striking allelic imbalance with 5.4-fold lower PPARG2 mRNA expression from the C risk allele (p = 6.0 × 10−4) (Figure 4E). Given the role of PPARG2 in adipogenesis, we then tested whether the rs4684847 C risk allele might affect PPARG2 mRNA expression during adipogenesis. The allele-specific primer extension analyses in hASCs from heterozygous risk allele carriers revealed that the risk allele-dependent suppression of PPARG2 mRNA diminished with progression of adipogenesis (p< 0.001) (Figure S4C). These data suggest a highly temporal context-specific effect of the risk allele on PPARG2 suppression in the undifferentiated state. Given the availability of cell-stage-dependent open chromatin data in hASCs reported by Mikkelsen et al. (2010), we sought supportive evidence for rs4684847 as the variant underlying the cell-stage-dependent allelic PPARG2 expression. We integrated all six variants in complex regions at the PPARG locus with genome-wide temporal regulatory annotations estimated by H3K27ac data. Among those six, only the flanking region rs4684847 (C/T) showed consistent cell stage-dependent H3K27ac density distributions (Figure S4D). Thus, the rs4684847-specific match with the T2D homeobox TFBS clustering, informed by conserved TFBS pattern analysis, could be confirmed by cell-stage-dependent regulatory regions estimated by chromatin state data.
Finally, we performed a host of in vitro and in vivo analyses to prove that the rs4684847 risk allele (C allele) mediates the suppression of PPARG2 mRNA expression via the transcriptional regulator PRRX1. By affinity chromatography and liquid chromatography-tandem mass spectrometry (LC-MS/MS), we could demonstrate a 2.3-fold increased binding of PRRX1 to the rs4684847 risk relative to nonrisk allele (Extended Experimental Procedures). Moreover, by EMSA we found rs4684847 risk allele-specific DNA-protein binding (Figure 4F), and competition EMSA and supershift experiments confirmed that PRRX1 was responsible for this allele-specific DNA-protein binding (Figure 4G). Furthermore, consistent with the GWAS signal for insulin resistance rather than insulin secretion (Voight et al., 2010), in luciferase reporter assays we observed rs4684847 cell type-specific effects in 3T3-L1 adipose cells, C2C12 myocytes and Huh7 hepatocytes, whereas pancreatic INS-1 β-cells and 293T cells lacked allelic activity (Figure S4E). Luciferase activity in 3T3-L1 preadipocytes was 5.2-fold lower for the C risk allele (p = 1.0 × 10−4, Figure 4H). This repressive effect was independent of 5′-versus 3′-orientation to the reporter gene (p = 0.03) and forward-reverse orientation (p = 0.03) (Figure S4F), suggesting enhancer function for the nonrisk allelic complex region. Importantly, perturbing the PRRX1 consensus sequence without affecting the SNP position itself fully abrogated the C risk allelic repression of reporter gene activity (Figure 4H), whereas overexpressing PRRX1 enhanced it (p = 2.0 × 10−4; Figure 4I).
We then sought proof that the rs4684847 risk allele—independent of correlated sequence variants—causes the suppression of endogenous PPARG2 expression. We used an adopted CRISPR/Cas homology-directed repair genome editing approach (Wang et al., 2013a) to introduce the rs4684847 non-risk allele in human Simpson-Golabi-Behmel syndrome (SGBS) preadipocytes, replacing the endogenous risk allele. Notably, the rs4684847 nonrisk allele was sufficient to increase PPARG2 transcript levels 5.4-fold (p = 0.005) (Figure 4J, left) (PPARG1 unaffected) (Figure S4G). In parallel experiments, we performed PRRX1 knockdown and confirmed that (1) risk allele-driven suppression of PPARG2 expression was reversed by PRRX1 silencing (p = 0.005), and (2) PRRX1 silencing did not affect PPARG2 expression in nonrisk allele cells (Figure 4J, right).
rs4684847 via PRRX1 Binding Affects FFA Homeostasis and Insulin Sensitivity
The SNP rs1801282 (Pro12Ala) in PPARG associates with BMI, fasting insulin, and insulin sensitivity (Deeb et al., 1998; Voight et al., 2010). rs4684847 is located 6.5 kb upstream of the PPARG2-specific promoter and is in complete LD (r2 = 1.0) with rs1801282. Via PMCA, we found that PRRX1 binds at the rs4684847 C risk allele and thus inhibits PPARG2 expression. On the other hand, the T allele of rs4684847 (minor allele frequency 6.5% in Caucasians) reduces the binding ability of PRRX1 and thus maintains a higher level of PPARG2 expression. Further in vivo evidence was obtained in primary human adipose stromal cells (hASCs) isolated from BMI-matched subjects, showing rs4684847-dependent PPARG2 mRNA expression (p = 1.4 × 10−20, n = 32). PPARg2 is crucial for maintaining insulin sensitivity: adipose-specific Pparg2 knockout mice develop insulin resistance independently of affecting body weight (Medina-Gomez et al., 2005), and PPARg is target of the thiozo-lidinedione (TZD) class of insulin-sensitizing drugs such as Rosi-glitazone (Rosi) (Lehmann et al., 1995). Indeed, we observed rs4684847-dependent association with lower T2D risk (Voight et al., 2010) (OR = 0.89, 95% CI = 0.86-0.92, p = 3.75 × 10−11, n = 80,648). Further, in hASCs we found rs4684847-dependent increase in adipocyte insulin sensitivity (p = 1.5 × 10−7, ratio insulin-stimulated/basal 2-deoxyglucose uptake, Pearson’s correlation, n = 32). We confirmed a significant interaction between the rs4684847 risk allele and adipose PRRX1 mRNA levels to HOMA-IR, independent of BMI (p = 0.044, n = 38, interaction model; Extended Experimental Procedures). In addition, we observed rs4684847-dependent correlations of PRRX1 mRNA levels with BMI, TG/HDL ratio, and BMI-adjusted HOMA-IR and with glucose infusion rate (GIR) measured by euglycemic hyperinsulinemic clamp in a cohort of 67 BMI- and body fat-matched obese patients (Table 1; Figure S4H).
Table 1. Correlation of Adipose Tissue PRRX1 mRNA Expression with T2D Traits in rs4684847 Risk Allele Carriers.
| PRRX1 mRNA | PRRX1 mRNA | PRRX1 mRNA | |||||
|---|---|---|---|---|---|---|---|
| All | CC | CT and TT | |||||
| rs4684847 genotypes | β | p | β | p | B | p | |
| A | n = 38 | n = 20 | n = 18 | ||||
| log(BMI) | — | 1.32 | 0.05 | 1.23 | 0.19 | 1.43 | 0.23 |
| age | 1.45 | 0.03 | 1.23 | 0.19 | 1.96 | 0.09 | |
| log(TG/HDL) | — | 6.92 | 7.54 × 10−4 | 6.40 | 0.02 | 6.35 | 0.07 |
| age | 6.97 | 7.36 × 10−4 | 6.14 | 0.02 | 6.81 | 0.07 | |
| age/BMI | 4.86 | 8.3 × 10−3 | 5.00 | 0.07 | 2.64 | 0.33 | |
| log(HOMAIR) | — | 2.77 | 3.52 × 10−3 | 3.13 | 8.3 × 10−3 | 1.80 | 0.29 |
| age | 2.77 | 3.77 × 10−3 | 3.12 | 8.6 × 10−3 | 1.70 | 0.34 | |
| age/BMI | 1.41 | 0.028 | 2.1 | 4.6 × 10−3 | -0.55 | 0.63 | |
| B | n = 67 | n = 54 | n = 13 | ||||
| log(GIR) | age/BMI | -0.51 | 1.83 × 10−7 | -0.78 | 3.30 × 10−8 | -0.38 | 0.28 |
| log(FFA) | age/BMI | 0.25 | 0.014 | 0.27 | 0.015 | -0.009 | 0.99 |
Gene expression and phenotypes were measured in (A) adipose tissue from a lean/obese patient cohort (mean ± SD 24.2 ± 9.1 kg/m2), and (B) adipose tissue samples from BMI-matched obese patients (mean ± SD 43.2 ± 3.1 kg/m2) characterized by hyperinsulinemic euglycemic clamp. rs4684847 risk allele and nonrisk allele genotypes were determined by Sequenom-assay. p values and b-estimates from linear regression analysis of PRRX1 mRNA expression levels with phenotype measures are shown. BMI, body mass index; FFA, free fatty acids; GIR, glucose infusion rate of hyperinsulinemic euglycemic clamp; HDL, high density lipoprotein; HOMA-IR, homeostasis model assessment of insulin resistance; TG, triglyceride.
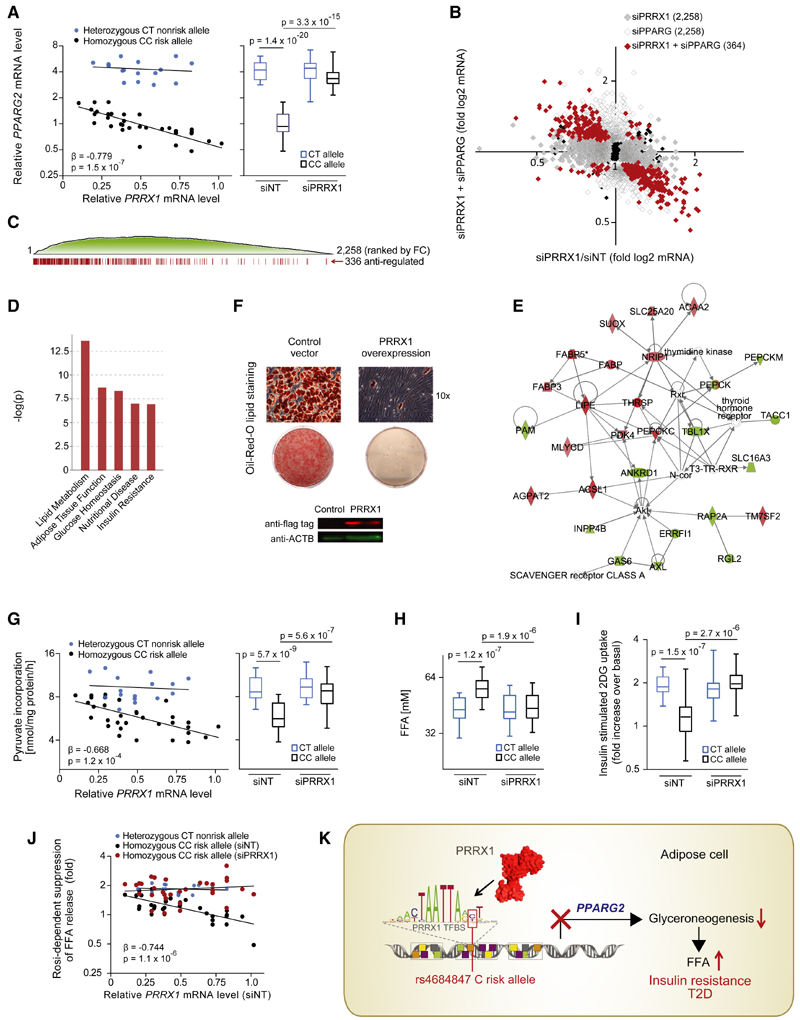
To further examine PRRX1 as mediator of the repressive rs4684847 risk allele (C allele) effect on PPARG2 expression, we performed knockdown of PRRX1 in primary hASCs and found that PRRX1 silencing was sufficient to revert the risk allelic suppression (p = 3.3 × 10−15) (Figure 5A; Table 2). Then, to inform on the cellular processes by which PRRX1 may contribute to T2D, we studied the impact of PRRX1 on PPARγ-regulated genes in hASCs from homozygous rs4684847 CC risk allele carriers by microarray analysis (n = 9). We found 2,258 transcripts regulated by PRRX1 knockdown (q < 0.2), 336 of which were reversely regulated by concomitant PPARG knockdown (Figure 5B). Gene set enrichment analysis (GSEA) highlighted an enrichment of those antiregulated genes among the most differentially expressed genes after PRRX1 knockdown (FDR = 0, Figure 5C), revealing that PPARg2 mediated the primary PRRX1 effect on global gene expression. Ingenuity pathway analysis (IPA) showed the strongest enrichment for lipid metabolism (p = 2.81 × 10−14) followed by adipose tissue function, glucose homeostasis, nutritional disease, and insulin resistance (Figure 5D). Accordingly, an inverse relationship between PRRX1 and adipocyte triglyceride (TG) accumulation was observed in PRRX1-overexpressing SGBS adipocytes (Figure 5E).
Figure 5. Binding of PRRX1 at the rs4684847 Risk Allele in Human Adipose Cells Affects Lipid Metabolism and Insulin Sensitivity.
(A) rs4684847-dependent PPARG2 and PRRX1 mRNA levels measured by qPCR (standardized to HPRT) in hASC from BMI-matched rs4684847 CT (n = 16) and CC (n = 32) risk allele carriers. siPRRX1 and siNT transfected concurrent with induction of adipogenic differentiation for 72 hr. Left: Pearson’s correlation in the siNT set. Right: box-whisker plot comparing PPARG2 mRNA in siNT- versus siPRRX1-treated cells (t test). FC, fold change.
(B and C) Global gene expression profiling by Illumina microarrays (q < 0.2) in hASCs from rs4684847 CC risk allele carriers transfected with siPRRX1 (n = 9, gray dots) and cotransfected with siPRRX1 and siPPARG (n = 4, red dots) for 72 hr after induction of adipogenic differentiation (B). Distribution of siPRRX1/siPPARG antiregulated genes among all regulated genes ranked by fold change (C).
(D and E) Biological pathways associated with siPRRX1/siPPARG antiregulated genes (D) and top scoring interaction network (E) from ingenuity pathway analysis.
(F) Oil Red O lipid staining of human SGBS cells with lentiviral-overexpressed flag-tagged PRRX1 (or control vector) 12 days after induction of adipocyte differentiation. Protein expression with aflag (PRRX1) and aACTB antibodies.
(G and H) rs4684847-dependent glyceroneogenesis rate measured by [1-14C]-pyruvate incorporation (G) and FFA release (H) in hASCs from BMI-matched rs4684847CT(n=16) and CC (n = 32) risk allele carriers aftersilencing ofPRRX1. (G) Left: Pearson’s correlation inthe siNT set. Right: box-whisker plot comparing siNT- versus siPRRX1-treated cells, t test. FFA, free fatty acids.
(I) rs4684847-dependent increase of [3H]-2-deoxyglucose ([3H]-2DG) uptake following insulin stimulation in hASCs. Box-whisker plot comparing siNT- versus siPRRX1-treated cells; t test.
(J) rs4684847-dependent rosiglitazone-mediated suppression of FFA-release during glyceroneogenesis. Pearson’s correlation comparing siNT versus siPRRX1. Mean ± SD, t test. See also Figures S4G and S4H; Tables 1 and 2.
(K) The rs4684847 risk allele (C allele) promotes PRRX1 binding 6.5 kb upstream of the PPARG2-specific promoter, leading to suppression of PPARG2 mRNA expression and perturbated lipid handling in adipose cells, increased circulating FFA levels, insulin resistance, and risk of T2D.
Table 2. Genotype-PRRX1-Dependent Regulation of PRXX1/PPARG Antiregulated Genes in hASCs.
| siNT | siPRRX1 | siPRRX1/siNT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hetero | Homo | Hetero/Homo | Hetero | Homo | Hetero/Homo | Hetero | Homo | |||||
| Mean ± SD | Mean ± SD | FC | p | Mean ± SD | Mean ± SD | FC | p | FC | p | FC | p | |
| PRRX1 | 0.52 ± 0.18 | 0.51 ± 0.19 | 1.01 | 0.92 | 0.11 ± 0.05 | 0.12 ± 0.06 | 0.90 | 0.56 | 0.25 | 2.83 × 10−7 | 0.22 | 4.02 × 10−8 |
| PPARG2 | 4.32 ± 1.07 | 0.79 ± 0.08 | 0.18 | 2.46 × 10−11 | 4.34 ± 1.47 | 3.37 ± 1.04 | 0.77 | 0.08 | 1.00 | 0.96 | 4.29 | 7.24 × 10−11 |
| PPARG1 | 1.07 ± 0.26 | 1.04 ± 0.33 | 1.03 | 0.79 | 1.18 ± 0.35 | 1.20 ± 0.49 | 0.98 | 0.90 | 1.15 | 0.35 | 1.10 | 0.41 |
| PEPCKC | 2.83 ± 0.58 | 1.03 ± 0.20 | 2.76 | 1.62 × 10−10 | 2.66 ± 0.50 | 2.98 ± 0.42 | 0.89 | 0.09 | 0.94 | 0.43 | 2.90 | 8.77 × 10−4 |
| PDK4 | 2.01 ± 0.88 | 0.74 ± 0.18 | 2.73 | 3.19 × 10−5 | 2.00 ± 0.60 | 1.73 ± 0.61 | 1.15 | 0.27 | 0.99 | 0.97 | 2.35 | 8.01 × 10−6 |
| LIPE | 1.37 ± 0.64 | 0.68 ± 0.32 | 2.01 | 2.00 × 10−3 | 1.30 ± 0.32 | 1.21 ± 0.45 | 1.08 | 0.56 | 0.95 | 0.74 | 1.77 | 2.03 × 10−3 |
| ADIPOQ | 1.89 ± 0.32 | 0.95 ± 0.31 | 1.98 | 7.92 × 10−8 | 1.85 ± 0.44 | 1.75 ± 0.61 | 1.05 | 0.66 | 0.98 | 0.81 | 1.84 | 2.84 × 10−4 |
| OPG | 0.78 ± 0.36 | 1.67 ± 0.53 | 0.47 | 3.91 × 10−5 | 0.84 ± 0.28 | 1.09 ± 0.38 | 0.77 | 0.07 | 1.08 | 0.61 | 0.65 | 4.10 × 10−3 |
| TIMP3 | 0.61 ± 0.21 | 1.50 ± 0.52 | 0.41 | 6.45 × 10−6 | 0.83 ± 0.33 | 1.00 ± 0.39 | 0.83 | 0.23 | 1.36 | 0.06 | 0.67 | 0.01 |
| BBOX1 | 2.16 ± 0.48 | 0.96 ± 0.30 | 2.26 | 8.04 × 10−8 | 1.84 ± 0.37 | 2.14 ± 0.44 | 0.86 | 0.07 | 0.85 | 0.07 | 2.23 | 3.09 × 10−8 |
| GLUT4 | 1.57 ± 0.35 | 0.99 ± 0.24 | 1.58 | 6.15 × 10−5 | 1.62 ± | 1.50 ± 0.31 | 1.09 | 0.26 | 1.03 | 0.67 | 1.50 | 1.08 × 10−4 |
| THRSP | 0.99 ± 0.28 | 1.61 ± 0.39 | 0.61 | 8.18 × 10−5 | 1.53 ± 0.33 | 1.60 ± 0.32 | 0.95 | 0.57 | 1.55 | 1.38 × 10−4 | 0.99 | 0.93 |
PRRX1/PPARG antiregulated genes were identified by Illumina microarray analysis in samples with PRRX1 knockdown and simultaneous PRRX1 and PPARG knockdown during adipogenic differentiation (Figure 5E). Confirmatory qRT-PCR was performed for these representative top regulated genes in hASC from BMI-matched heterozygous (hetero, n = 16) and homozygous (homo, n = 32) risk allele carriers (genotyped for the PPARG locus cis-regulatory variant rs4684847 and the tagSNP rs1801282 Pro12Ala). ADIPOQ, adiponectin, C1Q and collagen domain containing; BBOX1, butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase); FC, fold change; GLUT4, Glucose Transporter Type 4; LIPE, lipase, hormone-sensitive; OPG, Osteoprotegerin; p, p value from unpaired t test; PDK4, pyruvate dehydrogenase kinase, isozyme 4; PEPCKC, Phosphoenolpyruvate carboxylase cytosolic; PPARG, peroxisome proliferator-activated receptor gamma; PRRX1, paired-related homeobox 1; THRSP, thyroid hormone responsive Spot 14 Protein; TIMP3, TIMP metallopeptidase inhibitor 3.
By qPCR, we confirmed rs4684847 allele-dependent dysregulation of genes in the identified biological pathways. Notably, the gene with the strongest risk allele-dependent decrease in mRNA levels was PEPCKC (Table 2). The top scoring IPA interaction network reinforced a central role for PEPCKC (Figure 5F). PEPCK-C is the enzyme controlling the first committed step of glyceroneogenesis, a crucial metabolic process in adipocytes regulating the re-esterification of free fatty acids (FFA) to TG (Ballard et al., 1967). Glyceroneogenesis limits FFA release from adipocytes in the fasting state thereby controlling systemic FFA homeostasis and insulin sensitivity (Millward et al., 2010). In the 67 BMI- and body fat-matched obese subjects, we confirmed rs4684847 risk allele association with increased serum FFAs levels (p = 0.049) and risk allele-dependent association of PRRX1 mRNA with FFA levels (p = 0.015, Table 1). To prove that rs4684847, by determining PRRX1 binding, affects glyceroneogenesis and subsequent FFA release, we monitored pyruvate incorporation in TG (Ballard et al., 1967). We confirmed a PRRX1-dependent suppression of glyceroneogenesis in CC risk allele carriers, marked by a robust correlation with PRRX1 mRNA levels (Figure 5G) and a risk allele-dependent increase in FFA release (Figure 5H). In a parallel experiment, we also found that PRRX1 silencing was sufficient to restore cellular insulin sensitivity in risk allele carriers (Figure 5I). Importantly, the PPARg ligand Rosi pharmacologically promotes insulin sensitivity largely via control of FFA homeostasis through glyceroneo-genesis (Cadoudal et al., 2007), and Kang et al. (2005) reported impaired Rosi response in risk haplotype carriers. In our analysis of glyceroneogenesis in hASCs, we observed an impaired response to Rosi-mediated suppression of FFA release dependent on the risk allele (Figure 5J). Strikingly, PRRX1 silencing in CC risk allele patient samples was sufficient to abolish the reduced Rosi responsiveness, making PRRX1 a potential target for pharmacological T2D intervention.
In summary, by PMCA we demonstrate a clustering of specific homeobox TFBS at T2D risk SNPs. We specifically unveil a role of the homeobox TF PRRX1 as a repressor of PPARG2 via its enhanced binding at the rs4684847 C risk allele, thereby provoking dysregulation of FFA turnover and glucose homeostasis (Figure 5K).
Discussion
We have developed a bioinformatics approach, PMCA, which enables the extraction of cis-regulatory variants that may mechanistically contribute to human disease by dysregulation of gene expression. In line with our approach to exploit conservation in terms of co-occurring TFBS patterns, (Visel et al., 2013) has recently shown that combination of TFBS, rather than single TFBS, via combinatorial TF binding governs spatial enhancer activity in the developing telencephalon. Further, tissue-specific enhancers were accurately detected by in vivo mapping of the enhancer-associated proteins p300, in addition to comparative genomics approaches (Visel et al., 2009a; Blow et al., 2010).
Using T2D as a showcase, we demonstrate the utility of PMCA for the generic prediction of distinct homeobox TFBS at T2D risk SNPs, which is important for understanding disease regulatory circuits when we consider that interactions in a regulatory network involve numerous genes and a rather small set of TFs (Califano et al., 2012). Pursuing the results emerging from our comprehensive T2D analysis, we show that identification of the cis-regulatory variant rs4684847 at the PPARG locus enabled linking the molecular upstream factor PRRX1 to aberrant downstream mechanisms of impaired lipid handling and insulin sensitivity, explaining the GWAS association with T2D. Notably, PRRX1 was recently implicated in adipogenesis (Du et al., 2013), yet the regulated genes remain elusive.
Here, we restricted the analysis to SNPs in LD with GWAS SNPs. However, the approach could be applied to any other kind of variability, such as somatic mutations in cancer, without loss of generality. Certain issues will require consideration, e.g., analyzing genomes of closely related species to refine scoring criteria, and extending our analysis to whole genome sequencing studies, including rare variants data, should further inform on the genetic underpinnings of phenotypic diversity in humans. Our in silico scoring results predict varying numbers of regulatory SNPs per LD block. Studies have now found evidence for allelic heterogeneity (Maller et al., 2012; Schaub et al., 2012), yet the number of causal variants within a disease locus is elusive. We propose an integrative framework where computational TFBS modularity analysis may be synergistically combined with functional genomics and population genetics data.
In sum, our results demonstrate that the extension of sequence analysis to functional conservation integrates biological data with statistical signals, and our method should help to clarify the role of inherited and somatic variability in altering gene regulatory networks, in both mendelian and common human diseases.
Experimental Procedures
See the Extended Experimental Procedures for details.
LD Block Definition
SNPs in close LD (r2 ≥ 0.7) to GWAS tagSNPs (references in Tables S1, S5, S7, and S13) from 1000 Genomes Project, Pilot 1, CEU data (http://www.1000genomes.org/).
PMCA
PMCA analyzes the occurrence of conserved patternsof TFBS in a CRM within the genomic region flanking a noncoding variant, to predict its cis-regulatory functionality. For each variant the PMCA method provides a classification of the region surrounding the variant as being either complex or noncomplex. Complex regions are defined as being significantly enriched in conserved co-occurring TFBS (TFBS modules) according to the scoring scheme described in Extended Experimental Procedures.
Positional Bias Analysis
Complex and noncomplex regions (SNP ± 500 bp) were scanned for presence of TFBS family matches at SNP positions. Positional bias of TFBS families was calculated using overlapping 50 bp sliding windows in steps of 10 bp. Positional bias (p) was calculated as binomial p value for each TFBS family and each window.
Correlation with Evolutionary Constraint, DHSseq, and ChIP-Seq Regions
Complex/noncomplex SNP regions (SNP ± 60 bp) were correlated to constrained regions or DHSseq and ChIP-seq peaks. From midpoint of constrained regions (±500 bp), DHSseq (±1,000 bp), or ChIP-seq peaks (±1,000 bp), the overlapping positions (correlation) with complex/noncomplex regions were counted and plotted versus position relative to anchor. For the calculation of enrichment of DHS and ChIP-seq peak overlaps to complex/noncomplex SNPs only those SNPs were considered where an overlap was detected within ± 20 bp from SNP positions.
Primary Human Tissue and hASC
Human islets and adipose tissue were obtained with informed consent from each subject. The studies were approved by the local ethics committees of the Technische Universität München (Germany), the Haukeland University Hospital (Norway) and the Lund University (Sweden). Primary hASCs were isolated from subcutaneous adipose tissue and differentiated in vitro. Genotyping was done by MassARRAY (Sequenom), Omni express (Illumina), or Sanger Sequencing.
RNA Preparation and Expression Analysis
Total RNA was prepared by TRIzol (Invitrogen) or RNeasy Lipid Tissue Mini Kit (QIAGEN), and gene expression was measured by qPCR or microarrays (Affymetrix, Illumina). Allele-specific primer extension was performed with SNaPshotKit (ABI Prism).
Cell Culture and Reporter Assays
Huh7, INS-1, 293T, C2C12, 3T3-L1, and SGBS cells were cultured using standard protocols. Genomic sequences surrounding SNPs were synthesized (MWG), cloned in pGL4.22-TK-promoter (Promega) and transfected in cells by Lipofectamine (Invitrogen). Luciferase activity was measured by Luminoscan-Ascent (Thermo).
Gene Knockdown by Small Interfering RNA
All knockdowns were performed with ON-TARGETplus SMARTpool small interfering RNA (siRNA) (Dharmacon) and HiPerFect (QIAGEN).
CRISPR/Cas Genome Editing
HDR genome editing was performed in human SGBS preadipocytes by trans-fection of CRISPR/Cas9 and single guide RNA (sgRNA) expression vectors (sgRNA targeting a NGG PAM sequence 50 of rs4684847, R. Kühn, Munich) and rs4684847 DNA donor vectors (T allele to replace endogenous allele, C allele control). Cell enrichment by MACS selected transfected cell selection kit (Miltenyi). rs4684847 genome editing was confirmed by Sanger sequencing.
EMSA
Forty-two base pairs of allelic Cy5-labeled-DNAs (MWG) and nuclear protein were used for EMSA. Supershift experiments were performed with aPRRX1 or IgG, competition with excess unlabeled probe, and protein from pCMV-PRRX1-flag transfected 293T.
DNA-Protein Affinity Chromatography LC-MS/MS
DNA-protein affinity chromatography was performed with streptavidin magnetic beads (Invitrogen) and allelic biotinylated DNA-probes (MWG) and Ultimate3000nano HPLC (Dionex) LC-MS/MS coupled to LTQ OrbitrapXL (Thermo Fisher Scientific). Data were analyzed with Progenesis software v2.5.
Statistical Analysis
Statistical analyses were done using Graph Pad Prism v5.02, R Software v2.14.2 or Perl scripts.
Supplementary Material
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, 4 figures, and 17 tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.10.058.
Acknowledgments
This work was funded by the Else Kröner-Fresenius Foundation, Bad Homburg v.d.H, Germany; the grant Virtual Institute “Molecular basis of glucose regulation and type 2 diabetes” received from the Helmholtz Zentrum München, München-Neuherberg, Germany; the grant Clinical Cooperation Group “Nutrigenomics and type 2 diabetes” received from the Helmholtz Zentrum München, München-Neuherberg, Germany, and the Technische Universität München; the Helmholtz Graduate School for Environmental Health, HELENA; a grant from the German Federal Ministry of Education and Research to the German Centre for Diabetes Research (DZD e.V.); the Competence Network Obesity (German Obesity Biomaterial Bank; FKZ 01GI1128), and the University Duisburg-Essen (01KU1216E); the KG Jebsen Center for Diabetes Research, University of Bergen, Norway and the Western Norway Regional Health Authority, Norway; by grants from the Swedish Research Council, including strategic research area grant EXODIAB (2009-1039), Linnaeus grant (349-2006-237), Collaborative Grant (2011-3315), and Project Grant (521-2010-3490) and by an ERC Advanced Researcher Grant GENETARGET-T2D (GA 269045); and by “Biomedical Research Program” funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation. We thank Karl-Fredrik Eriksson and Targ Elgzyri for providing human fat biopsy material and Charlotte Ling for supporting generation of microarray data (Lund University). We are grateful to Andrea Califano (Columbia University, New York, NY) for critical review and constructive scientific comments on the manuscript. We further thank Bernd Baumann, Vidar M. Steen, and Jørn V. Sagen for expert advice, and Elisabeth Hofmair, Manuela Hubersberger, Margit Solsvik, Linn Jeanette Waagbø, Tone Nygaard Flølo, Jan-Inge Bjune, Zina Fandalyuk, Vivian Veum, and Christine Haugen for excellent technical assistance. We thank Ralf Kühn (Helmholtz Zentrum München, München-Neuherberg) for providing CRISPR/Cas vectors. We thank Christine Stansberg, Rita Holdhus, and Kjell Petersen at the Norwegian Microarray Consortium (NMC) (Bergen node, Norway), Douglas P. Kiel and David Karasik (Hebrew SeniorLife Institute for Aging Research, Harvard Medical School, Boston, MA), and Michael Molla (Joslin Diabetes Center, Harvard Medical School, Boston, MA) for expert assistance. We thank surgeonHans JørgenNielsen and colleagues (Voss Hospital, Norway), surgeons Barbara Auras Jaatun, Inge Glambæk, and colleagues (Haraldsplass Deaconess Hospital, Bergen), surgeon Christian Busch (Klinikk Bergen, Nesttun), and all the patients for providing human adipose tissue. We thank Reiner Schroeer and Dorothy Dankel for critical reading of the manuscript. The authors have filed a patent related to this work.
Footnotes
ACCESSION NUMBERS
Microarray data for hASC are available in ArrayExpress (E-MTAB-1906). The Gene Expression Omnibus (GEO) accession number for the adipose tissue microarray analysis reported in this paper is GSE25402.
Contributor Information
DIAGRAM+Consortium:
Benjamin F. Voight, Laura J. Scott, Valgerdur Steinthorsdottir, Andrew P. Morris, Christian Dina, Ryan P. Welch, Eleftheria Zeggini, Cornelia Huth, Yurii S. Aulchenko, Gudmar Thorleifsson, Laura J. McCulloch, Teresa Ferreira, Harald Grallert, Najaf Amin, Guanming Wu, Cristen J. Willer, Soumya Raychaudhuri, Steve A. McCarroll, Claudia Langenberg, OliverM. Hofmann, Josée Dupuis, Lu Qi, Ayellet V. Segrè, Mandy van Hoek, Pau Navarro, Kristin Ardlie, Beverley Balkau, Rafn Benediktsson, Amanda J. Bennett, Roza Blagieva, Eric Boerwinkle, Lori L. Bonnycastle, Kristina Bengtsson Boström, Bert Bravenboer, Suzannah Bumpstead, Noël P. Burtt, Guillaume Charpentier, Peter S. Chines, Marilyn Cornelis, David J. Couper, Gabe Crawford, Alex S.F. Doney, Katherine S. Elliott, Amanda L. Elliott, Michael R. Erdos, Caroline S. Fox, Christopher S. Franklin, Martha Ganser, Christian Gieger, Niels Grarup, Todd Green, Simon Griffin, Christopher J. Groves, Candace Guiducci, Samy Hadjadj, Neelam Hassanali, Christian Herder, Bo Isomaa, Anne U. Jackson, Paul R.V. Johnson, Torben Jørgensen, Wen H.L. Kao, Norman Klopp, Augustine Kong, Peter Kraft, Johanna Kuusisto, Torsten Lauritzen, Man Li, Aloysius Lieverse, Cecilia M. Lindgren, Valeriya Lyssenko, Michel Marre, Thomas Meitinger, Kristian Midthjell, Mario A. Morken, Narisu Narisu, Peter Nilsson, Katharine R. Owen, Felicity Payne, John R.B. Perry, Ann-Kristin Petersen, Carl Platou, Christine Proença, Inga Prokopenko, Wolfgang Rathmann, N. William Rayner, Neil R. Robertson, Ghislain Rocheleau, Michael Roden, Michael J. Sampson, Richa Saxena, Beverley M. Shields, Peter Shrader, Gunnar Sigurdsson, Thomas Sparsø, Klaus Strassburger, Heather M. Stringham, Qi Sun, Amy J. Swift, Barbara Thorand, Jean Tichet, Tiinamaija Tuomi, Rob M. van Dam, Timon W. van Haeften, Thijs van Herpt, Jana V. van Vliet-Ostaptchouk, G. Bragi Walters, Michael N. Weedon, Cisca Wijmenga, Jacqueline Witteman, Richard N. Bergman, Stephane Cauchi, Francis S. Collins, Anna L. Gloyn, Ulf Gyllens-ten, Torben Hansen, Winston A. Hide, Graham A. Hitman, Albert Hofman, David J. Hunter, Kristian Hveem, Markku Laakso, Karen L. Mohlke, Andrew D. Morris, Colin N.A. Palmer, Peter P. Pramstaller, Igor Rudan, Eric Sijbrands, Lincoln D. Stein, Jaakko Tuomilehto, Andre Uitterlinden, Mark Walker, Nicholas J. Wareham, Richard M. Watanabe, Goncalo R. Abecasis, Bernhard O. Boehm, Harry Campbell, Mark J. Daly, Andrew T. Hattersley, Frank B. Hu, James B. Meigs, James S. Pankow, Oluf Pedersen, H.-Erich Wichmann, Inês Barroso, Jose C Florez, Timothy M. Frayling, Leif Groop, Rob Sladek, Unnur Thorsteinsdottir, James F. Wilson, Thomas Illig, Philippe Froguel, Cornelia M. van Duijn, Kari Stefansson, David Altshuler, Michael Boehnke, and Mark I. McCarthy
References
- Alberobello AT, Congedo V, Liu H, Cochran C, Skarulis MC, Forrest D, Celi FS. An intronic SNP in the thyroid hormone receptor β gene is associated with pituitary cell-specific over-expression of a mutant thyroid hormone receptor b2 (R338W) in the index case of pituitary-selective resistance to thyroid hormone. J Transl Med. 2011;9:144. doi: 10.1186/1479-5876-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Ballard FJ, Hanson RW, Leveille GA. Phosphoenolpyruvate carboxykinase and the synthesis of glyceride-glycerol from pyruvate in adipose tissue. J Biol Chem. 1967;242:2746–2750. [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M, Shiota M, Nicholson WE, Gannon M, Knobel SM, Piston DW, Wright CVE, Powers AC. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J Biol Chem. 2002;277:11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- Cadoudal T, Blouin JM, Collinet M, Fouque F, Tan GD, Loizon E, Beale EG, Frayn KN, Karpe F, Vidal H, et al. Acute and selective regulation of glyceroneogenesis and cytosolic phosphoenolpyruvate carbox-ykinase in adipose tissue by thiazolidinediones in type 2 diabetes. Diabetologia. 2007;50:666–675. doi: 10.1007/s00125-006-0560-5. [DOI] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Kang MJ, Homer RJ, Kang HR, Zhang X, Lee PJ, Elias JA, Lee CG. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem. 2006;281:8161–8168. doi: 10.1074/jbc.M506770200. [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- Doria A, Patti M-E, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B, Cawthorn WP, Su A, Doucette CR, Yao Y, Hemati N, Kampert S, McCoin C, Broome DT, Rosen CJ, et al. The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-b (TGFb) signaling. J Biol Chem. 2013;288:3036–3047. doi: 10.1074/jbc.M112.440370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001046. e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- Fajas L, Fruchart JC, Auwerx J. PPARgamma3 mRNA: a distinct PPARgamma mRNA subtype transcribed from an independent promoter. FEBS Lett. 1998;438:55–60. doi: 10.1016/s0014-5793(98)01273-3. [DOI] [PubMed] [Google Scholar]
- FitzGerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen S, Argmann C, Feige JN, Koutnikova H, Champy M-F, Dali-Youcef N, Schadt EE, Laakso M, Auwerx J. The Pro12Ala PPARgamma2 variant determines metabolism at the gene-environment interface. Cell Metab. 2009;9:88–98. doi: 10.1016/j.cmet.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MC, Ahnfelt-Rønne J, Hald J, Madsen OD, Serup P, Hecksher-Sørensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EE. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kang ES, Park SY, Kim HJ, Kim CS, Ahn CW, Cha BS, Lim SK, Nam CM, Lee HC. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor gamma2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther. 2005;78:202–208. doi: 10.1016/j.clpt.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Laumen H, Saningong AD, Heid IM, Hess J, Herder C, Claussnitzer M, Baumert J, Lamina C, Rathmann W, Sedlmeier E-M, et al. Functional characterization of promoter variants of the adiponectin gene complemented by epidemiological data. Diabetes. 2009;58:984–991. doi: 10.2337/db07-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, Su Z, Howson JMM, Auton A, Myers S, Morris A, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez G, Virtue S, Lelliott C, Boiani R, Campbell M, Christodoulides C, Perrin C, Jimenez-Linan M, Blount M, Dixon J, et al. The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-g2 isoform. Diabetes. 2005;54:1706–1716. doi: 10.2337/diabetes.54.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward CA, Desantis D, Hsieh CW, Heaney JD, Pisano S, Olswang Y, Reshef L, Beidelschies M, Puchowicz M, Croniger CM. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/fatty acid cycle and development of insulin resistance in mice. J Lipid Res. 2010;51:1452–1463. doi: 10.1194/jlr.M005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO, et al. A large-scale, consortium-based genome wide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Science. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000895. e1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Sato H, Iida A, Mizuno H, Nakamura T, Miyamoto Y, Takahashi A, Tsunoda T, Ikegawa S, Kamatani N, et al. A functional SNP in PSMA6 confers risk of myocardial infarction in the Japanese population. Nat Genet. 2006;38:921–925. doi: 10.1038/ng1846. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintás-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR g is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott LJ, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR g 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009a;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009b;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013a;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012a;337:1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012b;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.